Abstract
The synaptonemal complex (SC) is a proteinaceous structure that mediates homolog engagement and genetic recombination during meiosis. In budding yeast, Zip-Mer-Msh (ZMM) proteins promote crossover (CO) formation and initiate SC formation. During SC elongation, the SUMOylated SC component Ecm11 and the Ecm11-interacting protein Gmc2 facilitate the polymerization of Zip1, an SC central region component. Through physical recombination, cytological, and genetic analyses, we found that ecm11 and gmc2 mutants exhibit chromosome-specific defects in meiotic recombination. CO frequencies on a short chromosome (chromosome III) were reduced, whereas CO and non-crossover frequencies on a long chromosome (chromosome VII) were elevated. Further, in ecm11 and gmc2 mutants, more double-strand breaks (DSBs) were formed on a long chromosome during late prophase I, implying that the Ecm11–Gmc2 (EG) complex is involved in the homeostatic regulation of DSB formation. The EG complex may participate in joint molecule (JM) processing and/or double-Holliday junction resolution for ZMM-dependent CO-designated recombination. Absence of the EG complex ameliorated the JM-processing defect in zmm mutants, suggesting a role for the EG complex in suppressing ZMM-independent recombination. Our results suggest that the SC central region functions as a compartment for sequestering recombination-associated proteins to regulate meiosis specificity during recombination.
INTRODUCTION
During meiosis, homologous chromosomes (‘homologs’) undergo dynamic structural changes and line up in pairs for recombination, which is initiated by the formation of programmed DNA double-strand breaks (DSBs) (1). In most sexually reproducing organisms, programmed DSB formation, which is catalyzed by the topoisomerase VI-like protein Spo11, occurs early in meiosis after DNA replication (2–4). The DSB ends subsequently undergo extensive nucleolytic resection to expose a 3′ single-stranded overhang of ∼800 nucleotides, which is required for homology searching (5–7). The ‘first’ DSB end undergoes strand exchange with the homolog chromatid through a process mediated by the RecA homologs Dmc1 and Rad51 and forms a nascent D-loop that is expanded into the single-end invasion (SEI) recombination intermediate (8–13). The ‘second’ DSB end is thought to engage with the displaced strand of the SEI to produce a double-Holliday junction (dHJ). Errors in meiotic DSB repair can lead to genome instability and nondisjunction of chromosomes (14). Multiple feedback control mechanisms regulate DSB frequency and distribution to maintain equilibrium, thereby managing recombination events (15–19).
In Saccharomyces cerevisiae, Zip-Mer-Msh (ZMM) proteins play a role in the feedback control of DSB formation (15,20). ZMM proteins, including Zip1–3, Spo22/Zip4, Mer3, Msh4, Msh5 and Spo16, are involved in SC assembly and act to stabilize nascent joint molecules (JMs), which are related to crossover (CO) formation (21–31). Mer3 and Msh4–Msh5 (MutSγ) bind nascent recombination intermediates to facilitate CO-specific recombination; Zip2–Zip4–Spo16 form a functional complex (the ZZS complex) to promote CO formation; and Zip1 contains a coiled-coil domain and a globular domain that form the transverse filament component of the synaptonemal complex (SC), a meiosis-specific zipper-like proteinaceous structures, comprising axial/lateral elements and a central element that interconnects these. The SC components are involved in the relocation of recombination complexes to SC central region (21,32–34).
In budding yeast, recombination intermediates are specifically designated to become interhomolog (IH) COs, whereas the remainders are designated as non-crossovers (NCOs) (22,35). Once CO/NCO differentiation has occurred in the early prophase, progression to the CO fate involves the production stable JMs, such as SEI and dHJ intermediates, in a ZMM-dependent manner (22,35). ZMM-dependent COs, often called ‘class I’ COs, exhibit a non-random distribution on chromosomes with positive interference. A fraction of the meiotic DSBs is repaired through a ZMM-independent pathway, which shows random resolution of the Holliday structures, yielding both CO and NCO products (32,36). The ZMM-independent COs are called ‘class II’ COs and do not exhibit interference (31,36). Additionally, dHJs may be processed into NCOs through dissolution, which involves branch migration of HJs (37–39). In normal meiosis in wild-type (WT), the majority of COs are formed via the ZMM-dependent pathway. In zmm mutants, NCOs occur at high frequencies, whereas CO-designated recombination is strongly defective at the HIS4LEU2 locus of chromosome III. In a zip1Δ mutant, CO recombination is not reduced on chromosome VII (20). The fact that zmm mutants demonstrate an elevated levels of DSB formation during late meiotic prophase I suggests that homolog engagement regulates the number and distribution of DSBs via the displacement of Spo11 accessory factors or axis proteins (Red1/Mek1/Hop1) (9,10,15,19,36,40–44). However, it is not clear whether late meiotic DSB formation in WT meiosis is directly downregulated by homolog engagement itself or by ZMM.
Several lines of evidence suggest that SUMOylation of Ecm11, which forms a complex with Gmc2, is important for Zip1 assembly between homologs and that the Ecm11–Gmc2 (EG) complex functions as a component of the SC central element (45–47). Gmc2 localizes to the synapsis initiation complex (SIC) in a SUMOylated Ecm11-dependent manner, whereas Ecm11 can localize to the SIC in a Gmc2-independent manner (45–47). Further, SUMOylated Ecm11 localizes to a discrete region of the central element domain that is assocated with the Zip1 N-terminus and limits excess MutSγ-mediated CO formation (48,49).
To better define the interplay between homolog engagement and recombination, we evaluated the regulatory roles of the EG complex in DSB formation and CO-designated DSB control through physical recombination, genetic, and cytological analyses in S. cerevisiae. We show that ecm11 and gmc2 deletion mutants exhibit increased DSB frequencies, particularly on a long chromosome, during late prophase I, suggesting that the EG complex is involved in the feedback control of DSB formation in a chromosome length-dependent manner. A zip3Δ mutant exhibited a severe reduction in CO-specific species and formed extra DSBs at late prophase. Meanwhile, the ecm11 and gmc2 mutation partially suppressed the formation of defects in meiotic recombination processing in the zip3Δ background. Together, these results reveal multiple roles for the EG complex in the control of late DSB formation and CO-specific events. The findings we present here provide new insights into the regulatory role of the EG complex in the SC central region.
MATERIALS AND METHODS
Strains
Detailed information on strain genotypes is presented in Supplementary Table S1.
Meiotic time course
Meiotic time course experiments were performed as previously described (9,41,50–52). Saccharomyces cerevisiae SK1 strains were patched onto YPG plates (2% bacteriological peptone, 1% yeast extract, 3% glycerol, 2% bacteriological agar) and incubated at 30°C for 12 h. Cells were inoculated onto YPD plates (2% bacteriological peptone, 1% yeast extract, 2% glucose, 2% bacteriological agar) and grown for two days to obtain single colonies. A single colony was inoculated into 2 ml of YPD liquid medium and grown to saturation at 30°C overnight. To synchronize cell cultures, overnight cultures were transferred to pre-warmed SPS medium (1% potassium acetate, 1% bacteriological peptone, 0.5% yeast extract, 0.17% yeast nitrogen base with ammonium sulfate and without amino acids, 0.5% ammonium sulfate, 0.05 M potassium biphthalate, adjusted to pH 5.5 with 10 N KOH) and grown for 17–18 h. Meiosis was induced by transferring the SPS-cultured cells to pre-warmed SPM medium (1% potassium acetate, 0.02% raffinose, two drops of Antifoam 204). SPM-cultured cells were harvested at 0, 2.5, 3.5, 4, 5, 6, 7, 8, 10 and 24 h after being transferred to the SPM medium and then cross-linked with psoralen under ultraviolet light at a wavelength of 360 nm. For the analysis of cells grown at 23°C, cells were cultured at 30°C for up to 2.5 h, and then shifted to 23°C.
Physical recombination analysis
Genomic DNA was extracted from cultured cells using a previously described guanidine-phenol extraction method (9,41,51,52). Cells were resuspended in spheroplasting buffer (1 M sorbitol, 50 mM potassium phosphate, zymolyase 100T, 1% β-mercaptoethanol, 10 mM EDTA, pH 7.5) and incubated at 37°C for 20 min. Spheroplasts were harvested and incubated in guanidine lysis buffer (4.5 M guanidine–HCl, 0.1 M EDTA, 0.15 M NaCl, 0.05% sodium lauroyl sarcosinate, adjusted to pH 8.0 with 50% NaOH) at 65°C for 15 min. The cell lysates were cooled on ice and precipitated in ethanol at –20°C overnight. The pellets were collected through centrifugation and then incubated in proteinase K solution at 65°C for 1 h. Genomic DNA was extracted twice with phenol/chloroform/isoamyl alcohol (25:24:1) solution and precipitated in ethanol. The DNA pellets were rinsed with 70% ethanol and dried at room temperature for 1 h.
XhoI restriction-site polymorphisms in the HIS4LEU2 hotspot on chromosome III were detected to obtain the DNA species for DSB, SEI, dHJ and CO products as previously described (9,41,51,52). To analyze DSBs and COs, genomic DNA (2 μg) was digested with 80 units of XhoI and subjected to one-dimensional (1D) gel electrophoresis using a 0.6% SeaKem agarose gel in TBE (89 mM Tris-borate, 2 mM EDTA, pH 8.3) and TBE gel running buffer at 2 V/cM for 24 h. XhoI restriction polymorphisms in the HIS4LEU2 locus allow the detection of total CO products (5.6 and 4.6 kb) from Mom and Dad fragments, which can be resolved through 1D gel electrophoresis of XhoI-digested DNA followed by Southern blotting. CO-1 and NCO-1 were distinguished using BamHI (Mom strain)/NgoMIV (Dad strain)-restriction enzyme sites inserted close to the DSB sites at the HIS4LEU2 locus. The XhoI/NgoMIV-digested genomic DNA (2 μg) was separated, and 1D gel electrophoresis was carried out in TBE gel running buffer. Southern blotting of XhoI/NgoMIV-digested DNA can resolve the presence of CO-1 (4.6 kb) and NCO-1 (4.3 kb), which are partial fragments of COs and NCOs from two parental species (Mom and Dad).
For native/native two-dimensional (2D) gel analysis, XhoI-digested DNA samples (2.5 μg per sample) were loaded onto a 0.4% SeaKem Gold agarose gel and electrophoresed at 1 V/cM for 21 h. The gels were stained with ethidium bromide (EtBr) (0.5 μg/ml in TBE), and the gel lanes containing the DNA of interest were cut. The gel strips were placed on 2D gel trays, and 0.8% SeaKem agarose containing EtBr was poured into the trays. 2D gel electrophoresis was performed in TBE buffer containing EtBr at 6 V/cm, at 4°C for 6 h. The gels were transferred to Biodyne B membranes (Pall, Port Washington, NY) for Southern hybridization.
For all physical analyses, radiolabeled probes were used to detect hybridizing DNA species. The probes were radiolabeled with α-32P-dCTP using a random priming kit. Hybridizations were carried out at 65°C in sodium phosphate buffer (0.5 M sodium phosphate, 1 mM EDTA, 7% SDS, pH 7.2) for 16 h. Blots were washed four times for 30 min each time with saline-sodium citrate buffer (150 mM sodium chloride, 15 mM trisodium citrate, 0.1% SDS, pH 7.0). The blots were then exposed to storage phosphor screens (GE Healthcare Life Sciences, Piscataway, NJ) in light-tight film cassettes. Hybridizing DNA species were visualized using a phosphoimager (Bio-Rad, Hercules, CA) and quantified using the QuantityOne software (Bio-Rad). For detection of the HIS4LEU2 locus by Southern blotting, probes were amplified from yeast genomic DNA using primers with the following sequences: 5′-ATATACCGGTGTTGGGCCTTT-3′ and 5′-ATATAGATCTCCTACAATATCAT-3′. The primer sequences of the DNA probes used to detect the ERG1 (SacII, SacII + SalI) locus were 5′-ATGGAAGATATAGAAGGATACGAACC-3′ and 5′-GCGACGCAAATTCGCCGATGGTTTG-3′. The sequences of the DNA probes used to detect the ERG1 (HindIII) locus were 5′-GGCAGCAACATATCTCAAGGCC-3′ and 5′-TCAATGTAGCCTGAGATTGTGGCG-3′.
Spore viability and genetic distance analyses
Spore viability and genetic distances between markers and CO interference were analyzed as previously described (28,53). Parental haploid strains (MSY4304 and MSY4988 derivatives, or S2921 and MSY5085 derivatives) were mated on YPAD plates for 3 h and then transferred to SPM plates. To exclude tetrads with mitotic COs, four independent crosses were analyzed. Map distances were calculated using the Perkins equation: cM = 100 (TT + 6NPD)/2(PD + TT + NPD). Standard errors were calculated using the Stahl Lab online tool (https://elizabethhousworth.com/StahlLabOnlineTools).
Chromosome spreading and immunofluorescence
Immunostaining of yeast meiotic chromosome spreads was performed as previously described (54). Stained samples were observed using an epifluorescence microscope (Zeiss Axioskop 2; Zeiss, Oberkochen, Germany) and a 100× objective (Zeiss AxioPlan, NA1.4). Images were captured using a CCD camera (Retiga; QImaging, British Columbia, Canada) and processed using iVision (BioVision Technologies, Exton, PA) and Photoshop (Adobe, San Jose, PA) software. Antibodies against Zip1 (rabbit and rat) (28), Zip3 (rabbit and rat) (28), Rad51 (guinea pig) (55), Dmc1 (rabbit) (55), Msh5 (rabbit) (28), and Red1 (chicken) (28) were generated using recombinant proteins purified from Escherichia coli.
Spo11-oligo assay
Spo11-oligo detection was performed according to previously described methods (56,57), with modifications. Spo11-oligo complexes were immunoprecipitated from 20 ml of synchronous meiotic yeast culture. After preparation of a cell extract using glass beads (Yasui Kikai Co Ltd., Osaka, Japan), Spo11-FLAG was immunoprecipitated using an anti-DYKDDDDK tag antibody (1E6, FUJIFILM Wako, Tokyo, Japan) and protein G-conjugated magnetic beads (Dynabeads, Veritas, Tokyo, Japan) in IP buffer (2% Triton X-100; 30 mM Tris-HCl, pH 8.0; 300 mM NaCl; 2 mM EDTA; 0.02% SDS). Immunoprecipitates were washed twice with IP buffer. Then, 10% volume of each sample was analyzed through western blotting, and the Spo11-FLAG protein levels in the precipitates were measured using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). The remaining samples (∼90%) were used for end-labeling reactions. For end-labeling of Spo11-oligo, immunoprecipitates were washed twice with NEBuffer #4 (New England Biolabs, Ipswich, MA). The beads were then suspended in TdT reaction buffer (NEBuffer #4, 0.25 mM CoCl2; 15 U TdT, Takara Bio, Shiga, Japan; 20 Ci α-32P-dCTP, 6000 Ci/mmol), and incubated at 37°C for 2 h. Radiolabeled Spo11-oligos were washed three times with IP buffer, separated through SDS-PAGE, visualized using a Phosphor imager BAS5000 (FUJIFILM), and quantified using the ImageQuant software (GE Healthcare).
Statistical analysis
Bar graphs and scatter plots for each tested mutant strain were generated using GraphPad Prism 5 software (GraphPad, San Diego, CA). Statistical details for each experiment are included in the figure legends. The data are presented as the means ± standard deviations (SDs) of at least three independent trials. Statistically significant differences from multiple experiments were assessed using an unpaired Student's t-test in Figures 2–4, and 6. The Mann−Whitney U-test was applied for statistical analysis in Figure 5G–I (****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, and ns indicates P > 0.05).
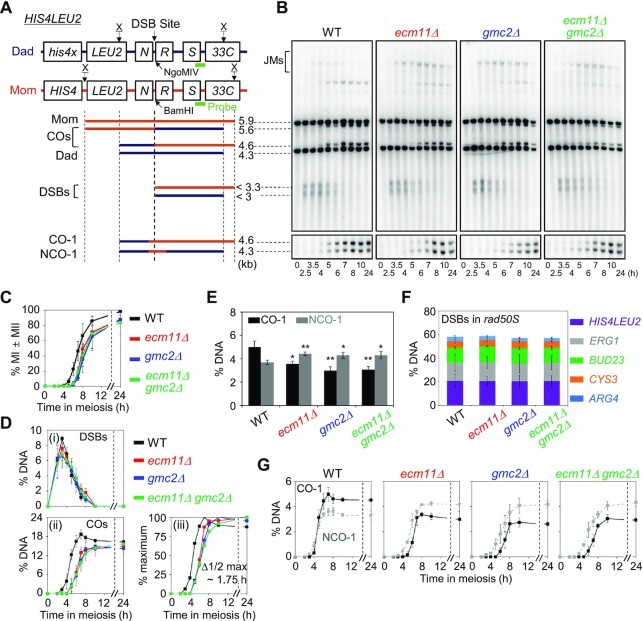
Figure 2.
Physical analysis of meiotic recombination in ecm11Δand gmc2Δ mutants. (A) Physical map of the HIS4LEU2 locus of chromosome III showing the XhoI (X) restriction endonuclease site and the position of the probes used for Southern hybridization. Maternal and paternal fragments were distinguished based on XhoI polymorphisms. For the analysis of CO-1 and NCO-1, DNA was digested with the XhoI and NgoMIV endonucleases. Mom, maternal species (5.9 kb); Dad, paternal species (4.3 kb); COs, crossovers (5.6 kb and 4.6 kb); DSBs, double-strand breaks (<3.3 and <3 kb); CO-1, crossover (4.6 kb); NCO-1, non-crossover (4.3 kb). (B) One-dimensional (1D) gel analysis of DSBs, COs and NCOs in WT, ecm11Δ, gmc2Δ, and ecm11Δ gmc2Δ strains. Gel analysis (1D) showing Mom, Dad, DSBs and CO species (top). CO-1 and NCO-1 of recombinants are displayed in the CO/NCO gel analysis (bottom). (C) Meiotic nuclear division for WT, ecm11Δ, gmc2Δ, and ecm11Δ gmc2Δ strains. Error bars indicate SDs based on three independent time-course experiments. (D) Quantitative analysis of DSBs and COs. (i) Quantification of DSBs. (ii) Quantification of COs. (iii) Normalized CO levels from the analysis shown in (ii). Δ1/2 max, the difference between the times of 50% of the maximum CO levels for WT and mutants. Data are the mean ± SD (N = 3). (E) Quantitative analysis of COs and NCOs in three independent meiotic time-course experiments (data are the mean ± SD). Significance was examined using an unpaired Student's t-test (**P < 0.01; *P < 0.05). (F) Quantitative analysis of DSBs at various loci in rad50S, rad50S ecm11Δ, rad50S gmc2Δ and rad50S ecm11Δ gmc2Δ strains. Data are mean ± SD (N = 3 for ARG4, BUD23, CYS3, and ERG1; N = 4 for HIS4LEU2). See also Supplementary Figure S3. (G) Quantitative analysis of CO-1 (black line) and NCO-1 (gray dashed line). Data are the mean ± SD (N = 3).
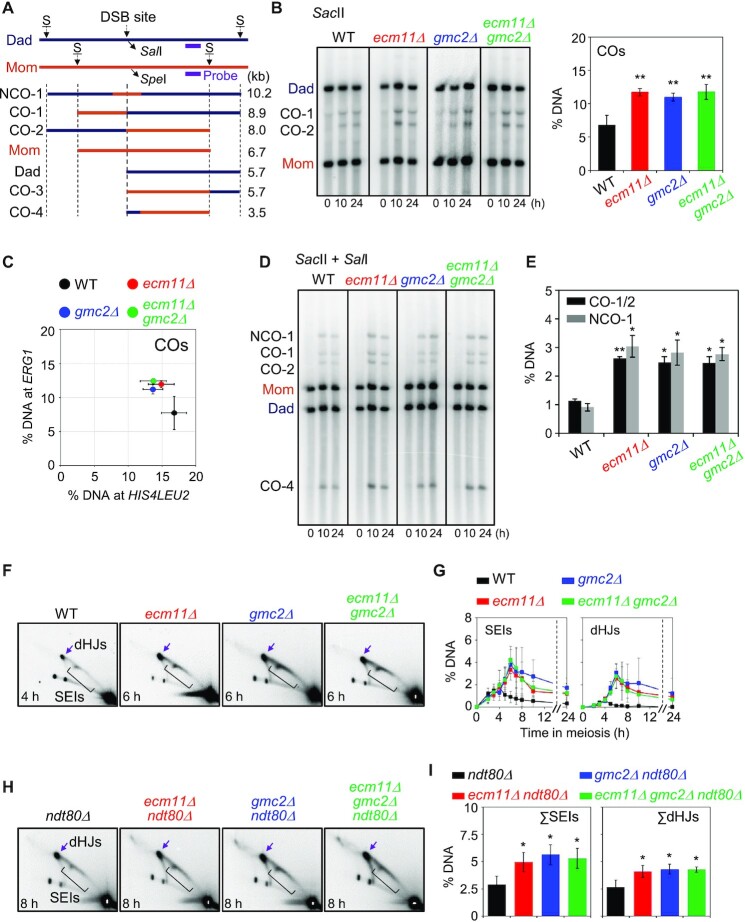
Figure 4.
Meiotic recombination analysis at the ERG1 locus in ecm11Δ, gmc2Δ and ecm11Δ gmc2Δ mutants. (A) Schematic diagram of the ERG1 locus showing restriction enzyme sites and the probe position (15). Parental chromosomes, Mom and Dad, were distinguished by restriction enzyme site polymorphisms (S = SacII). (B) Representative image of 1D gel analysis at the ERG1 locus in WT, ecm11Δ, gmc2Δ and ecm11Δ gmc2Δ strains. Quantitative 1D gel analysis of ERG1 locus in the WT, ecm11Δ, gmc2Δ, and ecm11Δ gmc2Δ strains. Maximum CO levels are shown. Data are the mean ± SD (N = 3). Significance was examined using an unpaired Student's t-test (**P < 0.01). (C) Comparison of CO levels at the HIS4LEU2 and ERG1 loci. Each colored circle indicates the HIS4LEU2 versus ERG1 CO. The data indicate the means ± SD (N = 3). (D) Representative image of CO and NCO analysis of WT, ecm11Δ, gmc2Δ, and ecm11Δ gmc2Δ strains. For CO and NCO gel analysis, the DNA samples were digested with SacII and SalI. (E) Quantitative analysis of CO and NCO. Data indicate the mean ± SD (N = 3). Significance was examined using unpaired Student's t-test (*P < 0.05, **P < 0.01). (F) Representative 2D gel analysis image of the ERG1 locus. (G) Quantitative analysis of SEIs and dHJs. The data indicate the mean ± SD (N = 3). (H) 2D gel analysis of the ERG1 locus in ndt80Δ, ndt80Δ ecm11Δ, ndt80Δ gmc2Δ and ndt80Δ ecm11Δ gmc2Δ strains. DNA samples were digested with HindIII and used for 2D analysis to detect JMs at the ERG1 locus via Southern blotting using an ERG1 probe (15). See also Supplementary Figure S6. (I) Quantification of SEIs and dHJs at the ERG1 locus. Gel images at t = 8 h were used to quantify DNA signals. Data are the mean ± SD (N = 3). Significance was examined using an unpaired Student's t-test (*P < 0.05).
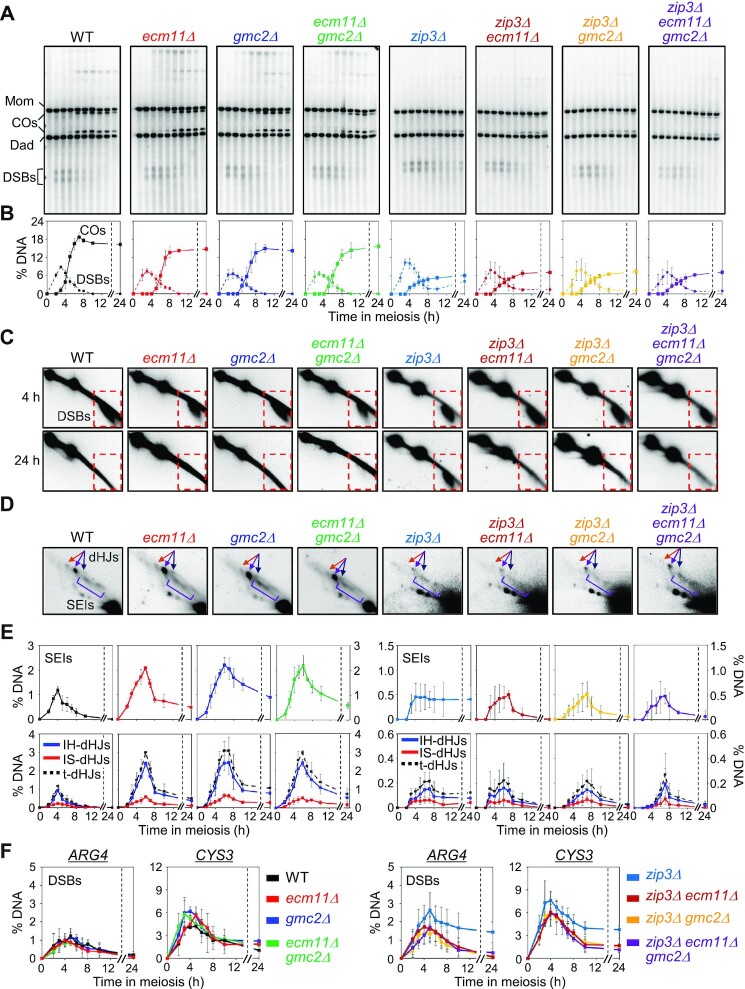
Figure 6.
Ecm11 and Gmc2 inhibit additional DSB formation in zip3Δ cells. (A) Representative Southern blot images of 1D gel analysis of WT, ecm11Δ, gmc2Δ, ecm11Δ gmc2Δ, zip3Δ, zip3Δ ecm11Δ, zip3Δ gmc2Δ, and zip3Δ ecm11Δ gmc2Δ mutants assessed at 0, 2.5, 3.5, 4, 5, 6, 7, 8, 10 and 24 h. (B) Quantitative analysis of DSBs and COs. The plots for WT, ecm11Δ, gmc2Δ, and ecm11Δ gmc2Δ are from Figure 2D. Data are the mean ± SD (N = 3). (C) 2-D gel detection of DSB formation at 4 h and 24 h. Dashed squares indicate DSB regions. (D) Representative images of 2D gel analysis. (E) Quantification of SEIs and dHJs. The plots for WT, ecm11Δ, gmc2Δ, and ecm11Δ gmc2Δ are from Figure 3B. Data are the means ± SD (N = 3 for WT, ecm11Δ, gmc2Δ, ecm11Δ gmc2Δ, zip3Δ ecm11Δ and zip3Δ ecm11Δ gmc2Δ; N = 4 for zip3Δ and zip3Δ gmc2Δ). IH-dHJ, interhomolog double-Holliday junction; IS-dHJ, intersister double-Holliday junction; SEI, single-end invasion; t-dHJs, total dHJs. (F) Quantitative analysis of DSB at the ARG4 and CYS3 loci. The data indicate the means ± SD (N = 3).
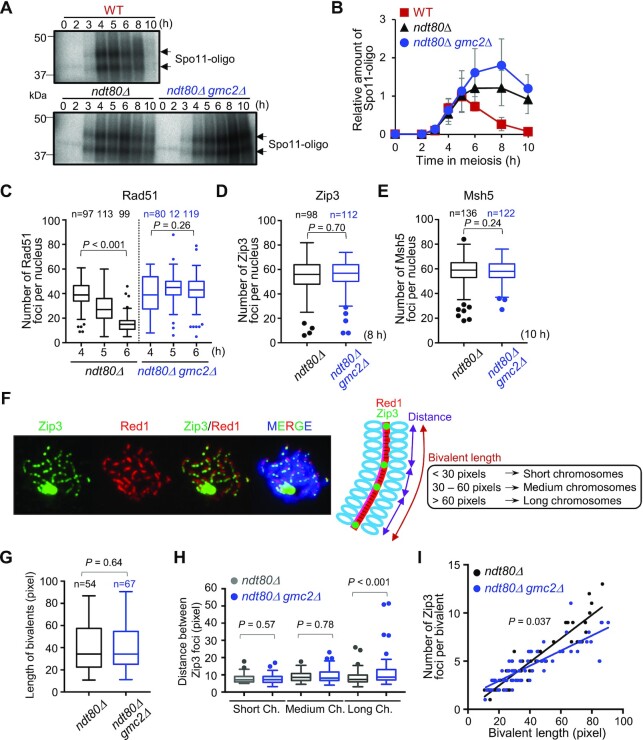
Figure 5.
DSB formation and Zip3 distribution in WT and gmc2Δ cells. (A) Representative image of 32P-labeled DNA fragments covalently bound to Spo11-3FLAG in immunoprecipitates from WT, ndt80Δ, and ndt80Δ gmc2Δcells at the indicated time points. (B) Relative DNA fragment signals at each time point. Relative amounts of Spo11-oligo complex were determined as described in the Materials and Methods section. Data are the mean ± SD (N = 3). (C) Average number of Rad51 foci per nucleus at the indicated time points in the ndt80Δ and ndt80Δ gmc2Δmutants. The number of nuclei at each time point is shown at the top. (D) Average number of Zip3 foci per nucleus in the ndt80Δ and ndt80Δ gmc2Δ mutants. The number of nuclei at each time point is shown at the top. (E) Average number of Msh5 foci per nucleus in the ndt80Δ and ndt80Δ gmc2Δ mutants. The number of nuclei counted at each time point is shown at the top. (F) Representative image of a meiotic nuclear spread from ndt80Δ cells at 8 h post-meiosis entry. The cells were co-stained for anti-Red1 (red) and anti-Zip3 (green). A schematic explanation of the classification into bivalent length categories is shown. (G) Comparison of the distribution of bivalent length between ndt80Δ and ndt80Δ gmc2Δmutants. (H) Distribution of distances between adjacent Zip3 foci on short, medium, and long bivalents. Data are the mean ± SD (N ≥ 3). The data were analyzed using the Mann−Whitney U-test, and the results are shown in panels G and H. Boxplots in C, D, E, G and H show medians and interquartile ranges (IQRs), and the whiskers indicate the values that are 1.5 times the IQR, based on the Tukey method. (I) Correlations between the total numbers of Zip3 foci on each bivalent and bivalent length in WT and gmc2Δ strains. P-values were determined using the Wald−Wolfowitz runs test.
RESULTS
A gmc2 mutant shows hyper-recombination on chromosome VII
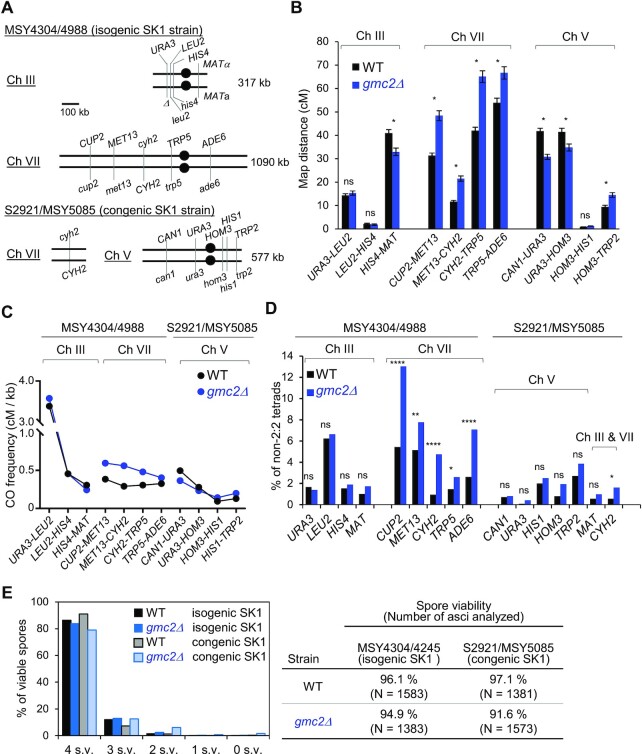
Previous studies have indicated that the EG complex is necessary for efficient Zip1 assembly, promoting synapsis of homologous chromosomes during meiotic prophase (45–47). Further, genetic analysis of ecm11Δ and gmc2Δ mutants of the yeast strain BR1919-8B revealed that CO frequencies increase within intervals of chromosomes III and VIII (48). Therefore, we first analyzed the frequencies of COs and non-Mendelian segregation on chromosome III (317 kb), including the HIS4LEU2 artificial hotspot, and chromosome VII (1090 kb) in the SK1 background (49), which shows some genetic differences from the BR strain. Similarly, we analyzed COs and non-Mendelian segregation on chromosome V (577 kb) in a congenic SK1 strain (Figure 1A). Consistent with the findings of Voelkel-Meiman et al. (49), gmc2Δ exhibited increased CO frequencies in four intervals of chromosome VII, but not any in interval of chromosome III (Figure 1B; Supplementary Table S2). In contrast, CO frequencies on intervals of chromosome III and on one interval of chromosome V in the gmc2Δ mutant were similar to those in the WT (Figure 1B). One interval on chromosome III and two intervals on chromosome V in the gmc2Δ mutant had significantly lower CO frequencies than their WT counterparts. The increase in CO frequencies along chromosome VII in the gmc2Δ mutant was more prominent than those on chromosomes III and V (Figure 1C). All five loci on chromosomes in the gmc2Δ mutant showed increased frequencies of non-Mendelian segregation, whereas none of the nine loci on chromosomes III and V did (Figure 1D; Supplementary Table S3). When CO interference was examined based on nonparental ditype (NPD) ratios, all intervals on chromosome VII of the gmc2Δ mutant showed decreased CO interference (increased NPD ratios) compared to those of the WT (Supplementary Figure S1). We observed similar results of weakened CO interference in the gmc2Δ mutant when we used the method described by Malkova et al. (58) to evaluate changes in CO interference (Supplementary Figure S2).
Figure 1.
The Ecm11–Gmc2 (EG) complex regulates meiotic recombination in a chromosome-dependent manner. (A) Schematic representation of the location of marker genes on chromosomes III and VII in the MSY4304/4998 diploid and chromosomes VII and V in the S2921/MSY5085 diploid. (B) Map distances within each indicated genetic interval of chromosomes III, VII, and V in the WT (black) and gmc2Δ (blue) strains determined using the Perkins equation. Error bars show standard errors (SEs). The SEs for map distances were calculated using the Stahl Lab online tool (https://elizabethhousworth.com/StahlLabOnlineTools/). The asterisks indicate significant differences between map distances in WT and gmc2Δ tetrads (see also Supplementary Table S2). ns, not significant. (C) CO frequencies (cM) per physical length (kb) of each genetic interval of chromosomes III, VII, and V in the WT and gmc2Δstrains. (D) Frequencies of non-Mendelian segregation of the indicated genetic loci in tetrads of WT and gmc2Δ strains. (E) Distribution of viable spores per tetrad (left) and spore viability in each strain (right).
We also determined the effect of gmc2Δ in spore viability of isogenic and congenic SK1 strains. The WT produced 96.1% and 97.1% viable spores for isogenic and congenic SK1, respectively. Spore viabilities were slightly reduced in the gmc2Δ mutant: 94.9% for isogenic SK1 and 91.6% for congenic SK1 (Figure 1E). Taken together, these results imply that the EG complex plays a role in regulating the frequencies and distribution of COs, which may be specific to chromosomal properties, such as length.
The EG complex is not required for DSB formation, but is necessary for CO-specific recombination at HIS4LEU2
To investigate the roles of the EG complex in meiotic recombination further, we used ecm11Δ/gmc2Δ single and double mutant strains to analyze recombination intermediates and products at the HIS4LEU2 locus on chromosome III, which contains a well-controlled single DSB site (Figure 2A and B). In WT cells, meiotic division was efficient, yielding ≥98% nuclei at 24 h (Figure 2C). The ecm11Δ and gmc2Δ mutants showed a substantial ∼2-h delay in the progression of meiotic division, with ∼85% of the cells undergoing meiosis at 24 h after the initiation of meiosis (Figure 2C).
In WT meiosis, DSBs appeared and disappeared, followed by the formation of CO products. The number of DSBs in the WT peaked at 3.5 h, and DSBs were completely processed after ∼8 h (Figure 2B and D). The frequencies of CO-I and NCO-I were ∼5.0% and 3.7%, respectively (Figure 2E and G). In the ecm11Δ and gmc2Δ mutants, meiotic DSBs at HIS4LEU2 formed at WT levels with normal post-DSB progression. DSB kinetics were similar between the WT and ecm11Δ/gmc2Δ strains, with respect to timing and peak levels (Figure 2D). The ecm11Δ and gmc2Δ single mutants and the ecm11Δ gmc2Δ double mutants formed COs (total COs in Figure 2D) at 14.8 ± 0.7%, 14.9 ± 1.2% and 15.6 ± 0.8%, respectively, whereas COs formed at a frequency of 18.6 ± 1.5% in the WT strain, indicating a modest reduction in CO frequency in the ecm11Δ and gmc2Δ mutant strains. Moreover, CO formation in the mutants exhibited a delay of ∼1.75 h compared to that in the WT (Figure 2D). However, NCO-1 levels in the ecm11Δ and gmc2Δ mutants were slightly higher than those in WT cells (∼4.4% in ecm11Δ and gmc2Δ mutants versus 3.7% in WT) (Figure 2E and G). Therefore, NCO formation progressed normally, but CO-fated DSB repair was defective in the mutants.
In a rad50S strain background, DSBs occurred in ∼20% of the chromatids at the HIS4LEU2 locus and failed to progress to recombination (Figure 2F; Supplementary Figure S3). In the rad50S background, the ecm11Δ and gmc2Δ mutants exhibited similar levels of DSBs at the HIS4LEU2 locus, which was also observed in a dmc1Δ background, in which meiotic DSB repair is blocked (Supplementary Figure S4). These findings indicate that the mutations do not affect early DSB formation at the HIS4LEU2 locus.
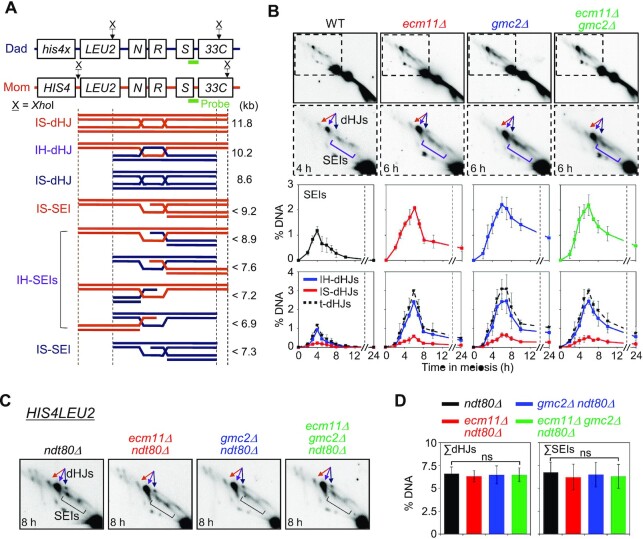
To determine whether the EG complex is required for JM formation, we analyzed SEIs and dHJs through native/native 2D gel electrophoresis followed by Southern hybridization. This analysis can also be used to distinguish between IH-JMs and intersister (IS)-JMs based on size differences (Figure 3A). In WT meiosis, SEIs and dHJs were detectable at 3.5 h and reached peak levels at 4 h, with an IH:IS dHJ ratio of ∼5:1. In the ecm11Δ and gmc2Δ mutants, SEIs and dHJs were observed at 3.5 h, similar to the WT, and peaked at 6 h, showing a 2-h delay in peak levels compared to the WT (Figure 3B; Supplementary Figure S5). Although steady-state levels of SEIs and dHJs were higher in the ecm11Δ, gmc2Δ and ecm11Δ gmc2Δ cells than in the WT between 5 and 8 h, the majority of these JMs disappeared after 8 h (Figure 3B). However, unresolved SEI and dHJ species persisted in the ecm11Δ and gmc2Δ mutants at later time points and may have caused a defect in CO formation. The ecm11Δ and gmc2Δ mutants exhibited an IH:IS dHJ ratio of ∼5.5:1, indicating normal IH bias in the mutants (Figure 3B). Overall, these results suggest that the ecm11Δ, gmc2Δ, and ecm11Δ gmc2Δ mutants show both normal DSB-SEI transition and normal IH-bias but may have defects in SEI-to-dHJ transition and/or dHJ resolution at the HIS4LEU2 locus. Alternatively, these mutants may have formed more SEIs and dHJs, possibly due to more frequent DSB formation than that observed in the WT.
Figure 3.
Two-dimensional (2D) gel analysis of ecm11Δ, gmc2Δ and ecm11Δ gmc2Δ in WT and ndt80Δ backgrounds. (A) Physical map of the HIS4LEU2 locus. IH-dHJ, interhomolog double-Holliday junction; IS-dHJ, intersister double-Holliday junction; SEI, single-end invasion. (B) Representative images of 2D analysis of WT, ecm11Δ, gmc2Δ, and ecm11Δ gmc2Δ strains (top). Quantitative analysis of SEIs and dHJs (bottom). Data are the mean ± SD (N = 3). Full images in Supplementary Figure S5. t-dHJs, total dHJs. (C) Representative 2D analysis images of the HIS4LEU2 locus in the ndt80Δ, ecm11Δ ndt80Δ, gmc2Δ ndt80Δ and ecm11Δ gmc2Δ ndt80Δ strains. (D) Quantitative analysis of dHJs and SEIs in an ndt80Δ background at the HIS4LEU2 locus. Gel images at t = 8 h were used to quantify DNA signals. Data are the mean ± SD (N = 3). Significance was examined using an unpaired Student's t-test (ns, not significant).
To distinguish between these two possibilities, we counted total dHJs in the ecm11Δ and gmc2Δ mutants in an ndt80Δ background, in which meiotic cells are arrested in middle pachytene, resulting in unresolved SEIs and dHJs (12,59). Total dHJ levels at the HIS4LEU2 locus in the ecm11Δ ndt80Δ, gmc2Δ ndt80Δ, and ecm11Δ gmc2Δ ndt80Δ mutants were similar to those in the ndt80Δ mutant (6.4 ± 0.8% in ndt80Δ; 6.2 ± 0.8% in ndt80Δ ecm11Δ; 6.5 ± 1.3% in ndt80Δ gmc2Δ; and 6.4 ± 1.0% in ndt80Δ ecm11Δ gmc2Δ; Figure 3C and D). Thus, we conclude that the EG complex plays a positive role in JM resolution, rather than in the regulation of DSB frequencies at the HIS4LEU2 locus.
The ecm11Δ and gmc2Δ mutants exhibit a locus-specific defect in DSB processing
Because the effects of ecm11Δ and gmc2Δ mutations on CO levels differed between chromosomes III and VII (Figure 1), we further analyzed meiotic recombination at the ERG1 locus, which is a natural hotspot on chromosome VII (15). In contrast to the HIS4LEU2 locus, the ecm11Δ and gmc2Δ mutants exhibited >2-fold increases in both CO and NCO frequencies at the ERG1 locus compared to the WT (Figure 4A−E). We then monitored JM formation at the ERG1 locus using 2D gel electrophoresis and quantified the levels of JM species from parallel ecm11Δ, gmc2Δ, and WT cultures. JMs appeared with normal timing in the ecm11Δ and gmc2Δ mutants; however, the peak levels of SEIs and dHJs were ∼3-fold higher in the ecm11Δ and gmc2Δ mutants than those in WT (Figure 4F and G; Supplementary Figure S6). We then further analyzed JM formation at the ERG1 locus in the ndt80Δ background. Interestingly, dHJ levels were increased from 2.9 ± 0.7% in ndt80Δ to 5.0 ± 0.8%, 5.6 ± 0.9% and 5.3 ± 0.9% in the ndt80Δ ecm11Δ, ndt80Δ gmc2Δ and ndt80Δ ecm11Δ gmc2Δ mutants, respectively (Figure 4H and I). Thus, in the ndt80Δ background, the ecm11Δ and gmc2Δ mutants showed ∼1.7–1.9-fold higher dHJ levels than the control. These findings indicate that more DSBs are formed at ERG1 in the ecm11Δ and gmc2Δ mutants (Supplementary Figure S7), but not at HIS4LEU2, resulting in higher JM levels at ERG1 in the ndt80Δ background. Furthermore, both hotspots exhibited increased JM steady-state levels at peak and late time points, indicating delays in second-end capture and/or dHJ resolution.
Early DSB formation is not affected by the absence of the EG complex
As suggested above, the elevated levels of meiotic recombination in the ecm11Δ and gmc2Δ mutants may have been caused by an increase in initiation events associated with DSB formation (Figure 4). Therefore, we investigated whether the increased recombination level was caused by the increased DSB formation rate in the ecm11Δ and gmc2Δ mutants. Through immunofluorescence analysis of chromosome spreads, we counted foci of recombination proteins, such as Rad51 and Dmc1, as well as the number of foci of ZMM proteins, such as Zip3 (Supplementary Figure S8). Foci formation by Rad51/Dmc1 and Zip3 in the ecm11Δ and gmc2Δ mutants began in a manner similar to that in the WT strain. However, the numbers of foci were higher at 5 h in the ecm11Δ and gmc2Δ mutants than in the WT strain. While this is consistent with a longer duration of DSB formation, we cannot distinguish whether this accumulation of Rad51/Dmc1 foci is caused by additional DSB formation or by inefficient turn over.
We also checked DSB levels at loci on chromosomes that are smaller than chromosome VII, including CYS3 (chromosome I), ARG4 (chromosome VIII), and BUD23 (chromosome III), in the rad50S and dmc1Δ backgrounds (Figure 2F; Supplementary Figures S3 and S4). The ecm11Δ and gmc2Δ mutants showed DSB levels similar to those in the control strains at these three loci. These findings suggest that the EG complex does not play a role in DSB formation in the early meiotic prophase I. Alternatively, the experimental approach may not have been appropriate to evaluate the homolog engagement-mediated suppression of DSB formation, because rad50S and dmc1Δ block recombination before the EG complex acts.
The EG complex restricts prolonged DSB formation independently of Ndt80
It has been reported that homolog engagement suppresses DSB formation in late prophase I (15). Additionally, Ndt80-mediated pachytene exit has been reported to down-regulate DSB formation, which is independent of homolog engagement suppression (15). We determined DSB levels in the absence of Ndt80 by quantifying Spo11-oligo complexes in the gmc2Δ background, which may be related to a homolog-engagement defect in this background (45–49). In WT, Spo11-oligos appeared with a peak at 5 h, and then disappeared (Figure 5A and B). The ndt80Δ mutant exhibited persistent Spo11-oligos at late time points, consistent with a previous report (15). Notably, ndt80Δ gmc2Δ cells had increased levels of Spo11-oligos, i.e. ∼1.7-fold level in the ndt80Δ single mutant at 8 h (Figure 5A and B). Consistent herewith, Keeney and colleagues reported increased steady-state levels of Spo11-oligos, particularly a later time points, in gmc2Δ and ecm11Δ mutants in a WT background (19). These findings suggest that DSB formation occurs at a higher rate in the late prophase I in gmc2Δ, which may be related to the homolog-engagement defect in this mutant, and this phenomenon appears to be independent of Ndt80-mediated pachytene exit.
Sgs1 is a RecQ family DNA helicase that reportedly suppresses aberrant crossing over (60). Mutations in sgs1 promote the joining of homolog axes in synapsis-defective mutants in a process called pseudo-synapsis (61). The sgs1-Δ200 truncation mutant lacks the helicase and RNase D C-terminal (HRDC) domain, but shows normal vegetative growth (62). We found that the sgs1-Δ200 mutation suppressed synapsis defects (Supplementary Figure S9A and B). We then evaluated whether pseudo-synapsis could suppress DSB formation in the gmc2Δ mutant. We found that the Spo11-oligo complex formation was increased by ∼1.7-fold in ndt80Δ gmc2Δ sgs1-Δ200 compared to that in the ndt80Δ gmc2Δ control (Supplementary Figure S9C). These results suggest that the role of the EG complex in the suppression of DSB formation could not be ameliorated by pseudo-synapsis.
The EG complex regulates COs on long chromosomes
The increase in Spo11-oligo levels observed in ndt80Δ gmc2Δ cells implied that the EG complex plays a role in late DSB formation in meiotic prophase I. To determine the roles of the EG complex on pachytene chromosomes, we analyzed CO formation and synapsis at meiotic prophase I in the absence of Ndt80 and/or Gmc2 by immunostaining of chromosome spreads. The number of Rad51 foci was slightly increased and was maintained at the late meiotic prophase in the ndt80Δ gmc2Δ mutant compared with that in the ndt80Δ mutant (Figure 5C). We also visualized Zip3 and Msh5 localization to detect CO-designated sites in the late prophase in the ndt80Δ background. Zip3 and Msh5 foci counts were similar in the ndt80Δ and the ndt80Δ gmc2Δ mutants in the late prophase (Figure 5D and E). Although more DSBs were produced in the gmc2Δ mutant in the late meiotic prophase I (Figure 5B), the increase in the number of DSBs did not contribute to the total number of Zip3/Msh5-dependent recombination intermediates in the ndt80Δ background. Thus, the additional DSBs in the mutant are not correlated with an increased number of Zip3 or Msh5 foci.
To examine the role of the EG complex in regulating CO control in a chromosome size-dependent manner, we measured bivalent length in the ndt80Δ background by staining Red1, which localized to the chromosome axis at prophase I (Figure 5F). The total bivalent length indicated by Red1 lines in a single spread in ndt80Δ gmc2Δ was similar to that in ndt80Δ (P = 0.64; Figure 5G). This observation implies that normal axis formation occurs in the absence of Gmc2. We then measured the distance between two adjacent Zip3 foci on a bivalent (Figure 5H) and counted the Zip3 foci per bivalent (Figure 5I). Chromosome length was classified according to detectable Red1 signals, as follows: (i) short chromosomes (<30 pixels), (ii) medium chromosomes (30–60 pixels) and (iii) long chromosomes (>60 pixels) (Figure 5H and I). The short- and medium-length chromosomes displayed similar distances between Zip3 foci and had comparable numbers of foci per bivalent in the ndt80Δ and ndt80Δ gmc2Δ mutants (P = 0.57 and P = 0.78; respectively). Importantly, long chromosomes exhibited increased inter-Zip3 distances in ndt80Δ gmc2Δ (more variation) compared with those in ndt80Δ (P < 0.001; Figure 5H). Furthermore, when the numbers of Zip3 foci per bivalent were plotted against chromosome length, the long chromosomes in ndt80Δ gmc2Δ showed reduced Zip3 counts compared to the control (Figure 5I). This reduction suggests that Zip3-dependent events are less frequent on long chromosomes than on chromosomes of other sizes in ndt80Δ gmc2Δ. Therefore, we hypothesize that the high levels of COs on long chromosomes were caused by additional DSB formation (20), which may be processed in a Zip3-focus-independent pathway.
Absence of the EG complex suppresses the DSB turnover defect in zip3 mutants
The EG complex extensively colocalizes with Zip3, a SIC component, between aligned chromosomes at early prophase I (45). To explore the role of the EG complex in regulating meiotic recombination further, DSB turnover and JM processing in the ecm11Δ and gmc2Δ mutants were determined in a zip3Δ background. In zip3Δ cells, some DSBs persisted at ∼40% of the maximum levels at 10–24 h at the HIS4LEU2 locus (Figure 6A−C), which is consistent with the findings of a previous report (22). 2D gel analysis revealed that the peak levels of both SEIs and dHJs in zip3Δ were ∼5-fold lower than those in the WT, irrespective of the ecm11Δ and gmc2Δ mutation status, and residual dHJs were detected at 24 h in zip3Δ (Figure 6D and E). In contrast, DSBs were detected at very low levels at 10–24 h and JMs were not detected in the ecm11Δ zip3Δ, gmc2Δ zip3Δ, and ecm11Δ gmc2Δ zip3Δ mutants at 24 h (Figure 6B and E). Consistent with the levels of JM processing, the CO levels were slightly higher in the ecm11Δ zip3Δ, gmc2Δ zip3Δ, and ecm11Δ gmc2Δ zip3Δ mutants than those in the zip3Δ single mutant (Figure 6B). The additional DSBs may not be processed into dHJs in zip3Δ, whereas they progressed to COs in the ecm11Δ and gmc2Δ mutants. Therefore, the EG complex seems to promote delays in DSB turnover and/or JM processing in the zip3Δ mutant. However, it is unclear whether in the ecm11Δ/gmc2Δ zip3Δ double mutants, fewer additional DSBs are formed and/or DSBs progress more efficiently to COs. Further research is required to distinguish between these possibilities.
The ecm11Δ and gmc2Δ mutants in the zip3Δ background exhibited more efficient meiotic progression than the zip3Δ single mutant (Supplementary Figure S10A), implying that the effects of zip3Δ are upstream of the effects of ecm11Δ and gmc2Δ. Thus, absence of the EG complex partly ameliorated the defect in recombination progression caused by the absence of Zip3. Similar suppression of delayed turnover of DSBs in the zip3Δ mutant by ECM11 and/or GMC2 deletion was observed at natural hotspots, including the ARG4 and CYS3 loci (Figure 6F; Supplementary Figure S10B and C). Taken together, these results indicate that the EG complex can suppress recombination in the absence of Zip3.
COs are reduced in mus81Δ and mus81Δ ecm11Δ cells
The XPF family endonuclease Mus81–Mms4 has been implicated in facilitating JM resolution to process recombination in the class II interference-independent pathway (30,31). To examine the combined roles of the EG complex and Mus81-Mms4 in CO formation, we compared ecm11Δ and/or mus81Δ single and double mutant strains. In both the ecm11Δ and mus81Δ mutants, meiotic division progression was delayed by ∼2 h (Supplementary Figure S11A). However, the ecm11Δ mus81Δ mutant exhibited a significant delay, with ∼70% of the cells undergoing the meiotic cell cycle (Supplementary Figure S11A). The ecm11Δ mus81Δ double mutant exhibited a synergistic defect in spore viability (Supplementary Figure S11B). Physical analysis showed that the maximum levels of CO were significantly reduced in ecm11Δ mus81Δ cells (76% of the WT level) compared to those in either single mutant (86% and 92% of the WT level in ecm11Δ and mus81Δ, respectively) (Supplementary Figure S11C and D). These findings indicate that the loss of the functions of both Ecm11 and Mus81 causes an additive reduction in the CO levels, suggesting that the EG complex and Mus81 are involved in CO formation via distinct pathways. Further, JM resolution was more delayed in the ecm11Δ mus81Δ mutant than in either single mutant (Supplementary Figure S11E and F), supporting that Mus81-dependent processing of JMs is predominant in the absence of the EG complex.
Low temperature suppresses the delay in recombination progression in ecm11Δ and gmc2Δ mutants
A previous study has shown that the effects of mutations in meiotic recombination are modulated by incubation temperature (22). Thus, we wondered whether temperature would affect the recombination defects in the ecm11Δ and gmc2Δ mutants and evaluated the mutant phenotypes at a low temperature of 23°C (Supplementary Figure S12). Similar to the findings at 30°C, the ecm11Δ and gmc2Δ mutants exhibited reduced CO levels at the HIS4LEU2 locus at 23°C, without affecting NCOs. At 23°C, JMs accumulated at high levels in the ecm11Δ and gmc2Δ mutants, but disappeared at later time points (Supplementary Figure S12). This indicates that low temperatures partially suppress the formation of JM processing-associated defects in these mutants. In other words, the kinetic barrier imposed by the EG complex is sensitive to temperature.
DISCUSSION
The EG complex is a component of the SC central region and plays a role in its initiation and elongation. In the current study, we characterized ecm11Δ and gmc2Δ mutants, whose phenotypes provided new insights into the control of DSB formation and the role of the SC central region in CO formation.
EG complex promotes the ZMM-dependent CO pathway
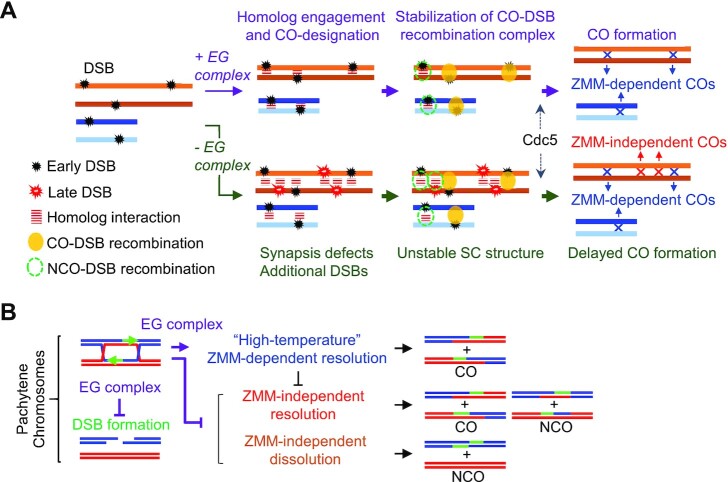
In normal meiosis, CO/NCO differentiation occurs at early meiotic prophase I (22,41). CO-designated DSBs are processed into JMs such as SEIs and dHJs. The dHJs are subsequently subjected to biased resolution into CO products. These JM-processing events are highly regulated in a meiosis-specific program, and are also associated with the following morphological changes in the chromosomes: (a) meiotic DSBs occur in tethered chromosome axis/loop complexes; (b) interaxis bridges formed by DSB/template interaction occur in the axis/loop complexes, suggesting that one DSB end is released from the axis to form a homology-searching DNA ‘tentacle’ and (c) the SC forms between aligned pairs of homologs (41,63,64). Meiosis-specific ZMM proteins play a major role in the formation of JM as a CO-specific precursor (22). Additionally, in meiotic cells, there are minor mitotic-processing pathways of JMs, which are resolved into either COs or NCOs, or dissolved into NCOs (65,66). Previous studies have suggested that the mitotic-like Sgs1-dependent resolution of JMs is induced at the time of pachytene exit (66,67). Physical analyses of meiotic recombination in this study showed that dHJ-to-CO transition was significantly delayed in the ecm11Δ and gmc2Δ mutants at 30°C, but not at 23°C. Interestingly, the DSB-JM transition in these mutants appeared normal, which indicates that the EG complex is not required for early ZMM-dependent JM processing (Figure 7A). This idea is supported by the fact that the ecm11Δ and gmc2Δ mutations do not affect NCOs, whose frequencies are indirectly affected by early ZMM functions that mediate D-loop stabilization to promote SEI formation (15,19,22). As such, the EG complex appears to be a positive modulator of late ZMM functions that promote the resolution of JMs to COs (Figure 7B).
Figure 7.
Roles of the EG complex in a feedback mechanism linked to DSB number and ZMM-dependent CO formation. (A) Proposed mechanism of chromosome synapsis-dependent feedback based on ecm11 and gmc2 mutant phenotypes. The EG complex facilitates Zip1 chromosomal assembly (45) and modulates the meiotic recombination frequency and distribution through chromosome synapsis-dependent feedback. Delay of JM resolution in the absence of the EG complex may be explained by a pathway in which unstable SC structures cause defective CO-fated recombination. The SC central region provides an environment for correct recombination processing through phase separation. (B) Model of the roles of the EG complex in feedback control of DSB formation and the ZMM-dependent crossover pathway.
We further showed that the ecm11Δ and gmc2Δ mutations suppressed the JM processing defect in the zip3Δ mutant. The CO levels in the ecm11Δ zip3Δ and gmc2Δ zip3Δ mutants increased slightly, compared to those in the zip3Δ single mutant. Given that the ecm11Δ zip3Δ and gmc2Δ zip3Δ mutants showed lower steady-state levels of JMs than either the ecm11Δ or the gmc2Δ mutant, most COs in the double mutants might not form through ZMM-mediated JMs. However, whether the persistence of a small fraction of dHJs in zip3Δ indicates a late ZMM function or is a consequence of progression along an unknown pathway remains to be determined. Moreover, JMs created through the ZMM pathway can be resolved by the mitotic endonucleases Mus81-Mms4/Yen1 to produce both COs and NCOs (30,65−67). In the absence of the EG complex in a WT background, even with delayed JM processing, about two-thirds of the JMs were resolved during late meiosis. This resolution in the mutants may be independent of the functions of ZMM. Indeed, we showed that ecm11Δ mus81Δ exhibited a significant delay in JM resolution compared to the single mutants; however, DSB turnover was similar among the WT strain, single mutants, and double mutants. Based on these results, we hypothesize that probable mitotic resolvases more actively process JMs via the ZMM-independent pathway in the absence of the EG complex than in its presence (Figure 7B). In other words, the EG complex may limit the activity of mitotic-like JM-processing enzymes not only in the absence of ZMM proteins but also in their presence. Further, it is noteworthy that the excess COs in ecm11Δ are MutSγ-dependent, implying that MutSγ-mediated COs may be limited by the presence of Ecm11 and Gmc2 (49).
The EG complex suppresses DSB formation in late prophase I
Genetic analysis revealed that CO frequencies on chromosome VII were increased in the gmc2Δ mutant, which is consistent with the results of a previous genetic analysis of chromosome VIII (49). This finding was supported by the results of physical analysis of the ERG1 locus on chromosome VII, which showed that the CO levels were increased in both the ecm11Δ and gmc2Δ mutants. The increase in recombination events may simply be due to increased instances of recombination initiation. At the HIS4LEU2 locus, reduced CO and normal NCO levels were observed in the ecm11Δ and gmc2Δ mutants. Importantly, the levels of JMs at the HIS4LEU2 locus in the ecm11Δ and gmc2Δ mutants in the ndt80Δ background were similar to those in parental ndt80Δ cells. Given that the mutants are defective in the processing of CO-designated DSBs, it is likely that the levels of ‘early’ DSBs on chromosome III in the mutants are similar or slightly increased compared to those in the WT. Indeed, when DSB levels at five loci on different chromosomes were measured in repair-deficient mutants, such as rad50S, we observed similar levels of DSBs between the WT, ecm11Δ, and gmc2Δ strains. This finding strongly suggests that the frequencies of early-forming DSBs are not affected in the absence of the EG complex (Figure 7A). We next analyzed the levels of the Spo11-oligo complex. This complex is a byproduct of Spo11-mediated DSB formation; thus, Spo11-oligo complex levels are proportional to DSB frequencies. We found that in ndt80Δ and ndt80Δ gmc2Δ mutants with pachytene arrest, the levels and kinetics of early Spo11-oligos were similar. In contrast, the increase in steady-state levels of Spo11-oligo at late meiotic prophase I was higher in the ndt80Δ gmc2Δ mutant than in the ndt80Δ mutant. This indicates that during late prophase I, more DSBs are formed in cells with defective SC central regions.
Homeostatic regulation of DSB formation
Previous studies have shown that Spo11-oligo levels are increased in mutants defective in synapsis (e.g. zmm mutants), suggesting that homolog engagement suppresses DSB formation as a negative feedback control (15,19,68). Phosphorylation of Zip1 at the C-terminal globular domain is required for chromosome synapsis and is known to suppress extra DSBs in a chromosome size-dependent manner (20). Our findings indicate that the loading of ZMM proteins on meiotic chromosomes is not directly involved in DSB suppression, as the ecm11Δ and gmc2Δ mutants showed normal Zip3/Msh5 foci formation. This indicates that Zip3/Msh5 recruitment to the SIC does not directly suppress DSB formation. Rather, SC elongation suppresses additional DSB formation in late prophase I as a feedback mechanism. In addition to the suppression by homolog engagement, DSB formation is negatively regulated by Ndt80 and recombination checkpoint kinases, such as Tel1ATM (15,69). We hypothesize that delayed JM processing in the ecm11Δ and gmc2Δ mutants induces recombination checkpoints to downregulate Ndt80-dependent pachytene exit. This is supported by the observation that in the ecm11Δ and gmc2Δ mutants, inducing the expression of Cdc5, which is activated by Ndt80 and triggers exit from pachytene, led to immediate resolution of JMs and a rapid increase in COs (MS Lee, unpublished data). As the Spo11-oligo levels were more elevated in the ndt80Δ gmc2Δ mutant than in the ndt80Δ mutant, EG complex-dependent suppression of late DSBs appears to work independently of Ndt80. Activation of Tel1ATM-dependent feedback control may explain the increased levels of DSBs in the ecm11Δ and gmc2Δ mutants (69,70). However, this is not very likely, as the levels of Spo11-oligos do not increase in the mutants in early meiosis, during which Tel1ATM is activated (69,70). Moreover, in the background of rad50S mutation, which robustly activates Tel1ATM kinase activity (69), no increase in DSB levels was observed in the ecm11Δ and gmc2Δ mutants.
During late prophase I, SC elongation mediates axis remodeling (18). The axis proteins Red1 and Hop1 are required for efficient DSB formation and are abundantly present on chromosomes in zmm mutants (41,42,44,45). Furthermore, we observed increased DSB levels in the ndt80Δ mutant, which forms a full-length SC with Red1/Hop1 (71) (Figure 5F). Potentially, Red1/Hop1 proteins persistently present on the chromosomes in the ecm11Δ and gmc2Δ mutants induce additional DSBs (45) (Figure 5F and G). Therefore, EG complex-dependent suppression of DSB formation is likely to occur through the removal of Red1/Hop1. Additionally, the meiotic DSB-forming machinery may be functionally suppressed in the context of a full-length SC and in the presence of the central regions.
Role of the SC central region in CO control
Synapsis-dependent suppression of DSB formation may explain the increased recombination levels observed on long chromosomes in the mutants. We speculate that late-forming DSBs along non-synaptic chromosomes are processed through a ZMM-independent recombination pathway that produces non-interfering COs and NCOs (Figure 7A). The gmc2Δ mutant showed reduced CO interference compared to the WT in the genetic assays. In contrast, these mutants appeared to produce WT-like levels of Zip3 foci on the chromosomes. The compromised CO interference in the mutants might be simply explained by the formation of non-interfering COs in addition to adequate levels of interfering COs. The fact that the gmc2Δ mutant retained significant CO interference with the WT number of Zip3 foci suggests that the establishment of CO interference is implemented in the absence of the matured SC central region, and consequently in the absence of SC elongation or polymerization. Thus, our results suggest that SC polymerization and/or the SC per se is not necessary for CO interference, which is consistent with previous reports (24,72,73).
CONCLUSION
Meiotic recombination plays important roles in chromosomal processes to mediate homolog pairing during prophase I and to ensure correct homolog segregation in the first meiotic division (63,64,74). Little is known about the role of the SC central region in meiotic recombination. In this study, by analyzing the role of the EG complex in meiotic recombination, we found that the EG complex in the SC central region is involved in multiple events that control recombination processes (Figure 7A), which ensure meiosis-specific properties, such as the regulated formation of interfering COs. The roles of the EG complex in controlling recombination include ZMM-dependent JM processing, JM resolution, and homeostatic regulation of meiotic DSB formation during late prophase I (Figure 7B). Based on our findings, we suggest that a compartment of the SC central region, which likely mediates phase separation (75), functions to sequester ZMM-dependent and -independent recombination proteins in the region. Further, we suggest that the SC central region allows shuttling of the DSB-forming machinery out of the region and that this process is mediated by the EG complex and transverse filament Zip1.
DATA AVAILABILITY
All data generated in this study are included in the published article and its supplementary data.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Scott Keeney and Hajime Murakami for personal communication of unpublished data. We are particularly thankful to Nancy Kleckner, Scott Keeney and Neil Hunter for providing the yeast strains.
Contributor Information
Min-Su Lee, Department of Life Science, Chung-Ang University, Seoul 06974, South Korea.
Mika T Higashide, Institute for Protein Research, Osaka University, Osaka 565-0871, Japan.
Hyungseok Choi, Department of Life Science, Chung-Ang University, Seoul 06974, South Korea.
Ke Li, Institute for Protein Research, Osaka University, Osaka 565-0871, Japan; Graduate School of Agriculture, Kindai University, Nara 631-8505, Japan.
Soogil Hong, Department of Life Science, Chung-Ang University, Seoul 06974, South Korea.
Kangseok Lee, Department of Life Science, Chung-Ang University, Seoul 06974, South Korea.
Akira Shinohara, Institute for Protein Research, Osaka University, Osaka 565-0871, Japan.
Miki Shinohara, Institute for Protein Research, Osaka University, Osaka 565-0871, Japan; Graduate School of Agriculture, Kindai University, Nara 631-8505, Japan; Agricultural Technology and Innovation Research Institute, Kindai University, Nara 631-8505, Japan.
Keun P Kim, Department of Life Science, Chung-Ang University, Seoul 06974, South Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea, funded by the Ministry of Science, ICT & Future Planning [2020R1A2C2011887, 2018R1A5A1025077 to K.P.K. and K.L.; 2018R1D1A1B07051054 to S.H]; Next-Generation BioGreen 21Program, Rural Development Administration, Republic of Korea [PJ015708012021 to K.P.K.]; Japan Society for the Promotion of Science (JSPS) KAKENHI [19K22402, 15H05973 to M.S.]; Hyogo Science and Technology Association (to M.S.). Funding for open access charge: National Research Foundation of Korea, funded by the Ministry of Science, ICT & Future Planning (to K.P.K.)
Conflict of interest statement. None declared.
REFERENCES
- 1. Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 2015; 7:a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lam I., Keeney S.. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 2014; 7:a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robert T., Nore A., Brun C., Maffre C., Crimi B., Bourbon H.M., de Massy B.. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science. 2016; 26:943–949. [DOI] [PubMed] [Google Scholar]
- 4. Vrielynck N., Chambon A., Vezon D., Pereira L., Chelysheva L., De Muyt A., Mézard C., Mayer C., Grelon M.. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science. 2016; 351:939–943. [DOI] [PubMed] [Google Scholar]
- 5. Garcia V., Phelps S.E., Gray S., Neale M.J.. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011; 479:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cannavo E., Cejka P., Kowalczykowski S.C.. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:e1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mimitou E.P., Yamada S., Keeney S.. A global view of meiotic double-strand break end resection. Science. 2017; 355:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cloud V., Chan Y.L., Grubb J., Budke B., Bishop D.K.. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012; 337:1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hong S., Sung Y., Yu M., Lee M., Kleckner N., Kim K.P.. The logic and mechanism of homologous recombination partner choice. Mol. Cell. 2013; 51:440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lao J.P., Cloud V., Huang C.C., Grubb J., Thacker D., Lee C.Y., Dresser M.E., Hunter N., Bishop D.K.. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013; 9:e1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Gaines W.A., Callender T., Busygina V., Oke A., Sung P., Fung J.C., Hollingsworth N.M.. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet. 2014; 10:e1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong S., Joo J.H., Yun H., Kim K.P.. The nature of meiotic chromosome dynamics and recombination in budding yeast. J. Microbiol. 2019; 57:221–231. [DOI] [PubMed] [Google Scholar]
- 13. Hunter N., Kleckner N.. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001; 106:59–70. [DOI] [PubMed] [Google Scholar]
- 14. Hassold T., Hall H., Hunt P.. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 2007; 16:203–208. [DOI] [PubMed] [Google Scholar]
- 15. Thacker D., Mohibullah N., Zhu X., Keeney S.. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014; 510:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cooper T.J., Garcia V., Neale M.J.. Meiotic DSB patterning: a multifaceted process. Cell Cycle. 2016; 15:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keeney S., Lange J., Mohibullah N.. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 2014; 48:187–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rockmill B., Lefrancois P., Voelkel-Meiman K., Oke A., Roeder G.S., Fung J.C.. High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity. PLoS Genet. 2013; 9:e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mu X., Murakami H., Mohibullah N., Keeney S.. Chromosome-autonomous feedback downregulates meiotic DSB competence upon synaptonemal complex formation. Genes Dev. 2020; 34:1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X., Suhandynata R.T., Sandhu R., Rockmill B., Mohibullah N., Niu H., Liang J., Lo H.C., Miller D.E., Zhou H.et al.. Phosphorylation of the synaptonemal complex protein Zip1 regulates the crossover/noncrossover decision during yeast meiosis. PLoS Biol. 2015; 13:e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agarwal S., Roeder G.S.. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000; 102:245–255. [DOI] [PubMed] [Google Scholar]
- 22. Börner G.V., Kleckner N., Hunter N.. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004; 117:29–45. [DOI] [PubMed] [Google Scholar]
- 23. Chua P.R., Roeder G.S.. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998; 93:349–359. [DOI] [PubMed] [Google Scholar]
- 24. Fung J.C., Rockmill B., Odell M., Roeder G.S.. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004; 116:795–802. [DOI] [PubMed] [Google Scholar]
- 25. Manhart C.M., Alani E.. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst.). 2016; 38:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pyatnitskaya A., Borde V., De Muyt A.. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma. 2019; 128:181–198. [DOI] [PubMed] [Google Scholar]
- 27. Tsubouchi T., Zhao H., Roeder G.S.. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev. Cell. 2006; 10:809–819. [DOI] [PubMed] [Google Scholar]
- 28. Shinohara M., Oh S.D., Hunter N., Shinohara A.. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genetics. 2008; 40:299–309. [DOI] [PubMed] [Google Scholar]
- 29. De Muyt A., Pyatnitskaya A., Andréani J., Ranjha L., Ramus C., Laureau R., Fernandez-Vega A., Holoch D., Girard E., Govin J.et al.. A meiotic XPF–ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 2018; 32:283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zakharyevich K., Tang S., Ma Y., Hunter N.. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012; 149:334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N.M.. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003; 164:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lynn A., Soucek R., Börner G.V.. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007; 15:591–605. [DOI] [PubMed] [Google Scholar]
- 33. Page S.L., Hawley R.S.. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004; 20:525–558. [DOI] [PubMed] [Google Scholar]
- 34. Dubois E., De Muyt A., Soyer J.L., Budin K., Legras M., Piolot T., Debuchy R., Kleckner N., Zickler D., Espagne E.. Building bridges to move recombination complexes. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:12400–12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allers T., Lichten M.. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001; 106:47–57. [DOI] [PubMed] [Google Scholar]
- 36. Anderson C.M., Oke A., Yam P., Zhuge T., Fung J.C.. Reduced crossover interference and increased ZMM-independent recombination in the absence of Tel1/ATM. PLoS Genet. 2015; 11:e1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cejka P., Plank J.L., Bachrati C.Z., Hickson I.D., Kowalczykowski S.C.. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1–Top3. Nat. Struct. Mol. Biol. 2010; 17:1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaur H., De Muyt A., Lichten M.. Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol. Cell. 2015; 57:583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martini E., Borde V., Legendre M., Audic S., Regnault B., Soubigou G., Dujon B., Llorente B.. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 2011; 7:e1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Callender T.L., Laureau R., Wan L., Chen X., Sandhu R., Laljee S., Zhou S., Suhandynata R.T., Prugar E., Gaines W.A.et al.. Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 2016; 12:e1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim K.P., Weiner B.M., Zhang L., Jordan A., Dekker J., Kleckner N.. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010; 143:924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith A.V., Roeder G.S.. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 1997; 136:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu L., Weiner B., Kleckner N.. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997; 11:106–118. [DOI] [PubMed] [Google Scholar]
- 44. Subramanian V.V., MacQueen A.J., Vader G., Shinohara M., Sanchez A., Borde V., Shinohara A., Hochwagen A.. Chromosome synapsis alleviates Mek1-dependent suppression of meiotic DNA repair. PLoS Biol. 2016; 14:e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Humphryes N., Leung W.K., Argunhan B., Terentyev Y., Dvorackova M., Tsubouchi H.. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet. 2013; 9:e1003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leung W.K., Humphryes N., Afshar N., Argunhan B., Terentyev Y., Tsubouchi T., Tsubouchi H.. The synaptonemal complex is assembled by a polySUMOylation-driven feedback mechanism in yeast. J. Cell Biol. 2015; 211:785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Voelkel-Meiman K., Taylor L.F., Mukherjee P., Humphryes N., Tsubouchi H., Macqueen A.J.. SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet. 2013; 9:e1003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Voelkel-Meiman K., Johnston C., Thappeta Y., Subramanian V.V., Hochwagen A., MacQueen A.J.. Separable crossover-promoting and crossover-constraining aspects of Zip1 activity during budding yeast meiosis. PLoS Genet. 2015; 11:e1005335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Voelkel-Meiman K., Cheng S.Y., Morehouse S.J., MacQueen A.J.. Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation. Genetics. 2016; 203:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oh S.D., Jessop L., Lao J.P., Allers T., Lichten M., Hunter N.. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol. Biol. 2009; 557:209–234. [DOI] [PubMed] [Google Scholar]
- 51. Yoon S.W., Lee M.S., Xaver M., Zhang L., Hong S.G., Kong Y.J., Cho H.R., Kleckner N., Kim K.P.. Meiotic prophase roles of Rec8 in crossover recombination and chromosome structure. Nucleic Acids Res. 2016; 44:9296–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hong S., Joo J.H., Yun H., Kleckner N., Kim K.P.. Recruitment of Rec8, Pds5 and Rad61/Wapl to meiotic homolog pairing, recombination, axis formation and S-phase. Nucleic Acids Res. 2019; 47:11691–11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shinohara M., Bishop D.K., Shinohara A.. Distinct functions in regulation of meiotic crossovers for DNA damage response clamp loader Rad24(Rad17) and Mec1(ATR) kinase. Genetics. 2019; 213:1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shinohara M., Gasior S.L., Bishop D.K., Shinohara A.. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:10814–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hayase A., Takagi M., Miyazaki T., Oshiumi H., Shinohara M., Shinohara A.. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004; 119:927–940. [DOI] [PubMed] [Google Scholar]
- 56. Neale M.J., Keeney S.. End-labeling and analysis of Spo11-oligonucleotide complexes in Saccharomyces cerevisiae. Methods Mol. Biol. 2009; 557:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Higashide M., Shinohara M.. Budding yeast SLX4 contributes to the appropriate distribution of crossovers and meiotic double-strand break formation on bivalents during meiosis. G3 (Bethesda). 2016; 6:2033–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Malkova A., Swanson J., German M., McCusker J.H., Housworth E.A., Stahl F.W., Haber J.E.. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics. 2004; 168:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu L., Ajimura M., Padmore R., Klein C., Kleckner N.. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell Biol. 1995; 15:6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oh S.D., Lao J.P., Hwang P.Y., Taylor A.F., Smith G.R., Hunter N.. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007; 130:259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rockmill B., Fung J.C., Branda S.S., Roeder G.S.. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 2003; 13:1954–1962. [DOI] [PubMed] [Google Scholar]
- 62. Mullen J.R., Kaliraman V., Brill S.J.. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000; 154:1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zickler D., Kleckner N.. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 2015; 7:a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hunter N. Aguilera A., Rothstein R.. Meiotic Recombination. Topics in Current Genetics, Molecular Genetics of Recombination. 2006; Heidelberg: Springer-Verlag; 381–442. [Google Scholar]
- 65. Dayani Y., Simchen G., Lichten M.. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 2011; 5:e1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., Dayani Y., Lichten M.. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell. 2012; 46:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tang S., Wu M.K., Zhang R., Hunter N.. Pervasive and essential roles of the Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol. Cell. 2015; 57:607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kauppi L., Barchi M., Lange J., Baudat F., Jasin M., Keeney S.. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 2013; 27:873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang L., Kim K.P., Kleckner N.E., Storlazzi A.. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc. Natl. Acad. Sci. U.S.A. 2011; 13:20036–20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mohibullah N., Keeney S.. Numerical and spatial patterning of yeast meiotic DNA breaks by Tel1. Genome Res. 2017; 27:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Joshi N., Barot A., Jamison C., Börner G.V.. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009; 5:e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang L., Liang Z., Hutchinson J., Kleckner N.. Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 2014; 10:e1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang L., Wang S., Yin S., Hong S., Kim K.P., Kleckner N.. Topoisomerase II mediates meiotic crossover interference. Nature. 2014; 511:551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Storlazzi A., Gargano S., Ruprich-Robert G., Falgue M., David M., Kleckner N., Zickler D.. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell. 2010; 141:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rog O., Köhler S., Dernburg A.. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. eLife. 2017; 6:e21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in the published article and its supplementary data.