Keywords: disease, gastrointestinal, microbiota, SARS-CoV-2, treatment
Abstract
COVID-19 represents a novel infectious disease induced by SARS-CoV-2. It has to date affected 24,240,000 individuals and killed 2,735,805 people worldwide. The highly infectious virus attacks mainly the lung, causing fever, cough, and fatigue in symptomatic patients, but also pneumonia in severe cases. However, growing evidence highlights SARS-CoV-2-mediated extrarespiratory manifestations, namely, gastrointestinal (GI) and hepatic complications. The detection of 1) the virus in the GI system (duodenum, colon, rectum, anal region, and feces); 2) the high expression of additional candidate coreceptors/auxiliary proteins to facilitate the virus entry; 3) the abundant viral angiotensin-converting enzyme 2 receptor; 4) the substantial expression of host transmembrane serine protease 2, necessary to induce virus-cell fusion; 5) the viral replication in the intestinal epithelial cells; and 6) the primarily GI disorders in the absence of respiratory symptoms lead to increased awareness of the risk of disease transmission via the fecal-oral route. The objectives of this review are to provide a brief update of COVID-19 pathogenesis and prevalence, present a critical overview of its GI and liver complications that affect clinical COVID-19 outcomes, clarify associated mechanisms (notably microbiota-related), define whether gut/liver disorders occur more frequently among critically ill patients with COVID-19, determine the impact of COVID-19 on preexisting gut/liver complications and vice versa, and discuss the available strategies for prevention and treatment to improve prognosis of the patients. The novel SARS-CoV-2 can cause gastrointestinal and hepatobiliary manifestations. Metagenomics studies of virobiota in response to SARS-CoV-2 infection are necessary to highlight the contribution of bacterial microflora to COVID-19 phenotype, which is crucial for developing biomarkers and therapeutics.
INTRODUCTION
From the very beginning of the pandemic, respiratory symptoms led us to naively assume that coronavirus disease 2019 (COVID-19) exclusively affects the respiratory system. Most of the efforts were channeled into this direction, but gradually, the information on extrapulmonary manifestations from severely hospitalized patients and from different centers has started reaching the medical world (Supplemental Fig. S1; see 10.6084/m9.figshare.14334044). A large proportion of patients exhibited anorexia, nausea, vomiting, diarrhea, and transaminases before or during the course of COVID-19, which pointed to gastrointestinal (GI) and hepatic perturbations (1, 2). The main objective of this review is to address the following questions: What is the incidence of GI and liver manifestations in subjects with laboratory-confirmed COVID-19? How does the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) affects the digestive and hepatic systems? Do patients with inflammatory bowel disease (IBD) or chronic liver disease have an increased risk of SARS-CoV-2 infection or could COVID-19 worsen preexisting conditions or even initiate these disorders? Are infected patients at a high risk of a complicated course of the illness? What are the mechanisms and to what extent are respiratory and intestinal microbiota implicated? This critical review is designed to help strengthen and optimize the knowledge and relationship between the GI/hepatic systems and COVID-19 and to stimulate the development of strategies for prevention measures, treatment, and guidelines.
Epidemiology and Pathogenesis of COVID-19
The world continues to be highly concerned and disquieted by the life-threatening SARS-CoV-2, causative of COVID-19, given its rapid contagiousness. As of today, date (March 2021), more than 178 million confirmed cases and 3.8 million deaths have been reported around the globe. The World Health Organization has declared the COVID-19 as a pandemic since the first confirmed infected patient (10/01/2020), and all the countries are fighting to stop the spread, and particularly with the identification of the new virulent SARS-CoV-2 variants (3). The outbreaks continue to progress by direct person-to-person transmission, mainly via respiratory droplets and the airborne route through small particles remaining in the air over time. So far, the precise contribution of this mode of transmission to the pandemic is not established (4, 5). The age-group of ≥60 yr seems the most affected, and rare are infected patients aged below 19 yr (6). In one study, severe cases constituted 74.5% of the total patients in the age-group of over 80 yr (7). Generally, the clinical presentation of patients with COVID-19 classically reflects an acute respiratory illness with symptoms, including fever, cough, and dyspnea (8–10). The proportions of these symptoms and others (chill, sore throat, and sneezing) are higher in severe cases than in mild patients. More comorbid diseases were found in the severe cases group (11–13). Radiological investigations focused on the lung and could document multifocal, subpleural ground-glass opacities with nodular consolidations bilaterally, bronchial thickening, and crazy-paving sign (14, 15). Importantly, GI and liver manifestations are not rare in patients with communicable COVID-19, but their prevalence might be affected by preexisting diseases.
GI and SARS-CoV-2 Infection
Evidence for the GI infection in subjects with COVID-19.
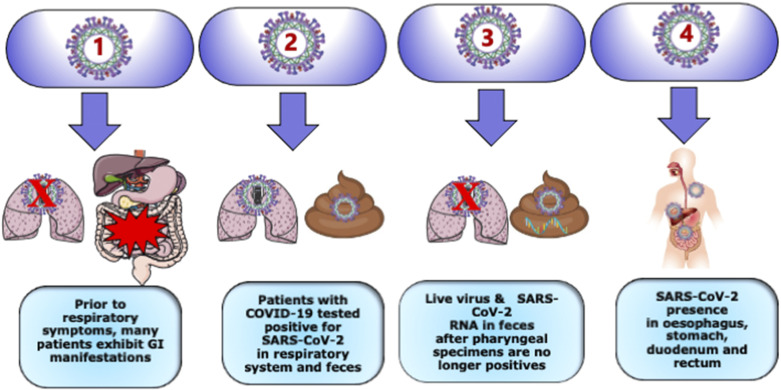
In addition to the detection of the viral presence in bronchoalveolar fluid, sputum, and blood specimens from patients with COVID-19, live virus and SARS-CoV-2 RNA were found in their feces (16). Persistent fecal viral shedding was also prominent in pediatric patients (17). Evidence was also obtained from anal testing in a limited number of confirmed cases (18). The esophagus, stomach, duodenum, and rectum displayed SARS-CoV-2 mRNA expression in severe patients (19). Unexpectedly, 64% of patients with COVID-19 remained positive for viral RNA in the feces (20) after the pharyngeal testing turned negative (Fig. 1). Further evidence using RT-qPCR validated fecal SARS-CoV-2 RNA content, whereas pharyngeal swabs were negative (21). Therefore, stools may represent a potential transmission route.
Figure 1.
The nonsimultaneous nature of SARS-CoV-2 infection characterizing the pulmonary system and the gastrointestinal tract. As the COVID-2 outbreak spreads worldwide, many individuals have experienced gastrointestinal distress, considered as the most common nonrespiratory symptoms of SARS-CoV-2 infection. Importantly, these subjects 1) have no respiratory symptoms, 2) tested positive for SARS-CoV-2 in respiratory system and feces, 3) exhibit live virus and SARS-CoV-2 RNA in feces even if pharyngeal specimens are no longer positive, and 4) display SARS-CoV-2 presence in various gastrointestinal sites such as esophagus, stomach, duodenum, and rectum. Therefore, caution is recommended, so as to not underestimate gastrointestinal symptoms in the absence of respiratory illness. Created with Servier Medical Art.
Histopathological findings indicate diffuse endothelial inflammation in the submucosal vessels of the small intestine from patients with COVID-19 (22). Accordingly, interstitial edema and infiltration of plasma cells and lymphocytes were noticed in lamina propria of the stomach, duodenum, and rectum of patients (23). These data underline microvascular small-bowel injuries, which likely contribute to severe disease progression.
Virus replication in the GI tract of animal models.
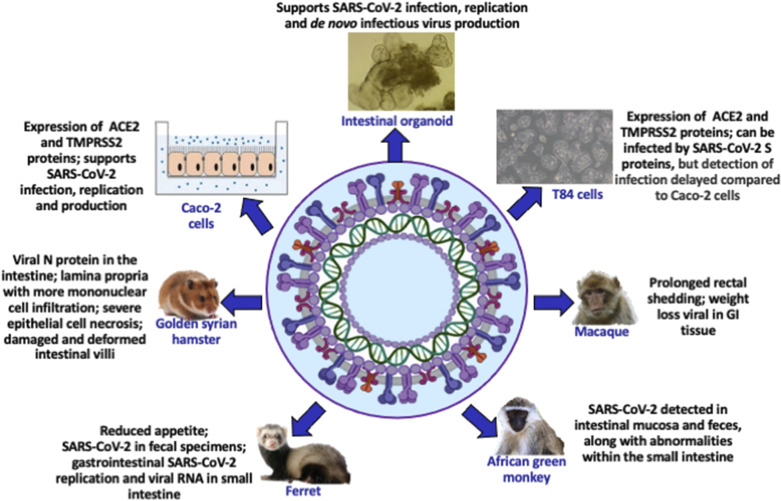
Various types of animals were tested to determine the susceptibility to SARS-CoV-2 (24) and to assess their utility for drug and vaccine development (Fig. 2). Replication of the virus was observed in not only the nasal turbinate, soft palate, and tonsils but also in the digestive tract and rectum of ferrets. Viral RNA was also noted in the small intestine of other animal models such as dogs and cats (25). Efficient transmission from SARS-CoV-2-infected ferrets and hamsters to naïve ferrets and hamsters was recorded concomitantly with simultaneous viral detection in fecal specimens (26, 27). Not only do animal models provide valuable data on viral infection and pathogenesis in extrapulmonary tissues, notably the GI tract, but they are also reliable for the development and preclinical evaluation of vaccines and therapeutics.
Figure 2.
Experimental models to comprehensively analyze gastrointestinal pathophysiology. As the mechanisms by which SARS-CoV-2 causes gastrointestinal infection remain largely unknown, long-term cultures of human cell lines and organoids are used, given their capacity to maintain lineage functions of the gut and to practically serve for coronavirus-associated transmission and multiplication. Similarly, animal models are essential to efficiently dissect SARS-CoV-2 life cycle steps and to understand detailed gastrointestinal pathophysiology. Created with BioRender.com.
Organoid modeling tool to evidence direct intestinal viral infection.
To improve our knowledge on SARS-CoV-2 entrance and replication in the human gut, live small intestinal organoids were cultured and examined three-dimensionally (3-D) (28) with high-resolution imaging (Fig. 2). Transmission electron microscopy, gene expression analysis, and live virus titration uncovered infection of enterocytes by SARS-CoV-2 virus (23). In experimental differentiated conditions, SARS-CoV-2 titers remained stable at 60 h postinfection concomitantly with upregulation of angiotensin-converting enzyme 2 (ACE2) mRNA (28). Although multiple disintegrated cells were noted in intestinal organoids, viral particles were produced and secreted into the lumen of the organoid at the basolateral and the apical side of enterocytes (28), confirming that the viral shedding is conceivable in humans with the high potential SARS-CoV-2 transmission via the fecal-oral route (29). Furthermore, mRNA sequence analysis showed increased cytokines and interferon-stimulated genes in response to SARS-CoV-2. These elegant studies support SARS-CoV-2 replication in the intestinal epithelium while demonstrating that the gut organoid model is effective in exploring coronavirus infection and biology. As additional workers reported SARS-CoV-2 mRNA detection and intracellular staining of viral nucleocapsid protein in gastric, duodenal, and rectal epithelia (23), it seems obvious that the intestine acts as a secondary site for coronavirus tropism and infection.
Life cycle of highly pathogenic SARS-CoV-2 in the host GI cells.
It is now well accepted that SARS-CoV-2 uses ACE2 as its receptor to enter human cells (30), though additional membrane proteins (alanyl aminopeptidase, glutamyl aminopeptidase, dipeptidyl peptidase-4) (31) may also assist virus entry (Supplemental Fig. S2; see 10.6084/m9.figshare.14334086). ACE2 is highly expressed in lung alveolar type-2 cells, myocardial cells, liver cholangiocytes, kidney proximal tubules, bladder urothelial cells, and GI epithelial cells (31, 32). According to in silico searches in publicly available databases, the highest ACE2 expression is found in the small intestine (33). The host transmembrane protease serine 2 (TMPRSS2), necessary to induce virus-cell fusion (34), is also expressed at high levels in both the small intestine and the colon (33). Vigorous SARS-CoV-2 replication was noted in this in vitro model. Collectively, these data indicate that intestinal cells might support natural infection by SARS-CoV-2, which can occur in patients with COVID-19. Nevertheless, research effort is required to determine the source of enteric infection in humans and answer the following questions: Is intestinal epithelium primarily infected with SARS-CoV-2 through the oral-fecal route? Is it the consequence of respiratory infection?
GI symptoms and diseases in association with SARS-CoV-2 infection.
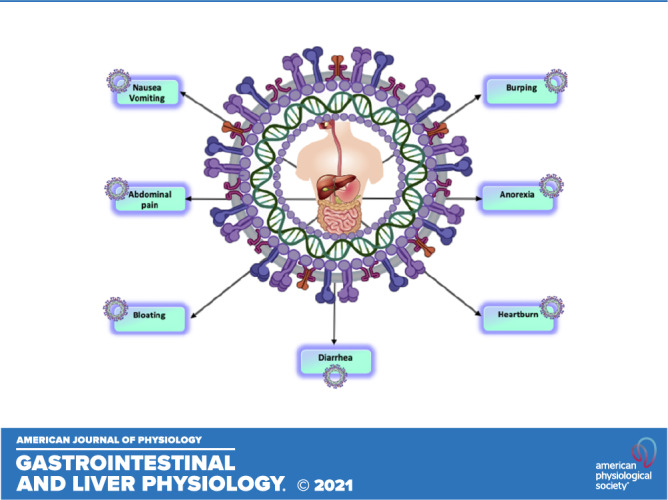
In patients with COVID-19, the typical intestinal mucosa can be disrupted by SARS-CoV-2, and this could result in GI symptoms (Supplemental Fig. S3; see 10.6084/m9.figshare.14334092). Anorexia, nausea, vomiting, diarrhea, and abdominal pain are present. Data from a meta-analysis comprising 60 investigations with 4,243 patients (35) revealed a pooled prevalence of GI symptoms (17.6%), with anorexia as the most commonly reported symptom (26.8%), followed by diarrhea (12.5%), nausea and vomiting (10.2%), and abdominal discomfort (9.2%). COVID-19 severity was often accompanied by GI symptoms (36). Interestingly, the occurrence of GI symptoms at presentation was related with a 70% augmented risk of SARS-CoV-2 infection (37). A plausible conclusion from these observations is that the orally fed patients may develop poor nutritional status, hence the need for enteral nutrition, which may exert diverse benefits in patients with COVID-19, e.g., intestinal mucosa stimulation and reduction of hospital stay length, costs, mortality rates, and septic complications (38).
Abdominal pain was more frequent in patients with COVID-19 who necessitated the intensive care unit (39). Therefore, these complications are likely to worsen the patients’ condition, which demands appropriate early special care and treatment. It is all the more critical given the observation describing GI signs in the absence of respiratory symptoms, obviously causing a delay in COVID-19 diagnosis (40). These observations raise larger questions about individuals with IBD: Do Crohn’s disease and ulcerative colitis subjects exhibit an increased risk of SARS-CoV-2 infection compared with the general population? Will this mean a significantly raised rate of morbidity and mortality in response to COVID-19 infection? What is the impact of SARS-CoV-2 infection on immunosuppressed patients with IBD?
Links between IBD and SARS-CoV-2 infection.
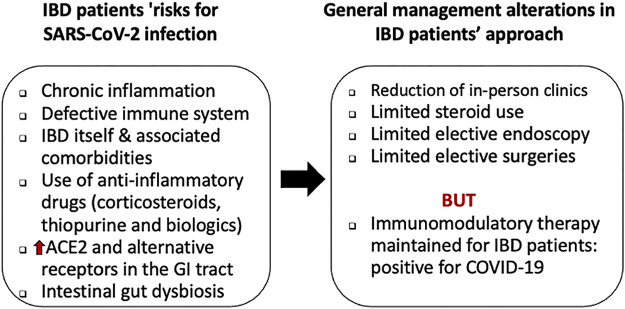
COVID-19 has been described to be more frequent in people with older age, coexisting pathologies, pregnancy, smoking tobacco, and weakened immune systems (41, 42). Consequently, the Centers for Disease Control and Prevention, as well as many countries, have stated that patients with IBD sharing these same characteristics would be prone to getting raised risks of more severe SARS-CoV-2 infections (43–46). However, to date, there has been a limited number of investigations (47, 48) (Fig. 3). At this point of time, all we can say, based on two independent studies from China and Italy, is that patients with IBD do not appear to have an increased risk of SARS-CoV-2 infection compared with the general population (49, 50). Nonetheless, the conclusions drawn from these studies suggest that patients with IBD appear to be underrepresented in those diagnosed with COVID-19 compared with what has been seen in the general populations (51).
Figure 3.
Risks of patients with IBD for SARS-CoV-2 infection and approach to management. Subjects with IBD as a preexisting chronic disease with immunosuppressive or biological treatment may be at a higher risk of developing severe forms of COVID-19 with gastrointestinal manifestations. Consequently, IBD specialists have been challenged to address specific aspects of SARS-CoV-2 infection, although patients with IBD do not appear to have an increased risk of being infected with SARS-CoV-2. IBD, inflammatory bowel disease.
As a large number of subjects with IBD receive long-term immunomodulatory and immunosuppressive treatments, there are fears that they are more susceptible to infections and associated complications. However, despite the use of immunosuppressive or immune-modifying therapies to control chronic inflammation, and notwithstanding the need to be present at medical facilities (possibly enhancing the risk of SARS-CoV-2 exposure), no baseline enhanced risk for viral infection has been detected in subjects with IBD (52). Nevertheless, one has to point out that bowel inflammation was not always assessed in patients, and fecal sample SARS-CoV-2 RNA lags behind that of respiratory tract samples, suggesting the possibility of extended duration of viral shedding in feces (53). In this case, safety measures should be taken to prevent transmission from hospitalized patients who have recovered from COVID-19, unless fecal samples are tested, while remaining aware about the progression of GI symptoms in patients with IBD. Without losing sight of IBD relapsing and remitting inflammatory condition of the bowel, and with due consideration for immunosuppression and consequent predisposition to infection in Crohn’s disease and ulcerative colitis, while simultaneously taking account of high intestinal SARS-CoV-2 infectivity, future research in this area is critically needed to uncover the pathological hallmarks. In fact, there is still much to learn on the impact of direct and indirect virus-induced GI damage by host immune-mediated damage.
Very recently, another piece of information concerning ACE2-expressing ileal biopsies has made the picture a little more nebulous (54). ACE2 was downregulated in Crohn’s disease compared with nondisease controls (54). These findings can logically lead to the conclusion that the entry and propagation of SARS-CoV-2 are limited, thereby lowering COVID-19 disease severity in patients with IBD. However, ACE2 activity plays a protective role since acute lung injury and respiratory distress syndrome are more severe in mice with ACE2 inactivation following SARS-CoV-2 infection (55). Obviously, after being infected, animals with total ACE2 deficiency exhibited worsened vascular permeability, lung edema, neutrophil accumulation, and lung dysfunction. Amelioration was recorded with the administration of catalytically active recombinant ACE2 proteins (56). Similar to the lungs, ACE2 expression was reduced in inflamed ileal tissue with a worse prognosis in both adult and pediatric cohorts (54). Although these data suggest a relationship between ACE2 downregulation and inflammation and worse outcomes in Crohn’s disease, further work is required to clarify the paradoxical function of ACE2 in IBD during COVID-19.
Enteric nervous system abnormalities and SARS-CoV-2 infection.
SARS-CoV-2 may gain access to the enteric nervous system as an alternative route to enter the central nervous system, given the high expression of ACE2 and TMPRSS2 in enteric neurons and glial cells of the small and large intestine, as well as in choroid plexus epithelial cells of postmortem in patients (57). Infection and replication of the SARS-CoV-2 in the gut, which was revealed by viral genetic material and endoscopy (19, 58), may explain GI symptoms, including diarrhea, vomiting, and abdominal pain. In addition to the lateral hypothalamic nuclei, the GI tract could also represent an entry strategy for SARS-CoV-2 to the brain, which may elucidate the underlying mechanism of nausea and anorexia symptoms (19, 59). Nevertheless, SARS-CoV-2-mediated intestinal inflammation may through the vagal nerve turn on a signal to the brain and affect cognitive functions, thereby causing nausea and anorexia (60–62). More studies are necessary to shed more light on the gut-brain axis mechanisms triggered by SARS-CoV-2 and leading to neurological and GI symptoms.
COVID-19 and Hepatobiliary Injury
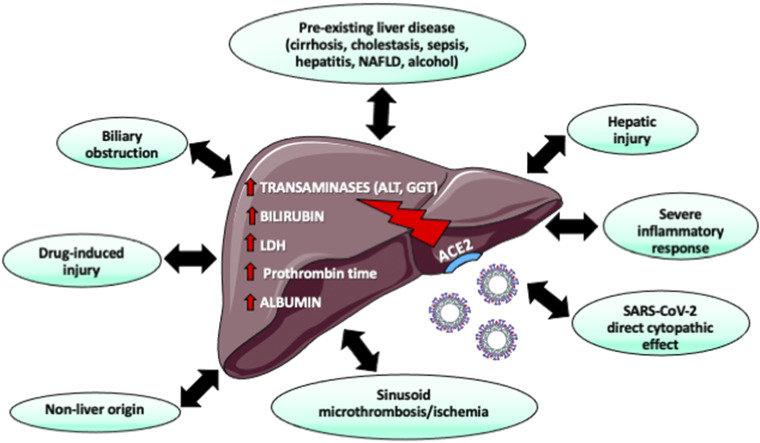
The liver is among the systemic and extrarespiratory organs affected by SARS-CoV-2 infection. As many reports have shown the wide distribution of ACE2 across human tissues, which also includes liver, when in fact the latter was suspected of being infected by SARS-CoV-2 via binding of host receptors before engaging the infection cycle (31, 32). Clinical and hepatic symptoms were associated with ACE2-expressing cells, such as liver cholangiocytes and hepatocytes (63). Indeed, determination of hepatic biomarkers in patients with COVID-19 revealed higher levels of transaminases and bilirubin, which may reveal the occurrence of liver injury, due to direct SARS-CoV-2 infection, systemic inflammation, or toxicity (64–66) from commonly used drugs (Fig. 4). Patients with GI symptoms have more elevated transaminase levels (36, 67). However, the abnormal liver function tests caused by the virus are transitory (64). Nevertheless, subjects with preexisting liver injury [e.g., nonalcoholic fatty liver disease (NAFLD) with fibrosis or cirrhosis] are at a high risk for severe COVID-19 since these patients express elevated levels of ACE2, thereby allowing the invasion of SARS-CoV-2 through liver cells (68). Although alkaline phosphatase and γ-glutamyl transferase levels have infrequently been reported, their elevation in subjects with NAFLD foreshadows a more severe course of the disease (69). Therefore, caution must be exercised to detect and treat liver impairment in patients with COVID-19, as depicted in supplemental data (Supplemental Fig. S4; see 10.6084/m9.figshare.14334281).
Figure 4.
Liver injury in SARS-CoV-2-infected patients. There is ample evidence that liver cells are equipped with ACE2 receptors. The marked affinity of SARS-CoV-2 for cholangiocytes and the high ACE2 expression in these biliary cells increase the risk of viral infection. Accordingly, numerous patients with SARS-CoV-2 had liver injury, reflected by raised transaminases, elevated bilirubin, and hypoalbuminemia. Although reduced albumin seems the result of inflammatory response, raised levels of gamma-glutamyl transpeptidase (GGT) and bilirubin could account for biliary damage. Further investigation is required to determine whether liver injury is due to direct viral damage, drug-induced liver injury, hypoxia, or microthromboses. ACE2, angiotensin-converting enzyme 2. Created with Servier Medical Art.
Many clinicians and scientists wonder what the mechanism is by which SARS-CoV-2 could deteriorate liver function. The detection of abundant ACE2 in bile duct cells may be indicative of the binding of the virus to cholangiocytes, which triggers dysregulation of liver function (70). Raised mitosis and ballooned hepatocytes reflected amplified apoptosis in the biopsies of patients with SARS-associated coronavirus infection (71). In line with these findings, the specific SARS-CoV-2 7a protein produces apoptosis through a caspase-dependent pathway in the liver and in cell lines representing different organs (72).
We cannot exclude the role of inflammation in the liver disorders characterizing patients with COVID-19 since 1) elevated circulating proinflammatory cytokines and chemokines were recorded with hepatic dysfunction (73) in line with various viral infections (74), and 2) a close association was generally observed between liver injury and inflammatory responses induced by SARS-CoV-2 infection (75). As mentioned previously, antiviral, antibiotic, and antipyretic drugs may contribute to liver injury in COVID-19 (76, 77).
SARS-CoV-2 infection and cholangiocyte damage.
Although biomarkers of hepatic abnormalities as a frequent manifestation of COVID-19 are considered transient and resolve with viral disease resolution, features similar to secondary sclerozing cholangitis are suggestive of direct hepatic injury from COVID-19 (78). Indeed, patients develop severe cholangiocyte injury and intrahepatic microangiopathy even during recovery from critical cardiopulmonary COVID-19. Some investigators infer the development of biliary obstructions, cholestatic liver injury, biliary cirrhosis, and end-stage liver disease to intravenous ketamine for sedation of patients with acute respiratory distress syndrome (79). In fact, arterial injuries supplying the bile ducts may result in ischemic lesions in response to ketamine, as well as mechanical ventilation for a long time, inspired oxygen fraction greater than 80%, high doses of vasopressors which further reduced splanchnic blood flow (80, 81). Therefore, these pathophysiological mechanisms constitute reasonable justifications for biliary lesions observed in patients with COVID-19. For its part, SARS-CoV-2 can bind to ACE2 receptors on cholangiocytes and move in the liver, which may lead to cholestatic injury in association with a higher likelihood of death (82). Cytokine storm and sepsis may represent an additional contributing mechanism for cholestasis etiology. Further studies are as well needed to clarify the limited occurrence of cholestatic liver injury in patients with COVID-19 (<1%), the increased mortality rate, and the long-term outlook of subjects with cholestatic liver injury (resolution versus chronic cholangiopathy).
COVID-19 and Liver Transplants
Theoretically, liver-transplant recipients may be at a higher risk of developing complications related to COVID-19 in view of their immunocompromised condition. This was the concern of clinical experts about immunocompromised patients facing the SARS-CoV-2 pandemic since immunosuppressive therapy provokes an impact on humoral, cell-mediated immunity, and neutrophil function, increasing the risk of severe infections caused by viral agents (83). The complex mechanisms underlying liver damage include hyperactivated immune responses and “cytokine storm,” systemic inflammation, severe hypoxemia and acute respiratory failure (especially in the critical phase), septic shock, multiple-organ dysfunction or failure, intestinal endotoxemia, drug toxicity, and progression of preexisting liver diseases (77, 84, 85). In the first studies in Italy, liver-transplant recipients were not at an increased risk of COVID-19 and did not develop clinical pulmonary disease despite testing positive for SARS-CoV-2 compared with the general population (86). Although the absence of worse prognosis was also confirmed by other studies (87, 88), but it is opposite to poorer outcomes reported by various groups (89–91). Liver injury is common in patients with COVID-19, but little is known about its clinical presentation and severity in the context of liver transplant. We describe a case of COVID-19 in a patient who underwent transplant 3 years ago for hepatocellular carcinoma. The patient came to the clinic with symptoms of respiratory disease, and pharyngeal swabs for severe acute respiratory syndrome COVID-19 were positive. His disease progressed rapidly from mild to critical illness and was complicated by several nosocomial infections and multiorgan failure. Despite multiple invasive procedures and rescue therapies, he died from the disease. The management of COVID-19 in the posttransplant setting presents complex challenges, emphasizing the importance of strict prevention strategies. In the latter cases, liver posttransplant patients who were infected by SARS-CoV-2 exhibited severe symptoms of respiratory disease, progressed rapidly to critical illness and multiorgan failure, and died despite aggressive therapeutic measures. This emphasizes the need of prevention strategies to avert fatal outcomes from opportunistic coronavirus infection. Therefore, there are still scarce data regarding the susceptibility of liver-transplant recipients to COVID-19 complications. To address important issues of ambiguity pertaining to donor and recipient liver-transplant candidates (e.g., practice patterns surrounding incidence, management, and outcome), the European Liver and Intestine Transplantation Association launched a survey on March 24, 2020, among the 149 active liver-transplant centers located in 30 European countries (92). Symptomatic COVID-19 was noted more frequently in liver-transplant candidates and recipients than in the general population, along with increased rate of admission to intensive care and mortality. In line with the recommendations of transplant societies, most of the centers performed PCR tests on nasopharyngeal swabs among both deceased donors and recipients for COVID-19 diagnosis. This survey revealed that several of the liver transplant centers in Europe temporarily halted liver-transplant activity due to the lack of donors and other collateral effects of the pandemic, whereas others restricted liver transplants to the most urgent candidates. Nevertheless, additional investigations are warranted to determine whether the progression of COVID-19 is more rapid and severe in immunocompromised hosts with liver transplants.
Intestinal Microbiota and COVID-19
Gut microbiota and viral respiratory infection.
The mammalian intestine is colonized by various types of microorganisms, including bacteria, viruses, fungi, and Archaea. The intestinal flora, currently named gut microbiota, exerts a significant impact on human health by providing metabolites and micronutrients, stimulating immunity, and promoting intestinal homeostasis (93). In physiological conditions, the gut microbiota maintains the homeostasis of the GI tract, allowing not only the full integrity of enteric digestive, nervous, endocrine, and immune systems but also the integrity of the whole organism. In fact, symbiotic or commensal flora have co-evolved with the human host and been revealed to be advantageous for host interaction such as involvement in regulating physiological processes required for metabolic and immune function, as well as digestion and nutrition (94).
However, dysbiosis of gut microbiota could induce or exacerbate GI and extra-GI diseases. It is particularly important to emphasize the predominant role of microbiota in the intestinal mucosal barrier, which acts as a first line of immunological defense against possible harmful luminal compounds and pathogenic microorganisms. Disruption of the homeostasis by several factors (e.g., dietary habits and lifestyles, medications, host genetics, and infections) between the microbiota and the epithelial layer leads to a leaky intestine, translocation of pathogens and lipopolysaccharide into systemic circulation, and the development of multiple pathologies such as obesity, cardiovascular diseases, carcinoma, and liver disease (95). Therefore, a better understanding of intestinal microbiota perturbations and mechanisms of impaired barrier function, under pathological conditions, may constitute nutritional and pharmacological therapeutic targets.
Disruptions of the normal microbial communities by an acute viral infection might contribute to the development of severe complications. One of the most striking examples is influenza infection, which causes modifications in intestinal microbiota, which in turn affect host immune responses and raise susceptibility to secondary bacterial infections (96) (Fig. 5). The key question is how to explain the effects of viral respiratory infection on distant gut microflora? Wang et al. (96) addressed this intriguing issue and discovered that lymphocytes originating from the respiratory system are able, in the aftermath of respiratory influenza infection, to precisely move toward intestinal mucosa. The guidance of lymphocytes to the gut is provided by the chemokine receptor 9 and its exclusive ligand, chemokine 25, whereas once arrived at the destination, lymphocytes perturb gut microbiota homeostasis, resulting in intestinal immune injury and inflammation. A second mechanism, through which influenza virus affects the gut, relies on the induction of type I interferon in the lungs, which alters intestinal microbial composition and suppresses host immunity to enhance a secondary Salmonella intestinal infection (97). Among gut microbiota perturbations, there were an increase in Bacteroidetes and a concomitant decrease in Firmicutes phyla abundance (98). Currently, in addition to the influenza virus, a close relationship was reported between respiratory infections with Burkholderia thailandensis, Streptococcus pneumoniae, Staphylococcus aureus, Aspergillus fumigatus, and Klebsiella pneumoniae (99, 100) and enteric microbiome composition (Fig. 6).
Figure 5.
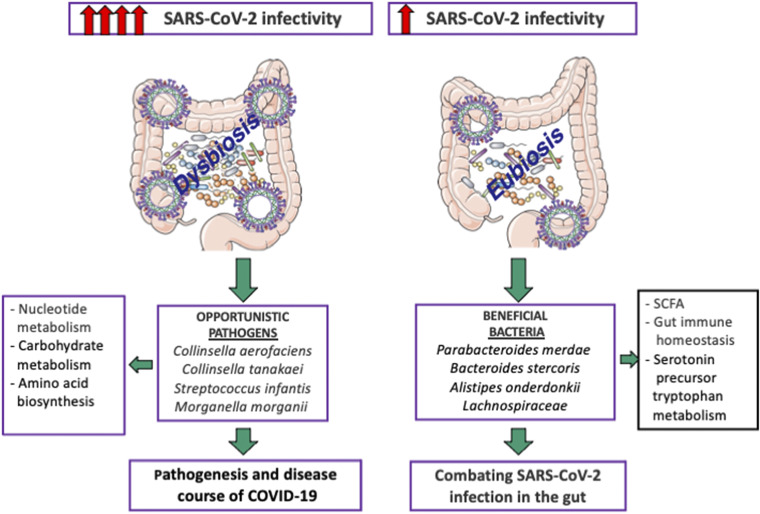
Evidence for active and prolonged “quiescent” GI infection even in the absence of GI manifestations and after recovery from respiratory infection of SARS-CoV-2. Gut microbiota of patients with active SARS-CoV-2 GI infection was characterized by enrichment of opportunistic pathogens, loss of salutary bacteria, and increased functional capacity for nucleotide and amino acid biosynthesis and carbohydrate metabolism. GI, gastrointestinal. Created with Servier Medical Art.
Figure 6.
Microbiota in eubiosis and dysbiosis conditions and cross talk with the pulmonary system. Created with Servier Medical Art.
If the respiratory microbiota, in response to influenza, can impact gut microbiota, the inverse is also true. Manipulating intestinal microbiota by fermentable fibers modified Firmicutes to Bacteroidetes ratio and gave rise to short-chain fatty acids resulting in protection from allergic airway inflammation, leading to curtailed inflammatory cell infiltration and diminished Th2 cytokines and total IgE in the lung (101). The administration of oral probiotics resulted in significantly reduced airway allergic sensitization and respiratory syncytial virus-induced pulmonary immunopathology (102).
Airway microbiota and viral infection.
The airway microbiota must take into account the two compartments: the upper and the lower respiratory tract. Although the lung and oral cavity share a number of bacterial communities, differences are observed in their microbiota composition (103). In particular, they share the Streptococcus, Prevotella, and Veillonella genera (104). However, the predominant phyla in the lungs are Bacteroidetes and Firmicutes, whereas the oropharynx is dominated by Firmicutes, Proteobacteria, and Bacteriodetes (103).
Viral infection and gut-lung axis.
The gut and lung microbiota intimately interact with the host immune system. Deterioration of one of them upsurges harmful modifications in the other microbial ecosystem, creating a vicious cycle, which aggravates diseases and underlines the gut-lung cross talk (105). The pathways and mechanisms carrying on the gut-lung axis are still not defined. Following infection with the influenza virus H1N1, mice exhibited pathological alterations in lung tissues (e.g., congestion, edema, and inflammatory exudation) along with intestinal injury (106). On the other hand, polysaccharide extract, as a functional food, lessened viral replication, raised the survival rate of H1N1-infected mice, and prevented lung (e.g., counteracting proinflammatory cytokines/chemokines, TLR4, and phospho-NFκB p65) and intestine (e.g., improvement of goblet cells and immune barrier) infection (106). Likely, the mutual interaction between gut and lung microbiota plays a mediatory role in the management of H1N1.
COVID-19 and microbiota.
Metatranscriptome sequencing of the bronchoalveolar lavage fluid from subjects with SARS-CoV-2 documented changes between patients and healthy controls (107). Although bronchoalveolar lavage fluid microbiota from patients with COVID-19 was enriched with pathogenic bacterial strains and commensal bacteria commonly found in the oral and the upper respiratory tract, its signature was quite similar to the imprinting left by other respiratory viruses such as influenza and respiratory syncytial virus (107). Abridged bacterial diversity was evident in COVID-19 microbiota, which was characterized by profuse opportunistic pathogens (e.g., Streptococcus, Rothia, Veillonella, and Actinomyces) and diminished beneficial symbionts (108) in association with the C-reactive protein and the index of bacterial infection (Fig. 5). Collectively, the changes in respiratory and intestinal microflora reflect gut-lung axis disturbances. Like other viral respiratory infections, these modifications are expected to cause intestinal injury by recruiting lung-derived CCR9+CD4+ T-cells to the small intestine and stimulating the production of interferon-γ by these cells (96). For its part, this intestinal milieu in full disruption, and already complicated by the binding of SARS-CoV-2 to ACE2 receptors leading to gastroenteritis-like symptoms, has the great potential to worsen the lung condition.
In a healthy condition, the gut and airway microbiota favorably regulate the immune system by blunting the formation of immune cells, including regulatory T-cells and innate lymphoid cells, which supports gut and lung homeostasis (109). Indeed, in physiological conditions, intestinal microbiota has the potential to trigger the inflammasome and recruit dendritic cells to initiate T-cell responses to viruses such as the influenza while strengthening antiviral responses in macrophages (110). These findings must be seriously taken into account to develop anti-COVID-19 therapy.
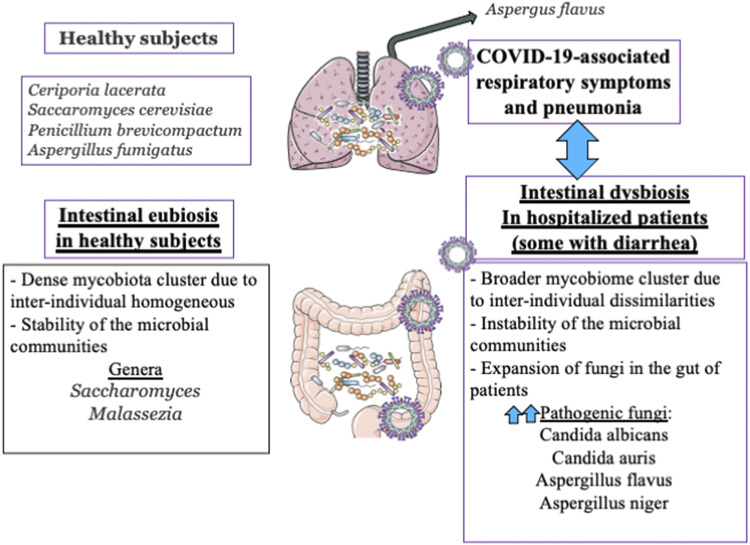
Various respiratory viral infections predispose to secondary bacterial infections, which may frequently present a more severe clinical course (111). It is well accepted that acute viral respiratory infections alter immune responses and result in dysbiosis of the respiratory and GI systems. Consequently, the immune function against secondary bacterial infection is inappropriate, thereby promoting the proliferation of potentially pathogenic bacterial species. As to SARS-CoV-2, the secondary infection (bacteria and fungi) rate may reach 10% and increase morbidity (112, 113) (Fig. 6). According to the extended descriptive study of Guo et al. (114), the bacterial cultures of patients with secondary infections included Acinetobacter baumannii, Klebsiella pneumoniae, Aspergillus flavus, Candida glabrata, and Candida albicans (115, 116). Although six additional studies reported the occurrence of bacterial coinfection with the copathogens Mycoplasma pneumoniae, Legionella pneumophila, Streptococcus pneumoniae, and Candida pneumonia, eight studies described viral coinfections with rhinovirus/enterovirus and influenza. Among the commonest copathogens with coronavirus, respiratory syncytial virus, parainfluenza, metapneumovirus, and influenza B virus were also stated (117). The mixed infections may lead to not only amplified complications in combination with dysbiosis but also derangements of the dynamics of the gut-lung axis. Diagnosis of the coinfections prompts both antiviral and antibiotic treatment.
COVID-19 and Virobiota
Although most of the studies focused on microbial community colonizing the GI tract, given its high abundance, there are also large communities of viruses that are also indigenous to the airway and GI tract, which suggests that virobiota could play a role in health and disease (118). At least, 109 virus‐like particles per gram are detected in human feces, and a great part of them are bacteriophages, not to mention, of course, viruses infecting eukaryotic host cells, and virus-derived genetic elements present in host chromosomes (119). Despite their pathogenicity, eukaryotic viruses are capable of establishing mutualistic relationships with the human host. Unfortunately, current knowledge is not yet sufficient concerning the mutualistic aspects of viral infection.
Bacteriophage populations (phageome), which dominates the virome, establish symbiotic relationships (known as lysogeny) with their bacterial hosts. In fact, the phageome regulates the complex bacterial network by turning on genes to increase defense or virulence (120). As noted, phages, as stable residents of the human body, have the potential to assist their bacterial hosts by killing related competing strains (121). Currently, scientists show a great interest in bacteriophages, given antibiotic-resistant bacteria (122). As an example, lytic phages can be used to destroy pathogenic bacteria (123). If bacteriophages infect the bacteria that stably colonize the human host, the picture is less obvious for eukaryotic viruses, chiefly because metagenomics faces difficulties with discriminating resident viruses from acute viruses (124). According to a recent investigation, 220 viruses are now able to infect humans, but only 100 are pathogenic since the remainder establishes commensal or symbiotic interactions with host (125). SARS-CoV-2 is not a prototypical, human commensal virus but, instead, a pathogen that replicates persistently and causes COVID-19. Currently, its effects on the whole virome and particularly on the phageome ecosystem are not defined. There is clearly a palpable lack of knowledge about the relationship/interaction between SARS-CoV-2, microbiota, and bacteriophage communities. At this point, metagenomics studies of virobiota in response to SARS-CoV-2 infection are necessary to highlight the contribution of bacterial and viral microflora to COVID-19 phenotype, which is crucial for developing future biomarkers and therapeutics. For the moment, we can only put forward the hypothesis that microbiota dysbiosis, including virobiota modifications, is implicated in symptomatic SARS-CoV-2 carriers and modulates disease severity. As phages are quite able to shape the genetic diversity and the species diversity of the microbiota, they therefore may have an impact on the host immune system.
From the concepts relative to virus-derived genetic elements present in host chromosomes, it appears that human endogenous retroviruses are able to repress the viral replication and prevent the horizontal transmission of infections between individuals (126). Depending on the host cell type and the physiological circumstances, various human endogenous retrovirus members are actively transcribed and differentially expressed, which makes exogenous viruses more or less invasive between individuals using epigenetic mechanisms (127). Whether virus-derived genetic elements present in human host chromosomes are behind the differences between symptomatic and asymptomatic individuals remains to be discovered.
Conclusions
The novel SARS-CoV-2 can cause GI and hepatobiliary manifestations in addition to severe respiratory pathology. The GI and hepatobiliary dissemination is probably the reflection of both SARS-CoV-2 replication and COVID-19 immunopathological sequelae. In fact, the co-expression of ACE2 and TMPRSS2 in the GI and hepatobiliary systems, the identification and replication of SARS-CoV-2 locally, and the gut and liver disorders in the absence of respiratory symptoms constitute indisputable evidence for direct viral infectivity and toxicity. Although it remains difficult to gauge the excess risk of death attributable to gut and liver failure, the GI tract and the hepatic system may represent possible routes of invasion and transmission of SARS-CoV-2. So far, the human physiological small intestinal and liver organoids and animal models have been instrumental to evidence that SARS-CoV-2 can infect and replicate in intestinal epithelial cells and hepatocytes, pointing out that the gut and the liver are viral target organs. Importantly, gut dysbiosis, featured by decreased beneficial bacterial communities and increased opportunistic pathogens, is correlated to poorer outcomes of COVID-19 infection, especially in hospitalized patients with severe preexisting GI and hepatic disorders. Metagenomics studies of virobiota in response to SARS-CoV-2 infection are necessary to highlight the contribution of bacterial and viral microflora to COVID-19 phenotype, which is crucial for developing future biomarkers and therapeutics.
SUPPLEMENTAL DATA
Supplemental Fig. S1: https://figshare.com/s/b08ec27b98182e5c5d08.
Supplemental Fig. S2: https://figshare.com/s/69e239e93d70daf890da.
Supplemental Fig. S3: https://figshare.com/s/3e0890a19d5425df54b3.
Supplemental Fig. S4: https://figshare.com/s/f0ad23e49559a50390fb.
GRANTS
This study was supported by the Canadian Institutes of Health Research Institute of Human Development, Child, and Youth Health Grant PJT 153113 and the J. A. DeSève Research Chair in Nutrition (E.L.).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
E.L. conceived and designed research; S.S. prepared figures; E.L. and S.S. drafted manuscript; E.L., A.S., A.C., Y.D., A.M., and S.S. edited and revised manuscript; E.L., A.S., A.C., Y.D., A.M., and S.S. approved final version of manuscript.
REFERENCES
- 1.Barrera FJ, Gonzalez GJG, Rodriguez GR. Gastrointestinal and liver involvement in patients with COVID-19. Lancet Gastroenterol Hepatol 5: 799, 2020. doi: 10.1016/S2468-1253(20)30209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boregowda U, Aloysius MM, Perisetti A, Gajendran M, Bansal P, Goyal H. Serum activity of liver enzymes is associated with higher mortality in COVID-19: a systematic review and meta-analysis. Front Med (Lausanne) 7: 431, 2020. doi: 10.3389/fmed.2020.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Oosterhout C, Hall N, Ly H, Tyler KM. COVID-19 evolution during the pandemic - implications of new SARS-CoV-2 variants on disease control and public health policies. Virulence 12: 507–508, 2021. doi: 10.1080/21505594.2021.1877066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klompas M, Baker MA, Rhee C. Airborne transmission of SARS-CoV-2: theoretical considerations and available evidence. JAMA 324: 441, 2020. doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- 5.Morawska L, Milton DK. It is time to address airborne transmission of coronavirus disease 2019 (COVID-19). Clin Infect Dis 71: 2311–2313, 2020. doi: 10.1093/cid/ciaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu C, Zhou M, Liu Y, Guo T, Ou C, Yang L, Li Y, Li D, Hu X, Shuai L, Wang B, Zou Z. Characteristics of asymptomatic COVID-19 infection and progression: a multicenter, retrospective study. Virulence 11: 1006–1014, 2020. doi: 10.1080/21505594.2020.1802194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JY, Hong SW, Hyun M, Park JS, Lee JH, Suh YS, Kim DH, Han SW, Cho CH, Kim HA. Epidemiological and clinical characteristics of coronavirus disease 2019 in Daegu, South Korea. Int J Infect Dis 98: 462–466, 2020. doi: 10.1016/j.ijid.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke RM, Killerby ME, Newton S, Ashworth CE, Berns AL, Brennan S, Bressler JM, Bye E, Crawford R, Harduar Morano L, Lewis NM, Markus TM, Read JS, Rissman T, Taylor J, Tate JE, Midgley CM; Case Investigation Form Working Group. Symptom profiles of a convenience sample of patients with COVID-19 - United States, January-April 2020. MMWR Morb Mortal Wkly Rep 69: 904–908, 2020. doi: 10.15585/mmwr.mm6928a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz P, Galar A, Catalan P, Valerio M, Aldamiz-Echevarria T, Colliga C, Bouza E; Gregorio Marañón Microbiology-ID COVID 19 Study Group. The first 100 cases of COVID-19 in a hospital in Madrid with a 2-month follow-up. Rev Esp Quimioter 33: 369–378, 2020. doi: 10.37201/req/072.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu P, Zhou Y, Wang F, Wang H, Zhang M, Pan X, Zhao Q, Liu J. Clinical characteristics, laboratory outcome characteristics, comorbidities, and complications of related COVID-19 deceased: a systematic review and meta-analysis. Aging Clin Exp Res 32: 1869–1878, 2020. doi: 10.1007/s40520-020-01664-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandorkar A, Coro A, Natori Y, Anjan S, Abbo LM, Guerra G, Mattiazzi AD, Mendez-Castaner LA, Morris MI, Camargo JF, Vianna R, Simkins J. Kidney transplantation during coronavirus 2019 pandemic at a large hospital in Miami. Transpl Infect Dis 22: e13416, 2020. doi: 10.1111/tid.13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habouzit V, Sanchez A, Dehbi S, Prevot N, Bonnefoy PB. Incidental finding of COVID-19 lung infection in 18F-FDG PET/CT: what should we do? Clin Nucl Med 45: 649–651, 2020. doi: 10.1097/RLU.0000000000003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maggiolo F, Zoboli F, Arosio M, Valenti D, Guarneri D, Sangiorgio L, Ripamonti D, Callegaro A. SARS-CoV-2 infection in persons living with HIV: a single center prospective cohort. J Med Virol 93: 1145–1149, 2021. doi: 10.1002/jmv.26352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, Li S, Shan H, Jacobi A, Chung M. Chest CT findings in coronavirus risease-19 (COVID-19): relationship to duration of infection. Radiology 295: 200463, 2020. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19). Radiology 295: 715–721, 2020. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323: 1843–1844, 2020. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J, Zhang H, Liu H, Xia H, Tang J, Zhang K, Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 26: 502–505, 2020. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, Wang YY, Xiao GF, Yan B, Shi ZL, Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 9: 386–389, 2020. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin L, Jiang X, Zhang Z, Huang S, Zhang Z, Fang Z, Gu Z, Gao L, Shi H, Mai L, Liu Y, Lin X, Lai R, Yan Z, Li X, Shan H. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 69: 997–1001, 2020. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, Yang Y, Liu B, Wang W, Wei C, Yang J, Ye G, Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol 92: 833–840, 2020. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Gao G, Xu Y, Pu L, Wang Q, Wang L, Wang W, Song Y, Chen M, Wang L, Yu F, Yang S, Tang Y, Zhao L, Wang H, Wang Y, Zeng H, Zhang F. SARS-CoV-2-positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann Intern Med 172: 832–834, 2020. doi: 10.7326/M20-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F, Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 395: 1417–1418, 2020. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 158: 1831–1833.e3, 2020. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahlmann R, Lode H. Medication for COVID-19-an overview of approaches currently under study. Dtsch Arztebl Int 117: 213–219, 2020. doi: 10.3238/arztebl.2020.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368: 1016–1020, 2020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MAB, Um J, Song MS, Jeong HW, Lai VD, Kim Y, Chin BS, Park JS, Chung KH, Foo SS, Poo H, Mo IP, Lee OJ, Webby RJ, Jung JU, Choi YK. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 27: 704–709.e2, 2020. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, Kaewpreedee P, Perera R, Poon LLM, Nicholls JM, Peiris M, Yen HL. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583: 834–838, 2020. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science 369: 50–54, 2020. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zang R, Gomez CM, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, Diamond MS, Ciorba MA, Whelan SPJ, Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 5: eabc3582, 2020. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273, 2020. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun 526: 135–140, 2020. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 14: 185–192, 2020. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419, 2015. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280.e8, 2020. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TWH, Tam AR, Yip CCY, Leung KH, Fung AY, Zhang RR, Lin Y, Cheng HM, Zhang AJX, To KKW, Chan KH, Yuen KY, Leung WK. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology 159: 81–95, 2020. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin X, Lian JS, Hu JH, Gao J, Zheng L, Zhang YM, Hao SR, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 69: 1002–1009, 2020. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nobel YR, Phipps M, Zucker J, Lebwohl B, Wang TC, Sobieszczyk ME, Freedberg DE. Gastrointestinal symptoms and coronavirus disease 2019: a case-control study from the United States. Gastroenterology 159: 373–375.e2, 2020. doi: 10.1053/j.gastro.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguila EJT, Cua IHY, Fontanilla JAC, Yabut VLM, Causing MFP. Gastrointestinal manifestations of COVID-19: impact on nutrition practices. Nutr Clin Pract 35: 800–805, 2020. doi: 10.1002/ncp.10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323: 1061–1069, 2020. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan L, Mu M, Yang P, Sun Y, Wang R, Yan J, Li P, Hu B, Wang J, Hu C, Jin Y, Niu X, Ping R, Du Y, Li T, Xu G, Hu Q, Tu L. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol 115: 766–773, 2020. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu AL, Guan M, Johannesen E, Stephens AJ, Khaleel N, Kagan N, Tuhlei BC, Wan XF. Placental SARS-CoV-2 in a pregnant woman with mild COVID-19 disease. J Med Virol 93: 1038–1044, 2020. doi: 10.1002/jmv.26386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramlall V, Thangaraj PM, Meydan C, Foox J, Butler D, Kim J, May B, De Freitas JK, Glicksberg BS, Mason CE, Tatonetti NP, Shapira SD. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat Med 26: 1609–1615, 2020. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashton JJ, Kammermeier J, Spray C, Russell RK, Hansen R, Howarth LJ, Torrente F, Deb P, Renji E, Muhammed R, Paul T, Kiparissi F, Epstein J, Lawson M, Hope B, Zamvar V, Narula P, Kadir A, Devadason D, Bhavsar H, Beattie RM. Impact of COVID-19 on diagnosis and management of paediatric inflammatory bowel disease during lockdown: a UK nationwide study. Arch Dis Child 105: 1186–1191, 2020. doi: 10.1136/archdischild-2020-319751. [DOI] [PubMed] [Google Scholar]
- 44.Rimondi A, Tontini GE, Mazza S, Caprioli F, Sangiovanni A, Lampertico P, Vecchi M. Fogging IBD management: an unusual case of IBD flare-up during the COVID-19 outbreak. Inflamm Bowel Dis 26: e128–e129, 2020. doi: 10.1093/ibd/izaa184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taxonera C, Alba C, Olivares D. What is the incidence of COVID-19 in patients with IBD in western countries? Gastroenterology 160: 1901–1902, 2021. doi: 10.1053/j.gastro.2020.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang YF, Qiu Y, He JS, Tan JY, Li XZ, Zhu LR, Chen Y, Liu ZJ, Iacucci M, Chen BL, He Y, Ben-Horin S, Shen B, Zeng ZR, Ghosh S, Chen MH, Mao R. Impact of COVID-19 outbreak on the care of patients with inflammatory bowel disease: a comparison before and after the outbreak in South China. J Gastroenterol Hepatol 36: 700–709, 2021. doi: 10.1111/jgh.15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baradaran Ghavami SH, Shahrokh SH, Hossein-Khannazer N, Shpichka A, Asadzadeh Aghdaei H, Timashev P, Vosough M. IBD patients could be silent carriers for novel coronavirus and less prone to its severe adverse events: true or false? Cell J 22: 151–154, 2020. doi: 10.22074/cellj.2020.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernstein CN, Ng SC, Banerjee R, Steinwurz F, Shen B, Carbonnel F, Hamid S, Sood A, Yamamoto-Furusho JK, Griffiths A, Benchimol EI, Travis S, Lopes S, Rubin DT, Kaplan GG, Armstrong D, Gearry R; IBD-Emerging Nations Consortium and the WGO IBD Task Force on COVID-19. Worldwide management of inflammatory bowel disease during the COVID-19 pandemic: an international survey. Inflamm Bowel Dis 27: 836–847, 2020. doi: 10.1093/ibd/izaa202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.An P, Ji M, Ren H, Su J, Ding NS, Kang J, Yin A, Zhou Q, Shen L, Zhao L, Jiang X, Xiao Y, Tan W, Lv X, Li J, Liu S, Zhou J, Chen H, Xu Y, Liu J, Chen M, Cao J, Zhou Z, Shen L, Tan S, Yu H, Dong W, Ding Y. Prevention of COVID-19 in patients with inflammatory bowel disease in Wuhan, China. Lancet Gastroenterol Hepatol 5: 525–527, 2020. doi: 10.1016/S2468-1253(20)30121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brenner EJ, Ungaro RC, Gearry RB, Kaplan GG, Kissous-Hunt M, Lewis JD, Ng SC, Rahier JF, Reinisch W, Ruemmele FM, Steinwurz F, Underwood FE, Zhang X, Colombel JF, Kappelman MD. Corticosteroids, but not TNF antagonists, are associated with adverse COVID-19 outcomes in patients with inflammatory bowel diseases: results from an International Registry. Gastroenterology 159: 481–491.e3, 2020. doi: 10.1053/j.gastro.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higgins PDR, Ng S, Danese S, Rao K. The risk of SARS-CoV-2 in immunosuppressed IBD patients. Crohns Colitis 360 2: otaa026, 2020. doi: 10.1093/crocol/otaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zingone F, Buda A, Savarino EV. Screening for active COVID-19 infection and immunization status prior to biologic therapy in IBD patients at the time of the pandemic outbreak. Dig Liver Dis 52: 604–605, 2020. doi: 10.1016/j.dld.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 5: 434–435, 2020. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potdar AA, Dube S, Naito T, Botwin G, Haritunians T, Li D, Yang S, Bilsborough J, Denson LA Daly M, Targan SR, Fleshner P, Braun J, Kugathasan S, Stappenbeck TS, McGovern DPB. Reduced expression of COVID-19 host receptor, ACE2 is associated with small bowel inflammation, more severe disease, and response to anti-TNF therapy in Crohn's disease [preprint]. medRxiv, 2020. doi: 10.1101/2020.04.19.20070995. [DOI] [Google Scholar]
- 55.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, Crackower MA, Fukamizu A, Hui CC, Hein L, Uhlig S, Slutsky AS, Jiang C, Penninger JM. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436: 112–116, 2005. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao H, Gao F, Xie G, Liu Z. Angiotensin-converting enzyme 2 inhibits apoptosis of pulmonary endothelial cells during acute lung injury shrough Suppressing MiR-4262. Cell Physiol Biochem 37: 759–767, 2015. doi: 10.1159/000430393. [DOI] [PubMed] [Google Scholar]
- 57.Deffner F, Scharr M, Klingenstein S, Klingenstein M, Milazzo A, Scherer S, Wagner A, Hirt B, Mack AF, Neckel PH. Histological evidence for the enteric nervous system and the choroid plexus as alternative routes of neuroinvasion by SARS-CoV2. Front Neuroanat 14: 596439, 2020. doi: 10.3389/fnana.2020.596439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian Q, Fan L, Liu W, Li J, Yue J, Wang M, Ke X, Yin Y, Chen Q, Jiang C. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin Infect Disciaa925, 2020. doi: 10.1093/cid/ciaa925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C, Zhou Y, Wang D, Miao X, Li Y, Hu B. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol 77: 683–690, 2020. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahima RS, Antwi DA. Brain regulation of appetite and satiety. Endocrinol Metab Clin North Am 37: 811–823, 2008. doi: 10.1016/j.ecl.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bostancıklıoğlu M. Temporal correlation between neurological and gastrointestinal symptoms of SARS-CoV-2. Inflamm Bowel Dis 26: e89–e91, 2020. doi: 10.1093/ibd/izaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Effenberger M, Grabherr F, Mayr L, Schwaerzler J, Nairz M, Seifert M, Hilbe R, Seiwald S, Scholl-Buergi S, Fritsche G, Bellmann-Weiler R, Weiss G, Muller T, Adolph TE, Tilg H. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut 69: 1543–1544, 2020. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203: 631–637, 2004. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garrido I, Liberal R, Macedo G. Review article: COVID-19 and liver disease-what we know on 1st May 2020. Aliment Pharmacol Ther 52: 267–275, 2020. doi: 10.1111/apt.15813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX; China Medical Treatment Expert Group for COVID-19, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382: 1708–1720, 2020. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Zheng L, Liu L, Zhao M, Xiao J, Zhao Q. Liver impairment in COVID-19 patients: a retrospective analysis of 115 cases from a single centre in Wuhan city. Liver Int 40: 2095–2103, 2020. doi: 10.1111/liv.14455. [DOI] [PubMed] [Google Scholar]
- 67.Zhang D, Li S, Wang N, Tan HY, Zhang Z, Feng Y. The cross-talk between gut microbiota and lungs in common lung diseases. Front Microbiol 11: 301, 2020. doi: 10.3389/fmicb.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Xu Q, Ma L, Wu D, Gao J, Chen G, Li H. Systematic profiling of ACE2 expression in diverse physiological and pathological conditions for COVID-19/SARS-CoV-2. J Cell Mol Med 24: 9478–9482, 2020. doi: 10.1111/jcmm.15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji D, Qin E, Xu J, Zhang D, Cheng G, Wang Y, Lau G. Non-alcoholic fatty liver diseases in patients with COVID-19: a retrospective study. J Hepatol 73: 451–453, 2020. doi: 10.1016/j.jhep.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertolini A, van de Peppel IP, Bodewes F, Moshage H, Fantin A, Farinati F, Fiorotto R, Jonker JW, Strazzabosco M, Verkade HJ, Peserico G. Abnormal liver function tests in patients with COVID-19: relevance and potential pathogenesis. Hepatology 72: 1864–1872, 2020. doi: 10.1002/hep.31480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chau TN, Lee KC, Yao H, Tsang TY, Chow TC, Yeung YC, Choi KW, Tso YK, Lau T, Lai ST, Lai CL. SARS-associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology 39: 302–310, 2004. doi: 10.1002/hep.20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan YJ, Fielding BC, Goh PY, Shen S, Tan TH, Lim SG, Hong W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol 78: 14043–14047, 2004. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hossain AN. Liver injury in severe COVID-19 infection: current insights and challenges. Expert Rev Gastroenterol Hepatol 14: 879–884, 2020. doi: 10.1080/17474124.2020.1794812. [DOI] [PubMed] [Google Scholar]
- 74.Adams DH, Hubscher SG. Systemic viral infections and collateral damage in the liver. Am J Pathol 168: 1057–1059, 2006. doi: 10.2353/ajpath.2006.051296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Portincasa P, Krawczyk M, Machill A, Lammert F, Di Ciaula A. Hepatic consequences of COVID-19 infection. Lapping or biting? Eur J Intern Med 77: 18–24, 2020. doi: 10.1016/j.ejim.2020.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boeckmans J, Rodrigues RM, Demuyser T, Pierard D, Vanhaecke T, Rogiers V. COVID-19 and drug-induced liver injury: a problem of plenty or a petty point? Arch Toxicol 94: 1367–1369, 2020. doi: 10.1007/s00204-020-02734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan Z, Chen L, Li J, Cheng X, Yang J, Tian C, Zhang Y, Huang S, Liu Z, Cheng J. Clinical features of COVID-19-related liver functional abnormality. Clin Gastroenterol Hepatol 18: 1561–1566, 2020. doi: 10.1016/j.cgh.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roth NC, Kim A, Vitkovski T, Xia J, Ramirez G, Bernstein D, Crawford JM. Post-COVID-19 cholangiopathy: a novel entity. Am J Gastroenterol 116: 1077–1082, 2021. doi: 10.14309/ajg.0000000000001154. [DOI] [PubMed] [Google Scholar]
- 79.Keta-Cov research group. Intravenous ketamine and progressive cholangiopathy in COVID-19 patients. J Hepatol 74: 1243–1244, 2021. doi: 10.1016/j.jhep.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gelbmann CM, Rummele P, Wimmer M, Hofstadter F, Gohlmann B, Endlicher E, Kullmann F, Langgartner J, Scholmerich J. Ischemic-like cholangiopathy with secondary sclerosing cholangitis in critically ill patients. Am J Gastroenterol 102: 1221–1229, 2007. doi: 10.1111/j.1572-0241.2007.01118.x. [DOI] [PubMed] [Google Scholar]
- 81.Leonhardt S, Veltzke-Schlieker W, Adler A, Schott E, Hetzer R, Schaffartzik W, Tryba M, Neuhaus P, Seehofer D. Trigger mechanisms of secondary sclerosing cholangitis in critically ill patients. Crit Care 19: 131, 2015. doi: 10.1186/s13054-015-0861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fu L, Fei J, Xu S, Xiang HX, Xiang Y, Hu B, Li MD, Liu FF, Li Y, Li XY, Zhao H, Xu DX. Liver dysfunction and its association with the risk of death in COVID-19 patients: a prospective cohort study. J Clin Transl Hepatol 8: 246–254, 2020. doi: 10.14218/JCTH.2020.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaltsas A, Sepkowitz K. Community acquired respiratory and gastrointestinal viral infections: challenges in the immunocompromised host. Curr Opin Infect Dis 25: 423–430, 2012. doi: 10.1097/QCO.0b013e328355660b. [DOI] [PubMed] [Google Scholar]
- 84.Singh S, Khan A. Clinical characteristics and outcomes of coronavirus disease 2019 among patients with preexisting liver disease in the United States: a multicenter research network study. Gastroenterology 159: 768–771.e3, 2020. doi: 10.1053/j.gastro.2020.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie H, Zhao J, Lian N, Lin S, Xie Q, Zhuo H. Clinical characteristics of non-ICU hospitalized patients with coronavirus disease 2019 and liver injury: a retrospective study. Liver Int 40: 1321–1326, 2020. doi: 10.1111/liv.14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D'Antiga L. Coronaviruses and immunosuppressed patients: the facts during the third epidemic. Liver Transpl 26: 832–834, 2020. doi: 10.1002/lt.25756. [DOI] [PubMed] [Google Scholar]
- 87.Bhoori S, Rossi RE, Citterio D, Mazzaferro V. COVID-19 in long-term liver transplant patients: preliminary experience from an Italian transplant centre in Lombardy. Lancet Gastroenterol Hepatol 5: 532–533, 2020. doi: 10.1016/S2468-1253(20)30116-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hayashi K, Ito Y, Yamane R, Yoshizaki M, Matsushita K, Kajikawa G, Kozawa T, Mizutani T, Shimizu Y, Nagano K, Tachi K, Yoshioka K, Goto H. The case of a liver-transplant recipient with severe acute respiratory syndrome coronavirus 2 infection who had a favorable outcome. Clin J Gastroenterol 14: 842–845, 2021. doi: 10.1007/s12328-021-01374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fava A, Cucchiari D, Montero N, Toapanta N, Centellas FJ, Vila-Santandreu A, Coloma A, Meneghini M, Manonelles A, Sellares J, Torres I, Gelpi R, Lorenzo I, Ventura-Aguiar P, Cofan F, Torregrosa JV, Perello M, Facundo C, Seron D, Oppenheimer F, Bestard O, Cruzado JM, Moreso F, Melilli E. Clinical characteristics and risk factors for severe COVID-19 in hospitalized kidney transplant recipients: a multicentric cohort study. Am J Transplant 20: 3030–3041, 2020. doi: 10.1111/ajt.16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garin E, Tselikas L, Guiu B, Chalaye J, Edeline J, de Baere T; DOSISPHERE-01 Study Group, et al. Personalised versus standard dosimetry approach of selective internal radiation therapy in patients with locally advanced hepatocellular carcinoma (DOSISPHERE-01): a randomised, multicentre, open-label phase 2 trial. Lancet Gastroenterol Hepatol 6: 17–29, 2021. doi: 10.1016/S2468-1253(20)30290-9. [DOI] [PubMed] [Google Scholar]
- 91.Huang JF, Zheng KI, George J, Gao HN, Wei RN, Yan HD, Zheng MH. Fatal outcome in a liver transplant recipient with COVID-19. Am J Transplant 20: 1907–1910, 2020. doi: 10.1111/ajt.15909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Polak WG, Fondevila C, Karam V, Adam R, Baumann U, Germani G, Nadalin S, Taimr P, Toso C, Troisi RI, Zieniewicz K, Belli LS, Duvoux C. Impact of COVID-19 on liver transplantation in Europe: alert from an early survey of European Liver and Intestine Transplantation Association and European Liver Transplant Registry. Transpl Int 33: 1244–1252, 2020. doi: 10.1111/tri.13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blacher E, Levy M, Tatirovsky E, Elinav E. Microbiome-modulated metabolites at the interface of host immunity. J Immunol 198: 572–580, 2017. doi: 10.4049/jimmunol.1601247. [DOI] [PubMed] [Google Scholar]
- 94.Yuan Y, Zallot R, Grove TL, Payan DJ, Martin-Verstraete I, Sepic S, Balamkundu S, Neelakandan R, Gadi VK, Liu CF, Swairjo MA, Dedon PC, Almo SC, Gerlt JA, de Crecy-Lagard V. Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens. Proc Natl Acad Sci USA 116: 19126–19135, 2019. doi: 10.1073/pnas.1909604116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Candela M, Biagi E, Maccaferri S, Turroni S, Brigidi P. Intestinal microbiota is a plastic factor responding to environmental changes. Trends Microbiol 20: 385–391, 2012. doi: 10.1016/j.tim.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 96.Wang J, Li F, Wei H, Lian ZX, Sun R, Tian Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J Exp Med 211: 2397–2410, 2014[Erratum inJ Exp Med211: 2683, 2014]. doi: 10.1084/jem.20140625. [25366965] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deriu E, Boxx GM, He X, Pan C, Benavidez SD, Cen L, Rozengurt N, Shi W, Cheng G. Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I interferons. PLoS Pathog 12: e1005572, 2016. doi: 10.1371/journal.ppat.1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Groves HT, Cuthbertson L, James P, Moffatt MF, Cox MJ, Tregoning JS. Respiratory disease following viral lung infection alters the murine gut microbiota. Front Immunol 9: 182, 2018. doi: 10.3389/fimmu.2018.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fagundes CT, Amaral FA, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, Teixeira MM, Souza DG. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol 188: 1411–1420, 2012. doi: 10.4049/jimmunol.1101682. [DOI] [PubMed] [Google Scholar]
- 100.Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, Hoogendijk AJ, de Beer R, de Vos A, Belzer C, de Vos WM, van der Poll T, Wiersinga WJ. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65: 575–583, 2016. doi: 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 20: 159–166, 2014. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 102.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, Boushey HA, Zoratti E, Ownby D, Lukacs NW, Lynch SV. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci USA 111: 805–810, 2014. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol 78: 481–504, 2016. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, Jablonski K, Kleerup E, Lynch SV, Sodergren E, Twigg H, Young VB, Bassis CM, Venkataraman A, Schmidt TM, Weinstock GM; Lung HIV Microbiome Project. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med 187: 1067–1075, 2013. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marsland BJ, Trompette A, Gollwitzer ES. The gut-lung axis in respiratory disease. Ann Am Thorac Soc 12 Suppl 2: S150–S156, 2015. doi: 10.1513/AnnalsATS.201503-133AW. [DOI] [PubMed] [Google Scholar]
- 106.Zhu H, Lu X, Ling L, Li H, Ou Y, Shi X, Lu Y, Zhang Y, Chen D. Houttuynia cordata polysaccharides ameliorate pneumonia severity and intestinal injury in mice with influenza virus infection. J Ethnopharmacol 218: 90–99, 2018. doi: 10.1016/j.jep.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 107.Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, Zhou Z, Yang J, Zhong J, Yang D, Guo L, Zhang G, Li H, Xu Y, Chen M, Gao Z, Wang J, Ren L, Li M. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis 71: 713–720, 2020. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, Guo F, Zhang X, Luo R, Huang C, Lu H, Zheng B, Zhang J, Yan R, Zhang H, Jiang H, Xu Q, Guo J, Gong Y, Tang L, Li L. Alterations of the gut microbiota in patients with COVID-19 or H1N1 influenza. Clin Infect Dis 71: 2669–2678, 2020. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, Withers DR, Hugues S, Farrar MA, Reith W, Eberl G, Baldassano RN, Laufer TM, Elson CO, Sonnenberg GF. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science 348: 1031–1035, 2015. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, Wherry EJ, Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37: 158–170, 2012. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huffnagle GB, Dickson RP, Lukacs NW. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol 10: 299–306, 2017. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395: 507–513, 2020. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395: 497–506, 2020. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guo L, Wei D, Zhang X, Wu Y, Li Q, Zhou M, Qu J. Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol 10: 2752, 2019. doi: 10.3389/fmicb.2019.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lescure FX, Bouadma L, Nguyen D, Parisey M, Wicky PH, Behillil S, Gaymard A, Bouscambert-Duchamp M, Donati F, Le HQ, Enouf V, Houhou-Fidouh N, Valette M, Mailles A, Lucet JC, Mentre F, Duval X, Descamps D, Malvy D, Timsit JF, Lina B, van-der-Werf S, Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis 20: 697–706, 2020. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 8: 475–481, 2020[Erratum inLancet Respir Med8: e26, 2020]. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lai CC, Wang CY, Hsueh PR. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J Microbiol Immunol Infect 53: 505–512, 2020. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Columpsi P, Sacchi P, Zuccaro V, Cima S, Sarda C, Mariani M, Gori A, Bruno R. Beyond the gut bacterial microbiota: the gut virome. J Med Virol 88: 1467–1472, 2016. doi: 10.1002/jmv.24508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell 138: 30–50, 2009. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 120.Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, Angly FE, McDole T, MariellaRP, Jr., Rohwer F, Haynes M. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci USA 108 Suppl 1: 4547–4553, 2011. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci USA 109: 17621–17626, 2012. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fernandes P. Antibacterial discovery and development–the failure of success? Nat Biotechnol 24: 1497–1503, 2006. doi: 10.1038/nbt1206-1497. [DOI] [PubMed] [Google Scholar]
- 123.Lu TK, Koeris MS. The next generation of bacteriophage therapy. Curr Opin Microbiol 14: 524–531, 2011. doi: 10.1016/j.mib.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 124.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol 14: 654–659, 2013. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Parker MT. An ecological framework of the human virome provides classification of current knowledge and identifies areas of forthcoming discovery. Yale J Biol Med 89: 339–351, 2016. [PMC free article] [PubMed] [Google Scholar]
- 126.Katzourakis A, Gifford RJ, Tristem M, Gilbert MT, Pybus OG. Macroevolution of complex retroviruses. Science 325: 1512, 2009. doi: 10.1126/science.1174149. [DOI] [PubMed] [Google Scholar]
- 127.Grandi N, Tramontano E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol 9: 2039, 2018. doi: 10.3389/fimmu.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]