Abstract
Background:
Metabolic alterations provide substrates that influence chromatin structure to regulate gene expression that determines cell function in health and disease. Heightened proliferation of smooth muscle cells (SMC) leading to the formation of a neointima is a feature of pulmonary arterial hypertension (PAH) and systemic vascular disease. Increased glycolysis is linked to the proliferative phenotype of these SMC.
Methods:
RNA Sequencing was applied to pulmonary arterial (PA) SMC from PAH patients with and without a BMPR2 mutation vs. control PASMC to uncover genes required for their heightened proliferation and glycolytic metabolism. Assessment of differentially expressed genes established metabolism as a major pathway, and the most highly upregulated metabolic gene in PAH PASMC was aldehyde dehydrogenase family 1 member 3 (ALDH1A3), an enzyme previously linked to glycolysis and proliferation in cancer cells and systemic vascular SMC. We determined if these functions are ALDH1A3-dependent in PAH PASMC, and if ALDH1A3 is required for the development of pulmonary hypertension in a transgenic mouse. Nuclear localization of ALDH1A3 in PAH PASMC led us to determine whether and how this enzyme coordinately regulates gene expression and metabolism in PAH PASMC.
Results:
ALDH1A3 mRNA and protein were increased in PAH vs control PASMC, and ALDH1A3 was required for their highly proliferative and glycolytic properties. Mice with Aldh1a3 deleted in SMC did not develop hypoxia-induced PA muscularization or pulmonary hypertension. Nuclear ALDH1A3 converted acetaldehyde to acetate to produce acetyl-CoA to acetylate H3K27, marking active enhancers. This allowed for chromatin modification at nuclear factor Y (NFY)A binding sites via the acetyltransferase KAT2B and permitted NFY mediated transcription of cell cycle and metabolic genes that is required for ALDH1A3-dependent proliferation and glycolysis. Loss of BMPR2 in PAH SMC with or without a mutation upregulated ALDH1A3, and transcription of NFYA and ALDH1A3 in PAH PASMC was β-catenin dependent.
Conclusions:
Our studies have uncovered a metabolic-transcriptional axis explaining how dividing cells use ALDH1A3 to coordinate their energy needs with the epigenetic and transcriptional regulation of genes required for SMC proliferation. They suggest that selectively disrupting the pivotal role of ALDH1A3 in PAH SMC, but not EC, is an important therapeutic consideration.
Keywords: ALDH1A3, metabolism, cell cycle, NFY, H3K27ac, SMC, glycolysis, proliferation, PAH
Introduction
Pulmonary arterial hypertension (PAH) is a progressive disease in which proliferation of smooth muscle cells (SMC) causes progressive obliteration of pulmonary arteries (PA) resulting in increased resistance to blood flow and culminating in right heart failure. Current therapies attempt to dilate the PA but do not address the mechanism of occlusive remodeling and are thus mainly effective in temporarily improving quality of life and survival. In familial PAH, 70–80% of those affected have a mutation in bone morphogenetic receptor 2 (BMPR2). These mutations also account for 20% of sporadic cases of idiopathic (I) PAH1, but a deficiency in BMPR2 expression or signaling is evident even in patients without a known mutation, or when PAH is a complication of other medical disorders2. While BMPR2 deficiency in PA endothelial cells (PAEC) can promote expression and release of growth factors to drive proliferation of PASMC, reduced BMPR2 signaling in PASMC also contributes directly to their enhanced proliferation3, 4.
The excessive growth and glycolytic nature of the PAH PASMC is similar to that of cancer cells. Suppressing the glycolytic metabolism of PAH PASMC or cancer cells can reduce their heightened proliferation5. Excessive glycolysis has been largely related to the fast turnover rate of ATP required by proliferating cells6. A number of studies, however, have shown that metabolites can change the landscape of chromatin to modulate gene expression in development and disease7–9. For example, in acute myeloid leukemia, isocitrate dehydrogenase 1/2 (IDH1/2) mutations result in DNA and histone hypermethylation as accumulation of (R)-2-hydroxyglutarate (R-2HG) inhibits the activity of TET-family DNA and JmjC-family histone demethylase enzymes10. In our previous studies we observed in PA endothelial cells (EC), that a by-product of PFKFB3 mediated glycolysis is acetyl-CoA, the acetyl source for histone acetylation11 of histone H3 lysine 27 (H3K27ac) enhancers at sites of Notch 1 intracellular domain-RBPJ and c-Myc regulated transcription9. Others12 have also shown that acetyl-CoA, via acetyltransferases such as p300, can mediate H3K27ac and facilitate gene transcription.
Aldehyde dehydrogenase family 1 member 3 (ALDH1A3) can convert acetaldehyde to acetate to produce nuclear acetyl-CoA13, 14, and pyruvate15 and citrate16 can also be sources of acetyl-CoA. ALDH1A3 has been described as a regulator of glycolysis linked to proliferation of cancer cells17 and more recently, SMC, in a rat carotid artery injury model18. Nuclear factor Y (NFY), also known as the CCAAT-binding factor CBF, is a transcription factor complex consisting of NFYA, NFYB and NFYC subunits19 that regulates cell cycle genes in proliferating cells20. The NFYA subunit binds to DNA and activates transcription at the proximal promotor21. Acetyltransferase KAT2B22 acts as a cofactor, that transfers acetyl groups to histones permitting transcription of target genes.
In this study, we show that active ß-catenin in PAH PASMC increases transcription of ALDH1A3 and NFYA. Nuclear ALDH1A3 generates acetyl-CoA. This metabolite is required together with the acetyltransferase KAT2B, to acetylate H3K27, producing active enhancers at NFYA transcription factor binding sites of cell cycle and metabolic genes that drive the PASMC proliferative and glycolytic phenotype. Thus, elevated ALDH1A3 can orchestrate changes in metabolism needed both for cell energy and for targeted epigenetic and transcriptional regulation of genes required by dividing cells.
Methods
The authors declare that all supporting data are available within the article and its files in the Data Supplement.
Cell culture.
De-identified primary human small PASMC were harvested from lungs explanted from patients with hereditary PAH with a BMPR2 mutation or idiopathic PAH, undergoing transplantation, or from unused donor lungs (as controls). Tables I-IV in the Data Supplement indicate demographics and other characteristics related to hemodynamic assessments and PAH medications.
PASMC and PAEC were isolated and cultured from small pulmonary arteries <1mm in diameter, and were used between passages 3–8, but at the same passage in each experiment. PASMC were isolated and cultured in Smooth Muscle Growth Medium-2 containing 5% fetal bovine serum (FBS) (Lonza, Indianapolis, IN), and PAEC were cultured in Endothelial Cell Medium (Sciencell, Carlsbad, CA). Isolated PASMC were cultured from the mice following removal of the adventitia and the EC layer. Cells were maintained in 95% air and 5% CO2 at 37°C. Starvation medium was basal medium plus 0.2% FBS. The cells were routinely tested for mycoplasma, and only mycoplasma negative cells were used.
Animal models.
SMC-specific Aldh1a3−/− mice were created by crossing non-inducible SM22-Cre mice (C57BL/6 background) with Aldh1a3−/− floxed mice. Wild-type littermates were used as controls. The number of mice per experiment is indicated in the figure legends. Male mice 7–8 weeks of age were housed in hypoxia (10% O2) for three weeks, or in room air for three weeks. RVSP, RVH, cardiac function and output, and pulmonary artery acceleration time (PAAT) were measured, all according to methods previously published by our group9. The heart and lungs were perfused with PBS, the left lung fixed, and sections embedded in paraffin for immunohistochemistry and immunofluorescence. The right lung was snap-frozen in liquid N2 and kept at −80°C.
Immunoblotting.
PASMC were synchronized by serum-starvation (0.2% FBS) for 48h, then cultured with 5% FBS for 72h. Protein concentration was determined using the BCA assay. Lysates were separated by SDS-PAGE and transferred to a PVDF membrane. The membranes were incubated with primary antibodies as listed below in 5% BSA. Appropriate secondary antibodies were used.
Nuclear and cytoplasmic fractions.
Nuclear and cytoplasmic fractions were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher Scientific, Waltham, MA).
Immunoprecipitation.
Nuclear extracts were diluted prior to immunoprecipitation. Equal protein concentrations were determined by BCA assay as described above. Diluted nuclear extracts or undiluted whole cell extracts were incubated with specific antibodies. Dynabeads Protein-G (Invitrogen) were added to cell extracts containing antibodies, and proteins were eluted in acid.
Immunofluorescence.
Formaldehyde-fixed, paraffin-embedded human or mouse lung tissue sections were deparaffined and rehydrated. Antigen retrieval was performed. Then the sections were incubated with primary antibodies against SM22α (1:400; Abcam, Cambridge, CA) and ALDH1A3 (1:100, Abgent, San Diego, CA) or PCNA(1:400, Cell Signaling Technology, Danvers MA), PKM2 (1:200, Cell Signaling Technology, Danvers MA), NFYA (1:100, Santa Cruz Technology, CA) and active ß-catenin (1:100, Millipore, Massachusetts, MA) overnight at 4°C. Sections were washed three times with PBS and then incubated with the secondary antibody Confocal analysis was performed using a confocal laser-scanning microscope (FV1000, Olympus, Center Valley, PA).
Proliferation assays.
Proliferation of PASMC was assessed by cell counts and MTT assays. Forty-eight hours following transfection in serum starvation (0.2% FBS), PASMC were exposed to 5% FBS for 72h, and cell growth was assessed in 48-well plates for the MTT cell proliferation assay (American Type Culture Collection (ATCC), Manassas, Virginia). Cell number was verified by cell count as shown in supplementary figures.
Reverse transcription qPCR (RT-qPCR).
Total RNA was extracted and purified using the Quick-RNA MiniPrep Kit (Zymo Research, Irvine, CA). The quantity and quality of RNA was determined by using a spectrophotometer. RNA was reverse transcribed using the High Capacity RNA to cDNA Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Primer sequences were designed using PrimerBank and are listed in the Supplementary Materials. Expression levels of genes was normalized to the expression level of β-actin. Primers are listed in Tables VI-VIII in the Data Supplement.
RNA-seq sample preparation and analysis.
RNA was extracted using the RNeasy Mini Kit (QIAGEN, Hilden, Germany). Libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit (Illumina, San Diego, CA) and subjected to sequencing on 1–2 Illumina HiSeq 2000 lanes to obtain an average of approximately 100–150 million uniquely mapped reads for each sample (Stanford Personalized Medicine Sequencing Core).
ChIP-Seq (chromatin immunoprecipitation followed by sequencing) and analysis.
Cells were trypsinized and cross-linked with 1% formaldehyde (EMD Millipore) and neutralized with 2M glycine (ThermoFisher Scientific). Nuclear pellets were immunoprecipitated with H3K27ac antibody (Cell signaling Technology). For the input sample, 100μl of sheared nuclear lysate was removed and stored. Protein A/G agarose beads (Millipore) were added to the chromatin-antibody complex and then the beads were eluted with SDS buffer (Santa Cruz Biotechnology). Supernatant containing ChIP-DNA was reverse cross-linked. ChIP-DNA was treated with RNase A (Qiagen) and proteinase K (ThermoFisher Scientific) and then purified. The ChIP-DNA samples were end repaired and A-tailed. The Illumina TruSeq adapters (Illumina, San Diego, CA) were ligated and size-selected from the gel before PCR amplification. PCR products were purified and size selected in the gel again. The final purified samples were sequenced on HiSeq4000 (Illumina). ChIP-seq data were aligned to the human genome (hg19). The pipeline for the preprocess is available online (https://github.com/ny-shao/chip-seq_preprocess).
ChIP-qPCR.
Samples were prepared as described above for ChIP-Seq with NFYA or active-β-catenin antibodies. Input DNA (before immunoprecipitation) and immunoprecipitated DNA samples were subjected to real-time PCR analysis. Primer sequences were designed using PrimerBank and are listed in the Supplementary Materials. Expression levels of bound sequences were normalized to the input DNA of those sequences.
Acetyl-CoA quantification.
Acetyl-CoA was measured by PicoProbe Acetyl-CoA Assay Kit (Abcam) following the manufacturer’s instructions: the assay was performed after 72h of serum stimulation preceded by 48 h serum starvation of PASMC with or without ALDH1A3 siRNA.
Acetaldehyde quantification.
Nuclear and cytosolic fractions were isolated as described above. The working reagent was added to the standards and sample wells, mixed briefly and thoroughly, then incubated for 30min at room temperature. Optical density was read at 565nm.
Statistics.
All data are expressed as arithmetical mean ±SEM. Multiple group comparisons were calculated using one-way ANOVA. Multiple group comparisons with multiple treatments were calculated using two-way ANOVA or repeated measures two-way ANOVA followed by Bonferroni analysis as indicated in the Figure legends. Statistical differences were assessed by either the unpaired two-tailed Student t test when two groups were independent, or by the paired Student t test when the same biological sample was being assessed at a different time or with a different treatment in the same experiment. A p-value of <0.05 was considered significant. The number of experiments, animals per group, and the statistical test used are indicated in the figure legends or in the appropriate text. Power calculations were based upon similar studies carried out by our group.
Study approval.
The Animal Care Committee of Stanford University approved all protocols, in keeping with the regulations of the American Physiological Society. Procurement of the tissues from human subjects is approved by the Administrative Panel on Human Subjects in Medical Research at Stanford University (IRB #350, Panel 6). Written informed consent was received from participants prior to inclusion in the study by the PHBI and all data were de-identified and accessed from the Data Coordinating Center at the University of Michigan.
Data and Software Availability.
RNAseq and ChIP-seq data have been deposited to GEO. The accession number is GEO: GSE168905.
Antibodies, primers, siRNA, and assays:
Detailed information is available under “Expanded Methods” in the Data Supplement.
Results
Transcriptomic analysis identifies increased ALDH1A3 in PASMC from PAH patients.
We carried out transcriptomic analyses using RNA sequencing to link altered gene expression to the hyperproliferative and glycolytic phenotype observed in PASMC from patients with PAH. PASMC were cultured from lungs removed at the time of transplant from 12 patients, including five with hereditary (H) PAH associated with a BMPR2 mutation and nine with idiopathic (I) PAH in whom no mutation was identified. These patients are collectively referred to as PAH. PASMC cultured from nine age and gender matched unused donor lungs served as controls. Other demographic and phenotypic data are shown as Tables I-IV in the Data Supplement.
Of 181 genes differentially expressed in PAH PASMC (FDR <0.1), 80 were upregulated as shown in the heatmap (Figure 1A) and the volcano plot (Supplemental Figure 1A). Among the highly upregulated transcripts, was ALDH1A3, which encodes an aldehyde dehydrogenase linked to glycolysis and hyper-proliferation of highly invasive cancer cells17, 23, as well as proliferation of atrial appendage progenitor cells23 and vascular SMC that form a neointima in balloon-injured rat carotid arteries18. ALDH1A3, was the only ALDH isoenzyme of the 19 described24 that was significantly changed in PASMC in this transcriptomic analysis. It was represented in 6/10 KEGG analysis pathways related to differentially expressed genes (Figure IB in the Data Supplement and gene list in Excel File I in the Data Supplement). We found no differences in the expression of ALDH1A3 when comparing IPAH vs. HPAH patients, and our subsequent PAH analyses reflected a combination of patients from both subgroups. Table V in the Data Supplement indicates the patient or control source of the PASMC or tissue used in each experiment.
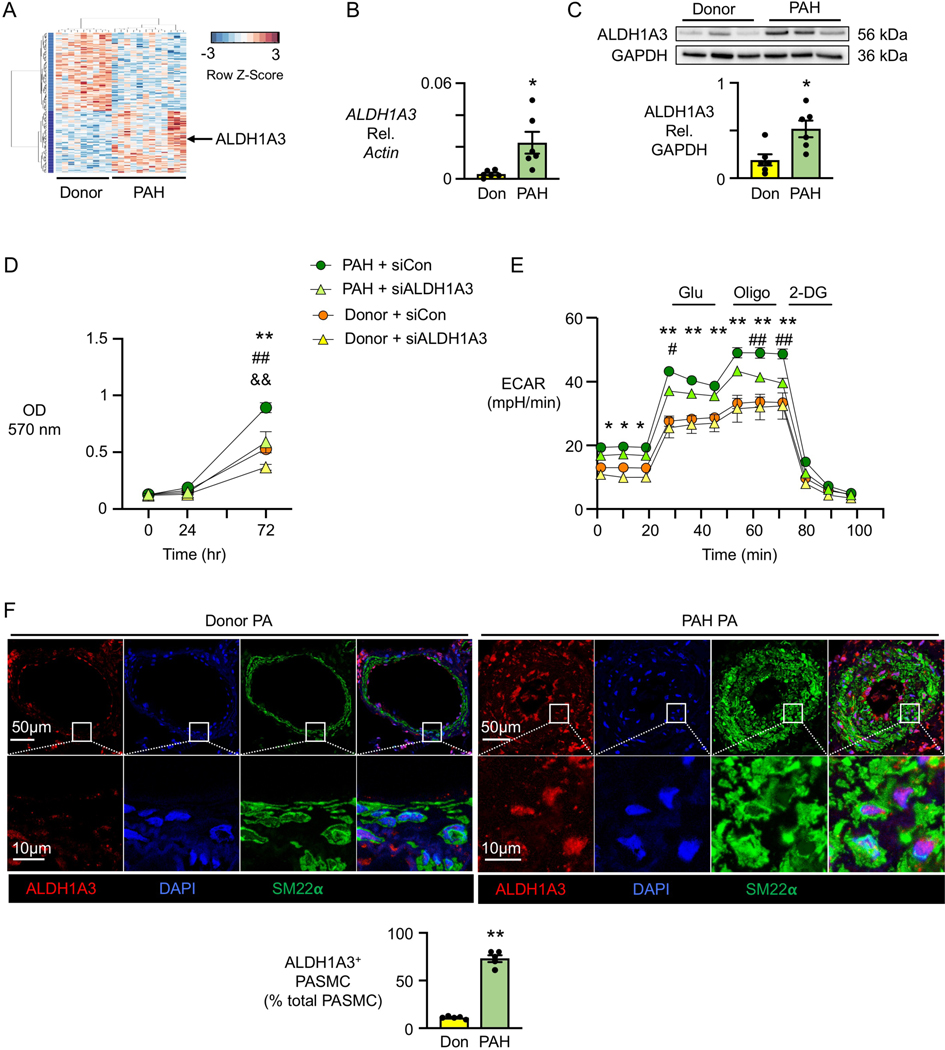
Figure 1. ALDH1A3 is induced in PAH PASMC.
(A) Heatmap of gene expression in PASMC from nine Donor controls vs. 12 PAH patients.
(B) ALDH1A3 mRNA expression in six donor controls (Don) and six PAH patients by qPCR under 48h serum starvation (0.2% FBS) followed by 72h serum stimulation (5% FBS).
(C) Representative immunoblot of ALDH1A3 protein expression relative to GAPDH in three donor controls and three PAH patients, with quantification, in PASMC from six donor controls and six PAH patients under the conditions of B. In (B, C), n=6. *p<0.05 by unpaired Student t test and Welch t-test in B since the variance was unequal.
(D) MTT assay (OD 570 nm) as a measure of cell proliferation using PASMC from three donor controls and three PAH patients transfected with non-targeting control (Con) siRNA or ALDH1A3 siRNA. Values are given after 48h of serum starvation (0 hour) followed by serum stimulation at 24, 48 and 72 hours.
(E) Extracellular acidification rate (ECAR) in response to glucose (Glu), oligomycin (Oligo), and 2-deoxyglucose (2DG). The measurements were normalized to cell numbers. In (D, E), n=3. *p<0.05, **p<0.01, Donor siCon vs PAH siCon; #p<0.05, ##p<0.01 PAH siCon vs PAH siALDH1A3, &&p<0.01 Donor siCon vs siALDH1A3, at the indicated time point, by repeated measures two-way ANOVA followed by Bonferroni analysis.
(F) Representative immunofluorescent staining of ALDH1A3 and SM22⍺ in pulmonary arteries of five donor controls and five PAH patients. Quantification of percent fluorescence of ALDH1A3 in five donor and five PAH lung tissues, three PA in each sample averaged. n=5. **p<0.01 by unpaired Student t test.
Elevation in ALDH1A3 mRNA in PAH vs donor PASMC was first confirmed by qPCR (Figure IC in the Data Supplement). We then related the elevation in ALDH1A3 more directly to the hyperproliferative and glycolytic phenotype of PAH vs. donor PASMC. First, we documented that ALDH1A3 mRNA (Figure 1B) and ALDH1A3 protein (Figure 1C) were elevated in PAH vs. donor PASMC in which proliferation was induced by first synchronizing the cells by 48h serum starvation, then followed during 72h of serum stimulation. We next determined whether ALDH1A3 is required for the heightened PASMC proliferation that is linked to glycolysis. We reduced ALDH1A3 by siRNA in PAH and donor PASMC and assessed cell proliferation by the MTT assay and cell count under the conditions described above. PASMC proliferation in PAH cells transfected with ALDH1A3 siRNA was reduced to donor control levels, and a further decrease in proliferation was evident in donor cells with reduced ALDH1A3. Figure 1D shows the MTT assay results and Supplemental Figure ID, shows the efficient ALDH1A3 knockdown and cell count results. Furthermore, heightened glycolysis, as measured by the extracellular acidification rate (ECAR), was reduced in PAH PASMC by decreasing ALDH1A3 (Figure 1E). Oxygen Consumption Rate (OCR) was significantly increased by ALDH1A3 siRNA in donor control PASMC, but not in PAH PASMC although a trend was apparent (Figure IE in the Data Supplement). In these Seahorse assays, cells were seeded at equal density 8h before the assay and verified as unchanged after the assay. We also showed that PAH PASMC were resistant to apoptosis compared to donor control PASMC, as detected by Caspase assay, and the resistance to apoptosis was negated by reducing ALDH1A3 in PAH PASMC (Figure IF in the Data Supplement).
The increase in ALDH1A3 associated with PASMC proliferation in cultured PAH vs. donor PASMC was then linked to the expanded SMC compartment of the media and neointima of the PA in PAH lung tissue sections. ALDH1A3 protein expression was abundant in PASMC in PAH compared to donor PA and was specifically localized to the nucleus as assessed by confocal microscopic analysis (Figure 1F), although it appeared to also be increased in some endothelial cells lining the vessel lumen as well as in some of the perivascular cells, where it was mostly evident in the cytoplasm. Immunoperoxidase staining substantiated an increase in ALDH1A3 positive cells particularly in the intima and media in PA of all sizes and severity of lesions including those characterized by medial hypertrophy, an occlusive neointima, or plexiform features (Figure IG-J in the Data Supplement).
As we observed ALDH1A3 in PAEC in the tissue sections, we also investigated the role of ALDH1A3 in those cells. Indeed, we found that ALDH1A3 was increased in PAH PAEC but in contrast to PASMC, there was a similar rate of proliferation in PAH and control PAEC. Moreover, reducing ALDH1A3 resulted in only a trend toward a reduction in glycolysis or proliferation of both cell types (Figure IK in the Data Supplement) but in a significant increase in the propensity to apoptosis in the PAH PAEC (Figure IL in the Data Supplement). While ALDH1A3 immunoreactivity of some cells in the intima and adventitia was quite strong, our isolation technique included both intimal and medial SMC. Adventitial cells were not studied.
Depleting SMC Aldh1A3 in mice prevents hypoxia induced pulmonary hypertension.
Hypoxia alone can cause SMC proliferation and pulmonary hypertension25. To determine whether hypoxia also increases ALDH1A3, we subjected donor PASMC to hypoxia (2% O2 for 72 hours) and observed elevated ALDH1A3 compared to PASMC maintained in room air (Figure 2A). Similarly, we observed elevated ALDH1A3 in the nuclei of SMC in PA with increased muscularization in lung sections from mice that were exposed to three weeks of chronic hypoxia (10% O2) compared to the SMC in PA from mice maintained in normoxia (21% O2) (Figure 2B). Elevated ALDH1A3 was also apparent in some of the perivascular cells in the vessel wall of the mice with chronic hypoxia induced pulmonary hypertension. We therefore determined whether elevated ALDH1A3 in SMC was required for the proliferation of PASMC and the development of disease. SM22α Cre mice were bred with floxed Aldh1a3 mice (a kind gift from Dr. Norbert B. Ghyselinck of Institut de Génétique et de Biologie Moléculaire et Cellulaire, France) to generate SM22α-Aldh1a3−/− (KO) mice and littermate controls (SM22α-Aldh1a3+/+, WT). We verified knockout of Aldh1a3 in SMC by PCR analysis of tail genomic DNA shown in Figure IIA in the Data Supplement. Loss of ALDH1A3 protein was evident by absence of fluorescent staining in the PASMC in lungs from KO vs WT mice under hypoxia (Figure IIB in the Data Supplement). SM22α-Aldh1a3−/− mice showed no visible developmental defects in room air judged by body weight, heart rate and cardiac output when compared to littermate controls (Figure IIC in the Data Supplement). At eight to ten weeks of age male SM22α-Aldh1a3−/− mice and littermate controls were randomly selected for exposure to three weeks of chronic hypoxia (10% O2) or were maintained in room air. In contrast to their control (WT) littermates, SM22α-Aldh1a3−/− mice did not develop PH in hypoxia, as judged by right ventricular systolic pressure (RVSP) (Figure 2C), right ventricular acceleration to ejection time (AT/ET) determined by echocardiography (Figure 2D), and right ventricular hypertrophy (RVH) measured as the ratio of the weight of the right ventricle/left ventricle + septum (RV/LV+S) (Figure 2E). Consistent with this, there was a reduction in the hypoxia-induced muscularization of peripheral PA in SM22α-Aldh1a3−/− mice vs. littermate controls as assessed using an anti-SM22a antibody (Figure 2F).
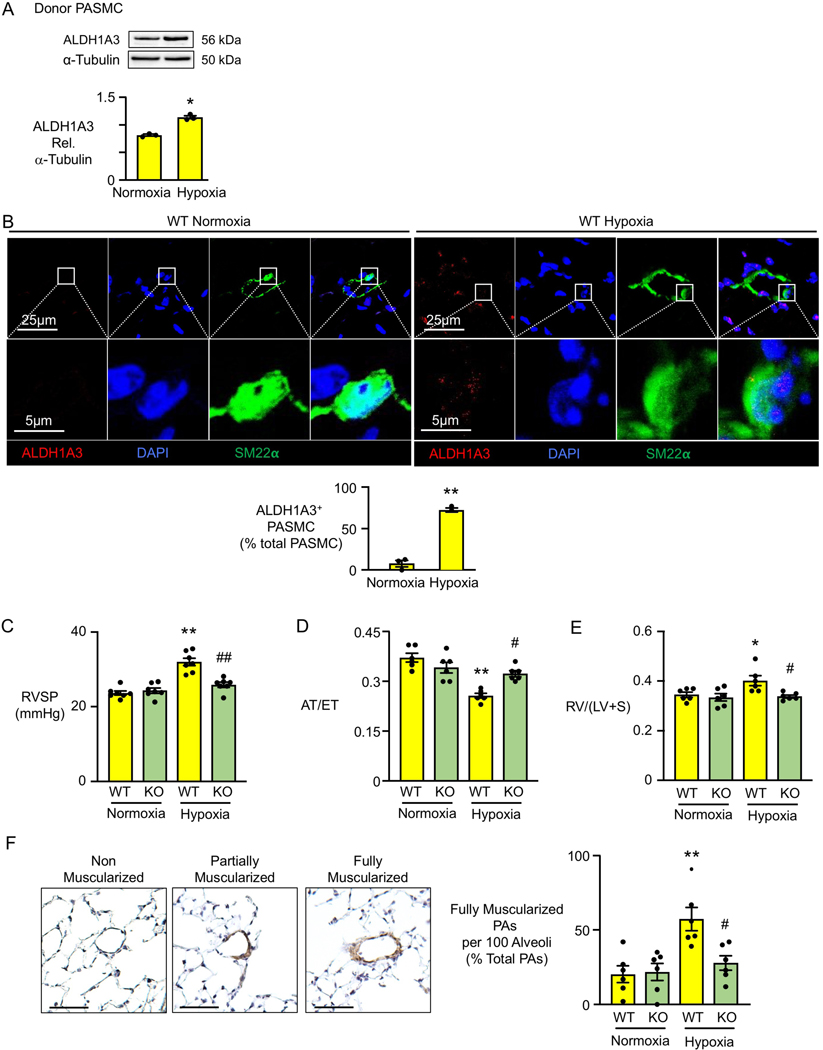
Figure 2. Mice with Deleted Aldh1a3 in SMC Do Not Develop Pulmonary Hypertension.
(A) Representative immunoblot of ALDH1A3 expression relative to α-tubulin with quantification in PASMC of three Donor controls, under normoxia (21% O2) or hypoxia (2% O2) for 72h. *p<0.05 by paired Student t test.
(B) Representative immunofluorescent staining of ALDH1A3 and SM22⍺ with quantification in pulmonary arteries of C57 wild type mice under three weeks of normoxia or hypoxia (10% O2). Scale bar =20μM. Average of five PA per mouse, n=3 mice, **p<0.01, by unpaired Student t test.
(C) Right ventricular systolic pressure (RVSP) of SM22-Aldh1a3−/− (KO) and wild type littermates (WT) after three weeks of hypoxia or normoxia.
(D) the ratio of right ventricular acceleration to ejection time (AT/ET)
(E) right ventricular hypertrophy (right ventricle/(left ventricle+septum) (RVH).
(F) Percentage of fully muscular arteries at alveolar duct and wall level is quantified per field of 100 alveoli in three fields in each mouse. Representative histology of non-muscularized, partially muscularized and fully muscularized arteries on the left. In (C, D, E): n=6 mice per group. *p<0.05, **p<0.01, WT normoxia vs. WT hypoxia; #p<0.05, ##p<0.01 hypoxia: WT vs KO, by two-way ANOVA followed by Bonferroni analysis.
We related our observations in the SM22α-Aldh1a3−/− mice to heightened SMC proliferation and glycolytic metabolism. First PASMC and aortic SMC were isolated from three WT and three KO male mice and we verified by qPCR only trivial expression of Aldh1a3 in the KO compared to WT mice (Figure IID in the Data Supplement). We observed decreased proliferation by cell count and glycolysis by ECAR in PAMSC from KO vs. WT mice, but OCR was not different (Figure IIE in the Data Supplement). Consistent with this, in the lung tissue sections from the mice, PCNA and PKM2 were increased in WT compared to KO PASMC of mice under hypoxia, supporting suppression of proliferation and glycolysis by inhibition of ALDH1A3 (Figure IIF in the Data Supplement). We used male mice in this study because female mice show only a minimal elevation in RVSP in response to hypoxia, a feature described in previous studies26 and verified by our data in mice in this experiment (Figure IIG in the Data Supplement).
ALDH1A3 regulates mRNA expression of cell cycle and metabolic genes in PAH PASMC.
To determine gene expression changes linked to the elevated level of ALDH1A3 in PAH vs. donor PASMC, we carried out transcriptomic analysis comparing PAH PASMC transfected with ALDH1A3 siRNA or non-targeting control siRNA. We found 243 differentially expressed genes with an FDR <0.01 (Figure 3A). KEGG enrichment analyses of genes (Excel File II in the Data Supplement) downregulated when ALDH1A3 was reduced in PAH PASMC indicated an impact on cell cycle genes (Figure IIIA in the Data Supplement). This was evident by decreased expression of transcripts such as cyclin B1 (CCNB1) and cyclin A2 (CCNA2) that control cell cycle transition27, cell division cycle 20 (CDC20) that activates the anaphase promoting complex (APC/C) to initiate chromatid separation and entrance into anaphase28; monopolar spindle 1 kinase (TTK), a key spindle assembly checkpoint protein that regulates proper chromosomal alignment and segregation during mitosis29; and mitotic arrest deficient 2 like 1 (MAD2L1), a component of the spindle-assembly checkpoint that prevents the onset of anaphase until all chromosomes are properly aligned at the metaphase plate30. Using qPCR, we confirmed a reduction in these transcripts when ALDH1A3 is decreased by siRNA in PAH PASMC (Figure 3B), and an increase in these cell cycle genes in PAH vs. donor PASMC (Figure 3C). Protein levels of these genes were similarly reduced in PAH PASMC transfected with ALDH1A3 siRNA vs. control siRNA; CCNB1 and TTK are shown as examples (Figure IIIB in the Data Supplement). Flow Cytometry cell cycle analysis, using propidium iodide DNA staining in PAH PASMC following transfection of ALDH1A3 siRNA vs. control siRNA, revealed a reduction in the G2/M phase of the cell cycle in the PAH PASMC with reduced ALDH1A3 (Figure IIIC in the Data Supplement).
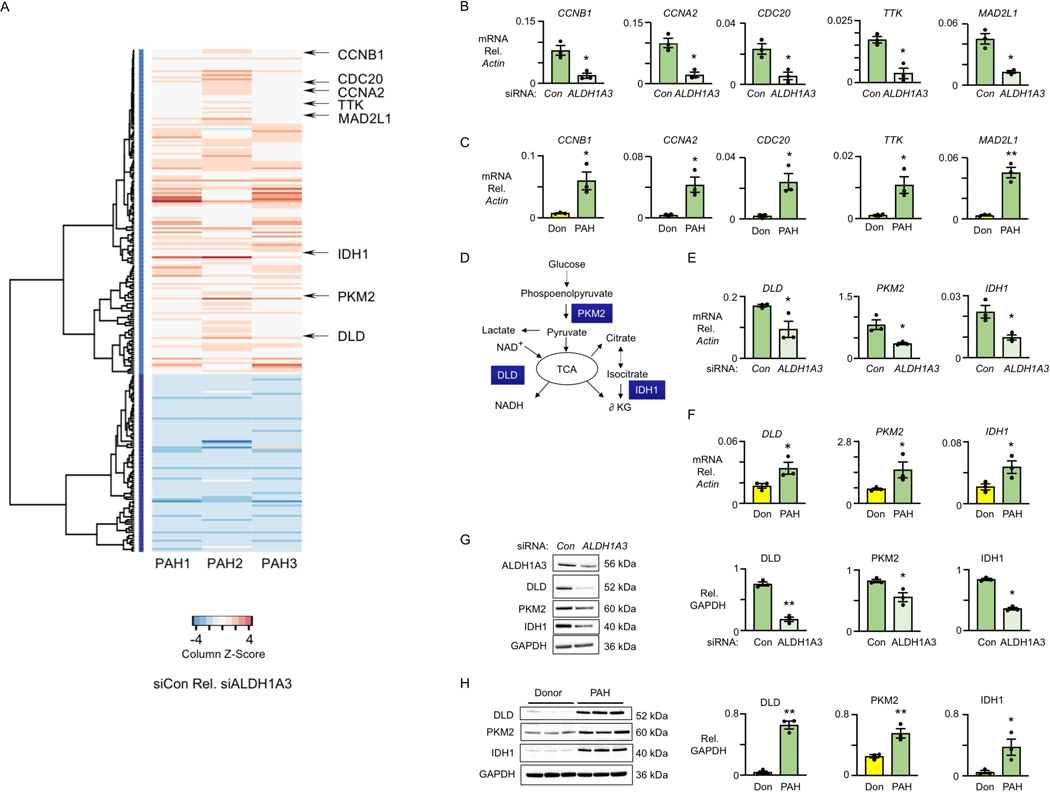
Figure 3. ALDH1A3 Increases Cell Cycle and Metabolic Genes in PAH PASMC.
(A) Heatmap of significantly changed genes in three PAH patients transfected with non- targeting (Con) siRNA vs. ALDH1A3 siRNA, n=3, FDR corrected p-value <0.01, FC>1.2. Red indicates genes upregulated in siCon vs siALDH1A3 and Blue indicates downregulated genes.
(B) Representative cell cycle genes - CCNB1, CCNA2, CDC20, TTK and MAD2L1 - by PCR of PASMC of three PAH patients with Con vs ALDH1A3 siRNA. *p<0.05 and **p<0.01 by paired Student t test.
(C) The same analysis in PASMC of three donor controls (Don) and three PAH patients. *p<0.05 and **p<0.01 by unpaired Student t test.
(D) Schema depicting the metabolic roles of DLD, PKM2 and IDH1 in glucose metabolism.
(E) Quantification of the mRNA level of DLD, IDH1 and PKM2 relative to β-actin in three PAH PASMC with Con or ALDH1A3 siRNA. *p<0.05 by paired Student t test.
(F) The same analysis in PASMC of three donor controls and three PAH patients. *p<0.05 by unpaired Student t test.
(G) Representative immunoblots of DLD, IDH1 and PKM2 relative to GAPDH in three PAH PASMC with Con vs ALDH1A3 siRNA with quantification. **p<0.01. by paired Student t test.
(H) The same analysis in PASMC of three donor controls and three PAH patients. **p<0.01 by unpaired Student t test.
Also evident in the heatmap (Figure 3A) and KEGG enrichment analysis (Figure IIIA in the Data Supplement and Supplement Excel File II), was a decrease in genes related to glycolysis and pyruvate metabolism. For example, there were reductions in dihydrolipoyl dehydrogenase (DLD), the E3 component of the pyruvate dehydrogenase complex that catalyzes nicotinamide adenine dinucleotide (NAD+) to NADH; and pyruvate kinase M2 (PKM2) that produces pyruvate from phospoenolpyruvate. PKM2 and DLD are involved in glucose metabolism and contribute to the TCA cycle (Figure 3D). Isocitrate dehydrogenase 1 (IDH1), which catalyzes the conversion of the citrate isomer isocitrate to α-ketoglutarate was also decreased (Figure 3D). Using qPCR, we verified the reduction in mRNA levels of PKM2, DLD and IDH1 in response to ALDH1A3 vs. control siRNA in PAH PASMC (Figure 3E), and an increase in these transcripts in PAH vs. donor PASMC (Figure 3F). Consistent with these findings, protein levels of DLD, PKM2 and IDH1 were reduced in PAH PASMC with ALDH1A3 vs. control siRNA (Figure 3G) and were elevated in PAH PASMC relative to those of donors (Figure 3H). Taken together, these findings suggest that ALDH1A3 coordinates the regulation of cell cycle and metabolic enzymes important in PASMC proliferation.
ALDH1A3 increases cell cycle and metabolic genes in PAH PASMC via H3K27ac.
ALDH1A3 catalyzes the conversion of acetaldehyde to acetate, a source of nuclear acetyl-CoA that can acetylate histones such as histone 3 lysine 27 (H3K27ac), a mark of active gene enhancers (Figure 4A). We confirmed by immunoblot analyses that ALDH1A3 is present in the nucleus as well as the cytoplasm of PAH PASMC (Figure IVA in the Data Supplement). Acetaldehyde is derived from pyruvate in mammalian cells13, and we detected higher levels of acetaldehyde in the nuclear fraction of PAH vs donor PASMC (Figure 4B) but no significant change in the cytoplasmic fraction (Figure IVB in the Data Supplement). Immunoblot analyses indicate that DLD, like ALDH1A3, is present in both the cytoplasm and nucleus of PAH PASMC, whereas PKM2 and IDH1 are mostly present in the cytoplasm (Figure IVC in the Data Supplement). As PKM2 and IDH1 are involved in generating citrate via the TCA cycle, they may also be responsible for the production of nuclear acetyl-CoA from citrate via the citrate transport protein (CTP)9 (Figure 4C).
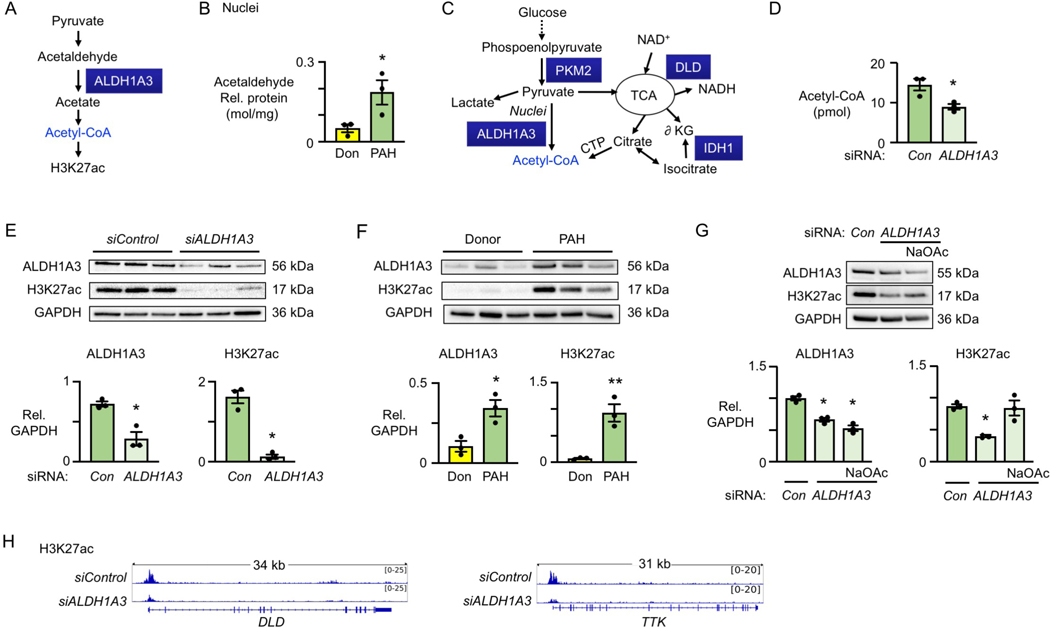
Figure 4. ALDH1A3 increases cell cycle and metabolic genes in PAH PASMC via H3K27ac.
(A) Schema of ALDH1A3 catalyzing acetaldehyde to acetate to acetyl-CoA, increasing H3K27ac.
(B) Concentration of acetaldehyde in nuclei from donor control and PAH PASMC normalized to nuclear protein (mol/mg), n=3, *p<0.05 by unpaired Student t test.
(C) Schema of DLD, PKM2 and IDH1 generating acetyl-CoA in cell cytoplasm and/or nucleus.
(D) Acetyl-CoA levels in nuclei per 1×106 PAH PASMC by fluorometric assay kit, comparing PAH PASMC transfected with nontargeting (Con) siRNA or ALDH1A3 siRNA. n=3, *p<0.05 by paired Student t test.
(E) Immunoblot and quantification of ALDH1A3 and H3K27ac relative to GAPDH in three PAH patients with Con and ALDH1A3 siRNA. **p<0.01 by paired Student t test.
(F) Immunoblot and quantification of ALDH1A3 and H3K27ac in three donor control vs. three PAH PASMC, Unpaired Student t test, **p<0.01.
(G) Immunoblot and quantification of ALDH1A3 and H3K27ac in three PAH patients with Con siRNA, ALDH1A3 siRNA or ALDH1A3 siRNA plus 5mM sodium acetate (NaOAc) for 8h. *p<0.05 by one-way ANOVA.
(H) H3K27ac peak density of DLD and TTK in PAH PASMC with Con vs. ALDH1A3 siRNA visualized in the IGV genome browser.
We showed that nuclear acetyl-CoA (Figure 4D) and H3K27ac (Figure 4E) are reduced when levels of ALDH1A3 are decreased in PAH PASMC by ALDH1A3 vs. control siRNA. Moreover, H3K27ac is elevated in PAH vs. donor PASMC (Figure 4F). To relate ALDH1A3 mediated acetyl-CoA to H3K27ac in PAH PASMC, we showed an increase in H3K27ac in PASMC despite ALDH1A3 siRNA when we added exogenous acetate (Figure 4G). It is also evident that DLD, PKM2 and IDH1 that are regulated by ALDH1A3 can contribute to production of acetyl-CoA since reducing levels of each of these enzymes also decreased H3K27ac in PAH PASMC (Figure IVD in the Data Supplement).
To determine whether ALDH1A3 mediated H3K27ac was increased in ALDH1A3-dependent transcripts, we carried out H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) in PASMC from three PAH patients (± ALDH1A3 siRNA) that we used for the transcriptomic analyses in Figure 3. More than 2/3 of the H3K27ac ChIP peaks were distributed within 3kb of the proximal promoter of genes in which expression was reduced by ALDH1A3 siRNA (Figure IVE in the Data Supplement). ALDH1A3-dependent H3K27ac was evident in the ALDH1A3-dependent metabolic genes (DLD, IDH1, PKM2) and cell cycle genes (CCNA2, CCNB1, TTK and CDC20). DLD and TTK tracks are shown in Figure 4H as examples and the others are in Figure IVF in the Data Supplement.
ALDH1A3 targets H3K27ac to NFY binding sites via KAT2B.
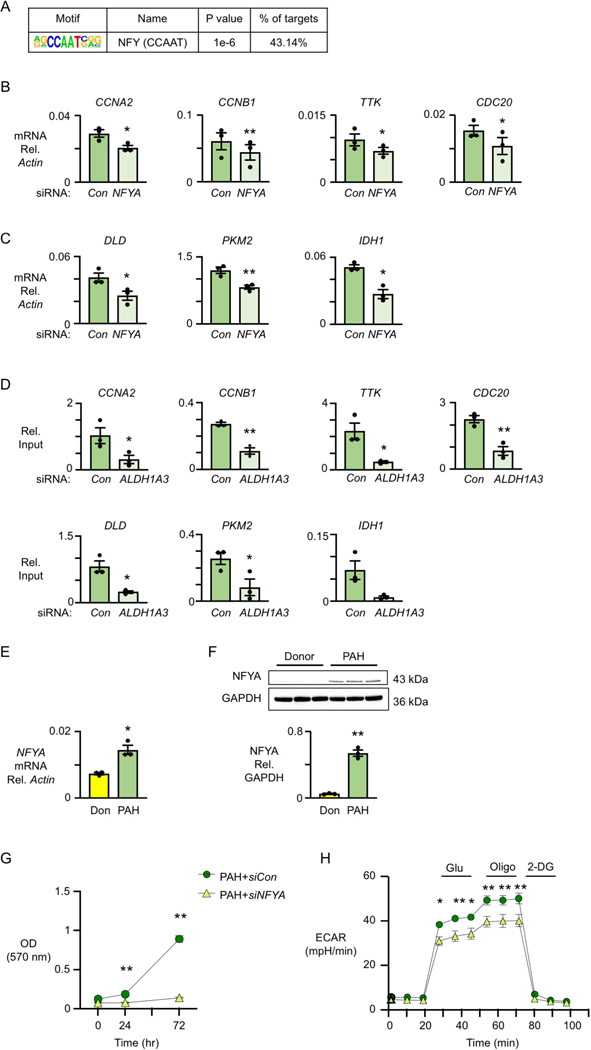
We next investigated whether ALDH1A3 mediates H3K27ac at specific transcription factor binding sites that impact gene expression. Motif enrichment analyses of transcriptomic datasets in PAH PASMC impacted by ALDH1A3 siRNA identified NFY as a putative transcription factor in 40% of the significantly changed transcripts with a p value of 1e-6 (Figure 5A). The cell cycle genes CCNA2, CCNB1, CDC20 and TTK, and the metabolic genes DLD, PKM2 and IDH1 all have NFYA binding sites. To verify these as NFY target genes, we decreased NFYA, the subunit that binds to DNA, by siRNA in PAH PASMC and observed reduced cell cycle gene transcripts as assessed by qPCR (Figure 5B) and a reduction in representative proteins as measured by immunoblots (Figure VA in the Data Supplement). Similarly, mRNA expression of the metabolic enzymes, DLD, PKM2 and IDH1 is NFYA-dependent (Figure 5C), and protein levels related to all three transcripts are decreased in PAH PASMC with siNFYA treatment (Figure VB in the Data Supplement). These findings were validated by NFYA ChIP-qPCR (Figure 5D), confirming the binding of NFYA to all target genes with the exception of IDH1 where a strong trend, with p value of 0.07, is evident.
Figure 5. Cell cycle and metabolic genes are regulated by NFYA in PAH PASMC.
(A) Transcription factor motif enrichment analysis of RNA-seq data in PAH PASMC transfected with nontargeting (Con) vs. ALDH1A3 siRNA. The analysis shows that there is an NFY motif in 43% of genes downregulated in PAH PASMC with ALDH1A3 siRNA.
(B) mRNA expression of CCNB1, TTK, CCNA2 and CDC20 relative to β-actin by qPCR in PAH PASMC with nontargeting (Con) or NFYA siRNA.
(C) mRNA expression of DLD, PKM2 and IDH1 relative to β-actin by qPCR in PAH PASMC with Con or NFYA siRNA. In (B, C): n=3, *p<0.05, **p<0.01 by paired Student t test.
(D) NFYA ChIP-qPCR of CCNA2, CCNB1, TTK, CDC20, DLD, PKM2 and IDH1 normalized to input in PAH PASMC with Con and ALDH1A3 siRNA. n=3, *p<0.05, **p<0.01 by paired Student t test.
(E) Expression of NFYA relative to β-actin in Donor vs PAH PASMC. (F) Immunoblots of NFYA relative to GAPDH and quantification in Donor and PAH PASMC. In (E, F), n=3, *p<0.05, **p<0.01 by unpaired Student t test.
(G) MTT assay (OD 570nm) as a measure of cell proliferation using PASMC from three PAH patients transfected with non-targeting (Con) siRNA or NFYA siRNA. Values given after 48h of serum starvation (0 hour) vs serum stimulation at 24, 48 and 72 hours. n=3, **p<0.01 by repeated measures two-way ANOVA, followed by Bonferroni analysis.
(H) Glycolytic function of control and PAH PASMC with siCon vs. siNFYA (n=3). *p<0.05, **p<0.01, by repeated measures two-way ANOVA, followed by Bonferroni analysis.
Moreover, an elevation in NFYA mRNA (Figure 5E) and protein (Figure 5F) was observed in PAH vs donor PASMC. Consistent with its function as a transcription factor for ALDH1A3-dependent metabolic and cell cycle genes, depleting NFYA by siRNA resulted in decreased PAH PASMC proliferation as assessed by MTT assay (Figure 5G) and cell counts (Figure VC in the Data Supplement) and glycolysis determined by ECAR (Figure 5H). However, the OCR was unchanged by NFYA siRNA in PAH PASMC (Figure VD in the Data Supplement). The apoptosis resistant phenotype of the PAH PASMC was negated by a reduction in NFYA (Figure VE in the Data Supplement).
We also carried out gain of function experiments by transfecting control PASMC with a plasmid expressing NFYA and ALDH1A3 but, despite marked overexpression of both, we did not observe an increase in cell number or in glycolytic function (Figure VF in the Data Supplement), suggesting that while ALDH1A3 and NFYA are necessary, they are not sufficient to reproduce the phenotype. Other factors, perhaps other NFY family members, other transcription factors or chromatin remodelers may be required.
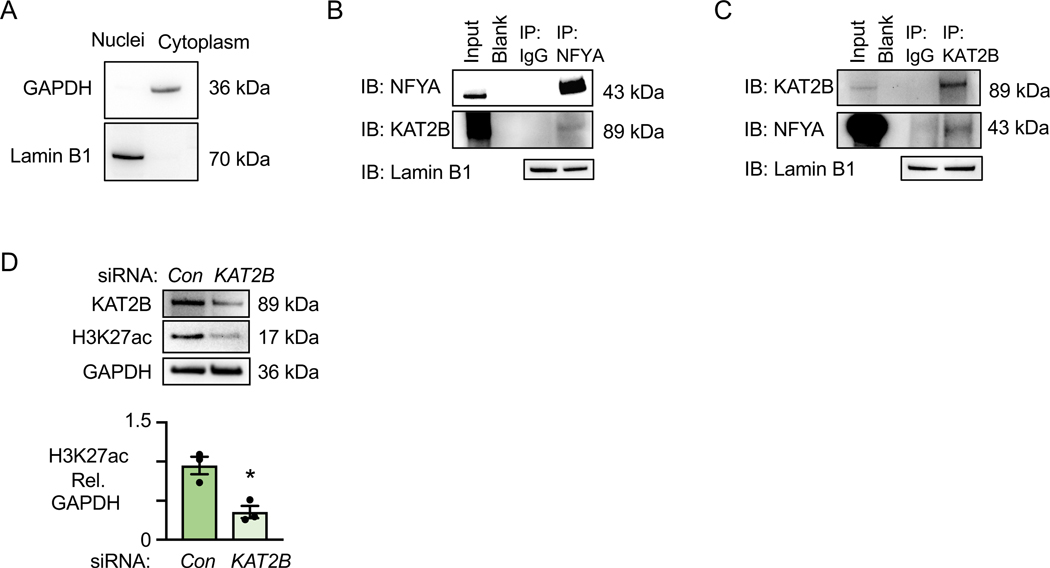
KAT2B is an acetyltransferase known to bind NFYA31. We hypothesized that KAT2B transfers acetyl-CoA to acetylate H3K27 at NFYA binding sites. The cytosolic marker GAPDH shows no cytosolic contamination of the nuclear fraction marked by Lamin B1 (Figure 6A). Co-immunoprecipitation studies showed an interaction with KAT2B when NFYA was immunoprecipitated (Figure 6B) and an interaction with NFYA when KAT2B is immunoprecipitated (Figure 6C). To confirm that KAT2B acts as the acetyltransferase at NFYA binding sites contributing to H3K27ac, we reduced KAT2B by siRNA and observed a decrease in H3K27ac (Figure 6D).
Figure 6. NFYA binding to KAT2B Regulates H3K27ac in PAH PASMC.
(A) Immunoblots of GAPDH (cytoplasmic marker) and Lamin B1 (nuclear marker) in nuclear and cytoplasm fractions of PAH PASMC.
(B) Immunoblots of KAT2B immunoprecipitated with normal IgG or NFYA antibody in PAH PASMC nuclear lysates. Lamin B1 is the loading control for the nuclear fraction.
(C) Immunoblots of NFYA immunoprecipitated with normal IgG or KAT2B antibody in nuclear lysates of PAH PASMC. Lamin B1 is the loading control for the nuclear fraction.
(D) Immunoblots and quantification of KAT2B and H3K27ac in PASMC of PAH patients transfected with nontargeting (Con) or KAT2B siRNA. n=3. *p<0.05 by paired Student t test.
β-catenin coordinately regulates ALDH1A3 and NFYA gene expression in PAH PASMC.
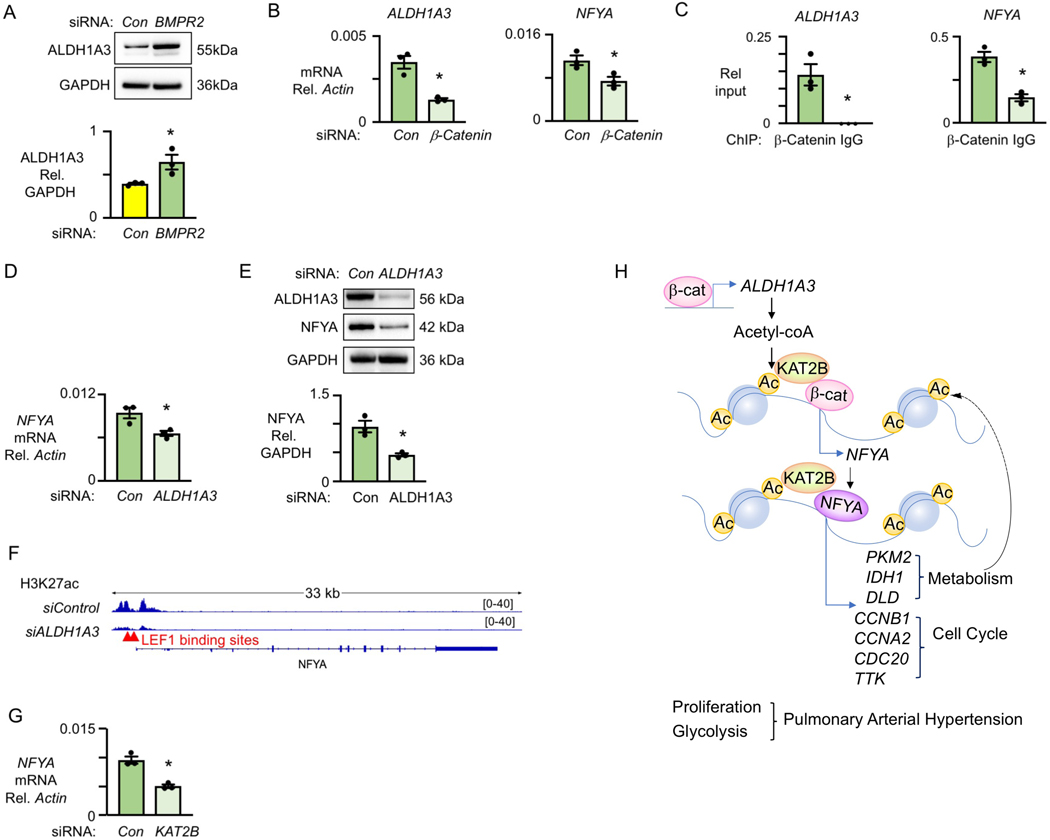
The findings above led us to investigate how ALDH1A3 and NFYA are coordinately upregulated in PAH PASMC. Decreasing BMPR2 by siRNA in donor PASMC induced ALDH1A3 (Figure 7A). Previous studies from our laboratory4 and those of others32 demonstrated that active β-catenin is a candidate transcription factor that drives PASMC proliferation, and both ALDH1A3 and NFYA are predicted to have β-catenin/TCF-LEF1 binding sites. When β-catenin was reduced by siRNA in PAH PASMC, both ALDH1A3 and NFYA was decreased (Figure 7B). β-catenin ChIP-qPCR confirmed β-catenin binding sites in NFYA and ALDH1A3 (Figure 7C).
Figure 7. ALDH1A3 targets NFY through β-catenin.
Cells were synchronized by culture for 48h under serum starvation, then cultured under serum stimulation and assessed at 72h.
(A) Representative immunoblots of and quantification of ALDH1A3 protein normalized to GAPDH in donor PASMC with nontargeting (Con) and BMPR2 siRNA.
(B) Expression of ALDH1A3 and NFYA relative to β-actin in PAH PASMC with nontargeting (Con) and β-catenin siRNA detected by qPCR.
(C) β-catenin and IgG ChIP-qPCR of ALDH1A3 and NFYA relative to input in PAH PASMC.
(D) mRNA expression, by qPCR, of NFYA relative to β-actin in PAH PASMC treated with Con or ALDH1A3 siRNA.
(E) Protein expression by immunoblots of NFYA relative to GAPDH in three PAH PASMC treated with Con or ALDH1A3 siRNA.
(F) Representative LEF1 binding site of NFYA and H3K27ac histone mark in PAH PASMC treated with Con vs. ALDH1A3 siRNA visualized in the IGV genome browser.
(G) mRNA expression by qPCR of NFYA relative to β-actin in PAH PASMC treated with Con vs. KAT2B siRNA. (H) Model summarizing all data. n=3. *p<0.05**p<0.01, by paired Student t test.
We also found evidence that NFYA is regulated by β-catenin in an ALDH1A3-dependent manner. Expression of NFYA mRNA (Figure 7D) and NFYA protein (Figure 7E) were reduced when we decreased ALDH1A3 in PAH PASMC. NFYA is also reduced in PASMC from KO mice compared to WT mice under hypoxia as shown by immunofluorescent staining (Figure VIA in the Data Supplement). Moreover, the peak density of H3K27ac on the LEF1 binding sites of the NFYA promoter was reduced in PAH PASMC by ALDH1A3 siRNA (Figure 7F). Since KAT2B is known to interact with β-catenin33, we hypothesized that KAT2B is the acetyltransferase responsible for acetylating histones at the LEF1 binding site. This is supported by our finding that reducing KAT2B by siRNA decreases NFYA mRNA (Figure 7G). Consistent with these data, increased active β-catenin is observed in the PASMC in the lung tissue sections from WT vs. KO mice (Figure VIB in the Data Supplement).
Taken together, our studies produce a model (Figure 7H) whereby in PAH PASMC, an increase in ALDH1A3 gene expression results from elevated levels of β-catenin binding to the TCF/LEF1 site of the ALDH1A3 gene. Then, elevated levels of ALDH1A3 increase NFYA gene expression owing to the production of acetyl-CoA and acetylation of H3K27 in conjunction with KAT2B at the β-catenin TCF/LEF1 binding site of the NFYA gene. The increase in NFYA together with ALDH1A3- mediated acetylation of H3K27 via KAT2B at NFYA binding sites promotes transcription of cell cycle genes that regulate proliferation and metabolic enzymes that fuel glycolysis, processes that drive the pathology of PAH.
Discussion
This study demonstrates the mechanism by which, ALDH1A3, a metabolic enzyme previously implicated in proliferation and glycolysis of cancer cells as well as rat vascular SMC, is increased in the nucleus of PAH PASMC and regulates H3K27 acetylation at NFYA binding sites in transcripts related to cell cycle and metabolism. Our findings further support the concept that altered metabolism regulates chromatin remodeling and transcription to impact cell function.
ALDH1A3 is one of 19 ALDH isoenzymes, but the only one observed to be significantly upregulated in transcriptomic analyses in PASMC from PAH vs. donor controls. We initially focused on the PASMC because previous transcriptomic analyses in PAH vs control PAEC from our laboratory did not show a significant elevation in this transcript or in other ALDH isoforms34,35. Moreover, deleting ALDH1A3 in murine SMC was sufficient to prevent hypoxia induced pulmonary hypertension. However, our immunofluorescent studies in PAH vs. control lung tissue sections, turned our attention to evaluating the role of ALDH1A3 in PAH vs. control PAEC as described below.
Our studies focused on the role of ALDH1A3 upregulation in inducing the proliferative and glycolytic phenotype of PAH PASMC. While there is considerable heterogeneity of SMC36 in the vessel wall the increase in nuclear ALDH1A3 was uniformly seen in the media and adventitia at least as judged by co-staining with SM22α. There are other phenotypic characteristics of PAH PASMC related to increased migration, reduced contractility and production of extracellular matrix37, and altered mitochondrial biology38 that we did not investigate and could be of interest, as well as others reflected in pathway analysis as ALDH1A3 dependent. ALDH isoforms are differentially regulated by p53 in different cancer cell types39 but as deletion of p53 in SMC in mice does not worsen PH40, it is unlikely that ALDH1A3 is p53 dependent in PAH PASMC.
Nuclear expression of ALDH1A3 suggested that it could be regulating proliferation by a mechanism independent of glycolysis. Before investigating this mechanism, we found further evidence supporting the functional importance of elevated ALDH1A3 in PAH. There was increased expression of nuclear ALDH1A3 in PASMC in PAH lung tissue sections and a transgenic mouse, in which Aldh1a3 was depleted in SMC, failed to show proliferation of PASMC in peripheral arteries and the associated pulmonary hypertension after exposure to chronic hypoxia. Unfortunately, all experimental interventions to block ALDH1A3 have been carried out with disulfiram, a non-selective ALDH inhibitor. However, when selective inhibitors become available26, particularly if they can be modified in a way that specifically targets PASMC and not PAEC, it would be of interest to determine whether inhibiting ALDH1A3 can also induce regression of severe pulmonary hypertension induced in more severe models of pulmonary hypertension such as in the rat heterozygous for BMPR2 administered 5-lipoxygenase41.
To elucidate mechanisms accounting for ALDH1A3-mediated PAH PASMC proliferation, we compared the transcriptome of PAH PASMC under basal conditions and when ALDH1A3 was reduced. We found ALDH1A3-dependent regulation of mRNA levels of genes related to cell cycle and metabolism. DLD is the oxidized lipoamide cofactor in pyruvate and α-ketoglutarate-dehydrogenase complexes that generates NADH and participates in ATP production. DLD also functions as a diaphorase, using NADH to generate reactive oxygen species (ROS)42. PKM2 has been associated with both aerobic glycolysis and anabolic metabolism in cancer cells43. PKM2 also regulates mitosis in cancer cells by increasing deoxynucleotide levels for DNA replication, as well as ribose production via the oxidative pentose phosphate pathway44. IDH1 promotes aggressive growth in primary glioblastoma by shifting metabolic adaptation to macromolecular synthesis, whereas inactivating IDH1 decreases glioblastoma cell growth and promotes a more differentiated tumor cell state by enhancing methylation and differentiation marker gene expression45. Taken together, DLD, PKM2, and IDH1 contribute to the metabolic shift initiated by ALDH1A3 in PAH PASMC and also impact nucleotide synthesis via PKM244 and cell cycle genes required for cell proliferation.
H3K27ac is found at active promoters and distal enhancers in association with open chromatin46. Increased H3K27ac mediated by acetyl-CoA is evident in hyperproliferative yeast and mammalian cells47, 48. Our previous work showed a global increase in H3K27ac related to acetyl-CoA produced by PFKFB3, that was required for PAEC proliferation and regeneration in response to injury9. It would be of interest to determine whether there is differential expression in donor and PAH PASMC of other histone marks and methylation changes.
While the role of NFYA in regulating cell cycle genes is described in cancer cells, we have shown that NFYA also regulates metabolic genes. KAT2B is a candidate acetyltransferase that interacts with NFYA22 and we confirmed the interaction in PAH PASMC. KAT2B is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation49. It is also known that KAT2B regulates the accessibility of β-catenin forming a complex at the promoter region of gene targets in association with myoblast proliferation50.
In this study, we linked elevated ALDH1A3, with transcription of NFYA by active β-catenin, to promote cell cycle and metabolic genes in PAH PASMC, causing a proliferative and glycolytic phenotype. Increased ALDH1A3 is also observed in PAH PAEC, but reducing ALDH1A3 worsened the apoptosis vulnerability of PAH PAEC. Inhibitors of some ALDH enzymes such as ALDH2 are in clinical use and have been tested experimentally in PDGF mediated intimal hyperplasia in models of systemic vascular disease18. Small molecule inhibitors could be developed for the ALDH1A3 isoenzyme. Our findings related to ALDH1A3 merit its future consideration as a selective therapeutic target in PAH, but because of the adverse effects on PAEC, our studies indicate that selective inhibition of ALDH1A3 in PASMC would be important.
Supplementary Material
Clinical Perspective.
What’s new?
An increase in an aldehyde dehydrogenase, ALDH1A3, underlies the heightened proliferation and glycolysis of pulmonary arterial smooth muscle cells in patients with idiopathic and hereditary pulmonary arterial hypertension, while promoting survival of their endothelial cells under stress.
ALDH1A3 has the dual function of providing energy to fuel smooth muscle cell proliferation and acetyl CoA to acetylate histones (H3K27) at enhancer sites to increase expression of genes that control metabolism and proliferation and are regulated by the transcription factor, NFYA.
Transgenic mice with ALDH1A3 deleted in smooth muscle cells do not develop chronic hypoxia induced pulmonary hypertension.
What are the clinical implications?
Agents that target the excessive production of ALDH1A3 in vascular smooth muscle cells could be highly effective in treating and potentially reversing pulmonary arterial hypertension.
The same therapeutic target may have adverse effects in one cell type and beneficial effects in another cell type in the vessel wall.
Altering metabolic processes in cardiovascular cells can have profound effects on gene regulation as the two are coordinately regulated.
Acknowledgments
D. L. conceived and performed the experiments, interpreted data and wrote the manuscript. N-Y. S. performed the analyses of RNA-seq and ChIP-seq. J. R. M. helped with the immunofluorescence imaging. Z. Z. helped in the preparation of libraries and analyses for RNA Seq. M. S. helped in the preparation of the libraries for ChIP-seq. S. O. performed the animal experiments. L. W. bred the mice and helped with genotyping and physiologic studies. T. N. performed quantification of muscularity in tissue sections from mice in a blinded manner and. E. Y. performed quantification of ALDH1A3 staining in tissue sections from humans and mice, also in a blinded manner. C. G. L., D. P. M., K. C., and J. R. M contributed to the experimental design, editing and critical review of the manuscript. J. C. W. and M. P. S were responsible for overseeing the sequencing studies and analyses. M.R. oversaw study design, data acquisition and analysis, and manuscript preparation and editing.
We thank Dr. Norbert B. Ghyselinck (Institut de Génétique et de Biologie Moléculaire et Cellulaire, IllKirch, France) for providing the floxed Aldh1a3 mice embryos; Dr. Amato Giaccia (Department of Radiation Oncology, Stanford University) for providing the hypoxia chamber; Dr. Tushar Desai (Stanford University) for the use of the Leica confocal microscope, Drs. Daria Mochly-Rosen and Che-Hong Chen (Department of Chemical and System Biology, Stanford University) for suggestions regarding ALDH inhibition; Dr. Jaecheol Lee (Stanford University) for the advice on ChIP-qPCR; Dr. Yinhua Jin in Prof. Roeland Nusse laboratory (Stanford University) for processing the flow cytometry related to cell cycle analysis; Drs. Chengkun Wang and Chen Chen for helping with plasmid design; Drs. Sarasa Isobe and Tsutomu Shinohara for helping with confocal microscopy; Drs. Jan K Hennigs and Aiqin Cao for help in the isolation and culture of primary PASMC; Ms. Patricia A. del Rosario (Stanford University) for providing clinical information related to the cell and tissue samples. We greatly appreciate the editorial and technical assistance of Dr. Michal Bental Roof in preparing both the figures and the scientific editing of the text, and the administrative help of Ms. Michelle Fox. We are indebted to the Pulmonary Hypertension Breakthrough Initiative (PHBI), as the source of cells and tissues from PAH patients and unused donor controls. Deidentified demographic and clinical data were supplied by the Data Coordinating Center at the University of Michigan.
Sources of Funding
This work was supported by the NIH/NHLBI R01 HL122887 (Marlene Rabinovitch and Michael P. Snyder) and R01 HL074186 (Marlene Rabinovitch). Dr. Dan Li was supported by an American Lung Association Senior Research Training Fellowship RT-509274. The PHBI is funded by NIH/NHLBI R24 HL123767 and the Cardiovascular Medical Research and Education Fund (CMREF) UL1RR024986. Dr. Marlene Rabinovitch is also supported by the Dwight and Vera Dunlevie Chair in Pediatric Cardiology at Stanford University.
Non-standard Abbreviations and Acronyms
- ALDH1A3
aldehyde dehydrogenase family 1 member 3
- PAH
pulmonary arterial hypertension
- PA
pulmonary arteries
- SMC
smooth muscle cells
- BMPR2
bone morphogenetic receptor 2
- PAEC
pulmonary artery endothelial cells
- IDH1/2
isocitrate dehydrogenase 1/2
- R-2HG
(R)-2-hydroxyglutarate
- H3K27ac
histone H3 lysine 27
- NFY
nuclear factor Y
- IPAH
idiopathic PAH
- HPAH
hereditary PAH
- ECAR
extracellular acidification rate
- OCR
oxygen consumption rate
- KO
Knock-out (SM2α-Aldh1a3−/−)
- WT
Wild-type (SM22α-Aldh1a3+/+)
- RVSP
right ventricular systolic pressure
- AT/ET
right ventricular acceleration time relative to ejection time
- RVH
right ventricular hypertrophy
- RV/LV+S
right ventricle/left ventricle + septum (weight ratio)
- CCNB1
cyclin B1
- CCNA2
cyclin A2
- CDC20
cell division cycle 20
- APC/C
anaphase promoting complex
- TTK
monopolar spindle 1 kinase
- DLD
dihydrolipoyl dehydrogenase
- NAD+
nicotinamide adenine dinucleotide
- PKM2
pyruvate kinase M2
- ChIP-seq
chromatin immunoprecipitation sequencing
- ROS
reactive oxygen species
- FBS
fetal bovine serum
- PAAT
pulmonary artery acceleration time
- RT-qPCR
reverse transcription qPCR
Footnotes
Disclosures
None
References
- 1.Machado RD, Pauciulo MW, Thomson JR, Lane KB, Morgan NV, Wheeler L, Phillips JA, Newman J, Williams D, Galie N, et al. BMPR2 haploinsufficiency as the inherited molecular mechanism for primary pulmonary hypertension. Am J Hum Genet. 2001;68:92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson C, Stewart S, Upton PD, Machado R, Thomson JR, Trembath RC and Morrell NW. Primary pulmonary hypertension is associated with reduced pulmonary vascular expression of type II bone morphogenetic protein receptor. Circulation. 2002;105:1672–1678. [DOI] [PubMed] [Google Scholar]
- 3.Hansmann G, de Jesus Perez VA, Alastalo TP, Alvira CM, Guignabert C, Bekker JM, Schellong S, Urashima T, Wang L, Morrell NW, et al. An antiproliferative BMP-2/PPARgamma/apoE axis in human and murine SMCs and its role in pulmonary hypertension. J Clin Invest. 2008;118:1846–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez VA, Ali Z, Alastalo TP, Ikeno F, Sawada H, Lai YJ, Kleisli T, Spiekerkoetter E, Qu X, Rubinos LH, et al. BMP promotes motility and represses growth of smooth muscle cells by activation of tandem Wnt pathways. J Cell Biol. 2011;192:171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S, Harry G, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J and Thompson CB. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol. 2019;20:436–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Egervari G, Wang Y, Berger SL and Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol. 2018;19:563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinnaird A, Zhao S, Wellen KE and Michelakis ED. Metabolic control of epigenetics in cancer. Nat Rev Cancer. 2016;16:694–707. [DOI] [PubMed] [Google Scholar]
- 9.Miyagawa K, Shi M, Chen PI, Hennigs JK, Zhao Z, Wang M, Li CG, Saito T, Taylor S, Sa S, et al. Smooth Muscle Contact Drives Endothelial Regeneration by BMPR2-Notch1-Mediated Metabolic and Epigenetic Changes. Circ Res. 2019;124:211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F and Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–821. [DOI] [PubMed] [Google Scholar]
- 12.Ogryzko VV, Schiltz RL, Russanova V, Howard BH and Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Cooper DE, Cluntun AA, Warmoes MO, Zhao S, Reid MA, Liu J, Lund PJ, Lopes M, Garcia BA, et al. Acetate Production from Glucose and Coupling to Mitochondrial Metabolism in Mammals. Cell. 2018;175:502–513 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E and Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. [DOI] [PubMed] [Google Scholar]
- 16.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR and Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie X, Urabe G, Marcho L, Stratton M, Guo LW and Kent CK. ALDH1A3 Regulations of Matricellular Proteins Promote Vascular Smooth Muscle Cell Proliferation. iScience. 2019;19:872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maity SN and de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. [DOI] [PubMed] [Google Scholar]
- 20.Benatti P, Dolfini D, Vigano A, Ravo M, Weisz A and Imbriano C. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res. 2011;39:5356–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, Yellaboina S and Jothi R. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol Cell. 2014;55:708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolfini D, Gatta R and Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47:29–49. [DOI] [PubMed] [Google Scholar]
- 23.Puttini S, Plaisance I, Barile L, Cervio E, Milano G, Marcato P, Pedrazzini T and Vassalli G. ALDH1A3 Is the Key Isoform That Contributes to Aldehyde Dehydrogenase Activity and Affects in Vitro Proliferation in Cardiac Atrial Appendage Progenitor Cells. Front Cardiovasc Med. 2018;5:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang PM, Chen CH, Yeh CC, Lu HJ, Liu TT, Chen MH, Liu CY, Wu ATH, Yang MH, Tai SK, et al. Transcriptome analysis and prognosis of ALDH isoforms in human cancer. Sci Rep. 2018;8:2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenmark KR, Fagan KA and Frid MG. Hypoxia-induced pulmonary vascular remodeling: cellular and molecular mechanisms. Circ Res. 2006;99:675–691. [DOI] [PubMed] [Google Scholar]
- 26.Spiekerkoetter E, Tian X, Cai J, Hopper RK, Sudheendra D, Li CG, El-Bizri N, Sawada H, Haghighat R, Chan R, et al. FK506 activates BMPR2, rescues endothelial dysfunction, and reverses pulmonary hypertension. J Clin Invest. 2013;123:3600–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanchard JM. Cyclin A2 transcriptional regulation: modulation of cell cycle control at the G1/S transition by peripheral cues. Biochem Pharmacol. 2000;60:1179–1184. [DOI] [PubMed] [Google Scholar]
- 28.Kapanidou M, Curtis NL and Bolanos-Garcia VM. Cdc20: At the Crossroads between Chromosome Segregation and Mitotic Exit. Trends Biochem Sci. 2017;42:193–205. [DOI] [PubMed] [Google Scholar]
- 29.Pachis ST and Kops G. Leader of the SAC: molecular mechanisms of Mps1/TTK regulation in mitosis. Open Biol. 2018;8 (8): 180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gisselsson D, Hakanson U, Stoller P, Marti D, Jin Y, Rosengren AH, Stewenius Y, Kahl F and Panagopoulos I. When the genome plays dice: circumvention of the spindle assembly checkpoint and near-random chromosome segregation in multipolar cancer cell mitoses. PLoS One. 2008;3:e1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Currie RA. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. [DOI] [PubMed] [Google Scholar]
- 32.DiRenzo DM, Chaudhary MA, Shi X, Franco SR, Zent J, Wang K, Guo LW and Kent KC. A crosstalk between TGF-beta/Smad3 and Wnt/beta-catenin pathways promotes vascular smooth muscle cell proliferation. Cell Signal. 2016;28:498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge X, Jin Q, Zhang F, Yan T and Zhai Q. PCAF acetylates {beta}-catenin and improves its stability. Mol Biol Cell. 2009;20:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhodes CJ, Im H, Cao A, Hennigs JK, Wang L, Sa S, Chen PI, Nickel NP, Miyagawa K, Hopper RK, et al. RNA Sequencing Analysis Detection of a Novel Pathway of Endothelial Dysfunction in Pulmonary Arterial Hypertension. Am J Respir Crit Care Med. 2015;192:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes-Palomares A, Gu M, Grubert F, Berest I, Sa S, Kasowski M, Arnold C, Shuai M, Srivas R, Miao S, et al. Remodeling of active endothelial enhancers is associated with aberrant gene-regulatory networks in pulmonary arterial hypertension. Nat Commun. 2020;11:1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frid MG, Aldashev AA, Dempsey EC and Stenmark KR. Smooth muscle cells isolated from discrete compartments of the mature vascular media exhibit unique phenotypes and distinct growth capabilities. Circ Res. 1997;81:940–952. [DOI] [PubMed] [Google Scholar]
- 37.Saygin D, Tabib T, Bittar HET, Valenzi E, Sembrat J, Chan SY, Rojas M and Lafyatis R. Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension. Pulm Circ. 2020;10: 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen KH, Dasgupta A, Lin J, Potus F, Bonnet S, Iremonger J, Fu J, Mewburn J, Wu D, Dunham-Snary K, et al. Epigenetic Dysregulation of the Dynamin-Related Protein 1 Binding Partners MiD49 and MiD51 Increases Mitotic Mitochondrial Fission and Promotes Pulmonary Arterial Hypertension: Mechanistic and Therapeutic Implications. Circulation. 2018;138:287–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gui S, Xie X, O’Neill WQ, Chatfield-Reed K, Yu JG, Teknos TN and Pan Q. p53 functional states are associated with distinct aldehyde dehydrogenase transcriptomic signatures. Sci Rep. 2020;10:1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wakasugi T, Shimizu I, Yoshida Y, Hayashi Y, Ikegami R, Suda M, Katsuumi G, Nakao M, Hoyano M, Kashimura T, et al. Role of smooth muscle cell p53 in pulmonary arterial hypertension. PLoS One. 2019;14:e0212889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian W, Jiang X, Sung YK, Shuffle E, Wu TH, Kao PN, Tu AB, Dorfmuller P, Cao A, Wang L, et al. Phenotypically Silent Bone Morphogenetic Protein Receptor 2 Mutations Predispose Rats to Inflammation-Induced Pulmonary Arterial Hypertension by Enhancing the Risk for Neointimal Transformation. Circulation. 2019;140:1409–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solmonson A and DeBerardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. J Biol Chem. 2018;293:7522–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo W and Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23:560–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo PN, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calvert AE, Chalastanis A, Wu Y, Hurley LA, Kouri FM, Bi Y, Kachman M, May JL, Bartom E, Hua Y, et al. Cancer-Associated IDH1 Promotes Growth and Resistance to Targeted Therapies in the Absence of Mutation. Cell Rep. 2017;19:1858–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai L, Sutter BM, Li B and Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Reyes I and Chandel NS. Acetyl-CoA-directed gene transcription in cancer cells. Genes Dev. 2018;32:463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malatesta M, Steinhauer C, Mohammad F, Pandey DP, Squatrito M and Helin K. Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res. 2013;73:6323–6233. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki A, Minamide R and Iwata J. The role of acetyltransferases for the temporal-specific accessibility of beta-catenin to the myogenic gene locus. Sci Rep. 2018;8:15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diebold I, Hennigs JK, Miyagawa K, Li CG, Nickel NP, Kaschwich M, Cao A, Wang L, Reddy S, Chen PI, et al. BMPR2 preserves mitochondrial function and DNA during reoxygenation to promote endothelial cell survival and reverse pulmonary hypertension. Cell Metab. 2015;21:596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng G, Xu J, Liu R, Fu Z, Li S, Hong W, Chen J, Li B and Ran P. Isolation, Culture and Identification of Pulmonary Arterial Smooth Muscel Cells from Rat Distal Pulmonary Arteries. Cytotechnology. 2017;69:831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y, Haghighat L, Spiekerkoetter E, Sawada H, Alvira CM, Wang L, Acharya S, Rodriguez-Colon G, Orton A, Zhao M, et al. Neutrophil Elastase is Produced by Pulmoanry Artery Smooth Muscle Cells and is Linked to Neointimal Lesions. Am J Pathol. 2011;179:1560–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu M, Shao NY, Sa S, Li D, Termglinchan V, Ameen M, Karakikes I, Sosa G, Grubert F, Lee J, et al. Patient-Specific iPSC-Derived Endothelial Cells Uncover Pathways that Protect against Pulmonary Hypertension in BMPR2 Mutation Carriers. Cell Stem Cell. 2017;20:490–504 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pertea M, Kim D, Pertea GM, Leek JT and Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y, Smyth GK and Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- 57.Love MI, Huber W and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H and Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Trapnell C, Pop M and Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R and Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen L, Shao NY, Liu X, Maze I, Feng J and Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8:e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorvaldsdottir H, Robinson JT and Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen L, Shao N, Liu X and Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.