Abstract
Transposable elements (TEs) are a major source of genetic and regulatory variation in their host genome and are consequently thought to play important roles in evolution. Many fungal and oomycete plant pathogens have evolved dynamic and TE-rich genomic regions containing genes that are implicated in host colonization and adaptation. TEs embedded in these regions have typically been thought to accelerate the evolution of these genomic compartments, but little is known about their dynamics in strains that harbor them. Here, we used whole-genome sequencing data of 42 strains of the fungal plant pathogen Verticillium dahliae to systematically identify polymorphic TEs that may be implicated in genomic as well as in gene expression variation. We identified 2,523 TE polymorphisms and characterize a subset of 8% of the TEs as polymorphic elements that are evolutionary younger, less methylated, and more highly expressed when compared with the remaining 92% of the total TE complement. As expected, the polyrmorphic TEs are enriched in the adaptive genomic regions. Besides, we observed an association of polymorphic TEs with pathogenicity-related genes that localize nearby and that display high expression levels. Collectively, our analyses demonstrate that TE dynamics in V. dahliae contributes to genomic variation, correlates with expression of pathogenicity-related genes, and potentially impacts the evolution of adaptive genomic regions.
Keywords: transposable element, transposon, structural variation, genome evolution, plant pathogen
Introduction
Repetitive sequences such as TEs are ubiquitous components of prokaryote and eukaryote genomes (Bowen and Jordan 2002; Wicker et al. 2007). In eukaryotes, the repeat content can differ between species and range from only ∼5.6% in the fish Tetraodon nigroviridis (Jaillon et al. 2004) to up to 85% in the maize genome (Schnable et al. 2009; Jiao et al. 2017). TEs can transpose to different locations within the host genome, and thus are often considered “mobile genetic elements” (McClintock 1950). TEs have been traditionally classified into two classes based on their mode of transposition: class I retrotransposons transpose via a copy-and-paste mechanism, whereas class II DNA transposons mainly mobilize via a cut-and-paste system (Wicker et al. 2007; Piégu et al. 2015). TEs can impact host genome organization and gene function by disrupting protein-coding regions, modifying gene expression, or causing large-scale chromosomal rearrangements (Eichler and Sankoff 2003; Feschotte 2008; Bennetzen and Wang 2014; Le et al. 2015; Mita and Boeke 2016). Consequently, TEs and their activities have been considered to generally decrease fitness, and TEs and TE-induced mutations are typically rapidly purged from a population. Furthermore, TE activities are generally suppressed by host defense processes. These processes include DNA methylation (Selker et al. 2003; Zemach et al. 2010), histone modifications associated with highly condensed heterochromatin (Slotkin and Martienssen 2007; Hocher et al. 2018; Hocher and Taddei 2020), targeted processes such as RNA-silencing (Lippman and Martienssen 2004; Nicolas et al. 2013; Yadav et al. 2018), or repeat-induced point (RIP) mutation, a fungal specific process in in which duplicated or repetitive sequences are targeted by C to T mutations (Cambareri et al. 1989; Galagan and Selker 2004; Fudal et al. 2009; Wang et al. 2020).
Over the last decades, besides their detrimental effects, TEs have been increasingly recognized to play important roles in adaptive genome evolution (Venner et al. 2009; Hua-Van et al. 2011; Lynch et al. 2011; Bourque et al. 2018). For example, TEs can be co-opted as a source of cis-regulatory elements (Chuong et al. 2016; Kitano et al. 2018) and mediate swift changes in the transcriptional landscape under biotic and abiotic stresses (Sinzelle et al. 2009; Bourque et al. 2018; Muszewska et al. 2019). A classic example is the dark pigmentation of the peppered moth in response to industrialization that was caused by a single TE insertion into the first intron of a cell-cycle regulator gene, changing the transcript abundance of this gene and allowing the insect to camouflage on polluted surfaces (Cook and Saccheri 2013; Hof et al. 2016). Changes in mutation rates in genes in proximity TEs can be caused by leakage of TE silencing mechanisms (e.g., RIP) or DNA repair following TE insertion (Fudal et al. 2009; Rouxel et al. 2011; Yoshida et al. 2016; Wang et al. 2020). Furthermore, abundant TEs and other repetitive elements can act as an ectopic substrate for double-strand break repair pathways, thereby fostering chromosomal rearrangements (structural variants; SVs), such as chromosomal translocations and inversions as well as large-scale duplications and deletions (Chan and Kolodner 2011; Faino et al. 2016; Bourque et al. 2018). For example, TE expansions in crops such as tomato, rice, wheat, and soybean, have been shown to foster an increased accumulation of SVs associated with important traits during domestication (Fuentes et al. 2019; Alonge et al. 2020; Liu et al. 2020; Walkowiak et al. 2020). Interestingly, as TEs and SVs colocalize, they tend to form clusters in discrete regions in many eukaryotic organisms (Hastings et al. 2009; Lin and Gokcumen 2019; Marand et al. 2019). This colocalization, and its consequences for genome variation and organization, are important to understand the genome evolution of eukaryotic organisms (Kidwell and Lisch 2001; Wright and Finnegan 2001; Lynch and Conery 2003; Hurst et al. 2004; Lynch 2007). Remarkably, these TE-rich regions are often enriched in environmentally responsive genes, for example, in genes associated with roles in immunity or in secondary metabolism in plants (Sakamoto et al. 2004; Kawakatsu et al. 2016; Seidl et al. 2016; van Wersch and Li 2019). Therefore, TE-rich compartments are considered relevant for contribution to adaptation to various abiotic and biotic stresses (Crombach and Hogeweg 2007; Knibbe et al. 2007).
TE-rich compartments play important roles in the coevolutionary “arms-race” between pathogens and their hosts. Pathogens secrete molecules known as effectors to establish a successful parasitic relationship on their hosts (Rovenich et al. 2014; Sánchez-Vallet et al. 2018). In turn, hosts evolve receptors to activate immunity and halt pathogen colonization (Cook et al. 2015; Han 2019). Thus, in order to overcome recognition by host immune systems, pathogens have to purge or diversify effector genes once their gene products become recognized (Moller and Stukenbrock 2017; Fouche et al. 2018; Badet and Croll 2020). In many filamentous pathogens, effector genes are enriched in TE-rich and gene-sparse compartments, whereas they are typically underrepresented in TE-poor and gene-dense genomic regions that typically harbor housekeeping genes (Seidl and Thomma 2014; Dong et al. 2015; Frantzeskakis et al. 2020). TE-rich compartments are often characterized by increased substitution rates and increased occurrence of SVs and presence/absence polymorphisms (Raffaele et al. 2010; Croll and McDonald 2012; de Jonge et al. 2012, 2013; Dutheil et al. 2016; Hartmann and Croll 2017; Hartmann et al. 2017; Wang et al. 2017; Fokkens et al. 2018; Plissonneau et al. 2018; Grandaubert et al. 2019; Gan et al. 2021; Wyka et al. 2021). Notably, a similar association of TEs with genes involved in immune responses has also been observed in plant hosts (Leister 2004; Kawakatsu et al. 2016; Mascher et al. 2017; Seidl and Thomma 2017). Therefore, TE-rich compartments have been suggested to facilitate the generation of variation that is necessary to fuel the “arms-race” between pathogens and their hosts (Seidl and Thomma 2017; Frantzeskakis et al. 2019, 2020; Schrader and Schmitz 2019; Torres et al. 2020).
Verticillium dahliae is an asexual soil-borne fungal plant pathogen that colonizes the xylem of hundreds of susceptible host plants (Fradin and Thomma 2006; Klosterman et al. 2009, 2011; Inderbitzin and Subbarao 2014). Genomic comparisons between V. dahliae strains revealed that their genomes evolved by extensive chromosomal rearrangements (de Jonge et al. 2013; Faino et al. 2016). These rearrangements include chromosomal translocations, inversions, large-scale deletions, and duplications that collectively contributed to the formation of adaptive genomic compartments that originally have been referred to as lineage-specific (LS) regions, as these are variable between V. dahliae strains (Klosterman et al. 2011; de Jonge et al. 2012, 2013; Faino et al. 2016; Depotter et al. 2019). Initially, LS regions were solely defined by presence/absence polymorphisms among sequenced strains (de Jonge et al. 2013; Faino et al. 2016). However, subsequent work on the chromatin landscape in V. dahliae has revealed that these LS regions display a unique chromatin profile (Cook et al. 2020). Moreover, it was shown that a substantial number of additional genomic regions share chromatin characteristics with LS regions and, accordingly, that the amount of LS DNA has been underestimated (Cook et al. 2020). The originally identified LS regions, together with the additional genomic regions that share a similar chromatin profile, are now referred to as adaptive genomic regions (Cook et al. 2020). Importantly, these adaptive genomic regions are enriched for in planta-induced effector genes that contribute to host colonization (Klosterman et al. 2011; de Jonge et al. 2012, 2013; Kombrink et al. 2017; Gibriel et al. 2019; Cook et al. 2020; Chavarro-Carrero et al. 2021). Furthermore, these regions are enriched in relatively young and transcriptionally active TEs (Faino et al. 2016; Cook et al. 2020). Although a key role of TEs to drive formation and maintenance of adaptive genomic regions has been revealed (Amyotte et al. 2012; Faino et al. 2015, 2016; Shi-Kunne et al. 2018; Depotter et al. 2019; Cook et al. 2020; Seidl et al. 2020), it remains unclear what the impact of TE variation is on the biology of individual strains of V. dahliae. Changes in the TE landscape between V. dahliae strains, involving novel insertions as well as deletions, can cause structural as well as gene expression variation. Here, we exploited a collection of V. dahliae strains to analyze their TE landscape and systematically identify TE polymorphisms. Furthermore, we analyzed the polymorphic TEs in detail to study its impact on genome function and organization. Our analysis extends previous hypotheses on the relevance of TE dynamics in adaptive genomic regions (Faino et al. 2016; Cook et al. 2020), and further characterizes a TE subset implicated in genomic variation and gene expression in V. dahliae.
Results
Structural Variants Are Common in V. dahliae
Previous genomic comparisons among a narrow selection of V. dahliae strains have revealed extensive chromosomal rearrangements, which were proposed to have contributed to the establishment of adaptive genomic regions (de Jonge et al. 2013; Faino et al. 2016). To systematically assess their prevalence and abundance in a considerably larger collection of V. dahliae strains, we used paired-end sequencing reads for de novo prediction of SVs in 42 V. dahliae strains (Cook et al. 2020; Chavarro-Carrero et al. 2021) together with the gapless genome assembly of strain JR2 as a reference (Faino et al. 2015). With a combination of four commonly used SV-callers (supplementary fig. 1, Supplementary Material online), 919 high-confidence SVs were identified in total, comprising 410 deletions, 49 inversions, 24 duplications, and 436 translocations (fig. 1A and supplementary fig. 1, Supplementary Material online). Importantly, we successfully recalled all 19 chromosomal rearrangements that were previously manually identified between V. dahliae strains JR2 and VdLs17 (Faino et al. 2016). To investigate the occurrence of the 919 SVs across the V. dahliae strains, we calculated the frequency of each SV over all the strains included in the analysis. We observed that 87% (n = 803) of the SVs is shared by <20% of the strains, that is, they occur in less than eight strains (fig. 1B). Among the SVs, deletions occur at the highest frequency (fig. 1A and B; P < 2 × 10−16, Kruskal–Wallis test). Interestingly, the number of SVs per strain does not correspond with the phylogenetic relationship among the V. dahliae strains (fig. 1C and D). Collectively, our data confirm that SVs occur commonly in V. dahliae, but that individual SVs are typically only shared by a small subset of strains.
Fig. 1.
Structural variants (SVs) in a collection of 42 Verticillium dahliae strains. (A) The number of SVs is depicted per type for each V. dahliae strain included in the analysis; the color-code for each variant type as indicated in panel (B); (B) Frequency distribution of the 919 SVs in the collection of V. dahliae strains; (C) Unrooted neighbor-joining tree based on presence–absence of 919 high confident SVs relative to the reference genome assembly of V. dahliae strain JR2 (red), color code depicts different groups; (D) Unrooted neighbor-joining tree of the collection of V. dahliae strains based on 287,251 high-confident SNVs relative to the reference genome assembly of V. dahliae strain JR2 (red); the color of the strain names corresponds to the colors represented in (C); (E) Circular plot displaying the genomic distribution of the 919 SVs along the eight chromosomes of the V. dahliae strain JR2 reference genome assembly. The tracks are shown in the following order from outside to inside: centromeric regions (Seidl et al. 2020), gene density in 10 kb windows, TE distribution, adaptive genomic regions distribution (Cook et al. 2020), 410 deletions (light blue), 49 inversions (yellow), 24 duplications (red), and 436 translocations (blue lines). The color intensity of the lines for an individual translocation is based on its frequency in the V. dahliae collection; (F) Number of TEs around SV breakpoints is displayed in 10 kb windows; (G) The distribution of 10,000 random permutations of distances between SVs and TEs in adaptive genomic regions. The vertical dashed line represents the mean number of random occurrences, the yellow line indicates the average distance at 4,681.85 bp, significance P = 9.99 × 10−5, z score = −2.32.
Previously, we have demonstrated that SVs cluster at adaptive genomic regions in V. dahliae (de Jonge et al. 2013; Faino et al. 2016). Here, we similarly observed distinct genomic regions comprising multiple nonrandomly distributed SVs (P < 2 × 10−16, Kolmogorov–Smirnov test, supplementary fig. 2, Supplementary Material online), enriched in 17 regions (Pignatelli et al. 2009) (hypergeometric test; P < 0.05 after Benjamini–Hochberg correction; supplementary table 2, Supplementary Material online), and colocalized with the previously identified adaptive genomic regions (one-sided Fisher’s exact test, P = 0.00049; supplementary fig. 3, Supplementary Material online). Interestingly, SVs occur largely independently of nucleotide mutations in adaptive genomic regions (supplementary table 3, Supplementary Material online). We observed that only 0.06% of the nucleotides in adaptive genomic regions are affected by single-nucleotide variations, whereas 14.8% are affected by SVs. Thus, we conclude that SVs are a more likely phenomenon to increase variation in adaptive genomic regions than nucleotide mutations, especially when considering the previously reported high levels of sequence conservation in adaptive genomic regions (de Jonge et al. 2013; Faino et al. 2016; Shi-Kunne et al. 2018; Depotter et al. 2019).
A Subset of TEs in V. dahliae Is Polymorphic
TEs and other repetitive elements are often considered to drive the formation of SVs (Bourque et al. 2018). In V. dahliae, TEs have been proposed to mediate genomic rearrangements (Faino et al. 2016), and TEs and repetitive elements encompass 12.3% of the V. dahliae strain JR2 genome (Faino et al. 2015). Class I long-terminal repeat (LTR) retroelements are the most abundant, whereas Class II DNA is less abundant (supplementary table 4, Supplementary Material online). In line with previous observations (Klosterman et al. 2011; de Jonge et al. 2013; Faino et al. 2015, 2016; Cook et al. 2020), TEs are significantly enriched (3.5-fold) in adaptive genomic regions when compared with the core genome and colocalize with SVs (fig. 1F and G; supplementary fig. 4, Supplementary Material online). As TE dynamics may promote the formation of SVs, insertions, and deletions of TEs (i.e., polymorphic TEs) may directly impact host genome organization. We identified TE insertion and deletion polymorphisms using TEPID (Stuart et al. 2016) by querying paired-end sequencing data for 42 V. dahliae strains relative to the V. dahliae strain JR2 reference genome; deletions are defined as TEs that are present in the reference genome but absent in other strains, whereas insertions are those present in at least one other strain and absent from the syntenic region in the reference genome. Our approach uses a single V. dahliae strains as a reference, and thus we will be limited by the reference TEs annotation when describing TE polymorphisms. For example, two DNA/Tc1-Mariner elements annotated in V. dahliae strain VdLs17 (Amyotte et al. 2012) are absent in the reference strain JR2 (Faino et al. 2015), and consequently these two elements would have not been considered in our analysis. In total, we identified 135 and 30 unique loci that underwent TE deletions and insertions, respectively; these loci are responsible for a total of 2,387 deletions and 136 insertions we identified across the 42 V. dahliae strains (fig. 2B). In the case of insertions, we decided to keep only elements that belong to designated TE superfamilies, removing 12 “unspecified” elements that do not show association to a specific TE superfamily or the reference sequence length is incomplete. Only around 8% of all TEs are polymorphic, that is, they show insertion or deletion events in different V. dahliae strains compared with the reference strain JR2 (fig. 2A). We observed that polymorphic TEs are significatively larger than nonpolymorphic TEs (supplementary fig. 6, Supplementary Material online). To further analyze the polymorphic TEs, we first calculated their frequency in the V. dahliae strains relative to the reference strain JR2. This revealed that TE deletions occurred more frequently than insertions (P = 0.0349, one-sided Fisher’s exact test; fig. 2C); a similar frequency distribution was also observed for SV deletions (fig. 1C). Only five TEs display both deletions and insertions, of which one belongs to the LTR/Gypsy and four to the LTR/Copia superfamily. We observed that 33 polymorphic TEs (37% of which belong to the LTR/Copia superfamily) overlap protein-coding genes, 22 of which overlap introns and 11 exons. Thus, only a limited subset of TEs in the V. dahliae genome is polymorphic, whereas the vast majority is shared by all strains analyzed.
Fig. 2.
Transposable element polymorphisms in a collection of 42 Verticillium dahliae strains. (A) The percentage of polymorphic (shades of blue) and nonpolymorphic (gray) TEs in V. dahliae strain JR2 is depicted as a pie chart. The percentages of polymorphic TEs separated by superfamily relative to the TE annotation of the V. dahliae strain JR2 is shown in blue, and the percentage of polymorphic TEs within each superfamily is shown in red; (B) The proportion of identified TE polymorphisms is shown for every V. dahliae strain, depicting deletions (blue) and insertions (red) per superfamily; (C) Frequency distribution of 165 polymorphic TEs in the V. dahliae strain collection, depicted by TE variant type.
Polymorphic TEs Are Dynamic in V. dahliae
Fungi evolved a range of mechanisms to suppress TE activity and avoid TE expansions (Castanera et al. 2016). However, we previously observed that a fraction of TEs is transcriptionally active in V. dahliae, suggesting it comprises TEs that are potentially able to multiply and accumulate in the V. dahliae genome (Faino et al. 2016; Cook et al. 2020). Therefore, we hypothesized that polymorphic TEs are not suppressed and still able to multiply. To assess if polymorphic TEs lack typical signatures of suppression when compared with nonpolymorphic TEs in V. dahliae strain JR2, we assessed DNA methylation levels (in the three methylation contexts; mCG, mCHG, and mCHH), RIP signature (Composite Repeat-Induced-Mutations Index; CRI), Kimura distance (divergence from the TE consensus sequence), GC content, and expression levels in vitro (counts per million; CPM) by quantifying the abundance of TE-derived reads across all different TE copies in the genome using TEtranscripts (Jin et al. 2015; Jin and Hammell 2018). Dimensional reduction by principal component analysis (PCA) revealed that polymorphic TEs cluster and are largely separated from the nonpolymorphic TEs (fig. 3A). On the first dimension of the PCA, explaining 31.6% of the variation, polymorphic and nonpolymorphic TEs is separated based on CRI, DNA methylation, and GC content (supplementary fig. 5, Supplementary Material online). This observation can be explained by the association of TEs with strictly heterochromatic regions, in which TEs are characterized by high CRI and DNA methylation levels as well as by low GC content (Cook et al. 2020). The second dimension of the PCA, explaining 14.5% of the variation, revealed a more distinct separation between polymorphic and nonpolymorphic TEs mainly based on Kimura distance, DNA methylation, and transcriptional activity in vitro (supplementary table 5, Supplementary Material online). Specifically, we observed that the sequence of polymorphic TEs is more similar to the consensus sequence of the TE family (Kimura distance) and is characterized by a higher GC content when compared with nonpolymorphic TEs, indicating that polymorphic TEs are evolutionary younger when compared with nonpolymorphic TEs (fig. 3B). Notably, the CRI of polymorphic TEs is lower than nonpolymorphic TEs and resembles that of genomic regions located in the core genome (supplementary fig. 6, Supplementary Material online). Together with lower CG methylation levels in polymorphic TEs, our data suggest that polymorphic TEs are not silenced and are likely more active, as proxied by an increased transcriptional activity when compared with other TEs (fig. 3B). As expected, polymorphic TEs are expressed at higher levels in vitro and in planta when compared with nonpolymorphic TEs (fig. 3B and supplementary fig. 6, Supplementary Material online). Collectively, our analyses identified a subset of TEs as polymorphic among strains, and these tend to be evolutionary younger, less suppressed by RIP and methylation, as well as more highly expressed in V. dahliae.
Fig. 3.

The TE landscape across the Verticillium dahliae genome is dynamic. (A) Principal component analysis of TEs in V. dahliae strain JR2 based on methylation (mCG, mCHG, mCHH), GC proportion, Kimura distance, Composite Repeat-Induced Point Mutation index (CRI), frequency, and in vitro expression. Each point represents a single TE and colored ellipses represent the confidence interval for the polymorphic and nonpolymorphic TEs; (B) Polymorphic (n = 165) and nonpolymorphic TEs (n = 1,956) were compared based on GC proportion, the Kimura distance to the TE family consensus sequence, CRI, cytosine methylation, and in vitro expression (CPM). Statistical significance was assessed using one-sided Wilcoxon rank-sum test; (C) The association between polymorphic TEs and adaptive genomic regions was tested using a permutation test. The vertical dashed line shows the mean number of polymorphic TEs overlaps expected at 10,000 random permutations and the blue line indicates the empirical number of overlaps, significance P = 9.99 × 10−5, z score = 3.2927; (D) Circular plot displays the genomic distribution of 165 polymorphic TEs along the eight chromosomes of V. dahliae strain JR2. The tracks display the centromeric regions, gene density (in 10 kb windows), TE annotation, adaptive genomic regions, TE deletions, and TE insertions (with arrows indicating the insertion direction) from outside to inside.
Adaptive genomic regions in V. dahliae have been previously shown to be enriched for transcriptionally active TEs (supplementary fig. 4, Supplementary Material online; Faino et al. 2016; Cook et al. 2020). We hypothesized adaptive genomic regions are enriched for polymorphic TEs. Therefore, we analyzed the distribution of polymorphic TEs in V. dahliae strain JR2 (fig. 3D), leading to the identification of clusters of polymorphic TEs that colocalize with adaptive genomic regions (P = 0.00024, one-sided Fisher’s exact test; supplementary table 6, Supplementary Material online; fig. 3C). Interestingly, polymorphic TEs do not localize closer to SVs than nonpolymorphic TEs, suggesting that both types of TEs can contribute to the formation of SVs. Although deleted TEs were enriched in adaptive genomic regions, we could not observe a similar enrichment for TE insertions in the JR2 strain (supplementary fig. 7, Supplementary Material online). Polymorphic TEs belonging to LTR/Copia superfamilies or unspecified repetitive elements are highly enriched in adaptive regions (P = 8.24 × 10−9 and P = 1.36 × 10−5 respectively, one-sided Fisher’s exact test), and for instance, LTR/Copia represents 51.1% of all polymorphic TEs in adaptive regions (supplementary table 7, Supplementary Material online). Therefore, our results suggest that the LTR superfamily is an important component of the dynamic TE landscape.
Polymorphic TEs Impact the Functional Genome of V. dahliae
Next to their association with SVs, TE presence and activity have been suggested to directly or indirectly impact the functional genome, for example by modifying the expression of genes in their vicinity or by inducing changes in gene structure (Bourque et al. 2018; Schrader and Schmitz 2019). To establish to which extent TEs may affect gene expression in V. dahliae strain JR2, we first summarized the occurrence of TEs up to 5 kb upstream and downstream of all protein-coding genes (supplementary fig. 8, Supplementary Material online). The majority of annotated TEs are located within 5 kb range of a gene (67% of total TEs, n = 1,418; P = 0.0094, one-sided Fisher’s exact test), with an overrepresentation of polymorphic TEs belonging to the LTR/Copia, LINE/I, and DNA/Tc-1-Mariner superfamilies (fig. 4A and supplementary fig. 8, Supplementary Material online). By assessing the expression of TEs nearby genes (n = 1,418) as a proxy for their activity, we observed that many TE families are expressed in vitro and typically at higher levels during infection (P < 0.05, fig. 4B; supplementary fig. 9, Supplementary Material online). As expected, these families are abundant in polymorphic TEs (fig. 4B), suggesting that environmental changes induce the expression of TEs.
Fig. 4.
The presence of polymorphic transposable elements is correlated with expression of genes that localize nearby. (A) Ridgeline shows the density distribution of polymorphic TEs, classified by TE superfamily, upstream and downstream of genes; (B) Unsupervised clustering of TE expression per TE family and superfamily, color-coded based on the mean-per-family expression value (log2(CPM + 1)). The column depicts the abundance of polymorphic TEs within each group (yellow coded). Statistical significance for comparisons between in vitro (growth in MS media) and in planta (colonization of Arabidopsis thaliana at 28 days postinoculation) was assessed using a one-sided Wilcoxon rank-sum test *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001; (C) Relationship of gene expression ranks in vitro (blue) and in planta (red) and the number of TEs. Low rank = low expression and high rank = high expression. The plots display linear regression (dark line) and confidence interval (light gray) as well as the R and P values after linear regression. The histograms on top of the graph show the number of polymorphic TEs per expression rank and the dot-size reflects the proportion of polymorphic TEs relative to the total number of TEs per expression rank; (D) Relationship of gene expression rank and mean distance to TEs (bp) with annotations as for (C); (E) Gene expression in relation to the TE context (TEs within 5 kb); no TE in proximity = Distal (n = 8,742), or TEs flanked upstream (n = 1,098), downstream (n = 1,051), or in between (n = 546; blue). Significant differences were assessed using one-sided Wilcoxon rank-sum test; (F) TE distance to genes in 5 kb windows; TEs in upstream (n = 1,609), downstream (n = 1,511) or in between (n = 1,582). Statistical differences were assessed using one-sided Wilcoxon rank-sum test.
To further assess the correlation between the proximity of TEs and the expression of nearby genes, we compared the expression of genes located within 5 kb windows from TEs under in vitro and in planta conditions. We observed that 23% of all annotated genes reside within 5 kb of a TE (n = 2,696). Genes in proximity to TEs are transcribed at significantly lower levels when compared with genes that do not reside in close proximity to TEs (P < 2 × 10−16, in vitro, P = 1.9 × 10−9 in planta; Kruskal–Wallis test; fig. 4E, supplementary fig. 10, Supplementary Material online). Therefore, we hypothesized that the presence of polymorphic TEs correlates with expression in V. dahliae. To further investigate this, we aggregated genes and TEs within 5 kb by their expression rank and obtained the number of TEs in each rank. We observed only a moderate negative correlation between the number of polymorphic TEs and gene expression ranks in vitro (fig. 4C; R = –0.21, P = 0.034 linear regression). Conversely, the distance between genes and polymorphic TEs is moderately positively correlated with higher expression ranks (fig. 4D; R = 0.16, P = 0.013 linear regression). These results suggest that polymorphic TEs are associated, albeit weakly, with genes expressed at low levels under in vitro conditions. However, we observed a different pattern when considering host colonization, as gene expression ranks in planta did not correlate with TE density (fig. 4C;R = 0.025, P = 0.8 linear regression) whereas, similar to in vitro conditions, TE distance and expression are moderately positively correlated (fig. 4D;R = 0.19, P = 0.06 linear regression). Collectively, our data suggest the polymorphic TE density nearby protein-coding genes correlates only weakly with gene expression in different conditions.
To further assess the relationship between TEs and gene expression, we categorized genes into four categories based on the relative position of TEs; genes with no TEs in proximity (n = 8,742), genes with TEs localized upstream (n = 1,098), downstream (n = 1,051), or genes flanked by TEs on both sides (n = 546). The distance between TEs and genes is significantly shorter when genes are flanked by TEs, when compared with genes having TEs only upstream or downstream (fig. 4F). Importantly, genes flanked by TEs show reduced expression in vitro when compared with genes in the proximity of TEs located either upstream or downstream (fig. 4E), a pattern we similarly observed during host colonization. We similarly observed this pattern when only considering the subset of polymorphic TEs (supplementary figs. 10 and 11, Supplementary Material online). Interestingly, during host colonization genes in proximity to polymorphic TEs do not show a significant decrease in expression (supplementary fig. 10, Supplementary Material online). Thus, genes in proximity to nonpolymorphic TEs showed lower expression levels when compared with genes in proximity to polymorphic TEs.
Pathogenicity-Related Genes Colocalize with Polymorphic TEs
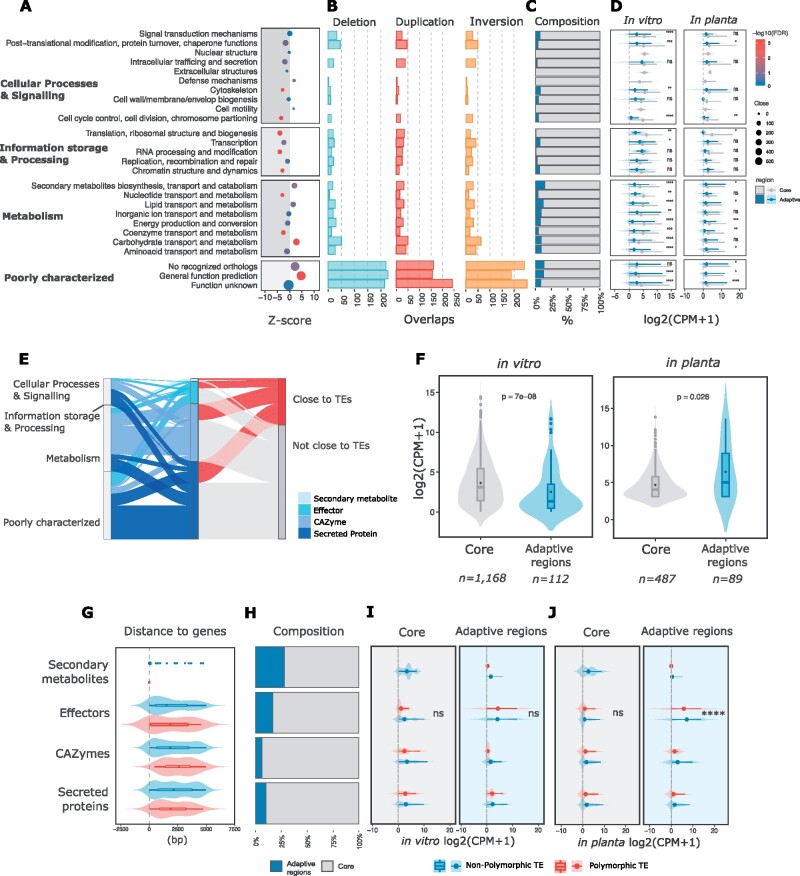
To understand which types of genes are enriched in proximity to TEs, we performed functional annotation of the V. dahliae strain JR2 gene catalog using a cluster for orthologs groups (COG) approach and subsequently tested for enrichment of genes with specific COG categories. No significant enrichment of genes in proximity to TEs could be observed for most of the categories belonging to “cellular processes” or to “information processing.” In contrast, genes annotated as “metabolism” and “poorly characterized” are significantly enriched (fig. 5A). Especially, genes belonging to the subcategories “defense mechanisms” or “carbohydrate metabolism” are highly enriched (fig. 5A). A total of 39% of genes that reside in proximity to TEs fall in the category “function unknown” (n = 579) or “not categorized” (n = 482), which refers to orthologs present in other organisms but without known function. Furthermore, we could not find orthologs for 14.9% of the genes that reside in proximity to TEs (n = 402), described in this work in the “not recognized orthologs” category, which can probably be explained as gene content specific to the V. dahliae lineage (fig. 5A and supplementary table 8, Supplementary Material online). Interestingly, most of these gene categories are associated with TEs (fig. 5A) and overlap with various SVs (fig. 5B). Subsequently, we annotated all protein-coding genes for functions that have previously been associated with pathogenicity, such as genes encoding secreted proteins (n = 672), carbohydrate-active enzymes (CAZymes, n = 498), secondary metabolites (n = 25), or effector candidates (n = 193). These genes can be broadly classified as “pathogenicity-related” and are enriched in the “poorly characterized” functional category, especially secreted proteins and effector candidates (P = 3.49 × 10−15 and n = 390, P = 2.85 × 10−6 and n = 127, respectively, one-sided Fisher’s exact test; supplementary table 9, Supplementary Material online). Genome-wide, we observed that 40.72% (n = 401) of predicted pathogenicity-related genes have a TE localized within 5 kb (fig. 5E). Polymorphic TEs are enriched in proximity (5 kb) to effector candidates (Z score = 3.4838, P = 0.0001; 5,000 random permutations), even though we did not observe that these genes and other pathogenicity-related genes are significantly closer to polymorphic TEs (fig. 5G). Thus, commonly considered pathogenicity-associated genes reside in proximity to TEs, yet only some genes are in proximity to polymorphic TEs.
Fig. 5.
Polymorphic transposable elements correlate with expression of a subset of genes. (A) Enrichment of TEs in proximity (within 5 kb range) to genes in different functional categories. Functional annotation was based on Clustering for Orthologous Groups (COGs) categories. The y axis shows the different functional categories and the x axis the z score after 5,000 random permutations; the gray shadow represents values with a negative z score indicating depletion rather than enrichment, and the −log10(P-value) after Benjamini–Hochberg correction for false-discovery rate is color-coded. The size is relative to the number of genes associated with TEs in each category; (B) The number of genes overlapping deletions (blue) and duplications (red), and inversions (yellow); (C) The relative proportions of genes localized in the adaptive genomic regions (blue) and core (gray) genomic regions; (D) Gene expression distribution in two conditions (in vitro and in planta; log2(CPM + 1)) and separated by core and adaptive genomic regions; (E) Predicted pathogenicity-related genes (n = 1,388) overlapping with COG categories and depicting the proportion of genes with TEs in proximity (5 kb); (F) Gene expression of pathogenicity-related genes in two conditions (in vitro and in planta; log2(CPM + 1)) and separated by core and adaptive genomic regions; (G) Distance between genes and TEs separated by polymorphic and nonpolymorphic TEs. Statistical significance was assessed using Wilcoxon rank-sum test; (H) The composition of pathogenicity-related gene categories separated in adaptive (blue) and core (gray) genomic regions; Pathogenicity-related gene expression in vitro (I) and in planta (J) separated by polymorphic and nonpolymorphic TEs. Statistical significance was assessed by a Kruskal–Wallis rank-sum test; ****P ≤ 0.0001.
When compared with the core genome, adaptive genomic regions in V. dahliae are relatively gene-poor (fig. 5C) and SV-rich (supplementary fig. 3, Supplementary Material online), as well as enriched for in planta-expressed genes (de Jonge et al. 2012, 2013; Kombrink et al. 2017; Cook et al. 2020; Chavarro-Carrero et al. 2021) (fig. 5D). Although a broad range of functional categories can be observed in adaptive genomic regions (fig. 5C), these are enriched for genes encoding secreted proteins and candidate effector genes (fig. 5H; Cook et al. 2020; P = 0.032 and P = 0.014, respectively; one-sided Fisher’s exact test). Since we have shown that gene expression levels correlate with the presence of both polymorphic as well as nonpolymorphic TEs (fig. 4), we queried to which extent polymorphic TEs in proximity of pathogenicity-related genes correlate with gene expression patterns in the adaptive genomic regions. We observed a significant enrichment of polymorphic TEs colocalizing with highly expressed genes in vitro and in planta (P = 0.0116 and P = 0.0010 for 50% and 75% upper quartiles for expression, respectively, after one-sided Fisher’s exact test; supplementary table 10, Supplementary Material online). Under in vitro conditions, expressed pathogenicity-related genes located in the core genome are expressed significantly higher than genes located in adaptive genomic regions (fig. 5F). As expected, we observed the opposite trend in planta (fig. 5F), where expressed pathogenicity-related genes that are localized in adaptive genomic regions are highly expressed during host colonization (de Jonge et al. 2012, 2013). Furthermore, we observed an enrichment of polymorphic TEs located within 5 kb of pathogenicity-related genes in adaptive genomic regions (P = 0.0062 after one-sided Fisher’s exact test). As expected, a higher proportion of these highly expressed genes are effector candidates (fig. 5I and J). These results suggest that polymorphic TEs occur more likely in close proximity to highly expressed pathogenicity-related genes in adaptive genomic regions.
Discussion
Transposable elements can contribute to genomic and transcriptomic variation (Bourque et al. 2018) and, consequently, TEs have been often considered to play crucial roles in genome evolution and function (Feschotte 2008). Here, we analyzed the TE landscape in the plant pathogen V. dahliae. We show that the presence of TEs is associated with abundant SVs that emerged independently in individual V. dahliae strains. The SVs form discrete clusters and colocalize within adaptive genomic regions. Importantly, we demonstrate that these SVs colocate with TEs, many of them polymorphic, and typically evolutionary younger, less silenced, and higher expressed. Furthermore, we show that the presence of polymorphic TEs is associated with gene expression of genes that can be assigned to diverse functional categories, particularly with host–pathogen interaction genes. Collectively, our results provide evidence for the hypothesis that polymorphic TEs contribute to increased genomic diversity, correlate with the expression of pathogenicity-related genes, and play important roles in the evolution of adaptive genomic regions.
TEs occur commonly in eukaryote genomes, yet there is an enormous variation in the TE content and diversity between different organisms (Wicker et al. 2007; Huang et al. 2012; Dietrich et al. 2013). Typically, TE expansions are suppressed by various genome defense mechanisms, but some TE copies can escape suppression and remain active and mobile. Polymorphic TEs have been associated with recent transposition activity (Huang et al. 2012), and in V. dahliae, a subset of TEs shares these characteristics. Only a subset of all predicted TEs is young and dynamic, which is in contrast to some other fungi in which there is a considerable amount of young TEs associated with recent TE expansions, such as in Zymoseptoria tritici (Badet et al. 2020; Lorrain et al. 2021; Oggenfuss et al. 2020), Leptosphaeria maculans (Grandaubert et al. 2014), Blumeria graminis (Frantzeskakis et al. 2018), and Magnaporthe oryzae (Kang et al. 2016). In many plants and fungi, TE expansions are mainly driven by LTR retrotransposons (Vitte and Bennetzen 2006; Tsukahara et al. 2009; Muszewska et al. 2011; Huang et al. 2012; Amselem et al. 2015; Donnart et al. 2017; Galindo-Gonzalez et al. 2017; Lorrain et al. 2021). We observed an enrichment of LTRs in facultative heterochromatic regions and at the centromeres in V. dahliae (Cook et al. 2020; Seidl et al. 2020). Similarly, LTRs are an important fraction of the polymorphic TEs in V. dahliae that are localized in the adaptive genomic regions. TE expansions have occurred at least twice during the evolution of V. dahliae, once during Verticillium diversification, and then right after V. dahliae speciation (Faino et al. 2016). The active polymorphic TEs we identified are likely the product of the most recent TE expansion in V. dahliae, and likely contributed to the formation of the adaptive genomic compartments.
Polymorphic TEs are a small fraction of the total TE repertoire, which suggests that most TEs are suppressed. Three main processes have been associated with TE suppression in fungi: RIP mutation, DNA methylation, and RNA-mediated silencing. RIP is usually associated with sexual fungal organisms (Galagan and Selker 2004), yet RIP mutations have been also observed in V. dahliae that is considered asexual (Clutterbuck 2011; Klosterman et al. 2011; Amyotte et al. 2012; Cook et al. 2020). RIP mutations have been associated with meiosis, but alternative mechanisms during vegetative propagation could lead to RIP-like mutations (Clutterbuck 2011). This has been recently observed in Z. tritici and N. crassa in which propagation through mitotic divisions generate RIP-like mutations (Möller et al. 2021; Wang et al. 2020). Intriguingly, polymorphic TEs are depleted in RIP mutations. Consequently, we consider different RIP scenarios, such as that RIP activity was lost during an evolutionary transition to asexuality in V. dahliae. In such a scenario, the mutations we observed in most TEs are remains of ancestral RIP activity. A second scenario is that V. dahliae is not strictly asexual, but rare sexual activity occurs in the population. In this scenario, the RIP machinery may be active during the occasional meiosis. Thirdly, a RIP-like mechanism could act outside the typical meiotic cycles and might be active in V. dahliae. These hypotheses are not mutually exclusive, and it remains challenging to elucidate the mechanistic origin of RIP mutations in V. dahliae.
Next to RIP, DNA methylation targets repetitive elements in many fungi (Nakayashiki et al. 1999; Zemach et al. 2010; Montanini et al. 2014; Bewick et al. 2019). We have previously demonstrated that the majority of TEs in V. dahliae is methylated (Cook et al. 2020; Seidl et al. 2020). We now show that polymorphic TEs are typically less methylated and not affected by RIP, collectively indicating that these elements are not suppressed. In the model plant Arabidopsis and the fruit-fly Drosophila, polymorphic and active TEs occur in discrete regions where novel insertions are not efficiently suppressed, whereas TE insertions outside of these regions are mostly suppressed by methylation (Hollister and Gaut 2009; Tsukahara et al. 2009; Stuart et al. 2016; Lee and Karpen 2017). We observed that TEs in adaptive genomic regions display low methylation levels (Cook et al. 2020). Whereas effective TE suppression outside adaptive regions might be an important mechanism to restrict TE expansions in the core genome, the activity TEs in adaptive regions suggests that that these regions are prone to TE-mediated variation.
Environmental changes can alter suppression mechanisms and drive a derepression of TEs (Bouvet et al. 2008; Carr et al. 2010; Fouche et al. 2020). We observed TE expression upon changes in environmental conditions, that is, different in vitro growth media as well as in planta growth. In addition, TE expression has been observed previously in different abiotic conditions in V. dahliae (Amyotte et al. 2012; Faino et al. 2016; Cook et al. 2020). For example, some Tc1/Mariner and Mutator elements are induced upon heat stress (Amyotte et al. 2012), whereas we observed that some LTR/Copia and LTR/Gypsy families highly expressed in planta. However, the function of TE derepression under changing environmental conditions remains unknown (Galhardo et al. 2007; Slotkin and Martienssen 2007; Koonin and Wolf 2009). Modifications in suppression patterns induced by changes in the environment could imply a trade-off between the functional TE derepression and increased probability of TE expansions and TE mobilization on nearby genes, or even changes in the genome structure (Seidl et al. 2016). In Schizosaccharomyces pombe, for instance, changes in temperature and nutrient availability induce TE insertions in promoters of stress-dependent genes, and these insertions have been associated with higher expression of the targeted genes (Feng et al. 2013; Maxwell 2020). In the plant-pathogen Z. tritici, changes in nutrient composition led to increased TE expression, and a TE insertion in proximity of an effector gene contribute to reduced expression and virulence (Fouche et al. 2020). Although the mechanisms in which the environment and TE derepression influence genome function and structure remain unclear, the expression of polymorphic TEs induced by changes in the environment could be similarly an important driver of the genome dynamics in V. dahliae.
Changes in the TE landscape have often been associated with gene expression changes (Goubert et al. 2020). Insertion of TEs within or near genes usually reduce gene expression, for example by spreading of silencing marks, such as DNA methylation or inducing changes in chromatin conformation (Castanera et al. 2016; Winter et al. 2018). TE insertions are associated with gene silencing and mostly have negative effects, yet also examples for an adaptive advantage exist. For example, in Z. tritici, the insertion of a TE in the promoter of a melanin biosynthesis gene causes reduced melanin synthesis and quicker growth, which would be beneficial for colonization (Krishnan et al. 2018). We observed that genes in proximity to TEs were typically less expressed during in vitro and in planta conditions, which was particularly clear for genes flanked by TEs. However, we observed that genes located in TE-rich regions are highly expressed in planta, raising a conundrum to link these opposing observations. Studies in adaptive genomic regions in V. dahliae and analogous regions in other fungi revealed that chromatin represents an additional layer of genomic regulation that creates a chromatin state allowing for accessibility to the transcriptional machinery (Schotanus et al. 2015; Fokkens et al. 2018; Soyer et al. 2019; Cook et al. 2020). We observed an enrichment of polymorphic TEs in close proximity of these highly expressed genes in adaptive regions. This observation raises the possibility that polymorphic TEs form part of the regulation of genes in the vicinity as cis-regulatory sequences or as a consequence of changes in the chromatin conformation (Le Rouzic et al. 2007; Hollister and Gaut 2009; Huang et al. 2012; Szitenberg et al. 2016; Choi and Lee 2020). For instance, TEs and TE-derived sequences can be co-opted in promoter sequences and contribute to the control of gene expression (Sundaram and Wysocka 2020; Cosby et al. 2021), as for example, Tf1 retrotransposon insertions in S. pombe occur in promoters and induce consistently higher expression of neighboring genes (Leem et al. 2008; Guo and Levin 2010; Maxwell 2020). Furthermore, this could explain the maintenance of polymorphic TEs in V. dahliae, as the TE suppression of these elements could negatively affect the expression levels of neighboring genes, favoring their colocalization.
Genome structure can differ significantly between species and even between strains of the same species (Lynch 2007). Here, we demonstrated that even closely related V. dahliae strains are characterized by their unique SV repertoire and colocalization of polymorphic TEs and SVs in adaptive regions. As previously reported, we similarly observed high levels of sequence conservation in adaptive regions (de Jonge et al. 2013; Faino et al. 2016; Shi-Kunne et al. 2018; Depotter et al. 2019). Recently, adaptive regions were characterized by an intermediate chromatin state between heterochromatin and euchromatin (Cook et al. 2020), which could indicate that this unique chromatin organization can allow for lower mutation rates in these adaptive regions (Prendergast et al. 2007). We observed that SVs emerge rapidly due to their association with polymorphic TEs, and they represent a relevant mechanism to generate variation, rather than single-nucleotide changes in adaptive regions. The colocalization of TEs and SVs in distinct regions has been reported previously associated with genome compartmentalization in diverse fungi (Schotanus et al. 2015; Faino et al. 2016; Plissonneau et al. 2016, 2018; Shi-Kunne et al. 2018; Ola et al. 2020; Torres et al. 2020). Relaxed selection has been proposed to explain the association of variation and TEs in genomic compartments, for example, the TE-rich accessory chromosomes in Z. tritici (Croll and McDonald 2012; Hartmann et al. 2017; Grandaubert et al. 2019). Conversely, adaptive regions in V. dahliae do not show signs of relaxed or strong positive selection (Depotter et al. 2019). Since V. dahliae is an asexual fungus and consequently has a low effective population size, genetic drift could represent an important evolutionary force that maintains SVs in population (Kelkar and Ochman 2012; Cvijovic et al. 2015), and these can therefore contribute to genome dynamics.
Compartmentalized genomes have been observed in diverse organisms (Torres et al. 2020). In filamentous plant pathogens, this genome organization is described in the “two-speed” genome model (Raffaele et al. 2010) in which dynamic genome compartments serve as cradles of variation (Croll and McDonald 2012; Frantzeskakis et al. 2019; Torres et al. 2020). Additionally, changes in chromatin organization have been shown to be relevant for compartmentalized genomes to further increase or facilitate genomic variation (Schotanus et al. 2015; Moller et al. 2019; Cook et al. 2020). Thus, further insights into chromatin organization and TE dynamics are necessary to further disentangle the impact of chromatin modifications on the emergence of genetic variation and unravel how these facilitate the evolution of virulence in plant pathogens.
Materials and Methods
Verticillium dahliae JR2 Genome, Repetitive Element, and Functional Gene Annotation
Repetitive element annotation of the chromosome-level genome assembly of V. dahliae strain JR2 (Faino et al. 2015) was based on available annotation (Faino et al. 2015; Cook et al. 2020; Seidl et al. 2020). Briefly, repetitive elements were annotated by using a combination of LTRharvest (Ellinghaus et al. 2008), LTRdigest (Steinbiss et al. 2009), and RepeatModeler. The repeat predictions were further curated and classified (Wicker et al. 2007) by a combination of PASTEC (Hoede et al. 2014) and sequence similarity to known TEs. The genome-wide occurrence of repeats was determined with RepeatMasker v 4.0.9, and the output was further processed using “one code to find them all” (Bailly-Bechet et al. 2014). Simple repeats and low-complexity regions were excluded (Cook et al. 2020; Seidl et al. 2020). GC-content, Kimura distance from a consensus TE family, and weighted DNA methylation data were previously summarized (Cook et al. 2020; Seidl et al. 2020). The CRI per TE was calculated by obtaining the RIP substrate and the RIP product index as defined by nucleotide frequencies: RIP product index = TpA/ApT and the RIP substrate index= (CpA + TpG)/(ApC + GpT). We also calculated the genome-wide RIP index (CRI) using RIPper (van Wyk et al. 2019) using sliding windows (1 kb, 500 bp slide).
Functional gene annotation was based on the available V. dahliae JR2 gene annotation (Faino et al. 2015) and was performed using eggNOG 5.0 (Huerta-Cepas et al. 2019) based on an hierarchical nonsupervised orthology search, restricted to a fungal database with e-values ≥0.001 and query coverage ≥50%. To define the V. dahliae secretome, we predicted N-terminal signal peptides using SignalP v.4.1 (Nielsen 2017). Subsequently, we predicted putative effectors with EffectorP v.2.0 (Sperschneider et al. 2018), using default parameters. Secondary metabolite biosynthetic genes were previously predicted using antiSMASH fungal version 4.0.2 (Weber et al. 2015; Shi-Kunne et al. 2018). Similarly, we used previously predicted CAZymes that cover signatures of glycoside hydrolases, polysaccharide lyases, carbohydrate esterases, glycosyltransferases, and carbohydrate-binding molecules (Seidl et al. 2015; Shi-Kunne et al. 2018). For further analysis, we considered the groups of “secreted proteins” as the secreted proteomes excluding candidate effectors, secondary metabolite genes, and CAZymes.
Gene and TE RNA-Sequencing Analysis
Transcriptional activity of genes and repetitive elements in V. dahliae strain JR2 was assessed using previously generated transcriptome data (Cook et al. 2020) of three in vitro conditions, (Potato Dextrose Broth, Murashige-Skoog, and Czapeck-Dox media) and of in planta colonization (Arabidopsis thaliana; 28 dpi). For further analysis, we considered only MS media for in vitro and in planta comparisons. Single-end sequencing reads of three biological replicates per condition were mapped to V. dahliae strain JR2 genome assembly (Faino et al. 2015) using STAR v.2.4.2.a, allowing multiple mapped reads with the following settings: –outFilterMultimapNmax 100 –winAnchorMultimapNmax 200 –outSAMtype BAM Unsorted –outFilterMismatchNmax 3 (Jin et al. 2015; Jin and Hammell 2018; Fouche et al. 2020). The bam files were sorted by read name with samtools v.1.2 and the transcriptional activity level was quantified using TEcount from the TEtrancripts package 2.2.1 (Jin et al. 2015), with the following parameters: –stranded no –mode multi, -iteration 1,000. TEcount considers multiple-mapped reads aligned to genes and TE regions to determine transcript abundance per condition/replicate. Furthermore, TEcount only considers reads that map in its entirety to TEs and reads mapping to only a fraction were discarded (Jin et al. 2015; Jin and Hammell 2018).
Sequencing reads summarized over TEs and genes were normalized between three replicates in the different conditions using R/Bioconductor package EdgeR v.3.8 (Robinson et al. 2010; Robinson and Oshlack 2010). For normalization, we only considered genes and TEs ≥1 reads in all samples (Anders et al. 2013). TEs with read count 0 were assumed to have no transcriptional activity. Libraries were normalized with TMM method (Robinson and Oshlack 2010), and converted to CPM-mapped reads using R/Bioconductor package EdgeR v.3.8 (Robinson et al. 2010). Comparisons between transcriptional levels were computed in R 3.6.3 (R Core Team 2019).
Single-Nucleotide Variant Detection and Analysis
Single-nucleotide variants were detected using paired-end sequencing reads of each 42 previously sequenced V. dahliae strains (supplementary table 1, Supplementary Material online). Each strain was mapped independently to the reference genome V. dahliae JR2 using BWA -MEM v.0.7 with default options (Li and Durbin 2010). Library artifacts were marked and removed using Picard tools v.2.18 with -MarkDuplicates followed by -SortSam to sort the reads (http://broadinstitute.github.io/picard/). Single-nucleotide variants were identified using the -HaplotypeCaller of the Genome Analysis Toolkit (GATK) v.4.0 (Poplin et al. 2018). Variations were detected individually per strain, using -ploidy 1 and -emitRefConfidence GVCF. Then, all strains were joined by -Jointgenotyping with -maxAltAlleles 2, and all non-SNVs were removed using SelectVariants -selectType SNP. To obtain high-quality SNVs, we applied the following cut-off filters: QUAL < 250, MQ < 40, QD < 20, FS > 60, SOR > 3.0, ReadPosRankSum < −5.0, MQRankSum between −2 and 2. Finally, we excluded variants with missing genotype calls in 10% of the strains. To explore the similarity between strains, an unrooted phylogenetic tree was constructed using a Neighbor-Joining approach based on the final single-nucleotide variance set, using distance function in R 3.6.3 (R Core Team 2019).
To further assess the sequence diversity between V. dahliae strains, we calculated the nucleotide diversity (π) based on Nei and Jin (1989) using a sliding window of 1 kb (500 bp sliding) as implemented in the PopGenome package (Pfeifer et al. 2014) in R. Single-nucleotide variants were annotated using SNPeff v.3.2 (Cingolani et al. 2012) using the refined annotation of V. dahliae JR2 strain. Based on the SNPeff prediction, we calculated the number of synonymous and nonsynonymous mutations.
Structural Variant Calling and Analysis
To predict SVs, we used the “sv-callers” workflow with few modifications that enabled parallel execution of multiple SV callers (Kuzniar et al. 2020), an approach that is considered optimal as it exploits often complementary information to predict SVs (Goerner-Potvin and Bourque 2018; Cameron et al. 2019). Briefly, the workflow takes every strain and maps the genomic reads to the reference genome V. dahliae JR2 using BWA-MEM v.0.7 with default options (Li and Durbin 2010). Then, library artifacts were marked and removed using Picard tools v.2.18 with -MarkDuplicates followed by -SortSam to sort the reads (http://broadinstitute.github.io/picard/). We used four different callers: DELLY v.0.8.1 (Rausch et al. 2012), LUMPY v.2.13 (Layer et al. 2014), Manta v.1.6.0 (Chen et al. 2016), and Wham v.1.0 (Kronenberg et al. 2015) in single-sample mode, using each 42 previously sequenced V. dahliae strains independently (supplementary table 1, Supplementary Material online). These callers incorporate a diverse range of information for SV detection such as split reads (DELLY, LUMPY, Wham), discordant read pairs (DELLY, LUMPY, Wham), read depth signal (LUMPY, Wham), short-read assembly (Manta, Wham), and soft-clipping detection (Wham). The four SV callers were used with default parameters, except for DELLY in which we used >1 as minimum quality for further processing. All outputs were first filtered using bcftools -filter v.1.3.2 with default settings used in the “sv-callers” workflow (Li 2011), except for Manta since the score model for postprocessing assumes a diploid genome (Chen et al. 2016). Therefore, we used the unscored Manta predictions for further processing.
For final filtering, the “sv-callers” workflow postprocesses the vcfs. Briefly, the results of the four different callers per strain were merged using SURVIVOR v.1.0.6 (Jeffares et al. 2017), keeping SVs predicted by at least three callers, allowing 1,000 bp as the maximum distance from breakpoints predicted by the different tools and considering only same SV types. Subsequently, only SVs with minimum size >50 bp and maximum size 1 Mb as well as localization outside of a low-quality region, defined as MQ = 0 and read support <10 for every strain, were kept. Finally, the 42 independent data sets we combined into a single vcf file using SURVIVOR by merging SVs up to 1,000 bp apart of the same SV type.
Transposable Element Polymorphism Prediction and Analysis
Transposable element presence/absence polymorphisms were analyzed using TEPID v.2.0 (Stuart et al. 2016), using paired-end short reads of 42 previously sequenced V. dahliae strains (supplementary table 1, Supplementary Material online). TEPID integrates split and discordant read mapping, mapping quality, sequencing breakpoints, and local variations in coverage to identify variants with respect to a reference TE annotation (Stuart et al. 2016). We mapped each of the 42 V. dahliae strains individually to V. dahliae JR2 reference genome, using tepid-map (average insert size -s 500), which uses Bowtie2 v.2.2.5 (Langmead and Salzberg 2012). TE variant discovery was computed using tepid-discover, considering insertion and deletion prediction and a conservative discovery through –strict option. We subsequently refined the discovered variants using tepid-refine to reduce false-negative calls within the group of 42 strains.
Statistical Analysis and Visualization
The distribution of SVs over the V. dahliae strain JR2 genome was determined considering the breakpoints (±1 bp) overlapping in 10 kb nonoverlapping windows. We predicted an expected probability distribution assuming a Poisson distribution, using λ = 0.77 (mean number of SVs breakpoints per 10 kb windows). To investigate if SVs colocalize, we performed a clustering analysis using CROC (Pignatelli et al. 2009) based on a hypergeometric distribution test and posterior multiple-testing correction using Benjamini–Hochberg; CROC scans every chromosome with a sliding window (SV-breakpoints per window = 10, slide = 1, >3 SV-breakpoints as a minimum). We computed the same test for TE clustering using TEs per window = 10, slide = 1, >3 polymorphic TEs as a minimum. Permutation tests were computed using R/Bioconductor regionR v.1.18.1 package (Gel et al. 2016) and performed 10,000 iterations, using the mean distance to evaluate the closest relationship (bp distance) between SV-breakpoints and TE elements, and circular randomization to maintain the order and distance of the regions in the chromosomes. We assumed the same parameters to evaluate the association of TEs and adaptive regions but considering the number of overlaps instead of distance. Finally, we computed enrichment association tests using bedtools -fisher v.2.25.0 based on a one-sided Fisher’s test (Quinlan and Hall 2010) and random association permutation tests using R/Bioconductor region v.1.18.1 (Gel et al. 2016), performing 5,000 iterations with a resampling randomization.
Principle component analyses were performed in R v.3.6.3, using packages FactoMineR v.1.42 and factoextra v.1.0.5 (Lê et al. 2008). The used variables for each TE were methylation (mCG, mCHG, mCHH), GC content, Kimura distance, CRI, Frequency, and in vitro (MS media) expression. To further investigate the relationship of TEs with gene expression, we summarized TEs in 5 kb windows upstream and downstream genes, using bedtools -window (Quinlan and Hall 2010). We classified genes in “upstream” or “downstream” if ≥1 TE is located in the position in relation to the gene. We considered genes to be located in “between” TEs if these had ≥1 TEs in both positions (upstream and downstream), and we excluded them from the other categories. We ranked each gene near TE based on their expression profile and counted the number of TEs associated with each independent rank. These rank counts were then binned to obtain the distribution of ranks and fit a linear model for the data and calculated the R2 and P value for the fit of the model in R v.3.6.3 (R Core Team 2019). All statistical analyses and comparison tests were performed in R v.3.6.3 (R Core Team 2019), and visualization with R packages ggplot2 and circlize (Gu et al. 2014; Wickham et al. 2016).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Consejo Nacional de Ciencia y Tecnología, México and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2048/1—Project ID: 390686111.
Data Availability
The data underlying this article are accessible at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject PRJNA592220 (see supplementary table 1, Supplementary Material online).
Literature Cited
- Alonge M, et al. 2020. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182(1):145–161.e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J, Lebrun MH, Quesneville H.. 2015. Whole genome comparative analysis of transposable elements provides new insight into mechanisms of their inactivation in fungal genomes. BMC Genomics 16:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyotte SG, et al. 2012. Transposable elements in phytopathogenic Verticillium spp.: insights into genome evolution and inter- and intra-specific diversification. BMC Genomics 13:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, et al. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 8(9):1765–1786. [DOI] [PubMed] [Google Scholar]
- Badet T, Croll D.. 2020. The rise and fall of genes: origins and functions of plant pathogen pangenomes. Curr Opin Plant Biol. 56:65–73. [DOI] [PubMed] [Google Scholar]
- Badet T, Oggenfuss U, Abraham L, McDonald BA, Croll D.. 2020. A 19-isolate reference-quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici. BMC Biol. 18(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M, Haudry A, Lerat E.. 2014. “One code to find them all”: a perl tool to conveniently parse RepeatMasker output files. Mobile DNA. 5(1):13. [Google Scholar]
- Bennetzen JL, Wang H.. 2014. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol. 65:505–530. [DOI] [PubMed] [Google Scholar]
- Bewick AJ, et al. 2019. Diversity of cytosine methylation across the fungal tree of life. Nat Ecol Evol. 3(3):479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet GF, Jacobi V, Plourde KV, Bernier L.. 2008. Stress-induced mobility of OPHIO1 and OPHIO2, DNA transposons of the Dutch elm disease fungi. Fungal Genet Biol. 45(4):565–578. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Jordan IK.. 2002. Transposable elements and the evolution of eukaryotic complexity. Curr Issues Mol Biol. 4(3):65–76. [PubMed] [Google Scholar]
- Cambareri EB, Jensen BC, Schabtach E, Selker EU.. 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science 244(4912):1571–1575. [DOI] [PubMed] [Google Scholar]
- Cameron DL, Di Stefano L, Papenfuss AT.. 2019. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat Commun. 10(1):3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr PD, et al. 2010. The transposon impala is activated by low temperatures: use of a controlled transposition system to identify genes critical for viability of Aspergillus fumigatus. Eukaryot Cell. 9(3):438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanera R, et al. 2016. Transposable elements versus the fungal genome: impact on whole-genome architecture and transcriptional profiles. PLoS Genet. 12(6):e1006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JE, Kolodner RD.. 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet. 7(5):e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarro-Carrero EA, Vermeulen JP, E., Torres D, Usami T, Schouten HJ, Bai Y, Seidl MF, Thomma BPHJ.. 2021. Comparative genomics reveals the in planta-secreted Verticillium dahliae Av2 effector protein recognized in tomato plants that carry the V2 resistance locus. Environ Microbiol. 23:1941–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, et al. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32(8):1220–1222. [DOI] [PubMed] [Google Scholar]
- Choi JY, Lee YCG.. 2020. Double-edged sword: the evolutionary consequences of the epigenetic silencing of transposable elements. PLoS Genet. 16(7):e1008872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C.. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351(6277):1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118. Fly (Austin) 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck AJ. 2011. Genomic evidence of repeat-induced point mutation (RIP) in filamentous ascomycetes. Fungal Genet Biol. 48(3):306–326. [DOI] [PubMed] [Google Scholar]
- Cook DE, Kramer HM, Torres DE, Seidl MF, Thomma BP.. 2020. A unique chromatin profile defines adaptive genomic regions in a fungal plant pathogen. Elife 9:e62208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Mesarich C, Thomma BPHJ.. 2015. Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol. 53:541–563. [DOI] [PubMed] [Google Scholar]
- Cook LM, Saccheri IJ.. 2013. The peppered moth and industrial melanism: evolution of a natural selection case study. Heredity (Edinb). 110(3):207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosby RL, et al. 2021. Recurrent evolution of vertebrate transcription factors by transposase capture. Science 371(6531):eabc6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, McDonald BA.. 2012. The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog. 8(4):e1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombach A, Hogeweg P.. 2007. Chromosome rearrangements and the evolution of genome structuring and adaptability. Mol Biol Evol. 24(5):1130–1139. [DOI] [PubMed] [Google Scholar]
- Cvijovic I, Good BH, Jerison ER, Desai MM.. 2015. Fate of a mutation in a fluctuating environment. Proc Natl Acad Sci U S A. 112(36):E5021–E5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, et al. 2012. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci U S A. 109(13):5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, et al. 2013. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23(8):1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depotter JRL, et al. 2019. Dynamic virulence-related regions of the plant pathogenic fungus Verticillium dahliae display enhanced sequence conservation. Mol Ecol. 28(15):3482–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Kuo S, Philippsen P.. 2013. Genomes of Ashbya fungi isolated from insects reveal four mating-type loci, numerous translocations, lack of transposons, and distinct gene duplications. G3 (Bethesda) 3(8):1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Raffaele S, Kamoun S.. 2015. The two-speed genomes of filamentous pathogens: waltz with plants. Curr Opin Genet Dev. 35:57–65. [DOI] [PubMed] [Google Scholar]
- Donnart T, Piednoel M, Higuet D, Bonnivard E.. 2017. Filamentous ascomycete genomes provide insights into Copia retrotransposon diversity in fungi. BMC Genomics 18(1):410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutheil JY, et al. 2016. A tale of genome compartmentalization: the evolution of virulence clusters in smut fungi. Genome Biol Evol. 8(3):681–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Sankoff D.. 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301(5634):793–797. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U.. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino L, et al. 2015. Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome. mBio 6(4):e00936–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino L, et al. 2016. Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 26(8):1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Leem Y-E, Levin HL.. 2013. Transposon integration enhances expression of stress response genes. Nucleic Acids Res. 41(2):775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 9(5):397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkens L, Shahi S, Connolly LR, Stam R, Schmidt SM, Smith KM, Freitag M, Rep M.. 2018. The multi-speed genome of Fusarium oxysporum reveals association of histone modifications with sequence divergence and footprints of past horizontal chromosome transfer events. bioRxiv.
- Fouche S, et al. 2020. Stress-driven transposable element de-repression dynamics and virulence evolution in a fungal pathogen. Mol Biol Evol. 37(1):221–239. [DOI] [PubMed] [Google Scholar]
- Fouche S, Plissonneau C, Croll D.. 2018. The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr Opin Microbiol. 46:34–42. [DOI] [PubMed] [Google Scholar]
- Fradin EF, Thomma BP.. 2006. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol. 7(2):71–86. [DOI] [PubMed] [Google Scholar]
- Frantzeskakis L, et al. 2018. Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genomics 19(1):381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantzeskakis L, et al. 2020. Rapid evolution in plant-microbe interactions – a molecular genomics perspective. New Phytol. 225(3):1134–1142. [DOI] [PubMed] [Google Scholar]
- Frantzeskakis L, Kusch S, Panstruga R.. 2019. The need for speed: compartmentalized genome evolution in filamentous phytopathogens. Mol Plant Pathol. 20(1):3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudal I, et al. 2009. Repeat-induced point mutation (RIP) as an alternative mechanism of evolution toward virulence in Leptosphaeria maculans. Mol Plant Microbe Interact. 22(8):932–941. [DOI] [PubMed] [Google Scholar]
- Fuentes RR, et al. 2019. Structural variants in 3000 rice genomes. Genome Res. 29(5):870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Selker EU.. 2004. RIP: the evolutionary cost of genome defense. Trends Genet. 20(9):417–423. [DOI] [PubMed] [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM.. 2007. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 42(5):399–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-Gonzalez L, Mhiri C, Deyholos MK, Grandbastien MA.. 2017. LTR-retrotransposons in plants: engines of evolution. Gene 626:14–25. [DOI] [PubMed] [Google Scholar]
- Gan P, et al. 2021. Telomeres and a repeat-rich chromosome encode effector gene clusters in plant pathogenic Colletotrichum fungi. Environ Microbiol. 10.1111/1462-2920.15490. [DOI] [PubMed]
- Gel B, et al. 2016. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32:289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibriel H, Li J, Zhu L, Seidl MF, Thomma BPHJ.. 2019. Verticillium dahliae strains that infect the same host plant display highly divergent effector catalogs. bioRxiv:528729.
- Goerner-Potvin P, Bourque G.. 2018. Computational tools to unmask transposable elements. Nat Rev Genet. 19(11):688–704. [DOI] [PubMed] [Google Scholar]
- Goubert C, Zevallos NA, Feschotte C.. 2020. Contribution of unfixed transposable element insertions to human regulatory variation. Philos Trans R Soc B. 375(1795):20190331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandaubert J, Dutheil JY, Stukenbrock EH.. 2019. The genomic determinants of adaptive evolution in a fungal pathogen. Evol Lett. 3(3):299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandaubert J, et al. 2014. Transposable element-assisted evolution and adaptation to host plant within the Leptosphaeria maculans-Leptosphaeria biglobosa species complex of fungal pathogens. BMC Genomics 15(1):891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B.. 2014. circlize implements and enhances circular visualization in R. Bioinformatics 30(19):2811–2812. [DOI] [PubMed] [Google Scholar]
- Guo Y, Levin HL.. 2010. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res. 20(2):239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G-Z. 2019. Origin and evolution of the plant immune system. New Phytol. 222(1):70–83. [DOI] [PubMed] [Google Scholar]
- Hartmann FE, Croll D.. 2017. Distinct trajectories of massive recent gene gains and losses in populations of a microbial eukaryotic pathogen. Mol Biol Evol. 34(11):2808–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FE, Sanchez-Vallet A, McDonald BA, Croll D.. 2017. A fungal wheat pathogen evolved host specialization by extensive chromosomal rearrangements. ISME J. 11(5):1189–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G.. 2009. Mechanisms of change in gene copy number. Nat Rev Genet. 10(8):551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocher A, et al. 2018. Expanding heterochromatin reveals discrete subtelomeric domains delimited by chromatin landscape transitions. Genome Res. 28(12):1867–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocher A, Taddei A.. 2020. Subtelomeres as specialized chromatin domains. Bioessays 42(5):e1900205. [DOI] [PubMed] [Google Scholar]
- Hoede C, et al. 2014. PASTEC: an automatic transposable element classification tool. PLoS One 9(5):e91929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof AEvt, et al. 2016. The industrial melanism mutation in British peppered moths is a transposable element. Nature 534(7605):102–105. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS.. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19(8):1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Burns KH, Boeke JD.. 2012. Active transposition in genomes. Annu Rev Genet. 46:651–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A, Le Rouzic A, Boutin TS, Filée J, Capy P.. 2011. The struggle for life of the genome’s selfish architects. Biol Direct. 6(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47(D1):D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pál C, Lercher MJ.. 2004. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 5(4):299–310. [DOI] [PubMed] [Google Scholar]
- Inderbitzin P, Subbarao KV.. 2014. Verticillium systematics and evolution: how confusion impedes Verticillium wilt management and how to resolve it. Phytopathology 104(6):564–574. [DOI] [PubMed] [Google Scholar]
- Jaillon O, et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431(7011):946–957. [DOI] [PubMed] [Google Scholar]
- Jeffares DC, et al. 2017. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 8:14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, et al. 2017. Improved maize reference genome with single-molecule technologies. Nature 546(7659):524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Hammell M.. 2018. Analysis of RNA-Seq data using TEtranscripts. Methods Mol Biol. 1751:153–167. [DOI] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, Hammell M.. 2015. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31(22):3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, et al. 2016. A novel method for identifying polymorphic transposable elements via scanning of high-throughput short reads. DNA Res. 23(3):241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, et al. 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166(2):492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar YD, Ochman H.. 2012. Causes and consequences of genome expansion in fungi. Genome Biol Evol. 4(1):13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR.. 2001. Perspective: transposable elements, parasitic DNA and genome evolution. Evolution 55(1):1–24. [DOI] [PubMed] [Google Scholar]
- Kitano S, Kurasawa H, Aizawa Y.. 2018. Transposable elements shape the human proteome landscape via formation of cis-acting upstream open reading frames. Genes Cells. 23(4):274–284. [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV.. 2009. Diversity, pathogenicity, and management of Verticillium species. Annu Rev Phytopathol. 47:39–62. [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, et al. 2011. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 7(7):e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibbe C, Coulon A, Mazet O, Fayard JM, Beslon G.. 2007. A long-term evolutionary pressure on the amount of noncoding DNA. Mol Biol Evol. 24(10):2344–2353. [DOI] [PubMed] [Google Scholar]
- Kombrink A, et al. 2017. Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol Plant Pathol. 18(4):596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI.. 2009. Is evolution Darwinian or/and Lamarckian? Biol Direct. 4(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan P, et al. 2018. Transposable element insertions shape gene regulation and melanin production in a fungal pathogen of wheat. BMC Biol. 16(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg ZN, et al. 2015. Wham: identifying structural variants of biological consequence. PLoS Comput Biol. 11(12):e1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniar A, et al. 2020. sv-callers: a highly portable parallel workflow for structural variant detection in whole-genome sequence data. PeerJ 8:e8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM.. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15(6):R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A, Boutin TS, Capy P.. 2007. Long-term evolution of transposable elements. Proc Natl Acad Sci U S A. 104(49):19375–19380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F.. 2008. FactoMineR: an R package for multivariate analysis. J Stat Softw. 25(1):18. [Google Scholar]
- Le TN, Miyazaki Y, Takuno S, Saze H.. 2015. Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana. Nucleic Acids Res. 43(8):3911–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Karpen GH.. 2017. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. Elife 6:e25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem Y-E, et al. 2008. Retrotransposon Tf1 is targeted to Pol II promoters by transcription activators. Mol Cell. 30(1):98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. 2004. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 20(3):116–122. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Gokcumen O.. 2019. Fine-scale characterization of genomic structural variation in the human genome reveals adaptive and biomedically relevant hotspots. Genome Biol Evol. 11(4):1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Martienssen R.. 2004. The role of RNA interference in heterochromatic silencing. Nature 431(7006):364–370. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. 2020. Pan-genome of wild and cultivated soybeans. Cell 182(1):162–176.e113. [DOI] [PubMed] [Google Scholar]
- Lorrain C, Feurtey A, Möller M, Haueisen J, Stukenbrock E. 2021. Dynamics of transposable elements in recently diverged fungal pathogens: lineage-specific transposable element content and efficiency of genome defenses. G3 (Bethesda). 11(4):jkab068. [DOI] [PMC free article] [PubMed]
- Lynch M. 2007. The origins of genome architecture. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Lynch M, Bobay LM, Catania F, Gout JF, Rho M.. 2011. The repatterning of eukaryotic genomes by random genetic drift. Annu Rev Genomics Hum Genet. 12:347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 302(5649):1401–1404. [DOI] [PubMed] [Google Scholar]
- Marand AP, et al. 2019. Historical meiotic crossover hotspots fueled patterns of evolutionary divergence in rice. Plant Cell 31(3):645–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, et al. 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544(7651):427–433. [DOI] [PubMed] [Google Scholar]
- Maxwell PH. 2020. Diverse transposable element landscapes in pathogenic and nonpathogenic yeast models: the value of a comparative perspective. Mob DNA. 11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A. 36(6):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P, Boeke JD.. 2016. How retrotransposons shape genome regulation. Curr Opin Genet Dev. 37:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller M, et al. 2019. Destabilization of chromosome structure by histone H3 lysine 27 methylation. PLoS Genet. 15(4):e1008093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller M, et al. 2021. Recent loss of the Dim2 DNA methyltransferase decreases mutation rate in repeats and changes evolutionary trajectory in a fungal pathogen. PLoS Genet. 17(3):e1009448. [DOI] [PMC free article] [PubMed]
- Moller M, Stukenbrock EH.. 2017. Evolution and genome architecture in fungal plant pathogens. Nat Rev Microbiol. 15(12):756–771. [DOI] [PubMed] [Google Scholar]
- Montanini B, et al. 2014. Non-exhaustive DNA methylation-mediated transposon silencing in the black truffle genome, a complex fungal genome with massive repeat element content. Genome Biol. 15(8):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszewska A, Hoffman-Sommer M, Grynberg M.. 2011. LTR retrotransposons in fungi. PLoS One 6(12):e29425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszewska A, Steczkiewicz K, Stepniewska-Dziubinska M, Ginalski K.. 2019. Transposable elements contribute to fungal genes and impact fungal lifestyle. Sci Rep. 9(1):4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki H, Nishimoto N, Ikeda K, Tosa Y, Mayama S.. 1999. Degenerate MAGGY elements in a subgroup of Pyricularia grisea: a possible example of successful capture of a genetic invader by a fungal genome. Mol Gen Genet. 261(6):958–966. [DOI] [PubMed] [Google Scholar]
- Nei M, Jin L.. 1989. Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol. 6(3):290–300. [DOI] [PubMed] [Google Scholar]
- Nicolas FE, Torres-Martinez S, Ruiz-Vazquez RM.. 2013. Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 9:e1003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H. 2017. Predicting secretory proteins with SignalP. Springer. p. 59–73. [DOI] [PubMed] [Google Scholar]
- Oggenfuss U, Badet T, Wicker T, Hartmann FE, Singh NK, Abraham LN, Karisto P, Vonlanthen T, Mundt CC, McDonald BA, et al. 2020. A population-level invasion by transposable elements in a fungal pathogen. bioRxiv. [DOI] [PMC free article] [PubMed]
- Ola M, et al. 2020. Polymorphic centromere locations in the pathogenic yeast Candida parapsilosis. Genome Res. 30(5):684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ.. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 31(7):1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piégu B, Bire S, Arensburger P, Bigot Y.. 2015. A survey of transposable element classification systems–a call for a fundamental update to meet the challenge of their diversity and complexity. Mol Phylogenet Evol. 86:90–109. [DOI] [PubMed] [Google Scholar]
- Pignatelli M, Serras F, Moya A, Guigó R, Corominas M.. 2009. CROC: finding chromosomal clusters in eukaryotic genomes. Bioinformatics 25(12):1552–1553. [DOI] [PubMed] [Google Scholar]
- Plissonneau C, Hartmann FE, Croll D.. 2018. Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome. BMC Biol. 16(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plissonneau C, Sturchler A, Croll D.. 2016. The evolution of orphan regions in genomes of a fungal pathogen of wheat. mBio. 7(5):e01231–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin R, et al. 2018. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. 36(10):983–987. [DOI] [PubMed] [Google Scholar]
- Prendergast JGD, et al. 2007. Chromatin structure and evolution in the human genome. BMC Evol Biol. 7:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2019. R: a language and environment for statistical computing, Version 3.5.3. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Raffaele S, et al. 2010. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330(6010):1540–1543. [DOI] [PubMed] [Google Scholar]
- Rausch T, et al. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28(18):i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A.. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11(3):R25–R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouxel T, et al. 2011. Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nat Commun. 2:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovenich H, Boshoven JC, Thomma BP.. 2014. Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr Opin Plant Biol. 20:96–103. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, et al. 2004. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134(4):1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vallet A, et al. 2018. The genome biology of effector gene evolution in filamentous plant pathogens. Annu Rev Phytopathol. 56:21–40. [DOI] [PubMed] [Google Scholar]
- Schnable PS, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115. [DOI] [PubMed] [Google Scholar]
- Schotanus K, et al. 2015. Histone modifications rather than the novel regional centromeres of Zymoseptoria tritici distinguish core and accessory chromosomes. Epigenet Chromatin. 8:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Schmitz J.. 2019. The impact of transposable elements in adaptive evolution. Mol Ecol. 28(6):1537–1549. [DOI] [PubMed] [Google Scholar]
- Seidl MF, Cook DE, Thomma BPHJ.. 2016. Chromatin biology impacts adaptive evolution of filamentous plant pathogens. PLoS Pathog. 12(11):e1005920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl MF, et al. 2015. The genome of the saprophytic fungus Verticillium tricorpus reveals a complex effector repertoire resembling that of its pathogenic relatives. Mol Plant Microbe Interact. 28(3):362–373. [DOI] [PubMed] [Google Scholar]
- Seidl MF, et al.. 2020. Repetitive elements contribute to the diversity and evolution of centromeres in the fungal genus Verticillium. mBio. 11(5):e01714–20. [DOI] [PMC free article] [PubMed]
- Seidl MF, Thomma B.. 2017. Transposable elements direct the coevolution between plants and microbes. Trends Genet. 33(11):842–851. [DOI] [PubMed] [Google Scholar]
- Seidl MF, Thomma BP.. 2014. Sex or no sex: evolutionary adaptation occurs regardless. Bioessays 36(4):335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, et al. 2003. The methylated component of the Neurospora crassa genome. Nature 422(6934):893–897. [DOI] [PubMed] [Google Scholar]
- Shi-Kunne X, Faino L, van den Berg GCM, Thomma B, Seidl MF.. 2018. Evolution within the fungal genus Verticillium is characterized by chromosomal rearrangement and gene loss. Environ Microbiol. 20(4):1362–1373. [DOI] [PubMed] [Google Scholar]
- Sinzelle L, Izsvák Z, Ivics Z.. 2009. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci. 66(6):1073–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R.. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 8(4):272–285. [DOI] [PubMed] [Google Scholar]
- Soyer JL, Grandaubert J, Haueisen J, Schotanus K, Stukenbrock EH.. 2019. In planta chromatin immunoprecipitation in Zymoseptoria tritici reveals chromatin-based regulation of putative effector gene expression. bioRxiv.
- Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM.. 2018. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol. 19(9):2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S.. 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37(21):7002–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, et al. 2016. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. Elife 5:e20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Wysocka J.. 2020. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos Trans R Soc B. 375(1795):20190347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szitenberg A, et al. 2016. Genetic drift, not life history or RNAi, determine long-term evolution of transposable elements. Genome Biol Evol. 8(9):2964–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres DE, Oggenfuss U, Croll D, Seidl MF.. 2020. Genome evolution in fungal plant pathogens: looking beyond the two-speed genome model. Fungal Biol Rev. 34(3):136–143. [Google Scholar]
- Tsukahara S, et al. 2009. Bursts of retrotransposition reproduced in Arabidopsis. Nature 461(7262):423–426. [DOI] [PubMed] [Google Scholar]
- van Wersch S, Li X.. 2019. Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci. 24(8):688–699. [DOI] [PubMed] [Google Scholar]
- van Wyk S, et al. 2019. The RIPper, a web-based tool for genome-wide quantification of repeat-induced point (RIP) mutations. PeerJ 7:e7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venner S, Feschotte C, Biemont C.. 2009. Dynamics of transposable elements: towards a community ecology of the genome. Trends Genet. 25(7):317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL.. 2006. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci U S A. 103(47):17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkowiak S, et al. 2020. Multiple wheat genomes reveal global variation in modern breeding. Nature 588(7837):277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, et al. 2017. Characterization of the two-speed subgenomes of Fusarium graminearum reveals the fast-speed subgenome specialized for adaption and infection. Front Plant Sci. 8:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. 2020. Repeat-induced point mutation in Neurospora crassa causes the highest known mutation rate and mutational burden of any cellular life. Genome Biol. 21(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, et al. 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43(W1):W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8(12):973–982. [DOI] [PubMed] [Google Scholar]
- Wickham H, Chang W, Wickham MH. 2016. Package ‘ggplot2’. Create Elegant Data Visualisations Using the Grammar of Graphics. Version, 2(1):1–189. Oxford University Press; [Google Scholar]
- Winter DJ, et al. 2018. Repeat elements organise 3D genome structure and mediate transcription in the filamentous fungus Epichloe festucae. PLoS Genet. 14(10):e1007467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S, Finnegan D.. 2001. Genome evolution: sex and the transposable element. Curr Biol. 11(8):R296–R299. [DOI] [PubMed] [Google Scholar]
- Wyka SA, et al. 2021. Whole-Genome Comparisons of Ergot Fungi Reveals the Divergence and Evolution of Species within the Genus Claviceps Are the Result of Varying Mechanisms Driving Genome Evolution and Host Range Expansion. Genome Biol Evol. 13(2):evaa267. [DOI] [PMC free article] [PubMed]
- Yadav V, et al. 2018. RNAi is a critical determinant of centromere evolution in closely related fungi. Proc Natl Acad Sci U S A. 115(12):3108–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, et al. 2016. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics 17:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D.. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328(5980):916–919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are accessible at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject PRJNA592220 (see supplementary table 1, Supplementary Material online).