Abstract
Accurate segmentation of the jaw (i.e., mandible and maxilla) and the teeth in cone beam computed tomography (CBCT) scans is essential for orthodontic diagnosis and treatment planning. Although various (semi)automated methods have been proposed to segment the jaw or the teeth, there is still a lack of fully automated segmentation methods that can simultaneously segment both anatomic structures in CBCT scans (i.e., multiclass segmentation). In this study, we aimed to train and validate a mixed-scale dense (MS-D) convolutional neural network for multiclass segmentation of the jaw, the teeth, and the background in CBCT scans. Thirty CBCT scans were obtained from patients who had undergone orthodontic treatment. Gold standard segmentation labels were manually created by 4 dentists. As a benchmark, we also evaluated MS-D networks that segmented the jaw or the teeth (i.e., binary segmentation). All segmented CBCT scans were converted to virtual 3-dimensional (3D) models. The segmentation performance of all trained MS-D networks was assessed by the Dice similarity coefficient and surface deviation. The CBCT scans segmented by the MS-D network demonstrated a large overlap with the gold standard segmentations (Dice similarity coefficient: 0.934 ± 0.019, jaw; 0.945 ± 0.021, teeth). The MS-D network–based 3D models of the jaw and the teeth showed minor surface deviations when compared with the corresponding gold standard 3D models (0.390 ± 0.093 mm, jaw; 0.204 ± 0.061 mm, teeth). The MS-D network took approximately 25 s to segment 1 CBCT scan, whereas manual segmentation took about 5 h. This study showed that multiclass segmentation of jaw and teeth was accurate and its performance was comparable to binary segmentation. The MS-D network trained for multiclass segmentation would therefore make patient-specific orthodontic treatment more feasible by strongly reducing the time required to segment multiple anatomic structures in CBCT scans.
Keywords: facial bones, dentofacial deformities, diagnostic imaging, image processing, neural networks, artificial intelligence
Introduction
Cone beam computed tomography (CBCT) is being increasingly used in orthodontics because of its low cost and low radiation dose (Carter et al. 2016). The 3-dimensional (3D) information embedded in CBCT scans allows orthodontists to accurately assess complex dental and skeletal malocclusions, which helps to substantially improve diagnosis and treatment planning (Kapila and Nervina 2015; Abdelkarim 2019). An essential step in the diagnosis of dentofacial abnormalities and deformities is the conversion of CBCT scans into virtual 3D models of relevant anatomic regions of interest, such as the mandible, the maxilla, and the teeth. Orthodontic changes can be visually and quantitatively assessed by superimposing 3D models of a patient at different time points. Moreover, these 3D models can be used to simulate stress distribution in alveolar bone via finite element analysis (Likitmongkolsakul et al. 2018).
Currently, the most challenging step in creating 3D models for orthodontics is CBCT image segmentation—that is, the partitioning of CBCT scans into various anatomic regions of interest. For example, it is laborious to segment the teeth due to the similar intensity of tooth roots and their surrounding alveolar bone. In addition, because of high noise levels, limited image resolution, and cone beam artifacts (Schulze et al. 2011), it is difficult to accurately segment bony structures—for example, the condyle and the ramus. Consequently, bony structures are often erroneously labeled, which leads to cavities and gaps when converted into 3D models, subsequently compromising the quality of the treatment planning and finite element analysis.
In the last decades, several methods have been proposed to (semi)automatically segment various anatomic structures in CBCT scans. Such automatic approaches include edge detection, watershed segmentation, region seed growing, clustering methods, morphologic snakes, random forests, and statistical shape models (Khan 2014; Mustafa et al. 2016; van Eijnatten et al. 2018). Although these methods are capable of segmenting CBCT scans, accurate segmentation of the mandible, the maxilla, and the teeth remains challenging. Therefore, laborious manual correction remains necessary in clinical settings to achieve accurate segmentation. Hence, new methods for automatic image segmentation are sought.
Deep convolutional neural networks (CNNs) have recently become increasingly utilized in medical image segmentation (Litjens et al. 2017; Altaf et al. 2019) and have achieved state-of-the-art performances (Minnema et al. 2018; Casalegno et al. 2019; Nguyen et al. 2020). The success of CNNs can be mainly attributed to their ability of learning nonlinear spatial features in input images. Several research groups have used CNNs to segment the mandible or the teeth (i.e., binary segmentation) and demonstrated that CNNs were able to perform accurate segmentation tasks (Egger et al. 2018; Qiu et al. 2018; Cui et al. 2019; Lee et al. 2020). However, no studies have been published in which CNNs were applied to simultaneously segment the jaw (i.e., mandible and maxilla) and the teeth in CBCT scans, also known as multiclass segmentation. As compared with binary segmentation, multiclass segmentation approaches require training only a single network to segment jaw and teeth, thus reducing the overall training time. Furthermore, multiclass segmentation does not suffer from conflicting segmentation labels. Such conflicting labels are caused when 1 binary segmentation network classifies a pixel as jaw and the other classifies it as teeth.
A novel CNN architecture, namely the mixed-scale dense (MS-D) CNN, has recently shown promising segmentation performances (Pelt and Sethian 2018; Minnema et al. 2019). This MS-D network allows for accurate and automatic segmentation of different bony structures. To reduce the time-consuming and costly manual labor required to create 3D models for patient-specific orthodontic treatment, our aim was to train the MS-D network to simultaneously segment the jaw and the teeth in CBCT scans.
Materials and Methods
CBCT scan information, CNN architecture, implementation and training details, and CNN performance evaluation are provided in the Appendix.
Data Acquisition and Preprocessing
Thirty dental CBCT scans were acquired from patients (age, 11 to 24 y; mean ± SD, 14.2 ± 3.4 y; 19 females and 11 males) who previously underwent orthodontic treatment at the Shanghai Xuhui Dental Center. The CBCT scans used in this study were obtained before the orthodontic treatment, and none of the patients had fillings, dental implants, or crowns. Thus, the CBCT scans were free of metal artifacts. Moreover, no patients had missing teeth, and wisdom teeth were not yet erupted in most patients (n = 22). Informed consent was signed by each patient and at least 1 parent. The use of patient data was approved by the medical ethics committee of the Shanghai Xuhui Dental Center (No. 20193).
Since this study focused on segmenting the jaw and the teeth, we cropped all CBCT scans to those anatomic regions, resulting in scans with axial dimensions ranging between 255 to 384. In total, 9507 slices were obtained from the 30 CBCT scans.
To acquire gold standard labels, all 30 CBCT scans were segmented into 3 classes: jaw, teeth, and background. The manual segmentation was carried out by 4 dentists with at least 2 y of working experience in dental clinics. The 4 dentists were well instructed and practiced extensively until they could accurately annotate jaw and teeth in CBCT scans. After that, the 30 CBCT scans were distributed among the 4 annotators, and each CBCT scan was segmented only once by a single annotator. This segmentation was performed with global thresholding, followed by manual correction—that is, removing noise, artifacts, and unrelated parts, as well as adding missing thin bony structures and filling erroneous cavities in the segmented scans through Mimics 21.0 software (Materialise). The resulting segmentation labels were used as the gold standard.
CNN Architecture
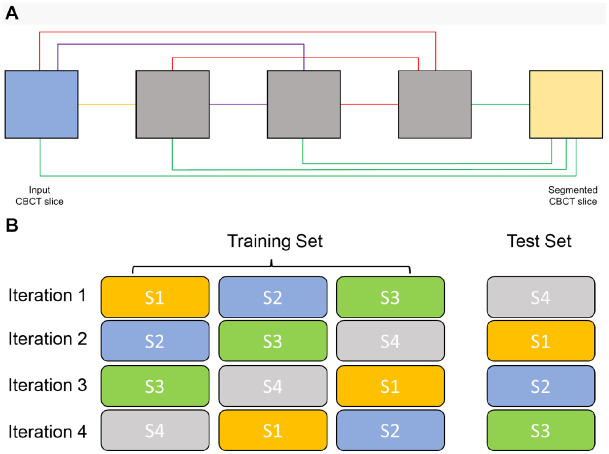
In this study, we employed an MS-D network that was developed by Pelt and Sethian (2018). A schematic overview of an MS-D network with a depth of 3 and a width of 1 is presented in Figure 1A.
Figure 1.
MS-D network architecture and 4-fold cross-validation scheme. (A) Schematic representation of an MS-D network with 3 convolutional layers and a width of 1; (B) 28 CBCT scans were divided into 4 subsets (S1, S2, S3, and S4), each containing 7 CBCT scans. For each iteration, 3 subsets were used for training and 1 for testing. CBCT, cone beam computed tomography; MS-D, mixed-scale dense.
Implementation and Training Details
Three experiments were designed to evaluate the MS-D network’s segmentation performance. The first experiment was multiclass segmentation, in which the MS-D network was trained to simultaneously segment 3 labels: jaw, teeth, and background. The second and third experiments were binary segmentation, where the MS-D network segmented jaw or teeth, respectively.
Twenty-eight CBCT scans were divided into 4 subsets (S1, S2, S3, and S4), each containing 7 scans. Each experiment followed a 4-fold cross-validation scheme (Anguita et al. 2012), which means that 3 subsets were used for training and 1 for testing. This process was repeated 4 times such that each CBCT scan was used for testing exactly once (Fig. 1B). The 2 CBCT scans that were not included in the 4-fold cross-validation scheme were used to determine the optimal number of epochs for training.
CNN Performance Evaluation
The segmentation performance of the MS-D network was evaluated with the Dice similarity coefficient (DSC; Zou et al. 2004). DSCs were calculated on the patient level, which means that a single DSC was calculated for each segmented CBCT volume.
Surface deviations between the MS-D network–based 3D models and the gold standards were calculated to evaluate the accuracy of the MS-D segmentation around the edges of bony structures. Additionally, mean absolute deviations (MADs) were calculated between the MS-D network–based 3D models and the gold standards.
After the 4 iterations of the cross-validation scheme, the performance of the MS-D network was averaged over the 28 CBCT scans.
Results
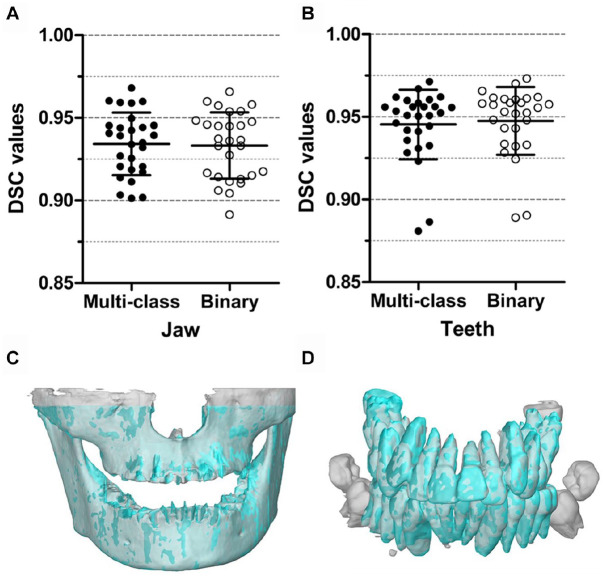
The multiclass and binary segmentation approaches achieved similar segmentation accuracies (Fig. 2A, B). The former approach resulted in DSCs of jaw between 0.901 (patient 3) and 0.968 (patient 28), with a mean of 0.934 ± 0.019. The DSCs of teeth ranged between 0.881 (patient 2) and 0.971 (patient 28), with a mean of 0.945 ± 0.021. For the binary segmentation, the DSCs of jaw ranged from 0.892 (patient 3) to 0.966 (patient 28), with a mean of 0.933 ± 0.020. The DSCs of teeth ranged from 0.889 (patient 2) to 0.973 (patient 28), with a mean of 0.948 ± 0.021. The lowest DSC of jaw from patient 3 was due to the larger excluded region of maxilla in its gold standard while this excluded region was segmented by the MS-D (Fig. 2C). The lowest DSC of teeth from patient 2 was attributed to the unerupted teeth not being included in the gold standard while the MS-D segmented these teeth (Fig. 2D). Comparison of the DSCs obtained with the multiclass and binary segmentation approaches showed 90% CIs of difference of −0.001 to 0.003 for jaw segmentation and −0.004 to −0.001 for teeth segmentation, which indicated that the 2 approaches resulted in equivalent segmentation performances with a confidence of 95%.
Figure 2.
Comparison of DSCs obtained using the multiclass and binary segmentation approaches. For jaw segmentation (A) and teeth segmentation (B), the multiclass segmentation and the binary segmentation were equivalent. In addition, 3D models of the jaw (C) and the teeth (D) are presented, which were acquired by using the CBCT scans that resulted in the lowest DSCs. 3D, 3-dimensional; CBCT, cone beam computed tomography; DSC, Dice similarity coefficient.
Examples of the multiclass segmentation of the CBCT scan from patient 9 are presented in Figure 3. Five axial CBCT slices representing different skull anatomies were selected. The difference maps show that the errors mainly occurred at the edges with deviations around 1 pixel (Fig. 3A, B). Some thin bony structures around the maxillary sinus were not segmented by the MS-D network as compared with the gold standard (Fig. 3E).
Figure 3.
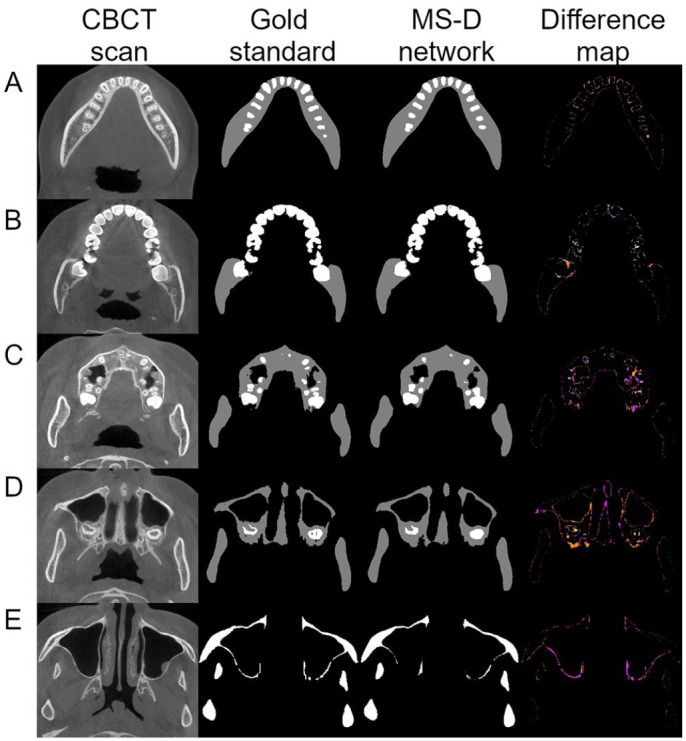
Example of jaw and teeth segmentation from 5 axial CBCT slices of patient 9 with the multiclass segmentation approach. The first column contains 5 axial CBCT slices, which represent different skull anatomies. The second and third columns show the gold standard segmentation and the MS-D segmentation, respectively. The last column indicates the difference between the gold standard and the MS-D segmentations. False negatives of the jaw are marked in fuchsia, and false positives are marked in ultra pink and gamboge. False negatives of the teeth are marked in pink, and false positives are marked in wheat and yellow. CBCT, cone beam computed tomography; MS-D, mixed-scale dense.
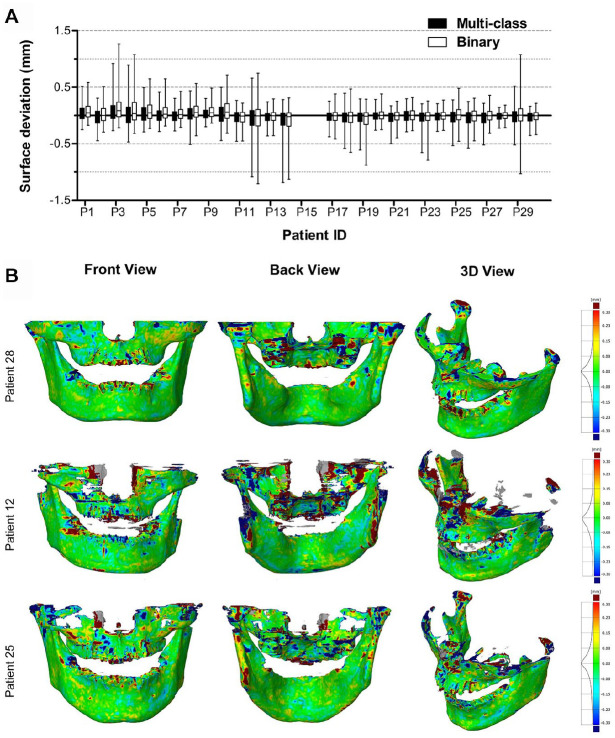
Figure 4A shows the surface deviations of all 3D jaw models obtained from the multiclass and binary segmentations. Figure 4B illustrates 3 jaw models from the multiclass segmentation. Patient 28 and 12 corresponded to the minimum and maximum MADs, respectively. Patient 25 had an MAD close to the mean MAD. All MAD values of jaw models are presented in the Appendix Table. When analyzing jaw models, the multiclass segmentation resulted in surface deviations from −0.191 ± 1.095 mm (patient 14) to 0.185 ± 1.011 mm (patient 3) and a mean MAD of 0.390 ± 0.093 mm. The binary segmentation resulted in surface deviations from −0.180 ± 1.069 mm (patient 14) to 0.252 ± 1.058 mm (patient 3) and a mean MAD of 0.410 ± 0.103 mm.
Figure 4.
Surface deviations between the MS-D network–based 3D models of the jaw and the corresponding gold standard 3D models. The CBCT scans from patients 15 and 16 were used for validation and therefore not included in the analysis. (A) Box and whisker plot of the surface deviations. The boxes represent the interquartile range, and the whiskers represent the 10th and 90th percentiles of the surface deviations. (B) The front, back, and isotropic views of color maps of surface deviations are presented for 3 jaw models. Patients 28 and 12 corresponded to the minimum and maximum MADs, respectively. Patient 25 had an MAD close to the mean MAD. 3D, 3-dimensional; CBCT, cone beam computed tomography; MAD, mean absolute deviation.
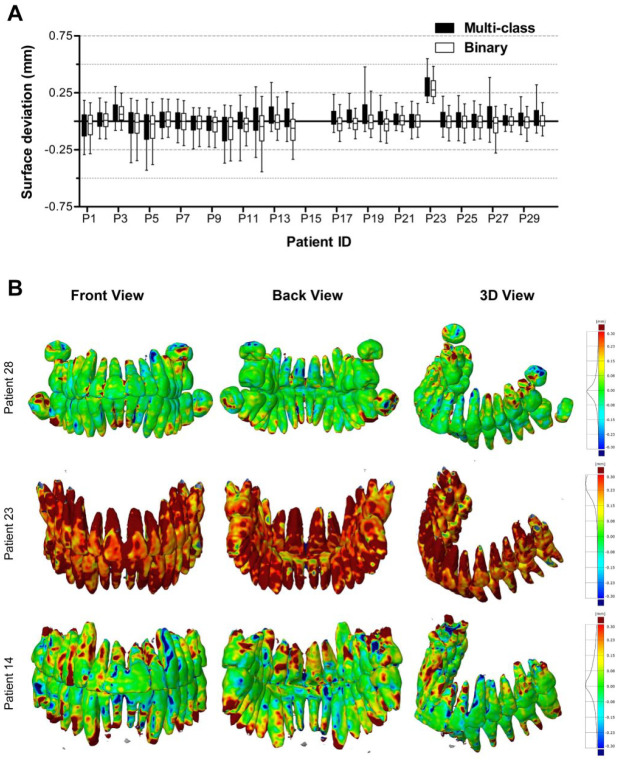
Figure 5A shows the surface deviations of all 3D teeth models obtained from the multiclass and binary segmentations. Figure 5B presents 3 teeth models from the multiclass segmentation. Patients 28 and 23 corresponded to the minimum and maximum MADs, respectively. Patient 14 had an MAD close to the mean MAD. All MAD values of teeth models are presented in the Appendix Table. When analyzing teeth models, the multiclass segmentation resulted in surface deviations from −0.107 ± 0.546 mm (patient 5) to 0.318 ± 0.347 mm (patient 23) and a mean MAD of 0.204 ± 0.061 mm. The binary segmentation resulted in surface deviations from −0.116 ± 0.534 mm (patient 12) to 0.290 ± 0.272 mm (patient 23) and a mean MAD of 0.163 ± 0.051 mm.
Figure 5.
Surface deviations between the MS-D network–based 3D models of the teeth and the corresponding gold standard models. The CBCT scans from patients 15 and 16 were used for validation and therefore not included in the analysis. (A) Box and whisker plot of the surface deviations. The boxes represent the interquartile range, and the whiskers represent the 10th and 90th percentiles of the surface deviations. (B) The front, back, and isotropic views of color maps of surface deviations are presented for 3 teeth models. Patients 28 and 23 corresponded to the minimum and maximum MADs, respectively. Patient 14 had an MAD close to the mean MAD. 3D, 3-dimensional; CBCT, cone beam computed tomography; MAD, mean absolute deviation; MS-D, mixed-scale dense.
Discussion
CBCT is increasingly utilized to create virtual 3D models for quantitative evaluation of orthodontic changes such as tooth resorption, condyle growth, and movement of the chin and teeth. Creating these 3D models requires accurate segmentation of jaw (i.e., mandible and maxilla) and teeth. However, manually segmenting these 2 anatomies is time-consuming, laborious, and expensive. In this study, we trained a novel MS-D network to simultaneously segment jaw and teeth in CBCT scans (i.e., multiclass segmentation). The jaw and teeth segmented by the MS-D network demonstrated high DSCs, and their 3D models showed minor surface deviations as compared with the gold standard. The MS-D network took approximately 25 s to segment jaw and teeth in 1 CBCT scan, thus markedly reducing the time required for segmentation. Therefore, the MS-D network trained for multiclass segmentation has promising potential to accurately and automatically segment multiple anatomies of interest for orthodontic diagnosis and treatment.
Multiclass segmentation has been considered challenging since it faces class data imbalance and interclass feature similarity problems (Chen et al. 2018; Novikov et al. 2018; Jafari et al. 2019). As compared with multiclass strategies, binary strategies are generally more robust and achieve higher accuracy but come at the cost of increased training time (Berstad et al. 2018; Gómez et al. 2020). In this study, the MS-D network trained for multiclass segmentation was able to accurately segment jaw and teeth in CBCT scans, achieving comparable accuracy as binary segmentation. This indicates that the MS-D network can be trained with 3 classes without losing segmentation accuracy as compared with binary segmentation. Moreover, multiclass segmentation has 2 important advantages over binary segmentation. The first is that multiclass segmentation requires training only a single CNN for segmentation of jaw and teeth, which was twice as fast as training the 2 CNNs needed for binary segmentation. Specifically, training an MS-D network took about 20 h (1 h per epoch), and segmentation of 1 CBCT scan took approximately 25 s. Nevertheless, it must be noted that the segmentation time of both deep learning approaches is still significantly less than that of manual segmentation, which took around 5 h per CBCT scan. The second advantage is that multiclass segmentation does not generate conflicting labels, as opposed to binary segmentation. These conflicting labels are caused when 1 binary segmentation network classifies a pixel as jaw and the other classifies it as teeth (Appendix Figure).
The MS-D networks trained in this study resulted in DSCs that are comparable to those presented in literature. For mandible segmentation, Qiu et al. (2018) obtained a mean DSC of 0.896 by training 3 CNNs using CBCT slices from axial, sagittal, or coronal planes and then combining the segmentation results from all 3 CNNs. For maxilla segmentation, a lower mean DSC of 0.800 ± 0.029 was found by S. Chen et al. (2020), who used a learning-based multisource integration framework. For teeth segmentation, Lee et al. (2020) applied a multiphase strategy to train a U-Net–based architecture, which resulted in DSCs ranging from 0.910 to 0.918. Furthermore, Cui et al. (2019) employed a 2-stage network consisting of a tooth edge map extraction network and a region proposal network and reported a mean DSC of 0.926. In comparison with the aforementioned studies, the MS-D network employed in this study achieved comparable DSCs. However, the reviewed studies used different data sets to evaluate their methods, which means that differences among the DSCs should be interpreted with caution.
All MS-D network–based 3D models closely resembled the corresponding gold standard 3D models. The surface deviations found in our study were generally lower than those obtained by Wang et al. (2016), who developed a random forest method to segment the mandible and the teeth in CBCT scans. When segmenting the mandible, their method resulted in a surface deviation of 0.420 ± 0.150 mm, whereas the segmentation of the upper and lower teeth resulted in surface deviations of 0.312 ± 0.103 mm and 0.346 ± 0.154 mm, respectively. Our MS-D network segmentation also resulted in lower surface deviations of teeth than the multitask fully CNN developed by Y. Chen et al. (2020). Their network was trained for individual tooth segmentation, which resulted in an average surface deviation of 0.363 ± 0.145 mm.
The segmentation errors of the MS-D network trained in this study mainly occurred at the edges of the bony structures (Fig. 3). These segmentation errors are likely due to the partial volume effect. This occurs when tissues with different densities are encompassed within the same voxel, which is typically the case at the border of 2 anatomic regions (e.g., bone and soft tissue). As a consequence, it is extremely hard to exactly define the edge regions of bony structures. This phenomenon might also explain why some thin bony structures in the maxillae were not correctly segmented by the MS-D network (Fig. 3E). The quality of CBCT scans also affects the accuracy of segmentation. For example, the ramus and the condyle were poorly segmented in the CBCT scan of patient 12 (Fig. 4B) because these regions were affected by movement artifacts.
One challenge for deep learning in medicine and dentistry is to obtain an accurate gold standard (Schwendicke et al. 2020). The gold standard is usually created by human annotators, which contains intrinsic inter- and intraobserver variability. However, deep learning can learn from large training data sets and therefore is able to overcome this variability (Naylor 2018). In this study, the gold standard segmentation labels were annotated by 4 dentists, which introduced subjective variability in the gold standard. For example, the unerupted teeth from 1 patient were not included in the gold standard. Because the MS-D network was able to learn from all segmented CBCT images, the unerupted teeth in the CBCT scans were all correctly segmented by the MS-D network. These findings indicate that the MS-D network can reduce the influence of subjective variability. If the inaccurate gold standard labels are included in the test set, this can affect the evaluation of the network, particularly for small data sets. However, since the performance of the MS-D network was evaluated on the 28 CBCT scans, the influence of 1 inaccurate gold standard scan in the test set can be minimized.
In the present study, the MS-D network was adopted to evaluate the multiclass segmentation performance. This network was chosen because it has relatively few parameters, making it easier to train and apply than other CNNs (Pelt and Sethian 2018). The multiclass segmentation performance of the MS-D network was compared with that of the U-Net, demonstrating that it was able to achieve similar segmentation accuracy as the U-Net (Pelt and Sethian 2018). Nevertheless, the MS-D network is not the only CNN architecture capable of performing multiclass segmentation. Several other CNN architectures have been implemented to perform multiclass segmentation of anatomies in brain (Chen et al. 2018; Jafari et al. 2019) and lung (Novikov et al. 2018; Saood and Hatem 2020).
In this study, all CBCT scans were obtained from patients without dental fillings, implants, or orthodontic devices to avoid the influence of metal artifacts. In daily clinical practice, there may be such artifacts, so caution should be taken to apply the current findings in clinics. Further studies shall be performed for CBCT image segmentation with complicated dental status.
To facilitate CNN training, the maxilla and mandible were considered a single class, and the upper and lower teeth were considered another class. However, one may wish to automatically separate the mandible from the maxilla and classify individual teeth during segmentation. The mandible and the maxilla can be easily separated by using region growing methods available in most image processing software, but individual teeth segmentation still requires postprocessing. To make the automatic segmentation of individual teeth possible, we aim to include individual labels of different teeth during the training of the MS-D network in future work.
Conclusion
This study applied a novel MS-D network to segment CBCT scans into jaw, teeth, and background. Multiclass segmentation achieved comparable segmentation accuracy as binary segmentation. In addition, the MS-D network–based 3D models closely resembled the gold standard 3D models. These results demonstrate that deep learning has the potential to accurately and simultaneously segment jaw and teeth in CBCT scans. This will substantially reduce the amount of time and effort spent in clinical settings, thereby facilitating patient-specific orthodontic treatment.
Author Contributions
H. Wang, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; J. Minnema, contributed to design, data analysis, and interpretation, critically revised the manuscript; K.J. Batenburg, F.J. Hu, contributed to data interpretation, critically revised the manuscript; T. Forouzanfar, contributed to conception, critically revised the manuscript; G. Wu, contributed to conception and data acquisition, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211005338 for Multiclass CBCT Image Segmentation for Orthodontics with Deep Learning by H. Wang, J. Minnema, K.J. Batenburg, T. Forouzanfar, F.J. Hu and G. Wu in Journal of Dental Research
Acknowledgments
We thank orthodontist Xiaoqing Ma for collecting CBCT scans and dentists Gaoli Xu, Yan Zhang, Zhennan Deng, and Danni Wu for helping to manually label jaw and teeth as the gold standard. We also thank Allard Hendriksen for his advice on using the MS-D network in this study. We further thank Dr. René van Oers for proofreading and discussing the manuscript.
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported partly by Nederlandse Organisatie voor Wetenschappelijk Onderzoek (040.20.009/040.20.010).
References
- Abdelkarim A. 2019. Cone-beam computed tomography in orthodontics. Dent J. 7(3):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf F, Islam SM, Akhtar N, Janjua NK. 2019. Going deep in medical image analysis: concepts, methods, challenges, and future directions. IEEE Access. 7:99540–99572. [Google Scholar]
- Anguita D, Ghelardoni L, Ghio A, Oneto L, Ridella S. 2012. The “k” in k-fold cross validation. Paper presented at: ESANN 2012 Proceedings, European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning; April 25–27, 2012; Bruges, BE, p. 441–446. [Google Scholar]
- Berstad TJD, Riegler M, Espeland H, de Lange T, Smedsrud PH, Pogorelov K, Stensland HK, Halvorsen P. 2018. Tradeoffs using binary and multiclass neural network classification for medical multidisease detection. In: 2018 IEEE International Symposium on Multimedia. Washington (DC): IEEE Computer Society. p. 1–8. [Google Scholar]
- Carter JB, Stone JD, Clark RS, Mercer JE. 2016. Applications of cone-beam computed tomography in oral and maxillofacial surgery: an overview of published indications and clinical usage in United States academic centers and oral and maxillofacial surgery practices. J Oral Maxillofac Surg. 74(4):668–679. [DOI] [PubMed] [Google Scholar]
- Casalegno F, Newton T, Daher R, Abdelaziz M, Lodi-Rizzini A, Schurmann F, Krejci I, Markram H. 2019. Caries detection with near-infrared transillumination using deep learning. J Dent Res. 98(11):1227–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang L, Li G, Wu TH, Diachina S, Tejera B, Kwon JJ, Lin FC, Lee YT, Xu T, et al. 2020. Machine learning in orthodontics: introducing a 3D auto-segmentation and auto-landmark finder of CBCT images to assess maxillary constriction in unilateral impacted canine patients. Angle Orthod. 90(1):77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liew JH, Xiong W, Chui CK, Ong SH. 2018. Focus, segment and erase: an efficient network for multi-label brain tumor segmentation. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y, editors. Computer vision—ECCV 2018. Lecture notes in computer science. Vol. 11217. Cham (Switzerland): Springer. [Google Scholar]
- Chen Y, Du H, Yun Z, Yang S, Dai Z, Zhong L, Feng Q, Yang W. 2020. Automatic segmentation of individual tooth in dental CBCT images from tooth surface map by a multi-task FCN. IEEE Access. 8:97296–97309. [Google Scholar]
- Cui Z, Li C, Wang W. 2019. ToothNet: automatic tooth instance segmentation and identification from cone beam CT images. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Washington (DC): IEEE Computer Society. p. 6361–6370. [Google Scholar]
- Egger J, Pfarrkirchner B, Gsaxner C, Lindner L, Schmalstieg D, Wallner J. 2018. Fully convolutional mandible segmentation on a valid ground-truth dataset. Annu Int Conf IEEE Eng Med Biol Soc. 2018:656–660. [DOI] [PubMed] [Google Scholar]
- Gómez O, Mesejo P, Ibáñez O, Valsecchi A, Cordón O. 2020. Deep architectures for high-resolution multi-organ chest X-ray image segmentation. Neural Comput Appl. 32:15949–15963. [Google Scholar]
- Jafari M, Li R, Xing Y, Auer D, Francis S, Garibaldi J, Chen X. 2019. FU-net: multi-class image segmentation using feedback weighted U-net. ICIGP. 2019:529–537. [Google Scholar]
- Kapila SD, Nervina JM. 2015. CBCT in orthodontics: assessment of treatment outcomes and indications for its use. Dentomaxillofac Radiol. 44(1):20140282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MW. 2014. A survey: image segmentation techniques. IJFCC. 3(2):89–93. [Google Scholar]
- Lee S, Woo S, Yu J, Seo J, Lee J, Lee C. 2020. Automated CNN-based tooth segmentation in cone-beam CT for dental implant planning. IEEE Access. 8:50507–50518. [Google Scholar]
- Likitmongkolsakul U, Smithmaitrie P, Samruajbenjakun B, Aksornmuang J. 2018. Development and validation of 3D finite element models for prediction of orthodontic tooth movement. Int J Dent. 2018:4927503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sanchez CI. 2017. A survey on deep learning in medical image analysis. Med Image Anal. 42:60–88. [DOI] [PubMed] [Google Scholar]
- Minnema J, van Eijnatten M, Hendriksen AA, Liberton N, Pelt DM, Batenburg KJ, Forouzanfar T, Wolff J. 2019. Segmentation of dental cone-beam CT scans affected by metal artifacts using a mixed-scale dense convolutional neural network. Med Phys. 46(11):5027–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnema J, van Eijnatten M, Kouw W, Diblen F, Mendrik A, Wolff J. 2018. CT image segmentation of bone for medical additive manufacturing using a convolutional neural network. Comput Biol Med. 103:130–139. [DOI] [PubMed] [Google Scholar]
- Mustafa ID, Hassan MA, Mawia A. 2016. A comparison between different segmentation techniques used in medical imaging. Am J Biomed Eng. 6(2):59–69. [Google Scholar]
- Naylor CD. 2018. On the prospects for a (deep) learning health care system. JAMA. 320(11):1099–1100. [DOI] [PubMed] [Google Scholar]
- Nguyen KCT, Duong DQ, Almeida FT, Major PW, Kaipatur NR, Pham TT, Lou EHM, Noga M, Punithakumar K, Le LH. 2020. Alveolar bone segmentation in intraoral ultrasonographs with machine learning. J Dent Res. 99(9):1054–1061. [DOI] [PubMed] [Google Scholar]
- Novikov AA, Lenis D, Major D, Hladůvka J, Wimmer M, Bühler K. 2018. Fully convolutional architectures for multi-class segmentation in chest radiographs. IEEE Trans Med Imaging. 37(8):1865–1876. [DOI] [PubMed] [Google Scholar]
- Pelt DM, Sethian JA. 2018. A mixed-scale dense convolutional neural network for image analysis. Proc Natl Acad Sci U S A. 115(2):254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu B, Guo J, Kraeima J, Borra R, Witjes M, van Ooijen P. 2018. 3D segmentation of mandible from multisectional CT scans by convolutional neural networks. arXiv. https://arxiv.org/abs/1809.06752
- Saood A, Hatem I. 2020. COVID-19 lung CT image segmentation using deep learning methods: UNET vs. SegNET. Research Square. doi: 10.21203/rs.3.rs-56882/v3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze R, Heil U, Gross D, Bruellmann DD, Dranischnikow E, Schwanecke U, Schoemer E. 2011. Artefacts in CBCT: a review. Dentomaxillofac Radiol. 40(5):265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendicke F, Samek W, Krois J. 2020. Artificial intelligence in dentistry: chances and challenges. J Dent Res. 99(7):769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijnatten M, van Dijk R, Dobbe J, Streekstra G, Koivisto J, Wolff J. 2018. CT image segmentation methods for bone used in medical additive manufacturing. Med Eng Phys. 51:6–16. [DOI] [PubMed] [Google Scholar]
- Wang L, Gao Y, Shi F, Li G, Chen KC, Tang Z, Xia JJ, Shen D. 2016. Automated segmentation of dental CBCT image with prior-guided sequential random forests. Med Phys. 43(1):336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou KH, Warfield SK, Bharatha A, Tempany CM, Kaus MR, Haker SJ, Wells WM, III, Jolesz FA, Kikinis R. 2004. Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports. Acad Radiol. 11(2):178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211005338 for Multiclass CBCT Image Segmentation for Orthodontics with Deep Learning by H. Wang, J. Minnema, K.J. Batenburg, T. Forouzanfar, F.J. Hu and G. Wu in Journal of Dental Research