Abstract
G-quadruplex, a unique DNA quartet motif with a pivotal role in regulation of the gene expression, has been established as a potent therapeutic target for the treatment of cancer. Small-molecule-mediated stabilization of the G-quadruplex and thus inhibition of the expression from the oncogene promoter and telomere region may be a promising anticancer strategy. Aloe vera-derived natural compounds like aloe emodin, aloe emodin-8-glucoside, and aloin have significant anticancer activity. Comparative binding studies of these three molecules with varieties of G-quadruplex sequences were carried out using different biophysical techniques like absorption spectral titration, fluorescence spectral titration, dye displacement, ferrocyanide quenching assay, and CD and DSC thermogram studies. Overall, this study revealed aloe emodin and aloe emodin-8-glucoside as potent quadruplex-binding molecules mostly in the case of c-KIT and c-MYC sequences with a binding affinity value of 105 order that is higher than their duplex DNA binding ability. This observation may be correlated to the anticancer activity of these aloe-active compounds and also be helpful in the potential therapeutic application of natural compound-based molecules.
Introduction
G-quadruplex, a unique secondary nucleic acid structure, is widespread in the human genome including telomere as well as promoter region of numerous oncogenes suggesting their significant biological relevance.1−3 The formation of this tetrad planar structures in guanine-rich sequences is associated to the interaction of four guanine bases through Hoogsteen and Watson-Crick faces of the neighboring guanine residues in the presence of a central monovalent metal ion.4,5 The extensively characterized TTAGGG repeats which are involved in the formation of the G-quadruplex is evident in the telomeric region of the human genome and plays a crucial role in regulation of uncontrolled cell division.6,7 Most interestingly, abundance of the G-quadruplex structure in several oncogene promoter regions such as c-KIT,8,9c-MYC,10,11BCL-2,12,13KRAS,14 and VEGF(15) clearly indicates their exclusive role in modulation of gene regulation. Due to its wide varieties throughout the different regulatory parts of the human genome, the quadruplex structure has been targeted for novel anticancer strategy.3,16−18 Selective targeting by small molecules can potentially result in stabilization of G-quadruplex structures and subsequently silencing or suppression in oncogene transcription.19 From this prediction, ligand-directed selective targeting of the G-quadruplex has been a new direction in the chemotherapeutic aspect. Several investigations have been carried out by small molecules like acridine,20−22 anthraquinone,23,24 telomestatin,25,26 TMPyP4,27 quindoline,28 phenanthridine,29 and napthoquinone30 derivatives to establish their ability to bind with different quadruplex structures.
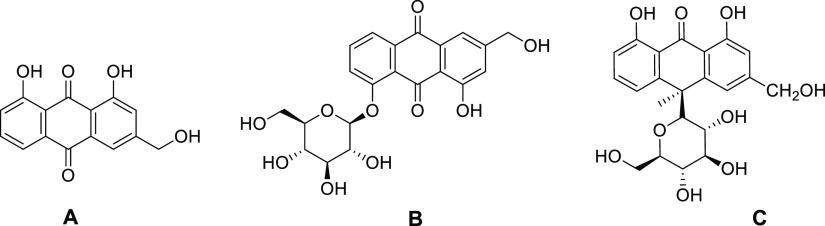
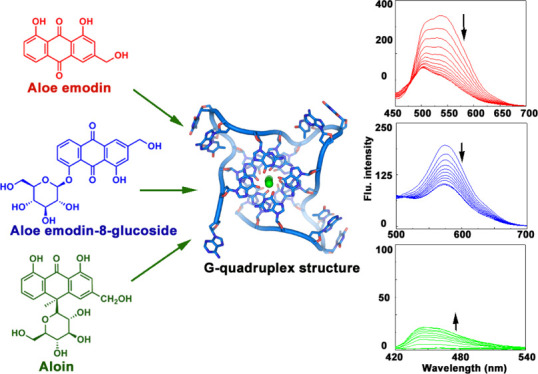
Natural compounds are gaining more importance day by day as an infinite resource of pharmaceutical drug development for diseases like cancer. Traditional plant-extracted molecules with potent anticancer activity have been regarded as important lead compounds for chemotherapeutic drug development.31−34 Anthraquinones are one of the most biologically active natural compounds with a wide variety of structural diversity as well as promising therapeutic activity. Many anthraquinone derivatives like daunomycin, doxorubicin, and mitoxantrone are used as potent chemotherapeutic agents due to their ability of targeting at the molecular DNA level.35−37Aloe vera-extracted anthraquinone compounds like aloe emodin, emodin, and aloin are well established therapeutic natural molecules with significant anticancer activity.38−42 Several reports have suggested these compounds having effective antiproliferative activity with different cancer cell lines.38−42 Anthraquinone glycosides and aloe emodin-8-glucoside are reported as antidiabetic molecules with potential activity in insulin-resistant cell line.43 In this paper, our perspective is targeting some biologically significant aloe active compounds to well establish biomolecular target G-quadruplex sequences. Here, aloe emodin, aloe emodin-8-glucoside, and aloin [Figure 1] have been chosen to study the binding interaction with a variety of quadruplex sequences like c-KIT, c-MYC, HUMTEL, BCL-2, KRAS, and VEGF. Moreover, these compounds are reported to moderately bind with natural DNA.44 Contextually, it is noteworthy to mention that some aloe compounds are reported as the telomeric G-quadruplex structure stabilizer.45,46 Based on the previous reports, our present work is an attempt for understanding of the comparative quadruplex recognition ability of these aloe active compounds. Overall, this rigorous analysis may highlight the aloe active compounds as promising candidatures in rational designing of the natural product-based chemotherapeutic agent.
Figure 1.
Chemical structures of [A] aloe emodin (ALM), [B] aloe emodin-8-glucoside (ALMG), and [C] aloin (ALN).
Results and Discussion
Equilibrium Binding Study from Absorption Titration Data
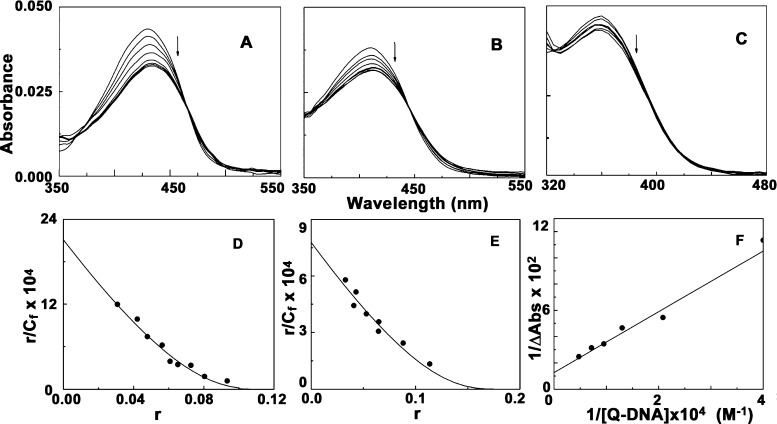
Quadruplex DNA binding behavior of the Aloe vera compounds (Figure 1) was studied by performing UV-spectroscopic titration with six sets of quadruplex DNA sequences of different proto-oncogenes and human telomeric sequences. Figure S1 shows that ALM, ALMG, and ALN when titrated with six different sets of quadruplexes c-KIT, c-MYC, HUMTEL, BCL-2, KRAS, and VEGF, resulted in different extent of hypochromic and bathochromic shifts of characteristic absorption spectra of respective compounds. Table S1 shows the characteristic changes in the absorption profile of ALM, ALMG, and ALN titrated with different quadruplex DNA sequences. ALM and ALMG both reached saturation in all cases with significant hypochromic effects and bathochromic shifts, though ALN showed comparatively less significant changes in absorption spectra and no clear saturations were obtained in all six set of quadruplex oligos. ALM showed the highest binding affinity compared to ALMG and ALN with all quadruplex sequences. ALM showed a characteristic isosbestic point around 460 nm in all cases clearly stating equilibrium condition in complex formation. ALM treating with c-KIT produced hypochromicity up to 24.53% along with a redshift of λmax up to 4 nm (Figure 2A). Among the six sets of quadruplexes, the c-KIT sequence has showed the strongest binding affinity value with ALM indicating c-KIT quadruplex selectivity of this aloe compounds. ALMG also exhibited significant changes upon interaction with these structures. ALN showed very poor quadruplex binding ability in comparison to ALM and ALMG. The spectrophotometric titration data were fitted into the Scatchard plots of r/Cf versus r, where r is the number of moles of ligand per mole of the DNA nucleotide.47 Representative binding isotherms for complexation are depicted in Figure 2D,E from which Ki, the intrinsic binding constant, and n, the number of nucleotide phosphates excluded by ligand molecule, were obtained (Table S1).
Figure 2.
Representative absorption spectral titration of [A] ALM (5 μM) treated with c-KIT (0–135 μM), [B] ALMG (5 μM) treated with c-MYC (0–180 μM), and [C] ALN (5 μM) treated with BCL-2 (0–250 μM). Scatchard plots of [D] ALM-c-KIT complexation and [E] ALMG-c-MYC complexation. The solid lines represent the non-linear least square best fit of the experimental points to the neighbor exclusion model. [F] Benesi–Hildebrand plot for ALN- BCL-2 complexation.
ALM showed the highest binding in all cases. Among the six set of quadruplexes, the highest binding of ALM was observed with the c-KIT quadruplex sequence with binding affinity value (2.11 ± 0.33) × 105 M–1 that indicates significantly preferential binding of ALM with the c-KIT quadruplex structure. The affinity toward DNA varied as c-KIT > c-MYC > BCL-2> HUMTEL > KRAS > VEGF. ALMG showed a comparatively lower but significant quadruplex binding property which was also most prominent in case of the c-KIT sequence. Spectral changes for ALN with all quadruplex DNA neither reached any saturation nor exhibited any isosbestic point and were analyzed by the Benesi–Hildebrand plot (Figure 2C,F).48 All the analyzed data are presented in Table S1. Alteration of the absorption spectra of ALM and ALMG upon interaction with quadruplex DNA indicated a π–π stacking interaction between DNA and ligand molecules. It is noteworthy to mention here that ALM and ALMG can also bind with ctDNA but with lower binding affinity.44 The equilibrium constants of binding to ctDNA are reported to be (6.02 ± 0.10) × 104 M–1 and (4.90 ± 0.11) × 104 M–1 at 298.15 K, for ALM and ALMG, respectively.44 This observation suggests ALM and ALMG as a potent quadruplex binding molecule though ALN is not that much capable of targeting the quadruplex sequences.
Fluorescence Titration Study
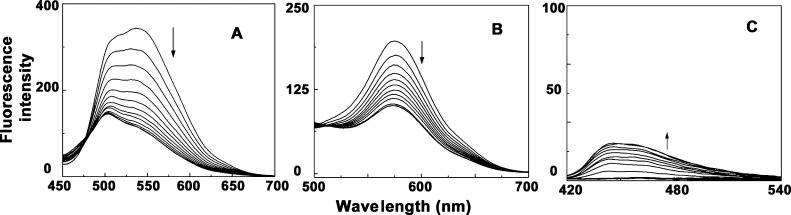
ALM and ALMG being fluorescent molecules, steady-state fluorometric titration was also performed for characterization of their binding with different quadruplex-forming sequences. The fluorescence intensity of ALM (emission maxima at 535 when excited at 430 nm) and ALMG (emission maxima at 576 nm when excited at 410 nm) was significantly quenched upon binding with quadruplex DNA (Figure 3). The details of parameters obtained from the fluorescence titration study are represented in Table S2 and Figure S2. In the case of ALM, fluorescence intensity at 536 nm was significantly quenched to different extents with the interaction with different sets of quadruplex structures suggesting effective interactions between the non-covalently bound Aloe-active molecules with the quadruplex DNA. Most significant change was observed for ALM binding with the c-KIT quadruplex with maximum quenching of 67% at saturation with a large blueshift of 32 nm at λmax (Figure 3). The fluorescence intensity of ALMG also quenched at 576 nm with increasing concentration of quadruplex DNA and finally saturated at higher concentrations (Figure S2). ALN did not show any significant fluorescence property. With the addition of different sets of DNA, changes in the fluorescence intensity were observed in different manners which is represented in Figure S2. Slight increases in fluorescence intensity were observed when ALN formed complexation with c-KIT, c-MYC, HUMTEL, and BCL-2 quadruplexes, whereas KRAS and VEGF did not significantly affect the fluorescence property of the free ALN. The data (Table S2) obtained from the spectrofluorimetric titration study were in full agreement with the absorption titration data. Fluorescence data also revealed that both ALM and ALMG most effectively binds to the c-KIT structure which is supposed to indicate preferential binding selectivity of the aloe active compounds to c-KIT DNA over a large set of quadruplex sequences. Comparing the previous report on the binding study of the aloe-active compound to ctDNA with the current study,44 both ALM and ALMG can be suggested as better quadruplex-binding molecules.
Figure 3.
Representative fluorescence titration of [A] ALM (4 μM) with c-KIT (0–112 μM), [B] ALMG (4 μM) with c-MYC (0–152 μM), and [C] ALN (4 μM) with BCL-2 (0–320 μM).
Fluorescence Quenching Studies
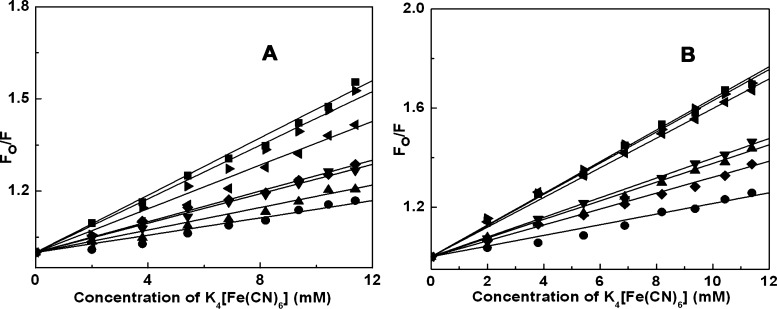
Fluorescence quenching phenomena were studied to understand the accessibility of ALM and ALMG to the quencher ion in the presence of G-quadruplex structures.49Ksv, the Stern–Volmer constant values for free ALM, was 46.64 M–1 which had been changed to 14.07, 18.29, 23.89, 25.03, 35.58, and 43.65 M–1, in quadruplex-bound condition with c-KIT, c-MYC, HUMTEL, BCL-2, KRAS and VEGF, respectively (Figure 4A). In the case of ALMG, ksv for the free drug was found to be 63.93 M–1 and 21.53, 37.75, 39.75, 32.12, 59.82, and 62.97 M–1, for complexation with c-KIT, c-MYC, HUMTEL, BCL-2, KRAS, and VEGF, respectively (Figure 4B). The decrease in the Ksv value after complex formation gives a clear indication that ALM and ALMG in the free state were readily accessible to the quencher, while in the complex-forming condition, they may be between base quartets of quadruplex structures, thus not easily available for the quencher. This observation also supported the binding of ALM and ALMG to the G-quartets structure, where the anionic ion is prevented from entering due to the repulsion from the negatively charged phosphate groups.
Figure 4.
Stern–Volmer plots for the quenching of (A) ALM and (B) ALMG by K4[Fe(CN)6] in free (■) and complexation with c-KIT (●), c-MYC (▲), HUMTEL (▼), BCL-2 (◆), KRAS (◀), and VEGF (▶), respectively.
Thiazole Orange Displacement Assay
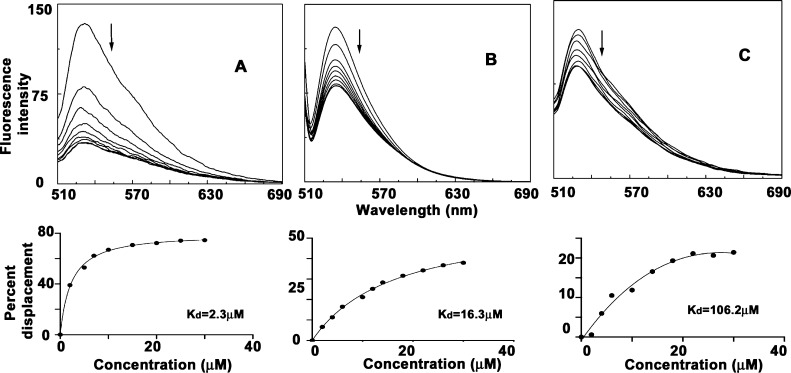
Thiazole orange (TO) is a well-established dye which binds with high affinity with the external quartets of a G-quadruplex by end-stacking.50 Thiazole orange upon forming a complex with the quadruplex DNA structures increases the fluorescence signal by thousand fold and can thus act as useful G-quadruplex binding fluorescent markers. An externally added quadruplex binding molecule can cause competitive displacement of TO from the quadruplex structure and as a result decrease the fluorescent intensity of the Thiazole orange-G4 DNA complex. The extent of quadruplex binding ability of the molecule can be correlated by TO displacement. TO displacement by ALM, ALMG, and ALN are represented in Figure S3. In the case of ALM interaction with c-KIT quadruplex, maximum quenching in fluorescence was observed which indicates significant binding affinity of ALM with the c-KIT quadruplex sequence (Figure 5A). Representative displacement of aloe compounds is depicted in Figure 5A–5C. TO displacement by aloe active compounds are plotted by percentage displacement with increased concentration of aloe compounds, and kD values were calculated for these fits (lower panel of Figure 5) which were in full support with other spectroscopic data (Table S3).
Figure 5.
Representative thiazole orange displacement assay for [A] ALM-c-KIT, [B] ALMG-VEGF, and [C] ALN-HUMTEL complexation. Bottom panel: KD value obtained from the corresponding fit.
Circular Dichroism Spectroscopy Study
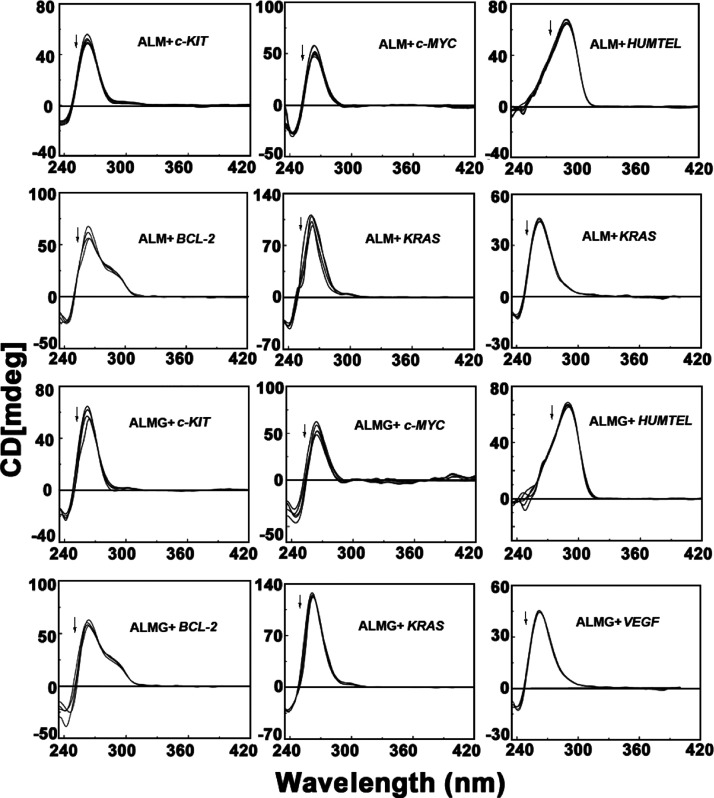
CD is an important technique to understand the structural perturbation induced in the nucleic acid structures by drug or ligand molecules. In the presence of the [K+] ion, c-KIT, c-MYC, BCL-2, VEGF, and KRAS, all oncogene G-quadruplex sequences show characteristic peaks (positive band at 262 nm and a negative band at 245 nm) which resembles a parallel G-quadruplex topology.51,52 In [K+] solution, the G-quadruplex of the telomeric DNA is reported to adopt a mixed parallel/anti-parallel structure ([3 + 1] hybrid structure) with a strong positive peak at 290 nm, a smaller hump at 265 nm and followed by a small negative peak at 240 nm.53−55
ALM and ALMG both resulted in slight decrease in the molar ellipticity value at the characteristic positive peaks of quadruplexes due to base stacking complexation though no significant conformational change in overall parallel topology of quadruplexes was observed (Figure 6). The overall secondary conformation of these quadruplex structures remains unaltered by the aloe-active compounds.
Figure 6.
Circular dichroism spectra for the interaction of quadruplex DNAs (4 μM) in the presence of aloe active compounds.
DSC Study
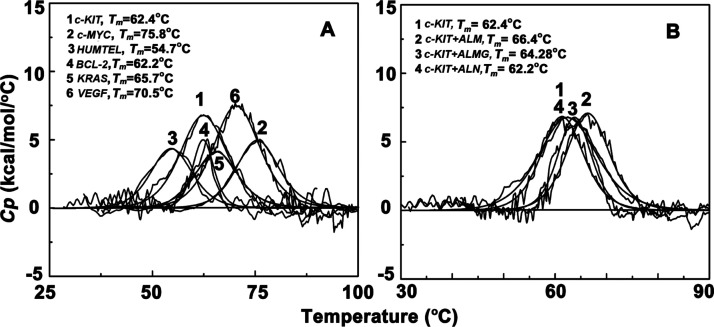
The DSC experiment was used to identify the characteristic thermal melting temperature of each quadruplex DNAs. From the DSC thermogram, the melting temperatures obtained for six set of quadruplex DNA were 62.4 °C for c-KIT, 75.8 °C for c-MYC, 54.7 °C for HUMTEL, 62.2 °C for BCL-2, 65.7 °C for KRAS, and 70.5 °C for VEGF, respectively (Figure 7). ALM after complexation with c-KIT and c-MYC increased the melting temperature by 4 °C, BCL-2 by 2 °C, HUMTEL by 1 °C, and KRAS and VEGF remain unaltered. ALMG increased the c-KIT and c-MYC melting temperature by 2 °C. Aloin did not show any stabilizing effect on the thermal melting of the quadruplex sequences. Increase in melting temperature clearly indicates that ALM and ALMG leads to enhanced structural stability of some quadruplex structures as they can form thermodynamically more stable complexation with quadruplex sequences predominantly with c-KIT.
Figure 7.
Differential scanning calorimetry thermograms of [A] different quadruplex DNAs (10 μM) and [B] c-KIT complex with aloe-active compounds.
Conclusions
In summary, significant binding and stabilization of a series of G-quadruplex structures by ALM and ALMG were confirmed by various biophysical techniques like absorption, fluorescence, circular dichroism, and differential scanning calorimetric studies. All the studies were in full agreement that ALM and ALMG both exhibit preferential binding selectivity toward to quadruplex DNA structures with the binding affinity value significantly more than normal duplex form of DNA, and this property of aloe compounds may be correlated to their established anticancer properties. Among three aloe compounds ALM was the most effective G-quadruplex binding ligand and both the drugs ALM and ALMG showed highest binding affinity toward the c-KIT quadruplex sequence. ALN also have weak binding affinity to these quadruplex-forming sequences. As the binding of ALM and ALMG progressed, significant changes in absorption and fluorescence spectra with moderate decrease in the molar ellipticity value indicate most likely the base stacking interactions of these two molecules. ALM, ALMG, and ALN, all these three molecules are neutral in charge. Though cationic charges in the DNA binding ligand facilitate their interaction by the electrostatic contact between the molecule and negatively charged phosphate group of DNA, it is not always the sole determining factor in the binding phenomena. The presence of the planar anthraquinone moiety in ALM and ALMG lead them to hydrophobic interaction with quadruplex structures by accumulation of base stacking forces due to aromatic π–π interaction. Overall, this study may be helpful for natural compound-based therapeutics approaches of targeting the G-quadruplex-dependent biological pathway.
Materials and Methods
Materials
Aloe vera-extracted compounds, aloe emodin (hereafter ALM) and aloin (hereafter ALN), were purchased from Cayman Chemical, USA, and aloe emodin-8-glucoside (hereafter ALMG) was purchased from Santa Cruz Biotechnology, USA. All compounds had purity >98%. All three aloe active compounds are DMSO-soluble. So, stock solutions were prepared by dissolving the compounds in 100% DMSO and were kept protected from light.44 All six sets of quadruplex-forming oligos c-KIT, c-MYC, telomeric HUMTEL, BCL-2, VEGF, KRAS, were obtained from Sigma-Aldrich and the sequences are as follows:
c-KIT d 5′[GGGAGGGCGCTGGGAGGGAGGG]3′
c-MYC d 5′[TGAGGGTGGGTAGGGTGGGTAA]3′
telomeric HUMTEL d 5′[AGGGTTAGGGTTAGGGTTAGGG]3′
BCL-2 d 5′[GGGCGCGGGAGGAATTGGGCGGG]3′
VEGF d 5′[GGG GCGGGCCGGGGGCGGGG]3′
KRAS d 5′[AGGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG]3′
Oligos were dissolved in phosphate buffer of pH 7.0 containing a potassium ion concentration of 100 mM. Quadruplex structures of these oligos were prepared by heating the solution at 95 °C for 10 min followed by very slow cooling to 5 °C and then equilibrating for 48 h at 5 °C. The quadruplex formation was confirmed by optical melting studies at 295 nm and also from the DSC thermogram study which revealed characteristic thermal melting at 62.4 °C for c-KIT, 75.8 °C for c-MYC, 54.7 °C for HUMTEL, 62.2 °C for BCL-2, 65.7 °C for KRAS, and 70.5 °C for VEGF, respectively. All the experiments throughout the study were performed in phosphate buffer of pH 7.0 containing a potassium ion concentration of 100 mM with additional 1% DMSO.
Evaluation of the Binding Affinity by Absorption Titration Study
Absorption titration of aloe compounds with variety of quadruplexes was performed in the Jasco V660 spectrophotometer (Jasco International Co Ltd, Hachioji, Japan). The changes in absorbance of free molecules were monitored upon gradual addition of quadruplex solutions, at respective characteristic absorption peaks ∼430 nm for ALM, 410 nm for ALMG, and ∼350 nm for ALN. Titration was carried out till saturation, at 25 ± 1.0 °C under conditions of constant stirring. The obtained spectral data of ALM and ALMG were fit into Scatchard plots of r/Cf versus r and were analyzed by McGhee–von Hippel equation, r/Cf = Ki(1 – nr)[(1 – nr)/{1 – (n – 1)r}](n – 1), where r denotes the moles of the bound ligand per mole of DNA, Cf is the concentration of the free ligand in solution, Ki is the intrinsic binding constant, and n is the number of DNA base pairs occupied by single ligand molecule.47 Spectral data of ALN were analyzed by the Benesi–Hildebrand equation.48
Binding Studies by Fluorescence Titration
ALM and ALMG both have strong fluorescence signal with emission maxima at 535 nm and 576 nm when at 430 and 410 nm, respectively, though ALN does not show any fluorescence property. Titration of free aloe molecules with increasing concentration of quadruplexes was carried out in a quartz cuvette (fluorescence free) of 1 cm path length by monitoring the fluorescence intensity at respective emission maxima on a Hitachi-F4010 fluorimeter (Hitachi, Tokyo, Japan). The data obtained were analyzed by the same method as mentioned in the absorption titration study.
Fluorescence Quenching Studies
Anionic quenchers ferrocyanide [Fe(CN)6]4– was used in increasing concentration to the solution of aloe-active molecules and its complex with quadruplex DNA, and changes in the fluorescence intensity were monitored. The data were plotted as Stern–Volmer equation, Fo/F = 1 + Ksv[Q], where [Q] is the quencher concentration, Fo and F are the fluorescence intensities in the absence and in the presence of the quencher. The Ksv value obtained from the slope of the equation can be correlated to the accessibility of the DNA-bound drug molecules by the quencher.49
Thiazole Orange Displacement Assay
As Thiazole orange (TO) is a well-known quadruplex binding dye, its displacement from the complex by some other molecules indicates the quadruplex binding ability of the molecules. Displacement assay was performed by monitoring fluorescence emission by exciting at 501 nm with gradual addition of aloe-active compounds on the prefolded G-quadruplex structure (0.25 μM) and TO (0.50 μM) complex. The decrease in fluorescence intensity of the TO-quadruplex complex by aloe-active compounds was plotted as percentage displacement, PD = 100 – ([FA/FAo] × 100), (FAo and FA are fluorescence intensities at 530 nm before and after the drug addition), against the concentration of the added aloe compound, and the KD value was calculated by curve fitting.50
Circular Dichroism Study
The Jasco J815 spectropolarimeter attached with a temperature control system (model PTC348WI) was used for the CD study. CD scans of quadruplex solution and their complex with aloe molecules were recorded with a scan speed of 50 nm/min at a bandwidth of 1 nm using a 1 cm path length rectangular cuvette. Each spectrum was represented after baseline correction, averaged from five successive accumulations and smoothing within permissible limits by the software.
Differential Scanning Calorimetry
Stabilization of the nucleic acid structure by ligand binding can be evaluated by the DSC thermogram study. Characteristic thermal denaturation of the quadruplex structure may be altered by the influence of aloe-active compound binding which was monitored by the Micro Cal, Inc, differential scanning calorimeter (DSC). Initially both the sample and the reference cells of the system were equilibrated with the degassed buffer solution at 30 °C for 15 min and then scanned from 35 to 100 °C with a rate of 60 °C/h several times until a stable overlapping baseline was achieved. As the stable overlapping baseline was obtained, the buffer solution was removed from the sample cell only within the cooling cycle and then was filled with different quadruplex solutions and their complex with aloe compounds. Each sample was scanned from 35 to 100 °C to get the DSC melting thermogram of quadruplex structures. The plot of excess heat capacity versus temperature obtained from DSC was analyzed by Origin 7.0 software to determine the transition temperature (Tm) as the sharp peak.
Acknowledgments
The authors acknowledge the support for this work by grants from Department of Science and Technology [DST, Women Scientists Scheme (WOS A) (file no. SR/WOS-A/CS-34/2017(G)] and also the Director, CSIR-IICB for laboratory facilities. S.D. acknowledges DST SERB grant no.: EMR/2017/000659.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c02207.
Absorption and fluorescence titration and thiozole orange displacement assay of aloe-active compounds with quadruplex DNAs and absorption titration, fluorescence spectral, and thiazole orange displacement data for the interaction of aloe-active compounds with G-quadruplex DNAs (PDF)
Author Contributions
A.D. has designed and carried out the research experiments. A.D. and S.D. have contributed to writing of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Todd A. K.; Johnston M.; Neidle S. Highly Prevalent Putative Quadruplex Sequence Motifs in Human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. Prevalence of Quadruplexes in The Human Genome. Nucleic Acids Res. 2005, 33, 2908–2916. 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.; Neidle S. G-Quadruplex Nucleic Acids as Therapeutic Targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell N. H.; Neidle S. G-Quadruplexes and Metal Ions. Met. Ions Life Sci. 2012, 10, 119–134. 10.1007/978-94-007-2172-2_4. [DOI] [PubMed] [Google Scholar]
- Sen D.; Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 1990, 344, 410–414. 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- Parkinson G. N.; Lee M. P. H.; Neidle S. Crystal Structure of Parallel Quadruplexes from Human Telomeric DNA. Nature 2002, 417, 876–880. 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Beginning to Understand the End of the Chromosome. Cell 2004, 116, 273–279. 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- Todd A. K.; Haider S. M.; Parkinson G. N.; Neidle S. Sequence Occurrence and Structural Uniqueness of a G-Quadruplex in the Human C-kit Promoter. Nucleic Acids Res. 2007, 35, 5799–5808. 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T.; Kuryavyi V.; Burge S.; Neidle S.; Patel D. J. Structure of an Unprecedented G-Quadruplex Scaffold in the Humanc-kitPromoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A.; Grand C. L.; Bearss D. J.; Hurley L. H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 11593–11598. 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.; Hurley L. H. Structure of the Biologically Relevant G-Quadruplex in the C-MYC Promoter. Nucleos Nucleot. Nucleic Acids 2006, 25, 951–968. 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- Dai J.; Chen D.; Jones R. A.; Hurley L. H.; Yang D. NMR Solution Structure of the Major G-Quadruplex Structure Formed in the Human BCL2 Promoter Region. Nucleic Acids Res. 2006, 34, 5133–5144. 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer T. S.; Sun D.; Hurley L. H. Deconvoluting the Structural and Drug-Recognition Complexity of the G-Quadruplex-Forming Region Upstream of thebcl-2P1 Promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi S.; Xodo L. E. G-Quadruplex Formation within the Promoter of the KRAS Proto-Oncogene and its Effect on Transcription. Nucleic Acids Res. 2006, 34, 2536–2549. 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Guo K.; Rusche J. J.; Hurley L. H. Facilitation of a Structural Transition in the Polypurine/Polypyrimidine Tract within the Proximal Promoter Region of the Human VEGF Gene by the Presence of Potassium and G-Quadruplex-Interactive Agents. Nucleic Acids Res. 2005, 33, 6070–6080. 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G. Panyutin I.; I. Onyshchenko M.; A. Englund E.; H. Appella D.; D. Neumann R. Targeting DNA G-Quadruplex Structures with Peptide Nucleic Acids. Curr. Pharm. Des. 2012, 18, 1984–1991. 10.2174/138161212799958440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsel-Hertsch R.; Di Antonio M.; Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- Rigo R.; Palumbo M.; Sissi C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta 2017, 1861, 1399–1413. 10.1016/j.bbagen.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Mazzini S.; Gargallo R.; Musso L.; De Santis F.; Aviñó A.; Scaglioni L.; Eritja R.; Di Nicola M.; Zunino F.; Amatulli A.; Dallavalle S. Stabilization of c-KIT G-Quadruplex DNA Structures by the RNA Polymerase I Inhibitors BMH-21 and BA-41. Int. J. Mol. Sci. 2019, 20, 4927. 10.3390/ijms20194927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C.; Gunaratnam M.; Stuart J.; Makwana V.; Greciano O.; Reszka A. P.; Kelland L. R.; Neidle S. Structure-Based Design of Benzylamino-Acridine Compounds as G-Quadruplex DNA Telomere Targeting Agents. Bioorg. Med. Chem. Lett. 2007, 17, 2293–2298. 10.1016/j.bmcl.2007.01.056. [DOI] [PubMed] [Google Scholar]
- Ferreira R.; Artali R.; Benoit A.; Gargallo R.; Eritja R.; Ferguson D. M.; Sham Y. Y.; Mazzini S. Structure and Stability of Human Telomeric G-Quadruplex with Preclinical 9-Amino Acridines. PLoS One 2013, 8, e57701 10.1371/journal.pone.0057701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero E.; Richter S. N. G-Quadruplexes and G-Quadruplex Ligands: Targets and Tools in Antiviral Therapy. Nucleic Acids Res. 2018, 46, 3270–3283. 10.1093/nar/gky187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Thompson B.; Cathers B. E.; Salazar M.; Kerwin S. M.; Trent J. O.; Jenkins T. C.; Neidle S.; Hurley L. H. Inhibition of Human Telomerase by a G-Quadruplex-Interactive Compound. J. Med. Chem. 1997, 40, 2113–2116. 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- Scaglioni L.; Mondelli R.; Artali R.; Sirtori F. R.; Mazzini S. Nemorubicin and doxorubicin bind the G-quadruplex sequences of the human telomeres and of the c-MYC promoter element Pu22. Biochim. Biophys. Acta 2016, 1860, 1129–1138. 10.1016/j.bbagen.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Tauchi T.; Shin-ya K.; Sashida G.; Sumi M.; Okabe S.; Ohyashiki J. H.; Ohyashiki K. Telomerase Inhibition with a Novel G-Quadruplex-Interactive Agent, Telomestatin: in Vitro and in Vivo Studies in Acute Leukemia. Oncogene 2006, 25, 5719–5725. 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- Tahara H.; Shin-Ya K.; Seimiya H.; Yamada H.; Tsuruo T.; Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene 2006, 25, 1955–1966. 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- Arora A.; Maiti S. Effect of Loop Orientation on Quadruplex–TMPyP4 Interaction. J. Phys. Chem. B 2008, 112, 8151–8159. 10.1021/jp711608y. [DOI] [PubMed] [Google Scholar]
- Funke A.; Weisz K. Comprehensive Thermodynamic Profiling for the Binding of a G-Quadruplex Selective Indoloquinoline. J. Phys. Chem. B 2017, 121, 5735–5743. 10.1021/acs.jpcb.7b02686. [DOI] [PubMed] [Google Scholar]
- Jarosova P.; Paroulek P.; Rajecky M.; Rajecka V.; Taborska E.; Eritja R.; Aviñó A.; Mazzini S.; Gargallo R.; Taborsky P. Naturally occurring quaternary benzo[c]phenanthridine alkaloids selectively stabilize G-quadruplexes. Phys. Chem. Chem. Phys. 2018, 20, 21772–21782. 10.1039/c8cp02681e. [DOI] [PubMed] [Google Scholar]
- Riva B.; Ferreira R.; Musso L.; Artali R.; Scaglioni L.; Mazzini S. Molecular recognition in naphthoquinone derivatives - G-quadruplex complexes by NMR. Biochim. Biophys. Acta 2015, 1850, 673–680. 10.1016/j.bbagen.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Williams D. H.; Stone M. J.; Hauck P. R.; Rahman S. K. Why Are Secondary Metabolites (Natural Products) Biosynthesized?. J. Nat. Prod. 1989, 52, 1189–1208. 10.1021/np50066a001. [DOI] [PubMed] [Google Scholar]
- Cragg G. M.; Kingston D. G. I.; Newman D. J.. Anticancer Agents from Natural Products; CRC Press, Taylor & Francis: Boca Raton, FL, USA, 2005. [Google Scholar]
- Newman D. J.; Shapiro S. Microbial Prescreens for Anticancer Activity. SIM News 2008, 58, 132–150. [Google Scholar]
- Kinghorn A. D.; Chin Y.-W.; Swanson S. M. Discovery of Natural Product Anticancer Agents from Biodiverse Organisms. Curr. Opin. Drug Discov. Dev 2009, 12, 189–196. [PMC free article] [PubMed] [Google Scholar]
- Johnson R. K.; Zee-Cheng R. K. Y.; Lee W. W.; Acton E. M.; Henry D. W.; Cheng C. C. The Experimental Antitumor Activity of Aminoanthraquinones. Canc. Treat Rep. 1979, 63, 425–439. [PubMed] [Google Scholar]
- Chaires J. B. Biophysical chemistry of the daunomycin-DNA interaction. Biophys. Chem. 1990, 35, 191–202. 10.1016/0301-4622(90)80008-u. [DOI] [PubMed] [Google Scholar]
- Minotti G.; Menna P.; Salvatorelli E.; Cairo G.; Gianni L. Anthracyclines: Molecular Advances and Pharmacologic Developments in Antitumor Activity and Cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- Lee H.-Z. Protein kinase C involvement in aloe-emodin- and emodin-induced apoptosis in lung carcinoma cell. Br. J. Pharmacol. 2001, 134, 1093–1103. 10.1038/sj.bjp.0704342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q.; Pan H.; Lou H.; Xu Y.; Tian L. Inhibition of The Angiogenesis and Growth of Aloin in Human Colorectal Cancer in Vitro and in Vivo. Canc. Cell Int. 2013, 13, 69. 10.1186/1475-2867-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-H.; Huang C.-Y.; Chen M.-C.; Lee Y.-T.; Yue C.-H.; Wang H.-Y.; Lin H. Emodin and Aloe Emodin Supress Breast Cancer Cell Proliferation Through Erα Inhibition. Evidence-Based Complementary Altern. Med. 2013, 2013, 376123. 10.1155/2013/376123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özenver N.; Saeed M.; Demirezer L. Ö.; Efferth T. Aloe-emodin as drug candidate for cancer therapy. Oncotarget 2018, 9, 17770–17796. 10.18632/oncotarget.24880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niciforovic A.; Adzic M.; Spasic S. D.; Radojcic M. B. Antitumor Effects of a Natural Anthracycline Analog (Aloin) Involve Altered Activity of Antioxidant Enzymes in HeLaS3 cells. Cancer Biol. Ther. 2007, 6, 1211–1216. 10.4161/cbt.6.8.4383. [DOI] [PubMed] [Google Scholar]
- Anand S.; Muthusamy V. S.; Sujatha S.; Sangeetha K. N.; Bharathi Raja R.; Sudhagar S.; Poornima Devi N.; Lakshmi B. S. Aloe Emodin Glycosides Stimulates Glucose Transport and Glycogen Storage Through PI3K Dependent Mechanism in L6 Myotubes and Inhibits Adipocyte Differentiation in 3T3L1 Adipocytes. FEBS Lett. 2010, 584, 3170–3178. 10.1016/j.febslet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Das A.; Kumar G. S.; Dutta S. Interaction of Aloe Active Compounds with Calf Thymus DNA. J. Mol. Recognit. 2019, 32, e2789 10.1002/jmr.2786. [DOI] [PubMed] [Google Scholar]
- Jin Y.; Chen G.; Wang Y. Gold nanorod-based FRET assay for selection of G-quadruplex-binding ligands. Gold Bull. 2011, 44, 163–169. 10.1007/s13404-011-0022-9. [DOI] [Google Scholar]
- Wang S.; Yan W.-W.; He M.; Wei D.; Long Z.-J.; Tao Y.-M. Aloe emodin inhibits telomerase activity in breast cancer cells: transcriptional and enzymological mechanism. Pharmacol. Rep. 2020, 72, 1383–1396. 10.1007/s43440-020-00062-w. [DOI] [PubMed] [Google Scholar]
- McGhee J. D.; Von Hippel P. H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Benesi H. A.; Hildebrand J. H. A Spectrophotometric Investigation of the Interaction of Iodine with Aromatic Hydrocarbons. J. Am. Chem. Soc. 1949, 71, 2703–2707. 10.1021/ja01176a030. [DOI] [Google Scholar]
- Das A.; Suresh Kumar G. Probing the binding of two sugar bearing anticancer agents aristololactam-β-d-glucoside and daunomycin to double stranded RNA polynucleotides: a combined spectroscopic and calorimetric study. Mol. BioSyst. 2012, 8, 1958–1969. 10.1039/c2mb25080b. [DOI] [PubMed] [Google Scholar]
- Monchaud D.; Allain C.; Teulade-Fichou M.-P. Thiazole Orange: A Useful Probe for Fluorescence Sensing of G-Quadruplex-Ligand Interactions. Nucleos Nucleot. Nucleic Acids 2007, 26, 1585–1588. 10.1080/15257770701548212. [DOI] [PubMed] [Google Scholar]
- Fernando H.; Reszka A. P.; Huppert J.; Ladame S.; Rankin S.; Venkitaraman A. R.; Neidle S.; Balasubramanian S. A Conserved Quadruplex Motif Located in a Transcription Activation Site of the Human C-kit Oncogene. Biochemistry 2006, 45, 7854–7860. 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y.; Hurley L. H. Structures, Folding Patterns, and Functions of Intramolecular DNA G-Quadruplexes Found in Eukaryotic Promoter Regions. Biochimie 2008, 90, 1149–1171. 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrus A.; Chen D.; Dai J.; Bialis T.; Jones R. A.; Yang D. Human Telomeric Sequence Forms a Hybrid-Type Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel Strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renciuk D.; Kejnovská I.; Skoláková P.; Bednárová K.; Motlová J.; Vorlícková M. Arrangements of Human Telomere DNA Quadruplex in Physiologically Relevant K+ Solutions. Nucleic Acids Res. 2009, 37, 6625–6634. 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Wang X.; Fu M.; Ren J.; Qu X. Chiral Metallo-Supramolecular Complexes Selectively Recognize Human Telomeric G-Quadruplex DNA. Nucleic Acids Res. 2008, 36, 5695–5703. 10.1093/nar/gkn569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.