Abstract
In every cell cycle, billions of nucleotides need to be duplicated within hours, with extraordinary precision and accuracy. The molecular mechanism by which cells regulate the replication event is very complicated, and the entire process begins way before the onset of S phase. During the G1 phase of the cell cycle, cells prepare by assembling essential replication factors to establish the pre-replicative complex at origins, sites that dictate where replication would initiate during S phase. During S phase, the replication process is tightly coupled with the DNA repair system to ensure the fidelity of replication. Defects in replication and any error must be recognized by DNA damage response and checkpoint signaling pathways in order to halt the cell cycle before cells are allowed to divide. The coordination of these processes throughout the cell cycle is therefore critical to achieve genomic integrity and prevent diseases. In this review, we focus on the current understanding of how the replication initiation events are regulated to achieve genome stability.
1. Pre-replication complex and Origin licensing
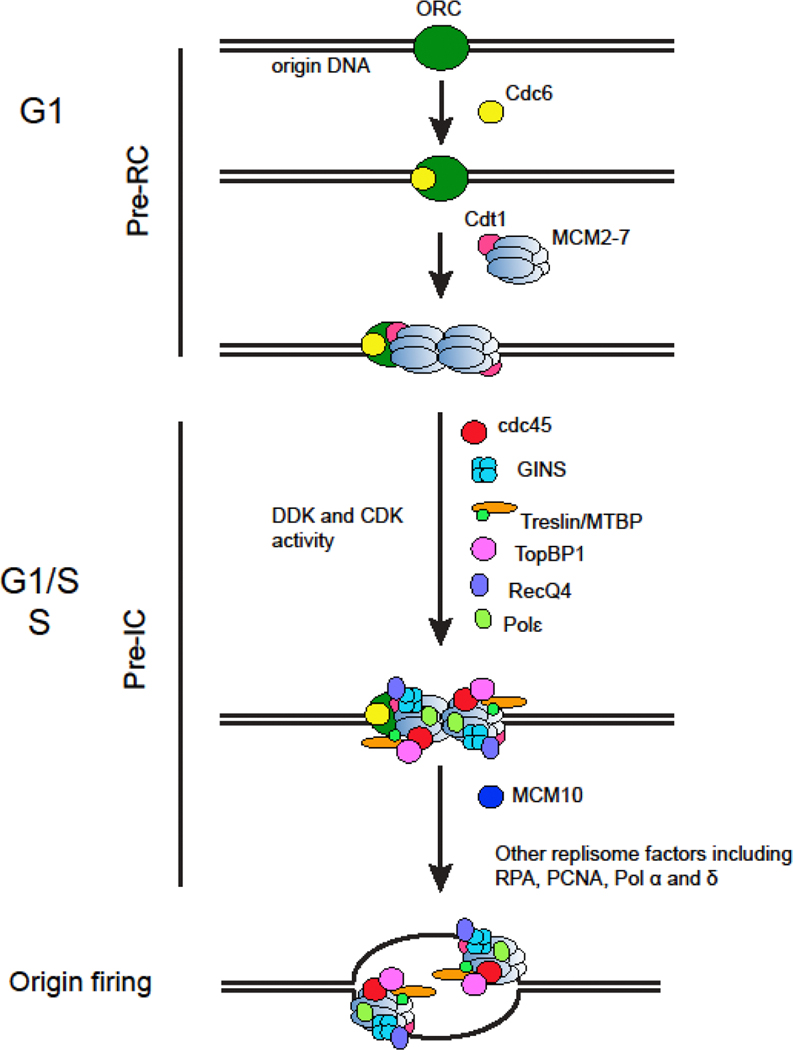
Accurate DNA replication is critical for achieving genome integrity and cell survival. The assembly of the pre-replication complex (pre-RC), which is the very first step required for DNA replication, is a stepwise event that starts with the binding of ORC (Origin Recognition Complex) to the origins on the DNA during G1 phase [1, 2]. ORC then recruits Cdc6, Cdt1, and finally the MCM (minichromosome maintenance) 2–7 complex, component of the DNA replicative helicase, to the origin. An origin is considered “licensed” once the MCM2-7 double hexamer is loaded, which can later fire to initiate replication during S phase (Figure 1) [3–5].
Fig. 1.
Overview of origin licensing and origin firing.
Origin Recognition Complex (ORC) is a heterohexameric protein complex that recognizes DNA replication origins and serves as the initiator of DNA replication [6]. Six subunits of ORC were named Orc1 through Orc6 in descending order of their molecular mass in S. cerevisiae, where they were first identified. Since their discovery in S. cerevisiae, the orthologs of all six subunits have been identified in many other eukaryotic organisms, and ORC’s function as the replication initiating factor is highly conserved across species [7–15]. Orc1-5 belong to AAA+ (ATPases Associated with diverse cellular Activities) family proteins with conserved AAA+ fold, including Walker A and Walker B motifs, suggesting their ATP binding ability and ring-shaped protein complex formation [16–18]. Orc1-5 also contain winged-helix (WH) domain at their C-termini. Orc6, albeit a member of ORC, does not contain such structural features. Structural and sequence analysis in Drosophila and human revealed that N-terminus of Orc6 contains two cyclin-box folds that are homologous to the DNA binding domain of transcription factor TFIIB [19–21]. The C-terminus contains a helix region that is required for Orc6 binding to ORC complex [22, 23].
The role of ORC proteins in establishing preRC at the origins is critical for the maintenance of genome integrity, as the distribution and density of origins must be sufficient to support the replication of the entire genome so that no region is left un-replicated. However, the origin selection program, especially in higher eukaryotes, is very complicated and remains an active field of research [24]. Early studies in S. cerevisiae for replication initiation sites revealed a conserved DNA sequence called autonomously replicating sequence (ARS) [25]. The ARS comprises one A element and three B elements (B1, B2, B3), which are essential for origin function [26]. The cryo-EM structure of ORC binding to ARS explained sequence-specific DNA recognition of ORC [27]. It was further revealed that this specificity lies within a 19-amino acids helix of Orc4 [28]. However, S. cerevisiae appears to be the only eukaryote with a well-defined DNA origin consensus sequence. In S. pombe, origins are AT-rich clustered stretches [29–31]. Origins in higher eukaryotes do not show any features at the level of a defined DNA sequence. Metazoan ORC binds to DNA promiscuously and replication can initiate from random sequences [32–34], suggesting that origins may be defined by 3-dimensional structures of DNA, chromatin environments, or interactions with other proteins. Before the publication of the first genome-scale study in 2008, only about 20 origins had been identified and categorized in the entire human genome [35, 36]. Certain determinants in metazoan origin selection have been revealed by genome-wide studies in the past decade [37]. Nucleosome positioning is an important factor, as metazoan origins, like in yeast, are maintained as nucleosome-free regions [38–41]. In human cells, it was demonstrated that origins correlate with DNase I-hypersensitive regions [42]. Similarly, genome-wide mapping of origins also discovered that origins are located near transcription start sites (TSSs), active promoters, CpG islands, and G-quadruplexes in metazoan [35, 43–49]. However, none of these factors is strictly required for origin selection. A recent study also defined “core origins” in human and mouse genomes and showed that they shared a G-rich sequence signature [50]. The licensing of origins in different chromatin regions, especially heterochromatin, has been an important area of investigation, as cells must be able to license origins across the entire genome regardless of the chromatin status. It is known that some of the ORC subunits localize to specific heterochromatin regions such as centromere and telomere [51, 52]. In terms of origin licensing, other proteins may also be required in facilitating the licensing in certain heterochromatin regions. TRF2 (telomeric repeat-binding factor 2) is known for protecting telomere as one of the shelterin complex subunits. TRF2 also recruits ORC to telomeric regions for facilitating licensing through the interaction with Orc2 [53]. Another well-known example is ORCA (ORC-associated, also known as LRWD1), which can recruit ORC to chromatin [54]. ORCA facilitates the licensing in certain heterochromatin regions [55]. Moreover, ORCA is also critical for heterochromatin organization as well as homologous recombination at telomeric regions [56, 57]. Thus, ORC and its associated proteins are equally important for the maintenance of genome integrity.
In recent years, a debate about whether ORC is an essential factor for origin licensing has been brought up by a study showing that human cells remain viable with no dramatic defect in replication after the depletion of Orc1 or Orc2 by CRISPR-mediated knockout [58]. This study challenged the long-standing view that ORC is the essential initiator of DNA replication. It is possible that partial ORC rings are still functional and able to load MCMs for proper replication; it is also possible that there exists an ORC-independent licensing system, but there is no evidence supporting this idea [59]. A follow-up study depleting another ORC subunit Orc5 or co-depleting Orc2/Orc5 showed that ORC is dispensable for replication [60]. However, other studies continue to suggest the essentiality of ORC using genome-wide or ORC-specific CRISPR screens [61–63]. It is also important to note that these different studies used different approaches and experimental systems to deplete and examine ORC’s role. Future studies are necessary to address this critical question.
After ORC binds to the origin, Cdc6 is the next factor to associate with ORC. Cdc6 is part of the pre-RC, that functions downstream of ORC and is important for the loading of MCM onto the chromatin [64–68]. Like Orc1-5, Cdc6 belongs to the AAA+ family and has ATPase activity [16, 69]. Evidence from yeast to human, all point that Cdc6’s ATPase activity is critical for the regulation of its pre-RC function [70–73]. It has the most extensive sequence similarity with Orc1 and they are suggested to be paralogs [18, 74]. Cdc6 directly interacts with ORC in vivo and in vitro [75, 76]. Cdc6 associates with chromatin-bound ORC in an ATP-dependent manner [71, 77, 78]. After ORC binds to the DNA, Cdc6 is recruited and docks into the Orc1/Orc2 gap, forming the ORC-Cdc6 complex and closing the ring to encircle DNA [79–81]. Therefore, Cdc6 provides the sixth AAA+ fold protein to the pentameric Orc1-5, establishing the classic hexameric AAA+ complex toroid configuration.
After the formation of the ORC-Cdc6 complex on origin DNA, MCM2-7 (Minichromosome maintenance 2–7) hexamer can then be loaded with the help of Cdt1 (Cdc10-dependent transcript 1), which acts as a molecular chaperone for MCM [82]. Cdt1 was recognized as an initiator of replication due to its ability to induce re-replication [83]. Cdt1 is conserved in eukaryotes from yeast to human and is required for MCM2-7 loading in all eukaryotes tested [84–87]. Cdt1 forms a stable complex with MCM2-7, primarily by interacting with MCM6 subunit [88–91], and helps maintain the integrity of MCM2-7 [91–95]. Moreover, structural studies in S. cerevisiae suggested that the binding of Cdt1 to MCM2-7 prevents MCM2/5 gate closure [96–98] which in turn allows the DNA to pass and enter the central channel. In S. cerevisiae, Cdt1 has also been reported to be critical for MCM loading to ORC-Cdc6 through its interaction with Orc6 [99–102]. However, it is unknown if this interaction occurs in metazoans. Other reports suggest that in S. cerevisiae MCM2-7 itself directly engages with ORC-Cdc6 [95, 103], but Cdt1 helps to overcome an MCM6-dependent autoinhibitory mechanism which would otherwise sterically hinder the MCM docking to ORC-Cdc6 [103]. Together, these studies demonstrated the critical role of Cdt1 in origin licensing.
MCM2-7 hexamer loading marks the final step of the pre-RC assembly on the chromatin [104]. MCM2-7 loading requires the coordinated function of ORC, Cdc6, and Cdt1. MCM proteins were first identified in genetic screens designed to uncover genes that are required for replication in S. cerevisiae [105]. The orthologs were subsequently identified in other eukaryotes and shown to have an essential function in replication [106–108]. Biochemical characterization soon revealed that MCM2-7 assemble into a hexameric complex in vivo and in vitro [109–111]. Similar to Orc1-5 and Cdc6, MCM2-7 belongs to AAA+ family [112]. MCM hexamer is formed in the order of MCM5-MCM3-MCM7-MCM4-MCM6-MCM2, with an open gate between MCM2 and 5 to allow the passage of DNA [113, 114]. As discussed above, Cdt1 helps to stabilize MCM integrity and prevent gate closure. The interaction of Cdc6 to the C-terminus of MCM3 has also been reported to be critical for MCM loading [95]. Following the loading of MCM, both Cdc6 and Cdt1 are ejected in an ATP hydrolysis-dependent manner. An ORC-Cdc6-Cdt1-MCM (OCCM) intermediate can be stabilized by using a nonhydrolyzable ATP analog [81], in a state prior to the loading of the next MCM2-7 hexamer.
The defining step of origin licensing is the formation of stable head-to-head MCM2-7 double hexamer around DNA [93, 115]. The formation of double hexamer has been extensively examined using S. cerevisiae proteins [116]. However, there were conflicting results observed using different methods regarding how the loading of the second MCM is achieved. Biochemical studies suggested that two hexamers are loaded independently on two inverted ORC binding sites near each other [95, 117]. In contrast, single molecular experiments showed that two single hexamers are loaded in a sequential way [78], and the loading of the second MCM requires a distinct Cdc6 molecule [96]. The latest cryo-EM experiments identified a key intermediate after OCCM, called MCM-ORC (MO), demonstrating that double hexamer loading is indeed coupled and sequential [118]. In OCCM, the MCM C-terminal domains are in contact with ORC. In the newly observed MO structure, however, ORC binds in an inverted configuration with N-terminal sides of MCM through Orc6 N-terminus. Therefore, it was concluded that the first MCM hexamer loading, coupled with the release of the first set ORC-Cdc6 and Cdt1, creates a distinct binding site and allows the second ORC binding in an inverted orientation. This configuration then recruits the second Cdc6 and allows for the loading of the second Cdt1-MCM2-7 using the same mechanism.
2. Pre-initiation complex and origin firing
Origins are licensed in G1 phase and that makes them ready to fire in S phase. At the onset of S phase, a wave of protein modifications and sequential recruitment regulate the conversion of inactive MCM2-7 complex to active CMG (Cdc45, MCM2-7, GINS) helicase. The CMG helicase, together with polymerases, PCNA, RPA (Replication Protein A), and other essential proteins, initiates DNA unwinding and replication fork establishment (Figure 1). The activation of helicase, like pre-RC assembly, is regulated by a series of events that is highly conserved in eukaryotes.
MCM2-7 hexamer itself has very limited helicase activity. It was later identified that the full helicase activity requires the assembly of Cdc45 and GINS (from the Japanese go-ichi-ni-san that stands for 5-1-2-3, representing Sld5, Psf1, Psf2, and Psf3 subunits) complex with MCM2-7, forming CMG helicase [119–122]. CMG contains a copy of MCM2-7, a single Cdc45, and one GINS tetramer forming a stable 11 subunits helicase. The assembly begins at the G1 to S transition, where MCM double hexamer serves as a platform to recruit DDK (Dbf4-dependent Cdc7 kinase) via interaction by MCM2 and MCM4 [123, 124]. DDK is a cell cycle regulated kinase [125, 126]. Together with S-phase CDK, they are two essential kinases that regulate the assembly of other initiation factors to MCM2-7. DDK phosphorylates multiple MCM subunits including MCM2, 4 and 6 [127–131]. Notably, DDK only targets chromatin bound MCM in the context of a double hexamer but not a single hexamer [132–136], suggesting only the double hexamer configuration is allowed to initiate productive replication. The phosphorylated MCMs then recruit Cdc45 [128]. The recruitment process is facilitated by Sld3 and Sld7 in S. cerevisiae, or their functional homologs Treslin and MTBP in humans [137–139]. In addition to Cdc45, GINS is also associated with MCM2-7. The assembly of GINS complex into MCM requires CDK activity and several chaperone proteins. Sld3 and Dpb11 (TopBP1 in vertebrates) facilitate a complex containing GINS, Sld2 (RecQ4 in vertebrates), and Polɛ to bind to MCM [140–142]. In this process, CDK phosphorylates Cdc45, Sld2, Sld3 and Sld7 to facilitate protein interaction [142, 143]. The protein complex formed at this point is defined as a pre-initiation complex (pre-IC) [144]. In the pre-IC, MCM double hexamer splits into two single hexamers, suggesting they are ready to form bi-directional replication forks [145]. MCM10 is also a critical protein required for origin firing. Although tightly coupled, the pre-IC formation can be separated from the subsequent firing event in vivo in cells lacking MCM10 [145], or when MCM10 is omitted in the in vitro reconstitution reactions [146].
After the pre-IC formation, MCM10 is recruited by interacting with CMG [147–149]. MCM10 is essential for origin firing not only for activating CMG but also stabilizing the replisome [150–152]. Meanwhile, the single strand DNA binding ability of budding yeast MCM10 is important for stabilizing the origin melting reaction [153]. A recent study further suggested that MCM10 is required for CMG to transit between dsDNA and ssDNA [154]. Importantly, in addition to its role in helicase activation, MCM10 also travels along at the replication fork and stimulates replication elongation [155, 156]. On the other hand, extensive structural and single-molecule studies have elucidated that active CMG translocates on DNA with its N-terminal domain in front and C-terminal motor domain pushing from behind, and ssDNA passing through the middle channel making contact with several MCM subunits [157–160].
3. Dormant origins
An important concept of DNA replication regarding the maintenance of genome stability is that in each G1 phase, there are way more origins that are licensed than the ones that actually fire in the subsequent S phase [161]. These origins, which are inactive during normal replication, are called “dormant origins”. It has been known for a long time that MCM is loaded in excess onto chromatin. Studies from yeast to human pointed out that MCM can be loaded up to 20-fold in excess over the numbers of loaded ORC or replication origins [162–164]. Indeed, it has been shown that DNA replication can occur normally with significantly reduced levels of MCM proteins in the cells [162, 165, 166]. Meanwhile, it was also found that though the replication efficiency is maintained in these MCM-depleted cells, they showed defects in the S-phase checkpoint. Critically, the MCM-depleted cells became hypersensitive to low levels of replication stress that would otherwise be tolerated by normal cells [163, 167]. Thus, the role of these excess licensed dormant origins was demonstrated as a safeguard system when cells are facing replication stress, and the dormant origins only fire when other replication forks fail to finish replication. The amount of dormant origins, or level of origin licensing, is not only correlated to the tolerability of cells to replication stress, but also shown to be critical for the maintenance of stem cell pluripotency [168].
Although the importance of dormant origins is well appreciated, the selection mechanism regarding which subset of licensed origins should be activated while others should stay dormant is poorly understood. It was suggested that a fraction of pre-RC is multi-mono-ubiquitinated on Orc3/Orc5 by an E3 ligase OBI1, which in turn marks those pre-RCs (out of all available pre-RC) for activation [169]. However, the mechanism of how OBI1 selects its substrate is still unknown. Recently, one study made an interesting observation that the parental MCMs inherited from the previous cell cycle have a distinct function from the nascent MCMs [170]. It was shown that the parental MCMs are preferred for forming active CMGs even though both parental and nascent MCMs can form pre-RC on chromatin. On the other hand, nascent MCMs serve to adjust the pace of active CMGs, possibly by acting as physical resistance of replication fork progression. It was therefore proposed that the surplus of licensed MCMs not only provides a backup system upon replication stress, but also acts to manage replication fork speed to actively prevent replication stress-associated DNA damage. Nevertheless, the mechanism that differentiates these two groups of MCMs remains to be elucidated.
4. Licensing checkpoint
As mentioned above, to prevent genome instability, cells should only enter S phase with a sufficient number of licensed origins. It was suggested that there is a “licensing checkpoint” that senses the number of licensed origins and delays or stops S phase entry if there is an insufficient number of origins licensed [171–173]. In untransformed cells, reducing MCM loading by different methods causes the cells to arrest at G1 phase. Importantly, the licensing checkpoint is often defective in cancer cells; where the inhibition of origin licensing leads to genome instability and apoptosis in several cancer cell lines due to their inability to halt from entering S phase [174–176]. Several of these reports link licensing checkpoint to the p53 status as well as cyclinE/CDK2 activity. However, the detailed mechanism of how cells sense the number of licensed origins remains elusive and requires further study. Recently, a study focusing on the re-entry of the cell cycle from quiescence, or G0, showed that MCM loading is slow and reduced in the first G1 phase, suggesting the licensing checkpoint is largely inactive when cells re-enter cell cycle [177]. Further, the chromatin environment was shown to impact origin utilization when quiescent cells reenter the cell cycle [178]. This under licensed condition in the first G1 phase leads to naturally occurring replication stress, as well as hypersensitivity towards genotoxic drugs. Together, the licensing checkpoint is critical for cells to maintain genome integrity and aberrant licensing can lead to cancer development.
5. Mechanisms preventing re-replication
One of the major challenges of DNA replication is to ensure that replication happens once and only once per cell cycle. Refiring of origins during the same S phase can result in re-replication leading to genome instability and chromosomal breakage during the following mitosis. Cells must exert elaborate regulatory events to prevent re-replication from happening, and the major regulatory point is to prevent re-licensing during S phase [179].
In budding yeast, all six ORC subunits remain chromatin-associated throughout the cell cycle [180]. However, in mammalian cells, Orc1 is released from chromatin upon S phase entry in a CDK-dependent manner, gets ubiquitinated by SCFSkp2 and degraded via ubiquitin-mediated proteasomal degradation [181–184]. This prevents ORC from executing a second round of licensing. Only at M phase to G1 transition is Orc1 able to re-associate with chromatin and enable origin licensing [185–188]. The cell cycle dependent regulation of Orc1 proves to be one of many mechanisms that allow replication to occur once and only once per cell cycle [189]. As mentioned previously, Orc3 and Orc5 are also ubiquitinated in a cell cycle dependent manner that is critical for origin function. The role of other E3 ligases in regulating pre-RC/pre-IC function, origin firing, and maintenance of genome stability are an area of intense research [190, 191]. In addition, a recent report suggested that budding yeast ORC dimerizes in a cell cycle dependent manner to control licensing [192]. It remains to be seen if human ORC is regulated in a similar way.
S. cerevisiae Cdc6 and its ortholog, Cdc18 in S. pombe, are rapidly degraded when cells enter S phase via ubiquitin-mediated proteasomal degradation, after being phosphorylated by S and M-phase CDKs [193–197]. Human Cdc6 is also regulated by CDK but through a different mechanism. Human Cdc6 is targeted for degradation by APC/C-dependent proteolysis [198]. During G1 phase where licensing takes place, Cdc6 is protected from destruction by cyclinE/Cdk2-dependent phosphorylation [199]. In S phase, it was proposed that Cdc6 is phosphorylated by cyclinA/Cdk2 and exported from the nucleus [75, 200, 201]. However, this model was challenged by other studies showing that a fraction of Cdc6 remains chromatin-bound even in S phase [202–204]. In addition, a more recent study showed that Cdc6 is also targeted for degradation by SCFcyclinF complex late in the cell cycle [205]. Therefore, the detailed mechanism regulating Cdc6 during cell cycle remains to be delineated.
The cell cycle-dependent regulation of Cdt1 is the major regulatory point for preventing re-replication. S. cerevisiae Cdt1 is controlled by CDK phosphorylation that inhibits its interaction with Orc6 [101], and subject to nuclear export during S phase [206]. In other eukaryotes including humans, however, Cdt1 protein is degraded upon S phase entry [207–209]. Cdt1 is controlled by two independent E3 ligase pathways to ensure its destruction after S phase entry. First, Cdt1 is phosphorylated by CDKs, which leads to its recognition by SCFSkp2 E3 ligase for ubiquitination and degradation [210–213]. Second, degradation of Cdt1 is further restricted to S phase by “replication-coupled destruction” through interacting with chromatin-bound PCNA (Proliferating Cell Nuclear Antigen), which is the processivity factor for DNA polymerase at the replication fork [214, 215]. Cdt1 contains a conserved PCNA-interacting protein (PIP) box that mediates its interaction with PCNA; the interaction, in turn, triggers Cdt1 ubiquitination and degradation by CRL4Cdt2 E3 ligase [216]. In addition, metazoan Cdt1 is inhibited by direct binding of another cell cycle-oscillating protein, Geminin [217–221]. Geminin levels are elevated in S phase and drastically reduced upon entry in M phase due to degradation by APC/C [222], thus allowing Cdt1 to be active only during G1. Moreover, a recent study uncovered that Cdt1 is also hyperphosphorylated in G2 phase by Cyclin A/CDK1, which inhibits the loading of MCM during G2 phase [223]. This finding provided yet another independent mechanism to prevent re-replication within one cell cycle and maintain genome integrity.
6. Replication initiation and replication stress
A significant threat to genomic and chromosomal stability comes from replication stress. Replication stress is referred to conditions that cause the slowing or stalling of replication fork progression and perturbation of the dynamics of DNA synthesis [224, 225]. Replication stress can be induced by exogenous or endogenous sources. Exogenous causes include ionizing radiation (IR), ultraviolet (UV) irradiation, and chemotherapeutic drugs that lead to DNA damage, interstrand crosslinks, and DNA breaks. These DNA lesions block CMG unwinding or polymerases, causing the fork to stall. Chemical compounds such as hydroxyurea (HU) that result in dNTP depletion/imbalance also induce replication stress by causing uncoupling of DNA polymerase with CMG helicase [226]. Similarly, endogenous DNA damage such as naturally occurring depurination or oxidation can lead to replication stress. A great fraction of endogenous causes of replication stress has resulted from sequence or chromatin features that make replication intrinsically difficult. These include short tandem repeats and microsatellites regions [227], secondary structures such as hairpins or G-quadruplexes [228, 229], centromeres and telomeres regions [230, 231]. Collisions of replication and transcription machinery and R-loop, the three-stranded structures containing a DNA-RNA duplex and the excluded ssDNA, also impede replication progression [232, 233]. Common fragile sites, the chromosomal regions prone to experience replication stress and break upon replication inhibition, usually contain one or several of the above features [234–236]. Importantly, common fragile sites also correlate with origin-poor and ORC-poor regions, indicating that inefficient origin activity and reduced dormant origins cause genome instability [237, 238].
A more direct relationship between replication initiation and replication stress lies in the context of oncogene activation or overexpression-induced replication stress [239, 240]. Early studies identified that the overexpression of cyclin E induces chromosomal instability due to defects in pre-RC chromatin loading [241, 242]. This could be due to short G1/premature S phase entry before sufficient origins are licensed. As discussed above, a reduced number of dormant origins due to insufficient licensing makes cells more vulnerable to replication stress. The common fragile sites represent regions where dormant origins are either inefficiently licensed/activated resulting in replication fork failure and increased susceptibility to breakage [243]. Moreover, overexpression of oncogenes, including cyclin E, Ras, and Myc, have been proven to drive excessive usage of origins that can lead to acute depletion of dNTPs in the cells, resulting in replication stress and genome instability [244, 245]. Increased replication initiation by oncogenes also increased the chance of replication-transcription collision [246]. A recent study using genome-wide mapping further revealed that oncogene overexpression induces a subset of intragenic origins to be fired early due to short G1/premature S phase entry, which greatly increases the replication-transcription collision and DNA breakage [247]. The regulation of transcription activity at origins is also critical for genomic integrity [248].
One direct consequence of replication stress is the formation of exposed ssDNA due to fork stalling. Thus, replication stress is tightly associated with the activation of ATR pathway of the DDR. The activation of ATR starts from RPA loading onto exposed ssDNA, which can be generated by end-resection of DSBs, replication stress, or intermediates during the DNA repair processes [249–255]. RPA-ssDNA then serves as a platform to recruit ATR via its partner protein ATRIP (ATR interacting protein) [256]. Two independent mechanisms then function to activate ATR. The binding of Rad17/RFC and Rad9-Rad1-Hus1 (9-1-1) complexes to the junction of RPA-ssDNA and dsDNA recruits TopBP1, the first ATR-activating domain (AAD)-containing protein, to activate ATR kinase activity [257–260]. On the other hand, ETAA1, the second protein identified with an AAD, binds to RPA directly and activates ATR in parallel to TopBP1 [261–263]. Subsequently, the activated ATR phosphorylates its downstream effector kinase Chk1 [264]. Like the ATM-Chk2 axis, ATR-Chk1 controls many cellular events during DNA damage. One proteomic study identified more than 700 ATM/ATR downstream substrates in human cells, highlighting the complexity of the DDR network [265]. Importantly, many pre-RC and pre-IC proteins have been identified in this study as ATM/ATR substrates, suggesting the existence of intricate regulatory mechanisms in replication initiation or perhaps playing other non-canonical roles. DNA Polymerase epsilon playing a critical role in checkpoint pathway during S phase to sense stalled replication confirms that a link between DNA replication machinery and S-Phase checkpoint exists in eukaryotes [266–268]. Further, Pol ϵ binds replication origins early in S phase and the essential function of Pol ϵ was not dependent on its DNA synthesis activity [269].
ATR signaling also has critical roles in the regulation of replication during S phase. Under genotoxic conditions, checkpoints are activated and the cell cycle is halted. It is well established that replication initiation is targeted for repression by ATR-Chk1 pathway in order to prevent S phase progression and further damage to the DNA until DNA damages are repaired [270–273]. The repression of origin firing by checkpoint also prevents DNA topological stress [274]. Interestingly, the ATR-Chk1 activation inhibits the global origin firing, yet promotes local origins to initiate in order to support the completion of those regions of DNA where replication forks are stalled [275]. However, the detailed mechanism remains elusive. On the contrary, even without exogenous sources of DNA damage, the basal level of ATR-Chk1 activity is critical to control origin activity. Many studies discovered that ATR or Chk1 inhibition causes increased global origin firing and abnormal early activation of late origins [276–281], suggesting an essential function of ATR pathway in regulating unperturbed S phase progression and its importance in genome integrity. On the other hand, deregulation of many pre-RC and pre-IC proteins directly affects ATR signaling pathway activation. Cdc6 has been shown to directly interact with ATR and activate replication-checkpoint [282]. MCM7 reduction leads to ATR activation defects [165]. Further, TopBP1, one of the pre-IC proteins, is also an essential protein for ATR activation [257]. The regulation of DNA replication initiation events and checkpoint signaling pathways remains an active field of study.
Highlights.
This review provides insights on the following topics:
Pre-replication complex and Origin licensing
Pre-initiation complex and origin firing
Dormant origins
Licensing checkpoint
Mechanisms preventing re-replication
Replication initiation and replication stress
Acknowledgements
The authors would like to thank Jay Sonalkar for his comments. FUNDING: SGP Lab is supported by. NSF (1243372 and 1818286) and NIH (R01GM125196) awards.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bell SP, Dutta A, DNA Replication in Eukaryotic Cells, Annual Review of Biochemistry, 71 (2002) 333–374. [DOI] [PubMed] [Google Scholar]
- [2].Bell SP, The origin recognition complex: from simple origins to complex functions, Genes Dev, 16 (2002) 659–672. [DOI] [PubMed] [Google Scholar]
- [3].Parker MW, Botchan MR, Berger JM, Mechanisms and regulation of DNA replication initiation in eukaryotes, Critical reviews in biochemistry and molecular biology, 52 (2017) 107–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Limas JC, Cook JG, Preparation for DNA replication: the key to a successful S phase, FEBS Lett, 593 (2019) 2853–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bleichert F, Mechanisms of replication origin licensing: a structural perspective, Current opinion in structural biology, 59 (2019) 195–204. [DOI] [PubMed] [Google Scholar]
- [6].Bell SP, Stillman B, ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex, Nature, 357 (1992) 128–134. [DOI] [PubMed] [Google Scholar]
- [7].Gavin KA, Hidaka M, Stillman B, Conserved Initiator Proteins in Eukaryotes, Science (New York, N.Y.), 270 (1995) 1667–1671. [DOI] [PubMed] [Google Scholar]
- [8].Gossen M, Pak DTS, Hansen SK, Acharya JK, Botchan MR, A Drosophila Homolog of the Yeast Origin Recognition Complex, Science (New York, N.Y.), 270 (1995) 1674–1677. [DOI] [PubMed] [Google Scholar]
- [9].Carpenter PB, Mueller PR, Dunphy WG, Role for a Xenopus Orc2-related protein in controlling DNA replication, Nature, 379 (1996) 357–360. [DOI] [PubMed] [Google Scholar]
- [10].Takahara K, Bong M, Brevard R, Eddy RL, Haley LL, Sait SJ, Shows TB, Hoffman GG, Greenspan DS, Mouse and human homologues of the yeast origin of replication recognition complex subunit ORC2 and chromosomal localization of the cognate human gene ORC2L, Genomics, 31 (1996) 119–122. [DOI] [PubMed] [Google Scholar]
- [11].Dhar SK, Dutta A, Identification and characterization of the human ORC6 homolog, J Biol Chem, 275 (2000) 34983–34988. [DOI] [PubMed] [Google Scholar]
- [12].Quintana DG, Thome KC, Hou ZH, Ligon AH, Morton CC, Dutta A, ORC5L, a new member of the human origin recognition complex, is deleted in uterine leiomyomas and malignant myeloid diseases, J Biol Chem, 273 (1998) 27137–27145. [DOI] [PubMed] [Google Scholar]
- [13].Quintana DG, Hou Z, Thome KC, Hendricks M, Saha P, Dutta A, Identification of HsORC4, a member of the human origin of replication recognition complex, J Biol Chem, 272 (1997) 28247–28251. [DOI] [PubMed] [Google Scholar]
- [14].Tugal T, Zou-Yang XH, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B, The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p, J Biol Chem, 273 (1998) 32421–32429. [DOI] [PubMed] [Google Scholar]
- [15].Yu G, Wu JR, Gilbert DM, Analysis of mammalian origin specification in ORC-depleted Xenopus egg extracts, Genes to cells : devoted to molecular & cellular mechanisms, 3 (1998) 709–720. [DOI] [PubMed] [Google Scholar]
- [16].Speck C, Chen Z, Li H, Stillman B, ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA, Nat Struct Mol Biol, 12 (2005) 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Giordano-Coltart J, Ying CY, Gautier J, Hurwitz J, Studies of the properties of human origin recognition complex and its Walker A motif mutants, Proc Natl Acad Sci U S A, 102 (2005) 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B, The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing, Cell, 83 (1995) 563–568. [DOI] [PubMed] [Google Scholar]
- [19].Liu S, Balasov M, Wang H, Wu L, Chesnokov IN, Liu Y, Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB, Proc Natl Acad Sci U S A, 108 (2011) 7373–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chesnokov IN, Chesnokova ON, Botchan M, A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity, Proc Natl Acad Sci U S A, 100 (2003) 9150–9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Balasov M, Huijbregts RP, Chesnokov I, Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster, Mol Cell Biol, 27 (2007) 3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bleichert F, Balasov M, Chesnokov I, Nogales E, Botchan MR, Berger JM, A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation, Elife, 2 (2013) e00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bleichert F, Botchan MR, Berger JM, Crystal structure of the eukaryotic origin recognition complex, Nature, 519 (2015) 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ganier O, Prorok P, Akerman I, Méchali M, Metazoan DNA replication origins, Current opinion in cell biology, 58 (2019) 134–141. [DOI] [PubMed] [Google Scholar]
- [25].Newlon CS, Yeast chromosome replication and segregation, Microbiological reviews, 52 (1988) 568–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Marahrens Y, Stillman B, A yeast chromosomal origin of DNA replication defined by multiple functional elements, Science (New York, N.Y.), 255 (1992) 817–823. [DOI] [PubMed] [Google Scholar]
- [27].Li N, Lam WH, Zhai Y, Cheng J, Cheng E, Zhao Y, Gao N, Tye BK, Structure of the origin recognition complex bound to DNA replication origin, Nature, 559 (2018) 217–222. [DOI] [PubMed] [Google Scholar]
- [28].Lee CSK, Cheung MF, Li J, Zhao Y, Lam WH, Ho V, Rohs R, Zhai Y, Leung D, Tye BK, Humanizing the yeast origin recognition complex, Nat Commun, 12 (2021) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Okuno Y, Satoh H, Sekiguchi M, Masukata H, Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin, Mol Cell Biol, 19 (1999) 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dai J, Chuang RY, Kelly TJ, DNA replication origins in the Schizosaccharomyces pombe genome, Proc Natl Acad Sci U S A, 102 (2005) 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Segurado M, de Luis A, Antequera F, Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe, EMBO Rep, 4 (2003) 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC, Sequence-independent DNA binding and replication initiation by the human origin recognition complex, Genes Dev, 17 (2003) 1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Méchali M, Kearsey S, Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast, Cell, 38 (1984) 55–64. [DOI] [PubMed] [Google Scholar]
- [34].Hyrien O, Méchali M, Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos, Embo j, 12 (1993) 4511–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN, Genome-wide studies highlight indirect links between human replication origins and gene regulation, Proc Natl Acad Sci U S A, 105 (2008) 15837–15842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Aladjem MI, Replication in context: dynamic regulation of DNA replication patterns in metazoans, Nat Rev Genet, 8 (2007) 588–600. [DOI] [PubMed] [Google Scholar]
- [37].Aladjem MI, Redon CE, Order from clutter: selective interactions at mammalian replication origins, Nat Rev Genet, 18 (2017) 101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM, Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading, Genome Res, 20 (2010) 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Karnani N, Taylor CM, Malhotra A, Dutta A, Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection, Mol Biol Cell, 21 (2010) 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM, Chromatin signatures of the Drosophila replication program, Genome Res, 21 (2011) 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lubelsky Y, Sasaki T, Kuipers MA, Lucas I, Le Beau MM, Carignon S, Debatisse M, Prinz JA, Dennis JH, Gilbert DM, Pre-replication complex proteins assemble at regions of low nucleosome occupancy within the Chinese hamster dihydrofolate reductase initiation zone, Nucleic Acids Res, 39 (2011) 3141–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mesner LD, Valsakumar V, Cieslik M, Pickin R, Hamlin JL, Bekiranov S, Bubble-seq analysis of the human genome reveals distinct chromatin-mediated mechanisms for regulating early- and late-firing origins, Genome Res, 23 (2013) 1774–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM, Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs, Nat Struct Mol Biol, 19 (2012) 837–844. [DOI] [PubMed] [Google Scholar]
- [44].Sequeira-Mendes J, Díaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gómez M, Transcription initiation activity sets replication origin efficiency in mammalian cells, PLoS Genet, 5 (2009) e1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bartholdy B, Mukhopadhyay R, Lajugie J, Aladjem MI, Bouhassira EE, Allele-specific analysis of DNA replication origins in mammalian cells, Nat Commun, 6 (2015) 7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Martin MM, Ryan M, Kim R, Zakas AL, Fu H, Lin CM, Reinhold WC, Davis SR, Bilke S, Liu H, Doroshow JH, Reimers MA, Valenzuela MS, Pommier Y, Meltzer PS, Aladjem MI, Genome-wide depletion of replication initiation events in highly transcribed regions, Genome Res, 21 (2011) 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Petryk N, Kahli M, d’Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen CL, Hyrien O, Replication landscape of the human genome, Nat Commun, 7 (2016) 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau JC, van Helden J, Méchali M, The chromatin environment shapes DNA replication origin organization and defines origin classes, Genome Res, 25 (2015) 1873–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Prorok P, Artufel M, Aze A, Coulombe P, Peiffer I, Lacroix L, Guédin A, Mergny JL, Damaschke J, Schepers A, Ballester B, Méchali M, Involvement of G-quadruplex regions in mammalian replication origin activity, Nat Commun, 10 (2019) 3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Akerman I, Kasaai B, Bazarova A, Sang PB, Peiffer I, Artufel M, Derelle R, Smith G, Rodriguez-Martinez M, Romano M, Kinet S, Tino P, Theillet C, Taylor N, Ballester B, Méchali M, A predictable conserved DNA base composition signature defines human core DNA replication origins, Nat Commun, 11 (2020) 4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B, Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance, Embo j, 23 (2004) 2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tatsumi Y, Ezura K, Yoshida K, Yugawa T, Narisawa-Saito M, Kiyono T, Ohta S, Obuse C, Fujita M, Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres, Genes to cells : devoted to molecular & cellular mechanisms, 13 (2008) 1045–1059. [DOI] [PubMed] [Google Scholar]
- [53].Drosopoulos WC, Deng Z, Twayana S, Kosiyatrakul ST, Vladimirova O, Lieberman PM, Schildkraut CL, TRF2 Mediates Replication Initiation within Human Telomeres to Prevent Telomere Dysfunction, Cell Rep, 33 (2020) 108379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, Prasanth SG, A WD-repeat protein stabilizes ORC binding to chromatin, Mol Cell, 40 (2010) 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang Y, Khan A, Marks AB, Smith OK, Giri S, Lin YC, Creager R, MacAlpine DM, Prasanth KV, Aladjem MI, Prasanth SG, Temporal association of ORCA/LRWD1 to late-firing origins during G1 dictates heterochromatin replication and organization, Nucleic Acids Res, 45 (2017) 2490–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Giri S, Aggarwal V, Pontis J, Shen Z, Chakraborty A, Khan A, Mizzen C, Prasanth KV, Ait-Si-Ali S, Ha T, Prasanth SG, The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin, Elife, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hsu RYC, Lin YC, Redon C, Sun Q, Singh DK, Wang Y, Aggarwal V, Mitra J, Matur A, Moriarity B, Ha T, Aladjem MI, Prasanth KV, Prasanth SG, ORCA/LRWD1 Regulates Homologous Recombination at ALT-Telomeres by Modulating Heterochromatin Organization, iScience, 23 (2020) 101038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Shibata E, Kiran M, Shibata Y, Singh S, Kiran S, Dutta A, Two subunits of human ORC are dispensable for DNA replication and proliferation, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bell SP, Rethinking origin licensing, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shibata E, Dutta A, A human cancer cell line initiates DNA replication normally in the absence of ORC5 and ORC2 proteins, J Biol Chem, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, Meyers RM, Ali L, Goodale A, Lee Y, Jiang G, Hsiao J, Gerath WFJ, Howell S, Merkel E, Ghandi M, Garraway LA, Root DE, Golub TR, Boehm JS, Hahn WC, Defining a Cancer Dependency Map, Cell, 170 (2017) 564–576.e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A, Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells, Nat Genet, 49 (2017) 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chou HC, Bhalla K, Demerdesh OE, Klingbeil O, Hanington K, Aganezov S, Andrews P, Alsudani H, Chang K, Vakoc CR, Schatz MC, McCombie WR, Stillman B, The human origin recognition complex is essential for pre-RC assembly, mitosis, and maintenance of nuclear structure, Elife, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liang C, Weinreich M, Stillman B, ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome, Cell, 81 (1995) 667–676. [DOI] [PubMed] [Google Scholar]
- [65].Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF, An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast, Nature, 379 (1996) 180–182. [DOI] [PubMed] [Google Scholar]
- [66].Donovan S, Harwood J, Drury LS, Diffley JF, Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast, Proc Natl Acad Sci U S A, 94 (1997) 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tanaka T, Knapp D, Nasmyth K, Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs, Cell, 90 (1997) 649–660. [DOI] [PubMed] [Google Scholar]
- [68].Schmidt JM, Bleichert F, Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6, Nat Commun, 11 (2020) 4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM, Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control, Mol Cell, 6 (2000) 637–648. [DOI] [PubMed] [Google Scholar]
- [70].Herbig U, Marlar CA, Fanning E, The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells, Mol Biol Cell, 10 (1999) 2631–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Perkins G, Diffley JF, Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders, Mol Cell, 2 (1998) 23–32. [DOI] [PubMed] [Google Scholar]
- [72].Weinreich M, Liang C, Stillman B, The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin, Proc Natl Acad Sci U S A, 96 (1999) 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wang B, Feng L, Hu Y, Huang SH, Reynolds CP, Wu L, Jong AY, The essential role of Saccharomyces cerevisiae CDC6 nucleotide-binding site in cell growth, DNA synthesis, and Orc1 association, J Biol Chem, 274 (1999) 8291–8298. [DOI] [PubMed] [Google Scholar]
- [74].Giraldo R, Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives, FEMS microbiology reviews, 26 (2003) 533–554. [DOI] [PubMed] [Google Scholar]
- [75].Saha P, Chen J, Thome KC, Lawlis SJ, Hou ZH, Hendricks M, Parvin JD, Dutta A, Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase, Mol Cell Biol, 18 (1998) 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mizushima T, Takahashi N, Stillman B, Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro, Genes Dev, 14 (2000) 1631–1641. [PMC free article] [PubMed] [Google Scholar]
- [77].Evrin C, Fernández-Cid A, Zech J, Herrera MC, Riera A, Clarke P, Brill S, Lurz R, Speck C, In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2-7 hexamer dimerization, Nucleic Acids Res, 41 (2013) 3162–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP, Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading, Cell, 161 (2015) 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H, Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy, Structure (London, England : 1993), 20 (2012) 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, Speck C, Li H, Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1, Nat Struct Mol Biol, 24 (2017) 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sun J, Evrin C, Samel SA, Fernandez-Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H, Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA, Nat Struct Mol Biol, 20 (2013) 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Pozo PN, Cook JG, Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability, Genes, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Nishitani H, Lygerou Z, Nishimoto T, Nurse P, The Cdt1 protein is required to license DNA for replication in fission yeast, Nature, 404 (2000) 625–628. [DOI] [PubMed] [Google Scholar]
- [84].Devault A, Vallen EA, Yuan T, Green S, Bensimon A, Schwob E, Identification of Tah11/Sid2 as the ortholog of the replication licensing factor Cdt1 in Saccharomyces cerevisiae, Current biology : CB, 12 (2002) 689–694. [DOI] [PubMed] [Google Scholar]
- [85].Whittaker AJ, Royzman I, Orr-Weaver TL, Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts, Genes Dev, 14 (2000) 1765–1776. [PMC free article] [PubMed] [Google Scholar]
- [86].Maiorano D, Moreau J, Méchali M, XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis, Nature, 404 (2000) 622–625. [DOI] [PubMed] [Google Scholar]
- [87].Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A, Inhibition of eukaryotic DNA replication by geminin binding to Cdt1, Science (New York, N.Y.), 290 (2000) 2309–2312. [DOI] [PubMed] [Google Scholar]
- [88].Ferenbach A, Li A, Brito-Martins M, Blow JJ, Functional domains of the Xenopus replication licensing factor Cdt1, Nucleic Acids Res, 33 (2005) 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Liu C, Wu R, Zhou B, Wang J, Wei Z, Tye BK, Liang C, Zhu G, Structural insights into the Cdt1-mediated MCM2-7 chromatin loading, Nucleic Acids Res, 40 (2012) 3208–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wei Z, Liu C, Wu X, Xu N, Zhou B, Liang C, Zhu G, Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6, J Biol Chem, 285 (2010) 12469–12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wu R, Wang J, Liang C, Cdt1p, through its interaction with Mcm6p, is required for the formation, nuclear accumulation and chromatin loading of the MCM complex, J Cell Sci, 125 (2012) 209–219. [DOI] [PubMed] [Google Scholar]
- [92].Kawasaki Y, Kim HD, Kojima A, Seki T, Sugino A, Reconstitution of Saccharomyces cerevisiae prereplicative complex assembly in vitro, Genes to cells : devoted to molecular & cellular mechanisms, 11 (2006) 745–756. [DOI] [PubMed] [Google Scholar]
- [93].Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF, Concerted loading of MCM2-7 double hexamers around DNA during DNA replication origin licensing, Cell, 139 (2009) 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Takara TJ, Bell SP, Multiple Cdt1 molecules act at each origin to load replication-competent MCM2-7 helicases, Embo j, 30 (2011) 4885–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Frigola J, Remus D, Mehanna A, Diffley JF, ATPase-dependent quality control of DNA replication origin licensing, Nature, 495 (2013) 339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ticau S, Friedman LJ, Champasa K, Corrêa IR Jr., Gelles J, Bell SP, Mechanism and timing of MCM2-7 ring closure during DNA replication origin licensing, Nat Struct Mol Biol, 24 (2017) 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhai Y, Cheng E, Wu H, Li N, Yung PY, Gao N, Tye BK, Open-ringed structure of the Cdt1-MCM2-7 complex as a precursor of the MCM double hexamer, Nat Struct Mol Biol, 24 (2017) 300–308. [DOI] [PubMed] [Google Scholar]
- [98].Frigola J, He J, Kinkelin K, Pye VE, Renault L, Douglas ME, Remus D, Cherepanov P, Costa A, Diffley JFX, Cdt1 stabilizes an open MCM ring for helicase loading, Nat Commun, 8 (2017) 15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Asano T, Makise M, Takehara M, Mizushima T, Interaction between ORC and Cdt1p of Saccharomyces cerevisiae, FEMS yeast research, 7 (2007) 1256–1262. [DOI] [PubMed] [Google Scholar]
- [100].Chen S, de Vries MA, Bell SP, Orc6 is required for dynamic recruitment of Cdt1 during repeated MCM2-7 loading, Genes Dev, 21 (2007) 2897–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chen S, Bell SP, CDK prevents MCM2-7 helicase loading by inhibiting Cdt1 interaction with Orc6, Genes Dev, 25 (2011) 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Semple JW, Da-Silva LF, Jervis EJ, Ah-Kee J, Al-Attar H, Kummer L, Heikkila JJ, Pasero P, Duncker BP, An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes, Embo j, 25 (2006) 5150–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Fernández-Cid A, Riera A, Tognetti S, Herrera MC, Samel S, Evrin C, Winkler C, Gardenal E, Uhle S, Speck C, An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly, Mol Cell, 50 (2013) 577–588. [DOI] [PubMed] [Google Scholar]
- [104].Yuan Z, Schneider S, Dodd T, Riera A, Bai L, Yan C, Magdalou I, Ivanov I, Stillman B, Li H, Speck C, Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6, Proc Natl Acad Sci U S A, 117 (2020) 17747–17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Maine GT, Sinha P, Tye BK, Mutants of S cerevisiae defective in the maintenance of minichromosomes, Genetics, 106 (1984) 365–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Treisman JE, Follette PJ, O’Farrell PH, Rubin GM, Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant, Genes Dev, 9 (1995) 1709–1715. [DOI] [PubMed] [Google Scholar]
- [107].Madine MA, Khoo CY, Mills AD, Laskey RA, MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells, Nature, 375 (1995) 421–424. [DOI] [PubMed] [Google Scholar]
- [108].Todorov IT, Attaran A, Kearsey SE, BM28, a human member of the MCM2–3-5 family, is displaced from chromatin during DNA replication, J Cell Biol, 129 (1995) 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chong JP, Mahbubani HM, Khoo CY, Blow JJ, Purification of an MCM-containing complex as a component of the DNA replication licensing system, Nature, 375 (1995) 418–421. [DOI] [PubMed] [Google Scholar]
- [110].Adachi Y, Usukura J, Yanagida M, A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast, Genes to cells : devoted to molecular & cellular mechanisms, 2 (1997) 467–479. [DOI] [PubMed] [Google Scholar]
- [111].Thömmes P, Kubota Y, Takisawa H, Blow JJ, The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides, Embo j, 16 (1997) 3312–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Koonin EV, A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication, Nucleic Acids Res, 21 (1993) 2541–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM, The structural basis for MCM2-7 helicase activation by GINS and Cdc45, Nat Struct Mol Biol, 18 (2011) 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Davey MJ, Indiani C, O’Donnell M, Reconstitution of the MCM2-7p heterohexamer, subunit arrangement, and ATP site architecture, J Biol Chem, 278 (2003) 4491–4499. [DOI] [PubMed] [Google Scholar]
- [115].Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C, A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication, Proc Natl Acad Sci U S A, 106 (2009) 20240–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Champasa K, Blank C, Friedman LJ, Gelles J, Bell SP, A conserved Mcm4 motif is required for MCM2-7 double-hexamer formation and origin DNA unwinding, Elife, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Coster G, Diffley JFX, Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading, Science (New York, N.Y.), 357 (2017) 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Miller TCR, Locke J, Greiwe JF, Diffley JFX, Costa A, Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM, Nature, 575 (2019) 704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Ilves I, Petojevic T, Pesavento JJ, Botchan MR, Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins, Mol Cell, 37 (2010) 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Moyer SE, Lewis PW, Botchan MR, Isolation of the Cdc45/MCM2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase, Proc Natl Acad Sci U S A, 103 (2006) 10236–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC, Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication, Mol Cell, 21 (2006) 581–587. [DOI] [PubMed] [Google Scholar]
- [122].Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K, GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks, Nat Cell Biol, 8 (2006) 358–366. [DOI] [PubMed] [Google Scholar]
- [123].Ramer MD, Suman ES, Richter H, Stanger K, Spranger M, Bieberstein N, Duncker BP, Dbf4 and Cdc7 proteins promote DNA replication through interactions with distinct MCM2-7 protein subunits, J Biol Chem, 288 (2013) 14926–14935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Sheu YJ, Stillman B, The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4, Nature, 463 (2010) 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Cheng L, Collyer T, Hardy CF, Cell cycle regulation of DNA replication initiator factor Dbf4p, Mol Cell Biol, 19 (1999) 4270–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Oshiro G, Owens JC, Shellman Y, Sclafani RA, Li JJ, Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability, Mol Cell Biol, 19 (1999) 4888–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK, CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein, Proc Natl Acad Sci U S A, 103 (2006) 11521–11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, Tamai K, Ohtani K, Arai K, Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin, J Biol Chem, 281 (2006) 39249–39261. [DOI] [PubMed] [Google Scholar]
- [129].Sheu YJ, Stillman B, Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression, Mol Cell, 24 (2006) 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP, Mec1 is one of multiple kinases that prime the MCM2-7 helicase for phosphorylation by Cdc7, Mol Cell, 40 (2010) 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Tsuji T, Ficarro SB, Jiang W, Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells, Mol Biol Cell, 17 (2006) 4459–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, Li H, Structural and mechanistic insights into MCM2-7 double-hexamer assembly and function, Genes Dev, 28 (2014) 2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP, Incorporation into the prereplicative complex activates the MCM2-7 helicase for Cdc7-Dbf4 phosphorylation, Genes Dev, 23 (2009) 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kang S, Warner MD, Bell SP, Multiple functions for MCM2-7 ATPase motifs during replication initiation, Mol Cell, 55 (2014) 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Costa A, Renault L, Swuec P, Petojevic T, Pesavento JJ, Ilves I, MacLellan-Gibson K, Fleck RA, Botchan MR, Berger JM, DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome, Elife, 3 (2014) e03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Li N, Zhai Y, Zhang Y, Li W, Yang M, Lei J, Tye BK, Gao N, Structure of the eukaryotic MCM complex at 3.8 Å, Nature, 524 (2015) 186–191. [DOI] [PubMed] [Google Scholar]
- [137].Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H, Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing, Current biology : CB, 21 (2011) 2055–2063. [DOI] [PubMed] [Google Scholar]
- [138].Boos D, Yekezare M, Diffley JF, Identification of a heteromeric complex that promotes DNA replication origin firing in human cells, Science (New York, N.Y.), 340 (2013) 981–984. [DOI] [PubMed] [Google Scholar]
- [139].Deegan TD, Yeeles JT, Diffley JF, Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation, Embo j, 35 (2016) 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H, GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast, Genes Dev, 17 (2003) 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Tanaka S, Komeda Y, Umemori T, Kubota Y, Takisawa H, Araki H, Efficient initiation of DNA replication in eukaryotes requires Dpb11/TopBP1-GINS interaction, Mol Cell Biol, 33 (2013) 2614–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H, CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast, Genes Dev, 24 (2010) 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP, Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases, Cell, 146 (2011) 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Zou L, Stillman B, Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin, Science (New York, N.Y.), 280 (1998) 593–596. [DOI] [PubMed] [Google Scholar]
- [145].Miyazawa-Onami M, Araki H, Tanaka S, Pre-initiation complex assembly functions as a molecular switch that splits the MCM2-7 double hexamer, EMBO Rep, 18 (2017) 1752–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF, Regulated eukaryotic DNA replication origin firing with purified proteins, Nature, 519 (2015) 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK, Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins, Genes Dev, 14 (2000) 913–926. [PMC free article] [PubMed] [Google Scholar]
- [148].Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC, Xenopus Mcm10 binds to origins of DNA replication after MCM2-7 and stimulates origin binding of Cdc45, Mol Cell, 9 (2002) 233–240. [DOI] [PubMed] [Google Scholar]
- [149].Douglas ME, Diffley JF, Recruitment of Mcm10 to Sites of Replication Initiation Requires Direct Binding to the Minichromosome Maintenance (MCM) Complex, J Biol Chem, 291 (2016) 5879–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Ricke RM, Bielinsky AK, Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha, Mol Cell, 16 (2004) 173–185. [DOI] [PubMed] [Google Scholar]
- [151].van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K, Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation, Embo j, 31 (2012) 2195–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK, Assembly of the Cdc45-MCM2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins, Proc Natl Acad Sci U S A, 106 (2009) 15628–15632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Perez-Arnaiz P, Kaplan DL, An Mcm10 Mutant Defective in ssDNA Binding Shows Defects in DNA Replication Initiation, Journal of molecular biology, 428 (2016) 4608–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Wasserman MR, Schauer GD, O’Donnell ME, Liu S, Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase, Cell, 178 (2019) 600–611.e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Lõoke M, Maloney MF, Bell SP, Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase, Genes Dev, 31 (2017) 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A, Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components, Nat Cell Biol, 16 (2014) 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Yuan Z, Georgescu R, Bai L, Zhang D, Li H, O’Donnell ME, DNA unwinding mechanism of a eukaryotic replicative CMG helicase, Nat Commun, 11 (2020) 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Burnham DR, Kose HB, Hoyle RB, Yardimci H, The mechanism of DNA unwinding by the eukaryotic replicative helicase, Nat Commun, 10 (2019) 2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Meagher M, Epling LB, Enemark EJ, DNA translocation mechanism of the MCM complex and implications for replication initiation, Nat Commun, 10 (2019) 3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].O’Donnell ME, Li H, The ring-shaped hexameric helicases that function at DNA replication forks, Nat Struct Mol Biol, 25 (2018) 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Courtot L, Hoffmann JS, Bergoglio V, The Protective Role of Dormant Origins in Response to Replicative Stress, International journal of molecular sciences, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC, MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts, J Biol Chem, 277 (2002) 33049–33057. [DOI] [PubMed] [Google Scholar]
- [163].Woodward AM, Göhler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ, Excess MCM2-7 license dormant origins of replication that can be used under conditions of replicative stress, J Cell Biol, 173 (2006) 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Tye BK, MCM proteins in DNA replication, Annu Rev Biochem, 68 (1999) 649–686. [DOI] [PubMed] [Google Scholar]
- [165].Cortez D, Glick G, Elledge SJ, Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases, Proc Natl Acad Sci U S A, 101 (2004) 10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Tsao CC, Geisen C, Abraham RT, Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling, Embo j, 23 (2004) 4660–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Ibarra A, Schwob E, Méndez J, Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication, Proc Natl Acad Sci U S A, 105 (2008) 8956–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Matson JP, Dumitru R, Coryell P, Baxley RM, Chen W, Twaroski K, Webber BR, Tolar J, Bielinsky AK, Purvis JE, Cook JG, Rapid DNA replication origin licensing protects stem cell pluripotency, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Coulombe P, Nassar J, Peiffer I, Stanojcic S, Sterkers Y, Delamarre A, Bocquet S, Méchali M, The ORC ubiquitin ligase OBI1 promotes DNA replication origin firing, Nat Commun, 10 (2019) 2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Sedlackova H, Rask MB, Gupta R, Choudhary C, Somyajit K, Lukas J, Equilibrium between nascent and parental MCM proteins protects replicating genomes, Nature, (2020). [DOI] [PubMed] [Google Scholar]
- [171].McIntosh D, Blow JJ, Dormant origins, the licensing checkpoint, and the response to replicative stresses, Cold Spring Harbor perspectives in biology, 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Liu P, Slater DM, Lenburg M, Nevis K, Cook JG, Vaziri C, Replication licensing promotes cyclin D1 expression and G1 progression in untransformed human cells, Cell cycle (Georgetown, Tex.), 8 (2009) 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Machida YJ, Teer JK, Dutta A, Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation, J Biol Chem, 280 (2005) 27624–27630. [DOI] [PubMed] [Google Scholar]
- [174].Shreeram S, Sparks A, Lane DP, Blow JJ, Cell type-specific responses of human cells to inhibition of replication licensing, Oncogene, 21 (2002) 6624–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Feng D, Tu Z, Wu W, Liang C, Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells, Cancer research, 63 (2003) 7356–7364. [PubMed] [Google Scholar]
- [176].Nevis KR, Cordeiro-Stone M, Cook JG, Origin licensing and p53 status regulate Cdk2 activity during G(1), Cell cycle (Georgetown, Tex.), 8 (2009) 1952–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Matson JP, House AM, Grant GD, Wu H, Perez J, Cook JG, Intrinsic checkpoint deficiency during cell cycle re-entry from quiescence, J Cell Biol, 218 (2019) 2169–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Lee PH, Osley MA, Chromatin structure restricts origin utilization when quiescent cells re-enter the cell cycle, Nucleic Acids Res, 49 (2021) 864–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Grant GD, Cook JG, The Temporal Regulation of S Phase Proteins During G(1), Advances in experimental medicine and biology, 1042 (2017) 335–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Liang C, Stillman B, Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants, Genes Dev, 11 (1997) 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Li CJ, DePamphilis ML, Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle, Mol Cell Biol, 22 (2002) 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Méndez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B, Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication, Mol Cell, 9 (2002) 481–491. [DOI] [PubMed] [Google Scholar]
- [183].Kreitz S, Ritzi M, Baack M, Knippers R, The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells, J Biol Chem, 276 (2001) 6337–6342. [DOI] [PubMed] [Google Scholar]
- [184].Li CJ, Vassilev A, DePamphilis ML, Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex’s largest subunit (Orc1) from binding to chromatin during mitosis, Mol Cell Biol, 24 (2004) 5875–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A, Localization of proteins bound to a replication origin of human DNA along the cell cycle, Embo j, 22 (2003) 4294–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Ohta S, Tatsumi Y, Fujita M, Tsurimoto T, Obuse C, The ORC1 cycle in human cells: II. Dynamic changes in the human ORC complex during the cell cycle, J Biol Chem, 278 (2003) 41535–41540. [DOI] [PubMed] [Google Scholar]
- [187].Kara N, Hossain M, Prasanth SG, Stillman B, Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning during G1 Phase and Assembly of the Origin Recognition Complex in Human Cells, J Biol Chem, 290 (2015) 12355–12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Fujita M, Ishimi Y, Nakamura H, Kiyono T, Tsurumi T, Nuclear organization of DNA replication initiation proteins in mammalian cells, J Biol Chem, 277 (2002) 10354–10361. [DOI] [PubMed] [Google Scholar]
- [189].DePamphilis ML, Cell cycle dependent regulation of the origin recognition complex, Cell cycle (Georgetown, Tex.), 4 (2005) 70–79. [DOI] [PubMed] [Google Scholar]
- [190].Lin YC, Wang Y, Hsu R, Giri S, Wopat S, Arif MK, Chakraborty A, Prasanth KV, Prasanth SG, PCNA-mediated stabilization of E3 ligase RFWD3 at the replication fork is essential for DNA replication, Proc Natl Acad Sci U S A, 115 (2018) 13282–13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Hsu RYC, Giri S, Wang Y, Lin YC, Liu D, Wopat S, Chakraborty A, Prasanth KV, Prasanth SG, The E3 ligase RFWD3 stabilizes ORC in a p53-dependent manner, Cell cycle (Georgetown, Tex.), 19 (2020) 2927–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Amin A, Wu R, Cheung MH, Scott JF, Wang Z, Zhou Z, Liu C, Zhu G, Wong CK, Yu Z, Liang C, An Essential and Cell-Cycle-Dependent ORC Dimerization Cycle Regulates Eukaryotic Chromosomal DNA Replication, Cell Rep, 30 (2020) 3323–3338.e3326. [DOI] [PubMed] [Google Scholar]
- [193].Luo KQ, Elsasser S, Chang DC, Campbell JL, Regulation of the localization and stability of Cdc6 in living yeast cells, Biochemical and biophysical research communications, 306 (2003) 851–859. [DOI] [PubMed] [Google Scholar]
- [194].Piatti S, Lengauer C, Nasmyth K, Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae, Embo j, 14 (1995) 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Drury LS, Perkins G, Diffley JF, The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast, Embo j, 16 (1997) 5966–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Perkins G, Drury LS, Diffley JF, Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis, Embo j, 20 (2001) 4836–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Mimura S, Seki T, Tanaka S, Diffley JF, Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control, Nature, 431 (2004) 1118–1123. [DOI] [PubMed] [Google Scholar]
- [198].Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Lazzerini Denchi E, Gieffers C, Matteucci C, Peters JM, Helin K, Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1, Genes Dev, 14 (2000) 2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Mailand N, Diffley JF, CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis, Cell, 122 (2005) 915–926. [DOI] [PubMed] [Google Scholar]
- [200].Jiang W, Wells NJ, Hunter T, Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6, Proc Natl Acad Sci U S A, 96 (1999) 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Petersen BO, Lukas J, Sørensen CS, Bartek J, Helin K, Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization, Embo j, 18 (1999) 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Alexandrow MG, Hamlin JL, Cdc6 Chromatin Affinity Is Unaffected by Serine-54 Phosphorylation, S-Phase Progression, and Overexpression of Cyclin A, Molecular and Cellular Biology, 24 (2004) 1614–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Fujita M, Yamada C, Goto H, Yokoyama N, Kuzushima K, Inagaki M, Tsurumi T, Cell cycle regulation of human CDC6 protein. Intracellular localization, interaction with the human mcm complex, and CDC2 kinase-mediated hyperphosphorylation, J Biol Chem, 274 (1999) 25927–25932. [DOI] [PubMed] [Google Scholar]
- [204].Coverley D, Pelizon C, Trewick S, Laskey RA, Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process, J Cell Sci, 113 ( Pt 11) (2000) 1929–1938. [DOI] [PubMed] [Google Scholar]
- [205].Walter D, Hoffmann S, Komseli ES, Rappsilber J, Gorgoulis V, Sørensen CS, SCF(Cyclin F)-dependent degradation of CDC6 suppresses DNA re-replication, Nat Commun, 7 (2016) 10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Tanaka S, Diffley JF, Interdependent nuclear accumulation of budding yeast Cdt1 and MCM2-7 during G1 phase, Nat Cell Biol, 4 (2002) 198–207. [DOI] [PubMed] [Google Scholar]
- [207].Nishitani H, Taraviras S, Lygerou Z, Nishimoto T, The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase, J Biol Chem, 276 (2001) 44905–44911. [DOI] [PubMed] [Google Scholar]
- [208].Zhong W, Feng H, Santiago FE, Kipreos ET, CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing, Nature, 423 (2003) 885–889. [DOI] [PubMed] [Google Scholar]
- [209].Nishitani H, Lygerou Z, Nishimoto T, Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region, J Biol Chem, 279 (2004) 30807–30816. [DOI] [PubMed] [Google Scholar]
- [210].Li X, Zhao Q, Liao R, Sun P, Wu X, The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation, J Biol Chem, 278 (2003) 30854–30858. [DOI] [PubMed] [Google Scholar]
- [211].Liu E, Li X, Yan F, Zhao Q, Wu X, Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation, J Biol Chem, 279 (2004) 17283–17288. [DOI] [PubMed] [Google Scholar]
- [212].Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, Lygerou Z, Nishimoto T, Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis, Embo j, 25 (2006) 1126–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Sugimoto N, Tatsumi Y, Tsurumi T, Matsukage A, Kiyono T, Nishitani H, Fujita M, Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding, J Biol Chem, 279 (2004) 19691–19697. [DOI] [PubMed] [Google Scholar]
- [214].Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A, PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination, J Biol Chem, 281 (2006) 6246–6252. [DOI] [PubMed] [Google Scholar]
- [215].Arias EE, Walter JC, PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication, Nat Cell Biol, 8 (2006) 84–90. [DOI] [PubMed] [Google Scholar]
- [216].Havens CG, Walter JC, Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2, Mol Cell, 35 (2009) 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Cook JG, Chasse DA, Nevins JR, The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells, J Biol Chem, 279 (2004) 9625–9633. [DOI] [PubMed] [Google Scholar]
- [218].Tada S, Li A, Maiorano D, Méchali M, Blow JJ, Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin, Nat Cell Biol, 3 (2001) 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Quinn LM, Herr A, McGarry TJ, Richardson H, The Drosophila Geminin homolog: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis, Genes Dev, 15 (2001) 2741–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Lutzmann M, Maiorano D, Méchali M, A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus, Embo j, 25 (2006) 5764–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, Lygerou Z, Medema RH, Blow JJ, Svergun DI, Taraviras S, Perrakis A, Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing, Proc Natl Acad Sci U S A, 106 (2009) 19807–19812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].McGarry TJ, Kirschner MW, Geminin, an inhibitor of DNA replication, is degraded during mitosis, Cell, 93 (1998) 1043–1053. [DOI] [PubMed] [Google Scholar]
- [223].Zhou Y, Pozo PN, Oh S, Stone HM, Cook JG, Distinct and sequential re-replication barriers ensure precise genome duplication, PLoS Genet, 16 (2020) e1008988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Zeman MK, Cimprich KA, Causes and consequences of replication stress, Nat Cell Biol, 16 (2014) 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Gaillard H, García-Muse T, Aguilera A, Replication stress and cancer, Nature reviews. Cancer, 15 (2015) 276–289. [DOI] [PubMed] [Google Scholar]
- [226].Muñoz S, Méndez J, DNA replication stress: from molecular mechanisms to human disease, Chromosoma, 126 (2017) 1–15. [DOI] [PubMed] [Google Scholar]
- [227].Kim JC, Mirkin SM, The balancing act of DNA repeat expansions, Current opinion in genetics & development, 23 (2013) 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Bochman ML, Paeschke K, Zakian VA, DNA secondary structures: stability and function of G-quadruplex structures, Nat Rev Genet, 13 (2012) 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA, Pif1 family helicases suppress genome instability at G-quadruplex motifs, Nature, 497 (2013) 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Higa M, Fujita M, Yoshida K, DNA Replication Origins and Fork Progression at Mammalian Telomeres, Genes, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Black EM, Giunta S, Repetitive Fragile Sites: Centromere Satellite DNA As a Source of Genome Instability in Human Diseases, Genes, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].García-Muse T, Aguilera A, R Loops: From Physiological to Pathological Roles, Cell, 179 (2019) 604–618. [DOI] [PubMed] [Google Scholar]
- [233].Gómez-González B, Aguilera A, Transcription-mediated replication hindrance: a major driver of genome instability, Genes Dev, 33 (2019) 1008–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Voutsinos V, Munk SHN, Oestergaard VH, Common Chromosomal Fragile Sites-Conserved Failure Stories, Genes, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Feng W, Chakraborty A, Fragility Extraordinaire: Unsolved Mysteries of Chromosome Fragile Sites, Advances in experimental medicine and biology, 1042 (2017) 489–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Brison O, El-Hilali S, Azar D, Koundrioukoff S, Schmidt M, Nähse V, Jaszczyszyn Y, Lachages AM, Dutrillaux B, Thermes C, Debatisse M, Chen CL, Transcription-mediated organization of the replication initiation program across large genes sets common fragile sites genome-wide, Nat Commun, 10 (2019) 5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Miotto B, Ji Z, Struhl K, Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers, Proc Natl Acad Sci U S A, 113 (2016) E4810–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Sugimoto N, Maehara K, Yoshida K, Ohkawa Y, Fujita M, Genome-wide analysis of the spatiotemporal regulation of firing and dormant replication origins in human cells, Nucleic Acids Res, 46 (2018) 6683–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Primo LMF, Teixeira LK, DNA replication stress: oncogenes in the spotlight, Genetics and molecular biology, 43 (2019) e20190138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Petropoulos M, Champeris Tsaniras S, Taraviras S, Lygerou Z, Replication Licensing Aberrations, Replication Stress, and Genomic Instability, Trends Biochem Sci, 44 (2019) 752–764. [DOI] [PubMed] [Google Scholar]
- [241].Spruck CH, Won KA, Reed SI, Deregulated cyclin E induces chromosome instability, Nature, 401 (1999) 297–300. [DOI] [PubMed] [Google Scholar]
- [242].Ekholm-Reed S, Méndez J, Tedesco D, Zetterberg A, Stillman B, Reed SI, Deregulation of cyclin E in human cells interferes with prereplication complex assembly, J Cell Biol, 165 (2004) 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B, Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites, Mol Cell, 43 (2011) 122–131. [DOI] [PubMed] [Google Scholar]
- [244].Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B, Nucleotide deficiency promotes genomic instability in early stages of cancer development, Cell, 145 (2011) 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R, Non-transcriptional control of DNA replication by c-Myc, Nature, 448 (2007) 445–451. [DOI] [PubMed] [Google Scholar]
- [246].Jones RM, Mortusewicz O, Afzal I, Lorvellec M, García P, Helleday T, Petermann E, Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress, Oncogene, 32 (2013) 3744–3753. [DOI] [PubMed] [Google Scholar]
- [247].Macheret M, Halazonetis TD, Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress, Nature, 555 (2018) 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Topal S, Van C, Xue Y, Carey MF, Peterson CL, INO80C Remodeler Maintains Genomic Stability by Preventing Promiscuous Transcription at Replication Origins, Cell Rep, 32 (2020) 108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Branzei D, Foiani M, Maintaining genome stability at the replication fork, Nature reviews. Molecular cell biology, 11 (2010) 208–219. [DOI] [PubMed] [Google Scholar]
- [250].Cejka P, DNA End Resection: Nucleases Team Up with the Right Partners to Initiate Homologous Recombination, J Biol Chem, 290 (2015) 22931–22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Marechal A, Zou L, RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response, Cell Res, 25 (2015) 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Chapman JR, Taylor MR, Boulton SJ, Playing the end game: DNA double-strand break repair pathway choice, Mol Cell, 47 (2012) 497–510. [DOI] [PubMed] [Google Scholar]
- [253].Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA, Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint, Genes Dev, 19 (2005) 1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Li Z, Pearlman AH, Hsieh P, DNA mismatch repair and the DNA damage response, DNA Repair (Amst), 38 (2016) 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Novarina D, Amara F, Lazzaro F, Plevani P, Muzi-Falconi M, Mind the gap: keeping UV lesions in check, DNA Repair (Amst), 10 (2011) 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Zou L, Elledge SJ, Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes, Science (New York, N.Y.), 300 (2003) 1542–1548. [DOI] [PubMed] [Google Scholar]
- [257].Kumagai A, Lee J, Yoo HY, Dunphy WG, TopBP1 activates the ATR-ATRIP complex, Cell, 124 (2006) 943–955. [DOI] [PubMed] [Google Scholar]
- [258].Mordes DA, Glick GG, Zhao R, Cortez D, TopBP1 activates ATR through ATRIP and a PIKK regulatory domain, Genes Dev, 22 (2008) 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM, The Rad9-Hus1-Rad1 (9–1-1) clamp activates checkpoint signaling via TopBP1, Genes Dev, 21 (2007) 1472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Lee J, Kumagai A, Dunphy WG, The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR, J Biol Chem, 282 (2007) 28036–28044. [DOI] [PubMed] [Google Scholar]
- [261].Bass TE, Luzwick JW, Kavanaugh G, Carroll C, Dungrawala H, Glick GG, Feldkamp MD, Putney R, Chazin WJ, Cortez D, ETAA1 acts at stalled replication forks to maintain genome integrity, Nature Cell Biology, 18 (2016) 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Haahr P, Hoffmann S, Tollenaere MA, Ho T, Toledo LI, Mann M, Bekker-Jensen S, Raschle M, Mailand N, Activation of the ATR kinase by the RPA-binding protein ETAA1, Nat Cell Biol, 18 (2016) 1196–1207. [DOI] [PubMed] [Google Scholar]
- [263].Lee YC, Zhou Q, Chen J, Yuan J, RPA-Binding Protein ETAA1 Is an ATR Activator Involved in DNA Replication Stress Response, Current biology : CB, 26 (2016) 3257–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Zhao H, Piwnica-Worms H, ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1, Mol Cell Biol, 21 (2001) 4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ, ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage, Science (New York, N.Y.), 316 (2007) 1160–1166. [DOI] [PubMed] [Google Scholar]
- [266].Araki H, Leem SH, Phongdara A, Sugino A, Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint, Proc Natl Acad Sci U S A, 92 (1995) 11791–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Navas TA, Zhou Z, Elledge SJ, DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint, Cell, 80 (1995) 29–39. [DOI] [PubMed] [Google Scholar]
- [268].Navas TA, Sanchez Y, Elledge SJ, RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae, Genes Dev, 10 (1996) 2632–2643. [DOI] [PubMed] [Google Scholar]
- [269].Feng W, Rodriguez-Menocal L, Tolun G, D’Urso G, Schizosacchromyces pombe Dpb2 binds to origin DNA early in S phase and is required for chromosomal DNA replication, Mol Biol Cell, 14 (2003) 3427–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Lopez-Mosqueda J, Maas NL, Jonsson ZO, Defazio-Eli LG, Wohlschlegel J, Toczyski DP, Damage-induced phosphorylation of Sld3 is important to block late origin firing, Nature, 467 (2010) 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J, An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication, Mol Cell, 11 (2003) 203–213. [DOI] [PubMed] [Google Scholar]
- [272].Zegerman P, Diffley JF, Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation, Nature, 467 (2010) 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Chen YH, Jones MJ, Yin Y, Crist SB, Colnaghi L, Sims RJ 3rd, Rothenberg E, Jallepalli PV, Huang TT, ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress, Mol Cell, 58 (2015) 323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Morafraile EC, Hänni C, Allen G, Zeisner T, Clarke C, Johnson MC, Santos MM, Carroll L, Minchell NE, Baxter J, Banks P, Lydall D, Zegerman P, Checkpoint inhibition of origin firing prevents DNA topological stress, Genes Dev, 33 (2019) 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Ge XQ, Blow JJ, Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories, J Cell Biol, 191 (2010) 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Shechter D, Costanzo V, Gautier J, ATR and ATM regulate the timing of DNA replication origin firing, Nat Cell Biol, 6 (2004) 648–655. [DOI] [PubMed] [Google Scholar]
- [277].Guo C, Kumagai A, Schlacher K, Shevchenko A, Shevchenko A, Dunphy WG, Interaction of Chk1 with Treslin negatively regulates the initiation of chromosomal DNA replication, Mol Cell, 57 (2015) 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Moiseeva TN, Yin Y, Calderon MJ, Qian C, Schamus-Haynes S, Sugitani N, Osmanbeyoglu HU, Rothenberg E, Watkins SC, Bakkenist CJ, An ATR and CHK1 kinase signaling mechanism that limits origin firing during unperturbed DNA replication, Proc Natl Acad Sci U S A, 116 (2019) 13374–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Marheineke K, Hyrien O, Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint, J Biol Chem, 279 (2004) 28071–28081. [DOI] [PubMed] [Google Scholar]
- [280].Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA, Chk1 regulates the density of active replication origins during the vertebrate S phase, Embo j, 26 (2007) 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M, Cyclin A-Cdk1 regulates the origin firing program in mammalian cells, Proc Natl Acad Sci U S A, 106 (2009) 3184–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Yoshida K, Sugimoto N, Iwahori S, Yugawa T, Narisawa-Saito M, Kiyono T, Fujita M, CDC6 interaction with ATR regulates activation of a replication checkpoint in higher eukaryotic cells, J Cell Sci, 123 (2010) 225–235. [DOI] [PubMed] [Google Scholar]