ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) is a multidrug-resistant pathogen of acute clinical importance. Combination treatment with an FtsZ inhibitor potentiates the activity of penicillin binding protein (PBP)-targeting β-lactam antibiotics against MRSA. To explore the mechanism underlying this synergistic behavior, we examined the impact of treatment with the FtsZ inhibitor TXA707 on the spatial localization of the five PBP proteins expressed in MRSA. In the absence of drug treatment, PBP1, PBP2, PBP3, and PBP4 colocalize with FtsZ at the septum, contributing to new cell wall formation. In contrast, PBP2a localizes to distinct foci along the cell periphery. Upon treatment with TXA707, septum formation becomes disrupted, and FtsZ relocalizes away from midcell. PBP1 and PBP3 remain significantly colocalized with FtsZ, while PBP2, PBP4, and PBP2a localize away from FtsZ to specific sites along the periphery of the enlarged cells. We also examined the impact on PBP2a and PBP2 localization of treatment with β-lactam antibiotic oxacillin alone and in synergistic combination with TXA707. Significantly, PBP2a localizes to the septum in approximately 15% of the oxacillin-treated cells, a behavior that likely contributes to the β-lactam resistance of MRSA. Combination treatment with TXA707 causes both PBP2a and PBP2 to localize in malformed septum-like structures. Our collective results suggest that PBP2, PBP4, and PBP2a may function collaboratively in peripheral cell wall repair and maintenance in response to FtsZ inhibition by TXA707. Cotreatment with oxacillin appears to reduce the availability of PBP2a to assist in this repair, thereby rendering the MRSA cells more susceptible to the β-lactam.
IMPORTANCE MRSA is a multidrug-resistant bacterial pathogen of acute clinical importance, infecting many thousands of individuals globally each year. The essential cell division protein FtsZ has been identified as an appealing target for the development of new drugs to combat MRSA infections. Through synergistic actions, FtsZ-targeting agents can sensitize MRSA to antibiotics like the β-lactams that would otherwise be ineffective. This study provides key insights into the mechanism underlying this synergistic behavior as well as MRSA resistance to β-lactam drugs. The results of this work will help guide the identification and optimization of combination drug regimens that can effectively treat MRSA infections and reduce the potential for future resistance.
KEYWORDS: FtsZ inhibitor, MRSA, PBP2a, anti-PBP2a antibody, antibacterial synergy, fluorescence microscopy, oxacillin, penicillin-binding proteins, protein localization, transmission electron microscopy
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is a multidrug-resistant bacterial pathogen that infects more than 120,000 individuals and causes nearly 20,000 deaths each year in the United States (1). Growing resistance to current standard-of-care antibiotics (2–4) has highlighted a critical need to develop new antibiotics that can address the MRSA threat (5–7). Toward this goal, recent efforts have been focused on the development of novel small-molecule chemotypes that target bacterial cell division (8–12). These efforts have resulted in the identification and development of lead compounds that target the essential cell division protein, FtsZ (11, 13–23). Treatment of MRSA with such FtsZ inhibitors disrupts bacterial cell division and induces cell death (14–18, 20, 23–26).
In almost all bacterial species, FtsZ plays a critical role in cell division by acting as a scaffold for recruitment of other components of the cell division machinery (the divisome) (27–29). FtsZ forms this scaffold by polymerizing at midcell and adopting a dynamic ring structure termed the Z-ring (30–32). Upon formation of the Z-ring, the divisome machinery assembles with the penicillin binding proteins (PBPs) being among the key proteins that are recruited. These proteins are involved in the synthesis of new cell walls at the septum (33). Methicillin-sensitive S. aureus (MSSA) expresses a total of four PBP proteins (PBP1, PBP2, PBP3, and PBP4), of which PBP1 and PBP2 have been identified as essential proteins (34). The PBP proteins are targeted by β-lactam antibiotics, which disable the transpeptidase domains of the proteins and thereby interfere with PBP-induced cross-linking of the bacterial cell wall (35, 36). MRSA expresses an additional PBP (PBP2a) that underlies the resistance of MRSA to most β-lactam antibiotics (37). The affinity of PBP2a for these β-lactam antibiotics is significantly reduced compared to the other PBPs (38, 39). Cotreatment with FtsZ inhibitors can repurpose β-lactam antibiotics for use against MRSA (13, 15, 25, 40, 41), with such combination treatments being associated with the greatest degree of synergy when the β-lactam targets PBP2 with high affinity (41).
The mechanism underlying the potentiation of β-lactam activity against MRSA by FtsZ inhibitors is unclear. Here, we examine the impact of treatment with the FtsZ inhibitor TXA707 (18) on the localization of the five PBPs expressed in MRSA. Toward this goal, we generated a series of MRSA strains expressing fluorescent fusion forms of FtsZ and either PBP1, PBP2, PBP3, or PBP4. We also developed an immunofluorescence approach to monitoring the localization of PBP2a. Our results shed light on how FtsZ inhibitors potentiate β-lactam antibiotic activity versus MRSA as well as on the mechanism by which PBP2a facilitates MRSA survival in response to β-lactam antibiotic exposure.
RESULTS
Impact of the genetic modifications on the growth and antibiotic sensitivity profile of MRSA LAC.
After constructing our genetically modified MRSA LAC strains, we first sought to explore whether our genetic alterations affected cell fitness and antibiotic sensitivity. To this end, we monitored the time-dependent growth of each genetically modified strain and compared the resulting growth curves to that of the wild-type LAC strain. Significantly, the growth rates of the genetically modified strains were similar to that of the wild-type strain (see Fig. S1 in the supplemental material), confirming that our genetic alterations did not alter the fitness of the MRSA cells. We also examined the sensitivity of our genetically modified strains to treatment with TXA707, vancomycin, and oxacillin, as well as with the cephalosporins ceftriaxone, cefotaxime, and cephalexin. All six agents were associated with similar MICs against wild-type MRSA LAC and strains expressing FtsZ-mCherry (LAC-FCh) alone or both FtsZ-mCherry and either superfolder green fluorescent protein (sfGFP)-PBP1 (LAC-FChP1GFP), sfGFP-PBP2 (LAC-FChP2GFP), sfGFP-PBP3 (LAC-FChP3GFP), or PBP4-sfGFP (LAC-FChP4GFP) (see Table S1), indicating that these genetic alterations had no impact on antibiotic sensitivity. In contrast, MRSA LAC cells expressing either PBP2a-mCherry (LAC-P2ACh-1) or mCherry-PBP2a (LAC-P2ACh-2) lost their resistance to oxacillin, ceftriaxone, cefotaxime, and cephalexin, with the MICs for oxacillin, ceftriaxone, cefotaxime, and cephalexin against these strains being 0.125, 4, 2, and 4 μg/ml, respectively (compared to corresponding MIC values of 64, 512, 128, and 128 μg/ml, respectively, against the wild-type strain). Significantly, the MIC values determined for oxacillin, ceftriaxone, cefotaxime, and cephalexin against the MRSA LAC-P2ACh-1 and LAC-P2ACh-2 strains are comparable to those determined for the four β-lactam antibiotics against the MSSA RN4220 strain (Table S1). Thus, the PBP2a-mCherry and mCherry-PBP2a proteins were nonfunctional, transforming the MRSA strains expressing these conjugate proteins into MSSA strains. These observations are consistent with previous studies by the Pinho group indicating that appending amino acids to the C or N termini of PBP2a was deleterious to the localization and function of the protein (42). We therefore used immunofluorescence for visualizing the localization of PBP2a (as detailed below).
Impact of TXA707 treatment on MRSA cell morphology and the localization of FtsZ.
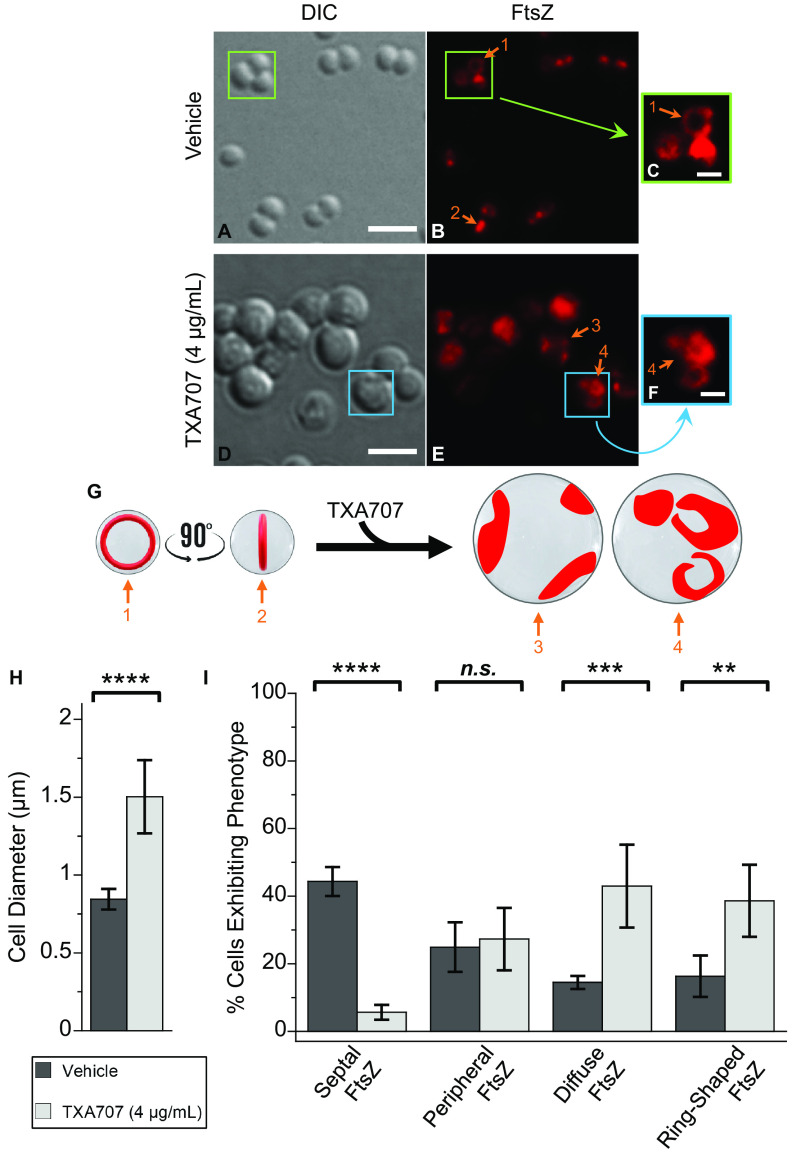
We sought to determine the impact of TXA707 treatment on FtsZ localization in MRSA LAC-FCh cells expressing FtsZ-mCherry. For this purpose, cells were treated with either vehicle (dimethyl sulfoxide [DMSO]) or 4 μg/ml TXA707 (4× MIC) for 3 h prior to visualization by total internal reflection fluorescence (TIRF) microscopy. Vehicle-treated cells (n = 641) maintained a normal average diameter of approximately 0.84 ± 0.07 μm (Fig. 1A and H). Among the 641 vehicle-treated cells analyzed, FtsZ is localized to the septum at midcell in 44% of them (Fig. 1I). In 16% of the analyzed cells, FtsZ forms visible Z-rings (as shown in Fig. 1B and C and schematically depicted in Fig. 1G). The remainder of the analyzed cells have FtsZ localized to the cell periphery (25%) or in a diffuse pattern throughout the cell (14%).
FIG 1.
Differential interference contrast (DIC) and fluorescence micrographs of MRSA LAC-FCh cells treated for 3 h with either DMSO vehicle (A to C) or 4 μg/ml (4× MIC) TXA707 (D to F) just prior to visualization. (C and F) Enlargements of the regions enclosed by the green and cyan boxes in panels B and E, respectively. (G) Localization of FtsZ (red) is schematically depicted, with the numbered arrows in the schematic depiction reflecting the correspondingly numbered arrows in the fluorescence micrographs. The scale bars for panels A to B and D to E represent 2 μm, while those for panels C and F represent 0.5 μm. (H) Bar graph showing the average diameter of the vehicle-treated cells (n = 641) as well as the TXA707-treated cells (n = 349). (I) Bar graph showing the prevalence of the various FtsZ phenotypes observed in both vehicle- and TXA707-treated cells. Each percentage reflects an average of 5 different fields of view, with the number of cells in each field of view ranging from 56 to 171. In both panels H and I, the indicated error bars reflect the standard deviation from the mean. The statistical significance of differences in cell diameter and FtsZ phenotype were analyzed using a one-way analysis of variance (ANOVA) test. For all figures: ****, P < 0.0001; ***, 0.0001 < P < 0.001; **, 0.001 < P < 0.01; *, 0.01 < P < 0.1.; n.s., not significant, as reflected by a P value of >0.1.
In marked contrast to vehicle-treated cells, cells treated with TXA707 (n = 349) almost double in size, with an average diameter of 1.50 ± 0.24 μm (Fig. 1D and H). Moreover, FtsZ is no longer predominantly localized to the septum at midcell, with only 6% of the analyzed cells exhibiting this phenotype compared to 44% of vehicle-treated cells (Fig. 1I). Instead, 43% of the TXA707-treated cells have FtsZ localized in a diffuse pattern throughout the cell (compared to only 14% of vehicle-treated cells with this phenotype). In addition, 39% of the 349 TXA707-treated cells analyzed exhibit a phenotype in which FtsZ adopts multiple ring-shaped structures inside the cell (shown in Fig. 1E and F and schematically depicted in Fig. 1G), with an average diameter of 0.52 ± 0.12 μm. Thus, treatment with TXA707 increases the percentage of cells in which FtsZ has formed visible ring-shaped structures by 2.4-fold (from 16% to 39%) relative to vehicle treatment (Fig. 1I). Relative to vehicle, treatment with TXA707 does not induce a significant change in the percentage of cells in which FtsZ is localized to the cell periphery (from 25% to 27%).
Impact of TXA707 treatment on the localization of FtsZ relative to that of PBP1, PBP2, PBP3, PBP4, or PBP2a in MRSA.
To understand the impact of FtsZ inhibition on the localization of the five PBPs in MRSA, we generated MRSA LAC strains expressing both FtsZ-mCherry as well as sfGFP fusion proteins of PBP1, PBP2, PBP3, and PBP4 (LAC-FChP1GFP, LAC-FChP2GFP, LAC-FChP3GFP, and LAC-FChP4GFP, respectively). In addition, we also developed an immunofluorescence approach to monitoring the impact of FtsZ inhibition on the localization of PBP2a in the LAC-FCh strain. The sections that follow describe our resulting characterizations.
(i) PBP1.
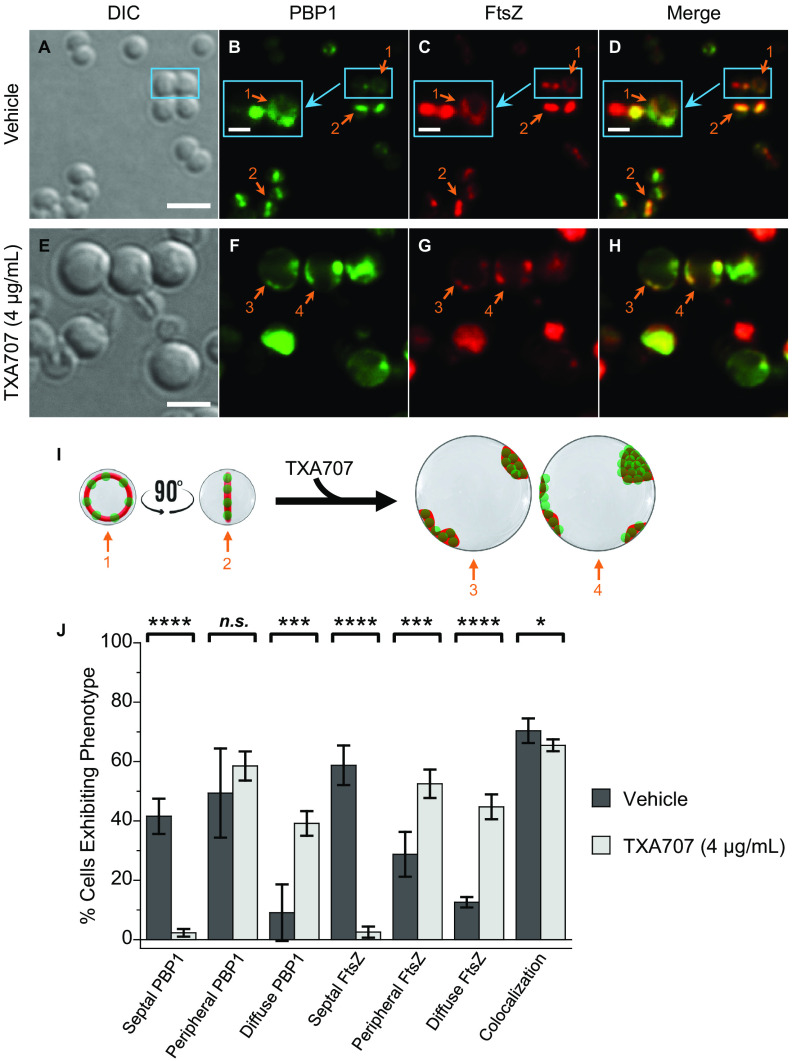
Our initial characterizations explored the impact of TXA707 treatment on MRSA LAC cells expressing FtsZ-mCherry and sfGFP-PBP1 (LAC-FChP1GFP) at its native locus. In vehicle-treated LAC-FChP1GFP cells (n = 919), PBP1 and FtsZ are localized to the septum at midcell in 42% and 59% of the cells, respectively (see Fig. 2B, D, and I to J). Most of the nonseptally localized PBP1 and FtsZ are localized to the cell periphery, with these phenotypes occurring in 49% and 29% of the analyzed cells, respectively (Fig. 2J). The remainder of the vehicle-treated cells have PBP1 and FtsZ localized in a more diffuse pattern (9% of the cells in the case of PBP1 and 13% of cells in the case of FtsZ). Significantly, PBP1 and FtsZ colocalize in the majority (70%) of the vehicle-treated LAC-FChP1GFP cells (Fig. 2J), with much of this colocalization occurring at the septum and FtsZ Z-ring structures (as depicted in Fig. 2D and I).
FIG 2.
(A to H) DIC and fluorescence micrographs of MRSA LAC-FChP1GFP cells treated for 3 h with either DMSO vehicle (A to D) or 4 μg/ml (4× MIC) TXA707 (E to H) just prior to visualization. (B to D) Insets depict enlargements of the regions enclosed by the small cyan boxes. (I) Localization of PBP1 (green) and FtsZ (red) is schematically depicted, with the numbered arrows in the scheme reflecting the correspondingly numbered arrows in the florescence micrographs. Scale bars for panels A to H represent 2 μm, while those for the insets in panels Bto D represent 0.5 μm. (J) Bar graph showing the prevalence of the various FtsZ and PBP1 phenotypes observed in both vehicle-treated cells (n = 919) and TXA707-treated cells (n = 535). Each percentage reflects an average of 5 different fields of view, with the number of cells in each field of view ranging from 98 to 256. The indicated error bars reflect the standard deviation from the mean. The statistical significance of differences in the FtsZ and PBP1 phenotypes were analyzed as described in the legend to Fig. 1. n.s., not significant.
Upon treatment of LAC-FChP1GFP cells (n = 535) with TXA707, the average cell diameter increases approximately 2.1-fold from 0.84 ± 0.09 to 1.77 ± 0.27 μm (Fig. 2A and E and Fig. S2), consistent with our observations described above for LAC-FCh cells. Moreover, TXA707 treatment markedly reduces the percentage of LAC-FChP1GFP cells exhibiting septally localized PBP1 from 49% to 2% (Fig. 2J). Instead, PBP1 localizes to distinct foci across the cell periphery in the majority (59%) of the TXA707-treated cells analyzed (Fig. 2F and J). Interestingly, FtsZ appears to colocalize to those same peripheral foci (Fig. 2G to I). Relative to vehicle, TXA707 treatment is also associated with a significant increase in the percentage of cells exhibiting a diffuse localization phenotype for both PBP1 (from 9% to 39%) and FtsZ (from 13% to 45%) (Fig. 2J). In all, PBP1 and FtsZ colocalize in 65% of the TXA707-treated cells, a behavior consistent with the two proteins undergoing a similar pattern of mislocalization in response to the FtsZ inhibitor.
(ii) PBP3.
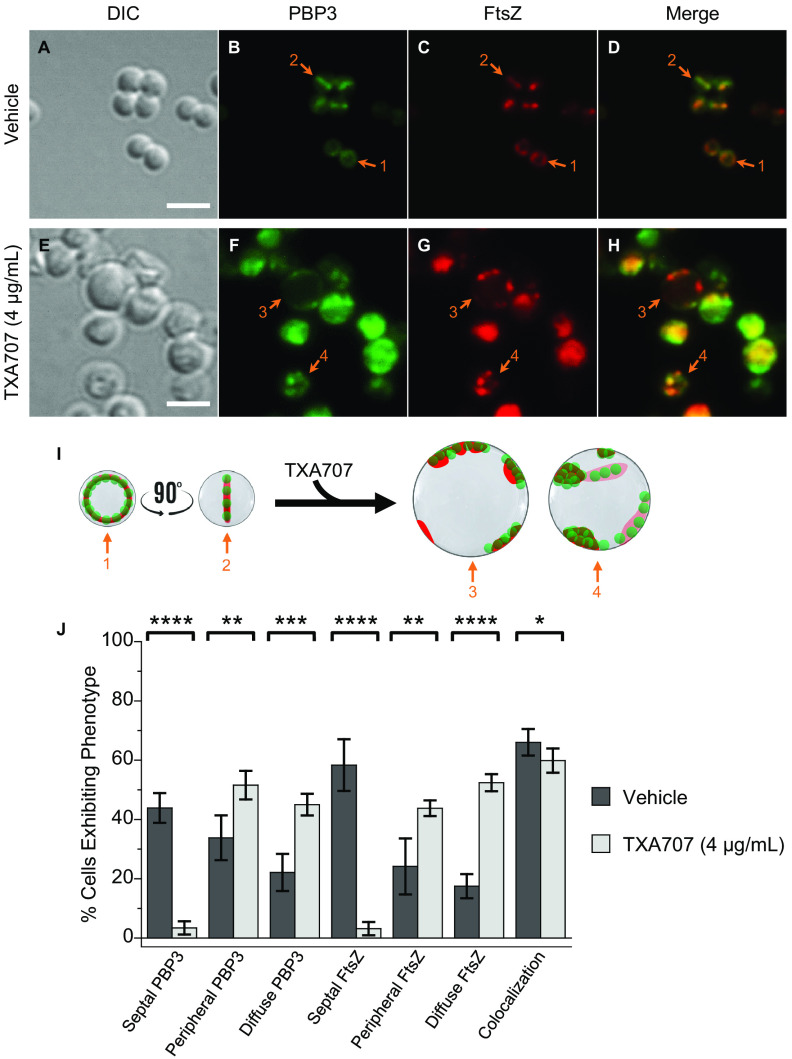
We next explored the impact of TXA707 treatment on MRSA LAC cells expressing FtsZ-mCherry and sfGFP-PBP3 (LAC-FChP3GFP) at its native locus. A total of 545 vehicle-treated and 326 TXA707-treated LAC-FChP3GFP cells were analyzed. This analysis revealed similar effects on cell diameter (Fig. S2) as well as on PBP3 and FtsZ localization phenotype (Fig. 3) to those observed in response to TXA707 treatment of LAC-FChP1GFP cells. This behavior included marked TXA707-induced reductions in the septal localization of both PBP3 (from 44% to 3% of cells) and FtsZ (from 58% to 3% of cells), coupled with significant colocalization (in 60% of TXA707-treated cells) of both PBP3 and FtsZ present in diffuse and peripheral localization phenotypes (Fig. 3H to J). These collective results suggest that the FtsZ inhibitor induces a similar pattern of mislocalization between FtsZ and not only PBP1 but also PBP3.
FIG 3.
(A to H) DIC and fluorescence micrographs of MRSA LAC-FChP3GFP cells treated as described in the legend for panels A to H in Fig. 2. (I) Localization of PBP3 (green) and FtsZ (red) is schematically depicted, with the numbered arrows in the scheme reflecting the correspondingly numbered arrows in the florescence micrographs. Scale bars for panels A to H represent 2 μm. (J) Bar graph showing the prevalence of the various FtsZ and PBP3 phenotypes observed in both vehicle-treated cells (n = 545) and TXA707-treated cells (n = 326). Each percentage reflects an average of 5 different fields of view, with the number of cells in each field of view ranging from 48 to 136. The indicated error bars reflect the standard deviation from the mean. The statistical significance of differences in the FtsZ and PBP3 phenotypes were analyzed as described in the legend to Fig. 1.
(iii) PBP2.
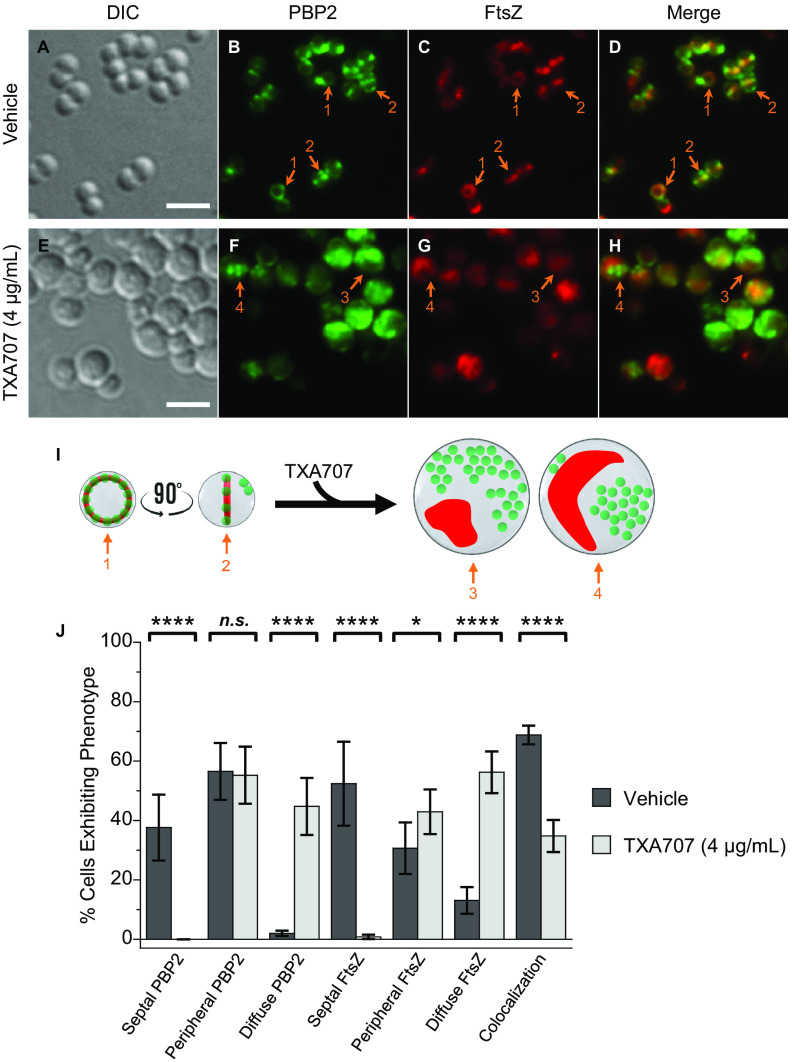
We also examined the impact of TXA707 treatment on MRSA LAC cells expressing FtsZ-mCherry and sfGFP-PBP2 (LAC-FChP2GFP) at its native locus, with a total of 1,071 vehicle-treated cells and 368 TXA707-treated cells being analyzed. The patterns of observed changes in cell diameter (Fig. 4A and E and Fig. S2) and FtsZ localization (Fig. 4C, G, I, and J) in response to TXA707 treatment were similar to those described above in our studies of LAC-FChP1GFP and LAC-FChP3GFP cells. In vehicle-treated LAC-FChP2GFP cells, PBP2 localizes to the septum at midcell in 38% of the analyzed cells and the cell periphery in 57% of the cells (Fig. 4B, F, and J). Very few (2%) of the vehicle-treated cells analyzed exhibit a phenotype in which PBP2 localizes in a diffuse pattern (Fig. 4J). PBP2 colocalizes with FtsZ in just over 2/3 (69%) of the vehicle-treated cells (Fig. 4D, I, and J). Treatment with TXA707 induces a significant decrease (from 38% to 0.2%) in cells exhibiting septally localized PBP2 and a concomitant increase (from 2% to 45%) in cells exhibiting a diffuse PBP2 phenotype in which multiple foci of PBP2 are distributed throughout large regions of the cell (Fig. 4F, I, and J). The percentage of cells in which PBP2 is localized to foci along the cell periphery does not change significantly upon TXA707 treatment (Fig. 4J). While PBP2 and FtsZ colocalize in a significant percentage (69%) of vehicle-treated cells, the two proteins colocalize in only 35% of TXA707-treated cells (Fig. 4D, H, to J). Thus, contrary to our observations described above for PBP1 and PBP3, the mislocalization of PBP2 in response to TXA707 treatment appears independent of FtsZ mislocalization. A similar behavior for PBP2 was previously reported by Tan et al. upon treatment with the FtsZ inhibitor PC190723 (25).
FIG 4.
(A to H) DIC and fluorescence micrographs of MRSA LAC-FChP2GFP cells treated as described in the legend for panels A to H in Fig. 2. (I) Localization of PBP2 (green) and FtsZ (red) is schematically depicted, with the numbered arrows in the scheme reflecting the correspondingly numbered arrows in the florescence micrographs. Scale bars for panels A to H represent 2 μm. (J) Bar graph showing the prevalence of the various FtsZ and PBP2 phenotypes observed in both vehicle-treated cells (n = 1,071) and TXA707-treated cells (n = 368). Each percentage reflects an average of 5 different fields of view, with the number of cells in each field of view ranging from 44 to 247. The indicated error bars reflect the standard deviation from the mean. The statistical significance of differences in the FtsZ and PBP2 phenotypes were analyzed as described in the legend to Fig. 1. n.s., not significant.
(iv) PBP4.
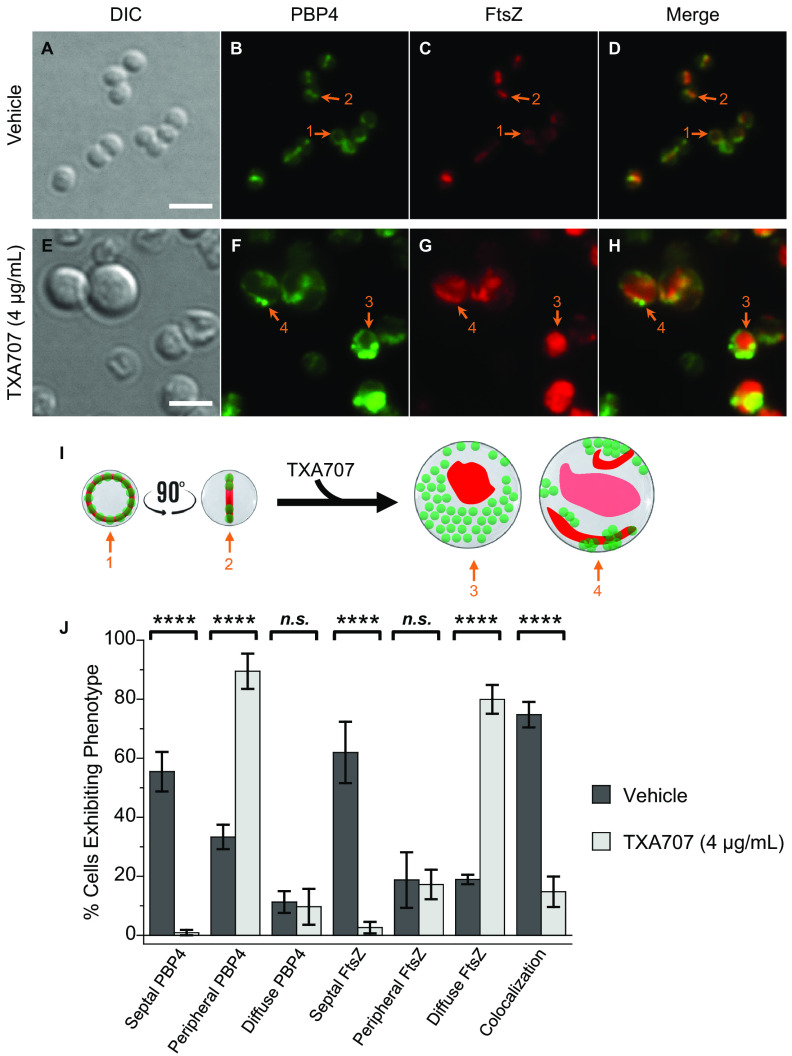
We next characterized the effects of vehicle relative to TXA707 treatment on MRSA LAC cells (n = 300 and 326, respectively) expressing FtsZ-mCherry and sfGFP-PBP4 (LAC-FChP4GFP) at its native locus. The overall impact of TXA707 treatment on LAC-FChP4GFP cell diameter (Fig. 5A and E and Fig. S2) and FtsZ localization (Fig. 5C, G, I, and J) was similar to that observed in LAC-FChP1GFP, LAC-FChP2GFP, and LAC-FChP3GFP cells. Like PBP1, PBP2, and PBP3, PBP4 localizes to the septum in a significant percentage (55%) of vehicle-treated cells while localizing in a more diffuse pattern in a much smaller percentage (11%) of cells (Fig. 5B, I, and J). PBP4 localizes to the cell periphery in approximately 1/3 (33%) of the vehicle-treated cells (Fig. 5J). The extent to which PBP4 colocalizes with FtsZ in vehicle-treated cells is significant, with 75% of the analyzed cells exhibiting this phenotype (Fig. 5D, I, and J).
FIG 5.
(A to H) DIC and fluorescence micrographs of MRSA LAC-FChP4GFP cells treated as described in the legend for panels A to H in Fig. 2. (I) Localization of PBP4 (green) and FtsZ (red) is schematically depicted, with the numbered arrows in the scheme reflecting the correspondingly numbered arrows in the florescence micrographs. Scale bars for panels A to H represent 2 μm. (J) Bar graph showing the prevalence of the various FtsZ and PBP4 phenotypes observed in both vehicle-treated cells (n = 300) and TXA707-treated cells (n = 326). Each percentage reflects an average of 3 to 5 different fields of view, with the number of cells in each field of view ranging from 56 to 162. The indicated error bars reflect the standard deviation from the mean. The statistical significance of differences in the FtsZ and PBP4 phenotypes were analyzed as described in the legend to Fig. 1. n.s., not significant.
Upon TXA707 treatment, PBP4 relocalizes away from the septum to distinct foci along the cell periphery (Fig. 5F, I, and J). In this connection, the percentage of cells exhibiting the septal PBP4 phenotype decreases from 55% to 0.9%, while the percentage of cells exhibiting the peripheral PBP4 phenotype increases from 33% to 89% (Fig. 5J). TXA707 treatment is also associated with a marked reduction in the observed colocalization of PBP4 and FtsZ, with the percentage of cells exhibiting this phenotype decreasing from 75% in vehicle-treated cells to 15% in TXA707-treated cells. Viewed as a whole, our characterizations of LAC-FChP1GFP, LAC-FChP2GFP, LAC-FChP3GFP, and LAC-FChP4GFP suggest that the mislocalization of FtsZ in response to treatment with the FtsZ inhibitor coincides with the mislocalization of PBP1 and PBP3 but appears independent of the mislocalization of PBP2 and PBP4.
(v) PBP2a.
We next explored the impact of TXA707 treatment on the relative localization of PBP2a and FtsZ in MRSA LAC cells expressing FtsZ-mCherry (LAC-FCh). We used an anti-MRSA monoclonal antibody and immunofluorescence to visualize PBP2a in these studies since fluorescent fusion forms of PBP2a were nonfunctional. Prior to utilizing the antibody in our immunofluorescence microscopy experiments, we verified its selectivity for PBP2a by Western blotting using purified PBP2a as well as cell lysates of MRSA LAC, MSSA RN4220, and Escherichia coli BL21(DE3) engineered to express S. aureus PBP1, PBP2, PBP3, PBP4, or PBP2a. Significantly, this analysis (the results of which are shown in Fig. S3) confirmed the selectivity of the anti-MRSA antibody for PBP2a, with no observed cross-reactivity with PBP1, PBP2, PBP3, or PBP4.
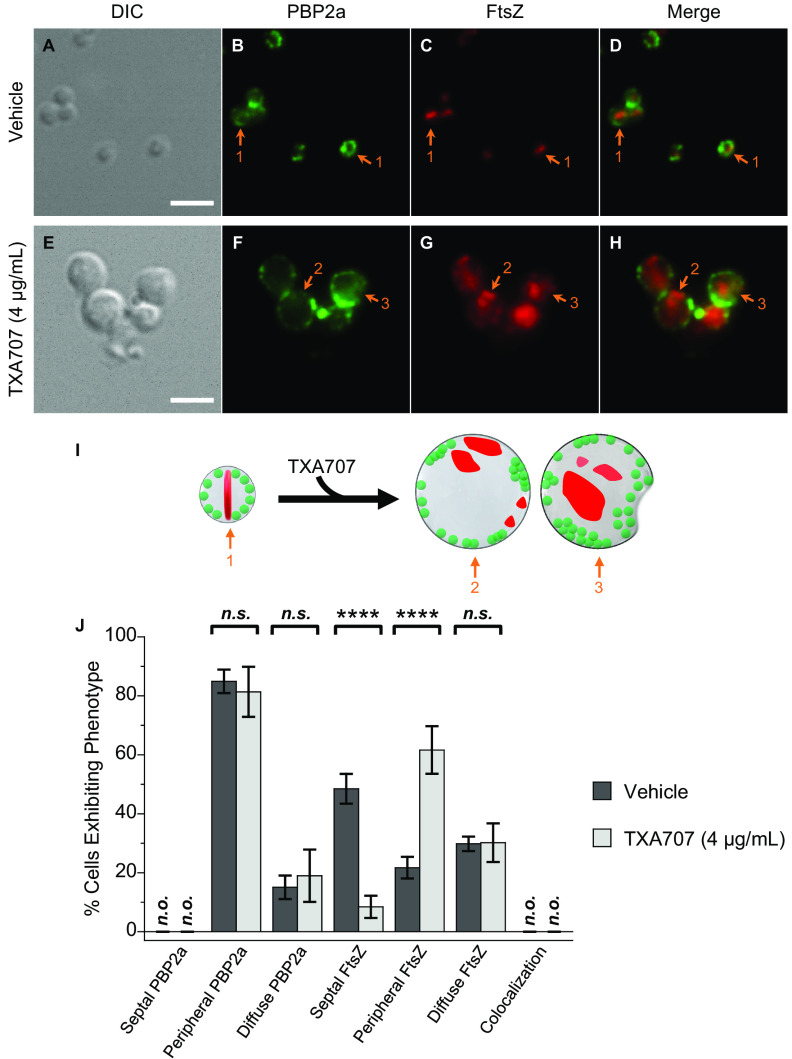
Figure 6 shows the results of our immunofluorescence microscopy studies probing the impact of TXA707 on the localization of PBP2a and FtsZ in LAC-FCh cells (n = 536 for vehicle treatment and 315 for TXA707 treatment). In marked contrast to the behavior of PBP1, PBP2, PBP3, and PBP4 in vehicle-treated cells, PBP2a does not localize to the septum at midcell, with none of the analyzed cells exhibiting this phenotype (Fig. 6B, I, and J). Instead, PBP2a localizes to distinct foci across the cell periphery in the vast majority (85%) of vehicle-treated cells and in a more diffuse pattern in the remaining cells. As expected, FtsZ localizes to the septum in a significant percentage (48%) of vehicle-treated cells while localizing to foci in the cell periphery or in a diffuse pattern in 22% and 30% of the cells, respectively (Fig. 6C, I, and J). Significantly, none of the 536 vehicle-treated cells analyzed exhibit a phenotype in which FtsZ and PBP2a are colocalized (Fig. 6D, I, and J).
FIG 6.
(A to H) DIC and fluorescence micrographs of MRSA LAC-FCh cells treated for 3 h with either DMSO vehicle (A to D) or 4 μg/ml (4× MIC) TXA707 (E to H), followed by immunostaining using a PBP2a-specific monoclonal mouse antibody and a goat anti-mouse Alexa Fluor 488-conjugated secondary antibody prior to visualization. (I) Localization of PBP2a (green) and FtsZ (red) is schematically depicted, with the numbered arrows in the scheme reflecting the correspondingly numbered arrows in the florescence micrographs. Scale bars for panels A to H represent 2 μm. (J) Bar graph showing the prevalence of the various FtsZ and PBP2a phenotypes observed in both vehicle-treated cells (n = 536) and TXA707-treated cells (n = 315). Each percentage reflects an average of 5 different fields of view, with the number of cells in each field of view ranging from 29 to 116. The indicated error bars reflect the standard deviation from the mean. The statistical significance of differences in the FtsZ and PBP2a phenotypes were analyzed as described in the legend to Fig. 1. n.s., not significant; n.o., none observed.
In response to TXA707 treatment, PBP2a localizes to an increased number of foci along the periphery of the enlarged cell (Fig. 6F and I), with the overall percentage of cells exhibiting peripherally localized PBP2a (81%) being similar to the 85% observed with vehicle treatment (Fig. 6J). As expected, the prevalence of cells in which FtsZ is localized to the septum decreases markedly (from 48% to 8%) with TXA707 treatment (Fig. 6C, G, I, and J). TXA707 treatment is also associated with FtsZ localization to foci in the cell periphery in 62% of the cells (Fig. 6J). As observed with vehicle treatment, none of the 315 TXA707-treated cells analyzed exhibit a phenotype in which FtsZ and PBP2a are colocalized (Fig. 6H to J). Even when both proteins are localized to the cell periphery, FtsZ and PBP2a appear to form foci distinct from one another (as exemplified by the cell denoted with arrow 2 in Fig. 6F to I). Like the behavior noted above for PBP2 and PBP4, the relocalization of PBP2a resulting from TXA707 treatment appears to be independent of the corresponding relocalization of FtsZ.
Impact of treatment with the synergistic combination of TXA707 and oxacillin on cell morphology as well as on the localization of FtsZ, PBP2a, and PBP2.
FtsZ inhibitors have been shown to sensitize MRSA to β-lactam antibiotics, resulting in a synergistic antibacterial effect (13, 15, 25, 40, 41). We sought to examine how treatment of MRSA LAC-FCh cells with a sub-MIC of TXA707 alone, oxacillin alone, or a combination of both TXA707 and oxacillin impacts cell morphology as well as the localization of FtsZ and PBP2a.
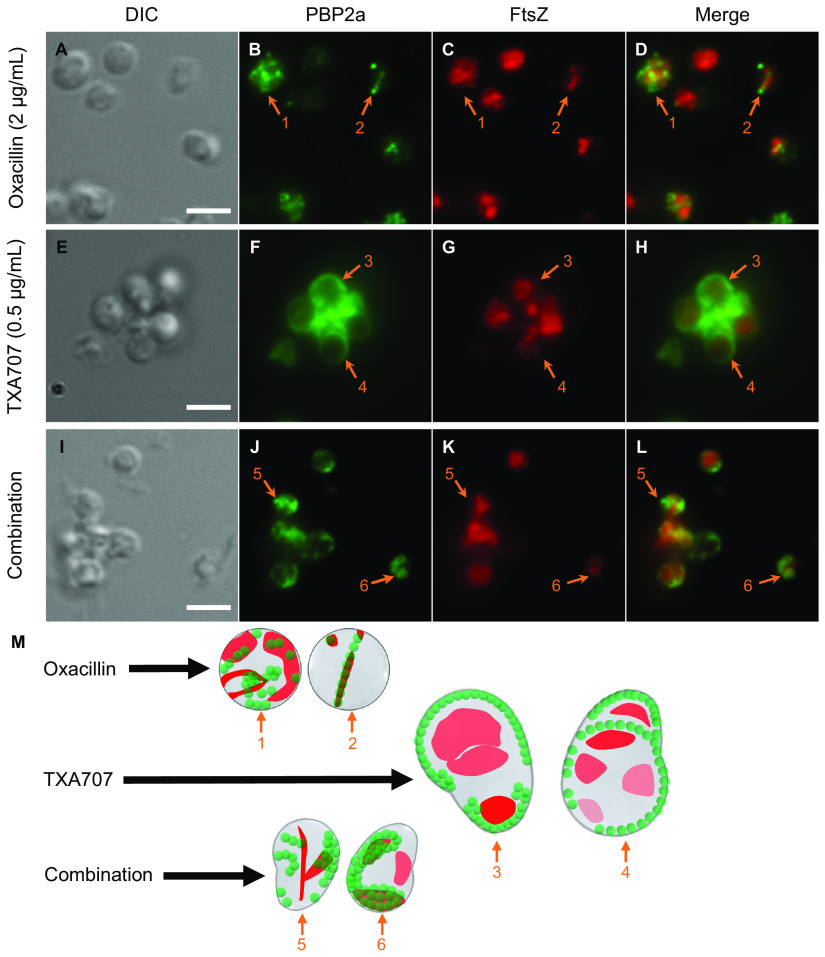
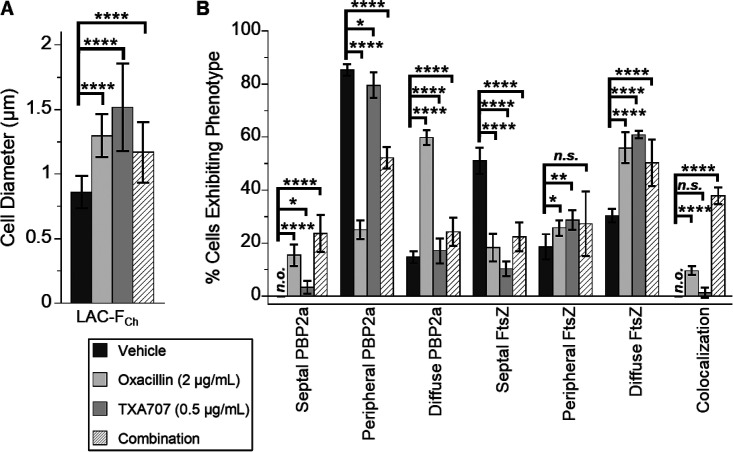
We first compared cells treated with 2 μg/ml (1/32× MIC) oxacillin alone (n = 835) relative to cells treated with vehicle (n = 535). Oxacillin-treated cells are associated with an average diameter of 1.30 ± 0.17 μm, a 1.5-fold increase relative to that associated with vehicle-treated cells (0.86 ± 0.13 μm) (Fig. 7A and Fig. 8A). In response to oxacillin treatment, PBP2a localizes to the septum at midcell (as exemplified by the cell denoted with arrow 2 in Fig. 7B and M) in 15% of the 835 cells analyzed (Fig. 8B). Significantly, this behavior markedly contrasts that associated with vehicle treatment, where none of the 535 analyzed cells exhibit septally localized PBP2a (Fig. 8B). Thus, treatment with oxacillin appears to induce the relocalization of PBP2a to the septum in a statistically significant percentage of cells. In addition, we observe oxacillin-treated cells in which PBP2a is localized to specific sites around the cell periphery (as indicated by the cell denoted with arrow 1 in Fig. 7B and M), although the prevalence of this PBP2a phenotype is markedly reduced (from 85% to 25%) relative to vehicle-treated cells (Fig. 8B). PBP2a localizes in a more diffuse pattern in 60% of oxacillin-treated cells, a rise of 45% over the vehicle treatment condition (Fig. 8B). FtsZ localization to the septum is reduced in response to oxacillin treatment, with a concomitant increase in the diffuse FtsZ phenotype (Fig. 7C and M and Fig. 8B). Significantly, in striking contrast to the absence of colocalization in vehicle-treated cells, FtsZ colocalizes with PBP2a in 10% of the 835 oxacillin-treated cells analyzed (Fig. 8B), with this colocalization occurring almost exclusively in cells where both PBP2a and FtsZ are septally localized (as indicated by the cell denoted with arrow 2 in Fig. 7D and M).
FIG 7.
(A to L) DIC and fluorescence micrographs of MRSA LAC-FCh cells treated for 3 h with either 2 μg/ml (1/32× MIC) oxacillin (A to D), 0.5 μg/ml (1/2× MIC) TXA707 (E to H), or a combination of 2 μg/ml oxacillin and 0.5 μg/ml TXA707 (I to L), followed by immunostaining of PBP2a as described in the legend to Fig. 6. (M) Localization of PBP2a (green) and FtsZ (red) is schematically depicted, with the numbered arrows in the scheme reflecting the correspondingly numbered arrows in the florescence micrographs. Scale bars for panels A to L represent 2 μm.
FIG 8.
Quantification of the cell diameter, FtsZ phenotype, and PBP2a phenotype results of the microscopy experiments depicted in Fig. 7. (A) Bar graph showing the average diameter of the vehicle-treated cells (n = 535), oxacillin-treated cells (n = 835), TXA707-treated cells (n = 270), and the cells treated with a combination of both oxacillin and TXA707 (n = 239). (B) Bar graph showing the prevalence of the various FtsZ and PBP2a phenotypes observed in the different treatment groups. Each percentage reflects an average of 5 to 6 different fields of view, with the number of cells in each field of view ranging from 28 to 325. In both panels A and B, the indicated error bars reflect the standard deviation from the mean. The statistical significance of differences in cell diameter, FtsZ phenotype, and PBP2a phenotype were analyzed as described in the legend to Fig. 1. n.s., not significant; n.o., none observed.
Treatment with 0.5 μg/ml (1/2× MIC) TXA707 alone results in enlarged cells (average diameter, 1.52 ± 0.34 μm) (Fig. 8A), some with oblong shapes (Fig. 7E and M), a behavior similar to that observed upon treatment with 4 μg/ml TXA707 (Fig. 6E and M). Interestingly, Pinho and coworkers observed a similar oblong-shaped morphology in untreated MRSA cells expressing an FtsZ variant containing the G193D mutation (43). The localization patterns for FtsZ and PBP2a observed upon treatment with 0.5 μg/ml TXA707 (Fig. 7F to H) were comparable to those observed with 4 μg/ml TXA707 treatment, with very few cells exhibiting a septal PBP2a phenotype or colocalization between PBP2a and FtsZ (Fig. 8B).
Remarkably, cotreatment with a combination of both 2 μg/ml oxacillin and 0.5 μg/ml TXA707 yields unique morphological changes and localization patterns for both FtsZ and PBP2a relative to those observed upon treatment with either agent alone. Specifically, the combination treatment causes many cells to adopt oblong shapes (Fig. 7I to M), though smaller in size (average diameter, 1.17 ± 0.24 μm) than those observed with TXA707 treatment alone (Fig. 8A). The smaller size of the combination-treated cells relative to the cells treated with 0.5 μg/ml TXA707 alone may reflect the synergistic bactericidal activity of oxacillin in combination with TXA707, with this combination inducing a greater rate of kill than either agent alone (41). This enhanced bactericidal activity may preclude the combination-treated cells from ever attaining the size induced by treatment with TXA707 alone. In 52% of the 239 combination-treated cells analyzed (Fig. 8B), PBP2a appears to localize into distinct curved structures across the cell periphery (as indicated by the cells denoted with arrows 5 and 6 in Fig. 7J and M) while also localizing to the septum in 24% of the combination-treated cells. FtsZ colocalizes with PBP2a in many (38%) of these cells (Fig. 7L and M and Fig. 8B).
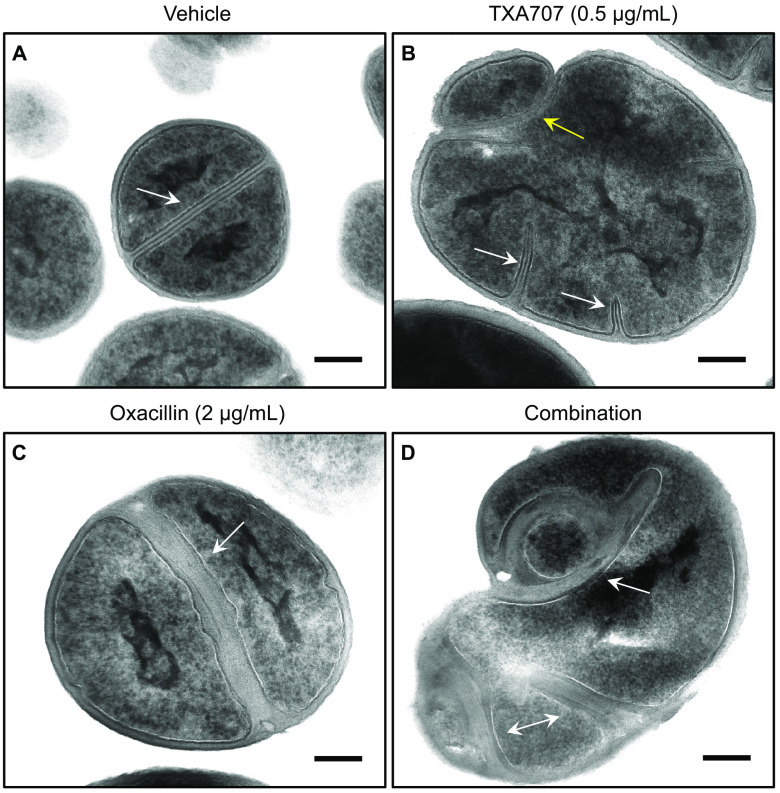
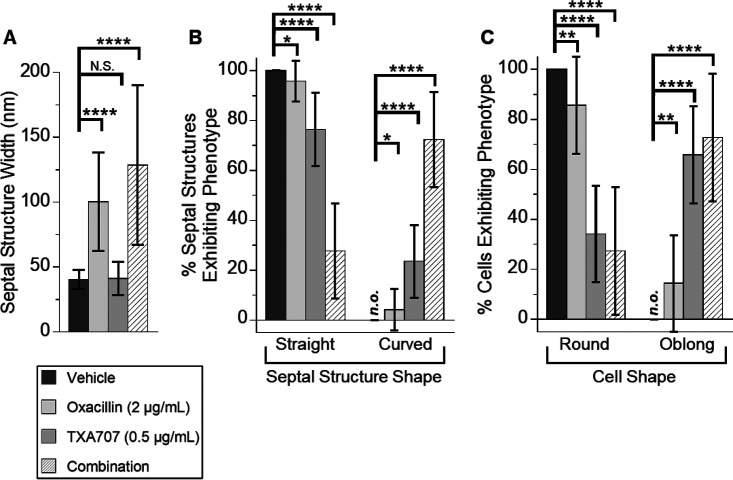
To gain further insight into the behavior of MRSA LAC-FCh cells in response to cotreatment with the synergistic combination of oxacillin and TXA707, we sought to correlate our DIC and fluorescence microscopy results shown in Fig. 7 with corresponding transmission electron microscopy (TEM) studies. In these studies, the total number of cells analyzed in each treatment condition were 55 for DMSO vehicle, 42 for TXA707 alone at 0.5 μg/ml (1/2× MIC), 56 for oxacillin alone at 2 μg/ml (1/32× MIC), and 53 for the combination of 2 μg/ml oxacillin and 0.5 μg/ml TXA707. Cells treated with vehicle are round and divide normally, forming well-defined straight septa at midcell averaging approximately 40.5 nm in width (Fig. 9A and 10). Treatment with TXA707 alone causes the cells to enlarge significantly and disrupts the ability of the cells to generate a normal septum (Fig. 9B). Approximately 48% of the TXA707-treated cells analyzed appear to undergo multiple attempts at generating a septal structure but are unable to complete the fully formed septum. The incomplete septal structures induced by TXA707 treatment (highlighted by the white arrows in Fig. 9B) are similar in width (averaging 41.2 nm) to the septa formed in the presence of vehicle (Fig. 10A), with 76% of these structures being straight and 24% being curved (Fig. 10B). Aberrant attempts at division in TXA707-treated cells can also result in the formation of blebs (as highlighted by the yellow arrow in Fig. 9B) and a significant percentage (66%) of oblong-shaped cells (Fig. 10C). Eighty-six percent of the cells treated with oxacillin alone were round (Fig. 9C and 10C), though they appear larger in size than vehicle-treated cells. The oxacillin-treated cells are still able to form complete straight septa 96% of the time (Fig. 9C and 10B). However, the septa in oxacillin-treated cells are approximately 2.5 times thicker than those in vehicle-treated cells (compare Fig. 9A and C), averaging 100.4 nm in width (Fig. 10A). Cotreatment with the combination of TXA707 and oxacillin yields a significant percentage (73%) of blebbed and oblong-shaped cells (Fig. 9D and 10C), with 66% of the cells exhibiting multiple thick septum-like structures (as highlighted by the arrows in Fig. 9D) averaging 128.7 nm in width (Fig. 10A). Seventy-two percent of these septum-like structures are highly curved and irregular (Fig. 9D and 10B), similar in nature to the curved PBP2a structures observed in our immunofluorescence micrographs of MRSA LAC-FCh cells treated with the same combination of agents (Fig. 7J and M).
FIG 9.
Transmission electron micrographs of MRSA LAC-FCh cells treated for 3 h with either vehicle (A), 0.5 μg/ml TXA707 alone (B), 2 μg/ml oxacillin alone (C), or 0.5 μg/ml TXA707 in combination with 2 μg/ml oxacillin (D). Scale bars for represent 0.2 μm. The white arrows in panels A and C highlight septa in dividing cells, while the white arrows in panels B and D highlight incomplete or aberrant septal structures. The yellow arrow in panel B highlights a bleb resulting from an aberrant attempt at division.
FIG 10.
Quantification of septal structure width (A), septal structure phenotype (B), and cell shape (C) for the results of the TEM experiments depicted in Fig. 9. (A) Bar graph showing the average septal structure width with vehicle treatment (n = 55), oxacillin treatment (n = 72), TXA707 treatment (n = 78), or treatment with a combination of both oxacillin and TXA707 (n = 106). (B) Bar graph showing the prevalence of the septal structure phenotype (straight versus curved) observed in the different treatment groups. (C) Bar graph showing the prevalence of the cell shape (round versus oblong) observed in the different treatment groups (n = 55 for vehicle, n = 56 for oxacillin alone, n = 42 for TXA707 alone, and n = 53 for the combination). In both panels B and C, each percentage reflects an average of 14 to 18 different fields of view, with the indicated error bars reflecting the standard deviation from the mean. The statistical significance of differences in septal structure width, septal structure phenotype, and cell shape were analyzed as described in the legend to Fig. 1. n.s., not significant; n.o., none observed.
We also examined the impact of treatment with 2 μg/ml oxacillin alone or in combination with 0.5 μg/ml TXA707 on the relative localization of PBP2 and FtsZ in MRSA LAC-FChP2GFP cells. Both treatment conditions result in FtsZ localization patterns (Fig. S4) similar to those observed in our immunofluorescence studies of MRSA LAC-FCh cells (Fig. 7 and 8). PBP2 localizes to the septum in 33% of the cells (n = 406) treated with oxacillin alone (Fig. S4B, I, and K). This behavior is consistent with that previously observed by Pinho and Errington, who demonstrated that β-lactam treatment yields septal localization of PBP2 in MRSA but not MSSA cells (44). PBP2 also localizes to foci in the cell periphery in 54% of the oxacillin-treated cells analyzed, with the prevalence of both peripheral and septal PBP2 being similar in oxacillin-treated relative vehicle-treated cells (Fig. S4). PBP2 colocalizes with FtsZ in 51% of the oxacillin-treated cells (Fig. S4D and K), compared to 69% in the vehicle treatment condition. Relative to treatment with oxacillin alone, treatment of MRSA LAC-FChP2GFP cells (n = 432) with a combination of both 2 μg/ml oxacillin and 0.5 μg/ml TXA707 results in a significant reduction (from 33% to 11%) in the prevalence of PBP2 localized to the septum coupled with an increase (from 54% to 72%) in the prevalence of PBP2 localized to foci in the cell periphery (Fig. S4). Colocalization of PBP2 and FtsZ is also markedly reduced (from 51% to 28%) with combination relative to oxacillin treatment (Fig. S4D, H, I, and K).
DISCUSSION
FtsZ and the PBPs are key components of the divisome machinery that play important roles in septum formation and cell division (32, 33, 45). FtsZ inhibitors have been shown to act synergistically with PBP-targeting β-lactam antibiotics against MRSA, effectively resensitizing MRSA to the β-lactams (13, 15, 25, 40, 41). To further our understanding of the basis for this behavior, we examined the impact of the FtsZ inhibitor TXA707 on the relative localization of FtsZ and the five PBPs in MRSA (PBP1, PBP2, PBP3, PBP4, and PBP2a). To this end, we genetically engineered MRSA LAC to express an FtsZ-mCherry fusion protein and an sfGFP fusion form of PBP1, PBP2, PBP3, or PBP4. We also developed an immunofluorescence approach for monitoring the localization of PBP2a in MRSA LAC expressing FtsZ-mCherry.
In MRSA cells treated with DMSO vehicle, PBP1, PBP2, PBP3, and PBP4 are recruited to FtsZ Z-rings formed at the midcell septum (Fig. 2 and 5), consistent with previous reports suggesting that all four PBPs are involved in cell wall synthesis at the septum during cell division (46–51). In striking contrast, PBP2a does not localize to the septum but, rather, to distinct foci around the cell periphery (Fig. 6). This observation suggests that under vehicle-treated conditions, PBP2a is not involved in new cell wall synthesis at the septa of MRSA cells. Instead, it may be functioning in cell wall maintenance and repair around the cell periphery. García-Fernández et al. have demonstrated that PBP2a is localized in lipid rafts in MRSA cell membranes (52). This observation, coupled with our results, suggests that the PBP2a may be localized in depots around the cell periphery where it is available to act in response to conditions encountered by the cell that compromise the cell wall.
Upon treatment with TXA707, septum formation and cell division are disrupted, with FtsZ and PBP1, PBP2, PBP3, and PBP4 being mislocalized away from midcell (Fig. 2 and 5). PBP1 and PBP3 relocalize to distinct foci across the cell periphery, with FtsZ colocalizing to these same foci in 60% to 65% of the cells analyzed (Fig. 2 and 3). This colocalization suggests that PBP1 and PBP3 may function in maintaining septal integrity through direct or indirect linkage with FtsZ. In contrast to the behavior of PBP1 and PBP3, PBP2 and PBP4 do not significantly colocalize with FtsZ in response to FtsZ inhibition with TXA707, exhibiting a colocalization phenotype in only 15% to 35% of the analyzed cells. Instead, PBP2 and PBP4 appear to localize in foci across the cell periphery that are distinct from those in which FtsZ has localized (Fig. 4 and 5). Pinho and Errington observed a similar behavior for PBP2 in methicillin-sensitive S. aureus (MSSA) cells in which cell division had been disrupted through depletion of FtsZ (53). The authors speculated that PBP2 plays a major role in the cell wall remodeling and repair necessitated by the enlarged cell resulting from the block of cell division. Our results suggest that both PBP2 and PBP4 are involved in such a cell wall remodeling and repair process induced by FtsZ inhibition with TXA707. Interestingly, this process does not appear to be coordinated by FtsZ.
In response to TXA707 treatment, PBP2a localizes to an increased number of foci around the periphery of the enlarged cell (Fig. 6). This behavior suggests that PBP2a may play a similar role to PBP2 and PBP4 in response to FtsZ inhibition, a function geared toward cell wall remodeling, maintenance, and repair necessitated by a disruption of cell division. Łęski and Tomasz have previously demonstrated that PBP2, PBP4, and PBP2a act cooperatively during cell wall synthesis in MRSA (47). The similar behaviors we observe for PBP2, PBP4, and PBP2a upon FtsZ inhibition with TXA707 are consistent with the three PBPs having cooperative roles in cell wall maintenance and repair in the enlarged cells.
To further explore the mechanism of synergy between FtsZ inhibitors and β-lactam antibiotics against MRSA, we investigated the impact of treatment with sub-MICs of TXA707 alone, oxacillin alone, or a combination of both agents on the localization of FtsZ and PBP2a. Upon treatment with oxacillin alone at 1/32× MIC, PBP2a relocalizes to the septum at midcell (Fig. 7 and 8), suggesting that PBP2a can indeed participate in new cell wall formation at the septum, but only in the presence of a β-lactam antibiotic. Although this behavior was observed in only 15% of the cells analyzed, its prevalence would be more than sufficient to confer MRSA with resistance to β-lactams. In addition to this localization phenotype, PBP2a also localizes to multiple locations across the cell periphery, suggesting that it is also involved in peripheral cell wall maintenance in response to β-lactam treatment.
Treatment with a synergistic combination of TXA707 and oxacillin yields a unique phenotype in which PBP2a now forms discrete curved structures across the cell periphery that colocalize with FtsZ in 38% of the cells analyzed (Fig. 7J to M and Fig. 8B). This behavior differs from that observed upon treatment with vehicle, TXA707 alone, or oxacillin alone. We further explored this behavior using TEM. TEM micrographs of MRSA cells treated with oxacillin alone revealed a substantial thickening of the septa (Fig. 9C and Fig. 10A). Recent studies by Müller et al. have indicated that PBP2a is associated with weak transpeptidase activity, making it a poor cross-linker of peptidoglycans in the bacterial cell wall (54). The thickening of the septa we observe in response to oxacillin treatment may reflect the poor cross-linking activity of PBP2a, which would result in a septal cell wall with reduced structural density. Additional layers of peptidoglycan would have to be incorporated into the septal cell wall to compensate for the reduced structural integrity, thereby resulting in a thickening of the septum. As both PBP2 and PBP2a are recruited to the septum upon treatment with oxacillin (Fig. 7 and 8; see Fig. S4 in the supplemental material), it is likely that the two proteins act collaboratively in the synthesis of new septal cell wall. This hypothesis is supported by previous studies suggesting that PBP2a complements the loss of PBP2 transpeptidase activity upon acylation by a β-lactam antibiotic (39, 44, 47).
TEM micrographs of MRSA cells treated with a synergistic combination of oxacillin and TXA707 (Fig. 9 and 10) reveal a similar pattern to that observed in our immunofluorescence studies (Fig. 7). In these TEM micrographs, we observe the formation of predominantly oblong-shaped cells with thick, curved, septum-like structures (Fig. 9D and Fig. 10) that are largely absent in cells treated with either vehicle, oxacillin alone, or TXA707 alone (Fig. 10B). As noted above, the thick septa observed in the presence of oxacillin may reflect a cell wall with reduced structural density resulting from the poor cross-linking activity of PBP2a. Such septal structures may thus be predisposed to adopt curvature in response to combination treatment with both oxacillin and TXA707.
The unique behavior associated with the response to the combination treatment provides insight into how FtsZ inhibitors can sensitize MRSA to β-lactam antibiotics. With PBP2a now localizing in the curved septal-like structures that arise in response to the combination treatment, it is less available to function in peripheral cell wall maintenance and repair in cooperation with PBP2, which can be targeted, to at least some degree, by the β-lactam itself. As a result, the cell would become sensitive to the β-lactam, particularly β-lactams that target PBP2 with a high affinity. This hypothesis is consistent with that previously suggested by Tan et al. (25), as well as with our previous studies demonstrating that FtsZ inhibitors synergize to the greatest extent with β-lactams like oxacillin that target PBP2 with a high affinity (41). By simultaneously interfering with the ability of critical PBPs (like PBP2 and PBP2a) to function in cell wall maintenance and repair in response to disrupted cell division, combinations of β-lactams and FtsZ inhibitors render MRSA cells more susceptible to the bactericidal effects of the β-lactams (25).
MATERIALS AND METHODS
Bacterial strains and other reagents.
S. aureus RN4220 (MSSA) and MRSA LAC were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, and NIH. E. coli NEB5α was obtained from New England Biolabs. All the MRSA LAC strains expressing fluorescent fusion forms of FtsZ and different PBPs (listed in Table 1) were generated as described below. The PBP2a-specific monoclonal antibody mouse anti-MRSA was obtained from RayBiotech (product code 130-10096-20), and the goat anti-mouse Alexa Fluor 488-conjugated antibody was from Thermo Fisher. Tryptic soy broth (TSB), tryptic soy agar (TSA), cation-adjusted Mueller-Hinton (CAMH) medium, and Luria-Bertani (LB) medium were obtained from Becton, Dickinson. Ampicillin sodium salt, oxacillin sodium salt, cephalexin monohydrate, erythromycin, chloramphenicol, isopropyl-β-d-1-thiogalactopyranoside (IPTG), Tween 20, bovine serum albumin (BSA), and high-resolution agarose were from Sigma. Cefotaxime and ceftriaxone sodium salts were from Toku-E. Phosphate-buffered saline (PBS) was obtained from Lonza. TXA707 was synthesized as previously described (18).
TABLE 1.
List of genetically modified MRSA LAC strains expressing fluorescence fusion forms of FtsZ and different PBPs
| Strain | Relevant characteristicsa |
|---|---|
| LAC-FCh | LAC attB::[pLL39 Pspac-ftsZ-mCherry lacI Tetr] pCM11-lacI Ermr |
| LAC-FChP1GFP | LAC-FCh ΔpbpA::sfgfp-pbpA |
| LAC-FChP2GFP | LAC-FCh ΔpbpB::sfgfp-pbpB |
| LAC-FChP3GFP | LAC-FCh ΔpbpC::sfgfp-pbpC |
| LAC-FChP4GFP | LAC-FCh ΔpbpD::pbpD-sfgfp |
| LAC-P2ACh-1 | LAC ΔmecA::mecA-mCherry |
| LAC-P2ACh-2 | LAC ΔmecA::mCherry-mecA |
Erm, erythromycin; Tet, tetracycline.
Generation of MRSA LAC strains expressing fluorescent fusion forms of FtsZ and different PBPs.
Fluorescent fusion proteins were cloned in MRSA LAC using the general strategy outlined below. The plasmids listed in Table S2 in the supplemental material were propagated in E. coli NEB5α in the presence of the appropriate antibiotic for each given plasmid. The plasmids were then isolated and purified using the Monarch plasmid miniprep kit (New England Biolabs) and subsequently introduced into electrocompetent S. aureus RN4220 cells as previously described (55). Bacteriophage 80α was used to transduce (56) the plasmids from S. aureus RN4220 into MRSA LAC-FCh, an MRSA strain that we previously generated (57) containing an ectopic ftsZ-mCherry fusion gene under the control of an IPTG-inducible promoter. The MRSA LAC-FCh strains expressing the fluorescent fusion form of sfGFP-PBP1, sfGFP-PBP2, sfGFP-PBP3, or PBP4-sfGFP (LAC-FChP1GFP, LAC-FChP2GFP, LAC-FChP3GFP, and LAC-FChP4GFP, respectively) were constructed by allelic replacement utilizing the pJB38 vector (58). In each case, DNA fragments P1, P2, and P3 (see Table S3) that contain complementary overhangs were PCR amplified using Q5 High-Fidelity DNA polymerase (New England Biolabs). The resulting amplified fragments were then combined in equimolar quantities with the pJB38 vector, previously linearized by PCR using primers pJB38-F and pJB38-R (Table S4). All four DNA fragments were then joined using the NEBuilder HiFi DNA assembly kit (New England Biolabs), with each resulting construct being verified by sequencing.
The integration of the pJB38 constructs into the MRSA LAC-FCh chromosome and subsequent excision were achieved through a double recombination process, leading to an allelic exchange (58, 59). Briefly, transductants containing the pJB38-derived constructs were maintained at the replication-permissive temperature of 30°C for plasmid maintenance and confirmation and then frozen down. To initiate recombination, frozen stocks of the transductants were streaked onto TSA plates supplemented with 30 μg/ml chloramphenicol and 10 μg/ml erythromycin and then incubated overnight at 44°C. Large colonies reflecting clones that had undergone a single recombination event were restreaked onto TSA plates supplemented with 30 μg/ml chloramphenicol and 10 μg/ml erythromycin and then incubated overnight at 44°C. These single recombinants were then inoculated into 5 ml of TSB and incubated at 30°C in the absence of chloramphenicol to promote a second round of recombination and subsequent plasmid loss. After consecutive passages over 5 days at 30°C, the cultures were then serially diluted and plated onto TSA plates supplemented with 100 ng/ml anhydrotetracycline and 10 μg/ml erythromycin. To identify cells that had undergone a second recombination event and subsequent loss of the plasmid, the resulting colonies were replica patched onto TSA plates supplemented with 10 μg/ml erythromycin alone, as well as onto TSA plates supplemented with both 10 μg/ml erythromycin and 30 μg/ml chloramphenicol. Chloramphenicol-sensitive colonies were screened by PCR to verify the presence of the appropriate allele utilizing the forward primer for the P1 DNA fragment and the reverse primer for the P3 DNA fragment (listed in Table S3 for each denoted strain). The MRSA LAC strains expressing the fluorescent fusion form of PBP2a-mCherry or mCherry-PBP2a (LAC-P2ACh-1 and LAC-P2ACh-2, respectively) were generated using a similar strategy to that described above, with the exception that wild-type MRSA LAC was used as the recipient strain instead of MRSA LAC-FCh.
Time-dependent growth assay.
An exponentially growing culture of each MRSA strain was diluted in CAMH broth to a final count of 5 × 105 CFU/ml. The CFU/ml of each culture at time zero was verified by plating serial dilutions in duplicate on TSA plates. The cultures were then incubated at 37°C with shaking in the presence of 100 nM IPTG to induce expression of the ftsZ-mCherry gene when present. The CFU/ml in each culture was determined over time by withdrawing samples at time points ranging from 3 to 24 h and then plating appropriate serial dilutions onto TSA plates. All TSA plates were incubated at 37°C for 24 h and the CFU/ml at each time point determined.
MIC assay.
MIC assays were conducted in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines for broth microdilution (60). Briefly, log-phase MRSA cells were added to 96-well microtiter plates (at 5 × 105 CFU/ml) containing 2-fold serial dilutions of TXA707, oxacillin, ceftriaxone, cefotaxime, cephalexin, or vancomycin in CAMH broth, with each concentration of antibacterial agent being present in duplicate. The final volume in each well was 0.1 ml, and the microtiter plates were incubated aerobically for 18 h at 37°C. Bacterial growth was monitored by measuring the optical density at 600 nm (OD600) using a SpectraMax M2 plate reader (Molecular Devices, Inc.), with the MIC being defined as the lowest compound concentration at which growth was ≥90% inhibited.
Differential interference contrast and fluorescence microscopy.
Each MRSA strain was grown to log phase in 5 ml of TSB supplemented with 10 μg/ml erythromycin and then diluted to an OD600 of 0.1 in 5 ml of TSB supplemented with 10 μg/ml erythromycin. The log-phase cultures were then treated with DMSO vehicle, 4 μg/ml TXA707 (4× MIC), and 10 nM IPTG for 3 h at 37°C. Each culture was then centrifuged at 15,000 × g for 1 min and washed twice with 1 ml of PBS. Cells were then resuspended in 200 μl of PBS. Eight microliters of this final cell suspension were spread on a 0.25-mm layer of 1.5% high-resolution agarose in PBS, which was mounted on a petrographic 27- by 46- by 1.2-mm microscope slide (Ward’s Natural Science) using a 1.7- by 2.8- by 0.025-cm gene frame (Thermo Fisher). A 24- by 40-mm coverslip (Azer Scientific) was then applied to the agarose pad in preparation for microscopic visualization.
All differential interference contrast (DIC) and fluorescence microscopy experiments were conducted using a total internal reflection fluorescence (TIRF) microscope that was custom-built on the basis of a commercial inverted microscope (Ti-E; Nikon) (61). The microscope was equipped with a high-numerical-aperture (NA) lens objective (chrome-free infinity [CFI] Apo 100×; NA, 1.49; Nikon), a scientific complementary metal oxide semiconductor (sCMOS) camera (Zyla 4.2; Andor), and the 488-nm Genesis MX488-1000 STM and 561 nm Genesis MX561-1000 STM excitation lasers (Coherent). For DIC imaging, a white light LED (LDB101F; Prior) was used along with Nikon’s DIC modules. sfGFP was imaged using 48-nm laser excitation and a green emission band-pass filter (ET525/50m; Chroma), while mCherry was imaged using 561-nm laser excitation and an orange emission band-pass filter (ET605/52m; Chroma). Multichannel images were obtained by triggered acquisition schemes, using acousto-optic tunable filter (AOTF) (AOTFnC-400.650-TN; Quanta-Tech), transistor-transistor logic signal out of the sCMOS camera, a data acquisition card (PCIe-7852R; NI), and the Nikon NIS-Elements software. Extra magnification was achieved using a built-in ×1.5 intermediate magnification changer on the microscope. MRSA samples were first inspected in the DIC channel and then switched to the fluorescence channel to adjust the focus as well as the excitation laser tilt angle to the optimal TIRF imaging condition. For imaging sfGFP, the 48-nm laser was used at power settings in the range of 2 to 15 mW, coupled with exposure times ranging from 80 to 400 ms. For imaging mCherry, the 561-nm laser was used at power settings in the range of 20 to 40 mW, coupled with exposure times ranging from 200 to 700 ms. The Perfect Focus system (Nikon) was used to actively stabilize focus drift while acquiring images.
Characterization of the specificity of the mouse anti-MRSA monoclonal antibody.
E. coli BL21(DE3) strains expressing recombinant forms of PBP1, PBP2, PBP3, or PBP4 from S. aureus (SaPBP1, SaPBP2, SaPBP3, and SaPBP4, respectively) were generated as described previously (41). S. aureus PBP2a (SaPBP2a) was cloned and expressed in E. coli BL21(DE3) as detailed in the supplemental material. For the expression of recombinant SaPBP1, SaPBP2, SaPBP3, SaPBP4, and SaPBP2a, each E. coli strain was cultured overnight at 37°C in LB broth and then diluted 1:100 into 10 ml of LB broth. These cultures were then grown at 37°C to an OD600 of 0.3, whereupon PBP expression was induced by the addition of 1 mM IPTG and subsequent incubation for 3 h at 37°C. Overnight cultures of MRSA LAC and MSSA RN4220 cells were diluted 1:10 into in TSB and grown for 3 h at 37°C. The MRSA and MSSA cells were then lysed by addition of lysostaphin (Sigma) at a concentration of 1 mg/ml and subsequent incubation for 1 h at 37°C.
All the E. coli, MRSA, and MSSA samples were then washed twice with 500 μl of PBS and resuspended in 2 ml of PBS. The cells were sonicated for 5 min at 0°C using a Qsonica Q500 sonicator (equipped with a 1/8-inch probe) with an on/off cycle of 10 s and amplitude set at 10%. The total protein concentration in each cell lysate was then quantified using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher). A 12% SDS-PAGE gel (Bio-Rad) was loaded with 5 μl of 0.5 μg/ml purified recombinant SaPBP2a and 5 μl of 500 μg total protein/ml from each cell lysate. Western blotting was conducted using standard procedures, followed by incubation with the mouse anti-MRSA antibody (RayBiotech) at a 1:1,000 dilution. Secondary antibody treatment was performed with goat anti-mouse IgG–horseradish peroxidase (HRP) (Thermo Fisher) at a 1:100,000 dilution. The Western blotting was developed using SuperSignal West Pico Plus (Thermo Fisher) solutions and visualized by chemiluminescence using an Amersham Imager 680.
Immunofluorescence microscopy.
MRSA LAC-FCh cells were grown to log phase in 5 ml of TSB supplemented with 10 μg/ml erythromycin and then diluted to an OD600 of 0.1 in 5 ml of TSB supplemented with 10 μg/ml erythromycin. The log-phase cultures were then supplemented with 10 nM IPTG and treated for 3 h at 37°C with either DMSO vehicle, 0.5 μg/ml TXA707 (1/2× MIC), 4 μg/ml TXA707 (4× MIC), 2 μg/ml oxacillin (1/32× MIC), or a combination of 0.5 μg/ml TXA707 and 2 μg/ml oxacillin. Each culture was then centrifuged at 15,000 × g for 1 min and washed twice with 1 ml of PBS. Cells were then resuspended in 500 μl of PBS containing 2.4% (vol/vol) formaldehyde, followed by incubation for 15 min at room temperature and then 25 min on ice. Each culture was then centrifuged at 15,000 × g for 1 min and washed twice with 1 ml of PBS containing 0.01% Tween 20. Cells were then resuspended in 500 μl of GTE buffer containing 50 mM glucose, 20 mM Tris-HCl (pH 7.6), and 10 mM EDTA. We then mounted 100 μl of the resulting suspension on a poly-l-lysine-coated, 18- by 18-mm coverslip (VWR). Two hundred microliters of 30 ng/μl lysostaphin were then applied and allowed to equilibrate for 1 min. Cells were then washed three times with GTE buffer, air dried, and subsequently rehydrated by applying 200 μl of PBS. After equilibration for 5 min, the cells were then blocked for 45 min using 300 μl of 2% (wt/vol) BSA. PBP2a was detected via immunofluorescence using the PBP2a-specific mouse anti-MRSA monoclonal antibody (RayBiotech). Serial 2-fold dilutions of antibody (ranging from 1:100 to 1:1,600) were added to each sample and incubated overnight at 4°C. The cells were then washed 8 times with PBS and incubated in the dark for 2 h with a goat anti-mouse Alexa Fluor 488-conjugated antibody (Thermo Fisher) diluted 3:1,000 with PBS containing 2% (wt/vol) BSA. Cells were again washed 8 times with PBS. Aqua-Poly/Mount medium (Polysciences) was then added, and the cells were mounted on a petrographic 27- by 46- by 1.2-mm microscope slide (Ward’s Natural Science). The slides were then visualized by DIC and fluorescence microscopy as described above.
Transmission electron microscopy.
Log-phase MRSA LAC-FCh cells were diluted to an OD600 of 0.1 in 5 ml of TSB and then cultured at 37°C for 3 h in the presence of DMSO vehicle, 0.5 μg/ml TXA707 (1/2× MIC), 2 μg/ml oxacillin (1/32× MIC), or a combination of 0.5 μg/ml TXA707 and 2 μg/ml oxacillin. The cultures were then centrifuged at 16,000 × g for 3 min at room temperature. The resulting bacterial pellets were washed with 1 ml of PBS and then resuspended in 500 μl of 100 mM cacodylate buffer (pH 7.2) containing 2.5% (vol/vol) glutaraldehyde and 4% (vol/vol) paraformaldehyde. The samples were then prepared for TEM acquisition and the micrographs acquired as previously described (41).
ACKNOWLEDGMENTS
This work was supported by NIAID grant 1RO1 AI118874-05 (to D.S.P.) and Department of Energy grant DE-SC0019313 (to S.-H.L.). The Boyd lab is funded by NIAID grant 1R01 AI139100-01 and USDA MRF project NE-1028.
Footnotes
Supplemental material is available online only.
Contributor Information
Sang-Hyuk Lee, Email: shlee@physics.rutgers.edu.
Daniel S. Pilch, Email: pilchds@rwjms.rutgers.edu.
Michael J. Federle, University of Illinois at Chicago
REFERENCES
- 1.Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, Nadle J, Kainer MA, Dumyati G, Petit S, Ray SM, Ham D, Capers C, Ewing H, Coffin N, McDonald LC, Jernigan J, Cardo D, Emerging Infections Program MRSA Author Group. 2019. Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections - United States. MMWR Morb Mortal Wkly Rep 68:214–219. 10.15585/mmwr.mm6809e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Freire-Moran L, Aronsson B, Manz C, Gyssens IC, So AD, Monnet DL, Cars O, Group E-EW, ECDC-EMA Working Group. 2011. Critical shortage of new antibiotics in development against multidrug-resistant bacteria—time to react is now. Drug Resist Updat 14:118–124. 10.1016/j.drup.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Cars O, Hedin A, Heddini A. 2011. The global need for effective antibiotics—moving towards concerted action. Drug Resist Updat 14:68–69. 10.1016/j.drup.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Craft KM, Nguyen JM, Berg LJ, Townsend SD. 2019. Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. Medchemcomm 10:1231–1241. 10.1039/c9md00044e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vestergaard M, Frees D, Ingmer H. 2019. Antibiotic resistance and the MRSA problem. Microbiol Spectr 7. 10.1128/microbiolspec.GPP3-0057-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sass P, Brotz-Oesterhelt H. 2013. Bacterial cell division as a target for new antibiotics. Curr Opin Microbiol 16:522–530. 10.1016/j.mib.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Kusuma KD, Payne M, Ung AT, Bottomley AL, Harry EJ. 2019. FtsZ as an antibacterial target: status and guidelines for progressing this avenue. ACS Infect Dis 5:1279–1294. 10.1021/acsinfecdis.9b00055. [DOI] [PubMed] [Google Scholar]
- 10.Hurley KA, Santos TM, Nepomuceno GM, Huynh V, Shaw JT, Weibel DB. 2016. Targeting the bacterial division protein FtsZ. J Med Chem 59:6975–6998. 10.1021/acs.jmedchem.5b01098. [DOI] [PubMed] [Google Scholar]
- 11.Tripathy S, Sahu SK. 2019. FtsZ inhibitors as a new genera of antibacterial agents. Bioorg Chem 91:103169. 10.1016/j.bioorg.2019.103169. [DOI] [PubMed] [Google Scholar]
- 12.Casiraghi A, Suigo L, Valoti E, Straniero V. 2020. Targeting bacterial cell division: a binding site-centered approach to the most promising inhibitors of the essential protein FtsZ. Antibiotics (Basel) 9:69. 10.3390/antibiotics9020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan FY, Sun N, Leung YC, Wong KY. 2015. Antimicrobial activity of a quinuclidine-based FtsZ inhibitor and its synergistic potential with β-lactam antibiotics. J Antibiot (Tokyo) 68:253–258. 10.1038/ja.2014.140. [DOI] [PubMed] [Google Scholar]
- 14.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, Heal JR, Sheridan JM, Aiwale ST, Chauhan PK, Srivastava A, Taneja A, Collins I, Errington J, Czaplewski LG. 2008. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321:1673–1675. 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 15.Kaul M, Mark L, Parhi AK, LaVoie EJ, Pilch DS. 2016. Combining the FtsZ-targeting prodrug TXA709 and the cephalosporin cefdinir confers synergy and reduces the frequency of resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 60:4290–4296. 10.1128/AAC.00613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaul M, Mark L, Zhang Y, Parhi AK, Lavoie EJ, Pilch DS. 2013. An FtsZ-targeting prodrug with oral antistaphylococcal efficacy in vivo. Antimicrob Agents Chemother 57:5860–5869. 10.1128/AAC.01016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaul M, Mark L, Zhang Y, Parhi AK, LaVoie EJ, Pilch DS. 2013. Pharmacokinetics and in vivo antistaphylococcal efficacy of TXY541, a 1-methylpiperidine-4-carboxamide prodrug of PC190723. Biochem Pharmacol 86:1699–1707. 10.1016/j.bcp.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Kaul M, Mark L, Zhang Y, Parhi AK, Lyu YL, Pawlak J, Saravolatz S, Saravolatz LD, Weinstein MP, LaVoie EJ, Pilch DS. 2015. TXA709, an FtsZ-targeting benzamide prodrug with improved pharmacokinetics and enhanced in vivo efficacy against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 59:4845–4855. 10.1128/AAC.00708-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margalit DN, Romberg L, Mets RB, Hebert AM, Mitchison TJ, Kirschner MW, RayChaudhuri D. 2004. Targeting cell division: small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc Natl Acad Sci U S A 101:11821–11826. 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stokes NR, Baker N, Bennett JM, Berry J, Collins I, Czaplewski LG, Logan A, Macdonald R, Macleod L, Peasley H, Mitchell JP, Nayal N, Yadav A, Srivastava A, Haydon DJ. 2013. An improved small-molecule inhibitor of FtsZ with superior in vitro potency, drug-like properties, and in vivo efficacy. Antimicrob Agents Chemother 57:317–325. 10.1128/AAC.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stokes NR, Baker N, Bennett JM, Chauhan PK, Collins I, Davies DT, Gavade M, Kumar D, Lancett P, Macdonald R, Macleod L, Mahajan A, Mitchell JP, Nayal N, Nayal YN, Pitt GR, Singh M, Yadav A, Srivastava A, Czaplewski LG, Haydon DJ. 2014. Design, synthesis and structure-activity relationships of substituted oxazole-benzamide antibacterial inhibitors of FtsZ. Bioorg Med Chem Lett 24:353–359. 10.1016/j.bmcl.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Haydon DJ, Czaplewski LG, Stokes NR, Davies D, Collins I, Palmer JT, Mitchell JP, Pitt GRW, Offermann D. 6 December 2016. Aromatic amides and uses thereof. U.S. patent 9,511,073.
- 23.Knudson SE, Awasthi D, Kumar K, Carreau A, Goullieux L, Lagrange S, Vermet H, Ojima I, Slayden RA. 2014. A trisubstituted benzimidazole cell division inhibitor with efficacy against Mycobacterium tuberculosis. PLoS One 9:e93953. 10.1371/journal.pone.0093953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsen NL, Lu J, Parthasarathy G, Reid JC, Sharma S, Soisson SM, Lumb KJ. 2012. Mechanism of action of the cell-division inhibitor PC190723: modulation of FtsZ assembly cooperativity. J Am Chem Soc 134:12342–12345. 10.1021/ja303564a. [DOI] [PubMed] [Google Scholar]
- 25.Tan CM, Therien AG, Lu J, Lee SH, Caron A, Gill CJ, Lebeau-Jacob C, Benton-Perdomo L, Monteiro JM, Pereira PM, Elsen NL, Wu J, Deschamps K, Petcu M, Wong S, Daigneault E, Kramer S, Liang L, Maxwell E, Claveau D, Vaillancourt J, Skorey K, Tam J, Wang H, Meredith TC, Sillaots S, Wang-Jarantow L, Ramtohul Y, Langlois E, Landry F, Reid JC, Parthasarathy G, Sharma S, Baryshnikova A, Lumb KJ, Pinho MG, Soisson SM, Roemer T. 2012. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci Transl Med 4:126ra35. 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- 26.Knudson SE, Awasthi D, Kumar K, Carreau A, Goullieux L, Lagrange S, Vermet H, Ojima I, Slayden RA. 2015. Cell division inhibitors with efficacy equivalent to isoniazid in the acute murine Mycobacterium tuberculosis infection model. J Antimicrob Chemother 70:3070–3073. 10.1093/jac/dkv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goehring NW, Beckwith J. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol 15:R514-26. 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 28.den Blaauwen T, Hamoen LW, Levin PA. 2017. The divisome at 25: the road ahead. Curr Opin Microbiol 36:85–94. 10.1016/j.mib.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margolin W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev 24:531–548. 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 30.Ortiz C, Natale P, Cueto L, Vicente M. 2016. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev 40:57–67. 10.1093/femsre/fuv040. [DOI] [PubMed] [Google Scholar]
- 31.Coltharp C, Xiao J. 2017. Beyond force generation: why is a dynamic ring of FtsZ polymers essential for bacterial cytokinesis? Bioessays 39:1–11. 10.1002/bies.201600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McQuillen R, Xiao J. 2020. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu Rev Biophys 49:309–341. 10.1146/annurev-biophys-121219-081703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 34.Reed P, Atilano ML, Alves R, Hoiczyk E, Sher X, Reichmann NT, Pereira PM, Roemer T, Filipe SR, Pereira-Leal JB, Ligoxygakis P, Pinho MG. 2015. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog 11:e1004891. 10.1371/journal.ppat.1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandes R, Amador P, Prudêncio C. 2013. β-Lactams: chemical structure, mode of action and mechanisms of resistance. Rev Med Microbiol 24:7–17. 10.1097/MRM.0b013e3283587727. [DOI] [Google Scholar]
- 36.Kong KF, Schneper L, Mathee K. 2010. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS 118:1–36. 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peacock SJ, Paterson GK. 2015. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem 84:577–601. 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 38.Fishovitz J, Hermoso JA, Chang M, Mobashery S. 2014. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 66:572–577. 10.1002/iub.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinho MG, de Lencastre H, Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci U S A 98:10886–10891. 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lui HK, Gao W, Cheung KC, Jin WB, Sun N, Kan JWY, Wong ILK, Chiou J, Lin D, Chan EWC, Leung YC, Chan TH, Chen S, Chan KF, Wong KY. 2019. Boosting the efficacy of anti-MRSA β-lactam antibiotics via an easily accessible, non-cytotoxic and orally bioavailable FtsZ inhibitor. Eur J Med Chem 163:95–115. 10.1016/j.ejmech.2018.11.052. [DOI] [PubMed] [Google Scholar]
- 41.Ferrer-González E, Kaul M, Parhi AK, LaVoie EJ, Pilch DS. 2017. β-Lactam antibiotics with a high affinity for PBP2 act synergistically with the FtsZ-targeting agent TXA707 against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 61:e00863-17. 10.1128/AAC.00863-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorge AMR. 2012. Insights into cell wall synthesis and cell division in Staphylococcus aureus. PhD thesis. Universidade NOVA de Lisboa, Lisbon, Portugal. [Google Scholar]
- 43.Pereira AR, Hsin J, Król E, Tavares AC, Flores P, Hoiczyk E, Ng N, Dajkovic A, Brun YV, VanNieuwenhze MS, Roemer T, Carballido-Lopez R, Scheffers DJ, Huang KC, Pinho MG. 2016. FtsZ-dependent elongation of a coccoid bacterium. mBio 7:e00908-16. 10.1128/mBio.00908-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinho MG, Errington J. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol Microbiol 55:799–807. 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- 45.Lund VA, Wacnik K, Turner RD, Cotterell BE, Walther CG, Fenn SJ, Grein F, Wollman AJ, Leake MC, Olivier N, Cadby A, Mesnage S, Jones S, Foster SJ. 2018. Molecular coordination of Staphylococcus aureus cell division. Elife 7:e32057. 10.7554/eLife.32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira SF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. 2009. Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol Microbiol 72:895–904. 10.1111/j.1365-2958.2009.06687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Łeski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 187:1815–1824. 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinho MG, de Lencastre H, Tomasz A. 2000. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J Bacteriol 182:1074–1079. 10.1128/jb.182.4.1074-1079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reichmann NT, Tavares AC, Saraiva BM, Jousselin A, Reed P, Pereira AR, Monteiro JM, Sobral RG, VanNieuwenhze MS, Fernandes F, Pinho MG. 2019. SEDS-bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat Microbiol 4:1368–1377. 10.1038/s41564-019-0437-2. [DOI] [PubMed] [Google Scholar]
- 50.Monteiro JM, Fernandes PB, Vaz F, Pereira AR, Tavares AC, Ferreira MT, Pereira PM, Veiga H, Kuru E, VanNieuwenhze MS, Brun YV, Filipe SR, Pinho MG. 2015. Cell shape dynamics during the staphylococcal cell cycle. Nat Commun 6:8055. 10.1038/ncomms9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, Veiga H, Tavares AC, Santos M, Ferreira MT, Macario V, VanNieuwenhze MS, Filipe SR, Pinho MG. 2018. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554:528–532. 10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.García-Fernández E, Koch G, Wagner RM, Fekete A, Stengel ST, Schneider J, Mielich-Süss B, Geibel S, Markert SM, Stigloher C, Lopez D. 2017. Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 171:1354–1367.e20. 10.1016/j.cell.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinho MG, Errington J. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol 50:871–881. 10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- 54.Müller S, Wolf AJ, Iliev ID, Berg BL, Underhill DM, Liu GY. 2015. Poorly cross-linked peptidoglycan in MRSA due to mecA induction activates the inflammasome and exacerbates immunopathology. Cell Host Microbe 18:604–612. 10.1016/j.chom.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneewind O, Missiakas D. 2014. Genetic manipulation of Staphylococcus aureus. Curr Protoc Microbiol 32:Unit 9C.3. 10.1002/9780471729259.mc09c03s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol 204:587–636. 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 57.Ferrer-González E, Fujita J, Yoshizawa T, Nelson JM, Pilch AJ, Hillman E, Ozawa M, Kuroda N, Al-Tameemi HM, Boyd JM, LaVoie EJ, Matsumura H, Pilch DS. 2019. Structure-guided design of a fluorescent probe for the visualization of FtsZ in clinically important Gram-positive and Gram-negative bacterial pathogens. Sci Rep 9:20092. 10.1038/s41598-019-56557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosario-Cruz Z, Chahal HK, Mike LA, Skaar EP, Boyd JM. 2015. Bacillithiol has a role in Fe-S cluster biogenesis in Staphylococcus aureus. Mol Microbiol 98:218–242. 10.1111/mmi.13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed. CLSI standard M07. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 61.Lee S-H. 2018. Optimal integration of wide field illumination and holographic optical tweezers for multimodal microscopy with ultimate flexibility and versatility. Opt Express 26:8049–8058. 10.1364/OE.26.008049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4, Tables S1 to S4, and supplemental text. Download JB00204-21_Supp_1_seq1.pdf, PDF file, 0.8 MB (854.1KB, pdf)