Abstract
Autism spectrum disorder (ASD) is a highly heterogeneous neurodevelopmental condition characterized by atypical social interaction and communication together with repetitive behaviors and restricted interests. The prevalence of ASD has been increased these years. Compelling evidence has shown that genetic factors contribute largely to the development of ASD. However, knowledge about its genetic etiology and pathogenesis is limited. Broad applications of genomics studies have revealed the importance of gene mutations at protein-coding regions as well as the interrupted non-coding regions in the development of ASD. In this review, we summarize the current evidence for the known molecular genetic basis and possible pathological mechanisms as well as the risk genes and loci of ASD. Functional studies for the underlying mechanisms are also implicated. The understanding of the genetics and genomics of ASD is important for the genetic diagnosis and intervention for this condition.
Keywords: Autism spectrum disorder, genetic basis, genomic structural variation, neurodevelopment, whole-genome sequencing
Introduction
Autism spectrum disorder (ASD) is defined as social interaction and communication deficits, restrictive repetitive behaviors across a phenotypic spectrum, with onset during early childhood [1]. Complications often occur including intellectual disability, epilepsy, motor deficits (hypotonia, apraxia or motor delay), gastrointestinal disturbances, and sleep abnormalities. ASD is a common neurodevelopmental disorder occurring in approx. 1% of individuals worldwide. The affected number has been increased rapidly in these years. According to the Centers for Disease Control and Prevention (U.S.A.), the prevalence of ASD is approx. 1:54, with a significantly higher proportion of males affected compared with females [2].
ASD is a complex and highly inheritable disease. Its clinical presentation is highly heterogeneous by encompassing a wide range of cognitive and adaptive abilities. The degree of heritability of ASD has been estimated as 40–90% [3,4], but a significant proportion of genetic risk factors remains undefined.
As the technology of molecular genetics and genomics developed rapidly, inherited risk factors of ASD have been identified through big genomics data in large patient cohort studies. Functional experiments are conducted to explain how these genomic variations play a role in molecular, cellular, or brain functions to uncover the potential pathogenic mechanisms. In this review, we highlight the recent findings of molecular genetics and genomics of ASD, shading light on how genetic risk factors affect cellular functions and clinical phenotypes, to direct the precision diagnosis and intervention of ASD.
Genetic basis of ASD
Genetic or environmental risk factors of prenatal, perinatal or postnatal period, could cause ASD alone or together [5]. Environmental factors such as exposure to heavy metal, deficiency of vitamin D, advanced parental age, and complications of pregnancy or birth have been shown to be risk factors of ASD [5–7]. According to the twin studies over 50 years, ASD concordance is 50–90% in monozygotic twins, while the concordance in dizygotic twins is 30% [8–11]. It is now accepted that ASD is a highly inheritable condition, the risk becomes higher with a closer kinship of affected individuals [8]. Known genetic causes include (but not limited to) copy number variants (CNVs), de novo single nucleotide variants (SNVs), common genetic variants, mosaicism, non-coding and regulatory pathogenic variations, and inherited recessive variants. However, to date, approx. 70% of the affected individuals have no genetic etiology identified [12].
De novo CNVs
CNVs refer to large deletions or duplications often involving in several genes. The association of phenotype with gene dosage exists, but the confirmation of relationship is often difficult. In 2007, comparative genomic hybridization was used to establish a significant association between de novo submicroscopic structural variation and autism [13]. From then on, more CNVs related to autism have been identified. The curated CNVs with ASD from SFARI Gene (https://gene.sfari.org/) are shown in Table 1.
Table 1. Known CNVs in ASD.
| Location | CNV type | Syndrome | Spanning range* | Associated/candidate genes |
|---|---|---|---|---|
| 1q21.1 | Deletion/Duplication | 1q21.1 deletion/duplication syndrome | Chr1:145900678-147965543 | - |
| 2p16.3 | Deletion/Duplication | NRXN1 deletion syndrome | Chr2:47600165- 53040270 | FBXO11, NRXN1 |
| 2q11.2 | Deletion/Duplication | 2q11.2 deletion syndrome | Chr2:97739057- 98115695 | - |
| 3q29 | Deletion/Duplication | 3q29 deletion/duplication syndrome | Chr3:195676676-197366632 | DLG1, PAK2, TM4SF19 |
| 5q35 | Duplication | 5q35 duplication | Chr5:178554060-179589550 | - |
| 7q11.23 | Deletion/Duplication | Williams–Beuren syndrome/Williams–Beuren duplication syndrome | Chr7:72311894- 74129587 | CLIP2, GTF2I, STX1A |
| 8p23.1 | Deletion/Duplication | 8p23.1 deletion/8p23.1 duplication syndrome | Chr8:8123460- 11384691 | - |
| 15q13.3 | Deletion/Duplication | 15q13.3 deletion syndrome/15q13.3 duplication | Chr15:30938215-32914140 | ARHGAP11A, CHRNA7, FAN1, OTUD7A, TRPM1 |
| 16p11.2 | Deletion/Duplication | 16p11.2 deletion/duplication syndrome | Chr16:29692499-30792499 | CORO1A, KCTD13, MAPK3, SEZ6L2, SRCAP |
| 16p12.2 | Deletion/Duplication | 16p12.2 deletion/duplication | Chr16:21356420-21577433 | - |
| 16p13.11 | Deletion/Duplication | 16p13.11 microdeletion syndrome/16p13.11 microduplication | Chr16:14972499-16522499 | - |
| 16p13.3 | Deletion/Duplication | 16p13.3 deletion syndrome/16p13.3 duplication | Chr16:3392370- 5752860 | CREBBP |
| 17p11.2 | Deletion/Duplication | Smith–Magenis syndrome/Potocki–Lupski syndrome | Chr17:16532736-20464365 | RAI1 |
| 17q11.2 | Deletion/Duplication | 17q11.2 deletion syndrome/17q11.2 duplication | Chr17:29015932-29149664 | - |
| 17q12 | Deletion/Duplication | 17q12 deletion/duplication syndrome | Chr17:37228545-39077997 | CACNB1, KRT26, NR1D1, THRA |
| 22q11.2 | Duplication | 22q11.2 duplication syndrome | Chr22:21031117-21651381 | LZTR1 |
| 22q13.3 | Deletion | 22q13.3 deletion syndrome | Chr22:41122568-49565875 | EP300, TCF20, XRCC6 |
Genomic location is referred to Human Genome GRCh37/hg19.
De novo SNVs
The clinical implementation of trio exome sequencing has shown a significant contribution to the discovery of de novo SNVs to autism risk [14–16]. As these variants usually affect a single gene, it is particularly important in emphasizing the underlying neurobiology of de novo SNVs associated with autism.
Enhanced bioinformatics analyses integrate evolutionary constraints to identify risk genes with a false discovery rate less than or equal to 0.1. In addition to utilizing probability of loss of function (pLI), missense badness, PolyPhen-2 constraint score, researchers are able to identify variants affecting gene functions by predicted impact [17]. These analyses not only confirm enrichment of de novo loss-of-function mutations which affect highly constrained genes, but also identify pathogenic missense mutations. Besides, functional experiments are crucial for these validations to better understand the mechanism of pathogenicity.
The SFARI gene database (https://gene.sfari.org/) has comprehensive and updated information on ASD-associated genes [18]. In the released 2020 Q4 database (updated on 13 January 2021), 1003 genes are divided into score 1 (High Confidence), 2 (Strong Candidate), or 3 (Suggestive Evidence) due to the current evidence to support the function of a certain risk gene in ASD development. According to the gene list, the risk genes have a bias of distribution on each chromosome, for instance, high confidence ASD-associated genes (score 1) mainly clustered on the chromosome X. This bias has confirmed the male-to-female ASD ratio which is approx. 4 to 1 [19]. Transcriptome and whole-genome sequencing (WGS) suggest that ASD risk genes may be involved in dysregulation in specific molecular processes, including chromatin modifications, RNA splicing, signaling pathways, gene expression regulation, neuronal communication, cytoskeletal organization, and cell cycling [20–22]. The curated 889 ASD risk genes with SNVs from SFARI Gene (https://gene.sfari.org/) are listed in Supplementary Table S1 supported by evidence from literature.
Common genetic variants
Common genetic variants are those variants with higher allele frequencies (usually greater than 0.05). Each of the variants has a small effect on ASD, or together with environmental factors, resulting in an individual bypassing a risk threshold to develop into the disease [4,23,24]. This is also called a polygenic model. Polygenic models are supported by the following multiple lines of evidence: (1) the genetic factors are highly and repeatedly inherited in ASD families [10,25]. (2) The proportion of related phenotypes such as social and behavioral problem is higher in the first-degree relatives of children with ASD compared with the general population [26,27]. (3) Through analyzing the single-nucleotide polymorphism (SNP) data, inherited common variants (minor allele frequency > 0.05) and variants marked by common genetic variants account for a large proportion of the ASD risk in total [3,4,28].
Due to the limited sample size of ASD individuals, specific common variants had not been identified until 2019 by the large-scale genome-wide association study (GWAS) in autism study [29]. Several significant common risk loci delineating the genetic heterogeneity of phenotypic subgroups have been found to reveal that common variants play a large role in high-functioning autism. It has shown that common variants in autism are enriched in regulatory elements, which are predicted to influence the development of the human cortex by utilizing Hi-C data from the developing fetal brain [29]. With increasing GWAS sample size and power, as well as functional experiments, more common risk loci will be identified, which will provide more genetic data for further investigation. The 316 curated known genes with common variants of ASD from SFARI Gene (https://gene.sfari.org/) are summarized in Supplememtary Table S2 supported by evidence from literature.
Mosaicism
Mosaic mutations are de novo variants occurring after fertilization, only involving some cell lineages of the body. The proportion of autism cases affected by somatic variants is unknown, while recent studies revealed that 0–7.5% of de novo mutations in autism were postzygotic mosaic mutations [30,31]. Despite the limitation of small available samples and the absence of parental samples, studies of mosaic mutations in postmortem brain tissue suggest the presence of damaging mosaic mutations in some autism brains. Targeted sequencing and WGS in postmortem autism brain identified potentially risk-modifying somatic mutations present in brain DNA. Somatic mutations may also contribute to the risk of ASD by disrupting the gene regulatory elements [32,33].
Research on mosaic mutations in autism provides another approach to understand cells and circuits critical for the underlying neurobiology, which may help to explain a fraction of cases without genetic causes identified. It is worth noting that mutations at low cell proportion in the brain might not be detectable in peripheral DNA, while peripherally detected somatic mutations may be present in different cell types and distributions within the brain [12]. Single-cell sequencing (scSeq) provides a new solution to identify somatic mutations in different cell lineages.
Non-coding and regulatory pathogenic variants
De novo and inherited non-coding variants have shown to be involved in autism risk [34–38]. Due to lack of robust functional categorization of the non-coding genomic structures, bioinformatics and experimental approaches are often adopted to pursue this issue. Comparative genomics technology is used to identify regions of the human genome with accelerated divergence, or human accelerated regions (HARs), from evolutionarily conserved sequences, specifically reflecting critical function in the human beings, of which many regions are predicted as regulatory function in brain development. A significant amount of both de novo CNVs and biallelic SNVs in individuals with autism have been identified through HAR analyses [35,36]. WGS has detected multiple smaller and gene-disruptive CNVs involving dosage sensitivity. Many neurodevelopmental genes such as ARID1B, SCN2A, NR3C2, PRKCA DSCAM, DISC1, WNT7A, RBFOX1, MBD5, CANX, SAE1, and PIK3CA are associated with ASD by affecting putative regulatory elements of these genes [37]. Further functional experiments (in vitro cellular reporter assays and in vivo mouse models) provide more evidence with respect to the impact of the identified variants, locating at the active enhancers of CUX1, PTBP2, GPC4, CDKL5, therefore such ASD or neural function linked biallelic HAR mutations have been revealed [36]. In addition, it has been shown that paternally inherited cis-regulatory structural variants (SVs), including but not limited to CNTN4, LEO1, RAF1, and MEST, are preferentially transmitted to affected offspring [35].
By utilizing a deep learning method in combination with extensive experimental data, de novo variants in probands in the Simons Simplex Collection were annotated [39]. They demonstrated that regulatory de novo mutations in probands had a significantly higher predicted functional impact than those in unaffected siblings, while some of the variants were involved in the regulation of previously identified biological pathways, suggesting that both non-coding and coding variations may have effects on the risk of ASD.
Another creative approach using rigorous and unbiased genome-wide category-based de novo risk score is adopted to reveal that de novo mutations at distal conserved promotors could increase autism risk [38]. Larger samples are required to determine the nuances of how non-coding and regulatory variations affect risk of ASD.
Inherited recessive variants
It was predicted that inherited recessive variants contribute to autism risk in 1985 [40]. Whole-exome sequencing (WES) has identified rare de novo heterozygous mutations, as well as rare recessive mutations inherited from consanguineous families [41,42], indicating that inherited recessive variants play a role in autism liability [40,41].
A study in 2019 has estimated that approx. 5% of total ASD cases are caused by biallelic loss-of-function or damaging missense mutations. An excess of damaging biallelic missense variation was significantly enriched in cases than in controls. This study agreed with the conclusion of earlier studies that females have protective effect for rare recessive mutations [43]. We curated 207 known ASD recessive inherited risk genes from SFARI Gene (https://gene.sfari.org/) in Supplementary Table S3 supported by evidence from literature.
In conclusion, current studies have mostly focused on protein-truncating variants and recurrent CNVs, however, the contribution of non-coding variants is largely unknown or underestimated. In fact, non-coding variants account for a considerable amount of ASD cases with known molecular etiology. The percentages of ASD individuals harboring known mutations are syndromic (3.4%), de novo SNVs (1.34%), and CNVs (1.28%). The heritability of autism in addictive genetic effect (rare inherited and common inherited) is estimated to be 52%, and the non-additive genetic effect is approx. 7% including de novo mutations and non-additive effects [4,44]. To ascertain the genetic basis of ASD, more attention should be paid on the non-coding regions of the genome structure. Non-coding variants contribute to ASD development at a complicated mode. It has been revealed that disrupted non-coding RNAs, regulatory elements, or 3D chromatin conformation have profound effects on ASD and neurodevelopment [45].
ASD and big genomics data
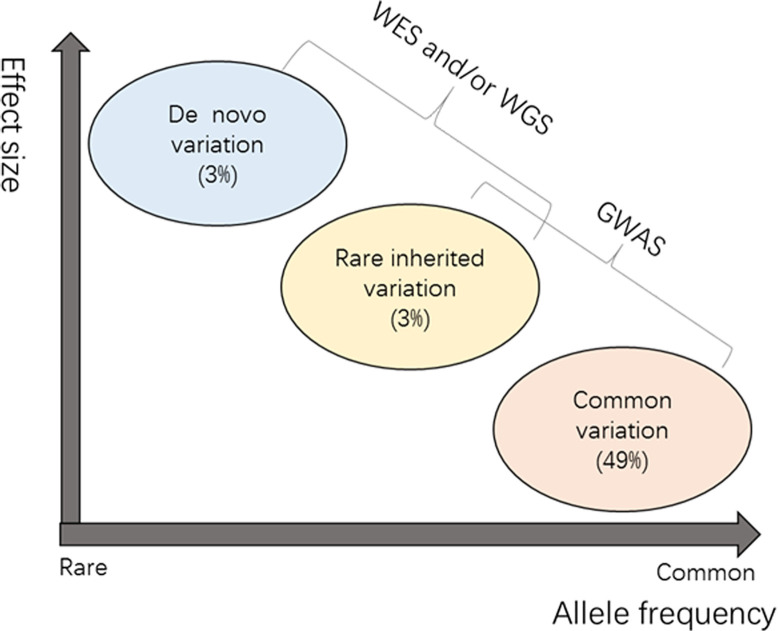
Family and twin studies on ASD have shown the importance of common variants in heritability, as well as the large effects of rare and de novo variants in individuals [4] (Figure 1). Genomic studies of ASD we refer here mainly include results from GWAS, WES, and WGS. Comparison of advantages and limitations of these genomics studies is summarized in Table 2.
Figure 1. Genetic architecture of ASD.
This is a sketch map showing liabilities in three mutation classes, namely common variation (minor allele frequency (MAF) > 5%) and rare inherited variation (MAF < 1%) and de novo variation [4]. The x-axis represents the allele frequency from rare to common. The effect size is increasing from bottom to top. The liabilities in different mutation classes are shown in brackets.
Table 2. Comparison of advantages and limitations of genomic studies in ASD research.
| Approach | Advantages | Disadvantages |
|---|---|---|
| GWAS | 1) Relatively easy to perform technically 2) Meta-analysis can be used to increase the statistical power 3) Common variant in common disease model is often adopted |
1) Sample size limitation is common in association studies 2) Population stratification or selection criteria for cases and controls are confounders in GWAS or meta-analysis 3) Replication of risk loci in different populations or labs is rarely seen 4) Biological relevance of risk loci is often very difficult to validate, especially for those SNPs located in the intergenic region 5) Rare pathogenic allele is often missing |
| WES | 1) Sample collection is easy to obtain in a single center 2) Trio-WES is often adopted to analyze allele transmission and de novo variants 3) Candidate genes are easily selected from pathogenic or likely pathogenic variants 4) Rare variants are often focused 5) Cost-effective in identifying coding variants or de novo variants |
1) Variants are limited to the exonic regions or exon/intron boundaries 2) VUSs are difficult to interpretate without functional assays 3) Multicenter validation is often limited |
| WGS | 1) Sample collection is easy 2) SNV, CNV, SV can be simultaneously analyzed at the genomic level 3) De novo variants can be found in Trio-WGS 4) Variants from both coding and non-coding regions are covered |
1) Relatively expensive for one sample 2) Length and depth of sequencing reads are important for the quality in expense of the costs 3) Variants in non-coding regions are difficult to replicate in animal models 4) Accuracy of prediction models for SVs needs to be improved |
Common variants in GWAS
GWAS is a widely adopted approach to associate common variants with complex diseases. From 2009 on, many GWASs of ASD have been conducted [3,46–49]. However, no variant has been robustly replicated in these GWASs due to sample size limitation. One way of increasing sample size is to perform meta-analysis study by integrating individuals from different ASD cohorts. Recently, a meta-analysis based GWAS with 18381 ASD cases and 27969 controls including iPSYCH samples, Psychiatric Genomic Consortium samples, and five follow-up samples identified five genome-wide significant loci at LINC02790, AC120036.1, AC090987.1, AC025839.1, RSU1 [29].
Another way is to take advantage of phenotypic similarity to boost statistical power. A cross-trait meta-analysis was conducted based on GWAS data which included 65967 schizophrenia, 41653 bipolar disorder, 46350 ASD, 55374 attention deficit hyperactivity disorder, and 688809 depressions [50]. This study identified ten ASD associated genomic loci at genes RSRC1, AC099520.1, GALNT10, AC003044.1, SOX7, SORCS3, RBFOX1, DCC, MACROD2, ZNF877P. Cross-Disorder Group of the Psychiatric Genomics Consortium analyzed 232964 cases and 494162 controls from GWAS of anorexia nervosa, attention-deficit/hyperactivity disorder, ASD, bipolar disorder, major depression, obsessive-compulsive disorder, schizophrenia, and Tourette syndrome [51]. This meta-analysis detected 109 loci across eight disorders, where 23 genomic loci are commonly associated with four or more disorders including genomic loci at the transcription start sites of DCAF4L1, CTNND1, MRPS33, DFNA5 and at the gene bodies of DCC, RBFOX1, SORCS3, PLCL1, RGS6, CHADL, KCNB1, SOX5. In addition to increased sample size, functional annotations on common variants tend to incorporate with multiomics datasets, i.e. expression data from brain [52], HiC seq [29], in order to explain the effects of common variants on their molecular pathways. It should be noticed that GWAS has largely conducted in samples of European origin, which is inefficient to capture population-specific signals.
Rare and de novo variants from WES
The remarkable work on the identification of rare and de novo mutations in ASD is from the applications in high-throughput sequencing as well as large family-based cohort. At coding regions, the functional roles of rare and de novo mutations are assessed by the potential impact on protein function and structure. After the initial step, discovery and prioritization of ASD-related variants can be done by group-wise tests, e.g. Transmission and De Novo Association test (TDNA), giving the low frequency of rare and de novo mutations. Current evidence of ASD genes highlights the potential pathogenic role of genes carrying protein truncating variants and probably damaging missense variants. TDNA evaluates mutation burden in a gene-based model with weighted categories of above-mentioned two types of mutations. Researchers applied this model to 3871 cases and 9937 controls, where 22 autosomal genes were implicated including ADNP, ANK2, ARID1B, CHD8, CUL3, DYRK1A, GRIN2B, KATNAL2, POGZ, SCN2A, SUV420H1, SYNGAP1, TBR1, ASXL3, BCL11A, CACNA2D3, MLL3, CTTNBP2, GABRB3, PTEN, RELN, MIB1 [53]. Along with the growing sample size and heterogeneous insight of ASD, several refined versions are developed by either multiple population data [54], or probable intolerance of loss-of-function variation score (pLI) as a continuous metric to weight mutations [21].
As such, the TDNA model and other group-wise tests serve as a basis in prioritizing ASD genes and modified models could add new insights into ASD etiology, but the gene list generated by group-wise tests could be large and further refinement may be required. Enrichment analysis, protein–protein interaction (PPI) networks are useful approaches in refining the gene list [55]. The key to these approaches is either to find out the significant depleted or enriched groups of genes based on known knowledge, or to prioritize novel genes or weakly associated genes via interaction maps.
Another commonly applied method in prioritizing ASD genes is network analysis. For example, Detecting Association With Networks (DAWN) is a Hidden Markov Model (HMM)-based approach which models two sets of data, rare variants, and gene co-expression. Combining TADA scores and BrainSpan gene co-expression data, DAWN identified 102 genes through complex gene networks from 35584 individuals with 11986 ASD [54]. Majority of these genes are expressed in brain, where 53 are ASD predominant genes (e.g. ASH1L, CHD8, KMT5B, DEAF1, KDM6B, ANK2, SHANK3, PTEN, DSCAM), 49 are ASD and neurodevelopmental delay genes (e.g. ADNP, ANKRD11, ARID1B, MED13L, CHD2,TLK2, CTNNB1, POGZ, FOXP1, SLC6A1, SYNGAP1, GRIN2B, SCN2A, DYRK1A).
One important aspect is that all these approaches alone are useful in prioritizing candidate genes, but an increasing number of studies have applied combined or refined approaches to boost discovery power giving the complexity of ASD genetics. Moreover, multidimensional data are used to understand the underlying molecular mechanisms of ASD in recent years.
Rare and de novo variants from WGS
The basic ide a of prioritizing variants from WGS is very similar to WES, in which functional annotations and grouping are usually the initial step. By doing so, rare and de novo variants can be analyzed collectively in statistical models. However, WGS generates comprehensive types of variants encompassing SNVs, InDels and SVs, as well as vast number of variants locating at non-coding regions compared with WES, where methods have to be modified. The Category-Wide Association Study (CWAS), akin to GWAS, with SNP substituted for annotation categories, can be applied to variants generated from WGS. For example, CWAS was applied to all kinds of variants by grouping variants into various functional regions, such as gene-coding regions, conserved regions, with modest findings only [38]. Like GWAS, a big sample size increases the power of detection. Within 1902 quartet families (parents and one ASD-affected child, one healthy sibling), indications of de novo mutations at promoter regions by CWAS were found [34].
Focusing on only SVs, enrichment analysis and group-wise tests are commonly used. Brandler et al. applied enrichment analysis over several functional catalogs in total of 9274 subjects from 2600 families [35]. Burden test, a type of group-wise test, is applied for the comparisons of variant frequencies between cases and controls where novel loci and known tandem repeat expansions were found [56]. The identified SVs often need additional methods for further refinement. A method that adopted the idea that SVs lead to long-range interactions has been developed in our lab [57]. A similar method that took advantage of known disease-related SVs is used to refine the list of candidate SVs [58]. Furthermore, giving the complexity of SVs and a variety of source data providers, many efforts have been done to integrate SVs into a clean dataset. A unified workflow is necessary to accelerate the study on SVs in ASD.
Together, genomics studies established the importance of de novo mutations at protein-coding regions in ASD development, as well as with growing evidence for the modest effects of non-coding regions. Multidimensional data may also leverage our knowledge on molecular etiology of ASD, such as expression data [59–61] and epigenetics data [62].
ASD models
Cell models
Successful reprogramming of adult somatic cells transforms differentiated cells into induced pluripotent stem cells (iPSCs) [63]. The main features of iPSCs include the self-renewal capability and differentiation potential. As human iPSCs can differentiate into a variety of cell types, somatic cells obtained directly from patients could be induced into specialized in vitro cell models to study the disease mechanisms. Compared with other in vitro cell models, iPSC models directly derived from patients keep the genetic background of the ASD patients [64,65]. With the cellular models obtained from patients, the biological basis or molecular mechanism of the disorders is investigated to validate the association between the genotype and phenotype, and to develop new cell or pharmacological therapeutic approaches [64,65]. In recent years, numerous in vitro cell models have been utilized to study the mechanism of ASD (Supplementary Table S4). Challenge of in vitro cellular models is how to model the cellular and physiological phenotypes which are most relevant to ASD patients. Although there are a lot of advantages of in vitro cell models, cells in culture can not fully recapitulate all the complex mechanism of ASD.
Animal models
Flourishing genetic achievements has accelerated the generation and characterization of different types of animal models of ASD. The commonly used animal models include zebrafish, mouse, rat, and non-human primates (Supplementary Table S5). Mouse or rat genes are highly homologous to human genes, compared with other animal models. Mouse models provide an experimental platform to study molecular mechanisms, cellular pathways, circular disturbances and behavioral analyses of ASD, offering the opportunity to explore whether the behavioral abnormalities could be reversed by potential therapeutics before translating them to humans [66]. However, there are still some drawbacks of mouse models. Neuropsychiatric behaviors assessed by psychiatrists are difficult to be measured or recorded in mice (e.g. language). Comorbidities such as sensory dysfunction, learning deficits, locomotor dysfunction, fear and anxiety could confound with core ASD phenotypes in human during the assessment. Primate models or invertebrates are complementary to mouse models [67]. Non-human primates models could simulate the complex behaviors and higher cortical functions of human, whereas zebrafish and invertebrates could be efficiently manipulated in large-scale parallel experiments [44,67,68]. However, due to the non-conserved non-coding regions in animals, it is difficult to model those SNVs, CNVs, or SVs in the non-coding regions.
Pathogenic mechanisms of ASD development
There are plenty of hypothetical pathophysiological mechanisms of ASD, which have been tested by studies in humans or model systems from different aspects. Most of these mechanisms demand more work to figure out the exact molecular pathways. Furthermore, these models overlap at some extent. Different stages of brain development may share the same genes or molecular pathways. More interestingly, how early developmental disruption correlates to phenotypes after birth or at later ages remains unknown. According to recent studies, it is shown that the abnormal development of ASD may begin in prenatal period [69]. ASD risk genes are expressed in prenatal and postnatal stages, most of which are expressed broadly as regulatory genes in a variety of biological processes in brain development. The aberrations of ASD risk genes could cause abnormalities through disrupting regulatory networks and dysregulating key signaling pathways such as PI3K/AKT, RAS/ERK, Notch, and Wnt/ β-catenin.
From the first to the third trimesters, namely Epoch-1, with the combination of broadly expressed regulatory risk genes and brain-specific risk genes, many embryonic development processes (e.g. cell proliferation, neurogenesis, cell fate determination, and migration) are disrupted. In the third trimester and early postnatal period, namely Epoch-2, there may exist dysregulation of cortical wiring (including neurite outgrowth, neural network organization, synaptogenesis) due to a different set of genes.
Here we summarize the main ASD-related genes of different brain developmental stages in Table 3 [70–85], and the prevailing hypotheses of ASD development [44] are discussed below.
Table 3. ASD risk genes and associated affected developmental processes.
| Affected developmental process | Gene symbol |
|---|---|
| Neuron migration | ASTN2, AUTS2, CHD8, CNTNAP2, DLX1/2, FOXP1, LIS1, NCAM2, NCKAP1, NDE1, RELN, TBR1, TCF4 |
| Cell–cell adhesion | ASTN2, CHD8,CHD9, CHD10, CHD13, NRLG1/2/3/4/4Y, NRXN1/2/3 |
| Neurite growth | AUTS2, CSDE1, CTNND2, DOCK4, KIAA2022/NEXMIF, MECP2, NF1, PTEN, RELN, TAOK2, TSC1, TSC2, UBE3A, |
| Synapse formation | CNTNAP2, CTTNBP2, FMR1, TAOK2 |
| Synaptic function | CNTNAP2, DIP2A, NRXN1/2/3, SYN1/2/3 |
| Synaptogenesis | DLG4, GPHN, MECP2, NRLG1/2/3/4/4Y, NRXN1/2/3, PTEN, SHANK1/2/3, SYN1/2/3, TSC1, TSC2 |
| Synaptic plasticity | FMR1, MECP2, SHANK1/2/3, TSC1, TSC2, UBE3A |
| Translation | AGO1, CNOT3, DYRK1A, eEF1A2, eEF2, eIF3g, eIF4B, eIF4E, FMR1, JAKMIP1, PABPC1, PTEN, RPL10, RPS6, TNRC6B, UPF3B |
| Intracellular transport | CYFIP1, LIS1, NDE1,WDFY3 |
| Neurogenesis | LIS1, NDE1, PTEN, WDFY3 |
| Transcription | MECP2, TCF4 |
Dysregulation of fetal cortical development
Multiple lines of evidence from human genetic studies and postmortem studies support the notion that dysregulation of fetal cortical development could result in ASD [86–88]. Neuropathological studies have revealed a number of cortical developmental errors such as smaller neuron size, more neuron number, mislocated neurons, misoriented pyramidal neurons, disrupted lamination, reduced white matter tracks, and abnormal dendrites in ASD patients [86]. Other studies showed that the cortical minicolumn, which is a basic processing unit of cortical circuits, is more narrow and densely packed [89]. Patches of cortical cells could not be laminated regularly due to lack of specific laminar markers [90]. The brain size is reduced at birth but overgrown during childhood in individuals affected with ASD [91].
It has been reported that the target genes involved in the mTOR pathway harbor more ASD-related variants, and may affect the regulation of processes of cell proliferation, cell growth, and neuronal morphogenesis [92]. The target genes involved in the Wnt pathway may regulate the courses of radial glia self-renewal, neuronal differentiation, and brain dorsoventral pattern [93]. We found that BLOS2 interacts with Notch1 to mediate the endolysosomal trafficking of Notch1. Loss of BLOS2 leads to elevated Notch signaling, which consequently increases the proliferation of neural progenitor cells and inhibits neuronal differentiation during cortical development [94]. Target genes involved in the BAF complex (a multisubunit complex mediating chromatin remodeling) may regulate neurogenesis and neuronal morphogenesis [95]. Mutations of the genes involved in these pathways are considered to play a role in cortical development.
Synaptic dysfunction
Neuropathological studies have provided strong genetic evidence for synaptic dysfunction. Mutations in genes encoding synaptic cell-adhesion molecules, excitatory and inhibitory synaptic scaffolding molecules, the excitatory glutamatergic receptor, inhibitory GABAergic receptor subunits, inhibitory synaptic scaffolding molecule gephyrin and neurotransmitter release regulators are associated with ASD in numerous studies. Namely, these molecules include neurexins [53,96,97], neuroligins [98], the SH3 and multiple ankyrin-repeat domain (SHANK) proteins [53,97], GRIN2B, GABAR [53,96], GPHN [53], the synaptotagmins [16,53], and synapsins [53,99].
The dysregulation in synaptogenesis and synaptic transmission have effects on ASD [100]. Meanwhile, the glutamatergic and GABAergic synaptic dysfunction raise the hypothesis that disruption in the excitatory/inhibitory (E/I) balance leads to ASD [101]. However, E/I imbalance is also frequently observed in other neuropsychiatric disorders such as epilepsy, Alzheimer’s disease, and schizophrenia [102]. Thus, how E/I imbalance has an effect on ASD pathophysiology demands dissecting its spatiotemporal dynamics, which means we need to figure out whether there is a key period or whether it is circuit specific that an E/I imbalance leads to ASD-relevant behavior in numerous ASD models. Moreover, an E/I imbalance could result from either synaptic physiology changes or altered cell fates that result in shifted ratio of inhibitory and excitatory synaptic neurons [103].
Abnormalities of gene transcription and translation
Neuronal activity could dynamically regulate gene transcription and protein translation in neurons, to ensure that specific gene could expressed spatially or contextually within subcellular partition [104]. Many studies have shown that disruption of this activity-dependent gene transcription and translation may cause ASD. Mutations of risk genes or risk loci such as TSC1, TSC2, FMR1 and dup15q11-q13 suggest that ASD patients may result from dysregulated neuronal translation [92,105]. Studies in mice emphasize the viewpoint of molecular enrichment between synaptic function, synaptic plasticity, and translational regulation [106]. Synaptic pruning and stability are also regulated though activity-dependent transcription and translation [107]. As a supporting evidence, it has been reported that ASD patients present increased dendritic spine density in the temporal lobe [108].
Many ASD risk genes are confirmed to have an increased risk for ASD, these genes could be transcriptionally co-regulated (such as MEF2A, MEF2C, and SATB1) as well as translationally regulated (e.g. FMR1), suggesting that a potential convergent mechanism in ASD development is activity-dependent gene regulation [109]. It is critical to explore the link between ASD risk gene-related changes in synapse dynamics and the specific phenotypes presented in ASD models and patients. There is an assumption that small differences in synaptic function and timing will dysregulate the linkage between higher order association regions, including the frontal–parietal, frontal–temporal, and frontal–striatal circuits which mediate social behaviors [110]. To investigate the linkage between synaptic dysfunction and multiple heterogeneous phenotypes in ASD patients, it is necessary to study transcriptional and translational regulation related to spatiotemporal dynamics and the differences in micro- and macro-circuit connection [111].
Altered neural circuitry
Studies of neuroimaging and neuropathology in ASD patients imply that within the cortex and in cortico–striatal circuits, there exists disruption of resting state network activity as well as altered macrocircuit connectivity [112,113]. By utilizing systematic imaging in ASD-like mouse models, studies illustrated that the parieto–temporal lobe, the cerebellar cortex, the frontal lobe, the hypothalamus, and the striatum are the most commonly affected regions, shared by all the 26 mouse models [114]. Another candidate ASD region is the amygdala, which plays a critical role in modulating the emotion of fear as well as social behaviors [113,115]. Besides, striatal dysfunction is likely the neural basis for repetitive behavior as well as motor routine learning both in mice and in humans [116].
Except for the frontal circuits, cerebellar function also plays a role in social behavior. Individuals with ASD have deficient processing of abstract animations [117] and body motion [100]. A recent study showed that both degenerative cerebellar disease and autistic patients could not handle the immediate perceptual component of the mental state recognition (for example, to distinguish other people’s psychosis from their eye expression) and superior conceptual level of mentalization (for example, to tell a false opinion) [118]. Additionally, functional MRI (fMRI) studies illustrated that social impairment typically observed in ASD patients may link the dysregulation of cerebellar outputs to default network brain areas [119]. Differences in cerebellar volume [120] and decreased gray matter volumes in given cerebellar areas [121] have been recognized from the earlier neuroimaging studies of ASD. Early cerebellar damage is responsible for increased internalizing behaviors, emotional and attentional deficits, and social contact disorder [122], suggesting that the autistic characteristic behaviors could derive from atypical cerebellar development [123].
From cells to systems, animal models have also shown how the cerebellum is involved in autism [124,125]. All of the 26 autism mouse models revealed cerebellar abnormalities through the clustering analysis [126]. Knockout of Tsc1 in mouse cerebellar Purkinje cells could cause core ASD-like behaviors [127], suggesting that social deficits of autism could be derived from cerebellar dysfunction in mice. As in humans, disrupted cerebellar development in early stage in rodents may result in ASD-like phenotypes [128]. However, how to correlate the phenotypes of mouse to human circuits is still a challenging task. Moreover, many relevant brain areas in humans, like the frontal and temporal lobes, have changes dramatically during the evolution of primates [129]. Together with mouse models, primate models can be a complement to utilize comparative studies, which may reveal new candidate brain circuits related to ASD pathogenesis.
Studies of cerebellar structural and functional connectivity in ASD patients provide evidence for the involvement of cerebellar in autism [130]. Decreased cerebellar white matter density [131] and larger cerebellar white matter volume [132] in autism has been reported. Diffusion imaging studies showed that the cerebellum and the cerebral cortex connected pathways change its integrity [133], suggesting that in the middle and superior cerebellar peduncles, fractional anisotropy (FA) could be decreased, while mean diffusivity increased. Thus, cerebellar white matter volume could be utilized as a predictor of future autism diagnosis [134], while decreased cerebellar FA correlates with autism severity [135].
The findings of resting-state functional connectivity implied atypical cerebro–cerebellar networks of ASD, presenting decreased connectivity within settled networks, most of which worked for social intercourse. Besides, in autism there are increased connectivity between cerebellum non-motor areas and sensori-motor cerebral cortical areas, implying atypical linkage between sensori-motor and non-motor cerebro–cerebellar circuits [136].
Dysregulated neuron-glia signaling and neuroinflammation
It is reported that there are numerous activated microglia and astrocytosis in multiple brain areas in brains from individuals with ASD. Imaging studies utilizing positron emission tomography (PET) [137] and studies on postmortem brains [88] show a large number of activated microglial and astrocytosis cells in the brains of ASD affected individuals [138]. Synaptic dysfunction in ASD patients, which causing abnormal synapses numbers, functions, and E/I balance which finally result in autistic phenotype, could be caused by dysregulated synaptic pruning and homeostasis due to the vicious cycle of up-regulated microglia and astrocytes [139,140]. As synaptic development and pruning could be regulated by astrocytes and microglia [141,142], it could provide another clue for therapeutic targets [143].
There is a gender bias in ASD incidence: the affected males are four-times higher than the affected females. In current studies, the properties of microglia have a discrepancy in different sex. Compared with males, the cultured microglial cells of females have stronger ability of phagocytosis [144]. According to a microglia-specific eIF4E overexpressing mice experiment, only male mice showed the phenotype of abnormal behaviors and microglial morphologies ability [145]. Abnormal phagocytic ability of microglia may be the mechanism of activated microglia in ASD brain: the accumulation of degenerated cells and materials caused by impaired phagocytic ability enhancing activation and proliferation of microglia via damage-associated molecular patterns, while the damaged phagocytosis is unaffected [146].
The dramatically increased incidence of ASD may not be simply considered as changes in diagnostic criteria and/or methods [147]. It has shown that environmental factors which affecting immunological responses in human brain may lead to autism. Microglia in fetal brains could be activated by environmental factors through maternal immune activation (MIA), which consequently lead to ASD. In a study of mouse model, infected with microorganisms like herpes simplex virus during pregnancy caused MIA, hyperactivating fetal brain microglia, and finally resulting in autistic behaviors in the offspring [148]. The mechanism of how MIA-induced activated microglia impact the synaptic transmission, causing autistic symptoms is still not fully clear.
Impaired adult neurogenesis
Neurodevelopment continues after birth. Circuitry maturation or plasticity is further established upon adult neurogenesis. Adult neurogenesis is subject to epigenetic changes. Our results have shown that dysbindin-1C, an isoform of schizophrenia susceptibility gene DTNBP1, is involved in the maturation of adult newborn neurons in the dentate gyrus (DG) by regulating the survival of hilar mossy cells [149,150]. Similarly, FMR1 is involved in the cell survival at the ventral subregion of the DG [151]. FMRP plays an important role in adult hippocampal neurogenesis and hippocampus-dependent learning by regulating the adult neural stem cell (aNSC) fate through the translational regulation of GSK3β [152]. Loss of FMRP compromises the differentiation of aNSCs by impacting many mitosis and neurogenesis genes at transcription or translation level. In addition, knockdown of necdin, an FMRP-repressed transcriptional factor, rescues aNSC differentiation [153]. Another mechanism related to loss of FMRP is the increased protein synthesis of histone acetyltransferase EP300 and ubiquitination-mediated degradation of histone deacetylase HDAC1 in aNSCs [154]. As adult hippocampal neurogenesis converges several pathways, other ASD risk genes may be involved in this process.
Epigenetic and transcriptomic differences
Except for the FMR1 gene in regulating histone modifications as mentioned above [154], it has been shown that DNA methylation differences in genomic regions linked with immunity and neuronal regulation in ASD brain [155,156]. Histone H3K27 acetylation is also clarified to have an effect on genes involved in synaptic transmission and morphogenesis [157]. By integrating omics studies of mRNA expression, miRNA expression, DNA methylation, and histone acetylation from ASD and control brains, researchers have dissected a convergent molecular subtype of ASD with shared dysregulation across both the epigenome and transcriptome. They expanded the repertoire of differentially expressed genes in ASD as well as identified a component of up-regulated immune processes which is related to hypomethylation. By utilizing eQTL and chromosome conformation datasets, differentially acetylated regions with their cognate genes could imply an enrichment of ASD genetic risk variants in hyperacetylated non-coding regulatory regions linked to neuronal genes [158]. Besides, the expression differences in one-carbon metabolites transcript of TGR-AS1, SQSTM1, HLA-C, and RFESD were identified to be related to ASD through differential expression analysis [159].
Conclusion and perspectives
ASD has a complex origin, arising from both genetic risks and environmental exposures. As the increase in sample sizes and development of statistical and biological approaches, there are more and more evidence to uncover diverse genetic mechanisms and biological pathways of ASD. Big genomics data and bioinformatics and experimental innovation accelerate the investigation of ASD genetic risk factors, especially the common variants and those variants in non-coding and regulatory regions.
A challenge in the omics era is how to integrate the meaningful link from multiomics data. Meaningful genomic variants can be evidenced by transcriptomic and proteomic data. scSeq is now a trend to explore the abnormalities in spatiotemporal ASD brains and in inflammatory cells of both brain regions and bloodstream that are potential features or biomarkers of ASD.
In future, along with the understanding of the genetics and genomics of ASD, studies should try to integrate the intricate connections among different genetic sources, biological pathways and brain connectomes, exploiting potential biomarkers and therapeutics for individuals affected with ASD. It is well accepted that ASD can be categorized into different subtypes by inputting DSM-V scores, NMR images, and omics data. Uncovering these subtypes will be another challenge in ASD research. Combined with new science and technology such as artificial intelligence (AI), it could be possible to improve the genetic diagnosis and intervention for this condition.
Supplementary Material
Abbreviations
- aNSC
adult neural stem cell
- ASD
autism spectrum disorder
- CNV
copy number variant
- CWAS
category-wide association study
- DAWN
Detecting Association With Networks
- DG
dentate gyrus
- E/I
excitatory/inhibitory
- eQTL
expression quantitative trait loci
- FA
fractional anisotropy
- GWAS
genome-wide association study
- HAR
human accelerated region
- InDel
insertion and deletion
- iPSC
induced pluripotent stem cell
- MIA
maternal immune activation
- scSeq
single-cell sequencing
- SNP
single-nucleotide polymorphism
- SNV
single nucleotide variant
- SV
structural variant
- TDNA
Transmission and De Novo Association test
- WES
whole-exome sequencing
- WGS
whole-genome sequencing
Contributor Information
Xin Ni, Email: nixin@bch.com.cn.
Wei Li, Email: liwei@bch.com.cn.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was partially supported by the Ministry of Science and Technology of China [grant number 2016YFC1000306]; the National Natural Science Foundation of China [grant number 31830054]; and the Beijing Municipal Health Commission [grant number JingYiYan 2018-5].
References
- 1.American Psychiatric Association (2013) Diagnostic and Statistical Manual of Mental Disorders, 5th, ed., Arlington: American Psychiatric Association, Washington, DC [Google Scholar]
- 2.Maenner M.J.et al. (2020) Prevalence of autism spectrum disorder among children aged 8 years — Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 69, 1–12 10.15585/mmwr.ss6904a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cross-Disorder Group of the Psychiatric Genomics Consortium (2013) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 45, 984–994 10.1038/ng.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaugler T.et al. (2014) Most genetic risk for autism resides with common variation. Nat. Genet. 46, 881–885 10.1038/ng.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lord C.et al. (2020) Autism spectrum disorder. Nat. Rev. Dis. Primers 6, 5 10.1038/s41572-019-0138-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modabbernia A.et al. (2017) Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses. Mol. Autism 8, 13 10.1186/s13229-017-0121-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu S.et al. (2017) Advanced parental age and autism risk in children: a systematic review and meta-analysis. Acta Psychiatr. Scand. 135, 29–41 10.1111/acps.12666 [DOI] [PubMed] [Google Scholar]
- 8.Sandin S.et al. (2017) The heritability of autism spectrum disorder. J. Am. Med. Assoc. 318, 1182–1184 10.1001/jama.2017.12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tick B.et al. (2016) Heritability of autism spectrum disorders: a meta-analysis of twin studies. J. Child. Psychol. Psychiatry 57, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallmayer J.et al. (2011) Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 68, 1095–1102 10.1001/archgenpsychiatry.2011.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg R.E.et al. (2009) Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch. Pediatr. Adolesc. Med. 163, 907–914 10.1001/archpediatrics.2009.98 [DOI] [PubMed] [Google Scholar]
- 12.Dias C.M.et al. (2020) Recent advances in understanding the genetic architecture of autism. Annu. Rev. Genomics Hum. Genet. 21, 289–304 10.1146/annurev-genom-121219-082309 [DOI] [PubMed] [Google Scholar]
- 13.Sebat J.et al. (2007) Strong association of de novo copy number mutations with autism. Science 316, 445–449 10.1126/science.1138659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iossifov I.et al. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neale B.M.et al. (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 10.1038/nature11011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders S.J.et al. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 10.1038/nature10945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lek M.et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abrahams B.S.et al. (2013) SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 36 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loomes R.et al. (2017) What is the male-to-female ratio in autism spectrum disorder? A systematic review and meta-analysis J. Am. Acad. Child. Adolesc. Psychiatry 56, 466–474 10.1016/j.jaac.2017.03.013 [DOI] [PubMed] [Google Scholar]
- 20.Ruzzo E.K.et al. (2019) Inherited and de novo genetic risk for autism impacts shared networks. Cell 178, 850.e26–866.e26 10.1016/j.cell.2019.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satterstrom F.K.et al. (2020) Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568.e23–584.e23 10.1016/j.cell.2019.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian Y.et al. (2014) Alteration in basal and depolarization-induced transcriptional network in iPSC-derived neurons from Timothy syndrome. Genome Med. 6, 75 10.1186/s13073-014-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulik-Sullivan B.K.et al. (2015) LD score regression distinguishes confounding from poly-genicity in genome-wide association studies. Nat. Genet. 47, 291–295 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wray N.R. and Visscher P.M. (2015) Quantitative genetics of disease traits. J Anim Breed Genet. 132, 198–203 10.1111/jbg.12153 [DOI] [PubMed] [Google Scholar]
- 25.Sandin S.et al. (2014) The familial risk of autism. J. Am. Med. Assoc. 311, 1770–1777 10.1001/jama.2014.4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Virkud Y.V.et al. (2009) Familial aggregation of quantitative autistic traits in multiplex versus simplex autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 150B, 328–334 10.1002/ajmg.b.30810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyall K.et al. (2014) Parental social responsiveness and risk of autism spectrum disorder in offspring. JAMA Psychiatry 71, 936–942 10.1001/jamapsychiatry.2014.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klei L.et al. (2012) Common genetic variants, acting additively, are a major source of risk for autism. Mol. Autism 3, 9 10.1186/2040-2392-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grove J.et al. (2019) Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 10.1038/s41588-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed D. and Pevsner J. (2016) The contribution of mosaic variants to autism spectrum disorder. PLoS Genet. 12, e1006245 10.1371/journal.pgen.1006245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim E.T.et al. (2017) Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 20, 1217–1224 10.1038/nn.4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Gama A.M.et al. (2015) Targeted DNA sequencing from autism spectrum disorder brains implicates multiple genetic mechanisms. Neuron 88, 910–917 10.1016/j.neuron.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodin R.E.et al. (2021) The landscape of somatic mutation in cerebral cortex of autistic and neurotypical individuals revealed by ultra-deep whole-genome sequencing. Nat. Neurosci. 24, 176–185 10.1038/s41593-020-00765-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An J.Y.et al. (2018) Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science 362, eaat6576 10.1126/science.aat6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandler W.M.et al. (2018) Paternally inherited cis-regulatory structural variants are associated with autism. Science 360, 327–331 10.1126/science.aan2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doan R.N.et al. (2016) Mutations in human accelerated regions disrupt cognition and social behavior. Cell 167, 341.e12–354.e12 10.1016/j.cell.2016.08.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner T.N.et al. (2016) Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 98, 58–74 10.1016/j.ajhg.2015.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werling D.M.et al. (2018) An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat. Genet. 50, 727–736 10.1038/s41588-018-0107-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J.et al. (2019) Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat. Genet. 51, 973–980 10.1038/s41588-019-0420-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritvo E.R.et al. (1985) Evidence for autosomal recessive inheritance in 46 families with multiple incidences of autism. Am. J. Psychiatry 142, 187–192 10.1176/ajp.142.2.187 [DOI] [PubMed] [Google Scholar]
- 41.Morrow E.M.et al. (2008) Identifying autism loci and genes by tracing recent shared ancestry. Science 321, 218–223 10.1126/science.1157657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu T.W.et al. (2013) Using whole-exome sequencing to identify inherited causes of autism. Neuron 77, 259–273 10.1016/j.neuron.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doan R.N.et al. (2019) Recessive gene disruptions in autism spectrum disorder. Nat. Genet. 51, 1092–1098 10.1038/s41588-019-0433-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de la Torre-Ubieta L.et al. (2016) Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 22, 345–361 10.1038/nm.4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'haene E. and Vergult S. (2021) Interpreting the impact of noncoding structural variation in neurodevelopmental disorders. Genet. Med. 23, 34–46 10.1038/s41436-020-00974-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anney R.et al. (2010) A genome-wide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 19, 4072–4082 10.1093/hmg/ddq307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X.et al. (2016) Genome-wide association study of autism spectrum disorder in the East Asian populations. Autism Res. 9, 340–349 10.1002/aur.1536 [DOI] [PubMed] [Google Scholar]
- 48.Weiss L.A.et al. (2009) A genome-wide linkage and association scan reveals novel loci for autism. Nature 461, 802–808 10.1038/nature08490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia K.et al. (2014) Common genetic variants on 1p13.2 associate with risk of autism. Mol. Psychiatry 19, 1212–1219 10.1038/mp.2013.146 [DOI] [PubMed] [Google Scholar]
- 50.Wu Y.et al. (2020) Multi-trait analysis for genome-wide association study of five psychiatric disorders. Transl. Psychiatry 10, 209 10.1038/s41398-020-00902-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cross-Disorder Group of the Psychiatric Genomics Consortium (2019) Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469.e11–1482.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matoba N.et al. (2020) Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism. Transl. Psychiatry 10, 265 10.1038/s41398-020-00953-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Rubeis S.et al. (2014) Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen H.T.et al. (2017) Integrated Bayesian analysis of rare exonic variants to identify risk genes for schizophrenia and neurodevelopmental disorders. Genome Med. 9, 114 10.1186/s13073-017-0497-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanders S.J.et al. (2015) Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trost B.et al. (2020) Genome-wide detection of tandem DNA repeats that are expanded in autism. Nature 586, 80–86 10.1038/s41586-020-2579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X.et al. (2020) Prioritizing long range interactions in noncoding regions using GWAS and deletions perturbed TADs. Comp. Structr. Biotechnol. J. 18, 2945–2952 10.1016/j.csbj.2020.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geoffroy V.et al. (2018) AnnotSV: an integrated tool for structural variations annotation. Bioinformatics 34, 3572–3574 10.1093/bioinformatics/bty304 [DOI] [PubMed] [Google Scholar]
- 59.Lee C.et al. (2019) Profiling allele-specific gene expression in brains from individuals with autism spectrum disorder reveals preferential minor allele usage. Nat. Neurosci. 22, 1521–1532 10.1038/s41593-019-0461-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tran S.S.et al. (2019) Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 22, 25–36 10.1038/s41593-018-0287-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velmeshev D.et al. (2019) Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 10.1126/science.aav8130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Massrali A.et al. (2019) Integrated genetic and methylomic analyses identify shared biology between autism and autistic traits. Mol. Autism 10, 31 10.1186/s13229-019-0279-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi K. and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 64.Shi Y.et al. (2017) Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 16, 115–130 10.1038/nrd.2016.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juopperi T.A.et al. (2011) Modeling neurological diseases using patient-derived induced pluripotent stem cells. Future Neurol. 6, 363–373 10.2217/fnl.11.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silverman J.L.et al. (2010) Behavioral phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 11, 490–502 10.1038/nrn2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watson K.K. and Platt M.L. (2012) Of mice and monkeys: using nonhuman primate models to bridge mouse- and human-based investigations of autism spectrum disorders. J. Neurodev. Disord. 4, 21 10.1186/1866-1955-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dolmetsch R. and Geschwind D.H. (2011) The human brain in a dish: the promise of iPSC derived neurons. Cell 145, 831–834 10.1016/j.cell.2011.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Courchesne E.et al. (2020) Prenatal origins of ASD: The when, what, and how of ASD development. Trends Neurosci. 43, 326–342 10.1016/j.tins.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gilbert J. and Man H.Y. (2017) Fundamental elements in autism: from neurogenesis and neurite growth to synaptic plasticity. Front. Cell Neurosci. 11, 359 10.3389/fncel.2017.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H.et al. (2019) Disruption of TCF4 regulatory networks leads to abnormal cortical development and mental disabilities. Mol. Psychiatry 24, 1235–1246 10.1038/s41380-019-0353-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Q.et al. (2018) Autism-associated CHD8 deficiency impairs axon development and migration of cortical neurons. Mol. Autism 9, 65 10.1186/s13229-018-0244-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parcerisas A.et al. (2020) NCAM2 regulates dendritic and axonal differentiation through the cytoskeletal proteins MAP2 and 14-3-3. Cereb. Cortex 30, 3781–3799 10.1093/cercor/bhz342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo H.et al. (2020) NCKAP1 disruptive variants lead to a neurodevelopmental disorder with core features of autism. Am. J. Hum. Genet. 107, 963–976 10.1016/j.ajhg.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X.et al. (2019) An autism-related, nonsense foxp1 mutant induces autophagy and delays radial migration of the cortical neurons. Cereb. Cortex 29, 3193–3208 10.1093/cercor/bhy185 [DOI] [PubMed] [Google Scholar]
- 76.Guo H.et al. (2019) Disruptive variants of CSDE1 associate with autism and interfere with neuronal development and synaptic transmission. Sci. Adv. 5, eaax2166 10.1126/sciadv.aax2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang M.et al. (2020) Two autism/dyslexia linked variations of DOCK4 disrupt the gene function on rac1/rap1 activation, neurite outgrowth, and synapse development. Front. Cell. Neurosci. 13, 577 10.3389/fncel.2019.00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shih P.et al. (2020) CTTNBP2 controls synaptic expression of zinc-related autism-associated proteins and regulates synapse formation and autism-like behaviors. Cell Rep. 31, 107700 10.1016/j.celrep.2020.107700 [DOI] [PubMed] [Google Scholar]
- 79.Wang J.et al. (2018) Neurexin gene family variants as risk factors for autism spectrum disorder. Autism Res. 11, 37–43 10.1002/aur.1881 [DOI] [PubMed] [Google Scholar]
- 80.Richter M.et al. (2019) Altered TAOK2 activity causes autism-related neurodevelopmental and cognitive abnormalities through RhoA signaling. Mol. Psychiatry 24, 1329–1350 10.1038/s41380-018-0025-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma J.et al. (2019) Autism candidate gene DIP2A regulates spine morphogenesis via acetylation of cortactin. PLoS Biol. 17, e3000461 10.1371/journal.pbio.3000461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y.et al. (2019) Dysregulated translation in neurodevelopmental disorders: an overview of autism-risk genes involved in translation. Dev. Neurobiol. 79, 60–74 10.1002/dneu.22653 [DOI] [PubMed] [Google Scholar]
- 83.Napoli E.et al. (2018) Beyond autophagy: a novel role for autism-linked Wdfy3 in brain mitophagy. Sci. Rep. 8, 11348 10.1038/s41598-018-29421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanellopoulos A.K.et al. (2020) Aralar sequesters GABA into hyperactive mitochondria, causing social behavior deficits. Cell 180, 1178.e20–1197.e20 10.1016/j.cell.2020.02.044 [DOI] [PubMed] [Google Scholar]
- 85.Forrest M.P.et al. (2018) The psychiatric risk gene transcription factor 4 (TCF4) regulates neurodevelopmental pathways associated with schizophrenia, autism, and intellectual disability. Schizophr. Bull. 44, 1100–1110 10.1093/schbul/sbx164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen J.A.et al. (2015) The emerging picture of autism spectrum disorder: genetics and pathology. Annu. Rev. Pathol. 10, 111–144 10.1146/annurev-pathol-012414-040405 [DOI] [PubMed] [Google Scholar]
- 87.Willsey A.J.et al. (2013) Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 10.1016/j.cell.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Voineagu I.et al. (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 10.1038/nature10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Casanova M.F.et al. (2006) Minicolumnar abnormalities in autism. Acta Neuropathol. 112, 287–303 10.1007/s00401-006-0085-5 [DOI] [PubMed] [Google Scholar]
- 90.Stoner R.et al. (2014) Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 370, 1209–1219 10.1056/NEJMoa1307491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Redcay E. and Courchesne E. (2005) When is the brain enlarged in autism? A meta-analysis of all brain size reports Biol. Psychiatry 58, 1–9 10.1016/j.biopsych.2005.03.026 [DOI] [PubMed] [Google Scholar]
- 92.Lipton J.O. and Sahin M. (2014) The neurology of mTOR. Neuron 84, 275–291 10.1016/j.neuron.2014.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hansen D.V.et al. (2011) Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron 70, 645–660 10.1016/j.neuron.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou W.et al. (2016) BLOS2 negatively regulates Notch signaling during neural and hematopoietic stem and progenitor cell development. Elife 5, e18108 10.7554/eLife.18108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ronan J.L.et al. (2013) From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 14, 347–359 10.1038/nrg3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krumm N.et al. (2015) Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 10.1038/ng.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pinto D.et al. (2014) Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 94, 677–694 10.1016/j.ajhg.2014.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jamain S.et al. (2003) Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 34, 27–29 10.1038/ng1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corradi A.et al. (2014) SYN2 is an autism predisposing gene: loss-of-function mutations alter synaptic vesicle cycling and axon outgrowth. Hum. Mol. Genet. 23, 90–103 10.1093/hmg/ddt401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Südhof T.C. (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yizhar O.et al. (2011) Neocortical excitation-inhibition balance in information processing and social dysfunction. Nature 477, 171–178 10.1038/nature10360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Palop J.J.et al. (2007) Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer’s disease. Neuron 55, 697–711 10.1016/j.neuron.2007.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mariani J.et al. (2015) FOXG1-dependent dysregulation of GABA-glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390 10.1016/j.cell.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Buffington S.A.et al. (2014) Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 37, 17–38 10.1146/annurev-neuro-071013-014100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nishimura Y.et al. (2007) Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum. Mol. Genet. 16, 1682–1698 10.1093/hmg/ddm116 [DOI] [PubMed] [Google Scholar]
- 106.Darnell J.C.et al. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cohen S. and Greenberg M.E. (2008) Communication between the synapse and the nucleus in neuronal development, plasticity and disease. Annu. Rev. Cell Dev. Biol. 24, 183–209 10.1146/annurev.cellbio.24.110707.175235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tang G.et al. (2014) Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83, 1131–1143 10.1016/j.neuron.2014.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Parikshak N.N.et al. (2013) Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 10.1016/j.cell.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abrahams B.S. and Geschwind D.H. (2010) Connecting genes to brain in the autism spectrum disorders. Arch. Neurol. 67, 395–399 10.1001/archneurol.2010.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rudie J.D.et al. (2012) Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron 75, 904–915 10.1016/j.neuron.2012.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Minshew N.J. and Williams D.L. (2007) The new neurobiology of autism: cortex, connectivity and neuronal organization. Arch. Neurol. 64, 945–950 10.1001/archneur.64.7.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Amaral D.G.et al. (2008) Neuroanatomy of autism. Trends Neurosci. 31, 137–145 10.1016/j.tins.2007.12.005 [DOI] [PubMed] [Google Scholar]
- 114.Ellegood J.et al. (2015) Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol. Psychiatry 20, 118–125 10.1038/mp.2014.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Amaral D.G. (2003) The amygdala, social behavior and danger detection. Ann. N.Y. Acad. Sci. 1000, 337–347 10.1196/annals.1280.015 [DOI] [PubMed] [Google Scholar]
- 116.Langen M.et al. (2011) The neurobiology of repetitive behavior: …and men. Neurosci. Biobehav. Rev. 35, 356–365 10.1016/j.neubiorev.2010.02.005 [DOI] [PubMed] [Google Scholar]
- 117.Fatemi S.H.et al. (2002) Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol. Psychiatry 52, 805–810 10.1016/S0006-3223(02)01430-0 [DOI] [PubMed] [Google Scholar]
- 118.Van Overwalle F.et al. (2019) The sequencing process generated by the cerebellum crucially contributes to social interactions. Med. Hypotheses 128, 33–42 10.1016/j.mehy.2019.05.014 [DOI] [PubMed] [Google Scholar]
- 119.Stessman H.A.et al. (2014) A genotype-first approach to defining the subtypes of a complex disease. Cell 156, 872–877 10.1016/j.cell.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tyzio R.et al. (2014) Oxytocin-mediated GABA inhibition during delivery attenuates autism pathogenesis in rodent offspring. Science 343, 675–679 10.1126/science.1247190 [DOI] [PubMed] [Google Scholar]
- 121.Owen S.F.et al. (2013) Oxytocin enhances hippocampal spike transmission by modulating fast-spiking interneurons. Nature 500, 458–462 10.1038/nature12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Han S.et al. (2012) Autistic-like behaviour in Scn1a+/- mice and rescue by enhanced GABA-mediated neurotransmission. Nature 489, 385–390 10.1038/nature11356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ehninger D.et al. (2008) Reversal of learning deficits in a Tsc2+/- mouse model of tuberous sclerosis. Nat. Med. 14, 843–848 10.1038/nm1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ru W.et al. (2012) A role of the mammalian target of rapamycin (mTOR) in glutamate-induced downregulation of tuberous sclerosis complex proteins 2 (TSC2). J. Mol. Neurosci. 47, 340–345 10.1007/s12031-012-9753-1 [DOI] [PubMed] [Google Scholar]
- 125.Chang Q.et al. (2006) The disease progression of Mecp2-mutant mice is affected by the level of BDNF expression. Neuron 49, 341–348 10.1016/j.neuron.2005.12.027 [DOI] [PubMed] [Google Scholar]
- 126.Bernardet M. and Crusio W.E. (2006) Fmr1 KO mice as a possible model of autistic features. Sci. World J. 6, 1164–1176 10.1100/tsw.2006.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsai P.T.et al. (2012) Autistic-like behavior and cerebellar dysfunction in Purkinje cell Tsc1-mutant mice. Nature 488, 647–651 10.1038/nature11310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mellios N.et al. (2014) β2-adrenergic receptor agonist ameliorates phenotype and corrects microRNA-mediated IGF1 deficits in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 111, 9947–9952 10.1073/pnas.1309426111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Geschwind D.H. and Rakic P. (2013) Cortical evolution: judge the brain by its cover. Neuron 80, 633–647 10.1016/j.neuron.2013.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.D'Mello A.M. and Stoodley C.J. (2015) Cerebro-cerebellar circuits in autism spectrum disorder. Front. Neurosci. 9, 408 10.3389/fnins.2015.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McAlonan G.M. (2004) Mapping the brain in autism. A voxel-based MRI study of volumetric differences and intercorrelations in autism. Brain 128, 268–276 10.1093/brain/awh332 [DOI] [PubMed] [Google Scholar]
- 132.Courchesne E.et al. (2001) Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology 57, 245–254 10.1212/WNL.57.2.245 [DOI] [PubMed] [Google Scholar]
- 133.Hanaie R.et al. (2013) Altered microstructural connectivity of the superior cerebellar peduncle is related to motor dysfunction in children with autistic spectrum disorders. Cerebellum 12, 645–656 10.1007/s12311-013-0475-x [DOI] [PubMed] [Google Scholar]
- 134.Akshoomoff N.et al. (2004) Outcome classification of preschool children with autism spectrum disorders using MRI brain measures. J. Am. Acad. Child. Adolesc. Psychiatry 43, 349–357 10.1097/00004583-200403000-00018 [DOI] [PubMed] [Google Scholar]
- 135.Catani M.et al. (2008) Altered cerebellar feedback projections in Asperger syndrome. Neuroimage 41, 1184–1191 10.1016/j.neuroimage.2008.03.041 [DOI] [PubMed] [Google Scholar]
- 136.Khan A.J.et al. (2015) Cerebro-cerebellar resting-state functional connectivity in children and adolescents with autism spectrum disorder. Biol. Psychiatry 78, 625–634 10.1016/j.biopsych.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Suzuki K.et al. (2013) Microglial activation in young adults with autism spectrum disorder. JAMA Psychiatry 70, 49–58 10.1001/jamapsychiatry.2013.272 [DOI] [PubMed] [Google Scholar]
- 138.Tetreault N.A.et al. (2012) Microglia in the cerebral cortex in autism. J. Autism Dev. Disord. 42, 2569–2584 10.1007/s10803-012-1513-0 [DOI] [PubMed] [Google Scholar]
- 139.Gatto C.L. and Broadie K. (2010) Genetic controls balancing excitatory and inhibitory synaptogenesis in neurodevelopmental disorder models. Front. Synap. Neurosci. 2, 4, eCollection 2010 10.3389/fnsyn.2010.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Koyama R. and Ikegaya Y. (2015) Microglia in the pathogenesis of autism spectrum disorders. Neurosci. Res. 100, 1–5 10.1016/j.neures.2015.06.005 [DOI] [PubMed] [Google Scholar]
- 141.Chung W.S.et al. (2013) Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 10.1038/nature12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schafer D.P.et al. (2012) Microglia sculpt postnatal neural circuits in an activity- and complement-dependent manner. Neuron 74, 691–705 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Andoh M.et al. (2019) Microglia as possible therapeutic targets for autism spectrum disorders. Prog. Mol. Biol. Transl. Sci. 167, 223–245 10.1016/bs.pmbts.2019.06.012 [DOI] [PubMed] [Google Scholar]
- 144.Bordt E.A.et al. (2020) Microglia and sexual differentiation of the developing brain: a focus on ontogeny and intrinsic factors. Glia 68, 1085–1099 10.1002/glia.23753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu Z.X.et al. (2020) Elevated protein synthesis in microglia causes autism-like synaptic and behavioral aberrations. Nat. Commun. 11, 1797 10.1038/s41467-020-15530-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Miyanishi K.et al. (2020) Synaptic elimination by microglia and disturbed higher brain functions. Neurochem. Int. 142, 104901 10.1016/j.neuint.2020.104901 [DOI] [PubMed] [Google Scholar]
- 147.Pérez-Crespo L.et al. (2019) Temporal and geographical variability of prevalence and incidence of autism spectrum disorder diagnoses in children in Catalonia. Spain Autism Res. 12, 1693–1705 10.1002/aur.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Malkova N.V.et al. (2012) Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain Behav. Immun. 26, 607–616 10.1016/j.bbi.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang H.et al. (2014) Dysbindin-1C is required for the survival of hilar mossy cells and the maturation of adult newborn neurons in dentate gyrus. J. Biol. Chem. 289, 29060–29072 10.1074/jbc.M114.590927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yuan Y.et al. (2015) Impaired autophagy in hilar mossy cells of the dentate gyrus and its implication in schizophrenia. J. Genet. Genomics 42, 1–8 10.1016/j.jgg.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 151.Eadie B.D.et al. (2009) Fmr1 knockout mice show reduced anxiety and alterations in neurogenesis that are specific to the ventral dentate gyrus. Neurobiol. Dis. 36, 361–373 10.1016/j.nbd.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 152.Guo W.et al. (2012) Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum. Mol. Genet. 21, 681–691 10.1093/hmg/ddr501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu B.et al. (2018) Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, E11397–E11405 10.1073/pnas.1809588115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li Y.et al. (2018) Reducing histone acetylation rescues cognitive deficits in a mouse model of Fragile X syndrome. Nat. Commun. 9, 2494 10.1038/s41467-018-04869-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Saffari A.et al. (2019) RNA sequencing of identical twins discordant for autism reveals blood-based signatures implicating immune and transcriptional dysregulation. Mol. Autism 10, 38 10.1186/s13229-019-0285-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wu Y.E.et al. (2016) Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat. Neurosci. 19, 1463–1476 10.1038/nn.4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Castel D.et al. (2018) Transcriptomic and epigenetic profiling of ‘diffuse midline gliomas, H3 K27M-mutant’ discriminate two subgroups based on the type of histone H3 mutated and not supratentorial or infratentorial location. Acta Neuropathol. Commun. 6, 117 10.1186/s40478-018-0614-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ramaswami G.et al. (2020) Integrative genomics identifies a convergent molecular subtype that links epigenomic with transcriptomic differences in autism. Nat. Commun. 11, 4873 10.1038/s41467-020-18526-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu Y.et al. (2021) Expression changes in epigenetic gene pathways associated with one-carbon nutritional metabolites in maternal blood from pregnancies resulting in autism and non-typical neurodevelopment. Autism Res. 14, 11–28 10.1002/aur.2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.