Abstract
Early responses to vaccination are important for shaping both humoral and cellular protective immunity. Dissecting innate vaccine signatures may predict immunogenicity to help optimize the efficacy of mRNA and other vaccine strategies. Here, we characterize the cytokine and chemokine responses to the 1st and 2nd dose of the BNT162b2 mRNA (Pfizer/BioNtech) vaccine in antigen-naive and in previously coronavirus disease 2019 (COVID-19)-infected individuals (NCT04743388). Transient increases in interleukin-15 (IL-15) and interferon gamma (IFN-γ) levels early after boost correlate with Spike antibody levels, supporting their use as biomarkers of effective humoral immunity development in response to vaccination. We identify a systemic signature including increases in IL-15, IFN-γ, and IP-10/CXCL10 after the 1st vaccination, which were enriched by tumor necrosis factor alpha (TNF-α) and IL-6 after the 2nd vaccination. In previously COVID-19-infected individuals, a single vaccination results in both strong cytokine induction and antibody titers similar to the ones observed upon booster vaccination in antigen-naive individuals, a result with potential implication for future public health recommendations.
Keywords: SARS-CoV-2, mRNA vaccine, innate immunity, IL-15, IFN-γ, cytokines, IP10/CXCL10, Spike antibody
Graphical abstract
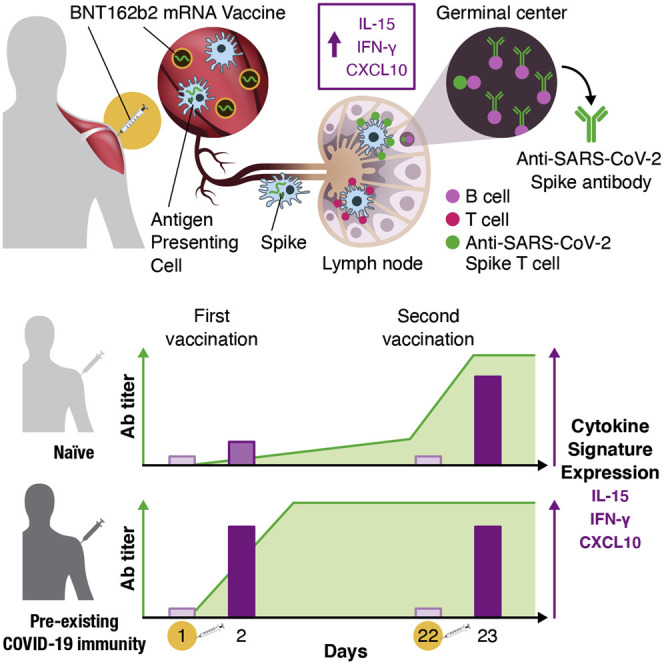
Bergamaschi et al. find that the SARS-CoV-2 BNT162b2 mRNA vaccine induces a distinct transient cytokine response featuring IL-15, IFN-γ, and IP-10/CXCL10. mRNA-vaccine-induced IFN-γ and IL-15 correlate with spike antibody response. A single vaccination of convalescent persons leads to both robust cytokine signature and antibody response.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 131 million individuals worldwide and is responsible for more than 2.8 million deaths to date (https://www.coronavirustraining.org/live-map). Infection or vaccination is associated with the development of variable levels of antibodies with neutralizing activity that can protect against infection and/or disease development in animal models (reviewed in Muñoz-Fontela et al., 2020). Administration of anti-Spike neutralizing antibodies (NAbs) was shown to provide strong protection from disease in animal models and humans (Baum et al., 2020; Chen et al., 2021; Gottlieb et al., 2021; Ledford, 2021; Weinreich et al., 2021). Several SARS-CoV-2 vaccines currently tested induced potent antibody responses in humans, which led to the advancement of several candidates to the clinic under Emergency Use Authorization or Conditional Marketing Authorization and demonstrated protective efficacy (Baden et al., 2021; Barrett et al., 2021; Folegatti et al., 2020; Polack et al., 2020; Sadoff et al., 2021; Stephenson et al., 2021; van Doremalen et al., 2020; Walsh et al., 2020; Widge et al., 2021). SARS-CoV-2 vaccines showed real-world effectiveness as reported in Israel (Chodick et al., 2021) as well as by CDC (2021) and Public Health England (2021).
Cytokines and chemokines are important drivers of inflammation and innate immunity and have a pivotal role in the development and maintenance of adaptive immunity, in response to both infection and vaccination. Identification of a robust signature of cytokine induction leading to successful vaccination would be important for further vaccine development and optimization (Arunachalam et al., 2020; Fourati et al., 2019; Hagan and Pulendran, 2018; Kuri-Cervantes et al., 2016). Immune signatures in vaccine recipients receiving yellow fever, HIV-Ade5, or HIV canary pox virus vaccine (ALVAC) vaccines have been described (Andersen-Nissen et al., 2021; Gaucher et al., 2008; Querec et al., 2009; Zak et al., 2012).
To identify markers associated with vaccination resulting in beneficial antibody development, we studied cytokines and chemokines triggered by prime and boost vaccination by the Pfizer/BioNtech BNT162b2 mRNA vaccine at various times after the 1st and 2nd dose. Such analytes could support the identification of pathways leading to efficient vaccination (reviewed in Cagigi and Loré, 2021) and could be used as biomarkers predicting successful application of mRNA vaccines.
Results
SARS-CoV-2 anti-Spike antibody titers detected in BNT162b2 mRNA (Pfizer/BioNTech) vaccine recipients
A cohort of 63 health-care workers (Table 1 ) received the BNT162b2 mRNA vaccine and was monitored for the development of anti-Spike-receptor-binding domain (RBD) immunoglobulin G (IgG) and antibodies recognizing full-length trimeric Spike (Figures 1A and 1B). Sera were analyzed on the day of vaccination (day 1), 1 and 3 weeks later (day 8, day 22), and 2 and 4 weeks after the 2nd vaccination (day 36 and day 50). We found strong correlations (Figures S1A–S1C) between anti-Spike-RBD (Figure 1A) and Spike (Figure 1B) antibodies, which is in agreement with our previous reports using sera from SARS-CoV-2 convalescent patients and Spike-DNA-vaccinated macaques (Rosati et al., 2021; Terpos et al., 2020, 2021a) and supports the notion that RBD is the major antibody target.
Table 1.
Description of vaccine recipients
| Parameter | N |
|---|---|
| Naive vaccine recipients | 58 |
| Vaccine recipients with pre-existing immunity | 5 |
| Sex | |
| Male | 27 |
| Female | 36 |
| Age (years) | |
| <50 | 30 |
| >50 | 33 |
| Medical history | |
| None | 37 |
| Yes | 26 |
| Adverse vaccine effects | |
| 1stDose | |
| No | 22 |
| Yes | 41 |
| 2ndDose | |
| No | 13 |
| Yes | 50 |
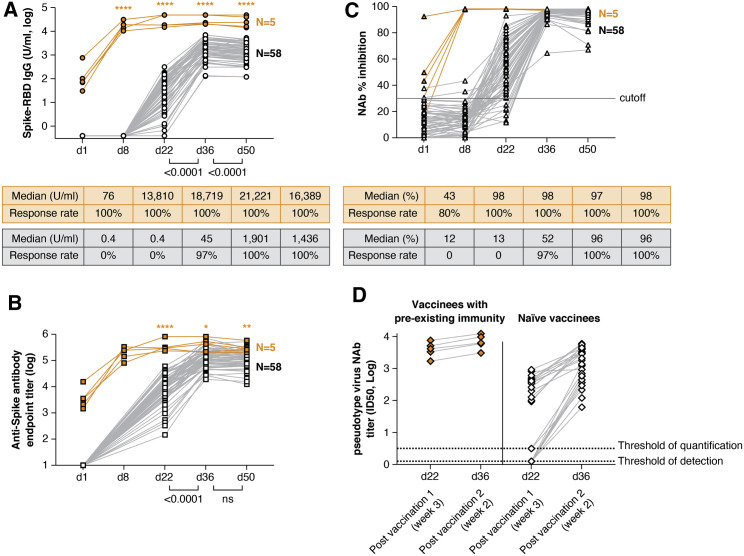
Figure 1.
Anti-SARS-CoV2 antibody development upon BNT162b2 mRNA vaccination
Vaccine recipients were monitored after the 1st vaccination (day 1, day 8, and day 22) and at weeks 2 and 4 after the 2nd vaccination (day 36 and day 50). Responses are shown for recipients with pre-existing immunity due to prior infection (orange symbols) and vaccine recipients naive to SARS-CoV-2 (black symbols).
(A and B) Over time, an analysis of binding antibodies recognizing anti-Spike-RBD IgG (ROCHE, U/ml in log) (A) and the full-length trimeric Spike (ELISA, endpoint titer in log) (B) are shown. The Spike-RBD IgG antibody assay (ROCHE) has a range of >0.4 −2,500 U/ml and was run with serial dilutions for some samples reaching >50,000 U/ml.
(C and D) Neutralizing antibodies (NAbs) were assessed by a surrogate virus neutralization test (GenScript) (C) and pseudotype NAb assays (D) using HIV-1NLΔEnv-NanoLuc-derived pseudotype virus carrying Wuhan-Hu-1 Spike. Pseudotype neutralization was performed in sera from 5 vaccinees with pre-existing immunity and a subset of samples selected to cover a range of low to high responses (n = 25 naive vaccinees; black symbols) at day 22 and day 36. Sera from vaccinees with pre-existing immunity showed pseudotype NAb titers (50% inhibitory dose, ID50) with a median 3.6 log (range, 3.2–3.9) upon a single vaccination with similar levels after the booster vaccination (median, 3.8 log; range, 3.5–4.1). Naive vaccinees showed NAb ID50 titers ranging from 0.1 to 2.94 log at day 22 and from 1.79 to 3.78 log at day 36. The surrogate virus inhibition assay showed median 96% inhibition levels after the 2nd dose (>90% inhibition by 95% of day 36 sera and by 83% of day 50 sera, respectively). (A and C) Median Spike-RBD antibody and % inhibition and response rate (%) are listed. Orange asterisks indicate significant difference between vaccine recipient with or without prior immunity to SARS-CoV-2 (Mann-Whitney test). See also Figures S1 and S2 and Table S1.
We noted that the recipients could be separated into 2 groups; 58 recipients showed responses first detected 3 weeks after the 1st dose (day 22), which was followed by a significant increase after the 2nd dose by day 36 (Figures 1A and 1B, black symbols). In contrast, the 5 recipients (Figure 1, orange symbols) with pre-existing SARS-CoV-2 immunity (Table S1) showed antibody responses to Spike-RBD (Figure 1A) and trimeric Spike (Figure 1B) at the day of vaccination, followed by an immediate strong anamnestic response after the 1st dose (day 8). The antibody responses did not further increase upon the 2nd vaccination and remained significantly higher than those in the SARS-CoV-2-naive vaccine recipients (Figure 1, orange asterisks).
An analysis of the kinetics of Spike-RBD and Spike antibody development in the 58 naive vaccine recipients showed a maximal level reached after the 2nd vaccination. The Spike-RBD (Figure 1A), but not the anti-Spike (Figure 1B), antibody levels contracted significantly by day 50 in these recipients. The difference could be explained by the different antibody half-lives and is corroborated by our report that antibodies to Spike-RBD have a shorter half-life than antibodies to complete Spike (Terpos et al., 2021a). The observed differences reflect the nature of these antibodies not only in convalescent patients (Terpos, 2021) but also in mRNA vaccinated persons; therefore, this is a general feature of the antibody specificities.
The neutralizing ability of these antibodies was measured using a surrogate virus neutralization test (GenScript; Figure 1C), an assay that reached a median of 96% inhibition after the 2nd dose, making any further comparisons uncertain, and a Spike pseudotyped Nanoluc reporter virus assay, performed in a subset of samples selected to cover a range of low to highest responses (Figure 1D). Overall NAb levels followed those of the binding antibodies. The pseudotype NAb levels correlated significantly with the anti-Spike-RBD (Figure S2A) and anti-Spike antibodies (Figure S2B), as expected from our studies of convalescent patients and of vaccinated macaques (Rosati et al., 2021; Terpos et al., 2020, 2021a), supporting that antibody measurements (Spike and Spike-RBD) can serve as surrogates for NAb measurements.
For the 5 recipients with pre-existing coronavirus disease 2019 (COVID-19) immunity, 1 single vaccination induced potent recall responses with maximal antibody levels reached by day 8 for both Spike-RBD antibodies and NAb (Figure 1), which is similar to recent findings reported while this report was in preparation (Ebinger et al., 2021; Gobbi et al., 2021; Manisty et al., 2021). These levels did not decline within the 4-week follow-up after the 2nd vaccination (day 22 or day 50), which is in contrast to the decline found in 86% of the naive vaccine recipients (49 of 57 paired samples). Thus, a single vaccination in the presence of pre-existing immunity due to CoV-2 infection induced faster and more durable immune responses. Based on these results, a subsequent analysis of serum cytokines and chemokines was performed in SARS-CoV-2 naive (n = 58) versus previously infected (n = 5) vaccine recipients.
Serum cytokine and chemokine profile induced by the BNT162b2 mRNA vaccine
Sera collected on the day of and after the 1st vaccination (day 1, day 2, and day 8) and the 2nd vaccination (day 22 and day 23) were subjected to cytokine/chemokine analysis by using the MSD (Meso Scale Discovery) platform that analyzed 41 analytes (Table S2). Of these analytes, 19 showed significant changes, 8 showed none or marginal changes, and 14 were below the threshold of detection.
The chemokine/cytokine levels showing changes (Figure 2 ; Figure S3) include molecules released in response to inflammation with both a pro-inflammatory role (interleukin-6 [IL-6], VEGF-A, and acute phase proteins SAA and CRP) (Hunter and Jones, 2015; Mangalmurti and Hunter, 2020) and anti-inflammatory function (IL-1Ra) (Dinarello, 2018; Mantovani et al., 2019); chemokines involved in lymphocyte, monocyte/macrophage, and granulocyte recruitment (IP-10/CXCL10, IL-8, IL-16, MIP-1α/CCL3, MIP-1β/CCL4, MCP-1/CCL2, MDC/CCL22, and Eotaxin) (Griffith et al., 2014); and cytokines that promote innate and adaptive immune response (interferon gamma [IFN-γ], IL-15, IL-12/IL-23p40, tumor necrosis factor alpha [TNF-α], IL-3, and IL-7) (Hu and Ivashkiv, 2009; Leonard et al., 2019).
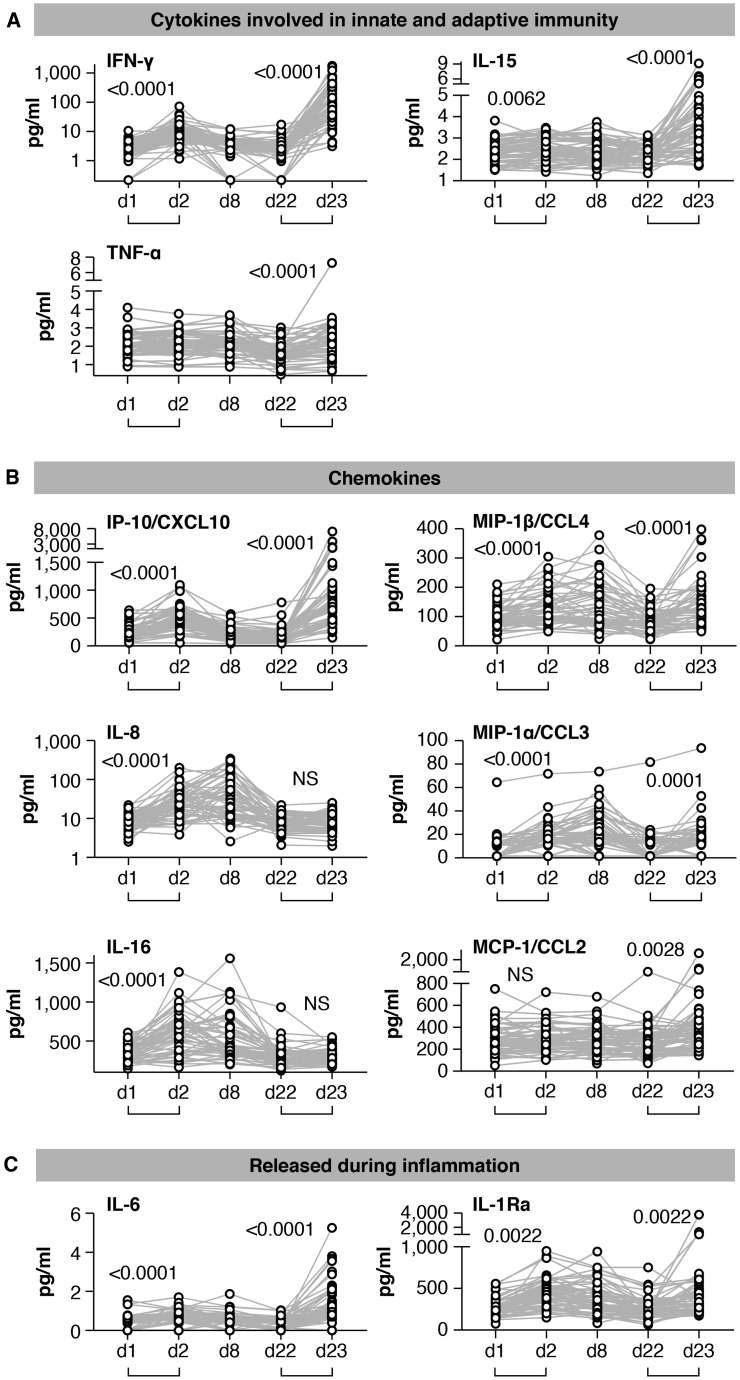
Figure 2.
Serum cytokine and chemokine levels after the 1st and 2nd vaccination in COVID-19-naive vaccine recipients
Cytokine and chemokine levels were measured over time using the MSD assay after the 1st vaccination (day 1, day 2, day 8, and day 22) and at 1 day after the 2nd vaccination (day 23) in the 58 COVID-19-naive vaccine recipients.
(A–C) Serum levels of 11 selected analytes among 19 analytes showing changes upon vaccination are plotted over time. (A) Cytokines involved in both innate and adaptive immunity. (B) Chemokines. (C) Molecules released during inflammation. See also Figure S3 and Tables S2 and S3. p values are from paired t test.
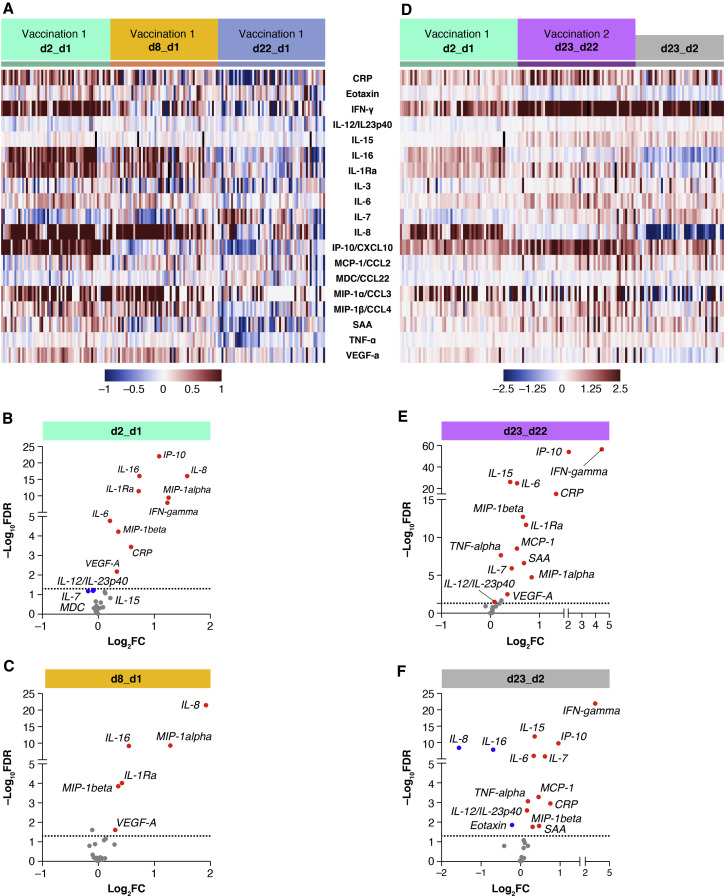
The cytokine/chemokine profile induced by the 1st and 2nd vaccination and the comparison between the effects induced by each dose for the individual recipients are represented in heatmaps (Figures 3A and 3D).
Figure 3.
Comparison of serum cytokine and chemokine levels
Cytokine and chemokine levels were measured using the MSD assay after the 1st and 2nd vaccination in 58 COVID-19 naive recipients (as described for Figure 2).
(A and D) Heatmaps depicted log2 fold changes in 19 analytes upon the 1st vaccination (green: d2_d1; orange: d8_d1; blue: d22_d1) (A) and after both the 1st (green: d2_d1) and 2nd vaccinations (purple: d23_d22) (D). The comparison of the effects induced by the 2nd vaccination over the 1st (d23_d22 over d2_d1) is shown in gray in (D). Different scales are used in (A) and (D) to better visualize the distinct changes upon the 1st and 2nd vaccination.
(B, C, E, and F) Volcano plots of data shown in (A) and (D) depict differentially expressed analytes upon the 1st vaccination at day 2 in comparison to day 1 (B) and at day 8 in comparison to day 1 (C) and after the 2nd vaccination at day 23 in comparison to day 22 (E). (F) Differentially affected analytes after the 2nd vaccination in comparison to the 1st vaccination (1 day after each vaccine dose). Red dots indicate significant upregulation; blue dots indicate significant downregulation (FDR < 0.05 represented by the broken horizontal line). See also Figure S4 and Tables S2 and S3.
After the 1st vaccination, the 58 naive vaccine recipients showed a highly significant but transient increase of IFN-γ (∼2.5×; Figure 2A), IP-10/CXCL10 (∼2×; Figure 2B), and IL-6 (1.5×; Figure 2C) at day 2 followed by rapid downregulation close to baseline levels by day 8. IL-15 also showed a small but significant upregulation (Figure 2A). Other analytes including IL-8 (∼3×; Figure 2B), IL-16 (∼1.5×; Figure 2B), MIP-1α/CCL3 (∼2.5×; Figure 2B), MIP-1β/CCL4 (∼1.5×; Figure 2B), and IL-1Ra (∼2×; Figure 2C) were significantly upregulated over baseline both at day 2 and day 8, indicating longer-lasting vaccine effects (1 week after administration). No significant differences after the 1st vaccination were observed for TNF-α (Figure 2A), MCP-1/CCL2 (Figure 2B), and IL-3 (Figure S3). A slight decrease in IL-12/IL-23p40, MDC/CCL22, and IL-7 levels (Figure S3) was also observed, whereas CRP, Eotaxin, SAA, and VEGF-A were all increased (Figure S3) as consequence of the inflammation process. For most of the analytes, serum levels at day 22, prior to the 2nd vaccination, were comparable to pre-vaccination levels, as shown in the heatmap (Figure 3A).
The cytokine/chemokine response pattern was different at day 23 (1 day after the 2nd vaccination; Figures 2, S3, and 3D–3F). IFN-γ, IL-15, IP-10/CXCL10, and IL-6 showed elevated levels at day 23 that were significantly higher than those at day 2 (Figure 2). Remarkably, IFN-γ and IP-10/CXCL10 levels increased up to ∼20× and ∼4× over baseline after the 2nd vaccination, respectively. About 2× higher IL-15 and IL-6 peaks were detected after the 2nd vaccination (Figure 2). A similar effect was also observed for MIP-1β/CCL4 (Figure 2B), CRP, and SAA (Figure S3). IL-16, IL-8 (Figure 2B), MDC/CCL22, and VEGF-A (Figure S3) were not affected, whereas MIP-1α/CCL3 and IL-1Ra behave similarly after each vaccine dose (Figures 2B and 2C). Eotaxin showed a slight downregulation at day 23 (Figure S3). Importantly, TNF-α (Figure 2A), MCP-1/CCL2 (Figure 2B), IL-7 (Figure S3), IL-3 (Figure S3), and IL-12/IL-23p40 (Figure S3) showed significant increases only after the 2nd vaccination (day 23; Figure 2; Figure S3).
To confirm our results, we also performed differential expression analysis comparing mean cytokine levels for the 58 naive vaccine recipients at day 2 to day 1 (Figure 3B), day 8 to day 1 (Figure 3C), day 23 to day 22 (Figure 3E), and day 23 to day 2 (Figure 3F), by setting significance at a strict cut-off of a false discovery rate (FDR) of <0.05. Indeed, the 1st vaccination induced both acute and durable effects on the levels of the analyzed cytokines and chemokines (Figures 3B and 3C). A significant upregulation of IFN-γ, IP-10/CXCL10, IL-6, and CRP was detected only at day 2 (Figure 3B, red dots). Similarly, a marginal downregulation was observed for IL-12/IL-23p40 and IL-7 (Figure 3B, blue dots). In contrast, the significant positive effects induced by the 1st vaccination on IL-8, IL-16, MIP-1α/CCL3, MIP-1β/CCL4, IL-1Ra, and VEGF-A were maintained both at day 2 and day 8 (Figure 3C). These results allow us to distinguish vaccine-induced transient effects from longer lasting ones (1 week after vaccine administration). Our analysis at day 23 showed that the 2nd vaccination induced effects much broader and greater in magnitude on the cytokine/chemokine profile (Figure 3D). Newly induced analytes upon 2nd vaccination include TNF-α, MCP-1/CCL2, IL-7, and IL-12/IL-23p40 (Figure 3E; see also Figures 2 and S3). IL-15 upregulation also reached significance using an FDR of <0.05 (Figure 3E). For several analytes (IFN-γ, IP-10/CXCL10, IL-15, IL-6, CRP, and MIP-1β), the log2 fold changes were higher after the 2nd vaccination (Figure 3E). The differential outcome of the 2nd vaccination versus 1st vaccination is represented in a volcano plot (Figure 3F).
Vaccine recipient clustering did not reveal a differential effect of the vaccination on the cytokine/chemokine profile based on age (cutoff age of 50) in this cohort. However, we found a stronger induction in female versus male vaccine recipients (Figure S4) of IFN-γ, IL-15, IL-6, and IP-10/CXCL10 upon the 2nd vaccination. Because the dominant adverse effect (AE) upon the vaccinations (Table S3) was pain at site of injection as reported by 80% and 76% of vaccinees, respectively, this precluded further dissection of AE and cytokine/chemokine changes.
Serum cytokine and chemokine profile induced by the BNT162b2 mRNA vaccine in recipients with pre-existing anti-COVID-19 immunity
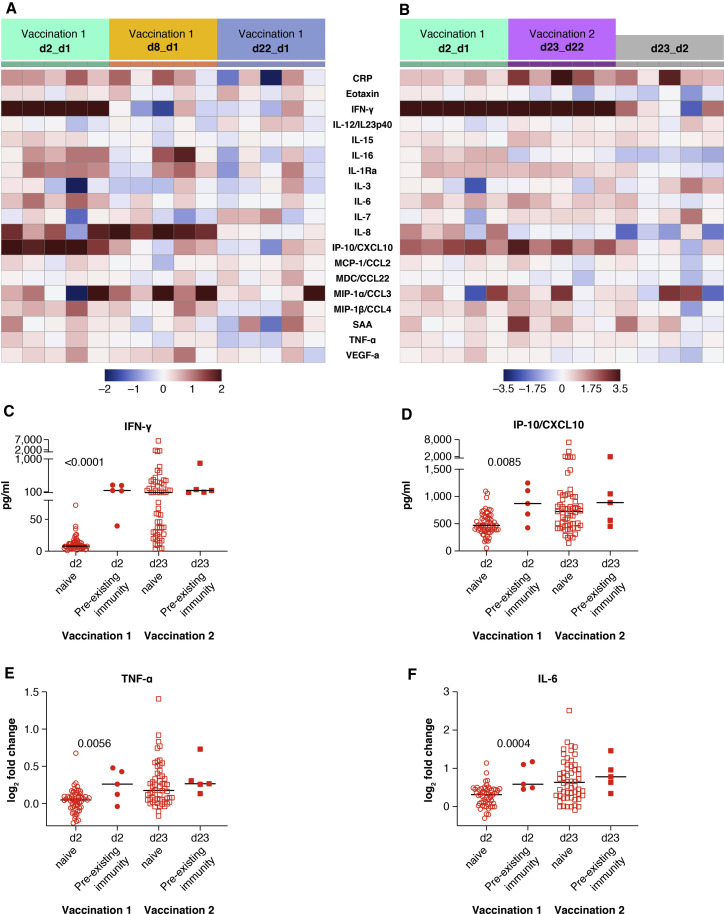
A similar chemokine/cytokine analysis was performed in the 5 vaccine recipients with pre-existing SARS-CoV-2 immunity. The cytokine/chemokine signature upon the 1st and 2nd vaccination is depicted in heatmaps (Figures 4A and 4B). The effects of the 1st vaccination were also compared among the 2 vaccine groups (Figures 4C to 4F). In recipients with pre-existing CoV-2 immunity, the 1st vaccination induced a much stronger upregulation of IFN-γ, IP-10/CXCL10, TNF-α, and IL-6. At day 2, high levels of IFN-γ and IP-10/CXCL10 were detected that were comparable to the levels achieved at 1 day after the 2nd vaccination in CoV-2 naive recipients (Figures 4C and 4D). Similarly, a greater increase of TNF-α and IL-6 was found after the 1st vaccination in individuals with pre-existing COVID-19 immunity (Figures 4E and 4F). Both vaccine groups showed similar levels for these analytes after the 2nd vaccination.
Figure 4.
Serum cytokine and chemokine levels after the 1st and 2nd vaccination in COVID-19 vaccine recipients with pre-existing immunity
Cytokine and chemokine levels were measured using the MSD assay after the 1st and 2nd vaccination in 5 recipients with pre-existing immunity (as described for Figure 2).
(A and B) Heatmaps representing the 19 analytes that showed significant changes upon the 1st (A) and the 1st and 2nd vaccinations (B), and comparison of both vaccinations are shown. Different scales are used in (A) and (B) to better visualize the distinct changes upon the 1st and 2nd vaccination.
(C–F) Comparison of changes between the 58 COVID-19-naive individuals and the 5 individuals with prior COVID-19 infection in serum levels (pg/ml) of IFN-γ (C) and IP-10/CXCL10 (D) and in log2 fold changes for TNF-α (E) and IL-6 (F). p values are from unpaired non-parametric t test (Mann-Whitney). See also Tables S2 and S3.
On the contrary, IL-15, IL-8, IL-16, MIP-1α, MIP-1β, IL-1Ra, and MCP-1 molecules released as result of inflammation showed a similar pattern over time with no significant differences observed among both groups of vaccinees, regardless of their SARS-CoV-2 serological status.
Overall, these data showed that BNT162b2 mRNA vaccination is accompanied by the rapid release in the blood of inflammatory markers, chemokines, and cytokines. In particular, the vaccination resulted in a strong response driven by IL-15, IFN-γ, and IP-10/CXCL10. Booster vaccination in naive individuals or one single vaccine dose in previously SARS-CoV-2-infected individuals induced anamnestic responses and high levels of cytokines critical for the rapid recruitment and stimulation of virus-specific effector immune cells.
Correlation between cytokine changes induced by vaccination
mRNA vaccination results in an innate signature characterized by the co-expression of several cytokines and chemokines. In order to assess the inter-relationship of the vaccine-induced effects on different serum cytokines and chemokines, we performed a pairwise correlation analysis by using the log2 fold change at day 2 (1st vaccination) and day 23 (2nd vaccination).
Effects at day 2 (upon 1st vaccination) identified positive associations in a cluster featuring IL-15, IFN-γ, IP-10/CXCL10, and IL-6 (Figure S5). However, these associations (Spearman r between 0.25 and 0.42; Figure S5) were below our cutoff for a Spearman correlation coefficient corresponding to an adjusted p value of <0.05. The cytokine signature at day 2 suggested a rapid co-expression of molecules promoting inflammation and priming of adaptive immunity. Indeed, a role of IL-15 in inducing both IFN-γ directly and IP-10/CXCL10 through the IFN type 2 pathway has been previously reported (Bergamaschi et al., 2020). Additionally, in autoimmune conditions such as rheumatoid arthritis, localized inflammatory responses are characterized by the concerted release of both IL-15 and IL-6 that are maintained by positive feedback loops and result in systemic disorders (McInnes and Schett, 2011).
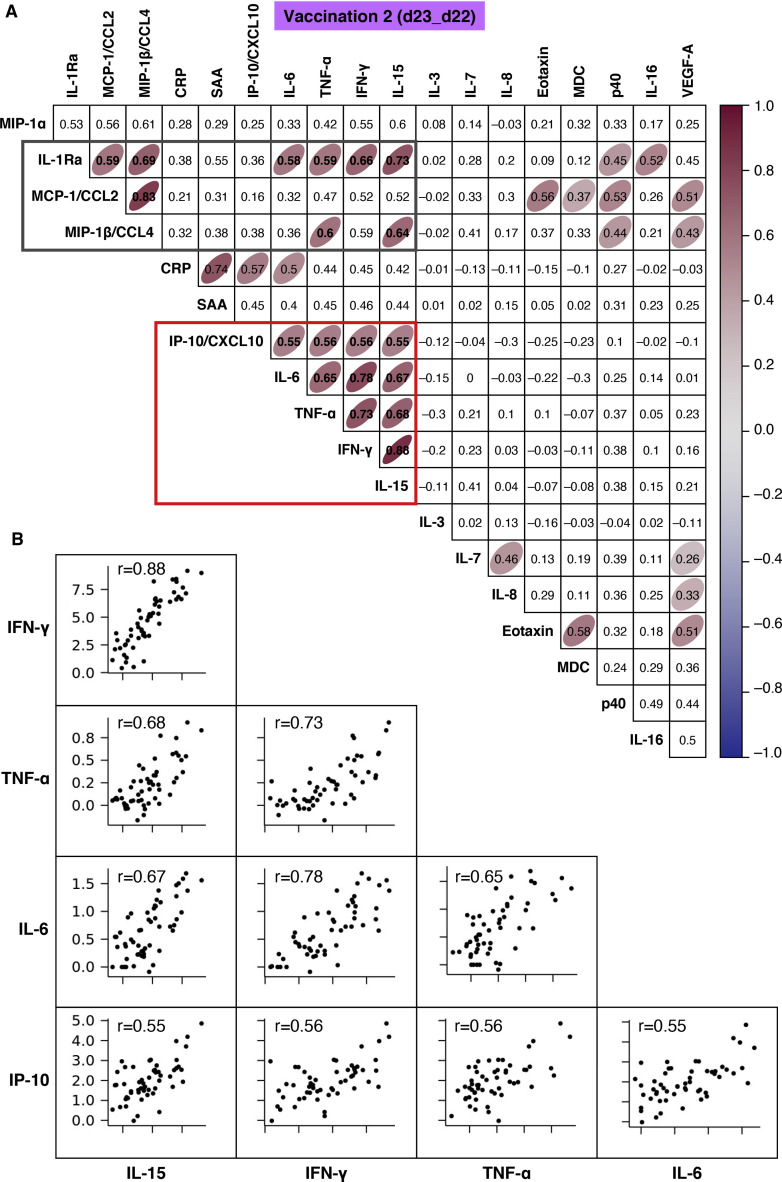
A correlation matrix of the measurements after the 2nd vaccination was also calculated (Figure 5 A). Several cytokines were co-expressed upon the 2nd vaccination, and individual correlation plots of cytokines within the red and gray boxes in the heatmap are shown in Figure 5B and Figure S6, respectively. A concerted and highly significant effect on IL-15, IFN-γ, TNF-α, IL-6, and IP-10/CXCL10 was observed (Figure 5A, red box), suggesting an amplification of the responses already induced at day 2 upon the 1st vaccination (Figure S5). Among the cytokine pairs that correlated highly, we identified IL-15 and IFN-γ, IL-15 and TNF-α, IL-15 and IL-6, IL-15 and IP-10/CXCL10, IFN-γ and TNF-α, IFN-γ and IL-6, IFN-γ and IP-10/CXCL10, TNF-α and IL-6, TNF-α and IP-10/CXCL10, and IL-6 and IP-10/CXCL10 (Figure 5B). These results are consistent with a coordinate role of these cytokines in supporting both innate and adaptive immunity and in the recall of immune memory response.
Figure 5.
Correlation of chemokine and cytokine changes
Pairwise correlations were calculated among the log2 fold changes at day 23 (after the 2nd vaccination) for the 19 biomarkers that were affected by the vaccination by using the Spearman correlation coefficient (adjusted p < 0.05). The analysis was performed for the 58 COVID-19-naive vaccine recipients.
(A) Correlation matrix for the 2nd vaccination is plotted as a heatmap. Spearman r values of correlations are indicated in the grid cells, and ellipses identified significant correlations. The color and shape of ellipses correspond to the value of the Spearman correlation coefficient, with red color indicating a positive correlation. The red box identifies the cluster of positive associations featuring IFN-γ, IL-15, TNF-α, IL-6, and IP-10/CXCL10. The gray box data are described in Figure S6.
(B) Correlation plots for the selected analytes from (A) (red box). Each dot represents a single vaccine recipient response. r is shown in plots; all correlations are characterized by an adjusted p < 0.05. See also Figure S5 and Tables S2 and S3.
Additionally, correlations suggesting a generic pattern of cytokine/chemokine co-expression as a consequence of the inflammation process were also identified. The day 23 IL-15 log2 fold change significantly correlated with the day 23 log2 fold change for MIP-1β/CCL4 and IL-1Ra (Figure 5A, gray box; Figure S6). Positive associations were also found for the pairs IL-6 and IL-1Ra, TNF-α and IL-1Ra, TNF-α and MIP-1β/CCL4, and IFN-γ and IL-1Ra (Figure 5A, gray box; Figure S6). Other chemokines had significant positive correlations with a Spearman r of >0.55, namely, IL-1Ra and MCP-1/CCL2, IL-1Ra and MIP-1β/CCL4, and MIP-1β/CCL4 and MCP-1/CCL2 (Figure 5A, gray box; Figure S6). The booster vaccination resulted in a coordinated release of the chemokines MCP-1/CCL2, MIP-1β/CCL4, Eotaxin, and MDC/CCL22, as well of the anti-inflammatory molecule IL-1Ra. The concerted chemokine response is likely responsible for the recruitment and mobilization of different immune cell subsets, supporting regulated priming and activation of immune responses. Given the anti-inflammatory role of IL-1Ra (Dinarello, 2018; Mantovani et al., 2019), these relationships may also suggest a self-modulatory vaccine effect. The tissues and cells participating in these processes remain to be identified by further experiments.
Identification of biomarkers of successful vaccination resulting in efficient antibody development
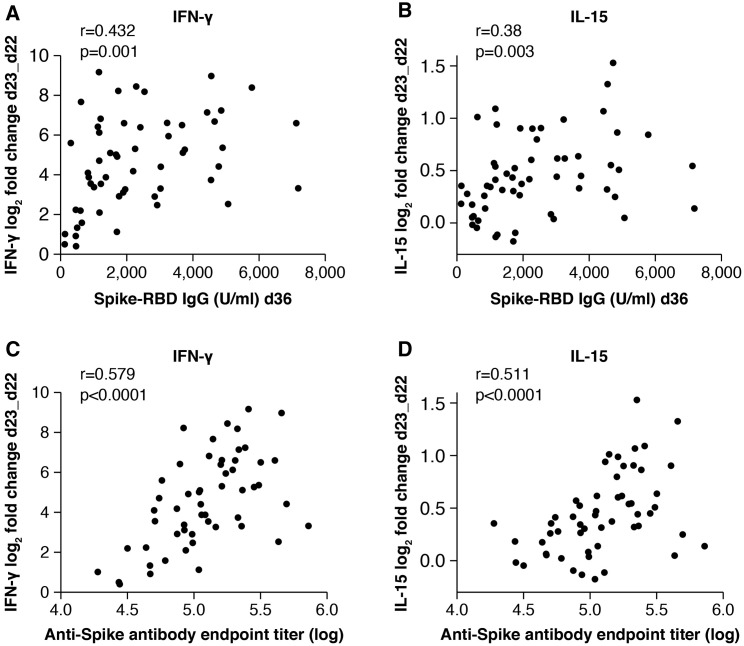
Our analysis demonstrated a vaccine-induced cytokine signature featuring IL-15, IFN-γ, and IP-10/CXCL10. Systemic levels of several other cytokines and chemokines were also affected by the vaccination. We therefore examined the relationships between alterations in these cytokines and the levels of anti-Spike antibodies detected at peak (day 36) to identify biomarkers of efficient humoral responses to vaccination. Indeed, both IFN-γ and IL-15 log2 fold changes at day 23 positively correlated with the anti-Spike-RBD antibody levels detected at day 36 (IFN-γ and anti-Spike antibody day 36: r = 0.43, p = 0.001; IL-15 and anti-Spike antibody day 36: r = 0.38, p = 0.003; Figures 6A and 6B, respectively). Similar correlations were also found at day 50. These results suggest that the IL-15/IFN-γ signature could be used as an early immune biomarker of effective development of vaccine-induced humoral responses. Additional correlations were identified between anti-Spike-RBD antibody at day 36 and changes in the chemokines MIP-1α/CCL3, MIP-1β/CCL4, and MDC/CCL22 and inflammatory markers IL-12/IL-23p40 and IL-1Ra and marginally with VEGF-A and SAA, supporting the role of leukocyte recruitment and self-limiting inflammation in the priming and recall of humoral responses (Table S4). Interestingly, significant correlations include several cytokines belonging to clusters of co-expression as reported in Figure 5. These results suggest a coordinated response to the vaccine and highlight the important role of innate responses to vaccination in shaping adaptive immunity.
Figure 6.
Biomarkers of effective vaccination
Correlations of log2 fold changes after the 2nd vaccination (d23_d22) of IFN-γ (A and C) and IL-15 (B and D) and levels of antibodies against Spike-RBD (U/ml) (A and B) and trimeric Spike (ELISA, endpoint titer) (C and D). The analysis was performed at 2 weeks after the 2nd vaccination (day 36). Spearman r and p values are given (GraphPad Prism). See also Table S4.
We found the same correlation of biomarkers with anti-Spike antibody responses, when we performed a similar analysis by using the antibody titers elicited against full-length trimeric Spike upon the 2nd vaccination (day 36). We confirmed a positive association with the log2 fold changes at day 23 for both IFN-γ (r = 0.58, p < 0.0001) and IL-15 (r = 0.51, p < 0.0001) (Figures 6C and 6D, respectively). These data are expected based on the strong correlations between Spike-RBD and Spike antibody levels (Figure S1).
Together, these results suggest a coordinated response to the vaccine and highlight the important role of innate responses to vaccination in shaping adaptive immunity.
Discussion
The field of vaccination against infectious diseases has witnessed rapid advances during the COVID-19 pandemic, with the clinical introduction of novel platforms and especially mRNA-based vaccines. Such novel vaccine technologies as the BNT162b2 mRNA COVID-19 vaccine elicit a range of responses, but the mechanisms that determine the quality and quantity of these responses are largely uncharacterized (Teijaro and Farber, 2021). In the present study, we applied systems serology to study the effects of the BNT162b2 mRNA COVID-19 vaccine to identify immunological parameters predictive of beneficial response to mRNA-based vaccination. Our analysis on the circulating levels of cytokines, chemokines, and inflammation markers as well as on the generation of anti-Spike-RBD antibodies suggests that cytokine modulation could indeed be a biomarker of successful vaccination resulting in efficient antibody development.
In antigen-naive individuals, the 1st vaccination resulted in both acute and more persistent effects on serum cytokine/chemokine levels (up to 1 week after dose administration), which were a result of inflammation and innate immune system activation. Broader and greater cytokine changes were observed after 2nd vaccination, which also suggests stimulation of anamnestic responses. Indeed, BNT162b2 mRNA vaccine administration induced a systemic cytokine/chemokine signature featuring IL-15, IFN-γ, and IP-10/CXCL10, which are molecules with a pivotal role in eliciting innate immune responses as well as in shaping adaptive immunity and leading to immunological memory. Importantly, changes in the level of IFN-γ and IL-15 positively correlated with antibody titers against SARS-CoV-2 Spike-RBD. Several associations between cytokine alterations were also identified, suggesting a coordinate response to the vaccine.
Immune signatures in vaccine recipients receiving yellow fever, HIV-Ade5, or HIV ALVAC vaccines have been described (Andersen-Nissen et al., 2021; Gaucher et al., 2008; Querec et al., 2009; Zak et al., 2012). These studies underscored the importance of analysis within 24 h after vaccination to determine innate signatures and early biomarkers that shape and predict protective adaptive responses elicited by different vaccine platforms. Analyzing the effect of a non-replicating HIV-ALVAC vaccine, Andersen-Nissen et al. (2021), reported a signature of plasma serum cytokines featuring IFN-γ, IL-15, and IP-10/CXCL10, which is similar to our findings on the BTN162b2 mRNA COVID-19 vaccine. In our study, we further found a correlation of systemic IL-15 and IFN-γ changes and anti-Spike antibody responses, supporting their identification as biomarkers of successful vaccination resulting in the development of effective humoral responses. Studies analyzing the effect of the Moderna and Curevac COVID-19 mRNA vaccines will shed more light on similarities and differences of immune signatures induced by the different mRNA platforms. It will also be important to identify early predictors of induction of humoral and cellular immunity because different vaccine methods engage the immune system differently. The identification of biomarkers measured early after vaccination (i.e., within 24 h) that correlate with immunogenicity to the full vaccine regimen (i.e., 2 weeks after booster vaccine dose) is important for vaccine clinical development and public health management. Such biomarkers could be used as surrogates of vaccine-induced protective responses, allowing for much faster decisions in trial planning and execution. Biomarkers could also help the refinement of regimens to increase vaccine efficacy, applicability, and distribution, through the identification of individuals with supra- or sub-optimal responses, especially during an outbreak. In our study, the early cytokine profile and the antibody titers induced by one mRNA vaccination in persons with pre-existing COVID-19 immunity mimic the response to the booster vaccination in COVID-naive persons. These findings support the proposal of administering a single vaccine dose to individuals with pre-existing SARS-CoV-2 immunity, which is useful during the present period of limited vaccine supply. This conclusion is in agreement with recent publications (Ebinger et al., 2021; Gobbi et al., 2021; Manisty et al., 2021).
IL-15 is a heterodimeric cytokine, comprising the IL-15 and IL-15 receptor alpha chains, termed hetIL-15 (Bergamaschi et al., 2008, 2012, 2021; Chertova et al., 2013). It affects both the innate and adaptive immune system, by supporting proliferation, survival, and function of many lymphocytes (Berard et al., 2003; Carson et al., 1994; Ma et al., 2006; Picker et al., 2006; Zhang et al., 1998). IL-15 possesses a non-redundant role in supporting long-lasting immune responses (Li et al., 2015; Rubinstein et al., 2008; Schluns et al., 2002) and in stimulating cytotoxic activity of immune cells (Bergamaschi et al., 2020; Ng et al., 2017; Watson et al., 2018). In a humanized mouse model, IL-15 treatment resulted in the development of T-cell-dependent antigen-specific B responses, following immunization (Huntington et al., 2011). These functions provided the rationale for exploring the use of IL-15 in conjunction with vaccination and evaluating its role in promoting immunogenicity. Indeed, several studies demonstrated an enhanced immune response to different vaccine platforms by IL-15 (Moore et al., 2002; Oh et al., 2003). We have also previously shown that the use of hetIL-15 as a molecular adjuvant in the therapeutic vaccination of simian immunodeficiency virus (SIV)-infected macaques resulted in robust induction of SIV-specific effector memory cells and virological benefit with strong reduction of viremia (Valentin et al., 2010).
Both IFN-γ and IP-10/CXCL10 play a role in the IL-15 effects on the immune system. IL-15 directly stimulates lymphocytes to produce IFN-γ. Both type I and type II IFN responses, in conjunction with IL-15, often represent the first innate barrier against pathogens (Perera et al., 2012). In addition, IFN-γ is critical for the development and maintenance of type 1 and antiviral immune responses (reviewed in Lin and Young, 2014; Schroder et al., 2004). An involvement of IFN-γ in shaping humoral responses by controlling Ig isotypes produced by B cells and supporting long-lived antibody-secreting cells has also been documented (Baumgarth, 2021; Stone et al., 2019).
The chemokine IP-10/CXCL10 is often released in the context of inflammation by many cells including leukocytes, neutrophils, eosinophils, monocytes, and stromal cells, in response to IFN-γ. IP-10/CXCL10 promotes the chemotaxis of CXCR3+ cells, which are mainly activated T and B lymphocytes (reviewed in Griffith et al., 2014; Liu et al., 2011). A recent study proposed a mechanism by which IL-15 indirectly acts on dendritic cells and macrophage/monocytes to induce the secretion of IP-10/CXCL10, by IFN-γ (Bergamaschi et al., 2020). Recent studies identified early innate immune responses to both flu and Ebola virus vaccines. Serum IP-10/CXCL10 levels, an innate signature linked to IP-10/CXCL10 and IFN-related genes, were associated with higher vaccine-induced antibody titers (Gonçalves et al., 2019; Rechtien et al., 2017). IP-10/CXCL10 was also described to drive activated B cells to differentiate into plasma cells (Xu et al., 2012).
Given their action, IL-15, IFN-γ, and IP-10/CXCL10 have emerged as critical components of an immune response against viral infections. In our study, the role of these cytokines upon vaccination became more apparent after the 2nd vaccination, which also induced TNF-α and IL-6. Importantly, a similar cytokine/chemokine pattern of expression at 24 h post-vaccination was found between vaccine recipients with pre-existing SARS-CoV-2 immunity who received the 1st vaccine dose and antigen-naive individuals after 2nd vaccine dose, suggesting induction of anamnestic responses, with higher levels of IFN-γ, IP-10/CXCL10, IL-6, and TNF-α for the rapid recruitment and stimulation of effector immune cells.
Many studies have shown that uncontrolled inflammation and cytokine storm syndrome contribute to the severity of COVID-19 disease. Patients with severe disease are characterized by high levels of inflammatory markers, including CRP, ferritin, and D-dimer and high levels of chemokines, such as granulocyte colony-stimulating factor (G-CSF), MCP-1/CCL2, MIP-1α/CCL3, IL-8, and IP-10/CXCL10, resulting in inflammatory cell infiltration and tissue damage in the lungs and in a high neutrophil-to-lymphocyte ratio (Mehta et al., 2020; Merad and Martin, 2020). A systemic increase in the levels of IL-2, IL-7, IL-10, IL-6, and TNF-α has also been reported (Huang et al., 2020). In particular, IL-6, IL-8, and TNF-α serum levels are significant predictors of disease severity and death (Del Valle et al., 2020). In contrast, early activation of the IFN type I pathway was associated with the prevention of disease progression (Bastard et al., 2020; Zhang et al., 2020). Although several of the cytokines and chemokines induced by viral infection were also elevated after mRNA vaccination, important differences are to be highlighted. Upon vaccination, we observed an early but transient inflammatory cytokine response. IFN-γ, IP-10/CXCL10, IL-6, and CRP increased acutely at day 2 and returned to baseline levels by day 8 after vaccine administration. More durable effects lasting up to day 8 were observed for the chemokines IL-8, IL-16, MIP-1α/CCL3, and MIP-1β/CCL4. In contrast, in COVID-19 patients, systemic levels of IP-10/CXCL10 and IL-6 remain elevated throughout the COVID-19 infection response (Buszko et al., 2021). Additionally, the vaccine-induced upregulation of the anti-inflammatory molecule IL-1Ra may also indicate a self-modulatory and limiting inflammatory effect of the vaccine. Importantly, our data suggest that mRNA vaccination is associated with a cytokine signature featuring IL-15, IFN-γ, and IP-10/CXCL10, which results in an efficient anti-viral immune response that is usually weakened in COVID-19 patients.
In conclusion, this study highlights important associations of several immunoregulatory molecules induced by vaccination with innate and adaptive immune responses elicited by an mRNA-based vaccine. The early cytokine/chemokine signature featuring IL-15, IFN-γ, and IP-10/CXCL10 may be used to monitor effective vaccination and as a guide to optimize the efficacy of mRNA vaccination strategies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| sera from BNT162b2 vaccinated persons | NCT04743388 | N/A |
| mouse anti-human IgGFc-HRP | Southern Biotech | 9040-05; RRID:AB_2687484 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 Spike | D. Esposito, NCI | N/A |
| Critical commercial assays | ||
| V-PLEX Human Biomarker Assay kit | Meso Scale Diagnostics, Maryland, USA | K15248D-2 |
| Elecsys Anti-SARS-CoV-2 S assay | Roche Diagnostics GmbH, Mannheim, Germany | N/A |
| SARS-CoV-2 Surrogate Virus Neutralization Test | GenScript, Piscataway, NJ, USA | N/A |
| Nano-Glo® Luciferase Assay System | Promega, USA | N1130 |
| Luciferase Cell Culture Lysis 5X Reagent | Promega, USA | E1531 |
| Deposited data | ||
| Cytokine expression data and analysis code | This paper | https://github.com/NCI-VB/felber_covid_vaccine |
| Experimental models: Cell lines | ||
| HEK293T/ACE2wt | T. Hatziioanou, Rockefeller University | N/A |
| Recombinant DNA | ||
| pHIVNLDEnv-Nanoluc | T. Hatziioanou, Rockefeller University | N/A |
| Spike Wuhan1253 | T. Hatziioanou, Rockefeller University | N/A |
| Software and algorithms | ||
| GraphPad Prism version 9.0.2 for MacOS X | GraphPad Software, Inc, La Jolla, CA | N/A |
| R | this paper | https://www.r-project.org/ v3.5.1 |
| edgeR | this paper | https://bioconductor.org/packages/release/bioc/html/edgeR.html v3.24.3 |
| limma | this paper | https://bioconductor.org/packages/release/bioc/html/limma.html v3.38.3 |
| corrplot | this paper | https://cran.r-project.org/web/packages/corrplot/index.html v0.84 |
| ggpubr | this paper | https://cran.r-project.org/web/packages/ggpubr/index.html v0.2.5 |
| NIH Integrated Data Analysis Platform | this paper | https://nidap.nih.gov/workspace |
| Antibody endpoint titer: R functions SSasymp() and nls() from the “stats” R package | this paper | https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/nls |
| Cytokine expression data and analysis code | this paper | https://github.com/NCI-VB/felber_covid_vaccine |
| Other | ||
| SpectraMax M3 | Molecular Devices, LLC | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Barbara K. Felber (Barbara.felber@nih.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Study Design
This is an ongoing prospective study (NCT04743388) that evaluates the kinetics of antibodies against SARS-CoV-2 as well as the kinetics of serum cytokines and chemokines associated with the immune response and of B- and T cell subpopulations in volunteers receiving the BNT162b2 vaccine against SARS-Cov-2 (Comirnaty™) initiated on January 4, 2021, in Greece. This analysis reports the results regarding the alterations of cytokines and chemokines in volunteer donors, which is a secondary endpoint of the study.
Inclusion/Exclusion Criteria
Major inclusion criteria for participation in this study included: (i) age above 18 years; (II) ability to sign the informed consent form, and (iii) eligibility for vaccination, according to the national (Greek) program for COVID-19 vaccination (i.e., individuals who had no serious allergy problem and especially they have not been hospitalized due to a serious allergic reaction (anaphylaxis). Major exclusion criteria included the presence of: (i) autoimmune disorder under immunosuppressive therapy; (ii) active malignant disease and (iii) end-stage renal disease, as previously described (Terpos et al., 2021b).
Volunteer donors between the ages of 20-78 (Male = 27 and Female = 36) were tested in the period January 4 to April 1, 2021, in the vaccine center of Alexandra General Hospital in Athens, Greece. All study procedures were carried out in accordance with the declaration of Helsinki (18th World Medical Association Assembly), its subsequent amendments, the Greek regulations and guidelines, as well as the good clinical practice guidelines (GCP) as defined by the International Conference of Harmonization. The study was also approved by the local ethic committee of Alexandra General Hospital (no 15/23 December 2020).
Method details
Detection of antibodies against SARS-CoV-2
Serum was collected at day of vaccination (d1) and 1 and 3 weeks later (d8, d22) and 2 and 4 weeks after the 2nd vaccination dose (d36 and d50). After vein puncture, serum was separated within 4 hours from blood collection and stored at −80°C until the day of measurement. Stored samples from different time points of the same donor were measured in parallel assays.
Anti-Spike-RBD IgG antibodies and neutralizing antibodies (NAbs) against SARS-CoV-2 were measured using FDA approved methods, i.e., the Elecsys Anti-SARS-CoV-2 S assay (Roche Diagnostics GmbH, Mannheim, Germany) (Higgins et al., 2021) and the cPass SARS-CoV-2 NAbs Detection Kit (GenScript, Piscataway, NJ, USA) (Tan et al., 2020), respectively; the latter allows the indirect detection of potential SARS-CoV-2 NAbs in the blood, by assaying the antibody (independent of class)-mediated inhibition of SARS-CoV-2 RBD binding to human host receptor angiotensin converting enzyme 2 (ACE2).
An in-house ELISA assay described elsewhere (Terpos et al., 2020, 2021a) was used to detect the complete trimeric Spike using mammalian Expi293-cells produced proteins (Esposito et al., 2020) measuring eight 4-fold serial dilutions starting at 1:50. Antibody levels were expressed as endpoint titers using a model fit approach conducted in R to model the curve to more accurately define endpoint titers (Terpos et al., 2021a).
The Spike pseudotyped pHIVNLΔEnv-Nanoluc assay (Robbiani et al., 2020; Schmidt et al., 2020) was performed as previously described (Terpos et al., 2020, 2021a). Spike pseudotyped pHIVNLΔEnv-Nanoluc was tested with 8 dilutions of heat-inactivated sera (1:40 to 1: 655,360) in triplicates in HEK293T/ACE2wt cells and luciferase levels in the cell lysates were measured. The 50% Inhibitory dose (ID50) was calculated using GraphPad Prism version 9.0.2 for MacOS X (GraphPad Software, Inc, La Jolla, CA) with nonlinear regression curve fit using inhibitor versus responses variable slope (four parameters). The NAb ID50 threshold of quantification is 0.5 log and the threshold of detection is 0.1 log in this assay.
Cytokine/chemokine analysis
Serum cytokine/chemokine concentrations were measured with the V-PLEX Human Biomarker Assay kit (Meso Scale Diagnostics, Maryland, US) according to manufacturer’s recommendations. This allowed for the concurrent measurement of the following cytokines and chemokines (see also Table S2). For analysis, biomarkers falling below the detection limit/standard range were removed if absent in more than 50% of the samples or adjusted to 0.5 of the lowest standard point/detection limits.
Bioinformatics
Biomarker analysis was performed with a workflow written in R and through a user interface developed on the Foundry Platform (Palantir Technologies). The limma package was used to compare biomarker changes between time points, setting significance for False Discovery Rate (FDR) < 0.05. Heatmaps were represented as log2 fold change over day 1 (1st vaccination) or day 22 (2nd vaccination). A comparison of the effects after 2nd vaccination over the ones induced by the 1st vaccination was also performed. Pairwise correlations were performed among the log2 fold-changes in concentration at day 2 and 23 for the 19 biomarkers that were affected by the vaccination, using an adjusted Spearman p value < 0.05. (Link: https://github.com/NCI-VB/felber_covid_vaccine).
Quantification and statistical analysis
Software
All software is freely or commercially available and is listed in the STAR Methods description and Key resources table.
Acknowledgments
We thank R. Burns for assistance; Y. Wang and J. Inglefield, Clinical Services Program, Frederick National Laboratory for Cancer Research for technical assistance; D. Donohue (Data Management Service, NCI) for assistance with data analysis; T. Hatziioanou (Rockefeller University) for pseudotype virus reagents and discussions; G. Rentziou, I. Charitaki, C.Y. Liacos, N. Mavrianou, N.A. Kokkali, and S. Skourti for administrative, technical, and/or material support; X. Hu for additional antibody assays; D. Esposito (Protein Expression Laboratory, FNLCR) for materials; and members of the Felber and Pavlakis labs for discussions; C.A. Stewart for discussion; and T. Jones for editorial assistance. This research was supported in part by the Intramural Research Program of the NIH, NCI to G.N.P. and B.K.F.
Author contributions
Conceptualization, G.N.P., B.K.F., E.T., and M.A.D.; data curation, M.A., E.T., C.B., G.N.P., and B.K.F.; formal analysis, C.B., M.A., G.N.P., and B.K.F.; funding acquisition, G.N.P., B.K.F., and M.A.D.; investigation, J.B., E.T., M.A., S.G., I.P.T., F.A., T.B., D.P., M.R., D.S., and S.K.; visualization, C.B., M.A., and B.K.F.; writing – original draft, C.B., G.N.P., and B.K.F.; writing – review & editing, C.B., G.N.P., B.K.F., E.T., M.A.D., and all coauthors reviewed the paper.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: August 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109504.
Supplemental Information
Data and code availability
-
•
Data and analysis generated during the study are available at https://github.com/NCI-VB/felber_covid_vaccine.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Andersen-Nissen E., Fiore-Gartland A., Ballweber Fleming L., Carpp L.N., Naidoo A.F., Harper M.S., Voillet V., Grunenberg N., Laher F., Innes C., et al. Innate immune signatures to a partially-efficacious HIV vaccine predict correlates of HIV-1 infection risk. PLoS Pathog. 2021;17:e1009363. doi: 10.1371/journal.ppat.1009363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.R., Belij-Rammerstorfer S., Dold C., Ewer K.J., Folegatti P.M., Gilbride C., Halkerston R., Hill J., Jenkin D., Stockdale L., et al. Oxford COVID Vaccine Trial Group Phase 1/2 trial of SARS-CoV-2 vaccine ChAdOx1 nCoV-19 with a booster dose induces multifunctional antibody responses. Nat. Med. 2021;27:279–288. doi: 10.1038/s41591-020-01179-4. [DOI] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N. The Shaping of a B Cell Pool Maximally Responsive to Infections. Annu. Rev. Immunol. 2021;39:103–129. doi: 10.1146/annurev-immunol-042718-041238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berard M., Brandt K., Bulfone-Paus S., Tough D.F. IL-15 promotes the survival of naive and memory phenotype CD8+ T cells. J. Immunol. 2003;170:5018–5026. doi: 10.4049/jimmunol.170.10.5018. [DOI] [PubMed] [Google Scholar]
- Bergamaschi C., Rosati M., Jalah R., Valentin A., Kulkarni V., Alicea C., Zhang G.M., Patel V., Felber B.K., Pavlakis G.N. Intracellular interaction of interleukin-15 with its receptor alpha during production leads to mutual stabilization and increased bioactivity. J. Biol. Chem. 2008;283:4189–4199. doi: 10.1074/jbc.M705725200. [DOI] [PubMed] [Google Scholar]
- Bergamaschi C., Bear J., Rosati M., Beach R.K., Alicea C., Sowder R., Chertova E., Rosenberg S.A., Felber B.K., Pavlakis G.N. Circulating IL-15 exists as heterodimeric complex with soluble IL-15Rα in human and mouse serum. Blood. 2012;120:e1–e8. doi: 10.1182/blood-2011-10-384362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi C., Pandit H., Nagy B.A., Stellas D., Jensen S.M., Bear J., Cam M., Valentin A., Fox B.A., Felber B.K., Pavlakis G.N. Heterodimeric IL-15 delays tumor growth and promotes intratumoral CTL and dendritic cell accumulation by a cytokine network involving XCL1, IFN-γ, CXCL9 and CXCL10. J. Immunother. Cancer. 2020;8:e000599. doi: 10.1136/jitc-2020-000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi C., Stravokefalou V., Stellas D., Karaliota S., Felber B.K., Pavlakis G.N. Heterodimeric IL-15 in Cancer Immunotherapy. Cancers (Basel) 2021;13:837. doi: 10.3390/cancers13040837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszko M., Nita-Lazar A., Park J.H., Schwartzberg P.L., Verthelyi D., Young H.A., Rosenberg A.S. Lessons learned: new insights on the role of cytokines in COVID-19. Nat. Immunol. 2021;22:404–411. doi: 10.1038/s41590-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagigi A., Loré K. Immune Responses Induced by mRNA Vaccination in Mice, Monkeys and Humans. Vaccines (Basel) 2021;9:61. doi: 10.3390/vaccines9010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson W.E., Giri J.G., Lindemann M.J., Linett M.L., Ahdieh M., Paxton R., Anderson D., Eisenmann J., Grabstein K., Caligiuri M.A. Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J. Exp. Med. 1994;180:1395–1403. doi: 10.1084/jem.180.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2021. CDC COVID-19 Study Shows mRNA Vaccines Reduce Risk of Infection by 91 Percent for Fully Vaccinated People.https://www.cdc.gov/media/releases/2021/p0607-mrna-reduce-risks.html?fbclid=IwAR2jtHTZO2XxwMHHvwWReHrDg59qfTo2ZntZaZmkDy-7xPEJSC0b8q_X8ng [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. BLAZE-1 Investigators SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertova E., Bergamaschi C., Chertov O., Sowder R., Bear J., Roser J.D., Beach R.K., Lifson J.D., Felber B.K., Pavlakis G.N. Characterization and favorable in vivo properties of heterodimeric soluble IL-15·IL-15Rα cytokine compared to IL-15 monomer. J. Biol. Chem. 2013;288:18093–18103. doi: 10.1074/jbc.M113.461756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodick G., Tene L., Rotem R.S., Patalon T., Gazit S., Ben-Tov A., Weil C., Goldshtein I., Twig G., Cohen D., Muhsen K. The effectiveness of the TWO-DOSE BNT162b2 vaccine: analysis of real-world data. Clin. Infect. Dis. 2021:ciab438. doi: 10.1093/cid/ciab438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 2018;281:8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinger J.E., Fert-Bober J., Printsev I., Wu M., Sun N., Prostko J.C., Frias E.C., Stewart J.L., Van Eyk J.E., Braun J.G., et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat. Med. 2021;27:981–984. doi: 10.1038/s41591-021-01325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D., Mehalko J., Drew M., Snead K., Wall V., Taylor T., Frank P., Denson J.P., Hong M., Gulten G., et al. Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein Expr. Purif. 2020;174:105686. doi: 10.1016/j.pep.2020.105686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., et al. Oxford COVID Vaccine Trial Group Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourati S., Ribeiro S.P., Blasco Tavares Pereira Lopes F., Talla A., Lefebvre F., Cameron M., Kaewkungwal J., Pitisuttithum P., Nitayaphan S., Rerks-Ngarm S., et al. Integrated systems approach defines the antiviral pathways conferring protection by the RV144 HIV vaccine. Nat. Commun. 2019;10:863. doi: 10.1038/s41467-019-08854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher D., Therrien R., Kettaf N., Angermann B.R., Boucher G., Filali-Mouhim A., Moser J.M., Mehta R.S., Drake D.R., III, Castro E., et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbi F., Buonfrate D., Moro L., Rodari P., Piubelli C., Caldrer S., Riccetti S., Sinigaglia A., Barzon L. Antibody Response to the BNT162b2 mRNA COVID-19 Vaccine in Subjects with Prior SARS-CoV-2 Infection. Viruses. 2021;13:422. doi: 10.3390/v13030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves E., Bonduelle O., Soria A., Loulergue P., Rousseau A., Cachanado M., Bonnabau H., Thiebaut R., Tchitchek N., Behillil S., et al. Innate gene signature distinguishes humoral versus cytotoxic responses to influenza vaccination. J. Clin. Invest. 2019;129:1960–1971. doi: 10.1172/JCI125372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb R.L., Nirula A., Chen P., Boscia J., Heller B., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J.W., Sokol C.L., Luster A.D. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- Hagan T., Pulendran B. Will Systems Biology Deliver Its Promise and Contribute to the Development of New or Improved Vaccines? From Data to Understanding through Systems Biology. Cold Spring Harb. Perspect. Biol. 2018;10:a028894. doi: 10.1101/cshperspect.a028894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins V., Fabros A., Kulasingam V. Quantitative Measurement of Anti-SARS-CoV-2 Antibodies: Analytical and Clinical Evaluation. J. Clin. Microbiol. 2021;59:e03149-20. doi: 10.1128/JCM.03149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Ivashkiv L.B. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Alves N.L., Legrand N., Lim A., Strick-Marchand H., Mention J.J., Plet A., Weijer K., Jacques Y., Becker P.D., et al. IL-15 transpresentation promotes both human T-cell reconstitution and T-cell-dependent antibody responses in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:6217–6222. doi: 10.1073/pnas.1019167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Fourati S., Canderan G., Sekaly R.P. Systems biology and the quest for correlates of protection to guide the development of an HIV vaccine. Curr. Opin. Immunol. 2016;41:91–97. doi: 10.1016/j.coi.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Ledford H. COVID antibody treatments show promise for preventing severe disease. Nature. 2021;591:513–514. doi: 10.1038/d41586-021-00650-7. [DOI] [PubMed] [Google Scholar]
- Leonard W.J., Lin J.X., O’Shea J.J. The γc Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity. 2019;50:832–850. doi: 10.1016/j.immuni.2019.03.028. [DOI] [PubMed] [Google Scholar]
- Li J., Valentin A., Ng S., Beach R.K., Alicea C., Bergamaschi C., Felber B.K., Pavlakis G.N. Differential effects of IL-15 on the generation, maintenance and cytotoxic potential of adaptive cellular responses induced by DNA vaccination. Vaccine. 2015;33:1188–1196. doi: 10.1016/j.vaccine.2014.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.C., Young H.A. Interferons: Success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 2014;25:369–376. doi: 10.1016/j.cytogfr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Guo S., Hibbert J.M., Jain V., Singh N., Wilson N.O., Stiles J.K. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011;22:121–130. doi: 10.1016/j.cytogfr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., Koka R., Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu. Rev. Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- Mangalmurti N., Hunter C.A. Cytokine Storms: Understanding COVID-19. Immunity. 2020;53:19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manisty C., Otter A.D., Treibel T.A., McKnight Á., Altmann D.M., Brooks T., Noursadeghi M., Boyton R.J., Semper A., Moon J.C. Antibody response to first BNT162b2 dose in previously SARS-CoV-2-infected individuals. Lancet. 2021;397:1057–1058. doi: 10.1016/S0140-6736(21)00501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Dinarello C.A., Molgora M., Garlanda C. Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity. Immunity. 2019;50:778–795. doi: 10.1016/j.immuni.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes I.B., Schett G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., HLH Across Speciality Collaboration, UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A.C., Kong W.P., Chakrabarti B.K., Nabel G.J. Effects of antigen and genetic adjuvants on immune responses to human immunodeficiency virus DNA vaccines in mice. J. Virol. 2002;76:243–250. doi: 10.1128/JVI.76.1.243-250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.S.M., Nagy B.A., Jensen S.M., Hu X., Alicea C., Fox B.A., Felber B.K., Bergamaschi C., Pavlakis G.N. Heterodimeric IL-15 treatment enhances tumor infiltration, persistence and effector functions of adoptively transferred tumor-specific T cells in the absence of lymphodepletion. Clin. Cancer Res. 2017;23:2817–2830. doi: 10.1158/1078-0432.CCR-16-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S., Berzofsky J.A., Burke D.S., Waldmann T.A., Perera L.P. Coadministration of HIV vaccine vectors with vaccinia viruses expressing IL-15 but not IL-2 induces long-lasting cellular immunity. Proc. Natl. Acad. Sci. USA. 2003;100:3392–3397. doi: 10.1073/pnas.0630592100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera P.Y., Lichy J.H., Waldmann T.A., Perera L.P. The role of interleukin-15 in inflammation and immune responses to infection: implications for its therapeutic use. Microbes Infect. 2012;14:247–261. doi: 10.1016/j.micinf.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picker L.J., Reed-Inderbitzin E.F., Hagen S.I., Edgar J.B., Hansen S.G., Legasse A., Planer S., Piatak M., Jr., Lifson J.D., Maino V.C., et al. IL-15 induces CD4 effector memory T cell production and tissue emigration in nonhuman primates. J. Clin. Invest. 2006;116:1514–1524. doi: 10.1172/JCI27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England. 2021. PHE monitoring of the effectiveness of COVID-19 vaccination.https://www.gov.uk/government/publications/phe-monitoring-of-the-effectiveness-of-covid-19-vaccination [Google Scholar]
- Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtien A., Richert L., Lorenzo H., Martrus G., Hejblum B., Dahlke C., Kasonta R., Zinser M., Stubbe H., Matschl U., et al. VEBCON Consortium Systems Vaccinology Identifies an Early Innate Immune Signature as a Correlate of Antibody Responses to the Ebola Vaccine rVSV-ZEBOV. Cell Rep. 2017;20:2251–2261. doi: 10.1016/j.celrep.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati M., Agarwal M., Hu X., Devasundaram S., Stellas D., Chowdhury B., Bear J., Burns R., Donohue D., Pessaint L., et al. Control of SARS-CoV-2 infection after Spike DNA or Spike DNA+Protein co-immunization in rhesus macaques. bioRxiv. 2021 doi: 10.1101/2021.06.11.448032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M.P., Lind N.A., Purton J.F., Filippou P., Best J.A., McGhee P.A., Surh C.D., Goldrath A.W. IL-7 and IL-15 differentially regulate CD8+ T-cell subsets during contraction of the immune response. Blood. 2008;112:3704–3712. doi: 10.1182/blood-2008-06-160945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluns K.S., Williams K., Ma A., Zheng X.X., Lefrançois L. Cutting edge: requirement for IL-15 in the generation of primary and memory antigen-specific CD8 T cells. J. Immunol. 2002;168:4827–4831. doi: 10.4049/jimmunol.168.10.4827. [DOI] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Hertzog P.J., Ravasi T., Hume D.A. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Stephenson K.E., Le Gars M., Sadoff J., de Groot A.M., Heerwegh D., Truyers C., Atyeo C., Loos C., Chandrashekar A., McMahan K., et al. Immunogenicity of the Ad26.COV2.S Vaccine for COVID-19. JAMA. 2021;325:1535–1544. doi: 10.1001/jama.2021.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Peel J.N., Scharer C.D., Risley C.A., Chisolm D.A., Schultz M.D., Yu B., Ballesteros-Tato A., Wojciechowski W., Mousseau B., et al. T-bet Transcription Factor Promotes Antibody-Secreting Cell Differentiation by Limiting the Inflammatory Effects of IFN-γ on B Cells. Immunity. 2019;50:1172–1187.e7. doi: 10.1016/j.immuni.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I., Tiu C., Hu Z., Chen V.C., Young B.E., Sia W.R., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Teijaro J.R., Farber D.L. COVID-19 vaccines: modes of immune activation and future challenges. Nat. Rev. Immunol. 2021;21:195–197. doi: 10.1038/s41577-021-00526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpos E., Politou M., Sergentanis T.N., Mentis A., Rosati M., Stellas D., Bear J., Hu X., Felber B.K., Pappa V., et al. Anti-SARS-CoV-2 Antibody Responses in Convalescent Plasma Donors Are Increased in Hospitalized Patients; Subanalyses of a Phase 2 Clinical Study. Microorganisms. 2020;8:E1885. doi: 10.3390/microorganisms8121885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpos E., Stellas D., Rosati M., Sergentanis T.N., Hu X., Politou M., Pappa V., Ntanasis-Stathopoulos I., Karaliota S., Bear J., et al. SARS-CoV-2 antibody kinetics eight months from COVID-19 onset: Persistence of spike antibodies but loss of neutralizing antibodies in 24% of convalescent plasma donors. Eur. J. Intern. Med. 2021;89:87–96. doi: 10.1016/j.ejim.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpos E., Trougakos I.P., Apostolakou F., Charitaki I., Sklirou A.D., Mavrianou N., Papanagnou E.-D., Liacos C.-I., Gumeni S., Rentziou G., et al. Age-dependent and gender-dependent antibody responses against SARS-CoV-2 in health workers and octogenarians after vaccination with the BNT162b2 mRNA vaccine. Am. J. Hematol. 2021;96:E257–E259. doi: 10.1002/ajh.26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin A., von Gegerfelt A., Rosati M., Miteloudis G., Alicea C., Bergamaschi C., Jalah R., Patel V., Khan A.S., Draghia-Akli R., et al. Repeated DNA therapeutic vaccination of chronically SIV-infected macaques provides additional virological benefit. Vaccine. 2010;28:1962–1974. doi: 10.1016/j.vaccine.2009.10.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D.C., Moysi E., Valentin A., Bergamaschi C., Devasundaram S., Fortis S.P., Bear J., Chertova E., Bess J., Jr., Sowder R., et al. Treatment with native heterodimeric IL-15 increases cytotoxic lymphocytes and reduces SHIV RNA in lymph nodes. PLoS Pathog. 2018;14:e1006902. doi: 10.1371/journal.ppat.1006902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. mRNA-1273 Study Group Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Joo H., Clayton S., Dullaers M., Herve M.C., Blankenship D., De La Morena M.T., Balderas R., Picard C., Casanova J.L., et al. Macrophages induce differentiation of plasma cells through CXCL10/IP-10. J. Exp. Med. 2012;209:1813–1823. doi: 10.1084/jem.20112142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak D.E., Andersen-Nissen E., Peterson E.R., Sato A., Hamilton M.K., Borgerding J., Krishnamurty A.T., Chang J.T., Adams D.J., Hensley T.R., et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8+ T-cell responses but is attenuated by preexisting Ad5 immunity. Proc. Natl. Acad. Sci. USA. 2012;109:E3503–E3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Sun S., Hwang I., Tough D.F., Sprent J. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity. 1998;8:591–599. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. COVID-STORM Clinicians. COVID Clinicians. Imagine COVID Group. French COVID Cohort Study Group. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort. NIAID-USUHS/TAGC COVID Immunity Group Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data and analysis generated during the study are available at https://github.com/NCI-VB/felber_covid_vaccine.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.