Abstract
A machine learning approach is presented for analyzing complex two-dimensional hyperfine sublevel correlation electron paramagnetic resonance (HYSCORE EPR) spectra with the proficiency of an expert spectroscopist. The computer vision algorithm requires no training on experimental data; rather, all of the spin physics required to interpret the spectra are learned from simulations alone. This approach is therefore applicable even when insufficient experimental data exist to train the algorithm. The neural network is demonstrated to be capable of utilizing the full information content of two-dimensional 14N HYSCORE spectra to predict the magnetic coupling parameters and their underlying probability distributions that were previously inaccessible. The predicted hyperfine (a, T) and 14N quadrupole (K, η) coupling constants deviate from the previous manual analyses of the experimental spectra on average by 0.11 MHz, 0.09 MHz, 0.19 MHz, and 0.09, respectively.
Graphical Abstract
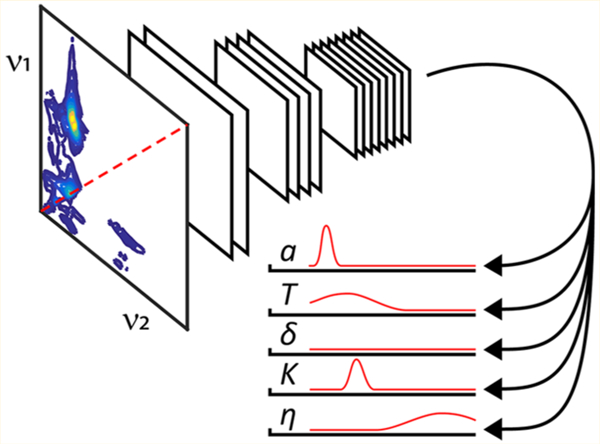
Recent advances in machine learning provide computers with an unprecedented visual capacity to match or even outperform humans in tasks ranging from everyday image recognition1–3 to disease diagnosis,4,5 yet only in a handful of cases have these types of neural network architectures been applied to problems in spectroscopy. Central to these computer vision algorithms is the convolutional neural network (CNN), which has allowed automated assignment of Raman spectra even in the presence of significant baseline distortion artifacts6 and has demonstrated a high performance level for automatic spectral assignments in multidimensional nuclear magnetic resonance (NMR) spectra.7,8 In this Letter, we describe a general purpose CNN architecture for extracting parameters of interest directly from two-dimensional (2D) spectra. The spectrum and experimental settings are the input to the algorithm, and the predictions and their associated confidence intervals are returned as output. As a case study, this neural network’s capabilities are demonstrated for the pulsed electron paramagnetic resonance (EPR) experiment HYSCORE (Hyperfine Sublevel Correlation), with the pulse sequence shown in Figure 1.9 Specifically, HYSCORE is used to differentiate all the connectivities of the hyperfine levels and the coupling parameters between electron spins (S = 1/2) and their surrounding nuclear spin environments. For spin I = 1/2 nuclei such as 1H, 13C, and 15N, the hyperfine coupling with the electron spin gives rise to simple cross-ridge lineshapes in HYSCORE spectra of powder samples that allow for the magnetic coupling parameters to be extracted by simple least-squares fitting.10,11 For 14N nuclei (I = 1) considered in this work, the situation is complicated by the nonequivalent splitting of the nuclear levels into the mI = −1, 0, and 1 spin states because of the quadrupolar interaction. The HYSCORE spectrum for even a single 14N spin is generally a multiline pattern with up to nine cross-peak pairs, and the cross-ridge intensities are a complicated function of the relative hyperfine and nuclear quadrupole couplings and the experimental parameters with which the measurement was performed.12 Spectral artifacts from τ-suppression11 and the appearance of diagonal peaks from incomplete inversion by the π-pulse can further contribute to complicating the spectral analysis.13 These challenges make 2D 14N HYSCORE an appealing case study for using machine learning to extract information convolved with these spectral artifacts.
Figure 1.
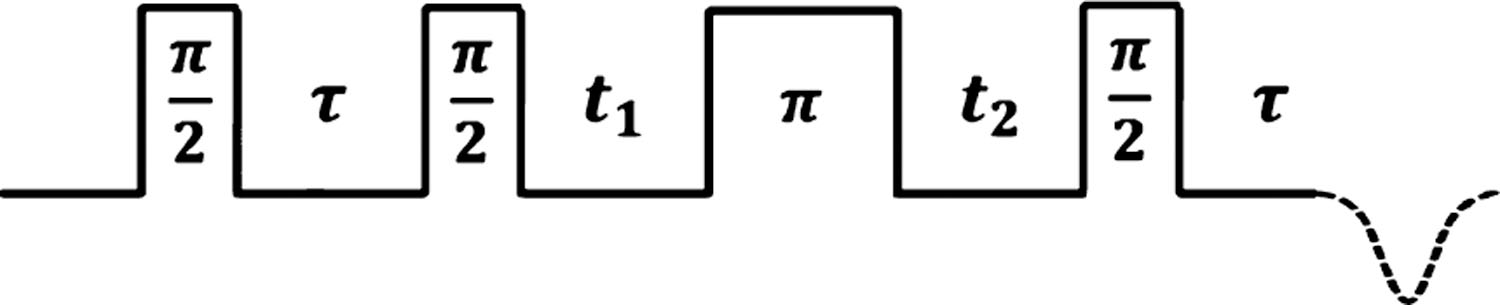
HYSCORE pulse sequence investigated in this work. The delay between the first and second π/2-pulses is held at a fixed value τ. The intensity of the inverted echo (dashed) is acquired as a function of t1 and t2, resulting in a 2D time-domain pattern correlating the change in nuclear resonant frequencies as the electron spin is flipped between its +1/2 and −1/2 spin states. All spectra were acquired with a 4-step phase cycle.14
However, an inherent problem with HYSCORE, as for most multidimensional spectroscopies in magnetic resonance, is a paucity of experimental data available to train the neural network. Here we present a simple, yet novel solution where a CNN is trained on simulations in order to teach it to analyze experimental spectra. A similar strategy of training a neural network on simulations was successfully implemented recently to process double electron–electron resonance data.15 The result described here is a neural network that solves the inverse problem of extracting magnetic coupling parameters from a HYSCORE spectrum, revealing the full probability distributions of these parameters for the first time.
Hyperfine and Nuclear Quadrupole Couplings.
The goal of analyzing an 14N HYSCORE spectrum is the determination of the hyperfine and nuclear quadrupole tensors, which provide a quantum mechanical description of the couplings between the nitrogen spin and its nearby electron spin. The principal components of the hyperfine tensor (AXX, AYY, and AZZ) can be represented in spherical form as a − T(1 + δ), a − T(1 − δ), and a + 2T, respectively, where a is the isotropic constant, T the anisotropic constant, and δ the unitless rhombicity factor that varies between 0 and 1. The tensor for the nuclear quadrupole interaction is traceless, and therefore, its principal components (QXX, QYY, and QZZ) can be represented with only two values, the quadrupole coupling constant K = e2Qq/h and the unitless asymmetry parameter η = (QXX − QYY)/QZZ.13,16
Under most circumstances, currently available analytical expressions allow only for a and K2(3 + η2) to be directly estimated from 14N spectra (valid only when the hyperfine anisotropy is weak, i.e. T ≈ 0).16,17 In the special case of the cancellation condition, where the nuclear Larmor frequency (ν14N) exactly matches the hyperfine interaction (|ν14N − |a|/2| ≈ 0), direct determinations of K and η may also be possible.16,17 In order to determine the remaining parameters, 14N HYSCORE simulations are performed by optimizing a vast parameter space of unknown variables. These simulations are often a tedious trial-and-error task prone to operator bias.
Machine Learning Algorithm.
A CNN architecture was designed and trained on EasySpin v5.2.1320 simulations to extract a, T, δ, K, and η directly from the raw X-band (∼9 GHz) HYSCORE time-domain patterns (Figure 2). For spectra with multiple nitrogen couplings, the cross-peaks were manually separated such that HYSCORE spectra corresponding to a single 14N served as input to the neural network. The machine learning algorithm’s performance was validated on our previously analyzed experimental HYSCORE spectra21–25 that exhibit well-resolved 14N couplings. The outputs of the CNN are unnormalized probability distributions for each parameter, where the amplitudes of the distributions indicate the neural network’s confidence in making its predictions (a flat baseline of zeros indicates no confidence in making a prediction). The CNN outputs are compared with the literature values in Figure 3. The chemical structures of the spin systems considered in this work are shown in Figure 4. Full details on the machine learning architectural design and training are provided in the Supporting Information.
Figure 2.
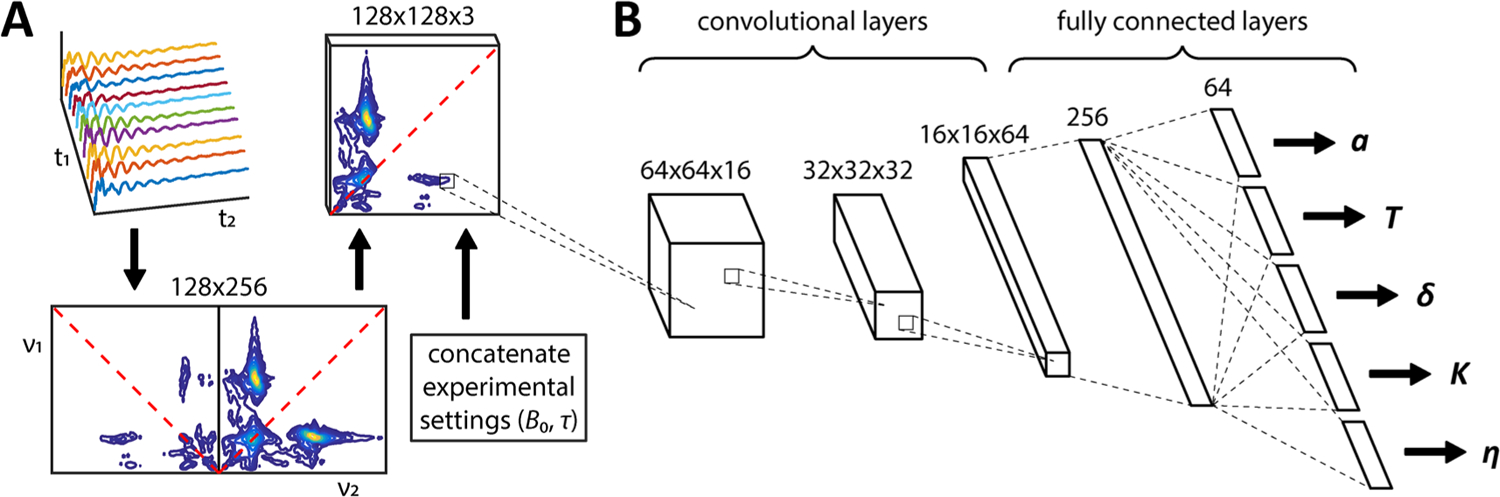
Machine learning architecture for predicting the 14N hyperfine and nuclear quadrupole coupling constants directly from the raw HYSCORE data. (A) Spectral preprocessing involves 2D Fourier transformation of the time-domain data, followed by averaging and rearranging of the nearly symmetrical triangular halves into a compact square matrix representation. The experimental settings (in this case the magnetic field B0 and pulse delay time τ) are then concatenated onto the back of the HYSCORE spectrum. (B) Preprocessed HYSCORE spectra are passed through 3 convolutional layers with 16, 32, and 64 channels (kernel size = 3 × 3, stride = 2), and the output is flattened along the image dimensions. After passage through one fully connected layer of size 256, the network branches into 5 parallel fully connected layers each of size 64 for predicting the values of a, T, δ, K, and η. ReLU18 activations are applied to all layer outputs except for the five final classification layers, where the SoftPlus19 activation ln(1 + ex) is used instead. The full details of the spectral preprocessing, CNN architectural design, and network training are provided in the Supporting Information.
Figure 3.
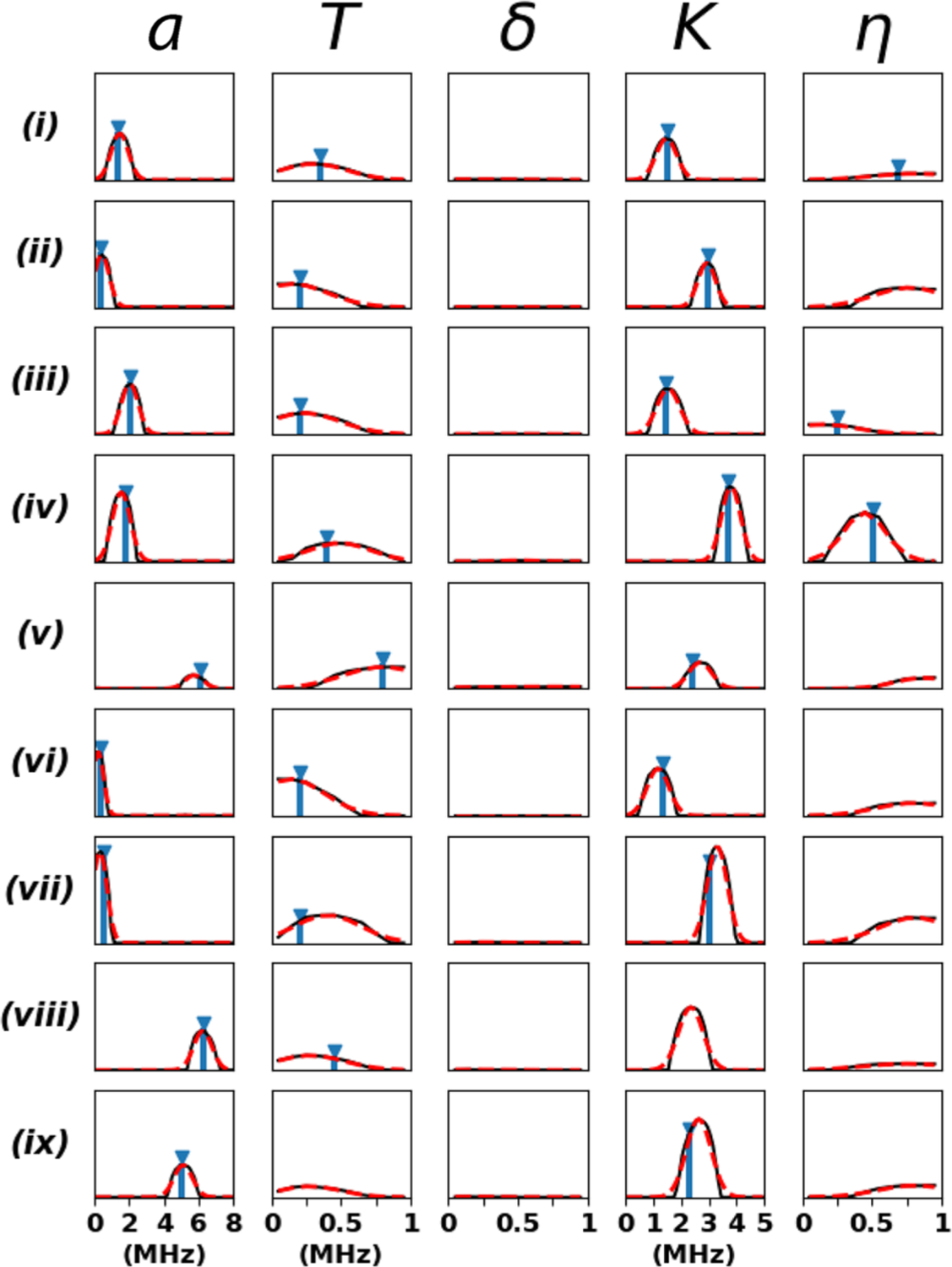
Comparison of the CNN probability distributions for a, T, δ, K, and η (black) with the previously reported literature values (blue) where available: (i) SQB RC (L-H190-Nδ) – semiquinone (SQ) in the QB ubiquinone binding site of the reaction center from Rhodobacter sphaeroides interacting with Nδ of residue L-H190;21 (ii) SQB RC (L-G225-Np) – same as previous for Np of L-G225;21 (iii) SQH cyt aa3 (H70-Nε) – SQ in the QH high-affinity menaquinone binding site of the R70H mutant of cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis interacting with Nε of residue H70;22 (iv) SQH cyt bo3 (R71-Nε) – semiquinone in the QH high-affinity ubiquinone binding site of the cytochrome bo3 ubiquinol oxidase from Escherichia coli interacting with Nε of residue R71;23 (v) [2Fe-2S] rat mitoNEET (H87-Nδ) – reduced [2Fe-2S](Cys)3(His)1 cluster in rat mitoNEET interacting with Nδ of residue H87;24 (vi) [2Fe-2S] rat mitoNEET (H87-Nε) – same as previous for Nε of residue H87;24 [2Fe-2S] rat mitoNEET (Np) – same as previous for Np in the cluster environment;24 (viii) VO2+ (Imidazole) – VO2+ ion interaction with coordinating imidazole ligand nitrogens;25 and (ix) VO2+ (Histidine-Nα)2 – VO2+ ion interaction with coordinating NαH2 of histidine ligands.25 The final predictions and their standard deviation errors are determined by a Gaussian fit to the probability distributions (dashed red). It took ∼3 s for the machine learning algorithm to process the time-domain patterns and generate this figure on a desktop equipped with an Intel Core i5–4570 processor (3.2 GHz) and 16 GB of RAM.
Figure 4.
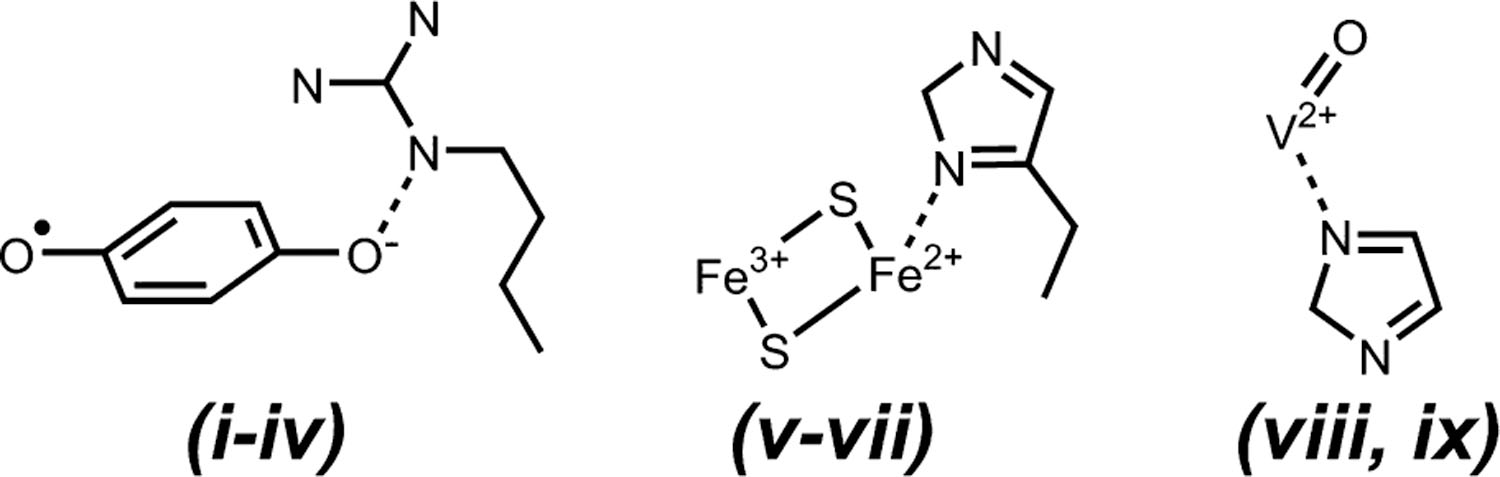
Chemical structures of the three major electron spin systems studied in this work (each shown in complex with an example of one of the 14N ligands from this Letter): (left) semiquinone hydrogen bonded to Nε of arginine, (middle) reduced [2Fe-2S] cluster in coordination with histidine, and (right) VO2+ ion coordinated to an imidazole. The spin systems in Figure 3 belonging to each class are listed at the bottom.
The predictions and their associated errors were determined by fitting a Gaussian to the CNN output probability distributions (Table S1 in the Supporting Information). The machine learning algorithm in Figure 3 utilizes the HYSCORE spectra along with the experimental settings B0 and τ to predict a and K to within an average deviation of 0.10 and 0.17 MHz from the manually assigned values, respectively. These errors approach the spectral frequency resolution of 0.1 MHz used in the HYSCORE preprocessing and analysis. Remarkably, the neural network also provides quantitative estimates for the hyperfine anisotropy T, for which no algorithm exists to directly extract this parameter from the spectrum. While T is best estimated from an analysis of 15N HYSCORE spectra because they lack contributions from the nuclear quadrupole coupling (all of the literature values of T in Table S1 are based on analyses of 15N-enriched samples), the machine learning algorithm is apparently able to extract this information directly from the natural abundance 14N spectrum. The error in the prediction for T is on average 0.08 MHz with respect to the manually assigned values. The nuclear quadrupole asymmetry parameter, η, can be determined only in certain cases, but this is understandable as the single-quantum transitions necessary to quantify this parameter appear in HYSCORE spectra only when the cancellation condition is satisfied.12,16,17 For the spin systems sufficiently near the cancellation condition (SQB RC L-H190-Nδ, SQH cyt aa3 H70-Nε, and SQH cyt bo3 R71-Nε),21–23 an average prediction error for η of 0.10 with respect to manual analysis is obtained. We note SQB RC L-H190-Nδ satisfies the cancellation condition poorly at X-band, and S-band measurements were required for an accurate determination of η.21 Despite this, the machine learning algorithm presented here is able to provide a reasonable estimate of η even without the low-frequency measurements. Finally, the CNN is not able to predict the hyperfine rhombicity parameter, δ. This is consistent with the previous inability to determine δ accurately for these spin systems because of insufficient spectral resolution. The failure of the machine learning algorithm to extract δ suggests the information required for specifying this parameter simply does not exist in these 14N HYSCORE spectra.
Comparison of neural network models trained with and without various combinations of B0 and τ shows that only B0 provides a small but significant improvement to the fit (Table 1). This suggests this neural network is able to infer the magnetic field (within the range of 300–400 mT used in the training set) directly from the spectrum alone. Indeed, when this CNN is repurposed to extract B0 from the spectrum instead of a, T, δ, K, and η, it does so with an average deviation of 20 mT from the experimental truth value. Therefore, the magnetic field of the spectrum is, to some degree of accuracy, encoded into the pattern of peaks and cross-ridges. Additional performance benchmarks of the neural network as a function of various simulations parameters are provided in the Supporting Information: prediction accuracy does not show any dependence on magnetic field (Figure S3) or τ-value (Figure S4) within the intervals the neural network was trained, performance degrades as the spectrum becomes dominated by diagonal peak intensity (Figure S5), and the machine learning algorithm exhibits robust performance for signal-to-noise ratios above ∼4 (Figure S6).
Table 1.
Comparison of CNN Model Prediction Errorsa
| model | Δa (MHz) | ΔT (MHz) | ΔK (MHz) | Δη |
|---|---|---|---|---|
| HYnetb | 0.13 (±0.02) | 0.09 (±0.04) | 0.26 (±0.04) | 0.14 (±0.04) |
| HYnet-B0c | 0.12 (±0.01) | 0.08 (±0.01) | 0.19 (±0.01) | 0.11 (±0.02) |
| HYnet-τd | 0.13 (±0.03) | 0.09 (±0.01) | 0.25 (±0.08) | 0.11 (±0.01) |
| HYnet-B0τe | 0.11 (±0.03) | 0.09 (±0.01) | 0.19 (±0.02) | 0.09 (±0.01) |
Prediction error is calculated as the absolute difference between the CNN output and the literature values. For each model, the prediction error was averaged over five training runs with the standard deviation in parentheses.
CNN model not given any information on the experimental settings at which the HYSCORE spectrum was measured.
Only the magnetic field of the experiment (B0) is provided to the model.
Only the τ-value of the experiment is provided to the model.
Both B0 and the τ-value are provided to the model.
Here we describe a CNN architecture capable of utilizing and interpreting the full information content of 2D EPR data and apply this approach for the first time to the analysis of 14N HYSCORE spectroscopy. The algorithm is trained only on spectral simulations and is robust to variations in diagonal peak intensity, noise, τ-suppression, and variations in t1 and t2 step size. However, the training set includes only narrow-line radicals with a maximum g-tensor span of 0.1 and no resolvable splittings in the EPR spectrum. As such, the CNN exhibits instability for cases of complex EPR spectra, such as the VO2+ spin system (strong hyperfine interactions with the I = 7/2 vanadium, Figure S7) and to a lesser degree the rat mitoNEET spin system (g-tensor span of 0.11 with small splittings in the EPR spectrum, Figure S8). Accounting for these complex patterns in the EPR spectrum is outside the scope of the present work and is currently not supported in the EasySpin simulation package.20 However, the SPINACH package can model these features and may help generalize the neural network to handle arbitrary EPR spectral patterns.26 An additional limitation of the current approach is that it can only analyze spectra corresponding to a single 14N nucleus, and therefore, manual intervention is required for analyzing multinuclear spectra. The solution to this problem will likely involve semantic image segmentation, unsupervised clustering, or a combination of the two for automatically separating the spectrum into its different components before being fed into the neural network. Despite the limitations of the present algorithm, it can provide a useful starting point for setting up HYSCORE simulations bounded by confidence intervals that are bias free (Figure S9). The code is provided as an open source package and is available online to download (https://griffingroup.mit.edu/EPR_Spectroscopy).
Thus, the approach outlined here proposes an alternative paradigm to the typical trial-and-error simulation process for interpreting multidimensional spectra. Machine learning has demonstrated the incredible ability to extract the magnetic coupling parameters of interest directly from the multidimensional spectra by treating it as an image classification problem. In the case of HYSCORE spectroscopy, the CNN derives an entirely new algorithm for estimating the hyperfine anisotropy parameter T directly from the spectrum. Importantly, the neural network extracts the full underlying probability distributions of each parameter for the first time, allowing for error estimates free from bias. Future efforts will be applied toward adapting these neural network architectures to the three- and four-dimensional spectra often encountered in NMR, where automation of structure calculations directly from the raw spectra may eventually become a reality.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the National Institutes of Biomedical Imaging and Bioengineering (EB002804, EB001960, and EB002026) to R.G.G.; a grant from the Chemical Sciences, Geosciences and Biosciences Division of the Office of Basic Energy Sciences at the U.S. Department of Energy (DE-FG02–08ER15960, for pulsed EPR work) to S.A.D.; an NIH F32 Fellowship (GM 123596) to A.T.T.; and an NSF Graduate Research Fellowship (#112237) and the Martin Family Society of Fellows for Sustainability to E.D.E.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jp-clett.8b03797.
Full description of the training set simulation and preprocessing and the neural network architecture, training, and design considerations (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Krizhevsky A; Sutskever I; Hinton GE ImageNet classification with deep convolutional neural networks. Proceedings of the 25th International Conference on Neural Information Processing Systems 2012, 1, 1097–1105. [Google Scholar]
- (2).Szegedy C; Liu W; Jia Y; Sermanet P; Reed S; Anguelov D; Erhan D; Vanhoucke V; Rabinovich A Going Deeper with Convolutions. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR);2015.
- (3).He K; Zhang X; Ren S; Sun J Deep Residual Learning for Image Recognition. arXiv:1512.03385 2015.
- (4).Esteva A; Kuprel B; Novoa RA; Ko J; Swetter SM; Blau HM; Thrun S Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kermany DS; Goldbaum M; Cai W; Valentim CCS; Liang H; Baxter SL; McKeown A; Yang G; Wu X; Yan F; Dong J; Prasadha MK; Pei J; Ting M; Zhu J; Li C; Hewett S; Dong J; Ziyar I; Shi A; Zhang R; Zheng L; Hou R; Shi W; Fu X; Duan Y; Huu VAN; Wen C; Zhang ED; Zhang CL; Li O; Wang X; Singer MA; Sun X; Xu J; Tafreshi A; Lewis MA; Xia H; Zhang K Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131. [DOI] [PubMed] [Google Scholar]
- (6).Liu J; Osadchy M; Ashton L; Foster M; Solomon CJ; Gibson SJ Deep convolutional neural networks for Raman spectrum recognition: a unified solution. Analyst 2017, 142, 4067–4074. [DOI] [PubMed] [Google Scholar]
- (7).Klukowski P; Augoff M; Zieba M; Drwal M; Gonczarek A; Walczak MJ NMRNet: A deep learning approach to automated peak picking of protein NMR spectra. Bioinformatics 2018, 34, 2590–2597. [DOI] [PubMed] [Google Scholar]
- (8).Klukowski P; Walczak MJ; Gonczarek A; Boudet J; Wider G Computer vision-based automated peak picking applied to protein NMR spectra. Bioinformatics 2015, 31, 2981–2988. [DOI] [PubMed] [Google Scholar]
- (9).Höfer P; Grupp A; Nebenführ H; Mehring M Hyperfine sublevel correlation (HYSCORE) spectroscopy: A 2D ESR investigation of the squaric acid radical. Chem. Phys. Lett 1986, 132, 279–282. [Google Scholar]
- (10).Dikanov SA; Bowman MK Cross-Peak Lineshape of Two-Dimensional ESEEM Spectra in Disordered S = 1/2, I = 1/2 Spin Systems. J. Magn. Reson., Ser. A 1995, 116, 125–128. [Google Scholar]
- (11).Dikanov SA; Tyryshkin AM; Bowman MK Intensity of Cross-Peaks in HYSCORE Spectra of S = 1/2, I = 1/2 Spin Systems. J. Magn. Reson 2000, 144, 228–242. [DOI] [PubMed] [Google Scholar]
- (12).Dikanov SA; Xun L; Karpiel AB; Tyryshkin AM; Bowman MK Orientationally-Selected Two-Dimensional ESEEM Spectroscopy of the Rieske-Type Iron–Sulfur Cluster in 2,4,5-Trichlorophenoxyacetate Monooxygenase from Burkholderia cepacia AC1100. J. Am. Chem. Soc 1996, 118, 8408–8416. [Google Scholar]
- (13).Schweiger A; Jeschke G Principles of Pulse Electron Paramagnetic Resonance; Oxford University Press: Oxford, U.K., 2001. [Google Scholar]
- (14).Gemperle C; Aebli G; Schweiger A; Ernst RR Phase cycling in pulse EPR. J. Magn. Reson 1990, 88, 241–256. [Google Scholar]
- (15).Worswick SG; Spencer JA; Jeschke G; Kuprov I Deep neural network processing of DEER data. Sci. Adv 2018, 4, No. eaat5218. [DOI] [PMC free article] [PubMed]
- (16).Dikanov SA; Tsvetkov YD; Bowman MK; Astashkin AV Parameters of quadrupole coupling of 14N nuclei in chlorophyll a cations determined by the electron spin echo method. Chem. Phys. Lett 1982, 90, 149–153. [Google Scholar]
- (17).Flanagan HL; Singel DJ Analysis of 14N ESEEM patterns of randomly oriented solids. J. Chem. Phys 1987, 87, 5606–5616. [Google Scholar]
- (18).Nair V; Hinton GE Rectified Linear Units Improve Restricted Boltzmann Machines. In 27th International Conference on Machine Learning, 2010; pp 807–814.
- (19).Dugas C; Bengio Y; Beĺisle F; Nadeau C; Garcia R Incorporating Second-Order Functional Knowledge for Better Option Pricing. 13th International Conference on Neural Information Processing Systems, 2000.
- (20).Stoll S; Schweiger A EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson 2006, 178, 42–55. [DOI] [PubMed] [Google Scholar]
- (21).Taguchi AT; O’Malley PJ; Wraight CA; Dikanov SA Nuclear hyperfine and quadrupole tensor characterization of the nitrogen hydrogen bond donors to the semiquinone of the QB site in bacterial reaction centers: a combined X- and S-band 14,15N ESEEM and DFT study. J. Phys. Chem. B 2014, 118, 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yi SM; Taguchi AT; Samoilova RI; O’Malley PJ; Gennis RB; Dikanov SA Plasticity in the High Affinity Menaquinone Binding Site of the Cytochrome aa3–600 Menaquinol Oxidase from Bacillus subtilis. Biochemistry 2015, 54, 5030–5044. [DOI] [PubMed] [Google Scholar]
- (23).Lin MT; Baldansuren A; Hart R; Samoilova RI; Narasimhulu KV; Yap LL; Choi SK; O’Malley PJ; Gennis RB; Dikanov SA Interactions of intermediate semiquinone with surrounding protein residues at the QH site of wild-type and D75H mutant cytochrome bo3 from Escherichia coli. Biochemistry 2012, 51, 3827–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Iwasaki T; Samoilova RI; Kounosu A; Ohmori D; Dikanov SA Continuous-wave and pulsed EPR characterization of the [2Fe-2S](Cys)3(His)1 cluster in rat MitoNEET. J. Am. Chem. Soc 2009, 131, 13659–13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dikanov SA; Samoilova RI; Smieja JA; Bowman MK Two-Dimensional ESEEM Study of VO2+ Complexes with Imidazole and Histidine: Histidine Is a Polydentante Ligand. J. Am. Chem. Soc 1995, 117, 10579–10580. [Google Scholar]
- (26).Hogben HJ; Krzystyniak M; Charnock GT; Hore PJ; Kuprov I Spinach–a software library for simulation of spin dynamics in large spin systems. J. Magn. Reson 2011, 208, 179–194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


