ABSTRACT
Fanconi anemia (FA) is a rare genetic disease characterized by increased risk for bone marrow failure and cancer. The FA proteins function together to repair damaged DNA. A central step in the activation of the FA pathway is the monoubiquitination of the FANCD2 and FANCI proteins, which occurs upon exposure to DNA-damaging agents and during the S phase of the cell cycle. The regulatory mechanisms governing S-phase monoubiquitination, in particular, are poorly understood. In this study, we have identified a cyclin-dependent kinase (CDK) regulatory phosphosite (S592) proximal to the site of FANCD2 monoubiquitination. FANCD2 S592 phosphorylation was detected by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and by immunoblotting with an S592 phospho-specific antibody. Mutation of S592 leads to abrogated monoubiquitination of FANCD2 during the S phase. Furthermore, FA-D2 (FANCD2−/−) patient cells expressing S592 mutants display reduced proliferation under conditions of replication stress and increased mitotic aberrations, including micronuclei and multinucleated cells. Our findings describe a novel cell cycle-specific regulatory mechanism for the FANCD2 protein that promotes mitotic fidelity.
KEYWORDS: Fanconi anemia, FANCD2, CDK phosphorylation, cell cycle, chromosome stability, ubiquitination, cell cycle, ubiquitin
INTRODUCTION
Protection of the integrity of our genome depends on the concerted activities of several DNA repair pathways that ensure the timely and accurate repair of damaged DNA. These DNA repair pathways must be tightly regulated upon cellular exposure to exogenous DNA-damaging agents, as well as during the cell cycle. Somatic disruption of the DNA damage response leads to mutation, genome instability, and cancer. In addition, germline mutations in DNA repair genes are associated with hereditary diseases characterized by increased cancer risk and other clinical manifestations, examples of which include Fanconi anemia (FA) and xeroderma pigmentosum (XP). FA is a rare genetic disease characterized by congenital abnormalities, increased risk for bone marrow failure and cancer, and accelerated aging (1). FA is caused by germline mutations in any 1 of 23 genes. The FA proteins function together in a pathway to repair damaged DNA and to maintain genome stability. A major role for the FA pathway in the repair of DNA interstrand cross-links (ICLs) has been described (2).
The main activating step of the FA pathway is the monoubiquitination of the FANCD2-FANCI heterodimer (ID2), which is catalyzed by the FA core complex, comprising FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, UBE2T/FANCT, and the FA-associated proteins. Monoubiquitination occurs following exposure to DNA-damaging agents and during the S phase of the cell cycle (3, 4). Recently, it was discovered that the monoubiquitination of FANCD2 and FANCI leads to the formation of a closed ID2 clamp that encircles DNA (5, 6). Moreover, ubiquitinated ID2 (ID2-Ub) assembles into nucleoprotein filament arrays on double-stranded DNA (7). The monoubiquitination of FANCD2 is necessary for efficient ICL repair, the maintenance of common fragile site stability, and faithful chromosome segregation, events that are all crucial for genomic stability (8, 9).
A DNA damage-independent role for the FA pathway during the cell cycle has been implicated in several studies. For example, FANCD2 promotes replication fork protection during the S phase and ensures the timely and faithful replication of common chromosome fragile sites (CFSs) (8, 10, 11). FANCD2 has also been shown to be involved in mitotic DNA synthesis (MiDAS) during prophase and is present on the terminals of anaphase ultrafine bridges (12, 13). During the cell cycle, FANCD2 monoubiquitination is maximal during the S phase and minimal during the M phase (4). FANCD2 and FANCI are phosphorylated by the ATR and ATM kinases following exposure to DNA-damaging agents, promoting their monoubiquitination (14–17). However, in general, the function and regulation of FANCD2 and FANCI during the cell cycle remain poorly understood.
Cyclin-dependent kinases (CDKs) play a major role in regulating cell cycle progression, with CDK-mediated hyperphosphorylation of the retinoblastoma protein (pRb) being the primary mechanism of cell cycle regulation. CDKs are also known to regulate DNA repair during the cell cycle. Several protein components of the homologous recombination repair (HR), translesion DNA synthesis (TLS), nonhomologous end joining (NHEJ), and telomere maintenance pathways are CDK substrates (18). For example, the BRCA2 protein is phosphorylated by CDK1 as cells progress toward mitosis. CDK-mediated phosphorylation of BRCA2 blocks interaction with RAD51 and thereby restricts HR DNA repair during the M phase (19). Conversely, CDK1/2 phosphorylate the EXO1 nuclease to promote DNA strand resection and HR repair during the S phase (20).
In this study, we have examined the regulation of the FANCD2 protein by phosphorylation, specifically during the cell cycle. We show that FANCD2 is phosphorylated on S592, a putative CDK site, during the S phase but not during the M phase and is phosphorylated by CDK2 in vitro. Mutation of S592, as well as CDK inhibition, disrupts S-phase FANCD2 monoubiquitination. Furthermore, we demonstrate that FA-D2 (FANCD2−/−) patient cells stably expressing FANCD2 mutated at S592 display reduced growth under conditions of replication stress, an altered cell cycle profile, and increased genomic instability manifested as increased micronuclei, nucleoplasmic bridges, and aneuploidy. Our results uncover important mechanistic insight into the regulation of a key DNA repair/genome maintenance pathway under unperturbed cellular conditions.
RESULTS
FANCD2 is phosphorylated under unperturbed conditions and during the S phase of the cell cycle.
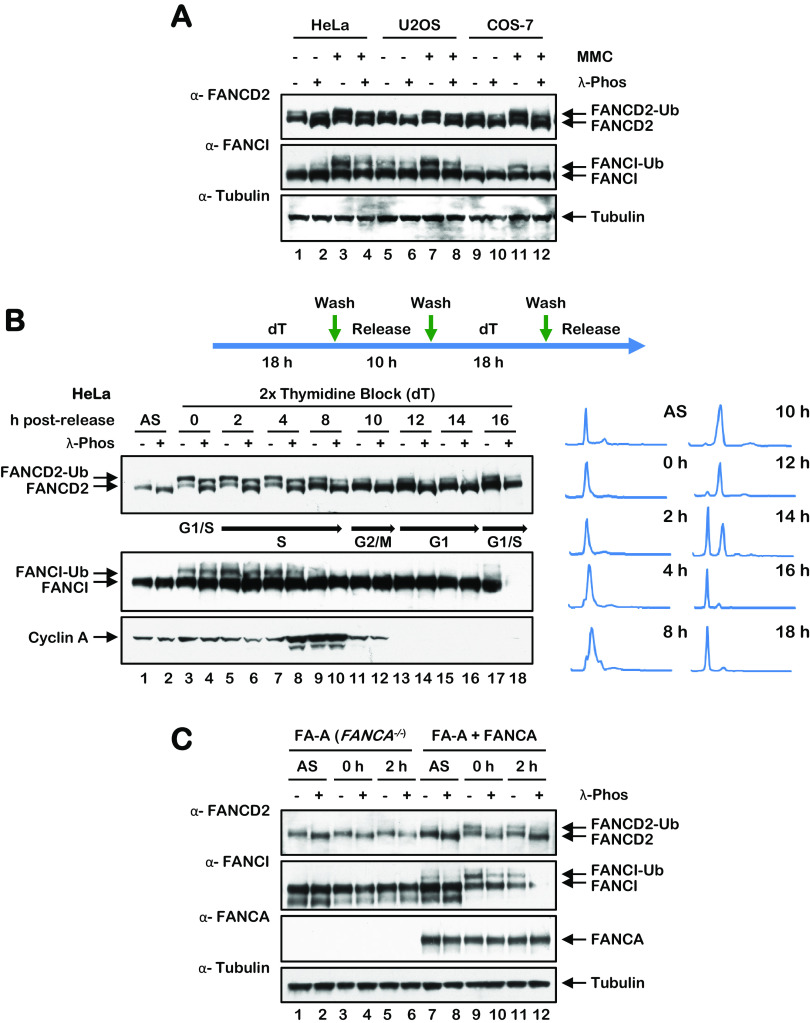
To begin to study the phosphorylation of the FANCD2 protein in the absence and presence of DNA-damaging agents, we incubated cells in the absence or presence of the DNA cross-linking agent mitomycin C (MMC), treated whole-cell lysates with or without lambda phosphatase, and analyzed FANCD2 via immunoblotting. Notably, we observed a large increase in FANCD2 mobility following incubation of lysates with lambda phosphatase, even in the absence of MMC (Fig. 1A). These results suggest that FANCD2 is subject to extensive phosphorylation even in the absence of exogenous DNA damage. A similar change in protein mobility was not observed for FANCI (Fig. 1A). To determine if FANCD2 is subject to phosphorylation during the cell cycle, we arrested cells in the early S phase using double-thymidine synchronization and analyzed phosphorylation at regular time points following release. We observed maximal phosphorylation of FANCD2 during the S phase of the cell cycle, with less phosphorylation observed as cells progressed through G2/M and G1 phases of the cell cycle (Fig. 1B). Again, we observed no appreciable change in FANCI mobility under the same conditions, suggesting that FANCI is not subject to the same levels of phosphorylation as FANCD2 during the cell cycle (Fig. 1B). Similar findings were observed with U2OS cells as well as following synchronization with nocodazole (results not shown). The FANCA protein is a central component of the FA core complex, a multisubunit ubiquitin ligase that catalyzes the monoubiquitination of FANCD2 (3, 21). To determine if FANCD2 phosphorylation was dependent on its monoubiquitination, we performed a lambda phosphatase assay with asynchronous and early S-phase-synchronized FA-A (FANCA−/−) and FANCA-complemented FA-A cells. S-phase FANCD2 phosphorylation was observed in the absence of FANCA, albeit to a slightly lesser extent than in FANCA-complemented FA-A cells (Fig. 1C). These results suggest that S-phase phosphorylation is coupled to, but not dependent upon, the monoubiquitination of FANCD2.
FIG 1.
FANCD2 is phosphorylated under nonstressed conditions and during the S phase of the cell cycle. (A) HeLa, U2OS, and COS-7 cells were incubated with or without 200 nM MMC for 24 h. Cells were harvested and lysed in lambda phosphatase lysis buffer and incubated in the absence or presence of lambda phosphatase and proteins analyzed by immunoblotting. (B) HeLa cells were synchronized at the G1/S boundary by a double-thymidine block and released into thymidine-free media. Cells were lysed in lambda phosphatase buffer and incubated in the absence or presence of lambda phosphatase and lysates analyzed by immunoblotting. For cell cycle stage analysis, cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. (C) FA-A (FANCA−/−) and FANCA-complemented FA-A cells were synchronized at the G1/S boundary by a double-thymidine block and released into thymidine-free media. Cells were harvested, lysed in lambda phosphatase buffer, incubated in the presence or absence of lambda phosphatase, and analyzed by immunoblotting.
FANCD2 is a CDK substrate.
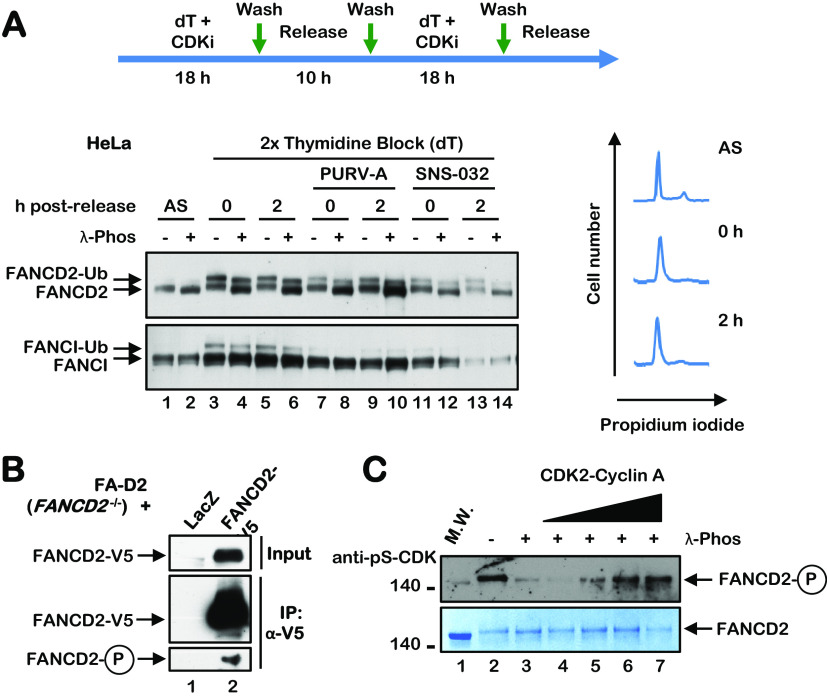
Phosphorylation of FANCD2 during the cell cycle suggested a role for cyclin-dependent kinases (CDKs) in the regulation of FANCD2. CDKs phosphorylate serine or threonine residues in the consensus sequence [S/T*]PX[K/R], and several DNA repair proteins are known CDK substrates, including BRCA2, FANCJ, and CtIP (19, 22, 23). Both FANCD2 and FANCI have multiple putative CDK phosphorylation sites with various degrees of conservation. To begin to assess the role of CDKs in the regulation of FANCD2, we performed a double-thymidine block in the absence and presence of the CDK inhibitors purvalanol A and SNS-032. Treatment with both inhibitors resulted in a significant reduction in the levels of FANCD2 and FANCI monoubiquitination upon double-thymidine arrest and a modest reduction in the FANCD2 phosphorylation mobility shift (Fig. 2A). These results suggest that CDK phosphorylates FANCD2 during the cell cycle but also indicate that CDK is not the sole cell cycle FANCD2 kinase. To determine if FANCD2 is a CDK substrate, we immunoprecipitated FANCD2 from FA-D2 (FANCD2−/−) patient cells stably expressing V5-tagged LacZ or FANCD2, and probed immune complexes with a pan anti-pS/T-CDK antibody. An immune-reactive band was detected in immune complexes from cells expressing FANCD2 and not LacZ (Fig. 2B), suggesting that FANCD2 is a CDK substrate. We also performed an in vitro CDK kinase assay with CDK2-cyclin A and full-length FANCD2 purified from High Five insect cells (24). CDK-mediated FANCD2 phosphorylation was already evident upon purification from insect cells (Fig. 2C). Following dephosphorylation with lambda phosphatase, we observed a concentration-dependent increase in FANCD2 phosphorylation upon incubation with CDK2-cyclin A (Fig. 2C). These results indicate that FANCD2 is a CDK substrate and suggest that FANCD2 monoubiquitination may be coupled to CDK phosphorylation.
FIG 2.
FANCD2 is a CDK substrate. (A) HeLa cells were synchronized at the G1/S boundary by a double-thymidine block performed in the absence or presence of the CDK inhibitors purvalanol A (PURV-A) or SNS-032. Cells were lysed in lambda phosphatase buffer, incubated in the absence or presence of lambda phosphatase, and analyzed by immunoblotting. Cells were also fixed, stained with propidium iodide, and analyzed by flow cytometry to determine cell cycle stage. (B) FA-D2 (FANCD2−/−) cells stably expressing LacZ-V5 or FANCD2-V5 were immunoprecipitated with anti-V5 agarose, and immune complexes were immunoblotted with anti-FANCD2 and pan-anti-pS/T-CDK antibodies. (C) Purified FANCD2 was incubated in the absence (−) or presence (+) of lambda phosphatase followed by incubation with increasing concentrations of CDK2-cyclin A and ATP at 30°C for 30 min. Samples were immunoblotted with a pan anti-pS-CDK antibody or stained with SimplyBlue SafeStain.
FANCD2 is phosphorylated on S592 during the S phase of the cell cycle.
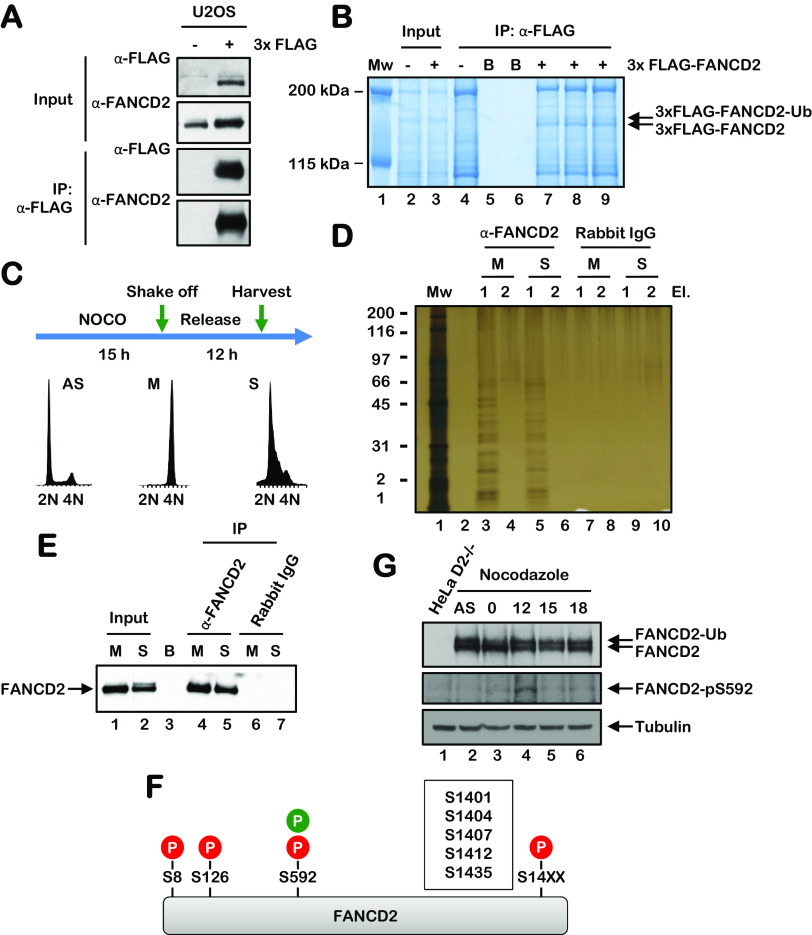
To map the sites of FANCD2 phosphorylation, we immunoprecipitated FANCD2 from asynchronous U2OS cells stably expressing 3×FLAG-FANCD2 under stringent conditions, and we performed phosphoproteomic analysis using liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 3A and B). Under these conditions, we observed the phosphorylation of multiple FANCD2 sites, including the previously detected ATM/ATR phosphorylation sites S1401 and S1404 (Table 1) (17). We also detected phosphorylation of the putative CDK site S592 (Table 1). To analyze the phosphorylation of FANCD2 during the cell cycle, we synchronized HeLa cells in the M phase using nocodazole, immunoprecipitated FANCD2 immediately upon release and at 15 h postrelease, and performed phosphoproteomic analysis using LC-MS/MS (Fig. 3C to F). Under these conditions, we also detected phosphorylation of FANCD2 on S592, specifically during the S phase and not during the M phase. To study FANCD2 S592 phosphorylation more closely, we generated an S592 phospho-specific antibody. We arrested cells in the M phase using nocodazole and analyzed FANCD2 S592 phosphorylation upon release from nocodazole block. A FANCD2 pS592 immunoreactive band was observed at 12 h (S phase) following release (Fig. 3G), consistent with the results of our LC-MS/MS phosphoproteomic analysis. Taken together, our results demonstrate that FANCD2 is phosphorylated on S592 during the S phase of the cell cycle and suggest that S592 phosphorylation might play an important regulatory function during the S phase (Fig. 3F).
FIG 3.
FANCD2 is phosphorylated on S592 during S-phase. (A and B) FANCD2 was immunoprecipitated from U2OS cells stably expressing 3×FLAG-FANCD2 under stringent conditions using anti-FLAG–agarose. Immune complexes were analyzed by immunoblotting using anti-FLAG and anti-FANCD2 antibodies (A) and by staining with SimplyBlue SafeStain (B). Immunoprecipitated FANCD2 bands were combined and subjected to phosphoproteomic analysis using LC-MS/MS. (C) HeLa cells were synchronized in the M phase by nocodazole block. Mitotic cells were harvested or released into nocodazole-free medium for 12 h (S phase) prior to harvesting. (D and E) FANCD2 was immunoprecipitated from M-phase- and S-phase-synchronized cells using an anti-FANCD2 antibody and visualized using silver staining (D) or by immunoblotting (E). (F) Schematic of the FANCD2 protein indicating the various phosphorylation sites identified by LC-MS/MS in this study; red represents sites identified in asynchronous U2OS cells, and green represents sites identified in synchronous HeLa cells. (G) HeLa cells were synchronized in the M phase by nocodazole block. Samples were harvested and analyzed by immunoblotting using an anti-FANCD2 antibody and an anti-FANCD2 pS592 phospho-specific antibody.
TABLE 1.
Immunoprecipitation mass spectrometry analysis of FANCD2
| Protein | Peptide sequence | P site(s) | Putative kinase(s)a | Previously detected (reference no.) |
|---|---|---|---|---|
| FANCD2 | R.LS*KSEDKESLTEDASK.T | S8 | PKACa | Yes (41, 42) |
| FANCD2 | R.LQDEEAS*M#GASYSK.S | S126 | RSK2 | No |
| FANCD2 | R.SES*PSLTQER.A | S592 | P38/MAPK, CDK1, CDK2 | Yes (41, 43) |
| FANCD2 | R.DLQGEEIKS*QNSQESTADES*EDDM#SSQASK.S | S1401, S1412 | ATR, DNA-PK, BARK1 | Yes (17, 43, 44) |
| FANCD2 | K.SQNS*QESTADESEDDM#SSQASK.S | S1404 | ATR, DNA-PK | Yes (17) |
| FANCD2 | K.SQNS*QESTADES*EDDM#SSQASK.S | S1404, S1412 | ATR, DNA-PK, BARK1 | Yes (17, 43, 44) |
| FANCD2 | K.SQNSQES*TADES*EDDM#SSQASK.S | S1407, S1412 | CK1a1, BARK1 | Yes (43, 45) |
| FANCD2 | K.SKATEDGEEDEVS*AGEK.E | S1435 | CKII, CKI | Yes (43) |
| FANCD2 | K.ATEDGEEDEVS*AGEK.E | S1435 | CKII, CKI | Yes (43) |
| FANCD2 | R.SES*PSLTQER.A | S592 | P38/MAPK, CDK1, CDK2 | Yes (41, 43) |
MAPK, mitogen-activated protein kinase.
Mutation of S592 disrupts S-phase FANCD2 monoubiquitination.
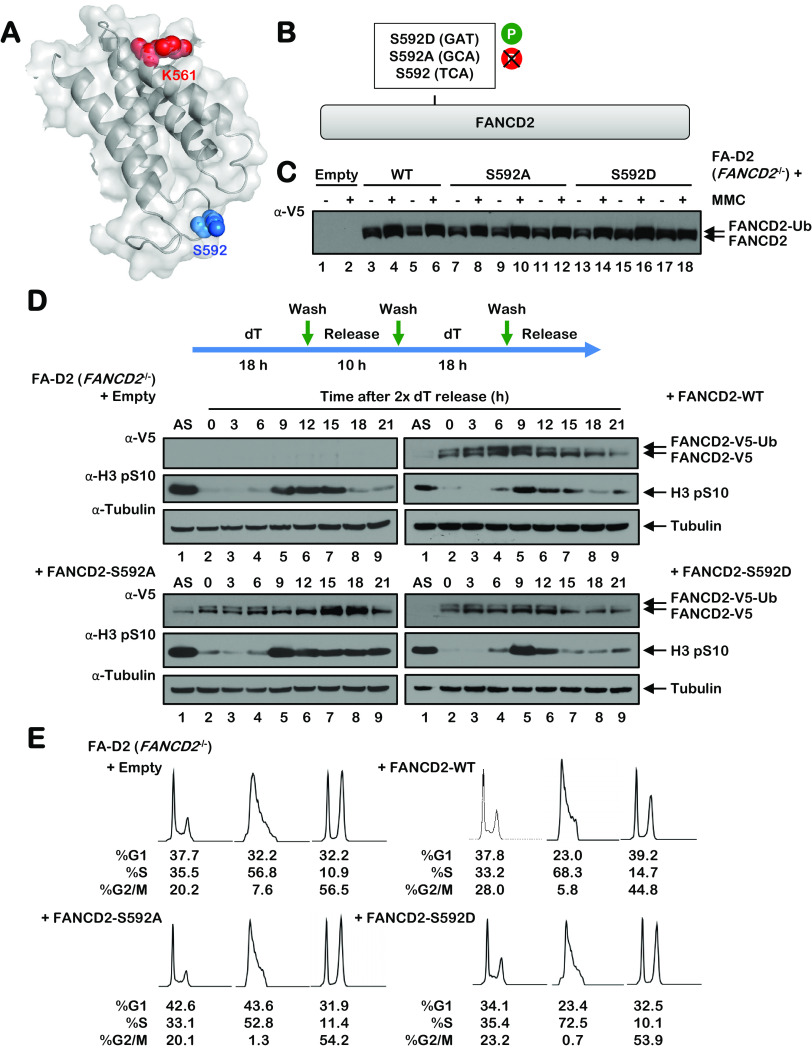
FANCD2 S592 is predicted to localize to a flexible loop proximal to K561, the site of monoubiquitination, suggesting a potential regulatory function for S592 phosphorylation (Fig. 4A). To begin to characterize the role of S592 phosphorylation, we generated phospho-dead (S592A) and phospho-mimetic (S592D) variants and stably expressed these in FA-D2 (FANCD2−/−) patient-derived cells (Fig. 4B and C). Mutation of S592 did not adversely impact the stability or overall structure of FANCD2, as both variants were expressed at comparable levels to wild-type FANCD2 and were subject to similar levels of monoubiquitination following MMC exposure (Fig. 4C). Previous studies have shown that FANCD2 undergoes monoubiquitination as cells progress through the S phase of the cell cycle (4). To analyze the effects of S592 mutation on S-phase FANCD2 monoubiquitination, cells were subject to a double-thymidine block, and FANCD2 monoubiquitination was analyzed upon release. Consistent with previous studies, we observed an increase in the monoubiquitination of wild-type FANCD2 as cells progressed through the S phase (Fig. 4D). In contrast, S-phase monoubiquitination was markedly attenuated for both FANCD2-S592A and FANCD2-S592D (Fig. 4D). We also analyzed levels of the mitotic marker H3 pS10 in these cells. Compared to cells expressing wild-type FANCD2, we observed persistent levels of H3 pS10 in FA-D2 cells expressing empty vector and the FANCD2-S592A mutant (Fig. 4D). We observed a similar phenotype of elevated H3 pS10 in HeLa FANCD2−/− cells generated by CRISPR-Cas9 gene editing (results not shown) (25). In contrast to cells expressing empty vector and the FANCD2-S592A mutant, we observed a more rapid disappearance of H3 pS10 in cells expressing FANCD2-S592D (Fig. 4D). Furthermore, fluorescence-activated cell sorter (FACS) analysis of FA-D2 patient cells expressing empty vector and the S592 variants upon release from double-thymidine block indicated a higher percentage of cells in G2/M at earlier time points than cells expressing FANCD2 wild type (FANCD2-WT) (Fig. 4E). Taken together, these results demonstrate that mutation of FANCD2 S592 disrupts S-phase monoubiquitination and leads to aberrant G2-M cell cycle progression.
FIG 4.
Mutation of S592 disrupts FANCD2 monoubiquitination during the S phase and following exposure to DNA-damaging agents. (A) Shown is a partial model of the human FANCD2 protein, modeled on the mouse Fancd2 structure (PDB ID 3S4W), illustrating the site of monoubiquitination K561 in red and S592 in blue. (B) Site-directed mutagenesis was used to generate phospho-dead (S592A) and phospho-mimetic (S592D) FANCD2 S592 variants. (C) Polyclonal populations of FA-D2 (FANCD2−/−) patient cells expressing empty vector or V5-tagged FANCD2-WT, FANCD2-S592A, or FANCD2-S592D were incubated in the absence (−) or presence (+) of 200 nM mitomycin C (MMC) for 24 h, and whole-cell lysates were analyzed by immunoblotting. (D) FA-D2 cells stably expressing empty vector or V5-tagged FANCD2-WT, FANCD2-S592A, or FANCD2-S592D were synchronized at the G1/S boundary by double-thymidine block. Cells were released into thymidine-free medium, and whole-cell lysates were analyzed by immunoblotting at the indicated times postrelease. (E) FA-D2 cells stably expressing empty vector or V5-tagged FANCD2-WT, FANCD2-S592A, or FANCD2-S592D were synchronized at the G1/S boundary by double-thymidine block (2× deoxyribosylthymine [dT]) and then released into thymidine-free medium. At the indicated time points, cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. Cell cycle stage analysis was performed using FlowJo v10.2 software.
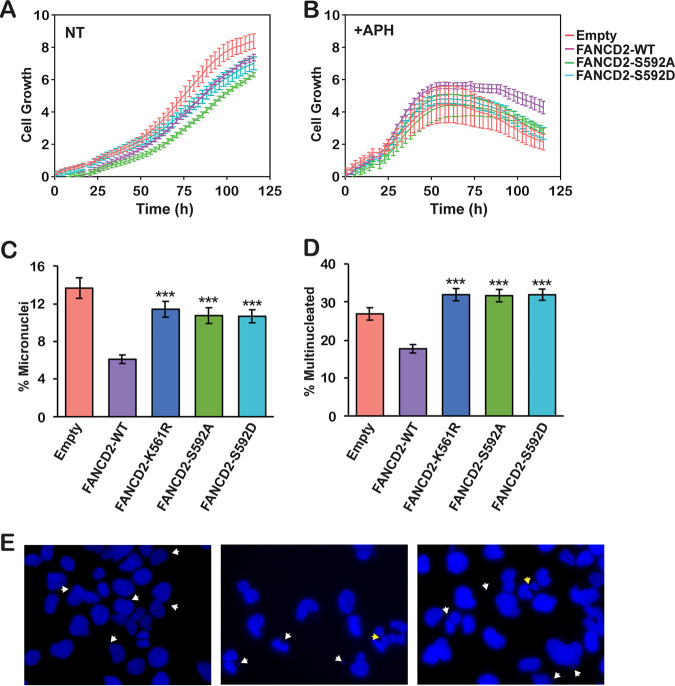
Mutation of FANCD2 S592 leads to decreased proliferative capacity and increased mitotic defects.
To assess the functional impacts of mutation of FANCD2 S592, we monitored the proliferation of FA-D2 patient cells stably expressing FANCD2-WT and the S592 variants in the absence or presence of low concentrations of the DNA polymerase inhibitor aphidicolin (APH) for prolonged periods using the xCELLigence real-time cell analysis system. In the absence of APH, cells expressing FANCD2-S592D displayed similar levels of proliferation to cells expressing wild-type FANCD2, while cells expressing FANCD2-S592A exhibited decreased proliferation compared to all other cells (Fig. 5A). When cultured in the presence of APH, cells expressing empty vector and the S592 variants exhibited decreased proliferation compared to cells expressing wild-type FANCD2 (P < 0.05 for all time points from 90 to 120 h) (Fig. 5B). In contrast, mutation of FANCD2 S592 had no discernible impact on growth in the presence of low concentrations of mitomycin C (MMC) (results not shown). Next, we examined the effects of mutation of S592 on endogenous levels of mitotic aberrations. Compared to FA-D2 cells expressing wild-type FANCD2, we observed increased endogenous levels of micronuclei and bi- and multinucleated cells in FA-D2 cells expressing the S592 variants (Fig. 5C and D). Elevated levels of mitotic aberrations were also observed in FA-D2 cells expressing FANCD2-K561R, which cannot be monoubiquitinated (Fig. 5C and D). Taken together, these results indicate that S592 phosphorylation is functionally necessary to ensure mitotic fidelity and chromosome stability under nonstressed conditions.
FIG 5.
Mutation of FANCD2 S592 leads to decreased proliferation under conditions of replicative stress and increased mitotic defects. (A and B) FA-D2 cells stably expressing empty vector or V5-tagged FANCD2-WT, FANCD2-S592A, or FANCD2-S592D were incubated in the absence (NT) (A) or presence of 0.4 μM aphidicolin (+APH) (B). Cellular proliferation was monitored by measuring electrical impedance every 15 min over a 120-h period using the xCELLigence real-time cell analysis system. Error bars represent the standard deviations of quadruplicate measurements, and this experiment was repeated three times with similar results. P < 0.05 between cells expressing wild-type FANCD2 and those expressing empty vector or the S592 variants for all time points between 90 and 120 h. (C and D) FA-D2 cells stably expressing empty vector or V5-tagged FANCD2-WT, FANCD2-K561R, FANCD2-S592A, or FANCD2-S592D were incubated in the absence of any DNA-damaging agents and mitotic aberrations, including micronuclei (C) and multinucleated (>2) cells (D), were scored. At least 400 cells per group were scored, and the combined results from two independent experiments are shown. ***, P < 0.001. (E) Representative images of micronuclei, nucleoplasmic bridges, and multinucleated cells, indicated by white arrows, from FA-D2 cells expressing FANCD2-S592A or FANCD2-S592D.
DISCUSSION
In this study, we have investigated the posttranslational regulation of the central FA pathway protein FANCD2 largely under unperturbed conditions. The majority of studies to date have focused on the posttranslational regulation of FANCD2 and FANCI following exposure to DNA-damaging agents (15, 16, 26). Here, we have observed that FANCD2 undergoes extensive phosphorylation during the cell cycle and, in particular, during the S phase of the cell cycle. Notably, in contrast, FANCI—a FANCD2 paralog—does not appear to be subject to the same degree of phosphorylation during the cell cycle. We have also determined that FANCD2 is phosphorylated on S592 by CDK2-cyclin A during the S phase using several approaches, including phosphoproteomic analysis of FANCD2 immune complexes from synchronized cell populations and immunoblotting with an S592 phospho-specific antibody. FANCD2 S592 is contained within a weak CDK consensus sequence, S/T-P, as opposed to the strong S/T-P-X-R/K consensus sequence, similar to that of other DNA repair proteins, including BRCA2, CtIP, and EXO1 (19, 20, 22). However, S592 is one of several putative CDK sites located proximal to FANCD2 K561, consistent with the known clustering of CDK sites in CDK substrates (27). Previous studies have shown that FANCD2 is monoubiquitinated as cells traverse the S phase, and this has been shown to contribute to the protection of stalled replication forks from degradation (4, 11). Here, we show that mutation of S592, or CDK inhibition, abrogates S-phase monoubiquitination, strongly suggesting that S592 phosphorylation primes FANCD2 for ubiquitination during the S phase. Recent studies have established that the FANCL RING E3 ubiquitin ligase allosterically alters the active site of the UBE2T E2 ubiquitin-conjugating enzyme to promote site-specific monoubiquitination of FANCD2 (28). Specifically, FANCL binding to UBE2T exposes a basic triad of the UBE2T active site, promoting favorable interactions with a conserved acidic patch proximal to K561, the site of ubiquitination (28). We speculate that S592 phosphorylation may augment interaction with the basic active site of UBE2T. Alternatively, S592 phosphorylation may inhibit FANCD2 deubiquitination by USP1 in a manner similar to that previously reported for the FANCI S/TQ cluster (15).
Our studies further emphasize the critical nature of the coordinated posttranslational modification of FANCD2. Several studies have previously established intricate dependent and independent relationships between FANCD2 monoubiquitination and phosphorylation. For example, the ATM kinase phosphorylates FANCD2 on several S/TQ motifs following exposure to ionizing radiation, e.g., S222, S1401, S1404, and S1418 (17)—phosphorylation of S1401 and S1404 were also detected under unperturbed conditions in this study. While phosphorylation of S222 promotes the establishment of the ionizing radiation (IR)-inducible S-phase checkpoint, S222 phosphorylation and K561 monoubiquitination appear to function as independent events (17). In contrast, phosphorylation of FANCD2 on T691 and S717 by the ATR kinase is required for efficient FANCD2 monoubiquitination (16, 29). CHK1-mediated phosphorylation of FANCD2 S331 also promotes DNA damage-inducible monoubiquitination and its interaction with FANCD1/BRCA2 (30). Conversely, CK2-mediated phosphorylation of FANCD2 inhibits its monoubiquitination and ICL recruitment (31). Our results suggest that CDK-mediated phosphorylation of S592 primes FANCD2 for monoubiquitination during the S phase in particular. It remains to be determined if other posttranslational modifications, including phosphorylation at other sites, e.g., S1401 and S1404 detected in this study, also contribute to the regulation of FANCD2 during the cell cycle.
Our work aligns with several previous studies describing the coordination of the DNA damage response with cell cycle progression. For example, BRCA2 carboxy-terminal phosphorylation by CDK2 precludes the binding of RAD51, thereby inactivating HR prior to the onset of mitosis (19). In contrast, CDK1/2 phosphorylation of EXO1 endonuclease positively regulates DNA strand resection and HR repair during the S phase (20). Similarly, CDK-mediated phosphorylation of CtIP allosterically stimulates its phosphorylation by ATM, resulting in the promotion of strand resection and HR during the S phase (32). While a role for FANCD2 in HR has been established (33), it remains to be determined if CDK-mediated FANCD2 S592 phosphorylation promotes HR or other related functions. For example, FANCD2 has been shown to have a DNA repair-independent role in protecting stalled replication forks from MRE11-mediated degradation (11). We have also previously established an important role for FANCD2 in the maintenance of CFS stability: in the absence of FANCD2, CFS replication forks stall at a greater frequency, a likely consequence of persistent toxic R-loop structures, leading to visible gaps and breaks on metaphase chromosomes (8, 10, 34, 35). Under conditions of replication stress, CFSs can remain unreplicated until mitosis, where replication can be completed through a process referred to as mitotic DNA synthesis (MiDAS), a POLD3- and RAD52-dependent process that shares features with break-induced DNA replication (BIR) (36, 37). FANCD2 has been shown to play a role in MiDAS (12, 13). Defects in—or an overreliance upon—this process are likely to lead to chromosome missegregation and mitotic defects, as we have observed upon mutation of S592. Increased mitotic aberrations have previously been observed in primary murine FA pathway-deficient hematopoietic stem cells and FA-patient-derived bone marrow stromal cells (9). Taken together, our results support a model whereby CDK-mediated S592 phosphorylation promotes efficient S-phase monoubiquitination, ensuring the efficient and timely completion of CFS replication and mitotic chromosome stability. A greater understanding of the function and regulation of the FA pathway under physiologically relevant nonstressed conditions will provide greater insight into the pathophysiology of this disease and ultimately lead to improved therapeutic options for FA patients.
MATERIALS AND METHODS
Cell culture and generation of mutant cell lines.
HeLa cervical carcinoma and U2OS osteosarcoma cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS), l-glutamine, and penicillin-streptomycin. 293FT viral producer cells (Invitrogen) were cultured in DMEM containing 12% (vol/vol) FBS, 0.1 mM nonessential amino acids (NEAA), 1% (vol/vol) l-glutamine, 1 mM sodium pyruvate, and 1% (vol/vol) penicillin-streptomycin. PD20 FA-D2 (FANCD2−/−) patient cells were purchased from Coriell Cell Repositories (catalog no. GM16633). These cells harbor a maternally inherited A-G change at nucleotide 376 that leads to the production of a severely truncated protein and a paternally inherited missense hypomorphic mutation leading to an R1236H change. PD20 FA-D2 cells were stably infected with pLenti6.2/V5-DEST (Invitrogen) harboring wild-type or mutant FANCD2 cDNAs. Stably infected cells were grown in DMEM complete medium supplemented with 2 μg/ml blasticidin.
Immunoprecipitation.
FA-D2 cells stably expressing LacZ or V5-tagged FANCD2, U2OS, and U2OS 3×FLAG FANCD2 cells were lysed in Triton X-100 lysis buffer (50 mM Tris-HCl [pH 7.5], 1% [vol/vol] Triton X-100, 200 mM NaCl, 5 mM EDTA, 2 mM Na3VO4, protease inhibitors [Roche], and 40 mM β-glycerophosphate) on ice for 15 min followed by sonication for 10 s at 10% amplitude using a Fisher Scientific model 500 Ultrasonic Dismembrator. Anti-V5 or anti-FLAG–agarose were washed and blocked with NETN100 buffer (20 mM Tris-HCl [pH 7.5], 0.1% NP-40, 100 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, and protease inhibitors [Roche]) plus 1% bovine serum albumin (BSA), and the final wash and resuspension were done in Triton X-100 lysis buffer. Lysates were incubated with agarose beads at 4°C for 2 h with nutating. Agarose beads were then washed in Triton X-100 lysis buffer and boiled in 1× NuPAGE buffer (Invitrogen) and analyzed for the presence of proteins by SDS-PAGE and immunoblotting or stained using Colloidal blue staining kit (Invitrogen) for mass spectrometry.
Cell cycle FANCD2 immunoprecipitation.
S-phase and M-phase synchronized populations of HeLa cells were lysed in lysis buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 400 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.1 mM dithiothreitol [DTT], 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], and complete protease inhibitors). We added 100 units of benzonase per 100 μl of lysis buffer. Protein G magnetic beads (Dynabeads; Novex) were cross-linked with anti-FANCD2 (catalog no. NB100-182; Novus Biologicals) antibody. An equal volume of no-salt buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.2 mM KCl, 0.2 mM EDTA, 0.1 mM DTT, and 0.1% NP-40, plus complete protease inhibitors) was added to 2 mg of whole-cell lysate. Samples were resuspended in anti-FANCD2-bound beads rotating at 4°C for 4 h. Beads were washed in wash buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.2 mM KCl, 300 mM KCl, 10% glycerol, 0.2 mM EDTA, 0.1 mM DTT, and 0.1% NP-40, plus complete protease inhibitor). The samples were eluted in urea elution buffer (8 M urea, 1 mM Na3VO4, 2.5 mM Na2H2P2O, 1 mM β-glycerophosphate, and 25 mM HEPES, pH 8.0). Silver staining was performed to visualize the immunoprecipitation (IP), and FANCD2 pulldown was confirmed by Western blot analysis. Samples were sent for LC-MS/MS analysis at the COBRE Center for Cancer Research Development Proteomics Core at Rhode Island Hospital.
Sample preparation and phosphopeptide enrichment for LC-MS/MS analysis.
Cell pellets were lysed in 8 M urea, 1 mM sodium orthovanadate, 20 mM HEPES, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate (pH 8.0, 20 min, 4°C); sonicated; and cleared by centrifugation (14,000 × g, 15 min, 4°C). A total of 100 μg of protein per sample was subject to trypsin digestion, and tryptic peptides were desalted using C18 Sep-Pak Plus cartridges (Waters, Milford, MA) and lyophilized for 48 h to dryness. Phosphopeptides were enriched using Titansphere Phos-TiO tips (GL Sciences, Tokyo, Japan) following the manufacturer’s protocol with minor modifications. Dried eluted phosphopeptides were analyzed by LC-MS/MS using a fully automated proteomic technology platform that includes an Agilent 1200 series Quaternary high-performance liquid chromatography (HPLC) system (Agilent Technologies, Santa Clara, CA) connected to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA) using conditions as described previously (38–40). Peptide spectrum matching of MS/MS spectra of each file was searched against the UniProt database using Mascot v2.4 (Matrix Science, Ltd., London, UK). The RAW data files associated with LC/MS-MS phosphoproteomic analysis have been deposited to figshare.
Lambda phosphatase assay.
Cells were harvested, and cell pellets were split into two, lysed in either lambda phosphatase lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1 mM EGTA, 1 mM dithiothreitol, 2 mM MnCl2, 0.01% Brij35, 0.5% NP-40, and protease inhibitor) or lambda phosphatase buffer with the addition of phosphatase inhibitors, 2 mM Na3VO4 and 5 mM NaF, for 15 min at 4°C followed by sonication for 10 s at 10% amplitude using a Fisher Scientific model 500 Ultrasonic Dismembrator. Lysates were incubated with or without 30 U of lambda phosphatase 1 h at 30°C.
Immunoblotting.
For immunoblotting analysis, cell pellets were washed in phosphate-buffered saline (PBS) and lysed in 2% (wt/vol) SDS, 50 mM Tris-HCl, and 10 mM EDTA followed by sonication for 10 s at 10% amplitude. Proteins were resolved on NuPAGE 3 to 8% (wt/vol) Tris-acetate or 4 to 12% (wt/vol) Bis-Tris gels (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membranes. The following antibodies were used: rabbit polyclonal antisera against FANCD2 (catalog no. NB100-182; Novus Biologicals), FANCI (catalog no. A301-254A; Bethyl), FLAG (product no. F7425; Sigma), phospho-histone H3 pS10 (product no. 9701; Cell Signaling), pan pS/T-CDK (product no. 9477; Cell Signaling), and V5 (product no. 13202; Cell Signaling) and mouse monoclonal antisera against α-tubulin (catalog no. MS-581-PO; Neomarkers). The FANCD2 S592 phospho-specific antibody was generated against the following 14-amino-acid (aa) peptide: FANCD2 aa587-599 ADRSE-pS-PSLTQER-Cys (Pacific Immunology).
Plasmids.
The FANCD2 S592A and S592D cDNAs were generated by site-directed mutagenesis of the wild-type FANCD2 cDNA using the QuikChange site-directed mutagenesis kit (Stratagene). The forward (FP) and reverse (RP) oligonucleotide sequences used are as follows: S592A FP, 5′- CGGCAGACAGAAGTGAAGCACCTAGTTTGACCCAAG-3′; S592A RP, 5′-CTTGGGTCAAACTAGGTGCTTCACTTCTGTCTGCCG-3′; S592D FP, 5′-GGCGGCAGACAGAAGTGAAGATCCTAGTTTGACCCAAGAG-3′; and S592D RP, 5′-CTCTTGGGTCAAACTAGGATCTTCACTTCTGTCTGCCGCC-3′. The full-length FANCD2 cDNA sequences were TOPO cloned into the pENTR/d-TOPO (Invitrogen) entry vector and subsequently recombined into the pLenti6.2/V5-DEST (Invitrogen) destination vector and used to generate lentivirus for the generation of stable cell lines.
Cell cycle synchronization.
For early S-phase arrest, cells were synchronized by the double-thymidine block method. Cells were seeded in 10-cm2 dishes and treated with 2 mM thymidine (catalog no. 226740050; Acros Organics) for 18 h. Cells were washed with PBS and released into thymidine-free media for 10 h following by a second incubation in 2 mM thymidine for 18 h. Cells were washed twice with PBS and released into thymidine-free media. For M-phase arrest, cells were synchronized using the mitotic shake-off method. HeLa cells were seeded in 10-cm2 dishes. Cells were treated with 100 ng/ml of nocodazole (catalog no. SML1665; Sigma) at 80 to 90% confluence for 15 h. Mitotic cells were collected by the shake-off method and replated in nocodazole-free media for cell cycle progression analysis.
Cell cycle analysis by FACS.
Cells were fixed in ice-cold methanol, washed in PBS, and incubated in 50 μg/ml propidium iodide (PI) (Sigma) and 30 U/ml RNase A for 10 min at 37°C, followed by analysis using a BD FACSVerse flow cytometer. The percentages of cells in G1, S, and G2/M were determined by analyzing PI histograms with FlowJo v10.2 software.
In vitro CDK phosphorylation assay.
Purified FANCD2 CDK2-cyclin A proteins were a generous gift from Andrew Deans at the University of Melbourne. In order to remove any previous phosphorylation, 2 μg of protein were incubated with 100 U of lambda phosphatase, 10 mM protein metallophosphatases (PMP), and 10 mM MnCl2 at 30°C for 30 min. For the phosphorylation reaction, each reaction tube was composed of the following reagents in this specific order: 10× CDK kinase buffer (250 mM Tris [pH 7.5], 250 mM glycerophosphate, 50 mM EGTA, and 100 mM MgCl2), 2 μg of protein, 15 nM CDK2-cyclin A, and 20 μM ATP. Samples were incubated at 30°C for 30 min, and the reaction was stopped by adding 10 μl of 10% β-mercaptoethanol in 4× lithium dodecyl sulfate (LDS) buffer.
Cell proliferation assay.
For cell proliferation assays, we performed electrical impedance analysis using the xCELLigence RTCA DP system from Acea Biosciences. Cells were seeded in polyethylene terephthalate (PET) E-plates (part no. 300600890; Acea Biosciences). Cells were incubated in the absence or presence of 0.4 μM aphidicolin (APH) or 20 nM mitomycin C (MMC) for 120 h. Electrical impedance measurements were taken every 15 min over the course of the incubation. Statistical analysis was performed using R.
ACKNOWLEDGMENTS
We thank members of the Howlett, Camberg, and Dutta laboratories for critical discussions. We thank Andrew A. Deans and Winnie Tan at the University of Melbourne for purified proteins and Martin A. Cohn at the University of Oxford for HeLa FANCD2−/− cells.
N.G.H. designed the research, supervised the experiments, analyzed the data, and wrote the paper. J.A.C.-V. performed the majority of experiments, analyzed the data, and wrote the paper with N.G.H. J.L.B., D.A.V., R.A.B., J.L.G., and B.P. performed experiments and analyzed data. W.T. and A.J.D. contributed vital reagents and advice.
We declare that we have no conflicts of interest.
This work was supported by NIH/NHLBI grant R01HL149907 ( to N.G.H.), an American Society of Hematology Bridge grant (to N.G.H.), Rhode Island IDeA Network of Biomedical Research Excellence (RI-INBRE) grant P20GM103430 from the National Institute of General Medical Science, and Rhode Island Experimental Program to Stimulate Competitive Research (RI-EPSCoR) grant no. 1004057 from the National Science Foundation.
REFERENCES
- 1.Fanconi Anemia Research Fund, Inc. 2014. Fanconi Anemia: guidelines for Diagnosis and Management, Fourth ed Fanconi Anemia Research Fund, Inc., Eugene, OR. [Google Scholar]
- 2.Kottemann MC, Smogorzewska A. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493:356–363. 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7:249–262. 10.1016/S1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100:2414–2420. 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 5.Alcón P, Shakeel S, Chen ZA, Rappsilber J, Patel KJ, Passmore LA. 2020. FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair. Nat Struct Mol Biol 27:240–248. 10.1038/s41594-020-0380-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, Wang S, Dhar A, Peralta C, Pavletich NP. 2020. DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex. Nature 580:278–282. 10.1038/s41586-020-2110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan W, van Twest S, Leis A, Bythell-Douglas R, Murphy VJ, Sharp M, Parker MW, Crismani W, Deans AJ. 2020. Monoubiquitination by the human Fanconi anemia core complex clamps FANCI:FANCD2 on DNA in filamentous arrays. Elife 9:e54128. 10.7554/eLife.54128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. 2005. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet 14:693–701. 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 9.Vinciguerra P, Godinho SA, Parmar K, Pellman D, D'Andrea AD. 2010. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest 120:3834–3842. 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madireddy A, Kosiyatrakul ST, Boisvert RA, Herrera-Moyano E, Garcia-Rubio ML, Gerhardt J, Vuono EA, Owen N, Yan Z, Olson S, Aguilera A, Howlett NG, Schildkraut CL. 2016. FANCD2 facilitates replication through common fragile sites. Mol Cell 64:388–404. 10.1016/j.molcel.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22:106–116. 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan KL, Hickson ID. 2011. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol 22:906–912. 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11:753–760. 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 14.Chen YH, Jones MJ, Yin Y, Crist SB, Colnaghi L, Sims RJ, III, Rothenberg E, Jallepalli PV, Huang TT. 2015. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol Cell 58:323–338. 10.1016/j.molcel.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung RS, Castella M, Abeyta A, Gafken PR, Tucker N, Taniguchi T. 2017. Ubiquitination-linked phosphorylation of the FANCI S/TQ cluster contributes to activation of the Fanconi anemia I/D2 complex. Cell Rep 19:2432–2440. 10.1016/j.celrep.2017.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho GP, Margossian S, Taniguchi T, D'Andrea AD. 2006. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol 26:7005–7015. 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD. 2002. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109:459–472. 10.1016/S0092-8674(02)00747-X. [DOI] [PubMed] [Google Scholar]
- 18.Wohlbold L, Fisher RP. 2009. Behind the wheel and under the hood: functions of cyclin-dependent kinases in response to DNA damage. DNA Repair (Amst) 8:1018–1024. 10.1016/j.dnarep.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. 2005. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434:598–604. 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 20.Tomimatsu N, Mukherjee B, Catherine Hardebeck M, Ilcheva M, Vanessa Camacho C, Louise Harris J, Porteus M, Llorente B, Khanna KK, Burma S. 2014. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat Commun 5:3561. 10.1038/ncomms4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Higuera I, Kuang Y, Denham J, D'Andrea AD. 2000. The Fanconi anemia proteins, FANCA and FANCG, stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood 96:3224–3230. 10.1182/blood.V96.9.3224. [DOI] [PubMed] [Google Scholar]
- 22.Huertas P, Jackson SP. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284:9558–9565. 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X, Chini CC, He M, Mer G, Chen J. 2003. The BRCT domain is a phospho-protein binding domain. Science 302:639–642. 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 24.van Twest S, Murphy VJ, Hodson C, Tan W, Swuec P, O'Rourke JJ, Heierhorst J, Crismani W, Deans AJ. 2017. Mechanism of ubiquitination and deubiquitination in the Fanconi anemia pathway. Mol Cell 65:247–259. 10.1016/j.molcel.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Liang CC, Li Z, Lopez-Martinez D, Nicholson WV, Venien-Bryan C, Cohn MA. 2016. The FANCD2-FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat Commun 7:12124. 10.1038/ncomms12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, Tashiro S, Elledge SJ, Takata M. 2008. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol 15:1138–1146. 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moses AM, Hériché JK, Durbin R. 2007. Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol 8:R23. 10.1186/gb-2007-8-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaugule VK, Arkinson C, Rennie ML, Kamarainen O, Toth R, Walden H. 2020. Allosteric mechanism for site-specific ubiquitination of FANCD2. Nat Chem Biol 16:291–301. 10.1038/s41589-019-0426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreassen PR, D'Andrea AD, Taniguchi T. 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev 18:1958–1963. 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhi G, Wilson JB, Chen X, Krause DS, Xiao Y, Jones NJ, Kupfer GM. 2009. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for Fanconi anemia pathway function and BRCA2 interaction. Cancer Res 69:8775–8783. 10.1158/0008-5472.CAN-09-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Martinez D, Kupculak M, Yang D, Yoshikawa Y, Liang CC, Wu R, Gygi SP, Cohn MA. 2019. Phosphorylation of FANCD2 inhibits the FANCD2/FANCI complex and suppresses the Fanconi anemia pathway in the absence of DNA damage. Cell Rep 27:2990–3005.e2995. 10.1016/j.celrep.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Shi LZ, Wong CC, Han X, Hwang PY, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, Yates JR, III, Chen L, Wu X. 2013. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet 9:e1003277. 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A 102:1110–1115. 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Rubio ML, Perez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, Aguilera A. 2015. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet 11:e1005674. 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, Cohn MA, Gibbons RJ, Deans AJ, Niedzwiedz W. 2015. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell 60:351–361. 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhowmick R, Minocherhomji S, Hickson ID. 2016. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol Cell 64:1117–1126. 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 37.Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID. 2015. Replication stress activates DNA repair synthesis in mitosis. Nature 528:286–290. 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- 38.Ahsan N, Belmont J, Chen Z, Clifton JG, Salomon AR. 2017. Highly reproducible improved label-free quantitative analysis of cellular phosphoproteome by optimization of LC-MS/MS gradient and analytical column construction. J Proteomics 165:69–74. 10.1016/j.jprot.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu K, Salomon AR. 2009. PeptideDepot: flexible relational database for visual analysis of quantitative proteomic data and integration of existing protein information. Proteomics 9:5350–5358. 10.1002/pmic.200900119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu K, Salomon AR. 2010. HTAPP: high-throughput autonomous proteomic pipeline. Proteomics 10:2113–2122. 10.1002/pmic.200900159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, Clauser KR, Qiao JW, Gritsenko MA, Moore RJ, Levine DA, Townsend R, Erdmann-Gilmore P, Snider JE, Davies SR, Ruggles KV, Fenyo D, Kitchens RT, Li S, Olvera N, Dao F, Rodriguez H, Chan DW, Liebler D, White F, Rodland KD, Mills GB, Smith RD, Paulovich AG, Ellis M, Carr SA. 2014. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics 13:1690–1704. 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3:ra3. 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 43.Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, Kawaler E, Mundt F, Krug K, Tu Z, Lei JT, Gatza ML, Wilkerson M, Perou CM, Yellapantula V, Huang KL, Lin C, McLellan MD, Yan P, Davies SR, Townsend RR, Skates SJ, Wang J, Zhang B, Kinsinger CR, Mesri M, Rodriguez H, Ding L, Paulovich AG, Fenyö D, Ellis MJ, Carr SA, NCI CPTAC. 2016. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534:55–62. 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruse CI, McClatchy DB, Lu B, Cociorva D, Motoyama A, Park SK, Yates JR, III, 2008. Motif-specific sampling of phosphoproteomes. J Proteome Res 7:2140–2150. 10.1021/pr800147u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. 2011. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal 4:rs5. 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]