Abstract
Sepsis is a major cause of mortality among hospitalized patients worldwide. Shorter time to administration of broad‐spectrum antibiotics is associated with improved outcomes, but early recognition of sepsis remains a major challenge. In a two‐center cohort study with prospective sample collection from 1400 adult patients in emergency departments suspected of sepsis, we sought to determine the diagnostic and prognostic capabilities of a machine‐learning algorithm based on clinical data and a set of uncommonly measured biomarkers. Specifically, we demonstrate that a machine‐learning model developed using this dataset outputs a score with not only diagnostic capability but also prognostic power with respect to hospital length of stay (LOS), 30‐day mortality, and 3‐day inpatient re‐admission both in our entire testing cohort and various subpopulations. The area under the receiver operating curve (AUROC) for diagnosis of sepsis was 0.83. Predicted risk scores for patients with septic shock were higher compared with patients with sepsis but without shock (p < 0.0001). Scores for patients with infection and organ dysfunction were higher compared with those without either condition (p < 0.0001). Stratification based on predicted scores of the patients into low, medium, and high‐risk groups showed significant differences in LOS (p < 0.0001), 30‐day mortality (p < 0.0001), and 30‐day inpatient readmission (p < 0.0001). In conclusion, a machine‐learning algorithm based on electronic medical record (EMR) data and three nonroutinely measured biomarkers demonstrated good diagnostic and prognostic capability at the time of initial blood culture.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Sepsis represents significant morbidity, mortality, and cost in modern health care. Timely treatment with antibiotics improves outcomes, but it can be difficult to identify patients with sepsis early on in the clinical course.
WHAT QUESTION DID THIS STUDY ADDRESS?
Can a machine‐learning algorithm incorporating basic clinical data and nonroutinely measured biomarkers accurately predict sepsis and other related secondary outcomes?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
A machine‐learning algorithm incorporating basic clinical data and nonroutinely measured biomarkers accurately identify sepsis. Meanwhile, a higher score outputted by the algorithm predicts less favorable outcomes with respect to discharge time, 30‐day mortality, and 30‐day inpatient re‐admission.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Earlier treatment of patients who are on a course for poor outcomes has the potential to significantly improve those outcomes. This study suggests that a machine‐learning‐based score may assist clinicians in identifying such patients.
INTRODUCTION
Sepsis, a dysregulated immune response to infection resulting in organ dysfunction, is responsible for significant morbidity and mortality worldwide. 1 Early therapy—particularly antibiotics—leads to improved outcomes. 2 , 3 , 4 , 5 , 6 However, vague presenting symptoms make the recognition of sepsis difficult and lead to increased mortality. 7 The initial recognition and treatment of sepsis often occur in the emergency departments that can be chaotic and understaffed, complicating recognition of this syndrome.
The timely treatment of sepsis remains a widespread challenge. In 2015, the Centers for Medicare and Medicaid Services (CMS) established a sepsis quality measure (SEP‐1) that aims to improve outcomes through 3‐h and 6‐h treatment bundles, including serum lactate measurement, blood culture collection, broad‐spectrum antibiotic administration, and intravenous fluids and vasopressors when indicated. Compliance with these goals has varied widely among 2851 hospitals reporting SEP‐1 data with a mean of only 48.9% of patients receiving all bundle components in the designated timeframe. 8 Failure to meet these quality measures represents suboptimal treatment that likely results in increased sepsis morbidity and mortality. 9
Multiple factors contribute to delayed interventions in sepsis. In patients presenting with vague symptoms, the need for antibiotics may go unrecognized. Meanwhile, antibiotics cannot be administered indiscriminately as potential benefits must be weighed against risks, including direct adverse effects of antibiotic agents and the growing problem of antimicrobial resistance. 10 Therefore, each patient requires evaluation by a healthcare provider to assess for the appropriateness of antibiotics.
In this context, multiple factors contribute to delayed interventions in sepsis. In patients presenting with vague symptoms, the need for antibiotics may go unrecognized. Meanwhile, treatment may get delayed while waiting for an initial evaluation by a primary provider, processing laboratory tests, or as a result of distraction by other urgent cases in overburdened emergency departments. Even after adequate data are available and a presumptive diagnosis is made, alerting everyone on the healthcare team (e.g., nurses and pharmacists) to prioritize treatment may reduce time to intervention relative to usual care. Each step in the healthcare team’s workflow (examples depicted in Figure S1) is a potential opportunity for swifter action relative to when occult sepsis is unrecognized.
The prevailing wisdom has been that more reliable recognition of sepsis may lead to earlier treatment and improved outcomes. Numerous methods have been described to screen for sepsis and facilitate early response—particularly timely antibiotic administration. These range from simple scoring systems to complex multivariable algorithms. The availability of large volumes of electronic medical record (EMR) data has led to the development of machine‐learning methods for identifying these patients. 11 , 12 , 13 Although these tools may provide the ability to identify the patients in whom sepsis should be strongly considered, they do little else to inform the risks‐benefits dilemma the clinician must weigh when deciding to initiate antibiotics.
Tools that solely dichotomize patients (sepsis vs. not) fail to embrace the reality that sepsis encompasses a heterogeneous group of poorly defined disorders exhibiting a complicated spectrum of severity. 14 , 15 No tool currently identifies or predicts where a patient falls on this spectrum; yet, such information, if reliable, may be extremely valuable for the clinician considering the risks and benefits of initiating antimicrobial therapy and for prioritizing actions leading to the administration of antibiotics in patients who need them most. For example, lower severity of illness may afford more judicious use of antibiotics, whereas a bleak prognosis may not only inspire earlier antimicrobial therapy but also prompt the multidisciplinary healthcare team to swifter action.
We previously applied machine‐learning using EMR data and 15 novel biomarkers to identify patients with sepsis and demonstrated improved diagnostic performance with the addition of novel biomarker measurements to standard clinical data. 16 Given that diagnostic performance alone does not address the entire dilemma facing healthcare providers today, we now present a more holistic analysis of a subset of NOSIS, a large, multi‐center, novel data set comprised of three plasma proteins (procalcitonin [PCT], interleukin‐6 [IL‐6], and C‐reactive protein [CRP]) and routinely measured EMR parameters. We specifically selected these three biomarkers because numerous studies, including one we previously conducted, have consistently demonstrated that PCT, IL‐6, and CRP have strong predictive power with regard to sepsis and sepsis‐related outcomes. 16 , 17 , 18 , 19 , 20 , 21 Furthermore, they are readily availability in commercial immunoanalyzers, suggesting that a model incorporating such parameters can be readily translatable.
METHODS
Participants and source of data
We performed a prospective observational cohort study of all adult inpatients with a blood culture ordered at Carle Foundation Hospital (CFH), a 413‐bed regional hospital in central Illinois, and OSF Saint Francis Medical Center, a 616‐bed regional hospital also in central Illinois. Data and specimens were collected between February 2018 and September 2019 according to standards approved by the CFH institutional review board (IRB) and OSF IRB with informed consent, when required. De‐identified clinical data from the EMRs were extracted by data engineers.
Clinical samples
Plasma samples were obtained from remnant clinical blood specimens prior to disposal. At both sites, samples were obtained from Lithium Heparin PST tubes used in routine clinical testing.
Sample pre‐processing steps
Lithium heparin plasma samples included in the study were aliquoted into a 2 ml microcentrifuge tube and centrifuged at 1200 g for 10 min at 4°C. After centrifugation, the supernatant was aliquoted into a new 1.5 ml microcentrifuge tube and vortexed for 5 s to ensure the sample was homogenous. Then up to 600 µL of each sample was aliquoted into four 150 µL aliquots and the remainder of the sample was left in the 1.5 ml microcentrifuge tube. After processing, all aliquots were stored at −80°C before being transported on dry ice to Prenosis’ central laboratory where they were immediately placed into −80°C storage.
Biomarker measurement process
Plasma protein biomarkers were measured using the Magnetic Luminex Assay technology, a bead‐based multiplex assay. For each assay, 37 patient plasma samples, 3 quality controls (QCs; high, mid, and low), and a 7‐point calibration curve were prepared in duplicate and run on a 96‐well plate. Appropriate calibrator diluent served as the blank. Prior to running the assay, all samples including the QCs were diluted using appropriate calibrator diluent. Optimal dilutions for each plasma protein were determined according to the US Food and Drug Administration (FDA)’s fit‐for‐purpose guidelines for ligand binding assays. Each plasma protein was measured using the Luminex MAGPIX CCD Imager.
Biomarker assay validation
Biomarker assay validation was conducted following the principles and procedures of the FDA’s Bioanalytical Method Validation guidance document. The validation consisted of pre‐analytical validation (i.e., sample collection, handling, and storage) and analytical validation (i.e., accuracy, precision, and reproducibility of biomarker measurements). Incurred sample re‐analysis and multilevel QC monitoring were used to identify erroneous measurements.
Sepsis definitions
We used criteria for sepsis grounded in the Sepsis‐3 framework, which formally defines sepsis as life‐threatening organ dysfunction caused by a dysregulated host response to infection. 1 Strengths of the Sepsis‐3 definition rest on its ability to provide a standardized conceptual framework, capture current consensus in sepsis pathophysiology, and predict outcomes relevant in the context of sepsis, such as intensive care unit (ICU) admission or death. In this study, we defined a patient as septic if they had a suspected infection and life‐threatening organ dysfunction.
Life‐threatening organ dysfunction was defined as an acute change in total Sequential Organ Failure Assessment (SOFA) score by two or more points consequent to infection. 1 Baseline SOFA was adjudicated by two physicians. A patient was considered to have suspected infection if a blood culture was obtained (all of study cohort) and more than 4 qualifying antimicrobial days (QADs) were given within ± 2 days of blood culture. 22 We utilized the list of antimicrobials in the Centers for Disease Control and Prevention (CDC)’s Hospital Toolkit for Adult Sepsis Surveillance to define QAD. 23 The methodology was validated by an adjudication process on a subset of patients. Detailed methodology of the adjudication is presented in Supplementary Text S1.
A patient was considered in septic shock if they satisfied our criteria for sepsis and had a lactate greater than 2.0 mmol/L and were administered norepinephrine, dopamine, epinephrine, phenylephrine, or vasopressin at any time during their hospitalization.
Because we independently defined life‐threatening organ dysfunction and suspected infection, a patient can be categorized into one of four categories: (1) no organ dysfunction and no infection, (2) infection without organ dysfunction, (3) organ dysfunction without infection, (4) and organ dysfunction present alongside infection. The presence or absence of each event is determined in the context of the patient’s entire hospital stay.
Subpopulation definitions
To examine the pragmatic utility of this risk score, subgroup analyses were performed among clinically interesting subpopulations. Specifically, patients who did not meet at least two of four systemic inflammatory response syndrome (SIRS) criteria at the time of blood culture collection (SIRS‐negative patients), and patients whose SOFA score was greater than or equal to two with respect to baseline at the time of blood culture collection were examined (SOFA‐positive patients).
Algorithms and predictors
Features
Three biomarkers (not routinely measured in standard sepsis clinical care) were included in the machine‐learning analysis: PCT, IL‐6, and CRP. Biomarkers were log‐transformed prior to being input into the model.
The following EMR parameters were included in the machine‐learning analysis: patient age, sex, Glasgow Coma Scale, vital signs, and standard laboratory measurements. Vital signs included systolic blood pressure, diastolic blood pressure, temperature, respiratory rate, heart rate, and blood oxygen saturation. Hematology parameters included white blood cell count, absolute monocyte count, absolute neutrophil count, and platelet count. Chemistry parameters included plasma albumin, blood urea nitrogen, creatinine, potassium, lactate, glucose, sodium, and total bilirubin. For each EMR parameter, the value used was the one that was drawn nearest to the draw time of the sample associated with the measurement of the three biomarkers within a certain timeframe. For vitals and assessments, the timeframe was any time prior to the draw time of the sample and 30 min after the draw time of the sample. For laboratories, the timeframe was any time prior to the draw time of the sample and 1 h after the draw time of the sample.
Inclusion criteria
Patients were included in the analysis if (1) a blood culture was drawn during their hospital stay and (2) a blood sample for a basic or complete metabolic panel was drawn within 3 h of their first culture being ordered and prior to the initial administration of antimicrobials; if multiple samples satisfied this condition, the sample drawn nearest to the time of the ordering of the first culture was used. The latter condition was imposed because we wanted the sample that served as the source of our biomarker measurements to be drawn at a clinically relevant timepoint. In Figure S2, we report the distribution of the sample draw time minus the first blood culture order time.
Train and test split
We performed a 2:1 split of the data that was conducted as follows: we ordered our patients sequentially in terms of their inclusion date in our study on a site‐by‐site basis and then assigned every third patient to be in the testing set while assigning the remaining patients to be in the training set.
Based on our inclusion criteria, 1400 patients were included in the analysis, 933 of which were in the training set and 467 of which were in the testing set.
Label
A patient was assigned a positive label if they were administered four of more qualifying antimicrobial days (a surrogate for suspected infection) and exhibited an increase in SOFA score by two points or greater from baseline within 12 h of emergency department presentation. These criteria serve as our surrogate for a patient having sepsis, which is grounded in the Sepsis‐3 framework.
Algorithm
We utilized random forests to build our predictive models using the R ranger package. 24 Random forest modeling was used due to its ability to model nonlinear relationships and its inherent mechanisms of random sampling and ensemble strategies that tend to enable better generalization performance. 25 To demonstrate the relative value of random forests, we also provide results derived from a logistic regression model for comparison.
Missing data
Biomarker measurements were missing either due to inadequate volume from the plasma sample or if a measurement did not pass our quality control criteria. EMR parameters were missing if they were not available in their respective, requisite timeframe (specified in the “Features” description). We report missing data statistics for all parameters in Figure S3 for both the training and test sets. In the test set, the biomarkers all have less than 7% missing measurements. If the value of a subject’s covariate was missing, it was imputed with its median value from the training cohort.
Hyperparameters
Using the R caret package, 26 the number of variables randomly sampled at each split was optimized via 10 repeats of 5‐fold cross‐validation in the training cohort. The optimized value of this parameter was two. The number of trees used was set to a value of 1000.
Output
A random forest model was fit on the training dataset using fixed hyperparameters derived as above. The model was then evaluated on the testing cohort, yielding a probability ranging between 0.0 and 1.0 per patient. Details regarding how predictions were generated for patients in the training cohort are provided in Supplementary Text S2.
Feature importance
Feature importance was determined according to a permutation‐based method, outlined by Altman et al. 27 Briefly, this method calculates the distribution of each feature’s importance under the null hypothesis of no association to the response by repeatedly permuting the outcome.
Risk category determination
To assess the prognostic power of the score, the score was discretized into three risk groups (low, medium, and high). To define the first threshold to categorize a patient as low‐risk, the threshold closest to the top‐left of the receiver operating characteristic (ROC) plot in the training cohort was used (defined as the threshold satisfying min((1‐sensitivities)2 +(1‐specificities)2)), as it provides a balance between optimal sensitivity and specificity with respect to a patient being septic or not. The second threshold used to define a patient as medium/high‐risk was defined based on the prevalence of septic shock in the training cohort. Specifically, if the prevalence of septic shock in our dataset is defined as p shock, the (1‐p shock)th percentile of the scores of all patients was used as the threshold.
Survival analysis
To evaluate prognostic capabilities of the algorithm, survival curves were generated for low, medium, and high‐risk groups with respect to length of stay (LOS), 30‐day mortality, and 30‐day inpatient re‐admission. Survival estimates were based on the Kaplan‐Meier method, and comparisons of survival distributions were based on the log rank test. Details regarding the construction of censored outcomes are presented in Supplementary Text S3.
Probability distribution group comparison
Comparisons of probability distributions between the two groups were performed using the Wilcoxon rank‐sum test, and multigroup comparisons were performed using the Kruskal‐Wallis test.
RESULTS
Patients
Twenty‐nine percent of the 1400 patients in our analysis met the primary diagnostic outcome of sepsis based on the QAD/SOFA definition. Demographics and relevant clinical characteristics of the entire cohort are presented in Table 1. Demographics and relevant clinical characteristics are also presented for the training and testing cohort (Tables S1 and S2).
TABLE 1.
Baseline data of the entire cohort
| All patients | No sepsis | Sepsis | Septic shock | |
|---|---|---|---|---|
| N | 1400 | 990 | 350 | 60 |
| Age (median, IQR) | 65 (54–75) | 65 (53–75) | 67 (55–76) | 64 (55–76) |
| Gender (male) | 51% | 50% | 55% | 55% |
| Race | ||||
| White | 85.5% | 85.7% | 83.9% | 91.7% |
| Black | 10.5% | 10.8% | 10.6% | 6.7% |
| Other | 2.9% | 2.7% | 4.0% | 0% |
| Patients from site 1 (Carle), patients from Site 2 (OSF) | 59%, 41% | 56%, 44% | 67%, 33% | 58%, 42% |
| Blood culture order time [minutes] (median, IQR) | 35 (17–87) | 35 (17–93) | 34 (17–76) | 25 (10–81) |
| Discharge time [days] (median, IQR) | 4.17 (2.67,7.25) | 3.14 (2.11, 5.35) | 6.85 (4.92, 9.96) | 8.65 (5.14, 14.36) |
| Patients with >= 1 30‐day inpatient re‐admission | 30.1% | 26.0% | 40.9% | 33.3% |
| 30‐day mortality | 5.1% | 2.9% | 6.0% | 36.7% |
| Comorbidities | ||||
| Diabetes | 38% | 37% | 38% | 40% |
| COPD | 20% | 21% | 19% | 12% |
| Congestive heart failure | 16% | 15% | 16% | 27% |
| Chronic kidney disease | 19% | 18% | 23% | 15% |
| Chronic liver disease | 5% | 4% | 7% | 7% |
| Cancer | 3% | 2% | 3% | 3% |
| Presenting features at time of blood culture | ||||
| SIRS | 1.0 (1.0–2.0) | 1.0 (1.0–2.0) | 2.0 (1.0–2.0) | 2.0 (1.0–2.0) |
| SOFA | 1.0 (0.0–3.0) | 1.00 (0.0–2.0) | 3.0 (2.0–4.0) | 5.0 (2.0–8.0) |
| Lactate >= 2 | 41% | 37% | 43% | 100% |
| SOFA‐positive | 48% | 34% | 80% | 82% |
| SIRS‐negative | 53% | 58% | 43% | 33% |
Demographic characteristics, comorbidity information, and statistics of relevant features at the time of blood culture are presented for the entire population, non‐septic patients, patients with sepsis without shock, and patients with septic shock.
Abbreviations: COPD, chronic obstructive pulmonary disease; IQR, interquartile range; OSF, OSF Saint Francis Medical Center; SIRS, systemic inflammatory response syndrome; SOFA, Sequential Organ Failure Assessment.
Diagnostic performance
Diagnostic performance of the algorithm in the testing cohort with regard to sepsis is described in Figure 1 and Figure 2. The 12‐h constraint on the positive sepsis label was imposed to enable a more standardized comparison to other machine‐learning based sepsis prediction algorithms discussed in the literature. 13 , 28 Among the testing cohort the area under the ROC curve (AUROC) was 0.83 (Figure 1a) and the area under the precision recall curve (AUPR) was 0.61 (Figure 2a).
FIGURE 1.
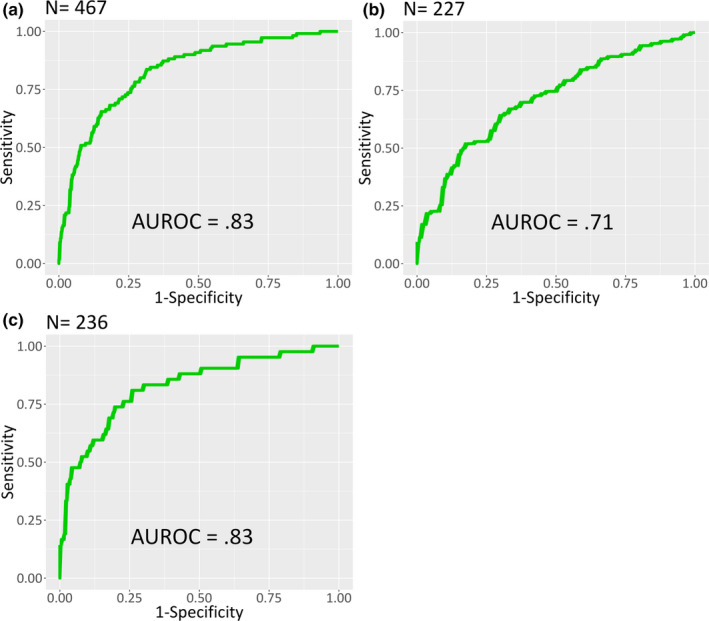
ROC curves of the algorithm in the testing cohort for (a) all patients, (b) SOFA‐positive patients, and (c) SIRS‐negative patients. In all subpopulations, the algorithm demonstrates a strong ability to differentiate patients who satisfied the criteria for sepsis within 12 h of emergency department presentation from those who did not. AUROC, area under the receiver operating curve; ROC, receiver operating characteristic; SIRS, systemic inflammatory response syndrome; SOFA, Sequential Organ Failure Assessment
FIGURE 2.
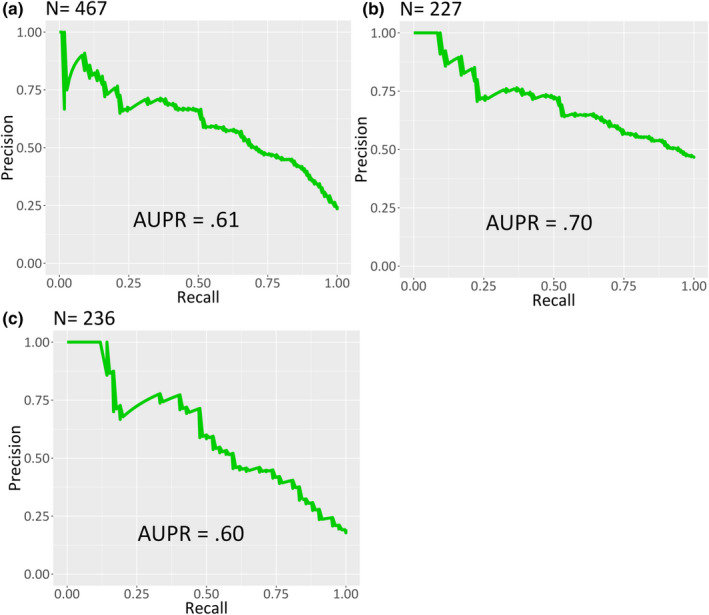
PR curves of the algorithm in the testing cohort for (a) all patients, (b) SOFA‐positive patients, and (c) SIRS‐negative patients. Recall (also known as sensitivity) is displayed on the x‐axis and precision (also known as positive predictive value) is displayed on the y‐axis. PR, precision recall; SIRS, systemic inflammatory response syndrome; SOFA, Sequential Organ Failure Assessment
Subgroup analyses for previously defined, clinically challenging patient populations are presented in Figure 1b,c and Figure 2b,c. Among 227 SOFA‐positive patients at the time of culture, the algorithm achieved an AUROC of 0.71 (Figure 1b) and an AUPR of 0.70 (Figure 2b). Among 236 patients not meeting at least 2 of 4 SIRS criteria, the algorithm achieved an AUROC of 0.83 (Figure 1c) and an AUPR of 0.60 (Figure 2c).
Sensitivity, specificity, positive predictive value, and negative predictive value at the threshold used to categorize a patient as low risk (see Methods) is presented in Table S3. AUROC, AUPR, and F1 score for each population is presented in Table S4.
Importance designated to each feature by random forest placed PCT and IL‐6 among the top three features. Figure 3 displays a ranking of all incorporated features.
FIGURE 3.
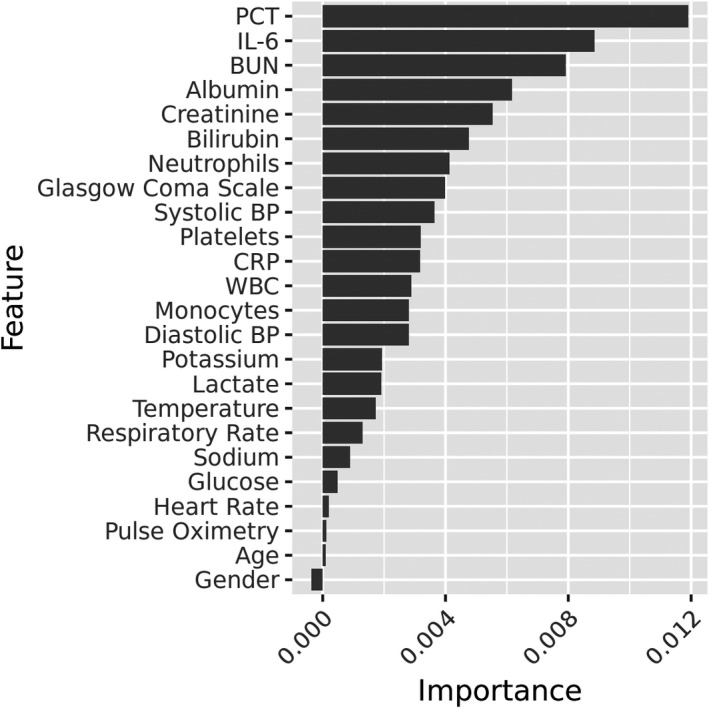
Feature importance outputted by random forest. PCT and IL‐6 emerge as the most important features
Figure 4 depicts algorithm output (probability of sepsis) for subgroups of the primary outcome. In the comparison of nonseptic patients, patients with sepsis without shock, and patients with septic shock, median probability was 0.22, 0.44, and 0.53, respectively. In the comparison of patients with no organ dysfunction and no infection, organ dysfunction only, infection only, and organ dysfunction and infection, median probability was 0.16, 0.23, 0.31, and 0.43, respectively. For the comparison among patients who were nonseptic, septic, and with septic shock, results were statistically significant for all pairwise comparisons (p < 0.0001, Wilcoxon) and the aggregate groupwise comparison (p < 0.0001, Kruskal‐Wallis). For the comparison between patients with/without organ dysfunction and/or with/without infection, results were statistically significant for all pairwise comparisons (p < 0.0001, Wilcoxon) and the aggregate groupwise comparison (p < 0.0001, Kruskal‐Wallis).
FIGURE 4.
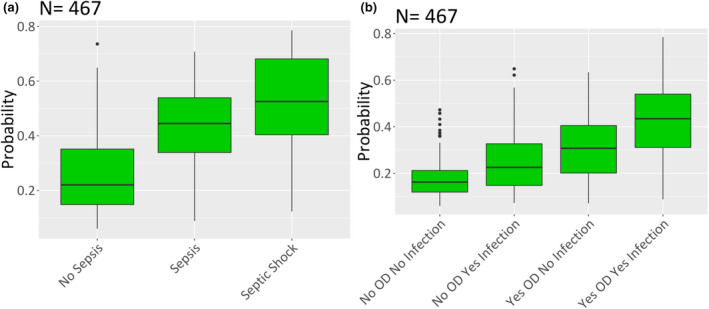
Algorithm‐determined probability of sepsis for specific population subgroups in the testing cohort. (a) Subgroups of sepsis based on the Sepsis‐3 definition. (b) Subgroups of nonseptic patients based on the presence of infection and/or organ dysfunction (OR). In both subgroups, a trend of increasing probability with increasing disease severity emerges
Analogous diagnostic results for the training cohort are presented in Figures S4–S6 and Tables S5 and S6. For reference, we also report the distributions of the IL‐6, PCT, and CRP among nonseptic patients, patients with sepsis without shock, and patients with septic shock in the training cohort (Figure S7). Last, Figures S8 and S9 display analogous diagnostic results for the testing cohort derived from a logistic regression model. We observe that the random forest model consistently outperforms the logistic regression model.
Prognostic performance
Secondary outcomes were compared among low, medium, and high‐risk groups in the testing cohort and the entire cohort. Survival curves for LOS, 30‐day mortality, and 30‐day inpatient re‐admission are depicted for the testing cohort in Figure 5 and for the entire cohort in Figure S10. In the testing cohort, median LOS was 3.2 days for 273 patients in the low‐risk group, 5.0 days for 164 patients in the moderate‐risk group, and 8.5 days for 30 patients in the high‐risk group (p < 0.0001). Mortality rates were higher among higher severity risk groups as a function of time within a 30‐day window (p < 0.0001). Re‐admission rates were higher among higher severity risk groups as a function of time within a 30‐day window (p < 0.0001). Detailed statistics for each risk category and secondary outcome in the testing cohort and the entire cohort are provided in Table 2.
FIGURE 5.
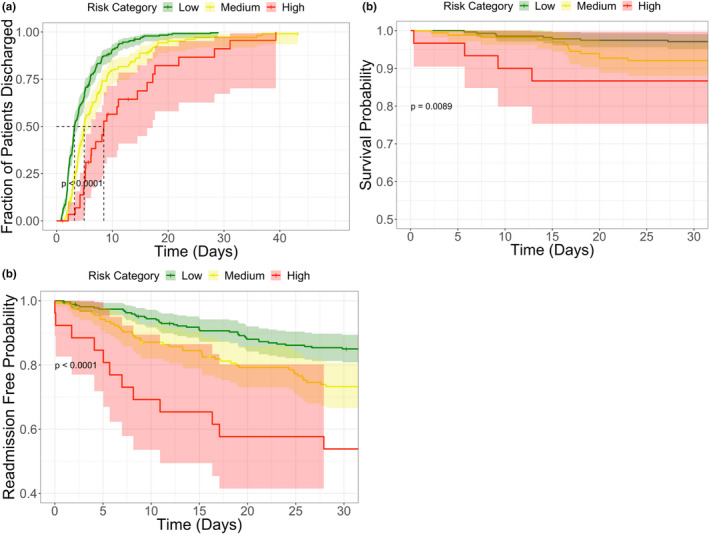
Risk group analysis for three outcomes in the testing cohort: (a) length of hospital stay, (b) 30‐day mortality, (c) 30‐day inpatient readmission. For each risk group and outcome, survival estimates were generated based on the Kaplan‐Meier method, and comparisons of survival distributions were based on the log‐rank test. Statistically significant differences between each of the risk groups are observed for all three outcomes
TABLE 2.
Prognostic characteristics for the testing cohort and entire cohort
| Risk category | Cohort | N | Median LOS | 30‐day Mortality rate | 30‐day Re‐admission rate |
|---|---|---|---|---|---|
| Low | Testing | 273 | 3.2 | 2.9% | 23% |
| Medium | Testing | 164 | 5.0 | 7.9% | 36% |
| High | Testing | 30 | 8.5 | 13.3% | 57% |
| Low | Entire | 859 | 3.4 | 2.2% | 22% |
| Medium | Entire | 464 | 5.3 | 6.9% | 41% |
| High | Entire | 77 | 8.1 | 27.3% | 60% |
Median length of hospital stay, 30‐day mortality rate, and 30‐day inpatient readmission rate are presented as a function of risk category for each cohort a function of risk category for each cohort.
Abbreviation: LOS, length of stay.
DISCUSSION
The challenge facing timely treatment of sepsis lies at many levels of the clinical care process. Some patients with sepsis may be missed early in their presentation due to vague signs and symptoms. Other patients, even with concern for infection, may not manifest illness in ways that sway the obligatory risk‐benefit analysis toward an immediate commitment to broad‐spectrum antibiotics. Even after commitment to broad‐spectrum antibiotics, some cases may harbor an occult risk for decompensation which, if revealed with an adequate tool, would compel providers toward swift action. We have described an approach, evaluated at two separate clinical sites, which uniquely incorporates a large volume of clinical data and three nonroutinely measured plasma biomarkers in a machine‐learning model to rapidly identify patients with sepsis and stratify them based on severity at the time a first clinical specimen is acquired.
One of the largest barriers in developing predictive models for sepsis lies in the lack of a gold standard. 29 Fully appreciating the difficulty (and arguably futility) of defining a gold‐standard for this syndrome, the Third International Consensus Definitions Task Force defined sepsis in a manner that would enable standardization, capture our current conceptual understanding of the disease, and have high predictive validity with respect to clinically relevant end points. When decomposing each aspect of the Sepsis‐3 definition, it becomes clear that some lend themselves to a more robust quantification more than others. Life‐threatening organ dysfunction, defined as an acute change in total SOFA score by 2 or more points consequent to infection, can be accurately quantified through a combination of curating relevant parameters in the EMR and a targeted physician adjudication. Meanwhile, no tool exists which is broadly sensitive and specific for identifying infection and we rely on the judgment of clinicians as indicated by the surrogate developed by Rhee et al. 22 and embraced by the CDC. Utilizing this surrogate for infection and SOFA‐based definition of organ dysfunction, our final sepsis label exhibited a false‐negative rate around 30% and a false‐positive rate around 15% with respect to an adjudication conducted by three physicians on a subset of patients in our analysis (Supplementary Text S1).
Despite this error in our label, our intention is not to build a tool which correlates to the imperfect “gold‐standard” for sepsis but rather an acuity score demonstrating a signal with respect to a wide variety of outcomes. An imperfect label does not preclude one from building a useful score based on that label, but it does likely impose a ceiling on diagnostic performance 30 and impose limitations on the set of actions that can take place based on the score (e.g., deciding whether or not to prescribe antibiotics for a patient).
Despite the noise in our label, we demonstrate good prospective diagnostic performance in a heterogeneous population with respect to the Sepsis‐3 surveillance definition. Robust diagnostic performance in the subgroup with vague or confounding presentation is a novel aspect of these results: the algorithm is no less capable of identifying sepsis in those patients who lack obvious features of infection (i.e., SIRS‐negative) or exhibit features potentially caused by an infection (i.e., SOFA positive) on initial presentation. This tool may therefore be capable of effectively identifying the problematic patient population with vague presentation in need of prompt broad‐spectrum antibiotics earlier than clinical gestalt. Meanwhile, the comparison of subgroups based on the presence or absence of organ dysfunction and infection suggest that the algorithm distinguishes septic patients (i.e., patients with both infection and organ dysfunction) from patients with either feature present independently. This suggests the approach is less likely to misclassify patients who either have uncomplicated infection or have organ dysfunction due to a noninfectious process.
We also present evidence that the score is reflective of disease severity. The analysis stratifying patients into subgroups with and without shock shows a significantly higher probability score produced by the algorithm corresponding to patients with shock. This pattern was observed without providing the algorithm a label for shock. This suggests that the algorithm output may not simply correspond to the dichotomy of sepsis versus not sepsis, but rather a higher score may reflect greater severity of illness. We were similarly interested in how the model output would relate to LOS—a surrogate for severity of illness and major determinant of cost of care, 30‐day mortality, and 30‐day inpatient readmission. Our hypothesis was that higher probability scores will correspond to greater severity of illness, which drive longer hospital stays, greater mortality rates, and greater re‐admission rates. Figure 5 and Figure S10 support this conclusion and provide evidence for prognostic capabilities of the model. Again, this pattern was observed without providing the algorithm any of this direct information.
PCT and IL‐6 are among the most important features in the model, suggesting these biomarkers are useful in differentiating patients with organ dysfunction and infection from those satisfying either condition. We note that this is a departure from the more “Sepsis‐2 grounded” claim that host response markers are useful in differentiating inflammatory responses from noninfectious and infectious stimuli.
There are multiple limitations to this study. In the testing cohort, the analysis would benefit from larger sample sizes when looking at diagnostic results in subpopulations and prognostic results in the high‐risk group for 30‐day mortality and 30‐day inpatient re‐admission. With that said, we do provide subpopulation diagnostic results in the training cohort and prognostic results in the entire cohort for reference. Another limitation of this study is that it includes only two level‐one trauma clinical centers in the central Illinois region. Including more geographically and socioeconomically diverse sites is crucial to better capture the heterogeneity of the suspected sepsis population.
A final potential limitation to this study is that it is restricted to patients for whom blood cultures were ordered in the inpatient or emergency department environment. It is our experience that blood cultures indicate a high clinical suspicion for infection, even if a commitment to broad‐spectrum antibiotic therapy has not yet been made by the clinician. It is clear from our data that even among these patients, who have a higher pre‐test probability for sepsis, a significant portion still do not receive broad‐spectrum antibiotics within 3 h. Therefore, there is clinical value to be gained from implementation of an algorithm which quantifies appropriateness of broad‐spectrum antibiotics in the “blood cultured” population. Tools to identify sepsis among patients for whom concern for infection has been completely missed are needed. However, studies that focus on broad patient populations with low prevalence of disease face methodological challenges as traditional machine‐learning algorithms optimize for overall error rate, weigh positives, and negatives equally, and generally lead to a low positive predictive value. 31 , 32
On the other hand, blood cultures represent a simple inclusion criteria that leverages clinical expertise yet is still broader than commonly applied criteria such as SIRS‐positive 33 or ICU‐only patients. 34 Roughly 50% of our patient population is SIRS‐negative (25% of which were septic) and the majority of our patients (>85%) were never transferred to the ICU, so more restrictive inclusion criteria would severely limit the population our tool could be useful for.
Methods for identifying patients in need of broad‐spectrum antibiotic therapy and facilitating prompt initiation of that therapy are among the greatest immediate needs for the reduction of morbidity, mortality, and healthcare costs in the United States. The challenge of sepsis, which can present with vague and confounding signs and symptoms, is that a decision to administer antibiotics is typically made with a great deal of uncertainty. A tool identifying patients with—or soon to develop—sepsis may facilitate appropriate treatment significantly. We have demonstrated the ability of a machine‐learning based score using clinical data from the EMR and three nonroutinely measured biomarkers trained on a surveillance definition of sepsis to both differentiate between various strata of sepsis and reflect severity of illness when discretized into three risk categories. An early identification tool based on this approach may be used to guide clinicians and staff across the care team and facilitate prioritization of broad‐spectrum antibiotic administration for medium or high‐risk category patients, leading to improved clinical outcomes.
CONFLICT OF INTEREST
I.T., G.L.D., C.L.E., S.K., L.S., S.S.T., L.Q., S.M., B.R.J., and R.B. have financial interests in Prenosis Inc. All other authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
I.T., G.L.D., C.L.E., and B.R.J. wrote the manuscript. B.R.J. and R.B. designed the research. S.K., L.S., S.S.T., L.Q., S.M., E.V., K.W., J.K., S.M., A.V., L.Y., G.L.D., J.S., M.D., S.A., R.M., R.U., A.S., T.B., J.D., C.H., and G.P. performed the research. I.T., C.L.E., S.D.Z., R.Z., and B.R.J. analyzed the data.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Vasantha Reddi, Nandini Goswami, Ali Moll, Debby Vannoy, and Jennifer Eardley from Carle Foundation Hospital and Savannah Cranford, Susan Peterson, Kimberly Hartwig, and Sara Riggenbach from OSF Healthcare for their help in conducting the clinical studies.
Ishan Taneja and Gregory L. Damhorst contributed equally to this work.
Funding information
No funding was received for this work.
REFERENCES
- 1. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis‐3). JAMA. 2016;315:801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferrer R, Martin‐Loeches I, Phillips G, et al. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour. Crit Care Med. 2014;42:1749‐1755. [DOI] [PubMed] [Google Scholar]
- 3. Gaieski DF, Mikkelsen ME, Band RA, et al. Impact of time to antibiotics on survival in patients with severe sepsis or septic shock in whom early goal‐directed therapy was initiated in the emergency department. Crit Care Med. 2010;38:1045‐1053. [DOI] [PubMed] [Google Scholar]
- 4. Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589‐1596. [DOI] [PubMed] [Google Scholar]
- 5. Puskarich MA, Trzeciak S, Shapiro NI, et al. Association between timing of antibiotic administration and mortality from septic shock in patients treated with a quantitative resuscitation protocol. Crit Care Med. 2011;39:2066‐2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu VX, Fielding‐Singh V, Greene JD, et al. The timing of early antibiotics and hospital mortality in sepsis. Am J Respir Crit Care Med. 2017;196:856‐863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Filbin MR, Lynch J, Gillingham TD, et al. Presenting symptoms independently predict mortality in septic shock: importance of a previously unmeasured confounder. Crit Care Med. 2018;46:1592‐1599. [DOI] [PubMed] [Google Scholar]
- 8. Barbash IJ, Davis B, Kahn JM. National performance on the Medicare SEP‐1 sepsis quality measure. Crit Care Med. 2018;47:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seymour CW, Gesten F, Prescott HC, et al. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med. 2017;376:2235‐2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pulia MS, Redwood R, Sharp B. Antimicrobial stewardship in the management of sepsis. Emerg Med Clin North Am. 2017;35:199‐217. [DOI] [PubMed] [Google Scholar]
- 11. Delahanty RJ, Alvarez J, Flynn LM, Sherwin RL, Jones SS. Development and evaluation of a machine learning model for the early identification of patients at risk for sepsis. Ann Emerg Med. 2019;73(4):334‐344. [DOI] [PubMed] [Google Scholar]
- 12. Henry KE, Hager DN, Pronovost PJ, Saria S. A targeted real‐time early warning score (TREWScore) for septic shock. Sci Transl Med. 2015;7:299ra122. [DOI] [PubMed] [Google Scholar]
- 13. Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med. 2018;46(4):547‐553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hotchkiss RS, Moldawer LL, Opal SM, Reinhart K, Turnbull IR, Vincent JL. Sepsis and septic shock. Nature Reviews Disease Primers. 2016;2(1):16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kesselmeier M, Scherag A. Commentary: arguing for adaptive clinical trials in sepsis. Front Immunol. 2018;9:2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taneja I, Reddy B, Damhorst G, et al. Combining biomarkers with EMR data to identify patients in different phases of sepsis. Sci Rep. 2017;7:10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bloos F, Reinhart K. Rapid diagnosis of sepsis. Virulence. 2014;5:154‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma L, Zhang H, Yin Y, et al. Role of interleukin‐6 to differentiate sepsis from non‐infectious systemic inflammatory response syndrome. Cytokine. 2016;88:126‐135. [DOI] [PubMed] [Google Scholar]
- 19. Lin KH, Wang FL, Wu MS, et al. Serum procalcitonin and C‐reactive protein levels as markers of bacterial infection in patients with liver cirrhosis: a systematic review and meta‐analysis. Diagn Microbiol Infect Dis. 2014;80:72‐78. [DOI] [PubMed] [Google Scholar]
- 20. Schuetz P, Birkhahn R, Sherwin R, et al. Serial procalcitonin predicts mortality in severe sepsis patients: Results from the multicenter procalcitonin monitoring SEpsis (MOSES) Study. Crit Care Med. 2017;45:781‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ryoo SM, Han KS, Ahn S, et al. The usefulness of C‐reactive protein and procalcitonin to predict prognosis in septic shock patients: a multicenter prospective registry‐based observational study. Sci Rep. 2019;9(1):6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rhee C, Dantes R, Epstein L, et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009–2014. JAMA. 2017;318:1241‐1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. CDC . Hospital Toolkit for Adult Sepsis Surveillance. CDC; 2018. https://www.cdc.gov/sepsis/pdfs/Sepsis‐Surveillance‐Toolkit‐Aug‐2018_508.pdf [Google Scholar]
- 24. Wright MN, Ziegler A. Ranger: a fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw. 2017;77:1‐17. [Google Scholar]
- 25. Breiman L. Random forests. Mach Learn. 2001;45:5‐32. [Google Scholar]
- 26. Kuhn M. caret: Classification and Regression Training. 2018. https://cran.r‐project.org/web/packages/caret/index.html
- 27. Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26:1340‐1347. [DOI] [PubMed] [Google Scholar]
- 28. Crouser ED, Parrillo JE, Seymour CW, et al. Monocyte distribution width: a novel indicator of sepsis‐2 and sepsis‐3 in high‐risk emergency department patients. Crit Care Med. 2019;47:1018‐1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis for the third international consensus definitions for sepsis and septic shock (Sepsis‐3). JAMA. 2016;315:762‐774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McHugh LC, Snyder K, Yager TD. The effect of uncertainty in patient classification on diagnostic performance estimations. PLoS One. 2019;14:1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saito T, Rehmsmeier M. The precision‐recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10:1‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen C, Liaw A, Breiman L. Using random forest to learn imbalanced data. Discovery 1–12. https://statistics.berkeley.edu/sites/default/files/tech‐reports/666.pdf
- 33. Miller RR, Lopansri BK, Burke JP, et al. Validation of a host response assay, SeptiCyte LAB, for discriminating sepsis from systemic inflammatory response syndrome in the ICU. Am J Respir Crit Care Med. 2018;198:903‐913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Desautels T, Calvert J, Hoffman J, et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Informatics. 2016;4:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


