Abstract
Background
The outbreak of Coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 infection has become a global health emergency. We aim to decipher SARS-CoV-2 infected cell types, the consequent host immune response and their interplay in lung of COVID-19 patients.
Methods
We analyzed single-cell RNA sequencing (scRNA-seq) data of bronchoalveolar lavage fluid (BALF) samples from 10 healthy donors, 6 severe COVID-19 patients and 3 mild recovered patients. The expressions of SARS-CoV-2 receptors (ACE2 and TMPRSS2) were examined among different cell types. The immune cells infiltration patterns, their expression profiles, and interplays between immune cells and SARS-CoV-2 target cells were further investigated.
Findings
Compared to healthy controls, ACE2 and TMPRSS2 expressions were significantly higher in lung epithelial cells of COVID-19 patients, in particular club and ciliated cells. SARS-CoV-2 activated pro-inflammatory genes and interferon/cytokine signaling in these cells. In severe COVID-19 patients, significantly higher neutrophil, but lower macrophage in lung was observed along with markedly increased cytokines expression compared with healthy controls and mild patients. By contrast, neutrophil and macrophage returned to normal level whilst more T and NK cells accumulation were observed in mild patients. Moreover, SARS-CoV-2 infection altered the community interplays of lung epithelial and immune cells: interactions between the club and immune cells were higher in COVID-19 patients compared to healthy donors; on the other hand, immune-immune cells interactions appeared the strongest in mild patients.
Interpretation
SARS-CoV-2 could infect lung epithelium, alter communication patterns between lung epithelial cells and immune system, and drive dysregulated host immune response in COVID-19 patients.
Keywords: COVID-19, SARS-CoV-2, scRNA-seq, lung epithelium, Dysregulated immune response
Research in context.
Evidence before this study
The Coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), becomes a global threat to humanity. In severe COVID-19 patients, excessive proinflammatory responses and impaired host immune system are observed which could result in the progression of acute respiratory distress syndrome (ARDS) and even death. In-depth investigation of SARS-CoV-2 infected cell types, the consequent host immune response, and the communication patterns between infected cells and immune system could facilitate the identification of COVID-19 patients and their optimal treatments.
Added value of this study
By analyzing single-cell RNA sequencing data of bronchoalveolar lavage fluid samples from COVID-19 patients and healthy donors, our study revealed high expression of SARS-CoV-2 receptor ACE2 and TMPRSS2 in lung epithelial cells of COVID-19 patients, in particular club and ciliated cells. SARS-CoV-2 infection induced pro-inflammatory genes expression and interferon/cytokine signaling in these cells. In severe COVID-19 patients, remarkably high neutrophil but low macrophage cells in lung were observed along with excessive expressions of cytokines. Moreover, SARS-CoV-2 infection altered the community interplays among lung epithelial and immune cells, and the dysregulated cytokine/receptor interactions were correlated with severity of COVID-19.
Implication of all the available evidence
Our study identified critical cell type, dysregulated immunity and their interactions in the lung of SARS-CoV-2 infection, thus provided new insight into the mechanisms underlying of SARS-CoV-2 infection in COVID-19 patients.
Alt-text: Unlabelled box
1. Introduction
The Coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), poses a tremendous global challenge. SARS-CoV-2 belongs to coronaviruses family which are single-stranded and positive-sense RNA viruses characterized by club-like spike on their surface [1]. SARS-CoV-2 binds to the surface expressed proteins, angiotensin-converting enzyme 2 (ACE2), to entry into cells which is similar as SARS-CoV [2], [3], [4]. In addition to ACE2, the expression of serine protease TMPRSS2 on target cells is required for activation of viral spike (S) proteins to facilitate viral entry [4]. It is thus important to investigate ACE2- and TMPRSS2- expressing cell types and their expression level in the lung of COVID-19 patients.
Although SARS-CoV-2 could be recognized by the host immune system to mount an antiviral response [3,5], imbalanced immune responses have been observed in most patients, as exemplified by high neutrophil to lymphocyte ratio [6], [7], [8], [9]. Moreover, a large number of severe COVID-19 patients suffered cytokine storm with markedly release of proinflammatory cytokines such as interleukin 6 (IL-6), interleukin 10 (IL-10) and tumor necrosis factor (TNF)-α, leading to the progression of acute respiratory distress syndrome (ARDS) and potentially death [6,10]. However, it is still unclear how SARS-CoV-2 infection contributes to dysregulated immune response in the lung of COVID-19 patients.
In this study, we comprehensively evaluated the single cell sequencing data from bronchoalveolar lavage fluid (BALF) samples of 19 subjects (6 severe COVID-19 patients, 3 recovered COVID-19 patients with mild symptoms and 10 healthy donors) to uncover cell types with ACE2 and TMPRSS2 expression in the lung infected with SARS-CoV-2. We further investigated the patterns of immune cells infiltration and their expression profiles across the different severity of infected patients as compare to the healthy controls. We finally evaluated the crosstalk between lung epithelial cells and immune cells.
2. Methods
2.1. Data availability
Single cell RNA sequence (scRNA-Seq) data were retrieved from published resources, including bronchoalveolar lavage fluid (BALF) from 6 severe and 3 moderate COVID-19 patients [11], and 10 healthy transplant donors. Bulk RNA-Seq data in three SARS-CoV-2 treated cell lines were obtained for validation purpose, including primary human bronchial epithelial cells (NHBE), Calu-3 and A549-ACE2 (with vector expressing human ACE2) [12]. All relevant data were downloaded from Gene Expression Omnibus under the accession number GSE145926, GSE151928 and GSE147507.
2.2. scRNA-Seq data analysis
We re-analyzed the data from a count quantification matrix due to the un-available per-cell annotation. Cells with mitochondrial gene proportion higher than 15% were filtered out. For each individual dataset, raw count matrix was first normalized and the top 2000 most variable genes were chosen. For each cell, we divided the gene counts by the total counts and multiplied by 10,000, followed by natural-log transformation. High variable genes were determined using FindVariableFeatures in Seurat pipeline [13]. To remove batch effects, following recommended batch-effect correction workflow of Seurat (v3.2.2), we scaled each dataset, selected 2000 high-variable genes as input to compute the integration anchors (FindIntegrationAnchors), and then integrated (IntegrateData) the batches using the anchors. Multiple datasets were then integrated via searching the “anchors” among them [13], enabling us to explore shared cell types presented across different datasets and conditions. The integrated data were scaled followed by principal component analysis (PCA), and we retained the top 30 principal components (PCs) for further analysis. To visualize the cells, we applied the t-distributed stochastic neighbor embedding (t-SNE) on the top PCs. The selected PCs were also used for computing nearest-neighbour graphs and for clustering the cells. To re-annotate the cells, we identified the conserved markers for each cluster across different conditions, by comparing with all remaining clusters using FindConservedMarkers method in Seurat pipeline. Markers specific for major cell types were listed below: EPCAM (epithelial), PTPRC (immune), PECAM1 (Endothelia), and PDGFRA (Firboblasts). To identify different immune cell types, we re-clustered non-epithelial cells into 22 subclusters. For epithelial and non-epithelial clusters, we annotated cell identity by the expressions of known marker genes [14]. MAST [15] algorithm was used to identify the altered genes under SARS-CoV-2 infection for the epithelial-related (EPCAM+) and immune-related (CD45+) clusters, respectively.
2.3. Regulatory network inference
We constructed gene regulatory networks using SCENIC (pySCENIC, v0.11.2) [16] to predict transcription factors (TFs) activities for scRNA-seq data. Briefly, TFs and target genes co-expression modules were firstly inferred via regression-based network inference using GENIE3. Subsequently, those modules were pruned via motif enrichment analysis by cisTarget. After the putative modules (regulon) were defined, the method AUCell was used to score individual cell for the relative biological activity of regulon.
2.4. Cell-cell interaction analysis using CellPhoneDB
A systematic analysis of cell-cell communication was performed by CellPhoneDB package (www.cellphonedb.org), which explores the ligand-receptor interactions based on expression of ligand / receptor between different cell types (clusters). We chose the receptors and ligands expressed in more than 10% of the cells in the specific cluster for subsequent analysis. To minimize the effects with unequal cell numbers, all cell clusters were randomly down-sampled to the size of 100, which was closed to cell number of the smallest cluster. This procedure was repeated 100 times and the average receptor expression level of a cluster and the average ligand expression level of the interacting cluster were calculated. P < 0.05 was considered as significant cell–cell interaction.
3. Functional analysis
Functional enrichment analysis was used to identify classes of molecules (genes or proteins) that were over-represented in a set of pre-defined molecules and predicted its association with disease phenotypes. We performed this method to uncover potential biological function shift under SARS-CoV-2 infection through mapping the molecules into known molecule sets by WebGestalt [17].
4. Bulk RNA-Seq data analysis
RNA-seq reads were mapped onto the human reference (GRCh38 with gene annotations GENCODE v30) by HISAT2 (version 2.1.0) with the default options. The number of reads mapped to each of genes was counted by using featureCount (version v1.6.4). Gene expression levels were calculated as FPKM (Fragments per Kilobase of transcript per Million mapped reads) by rpkm method in edgeR. Differentially expressed genes (DEGs) were determined using DESeq2.
5. Statistics
The statistical analyses used have been described in each subsection. SCOPIT [18] was carried out to determine the minimum number of cells that were required for analysis. Randomisation, blinding, inclusion/exclusion criteria were not applicable for this study, and available single cell RNA sequence (scRNA-Seq) data of BALF samples from COVID-19 patients and healthy donors were retrieved from published resources for analysis. In this study, we applied a flexible generalized linear model, MAST [15], to characteristic bimodal expression distributions in the scRNA-seq data. We detected the conserved markers among t-SNE clusters (FindConservedMarkers) and differentially expressed genes among groups (Healthy, Severe and Mild) for different cell cluster(s) (FindMarkers). To account for multiple testing, P values were adjusted using bonferroni correction.
6. Role of funding source
Funders of this study had no role in study design, data collection, data analyses, interpretation, or writing of the report.
7. Results
7.1. Lung epithelial cells express higher ACE2 and TMPRSS2 in COVID-19 patients
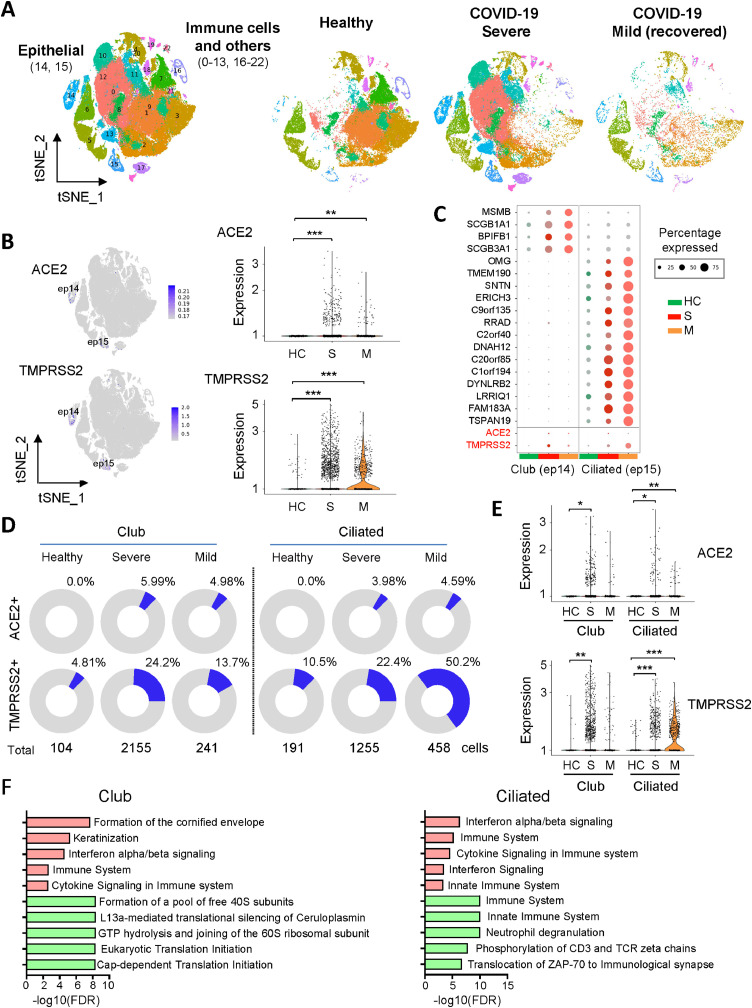
To examine the expressions of SARS-CoV-2 entry genes, ACE2 and TMPRSS2, in different cell types of human lung after SARS-CoV-2 infection, single-cell RNA sequencing (scRNA-seq) data of lung bronchoalveolar lavage fluid (BALF) from 3 recovered mild cases and 6 severe cases (GSE145926) [11], as well as 10 healthy donors (GSE151928) [19] were retrieved from NCBI database. Unsupervised analysis identified 23 distinct cell clusters (Fig. 1A and Figure S1), including epithelial (EPCAM+) and immune (PTPRC+) cell populations (Figure S2). ACE2 and TMPRSS2 were primarily expressed in lung epithelial cells (Fig. 1B), in line with other studies [20,21]. Among lung epithelial populations, a relative high percentage of ACE2 or TMPRSS2 positive cells were shown in club and ciliated cells which may act as primary target cells of SARS-CoV-2 infection (Fig. 1C). Notably, the percentages of ACE2 or TMPRSS2 positive cells among these two types of lung epithelial cells were all significantly higher in BALF samples from either severe or mild COVID-19 patients as compared to healthy controls (Fig. 1D). In keeping with this, the ACE2 or TMPRSS2 mRNA expression level was significantly higher in COVID-19 patients compared to healthy controls in club and ciliated cells (Fig. 1E). However, the correlation between increased ACE2 and TMPRSS2 expressions in the lung epithelial cells from COVID-19 patients and SARS-CoV-2 infection needs further in-depth investigation, considering the small sample size in this study and the treatment administrated to these patients.
Fig. 1.
High ACE2 and TMPRSS2 expression in lung epithelial cells from COVID-19 patients. (A) The t-SNE plot displayed the major cell types (epithelial, immune and others) in 23 clusters for bronchoalveolar lavage fluid (BALF) samples from 6 severe (S) and 3 recovered mild COVID-19 patients (M), as well as 10 healthy controls (HC). (B) The t-SNE plot displayed RNA expression of ACE2 or TMPRSS2. Right panel shows ACE2 or TMPRSS2 expression in lung epithelial cells from different groups using violin plot. (C) Dot plot of ACE2 or TMPRSS2 expression for each cell-type of lung epithelial cells from different groups. Dot size represents the percentage of cells in individual clusters expressing a given gene. (D) The pie chart shows the percentages of ACE2- or TMPRSS2- positive cells in club (cluster 14) and ciliated (cluster 15) cells. (E) Violin plot of expression values of ACE2 or TMPRSS2 in different cell types of lung epithelial cells from different group. (F) The top 5 enriched signaling pathways of up-regulated (red) or down-regulated (green) genes in lung epithelial cells after SARS-CoV-2 infection (severe vs. health). * P < 0.05; ** P < 0.01; *** P < 0.001.
7.2. SARS-CoV-2 leads to cellular transcriptome alterations in lung epithelial cells
We next investigated cellular transcriptome alterations of lung epithelial cells in response to SARS-CoV-2 infection. Profoundly altered gene transcriptional expressions in club and ciliated cells were present in severe COVID-19 patients (Figure S3). In club cells, we detected 107 up-regulated and 65 down-regulated transcripts in severe COVID-19 patients as compared to healthy control (adjusted p ≤ 0.01 and |log2Fold change (FC)| ≥ 1) (Table S1). On the other hand, 162 up-regulated and 138 down-regulated transcripts (adjusted p ≤ 0.01 and |log2Fold change (FC)| ≥ 1) after SARS-CoV-2 infection were identified in ciliated cells (Table S1). Over-representation analysis of these candidate genes revealed that SARS-CoV-2 infection induced interferon pathway and cytokine signaling in the lung epithelial cells of severe COVID-19 patients (Fig. 1F). On the other hand, SARS-CoV-2 was capable to suppress host protein translation in club cells (Fig. 1F). Furthermore, comparative analysis of lung epithelial cells transcriptome between mild and severe COVID-19 patients indicated that T cell activation signaling such as MHC class II antigen presentation and Phosphorylation of CD3 and TCR zeta chains were induced but the cytokine signaling was inhibited in mild patients as compared to severe patients (Figure S4). Intriguingly, major histocompatibility complex (MHC) class II genes, including HLA-DR, HLA-DQ, HLA-DP and HLA-DM, were found significantly decreased in lung epithelial cells of severe COVID-19 patients as compared to healthy controls (Figure S3 and Table S1). This may partially explain the failure of T cells induction and poor patient outcome. On the other hand, expressions of MHC class II genes were restored in mild recovered COVID-19 patients (Table S2).
7.3. SARS-CoV-2 infection drives lung immune response
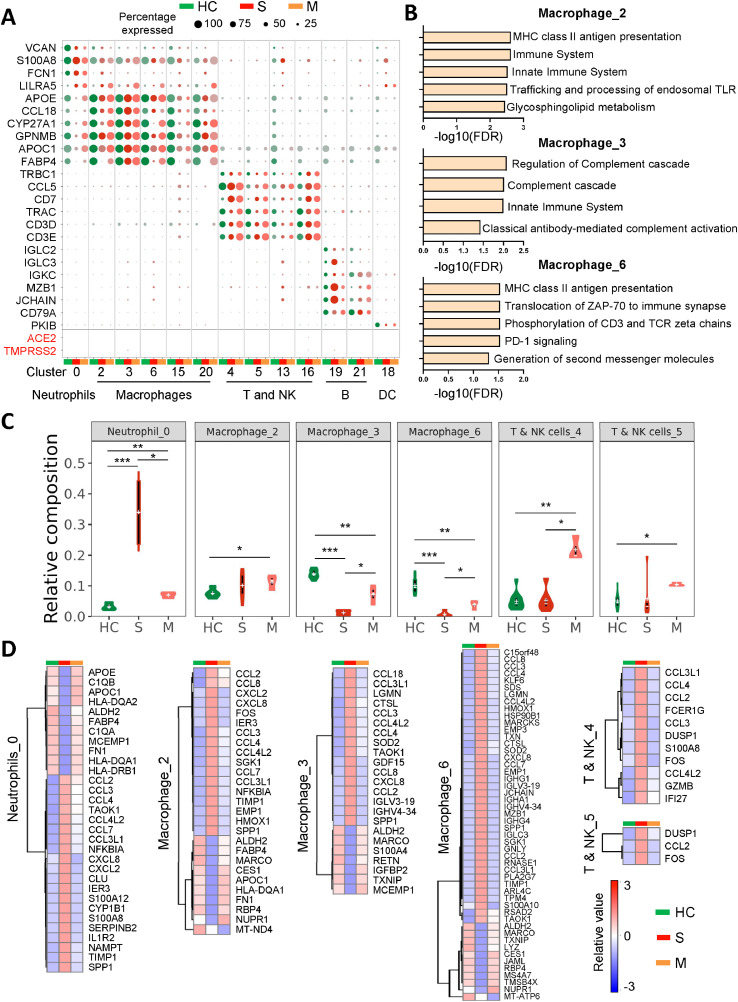
We further studied the specification of immune cells fates in response to SARS-CoV-2 infection. The non-epithelial cells were re-clustered into 22 subclusters and different immune cell types were identified (Figure S5), based on their typical markers [14]. As shown in Fig. 1B and Fig. 2A, ACE2 or TMPRSS2 was almost not expressed in the immune cells of BALF samples from COVID-19 patients, implying that the immune cells might be not susceptible to SARS-CoV-2 infection. We next studied three macrophage subclusters (cluster 2, 3 and 6), two T & NK cells subclusters (cluster 4 and 5), as well as one neutrophils subcluster (cluster 0) which contained sufficient cell number for analysis (Figure S6). Over-representation analysis of subcluster-specific genes revealed activation of MHC class II antigen presentation signaling in Macrophage_2 and Macrophage_6; in contrast, Macrophage_3 could carry out complement effector function (Fig. 2B). We also investigated differential regulon activities of transcription factors (TFs) among different epithelial and immune cell subtypes using SCENIC, and multiple TFs associated with specific cell types showed enriched regulon activity including TCF7L2 in macrophage subclusters, and GATA3 and NFATC2 for T and NK subclusters (Figure S7). By comparing different immune cell populations between COVID-19 patients and healthy controls, we found a dysregulated immune response in the lungs after SARS-CoV-2 infection (Fig. 2C and S8). A massive increase of neutrophils was observed in severe COVID-19 patients as compared with healthy controls, while the level was restored to normal after the patients recovered (Fig. 2C). Whilst macrophage number (cluster 3 and 6) was significantly lower in severe COVID-19 patients compared to healthy controls, they were restored in recovered patients (Fig. 2C). Meanwhile, T/NK cells were only induced in recovered but not severe COVID-19 patients as compared to healthy controls (Fig. 2C).
Fig. 2.
SARS-CoV-2 infection induced imbalanced host immune response in severe COVID-19 patients. (A) Dot plot of ACE2 or TMPRSS2 expression for each cell-type of lung immune cells from different groups. Dot size represents the percentage of cells in individual clusters expressing a given gene. (B) Top 5 enriched signaling pathways of markers genes for macrophage (cluster 2, 3 and 6). (C) The percentages of different immune cell types of all CD45+ cells in BALF of severe (S), recovered mild COVID-19 patients (M), and healthy controls (HC). (D) Heatmaps of transcript level of candidate genes in different immune cell types. * P < 0.05; ** P < 0.01; *** P < 0.001.
We then explored the differential gene expression profiling of immune cells in the lung between COVID-19 patients and healthy controls. Differential gene expression patterns of neutrophil, macrophage and T/NK cells were demonstrated in severe COVID-19 patients, mild recovered COVID-19 patients and healthy controls (Fig. 2D and S9). In severe COVID-19 patients, we identified a variety of cytokines which were markedly increased in neutrophil, macrophage, and T/NK cells (CCL2, CCL3, CCL3L1, CCL4 and CCL4L2), and in neutrophil and macrophage (CCL7 and CXCL8) (Fig. 2D). The concomitant high expression of these cytokines derived from dysregulated immune cells attracted by SARS-CoV-2 infection suggested occurrence of cytokine storm in these patients. Of note, FABP4 was highly expressed in macrophage_2 of healthy controls and mild patients as compared to severe COVID-19 patients; by contrast, SPP1 were highly expressed in macrophages (cluster 2, 3 and 6) of severe COVID-19 patients (Fig. 2D). These observations were in line with previous study [11]. Moreover, MARCO expression in macrophages was suppressed in severe COVID-19 patients, implying that the lung alveolar macrophages may fail to clean up neutralized viruses after SARS-CoV-2 infection (Fig. 2D).
7.4. SARS-CoV-2 caused abnormal epithelial-immune cell interaction in lung
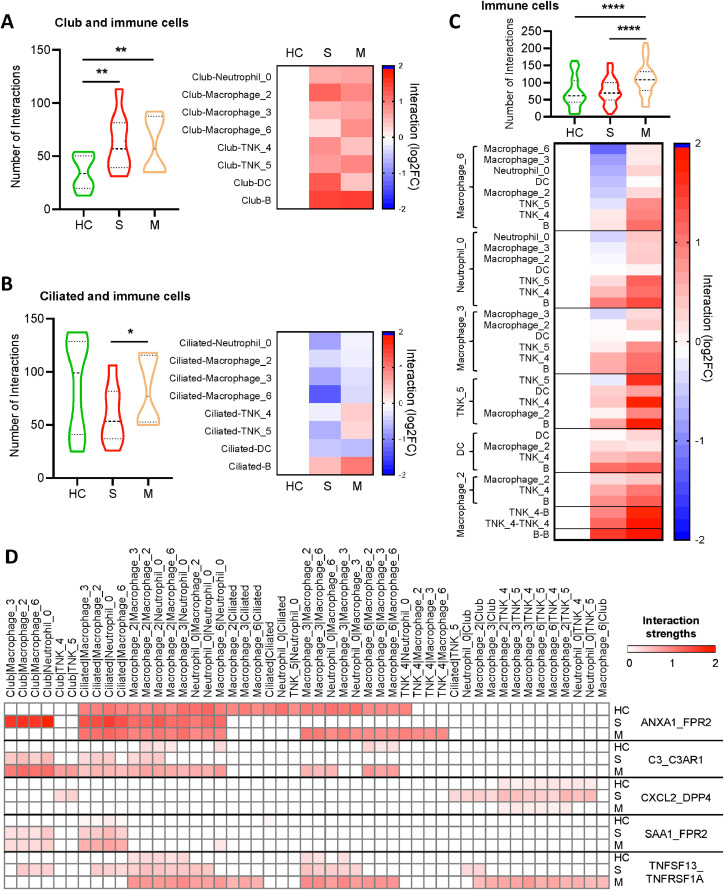
We next explored the intercellular communications within lung based on expression of ligand–receptor pairs among different epithelial and immune cell types by CellPhoneDB [22]. The number of significant cell interactions (P < 0.05) was calculated. We found club-immune cells interactions (Fig. 3A), but not ciliated-immune cells interactions (Fig. 3B), were significantly increased in COVID-19 patients as compared to healthy controls. As to immune-immune cells communications, BALF samples of mild patients presented more cells interactions than those of severe patients and healthy donors (Fig. 3C). To understand whether these interactions correlated with COVID-19 severity, we further looked into specific cytokine/receptor interactions among epithelial and immune cells. We found occurrence of ANXA1/FPR2 and TNFSF13/TNFRSF1A interactions between club and macrophage (or Neutrophil) cells, and CXCL2/DPP4 interaction between club and T/NK cells in severe COVID-19 patients, but not in mild patients or healthy controls, indicating that cytokine/receptor interactions between lung epithelial and immune cells are associated with severity of COVID-19 infection (Fig. 3D). Intriguingly, SARS-CoV-2 might gain entry into host cells through DPP4, and inhibition of DPP4 potentially benefits diabetes patients with COVID-19 [23]. Furthermore, we found that SARS-CoV-2 infection induced both C3/C3AR1 and SAA1/FPR2 interactions between lung epithelial cells (club and cilliated) with macrophage (or Neutrophil); loss of ANXA1/FPR2 interactions among macrophages were observed in severe COVID-19 patients; and TNFSF13/TNFRSF1A interaction appeared between macrophages and T/NK cells in mild patients, but not in severe patients or healthy donors (Fig. 3D). We further validated ANXA1, C3, CXCL2, SAA1 and TNFSF13 expressions in lung epithelial cells after SARS-CoV-2 infection. The result demonstrated that the expression of ANXA1, C3 and SAA1 were all significantly higher in lung epithelial cells of severe COVID-19 patients as compared to healthy controls (Figure S10). An increase trend was observed for CXCL2, but not TNFSF13, in lung epithelial cells after SARS-CoV-2 infection (Figure S10). In supporting this, SARS-CoV-2 infection significantly increased the mRNA expression of C3, CXCL2 and SAA1 in human lung cancer cell line A549 overexpressing ACE2, lung cancer cell line Calu-3, and human normal bronchial epithelial cells (NHBE) (Figure S10). Our findings suggest that the specific networks between epithelial cells and immune cells were formed in lung after SARS-CoV-2 infection which might contribute to severity of COVID-19 infection.
Fig. 3.
SARS-CoV-2 infection resulted in abnormal epithelial-immune cell interaction in lung. (A-C) We evaluated intercellular communications based on expression of ligand–receptor pairs among different cell types by CellPhoneDB. Club-immune cells (A), ciliated-immune cells (B), and immune-immune communications (C) in severe (S, n=6), recovered mild (M, n=3) COVID-19 patients, and healthy controls (HC, n=10) were shown. Only significant interactions were calculated, and the number of interactions was depicted in violin plot, or normalized against those of healthy controls in heatmap. (C) Heat map depicting different ligand–receptor interactions among different cell types. Interaction strengths are color coded. [One-way Analysis of Variance (ANOVA) followed by Tukey multiple comparisons (A-C)]. * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001.
8. Discussion
In this study, we identified high expressions of ACE2 and TMPRSS2 in ciliated and club cells of lung epithelium in COVID-19 patients. ACE2 and TMPRSS2 are two critical entry genes required for SARS-CoV-2 infection [4]. The expression of ACE2 has been reported in a variety of tissues including respiratory tract and gastrointestinal mucosa [24]. ACE2 and TMPRSS2 mRNA were expressed in lung type II pneumocytes, ileal absorptive enterocytes, and nasal goblet secretory cells [21], and their protein expressions were detected in nasal and bronchial epithelium [25]. Our study provides some novel information on the cell types of lung epithelial cells expressing ACE2 and TMPRSS2, and their expression levels in COVID-19 patients based on single cell sequencing analyses. However, the expressions of ACE2 and TMPRSS2 were not found in immune cells in the BALF of COVID-19 patients. In keeping this, no SARS-CoV-2 viral gene expression was detected in peripheral blood mononuclear cell in three SARS-CoV-2 patients [26]. Thus, lung epithelial cells are more susceptible to SARS-CoV-2 infection as compared to immune cells.
We further demonstrated that SARS-CoV-2 led to host cellular transcriptome alterations in club and ciliated cells of COVID-19 patients, resulting in activation of interferon pathway and cytokine signaling. These secreted signaling molecules serve to initiate host immune response by recruiting various immune cells [27]. Intriguingly, reduced expressions of MHC class II genes such as HLA-DR, HLA-DQ, HLA-DP and HLA-DM were identified in lung epithelial cells of severe COVID-19 patients, and their expressions were restored in mild recovered patients. MHC class II molecules play a key role in adaptive immune responses, and are responsible for presenting pathogen-derived peptide to T cells [28]. As a consequence, massive infiltration of neutrophils, but not T cells, was observed in severe COVID-19 patients. Moreover, the accumulated immune cells (neutrophils, macrophage and T/NK cells) in the lung of severe COVID-19 patients expressed significantly higher levels of cytokines including CCL2, CCL3, CCL3L1, CCL4, CCL4L2, CCL7 and CXCL8, and finally formed a cytokine "hurricane". Consistently, a recent study reported that CCL2 and CCL3 cytokines released by the lung may contribute to lung tissue damage [29]. Thus targeting cytokines could benefit severe COVID-19 patients. We also noticed that macrophages of severe COVID-19 patients expressed less MARCO. MARCO is expressed on alveolar macrophages and thought to promote protective innate immunity against infection [30]. Thus the immune system of severe COVID-19 patients failed to clean up SARS-CoV-2. Taken together, SARS-CoV-2 infection could induce aberrant genes expressions, enriched pro-inflammation signaling, and the dysregulated host immune response in the lung of COVID-19 patients.
We went further to look into the intercellular communications within lung using CellPhoneDB [22]. The interaction networks between epithelial-immune cells or immune-immune cells were quite different among healthy donors, severe and mild patients. We revealed more club-immune cells interactions in lung of COVID-19 patients as compared to healthy controls. In addition, BALF samples of mild patients presented more immune-immune cells communications than those of severe patients and healthy donors. In particular, ANXA1/FPR2 interaction occurred between club and macrophage (or neutrophil) cells, but diminished between macrophage and neutrophils in severe COVID-19 patients. ANXA1 is commonly highly expressed in immune cells such as neutrophils and macrophages. ANXA1 could bind to and activate FPR2 receptor, and exert anti-inflammatory effects [31,32]. Therefore, the ANXA1/FPR2 interaction switch, or loss of ANXA1/FPR2 interaction in macrophage/neutrophil, may result in hyperinflammatory response in severe COVID-19 patients. Moreover, CXCL2/DPP4 interaction occurred between club and T/NK cells in severe COVID-19 patients, but not in mild patients or healthy controls. DPP4 is highly expressed on T cells and inhibition of DPP-4 promotes accumulation of anti-inflammatory T cells and anti-inflammatory cytokines [33]. Targeting DPP4 may ameliorate severity of SARS-CoV-2 infection given that DPP4 could favor SARS-CoV-2 entry into cells [23]. Our findings suggest that dysregulated cytokine/receptor interactions in the lung could result in severity of COVID-19 infection.
Our study has some limitations. First, the sample size used was relatively small. Therefore we could not examine the correlation between the patients’ demographic characteristics such as age, gender and ethnicity, with aberrant gene expression profiling and dysregulated lung immune response in the lung of COVID-19 patients. Second, although our analysis indicated that SARS-CoV-2 infection induced aberrant gene expression profiling of lung epithelial and immune cells, and disrupted their interplays, validation studies using cell line and animal models are needed in future.
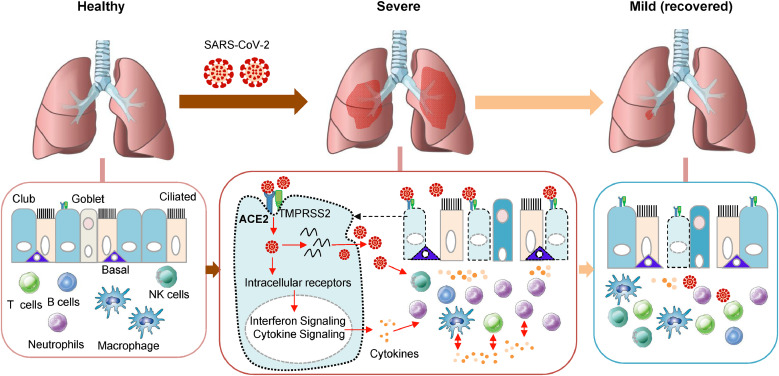
In conclusion, SARS-CoV-2 infection induces aberrant gene expression profiling and activation of pro-inflammatory signaling of lung epithelium cells (ciliated and club) that expressing high levels of ACE2 and TMPRSS2. Moreover, SARS-CoV-2 infection causes dysregulated lung immune response and massive production of pro-inflammatory cytokines and disrupts the interplays of epithelial cells and immune cells. All these contribute to severity of COVID-19 infection (Fig. 4).
Fig. 4.
SARS-CoV-2 infection and host immune response in COVID-19 patients. In COVID-19 patients, the SARS-CoV-2 may infect ciliated cells, club cells, and basal cells expressing ACE2 and TMPRSS2 in lung epithelium and actively replicate in host cells. This could lead to activation of pro-inflammatory signaling and production of pro-inflammatory cytokines which subsequently attract both innate and adaptive immune cells including neutrophils, macrophages and T cells to the infection site to fight virus and virus-infected cells. Besides, the immune cells also release cytokines to attract more immune cells, creating a positive feedback loop of cytokine creation. Massive accumulation of pro-inflammatory cytokines producing-immune cells in the lungs could increase the severity of COVID-19 patients.
Author Contributions
HRC and WXL designed the study; WXL and HRC performed data analysis; HRC, WXL, YFW, DBL and LYZ contributed to the preparation of the manuscript; JY designed, supervised the study and revised the paper.
Data Sharing Statement
All relevant data is available online at Gene Expression Omnibus under the accession number GSE145926, GSE151928 and GSE147507.
Declaration of Competing Interest
The authors declared no conflict of interest.
Funding
This project was supported by National Key R&D Program of China (No. 2018YFC1315000/2018YFC1315004), Science and Technology Program Grant Shenzhen (JCYJ20170413161534162), HMRF Hong Kong (17160862), RGC-CRF Hong Kong (C4039-19G), RGC-GRF Hong Kong (14163817), Vice-Chancellor's Discretionary Fund CUHK and CUHK direct grant, Shenzhen Virtual University Park Support Scheme to CUHK Shenzhen Research Institute.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103500.
Appendix. Supplementary materials
Reference
- 1.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26(4):453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan M, Liu Y, Zhou R, Deng X, Li F, Liang K. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160(3):261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Investig. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 12.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019;25(7):1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 15.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y, Wang J, Jaehnig EJ, Shi Z, WebGestalt ZB. 2019: gene set analysis toolkit with revamped UIs and APIs. Nucl Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis A, Gao R, Navin NE. SCOPIT: sample size calculations for single-cell sequencing experiments. BMC Bioinform. 2019;20(1):566. doi: 10.1186/s12859-019-3167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mould KJ, Moore CM, McManus SA, McCubbrey AL, McClendon JD, Griesmer CL. Airspace macrophages and monocytes exist in transcriptionally distinct subsets in healthy adults. Am J Respir Crit Care Med. 2021;203(8):946–956. doi: 10.1164/rccm.202005-1989OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protocols. 2020;15(4):1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 23.Scheen AJ. DPP-4 inhibition and COVID-19: from initial concerns to recent expectations. Diabetes Metab. 2021;47(2) doi: 10.1016/j.diabet.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532(1-2):107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 25.Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PloS One. 2012;7(4):e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberholzer A, Oberholzer C, Moldawer LL. Cytokine signaling–regulation of the immune response in normal and critically ill states. Crit Care Med. 2000;28(4):N3–12. doi: 10.1097/00003246-200004001-00002. Suppl. [DOI] [PubMed] [Google Scholar]
- 28.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–S33. doi: 10.1016/s0092-8674(02)00696-7. Suppl. [DOI] [PubMed] [Google Scholar]
- 29.Szabo PA, Dogra P, Gray JI, Wells SB, Connors TJ, Weisberg SP. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54(4):797–814. doi: 10.1016/j.immuni.2021.03.005. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanno S, Hirano S, Sakamoto T, Furuyama A, Takase H, Kato H. Scavenger receptor MARCO contributes to cellular internalization of exosomes by dynamin-dependent endocytosis and macropinocytosis. Sci Rep. 2020;10(1):21795. doi: 10.1038/s41598-020-78464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McArthur S, Juban G, Gobbetti T, Desgeorges T, Theret M, Gondin J. Annexin A1 drives macrophage skewing to accelerate muscle regeneration through AMPK activation. J Clin Investig. 2020;130(3):1156–1167. doi: 10.1172/JCI124635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scannell M, Flanagan MB, deStefani A, Wynne KJ, Cagney G, Godson C. Annexin-1 and peptide derivatives are released by apoptotic cells and stimulate phagocytosis of apoptotic neutrophils by macrophages. J Immunol. 2007;178(7):4595–4605. doi: 10.4049/jimmunol.178.7.4595. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Zheng P, Huang G, Yang L, Zhou Z. Dipeptidyl peptidase-4(DPP-4) inhibitors: promising new agents for autoimmune diabetes. Clin Exp Med. 2018;18(4):473–480. doi: 10.1007/s10238-018-0519-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell RNA sequence (scRNA-Seq) data were retrieved from published resources, including bronchoalveolar lavage fluid (BALF) from 6 severe and 3 moderate COVID-19 patients [11], and 10 healthy transplant donors. Bulk RNA-Seq data in three SARS-CoV-2 treated cell lines were obtained for validation purpose, including primary human bronchial epithelial cells (NHBE), Calu-3 and A549-ACE2 (with vector expressing human ACE2) [12]. All relevant data were downloaded from Gene Expression Omnibus under the accession number GSE145926, GSE151928 and GSE147507.