Abstract
A pilot study was conducted from late October 2020 until mid-April 2021, aiming to examine the association between SARS-CoV-2 RNA concentrations in untreated wastewater and recorded COVID-19 cases in two Greek municipalities. A population of Random Forest and Linear Regression Machine Learning models was trained and evaluated incorporating the concentrations of SARS-CoV-2 RNA in 111 wastewater samples collected from the inlets of two Wastewater Treatment Plants, along with physicochemical parameters of the wastewater influent. The model's predictions were adequately associated with the 7-day cumulative cases with the correlation coefficients (after 5-fold cross validation) ranging from 0.754 to 0.960 while the mean relative errors ranged from 30.42% to 59.46%. Our results provide indications that wastewater-based predictions can be applied in diverse settings and in prolonged time periods, although the accuracy of these predictions may be mitigated. Wastewater-based epidemiology can support and strengthen epidemiological surveillance.
Keywords: Wastewater-based epidemiology (WBE), COVID-19, SARS-CoV-2, Machine learning, RNA, RT-PCR
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped beta coronavirus responsible for the ongoing COVID-19 pandemic. Transmission of SARS-CoV-2 occurs predominantly through direct or indirect contact with infected individuals, when respiratory particles are inhaled or deposited onto exposed mucous membranes (Cevik et al., 2020). Upon exposure, the virus binds to the angiotensin-converting enzyme 2 (ACE-2) receptor, which in the respiratory system is mainly expressed on type II alveolar epithelial cells (Ni et al., 2020). Within infected cells viral RNA is replicated and translated and new viral particles are released infecting adjacent cells. The infection of the type II alveolar epithelial cells can result in various pathological findings, including the development of the Acute Respiratory Distress Syndrome (ARDS) which occurs either through mechanisms involving the release of inflammatory cytokines (cytokine storm) or by apoptosis pf the host's pneumocytes (Parasher, 2021). In addition to the respiratory system, the ACE-2 receptor has also been found to be expressed in gastrointestinal cells, and it has been hypothesized that SARS-CoV-2 can infect and replicate in the gastrointestinal tract (Ng and Tilg, 2020, Wong et al., 2020). Until present, numerus studies have confirmed the presence of SARS CoV-2 RNA in stool samples, in a significant proportion of infected individuals (Jones et al., 2020).
Wastewater-based epidemiology has been gaining increasing attention in the era of COVID-19 pandemic, as a supplementary tool for monitoring the epidemiological burden in communities and detecting trends in the dynamics of virus spread. The concept is based on the fact that infected individuals excrete SARS-CoV-2 RNA, mainly through feces (Jones et al., 2020), which is then carried through the sewerage system to the Wastewater Treatment Plants (WWTPs), where it can be detected in untreated wastewater samples. Consequently, wastewater samples acquired from the inlet of treatment plants can be considered as representative samples of the population residing in the entire catchment area. Monitoring SARS-CoV-2 spread through wastewater measurements may constitute a cost-effective approach which is not affected by biases and limitations occurring in conventional surveillance practices (e.g spatial and temporal differences in health seeking behaviors, sampling rate, contact tracing, screening etc) (Larsen and Wigginton, 2020).
It has been claimed that active carriers of the virus in a community served by a particular WWTP can be back-calculated from sewage measurements through a function incorporating the concentration of SARS-CoV-2 RNA detected in wastewater samples along with the catchment area population, the wastewater flow, the decay ratio of SARS-CoV-2 RNA in wastewater and the excretion rate of SARS-CoV-2 RNA from infected people (Ahmed et al., 2020, Li et al., 2021). However, the accurate estimation of abovementioned parameters is complex and characterized by a considerable degree of uncertainty recently reviewed by Li et al. (2021). Shading dynamics have been shown to have high inter-individual and temporal variability (Miura et al., 2021), while analytical uncertainties concerning the absolute quantitation of SARS-CoV-2 RNA in wastewater should also be kept in mind. Nonetheless, there are several studies reporting associations between SARS-CoV-2 levels in wastewater and indicators related to virus's spread. The current state of knowledge concerning this association suggests that quantitative estimates of viral load in sewage can be linked to epidemiological indicators such as cumulative incidence, estimated period prevalence and hospitalization rates (Peccia et al., 2020; Medema et al., 2020a, 2020b; Vallejo et al., 2020). The determination of magnitude, the temporal consistency and the generalizability of these associations in diverse settings, remains a critical issue to be clarified in the evaluation of WBE as a credible surveillance tool. Moreover, some reports indicate that sewage surveillance can function as a crucial early warning tool (Róka et al., 2021, Wu et al., 2020). In the present study we examined the association between SARS-CoV-2 RNA concentrations in untreated wastewater and 7-day cumulative cases in two Greek municipalities of ~150.000 residents each, during a 5 month period, from late October 2020 until mid-April 2021, when two distinctive peaks of the epidemic occurred.
2. Methods
2.1. Study settings and sampling
Wastewater samples were acquired from two different Waste Water Treatment Plants located in the cities of Larissa and Volos, Central Greece. Sixty-three wastewater samples were collected from the WWTP of Larissa between October 29, 2020 and April 14, 2021 and 48 samples were collected from WWTP of Volos between November 9, 2020 and April 14, 2021. The sewerage network in the municipality of Larissa has a length of 516 Km, and receives only municipal wastewater. It currently serves approximately 150,000 residents. Samples were obtained from the sewage inlet of the treatment plant. The vast majority of samples (58/63) were taken with the use of a Sigma SD900 portable sampler, (HACH Company, US), while the first 5 samples were collected by merging 4 different grab samples collected at 2-h intervals. The composite 24-h samples were obtained with a sampling rate of 150 ml/hour. The sewerage network in the municipality of Volos has a length of 775Km, and receives mainly municipal wastewater and a small proportion of industrial wastewater (≈5%). It currently serves approximately 155,000 residents. Samples were obtained from the sewage inlet of the treatment plant with the use of an AS950 portable sampler (HACH Company, US). Composite 24-h samples were obtained, with a sampling rate of 100ml/10min. In both WWTPs, at the end of the 24-h period, wastewater was transfused to 1.5 l containers which were immediately transported to the laboratory for analysis within isothermal boxes at 2–8 °C.
2.2. Laboratory analyses
For SARS-CoV-2 detection in wastewater, a concentration protocol based on polyethylene glycol precipitation of the virus from 105 mL of primary effluent, followed by high-speed centrifugation was applied. For each sample, a standard concentration of poliovirus Sabin 1 vaccine strain was added, as a process control virus, in order to control concentration and extraction efficiency. RNA extraction was performed on a KingFisher Flex System (ThermoFisher Scientific) using the MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit (Applied Biosystems™). Finally, for the real-time reverse transcription polymerase chain reaction (RT-PCR) the TaqPath™ COVID-19 CE-IVD RT-PCR Kit (Applied Biosystems™) (targets three different SARS-CoV-2 specific genomic regions; ORF1ab, the Spike ORF and the Nucleocapsid ORF) was used following the manufacturer's instructions, on a validated QuantStudio™ 5 Real-Time PCR System (ThermoFisher Scientific). Furthermore, for each sample a 10-1 dilution was also analyzed, and both samples and their dilutions were analyzed in duplicate. Subsequently, viral load in each sample was calculated as shown below:
| [1] |
Ct: Cycle threshold value measured for the unknown sample
b: y-intercept of the standard curve
m: slope of the standard curve
| [2] |
rnatotal: Total volume of RNA eluted from magnetic bead extraction
rnaPCR: Volume of purified RNA tested in PCR
concentratetotal: Total volume of wastewater concentrate
concentrateextracted: Volume of wastewater concentrate from which RNA was extracted
wastewater: volume of the original wastewater sample processed with PEG procedure.
In each sample, pH, electrical conductivity, total suspended solid (TSS), Biochemical Oxygen Demand (BOD), Chemical Oxygen Demand (COD), Total Nitrogen (N), Ammonium-Nitrogen (NH4+-N), total phosphorus (P) and chlorides (Cl−) were determined, according to in-house methods based on APHA et al., 2017 (APHA/AWWA/WEF, 2017).
2.3. Acquisition and processing of epidemiological and sewage data
Daily COVID-19 cases from the municipalities served by each WWTP, were obtained from the Hellenic National Public Health Organization, in collaboration with the Region of Thessaly. In the developed dataset, for each date cases were attributed based on the sampling date, not the reporting date. Epidemiological data were smoothed using cubic spline interpolation after adjusting for the week day effect. More specifically, two time points per week were used for spline interpolation; the first data point within a given week was the average value of cases from Sunday until Wednesday and the second data point within a given week was the average value of cases from Thursday until Saturday. Based on these two data points per week, the cubic spline interpolated the cases for each day of the week. This procedure was performed to reduce the “noise” and correct the systematic variation of sampling rate occurring within the same week. Cumulative cases of the previous 7 days were then calculated for each date. A similar approach was performed for smoothing of wastewater RNA concentrations to reduce the within week variability between measurements. In particular, the weekly average was appointed to the mid-day of the week (Thursday) and then cubic spline interpolation was performed. In Supplementary Figures (SF1-SF4), the actual and smoothed/adjusted daily cases and wastewater SARS-CoV-2 RNA concentrations are presented for both municipalities.
2.4. Machine learning models to determine the association between RNA concentrations in sewage water and epidemiological data
To examine the correlation between wastewater measurements and the 7-day cumulative cases, Linear Regression (LR) and Random Forest (RF) models were trained and tested with the machine learning Waikato Environment for Knowledge Analysis (Weka), using default parameters. Concerning RFs, 100 random trees were estimated, whereas the depth of each tree was not set to a certain threshold, but was allowed to be unlimited (all other parameters were default). All models were evaluated with 5- fold cross-validation. Machine learning models were trained and tested using as predictors/features the following expressions of SARS-CoV-2 concentrations: i) RNA copies/ml, ii) RNA copies/ml normalized for COD, iii) RNA copies/ml normalized for BOD, iv) RNA copies/ml normalized for TSS, v) RNA copies/ml multiplied with total flow (m3/day). Next, we evaluated the various models’ performance incorporating different mathematical expressions of the normalized values namely decimal logarithm (log10), natural logarithm (ln) and square root as well as non-transformed values. The performance of the models was evaluated by calculating the correlation coefficients, mean absolute errors and root mean squared error.
Feature selection was performed within WEKA by implementing the WrapperSubsetEval method for LR and RFs. When a certain subset of features was selected as optimal for a certain combination of training set (municipality of Larissa, or municipality of Volos or both municipality together) and for a certain algorithm (LR or RF), this feature set was also used to develop models of the other combinations of cities/algorithm. For example, based on feature selection, a certain subset of features was selected to develop a LR model of Larissa. This same subset of features was used to develop five other models, i) a LR model of Volos, ii) a RF model of Larissa, iii) a RF model of Volos, iv) a LR model of the combined data of Larissa and Volos, and v) a RF model of the combined data of Larissa and Volos. The same procedure was followed for each municipality/model combination. Thus, a large population of models was assessed and from this population, the best performing model was selected for each of the six combinations of city (Larissa, Volos or both municipalities together) and algorithm (LR or RF). In addition, Larissa and Volos specific LR and RF models were also developed by using only the square root of SARS-CoV-2 RNA concentrations or the square root of COD normalized concentrations. These 8 additional simple models were developed as a baseline to estimate the improvement of the model's performance when using the extra features. For all models incorporating more than one feature the Variance Inflation Factors (VIFs) for each feature were computed to assess multicollinearity, by using IBM SPSS Statistics, Version 22.0 (IBM Corp., Armonk, NY, USA).
3. Results
Regarding the chemical composition and properties of wastewater in the examined municipalities, the values of the physicochemical parameters measured in wastewater samples differed significantly between the two investigated WWTPs. Major differences were observed in conductivity, chlorides, total nitrogen, ammonium Nitrogen and phosphorus. The mean values for each parameter are presented in Supplementary Table 1.
Concerning the smoothing and normalization processes, we observed that the cubic-spline corrected SARS-CoV-2 concentrations were better correlated to the seven day cumulative cases than the raw wastewater RNA measurements. In addition, the correlation was further increased when the cubic-spline corrected concentrations were converted to their square-root values. For the municipalities of Larissa and Volos separately, the square-root of the viral load achieved the best correlation, whereas when the data of both municipalities were merged, the square-root of the RNA concentrations normalized for COD achieved slightly better performance. Concentrations normalized for BOD, TSS or multiplied by total flow did not demonstrate a higher correlation.
Subsequently, 14 different machine learning models were developed from different training datasets. The VIFs for each feature included in the models are presented in Supplementary Table 2. In models 1, 3 and 4 (Table 1 ) multicollinearity was observed with Total nitrogen (N) and ammonium nitrogen (NH4–N) having very high VIFs. For the main predictor (SARS-CoV-2 RNA concentration in wastewater) the VIFs were low (<2) in all models.
Table 1.
Association between sewage measurements and cumulative cases by different machine learning models.
| Model description | Evaluation set | Correlation coefficient | Mean Absolute Error % | |
|---|---|---|---|---|
| 1 | Method: Linear Regression | Dataset 1 (Larissa), 5-fold CV | 0.8814 | 42.27 |
| Train set: Dataset 1 (Larissa) | Dataset 2 (Volos) | 0.9077 | 343.95 | |
| Features: , P, NH4–N, N |
Dataset 1 (Larissa) |
0.8934 |
37.02 |
|
| 2 | Method: Random Forest | Dataset 1 (Larissa), 5-fold CV | 0.8878 | 38.85 |
| Train set: Dataset 1 (Larissa) | Dataset 2 (Volos) | 0.7984 | 93.6 | |
| Features: , P, NH4–N |
Dataset 1 (Larissa) |
0.9897 |
11.64 |
|
| 3 | Method: Linear Regression | Dataset 1&2(All data), 5-fold CV | 0.8646 | 43.81 |
| Train set: Dataset 1&2 (All data) | Dataset 1 (Larissa) | 0.8622 | 43.82 | |
| Features: , M3, pH, P, NH4–N |
Dataset 2(Volos) |
0.9074 |
37.15 |
|
| 4 | Method: Random Forest | Dataset 1&2(All data), 5-fold CV | 0.9188 | 33.72 |
| Train set: Dataset 1&2 (All data) | Dataset 1 (Larissa) | 0.9921 | 9.25 | |
| Features: , Cl−, P, NH4–N, N |
Dataset 2 |
0.9923 |
11.28 |
|
| 5 | Method: Linear Regression | Dataset 2 (Volos), 5-fold CV | 0.9377 | 35.89 |
| Train set: Dataset 2 (Volos) | Dataset 1 (Larissa) | 0.7281 | 188.15 | |
| Features: , P, NH4–N |
Dataset 2 (Volos) |
0.9511 |
24.29 |
|
| 6 | Method: Random Forest | Dataset 2 (Volos), 5-fold CV | 0.9602 | 31.56 |
| Train set: Dataset 2 (Volos) | Dataset 1 (Larissa) | 0.7371 | 49.87 | |
| Features: , Cl, NH4–N |
Dataset 2 (Volos) |
0.9956 |
8.25 |
|
| 7 | Method: Linear Regression | Dataset 1 (Larissa), 5-fold CV | 0.7741 | 59.46 |
| Train set: Dataset 1 (Larissa) | Dataset 2 (Volos) | 0.9244 | 43.88 | |
| Features: |
Dataset 1 (Larissa) |
0.7904 |
55.22 |
|
| 8 | Method: Random Forest | Dataset 1 (Larissa), 5-fold CV | 0.7543 | 53.88 |
| Train set: Dataset 1 (Larissa) | Dataset 2 (Volos) | 0.8347 | 62.97 | |
| Features: |
Dataset 1 (Larissa) |
0.9674 |
18.79 |
|
| 9 | Method: Linear Regression | Dataset 2 (Volos), 5-fold CV | 0.9107 | 45.47 |
| Train set: Dataset 2 (Volos) | Dataset 1 (Larissa) | 0.7904 | 49.13 | |
| Features: |
Dataset 2 (Volos) |
0.9244 |
31.86 |
|
| 10 | Method: Random Forest | Dataset 2 (Volos), 5-fold CV | 0.8721 | 52.58 |
| Train set: Dataset 2 (Volos) | Dataset 1 (Larissa) | 0.7359 | 50.69 | |
| Features: |
Dataset 2 (Volos) |
0.9871 |
13.20 |
|
| 11 | Method: Linear Regression | Dataset 1 (Larissa), 5-fold CV | 0.8157 | 55.58 |
| Train set: Dataset 1 (Larissa) | Dataset 2 (Volos) | 0.9373 | 51.77 | |
| Features: |
Dataset 1 (Larissa) |
0.8249 |
57.77 |
|
| 12 | Method: Random Forest | Dataset 1 (Larissa), 5-fold CV | 0.7792 | 51.79 |
| Train set: Dataset 1 (Larissa) | Dataset 2 (Volos) | 0.8492 | 84.60 | |
| Features: |
Dataset 1 (Larissa) |
0.9739 |
17.21 |
|
| 13 | Method: Linear Regression | Dataset 2 (Volos), 5-fold CV | 0.9261 | 40.06 |
| Train set: Dataset 2 (Volos) | Dataset 1 (Larissa) | 0.8249 | 44.81 | |
| Features: |
Dataset 2 (Volos) |
0.9373 |
29.56 |
|
| 14 | Method:Random Forest | Dataset 2 (Volos), 5-fold CV | 0.9547 | 30.42 |
| Train set: Dataset 2 (Volos) | Dataset 1 (Larissa) | 0.7801 | 47.17 | |
| Features: | Dataset 2 (Volos) | 0.9939 | 7.87 |
Features: Cw: SARS-COV-2 RNA concentrations in wastewater (RNA Copies/ml), COD: Chemical Oxygen Demand(mg/l), NH4–N: Ammonium-nitrogen(mg/l), P: total Phosphorus(mg/l), N: total nitrogen(mg/l), Cl: Chlorides(mg/l).F: Wastewater flow (m3/day) Depended Variable: Cumulative reported cases of the previous 7 days Dataset 1: Waste water measurements and epidemiological data from the municipality of Larissa(63 data points), Dataset 2: Waste water measurements and epidemiological data from the municipality of Volos(48 data points), 5-fold CV: 5- fold cross-validation.
Table 1 presents the association between sewage measurements and cumulative cases, as determined by the various machine learning models applied. Random Forest and Linear Regression models trained with all available data, resulted in correlation coefficients of 0.919 (model 4) and 0.865 (model 3) respectively after evaluation with 5-fold cross validation. The highest correlations were observed for models trained and evaluated with data from the municipality of Volos where the correlation coefficients ranged from 0.872 (model 10) to 0.96 (model 6). For models trained and applied to data from the municipality of Larissa, the corresponsive coefficients ranged from 0.754 (model 8) to 0.89 (model 2). When a municipality-specific model was tested against the other municipality meaning that the models were trained by data form one municipality and then evaluated with data from the other, the models still performed well although the associations were weaker. In particular, for models trained with the Larissa dataset and evaluated with data from Volos the correlation ranged from 0.798 to 0.937, while when the procedure was implemented in reverse (train set: Volos, Evaluation set: Larissa) the correlations ranged from 0.728 (model 5) to 0.825 (model 13). It is noteworthy that in cross-municipality evaluation, simpler models not including physicochemical measurements performed better. The mean absolute error for 5-fold cross-validation evaluations ranged from 30.42% to 59.46%. In cross-municipality evaluations, linear regression models based on non-normalized concentrations were characterized with very high errors (model 1 : 343.95%, model 5: 188.15%), a phenomenon that was significantly mitigated when concentrations normalized for COD were included (model 7: 43.88%, model 9 49.13%). These extreme error levels were not observed in any of the RF models.
Finally, as expected, 5-fold cross-validation of the data demonstrated significantly lower performance compared to non cross-validated evaluations, where the model's estimations were very accurate (e.g model 14, Correlation coefficient:0.99, mean absolute error: 7.87%).
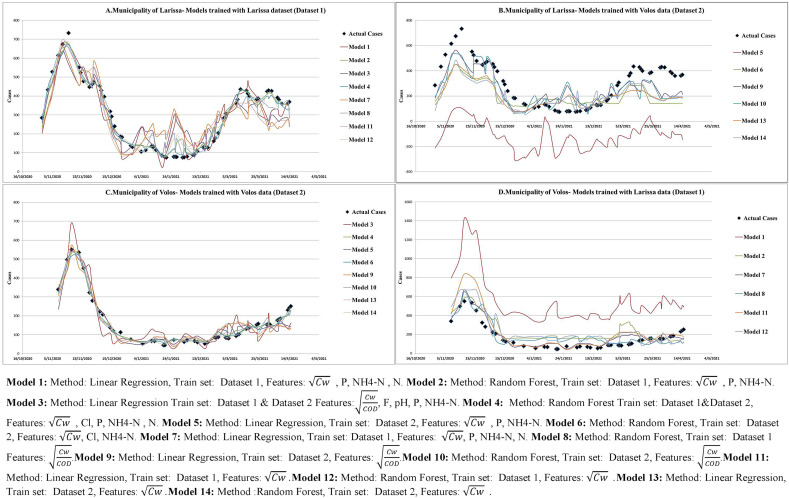
Fig. 1 provides a graphical representation of actual and predicted cases with 14 different Random Forest and linear regression models. In general, wastewater based predictions seem to capture short term changes in disease incidence and resembled the epidemic curve in both municipalities. However, we identified specific periods (e.g, January in Larissa) were considerable deviations between actual and predicted case were observed. On the other hand, very high accuracy was observed during the first epidemic wave in both municipalities (from October until December). When training and test sets were heterogeneous (the model was trained in one municipality and then tested on the other) the course of the epidemic was still captured satisfactorily (Fig. 1B and D).
Fig. 1.
Actual and predicted cases estimated by wastewater measurements using different Machine learning models in the Municipalities of Larissa and Volos
Model 1: Method: Linear Regression, Train set: Dataset 1, Features: , P, NH4–N, N. Model 2: Method: Random Forest, Train set: Dataset 1, Features: , P, NH4–N. Model 3: Method: Linear Regression Train set: Dataset 1 & Dataset 2 Features: , F, pH, P, NH4–N. Model 4: Method: Random Forest Train set: Dataset 1&Dataset 2, Features: , Cl, P, NH4–N, N. Model 5: Method: Linear Regression, Train set: Dataset 2, Features: , P, NH4–N. Model 6: Method: Random Forest, Train set: Dataset 2, Features: , Cl, NH4–N. Model 7: Method: Linear Regression, Train set: Dataset 1, Features: , P, NH4–N, N. Model 8: Method: Random Forest, Train set: Dataset 1 Features: .Model 9: Method: Linear Regression, Train set: Dataset 2, Features: .Model 10: Method: Random Forest, Train set: Dataset 2, Features: .Model 11: Method: Linear Regression, Train set: Dataset 1, Features: .Model 12: Method: Random Forest, Train set: Dataset 1, Features: .Model 13: Method: Linear Regression, Train set: Dataset 2, Features: .Model 14: Method:Random Forest, Train set: Dataset 2, Features: .
4. Discussion
Sewage water surveillance for monitoring SARS-COV-2 RNA concentrations constitutes a low-cost and feasible approach for tracking community virus spread. In this study, we demonstrated that wastewater monitoring can function as a supplementary surveillance tool and can reflect short-term changes of COVID-19 incidence during localized epidemics. Our results also suggest that when sewage monitoring data are supported by machine learning models the derived estimations of cumulative cases can be very accurate. It is very encouraging that various models performed relatively well in heterogeneous environments, in two WWTPs with different technical characteristics.
In general, the population of trained models yielded satisfactory correlations between actual and predicted cases. These findings indicate that wastewater-based SARS-CoV-2 surveillance can be used for monitoring the trends and dynamics of the virus's spread. More caution is suggested when quantitative estimates of incidence are attempted based on wastewater data, since the mean errors of the predictions are not insignificant. The fact that we could not identify a single model as the overall best performing in terms of correlation, accuracy and generalizability in both municipalities, underlines the complexity of the relationship between sewage measurements and disease spread. The interpretation can be sought in the wide variety of factors potentially influencing the investigated relationship which include differences in technical characteristics of the WWTP, chemical composition and physicochemical properties of the wastewater samples, environmental parameters, analytical issues, sampling procedures, changes in surveillance practices and alteration in the virus's properties. Another issue to be discussed is that the strength association between actual and predicted cases varied over time. In the period of the first epidemic wave that peaked in November 2020, the model's predictions were very accurate in both municipalities, and both the increase and decline in disease incidence was captured satisfactorily. However, in the municipality of Larissa we observed a period (January 2021) when a clear increase in SARS-CoV-2 RNA concentrations was not confirmed by epidemiological data, since during this specific period the reported cases were stable. The second epidemic wave was also resembled by the model's predictions, although the relationship seems altered (the ratio of sewage RNA measurements to reported cases declined in comparison to the first wave). There is no ready explanation for this observation, but it should be noted that the two epidemic waves differed in terms of prevalence of different SARS-COV-2 variants, climatological parameters, testing rate and presumably health seeking behaviors.
Our analysis was focused on the relationship between SARS-CoV-2 RNA concentrations and 7-day cumulative cases, although a larger time window would also reflect a plausible scenario. SARS-CoV-2 RNA can enter the sewage system through wastewater discharged from households and other establishments frequently inhabited by active carriers, but also from hospitals and isolation facilities (Giacobbo et al., 2021) and may occur in several forms in wastewater which include at least i) infectious protected, ii) non-infectious protected and iii) non-protected forms (Wurtzer et al., 2021). It has been shown that infected persons have been found to excrete the virus for prolonged periods (Cevik et al., 2021, Zhang et al., 2020). Thus, wastewater RNA concentrations are obviously affected from the accumulated cases reported earlier than the 7 day period. However, the period that infected individuals excrete SARS-Cov-2 genetic material varies from person to person. From a practical perspective monitoring cumulative incidence within larger periods during an ongoing epidemic, is of limited importance for policy makers as it mostly provides retrospective insights at the time of measurement. Our results indicate that estimating cumulative cases choosing a 7-day window is feasible. We found no clear evidence that wastewater measurements can foreshadow reported cases. However, in the present report, samples were taken during epidemics in the examined municipalities, when significant circulation of the virus was present. More data of systematic wastewater monitoring covering periods preceding the beginning of outbreaks are essential to test the hypothesis. It still remains unclear if wastewater based epidemiology can address a critical limitation of epidemiological surveillance which is to capture silent SARS-CoV-2 transmission from asymptomatic and pre-symptomatic cases, a factor that substantially contributes to the emergence of COVID-19 outbreaks (Huff and Singh, 2020).
Some limitations must be taken into account during result interpretation. In some instances, a non-negligent degree of variance was observed in repeated wastewater measurements from the same WWTP in short time intervals. This variance cannot be entirely attributed to changes in the epidemiological burden and it is logical to assume that they are a consequence of changes in environmental, physicochemical or biological factors. Thankfully, the methods concerning the analysis and interpretation of the presence of SARS-CoV-2 in sewage water are constantly advancing. Promising efforts to reduce the associated uncertainties by rationalizing the measurements and thus providing more reliable data are in progress (Petala et al., 2021). A deeper understanding of how different parameters affect the measured concentrations can lead to the development of more valid predictions and upgrade the role of wastewater epidemiology as an epidemiological surveillance tool. In addition, standardization of sampling procedures and optimization of the analytical protocols will not only increase reliability but also reproducibility and comparability of studies conducted in various settings (Alygizakis et al., 2021). Finally, the sample size used in this study was relatively small and a larger dataset could have improved the performance of machine learning models yielding lower error levels (Breiman, 2001).
5. Conclusions
The strong associations between wastewater-based based estimations and actual COVID-19 cases observed in the present investigation, indicate that wastewater monitoring can be exploited by Public Health Authorities to increase the confidence in the results of conventional surveillance practices and can identify temporal trends of the disease's spread. It is encouraging that the developed models performed well in heterogeneous environments in two different WWTPs, and for a prolonged time period. Future studies should focus on the mechanisms by which diverse factors (physicochemical, analytical, environmental, biological) can affect wastewater SARS-CoV-2 RNA levels, in order to develop novel methodologies that yield more robust and accurate estimations.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like thank the Municipal Water Supply and Sewerage Companies of Larissa and Volos that participated in the study and especially Mr. Vaios Zembeoglou. Moreover, the authors would like to thank the region of Thessaly and the National Public Health Organization for providing surveillance data on COVID-19.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envres.2021.111749.
Author’s contributions
Michalis Koureas: Conceptualization, Methodology, Formal analysis, Writing – original draft. Grigoris D. Amoutzias: Conceptualization, Software, Formal analysis, Data curation, Writing. Alexandros Vontas: Investigation, Validation. Maria Kyritsi: Investigation, Supervision. Ourania Pinaka: Resources, Investigation. Argyrios Papakonstantinou: Resources, Investigation. Katerina Dadouli: Resources, Data curation. Marina Hatzinikou: Investigation. Anastasia Koutsolioutsou: Project administration, Supervision. Varvara A. Mouchtouri: Methodology, Writing – review & editing. Matthaios Speletas: Supervision, Writing – review & editing. Sotirios Tsiodras: Supervision, Conceptualization. Christos Hadjichristodoulou: Conceptualization, Writing – review & editing, Supervision, Project administration.
Funding
The study was partially funded by the Hellenic National Public Health Organization (NPHO) under the operation of the National Network of Wastewater Surveillance.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmed W., Angel N., Edson J., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alygizakis N., Markou A.N., Rousis N.I., et al. Analytical methodologies for the detection of SARS-CoV-2 in wastewater: protocols and future perspectives. Trends Anal. Chem. : TRAC. 2021;134:116125. doi: 10.1016/j.trac.2020.116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA/AWWA/WEF . 23rd Edition. American Public Health Association, American Water Works Association, Water Environment Federation; Denver: 2017. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Cevik M., Kuppalli K., Kindrachuk J., et al. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;371:m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- Cevik M., Tate M., Lloyd O., et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. The Lancet Microbe. 2021;2(1):e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacobbo A., Rodrigues M.A.S., Zoppas Ferreira J., et al. A critical review on SARS-CoV-2 infectivity in water and wastewater. What do we know? Sci. Total Environ. 2021;774:145721. doi: 10.1016/j.scitotenv.2021.145721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff H.V., Singh A. Asymptomatic transmission during the coronavirus disease 2019 pandemic and implications for public health strategies. Clin. Infect. Dis. 2020;71(10):2752–2756. doi: 10.1093/cid/ciaa654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749:141364. doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38(10):1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang S., Shi J., et al. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021;415:129039. doi: 10.1016/j.cej.2021.129039. Lausanne, Switzerland : 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., et al. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Current Opinion Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., et al. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Miura F., Kitajima M., Omori R. Duration of SARS-CoV-2 viral shedding in faeces as a parameter for wastewater-based epidemiology: Re-analysis of patient data using a shedding dynamics model. Sci. Total Environ. 2021;769:144549. doi: 10.1016/j.scitotenv.2020.144549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.C., Tilg H. COVID-19 and the gastrointestinal tract: more than meets the eye. Gut. 2020;69(6):973. doi: 10.1136/gutjnl-2020-321195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Yang X., Yang D., et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24(1) doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasher A. COVID-19: current understanding of its pathophysiology, clinical presentation and treatment. Postgrad. Med. 2021;97(1147):312–320. doi: 10.1136/postgradmedj-2020-138577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petala M., Dafou D., Kostoglou M., et al. A physicochemical model for rationalizing SARS-CoV-2 concentration in sewage. Case study: the city of Thessaloniki in Greece. Sci. Total Environ. 2021;755(Pt 1):142855. doi: 10.1016/j.scitotenv.2020.142855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Róka E., Khayer B., Kis Z., et al. Ahead of the second wave: early warning for COVID-19 by wastewater surveillance in Hungary. Sci. Total Environ. 2021:786. doi: 10.1016/j.scitotenv.2021.147398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo J.A., Rumbo-Feal S., Conde-Pérez K., et al. Predicting the number of people infected with SARS-COV-2 in a population using statistical models based on wastewater viral load. medRxiv. 2020:20144865. doi: 10.1101/2020.07.02.20144865. 2020.07.02. [DOI] [Google Scholar]
- Wong S.H., Lui R.N.S., Sung J.J.Y. Covid-19 and the digestive system. J. Gastroenterol. Hepatol. 2020;35(5):744–748. doi: 10.1111/jgh.15047. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., et al. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. medRxiv. 2020 doi: 10.1101/2020.06.15.20117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Ferrier-Rembert A., et al. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: implication for wastewater-based epidemiology and risk assessment. Water Res. 2021;198:117183. doi: 10.1016/j.watres.2021.117183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Gong Y., Meng F., et al. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. medRxiv. 2020:20043059. doi: 10.1101/2020.03.28.20043059. 2020.03.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.