Abstract
We describe three cases with viral strains that demonstrate impaired N2-gene detection on the Cepheid Xpert Xpress SARS-CoV-2 assay, with two previously undescribed single nucleotide polymorphisms (SNPs): C29197T and G29227T. We propose that these SNPs are likely responsible since they are in close proximity to the previously described C29200T/C29200A SNPs, already shown to abolish N2-gene detection by the Xpert assay. Whether these SNPs abolish N2-gene detection by the Xpert assay individually or only in combination requires more work to elucidate.
Keywords: COVID-19, SARS-CoV-2, PCR, Xpert, Mutations, Single nucleotide polymorphisms
Introduction
The gold standard for diagnosis of coronavirus disease-2019 (COVID-19) is detection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) ribonucleic acid (RNA) by real time reverse transcription polymerase chain reaction (RT-PCR). One of the most widely used commercial RT-PCR assays is the Cepheid Xpert Xpress SARS-CoV-2 test (Xpert), which detects SARS-CoV-2 E- and N2-gene targets. Single nucleotide polymorphisms (SNPs) in the N2-gene which abolish detection by the Xpert assay have been reported [1,2]. Here we describe our experience of three clinical cases and novel SNPs which may impact the Xpert assay.
Methods
Nasopharyngeal swabs (NPSs) collected from patients underwent routine diagnostic testing at Middlemore Hospital and LabPLUS Auckland City Hospital laboratories, Auckland, New Zealand. Positive samples were frozen and underwent repeat testing on alternative assays. Note that the freeze-thaw process can result in RNA degradation leading to inter-assay variation in cycle threshold (Ct) values. For assays with separate extraction and amplification steps, RNA was extracted from samples using either MagNA Pure 96 or MagNA Pure 24 Systems (Roche Diagnostics, Basel, Switzerland). Samples were tested using combinations of the following assays: Xpert Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA, USA); E-gene RT-PCR laboratory developed test (LDT) [3] on Panther Fusion (Hologic, Marlborough, MA, USA); TaqPath COVID-19 RT-PCR Kit (Thermo Fisher Scientific, Waltham, MA, USA) on LightCycler 480 (Roche Diagnostics, Basel, Switzerland); MiRXES Fortitude Kit 2.1 (MiRXES, Singapore) on LightCycler 480; BioFire FilmArray Respiratory 2.1 panel (BioFire Diagnostics, Salt Lake City, UT, USA); Aptima SARS-CoV-2 Assay (Hologic, Marlborough, MA, USA) on Panther Fusion. Whole-genome sequencing was undertaken using a 1200-bp amplicon protocol and Oxford Nanopore Rapid barcoding R9.0 sequencing method [4].
Cases
New Zealand’s borders have been closed to all but citizens and residents since 25th March 2020, when the government declared a state of national emergency in response to the COVID-19 pandemic, and the country entered ‘alert level 4’ comprising a stringent nationwide lockdown. Since this time there have been fluctuations in local alert levels but border restrictions have remained in place. Returning travellers must complete 14 days quarantine in a managed isolation facility [5]. Whilst in managed isolation, asymptomatic individuals undergo mandatory SARS-CoV-2 RT-PCR testing via NPS on days 0, 3 and 12 following arrival. Symptomatic individuals are tested as soon as possible following symptom onset. Additional testing, including serology, is conducted at the discretion of public health and the facility physician.
Case A is a 29-year-old male who departed New Delhi on 5th January 2021, transiting via Dubai, and arriving in Auckland on 7th January. On 9th January he developed fever, headache and arthralgia, and tested positive for SARS-CoV-2 the following day.
Case B is a 24-year-old female who tested positive for SARS-CoV-2 in the United Kingdom on 2nd November 2020 when symptomatic with sore throat, fever and cough. She departed London on 5th January 2021, transiting via Dubai. From Dubai to Auckland she was on the same flight as Case A, sitting in seat 19C whilst Case A sat five rows behind in seat 24K. Following her arrival in Auckland on 7th January, two RT-PCR tests were negative. She developed cough, sore throat, anosmia, myalgia, headache and nasal congestion on 8th January, and an NPS collected the following day tested negative. She was tested again on 13th and 15th January with positive results.
Case C is a 15-year-old female who departed Los Angeles on 27th January 2021 and arrived in Auckland on 28th January. She developed cough and sore throat on arrival on 28th January, noted at a border health check, and tested positive the same day. SARS-CoV-2 RT-PCR results for each case are summarised in Table 1.
Table 1.
Summary of SARS-CoV-2 RT-PCR results for each case.
| Case | Sample Collection Date (days since arrival) | Result | Assay/Platform | Target: Ct Value | Sequencing Results |
|---|---|---|---|---|---|
| A | 10/01/21 (Day 3) | POSITIVE | Xpert Xpress SARS-CoV-2 | E-gene: 26.1 | Lineage:
|
| N2-gene: ND | |||||
| E-gene LDT / Panther Fusion | E-gene: 26.4 | ||||
| TaqPath COVID-19 RT-PCR Kit / LightCycler 480 | Orf1ab: 25.03 | ||||
| N-gene: 25.9 | |||||
| S-gene: 24.0 | |||||
| B | 07/01/21 (Day 0) | Negative | E-gene LDT / Panther Fusion | E-gene: ND | Lineage:
|
| 09/01/21 (Day 2) | Negative | MiRXES Fortitude Kit 2.1 / LightCycler 480 | Orf1ab Region 1/2: ND | ||
| 13/01/21 (Day 6) | POSITIVE | Xpert Xpress SARS-CoV-2 | E-gene: Ct 15.4 | ||
| N2-gene: Ct 39.5 | |||||
| E-gene LDT / Panther Fusion | E-gene: 16.7 | ||||
| TaqPath COVID-19 RT-PCR Kit / LightCycler 480 | Orf1ab: 29.49 | ||||
| N-gene: 30.73 | |||||
| S-gene: 26.93 | |||||
| 15/01/21 (Day 8) | POSITIVE | Xpert Xpress SARS-CoV-2 | E gene: 16.0 | ||
| N2 gene: ND | |||||
| BioFire FilmArray Respiratory 2.1 panel | Detected | ||||
| Aptima SARS-CoV-2 Assay / Panther Fusion | RLU 1203 | ||||
| C | 28/01/21 (Day 0) | POSITIVE | Xpert Xpress SARS-CoV-2 | E gene: 18.2 | Lineage:
|
| N2 gene: ND | |||||
| BioFire FilmArray Respiratory 2.1 panel | Detected | ||||
| Aptima SARS-CoV-2 Assay / Panther Fusion | RLU 1258 |
Key: Ct, cycle threshold; ND, not detected; LDT, laboratory developed test; RLU, relative light units; SNP, single nucleotide polymorphism.
All positive samples from the three cases were tested on the Xpert assay, which failed to detect the N2-gene target but detected the E-gene target with low-moderate Ct values ranging from 15 to 26. The exception to this was the first positive sample from Case B, from 13th January, which yielded an Xpert E-gene result with a low Ct of 15.4 and a discrepant N2-gene result with a high Ct of 39.5. Following these findings on the Xpert assay, the samples were tested on alternative assays to confirm the positive results and assess for amplification failure of the N-gene target on other assays. These samples underwent whole genome sequencing, with findings summarised in Table 1. Viral culture was attempted on the positive samples from Cases B and C, with the sample from Case B yielding live virus.
Discussion
We describe three cases that demonstrate impaired N2-gene detection on the Xpert assay. In this assay, detection of the E-gene target alone gives rise to a ‘presumptive positive’ result [6]. This is because the E-gene target is present in other members of the Sarbecovirus subgenus of coronaviruses, including SARS-CoV-1, and thus technically E-gene detection alone cannot distinguish between SARS-CoV-1 and SARS-CoV-2, [6,7] although SARS-CoV-1 is not known to be circulating after the end of the SARS outbreak in 2003 [8]. The N-gene of SARS-CoV-2 is the least conserved of the commonly used RT-PCR gene targets [9,10]. There have been reports of SNPs which impair N-gene target assay performance, such as G29140U, which abolished detection by a Japanese N-gene assay [11].
Cases A and B were found to have genomically identical viruses belonging to lineage B.1.1.281. The C29200T SNP common to Cases A and B has already been described to cause loss of amplification of the N2-gene target of the Xpert assay [1]. Furthermore, a different SNP at the same position (C29200A) also causes failure of amplification of the Xpert N2-gene target [2].
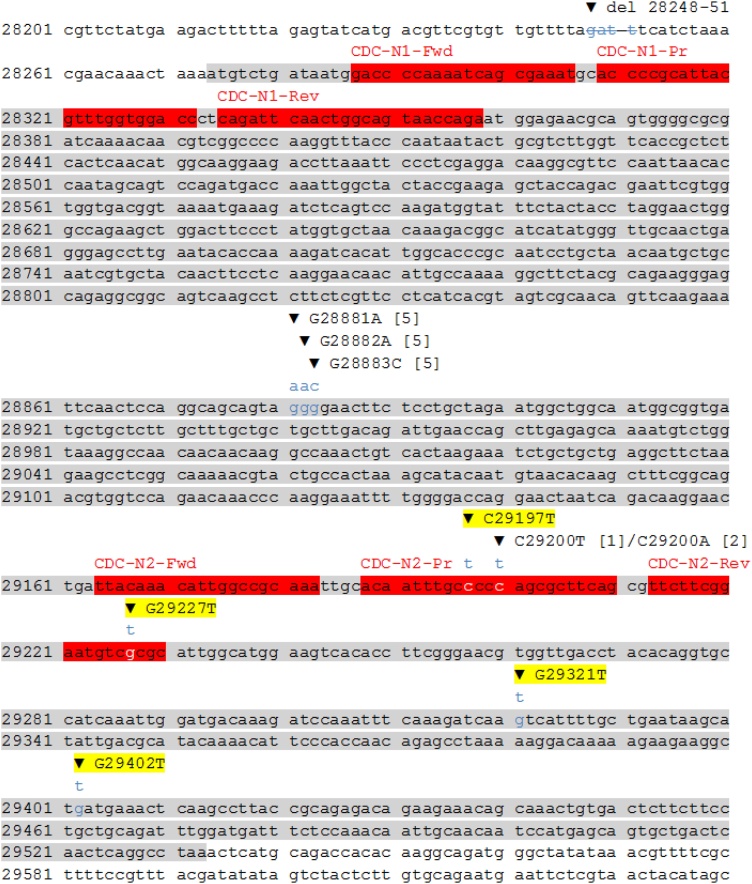
The virus isolated from Case C belonged to lineage B.1.1.519. Six SNPs were identified in the N-gene of this virus (Table 1). The SNPs G28881A, G28882A and G28883C have been previously described, occurring at the 3′ end of the Chinese CDC N-gene forward primer [10]. They have minimal effect on the Chinese CDC assay [10] and have not been reported to affect the Xpert assay. The remaining three SNPs found in the virus from Case C, C29197T, G29227T and G29321T, are not previously known to cause loss of amplification of the Xpert N2-gene target. To the best of our knowledge this is the first report of these mutations potentially impacting this assay. Whilst the primers and probes utilised in the Xpert assay are proprietary, we postulate that the C29197T and G29227T SNPs are most likely to be responsible for loss of amplification of the N2-gene target since these SNPs are in close proximity to the previously described C29200T/C29200A SNPs [1,2], already shown to abolish N2-gene detection by the Xpert assay (Fig. 1). That these SNPs also fall within the US CDC N2-gene primer-probe sequences [12] suggests the US CDC sequences may overlap with or form the basis for those used in the Xpert assay. Whether these SNPs abolish N2-gene detection by the Xpert assay individually or only in combination requires more work to elucidate.
Fig. 1.
Location of SNPs identified in Cases A, B and C in relation to the N-gene and the CDC N1- and N2-gene primers and probes. The N-gene has been highlighted in grey and the US CDC primers and probes highlighted in red and labelled above. SNPs are shown with the base substitution above and labelled, with previously described SNPs referenced [square brackets] and the novel SNPs described here highlighted. Strikethrough delineates the 4 base deletion seen in Cases A and B. Key: del, deletion; CDC, US Centers for Disease Control and Prevention; Fwd, forward primer; Pr, probe; Rev, reverse primer.
The more recently released Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV assay (XpertSFR) also detects SARS-CoV-2 E- and N2-gene targets [13]. Assuming that the same primer-probe combinations used for the Xpert are also utilised in the XpertSFR, it would be expected that the novel mutations described here would also impact N2-gene detection by the newer XpertSFR assay. It should be noted however, that in the XpertSFR the E- and N2-targets are combined in the same optical detection channel, and thus separate results for each targets are not provided, meaning mutations affecting only one target may not be evident.
Of additional interest, it is possible that Cases A and B, who had genomically identical viruses and were seated in close proximity on the Dubai-Auckland flight, represent an episode of in-flight transmission. Case A was mostly likely infectious during the Dubai-Auckland flight as he developed symptoms on day 2 after arrival. His first NPS was taken on day 3 after arrival whereas Case B tested negative twice after arrival.
The FDA Policy for Evaluating Impact of Viral Mutations on COVID-19 Tests provides guidance for evaluating the potential impact of novel mutations on existing assays. The FDA notes that two independent single point mutations reduce the Xpert’s sensitivity for detecting the N2-gene target [14]. The E-gene target is still detected when enough virus is present leading to a ‘presumptive positive’ result, as described above [6]. Case C provides initial evidence for other putative SNPs in circulating viruses which may cause failure of amplification of the N2-gene target.
The three cases described here highlight the impact of SNPs on the widely used Cepheid Xpert Xpress SARS-CoV-2 assay, and we report two potentially novel SNPs which abolish N2-gene target detection by this test. This underscores the importance of utilising RT-PCR assays with multiple gene targets to prevent false-negative results due to mutations affecting a single gene target.
Declaration of Competing Interest
The authors report no declarations of interest.
Funding
The authors received no specific funding for this work.
Ethical approval
Not applicable.
Consent
Written informed consent was obtained from the patients for publication of this case series. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request.
CRediT authorship contribution statement
Shivani Fox-Lewis: Conceptualization, Writing - original draft, Writing - review & editing. Andrew Fox-Lewis: Conceptualization, Writing - original draft, Writing - review & editing. Jay Harrower: Investigation, Writing - original draft. Richard Chen: Investigation, Writing - original draft. Jing Wang: Investigation, Writing - original draft. Joep de Ligt: Investigation, Writing - original draft. Gary McAuliffe: Writing - original draft, Writing - review & editing, Supervision. Susan Taylor: Writing - original draft, Supervision. Erasmus Smit: Conceptualization, Writing - original draft, Writing - review & editing, Supervision.
Acknowledgements
We would like to thank the three individuals reported here for proving written consent to publish their anonymised clinical cases.
References
- 1.Zeigler K., Steininger P., Ziegler R., Steinmann J., Korn K., Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25(39):2001650. doi: 10.2807/1560-7917.ES.2020.25.39.2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasan M.R., Sundararaju S., Manickam C., Mirza F., Al-Hail H., Lorenz S. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by reverse transcription-quantitative PCR. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.03278-20. e03278-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swadi T., Geoghegan J.L., Devine T., McElnay C., Sherwood J., Shoemack P. Genomic evidence of in-flight transmission of SARS-CoV-2 despite predeparture testing. Emerg Infect Dis. 2021;27(March (3)):687–693. doi: 10.3201/eid2703.204714. Epub 2021 Jan 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.2021. Managed isolation and quarantine: arriving in New Zealand.https://www.miq.govt.nz/travel-to-new-zealand/arriving-in-nz/ [Accessed 8 April 2021] [Google Scholar]
- 6.Cepheid . 2021. SARS-CoV-2 test information - Xpert® Xpress-SARS-CoV-2 EUA package insert (English)https://www.cepheid.com/en_US/package-inserts/1615 [Accessed 16 July 2021] [Google Scholar]
- 7.Loeffelholz M.J., Alland D., Butler-Wu S.M., Pandey U., Perno C.F., Nava A. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2 test. J Clin Microbiol. 2020;58(July (8)) doi: 10.1128/JCM.00926-20. e00926-20 Print 2020 Jul 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis. 2020;20(September (9)):e238–e244. doi: 10.1016/S1473-3099(20)30484-9. Epub 2020 Jul 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R., Hozumi Y., Yin C., Wei G.W. Mutations on COVID-19 diagnostic targets. Genomics. 2020;112(6):5204–5213. doi: 10.1016/j.ygeno.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore C., Davies L., Rees R., Gifford L., Lewis H. Localised community circulation of SARS-CoV-2 viruses with an increased accumulation of single nucleotide polymorphisms that adversely affect the sensitivity of real-time reverse transcription assays targeting. Nucleocapsid Protein. 2021 https://www.medrxiv.org/content/10.1101/2021.03.22.21254006v1 Preprint at. [Google Scholar]
- 11.Vanaerschot M., Mann S.A., Webber J.T., Kamm J., Bell S.M., Bell J. Identification of a polymorphism in the N gene of SARS-CoV-2 that adversely impacts detection by reverse transcription-PCR. J Clin Microbiol. 2020;59(1) doi: 10.1128/JCM.02369-20. e02369-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention . 2021. Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primers and probes.https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html [Accessed 10 May 2021] [Google Scholar]
- 13.Mostafa H.H., Carroll K.C., Hicken R., Berry G.J., Manji R., Smith E. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV test. J Clin Microbiol. 2021;59(February (3)) doi: 10.1128/JCM.02955-20. e02955-20 Print 2021 Feb 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.U.S. Food and Drug Administration . 2021. SARS-CoV-2 viral mutations: impact on COVID-19 tests.https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests#xpert [Accessed 10 May 2021] [Google Scholar]