Abstract
The mandibular and hyoid arches collectively make up the facial skeleton, also known as the viscerocranium. Although all three germ layers come together to assemble the pharyngeal arches, the majority of tissue within viscerocranial skeletal components differentiates from the neural crest. Since nearly one third of all birth defects in humans affect the craniofacial region, it is important to understand how signalling pathways and transcription factors govern the embryogenesis and skeletogenesis of the viscerocranium. This review focuses on mouse and zebrafish models of craniofacial development. We highlight gene regulatory networks directing the patterning and osteochondrogenesis of the mandibular and hyoid arches that are actually conserved among all gnathostomes. The first part of this review describes the anatomy and development of mandibular and hyoid arches in both species. The second part analyses cell signalling and transcription factors that ensure the specificity of individual structures along the anatomical axes. The third part discusses the genes and molecules that control the formation of bone and cartilage within mandibular and hyoid arches and how dysregulation of molecular signalling influences the development of skeletal components of the viscerocranium. In conclusion, we notice that mandibular malformations in humans and mice often co-occur with hyoid malformations and pinpoint the similar molecular machinery controlling the development of mandibular and hyoid arches.
Keywords: neural crest cells, craniofacial development, pharyngeal arches, jaw development, hyoid bone, patterning, cartilage, bone, chondrogenesis, osteogenesis
1. Introduction
The primary function of the gnathostome facial skeleton is to encase the openings to the mouth and airways and to accommodate several sensory organs (such as vision, smell, or taste). This ancestral function of facial skeleton is shared among all of the gnathostome species and may have played a central role in their evolution. The facial skeleton of gnathostomes, also known as the viscerocranium, is composed of bone and cartilage that collectively form the skeleton of the face and throat. The membranous viscerocranium is formed by a process of intramembranous ossification, whereas the cartilaginous viscerocranium utilizes the endochondral ossification to form bones. Since the last common ancestor of mammals and teleosts roamed the Earth ≈450 million years ago, the composition of murine and zebrafish viscerocrania is vastly different [1]. However, studies show that genetic regulation of craniofacial morphogenesis between the mouse and the zebrafish is similar, indicating a common regulatory circuit during facial development among gnathostomes. This relative similarity means that the zebrafish is complementary to the mouse in the research of craniofacial defects. Currently, the research of zebrafish craniofacial development is growing in intensity, since the genetic machinery controlling PA development is similar among zebrafish, mice, and humans. Some researchers even use a zebrafish model to study human craniofacial diseases, such as CATSHL syndrome (tall stature and hearing loss) and cleft lip/palate [2,3,4]. The adult viscerocranium is composed of many individually distinct elements and requires a coordinated integration of various tissues. Intricate molecular signals and transcription factors among cranial tissues regulate the patterning of the prospective face, which ensures the formation of heterogenous bone and cartilage. Perturbation and impaired regulation of craniofacial development results in dysmorphy of bones and cartilage, which collectively accounts for at least a third of all birth defects in humans [5]. Understanding the precise mechanism of how bony and cartilaginous structures arise and attain their distinct shape may improve treatment and reduce the impact of certain craniofacial birth defects on human patients. During the embryonic development in amniotes, transient embryonic structures known as pharyngeal arches (PAs) undergo extensive growth and differentiation to create the adult viscerocranium. Pharyngeal arches are a series of paired, bilaterally symmetrical outgrowths on both sides of the developing pharynx. Cells from all germ layers take part in assembling the PAs. Each PA consists primarily of two robust mesenchymal populations, the neural crest-derived mesenchyme (also known as “ectomesenchyme”) and the paraxial mesoderm [6]. The oral surface of arches is coated with the ectodermal epithelium, whereas the pharyngeal surface is lined with the endodermal epithelium. The neural crest (NC), sometimes colloquially termed as the fourth germ layer, is a multipotent embryonic population of cells that arises at the lateral border of the neural plate, from which it subsequently delaminates and undergoes extensive migration into distant parts of the body [7]. Neural crest cells (NCCs) are regarded as multipotent because they have the capacity to differentiate into plethora of cell types—osteoblasts, chondroblasts, fibroblasts, neurons, and glia, among many others [8]. A subpopulation of NCCs coming from the level of the future brain—named the cranial neural crest—gives rise to many tissues, including the viscerocranium, the connective tissues, and part of the neurosensory ganglia of the cranium [9,10]. On the other hand, the cranial paraxial mesoderm within PAs forms the muscles and blood vessels of the face, neck, and throat [11].
The segmentation of the pharyngeal region appears to be driven by the endoderm and is independent of NCCs that migrate into PAs [12]. Moreover, the pharyngeal endoderm provides positional clues for the mesenchyme within PAs and is also responsible for the formation of particular arch components. Via interaction with migrating NCCs, the mesoderm actively participates in the formation of PAs [13]. The cranial paraxial mesoderm proliferates ahead of the neural crest migratory front, thus prior to the migration of the neural crest. Like the NC, the mesoderm is inherently motile. Proliferating mesodermal cells commence the PA formation by driving outgrowth in the lateral direction. After the initiation of NC migration, a portion of mesodermal cells freely intermingles with NCCs, while others are displaced by migrating NCCs. After NCCs invade the nascent arches, they actively proliferate in order to stay in pace with the mesoderm. Thus, the mesoderm is the main driver of PA growth, and PAs can form even the absence of NC. The fact that mouse mutants lacking specific NC streams will still form normal PAs supports the notion that NCCs are not required during the initial stages of PA formation. Thus, the formation of the PA template precedes the appearance of NCCs in the pharyngeal region [14,15]. This narrative review focuses on the morphogenesis and skeletogenesis of the first two PAs in mice and zebrafish, since both these models are used in the research of craniofacial diseases. Tight control of temporospatial cell specification and differentiation along the anatomical axes is crucial for the embryonic formation of various structures. The authors present an overview of signalling pathways and regulatory networks involved in this process in the mandibular and hyoid arches. Furthermore, we outline that the mandibular and hyoid arches are collectively governed by a shared gene regulatory network.
2. Anatomy and Fate of Pharyngeal Arches
In amniotes, including humans, there are five PAs, numbered first, second, third, fourth, and fifth. Previously, the terminal arch used to be labelled as the sixth, while the fifth was considered rudimentary, disappearing almost as soon as it has formed. However, new analyses show that there is no evidence from amniote embryology for the existence of a transient, rudimentary fifth arch [16]. Collectively, the abnormal development of PAs is linked to several major groups of birth defects in humans [17]. The first two PAs are called the mandibular and the hyoid and have been named according to the anatomical structures they turn into in the adult organism. The third, fourth, and fifth PAs are collectively known as the posterior pharyngeal arches. After formation of the mandibular arch, the first PA is split into upper maxillary and lower mandibular processes. Cartilage elements and endochondral bone originating from PAs collectively make up the splanchnocranium. In the mandibular arch, two cartilaginous elements arise—a rod shaped, oblongate Meckel’s cartilage in the mandibular process and subtler palatoquadrate cartilage in the maxillary process. During craniofacial morphogenesis, palatoquadrate cartilage undergoes endochondral ossification to form a portion of orbital and lateral skull wall, the alisphenoid, and the second middle ear bone, the incus [18,19,20]. In contrast, a fraction of NCCs encasing the splanchnocranium differentiates directly into functional osteoblasts without a cartilaginous intermediate by the process of intramembranous ossification. In the maxillary process, NCCs surrounding the palatoquadrate cartilage form the maxilla, zygomatic, and squamous part of the temporal bone [21]. Facial bones, which are created around splanchnocranial cartilages, collectively comprise a membranous viscerocranium and serve as the functional jaws in mammals. Interestingly, in the mouse, only NCCs in the first PA have the potential to generate osteoblasts that undergo intramembranous ossification.
In all gnathostomes, Meckel’s cartilage represents a strut of the lower jaw during embryonic development. Meckel’s cartilage initially consists of a pair of continuous rods of cartilage, which subsequently elongate anteriorly and later fuse in the distal midline to form a V-shaped structure outlining the forming lower jaw in mice. In mammals, Meckel’s cartilage can be divided into three parts according to the fate of each region: anterior/distal, intermediate/central, and posterior/proximal [22,23,24,25]. In humans, the distal part of Meckel’s cartilage undergoes endochondral ossification and forms a portion of dentary bone extending from the mental foramen to the midline. However, isolated cartilaginous nodules originating from Meckel’s cartilage can be found on the dorsal surface of the mandibular symphysis [26]. The most proximal part of Meckel’s cartilage turns to bone and forms the first middle ear bone—the malleus. Even though the intermediate part of Meckel’s cartilage initially serves as a template during the development of the lower jaw, it later degenerates, and a dentary bone emerges in its place, also known as the jawbone or the mandible. Although chondrocytes in Meckel’s cartilage have been shown to be able to transdifferentiate into osteogenic cells, evidence for the ossification of the intermediate part of Meckel’s cartilage in vivo is currently limited [27,28,29,30,31,32]. Most importantly, the cartilaginous matrix of Meckel’s cartilage is removed during the mandibular development. Nonetheless, two separate parts of the intermediate region of Meckel’s cartilage, one at the base of skull and the other just at the periphery of the mandibular foramen, ultimately undergo endochondral ossification and turn into the spine of the sphenoid and the lingula of the mandible, respectively. In the adult organism, the dentary bone and the malleus are interconnected by ligaments. Parts of Meckel’s cartilage connecting the spine and lingula are thought to transdifferentiate to become the anterior ligament of the malleus and sphenomandibular ligament [33,34]. The sphenomandibular ligament connects the lingula of the mandible, situated at the periphery of the mandibular foramen and the spine of the sphenoid, hanging from the cranial base, from which it continues as an anterior ligament of the malleus to the middle ear cavity and attaches itself to the malleus. In adulthood, the connection between the mandible and the middle ear is still apparent, as trauma to the jaw joint can potentially cause dislocation of ear bones [35]. After the dentary bone undergoes intramembranous ossification, secondary ossification centres appear in the key points of articulation and mechanical force—in the condylar, coronoid, and angular processes of the mandible—where they initiate endochondral ossification. Since Meckel’s cartilage acts as a template for later formation of the lower jaw bones, its defects lead to anomalies in the pattern and size of the lower jaw in both mouse and human embryos [36]. In summary, Meckel’s cartilage turns into diverse structures along the proximal-distal axis: the malleus; ligaments, replaced by the dentary; and mandibular symphyseal cartilage [25].
Generation of bone in the second PA generally involves endochondral ossification [37]. In the mammalian hyoid arch, several separate cartilaginous elements arise, i.e., anlage of the third middle ear bone—the stapes, and Reichert’s cartilage. Unlike Meckel’s cartilage, Reichert’s cartilage is not a continuous structure [38]. The cranial portion of Reichert’s cartilage is continuous with the ear capsule and undergoes endochondral ossification to form a bony projection of the temporal bone, termed the styloid process. The smaller caudal segment of Reichert’s cartilage develops in close relation to the oropharynx and undergoes endochondral ossification to form lesser horns of the hyoid. No cartilage connection between these segments exists, although they are temporarily linked by a mesenchymal band, which is thought to differentiate into muscles and ligaments [39]. The cartilage element of the third PA does not bear any eponymous name and contributes to the development of greater horns of the hyoid and possibly to superior horns of the thyroid [40]. The body of the hyoid bone originates from a single growth centre, without overt contributions from the second PA and third cartilage elements. In mammals, posterior PAs probably bear a miniscule importance. Analyses of chondrogenesis and myogenesis in the chick and mouse, as well as three-dimensional analysis of human embryos, revealed that cartilage formation does not occur within the fourth and fifth PAs [41,42]. Laryngeal cartilages, previously considered to be derived from the posterior PAs, likely develop as new mesenchymal condensations in the throat region [41].
Interestingly, reports of abnormalities in the hyoid arch in humans are uncommon in the literature. However, severe hyoid abnormalities associated with swallowing dysfunction occur in patients with Pierre Robin sequence [43], and infants with cleft lip and palate occasionally exhibit delayed ossification of the hyoid bone, as well as a significantly lower position of the hyoid bone relative to the cervical vertebrae [44]. Conversely, the hyoid bone has been shown to have a more superior and posterior position in patients with hyperdivergent vertical facial growth [45]. In a 15-year-old boy patient with cleidocranial dysplasia, Yoshida et al. reported a unique case of abnormal ossification of the hyoid bone [46]. Cephalometry of children with 22q11.2 deletion syndrome revealed a reduction of hyoid bone lengths, and hyoidal gaps, which reflect the fusion of the hyoidal segments, the greater horns, and the body, were larger than those of the controls [47]. This finding indicates that the ossification of the hyoid bone is delayed in children with 22q11.2 deletion syndrome. In accordance with this, the delayed ossification of the hyoid bone was suggested to be a useful tool in the diagnosis of DiGeorge syndrome during the first postnatal months, before the diagnostic use of the FISH hybridization techniques [48]. Moreover, autopsied infants with DiGeorge syndrome, tetralogy of Fallot, and interrupted aortic arch showed a significantly low incidence of visible hyoid ossification centre [49]. Since the hyoid bone has an important role in respiration, deglutition, and speech, delayed development of the hyoid bone in children with 22q11.2 deletion syndrome may be related to hypotonia of the velopharyngeal muscles and nasal speech.
In teleosts, seven PAs, numbered first, second, third, fourth, fifth, sixth, and seventh, have been described. As a hallmark of gnathostomes, the first PA/the mandibular arch in teleosts transforms into the jaws during embryogenesis. The second PA, the hyoid arch, mainly provides the attachment of the jaws to the base of neurocranium. The remaining five PAs, also known as branchial/gill arches, provide a gill-supporting function. The mandibular arch is divided into two clearly distinguishable cartilaginous bars, Meckel’s cartilage and palatoquadrate cartilage. While Meckel’s cartilage is the precursor of the lower jaw, dorsally situated palatoquadrate cartilage precedes the appearance of individual bones in the upper jaw. Similar to the mandibular, the hyoid arch is divided into a dorsal region, represented by hyosymplectic cartilage, and a ventral region, represented by ceratohyal and basihyal cartilages. The hyosymplectic cartilage in the dorsal region provides the attachment of jaw to the neurocranium, whereas the ventrally situated ceratohyal and basihyal cartilages act to stabilize the jaw and support the neck region. This primary setting of the teleost viscerocranium represents a base for further development, which is the bone formation. The mechanism of ossification in teleosts slightly differs from that in mammals. At first, endochondral bone usually goes through a perichondral ossification, meaning that cartilage ossification initially occurs within the perichondrium and then continues progressively from outside to inside. It is important to note that the nascent bone still remains a rod-like shaped cartilage in the centre [49].
In the zebrafish lower jaw, the perichondral ossification of Meckel’s cartilage is initiated on the anterior, labial side. The bulkiest bone arising from Meckel’s cartilage via perichondral ossification is the anguloarticular [49]. Nonetheless, most parts of Meckel’s cartilage are encased within intramembranous bone, such as the dentary bone. In the upper jaw, palatoquadrate cartilage turns to bone and in the posterior region gives rise to the endochondral bone, the quadrate, which articulates with the anguloarticular in the jaw hinge region. Conversely, the upper jaw is composed of two dermal bones in the anterior region, premaxilla and maxilla [50]. In contrast with teleosts, there is an evolutionary trend towards the reduction and/or fusion of skeletal elements within mandibular and hyoid arches in mammals. Teleost jaws are composed of a large amount of individual bones which are mostly clearly identifiable.
Similar to Meckel’s, the ossification of ceratohyal cartilage in the ventral region of the hyoid arch starts within the perichondrium, progressing in the anteroposterior direction. In the dorsal region of the hyoid arch, the hyosymplectic cartilage ossifies into the hyomandibular bone. The hyomandibula is fused with the symplectic bone, which itself derives from the hyosymplectic cartilage in the middle region. Moreover, the hyomandibula articulates with the opercular series, which is composed of several intramembranous bones, including the opercle. Collectively, the intramembranous bones of the opercular series serve as a protection of gill slits. Meanwhile, subsequent branchial arches also ossify perichondrally [50] Opercular intramembranous bones are in sharp contrast with the mammalian hyoid arch, as NCCs in the murine hyoid arch are incompetent at forming intramembranous bone under normal conditions. A summary of skeletal derivates of PAs in the mouse and zebrafish can be found in Table 1.
Table 1.
Skeletal derivatives of PAs in the mouse and zebrafish.
| Viscerocranium | Mouse | Zebrafish | ||
|---|---|---|---|---|
| Cartilaginous | Membranous | Cartilaginous | Membranous | |
| First pharyngeal arch (the mandibular) |
Palatoquadrate cartilage: Alisphenoid Incus |
Premaxilla Maxilla Zygomatic bone Temporal squama |
Palatoquadrate cartilage: Quadrate Metapterygoids Palatines |
Premaxilla Maxilla Ectopterygoid Entopterygoid |
| Meckel’s cartilage: Mandibular symphysis Lingula of mandible Sphenomandibular ligament Spine of sphenoid Anterior ligament of malleus Malleus |
Dentary bone | Meckel’s cartilage: Retroarticular |
Dentary Anguloarticular Coronomeckelian |
|
| Second pharyngeal arch (the hyoid) | Stapes Styloid process of the temporal bone Stylohyoid ligament Lesser horns of the hyoid bone |
Basihyal Ceratohyal Epihyal Hypohyal Hyomandibula Interhyal Symplectic |
Urohyals Branchiostegal rays Interopercle Opercles Preopercles Subopercles |
|
| Third pharyngeal arch | Greater horns of the hyoid bone | Basibranchials Ceratobranchials Epibranchials Hypobranchials Pharyngobranchials |
||
In teleosts, the cranial skeleton is highly developed, and the function of pharyngeal cartilage is more akin to the ancestral gnathostome state in comparison with amniotes, as each PA-derived element reflects the adaption to aquatic life [51]. The number of PAs in aquatic species is usually higher, since the gills are relatively inefficient filters. There appears to be a general trend towards the reduction of PAs during evolution. Fossil fish have high numbers of PAs, and there have even been ostracoderm fossils with as many as 30 arches [52]. One reason for the general decrease in PA number in amniotes could be the transition from an aquatic to land dwelling lifestyle. However, it is important to note that the anterior–posterior and dorsal–ventral PA identity and polarity has largely been conserved among all gnathostomes [21].
3. Specification of Pharyngeal Arches by the Hox Code
The anatomical identity of individual PAs is dependent on their position along the anterior-posterior axis. The axial identity of PAs is determined by the expression of Hox genes in the hindbrain and in migrating NCCs [53,54]. However, even crestless PAs have a sense of individual identity [15]. Hox genes control the segmentation of the hindbrain by the principle of collinearity, meaning that they are organized in clusters in the chromosomes in the same order, as is their expression along the anterior–posterior axis [55]. Cranial NCCs populate PAs in distinct segregated streams, which are defined by the spatiotemporal expression of Hox genes in the hindbrain [56]. The neuroepithelium of the hindbrain is transiently subdivided into a series of eight metameric segments, called rhombomeres (r1–r8) [57]. NCCs arising at the level of rhombomeres colonize PAs, which are worth to note also metameric [9]. While NCCs migrating from the level of the forebrain, midbrain, and anterior hindbrain do not express Hox genes, those that arise at the level of r3–r8 are Hox-positive. According to this anterior–posterior specification, NCCs colonizing the prospective face and the mandibular arch are Hox-negative, whereas the hyoid and posterior PAs are Hox-positive [58]. In humans and mice, four Hox paralogue groups, numbered Hox1, Hox2, Hox3, and Hox4, are expressed in the head and neck. Each paralogue group contains Hox genes from at least two Hox clusters—Hoxa, Hoxb, Hoxc, and Hoxd. For example, Hox1a and Hox1b collectively form one paralogue group, and they both come from two distinct clusters. Due to teleost-specific duplication, as much as seven hox clusters appear in the zebrafish genome—hoxaa/hoxbb, hoxba/hoxbb, hoxca/hoxcb, and hoxda/hoxdb [59]. Generally, each PA is governed by one Hox group—the second PA is controlled by Hox2, the third PA by Hox3, and the fourth PA by Hox4 [60,61]. Hoxa1 itself is not expressed in migrating NCCs but solely in their precursors at the neural plate prior to NCC delamination, and Hoxa1 lineage gives rise to all NCCs that emanate from r4 [62,63]. Likewise, the expression of Hoxb1 is apparent only in the neuroepithelium and is very temporary in the mouse [56]. In the zebrafish, hoxb1 in conjunction with other transcription factors modulates NCC activity in streams migrating from r4 [56]. Mice lacking Hoxa1 show a significant decrease in migratory NCCs in the second PA and the reduction of the NCC number is even stronger in Hoxa1 and Hoxb2 double-null embryos, which lack any NCCs from r4, a major site of origin of the second PA neural crest [14]. In zebrafish, overexpression of hoxa1 results in robust and partially duplicated ceratohyal cartilages, while the remaining PAs, including the mandibular arch, are underdeveloped [64]. Interestingly, single Hoxb1-null mouse embryos display no discernible defects in NCCs [14,65].
In contrast, Hoxa2 has a more direct effect on the craniofacial morphogenesis, since Hoxa2 is expressed in NCCs emanating to the second, third, and fourth PAs. Strictly speaking, Hoxa2 is a key determinant of the second PA fate in the mouse [66]. Hoxa2-null mice exhibit a homeotic transformation of the first arch derivatives into the second arch skeletal elements [67,68]. Although not studied in the mouse, the ectopic activation of Hoxa2 in the mandibular arch of fish, frog, and chick transforms the identity of the first PA elements into that of the second arch [69,70,71]. Phenotypic changes in Hoxa2 mutants suggest that Hox genes are incompatible with the mandibular arch development, and this idea is further supported by mutants missing the entire Hoxa cluster. In these mouse mutants, the individual identities of the second, third, and fourth PAs are diminished and all are transformed the into rudimentary first PA elements, while posterior PA derivatives do not develop altogether [72]. Nonetheless, this is not firm evidence that the first PA represents a ground state, and the formation of successive PAs requires the Hoxa cluster. Akin to Hoxb1, mouse Hoxb2 mutants have only mild craniofacial defects, and their pharyngeal skeletal elements appear normal. In zebrafish, hoxb2 is expressed only in NCCs emanating from r4 to the second PA, and its individual function is not necessary for hyoid arch development. However, it is important to note that hoxa2 alone does not drive the development of second PA derivatives in the zebrafish [70]. During the second PA morphogenesis in zebrafish, the combined action of hoxa2 and hoxb2 patterns the nascent hyoid arch, as hoxa2/hoxb2 double knockdown changes the morphology of the second PA derivates so that they appear similar to the mandibular arch-derived elements. Altogether, data from Hox mutants suggest that the Hoxa gene cluster has a primary role in the specification of the axial identity of the PAs, whereas Hoxb cluster may serve as a fine tuner of the nascent PA morphology.
4. Specification of Mandibular and Hyoid Arches by the MEIS/PBX Complex
MEIS and PBX transcription factors are regulatory proteins containing TALE (three-amino-acid-loop extension) homeodomain. MEIS binds PBX, among other transcription factors, and they collectively form a complex that binds to a DNA via respective MEIS- and PBX-consensus binding sites [73]. Mice and humans possess three Meis paralogues—Meis1, Meis2, and Meis3. In zebrafish, meis1 and meis2 genes were duplicated during teleost evolution, and its genome contains meis1a/meis1b and meis2a/meis2b. When it comes to Pbx, mice and humans have four Pbx genes Pbx1, Pbx2, Pbx3, and Pbx4, whereas the pbx family in the zebrafish genes consists of pbx1a, pbx2, pbx3, and pbx4, whose function is more akin to murine Pbx1 [74].
MEIS and PBX transcription factors serve important roles by interaction with HOX proteins during development of the hindbrain and NC. MEIS factors bind to the PBX–HOX complex, therefore forming a stable trimeric complex, allowing the modulation of Hox expression [73,75]. Crosstalk between MEIS and HOX is likely required for the determination of PA identity. In zebrafish, the Meis–Pbx–Hox complex regulates chromatin accessibility in hoxb1a and hoxb2a gene loci, thereby regulating their expression in the second PA [76]. Correspondingly, the murine MEIS–HOXA2 complex regulates the identity of the second PA by controlling the expression of second PA-specific genes [77]. The elimination of Meis2 specifically in NCCs results in extensive craniofacial defects [78]. Furthermore, NC-specific Meis2 embryonic mutants have elevated osteogenesis in the mandibular and hyoid arch at the expense of disrupted tongue development [79]. Craniofacial defects in the maxillary and mandibular processes of Meis2-deficient embryos thus reveal the Hox-independent function of Meis2. Furthermore, altered osteogenesis within the hyoid arch also results in various defects of the hyoid apparatus in Meis2-deficient embryos [78,79]. The MEIS–PBX–HOX regulatory circuit seems to be evolutionary conserved. In a clinical setting, human patients with heterozygous mutations in MEIS2 are afflicted by craniofacial and cardiac defects, in addition to intellectual disabilities [80,81,82,83,84,85]. In both lamprey (cyclostomes) and zebrafish (gnathostomes), the deletion of evolutionary conserved hoxa2 and hoxb2 enhancers results in loss of hox expression in the second migratory stream of NCCs, which contains precursors of second PA cartilage [86]. Intriguingly, the targeted deletion of conserved meis- and pbx-binding sites in these hox enhancers leads to the same result. The combined knockdown of meis1 and meis2 leads to malformations of craniofacial cartilage, e.g., a fusion of viscerocranial cartilages, demonstrating the importance of meis in cranial NCCs [87]. This indicates the improper specification of PAs at the earliest stages, which may affect subsequent steps of cartilage formation.
Zebrafish Pbx4, the functional equivalent of mammalian PBX1, cooperates with HOX in PA segment setting, as pbx4 mutants exhibit hypoplastic jaws and the fusion of first and second PA skeletal derivatives [88]. In contrast to zebrafish, murine Pbx1 is expressed in the ectomesenchyme and ectoderm of the second arch, while maxillary and mandibular processes show much lower expression. Nonetheless, Pbx-null and Pbx2 heterozygous mutants have been reported to exhibit mandibular hypoplasia [89]. Compound Pbx1/Pbx2 mutant mice show abnormal forebrain development, hindbrain segmentation, and hypoplasia of posterior PAs [90]. Mice with systemic elimination of Pbx1 have morphological alternations of splanchnocranial cartilage derived from the second PA, which mimics the homeotic transformation in Hoxa2-null mice. Both lesser horns of the hyoid and styloid processes of the temporal bone develop elongated outgrowths that are fused together. This newly formed cartilaginous structure is oddly reminiscent of the hyoid apparatus of certain nonhuman mammals and of Eagle’s syndrome in humans. Additionally, Pbx1 mutants lack stapes, another skeletal element derived from the second PA [91]. Elongation of the styloid process or calcification of the stylohyoid ligament above a specific threshold is a medical condition called Eagle’s syndrome. Alongside Hoxa2 and Pbx1-null mutants, calcification or chondrification of the stylohyoid ligament resembling human Eagle’s syndrome can also be observed in Meis2 and Prrx2 mutants [79,92]. Calcified stylohyoid ligament conspicuously resembles the hyoid apparatus of some nonhuman mammals, in which it may consist of more parts than in humans. It has been hypothesized that the elongated styloid process in humans is evolutionary coded and represents a form of atavism of the bony hyoid apparatus of our evolutionary ancestors. Clinically, the condition is characterized mostly by pain in the head and neck due to compression of the surrounding structures either by elongation or angulation of enlarged styloid process [93]. Multiple aetiologies of Eagle’s syndrome have been suggested in the literature, ranging from genetic, developmental, endocrine, traumatic, degenerative, and metaplastic. To summarize, the MEIS/PBX complex regulates cell specification within the mandibular arch, whereas the trimeric complex MEIS/PBX/HOX determines cell identity within the hyoid arch.
5. Endothelin–Dlx–Hand Gene Regulatory Network Controlling Anatomical Axes in Mandibular and Hyoid Arches
5.1. Mouse
Already at the onset of PA formation, molecular signals determine the pattern and polarity of the respective arch. The mandibular arch contains Hox-free NCCs, so its molecular determination is dependent on distinct signalling cascades, primarily on the Endothelin–Dlx–Hand regulatory network. Endothelin1 (EDN1) is a peptide ligand that binds to G protein-coupled receptor EDNRA and together with its downstream components, DLX and HAND, governs the patterning of jaws [94,95,96]. Both EDN1 and its receptor EDNRA are required for the induction of Dlx and Hand expression in the mandibular arch [97,98,99]. Edn1 is expressed in the epithelium, in the paraxial mesoderm, and in the aortic arch vessel endothelium of the mandibular arch, whereas Endra is extensively expressed in the ectomesenchyme of the head [100,101]. Edn1- and Ednra-null mouse embryos exhibit a homeotic transformation of the lower jaw to an upper jaw identity [101,102]. Similarly, ventral structures of the hyoid arch (lesser horns) appear more severely affected in comparison to the dorsal structures (stapes) in Edn1 mutant mice [103]. Moreover, Edn1-null mouse mutants display the absence of the styloid process, and the hyoid bone is largely deformed and fused to the pterygoid process. Conversely, ectopic activation of Ednra in the cranial NCCs leads to homeotic transformation of the maxilla into the mandible-like structure [104]. In line with this, misexpression of Edn1 in the maxilla induces the ectopic dentary bone in the upper jaw region, again demonstrating the reversal of the molecular switch [104]. Cranial NCCs within the mandibular arch are competent at forming both maxilla and the mandible, and Edn1 is a molecular switch responsible for the choice of the mandibular-specific morphogenetic program [104]. Intriguingly, even indirectly induced ectopic Edn1 signalling in Six1-null mice present in the proximal end of the mandibular arch leads to the formation of rod-shaped bone at the zygomatic arch with a cartilaginous tip [105].
A close relationship among Edn1 and its downstream targets Dlx and Hand has been proposed in several loss-of-function studies. The combined loss of Dlx5 and Dlx6 causes the homeotic transformation of the lower jaw into the maxilla-like structure, which essentially phenocopies Edn1 knockout [102,104,106]. Furthermore, misexpression of Hand2 in the Ednra domain of the cranial NCCs causes similar transformation to ectopic Ednra activation [107]. Altogether, the EDN–DLX–HAND regulatory network is a prime regulator of anterior–posterior (synonymous with ventral–dorsal in the zebrafish) patterning of the mandibular arch and, in that sense, upper and lower jaw identity [104].
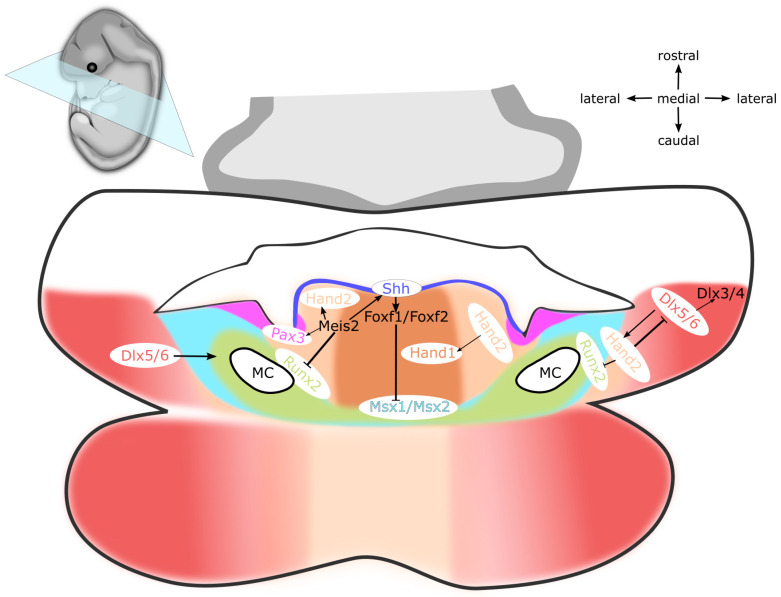
Dlx genes are homeodomain transcription factors that control the intra-arch polarity of pharyngeal arches and anterior–posterior and proximal–distal patterning [106]. In mice, they are organized as three bigene pairs, namely Dlx1/Dlx2, Dlx3/Dlx4, and Dlx5/Dlx6, in the proximity of Hox genes in the chromosomes [108,109]. In both mice and zebrafish, there are six Dlx genes that are expressed in the ectomesenchyme of the mandibular and hyoid arches. While Dlx1/Dlx2 are expressed almost throughout the entire first two arches, Dlx3/Dlx4 and Dlx5/Dlx6 show more restricted domains [110]. Dlx5/Dlx6 are expressed solely in the mandibular process and hyoid arch (see Figure 1), in the nested domains within Dlx1/Dlx2 territory, whereas Dlx3/Dlx4 are expressed only in the most distal part of the mandibular process and hyoid arch, within Dlx5/Dlx6 territory.
Figure 1.
Schematics of a frontal section through developing oral cavity of a mouse embryo at E12. Dlx5/Dlx6 are expressed in ectomesenchyme of the mandibular process and the hyoid arch. Hand1/Hand2 and Meis2 are expressed in ectomesenchyme in the medial region of the mandibular process and the hyoid arch, which also includes the lingual ectomesenchyme. Msx1 and Runx2 are expressed within the primordium of prospective dentary bone. Pax3 is expressed in the ectomesenchyme around the alveolingual sulcus, which represents an anatomical boundary between the dentary bone and tongue. Shh is expressed in the lingual epithelium, whereas Foxf1/Foxf2 are expressed in the lingual ectomesenchyme. Abbreviations: E, embryonic day; MC, Meckel’s cartilage. Expression domains of genes written in black text are not highlighted in the figure.
Single and compound Dlx1/Dlx2 mouse mutants display malformations selectively in the upper jaw and upper hinge region, with barely any effect in the lower jaw [110,111,112]. Many anterior first PA elements, such as alisphenoid and incus, are malformed in the Dlx2-null mice, whereas Dlx1-null mutants exhibit a similar phenotype, although much milder. However, compound double-null Dlx1/Dlx2 mutants develop more severe defects that are not present in either of the single-null mutants [111,112], suggesting their functional redundancy for the development of maxilla. Furthermore, Dlx1/Dlx2 may be dispensable for the development of the lower jaw, as there are barely any malformations in the lower jaw associated with Dlx1/Dlx2 single or compound mutations. Single Dlx2 and compound Dlx1/Dlx2 mouse mutants exhibit no abnormalities in the ventral region of the hyoid arch, although they display cleft hyoid bodies and fusion of the greater horns to the superior horns of the thyroid cartilage [110]. Interestingly, although the expression of Dlx2 is unaltered in the mandibular process of Edn1 mutants, it is slightly diminished in the hyoid arch [103]. On the other hand, mice with targeted deletion of Dlx5 have lower jaw defects, particularly hypotrophy and dysmorphy of Meckel’s cartilage [113,114]. In double-null Dlx5/Dlx6 mice, the shape of maxillary and mandibular processes is identical during embryogenesis, and the lower jaw never develops Meckel‘s cartilage, but mouse whiskers arise on its surface [110,115,116]. Concomitantly, compound Dlx5/Dlx6 mouse mutants exhibit the truncated styloid process with an ectopic process extending towards it from the hyoid bone and lesser horns projecting towards the neurocranial base [110]. The forced expression of Dlx5 in NCCs in the maxillary process leads to upregulation of mandibular-specific genes and appearance of several phenotypic hallmarks of the mandible in the maxilla region [106]. This represents the aforementioned homeotic transformation of the lower jaw into the upper jaw-like structure, which suggests that the default state of the jaw is maxillary, and EDN–DLX–HAND is required to initiate the lower jaw development programme. The specific function of Dlx3/Dlx4 during the development of the mandibular arch remains elusive, since no craniofacial phenotype has been described in Dlx3-null mice, and Dlx4-null mice have not been reported yet [117]. However, Dlx3/Dlx4 is induced by Dlx5/Dlx6 [116,118], and their functional redundancy cannot be excluded. In summary, combinatorial Dlx expression domains within PAs make up a prerequisite for intra-arch identity of individual skeletal elements along the proximal–distal axis [37,110].
Hand basic helix-loop-helix transcription factors are expressed in the distal region of the mandibular process, where they act to specify the so-called distal tip. Hand2 is regulated by Dlx5/Dlx6, which are induced by EDN1 signalling (END–DLX–HAND), therefore specifying the mandibular identity (see Figure 1). The view that Hand2 expression is not compatible with maxillary development is further supported by Sato et al., who show that ectopic Hand2 expression transforms the maxilla into the mandible [104]. Of note, Dlx5/Dlx6 and Hand2 are severely reduced in the hypoplastic mandibular process of Mef2c NC-specific mutants, which links Mef2c to the Edn–Dlx–Hand regulatory network [119]. Contrary to Edn1 induced by the expression of Hand2, the expression of Hand1 requires BMP signalling. Moreover, HAND2 acts synergistically with BMP to regulate the expression of Hand1 [120,121], since Hand1 expression is markedly downregulated in Hand2 mutants. Hand1 and Hand2 are expressed in the distal tip domain of the mandibular process and hyoid arch, which is mutually exclusive with the more proximal expression domain of Dlx5/Dlx6. Tissue-specific inactivation of Hand2 in NCCs leads to ectopic ossification in the distal tip of the mandible, heterotopic bone in the symphysis, and tongue hypoplasia [120,122]. Similarly, the deletion of the branchial enhancer of Hand2 in the mandibular arch leads to the hypoplasia of the mandible and cartilage malformations, such as truncation of Meckel’s cartilage and abnormal projections of the malleus and lesser horns of the hyoid [107]. Multiple defects of the hyoid apparatus have been reported to occur in Hand mutants, including poor ossification of the hyoid bone and lesser horns, deformation of the hyoid body in the midline, fusion of the hyoid body and thyroid cartilage in the midline, fusion of lesser horns and palatine bones, and aberrant articulation of the styloid process with greater horns [120,123,124,125]. Meis2 appears to act upstream of Hand2 because NC-specific Meis2 mutants exhibit decreased Hand2 expression in the first and second PAs. In Wnt1-Cre2-driven, tissue-specific deletion of either Meis2 or Hand2 in NCCs, mutant mice show comparable tongue hypoplasia, mandibular retrognathia, and symphyseal ossification [79,120]. The expression of Hand genes in the distal tip of the mandibular process thus restricts osteogenesis in the prospective tongue region, while Dlx genes ensure the development of individual pharyngeal skeletal elements in the proximal region [121]. Altogether, BMP simultaneously with EDN1 acts to divide the nascent mandibular process into the tongue-forming Hand-positive nested domain and the bone-forming Dlx-positive nested domain.
5.2. Zebrafish
In the zebrafish, edn1 is expressed in the pharyngeal ectoderm, mesoderm, and endoderm. However, only ectodermal edn1 seems to control the fate of NCCs during the formation of the intermediate-ventral region of PAs (see Figure 2). Thus, mutations in edn1 lead to hypoplasia of the Meckel’s cartilage and its fusion with the palatoquadrate cartilage [126]. In addition, both edn1 mutants and edn1 morphants have malformed intramembranous bones within the mandibular and hyoid arches [127]. The mutant phenotype is also reflected in the alteration of the molecular imprint, as edn1 mutants have decreased expression of hand2, dlx2a, msxE, and gsc, especially in the ventral region of PAs [128], while nkx3.2 in the jaw hinge region is also reduced [129]. The essential role of edn1 during patterning of the ventral region of PAs was confirmed by heat-shock experiments, resulting in disrupted expression of edn1, the reduction of hand2, and the simultaneous expansion of dlx3b, dlx5a, dlx6a, and nkx3.2 [130]. Unlike in the mouse, edn1 does not recognize one but two paralogue receptors, Ednra1 and Ednra2. Ednra1 is expressed in the migratory and early postmigratory NCCs within PAs, whereas endra2 is expressed in the late postmigratory NCCs [126]. Ednra1 knockdowns display the fusion of joints in hinge regions of the mandibular and hyoid arches, as well as retrognathia. Unlike ednra1, knockdown of ednra2 does not affect PA development. Moreover, ednra1/2 double knockdown mutants miss the lower jaw and ceratohyal cartilage, similar to edn1 mutants [126]. Thus, Edn1 signalling via Ednra1 and Ednra2 is important during development of the ventral region of PAs. [131]. Collectively, data from mice and zebrafish suggest the evolutionarily conserved function of edn1 in postmigratory NCCs and during the development of ventral pharyngeal cartilages in gnathostomes.
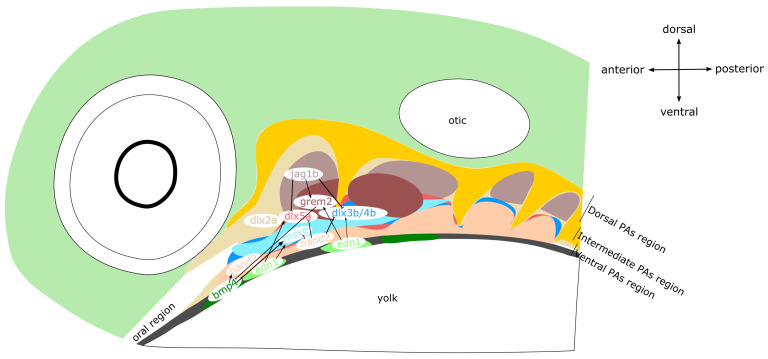
Figure 2.
Schematic model of PA patterning along the dorsal–ventral axis in zebrafish at 30–36 hpf (hours post-fertilization). Lateral view, head to the left, pharyngeal endoderm in dark yellow, pharyngeal ectoderm in dark grey. The dorsal region of PAs is established by co-operative action of jag1b and grem2 and characterized by the expression of dlx2a. The intermediate region of PAs is controlled by dlx3b/4b/5a and msxe, which are downregulated by edn1 in the pharyngeal ectoderm. The ventral region of PAs is dominated by the expression of bmp4-hand2.
The craniofacial phenotypes of edn1 and hand2 mutants appear to be similar. Hand2 zebrafish mutants lack the lower jaw and ventral set of second pharyngeal cartilages [129]. In fact, edn1 positively regulates hand2 during the development of ventral pharyngeal cartilages. Upon the early NC migration, hand2 restricts cell proliferation during the anterior-ventral protrusion of NCCs, which is under the control of edn1. However, at later stages of development, the function of hand2 shifts, and it eventually promotes the cell proliferation [131]. Additionally, hand2 also influences the cell movement within the mandibular arch, but apparently independently of edn1 [131]. Nkx3.2 is expressed in ectomesenchyme of the lower jaw primordium, and during the chondrification, its expression becomes localized within and around the jaw joint. In keeping with this, hand2 regulates development of the jaw joint via modulation of nkx3.2 expression [129,132]. Therefore, nkx3.2 is involved in specification of the intermediate region of PAs, the hinge region, and is expressed ventrally to dlx2a and dorsally to hand2 [129]. The expression of nkx3.2 in the presumptive jaw hinge region is regulated by Hand2 via gsc and dlx3b/4b/5a. Hand2 activates the expression of gsc, which in turn represses nkx3.2. Meanwhile, dlx3b/4b/5a repress gsc and activate nkx3.2 [132].
Dlx genes are under the control of Edn1 and Bmp signalling [133]. The genome of zebrafish contains four bigene dlx pairs—dlx1a/dlx2a, dlx3b/dlx4b, dlx5a/dlx6a, and dlx2b/dlx4a [134]. Along the dorsoventral axis of PAs, dlx3b/dlx4b and dlx4a can be detected in the intermediate region. While dlx2a is expressed in the dorsal region of PAs, dlx5a/dlx6a are found in the ventral region and the intermediate region of PAs (see Figure 2). The expression of dlx2b is excluded from the first two PAs. In the ventral region of PAs, dlx genes are repressed by hand2 [132]. Double knockdown of dlx1a/dlx2a causes defects in the dorsal pharyngeal cartilages (palatoquadrate and hyosymplectic cartilages), bearing similarities to murine Dlx1/Dlx2 mutants [110,111,112]. The patterning of mandibular and hyoid arch hinge regions, the opercle, and branchiostegal rays is influenced by dlx5a, dlx3b, and dlx4b. Interestingly, single knockdown of dlx5a, dlx3b, or dlx4b does not produce any changes in the pattern of expression. Conversely, simultaneously knocking down all of them leads to the loss of hinge region joints and fusion of the opercle with branchiostegal rays [132]. Taken together, dlx1a and dlx2a control the patterning of the dorsal region in the mandibular and hyoid arches, whereas both dlx3b/dlx4b and dlx5a/dlx6a regulate the development of intermediate region. The expression of dlx genes in the zebrafish is in accordance with the mouse, as Dlx1/Dlx2 govern morphogenesis in the dorsal region of PAs, which equals the upper jaw region, the styloid process, and the stapes, whilst Dlx3/Dlx4 and Dlx5/Dlx6 establish the intermediate and ventral region of PAs, which comprises the presumptive lower jaw and lesser horns of the hyoid. Therefore, the patterning along the anterior–posterior and dorsal–ventral axes in mice and zebrafish is under the control of a common regulatory cascade, EDN–HAND–DLX (see Figure 1 and Figure 2).
6. Combinatorial Action of FGF8, BMP4, and SHH Signalling Pathways during Morphogenesis of Mandibular and Hyoid Arches
6.1. Mouse
Alongside transcription factors, numerous protein ligands also serve essential functions during the patterning of mandibular and hyoid arches. Ffg8 is expressed in the oral epithelium from which it diffuses into the underlying ectomesenchyme. Fgf8 is a key survival factor of the NC because its ablation in the oral ectoderm leads to massive apoptosis in the mandibular arch, as well as to complete loss of the proximal mandibular structures [135]. Moreover, Fgf8 determines the rostral–caudal axis of the mandibular arch. The expression of Fgf8 in the oral surface ectoderm induces the expression of transcription factors Lhx6/Lhx8 in the rostral mandibular mesenchyme. Concomitantly, this results in the restriction of Goosecoid (Gsc) expression in the caudal mandibular mesenchyme, therefore establishing the subdivision of the mandibular process into the rostral and caudal domain (see Figure 3 and Figure 4) [136]. At the same time, FGF8 acts together with BMP4 to specify the proximal–distal axis by regulating the expression of specific homeodomain-containing transcription factors in the ectomesenchyme, which subsequently defines the positional identity of individual teeth. Barx1 induced by FGF8 in the proximal region determines the molar identity, whereas BMP4-regulated Msx1 in the distal aboral region specifies the prospective incisors. Intriguingly, early mandibular epithelium can organize dental mesenchyme and dental papilla in the mouse hyoid arch, indicating a common regulatory circuit between the mandibular and hyoid arches during the early stages of PA development [137]. In the mandibular process, maintenance of Fgf8 expression is ensured by transcription factor PITX2, which simultaneously represses Bmp4 expression. Consistently, the expression of Fgf8 and its target genes, such as Barx1 and Pitx1, is severely reduced in Pitx2-null mutants, whereas the expression of Gsc in the mandibular process is expanded rostrally. Moreover, since high doses of Pitx2 are required for repression of BMP signalling, the expression of Bmp4, Msx1, and Msx2 is expanded as well. As a result, disrupted signalling in the mandibular arch due to the mutation in either Pitx1 or Pitx2 leads to a severe micrognathia, while single Pitx1 mutants also suffer from the bifurcation of the tongue and a novel bone deposition around Meckel’s cartilage [138,139,140].
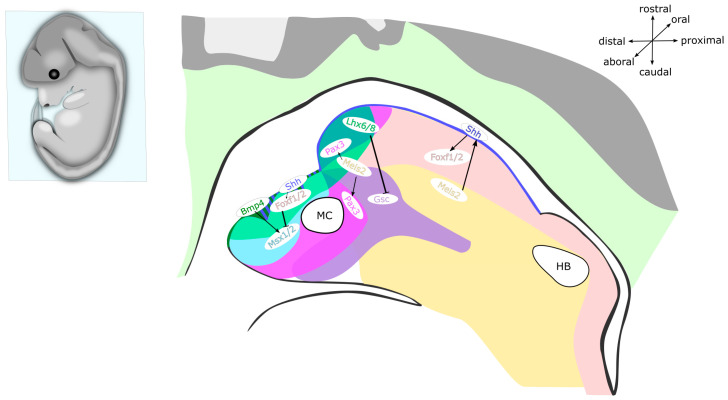
Figure 3.
Schematics of a mid-sagittal section through the mandibular and hyoid arches of a mouse embryo at E12. Bmp4 is expressed in the distal mandibular epithelium, at the site of presumptive incisors, while Msx1 is expressed in the distal mandibular ectomesenchyme, surrounding the incisor primordia. Shh is expressed in the vestibular lamina and dental epithelium, as well as in the lingual epithelium, while Foxf1/Foxf2 are expressed in ectomesenchyme in the medial region of the mandibular process, surrounding the incisor primordium and inside the nascent tongue. Pax3 is expressed in the ectomesenchyme of distal tip of the mandibular process and nascent tongue. Lhx6/Lhx8 are expressed in ectomesenchyme on the rostral side, while Gsc is expressed on the caudal side of the mandibular process. Meis2 is expressed in the medial proximal region of the mandibular process and in the medial region of the hyoid arch. Abbreviations: E, embryonic day; MC, Meckel’s cartilage; HB, cartilage primordium of the hyoid bone. Expression domains of genes written in black text are not highlighted in the figure.
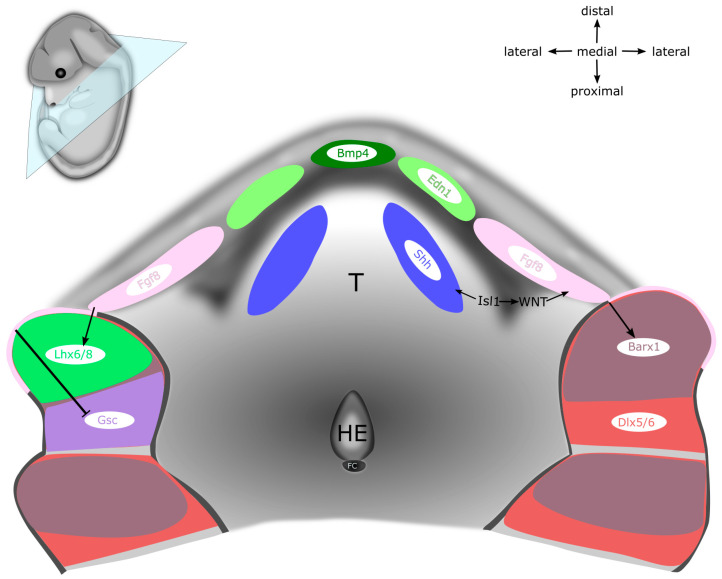
Figure 4.
Schematics of the mandibular and hyoid regions of a mouse embryo at E12, superior view at the nascent tongue from inside of the alimentary canal. Bmp4 and Edn1 are expressed in the distal mandibular epithelium, while Bmp4 also marks the site of presumptive incisors. Shh is expressed in lingual epithelium of the tongue primordium. Fgf8 is expressed in the proximal mandibular epithelium, at the site of presumptive molars, while Barx1 is expressed in the proximal mandibular ectomesenchyme, surrounding the molar primordia, and in ectomesenchyme of the hyoid arch. Dlx5/Dlx6 are expressed in ectomesenchyme of the mandibular process and the hyoid arch. Lhx6/Lhx8 are expressed in the ectomesenchyme on the rostral side, while Gsc is expressed on the caudal side of the mandibular process. Abbreviations: E, embryonic day; T, tongue primordium; HE, hypobranchial eminence (second PA); FC, foramen caecum. Expression domains of genes written in black text are not highlighted in the figure.
The rostral–caudal axis, which is defined by the complementary expression of Lhx6/Lhx8 and Gsc, is actually distinct from the oral–aboral axis (see Figure 3 and Figure 4) [141]. Corresponding to the expression pattern of Sonic hedgehog (Shh) in the oropharyngeal epithelium, the downstream targets and mediators of Hedgehog (HH) signalling, Foxf1 and Foxf2 are expressed in the subjacent mandibular mesenchyme [141,142]. Complementary to the expression of Shh on the oral side of the mandibular arch, Bmp4 is expressed in the complementary subdomain on the aboral side (the same domain that is important for the development of incisors) [141,143]. The expression of Foxf1/Foxf2 genes in the mandibular mesenchyme antagonizes the expression of Msx1/Msx2 induced by BMP4, thereby preventing the osteogenesis in the prospective tongue region. Upon ablation of either Smo or Foxf1/Foxf2 in NCCs via Wnt1-Cre2 recombination, the Bmp4 expression domain expands to the oral side of the mandibular arch, which leads to the formation of heterotopic bone on the oral side of the mandible [141]. Altogether, this shows that HH signalling in the mandibular arch is required for patterning the oral–aboral axis of the mandible.
Transcription factor MEIS2 modulates SHH activity in the mandibular process and determines its medial–lateral axis [79]. The targeted deletion of Meis2 in the NC using Wnt1-Cre2 driver leads to the downregulation of Shh and Ptc1 expression on the oral side of the mandibular process. Furthermore, the expression of Hand1/Hand2 in the distal tip of the medial region of mandibular and hyoid arches is reduced, while the gradient of Dlx5 and Barx1 expands from the lateral to medial regions. This patterning shift along the medial–lateral axis leads to the loss of molecular identity of NCCs in the prospective tongue. When Meis2 is deleted within NCCs in Wnt1-Cre2; Meis2 fl/fl mutants, the levels of PAX3 around the alveolingual sulcus (anatomical boundary between dentary bone and tongue) are markedly reduced and replaced with RUNX2, which subsequently leads to the formation of ectopic bone in the same region. As a result, the tongue is severely hypoplastic and its lateral edges are invaded by heterotopic bone. Altogether, the determination of the oral–aboral and medial–lateral axes in the mandibular process by the coordinated interaction of SHH, BMP, and the EDN–DLX–HAND regulatory cascade may be linked to the MEIS2 regulatory network, since its ablation in the NCCs leads to the downregulation of both Shh and Hand2.
Shortly after the colonization of PAs by NCCs, Shh is expressed in the oropharyngeal epithelium, from which it maintains the survival, proliferation, and patterning of the underlying mandibular mesenchyme. Both epithelial and mesenchymal cells in the mandibular arch express receptors Smo and Ptch1 and are therefore able to respond to SHH ligand. As development of the mandibular arch proceeds, spatially restricted centres of Shh induce the formation of numerous oral structures, including tongue, teeth, palate, and salivary glands (see Figure 1, Figure 3 and Figure 4) [144,145]. The elimination of SHH activity in either oropharyngeal epithelium via Nkx2.1-Cre,Shhflox or SHH responsiveness in the ectomesenchyme using Wnt1-Cre2;Smoflox causes extensive apoptosis of NCCs and results in mandible and tongue defects [141,142,146,147,148]. At the midline of the mandibular process, the expression of Shh specifies NCCs in the tongue primordium, thereby establishing the oral–aboral and medial–lateral axes. Moreover, Shh in this region allows the invading myogenic progenitors to permeate the nascent tongue primordium, thereby promoting the tongue development and preventing osteogenic differentiation in the midline [79,141]. Thus far, Shh has not been reported to exert any patterning activity in the second PA [149,150].
In the early pharyngula, signalling centres expressing Fgf8 and Shh are set up by Islet1 (ISL1). Isl1 is a member of the Lhx family that encodes transcription factors containing two LIM domains and a homeodomain. In the PA development, ISL1 acts as an epithelial ligand expressed in the oral ectoderm of the first PA and the endoderm of other arches [151]. Loss of Isl1 in β-catenin expressing cells leads to agnathia, a complete absence of the lower jaw [151]. When Isl1 is inactivated in the mandibular epithelium, specifically in Shh-expressing cells, the aberrant bony fusion of the distal tip of the dentary bone occurs, similar to Hand2 and Meis2 mouse mutants [79,123]. Both Fgf8 and Shh are missing in the oropharyngeal epithelium of the early pharyngula in Isl1 mutants [151,152]. Canonical WNT signalling is known to be upstream of Fgf8 in the first PA epithelium, and WNT signalling is disrupted in the first PA of Isl1 mutants, indicating a regulatory circuit of Isl1-Wnt-Fgf8 [151,153]. ISL1 may activate epithelial β-catenin signalling via repression of WNT antagonist. Intriguingly, reactivation of β-catenin in the mandibular epithelium of Isl1 mutants rescued the mandibular morphogenesis through SHH signalling to the mandibular ectomesenchyme. Furthermore, overexpression of Shh in the first PA epithelium partially restored the morphologic defect in Isl1 mutants and led to successful outgrowth of the dentary bone [154].
6.2. Zebrafish
In the zebrafish, fgf8 in concert with fgf3 establishes the segmentation of the pharyngeal endoderm within PAs during the early pharyngula stage [155]. Together, fgf8 and fgf3 control the survival of NCCs during the formation of pharyngeal cartilages. Therefore, loss of fgf8 leads to hypoplasia of the mandibular cartilage [2,155]. Furthermore, fgf3 knockdown results in misshapen ceratohyal and lack of ceratobranchial cartilages. The complementary function of fgf8 and fgf3 is strongly supported by severe pharyngeal malformations in compound fgf8 and fgf3 knockdown mutants, which results in loss of all ceratobranchial and hyoid arch cartilages, accompanied by significant size reduction of mandibular arch cartilages [155]. Later in development, fgf8 is essential for the proper expression of the osteogenic genes runx2 and sp7 during the craniofacial ossification [2]. Together with Bmp, Fgf signalling controls the expression of barx1. Barx1 is expressed in migratory NCCs and also later in the ectomesenchyme within PAs, where it maintains the chondrogenic cell fate and negatively regulates the development of the jaw joint, and its loss initiates osteogenic differentiation within chondrocytes [156].
In conjunction with Edn1, Bmp signalling patterns the dorsal–ventral axis of PAs (see Figure 2) [130,133]. Lack of Bmp signalling in PAs leads to either reduction or even loss of ventral pharyngeal cartilages, such as Meckel’s cartilage and ceratohyal cartilage, and intermediate pharyngeal cartilages, such as joints, interhyal cartilage, and the ventral part of symplectic cartilage [133]. Conversely, bmp overexpression transforms and fuses hyosymplectic cartilage into a structure reminiscent of ceratohyal cartilage, and joints within mandibular, hyoid arches, as well as the ventral part of hyosymplectic cartilage, are lost. Moreover, palatoquadrate cartilage is also transformed into a structure resembling Meckel’s cartilage [130]. Akin to the loss of Bmp signalling, edn1 overexpression results in similar defects in dorsal pharyngeal cartilages of the mandibular and hyoid arches, except for the joints.
In the early pharyngula, the inhibition of Bmp signalling causes the downregulation of Edn1 signalling, as well as downregulation of hand2 and dlx6a in ventral pharyngeal cartilages [133]. At first, Bmp induces Edn1 signalling and restricts the expression of jag1b in the dorsal region of nascent PAs. In addition, the joint action of Bmp and Edn1 activates hand2 via dlx5a/dlx6a in the ventral region of PAs. After the NC migration, Bmp controls the ventral fate of PAs in an independent manner, whereas Edn1 regulates the intermediate region of PAs. During this stage, hand2 in the ventral region is under the exclusive control of Bmp. Furthermore, Hand2 represses intermediate-region-specific genes, such as nkx3.2, as well as ventral-region-specific dlx3b/dlx4b/dlx5a [130,133]. The restriction of bmp in the ventral region of PAs is mediated by the dorsal-intermedial expression of grem2, an antagonist of Bmp signalling, induced by edn1 and jag1b (see Figure 2). In the ventral region of PAs, Bmp inhibits grem2 expression [130], whereas the intermediate region of PAs is established by the collective action of dlx3b, msxe, and nkx3.2 [133]. While msxe expression is coregulated by both Bmp and Edn1, dlx3b expression is driven solely by Edn1. To sum up, the grem2-mediated repression of bmp restricts hand2 to the ventral region of PAs, where Hand2 acts to inhibit the expression of dlx3b, dlx5a, dlx6a, and nkx3.2 (see Figure 2) [130,133]. Thus, during pharyngeal chondrogenesis, Bmp signalling governs the specification of ventral cartilages, whereas Edn1 regulates the development of intermediate cartilages [130,133].
In contrast to zebrafish, FGF, BMP, and SHH set up the position of the prospective tongue and teeth within the murine oral cavity. In zebrafish, these molecules do not play a complementary role, as teeth in zebrafish grow inside the pharynx, not within the oral cavity, and they are not heterogenous, meaning they do not have incisor/molar identity. Moreover, a tongue-like structure in zebrafish is not homologous with the muscular tongue of tetrapods, so SHH signalling at the midline of murine embryos is not readily comparable to that in the zebrafish [157]. However, in both mice and zebrafish, Bmp signalling controls the development of the ventral pharyngeal region, as loss of bmp in the zebrafish leads to the lack of ventral pharyngeal cartilages, and ectopic expression of Bmp in the mouse results in the duplication of the dentary bone.
7. Molecular Regulation of Osteochondrogenesis in the Mandibular and Hyoid Arches
7.1. Mouse
During the morphogenesis of PA-derived skeletal elements, the differentiation of osteoblasts and chondroblasts from a common osteochondral progenitor represents a critical step towards the formation of bone and cartilage. In regions of prospective cartilage and bone, these osteochondral progenitors aggregate and condense. Both intramembranous and endochondral ossification start from mesenchymal condensations, but the processes themselves are different: during intramembranous ossification, mesenchymal progenitors can differentiate exclusively into osteoblasts, whereas endochondral ossification encompasses the differentiation of both osteoblasts and chondroblasts. The key difference is that chondroblast differentiation precedes the formation of endochondral bone. Osteochondroblastic differentiation and maturation are regulated by three master transcription factors, SOX9, RUNX2, and SP7 (also known as Osterix, OSX). Osteochondral progenitors in early mesenchymal condensation have dual differentiation potential, as they coexpress Sox9 and Runx2 [158,159,160,161].
During skeletogenic differentiation, WNT signalling is a key regulator of chondroblast versus osteoblast cell fate choice in NCCs. The tissue-specific conditional deletion of β-catenin (Ctnnb1), the effector of canonical WNT signalling, results in the complete agenesis of cranial bones [162]. Concomitantly with WNT signalling the inactivation and agenesis of cranial bone, osteogenic progenitors are diverted into the chondrogenic fate, and an ectopic cartilage forms [163,164]. An alternative hypothesis is that RUNX2 and SP7 are intrinsic factors which are not only required for the determination of osteoblastic cell type, but they also play a role in suppressing the differentiation program that leads to chondroblastic cell fate. Cell fate at early stages of differentiation is seemingly still flexible because Runx2-expressing osteoblasts still maintain some cell fate plasticity. Full differentiation along the osteoblast lineage is likely ensured by Sp7, since in mouse mutants with inactivated Sp7, ectopic chondrocytes form at the expense of osteoblasts in some areas where intramembranous bone should form [165].
Transcription factor SOX9 is a master regulator of chondrogenesis, and its expression in NCCs is necessary for the formation of craniofacial cartilage. SOX9 probably regulates chondrogenesis by upregulating the expression of Col2a1 and Col11a2, types of collagen found predominantly in the cartilage [158,166]. The tissue-specific deletion of Sox9 in the NCCs results in loss of all cartilage elements derived from the cranial neural crest. Intriguingly, although the dentary is smaller in Sox9-deficient mice, the gross morphology and bone formation are not severely affected [167]. Furthermore, inactivation of Sox9 in cranial NCCs also results in upregulation of osteoblast marker genes such as Runx2, Sp7, and Col1a1 [167]. This further supports the notion that the osteoblastic differentiation programme plays a role in suppressing chondroblastic cell fate, and vice versa. When it comes to the hyoid arch, the specifier of second arch fate HOXA2 regulates the expression of Sox9. Under normal circumstances, HOXA2 prevents chondrogenesis in the second PA by suppressing the expression of Sox9 [66]. When HOXA2 is absent, chondrogenesis is activated ectopically and a duplicated set of first PA cartilages appear in the Hoxa2 expression domain of the hyoid arch. Thus, the expression of Sox9 in NCCs is required for the differentiation of common osteochondrogenic progenitors into chondroblasts and for the formation of all craniofacial cartilages.
RUNX2 is a transcription factor that controls the differentiation of mesenchymal progenitors (preosteoblasts) into osteoblasts and is expressed in early osteoblasts, hypertrophic Meckel’s cartilage, and mineralized bone [168]. RUNX2 is also a positive regulator of hypertrophic differentiation, as Runx2-null mice lack hypertrophic cartilage whatsoever. Systemic deletion of Runx2 in mice shows that it is important for both intramembranous and endochondral ossification [169]. Loss of Runx2 in mice leads to total agenesis of bone and a complete loss of expression of osteocalcin and osteopontin, two major non-collagenous proteins in the bone matrix [169]. In the absence of Runx2 solely in the neural crest, loss of frontal, zygomatic, squamous temporal bone occurs, whereas the dentary, maxilla, premaxilla, and nasal bones are severely hypoplastic and hypomineralized [170]. However, deficiency of Runx2 in mice not only affects bone but also both the primary and secondary cartilage, as mutant mice lack the condylar cartilage and have deformed Meckel’s cartilage [171]. Runx2 is controlled by DLX5 and both are essential in driving the differentiation of mesenchymal precursors into osteoblasts. In the prospective tongue region in the mandibular arch, Hand2 plays a major role in establishing a negative feedback loop in the DLX5/DLX6-RUNX2 circuit. Furthermore, ossification defects in Runx2-deficient mice reach beyond the mandibular arch, as the mineralization of the hyoid body is impaired as well. The transition of preosteoblasts into mature osteoblasts is regulated by SP7, a major downstream target of RUNX2. All osteoblasts and even hypertrophic chondrocytes express Sp7. Although deficiency of Sp7 in mice leads to the loss of dentary bone, the development of Meckel’s cartilage is seemingly not affected at all [165]. Interestingly, when Sp7 is lost exclusively in the neural crest, the dentary bone forms but ends up tiny and rudimentary [172]. To summarize, Runx2 expression within PAs gives NCCs the potency to form bone, while Sp7 is required for full commitment to osteoblastic lineage.
Muscle segment homeobox transcription factors (Msx1 and Msx2) are initially expressed together with Sox9 in the migrating cranial NCCs. Upon complete colonization of PAs, expression domains of Msx and Sox9 become separate [173]. Until cranial NCC migration within the mandibular process is completed, MSX2 inhibits chondrogenic differentiation of Sox9-positive NCCs. In mice and humans, both single Msx2 and compound Msx1 and Msx2 mutations lead to cleidocranial dysplasia with enlarged parietal foramina [174]. This rare genetic condition is characterized by disrupted osteoblast differentiation that clinically presents with hypoplasia of jaw and tooth abnormalities, among many other symptoms. Loss of Tbx1 specifically in murine NCCs induces a similar phenotype to cleidocranial dysplasia and results in a lack of the hyoid body and fusion to the thyroid cartilage [175]. Generally, the genetic cause of classical cleidocranial dysplasia in humans is heterozygous loss of RUNX2, not MSX2. However, the hyoid phenotypes of Runx2+/− and Tbx1−/− are different, indicating that Tbx1 might have a primary role in early patterning and perichondral ossification in the hyoid bone [175]. As mentioned before, hyoid anomalies occur in human patients with 22q11.2 deletion syndrome, the clinical picture of which is thought to be caused by loss of the TBX1 gene. Alongside the cleidocranial dysplasia, a mutation of the MSX2 gene in humans can cause craniosynostosis and enlarged parietal foramina, whereas haploinsufficiency can lead to midline cranial defects [176,177,178]. On the other hand, mutations in MSX1 are connected predominantly with dental abnormalities in humans, such as Witkop syndrome and tooth agenesis [179]. Mutations of Msx genes in mice also encompass a wide variety of first and second arch malformations. Overexpression of either wild-type or mutant human MSX2 in transgenic mice causes mandibular hypoplasia, cleft palate, and decreased ossification of the hyoid, etc. [180]. Single Msx1-null mouse mutants display an anomalous malleus, loss of alveolar dentary bone and maxilla, and failure of tooth development. [181]. The combined loss of Msx1 and Msx2 in mice results in severe defects such as cleft palate, truncated mandibular process, and decreased volume of trigeminal ganglia [182]. To summarize, during early craniofacial development, Msx genes influence the suppression of chondrogenesis and later control the skeletogenic differentiation, as overexpression, misexpression, or deficiency of Msx impedes the osteoblastic differentiation and results in craniofacial bone, cartilage, and tooth defects.
PAX3 is a transcription factor that is robustly expressed in cranial NCCs that make up the entire palatal, lingual, and mandibular mesenchyme, where it possibly keeps mesenchymal NCCs in an undifferentiated state [183]. Later in development, the mesenchymal expression localizes to the distal tip of tongue and the mandible [79,183]. Pax3 mutants with persistent Pax3 overexpression in the entire mandibular arch, including the tongue, display defects in osteogenesis. In NCCs, PAX3 directly regulates the expression of a soluble inhibitor Sotdc1, which diminishes responsiveness to BMP and decreases the expression of Runx2 [183]. In Meis2 NC-specific conditional mutants, the expression domain of Pax3 in the tongue is dramatically reduced, whereas the Runx2 expression domain is expanded in the medial mandibular region, which leads to heterotopic ossification in the lingual mesenchyme [79]. In Pax3-deficient Splotch mice, the hyoid bone is often split and partially fused to the thyroid cartilage [184]. In NCCs, Pax3 seems to be colocalized with Goosecoid (Gsc) in the postotic NC, frontonasal prominence, mandibular arch, and hyomandibular cleft [185]. Gsc encodes a highly conserved homeodomain transcription factor. During cranial morphogenesis, Gsc is initially expressed in the undifferentiated tissue of first and second PAs. During PA formation in mouse, Gsc expression persists in the nascent lower jaw and tongue, as well as in the hyomandibular cleft [186]. Among many skeletal malformations, Gsc-null mice exhibit malformations of malleus, palatine, maxillary, alisphenoid, pterygoid, coronoid, and angular processes [187,188]. Since Gsc is also expressed in the hyomandibular cleft, its inactivation results in auditory canal atresia and loss of tympanic rings. In humans, heterozygous loss of GSC results in SAMS syndrome (short stature, auditory canal atresia, mandibular hypoplasia, and skeletal abnormalities), which further confirms the role of Gsc in craniofacial and joint development. Gsc is possibly a downstream effector gene of regulatory networks that defines the specification and cell fate of neural crest and mesodermal lineages. Therefore, dysregulation of GSC-mediated gene expression in the connective tissue results in pathological differentiation and adaptation of new cartilaginous or osseous fate [185].
Skeletogenesis in the medial region of the mandibular process is regulated by Prrx transcription factors. Prrx1 and Prrx2 are expressed in overlapping domains throughout the ectomesenchyme of PAs, with the strongest expression in the mandibular and hyoid arches [189,190]. Of note, the domains of Prrx1 expression in the ventral regions of the mandibular and hyoid arches are similar to the expression domains of Hand2. Single Prrx1 knockout mice display extensive malformations of the viscerocranium derived from the mandibular and hyoid arches, including fusion of the incus to palatoquadratum and fusion of the stapes to Reichert’s cartilage [191]. Single Prrx2 knockout does not result in any discernible abnormalities in the craniofacial skeleton, suggesting that Prrx1 compensates for the loss of Prrx2. However, compound Prrx1 and Prrx2 knockout mice have amplification of the craniofacial phenotype found in single Prrx1 mutants [192]. As a result, the lower jaw is micrognathic, fused at its anterior tip and often has only a single incisor in the midline. The cause of this defect may have several explanations [193]. Firstly, the downregulation of Shh in the oral epithelium of compound Prrx1/Prrx2 mutants can lead to reduced cell proliferation, resulting in the mandibular hypoplasia. Secondly, Prrx1/Prrx2 double mutants contain a large population of Runx2-positive cells in the middle and rostral region, indicating precocious or accelerated osteogenesis in the mandibular process. Only remnants of Meckel’s cartilage in the rostral region of the mandibular process are preserved in Prrx1/Prrx2 double mutants. Loss of the main body of Meckel’s cartilage and increased osteogenesis may be related to changes in the mesenchymal precursors from chondrogenic to osteogenic fate. Thirdly, the expanded domain of Runx2 expression in mutants may be a result of increased proliferation or decreased apoptosis of osteogenic mesenchyme and/or the recruitment of additional osteoprogenitor cells to compensate for the loss of Meckel’s cartilage. Unlikely, but still possible, the expanded domain of expression of Runx2 may reflect the fusion of multiple osteogenic condensations. Similar to mice, the homozygous or dominant heterozygous loss of PRRX1 in humans has been associated with loss of the lower jaw [194,195,196,197,198]. Prrx1/Prrx2 mouse mutants also display abnormalities in the dorsal hyoid arch. The stylohyoid ligament is not formed completely in the Prrx1-null mutant, and a part of it develops as a cartilaginous element [191]. In contrast, in compound Prrx1 and Prrx2 mutants, the entire stylohyoid ligament chondrifies and forms a continuous structure stretching from the stapes and styloid process to the lesser horns of the hyoid bone. Moreover, the chondrification of the stylohyoid ligament also occurs in Prrx1+/− Prrx2−/− mice [92]. The accelerated osteogenesis in the Prrx1/Prrx2 double mutants results in similar abnormalities as those in the Hand compound mutants and abnormalities in mice with the deletion of PA-specific enhancer dHand (Hand2), which also exhibited accelerated osteoblast differentiation [122,123]. Of note, the expanded Dlx5- and Runx2-positive domains and downregulation of Shh in the medial and oral region of the mandibular process are also found in Meis2 NC-specific mutants [79]. The second PA phenotype of Prrx1/Prrx2 double null mice is astoundingly similar to the hyoid abnormalities in Meis2-null mice, which suggests that there might be a link between Prrx1/Prrx2 and Meis2 during the hyoid arch development [92]
During the pharyngula stage, Shh influences the development of Meckel’s cartilage. Tissue-specific inactivation of Shh in the oropharyngeal epithelium in Nkx2.1-Cre; Shhflox leads to a complete lack of Meckel’s cartilage formation in the mouse embryos. In contrast, when SHH responsiveness is deleted from the ectomesenchyme using Wnt1-Cre2; Smoflox, Meckel’s cartilage still develops, albeit truncated. Either SHH acts through an SMO-independent mechanism or its effect on Meckel’s cartilage is indirect through another signalling molecule, such as FGF8.
Another member of the Hedgehog family, Indian Hedgehog (IHH), is a signalling molecule widely recognized as a regulator of skeletal development that is expressed in the prehypetrophic chondrocytes and early hypertrophic chondrocytes. In craniofacial morphogenesis, the expression of Ihh has traditionally been associated with the secondary cartilage—the mandibular symphysis, angular, coronoid, and condylar processes. In Ihh-null mice, the development of mandibular symphysis is defective due to precocious chondrocyte maturation and reduced proliferation of the chondroblast progenitors. However, this phenotype can be rescued upon ablation of Gli3, which thus acts as a negative regulator of symphyseal development [199,200]. Furthermore, IHH signalling during embryogenesis promotes the expression of parathyroid hormone-related protein (Pthrp) at the apical end of the presumptive condylar cartilage, thereby increasing numbers of presumptive chondroprogenitor cells. In keeping with this, the articular disc and temporomandibular joint are absent in Ihh-null mice, and the condylar process directly opposes the glenoid fossa. Interestingly, the disc phenotype of Ihh-null mice is not rescued in the concurrent absence of Gli3 [199]. Conversely, tissue-specific augmentation of Ihh expression in the NCCs leads to severe craniofacial abnormalities, including a complete loss of the glenoid fossa [201]. In mice, IHH signalling via PTC1 controls the proliferation and differentiation of mesenchymal cells into chondrocytes during growth of the mesial alveolar process of dentary bone. Furthermore, Ihh-null newborn mice have the overall length of the mandibular body reduced by as much as a third, including secondary cartilages [202]. Likewise, in humans, patients carrying a mutation in GLI2 exhibit a range of facial defects, including mandibular hypoplasia [203]. To summarize, IHH regulates a myriad of processes during craniofacial morphogenesis—including the proliferation and maturation rate of chondrocytes, endochondral ossification, the expression of Pthrp in periarticular tissue, articular disc formation, and synovial cavity formation.
Transforming growth factor beta (TGF-β) is a protein ligand that acts as a stimulant in cranial NCCs, increasing the proliferation of chondrocytes and the production of cartilage extracellular matrix [204]. Tissue-specific loss of Tgfbr1 in NCCs causes delayed tooth initiation and mandibular defects, particularly in the proximal region, including loss of secondary mandibular cartilages [205]. Moreover, Tgfbr1 mutants also exhibit a lack of the stapes and severe malformations of second PA cartilage with differentiation of multiple novel ectopic elements derived from the NC. Viscerocranial phenotypes of NC-specific Tgfbr2 mouse mutants are less severe but comparable to those in Tgfbr1 mutants [205,206]. Tissue-specific deletion of Tgfbr2 in NCCs particularly affects the lower jaw and palate [207,208,209]. Explicitly, the elimination of Tgfbr2 causes micrognathia and loss of secondary mandibular cartilages.
Signalling proteins from the BMP subfamily are major factors influencing the development of dentary bone, as ectopic expression of Bmp on the oral side of the mandibular process results in the formation of mirror-image dentary bone. The absence of two dedicated BMP4 antagonists, Chordin and Noggin, in the distal mandibular epithelium during the mandibular process patterning results in elevated levels of BMP4 at the expense of FGF8, increasing cell death and leading to the spectrum of mandibular hypoplasia, culminating in almost total agnathia [210]. In contrast, the absence of a single allele of BMP4 antagonist Noggin leads to significantly thicker cartilage along with increased pSmad1/5/8 expression, leading to ossification rather than degeneration of Meckel’s cartilage [211]. Similarly, in mutant mice with a complete lack of Noggin, the hyoid body is greatly enlarged, and its shape changes due to significantly shorter and wider horns [212]. Ectopic Bmp4 expression in NCCs leads to bony fusion of the dentary and maxilla, which is reminiscent of the syngnathia birth defect in humans [213]. Upon ablation of Bmp2 specifically in NCCs, the expression of Sox9 is downregulated in both the mandibular process and Meckel’s cartilage, which results in micrognathia and cleft palate, characteristic features of Pierre Robin malformation sequence [214]. Compound loss of Bmp2 and Bmp4 in NCCs results in more severe shortening of mandible then in either of single mutants [215]. In mice with homozygous mutation in Bmp5, loss of the lesser horns and shortening of the greater horns of the hyoid occurs [216]. Mice with a deficiency of Bmp7 exhibit shortening of maxilla and mandible, as well as a failure of Meckel’s cartilage fusion at the anterior tip [217].
Even though mutations in Fgfs and genes coding FGF receptors (FGFRs) are usually associated with craniosynostoses, these signalling molecules exert their influence over PAs as well. Fgfr1 is expressed in the pharyngeal epithelium and is necessary to create a permissive environment for the immigration of NCCs. Mice homozygous for a hypomorphic allele of Fgfr1 display reduced Fgf3 expression in the pharyngeal epithelium, which prevents NCCs from entering the second PA and induces apoptosis instead [218]. In Ffgr1 hypomorphs, craniofacial skeletal malformations occur mainly within the second PA and include a deficient or missing stapes and a missing proximal part of the styloid process. In general, the abnormalities in pharyngeal development in embryos in which FGF signalling has been disrupted are strikingly similar to each other, with the most consistent phenotypic feature being the hyoid arch hypoplasia [218,219,220,221,222]. Elimination of Fgfr1 specifically in NCCs causes orofacial dysformation, tooth bud defects, and micrognathia [223]. One diagnostic feature of craniosynostosis syndromes in humans is mandibular dysgenesis. Gain-of-function mutation in Fgfr2 leads to Ffgr2-related craniosynostosis and mandibular dysmorphogenesis, demonstrating that Fgfr2 influences cartilage and intramembranous bone formation [224]. Additionally, in a mouse model of achondroplasia, gain-of-function mutation in Fgfr3 leads to structural anomalies of Meckel’s cartilage and secondary mandibular cartilages, resulting in mandibular hypoplasia and dysmorphogenesis [225]. The formation of Meckel’s cartilage is critically dependent on the FGF8 molecule. Exogenous FGF8 is able to rescue Meckel’s cartilage in mouse mandibular explant cultures treated with the antagonist of HH signalling [226]. In the chick mandibular process, implanting SHH-soaked beads into the tissue leads to the development of supernumerary Meckel’s cartilage and an ectopic expression of Fgf8. Conversely, a significant reduction of Fgf8 in the proximal region of the mandibular process is seen in mouse mutants with tissue-specific ablation of Shh in the oropharyngeal epithelium, which lack Meckel’s cartilage altogether [146]. Intriguingly, overexpression of Fgf8 in NCCs leads to severe craniofacial malformations, including exencephaly, maxilla, and dentary bone agenesis [227]. In Fgf8 hypomorphic mutants, both the mandibular and hyoid arches are obviously smaller, showing a reduction in total size. Moreover, these mutants have either absent or severely hypoplastic Meckel’s cartilage, absent malleus and incus; severely defective dentary bone and tympanic ring; and reduced or absent alisphenoid, presphenoid, squamous temporal bone, pterygoid, palatine, and ala temporalis bone. In Fgf8 hypomorphic embryos, the stapes is normal or slightly smaller, the styloid process is thickened and/or shortened, and in a subset of mutants, the hyoid bone is mildly defective, which is surprising given the severity of defects noted in the early hyoid arch development [228]. Fgf10 modulates the early morphogenesis of Meckel’s cartilage by controlling cell differentiation in the mandibular process. Overexpression of Fgf10 in rat mandibular explants results in deformation of Meckel’s cartilage and a significant increase in its size, whilst also inducing the upregulation of cartilage specific genes, such as Col2a1 and Sox9 [229]. Accordingly, genetic polymorphism in FGF10 has been linked with mandibular prognathism in humans [230]. The FGF pathway is overreactive in mice with mutations of Sprouty genes (Spry), which encode for inhibitors of receptor tyrosine kinases that are crucial for regulation of FGF downstream signalling. NC-specific deletion of Spry1 results in malformed and incomplete maxilla, as well as a smaller mandible. Single Spry4 loss or simultaneous loss of both Spry2 and Spry4 leads to micrognathia and growth retardation of the mandible, as well as incisor anomalies [231]. In summary, the combined action of the TGB-β, BMP, and FGF signalling pathways collectively controls the proliferation, maintenance, and cell fate specification of ectomesenchymal cells during osteogenic and chondrogenic differentiation. Furthermore, several mouse mutants with anomalies in the mandibular and hyoid arches have supernumerary pharyngeal skeletal elements, indicating that the ectomesenchyme in the head indeed retains an ability to form a spectrum of novel structures in response to either loss of cell signalling or ectopic cell signalling. These novel structures may represent skeletal atavisms and could be caused by a reactivation of a dormant developmental programme.
7.2. Zebrafish
Similar to the mouse, Wnt signalling is important for chondrogenic cell fate during craniofacial development in the zebrafish. The Wnt–Frizzled (Frz) complex modulates the jaw and ethmoid plate development [232]. Knockdown of frzb and frzd7a in zebrafish morphants results in lack of the lower jaw and ceratobranchial cartilage, as well as the loss of chondrocytes within the ethmoid plate. Altogether, the interaction between Wnt9a/Frzb/Frzd7a is crucial for the chondrogenic proliferation and cell fate within the ethmoid plate. Moreover, Wnt9/Frzb/Frzd7 also drive the development of the lower jaw. Ergo, Wnt–Frz complex regulates both canonical and planar-cell-polarity pathways during craniofacial chondrogenesis [233].
Wnt9a drives the expression of orthologue sox9a during early chondrogenic differentiation. Zebrafish sox9a mutants lack almost the entire set of cranial cartilage, except for the ceratohyal cartilage, which eventually leads to a reduction in a number of cranial intramembranous bones, including the dentary bone, maxilla, and the opercle [234,235]. On the other hand, sox9b knockdowns reveal a mild reduction of cartilages within mandibular and hyoid arches of morphants, which is in striking comparison with sox9a mutants [235]. Furthermore, sox9b knockdown mutants lack nearly all cranial bones, including most intramembranous bones, except for the cleithrum and the opercle [235]. However, the craniofacial phenotype remains unaffected in sox9b-null mutants [234]. All in all, analysis of sox9a zebrafish knockout mutants shows that sox9a is important during chondrogenic differentiation [235,236].
Two runx2 orthologues, runx2a and runx2b, can be found within the zebrafish genome [236]. Runx2b is expressed throughout the mesenchyme of presumptive pharyngeal cartilages even before chondrogenic differentiation [236,237]. In contrast to murine Runx2, runx2b in zebrafish is expressed in all chondrocytes and is downregulated in sox9b mutants but remains unaffected in sox9a mutants [235]. Runx2a is expressed predominantly in the mandibular arch, while being expressed only weakly in the hyoid cartilages [236,237]. While runx2b morphants lack all pharyngeal cartilage, runx2a knockdown has barely any effect on pharyngeal chondrogenesis. Indeed, runx2b can possibly compensate for the lack of runx2a during pharyngeal skeletogenesis [237]. In contrast with mice, the expression of runx2a/runx2b is not affected by canonical Wnt and Fgf signalling during the early osteogenic differentiation [238]. Rather, Runx2b is induced by Runx3 emanating from the endoderm [237]. Following the induction of Runx3, Egr1 is activated in the endoderm, which in turn downregulates the expression of sox9b and follistatin A (fsta). Together, Runx3/Egr1/Sox9b/Fsta enable Bmp signalling during cranial cartilage development via inhibition of Bmp antagonists [239]. In addition, both Sox9 and Runx2 are affected by Foxe1 during osteochondrogenesis. Foxe1 suppresses Fgfr2, which in turn enables development of the cartilage within PAs [240]. Taken together, Wnt signalling and SOX9/Sox9a transcription factors are master regulators of chondrogenesis in both mice and zebrafish. Moreover, Runx2 participates in chondrogenesis in both species. However, the key difference is that cartilage within PAs cannot form without runx2b in zebrafish, whilst in mice, it can form even when the expression of Runx2 is lost.
In contrast with runx2, sp7 expression in zebrafish is regulated by canonical Wnt signalling, which acts in concert with Fgf to modulate osteogenic differentiation [238]. Further support for Wnt-mediated control of osteogenesis during craniofacial development in zebrafish can be found within developing intramembranous bones, which express a mediator of canonical Wnt signalling, tcf7 [241]. Delayed ossification of maxilla occurs in sp7 mutants, while other cranial intramembranous bones (cleithrum, brachiostegal rays, opercle, parasphenoid) are misshapen. In general, intramembranous bones of sp7 mutants show reduced ossification, whereas the cartilage development appears unchanged, as the expression of chondrocyte differentiation markers sox9a, sox9b, runx2a, and runx2b is unaltered. However, the expression of osteogenesis-related markers, such as bglap, spp1, col1a1a, and col1a1b, is decreased. Of note, sp7 is a driver of col10a1a expression in osteoblasts, and the role of col10a1a during osteoblastogenesis has only been described in zebrafish so far [241,242,243].
In the zebrafish neurula, Shh signalling radiates from the ventral brain primordium into the presumptive stomodeal area. When NCCs colonize the mandibular arch, a signal from the stomodeum diffuses into anterior NCCs, initiating the mesenchymal condensation and giving rise to the pterygoid process of palatoquadrate cartilage and the neurocranium. Double knockdown of shh and twhh, two members of the hh family, leads to the loss of ectomesenchymal condensations and the prospective anterior cranial skeleton [244]. In addition, Hh signalling influences development of the jaw joint via regulation of nkx3.2 and gd5 [245].
Later in the craniofacial development, Hh signalling via Ptc1 and Ptc2 plays a role during the differentiation of osteoblasts in the perichondrium of ceratohyal and hyosymplectic cartilages [246]. In ihha-null zebrafish, reduced proliferation of chondrocytes is reflected in the loss of mineralized endochondral bones in the cranium. Depending on the concentration, ihha regulates chondrogenic and osteogenic proliferation via Gli1 and Gli3 transcription factors [247]. In addition, Hh signalling drives the proliferation of preosteoblasts during the intramembranous osteogenesis [248]. During the endochondral bone formation, Hh signalling influences the expression of runx2a, runx2b, sp7, and colX in ossification centres [246]. Not unlike ihh, shh also plays a role during the ossification and mineralization, as it upregulates the expression of bmp2, sp7, and col10a1a. Moreover, Hh signalling has also been discovered to downregulate autophagy during osteoblastogenesis [249].
In contrast with mice, Meckel’s cartilage is affected only mildly in zebrafish prrx1a/prrx1b mutants, whereas dorsal pharyngeal cartilages are affected more significantly. During the chondrogenic differentiation, prrx1a/prrx1b are negatively regulated by Edn1 in ventral pharyngeal cartilages. Conversely, Jag1b-Notch signalling in concert with Prrx1a/Prrx1b sets up dorsal pharyngeal cartilages via inhibition of barx1 [250]. Intriguingly, both zebrafish and mice exhibit ectopic and abnormal cartilage in the dorsal region of mandibular and hyoid arches; however, loss of prrx1a and prrx1b in the zebrafish produces very minor defects in ventral cartilages. In contrast, knockout of Prrx1 in mice produces extensive malformations within the mandibular process and both ventral and dorsal hyoid arches.
Knockdown of tgfbr2 in zebrafish causes the shortening of jaws and misshapen palate [251], which is similar to findings in mouse mutants. In the zebrafish, tgfβ-3 governs the generation and survival of the cranial NC from premigratory to migratory stages, and its inhibition leads to the increased apoptosis of NCCs, resulting in malformations of the palatoquadrate and other cranial cartilages [252]. Zebrafish fgfr3 knockout mutants display malformations of the mandible and delayed ossification of the craniofacial skeleton. Both early stage osteoblast markers, such as col10a1a, and late-stage osteoblast markers, such as spp1, osn, and col1a2, exhibit decreased expression. Additionally, upregulation of the chondrogenic proliferation and irregular directional orientation of chondrocytes can be observed in fgfr3 mutant fish, accompanied by an upregulation of Ihh and canonical Wnt signalling [3]. Thus, fgfr3 downregulates Wnt/ß-catenin and Ihh signalling. The importance of Fgf signalling during zebrafish chondrogenesis is further supported by knockdown experiments of fgf10a, which cause the reduction of Meckel’s cartilage and deformation of the palatoquadrate cartilage [251]. Bmp is another signalling pathway that is crucial for ossification of the cranial skeleton in the zebrafish. It is mediated via the production of nitric oxide [253] Knockdown of zebrafish orthologues msxB, msxC, and msxE leads to the loss of jaws, including other ventral pharyngeal cartilages, as a result of arch ventralization [254].
8. Conclusions
The pharyngeal apparatus is one of the hallmarks of gnathostome embryogenesis and evolution. The neural crest gives rise to most of the tissue within PA-derived skeletal elements, which collectively form the viscerocranium. The expression of Hox genes in the hindbrain and NCCs specifies the identity of individual PAs. Cell identity in the mandibular arch is regulated by the MEIS/PBX complex, whereas in the hyoid arch, a trimeric complex of HOX/MEIS/PBX specifies the second PA fate. The skeletal polarity within the individual PA is governed by the EDN–DLX–HAND regulatory cascade. Numerous signalling pathways operating within PAs, including FGF, BMP, and SHH, pattern the nascent arch and thus ensure the genesis of heterogenous structures, including teeth, skeletal components, and the tongue. In this review, we have discussed the role of transcription factors SOX9, RUNX2, SP7, PAX3, GSC, MSX, and PRRX and the signalling pathways WNT and HH during mandibular and hyoid arch skeletogenesis. Within the PAs, these molecules influence chondrogenic and osteoblastic differentiation, the transition from chondroblastic to osteoblastic fate, and the choice between osteoblastic and chondroblastic cell fate. In addition, multiple ligands from the TGF-β, BMP, and FGF families control the proliferation, maintenance, and differentiation of NCCs in conjunction with the aforementioned transcription factors. Disruption of these signalling pathways in the ectomesenchyme within the mandibular and hyoid arches results in malformations and dysmorphies of the viscerocranium.
In contrast with zebrafish, the hyoid arch is rarely a subject of interest in mice and humans. Certainly, one reason for this is a scarcity of patients with hyoid arch abnormalities in a clinical setting. As the mandibular arch has a much larger impact on development of the viscerocranium than the hyoid arch, morphogenesis of the hyoid arch has naturally been less studied by researchers. Nonetheless, malformations in the mandibular arch often co-occur with hyoid arch malformations in mouse genetic mutants. Although hyoid abnormalities have not been casually reported in human patients with first arch syndromes, they have been described to co-occur with Pierre Robins sequence, 22q11.2 deletion syndrome, and cleft lip/cleft palate, which highlights the importance of the management of hyoid abnormalities in patients with first arch anomalies. In a clinical setting, symptomatic anatomical variants of the hyoid–larynx complex can often be overlooked by physicians. Nonetheless, it seems plausible that the first arch anomalies are often accompanied with hyoid anomalies in humans but remain unnoticed or underreported.
It is important to note that signalling molecules and transcription factors governing PA development are similar between the mandibular and hyoid arches, which is in fact already known from studies in zebrafish. In both mice and zebrafish, the Edn1, Dlx5/Dlx6, and Hand2 genes control cell fate in the ventral regions of the mandibular and hyoid arches—the mandibular process and the ventral hyoid arch—and, accordingly, a mutation in any of them results in malformations particularly in the lower jaw and lesser horns of the hyoid. Dlx1/Dlx2 specify dorsal regions of the mandibular and hyoid arches, and their absence does not result in malformations of the ventral PA derivatives. In mice, the expression of the Meis and Pbx genes in the mandibular and hyoid arches largely overlaps with Hand2, and both Meis and Pbx mutants display malformations of the mandibular process and the hyoid bone. Therefore, Meis, Pbx, Hand2, and Prrx1 share similar temporospatial patterns of expression in PAs—weak expression in lateral regions, while being abundant in medial regions. This pattern of expression in medial domains of the mandibular and hyoid arches suggests a tight link between the medial structures of first two PAs. Although meis and pbx govern both mandibular and hyoid arches, the precise link between pbx/meis and hand is not well studied in zebrafish. In both zebrafish and mice, Prrx1 is involved in the specification of the dorsal fate of the mandibular and hyoid arches, whereas in mice, Prrx1 participates in the specification of both ventral and dorsal fates of the mandibular and hyoid arches. In conclusion, we presume that the development of mandibular and hyoid arches involves a common regulatory network involving MEIS, PBX, EDN1, DLX5/6, HAND2, and PRRX1 that is shared among gnathostomes.
Author Contributions
J.F., V.P. and O.M. conceived this review article and wrote it. V.P. prepared the figures. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Grant Agency of Charles University (grants #1034120 and 340321).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kumar S., Hedges S.B. A Molecular Timescale for Vertebrate Evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 2.Gebuijs I.G.E., Raterman S.T., Metz J.R., Swanenberg L., Zethof J., Van den Bos R., Carels C.E.L., Wagener F.A.D.T.G., Von den Hoff J.W. Fgf8a Mutation Affects Craniofacial Development and Skeletal Gene Expression in Zebrafish Larvae. Biol. Open. 2019;8:bio039834. doi: 10.1242/bio.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun X., Zhang R., Chen H., Du X., Chen S., Huang J., Liu M., Xu M., Luo F., Jin M., et al. Fgfr3 Mutation Disrupts Chondrogenesis and Bone Ossification in Zebrafish Model Mimicking CATSHL Syndrome Partially via Enhanced Wnt/β-Catenin Signaling. Theranostics. 2020;10:7111–7130. doi: 10.7150/thno.45286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S., Max S.R., Panny S.R., Cohen M.M. Deletions of Proximal 15q and Non-Classical Prader-Willi Syndrome Phenotypes. Am. J. Med. Genet. 1985;20:255–263. doi: 10.1002/ajmg.1320200208. [DOI] [PubMed] [Google Scholar]
- 5.Jones N.C., Trainor P.A. The Therapeutic Potential of Stem Cells in the Treatment of Craniofacial Abnormalities. Expert Opin. Biol. Ther. 2004;4:645–657. doi: 10.1517/14712598.4.5.645. [DOI] [PubMed] [Google Scholar]
- 6.Noden D.M., Trainor P.A. Relations and Interactions between Cranial Mesoderm and Neural Crest Populations. J. Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad M.S., Charney R.M., García-Castro M.I. Specification and Formation of the Neural Crest: Perspectives on Lineage Segregation. Genesis. 2019;57:e23276. doi: 10.1002/dvg.23276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauka-Spengler T., Bronner-Fraser M. A Gene Regulatory Network Orchestrates Neural Crest Formation. Nat. Rev. Mol. Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 9.Kontges G., Lumsden A. Rhombencephalic Neural Crest Segmentation Is Preserved throughout Craniofacial Ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- 10.D’Amico-Martel A., Noden D.M. Contributions of Placodal and Neural Crest Cells to Avian Cranial Peripheral Ganglia. Am. J. Anat. 1983;166:445–468. doi: 10.1002/aja.1001660406. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida T., Vivatbutsiri P., Morriss-Kay G., Saga Y., Iseki S. Cell Lineage in Mammalian Craniofacial Mesenchyme. Mech. Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Graham A. Deconstructing the Pharyngeal Metamere. J. Exp. Zool. B Mol. Dev. Evol. 2008;310:336–344. doi: 10.1002/jez.b.21182. [DOI] [PubMed] [Google Scholar]
- 13.McKinney M.C., McLennan R., Giniunaite R., Baker R.E., Maini P.K., Othmer H.G., Kulesa P.M. Visualizing Mesoderm and Neural Crest Cell Dynamics during Chick Head Morphogenesis. Dev. Biol. 2020;461:184–196. doi: 10.1016/j.ydbio.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavalas A., Trainor P., Ariza-McNaughton L., Krumlauf R. Synergy between Hoxa1 and Hoxb1: The Relationship between Arch Patterning and the Generation of Cranial Neural Crest. Development. 2001;128:3017–3027. doi: 10.1242/dev.128.15.3017. [DOI] [PubMed] [Google Scholar]
- 15.Veitch E., Begbie J., Schilling T.F., Smith M.M., Graham A. Pharyngeal Arch Patterning in the Absence of Neural Crest. Curr. Biol. 1999;9:1481–1484. doi: 10.1016/S0960-9822(00)80118-9. [DOI] [PubMed] [Google Scholar]
- 16.Graham A., Poopalasundaram S., Shone V., Kiecker C. A Reappraisal and Revision of the Numbering of the Pharyngeal Arches. J. Anat. 2019;235:1019–1023. doi: 10.1111/joa.13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passos-Bueno M.R., Ornelas C.C., Fanganiello R.D. Syndromes of the First and Second Pharyngeal Arches: A Review. Am. J. Med. Genet. Part A. 2009;149A:1853–1859. doi: 10.1002/ajmg.a.32950. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Vázquez J.F., Yamamoto M., Abe S., Katori Y., Murakami G. Development of the Human Incus with Special Reference to the Detachment From the Chondrocranium to Be Transferred into the Middle Ear. Anat. Rec. 2018;301:1405–1415. doi: 10.1002/ar.23832. [DOI] [PubMed] [Google Scholar]
- 19.Woronowicz K.C., Schneider R.A. Molecular and Cellular Mechanisms Underlying the Evolution of Form and Function in the Amniote Jaw. EvoDevo. 2019;10:17. doi: 10.1186/s13227-019-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaupp E. Über Die Ala Temporalis Des Säugerschädels Und Die Regio Orbtailis Einiger Anderer Wirbeltierschädel. Anat. Hefte. 1902;19:155–230. doi: 10.1007/BF02169846. [DOI] [Google Scholar]
- 21.Frisdal A., Trainor P.A. Development and Evolution of the Pharyngeal Apparatus. Wiley Interdiscip. Rev. Dev. Biol. 2014;3:403–418. doi: 10.1002/wdev.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhaskar S.N., Weinmann J.P., Schour I. Role of Meckel’s Cartilage in the Development and Growth of the Rat Mandible. J. Dent. Res. 1953;32:398–410. doi: 10.1177/00220345530320031401. [DOI] [PubMed] [Google Scholar]
- 23.Ito Y., Bringas P., Mogharei A., Zhao J., Deng C., Chai Y. Receptor-Regulated and Inhibitory Smads Are Critical in Regulating Transforming Growth Factorβ–Mediated Meckel’s Cartilage Development. Dev. Dyn. 2002;224:69–78. doi: 10.1002/dvdy.10088. [DOI] [PubMed] [Google Scholar]
- 24.Shimo T., Kanyama M., Wu C., Sugito H., Billings P.C., Abrams W.R., Rosenbloom J., Iwamoto M., Pacifici M., Koyama E. Expression and Roles of Connective Tissue Growth Factor in Meckel’s Cartilage Development. Dev. Dyn. 2004;231:136–147. doi: 10.1002/dvdy.20109. [DOI] [PubMed] [Google Scholar]
- 25.Svandova E., Anthwal N., Tucker A.S., Matalova E. Diverse Fate of an Enigmatic Structure: 200 Years of Meckel’s Cartilage. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez-Vázquez J.F., Mérida-Velasco J.R., Mérida-Velasco J.A., Sánchez-Montesinos I., Espín-Ferra J., Jiménez-Collado J. Development of Meckel’s Cartilage in the Symphyseal Region in Man. Anat. Rec. 1997;249:249–254. doi: 10.1002/(SICI)1097-0185(199710)249:2<249::AID-AR12>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 27.Eames B.F., Sharpe P.T., Helms J.A. Hierarchy Revealed in the Specification of Three Skeletal Fates by Sox9 and Runx2. Dev. Biol. 2004;274:188–200. doi: 10.1016/j.ydbio.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Harada Y., Ishizeki K. Evidence for Transformation of Chondrocytes and Site-Specific Resorption during the Degradation of Meckel’s Cartilage. Anat. Embryol. 1998;197:439–450. doi: 10.1007/s004290050155. [DOI] [PubMed] [Google Scholar]
- 29.Ishizeki K. Imaging Analysis of Osteogenic Transformation of Meckel’s Chondrocytes from Green Fluorescent Protein-Transgenic Mice during Intrasplenic Transplantation. Acta Histochem. 2012;114:608–619. doi: 10.1016/j.acthis.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Ishizeki K., Takigawa M., Nawa T., Suzuki F. Mouse Meckel’s cartilage chondrocytes evoke bone-like matrix and further transform into osteocyte-like cells in culture. Anat. Rec. 1996;245:25–35. doi: 10.1002/(SICI)1097-0185(199605)245:1<25::AID-AR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 31.Ishizeki K., Saito H., Shinagawa T., Fujiwara N., Nawa T. Histochemical and Immunohistochemical Analysis of the Mechanism of Calcification of Meckel’s Cartilage during Mandible Development in Rodents. Pt 2J. Anat. 1999;194:265–277. doi: 10.1046/j.1469-7580.1999.19420265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishizeki K., Kagiya T., Fujiwara N., Otsu K., Harada H. Expression of Osteogenic Proteins during the Intrasplenic Transplantation of Meckel’s Chondrocytes: A Histochemical and Immunohistochemical Study. Arch. Histol. Cytol. 2009;72:1–12. doi: 10.1679/aohc.72.1. [DOI] [PubMed] [Google Scholar]
- 33.Anthwal N., Joshi L., Tucker A.S. Evolution of the Mammalian Middle Ear and Jaw: Adaptations and Novel Structures. J. Anat. 2013;222:147–160. doi: 10.1111/j.1469-7580.2012.01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishizeki K., Takahashi N., Nawa T. Formation of the Sphenomandibular Ligament by Meckel’s Cartilage in the Mouse: Possible Involvement of Epidermal Growth Factor as Revealed by Studies in Vivo and in Vitro. Cell Tissue Res. 2001;304:67–80. doi: 10.1007/s004410100354. [DOI] [PubMed] [Google Scholar]
- 35.Cheynet F., Guyot L., Richard O., Layoun W., Gola R. Discomallear and Malleomandibular Ligaments: Anatomical Study and Clinical Applications. Surg. Radiol. Anat. 2003;25:152–157. doi: 10.1007/s00276-003-0097-y. [DOI] [PubMed] [Google Scholar]
- 36.Amano O., Doi T., Yamada T., Sasaki A., Sakiyama K., Kanegae H., Kindaichi K. Meckel’s Cartilage: Discovery, Embryology and Evolution: —Overview of the Specificity of Meckel’s Cartilage—. J. Oral Biosci. 2010;52:125–135. doi: 10.1016/S1349-0079(10)80041-6. [DOI] [Google Scholar]
- 37.Dash S., Trainor P.A. The Development, Patterning and Evolution of Neural Crest Cell Differentiation into Cartilage and Bone. Bone. 2020;137:115409. doi: 10.1016/j.bone.2020.115409. [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez-Vázquez J.F., Mérida-Velasco J.R., Verdugo-López S., Sánchez-Montesinos I., Mérida-Velasco J.A. Morphogenesis of the Second Pharyngeal Arch Cartilage (Reichert’s Cartilage) in Human Embryos. J. Anat. 2006;208:179–189. doi: 10.1111/j.1469-7580.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodríguez-Vázquez J.F., Verdugo-López S., Abe H., Murakami G. The Origin of the Variations of the Hyoid Apparatus in Human. Anat. Rec. 2015;298:1395–1407. doi: 10.1002/ar.23166. [DOI] [PubMed] [Google Scholar]
- 40.Rodríguez-Vázquez J.F., Kim J.H., Verdugo-López S., Murakami G., Cho K.H., Asakawa S., Abe S.-I. Human Fetal Hyoid Body Origin Revisited. J. Anat. 2011;219:143–149. doi: 10.1111/j.1469-7580.2011.01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bakker B.S., de Bakker H.M., Soerdjbalie-Maikoe V., Dikkers F.G. The Development of the Human Hyoid-Larynx Complex Revisited. Laryngoscope. 2018;128:1829–1834. doi: 10.1002/lary.26987. [DOI] [PubMed] [Google Scholar]
- 42.Poopalasundaram S., Richardson J., Scott A., Donovan A., Liu K., Graham A. Diminution of Pharyngeal Segmentation and the Evolution of the Amniotes. Zool. Lett. 2019;5:6. doi: 10.1186/s40851-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El Amm C.A., Denny A. Hyoid Bone Abnormalities in Pierre Robin Patients. J. Craniofacial Surg. 2008;19:259–263. doi: 10.1097/scs.0b013e31815c9460. [DOI] [PubMed] [Google Scholar]
- 44.Rajion Z.A., Townsend G.C., Netherway D.J., Anderson P.J., Hughes T., Shuaib I.L., Halim A.S., Samsudin A.R., McLean N.R., David D.J. The Hyoid Bone in Malay Infants with Cleft Lip and Palate. Cleft Palate-Craniofacial J. 2006;43:532–538. doi: 10.1597/05-085. [DOI] [PubMed] [Google Scholar]
- 45.Erdinc A.M.E., Dincer B., Sabah M.E. Evaluation of the Position of the Hyoid Bone in Relation to Vertical Facial Development. J. Clin. Pediatr. Dent. 2003;27:347–352. doi: 10.17796/jcpd.27.4.v619q30222674w30. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida K., Yokoi T., Mori S., Achiwa M., Kuroiwa Y., Kurita K. Abnormal Ossification of the Hyoid Bone in Cleidocranial Dysplasia Rare Case and Literature Review. Int. J. Oral Maxillofac. Surg. 2017;46:375–376. doi: 10.1016/j.ijom.2017.02.1263. [DOI] [Google Scholar]
- 47.Heliövaara A., Hurmerinta K. Craniofacial Cephalometric Morphology in Children with CATCH 22 Syndrome. Orthod. Craniofacial Res. 2006;9:186–192. doi: 10.1111/j.1601-6343.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 48.Wells T.R., Gilsanz V., Senac M.O., Landing B.H., Vachon L., Takahashi M. Ossification Centre of the Hyoid Bone in DiGeorge Syndrome and Tetralogy of Fallot. Br. J. Radiol. 1986;59:1065–1068. doi: 10.1259/0007-1285-59-707-1065. [DOI] [PubMed] [Google Scholar]
- 49.Milligan B., Harris N., Franz-Odendaal T.A. Understanding Morphology: A Comparative Study on the Lower Jaw in Two Teleost Species: Lower Jaw Morphology. J. Appl. Ichthyol. 2012;28:346–352. doi: 10.1111/j.1439-0426.2012.01998.x. [DOI] [Google Scholar]
- 50.Cubbage C.C., Mabee P.M. Development of the Cranium and Paired Fins in the Zebrafish Danio Rerio (Ostariophysi, Cyprinidae) J. Morphol. 1996;229:121–160. doi: 10.1002/(SICI)1097-4687(199608)229:2<121::AID-JMOR1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 51.Graham A. The Development and Evolution of the Pharyngeal Arches. J. Anat. 2001;199:133–141. doi: 10.1046/j.1469-7580.2001.19910133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janvier P., Desbiens S., Willett J.A., Arsenault M. Lamprey-like Gills in a Gnathostome-Related Devonian Jawless Vertebrate. Nature. 2006;440:1183–1185. doi: 10.1038/nature04471. [DOI] [PubMed] [Google Scholar]
- 53.Santagati F., Rijli F.M. Cranial Neural Crest and the Building of the Vertebrate Head. Nat. Rev. Neurosci. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- 54.Trainor P.A., Krumlauf R. Hox Genes, Neural Crest Cells and Branchial Arch Patterning. Curr. Opin. Cell Biol. 2001;13:698–705. doi: 10.1016/S0955-0674(00)00273-8. [DOI] [PubMed] [Google Scholar]
- 55.Pearson J.C., Lemons D., McGinnis W. Modulating Hox Gene Functions during Animal Body Patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 56.Parker H.J., Pushel I., Krumlauf R. Coupling the Roles of Hox Genes to Regulatory Networks Patterning Cranial Neural Crest. Dev. Biol. 2018;444:S67–S78. doi: 10.1016/j.ydbio.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 57.Lumsden A. Segmentation and Compartition in the Early Avian Hindbrain. Mech. Dev. 2004;121:1081–1088. doi: 10.1016/j.mod.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 58.Couly G., Grapin-Botton A., Coltey P., Ruhin B., Le Douarin N.M. Determination of the Identity of the Derivatives of the Cephalic Neural Crest: Incompatibility between Hox Gene Expression and Lower Jaw Development. Development. 1998;125:3445–3459. doi: 10.1242/dev.125.17.3445. [DOI] [PubMed] [Google Scholar]
- 59.Jozefowicz C., McClintock J., Prince V. The fates of zebrafish Hox gene duplicates. In: Meyer A., Van de Peer Y., editors. Genome Evolution. Springer; Dordrecht, The Netherlands: 2003. pp. 185–194. [PubMed] [Google Scholar]
- 60.Hunt P., Krumlauf R. Hox Genes Coming to a Head. Curr. Biol. 1991;1:304–306. doi: 10.1016/0960-9822(91)90092-B. [DOI] [PubMed] [Google Scholar]
- 61.Hunt P., Krumlauf R. Deciphering the Hox Code: Clues to Patterning Branchial Regions of the Head. Cell. 1991;66:1075–1078. doi: 10.1016/0092-8674(91)90029-X. [DOI] [PubMed] [Google Scholar]
- 62.Makki N., Capecchi M.R. Hoxa1 Lineage Tracing Indicates a Direct Role for Hoxa1 in the Development of the Inner Ear, the Heart, and the Third Rhombomere. Dev. Biol. 2010;341:499–509. doi: 10.1016/j.ydbio.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murphy P., Hill R.E. Expression of the Mouse Labial-like Homeobox-Containing Genes, Hox 2.9 and Hox 1.6, during Segmentation of the Hindbrain. Development. 1991;111:61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- 64.Alexandre D., Clarke J.D., Oxtoby E., Yan Y.L., Jowett T., Holder N. Ectopic Expression of Hoxa-1 in the Zebrafish Alters the Fate of the Mandibular Arch Neural Crest and Phenocopies a Retinoic Acid-Induced Phenotype. Development. 1996;122:735–746. doi: 10.1242/dev.122.3.735. [DOI] [PubMed] [Google Scholar]
- 65.Gavalas A., Studer M., Lumsden A., Rijli F.M., Krumlauf R., Chambon P. Hoxa1 and Hoxb1 Synergize in Patterning the Hindbrain, Cranial Nerves and Second Pharyngeal Arch. Development. 1998;125:1123–1136. doi: 10.1242/dev.125.6.1123. [DOI] [PubMed] [Google Scholar]
- 66.Kanzler B., Kuschert S.J., Liu Y.H., Mallo M. Hoxa-2 Restricts the Chondrogenic Domain and Inhibits Bone Formation during Development of the Branchial Area. Development. 1998;125:2587–2597. doi: 10.1242/dev.125.14.2587. [DOI] [PubMed] [Google Scholar]
- 67.Gendron-Maguire M., Mallo M., Zhang M., Gridley T. Hoxa-2 Mutant Mice Exhibit Homeotic Transformation of Skeletal Elements Derived from Cranial Neural Crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- 68.Rijli F.M., Mark M., Lakkaraju S., Dierich A., Dollé P., Chambon P. A Homeotic Transformation Is Generated in the Rostral Branchial Region of the Head by Disruption of Hoxa-2, Which Acts as a Selector Gene. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- 69.Grammatopoulos G.A., Bell E., Toole L., Lumsden A., Tucker A.S. Homeotic Transformation of Branchial Arch Identity after Hoxa2 Overexpression. Development. 2000;127:5355–5365. doi: 10.1242/dev.127.24.5355. [DOI] [PubMed] [Google Scholar]
- 70.Hunter M.P., Prince V.E. Zebrafish Hox Paralogue Group 2 Genes Function Redundantly as Selector Genes to Pattern the Second Pharyngeal Arch. Dev. Biol. 2002;247:367–389. doi: 10.1006/dbio.2002.0701. [DOI] [PubMed] [Google Scholar]
- 71.Pasqualetti M., Ori M., Nardi I., Rijli F.M. Ectopic Hoxa2 Induction after Neural Crest Migration Results in Homeosis of Jaw Elements in Xenopus. Development. 2000;127:5367–5378. doi: 10.1242/dev.127.24.5367. [DOI] [PubMed] [Google Scholar]
- 72.Minoux M., Antonarakis G.S., Kmita M., Duboule D., Rijli F.M. Rostral and Caudal Pharyngeal Arches Share a Common Neural Crest Ground Pattern. Development. 2009;136:637–645. doi: 10.1242/dev.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schulte D., Geerts D. MEIS Transcription Factors in Development and Disease. Development. 2019;146 doi: 10.1242/dev.174706. [DOI] [PubMed] [Google Scholar]
- 74.Longobardi E., Penkov D., Mateos D., Florian G.D., Torres M., Blasi F. Biochemistry of the Tale Transcription Factors PREP, MEIS, and PBX in Vertebrates. Dev. Dyn. 2014;243:59–75. doi: 10.1002/dvdy.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waskiewicz A.J., Rikhof H.A., Hernandez R.E., Moens C.B. Zebrafish Meis Functions to Stabilize Pbx Proteins and Regulate Hindbrain Patterning. Development. 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- 76.Choe S.-K., Lu P., Nakamura M., Lee J., Sagerström C.G. Meis Cofactors Control HDAC and CBP Accessibility at Hox-Regulated Promoters during Zebrafish Embryogenesis. Dev. Cell. 2009;17:561–567. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amin S., Donaldson I.J., Zannino D.A., Hensman J., Rattray M., Losa M., Spitz F., Ladam F., Sagerström C., Bobola N. Hoxa2 Selectively Enhances Meis Binding to Change a Branchial Arch Ground State. Dev. Cell. 2015;32:265–277. doi: 10.1016/j.devcel.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Machon O., Masek J., Machonova O., Krauss S., Kozmik Z. Meis2 Is Essential for Cranial and Cardiac Neural Crest Development. BMC Dev. Biol. 2015;15:40. doi: 10.1186/s12861-015-0093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fabik J., Kovacova K., Kozmik Z., Machon O. Neural Crest Cells Require Meis2 for Patterning the Mandibular Arch via the Sonic Hedgehog Pathway. Biol. Open. 2020;9 doi: 10.1242/bio.052043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crowley M.A., Conlin L.K., Zackai E.H., Deardorff M.A., Thiel B.D., Spinner N.B. Further Evidence for the Possible Role of MEIS2 in the Development of Cleft Palate and Cardiac Septum. Am. J. Med. Genet. A. 2010;152A:1326–1327. doi: 10.1002/ajmg.a.33375. [DOI] [PubMed] [Google Scholar]
- 81.Douglas G., Cho M.T., Telegrafi A., Winter S., Carmichael J., Zackai E.H., Deardorff M.A., Harr M., Williams L., Psychogios A., et al. De Novo Missense Variants in MEIS2 Recapitulate the Microdeletion Phenotype of Cardiac and Palate Abnormalities, Developmental Delay, Intellectual Disability and Dysmorphic Features. Am. J. Med. Genet. A. 2018;176:1845–1851. doi: 10.1002/ajmg.a.40368. [DOI] [PubMed] [Google Scholar]
- 82.Erdogan F., Ullmann R., Chen W., Schubert M., Adolph S., Hultschig C., Kalscheuer V., Ropers H.-H., Spaich C., Tzschach A. Characterization of a 5.3 Mb Deletion in 15q14 by Comparative Genomic Hybridization Using a Whole Genome “Tiling Path” BAC Array in a Girl with Heart Defect, Cleft Palate, and Developmental Delay. Am. J. Med. Genet. A. 2007;143A:172–178. doi: 10.1002/ajmg.a.31541. [DOI] [PubMed] [Google Scholar]
- 83.Giliberti A., Currò A., Papa F.T., Frullanti E., Ariani F., Coriolani G., Grosso S., Renieri A., Mari F. MEIS2 Gene Is Responsible for Intellectual Disability, Cardiac Defects and a Distinct Facial Phenotype. Eur J. Med. Genet. 2019 doi: 10.1016/j.ejmg.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 84.Johansson S., Berland S., Gradek G.A., Bongers E., de Leeuw N., Pfundt R., Fannemel M., Rødningen O., Brendehaug A., Haukanes B.I., et al. Haploinsufficiency of MEIS2 Is Associated with Orofacial Clefting and Learning Disability. Am. J. Med. Genet. Part A. 2014;164:1622–1626. doi: 10.1002/ajmg.a.36498. [DOI] [PubMed] [Google Scholar]
- 85.Verheije R., Kupchik G.S., Isidor B., Kroes H.Y., Lynch S.A., Hawkes L., Hempel M., Gelb B.D., Ghoumid J., D’Amours G., et al. Heterozygous Loss-of-Function Variants of MEIS2 Cause a Triad of Palatal Defects, Congenital Heart Defects, and Intellectual Disability. Eur. J. Hum. Genet. 2019;27:278–290. doi: 10.1038/s41431-018-0281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parker H.J., De Kumar B., Green S.A., Prummel K.D., Hess C., Kaufman C.K., Mosimann C., Wiedemann L.M., Bronner M.E., Krumlauf R. A Hox-TALE Regulatory Circuit for Neural Crest Patterning Is Conserved across Vertebrates. Nat. Commun. 2019;10:1189. doi: 10.1038/s41467-019-09197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Melvin V.S., Feng W., Hernandez-Lagunas L., Artinger K.B., Williams T. A Morpholino-Based Screen to Identify Novel Genes Involved in Craniofacial Morphogenesis. Dev. Dyn. 2013;242:817–831. doi: 10.1002/dvdy.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pöpperl H., Rikhof H., Chang H., Haffter P., Kimmel C.B., Moens C.B. Lazarus Is a Novel Pbx Gene That Globally Mediates Hox Gene Function in Zebrafish. Mol. Cell. 2000;6:255–267. doi: 10.1016/S1097-2765(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 89.Ferretti E., Li B., Zewdu R., Wells V., Hebert J.M., Karner C., Anderson M.J., Williams T., Dixon J., Dixon M.J., et al. A Conserved Pbx-Wnt-P63-Irf6 Regulatory Module Controls Face Morphogenesis by Promoting Epithelial Apoptosis. Dev. Cell. 2011;21:627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vitobello A., Ferretti E., Lampe X., Vilain N., Ducret S., Ori M., Spetz J.-F., Selleri L., Rijli F.M. Hox and Pbx Factors Control Retinoic Acid Synthesis during Hindbrain Segmentation. Dev. Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Selleri L., Depew M.J., Jacobs Y., Chanda S.K., Tsang K.Y., Cheah K.S.E., Rubenstein J.L.R., O’Gorman S., Cleary M.L. Requirement for Pbx1 in Skeletal Patterning and Programming Chondrocyte Proliferation and Differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- 92.ten Berge D., Brouwer A., Korving J., Martin J.F., Meijlink F. Prx1 and Prx2 in Skeletogenesis: Roles in the Craniofacial Region, Inner Ear and Limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- 93.Czako L., Simko K., Thurzo A., Galis B., Varga I. The Syndrome of Elongated Styloid Process, the Eagle’s Syndrome—From Anatomical, Evolutionary and Embryological Backgrounds to 3D Printing and Personalized Surgery Planning. Report of Five Cases. Medicina. 2020;56:458. doi: 10.3390/medicina56090458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arai H., Hori S., Aramori I., Ohkubo H., Nakanishi S. Cloning and Expression of a CDNA Encoding an Endothelin Receptor. Nature. 1990;348 doi: 10.1038/348730a0. [DOI] [PubMed] [Google Scholar]
- 95.Sakurai T., Yanagisawa M., Takuwa Y., Miyazaki H., Kimura S., Goto K., Masaki T. Cloning of a CDNA Encoding a Non-Isopeptide-Selective Subtype of the Endothelin Receptor. Nature. 1990;348:732–735. doi: 10.1038/348732a0. [DOI] [PubMed] [Google Scholar]
- 96.Yanagisawa M. The Endothelin System. A New Target for Therapeutic Intervention. Circulation. 1994;89:1320–1322. doi: 10.1161/01.CIR.89.3.1320. [DOI] [PubMed] [Google Scholar]
- 97.Clouthier D.E., Williams S.C., Yanagisawa H., Wieduwilt M., Richardson J.A., Yanagisawa M. Signaling Pathways Crucial for Craniofacial Development Revealed by Endothelin-A Receptor-Deficient Mice. Dev. Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- 98.Fukuhara S., Kurihara Y., Arima Y., Yamada N., Kurihara H. Temporal Requirement of Signaling Cascade Involving Endothelin-1/Endothelin Receptor Type A in Branchial Arch Development. Mech. Dev. 2004;121:1223–1233. doi: 10.1016/j.mod.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 99.Thomas T., Kurihara H., Yamagishi H., Kurihara Y., Yazaki Y., Olson E.N., Srivastava D. A Signaling Cascade Involving Endothelin-1, DHAND and Msx1 Regulates Development of Neural-Crest-Derived Branchial Arch Mesenchyme. Development. 1998;125:3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- 100.Clouthier D.E., Hosoda K., Richardson J.A., Williams S.C., Yanagisawa H., Kuwaki T., Kumada M., Hammer R.E., Yanagisawa M. Cranial and Cardiac Neural Crest Defects in Endothelin-A Receptor-Deficient Mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- 101.Kurihara Y., Kurihara H., Suzuki H., Kodama T., Maemura K., Nagai R., Oda H., Kuwaki T., Cao W.H., Kamada N. Elevated Blood Pressure and Craniofacial Abnormalities in Mice Deficient in Endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- 102.Ruest L.-B., Xiang X., Lim K.-C., Levi G., Clouthier D.E. Endothelin-A Receptor-Dependent and -Independent Signaling Pathways in Establishing Mandibular Identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ozeki H., Kurihara Y., Tonami K., Watatani S., Kurihara H. Endothelin-1 Regulates the Dorsoventral Branchial Arch Patterning in Mice. Mech. Dev. 2004;121:387–395. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 104.Sato T., Kurihara Y., Asai R., Kawamura Y., Tonami K., Uchijima Y., Heude E., Ekker M., Levi G., Kurihara H. An Endothelin-1 Switch Specifies Maxillomandibular Identity. Proc. Natl. Acad. Sci. USA. 2008;105:18806–18811. doi: 10.1073/pnas.0807345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tavares A.L.P., Cox T.C., Maxson R.M., Ford H.L., Clouthier D.E. Negative Regulation of Endothelin Signaling by SIX1 Is Required for Proper Maxillary Development. Development. 2017;144:2021–2031. doi: 10.1242/dev.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shimizu M., Narboux-Nême N., Gitton Y., de Lombares C., Fontaine A., Alfama G., Kitazawa T., Kawamura Y., Heude E., Marshall L., et al. Probing the Origin of Matching Functional Jaws: Roles of Dlx5/ 6 in Cranial Neural Crest Cells. Sci. Rep. 2018;8:14975. doi: 10.1038/s41598-018-33207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Panganiban G., Rubenstein J.L.R. Developmental Functions of the Distal-Less/Dlx Homeobox Genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 108.Stock D.W., Ellies D.L., Zhao Z., Ekker M., Ruddle F.H., Weiss K.M. The Evolution of the Vertebrate Dlx Gene Family. Proc. Natl. Acad. Sci. USA. 1996;93:10858–10863. doi: 10.1073/pnas.93.20.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Depew M.J., Simpson C.A., Morasso M., Rubenstein J.L. Reassessing the Dlx Code: The Genetic Regulation of Branchial Arch Skeletal Pattern and Development. J. Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qiu M., Bulfone A., Martinez S., Meneses J.J., Shimamura K., Pedersen R.A., Rubenstein J.L. Null Mutation of Dlx-2 Results in Abnormal Morphogenesis of Proximal First and Second Branchial Arch Derivatives and Abnormal Differentiation in the Forebrain. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- 111.Qiu M., Bulfone A., Ghattas I., Meneses J.J., Christensen L., Sharpe P.T., Presley R., Pedersen R.A., Rubenstein J.L. Role of the Dlx Homeobox Genes in Proximodistal Patterning of the Branchial Arches: Mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 Alter Morphogenesis of Proximal Skeletal and Soft Tissue Structures Derived from the First and Second Arches. Dev. Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 112.Acampora D., Merlo G.R., Paleari L., Zerega B., Postiglione M.P., Mantero S., Bober E., Barbieri O., Simeone A., Levi G. Craniofacial, Vestibular and Bone Defects in Mice Lacking the Distal-Less-Related Gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 113.Depew M.J., Liu J.K., Long J.E., Presley R., Meneses J.J., Pedersen R.A., Rubenstein J.L. Dlx5 Regulates Regional Development of the Branchial Arches and Sensory Capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- 114.Beverdam A., Merlo G.R., Paleari L., Mantero S., Genova F., Barbieri O., Janvier P., Levi G. Jaw Transformation with Gain of Symmetry after Dlx5/Dlx6 Inactivation: Mirror of the Past? Genesis. 2002;34:221–227. doi: 10.1002/gene.10156. [DOI] [PubMed] [Google Scholar]
- 115.Depew M.J., Lufkin T., Rubenstein J.L.R. Specification of Jaw Subdivisions by Dlx Genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- 116.Morasso M.I., Grinberg A., Robinson G., Sargent T.D., Mahon K.A. Placental Failure in Mice Lacking the Homeobox Gene Dlx3. Proc. Natl. Acad. Sci. USA. 1999;96:162–167. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jeong J., Li X., McEvilly R.J., Rosenfeld M.G., Lufkin T., Rubenstein J.L.R. Dlx Genes Pattern Mammalian Jaw Primordium by Regulating Both Lower Jaw-Specific and Upper Jaw-Specific Genetic Programs. Development. 2008;135:2905–2916. doi: 10.1242/dev.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Verzi M.P., Agarwal P., Brown C., McCulley D.J., Schwarz J.J., Black B.L. The Transcription Factor MEF2C Is Required for Craniofacial Development. Dev. Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barron F., Woods C., Kuhn K., Bishop J., Howard M.J., Clouthier D.E. Downregulation of Dlx5 and Dlx6 Expression by Hand2 Is Essential for Initiation of Tongue Morphogenesis. Development. 2011;138:2249–2259. doi: 10.1242/dev.056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vincentz J.W., Casasnovas J.J., Barnes R.M., Que J., Clouthier D.E., Wang J., Firulli A.B. Exclusion of Dlx5/6 Expression from the Distal-Most Mandibular Arches Enables BMP-Mediated Specification of the Distal Cap. Proc. Natl. Acad. Sci. USA. 2016;113:7563–7568. doi: 10.1073/pnas.1603930113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Funato N., Chapman S.L., McKee M.D., Funato H., Morris J.A., Shelton J.M., Richardson J.A., Yanagisawa H. Hand2 Controls Osteoblast Differentiation in the Branchial Arch by Inhibiting DNA Binding of Runx2. Development. 2009;136:615–625. doi: 10.1242/dev.029355. [DOI] [PubMed] [Google Scholar]
- 122.Yanagisawa H., Clouthier D.E., Richardson J.A., Charité J., Olson E.N. Targeted Deletion of a Branchial Arch-Specific Enhancer Reveals a Role of DHAND in Craniofacial Development. Development. 2003;130:1069–1078. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- 123.Barbosa A.C., Funato N., Chapman S., McKee M.D., Richardson J.A., Olson E.N., Yanagisawa H. Hand Transcription Factors Cooperatively Regulate Development of the Distal Midline Mesenchyme. Dev. Biol. 2007;310:154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Firulli B.A., Fuchs R.K., Vincentz J.W., Clouthier D.E., Firulli A.B. Hand1 Phosphoregulation within the Distal Arch Neural Crest Is Essential for Craniofacial Morphogenesis. Development. 2014;141:3050–3061. doi: 10.1242/dev.107680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Funato N., Kokubo H., Nakamura M., Yanagisawa H., Saga Y. Specification of Jaw Identity by the Hand2 Transcription Factor. Sci. Rep. 2016;6:28405. doi: 10.1038/srep28405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nair S., Li W., Cornell R., Schilling T.F. Requirements for Endothelin Type-A Receptors and Endothelin-1 Signaling in the Facial Ectoderm for the Patterning of Skeletogenic Neural Crest Cells in Zebrafish. Development. 2007;134:335–345. doi: 10.1242/dev.02704. [DOI] [PubMed] [Google Scholar]
- 127.Kimmel C.B., Ullmann B., Walker M., Miller C.T., Crump J.G. Endothelin 1-Mediated Regulation of Pharyngeal Bone Development in Zebrafish. Development. 2003;130:1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- 128.Miller C.T., Schilling T.F., Lee K., Parker J., Kimmel C.B. Sucker Encodes a Zebrafish Endothelin-1 Required for Ventral Pharyngeal Arch Development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- 129.Miller C.T., Yelon D., Stainier D.Y.R., Kimmel C.B. Two Endothelin 1 Effectors, Hand2 and Bapx1, Pattern Ventral Pharyngeal Cartilage and the Jaw Joint. Development. 2003;130:1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- 130.Zuniga E., Rippen M., Alexander C., Schilling T.F., Crump J.G. Gremlin 2 Regulates Distinct Roles of BMP and Endothelin 1 Signaling in Dorsoventral Patterning of the Facial Skeleton. Development. 2011;138:5147–5156. doi: 10.1242/dev.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sasaki M.M., Nichols J.T., Kimmel C.B. Edn1 and Hand2 Interact in Early Regulation of Pharyngeal Arch Outgrowth during Zebrafish Development. PLoS ONE. 2013;8:e67522. doi: 10.1371/journal.pone.0067522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Talbot J.C., Johnson S.L., Kimmel C.B. Hand2 and Dlx Genes Specify Dorsal, Intermediate and Ventral Domains within Zebrafish Pharyngeal Arches. Development. 2010;137:2507–2517. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Alexander C., Zuniga E., Blitz I.L., Wada N., Le Pabic P., Javidan Y., Zhang T., Cho K.W., Crump J.G., Schilling T.F. Combinatorial Roles for BMPs and Endothelin 1 in Patterning the Dorsal-Ventral Axis of the Craniofacial Skeleton. Development. 2011;138:5135–5146. doi: 10.1242/dev.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Quint E., Zerucha T., Ekker M. Differential Expression of Orthologous Dlx Genes in Zebrafish and Mice: Implications for the Evolution of the Dlx Homeobox Gene Family. J. Exp. Zool. 2000;288:235–241. doi: 10.1002/1097-010X(20001015)288:3<235::AID-JEZ4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 135.Trumpp A., Depew M.J., Rubenstein J.L.R., Bishop J.M., Martin G.R. Cre-Mediated Gene Inactivation Demonstrates That FGF8 Is Required for Cell Survival and Patterning of the First Branchial Arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tucker A.S., Yamada G., Grigoriou M., Pachnis V., Sharpe P.T. Fgf-8 Determines Rostral-Caudal Polarity in the First Branchial Arch. Development. 1999;126:51–61. doi: 10.1242/dev.126.1.51. [DOI] [PubMed] [Google Scholar]
- 137.Mina M., Kollar E.J. The Induction of Odontogenesis in Non-Dental Mesenchyme Combined with Early Murine Mandibular Arch Epithelium. Arch. Oral Biol. 1987;32:123–127. doi: 10.1016/0003-9969(87)90055-0. [DOI] [PubMed] [Google Scholar]
- 138.Lanctôt C., Moreau A., Chamberland M., Tremblay M.L., Drouin J. Hindlimb Patterning and Mandible Development Require the Ptx1 Gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- 139.Lu M.F., Pressman C., Dyer R., Johnson R.L., Martin J.F. Function of Rieger Syndrome Gene in Left-Right Asymmetry and Craniofacial Development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- 140.Liu W., Selever J., Lu M.-F., Martin J.F. Genetic Dissection of Pitx2 in Craniofacial Development Uncovers New Functions in Branchial Arch Morphogenesis, Late Aspects of Tooth Morphogenesis and Cell Migration. Development. 2003;130:6375–6385. doi: 10.1242/dev.00849. [DOI] [PubMed] [Google Scholar]
- 141.Xu J., Liu H., Lan Y., Adam M., Clouthier D.E., Potter S., Jiang R. Hedgehog Signaling Patterns the Oral-Aboral Axis of the Mandibular Arch. eLife. 2019;8:e40315. doi: 10.7554/eLife.40315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jeong J., Mao J., Tenzen T., Kottmann A.H., McMahon A.P. Hedgehog Signaling in the Neural Crest Cells Regulates the Patterning and Growth of Facial Primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tucker A.S., Matthews K.L., Sharpe P.T. Transformation of Tooth Type Induced by Inhibition of BMP Signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- 144.Dworkin S., Boglev Y., Owens H., Goldie S.J. The Role of Sonic Hedgehog in Craniofacial Patterning, Morphogenesis and Cranial Neural Crest Survival. J. Dev. Biol. 2016;4:24. doi: 10.3390/jdb4030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xavier G.M., Seppala M., Barrell W., Birjandi A.A., Geoghegan F., Cobourne M.T. Hedgehog Receptor Function during Craniofacial Development. Dev. Biol. 2016;415:198–215. doi: 10.1016/j.ydbio.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 146.Billmyre K.K., Klingensmith J. Sonic Hedgehog from Pharyngeal Arch 1 Epithelium Is Necessary for Early Mandibular Arch Cell Survival and Later Cartilage Condensation Differentiation. Dev. Dyn. 2015;244:564–576. doi: 10.1002/dvdy.24256. [DOI] [PubMed] [Google Scholar]
- 147.Millington G., Elliott K.H., Chang Y.-T., Chang C.-F., Dlugosz A., Brugmann S.A. Cilia-Dependent GLI Processing in Neural Crest Cells Is Required for Tongue Development. Dev. Biol. 2017;424:124–137. doi: 10.1016/j.ydbio.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yamagishi C., Yamagishi H., Maeda J., Tsuchihashi T., Ivey K., Hu T., Srivastava D. Sonic Hedgehog Is Essential for First Pharyngeal Arch Development. Pediatr. Res. 2006;59:349–354. doi: 10.1203/01.pdr.0000199911.17287.3e. [DOI] [PubMed] [Google Scholar]
- 149.Brito J.M., Teillet M.-A., Le Douarin N.M. Induction of Mirror-Image Supernumerary Jaws in Chicken Mandibular Mesenchyme by Sonic Hedgehog-Producing Cells. Development. 2008;135:2311–2319. doi: 10.1242/dev.019125. [DOI] [PubMed] [Google Scholar]
- 150.Haworth K.E., Wilson J.M., Grevellec A., Cobourne M.T., Healy C., Helms J.A., Sharpe P.T., Tucker A.S. Sonic Hedgehog in the Pharyngeal Endoderm Controls Arch Pattern via Regulation of Fgf8 in Head Ectoderm. Dev. Biol. 2007;303:244–258. doi: 10.1016/j.ydbio.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 151.Akiyama R., Kawakami H., Taketo M.M., Evans S.M., Wada N., Petryk A., Kawakami Y. Distinct Populations within Isl1 Lineages Contribute to Appendicular and Facial Skeletogenesis through the β-Catenin Pathway. Dev. Biol. 2014;387:37–48. doi: 10.1016/j.ydbio.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lin L., Bu L., Cai C.-L., Zhang X., Evans S. Isl1 Is Upstream of Sonic Hedgehog in a Pathway Required for Cardiac Morphogenesis. Dev. Biol. 2006;295:756–763. doi: 10.1016/j.ydbio.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 153.Sun Y., Teng I., Huo R., Rosenfeld M.G., Olson L.E., Li X., Li X. Asymmetric Requirement of Surface Epithelial β-Catenin during the Upper and Lower Jaw Development. Dev. Dyn. 2012;241:663–674. doi: 10.1002/dvdy.23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li F., Fu G., Liu Y., Miao X., Li Y., Yang X., Zhang X., Yu D., Gan L., Qiu M., et al. ISLET1-Dependent β-Catenin/Hedgehog Signaling Is Required for Outgrowth of the Lower Jaw. Mol. Cell. Biol. 2017;37 doi: 10.1128/MCB.00590-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Crump J.G., Maves L., Lawson N.D., Weinstein B.M., Kimmel C.B. An Essential Role for Fgfs in Endodermal Pouch Formation Influences Later Craniofacial Skeletal Patterning. Development. 2004;131:5703–5716. doi: 10.1242/dev.01444. [DOI] [PubMed] [Google Scholar]
- 156.Sperber S.M., Dawid I.B. Barx1 Is Necessary for Ectomesenchyme Proliferation and Osteochondroprogenitor Condensation in the Zebrafish Pharyngeal Arches. Dev. Biol. 2008;321:101–110. doi: 10.1016/j.ydbio.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Iwasaki S. Evolution of the Structure and Function of the Vertebrate Tongue. J. Anat. 2002;201:1–13. doi: 10.1046/j.1469-7580.2002.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bell D.M., Leung K.K., Wheatley S.C., Ng L.J., Zhou S., Ling K.W., Sham M.H., Koopman P., Tam P.P., Cheah K.S. SOX9 Directly Regulates the Type-II Collagen Gene. Nat. Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 159.Bi W., Deng J.M., Zhang Z., Behringer R.R., de Crombrugghe B. Sox9 Is Required for Cartilage Formation. Nat. Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 160.Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. Osf2/Cbfa1: A Transcriptional Activator of Osteoblast Differentiation. Cell. 1997;89:747–754. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 161.Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W.H., Beddington R.S.P., Mundlos S., Olsen B.R., et al. Cbfa1, a Candidate Gene for Cleidocranial Dysplasia Syndrome, Is Essential for Osteoblast Differentiation and Bone Development. Cell. 1997;89:765–771. doi: 10.1016/S0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 162.Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D.H., McMahon A.P., Sommer L., Boussadia O., Kemler R. Inactivation of the Beta-Catenin Gene by Wnt1-Cre-Mediated Deletion Results in Dramatic Brain Malformation and Failure of Craniofacial Development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 163.Day T.F., Guo X., Garrett-Beal L., Yang Y. Wnt/Beta-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis. Dev. Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 164.Goodnough L.H., DiNuoscio G.J., Ferguson J.W., Williams T., Lang R.A., Atit R.P. Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors. PLoS Genet. 2014;10:e1004152. doi: 10.1371/journal.pgen.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. The Novel Zinc Finger-Containing Transcription Factor Osterix Is Required for Osteoblast Differentiation and Bone Formation. Cell. 2002;108:17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 166.Lefebvre V., Huang W., Harley V.R., Goodfellow P.N., de Crombrugghe B. SOX9 Is a Potent Activator of the Chondrocyte-Specific Enhancer of the pro Alpha1(II) Collagen Gene. Mol. Cell Biol. 1997;17:2336–2346. doi: 10.1128/MCB.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mori-Akiyama Y., Akiyama H., Rowitch D.H., de Crombrugghe B. Sox9 Is Required for Determination of the Chondrogenic Cell Lineage in the Cranial Neural Crest. Proc. Natl. Acad. Sci. USA. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang Z., Wlodarczyk B.J., Niederreither K., Venugopalan S., Florez S., Finnell R.H., Amendt B.A. Fuz Regulates Craniofacial Development through Tissue Specific Responses to Signaling Factors. PLoS ONE. 2011;6:e24608. doi: 10.1371/journal.pone.0024608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.-H., Inada M., et al. Targeted Disruption of Cbfa1 Results in a Complete Lack of Bone Formation Owing to Maturational Arrest of Osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 170.Shirai Y., Kawabe K., Tosa I., Tsukamoto S., Yamada D., Takarada T. Runx2 Function in Cells of Neural Crest Origin during Intramembranous Ossification. Biochem. Biophys. Res. Commun. 2019;509:1028–1033. doi: 10.1016/j.bbrc.2019.01.059. [DOI] [PubMed] [Google Scholar]
- 171.Shibata S., Suda N., Yoda S., Fukuoka H., Ohyama K., Yamashita Y., Komori T. Runx2-Deficient Mice Lack Mandibular Condylar Cartilage and Have Deformed Meckel’s Cartilage. Anat. Embryol. 2004;208:273–280. doi: 10.1007/s00429-004-0393-2. [DOI] [PubMed] [Google Scholar]
- 172.Baek W.-Y., Kim Y.-J., de Crombrugghe B., Kim J.-E. Osterix Is Required for Cranial Neural Crest-Derived Craniofacial Bone Formation. Biochem. Biophys. Res. Commun. 2013;432:188–192. doi: 10.1016/j.bbrc.2012.12.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Semba I., Nonaka K., Takahashi I., Takahashi K., Dashner R., Shum L., Nuckolls G.H., Slavkin H.C. Positionally-Dependent Chondrogenesis Induced by BMP4 Is Co-Regulated by Sox9 and Msx2. Dev. Dyn. 2000;217:401–414. doi: 10.1002/(SICI)1097-0177(200004)217:4<401::AID-DVDY7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 174.Garcia-Miñaur S., Mavrogiannis L.A., Rannan-Eliya S.V., Hendry M.A., Liston W.A., Porteous M.E.M., Wilkie A.O.M. Parietal Foramina with Cleidocranial Dysplasia Is Caused by Mutation in MSX2. Eur. J. Hum. Genet. 2003;11:892–895. doi: 10.1038/sj.ejhg.5201062. [DOI] [PubMed] [Google Scholar]
- 175.Funato N., Nakamura M., Richardson J.A., Srivastava D., Yanagisawa H. Loss of Tbx1 Induces Bone Phenotypes Similar to Cleidocranial Dysplasia. Hum. Mol. Genet. 2015;24:424–435. doi: 10.1093/hmg/ddu458. [DOI] [PubMed] [Google Scholar]
- 176.Jabs E.W., Müller U., Li X., Ma L., Luo W., Haworth I.S., Klisak I., Sparkes R., Warman M.L., Mulliken J.B. A Mutation in the Homeodomain of the Human MSX2 Gene in a Family Affected with Autosomal Dominant Craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 177.Liu Y.H., Kundu R., Wu L., Luo W., Ignelzi M.A., Snead M.L., Maxson R.E. Premature Suture Closure and Ectopic Cranial Bone in Mice Expressing Msx2 Transgenes in the Developing Skull. Proc. Natl. Acad. Sci. USA. 1995;92:6137–6141. doi: 10.1073/pnas.92.13.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wilkie A.O.M., Tang Z., Elanko N., Walsh S., Twigg S.R.F., Hurst J.A., Wall S.A., Chrzanowska K.H., Maxson R.E. Functional Haploinsufficiency of the Human Homeobox Gene MSX2 Causes Defects in Skull Ossification. Nat. Genet. 2000;24:387–390. doi: 10.1038/74224. [DOI] [PubMed] [Google Scholar]
- 179.Jumlongras D., Bei M., Stimson J.M., Wang W.-F., DePalma S.R., Seidman C.E., Felbor U., Maas R., Seidman J.G., Olsen B.R. A Nonsense Mutation in MSX1 Causes Witkop Syndrome. Am. J. Hum. Genet. 2001;69:67–74. doi: 10.1086/321271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Winograd J., Reilly M.P., Roe R., Lutz J., Laughner E., Xu X., Hu L., Asakura T., vander Kolk C., Strandberg J.D., et al. Perinatal Lethality and Multiple Craniofacial Malformations in MSX2 Transgenic Mice. Hum. Mol. Genet. 1997;6:369–379. doi: 10.1093/hmg/6.3.369. [DOI] [PubMed] [Google Scholar]
- 181.Satokata I., Maas R. Msx1 Deficient Mice Exhibit Cleft Palate and Abnormalities of Craniofacial and Tooth Development. Nat. Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 182.Ishii M., Han J., Yen H.-Y., Sucov H.M., Chai Y., Maxson R.E., Jr. Combined Deficiencies of Msx1 and Msx2 Cause Impaired Patterning and Survival of the Cranial Neural Crest. Development. 2005;132:4937–4950. doi: 10.1242/dev.02072. [DOI] [PubMed] [Google Scholar]
- 183.Wu M., Li J., Engleka K.A., Zhou B., Lu M.M., Plotkin J.B., Epstein J.A. Persistent Expression of Pax3 in the Neural Crest Causes Cleft Palate and Defective Osteogenesis in Mice. J. Clin. Investig. 2008;118:2076–2087. doi: 10.1172/JCI33715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tremblay P., Dietrich S., Mericskay M., Schubert F.R., Li Z., Paulin D. A Crucial Role for Pax3 in the Development of the Hypaxial Musculature and the Long-Range Migration of Muscle Precursors. Dev. Biol. 1998;203:49–61. doi: 10.1006/dbio.1998.9041. [DOI] [PubMed] [Google Scholar]
- 185.Parry D.A., Logan C.V., Stegmann A.P.A., Abdelhamed Z.A., Calder A., Khan S., Bonthron D.T., Clowes V., Sheridan E., Ghali N., et al. SAMS, a Syndrome of Short Stature, Auditory-Canal Atresia, Mandibular Hypoplasia, and Skeletal Abnormalities Is a Unique Neurocristopathy Caused by Mutations in Goosecoid. Am. J. Hum. Genet. 2013;93:1135–1142. doi: 10.1016/j.ajhg.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gaunt S.J., Blum M., De Robertis E.M. Expression of the Mouse Goosecoid Gene during Mid-Embryogenesis May Mark Mesenchymal Cell Lineages in the Developing Head, Limbs and Body Wall. Development. 1993;117:769–778. doi: 10.1242/dev.117.2.769. [DOI] [PubMed] [Google Scholar]
- 187.Rivera-Perez J.A., Mallo M., Gendron-Maguire M., Gridley T., Behringer R.R. Goosecoid Is Not an Essential Component of the Mouse Gastrula Organizer but Is Required for Craniofacial and Rib Development. Development. 1995;121:3005–3012. doi: 10.1242/dev.121.9.3005. [DOI] [PubMed] [Google Scholar]
- 188.Yamada G., Ueno K., Nakamura S., Hanamure Y., Yasui K., Uemura M., Eizuru Y., Mansouri A., Blum M., Sugimura K. Nasal and Pharyngeal Abnormalities Caused by the Mouse Goosecoid Gene Mutation. Biophys. Res. Commun. 1997;233:161–165. doi: 10.1006/bbrc.1997.6315. [DOI] [PubMed] [Google Scholar]
- 189.Cserjesi P., Lilly B., Bryson L., Wang Y., Sassoon D.A., Olson E.N. MHox: A Mesodermally Restricted Homeodomain Protein That Binds an Essential Site in the Muscle Creatine Kinase Enhancer. Development. 1992;115:1087–1101. doi: 10.1242/dev.115.4.1087. [DOI] [PubMed] [Google Scholar]
- 190.Opstelten D.-J.E., Vogels R., Robert B., Kalkhoven E., Zwartkruis F., de Laaf L., Destrée O.H., Deschamps J., Lawson K.A., Meijlink F. The Mouse Homeobox Gene, S8, Is Expressed during Embryogenesis Predominantly in Mesenchyme. Mech. Dev. 1991;34:29–41. doi: 10.1016/0925-4773(91)90089-O. [DOI] [PubMed] [Google Scholar]
- 191.Martin J.F., Bradley A., Olson E.N. The Paired-like Homeo Box Gene MHox Is Required for Early Events of Skeletogenesis in Multiple Lineages. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- 192.ten Berge D., Brouwer A., Korving J., Reijnen M.J., van Raaij E.J., Verbeek F., Gaffield W., Meijlink F. Prx1 and Prx2 Are Upstream Regulators of Sonic Hedgehog and Control Cell Proliferation during Mandibular Arch Morphogenesis. Development. 2001;128:2929–2938. doi: 10.1242/dev.128.15.2929. [DOI] [PubMed] [Google Scholar]
- 193.Balic A., Adams D., Mina M. Prx1 and Prx2 Cooperatively Regulate the Morphogenesis of the Medial Region of the Mandibular Process. Dev. Dyn. 2009;238:2599–2613. doi: 10.1002/dvdy.22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Çelik T., Simsek P.O., Sozen T., Ozyuncu O., Utine G.E., Talim B., Yiğit Ş., Boduroglu K., Kamnasaran D. PRRX1 Is Mutated in an Otocephalic Newborn Infant Conceived by Consanguineous Parents. Clin. Genet. 2012;81:294–297. doi: 10.1111/j.1399-0004.2011.01730.x. [DOI] [PubMed] [Google Scholar]
- 195.Dasouki M., Andrews B., Parimi P., Kamnasaran D. Recurrent Agnathia–Otocephaly Caused by DNA Replication Slippage in PRRX1. Am. J. Med. Genet. Part A. 2013;161:803–808. doi: 10.1002/ajmg.a.35879. [DOI] [PubMed] [Google Scholar]
- 196.Donnelly M., Todd E., Wheeler M., Winn V.D., Kamnasaran D. Prenatal Diagnosis and Identification of Heterozygous Frameshift Mutation in PRRX1 in an Infant with Agnathia-Otocephaly. Prenat. Diagn. 2012;32:903–905. doi: 10.1002/pd.3910. [DOI] [PubMed] [Google Scholar]
- 197.Herman S., Delio M., Morrow B., Samanich J. Agnathia–Otocephaly Complex: A Case Report and Examination of the OTX2 and PRRX1 Genes. Gene. 2012;494:124–129. doi: 10.1016/j.gene.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 198.Sergi C., Kamnasaran D. PRRX1 Is Mutated in a Fetus with Agnathia-Otocephaly. Clin. Genet. 2011;79:293–295. doi: 10.1111/j.1399-0004.2010.01531.x. [DOI] [PubMed] [Google Scholar]
- 199.Shibukawa Y., Young B., Wu C., Yamada S., Long F., Pacifici M., Koyama E. Temporomandibular Joint Formation and Condyle Growth Require Indian Hedgehog Signaling. Dev. Dyn. 2007;236:426–434. doi: 10.1002/dvdy.21036. [DOI] [PubMed] [Google Scholar]
- 200.Sugito H., Shibukawa Y., Kinumatsu T., Yasuda T., Nagayama M., Yamada S., Minugh-Purvis N., Pacifici M., Koyama E. Ihh Signaling Regulates Mandibular Symphysis Development and Growth. J. Dent. Res. 2011;90:625–631. doi: 10.1177/0022034510397836. [DOI] [PubMed] [Google Scholar]
- 201.Yang L., Gu S., Ye W., Song Y., Chen Y. Augmented Indian Hedgehog Signaling in Cranial Neural Crest Cells Leads to Craniofacial Abnormalities and Dysplastic Temporomandibular Joint in Mice. Cell Tissue Res. 2016;364:105–115. doi: 10.1007/s00441-015-2306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bechtold T.E., Kurio N., Nah H.-D., Saunders C., Billings P.C., Koyama E. The Roles of Indian Hedgehog Signaling in TMJ Formation. Int. J. Mol. Sci. 2019;20:6300. doi: 10.3390/ijms20246300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bertolacini C.D.P., Ribeiro-Bicudo L.A., Petrin A., Richieri-Costa A., Murray J.C. Clinical Findings in Patients with GLI2 Mutations--Phenotypic Variability. Clin. Genet. 2012;81:70–75. doi: 10.1111/j.1399-0004.2010.01606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chai Y., Mah A., Crohin C., Groff S., Bringas P., Le T., Santos V., Slavkin H.C. Specific Transforming Growth Factor-Beta Subtypes Regulate Embryonic Mouse Meckel’s Cartilage and Tooth Development. Dev. Biol. 1994;162:85–103. doi: 10.1006/dbio.1994.1069. [DOI] [PubMed] [Google Scholar]
- 205.Zhao H., Oka K., Bringas P., Kaartinen V., Chai Y. TGF-Beta Type I Receptor Alk5 Regulates Tooth Initiation and Mandible Patterning in a Type II Receptor-Independent Manner. Dev. Biol. 2008;320:19–29. doi: 10.1016/j.ydbio.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dudas M., Kim J., Li W.-Y., Nagy A., Larsson J., Karlsson S., Chai Y., Kaartinen V. Epithelial and Ectomesenchymal Role of the Type I TGF-β Receptor ALK5 during Facial Morphogenesis and Palatal Fusion. Dev. Biol. 2006;296:298–314. doi: 10.1016/j.ydbio.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ito Y., Yeo J.Y., Chytil A., Han J., Bringas P., Nakajima A., Shuler C.F., Moses H.L., Chai Y. Conditional Inactivation of Tgfbr2 in Cranial Neural Crest Causes Cleft Palate and Calvaria Defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- 208.Iwata J., Parada C., Chai Y. The Mechanism of TGF-β Signaling during Palate Development. Oral Dis. 2011;17:733–744. doi: 10.1111/j.1601-0825.2011.01806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Oka K., Oka S., Hosokawa R., Bringas P., Brockhoff H.C., Nonaka K., Chai Y. TGF-β Mediated Dlx5 Signaling Plays a Crucial Role in Osteo-Chondroprogenitor Cell Lineage Determination during Mandible Development. Dev. Biol. 2008;321:303–309. doi: 10.1016/j.ydbio.2008.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Stottmann R.W., Anderson R.M., Klingensmith J. The BMP Antagonists Chordin and Noggin Have Essential but Redundant Roles in Mouse Mandibular Outgrowth. Dev. Biol. 2001;240:457–473. doi: 10.1006/dbio.2001.0479. [DOI] [PubMed] [Google Scholar]
- 211.Wang Y., Zheng Y., Chen D., Chen Y. Enhanced BMP Signaling Prevents Degeneration and Leads to Endochondral Ossification of Meckel′s Cartilage in Mice. Dev. Biol. 2013;381:301–311. doi: 10.1016/j.ydbio.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lana-Elola E., Tylzanowski P., Takatalo M., Alakurtti K., Veistinen L., Mitsiadis T.A., Graf D., Rice R., Luyten F.P., Rice D.P. Noggin Null Allele Mice Exhibit a Microform of Holoprosencephaly. Hum. Mol. Genet. 2011;20:4005–4015. doi: 10.1093/hmg/ddr329. [DOI] [PubMed] [Google Scholar]
- 213.He F., Hu X., Xiong W., Li L., Lin L., Shen B., Yang L., Gu S., Zhang Y., Chen Y. Directed Bmp4 Expression in Neural Crest Cells Generates a Genetic Model for the Rare Human Bony Syngnathia Birth Defect. Dev. Biol. 2014;391:170–181. doi: 10.1016/j.ydbio.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chen Y., Wang Z., Chen Y., Zhang Y. Conditional Deletion of Bmp2 in Cranial Neural Crest Cells Recapitulates Pierre Robin Sequence in Mice. Cell Tissue Res. 2019;376:199–210. doi: 10.1007/s00441-018-2944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bonilla-Claudio M., Wang J., Bai Y., Klysik E., Selever J., Martin J.F. Bmp Signaling Regulates a Dose-Dependent Transcriptional Program to Control Facial Skeletal Development. Development. 2012;139:709–719. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ho A.M., Marker P.C., Peng H., Quintero A.J., Kingsley D.M., Huard J. Dominant Negative Bmp5mutation Reveals Key Role of BMPs in Skeletal Response to Mechanical Stimulation. BMC Dev. Biol. 2008;8:35. doi: 10.1186/1471-213X-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kouskoura T., Kozlova A., Alexiou M., Blumer S., Zouvelou V., Katsaros C., Chiquet M., Mitsiadis T.A., Graf D. The Etiology of Cleft Palate Formation in BMP7-Deficient Mice. PLoS ONE. 2013;8:e59463. doi: 10.1371/journal.pone.0059463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Trokovic N., Trokovic R., Mai P., Partanen J. Fgfr1 Regulates Patterning of the Pharyngeal Region. Genes Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Trokovic N., Trokovic R., Partanen J. Fibroblast Growth Factor Signalling and Regional Specification of the Pharyngeal Ectoderm. Int. J. Dev. Biol. 2005;49:797–805. doi: 10.1387/ijdb.051976nt. [DOI] [PubMed] [Google Scholar]
- 220.Hoch R.V., Soriano P. Context-Specific Requirements for Fgfr1 Signaling through Frs2 and Frs3 during Mouse Development. Development. 2006;133:663–673. doi: 10.1242/dev.02242. [DOI] [PubMed] [Google Scholar]
- 221.Kameda Y., Ito M., Nishimaki T., Gotoh N. FRS2alpha Is Required for the Separation, Migration, and Survival of Pharyngeal-Endoderm Derived Organs Including Thyroid, Ultimobranchial Body, Parathyroid, and Thymus. Dev. Dyn. 2009;238:503–513. doi: 10.1002/dvdy.21867. [DOI] [PubMed] [Google Scholar]
- 222.Jackson A., Kasah S., Mansour S.L., Morrow B., Basson M.A. Endoderm-Specific Deletion of Tbx1 Reveals an FGF-Independent Role for Tbx1 in Pharyngeal Apparatus Morphogenesis. Dev. Dyn. 2014;243:1143–1151. doi: 10.1002/dvdy.24147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang C., Chang J.Y.F., Yang C., Huang Y., Liu J., You P., McKeehan W.L., Wang F., Li X. Type 1 Fibroblast Growth Factor Receptor in Cranial Neural Crest Cell-Derived Mesenchyme Is Required for Palatogenesis. J. Biol. Chem. 2013;288:22174–22183. doi: 10.1074/jbc.M113.463620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Motch Perrine S.M., Wu M., Stephens N.B., Kriti D., van Bakel H., Jabs E.W., Richtsmeier J.T. Mandibular Dysmorphology Due to Abnormal Embryonic Osteogenesis in FGFR2-Related Craniosynostosis Mice. Dis. Models Mech. 2019;12 doi: 10.1242/dmm.038513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Biosse Duplan M., Komla-Ebri D., Heuzé Y., Estibals V., Gaudas E., Kaci N., Benoist-Lasselin C., Zerah M., Kramer I., Kneissel M., et al. Meckel’s and Condylar Cartilages Anomalies in Achondroplasia Result in Defective Development and Growth of the Mandible. Hum. Mol. Genet. 2016;25:2997–3010. doi: 10.1093/hmg/ddw153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Melnick M., Witcher D., Bringas P., Carlsson P., Jaskoll T. Meckel’s Cartilage Differentiation Is Dependent on Hedgehog Signaling. Cells Tissues Organs. 2005;179:146–157. doi: 10.1159/000085950. [DOI] [PubMed] [Google Scholar]
- 227.Shao M., Liu C., Song Y., Ye W., He W., Yuan G., Gu S., Lin C., Ma L., Zhang Y., et al. FGF8 Signaling Sustains Progenitor Status and Multipotency of Cranial Neural Crest-Derived Mesenchymal Cells in Vivo and in Vitro. J. Mol. Cell Biol. 2015;7:441–454. doi: 10.1093/jmcb/mjv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Abu-Issa R., Smyth G., Smoak I., Yamamura K., Meyers E.N. Fgf8 Is Required for Pharyngeal Arch and Cardiovascular Development in the Mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 229.Terao F., Takahashi I., Mitani H., Haruyama N., Sasano Y., Suzuki O., Takano-Yamamoto T. Fibroblast Growth Factor 10 Regulates Meckel’s Cartilage Formation during Early Mandibular Morphogenesis in Rats. Dev. Biol. 2011;350:337–347. doi: 10.1016/j.ydbio.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 230.Cruz C.V., Mattos C.T., Maia J.C., Granjeiro J.M., Reis M.F., Mucha J.N., Vilella B., Ruellas A.C., Luiz R.R., Costa M.C., et al. Genetic Polymorphisms Underlying the Skeletal Class III Phenotype. Am. J. Orthod. Dentofac. Orthop. 2017;151:700–707. doi: 10.1016/j.ajodo.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 231.Taniguchi K., Ayada T., Ichiyama K., Kohno R.-I., Yonemitsu Y., Minami Y., Kikuchi A., Maehara Y., Yoshimura A. Sprouty2 and Sprouty4 Are Essential for Embryonic Morphogenesis and Regulation of FGF Signaling. Biochem. Biophys. Res. Commun. 2007;352:896–902. doi: 10.1016/j.bbrc.2006.11.107. [DOI] [PubMed] [Google Scholar]
- 232.Curtin E., Hickey G., Kamel G., Davidson A.J., Liao E.C. Zebrafish Wnt9a Is Expressed in Pharyngeal Ectoderm and Is Required for Palate and Lower Jaw Development. Mech. Dev. 2011;128:104–115. doi: 10.1016/j.mod.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 233.Kamel G., Hoyos T., Rochard L., Dougherty M., Kong Y., Tse W., Shubinets V., Grimaldi M., Liao E.C. Requirement for Frzb and Fzd7a in Cranial Neural Crest Convergence and Extension Mechanisms during Zebrafish Palate and Jaw Morphogenesis. Dev. Biol. 2013;381:423–433. doi: 10.1016/j.ydbio.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 234.Lin Q., He Y., Gui J.-F., Mei J. Sox9a, Not Sox9b Is Required for Normal Cartilage Development in Zebrafish. Aquac. Fish. 2021;6:254–259. doi: 10.1016/j.aaf.2019.12.009. [DOI] [Google Scholar]
- 235.Yan Y.-L., Willoughby J., Liu D., Crump J.G., Wilson C., Miller C.T., Singer A., Kimmel C., Westerfield M., Postlethwait J.H. A Pair of Sox: Distinct and Overlapping Functions of Zebrafish Sox9 Co-Orthologs in Craniofacial and Pectoral Fin Development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 236.Flores M.V., Tsang V.W.K., Hu W., Kalev-Zylinska M., Postlethwait J., Crosier P., Crosier K., Fisher S. Duplicate Zebrafish Runx2 Orthologues Are Expressed in Developing Skeletal Elements. Gene Expr. Patterns. 2004;4:573–581. doi: 10.1016/j.modgep.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 237.Flores M.V., Lam E.Y.N., Crosier P., Crosier K. A Hierarchy of Runx Transcription Factors Modulate the Onset of Chondrogenesis in Craniofacial Endochondral Bones in Zebrafish. Dev. Dyn. 2006;235:3166–3176. doi: 10.1002/dvdy.20957. [DOI] [PubMed] [Google Scholar]
- 238.Felber K., Elks P.M., Lecca M., Roehl H.H. Expression of Osterix Is Regulated by FGF and Wnt/β-Catenin Signalling during Osteoblast Differentiation. PLoS ONE. 2015;10:e0144982. doi: 10.1371/journal.pone.0144982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dalcq J., Pasque V., Ghaye A., Larbuisson A., Motte P., Martial J.A., Muller M. RUNX3, EGR1 and SOX9B Form a Regulatory Cascade Required to Modulate BMP-Signaling during Cranial Cartilage Development in Zebrafish. PLoS ONE. 2012;7:e50140. doi: 10.1371/journal.pone.0050140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Nakada C., Iida A., Tabata Y., Watanabe S. Forkhead Transcription Factor Foxe1 Regulates Chondrogenesis in Zebrafish. J. Exp. Zool. 2009;312B:827–840. doi: 10.1002/jez.b.21298. [DOI] [PubMed] [Google Scholar]
- 241.Li N., Felber K., Elks P., Croucher P., Roehl H.H. Tracking Gene Expression during Zebrafish Osteoblast Differentiation. Dev. Dyn. 2009;238:459–466. doi: 10.1002/dvdy.21838. [DOI] [PubMed] [Google Scholar]
- 242.Chen Z., Song Z., Yang J., Huang J., Jiang H. Sp7/Osterix Positively Regulates Dlx2b and Bglap to Affect Tooth Development and Bone Mineralization in Zebrafish Larvae. J. Biosci. 2019;44:127. doi: 10.1007/s12038-019-9948-5. [DOI] [PubMed] [Google Scholar]
- 243.Niu P., Zhong Z., Wang M., Huang G., Xu S., Hou Y., Yan Y., Wang H. Zinc Finger Transcription Factor Sp7/Osterix Acts on Bone Formation and Regulates Col10a1a Expression in Zebrafish. Sci. Bull. 2017;62:174–184. doi: 10.1016/j.scib.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 244.Eberhart J.K., Swartz M.E., Crump J.G., Kimmel C.B. Early Hedgehog Signaling from Neural to Oral Epithelium Organizes Anterior Craniofacial Development. Development. 2006;133:1069–1077. doi: 10.1242/dev.02281. [DOI] [PubMed] [Google Scholar]
- 245.Schwend T., Ahlgren S.C. Zebrafish Con/Disp1 Reveals Multiple Spatiotemporal Requirements for Hedgehog-Signaling in Craniofacial Development. BMC Dev. Biol. 2009;9:59. doi: 10.1186/1471-213X-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Felber K., Croucher P., Roehl H.H. Hedgehog Signalling Is Required for Perichondral Osteoblast Differentiation in Zebrafish. Mech. Dev. 2011;128:141–152. doi: 10.1016/j.mod.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 247.Hammond C.L., Schulte-Merker S. Two Populations of Endochondral Osteoblasts with Differential Sensitivity to Hedgehog Signalling. Development. 2009;136:3991–4000. doi: 10.1242/dev.042150. [DOI] [PubMed] [Google Scholar]
- 248.Huycke T.R., Eames B.F., Kimmel C.B. Hedgehog-Dependent Proliferation Drives Modular Growth during Morphogenesis of a Dermal Bone. Development. 2012;139:2371–2380. doi: 10.1242/dev.079806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hu Z., Chen B., Zhao Q. Hedgehog Signaling Regulates Osteoblast Differentiation in Zebrafish Larvae through Modulation of Autophagy. Biol. Open. 2019;8:bio040840. doi: 10.1242/bio.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Barske L., Askary A., Zuniga E., Balczerski B., Bump P., Nichols J.T., Crump J.G. Competition between Jagged-Notch and Endothelin1 Signaling Selectively Restricts Cartilage Formation in the Zebrafish Upper Face. PLoS Genet. 2016;12:e1005967. doi: 10.1371/journal.pgen.1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Swartz M.E., Sheehan-Rooney K., Dixon M.J., Eberhart J.K. Examination of a Palatogenic Gene Program in Zebrafish. Dev. Dyn. 2011;240:2204–2220. doi: 10.1002/dvdy.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cheah F.S.H., Winkler C., Jabs E.W., Chong S.S. Tgfβ3 Regulation of Chondrogenesis and Osteogenesis in Zebrafish Is Mediated through Formation and Survival of a Subpopulation of the Cranial Neural Crest. Mech. Dev. 2010;127:329–344. doi: 10.1016/j.mod.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 253.Windhausen T., Squifflet S., Renn J., Muller M. BMP Signaling Regulates Bone Morphogenesis in Zebrafish through Promoting Osteoblast Function as Assessed by Their Nitric Oxide Production. Molecules. 2015;20:7586–7601. doi: 10.3390/molecules20057586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Phillips B.T., Kwon H.-J., Melton C., Houghtaling P., Fritz A., Riley B.B. Zebrafish MsxB, MsxC and MsxE Function Together to Refine the Neural–Nonneural Border and Regulate Cranial Placodes and Neural Crest Development. Dev. Biol. 2006;294:376–390. doi: 10.1016/j.ydbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.