Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are two common betacoronaviruses, which are still causing transmission among the human population worldwide. The major difference between the two coronaviruses is that MERS-CoV is now causing sporadic transmission worldwide, whereas SARS-CoV-2 is causing a pandemic outbreak globally. Currently, different guidelines and reports have highlighted several diagnostic methods and approaches which could be used to screen and confirm MERS-CoV and SARS-CoV-2 infections. These methods include clinical evaluation, laboratory diagnosis (nucleic acid-based test, protein-based test, or viral culture), and radiological diagnosis. With the presence of these different diagnostic approaches, it could cause a dilemma to the clinicians and diagnostic laboratories in selecting the best diagnostic strategies to confirm MERS-CoV and SARS-CoV-2 infections. Therefore, this review aims to provide an up-to-date comparison of the advantages and limitations of different diagnostic approaches in detecting MERS-CoV and SARS-CoV-2 infections. This review could provide insights for clinicians and scientists in detecting MERS-CoV and SARS-CoV-2 infections to help combat the transmission of these coronaviruses.
Abbreviations: acpcPNA, pyrrolidinyl peptide nucleic acid; ALI, Acute lung injury; ASO, Antisense oligonucleotides; BSL, Biosafety level; CDC, Centers for Disease Control and Prevention; CoV, Coronavirus; COVID-19, Coronavirus disease of 2019; CRISPR, Clustered regularly interspaced short palindromic repeats; CT, Computed tomography; CXR, Chest radiography; ddNTPs, dideoxynucleotide triphosphates; DNA, Deoxyribonucleic acid; dPCR, Digital polymerase chain reaction; EIA, Enzyme immunoassay; ELISA, Enzyme-linked immunosorbent assay; EUA, Emergency use authorization; FDA, Food and Drug Administration; FIPV, Feline infectious peritonitis virus; Hel, Helicase; HIV, Human immunodeficiency virus; ICU, Intensive care unit; IFA, Immunofluorescence assay; LAMP, Loop-mediated isothermal amplification; LOD, Limit of detection; LOQ, Limit of quantification; MALDI-TOF, MassARRAY matrix-assisted laser desorption ionization-time of flight; MERS, Middle East respiratory syndrome; MN, Microneutralisation; MS, Mass spectrometry; NP, Nucleocapsid protein; NSP, Non-structural proteins; ORF, Open reading frame; RAD, Rapid antigen test; PCR, Polymerase chain reaction; PFU, Plaque forming units; POCT, Point of care testing; ppNT, Pseudoparticle neutralization test; PRNT, Plaque reduction neutralization test; qPCR, Quantitative or real-time polymerase chain reaction; qRT-PCR, Real-time reverse transcription polymerase chain reaction; RBD, Receptor binding domain; RdRp, RNA-dependent RNA polymerase; RFLP, Restriction fragment length polymorphism; RNA, Ribonucleic acid; RSV, Respiratory syncytial virus; RT-iiPCR, Reverse transcription-insulated isothermal polymerase chain reaction; RT-RPA, Reverse transcription recombinase polymerase amplification; SARS, Severe acute respiratory syndrome; SNP, Single nucleotide polymorphism; TCID, Tissue culture infectious dose; TRPMSS2, Type II transmembrane serine protease; upE, Regions upstream of gene E; VOC, Variants of concern; VOHC, Variant of high consequence; VOI, Variant of interest; VUM, Variant under monitoring; WGS, Whole-genome sequencing; WHO, World Health Organization; OSN-qRT-PCR, One-step single-tube nested (OSN)-qRT-PCR; 5’-UTR, 5’-untranslated region
Keywords: MERS-CoV, SARS-CoV-2, Clinical, Laboratory, Radiological, Diagnosis
1. Introduction
Human coronavirus was first discovered in the 1960s and it is characterized as a virus that is responsible for causing respiratory tract infections in the human population [1]. Since the year 2003, at least six different types of coronaviruses have been discovered and these include severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1], [2], [3]. The two coronaviruses which have caused massive transmission in the human population in the past ten years are MERS-CoV and SARS-CoV-2 [3], [4].
MERS-CoV infection was initially reported in the year 2012 in the Kingdom of Saudi Arabia [5], and till the year 2020, more than 2,500 MERS-CoV cases have been reported worldwide and more than 880 patients had passed away because of this coronavirus infection [3], [6]. The fatality rate of MERS-CoV infection is about 35% [6], [7], which is way higher than the SARS outbreak that happened in the year 2003, in which the fatality rate of SARS-CoV infection was around 11% [3]. The MERS-CoV outbreak peaked and declined in four years’ time, from the year 2012–2015 [8]. Regardless, the transmission of this coronavirus still happens in a sporadic pattern and the last case was reported in December 2020 [6]. Compared to the MERS-CoV outbreak, the SARS-CoV epidemic was peaked in the year 2002–2003 and the transmission of this coronavirus has stopped since then [3], [9].
SARS-CoV-2 was first reported in the Hubei province, China, at the end of the year 2019, and was declared a global pandemic crisis in early 2020 [4]. SARS-CoV-2 is responsible for causing coronavirus disease 2019 (COVID-19) [10]. Until today (17th July 2021), more than 180 million Covid-19 cases have been reported worldwide and more than four million people have passed away because of Covid-19 [11]. Even though SARS-CoV-2 is causing more massive transmission than SARS-CoV and MERS-CoV, the fatality rate of SARS-CoV-2 infection is estimated to be around 2–3% [12], much lower than that of both the SARS-CoV [3] and MERS-CoV [6].
One of the biggest challenges in detecting MERS-CoV and SARS-CoV-2 infections is that these coronaviruses would cause respiratory tract infections similar to bacteria and other common cold viruses such as influenza virus and rhinovirus [13], [14]. Therefore, it would be difficult to detect the presence of coronavirus infection in patients who are presented with common cold symptoms such as fever, cough, and sore throat [14]. Besides, MERS-CoV and SARS-CoV-2 are highly pathogenic coronaviruses that can cause acute lung injury (ALI), severe pneumonia, and severe acute respiratory distress syndrome (SARS) which could be life-threatening [13]. A previously published report [15] described that the impact which could be brought by coronavirus infection “is more than a common cold infection” as it has the potential to cause a global outbreak that is accompanied by high mortality and morbidity rate.
To date, numerous studies and guidelines have outlined and compared different diagnostic approaches to detect MERS-CoV and SARS-CoV infections, which include clinical assessment, molecular diagnosis, serological diagnosis, and radiological diagnostic approaches [16], [17], [18]. With the presence of multiple, different diagnostic approaches to confirm the presence of MERS-CoV and SARS-CoV infections, it could be confusing sometimes for the clinicians and diagnostic laboratories to decide the best strategies to detect MERS-CoV and SARS-CoV infections [17], [18]. Besides, it is also not easy to differentiate MERS-CoV and SARS-CoV-2 infections from each other, and from other common cold virus infections [3], [13], [14]. This review, therefore, aims to provide an up-to-date comparison of the advantages and limitations of different diagnostic strategies in detecting MERS-CoV and SARS-CoV infections. It is hoped that this review would provide fruitful insights for the clinicians and scientists in detecting and confirming MERS-CoV and SARS-CoV-2 infections to help fight against these coronaviruses.
2. Brief overview of MERS-CoV and SARS-CoV-2
Coronavirus is a type of enveloped, positive-sense, single-stranded RNA virus grouped under the Coronaviridae family [19]. This virus has the potential to infect vertebrates such as humans and animals and is divided into four different types: alpha, beta, gamma, and delta coronavirus [20]. Alpha and beta coronaviruses are by far the only viruses that could infect mammals, while gamma and delta coronaviruses only infect birds [20]. MERS-CoV and SARS-CoV are classified as betacoronaviruses in which MERS-CoV is believed to originate from the bat and was spread to humans via camel [3]. The origin of SARS-CoV-2 is still in debate and it is hypothesized that this virus was originated from bats, since there is no sufficient evidence to suggest that it is an intentionally engineered novel coronavirus [21].
Compared to other RNA viruses, coronavirus has the biggest RNA viral genome which ranges from 26,000 bp to 32,000 bp in length [19], [22]. It was reported that the sequence identity between SARS-CoV, SARS-CoV-2, and MERS-CoV is at least 80% [19]. However, in another report, it was said that SARS-CoV and SARS-CoV-2 share around 80% of the sequence similarity while the sequences between SARS-CoV-2 and MERS-CoV are only about 50% similar [23]. The International Committee on Taxonomy of Viruses (ICTV) recommends that viruses sharing more than 90% of sequence identity in the conserved replicase domains belong to the same species [24]. Thus, it was concluded that both MERS-CoV and SARS-CoV-2 are novel and distinct betacoronavirus [5], [23]. The genome of MERS-CoV contains around 30,110 nucleotides [19], and this genome size is slightly larger compared to the genome of SARS-CoV-2 which contains about 30,000 nucleotides [19], [25]. In terms of infectivity, SARS-CoV-2 was reported to be more infectious as compared to both SARS-CoV and MERS-CoV [19]. One of the possible reasons to explain this phenomenon is that some mammals could potentially act as intermediate hosts during SARS-CoV-2 transmission and the virus is believed to acquire a certain level of mutation in order for them to transmit to the human from the intermittent host [19].
The RNAs of both viruses are capped at the 5’ end and poly-A tail is found at their 3’ end [19], [22]. As SARS-CoV, MERS-CoV and SARS-CoV-2 have a certain level of similar sequence identity as reported previously [19], therefore, the genetic structures of these three viruses are also quite similar. Generally, the viral genomes contain open reading frame (ORF) 1a and 1b, spike (S) gene, envelope (E) gene, membrane (M) gene, and nucleocapsid (N) gene [18], [19], [22]. According to the 5’ to 3’ order of arrangement, ORF1a and ORF1b are situated towards the 5’ end, followed by S, E, M, and N genes [18], [19], [22]. All these genes play vital roles in virus virulence, virus entry, and survival in the targeted host cells [18], [19], [22]. The S gene encodes for spike protein which helps facilitate the binding and attachment of the virus to the host cell [26], whereas the M gene encodes M protein which maintains the virus shape and structure [22]. E gene encodes E protein that plays an essential role in viral assembly and production [22]. N protein coded by the N gene is important in binding to the viral RNA and this protein helps in regulating viral replication [19], [22]. Other than structural protein-encoding genes, both MERS-CoV and SARS-CoV-2 also carry genes that encode a number of non-structural proteins (NSPs) that are important in regulating various virus activities such as viral replication [3], [19], [22], [27].
3. Different diagnostic approaches to detect MERS-CoV and SARS-CoV-2 infections
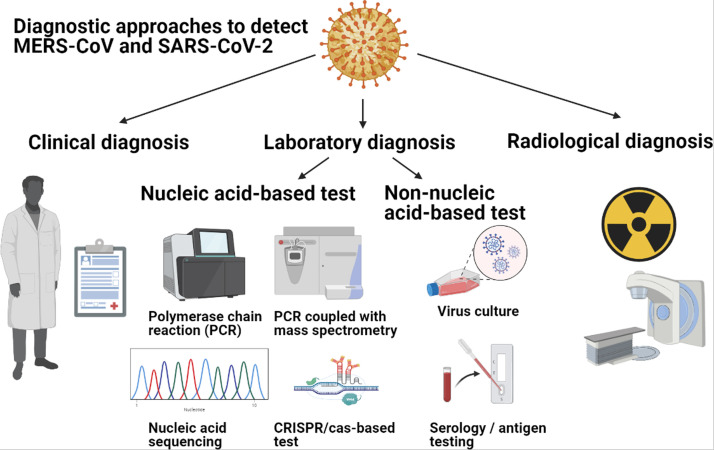
To date, there are several published methods and approaches ( Fig. 1) which could be used to screen and diagnose MERS-CoV and SARS-CoV-2 infections. These methods can be generally divided into three main categories, which refer to clinical diagnosis, laboratory diagnosis, and radiological diagnosis ( Table 1) [16], [17], [18]. Laboratory diagnosis can be further sub-divided into nucleic acid-based detection, protein-based detection, or viral culture to confirm the virus infections [16], [17], [18], [28].
Fig. 1.
Various diagnostic approaches which could be used to detect MERS-CoV and SARS-CoV infections. Clinical diagnosis relies on history taking and clinical assessment to determine whether an individual is at high risk of contracting the coronaviruses [6], [29], [30], [33]. Laboratory diagnostic tests can be divided into nucleic acid-based, protein-based, and virus-culture tests [16], [17]. Examples of nucleic acid-based diagnostic tests include polymerase chain reactions like qRT-PCR and dPCR, or RT-LAMP, RT-RPA, PCR-coupled with mass spectrometry (MS) and CRISPR/Cas-based detection test [16], [17], [99], [104], [105], [118], [121], [126], [185], [54], [57], [66], [74], [77], [95], [96], [97]. Protein-based diagnostic tests include virus serology, neutralization and virus antigen tests [2], [16], [17], [54], [57], [126], [180], [185], [257]. Radiological diagnosis involves the use of chest radiography or CT scan to assess the thoracic cavity of the individuals who are suspected to have pneumonia secondary to coronavirus infection [16], [277], [278], [279].
Table 1.
Overview and comparison of the strengths and limitations of different diagnostic approaches to detect MERS-CoV and SARS-CoV-2.
| Clinical diagnosis | Laboratory diagnosis |
Radiological diagnosis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Nucleic acid-based tests |
Non-nucleic acid-based tests |
||||||||
| PCR-based methods | Isothermal amplification-based detection methods | Nucleic acid sequencing | CRISPR/Cas-based method | Others: Mass spectrometry detection approach | Virus culture | Protein-based tests | |||
| Description | Assessment on the patient history and clinical features | Quantitative and qualitative detection of virus | Quantitative and qualitative detection of virus | Virus genome sequencing | Detection of specific viral genetic regions | MS approach to detect viral genetic targets | Live culture of virus | Antibody or antigen detection of the viral proteins | Radiological evaluation of the thoracic cavity of the individual |
| Advantages | Fast | Gold standard test, highly sensitive and specific | Might require shorter test time than qRT-PCR and do not need sophisticated instruments | Alternative test to detect the virus, differentiating different coronaviruses | Might require shorter test time | Highly sensitive and specific, faster test results | Could be used in vaccine development | Fast | Non-invasive |
| Limitations | Only clinicians could assess, less sensitive than molecular tests | Require specialized machine and trained staff to perform | Might be less sensitive and specific than qRT-PCR | Require specialized machine and trained staff to perform, expensive | Require trained staff to perform, expensive, multi-steps reactions might cause contamination | Require specialized machine and trained staff to perform, expensive | Require trained staff to perform, time-consuming | Risk of cross-reactivity with other pathogens | Require radiological facilities to perform the test, less sensitive than molecular tests |
| References | [29], [30], [31], [32], [33] | [16], [17], [54], [57], [60], [66], [67] | [95], [96], [97], [104], [105], [116], [118], [121] | [29], [34], [54], [57], [126] | [16], [144], [145], [148], [149] | [156], [157] | [16], [32], [35], [116], [118] | [26], [54], [57], [88], [107], [108], [110], [111], [132] | [16], [17], [277], [278], [279] |
3.1. Clinical diagnosis of MERS-CoV and SARS-CoV-2 infections
Assessing the patient history like a history of travel to coronavirus outbreak area and history of having close contact with confirmed cases are helpful to screen whether the patient is having a high risk to contract coronavirus [29], [30]. Besides, clinical evaluation of signs and symptoms which could be caused by MERS-CoV and SARS-CoV-2 infections is useful to rapidly screen for suspected cases at the triage area of the health facility [29], [31]. However, like other microorganisms or common cold viruses which could cause respiratory tract infections, both MERS-CoV and SARS-CoV-2 infections would also cause several common clinical signs and symptoms suggestive of respiratory system infection, such as cough, shortness of breath, respiratory distress, rhinorrhea, and hemoptysis [5], [29], [30], [32]. Besides, the coronavirus infections were also reported to show clinical symptoms in the gastrointestinal tract (vomiting and diarrhea), neurological system (headache and confusion), musculoskeletal system (muscle pain), and other general symptoms including fever [5], [9], [27], [29], [30], [32]. As most of these clinical signs and symptoms are non-specific to coronavirus infection, clinical evaluation alone could not be used to confirm the coronavirus infection [29], [30], [33], [34]. In addition, some of the patients infected with MERS-CoV and SARS-CoV-2 might be asymptomatic. Therefore, all suspected individuals who have recent contact with the confirmed MERS-CoV and SARS-CoV-2 patients, or had a history of travel to the outbreak areas, should be subjected to further laboratory testing to confirm the diagnosis of MERS-CoV and SARS-CoV-2 infections [29], [30], [33], [34].
In severe cases of MERS-CoV and SARS-CoV-2 infections, the patients might be presented with severe pneumonia, respiratory failure, or severe acute respiratory syndrome, in which these patients need to be supported in the intensive care unit (ICU) with mechanical ventilation support [12], [13], [35]. Regardless, none of these symptoms are exclusive to MERS-CoV and SARS-CoV-2 infections, therefore could not be used to differentiate MERS-CoV and SARS-CoV-2 infections from infections caused by other causative agents such as a respiratory syncytial virus (RSV) [36], [37].
3.2. Laboratory detection of MERS-CoV and SARS-CoV-2 infections
Generally, laboratory tests that could be employed to detect MERS-CoV and SARS-CoV-2 can be divided into the nucleic acid-based test, protein-based test, or viral culture, and each of these tests has its own advantages and limitations (Table 1). Nucleic acid-based detection methods which have been reported in detecting these coronaviruses include polymerase chain reaction (PCR), loop-mediated isothermal amplification methods (LAMP), next-generation sequencing (NGS), CRISPR/Cas-based detection test, and some other tests [16], [17], [18], [28]. Protein-based tests which could be used to detect MERS-CoV and SARS-CoV-2 infections comprise viral serology and antigen tests (Fig. 1) [16], [17], [18], [28].
3.2.1. Nucleic acid-based detection of MERS-CoV and SARS-CoV-2
Molecular diagnosis is important in the identification, prevention, and treatment of infectious diseases [38]. Molecular diagnosis is a rapidly growing discipline in laboratory medicine, with new methods and applications continually becoming available and improvements being made [38], [39]. Viral nucleic acids can be detected using molecular approaches such as polymerase chain reaction (PCR), reverse transcription loop-mediated isothermal amplification (RT-LAMP), and next-generation sequencing (NGS), and these methods are making their way into clinical laboratories [16], [18], [40]. Molecular tests enable the rapid detection of unculturable or fastidious microorganisms from clinical samples without the need for culture [41]. In addition, sequence analysis of amplified DNA from the infectious agents allows for identification and better characterization of the virus [42]. Besides, sequence analysis allows scientists to correlate the relationships of different pathogens based on their phylogenetic relationships [42]. The variation in the viral subspecies may affect infectivity, thus influence the disease prognosis [43]. Viral genome sequencing also enables the direct detection of genes or gene mutations in the virus which are responsible for contributing to its virulence and drug resistance [44]. On top of that, the molecular diagnostic technique is useful to quantify viral load [45]. With the advancement in molecular diagnostic technology, the presence of computerized, automated machines and handy software has allowed molecular diagnosis to be conducted more commonly with high precision [46]. In short, the detection of viral agents up to the nucleic acid level signifies a notable breakthrough in clinical microbiology.
3.2.1.1. Real-time reverse transcription PCR (qRT-PCR)
The nature and sequence of the first MERS-CoV isolate were uncovered using a random-amplification deep-sequencing approach [5]. Following that, in the year 2013, the first MERS-CoV isolate was patented, and many authorities were concerned such an act might restrict the progress on the development of viral diagnostics tests, viral vaccines, and anti-viral drugs [47]. Fortunately, the designated authorities granted virus isolate access to the related parties as long as the applicants follow the strict listed biosafety rules [47]. The molecular detection of MERS-CoV RNA started with the sequencing of conserved domains ORF 1a and 1b [48], [49]. Both ORFs help to identify the coronavirus species [48]. Thereafter, a sensitive molecular diagnostic approach using real-time reverse transcription PCR (qRT-PCR) was quickly defined, validated, and widely employed to diagnose MERS-CoV infection [27]. For SARS-CoV-2, the virus was first isolated from human airway epithelial cells from patients in Wuhan, China, at the end of the year 2019, and was subsequently subjected to genetic sequencing [50]. As SARS-CoV-2 was started to cause a global pandemic crisis in early 2020, many countries were then working hard to produce and validate different qRT-PCR diagnostic tests to detect the patients who carry the virus as part of the efforts to combat the transmission of SARS-CoV-2 [16].
Real-time PCR-based analyses combine the traditional PCR approaches and fluorescent-emitting compounds to measure the number of amplicons produced during the PCR amplification process in “real-time” [51]. By combining both real-time PCR and reverse transcription reaction, the gene expression level in the samples can be calculated [51]. With the introduction of qRT-PCR to detect both MERS-CoV and SARS-CoV, different health agencies and authorities like World Health Organization, US Food and Drug Administration (FDA), and US Centers for Disease Control and Prevention has sanctioned and revised the confirmed MERS and SARS-CoV-2 case definition in which individual who is tested positive for MERS-CoV or SARS-CoV-2 using qRT-PCR or serology test will be regarded as confirmed case, regardless of appearant clinical signs and symptoms ( Fig. 2) [16], [29], [30], [34], [52], [53], [54]. Subsequently, several MERS-CoV and SARS-CoV-2 diagnostic kits which consist of the assay signatures like ORF 1a, upE, and, S, N/ RdRp, and an appropriate positive control were developed [16], [18], [55], [56]. CDC and WHO suggest that a patient is potentially negative for active MERS-CoV infection following one negative qRT-PCR test on the tested viral genetic target as described (Fig. 2), but further testing is recommended to confirm the absence of MERS-CoV infection [29], [54]. Whereas, a patient diagnosed with MERS is required to have at least two consecutive negative qRT-PCR tests on all specimens to be considered as cleared for MERS-CoV infection [54]. For SARS-CoV-2, it is also recommended that patients who are symptomatic or had a history of travel to outbreak areas or had a history of close contact with the confirmed cases be subjected to a second qRT-PCR test if their first test result is negative [30], [57]. However, to discharge a patient who is confirmed to have SARS-CoV-2 infection, WHO has released updated guidelines which state that symptomatic patients could be discharged 10 days after the onset of symptoms while asymptomatic patients could be discharged 10 days after the molecular diagnosis of SARS-CoV-2 [58]. For both MERS-CoV and SARS-CoV-2, both CDC and WHO recommend that multiple specimens should be collected for molecular testings from various body sites like upper (nasopharyngeal and oropharyngeal swabs) and lower (bronchoalveolar lavage, sputum, and trachea) respiratory tracts, blood, and lower gastrointestinal tract (stool specimen) [29], [54], [57]. Nonetheless, numerous factors which might affect the rate of success and accuracy of qRT-PCR testing of coronavirus have been identified and these factors include type and quality of specimens, the expertise of laboratory staff, and laboratory environment (contamination) [26].
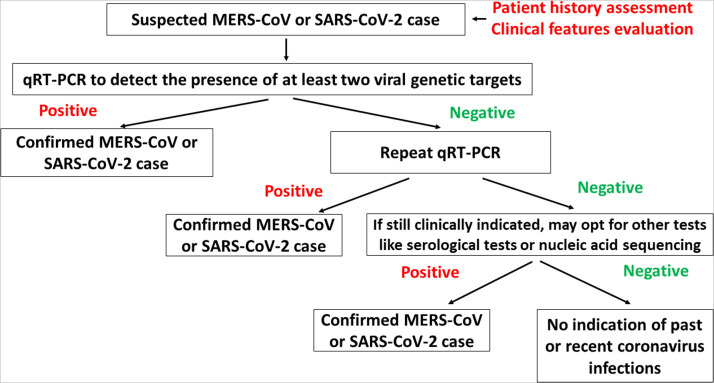
Fig. 2.
Diagnostic approaches to confirm the presence of MERS-CoV and SARS-CoV-2. For clinically suspicious cases, qRT-PCR remains as the gold standard molecular diagnostic test to confirm MERS-CoV and SARS-CoV infections [6], [29], [30], [33], [34], [57], [126]. For individuals who have negative qRT-PCR test results, a repeat test is highly recommended [29], [34], [54], [57]. In the case in which qRT-PCR results are inconclusive, further tests like nucleic acid sequencing and serological tests could be performed to confirm the coronavirus infection [29], [34], [54], [57]. For MERS-CoV, the guidelines state that the patients need to have at least two consecutive negative qRT-PCR results before they are allowed to be discharged from the isolation services [29], [54]. However, for SARS-CoV-2, the latest guideline recommends that the symptomatic patient is allowed to be discharged 10 days after the onset of symptoms while asymptomatic patients could be discharged 10 days after the molecular diagnosis of the virus. [58].
It has been recommended by WHO that qRT-PCR targeting both the upE and ORF 1a regions are highly sensitive to be used for the detection of MERS-CoV [59], [60]. A study had previously reported the use of a qRT-PCR test that was aimed to target the upE and ORF 1a regions of MERS-CoV and the test was able to detect 5 RNA copies per reaction with 100% specificity ( Table 2) [59]. A commercial test kit that could detect both UpE and ORF 1a regions of MERS-CoV was reported to have a sensitivity of 95% and specificity of 100% [61]. This suggested that targeting both UpE and ORF 1a could be useful to confirm the presence of MERS-CoV. Two other studies had reported that targeting UpE could be useful to specifically detect MERS-CoV infection in which the qPCR test sensitivity could be above 95% [62] and the limit of detection of such test could be 5–10 RNA copies per reaction [52]. On the other hand, another two studies reported the use of other targets like ORF 1b to confirm the presence of MERS-CoV and such qPCR tests could detect 4–64 RNA copies per reaction with a specificity of 100% [55], [60]. Compared to the previously mentioned genetic targets of MERS-CoV, the use of other targets like the N region might not be as sensitive as upE and ORF regions in confirming the presence of MERS-CoV. It was reported that the use of qRT-PCR to detect the N gene of MERS-CoV had a sensitivity that ranged from 55–100% and the specificity could vary from 33% to 100% [63], [64]. These findings suggested that the use of N gene detection to confirm MERS-CoV infection should be accompanied by tests that target other genetic regions like upE and ORF regions to avoid false-negative results. A group of Chinese researchers had previously reported the use of leader sequence at the 5’-untranslated regions (5’-UTR) of the MERS-CoV to confirm the presence of the virus and it was demonstrated that such qRT-PCR test could detect 5 RNA copies per reaction or 5.62 × 10–2 TCID50/mL with 100% specificity [65]. The findings from this study highlighted that targeting the 5’-UTR regions of the virus could also be useful to detect MERS-CoV [65].
Table 2.
Comparison of the sensitivity and specificity of qRT-PCR or qPCR in detecting different molecular targets of MERS-CoV and SARS-CoV-2.
| Virus type | Molecular targets | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | N | 54.5% | 33% | [64] |
| N, E | 60–100% | 100% | [63] | |
| N & upE | LOD: 5–10 RNA copies/reactions | 100% | [52] | |
| upE, ORF 1a | LOD: 0.5–0.9 RNA copies/μL (~95%) | 100% | [61] | |
| upE & ORF 1a | LOD: ~5 RNA copies/reaction for singleplex and multiplex qRT-PCR | 100% | [59] | |
| upE & ORF 1b | upE: LOD of 3.4 RNA copies/reaction | 100% | [60] | |
| ORF 1b: LOD of 64 RNA copies/reaction | ||||
| UpE, S | >98% | Unclear | [62] | |
| ORF 1b | ORF 1b: LOD of 4.1 RNA copies/reaction | 100% | [55] | |
| 5’-UTR | LOD: 5 RNA copies/reaction or 5.62 × 10–2 TCID50/mL | 100% | [65] | |
| SARS-CoV-2 | N | LOD: 0.0187 ng RNA compared to LOD of dPCR of 0.00187 ng. | Unclear | [66] |
| N | LOD: 5 RNA copies/reaction (>99%) | 100% | [318] | |
| N | LOD: <5 RNA copies/reaction (100%) | 100% | [71] | |
| N, ORF 1ab | LOD: 500–1000 RNA copies/mL (100%) | 95.3% | [73] | |
| N, ORF 1ab | LOD: 1–10 RNA copies/reaction | 100% | [75] | |
| N, ORF 1ab | ORF 1ab: LOD of 520.1 RNA copies/mL | 100% | [74] | |
| N: LOD of 528.1 RNA copies/mL | ||||
| Overall sensitivity to detect positive Covid-19 samples: 58.82% | ||||
| N, E, RdRp | ≥79% | 100% | [76] | |
| N, E, RdRp | LOD: 80–154 RNA copies/mL (90–100%) | 100% | [72] | |
| N, E, RdRp | LOD: ≥ 10 RNA copies/reaction (For N & E); LOD for RdRp could be 20-folds less sensitive | Unclear | [70] | |
| N, E. NSP, RdRp | LOD: 100 RNA copies/μL (~100%) | 100% | [88] | |
| N, S, RdRp/Hel, RdRp-P2 | N, S, RdRp/Hel: 1.8 × 100 TCID50/mL | 100% | [77] | |
| RdRp-P2: 1.8 × 101 TCID50 /mL | ||||
| N, E, ORF 1ab, RdRp | E: Detectable at 1:80 dilution | 100% | [93] | |
| N: Detectable at 1:160 dilution | ||||
| ORF 1ab: Detectable at 1:40 dilution | ||||
| RdRp: Detectable at 1:10 dilution | ||||
| N, E, S, ORF 1ab, RdRp | 60–97.7% (depending on targets) | Unclear (However, false negative ranged from 2% to 40%) | [319] | |
| N, E, S, ORF 1ab, RdRp | E: LOD95 of 0.91–4.8 RNA copy/mL | 100% | [67] | |
| N: LOD95 of 4.8 RNA copy/mL | ||||
| S: LOD95 of 3.8–4.3 RNA copy/mL | ||||
| ORF 1ab/RdRp: LOD95 of 3.1–23 RNA copy/mL | ||||
| N, E, S, ORF 1ab, RdRp | LOD: 0.5 RNA copies/μL to 12,500 copies/mL (94.1–100%) | 90–100% | [68], [78], [79] |
Compared to MERS-CoV-2, there are more studies which had described the use of qRT-PCR and qPCR in confirming the diagnosis of SARS-CoV-2 (Table 2) [66], [67], [68], [69], [70], [71]. Some studies had described the use of a single target like N gene to detect the presence of SARS-CoV-2 and such qRT-PCR test could detect around 5 RNA copies per reaction with a sensitivity of above 99% and specificity of 100% [71], [72]. Another two studies reported that when qPCR was used to detect N and ORF 1ab regions, the limit of detection could range from 500 to 1000 RNA copies per mL, and the test overall sensitivity was ranged from 58% to 100% with test specificity of 100% [73], [74]. However, a group of Chinese scientists had reported that the limit of detection of qRT-PCR that targets both N and ORF 1ab could be as low as 1–10 RNA copies per reaction [75] and this implied that the test sensitivities could vary between different research groups and experimental conditions [73], [74], [75]. On contrary, three research groups had employed three SARS-CoV-2 targets that include N, E, and RdRp in their qPCR testings, and the test sensitivities were varied between different published reports but the test specificities were quite consistent (100%) between the three published findings [70], [72], [76]. Compared to N and E genes, the detection of RdRp could be at least 20-folds less sensitive and this suggested that both N and E genes should be selected to be used in the qPCR test to confirm SARS-CoV-2 infection, followed by RdRp [70]. In terms of COVID-19 RdRp-qRT-PCR assay to detect SARS-CoV-2, a study has shown that when two different RdRp-based assays, namely RdRp/Hel and RdRp-P2 assays were used to detect SARS-CoV-2, the use of RdRp/Hel based assay was more sensitive than the other one in detecting the presence of SARS-CoV-2 [77]. In addition, the RdRp/Hel based assay was also found to have comparable sensitivity and specificity with N and S targets-based qPCR assay [77], suggesting that this RdRp/Hel based assay could be potentially used as an alternative test to detect SARS-CoV-2.
Apart from being widely reported in numerous original research articles, the use of qRT-PCR in detecting SARS-CoV-2 has also been expanded to commercial use, and to date, at least 20 different qRT-PCR commercial kits had been approved by the FDA, and examples of these kits include RealStar SARS-CoV-2 RT-PCR kit, Solaris Multiplex SARS-CoV-2 assay, SalivaDirect, ViroKey™ SARS-CoV-2 RT-PCR test and EURORealTime SARS-CoV-2 kit [68], [78], [79]. All these different commercial qRT-PCR or qPCR test kits could detect different molecular targets like N, E, S, RdRp, and ORF 1ab and the limit of detection of these test kits could be varied from 0.5 RNA copies per μL to 12,500 RNA copies per mL [68], [78], [79]. The sensitivities and specificities of these test kits generally ranged from 94% to 100%, and 90–100%, respectively [68], [78], [79]. The superior sensitivities and specificities of these different SARS-CoV-2 qPCR commercial test kits have helped the local health authorities to use them in detecting the virus without the need to use conventional qRT-PCR tests in which the laboratory scientists might need to undergo more tedious and lengthy steps to get the tests done [16], [17], [68].
3.2.1.2. Digital PCR (dPCR)
Other than qPCR, another type of PCR known as digital PCR (dPCR) can also be used to detect both MERS-CoV and SARS-CoV-2 ( Table 3) [66], [80], [81]. dPCR is a new nucleic acid amplification technology that is made commercially available since the year 2011 [82]. The main difference between dPCR and qPCR is that in dPCR, the PCR reaction is being segregated or partitioned into thousands of individual, independent reactions before the start of amplification and thus, this would give more precise data especially if the sample is present in minute quantity [82]. For MERS-CoV, a study has shown that the use of dPCR could detect 64–167 copies of MERS-CoV virus per reaction when primers or probes that targeted N, E, and ORF 1ab were used and the dPCR specificity was 100% [81]. Compared to the qRT-PCR test that targeted similar targets that had been reported to have lower LOD (<10 RNA copies per reaction) [52], [55], [59], [60], the sensitivity of dPCR in detecting MERS-CoV seems to be lower but the specificity of dPCR and qPCR are about the same.
Table 3.
Comparison of the sensitivity and specificity of dPCR in detecting different molecular targets of MERS-CoV and SARS-CoV-2.
| Virus type | Molecular targets | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | N, E, ORF 1ab | E: LOD of 167 RNA copies/reaction | 100% | [81] |
| N: LOD of 156 RNA copies/reaction | ||||
| ORF 1ab: LOD of 64 RNA copies/reaction | ||||
| SARS-CoV-2 | N | LOD and sensitivities of both qRT-PCR and dPCR are highly correlated. Mean detectable copies/samples using both methods are unclear | Unclear | [83] |
| N | LOD: 2.5 RNA copies/reaction as compared to 10 RNA copies/reaction of qRT-PCR (≥86%) | Unclear | [84] | |
| N | LOD: 0.00187 ng RNA compared to LOD of qRT-PCR of 0.0187 ng | 100% | [66] | |
| Could detect up to 0.08 virus copies/μL after 10-fold dilutions | ||||
| N | dPCR able to distinguish true positive and negative samples with low viral load (10-4 dilutions) while qRT-PCR was unable | 100% | [80] | |
| N | LOD: <2 RNA copies/μL for dPCR while qRT-PCR could not | Unclear | [86] | |
| RdRp | Median detection: 128 RNA copies/mL (~99%) | ~95% | [87] | |
| N, ORF 1ab | ORF 1ab: LOD of 401.8 RNA copies/mL | 100% | [74] | |
| N: LOD of 336.8 RNA copies/mL | ||||
| Overall sensitivity to detect positive Covid-19 samples: 67.65% |
Like qPCR, there were more reported studies (Table 3) that had described the use of dPCR in detecting SARS-CoV-2 than MERS-CoV [66], [83], [84], [85], [86]. When genetic targets like N, E, RdRp, and ORF 1ab were used in the SARS-CoV-2 dPCR test, the sensitivity of such tests was found to be ranging from 60% to 99% with the limit of detection that varied from 10 to 400 RNA copies per mL [74], [85], [87]. When compared to qRT-PCR tests that targeted similar targets that could achieve higher test sensitivity, for example, the sensitivity of almost 100% [71], [73], [88], the sensitivity of dPCR reported in these studies seems to be lower [74], [85], [87]. However, on the other hand, when another four studies were conducted to compare the efficiency, sensitivity, and specificity of qRT-PCR and dPCR in detecting the N gene of SARS-CoV-2, the findings showed that generally, dPCR has higher sensitivity and specificity than qRT-PCR (Table 2) [66], [80], [84], [86]. In another word, when the SARS-CoV-2 viral load is too low to be detected by qRT-PCR, dPCR could be utilized to detect the virus [66], [80], [84], [86]. Another Italian study showed that the LOD and sensitivities of qRT-PCR and dPCR in detecting N gene of SARS-CoV-2 are highly correlated and comparable and this further supported that dPCR could be employed as an alternative COVID-19 diagnostic test other than qRT-PCR [83].
Apart from being utilized widely to diagnose MERS-CoV and SARS-CoV-2 infections, PCR like qRT-PCR and dPCR are also being used to monitor viral load and disease progression [66], [80], [89], [90], [91]. Several studies have found a direct association between high MERS-CoV loads in clinical samples and worse clinical outcomes [89], [90], where the high abundance of virions could lead to increased lung damage via direct destruction of respiratory cells or initiate an exaggerated inflammatory response [92]. For SARS-CoV-2, it was proposed that high viremia is associated with a hyperinflammation state which will then lead to endothelial damage, perivascular inflammation, and systemic micro-and macrovascular complications [91]. To sum up, the introduction of different PCR methods like qRT-PCR and dPCR has helped in confirming coronavirus infection and also aided in the clinical management of the disease development.
3.2.1.3. Reverse transcription loop-mediated isothermal amplification (RT-LAMP)
To date, qRT-PCR remains one of the most sensitive and specific tests to detect both MERS-CoV and SARS-CoV-2 [59], [93]. However, researchers and laboratories are still looking for a simpler method that can amplify targeted viral genes with high sensitivity, specificity, and rapidity [94]. Because of this, several isothermal amplification methods have been developed to detect both MERS-CoV and SARS-CoV, which include reverse transcription loop-mediated isothermal amplification (RT-LAMP) [94], [95] and reverse transcription recombinase polymerase amplification (RT-RPA) assays [48], [96], [97].
RT-LAMP is a rapid and simple nucleic acid amplification assay that relies on Bst DNA polymerase large fragment for target amplification and could generate approximately 109 DNA copies in less than an hour [98]. Compared to RT-PCR, RT-LAMP reaction occurs at lower temperature (60–65 °C) [48], [98]. This means that it does not require sophisticated and expensive equipment for precise temperature control. Besides, some of the RT-LAMP assays can be completed within one hour [99] as compared to qRT-PCR which could usually take up to three to four hours to complete [100]. To detect the N gene of MERS-CoV, RT-LAMP was found to have LOD of 0.4 RNA copies per reaction in a Korean study [99] and LOD of 10–20 RNA copies per μL in a Chinese study [101], and the test specificities of both studies were 100% [99], [101]. When RT-LAMP was used to detect the presence of other MERS-CoV targets like ORF 1a and ORF 1b, the test sensitivity was found to varied from 2 to 120 RNA copies per reaction in which the test specificity was also 100% ( Table 4) [102], [103]. In addition, the test sensitivity of RT-LAMP was proven to be equivalent to qRT-PCR [103]. A similar study which was aimed to detect upE and ORF 1a of MERS-CoV was also found that both RT-LAMP and qRT-PCR have comparable and equivalent sensitivities, further suggesting that RT-LAMP could be used as an alternative diagnostic tool other than qRT-PCR [94]. The only study which has shown that RT-LAMP was less sensitive than qRT-PCR in detecting MERS-CoV was an American study that reported that RT-LAMP could be 2 to 200-folds less sensitive than qRT-PCR in detecting ORF 1ab of MERS-CoV to confirm the presence of the virus [48].
Table 4.
Comparison of the sensitivity and specificity of RT-LAMP in detecting different molecular targets of MERS-CoV and SARS-CoV-2.
| Virus type | Molecular targets | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | N | LOD: 1–2 × 101 RNA copies/μL of samples | 100% | [101] |
| N | LOD: 0.4 RNA copies/reaction | 100% | [99] | |
| ORF 1ab | 0.02–0.2 plaque forming units (PFU) | 100% | [48] | |
| 2–200-folds less sensitive than qRT-PCR | ||||
| N, ORF 1ab | LOD: 120 RNA copies/reaction | 100% | [102] | |
| N, ORF 1a | LOD: 2 RNA copies/reaction (same as qRT-PCR) | 100% | [103] | |
| upE & ORF 1a | upE: LOD of 1.6 RNA copies/reaction | ~100% | [94] | |
| ORF 1a: LOD of 3.4 RNA copies/reaction | ||||
| *Equivalent sensitivity to qRT-PCR | ||||
| SARS-CoV-2 | N | LOD: 100 RNA copies/reaction (10-folds lesser than qRT-PCR) | 100% | [106] |
| N | LOD: ~ 100 RNA copies/μL sample 86% (poorer than qRT-PCR) | 99.5% (poorer than qRT-PCR) | [95] | |
| N | LOD: 900 RNA copies/mL (+ve agreement: 94, -ve agreement: 98%) | 100% | [112] | |
| N | LOD: 20,000 RNA copies/mL (95%) | 100% | [114] | |
| ORF 1ab | LOD: 125 genomic equivalents/swab (96.6%) | 100% | [320] | |
| N, E | LOD: 25–75 RNA copies/mL (+ve agreement: 98%, –ve agreement: 100%) | Cross-react with SARS-CoV | [111] | |
| N, ORF 1ab | LOD: 2000 RNA copies/swab (100%) | 99% | [321] | |
| N, ORF 1ab | LOD: 1 RNA copy/μL | Unclear | [113] | |
| N, ORF 8 | LOD: 100 RNA copies/μL (at least 2-folds lower than qRT-PCR | 100% | [108] | |
| N, E, ORF 1ab | LOD: 0.75 RNA copies/μL (100%) | 100% | [322] | |
| N, S, ORF 1ab | LOD: 80 RNA copies/mL (Comparable sensitivities with qRT-PCR) | 100% | [104] | |
| N, E, RdRp, NSP | LOD: ~3 RNA copies/25 μL (44.8–82.8%) | <100% (few cross-reactivity with other pathogens) | [107] | |
| N, S, ORF 1a, ORF 8 | LOD: 0.75 RNA copies/μL (100%) | 100% | [109] | |
| N, S, NSP, ORF8 | LOD: ~ 100 RNA copies/reaction | 100% | [105] | |
| Unclear | LOD: 500 RNA copies/reaction | Unclear | [115] | |
| Unclear | LOD: 6.7 RNA copies/reaction (at Day 9) (92.8% compared to qRT-PCR) | 100% | [110] |
As RT-LAMP has been proven to be a sensitive diagnostic test to detect MERS-CoV, therefore, this method was also being tested and used to confirm SARS-CoV-2 infection when this virus started to cause a global public health crisis since the end of 2019 [95], [104], [105]. To date, several genetic targets of SARS-CoV-2 have been reported to be able to be detected by RT-LAMP (Table 4) and one of them was the N gene [95], [106]. RT-LAMP was able to detect 100 RNA copies per reaction [106] or 100 RNA copies in 1 μL of the tested sample [95]. Compared to qRT-PCR, RT-LAMP was found to have lesser sensitivity to detect N gene of SARS-CoV-2 even though both qRT-PCR and RT-LAMP could have comparable specificity (~100%) in detecting SARS-CoV-2 [95], [106]. The lower sensitivity of RT-LAMP in detecting SARS-CoV-2 was further shown in a study in which the authors reported that RT-LAMP has a sensitivity that ranged from 45% to 83% and specificity of less than 100% because of the possibility of the test to cross-react with other pathogens [107]. Besides, another study also reported that the sensitivity of RT-LAMP was at least 2-folds less than qRT-PCR in detecting genetic targets of SARS-CoV-2 like N and ORF 8 genes [108]. However, in a Chinese study [104], it was demonstrated that RT-LAMP could detect 80 RNA copies per mL when targets like N, S, and ORF 1ab regions were used in the assay and the test sensitivity was comparable to that of qRT-PCR, and the RT-LAMP assay specificity was 100%. This finding was against the four mentioned studies that had proposed that RT-LAMP was less sensitive than qRT-PCR in detecting SARS-CoV [95], [104], [106], [107]. An American study also reported that RT-LAMP could produce 100% sensitivity and specificity when genetic targets of SARS-CoV-2 like N, S, ORF 1a, and ORF 8 were targeted in the assay and the LOD of the assay could be as low as 0.75 RNA copies per μL [109]. Therefore, it could be summed up that compared to qRT-PCR, RT-LAMP could sometimes produce comparable sensitivity and specificity in detecting SARS-CoV-2 and so, it could serve as a potential diagnostic test for this viral infection when qRT-PCR results are inconclusive [95]. In terms of molecular targets, it seems like the sensitivity of RT-LAMP in detecting different molecular targets of SARS-CoV-2 could vary from one published report to the other [95], [104], [105], [107], [110], and thus, no target is said to be more reliable to be chosen as a molecular target for RT-LAMP.
As multiple studies have reported that RT-LAMP could have promising sensitivity and specificity in detecting SARS-CoV-2 [104], [105], [109], therefore, several developed RT-LAMP test kits were also being introduced to help diagnose COVID-19 across the world. Examples of such kits include MobileDetect BioBCC19 test kit, Lucira COVID-19 All-In-One test kit, AQ-TOP COVID-19 rapid detection kit plus, and SARS-CoV-2 RNA DETECTR assay [68], [111], [112], [113], [114]. All these RT-LAMP-based test kits were invented to detect various targets of SARS-CoV-2 like N, E, and ORF 1ab, and the sensitivity and specificity of these kits were all above 90% [68], [111], [112], [113], [114], [115]. This means that most of these test kits are specific enough to detect SARS-CoV-2 without cross-reacting with other pathogens from the respiratory tracts or other coronavirus species [68], [111], [112], [113], [114], [115].
3.2.1.4. Reverse transcription recombinase polymerase amplification (RT-RPA) and other isothermal amplification-based test
Another type of isothermal amplification-based assay which could be used to detect both MERS-CoV and SARS-CoV-2 is RT-RPA ( Table 5) [96], [97], [116]. Compared to qRT-PCR, RT-RPA assay could run at a lower temperature like 42 °C for 15 min (shorter test duration) and it can amplify the specific genetic targets after the enzyme recombinase and the oligonucleotide primers bind to the specific genetic regions [96], [116]. When RT-RPA was used to detect the N gene of MERS-CoV, it was shown that RT-RPA could detect as low as 1.2 RNA copies in 1 μL of a sample with 100% sensitivity [117] or 10 RNA molecules per reaction with comparable sensitivity to qRT-PCR [96]. Besides, RT-RPA was demonstrated to have 100% specificity in detecting MER-CoV [96], [117]. These findings suggested that RT-RPA could serve as a sensitive diagnostic tool to detect MERS-CoV by targeting the N gene [96], [117]. Compared to the N gene, RT-RPA was found to be slightly less sensitive to detect other molecular targets of MERS-CoV like UpE and ORF 1a [96], [117], [118]. The sensitivity of RT-RPA to detect UpE was ranged from 86% to 98.06% while the sensitivity of RT-RPA to detect ORF 1a was 99% [117], [118].
Table 5.
Comparison of the sensitivity and specificity of RT-RPA in detecting different molecular targets of MERS-CoV and SARS-CoV-2.
| Virus type | Molecular targets | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | N | LOD: 10 RNA molecules (as sensitive as qRT-PCR) | 100% | [96] |
| N, UpE | UpE: LOD of 12 RNA copies/μL (86%) | UpE: 100% | [117] | |
| N: 100% | ||||
| N: LOD of 1.2 RNA copies/μL (100%) | ||||
| UpE, ORF 1a | LOD: 3.7-1 PFU of MERS-CoV | 100% | [118] | |
| UpE: 98.06% (compared with qRT-PCR) | ||||
| ORF 1a: 99.03% (compared with qRT-PCR) | ||||
| SARS-CoV-2 | N | LOD: 7.8 RNA molecules/reaction (100% concordance to qRT-PCR) | 100% | [120] |
| N | LOD: 5 RNA copy/μL sample (98%) | 100% | [119] | |
| N, S | LOD: 0.05 RNA copy/μL sample | Unclear | [97] | |
| ORF 1ab | 96.8% compared to qRT-PCR | Unclear | [121] | |
| N, RdRp | 2.5 RNA copies/μL input | 100% | [122] | |
| For qRT-PCR: LOD was 1 RNA copies/μL input | ||||
| For RT-LAMP: LOD was 10 RNA copies/μL input | ||||
| N, E, ORF 1ab | LOD: 1 RNA copy/μL sample (97%) | 100% | [323] | |
| N, E, RdRp | LOD: 2–15 RNA molecules/reaction (65–94%) | 77–100% | [124] |
For SARS-CoV-2, RT-RPA has also been widely reported to detect genetic targets of the virus such as the N gene (Table 5) and the LOD of the assay could be 5 RNA copies in 1 μL of a sample with a sensitivity of 98% [119] or 7.8 RNA molecules per reaction with 100 sensitivity as compared to qRT-PCR [120]. Besides, the assay specificity could be as high as 100% [119], [120]. On the other hand, when other molecular targets like RdRp and ORF 1ab were used in RT-RPA to detect the presence of SARS-CoV-2, the assay sensitivity was lower [121], [122] and the LOD of the assay could be 10-folds less sensitive than qRT-PCR [122]. These findings implied that compared to other molecular targets, the N gene seems to be the target that is easier to be detected using the RT-RPA method [120], [123]. When compared to qRT-PCR, RT-RPA seems to have lower sensitivity in which its sensitivity could vary from 65% to 94% while its specificity could range from 77% to 100% [124]. Therefore, it is said that RT-RPA could still be less sensitive and specific than qRT-PCR in confirming SARS-CoV-2 infection [124].
On other hand, a newly developed reverse transcription-insulated isothermal PCR (RT-iiPCR) method targeting the ORF1a and upE genes of MERS-CoV was also developed to assist in the MERS case detection [118]. RT-iiPCR is a fluorescent probe-based nucleic acid detection technique that can be performed in a capillary tube at a single temperature (95 °C) [118]. It was demonstrated that RT-iiPCR assays could detect 3.7-1 PFU of MERS-CoV in infected cell culture supernatant and sputum samples, indicating the assay is highly sensitive [118]. The viral nucleic acids of human coronavirus (HCoV)-229E, HCoV-OC43, feline infectious peritonitis virus (FIPV), influenza type A and B virus strains showed no cross-reaction towards the assay and this suggested that RT-iiPCR assay is a highly specific assay that is less likely to produce false-positive test results [118]. RT-iiPCR has also been developed and validated to detect SARS-CoV-2, which has been proven to have comparable sensitivity and specificity to conventional qRT-PCR in confirming SARS-CoV-2 infection [121]. In addition, RT-iiPCR test results are available in less than 1.5 h, while qRT-PCR results might take 3–4 h to be available as it requires additional nucleic acid extraction step [121]. Therefore, RT-iiPCR could be a potential alternative test to confirm the presence of SARS-CoV-2 and MERS-CoV [121].
3.2.1.5. Nucleic acid sequencing
With the advancement in molecular diagnostic technology, nucleic acid sequencing has become one of the options used in diagnosing MERS-CoV and SARS-CoV-2 infections [26], [50], [94], [125]. Other than qRT-PCR, both WHO and CDC have recommended nucleic acid sequencing as an alternative option used in confirming the presence of MERS-CoV and SARS-CoV-2 (Fig. 2) [29], [34], [54], [57], [126]. As such, it is recommended that nucleic acid sequencing can be combined with nucleic acid amplification tests like qRT-PCR in confirming the diagnosis of MERS-CoV and SARS-CoV-2 [29], [30], [57], [126]. For example, to confirm the presence of MERS-CoV, at least two viral genetic targets need to be confirmed using qRT-PCR, but the diagnosis of MERS-CoV infection can be confirmed with only a single positive target detected using qRT-PCR while the second viral genetic target is confirmed using viral nucleic acid sequencing test [29]. For SARS-CoV-2 infection, a nucleic acid sequencing test can also be used to confirm the infection if the nucleic acid amplification test result is questionable or invalid [57].
However, compared to other molecular diagnostic tests like qRT-PCR, nucleic acid sequencing is more costly and time-consuming, and it requires trained personnel to perform and analyze the test findings [17]. In addition, nucleic acid sequencing like whole genome sequencing involves the complete sequencing of the viral genome, where the process is rather complicated and is less suitable for large-scale detection of the virus in a population [17]. Nevertheless, the use of this technique is essential for scientists to identify specific viral gene sequences in order to design suitable primers and probes for subsequent nucleic acid detection tests like qRT-PCR [127].
Besides, nucleic acid sequencing could be used to study the genetic sequences of both MERS-CoV and SARS-CoV-2, which is particularly important in differentiating coronaviruses based on their genetic sequences [5], [18], [23], [128]. On top of that, nucleic acid sequencing is vital to investigate the evolving sequence mutations of the virus which may affect the virus infectivity [17], [127], [129], [130]. A previously reported study has highlighted the unique amino acid substitutions in the ORF 1ab, N, and S proteins would increase MERS-CoV virulence [129]. The study employed whole-genome sequencing to study the viral genetic sequences of eight clinical sample isolates [129]. Similarly, nucleic acid sequencing was also being used to study the specific genetic mutations in SARS-CoV-2, such as D614G mutation, in which this unique nucleotide base substitution increases the transmission rate of the virus [131]. A genome-wide study that investigated over 2400 complete or near-complete genomes of SARS-CoV-2 reported in the GISAID database has found that specific mutations like AA mutations occurred at a higher frequency in the SARS-CoV-2 genomes reported in Europe, followed by Asia then North America [128]. This implied that SARS-CoV-2 that has been circulating worldwide is having different genetic mutations and is highly heterogeneous [128]. Therefore, nucleic acid sequencing could also be used to study the epidemiological distribution and the dynamic of the coronavirus infection [128], [132].
In terms of the sensitivity and specificity of nucleic acid sequencing in detecting MERS-CoV, it was reported that the sensitivity of the test could range from 92% to 100%, depending on the occurrence of events like single nucleotide polymorphism (SNP) or nucleotide/sequence mismatch in which these events would reduce the sequencing sensitivity [133]. The specificity of the test in confirming MERS-CoV infection, however, was not further described in the mentioned study [133]. For SARS-CoV-2, a number of protocols and kits were introduced to detect or sequence SARS-CoV-2 ( Table 6) [68]. An assay was introduced to sequence the N gene of SARS-CoV and the LOD of the assay was found to be 3000 RNA copies in 1 mL and the test specificity was 100% [134]. For the S gene, two different test flows were established to sequence this genetic region and the targeted genome sequencing could detect 125–250 RNA copies in a reaction with 100% sensitivity and specificity [135], [136]. Besides, some assays were introduced to sequence multiple genetic targets of SARS-CoV-2 and in an assay that could detect up to 21 targets of SARS-CoV-2, the LOD of the assay was found to be 2 RNA copies per μL and the assay specificity could range from 90% to 100% [137]. In another protocol that could detect up to multiple targets of SARS-CoV-2, the LOD of the assay was found to be 7–10 RNA copies in 1 mL and the test sensitivity was about 98% [138]. Other than assays that were designed to sequence targeted regions of SARS-CoV-2, some protocols were established to perform whole viral sequencing and the LOD of the assays could range from 3 to 20 RNA copies per assay with a test sensitivity of 90–100% [139], [140], [141]. Compared to single target sequencing which was found to have higher sensitivity and specificity (~100%) [135], [136], the use of nucleic acid sequencing technology to sequence multiple targets or whole viral genome was reported to have slightly lower sensitivity and specificity (90–100%) [137], [138], [141]. Some studies have reported that during the genome sequencing process, reads error might occur in one out of a certain number of bases [142], [143]. So, the sequencing of multiple targets or sequencing that covers longer genome regions might prone to have more errors that could reduce the test sensitivity and specificity.
Table 6.
Comparison of the sensitivity and specificity of nucleic acid sequencing in detecting different molecular targets of MERS-CoV and SARS-CoV-2.
| Virus type | Molecular targets | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | N, UpE, ORF 1ab | 92–100% (depending on SNP) | Unclear | [133] |
| SARS-CoV-2 | N | LOD: ~3000 RNA copies/mL | 100% | [134] |
| S | LOD: 125 RNA copies/mL (100%) | 100% | [135] | |
| S | LOD: 250 RNA copies (100%) | Unclear | [136] | |
| Whole virus sequence | LOD: 20 RNA copies/sample | Unclear | [139] | |
| Whole virus sequence | LOD: 3–5 RNA copies/assay | Unclear | [140] | |
| Whole virus sequence | LOD: 10 RNA copies/assay (>90%) | Unclear | [141] | |
| N, E, ORF 1a | LOD: 7–10 RNA copies/μL (98%) | 100% | [138] | |
| ~21 targets including N, S | LOD: 2 RNA copies/μL (100%) | 90–100% | [137] | |
| Various targets (up to 98 targets) | LOD: 1000 RNA copies/mL | Unclear | [324] |
3.2.1.6. CRISPR/Cas-based detection approach
CRISPR/Cas-based technology has recently emerged as one of the potential diagnostic tests to confirm the presence of coronaviruses like MERS-CoV and SARS-CoV-2 [144], [145]. The CRISPR/Cas-based assay aids in detecting a specific coronavirus by recognizing its specific genetic sequence and this is followed by the cutting and release of the reporter molecule into the reaction mixture to allow the presence of the virus to be identified [16]. This assay is said to be highly specific and sensitive to detect coronaviruses, and the virus confirmation could be done in as short as 10 min [16], [144]. Like the use dPCR to detect MERS-CoV, the use of CRISPR/Cas-based technology in detecting the presence of MERS-CoV is not widely reported, even though some study has incorporated such test to check for the presence of MERS-CoV when the test was originally designed to detect SARS-CoV-2 [144], [145]. The use of CRISPR/Cas-based diagnostic techniques was reported to be specific enough to differentiate SARS-CoV-2 from other respiratory pathogens like MERS-CoV [145], [146], [147], [148], [149].
To date, multiple studies have reported the use of CRISPR/Cas-based technology to detect SARS-CoV-2 ( Table 7) [16], [68]. Two studies were previously conducted to use the CRISPR/Cas-based method to detect the N gene of SARS-CoV-2 and it was found that the test could detect up to few RNA copies in a sample with test specificity of 100% [146], [150]. When compared the assay finding to that of qRT-PCR, it was shown that the CRISPR/Cas-based detection method results were 95% concordance to the qRT-PCR results [150]. In another study when the CRISPR/Cas-based method was used to detect the S gene, the LOD of the assay was found to be 10 RNA copies per reaction with 100% specificity and the test results were 96.23% consistent with RT-RPA results [151]. When both N and E regions of the virus were targeted in the CRISPR/Cas-based detection test [152], it was found that the assay sensitivity and specificity were 93.1% and 98.5%, respectively, when compared to the qRT-PCR test results. Combining the findings from the discussed studies, it was demonstrated that the CRISPR/Cas-based detection approach is slightly less sensitive than other methods like qRT-PCR and RT-RPA [146], [150], [151], [152]. However, a Chinese study reported that the use of a CRISPR/Cas12a-based method that was aimed to target the N and ORF 1ab could detect 1–10 RNA copies per reaction with test sensitivity and specificity of 100% as compared to qRT-PCR [149]. This proved that the CRISPR/Cas-based detection approach could also produce comparable test findings like other SARS-CoV-2 diagnostic tests [149].
Table 7.
Comparison of the sensitivity and specificity of CRISPR/Cas-based tests in detecting different molecular targets of MERS-CoV and SARS-CoV-2.
| Virus type | Molecular targets | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | N, E, S, M, ORF 1ab, RdRp | No data | 100% (no cross-reactivity with other pathogens like SARS-CoV-2) | [145], [146], [147], [148], [149] |
| SARS-CoV-2 | N | 95% concordance to qRT-PCR results | 100% | [150] |
| N | LOD: few RNA copies/sample | 100% | [146] | |
| S | LOD: 10 RNA copies/reaction(96.23% compared to RT-RPA) | 100% | [151] | |
| RdRp | LOD: 1 × 104 RNA copies/mL | 100% | [145] | |
| N, E | LOD: ~25 RNA copies/μL (93.1%) | 98.5% | [152] | |
| N, E | LOD: ~80 RNA copies/sample | <100% (minor cross-reactivity with other pathogens) | [155] | |
| N, ORF 1ab | LOD: 6.75 RNA copies/μL | 100% | [325] | |
| N, ORF 1ab | LOD: 1–10 RNA copies/reaction (100% consistent result with qRT-PCR) | 100% | [149] | |
| N, ORF 1ab | LOD: 7.5–25 RNA copies/μL | 100% | [326] | |
| E, ORF 1ab | LOD: 4 RNA copies/μL (100% -ve predictive agreement, 97.14% +ve predictive agreement) | 100% | [154] | |
| N, S, M | LOD: 0.1 RNA copies/μL | 100% | [153] | |
| Unclear | LOD: ~100 RNA copies/mL | 100% | [147] |
In terms of the test specificity, most of the test studies which had employed the CRISPR/Cas-based detection method reported a test specificity of 100%, regardless the test was targeting single or multiple genetic regions of SARS-CoV-2 [145], [146], [147], [149], [150], [151], [153], [154]. Only some studies had reported that the CRISPR/Cas-based detection assay might be causing some cross-reactivity with other respiratory pathogens and this could be affected by the choice of the molecular targets and cas enzyme [152], [155]. For example, a study has reported that the use of LwCas13 to detect ORF 1a, N, and S targets would have a sensitivity of 55% while the use of Cas3a and LbCas12a to detect the N gene would have a specificity of 95% [148]. Nevertheless, the CRISPR/Cas-based detection approach is still new compared to more established methods like qRT-PCR and it is believed that as time passes, more sensitive, reliable, and accurate CRISPR/Cas-based detection assay will be introduced. When compared between different molecular targets, numerous studies had shown that the CRISPR/Cas-based detection assay that was targeting N, S, E, and ORF 1ab could have lower LOD (<100 RNA copies per reaction or mL) [146], [149], [151], [154], [155]. Other targets like RdRp, when used in the CRISPR/Cas-based detection assay, however, were found to have a higher LOD which could be as high as 10,000 copies per mL [145]. Thus, in terms of molecular targets, genetic targets like N, E, S, and ORF 1ab should be selected for detection using CRISPR/Cas-based detection assay because these targets could be easier to be detected when the virus load is lower.
3.2.1.7. Other nucleic acid-related tests
In China, a study [156] was conducted to design and evaluate a multiplexed CoVs test coupled with mass spectrometry (MS) sequencing technique that was aimed to target the MERS-CoV N, upE, RdRp, and ORF 1b regions. Such a method was said to be able to detect and differentiate up to 6 known human coronaviruses including MERS-CoV and SARS with LOD of 10–100 RNA copies per reaction and assay specificity of 100% [156]. In this multiplexed coronavirus detection assay, the targeted sequences will be amplified using multiplex PCR, where dideoxynucleotide triphosphates (ddNTPs) are included in the reaction to produce amplification products of various lengths [156]. As the site-specific primers bind to the respective amplicons, it will extend the amplicons by a single base and MassARRAY matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS will be used to measure the masses of the extended primers [156].
For SARS-CoV-2, a PCR test that was coupled with MS was also widely used to detect this coronavirus ( Table 8) [157], [158], [159], [160]. In a Chinese study [159], a PCR test coupled with an MS test was used to detect the N and ORF 1ab genes of SARS-CoV and the detection rate of this assay was noted to be around 75%. On contrary, an American study reported that a similar assay would detect about 1563 RNA copies in mL when it was used to detect N and ORF 1ab genes of SARS-CoV-2 with test sensitivity that ranged from 90% to 100% as compared to qRT-PCR [160]. The drawback of this study was that the tendency of the assay to cross-react with other pathogens was unexplored and thus, the test specificity was unclear [160]. The test specificity of the PCR-MS-based test was also unspecified in another study [158], even though the study reported that such test could detect 400 RNA copies in 1 mL. To further compare the sensitivity and specificity of PCR-MS-based tests in detecting SARS-CoV-2 with other diagnostic methods, a study was conducted in Germany and it was shown that PCR-MS-based assay would produce test sensitivity and specificity that was comparable and concordance to qRT-PCR test findings [157]. This means the PCR-MS-based assay could have the potential to be further developed as an alternative diagnostic test for coronavirus detection [157]. Since PCR-MS test was shown to provide a promising outcome to detect SARS-CoV-2, a test assay that is dependent on the PCR-MS-based detection approach was approved for use under Emergency Use Authorization (EUA) and this test assay could be used to detect the N and ORF 1ab genes of SARS-CoV-2 [161]. This approved test could detect 0.34–110 RNA copies in 1 μL of sample and its specificity could vary from 90% to 100% because of its potential tendency to cross-react with other respiratory pathogens [161].
Table 8.
Comparison of the sensitivity and specificity of PCR-MS-based tests in detecting different molecular targets of MERS-CoV and SARS-CoV-2.
| Virus type | Molecular targets | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | N, UpE, RdRp, ORF 1b | LOD: 10–100 RNA copies/reaction | 100% | [156] |
| SARS-CoV-2 | N, ORF 1ab | Results 100% concordance to qRT-PCR | Specific and no false positive/negative results | [157] |
| N, ORF 1ab | LOD: 1562.5 RNA copies/mL (90–100%) | Unclear | [160] | |
| N, ORF 1ab | LOD: 400 RNA copies/mL | Unclear | [158] | |
| N, ORF 1ab | Detection rate: 75% | Unclear | [159] | |
| N, ORF 1ab | LOD: 0.34–110 RNA copies/μL | <100% (cross-reactive with other respiratory pathogens) | [161] |
On the other side, a paper-based colorimetric DNA assay which is depending on the detection of pyrrolidinyl peptide nucleic acid (acpcPNA)-induced nanoparticle aggregation was being reported as an alternative diagnostic test for MERS-CoV [162]. In the presence of targeted DNA sequences, a DNA-acpcPNA duplex will be formed and this would induce the dispersion of the AgNPs [162]. The nanoparticle dispersion would in turn induce color changes, which could then be observed on the specific paper for further analysis [162]. This paper-based DNA detection approach is a rapid test that could be employed as a Point of Care Testing (POCT) device to directly detect MERS-CoV from the sample itself in an automatic detection manner [162]. This type of assay has been reported to show no cross-reactivity with other CoVs, and its limit of detection for MERS-CoV was found to be 1.53 nM [162]. Thus, this assay could be potentially used as a low-cost diagnostic tool for the rapid screening of infectious diseases [162]. Similarly, the colorimetric assay based on the binding of the nanoparticles with viral nucleic acid was also being developed to detect SARS-CoV-2 [163]. In a study which was conducted in the USA, the scientists developed a type of gold nanoparticle capped with thiol-modified antisense oligonucleotides (ASO), of which the nanoparticle complex is specific to the N gene of the SARS-CoV [163]. When bound to the viral genetic targets, the nanoparticles complexes would form precipitation and the test results could be visualized using the naked eye in less than 10 min [163]. Besides, the assay can differentiate SARS-CoV-2 from other coronaviruses like MERS-CoV, where the detection limit of the test could be as low as 0.18 ng/uL of the RNA isolated from SARS-CoV-2 [163].
3.2.1.8. Rapid, extraction-free nucleic acid tests in detecting SARS-CoV-2
The rapid spreading of SARS-CoV-2 across the world has encouraged many scientists and sci-tech companies to develop and introduce different commercial kits to help in the diagnosis of SARS-CoV-2 [164], [165]. Among these commercial diagnostic kits, rapid and extraction-free nucleic acid-based detection assays have been introduced to speed up the diagnostic process without the need to extract RNA from the patient samples ( Table 9) [16], [68]. These extraction-free nucleic acid tests are currently mainly based on the RT-PCR detection technology, followed by other detection methods like RT-LAMP [68].
Table 9.
Comparison of the sensitivity and specificity of different rapid, extraction-free molecular test kits/protocols in detecting SARS-CoV.
| Name of test kits (manufacturer/country) | Detection technology | Targets | Sensitivity | Specificity | References |
|---|---|---|---|---|---|
| Advanta Dx SARS-CoV-2 RT-PCR assay (Fluidigm) | RT-PCR | N | LOD: 6.25 RNA copies/μL | 100% | [171] |
| qScript ® XLT qRT-PCR (Quantabio) | RT-PCR | N | LOD: 6–12 RNA/μL(sensitivity: 85%) | Unclear | [173] |
| VereRT™ ZeroPrep™ COVID-19 PCR Kit (Veredus) | RT-PCR | N | LOD: 2 RNA copies/reaction | Unclear | [172] |
| XFree™ COVID-19 qRT-PCR test (BioGX) | RT-PCR | N | >90% | >90% | [170] |
| FastPlex™ Triplex SARS-CoV-2 Detection Kits | RT-PCR | N | 97.9% | 95.7% | [174] |
| SalivaDirect (Yale) | RT-PCR | N | LOD: 1.5 RNA copies/μL | 100% | [167] |
| Xpert ® Xpress (Cepheid) | RT-PCR | N, E | LOD: 0.005–0.02 PFU/mL | 100% | [166] |
| SARS-CoV-2 SANSURE®BIOTECH Novel Coronavirus (Sansure). | RT-PCR | N, ORF 1ab | 69.9–94.6% | 100% | [169] |
| SwabExpress (USA) | RT-PCR | S, ORF 1b | LOD: 2–4 RNA/μL (sensitivity: 100%) | 99.4% | [168] |
| PrimeDirect® Probe RT-qPCR Mix (Takara) | RT-PCR | E, RdRp | 55.1–91.9% | 88% | [169] |
| PrimeScript®RT-PCR (Takara) | RT-PCR | E, RdRp | 69.6–89.2% | 100% | [169] |
| ViroReal® Kit RT-LAMP SARS-CoV-2 (Ingenetix) | RT-LAMP | ORF 1ab | LOD: 100 RNA copies/reaction (95%) | 99% | [175] |
| CASSPIT (India) | CRISPR/Cas | S, ORF 1ab | LOD: 100 RNA copies/reaction (~100%) | 100% | [176] |
| QSanger™-Covid-19 Assay (Swift Biosciences) | Nucleic acid sequencing | Unclear | LOD: 3200 RNA copies/mL | Unclear | [177] |
For RT-PCR-based extraction-free test kits, currently, the relevant health authorities like FDA, WHO, or CDC have approved at least ten types of such kits and examples of these kits include Xpert®Xpress [166], SalivaDirect [167], SwabExpress [168], PrimeDirect® qRT-PCR Mix [169], Prime® ScriptRT-PCR [169] and few more [16], [68]. These test assays or kits could be used to detect various genetic targets of SARS-CoV-2 such as N, E, S, RdRp, and ORF genes and the test sensitivity and specificity would vary greatly from one type of assay to another [166], [167], [169], [170], [171], [172]. In general, the test kits that are made to detect the N gene of the virus are found to have a better sensitivity (>80%) [168], [170], [173], [174] and this is higher compared to test kits that are specifically designed to detect other molecular targets of SARS-CoV-2 like RdRp and E (<70%) [169]. This suggested that the N target could be a better and more reliable target to be used for rapid detection of SARS-CoV-2 as the rapid diagnostic kits are more sensitive to detect this target. In terms of the test specificity, most of the introduced SARS-CoV-2 RT-PCR extraction-free test kits were reported to have high test specificity (>80%) [166], [167], [168], [169], [170], [171], except that the test specificity of some kits was unclear like VereRT™ ZeroPrep™ COVID-19 PCR kit [172] and qScript ® XLT qRT-PCR test kit [173]. However, both these test kits have generally low LOD (<12 RNA copies in 1 μL) and high test sensitivity (>85) [172], [173].
The increasing use of RT-LAMP in detecting SARS-CoV-2 has also led to the development of an extraction-free test kit that was based on this detection technology and an example of it is the ViroReal ® Kit RT-LAMP SARS-CoV-2 kit [175]. This kit was reported to have test sensitivity and specificity of 95% and 100%, respectively, and the test LOD is 100 RNA copies per reaction [175]. This kit was designed to detect ORF 1ab of SARS-CoV-2 and its superior sensitivity and specificity have made it a good candidate to be employed in rapid diagnosis of SARS-CoV-2 [175]. Next, an Indian company introduced an extraction-free test kit that was based on the CRISPR/Cas-based detection technology and this test assay was aimed to detect S and ORF 1ab of SARS-CoV-2 [176]. This test kit was reported to have a similar LOD as the ViroReal ® Kit RT-LAMP SARS-CoV-2 kit (100 RNA copies in a reaction) and the sensitivity and specificity of this CRISPR/Cas-based test kit was reported to be almost 100% [176]. On the other side, an extraction-free assay that was dependent on the nucleic acid sequencing technology was introduced and approved for use to detect SARS-CoV-2 and it was reported to be able to detect 3200 RNA copies in 1 mL [177]. However, the genetic targets that are targeted by this rapid diagnostic assay and its specificity were unclear [177]. By comparing the test sensitivity and specificity of the extraction-free diagnostic assays that are based on RT-LAMP, CRISPR/Cas, and nucleic acid sequencing [175], [176], [177], it is found that these assays have comparable good sensitivity and specificity to detect SARS-CoV-2 by targeting various molecular targets of the virus. Thus, more focus should also be given to extraction-free diagnostic tests that are based on the non-RT-PCR approach to increase the selectability and options for the users to choose and use.
In brief, molecular diagnostic techniques are the main approaches for the detection of both the MERS-CoV and SARS-CoV-2 [16], [59], [163], [60], [66], [96], [99], [105], [118], [156], [157]. The advancements in molecular biology and nucleic acids diagnostic technology have enhanced the epidemiological investigations of these viral communicable diseases by enabling the detection of the origins of the coronaviruses, and this also assists in controlling the spread of the infections by isolating the confirmed cases from the community [3], [21], [30], [52], [156], [178]. In time, it is believed that more sensitive, rapid, easy to perform and cheaper molecular diagnostic kits will be introduced to help detect the presence of human coronaviruses and prevent the spread of the viruses.
3.2.2. Protein-based tests to detect MERS-CoV and SARS-CoV-2 infections
Protein-based tests like serological tests and viral antigen detection have been reported as alternative diagnostic tests which could detect both MERS-CoV and SARS-CoV-2 [16], [17], [183], [184], [185], [186], [26], [32], [35], [123], [179], [180], [181], [182]. Generally, compared to nucleic acid-based detection tests, the protein-based test can be less sensitive as it does not involve target amplification [185], and there is a higher possibility of getting false-positive results [183].
Serological tests are applicable to individuals who might have been exposed to MERS-CoV and SARS-CoV-2 or suspected to have these coronaviruses infection since the presence of antibodies to the respective virus reveals that the individual has developed an immune response to the infection [26], [186]. Hence, one of the main purposes to conduct serological tests for MERS-CoV and SARS-CoV-2 cases is mainly meant for seroepidemiological study instead of for diagnostic purposes as the presence of virus antigens in infected individuals and the production of antibodies to both coronaviruses could be delayed up to 10 days after the illness onset or the arising of the clinical symptoms [26], [55], [126], [187], [188].
A few serological assays were introduced to detect betacoronaviruses, and each of these tests has its pros and cons [16], [17], [189], [190]. For both MERS-CoV and SARS-CoV-2, paired sampling at different time points is recommended for verification purposes, as well as to determine whether the patient is having past or recent acute infection [57], [126]. The first sample is to be collected during the illness onset, and the second sample should be ideally taken 21–28 days later, as the increase in the antibody titer could be greater than several-fold in that period [57], [126]. For MERS-CoV, a two-phase approach is recommended by both CDC and WHO. First, a rapid screening assay such as immunofluorescence assay (IFA) or enzyme-linked immunosorbent assay (ELISA) is performed to determine the presence of specific antibodies towards viral proteins in the serum samples, followed by a microneutralization (MN) test to confirm the presence of the respective antibodies [54], [126]. However for SARS-CoV-2, both CDC and WHO do not recommend the use of virus neutralization test as a routine diagnostic test for Covid-19, as the test requires highly skilled personnel to perform and it takes a longer time for the results to be available [57], [185]. Therefore, for SARS-CoV-2, only ELISA is conducted routinely to screen for recent or past infection by this coronavirus as it takes a shorter time [57], [185].
3.2.2.1. Diagnosis of MERS-CoV and SARS-CoV-2 using IFA
Anti-MERS-CoV IFA works by illuminating fluorescence of granular structures of viral particles in the cytoplasm of MERS-CoV infected cells, whereas the absence of illumination suggests that the cell is uninfected [191]. This method hinged on the MERS-CoV infected cells as the antigen platform to determine the presence of human anti-MERS-CoV IgG and IgM in the infected individual sera [5], [55], [192]. The test specificity of IFA was found to be generally low (<85%) and few studies had reported that the use of this assay would likely show cross-reaction with other respiratory pathogens ( Table 10) [193], [194]. This situation could be improved if the recombinant proteins are used as the antigens in IFA instead of using the whole-virus IFA approach [192]. Although the specificity of IFA is not significant as compared to other molecular diagnostic approaches, however, additional diagnostic information is still available from the unique fluorescent staining patterns in MERS-CoV infected cells as compared to other serological tests like enzyme immunoassay (EIA) [193]. In terms of the test sensitivity of IFA, even though a study has previously reported that IFA was sensitive enough to detect the presence of MERS-CoV in all tested patient samples [193], however, other studies had reported that IFA could have a low detection rate of below 40% [195] and its detection sensitivity would reduce when the samples infected with the virus was further diluted [194]. Another study also reported that when the S protein was employed as the assay target, IFA had a low ability to detect anti-MERS-CoV IgM (30%) and the ability of IFA to detect anti-MERS-CoV IgG was higher (90%). Combining the findings from various studies [192], [193], [194], [195], it is said that IFA might not be the best and highly sensitive and specific test to detect the presence of MERS-CoV.
Table 10.
Comparison of the sensitivity and specificity of IFA in detecting different targets of MERS-CoV and SARS-CoV-2.
| Virus type | Targets/Sources of samples | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | S | 3/10 (30%) for IgM compared to 9/10 (90%) compared to IgG | Varied due to cross-reactivity with other pathogens in sera of different patients (<100%) | [192]; |
| Whole virus | 100%; | 0–83% (depending on cross-reaction with different pathogens) | [193] | |
| Infected cells | ~97.9% at <1:20 dilution | 25% of SARS-CoV patients had anti-MERS-CoV antibody reactions | [194] | |
| Unclear | Detection rate of MERS-CoV = 38% compared to detection rate of MERS-CoV using ELISA = 30% (comparable results | Unclear | [195] | |
| SARS-CoV-2 | S | 85.7–96.3% (IgG) | High (unclear on the values) | [199] |
| Inactivated infected cells | Unclear | ~100% (IgG) | [197] | |
| Inactivated infected cells | 76.5–100% (IgG) | 86.4% (IgG) | [196] | |
| Inactivated infected cells | 41.9 (IgG), 35.5% (IgM) | Unclear | [183] | |
| Inactivated infected cells | 99% (IgG) | 100% (IgG) | [198] | |
| Inactivated infected cells | 41% (IgG) | 93% (Ig G), 98.5–100% (IgM/A) | [184] | |
| Virus from patient sera | 91.3% (IgG/A/M) (at Day 14 after onset of symptoms) | 98.9% (IgG/A/M) (at Day 14 after onset of symptoms) | [327] |
Like MERS-CoV, IFA has also been used in confirming the serological status of the patients infected with SARS-CoV-2 (Table 10) [183], [184]. Two studies had reported that the test sensitivity of IFA could be low (<50%) when the assay was used to detect IgG and IgM against SARS-CoV [183], [184]. Both these studies used inactivated infected cells for the assay [183], [184] and the test specificity of one of the studies was shown to be good (93–100%) in detecting different Ig against SARS-CoV-2 [184]. In another study where inactivated infected cells were used in the IFA [196], the test sensitivity and specificity ranged from 76% to 100%, and 86.4%, respectively, suggesting that the IFA test results might vary in different studies even though inactivated infected cells were used for the same assay. This hypothesis was further supported when another two studies that used inactivated infected cells showed good sensitivity and specificity in detecting IgG against SARS-CoV-2 (>95%) [197], [198]. In a Swiss study that used spike protein-based IFA (recombinant IFA) to detect IgG against SARS-CoV-2, the assay sensitivity was found to be high (>85) [199] and this suggested that the use of either inactivated infected cells or recombinant protein in IFA would produce comparable test findings to detect for the presence of SARS-CoV-2.
3.2.2.2. Diagnosis of MERS-CoV and SARS-CoV-2 using ELISA
Since the start of the MERS-CoV outbreak, an ELISA screening test that used recombinant N and S proteins had been introduced to detect the presence of antibodies specific against the N and S proteins of MERS-CoV [28], [200]. Both N and S proteins are highly immunogenic, especially S1 protein is a typical antigen that was used to explore the phylogenetic relationships of coronavirus and epidemiology of MERS-CoV infection [3]. The sensitivity and specificity of ELISA to detect antibodies against MERS-CoV N and S antigens could vary from 20% to 100% and the test findings were depending on the dilution factor of the test samples ( Table 11) [200]. Compared to IFA, ELISA was reported to have generally higher sensitivity and specificity in detecting the antibodies against MERS-CoV S protein in which ELISA could detect antibodies against S protein of MERS-CoV in all patient samples while IFA would unable to do so [192]. In another two studies [190], [201], ELISA was shown to have superior sensitivity (>90%) to detect IgG against S protein of MERS and the use of this test would not cause false-positive results by cross-reacting with other respiratory pathogens. Therefore, ELISA is said to be a more sensitive and specific test than IFA to evaluate the serological response of the MERS-CoV patients [192].
Table 11.
Comparison of the sensitivity and specificity of ELISA in detecting different targets of MERS-CoV and SARS-CoV-2.
| Virus type | Targets/Sources of samples | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | S | ~100% | Varied due to cross-reactivity with other pathogens in sera of different patients (<100%) | [192] |
| S | LOD: 0.04–5.9 ng/mL of S protein | Unclear (no significant intra-/inter-assay variability) | [328] | |
| S | 92.3% (IgG) | 100% (IgG) | [201] | |
| S | ~97% at day 21 and above (IgG) | Unclear | [190] | |
| N, S | varied from ~20–100%; | varied from 0% to 100% (depending on dilution factors) | [200] | |
| SARS-CoV-2 | N | LOD: 9.00 ng/μL | High (Unclear on the numerical value) | [329] |
| N | 94.9% (IgG) | 97.1% (IgG) | [202] | |
| N | 90.3% (IgG) | 97.9% (IgG) | [203] | |
| S | 88.9–92.9% (IgG) | High (unclear on the values) | [199] | |
| S | 64.1 (IgG), 74.3% (IgA) | 95.2 (IgG), IgA (84.2%) | [207] | |
| S | 95% (IgG) | 97.8% (IgG) | [205] | |
| S | 65 (IgG/A) | 96% (IgG/A) | [206] | |
| S | 95.6–100% (IgG) | 86.7–100% (IgG) | [204] | |
| S (RBD), inactivated virus | 53.3–92.1% (IgG) | ~99% | [209] | |
| S (RBD) | 92% (IgA), 96% (IgG), 98% (IgM) | 99.3% | [208] | |
| N, S | LOD: 5 pg/μL | 100% | [214] | |
| N, S | 88–100% (depending on Ig types) | 96.4–100% (depending on Ig types) | [230], [330] | |
| N, S | 58.8–96.2% (IgG) | ~95% (IgG) | [196] | |
| N, S | 56.3–81.6% (IgG) | 99% | [216] | |
| N, S | 74.3–82.2% (IgG), 77.1–80.4% (IgM) | 100% (IgG/M) | [213] | |
| N, S | 84.2% (IgG) | 100% (IgG) | [212] | |
| N, S | 93.1–98.3% (IgG/total antibody) | 86.3–96.4% (IgG/total antibody) | [211] | |
| N, S | 35.5–61.3% (IgG/M); | Unclear | [183] | |
| N, S | 86.4–87.9% (IgG) | 75.7–98.6% (IgG) | [215] | |
| N, S, total antibody | 92.7–99.1% (IgG/total antibody) | 98.9–99.9% (IgG/total antibody) | [210] | |
| Sera (unclear target) | 45% (IgG) | 97% (IgG) | [184] | |
| Sera (unclear target) | 69–80.6% (IgA), 48.8–72.9% (IgM), 79.8–83.7% (IgG); | 97.6–100% (IgA), 88.1–97.6% (IgM), 97.6–100% (IgG) | [182] |
For SARS-CoV-2, several studies had reported the use of ELISA to detect antibodies against N protein of SARS-CoV and the sensitivity and specificity of such test ranged from 90% to 94%, and, above 97%, respectively (Table 11) [202], [203]. Compared to ELISA assay that was aimed to detect antibodies like IgG against the N protein of SARS-CoV-2 [202], [203], the use of ELISA test that was focused to detect antibodies against S protein of SARS-CoV-2 showed wide range values of sensitivity and specificity, according to the findings reported from various studies [199], [204], [205], [206], [207]. Some studies showed that S protein-based ELISA test was less sensitive to detect IgG and IgA against SARS-CoV-2 (<70%) [206], [207] while few other studies reported that such test was highly sensitive (at least 90%) to detect IgG, IgA, or IgM against the virus S antigen [199], [204], [205], [208]. Therefore, N protein-based ELISA assay could be a better choice of ELISA test to predict serological response in the SARS-CoV-2 patients as the test results of this assay seem to be more consistent and vary less in different reported studies [202], [203] as compared to the S protein-based ELISA test [199], [204], [206], [207], [208], [209]. On the other hand, some studies had reported the use of ELISA test that was aimed to detect different antibodies against different proteins of SARS-CoV-2, and examples of these tests were tests that were focusing to detect antibodies against N and S protein of SARS-CoV-2 [196], [204], [209], [210], [211], [212], [213], [214], [215]. Compared to molecular detection using the qRT-PCR method that showed more consistent test sensitivity and specificity across different reported studies as an example [71], [72], [87], [88], the sensitivity and specificity of ELISA assays in detecting various antibodies (IgG, IgA, and IgM) against different antigen proteins of SARS-CoV-2 demonstrated a high degree of variations in which some studies reported low test sensitivity (<60%) [183], [196], [216] and specificity (as low as below 80%) [215] whereas some studies would report high test sensitivity and specificity (>85%) [210], [211]. Even though the ELISA test might be less sensitive and specific in detecting the presence of SARS-CoV-2 based on the findings from few studies [196], [209], [215], [217], however, this assay was reported to have higher sensitivity and specificity than other serological tests like IFA in detecting the presence of SARS-CoV-2 [183], [184].
To date, FDA has approved at least ten different types of commercial ELISA test kits under EUA use ( Table 12), and generally, these kits could be used for rapid detection of various antibodies against different antigenic targets of SARS-CoV-2 such as N, S, and M proteins [218], [219], [228], [220], [221], [222], [223], [224], [225], [226], [227]. Examples of these test kits include Anti-SARS-CoV-2 ELISA by EUROIMMUNE [221], Platelia SARS-CoV-2 Ab Assay by Bio-Rad [218], Wantai SARS-CoV-2 Ab ELISA by Beijing Wantai Co. Ltd [227], SARS-CoV-2 Detect IgG/M ELISA by InBios [220] and others. The test sensitivity and specificity of these approved ELISA test kits were noted to be generally good and ranged from 90% to 100%, and above 95%, respectively [218], [219], [230], [220], [221], [222], [225], [226], [227], [228], [229]. Therefore, these test kits would probably help to screen and detect SARS-CoV-2 infection in areas or places where molecular diagnostic tests like qRT-PCR are unavailable [16], [230].
Table 12.
Some of the selected examples of FDA EUA-approved commercial ELISA test kits to detect SARS-CoV-2.
| Name of test kits (manufacturer/country) | Antigen targets | Antibody | Sensitivity | Specificity | References |
|---|---|---|---|---|---|
| Platelia SARS-CoV-2 Ab Assay (Bio-Rad) | N | Total antibody | 92% | 99% | [218] |
| Anti-SARS-CoV-2 ELISA (EUROIMMUN) | S | IgG | 90% | 100% | [221] |
| Beijing Wantai SARS-CoV-2 Ab ELISA (Beijing Wantai Co. Ltd) | S | IgG | 96.7% | 97.5% | [227] |
| Kantaro Semi-Quantitative SARS-CoV-2 IgG Antibody Kit (Kantaro Biosciences, LLC) | S | IgG | 99.1% | 99.6% | [223] |
| Smybiotica COVID-19 Self-Collected Antibody Test System (Symbiotica. Inc) | S | IgG | 100% | 98% | [225] |
| COVID-19 ELISA pan-Ig Antibody Test (University of Arizona) | S | Total antibody | 97.5% | 99.1% | [222] |
| cPass SARS-CoV-2 Neutralization Antibody Kit (GenScript) | S | Total antibody | 100% | 100% | [229] |
| Dimension Vista SARS-CoV-2 Total Antibody Assay (Siemens Healthcare) | S | Total antibody | 100% | 100% | [219] |
| Mount Sinai Hospital COVID-19 ELISA Antibody Test (Mount Sinai Hospital) | S | Total antibody | 92.5% | 100% | [228] |
| OmniPath COVID-19 Total Antibody ELISA Test (ThermoFisher) | S | Total antibody | 96.7% | 97.5% | [224] |
| SARS-CoV-2 Detect IgG/M ELISA (InBios) | N, S | IgG, IgM | 96–100% | 98–100% | [220] |
| United Biomedical UBI SARS-CoV-2 ELISA (United Biomedical, Inc) | N, S, M | IgG | 89.7% | 100% | [226] |
3.2.2.3. Diagnosis of MERS-CoV and SARS-CoV-2 using virus micro-neutralization (MN) test
A virus neutralization test is a more definite assay that could be used to detect the presence of neutralizing antibodies specific to MERS-CoV and SARS-CoV-2 as these antibodies would not neutralize the other human coronaviruses [2], [57], [185], [231]. A total of three well-established virus neutralization tests have been published to detect coronaviruses, which are MN assay [35], [181], [232], pseudoparticle neutralization test (ppNT) [180], [233], and plaque reduction neutralization test (PRNT) [123], [193], [234]. For MN, specific anti-MERS-CoV antibodies were developed and it was used to determine the highest serum dilution required to inactivate the virus cytopathic effect in Vero cells [231]. This method is recognized as a gold standard test to detect specific anti-MERS-CoV as the neutralizing antibodies could be measured [231], [235]. However, the MN assay is laborious whereby three to five days of incubation is needed [188], [231]. In a study which was aimed to compare the efficiency of MN and ppNT in detecting the neutralizing antibodies against MERS-CoV, it was shown that MN could detect antibodies in 16 out of the 21 samples (76.2%) as compared to ppNT which could detect neutralizing antibodies in 20 out of 21 samples, indicating MN is less sensitive than ppNT in detecting antibodies against MERS-CoV ( Table 13) [231]. However, in another study that compared the efficacy between MN, ppNT, PRNT, and ELISA, it was shown that all these four tests were able to detect antibodies in an almost similar number of tested samples at different time intervals, suggesting that the efficiencies of all these four tests were highly correlated and comparable to each other in detecting antibodies against MERS-CoV [190].
Table 13.
Comparison of the sensitivity and specificity of micro-neutralization (MN) test in detecting different targets of MERS-CoV and SARS-CoV-2.
| Virus type | Targets/Sources of samples | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | Whole virus | 16/21 (76.2%) compared to 20/21 (95.2%) of ppNT; | 100% | [231] |
| Whole virus | 97% at day 21 and above (IgG); comparable efficiency with ppNT, PRNT and ELISA | Unclear | [190] | |
| SARS-CoV-2 | Whole virus | Sensitivity of three immunoassays to MN = 63.1–91.1% | Specificity of six immunoassays to MN = 100% | [239] |
| Whole virus | Varied and weaker compared to ELISA (most tests only detected virus at 1:10/1:20 dilution) | Unclear | [237] | |
| Whole virus | Able to confirm presence of SARS-CoV-2 when ELISA was unable | Unclear | [238] | |
| Whole virus | LOD for MN: 1/40 dilution Sensitivity of six immunoassays to MN = 43.8–87.8% | Specificity of six immunoassays to MN = 68.3–97.5% | [236] | |
| Whole virus | ≥50% compared to ELISA | Unclear | [331] | |
| Whole virus | Sensitivity of six immunoassays to MN = 69–100% | Unclear | [240] |
Similarly, an MN assay was also being developed to detect SARS-CoV-2 when the virus started to cause a global outbreak (Table 13) [181]. But, as mentioned previously, MN takes at least 3–5 days for the results to be available and it requires highly trained staff to perform the test [181], and thus, both WHO and CDC do not recommend the use of MN in detecting SARS-CoV-2 [57], [185]. The ability of MN to detect neutralizing antibodies against SARS-CoV-2 was reported to be highly dependent on the dilution factor used in a test [236], [237]. Some studies reported that MN could only detect antibodies against SARS-CoV-2 at a dilution factor of 1:20 and it would not be able to detect the antibodies when the testing reaction was diluted further [237]. In another study, it was reported that the LOD of MN was at a dilution factor of 1:40 [236], implying that the LOD of this assay might vary across different assay settings. Compared to other protein-based tests like ELISA, it was shown that MN could serve as an alternative test to confirm the presence of antibodies against SARS-CoV-2 when ELISA was unable to do so, suggesting MN has superior sensitivity than ELISA [238]. Thus, in the development and validation of the sensitivity and efficiency of various immunoassays or ELISA to detect antibodies against SARS-CoV-2, MN was always used as a standard test to be compared with [236], [239], [240].
3.2.2.4. Diagnosis of MERS-CoV and SARS-CoV-2 using pseudoparticle neutralization test (ppNT)
The main advantage of ppNT in MERS-CoV and SARS-CoV-2 diagnosis is that biosafety level (BSL)-3 containment is not compulsory, as no actual Risk Group 3 virus is being used [188], [190]. ppNT which utilizes HIV/MERS spike pseudoparticles to infect the Vero E6 cells was reported to have a shorter incubation time than MN assay [231]. For the time being, ppNT assay is mainly applied in the seroepidemiology study which involves animals and livestock, where the assay demonstrates a good correlation with the gold standard test, MN [2], [186], [231]. Besides, ppNT assay was demonstrated to produce a reliable testing outcome when it was used in human samples during the first six weeks of MERS-CoV infection [190]. Compared to other protein-based tests like MN and PRNT, ppNT was said to produce comparable test findings with these protein tests [190]. Therefore, when protein tests like ELISA and MN were unable to detect any antibodies against MERS-CoV, ppNT could be used as another optional test to confirm the presence of MERS-CoV [241], [242]. In terms of its LOD, a study has reported that ppNT was able to detect antibodies against MERS-CoV in more than 90% of the test samples when the dilution factor was set at 1:40 [233]. Compared to another study that has reported that MN could detect anti-SARS-CoV-2 at a dilution factor of 1:40 [236], ppNT was said to have a similar detection limit with that of MN when used to detect another coronavirus type. This further supported that ppNT has a sensitivity that is not inferior to MN ( Table 14). For SARS-CoV-2, a ppNT test that could be conducted in the BSL-2 facility was reported and the limit of detection was found to be 43 and 22 for mouse and human samples, when the cutoff values were set at 50 and 30, for mouse and human samples, respectively [180]. Besides, the ppNT assay was also shown to be highly reproducible with low intra- and inter-assay variations of 15.9% and 16.2%, respectively [180]. In the clinical setting, ppNT could be used to measure the neutralizing antibodies against SARS-CoV and this is important to evaluate the remaining antibody levels in the patient body post-infection [243], [244]. The findings from two ppNT-based studies (Table 14) reported that the neutralizing antibodies against the pseudoparticle virus would start to drop within 4–8 weeks post-infection [243], [244], suggesting that the patients might have a risk of being re-infected. In short, the introduction of ppNT has helped to reduce the risk of viral infection among the testing personnel as it does not require the handling of live, infectious viruses. [180], [188], [190]. Its good accuracy and versatility might ease the development of a viral vaccine or drug [180].
Table 14.
Comparison of the sensitivity and specificity of pseudoparticle neutralization test (ppNT) in detecting different targets of MERS-CoV and SARS-CoV-2.
| Virus type | Targets/Sources of samples | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | S protein pseudoparticle | MERS-CoV undetected by MN could be detected by ppNT (% sensitivity unclear) | Unclear | [241] |
| Virus pseudoparticle | 118/131 (90%) (at 1:40 dilution) | 100% | [233] | |
| Virus pseudoparticle | 94% at day 21 and above (IgG) | 100% | [190] | |
| Unclear | ppNT was used to confirm MERS-CoV infection when ELISA showed false +ve/-ve findings | Unclear | [242] | |
| SARS-CoV-2 | Virus pseudoparticle | Neutralizing antibody levels would drop after 8 weeks of infection but still measurable up to 8 months. Pseudovirion improved infectivity of SARS-CoV-2 neutralization assay | Unclear | [244] |
| Virus pseudoparticle | LOD: 1:100 dilution; LOD of 22.1 and 43.2 for human and mice sera when 120 negative samples were used | Low intra- and inter-assay variations of 15.9% and 16.2%, respectively | [180] | |
| Virus pseudoparticle | Unclear but 1/3 of the patients would lose the antibodies against pseudoparticle a month after onset of symptoms | Unclear | [243] |
3.2.2.5. Diagnosis of MERS-CoV and SARS-CoV-2 using plaque reduction neutralization test (PRNT)
PRNT is considered as another gold standard test to determine the neutralizing antibody titers against an infectious agent [123]. For MERS-CoV, PRNT exhibits slightly higher sensitivity in detecting the early antibody responses in MERS-CoV infection compared to MN and ppNT neutralization assays [190]. Hence, it is recommended that PRNT be conducted in patients with poor serological responses as it gives great sensitivity to other neutralization assays [190]. In another study [192], PRNT was reported to have comparable sensitivity with other protein-based tests such as IFA and ELISA in detecting the antibodies against MERS-CoV. However, in terms of its specificity, even though the authors suggested that PRNT could be the most specific test to detect MERS-CoV than other tests, but cross-reactivity with other pathogens cannot be ruled out [192]. Nevertheless, the same study has proven that the antibodies against MERS-CoV would fade over time in the human body post-infection and this suggested that the patient has a risk of re-infection with MERS-CoV in the future [192]. For SARS-CoV-2, several studies had been conducted to compare the sensitivity and efficacy of PRNT to detect antibodies against SARS-CoV-2 ( Table 15), and in one of the studies [217], it was shown that PRNT has comparable sensitivity with ELISA in detecting antibodies against SARS-CoV-2. In another study that compared the efficiency and sensitivity of three tests, namely, ELISA, MN, and PRNT, the authors reported that PRNT is more sensitive than MN and ELISA in detecting anti-SARS-CoV-2 [245]. The superiority of PRNT in detecting antibodies against SARS-CoV-2 was further shown when a Canadian study employed PRNT as a reference test to evaluate the sensitivity and specificity of the virus neutralization test [246]. Compared to PRNT, the virus neutralization test was demonstrated to have a slightly lower sensitivity of 89.8–99% whereas in terms of the test specificity, both neutralization test and PRNT were found to have similar and comparable specificity (~100%) [246]. In clinical use, like in the case of MERS-CoV, PRNT was also used to evaluate the levels of neutralizing antibodies in the bodies of the patients infected with SARS-CoV-2 and it was shown that patients who had more severe infections would have higher peak levels of PRNT90 and PRNT50 after months of infection, compared to patients with mild infection or asymptomatic patients [123]. This suggested that the levels of neutralizing antibodies were correlated to disease severity and SARS-CoV-2 is possible to create robust neutralizing antibodies in patients with severe infection [123]. A limitation that was not well-explored in the study [123] is that the authors did not specify whether other neutralization tests would still be able to detect the neutralizing antibody in the patients after two months of infection, and thus, it is inconclusive on whether PRNT is more superior and sensitive than other neutralizing tests in detecting the antibody responses towards SARS-CoV-2 after months of infection.
Table 15.
Comparison of the sensitivity and specificity of plaque reduction neutralization test (PRNT) in detecting different targets of MERS-CoV and SARS-CoV-2.
| Virus Type | Targets/Sources of samples | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | Whole virus | Comparable sensitivity with IFA and ELISA | Varied due to potential cross-reactivity with other pathogens in sera of different patients (<100%) | [192] |
| Whole virus | 97–100% at day 21 and above (IgG) | Unclear | [190] | |
| SARS-CoV-2 | Whole virus | 76.5–100% | Unclear | [196] |
| Whole virus | Over 99% of the sera remained seropositive for both 90% (PRNT90) and 50% (PRNT50) neutralization endpoints after 61 days of infection | Unclear | [123] | |
| Whole virus | Neutralizing activity was 93% for PRNT50 and 54% for PRNT90 at around day 33 after onset of symptoms | Unclear | [332] | |
| Whole virus | 100% | Unclear | [206] | |
| Whole virus | 100% compared to ELISA | Unclear | [217] | |
| Whole virus | PRNT > MN and ELISA (% of sensitivity unclear) | Unclear | [245] | |
| Whole virus | Virus neutralization test vs PRNT (89.8–99.0%) | Virus neutralization test vs PRNT (100%) | [246] |
3.2.2.6. Viral antigen detection for MERS-CoV and SARS-CoV-2
A viral antigen test aims to detect the specific antigen which is present on the virus surface. This type of assay could also be used to detect both MERS-CoV and SARS-CoV-2 [179], [247]. In a study that used 129 nasopharyngeal aspirates which were positive to various human respiratory viruses, it was found that the use of MERS-CoV-nucleocapsid protein (NP)-specific monoclonal antibodies were sensitive and specific in detecting MERS-CoV antigen, in which the test specificity was almost 100% [179]. Besides, the MERS-CoV-NP-specific monoclonal antibodies could also detect the presence of live MERS-CoV in simulated nasopharyngeal aspirates samples after rounds of serial dilutions, when the tissue culture infectious dose (TCID50) per 0.5 mL was at least 10 [179]. However, the sensitivity and validity of the virus antigen test reported in this study were not compared with other molecular tests like qRT-PCR [179]. In the year 2016, a similar group of researchers compared the sensitivity of viral antigen test to detect spike protein of MERS-CoV with the test sensitivity of ELISA to detect antibodies against MERS-CoV [100]. The report showed that the test sensitivity of antigen test was about 81% as compared to ELISA and it was estimated to be at least 25–100-folds less sensitive than ELISA to detect for the presence of MERS-CoV ( Table 16) [100]. However, the viral antigen test was proven to have high specificity to detect MERS-CoV and would produce results in less than half an hour [100].
Table 16.
Comparison of the sensitivity and specificity of virus antigen test in detecting different targets of MERS-CoV and SARS-CoV-2.
| Virus type | Targets/Sources of samples | Sensitivity (SN) | Specificity (SP) | References |
|---|---|---|---|---|
| MERS-CoV | N | +ve results for samples with 10 TCID50/100 μL; | 100% | [179] |
| S | 103.7–104.2 TCID50/mL (81%, ~100 times less sensitive than ELISA) | 100% | [100] | |
| SARS-CoV-2 | N | 98.33% | 98.73% | [257] |
| N | LOD95: 2·07 × 106 and 2·86 × 107 copies/swab | 98.5–100% | [256] | |
| N | 45.4–50.3% | 97.7–97.8% | [250] | |
| N, S | LOD: 0.31–102 TCID50/mL (84–97.7%) (depending on antigen type) | 93.9–100% | [230], [248] | |
| N, E, S, M | LOD: 100,000 RNA copies/mL (78.8%) | 99.7% | [253] | |
| Unclear | 96.52% | 99.68% | [255] | |
| Unclear | 11.1–45.7% (qRT-PCR>virus culture>antigen test) | ~100% (no cross-reactivity with other pathogens) | [249] | |
| Unclear | 81% | 99.1% | [254] | |
| Unclear | 69.86% | 99.61% | [251] | |
| Unclear | 72.5% | 99.4% | [252] | |
| Unclear | 88.9–100% | 46.2–100% | [258] |
On the other hand, a viral antigen test was also being used to detect the presence of SARS-CoV-2 (Table 16) [68], [248]. Like MERS-CoV, virus antigen test was generally reported to have lower sensitivity (<80%) in detecting the specific antigen of SARS-CoV-2, especially when this test was compared to the molecular diagnostic test like qRT-PCR [249], [250], [251], [252] In terms of the test specificity, antigen test was found to be most specific (>90%) to detect SARS-CoV-2 without creating cross-reaction when other respiratory pathogens were present [249], [250], [251], [252], [253], [254], [255], [256], [257]. However, there was also some study which had reported that the antigen test results could have low specificity (as low as 46.2%) and this suggested that antigen test could be used in rapid screening of the SARS-CoV-2 infection but the use of other confirmatory tests like qRT-PCR should be conducted to confirm the viral infection [258]. As for the molecular target that is frequently used in the virus antigen test, both N and S antigens of SARS-CoV-2 are the commonly employed antigenic targets in the virus antigen test, and the LOD to detect these antigenic targets could range from 100,000 RNA copies in 1 mL to 2.86 × 107 RNA copies in a single swab [253], [256]. Therefore, compared to molecular tests like qRT-PCR which could detect as low as few RNA copies in 1 μL [68], [78], it is said that more viral copies are needed to be present in the test samples for the virus to be detected by the viral antigen test [253], [256].
Rapid case detection is essential to identify subjects who have been infected with SARS-CoV-2 and this ensures the infected people will be isolated from other people to prevent the virus from spreading in a community [57], [58]. For rapid virus screening, multiple rapid viral antigen tests have been approved by FDA for EUA use ( Table 17) and examples of these tests include BinaxNOW COVID-19 Ag Card Home Test by Abbott Diagnostic Scarborough Inc [259], CareStart COVID-19 Antigen Test by Access Bio Inc [260], GenBody COVID-19 Antigen Test by GenBody Inc [261], InteliSwab COVID-19 RAPID Test Pro by OraSure Technologies Inc [262]. In general, these test kits were developed to detect N and S antigens of SARS-CoV-2 and the test sensitivity and specificity are ranging from 80% to 100%, and 90–100%, respectively [259], [260], [261], [263], [264], [265], [266], [267], [268]. In terms of rapid virus screening and detection, the sensitivity and specificity of these tests are considered to be good to enable the local medical personnel to screen for any suspected SARS-CoV-2 infection in a community [16], [17], [230]. For any indeterminate test finding, further test like qRT-PCR is recommended to confirm whether a subject has contracted the virus [57], [247].
Table 17.
Some of the selected examples of FDA EUA-approved commercial rapid antigen test kits to detect SARS-CoV-2.
| Name of test kits (manufacturer/country) | Antigen targets | Sensitivity | Specificity | References |
|---|---|---|---|---|
| BD Veritor System for Rapid Detection of SARS-CoV-2 (Becton, Dickinson and Company) | N | 84% | 100% | [265] |
| BinaxNOW COVID-19 Ag Card Home Test (Abbott Diagnostics Scarborough Inc) | N | 97.1% | 98.5% | [259] |
| CareStart COVID-19 Antigen Test (Access Bio, Inc) | N | 88% | 100% | [260] |
| Ellume COVID-19 Home Test (Ellume Limited) | N | 95% | 97% | [267] |
| GenBody COVID-19 Antigen Test (GenBody Inc) | N | 90% | 98% | [261] |
| InteliSwab COVID-19 RAPID Test Pro (OraSure Technologies, Inc.) | N | 85% | 99% | [262] |
| LumiraDx SARS-CoV-2 Ag Test (LumiraDx UK Ltd) | N | 97.6% | 97% | [268] |
| Omnia SARS-CoV-2 Antigen Test (Qorvo Biotechnologies, LLC.) | N | 89.5% | 100% | [333] |
| SCoV-2 Ag Detect™ Rapid Test (InBios International Lnc) | N | 86.67% | 100% | [264] |
| Sienna-Clarity COVID-19 Antigen Rapid Test Cassette (Salofa Oy) | N | 87.5% | 98.9% | [263] |
| Status COVID-19/Flu Test (Princeton BioMeditech) | N | 93.9% | 94% | [266] |
| COVID-19 Antigen MIA (Celltrion USA) | S | 94.4% | 100% | [334] |
To sum up, for this section, both ELISA and virus antigen tests can be done rapidly and both assays could be completed in a shorter time (<1 h) as compared to viral neutralization tests [17]. However, the virus antigen test is said to have lower accuracy, sensitivity, and is less reliable when compared to ELISA [17], [68]. In terms of invasive-ness, even though the use of nasopharyngeal swabs or aspirates for virus antigen detection sounds more user-friendly, but the nasopharyngeal samples collection process could be sometimes invasive and could lead to bleeding in the upper nasopharyngeal regions [269]. Thus, a different new method like the saliva specimen collection approach [269] has been introduced to screen for the presence of the virus antigen in the patient specimens. But, this new method was shown to have poorer sensitivity in detecting the coronaviruses, for example, when saliva specimen was used in the antigen testing for SARS-CoV-2, the sensitivity of the test was just about 21% as compared to the nasopharyngeal swab antigen test which could show a sensitivity of 52% [269].
3.2.3. Virus culture of MERS-CoV and SARS-CoV-2
The concept of virus-cell culture involves the use of cell lines to culture virus isolated from patients who are suspected to have MERS-CoV and SARS-CoV-2 infections [32], [35]. Examples of commonly used cell lines for virus-cell culture include Vero E6 and LLC-MK2 cells, in which the former is derived from the kidney of African green monkey while the latter is derived from the kidney of rhesus monkey [16], [270]. Following virus replication, the infected cells might die secondary to cellular apoptosis and the cells which survive after virus replication would show cellular morphology similar to healthy, uninfected cells, and the survived cells would continue to support the virus production [16]. The entry of both MERS-CoV and SARS-CoV-2 into host cells is mediated and enhanced by the proteolytic cleavage of the viral S protein by a host protein called type II transmembrane serine protease (TMPRSS2) [271], [272]. A group of Japanese researchers has engineered a Vero E2 cell line which expresses TMPRSS2, where the use of such cell lines would enhance the viral RNA production by at least ten-folds for both MERS-CoV and SARS-CoV-2 [271], [272]. Hence, such cell lines can be potentially used in diagnostic laboratories to speed up the virus identification process.
Apart from being used in virus identification, virus-cell culture has also been widely used in the anti-viral drug development, in which anti-viral agent will be administered to the cultured cells to determine whether it would help to inhibit virus replication [273], [274]. As such, virus-cell culture has been extensively employed to test the potential anti-viral agents against SARS-CoV-2 [273] and influenza virus [274]. The limitation of virus-cell culture is that the identification of virus using this method requires further tests like qRT-PCR or nucleotide sequencing to confirm the virus identity which is isolated from the clinical samples [275]. Besides, this method is time-consuming and insensitive, and it requires trained and specialized personnel to perform the procedure [35], [276].
3.3. Radiological diagnosis of MERS-CoV and SARS-CoV-2 infections
Chest radiography (CXR) and computed tomography (CT) scans are the commonly used radiological investigations that could be used to help identify chest infection caused by both MERS-CoV and SARS-CoV-2 [277], [278], [279]. For pneumonia caused by MERS-CoV, it was reported that both CXR and chest CT would show features of organizing pneumonia evidenced by the presence of multifocal ground glass opacities [278], [279]. Similarly, the CXR and chest CT in the patients infected with SARS-CoV-2 would also show features of ground-glass opacities [17], [277], suggesting chest infection caused by coronavirus infection is more commonly presented with ground-glass appearances under radiological imaging. A comparative review has reported that MERS-CoV infection is more prone to cause unilateral chest infection and pleural effusion, while SARS-CoV-2 infection is more likely to cause bilateral chest infection [277]. Therefore, the laterality involvement and the presence of pleural effusion could be potentially used to differentiate MERS-CoV and SARS-CoV-2 chest infection [277]. Another common radiological feature that could be found in the chest imaging of the patients infected with MERS-CoV and SARS-CoV-2 is the presence of signs of pulmonary fibrosis, where this predominant finding is obvious for patients who have recovered from coronavirus infection [277].
Even though chest infection caused by both MERS-CoV and SARS-CoV-2 would produce some typical features under radiological imaging, however, these radiological features are non-exclusive to the respective viruses. For example, pleural effusion has been being reported in 153 out of 473 patients (32.3%) infected with SARS-CoV-2 in China [280]. Besides, pneumonia with ground-glass opacity could also be caused by other viruses like cytomegalovirus [281], and pleural effusion could be present in pneumonia caused by other causative agents such as SARS-CoV [277] and bacteria like Pseudomonas aeruginosa [282]. Likewise, pulmonary fibrosis could also be caused by other infectious agents such as herpes and Epstein-Barr viruses [283]. In short, the presence of ground-glass opacity, pleural effusion, and pulmonary fibrosis under radiological imaging could not be used to confirm coronavirus chest infection.
4. Detections of different variants of SARS-CoV-2
The COVID-19 pandemic has hit mankind for more than 16 months since its first appearance at end of 2019 [284], [285], [286], [287]. During this period, the SARS-CoV-2 virus was found to undergo continuous mutations in their genetic sequences and different variants of SARS-CoV-2 have spread to many different countries [284], [285], [286], [287], [288], [289]. The appearance of these different SARS-CoV-2 variants has led to the increased virus transmissibility and reduced effectiveness of the currently available vaccines in curbing the spreading of SARS-CoV-2 [284], [285], [286], [287], [290], [291], [292], [293], [294], [295]. To ease the classification and grouping of the different virus variants, the relevant global and local health authorities have decided to divide the SARS-CoV-2 variants into four main groups ( Table 18), namely, variant of high consequence (VOHC), variant of concern (VOC), variant of interest (VOI) and variant under monitoring (VUM) [284], [287], [296].
Table 18.
Variant of concern (VOC) and variant of interest (VOI) of SARS-CoV-2.
| Variant of concern (VOC) |
Variant of interest (VOI) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variant type | B.1.1.7 (Alpha) | B.1.351 (Beta) | B.1.617.2 (Delta) | P.1 (Gamma) | B.1.427 and B.1.429 (Epsilon) | B.1.525 (Eta) | B.1.526 (Iota) | B.1.617.1 (Kappa) | P.2 (Zeta) |
| Origin | United Kingdom | South Africa | India | Brazil and Japan | California, United States of America | United Kingdom and Nigeria | New York, United States of America | India | Brazil |
| Protein changes/mutations | 69del, 70del, 144del, (E484Ka), (S494Pa), N501Y, A570D, D614G, P681H, T716I, S982A, D1118H (K1191N*) | D80A, D215G, 241del, 242del, 243del, K417N, E484K, N501Y, D614G, A701V | T19R, (G142Da), 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I | L452R, D614G, S13I, W152C | A67V, 69del, 70del, 144del, E484K, D614G, Q677H, F888L | L5F, (D80Ga), T95I, (Y144-a), (F157Sa), D253G, (L452Ra), (S477Na), E484K, D614G, A701V, (T859Na), (D950Ha), (Q957Ra) | (T95I), G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H | E484K, (F565La), D614G, V1176F |
| Effects on transmissibility | Increased by 50% | Increased by 50% | Increase (% unclear) | Unclear | Increased by 20% | Unclear | Unclear | Unclear | Unclear |
| Reduce neutralization by sera/antibody | Low impact | Significant reduced neutralization | Potential reduced neutralization | Significant reduced neutralization | Moderate reduced neutralization | Potential reduced neutralization | Reduced neutralization | Potential reduced neutralization | Potential reduced neutralization |
| References | [284], [285], [294], [295], [296], [297], [298], [299], [300], [335], [336], [337], [286], [338], [339], [340], [341], [342], [343], [287], [288], [289], [290], [291], [292], [293] | ||||||||
Amino acid changes were detected in some of the tested sequences but not all of the tested samples. Till date, there is no any variant that is classified as the variant of high consequence (VOHC). The variant list is not limited to the above variants and the list can be expanding from time to time.
VOHC is defined as a variant that would significantly reduce the effectiveness of the preventive measures and medical countermeasures that are designed to eradicate and prevent the transmission of the concerned virus [284], [287], [297]. To date, there is no any SARS-CoV-2 variant that is classified as VOHC [284], [287]. On the other hand, several SARS-CoV-2 variants have been grouped under (VOC) and these include alpha (B1.1.7), beta (B.1.351), delta (B.1.617.2), and gamma (P.1) [284], [287]. VOC is said to have increased transmissibility and the specific mutations which they have acquired would reduce the effectiveness of the currently available vaccines, and there is much scientific evidence to suggest that these variants would increase the disease severity [287], [288], [289], [297], [298], [299]. The spike protein mutations which the VOC could have acquired include E484K, D614G, N501Y, L452R, and many more, and these mutations could increase the virus ability to bind to the human cell receptor to increase their ability to enter and infect the human cells [284], [287], [297] VOI is a variant that is considered to be less virulent than VOC and they have the potential to cause increased transmission of the virus, worsened the disease severity and could probably reduce the virus neutralization by antibodies produced from past infection or vaccination [284], [287]. Examples of VOI include epsilon (B.1.427 and B.1.429), eta (B.1.525), iota (B.1.526), kappa (B.1.617.1), and zeta (P.2) [284], [287]. On contrary, VUM is a variant that was detected through some variant screening and they could have properties similar to VOC but there was limited scientific evidence to group them under VOC [287]. Currently, several variants are classified as VUM and these include lambda (B.1.351+E516Q or B.1.1.7+L452R or B.1.1.7+S494P or C.36+L452R or AT.1 or C.37), Iota (B.1.526.1 or B.1.526.2), and Zeta (B.1.1.519 or AV.1 or P.1+P681H or B.1.671.2+K417N) [287].
Detecting and understanding the variant type of SARS-CoV-2 is essential to help to contain the transmission of the virus in a specific region because certain variants like alpha and delta virus strains are known to accelerate the virus infection in a community and it is well-established that there is reduced in the effectiveness of the currently available vaccines to neutralize these viruses [289], [290], [291], [292], [297], [298], [300]. Currently, several types of detection tests could be performed to detect or identify different SARS-CoV-2 variants ( Table 19) [296]. One of them is RT-PCR in which a target failure or weak target detection could suggest the presence of a mutation like deletion [296], [301]. This technique, however, has a limitation because it cannot differentiate different VOC or VOI because mutations like N501Y are almost present in all variant types [296]. In terms of the test sensitivity, it was reported that the qRT-PCR method could detect virus variants at as low as 4–10 RNA copies per reaction [302], [303], while some studies reported that the LOD of RT-PCR could be ranging from 100 to 1000 RNA copies in 1 mL [301], [304]. By comparing to other detection methods like nucleic acid sequencing, qRT-PCR was shown to have comparable sensitivity and specificity in detecting the SARS-CoV-2 variant types [301], [305].
Table 19.
Detection of various SARS-CoV-2 variants using different methods.
| Methods | Types of detectable variants | Examples of detectable mutations | Key findings | References |
|---|---|---|---|---|
| qPCR/PCR | B.1.1.7 (Alpha) | N501Y | LOD: 4 RNA copies/reaction, Sensitivity and specificity: 100% (MC) | [303] |
| B.1.1.7 (Alpha), B.1.351 (Beta), | N501Y, E484K | LOD: 10 RNA copies/reaction | [302] | |
| B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma) | N501Y | LOD:5000 RNA copies/mL, sensitivity and specificity = 100% | [301] | |
| B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma) | K417N, N501Y | Sensitivity of PCR to detect VOC could be 10% better than whole genome sequencing | [305] | |
| Almost all variants | N501Y, E484K, K417N | RT-PCR and melt curve analyses were able to detect 80% of alpha, 5% of beta, 2% of gamma and 13% of non-variants of SARS-CoV-2 from 989 samples | [344] | |
| Almost all variants | N501Y, D614G | LOD: 100 RNA copies/mL, specificity unclear | [304] | |
| Almost all variants | N501Y, 69del, 70del, K417N, and E484K | Sensitivity and specificity unclear | [345] | |
| B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma) | E484K | LOD: 1–100 copies/μL | [69] | |
| Sensitivity and specificity: cannot differentiate all VOC/VOI | ||||
| Nucleic acid sequencing | B.1.1.7 (Alpha) | N501Y | LOD:18.5 RNA copies/reaction, sensitivity and specificity unclear | [308] |
| B.1.1.7 (Alpha) | P681H, E484K | Nucleic acid sequencing is able to differentiate B.1.1.7 (58%) and non-B1.1.7 infection (42%) | [306] | |
| B.1.1.7 (Alpha), B.1.1.162. B.1.1.267, B.1.1 | D614G, A222V, S982A, T716I, P681H, A570D, E583D | Nucleic acid sequencing was able to detect more than 60% of the SARS-CoV-2 variants from different samples collected from different infection waves. | [307] | |
| B.1.351 (Beta) | K417N, E484K, N501Y | Number of accumulated amino acid changes in whole genome = 341 | [338] | |
| PCR + Nucleic acid sequencing | B.1.1.7 (Alpha) | N501Y, S477N and D614G | LOD: Only samples with Cq value < 25 for N gene could be detected using WGS. Specificity: Unclear | [309] |
| Almost all variants | 69del, 70del, Y144del, N501Y and A570D | Compared to WGS, RT-PCR and sanger sequencing require shorter time and lower cost. LOD unclear | [310] | |
| SNP genotyping | Almost all variants | Unreported | >62% to be able to distinguish two variants of different genotypes | [311] |
| RT-LAMP | P.1 (Gamma), P.2 (Zeta) | N501Y, E484K/Q, K417N/T | Sensitivity: 97%, Specificity: 100% (based on N/E targets) | [312] |
| B.1.1.7 (Alpha) | 69del, 70del | LOD: 39–10,000 RNA copies/reaction | [313] | |
| CRISRP/Cas-based technique | B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma) | N501Y | LOD, sensitivity and specificity unclear | [346] |
| Almost all variants have D614G | D614G | LOD: 10 RNA copies/reaction | [314] | |
| Other variants | E174R/S542R/K548R, S254F | LOD: 50–1000 RNA copies/reaction, specificity: 100% | [315] | |
| Almost all variants have D614G | D614G | LOD: 82 RNA copies/reaction, % specificity unclear | [155] | |
| Antigen test | B.1.1.7 (Alpha), B.1.351 (Beta), | Unclear | LOD: 1.7 × 105 – 6.6 × 107 RNA copies/mL | [316] |
| Sensitivity and specificity: Unclear | ||||
| Antigen and antibody test | B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma) | Unclear | Detection rate of antigen/antibody tests to detect these three variants were reported to be >90% | [317] |
Nucleic acid sequencing remains an essential gold-standard method to detect different SARS-CoV-2 variants because this technique allows the detailed sequencing of the viral genome or specific gene to unravel any mutational changes [296]. Currently, few studies have reported the use of nucleic acid sequencing to identify and differentiate various VOC and VOI [296], [306], [307], and its LOD was reported to be around 19 RNA copies per reaction [308]. Compared to qRT-PCR, nucleic acid sequencing is said to be a more objective and unbiased test to detect the SARS-CoV-2 variant because the test user does not know about the virus information before the test [296]. Besides, some studies reported the use of qRT-PCR to screen for the presence of a specific target mutation, followed by nucleic acid sequencing tests like Sanger sequencing to confirm the mutation [309], [310]. This approach could help to speed up test time and reduce test cost as there is no need to run whole-genome sequencing of the virus to confirm its identity [310]. On contrary, SNP genotyping was also reported to be employed in detecting different SARS-CoV-2 variants and this assay was found to have the ability (>62%) to distinguish two virus variants of different genotypes [311].
Other than RT-PCR and nucleic acid sequencing, the RT-LAMP-based approach was also being reported to be used in the detection of different SARS-CoV-2 variants [312], [313]. This assay was used to identify various variants like Alpha (B.1.1.7) and Gamma (P.1) by detecting mutations like 69del, 70del, and N501Y [312], [313]. The LOD of this test approach was reported to be ranging from 39 to 10,000 RNA copies in a reaction [313], and the test sensitivity and specificity were shown to be around 97%, and 100%, respectively [312]. On the other side, a molecular diagnostic approach that is based on the CRISPR/Cas-based detection technique was also being employed in identifying different SARS-CoV-2 variants and the LOD of this test was reported to vary from 10 to 1000 RNA copies in a reaction [155], [314], [315]. The specific binding and interaction between the Cas enzyme and the RNA strand allow the specific recognition of a unique virus genome mutation [155]. In terms of the assay sensitivity and specificity, the use of the CRISPR/Cas-based detection technique was also shown to provide superior sensitivity and specificity in detecting the SARS-CoV-2 variant (~100%), making it another alternative tool to screen for virus mutation [315].
Although some published protocol has suggested that protein-based tests like viral antigen and the serological test could be potentially used to detect SARS-CoV-2 variant (Table 19), both these tests could not distinguish between different VOI and VOC and are said to have poorer sensitivity and specificity [296]. A study has previously reported the use of virus antigen test to try to identify different SARS-CoV-2 variants and the LOD was reported to be 1.7–6.6 × 107 RNA copies in 1 mL [316]. The test sensitivity and specificity in confirming the identity of different SARS-CoV-2 variants, however, was not described further by the authors [316]. In another study [317], it was demonstrated that antigen and the antibody-based test were sensitive (>80%) to identify three SARS-CoV-2 variants, namely, Alpha (B.1.1.7), Beta (B.1.351), and Gamma (P.1). This suggested that protein-based tests could be helpful to aid in screening identity of virus variants in a shorter test time [317]. However, sequence analysis is still recommended to be done to further confirm the screening test results [296].
5. Conclusion
This review summarizes the various approaches and methods, which could be used to confirm the diagnosis of both the MERS-CoV and SARS-CoV-2 infections. To date, qRT-PCR and viral nucleic acid sequencing serve as the two most commonly used and accurate diagnostic tests to confirm MERS-CoV and SARS-CoV-2 cases. Protein-based detection tests like viral serological tests could aid in determining whether a patient has past or recent acute infection by the coronavirus, but the test might be less sensitive to detect the presence of the antibodies towards the viruses if the individual is in the early stage of infection, or has long recovered from the infection. Virus cell culture and virus neutralization tests could be alternative tests to confirm the coronaviruses infections, but both of these tests require highly trained personnel to perform the tests, and that the test results take a longer time to be available. Clinical evaluation and radiological assessment of the thoracic cavity of the patients might help predict MERS-CoV and SARS-CoV-2 infections. However, confirmatory tests are still required. Compared to MERS-CoV which is now causing sporadic transmission worldwide, SARS-CoV-2 still causes a severe global pandemic crisis. By far qRT-PCR is still the most recommended test to screen for the transmission of SARS-CoV-2 worldwide while rapid antigen tests and serological tests like ELISA could help in screening for the presence of the virus when the qRT-PCR service is unavailable, especially in the remote regions. It is hoped that with the continuous development in molecular diagnostic technology, cheaper, faster, and highly sensitive diagnostic kits could be made available to allow massive screening of the coronavirus worldwide to help block the transmission chain, and thus, the world could be able to return to the normal state before the Covid-19 pandemic started.
Authors’ contributions
Swee Keong Yeap, Chean Yeah Yong, Wan Yong Ho and Stephanie Y.L. Ng conceptualized the idea, developed the structure of the article and guided the selection of references; Zhi Xiong Chong, Winnie Pui Pui Liew, Hui Kian Ong, Chong Seng Shit, Swee Keong Yeap, Chean Yeah Yong, Wan Yong Ho and Stephanie Y.L. Ng prepared, wrote and edited the draft; Zhi Xiong Chong prepared the figures/Tables. All authors have read and agreed to the published version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Xiamen University Research Fund, Malaysia (grant no. XMUMRF/2019-C4/ICAM/0006).
References
- 1.Kahn J.S., McIntosh K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005;24(11):223–227. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 2.Hemida M.G., Perera R.A., Wang P., Alhammadi M.A., Siu L.Y., Li M., Poon L.L., Saif L., Alnaeem A., Peiris M. Middle east respiratory syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Eurosurveillance. 2013;18(50):20659. doi: 10.2807/1560-7917.ES2013.18.50.20659. [DOI] [PubMed] [Google Scholar]
- 3.Masood N., Malik S.S., Raja M.N., Mubarik S., Yu C. Unraveling the epidemiology, geographical distribution, and genomic evolution of potentially lethal coronaviruses (SARS, MERS, and SARS CoV-2) Front. Cell. Infect. Microbiol. 2020;10:499. doi: 10.3389/fcimb.2020.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Quantitative analysis of RNA by HPLC and evaluation of RT-dPCR for coronavirus RNA quantification. New Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.WHO, Middle East respiratory syndrome, 2021. http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html (Accessed 7 March 2021).
- 7.WHO, Middle East respiratorysyndrome coronavirus (MERS- CoV), 2019. https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov) (Accessed 7 March 2021).
- 8.Chen X., Chughtai A.A., Dyda A., Macintyre C.R. Comparative epidemiology of Middle East respiratory syndrome coronavirus (MERS-CoV) in Saudi Arabia and South Korea. Emerg. Microbes Infect. 2017;6(6):51. doi: 10.1038/emi.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuen K.-S., Ye Z.-W., Fung S.-Y., Chan C.-P., Jin D.-Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO, Coronavirus (COVID-19) Dashboard, 2021. https://covid19.who.int/ (Accessed 18 July 2021).
- 12.Cao Y., Hiyoshi A., Montgomery S. COVID-19 case-fatality rate and demographic and socioeconomic influencers: worldwide spatial regression analysis based on country-level data. BMJ Open. 2020;10(11) doi: 10.1136/bmjopen-2020-043560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelrahman Z., Li M., Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A respiratory viruses. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg S.B. Update on human rhinovirus and coronavirus infections. Semin. Respir. Crit. Care Med. 2016;37(4):555–571. doi: 10.1055/s-0036-1584797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections—more than just the common cold. J. Am. Med. Assoc. 2020;323(8):707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 16.Asrani P., Eapen M.S., Chia C., Haug G., Weber H.C., Hassan M.I., Sohal S.S. Diagnostic approaches in COVID-19: clinical updates. Expert Rev. Respir. Med. 2021;15(2):197–212. doi: 10.1080/17476348.2021.1823833. [DOI] [PubMed] [Google Scholar]
- 17.Benzigar M.R., Bhattacharjee R., Baharfar M., Liu G. Current methods for diagnosis of human coronaviruses: pros and cons. Anal. Bioanal. Chem. 2020:1–20. doi: 10.1007/s00216-020-03046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30(3) doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys. Acta Mol. Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue L., Li J., Wei L., Ma C. A quick look at the latest developments in the COVID-19 pandemic. J. Int. Med. Res. 2020;48(9) doi: 10.1177/0300060520943802. [DOI] [Google Scholar]
- 21.Lundstrom K., Seyran M., Pizzol D., Adadi P., Mohamed Abd El-Aziz T., Hassan S.S., Soares A., Kandimalla R., Tambuwala M.M., Aljabali A.A.A., Kumar Azad G., Pal Choudhury P., Uversky V.N., Sherchan S.P., Uhal B.D., Rezaei N., Brufsky A.M. The importance of research on the origin of SARS-CoV-2. Viruses. 2020;12(11):1203. doi: 10.3390/v12111203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatmal M.M., Alshaer W., Al-Hatamleh M.A.I., Hatmal M., Smadi O., Taha M.O., Oweida A.J., Boer J.C., Mohamud R., Plebanski M. Comprehensive structural and molecular comparison of spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their interactions with ACE2. Cells. 2020;9(12):2638. doi: 10.3390/cells9122638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ICTV, Coronaviridae, 2011. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/222/coronaviridae (Accessed 21 February 2021).
- 25.Wang Y., Mao J.-M., Wang G.-D., Luo Z.-P., Yang L., Yao Q., Chen K.-P. Human SARS-CoV-2 has evolved to reduce CG dinucleotide in its open reading frames. Sci. Rep. 2020;10:12331. doi: 10.1038/s41598-020-69342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdel-Moneim A.S. Middle East respiratory syndrome coronavirus (MERS-CoV): evidence and speculations. Arch. Virol. 2014;159(7):1575–1584. doi: 10.1007/s00705-014-1995-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowlatshahi S., Shabani E., Abdekhodaie M.J. Serological assays and host antibody detection in coronavirus-related disease diagnosis. Arch. Virol. 2021;166(3):715–731. doi: 10.1007/s00705-020-04874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WHO, Middle East respiratory syndrome Case definition for reporting to WHO Interim case definition, 2018. https://www.who.int/csr/disease/coronavirus_infections/case_definition/en/%0Ahttp://www.who.int/csr/disease/coronavirus_infections/mers-interim-case-definition.pdf?ua=1 (Accessed 14 February 2021).
- 30.WHO, COVID-19 case definition, 2020. https://www.who.int/publications/i/item/WHO-2019-nCoV-Surveillance_Case_Definition-2020.2 (Accessed 13 March 2021).
- 31.CDC, Standard operating procedure (SOP) for triage of suspected COVID-19 patients in non-US healthcare settings: Early identification and prevention of transmission during triage, 2020. https://www.cdc.gov/coronavirus/2019-ncov/hcp/non-us-settings/sop-triage-prevent-transmission.htmln.html (Accessed 14 March 2021).
- 32.Wu Y.-C., Chen C.-S., Chan Y.-J. The outbreak of COVID-19: an overview. J. Chin. Med. Assoc. 2020;83(3):217–220. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.CDC, MERS clinical features, 2020. 10.7326/m13-2486. [DOI]
- 34.CDC, Overview of testing forSARS-CoV-2 (COVID-19), 2020. https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html (Accessed 13March 2021).
- 35.Chafekar A., Fielding B.C. MERS-CoV: understanding the latest human coronavirus threat. Viruses. 2018;10(2):93. doi: 10.3390/v10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nye S., Whitley R.J., Kong M. Viral infection in the development and progression of pediatric acute respiratory distress syndrome. Front. Pediatr. 2016;4:128. doi: 10.3389/fped.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bauer T.T., Ewig S., Rodloff A.C., Müller E.E. Acute respiratory distress syndrome and pneumonia: a comprehensive review of clinical data. Clin. Infect. Dis. 2006;43(6):748–756. doi: 10.1086/506430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y., Huang Y.-Y., Liu X., Zhang X., Ferrari M., Qin L. Point-of-care technologies for molecular diagnostics using a drop of blood. Trends Biotechnol. 2014;32(3):132–139. doi: 10.1016/j.tibtech.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Connelly J.T., Rolland J.P., Whitesides G.M. “Paper Machine” for molecular diagnostics. Anal. Chem. 2015;87(15):7595–7601. doi: 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- 40.Kohl C., Brinkmann A., Dabrowski P.W., Radonic A., Nitsche A., Kurth A. Protocol for metagenomic virus detection in clinical specimens. Emerg. Infect. Dis. 2015;21(1):48–57. doi: 10.3201/eid2101.140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bursle E., Robson J. Non-culture methods for detecting infection. Aust. Prescr. 2016;39(5):171–175. doi: 10.18773/austprescr.2016.059. https://doi.org/http://dx.doi.org/10.18773/ austprescr.2016.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang P., Croxen M.A., Hasan M.R., Hsiao W.W.L., Hoang L.M. Infection control in the new age of genomic epidemiology. Am. J. Infect. Control. 2017;45(2):170–179. doi: 10.1016/j.ajic.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Eastwood J.R., Ribot R.F.H., Rollins L.A., Buchanan K.L., Walder K., Bennett A.T.D., Berg M.L. Host heterozygosity and genotype rarity affect viral dynamics in an avian subspecies complex. Sci. Rep. 2017;7:13310. doi: 10.1038/s41598-017-13476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adedeji A.O., Sarafianos S.G. Future treatment strategies for novel Middle East respiratory syndrome coronavirus infection. Future Med. Chem. 2013;5(18):2119–2122. doi: 10.4155/fmc.13.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Memish Z.A., Al-Tawfiq J.A., Makhdoom H.Q., Assiri A., Alhakeem R.F., Albarrak A., Alsubaie S., Al-Rabeeah A.A., Hajomar W.H., Hussain R., Kheyami A.M., Almutairi A., Azhar E.I., Drosten C., Watson S.J., Kellam P., Cotton M., Zumla A. Respiratory tract samples, viral load, and genome fraction yield in patients with middle east respiratory syndrome. J. Infect. Dis. 2014;210(10):1590–1594. doi: 10.1093/infdis/jiu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner S.A., Tsongalis G.J. Automation of the molecular diagnostic Laboratory. Diagn. Mol. Pathol. 2017:35–46. doi: 10.1016/b978-0-12-800886-7.00004-2. [DOI] [Google Scholar]
- 47.Butler D. Tensions linger over discovery of coronavirus. Nat. N. 2013;493 doi: 10.1038/nature.2012.12108. [DOI] [Google Scholar]
- 48.Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raj V.S., Osterhaus A.D.M.E., Fouchier R.A.M., Haagmans B.L. MERS: emergence of a novel human coronavirus. Curr. Opin. Virol. 2014;5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park W.B., Kwon N.-J., Choi S.-J., Kang C.K., Choe P.G., Kim J.Y., Yun J., Lee G.-W., Seong M.-W., Kim N.J., Seo J.-S., Oh M. Virus isolation from the first patient with SARS-CoV-2 in Korea. J. Korean Med. Sci. 2020;35(7):84. doi: 10.3346/jkms.2020.35.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith C.J., Osborn A.M. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 2009;67(1):6–20. doi: 10.1111/j.1574-6941.2008.00629.x. [DOI] [PubMed] [Google Scholar]
- 52.Lu X., Whitaker B., Sakthivel S.K.K., Kamili S., Rose L.E., Lowe L., Mohareb E., Elassal E.M., Al-sanouri T., Haddadin A., Erdman D.D. Real-time reverse transcription-PCR assay panel for middle east respiratory syndrome coronavirus. J. Clin. Microbiol. 2014;52(1):67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.WHO, Contacts of Middle East respiratory syndrome Coronavirus (MERS-CoV) patients, 2013. https://www.who.int/csr/disease/coronavirus_infections/WHO_Contact_Protocol_MERSCoV_19_November_2013.pdf (Accessed 15 February 2021).
- 54.CDC, CDC laboratory testing for Middle East Respiratory Syndrome Coronavirus (MERS-CoV), 2013, pp. 1–2. http://www.cdc.gov/coronavirus/mers/lab/lab-testing.html (Accessed 14 February 2021).
- 55.Corman V.M., Müller M.A., Costabel U., Timm J., Binger T., Meyer B., Kreher P., Lattwein E., Eschbach-Bludau M., Nitsche A., Bleicker T., Landt O., Schweiger B., Drexler J.F., Osterhaus A.D., Haagmans B.L., Dittmer U., Bonin F., Wolff T., Drosten C. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveillance. 2012;17(49):20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 56.Corman V.M., Olschlager S., Wendtner C.-M., Drexler J.F., Hess M., Drosten C. Performance and clinical validation of the RealStar® MERS-CoV Kit for detection of Middle East respiratory syndrome coronavirus RNA. J. Clin. Virol. 2014;60(2):168–171. doi: 10.1016/j.jcv.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.WHO, Diagnostic testing forSARS-CoV-2, 2020. https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2 (Accessed 21 March 2021).
- 58.WHO, Criteria for releasing COVID-19 patients from isolation, 2020. https://www.who.int/publications/i/item/criteria-for-releasing-covid-19-patients-from-isolation (Accessed 21 March 2021). [DOI] [PMC free article] [PubMed]
- 59.Shirato K., Nao N., Matsuyama S., Kageyama T. Ultra-rapid real-time RT-PCR method for detecting Middle East respiratory syndrome coronavirus using a mobile PCR device, PCR1100. Jpn. J. Infect. Dis. 2020;73(3):181–186. doi: 10.7883/yoken.JJID.2019.400. [DOI] [PubMed] [Google Scholar]
- 60.Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T.M., Muth D., Müller M.A., Drexler J.F., Zambon M., Osterhaus A.D., Fouchier R.M., Drosten C. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance. 2012;17(39):20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 61.RealStar ® MERS RT-PCR Kit 1.0 07/2017, 2017. https://www.altona-diagnostics.com/en/products/reagents-140/reagents/realstar-real-time-pcr-reagents/realstar-mers-cov-rt-pcr-kit-ce.html (Accessed 12 July 2021).
- 62.Lee J.-S., Ahn J.S., Yu B.S., Cho S.I., Kim M.J., Choi J.M., Seo S.H., Park S.S., Seong M.-W. Evaluation of a real-time reverse transcription-PCR (RT-PCR) assay for detection of middle east respiratory syndrome coronavirus (MERS-COV) in clinical samples from an outbreak in South Korea in 2015. J. Clin. Microbiol. 2017;55(8):2554–2555. doi: 10.1128/JCM.00667-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frans G., Beuselinck K., Peeters B., Van Ranst M., Saegeman V., Desmet S., Lagrou K. Migrating a lab-developed MERS-CoV real-time PCR to 3 “Sample to Result” systems: experiences on optimization and validation. Diagn. Microbiol. Infect. Dis. 2019;94(4):349–354. doi: 10.1016/j.diagmicrobio.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hecht L.-S., Jurado-Jimenez A., Hess M., El Halas H., Bochenek G., Mohammed H., Alzahrani F., Asiri M.O., Hasan R., Alamri A., Alotaibi S. Verification and diagnostic evaluation of the RealStar® Middle East respiratory syndrome coronavirus (N gene) reverse transcription-PCR kit 1.0. Future Microbiol. 2019;14(11):941–948. doi: 10.2217/fmb-2019-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan J.F.-W., Choi G.K.-Y., Tsang A.K.-L., Tee K.-M., Lam H.-Y., Yip C.C.-Y., To K.K.-W., Cheng V.C.-C., Yeung M.-L., Lau S.K.-P., Woo P.C.-Y., Chan K.-H., Tang B.S.-F., Yuen K.-Y. Development and evaluation of novel real-time reverse transcription-PCR assays with locked nucleic acid probes targeting leader sequences of human-pathogenic coronaviruses. J. Clin. Microbiol. 2015;53(8):2722–2726. doi: 10.1128/JCM.01224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falzone L., Musso N., Gattuso G., Bongiorno D., Palermo C.I., Scalia G., Libra M., Stefani S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020;46(3):957–964. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shaffaf T., Ghafar-Zadeh E. Covid-19 diagnostic strategies. Part I: Nucleic acid-based technologies. Bioengineering. 2021;8(4):49. doi: 10.3390/bioengineering8040049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogels C.B.F., Breban M.I., Ott I.M., Alpert T., Petrone M.E., Watkins A.E., Kalinich C.C., Earnest R., Rothman J.E., de Jesus J.G., Claro I.M., Ferreira G.M., Crispim M.A.E., Brazil-UK CADDE Genomic Network, Singh L., Tegally H., Anyaneji U.J., Network for Genomic Surveillance in South Africa, Hodcroft E.B., Mason C.E., Khullar G., Metti J., Dudley J.T., MacKay M.J., Nash M., Wang J., Liu C., Hui P., Murphy S., Neal C., Laszlo E., Landry M.L., Muyombwe A., Downing R., Razeq J., de Oliveira T., Faria N.R., Sabino E.C., Neher R.A., Fauver J.R., Grubaugh N.D. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19(5) doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reijns M.A.M., Thompson L., Acosta J.C., Black H.A., Sanchez-Luque F.J., Diamond A., Parry D.A., Daniels A., O’Shea M., Uggenti C., Sanchez M.C., O’Callaghan A., McNab M.L.L., Adamowicz M., Friman E.T., Hurd T., Jarman E.J., Chee F.L.M., Rainger J.K., Walker M., Drake C., Longman D., Mordstein C., Warlow S.J., McKay S., Slater L., Ansari M., Tomlinson I.P.M., Moore D., Wilkinson N., Shepherd J., Templeton K., Johannessen I., Tait-Burkard C., Haas J.G., Gilbert N., Adams I.R., Jackson A.P. A sensitive and affordable multiplex RT-qPCR assay for SARS-CoV-2 detection. PLoS Biol. 2020;18:1–20. doi: 10.1371/journal.pbio.3001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petrillo S., Carrà G., Bottino P., Zanotto E., De Santis M.C., Margaria J.P., Giorgio A., Mandili G., Martini M., Cavallo R., Barberio D., Altruda F. A novel multiplex qRT-PCR assay to detect sars-cov-2 infection: High sensitivity and increased testing capacity. Microorganisms. 2020;8(7):1064. doi: 10.3390/microorganisms8071064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hur K.H., Park K., Lim Y., Jeong Y.S., Sung H., Kim M.N. Evaluation of four commercial kits for SARS-CoV-2 real-time reverse-transcription polymerase chain reaction approved by emergency-use-authorization in Korea. Front Med (Lausanne) 2020;7:521. doi: 10.3389/fmed.2020.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freire-Paspuel B., Garcia-Bereguiain M.A. Clinical performance and analytical sensitivity of three SARS-CoV-2 nucleic acid diagnostic tests. Am. J. Trop. Med. Hyg. 2021;104(4):1516–1518. doi: 10.4269/ajtmh.20-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y., Dai C., Wang H., Gao Y., Li T., Fang Y., Shen Z., Chen L., Chen Z., Ma X., Li M. Analysis and validation of a highly sensitive one-step nested quantitative real-time polymerase chain reaction assay for specific detection of severe acute respiratory syndrome coronavirus 2. Virol. J. 2020;17(1):197. doi: 10.1186/s12985-020-01467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J., Cai K., Zhang R., He X., Shen X., Liu J., Xu J., Qiu F., Lei W., Wang J., Li X., Gao Y., Jiang Y., Xu W., Ma X. Novel one-step single-tube nested quantitative real-time PCR assay for highly sensitive detection of SARS-CoV-2. Anal. Chem. 2020;92(13):9399–9404. doi: 10.1021/acs.analchem.0c01884. [DOI] [PubMed] [Google Scholar]
- 76.Clerici B., Muscatello A., Bai F., Pavanello D., Orlandi M., Marchetti G.C., Castelli V., Casazza G., Costantino G., Podda G.M. Sensitivity of SARS-CoV-2 detection with nasopharyngeal swabs. Front. Public Heal. 2021;8:1–5. doi: 10.3389/fpubh.2020.593491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H., Fung A.Y.-F., Ng A.C.-K., Zou Z., Tsoi H.-W., Choi G.K.-Y., Tam A.R., Cheng V.C.-C., Chan K.-H., Tsang O.T.-Y., Yuen K.-Y. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel, 2020. https://www.fda.gov/media/134922/download (Accessed 10 July 2021). [DOI] [PMC free article] [PubMed]
- 79.Labcorp’s COVID-19 RT-PCR testEUA summary, 2021. 10.31857/s0026898421010080. [DOI]
- 80.Liu X., Feng J., Zhang Q., Guo D., Zhang L., Suo T., Hu W., Guo M., Wang X., Huang Z., Xiong Y., Chen G., Chen Y., Lan K. Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microbes Infect. 2020;9(1):1175–1179. doi: 10.1080/22221751.2020.1772679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Niu C., Dong L., Gao Y., Zhang Y., Wang X., Wang J. Quantitative analysis of RNA by HPLC and evaluation of RT-dPCR for coronavirus RNA quantification. Talanta. 2021;228 doi: 10.1016/j.talanta.2021.122227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taylor S.C., Laperriere G., Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 2017;7(1):2409. doi: 10.1038/s41598-017-02217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brandolini M., Taddei F., Marino M.M., Grumiro L., Scalcione A., Turba M.E., Gentilini F., Fantini M., Zannoli S., Dirani G., Sambri V. Correlating qRT-PCR, dPCR and viral titration for the identification and quantification of sars-cov-2: a new approach for infection management. Viruses. 2021;13(6):1022. doi: 10.3390/v13061022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duong K., Ou J., Li Z., Lv Z., Dong H., Hu T., Zhang Y., Hanna A., Gordon S., Crynen G., Head S.R., Ordoukhanian P., Wang Y. Increased sensitivity using real-time dPCR for detection of SARS-CoV-2. Biotechniques. 2020;70(1):7–20. doi: 10.2144/BTN-2020-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mio C., Cifù A., Marzinotto S., Marcon B., Pipan C., Damante G., Curcio F. Validation of a one-step reverse transcription-droplet digital PCR (RT-ddPCR) approach to detect and quantify SARS-CoV-2 RNA in nasopharyngeal swabs. Dis. Markers. 2021;2021 doi: 10.1155/2021/8890221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park C., Lee J., ul Hassan Z., Ku K.B., Kim S.-J., Kim H.G., Park E.C., Park G.-S., Park D., Baek S.-H., Park D., Lee J., Jeon S., Kim S., Lee C.-S., Yoo H.M., Kim S. Comparison of digital PCR and quantitative PCR with various sars-cov-2 primer-probe sets. J. Microbiol. Biotechnol. 2021;31(3):358–367. doi: 10.4014/JMB.2009.09006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alteri C., Cento V., Antonello M., Colagrossi L., Merli M., Ughi N., Renica S., Matarazzo E., Di Ruscio F., Tartaglione L., Colombo J., Grimaldi C., Carta S., Nava A., Costabile V., Baiguera C., Campisi D., Fanti D., Vismara C., Fumagalli R., Scaglione F., Epis O.M., Puoti M., Perno C.F. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One. 2020;15(9) doi: 10.1371/journal.pone.0236311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O.-E., Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alsolamy S., Arabi Y.M. Infection with Middle East respiratory syndrome coronavirus. Can. J. Respir. Ther. 2015;51(4):102–103. [PMC free article] [PubMed] [Google Scholar]
- 90.Feikin D.R., Alraddadi B., Qutub M., Shabouni O., Curns A., Oboho I.K., Tomczyk S.M., Wolff B., Watson J.T., Madani T.A. Association of higher MERS-CoV virus load with severe disease and death, Saudi Arabia, 2014. Emerg. Infect. Dis. 2015;21(11):2029–2035. doi: 10.3201/eid2111.150764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka-Trocha A., Atyeo C., Fischinger S., Chan A., Flaherty K.T., Hall K., Dougan M., Ryan E.T., Gillespie E., Chishti R., Li Y., Jilg N., Hanidziar D., Baron R.M., Baden L., Tsibris A.M., Armstrong K.A., Kuritzkes D.R., Alter G., Walker B.D., Yu X., Li J.Z. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Prescott J., Falzarano D., de Wit E., Hardcastle K., Feldmann F., Haddock E., Scott D., Feldmann H., Munster V.J. Pathogenicity and viral shedding of MERS-CoV in immunocompromised rhesus macaques. Front. Immunol. 2018;9:205. doi: 10.3389/fimmu.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou Y., Pei F., Ji M., Wang L., Zhao H., Li H., Yang W., Wang Q., Zhao Q., Wang Y. Sensitivity evaluation of 2019 novel coronavirus (SARS-CoV-2) RT-PCR detection kits and strategy to reduce false negative. PLoS One. 2020;15(11) doi: 10.1371/journal.pone.0241469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shirato K., Yano T., Senba S., Akachi S., Kobayashi T., Nishinaka T., Notomi T., Matsuyama S. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol. J. 2014;11:139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P.M., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Kräusslich H.-G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12(556):eabc7075. doi: 10.1126/SCITRANSLMED.ABC7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.El Wahed A.A., Patel P., Heidenreich D., Hufert F.T., Weidmann M. Reverse transcription recombinase polymerase amplification assay for the detection of middle east respiratory syndrome coronavirus. PLoS Curr. 2013;5 doi: 10.1371/currents.outbreaks.62df1c7c75ffc96cd59034531e2e8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xia S., Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT–RPA. Cell Discov. 2020;6:37. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee S.H., Baek Y.H., Kim Y.-H., Choi Y.-K., Song M.-S., Ahn J.-Y. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front. Microbiol. 2016;7:2166. doi: 10.3389/fmicb.2016.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Y., Chan K.H., Hong C., Kang Y., Ge S., Chen H., Wong E.Y.M., Joseph S., Patteril N.G., Wernery U., Xia N., Lau S.K.P., Woo P.C.Y. A highly specific rapid antigen detection assay for on-site diagnosis of MERS. J. Infect. 2016;73(1):82–84. doi: 10.1016/j.jinf.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang P., Wang H., Cao Z., Jin H., Chi H., Zhao J., Yu B., Yan F., Hu X., Wu F., Jiao C., Hou P., Xu S., Zhao Y., Feng N., Wang J., Sun W., Wang T., Gao Y., Yang S., Xia X. A rapid and specific assay for the detection of MERS-CoV. Front. Microbiol. 2018;9:1101. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park S., Lee J.W. Detection of coronaviruses using RNA toehold switch sensors. Int. J. Mol. Sci. 2021;22(4):1772. doi: 10.3390/ijms22041772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shirato K., Semba S., El-kafrawy S.A., Hassan A.M., Tolah A.M., Takayama I., Kageyama T., Notomi T., Kamitani W., Matsuyama S., Azhar E.I. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J. Virol. Methods. 2018;258:41–48. doi: 10.1016/j.jviromet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I.L., Kim B.-T., Maeng J.-S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baek Y.H., Um J., Antigua K.J.C., Park J.-H., Kim Y., Oh S., Kim Y., Choi W.-S., Kim S.G., Jeong J.H., Chin B.S., Nicolas H.D.G., Ahn J.-Y., Shin K.S., Choi Y.K., Park J.-S., Song M.-S. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 2020;9(1):998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dong Y., Wu X., Li S., Lu R., Li Y., Wan Z., Qin J., Yu G., Jin X., Zhang C. Comparative evaluation of 19 reverse transcription loop-mediated isothermal amplification assays for detection of SARS-CoV-2. Sci. Rep. 2021;11:2936. doi: 10.1038/s41598-020-80314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mautner L., Baillie C.-K., Herold H.M., Volkwein W., Guertler P., Eberle U., Ackermann N., Sing A., Pavlovic M., Goerlich O., Busch U., Wassill L., Huber I., Baiker A. Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol. J. 2020;17:160. doi: 10.1186/s12985-020-01435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., Stewart de Ramirez S.A., Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117(37):22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Inaba M., Higashimoto Y., Toyama Y., Horiguchi T., Hibino M., Iwata M., Imaizumi K., Doi Y. Diagnostic accuracy of LAMP versus PCR over the course of SARS-CoV-2 infection. Int. J. Infect. Dis. 2021;107:195–200. doi: 10.1016/j.ijid.2021.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.MobileDetect-BIO BCC19 testkit-FDA, 2020. https://www.accessdata.fda.gov/scripts/medwatch/index.cfm?action=reporting.home (Accessed 10 July 2021).
- 112.Lucira COVID-19 All-in-one testkit instruction for use-FDA, 2020. https://www.fda.gov/media/143808/download (Accessed 10 July 2021).
- 113.AQ-TOPTM COVID-19 rapid detection kit, 2020. https://www.fda.gov/media/138307/download (Accessed 11 July 2021).
- 114.SARS-CoV-2 RNA DETECTR Assay, 2020. https://www.fda.gov/media/139937/download (Accessed 10 July 2021).
- 115.CapitalBio’s Isothermal Microfludic Chip Analyzer gains CE certification, 2020. http://en.thholding.com.cn/2020-04/28/c_488807.htm (Accessed 11 July 2021).
- 116.Moore M.D., Jaykus L.-A. Recombinase polymerase amplification: a promising point-of-care detection method for enteric viruses. Future Virol. 2017;12(8):421–429. doi: 10.2217/fvl-2017-0034. [DOI] [Google Scholar]
- 117.Huang P., Jin H., Zhao Y., Li E., Yan F., Chi H., Wang Q., Han Q., Mo R., Song Y., Bi J., Jiao C., Li W., He H., Wang H., Ma A., Feng N., Wang J., Wang T., Yang S., Gao Y., Xia X., Wang H. Nucleic acid visualization assay for middle east respiratory syndrome coronavirus (Mers-cov) by targeting the upE and N gene. PLoS Negl. Trop. Dis. 2021;15(3) doi: 10.1371/journal.pntd.0009227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Go Y.Y., Kim Y.-S., Cheon S., Nam S., Ku K.B., Kim M., Cho N.H., Park H., Lee P.-Y.A., Tsai Y.-C., Tsai Y.-L., Wang H.-T.T., Balasuriya U.B.R. Evaluation and clinical validation of two field-deployable reverse transcription-insulated isothermal PCR assays for the detection of the Middle East respiratory syndrome-coronavirus. J. Mol. Diagnos. 2017;19(6):817–827. doi: 10.1016/j.jmoldx.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lau Y.L., binti Ismail I., binti Mustapa N.I., Lai M.Y., Soh T.S.T., Hassan A.H., Peariasamy K.M., Lee Y.L., Kahar M.K.B.A., Chong J., Goh P.P. Development of a reverse transcription recombinase polymerase amplification assay for rapid and direct visual detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) PLoS One. 2021;16(1) doi: 10.1371/journal.pone.0245164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Behrmann O., Bachmann I., Spiegel M., Schramm M., Abd El Wahed A., Dobler G., Dame G., Hufert F.T. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (Exo-IQ) Clin. Chem. 2020;66(8):1047–1054. doi: 10.1093/clinchem/hvaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chang P.-L., Lin C.-Y., Chen C.-P., Lin Y.-C., Hu H.-C., Cheng S.-H., Cheng C.-Y. Clinical validation of an automated reverse transcription-insulated isothermal PCR assay for the detection of severe acute respiratory syndrome coronavirus 2. J. Microbiol. Immunol. Infect. 2021;54(3):522–526. doi: 10.1016/j.jmii.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sun Y., Yu L., Liu C., Ye S., Chen W., Li D., Huang W. One‑tube SARS‑CoV‑2 detection platform based on RT‑RPA and CRISPR/Cas12a. J. Transl. Med. 2021;19:74. doi: 10.1186/s12967-021-02741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lau E.H.Y., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.W., Chan W., Chiu S.S., Ko R.L.W., Chan K.H., Cheng S.M.S., Perera R.A.P.M., Cowling B.J., Poon L.L.M., Peiris M. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021;12:63. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.El Wahed A.A., Patel P., Maier M., Pietsch C., Rüster D., Böhlken-Fascher S., Kissenkötter J., Behrmann O., Frimpong M., Diagne M.M., Faye M., Dia N., Shalaby M.A., Amer H., Elgamal M., Zaki A., Ismail G., Kaiser M., Corman V.M., Niedrig M., Landt O., Faye O., Sall A.A., Hufert F.T., Truyen U., Liebert U.G., Weidmann M. Suitcase lab for rapid detection of SARS-CoV-2 based on recombinase polymerase amplification assay. Anal. Chem. 2021;93(4):2627–2634. doi: 10.1021/acs.analchem.0c04779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kandeil A., Shehata M.M., El Shesheny R., Gomaa M.R., Ali M.A., Kayali G. Complete genome sequence of Middle East respiratory syndrome coronavirus isolated from a dromedary camel in Egypt. Genome Announc. 2016;4(2):e00309–e00316. doi: 10.1128/genomeA.00309-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.WHO, Laboratory testing for Middle East Respiratory Syndrome Coronavirus, 2018. https://www.who.int/csr/disease/coronavirus_infections/mers-laboratory-testing/en/ (Accessed 14 February 2021).
- 127.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 128.Islam M.R., Hoque M.N., Rahman M.S., Alam A.S.M.R.U., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10:14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.AlBalwi M.A., Khan A., AlDrees M., GK U., Manie B., Arabi Y., Alabdulkareem I., AlJohani S., Alghoribi M., AlAskar A., AlAjlan A., Hajeer A. Evolving sequence mutations in the Middle East respiratory syndrome coronavirus (MERS-CoV) J. Infect. Public Health. 2020;13(10):1544–1550. doi: 10.1016/j.jiph.2020.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yassim A.S.M., Asras M.F.F., Gazali A.M., Marcial-Coba M.S., Zainulabid U.A., Ahmad H.F. COVID-19 outbreak in Malaysia: decoding D614G mutation of SARS-CoV-2 virus isolated from an asymptomatic case in Pahang. Mater. Today Proc. 2021 doi: 10.1016/j.matpr.2021.02.387. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Speranza E., Williamson B.N., Feldmann F., Sturdevant G.L., Perez-Perez L., Meade-White K., Smith B.J., Lovaglio J., Martens C., Munster V.J., Okumura A., Shaia C., Feldmann H., Best S.M., de Wit E. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci. Transl. Med. 2021;13(578) doi: 10.1126/scitranslmed.abe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Furuse Y., Okamoto M., Oshitani H. Conservation of nucleotide sequences for molecular diagnosis of Middle East respiratory syndrome coronavirus. Int. J. Infect. Dis. 2015;40(2015):25–27. doi: 10.1016/j.ijid.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Instructions for use: qSanger-COVID-19 assay, 2020. https://www.fda.gov/media/141935/download (Accessed 11 July 2021).
- 135.Helix COVID-19 NGS Test, 2020. https://www.fda.gov/media/140917/download#:~:text=The Helix COVID-19 NGS Test is an amplicon based,anterior nasal swabs) from individuals (Accessed 10 July 2021).
- 136.UCLA Swabseq COVID-19 diagnostic platform-FDA, 2020. https://www.fda.gov/media/142805/download (Accessed 10 July 2021).
- 137.Clear Dx TM SARS-CoV-2 Test, 2021. https://www.fda.gov/media/142805/download (Accessed 10 July 2021).
- 138.IVD testing for COVID-19 using LamPORE, 2021. https://nanoporetech.com/covid-19/lampore (Accessed 10 July 2021).
- 139.Ion AmpliSeq SARS-CoV-2 Insight research assay, 2020. https://www.thermofisher.com/my/en/home/life-science/sequencing/dna-sequencing/microbial-sequencing/microbial-identification-ion-torrent-next-generation-sequencing/viral-typing/coronavirus-research.html (Accessed 11 July 2021).
- 140.CleanPlex ® for MGI SARS-CoV-2 Panel, 2020. https://www.paragongenomics.com/product/cleanplex-for-mgi-sars-cov-2-panel/ (Accessed 11 July 2021).
- 141.Tools Address. SARS-CoV-2 Virus, 2020. https://www.twistbioscience.com/productchild/tools-addressing-sars-cov-2-virus (Accessed 11 July 2021).
- 142.Pfeiffer F., Gröber C., Blank M., Händler K., Beyer M., Schultze J.L., Mayer G. Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Sci. Rep. 2018;8:10950. doi: 10.1038/s41598-018-29325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.International Human Genome Sequencing Consortium Erratum: Initial sequencing and analysis of the human genome: International Human Genome Sequencing Consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 144.Javalkote V.S., Kancharla N., Bhadra B., Shukla M., Soni B., Sapre A., Goodin M., Bandyopadhyay A., Dasgupta S. CRISPR-based assays for rapid detection of SARS-CoV-2. Methods. 2020 doi: 10.1016/j.ymeth.2020.10.003. (Article in Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R.R., Zhou Q., Li W. SARS-CoV-2 detection with CRISPR diagnostics. Cell Disco. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fozouni P., Son S., Díaz de León Derby M., Knott G.J., Gray C.N., D’Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.-L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.-Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184(2):323–333.e9. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Palaz F., Kalkan A.K., Tozluyurt A., Ozsoz M. CRISPR-based tools: alternative methods for the diagnosis of COVID-19. Clin. Biochem. 2021;89:1–13. doi: 10.1016/j.clinbiochem.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xiong D., Dai W., Gong J., Li G., Liu N., Wu W., Pan J., Chen C., Jiao Y., Deng H., Ye J., Zhang X., Huang H., Li Q., Xue L., Zhang X., Tang G. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PLoS Biol. 2020;18(12) doi: 10.1371/journal.pbio.3000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brandsma E., Verhagen H.J.M.P., Van De Laar T.J.W., Claas E.C.J., Cornelissen M., Van Den Akker E. Rapid, sensitive, and specific severe acute respiratory syndrome coronavirus 2 detection: a multicenter comparison between standard quantitative reverse-transcriptase polymerase chain reaction and CRISPR-Based DETECTR. J. Infect. Dis. 2021;223(2):206–213. doi: 10.1093/infdis/jiaa641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mayuramart O., Nimsamer P., Rattanaburi S., Chantaravisoot N., Khongnomnan K., Chansaenroj J., Puenpa J., Suntronwong N., Vichaiwattana P., Poovorawan Y., Payungporn S. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp. Biol. Med. 2021;246(4):400–405. doi: 10.1177/1535370220963793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Joung J., Ladha A., Saito M., Kim N.-G., Woodley A.E., Segel M., Barretto R.P.J., Ranu A., Macrae R.K., Faure G., Ioannid E.I., Krajeski R.N., Bruneau R., Huang M.-L.W., Yu X.G., Li J.Z., Walker B.D., Hung D.T., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsou J.H., Liu H., Stass S.A., Jiang F. Rapid and sensitive detection of SARS-CoV-2 using clustered regularly interspaced short palindromic repeats. Biomedicines. 2021;9(3):1–11. doi: 10.3390/BIOMEDICINES9030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xiong E., Jiang L., Tian T., Hu M., Yue H., Huang M., Lin W., Jiang Y., Zhu D., Zhou X. Simultaneous dual-gene diagnosis of SARS-CoV-2 based on CRISPR/Cas9-mediated lateral flow assay. Angew. Chem. Int. Ed. 2021;60(10):5307–5315. doi: 10.1002/anie.202014506. [DOI] [PubMed] [Google Scholar]
- 155.Wang Y., Zhang Y., Chen J., Wang M., Zhang T., Luo W., Li Y., Wu Y., Zeng B., Zhang K., Deng R., Li W. Detection of SARS-CoV-2 and Its mutated variants via CRISPR-Cas13-based transcription amplification. Anal. Chem. 2021;93(7):3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- 156.Xiu L., Zhang C., Wu Z., Peng J. Establishment and application of a universal coronavirus screening method using MALDI-TOF mass spectrometry. Front. Microbiol. 2017;8:1510. doi: 10.3389/fmicb.2017.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wandernoth P., Kriegsmann K., Groh-Mohanu C., Daeumer M., Gohl P., Harzer O., Kriegsmann M., Kriegsmann J. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by mass spectrometry. Viruses. 2020;12(8):849. doi: 10.3390/v12080849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rybicka M., Miłosz E., Bielawski K.P. Superiority of MALDI-TOF mass spectrometry over real-time PCR for SARS-CoV-2 RNA detection. Viruses. 2021;13(5):730. doi: 10.3390/v13050730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang J., Zhang D., Chen Q., Yu J., Yang Y., Qiao L., Sun Y. Establishment and application of PCR-time-of-flight mass spectrometer for SARS-CoV-2 nucleic ncid detection, Chinese. Gen. Pract. 2020;23(35):4430–4435. [Google Scholar]
- 160.Hernandez M.M., Banu R., Shrestha P., Patel A., Chen F., Cao L., Fabre S., Tan J., Lopez H., Chiu N., Shifrin B., Zapolskaya I., Flores V., Lee P.Y., Castañeda S., Ramírez J.D., Jhang J., Osorio G., Gitman M.R., Nowak M.D., Reich D.L., Cordon-Cardo C., Sordillo E.M., Paniz-Mondolfi A.E. RT-PCR/MALDI-TOF mass spectrometry-based detection of SARS-CoV-2 in saliva specimens. J. Med. Virol. 2021:1–6. doi: 10.1002/jmv.27069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.SARS-COV-2 Massarray test, 2020. https://www.fda.gov/media/142548/download (Accessed 11 July 2021).
- 162.Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal. Chem. 2017;89(10):5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19(3):171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lee S. Steering the private sector in COVID-19 diagnostic test kit development in South Korea. Front Public Health. 2020;8 doi: 10.3389/fpubh.2020.563525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xpert® Xpress SARS-CoV-2, 2021 . https://www.fda.gov/media/136314/download (Accessed 15 July 2021).
- 167.SARS-CoV-2 RT-PCR Assay (SalivaDirect Assay), 2021. 10.31857/s0026898421010080. [DOI]
- 168.Srivatsan S., Heidl S., Pfau B., Martin B.K., Han P.D., Zhong W., van Raay K., McDermot E., Opsahl J., Gamboa L., Smith N., Truong M., Cho S., Barrow K.A., Rich L.M., Stone J., Wolf C.R., McCulloch D.J., Kim A.E., Brandstetter E., Sohlberg S.L., Llcisin M., Geyer R.E., Chen W., Gehring J., Seattle Flu Study Investigators, Kosuri S., Bedford T., Rieder M.J., Nickerson D.A., Chu H.Y., Konnick E.Q., Debley J.S., Shendure J., Lockwood C.M., Starita L.M. SwabExpress: an end-to-end protocol for extraction-free COVID-19 testing. BioRxiv. 2021 doi: 10.1101/2020.04.22.056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Visseaux B., Collin G., Houhou-Fidouh N., Le Hingrat Q., Ferr V.M., Damond F., Ichou H., Descamps D., Charpentier C. Evaluation of three extraction-free SARS-CoV-2 RT-PCR assays: a feasible alternative approach with low technical requirements. J. Virol. Methods. 2021;291 doi: 10.1016/j.jviromet.2021.114086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xfree TM COVID-19 qRT-PCR test, 2021. https://biogx.com/wp-content/uploads/2021/04/BioGX-XfreeTM-COVID-19-Direct-RT-PCR-Product-Overview-Rev.-04.pdf (Accessed 15 July 2021).
- 171.Advanta Dx SARS-CoV-2 RT-PCRassay, 2020. https://www.fda.gov/media/141541/download (Accessed 15 July 2021).
- 172.VereRTTM ZeroPrepTMCOVID-19 PCR Kit, 2020. https://www.hsa.gov.sg/docs/default-source/hprg-mdb/veredus_verert-zeroprep-_provisional-authorisation-for-covid-19-tests_07122020.pdf (Accessed 15 July 2021).
- 173.Quantabio’s RT-qPCR ToughMixes, 2020. https://www.quantabio.com/media/contenttype/MK-AN-0007_REV01_COVID19_Testing_0321.pdf (Accessed 15 July 2021).
- 174.FastPlex TM Triplex SARS-CoV-2 detection kits, 2020. https://www.precigenome.com/coronavirus-covid-19-pcr-kit-qpcr (Accessed 15 July 2021).
- 175.ViroReal ® Kit RT-LAMP SARS-CoV-2, n.d. https://www.ingenetix.com/wp-content/uploads/2021/01/ViroReal-KIT-LAMP-PCR-SARS-CoV-2_Description_IVD_v1–1en.pdf (Accessed 15 July 2021).
- 176.Azmi I., Faizan M.I., Kumar R., Raj Yadav S., Chaudhary N., Kumar Singh D., Butola R., Ganotra A., Datt Joshi G., Deep Jhingan G., Iqbal J., Joshi M.C., Ahmad T. A saliva-based RNA extraction-free workflow integrated with Cas13a for SARS-CoV-2 detection. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.632646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.QSANGER TM -COVID-19 ASSAY, 2021. https://swiftbiosci.com/rna-extraction-free-qsanger-covid-19-test-kit/ (Accessed 15 July 2021).
- 178.WHO, Middle East respiratory syndrome coronavirus (MERS-CoV), 2018. https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1 (Accessed 14 February 2021).
- 179.Chen Y., Chan K.-H., Kang Y., Chen H., Luk H.K.H., Poon R.W.S., Chan J.F.W., Yuen K.-Y., Xia N., Lau S.K.P., Woo P.C.Y. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerg. Microbes Infect. 2015;4(4):26. doi: 10.1038/emi.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Fan C., Huang W., Xu M., Wang Y. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9(1):680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Amanat F., White K.M., Miorin L., Strohmeier S., McMahon M., Meade P., Liu W.-C., Albrecht R.A., Simon V., Martinez-Sobrido L., Moran T., García-Sastre A., Krammer F. An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr. Protoc. Microbiol. 2020;58(1) doi: 10.1002/cpmc.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Padoan A., Zuin S., Cosma C., Basso D., Plebani M. Clinical performances of an ELISA for SARS-CoV-2 antibody assay and correlation with neutralization activity. Clin. Chim. Acta. 2020;510:654–655. doi: 10.1016/j.cca.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Michel M., Bouam A., Edouard S., Fenollar F., Di Pinto F., Mège J.-L., Drancourt M., Vitte J. Evaluating ELISA, immunofluorescence, and lateral flow assay for SARS-CoV-2 serologic assays. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.597529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Edouard S., Colson P., Melenotte C., de Pinto F., Thomas L., la Scola B., Million M., Tissot-Dupont H., Gautret P., Stein A., Brouqui P., Parola P., Lagier J.-C., Raoult D., Drancourt M. Evaluating the serological status of COVID-19 patients using an indirect immunofluorescent assay, France. Eur. J. Clin. Microbiol. Infect. Dis. 2020;40:361–371. doi: 10.1101/2020.05.05.20092064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.CDC, Interim guidelines for COVID-19 antibody testing., 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html (Accessed 28 March 2021).
- 186.Hemida M.G., Al-Naeem A., Perera R.A.P.M., Chin A.W.H., Poon L.L.M., Peiris M. Lack of middle east respiratory syndrome coronavirus transmission from infected camels. Emerg. Infect. Dis. 2015;21(4):699–701. doi: 10.3201/eid2104.141949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Müller M.A., Meyer B., Corman V.M., Al-Masri M., Turkestani A., Ritz D., Sieberg A., Aldabbagh S., Bosch B.-J., Lattwein E., Alhakeem R.F., Assiri A.M., Albarrak A.M., Al-Shangiti A.M., Al-Tawfiq J.A., Wikramaratna P., Alrabeeah A.A., Drosten C., Memish Z.A. Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infect. Dis. 2015;15(5):559–564. doi: 10.1016/S1473-3099(15)70090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Al Johani S., Hajeer A.H. MERS-CoV diagnosis: an update. J. Infect. Public Health. 2016;9(3):216–219. doi: 10.1016/j.jiph.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Park S.W., Perera R.A.P.M., Choe P.G., Lau E.H.Y., Choi S.J., Chun J.Y., Oh H.S., Song K., Bang J.H., Kim E.S., Kim H.B., Park W.B., Kim N.J., Poon L.L.M., Peiris M., Oh M. Comparison of serological assays in human Middle East respiratory syndrome (MERS)-coronavirus infection. Eurosurveillance. 2015;20(41):30042. doi: 10.2807/1560-7917.ES.2015.20.41.30042. [DOI] [PubMed] [Google Scholar]
- 191.Nakagawa K., Narayanan K., Wada M., Makino S. Inhibition of stress granule formation by Middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J. Virol. 2018;92(20):e00902–e00918. doi: 10.1128/jvi.00902-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Drosten C., Meyer B., Müller M.A., Corman V.M., Al-Masri M., Hossain R., Madani H., Sieberg A., Bosch B.J., Lattwein E., Alhakeem R.F., Assiri A.M., Hajomar W., Albarrak A.M., Al-Tawfiq J.A., Zumla A.I., Memish Z.A. Transmission of MERS-coronavirus in household contacts. New Engl. J. Med. 2014;371(9):828–835. doi: 10.1056/nejmoa1405858. [DOI] [PubMed] [Google Scholar]
- 193.Aburizaiza A.S., Mattes F.M., Azhar E.I., Hassan A.M., Memish Z.A., Muth D., Meyer B., Lattwein E., Müller M.A., Drosten C. Investigation of anti-Middle East respiratory syndrome antibodies in blood donors and slaughterhouse workers in Jeddah and Makkah, Saudi Arabia, Fall 2012. J. Infect. Dis. 2014;209(2):243–246. doi: 10.1093/infdis/jit589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chan K.-H., Chan J.F.-W., Tse H., Chen H., Lau C.C.-Y., Cai J.-P., Tsang A.K.-L., Xiao X., To K.K.-W., Lau S.K.-P., Woo P.C.-Y., Zheng B.-Ji, Wang M., Yuen K.-Y. Cross-reactive antibodies in convalescent SARS patients’ sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J. Infect. 2013;67(2):130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bold D., Van Doremalen N., Myagmarsuren O., Zayat B., Munster V.J., Richt J.A. Middle East respiratory syndrome-coronavirus seropositive Bactrian camels, Mongolia. Vector-Borne Zoonotic Dis. 2021;21(2):128–131. doi: 10.1089/vbz.2020.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Clinical performance of different SARS-CoV-2 IgG antibody tests. J. Med. Virol. 2020;92(10):2243–2247. doi: 10.1002/jmv.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nguyen D., Skelly D., Goonawardane N. A novel immunofluorescence assay for the rapid serological detection of SARS-CoV-2 infection. Viruses. 2021;13(5):747. doi: 10.3390/v13050747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang D., He S., Wang X., Yan Y., Liu J., Wu S., Liu S., Lei Y., Chen M., Li L., Zhang J., Zhang L., Hu X., Zheng X., Bai J., Zhang Y., Zhang Y., Song M., Tang Y. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4(12):1150–1158. doi: 10.1038/s41551-020-00655-z. [DOI] [PubMed] [Google Scholar]
- 199.Andrey D.O., Cohen P., Meyer B., Torriani G., Yerly S., Mazza L., Calame A., Arm-Vernez I., Guessous I., Stringhini S., Roux-Lombard P., Fontao L., Agoritsas T., Stirnemann J., Reny J.-L., Siegrist C.-A., Eckerle I., Kaiser L., Vuilleumier N. Head-to-head accuracy comparison of three commercial COVID-19 IgM/IgG serology rapid tests. JCM. 2020;9(8):2369. doi: 10.3390/jcm9082369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Trivedi S., Miao C., Al-Abdallat M.M., Haddadin A., Alqasrawi S., Iblan I., Nsour M.A., Alsanouri T., Ali S.S., Rha B., Gerber S.I., Payne D.C., Tamin A., Thornburg N.J. Inclusion of MERS-spike protein ELISA in algorithm to determine serologic evidence of MERS-CoV infection. J. Med. Virol. 2018;90(2):367–371. doi: 10.1002/jmv.24948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Okba N.M.A., Raj V.S., Widjaja I., GeurtsvanKessel C.H., de Bruin E., Chandler F.D., Park W.B., Kim N.-J., Farag E.A.B., Al-Hajri M., Bosch B.-J., Oh M., Koopmans M.P.G., Reuskan C.B.E.M., Haagmans B.L. Sensitive and specific detection of low-level antibody responses in mild Middle East respiratory syndrome coronavirus infections. Emerg. Infect. Dis. 2019;25(10):1868–1877. doi: 10.3201/eid2509.190051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sil B.K., Jahan N., Haq M.A., Oishee M.J., Ali T., Khandker S.S., Kobatake E., Mie M., Khondoker M.U., Jamiruddin M.R., Adnan N. Development and performance evaluation of a rapid in-house ELISA for retrospective serosurveillance of SARS-CoV-2. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0246346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tozetto-Mendoza T.R., Kanunfre K.A., Vilas-Boas L.S., Espinoza E.P.S., Paiao H.G.O., Rocha M.C., de Paula A.V., de Oliveira M.S., Zampelli D.B., Jr J.M.V., Buss L., Costa S.F., Sabino E.C., Witkin S.S., Okay T.S., Mendes-Correa M.C. Nucleoprotein-based ELISA for detection of SARS-COV-2 IgG antibodies: could an old assay be suitable for serodiagnosis of the new coronavirus? J. Virol. Methods. 2021;290 doi: 10.1016/j.jviromet.2021.114064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rongqing Z., Li M., Song H., Chen J., Ren W., Feng Y., Gao G.F., Song J., Peng Y., Su B., Guo X., Wang Y., Chen J., Li J., Sun H., Bai Z., Cao W., Zhu J., Zhang Q., Sun Y., Sun S., Mao X., Su J., Chen X., He A., Gao W., Jin R., Jiang Y., Sun L. Early detection of severe acute respiratory syndrome coronavirus 2 antibodies as a serologic marker of infection in patients with coronavirus disease 2019. Clin. Infect. Dis. 2020;71(16):2066–2072. doi: 10.1093/cid/ciaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Krahling V., Halwe S., Rohde C., Becker D., Berghofer S., Dahlke C., Eickmann M., Ercanoglu M.S., Gieselmann L., Herwig A., Kupke A., Muller H., Neubauer-Radel P., Klein F., Keller C., Becker S. Development and characterization of an indirect ELISA to detect SARS-CoV-2 spike protein-specific antibodies. J. Immunol. Methods. 2021;490 doi: 10.1016/j.jim.2021.112958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Okba N.M.A., Müller M.A., Li W., Wang C., Geurtsvankessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., De Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gededzha M.P., Mampeule N., Jugwanth S., Zwane N., David A., Burgers W.A., Blackburn J.M., Grove J.S., George J.A., Sanne I., Scott L., Stevens W., Mayne E.S. Performance of the EUROIMMUN anti-SARSCoV-2 ELISA assay for detection of IgA and IgG antibodies in South Africa. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0252317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Peterhoff D., Glück V., Vogel M., Schuster P., Schütz A., Neubert P., Albert V., Frisch S., Kiessling M., Pervan P., Werner M., Ritter N., Babl L., Deichner M., Hanses F., Lubnow M., Müller T., Lunz D., Hitzenbichler F., Audebert F., Hähnel V., Offner R., Müller M., Schmid S., Burkhardt R., Glück T., Koller M., Niller H.H., Graf B., Salzberger B., Wenzel J.J., Jantsch J., Gessner A., Schmidt B., Wagner R. A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization. Infection. 2021;49(1):75–82. doi: 10.1007/s15010-020-01503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mehdi F., Chattopadhyay S., Thiruvengadam R., Yadav S., Kumar M., Sinha S.K., Goswami S., Kshetrapal P., Wadhwa N., Chandramouli Natchu U., Sopory S., Koundinya Desiraju B., Pandey A.K., Das A., Verma N., Sharma N., Sharma P., Bhartia V., Gosain M., Lodha R., Lamminmäki U., Shrivastava T., Bhatnagar S., Batra G. Development of a fast SARS-CoV-2 IgG ELISA, based on receptor-binding domain, and its comparative evaluation using temporally segregated samples from RT-PCR positive individuals. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.618097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ainsworth M., Andersson M., Auckland K., Baillie J.K., Barnes E., Beer S., Beveridge A., Bibi S., Blackwell L., Borak M., Bown A., Brooks T., Burgess-Brown N.A., Camara S., Catton M., Chau K.K., Christott T., Clutterbuck E., Coker J., Cornall R.J., Cox S., Crawford-Jones D., Crook D.W., D’Arcangelo S., Dejnirattsai W., Dequaire J., Dimitriadis S., Dingle K.E., Doherty G., Dold C., Dong T., Dunachie S.J., Ebner D., Emmenegger M., Espinosa A., Eyre D.W., Fairhead R., Fassih S., Feehily C., Felle S., Fernandez-Cid A., Fernandez Mendoza M., Foord T.H., Fordwoh T., Fox McKee D., Frater J., Gallardo Sanchez V., Gent N., Georgiou D., Groves C.J., Hallis B., Hammond P.M., Hatch S.B., Harvala H.J., Hill J., Hoosdally S.J., Horsington B., Howarth A., James T., Jeffery K., Jones E., Justice A., Karpe F., Kavanagh J., Kim D.S., Kirton R., Klenerman P., Knight J.C., Koukouflis L., Kwok A., Leuschner U., Levin R., Linder A., Lockett T., Lumley S.F., Marinou S., Marsden B.D., Martinez J., Martins Ferreira L., Mason L., Matthews P.C., Mentzer A.J., Mobbs A., Mongkolsapaya J., Morrow J., Mukhopadhyay S., Neville M.J., Oakley S., Oliveira M., Otter A., Paddon K., Pascoe J., Peng Y., Perez E., Perumal P.K., Peto T., Pickford H., Ploeg R.J., Pollard A.J., Richardson A., Ritter T.G., Roberts D.J., Rodger G., Rollier C.S., Rowe C., Rudkin J.K., Screaton G., Semple M.G., Sienkiewicz A., Silva-Reyes L., Skelly D.T., Sobrino Diaz A., Stafford L., Stockdale L., Stoesser N., Street T., Stuart D.I., Sweed A., Taylor A., Thraves H., Tsang H.P., Verheul M.K., Vipond R., Walker T.M., Wareing S., Warren Y., Wells C., Wilson C., Withycombe K., Young R.K. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect. Dis. 2020;20(12):1390–1400. doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Manthei D.M., Whalen J.F., Schroeder L.F., Sinay A.M., Li S.-H., Valdez R., Giacherio D.A., Gherasim C. Differences in performance characteristics among four high-throughput assays for the detection of antibodies against SARS-CoV-2 using a common set of patient samples. Am. J. Clin. Pathol. 2021;155(2):267–279. doi: 10.1093/ajcp/aqaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.MacMullan M.A., Ibrayeva A., Trettner K., Deming L., Das S., Tran F., Moreno J.R., Casian J.G., Chellamuthu P., Kraft J., Kozak K., Turner F.E., Slepnev V.I., Page L.M. Le. ELISA detection of SARS-CoV-2 antibodies in saliva. Sci. Rep. 2020;10(1):20818. doi: 10.1038/s41598-020-77555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58(6):e00461–20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Di Domenico M., De Rosa A., Boccellino M. Detection of SARS-COV-2 proteins using an ELISA test. Diagnostics. 2021;11(4):698. doi: 10.3390/diagnostics11040698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yassine H.M., Al-Jighefee H., Al-Sadeq D.W., Dargham S.R., Younes S.N., Shurrab F., Marei R.M., Hssain A.A., Taleb S., Alhussain H., Al-Nesf M.A., Al-Khal A., Qotba H., Althani A.A., Tang P., Abu-Raddad L.J., Nasrallah G.K. Performance evaluation of five ELISA kits for detecting anti-SARS-COV-2 IgG antibodies. Int. J. Infect. Dis. 2021;102:181–187. doi: 10.1016/j.ijid.2020.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lagerqvist N., Maleki K.T., Verner-Carlsson J., Olausson M., Dillner J., Wigren Byström J., Monsen T., Forsell M., Eriksson J., Bogdanovic G., Muschiol S., Ljunggren J., Repo J., Kjerstadius T., Muradrasoli S., Brytting M., Szekely Björndal Å., Åkerlund T., Nilsson C., Klingström J. Evaluation of 11 SARS-CoV-2 antibody tests by using samples from patients with defined IgG antibody titers. Sci. Rep. 2021;11(1):7614. doi: 10.1038/s41598-021-87289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Padoan A., Bonfante F., Pagliari M., Bortolami A., Negrini D., Zuin S., Bozzato D., Cosma C., Sciacovelli L., Plebani M. Analytical and clinical performances of five immunoassays for the detection of SARS-CoV-2 antibodies in comparison with neutralization activity. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Platelia SARS-CoV-2 Total Ab, 2020. https://www.fda.gov/media/138810/download (Accessed 10 July 2021).
- 219.Dimension Vista SARS-CoV-2 Total Antibody Assay, 2021. https://www.fda.gov/media/145015/download (Accessed 20 July 2021).
- 220.InBios SCoV-2 DetectTM IgG/M ELISA, 2021. https://www.fda.gov/media/138810/download (Accessed 20 July 2021).
- 221.Anti-SARS-CoV-2 ELISA (IgG), 2020. https://www.fda.gov/media/139053/download (Accessed 20 July 2021).
- 222.COVID-19 ELISA Pan-Ig Antibody Test, 2020. https://www.fda.gov/media/141777/download (Accessed 20 July 2021).
- 223.COVID-SeroKlir, Kantaro Semi-Quantitative SARS-CoV-2 IgG Antibody Kit, 2020. https://www.fda.gov/media/144010/download (Accessed 20 July 2021).
- 224.OmniPATH COVID-19 Total Antibody ELISA test, 2020. https://www.fda.gov/media/142700/download (Accessed 20 July 2021).
- 225.COVID-19 Self-Collected Antibody Test System, 2021. https://www.fda.gov/media/147368/download (Accessed 20 July 2021).
- 226.UBI SARS-CoV-2 ELISA, 2021. https://www.fda.gov/media/145277/download (Accessed 20 July 2021).
- 227.WANTAI SARS-CoV-2 Ab ELISA Kit, 2020. https://www.fda.gov/media/140929/download (Accessed 20 July 2020).
- 228.COVID-19 ELISA IgG Antibody Test – Mount Sinai, 2020. https://www.fda.gov/media/137029/download (Accessed 20 July 2021).
- 229.GenScript cPassTM SARS-CoV-2 Neutralization Antibody Detection Kit, 2020. https://www.genscript.com/cpass-sars-cov-2-neutralization-antibody-detection-Kit.html (Accessed 20 July 2021).
- 230.Shaffaf T., Ghafar-Zadeh E. COVID-19 diagnostic strategies part II: Protein-based technologies. Bioengineering. 2021;8(5):54. doi: 10.3390/bioengineering8050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Perera R.A., Wang P., Gomaa M.R., El-Shesheny R., Kandeil A., Bagato O., Siu L.Y., Shehata M.M., Kayed A.S., Moatasim Y., Li M., Poon L.L., Guan Y., Webby R.J., Ali M.A., Peiris J.S., Kayali G. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Eurosurveillance. 2013;18(36):20574. doi: 10.2807/1560-7917.ES2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- 232.Mackay I.M., Arden K.E. MERS coronavirus: diagnostics, epidemiology and transmission. Virol. J. 2015;12:222. doi: 10.1186/s12985-015-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hemida M., Perera R., Al Jassim R., Kayali G., Siu L., Wang P., Chu D., Perlman S., Ali M., Alnaeem A., Poon L., Saif L., Peiris M. Seroepidemiology of Middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Eurosurveillance. 2014;19(23):20828. doi: 10.2807/1560-7917.es2014.19.23.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Reusken C.B., Ababneh M., Raj V.S., Meyer B., Eljarah A., Abutarbush S., Godeke G.J., Bestebroer T.M., Zutt I., Müller M.A., Bosch B.J., Rottier P.J., Osterhaus A.D., Drosten C., Haagmans B.L., Koopmans M.P. Middle east respiratory syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Eurosurveillance. 2013;18(50):20662. doi: 10.2807/1560-7917.ES2013.18.50.20662. [DOI] [PubMed] [Google Scholar]
- 235.Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C.Y., Lu L., Liu Q., Wang L., Feng Y., Wang Y., Zheng B.-J., Yuen K.-Y., Jiang S., Dimitrov D.S. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88(14):7796–7805. doi: 10.1128/jvi.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Jääskeläinen A., Kuivanen S., Kekalainen E., Ahava M., Loginov R., Kallio-Kokko H., Vapalahti O., Jarva H., Kurkela S., Lappalainen M. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bennett R.S., Postnikova E.N., Liang J., Gross R., Mazur S., Dixit S., Kocher G., Yu S., Georgia-Clark S., Gerhardt D., Cai Y., Marron L., Lukin V.V., Holbrook M.R. Scalable, micro-neutralization assay for assessment of sars-cov-2 (Covid-19) virus-neutralizing antibodies in human clinical samples. Viruses. 2021;13(5):893. doi: 10.3390/v13050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bonelli F., Blocki F.A., Bunnell T., Chu E., Arriel De La O., Grenache D.G., Marzucchi G., Montomoli E., Okoye L., Pallavicini L., Streva V.A., Torelli A., Wagner A., Zanin D., Zierold C., Wassenberg J.J. Evaluation of the automated LIAISON® SARS-CoV-2 TrimericS IgG assay for the detection of circulating antibodies. Clin. Chem. Lab. Med. 2021;59(8):1463–1467. doi: 10.1515/cclm-2021-0023. [DOI] [PubMed] [Google Scholar]
- 239.Barchuk A., Shirokov D., Sergeeva M., Tursunzade R., Dudkina O., Tychkova V., Barabanova L., Skougarevskiy D., Danilenko D. Evaluation of the performance of SARS-CoV-2 antibody assays for a longitudinal populationbased study of COVID-19 spread in St. Petersburg, Russia. J. Med. Virol. 2021 doi: 10.1002/jmv.27126. Article in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Walker G.J., Naing Z., Stella A.O., Yeang M., Caguicla J., Ramachandran V., Isaacs S.R., Agapiou D., Bull R.A., Stelzer-Braid S., Daly J., Gosbell I.B., Hoad V.C., Irving D.O., Pink J.M., Turville S., Kelleher A.D., Rawlinson W.D. Sars coronavirus-2 microneutralisation and commercial serological assays correlated closely for some but not all enzyme immunoassays. Viruses. 2021;13(2):247. doi: 10.3390/v13020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lester S., Harcourt J., Whitt M., Al-Abdely H.M., Midgley C.M., Alkhamis A.M., Aziz Jokhdar H.A., Assiri A.M., Tamin A., Thornburg N. Middle East respiratory coronavirus (MERS-CoV) spike (S) protein vesicular stomatitis virus pseudoparticle neutralization assays offer a reliable alternative to the conventional neutralization assay in human seroepidemiological studies. Access Microbiol. 2019;1(9) doi: 10.1099/acmi.0.000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Al Kahlout R.A., Nasrallah G.K., Farag E.A., Wang L., Lattwein E., Müller M.A., El Zowalaty M.E., Al Romaihi H.E., Graham B.S., Al Thani A.A., Yassine H.M. Comparative serological study for the prevalence of anti-MERS coronavirus antibodies in high- and low-risk groups in Qatar. J. Immunol. Res. 2019;2019 doi: 10.1155/2019/1386740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.-H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Dispinseri S., Secchi M., Pirillo M.F., Tolazzi M., Borghi M., Brigatti C., De Angelis M.L., Baratella M., Bazzigaluppi E., Venturi G., Sironi F., Canitano A., Marzinotto I., Tresoldi C., Ciceri F., Piemonti L., Negri D., Cara A., Lampasona V., Scarlatti G. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat. Commun. 2021;12(1):2670. doi: 10.1038/s41467-021-22958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Perera R.A., Mok C.K., Tsang O.T., Lv H., Ko R.L., Wu N.C., Yuan M., Leung W.S., Chan J.M., Chik T.S., Choi C.Y., Leung K., Chan K.H., Chan K.C., Li K.-C., Wu J.T., Wilson I.A., Monto A.S., Poon L.L., Peiris M. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Eurosurveillance. 2020;25(16) doi: 10.2807/1560-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Valcourt E.J., Manguiat K., Robinson A., Chen J.C.-Y., Dimitrova K., Philipson C., Lamoureux L., McLachlan E., Schiffman Z., Drebot M.A., Wood H. Evaluation of a commercially-available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) Diagn. Microbiol. Infect. Dis. 2021;99(4) doi: 10.1016/j.diagmicrobio.2020.115294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.CDC, Interim guidance for rapid antigen testing for SARS-CoV-2, 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html (Accessed 28 March 2021).
- 248.Coronavirus (COVID-19) update: FDA authorizes antigen test as first over-the-counter fully at-home diagnostic test for COVID-19, 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-antigen-test-first-over-counter-fully-home-diagnostic (Accessed 10 July 10).
- 249.Mak G.C.K., Cheng P.K.C., Lau S.S.Y., Wong K.K.Y., Lau C.S., Lam E.T.K., Chan R.C.W., Tsang D.N.C. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Osterman A., Baldauf H.-M., Eletreby M., Wettengel J.M., Afridi S.Q., Fuchs T., Holzmann E., Maier A., Döring J., Grzimek-Koschewa N., Muenchhoff M., Protzer U., Kaderali L., Keppler O.T. Evaluation of two rapid antigen tests to detect SARS-CoV-2 in a hospital setting. Med. Microbiol. Immunol. 2021;210(1):65–72. doi: 10.1007/s00430-020-00698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Peña M., Ampuero M., Garcés C., Gaggero A., García P., Velasquez M.S., Luza R., Alvarez P., Paredes F., Acevedo J., Farfán M.J., Solari S., Soto-Rifo R., Valiente-Echeverría F. Performance of SARS-CoV-2 rapid antigen test compared with real-time RT-PCR in asymptomatic individuals. Int. J. Infect. Dis. 2021;107:201–204. doi: 10.1016/j.ijid.2021.04.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Salvagno G.L., Gianfilippi G., Bragantini D., Henry B.M., Lippi G. Clinical Assessment of the Roche SARS-CoV-2 rapid antigen test. Diagnosis. 2021 doi: 10.1515/dx-2020-0154. Article in press. [DOI] [PubMed] [Google Scholar]
- 253.Peto T., UK COVID- Lateral Flow Oversight T. COVID-19: rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine. 2021;36 doi: 10.1016/j.eclinm.2021.100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Nsoga M.T.N., Kronig I., Rodriguez F.J.P., Sattonnet-Roche P., Da Silva D., Helbling J., Sacks J.A., de Vos M., Boehm E., Gayet- Ageron A., Berger A., Jacquerioz-Bausch F., Chappuis F., Kaiser L., Schibler M., Renzoni A., Eckerle I. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0253321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.K K.K., UC S., Maganthy V., Prakash A.A., Basumatary J., Adappa K., S C., TG N., S G.P.N., B P., Gangasagara S.B., L S.R.B. Pre-operative SARS CoV-2 rapid antigen test and reverse transcription polymerase chain reaction: a conundrum in surgical decision making. Indian J. Ophthalmol. 2021;69(6):1560–1562. doi: 10.4103/ijo.IJO_430_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Corman V.M., Haage V.C., Bleicker T., Schmidt M.L., Mühlemann B., Zuchowski M., Jo W.K., Tscheak P., Möncke-Buchner E., Müller M.A., Krumbholz A., Drexler J.F., Drosten C. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. Lancet Microbe. 2021;2:e311–319. doi: 10.1016/S2666-5247(21)00056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Chaimayo C., Kaewnaphan B., Tanlieng N., Athipanyasilp N., Sirijatuphat R., Chayakulkeeree M., Angkasekwinai N., Sutthent R., Puangpunngam N., Tharmviboonsri T., Pongraweewan O., Chuthapisith S., Sirivatanauksorn Y., Kantakamalakul W., Horthongkham N. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol. J. 2020;17(1):177. doi: 10.1186/s12985-020-01452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Van Honacker E., Van Vaerenbergh K., Boel A., De Beenhouwer H., Leroux-Roels I., Cattoir L. Comparison of five SARS-CoV-2 rapid antigen detection tests in a hospital setting and performance of one antigen assay in routine practice: a useful tool to guide isolation precautions. J. Hosp. Infect. 2021;S0195–6701(21):00142-0–0. doi: 10.1016/j.jhin.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.BinaxNOW COVID-19 Ag Card, 2020. https://www.fda.gov/media/141567/download (Accessed 20 July 2021).
- 260.CareStart COVID-19 Antigen Test Kit, 2021. https://www.fda.gov/media/142916/download (Accessed 20 July 2021).
- 261.GenBody COVID-19 Antigen Test, 2021 . http://diagnostici.eurospital.it/FILES/files/pdf-covid-2020/WIN 004Rev0 COVID19 GenBody EN.pdf (Accessed 20 July 2021).
- 262.InteliSwab COVID-19 RAPID Test Pro, 2021.
- 263.Sienna-Clarity COVID-19 Antigen Rapid Test Cassette, 2021. https://www.fda.gov/media/149055/download (Accessed 20 July 2021).
- 264.SCoV-2 Ag Detect TM Rapid Test, 2020. https://inbios.com/scov-2-ag-detecttm-rapid-test/ (Accessed 20 July 2021).
- 265.Veritor TM System For Rapid Detection of SARS-CoV-2, 2021. https://bdveritor.bd.com/content/dam/bdveritor/pdfs/BD-Veritor-IFU.pdf (Accessed 20 July 2021).
- 266.Status COVID-19/Flu, 2021. https://www.fda.gov/media/145694/download (Accessed 20 July 2021).
- 267.ellume.lab COVID Antigen Test, 2021. https://www.fda.gov/media/150684/download (Accessed 20 July 2021).
- 268.LumiraDx SARS-CoV-2 Ag Test EUA, 2021. https://www.fda.gov/media/141301/download (accessed 20 July 2021).
- 269.Saeed U., Uppal S.R., Piracha Z.Z., Rasheed A., Aftab Z., Zaheer H., Uppal R. Evaluation of SARS-CoV-2 antigen-based rapid diagnostic kits in Pakistan: formulation of COVID-19 national testing strategy. Virol. J. 2021;18:34. doi: 10.1186/s12985-021-01505-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Gillim-Ross L., Taylor J., Scholl D.R., Ridenour J., Masters P.S., Wentworth D.E. Discovery of novel human and animal cells infected by the severe acute respiratory syndrome coronavirus by replication-specific multiplex reverse transcription-PCR. J. Clin. Microbiol. 2004;42(7):3196–3206. doi: 10.1128/JCM.42.7.3196-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87(23):12552–12561. doi: 10.1128/jvi.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kumar S., Sarma P., Kaur H., Prajapat M., Bhattacharyya A., Avti P., Sehkhar N., Kaur H., Bansal S., Mahendiratta S., Mahalmani V.M., Singh H., Prakash A., Kuhad A., Medhi B. Clinically relevant cell culture models and their significance in isolation, pathogenesis, vaccine development, repurposing and screening of new drugs for SARS-CoV-2: a systematic review. Tissue Cell. 2021;70 doi: 10.1016/j.tice.2021.101497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Barrett P.N., Berezuk G., Fritsch S., Aichinger G., Hart M.K., El-Amin W., Kistner O., Ehrlich H.J. Efficacy, safety, and immunogenicity of a Vero-cell-culture-derived trivalent influenza vaccine: a multicentre, double-blind, randomised, placebo-controlled trial. Lancet. 2011;377(9767):751–759. doi: 10.1016/S0140-6736(10)62228-3. https://doi.org/doi:10.1016/S0140-6736(10)62228-3. [DOI] [PubMed] [Google Scholar]
- 275.Jefferson T., Spencer E.A., Brassey J., Heneghan C. Viral cultures for COVID-19 infectious potential assessment – a systematic review. Clin. Infect. Dis. 2020:ciaa1764. doi: 10.1093/cid/ciaa1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Leland D.S., Ginocchio C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007;20(1):49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Franquet T., Jeong Y.J., Lam H.Y.S., Wong H.Y.F., Chang Y.-C., Chung M.J., Lee K.S. Imaging findings in coronavirus infections: SARS-CoV, MERS-CoV, and SARS-CoV-2. Br. J. Radiol. 2020;93(1112) doi: 10.1259/bjr.20200515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ajlan A.M., Ahyad R.A., Jamjoom L.G., Alharthy A., Madani T.A. Middle East respiratory syndrome coronavirus (MERS-CoV) infection: Chest CT findings. Am. J. Roentgenol. 2014;203(4):782–787. doi: 10.2214/AJR.14.13021. [DOI] [PubMed] [Google Scholar]
- 279.Hamimi A. MERS-CoV: Middle East respiratory syndrome corona virus: can radiology be of help? Initial single center experience. Egypt. Soc. Radiol. Nucl. Med. 2016;47(1):95–106. doi: 10.1016/j.ejrnm.2015.11.004. [DOI] [Google Scholar]
- 280.Zhan N., Guo Y., Tian S., Huang B., Tian X., Zou J., Xiong Q., Tang D., Zhang L., Dong W. Clinical characteristics of COVID-19 complicated with pleural effusion. BMC Infect. Dis. 2021;21(1):176. doi: 10.1186/s12879-021-05856-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Du C.-J., Liu J.-Y., Chen H., Yan S., Pu L., Xiong H.-F., Xiang P., Li C.-S., Zhang M., Xie R.-M., Chen B.-D., Li A. Differences and similarities of high-resolution computed tomography features between pneumocystis pneumonia and cytomegalovirus pneumonia in AIDS patients. Infect. Dis. Poverty. 2020;9(1):149. doi: 10.1186/s40249-020-00768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Okada F., Ono A., Ando Y., Nakayama T., Ishii R., Sato H., Kira A., Tokimatsu I., Kadota J., Mori H. Thin-section CT findings in Pseudomonas aeruginosa pulmonary infection. Br. J. Radiol. 2012;85(1020):1533–1538. doi: 10.1259/bjr/54468236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chioma O.S., Drake W.P. Role of microbial agents in pulmonary fibrosis, Yale. J. Biol. Med. 2017;90(2):219–227. [PMC free article] [PubMed] [Google Scholar]
- 284.CDC, SARS-CoV-2 variant classifications and definitions, 2021. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html%0Ahttps://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html%0Ahttps://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-in (Accessed 4 July 2021).
- 285.FDA, Fact sheet for health care providers emergency use authorization (EUA) of REGEN-COV (casirivimab and imdevimab), 2021. https://pi.lilly.com/eua/bamlanivimab-eua-factsheet-hcp.pdf (Accessed 4July 2021).
- 286.FDA, Fact sheet for health care providers emergency use authorization (EUA) of bamlanivimab and etesevimab, 2021. https://pi.lilly.com/eua/bamlanivimab-eua-factsheet-hcp.pdf (Accessed 4 July 2021).
- 287.ECDPC, SARS-CoV-2 variants of concern as of 1 July, 2021. https://www.ecdc.europa.eu/en/covid-19/variants-concern (Accessed 3 July 2021).
- 288.Johnson & Johnson, Johnson & Johnson COVID-19 vaccine authorized by U.S. FDA for emergency use - First single-shot vaccine in fight against global pandemic, 2021. https://www.jnj.com/johnson-johnson-covid-19-vaccine-authorized-by-u-s-fda-for-emergency-usefirst-single-shot-vaccine-in-fight-against-global-pandemic (Accessed 4 July 2021).
- 289.Novavx, Novavax COVID-19 vaccine demonstrates 89.3% efficacy in UK phase 3 trial, 2021. 〈https://mvec.mcri.edu.au/novavax-covid-19-vaccine-demonstrates-89–3-efficacy-in-uk-phase-3-trial/〉 (Accessed 4 July 2021).
- 290.Annavajhala M.K., Mohri H., Wang P., Nair M., Zucker J.E., Sheng Z., Gomez-Simmonds A., Kelley A.L., Tagliavia M., Huang Y., Bedford T., Ho D.D., Uhlemann A.-C. A novel SARS-CoV-2 variant of concern, B.1.526, identified in New York. MedRxiv. 2021 doi: 10.1101/2021.02.23.21252259. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Garcia-Beltran W.F., Lam E.C., Denis K., St., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., Sigal A., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity | Elsevier Enhanced Reader. Cell. 2021;184(9):2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S., Reddy N.P., Sanchez San Martin C., Federman S., Cheng J., Balcerek J., Taylor J., Streithorst J.A., Miller S., Sreekumar B., Chen P.-Y., Schulze-Gahmen U., Taha T.Y., Hayashi J.M., Simoneau C.R., Kumar G.R., McMahon S., Lidsky P.V., Xiao Y., Hemarajata P., Green N.M., Espinosa A., Kath C., Haw M., Bell J., Hacker J.K., Hanson C., Wadford D.A., Anaya C., Ferguson D., Frankino P.A., Shivram H., Lareau L.F., Wyman S.K., Ott M., Andino R., Chiu C.Y. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184(13):3426–3437.e8. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Yadav P.D., Sapkal G.N., Abraham P., Ella R., Deshpande G., Patil D.Y., Nyayanit D.A., Gupta N., Sahay R.R., Shete A.M., Panda S., Bhargava B., Mohan K. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin. Infect. Dis. 2021:ciab411. doi: 10.1093/cid/ciab411. [DOI] [PubMed] [Google Scholar]
- 294.Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S., Carfi A., Corbett K.S., Seder R.A., Edwards D.K. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. BioRxiv. 2021 doi: 10.1101/2021.01.25.427948. (preprint) [DOI] [Google Scholar]
- 295.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 296.WHO, Methods for the detection and identification of SARS-CoV-2 variants (Tecnical report) 2021. https://www.who.int/publications/i/item/diagnostic.
- 297.WHO, Tracking SARS-CoV-2 variants, 2021. https://www.cdph.ca.gov/Programs/CID/DCDC/Pages/COVID-19/COVID-Variants.aspx (Accessed 29 June 2021).
- 298.Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Personalized Virology Initiative Study Group, Krammer F., Simon V., Martinez-Sobrido L., Garcia-Sastre A., Schotsaert M. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2(7):E283–E284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.NERVTAG, NERVTAG: Update note on B.1.1.7 severity, 11 February, 2021. https://www.gov.uk/government/publications/nervtag-update-note-on-b117-severity-11-february-2021 (Accessed 3 July 2021).
- 300.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS- CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Korukluoglu G., Kolukirik M., Bayrakdar F., Ozgumus G.G., Altas A.B., Cosgun Y., Kolukirik C.Z. Ketre. 40 min RT-qPCR assay for screening Spike N501Y and HV69-70del mutations. BioRxiv. 2021 doi: 10.1101/2021.01.26.428302. (preprint) [DOI] [Google Scholar]
- 302.Barua S., Hoque M., Kelly P.J., Bai J., Hanzlicek G., Noll L., Walz H., Johnson C., Kyriakis C., Wang C. High‐resolution melting curve FRET‐PCR rapidly identifies SARS‐CoV‐2 mutations. J. Med. Virol. 2021;93:5588–5593. doi: 10.1002/jmv.27139. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Banada P., Green R., Banik S., Chopoorian A., Streck D., Jones R., Chakravorty S., Alland D. A simple RT-PCR melting temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. MedRxiv. 2021 doi: 10.1101/2021.03.05.21252709. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Shen S., Fu A.Y., Jamba M., Li J., Powell M.J., Zhang A., Lu C.M., Sha M.Y. A flexible, pan-species, multi-antigen platform for the detection and monitoring of SARS-CoV-2-specific antibody responses. MedRxiv. 2021 doi: 10.1101/2021.04.01.21254484. (preprint) [DOI] [Google Scholar]
- 305.Matic N., Lowe C.F., Ritchie G., Stefanovic A., Lawson T., Jang W., Young M., Dong W., Brumme Z.L., Brumme C.J., Leung V., Romney M.G. Rapid detection of SARS-CoV-2 variants of concern, including B.1.1.28/P.1, British Columbia, Canada. Emerg. Infect. Dis. 2021;27(6):1673–1676. doi: 10.3201/eid2706.210532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., Scott R., Sconza R., Price J., Margaritis M., Bergstrom M., Spyer M.J., Miralhes P.B., Grant P., Kirk S., Valerio C., Mangera Z., Prabhahar T., Moreno-Cuesta J., Arulkumaran N., Singer M., Shin G.Y., Sanchez E., Paraskevopoulou S.M., Pillay D., McKendry R.A., Mirfenderesky M., Houlihan C.F., Nastouli E. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021;3099(21):1–11. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.P.W.G M., F C., G G., W T., A.A G.L., M M., de B.E., O Y., J.S L., C.J W., J.G K., E F., M C., P D., P.D C. Whole-genome sequencing of SARS-CoV-2 in the Republic of Ireland during waves 1 and 2 of the pandemic. MedRxiv. 2021 doi: 10.1101/2021.02.09.21251402. [DOI] [Google Scholar]
- 308.Bar-Or I., Weil M., Indenbaum V., Bucris E., Bar-Ilan D., Elul M., Levi N., Aguvaev I., Cohen Z., Shirazi R., Erster O., Sela-Brown A., Sofer D., Mor O., Mendelson E., Zuckerman N.S. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci. Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.148002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Bal A., Destras G., Gaymard A., Stefic K., Marlet J., Eymieux S., Regue H., Semanas Q., D’Aubarede C., Billaud G., Laurent F., Gonzalez C., Mekki Y., Valette M., Bouscambert M., Gaudy-Graffin C., Lina B., Morfin F., Josset L. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69–V70, France, August to December 2020. Euro Surveill. 2021;26(3) doi: 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.La Rosa G., Mancini P., Ferraro G.B., Veneri C., Iaconelli M., Lucentini L., Bonadonna L., Brusaferro S., Brandtner D., Fasanella A., Pace L., Parisi A., Galante D., Suffredini E. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Harper H., Burridge A., Winfield M., Finn A., Davidson A., Matthews D., Hutchings S., Vipond B., Jain N. Detecting SARS-CoV-2 variants with SNP genotyping. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0243185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Alves P.A., de Oliveira E.G., Franco-Luiz A.P.M., Almeida L.T., Goncalves A.B., Borges I.A., de F., Rocha S., Rocha R.P., Bezerra M.F., Miranda P., Capanema F.D., Martins H.R., Weber G., Teixeira S.M.R., Wallau G.L., do Monte-Neto R.L. Clinical validation of colorimetric RT-LAMP a fast highly sensitive and specific COVID-19 molecular diagnostic tool that is robust to detect SARS-CoV-2 variants of concern. MedRxiv. 2021 doi: 10.1101/2021.05.26.21257488. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Sherrill-Mix S., Van Duyne G.D., Bushman F.D. Molecular beacons allow specific RT-LAMP detection of B.1.1.7 variant SARS-CoV-2. MedRxiv. 2021 doi: 10.1101/2021.03.25.21254356. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Meng Q., Wang X., Wang Y., Dang L., Liu X., Ma X., Chi T., Wang X., Zhao Q., Yang G., Liu M., Huang X., Ma P. Detection of the SARS-CoV-2 D614G mutation using engineered Cas12a guide RNA. Biotechnol. J. 2021;16(6) doi: 10.1002/biot.202100040. [DOI] [PubMed] [Google Scholar]
- 315.Ooi K.H., Liu M.M., Tay J.W.D., Teo S.Y., Kaewsapsak P., Jin S., Lee C.K., Hou J., Maurer-Stroh S., Lin W., Yan B., Yan G., Gao Y.-G., Tan M.H. An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 2021;12:1739. doi: 10.1038/s41467-021-21996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Jungnick S., Hobmaier B., Mautner L., Hoyos M., Haase M., Baiker A., Lahne H., Eberle U., Wimmer C., Hepner S., Sprenger A., Berger C., Dangel A., Wildner M., Liebl B., Ackermann N., Sing A., Fingerle V. Detection of the new SARS-CoV-2 variants of concern B.1.1.7 and B.1.351 in five SARS-CoV-2 rapid antigen tests (RATs), Germany, March 2021. Eurosurveillance. 2021;26(16) doi: 10.2807/1560-7917.ES.2021.26.16.2100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Rodgers M.A., Batra R., Snell L.B., Daghfal D., Roth R., Huang S., Kovacs S., Nebbia G., Douthwaite S., Cloherty G.A. Detection of SARS-CoV-2 variants by Abbott molecular, antigen, and serological tests. MedRxiv. 2021 doi: 10.1016/j.jcv.2022.105080. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(8):1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Alcoba-Florez J., Gil-Campesino H., de Artola D.G.-M., González-Montelongo R., Valenzuela-Fernández A., Ciuffreda L., Flores C. Sensitivity of different RT-qPCR solutions for SARS-CoV-2 detection. Int. J. Infect. Dis. 2020;99:190–192. doi: 10.1016/j.ijid.2020.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Pro-Lab Diagnostics – Pro-AmpRTSARS-CoV-2 test EUA Summary, 2020. 10.1101/2020.02.20.20025874. [DOI]
- 321.C.-D. Kit, iAMP-COVID19 detection kit, 2020. https://www.fda.gov/media/136870/download (Accessed 10 July 2021).
- 322.Color SARS-CoV-2 RT-LAMP diagnostic assay, 2021. https://www.fda.gov/media/138249/download (Accessed 10 July 2021).
- 323.Liu D., Shen H., Zhang Y., Shen D., Zhu M., Song Y., Zhu Z., Yang C. A microfluidic-integrated lateral flow recombinase polymerase amplification (MI-IF-RPA) assay for rapid COVID-19 detection. Lab Chip. 2021;21(10):2019–2026. doi: 10.1039/d0lc01222j. [DOI] [PubMed] [Google Scholar]
- 324.Comprehensive workflow for detecting coronavirus using Illumina benchtop systems, 2020. https://www.illumina.com/content/dam/illumina-marketing/documents/products/appnotes/ngs-coronavirus-app-note-1270-2020-001.pdf (Accessed 11 July 2021).
- 325.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Innovative solutions for SARS-CoV-2 testing, 2021. https://www.caspr.bio/pdf/Brochure.pdf (Accessed 10 July 2021).
- 327.Hueston L., Kok J., Guibone A., McDonald D., Hone G., Goodwin J., Carter I., Basile K., Sandaradura I., Maddocks S., Sintchenko V., Gilroy N., Chen S., Dwyer D.E., O’Sullivan M.V.N. The antibody response to SARS-CoV-2 infection. Open Forum Infect. Dis. 2020;7(9):1–8. doi: 10.1093/ofid/ofaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Lee K., Ko H.L., Lee E.-Y., Park H.-J., Kim Y.S., Kim Y.-S., Cho N.-H., Park M.-S., Lee S.-M., Kim J., Kim H., Seong B.L., Nam J.-H. Development of a diagnostic system for detection of specific antibodies and antigens against Middle East respiratory syndrome coronavirus. Microbiol. Immunol. 2018;62(9):574–584. doi: 10.1111/1348-0421.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kasetsirikul S., Umer M., Soda N., Sreejith K.R., Shiddiky M.J.A., Nguyen N.-T. Detection of the SARS-CoV-2 humanized antibody with paper-based ELISA. Analyst. 2020;145(23):7680–7686. doi: 10.1039/d0an01609h. [DOI] [PubMed] [Google Scholar]
- 330.EUA authorized serology test performance, 2020. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance (Accessed 10 July 2021).
- 331.Manenti A., Maggetti M., Casa E., Martinuzzi D., Torelli A., Trombetta C.M., Marchi S., Montomoli E. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J. Med. Virol. 2020;92(10):2096–2104. doi: 10.1002/jmv.25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Lee W.T., Girardin R.C., Dupuis A.P., II, Kulas K.E., Payne A.F., Wong S.J., Arinsburg S., Nguyen F.T., Mendu D.R., Firpo-Betancourt A., Jhang J., Wajnberg A., Krammer F., Cordon-Cardo C., Amler S., Montecalvo M., Hutton B., Taylor J., Mcdonough K.A. Neutralizing antibody responses in COVID-19 convalescent Sera. J. Infect. Dis. 2021;223(1):47–55. doi: 10.1093/infdis/jiaa673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Qorvo Biotechnologies OmniaTM SARS-CoV-2 Antigen Test, 2021. https://www.fda.gov/media/147578/download (Accessed 20 July 2021).
- 334.Celltrion DiaTrustTM COVID-19 Ag Rapid Test, 2021. https://www.fda.gov/media/147691/download (Accessed 20 July 2021).
- 335.Edara V.-V., Lai L., Sahoo M.K., Floyd K., Sibai M., Solis D., Flowers M.W., Hussaini L., Ciric C.R., Bechnack S., Stephens K., Mokhtari E.B., Mudvari P., Creanga A., Pegu A., Derrien-Colemyn A., Henry A.R., Gagne M., Graham B.S., Wrammert J., Douek D.C., Boritz E., Pinsky B.A., Suthar M.S. Infection and vaccine-induced neutralizing antibody responses to the SARS-CoV-2 B.1.617.1 variant. BioRxiv. 2021 doi: 10.1101/2021.05.09.443299. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R., Cutland C.L., Darton T.C., Dheda K., Duncan C., Emary K., Ewer K.J., Fairlie L., Faust S.N., Feng S., Ferreira D.M., Finn A., Goodman A.L., Green C.M., Green C.A., Heath P.T., Hill C., Hill H., Hirsch I., Hodgson S., Izu A., Jackson S., Jenkin D., Joe C., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A.M., Lelliott A., Libri V., Lillie P.J., Mallory R., Mendes A., Milan E.P., Minassian A.M., McGregor A., Morrison H., Mujadidi Y.F., Nana A., O’Reilly P.J., Padayachee S.D., Pittella A., Plested E., Pollock K.M., Ramasamy M.N., Rhead S., Schwarzbold A.V., Singh N., Smith A., Song R., Snape M.D., Sprinz E., Sutherland R.K., Tarrant R., Thomson E.C., Török M.E., Toshner M., Turner D., Vekemans J., Villafana T.L., Watson M., Williams C.J., Douglas A.D., Hill A., Lambe T., Gilbert S.C., Pollard A.J., G.Oxford COVID Vaccine Trial Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., Briner C., Kwatra G., Ahmed K., Aley P., Bhikha S., Bhiman J.N., Bhorat A.E., du Plessis J., Esmail A., Groenewald M., Horne E., Hwa S.H., Jose A., Lambe T., Laubscher M., Malahleha M., Masenya M., Masilela M., McKenzie S., Molapo K., Moultrie A., Oelofse S., Patel F., Pillay S., Rhead S., Rodel H., Rossouw L., Taoushanis C., Tegally H., Thombrayil A., van Eck S., Wibmer C.K., Durham N.M., Kelly E.J., Villafana T.L., Gilbert S., Pollard A.J., de Oliveira T., Moore P.L., Sigal A., Izu A., NGS-SA G., G.Wits-VIDA COVID Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384(20):1885–1898. doi: 10.1056/nejmoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N., Korsman S., Davies M.-A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Kosakovsky Pond S.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 339.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Emary K.R.W., Golubchik T., Aley P.K., Ariani C.V., Angus B., Bibi S., Blane B., Bonsall D., Cicconi P., Charlton S., Clutterbuck E.A., Collins A.M., Cox T., Darton T.C., Dold C., Douglas A.D., Duncan C., Ewer K.J., Flaxman A.L., Faust S.N., Ferreira D.M., Feng S., Finn A., Folegatti P.M., Fuskova M., Galiza E., Goodman A.L., Green C.M., Green C.A., Greenland M., Hallis B., Heath P.T., Hay J., Hill H.C., Jenkin D., Kerridge S., Lazarus R., Libri V., Lillie P.J., Ludden C., Marchevsky N.G., Minassian A.M., McGregor A.C., Mujadidi Y.F., Phillips D.J., Plested E., Pollock K.M., Robinson H., Smith A., Song R., Snape M.D., Sutherland R.K., Thomson E.C., Toshner M., Turner D., Vekemans J., Villafana T.L., Williams C.J., Hill A., Lambe T., Gilbert S.C., Voysey M., Ramasamy M.N., Pollard A.J., c.COVID- Genomics UK, AMPHEUS P., G.Oxford COVID- Vaccine Trial Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397(10282):1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., Bianchi S., Tortorici M.A., Bowen J., Culap K., Jaconi S., Cameroni E., Snell G., Pizzuto M.S., Pellanda A.F., Garzoni C., Riva A., T.C.-N.B.C.-19 Collaboration, Elmer A., Kingston N., Graves B., McCoy L.E., Smith K.G.C., Bradley J.R., Temperton N., Ceron-Gutinerrez L., Barcenas-Morales G., T.C.-19 G.U. (COG-U. Consortium, Harvey W., Virgin H.W., Lanzavecchia A., Piccoli L., Doffinger R., Wills M., Veesler D., Corti D., Gupta R.K. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593(7857):136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Edara V.V., Hudson W.H., Xie X., Ahmed R., Suthar M.S. An mRNA vaccine against SARS-CoV-2 variants after infection and vaccination. New Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1001/jama.2021.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O., Gnanakaran S., Hengartner N., Pajon R., Smith G., Glenn G.M., Korber B., Montefiori D.C. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29(4):529–539.e4. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Ong D.S.Y., Koeleman J.G.M., Vaessen N., Breijer S., Paltansing S., de Man P. Rapid screening method for the detection of SARS-CoV-2 variants of concern. J. Clin. Virol. 2021;141 doi: 10.1016/j.jcv.2021.104903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Vega-Magaña N., Sánchez-Sánchez R., Hernández-Bello J., Venancio-Landeros A.A., Peña-Rodríguez M., Vega-Zepeda R.A., Galindo-Ornelas B., Díaz-Sánchez M., García-Chagollán M., Macedo-Ojeda G., García-González O.P., Muñoz-Valle J.F. RT-qPCR assays for rapid detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 mutations: a screening strategy to identify variants with clinical impact. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Kumar M., Gulati S., Ansari A.H., Phutela R., Acharya S., Kathpalia P., Kanakan A., Maurya R., Vasudevan J.S., Murali A., Pandey R., Maiti S., Chakraborty D. RAY: CRISPR diagnostic for rapid and accurate detection of SARS-CoV2 variants on a paper strip. MedRxiv. 2021 doi: 10.1101/2021.02.01.21250900. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]