Abstract
Fluorescence in situ hybridization (FISH) has proven to be particularly useful to describe the microbial composition and spatial organization of mixed microbial infections, as it happens in periodontitis. This scoping review aims to identify and map all the documented interactions between microbes in periodontal pockets by the FISH technique. Three electronic sources of evidence were consulted in search of suitable articles up to 7 November 2020: MEDLINE (via PubMed), Scopus (Elsevier: Amsterdam, The Netherlands), and Web of Science (Clarivate Analytics: Philadelphia, PA, USA) online databases. Studies that showed ex vivo and in situ interactions between, at least, two microorganisms were found eligible. Ten papers were included. Layered or radially ordered multiple-taxon structures are the most common form of consortium. Strict or facultative anaerobic microorganisms are mostly found in the interior and the deepest portions of the structures, while aerobic microorganisms are mostly found on the periphery. We present a model of the microbial spatial organization in sub- and supragingival biofilms, as well as how the documented interactions can shape the biofilm formation. Despite the already acquired knowledge, available evidence regarding the structural composition and interactions of microorganisms within dental biofilms is incomplete and large-scale studies are needed.
Keywords: periodontal diseases, fluorescence in situ hybridization, dental biofilm, imaging, microbes, oral microbiota
1. Introduction
Periodontitis is a chronic inflammatory disease that results in the loss of the tooth-supporting tissues, which can lead to tooth loss when untreated [1]. It is the consequence of the imbalance between the polymicrobial microbiota, that colonizes the tooth surfaces in form of biofilms, the immune and inflammatory host response within the gingival tissues [1,2,3] and the individual variations in the stock of these taxa [4]. This imbalance, in susceptible individuals, results in the loss of clinical attachment, triggering the formation of periodontal pockets [5,6]. As with other chronic diseases, periodontitis requires supportive care to avoid its recurrence [7]. Furthermore, mounting evidence suggests that many chronic disorders, such as diabetes and cardiovascular diseases, are related to periodontitis via systemic inflammation caused by periodontal bacteria [8].
A total of 2074 genomes and 529 taxa of microbes are estimated to inhabit the oral ecosystem [9]. While most of these microorganisms are not directly associated with periodontitis, they may create the necessary conditions (e.g., nutrient supply or oxygen depletion) for other microorganisms to grow and disrupt the periodontal balance between host and microbes. Microbes’ spatial organization, the understanding of the interactions between microbes in supra- and subgingival biofilms, and how it influences the establishment, development, and the outcome of the periodontal diseases have been emerging subjects in periodontal microbiology.
The development of culture-independent methods has allowed the identification of periodontitis-associated uncultured and fastidious species, providing a more detailed look at the bacterial communities in periodontal tissues. Fluorescence in situ hybridization (FISH) has proven to be particularly useful to describe the microbial composition and spatial organization of mixed microbial infections, in time and space within their natural context. This molecular technique relies on the hybridization of single-stranded, fluorescently labeled DNA-, RNA-, or nucleic acid mimics-targeted oligonucleotides probes with fluorescent molecules (e.g., fluorochromes) that hybridize to its complementary conserved 16S or 23S rRNA sequences in the microorganism [10,11,12].
Even in simple samples where only two or three fluorochromes are used, spectral overlap and crosstalk seem difficult to eradicate when performing standard FISH in multiplex experiments. Additionally, the use of bandpass filters in fluorescence image acquisition restricts the number of fluorochromes that can be simultaneously distinguished, making it problematic to recognize one signal from another simultaneously with certainty and consequently restricting the microscopic identification of various taxa of microbes in single samples [13,14].
To avoid spectral overlap and crosstalk, Valm, A.M. et al. (2012) [14,15] developed a strategy—the Combinatorial Labeling and Spectral Imaging (CLASI)—and blended it with FISH (CLASI-FISH). This technique relies on the labeling of microbes of interest with two or more combinations of fluorochromes [14], increasing spectral discrimination of the fluorochromes that have a propensity to overlap in excitation and emission spectra. CLASI-FISH is currently capable to distinguish unambiguously 120 differently labeled organisms [16], resulting in exclusive mixed colors that are distinguished by the application of linear unmixing algorithms.
This scoping review has the purpose to identify and map the existing evidence about the in situ and ex vivo interactions between microbes in periodontal pockets, identified with the FISH technique, as well as to identify and analyze any knowledge gaps that can point to further research directions.
2. Materials and Methods
2.1. Focused Question
This scoping review was conducted following the guidelines of the Transparent Reporting of Systematic Reviews and Meta-Analyses extension for scoping reviews (PRISMA-ScR) [17], to answer the focused (PCC—Population, Concept, Context) question: “What are the identified interactions between microbes in periodontal pockets as evaluated by the FISH technique?”
2.2. Search Strategy and Information Sources
Three electronic sources of evidence were consulted in search of suitable articles that matched the aim of this review: MEDLINE (via PubMed), Scopus (Elsevier: Amsterdam, The Netherlands), and Web of Science (Clarivate Analytics: Philadelphia, PA, USA) online databases. The databases were consulted up to the 7 November 2020.
Papers were searched using the following keywords: “Periodontal disease”; “Periodont *”; “Periodontal pockets”; “Microbes”; “Oral biofilm”; “Micro *”; “Oral micro *”; “Bacteria”; “Imaging”; “FISH”; “FISH technique”; “fluorescence in situ hybridization” and “hybridization”. The keywords were combined with the Boolean operators “AND” or “OR” with the proximity operators [“” and ()] and with the truncation operator (*) used whenever appropriate. The search strategy was personalized according to the different databases (Table S1).
The electronic database search was supplemented with a hand search across the references of all included papers. The authors of the included papers were contacted to find additional unpublished images that could fulfill our inclusion criteria and were asked permission to reproduce images in this scoping review. When required, additional permission of all reproduced images was granted by the Copyright Clearance Center (Danvers, MA, USA).
2.3. Eligibility Criteria
All studies that described ex vivo and in situ interactions between, at least, two microorganisms were found eligible. We only incorporated articles applied in humans.
The exclusion criteria detached experiments using other molecular cytogenetic techniques rather than the FISH technique, articles with no images, or experiments that used manufactured bovine enamel/dentin slabs, acrylic, or epoxy resin appliances to extract dental biofilm. Restrictions were also made to article type excluding reviews, case reports, or letters.
2.4. Screening and Selection of Sources of Evidence
Two independent reviewers (G.M.E., L.M.) selected papers by evaluating their titles and their abstracts information. Any disagreements in the acquired results were resolved upon discussion with a third reviewer (A.S.A.). Furthermore, the selected articles were then read in full and were not included if did not fulfill the inclusion criteria or if any of the exclusion criteria was detected. Using the Cohen’s Kappa method and IBM SPSS (Version 26) program, the interrater reliability (IRR) was calculated.
2.5. Data Extraction and Analysis
Data from the included articles were processed for analysis. Information regarding the year of publication, study design, FISH conditions, probes’ names and sequences, images, microorganisms found, their location, and their interactions within the biofilm were collected in parallel by G.M.E. and L.M. The data’s interpretation and analysis were debated until a consensus was reached.
2.6. Synthesis of Results
The studies were categorized based on the collected data. Evidence is reported in a table and a visual representation, that incorporates all the images in the finest resolution acquired from the FISH experiments performed on the included papers.
3. Results
3.1. Selection of Sources of Evidence
The initial electronic search resulted in 2090 studies, of which 832 located in PubMed, 625 in Web of Science (Clarivate Analytics: Philadelphia, PA, USA), and 633 in Scopus (Elsevier: Amsterdam, The Netherlands). After removing 1144 duplicated studies, 905 studies were rejected after screening articles by title and abstract. The remaining 41 studies were obtained and analyzed. After full-text reading, 8 studies met the inclusion criteria. Additional hand searching of the reference lists of the selected papers retrieved 10 additional studies for full-text reading, of which 2 papers met the inclusion criteria. As such, 10 papers [3,18,19,20,21,22,23,24,25,26] were included in the present scoping review.
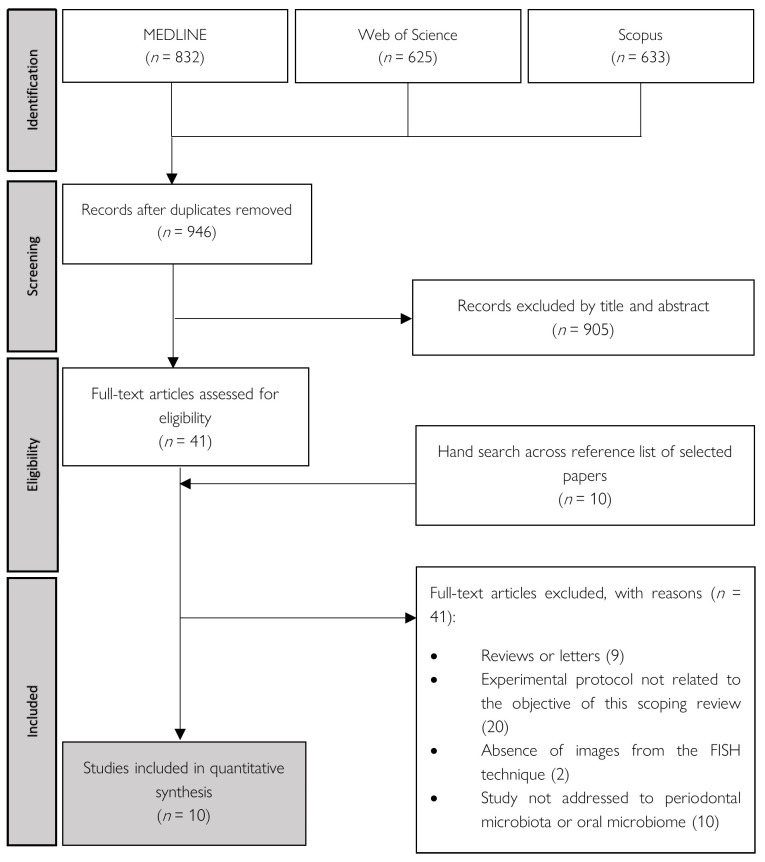
The Cohen’s Kappa method was used to calculate the interrater reliability (IRR) in the selection process by titles and abstracts, which yielded a value of 0.965. The PRISMA flow diagram (Figure 1) demonstrates the selection process.
Figure 1.
Flowchart of literature search and study selection.
3.2. Characteristics of Sources of Evidence
The general characteristics of the six case series, the three case-control studies, and the one cross-sectional study are presented in this section (Table 1). We divided certain images into three panels to make the analysis of the gathered images simpler (Figure 2, Figure 3 and Figure 4). Due to the well-defined approach, we paid special attention to the results of one particular study (Mark Welch, J.L., et al.).
Table 1.
Characteristics of the included studies, such as location of the sampled dental biofilm, exclusion criteria, characteristics of the controls and/or cases, and the purpose of each study. The abbreviations can be found at the bottom of the table.
| Case Series (6) | |||||||
| Authors (Year) | Sample | Exclusion Criteria | Cases | Aim | |||
| Characteristics | n | ||||||
| Wecke, J., et al. (2000) | Subgingival | Chronic diseases; antimicrobial therapy within the past 6 months | PPD means of 8.07 ± 1.63 mm | 52 | Assess the frequency and spatial organization of specific treponemes in RPP | ||
| Gmür, R., et al. (2004) | Supragingival | Chronic diseases; Antimicrobial therapy within the past 3 months | Gingival pain, gingival ulcers or necrosis, pseudomembranes, fetid odor, loss of gingival papillae, PPD ≤ 4 mm, having at least 20 natural teeth, BOP, and plaque index | 42 | Compare the distribution of periodontal pathogens in gingivitis and NUG | ||
| Gmür, R. and Lüthi-Schaller, H. (2007) | Subgingival | NAI | Patients with ACP | NAI | Develop an IF-FISH protocol | ||
| Schlafer, S., et al. (2010) | Subgingival | Chronic diseases; Antimicrobial/anti-inflammatory therapy within the past 6 months; pregnant or lactating women | GAP: having a disease onset estimated at <30 years and PPD ≥ 6 mm at more than 3 permanent teeth (other than first molars or incisors) | 11 | Study the architectural function of Filifactor alocis in GAP | ||
| Zjinge, V., et al. (2010) | Supra- and subgingival | Chronic diseases; antimicrobial therapy within the past 3 months | PPD > 6 mm RBL > 30% |
10 | Study the biofilm architecture of predominant periodontal taxa | ||
| Mark Welch, J.L., et al. (2016) | Supragingival | Subjects suffering from chronic diseases | Subjects refrained from oral hygiene for 12 to 48 h before sample collection | 22 | |||
| Case-Control Studies (3) | |||||||
| Authors (Year) | Sample | Exclusion Criteria | Cases | Controls | Aim | ||
| Characteristics | n | Characteristics | n | ||||
| Moter, A., et al. (1998) | Subgingival | Chronic diseases; antimicrobial/anti-inflammatory therapy within the past 6 months | PPD ≥ 6 mm BOP |
200 | Site clinically not affected | 44 | Assess the frequency of specific treponemes in RPP |
| Lepp, P.W., et al. (2004) | Subgingival | Chronic diseases; antimicrobial therapy within the past 3 months; pregnant or lactating women; diabetes or HIV-positive | Gingivitis (BOP; CAL ≤ 1 mm; PPD ≤ 4 mm); Slight periodontitis (BOP; CAL 2–3 mm; PPD ≥ 4 mm); Moderate periodontitis (BOP; CAL 4–5 mm; PPD ≥ 4 mm); Severe periodontitis (BOP; CAL ≥ 6 mm; PPD ≥ 4 mm) | 167 | Healthy (BOP; CAL ≤ 1 mm; PPD ≤ 3 mm) | 67 | Identify populations of Archaea in periodontal pockets |
| Drescher, J., et al. (2010) | Subgingival | Chronic diseases; antimicrobial/anti-inflammatory therapy within the past 6 months; pregnant or lactating women | GAP: having a disease onset estimated at <30 years and PPD ≥ 6 mm at more than 3 permanent teeth (other than first molars or incisors); CP: PPD of ≥4 mm at 30% or more of the residual teeth | 144 | PR subjects: (age ≥ 65 years; at least 20 natural teeth; CAL ≤ 2 mm; PPD ≤ 5 mm) | 19 | Study the spatial organization of Selenomonas sp. in GAP |
| Cross-Sectional Study (1) | |||||||
| Authors (Year) | Sample | Exclusion Criteria | Subjects ’ Condition | n | Aim | ||
| Machado, F.C., et al. (2012) | Subgingival | Chronic diseases; antimicrobial/psychotropic/anticonvulsant therapy within the past 3 months; professional tooth-cleaning in the previous 6 months or were receiving orthodontic treatment | Pregnant | 20 | Study the qualitative/quantitative differences of eight periodontal pathogens | ||
| Non-pregnant | 20 | ||||||
ACP: advanced chronic periodontitis; BOP: bleeding on probing; CAL: clinical attachment loss; CP: chronic periodontitis; GAP: generalized aggressive periodontitis; IF-FISH: immunofluorescence-fluorescence in situ hybridization; NAI: no available information; NUG: necrotizing ulcerative gingivitis; PPD: probing pocket depth; PR: periodontitis-resistant; RBL: radiographic bone loss; RPP: rapidly progressive periodontitis.
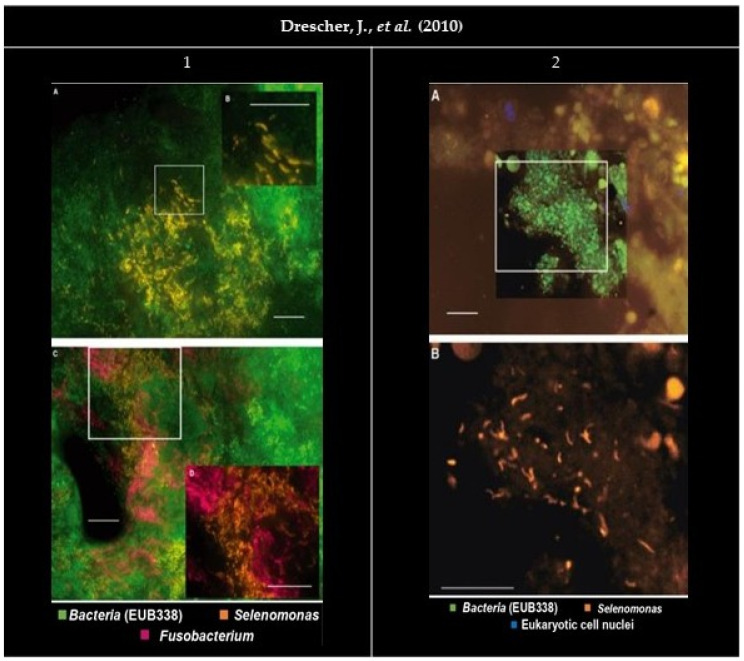
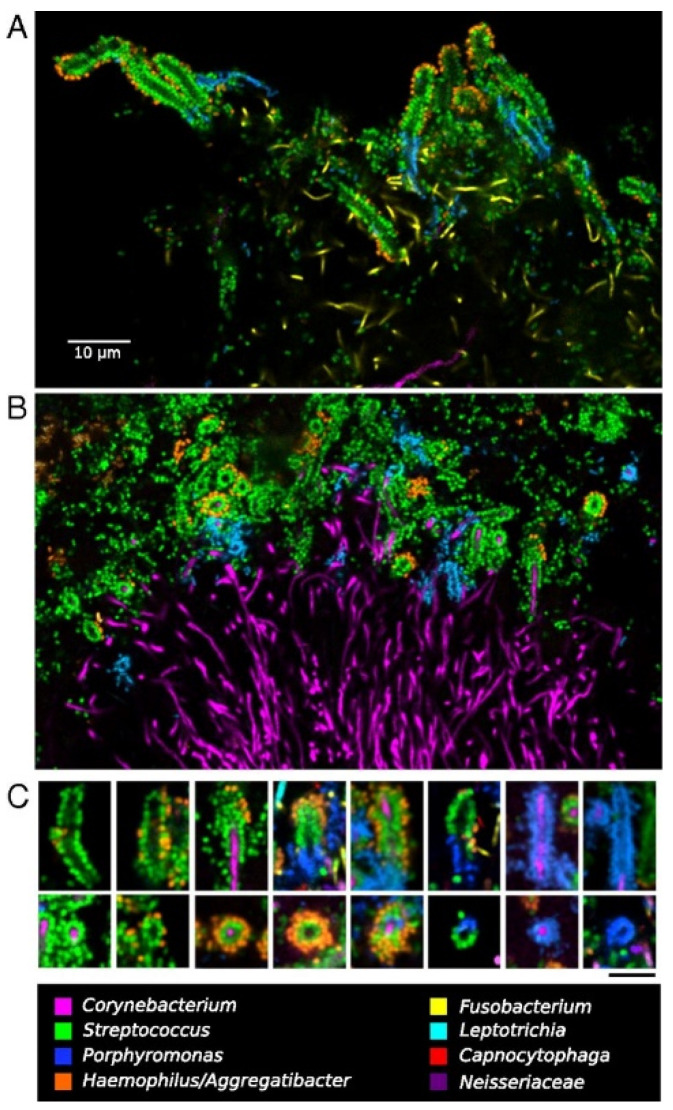
Figure 2.
Panel of the gathered images. (1A,C) Organisms stained by the probe SELE appear as densely packed groups in the cervical portion of the subgingival biofilm. (1B) Higher magnification shows a crescent-shaped morphology. (1C,D) EUB338FITC (Fluorescein isothiocyanate) (green), SELECy3 (Cyanine 3) (bright orange) and FUSOCy5 (Cyanine 5) (magenta). (1D) Higher magnification, with EUB338FITC filter removed for better interpretation. Bars indicate 10 µm. (2) FISH (Fluorescence in situ hybridization) performed on a gingival biopsy. (2A) EUB338FITC (green), SELECy3 (orange), and eukaryotic cell nuclei stained with DAPI (4′, 6-diamino-2-phenylindole) (blue). (2B) Higher magnification took with the Cy3 filter set only. Bars indicate 10 µm. All images were reprinted and adapted with the publisher’s permission.
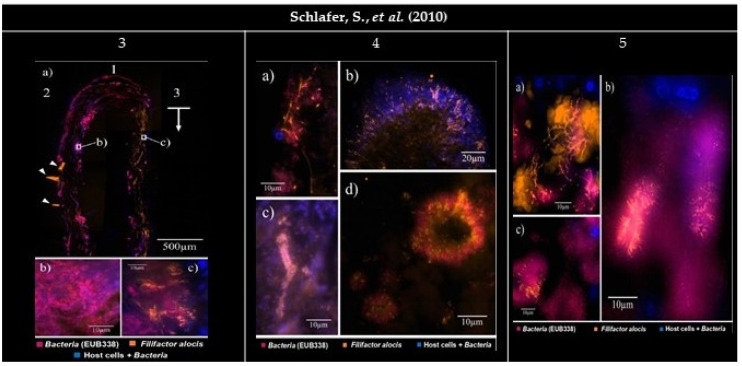
Figure 3.
Panel of the gathered images. (3) Subgingival biofilm visualized by FISH (Fluorescence in situ hybridization). EUB338Cy5 (Cyanine 5) (magenta), FIALCy3 (Cyanine 3) (bright orange), and DAPI (4′, 6-diamino-2-phenylindole) staining (blue). DAPI stains both host cell nuclei and bacteria. (3a1) The carrier tip. (3a2) The carrier side facing the tooth. (3a3) The carrier side facing the pocket epithelium. (3a1,2) Little or no presence of Filifactor alocis. (3a3) Presence of a bright orange signal, indicating a vindicated presence of F. alocis (arrow). The image show artifacts caused by the folding of the embedded carriers (arrowheads). (3b,c) Higher magnification. (3b) Rare colonization of F. alocis amongst the bacteria. (3c) F. alocis in densely packed groups among the organisms on the carrier side facing the soft tissues and host cell nuclei (blue). (4) Establishment of Filifactor alocis in a subgingival biofilm. (4a) Overlay of FIALCy3, EUB338Cy5, and DAPI filter sets. In some parts of the biofilm F. alocis rods can reach a considerable length. (4b,c) Overlay of FIALCy3 and DAPI filter sets. (4b) Radial orientation of F. alocis towards the exterior of a mushroom-like protuberance. (4c) Test-tube-brush formations of F. alocis around signal-free channels. (4d) Overlay of FIALCy3 and EUB338Cy5 filter sets. F. alocis and fusiform bacteria form concentrical structures. (5) Establishment of Filifactor alocis in periodontal tissue. (5a) F. alocis forming tree-like structures among coccoid and fusiform bacteria and autofluorescent erythrocytes. (5b) F. alocis forming palisades with fusiform bacteria around large rod-shaped eubacterial organisms. (5c) F. alocis being part of concentrical bacterial aggregations such as those found in (4d). All images were reprinted and adapted with the publisher’s permission.
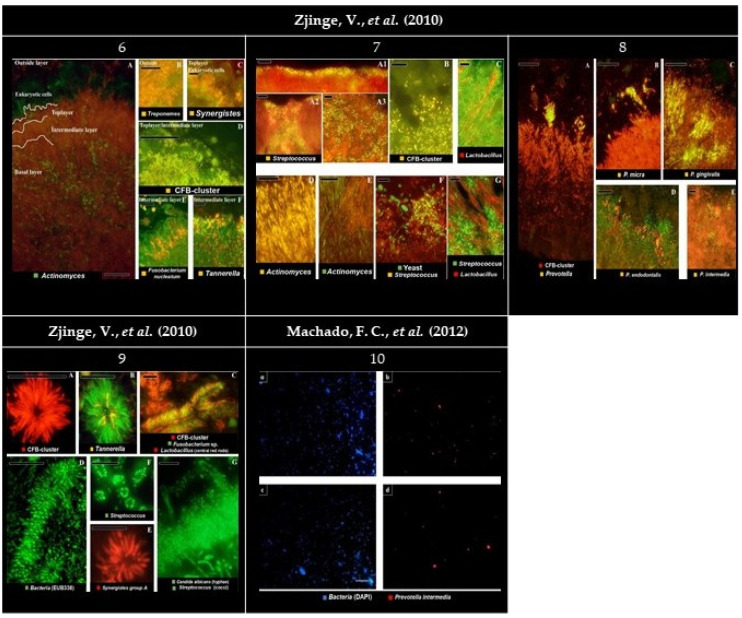
Figure 4.
Panel of the gathered images. (6) Localization of the most abundant species in subgingival biofilms. (6A) Overview of the four layers of subgingival biofilm. Actinomyces sp. (green), bacteria (red), and eukaryotic cells (large green cells on top). (6B) Spirochaetes (yellow) outside the biofilm, without clear organization. (6C) Detail of Synergistetes (yellow) in the top layer near PMN’s (Polymorphonuclear leukocytes) (green). (6D) Presence of the Cytophaga-Flavobacterium-Bacteroides cluster (CFB-cluster) (yellow) in the top and intermediate layer. (6E) Fusobacterium nucleatum in the intermediate layer. (6F) Tannerella sp. (yellow) in the intermediate layer. Each panel is double-stained with probe EUB338 labeled with FITC (Fluorescein isothiocyanate) or Cy3 (Cyanine 3). Bars indicate 10 µm. (7) Localization of the most abundant species in supragingival biofilms. (7A1–C) The second layer. (7D–G) Basal layer. (7A1–A3) Streptococcus sp. disposed of in different ways on the second layer. (7B) Cells from the CFB-cluster stained in the top layer of the biofilm. (7C) Lactobacillus sp. (red) through the top layer. (7D) On the basal layer, Actinomyces sp. cells (yellow). (7E) Actinomyces sp. (green) and chains of cocci. (7F) Colonies of Streptococcus sp. (yellow) all-around yeast cells (green) and bacteria unidentified (red). (7G) Streptococcus sp. (green) growing closely to Lactobacillus sp. (red). Black holes might be channels through the biofilm. Panels (A–C,E,F) are double stained with probe EUB338 labeled with FITC or Cy3. Bars indicate 10 µm. (8) Localization of presumptive species associated with periodontitis in subgingival biofilms. (8A) Colonization of the subgingival biofilm by the CFB-cluster species (red) and Prevotella sp. (yellow). Since Prevotella sp. are part of the CFB-cluster of bacteria, cells appear in yellow. (8B) Top of the biofilm with a micro-colony of Parvimonas micra (yellow). (8C,D) Micro-colonies of Porphyromonas gingivalis (yellow) and Porphyromonas endodontalis (yellow) in the top layer, respectively. (8E) Micro-colonies of Prevotella intermedia in the top layer. Panels B, C, D, and E are double stained with probe EUB338 labeled with FITC or Cy3. Bars indicate 10 µm. (9) Bacterial aggregates detected in both sub- and supragingival plaque. (9A) Filamentous cells from the CFB-cluster in the fourth layer of the subgingival plaque. (9B) Tannerella sp. (yellow) in a test-tube brush. (9C) Test-tube brush with Lactobacillus sp. (red rods) as central structures. Fusobacterium nucleatum (green) and CFB-cluster filaments (red), morphologically identical to Tannerella forsythia, perpendicularly radiating around lactobacilli. (9D) Test tube brush stained with the eubacterial probe. (9E) Synergistetes group A species forming aggregates solely with themselves. (9F) Streptococcus sp. (green) aggregation around a central cell (not stained) in supragingival plaque. (9G) Supragingival plaque with Streptococcus sp. (green cocci) adhering to a central axis of yeast cells or hyphae, such as Candida albicans (green hyphae). Bars indicate 10 µm. (10a,c) Plaque specimen stained for total bacterial cells with DAPI (4′, 6-diamino-2-phenylindole) in pregnant and non-pregnant women, respectively. (10b,d) Plaque specimen stained for Prevotella intermedia (with probe Pint649) in pregnant and non-pregnant women, respectively. Bars indicate 20 µm. All images were reprinted and adapted with the publisher’s permission.
In Mark Welch, J.L., et al. (2016) [18] only 13 out of 57 genera tested had at least 3% mean abundance and were also prevalent, being identified in more than 90% of supragingival specimens (Corynebacterium sp., Capnocytophaga sp., Fusobacterium sp., Leptotrichia sp., Actinomyces sp., Streptococcus sp., Neisseria sp., Haemophilus/Aggregatibacter sp., Porphyromonas sp., Rothia sp., Lautropia sp., Veilonella sp., and Prevotella sp.).
Corynebacterium sp. was remarkably specific to supragingival (12%) and subgingival (8%) plaque. By contrast, genera such as Streptococcus sp., Veillonella sp., and Haemophilus sp. occupied a wide range of substrates in oral ecosystems.
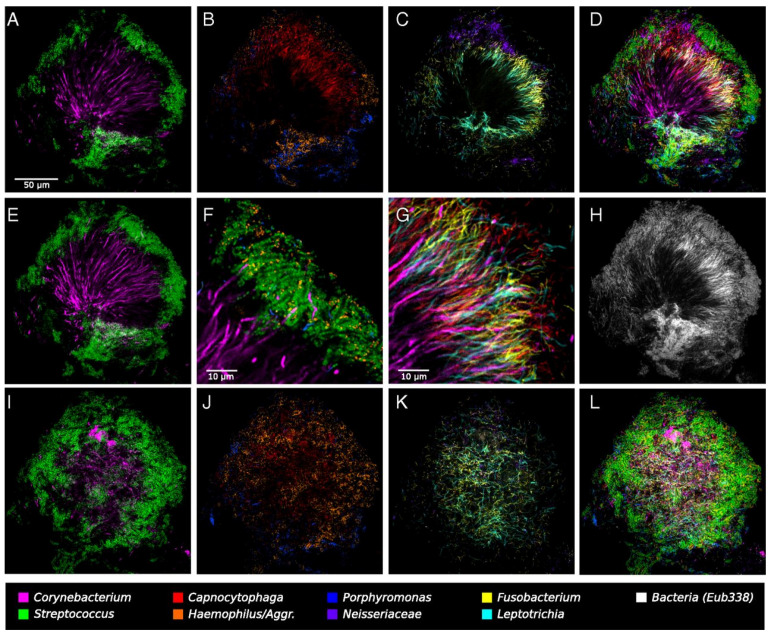
A complex microbial consortium in a hedgehog-shaped structure was observed, showing the spatial organization of the plaque microbiota. In short, these hedgehog structures were radially organized, with a multi-taxa consortium composed of a skeleton mainly of Corynebacterium sp. with Streptococcus sp. cells arranged around the distal tips, a multi-genus filament-rich halo composed of Fusobacterium sp., Leptotrichia sp., and Capnocytophaga sp. cells, and a periphery of corncobs structures composed by a filamentous core bordered primarily with Streptococcus sp. cells, Porphyromonas sp. and Haemophilus/Aggregatibacter sp. (Figure 5).
Figure 5.
Hedgehog-shape structure. (A–D,F–H) represents a single focal plane nearby the middle part of the structure. (A) Corynebacterium sp. filaments radiating outward from near the center of the image and coccoid Streptococcus sp. cells arranged around the distal tips of the Corynebacterium sp. filaments. (B) Cells of Haemophilus/Aggregatibacter sp. and Porphyromonas sp. sited at the structure’s periphery, in the same region as the Streptococcus sp. cells. Capnocytophaga sp. also inhabited a wideband inside the periphery. (C) Fusobacterium sp. and Leptotrichia sp. occupying this band called “filament-rich halo”. Neisseriaceae establishing constellations in and nearby the periphery. Actinomyces sp. represented by a small number of cells located near the structure’s base. (D) All taxa overlayed. Many cells from the “filament-rich annulus” overlapped the exterior of the consortia. (E–G) Detail of the Corynebacterium’s arrangement relatively to other taxa. (E) The maximum intensity projection of three adjacent optical sections shows that these filaments are continuous from the center to the periphery of the structure for more than 50 µm. Some filaments persist visible within the Streptococcus sp. cells. (F) Detail of the periphery. Corncob structures are composed of a filamentous core (sometimes visualized as Corynebacterium filaments but often not stained) bordered primarily by Streptococcus sp. cells but also by Porphyromonas sp. and Haemophilus/Aggregatibacter sp., both in proximal contact with Streptococcus sp. cells. (G) On the periphery of these corncob structures, Corynebacterium sp. filaments pass through the halo that is highly densely colonized with elongated rods of Fusobacterium sp., Leptotrichia sp., and Capnocytophaga species. (H) The fluorescent signal of the universal probe EUB338. (I–L) The exterior of the hedgehog structure, composed mainly of corncobs. (K) The edge of the Fusobacterium-Leptotrichia sp. halo. Reprinted with the publisher’s permission.
The corncobs at the periphery showed that “kernels” (coccoid cells) were composed of different taxonomic types and could be either single or double layered (Figure 6). Single-layer corncobs had coccoid cells of both Streptococcus sp. or Porphyromonas sp. (in some cases Porphyromonas sp. kernels coexisted with Streptococcus sp. around the same filament), whereas double-layer kernels consisted of a combination of Streptococcus sp. in the inner layer and Haemophilus/Aggregatibacter sp. in the outer layer.
Figure 6.
Dense corncob structures in supragingival plaque. (A) The central filament lacked hybridization in whole-mount preparation. (B) The central filament was well visualized in methacrylate-embedded and sectioned preparations. (C) Representative images of types of corncobs found. Bar indicates 5 µm in (C). Reprinted with the publisher’s permission.
The most common type of kernels visualized were the ones that had a single layer of Streptococcus sp. cells surrounded by a partial or complete layer of Haemophilus/Aggregatibacter species. Porphyromonas sp. cells were only observed organized in single-layer corncobs. On contrary, cells of the genus Haemophilus/Aggregatibacter sp. were only found forming double-layer structures, exclusively with Streptococcus sp. cells, demonstrating an undoubtedly specific relationship. Competitive, exploitative, or mutualistic interactions between these taxa were detected in this specific biofilm architecture between single- and double-layers corncobs.
Within hedgehogs, bacteria do not form broad single-taxon groups; rather, cells were observed intermingling with at least four different taxa. The authors only did not find this type of interaction between Actinomyces sp. and Corynebacterium sp. cells, where were visualized irregular clumps in the base of the hedgehogs or nearby the hedgehogs, rather than intermixed within this structure.
Hedgehogs were the most observed type of structure. Some samples had multiple hedgehogs’ structures adjacent to each other (Figure S1), especially on the tooth surface on the buccal side and plaque from the gingival margin. Another kind of consortia was also found: a cauliflower structure in plaque (Figure S2) constituted by Lautropia sp., forming the center of the structure, surrounded by Streptococcus sp., Haemophilus/Aggregatibacter sp., and Veilonella species. Dispersed cells of Prevotella sp., Rothia sp., and Capnocytophaga sp. were also visible.
The findings collected from the included articles in this scoping review are summarized in Table 2. We also included a supplemental table, with details on all the oligonucleotide probes and the FISH conditions used in each microorganisms’ analysis (Table S2).
Table 2.
A summary in terms of the location where the dental biofilm was collected, the microorganisms detected, their shape and spatial organization within the biofilm, the most relevant interactions detected, and the microscopic technique used in each study. The abbreviations can be found at the bottom of the table.
| Authors (Year) | Site | Microorganisms | Shape and Spatial Organization of Microorganisms | Relevant Interactions | Microscopic Technique |
|---|---|---|---|---|---|
| Moter, A., et al. (1998) | SUBG | Group I of oral treponemes | Large and dense | These organisms are present in high proportions in subgingival plaque samples and thus represent the predominant flora. Group I treponemes outnumbered group II treponemes. All the treponemes identified predominated at diseased sites but were found infrequently at periodontally stable sites | Dark-field |
| Group II of oral treponemes | Thin, slender, wavily | ||||
| Wecke, J., et al. (2000) | SUBG | Group I of oral treponemes | Large and undulated | Treponemes appeared spread between Gram-negative bacteria in the deepest parts of the periodontal pockets. Gram-positive cocci were located on the most coronal section of the specimens | CLSM |
| Bacteria | Rods and coccoid | ||||
| Lepp, P.W., et al. (2004) | SUBG | Methanobrevibacter oralis | Diplococcobacilli | Treponemal rDNA was found in significantly lower abundance in sites with archaeal rDNA than in sites without archaeal rDNA | CLSM |
| Gmür, R., et al. (2004) | SUPG | Leptotrichia buccalis | Wide and segmented fusiform rods | There was a significantly increased total abundance of periodontal pathogens in the NUG group compared with the gingivitis group | Dark-field |
| Indistinguishable Fusobacterium nucleatum, Capnocytophaga sp., and Fusobacterium periodonticum | Smaller, thin, spindle-shaped, and dotted fusiform rods | ||||
| Gmür, R. and Lüthi-Schaller, H. (2007) | SUBG | Tannerella forsythia | Clumps | Tannerella forsythia was detected in the deepest zones of the periodontal pockets | Epifluorescence |
| Drescher, J., et al. (2010) | SUBG | Selenomonas sp. | Densely packed groups with a crescent-shaped structure from the cervical section to the biofilm portion derived from the pocket’s depth and both on the side facing the tooth and the side facing the soft tissue | These genera appeared spatially related and tangled with each other | Epifluorescence |
| Fusobacterium sp. | Densely groups with a fusiform shape | ||||
| Schlafer, S., et al. (2010) | SUBG | Filifactor alocis | (i) Short rod clustered in radial-orientated structures nearby fusiform bacteria on mushroom-shaped biofilms; (ii) Test-tube brush shapes; (iii) Branch-like structures in gingival tissue; (iv) Palisades structures nearby fusiform bacteria and eubacterial organisms | F. alocis was present in areas that corresponded to the depth of the pockets, but very occasionally in areas that corresponded to the cervical portion and the carrier’s very tip. F. alocis colonized the carrier side facing the soft tissue in most cases and was present in small numbers or not at all on the carrier side facing the root | Epifluorescence |
| Zjinge, V., et al. (2010) | SUBG-FL | Actinomyces sp | Rod-shaped | Display little fluorescence | Epifluorescence |
| SUBG-IL | Fusobacterium nucleatum | Fusiform cells | TL and a portion of the IL were mostly made of filamentous, rod-shaped, or even coccoid bacteria from the CFB-cluster | ||
| Tannerella forsythia | |||||
| Tannerella sp. | |||||
| CFB-cluster | Filamentous, rod-shaped, coccoid | ||||
| SUBG-TL | CFB-cluster | Filamentous, rod-shaped, coccoid, micro-colonies (Prevotella) | |||
| Synergistetes group A | Wide cigar-like bacteria in a palisade lining | ||||
| Parvimonas micra | Micro-colonies | ||||
| SUBG-OL | Treponemes | Test-tube brush shapes | CFB-cluster cells were found perpendicularly arranged around Lactobacillus sp. in test-tube brush shape. Test-tube brushes shapes were also found in a complex mixture of cells | ||
| CFB-cluster | |||||
| Lactobacillus sp. | Rod-shaped | ||||
| Porphyromonas gingivalis | Micro-colonies | ||||
| Porphyromonas endodontalis | |||||
| Tannerella forsythia | Test-tube brush shapes | ||||
| Campylobacter sp. | |||||
| Parvimonas micra | |||||
| Fusobacterium sp. | |||||
| Synergistetes group A | |||||
| SUPG-BL | Actinomyces sp. | Rod-shaped | Bacterial deposits made up of early colonizers, growing perpendicularly to the tooth surface | ||
| Actinomyces sp. + chains of cocci | Rod-shaped and coccoid | ||||
| Streptococcus sp. + yeast and not identified bacteria | Filamentous | ||||
| Streptococcus sp. + Lactobacillus sp. | |||||
| SUPG-SL | Streptococcus sp. | (i) Thin coat; (ii) Colonizing biofilm’s cracks; (iii) Without clear organization | Corncob structures consisting of Streptococcus sp. adhering to a central axis of yeast/hyphae cells | ||
| CFB-cluster | Heterogenous and without clear organization | ||||
| Lactobacillus sp. | Long string-shape | ||||
| Machado, F.C., et al. (2012) | SUBG | Prevotella intermedia | Patchy groups | P. intermedia was frequently found in the plaque of pregnant women | Epifluorescence |
| Mark Welch, J.L., et al. (2016) | SUPG | Corynebacterium sp. | Continuous filaments from the base to the periphery of the structure | Corynebacterium sp. filaments were crusted at their distal tips by brilliant cocci | CLSM |
| Streptococcus sp. | Coccoid | ||||
| Capnocytophaga sp. | Filamentous | Part of a multi-genus halo | |||
| Fusobacterium sp. | |||||
| Leptotrichia sp. | |||||
| Actinomyces sp. | Patchy groups | Observed in the base of the hedgehogs’ structures | |||
|
Haemophilus/ Aggregatibacter sp. |
Filamentous | Built a periphery of corncobs structures in addiction with Streptococcus sp. cells | |||
| Porphyromonas sp. | |||||
| Rothia sp. | Cells of at least four different taxa interact with one another at a micron scale | ||||
| Lautropia sp. | |||||
| Veilonella sp. | |||||
| Prevotella sp. | |||||
| Neisseria sp. |
CLSM: confocal laser scanning microscopy; CP: chronic periodontitis; GAP: generalized aggressive periodontitis; NUG: necrotizing ulcerative gingivitis; SUBG-FL: subgingival first layer; SUBG-IL: subgingival intermediate layer; SUBG-OL: subgingival outside layer; SUBG-TL: subgingival top layer; SUBG: subgingival; SUPG-BL: supragingival basal layer; SUPG-SL: supragingival second layer; SUPG: supragingival.
4. Discussion
4.1. Summary of Evidence
Our findings from the obtained images indicate a paucity of research focusing specifically on the study of the spatial organization of periodontal pathogens within oral biofilms from the Bacteria domain and its etiologic significance.
In our results, the biofilm’s fluorescent intensity and variety of labeled microorganisms increased as images drifted from the tooth to the epithelium side, and from the pocket’s depth to the coronal surface, indicating differences in the physiological activity of the cells. Subsequently, we present three possible explanations: (i) the cell structures and morphologies of supragingival biofilms are much more diverse than those of subgingival biofilms; (ii) the unidentified microorganisms may belong to species for which there are no probes available (iii) prior stages of the biofilm that have been shielded from nutrients, comprising dead/inactive cells with low fluorescence activity, are located in the basal layers [26].
Although the stock of the oral microbiota has a lot of inter-and intra-individual variation, about 16 genera are almost universally found in supragingival dental biofilm, including Corynebacterium sp., Capnocytophaga sp., Fusobacterium sp., Leptotrichia sp., Actinomyces sp., Campylobacter sp., Streptococcus sp., Neisseria sp., Selenomonas sp., Haemophilus/Aggregatibacter sp., Porphyromonas sp., Rothia sp., Lautropia sp., Veilonella sp., and Prevotella sp. [18,23,26,27,28]. Bacteria that thrive below the gumline are cut off from high oxygen stress, salivary and dietary nutrients, relying on gingival crevicular fluid (GCF) for nutrition. As a result, strict or facultative anaerobic proteolytic bacteria become more prevalent in the subgingival microbiota, such as Filifactor sp., Fusobacterium sp., Parvimonas sp., Porphyromonas sp., Prevotella sp., Tannerella sp., and Treponema sp. [3,24,25,26,29,30], particularly in individuals with periodontitis.
In healthy people, the fungal load is thought to be lower than the bacterial load. However, the size and morphology of fungal cells, as well as their synergistic interactions with bacteria, indicate that these species play an important role in the formation of dental biofilms [26,31,32].
The adsorbing of salivary proteins and glycoproteins to the tooth’s surface triggers the initial plaque establishment, forming the acquired enamel pellicle (AEP)—a conditioning layer that covers all teeth present in the oral cavity and acts as a substrate for bacterial attachment [33,34]. The AEP is formed during the early stages of teeth eruption when saliva contacts the tooth’s surface [34] and is never fully removed even if professional dental hygiene is performed.
Planktonic cells, aggregates of cells, and early colonizers (e.g., Streptococcus sp., Lactobacillus sp., Actinomyces sp., and Candida sp.) adhere to this pellicle via specialized adhesins on the bacterial cell surface [35,36]. These species do not promiscuously bind to any filament available, but rather engage in a highly specific interaction with already adhered cells, such as Corynebacterium sp., Actinomyces sp., [18], or yeast/hyphae cells forming the first layer of supragingival biofilm [26,31].
The role of Actinomyces sp. as a primary colonizer has already been proved due to its significant role in gingivitis [37]. Many Actinomyces species, such as A. naeslundii, A. oris, and A. johnsonii [38] have been found in supra- and subgingival biofilms’ specimens from diseased patients, suggesting that these are the most significant biofilm initial formers among the Actinomyces genus. Actinomyces sp. can store intracellular glycogen or search for biofilm material such as extracellular polymeric substances and compounds from dead bacterial cells [26,39], which can create an important advantage in surviving in the deepest layers of the biofilm community. Actinomyces sp. were found in the base of the hedgehogs’ structures [18], which also suggests that Corynebacterium sp. cells do not directly colonize the tooth surface but on a previous and established biofilm containing Actinomyces species.
Possibly, these already adhered microorganisms may create a microenvironment favorable to other colonizers to grow. Therefore, biofilm maturation occurs by the coaggregation of planktonic bacteria to an already adhered biofilm [40].
Early colonizers of supragingival dental surfaces are usually facultative anaerobic bacteria, such as Streptococcus sp. These species produce carbon dioxide (CO2), lactate, and acetate, containing hydrogen peroxide (H2O2) by consuming oxygen (O2). The redox potential is lowered, allowing strict anaerobes (e.g., Fusobacterium sp., Leptotrichia sp., and Capnocytophaga sp.) to settle and multiply in the biofilm.
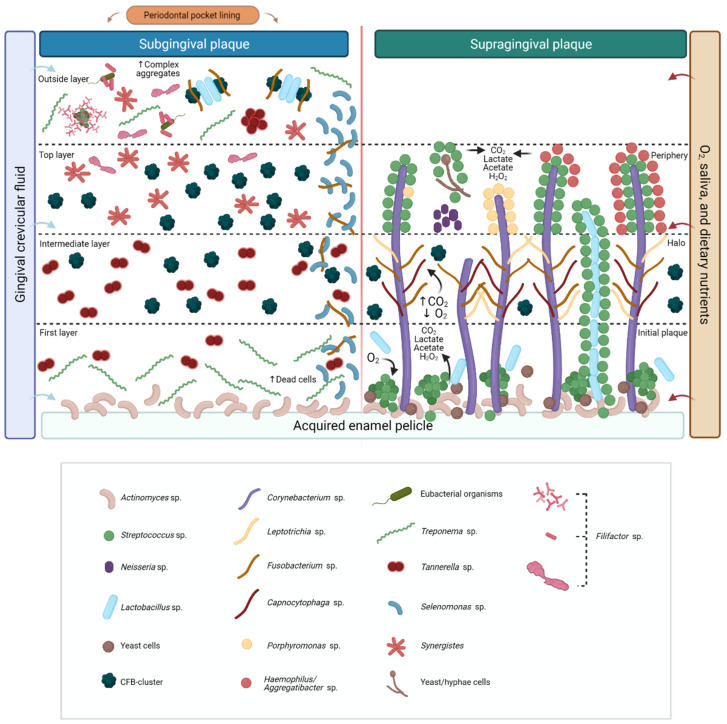
The presence of Streptococcus sp. and bacteria from the Cytophaga-Flavobacterium-Bacteroides cluster (CFB-cluster) in the second layer of supragingival biofilm [26] could indicate a critical transition of a supragingival biofilm made of predominantly Gram-positive saccharolytic bacteria to a Gram-negative proteolytic subgingival biofilm, which could be caused by nutrient availability (e.g., dietary sugars in the supragingival biofilm, or proteins from saliva and GCF in the subgingival biofilm). Additionally, the presence of Streptococcus sp. in the basal and second layers of biofilm also allows us to infer that these organisms can adapt to a wide variety of environments, settling first as early colonizers but with the ability to expand and prosper. Our hypothesis is summarized in Figure 7.
Figure 7.
Summary hypothesis for the interpretation of the gathered evidence. Microorganisms that thrive in the subgingival environment, in contrast to bacteria in supragingival biofilm, are cut off from high oxygen stress, salivary, and dietary nutrients, depending on gingival crevicular fluid (GCF) for nutrition. The adsorbing of salivary proteins and glycoproteins to the tooth’s surface triggers the initial plaque establishment, forming the acquired enamel pellicle (AEP)—a conditioning layer for bacterial attachment. In the supragingival biofilm, Corynebacterium sp. filaments bind to an existing biofilm containing Streptococcus sp., Actinomyces sp., yeast cells, and Lactobacillus sp. These species produce carbon dioxide (CO2), lactate, and acetate, containing hydrogen peroxide (H2O2) by consuming oxygen (O2). The redox potential is lowered, allowing strict anaerobes (such as Fusobacterium sp., Leptotrichia sp., and Capnocytophaga sp.) to settle and multiply in the CO2-requiring “halo” section. This environment is also favorable to the growth and development of microorganisms from the Cytophaga-Flavobacterium-Bacteroides cluster (CFB-cluster), especially, Prevotella sp. In the periphery, cells shape single- or double-layered corncobs structures. Streptococcus sp. adheres to a central axis of yeast/hyphae, Corynebacterium sp., or Lactobacillus sp. cells. Subgingival biofilm is divided into four layers: (i) the first layer, which is made of early colonizers displaying few fluorescences; (ii) the intermediate and (iii) the top layers are composed of a variety of microorganisms from the CFB-cluster, Treponema sp., Tannerella sp., Synergistetes, and Fusobacterium sp. tangled with Selenomonas sp. in the cervical portion; (iv) the outside layer, made of Treponemes nearby a complex mixture of CFB-cluster, Lactobacillus sp. and Fusobacterium sp. cells coaggregated in fine test-tube brushes. Tannerella sp. and Synergistetes formed aggregates solely with themselves. In this last layer, Filifactor sp. form a variety of concentrical-orientated structures, tree-like shapes among coccoid cells, palisades around eubacterial organisms, or remote test-tube-brush shapes.
The presence and abundance of Corynebacterium sp. in hedgehog structures in a bush-like skeleton, anchored from the presumed tooth surface [18,41] may also indicate that this genus plays a key role in the biofilm community, while its biofilm specificity indicates that it occupies a niche that is influenced by tooth surface and/or GCF properties.
By reducing the flow of GCF with the use of anti-inflammatory agents the outgrowth of proteolytic bacteria would be prevented. Another alternative would be to use oxygenating or redox agents to make the gingival crevice less anaerobic, selectively inhibiting the growth of obligate anaerobes [42]. Therefore, the conventional periodontal treatment approaches that involve mechanical removal of biofilm, physical disruption of biofilm structure, or antibiotic therapies could be enhanced.
4.2. Microbes’ Herd Mentality Behavior
Taxa that are present primarily or exclusively in one site may provide clues to the distinctive features of the habitat and the role that those taxa contribute to the site but it’s still unclear if pathogens found at the control sites are part of the local/commensal flora or are originated from adjacent periodontal lesion sulcus crevicular fluid [24].
Depending on which partner the microorganism was found coaggregated with, F. alocis shaped several conformations, especially with fusiform bacteria, which formed concentrical radial-orientated structures in mushroom-shaped biofilms or palisades structures when the coaggregation also occurred with eubacterial organisms [3]. When isolated, F. alocis formed test-tube brushes shapes around signal-free channels. This pattern and degree of organization can play a role in the events that occur during biofilm growth and maturation and be strongly linked to biofilm formation.
Our findings propose that other microorganisms adopted different spatial arrangments influenced by their peers in different biofilm areas and tissues. Test-tube brushes shapes were once again found in a complex mixture of cells containing T. forsythia, Campylobacter sp., P. micra, Fusobacterium sp., and Synergistetes group A in the outside layer of subgingival biofilm [26]. On the other hand, Synergistetes group A adopted a wide cigar-like shape in a palisade lining when found isolated, forming aggregates exclusively with themselves. The CFB-cluster also manifested different shapes, especially Prevotella sp. that formed micro-colonies in the top layer of subgingival biofilm but were observed as filamentous, rod-shaped, and coccoid on the intermediate layer.
Depending on which microorganisms they are coaggregated with, some microorganisms appear to “metamorphose”. We call this process microbes’ “herd mentality behavior” and we believe that might be an important advantage of periodontal pathogens. To verify our hypothesis, future research should focus on the spatial organization of periodontal microbiota. The studies should be performed considering sample collection and preparation that preserves as much spatial organization as possible, such as whole-mount preparations that permit the imaging of 3D structures [18].
4.3. Limitations
None of the included articles classified periodontal diseases according to current diagnostic criteria. In June 2018, the American Academy of Periodontology (AAP) released a series of reviews in partnership with the European Federation of Periodontology (EFP) [7,43,44,45,46] suggesting significant updates in periodontal diagnose, such as combining chronic and aggressive periodontitis in unique-entity periodontitis with various phenotypes. Another major improvement was the association of the periodontal disease’s classification with its progression rate and prognostic factors in a more accurate and reliable staging classification [47].
The results demonstrate that the available evidence relies on a limited number of observations. Assuming that these observable interactions are somehow representative of the periodontal microbiota in health and disease, is a leap of faith that needs confirmation in large-scale studies. Some images catch our eye, but in the absence of meticulous cell counting, it may contribute to an overestimation of microbes’ population in biofilm specimens, contradicting quantitative findings. This raises the question of whether all FISH studies should involve quantitative cell counting or dynamic time lap observations.
We did not include the identification and screening of gray literature in the scoping review process. However, all authors of the included publications in this review were requested to submit unpublished images and/or articles that matched the review’s goal.
5. Conclusions and Future Perspectives
Supragingival biofilm seems to present a radially ordered multiple-taxon structure, backboned by facultative anaerobic long rods, such as Corynebacterium sp. and Lactobacillus sp., bordered by coccoid and filamentous cells. On the other hand, subgingival biofilm seems to present a more layered multiple-taxon structure from the tooth’s surface to the pocket’s epithelium. Anaerobic microorganisms dominate the subgingival environment, especially in the deepest portions. Outside layers with increased microbial diversity were found in both sub- and supragingival biofilms when compared with the inner layers, and the largest contrast was observed in the subgingival biofilm. Consumers and producers of certain metabolites tend to be spatially related; some microorganisms exhibited the ability to shape various structures influenced by their peers in different biofilm areas. We referred to this last phenomenon as “microbes’ herd mentality behavior” and we believe that it may represent an imperative benefit of certain members of the periodontal microbiota.
In vitro studies or studies performed in animals provided the foundations for the molecular pathology reasoning discussed in this scoping review. We suggest that future research should focus on studying species networks in a more realistic and holistic setting, using complex community-like structures as a model.
In conclusion, and in response to the previous query, the FISH technique was able to detect interactions between microorganisms from the three domains of life, enabling the construction of a possible hypothesis for understanding the influence of these interactions in the establishment and development of periodontal diseases. However, the evidence about the structural composition and microorganisms’ interactions in supra- and subgingival biofilms is scarce and no definitive conclusion can be drawn. The enhanced FISH-based techniques provide significant information about the spatial organization of a complex natural polymicrobial community, allowing researchers and dental medicine doctors to better understand how the individual taxa interact within a community in periodontal diseases, and how their relations compromise the whole assemblage.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9071504/s1: Figure S1: Multiple hedgehogs’ structures adjacent to each other; Figure S2: A cauliflower structure in plaque; Table S1: Database search strategy; Table S2: Oligonucleotide probes’ names and sequences, the microorganisms targeted, and the FISH conditions used in the studies included in this scoping review.
Author Contributions
Conceptualization, L.M. and G.M.E.; methodology, L.M., A.S.A. and G.M.E.; data curation, G.M.E. and L.M.; writing—original draft preparation, G.M.E., L.M. and A.S.A.; writing—review and editing, G.M.E., L.M., A.S.A., N.F.A., J.A.P.; supervision, L.M. and A.S.A.; funding acquisition, A.S.A. and N.F.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by: Base Funding—UIDB/00511/2020 of the Laboratory for Process Engineering, Environment, Biotechnology and Energy—LEPABE—funded by national funds through the FCT/MCTES (PIDDAC); Project POCI-01-0145-FEDER-030431 (CLASInVivo), funded by FEDER funds through COMPETE2020—Programa Operacional Competitividade e Internacionalização (POCI) and by national funds (PIDDAC) through FCT/MCTES.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vartoukian S.R., Palmer R.M., Wade W.G. Diversity and morphology of members of the phylum “synergistetes” in periodontal health and disease. Appl. Environ. Microbiol. 2009;75:3777–3786. doi: 10.1128/AEM.02763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bostanci N., Bao K., Greenwood D., Silbereisen A., Belibasakis G.N. Advances in Clinical Chemistry. Volume 93. Elsevier; Amsterdam, The Netherlands: 2019. Periodontal disease: From the lenses of light microscopy to the specs of proteomics and next-generation sequencing; pp. 263–290. [DOI] [PubMed] [Google Scholar]
- 3.Schlafer S., Riep B., Griffen A.L., Petrich A., Hübner J., Berning M., Friedmann A., Göbel U.B., Moter A. Filifactor alocis—Involvement in periodontal biofilms. BMC Microbiol. 2010;10:66. doi: 10.1186/1471-2180-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis M.A., Diaz P.I., Van Dyke T.E. The role of the microbiota in periodontal disease. Periodontology 2000. 2020;83:14–25. doi: 10.1111/prd.12296. [DOI] [PubMed] [Google Scholar]
- 5.Mendes L., Rocha R., Azevedo A.S., Ferreira C., Henriques M., Pinto M.G., Azevedo N.F. Novel strategy to detect and locate periodontal pathogens: The PNA-FISH technique. Microbiol. Res. 2016;192:185–191. doi: 10.1016/j.micres.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Hajishengallis G., Lamont R.J. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caton J.G., Armitage G., Berglundh T., Chapple I.L., Jepsen S., Kornman K.S., Mealey B.L., Papapanou P.N., Sanz M., Tonetti M.S. A New Classification Scheme for Periodontal and Peri-Implant Diseases and Conditions—Introduction and Key Changes from the 1999 Classification. Wiley Online Library; Hoboken, NJ, USA: 2018. [DOI] [PubMed] [Google Scholar]
- 8.Sanz M., Marco del Castillo A., Jepsen S., Gonzalez-Juanatey J.R., D’Aiuto F., Bouchard P., Chapple I., Dietrich T., Gotsman I., Graziani F. Periodontitis and cardiovascular diseases: Consensus report. J. Clin. Periodontol. 2020;47:268–288. doi: 10.1111/jcpe.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escapa I.F., Chen T., Huang Y., Gajare P., Dewhirst F.E., Lemon K.P. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): A resource for the microbiome of the human aerodigestive tract. Msystems. 2018;3:e00187-18. doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moter A., Göbel U.B. Fluorescence In Situ Hybridization (FISH) for Direct Visualization of Microorganisms. Elsevier; Amsterdam, The Netherlands: 2000. [DOI] [PubMed] [Google Scholar]
- 11.Levsky J.M., Singer R.H. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 12.Azevedo A.S., Sousa I.M., Fernandes R.M., Azevedo N.F., Almeida C. Optimizing locked nucleic acid/2′-O-methyl-RNA fluorescence in situ hybridization (LNA/2′OMe-FISH) procedure for bacterial detection. PLoS ONE. 2019;14:e0217689. doi: 10.1371/journal.pone.0217689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickinson M., Bearman G., Tille S., Lansford R., Fraser S. Multi-spectral imaging and linear unmixing add a whole new dimension to laser scanning fluorescence microscopy. Biotechniques. 2001;31:1272–1278. doi: 10.2144/01316bt01. [DOI] [PubMed] [Google Scholar]
- 14.Valm A.M., Welch J.L.M., Borisy G.G. CLASI-FISH: Principles of combinatorial labeling and spectral imaging. Syst. Appl. Microbiol. 2012;35:496–502. doi: 10.1016/j.syapm.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valm A.M., Welch J.L.M., Rieken C.W., Hasegawa Y., Sogin M.L., Oldenbourg R., Dewhirst F.E., Borisy G.G. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. USA. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valm A.M., Oldenbourg R., Borisy G.G. Multiplexed spectral imaging of 120 different fluorescent labels. PLoS ONE. 2016;11:e0158495. doi: 10.1371/journal.pone.0158495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tricco A.C., Lillie E., Zarin W., O’Brien K.K., Colquhoun H., Levac D., Moher D., Peters M.D., Horsley T., Weeks L. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann. Intern. Med. 2018;169:467–473. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 18.Mark Welch J.L., Rossetti B.J., Rieken C.W., Dewhirst F.E., Borisy G.G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gmür R., Lüthi-Schaller H. A combined immunofluorescence and fluorescent in situ hybridization assay for single cell analyses of dental plaque microorganisms. J. Microbiol. Methods. 2007;69:402–405. doi: 10.1016/j.mimet.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Machado F.C., Cesar D.E., Assis A.V., Diniz C.G., Ribeiro R.A. Detection and enumeration of periodontopathogenic bacteria in subgingival biofilm of pregnant women. Braz. Oral Res. 2012;26:443–449. doi: 10.1590/S1806-83242012000500011. [DOI] [PubMed] [Google Scholar]
- 21.Gmür R., Wyss C., Xue Y., Thurnheer T., Guggenheim B. Gingival crevice microbiota from Chinese patients with gingivitis or necrotizing ulcerative gingivitis. Eur. J. Oral Sci. 2004;112:33–41. doi: 10.1111/j.0909-8836.2004.00103.x. [DOI] [PubMed] [Google Scholar]
- 22.Lepp P.W., Brinig M.M., Ouverney C.C., Palm K., Armitage G.C., Relman D.A. Methanogenic Archaea and human periodontal disease. Proc. Natl. Acad. Sci. USA. 2004;101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drescher J., Schlafer S., Schaudinn C., Riep B., Neumann K., Friedmann A., Petrich A., Göbel U.B., Moter A. Molecular epidemiology and spatial distribution of Selenomonas spp. in subgingival biofilms. Eur. J. Oral Sci. 2010;118:466–474. doi: 10.1111/j.1600-0722.2010.00765.x. [DOI] [PubMed] [Google Scholar]
- 24.Moter A., Hoenig C., Choi B.-K., Riep B., Göbel U.B. Molecular epidemiology of oral treponemes associated with periodontal disease. J. Clin. Microbiol. 1998;36:1399–1403. doi: 10.1128/JCM.36.5.1399-1403.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wecke J., Kersten T., Madela K., Moter A., Göbel U.B., Friedmann A., Bernimoulin J. A novel technique for monitoring the development of bacterial biofilms in human periodontal pockets. FEMS Microbiol. Lett. 2000;191:95–101. doi: 10.1111/j.1574-6968.2000.tb09324.x. [DOI] [PubMed] [Google Scholar]
- 26.Zijnge V., van Leeuwen M.B., Degener J.E., Abbas F., Thurnheer T., Gmür R., Harmsen H.J. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X., He J., Xue J., Wang Y., Li K., Zhang K., Guo Q., Liu X., Zhou Y., Cheng L. Oral cavity contains distinct niches with dynamic microbial communities. Environ. Microbiol. 2015;17:699–710. doi: 10.1111/1462-2920.12502. [DOI] [PubMed] [Google Scholar]
- 28.Bik E.M., Long C.D., Armitage G.C., Loomer P., Emerson J., Mongodin E.F., Nelson K.E., Gill S.R., Fraser-Liggett C.M., Relman D.A. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge X., Rodriguez R., Trinh M., Gunsolley J., Xu P. Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS ONE. 2013;8:e65520. doi: 10.1371/journal.pone.0065520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., He J., He Z., Zhou Y., Yuan M., Xu X., Sun F., Liu C., Li J., Xie W. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 2014;8:1879–1891. doi: 10.1038/ismej.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koo H., Andes D.R., Krysan D.J. Candida–streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. 2018;14:e1007342. doi: 10.1371/journal.ppat.1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valm A.M. The structure of dental plaque microbial communities in the transition from health to dental caries and periodontal disease. J. Mol. Biol. 2019;431:2957–2969. doi: 10.1016/j.jmb.2019.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakubovics N.S. Saliva as the sole nutritional source in the development of multispecies communities in dental plaque. Metab. Bact. Pathog. 2015:263–277. doi: 10.1128/9781555818883.ch12. [DOI] [PubMed] [Google Scholar]
- 34.Chawhuaveang D.D., Yu O.Y., Yin I.X., Lam W.Y.-H., Mei M.L., Chu C.-H. Acquired salivary pellicle and oral diseases: A literature review. J. Dent. Sci. 2020;16:523–529. doi: 10.1016/j.jds.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katharios-Lanwermeyer S., Xi C., Jakubovics N., Rickard A. Mini-review: Microbial coaggregation: Ubiquity and implications for biofilm development. Biofouling. 2014;30:1235–1251. doi: 10.1080/08927014.2014.976206. [DOI] [PubMed] [Google Scholar]
- 36.Borisy G.G., Valm A.M. Spatial scale in analysis of the dental plaque microbiome. Periodontology 2000. 2021;86:97–112. doi: 10.1111/prd.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Socransky S., Haffajee A., Cugini M., Smith C., Kent R., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 38.Vielkind P., Jentsch H., Eschrich K., Rodloff A.C., Stingu C.-S. Prevalence of Actinomyces spp. in patients with chronic periodontitis. Int. J. Med Microbiol. 2015;305:682–688. doi: 10.1016/j.ijmm.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi N., Yamada T. Glucose and lactate metabolism by Actinomyces naeslundii. Crit. Rev. Oral Biol. Med. 1999;10:487–503. doi: 10.1177/10454411990100040501. [DOI] [PubMed] [Google Scholar]
- 40.Palmer R.J., Jr., Gordon S.M., Cisar J.O., Kolenbrander P.E. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 2003;185:3400–3409. doi: 10.1128/JB.185.11.3400-3409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diaz P., Valm A. Microbial interactions in oral communities mediate emergent biofilm properties. J. Dent. Res. 2020;99:18–25. doi: 10.1177/0022034519880157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsh P.D. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 43.Chapple I.L., Mealey B.L., Van Dyke T.E., Bartold P.M., Dommisch H., Eickholz P., Geisinger M.L., Genco R.J., Glogauer M., Goldstein M. Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: Consensus report of workgroup 1 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018;89:S74–S84. doi: 10.1002/JPER.17-0719. [DOI] [PubMed] [Google Scholar]
- 44.Papapanou P.N., Sanz M., Buduneli N., Dietrich T., Feres M., Fine D.H., Flemmig T.F., Garcia R., Giannobile W.V., Graziani F. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018;89:S173–S182. doi: 10.1002/JPER.17-0721. [DOI] [PubMed] [Google Scholar]
- 45.Jepsen S., Caton J.G., Albandar J.M., Bissada N.F., Bouchard P., Cortellini P., Demirel K., de Sanctis M., Ercoli C., Fan J. Periodontal manifestations of systemic diseases and developmental and acquired conditions: Consensus report of workgroup 3 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018;45:S219–S229. doi: 10.1111/jcpe.12951. [DOI] [PubMed] [Google Scholar]
- 46.Berglundh T., Armitage G., Araujo M.G., Avila-Ortiz G., Blanco J., Camargo P.M., Chen S., Cochran D., Derks J., Figuero E. Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018;89:S313–S318. doi: 10.1002/JPER.17-0739. [DOI] [PubMed] [Google Scholar]
- 47.Fine D.H., Patil A.G., Loos B.G. Classification and diagnosis of aggressive periodontitis. J. Clin. Periodontol. 2018;45:S95–S111. doi: 10.1111/jcpe.12942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.