Significance
Antimalarial resistance has emerged and spread with every antimalarial deployed to date. Currently, parasite genotypes associated with reduced artemisinin and partner drug sensitivity have been reported in Asia, South America, and, most recently, Africa. Analyzing spatial-temporal trends in molecular markers can help policymakers choose efficacious partner drugs and slow the emergence of artemisinin resistance and spread of multidrug-resistant parasites. We display evidence of a continent-wide increase in molecular markers associated with reduced lumefantrine susceptibility, the partner drug of the most widely used artemisinin-based combination therapy in sub-Saharan Africa. We also generate hypotheses for large-scale demographic and environmental risk factors implicated in the spread of antimalarial resistance. Our results can help identify regions of developing parasite resistance that may require enhanced surveillance.
Keywords: malaria, drug resistance, surveillance
Abstract
Resistance to artemisinin-based combination therapies (ACTs) threatens the global control of Plasmodium falciparum malaria. ACTs combine artemisinin-derived compounds with partner drugs to enable multiple mechanisms of clearance. Although ACTs remain widely effective in sub-Saharan Africa, long-standing circulation of parasite alleles associated with reduced partner drug susceptibility may contribute to the development of clinical resistance. We fitted a hierarchical Bayesian spatial model to data from over 500 molecular surveys to predict the prevalence and frequency of four key markers in transporter genes (pfcrt 76T and pfmdr1 86Y, 184F, and 1246Y) in first-level administrative divisions in sub-Saharan Africa from the uptake of ACTs (2004 to 2009) to their widespread usage (2010 to 2018). Our models estimated that the pfcrt 76T mutation decreased in prevalence in 90% of regions; the pfmdr1 N86 and D1246 wild-type genotypes increased in prevalence in 96% and 82% of regions, respectively; and there was no significant directional selection at the pfmdr1 Y184F locus. Rainfall seasonality was the strongest predictor of the prevalence of wild-type genotypes, with other covariates, including first-line drug policy and transmission intensity more weakly associated. We lastly identified regions of high priority for enhanced surveillance that could signify decreased susceptibility to the local first-line ACT. Our results can be used to infer the degree of molecular resistance and magnitude of wild-type reversion in regions without survey data to inform therapeutic policy decisions.
Artemisinin-based combination therapies (ACTs) are recommended as the first-line treatment for uncomplicated Plasmodium falciparum malaria worldwide (1). ACTs combine potent artemisinin-derived compounds and partner drugs with longer half-lives, allowing for dual mechanisms of action and prolonged antiparasitic action. Of the ACTs currently recommended by the World Health Organization, artemether-lumefantrine (AL) and artesunate-amodiaquine (AS-AQ) are the most commonly adopted first-line ACTs in sub-Saharan Africa (1). While efficacy remains high, recent reports of mutations associated with reduced artemisinin sensitivity in Rwanda and long-standing circulation of alleles associated with bidirectional impacts on partner drug susceptibility raise concern that ACT resistance may arise and spread in sub-Saharan Africa (2–4).
Molecular markers are single nucleotide polymorphisms (SNPs), deletions, or copy number variations in the parasite genome that are associated with reduced susceptibility to antimalarial drugs. SNPs along two key transporter genes, P. falciparum chloroquine resistance transporter (pfcrt) and P. falciparum multidrug resistance 1 (pfmdr1), were first discovered to confer resistance to antimalarial drugs in the 1990s, primarily pfcrt 76T to chloroquine, and have since been implicated in impacting susceptibility to AL and AS-AQ (5). Notably, the partner drugs lumefantrine and AQ exert opposing selective pressures: parasites with genotype pfmdr1 86Y, Y184, 1246Y, and pfcrt 76T have reduced susceptibility to AQ, while pfmdr1 N86, 184F, D1246, and pfcrt K76 confer reduced susceptibility to lumefantrine (5–7). While data are strongest for partner drugs, artemisinins may also directly exert a selective pressure at pfmdr1 and other loci (8, 9).
In southeast Asia, partner drug resistance arose rapidly in the wake of, and in some regions independently of, artemisinin resistance (10). In sub-Saharan Africa, it has been suggested that partner drug failure is expected to result in greater increases in malaria morbidity than would be observed from artemisinin resistance alone (4). The population prevalence of markers in pfcrt and pfmdr1 can indicate the level of clinical resistance or tolerance to the partner drugs AQ and lumefantrine (11). When used as a surveillance tool, molecular marker data can rapidly provide data to support modifications of local partner drugs and to sustain artemisinin-based treatment and prevention options. Despite broad consensus to scale up molecular surveillance for antimalarial resistance, few studies have proposed systematically designed approaches such as where sentinel sites should be located or how often sampling should be conducted (12, 13).
We previously developed a database of studies assessing prevalence of partner drug resistance–associated markers in sub-Saharan Africa, supplementing an existing database by the Worldwide Antimalarial Resistance Network (14, 15). That work revealed important gaps in the geographic coverage of surveillance. We hypothesized that the geographic landscape of mutations associated with partner drug resistance exhibit spatial patterns that can be mapped and leveraged to determine sites needing additional surveillance. In this study, we use a hierarchical Bayesian spatial method to map the geographical distribution of key molecular markers in pfcrt and pfmdr1 in sub-Saharan Africa since the uptake of ACTs and analyze potential demographic and environmental determinants of resistance landscapes. We also identify potential regions for enhanced surveillance based on the magnitude and direction of changes in genotypes over time, suggesting areas that may pose an increased risk of artemisinin emergence or spread (16). Our work will assist in strategic surveillance efforts to sustain first-line ACT partner drug efficacy in sub-Saharan Africa and inform future surveillance as new antimalarials are deployed and additional molecular markers are identified.
Results
Geographic Distribution of Mutant Genotypes.
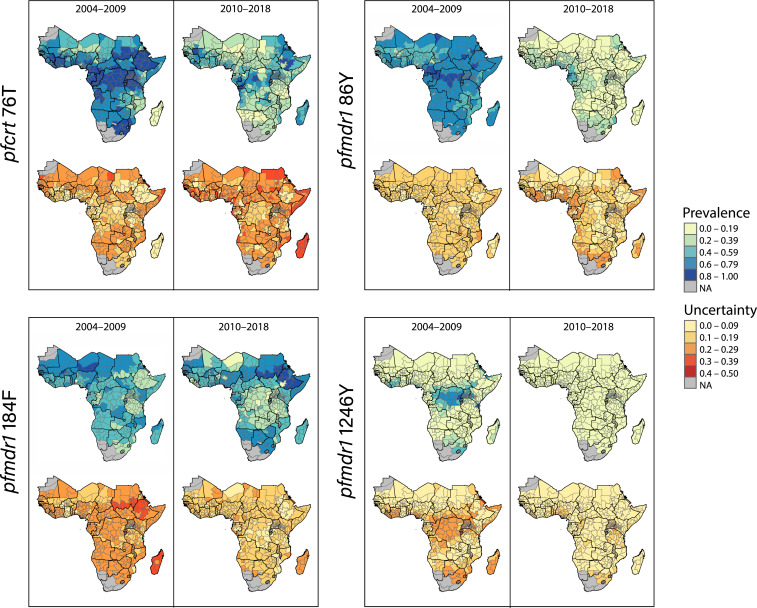
We generated maps of marker prevalence and uncertainty interpolated across the malaria-endemic subcontinent from 2004 to 2009 and from 2010 to 2018 (Fig. 1). The mutations at pfcrt 76T and pfmdr1 86Y were extensively prevalent in 2004 to 2009 and much more sparsely present in 2010 to 2018 (Table 1), with pfmdr1 86Y prevalent only in the Gulf of Guinea region. The pfmdr1 1246Y genotype exhibited the highest degree of prevalence in central and east Africa, as well as Côte d'Ivoire and South Africa, compared to all other regions from 2004 to 2009, but low prevalence across the continent in the latter time period. Meanwhile, pfmdr1 184F exhibited more randomness over time and space than the other markers, concentrated in the northwest from 2004 to 2009 and the northeast from 2010 to 2018 (Table 1). Pfcrt 76T estimates displayed the highest levels of uncertainty compared to other markers in the later time period, with isolated hotspots of uncertainty on the mainland and high uncertainty in Madagascar. Because of the similarity in regional trends and contributions of covariates between frequency and prevalence estimates (SI Appendix, Figs. S4 and S5) and superior prevalence data coverage, additional analyses involved prevalence estimates only.
Fig. 1.
Maps displaying model estimates and predictions for molecular marker prevalence in first-level administrative divisions from 2004 to 2009 and 2010 to 2018 and respective posterior SDs representing uncertainty.
Table 1.
Fitted and predicted estimates of molecular marker prevalence
| Marker | Time period | East (mean, SD) | West (mean, SD) | Central (mean, SD) | Southern (mean, SD) | Aggregate prevalence | No. of ADs with decreases in mutation prevalence (%) |
| pfcrt 76T | 2004 to 2009 | 0.76 (0.19) | 0.67 (0.17) | 0.74 (0.15) | 0.50 (0.27) | 0.75 (0.21) | 544 (89.5%) |
| 2010 to 2018 | 0.52 (0.19) | 0.44 (0.14) | 0.45 (0.18) | 0.33 (0.13) | 0.45 (0.18) | ||
| pfmdr1 86Y | 2004 to 2009 | 0.70 (0.09) | 0.54 (0.12) | 0.69 (0.12) | 0.58 (0.12) | 0.62 (0.15) | 585 (96.2%) |
| 2010 to 2018 | 0.34 (0.11) | 0.25 (0.14) | 0.24 (0.17) | 0.18 (0.14) | 0.23 (0.16) | ||
| pfmdr1 184F | 2004 to 2009 | 0.43 (0.13) | 0.62 (0.11) | 0.55 (0.12) | 0.49 (0.11) | 0.52 (0.21) | 313 (51.4%) |
| 2010 to 2018 | 0.54 (0.19) | 0.55 (0.16) | 0.49 (0.15) | 0.49 (0.16) | 0.51 (0.17) | ||
| pfmdr1 1246Y | 2004 to 2009 | 0.36 (0.26) | 0.14 (0.16) | 0.21 (0.20) | 0.13 (0.13) | 0.23 (0.15) | 496 (81.6%) |
| 2010 to 2018 | 0.09 (0.11) | 0.07 (0.07) | 0.05 (0.04) | 0.05 (0.03) | 0.08 (0.08) |
The table shows the average fitted/estimated prevalence and SD for all administrative divisions in the respective time period and region of sub-Saharan Africa, the overall average for all administrative divisions (ADs), and the overall number of ADs with decreased prevalence of mutant alleles in the latter time period.
Temporal Reversion to Wild-Type Genotypes.
The mean estimated prevalence of mutant genotypes decreased significantly from 2004 to 2009 to 2010 to 2018 for pfmdr1 86Y and 1246Y and pfcrt 76T [1.0 for all three markers (SI Appendix, Fig. S6)], with the greatest magnitude of change for pfmdr1 86Y (Table 1). West and southern Africa exhibited the smallest estimated changes in prevalence of all four markers. There were noticeable exceptions to wild-type reversion: For example, pfcrt 76T prevalence increased, according to our predictions, in parts of Niger, Nigeria, Djibouti, and Madagascar, although such areas tended to exhibit higher uncertainty. The distribution of pfmdr1 184F changed markedly over the two time periods, yet we found no evidence of directional selection toward the wild-type pfmdr1 Y184, suggesting more random and less spatially explicit patterns of selection [P]. The magnitude and direction of temporal changes for each administrative division can be visualized in SI Appendix, Fig. S6. The prevalence of pfcrt 76T was positively correlated with pfmdr1 86Y, which, in turn, was associated with pfmdr1 Y184 and pfmdr1 1246Y. Correlations between changes in prevalence for each pair of markers can be found in SI Appendix, Fig. S7.
Contributions of Covariates to Genotypic Variability.
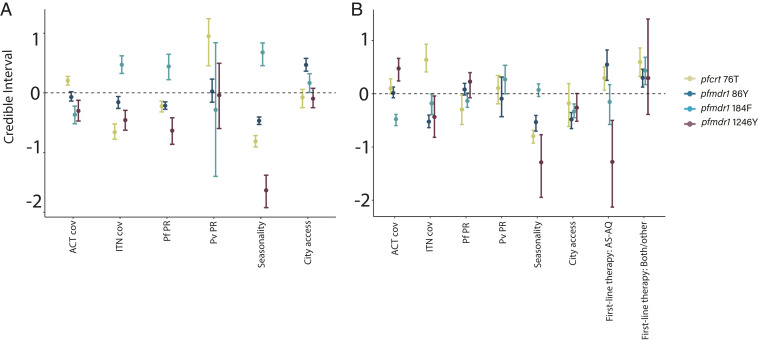
Seasonality was consistently the strongest variable associated with the presence of wild-type alleles pfcrt K76, pfmdr1 N86, and pfmdr1 D1246 and of the mutant allele pfmdr1 184F (Fig. 2 and SI Appendix, Fig. S4) in both time periods. From 2004 to 2009, transmission intensity of P. falciparum was associated with prevalence of markers pfcrt K76 and pfmdr1 N86, 184F, and D1246, while the transmission intensity of Plasmodium vivax was associated with pfcrt 76T. From 2010 to 2018, city accessibility and first-line drug policy were correlated with the prevalence of pfcrt K76, pfmdr1 N86, pfmdr1 D1246, and pfmdr1 184F alleles, with AS-AQ and both/other drug regimens generally associated with the presence of opposing alleles compared to AL (Fig. 2).
Fig. 2.
Contributions of covariates in the periods of 2004–2009 (A) and 2010–2018 (B). Posterior regression parameter estimates (mean, points, and 95% credible interval, whiskers) for covariates used in spatial models estimating the prevalence of individual molecular markers and time periods. Continuous variables are scaled for ease of comparison. The reference category for first-line therapy was AL. First-line drug policies were included only as covariates in the latter time period because they were assumed to have little effect during the period of ACT uptake (details in SI Appendix, Table S1).
We also estimated the impacts of covariates on the magnitude and direction of changes in marker prevalence over time (SI Appendix, Fig. S8). Areas with higher rainfall seasonality exhibited smaller changes in prevalence over time for all four markers after controlling for first-line drug policy and other variables. First-line drug policy was associated with the magnitude of change of most markers, although with much greater bounds of uncertainty: Compared to AL, AS-AQ was associated with a greater magnitude of change toward mutant genotypes pfcrt 76T and pfmdr1 86Y, and the use of multiple or other first-line therapies was associated with smaller changes toward wild-type genotypes pfmdr1 Y184 and pfmdr1 D1246. When stratifying the results by first-line drug regimen, trends were more difficult to parse out (SI Appendix, Table S2 and Fig. S9).
Regions for Enhanced Surveillance.
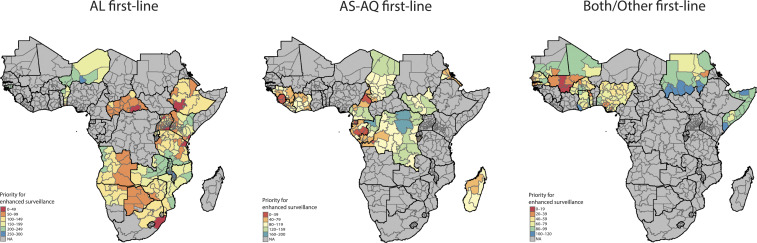
We identified numerous regions that could be prioritized for enhanced surveillance (Fig. 3) based on the relative strength and direction of change in prevalence over time and the degree of uncertainty in model predictions. Higher-priority regions were located in parts of the Central African Republic, Ethiopia, Tanzania, Gabon, Equatorial Guinea, Sierra Leone, Nigeria, Togo, and Mali. A specific set of administrative divisions demonstrated the highest posterior probability of selection (surveillance prioritization) across all model iterations, suggesting greater certainty in the relative strength of directional selection over time (SI Appendix, Fig. S10). These administrative divisions include Luapula Province, Zambia; Litoral Province, Equatorial Guinea; Southern Nations, Nationalities, and People’s Region, Ethiopia; Pointe-Noire Department, Republic of the Congo; and Edo State, Nigeria, with Pointe-Noire having the highest probability of selection for enhanced surveillance compared to any other region. These results may be partly reflective of higher sampling density in these subregions.
Fig. 3.
Prioritized sites for enhanced molecular surveillance. First-level administrative divisions were comparatively ranked according to the relative magnitude of change over time, dependent on first-line drug regimen to account for directional selection, after accounting for uncertainty in model results. Red regions are ranked highest priority for surveillance; blue regions are lower priority.
Discussion
Our study demonstrates the striking change in prevalence of key partner drug susceptibility–associated molecular markers in sub-Saharan Africa since the introduction of ACTs and provides an attempt to identify focal regions to monitor for decreased partner drug susceptibility. We found a strong wild-type resurgence of pfcrt K76, likely reflecting selection due to the combined impacts of the uptake of AL and withdrawal of chloroquine monotherapy, as evidenced by many studies at smaller spatial scales or in imported parasites (17, 18). We also developed spatially smoothed maps demonstrating wild-type selection of pfmdr1 N86 and D1246 across the continent, supporting a study by Okell et al. (19) that also found evidence of widespread wild-type reversion of these markers. Together, our results suggest that surveillance can be more effective if not only coordinated at a national scale but also taking into account regional spatial dependencies.
We identified intriguing areas of remaining pfcrt 76T and pfmdr1 86Y genotype presence, particularly in Madagascar, Ethiopia, the Gulf of Guinea, and parts of West Africa. One potential reason for the persistence in Madagascar and Ethiopia may be related to the usage of chloroquine to treat P. vivax where the species remains endemic (1, 20). In the Gulf of Guinea, a region where seasonal malaria chemoprevention (SMC) with AQ and sulfadoxine-pyrimethamine (SP) is used in children and higher rates of AS-AQ are used for treatment than in many other parts of Africa, the added pressure of AQ may explain the higher prevalence of these alleles (1). However, because the widespread administration of SMC began in the latter half of the second time interval, SMC was not included as a covariate. In all of these regions, the continued use of chloroquine to treat P. falciparum may also drive mutation persistence, but rigorous data on consumption of chloroquine are not available at this scale (21–23).
We also assessed the contribution of socioecological factors that may help explain drug resistance heterogeneity, aided by the additional power in scale of this analysis. Wild-type reversion of pfcrt K76, pfmdr1 N86, and pfmdr1 D1246 occurred across sub-Saharan Africa regardless of first-line therapy, yet countries using AS-AQ or multiple first-line therapies tended to exhibit a smaller magnitude of reversion overall. Areas with high seasonality strongly favored wild-type genotypes, but again, the extent of the reversion to wild type was partially lessened by the use of AQ. Our results support strong evidence that pfcrt 76T confers a fitness cost to the parasite and add to a growing body of evidence of a fitness cost of pfmdr1 86Y and 1246Y (24–26). In the dry season, these mutations may lose their competitive advantage in the absence of sustained drug pressure (27, 28). We also found that higher transmission intensity favored pfcrt K76, pfmdr1 N86, and pfmdr1 D1246 in the earlier time period, which may be related to higher rates of recombination in circulating parasites, increased levels of immunity in high transmission regions, and heterogeneity in the proportion of infections that encounter treatments (29–31). Our results ultimately suggest that environmental and epidemiologic factors may be of increased importance, in addition to national policy, in delineating factors to consider when conducting surveillance.
The covariates in our models and their relationship to drug resistance have important caveats. Our results are subject to ecological fallacy due to the large geographic scale of the analysis. In particular, because of limited data on antimalarial consumption patterns available across the subcontinent, multiple variables acted as potential proxies for detailed drug consumption patterns at the level of the administrative division, including first-line drug policy, ACT coverage, insecticide-treated bed net (ITN) coverage, and city accessibility. For example, we found that ITN coverage and city accessibility were correlated with pfmdr1 N86, 184F, and D1246, but these factors likely acted as proxies for drug quality, access, and mobility patterns (32). More studies are needed to determine the explicit association between these covariates, drug consumption patterns, and molecular marker prevalence (21, 22, 32). Our modeling approach cannot speak to many of the biological aspects of malaria transmission or mechanisms by which these covariates impact drug resistance. Future field and experimental studies, perhaps at more focal scales at which biological-level data can be obtained, are also needed.
Our analyses are also limited by gaps in spatial, temporal, and genetic coverage of data. To increase the predictive capacity of our models, we aggregated data into two time units, and our results may not precisely match localized reports of resistance or trends where more continuous data are available. To develop the largest possible dataset, we examined four well-known, commonly assessed molecular markers, but we were not able to analyze haplotypes or incorporate more detailed sequencing data. Although copy number variation in pfmdr1 has been found in parts of Africa, limited reporting and large heterogeneity in polygenomic infections made these data challenging to model and interpret (15, 33). Our framework allows for incorporation of existing markers as well as novel mutations as they become available and more widely assessed, such as those conferring phenotypic resistance to artemisinin in Africa, as was the case in southeast Asia (34). Finally, systematic acquisition and storage of samples can also serve as a repository for retrospective studies of newly identified markers of interest.
A number of previous studies have mapped molecular markers associated with antimalarial resistance at regional scales. Geospatial maps were first developed for markers implicated in resistance to SP, a former first-line P. falciparum treatment, in sub-Saharan Africa (13, 35–37). Additional studies by Tun et al. (38) and Grist et al. (39) mapped polymorphisms in pfK13 conferring artemisinin resistance in southeast Asia. Finally, Okell et al. (19) analyzed trends in pfmdr1 markers using point prevalence surveys across sub-Saharan Africa. We add to this body of work by including another critical locus, pfcrt K76T, and by developing spatially smoothed maps of partner drug resistance prevalence and uncertainty. These maps enable researchers to estimate marker prevalence in areas without sampling and to leverage uncertainty for strategic surveillance (14). This study also helps determine continent-wide geographic and temporal trends in key molecular markers and their association with environmental and policy landscapes.
In the final section of this study, we provide maps (Fig. 3) delineating regions undergoing potentially strong selection toward genotypes associated with reduced drug susceptibility, providing a genetic background within which artemisinin resistance may be more likely to emerge or spread (16). We recommend enhanced surveillance in these regions, which could entail more frequent surveys to establish the real-time frequency of molecular markers as well as patterns of drug consumption and quality. Additional focused studies in these areas may include treatment efficacy and pharmacokinetic studies as well as deep sequencing of local parasite strains to delineate haplotypes and novel variants (11, 12, 40, 41). The utility of our surveillance maps will improve with additional data coverage from surveillance conducted throughout a country/region, ideally under the guidance of national malaria control programs, and future versions should be optimized to support local research and decision making (42, 43). We intend for this work to help catalyze global agencies and regional networks alike to allocate resources urgently and strategically for drug resistance surveillance.
Materials and Methods
Prevalence Data Assembly and Aggregation.
We aggregated the results of 254 published studies in 35 countries in sub-Saharan Africa and Sudan, as previously described, encompassing 501 surveys assessing molecular markers in a specific location and year (14). In brief, all surveys assessed P. falciparum samples collected between 2004 and 2018 in sub-Saharan Africa for genotypes pfmdr1 N86Y, Y184F, and D1246Y and/or pfcrt K76T. We excluded studies that reported prevalence data aggregated across multiples sites more than 300 km apart and whose results could not be disaggregated (n = 23 surveys) (35). From each survey, we extracted the midpoint year of sampling, geographic coordinates of the study site, the number of samples tested at each locus, and the number of samples with wild-type, mutant, or mixed genotypes (in the case of polygenomic infections) at each locus. Prevalence was calculated as the number of individuals with mutant or mixed infections out of the total surveyed (35).
The study region was divided into the 608 first-level administrative divisions having endemic malaria transmission during the study period (44); the island nation of Comoros was excluded because administrative divisions did not have spatially contiguous neighbors, a requirement for our spatial models. The study period was divided into two time units representing the uptake of ACTs, 2004 to 2009, and widespread usage of ACTs, 2010 to 2018 (19). The observed prevalence of each marker in each administrative division was calculated as the weighted average of all respective survey estimates within each administrative division and time period (SI Appendix, Fig. S1).
While we define prevalence as the number of individuals with mutant or mixed infections out of the total number of P. falciparum–infected individuals surveyed, frequency is the proportion of parasite clones with the molecular marker in the parasite population, which may matter in highly endemic regions in which individuals often have polygenomic infections (35, 45). For example, say an individual is infected with four clones of P. falciparum, one of which has the mutant genotype of interest: If only that single person were surveyed, the frequency of the mutation would be 25%, whereas the prevalence would be 100%. Because the majority of studies (n = 209/254 studies) did not report the average number of clones within the surveyed population, we used the subset of data that reported the prevalence of mixed and mutant infections separately (n = 245/501 surveys) and inferred the proportion of multiclonal infections using the P. falciparum parasite rate obtained from the Malaria Atlas Project (44). We then applied methodology developed by Okell et al. (35) to estimate the observed frequency of each marker in each survey and aggregated surveys in the same method as described above.
Covariate Data Assembly and Selection.
Variables that we hypothesized to be associated with drug resistance were considered for inclusion (SI Appendix, Table S1 and Supplementary Methods) (12, 46). The final selected set of variables were P. falciparum parasite rate in children ages 2 to 10 y (P. falciparum parasite rate [PfPR]2–10), P. vivax parasite rate in individuals ages 0 to 99 y (P. vivax parasite rate [PvPR]0–99), seasonality of annual rainfall, ACT coverage, ITN coverage, city accessibility, and national first-line drug policy (AL, AS-AQ, or both/other; SI Appendix, Fig. S2) for the treatment of P. falciparum. For all covariates, excluding those only available at the national level, we averaged pixel values from rasterized data to generate administrative division-level estimates. Covariates were extracted for the midpoint year of both time periods, when possible, and for the equivalent study region (SI Appendix, Table S1 and Fig. S3).
Bayesian Hierarchical Spatial Model.
We used a logistic regression framework with spatially correlated random effects to identify factors that explain variability in the prevalence of molecular markers associated with partner drug resistance by first-level administrative divisions and to predict prevalence in administrative divisions without observed data. We previously found evidence of spatial autocorrelation in the data, suggesting utility of spatial analysis (14). Here, we specify a conditional autoregressive (CAR) model for the random effects which allows for the possibility that neighboring administrative divisions exhibit similar behavior, leading to localized smoothness of estimated prevalence, potentially improved predictions in unobserved regions, and accurate statistical inference for the regression associations of interest. The model for the aggregated survey data during a selected time period t (t = 1: 2004 to 2009, t = 2: 2010 to 2018) is given as
and
where is the number of mutant or mixed genotypes observed in administrative division k during time period t, is the total number of individuals tested, and is the true prevalence of resistance. Logistic regression is used to link with the covariates and random effect, where represents a vector of previously described covariates specific to administrative division k during time period t and is the spatially correlated random effect.
We specify the Leroux version of the CAR model for the spatial random effects (47), with an intuitive conditional form such that
where is the full vector of spatial random effects from time period t with removed and is equal to one if administrative division k and administrative division j are neighbors (i.e., share a common border) and is equal to zero otherwise; is equal to zero for all k (4). The CAR model includes two hyperparameters that control the total variability of the effects () and whether this variability is spatially smooth () or independent (), both of which are assigned prior distributions so that the data inform these quantities. This model specifies that the prior mean of a particular random effect is a weighted average of its neighbors’ random effect values. As approaches one, more emphasis is placed on the neighbors’ values, whereas the random effect becomes centered closer to zero as approaches zero.
Models were fit separately for each marker and time period. We selected weakly informative priors for model parameters to allow the data to drive the inference, such that (0, 1002), Uniform(0, 1) and Inverse gamma(0.01, 0.01). We collected 1,000,000 samples (after discarding 200,000 during a burn-in period) from the joint posterior distribution of the parameters using a Markov chain Monte Carlo sampling algorithm. We thinned these samples by a factor of 100 to reduce posterior autocorrelation, resulting in 10,000 nearly independent samples with which to make posterior inference. Convergence was assessed through visual inspection of trace plots and calculation of the Geweke diagnostic for all monitored parameters, with no obvious signs of nonconvergence across all fitted models. All analyses were conducted in R version 3.5.1. Spatial models were implemented with the CARBayes package (48).
Estimation of Marker Prevalence and Changes over Time.
In administrative divisions with observed data, we extracted the posterior mean of . In administrative divisions without observed data, we used the CAR structure of the spatial model to predict the prevalence and uncertainty by collecting samples from the corresponding posterior predictive distributions of the random effects. Hereafter, prevalence represents the posterior mean or SD of the predicted or fitted estimates for each administrative division. For each marker, we compared the change in prevalence over time by calculating the posterior probability that the prevalence from 2004 to 2009 was larger than the prevalence from 2010 to 2018. We also fitted a linear regression model with CAR random effects to determine which covariates explained variability in the estimated differences in prevalence over time while also accounting for spatial correlation in the data (SI Appendix, Supplementary Methods).
Identification of Potential Regions for Enhanced Surveillance.
We considered regions of high priority for enhanced surveillance to be those with large relative changes toward the genotypes that confer reduced susceptibility to the first-line regimen used in that region, or a large relative change in any direction in the case of multiple first-line therapies. For each administrative division, we calculated the relative change in marker prevalence over the two time periods for each posterior sample collected, yielding 10,000 difference estimates. We then subset administrative divisions by the first-line drug regimen used in that region (AL, AS-AQ, or both/other) to account for opposing selective pressures of drugs on markers in pfcrt and pfmdr1. Maps were created with the tmap package (49).
Supplementary Material
Acknowledgments
H.Y.E. was supported by the NIH National Institute of Allergy and Infectious Diseases (NIAID) Ruth L. Kirschstein National Research Service Award F31-AI150168; A.K.B. was supported by the NIH Fogarty International Center Grant K01-TW010496; D.M.W. and J.L.W. were supported by NIAID Grant R01-AI137093; and S.P. was supported by NIAID Grant R21-AI135477.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100685118/-/DCSupplemental.
Data Availability
Data, code, and maps generated by this study are publicly available in GitHub at https://github.com/hannaehrlich/maldrugres_SSA (50). Molecular marker survey data can also be viewed and deposited in the Worldwide Antimalarial Resistance Network at https://www.wwarn.org/tracking-resistance/act-partner-drug-molecular-surveyor. Please direct any requests for tailored mapping modifications to the corresponding author.
References
- 1.World Health Organization , World Malaria Report (World Health Organization, Geneva, Switzerland, 2020). [Google Scholar]
- 2.Uwimana A., et al., Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 26, 1602–1608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uwimana A., et al., Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: An open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect. Dis. S1473-3099(21)00142-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slater H. C., Griffin J. T., Ghani A. C., Okell L. C., Assessing the potential impact of artemisinin and partner drug resistance in sub-Saharan Africa. Malar. J. 15, 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatesan M., et al., Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: Parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am. J. Trop. Med. Hyg. 91, 833–843 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picot S., et al., A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar. J. 8, 89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arya A., Kojom Foko L. P., Chaudhry S., Sharma A., Singh V., Artemisinin-based combination therapy (ACT) and drug resistance molecular markers: A systematic review of clinical studies from two malaria endemic regions—India and sub-Saharan Africa. Int. J. Parasitol. Drugs Drug Resist. 15, 43–56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henriques G., et al., Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J. Infect. Dis. 210, 2001–2008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duraisingh M. T., Roper C., Walliker D., Warhurst D. C., Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol. Microbiol. 36, 955–961 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Imwong M., et al., The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: A molecular epidemiology observational study. Lancet Infect. Dis. 17, 491–497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nsanzabana C., Ariey F., Beck H.-P., et al., Molecular assays for antimalarial drug resistance surveillance: A target product profile. PLoS One 13, e0204347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talisuna A. O., et al., Mitigating the threat of artemisinin resistance in Africa: Improvement of drug-resistance surveillance and response systems. Lancet Infect. Dis. 12, 888–896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naidoo I., Roper C., Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol. 29, 505–515 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich H. Y., Jones J., Parikh S., Molecular surveillance of antimalarial partner drug resistance in sub-Saharan Africa: A spatial-temporal evidence mapping study. Lancet Microbe 1, e209–e217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otienoburu S. D., et al., An online mapping database of molecular markers of drug resistance in Plasmodium falciparum: The ACT Partner Drug Molecular Surveyor. Malar. J. 18, 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson O. J., et al., Pre-existing partner-drug resistance facilitates the emergence and spread of artemisinin resistance: A consensus modelling study. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.04.08.437876v1 (Accessed 25 April 2021). [DOI] [PMC free article] [PubMed]
- 17.Laufer M. K., et al., Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 355, 1959–1966 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Lu F., et al., Return of chloroquine sensitivity to Africa? Surveillance of African Plasmodium falciparum chloroquine resistance through malaria imported to China. Parasit. Vectors 10, 355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okell L. C., Reiter L. M., Ebbe L. S., et al., Emerging implications of policies on malaria treatment: Genetic changes in the Pfmdr-1 gene affecting susceptibility to artemether–lumefantrine and artesunate–amodiaquine in Africa. BMJ Glob. Health 3, e000999 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hailemeskel E., et al., Prevalence of Plasmodium falciparum Pfcrt and Pfmdr1 alleles in settings with different levels of Plasmodium vivax co-endemicity in Ethiopia. Int. J. Parasitol. Drugs Drug Resist. 11, 8–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frosch A. E., Venkatesan M., Laufer M. K., Patterns of chloroquine use and resistance in sub-Saharan Africa: A systematic review of household survey and molecular data. Malar. J. 10, 116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ocan M., et al., Persistence of chloroquine resistance alleles in malaria endemic countries: A systematic review of burden and risk factors. Malar. J. 18, 76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flegg J. A., et al., Trends in antimalarial drug use in Africa. Am. J. Trop. Med. Hyg. 89, 857–865 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ecker A., Lehane A. M., Clain J., Fidock D. A., PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 28, 504–514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayward R., Saliba K. J., Kirk K., pfmdr1 mutations associated with chloroquine resistance incur a fitness cost in Plasmodium falciparum. Mol. Microbiol. 55, 1285–1295 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Ochong E., Tumwebaze P. K., Byaruhanga O., Greenhouse B., Rosenthal P. J., Fitness consequences of Plasmodium falciparum pfmdr1 polymorphisms inferred from ex vivo culture of Ugandan parasites. Antimicrob. Agents Chemother. 57, 4245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babiker H. A., Gadalla A. A., Ranford-Cartwright L. C., The role of asymptomatic P. falciparum parasitaemia in the evolution of antimalarial drug resistance in areas of seasonal transmission. Drug Resist. Updat. 16, 1–9 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Ord R., et al., Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum imply reduced fitness of chloroquine-resistant parasites. J. Infect. Dis. 196, 1613–1619 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Klein E. Y., Smith D. L., Laxminarayan R., Levin S.. Superinfection and the evolution of resistance to antimalarial drugs. Proc. Biol. Sci. 279, 3834–3842 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ataide R., et al., Host immunity to Plasmodium falciparum and the assessment of emerging artemisinin resistance in a multinational cohort. Proc. Natl. Acad. Sci. U.S.A. 114, 3515–3520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenthal P. J., The interplay between drug resistance and fitness in malaria parasites. Mol. Microbiol. 89, 1025–1038 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rathmes G., et al., Global estimation of anti-malarial drug effectiveness for the treatment of uncomplicated Plasmodium falciparum malaria 1991–2019. Malar. J. 19, 374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gadalla N. B., et al., Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrob. Agents Chemother. 55, 5408–5411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariey F., et al., A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okell L. C., Griffin J. T., Roper C., Mapping sulphadoxine-pyrimethamine-resistant Plasmodium falciparum malaria in infected humans and in parasite populations in Africa. Sci. Rep. 7, 7389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flegg J. A., et al., Spatiotemporal mathematical modelling of mutations of the dhps gene in African Plasmodium falciparum. Malar. J. 12, 249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sridaran S., et al., Anti-folate drug resistance in Africa: Meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malar. J. 9, 247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tun K. M., et al., Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: A cross-sectional survey of the K13 molecular marker. Lancet Infect. Dis. 15, 415–421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grist E. P., et al., Optimal health and disease management using spatial uncertainty: A geographic characterization of emergent artemisinin-resistant Plasmodium falciparum distributions in Southeast Asia. Int. J. Health Geogr. 15, 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apinjoh T. O., Ouattara A., Titanji V. P. K., Djimde A., Amambua-Ngwa A., Genetic diversity and drug resistance surveillance of Plasmodium falciparum for malaria elimination: Is there an ideal tool for resource-limited sub-Saharan Africa? Malar. J. 18, 217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nsanzabana C., Resistance to artemisinin combination therapies (ACTs): Do not forget the partner Drug! Trop. Med. Infect. Dis. 4, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren J. L., Perez-Heydrich C., Yunus M., Bayesian spatial design of optimal deep tube well locations in Matlab, Bangladesh. Environmetrics 24, 377–386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diggle P. J., et al., Spatial modelling and the prediction of Loa loa risk: Decision making under uncertainty. Ann. Trop. Med. Parasitol. 101, 499–509 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Weiss D. J., et al., Mapping the global prevalence, incidence, and mortality of Plasmodium falciparum, 2000-17: A spatial and temporal modelling study. Lancet 394, 322–331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson T., Campino S. G., Auburn S., et al., Drug-resistant genotypes and multi-clonality in Plasmodium falciparum analysed by direct genome sequencing from peripheral blood of malaria patients. PLoS One 6, e23204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeffer D. A., et al., malariaAtlas: An R interface to global malariometric data hosted by the Malaria Atlas Project. Malar. J. 17, 352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leroux B. G., Lei X., Breslow N., “Estimation of disease rates in small areas: A new mixed model for spatial dependence” in Statistical Models in Epidemiology, the Environment, and Clinical Trials, Halloran M. E., Berry D., Eds. (Springer, New York, NY, 2000), pp. 179–191. [Google Scholar]
- 48.Lee D., CARBayes version 4.6: An R package for spatial areal unit modelling with conditional autoregressive priors. J. Stat. Softw. 55, 1–24 (2013). [Google Scholar]
- 49.Tennekes M., tmap: Thematic Maps in R. J. Stat. Softw. 84, 39 (2018). [Google Scholar]
- 50.Ehrlich H. Y., Sunil Parikh, maldrugres_SSA. GitHub. https://github.com/hannaehrlich/maldrugres_SSA. Deposited 27 June 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data, code, and maps generated by this study are publicly available in GitHub at https://github.com/hannaehrlich/maldrugres_SSA (50). Molecular marker survey data can also be viewed and deposited in the Worldwide Antimalarial Resistance Network at https://www.wwarn.org/tracking-resistance/act-partner-drug-molecular-surveyor. Please direct any requests for tailored mapping modifications to the corresponding author.