Abstract
The study investigated carbapenemase-producing Klebsiella pneumoniae (CPKP) isolates of patients in an intensive care unit (ICU) in a public hospital in the KwaZulu-Natal province, South Africa using whole-genome sequencing (WGS). Ninety-seven rectal swabs, collected from all consenting adult patients (n = 31) on days 1, 3, and 7 and then weekly, were screened for carbapenemase-production using Chrome-ID selective media. Antibiotic susceptibility was determined for the fourteen positive CPKP isolates obtained using the VITEK 2 automated system. All isolates (100%) were resistant to ertapenem and meropenem, and 71.4% (n = 10) were resistant to imipenem. All CPKP isolates were subjected to ERIC/PCR, and a sub-sample of isolates was selected for WGS based on their antibiograms and clonality. All sequenced isolates harbored the blaOXA-181 carbapenemase (100%) and co-carried other β-lactamase genes such as blaOXA-1, blaCTX-M-15, blaTEM-1B, and blaSHV-1. IncF, IncX3, and Col plasmid replicons groups and class I integrons (ln191 and ln27) were detected. All isolates belonged to the same sequence type ST307 and capsular serotypes (K102, O2v2). All the isolates carried the same virulence repertoire, reflecting the epidemiological relationship between isolates. blaOXA-181 was located on a multi-replicon plasmid similar to that of E. coli p010_B-OXA181, and isolates were aligned with several South African and international clades, demonstrating horizontal and vertical transboundary distribution. The findings suggest that blaOXA-181 producing K. pneumoniae is endemic in this ICU, colonizing the patients. CRE screening and enhanced infection prevention and control measures are urgently required.
Keywords: carbapenem resistance, Enterobacterales, Klebsiella pneumoniae, whole genome sequencing, South Africa, intensive care
1. Introduction
β-lactams are one of the most prescribed antibiotics globally [1], in part because they are flexible and can be structurally modified to result in several active forms, a malleability not possessed by other antibiotic classes [2]. Carbapenems, among the β-lactams, are effective against Gram-positive and Gram-negative bacteria, providing a broad spectrum of antibacterial activity. A particular molecular structure, along with the β-lactam ring, gives added stability to most β-lactamases, including extended-spectrum β-lactamases (ESBLs), making carbapenems “last-resort antibiotics” or “last-line agents” as treatments for critically ill patients suspected of drug-resistant infections [3]. The widespread use of carbapenems for multidrug-resistant bacterial infections has contributed to the development of resistance, most frequently mediated by β-lactamases, called carbapenemases [4]. Less common resistance mechanisms include altered target penicillin-binding proteins (PBPs) or reduced cell entry due to upregulated efflux pumps or modifications in the outer membrane proteins [5].
Carbapenemases belong to the Ambler class A (e.g., KPC, SME, IMI, and GES), class B (e.g., NDM, VIM, and IMP), and class D (OXA-48–like). The carbapenemases of class A and class D are serine β-lactamases, and the carbapenemases of class B are metallo-β-lactamases (MBLs) [6]. Class B carbapenemases, particularly NDM, are more potent than other classes and cannot be inhibited by β-lactamase inhibitors (BLIs) such as clavulanic acid, tazobactam, or sulbactam [7]. OXA carbapenemases, and specifically blaOXA-48, have a more restricted spectrum of β-lactam hydrolysis with clinically significant penicillin and imipenem hydrolysis and lower meropenem hydrolysis [5]. BLIs poorly inhibit OXA carbapenemases except for vaborbactam [8]. The blaOXA-181 gene, being a variant of blaOXA-48, was identified in different members of the Enterobacterales [9,10].
Carbapenem-resistant Enterobacterales (CRE), Pseudomonas aeruginosa, and Acinetobacter baumannii are listed by the World Health Organization (WHO) as high-priority pathogens for the research and development of new antibiotics [11]. Severe CRE infections have a higher risk of fatality than those caused by carbapenem-sensitive isolates [12]. This threat is considerably higher when the carbapenem resistance in Enterobacterales is due to carbapenemase production (CPEs; carbapenemase-producing Enterobacterales). Compared with the non-CPE infections (n = 37,45%), CPE infections (n = 46, 55%) were reported to cause four times the risk of death within 14 days among hospitalized patients in an academic hospital in the USA between March 2013 and April 2016 [13].
Carbapenemase resistance genes are associated with diverse mobile genetic elements (MGEs), including but not limited to plasmids, transposons, and integrons [14]. IncF plasmid replicons are the most common and are primarily reported to bear the blaKPC and blaNDM genes [15]. The transposon Tn125, harboring the insertion element ISAba125, is recognized as the primary vehicle for disseminating blaNDM-1 genes in Acinetobacter baumannii [16]. The successful propagation of blaOXA-48 is linked with different Tn1999 variants on highly transferable IncL plasmids, strengthened by the global distribution of specific high-risk clones such as Klebsiella pneumoniae ST307 [17].
Klebsiella pneumoniae is an invasive and virulent bacterium among Enterobacterales harboring multiple antibiotic-resistance genes (ARGs) [18]. K. pneumoniae easily assimilates ARGS by horizontal gene transfer (HGT) of MGEs [19]. The increase in multi-drug-resistant (MDR) K. pneumoniae and especially carbapenem-resistant K. pneumoniae (CRKP) is of significant medical concern. A recent meta-analysis approximated mortality associated with K. pneumoniae healthcare-associated infections (HAI) at 42% for CRKP compared with 21% for carbapenem-susceptible strains [20]. We used whole-genome sequencing (WGS) and bioinformatics analysis to characterize CPE implicated in the colonization of patients in an intensive care unit (ICU) of a public hospital in the uMgungundlovu District KwaZulu-Natal, South Africa.
2. Materials and Methods
2.1. Study Population
The study focused on the incidence and transmission of CPEs among ICU patients receiving carbapenem treatment at the uMgungundlovu District hospital from February 2020 to March 2020. This study’s participants were all consenting adult patients admitted over one month into the general ICU (medical and surgical).
2.2. Sample Collection and Screening for CPEs
Patients were screened for CPE using rectal swabs on days 1, 3, and 7 and weekly after that until transfer (to another ward or healthcare institution), discharge, or death.
Ninety-seven rectal swabs from 31 patients were screened for CPE using chromogenic screening media Chrom-ID CARBA SMART according to manufacturer instructions (BioMérieux, Marcy l’Étoile, France). Fourteen samples, positive for CPEs (14.4% of total samples) from five patients (16% of total patients enrolled), formed the final sample.
2.3. Antimicrobial Susceptibility Testing (AST)
Identification of the isolates to species levels and antibiotic susceptibility testing was conducted using the VITEK 2® automated system (BioMérieux-Vitek, Marcy-l’Étoile, France), and results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints. The antibiotic panel consisted of 18 antibiotics: ampicillin (AMP), amoxicillin-clavulanic acid (AMC), piperacillin-tazobactam (TZP), cefuroxime (CXM), cefuroxime-axetil (CXM-AX), cefoxitin (FOX), cefotaxime (CTX), ceftazidime (CAZ), cefepime (FEP), ertapenem (ERT), imipenem (IPM), meropenem (MEM), amikacin (AMK), gentamicin (GEN), ciprofloxacin (CIP), tigecycline (TGC), nitrofurantoin (NIT), and trimethoprim-sulfamethoxazole (SXT).
2.4. Genomic Characterization
2.4.1. DNA Extraction of CPKP Isolates
Genomic DNA was extracted using GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions for further molecular tests. Extracted DNA was checked for suitable concentrations and purity using Nanodrop 8000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).
2.4.2. Genomic Fingerprinting
Clonality was investigated using Enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR). The primers ERIC1 (5′-ATGTAAGCTCCTGGGATTCAC-3′) and ERIC2 (5′-AAGTAAGTGACTGGGTGAGCG-3′) (Inqaba Biotech (Pty.) Ltd., Pretoria, South Africa) were used to perform ERIC-PCR reactions on a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA) with a total volume of 25 μL consisting of 12.5 μL of Dream Taq Green PCR Master Mix (2X) (ThermoFisher Scientific, Waltham, MA, USA), 0.1 μL (100 μM) each of primers ERIC1 and ERIC2, 8.3 μL of nuclease-free H2O, and 4 μL of total genomic DNA. The PCR conditions were initial denaturation at 94 °C for 3 min, followed by 29 cycles denaturation at 94 °C (30 s), annealing at 50 °C (1 min), an extension at 65 °C (8 min), and a final extension at 65 °C (16 min). PCR amplicons were resolved horizontally by electrophoresis on 1.5% (w/v) on SeaKem® LE agarose gel at 90 V for two hours (Lonza, Rockland, ME, USA) followed by staining in 0.1 mg/mL of ethidium bromide for 15 min. The Quick-load®1-kb DNA ladder (New England BioLabs, Ipswich, MA, USA), a positive control (K. pneumoniae ATCC BAA-1705), and no-template control (NTC) were included in these reactions. The gel was visualized, and the images were captured under ultraviolet light using the Gel DocTM XR+ system (Bio-Rad, Hercules, CA, USA). Data was exported to BioNumerics software (version 7.6, Applied Maths, Austin, TX, USA) for cluster analysis. Strains were allocated to various clusters by evaluating the similarity coefficient from the homology matrix using the Jaccard approach. Dendrograms were assembled using the Unweighted Pair-Group Method (UPGMA). A 1% optimization and band tolerance were set, and 90% cut-off similarity was used to identify clusters.
2.4.3. Whole Genome Sequencing (WGS) of CPKP Isolates
A sub-sample of ten isolates, one per patient, were chosen based on similar antibiograms and ERIC profiles and subjected to whole-genome sequencing on the Illumina MiSeq platform. Using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA), multiplexed paired-end libraries (2300 bp) were prepared, and sequences were measured on 100-coverage. Utilizing CLC Genomics Workbench version 10, the resulting raw reads were screened for quality, trimmed, and de novo assembled into contigs (CLC, Bio-QIAGEN, Aarhus, Denmark). Two isolates were removed due to their poor DNA quality, and the remaining eight isolates were used for further downstream analysis.
2.4.4. Genomic Analyses and Annotation
The National Centre for Biotechnology Information (NCBI) prokaryotic genome annotation pipeline (PGAP) and RAST 2.0 server [21] were used to annotate assembled reads. All contiguous sequences were subsequently submitted to GenBank and assigned accession numbers under the Bioproject (PRJNA674742). ARGs were detected using ResFinder [22], and plasmids were delineated using PlasmidFinder [23]. The RAST SEED viewer [24] was employed to detect the genomic characteristics of the contigs and the presence of transposases flanking the β-lactamase genes (or carbapenemases). The INTEGRALL database was used to assess integrons (http://integrall.bio.ua.pt/). Insertion sequences (IS) residing in genomes were inferred by uploading contigs to the MobileElementFinder [25]. The BacWGSTdb and Pathogenwatch servers were employed to infer virulence genes and factors (http://bacdb.cn/BacWGSTdb/, https://pathogen.watch/ (accessed on 17 February 2021)). Kaptive-web, the reference online site tool for Klebsiella WGS data, was used to predict isolates’ serotypes (K types, wzc, and wzi allelic types, and O types) [26]. Multilocus sequence type (MLST) analyses were performed to determine sequence types (STs) of the study isolates on the PubMLST database (https://pubmlst.org/).
The carbapenemase genes and their flanking sequences retrieved from the RAST SEED viewer were searched on the NCBI microbial nucleotide basic local alignment search tool (BLAST). Fully sequenced plasmids with the closest synteny obtained from the BLAST search were used as a reference input to GView Server (https://server.gview.ca/) to visualize the presumed presence/absence of specific plasmid DNA.
2.4.5. Phylogenomics and Epidemiological Insights
Phylogenomic analysis was undertaken to determine how the study isolates compare to K. pneumoniae genomes from South Africa. All K. pneumoniae genomes reported in South Africa (n = 89) were downloaded from the PATRIC website (https://www.patricbrc.org/), annotated (Table S1), and included in the analysis. The phylogenetic tree was constructed based on the maximum likelihood method using the CSIPhylogeny (https://cge.cbs.dtu.dk/services/CSIPhylogeny/ (accessed on 23 February 2021)) [27], which performs single-nucleotide polymorphism (SNP) calling, filters the SNPs, and infers phylogeny based on the concatenated alignment of the high-quality SNPs, using the assembled contigs. The analysis was performed on the platform using default parameters as follows: minimum depth at SNP positions, 10X; minimum relative depth at SNP positions of 10%; minimum distance between SNPs (prune) at 10 bp; minimum SNP quality, 30; minimum read mapping quality of 25, and minimum Z-score of 1.96. The Escherichia coli ATCC 25922 was used as the outgroup strain (reference genome), facilitating the configuration of the phylogenetic distance between the isolates on the branches. The Figtree software (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 23 February 2021)) was used to visualize, edit, and annotate the generated phylogenetic tree.
3. Results
3.1. Population Demographics
Of the five consenting adult patients who screened positive for CPE, three were males, while two were females, aged between 26 and 64. Although they were admitted to the ICU with different diagnoses, almost all had the same invasive devices. The relevant patients’ data appear in Supplementary Table S2.
3.2. Antimicrobial Resistance Profiles
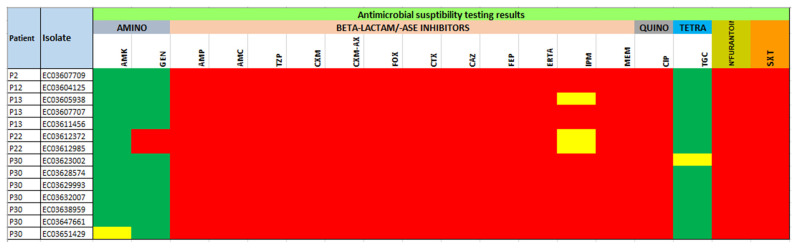
Isolates showed a similar resistance pattern to the panel of antibiotics. All isolates (100%) were resistant to 14 antibiotics, viz., ampicillin, amoxicillin-clavulanic acid, piperacillin-tazobactam, cefuroxime, cefuroxime-axetil, cefoxitin, cefotaxime, ceftazidime, cefepime, ertapenem, meropenem, ciprofloxacin, nitrofurantoin, and trimethoprim-sulfamethoxazole. However, they were fully susceptible to tigecycline and amikacin. Twelve (85.7%) isolates were susceptible to gentamycin, and four (28.6%) were susceptible to imipenem. All isolates were MDR, and the resistance phenotype was confirmed by the ARGs detected by WGS (Figure 1 and Table S3).
Figure 1.
Antibiotic susceptibility patterns of CPKP isolates (n = 14). Antibiotic susceptibility profiles are shown for the fourteen sequenced K. pneumonia isolates. Antibiotic resistance is displayed in red, intermediate resistance is displayed in yellow, and susceptibility is displayed in green.
3.3. Genomic Fingerprinting
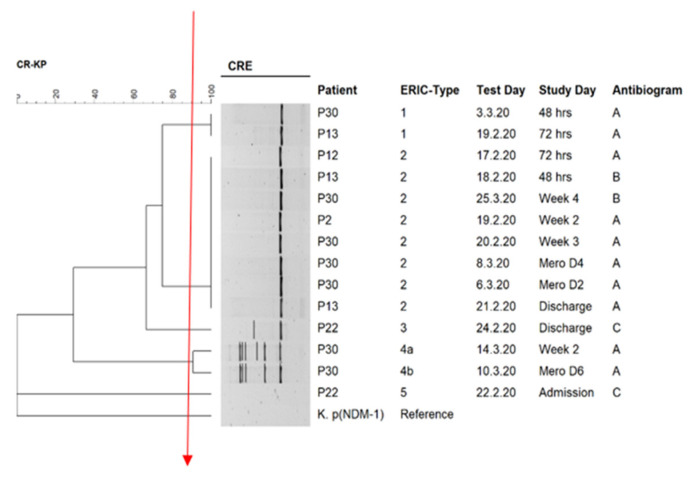
The ERIC dendrogram showed five main clusters. Isolates belonging to the same ERIC cluster generally had similar antibiograms, except for ERIC cluster 2, where isolates exhibited two antibiograms. Patients generating more than one CPE sample carried isolates belonging to 2–4 different ERIC clusters (e.g., isolates 13, 22, and 30) (Figure 2 and Table 1). All the isolates belonged to ST307 and harbored the blaOXA-181 carbapenemase and different combinations of other β-lactamases (Table 1).
Figure 2.
ERIC/PCR dendrogram for CPKP isolates (n = 14). Isolates clustered into five main clusters based on the similarity index of 90%. The solid red line indicates the ERIC-type cut-off. Antibiogram: A = AMP-AMC-TZP-CXM-CXM-AX-FOX-CTX-CAZ-FEP-ERTA-IPM-MEM-CIP-NIT-SXT; B = AMP-AMC-TZP-CXM-CXM-AX-FOX-CTX-CAZ-FEP-ERTA-MEM-CIP-NIT-SXT, C = AMP-AMC-TZP-CXM-CXM-AX-FOX-CTX-CAZ-FEP-ERTA-MEM-GEN-CIP-NIT-SXT; Mero D6 = 6th day of meropenem therapy; Mero D = 4th day of meropenem therapy and Mero D2 = 4th day of meropenem therapy.
Table 1.
Patient demographics, antibiograms, sequence types, carbapenemase, other β-lactamases, and ARGs in K. pneumoniae isolates.
| Isolate | Demographic Information | Resistome | Plasmid Incompatibility Group | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Gender | Date | Study Day | Antibiogram | ERIC Type | Other β-Lactamases | Other ARGs | ||
| EC03607709 | P2 | Male | 19 February 2020 | Week 2 | A | 2 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03605938 | P13 | Male | 18 February 2020 | 48 h | B | 2 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3′)-VIa, armA, qnrS1, oqxB, oqxA, dfrA14, fosA, msr(E), mph(E), catB3, tet(A), cmlA1, ARR-2 | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03607707 | P13 | Male | 19 February 2020 | 72 h | A | 1 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3′)-VIa, armA, qnrS1, oqxB, oqxA, dfrA14, fosA, msr(E), mph(E), catB3, tet(A), cmlA1 | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03612985 | P22 | Female | 24 February 2020 | Discharge | C | 5 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1C | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(pNDM-Mar), IncX3 |
| EC03623002 | P30 | Male | 3 March 2020 | 48 h | A | 1 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)- Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03629993 | P30 | Male | 8 March 2020 | Mero D4 | A | 2 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03632007 | P30 | Male | 10 March 2020 | Mero D6 | A | 4b | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(K), IncFII(K) |
| EC03638959 | P30 | Male | 14 March 2020 | Week 2 | A | 4a | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFII(K) |
Mero D6 = 6th day of meropenem therapy; Mero D4 = 4th day of meropenem therapy, A = AMP-AMC-TZP-CXM-CXM-AX-FOX-CTX-CAZ-FEP-ERTA-IPM-MEM-CIP-NIT-SXT; B = AMP-AMC-TZP-CXM-CXM-AX-FOX-CTX-CAZ-FEP-ERTA-MEM-CIP-NIT-SXT; C = AMP-AMC-TZP-CXM-CXM-AX-FOX-CTX-CAZ-FEP-ERTA-MEM-GEN-CIP-NIT-SXT.
3.4. Genome Characteristics of the Isolates
The draft genomes sizes of the eight isolates subjected to WGS and bioinformatics analysis were similar and ranged from 5.58 Mb to 5.72 Mb. The (G + C) content was similar for all isolates (57.1) except two that differed slightly in terms of median contigs lengths (N50, L50), contigs, RNAs present, and the number of coding sequences (Table S4).
3.5. In Silico ARGs Analysis
The genomic data (resistomes) confirmed the resistance phenotypes in most isolates (Table 1). Anomalies were detected for aminoglycosides where the presence of the aadA1 (n = 2), aadA2 (n = 1), aac(6′)-Ib-cr (n = 8), aph(3″)-Ib (n = 5), aph(3′)-Ia (n = 2), aph(3′)-VIa (n = 2), armA (n = 2), aph(6)-Id (n = 5), and aac(3)-Ia (n = 1) aminoglycoside ARGs did not translate into phenotypic aminoglycoside resistance on AST (Table S3). A single carbapenemase gene (blaOXA-181) was common to all isolates. Five other β-lactamase were detected (blaCTX-M-15, blaOXA-1, blaSHV-106) (n = 8), blaTEM-1B (n = 7), and blaTEM-1C (n = 1). Carbapenemases from classes A, B, and C were not detected. Other ARGs detected conferred resistance to tetracyclines (tet(A)) (n = 7), fluoroquinolones (aac(6′)-lb-cr, oqxA, oqxB, qnrB1, qnrS1) (n = 8), trimethoprim (dfrA12 (n = 1), dfrA14 (n = 7)), sulphonamides (sul1 (n = 1), sul2 (n = 5)), fosfomycin (fosA) (n = 8), macrolides (msr(E) (n = 2), mph(E) (n = 2)), phenicols (catB3 (n = 8), cmlA1 (n = 2)), rifamycin (ARR-2) (n = 1) and streptogramin b (msr(E) (n = 2)).
3.6. Sequence Types Analysis (MLST), In Silico Mobile Genetic Elements (Mobilome), Virulome and Capsular Serotypes
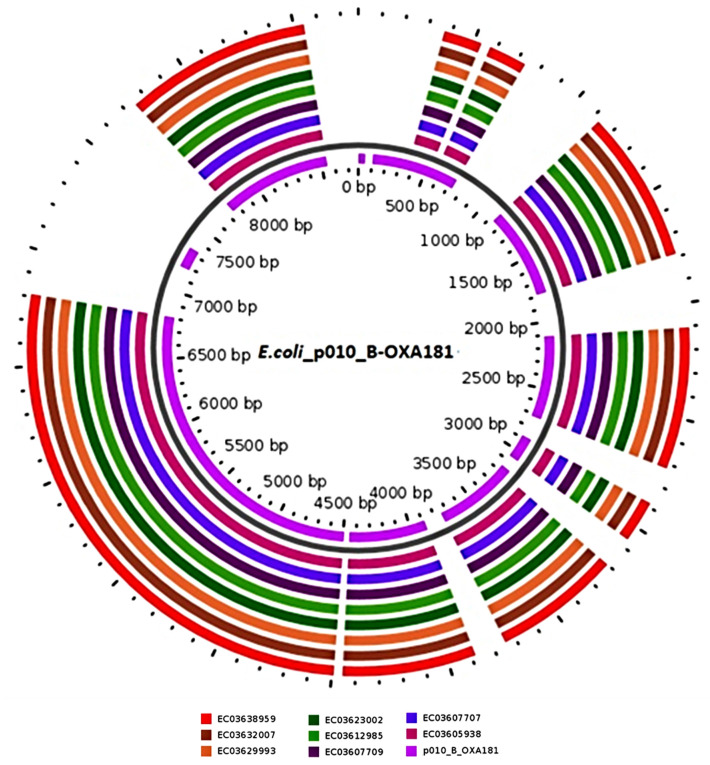
All isolates belonged to ST307 (Table 1). Plasmid analysis showed that each isolate harbored at least two replicon types simultaneously (Table 2). All isolates carried ColKP3 and 6, 7, and six isolates carried the IncX3, IncFIB (K), and IncFII (K) replicons, respectively. One isolate carried the IncFIB (pNDM-Mar) from the same group. The search for carbapenemase (OXA-181) and its flanking sequences using NCBI microbial nucleotide BLAST showed that it was located on a multi replicon plasmid E. coli p010-B-OXA181-like (accession no.: CP048332.1) in the CPKP isolates. Comparative analyses confirmed the presence of genome assemblies with similar genetic synteny and 99.98% coverage and identity to the E. coli p010 B-OXA181-like reference through the GView server (Figure 3) intimating the location of OXA-181 on a similar plasmid in all the CPKP isolates.
Table 2.
Plasmid replicon types, class 1 integrons, and gene cassettes found in K. pneumoniae isolates.
| Isolate | Plasmids Replicons | pMLST | Integron Class | Integron | Cassette Arrays | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| GC1 a | GC2 | GC3 | GC4 | GC5 | GC6 | |||||
| EC03607709 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — b | — | — | — | — |
| EC03638959 | ColKP3, IncFII(K) | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03632007 | ColKP3, IncFIB(K), IncFII(K) | [K7:A-:B-] | ND | ND | dfrA14 | — | — | — | — | — |
| EC03629993 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03607707 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03623002 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03605938 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03612985 | ColKP3, IncFIB(pNDM-Mar), IncX3 | [F-:A-:B-] | Integron integrase IntI1 | In27 | dfrA12 | gcuF | aadA2 | — | — | — |
Note: a GC denotes gene cassettes. b denotes the missing cassette arrays due to draft genomic sequences that were fragmented during the sequencing and assembling process into different contigs.
Figure 3.
Comparison of E. coli plasmid p010 B-OXA181 with genome assemblies containing OXA-181 in CPKP isolates (n = 8). The map was developed using the online server GView (https://server.gview.ca/. Accessed on 24 February2021). The concentric circles reflect genome assemblies containing OXA-181 in K. pneumoniae isolates for comparisons with E. coli p010-B-OXA181, starting with the inner circle. Color codes are assigned for every isolate with a plasmid sequence identity, ranging from 99–100%.
The determination of the location of ARGs and MGEs in synteny in the study isolates on plasmids and chromosomes on the NCBI databases revealed their location on the plasmids except for one β-lactamase gene, blaSHV-106, that was encoded on a chromosome (Table 3). It is noteworthy that the ISKpn19 insertion sequence containing the blaOXA-181 also harbored the EreA and QnrS1 genes in seven of the eight isolates together with recombinase and transposases. One isolate carried the same insertion sequence but was located on transposon (Tn3-like IS3000 transposase), found in the same plasmid and with the same companions (Table 3).
Table 3.
ARGs and MGEs in synteny and their plasmidic/chromosomal locations.
| Isolate | Contig | Synteny of Resistance Genes and MGEs | Plasmid/Chromosomal Sequence with the Closest Nucleotide Homology (Accession Number) |
|---|---|---|---|
| EC03607709 | 49 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 34 | IS5:IS5075 (IS110)::Sul2:aph(3″)-Ib:aph(6)-Id:IS91:TEM-1B:recombinase::IS1380 (ISEc9):blaCTX-M-15::Tn3 family:IS6 (IS6100) | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 54 | CatB3:bla OXA-1 :aac(6′)-Ib-cr5 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 15 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 50 | IS6 (IS6100)::dfrA14:IntI1 | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03638959 | 95 | transposase:QnrS1:recombinase::::recombinase:ISKra4 (ISKpn19)::EreA:blaOXA-181 | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 98 | blaTEM-1B:recombinase::IS1380 (ISEc9):blaCTX-M-15::Tn3 family:IS6 (IS6100) | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 109 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 24 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 102 | dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03632007 | 93 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 96 | Tn3 family::blaCTX-M-15:IS1380 (ISEc9)::recombinase:TEM-1B | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 112 | CatB3:blaOXA-1:aac(6′)-Ib-cr5 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 23 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 101 | dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03629993 | 47 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 35 | IS5:IS5075 (IS110)::Sul2:aph(3″)-Ib:aph(6)-Id:IS91:TEM-1B:recombinase::IS1380 (ISEc9):blaCTX-M-15::Tn3 family:IS6 (IS6100) | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 55 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 16 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 49 | IntI1:dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03607707 | 50 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 36 | IS6 (IS6100):Tn3 family::blaCTX-M-15:IS1380 (ISEc9)::recombinase:TEM-1B:IS91:aph(6)-Id:aph(3″)-Ib:Sul2::IS5075 (IS110):IS5 | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 56 | CatB3:blaOXA-1:aac(6′)-Ib-cr5 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 13 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 52 | IS6(IS6100)::dfrA14:IntI1 | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03623002 | 45 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 32 | IS5:IS5075 (IS110)::Sul2:aph(3″)-Ib:aph(6)-Id:IS91:TEM-1B:recombinase::IS1380 (ISEc9):blaCTX-M-15::Tn3 family:IS6 (IS6100) | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 54 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 16 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 47 | IntI1:dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03605938 | 48 | transposase:QnrS1:recombinase::::recombinase:ISKra4 (ISKpn19)::EreA:blaOXA-181 | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 34 | IS6 (IS6100):Tn3 family::blaCTX-M-15:IS1380 (ISEc9)::recombinase:TEM-1B:IS91:aph(6)-Id:aph(3″)-Ib:Sul2::IS5075 (IS110):IS5 | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 53 | aac(6′)-Ib-cr5:bla OXA-1 :CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 14 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 50 | IntI1:dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03612985 | 48 | Tn3-like IS3000:blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 54 | ::blaCTX-M-15:IS1380 (ISEc9)::recombinase:blaTEM-1C:: | E. coli str. 473 pRCS52 (99.9%), accession (LO017736.1) (plasmid) | |
| 61 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 2 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 49 | recombinase::IntI1:dfrA12:gcuF:aadA2::Sul1::::::::IS6 (IS6100)::::Mph(A) | K.p plasmid unnamed1 (99.99%), accession (CP060050.1) (plasmid) | |
| 59 | IS3:aac(3)-IIa | K.p pMS14393B (100%), accession (CP054305.1) (plasmid) |
We further identified blaSHV-106 on all isolates in parallel with other β-lactamase genes viz., blaOXA-1, blaCTX-M-15, blaTEM-1B, and blaTEM-1C in various permutations and combinations, the synteny of which was closely related to plasmid sequences already described in Genbank and summarized in Table 3. Across the isolates, blaCTX-M-15 (bracketed by Tn3 transposon, IS6100 and ISEc9); most blaTEM-1B genes (bracketed by IS91, and a recombinase); and most aph(6)-Id, aph(3”)-Ib, sul2 genes (bracketed by IS91, IS5070, and IS5) were found in close synteny on identical plasmids, except for one isolate (EC03612985) that encoded both blaCTX-M-1 and blaTEM-1C on one insertion sequence (ISEc9), showing a slightly different genetic environment (Table 3).
All isolates had trimethoprim and aminoglycoside resistance genes located on a class 1 integron (In191), with five isolates also containing the dfrA14 genes on gene cassettes. Another class 1 integron (In27) was detected in only one isolate and contained the dfrA12-gcuF-aadA2 cassette arrays. Only one isolate did not have any integrons with aminoglycosides or trimethoprim-resistance genes (Table 2).
All isolates carried an identical repertoire of 65 virulence genes and hence were clonal (Table S5). The virulence determinants belonged to the eight major virulence factor classes of Klebsiella: adherence, biofilm formation, efflux, immune evasion, iron uptake, nutritional factor, secretion system, and toxin. The virulence The O and K capsules (KL102 and O2v2) types in the strains were also highly clone specific.
3.7. Phylogenomic Analysis
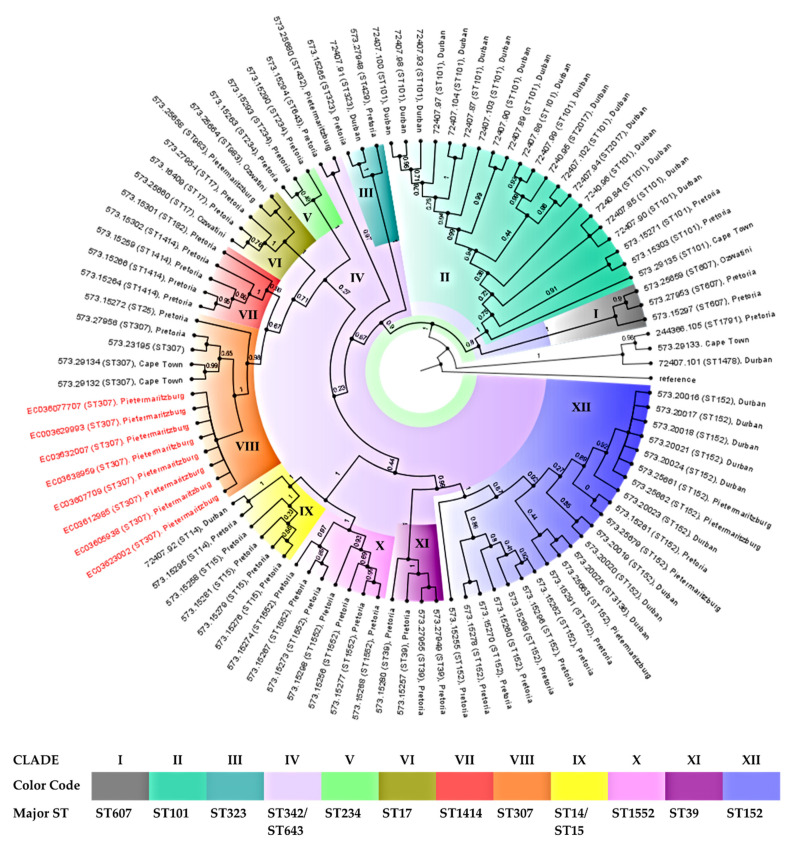
Although all eight isolates belonged to the same sequence type, ST307, small differences were evident from their phylogenetic tree (Figure 4). When compared to genomes of K. pneumoniae isolates from studies conducted in Durban, Pretoria, Pietermaritzburg, Ozwatini, and Cape Town (Table S1), the study isolates showed the greatest similarity to the clade of ST307 from Pretoria and Cape Town (clade VIII) with one ST25 clustered close to the ST307 clade (Figure 4). Of note, even though the isolates clustered according to their sequences, whole-genome phylogenetics showed higher resolution than the MLST typing scheme.
Figure 4.
Phylogeography and resistance profiles of K. pneumoniae. The phylogeographic relationship between the eight K. pneumoniae isolates with other South African K. pneumoniae isolates. The Escherichia coli ATCC 25922 was used as the outgroup strain (reference genome).
4. Discussion
The global spread of CPE presents a significant challenge to public health and clinical practice as these bacteria are resistant to last-resort antibiotics (carbapenems) and cause high fatality rates [28]. WGS data for eight CPKP strains isolated from patients at a general ICU of a regional hospital in South Africa were analyzed to delineate the genetic context of carbapenem resistome and associated mobilome together with the broader resistome virulome, clonality, and phylogeny.
K. pneumoniae is an established and significant pathogen in nosocomial infections. MDR and extremely drug-resistant (XDR) K. pneumoniae are closely linked to ARGs acquired via plasmids and other MGEs, resulting in a “super resistome” [29]. CRKP has been implicated in outbreaks in South Africa and appears to be endemic to certain hospitals and regions. In May 2012, a tertiary academic hospital in Cape Town reported one of the first laboratory-confirmed outbreaks of K. pneumoniae expressing OXA-181 in South Africa, linked to two patients admitted to the hematology ICU. Before then, laboratory records and the results of 340 rectal swabs or stool specimens collected from patients and staff members revealed an “outbreak” involving eight patients. Seven of them were admitted to the hematology unit during their hospitalization period. The remaining patient was epidemiologically linked to another patient because he was admitted to an adjacent bed [30].
The identification of ARGs from WGS data correlated with the phenotypic resistance observed in most instances in the study, with resistance ranging from 14% to 100%. However, some anomalies were observed. For example, the presence of aminoglycoside ARGs on WGS was not phenotypically evident, intimating transcriptionally silent or unexpressed ARGs. More so, the discrepancy between the aminoglycoside-modifying enzymes and phenotypes is not peculiar and has been widely reported in Carbapenemase- and ESBL-producing Enterobacter species [31,32,33]. This necessitates the urgent development of phenotypic assays that can accurately and rapidly predict aminoglycoside responses among Carbapenemase and ESBL-producing Enterobacter spp. to avoid treatment failures. All isolates belonged to the same ST type and the same phylogenetic clade, suggesting an endemic strain in the ICU.
The study findings showed that the CPKP hospital population was not diverse and belonged to a single sequence type (ST307) (Table 1), clade, and capsular serotype and contained identical repertoires of virulence genes. The phylogeography analysis with South African K. pneumonia isolates showed the isolates clustering in clade VIII to be similar to isolates from Pretoria and Cape Town (Figure 4). The Pretoria study characterized 56 CRKP with reduced susceptibility to carbapenems from six public hospitals and centers in the Tshwane District in 2018. The K. pneumoniae isolates belonged to ST307 and had similar resistance determinants, particularly blaOXA-181, along with blaOXA-1, blaTEM-1B, blaCTX-M-15, and blaSHV-28 genes. The similar plasmid (Inc) types (ColKP3, IncX3, IncFIB (k), IncFII (k)) and serotypes (KL102, O2v2) were evident [34], suggesting that the same strain is circulating in the Kwazulu-Natal area. More so, observation of imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae producing OXA-181 was not peculiar, as it has been reported in the literature [35].
Klebsiella pneumoniae ST307 is a relative newcomer to successful high-risk antibiotic-resistant strains contributing to a significant global public health burden. It possibly originated in Europe during the early to mid-1990s but was first published in the MLST database from The Netherlands in 2008 (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html. Accessed on 24 February2021). The earliest published reports came from the USA [36] and Pakistan [37] in 2013 and were followed by reports from different countries, including Russia [38], Spain [39], Brazil [40], and Japan [41] in 2016. The global distribution of K. pneumoniae ST307 has been recorded in all continents except Antarctica and has been implicated in many nosocomial [42] and long-term care center outbreaks. Findings from Colombia [43], Texas, the United States [44], Argentina [45], and Italy [46] have also shown that the incidence of ST307 among CPKP has been rising over the years, even replacing other high-risk antibiotic-resistant strains, including ST258 in regions such as Italy and Colombia [43,46].
A recent study examined major nationwide nosocomial outbreaks of blaOXA-181 and blaCTX-M-15 within several South African provinces. blaOXA-181 was reported in 471 K. pneumoniae isolates belonging to ST307, isolated from 1,247 unique clinical isolates from a private laboratory network repository between 2014 and 2016. Bayesian evolution analysis showed that ST307 isolates have evolved into six clades with clades I–IV restricted to Texas in the United States and clade VI confined to South Africa. Clade V is, in contrast, globally distributed. Clade VI in South Africa originated in 2013 and developed into two distinct lineages during 2014, spreading over 15 months across 23 cities/towns in six provinces of South Africa [47].
Several carbapenemases have been identified in K. pneumoniae in South Africa. Lowe and colleagues (2019) reported a surge in K. pneumoniae producing OXA-48-like strains, contrary to two studies in 2016 that detected the predominance of blaNDM-1 compared to blaOXA-48 [47]. In 2016, Perovic and colleagues reported data from private sector laboratories in South Africa, where a high prevalence of blaNDM-1 was observed in Gauteng province compared to blaOXA-48 through an evaluation of 9029 Gram-negative ESKAPE (K. pneumoniae, A. baumannii, P. aeruginosa, E. cloacae, and E. coli) isolates [48]. A review in the same year (2016), assessing carbapenem resistance-reporting publications from January 2000 and May 2016, showed that most common carbapenem-resistant isolates were isolated in Enterobacterales such as K. pneumoniae, E. cloacae, and S. marcescens with NDM (n = 860) and OXA-48 (n = 584) among the most reported carbapenemases [49]. A recent report described another increment in K. pneumoniae producing OXA-48-like strains, where blaOXA-48 (65%), followed by blaNDM-1 (29%), was the most common carbapenemases detected [34], in contrast to the study findings where blaOXA-181 was dominant. blaNDM-1 to blaOXA-48 have also been identified in Gauteng and the Eastern Cape [48].
The isolates carried carbapenemase blaOXA-181, other β-lactamases, plasmid-mediated quinolone resistance (PMQR) genes, and other ARGs conferring resistance to several antibiotic classes (Table 3). This suggests the potential co-selection of resistance genes and, more importantly, HGT [50] via plasmids and other MGEs [51,52].
blaCTX-M-15 was located on ISEc9 in all isolates and most of the blaTEM-1B on IS91 (Table 3). This globally observed pattern demonstrates the clonal and plasmid-mediated distribution of these ARGs within the same genetic context and on the same plasmid replicons in the same and different species. For example, IncF plasmids and ISEc9 have been shown to promote the global spread of blaCTX-M-15, alongside blaOXA-10, aac(6′)Ib-cr, and blaTEM across different species [52]. The presence of these genes in similar genetic contexts was reported in another South African study describing 20 consecutive MDR E. coli isolates collected from a referral laboratory serving two secondary and three tertiary academic hospitals in Gauteng province. The study reported ARGs mediating resistance to fluoroquinolones, aminoglycosides, and tetracyclines in E. coli isolates alongside β-lactam resistance mediated by OXA, CTX-M, and TEM-1B. Many of these resistance determinants were located on contigs containing multiple plasmid replicons and bracketed by composite transposons (Tn3), various ISs, and class 1 integrons. IncF was the most common plasmid replicon. Class 1 integrons were the only integron type identified, and blaCTX-M genes were frequently detected in ISEc9 [53], similar to the study findings.
The diverse mobilome consisting of different plasmid replicons (ColKP3, IncFIB(K), IncFII(K), IncFIB(pNDM-Mar), and IncX3) (Table 2) and insertion sequences (Table 3) intimates the potential mobility of this blaOXA-181 by HGT among isolates. Furthermore, the global dominance of the IncF plasmid, an HGT-associated MGE, is verified in previous studies. For instance, Pedersen and colleagues confirmed the role of MGEs in the potential distribution of carbapenemases. They investigated the molecular epidemiology of CRE clinical samples (n = 45) collected by a private laboratory in Durban from hospitalized patients in 10 separate private hospitals between 2012 and 2013. Focusing on the carbapenem-resistance encoding determinants and their genetic support, they demonstrated that patterns of dissemination of CPE gens (blaNDM-1, blaGES-5, blaOXA-232, and blaNDM-5) via MGEs (e.g., integrons and insertion sequences) embedded in five different plasmids have been revealed to mediate their horizontal transfer, together with clonal transmission, between various Enterobacterales species [54]. Although a small number of CRKP isolates were obtained, this study adds to the limited knowledge on the genomic insights into the resistance mechanisms, mobile genetic support, and phylogenetic relationship of isolates resistant to last-resort antibiotics such as carbapenems in Africa. Future studies should consider using larger sample sizes from diverse geographical locations and settings to understand the resistance mechanisms and transmission dynamics of CREs.
5. Conclusions
OXA-181-producing K. pneumoniae belonging to ST307 was potentially endemic in the hospital ICU environment of a public hospital in KwaZulu-Natal, South Africa. Many ARGs and/or MGEs in different permutations and combinations present challenges to clinical management and infection prevention and control measures. This necessitates a CRE screening programme and strict infection prevention and control measures to detect and eliminate this endemic clone.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12070951/s1, Table S1: Comparison of K. pneumoniae genomes with South African genomes, downloaded from NCBI and PATRIC servers; Table S2: Patients’ demographics; Table S3: Antibiotic susceptibility characteristics (MICs) of the carbapenemase-producing K. pneumoniae (CPKP) isolates collected from patients in an intensive care unit (ICU); Table S4: General genomic features of eight sequenced K. pneumoniae isolates; Table S5: Virulence analysis of the K. pneumoniae isolates showing the virulence genes repertoire (all isolates carried the same repertoire of 65 genes).
Author Contributions
Conceptualization, all; methodology, O.M.; validation, all; formal analysis, O.M., A.L.K.A., and D.G.A.; investigation, O.M.; resources, S.Y.E.; writing—original draft preparation, O.M.; writing—review and editing, all; supervision, S.Y.E., A.L.K.A., D.G.A., and J.R.; project administration, S.Y.E.; funding acquisition, S.Y.E. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa (Grant No. 98342), the National Research Foundation Competitive Grant for Rated Researchers (Grant no. 106063), jointly funded by the South African Medical Research Council (SAMRC), and UK Medical Research Council Newton Fund and the SAMRC Self-Initiated Research Grant. Any opinion, finding, and conclusion or recommendation expressed in this material is that of the authors, and neither the NRF nor the other funding bodies accept any liability in this regard.
Institutional Review Board Statement
This study was approved by the UKZN Biomedical Research Ethics Committee (BREC) (Ref No. BE709/18 and BREC/00001723/2020). Permission was granted by the KwaZulu-Natal Department of Health, the uMgungundlovu District Manager, the hospital Chief Executive Officer, and the ICU Manager. Voluntary informed consent was obtained from patients.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study, their next of kin, or proxy consent was obtained from the ICU manager.
Data Availability Statement
All analyzed data have been included in the manuscript. The nucleotide sequences of the eight isolates analyzed in this study were deposited in the NCBI GenBank database in the Bioproject number (PRJNA674742): under the Accession numbers; JADKPL000000000.1, JADOEH000000000.1, JADOEG000000000.1, JADKPK000000000.1, JADKPJ000000000.1, JADKPI000000000.1, JADKPH000000000.1, and JADKPG000000000.1.
Conflicts of Interest
S.Y.E. is chairperson of the Global Respiratory Infection Partnership and a member of the Global Hygiene Council, both sponsored by an unrestricted educational grant from Reckitt and Benckiser Ltd., UK. All other authors have no competing interests. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Klein E.Y., Van Boeckel T.P., Martinez E.M., Pant S., Gandra S., Levin S.A., Goossens H., Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA. 2018;115:E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K., Bradford P.A. β-lactams and β-lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2016;6:a025247. doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsamy Y., Mlisana K.P., Allam M., Amoako D.G., Abia A.L.K., Ismail A., Singh R., Kisten T., Han K.S., Muckart D.J.J., et al. Genomic analysis of carbapenemase-producing extensively drug-resistant Klebsiella pneumonia isolates reveals the horizontal spread of p18-43_01 plasmid encoding blandm-1 in South Africa. Microorganisms. 2020;8:137. doi: 10.3390/microorganisms8010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitout J.D.D., Nordmann P., Poirel L. Carbapenemase-producing klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 2015;59:5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K., Bradford P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020;33:e00047-19. doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui X., Zhang H., Du H. Carbapenemases in Enterobacteriaceae: Detection and antimicrobial therapy. Front. Microbiol. 2019;10:1823. doi: 10.3389/fmicb.2019.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Somboro A.M., Osei Sekyere J., Amoako D.G., Essack S.Y., Bester L.A. Diversity and proliferation of metallo-β-lactamases: A clarion call for clinically effective metallo-β-lactamase inhibitors. Appl. Environ. Microbiol. 2018;84:1–20. doi: 10.1128/AEM.00698-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomovskaya O., Sun D., Rubio-Aparicio D., Nelson K., Tsivkovski R., Griffith D.C., Dudley M.N. Vaborbactam: Spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in enterobacteriaceae. Antimicrob. Agents Chemother. 2017;61:61. doi: 10.1128/AAC.01443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brink A.J., Coetzee J., Corcoran C., Clay C.G., Hari-Makkan D., Jacobson R.K., Richards G.A., Feldman C., Nutt L., van Greune J. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J. Clin. Microbiol. 2013;51:369–372. doi: 10.1128/JCM.02234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel L., Potron A., Nordmann P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization WHO Priority Pathogens List for R&D of New Antibiotics. [(accessed on 3 September 2020)]; Available online: http://www.who.int/bulletin/volumes/94/9/16-020916.pdf.
- 12.Martin A., Fahrbach K., Zhao Q., Lodise T. Association between carbapenem resistance and mortality among adult, hospitalized patients with serious infections due to enterobacteriaceae: Results of a systematic literature review and meta-analysis. Open Forum Infect. Dis. 2018;5:ofy150. doi: 10.1093/ofid/ofy150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamma P.D., Goodman K.E., Harris A.D., Tekle T., Roberts A., Taiwo A., Simner P.J. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase- producing carbapenem-resistant enterobacteriaceae bacteremia. Clin. Infect. Dis. 2017;64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch T., Petkau A., Knox N., Graham M., Domselaar V. A primer on infectious disease bacterial genomics. Clin. Microbiol. Rev. 2016;29:881–913. doi: 10.1128/CMR.00001-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rozwandowicz M., Brouwer M.S.M., Fischer J., Wagenaar J.A., Gonzalez-Zorn B., Guerra B., Mevius D.J., Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 16.Khan A.U., Maryam L., Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017;17:101. doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitout J.D.D., Peirano G., Kock M.M., Strydom K.-A., Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 2019;33 doi: 10.1128/CMR.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partridge S.R. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 2011;35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 19.Mathers A.J., Peirano G., Pitout J.D.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant enterobacteriaceae. Clin. Microbiol. Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu L., Sun X., Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 2017;16:18. doi: 10.1186/s12941-017-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatusova T., Dicuccio M., Badretdin A., Chetvernin V., Nawrocki P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Microbiol. Resour. Announc. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., Møller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson M.H.K., Bortolaia V., Tansirichaiya S., Aarestrup F.M., Roberts A.P., Petersen T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021;76:101–109. doi: 10.1093/jac/dkaa390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyres K.L., Wick R.R., Gorrie C., Jenney A., Follador R., Thomson N.R., Holt K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:1–8. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheu C.-C., Chang Y.-T., Lin S.-Y., Chen Y.-H., Hsueh P.-R. Infections Caused by Carbapenem-Resistant Enterobacteriaceae: An Update on Therapeutic Options. Front. Microbiol. 2019;10:80. doi: 10.3389/fmicb.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson R.K., Manesen M.R., Moodley C., Smith M., Williams S., Nicol M., Bamford C. Molecular characterisation and epidemiological investigation of an outbreak of blaOXA-181 carbapenemase-producing isolates of Klebsiella pneumoniae in South Africa. S. Afr. Med. J. 2015;105:1030. doi: 10.7196/SAMJ.2015.v105i12.9926. [DOI] [PubMed] [Google Scholar]
- 31.Haidar G., Alkroud A., Cheng S., Churilla T.M., Churilla B.M., Shields R.K., Doi Y., Clancy C.J., Nguyen M.H. Association between the presence of aminoglycoside-modifying enzymes and in vitro activity of gentamicin, tobramycin, amikacin, and plazomicin against klebsiella pneumoniae carbapenemase- and extended-spectrum-β-lactamase-producing enterobacter species. Antimicrob. Agents Chemother. 2016;60:5208–5214. doi: 10.1128/AAC.00869-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almaghrabi R., Clancy C.J., Doi Y., Hao B., Chen L., Shields R.K., Press E.G., Iovine N.M., Townsend B.M., Wagener M.M., et al. Carbapenem-resistant klebsiella pneumoniae strains exhibit diversity in aminoglycoside-modifying enzymes, which exert differing effects on plazomicin and other agents. Antimicrob. Agents Chemother. 2014;58:4443–4451. doi: 10.1128/AAC.00099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler D.A., Rana A.P., Krapp F., Patel S.R., Huang Y., Ozer E.A., Hauser A.R., Bulman Z.P. Optimizing aminoglycoside selection for KPC-producing Klebsiella pneumoniae with the aminoglycoside-modifying enzyme (AME) gene aac(6′)-Ib. J. Antimicrob. Chemother. 2021;76:671–679. doi: 10.1093/jac/dkaa480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopotsa K., Mbelle N.M., Osei Sekyere J. Epigenomics, genomics, resistome, mobilome, virulome and evolutionary phylogenomics of carbapenem-resistant Klebsiella pneumoniae clinical strains. Microb. Genom. 2020;6:e000474. doi: 10.1099/mgen.0.000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayama S., Koba Y., Shigemoto N., Kuwahara R., Kakuhama T., Kimura K., Hisatsune J., Onodera M., Yokozaki M., Ohge H., et al. Imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae producing OXA-181 in Japan. Antimicrob. Agents Chemother. 2015;59:1379–1380. doi: 10.1128/AAC.04330-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castanheira M., Farrell S.E., Wanger A., Rolston K.V., Jones R.N., Mendes R.E. Rapid expansion of KPC-2-producing klebsiella pneumoniae isolates in two texas hospitals due to clonal Spread of ST258 and ST307 Lineages. Microb. Drug Resist. 2013;19:295–297. doi: 10.1089/mdr.2012.0238. [DOI] [PubMed] [Google Scholar]
- 37.Habeeb M.A., Haque A., Nematzadeh S., Iversen A., Giske C.G. High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum β-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int. J. Antimicrob. Agents. 2013;41:524–526. doi: 10.1016/j.ijantimicag.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 38.Lazareva I.V., Ageevets V.A., Ershova T.A., Zueva L.P., Goncharov A.E., Darina M.G., Svetlichnaya Y.S., Uskov A.N., Sidorenko S.V. Prevalence and antibiotic resistance of carbapenemase-producing gram-negative bacteria in Saint Petersburg and some other regions of the Russian Federation. Antibiot. Chemoterapy. 2016;61:28–38. [PubMed] [Google Scholar]
- 39.Ruiz-Garbajosa P., Hernández-García M., Beatobe L., Tato M., Méndez M.I., Grandal M., Aranzábal L., Alonso S., Lópaz M.Á., Astray J., et al. A single-day point-prevalence study of faecal carriers in long-term care hospitals in Madrid (Spain) depicts a complex clonal and polyclonal dissemination of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2016;71:348–352. doi: 10.1093/jac/dkv355. [DOI] [PubMed] [Google Scholar]
- 40.Dropa M., Lincopan N., Balsalobre L.C., Oliveira D.E., Moura R.A., Fernandes M.R., da Silva Q.M., Matté G.R., Sato M.I.Z., Matté M.H. Genetic background of novel sequence types of CTX-M-8- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae from public wastewater treatment plants in São Paulo, Brazil. Environ. Sci. Pollut. Res. 2016;23:4953–4958. doi: 10.1007/s11356-016-6079-5. [DOI] [PubMed] [Google Scholar]
- 41.Harada K., Shimizu T., Mukai Y., Kuwajima K., Sato T., Usui M., Tamura Y., Kimura Y., Miyamoto T., Tsuyuki Y., et al. Phenotypic and molecular characterization of antimicrobial resistance in Klebsiella spp. isolates from companion animals in Japan: Clonal dissemination of multidrug-resistant extended-spectrum β-lactamase-producing klebsiella pneumoniae. Front. Microbiol. 2016;7:1021. doi: 10.3389/fmicb.2016.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baek E.-H., Kim S.-E., Kim S., Lee S., Cho O.-H., In Hong S., Shin J.H., Hwang I. Successful control of an extended-spectrum beta-lactamase-producing Klebsiella pneumoniae ST307 outbreak in a neonatal intensive care unit. BMC Infect. Dis. 2020;20:166. doi: 10.1186/s12879-020-4889-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ocampo A.M., Chen L., Cienfuegos A.V., Roncancio G., Chavda K.D., Kreiswirth B.N., Jiménez J.N. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 Clones of carbapenem-resistant klebsiella pneumoniae with distinct clinical characteristics. Antimicrob. Agents Chemother. 2016;60:332–342. doi: 10.1128/AAC.01775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long S.W., Olsen R.J., Eagar T.N., Beres S.B., Zhao P., Davis J.J., Brettin T., Xia F., Musser J.M. Population Genomic Analysis of 1,777 Extended-spectrum beta-lactamase-producing klebsiella pneumoniae isolates, Houston, Texas: Unexpected abundance of clonal group 307. MBio. 2017;8:e00489-17. doi: 10.1128/mBio.00489-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cejas D., Elena A., Guevara Nuñez D., Sevillano Platero P., De Paulis A., Magariños F., Alfonso C., Berger M.A., Fernández-Canigia L., Gutkind G., et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: Emergence of hypermucoviscous ST25 and high-risk clone ST307. J. Glob. Antimicrob. Resist. 2019;18:238–242. doi: 10.1016/j.jgar.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Bonura C., Giuffrè M., Aleo A., Fasciana T., Di Bernardo F., Stampone T., Giammanco A., MDR-GN Working Group. Palma D.M., Mammina C. An update of the evolving epidemic of blaKPC carrying klebsiella pneumoniae in Sicily, Italy, 2014: Emergence of multiple non-ST258 clones. PLoS ONE. 2015;10:e0132936. doi: 10.1371/journal.pone.0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowe M., Kock M.M., Coetzee J., Hoosien E., Peirano G., Strydom K.-A., Ehlers M.M., Mbelle N.M., Shashkina E., Haslam D.B., et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014–2016. Emerg. Infect. Dis. 2019;25:739–747. doi: 10.3201/eid2504.181482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perovic O., Ismail H., Van Schalkwyk E., Lowman W., Prentice E., Senekal M., Govind C.N. Antimicrobial resistance surveillance in the South African private sector report for 2016. South. Afr. J. Infect. Dis. 2018;33:114–117. doi: 10.1080/23120053.2018.1482646. [DOI] [Google Scholar]
- 49.Osei Sekyere J. Current state of resistance to antibiotics of last-resort in South Africa: A review from a public health perspective. Front. Public Health. 2016;4:209. doi: 10.3389/fpubh.2016.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asante J., Osei Sekyere J. Understanding antimicrobial discovery and resistance from a metagenomic and metatranscriptomic perspective: Advances and applications. Environ. Microbiol. Rep. 2019;11:62–86. doi: 10.1111/1758-2229.12735. [DOI] [PubMed] [Google Scholar]
- 51.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31:1–61. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kopotsa K., Osei Sekyere J., Mbelle N.M. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: A review. Ann. N. Y. Acad. Sci. 2019;1457:61–91. doi: 10.1111/nyas.14223. [DOI] [PubMed] [Google Scholar]
- 53.Mbelle N.M., Feldman C., Osei Sekyere J., Maningi N.E., Modipane L., Essack S.Y. The resistome, mobilome, virulome and phylogenomics of multidrug-resistant escherichia coli clinical isolates from Pretoria, South Africa. Sci. Rep. 2019;9:16457. doi: 10.1038/s41598-019-52859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedersen T., Sekyere J.O., Govinden U., Moodley K., Sivertsen A., Samuelsen Ø., Essack S.Y., Sundsfjord A. Spread of plasmid-encoded NDM-1 and GES-5 carbapenemases among extensively drug-resistant and pandrug-resistant clinical enterobacteriaceae in Durban, South Africa. Antimicrob. Agents Chemother. 2018;62:e02178–17. doi: 10.1128/AAC.02178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyzed data have been included in the manuscript. The nucleotide sequences of the eight isolates analyzed in this study were deposited in the NCBI GenBank database in the Bioproject number (PRJNA674742): under the Accession numbers; JADKPL000000000.1, JADOEH000000000.1, JADOEG000000000.1, JADKPK000000000.1, JADKPJ000000000.1, JADKPI000000000.1, JADKPH000000000.1, and JADKPG000000000.1.