Highlights
-
•
The XPO1 inhibitor Selinexor inhibits growth of KRASmut lung cancer PDXs.
-
•
Selinexor is effective in G12C and non-G12C KRASmut lung cancers.
-
•
Selinexor Is more effective than the MEK inhibitor trametinib in KRASmut lung cancers.
-
•
Selinexor is less effective than kinase inhibitors in kinase-driven lung cancers.
Keywords: KRAS, XPO1, Lung adenocarcinoma, Patient-derived xenografts, Targeted therapeutics
Abstract
Gain-of-function Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations occur in 25% of lung adenocarcinomas, and these tumors are challenging to treat. Some preclinical work, largely based on cell lines, suggested KRASmut lung cancers are especially dependent on the nuclear export protein exportin-1 (XPO1), while other work supports XPO1 being a broader cancer dependency. To investigate the sensitivity of KRASmut lung cancers to XPO1 inhibition in models that more closely match clinical tumors, we treated 10 independently established lung cancer patient-derived tumor xenografts (PDXs) with the clinical XPO1 inhibitor, Selinexor. Monotherapy with Selinexor reduced tumor growth in all KRASmut PDXs, which included 4 different codon mutations, and was more effective than the clinical MEK1/2 inhibitor, Trametinib. Selinexor was equally effective in KRASG12C and KRASG12D tumors, with TP53 mutations being a biomarker for a weaker drug response. By mining genome-wide dropout datasets, we identified XPO1 as a universal cancer cell dependency and confirmed this functionally in two KRASWT PDX models harboring kinase drivers. However, targeted kinase inhibitors were more effective than Selinexor in these models. Our findings support continued investigation of XPO1 inhibitors in KRASmut lung adenocarcinoma, regardless of the codon alteration.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide [1,2], and the most common form is adenocarcinoma (LUAD) [2], the predominant form of non-small cell lung cancer (NSCLC). Several prominent molecular alterations have been identified in LUAD, including Kirsten rat sarcoma viral oncogene homolog (KRAS) point mutations in 20–25% of tumors [3,4]. Patients harboring KRAS-mutant tumors have historically been treated with standard-of-care chemotherapy, radiotherapy, and surgery [5]. However, inhibitors highly specific to KRASG12C have recently been developed and are currently in clinical trials [6,7] (NCT03600883; NCT03785249). One such inhibitor, Sotorasib, has just been approved for advanced stage NSCLC patients with KRASG12C-mutated tumors that have failed prior chemotherapy [8,9]. Unfortunately, KRASG12C mutations only comprise 40% of the total KRAS mutations in LUAD [10]. These G12C-specific inhibitors would be ineffective for the majority of the KRAS-mutant (KRASmut) LUAD patient population [6,7]. In addition, there is already evidence that patients treated with KRASG12C-specific inhibitors can acquire resistance mutations over time [11,12]. Thus, a great deal of research effort continues to focus on developing small-molecule inhibitors targeting pan-KRAS downstream effectors or uncovering pan-KRAS synthetic lethalities. Both approaches are hoped to identify therapeutic strategies that would be effective against all KRAS-point mutated tumors, regardless of their KRAS amino acid change.
Several KRAS synthetic lethal targets have been identified through short hairpin/interfering RNA (shRNA/siRNA) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas)9 screens [13]. Stemming from an siRNA screen, one group reported that the nuclear export protein, exportin-1 (XPO1), is a druggable vulnerability with enriched specificity for KRASmut NSCLC [14]. All eukaryotic cells require nuclear-cytoplasmic transport for normal functioning [15], and nuclear macromolecule export is orchestrated by the karyopherin family of proteins, which includes XPO1 [16]. There are over 350 cargoes which possess nuclear export signals, which XPO1 may transport from the nuclear to cytoplasmic compartments, including many tumor suppressors [17]. XPO1 inhibition leads to nuclear accumulation of many of these cargoes via slowly reversible binding to the cargo-binding groove in XPO1 [18]. Nuclear export may be exploited by multiple cancers as part of a general malignancy mechanism, as XPO1 is overexpressed in a number of cancers [19], and its overexpression in immortalized human bronchial epithelial cells or baby hamster kidney fibroblasts causes malignant transformation and alterations in cargo localization [20,21]. In the siRNA study which identified XPO1 as a druggability in KRASmut NSCLC, they observed a greater sensitivity to XPO1 inhibition in KRASmut rather than KRASWT LUAD cancer cells. However, this finding was largely based on cell line data and contrasts with other cell line work, which described sensitivity of KRASWT NSCLC cells both in vitro and in vivo [22,23]. Furthermore, additional work in other cancer types found the antitumor activity of XPO1 inhibitors to be independent of TP53 and RAS mutation status [24].
XPO1 inhibitors are currently in over 80 clinical trials covering a wide array of different cancers with varying frequencies of KRAS mutations and have been approved for relapsed/refractory multiple myeloma [25] and relapsed/refractory diffuse large B-cell lymphoma [26]. As very little work has been done to assess the effectiveness of XPO1 inhibitors towards KRASmut NSCLC in a more clinically relevant setting, we conducted an in vivo study with 10 independently established patient tumor-derived xenograft (PDX) models that retain the major histological and molecular features of patient disease [27,28]. We treated these models with Selinexor, an XPO1 inhibitor with clinical approval for two blood cancers [25,26]. Additionally, we compared the effectiveness of Selinexor to Trametinib, a mitogen-activated protein kinase kinase 1/2 (MEK1/2) inhibitor that is under investigation for KRASmut advanced stage NSCLC [29,30] (NCT02642042, NCT03704688). We now report that Selinexor is indeed effective in KRASmut NSCLC, with all 10 models showing a response, including those with non-G12C codon alterations. We further show that Selinexor is more effective than Trametinib and we provide evidence for XPO1 being a broader cancer target, regardless of KRAS mutation status. However, we did find that TP53 mutation status was associated with a weaker response to Selinexor. Our findings provide a compelling preclinical rationale to continue to study XPO1 inhibitors for the treatment of KRASmut NSCLC.
Materials and methods
Lung cancer PDX models
PDX models were established from resected LUAD as previously described and using the protocols approved by the Research Ethics Board (REB 09-0510-T) and Animal Care Committee (AUP#743) at the University Health Network [27,28]. To assure model authenticity, short tandem repeat DNA fingerprinting analysis was conducted to verify PDX passages to their matched patient tumor/normal reference using a 16 loci, 32-read AmpFLSTR™ Identifiler™ PCR Amplification Kit (Thermo Fisher Scientific, Waltham, MA). All models used in these experiments were passages 4-6.
PDX treatment protocol
Models were grown from cryopreserved PDX tumor tissue. Fragments were thawed, washed with fresh media, mixed with 0.1% Matrigel at 4 °C (BD Biosciences, San Jose, CA), and subcutaneously implanted into one to three 6 to 8-week-old male non-obese diabetic severe combined immunodeficient (NOD/SCID) mice. Once donor tumors grew and reached the humane endpoint of 1.5cm in diameter as defined in the Animal Care Committee (AUP#743), mice were euthanized, and tumors were harvested and cut into 3 mm pieces for serial expansion into treatment arms. Once tumors reached an average size of 150–250 mm [3], mice were assigned into three treatment arms based on their tumor volumes using a stratified randomization process for equal distribution of tumor sizes.
Selinexor (CAS 1393477–72–9), Trametinib (CAS 871700–17–3), erlotinib (CAS 183321–74–6), crizotinib (CAS 877399-52-5) were purchased from UHN Shanghai (Shanghai, China). All drugs were certified to be at least 98% pure. Trametinib (1mg/kg) was dissolved in 0.5% hydroxypropyl methylcellulose and 0.2% tween80 in sterile dH2O; Selinexor (10mg/kg) [14, 23] was dissolved in 0.6% pluronic F-68 (w/v) and 0.6% PVP K29/32 (w/v) in sterile dH2O; Erlotinib (50mg/kg) was dissolved in 6% captisol in sterile dH2O; Crizotinib (50mg/kg) was diluted in 0.5% hydroxypropyl methylcellulose and 0.4% tween80 in sterile dH2O. All drugs were orally administered by gavage. For chronic dosing, Trametinib, erlotinib and crizotinib were administered five times per week; Selinexor was administered 3 times per week (Monday, Wednesday, Friday). For acute dosing, Selinexor was administered once, and tumors were collected at 0, 24, 72 h post drug administration. Mice were weighed every day of treatment and tumor volumes measured twice per week using digital calipers.
Drug treatment was withheld from mice if their body weights fell 15% below that of the body weight at the time of treatment initiation, and only continued once weight was regained. Time to regain weight was noticeably longer in the Selinexor treatment group than that of Trametinib and vehicle groups. However, most mice treated with Selinexor whose weight fell below the 15% threshold only missed one or two doses in a 30-day dosing period (~12 doses total). Therefore, the dosing on average ranged between 2,3x/week for Selinexor.
DNA profiling and somatic mutation calling
The gSYNCTM DNA Extraction Kit (Geneaid, New Taipei City, Taiwan) was used to extract DNA from PDX models and their matched patient adjacent normal lung tissue, as per manufacturer's instructions. Library enrichment was performed using the Agilent SureSelect Human All Exon 50Mb + Cosmic capture kits (Santa Clara, CA). Paired-end sequence reads were generated using the Illumina HiSeq 2500 platform. Mouse stromal reads were filtered out by using Xenome(v1.0.1) [31] to align these reads to NOD/SCID mouse DNA. The remaining reads were aligned to the human reference genome (hg19) using Burrows-Wheeler Aligner (v0.7.12) [32]. Quality control, indexing, marking duplicates, indel local realignment, base quality score recalibration, and further data processing were performed using a standard Genome Analysis Toolkit (GATK) pipeline (v3.4) [33], samtools (v1.2) [34] and Picard (v1.140) (https://broadinstitute.github.io/picard/). Mutect v1.1.15 and Varscan v2.3.8 were used to call somatic mutation SNVs and indels, respectively. Samples without matched normal tissue underwent additional filtering using dbSNP138, ExAC03 and ESP6500. The calls were annotated using ANNOVAR [35]. Mutations were considered oncogenic or likely oncogenic according to PolyPhen-2 [36].
KRAS mutation verification
KRAS mutations called from PDX whole exome sequencing data were verified by Sanger sequencing. DNA was incubated with KRAS-specific primers (F: 5’-CATTTCGGACTGGGAGCGA-3’; R: 5’- AAAGAAAGCCCTCCCCAGT-3’) and underwent polymerase chain reaction using Taq polymerase (Lucigen, Middleton, WI) in a Biometra T3 thermocycler (Analytik Jena, Jena, Germany) (15 cycles: 94 °C 10m; 94 °C 30s; 57 °C 45s; 72 °C 45s. 20 cycles: 94 °C 30s; 50 °C 30s; 72 °C 45s. 72 °C 10m. Lower to 4 °C). Post-PCR amplicons were run on a 1% agarose gel and analyzed under UV light. Good amplicons were cleaned using ExoSAP-IT (Affymetrix, Santa Clara, CA) (37 °C 20m. 80 °C 15m. Lower to 4 °C) and were sent to be Sanger sequenced with KRAS-specific primers (F: 5’- CGCGGCGCAGGCACTGAA-3’; R: 5’- TCGAGAATATCCAAGAGAGAGGT-3’). Mutations and mutant allele frequency were confirmed from forward and reverse sequences.
Immunoblot analysis
PDX tumor tissues were snap frozen, banked in liquid nitrogen, then homogenized in RIPA lysis buffer (Sigma-Aldrich, St. Louis, MO) supplemented with PMSF, Na3VO4, and cOmplete, mini, EDTA-free protease inhibitor cocktail tablets (Roche, Basel, Switzerland). Protein concentration was determined using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA) as per manufacturer's instructions, in 96-well plates as triplicates. Samples had RIPA lysis buffer and 4X Laemmli sample buffer (Bio-Rad, Hercules, CA) added to them, then were boiled at 95°C for 5 min to denature protein. Protein was electrophoresed on a 4% to 20% mini-PROTEAN TGX gel (Bio-Rad, Hercules, CA) and transferred onto nitrocellulose membrane using a Trans-blot Turbo transfer machine (Bio-Rad, Hercules, CA). Membranes were blocked in 5% skim milk (Bio-Rad, Hercules, CA) in 1x TBST for 1hr at RT on a rocking table. The membranes were cut to isolate specific areas of the blots and incubated overnight at 4°C with monoclonal anti-XPO1 (1:1000, #46249 Cell Signaling Technology, Danvers, MA), anti-KRAS (1:200, #sc-030 Santa Cruz Biotechnology, Dallas, TX), anti-MEK1/2 (1:2000, #9122 Cell Signaling Technology, Danvers, MA), anti-pERK1/2 T202/Y204 (1:2000, #4370 Cell Signaling Technology, Danvers, MA), anti-ERK1/2 (1:2000, #4695 Cell Signaling Technology, Danvers, MA), and anti-β-Actin (1:2000, #A1978 Sigma-Aldrich, St. Louis, MO). All primary antibodies were diluted in 1x TBST with 5% bovine serum albumin (Wisent, Saint-Bruno, Canada). Membranes were washed with 1x TBST three times for 5 min at RT and later incubated with secondary anti-rabbit IgG HRP-linked antibody (1:10000, #7074 Cell Signaling Technology, Danvers, MA) or anti-mouse IgG HRP-linked antibody (1:10000, #7076 Cell Signaling Technology, Danvers, MA) dissolved in 5% skim milk for 1 h at RT. Membranes were washed with 1x TBST three times for 10 min at RT. Protein was visualized by Clarity ECL (Bio-Rad, Hercules, CA) and medical-grade X-ray film (Carestream, Rochester, NY).
DepMap analysis
CRISPR/Cas9 20Q4 and RNAi DEMETER2 v6 gene dependency data were downloaded from https://depmap.org/portal/ with files titled Achilles_gene_dependency.csv and D2_Achilles_gene_dep_scores.csv, respectively. CRISPR/Cas9 and RNAi data on XPO1 was available for 808 [37] and 597 [38] cancer cell lines, respectively. Data was compiled and input into Graphpad Prism 5.01. RNAi data for one NSCLC cell line was omitted from NSCLC KRASWT/KRASmut categories, as KRAS mutation status was unknown. Our NSCLC category did not include small-cell lung cancer, cases with unknown lung cancer pathologies, or mesotheliomas.
Statistical analyses
To quantify the difference in treatment effects, log linear mixed effects models [39] of tumor size were generated using R version 3.6.2 for each patient sample. Models include week, treatment, and a week/treatment interaction as main effects, as well as a random intercept and growth rate for each mouse Eq. (1). We reported the predicted change in weekly growth rates for treatment compared to the vehicle and associated p-value, which were adjusted for multiple comparisons using the false discovery rate approach. For example, a value of 0.9 would indicate the tumors given the treatment are growing at a rate of 0.9 times the rate of a tumors treated with a vehicle. Aggregate treatment effects across all patients were generated utilizing the same approach as per patient with the addition of a random patient intercept, and growth rate. To assess the treatment effect in the G12D and G12C KRAS codon alterations, the alteration type, and interactions with day, treatment, and day/treatment were included in model. The same approach was used to assess the TP53 mutations. P-values for Achilles data were generated using the Mann-Whitney test. P < 0.05 was considered to be statistically significant. For all figures, * = p < 0.05; ** = p < 0.01; *** = p < 0.005; **** = p < 0.001.
| (1) |
where
Where is the average log tumor volume at time zero of the control group, is the average logged growth rate for the control, is the difference in log tumor volume at time zero for the treated vs control is the log difference in growth rates for treated vs control, is mouse specific log volume at time zero, and is mouse specific log growth. represents the variation in starting tumor volume across mice, represent the tumor growth variation across mice, and is the variation within mice.
Results
KRAS-mutant lung cancer PDX models
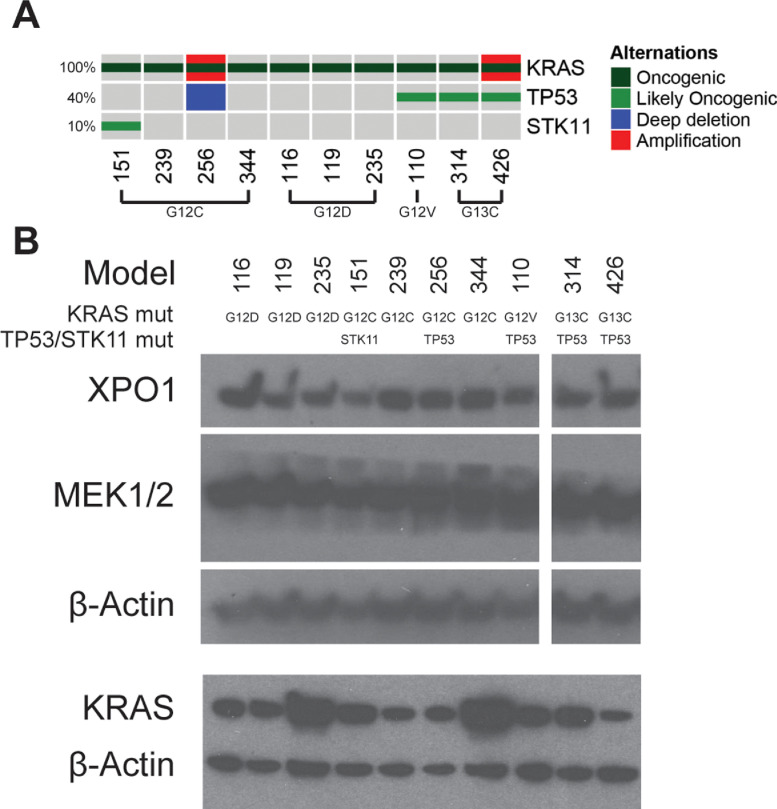
We previously reported establishment of a large number of PDXs from resected tumors of NSCLC patients [27,28]. Models that maintained tumorigenicity beyond passage three were considered stable and were profiled by whole-exome sequencing (WES) and for copy number variations. Among these PDX models, 27 harbored KRAS non-synonymous mutations. Ten models that cover the spectrum of the most common KRAS alterations found in the LUAD patient-population, including KRAS G12C, G12D, G12V, and G13C [40], were selected for further study and their KRAS mutations were verified by Sanger sequencing (Supplementary Table S1). These models were also annotated for STK11 and TP53 alterations, as it has been suggested that tumors with alterations in these genes in conjunction with KRAS mutations may define subgroups of tumors with distinct sensitivities to various therapeutic agents [41,42] (Fig. 1A, Supplementary Table S1). Western blotting confirmed expression of KRAS, XPO1, and MEK1/2 in these models (Fig. 1B, Supplementary Fig. S1).
Fig. 1.
Characterization of LUAD PDXs. (A) Oncoprint of KRAS, TP53, and STK11 alterations in the KRASmut PDX cohort. Mutations that were considered oncogenic or likely oncogenic according to PolyPhen-2 [36] were included, in addition to genetic amplifications and deep deletions. (B) Western blot of baseline expression of drug targets in LUAD PDXs used for drug screening.
Selinexor suppresses growth of KRAS-mutant lung cancer PDXs
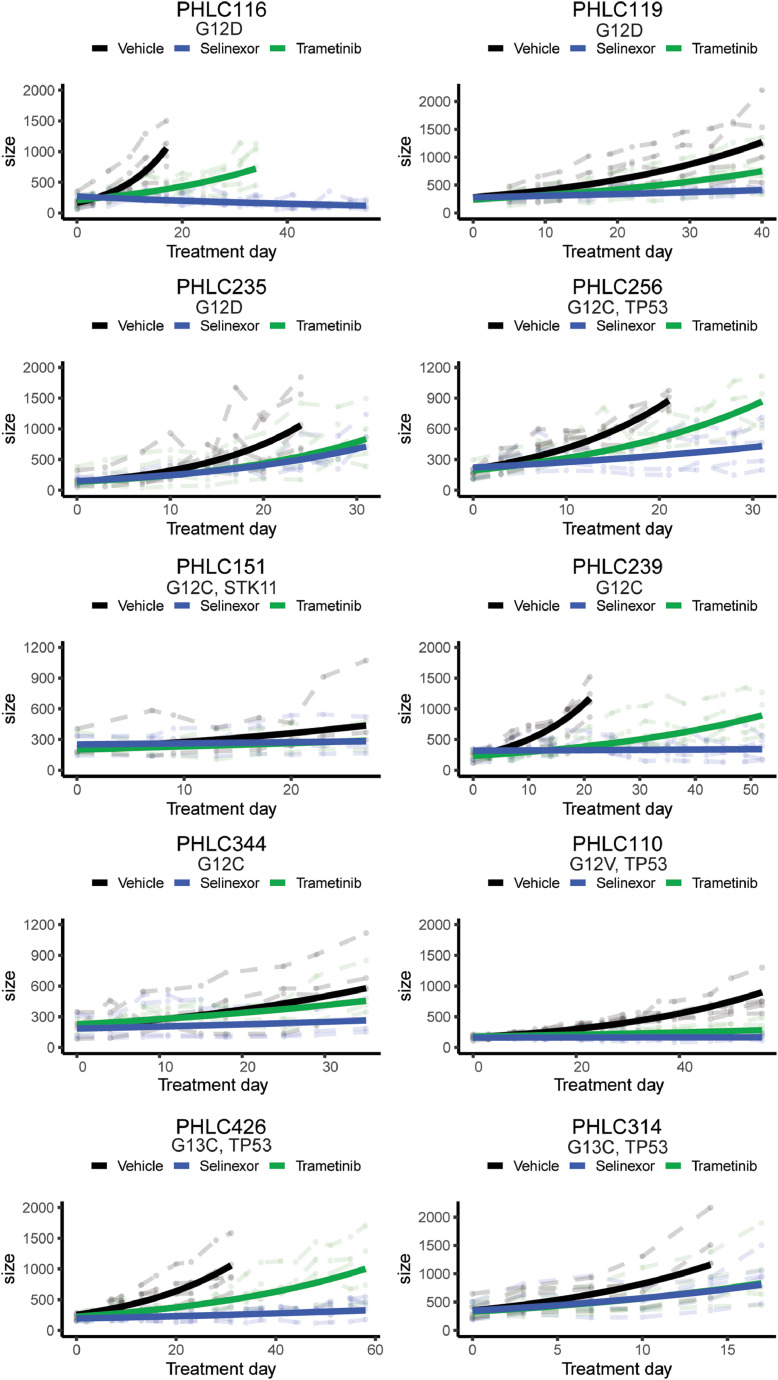
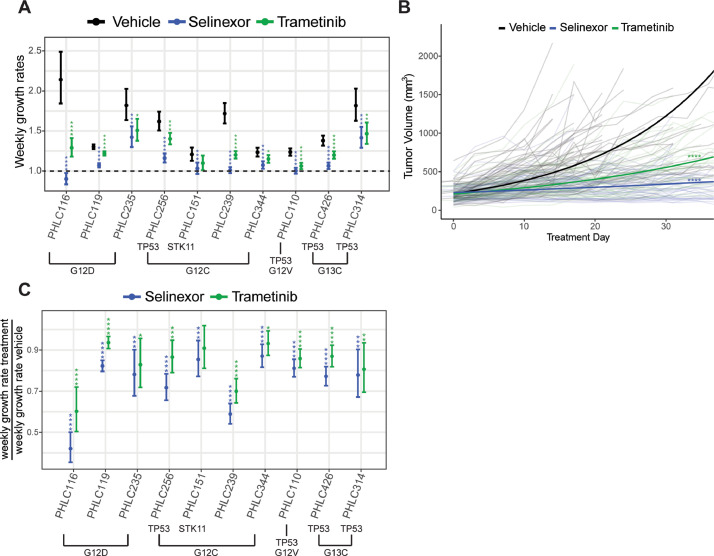
To investigate the potential anti-tumor effects of the XPO1 inhibitor Selinexor, and the MEK1/2 inhibitor Trametinib, on KRAS-mutant-driven lung cancer, the 10 PDX models were used in 3-arm studies that included vehicle and each of the two drugs. By inputting tumor measurements into log linear mixed effects models [39], we predicted the growth rates of these tumors (Figs. 2, 3A). When pooling data from all mice for all PDX models, Selinexor was found to reduce the overall weekly growth rate of KRAS-mutant-driven lung cancer by a factor of 0.73 or 27% (Fig. 3B, Supplementary Table S2). Considering that tumors grow exponentially, this effect on weekly growth rate translates to a much greater effect than 27% over time (Supplementary Table S2). We examined the potential effect of Selinexor on ERK activation in 8 models and found no evidence for impairment (Supplementary Fig. S2), suggesting Selinexor acts on a parallel pathway that cooperates with KRAS, but may not be under its direct control.
Fig. 2.
Growth rates and predicted growth rates of Selinexor and Trametinib treated KRASmut mice. Mice were treated with Trametinib (1mg/kg) 5x/week, Selinexor (10mg/kg) [14,23] 3x/week. Data are separated by the individual independently established KRASmut PDX models. PHLC116: n = 5/arm; PHLC119: n = 5/arm; PHLC235: n = 5/arm; PHLC151: n = 2/Trametinib, n = 3/vehicle, Selinexor; PHLC239: n = 5/arm; PHLC256: n = 5/arm; PHLC344: n = 4/arm; PHLC110: n = 5/arm; PHLC314: n = 5/arm; PHLC426: n = 5/arm. Faded dashed lines in the background are the measured tumor growth patterns of individual mice. Dark lines represent predicted growth curves generated from the log linear mixed effects models [39]. KRAS mutation is indicated, along with prevalence of STK11/TP53 mutations.
Fig. 3.
Weekly growth rate comparisons between different KRASmut PDXs treated with either Selinexor or Trametinib. (A) Weekly growth rates predicted by the log linear mixed effects models [39] are plotted for each treatment of every model. Values > 1.00 represent tumor growth; values < 1.00 represent tumor shrinkage. KRAS, TP53 andSTK11 mutation status is indicated. (B) Overall responses of pooled KRASmut models to either Selinexor or Trametinib. Day 0 was the start of treatment. Faded lines in the background are the measured growth data from individual mice, and dark lines represent the predicted weekly growth curve for each treatment. Overall, vehicle (n = 47), Trametinib (n = 46), Selinexor (n = 47) are plotted here. The p-values are comparing growth rate of treatments to vehicle using a log linear mixed effects model: * = p < 0.05; ** = p < 0.01; *** = p < 0.005; ****= p < 0.001. (C) Predicted weekly change in growth rates for treatment compared to vehicle, where the vehicle growth rate was normalized to a value of 1.00. Results are from log linear mixed effects models [39]. KRAS, TP53 andSTK11 mutation status is indicated. Plots are presented with confidence intervals and FDR adjusted p-values: * = p < 0.05; ** = p < 0.01; *** = p < 0.005; ****= p < 0.001.
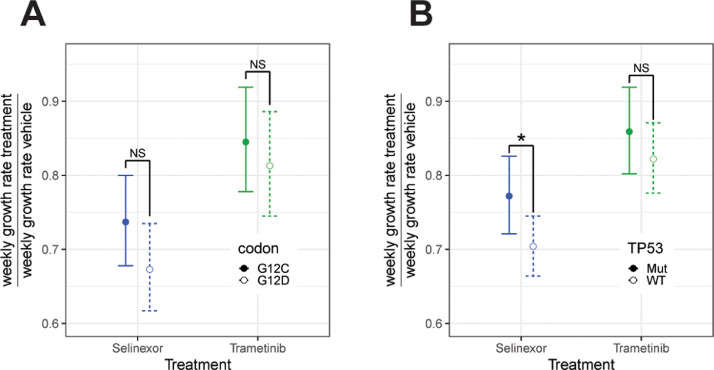
The overall growth suppression of KRASmut lung cancer by Selinexor was not due to selective effects on specific KRASmut models, but instead, reflected significant growth suppression of each of the 10 tested PDX models (Fig. 3A, C). Across these models, weekly tumor growth rates were reduced by a factor of 0.87 to 0.42 (13%–58%) (Fig. 3C, Supplementary Table S3). The most common codon alterations in our cohort, G12C and G12D, did not show differences in Selinexor responses between each other (Fig. 4A). However, TP53WT tumors responded better to Selinexor than TP53mut tumors (Fig. 4B).
Fig. 4.
Comparisons of Selinexor and Trametinib effectiveness depending on KRAS codon and TP53 alterations. (A, B) Predicted weekly change in growth rates for treatment compared to vehicle, where vehicle was normalized to equal a weekly growth rate of 1. Results are from a log linear mixed effects model testing the interaction between treatment, weekly growth rate, and KRAS codon or TP53 mutation [39]. (A) Comparison of predicted weekly growth rates after treatment with either Selinexor or Trametinib depending on presence of G12C (n = 4) or G12D (n = 3) codon alterations. There was no significant difference for either Selinexor (p = 0.15) or Trametinib (p = 0.52). (B) Comparison of predicted weekly growth rates after treatment with either Selinexor or Trametinib depending on TP53WT (n = 6) or TP53mut (n = 4) status. Predicted weekly change in growth rates for treatment compared to vehicle, where vehicle was normalized to equal a weekly growth rate of 1. There was no significant difference in the effect of Trametinib (p = 0.34), but Selinexor decreased the rate of growth significantly more in TP53WT compared to TP53mut models (p = 0.04).
Selinexor is more effective than Trametinib in KRAS-mutant lung cancer PDXs
Trametinib reduced the overall weekly growth rate of KRAS-mutant-driven lung cancer by 0.84 (16%) (Fig. 3B, Supplementary Table S2), which was 11% less effective than Selinexor (Supplementary Table S2). In addition, one of the 10 individual models, PHLC151, did not respond to Trametinib (Fig. 3A, C, Supplementary Table S3) and for 7 models, the response to Trametinib was significantly inferior to that of Selinexor (Trametinib 7%–43% faster growing than Selinexor) (Table 1). In no instance was the response to Trametinib greater than to Selinexor (Table 1). There was no significant difference in Trametinib response between lung cancers harboring KRASG12C versus KRASG12D mutations or depending on TP53 mutation status (Fig. 4).
Table 1.
Weekly overall growth rates of treatment arms versus Selinexor (normalized to a value of 1.00) based on log linear mixed effects models, and predicted tumor size at day 7, 14, and 21 if tumor treatment started at 200mm.
| Model | KRAS mutations (Whole exome Seq) | Treatment | Growth rate versus Selinexor (95% CI) | Adjusted p-value | Day 7 average tumor volume (mm3) | Day 14 average tumor volume (mm3) | Day 21 average tumor volume (mm3) |
|---|---|---|---|---|---|---|---|
| All KRASmut overall | Trametinib | 1.145(1.097,1.195) | < 0.001 | 250.8 | 314.5 | 394.3 | |
| PHLC116 | G12D | Trametinib | 1.431(1.267,1.616) | < 0.001 | 257.9 | 332.7 | 429 |
| G12D | Selinexor | - | - | 180.2 | 162.4 | 146.4 | |
| PHLC119 | G12D | Trametinib | 1.138(1.101,1.176) | < 0.001 | 243.9 | 297.4 | 362.6 |
| G12D | Selinexor | - | - | 214.4 | 229.7 | 246.2 | |
| PHLC235 | G12D | Trametinib | 1.061(0.931,1.209) | 0.4 | 301.6 | 455 | 686.2 |
| G12D | Selinexor | - | - | 284.4 | 404.4 | 575.1 | |
| PHLC151 | G12C | Trametinib | 1.064(0.949,1.193) | 0.32 | 219.5 | 241 | 264.6 |
| G12C | Selinexor | - | - | 206.3 | 212.9 | 219.6 | |
| PHLC239 | G12C | Trametinib | 1.188(1.125,1.255) | < 0.001 | 240.2 | 288.4 | 346.3 |
| G12C | Selinexor | - | - | 202.1 | 204.3 | 206.4 | |
| PHLC256 | G12C | Trametinib | 1.207(1.123,1.297) | < 0.001 | 280.4 | 393 | 550.9 |
| G12C | Selinexor | - | - | 232.2 | 269.7 | 313.2 | |
| PHLC344 | G12C | Trametinib | 1.07(1.006,1.139) | 0.041 | 230 | 264.5 | 304.1 |
| G12C | Selinexor | - | - | 214.8 | 230.8 | 247.9 | |
| PHLC110 | G12V | Trametinib | 1.058(1.004,1.116) | 0.041 | 212.4 | 225.6 | 239.7 |
| G12V | Selinexor | - | - | 200.7 | 201.4 | 202.1 | |
| PHLC314 | G13C | Trametinib | 1.036(0.906,1.185) | 0.61 | 293.2 | 429.7 | 629.9 |
| G13C | Selinexor | - | - | 283 | 400.5 | 566.7 | |
| PHLC426 | G13C | Trametinib | 1.126(1.067,1.189) | < 0.001 | 239.7 | 287.4 | 344.5 |
| G13C | Selinexor | - | - | 212.8 | 226.5 | 241 | |
| PHLC137 | WT | Erlotinib | 0.506(0.446,0.573) | < 0.001 | 114.9 | 66 | 37.9 |
| WT | Selinexor | - | - | 227.2 | 258.1 | 293.2 | |
| PHLC148 | WT | Crizotinib | 0.649(0.585,0.721) | < 0.001 | 127 | 80.6 | 51.2 |
| WT | Selinexor | - | - | 195.9 | 191.8 | 187.8 |
Selinexor and Trametinib monotherapy cause stable disease and rarely induce tumor regression
With one exception, Selinexor and Trametinib generally slowed tumor growth, rather than caused tumor regression (Fig. 3A). For Selinexor, treatment of 7 models resulted in weekly tumor growth rates close to 1.0, which indicates cessation of growth and is the equivalent of stable disease, while treatment of one model, PHLC116, reduced the weekly growth rate below 1.0, leading to tumor regression (Fig. 3A). By contrast, while Trametinib significantly slowed tumor growth across 9/10 models relative to vehicle, a weekly growth rate close to 1.0 was only achieved with one of those nine models (PHLC110) and no treated model showed tumor regression (Fig. 3A).
XPO1 is a common essential cancer gene
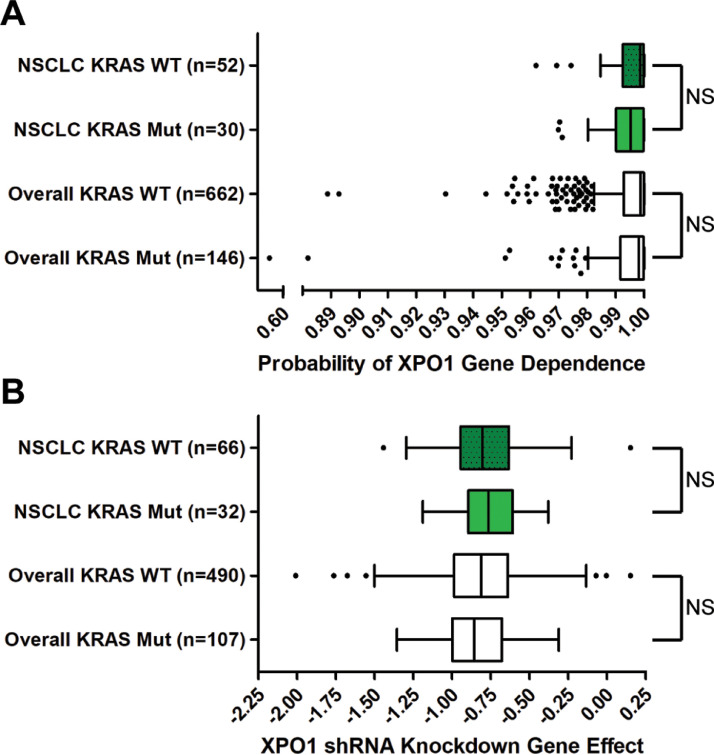
The sensitivity of all 10 tested KRASmut lung cancer PDXs to Selinexor could reflect a specific and greater dependency of KRAS-driven lung cancers on XPO1 activity, as suggested by some work [14], or could reflect a general dependency of cancer growth on XPO1, regardless of KRAS mutation status, as suggested by other studies [22], [23], [24]. To distinguish between these possibilities, we examined gene level data on XPO1 dependency across hundreds of cancer cell lines, as determined by CRISPR/Cas9 knockout or shRNA knockdown. In a genome-wide CRISPR/Cas9 dropout screen conducted in 808 cancer cell lines (Cancer Dependency Map Project) [37], XPO1 was found to be a dependency in at least 90% of the cell lines, falling into the classification of “common essential gene”. Consistent with XPO1 being a common essential gene, we found no difference in the gene dependency score when comparing cell lines with and without KRAS mutations (Fig. 5A). Similarly, the XPO1 gene dependency score was not different when specifically comparing KRASmut vs KRASWT NSCLC cell lines (Fig. 5A). Furthermore, the same results were obtained when examining the effect of knocking down XPO1 levels by shRNA in 597 cell lines [38]. In neither the overall dataset nor the data pertaining specifically to NSCLC, was the gene effect different between KRASmut vs KRASWT cell lines (Fig. 5B). These genetic data support the majority of the targeted preclinical studies that found that XPO1 is a broad dependency across multiple cancer types [[22], [23], [24],[43], [44], [45]].
Fig. 5.
XPO1 gene dependency across different cancer cell lines. (A) Box and whisker plots of the probabilities of cancer cell line XPO1 genetic dependency from CRISPR/Cas9 knockout screens, compiled from the Achilles Project 20Q4 [37]. Values were estimated previously and shifted and scaled per cell line so a score of 0 represents 0% dependence on the gene of interest per cell line; a score of 1 represents 100% dependence on the gene of interest per cell line [37]. XPO1 dependency scores of KRASWT and KRASmut NSCLC cell lines (p = 0.27) or all cell lines overall (not restricted to NSCLC) (p = 0.066) were not significantly different. (B) Box and whisker plots of the probabilities of cancer cell line XPO1 genetic dependencies from RNAi knockdown screens, compiled from the Achilles Project [38]. Values were estimated previously, with a lower value representing a greater dependence on XPO1 for survival, although no scale was explicitly defined [38]. XPO1 dependency scores of KRASWT and KRASmut NSCLC cell lines (p = 0.58) or all cell lines overall (not restricted to NSCLC) (p = 0.43) were not significantly different. Mann-Whitney tests were used to compare means of each group. Tukey whiskers are plotted. NS = not significant.
Selinexor inhibits growth of lung cancers driven by oncogenic kinases, but not as effectively as targeted kinase inhibitors
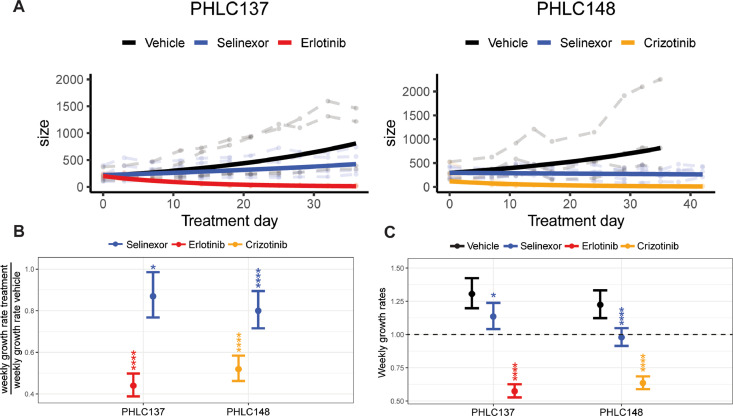
To further investigate whether KRASmut lung cancer PDX sensitivity to Selinexor reflects a general cancer dependency on XPO1 rather than a specific and greater dependency of KRAS-driven cancers, we also tested Selinexor in non-KRAS-driven lung cancer PDXs (Supplementary Fig. S3). PHLC137 and PHLC148 are lung cancer PDX models that harbor activating oncogenic EGFR mutations [46]. However, while PHLC137 is driven by mutant EGFR, PHLC148 is driven by a co-occurring amplification of the MET receptor tyrosine kinase [46]. Relative to the control vehicle, Selinexor comparably reduced the growth rates of these tumors by a factor of 0.87–0.80 (13–20%) (Fig. 6A, B, Supplementary Table S3), which was in the range of Selinexor effects observed in the KRASmut lung cancer PDXs (Fig. 3C, Supplementary Table S3). In fact, treatment of PHLC148 resulted in stable disease (weekly growth rate of 1.0, Fig. 6C), as seen in most of the KRASmut lung cancer PDXs (Fig. 3A). However, the approved targeted inhibitors for the EGFR and MET, erlotinib and crizotinib, respectively, were much more effective than Selinexor in these models, causing tumor regression to the point of the tumors becoming non-palpable (Fig. 6, Table 1, Supplementary Table S3).
Fig. 6.
Growth rates and predicted growth rates of KRASWT PDXs treated with either Selinexor or a tyrosine kinase inhibitor. Mice were treated with erlotinib (50mg/kg) 5x/week or crizotinib (50mg/kg) 5x/week. PHLC137: n = 4/arm; PHLC148: n = 4/arm. (A) Growth patterns over time. Faded lines in the background are the measured tumor growth patterns of individual mice and dark lines represent predicted growth curves generated from log linear mixed effects modeling [39]. (B) Predicted change in weekly growth rates for treatment compared to vehicle, where vehicle was normalized to equal a weekly growth rate of 1.00. (C) Raw predicted weekly growth rates. Values > 1.00 represent tumor growth; values < 1.00 represent tumor shrinkage. Plots are presented with confidence intervals and FDR adjusted p-values: * = p < 0.05; ** = p < 0.01; *** = p < 0.005; ****= p < 0.001.
Discussion
KRAS-mutant-driven lung cancers have generally been treated with platinum-based therapy [5,47]. Unfortunately, the five-year survival is poor [48], and only recently, has a targeted therapy become available for patients that specifically have KRASG12C-mutated tumors [8,9]. Thus, additional therapeutic strategies are still needed for KRASmut lung cancers, especially those harboring non-G12C mutations [6,7]. Using cell lines, a genetically engineered mouse model, and a single human lung cancer PDX, one study proposed that the XPO1 nuclear exporter is a specific vulnerability in KRAS-mutant-driven lung cancers [14]. This notion was further supported by the finding that pancreatic ductal carcinoma cell lines, PDXs, and patients, where KRAS mutations are common, also show sensitivity to Selinexor, either alone or in conjunction with chemotherapy [49]. However, other preclinical studies have highlighted XPO1 as a broader vulnerability across multiple cancer types [[22], [23], [24],[43], [44], [45]]. To better approximate the sensitivity of clinical lung cancer to XPO1 inhibition, we tested the sensitivity of ten KRASmut LUAD PDXs to Selinexor, an XPO1 inhibitor under clinical investigation for numerous cancers [25,26,50]. These PDX models generally recapitulate the genetic and phenotypic properties of their matched patient tumors with high fidelity [27,28,46], and thus, are considered reliable models of clinical disease. In addition, we compared the activity of Selinexor to that of a clinical MEK1/2 inhibitor, Trametinib, which is being investigated for the treatment of KRASmut lung cancer [29,30] (NCT02642042, NCT03704688).
Selinexor caused significant reductions in tumor growth rates of every tested KRASmut PDX model. Extended drug exposure led to maintenance of the initial tumor growth curve trajectory, suggesting sustained activity of the drug with our dosing regimen over periods lasting longer than 30 days, which was not previously documented in other mouse studies of Selinexor for lung cancer [14,22,23]. Overall, 80% of the models had at least stable disease, and one model regressed on Selinexor. We observed antitumor effectiveness across all tested KRAS codon alterations, including G12C, G12D, G12V, and G13C, which collectively span ~90% cases of KRASmut lung cancer [10]. Although there is evidence for some of these mutant KRAS proteins having distinct signaling properties and conferring different sensitivities to certain chemotherapeutics [51,52], our findings suggest that XPO1 is required for all mutant KRAS function, which is also consistent with XPO1 being broadly required for growth of cancer cells, in general. Trametinib significantly slowed tumor growth in 90% of the models, but responsiveness was inferior to that of Selinexor. With Trametinib, only one model achieved stable disease and no model showed regression.
We also investigated whether TP53 co-mutation impacted responsiveness to Selinexor. Somatic TP53 mutations are observed in 50% of LUAD [53,54] and prior work found that certain co-mutations, including in TP53, could subset KRASmut LUAD into distinct biological groups [41]. Moreover, TP53 co-mutation has sometimes been associated with differential sensitivity to certain therapies when compared to KRASmut/TP53WT cancer cells. For example, TP53 co-mutation increases sensitivity to anti-PD-1 therapy [42] but reduces sensitivity to adjuvant platinum-based chemotherapy [54]. In our KRASmut lung cancer PDX cohort, we found that TP53 co-mutation was significantly associated with a weaker response to Selinexor (relative to control vehicle), as compared to TP53WT tumors. By contrast, we did not detect any differential sensitivity between TP53mut and TP53WT tumors to Trametinib. Increased sensitivity of TP53WT cells to XPO1 inhibition relative to TP53mut cells has also been reported in other cancer types, even in those that do not have KRAS mutations such as mantle cell lymphoma [45]. One possible mechanistic explanation for the increased sensitivity of TP53WT tumors to XPO1 inhibition regardless of the driver could involve XPO1 regulation of TP53 trafficking and TP53’s ability to trigger cell cycle arrest. XPO1 inhibitors have been shown to promote nuclear accumulation and activation of TP53, which in turn may increase TP73 and TP21 levels to cause cell cycle arrest [55]. However, given that all our TP53mut lung cancer PDXs still had significant responses to Selinexor, XPO1 inhibitors must have additional modes of action to slow tumor growth besides promoting activation of TP53.
Since our finding that all ten KRASmut lung cancer PDXs responded to Selinexor could be consistent with either XPO1 being a specific dependency for KRASmut cancer cells, or a broadly acting dependency across many types of cancers, we examined genetic dependency of hundreds of different cancer cell lines on XPO1. This analysis was conducted by mining publicly available data from genome-wide shRNA and CRISPR/Cas9 dropout screenings [37,38]. From these data, we found that both among all cancers and specifically among lung cancer, there was no difference in the XPO1 dependency score between KRASmut and KRASWT cells, with nearly all cell lines being dependent on XPO1, leading to its classification as a common essential cancer gene. To specifically address the essentiality of XPO1 to KRASWT lung cancer cells in the context of PDX experiments, we also treated two wild-type KRAS lung cancer PDXs with Selinexor. These models were driven either by EGFR or MET alterations [46]. Selinexor was similarly effective in these models as in the KRASmut models, also being able to generate situations close to stable disease or weak regression. However, in both models, approved tyrosine kinase inhibitors generated superior responses. Notably, the MET-amplified, EGFR-TKI-resistant NCI-H1993 LUAD cell line [56] was reported to be somewhat resistant to Selinexor in vitro, with the drug being unable to induce caspase-dependent apoptosis [14]. By contrast, Selinexor induced weak regression in our EGFR-TKI-resistant/MET-amplified lung cancer PDX. This difference in responsiveness to Selinexor could reflect subtly different modulation of the drug response between in vitro and in vivo settings, as the genetic studies indicate the vast majority of cancer cells growing in vitro are still dependent on XPO1. Overall, both our PDX experiments and our genetic analyses support XPO1 inhibitors having anti-cancer effects in KRASmut and KRASWT cancer cells. These pan-cancer effects could be manifested due to distinct or common mechanisms. In addition to TP53 activation, XPO1 inhibition has also been suggested to promote killing in certain cancer cells such as those harboring KRAS mutations through nuclear accumulation of IκBα, which can inhibit NF-κB signaling [14]. However, since the export of over 350 cargoes could be dysregulated upon XPO1 inhibition [17], nuclear accumulation of any number of individual or combinations of cargoes could contribute to the broad anti-cancer effects of XPO1 inhibitors.
Selinexor is currently approved for relapsed/refractory multiple myeloma [25], which has a high frequency of KRAS mutation [57], and relapsed/refractory diffuse large B-cell lymphoma [26], where KRAS mutations are not common [58,59]. There are many ongoing clinical trials of XPO1 inhibitors, including two in NSCLC with Selinexor in combination with docetaxel (NCT04256707 and NCT03095612). Our data support continued trials of Selinexor in NSCLC, including both KRASWT and KRASmut subsets, without having to discriminate between the mutant KRAS alteration. Importantly, Selinexor could be an option for patients harboring non-G12C KRAS mutations. However, our results also suggest that Selinexor is unlikely to surpass promising and approved targeted therapies for non-KRAS-mutated NSCLC (e.g. EGFR mutations, ALK fusions, MET amplification), making the drug better suited for cancers without readily targetable driver alterations or for salvage therapy, and especially for patients with wild-type TP53. Although Selinexor is currently being investigated in combination with a standard of care chemotherapy, docetaxel, it may be challenging to adjust dosing to minimize toxicity. Over one-third of relapsed/refractory multiple myeloma patients treated with Selinexor experienced neutropenia (NCT02336815), a common toxicity also associated with docetaxel [60,61]. Thus, it may be worthwhile to also consider combining Selinexor with alternative treatments such as immunotherapy, where in a preclinical model of melanoma, Selinexor was shown to increase responses to several checkpoint inhibitors [62]. This combination may be especially beneficial for KRAS/TP53 double mutant NSCLC, which is thought to benefit from immunotherapy [41,42]. Other intriguing combinations could include Selinexor and KRASG12C inhibitors, as both preclinical and clinical trial studies indicate G12C inhibitor monotherapy can lead to emergence of resistant subpopulations of cancer cells [6,7,11,12,63]. This combination has recently been observed to reduce proliferation of MiaPaCa-2 pancreatic ductal adenocarcinoma and NCI-H2122 NSCLC cell lines, and disintegration of their respective spheroids [64].
In conclusion, Selinexor remains an intriguing drug for continued investigation in NSCLC.
CRediT authorship contribution statement
Joshua C. Rosen: Conceptualization, Methodology, Validation, Investigation, Writing – original draft, Writing – review & editing, Visualization. Jessica Weiss: Software, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Nhu-An Pham: Methodology, Writing – review & editing, Project administration. Quan Li: Software, Formal analysis. Sebastiao N. Martins-Filho: Conceptualization, Investigation. Yuhui Wang: Investigation. Ming-Sound Tsao: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition. Nadeem Moghal: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding support
This work is funded by the Canadian Institutes of Health Research (CIHR) Project Grant PJT-175190 (NM) and Foundation Grant FDN-148395 (MST), and the Princess Margaret Cancer Foundation. Joshua C. Rosen is funded by a University of Toronto Student Opportunity Trust Fund (OSOTF). Dr. Sebastiao Martins-Filho is supported by the Terry Fox Foundation Training grant for Clinician Scientist in Oncologic Pathology and Fellowship from the Ontario Molecular Pathology Research Network. M.-S. Tsao is the M. Qasim Choksi Chair in Lung Cancer Translational Research.
Acknowledgments
We thank Dr. Kugeng Huo for his aid in tumor measurement.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101179.
Appendix. Supplementary materials
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C., Dicker D., Pain A., Hamavid H., Moradi-Lakeh M. The global burden of cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barta J.A., Powell C.A., Wisnivesky J.P. Global epidemiology of lung cancer. Ann. Glob. Health. 2019;85:8. doi: 10.5334/aogh.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepherd F.A., Domerg C., Hainaut P., Jänne P.A., Pignon J.P., Graziano S. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 2013;31:2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Osta B., Behera M., Kim S., Berry L.D., Sica G., Pillai R.N. Characteristics and outcomes of patients with metastatic KRAS-mutant lung adenocarcinomas: the lung cancer mutation consortium experience. J. Thorac. Oncol. 2019;14:876–889. doi: 10.1016/j.jtho.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemjabbar-Alaoui H., Hassan O.U., Yang Y.W., Buchanan P. Lung cancer: biology and treatment options. Biochim. Biophys. Acta. 2015;1856:189–210. doi: 10.1016/j.bbcan.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 7.Hallin J., Engstrom L.D., Hargis L., Calinisan A., Aranda R., Briere D.M. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.FDA Approves First KRAS Inhibitor: Sotorasib. Cancer Discov 2021 June 22 (Epub ahead of print). [DOI] [PubMed]
- 9.Skoulidis F., Li B.T., Dy G.K., Price T.J., Falchook G.S., Wolf J. Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka N., Lin J.J., Li C., Ryan M.B., Zhang J., Kiedrowski L.A. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-21-0365. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awad M.M., Liu S., Rybkin I.I., Arbour K.C., Dilly J., Zhu V.W. Acquired resistance to KRAS(G12C) inhibition in cancer. N. Engl. J. Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguirre A.J., Hahn WC. Synthetic lethal vulnerabilities in KRAS-mutant cancers. Cold. Spring Harb. Perspect. Med. 2018;8 doi: 10.1101/cshperspect.a031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J., McMillan E., Kim H.S., Venkateswaran N., Makkar G., Rodriguez-Canales J. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538:114–117. doi: 10.1038/nature19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran E.J., King M.C., Corbett AH. Macromolecular transport between the nucleus and the cytoplasm: advances in mechanism and emerging links to disease. Biochim. Biophys. Acta. 2014;1843:2784–2795. doi: 10.1016/j.bbamcr.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Reilly A.J., Dacks J.B., Field MC. Evolution of the karyopherin-β family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS One. 2011;6:e19308. doi: 10.1371/journal.pone.0019308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu D., Grishin N.V., Chook YM. NESdb: a database of NES-containing CRM1 cargoes. Mol. Biol. Cell. 2012;23:3673–3676. doi: 10.1091/mbc.E12-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Q., Carrasco Y.P., Hu Y., Guo X., Mirzaei H., Macmillan J. Nuclear export inhibition through covalent conjugation and hydrolysis of leptomycin B by CRM1. Proc. Natl. Acad. Sci. U S A. 2013;110:1303–1308. doi: 10.1073/pnas.1217203110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birnbaum D.J., Finetti P., Birnbaum D., Mamessier E., Bertucci F. XPO1 expression is a poor-prognosis marker in pancreatic adenocarcinoma. J. Clin. Med. 2019;8:596. doi: 10.3390/jcm8050596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J., McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 21.Gao W., Lu C., Chen L., Keohavong P. Overexpression of CRM1: a characteristic feature in a transformed phenotype of lung carcinogenesis and a molecular target for lung cancer adjuvant therapy. J. Thorac. Oncol. 2015;10:815–825. doi: 10.1097/JTO.0000000000000485. [DOI] [PubMed] [Google Scholar]
- 22.Wang S., Han X., Wang J., Yao J., Shi Y. Antitumor effects of a novel chromosome region maintenance 1 (CRM1) inhibitor on non-small cell lung cancer cells in vitro and in mouse tumor xenografts. PLoS One. 2014;9:e89848. doi: 10.1371/journal.pone.0089848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun H., Hattori N., Chien W., Sun Q., Sudo M., GL E.L. KPT-330 has antitumour activity against non-small cell lung cancer. Br. J. Cancer. 2014;111:281–291. doi: 10.1038/bjc.2014.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arango N.P., Yuca E., Zhao M., Evans K.W., Scott S., Kim C. Selinexor (KPT-330) demonstrates anti-tumor efficacy in preclinical models of triple-negative breast cancer. Breast Cancer Res. 2017;19:93. doi: 10.1186/s13058-017-0878-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chari A., Vogl D.T., Gavriatopoulou M., Nooka A.K., Yee A.J., Huff C.A. Oral selinexor-dexamethasone for triple-class refractory multiple myeloma. N. Engl. J. Med. 2019;381:727–738. doi: 10.1056/NEJMoa1903455. [DOI] [PubMed] [Google Scholar]
- 26.Kalakonda N., Maerevoet M., Cavallo F., Follows G., Goy A., Vermaat J.S. Selinexor in patients with relapsed or refractory diffuse large B-cell lymphoma (SADAL): a single-arm, multinational, multicentre, open-label, phase 2 trial. Lancet Haematol. 2020;7:E511–E522. doi: 10.1016/S2352-3026(20)30120-4. [DOI] [PubMed] [Google Scholar]
- 27.John T., Kohler D., Pintilie M., Yanagawa N., Pham N.A., Li M. The ability to form primary tumor xenografts is predictive of increased risk of disease recurrence in early-stage non-small cell lung cancer. Clin. Cancer Res. 2011;17:134–141. doi: 10.1158/1078-0432.CCR-10-2224. [DOI] [PubMed] [Google Scholar]
- 28.Wang D., Pham N.A., Tong J., Sakashita S., Allo G., Kim L. Molecular heterogeneity of non-small cell lung carcinoma patient-derived xenografts closely reflect their primary tumors. Int. J. Cancer. 2017;140:662–673. doi: 10.1002/ijc.30472. [DOI] [PubMed] [Google Scholar]
- 29.Blumenschein G.R., Smit E.F., Planchard D., Kim D.W., Cadranel J., De Pas T. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)†. Ann. Oncol. 2015;26:894–901. doi: 10.1093/annonc/mdv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gadgeel S.M., Miao J., Riess J.W., Mack P.C., Gerstner G.J., Burns T.F. S1507: Phase II study of docetaxel and Trametinib in patients with G12C or non-G12C KRAS mutation positive (+) recurrent non-small cell lung cancer (NSCLC) J. Clin. Oncol. 2019;37(15_suppl):9021. doi: 10.1158/1078-0432.CCR-22-3947. –9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway T., Wazny J., Bromage A., Tymms M., Sooraj D., Williams E.D. Xenome-a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28:i172–i178. doi: 10.1093/bioinformatics/bts236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFarland J.M., Ho Z.V., Kugener G., Dempster J.M., Montgomery P.G., Bryan J.G. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 2018;9:4610. doi: 10.1038/s41467-018-06916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo S., Jiang X., Mao B., Li Q-X. The design, analysis and application of mouse clinical trials in oncology drug development. BMC Cancer. 2019;19:718. doi: 10.1186/s12885-019-5907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H.A., Sima C.S., Shen R., Kass S., Gainor J., Shaw A. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J. Thorac. Oncol. 2015;10:431–437. doi: 10.1097/JTO.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skoulidis F., Byers L.A., Diao L., Papadimitrakopoulou V.A., Tong P., Izzo J. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 43.Sexton R., Mahdi Z., Chaudhury R., Beydoun R., Aboukameel A., Khan H.Y. Targeting nuclear exporter protein XPO1/CRM1 in gastric cancer. Int. J. Mol. Sci. 2019;20:4826. doi: 10.3390/ijms20194826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y., Camacho S.C., Silvers T.R., Razak A.R., Gabrail N.Y., Gerecitano J.F. Inhibition of the nuclear export receptor XPO1 as a therapeutic target for platinum- resistant ovarian cancer. Clin. Cancer Res. 2017;23:1552–1563. doi: 10.1158/1078-0432.CCR-16-1333. [DOI] [PubMed] [Google Scholar]
- 45.Yoshimura M., Ishizawa J., Ruvolo V., Dilip A., Quintás-Cardama A., McDonnell T.J. Induction of p53-mediated transcription and apoptosis by exportin-1 (XPO1) inhibition in mantle cell lymphoma. Cancer Sci. 2014;105:795–801. doi: 10.1111/cas.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart E.L., Mascaux C., Pham N.A., Sakashita S., Sykes J., Kim L. Clinical utility of patient-derived xenografts to determine biomarkers of prognosis and map resistance pathways in EGFR-mutant lung adenocarcinoma. J. Clin. Oncol. 2015;33:2472–2480. doi: 10.1200/JCO.2014.60.1492. [DOI] [PubMed] [Google Scholar]
- 47.Kris M.G., Gaspar L.E., Chaft J.E., Kennedy E.B., Azzoli C.G., Ellis P.M. Adjuvant Systemic therapy and adjuvant radiation therapy for stage I to IIIA completely resected non-small-cell lung cancers: American society of clinical oncology/cancer care ontario clinical practice guideline update. J. Clin. Oncol. 2017;35:2960–2974. doi: 10.1200/JCO.2017.72.4401. [DOI] [PubMed] [Google Scholar]
- 48.Ma B., Geng Y., Meng F., Yan G., Song F. Identification of a sixteen-gene prognostic biomarker for lung adenocarcinoma using a machine learning method. J. Cancer. 2020;11:1288–1298. doi: 10.7150/jca.34585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azmi A.S., Khan H.Y., Muqbil I., Aboukameel A., Neggers J.E., Daelemans D. Preclinical assessment with clinical validation of selinexor with gemcitabine and nab-paclitaxel for the treatment of pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2020;26:1338. doi: 10.1158/1078-0432.CCR-19-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azizian N.G., Li Y. XPO1-dependent nuclear export as a target for cancer therapy. J. Hematol. Oncol. 2020;13:61. doi: 10.1186/s13045-020-00903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garassino M.C., Marabese M., Rusconi P., Rulli E., Martelli O., Farina G. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann. Oncol. 2011;22:235–237. doi: 10.1093/annonc/mdq680. [DOI] [PubMed] [Google Scholar]
- 52.Ihle N.T., Byers L.A., Kim E.S., Saintigny P., Lee J.J., Blumenschein G.R. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J. Natl. Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cancer genome atlas research network. comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shepherd F.A., Lacas B., Le Teuff G., Hainaut P., Jänne P.A., Pignon J.P. Pooled analysis of the prognostic and predictive effects of TP53 Comutation status combined with KRAS or EGFR mutation in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 2017;35:2018–2027. doi: 10.1200/JCO.2016.71.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang A.Y., Liu H. The past, present, and future of CRM1/XPO1 inhibitors. Stem. Cell Investig. 2019;6:6. doi: 10.21037/sci.2019.02.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubo T., Yamamoto H., Lockwood W.W., Valencia I., Soh J., Peyton M. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. Int. J. Cancer. 2009;124:1778–1784. doi: 10.1002/ijc.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu Y., Chen W., Wang J. Progress in the identification of gene mutations involved in multiple myeloma. Onco. Targ. Ther. 2019;12:4075–4080. doi: 10.2147/OTT.S205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lohr J.G., Stojanov P., Lawrence M.S., Auclair D., Chapuy B., Sougnez C. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA. 2012;109:3879. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pasqualucci L., Dalla-Favera R. The genetic landscape of diffuse large B-cell lymphoma. Semin. Hematol. 2015;52:67–76. doi: 10.1053/j.seminhematol.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kenmotsu H., Tanigawara Y. Pharmacokinetics, dynamics and toxicity of docetaxel: Why the Japanese dose differs from the western dose. Cancer Sci. 2015;106:497–504. doi: 10.1111/cas.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanna N., Shepherd F.A., Fossella F.V., Pereira J.R., De Marinis F., von Pawel J. Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy. J. Clin. Oncol. 2004;22:1589–1597. doi: 10.1200/JCO.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 62.Farren M.R., Hennessey R.C., Shakya R., Elnaggar O., Young G., Kendra K. The exportin-1 inhibitor selinexor exerts superior antitumor activity when combined with T-cell checkpoint inhibitors. Mol. Cancer Ther. 2017;16:417–427. doi: 10.1158/1535-7163.MCT-16-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xue J.Y., Zhao Y., Aronowitz J., Mai T.T., Vides A., Qeriqi B. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature. 2020;577:421–425. doi: 10.1038/s41586-019-1884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khan H.Y., MdH U., Zhang Y., Landesman Y., Sukari A., Nagasaka M. Abstract 1058: inhibition of nuclear transport protein XPO1 potentiates the effect of KRASG12C inhibitors. Cancer Res. 2021;81(13 Supplement):1058. Jul 1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.