ABSTRACT
Bacterial genomes encode various multidrug efflux pumps (MDR) whose specific conditions for fitness advantage are unknown. We show that the efflux pump MdtEF-TolC, in Escherichia coli, confers a fitness advantage during exposure to extreme acid (pH 2). Our flow cytometry method revealed pH-dependent fitness trade-offs between bile acids (a major pump substrate) and salicylic acid, a membrane-permeant aromatic acid that induces a drug resistance regulon but depletes proton motive force (PMF). The PMF drives MdtEF-TolC and related pumps such as AcrAB-TolC. Deletion of mdtE (with loss of the pump MdtEF-TolC) increased the strain’s relative fitness during growth with or without salicylate or bile acids. However, when the growth cycle included a 2-h incubation at pH 2 (below the pH growth range), MdtEF-TolC conferred a fitness advantage. The fitness advantage required bile salts but was decreased by the presence of salicylate, whose uptake is amplified by acid. For comparison, AcrAB-TolC, the primary efflux pump for bile acids, conferred a PMF-dependent fitness advantage with or without acid exposure in the growth cycle. A different MDR pump, EmrAB-TolC, conferred no selective benefit during growth in the presence of bile acids. Without bile acids, all three MDR pumps incurred a large fitness cost with salicylate when exposed at pH 2. These results are consistent with the increased uptake of salicylate at low pH. Overall, we showed that MdtEF-TolC is an MDR pump adapted for transient extreme-acid exposure and that low pH amplifies the salicylate-dependent fitness cost for drug pumps.
IMPORTANCE Antibiotics and other drugs that reach the gut must pass through stomach acid. However, little is known of how extreme acid modulates the effect of drugs on gut bacteria. We find that extreme-acid exposure leads to a fitness advantage for a multidrug pump that otherwise incurs a fitness cost. At the same time, extreme acid amplifies the effect of salicylate selection against multidrug pumps. Thus, organic acids and stomach acid could play important roles in regulating multidrug resistance in the gut microbiome. Our flow cytometry assay provides a way to measure the fitness effects of extreme-acid exposure to various membrane-soluble organic acids, including plant-derived nutrients and pharmaceutical agents. Therapeutic acids might be devised to control the prevalence of multidrug pumps in environmental and host-associated habitats.
KEYWORDS: AcrAB-TolC, Gad, MdtEF-TolC, bile acid, drug resistance evolution, extreme acid, flow cytometry, multidrug efflux pump, relative fitness, salicylate
INTRODUCTION
Bacterial multidrug resistance (MDR) efflux systems export diverse antibiotics, metals, and harmful products of metabolism (1–3). These MDR pumps also remove environmental or host-derived antimicrobials like bile acids (4) as well as toxic products of the bacterium’s own metabolism (5, 6). For pathogens, MDR pumps serve as a first-line defense against multiple antibiotics at low levels (7). Thus, MDR pumps in pathogens pose a major threat to human health (8–11). However, the metabolic and biochemical conditions in which they provide a fitness benefit are poorly understood.
The Escherichia coli K-12 genome contains genes for at least 36 multidrug efflux systems (5, 12). Only a few of these are understood in detail. The pump best known is AcrAB-TolC, a member of the resistance-nodulation-cell division (RND) superfamily (13). AcrAB-TolC exports bile acids (4, 14) as well as antimicrobial drugs, dyes, organic solvents, essential oils, and hormones (15, 16). AcrAB expression is upregulated by many proteins, including the global regulator MarA, which is activated by aspirin derivatives such as salicylate and benzoate (17, 18) (Fig. 1). The salicylate-induced Mar regulon intersects with the Gad acid fitness island, which includes mdtEF genes encoding components of MdtEF-TolC, an RND pump that is structurally similar to AcrAB-TolC (19, 20). MdtEF-TolC contributes to biofilm formation (21) and nitrate respiration (22). Other tripartite pumps similar to AcrAB-TolC include members of the major facilitator superfamily (MFS), such as EmrAB-TolC (23).
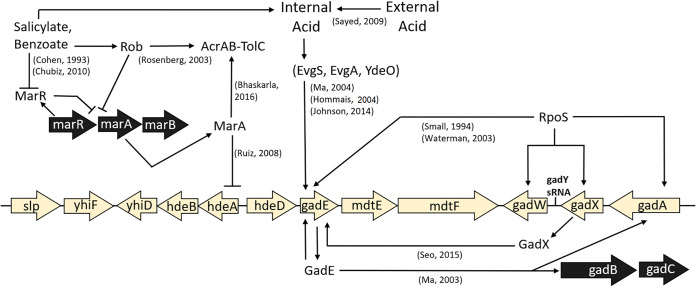
FIG 1.
The salicylate-inducible Mar drug resistance regulon intersects with the Gad acid resistance regulon. Selected components relevant to this work are shown. Cited references include 17, 31, 41, 49, 52, and 64–71. (Modified from reference 26.)
Clinical and environmental management of MDR-associated antibiotic resistance requires understanding the physiological trade-offs of such pumps, most of which spend substantial amounts of energy (12, 13). Our laboratory uses experimental evolution to explore conditions that could reverse the fitness benefit of drug efflux pumps and thus decrease their prevalence in microbial communities (24–26). Surprisingly, such conditions include the presence of salicylate or benzoate, which induce drug resistance regulons. In serial culture, these MDR inducers actually select against resistance to antibiotics, favoring mutants that have lost pumps such as MdtEF and EmrAB (24, 26, 27). Transcription profiles support a model in which genes responding to transient aromatic-acid stress actually decrease fitness over periods of chronic exposure (26).
Little is known regarding the effects of salicylates on the gut microbiome and their interactions with substances such as bile acids. The antimicrobial activity of bile acids plays an important role in structuring the gut microbiome (28–30), and bile acids induce AcrAB-TolC (31).
Most drug pumps power their efflux by spending proton motive force (PMF). For example, PMF drives pumps of the RND superfamily, such as AcrAB-TolC (15, 16), and the MFS superfamily pump EmrD (32). However, these pumps cannot function when the cell is exposed to PMF uncouplers, such as carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (12, 25). Full uncouplers penetrate the membrane in the protonated and unprotonated forms, whereas partial uncouplers cross mainly in the protonated form. Partial uncouplers include membrane-soluble aromatic acids, such as aspirin, salicylic acid, and ibuprofen, which belong to the class of nonsteroidal anti-inflammatory drugs (NSAIDs) (33, 34). Membrane-soluble acids and their derivatives are of interest for the human diet, as many are phytochemicals produced by plants, which control their microbiomes in ways that are poorly understood.
An important aspect of proton-driven drug pumps is their association with extreme-acid resistance (35). Extreme-acid resistance enables E. coli and other enteric bacteria to survive transient exposure at a range of pH 1 to 3, as found in the human stomach (36, 37). Moderate acid (pH 5.0 to 6.8) upregulates systems for extreme-acid survival such as those of the Gad acid fitness island (38, 39). The acid fitness island includes mdtEF, encoding the MdtEF-TolC pump (19, 20), and genes for Gad transcriptional regulators such as GadE (40). Gad-dependent acid resistance allows stationary-phase survival during exposure to extreme acid where E. coli cannot grow. The system raises intracellular pH by consuming protons through the decarboxylation of glutamate and glutamine by GadA and GadB, regulated by GadE. Despite the importance of this system, few studies of experimental evolution incorporate extreme-acid exposure. In one report, 20 cycles at pH 2.5 selected for mutations in the acid resistance regulator EvgS (41). Such an experiment, in effect, tests evolving mutants for relative death rates under an adverse condition.
We investigated the trade-offs between PMF-driven MDR pumps and the presence of salicylate and bile acids (24–26). We developed a method using flow cytometry (42) to measure the relative fitness contributions of MDR genes versus null alleles under various conditions, including the presence of uncouplers and of exportable substrates such as bile acids (cholic acid and deoxycholic acid). We modified the method to include cycles of extreme acid exposure. Using this approach, we showed that the MdtEF-TolC pump requires pH 2 exposure to confer positive relative fitness. This finding represents a novel case of an extreme-acid-dependent drug efflux pump. We also observed distinct patterns of relative fitness for the AcrAB-TolC and EmrAB-TolC efflux pumps.
RESULTS
Relative fitness measurement by flow cytometry of YFP and CFP.
We sought to measure the relative fitness contributions of MDR genes in the presence of various organic acids. For this purpose, we modified the flow cytometry assay of Gullberg et al. (42), which uses yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) markers (Fig. 2). In our assay, each strain had a gene of interest knocked out by replacement with kanR from alleles of the KEIO collection with the exception of the Δslp-gadX strain (43). The kanR gene constitutively expresses an aminoglycoside 3′-phosphotransferase (43, 44). In early competition trials, we found that the presence of kanR incurs a small fitness cost relative to the control strain, E. coli W3110. Therefore, in all assays involving knockout strains that expressed kanR, we used a control strain with the yhdN::kanR allele. For the single gene deletions used in this experiment (ΔacrA::kanR, ΔmdtE::kanR, ΔgadE::kanR, and ΔemrA::kanR), each allele was transduced into a strain of E. coli K-12 W3110 containing the fluorophore allele galK::yfp or galK::cfp, both of which are inducible via a lac promoter (42).
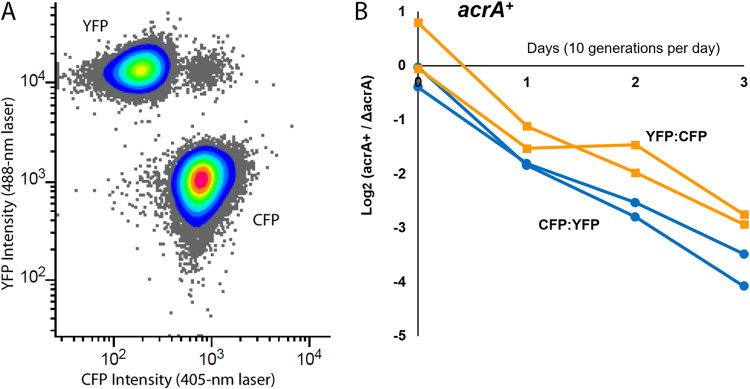
FIG 2.
Flow cytometry competition assays of strains expressing YFP or CFP. (A) Emission intensity indicates cells expressing cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) for two strains in coculture. The combined culture was diluted and incubated for 2 h with IPTG to induce fluorophore expression. (B) The slope of the log2 ratios for LBK-PIPES pH 6.8 control medium (n = 12) was compiled for day 0 through day 3 for all weeks tested. The absolute values of these ratios were taken from cell counts for ΔyhdN::kanR (acrA+)/ΔacrA::kanR strains. Each competition assay included an equal number of assays in which the ΔyhdN::kanR strain expressed YFP versus CFP. Cultures were diluted 1:1,000 daily and assayed by flow cytometry as described in Materials and Methods. Slopes were calculated over days 0 to 3 of testing. The flow cytometry threshold for percentage of each cell type was 0.01%.
The mixture of two strains was serially diluted 1,000-fold each day and observed over a total of 30 generations (doublings) from day 0 to day 3. This period was sufficient to permit accurate measurement of relative fitness but not long enough to make the rise of new mutations likely (42). All culture media were buffered at pH 6.8, a level that allows cytoplasmic pH depression by membrane-permeant acids of lower pKa. Each day, a parallel dilution with IPTG (isopropyl-β-d-thiogalactopyranoside) inducer was performed using LBK-PIPES pH 6.8 (see Materials and Methods) buffered medium without stressors. The IPTG-induced populations express YFP or CFP for flow cytometry, whereas the overnight 1,000-fold dilutions avoid energy-expensive fluorophore expression. This procedure enabled us to minimize the fitness effects of fluorophore expression during stress selection.
Figure 2A shows the appearance of a distinctive population of YFP-expressing cells showing high fluorescence intensity with 488-nm excitation and low-intensity fluorescence at 405 nm, versus a second population expressing CFP with low intensity at 488-nm excitation and high intensity at 405 nm. Figure 2B show results of a typical experiment in which the log2 ratios of YFP and CFP populations were reported for the tested mutant (W3110 acrA::kanR) and the control (W3110 yhdN::kanR). For each competition experiment, equal numbers of trials were performed with the gene knockout strain expressing YFP and the control strain expressing CFP, and vice versa.
Relative fitness was measured as the selection rate, defined as the daily change in log2 ratio of cocultured strain populations (45). A selection rate of 1 unit per day indicates a 2-fold increase in population of one strain compared to the competing strain’s population. This measure indicates the relative fitness of two strains cultured under a given stress condition.
The fitness benefit of MdtE requires extreme acid exposure (pH 2).
In a strain lacking AcrAB-TolC, overexpression of MdtEF-TolC confers resistance to antibiotics and bile acids (46). The range of substrates transported by an RND pump depends on its distal pocket, the periplasmic portion of its substrate-binding pocket (47). MdtE shares 55% amino acid sequence identity with AcrA (13), but its periplasmic substrate-binding pocket includes amino acid residues with a lower isoelectric point (pI 3.1 for MdtEF and pI 4.0 for AcrAB) (20). For this reason, we investigated roles for acids and acidic conditions in pump function.
We first tested the relative fitness contribution of mdtE by competition of an mdtE+ strain (W3110 ΔyhdN::kanR) against W3110 ΔmdtE::kanR in the presence of 6 mM salicylate and 0.15% bile acids (Fig. 3A). The mdtE deletion allele increased the relative fitness under all conditions—in the pH 6.8 control medium, as well as in 0.15% bile acids or in 6 mM salicylate (P < 0.001). Selection against the mdtE+ strain was further increased by the addition of 10 μM CCCP (P < 0.001), which depletes the PMF needed to drive efflux (Fig. 3A). The expenditure of limited PMF by the MdtEF-TolC could be one possible explanation for the pump’s fitness cost. All conditions tested led to a fitness cost or neutral selection for MdtEF-TolC.
FIG 3.
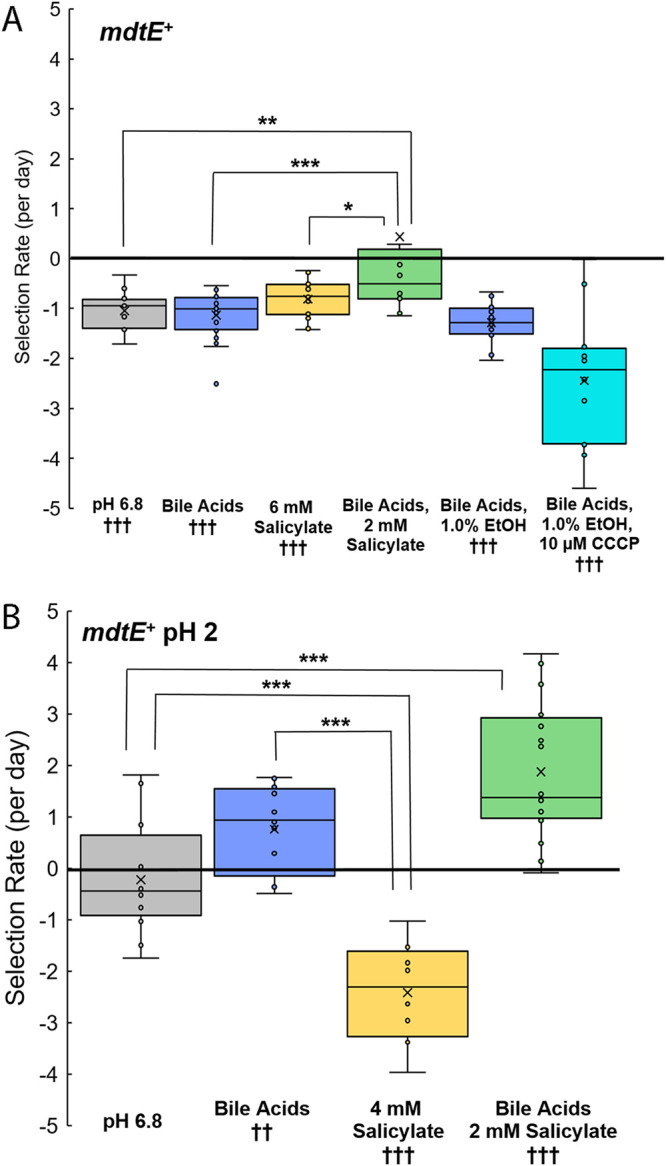
Selection for mdtE+ with or without pH 2 exposure. The selection rate is given by log2(ΔyhdN::kanR/ΔmdtE::kanR)/day. The slopes were calculated over days 0 to 3 of testing. (A) Conditions include LBK-PIPES pH 6.8 alone (n = 16), with 0.15% bile acids (n = 24), with 6 mM salicylate (n = 16), with 0.15% bile acids and 2 mM salicylate (n = 12), with 0.15% bile acids and 1.0% ethanol (n = 12), and with 0.15% bile acids, 1.0% ethanol, and 10 μM CCCP (n = 12). (B) Conditions include LBK-PIPES pH 6.8 alone (n = 16), with 0.15% bile acids (n = 16), with 4 mM salicylate (n = 8), and with 0.15% bile acids and 2 mM salicylate (n = 16). The daily growth cycle included 100-fold dilution in unbuffered LBK pH 2, with incubation for 2 h. Then the cultures were diluted 10-fold in the appropriate competition media adjusted to pH 7.0, yielding a final pH of 6.8. For each condition, ANOVA and post hoc Tukey tests were used to compare conditions to one another, shown with brackets (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Single-sample t tests were performed to compare each selection rate to a value of zero (††, P < 0.01; †††, P < 0.001). Statistical results are presented in File S1.
Under what growth conditions does MdtEF-TolC confer advantage? We tried including a period of pH 2 exposure, a condition for which the Gad regulon maintains extreme-acid survival dependent on stationary phase (35). This condition of stationary phase in extreme acid mimics the passage of bacteria through an acidic stomach, an adaptation that helps enteric E. coli gain access to the intestinal tract and may activate drug efflux pumps (19). Our serial cultures enter stationary phase for at least 12 h, so they are fully acid resistant before each dilution. Starting on day 0, the CFP/YFP strain mixture was diluted 100-fold in unbuffered LBK at pH 2.0. The acidified cell suspension, approximately pH 2, was incubated for 2 h. Under this condition, survival of E. coli K-12 strains is typically 50 to 100% (35, 48, 49). The acidified suspension was then diluted 10-fold in medium buffered with PIPES at pH 7.0, leading to a final pH of 6.8. The total dilution overall was 1,000-fold per day, comparable to our original relative fitness assay. This period of pH 2 exposure was repeated for each of days 1 to 3 of the assay.
The 2-h exposure to extreme acid eliminated the fitness cost of mdtE during coculture at pH 6.8. The mdtE allele conferred a fitness benefit in the presence of bile acids, with or without 2 mM salicylate (Fig. 3B). A higher concentration of 4 mM salicylate, however, incurred a large fitness cost for MdtE (P < 0.001). The highest salicylate concentration tested (6 mM) did not permit growth over 3 days. Note that after 100-fold dilution in LBK pH 2, there would still be a 0.06 mM concentration of salicylate. At external pH 2, where the E. coli internal pH is about 5 (49), the uptake of salicylic acid (with a pKa of 2.8) would be greatly increased by the transmembrane pH difference, a ΔpH of approximately 3 units. Thus, the low pH could amplify the fitness effect of a small salicylate concentration.
AcrA incurs fitness trade-offs.
AcrAB-TolC is the best known of the E. coli RND-type efflux pumps and is the most important for efflux of bile acids and other deleterious molecules (13). Nevertheless, we found that the acrA+ strain incurs a measurable fitness cost during culture at pH 6.8 (P < 0.01) (Fig. 4A). The fitness cost was seen with or without salicylate. The acrA+ allele conferred a fitness benefit only in the presence of bile acids (P < 0.001) (Fig. 4A). This result might show that the AcrAB-TolC efflux pump is worth the energy expenditure only when it is needed to export a deleterious substrate. The advantage conferred by bile acids was eliminated by CCCP, which would be consistent with pump dependence on PMF.
FIG 4.
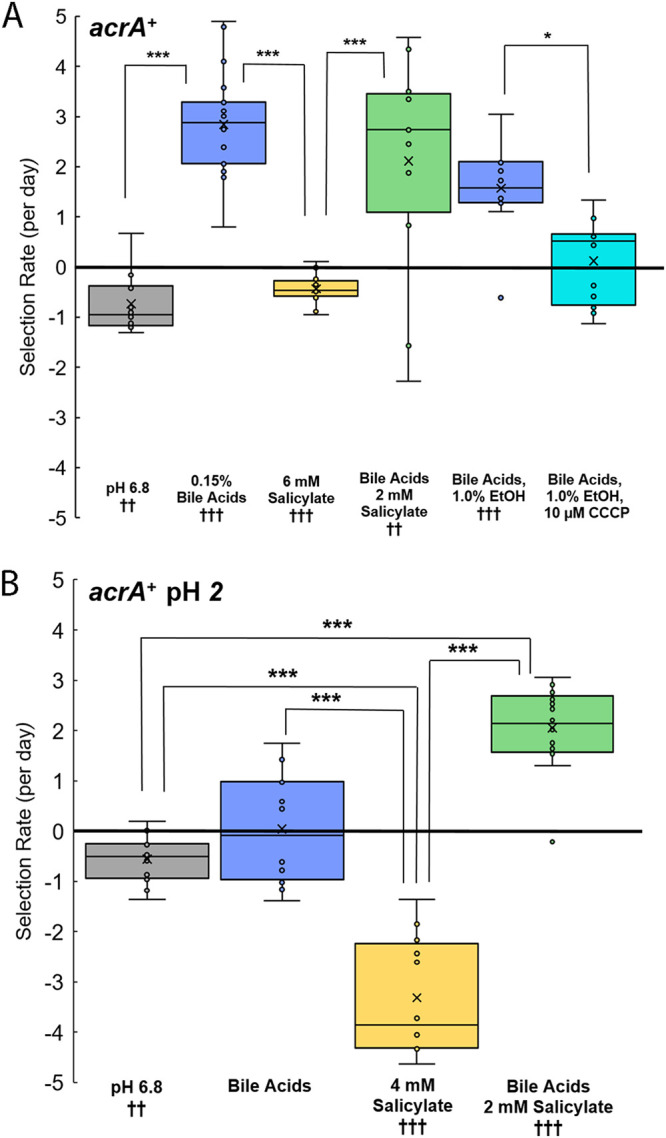
Selection for acrA+ in the presence of salicylate, bile acids, and CCCP. The selection rate is given by log2(ΔyhdN::kanR/ΔacrA::kanR)/day. The slopes were calculated over days 0 to 3 of testing. Significance was determined as for Fig. 3. (A) Conditions include LBK-PIPES pH 6.8 alone (n = 12), with 0.15% bile acids (n = 20), with 6 mM salicylate (n = 12), with 0.15% bile acids and 2 mM salicylate (n = 12), with 0.15% bile acids and 1.0% ethanol (n = 12), and with 0.15% bile acids, 1.0% ethanol (n = 12), and 10 μM CCCP (n = 12). (B) Conditions include LBK-PIPES pH 6.8 alone (n = 12), with 0.15% bile acids (n = 12), with 4 mM salicylate (n = 12), and with 0.15% bile acids and 2 mM salicylate (n = 16). The daily growth cycle included 100-fold dilution in unbuffered LBK pH 2, with incubation for 2 h. Then, the cultures were diluted 10-fold in the appropriate competition medium adjusted to pH 7.0, yielding a final pH of 6.8.
The daily growth cycle was adjusted to include a 2-h period in stationary phase at pH 2 (Fig. 4B). With this adjusted cycle, the acrA+ strain showed a small fitness cost (P < 0.01) and lost its fitness benefit in the presence of bile acids. A large fitness cost was incurred with 4 mM salicylate (P < 0.001), whereas the fitness advantage was restored with bile acids and a small amount of salicylate (P < 0.001) (Fig. 4B). The smaller amount of salicylate may tip the balance by inducing AcrAB expression during the pH 6.8 portion of the growth cycle (31). Thus, the bacteria’s ability to make AcrAB-TolC leads to very different trade-offs in the presence of bile acids and salicylate, dependent on pH 2 exposure.
EmrA carries a fitness cost.
The EmrAB-TolC efflux pump structurally resembles the AcrAB-TolC efflux pump, with EmrA providing a link between the outer membrane efflux pump component, TolC, and the inner membrane component, EmrB (50). The EmrAB-TolC efflux pump is activated by salicylate and confers resistance to some antibiotics as well as CCCP (23). As seen for mdtE, emrA incurred a fitness cost in the pH 6.8 medium under all conditions tested, with the exception of bile acids plus salicylate (Fig. 5A). The fitness cost was increased by the presence of CCCP (P < 0.01) (Fig. 5A). This is surprising, since the EmrAB-TolC efflux pump confers resistance to CCCP, but CCCP resistance may require emrA overexpression through mutations to the transcriptional repressor emrR (mprA) (25).
FIG 5.
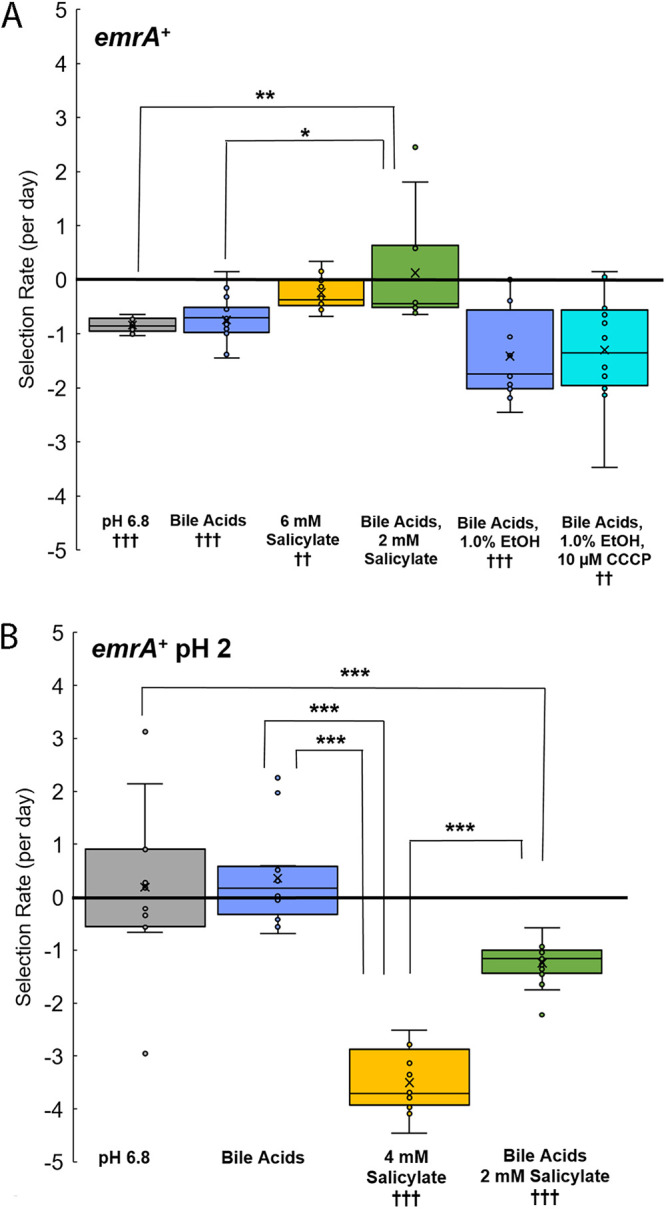
Selection for emrA+. Selection rate is given by log2(ΔyhdN::kanR/ΔemrA::kanR)/day. The slopes were calculated over days 0 to 3 of testing. Significance was determined as for Fig. 3. (A) Conditions include LBK-PIPES pH 6.8 alone (n = 16), with 0.15% bile acids (n = 16), with 6 mM salicylate (n = 16), 0.15% bile acids and 2 mM salicylate (n = 12), with 0.15% bile acids and 1.0% ethanol (n = 12), and with 0.15% bile acids, 1.0% ethanol, and 10 μM CCCP (n = 12). (B) Conditions include LBK-PIPES pH 6.8 alone (n = 12), with 0.15% bile acids (n = 12), with 4 mM salicylate (n = 16), and with 0.15% bile acids and 2 mM salicylate (n = 16). The daily growth cycle included 100-fold dilution in unbuffered LBK pH 2, with incubation for 2 h before a 10-fold dilution in the appropriate competition medium adjusted to pH 7.0, yielding a final pH of 6.8.
Inclusion of pH 2 exposure during the growth cycle did not increase relative fitness for emrA+ (Fig. 5B). The emrA+ strain was less fit in the presence of salicylate, with or without bile acids (P < 0.001). Unlike MdtEF-TolC or AcrAB-TolC, EmrAB-TolC did not show evidence of functional bile acid export under the conditions tested.
The Gad island shows fitness trade-offs with bile acids and salicylate.
In addition to the multidrug efflux pump components mdtEF (46), the Gad acid fitness island (Fig. 1) includes genes whose products counteract acid stress, such as the periplasmic acid chaperone genes hdeA and hdeB and the acid resistance regulator genes gadE and gadX (38). Nonetheless, this region of the genome often undergoes deletion during experimental evolution with acid stress (26, 51).
We investigated how the mdtE fitness effects compare to the fitness effects of the entire Gad island. Competition assays were conducted using a strain with most of the Gad island deleted by recombineering (Δslp-gadX) (26). In this assay, the parent strain W3110 had a small fitness advantage over the Δslp-gadX strain (Fig. 6A). This result differed from the mdtE+ competition (Fig. 3A) in which the mdtE deletion increased fitness. The advantage conferred by the Gad island was independent of the presence or absence of bile acids but was reversed by the presence of 6 mM sodium salicylate. Thus, the negative effect of salicylate overcomes whatever net fitness contribution is provided by components of the Gad island. The net effects on fitness are small compared to the fitness effects observed for individual knockout strains (Fig. 3 to 5).
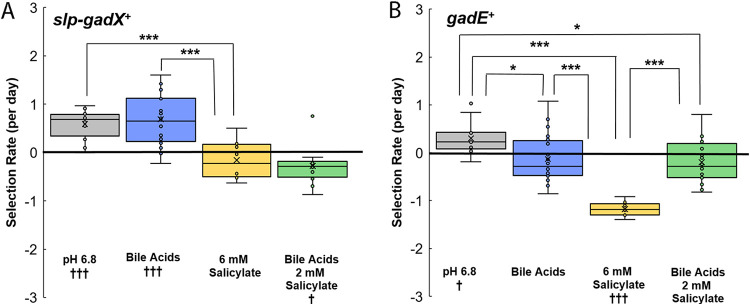
FIG 6.
Selection for Gad island and for gadE+. The slopes were calculated over days 0 to 3 of testing. Significance was determined as for Fig. 3 (†, P < 0.05). (A) slp-gadX+. Selection rate is given by log2(W3110/Δslp-gadX)/day. Conditions include LBK-PIPES pH 6.8 alone (n = 12), with 0.15% bile acids (n = 20), with 6 mM salicylate (n = 12), and with 0.15% bile acids and 6 mM salicylate (n = 12). (B) gadE+. Selection rate is given by log2(ΔyhdN::kanR/ΔgadE::kanR)/day. Conditions include LBK-PIPES pH 6.8 alone (n = 12), with 0.15% bile acids (n = 20), with 6 mM salicylate (n = 12), and with 0.15% bile acids and 6 mM salicylate (n = 16).
An important regulatory component of the glutamate-dependent acid resistance pathway is gadE (Fig. 1) (52, 53). GadE activates the decarboxylation of glutamate in the cell by the upregulation of gadA and gadB (48). We sought to determine if salicylate affects the fitness benefit of gadE+ similarly to that of the overall Gad island (slp-gadX). In pH 6.8 medium, the control strain had a fitness advantage over the ΔgadE::kanR strain, similar to the fitness advantage conferred by the Gad island as a whole (Fig. 6B). With bile acids, however, GadE conferred no selective benefit. The presence of 6 mM salicylate, in the absence of bile acids, incurred a fitness cost for gadE (P < 0.001) (Fig. 6B). These results suggest that some component regulated by GadE is sensitive to salicylate and that some unidentified trade-off exists with bile acids.
DISCUSSION
Previously, it was unknown why genes encoding the MdtEF pump reside in the Gad acid fitness island (Fig. 1) along with a complex set of regulators of acid resistance via glutamate decarboxylase and periplasmic chaperones (38). The MdtE fitness benefit with pH 2 exposure (Fig. 3) suggests that MdtEF-TolC exports bile acids during exposure to extreme acid, where the cell cannot grow but its death rate can be slowed by various acid resistance components of the Gad island. This would be consistent with the previously unexplained requirement of pump component TolC for Gad-related survival at pH 2 (54).
Our modified flow cytometry assay enables us to explore the fitness effects of extreme-acid exposure, which enteric pathogens as well as commensal strains must survive to reach the intestine (35, 48, 49). Our results may provide clues as to the relative fitness of drug-exporting strains during human uptake of aromatic acid medications such as aspirin. These efflux pumps consume PMF, and thus, their fitness contribution may be neutralized or reversed by the presence of the uncoupler CCCP (Fig. 3A, 4A, and 5A). We show that the efflux pump fitness contribution may also be reversed by a membrane-permeant aromatic acid such as salicylate during extreme-acid exposure (Fig. 3B, 4B, and 5B). The mechanisms of antimicrobial action of aromatic acids may involve depletion of PMF, as well as the ΔpH-amplified uptake of a molecule that disrupts the membrane (24, 26, 27, 55, 56).
Extreme-acid exposure had a surprising impact on the fitness effects of bile acids. Bile acids occur at highest concentration in the lumen of the small intestine, where they enhance lipid absorption (57–59). While bile does not normally reach the stomach, many patients exhibit chronic bile reflux gastritis (60), a condition associated with disorders such as carcinogenesis. Thus, pumps capable of extreme acid-dependent efflux of bile acids could be useful for bacteria that experience gastric transit.
The different MDR pumps we tested showed diverse effects of bile acids. AcrAB-TolC conferred a fitness benefit in the presence of bile acids under various conditions, whereas MdtEF-TolC conferred an advantage only when the growth cycle included exposure at pH 2. In contrast, EmrAB-TolC conferred no fitness advantage under any of our conditions tested. Thus, conditions favoring this pump’s action must involve substrates other than bile acids.
Although the intact mdtE gene decreased fitness under almost all conditions at pH 6.8, the overall Gad island (Fig. 6A) conferred a small selective benefit at pH 6.8. This advantage may derive from the net fitness contributions of Gad acid resistance genes other than mdtEF. The Gad regulon includes various components whose functions in acid resistance are poorly understood (26). The hdeAB acid stress operon encodes two periplasmic proteins that prevent protein aggregation at pH 2.0 (61). It may be more advantageous for the cell to lose one energetically expensive pump rather than its entire acid defense system. We will explore further the effect of acid exposure on fitness of other Gad components, as well as other MDR pumps.
Our interest in assessing fitness during acid exposure with salicylate has pharmaceutical implications. High levels of salicylate in the human stomach, whether through changes in diet or periodic exposure to drugs such as aspirin, can affect the relative survival of different bacteria that enter the stomach. Other pharmaceuticals likely interact with the microbiome in unexpected ways that influence clinical outcomes. Our relative fitness assay can reveal such interactions and explore their mechanisms.
MATERIALS AND METHODS
Strains and media.
All strains used in our experiments are derived from E. coli K-12 W3110 (Table 1). Strains were constructed by phage P1 transduction, with confirmation by PCR amplification across the sequence joint (54). The main growth medium was LBK broth (10 g/liter tryptone, 5 g/liter yeast extract, 7.45 g/liter potassium chloride) buffered at pH 6.8 with 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pKa= 6.80) using NaOH to adjust pH (26). This medium is designated LBK-PIPES pH 6.8. Sodium salicylate, bile acids (50/50 mixture of sodium cholate and sodium deoxycholate), and CCCP were all obtained from Millipore Sigma.
TABLE 1.
E. coli strains used in this study
| Strain | Genotype/description | Source |
|---|---|---|
| W3110 | E. coli K-12 F− λ− | Lab stock (72) |
| CH367 | Δlac::FRT ΔgalK::cfp-bla (AmpR) | 42 |
| CH372 | Δlac::FRT ΔgalK::yfp-bla (AmpR) | 42 |
| JLS1779 | W3110 ΔgalK::cfp-bla | This study |
| JLS1780 | W3110 ΔgalK::yfp-bla | This study |
| JLS1910 | W3110 ΔgalK::cfp-bla ΔyhdN::kanR | This study |
| JLS1911 | W3110 ΔgalK::yfp-bla ΔyhdN::kanR | This study |
| JLS1826 | W3110 ΔgalK::cfp-bla ΔacrA::kanR | This study |
| JLS1832 | W3110 ΔgalK::yfp-bla ΔacrA::kanR | This study |
| JLS1834 | W3110 ΔgalK::cfp-bla ΔmdtE::kanR | This study |
| JLS1835 | W3110 ΔgalK::yfp-bla ΔmdtE::kanR | This study |
| JLS1912 | W3110 ΔgalK::cfp-bla ΔemrA::kanR | This study |
| JLS1913 | W3110 ΔgalK::yfp-bla ΔemrA::kanR | This study |
| JLS1817 | W3110 ΔgalK::cfp-bla Δslp-gadX | This study |
| JLS1818 | W3110 ΔgalK::yfp-bla Δslp-gadX | This study |
| JLS1919 | W3110 ΔgalK::cfp-bla ΔgadE::kanR | This study |
| JLS1920 | W3110 ΔgalK::yfp-bla ΔgadE::kanR | This study |
Fluorescence-activated cell sorting (FACS) competition assays.
Relative fitness of cocultured strains was measured by flow cytometry of strains expressing yellow fluorescent protein (YFP) or cyan fluorescent protein (CFP) (42). Strains were cultured in 2 ml of LBK-PIPES pH 6.8 incubated in separate tubes at 37°C with rotation for 16 h. On the next day (day −1), 20 μl of each inoculated culture was pipetted into 2 ml of the appropriate competition medium for which the strains were competed and incubated for 24 h at 37°C with rotation. On day 0, for each experimental condition, 2 μl of a 1:1 mixture of the CFP and YFP strains to compete was added to 2 ml of the appropriate competition medium and incubated at 37°C with rotation for 24 h. From this culture, serial dilutions were repeated (2 μl into 2 ml) on days 1 and 2. Thus, each day led to a 1,000-fold dilution followed by approximately 10 doublings (generations), for a total of 30 doublings by day 3. During each daily cycle, the cells spent approximately 12 to 14 h in stationary phase (24, 26).
To include a period of pH 2 exposure, overnight cultures were inoculated in unbuffered LBK pH 2 for the first 2 h of incubation each day (starting in the morning, day 0 through day 2). At pH 2, the bacteria cannot grow but they remain viable via stationary-phase-induced Gad acid resistance (38, 40). Each day, 2 μl of the mixture of competing CFP and YFP strains was added to 200 μl of LBK pH 2 and incubated at 37°C for 2 h. After this incubation period, 1.8 ml of the competition medium buffered to pH 7 was added (restoring the pH to approximately 6.8), and the tubes were incubated for 24 h at 37°C.
For daily flow cytometry, a separate dilution of each CFP and YFP coculture was performed by adding 50 μl (1:40 dilution) of the 1:1 mixtures of labeled strains and 20 μl (1:100 dilution) of 100 mM IPTG to 2 ml of LBK-PIPES pH 6.8. This tube was incubated for 2 h at 37°C while rotating and then sampled using the BD FACSMelody cell sorter with a blue laser (488 nm) and violet laser (405 nm). A 545/20 filter was used for YFP emission and a 528/45 filter for CFP emission. Each competition mixture was diluted with 1× phosphate-buffered saline (PBS) to obtain 50,000 total events. The dilution was set such that the processed events were greater than 98% and the event rate was less than 10,000 events/second. The threshold value of counts for each tested strain in a coculture was set at 0.01%. Two technical replicates of each competition mixture were recorded and averaged. The percentages of cells with YFP or CFP fluorescence was recorded. This process was repeated each day for days 0 to 3 of testing. For each experimental condition, unless specified otherwise, 12 biological replicates were averaged, always with equal numbers of YFP-to-CFP and CFP-to-YFP competitions.
For each gene tested, the selection rate (s) was calculated as log2(Rt/R0)/t, where R represents the ratio of cell numbers for the control strain (W3110 ΔyhdN or W3110) to the knockout strain of interest and t represents time in days (with a daily dilution of 1:1,000, approximately 10 generations per day) (45, 62, 63). This rate gives a biological indication of the change in population distribution of cocultured genetic variants over time. For example, a selection rate of 1 unit per day (one doubling per 10 generations) means that each day, one of two cocultured strains increases its population advantage 2-fold over the competing strain.
Statistical analysis.
Analysis of variance (ANOVA) and post hoc Tukey tests were performed to compare trials under different conditions. Single sample t-tests were performed to compare the value of each selection rate to zero. Statistical results are presented in File S1 in the supplemental material.
ACKNOWLEDGMENTS
This work was supported by National Science Foundation awards MCB-1923077 and MRI-1725426.
We thank two anonymous reviewers for very helpful comments on the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Joan L. Slonczewski, Email: slonczewski@kenyon.edu.
Maia Kivisaar, University of Tartu.
REFERENCES
- 1.Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJV, Luisi BF. 2018. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16:523–539. 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 2.Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, DiDomenico B, Shaw KJ, Miller GH, Hare R, Shimer G. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother 45:1126–1136. 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tikhonova EB, Zgurskaya HI. 2004. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem 279:32116–32124. 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 4.Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518. 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teelucksingh T, Thompson LK, Cox G. 2020. The evolutionary conservation of Escherichia coli drug efflux pumps supports physiological functions. J Bacteriol 202:e00367-20. 10.1128/JB.00367-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosner JL, Martin RG. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol 191:5283–5292. 10.1128/JB.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolivos S, Cayron J, Dedieu A, Page A, Delolme F, Lesterlin C. 2019. Role of AcrAB-TolC multidrug efflux pump in drug-resistance acquisition by plasmid transfer. Science 364:778–782. 10.1126/science.aav6390. [DOI] [PubMed] [Google Scholar]
- 8.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 9.Malik B, Bhattacharyya S. 2019. Antibiotic drug-resistance as a complex system driven by socio-economic growth and antibiotic misuse. Sci Rep 9:15184. 10.1038/s41598-019-50846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spengler G, Kincses A, Gajdács M, Amaral L. 2017. New roads leading to old destinations: efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules 22:468. 10.3390/molecules22030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li XZ, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulsen IT, Brown MH, Skurray RA. 1996. Proton-dependent multidrug efflux systems. Microbiol Rev 60:575–608. 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anes J, McCusker MP, Fanning S, Martins M. 2015. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol 6:587. 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pos KM. 2009. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta 1794:782–793. 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515. 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Chen M, Yu Z, Bell JM, Wang H, Forrester I, Villarreal H, Jakana J, Du D, Luisi BF, Ludtke SJ, Wang Z. 2019. In situ structure and assembly of the multidrug efflux pump AcrAB-TolC. Nat Commun 10:4–9. 10.1038/s41467-019-10512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen SP, Levy SB, Foulds J, Rosner JL. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol 175:7856–7862. 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, Kunze C, Dunlop MJ. 2019. Salicylate increases fitness cost associated with MarA-mediated antibiotic resistance. Biophys J 117:563–571. 10.1016/j.bpj.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi A, Hirakawa H, Hirata T, Nishino K, Yamaguchi A. 2006. Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J Bacteriol 188:5693–5703. 10.1128/JB.00217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novoa D, Otakuye C-B. 2019. The anaerobic efflux pump MdtEF-TolC confers resistance to cationic biocides. bioRxiv 10.1101/570408. [DOI]
- 21.Matsumura K, Furukawa S, Ogihara H, Morinaga Y. 2011. Roles of multidrug efflux pumps on the biofilm formation of Escherichia coli K-12. Biocontrol Sci 16:69–72. 10.4265/bio.16.69. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Xiao M, Horiyama T, Zhang Y, Li X, Nishino K, Yan A. 2011. The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J Biol Chem 286:26576–26584. 10.1074/jbc.M111.243261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomovskaya O, Lewis K. 1992. emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A 89:8938–8942. 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Creamer KE, Ditmars FS, Basting PJ, Kunka KS, Hamdallah IN, Bush SP, Scott Z, He A, Penix SR, Gonzales AS, Eder EK, Camperchioli DW, Berndt A, Clark MW, Rouhier KA, Slonczewski JL. 2017. Benzoate- and salicylate-tolerant strains of Escherichia coli K-12 lose antibiotic resistance during laboratory evolution. Appl Environ Microbiol 83:e02736. 10.1128/AEM.02736-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffith JM, Basting PJ, Bischof KM, Wrona EP, Kunka KS, Tancredi AC, Moore JP, Hyman MRL, Slonczewski JL. 2018. Experimental evolution of Escherichia coli K-12 in the presence of proton motive force (PMF) uncoupler carbonyl cyanide m-chlorophenylhydrazone selects for mutations affecting PMF-driven drug efflux pumps. Appl Environ Microbiol 85:e02792-18. 10.1128/AEM.02792-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore JP, Li H, Engmann ML, Bischof KM, Kunka KS, Harris ME, Tancredi AC, Ditmars FS, Basting PJ, George NS, Bhagwat AA, Slonczewski JL. 2019. Inverted regulation of multidrug efflux pumps, acid resistance, and porins in benzoate-evolved Escherichia coli K-12. Appl Environ Microbiol 85:e00966-19. 10.1128/AEM.00966-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malla CF, Mireles NA, Ramírez AS, Poveda JB, Tavío MM. 2020. Aspirin, sodium benzoate and sodium salicylate reverse resistance to colistin in Enterobacteriaceae and Pseudomonas aeruginosa. J Antimicrob Chemother 75:3568–3575. 10.1093/jac/dkaa371. [DOI] [PubMed] [Google Scholar]
- 28.Martins A, Spengler G, Rodrigues L, Viveiros M, Ramos J, Martins M, Couto I, Fanning S, Pagès JM, Bolla JM, Molnar J, Amaral L. 2009. pH modulation of efflux pump activity of multi-drug resistant Escherichia coli: protection during its passage and eventual colonization of the colon. PLoS One 4:e6656. 10.1371/journal.pone.0006656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubert K, Olde Damink SWM, von Bergen M, Schaap FG. 2017. Interactions between bile salts, gut microbiota, and hepatic innate immunity. Immunol Rev 279:23–35. 10.1111/imr.12579. [DOI] [PubMed] [Google Scholar]
- 30.Kurdi P, Kawanishi K, Mizutani K, Yokota A. 2006. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol 188:1979–1986. 10.1128/JB.188.5.1979-1986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 48:1609–1619. 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 32.Lee J, Sands ZA, Biggin PC. 2016. A numbering system for MFS transporter proteins. Front Mol Biosci 3:21. 10.3389/fmolb.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardinale DA, Lilja M, Mandic M, Gustafsson T, Larsen FJ, Lundberg TR. 2017. Resistance training with co-ingestion of anti-inflammatory drugs attenuates mitochondrial function. Front Physiol 8:21. 10.3389/fmolb.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahmud T, Rafi SS, Scott DL, Wrigglesworth JM, Bjarnason I. 1996. Nonsteroidal antiinflammatory drugs and uncoupling of mitochondrial oxidative phosphorylation. Arthritis Rheum 39:1998–2003. 10.1002/art.1780391208. [DOI] [PubMed] [Google Scholar]
- 35.Foster JW. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2:898–907. 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 36.Dressman JB, Berardi RR, Dermentzoglou LC, Russell TL, Schmaltz SP, Barnett JL, Jarvenpaa KM. 1990. Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm Res 7:756–761. 10.1023/A:1015827908309. [DOI] [PubMed] [Google Scholar]
- 37.Beasley DE, Koltz AM, Lambert JE, Fierer N, Dunn RR. 2015. The evolution of stomach acidity and its relevance to the human microbiome. PLoS One 10:e0134116. 10.1371/journal.pone.0134116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mates AK, Sayed AK, Foster JW. 2007. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol 189:2759–2768. 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng Z, Shan Y, Pan Q, Gao X, Yan A. 2013. Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front Microbiol 4:e00194. 10.3389/fmicb.2013.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Z, Richard H, Tucker DL, Conway T, Foster JW. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J Bacteriol 184:7001–7012. 10.1128/JB.184.24.7001-7012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson MD, Bell J, Clarke K, Chandler R, Pathak P, Xia Y, Marshall RL, Weinstock GM, Loman NJ, Winn PJ, Lund PA. 2014. Characterization of mutations in the PAS domain of the EvgS sensor kinase selected by laboratory evolution for acid resistance in Escherichia coli. Mol Microbiol 93:911–927. 10.1111/mmi.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158. 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:e0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burghardt LT, Epstein B, Guhlin J, Nelson MS, Taylor MR, Young ND, Sadowsky MJ, Tiffin P. 2018. Select and resequence reveals relative fitness of bacteria in symbiotic and free-living environments. Proc Natl Acad Sci U S A 115:2425–2430. 10.1073/pnas.1714246115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishino K, Senda Y, Yamaguchi A. 2008. The AraC-family regulator GadX enhances multidrug resistance in Escherichia coli by activating expression of mdtEF multidrug efflux genes. J Infect Chemother 14:23–29. 10.1007/s10156-007-0575-Y. [DOI] [PubMed] [Google Scholar]
- 47.Vargiu AV, Nikaido H. 2012. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci U S A 109:20637–20642. 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richard H, Foster JW. 2004. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol 186:6032–6041. 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol 176:1729–1737. 10.1128/JB.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borges-Walmsley MI, Beauchamp J, Kelly SM, Jumel K, Candlish D, Harding SE, Price NC, Walmsley AR. 2003. Identification of oligomerization and drug-binding domains of the membrane fusion protein EmrA. J Biol Chem 278:12903–12912. 10.1074/jbc.M209457200. [DOI] [PubMed] [Google Scholar]
- 51.He A, Penix SR, Basting PJ, Griffith JM, Creamer KE, Camperchioli D, Clark MW, Gonzales AS, Erazo JSC, George NS, Bhagwat AA, Slonczewski JL. 2017. Acid evolution of Escherichia coli K-12 eliminates amino acid decarboxylases and reregulates catabolism. Appl Environ Microbiol 83:e00442-17. 10.1128/AEM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol 49:1309–1320. 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 53.Castanié-Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. 2010. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res 38:3546–3554. 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deininger KNW, Horikawa A, Kitko RD, Tatsumi R, Rosner JL, Wachi M, Slonczewski JL. 2011. A requirement of TolC and MDR efflux pumps for acid adaptation and GadAB induction in Escherichia coli. PLoS One 6:e18960. 10.1371/journal.pone.0018960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sundaramoorthy NS, Suresh P, Selva Ganesan S, GaneshPrasad AK, Nagarajan S. 2019. Restoring colistin sensitivity in colistin-resistant E. coli: combinatorial use of MarR inhibitor with efflux pump inhibitor. Sci Rep 9:19845. 10.1038/s41598-019-56325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosner JL, Chai TJ, Foulds J. 1991. Regulation of OmpF porin expression by salicylate in Escherichia coli. J Bacteriol 173:5631–5638. 10.1128/jb.173.18.5631-5638.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hofmann AF. 1999. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med 159:2647–2658. 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- 58.Hamilton JP, Xie G, Raufman JP, Hogan S, Griffin TL, Packard CA, Chatfield DA, Hagey LR, Steinbach JH, Hofmann AF. 2007. Human cecal bile acids: concentration and spectrum. Am J Physiol Gastrointest Liver Physiol 293:256–263. 10.1152/ajpgi.00027.2007. [DOI] [PubMed] [Google Scholar]
- 59.Hegyi P, Maléth J, Walters JR, Hofmann AF, Keely SJ. 2018. Guts and gall: bile acids in regulation of intestinal epithelial function in health and disease. Physiol Rev 98:1983–2023. 10.1152/physrev.00054.2017. [DOI] [PubMed] [Google Scholar]
- 60.Vere CC, Cazacu S, Comănescu V, Mogoantă L, Rogoveanu I, Ciurea T. 2005. Endoscopical and histological features in bile reflux gastritis. Rom J Morphol Embryol 46:269–274. [PubMed] [Google Scholar]
- 61.Kern R, Malki A, Abdallah J, Tagourti J, Richarme G. 2007. Escherichia coli HdeB is an acid stress chaperone. J Bacteriol 189:603–610. 10.1128/JB.01522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dykhuizen DE. 1990. Experimental studies of natural selection in bacteria. Annu Rev Ecol Syst 21:373–398. 10.1146/annurev.es.21.110190.002105. [DOI] [Google Scholar]
- 63.Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138:1315–1341. 10.1086/285289. [DOI] [Google Scholar]
- 64.Ma Z, Masuda N, Foster JW. 2004. Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J Bacteriol 186:7378–7389. 10.1128/JB.186.21.7378-7389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seo SW, Kim D, O'Brien EJ, Szubin R, Palsson BO. 2015. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat Commun 6:7970. 10.1038/ncomms8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waterman SR, Small PLC. 2003. Transcriptional expression of Escherichia coli glutamate-dependent acid resistance genes gadA and gadBC in an hns rpoS mutant. J Bacteriol 185:4644–4647. 10.1128/JB.185.15.4644-4647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chubiz LM, Rao CV. 2010. Aromatic acid metabolites of Escherichia coli K-12 can induce the marRAB operon. J Bacteriol 192:4786–4789. 10.1128/JB.00371-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhaskarla C, Das M, Verma T, Kumar A, Mahadevan S, Nandi D. 2016. Roles of Lon protease and its substrate MarA during sodium salicylate-mediated growth reduction and antibiotic resistance in Escherichia coli. Microbiology (Reading) 162:764–776. 10.1099/mic.0.000271. [DOI] [PubMed] [Google Scholar]
- 69.Ruiz C, McMurry LM, Levy SB. 2008. Role of the multidrug resistance regulator MarA in global regulation of the hdeAB acid resistance operon in Escherichia coli. J Bacteriol 190:1290–1297. 10.1128/JB.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sayed AK, Foster JW. 2009. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol Microbiol 71:1435–1450. 10.1111/j.1365-2958.2009.06614.x. [DOI] [PubMed] [Google Scholar]
- 71.Hommais F, Krin E, Coppée JY, Lacroix C, Yeramian E, Danchin A, Bertin P. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology (Reading) 150:61–72. 10.1099/mic.0.26659-0. [DOI] [PubMed] [Google Scholar]
- 72.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:2006.0007. 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data for Fig. 3 to 6. Download AEM.00724-21-s0001.xlsx, XLSX file, 0.04 MB (43.4KB, xlsx)