ABSTRACT
Enterococcus faecalis, a member of the commensal flora in the human gastrointestinal tract, has become a threatening nosocomial pathogen because it has developed resistance to many known antibiotics. More concerningly, resistance gene-carrying E. faecalis cells may transfer antibiotic resistance to resistance-free E. faecalis cells through their unique quorum sensing-mediated plasmid transfer system. Therefore, we investigated the role of probiotic bacteria in the transfer frequency of the antibiotic resistance plasmid pCF10 in E. faecalis populations to mitigate the spread of antibiotic resistance. Bacillus subtilis subsp. natto is a probiotic strain isolated from Japanese fermented soybean foods, and its culture fluid potently inhibited pCF10 transfer by suppressing peptide pheromone activity from chromosomally encoded CF10 (cCF10) without inhibiting E. faecalis growth. The inhibitory effect was attributed to at least one 30- to 50-kDa extracellular protease present in B. subtilis subsp. natto. Nattokinase of B. subtilis subsp. natto was involved in the inhibition of pCF10 transfer and cleaved cCF10 (LVTLVFV) into LVTL plus VFV fragments. Moreover, the cleavage product LVTL (L peptide) interfered with the conjugative transfer of pCF10. In addition to cCF10, faecalis-cAM373 and gordonii-cAM373, which are mating inducers of vancomycin-resistant E. faecalis, were also cleaved by nattokinase, indicating that B. subtilis subsp. natto can likely interfere with vancomycin resistance transfer in E. faecalis. Our work shows the feasibility of applying fermentation products of B. subtilis subsp. natto and L peptide to mitigate E. faecalis antibiotic resistance transfer.
IMPORTANCE Enterococcus faecalis is considered a leading cause of hospital-acquired infections. Treatment of these infections has become a major challenge for clinicians because some E. faecalis strains are resistant to multiple clinically used antibiotics. Moreover, antibiotic resistance genes can undergo efficient intra- and interspecies transfer via E. faecalis peptide pheromone-mediated plasmid transfer systems. Therefore, this study provided the first experimental demonstration that probiotics are a feasible approach for interfering with conjugative plasmid transfer between E. faecalis strains to stop the transfer of antibiotic resistance. We found that the extracellular protease(s) of Bacillus subtilis subsp. natto cleaved peptide pheromones without affecting the growth of E. faecalis, thereby reducing the frequency of conjugative plasmid transfer. In addition, a specific cleaved pheromone fragment interfered with conjugative plasmid transfer. These findings provide a potential probiotic-based method for interfering with the transfer of antibiotic resistance between E. faecalis strains.
KEYWORDS: Enterococcus faecalis, antibiotic resistance, pheromone-inducible conjugative plasmid transfer, probiotics, Bacillus subtilis subsp. natto, nattokinase
INTRODUCTION
Enterococcus faecalis is a hardy Gram-positive bacterium commonly found in the gastrointestinal tracts of humans and other mammals (1, 2). Most E. faecalis strains are considered harmless, and some can even be used as probiotics in feed additives to prevent diarrhea or to improve growth in animals (3, 4). However, this bacterium is also an opportunistic pathogen causing nosocomial bacteremia, surgical wound infection, endocarditis, and urinary tract infection (5–7). In recent decades, E. faecalis has gradually become a leading cause of health care-associated infections because it has developed resistance to multiple clinically used antibiotics, such as macrolides, tetracyclines, aminoglycosides, and glycopeptides, including vancomycin, which was previously used as the antibiotic of last resort for enterococcal infections (8–10). The emergence of vancomycin resistance has made the treatment of infections with E. faecalis a major challenge for clinicians, because it means that few or no treatment options are available (10, 11). Currently, in the United States, vancomycin-resistant enterococci (VRE) are the fourth most common pathogens causing death from antibiotic-resistant infection (11). It is estimated that more than 20,000 infections with VRE occur and that more than 1,300 patients die from these infections each year (11).
Even more concerningly, antibiotic resistance genes can undergo both intra- and interspecies transfer through pheromone-inducible conjugative plasmid transfer systems of E. faecalis (12). For example, transfer of pCF10 between E. faecalis bacteria is controlled by two counteracting peptide pheromones, where chromosomally encoded cCF10 secreted by pCF10-free E. faecalis (recipients) serves as a mating inducer to trigger conjugative plasmid transfer, and pCF10-encoded iCF10 acts as an inhibitor that prevents self-induction of pCF10-carrying E. faecalis (donors) (13, 14). As pCF10 is transferred, antibiotic resistance genes, such as the tetracycline resistance gene tet(M), may be carried and horizontally transferred to recipients concurrently (15). Such a system facilitates the dissemination of antibiotic resistance genes, which leads to ineffectiveness of antibiotics and considerable loss of life (16–18).
Discovering and developing new antibiotics is one way to effectively control and prevent the threat of such highly antibiotic-resistant E. faecalis strains. One drawback to this approach, however, is the unavoidable possibility of a new selection pressure favoring genetic variants with resistance to the new antibiotics, thus rendering the new antibiotics ineffective over time (19). Perhaps a more viable solution is to interfere with the pheromone-inducible conjugative plasmid transfer systems of E. faecalis. In addition to being involved in the dissemination of antibiotic resistance genes, these systems are also involved in the formation of aggregation substances that increase the adherence of E. faecalis to host tissues and enhance their ability for biofilm formation (14). By repressing pheromone-inducible conjugative plasmid transfer systems, we may simultaneously attenuate the fitness, virulence, and antibiotic resistance transfer of E. faecalis.
In recent decades, an increasing number of researchers have focused on preventing and treating gastrointestinal disorders or infections with probiotics, which are live microorganisms such as bacteria and yeast that could provide benefits to the host when administered in adequate amounts (20–22). Because of the long consumption history and assured safety of probiotics, probiotic-based therapies have been proposed as alternate, safe, and cost-effective treatments compared to ongoing therapeutic regimens (23). Studies have also revealed that many strains of probiotics play a key role in preventing the colonization and overgrowth of pathogens in the human intestine via actions such as competing for space or nutrients with pathogens, modulating the immune system of the host, or enhancing the intestinal epithelial barrier (24–26). Collectively, this evidence indicates that probiotic treatment may be a feasible strategy for combating E. faecalis.
Therefore, in the present study, six probiotics were selected and their effects on the transfer frequency of the tetracycline resistance plasmid pCF10 between E. faecalis bacteria were investigated. Plasmid pCF10 was selected because it has been used as a model system for analyzing pheromone-inducible conjugative plasmid transfer in enterococci for decades (27). Here, we conducted a modified E. faecalis conjugation assay in combination with treatment with probiotic supernatants to screen for potent pCF10 transfer-inhibiting probiotic strains. We further characterized the pCF10 transfer-inhibiting substances in the supernatants and attempted to clarify the mechanisms by which these substances affect the conjugative transfer of pCF10. Via this study, we aimed to identify promising probiotics and fermentation products that could be used clinically to combat E. faecalis by interfering with pheromone-inducible conjugative plasmid transfer to save the lives of people infected with multidrug-resistant E. faecalis.
RESULTS
Effect of probiotic spent culture supernatants on pCF10 transfer.
Each probiotic spent culture supernatant (SCS) was added to the mating culture of E. faecalis to examine its effect on the frequency of pCF10 transfer (as evaluated by the transconjugants per donor [T/D] ratio, an index for assessing bacterial conjugation efficiency). As shown in Fig. S1 and S2 in the supplemental material, the frequency of pCF10 transfer was markedly reduced by treatment with some probiotic supernatants. In addition to inhibiting pCF10 transfer, some supernatants also strongly inhibited the growth of E. faecalis donors and recipients. Even though the SCS of aerobically cultured Bacillus subtilis subsp. natto did not inhibit the growth of donors or recipients, both the concentration of transconjugants and the T/D ratio decreased, and the T/D ratio in this culture was 100-fold lower than that in the negative-control group to which no SCS was added. Based on its inhibitory effect on pCF10 transfer, the SCSs of aerobically cultured B. subtilis subsp. natto and aerobically cultured Bacillus coagulans, Lactobacillus reuteri, and Lactobacillus rhamnosus were selected for further investigation.
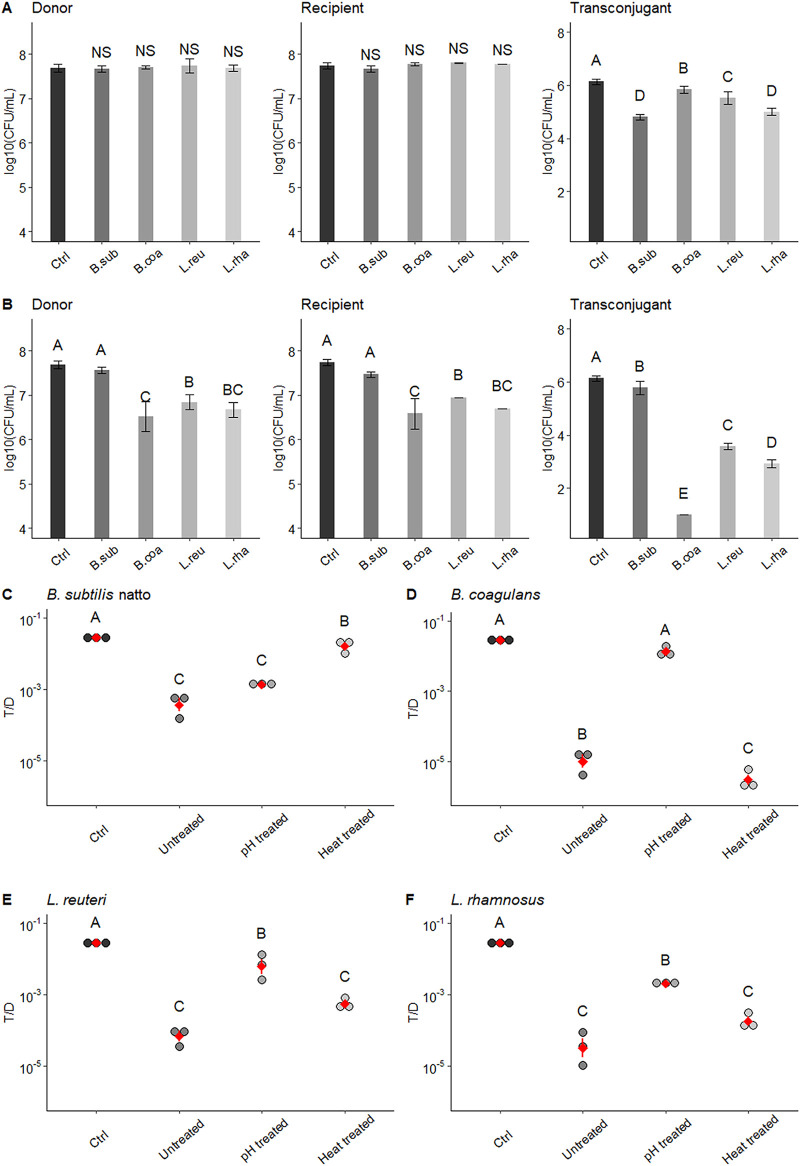
At the end of the culture period, the SCSs of these four probiotic bacteria had very different pH values (aerobically cultured B. subtilis subsp. natto, pH 6.19 ± 0.03; aerobically cultured B. coagulans, pH 4.68 ± 0.02; L. reuteri, pH 4.94 ± 0.04; and L. rhamnosus, pH 4.69 ± 0.05). To eliminate the possibility that the observed effect on the T/D ratio was caused by different pH values, the pH of these SCS was adjusted to an initial value of 6.5 with 1 N NaOH. After pH adjustment, the SCSs of aerobically cultured B. coagulans, L. reuteri, and L. rhamnosus lost their inhibitory effects on growth and pCF10 transfer. The concentrations of donors and recipients were not significantly different from those in the negative-control group to which no SCS was added (P > 0.05) (Fig. 1A). The T/D ratios were significantly increased compared with those in cultures exposed to untreated SCSs (P < 0.05) (Fig. 1D to F). However, the SCS of aerobically cultured B. subtilis subsp. natto lost its inhibitory effect on pCF10 transfer after heat treatment (80°C water bath for 30 min) but not after pH adjustment. As shown in Fig. 1B and C, both the concentration of transconjugants and the T/D ratio were markedly increased by the heat treatment. Thus, the pCF10 transfer-inhibiting substance(s) in the SCS of aerobically cultured B. subtilis subsp. natto is likely heat labile.
FIG 1.
Effect of pH-treated or heat-treated probiotic spent culture supernatants on growth and pCF10 transfer of E. faecalis. Spent culture supernatants (SCS) of aerobically cultured B. subtilis subsp. natto, B. coagulans, L. reuteri, and L. rhamnosus were first treated with pH adjustment (pH adjusted to 6.5 with NaOH) or heat (80°C water bath for 30 min) and were then added to the E. faecalis mating culture. After a 4-h mating assay, the concentrations of donors, recipients, and transconjugants were determined by plate counting. (A) Effect of pH-adjusted probiotic SCSs. (B) Effect of heat-treated probiotic SCSs. In panels A and B, the concentrations of donors, recipients, and transconjugants are presented as log10(CFU/ml) values. The data are presented as the means ± standard deviations (SDs) (n = 3). (C to F) Effects of untreated, pH-treated, or heat-treated probiotic SCSs on the number of transconjugants per donor (T/D ratios). Each dot in the figure represents a replication. The red diamonds with red lines indicate the means ± SDs (n = 3). Multiple comparisons were evaluated using Duncan’s multiple-range test. The different uppercase letters above the bars or dots indicate the significance of differences between groups (P < 0.05). Ctrl, negative control (the group treated with fresh M9B medium; no SCS treatment); B. sub, B. subtilis subsp. natto; B. coa, B. coagulans; L. reu, L. reuteri; L. rha, L. rhamnosus; NS, no significant difference.
At least one 30- to 50-kDa protein in B. subtilis subsp. natto supernatant inhibits pCF10 transfer.
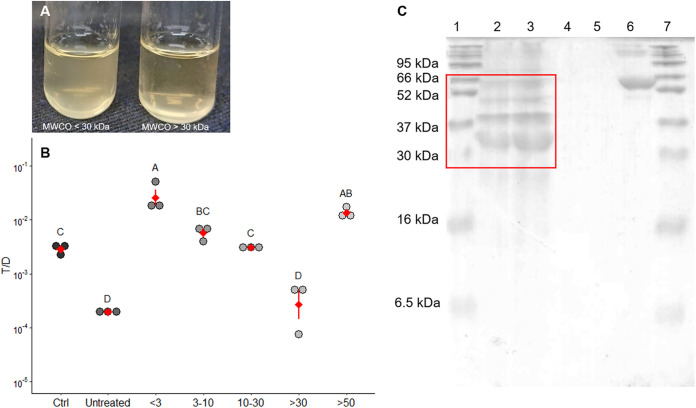
Since the factor that reduced the transfer of the conjugative plasmid pCF10 was heat labile, it was likely a protein molecule. The supernatant was fractionated using a series of membranes with molecular weight cutoffs (MWCOs) of 3, 5, 30, and 50 kDa. Fractions with different molecular weight (MW) ranges were then added to the E. faecalis mating culture. Only the fraction with a MW of greater than 30 kDa exerted an inhibitory effect on pCF10 transfer in E. faecalis (Fig. 2A and B). The T/D ratio was significantly lower (P < 0.05) than that in the negative-control group to which no SCS was added but similar to that in the group treated with unfractionated SCS. Moreover, the fraction with a MW of greater than 50 kDa clearly showed no inhibitory effect on pCF10 transfer in E. faecalis. Hence, the MW of the pCF10 transfer-inhibiting substances was likely greater than 30 kDa and less than 50 kDa. The total protein concentration in the SCS of B. subtilis subsp. natto was determined to be 162.0 ± 30 μg/ml, while that in fresh M9B medium (background) was 25.8 ± 1.8 μg/ml. Furthermore, several bands with MWs ranging from approximately 30 to 66 kDa were observed on Coomassie brilliant blue-stained gels (Fig. 2C). These results were consistent with the estimated MW of the pCF10 transfer-inhibiting proteins.
FIG 2.
At least one 30- to 50-kDa protein in B. subtilis subsp. natto supernatant inhibits pCF10 transfer. (A) Mating culture of E. faecalis treated with different MW fractions of B. subtilis subsp. natto supernatant. In this picture, the turbidity of the culture on the left, which was treated with the fraction with a MW less than 30 kDa appears higher than that of the culture on the right, which was treated with the fraction with a MW greater than 30 kDa. (B) Effect of different MW fractions of B. subtilis subsp. natto supernatant on the conjugative transfer of pCF10. The y axis indicates the T/D ratio, and the x axis indicates different MW (kDa) fractions of B. subtilis subsp. natto supernatant. Ctrl, negative control (the group treated with fresh M9B medium; no SCS treatment); untreated, the group treated with unfractionated B. subtilis subsp. natto supernatant. Each dot in the figure represents a replication. The red diamonds with red lines indicate the means ± SDs (n = 3). Multiple comparisons were evaluated using Duncan’s multiple-range test. The different uppercase letters above the dots indicate the significance of differences between groups (P < 0.05). (C) SDS-PAGE of B. subtilis subsp. natto supernatant. Lanes 1 and 7, protein marker; lane 2, B. subtilis subsp. natto supernatant (5 μl); lane 3, B. subtilis subsp. natto supernatant (10 μl); lane 4, M9B medium (5 μl); lane 5, M9B medium (10 μl); lane 6, BSA.
Inhibitory effect of B. subtilis subsp. natto supernatant on the conjugative transfer of pCF10.
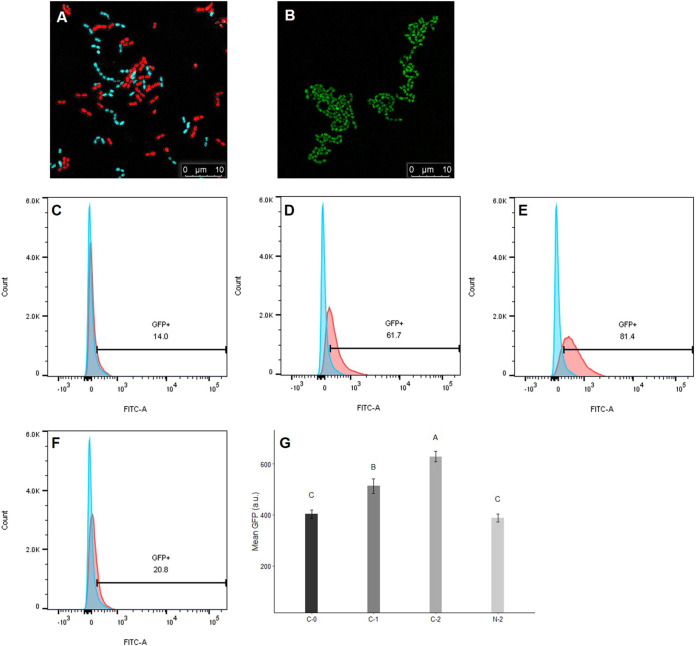
Under a confocal microscope, E. faecalis OG1RF::p23cfp (recipient), which constitutively expresses the cyan fluorescent protein (CFP) reporter, appears cyan, and E. faecalis OG1Sp/pCF10-iGFP-p23-tdTomato (donor), which constitutively expresses the tdTomato reporter, appears red (Fig. 3A). Upon induction with pheromone cCF10, donor cells expressed the green fluorescent protein (GFP) reporter, and green aggregates were observed (Fig. 3B). Here, the cCF10-inducible GFP reporter on pCF10 was used to evaluate the induction status of donor cells. Donor cells were incubated with 2.5 ng/ml cCF10 (more closely resembling the levels of cCF10 naturally produced by recipients [28]) for 120 min, and the expression of the GFP reporter was quantified using flow cytometry. As the induction time increased (0 to 120 min), the percentage of GFP-positive donor cells gradually increased from 14% to 81.0% (Fig. 3C to E). In addition, the mean GFP intensity increased over time (Fig. 3G). However, in the donor cell culture treated with the SCS of B. subtilis subsp. natto, both the percentage of GFP-positive donor cells and the mean GFP intensity were markedly decreased (Fig. 3F and G). The mean GFP intensity was not significantly different (P > 0.05) from that in the negative-control group (induced with 0 ng/ml cCF10 for 0 min) (Fig. 3G). Therefore, the SCS of B. subtilis subsp. natto can likely interfere with the ability of donor cells to sense cCF10 or can even affect the activity of the pheromone, thereby repressing the expression of the cCF10-inducible GFP reporter on pCF10.
FIG 3.
Influence of B. subtilis subsp. natto supernatant on the cCF10-inducible GFP reporter harbored on pCF10. (A) Under confocal microscopy, E. faecalis OG1Sp/pCF10-iGFP-p23-tdTomato (donor) appears red, and E. faecalis OG1RF::p23cfp (recipient) appears cyan. In the middle of the image, donor and recipient cells were close together, indicating that mating was likely happening. (B) When donor cells were induced with cCF10, they expressed the GFP reporter and exhibited green aggregates. (C to F) Donor cells were induced with 2.5 ng/ml cCF10 and harvested at 0, 60, and 120 min. Next, the expression of the GFP reporter was quantified using flow cytometry. Blue peak, wild-type E. faecalis OG1RF that would not express the GFP reporter, used as the negative control. Red peaks, donor cells induced with 0 ng/ml cCF10 for 0 min (C), 2.5 ng/ml cCF10 for 60 min (D), 2.5 ng/ml cCF10 for 120 min (E), or 2.5 ng/ml cCF10 plus B. subtilis subsp. natto SCS for 120 min (F). The numbers shown in the middle of panels C to F are the percentages of GFP-positive cells after gating. (G) Mean GFP intensity of GFP-positive donor cells after gating. Donor cells were induced with 0 ng/ml cCF10 for 0 min (C-0), 2.5 ng/ml cCF10 for 60 min (C-1), 2.5 ng/ml cCF10 for 120 min (C-2), or 2.5 ng/ml cCF10 plus B. subtilis subsp. natto SCS for 120 min (N-2). The data are presented as the means ± SDs (n = 3). Values with different uppercase letters were significantly different according to Duncan’s multiple-range tests (P < 0.05).
B. subtilis subsp. natto supernatant affects the activity of cCF10.
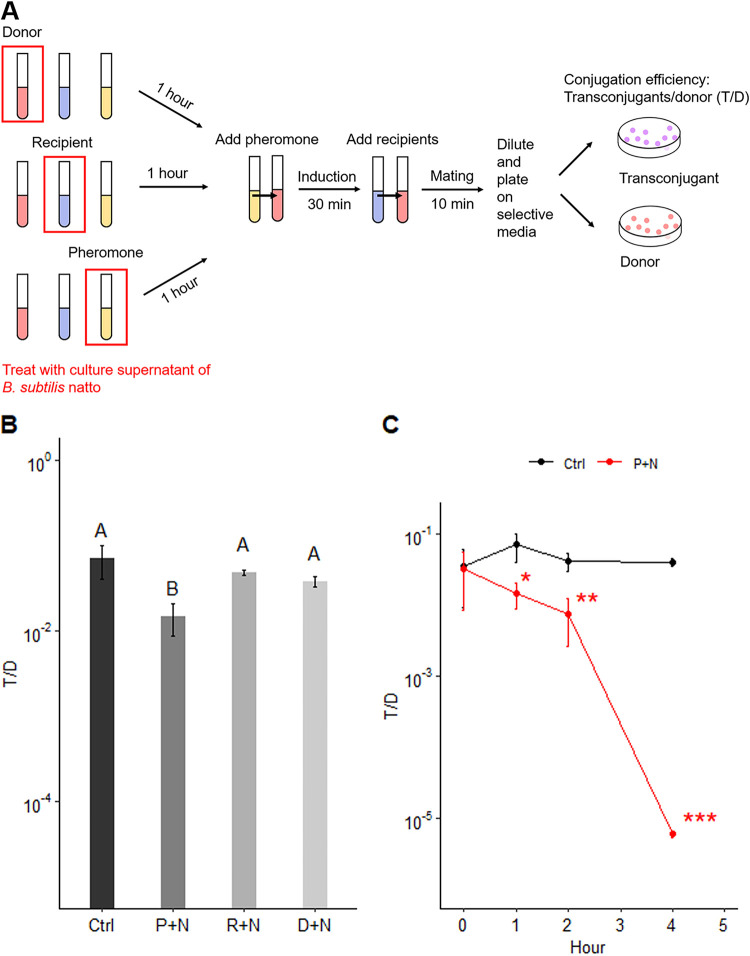
To clarify the factors affected by the SCS of B. subtilis subsp. natto, the supernatant was first treated separately with donor cells, recipient cells, or pheromone cCF10 at 37°C for 1 h. Next, donor cells were incubated with cCF10 at 37°C for 30 min. Then, recipient cells were added and incubated for 10 min to allow one round of conjugation before the entire culture was plated on selective medium for transconjugants and donors (Fig. 4A). The T/D ratio was only significantly decreased when cCF10 was treated with the SCS of B. subtilis subsp. natto compared to that in the negative-control group to which no SCS was added (P < 0.05) (Fig. 4B). Furthermore, the effect of the SCS of B. subtilis subsp. natto on cCF10 increased over time. At each tested time point (1, 2, and 4 h), the T/D ratio was significantly different from that in the negative-control group (1 h, P < 0.05; 2 h, P < 0.01; 4 h, P < 0.001) (Fig. 4C). Thus, the SCS of B. subtilis subsp. natto likely interferes with the conjugative transfer of pCF10 by affecting the activity of cCF10.
FIG 4.
B. subtilis subsp. natto supernatant affects the activity of cCF10. (A) Modified E. faecalis mating experiment. The SCS of B. subtilis subsp. natto was first treated separately with donor cells, recipient cells, or pheromone cCF10 at 37°C for 1 h. Next, donor cells were incubated with cCF10 at 37°C for 30 min and then mixed with recipient cells for one round of conjugation (10 min). Through this method, we were able to clarify the factor that is actually affected by the SCS of B. subtilis subsp. natto. (B) T/D ratios after separate treatment of pheromone cCF10, recipient cells, or donor cells with B. subtilis subsp. natto SCS. The y axis indicates the T/D ratio. Ctrl (control), not treated with SCS, P+N: pheromone cCF10 treated with B. subtilis subsp. natto SCS; R+N, recipient cells treated with B. subtilis subsp. natto SCS; D+N, donor cells treated with B. subtilis subsp. natto SCS. The data are presented as the means ± SDs (n = 3). Values with different uppercase letters were significantly different according to Duncan’s multiple-range tests (P < 0.05). (C) The effect of B. subtilis subsp. natto SCS on cCF10 increased over time. Donor cells were induced with cCF10 that was first treated with B. subtilis subsp. natto SCS for 0, 1, 2, and 4 h. Then, recipient cells were added and allowed one round of conjugation. The y axis indicates the T/D ratio. Ctrl (control), not treated with SCS; P+N, pheromone cCF10 treated with B. subtilis subsp. natto SCS. The data are presented as the means ± SDs (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with each negative control group (Student’s t test).
An extracellular protease of B. subtilis subsp. natto cleaves cCF10 and inhibits pCF10 transfer.
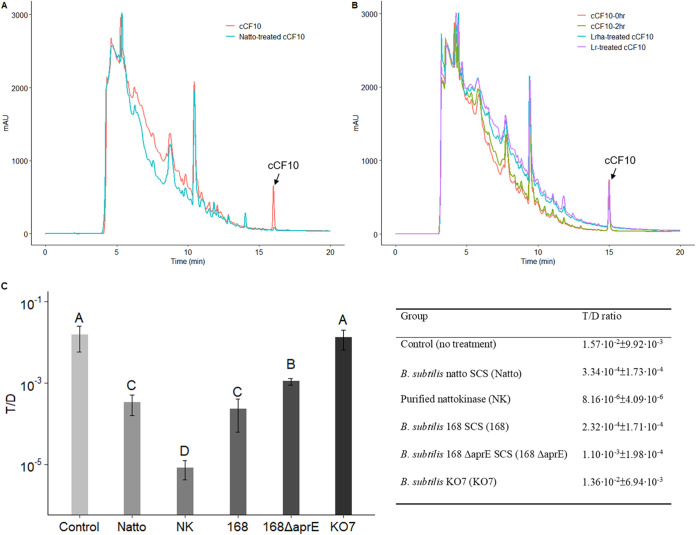
As mentioned above, we found that the pCF10 transfer-inhibiting substance(s) in the SCS of B. subtilis subsp. natto was one or more 30- to 50-kDa proteins and may directly affect the activity of the peptide pheromone cCF10. In addition, B. subtilis has been reported to be able to secrete several types of proteases into its growth medium at high concentrations (29, 30). Based on these findings, we hypothesized that the pCF10 transfer-inhibiting substance(s) was an extracellular hydrolase of B. subtilis subsp. natto with a MW of 30 to 50 kDa. To test this hypothesis, we surveyed the secreted proteins in the proteomes of Bacillus subtilis subsp. natto BEST195 (identifier [ID] UP000006805) and Bacillus subtilis subsp. natto CGMCC 2108 (ID UP000069510) obtained from UniProt. We identified two hydrolases: a poorly documented serine protease (34,887 Da) and subtilisin NAT (nattokinase) (30,697 Da). To examine whether cCF10 is cleaved after treatment with the SCS of B. subtilis subsp. natto, the treated peptides were analyzed using reverse-phase high-performance liquid chromatography (RP-HPLC). The results are shown in Fig. 5. As illustrated in the HPLC chromatogram, the peak of cCF10 (retention time [RT], 15.2 min) disappeared after treatment with the SCS of B. subtilis subsp. natto (Fig. 5A) but remained after treatment with fresh M9B medium or the SCS of L. rhamnosus or L. reuteri (Fig. 5B).
FIG 5.
An extracellular protease of B. subtilis subsp. natto cleaves cCF10 and inhibits pCF10 transfer. (A and B) HPLC chromatograms of probiotic supernatant-treated peptide pheromone cCF10. The synthetic peptide pheromone cCF10 (100 ng/ml) dissolved in M9B medium (150 μl total) was treated with 50 μl of probiotic SCSs or M9B medium at 37°C for 30 min. Next, the assay mixtures were analyzed using HPLC. The peak of cCF10 is at 15.2 min. (A) cCF10 was degraded when treated with B. subtilis subsp. natto SCS (Natto). (B) cCF10 was not degraded in M9B medium over time (0 to 2 h) or when treated with the SCS of L. rhamnosus (Lrha) or L. reuteri (Lr). (C) Conjugation assays with exogenously added B. subtilis 168 ΔaprE SCS, B. subtilis KO7 SCS, and purified nattokinase (final conc., 1 mg/ml). Donor and recipient cells were mixed 1:1 in M9B medium with and without treatment described above and allowed to grow for 4 h. The entire culture was then plated on selective medium for transconjugants and donors. The y axis indicates the T/D ratio. The data are presented as the means ± SDs (n = 3). Values with different uppercase letters were significantly different according to Duncan’s multiple-range tests (P < 0.05).
B. subtilis subsp. natto is closely related to well-known domesticated strain B. subtilis 168 (31). In 2010, Nishito et al. (32) conducted a genome comparison between B. subtilis subsp. natto BEST195 and B. subtilis 168. It was revealed that most predicted genes in B. subtilis subsp. natto BEST195 are one-to-one orthologous to genes in 168 (32). Although some genes are inserted or deleted in B. subtilis subsp. natto BEST195, these genes are mainly related to its ability to produce γ-poly-dl-glutamic acid (γ-PGA), polyketide, or plipastatin (32). According to the Natto Genome Project browser (http://natto-genome.org/), nattokinase-encoding gene aprN in B. subtilis subsp. natto is orthologous to aprE in B. subtilis 168, and these two genes show 99.2% similarity in nucleotide sequence. In addition to aprN, other genes encoding extracellular protease in B. subtilis subsp. natto BEST195 and their orthologous genes in 168 are listed in Table 1 (B. subtilis subsp. natto might lack nprB but has six other genes encoding extracellular protease in its genome). In B. subtilis 168, the extracellular proteases encoded by the seven genes listed in Table 1 account for more than 90% of its total extracellular protease activity (33). Herein, aprE-knockout strain B. subtilis BKK10300 (denoted B. subtilis 168 ΔaprE) and the seven-extracellular protease-deletion (aprE deficiency included) strain of B. subtilis KO7 were used to examine the role of nattokinase and other extracellular proteases of B. subtilis subsp. natto in the inhibition of E. faecalis pCF10 transfer.
TABLE 1.
Seven genes encoding extracellular protease in B. subtilis 168 and their orthologous genes in B. subtilis natto BEST195a
| B. subtilis 168 gene | B. subtilis subsp. natto BEST195 gene | Nucleotide sequence similarity (%) |
|---|---|---|
| nprE | nprE | 99.4 |
| aprE | aprN | 99.2 |
| epr | epr | 93 |
| mpr | mpr | 98.9 |
| nprB | No orthologous gene was found in the B. subtilis subsp. natto BEST195 genome | |
| vpr | vpr | 98.8 |
| bpr | bpr | 98.4 |
Natto Genome Project browser (http://natto-genome.org/).
A conjugation assay with exogenously added B. subtilis 168 ΔaprE SCS, B. subtilis KO7 SCS, and nattokinase that was purified from the culture supernatant of B. subtilis subsp. natto (a mass spectrometry [MS] analysis was conducted to ensure its purity and identity) (see Fig. S3 and Table S1) was performed. The results are shown in Fig. 5C. The T/D ratio in the group treated with 168 ΔaprE SCS was significantly higher (P < 0.05) than that in the group treated with wild type 168 SCS; the T/D ratio in the group treated with KO7 SCS showed no significant difference (P > 0.05) from the negative-control group to which no SCS was added. The pCF10 transfer-inhibiting effect of B. subtilis was decreased by approximately 80% after aprE was inactivated. Furthermore, after the seven genes encoding extracellular protease were all inactivated, B. subtilis nearly lost its overall pCF10 transfer-inhibiting effect (98%). In addition, the T/D ratio was significantly decreased (P < 0.001) when purified nattokinase (final concentration [conc.], 1 mg/ml) was exogenously added to E. faecalis mating cultures. The results described above indicated a key role for nattokinase of B. subtilis subsp. natto in the inhibition of E. faecalis pCF10 transfer. However, other extracellular proteases of B. subtilis subsp. natto might also contribute to the inhibitory effect, although to a lesser extent.
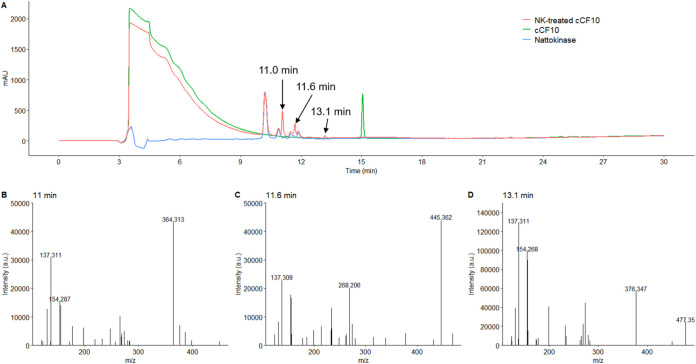
Furthermore, to identify the cleavage site on cCF10 (LVTLVFV), cCF10 was treated with nattokinase. The reaction product produced new peaks, as shown in the HPLC chromatogram (RT, 11.0, 11.6, and 13.1 min) (Fig. 6A). The peptides represented by the three peaks were analyzed using MS. The mass spectra of these three peaks are shown in Fig. 6B to D. The m/z (364.313) of the peak at 11.0 min was similar to the mass of fragment VFV (MW, 363.46), while the m/z (445.362) of the peak at 11.6 min was similar to that of the fragment LVTL (MW, 444.57). This pattern indicates a possible cleavage site between L and V, producing LVTL plus VFV fragments. Another detected cleavage product had an m/z of 477.352 (13.1 min), similar to the mass of the fragment LVFV (MW, 476.62). However, the intensity of this peak was relatively weak. These results show that cCF10 can be cleaved by nattokinase into LVTL+VFV and possibly also LVT+LVFV fragments (Table 2).
FIG 6.
HPLC chromatogram and mass spectra of nattokinase-treated cCF10. The synthetic peptide pheromone cCF10 (100 ng/ml) dissolved in 50 mM phosphate buffer containing 150 mM NaCl (pH 7.0) (150 μl total) was treated with 50 μl of 1 mg/ml B. subtilis nattokinase solution at 37°C for 30 min. Next, the assay mixtures were analyzed using HPLC and MS. (A) The peak of cCF10 is at 15.2 min. After cCF10 was treated with nattokinase (NK), the reaction product was found to produce three new peaks, as shown in the HPLC chromatogram (RTs, 11.0, 11.6, and 13.1 min). The mass spectra of these three new peaks are shown in panels B to D.
TABLE 2.
Possible cleavage sites in cCF10, faecalis-cAM373, and gordonii-cAM373
| Peptide name | Peptide sequence (mol wt) | m/z (nearby amino acid sequence) | Possible cleavage products of the peptide |
|---|---|---|---|
| cCF10 (secreted by E. faecalis) | LVTLVFV (790.01) | 364.313 (VFV) | LVTL + VFV; LVT + LVFV |
| 445.362 (LVTL) | |||
| 477.352 (LVFV) | |||
| faecalis-cAM373 (secreted by E. faecalis) | AIFILAS (733.91) | 576.393 (AIFIL) | AIFIL + AS |
| gordonii-cAM373 (secreted by S. gordonii) | SVFILAA (719.88) | 352.275 (SVF) | SVF + IL + AA |
| 578.370 (SVFIL) |
Small amounts of cCF10 fragments interfere with the conjugative transfer of pCF10.
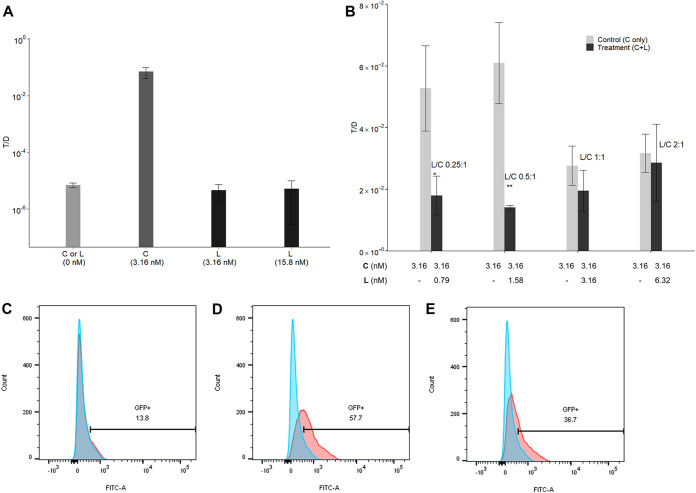
We found that cCF10 (LVTLVFV) was cleaved between L4 and V5 by nattokinase. In E. faecalis, the amino acid sequence of peptide pheromones, especially at the amino terminus, is crucial for its specific interactions with a conjugative plasmid-encoded sensing system (27). Therefore, we selected the fragment LVTL (denoted L peptide and comprising the first 4 amino acids at the amino terminus of cCF10) and investigated whether it exhibits activity to induce or interfere with the conjugative transfer of pCF10. L peptide was added alone or together with cCF10 to induce donor cells. Then, recipient cells were added to allow one round of conjugation before determining the T/D ratio. As shown in Fig. 7A, L peptide alone failed to exhibit cCF10-like pCF10 transfer-inducing activity at physiological (3.16 nM) or even higher (15.8 nM) concentrations. However, when L peptide was added to cCF10-inducing donor cells (3.16 nM), the T/D ratio was lower than that in the negative-control group to which no L peptide was added. Thus, L peptide can likely interfere with the conjugative transfer of pCF10. Next, different L peptide-to-cCF10 (L/C) ratios of 0.25:1, 0.5:1, 1:1, and 2:1 were also used to induce donor cells. At a low L/C ratio (0.5:1), the inhibitory effect of L peptide on the conjugative transfer of pCF10 was significant (P < 0.01) (Fig. 7B). Moreover, L peptide repressed the expression of the cCF10-inducible GFP reporter on pCF10. The percentage of GFP-positive donor cells decreased from 57.7% to 36.7% (Fig. 7C to E). Thus, L peptide likely affects the transport or sensing of cCF10 by donor cells, thus interfering with its conjugative transfer.
FIG 7.
Influence of L peptide on the conjugative transfer of pCF10. Donor cells were incubated with L peptide (L) (cCF10 fragment, sequence LVTL) alone or together with cCF10 (C) at 37°C for 30 min. Then, recipient cells were added and incubated for 10 min to allow one round of conjugation before the entire culture was plated on selective medium for transconjugants and donors. The conjugation frequency is represented by the number of transconjugants per donor (T/D ratio). (A) L was added at physiological (3.16 nM) or higher concentrations (15.8 nM) to induce donor cells. The groups induced with no peptide (L or C) or 3.16 nM C served as the negative-control and positive-control groups, respectively. The data are presented as the means ± SDs (n = 3). (B) Different volumes of 1 mg/ml L stock solution (dissolved in dimethyl sulfoxide [DMSO]) and a fixed volume of 1 mg/ml C stock solution (dissolved in DMSO; final conc., 3.16 nM) were added to achieve L/C ratios of 0.25:1, 0.5:1, 1:1, and 2:1 for induction of donor cells (treatment groups). The groups induced with 3.16 nM C served as the negative control. In addition, to eliminate the effect of DMSO, a volume of DMSO equal to that of the L stock solution was added to the negative control group. The data are presented as the means ± SDs (n = 3). *, P < 0.05; **, P < 0.01 compared with each negative control group (Student’s t test). (C to E) The expression of the cCF10-inducible GFP reporter harbored on pCF10 was quantified using flow cytometry. Blue peak, wild-type E. faecalis OG1RF that would not express the GFP reporter was used as the negative control. Red peaks, donor cells induced with no peptides for 0 min (C), 0.7 μl DMSO plus 3.16 nM C for 60 min (D), or 1.58 nM L plus 3.16 nM C (L/C = 1:2) for 60 min (E). The numbers shown in the middles of panels C to E are the percentages of GFP-positive cells after gating.
B. subtilis subsp. natto grows and secrets proteases under anaerobic conditions.
As shown in Fig. S1 and S2, B. subtilis subsp. natto grew slowly in M9B medium under anaerobic conditions (within 24 h of cultivation, the optical density at 600 nm [OD600] was 0.39 ± 0.05), and the T/D ratio of the group treated with anaerobically cultured B. subtilis subsp. natto SCS (M9B medium, 24 h) was not significantly different (P > 0.05) from that of the negative control to which no SCS was added.
Previous studies (34, 35) have shown that supplementing nitrate to the culture medium facilitates the growth and protease secretion of B. subtilis under anaerobic conditions. As reported, B. subtilis uses nitrate as the terminal electron acceptor for respiration in the absence of oxygen (34). In 2001, Espinosa-de-los-Monteros et al. found that B. subtilis grows anaerobically in Schaeffer’s sporulation medium supplemented with glucose and nitrate, and aprE expression is similar to that in bacteria cultured under aerobic conditions if sufficient glucose and nitrate are supplied (35). Therefore, B. subtilis subsp. natto was cultured in Schaeffer’s sporulation medium supplemented with glucose (4 g/liter) and potassium nitrate (20 mM) (denoted SGN medium). In Fig. 8A, B. subtilis subsp. natto was cultured alone or together with E. faecalis donors and recipients on a filter membrane on SGN agar supplemented with skim milk in the anaerobic jar. The clear zone that formed around the membrane indicated the presence of secreted protease. This phenomenon was observed in B. subtilis subsp. natto group and coculture group (B. subtilis subsp. natto and E. faecalis), but not in E. faecalis group within 24 h of cultivation. These results indicated that B. subtilis subsp. natto was able to grow and produce proteases anaerobically in the presence of E. faecalis.
FIG 8.
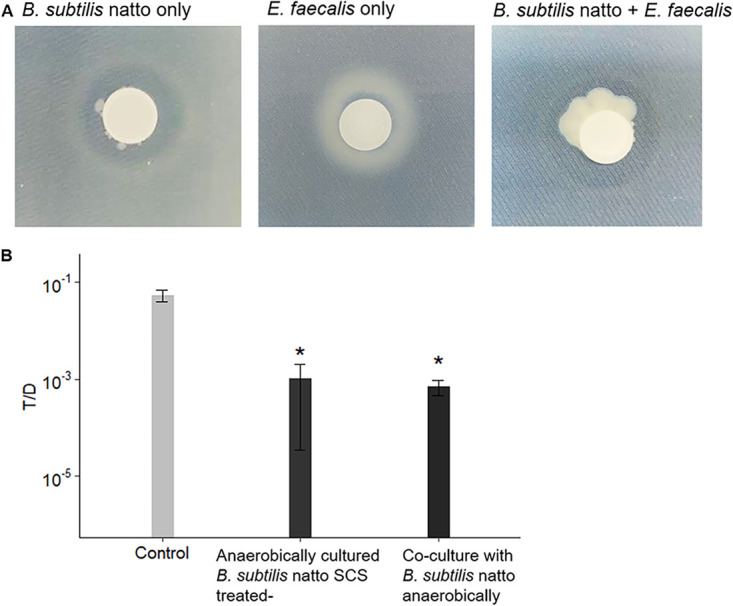
B. subtilis subsp. natto grows and secretes proteases under anaerobic conditions. (A) B. subtilis subsp. natto was cultured alone or together with E. faecalis donors and recipients on SGN (Schaeffer’s sporulation medium supplemented with glucose and nitrate) agar supplemented with skim milk. The amount of secreted protease was estimated from the clear zone that formed around the filter membrane. (B) Four-hour in vitro conjugation assays with exogenously added anaerobically cultured B. subtilis subsp. natto supernatant (SGN broth, 48 h) or B. subtilis subsp. natto (coculture). For bacterial coculture experiments, the cultures of B. subtilis subsp. natto were grown anaerobically in SGN broth to the early exponential phase and diluted according to the OD600. Next, the cultures were added to E. faecalis mating cultures and allowed to grow for 4 h. The entire culture was then plated on selective medium for E. faecalis transconjugants and donors. Fresh SGN broth was added to mating cultures to serve as the negative control group. The y axis indicates the T/D ratio. The data are presented as the means ± SDs (n = 3). *, P < 0.05 compared with negative control (Duncan’s multiple-range tests).
When B. subtilis subsp. natto was cultured in SGN broth supplemented with Oxyrase for broth for 48 h, the OD600 of the cultures was 0.93 ± 0.30, and nattokinase activity of its supernatant was 2.1 ± 0.58 fibrinolytic units (FU)/ml. The value of nattokinase activity was similar to that detected in 24-h aerobically cultured B. subtilis subsp. natto supernatant (2.4 ± 0.58 FU/ml). Conjugation assays with exogenously added anaerobically cultured B. subtilis subsp. natto supernatant (SGN medium, 48 h) were performed. The T/D ratio decreased by approximately 100-fold compared to that of negative control to which no SCS was added (Fig. 8B). Furthermore, B. subtilis subsp. natto was cocultured with E. faecalis donors and recipients in SGN broth anaerobically to examine whether B. subtilis subsp. natto itself could inhibit the transfer of pCF10. Similar to the effect of anaerobically cultured B. subtilis subsp. natto supernatant, the T/D ratio decreased by nearly 100-fold compared to that of negative control (Fig. 8B). These results supported the hypothesis that B. subtilis subsp. natto may grow and secrete nattokinase to inhibit E. faecalis pCF10 transfer under anaerobic conditions, such as those in the human intestinal tract, if appropriate substances were provided as terminal electron acceptors.
B. subtilis subsp. natto supernatant cleaves the mating inducer of vancomycin-resistant E. faecalis.
In addition to pCF10, several other families of pheromone-inducible conjugative plasmids have been identified in E. faecalis clinical isolates. Among them, pAM373 and its derivatives often carry vancomycin resistance genes, and their conjugative transfer can be induced with peptide pheromones secreted by E. faecalis and other bacterial species, including Staphylococcus aureus, Streptococcus gordonii, and Enterococcus hirae (36, 37). In addition to cAM373 secreted by E. faecalis (denoted faecalis-cAM373; sequence, AIFILAS), two other peptide pheromones, staph-cAM373 (AIFILAA) secreted by S. aureus and gordonii-cAM373 (SVFILAA) secreted by S. gordonii, can induce the conjugative transfer of pAM373 derivatives (38–40).
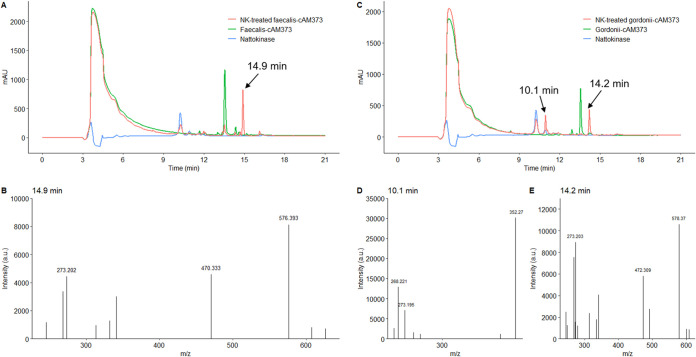
We investigated whether faecalis-cAM373 and gordonii-cAM373 can also be cleaved by the B. subtilis subsp. natto supernatant. Furthermore, degradation experiments using nattokinase were carried out, and the resulting peptides were analyzed by MS. As shown in Fig. 9A and C, both faecalis-cAM373 (RT, 13.5 min) and gordonii-cAM373 (RT, 13.6 min) were cleaved by nattokinase. Upon cleavage, the retention time of faecalis-cAM373 shifted to produce a new peak at 14.9 min with a major m/z of 576.393, corresponding to the mass of fragment AIFIL (MW, 575.75) (Fig. 9A and B). For gordonii-cAM373 (Fig. 9C to E), the two new peaks at 10.1 and 14.6 min had major m/z ratios of 352.275 and 578.370, respectively, corresponding to the masses of fragments SVF (MW, 351.40) and SVFIL (MW, 577.72). Hence, faecalis-cAM373 was cleaved into AIFIL plus AS fragments, while gordonii-cAM373 was cleaved into SVF plus IL plus AA fragments (Table 2). These results indicate that the protease, such as nattokinase, secreted by B. subtilis subsp. natto indeed cleaves intra- and interspecies mating pheromones possibly involved in the transfer of vancomycin resistance.
FIG 9.
HPLC chromatogram and mass spectra of nattokinase-treated faecalis-cAM373 and gordonii-cAM373. The synthetic peptide pheromone (faecalis-cAM373 or gordonii-cAM373) (100 ng/ml) dissolved in 50 mM phosphate buffer containing 150 mM NaCl (pH 7.0) (150 μl total) was treated with 50 μl of 1 mg/ml B. subtilis nattokinase solution at 37°C for 30 min. Next, the assay mixtures were analyzed using HPLC and MS. (A) The peak of faecalis-cAM373 (sequence AIFILAS) occurred at 13.5 min. After faecalis-cAM373 was treated with nattokinase (NK), the reaction product was found to produce a new peak, as shown in the HPLC chromatogram (RT, 14.9 min). The mass spectrum of this new peak is shown in panel B. (C) The peak of gordonii-cAM373 (sequence SVFILAA) occurred at 13.6 min. After gordonii-cAM373 was treated with nattokinase (NK), the reaction product was found to produce two new peaks, as shown in the HPLC chromatogram (RTs, 10.1 and 14.2 min). The mass spectra of these two new peaks are shown in panels D and E.
DISCUSSION
E. faecalis has attracted considerable attention from researchers in recent decades due to its pheromone-inducible conjugative plasmid transfer systems. These systems play a key role in facilitating the transfer of antibiotic resistance. Therefore, to search for potential probiotic strains or their fermentation products that can interfere with the transfer of antibiotic resistance between E. faecalis strains, we investigated whether probiotic SCSs have an inhibitory effect on the conjugative transfer of pCF10. Indeed, some tested probiotic SCSs showed a substantial inhibitory effect on pCF10 transfer in E. faecalis. However, after the pH of these probiotic SCSs was neutralized, only the SCS of aerobically cultured B. subtilis subsp. natto retained an inhibitory effect. Furthermore, our results indicated that at least one 30- to 50-kDa protein in the SCS of aerobically cultured B. subtilis subsp. natto cleaved the peptide pheromone cCF10, thus interfering with the conjugative transfer of pCF10. Thus, the pCF10 transfer-inhibiting protein(s) is likely an extracellular protease(s) of B. subtilis subsp. natto.
We found that B. subtilis lost approximately 80% of its pCF10 transfer-inhibiting activity after aprE was inactivated. Furthermore, after the seven genes encoding extracellular proteases were all inactivated, B. subtilis nearly lost its overall pCF10 transfer-inhibiting activity (98%). Therefore, we inferred that nattokinase of B. subtilis subsp. natto may play a key role in inhibiting E. faecalis pCF10 transfer. In addition to nattokinase, other extracellular proteases secreted by B. subtilis subsp. natto also contributed to inhibition, although their effects would be minor. As reported by Sloma et al., subtilisin (aprE gene) and neutral proteases (nprB and nprE genes) account for more than 90% of the total extracellular protease activity of B. subtilis, and the remaining activity is accounted by other minor extracellular proteases, such as bacillopeptidase F (bpr gene), Epr (epr gene), Mpr (mpr gene), and Vpr (vpr gene) (33).
Peptide pheromones, which are used as quorum sensing signals by Gram-positive bacteria, including E. faecalis, mediate conjugative plasmid transfer and many other fundamental behaviors, such as biofilm formation and virulence regulation (41). Our study showed that the conjugative transfer of pCF10 between E. faecalis strains was reduced by cleavage of cCF10. In addition, in previous studies, a cyclic peptide pheromone was found to be a potential target to inhibit the E. faecalis fsr quorum sensing system. In 2007, Nakayama et al. reported that siamycin I from actinomycete metabolites can attenuate fsr quorum sensing-mediated expression of virulence genes without inhibiting the growth of E. faecalis by inhibiting the production of gelatinase biosynthesis-activating pheromone (GBAP) (42). In 2009, the same group reported that ambuic acid from fungal butanol extracts can inhibit both GBAP production by E. faecalis and agr autoinducing peptide (AIP) production by S. aureus, thus affecting the quorum sensing signaling of these bacteria (43). These studies support the idea that various substances with pheromone-quenching activity likely exist among metabolites of microbial species and may be the key to combating E. faecalis and other pathogens.
In addition to identifying cleavage of cCF10, which attenuates quorum sensing signaling in E. faecalis, we also found that small amounts of the cleavage product L peptide, comprising the first 4 amino acids of the cCF10 amino terminus, can repress the conjugative transfer of pCF10. As reported by Dunny et al., the amino terminus of the E. faecalis peptide pheromone is crucial for its biological specificity and interactions with pheromone receptor proteins (27). Therefore, L peptide likely binds specifically to certain cCF10 receptor proteins of E. faecalis, such as the peptide-specific importer PrgZ or the transcriptional regulator PrgX, thereby interfering with the transport or regulatory effect of cCF10. In addition to L peptide, the pCF10-encoded inhibitor peptide iCF10 and membrane protein PrgY were shown to repress the conjugative transfer of pCF10 (44). iCF10 competes with cCF10 for binding to the cytoplasmic pheromone receptor PrgX, thus enhancing the repression of the promoter PQ that drives the expression of the prgQ operon on pCF10 (14), and PrgY potentially reduces pheromone activity by binding or degrading endogenous cCF10 (45). The role of L peptide is similar to that of iCF10. However, its amino acid sequence is very different from that of iCF10 (AITLIFI). More investigations are required to determine the mechanism by which L peptide interferes with the conjugative transfer of pCF10 and whether fragments of other E. faecalis peptide pheromones can also interfere with the conjugative transfer of specific plasmids.
We found that in addition to cCF10, faecalis-cAM373 and gordonii-cAM373, mating inducers of vancomycin-resistant E. faecalis, were also cleaved by nattokinase of B. subtilis subsp. natto. Conjugation experiments are necessary to examine whether the intra- and interspecies conjugative transfer of the vancomycin resistance gene-carrying plasmid pAM373 was indeed inhibited via the cleavage of the peptide pheromone faecalis-cAM373 or gordonii-cAM373. Our current data showed that nattokinase of B. subtilis subsp. natto might be useful to interfere with the transfer of vancomycin resistance to E. faecalis strains.
B. subtilis is a species of bacteria that can survive in the soil, which includes an abundance of anaerobic environments and anaerobic bacteria (46). In this study, we showed that B. subtilis subsp. natto grew and secreted nattokinase anaerobically in the presence of nitrate as a terminal electron acceptor. Therefore, B. subtilis subsp. natto likely inhibits E. faecalis pCF10 transfer under anaerobic conditions, such as those in the human intestinal tract. However, animal studies are further needed to evaluate the therapeutic potential of B. subtilis natto and nattokinase. In 2018, Piewngam et al. (47) found that mice fed B. subtilis spores exhibited a complete decolonization of methicillin-resistant Staphylococcus aureus (MRSA) in the feces and intestines. Furthermore, they demonstrated that B. subtilis can produce the lipopeptide fengycin to interfere with S. aureus agr quorum sensing signaling (47). This study supports the idea that B. subtilis may survive in the intestinal tract and produce functional substances to combat enteric pathogenic bacteria.
This work was an initial probiotic screening study that targeted the pheromone-inducible conjugative plasmid transfer system of E. faecalis. Our results showed that B. subtilis subsp. natto can secret nattokinase to degrade peptide pheromone cCF10, thereby interfering with the conjugative pCF10 transfer of E. faecalis. Moreover, we found that specific pheromone fragment L peptide was involved in the inhibition of conjugative pCF10 transfer. Based on these findings, we propose that nattokinase or specific peptide pheromone fragments might contribute to mitigating the spread of antibiotic resistance genes carried in the plasmid between E. faecalis populations.
MATERIALS AND METHODS
Bacterial strains, medium and growth conditions.
The E. faecalis strains and plasmids used in this study are listed in Table 3. All materials were obtained from the laboratory of Gary M. Dunny (University of Minnesota, USA). All E. faecalis strains were statically cultured at 37°C in M9 medium (3 g/liter yeast extract, 10 g/liter Casamino Acids, 36 g/liter glucose, 0.12 g/liter MgSO4, and 0.011 g/liter CaCl2 [12]) or in brain heart infusion (BHI) broth (BD Co., USA). If needed, antibiotics were added at the following concentrations: 10 μg/ml tetracycline, 200 μg/ml rifampicin, and 100 μg/ml spectinomycin (12). In the mating assay, OG1Sp carrying pCF10-iGFP-p23-tdTomato (denoted OG1Sp/pCF10-iGFP-p23-tdTomato) was used as the donor, and OG1RF::p23cfp was used as the recipient.
TABLE 3.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source | Reference(s) |
|---|---|---|---|
| E. faecalis strains | |||
| OG1RF | Rifr, Far | Gary M. Dunny | 61 |
| OG1Sp | Specr | Gary M. Dunny | 62 |
| OG1RF::p23cfp | OG1RF derivative with a constitutive CFP reporter fused to its genomic DNA | Gary M. Dunny | 63 |
| E. faecalis plasmids | |||
| pCF10 | Native E. faecalis conjugative plasmid; Tetr | Gary M. Dunny | 61, 64 |
| pCF10-iGFP-p23-tdTomato | pCF10 derivative that contains a constitutive tdTomato reporter and expresses GFP upon induction by the pheromone cCF10; tdTomato was inserted in the intergenic region between pcfR and Tn925; GFP was inserted in the intergenic region between prgC and prgD | Gary M. Dunny | 65 |
| Lactobacillus strains | |||
| L. plantarum KGW103 | National Chiayi University (Taiwan) | 66 | |
| L. rhamnosus ATCC 9595 | BCRC (Taiwan) | 67 | |
| L. reuteri ATCC 23272 | BCRC (Taiwan) | 68 | |
| Bacillus strains | |||
| B. coagulans ATCC 10545 | BCRC (Taiwan) | 69 | |
| B. subtilis subsp. natto NTU-18 (BCRC 80390) | Our laboratory | 48, 49 | |
| B. subtilis 168 (1A1) | BGSC (USA) | 70 | |
| B. subtilis BKK10300 | trpC2 ΔaprE::kan | BGSC (USA) | 71 |
| B. subtilis KO7 (1A1133) | ΔnprE ΔaprE Δepr Δmpr ΔnprB Δvpr Δbpr | BGSC (USA) | D. Zeigler, unpublished |
| Clostridium strain | |||
| C. butyricum MIYAIRI 588 | Miyarisan-BM (Miyarisan Pharmaceutical Co., Ltd., Japan) | 72 |
Rif, rifampicin; Fa, fusidic acid; Spec, spectinomycin; Tet, tetracycline.
Six probiotic strains (three lactic acid bacteria and three spore-forming bacteria) were selected, and their effects on pCF10 transfer between E. faecalis bacteria were examined (Table 3). The three lactic acid bacterial species are described below. Lactobacillus plantarum KGW103 was obtained from National Chiayi University (Taiwan). Lactobacillus rhamnosus (ATCC 9595) and Lactobacillus reuteri (ATCC 23272) were purchased from the Bioresource Collection and Research Center (BCRC, Taiwan). The three spore-forming bacterial species are listed below. Bacillus coagulans (ATCC 10545) was purchased from BCRC. Bacillus subtilis subsp. natto NTU-18 (BCRC 80390) isolated from the commercial product was maintained in our laboratory (48, 49). Clostridium butyricum MIYAIRI 588 was isolated from the product Miyarisan-BM (Miyarisan Pharmaceutical Co., Ltd., Japan). In addition, B. subtilis 168, B. subtilis BKK10300 (trpC2 ΔaprE::kan), and the seven-extracellular protease-deletion strain B. subtilis KO7 (ΔnprE ΔaprE Δepr Δmpr ΔnprB Δvpr Δbpr) were also used in this study to confirm the role of extracellular proteases of B. subtilis subsp. natto in the inhibition of E. faecalis pCF10 transfer (Table 3). The seven genes encoding extracellular protease in B. subtilis 168 and their orthologous genes in B. subtilis subsp. natto BEST195 (Natto Genome Project browser; http://natto-genome.org/) are listed in Table 1. These three strains were purchased from Bacillus Genetic Stock Center (BGSC, USA). All Lactobacillus spp. used in this study were anaerobically cultured at 37°C in De Man, Rogosa, and Sharpe (MRS) broth (BD Co., USA) supplemented with Oxyrase for broth (Oxyrase Inc., USA). All Bacillus spp. were cultured in LB broth (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter sodium chloride [50]) in orbital shakers at 37°C with shaking at 125 rpm, while C. butyricum MIYAIRI 588 was anaerobically cultured at 37°C in LB broth supplemented with Oxyrase for broth.
Preparation of probiotic spent culture supernatants.
The SCS of each probiotic bacterium was prepared using methods described by Lin et al. (51) and Wasfi et al. (52) with some modifications. In detail, all probiotic strains were cultured in M9 medium to eliminate growth differences arising from differences in culture media. Buffering agents (8.5 g/liter Na2HPO4·2H2O, 3 g/liter KH2PO4, 11.5 g/liter sodium acetate, and 1 ml/liter acetic acid) were added to the M9 medium to increase its buffering capacity (M9 medium containing the buffering agents is denoted M9B medium) and to prevent a sharp decrease in the pH of M9 medium during the growth of the probiotic bacteria. The initial pH value of M9B medium was 6.5 ± 0.1.
Furthermore, to ensure fully anaerobic culture conditions during the growth of the probiotic bacteria in M9B medium, anaerobic culture was conducted in Kimax culture tubes with screw caps and rubber liners (Kimble Inc., USA). In addition, Oxyrase for broth was added to fresh M9B medium to remove all dissolved oxygen before inoculation with the probiotics. Next, these culture tubes were placed in an AnaeroPack jar (Mitsubishi Gas Chemical Co., Japan) and incubated at 37°C in a growth chamber. Anaero-Indicator (bioMérieux Corp., France) and sodium resazurin solution (1 mg/liter) were used as oxygen indicators to assess the anaerobic status in both the atmosphere and medium during incubation.
Here, Lactobacillus spp. and C. butyricum MIYAIRI 588 were cultured under anaerobic conditions to collect their SCSs. On the other hand, for collection of the SCSs of B. subtilis subsp. natto and B. coagulans under different culture conditions, these bacteria were cultured under both anaerobic and aerobic conditions. Aerobic culture was performed in a Hinton flask with incubation in orbital shakers at 37°C with shaking at 125 rpm. After incubation for 24 h, the cultures were centrifuged (4,000 × g, 10 min) to remove all cells. Then, the supernatant was filter sterilized through 0.22-μm filters (Pall Co., USA) and stored as the untreated supernatant at 4°C.
In vitro mating assay with spent probiotic culture supernatants.
The in vitro mating assay with E. faecalis was performed as described by Breuer et al. (53) and Hirt et al. (54) with some modifications. In detail, cultures of E. faecalis OG1Sp/pCF10-iGFP-p23-tdTomato (donor) and E. faecalis OG1RF::p23cfp (recipient) were grown overnight at 37°C in M9 medium. One milliliter of each overnight culture was centrifuged, washed twice with phosphate-buffered saline (PBS) containing 2 mM EDTA, and resuspended in 400 μl of fresh M9B medium. Then, the cultures of the donor and recipient cells were mixed in a 1:1 ratio, and 30 μl of the mixed culture was inoculated into 3 ml (1% [vol/vol]) of mating mixture before anaerobic incubation at 37°C for 4 h. Here, the mating mixture was prepared by mixing the spent probiotic culture supernatants and fresh M9B medium supplemented 1:1 with Oxyrase for broth to generate anaerobic conditions.
After the 4-h in vitro mating assay, serial dilutions of these samples were plated on selective Todd-Hewitt broth (THB) agar medium containing 30 g/liter Bacto Todd-Hewitt broth (Neogen Corp., USA) and 15 g/liter agar to count the donors (spectinomycin and tetracycline resistant), recipients (rifampicin resistant), and transconjugants (rifampicin and tetracycline resistant). Antibiotics were added to THB agar at the following concentrations: 10 μg/ml tetracycline, 50 μg/ml rifampicin, and 630 μg/ml spectinomycin (54).
Characterization of pCF10 transfer-inhibiting substances in the spent probiotic culture supernatants.
The untreated supernatant of each probiotic culture was divided into equal portions for different assays. For the organic acid assay, the supernatant was adjusted to pH 6.5 ± 0.1 using 1 N NaOH (55, 56). For the protein or heat-labile compound assays, the supernatant was placed in an 80°C water bath for 30 min and immediately transferred to an ice bath. Treated and untreated supernatants were stored at 4°C.
Each supernatant subjected to different treatments was mixed with fresh M9B medium (1:1) as the mating mixture and used for the mating assay to examine the effect of treated probiotic supernatants on E. faecalis pCF10 transfer.
Supernatant fractionation through different molecular-weight-cutoff membranes.
Twelve milliliters of the supernatant were added to an Amicon Ultra-15 centrifugal filter device (Millipore Co., USA) with a MWCO of 30 kDa (30K) or 50 kDa (50K). The supernatant concentrate collected through the Amicon Ultra-15 30K and Amicon Ultra-15 50K filters was retained. The filtrate collected through the Amicon Ultra-15 30K filter was added to an Amicon Ultra-15 10K filter. Similarly, the concentrate was retained, and the filtrate was further added to an Amicon Ultra-15 3K filter. Finally, several fractions with different MWs (>50 kDa, >30 kDa, 10 to 30 kDa, 3 to 10 kDa, and <3 kDa) were obtained. All fractions were freeze-dried and redissolved in 4 ml of double-distilled water (ddH2O) to eliminate the effect of differences in concentration. Next, the effects of these concentrated (3×) fractions of supernatant on the transfer of plasmid pCF10 were examined.
Determination of the total protein concentration in the probiotic culture supernatant.
Fifty microliters of filter-sterilized culture supernatant were mixed with 200 μl of Coomassie brilliant blue G-250, and the absorbance was measured at 595 nm to determine the extracellular protein concentration in the probiotic culture supernatant. The concentration of total extracellular protein was calculated from the standard curve constructed from different concentrations of bovine serum albumin (BSA) (57).
SDS-PAGE of the probiotic culture supernatant was conducted using the procedure described by Cai et al. with some modifications (58). In detail, 1,800 μl of filter-sterilized culture supernatant was mixed with 200 μl of trichloroacetic acid (TCA) solution (6.1 N), maintained at 4°C overnight, and centrifuged at 13,000 × g for 10 min. The precipitate was washed twice with ethanol, dried, and redissolved in a solution containing 2 M thiourea and 8 M urea. The sample was then mixed with 5× SDS-PAGE sample buffer (4:1), and 5 or 10 μl of the solution was subjected to SDS-PAGE. A BSA solution (1 mg/ml) was used as the positive control. The gel was then stained with Coomassie brilliant blue R-250 solution.
GFP induction assay and analysis.
This assay was conducted using the method described by Breuer et al. (53), with some modifications. In detail, 1 ml of overnight cultures of E. faecalis OG1Sp/pCF10-iGFP-p23-tdTomato was centrifuged, washed twice with PBS containing 2 mM EDTA, and resuspended in fresh M9B medium. The cultures were then subcultured (1:4) in mating mixture and grown to the early exponential phase by incubating them for 1 h at 37°C. The cultures were then induced with 2.5 ng/ml cCF10, and cells were harvested at 0, 60, and 120 min. Cells were fixed immediately upon harvest by resuspending them in PBS containing 2% paraformaldehyde and incubated at 4°C overnight. After fixation, cells were collected by centrifugation at 14,000 × g for 2 min and subsequently declumped to break apart the bacterial aggregates before the expression of the GFP reporter was quantified using flow cytometry (BD FACSCanto II).
Analysis of peptide pheromones using HPLC and MS.
Synthetic pheromone (100 ng/ml) dissolved in M9B medium (150 μl total) was treated with 50 μl of the probiotic supernatant at 37°C for 30 min and immediately transferred to an ice bath. Next, the assay mixtures were injected into a reverse-phase high-performance liquid chromatography (RP-HPLC) system equipped with an Osaka Soda (Shiseido) CAPCELL PAK C18 column (particle size, 5 μm; column dimensions, 4.6 mm by 250 mm). A binary buffer system (solvent A, water containing 0.1% trifluoroacetic acid [TFA]; solvent B, acetonitrile containing 0.1% TFA) was used to establish the mobile phase gradient. Peptides were separated using a linear gradient of 0% to 100% solvent B for 30 min at a flow rate of 1 ml/min with detection at 220 nm.
For sample preparation for mass spectrometry (MS), synthetic pheromones were dissolved in 50 mM phosphate buffer containing 150 mM NaCl (pH 7.0) and cleaved with 1 mg/ml nattokinase solution. Nattokinase was purified from cultures of B. subtilis subsp. natto as described in our previous study (59). Briefly, the culture supernatant of B. subtilis subsp. natto was first fractionated according to the MWs of proteins. Only the fraction that presented nattokinase activity and a single band on 15% SDS-PAGE gels was used in the experiments. Nattokinase activity was determined using a chromogenic method with S2251 (H-d-Val-Leu-Lys-p-nitroanilide dihydrochloride) as the substrate (60). Purified nattokinase was further analyzed using MS to ensure its identity and purity. After the synthetic pheromone was treated with nattokinase, the new peaks appearing in the HPLC chromatogram were collected, purified, and analyzed using a matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer (Bruker AutoFlex III smartbeam TOF/TOF200).
Statistics.
All experiments were repeated at least three times with consistent results. R (version 4.0.0) was used for data analysis. One-way analysis of variance (ANOVA) was performed first. Next, multiple comparisons were evaluated using Duncan’s multiple-range test.
Data availability.
The data used to support the findings of this study are available from the corresponding author upon request.
ACKNOWLEDGMENTS
We thank the Technology Commons at the College of Life Science, NTU (Taiwan), for technical assistance with confocal laser scanning microscopy and the Genomic Core at the Institute of Molecular Biology, Academia Sinica (Taiwan), for assistance with the mass spectrometric analyses.
This work was funded by grants from the Ministry of Science and Technology (no. 107-2313-B-002-028 and 108-2313-B-002-056-MY3), Taiwan.
Yu-Chieh Lin wrote the manuscript. Kung-Ta Lee, Wei-Shou Hu, Gary M. Dunny, and Yu-Chieh Lin designed the experimental plan. Yu-Chieh Lin performed all experiments and analyzed the relevant data. Eric H.-L. Chen and Rita P.-Y. Chen assisted with peptide synthesis, HPLC analysis, and data interpretation. All authors contributed to the revision and final review of the manuscript.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Kung-Ta Lee, Email: ktlee@ntu.edu.tw.
Andrew J. McBain, University of Manchester
REFERENCES
- 1.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 2.Aakra A, Vebø H, Snipen L, Hirt H, Aastveit A, Kapur V, Dunny G, Murray BE, Murray B, Nes IF. 2005. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob Agents Chemother 49:2246–2259. 10.1128/AAC.49.6.2246-2259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanchi H, Mottawea W, Sebei K, Hammami R. 2018. The genus Enterococcus: between probiotic potential and safety concerns-an update. Front Microbiol 9:1791. 10.3389/fmicb.2018.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindenstrauss AG, Ehrmann MA, Behr J, Landstorfer R, Haller D, Sartor RB, Vogel RF. 2014. Transcriptome analysis of Enterococcus faecalis toward its adaption to surviving in the mouse intestinal tract. Arch Microbiol 196:423–433. 10.1007/s00203-014-0982-2. [DOI] [PubMed] [Google Scholar]
- 5.Jett BD, Huycke MM, Gilmore MS. 1994. Virulence of enterococci. Clin Microbiol Rev 7:462–478. 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormick JK, Tripp TJ, Dunny GM, Schlievert PM. 2002. Formation of vegetations during infective endocarditis excludes binding of bacterial-specific host antibodies to Enterococcus faecalis. J Infect Dis 185:994–997. 10.1086/339604. [DOI] [PubMed] [Google Scholar]
- 7.Carniol K, Gilmore MS. 2004. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J Bacteriol 186:8161–8163. 10.1128/JB.186.24.8161-8163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray BE. 1997. Vancomycin-resistant enterococci. Am J Med 102:284–293. 10.1016/S0002-9343(99)80270-8. [DOI] [PubMed] [Google Scholar]
- 9.Klevens RM, Edwards JR, Richards CL, Jr., Horan TC, Gaynes RP, Pollock DA, Cardo DM. 2007. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 122:160–166. 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arias CA, Contreras GA, Murray BE. 2010. Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect 16:555–562. 10.1111/j.1469-0691.2010.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frieden T. 2013. Antibiotic resistance threats in the United States. US Department of Health & Human Services, Washington, DC. [Google Scholar]
- 12.Bandyopadhyay A, O'Brien S, Frank KL, Dunny GM, Hu WS. 2016. Antagonistic donor density effect conserved in multiple enterococcal conjugative plasmids. Appl Environ Microbiol 82:4537–4545. 10.1128/AEM.00363-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunny GM. 2007. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc Lond B Biol Sci 362:1185–1193. 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breuer RJ, Hirt H, Dunny GM. 2018. Mechanistic features of the enterococcal pCF10 sex pheromone response and the biology of Enterococcus faecalis in its natural habitat. J Bacteriol 200:e00733-17. 10.1128/JB.00733-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akhtar M, Hirt H, Zurek L. 2009. Horizontal transfer of the tetracycline resistance gene tetM mediated by pCF10 among Enterococcus faecalis in the house fly (Musca domestica L.) alimentary canal. Microb Ecol 58:509–518. 10.1007/s00248-009-9533-9. [DOI] [PubMed] [Google Scholar]
- 16.Clewell DB, Francia MV, Flannagan SE, An FY. 2002. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 48:193–201. 10.1016/s0147-619x(02)00113-0. [DOI] [PubMed] [Google Scholar]
- 17.Manson JM, Hancock LE, Gilmore MS. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci U S A 107:12269–12274. 10.1073/pnas.1000139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willems RJ, Hanage WP, Bessen DE, Feil EJ. 2011. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev 35:872–900. 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 20.Saarela M, Lahteenmaki L, Crittenden R, Salminen S, Mattila-Sandholm T. 2002. Gut bacteria and health foods–the European perspective. Int J Food Microbiol 78:99–117. 10.1016/S0168-1605(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 21.Gourbeyre P, Denery S, Bodinier M. 2011. Probiotics, prebiotics, and synbiotics: impact on the gut immune system and allergic reactions. J Leukoc Biol 89:685–695. 10.1189/jlb.1109753. [DOI] [PubMed] [Google Scholar]
- 22.Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elshaghabee FMF, Rokana N, Gulhane RD, Sharma C, Panwar H. 2017. Bacillus as potential probiotics: status, concerns, and future perspectives. Front Microbiol 8:1490. 10.3389/fmicb.2017.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spinler JK, Taweechotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovic J. 2008. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe 14:166–171. 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macpherson AJ, Harris NL. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 4:478–485. 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 26.Bermudez-Brito M, Plaza-Diaz J, Munoz-Quezada S, Gomez-Llorente C, Gil A. 2012. Probiotic mechanisms of action. Ann Nutr Metab 61:160–174. 10.1159/000342079. [DOI] [PubMed] [Google Scholar]
- 27.Dunny GM, Antiporta MH, Hirt H. 2001. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response. Peptides 22:1529–1539. 10.1016/s0196-9781(01)00489-2. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. 1994. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol 176:7405–7408. 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling LF, Zi RX, Wei FL, Jiang BS, Ping L, Chun XH. 2007. Protein secretion pathways in Bacillus subtilis: implication for optimization of heterologous protein secretion. Biotechnol Adv 25:1–12. 10.1016/j.biotechadv.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Contesini FJ, Melo RR, Sato HH. 2018. An overview of Bacillus proteases: from production to application. Crit Rev Biotechnol 38:321–334. 10.1080/07388551.2017.1354354. [DOI] [PubMed] [Google Scholar]
- 31.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Dusterhoft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, Haiech J, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 32.Nishito Y, Osana Y, Hachiya T, Popendorf K, Toyoda A, Fujiyama A, Itaya M, Sakakibara Y. 2010. Whole genome assembly of a natto production strain Bacillus subtilis natto from very short read data. BMC Genomics 11:243. 10.1186/1471-2164-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sloma A, Rufo GA, Jr, Theriault KA, Dwyer M, Wilson SW, Pero J. 1991. Cloning and characterization of the gene for an additional extracellular serine protease of Bacillus subtilis. J Bacteriol 173:6889–6895. 10.1128/jb.173.21.6889-6895.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Priest FG. 1993. Systematics and ecology of Bacillus, p 1–16, In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and other Gram‐positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, DC. [Google Scholar]
- 35.Espinosa-de-los-Monteros J, Martinez A, Valle F. 2001. Metabolic profiles and aprE expression in anaerobic cultures of Bacillus subtilis using nitrate as terminal electron acceptor. Appl Microbiol Biotechnol 57:379–384. 10.1007/s002530100749. [DOI] [PubMed] [Google Scholar]
- 36.Showsh SA, De Boever EH, Clewell DB. 2001. Vancomycin resistance plasmid in Enterococcus faecalis that encodes sensitivity to a sex pheromone also produced by Staphylococcus aureus. Antimicrob Agents Chemother 45:2177–2178. 10.1128/aac.45.7.2177-2178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Boever EH, Clewell DB, Fraser CM. 2000. Enterococcus faecalis conjugative plasmid pAM373: complete nucleotide sequence and genetic analyses of sex pheromone response. Mol Microbiol 37:1327–1341. 10.1046/j.1365-2958.2000.02072.x. [DOI] [PubMed] [Google Scholar]
- 38.Clewell DB, An FY, Flannagan SE, Antiporta M, Dunny GM. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol Microbiol 35:246–247. 10.1046/j.1365-2958.2000.01687.x. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama J, Igarashi S, Nagasawa H, Clewell DB, An FY, Suzuki A. 1996. Isolation and structure of staph-cAM373 produced by Staphylococcus aureus that induces conjugal transfer of Enterococcus faecalis plasmid pAM373. Biosci Biotechnol Biochem 60:1038–1039. 10.1271/bbb.60.1038. [DOI] [PubMed] [Google Scholar]
- 40.Vickerman MM, Flannagan SE, Jesionowski AM, Brossard KA, Clewell DB, Sedgley CM. 2010. A genetic determinant in Streptococcus gordonii Challis encodes a peptide with activity similar to that of enterococcal sex pheromone cAM373, which facilitates intergeneric DNA transfer. J Bacteriol 192:2535–2545. 10.1128/JB.01689-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492. 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama J, Tanaka E, Kariyama R, Nagata K, Nishiguchi K, Mitsuhata R, Uemura Y, Tanokura M, Kumon H, Sonomoto K. 2007. Siamycin attenuates fsr quorum sensing mediated by a gelatinase biosynthesis-activating pheromone in Enterococcus faecalis. J Bacteriol 189:1358–1365. 10.1128/JB.00969-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama J, Uemura Y, Nishiguchi K, Yoshimura N, Igarashi Y, Sonomoto K. 2009. Ambuic acid inhibits the biosynthesis of cyclic peptide quormones in Gram-positive bacteria. Antimicrob Agents Chemother 53:580–586. 10.1128/AAC.00995-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandler JR, Dunny GM. 2008. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol 190:1172–1183. 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buttaro BA, Antiporta MH, Dunny GM. 2000. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J Bacteriol 182:4926–4933. 10.1128/jb.182.17.4926-4933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glaser P, Danchin A, Kunst F, Zuber P, Nakano MM. 1995. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J Bacteriol 177:1112–1115. 10.1128/jb.177.4.1112-1115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, Glose KA, Fisher EL, Hunt RL, Li B, Chiou J, Pharkjaksu S, Khongthong S, Cheung GYC, Kiratisin P, Otto M. 2018. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562:532–537. 10.1038/s41586-018-0616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuo LC, Cheng WY, Wu RY, Huang CJ, Lee KT. 2006. Hydrolysis of black soybean isoflavone glycosides by Bacillus subtilis natto. Appl Microbiol Biotechnol 73:314–320. 10.1007/s00253-006-0474-7. [DOI] [PubMed] [Google Scholar]
- 49.Kuo LC, Lee KT. 2008. Cloning, expression, and characterization of two beta-glucosidases from isoflavone glycoside-hydrolyzing Bacillus subtilis natto. J Agric Food Chem 56:119–125. 10.1021/jf072287q. [DOI] [PubMed] [Google Scholar]
- 50.Atlas RM. 2010. Handbook of microbiological media. CRC press, Boca Raton, FL. [Google Scholar]
- 51.Lin X, Chen X, Chen Y, Jiang W, Chen H. 2015. The effect of five probiotic lactobacilli strains on the growth and biofilm formation of Streptococcus mutans. Oral Dis 21:e128–e134. 10.1111/odi.12257. [DOI] [PubMed] [Google Scholar]
- 52.Wasfi R, Abd El-Rahman OA, Zafer MM, Ashour HM. 2018. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J Cell Mol Med 22:1972–1983. 10.1111/jcmm.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breuer RJ, Bandyopadhyay A, O'Brien SA, Barnes AMT, Hunter RC, Hu WS, Dunny GM. 2017. Stochasticity in the enterococcal sex pheromone response revealed by quantitative analysis of transcription in single cells. PLoS Genet 13:e1006878. 10.1371/journal.pgen.1006878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirt H, Greenwood-Quaintance KE, Karau MJ, Till LM, Kashyap PC, Patel R, Dunny GM. 2018. Enterococcus faecalis sex pheromone cCF10 enhances conjugative plasmid transfer in vivo. mBio 9:e00037-18. 10.1128/mBio.00037-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shokryazdan P, Sieo CC, Kalavathy R, Liang JB, Alitheen NB, Faseleh Jahromi M, Ho YW. 2014. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed Res Int 2014:927268. 10.1155/2014/927268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA. 2014. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog 10:e1003996. 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grintzalis K, Georgiou CD, Schneider YJ. 2015. An accurate and sensitive Coomassie brilliant blue G-250-based assay for protein determination. Anal Biochem 480:28–30. 10.1016/j.ab.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 58.Cai D, Wang H, He P, Zhu C, Wang Q, Wei X, Nomura CT, Chen S. 2017. A novel strategy to improve protein secretion via overexpression of the SppA signal peptide peptidase in Bacillus licheniformis. Microb Cell Fact 16:70. 10.1186/s12934-017-0688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu RL, Lee KT, Wang JH, Lee LY, Chen RP. 2009. Amyloid-degrading ability of nattokinase from Bacillus subtilis natto. J Agric Food Chem 57:503–508. 10.1021/jf803072r. [DOI] [PubMed] [Google Scholar]
- 60.Friberger P, Knos M, Gustavsson S, Aurell L, Claeson G. 1978. Methods for determination of plasmin, antiplasmin and plasminogen by means of substrate S-2251. Haemostasis 7:138–145. 10.1159/000214252. [DOI] [PubMed] [Google Scholar]
- 61.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 75:3479–3483. 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kristich CJ, Chandler JR, Dunny GM. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57:131–144. 10.1016/j.plasmid.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barnes AMT, Dale JL, Chen Y, Manias DA, Greenwood Quaintance KE, Karau MK, Kashyap PC, Patel R, Wells CL, Dunny GM. 2017. Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model. Virulence 8:282–296. 10.1080/21505594.2016.1208890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruhfel RE, Manias DA, Dunny GM. 1993. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J Bacteriol 175:5253–5259. 10.1128/jb.175.16.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Erickson RJB, Bandyopadhyay AA, Barnes AMT, O’Brien SA, Hu W-S, Dunny GM. 2020. Single-cell analysis reveals that the enterococcal sex pheromone response results in expression of full-length conjugation operon transcripts in all induced cells. J Bacteriol 202:e00685-19. 10.1128/JB.00685-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsieh CW, Chiu YC, Wang PS, Lin JH. 2016. Determining probiotic potential of lactic acid bacteria isolated from traditional Taiwan fermented vegetables. J Agric For Natl Chiayi Univ 13:81–93. [Google Scholar]
- 67.Davis GH. 1955. The classification of lactobacilli from the human mouth. J Gen Microbiol 13:481–493. 10.1099/00221287-13-3-481. [DOI] [PubMed] [Google Scholar]
- 68.Kandler O, Stetter K-O, Köhl R. 1980. Lactobacillus reuteri sp. nov., a new species of heterofermentative lactobacilli. Zentralblatt Für Bakteriologie: I Abt Originale C: Allgemeine Angewandte Und Ökologische Mikrobiologie 1:264–269. 10.1016/S0172-5564(80)80007-8. [DOI] [Google Scholar]
- 69.Gordon RE, Haynes WC, Pang CH-N. 1973. The genus Bacillus. Agricultural Research Service, US Department of Agriculture, Washington, DC. [Google Scholar]
- 70.Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190:6983–6995. 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koo BM, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, Wapinski I, Galardini M, Cabal A, Peters JM, Hachmann AB, Rudner DZ, Allen KN, Typas A, Gross CA. 2017. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst 4:291.e7–305.e7. 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seki H, Shiohara M, Matsumura T, Miyagawa N, Tanaka M, Komiyama A, Kurata S. 2003. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr Int 45:86–90. 10.1046/j.1442-200x.2003.01671.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S3, Table S1. Download AEM.00442-21-s0001.pdf, PDF file, 1.1 MB (1,008.9KB, pdf)
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.