ABSTRACT
Increasing resistance to antifungal therapy is an impediment to the effective treatment of fungal infections. Candida glabrata is an opportunistic human fungal pathogen that is inherently less susceptible to cost-effective azole antifungals. Gain-of-function mutations in the Zn-finger pleiotropic drug resistance transcriptional activator-encoding gene CgPDR1 are the most prevalent causes of azole resistance in clinical settings. CgPDR1 is also transcriptionally activated upon azole exposure; however, factors governing CgPDR1 gene expression are not yet fully understood. Here, we have uncovered a novel role for two FK506-binding proteins, CgFpr3 and CgFpr4, in the regulation of the CgPDR1 regulon. We show that CgFpr3 and CgFpr4 possess a peptidyl-prolyl isomerase domain and act redundantly to control CgPDR1 expression, as a Cgfpr3Δ4Δ mutant displayed elevated expression of the CgPDR1 gene along with overexpression of its target genes, CgCDR1, CgCDR2, and CgSNQ2, which code for ATP-binding cassette multidrug transporters. Furthermore, CgFpr3 and CgFpr4 are required for the maintenance of histone H3 and H4 protein levels, and fluconazole exposure leads to elevated H3 and H4 protein levels. Consistent with the role of histone proteins in azole resistance, disruption of genes coding for the histone demethylase CgRph1 and the histone H3K36-specific methyltransferase CgSet2 leads to increased and decreased susceptibility to fluconazole, respectively, with the Cgrph1Δ mutant displaying significantly lower basal expression levels of the CgPDR1 and CgCDR1 genes. These data underscore a hitherto unknown role of histone methylation in modulating the most common azole antifungal resistance mechanism. Altogether, our findings establish a link between CgFpr-mediated histone homeostasis and CgPDR1 gene expression and implicate CgFpr in the virulence of C. glabrata.
KEYWORDS: human fungal pathogens, histone modifications, histone chaperones, histone H3 lysine 36 methylation, FK506-binding protein, antifungal drug resistance, multidrug efflux pump expression, H3K36me3
INTRODUCTION
Successful treatment of fungal bloodstream infections (BSIs) is often restricted by the availability of a limited number of antifungal drugs and emerging resistance to existing antifungals (1–3). Polyenes, azoles, and echinocandins represent three major classes of antifungal drugs that are currently being used worldwide to treat fungal BSIs (4). Azoles block ergosterol biosynthesis by inhibiting the lanosterol 14-alpha-demethylase enzyme encoded by the ERG11 gene, while echinocandins impede the synthesis of 1,3-β-d-glucan in the fungal cell wall by targeting the β-glucan synthase catalytic subunit encoded by the FKS genes (4). The polyene antifungals bind to ergosterol in the fungal cell membrane and cause disruption of cell membrane integrity and/or ergosterol extraction from the cell membrane (4, 5). Resistance to azoles and echinocandin antifungal drugs is increasingly being reported across the world (1, 2, 6).
Candida species contribute substantially to fungal BSIs, with Candida albicans as the leading etiological agent (6–8). Candida glabrata is the second to fourth leading cause of candidemia around the world, whose prevalence has increased over the last 2 decades (6, 9–11). Many factors, including prior fluconazole exposure, older age, and geographical region, have been associated with the increased prevalence of C. glabrata (6, 9, 11). C. glabrata is intrinsically less susceptible to azole antifungals, and about 10% of C. glabrata isolates have been reported to be associated with fluconazole resistance in clinical settings (1, 6).
Azole resistance in clinical isolates of C. glabrata majorly arises from single-amino-acid substitution gain-of-function mutations in the CgPDR1 gene (12–14), which codes for a Zn2Cys6 binuclear zinc cluster domain-containing transcription factor (15–17). A variety of nonsynonymous mutations in CgPDR1 have been reported, which are associated with both azole resistance and increased virulence (12–14). CgPdr1 activates the expression of its target genes, including CgCDR1 and CgCDR2, by binding to pleiotropic drug response elements (PDREs) present in target gene promoters (16–18). CgCDR1 and CgCDR2 encode multidrug efflux pumps belonging to the ATP-binding cassette (ABC) transporter family (13, 19, 20). CgSNQ2 and EPA1, coding for an ABC transporter and an epithelial cell adhesin, are also target genes of CgPdr1 (18, 21, 22). Multidrug transporters mediate the efflux of azole drugs, thereby reducing the intracellular azole concentration and leading to drug resistance (13).
Echinocandin resistance in clinical isolates of C. glabrata has primarily been due to mutations in the CgFKS1 and CgFKS2 genes and is rising (1, 6, 23). Furthermore, up to 8% of azole-resistant isolates were found to display coresistance to echinocandins during the 2006–2016 SENTRY antifungal surveillance program (6). Recently, a new mechanism imparting multidrug resistance (MDR) in C. glabrata clinical isolates has been ascribed to mutations in the DNA mismatch repair gene CgMSH2 (24); however, CgMSH2 mutations were not always found to be associated with antifungal multidrug resistance (25–27).
Chromatin-dependent processes are increasingly being investigated for their role in fungal pathogenesis and antifungal drug resistance (28, 29). Acetylation and deacetylation of lysines in proteins, including histones, have been shown to be important for antifungal resistance (28, 30). Consistently, CgHst1 deacetylase in C. glabrata has been shown to negatively regulate CgPDR1 expression, with CgHST1 gene loss rendering cells azole resistant (31). Furthermore, HDAC (histone deacetylase) inhibitors have been reported to exhibit synergistic effects with azoles in the treatment of fungal infections (32).
In the current study, we have uncovered a novel role for two FK506-binding proteins (FKBPs), CgFpr3 and CgFpr4, which possess a peptidyl-prolyl cis-trans isomerase (PPIase) domain, in the regulation of histone H3 and H4 protein levels and azole antifungal resistance. Furthermore, while the Cgfpr3Δ4Δ mutant displayed increased basal expression of the CgPDR1 and CgCDR1 genes and consequent resistance to fluconazole, the loss of the histone demethylase CgRph1 resulted in increased susceptibility to fluconazole and diminished basal expression of the CgPDR1 and CgCDR1 genes. We also demonstrate for the first time that CgSet2 is a histone H3 lysine 36-specific methyltransferase, which negatively regulates azole resistance. Altogether, our findings unravel a whole new epigenetic layer of regulation of CgPDR1 and CgCDR1 gene expression that may be governed by the levels and posttranslational modifications (PTMs) of histones H3 and H4 in fungal cells.
RESULTS
CgFpr3 and CgFpr4 proteins contain a peptidyl-prolyl cis-trans isomerase domain at their C termini.
We have previously shown that the ability to remodel its chromatin contributes to the intracellular proliferation of C. glabrata, and mutants disrupted for genes involved in chromatin organization displayed decreased survival in human THP-1 macrophages (33). Of 7 chromatin organization-defective mutants identified in a screen for diminished survival/replication in macrophages, one carried a Tn7 insertion in the CgFPR4 (CAGL0M00638g) gene. The CgFPR4 gene is uncharacterized in C. glabrata; however, its ortholog in Saccharomyces cerevisiae codes for a nuclear FK506-binding protein, which possesses peptidyl-prolyl cis-trans isomerase (PPIase) activity (34, 35). Fpr4 in S. cerevisiae modulates lysine methylation of histones H3 and H4 by catalyzing the isomerization of proline residues in H3 and H4, acts as an acidic histone chaperone, and is implicated in histone homeostasis (36, 37). Since a reduced histone H4 gene dosage has recently been linked with elevated resistance to DNA damage (38), we sought to examine the role of the CgFPR4 gene in stress resistance and virulence in C. glabrata. In silico analysis identified the FKBP_C peptidyl-prolyl cis-trans isomerase and the nucleoplasmin-like domain (characteristic structural domain of the nucleoplasmin core domain superfamily proteins that bind to core histones and are pivotal to the assembly of nucleosomal arrays [39]) in the CgFpr4 protein (see Fig. S1A in the supplemental material). Since Fpr3 and Fpr4 constitute a paralog pair in S. cerevisiae and regulate the proteolysis of the centromeric histone H3 variant Cse4 (40), we next identified the ortholog of S. cerevisiae FPR3 in C. glabrata and found it to be the CAGL0L11484g open reading frame (ORF). The CgFPR3 and CgFPR4 gene loci are syntenic to the corresponding S. cerevisiae loci (Fig. S1B), and the CgFpr3 and CgFpr4 proteins showed 84% and 80% similarities to S. cerevisiae Fpr3 and Fpr4, respectively (Fig. S1C). Furthermore, similar to CgFpr4, CgFpr3 also contained the nucleoplasmin-like domain and the prolyl cis-trans isomerase domain at the N and C termini, respectively, with both proteins carrying a nuclear localization sequence in the midregion (Fig. S1A). This in silico analysis suggested that similar to their S. cerevisiae counterparts (36, 40, 41), CgFpr3 and CgFpr4 may have proline isomerase activity and may act as histone chaperones in C. glabrata.
CgFpr3 and CgFpr4 are required for maintenance of histone H3 and H4 protein levels.
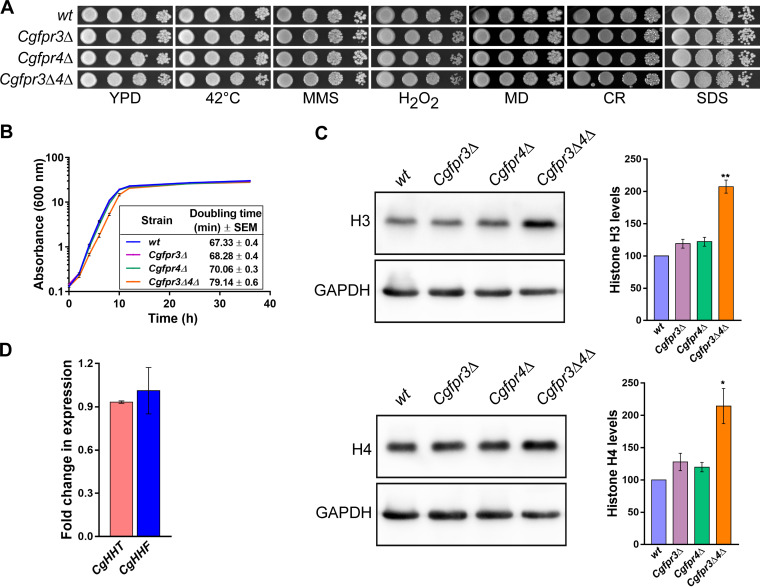
To study the role of CgFpr3 and CgFpr4 in histone homeostasis in C. glabrata, we created single-deletion strains lacking either the CgFPR3 or CgFPR4 gene as well as a double-deletion strain lacking both the CgFPR3 and CgFPR4 genes. Phenotypic characterization of the generated mutants revealed that CgFPR3 and CgFPR4 gene loss had no effect on the growth of C. glabrata under thermal (42°C), DNA damage (methyl methanesulfonate [alkylates DNA]), oxidative (hydrogen peroxide and menadione [produce reactive oxygen species]), cell wall (Congo red [inhibits the formation of two key components of the fungal cell wall, chitin and β-glucan]), and cell membrane (sodium dodecyl sulfate [SDS] [perturbs the plasma membrane]) stress conditions (Fig. 1A). Notably, the Cgfpr3Δ4Δ mutant exhibited a mild growth defect in rich yeast extract-peptone-dextrose (YPD) medium, with a doubling time of 79 min, compared to 67 min for the wild-type (wt) strain (Fig. 1B).
FIG 1.
The Cgfpr3Δ4Δ mutant contains elevated levels of histone H3 and H4 proteins. (A) Serial dilution spot analysis to assess the growth of the Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ mutants in the presence of different stressors. The indicated C. glabrata strains were grown overnight in YPD medium, and cultures were normalized to an OD600 of 1.0. Cultures were 10-fold serially diluted in PBS, and 3 μl of each dilution was spotted onto YPD medium lacking or containing the indicated compounds. Methyl methanesulfonate (MMS), hydrogen peroxide (H2O2), menadione (MD), Congo red (CR), and sodium dodecyl sulfate (SDS) were used at concentrations of 0.03%, 25 mM, 100 μM, 2 mg/ml, and 0.05%, respectively. All plates were incubated at 30°C unless indicated otherwise. Images were captured after 1 to 2 days of incubation. (B) Time course analysis of the wt, Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ strains. Cultures grown overnight in YPD medium were reinoculated into fresh YPD medium at an initial OD600 of 0.1 and incubated at 30°C. The absorbance of each culture was recorded at regular intervals until 36 h and plotted against time. Data represent means ± standard errors of the means (SEM) from 5 biological replicates. The growth period between 2 and 8 h, corresponding to the log phase of growth, was used to calculate the doubling time. Data represent means ± SEM from 5 biological replicates. Unpaired two-tailed Student’s t test was used to determine the statistical significance of doubling time differences between the wt and the Cgfpr3Δ4Δ mutant. ****, P ≤ 0.0001. (C) Representative immunoblots showing histone H3 and H4 levels in the indicated C. glabrata strains. Whole-cell extracts of log-phase cultures grown in YPD medium were prepared by the glass bead lysis method. Fifty micrograms of protein was resolved on a 15% SDS-PAGE gel and probed with anti-H3, anti-H4, and anti-GAPDH antibodies. CgGapdh was used as a loading control. For quantification, ImageJ densitometry software was used to measure the intensity of individual bands in 4 independent Western blots. The histone H3 and H4 signals were normalized to the corresponding CgGapdh signal. Data (means ± SEM; n = 4) represent percent changes in histone H3 and H4 levels in the CgfprΔ mutants compared to the wt strain (considered 100%) and are plotted as a bar graph on the right side of the blot images. *, P ≤ 0.05; **, P ≤ 0.01 (by paired two-tailed Student’s t test). (D) qPCR-based measurement of CgHHT (histone H3) and CgHHF (histone H4) transcript levels. Using the acid phenol method, total RNA was extracted from log-phase-grown wt and Cgfpr3Δ4Δ strains. Five hundred nanograms of the total RNA was used to set up real-time quantitative reverse transcriptase PCR. Transcript levels were quantified using the 2−ΔΔCT method. Data (means ± SEM; n = 3) were normalized against the CgTDH3 mRNA (which codes for GAPDH) control and represent fold changes in CgHHT and CgHHF expression in the Cgfpr3Δ4Δ mutant compared to the wt strain.
Next, we checked the levels of histones H3 and H4 in Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ mutants and found 2-fold-higher H3 and H4 levels in the Cgfpr3Δ4Δ mutant (Fig. 1C). Contrarily, H3 and H4 levels in single mutants were similar to those in the wt strain (Fig. 1C). These data suggest that CgFpr3 and CgFpr4 act redundantly to maintain the levels of two core histones, H3 and H4, in C. glabrata. Furthermore, to examine if CgFpr3 and CgFpr4 modulate the transcription of H3 and H4 genes, we performed quantitative real-time reverse transcriptase PCR (qPCR) to determine histone H3 (CgHHT) and H4 (CgHHF) transcript levels in log-phase cells. We found that the expression levels of both genes were similar in the Cgfpr3Δ4Δ and wt strains (Fig. 1D). These data suggest that CgFpr3 and CgFpr4 regulate histone H3 and H4 expression posttranscriptionally.
CgFPR3 and CgFPR4 genes are required for survival of C. glabrata in mice.
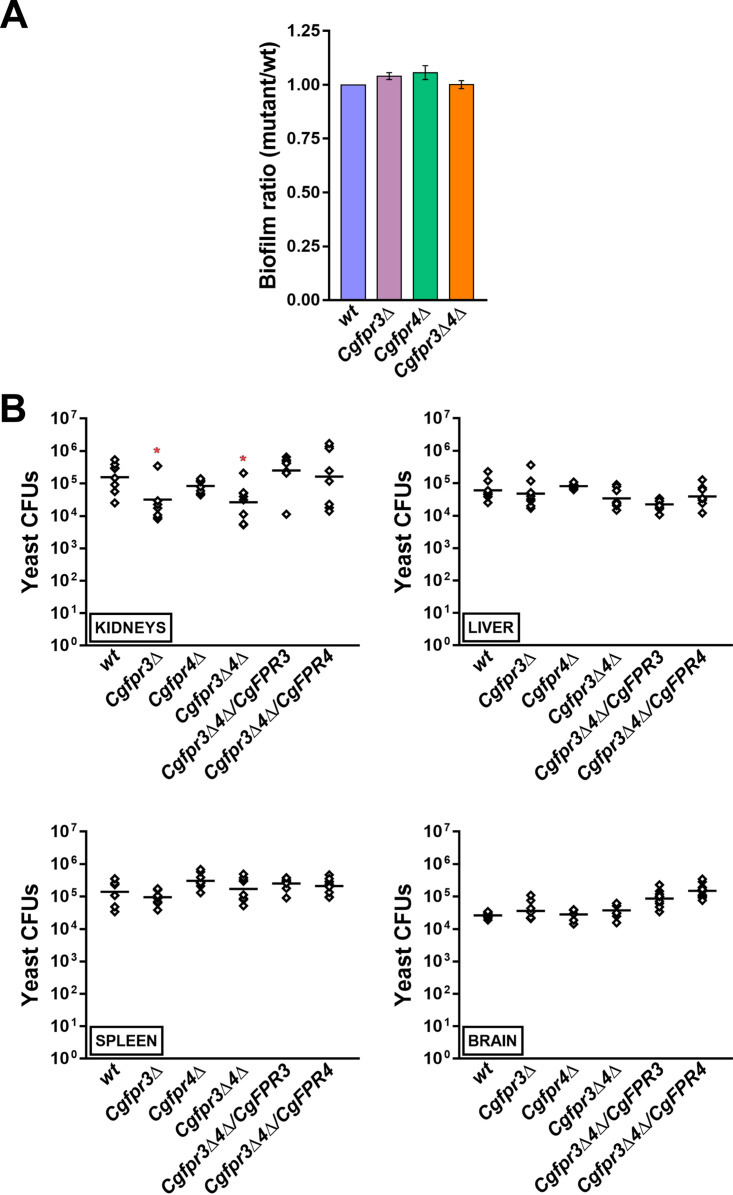
Reduced histone H4 levels have recently been associated with increased biofilm formation and diminished survival of C. glabrata in kidneys of mice (38). Since the Cgfpr3Δ4Δ mutant contained higher levels of histones H3 and H4 (Fig. 1C), we decided to determine the role of CgFpr3 and CgFpr4 in pathogenesis. For this, we studied two virulence-associated traits, viz., biofilm formation and survival, in the murine model of disseminated candidiasis. For the biofilm formation assay, we grew wt and Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ mutant cells in RPMI 1640 medium under static conditions and measured their ability to form biofilms on polystyrene-coated plates by a crystal violet stain-based assay. We found that all four strains, wt, Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ, produced similar amounts of biofilm (Fig. 2A), thereby suggesting that CgFpr3 and CgFpr4 are not required for biofilm formation in C. glabrata.
FIG 2.
CgFPR3 and CgFPR4 are required for virulence. (A) Biofilm formation assay. The wt, Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ strains were grown in RPMI 1640 medium containing 10% FBS in a 24-well polystyrene plate. After 48 h of incubation, the biofilm formed by yeast cells on polystyrene was stained with 0.4% crystal violet for 45 min, followed by three PBS washes. After destaining with 95% ethanol, the biofilm mass was measured by monitoring the absorbance at 595 nm. Data (means ± SEM; n = 3 to 4) represent biofilm ratios, which were calculated by dividing the absorbance units of mutants by those of the wt strain (considered 1.0). (B) Mouse infection assay. BALB/c mice were infected with the indicated C. glabrata strains intravenously and sacrificed 7 days after infection. Four organs, kidneys, liver, spleen, and brain, were harvested and homogenized in PBS. The homogenates were appropriately diluted and plated onto YPD medium containing penicillin and streptomycin. The CFU recovered from each organ of the individual mice are plotted. The individual mouse organ CFU are represented by diamonds, while bars mark the CFU geometric means (n = 8 to 9) for each organ. *, P < 0.05 (by a Mann-Whitney U test).
For in vivo virulence analysis, we infected BALB/c mice with the wt, Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ strains through tail vein injections and determined fungal survival in four target organs, kidneys, liver, spleen, and brain, by a CFU-based assay. We found similar organ fungal burdens in mice infected with the wt and Cgfpr4Δ strains (Fig. 2B). In contrast, the Cgfpr3Δ and Cgfpr3Δ4Δ mutants exhibited about 5-fold fewer CFU in the kidneys of infected mice than in those of wt-infected mice (Fig. 2B). Importantly, the ectopic expression of either CgFpr3 or CgFpr4 could rescue the survival defect of the Cgfpr3Δ4Δ mutant (Fig. 2B), indicating functional redundancy between CgFpr3 and CgFpr4 in vivo and implicating CgFpr3 and CgFpr4 in the virulence of C. glabrata in an organ-dependent manner. In light of these data and the previously reported link between reduced histone H4 gene dosage and diminished virulence (38), it is unlikely that the histone H4 levels per se contribute to the fitness of C. glabrata in vivo.
CgFPR3 and CgFPR4 gene loss led to elevated basal expression of multidrug transporter genes and azole resistance.
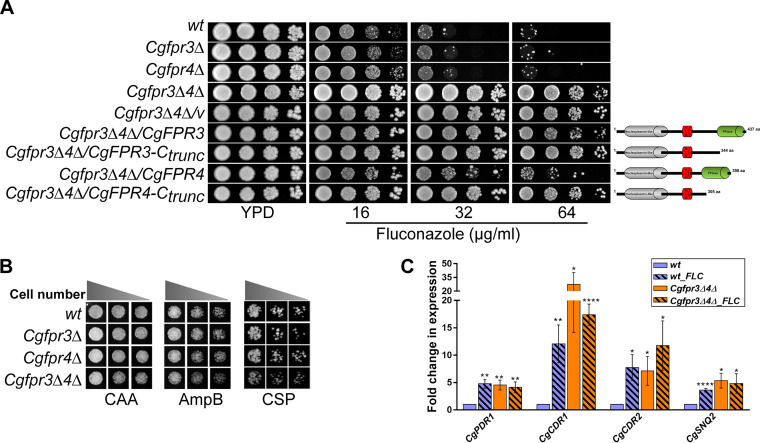
As mechanisms governing chromatin architecture and histone lysine acetylation have recently been implicated in resistance to antifungals (28, 29), we next investigated the role of CgFpr3 and CgFpr4 in antifungal drug resistance. For this, we checked the sensitivities of the Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ mutants to three classes of antifungal drugs, viz., azoles, echinocandins, and polyenes. We found that compared to wt cells, the Cgfpr3Δ and Cgfpr4Δ mutants grew slightly better in the presence of the azole antifungal fluconazole (Fig. 3A), while their growth in medium containing the drug amphotericin B (polyene) or caspofungin (echinocandin) was similar to that of the wt strain (Fig. 3B). Intriguingly, the Cgfpr3Δ4Δ double mutant exhibited a high level of azole resistance, with the mutant exhibiting robust growth in medium containing 64 μg/ml fluconazole (Fig. 3A). Notably, the MIC80s of fluconazole were found to be 16, 16, 16, and 64 μg/ml for the wt, Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ strains, respectively (Table S1), indicating that the deletion of the CgFPR3 or CgFPR4 gene individually had no effect on the fluconazole susceptibility of C. glabrata.
FIG 3.
The Cgfpr3Δ4Δ mutant is resistant to fluconazole. (A) Serial dilution spotting analysis indicating the fluconazole susceptibility of the indicated C. glabrata strains. The domain compositions of full-length and C-terminally truncated CgFpr3 and CgFpr4 proteins are shown schematically on the right side of the spotting image. NLS, nuclear localization signal. (B) Liquid medium-based growth analysis of the indicated C. glabrata strains to assess sensitivity to amphotericin B and caspofungin. wt and CgfprΔ mutant strains were grown in Casamino acids medium lacking (CAA) or containing amphotericin B (AmpB) (500 ng/ml) and caspofungin (CSP) (125 ng/ml) at 30°C. After 24 h of incubation, cultures were diluted in PBS, and 3 μl of 100-, 250-, and 500-fold-diluted cultures was spotted onto YPD medium. Images were captured after 1 day of growth at 30°C. (C) qPCR-based quantification of CgCDR1, CgCDR2, CgPDR1, and CgSNQ2 transcript levels. Log-phase wt and Cgfpr3Δ4Δ cells were either treated with 16 μg/ml fluconazole for 4 h or left untreated. RNA was extracted, and qPCR was set up using 500 ng of total RNA. Transcript levels were quantified using the 2−ΔΔCT method. Data (means ± SEM; n = 3 to 5) were normalized against the CgTDH3 mRNA as a control and represent fold changes in the expression of the CgCDR1, CgCDR2, CgPDR1, and CgSNQ2 genes in untreated Cgfpr3Δ4Δ and fluconazole (FLC)-treated wt and Cgfpr3Δ4Δ cells, compared to the untreated wt cells (taken as 1.0). *, P ≤ 0.0332; **, P ≤ 0.0021; ****, P ≤ 0.0001 (by multiple t tests).
Importantly, the ectopic expression of the CgFPR3 or CgFPR4 gene reverted the fluconazole resistance phenotype of the Cgfpr3Δ4Δ mutant slightly or substantially, respectively (Fig. 3A). The inability of the CgFpr3 and CgFpr4 proteins to fully complement the fluconazole resistance of the Cgfpr3Δ4Δ mutant could be due to their inadequate expression, functional alterations owing to the C-terminal fusion with green fluorescent protein (GFP), or both proteins being required for each other’s full activity. Next, to determine the importance of the PPIase domain of CgFpr3 and CgFpr4 in azole resistance, we generated C-terminally truncated CgFpr3 and CgFpr4 proteins that lacked the PPIase domain and first checked their expression, followed by their ability to complement the azole resistance phenotype of the Cgfpr3Δ4Δ mutant. Western analysis revealed that both the CgFpr3-Ctrunc and CgFpr4-Ctrunc proteins are expressed in the Cgfpr3Δ4Δ mutant (Fig. S2). However, a reversal of azole resistance in the Cgfpr3Δ4Δ mutant was not observed upon the expression of CgFpr3 and CgFpr4 proteins that lacked the C-terminal PPIase domain (Fig. 3A), thereby suggesting that the peptidyl-prolyl cis-trans isomerase activity of CgFpr3 and CgFpr4, catalyzing the isomerization between the cis and trans forms of peptide bonds, is pivotal to suppress azole resistance in the Cgfpr3Δ4Δ mutant. Collectively, these data suggest that CgFpr3 and CgFpr4 negatively regulate the response of C. glabrata to azole antifungals and are largely functionally redundant.
Furthermore, FK506 and fluconazole have previously been shown to act synergistically in C. glabrata (42). Therefore, we next checked the growth of the wt and the Cgfpr3Δfpr4Δ mutant in the presence of FK506 and fluconazole. Consistent with the previous report (42), we found a synergistic antifungal effect of fluconazole and FK506 on the growth of wt cells, while the growth of the Cgfpr3Δfpr4Δ mutant was only mildly retarded (Fig. S3). These results suggest that either the azole resistance in the Cgfpr3Δfpr4Δ mutant is refractory to the combinatorial inhibitory effect of fluconazole and FK506 or the CgFpr3 and CgFpr4 proteins are required for the synergistic effect of fluconazole and FK506.
Azole exposure in C. glabrata leads to the overexpression of the Zn2Cys6-type zinc finger motif-containing transcriptional regulator CgPdr1, which in turn activates the expression of the ATP-binding cassette multidrug transporter-encoding genes CgCDR1, CgCDR2, and CgSNQ2 (13). As expected, C. glabrata wt cells responded to fluconazole exposure by elevating the expression of the CgPDR1, CgCDR1, CgCDR2, and CgSNQ2 genes by 4- to 12-fold (Fig. 3C). Intriguingly, the Cgfpr3Δ4Δ mutant exhibited 4.5-fold-higher basal expression levels of the CgPDR1 gene (Fig. 3C). Consistent with this, basal transcript levels of the CgCDR1, CgCDR2, and CgSNQ2 genes, which are target genes of CgPdr1, were 5- to 27-fold higher in the Cgfpr3Δ4Δ mutant (Fig. 3C). Notably, fluconazole exposure led to no significant increase in CgPDR1, CgCDR1, CgCDR2, and CgSNQ2 gene expression in the Cgfpr3Δ4Δ mutant (Fig. 3C), underscoring the lack of fluconazole-induced MDR gene activation in the mutant cells. Altogether, the much higher constitutive levels of the CgPDR1, CgCDR1, CgCDR2, and CgSNQ2 genes in the Cgfpr3Δ4Δ mutant suggest that the CgFPR3 and CgFPR4 genes negatively regulate CgPdr1-dependent MDR gene expression under regular growth conditions.
Fluconazole exposure led to an increase in histone H3 and H4 protein levels.
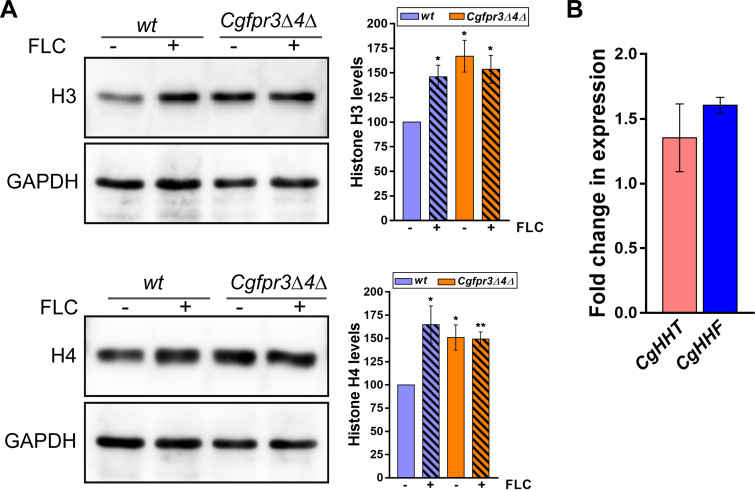
The Cgfpr3Δ4Δ mutant contained elevated histone H3 and H4 protein levels and exhibited higher MDR gene expression levels. Therefore, to further delineate the basis underlying the azole resistance phenotype of the Cgfpr3Δ4Δ mutant, we checked whether fluconazole exposure modulates histone protein levels in C. glabrata. Western analysis revealed that histone H3 and H4 levels were 50 to 60% higher in fluconazole-treated wt cells than in untreated wt cells (Fig. 4A). Contrarily, the Cgfpr3Δ4Δ mutant did not respond to fluconazole exposure by elevating H3 and H4 levels (Fig. 4A). Of note, histone H4 protein levels are known to be very tightly regulated in C. glabrata (38). Importantly, fluconazole treatment had no effect on the transcript levels of the histone H3 and H4 genes in wt cells (Fig. 4B), thereby ruling out fluconazole-responsive transcriptional regulation of H3 and H4 genes. Altogether, these data suggest that C. glabrata responds to fluconazole exposure by stabilizing histone H3 and H4 proteins, and the inability of the Cgfpr3Δ4Δ mutant to augment H3 and H4 upon fluconazole exposure could be due to already elevated H3 and H4 levels in the mutant. In this context, it is noteworthy that cells employ finely tuned regulatory mechanisms to ensure that histone H3 and H4 protein levels are tightly regulated (43).
FIG 4.
Histone H3 and H4 protein levels are elevated upon fluconazole treatment. (A) Representative immunoblots showing histone H3 and H4 protein levels in fluconazole-treated wt and Cgfpr3Δ4Δ cells. Log-phase wt and Cgfpr3Δ4Δ cells were either treated with 16 μg/ml fluconazole for 4 h or left untreated. Whole-cell lysates were prepared by glass bead lysis, and 50 μg protein was resolved on 15% SDS-PAGE gels and probed with anti-H3, anti-H4, and anti-GAPDH antibodies. CgGapdh was used as a loading control. The intensity of individual bands from 4 to 5 independent Western blots was quantified using ImageJ densitometry software. H3 and H4 intensity signals were normalized to the corresponding CgGapdh signal. Data (means ± SEM; n = 4 to 5) represent the percent changes in histone levels in untreated Cgfpr3Δ4Δ and fluconazole (FLC)-treated wt and Cgfpr3Δ4Δ cells, compared to the untreated wt cells (considered 100%), and are plotted as a bar graph on the right side of the blot image. *, P ≤ 0.05; **, P ≤ 0.01 (by paired two-tailed Student’s t test). (B) qPCR-based analysis of CgHHT and CgHHF gene expression in wt cells upon fluconazole exposure. Log-phase wt cells were either left untreated or treated with 16 μg/ml fluconazole for 4 h. Total RNA (500 ng) was used to set up qPCR, and transcript levels were quantified using the 2−ΔΔCT method. Data (means ± SEM; n = 3) were normalized against the CgTDH3 mRNA control and represent fold changes in CgHHT and CgHHF expression in fluconazole-treated wt cells compared to untreated wt cells (taken as 1.0).
The mutant lacking the histone demethylase CgRph1 displays diminished expression of MDR genes.
Since histone protein stability and functions are modulated by posttranslational modifications (PTMs) (44), we reasoned that CgFpr3 and CgFpr4 may contribute to the maintenance of H3 and H4 levels by regulating their PTMs and that there could be a link between histone PTMs and the azole resistance phenotype of the Cgfpr3Δ4Δ mutant. In this context, it is noteworthy that disruption of the CgSET2 (which encodes a putative histone methyltransferase) and CgRPH1 (which encodes a putative histone demethylase) genes has recently been reported to result in azole resistance and sensitivity, respectively (45, 46). Importantly, Set2 and Rph1 in S. cerevisiae are involved in the methylation and demethylation of histone H3 at lysine residue 36, respectively (47, 48). Furthermore, Fpr4-mediated isomerization of proline residue 38 of histone H3 has been reported to be inhibitory for Set2-mediated methylation of H3 at lysine residue 36 (36). Therefore, to examine the role of histone H3 methylation in azole resistance in C. glabrata, we analyzed CgRph1 and CgSet2 protein sequences for the presence of different domains and found that CgRph1 (CAGL0L11880p), a 980-amino-acid (aa) protein, possesses the JmjC domain, involved in histone demethylation reactions, at its N terminus (Fig. S4A). Furthermore, similar to its S. cerevisiae ortholog, the CgSet2 protein contained the SET domain, involved in the methylation of lysine residues, at its N terminus (Fig. S4B).
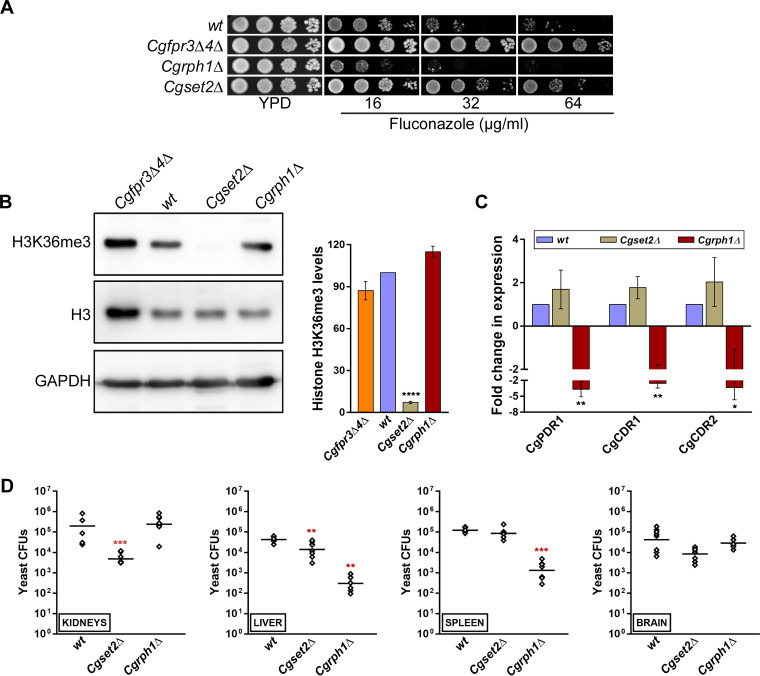
Next, we generated deletion strains for the CgSET2 and CgRPH1 genes, and phenotypic analysis revealed that the Cgrph1Δ and Cgset2Δ mutants were moderately sensitive and resistant, respectively, to fluconazole (Fig. 5A), in accordance with previous reports (45, 46). The MIC80s of fluconazole were found to be 8 and 32 μg/ml for the Cgrph1Δ and Cgset2Δ mutants, respectively (Table S2). We also checked the growth of the Cgrph1Δ and Cgset2Δ mutants under thermal stress (42°C), DNA damage (methyl methanesulfonate), oxidative stress (hydrogen peroxide and menadione), cell wall stress (Congo red), and cell membrane stress (sodium dodecyl sulfate) conditions and found that the Cgset2Δ mutant exhibited elevated susceptibility to SDS stress (Fig. S5). Since Fpr4 restrains the trimethylation of H3K36 in S. cerevisiae (36), we next examined the status of the H3K36me3 modification in the Cgset2Δ mutant. Western analysis showed that H3K36me3 was largely absent in the Cgset2Δ mutant (Fig. 5B), similar to its S. cerevisiae counterpart (47), thereby implicating CgSet2 in trimethylation at the H3K36 residue in C. glabrata. Of note, appreciable differences in H3K36me3 levels were not observed between the wt and Cgrph1Δ strains (Fig. 5B), thereby precluding a role for CgRph1 in controlling global levels of H3K36me3 modification.
FIG 5.
CgSet2 is a histone H3 lysine 36 methyltransferase. (A) Serial dilution spot analysis illustrating the fluconazole susceptibility of the Cgrph1Δ and Cgset2Δ mutants. Visible artifacts appear within the panels upon overexposure. (B) Representative immunoblots showing H3K36me3 modification levels in the indicated C. glabrata strains. Whole-cell lysates of YPD-grown log-phase cultures were prepared, and 50 μg protein was resolved on a 15% SDS-PAGE gel and probed with anti-H3K36me3, anti-H3, and anti-GAPDH antibodies. GAPDH serves as a loading control. For quantification, the intensity of individual bands from 3 to 4 independent Western blots was measured using ImageJ densitometry software. The histone H3K36me3 signal was normalized to the corresponding total histone H3 signal for each strain. Data (means ± SEM; n = 3 to 4) represent the percent changes in histone H3K36me3 levels in mutants compared to the wt strain (considered 100%) and are presented as a bar graph on the right side of the blot image. ****, P ≤ 0.0001 (by paired two-tailed Student’s t test). (C) qPCR-based analysis of CgPDR1, CgCDR1, and CgCDR2 transcript levels in the wt, Cgset2Δ, and Cgrph1Δ strains. Data (means ± SEM; n = 3 to 4) were normalized against the CgTDH3 mRNA as a control and represent fold changes in the expression of the CgPDR1, CgCDR1, and CgCDR2 genes in the Cgset2Δ and Cgrph1Δ mutants, compared to the wt strain (considered 1.0). *, P ≤ 0.05; **, P ≤ 0.01 (by paired two-tailed Student’s t test). (D) Organ fungal burden in BALB/c mice infected intravenously with wt, Cgset2Δ, or Cgrph1Δ C. glabrata cells. After 7 days of infection, the fungal burden in the indicated mouse organs was determined by organ collection and homogenization, followed by homogenate plating onto YPD medium containing penicillin and streptomycin. Diamonds and bars represent CFU recovered from organs of the individual mice and the CFU geometric mean (n = 6 to 8) for each organ, respectively. **, P ≤ 0.01; ***, P < 0.001 (by a Mann-Whitney U test).
Furthermore, to delineate the molecular basis underlying the differential azole susceptibilities of the Cgrph1Δ and Cgset2Δ mutants, we next quantified the CgPDR1, CgCDR1, and CgCDR2 transcript levels in the mutants. Quantitative real-time reverse transcriptase PCR revealed an ∼2.6- to 3.7-fold downregulation of the CgPDR1, CgCDR1, and CgCDR2 genes in the Cgrph1Δ mutant (Fig. 5C), which may account for the increased fluconazole susceptibility of the Cgrph1Δ mutant. However, despite the fluconazole resistance phenotype of the Cgset2Δ mutant, differences in CgPDR1, CgCDR1, and CgCDR2 transcript levels between the wt and Cgset2Δ strains were not statistically significant (Fig. 5C), suggesting that the global trimethylation at histone H3 lysine residue 36 does not regulate MDR gene expression appreciably in C. glabrata. Thus, the molecular basis underlying azole resistance in the Cgset2Δ mutant is yet to be elucidated.
The CgPdr1 transcriptional activator is also implicated in the expression regulation of virulence-related genes (12, 18, 22). Since the Cgrph1Δ mutant had lower CgPDR1 gene expression levels, we next examined the virulence potential of the Cgrph1Δ and Cgset2Δ mutants in the murine model of systemic candidiasis. We found 16- and 3-fold-lower fungal CFU in kidneys and liver, respectively, of Cgset2Δ mutant-infected mice than in wt-infected mice (Fig. 5D). In contrast, mice infected with the Cgrph1Δ mutant had 140- and 90-fold-lower fungal burdens in liver and spleen, respectively (Fig. 5D). Notably, no significant differences in C. glabrata CFU were observed in the brains of mice infected with the wt, Cgrph1Δ, or Cgset2Δ strain (Fig. 5D). Altogether, these data implicate for the first time the histone demethylase CgRph1 and the histone H3K36 methyltransferase CgSet2 in the survival of C. glabrata in mice.
CgCDR1 deletion led to reversal of fluconazole resistance in the Cgfpr3Δ4Δ mutant.
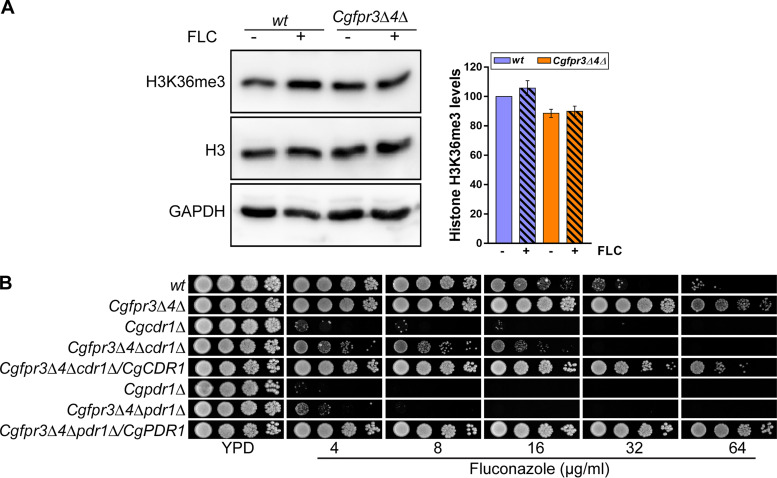
Next, to corroborate that global H3K36 trimethylation levels do not modulate CgPDR1-dependent fluconazole resistance, we checked the status of H3K36me3 modification in the wt and azole-resistant Cgfpr3Δ4Δ mutant strains. Notably, the mutant also had elevated basal expression of the CgPDR1 and CgCDR1 genes (Fig. 3C). Western analysis revealed similar basal levels of H3K36me3 in wt and Cgfpr3Δ4Δ cells (Fig. 6A). Importantly, fluconazole treatment led to no significant change in H3K36me3 levels in either strain (Fig. 6A). Collectively, these data suggest that the cellular response to fluconazole does not involve changes in the global trimethylation of histone H3 at lysine residue 36 in C. glabrata.
FIG 6.
CgPDR1 or CgCDR1 disruption reverses azole resistance in the Cgfpr3Δ4Δ mutant. (A) Representative immunoblots showing H3K36me3 modification levels in the indicated C. glabrata strains. Log-phase cultures of wt and Cgfpr3Δ4Δ cells were either treated with 16 μg/ml fluconazole for 4 h or left untreated. Whole-cell lysates (50 μg protein) were resolved on 15% SDS-PAGE gels and probed with anti-H3K36me3, anti-H3, and anti-GAPDH antibodies. GAPDH serves as a loading control. The intensity signal in each lane was enumerated using ImageJ densitometry software, and the H3K36me3 methylation signal was normalized to the total H3 signal. Data (means ± SEM; n = 3 to 4) are plotted as a bar graph on the right side of the blot image and represent the percent changes in H3K36me3 levels in untreated Cgfpr3Δ4Δ and fluconazole-treated wt and Cgfpr3Δ4Δ cells, compared to untreated wt samples (considered 100%). (B) Serial dilution spot analysis to assess the fluconazole susceptibility of the indicated C. glabrata strains.
Finally, to demonstrate that the higher basal levels of expression of the CgPDR1 and CgCDR1 genes contribute to azole resistance in the Cgfpr3Δ4Δ mutant, we generated two triple-deletion strains by deleting either the CgPDR1 or CgCDR1 gene in the Cgfpr3Δ4Δ mutant that lacked the genes encoding two FK506-binding histone chaperone proteins, CgFpr3 and CgFpr4, and checked their sensitivity to fluconazole. As a control, we used Cgpdr1Δ and Cgcdr1Δ mutants, which are supersensitive to fluconazole (Fig. 6B). The Cgfpr3Δ4Δ mutant showed robust growth in the presence of 64 μg/ml fluconazole, which was lost upon disruption of the CgPDR1 or CgCDR1 gene (Fig. 6B). Interestingly, while the Cgfpr3Δ4Δpdr1Δ triple mutant was unable to grow well even on 4 μg/ml fluconazole, the Cgfpr3Δ4Δcdr1Δ mutant exhibited some growth on medium containing 16 μg/ml fluconazole (Fig. 6B), thereby underscoring the contribution of other multidrug efflux pumps to CgPdr1-mediated azole resistance in the Cgfpr3Δ4Δ mutant. Importantly, the ectopic expression of CgPDR1 and CgCDR1 in the Cgfpr3Δ4Δpdr1Δ and Cgfpr3Δ4Δcdr1Δ triple mutants, respectively, restored the fluconazole resistance phenotype of the parental Cgfpr3Δ4Δ strain (Fig. 6B), indicating that CgPdr1 and CgCdr1 overexpression accounts for the azole resistance phenotype of the Cgfpr3Δ4Δ mutant. Altogether, these data unequivocally link the CgFpr3 and CgFpr4 proteins with the regulation of CgPDR1 and CgCDR1 gene expression in C. glabrata.
In conclusion, we have demonstrated a pivotal role for the first time for FK506-binding histone chaperones in azole resistance via regulation of CgPdr1-dependent MDR gene expression (schematically illustrated in Fig. 7) in C. glabrata. Additionally, our data point toward epigenetic control of azole antifungal resistance in clinical settings, with the overexpression of multidrug transporters being the most prevalent resistance mechanism.
FIG 7.
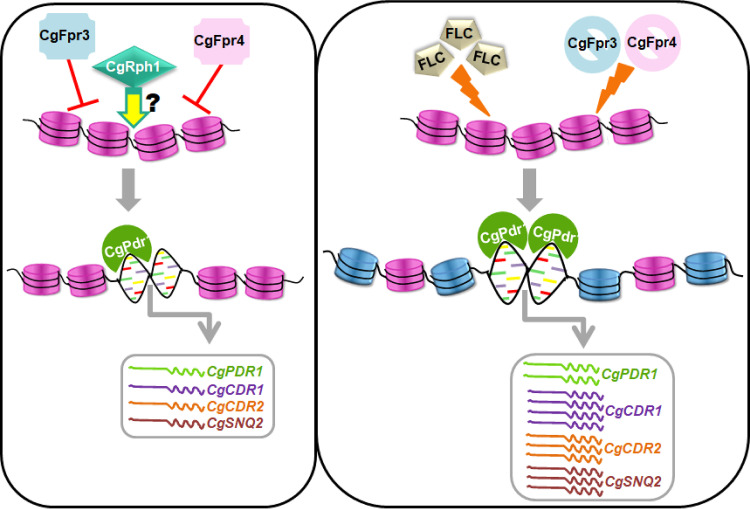
Schematic summarizing the key findings of the study. Under regular growth conditions, the FK506-binding proteins CgFpr3 and CgFpr4 maintain histone H3 and H4 levels and negatively regulate the CgPDR1 regulon, viz., the expression of the CgPDR1, CgCDR1, CgCDR2, and CgSNQ2 genes. Contrarily, CgRph1, a putative histone demethylase, positively regulates the CgPDR1 regulon. Of note, the underlying mechanism(s) is yet to be elucidated. Furthermore, in response to fluconazole exposure or upon the simultaneous deletion of the CgFPR3 and CgFPR4 genes, histone H3 and H4 protein levels are elevated, probably leading to differential acetylation and/or methylation of H3 and H4 (represented by blue nucleosomes), and the CgPDR1 regulon is activated, resulting in the increased expression of the CgPDR1, CgCDR1, CgCDR2, and CgSNQ2 genes. This activation of MDR genes is pivotal to the survival of fluconazole stress in C. glabrata. Of note, the loss of the CgFPR3 and CgFPR4 genes leading to an open chromatin state at the CgPDR1 promoter is yet to be demonstrated experimentally.
DISCUSSION
Invasive fungal infections are globally emerging as a significant challenge in hospitals worldwide (49). Treatment of C. glabrata infections is particularly difficult due to its intrinsic and acquired antifungal resistance (29, 49). As azole drugs still represent cost-effective, less toxic options for antifungal therapy, there is a compelling need to better understand azole resistance mechanisms and design resistance management strategies. The most clinically prevalent mechanism of azole resistance in C. glabrata involves the transcriptional activation of multidrug transporters through gain-of-function mutations in the master regulator-encoding gene CgPDR1 (12–14). Perturbation of chromatin modifications, including histone acetylation, has been proposed as a promising antifungal therapeutic strategy (28, 29), and HDAC inhibitors are known to act synergistically with azole antifungals (28, 29, 50). Despite this, the precise role of histones and histone modification proteins in antifungal resistance in C. glabrata remains to be explored (29). Toward this, here, we have elucidated the functions of two putative FK506-binding histone chaperones, CgFpr3 and CgFpr4, in the activation of CgPDR1 and its target genes CgCDR1, CgCDR2, and CgSNQ2. We show that CgFpr3 and CgFpr4 negatively regulate the expression of the CgPDR1 regulon and H3 and H4 protein levels. Overall, our work sheds light on the epigenetic networks that modulate the expression of the CgPDR1 gene, underscoring the multifaceted regulation of ABC transporter gene expression.
CgPdr1, the Cys6Zn2 DNA-binding motif-containing transcriptional regulator of pleiotropic drug resistance genes, is a major contributor to azole resistance via the control of both basal and azole-stimulated gene expression (16, 17). CgPdr1 is also known to regulate MDR gene expression by binding directly to azole drugs (51), and CgPdr1 loss results in azole susceptibility (16, 17). CgPdr1 is an autoactivator and regulates the expression of a spectrum of virulence genes, besides MDR genes (18, 22), which highlights its multifunctional roles. Consistent with this, CgPdr1 activity regulation is complex and multifaceted, with diminished CgERG11 levels and mitochondrial genome loss leading to increased CgPDR1 expression. Contrarily, the transcription factor CgStb5, the Hsp40 cochaperone CgJjj1, and the deubiquitinase subunit Bre5 act as negative regulators of CgPdr1 levels and/or functions (46, 52, 53). Additionally, the association of CgPdr1 with the CgGal11A subunit of the RNA polymerase II mediator complex also modulates its expression and activity (51, 54). Importantly, the NAD+-dependent histone deacetylase CgHst1 has been shown to be a repressor of CgPDR1 and CgCDR1 gene expression in C. glabrata (31). Our current findings add another regulatory layer of complexity to this circuitry and point toward histone chaperones also being key players in the CgPdr1-dependent cellular transcriptional response to fluconazole. Although how CgFpr3 and CgFpr4 govern CgPDR1 expression remains to be determined, one possible mechanism may include azole-stimulated differential recruitment of the regulatory mediator subunit complexes involving CgGal11A/Med15 to CgPDR1 target gene promoters (54).
An interesting aspect of our work is the increased and decreased susceptibility, arising from the loss of a putative histone demethylase, CgRph1, and H3K36 methyltransferase, CgSet2, respectively, to fluconazole. Although the nexus among CgRph1 and CgSet2 activity, H3K36me3 levels, CgFpr3/4-mediated histone homeostasis, and CgPDR1 regulon activation is yet to be established, the altered azole susceptibility of the Cgrph1Δ and Cgset2Δ mutants raises the possibility that cellular metabolism may govern azole stress survival. In this context, it is worth noting that a link between the availability of S-adenosylmethionine, which donates a methyl group during reactions catalyzed by methyltransferases, and histone methylation has been well established in higher eukaryotes (55, 56). Intriguingly, transcript levels of the S-adenosylmethionine synthetase-encoding gene SAM2 were found to be elevated in response to itraconazole treatment in C. albicans (57). Therefore, it is possible that the histone methylation-dependent regulation of the CgPDR1 gene is tightly intertwined with the cellular metabolic status that is perturbed upon azole exposure. Future studies will be designed to address this possibility as well as to examine other H3 and H4 methylation modifications in our fluconazole-sensitive (Cgrph1Δ) and -resistant (Cgset2Δ and Cgfpr3Δ4Δ) mutants, as current findings suggest that H3K36me3 is unlikely to be a pivotal determinant of azole resistance.
A multitude of histone modifications have been reported, and growing evidence points to an epigenetic regulation of drug resistance and virulence mechanisms in human-pathogenic fungi, with reversible lysine acetylation and methylation playing pivotal roles (28, 29). The loss of the catalytic subunit (NuB4) of the histone acetyltransferase complex Hat1 has previously been reported to result in elevated azole resistance (58), while the loss of the lysine acetyltransferase Gcn5 (catalytic subunit of the SAGA, SLIK, and ADA histone acetyltransferase complexes) had no effect on azole susceptibility in C. albicans (59). Furthermore, HDAC inhibitors are known to act synergistically with azole antifungals in C. albicans (50). In accordance, loss of the lysine deacetylases Rpd3 and Hda1 rendered C. albicans cells sensitive to azoles (60). However, compared to acetylation, histone methylation is an understudied PTM, and histone methyltransferases and demethylases are yet to be explored for their roles in azole resistance in human fungal pathogens. Of note, our finding that the Cgrph1Δ mutant (which lacks a putative histone demethylase) shows increased azole susceptibility raises the possibility of an analogous effect of histone acetylation and methylation on azole resistance. Furthermore, since cross talk between histone methylation and histone acetylation has previously been reported, with H3K36me3 stimulating the acetylation of histone H4 at the K16 residue (61), it is possible that the altered H3K36me3 levels may impact the status of histone acetylation in the Cgset2Δ mutant. Of note, histone methylation has also previously been associated with communicating the transcriptional memory of environmental stress responses through mitotic cell divisions in S. cerevisiae (62).
Finally, histone chaperones play an important role in chromatin homeostasis via direct binding to histone proteins and regulating the localization, protein levels, interaction, and DNA deposition of histone proteins (63). Histone chaperones belonging to the nucleoplasmin superfamily possess the 8-stranded beta barrel pentameric N-terminal core domain and bind to histones through a predominantly conserved mechanism (64). CgFpr3 and CgFpr4 represent nucleoplasmin-like proteins of this superfamily, and they contain a PPIase domain at their C termini (characteristic of the FKBP class of PPIase enzymes) and are yet to be functionally characterized in C. glabrata. S. cerevisiae Fpr4 has been shown to localize to the nucleus; bind to the H2B nuclear localization signal sequence; regulate ribosomal DNA (rDNA) silencing, lysine methylation, and gene expression; and act as an acidic histone chaperone in the assembly of nucleosomal arrays, with its PPIase domain inhibiting the histone chaperone activity (36, 37, 41, 65). Whereas the N-terminal tails of histones H3 and H4 are implicated in binding to Fpr4, the PPIase domain of Fpr4 is involved in the isomerization of proline residues 30 and 38 of H3 (36). Furthermore, S. cerevisiae Fpr3 is localized to the nucleolus, forms reversible aggregates upon thermal stress, and assists nucleosome assembly, and its PPIase domain serves as a transcriptional repressor (66–69). FPR3 and FPR4 gene deletion in S. cerevisiae is also known to result in the differential expression of a wide variety of genes, consistent with their products’ roles as histone chaperones (68). Although our data, taken together, suggest that CgFpr3 and CgFpr4 operate largely in a redundant manner and are likely to be functional orthologs of S. cerevisiae nuclear FKBPs, the mechanism(s) underlying the binding of CgFpr3 and CgFpr4 to histones H3 and H4 and the role of this binding in modulating azole resistance and the cellular status of histone PTMs are yet to be elucidated.
MATERIALS AND METHODS
Strains and media.
C. glabrata wild-type and mutant strains, which are derivatives of vaginal isolate BG2, were cultured in rich YPD or CAA (Casamino acids) medium at 30°C with shaking at 200 rpm. Bacterial strains were grown at 37°C in LB medium containing 60 μg/ml ampicillin. To obtain logarithmic-phase cells, C. glabrata strains were grown overnight in YPD or CAA medium and inoculated into fresh medium at an optical density at 600 nm (OD600) of 0.1. After 4 h of growth at 30°C, cultures were pelleted down, and cells were collected. The strains, plasmids, primers, and antibodies used are listed in Tables S3 to S6, respectively, in the supplemental material.
C. glabrata gene disruption and cloning.
Using the homologous-recombination-based approach, C. glabrata fpr3Δ, fpr4Δ, set2Δ, and rph1Δ strains were created with the nat1 gene (which confers nourseothricin resistance) as a selection marker, as described previously (70). Replacement of the disrupted ORF with the Flp recombination target (FRT)-nat1 cassette was confirmed by PCR. To create the double-deletion strain, the Cgfpr4Δ mutant was first transformed with the pRK70 plasmid, which expresses the flip recombinase (enzyme catalyzing recombination at FRT sites)-encoding gene. Transformants were selected for uracil prototrophy and screened for nourseothricin (200 μg/ml) sensitivity after colony purification. Since the nat1 gene was flipped out of nourseothricin-sensitive Cgfpr4Δ colonies, these colonies were grown in YPD medium for about 20 generations, followed by the selection of uracil auxotroph colonies. Disruption of the CgFPR3 gene in the ura−, nourseothricin-sensitive Cgfpr4Δ mutant strain was confirmed by PCR. To generate the triple-deletion strain Cgfpr3Δfpr4Δcdr1Δ, the cassette containing the nat1 gene flanked with the 5′ and 3′ untranslated regions (UTRs) of the CgCDR1 gene was amplified from genomic DNA of the Cgcdr1Δ mutant and transformed into the ura−, nourseothricin-sensitive Cgfpr3Δfpr4Δ mutant strain. For the generation of C-terminal GFP fusion proteins for mutant complementation studies, CgFPR3 (CAGL0L11484g) (1.31 kb) and CgFPR4 (CAGL0M00638g) (1.19 kb) were cloned into the pRK1018 plasmid between the PGK1 promoter and the GFP-encoding sequence in the XbaI/SpeI restriction sites. The clones were verified by PCR and restriction digestion, and the resultant plasmids were transformed into C. glabrata strains for complementation analyses.
Quantitative real-time PCR.
Log-phase cultures were inoculated at an OD600 of 0.1 in YPD medium lacking or containing fluconazole (16 μg/ml) and incubated for 4 h at 30°C in an incubator shaker. Cells were harvested and washed with ice-cold diethyl pyrocarbonate (DEPC)-treated water. Using the acid phenol extraction method, total RNA was extracted and treated with DNase I to remove any residual DNA. cDNA was synthesized by a reverse transcriptase enzyme (SuperScript III first-strand synthesis system for RT-PCR; Invitrogen) using 500 ng of DNase I-digested RNA. Quantitative real-time PCR (qPCR) was performed with the SYBR green qPCR master mix using primers specific for the CgCDR1, CgCDR2, CgPDR1, CgSNQ2, CgHHF, and CgHHT genes and the housekeeping gene CgTDH3. CgTDH3, which codes for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and whose expression was not altered upon fluconazole treatment, was used as the reference gene. CT (cycle threshold) values of the CgCDR1, CgCDR2, CgPDR1, CgSNQ2, CgHHF, and CgHHT genes were normalized against the corresponding CT value obtained for the CgTDH3 gene under similar conditions. The fold change in expression for fluconazole-treated samples compared to untreated samples was calculated by the comparative CT (2–ΔΔCT) method.
Stress susceptibility assays.
The susceptibility of C. glabrata strains to azole antifungals was evaluated in solid medium by a serial dilution spot assay. For serial dilution spotting analysis, cultures of C. glabrata strains grown overnight were normalized to an OD600 of 1.0 and serially diluted 10-fold in phosphate-buffered saline (PBS). Three microliters of each dilution was spotted onto YPD medium lacking or containing different concentrations of the azole antifungal fluconazole and other stressors. Plates were incubated at 30°C, and growth was recorded after 24 to 48 h. The liquid growth assay was performed in CAA medium lacking or containing amphotericin B and caspofungin in a 96-well plate. Each well was inoculated with a culture of C. glabrata strains grown overnight corresponding to an OD600 of 0.2 to a final volume of 100 μl and incubated for 24 h at 30°C. Next, 100-, 250-, and 500-fold culture dilutions were made, and 3 μl of each dilution was spotted onto YPD medium. The plates were incubated at 30°C, and images were captured between 16 and 48 h.
Growth curve analysis.
C. glabrata wild-type, Cgfpr3Δ, Cgfpr4Δ, and Cgfpr3Δ4Δ mutant strains were grown in 10 ml YPD broth for 16 h at 30°C, followed by inoculation at an OD600 of 0.1 in a 100 ml flask containing 20 ml YPD broth. Cultures were incubated in a shaker incubator at 30°C, and the absorbance was recorded at regular intervals until 36 h.
MIC determination.
The MIC was determined using the EUCAST method (71). Briefly, RPMI 1640 medium without sodium bicarbonate was prepared at a 2× concentration and supplemented with 2% glucose and 0.165 M morpholinepropanesulfonic acid (MOPS). The medium pH was adjusted to 7.0 with NaOH and filter sterilized. One hundred microliters of medium lacking or containing various fluconazole concentrations (4, 8, 16, 32, 64, and 128 μg/ml) was added to each well of a 96-well plate. C. glabrata wild-type and mutant strains were grown in YPD medium at 30°C at 200 rpm for 16 h and added at a density of 1 × 105 cells/well in the 96-well plate. After incubation at 37°C for 24 h in a moist container, the culture absorbance was recorded visually and measured at 530 nm in the SpectroMax multiplate reader. Endpoints were determined by comparing the OD530 values of C. glabrata cells grown in the absence and presence of fluconazole. The MIC80 of fluconazole was defined as the lowest drug concentration that inhibited 80% of a strain’s growth at 24 h, compared to the control without fluconazole.
Protein extraction and immunoblotting.
Log-phase cultures were inoculated at an OD600 of 0.1 in YPD medium lacking or containing fluconazole (16 μg/ml) and incubated for 4 h at 30°C in an incubator shaker. Cells were harvested, washed, and suspended in protein extraction buffer (50 mM Tris-HCl [pH 7.5], 2 mM EDTA, 2% glucose) containing 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium fluoride, 1 mM sodium orthovanadate, and a protease inhibitor mixture. Cells were lysed with glass beads using a Fastprep-24 instrument at maximum speed for 60 s five times and spun down at 13,000 rpm for 15 min at 4°C. The proteins were quantified using the bicinchoninic acid (BCA) protein assay kit, run on a 15% SDS-PAGE gel, and immunoblotted with the appropriate antibodies.
Biofilm formation assay.
C. glabrata cells were grown overnight in YPD medium, suspended in RPMI 1640 medium containing 10% fetal bovine serum (FBS), and seeded at a density of 1 × 107 cells per well in a 24-well polystyrene plate. After a 90 min incubation at 37°C, wells were washed twice with PBS, and fresh RPMI 1640 medium was added. The plate was incubated at 37°C for 48 h, with the removal of spent medium and the addition of RPMI 1640 medium after 24 h. The unbound C. glabrata cells were removed, and wells were washed three times with PBS. Crystal violet (0.4% [wt/vol]) stain was added to each well to stain the adherent C. glabrata cells. After 45 min, 95% ethanol was added for destaining purposes, followed by absorbance measurement of the destaining solution at 595 nm.
Mouse infection assay.
For animal infection, C. glabrata strains were grown overnight in YPD medium. After washing with PBS, a 100 μl cell suspension (4 × 107 cells) was injected into the tail vein of 6- to 8-week-old female BALB/c mice. On the 7th day postinfection, mice were sacrificed, and four organs, kidneys, liver, spleen, and brain, were harvested. After homogenizing organs in PBS, appropriate dilutions were plated on YPD medium containing penicillin and streptomycin, and the numbers of C. glabrata colonies that appeared after 2 days of incubation at 30°C were counted. Mouse infection procedures were designed to minimize animal suffering, performed at the Animal House Facility of the Centre for DNA Fingerprinting and Diagnostics (CDFD), Hyderabad, India, in accordance with guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals, Government of India; and approved by the Institutional Animal Ethics Committee (EAF/RK/CDFD/22).
ACKNOWLEDGMENTS
This work was supported by a DBT/Wellcome Trust India Alliance senior fellowship to R.K. (IA/S/15/1/501831 [www.indiaalliance.org/]) and grants from the Department of Biotechnology (BT/HRD/NBA/37/01/2014 [www.dbtindia.gov.in/]) and the Science and Engineering Research Board, Department of Science and Technology (EMR/2016/005375 [www.serb.gov.in/]), Government of India. K.K. is a recipient of a Shyama Prasad Mukherjee fellowship of the Council of Scientific and Industrial Research, New Delhi, India (www.csirhrdg.res.in/).
R.M. and K.K. conceived the idea. R.M., K.K., and R.K. designed the study. R.M. and K.K. performed experiments and acquired data. R.M., K.K., and R.K. analyzed data. R.M. and K.K. prepared figures. R.M., K.K., and R.K. wrote the manuscript.
We declare that we have no conflict of interest.
Footnotes
Supplemental material is available online only.
aac.02415-20-s000s1.pdf (4.3MB, pdf)
REFERENCES
- 1.Arendrup MC, Patterson TF. 2017. Multidrug-resistant candida: epidemiology, molecular mechanisms, and treatment. J Infect Dis 216:S445–S451. 10.1093/infdis/jix131. [DOI] [PubMed] [Google Scholar]
- 2.Kontoyiannis DP. 2017. Antifungal resistance: an emerging reality and a global challenge. J Infect Dis 216:S431–S435. 10.1093/infdis/jix179. [DOI] [PubMed] [Google Scholar]
- 3.Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. 2017. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis 17:e383–e392. 10.1016/S1473-3099(17)30316-X. [DOI] [PubMed] [Google Scholar]
- 4.Lewis RE. 2011. Current concepts in antifungal pharmacology. Mayo Clin Proc 86:805–817. 10.4065/mcp.2011.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson TM, Clay MC, Cioffi AG, Diaz KA, Hisao GS, Tuttle MD, Nieuwkoop AJ, Comellas G, Maryum N, Wang S, Uno BE, Wildeman EL, Gonen T, Rienstra CM, Burke MD. 2014. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol 10:400–406. 10.1038/nchembio.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. 2019. Twenty years of the SENTRY Antifungal Surveillance Program: results for Candida species from 1997-2016. Open Forum Infect Dis 6:S79–S94. 10.1093/ofid/ofy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arendrup MC, Dzajic E, Jensen RH, Johansen HK, Kjældgaard P, Knudsen JD, Kristensen L, Leitz C, Lemming LE, Nielsen L, Olesen B, Rosenvinge FS, Røder BL, Schønheyder HC. 2013. Epidemiological changes with potential implication for antifungal prescription recommendations for fungaemia: data from a nationwide fungaemia surveillance programme. Clin Microbiol Infect 19:E343–E353. 10.1111/1469-0691.12212. [DOI] [PubMed] [Google Scholar]
- 8.Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi 3:57. 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 10.Andes DR, Safdar N, Baddley JW, Alexander B, Brumble L, Freifeld A, Hadley S, Herwaldt L, Kauffman C, Lyon GM, Morrison V, Patterson T, Perl T, Walker R, Hess T, Chiller T, Pappas PG, TRANSNET Investigators. 2016. The epidemiology and outcomes of invasive Candida infections among organ transplant recipients in the United States: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Transpl Infect Dis 18:921–931. 10.1111/tid.12613. [DOI] [PubMed] [Google Scholar]
- 11.Astvad KMT, Johansen HK, Røder BL, Rosenvinge FS, Knudsen JD, Lemming L, Schønheyder HC, Hare RK, Kristensen L, Nielsen L, Gertsen JB, Dzajic E, Pedersen M, Østergård C, Olesen B, Søndergaard TS, Arendrup MC. 2018. Update from a 12-year nationwide fungemia surveillance: increasing intrinsic and acquired resistance causes concern. J Clin Microbiol 56:e01564-17. 10.1128/JCM.01564-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, Rohde B, Bauser C, Bader O, Sanglard D. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog 5:e1000268. 10.1371/journal.ppat.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whaley SG, Rogers PD. 2016. Azole resistance in Candida glabrata. Curr Infect Dis Rep 18:41. 10.1007/s11908-016-0554-5. [DOI] [PubMed] [Google Scholar]
- 14.Cavalheiro M, Costa C, Silva-Dias A, Miranda IM, Wang C, Pais P, Pinto SN, Mil-Homens D, Sato-Okamoto M, Takahashi-Nakaguchi A, Silva RM, Mira NP, Fialho AM, Chibana H, Rodrigues AG, Butler G, Teixeira MC. 2019. A transcriptomics approach to unveiling the mechanisms of in vitro evolution towards fluconazole resistance of a Candida glabrata clinical isolate. Antimicrob Agents Chemother 63:e00995-18. 10.1128/AAC.00995-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vermitsky JP, Edlind TD. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother 48:3773–3781. 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai HF, Krol AA, Sarti KE, Bennett JE. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob Agents Chemother 50:1384–1392. 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol 61:704–722. 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- 18.Caudle KE, Barker KS, Wiederhold NP, Xu L, Homayouni R, Rogers PD. 2011. Genomewide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryot Cell 10:373–383. 10.1128/EC.00073-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob Agents Chemother 43:2753–2765. 10.1128/AAC.43.11.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumikawa K, Kakeya H, Tsai H-F, Grimberg B, Bennett JE. 2003. Function of Candida glabrata ABC transporter gene, PDH1. Yeast 20:249–261. 10.1002/yea.962. [DOI] [PubMed] [Google Scholar]
- 21.Torelli R, Posteraro B, Ferrari S, La Sorda M, Fadda G, Sanglard D, Sanguinetti M. 2008. The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol Microbiol 68:186–201. 10.1111/j.1365-2958.2008.06143.x. [DOI] [PubMed] [Google Scholar]
- 22.Vale-Silva LA, Moeckli B, Torelli R, Posteraro B, Sanguinetti M, Sanglard D. 2016. Upregulation of the adhesin gene EPA1 mediated by PDR1 in Candida glabrata leads to enhanced host colonization. mSphere 1:e00065-15. 10.1128/mSphere.00065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallabhaneni S, Cleveland AA, Farley MM, Harrison LH, Schaffner W, Beldavs ZG, Derado G, Pham CD, Lockhart SR, Smith RM. 2015. Epidemiology and risk factors for echinocandin nonsusceptible Candida glabrata bloodstream infections: data from a large multisite population-based candidemia surveillance program, 2008-2014. Open Forum Infect Dis 2:ofv163. 10.1093/ofid/ofv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Healey KR, Zhao Y, Perez WB, Lockhart SR, Sobel JD, Farmakiotis D, Kontoyiannis DP, Sanglard D, Taj-Aldeen SJ, Alexander BD, Jimenez-Ortigosa C, Shor E, Perlin DS. 2016. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun 7:11128. 10.1038/ncomms11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bordallo-Cardona MÁ, Agnelli C, Gómez-Nuñez A, Sánchez-Carrillo C, Bouza E, Muñoz P, Escribano P, Guinea J. 2019. MSH2 gene point mutations are not antifungal resistance markers in Candida glabrata. Antimicrob Agents Chemother 63:e01876-18. 10.1128/AAC.01876-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh A, Healey KR, Yadav P, Upadhyaya G, Sachdeva N, Sarma S, Kumar A, Tarai B, Perlin DS, Chowdhary A. 2018. Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob Agents Chemother 62:e00195-18. 10.1128/AAC.00195-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arastehfar A, Lass-Flörl C, Garcia-Rubio R, Daneshnia F, Ilkit M, Boekhout T, Gabaldon T, Perlin DS. 2020. The quiet and underappreciated rise of drug-resistant invasive fungal pathogens. J Fungi 6:138. 10.3390/jof6030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuchler K, Jenull S, Shivarathri R, Chauhan N. 2016. Fungal KATs/KDACs: a new highway to better antifungal drugs? PLoS Pathog 12:e1005938. 10.1371/journal.ppat.1005938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Kane CJ, Weild R, Hyland EM. 2020. Chromatin structure and drug resistance in Candida spp. J Fungi 6:121. 10.3390/jof6030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarzmüller T, Ma B, Hiller E, Istel F, Tscherner M, Brunke S, Ames L, Firon A, Green B, Cabral V, Marcet-Houben M, Jacobsen ID, Quintin J, Seider K, Frohner I, Glaser W, Jungwirth H, Bachellier-Bassi S, Chauvel M, Zeidler U, Ferrandon D, Gabaldón T, Hube B, d’Enfert C, Rupp S, Cormack B, Haynes K, Kuchler K. 2014. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog 10:e1004211. 10.1371/journal.ppat.1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orta-Zavalza E, Guerrero-Serrano G, Gutiérrez-Escobedo G, Cañas-Villamar I, Juárez-Cepeda J, Castaño I, De Las Peñas A. 2013. Local silencing controls the oxidative stress response and the multidrug resistance in Candida glabrata. Mol Microbiol 88:1135–1148. 10.1111/mmi.12247. [DOI] [PubMed] [Google Scholar]
- 32.Pfaller MA, Messer SA, Georgopapadakou N, Martell LA, Besterman JM, Diekema DJ. 2009. Activity of MGCD290, a Hos2 histone deacetylase inhibitor, in combination with azole antifungals against opportunistic fungal pathogens. J Clin Microbiol 47:3797–3804. 10.1128/JCM.00618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rai MN, Balusu S, Gorityala N, Dandu L, Kaur R. 2012. Functional genomic analysis of Candida glabrata-macrophage interaction: role of chromatin remodeling in virulence. PLoS Pathog 8:e1002863. 10.1371/journal.ppat.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolinski K, Muir S, Cardenas M, Heitman J. 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 94:13093–13098. 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davey M, Hannam C, Wong C, Brandl CJ. 2000. The yeast peptidyl proline isomerases FPR3 and FPR4, in high copy numbers, suppress defects resulting from the absence of the E3 ubiquitin ligase TOM1. Mol Gen Genet 263:520–526. 10.1007/s004380051197. [DOI] [PubMed] [Google Scholar]
- 36.Nelson CJ, Santos-Rosa H, Kouzarides T. 2006. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126:905–916. 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 37.Xiao H, Jackson V, Lei M. 2006. The FK506-binding protein, Fpr4, is an acidic histone chaperone. FEBS Lett 580:4357–4364. 10.1016/j.febslet.2006.06.093. [DOI] [PubMed] [Google Scholar]
- 38.Kumar K, Moirangthem R, Kaur R. 2020. Histone H4 dosage modulates DNA damage response in the pathogenic yeast Candida glabrata via homologous recombination pathway. PLoS Genet 16:e1008620. 10.1371/journal.pgen.1008620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eirín-López JM, Frehlick LJ, Ausió J. 2006. Long-term evolution and functional diversification in the members of the nucleophosmin/nucleoplasmin family of nuclear chaperones. Genetics 173:1835–1850. 10.1534/genetics.106.058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohkuni K, Abdulle R, Kitagawa K. 2014. Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl cis-trans isomerase. Genetics 196:1041–1045. 10.1534/genetics.114.161224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shan X, Xue Z, Mélèse T. 1994. Yeast NPI46 encodes a novel prolyl cis-trans isomerase that is located in the nucleolus. J Cell Biol 126:853–862. 10.1083/jcb.126.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaur R, Castano I, Cormack BP. 2004. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob Agents Chemother 48:1600–1613. 10.1128/aac.48.5.1600-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunjan A, Paik J, Verreault A. 2006. The emergence of regulated histone proteolysis. Curr Opin Genet Dev 16:112–118. 10.1016/j.gde.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 44.O’Kane CJ, Hyland EM. 2019. Yeast epigenetics: the inheritance of histone modification states. Biosci Rep 39:BSR20182006. 10.1042/BSR20182006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhakt P, Shivarathri R, Choudhary DK, Borah S, Kaur R. 2018. Fluconazole-induced actin cytoskeleton remodeling requires phosphatidylinositol 3-phosphate 5-kinase in the pathogenic yeast Candida glabrata. Mol Microbiol 110:425–443. 10.1111/mmi.14110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whaley SG, Caudle KE, Simonicova L, Zhang Q, Moye-Rowley WS, Rogers PD. 2018. Jjj1 is a negative regulator of Pdr1-mediated fluconazole resistance in Candida glabrata. mSphere 3:e00466-17. 10.1128/mSphere.00466-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahl BD, Grant PA, Briggs SD, Sun Z-W, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22:1298–1306. 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klose RJ, Gardner KE, Liang G, Erdjument-Bromage H, Tempst P, Zhang Y. 2007. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Mol Cell Biol 27:3951–3961. 10.1128/MCB.02180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denning DW, Bromley MJ. 2015. How to bolster the antifungal pipeline. Science 347:1414–1416. 10.1126/science.aaa6097. [DOI] [PubMed] [Google Scholar]
- 50.Smith WL, Edlind TD. 2002. Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: correlation with reduction in CDR and ERG upregulation. Antimicrob Agents Chemother 46:3532–3539. 10.1128/aac.46.11.3532-3539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Näär AM. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604–609. 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 52.Noble JA, Tsai HF, Suffis SD, Su Q, Myers TG, Bennett JE. 2013. STB5 is a negative regulator of azole resistance in Candida glabrata. Antimicrob Agents Chemother 57:959–967. 10.1128/AAC.01278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paul S, McDonald WH, Moye-Rowley WS. 2018. Negative regulation of Candida glabrata Pdr1 by the deubiquitinase subunit Bre5 occurs in a ubiquitin independent manner. Mol Microbiol 110:309–323. 10.1111/mmi.14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishikawa JL, Boeszoermenyi A, Vale-Silva LA, Torelli R, Posteraro B, Sohn YJ, Ji F, Gelev V, Sanglard D, Sanguinetti M, Sadreyev RI, Mukherjee G, Bhyravabhotla J, Buhrlage SJ, Gray NS, Wagner G, Naar AM, Arthanari H. 2016. Inhibiting fungal multidrug resistance by disrupting an activator-mediator interaction. Nature 530:485–489. 10.1038/nature16963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gómez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, Nichenametla SN, Locasale JW. 2015. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab 22:861–873. 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serefidou M, Venkatasubramani AV, Imhof A. 2019. The impact of one carbon metabolism on histone methylation. Front Genet 10:764. 10.3389/fgene.2019.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Backer MD, Ilyina T, Ma XJ, Vandoninck S, Luyten WHML, Vanden Bossche H. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob Agents Chemother 45:1660–1670. 10.1128/AAC.45.6.1660-1670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tscherner M, Zwolanek F, Jenull S, Sedlazeck FJ, Petryshyn A, Frohner IE, Mavrianos J, Chauhan N, von Haeseler A, Kuchler K. 2015. The Candida albicans histone acetyltransferase Hat1 regulates stress resistance and virulence via distinct chromatin assembly pathways. PLoS Pathog 11:e1005218. 10.1371/journal.ppat.1005218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shivarathri R, Tscherner M, Zwolanek F, Singh NK, Chauhan N, Kuchler K. 2019. The fungal histone acetyl transferase Gcn5 controls virulence of the human pathogen Candida albicans through multiple pathways. Sci Rep 9:9445. 10.1038/s41598-019-45817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, Cai Q, Mei H, Zhou X, Shen Y, Li D, Liu W. 2015. The Rpd3/Hda1 family of histone deacetylases regulates azole resistance in Candida albicans. J Antimicrob Chemother 70:1993–2003. 10.1093/jac/dkv070. [DOI] [PubMed] [Google Scholar]
- 61.Li L, Wang Y. 2017. Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J Biol Chem 292:11951–11959. 10.1074/jbc.M117.788224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabrizio P, Garvis S, Palladino F. 2019. Histone methylation and memory of environmental stress. Cells 8:339. 10.3390/cells8040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burgess RJ, Zhang Z. 2013. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol 20:14–22. 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Namboodiri VMH, Akey IV, Schmidt-Zachmann MS, Head JF, Akey CW. 2004. The structure and function of Xenopus NO38-core, a histone chaperone in the nucleolus. Structure 12:2149–2160. 10.1016/j.str.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 65.Kuzuhara T, Horikoshi M. 2004. A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat Struct Mol Biol 11:275–283. 10.1038/nsmb733. [DOI] [PubMed] [Google Scholar]
- 66.Manning-Krieg UC, Henríquez R, Cammas F, Graff P, Gavériaux S, Movva NR. 1994. Purification of FKBP-70, a novel immunophilin from Saccharomyces cerevisiae, and cloning of its structural gene, FPR3. FEBS Lett 352:98–103. 10.1016/0014-5793(94)00927-9. [DOI] [PubMed] [Google Scholar]
- 67.Benton BM, Zang JH, Thorner J. 1994. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Saccharomyces cerevisiae is a proline rotamase localized to the nucleolus. J Cell Biol 127:623–639. 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park SK, Xiao H, Lei M. 2014. Nuclear FKBPs, Fpr3 and Fpr4 affect genome-wide genes transcription. Mol Genet Genomics 289:125–136. 10.1007/s00438-013-0794-0. [DOI] [PubMed] [Google Scholar]
- 69.Wallace EWJ, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, Airoldi EM, Pan T, Budnik BA, Drummond DA. 2015. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162:1286–1298. 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borah S, Shivarathri R, Kaur R. 2011. The Rho1 GTPase-activating protein CgBem2 is required for survival of azole stress in Candida glabrata. J Biol Chem 286:34311–34324. 10.1074/jbc.M111.264671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez-Tudela JL, Arendrup MC, Barchiesi F, Bille J, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Denning DW, Donnelly JP, Dromer F, Fegeler W, Lass-Flörl C, Moore C, Richardson M, Sandven P, Velegraki A, Verweij P. 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin Microbiol Infect 14:398–405. 10.1111/j.1469-0691.2007.01935.x. [DOI] [PubMed] [Google Scholar]