Abstract
Although MR-guided radiotherapy (MRgRT) is advancing rapidly, generating accurate synthetic CT (sCT) from MRI is still challenging. Previous approaches using deep neural networks require large dataset of precisely co-registered CT and MRI pairs that are difficult to obtain due to respiration and peristalsis. Here, we propose a method to generate sCT based on deep learning training with weakly paired CT and MR images acquired from an MRgRT system using a cycle-consistent GAN (CycleGAN) framework that allows the unpaired image-to-image translation in abdomen and thorax. Data from 90 cancer patients who underwent MRgRT were retrospectively used. CT images of the patients were aligned to the corresponding MR images using deformable registration, and the deformed CT (dCT) and MRI pairs were used for network training and testing. The 2.5D CycleGAN was constructed to generate sCT from the MRI input. To improve the sCT generation performance, a perceptual loss that explores the discrepancy between high-dimensional representations of images extracted from a well-trained classifier was incorporated into the CycleGAN. The CycleGAN with perceptual loss outperformed the U-net in terms of errors and similarities between sCT and dCT, and dose estimation for treatment planning of thorax, and abdomen. The sCT generated using CycleGAN produced virtually identical dose distribution maps and dose-volume histograms compared to dCT. CycleGAN with perceptual loss outperformed U-net in sCT generation when trained with weakly paired dCT-MRI for MRgRT. The proposed method will be useful to increase the treatment accuracy of MR-only or MR-guided adaptive radiotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13534-021-00195-8.
Introduction
In radiation therapy, radiation should be delivered accurately to the planning target volume (PTV) to eliminate cancer while simultaneously delivering minimal radiation to unwanted areas to prevent side effects. Therefore, accurate delineation of target and normal tissues is essential, and computed tomography (CT) is primarily used for target localization and organ contouring in radiotherapy planning.
Magnetic resonance imaging (MRI) is increasingly used for radiotherapy planning because of its superior soft-tissue contrast compared to CT, which facilitates tumor and organ-at-risk (OAR) delineation. However, dose estimation based solely on MR images is challenging because MR images do not provide direct information on electron density. Therefore, additional CT scans of patients are acquired and co-registered with MRI for the MR-guided radiotherapy (MRgRT) systems, including MR-Linac, which causes unnecessary radiation exposure and extra cost to patients. Dose estimation error due to spatial misregistration and temporal changes in anatomy between MRI and CT is another drawback of the co-registered CT-based approach [1].
Recently, considerable research interest has been directed toward the direct conversion of MRI into synthetic CT (sCT) based on deep learning (DL) approaches to overcome the drawbacks of co-registered CT. Previous studies using the DL method for MRI-to-CT translation employed supervised learning models that are easy to train but generally produce blurry output images [2–8]. The most commonly used neural network for MRI-to-CT translation is the U-net architecture, an encoder–decoder network with skip connections trained in a discriminative manner [9]. In contrast, the incorporation of generative models usually provides more realistic output images than discriminative models. One of the approaches to generative modeling is the generative adversarial network (GAN) in which generator and discriminator networks are trained simultaneously through min–max optimization [10]. In several recent studies on sCT generation from MR images, GANs that utilize adversarial feedback from a discriminator network have shown superior performance than the corresponding U-nets [11–14]. However, previous approaches using GANs were based on paired image-to-image translation, requiring a large dataset of precisely co-registered CT and MRI pairs that are difficult to obtain due to respiration and peristalsis in thorax and abdomen. In addition, there are a limited number of studies on generating MR-based sCT for MRgRT systems [15, 16]. The datasets used in previous studies are also limited to the head, neck, pelvis, and liver.
Therefore, in this study, we propose a method to generate sCT based on DL training from weakly paired CT and MR images acquired from the MRgRT system using a cycle-consistent GAN (CycleGAN) framework that allows unpaired image-to-image translation [17–20]. The proposed CycleGAN method has been applied to the head and neck or the pelvis area, but as far as we know it has not been applied to the abdomen and thorax [18, 21]. To improve the sCT generation performance, a perceptual loss that explores the discrepancy between high-dimensional representations of images extracted from a discriminator was incorporated into the CycleGAN. In addition, the weakly paired dataset included CT and MR images of the pelvis, thorax, and abdomen to prevent the overfitting of networks from being specialized in a specific area. The sCT generation performance of CycleGAN was compared with U-net, and the dose estimation accuracy was evaluated using the treatment planning system (TPS) of the MRgRT system.
Methods and materials
Data acquisition and preprocessing
Data from 90 cancer patients who underwent MRgRT were retrospectively used. The retrospective use of the scan data and waiver of consent were approved by the Institutional Review Board of our institute. The patient data were divided into three groups according to the treatment region: pelvis (n = 30), thorax (n = 30), and abdomen (n = 30). Detailed patient characteristics are provided in Supplementary Table 1.
CT simulation scans of all patients were acquired using the Brilliance Big Bore CT scanner (Philips, Cleveland, OH). Fifteen minutes after the CT scan, MR images were acquired using a 0.35 T MRI scanner combined with the radiation therapy unit of the MRIdian MRgRT system (ViewRay, Oakwood, OH). The MRI scans were performed in the same patient setup as the CT simulations using true fast imaging with steady-state precession (TrueFISP; TRUFI) sequence. Thereafter, the CT images were co-registered with the corresponding MR images using a deformable registration algorithm, provided by ViewRay TPS. The deformed CT (dCT) images were resampled to have the same dimensions as the MRI images (320 × 320 × 144). A dataset with mismatched body and organ boundaries on the co-registered CT and MRI were not included in the data of the 90 patients. The low-frequency intensity and non-uniformity present in MRI were corrected using the N4 bias field correction algorithm [22].
Of the 90 MR–dCT image pairs, 80% (72 pairs) were used for network training and the remaining 20% (18 pairs) were used for testing and validation. To avoid 3D discontinuities in sCT, 2.5D data consisting of three adjacent slices were provided to the network. The intensity of the images was normalized using the 95% percentile value of the intensity peak of each image to enable effective network training.
Network architecture
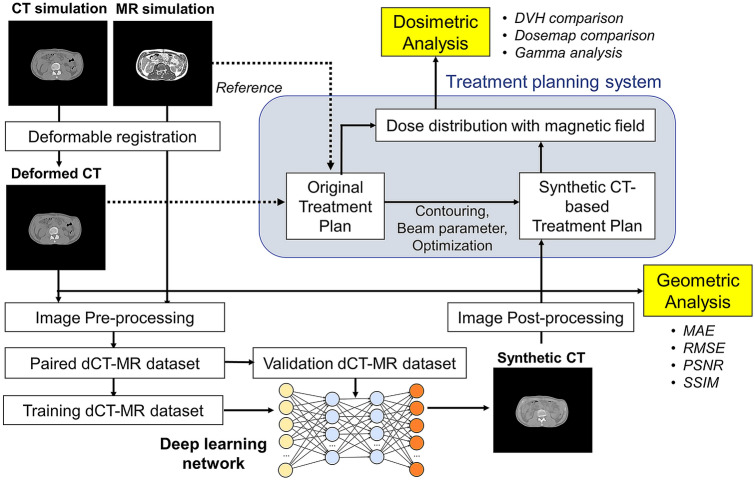
As mentioned previously, two different deep learning networks, U-net and CycleGAN, were used for sCT generation. Figure 1 shows the procedures for image processing/analysis and network training/testing, and Supplementary Fig. 1 shows the detailed structure of the deep learning networks. For U-net, the standard network architecture used in Noise2Noise paper [23] was modified. The input and output of the U-net are MRI and sCT, respectively, and the ground truth is dCT. The network consisted of 17 convolutional layers containing a convolution and a leaky rectified linear unit (ReLU) activation function, as shown in Supplementary Fig. 1.b. The filter numbers for these layers were 64, 64, 128, 256, 512, 1024, and 1024 in the encoder for down-sampling, and 1024, 1024, 512, 512, 256, 256, 128, 128, 64, and 64 in the decoder for up-sampling. The weights and biases in the layers were trained by minimizing L1 loss, the mean absolute error (MAE) between the sCT and dCT. The network was trained for 200 epochs using an adaptive stochastic gradient descent optimizer (Adam) with a learning rate of 0.0001.
Fig. 1.
Image analysis procedures
The CycleGAN consisted of two generators (G1 and G2) and two discriminators (D1 and D2), as shown in Supplementary Fig. 1.a. The U-net described above was used as the generator. The discriminators consisted of five residual neural network (ResNet) blocks [24] followed by global average pooling layer (Supplementary Fig. 1.b). The generators produce synthetic images, and discriminators determine whether the synthetic images are real or fake by minimizing the following loss functions:
where LGAN is the GAN loss that generates a real-like desired image, Lcyc is the cyclic loss enforcing that the output of the generator is similar to the input, and Lid is the identical loss that stabilizes the training.
Deformable registration is nearly accurate for bones, but it is not perfect for non-rigid structures areas. Therefore, we have complemented fidelity loss between the generator output and the dCT with perceptual loss in the discriminator’s view. Perception loss is used to minimize high-level differences, such as content and style discrepancies between the generated and input images. The sum of L2 norm of the features in ResNet blocks 1, 2, and 3 of the discriminators was calculated as the perceptual loss, and spectral normalization was applied to each layer to stabilize the discriminator training [25]. Accordingly, the perceptual loss and total loss can be described as follows:
where n is the batch size and is the i-th ResNet block used to calculate the perceptual loss, which was extracted from each discriminator.
The number of the batch size and learning rate of the generator were 1 and 0.0001, respectively. We used the two time-scale update rule to assign different learning rates to generators and discriminators [26]. The learning rate of the discriminators was four times higher than that of the generators. The Adam optimization algorithm (β1 = 0.5 and β2 = 0.99) was used to minimize the above losses. All models were implemented using TensorFlow and trained on an NVIDIA Geforce GTX 1080Ti with 11 GB of memory.
Image analysis and treatment planning
As previously mentioned, 18 of 90 MR–dCT image pairs were randomly selected as the test set. To assess the accuracy of the image translation for the test set, the MAE, root mean square error (RMSE), peak signal-to-noise ratio (PSNR), and structural similarity (SSIM) between the sCT generated from the pre-processed MRI using the deep neural networks and the dCT were calculated as follows:
where i is a voxel within the body, N is the total number of voxels, and MAX is the maximum voxel value of the reference image.
We also analyzed the dosimetric accuracy of the use of sCT images based on the MRIdian treatment planning system used for the Co-60 ViewRay system. The dose distribution was recalculated by replacing dCT with sCT images under the same beam parameters as the original dCT treatment plan. The Monte Carlo simulation with magnetic-field correction was performed to calculate the dose distribution with a calculation grid size of 3 mm. The prescription dose to the PTV was not the same for all patients. To evaluate the dose distribution under the same condition, the dose distribution map was scaled to correspond to the prescription dose of 70 Gy for the pelvis, 38.5 Gy for the thorax, and 50 Gy for the abdomen. To compare the dose distributions estimated using the sCT and dCT, several dose-volume histogram (DVH) parameters, such as minimum (Dmin), maximum (Dmax), and mean (Dmean) absorbed doses for the PTV and OARs, were calculated. For the PTV area, D98%, D2%, and V100% were also calculated to assess the homogeneity and conformity of the dose distribution. Spatial dose distributions were also compared using 3D gamma analysis under 2%/2 mm and 3%/3 mm (dose discrepancy/distance agreement) criteria with 10, 50, and 90% thresholds.
All statistical analyses were performed using SPSS software. Wilcoxon signed-rank tests were used to compare the performance of the U-net and the proposed CycleGAN models. P values less than 0.05 were considered statistically significant.
Results
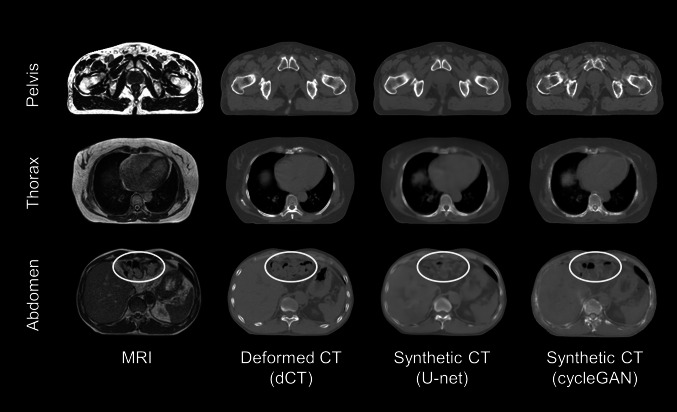
Figure 2 shows the representative case of the generated sCT slices exhibited with the corresponding MR and dCT slices in the pelvis, thorax, and abdomen. The sCT generated using CycleGAN (sCTCyGAN) showed sharper boundaries than the sCT generated using the U-net (sCTU-net). In addition, sCT showed better similarity to dCT in dense structures such as the pelvic bones and spine. The location and size of air pockets shown in the abdomen images are more accurate in sCT than in dCT (yellow ellipses).
Fig. 2.
Transverse slices of MRI, dCT, sCTU-net, and sCTcGAN in representative cases
The MAE, RMSE, PSNR, and SSIM between the dCT and sCT images calculated from the test set data are summarized in Table 1. In general, sCTCyGAN showed smaller errors (MAE and RMSE) and higher similarities (PSNR, SSIM) relative to dCT than sCTU-net. The error and similarity differences between CycleGAN and U-net were considerably large in the thorax and abdomen.
Table 1.
Errors (MAE and RMSE) and similarities (PSNR, SSIM) relative between sCT and dCT in the test group
| Pelvis (n = 6) | Thorax (n = 6) | Abdomen (n = 6) | Total (n = 18) | |
|---|---|---|---|---|
| U-net | ||||
| MAE | 56.3 ± 9.3 | 134 ± 27.8 | 150 ± 55.4 | 114 ± 54.7 |
| RMSE | 104 ± 7.0 | 168 ± 27.3 | 172 ± 22.9 | 148 ± 33.9 |
| PSNR | 27.9 ± 0.6 | 23.0 ± 1.7 | 22.9 ± 3.1 | 24.6 ± 3.1 |
| SSIM | 0.87 ± 0.04 | 0.89 ± 0.01 | 0.91 ± 0.01 | 0.89 ± 0.03 |
| CycleGAN | ||||
| MAE | 55.3 ± 5.5 | 63.7 ± 3.8 | 58.8 ± 4.4 | 59.2 ± 5.8 |
| RMSE | 118 ± 9.3 | 119 ± 8.4 | 113 ± 9.3 | 117 ± 9.4 |
| PSNR | 26.8 ± 0.7 | 25.9 ± 0.5 | 26.3 ± 0.7 | 26.3 ± 0.7 |
| SSIM | 0.89 ± 0.05 | 0.90 ± 0.02 | 0.91 ± 0.01 | 0.90 ± 0.03 |
MAE Mean absolute error, RMSE Root mean square error, PSNR Peak signal-to-noise ratio, SSIM Structural similarity
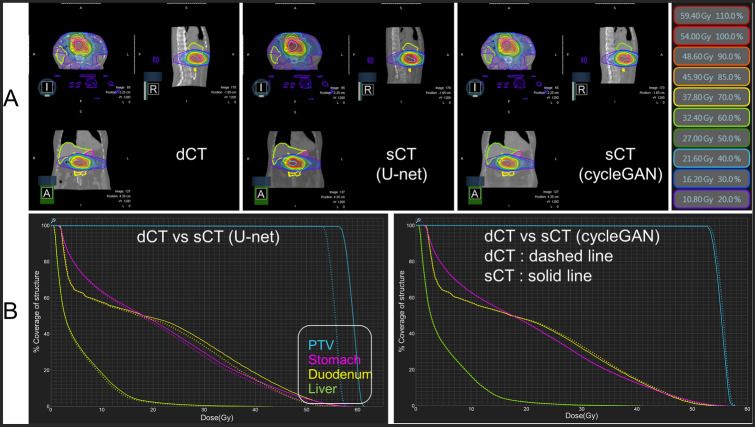
Figure 3 and Table 2 show the superiority of sCTCyGAN over sCTU-net in dose estimation for treatment planning. The treatment dose distribution and DVHs for the PTV and OARs generated using dCT and sCTs for a representative abdomen case are shown in Fig. 3. In this case, dCT and sCTs yield very similar isodose curves, as shown in Fig. 3a. The left column of Fig. 3b shows the significant DVH difference between dCT and sCTU-net, which is largest in the PTV. On the other hand, dCT and sCTCyGAN produce virtually identical DVHs, as shown in the right column of Fig. 3b. DVH parameters for the PTV and OARs in each region are summarized in Table 2.
Fig. 3.
Dosimetric comparison between dCT and sCTs in the abdomen. a Dose distribution and isodose curves. Each color contour shows the isodose line (see right legends). b Left: Comparison of dose volume histogram for PTV and OARs (stomach, duodenum, and liver) between dCT and sCT using U-net Right: Comparison of dose volume histogram for PTV and OARs between dCT and sCT using cycleGAN. Dashed line indicates the result of dCT and the solid line shows the result of sCT methods
Table 2.
Comparison of dose-Volume histogram (DVH) parameters between dCT and sCTs
| Pelvis | Thorax | Abdomen | |||||||
|---|---|---|---|---|---|---|---|---|---|
| dCT | sCTU-net | sCTcGAN | dCT | sCTU-net | sCTcGAN | dCT | sCTU-net | sCTcGAN | |
| PTV | |||||||||
| Dmean | 73.9 ± 0.7 | 75.6 ± 0.6* | 74.0 ± 0.7 | 40.1 ± 0.6 | 40.8 ± 0.8 | 40.2 ± 0.6 | 50.5 ± 2.1 | 54.1 ± 3.0** | 50.3 ± 1.5 |
| Dmin | 63.8 ± 2.1 | 66.3 ± 3.6 | 63.9 ± 2.3 | 35.3 ± 1.7 | 35.0 ± 2.0 | 35.2 ± 1.7 | 45.2 ± 2.2 | 48.7 ± 3.2 | 45.1 ± 2.5 |
| Dmax | 78.7 ± 1.3 | 80.3 ± 1.3* | 78.8 ± 1.5 | 42.5 ± 1.4 | 43.5 ± 1.7** | 42.6 ± 0.9 | 54.1 ± 1.2 | 57.5 ± 3.4** | 53.3 ± 0.9 |
| D98% | 68.6 ± 1.0 | 69.9 ± 1.3 | 68.6 ± 1.0 | 37.7 ± 0.9 | 37.8 ± 0.9 | 37.7 ± 1.0 | 49.3 ± 0.2 | 49.7 ± 3.4 | 48.8 ± 0.2* |
| D2% | 76.9 ± 1.1 | 78.5 ± 1.0* | 77.1 ± 1.0 | 41.9 ± 1.3 | 42.8 ± 1.6 | 41.9 ± 1.4 | 53.5 ± 0.6 | 56.4 ± 3.1 | 53.1 ± 0.6 |
| V100% | 77.5 ± 18.8 | 79.2 ± 19.0 | 77.8 ± 19.1 | 89.6 ± 27.8 | 92.5 ± 29.2 | 89.6 ± 27.6 | 36.1 ± 13.0 | 32.0 ± 12.4 | 34.2 ± 12.1 |
| OAR | Bladder | Heart | Duodenum | ||||||
| Dmean | 47.3 ± 3.7 | 47.9 ± 3.6 | 47.4 ± 3.7 | 3.1 ± 0.7 | 3.2 ± 0.8 | 3.0 ± 0.7 | 26.1 ± 8.9 | 27.4 ± 9.6 | 25.9 ± 8.9 |
| Dmin | 20.0 ± 10.5 | 20.1 ± 10.5 | 20.0 ± 10.4 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 | 5.4 ± 5.1 | 5.5 ± 5.3 | 5.5 ± 5.2 |
| Dmax | 75.8 ± 1.6 | 77.0 ± 2.1 | 75.7 ± 1.4 | 19.0 ± 10.9 | 19.3 ± 10.9 | 19.0 ± 10.9 | 49.4 ± 4.1 | 52.7 ± 5.1 | 46.7 ± 6.5 |
| OAR | Rectum | Left lung | Stomach | ||||||
| Dmean | 43.2 ± 6.1 | 43.7 ± 6.0 | 43.1 ± 6.2 | 1.6 ± 0.3 | 1.7 ± 0.3 | 1.6 ± 0.3 | 15.2 ± 5.1 | 15.7 ± 5.3 | 15.2 ± 5.1 |
| Dmin | 16.6 ± 8.0 | 16.5 ± 8.0 | 16.5 ± 7.9 | 0.2 ± 0.03 | 0.2 ± 0.04 | 0.2 ± 0.03 | 2.1 ± 1.6 | 2.2 ± 1.7 | 2.1 ± 1.7 |
| Dmax | 76.1 ± 1.7 | 76.9 ± 2.0 | 75.4 ± 1.3 | 9.1 ± 4.9 | 9.3 ± 4.7 | 8.9 ± 4.6 | 45.5 ± 5.2 | 47.1 ± 5.7 | 45.1 ± 5.4 |
| OAR | Femur head | Right lung | Liver | ||||||
| Dmean | 25.5 ± 6.7 | 25.8 ± 6.8 | 25.5 ± 6.7 | 6.4 ± 1.4 | 6.5 ± 1.4 | 6.3 ± 1.4 | 8.3 ± 5.3 | 8.7 ± 5.5 | 8.3 ± 5.3 |
| Dmin | 10.8 ± 6.0 | 11.1 ± 6.1 | 11.0 ± 6.1 | 0.2 ± 0.08 | 0.2 ± 0.07 | 0.2 ± 0.07 | 0.7 ± 0.3 | 0.8 ± 0.4 | 0.7 ± 0.4 |
| Dmax | 44.6 ± 7.4 | 45.0 ± 7.9 | 44.5 ± 7.9 | 40.1 ± 1.1 | 40.9 ± 0.8 | 39.9 ± 0.9 | 44.5 ± 8.9 | 47.4 ± 10.7 | 43.9 ± 8.8 |
The Wilcoxon test was used to compare the dose volume parameters between dCT and sCT. Significant differences were considered at p < 0.05* and p < 0.1**
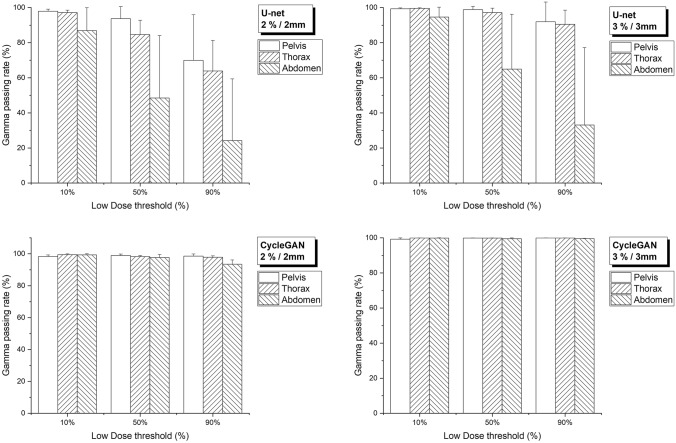
Figure 4 shows the average gamma passing rates between dCT and sCTs using 2%/2 mm and 3%/3 mm criteria at three different threshold levels. The gamma passing rate of cycleGAN exceeded 97%, except for 90% threshold in the abdomen. On the other hand, the U-net’s gamma passing rate exceeded 90% in all areas only at 3 mm/3% criteria with a 10% threshold, and was less than 90% when threshold increased or 2 mm/2% criteria was applied (see Supplementary Table 2 for detailed results).
Fig. 4.
Gamma passing rate according to two different criteria (2%/2 mm and 3%/3 mm) between dCT and sCTs
Discussion
Although MRgRT technology is advancing rapidly, generating accurate sCT from MRI is still an unsolved problem. Deep learning technology is also advancing rapidly, and image-to-image translation technology based on deep learning is being applied in various fields, including sCT generation from MRI [27–32]. However, most of the previous studies focused on generating sCT from diagnostic MRI, and in these studies, conventional TPS was used for dose calculation [3–5]. On the other hand, in our study, we generated sCT by applying deep learning models to low-field MR images used in the actual MRgRT system. The 0.35 T MRI images produced in the MRIdian MRgRT system exhibit different relaxation properties and image contrast than 1.5 T or 3 T MR images used for diagnostic purposes [33]. Furthermore, the MRIdian MRgRT system uses a TRUFI sequence that creates T2/T1 contrast that is different from the general T1 or T2 contrast [34].
Unlike most previous studies in which deep neural networks were trained using only data from a specific area, this study trained neural networks using all pelvis, thorax, and abdomen images to increase the performance and generality of the trained neural network. The main advantage of MRgRT is the application of real-time respiratory gating technology and the accurate identification of treatment targets based on the high soft-tissue contrast of MRI [35]. In this regard, it is particularly important to increase the accuracy of sCT generation in the thorax and abdomen areas that are heavily affected by respiration and inter- and intra-fraction movements [36]. However, there are limited studies that deal with the translation from MRI to CT of the thorax and abdomen because it is difficult to generate perfectly matched datasets due to changes in the shape and location of the non-rigid organs and air pockets.
U-net has shown superior performance in many image-to-image translation tasks [30, 31, 37–39]. However, this representative discriminative learning model has limited performance when trained with incomplete matched datasets, as shown in this study. To alleviate the incomplete matching problem, this study introduces CycleGAN that utilizes the flexible translation capabilities offered by the generative model structure [40]. The sCT generation performance was further improved by considering the perceptual loss that allows us to retain high-level features compared to pixel-wise loss, such as L1 or L2, which produce local smoothing [41] (Supplementary Fig. 2). Consequently, the CycleGAN using perceptual loss outperformed the U-net in terms of errors and similarities between sCT and dCT in the thorax and abdomen (Table 1). The dosimetric robustness of the proposed CycleGAN was also proven through gamma analysis (Fig. 4). In a previous study [14], conditional GAN with perceptual loss performed better than U-net in sCT generation from the MRI of patients with prostate cancer. However, the incorporation of perceptual loss in conditional GAN could not reduce MAE and improve the dose calculation accuracy, maybe because their dataset was limited to the pelvic area.
Recently, online MR-guided adaptive radiotherapy (MRgART) has been proposed to address anatomical changes in PTV and OARs during the treatment period [42]. In addition, a clinical trial called stereotactic MRI-guided on-table adaptive radiation therapy (SMART) for the pancreatic area is ongoing [43, 44]. However, changes in the air pocket position during MRgART cause significant differences in dose distribution [44]. To address this problem, in the current MRgART protocol, air pockets and body surfaces are recontoured in the dCT while referring to the MRI acquired daily, and correct electron densities are assigned to air and soft tissues, thereby increasing the time the patient is on the table. Therefore, improved sCT generation enabled by applying the proposed method will increase the accuracy of treatment planning and the convenience of patients in MRgART.
This study has some limitations, including a small number of datasets used for training and validating the network models. The accuracy of the sCT can be further improved by adding the MRI obtained daily for each fractionation and reducing the variability in MRI intensity through white stripe normalization [8, 45]. In addition, adding segmented bone regions from CT as input to networks will also improve the accuracy of sCT.
Conclusions
CycleGAN with perceptual loss outperformed U-net in sCT generation when trained with weakly paired dCT–MRI for MRgRT in abdomen and thorax. This approach will be useful to increase the treatment accuracy of MR-only radiotherapy and MRgART.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Grants from the Radiation Technology R&D program through the National Research Foundation of Korea funded by the Ministry of Science and ICT (2017M2A2A7A02020641, 2019M2A2B4095126, and 2020M2D9A109398911).
Data availability
Research data are not available at this time.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Seung Kwan Kang and Hyun Joon An have contributed equally to this work.
Contributor Information
Jong Min Park, Email: leodavinci@naver.com.
Jae Sung Lee, Email: jaes@snu.ac.kr.
References
- 1.Edmund JM, Nyholm T. A review of substitute CT generation for MRI-only radiation therapy. Radiat Oncol. 2017;12(1):28. doi: 10.1186/s13014-016-0747-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44(4):1408–1419. doi: 10.1002/mp.12155. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, et al. Technical Note: U-net-generated synthetic CT images for magnetic resonance imaging-only prostate intensity-modulated radiation therapy treatment planning. Med Phys. 2018;45(12):5659–5665. doi: 10.1002/mp.13247. [DOI] [PubMed] [Google Scholar]
- 4.Dinkla AM, et al. MR-only brain radiation therapy: dosimetric evaluation of synthetic CTs generated by a dilated convolutional neural network. Int J Radiat Oncol Biol Phys. 2018;102(4):801–812. doi: 10.1016/j.ijrobp.2018.05.058. [DOI] [PubMed] [Google Scholar]
- 5.Gupta D, et al. Generation of synthetic CT images From MRI for treatment planning and patient positioning using a 3-channel U-Net trained on sagittal images. Front Oncol. 2019;9:964. doi: 10.3389/fonc.2019.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neppl S, et al. Evaluation of proton and photon dose distributions recalculated on 2D and 3D Unet-generated pseudoCTs from T1-weighted MR head scans. Acta Oncol. 2019;58(10):1429–1434. doi: 10.1080/0284186X.2019.1630754. [DOI] [PubMed] [Google Scholar]
- 7.Fu J, et al. Deep learning approaches using 2D and 3D convolutional neural networks for generating male pelvic synthetic computed tomography from magnetic resonance imaging. Med Phys. 2019;46(9):3788–3798. doi: 10.1002/mp.13672. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez Andres E, et al. Dosimetry-driven quality measure of brain pseudo computed tomography generated from deep learning for mri-only radiation therapy treatment planning. Int J Radiat Oncol Biol Phys. 2020;108(3):813–823. doi: 10.1016/j.ijrobp.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer; 2015. pp. 234–41.
- 10.Goodfellow I, et al. Generative adversarial networks. In: Advances in neural information processing systems. 2014. pp. 2672–80.
- 11.Nie D, et al. Medical image synthesis with context-aware generative adversarial networks. Med Image Comput Comput Assist Interv. 2017;10435:417–425. doi: 10.1007/978-3-319-66179-7_48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isola P, et al. Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. pp. 1125–34.
- 13.Emami H, et al. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med Phys. 2018;45(8):3627–3636. doi: 10.1002/mp.13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Largent A, et al. Comparison of deep learning-based and patch-based methods for pseudo-CT generation in MRI-based prostate dose planning. Int J Radiat Oncol Biol Phys. 2019;105(5):1137–1150. doi: 10.1016/j.ijrobp.2019.08.049. [DOI] [PubMed] [Google Scholar]
- 15.Olberg S, et al. Synthetic CT reconstruction using a deep spatial pyramid convolutional framework for MR-only breast radiotherapy. Med Phys. 2019;46(9):4135–4147. doi: 10.1002/mp.13716. [DOI] [PubMed] [Google Scholar]
- 16.Fu J, et al. Generation of abdominal synthetic CTs from 0.35 T MR images using generative adversarial networks for MR-only liver radiotherapy. Biomedical Physics & Engineering Express. 2020;6(1):015033. doi: 10.1088/2057-1976/ab6e1f. [DOI] [PubMed] [Google Scholar]
- 17.Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. In: IEEE international conference on computer vision. 2017. pp. 2223–32.
- 18.Lei Y, et al. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med Phys. 2019;46(8):3565–3581. doi: 10.1002/mp.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shafai-Erfani G, et al. Dose evaluation of MRI-based synthetic CT generated using a machine learning method for prostate cancer radiotherapy. Med Dosim. 2019;44(4):e64–e70. doi: 10.1016/j.meddos.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. MRI-based treatment planning for liver stereotactic body radiotherapy: validation of a deep learning-based synthetic CT generation method. Br J Radiol. 2019;92(1100):20190067. doi: 10.1259/bjr.20190067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolterink JM, et al. Deep MR to CT synthesis using unpaired data. In: International workshop on simulation and synthesis in medical imaging. Springer; 2017. pp. 14–23.
- 22.Tustison NJ, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehtinen J, et al. Noise2noise: Learning image restoration without clean data. arXiv preprint https://arxiv.org/abs/1803.04189. 2018.
- 24.He K, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. pp. 770–8
- 25.Miyato T, et al. Spectral normalization for generative adversarial networks. arXiv preprint https://arxiv.org/abs/1802.05957. 2018.
- 26.Heusel M, et al. Gans trained by a two time-scale update rule converge to a local nash equilibrium. In: Advances in neural information processing systems. 2017. pp. 6629–40. https://proceedings.neurips.cc/paper/2017/hash/8a1d694707eb0fefe65871369074926d-Abstract.html.
- 27.Park J, et al. Computed tomography super-resolution using deep convolutional neural network. Phys Med Biol. 2018;63(14):145011. doi: 10.1088/1361-6560/aacdd4. [DOI] [PubMed] [Google Scholar]
- 28.Hwang D, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59(10):1624–1629. doi: 10.2967/jnumed.117.202317. [DOI] [PubMed] [Google Scholar]
- 29.Kang SK, et al. Adaptive template generation for amyloid PET using a deep learning approach. Hum Brain Mapp. 2018;39(9):3769–3778. doi: 10.1002/hbm.24210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MS, et al. Deep-dose: a voxel dose estimation method using deep convolutional neural network for personalized internal dosimetry. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-46620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang D, et al. Generation of PET attenuation map for whole-body time-of-flight 18F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019;60(8):1183–1189. doi: 10.2967/jnumed.118.219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JS. A review of deep Learning-based approaches for attenuation correction in positron emission tomography. IEEE Trans Radiat Plasma Med Sci. 2020;5:160–184. doi: 10.1109/TRPMS.2020.3009269. [DOI] [Google Scholar]
- 33.Korb JP, Bryant RG. Magnetic field dependence of proton spin-lattice relaxation times. Magn Reson Med Off J Int Soc Magn Reson Med. 2002;48(1):21–26. doi: 10.1002/mrm.10185. [DOI] [PubMed] [Google Scholar]
- 34.Klüter S. Technical design and concept of a 0.35 T MR-Linac. Clin Transl Radiat Oncol. 2019;18:98–101. doi: 10.1016/j.ctro.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JM, et al. Commissioning experience of tri-cobalt-60 MRI-guided radiation therapy system. Prog Med Phys. 2015;26(4):193–200. doi: 10.14316/pmp.2015.26.4.193. [DOI] [Google Scholar]
- 36.Henke L, et al. Magnetic resonance image-guided radiotherapy (MRIgRT): a 4.5-year clinical experience. Clin Oncol. 2018;30(11):720–727. doi: 10.1016/j.clon.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hegazy MA, et al. U-net based metal segmentation on projection domain for metal artifact reduction in dental CT. Biomed Eng Lett. 2019;9(3):375–385. doi: 10.1007/s13534-019-00110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comelli A, et al. Deep learning approach for the segmentation of aneurysmal ascending aorta. Biomed Eng Lett. 2020;11:1–10. doi: 10.1007/s13534-020-00179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J, et al. Measurement of glomerular filtration rate using quantitative SPECT/CT and deep-learning-based kidney segmentation. Sci Rep. 2019;9(1):1–8. doi: 10.1038/s41598-019-40710-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo J, Eom H, Choi YS. Image-to-image translation using a cross-domain auto-encoder and decoder. Appl Sci. 2019;9(22):4780. doi: 10.3390/app9224780. [DOI] [Google Scholar]
- 41.Wang C, et al. Perceptual adversarial networks for image-to-image transformation. IEEE Trans Image Process. 2018;27(8):4066–4079. doi: 10.1109/TIP.2018.2836316. [DOI] [PubMed] [Google Scholar]
- 42.Boldrini L, et al. Online adaptive magnetic resonance guided radiotherapy for pancreatic cancer: state of the art, pearls and pitfalls. Radiat Oncol. 2019;14(1):1–6. doi: 10.1186/s13014-019-1275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudra S, et al. Using adaptive magnetic resonance image-guided radiation therapy for treatment of inoperable pancreatic cancer. Cancer Med. 2019;8(5):2123–2132. doi: 10.1002/cam4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Placidi L, et al. On-line adaptive MR guided radiotherapy for locally advanced pancreatic cancer: Clinical and dosimetric considerations. Tech Innov Patient Support Radiat Oncol. 2020;15:15–21. doi: 10.1016/j.tipsro.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinohara RT, et al. Statistical normalization techniques for magnetic resonance imaging. NeuroImage Clin. 2014;6:9–19. doi: 10.1016/j.nicl.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Research data are not available at this time.