Abstract
Melanoma is an aggressive type of cancer originating from the skin that arises from neoplastic changes in melanocytes. Transforming growth factor-β (TGF-β) is a pleiotropic cytokine and is known to contribute to melanoma progression by inducing the epithelial-mesenchymal transition (EMT) program and creating an environment that favors tumor progression. There are three TGF-β isoforms, TGF-β1, TGF-β2 and TGF-β3, all of which engage in pro-tumorigenic activities by activating SMAD signaling pathways. All TGF-β isoforms activate signaling pathways by binding to their TGF-β type I (TβRI) and type II (TβRII) receptors. Thus, effective targeting of all TGF-β isoforms is of great importance. In the present study, chimeric proteins comprising the extracellular domains of TβRI and/or TβRII fused with the Fc portion of human immunoglobulin (IgG) were validated in the melanoma context. The Fc chimeric receptor comprising both TβRI and TβRII (TβRI-TβRII-Fc) effectively trapped all TGF-β isoforms. Conversely, TβRII-Fc chimeric receptor, that comprises TβRII only, was able to interact with TGF-β1 and TGF-β3 isoforms, but not with TGF-β2, which is a poor prognostic factor for melanoma patients. Accordingly, it was revealed that TβRI-TβRII-Fc chimeric receptor suppressed the EMT program in melanoma cells in vitro induced by any of the three TGF-β isoforms, as revealed by decreased expression of mesenchymal markers. Conversely, TβRII-Fc chimeric receptor inhibited the EMT program induced by TGF-β1 and TGF-β3. In addition, it was established that tumor growth in subcutaneous mouse melanoma was inhibited by TβRI-TβRII-Fc chimeric receptor indicating that Fc chimeric receptor could be applied to modify the tumor microenvironment (TME) of melanoma. Therefore, designing of Fc chimeric receptors targeting TGF-β signals that affect various components of the TME may result in the development of effective anti-melanoma agents.
Keywords: melanoma, TGF-β, tumor microenvironment, Fc chimeric receptor, EMT
Introduction
Malignant melanoma (melanoma) is the most aggressive skin cancer which arises from pigment-producing cells, melanocytes, or from dysplastic nevus cells. Despite various options basically involving enlarged surgical excision with or without anticancer drugs (chemotherapy) and various immune checkpoint inhibitors (1), there is a high demand for the development of new, more effective melanoma-targeted therapies.
The tumor microenvironment (TME) consists of tumor cells and tumor stroma including cancer-associated fibroblasts (CAFs), immune cells, and tumor endothelial cells (TECs). Cancer cells, CAFs, and TECs secrete transforming growth factor-β (TGF-β), a multifunctional cytokine, creating a favorable microenvironment that promotes tumor progression (2). There are three TGF-β isoforms that share structural similarity, TGF-β1, TGF-β2, and TGF-β3, and signal through the same type I (TβRI) and type II (TβRII) transmembrane receptors that are endowed with serine/threonine kinase activity (3). The formation of a complex between TGF-β and TβRII followed by binding of TβRI that results in activation of TβRI and phosphorylation of downstream components Smad2/3, which is followed by binding of Smad4 and translocation of the whole complex to the nucleus and transcription of direct target genes, plasminogen activator inhibitor-1 (PAI-1) (4) or transmembrane prostate androgen-induced protein (TMEPAI) (5).
TGF-β signaling has been implicated in the progression of various tumors by enhancing cell migration, promoting the invasion of cancer cells, and suppressing immune responses (6,7). The ability of TGF-β to induce epithelial-mesenchymal transition (EMT), which endows cancer cells of epithelial origin with the mesenchymal features, leading to loss of cell-cell contact and enhanced motility has also been widely studied (8). In the cells undergoing EMT, decreased expression of epithelial cell markers, E-cadherin or claudin-1 accompanied by increased expression of mesenchymal cell markers, smooth muscle protein 22α (SM22α), α-smooth muscle actin (αSMA), and fibronectin is observed (9). Although melanoma does not represent epithelial tumors, a decrease in E-cadherin expression and upregulated expression of EMT-related transcription factors, Snail, Slug, Twist, and Zeb1 have been revealed to be correlated with the enhanced invasion and acquisition of stem cell-like properties (10,11).
Elevated expression levels of TGF-β have been revealed to be associated with melanoma progression (12). Especially TGF-β2 can be detected in early as well as in advanced melanomas. A positive association between TGF-β2 expression level and tumor thickness (13) and high plasma levels of TGF-β2 have been observed in melanomas at advanced stages. TGF-β signals have been reported to stimulate melanoma cell dissemination from primary tumors (14). Previous findings indicated that activation of TGF-β signals in murine melanoma led to the upregulation of PAI-1 expression that resulted in tumor growth inhibition (15). Conversely, other studies demonstrated that activation of TGF-β signals facilitated progression of malignant melanoma (13,16) by stimulating cell proliferation in vivo and metastasis (17). In addition, TGF-β has also been revealed to affect stromal, immune, or endothelial cells and by this means exerts immunosuppressive and pro-angiogenic activities (18).
Current attempts in the development of melanoma-targeted therapies are based on the inhibition of proteins involved in the mitogen-activated protein kinase (MAPK) signal transduction pathway, B-Raf (BRAF) and MAPK kinase (1). In addition, the important role of interleukin-13 signals in melanoma progression has also been suggested (19). However, the previously approved drugs vemurafenib, dabrafenib, and trametinib which target the MAPK signaling pathway (20,21), may potentiate the risk of developing other skin cancers or formation of peripheral edema (22). As TGF-β has been associated with progression of melanomas, there have also been attempts to target TGF-β signals by using antisense oligonucleotides (trabedersen; API12009) (23), monoclonal anti-TGF-β antibodies (fresolimumab; GC1008) (24) or a low molecular weight compound (vactosertib; TEW-7197) that inhibits TβRI kinase activity (25). The efficacy of these approaches is still under evaluation; however, at present, various side effects such as cutaneous keratoacanthomas or cardiac toxicity have been observed (26).
Recently there have been attempts to develop chimeric proteins comprising the ligand-interacting ectodomains of TGF-β receptors fused with Fc domain of human immunoglobulin (IgG). The addition of the Fc region of IgG is known to prolong plasma half-life of such chimeric Fc receptors (27–29) and to affect their biological activities. As reviewed by Marotte and Cimaz, etanercept, a human tumor necrosis factor (TNF) receptor p75 fused with the Fc domain of human IgG has been revealed to have a prolonged half-life, resulting in an extended and more profound biological effect than its native form (30). A TβRI-TβRII-Fc chimeric receptor comprising extracellular domains of TGF-β type I and II receptors fused with the Fc portion of human IgG, which could effectively trap all TGF-β isoforms and inhibited EMT in oral squamous cell carcinoma cells as well as primary tumor growth, was previously designed by our research group (31). As all TGF-β isoforms facilitate melanoma progression, it was hypothesized that TβRI-TβRII-Fc chimeric receptor could potentially interfere with tumor-inducing TGF-β signals in melanomas. Thus, in the present study, using a melanoma syngeneic model and B16-F0 cell line, the effect of soluble TβRI-TβRII-Fc chimeric receptors on the induction of EMT and progression of melanoma were examined.
Materials and methods
Cell culture
The B16-F0 mouse melanoma cell line (cat. no. JCRB0202) was purchased from Japanese Collection of Research Bioresources (JCRB) Cell Bank and maintained in Eagle's Minimum Essential Medium (EMEM; FUJIFILM Wako Pure Chemical Corporation) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA), 100 U/ml penicillin and 100 µg/ml streptomycin (both from Nacalai Tesque, Inc.) under mycoplasma-free conditions. Clone M3 (Cloudman S91) melanoma cell line was obtained from European Collection of Authenticated Cell Cultures (ECACC) and cultured in Ham's F10 medium (FUJIFILM Wako Pure Chemical Corporation) supplemented with 15% FBS, 2 mM glutamine (Nacalai Tesque, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin. 293T and 293FT cells were obtained from Invitrogen; Thermo Fisher Scientific, Inc. HEK-Blue™ TGF-β cells were purchased from InvivoGen. 293T, 293FT and HEK-Blue™ TGF-β cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Nacalai Tesque, Inc.) supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. The cultured medium for 293FT cells was also supplemented with 1% non-essential amino acid solution (Nacalai Tesque, Inc.). All cell lines were cultured in a humidified incubator containing 5% CO2 at 37°C.
Reagents
TGF-β1 (PeproTech, Inc.), TGF-β2 (PeproTech, Inc.) and TGF-β3 (R&D Systems, Inc.) were used at concentrations of 1 ng/ml or 3 ng/ml depending on the experiment. SB431542 (FUJIFILM Wako Pure Chemical Corporation) was used at a concentration of 10 µM.
PrognoScan analysis
The correlation between the levels of expression of genes encoding all human TGF-β isoforms, TGFB1, TGFB2 or TGFB3 and overall survival of melanoma patients was performed using a public database, PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/) which comprises multiple cancer microarray datasets (32). TGFB1, TGFB2 and TGFB3 were used as queries. The analysis was conducted by minimum P-value approach, which allowed grouping of patients into two groups based on the expression levels of TGFB1, TGFB2 and TGFB3 at all possible cutoffs (cutoff points providing the best minimum corrected P-value were 0.76 for TGFB1, 0.87 for TGFB2 and 0.66 for TGFB3 analyses, respectively). The analysis results in the present study were based on the evaluation of TGFB1, TGFB2 and TGFB3 expression levels and survival of melanoma patients whose data was combined in dataset: GSE19234 (33). The log-rank test was used for statistical analysis.
Cell proliferation assay
The effect of expression of Fc chimeric receptors on proliferation of B16 melanoma cells was evaluated by WST-1 assay. The B16 cells (3.5×104) were seeded into a 12-well culture plate and incubated overnight at 37°C, 5% CO2. The following day the medium was refreshed, and the cells were cultured for 72 h. The assay was conducted using the WST-1 reagent (Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol. The colorimetric changes in the substrate were measured with a microplate reader (Model 680; Bio-Rad Laboratories, Inc.) at 450 nm. To evaluate the effect of TGF-β isoforms on the proliferation of B16 melanoma cells, B16 cells (7.5×104 cells/well) were seeded into 6-well culture plates and cultured overnight at 37°C in 5% CO2. The following day, the medium was replaced with 1 ml of EMEM and cells were treated with each TGF-β (3 ng/ml) isoform or SB431542 (10 µM) for 72 h followed by direct cell counting with Bürker-Türk hemocytometer (cat. no. 03-303-1; Erma, Inc.). The experiment was performed in triplicate and repeated twice.
Production of Fc chimeric receptors
Fc chimeric receptors were generated by transfection of 293T cells with respective Fc chimeric receptor expression vectors. The construction of expression vectors was performed as previously described (31). Briefly, to express Control-Fc protein, the Fc region of human IgG fused to the interleukin-2 signal peptide was inserted into pENTR201 vector (Invitrogen; Thermo Fisher Scientific, Inc.). For the expression of TβRII-Fc chimeric receptor the extracellular domain (ECD) of TβRII-Fc corresponding to the 184 amino acids (ECD1-184) was fused with the Fc region of human IgG and inserted into pENTR201 (Invitrogen; Thermo Fisher Scientific, Inc.). To express TβRI-TβRII-Fc chimeric receptor the ECD of TβRI corresponding to the 128 amino acids (ECD1-128) was fused with ECD of TβRII-Fc lacking signal peptide (ECD23-184) followed by the addition of the Fc region of human IgG and inserted into pENTR201 (Invitrogen; Thermo Fisher Scientific, Inc.). The Gateway Technology (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfer Control-Fc, TβRII-Fc and TβRI-TβRII-Fc cDNAs into pCSII-EF-RfA (a gift from Dr Hiroyuki Miyoshi, Keio University, deceased) to generate lentiviral expression vectors; pCSII-EF-RfA-Control-Fc, pCSII-EF-RfA-TβRII-Fc and pCSII-EF-RfA-TβRI-TβRII-Fc. 293T cells (9.0×106) were seeded into 10-cm tissue culture dishes and cultured overnight at 37°C and 5% CO2, followed by transfection with pCSII-EF-RfA vectors (20 µg/dish) expressing each chimeric receptor pCSII-EF-RfA-Control-Fc, pCSII-EF-RfA-TβRII-Fc and pCSII-EF-RfA-TβRI-TβRII-Fc, using Lipofectamine 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A total of 4 h post-transfection, the medium was replaced with serum-free Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) and the cells were incubated for 48 h to allow accumulation of secreted Fc chimeric receptors in culture medium. The accumulation of soluble chimeric receptors in the conditioned media was evaluated by immunoblotting with rabbit polyclonal anti-human IgG-Fc fragment (1:5,000; A80-105; Bethyl Laboratories, Inc.) as described in the Immunoblot analysis section. The concentration of each Fc chimeric receptor was assessed by enzyme-linked immunosorbent assay (ELISA) with the Human IgG ELISA Quantitation Set (E80-104; Bethyl Laboratories, Inc.). The collected conditioned media were aliquoted and stored at −80°C until use.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from B16 and Clone M3 cells. The extraction was performed using Sepasol (R)-RNA I Super G (Nacalai Tesque, Inc.) and reverse-transcribed to cDNA with ReverTra Ace qPCR-RT Master Mix (Toyobo Life Science) according to the manufacturer's protocol. Quantitative PCR analysis was performed using SYBR Green (Roche Applied Science) on a Step One Plus Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Inc.) under the following cycling conditions: 95°C, 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 30 sec with a final incubation at 95°C for 5 sec. The relative standard curve method was used to determine the relative expression of target genes (34). All expression data were normalized to the expression of β-actin. The genes and corresponding primer sequences are listed in Table SI.
Protein extraction
B16, Clone M3 and 293T cells were lysed using radioimmunoprecipitation assay buffer (RIPA Lysis Buffer System; Santa Cruz Biotechnology, Inc.) in the presence of a protease inhibitor (Sigma-Aldrich; Merck KGaA), followed by repeated freeze and thawing. Cell lysates were cleared by centrifugation at 16,400 × g for 30 min at 4°C, and the supernatants were collected. The protein concentration in obtained lysates was determined using a BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.).
Immunoblot analysis
Denatured cell lysates (30 µg of total proteins in Figs. 5A and S5B; 20 µg of proteins in Figs. 3D, 4C, S3B and S4B; or 10 µg of total protein in Fig. S1) were separated on 10.5% or 12% SDS-PAGE gel depending on the experiment, followed by transfer onto PVDF membranes (Merck KGaA). The membranes were then blocked with 3% bovine serum albumin (BSA; FUJIFILM Wako Pure Chemical Corporation) for 30 min at room temperature and incubated with appropriate primary antibodies diluted in 3% BSA (Nacalai Tesque, Inc.): Rabbit monoclonal anti-αSMA (1:1,000; product no. 19245; Cell Signaling Technology, Inc.), rabbit polyclonal anti-TAGLN/Transgelin (SM22α; 1:2,000; product code ab14106; Abcam), rabbit polyclonal anti-human IgG-Fc fragment (1:5,000), and rabbit polyclonal anti-α-tubulin (1:10,000; product code ab4074; Abcam) overnight at 4°C. The membranes were then incubated with goat anti-rabbit IgG HRP-linked antibody (1:5,000; product no. 7074S; Cell Signaling Technology, Inc.) for 1 h at room temperature. The target proteins were detected using an Enhanced Chemiluminescence Kit (ECL detection reagent; Cytiva) and visualized with a Fusion Solo S Imaging System (SOLO.6S.EDGE; Vilber Lourmat).
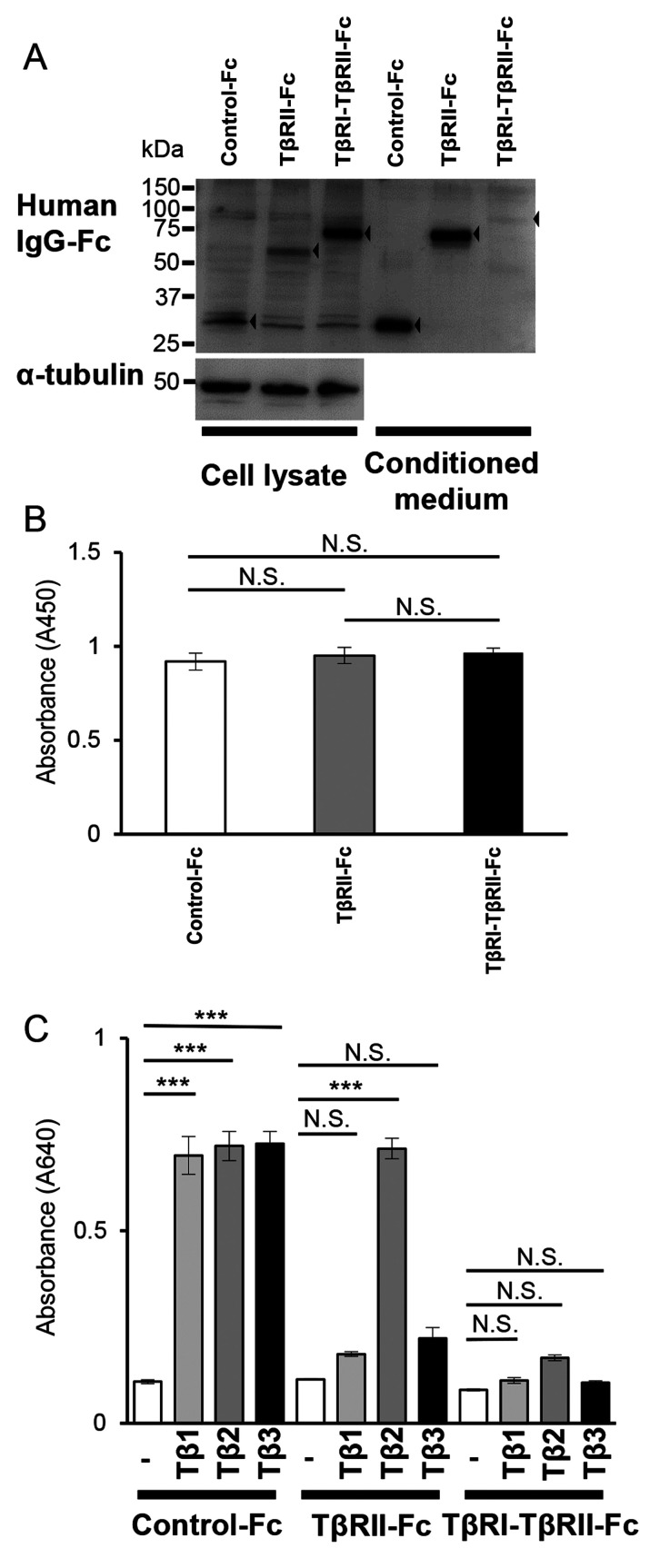
Figure 5.
TβRI-TβRII-Fc chimeric receptor expressed and secreted by B16 cells traps all TGF-β isoforms. B16 cells were infected with lentivirus expressing each Fc chimeric receptor (Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc). (A) The Fc chimeric receptors (black arrowhead; left, cell lysate; right, conditioned medium) were visualized by immunoblotting analysis using anti-human IgG-Fc antibody. α-tubulin was used as a loading control for the cell lysate. (B) The WST-1 cell proliferation assay of B16 cells expressing each Fc chimeric receptor. B16 cells were seeded into a 12-well tissue culture plate and allowed to grow for 72 h followed by the WST-1 assay. Error bars, SD. (C) HEK-Blue TGF-β reporter assay showing TGF-β-trapping ability of Fc chimeric receptors secreted by B16 cells. The values represent TGF-β signal activation corresponding to the colorimetric changes of the Quanti-Blue substrate by SEAP at 640 nm. Each experiment was performed in triplicate and repeated twice. Error bars, SD. ***P<0.001. TβRI, TGF-β type I receptor; TβRII, TGF-β type II receptor; TGF-β, transforming growth factor-β; NS, not significant; SEAP, secreted alkaline phosphatase; IgG, immunoglobulin G.
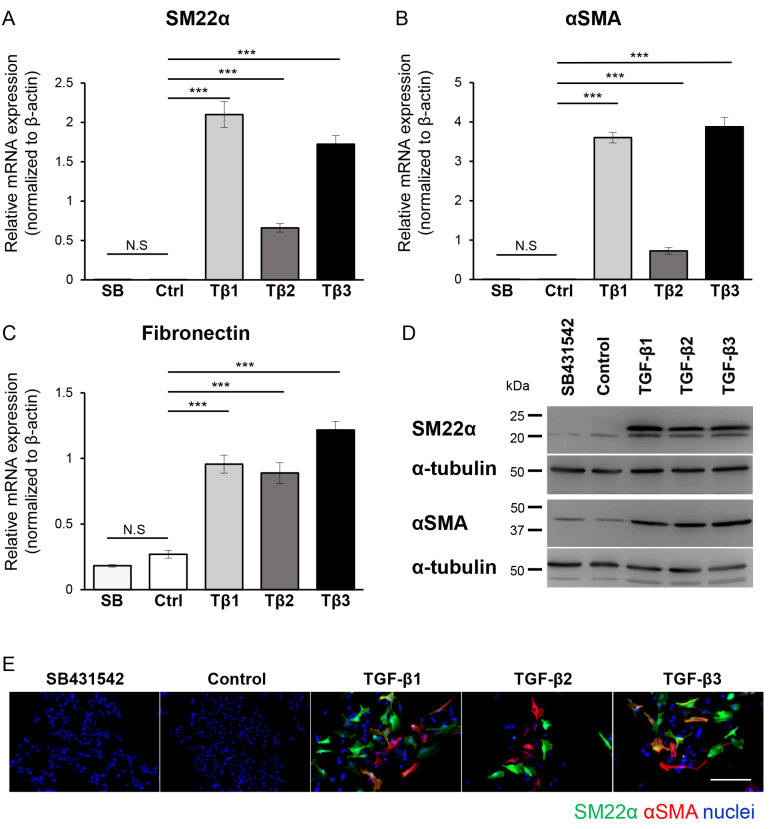
Figure 3.
EMT program in B16 melanoma cells is induced by all TGF-β isoforms. B16 cells were cultured in the absence (Ctrl or Control) or presence of TGF-β1 (Tβ1), TGF-β2 (Tβ2), or TGF-β3 (Tβ3) (3 ng/ml) or the TβRI kinase inhibitor, SB431542 (SB; 10 µM) for 72 h, followed by (A-C) RT-qPCR, (D) immunoblotting and (E) immunocytochemistry. Experiments were performed in triplicate and repeated twice. (A-C) The expression of mesenchymal markers (A) SM22α, (B) αSMA and (C) fibronectin were evaluated by RT-qPCR analyses. All RT-qPCR data were normalized to the β-actin expression. (D) The immunoblotting analysis with antibodies specific to SM22α, αSMA and α-tubulin (loading control). (E) Representative immunofluorescence images revealing staining of SM22α (green), αSMA (red) and nuclei (blue). Scale bar, 100 µm. Error bars, SD. ***P<0.001. EMT, epithelial-mesenchymal transition; TGF-β, transforming growth factor-β; TβRI, TGF-β type I receptor; RT-qPCR, reverse transcription-quantitative PCR; NS, not significant; SM22α, smooth muscle protein 22α; αSMA, α-smooth muscle actin.
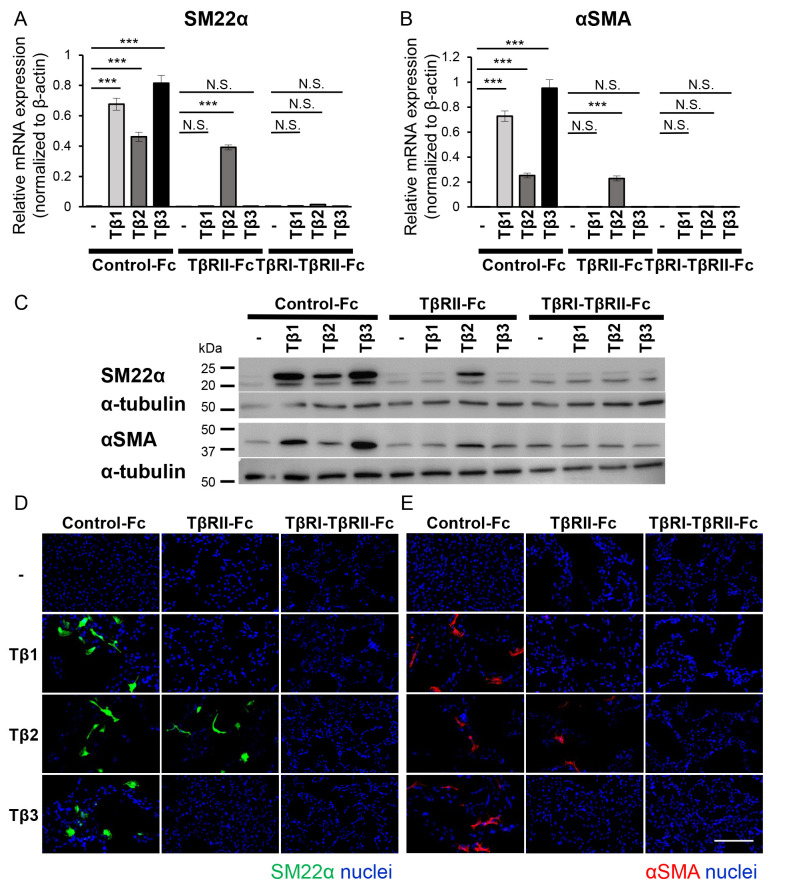
Figure 4.
TβRI-TβRII-Fc chimeric receptor inhibits the TGF-β-induced EMT program in B16 cells. B16 cells were incubated in the absence (−) or presence of TGF-β isoforms [TGF-β1 (Tβ1), TGF-β2 (Tβ2), or TGF-β3 (Tβ3)] (3 ng/ml) in combination with conditioned media from 293T cells containing Fc chimeric receptors (Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc). The activation of the EMT program, was estimated by (A and B) RT-qPCR analyses, (C) immunoblotting and (D and E) immunocytochemistry. Each experiment was performed in triplicate and repeated twice. (A and B) The RT-qPCR analysis for the expression of mesenchymal markers (A) SM22α and (B) αSMA. All data were normalized to the β-actin expression. (C) The immunoblotting analysis of the expression levels of SM22α, αSMA and α-tubulin (loading control). (D and E) Immunofluorescence staining of (D) SM22α (green) and (E) αSMA (red). The nuclei were stained with Hoechst (blue). Scale bar, 100 µm. Error bars, SD. ***P<0.001. TβRI, TGF-β type I receptor; TβRII, TGF-β type II receptor; TGF-β, transforming growth factor-β; EMT, epithelial-mesenchymal transition; RT-qPCR, reverse transcription-quantitative PCR; SM22α, smooth muscle protein 22α; αSMA, α-smooth muscle actin; NS, not significant.
Immunocytochemistry
B16 and Clone M3 cells (3.5×104 cells/well) were seeded on cover glasses placed into 12-well tissue culture plates and treated with TGF-β1, -β2, and -β3 in the presence or absence of Fc chimeric receptors for 72 h, at 37°C in 5% CO2. The cells were then fixed with methanol/acetone (1:1) for 20 sec on ice and blocked in PBS containing 1% BSA (FUJIFILM Wako Pure Chemical Corporation) for 30 min at room temperature and incubated with primary antibodies diluted in Blocking One buffer: Rabbit polyclonal anti-TAGLN/Transgelin (1:1,000), mouse monoclonal anti-actin, αSMA-Cy3™ (1:1,000; cat. no. C6198-2ML; Sigma-Aldrich; Merck KGaA) overnight at 4°C. To visualize SM22α and nuclei, samples were incubated for 1 h at room temperature with a mixture of donkey polyclonal anti-rabbit IgG (H+L) Alexa Fluor 488-conjugated secondary antibodies (1:1,000 in Blocking One buffer; cat. no. A-21206; Thermo Fisher Scientific, Inc.) and 500 ng/ml Hoechst33342 (Cell Signaling Technology, Inc.) for nuclear staining. The samples were then embedded in Fluoromount-G mounting medium (Cosmo Bio Co., Ltd.). Images were captured under a fluorescence microscope (BZ-X710; Keyence Corporation).
Treatment of B16 and Clone M3 cells with Fc chimeric receptors
B16 or Clone M3 cells (3.5×104 cells/well) were seeded into tissue culture plates (12-well plate or 6-well plate, depending on the experiment) and cultured overnight at 37°C in 5% CO2. The next day, the medium was replaced with 1 ml of serum-free Opti-MEM. The Fc chimeric receptor/ligand complexes were generated by mixing the conditioned medium from the 293T cells expressing Fc chimeric receptors containing the equal amount of Fc chimeric receptors (600 ng of Fc chimeric receptors in 500 µl of Opti-MEM) with TGF-β1, -β2, or -β3 at the concentration of 3 ng/ml. Samples were incubated for 2 h at 37°C to allow formation of the Fc chimeric receptor/ligand complexes and added to the B16 or Clone M3 cells. The cells were then incubated at 37°C and 5% CO2 with Fc chimeric receptor/ligand complexes for 4 or 72 h (depending on the experiment) and subjected to gene expression analysis by RT-qPCR, immunocytochemistry, or immunoblotting. Cells treated with each TGF-β ligand in serum-free Opti-MEM were used as controls for upregulation of the TGF-β signal while samples treated with the mixture of SB431542, a TβRI kinase inhibitor, and TGF-β in serum-free Opti-MEM were used as controls for inhibition of the TGF-β signal.
Lentivirus production and transduction of B16 cells
The lentiviral particles were produced as previously described (31). Briefly, the 293FT (8.0×106) cells were co-transfected with 5.5 µg of expression plasmids (pCSII-EF-RfA-Control-Fc, pCSII-EF-RfA-TβRII-Fc, and pCSII-EF-RfA-TβRI-TβRII-Fc) and packaging plasmids pCMV–VSV-G-RSV-Rev (3.25 µg; RIKEN BioResource Center) and pCAG-HIVgp (3.25 µg; RIKEN BioResource Center) using Lipofectamine 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in 5 ml Opti-MEM supplemented with 10% FBS. The control lentiviral particles expressing green fluorescent protein (GFP) were prepared by transfecting 293T cells with 5.5 µg of pCSII-EF-RfA-GFP and packaging plasmids pCMV–VSV-G-RSV-Rev (3.25 µg) and pCAG-HIVgp (3.25 µg). A total of 24 h post-transfection, the transfection medium was refreshed with 7.5 ml Opti-MEM, 10% FBS, and the recombinant lentiviruses were harvested 48 h later. The conditioned media containing viral particles were collected by centrifugation at 4°C for 5 min at 1,700 × g, and incubated at 4°C for 7 days on the rotary shaker with Lenti-X Concentrator (Takara Bio, Inc.). The viral particles were then centrifuged at 4°C for 45 min, at 1,500 × g, and resuspended in 140 µl of Opti-MEM. The 70-µl of concentrated lentiviral particles were used to infect B16 melanoma cells (3×105 cells/well in 12-well tissue culture plates). Transduction efficiency was evaluated using lentiviral particles expressing green GFP. The successful expression was examined by immunoblotting using rabbit polyclonal anti-human IgG-Fc antibody as described in the Immunoblot analysis section. The second generation of transduced B16 cells was used for further experiments.
Smad 2/3/4-responsive reporter assay (HEK-Blue reporter assay)
The ability of Fc chimeric receptors, secreted by transduced B16 cells, to trap TGF-β ligands was examined using the HEK-Blue TGF-β reporter system. The B16 cells (1×106 cells) expressing each of the Fc chimeric receptors were seeded into 10 cm tissue culture plates and incubated overnight at 37°C in 5% CO2. The following day, the medium was replaced with 5 ml of serum-free Opti-MEM and the cells were incubated for 48 h to allow the accumulation of Fc chimeric receptors in the culture supernatant. The conditioned media were collected and stored at −80°C until use.
HEK-Blue TGF-β reporter cells (1.0×105) were seeded into 96-well plates and incubated overnight at 37°C in 5% CO2. The following day, the medium was replaced with 200 µl of serum-free DMEM, and the cells were incubated for 3 h. The B16 cell-derived conditioned medium containing Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc chimeric proteins, was mixed with TGF-β ligands (1 ng/ml) and incubated for 2 h at 37°C to allow the formation of Fc chimeric receptor/ligand complexes. Next, Fc chimeric receptor/ligand complexes were added to the HEK-Blue TGF-β reporter cells, followed by incubation for 24 h, at 37°C. The activation of TGF-β/Smad signals was detected using QUANTI-Blue substrate (InvivoGen) following incubation for 30 min at 37°C. The colorimetric change of the substrate by the secreted alkaline phosphatase (SEAP) was quantified at 640 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Subcutaneous syngeneic tumor mouse model
A total of 62 female C57/BL6 mice (5–6 weeks old; average weight, 14–19 g) were purchased from Japan SLC, Inc. All animal experimental protocols were approved (approval no. R-02-017-1) by the Animal Experiment Committee of the Graduate School of Dentistry, Osaka University (Osaka, Japan). The mice were kept under a temperature of 23–24°C with 40–60% humidity and a 12-h light/dark cycle. Mice were provided with access to food and water ad libitum throughout the experiment. A total of 20 mice was used for injection of B16-Control-Fc cells, 21 for B16-TβRII-Fc cells, and 21 for B16-TβRI-TβRII-Fc cells. All animals underwent general anesthesia with mixture of medetomidine (0.3 mg/kg; Nippon Zenyaku Kogyo, Co., Ltd.) midazolam (4 mg/kg; Astellas Pharma, Inc.) and butorphanol (5 mg/kg; Meiji Seika Kaisha, Ltd.) by intraperitoneal administration (35,36). The B16 cells (5.0×105) expressing the Fc chimeric receptors, B16-Control-Fc, B16-TβRII-Fc, B16-TβRI-TβRII-Fc were suspended in 50 µl serum-free EMEM and subcutaneously injected into left flank region. Mice that did not develop any palpable tumor or did not survive until the endpoint of the experiment were excluded from the analysis. Tumor growth was monitored for 26 days. The tumor volume was measured twice per week and estimated using the following equation: Tumor volume (mm3) = [length (mm) × width (mm)2]/2. The size of developed tumors was selected as the humane endpoint. Mice were sacrificed when the size of the largest primary tumors started to exceed the 1,000 mm3. As large melanoma tumors often develop necrotic changes that would likely affect the experimental outcome, a total of 32 mice bearing primary tumors derived from Control-Fc (n=11), TβRII-Fc (n=9), and TβRI-TβRII-Fc (n=11) were thus euthanized on day 26 by intraperitoneal injection of the mixture of medetomidine (3 mg/kg), midazolam (40 mg/kg) and butorphanol (50 mg/kg).
Statistical analysis
Statistical analysis was carried out by EZR software (37). Results are presented as the mean ± standard deviation (SD) or standard error (SE). Each experiment was performed in triplicate and repeated twice. Comparisons of quantitative data were conducted using one-way ANOVA with post hoc Tukey's test or Mann-Whitney U test with post hoc Bonferroni test depending on experiment. P<0.05 was considered to indicate a statistically significant difference.
Results
TβRI-TβRII-Fc chimeric receptor efficiently suppresses TGF-β signals in melanoma cells
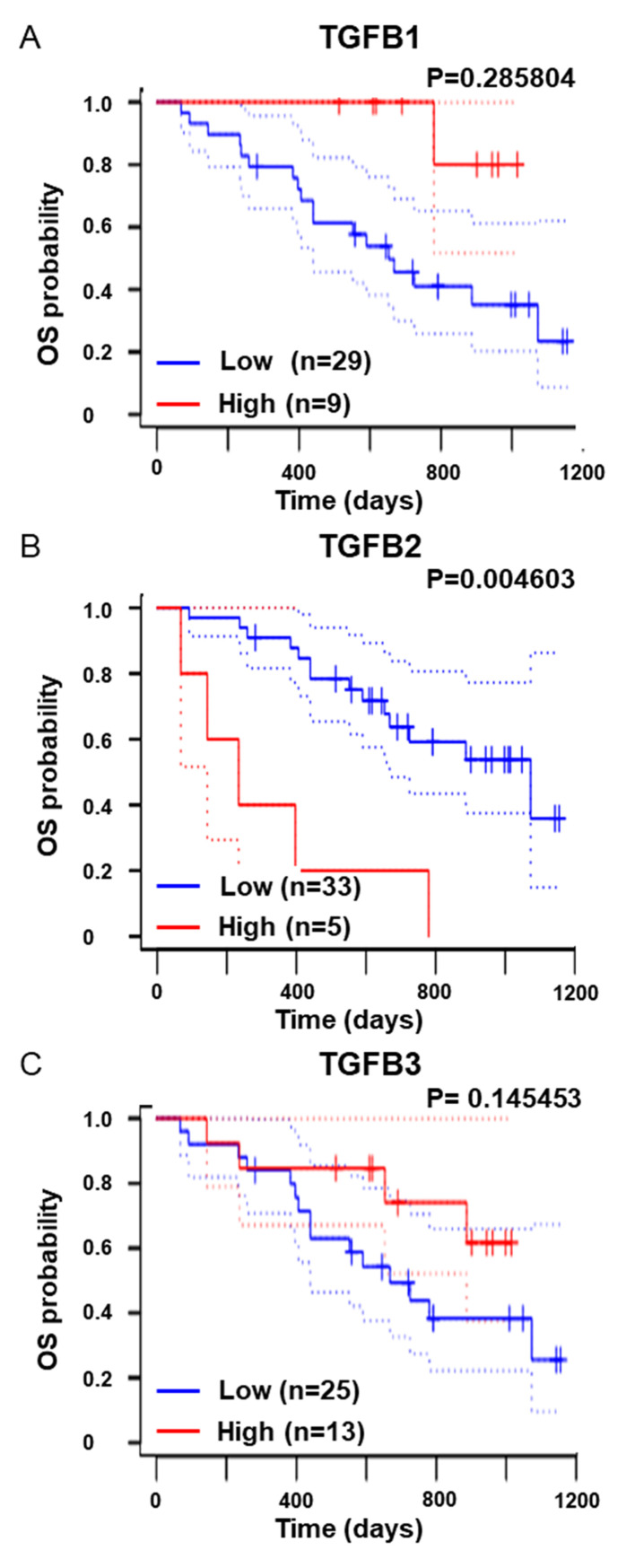
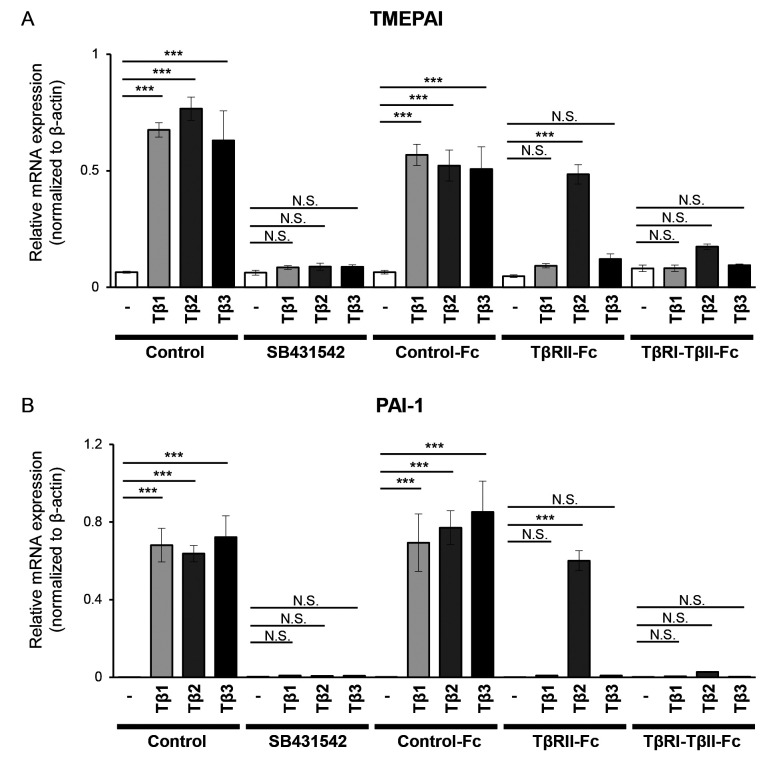
TGF-β has been revealed to promote invasiveness and progression of melanoma (13,16). Previous studies revealed that melanoma cells expressed all three TGF-β isoforms (12) and that an elevated level of TGF-β in melanoma patients was associated with metastatic outcomes (38). In addition, meta-analysis using a public database, PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/) (32) and dataset: GSE19234 (33), revealed that high expression levels of TGF-β2, but not that of TGF-β1 or TGF-β3, were associated with overall survival of melanoma patients (Fig. 1). Therefore, it was examined whether TβRI-TβRII-Fc chimeric receptor could be applied in the melanoma model. In our study, B16 melanoma cells were used, in which the EMT program can be activated in response to TGF-β. 293T cells were transfected with the vectors expressing Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc chimeric receptors and the accumulation of chimeric proteins in the conditioned media was confirmed by immunoblotting (Fig. S1). Such conditioned media were then used to examine the effect of soluble chimeric receptors on the activation of TGF-β signals. B16 cells were incubated in conditioned media of 293T cells expressing Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc chimeric receptors in an absence or presence of TGF-β1, -β2, or -β3, respectively, followed by an analysis of the expression of genes directly responding to TGF-β, TMEPAI, and PAI-1 by RT-qPCR. Incubation of B16 melanoma cells with any of the TGF-β isoforms upregulated the expression of both TMEPAI (Fig. 2A) and PAI-1 (Fig. 2B). As anticipated, SB431542, a TβRI kinase inhibitor, reduced the expression of both direct target genes to the background level (Fig. 2). Incubation of B16 melanoma cells with Control-Fc protein did not reduce the expression of TMEPAI, and PAI-1 induced by TGF-βs. Reduced expression of both genes in the presence of TβRII-Fc chimeric receptor was only observed when cells were treated with TGF-β1 and TGF-β3, but not by TGF-β2 (Fig. 2). Conversely, TβRI-TβRII-Fc chimeric receptor significantly inhibited the expression levels of TMEPAI and PAI-1 induced by all TGF-β isoforms indicating that TβRI-TβRII-Fc chimeric receptor effectively interfered with TGF-β signals also in the melanoma model. To generalize our findings, the same set of experiments were performed with another melanoma cell line, Clone M3. Clone M3 cells responded to all TGF-β isoforms as indicated by upregulated expression of TMEPAI (Fig. S2A) and PAI-1 (Fig. S2B). In addition, incubation with TβRI-TβRII-Fc decreased the expression of TMEPAI and PAI-1 induced by all TGF-βs when compared with the expression of both genes detected in the Clone M3 cells incubated with Control-Fc protein indicating that TβRI-TβRII-Fc chimeric receptor suppressed TGF-β signals in multiple types of melanoma cells.
Figure 1.
Elevated TGF-β2 expression is associated with poor prognosis in melanoma patients. Kaplan-Meier OS curves of melanoma patients (n=38) revealing the association with high (red) and low (blue) expression of (A) TGF-β1 (TGFB1), (B) TGF-β2 (TGFB2), and (C) TGF-β3 (TGFB3) using PrognoScan: A database for meta-analysis of the prognostic value of genes (http://dna00.bio.kyutech.ac.jp/PrognoScan/), and dataset: GSE19234. The statistical analysis reflects the log-rank P-values. The dotted lines represent 95% confidence intervals for each analyzed group. TGF-β, transforming growth factor-β; OS, overall survival.
Figure 2.
TβRI-TβRII-Fc chimeric receptor inhibits the expression of TGF-β direct target genes in B16 melanoma cells. B16 cells were incubated for 4 h in the absence (−) or presence of TGF-β isoforms [(TGF-β1 (Tβ1), TGF-β2 (Tβ2), or TGF-β3 (Tβ3) (3 ng/ml)] in combination with a vehicle (Control), 10 µM SB431542 or conditioned media from 293T cells containing Fc chimeric receptors (Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc). The expression levels of (A) TMEPAI and (B) PAI-1 were determined through reverse transcription-quantitative PCR analysis. Each experiment was performed in triplicate and repeated twice. All data are normalized to the expression of β-actin. Error bars, SD. ***P<0.001. TβRI, TGF-β type I receptor; TβRII, TGF-β type II receptor; TGF-β, transforming growth factor-β; NS, not significant; TMEPAI, transmembrane prostate androgen-induced protein; PAI-1, plasminogen activator inhibitor-1.
All TGF-β isoforms activate the EMT program in melanoma cells
Melanoma cells can activate the EMT program in response to TGF-β. Previous studies revealed that melanoma cells treated with TGF-β upregulated the expression of mesenchymal markers (14,17). Therefore, it was examined whether similar changes can be observed in B16 melanoma cells. B16 melanoma cells were cultured for 72 h in the absence or presence of each TGF-β isoform or SB431542, and the expression of various mesenchymal markers was determined using RT-qPCR. The treatment with any of TGF-β isoform resulted in upregulated expression of all mesenchymal markers; SM22α (Fig. 3A), αSMA (Fig. 3B) and fibronectin (Fig. 3C), while SB431542 did not exhibit any effect (Fig. 3A-C). These results were also confirmed at the protein level using immunoblotting and immunocytochemical analysis. A significant increase was observed in the band intensity corresponding to each mesenchymal marker, SM22α and αSMA (Fig. 3D), as well as an increase in a fluorescent signal related to the presence of SM22α-positive and αSMA-positive cells upon TGF-β treatment (Fig. 3E), indicating that B16 melanoma cells activated TGF-β-dependent EMT. The activation of the EMT program was also confirmed using Clone M3 cells. Treatment of the Clone M3 cells with any of the three TGF-β isoforms resulted in upregulated expression of SM22α as revealed by RT-qPCR (Fig. S3A), immunoblotting (Fig. S3B), and immunocytochemical analysis (Fig. S3C), supporting the findings that treatment with TGF-β upregulated the expression of SM22α in multiple types of melanoma cells.
EMT program, induced by all TGF-β isoforms in melanoma cells, is inhibited in the presence of TβRI-TβRII-Fc chimeric receptor
Our data indicated that the EMT program was activated upon treatment with any of the three TGF-β isoforms. In addition, as shown in Fig. 1B, high TGFB2 expression was a poor prognostic factor in overall survival in melanoma patients, indicating that inhibiting the EMT program may have beneficial effects on melanoma treatment. Therefore, in the following experiment, the effect of TβRI-TβRII-Fc chimeric receptor on activation of the EMT program was examined in B16 melanoma cells. The B16 melanoma cells were treated without or with TGF-β1, -β2 or -β3 in the presence of conditioned medium derived from 293T cells expressing Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc chimeric receptors for 72 h, followed by RT-qPCR analysis for the expression of mesenchymal markers, SM22α (Fig. 4A) and αSMA (Fig. 4B). The expression levels of both mesenchymal markers were upregulated when B16 melanoma cells were incubated in the conditioned medium containing Control-Fc protein. A significant suppression of SM22α (Fig. 4A) and αSMA (Fig. 4B) expression was observed in the cells stimulated with TGF-β1 or TGF-β3 in the presence of TβRII-Fc or TβRI-TβRII-Fc chimeric receptors. Conversely, the induction of the EMT program by TGF-β2 was inhibited only in the presence of TβRI-TβRII-Fc chimeric receptor (Fig. 4A and B), indicating that TβRI-TβRII-Fc chimeric receptor could modulate the response to TGF-β2 in the melanoma model. The aforementioned findings were also confirmed at the protein level using immunoblotting (Fig. 4C) and immunocytochemical analyses (Fig. 4D and E). In agreement with the RT-qPCR results, changes in the intensity of bands corresponding to upregulated expression of SM22α and αSMA proteins were observed (Fig. 4C) in response to all TGF-β isoforms, in the absence or presence of Fc chimeric receptors, as well as the number of SM22α-positive and αSMA-positive cells (Fig. 4D and E, respectively). Consistent with the RT-qPCR results, effective inhibition of the TGF-β2-induced EMT program was observed only in the presence of TβRI-TβRII-Fc chimeric receptor while Control-Fc and TβRII-Fc did not demonstrate such an effect (Fig. 4C-E). The suppressive effect of TβRI-TβRII-Fc chimeric receptor on EMT-associated changes in SM22α expression was also examined at both RNA and protein levels in Clone M3 cells. As anticipated, the expression of SM22α induced by any of the three TGF-β isoforms was inhibited only by TβRI-TβRII-Fc chimeric receptor (Fig. S4), indicating that TβRI-TβRII-Fc chimeric receptor could be potentially used for targeting all TGF-β isoforms present in the TME of melanoma tumors.
TβRI-TβRII-Fc chimeric receptor inhibits melanoma tumor growth in vivo
As our in vitro data revealed effective inhibition of the EMT program, examination of the effect of TβRI-TβRII-Fc chimeric receptor on melanoma tumor growth in vivo was performed. Therefore, B16 melanoma cells expressing Control-Fc, TβRII-Fc, and TβRI-TβRII-Fc chimeric proteins were established by infecting B16 melanoma cells with lentiviral vectors (Fig. S5). As revealed in Fig. 5A, all Fc chimeric receptors were expressed in B16 cells (Fig. 5A; cell lysate) and secreted into the culture media (Fig. 5A; conditioned medium); however, the amount of accumulated TβRI-TβRII-Fc chimeric receptor was lower when compared with the secreted amount of Control-Fc or TβRII-Fc chimeric receptors.
The anti-proliferative effect of TGF-β on normal epithelial cells has been previously reported (39). Moreover, in the early stage of melanoma progression, TGF-β has been revealed to inhibit cell growth (15). In agreement with these previous findings, B16 melanoma cells incubated in the presence of TGF-βs for 72 h demonstrated decreased proliferation when compared with the non-treated control cells, independently of the TGF-β isoform used (Fig. S6). The proliferation of B16 melanoma cells in the presence of SB431542, a TβRI kinase inhibitor, did not differ from the proliferation of control cells (Fig. S6). As B16 melanoma cells have been revealed to secrete active TGF-βs (40), there was a possibility that the expression of Fc chimeric receptors could alter their extracellular environment and affect cell proliferation. Therefore, the proliferation of B16 cells expressing each Fc chimeric receptor was examined and it was revealed that there was not any difference in the proliferation exhibited by B16 cells expressing TβRII-Fc and TβRI-TβRII-Fc chimeric receptors when compared with B16 cells expressing Control-Fc protein (Fig. 5B).
Next, the effect of TβRII-Fc and TβRI-TβRII-Fc chimeric receptors accumulated in conditioned media of B16 cells on TGF-β signaling was examined using HEK-Blue TGF-β reporter cells. HEK-Blue cells were cultured in the conditioned medium of B16 cells expressing Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc chimeric receptors in the absence or presence of TGF-β1, -β2, or -β3. As anticipated, stimulation of HEK-Blue cells with any TGF-β isoform in the presence of conditioned medium from B16 cells expressing Control-Fc protein resulted in upregulation of TGF-β signals (Fig. 5C). Conditioned medium derived from B16 cells expressing TβRII-Fc chimeric receptor significantly inhibited signals induced by TGF-β1 or TGF-β3 and had no effect on signals induced by TGF-β2 (Fig. 5C). Complete inhibition of TGF-β signals was observed only in the presence of conditioned medium derived from B16 cells expressing TβRI-TβRII-Fc chimeric receptor (Fig. 5C), indicating that TβRI-TβRII-Fc could trap all TGF-β isoforms.
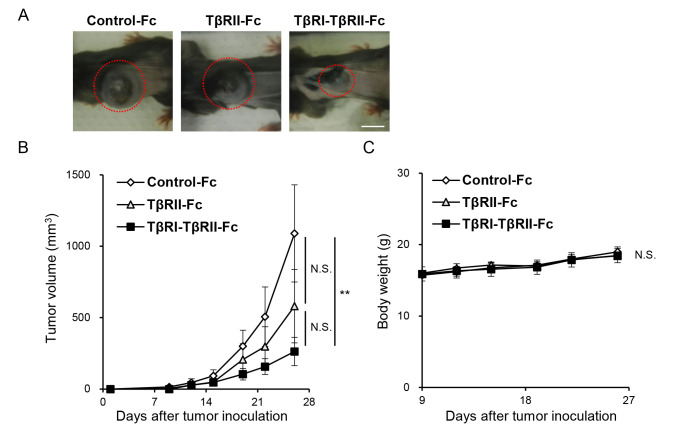
Finally, the effect of Fc chimeric receptors on melanoma tumor growth was examined in vivo. The B16 cells expressing Control-Fc, TβRII-Fc, or TβRI-TβRII-Fc chimeric receptors were subcutaneously inoculated in the left flank of C57/BL6 mice. The tumor growth and body weight were then monitored for 26 days. Expression of TβRI-TβRII-Fc chimeric receptor inhibited B16 melanoma tumor growth in vivo (Fig. 6A) when compared with Control-Fc. Moreover, the size of tumors originating from B16 cells expressing TβRI-TβRII-Fc chimeric receptor was significantly smaller than that developed from B16 cells expressing Control-Fc protein (Fig. 6B) indicating that it could effectively trap all TGF-β isoforms residing in the TME. Of note, no significant difference was observed between Control-Fc and TβRII-Fc or TβRII-Fc and TβRI-TβRII-Fc groups. In addition, no significant differences in body weight were observed between the three experimental groups (Fig. 6C).
Figure 6.
TβRI-TβRII-Fc chimeric receptor inhibits B16 melanoma tumor growth in vivo. B16 cells expressing Control-Fc, TβRII-Fc, and TβRI-TβRII-Fc chimeric receptors were subcutaneously inoculated into left flank region of C57/BL6 mice. The experiment was repeated twice. (A) Representative primary tumors formed by B16 melanoma cells expressing Control-Fc, TβRII-Fc and TβRI-TβRII-Fc chimeric receptors on day 26 (marked by red dashed circles). Scale bar, 10 mm. (B) Tumor growth was monitored for 26 days. Control-Fc (n=11), TβRII-Fc (n=9), and TβRI-TβRII-Fc (n=11). Error bars, SE. (C) The changes in body weight of the mice inoculated with B16 cells expressing Control-Fc, TβRII-Fc, and TβRI-TβRII-Fc chimeric receptors. Error bars, SE. **P<0.01. TβRI, TGF-β type I receptor; TβRII, TGF-β type II receptor; NS, not significant.
Discussion
TGF-β ligands facilitate progression of various types of cancer by affecting the components of the TME (6,7). Therefore, targeting of TGF-β signals will have an outcome in the development of effective agents. Recently, Fc chimeric receptors bearing the extracellular domains of various receptors and the Fc portion of IgG have attracted a significant amount of attention (28). The presence of Fc can extend the plasma half-life time of chimeric receptors and engage the immune response, thus being a favorable choice to develop effective agents (29). In the present study, it was demonstrated that previously developed TβRI-TβRII-Fc chimeric receptor could also trap all three TGF-β isoforms that resulted in inhibition of the EMT program in B16 melanoma cells in vitro. Our data also revealed that primary tumors originated from B16 melanoma cells expressing TβRI-TβRII-Fc chimeric receptor exhibited reduced growth in vivo, in a subcutaneous murine xenograft model, indicating that TβRI-TβRII-Fc chimeric receptor may represent a favorable strategy for the development of a novel drug for melanoma treatment.
Our previous study with human oral cancer cells, revealed that both TβRII-Fc and TβRI-TβRII-Fc chimeric receptors could significantly suppress tumor formation originated from oral cancer cells (31). However, in the present study, only the tumors formed by TβRI-TβRII-Fc-expressing cells appeared to be significantly smaller when compared with tumors formed by B16 cells expressing Control-Fc protein. In our study, TβRI-TβRII-Fc chimeric receptor was capable of interacting with all three isoforms indicating that TβRI-TβRII-Fc chimeric receptor could be used to control the level of TGF-β in the melanoma TME. Since the melanoma cells expressing the chimeric receptors in the present study were used, further experiments employing the recombinant soluble TβRI-TβRII-Fc chimeric receptor administered via blood system will shed light on the turnover of TβRI-TβRII-Fc chimeric receptor and allow its validation.
The exact mechanism by which TβRI-TβRII-Fc chimeric receptor inhibited melanoma tumor formation remains to be examined; however our data with the oral cancer cell model revealed that TβRI-TβRII-Fc chimeric receptor suppressed tumor formation by affecting tumor angiogenesis (31). As melanoma progression is tightly correlated with new vessel formation (41), it is possible that TβRI-TβRII-Fc chimeric receptor secreted by melanoma cells to the TME affected the angiogenic response of endothelial cells and resulted in reduced tumor size. The presence of TβRI-TβRII-Fc chimeric receptor in the TME that results in a local decrease in TGF-β level can also affect the formation of CAFs. Our group has previously revealed that growth of primary tumors derived from A375 human melanoma was stimulated by CAFs originated from TECs treated with TGF-β2, indicating that TGF-β2 conferred TECs with myofibroblastic properties leading to the formation of tumor-promoting CAFs (42). Therefore, TβRI-TβRII-Fc chimeric receptor present in the TME would likely trap TGF-β2, thus interfering with CAF formation and affecting tumor growth. Conversely, various studies have revealed the role of TGF-β in the regulation of immune responses, working both as an immunosuppressor of macrophages and various types of lymphocytes (43–45) or immune response inducer (7). Thus, depletion of TGF-β from the TME may also result in altered antitumor immunity.
TGF-β2 along with bone morphogenetic protein 7 (BMP7) has been reported to be expressed at high levels by proliferative and pro-invasive melanoma tumors (17). The aforementioned study indicated an important role of TGF-β superfamily members in melanoma development by regulating both melanoma invasion and proliferation. In addition, the important role of TGF-β in the induction of the EMT program in melanoma cells was also identified. A previous study has also revealed that melanoma cells undergoing EMT activate immunosuppressive regulatory T cells (Treg) (46). Kudo-Saito et al demonstrated that overexpression of EMT-related transcription factor, Snail, in mouse or human melanoma cells resulted in enhanced metastasis and immunoresistance of formed tumors (46). Moreover, treatment of human melanoma cells with TGF-β upregulated the expression of forkhead box P3 transcription factor (FOXP3), a marker of Treg. In our study, TGF-β2 induced the EMT program in melanoma cells leading to the myofibroblastic changes as revealed by upregulated αSMA expression. This effect was inhibited by TβRI-TβRII-Fc chimeric receptor, indicating that targeting TGF-β by the administration of TβRI-TβRII-Fc chimeric receptor could be potentially used to restore the immunocompetence in melanoma tumors.
Several chimeric receptors capable of inhibiting TGF-β have been designed and successfully tested to demonstrate the efficacy for the selective blockage of TGF-β family ligands in pathological conditions (47,48). Particularly, TβRII-Fc chimeric receptor has been applied in various studies (48,49). However, as revealed by Yung et al, TβRII-Fc therapeutic potential was isoform-selective, as it could trap only TGF-β1 and TGF-β3, but not TGF-β2 (49). A previous study has revealed that the elevated plasma expression levels of TGF-β2 detected in melanoma patients were associated with tumor progression, increased metastasis and poor prognosis (33). In addition, other approaches based on the small molecules targeting the kinase activity of TβRI kinase (25) or short hairpin RNA targeting TGF-β2 (23) revealed the involvement of TGF-β signals in melanoma progression (50). Therefore, effective trapping of all TGF-β isoforms may lead to improved clinical outcomes in treatment of melanoma patients.
TGF-β regulates melanoma cell plasticity and antitumor immunity by affecting the components of the TME (51). Therefore, targeting the TGF-β signals will be beneficial for the development of effective antitumor agents for melanoma. Considering the dual role of TGF-β and its tumor-suppressive activities, the complete inhibition of TGF-β signals may evoke tumorigenesis in normal epithelial cells or result in unwanted side effects. From this point of view, by adjusting the concentration of administered soluble Fc chimeric receptors, it may be possible to maintain the concentration of TGF-β at the level that exerts only tumor-suppressive effects without unwanted pro-tumorigenic responses.
Supplementary Material
Acknowledgements
The lentiviral vectors were kindly provided by Dr Hiroyuki Miyoshi (Keio University, deceased). The authors would like to thank the members at the Department of Biochemistry of Tokyo Medical and Dental University for critical discussion.
Glossary
Abbreviations
- BMP
bone morphogenetic protein
- BRAF
B-Raf
- BSA
bovine serum albumin
- CAFs
cancer-associated fibroblasts
- DMEM
Dulbecco's modified Eagle's medium
- ECD
extracellular domain
- ELISA
enzyme-linked immunosorbent assay
- EMEM
Eagle's minimum essential medium
- EMT
epithelial-mesenchymal transition
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- IgG
immunoglobulin G
- MAPK
mitogen-activated protein kinase
- PAI-1
plasminogen activator inhibitor-1
- RT-qPCR
reverse transcription-quantitative PCR
- SEAP
secreted alkaline phosphatase
- αSMA
α-smooth muscle actin
- SM22α
smooth muscle protein 22α
- TECs
tumor endothelial cells
- TGF-β
transforming growth factor-β
- TβRI
TGF-β type I receptor
- TβRII
TGF-β type II receptor
- TME
tumor microenvironment
- TMEPAI
transmembrane prostate androgen-induced protein
- Treg
regulatory T cells
Funding Statement
The present study was supported by a research program of the Japan Agency for Medical Research and Development (AMED) (grant no. 20cm0106253h0002 to TW). The present study was also supported in part by the Grant-in-Aid for Scientific Research (C) (grant nos. 17K11828 and 20K10111 to KAPI) and Grant-in-Aid for Early-Career Scientists (grant no. 19K19194 to TU) from the Japan Society for the Promotion of Science (JSPS).
Funding
The present study was supported by a research program of the Japan Agency for Medical Research and Development (AMED) (grant no. 20cm0106253h0002 to TW). The present study was also supported in part by the Grant-in-Aid for Scientific Research (C) (grant nos. 17K11828 and 20K10111 to KAPI) and Grant-in-Aid for Early-Career Scientists (grant no. 19K19194 to TU) from the Japan Society for the Promotion of Science (JSPS).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SK, KAPI, TU and TW conceived and designed the experiments. SK, TU, KK, HT, AS and KT performed the experiments. SK, KAPI, TU and TW analyzed and interpreted the data. TI, MK and ST interpreted the data. SK, TU and KK performed the data acquisition. SK, KAPI, TU and TW wrote the manuscript. SK, KAPI, TU, TI, MK, ST and TW conducted the manuscript revision/review. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experimental protocols were approved (approval no. R-02-017-1) by the Animal Experiment Committee of the Graduate School of Dentistry, Osaka University (Osaka, Japan). The molecular biology experimental procedures were approved (approval no. G2019-026C3) by the Genetically Modified Organisms Safety Committee of Tokyo Medical and Dental University (Tokyo, Japan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Falcone I, Conciatori F, Bazzichetto C, Ferretti G, Cognetti F, Ciuffreda L, Milella M. Tumor microenvironment: Implications in melanoma resistance to targeted therapy and immunotherapy. Cancers (Basel) 2020;12:2870. doi: 10.3390/cancers12102870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyazono K, Katsuno Y, Koinuma D, Ehata S, Morikawa M. Intracellular and extracellular TGF-beta signaling in cancer: Some recent topics. Front Med. 2018;12:387–411. doi: 10.1007/s11684-018-0646-8. [DOI] [PubMed] [Google Scholar]
- 3.Heldin CH, Moustakas A. Signaling receptors for TGF-β family members. Cold Spring Harb Perspect Biol. 2016;8:a022053. doi: 10.1101/cshperspect.a022053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe Y, Itoh S, Goto T, Ohnishi E, Inamitsu M, Itoh F, Satoh K, Wiercinska E, Yang W, Shi L, et al. TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol Cell. 2010;37:123–134. doi: 10.1016/j.molcel.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 6.Morikawa M, Derynck R, Miyazono K. TGF-beta and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 2016;8:a021873. doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsuno Y, Lamouille S, Derynck R. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol. 2013;25:76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 9.Moustakas A, Heldin CH. Mechanisms of TGFβ-induced epithelial-mesenchymal transition. J Clin Med. 2016;5:63. doi: 10.3390/jcm5070063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J, Hutchinson P, Tse G, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Heppt MV, Wang JX, Hristova DM, Wei Z, Li L, Evans B, Beqiri M, Zaman S, Zhang J, Irmler M, et al. MSX1-induced neural crest-like reprogramming promotes melanoma progression. J Invest Dermatol. 2018;138:141–149. doi: 10.1016/j.jid.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javelaud D, Alexaki VI, Mauviel A. Transforming growth factor-beta in cutaneous melanoma. Pigment Cell Melanoma Res. 2008;21:123–132. doi: 10.1111/j.1755-148X.2008.00450.x. [DOI] [PubMed] [Google Scholar]
- 13.Reed JA, McNutt NS, Prieto VG, Albino AP. Expression of transforming growth factor-beta 2 in malignant melanoma correlates with the depth of tumor invasion. Implications for tumor progression. Am J Pathol. 1994;145:97–104. [PMC free article] [PubMed] [Google Scholar]
- 14.Cantelli G, Orgaz JL, Rodriguez-Hernandez I, Karagiannis P, Maiques O, Matias-Guiu X, Nestle FO, Marti RM, Karagiannis SN, Sanz-Moreno V. TGF-β-induced transcription sustains amoeboid melanoma migration and dissemination. Curr Biol. 2015;25:2899–2914. doi: 10.1016/j.cub.2015.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramont L, Pasco S, Hornebeck W, Maquart FX, Monboisse JC. Transforming growth factor-beta1 inhibits tumor growth in a mouse melanoma model by down-regulating the plasminogen activation system. Exp Cell Res. 2003;291:1–10. doi: 10.1016/S0014-4827(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 16.Schlegel NC, von Planta A, Widmer DS, Dummer R, Christofori G. PI3K signalling is required for a TGFβ-induced epithelial-mesenchymal-like transition (EMT-like) in human melanoma cells. Exp Dermatol. 2015;24:22–28. doi: 10.1111/exd.12580. [DOI] [PubMed] [Google Scholar]
- 17.Tuncer E, Calçada RR, Zingg D, Varum S, Cheng P, Freiberger SN, Deng CX, Kleiter I, Levesque MP, Dummer R, Sommer L. SMAD signaling promotes melanoma metastasis independently of phenotype switching. J Clin Invest. 2019;129:2702–2716. doi: 10.1172/JCI94295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Díaz-Valdés N, Basagoiti M, Dotor J, Aranda F, Monreal I, Riezu-Boj JI, Borrás-Cuesta F, Sarobe P, Feijoó E. Induction of monocyte chemoattractant protein-1 and interleukin-10 by TGFbeta1 in melanoma enhances tumor infiltration and immunosuppression. Cancer Res. 2011;71:812–821. doi: 10.1158/0008-5472.CAN-10-2698. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto H, Yoshimatsu Y, Tomizawa T, Kunita A, Takayama R, Morikawa T, Komura D, Takahashi K, Oshima T, Sato M, et al. Interleukin-13 receptor α2 is a novel marker and potential therapeutic target for human melanoma. Sci Rep. 2019;9:1281. doi: 10.1038/s41598-019-39018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH, Jr, Kaempgen E, et al. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 22.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1203421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlingensiepen KH, Schlingensiepen R, Steinbrecher A, Hau P, Bogdahn U, Fischer-Blass B, Jachimczak P. Targeted tumor therapy with the TGF-beta 2 antisense compound AP 12009. Cytokine Growth Factor Rev. 2006;17:129–139. doi: 10.1016/j.cytogfr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Morris JC, Tan AR, Olencki TE, Shapiro GI, Dezube BJ, Reiss M, Hsu FJ, Berzofsky JA, Lawrence DP. Phase I study of GC1008 (fresolimumab): A human anti-transforming growth factor-beta (TGFβ) monoclonal antibody in patients with advanced malignant melanoma or renal cell carcinoma. PLoS One. 2014;9:e90353. doi: 10.1371/journal.pone.0090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin CH, Krishnaiah M, Sreenu D, Subrahmanyam VB, Rao KS, Lee HJ, Park SJ, Park HJ, Lee K, Sheen YY, Kim DK. Discovery of N-((4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-5-(6-methylpyridin-2-yl)-1H-imidazol-2-yl)methyl)-2-fluoroaniline (EW-7197): A highly potent, selective, and orally bioavailable inhibitor of TGF-β type I receptor kinase as cancer immunotherapeutic/antifibrotic agent. J Med Chem. 2014;57:4213–4238. doi: 10.1021/jm500115w. [DOI] [PubMed] [Google Scholar]
- 26.Colak S, Ten Dijke P. Targeting TGF-β signaling in cancer. Trends Cancer. 2017;3:56–71. doi: 10.1016/j.trecan.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Roopenian DC, Akilesh S. FcRn: The neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 28.Czajkowsky DM, Hu J, Shao Z, Pleass RJ. Fc-fusion proteins: New developments and future perspectives. EMBO Mol Med. 2012;4:1015–1028. doi: 10.1002/emmm.201201379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duivelshof BL, Murisier A, Camperi J, Fekete S, Beck A, Guillarme D, D'Atri V. Therapeutic Fc-fusion proteins: Current analytical strategies. J Sep Sci. 2021;44:35–62. doi: 10.1002/jssc.202000765. [DOI] [PubMed] [Google Scholar]
- 30.Marotte H, Cimaz R. Etanercept-TNF receptor and IgG1 Fc fusion protein: Is it different from other TNF blockers? Expert Opin Biol Ther. 2014;14:569–572. doi: 10.1517/14712598.2014.896334. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Akatsu Y, Podyma-Inoue KA, Matsumoto T, Takahashi H, Yoshimatsu Y, Koinuma D, Shirouzu M, Miyazono K, Watabe T. Targeting all transforming growth factor-β isoforms with an Fc chimeric receptor impairs tumor growth and angiogenesis of oral squamous cell cancer. J Biol Chem. 2020;295:12559–12572. doi: 10.1074/jbc.RA120.012492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogunovic D, O'Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, Darvishian F, Berman R, Shapiro R, Pavlick AC, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfaffl MW. Relative quantification. In: Dorak MT, editor. Real-time PCR. 1st edition. Taylor & Francis; London: 2006. [Google Scholar]
- 35.Kawai S, Takagi Y, Kaneko S, Kurosawa T. Effect of three types of mixed anesthetic agents alternate to ketamine in mice. Exp Anim. 2011;60:481–487. doi: 10.1538/expanim.60.481. [DOI] [PubMed] [Google Scholar]
- 36.Kirihara Y, Takechi M, Kurosaki K, Kobayashi Y, Kurosawa T. Anesthetic effects of a mixture of medetomidine, midazolam and butorphanol in two strains of mice. Exp Anim. 2013;62:173–180. doi: 10.1538/expanim.62.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krasagakis K, Thölke D, Farthmann B, Eberle J, Mansmann U, Orfanos CE. Elevated plasma levels of transforming growth factor (TGF)-beta1 and TGF-beta2 in patients with disseminated malignant melanoma. Br J Cancer. 1998;77:1492–1494. doi: 10.1038/bjc.1998.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Alexander PB, Wang XF. TGF-beta family signaling in the control of cell proliferation and survival. Cold Spring Harb Perspect Biol. 2017;9:a022145. doi: 10.1101/cshperspect.a022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penafuerte C, Galipeau J. TGF beta secreted by B16 melanoma antagonizes cancer gene immunotherapy bystander effect. Cancer Immunol Immunother. 2008;57:1197–1206. doi: 10.1007/s00262-008-0453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Streit M, Detmar M. Angiogenesis, lymphangiogenesis, and melanoma metastasis. Oncogene. 2003;22:3172–3179. doi: 10.1038/sj.onc.1206457. [DOI] [PubMed] [Google Scholar]
- 42.Akatsu Y, Takahashi N, Yoshimatsu Y, Kimuro S, Muramatsu T, Katsura A, Maishi N, Suzuki HI, Inazawa J, Hida K, et al. Fibroblast growth factor signals regulate transforming growth factor-β-induced endothelial-to-myofibroblast transition of tumor endothelial cells via Elk1. Mol Oncol. 2019;13:1706–1724. doi: 10.1002/1878-0261.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong D, Shi W, Yi SJ, Chen H, Groffen J, Heisterkamp N. TGFβ signaling plays a critical role in promoting alternative macrophage activation. BMC Immunol. 2012;13:31. doi: 10.1186/1471-2172-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W, Ten Dijke P. Immunoregulation by members of the TGFβ superfamily. Nat Rev Immunol. 2016;16:723–740. doi: 10.1038/nri.2016.112. [DOI] [PubMed] [Google Scholar]
- 45.Liu M, Kuo F, Capistrano KJ, Kang D, Nixon BG, Shi W, Chou C, Do MH, Stamatiades EG, Gao S, et al. TGF-β suppresses type 2 immunity to cancer. Nature. 2020;587:115–120. doi: 10.1038/s41586-020-2836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 47.Qin T, Barron L, Xia L, Huang H, Villarreal MM, Zwaagstra J, Collins C, Yang J, Zwieb C, Kodali R, et al. A novel highly potent trivalent TGF-β receptor trap inhibits early-stage tumorigenesis and tumor cell invasion in murine Pten-deficient prostate glands. Oncotarget. 2016;7:86087–86102. doi: 10.18632/oncotarget.13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muraoka RS, Dumont N, Ritter CA, Dugger TC, Brantley DM, Chen J, Easterly E, Roebuck LR, Ryan S, Gotwals PJ, et al. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J Clin Invest. 2002;109:1551–1559. doi: 10.1172/JCI0215234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yung LM, Nikolic I, Paskin-Flerlage SD, Pearsall RS, Kumar R, Yu PB. A selective transforming growth factor-β ligand trap attenuates pulmonary hypertension. Am J Respir Crit Care Med. 2016;194:1140–1151. doi: 10.1164/rccm.201510-1955OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohammad KS, Javelaud D, Fournier PG, Niewolna M, McKenna CR, Peng XH, Duong V, Dunn LK, Mauviel A, Guise TA. TGF-β-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 2011;71:175–184. doi: 10.1158/0008-5472.CAN-10-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Javelaud D, Alexaki VI, Dennler S, Mohammad KS, Guise TA, Mauviel A. The TGF-β/SMAD/GLI2 signaling axis in cancer progression and metastasis. Cancer Res. 2011;71:5606–5610. doi: 10.1158/0008-5472.CAN-11-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.