Abstract
Tyrosyl DNA phosphodiesterase 1 (TDP1) and DNA Ligase IIIα (LigIIIα) are key enzymes in single-strand break (SSB) repair. TDP1 removes 3′-tyrosine residues remaining after degradation of DNA topoisomerase (TOP) 1 cleavage complexes trapped by either DNA lesions or TOP1 inhibitors. It is not known how TDP1 is linked to subsequent processing and LigIIIα-catalyzed joining of the SSB. Here we define a direct interaction between the TDP1 catalytic domain and the LigIII DNA-binding domain (DBD) regulated by conformational changes in the unstructured TDP1 N-terminal region induced by phosphorylation and/or alterations in amino acid sequence. Full-length and N-terminally truncated TDP1 are more effective at correcting SSB repair defects in TDP1 null cells compared with full-length TDP1 with amino acid substitutions of an N-terminal serine residue phosphorylated in response to DNA damage. TDP1 forms a stable complex with LigIII170–755, as well as full-length LigIIIα alone or in complex with the DNA repair scaffold protein XRCC1. Small-angle X-ray scattering and negative stain electron microscopy combined with mapping of the interacting regions identified a TDP1/LigIIIα compact dimer of heterodimers in which the two LigIII catalytic cores are positioned in the center, whereas the two TDP1 molecules are located at the edges of the core complex flanked by highly flexible regions that can interact with other repair proteins and SSBs. As TDP1and LigIIIα together repair adducts caused by TOP1 cancer chemotherapy inhibitors, the defined interaction architecture and regulation of this enzyme complex provide insights into a key repair pathway in nonmalignant and cancer cells.
Keywords: DNA ligation, topoisomerase, phosphorylation, protein structure, small angle x-ray scattering, electron microscopy
Abbreviations: APTX, aprataxin; ATM, ataxia telangiectasia mutated; CPT, camptothecin; DBD, DNA-binding domain; DNA-PK, DNA-dependent protein kinase; DSB, double-strand break; MALS, multiangle light scattering; PARP1, poly (ADP-ribose) polymerase 1; PNKP, polynucleotide kinase phosphatase; ROS, reactive oxygen species; SAXS, small-angle X-ray scattering; SEC, size-exclusion chromatography; SSB, single-strand break; TDP1, tyrosyl DNA phosphodiesterase 1; TOP1, topoisomerase 1; XRCC1, X-ray cross complementing protein 1
DNA single-strand breaks (SSBs), which occur 1 to 2 orders of magnitude more frequently than DNA double-strand breaks (DSBs), are generated by multiple different mechanisms including directly by damaging agents, such as ionizing radiation and reactive oxygen species (ROS), and indirectly by DNA replication, DNA excision repair, and aberrant topoisomerase 1 (TOP1) activity (1, 2, 3, 4). Although SSBs are usually repaired rapidly, they are converted to cytotoxic one-ended DSBs when encountered by the replication fork (5, 6). This type of DSB, which is normally repaired by homology-dependent DSB repair, contributes to the cytotoxicity of inhibitors of poly (ADP-ribose) polymerase 1 (PARP1) and TOP1 that increase steady-state levels of SSBs in proliferating cells (7, 8, 9). Furthermore, the identification of mutations in genes encoding proteins involved in SSB repair, such as X-ray cross complementing protein 1 (XRCC1), polynucleotide kinase phosphatase (PNKP), aprataxin (APTX), and tyrosyl DNA phosphodiesterase 1 (TDP1) as the causative factors in inherited neurodegenerative diseases, indicates that unrepaired SSBs cause toxicity in terminally differentiated neuronal cells, presumably due to inhibition of transcription (10, 11, 12, 13, 14, 15).
The repair of SSBs is usually initiated by PARP1, an abundant nuclear protein that is activated to synthesize poly (ADP-ribose) chains on itself and other proteins using NAD+ as the substrate by binding to SSBs (16). Automodification results in the release of PARP1 from the SSB, thereby generating a high concentration of poly (ADP-ribose) in the vicinity of the SSB (16). Notably, automodified PARP1 molecules serve as the recruitment signal for SSB repair proteins, including TDP1 (17) and the DNA repair scaffold protein XRCC1 (18). A central BRCT within XRCC1 preferentially binds to poly (ADP-ribosylated) PARP1 (19, 20), thereby recruiting XRCC1 and other XRCC1 interacting proteins that include APTX, PNKP, AP endonuclease 1, DNA polymerase β, and DNA ligase IIIα (LigIIIα) to the vicinity of the SSB (18, 21, 22, 23, 24, 25, 26). While these enzymes are capable of processing and joining SSBs with a variety of different termini, they are unable to process 3′-phosphotyrosine adducts generated by proteolytic degradation of covalent TOP1-DNA complexes. The covalently linked tyrosine residues are predominantly removed by TDP1, which also acts upon other 3′-modifications including 3′-phosphoglycolates (27, 28, 29), whereas TDP2 resolves 5′ phosphotyrosyl DNA adducts but also has weak 3′ phosphotyrosyl phosphodiesterase activity (30, 31). In fission yeast unrepaired Top1-DNA adducts cause DNA damage, hyperrecombination, and checkpoint-mediated cell cycle arrest (32). In human cells the recruitment of TDP1 to SSBs resulting from aborted TOP1 reactions appears to be mediated, at least in part, via an interaction with PARP1 (17). Intriguingly, XRCC1 localization to SSBs induced by the TOP1 poison, camptothecin (CPT), is reduced in cells lacking TDP1 (17), suggesting that TDP1 contributes to the recruitment of XRCC1 and its partner proteins. Although TDP1 does not interact directly with XRCC1, it does interact with LigIIIα (11, 33), a constitutive partner protein of XRCC1, linking TDP1 with the other DNA repair enzymes that bind to XRCC1 and are required to complete repair of the SSB after removal of the 3′ tyrosine (18, 21, 22, 23, 24, 25, 26).
While the interactions of TDP1 with both PARP1 and LigIIIα are enhanced by DNA damage-dependent phosphorylation of the N-terminus of TDP1 by ataxia telangiectasia mutated (ATM) kinase and DNA-dependent protein kinase (DNA-PK) (33, 34), how they coordinate the repair of SSBs is not understood. Furthermore, since PARP1, TDP1, and LigIIIα occur in both nuclear and mitochondrial compartments, whereas XRCC1 is solely nuclear (27, 35, 36, 37, 38, 39), there are likely to be differences in the mechanisms by which SSBs resulting from aborted TOP1 reactions are repaired in these organelles. In this study, we have characterized the interaction between TDP1 and LigIIIα. These results support published studies showing that this interaction is regulated by phosphorylation of the noncatalytic N-terminal region of TDP1 but contradict the conclusion that it is mediated directly by the N-terminal 150 residues of TDP1 (33, 34). Specifically, we show that the TDP1 catalytic domain directly binds to the DNA-binding domain (DBD) within the catalytic region of LigIIIα, indicating that the interaction is indirectly regulated by posttranslational modification of the noncatalytic N-terminal region of TDP1. Furthermore, we find that TDP1 dephosphorylation induces large conformational changes in the N- but not the C-terminal region of TDP1. Following coexpression in insect cells, we purified and characterized LigIIIα-TDP1 and XRCC1-LigIIIα-TDP1 complexes using biochemical and biophysical approaches, including size-exclusion chromatography (SEC) and multiangle light scattering (MALS). Since XRCC1 is absent from mitochondria, the LigIIIα-TDP1 and XRCC1-LigIIIα-TDP1 complexes are likely to function in the repair of SSBs in the mitochondria and the nucleus, respectively (18, 23, 27, 35, 36, 37, 40, 41). Although refractory to atomic-resolution biophysical analysis because of high flexibility and aggregation tendencies, analysis of the LigIIIα-TDP1 and XRCC1-LigIIIα-TDP1 complexes by a combination of SEC-coupled small-angle X-ray scattering (SAXS) and negative stain electron microscopy (EM) to define flexible biomolecular interactions (42, 43) provided insights into their assembly architecture and conformational dynamicity.
Results
TDP1 interacts with LigIII DBD
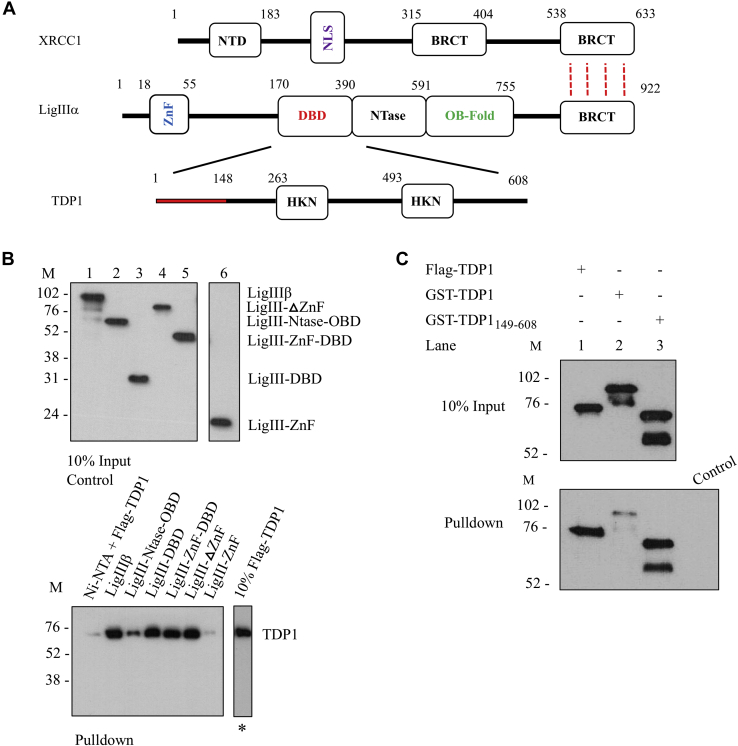
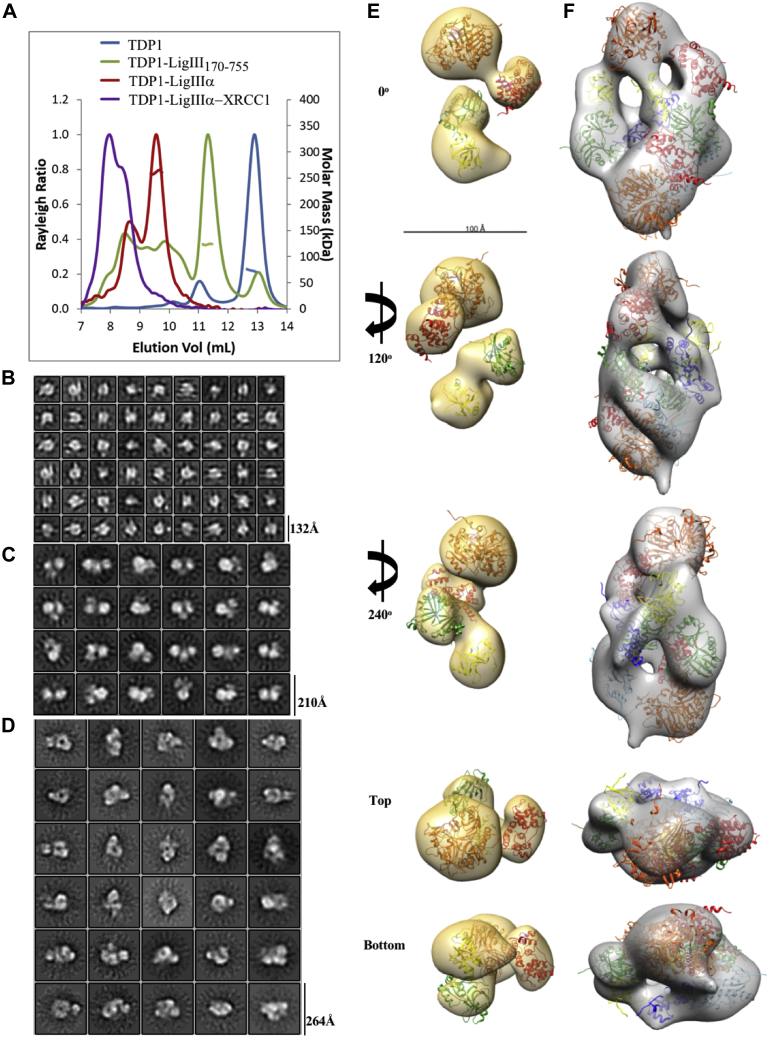
Previous mapping studies using the yeast two-hybrid assay concluded that the interaction between TDP1 and LigIIIα involves the N-terminal 150 residues of TDP1 and a fragment of LigIIIα (residues 173–862) that lacks the N-terminal zinc finger (ZnF) and the C-terminal BRCT domains (33) (Fig. 1A). To further refine the interacting region(s) of LigIII, we expressed His-tagged LigIIIβ, an alternatively spliced version of LigIII that lacks the C-terminal BRCT domain of LigIIIα (44) and different His-tagged truncations of LigIIIβ in E. coli. After purification (Fig. 1B, left panel), these proteins were examined for their ability to bind to purified insect-cell expressed FLAG-tagged TDP1 in pull-down assays with nickel beads. TDP1 was efficiently retained on beads liganded by all versions of LigIIIβ that contained the DBD (Fig. 1B, right panel), one of the three domains that constitute the catalytic region of LigIIIα (45, 46) (Fig. 1A). To confirm this and the behavior of the purified TDP1, we found that five different His-tagged versions of the DBD had similar TDP1-binding activity (Fig. S1A) and, in accord with published studies (17), TDP1 interacted with PARP1 but not XRCC1 (Fig. S1B). Thus, we conclude that the major interaction region for TDP1 resides within the DBD of LigIIIα.
Figure 1.
Mapping the region of LigIII that interacts with TDP1.A, domain architecture of human XRCC1, DNA Ligase III, and TDP1. B, Upper panel, input (10%) of full-length and different LigIII domains. Lanes 1 to 6 represent LigIIIβ full length (1–846), LigIII-Ntase-OBD (387–846), LigIII-DBD (152–390), LigIII-ΔZnF (170–846), LigIII-ZnF-DBD (1–390), and LigIII-ZnF (1–136), respectively. Immunoblotting with mouse anti-6His mAb (Abcam). Lower panel, binding of Flag-TDP1 (10 pmol) to Ni-beads liganded by LigIII domains (10 pmol of each). Lane (∗) represents 10% TDP1 input. Negative control reaction lane, Ni-NTA beads alone with Flag-TDP1. Immunoblotting with mouse anti-Flag mAb (Sigma M2). C, Upper panel, input (10%) of GST-TDP1 and Flag-TDP1. Ni-NTA beads liganded by LigIIIβ (10 pmol) were used to pull down full-length Flag-TDP1, GST-TDP1, and GST-TDP1149–608 (10 pmol of each). The control lane (Control) represents Ni-NTA beads alone incubated with GST-TDP1. Immunoblotting with rabbit anti-TDP1-C-term (Abcam).
TDP1 phosphorylation regulates interaction with LigIIIα
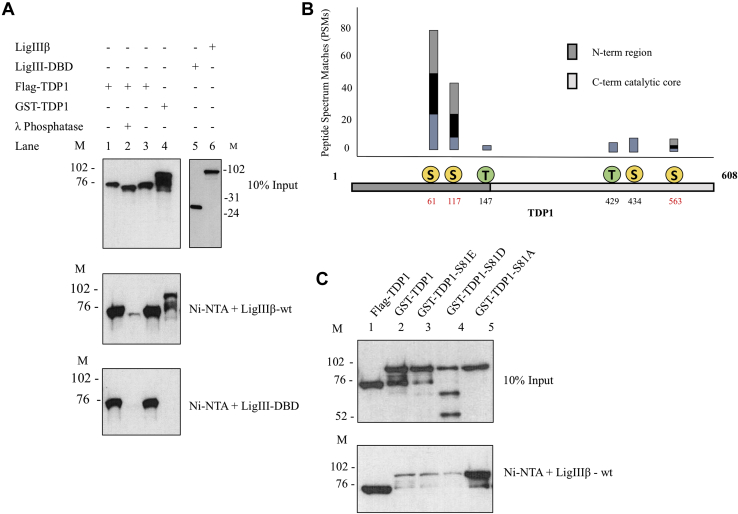
To map the region of TDP1 that interacts with LigIIIα, we initially purified a GST-tagged version of full-length TDP1 after expression in E. coli. In accord with studies indicating that the interaction between LigIIIα and TDP1 is enhanced by TDP1 phosphorylation (33, 34), the binding of unmodified GST-TDP1 to nickel beads liganded by either full-length LigIIIβ or the DBD was much less efficient than that of insect-cell-expressed Flag-tagged TDP1 that is assumed to be phosphorylated (Fig. 1C, compare lane 1 with lane 2). To confirm that insect-cell-expressed TDP1 is phosphorylated and that this modification enhances binding to LigIII, TDP1 was incubated with λ phosphatase, a serine-threonine phosphatase. As expected, treatment with λ phosphatase increased the mobility of purified TDP1 (Fig. 2A, compare lane 2 with lanes 1 and 3) and reduced TDP1 binding to nickel beads liganded by either full-length LigIIIβ or only its DBD (Fig. 2A, compare lane 2 with lanes 1 and 3).
Figure 2.
Phosphorylation status of TDP1 modulates its interaction with LigIII.A, Upper panel, input (10%) of Flag-TDP1, Flag-TDP1 incubated with λ phosphatase, Flag-TDP1 incubated with phosphatase buffer, GST-TDP1 (Lanes 1–4) and LigIII-DBD and LigIIIβ (lanes 5 and 6). Pull-down assay with Ni-NTA liganded by LigIIIβ (10 pmol, middle panel) and LigIII-DBD (10 pmol, lower panel). Lanes 1 to 4, immunoblotting with rabbit anti-TDP1-C-term (Abcam) and lanes 5 and 6, with mouse anti-6His mAb (Abcam). B, identification of phosphorylation sites in insect-cell-expressed TDP1 by mass spectrometry. In the bar diagram, peptide spectrum matches (PSM) represents the total number of times a peptide containing the indicated phosphorylated amino acid was identified. Sites with PSM values greater than 3 are shown. Amino acid residues indicated in red were substituted in subsequent studies. C, Upper panel, input (10%) of the indicated proteins (1 pmol of each). Lower panel, Ni-NTA beads liganded by LigIIIβ (10 pmol) were used to pull down the indicated proteins (10 pmol of each). Immunoblotting with rabbit anti-TDP1-C-term (Abcam).
Analysis of insect-cell-expressed TDP1 by mass spectrometry identified phosphorylated serine and threonine residues throughout the protein. Serine residues 61 and 117 were the most frequently modified residues as determined by the peptide spectrum match (PSM) score (Fig. 2B and Fig. S2A). Substitution of these residues with either Glu or Ala within the GST-TDP1 fusion protein marginally reduced the weak LigIII-binding activity of GST-TDP1 with the Glu substitution having a greater effect than the Ala (Fig. S3). While previous studies had reported that DNA-damage-dependent phosphorylation of Ser81 enhanced the interaction between TDP1 and LigIIIα (33, 34), we did not recover a peptide encompassing Ser81 despite identifying peptides covering 85% of TDP1 (Fig. S2A) and so cannot make any conclusions regarding the phosphorylation status of Ser81 and its contribution to the interaction with LigIIIα. To ask whether modification of Ser81 influences the binding of TDP1 with LigIIIα, Ser81 was replaced with either Asp or Glu to mimic phosphorylation or Ala in the E. coli expressed GST-TDP1 fusion protein. Surprisingly, while neither the Asp nor the Glu substitution exhibited enhanced binding, the retention of TDP1-S81A on the nickel beads liganded by LigIIIβ was markedly increased, reaching levels similar to the retention of phosphorylated TDP1 from insect cells (Fig. 2C).
The above studies using E. coli-expressed TDP1 examine the effect of a phosphomimetic amino acid substitution within the context of an otherwise unmodified protein, whereas TDP1 is phosphorylated at multiple sites in human cells. To examine the effects of substitution of Ser81 with either Glu or Ala on phosphorylation at other sites within TDP1, the mutant versions of TDP1 were expressed in and purified from insect cells. As with wild-type TDP1, serine residues 61 and 117 were the most frequently modified residues in TDP1-S81A and TDP1-S81E (Fig. S2A). While there were some differences in the minor phosphorylation sites detected, the overall patterns of phosphorylation in the wild-type and mutant versions of TDP1 were similar (Fig. S2A). The interactions of phosphorylated wild-type TDP1, TDP1-S81E, and TDP1-S81A with LigIII were similar (Fig. S4). Since pull-down assays are qualitative and the results may be influenced by the presence of the relatively large GST domain in the fusion proteins, we examined the interaction between His-tagged versions of LigIII and insect-cell-expressed TDP1 by surface plasmon resonance (Fig. S5). Under these conditions, TDP1 wild-type and TDP1-S81A have binding constants (KD) of 5.1 nM and 5.3 nM, respectively, whereas the binding constant for TDP1-S81E was 8.8 nM (Fig. S5D).
LigIIIα binds the catalytic C-terminal region of TDP1
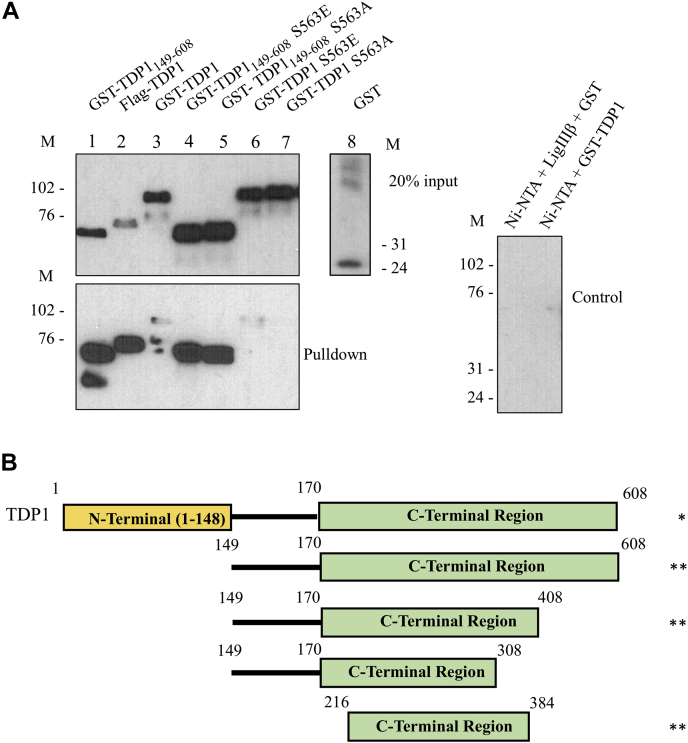
While the apparently paradoxical observations with the versions of TDP1 containing different residues at Ser81 could be due to the Asp and Glu substitutions failing to adequately mimic phosphorylation and/or disrupting protein structure, we also considered the possibility that N-terminus of TDP1 regulates the interaction between another region of TDP1 and LigIIIα. To test this, we initially examined the binding of a GST-TDP1 fusion protein lacking the N-terminal 148 residues of TDP1. Strikingly, the specific retention of truncated TDP1 on nickel beads liganded by LigIIIβ was much higher than full-length TDP1, reaching levels similar to the retention of phosphorylated TDP1 from insect cells (Figs. 1C and 3A). Further mapping studies identified residues 216 to 384 as the region within the TDP1 catalytic domain involved in the interaction interface (Fig. 3B and Fig. S6). Substitution of Ser563, the most frequently phosphorylated residue in the C-terminal region of wild-type TDP1 (Fig. 2B) with either Ala or Glu modestly reduced the low retention of full-length TDP1 (Fig. S3, compare lanes 3 and 4 with lane 2) but had no effect on the higher retention of N-terminally truncated TDP1 expressed in E. coli (Fig. 3A, compare lanes 4 and 5 with lane 1). Together, our results show that the catalytic regions of LigIII and TDP1 interact directly and that the noncatalytic N-terminal region of TDP1 negatively regulates this interaction.
Figure 3.
Mapping the region of TDP1 that interacts with LigIII.A, pull-down assay with N-terminal truncated (Δ148) single amino acid mutant TDP1. Upper panel, input (20%) of the indicated proteins. Immunoblotting with (left gel) rabbit anti-TDP1-C-term (Abcam) and (right gel) anti-GST (Santa Cruz Biotechnology). Lower panel (left gel), Ni-NTA beads liganded by LigIIIβ (10 pmol) were used to pull down the indicated proteins (10 pmol of each). Reaction controls (right gel) were Ni-NTA beads liganded by LigIIIβ incubated with GST and Ni-NTA beads alone incubated with GST-TDP1. Immunoblotting with rabbit anti-TDP1-C-term (Abcam). B, summary of pull-down assays with N- and C- terminally-truncated versions of TDP1. Asterisks represent the relative efficiency of the interaction.
Dephosphorylation causes a large conformational change in the noncatalytic N-terminal region of TDP1
Replacement of Ser81 with either Glu or Asp to mimic phosphorylation did not have the effect on in vitro binding to LigIII (Fig. 2C) predicted by published studies (33). However, in vitro binding to LigIII was affected by the change of Ser81 or other Ser residues phosphorylated in the N-terminal region of insect-cell-expressed TDP1 to Ala (Fig. S3), as well as dephosphorylation of insect-cell-expressed TDP1 (Fig. 2A). Taken together this suggests that the direct interaction between the TDP1 catalytic domain and the LigIII DBD is modulated by conformational changes in the unstructured N-terminal region of TDP1 induced by phosphorylation and/or alterations in amino acid sequence.
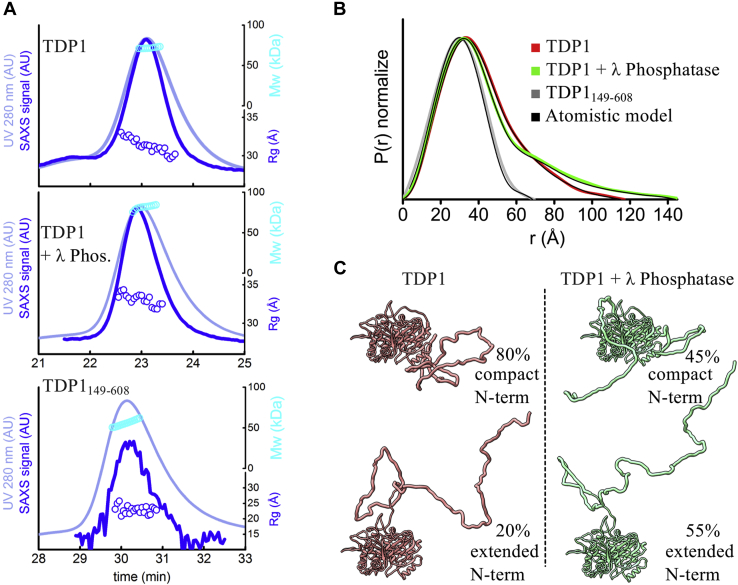
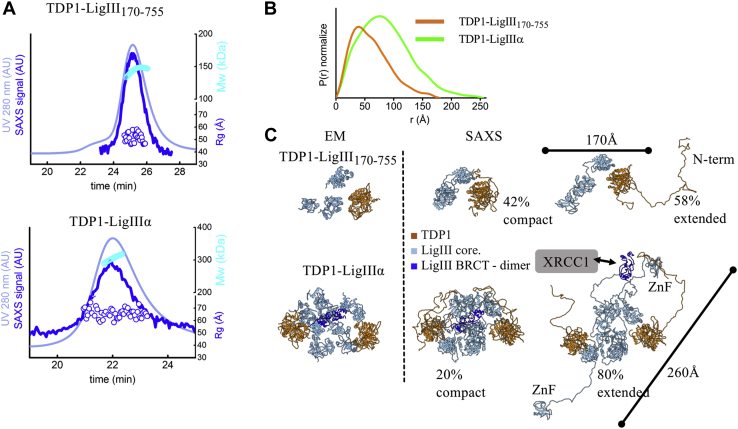
To visualize the conformational changes in the unstructured N-terminal region of purified insect-cell-expressed TDP1 (Fig. S7A, lane 8) induced by dephosphorylation, we used SAXS and MALS in line with SEC (SEC-SAXS-MALS) (43, 47, 48, 49). Both phosphorylated and dephosphorylated TDP1 eluted as a single asymmetric peak (Fig. 4A). MALS analysis of these peaks shows that mass across the peaks corresponded closely to the expected molecular mass of monomeric TDP1 (theoretical: 69 kDa, measured: ∼70 kDa). However, automated Guinier calculation of the radius of gyration (Rg) across the peak revealed a significant decrease in the Rg from ∼34 Å at the peak to ∼30 Å in the tail (Fig. 4A, Table 1). Taken together, these data suggest that TDP1 adopts a mixture of different conformational states that slowly interconvert over the time course of the chromatography. To further analyze the structure and dynamics of these conformational changes upon dephosphorylation of TDP1, we used merged SAXS curves limited to the peak of elution. As expected, the P(r) function observed for the C-terminal catalytic fragment of TDP1 (residues 149–608) was a good fit with the P(r) function calculated from the atomic resolution structure of the TDP1 catalytic domain (50) with a Dmax ∼65 Å (Fig. 4B). In contrast, the P(r) function for full-length phosphorylated TDP1 exhibited a long tail indicating an extended conformation with a Dmax ∼110 Å (Fig. 4B). The P(r) tail and Kratky Plot (Fig. S8) indicate the presence of unfolded protein regions and suggest that the TDP1 N-terminal region is partially unfolded. SAXS-based multistate modeling (48, 51) of full-length TDP1 further shows that the presence of unfolded N-terminal regions, in both the extended and compact arrangements (Fig. 4C), is required to fit the experimental SAXS curve (Fig. S9A). The compact conformer reflects the folded character of the N-terminal region, which is predicted to interact directly with the TDP1 catalytic core (Fig. 4C). Phosphatase treatment of full-length TDP1 resulted in a dramatic change in the P(r) function with a significant increase to a Dmax ∼145 Å, suggesting opening or further unfolding of the N-terminal region (Fig. 4B). In addition, multistate modeling predicted a higher frequency (55% versus 20%) of the extended N-terminal region following phosphatase treatment (Fig. 4C). As expected, the SAXS profile of phosphorylated TDP1-S81E was similar to that of phosphorylated wild-type TDP1 (Fig. S8), indicating a relatively compact N-terminal region. In contrast, despite a similar pattern of phosphorylation (Fig. S2A), TDPl-S81A behaved anomalously in SEC-SAXS-MALS experiments, suggesting that Ser81 is a structure-controlling residue and that its replacement with Ala results in unfolding and aggregation (Fig. S8B). Taken together, our studies show that the extent of folding and flexibility within the relatively unstructured noncatalytic N-terminal region of TDP1 is influenced by phosphorylation.
Figure 4.
TDP1 N-terminus adopts an open conformation upon protein dephosphorylation.A, SEC- SAXS-MALS chromatographs for TDP1, TDP1 with λ phosphatase, and TDP1149–608. Solid lines represent the UV 280 nm (light blue) or SAXS signal (blue) in arbitrary units, while symbols represent molecular mass determined by MALS (cyan) and Rg values for the principal SEC peak (blue) versus elution time. SEC-SAXS-MALS results show that TDP1 is a monomer. B, normalized pair distribution [P(r)] functions of TDP1 and TDP1 with λ phosphatase matching theoretical P(r) functions of atomistic models (black) shown in panel C. Normalized P(r) functions of TDP1149–608 matching theoretical P(r) functions of TDP1 crystal structure (PDB 1JY1). C, ensemble models of two conformers of TDP1 and λ phosphatase-treated TDP1 that were used to fit experimental SAXS curves shown in Figure S8 and P(r) functions shown in panel B.
Table 1.
Structural parameters from SAXS and MALS data
| SAXS sample | Dmax (Å) | Rg (Å) Guinier plot | Rg (Å) from P(r) | MW Seq. Calculated (kDa) | MW SAXS (kDa) | MW MALS (kDa) | Model fit x2 |
|---|---|---|---|---|---|---|---|
| TDP1149–698 | ∼70 | 23.1 ± 1.8 | 23.7 | 51 | 40 | 60 | 2.9 |
| TDP1 | ∼110 | 31.6 ± 0.3 | 32.3 | 68 | 73 | 70 | 1.2 |
| TDP1a | 68 | 72 | |||||
| TDP1 + λ phosphatase | ∼140 | 32.0 ± 0.7 | 33.32 | 68 | 64 | 70 | 1.1 |
| LigIII170–755-TDP1 | ∼160 | 45.0 ± 1.3 | 48.2 | 135 | 140 | 140 | 1.9 |
| LigIII170–755-TDP1a | 135 | 124 | |||||
| LigIIIα-TDP1 | ∼250 | 65.2 ± 3.0 | 68.0 | 172 | 340 | 340 | 1.8 |
| LigIIIα-TDP1a | 172 | 261 |
Molecular mass estimates from SEC and MALS using Superdex 200 column (see Experimental procedures).
LigIIIα forms stable complexes with TDP1 and XRCC1
While there is substantial evidence showing that LigIIIα forms a stable complex with XRCC1 both in vitro and in cells (21, 43, 52), it has not been shown that LigIIIα forms a stable complex with TDP1. To address this, His-tagged LigIIIα was coexpressed with Flag-tagged TDP1 in insect cells. Fractionation of the lysate by affinity chromatography with nickel beads resulted in copurification of TDP1 with His-tagged LigIIIα. These proteins were copurified through subsequent ion exchange and gel filtration columns (Fig. S7B), yielding a highly purified LigIIIα-TDP1 complex with an apparent stoichiometry of 1:1 (Fig. S7A, lane 3). In accord with the mapping data indicating that TDP1 interacts with the LigIIIα DBD (Fig. 1B), we were able to use the same approach to purify a complex of LigIIIα lacking the N-terminal ZnF and C-terminal BRCT domain (LigIII170–755) with TDP1 (Fig. S7A, lane 4). To confirm that the interactions of XRCC1 and TDP1 with LigIIIα are not mutually exclusive, His-tagged XRCC1 was coexpressed in insect cells with both LigIIIα and TDP1. Fractionation by affinity chromatography with nickel beads resulted in copurification of LigIIIα and TDP1 with His-tagged XRCC1. These proteins were copurified through subsequent gel filtration (Fig. S7C) and ion exchange columns, yielding a highly purified XRCC1-LigIIIα-TDP1 complex with an apparent stoichiometry of 1:1:1 (Fig. S7A, lane 6). The nick joining activities of the LigIIIα-TDP1, LigIII170–755-TDP1, and XRCC1-LigIIIα-TDP1 complexes were indistinguishable from that of purified LigIIIβ, LigIII170–755 and LigIIIα-XRCC1 (Figs. S7 and S10A). Similarly, the ability of TDP1 to remove the tyrosine residue from 3′-phosphotyrosine termini was not significantly changed by complex formation with LigIIIα (Figs. S7 and S10B).
Biophysical analysis reveals the architecture of LigIIIα-TDP1 complexes
Analysis of highly purified preparations of LigIIIα-TDP1, LigIII170–755-TDP1, and LigIIIα-XRCC1-TDP1 (Fig. S7A) by SEC revealed complex elution profiles, each with a single major peak that eluted earlier than expected when compared with globular protein standards (Fig. 5A). While this could be due to aggregation and/or the formation of multimers, it could also be due to the complexes having an elongated shape, particularly as this has been reported for XRCC1 (53), LigIII (46), and the LigIIIα-XRCC1 complex (43). We used SEC-MALS to determine the molecular mass, shape, and stoichiometry of the protein complexes (Fig. 5A). In comparing SEC with SEC-MALS (Table 1), LigIII170–755-TDP1 is less extended than full-length LigIIIα complexed with TDP1 but nonetheless still elutes earlier than expected for a globular complex of its mass (Fig. 5A, Table 1). While TDP1 alone was predominantly monomeric with a minor amount of dimerization, TDP1 and LigIII170–755 formed a stable heterodimer (Table 1). The stoichiometry deviates from a strict heterodimer when TDP1 forms a complex with full-length LigIIIα containing its ZnF and BRCT domains with multimer formation possibly mediated by BRCT-BRCT interactions (43, 52). Although a molecular mass could not be reliably estimated for the tripartite complex, it eluted even earlier than LigIIIα-TDP1 (Fig. 5A).
Figure 5.
SEC-MALS and EM of TDP1-LigIII170–755and TDP-LigIIIα.A, SEC-MALS analysis of TDP1 alone (blue) and in complex with LigIII170–755 (green), LigIIIα (red), or LigIIIα-XRCC1 (purple). The resultant molecular mass is plotted in the same color, where applicable. Molecular mass was not obtained for the tripartite complex due to its void volume elution. Negative-stain EM 2D classification of TDP1 alone (B), TDP1-LigIII170–755 (C), and TDP1-LigIIIα (D) shows homogeneous particles clustered in several orientations. The box size is indicated for each data set classification. The TDP1-LigIII170–755 (E) and TDP1-LigIIIα (F) 3D maps are shown rotated relative to 0°. Based on stoichiometry derived from molecular mass determination, crystal structures of TDP1149–608 (PDB 1JY1, orange) and LigIII170–755 (PDB 3L2P) were docked in a 1:1 ratio (E) or a 2:2 ratio (F). The DBD (red), NTase (green), and OB-fold (yellow) domains of LigIII170–755 were separated to fit into the EM map and the DBD was placed adjacent to TDP1149–608. For full-length LigIIIα, two ZnF domains (PDB 1UW0, light blue) and its BRCT homodimer (PDB 3PC8, navy blue) were also docked. The BRCT homodimer was placed in the center of the volume as it most likely mediates dimerization between two molecules of LigIIIα.
To gain insight into the shape and relative sizes of the LigIIIα-TDP1 complexes, the proteins were mildly chemically cross-linked and imaged using negative-stain EM. Using this high-contrast but low-resolution technique, we visualized TDP1 alone and in complex with either LigIII170–755 or full-length LigIIIα (Fig. S11). Individual particles were grouped into 2D classes (Fig. 5, B–D) and used to create 3D maps of the TDP1-containing complexes (Fig. 5, E and F). We readily identified the globular TDP1 (50) in complex with LigIII170–755 (gold map) and docked the LigIII catalytic core domains beginning with the interaction-mediating DBD. The 3D EM map of the LigIIIα-TDP1 complex (gray map) is consistent with a larger multimeric stoichiometry compared with LigIII170–755-TDP1, as indicated by SEC-MALS. However, the 3D map could accommodate a 2:1 or a 2:2 LigIIIα-TDP1 complex (the latter is modeled). Due to the modest resolution of 28 Å (Fig. S11F), we present here one possible spatial arrangement of the LigIIIα-TDP1 complex that represents the EM map when integrating with SAXS analysis (see below). We placed a LigIIIα BRCT dimer in the center of the map (43, 52) with two LigIIIα molecules filling out the central map density. Two molecules of TDP1 (50) were docked at the top and bottom parts of the map adjacent to the two LigIIIα DBDs. The assembled atomic coordinates were converted into low-resolution electron density maps and fit into their respective experimental negative-stain EM map. The map cross-correlations were 92% and 88% for TDP1 in complex with the catalytic fragment and full-length LigIIIα, respectively. Therefore, our docking is consistent with the experimental maps, although other arrangements are possible.
In parallel, we examined the solution structures of the LigIIIα-TDP1 complexes using SEC-SAXS-MALS (43, 47, 48, 49). In accord with the SEC-MALS data (Fig. 5A and Table 1), the molecular mass of the LigIII 170–755-TDP1 complex was determined to be 140 kDa, confirming a 1:1 complex between LigIII170–755 and TDP1 (Fig. 6A top). Based on the SAXS parameters and P(r) function, the complex has an extended conformation with maximal dimension of 160 Å (Fig. 6B and Table 1). To validate the LigIII170–755-TDP1 architecture in solution, we fitted the EM-derived models (Fig. 5, E and F) against solution SAXS data after the addition of missing LigIII linkers and unfolded regions in TDP1 to the EM-derived atomistic model (Fig. 6C) (see Experimental procedures). By means of rigid body modeling (43, 54), we explored movements of the NTase and OB-fold domains and unfolded TDP1 regions relative to the LigIIIα DBD-TDP1 interacting domains. The selected ensemble of two conformers (Fig. 6C) was a significantly better fit than a single conformer (x2= 1.9 versus 2.4, Fig. S11) to model the SAXS data, and the ensemble shows persistent flexibility in the ligase catalytic region as previously reported for LigIIIβ and LigIIIα-XRCC1 as well as the LigIV catalytic region (43, 46, 55, 56, 57).
Figure 6.
LigIIIα-TDP1 flexibility in solution.A, SEC-SAXS-MALS chromatographs for LigIII170–755- TDP1 and LigIIIα-TDP1. Solid lines represent the UV 280 nm (light blue) or SAXS signal (blue) in arbitrary units, while symbols represent molecular mass determined by MALS (cyan) and Rg values for the principal SEC peak (blue) versus elution time. SEC-SAXS-MALS results show that LigIII170–755-TDP1 is a homodimer and LigIII-TDP1 is heterotetramer. B, normalized P(r) functions of LigIII170–755-TDP1 and LigIIIα-TDP1. C, SAXS derived two-state model of LigIII170–755-TDP1 (top) and LigIIIα-TDP1 (bottom) are shown in comparison to the atomistic model derived by EM (shown in left panel). Weights of both SAXS conformers are indicated in percentage. Interaction between LigIII BRCT dimer (dark blue) and XRCC1 is highlighted. TDP1 and LigIII catalytic core are colored accordingly. Corresponding SAXS fits for the atomistic models are shown in Figure S8. Maximal dimension of the complexes (Dmax) determined from the P(r) functions (see panel B) is highlighted with the bars.
For full-length LigIIIα in complex with TDP1, the equal distribution of Rg values determined for individual SAXS frames across the elution peak (Fig. 6A bottom) is indicative of a homogeneous and stable complex. This prompted us to evaluate an overall LigIIIα-TDP1 domain arrangement for the putative tetrameric state (Fig. 6C) by fitting the EM-atomistic model against the solution SAXS data. First, we built an initial full-length LigIIIα-TDP1 model by connecting LigIII domains with the missing linkers and modeled terminal unfolded regions in TDP1 (see Experimental procedures). In the first rigid body modeling effort, we explored only the flexibility of the linker regions with the fixed domain arrangement as predicted by EM (Fig. 5F). The atomistic model that mimicked only linker flexibility did not match the SAXS data (x2= 3.5), suggesting that additional conformational variability of the domains is required to match the solution state. A multistate model, mimicking the flexibility of the LigIII ZnF and BRCT domains that has been reported previously (43, 46) (Fig. 6C bottom), significantly improved the fit to the SAXS data (x2= 1.8) (Fig. S9B). In the selected two-state model, one conformer adopts a compact arrangement similar to the EM map, whereas a second conformer displays an extended arrangement of the LigIIIα BRCT and ZnF domains (Fig. 6C). Taken together, the EM map of full-length LigIIIα-TDP1 agrees with the solution state model in which homodimerization of the LigIIIα BRCT domains links two LigIIIα-TDP1 heterodimers. In this relatively compact dimer of heterodimers, the two LigIII catalytic cores are positioned in the center, whereas two TDP1 molecules are located at the edges of the complex (Fig. 6C).
The noncatalytic TDP1 N-terminal region is not required for repair of CPT-induced single-strand breaks
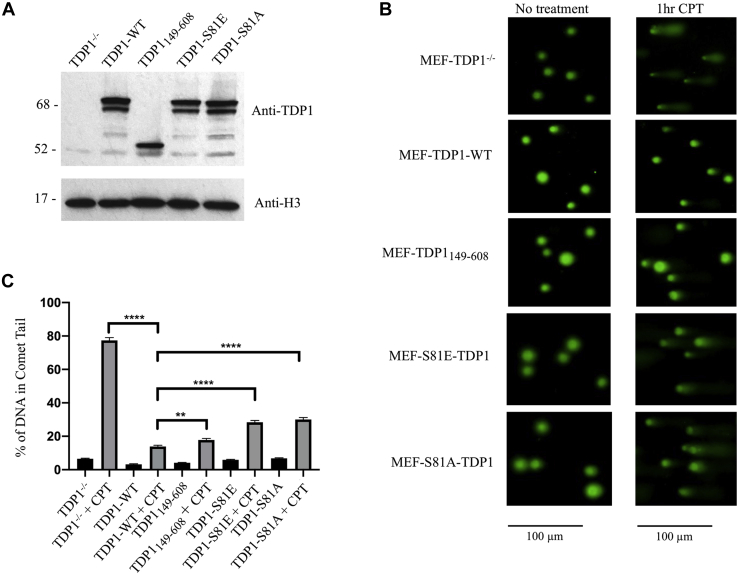
To examine the contribution of the TDP1 noncatalytic N-terminal domain and Ser81 phosphorylation to the cellular repair of CPT induced SSBs, wild-type TDP1, TDP1149–608, TDP1-S81E, and TDP1-S81A were expressed in TDP1−/−-p53−/− MEFs following transduction with recombinant lentiviruses. Although wild-type TDP1 and its mutant versions were expressed at similar levels (Fig. 7A), this expression was five- to ten-fold higher than that of endogenous TDP1 in p53−/− MEFs. As expected, expression of wild-type TDP1 in TDP1−/− p53−/− MEFs significantly enhanced the repair of CPT-induced SSBs (Fig. 7, B and C). A truncated version of TDP1 lacking the N-terminal 148 residues also significantly enhanced the repair of CPT-induced SSBs, achieving levels of repair that were close to but significantly lower than that in cells expressing wild-type TDP1 (Fig. 7, B and C). As reported by Chiang and colleagues (33), expression of full-length TDP1 with Ala in place of Ser81 enhanced the repair of CPT-induced SSBs but the extent of repair was significantly lower than that in cells expressing either wild-type TDP1 or TDP1 lacking the N-terminal region (Fig. 7, B and C). The repair of CPT-induced SSBs in cells expressing TDP1-S81E was similar to that in cells expressing TDP1-S81A (Fig. 7, B and C). Thus we conclude that, when overexpressed, neither the noncatalytic N-terminal region of TDP1 nor phosphorylation of Ser81 is required for the efficient repair of CPT-induced SSBs, but it is likely that posttranslational modification of the noncatalytic N-terminal region of TDP1 makes a larger contribution in determining the efficiency of SSB repair when TDP1 is present at endogenous steady-state levels.
Figure 7.
Alkaline comet assay of CPT-treated TDP1-transduced MEF-TDP1−/−cells.A, expression of TDP1-wt, TDP1149–608, TDP1-S81E, or TDP1-S81A in extracts of MEF-TDP1−/− cells after lentiviral transduction. Upper panel, immunoblotting with rabbit anti-TDP1-C-term (Abcam). Lower panel, immunoblotting with anti-histone H3 antibody (Cell Signaling Technology). B, alkaline comet assays of MEF-TDP1−/− cells and the indicated derivatives treated with CPT for 1 h were performed as described in Experimental procedures. C, comparison of the percentage of DNA in the comet tail MEF-TDP1−/− cells and the indicated derivatives. The results shown represent three independent experiments with at least 50 comet tail measurements for each sample. ∗∗ and ∗∗∗∗ represents p <0.0011 and <0.0001, respectively.
Discussion
Given the key role of TDP1 in repair of adducts caused by TOP1 inhibitors used in cancer chemotherapy, there is significant interest as to how this enzyme functions and is regulated in nonmalignant and cancer cells (58). Notably, the noncatalytic N-terminus of TDP1 has been implicated in interactions with both PARP1 and LigIIIα that are regulated by DNA-damage-dependent phosphorylation of Ser81 within TDP1 (11, 17, 33, 34). While supporting the conclusion that phosphorylation of the TDP1 N-terminus regulates its interaction with LigIIIα, our results reveal a direct interaction between the LigIII DBD and the TDP1 catalytic C-terminal domain rather than its noncatalytic N-terminal region as reported previously (11). Since TDP1 and LigIIIα are both present in the mitochondria as well as the nucleus (35, 37), it is likely that the interaction between these proteins contributes to the DNA repair in both these organelles. However, nuclear LigIIIα exists in a constitutive complex with the DNA repair scaffold protein XRCC1 that is not found in the mitochondria (21, 36, 59) suggesting that a TDP1-LigIIIα complex functions in mitochondrial DNA repair (35, 37), whereas TDP1 interacts with the LigIIIα-XRCC1 complex during nuclear DNA repair (11). Following coexpression in insect cells, we purified stable TDP1-LigIIIα and TDP1-LigIIIα-XRCC1 complexes that were predicted to form based on the mapping of protein–protein interactions described in this study and the published literature (21, 44, 60, 61). Here we describe the first structural insights into the complex formed between the catalytic regions of TDP1 and LigIIIα and the full-length TDP1-LigIIIα complex by complementary biophysical techniques (42).
DNA ligases are asymmetric, elongated, and flexible proteins (45). This is due in part to the catalytic fragment that undergoes a transition from an open, extended conformation to a closed, clamp-like conformation when it engages a DNA nick (46, 62), as well as to the noncatalytic regions that flank the catalytic fragment, such as the N-terminal ZnF region of LigIIIα (43, 46). While this complicates analysis of DNA ligase-containing complexes, we have employed SAXS and MALS in combination with gel filtration to gain insights into the heterogeneity, shape, and flexibility of complexes formed by LigIIIα and XRCC1 (43). Given the contribution of the LigIIIα N-terminal ZnF region to protein flexibility and asymmetry (43, 46) and the ability of the LigIIIα C-terminal BRCT domain to homodimerize (43, 52), we purified a TDP1-LigIIIα complex containing a version of LigIIIα lacking both the N-terminal ZnF and C-terminal BRCT domain in addition to the intact TDP1-LigIIIα complex. As expected, the truncated complex behaved as an asymmetric heterodimer, whereas the full-length complex exhibited evidence of oligomerization. Using a combination of SAXS and negative-stain EM together with atomic resolution structures of domains determined by X-ray crystallography (46, 50), integrated models for the structure and flexibility of truncated and full-length TDP1-LigIIIα complexes were developed in which the truncated complex is a heterodimer and the full-length complex is a dimer of heterodimers linked by an interaction between the C-terminal BRCT domains of LigIIIα (43, 52). It is likely that preferential binding to the C-terminal BRCT of XRCC1 would prevent the LigIIIα BRCT-BRCT homodimer interaction in the nucleus (43, 52). There may be an interaction between the LigIIIα BRCT and a mitochondrial protein that prevents formation of the dimer of heterodimers in this organelle. We were unable to confirm that XRCC1 blocks the BRCT-BRCT homodimer interaction because the TDP1-LigIIIα-XRCC1 complex was refractory to biophysical analysis by SAXS and EM. Notably, the Artemis nuclease interacts with the DBD of DNA ligase IV during V(D)J recombination and the repair of DNA DSBs by nonhomologous end joining (63, 64). This suggests that the binding of DNA repair enzymes to the flexible catalytic region of DNA ligases is a critical common feature that enables the catalytic regions of enzymes tethered within the same complex to dynamically engage with and, if appropriate, process DNA termini during strand break repair.
Posttranslational modifications, which regulate many protein–protein interactions, such as those involving DNA ligase I (65), often occur in unstructured protein regions, making it difficult to assess their effect on protein conformation and the protein–protein interaction interfaces. Notably, the flexibility of relatively unstructured regions makes them more amenable to large conformational changes induced by posttranslational modification compared with well-folded catalytic domains. As a relevant example, phosphorylation of a flexible XRCC1 region enhances its interaction with the end resection enzymes MRE11 and CtIP providing a possible switch between SSB repair and microhomology-mediated end joining (66) that is enhanced in BRCA2-deficient cells (67). Previously it has been shown that the phosphorylation status of the noncatalytic N-terminal domain of TDP1 regulates its interaction with LigIIIα (33). Our mass spectrometry and SAXS results show that the N-terminal domain of TDP1 is the major site of phosphorylation and that the phosphorylation status of this relatively unstructured region significantly alters its conformation, with dephosphorylation increasing the extension and dynamic flexibility of this region as well as reducing the interaction with LigIIIα. Substitutions of single amino acid residues corresponding to phosphorylation sites within the N-terminal region of TDP1 decreased the efficiency of binding to LigIII, whereas removal of the entire N-terminal region greatly enhanced the binding to LigIII. We therefore reason that certain conformations of the dynamic N-terminal region of TDP1 indirectly block the TDP1-LigIIIα interaction, most likely by occluding the interacting region in the catalytic C-terminal domain of TDP1. In this allosteric regulation, the relative distribution of the different conformations adopted by the N-terminal region of TDP1 is likely to be extremely sensitive to changes in amino acid sequence and/or charge with multiple phosphorylation events probably favoring conformations compatible with the TDP1-LigIIIα interaction.
While previous studies showed that DNA-damage-dependent phosphorylation of TDP1 Ser81 enhanced its interaction with LigIIIα (33), we were not able to ascertain the status of this residue in insect-cell-expressed TDP1. Although replacement of Ser81 with a phosphomimetic residue did not increase the binding of TDP1 to LigIIIα, it is possible that the amino acids substituted do not have the same effect on protein conformation as serine phosphorylation. Despite being highly overexpressed, versions of TDP1 in which Ser81 was replaced with either a nonphosphorylatable or a phosphomimetic residue as well as a version lacking the first 148 residues were not as effective as wild-type TDP1 in complementing the SSB repair defective MEF-TDP1−/− cells. This indicates that the noncatalytic N-terminal region of TDP1, while not essential, does contribute to the repair of SSBs in vivo. Notably, based on coimmunoprecipitation experiments with truncated versions of TDP1, it appears that its N-terminal region interacts with PARP1 (17), the key protein involved in sensing SSBs and generating the poly (ADP-ribose) polymers that serve as the recruitment factor for SSB repair factors, including XRCC1, the partner protein of LigIIIα (3). Interestingly, while XRCC1 binds to poly (ADP-ribosylated)-PARP1 (3, 67, 68), the recruitment of XRCC1 to SSB sites is diminished in cells lacking TDP1 even though TDP1 does not interact directly with XRCC1 (11, 17, 33, 69). Since the XRCC1 partner protein LigIIIα interacts with the C-terminal catalytic region of TDP1, it is possible that TDP1 can simultaneously engage with PARP1 and LigIIIα, thereby enhancing the recruitment of XRCC1 and partner proteins to SSBs resulting from aborted TOP1 reactions. Further work is needed to determine how the interactions of TDP1 with PARP1 and LigIIIα are regulated by posttranslational modifications and contribute to the repair of SSBs in the nucleus and mitochondria.
Experimental procedures
Plasmid constructs and proteins
cDNAs encoding full-length LigIIIα with or without a Strep-Tactin tag, LigIII170–755, TDP1 with a Flag tag and Flag-6His tag, and XRCC1 with a His-tag were subcloned into pFastBac (ThermoFisher). The resultant plasmids were converted into recombinant baculoviruses according to the manufacturer’s protocol. Insect Sf9 cells were infected either with a single baculovirus or a combination of baculoviruses (70). LigIII170–755 and TDP1 as well as LigIIIα-TDP1, LigIII170–755-TDP1, LigIIIα-XRCC1, and LigIIIα-XRCC1-TDP1 complexes were purified from baculovirus-infected insect cells (70). Cell pellets were resuspended in buffer containing 50 mM NaH2PO4, 10 mM Tris-HCl pH 8.0, 300 mM NaCl, 0.5% Igepal, and cOmplete Mini EDTA-free Protease Inhibitor Cocktail tablets (Roche) prior to sonication. Proteins were purified from clarified cell lysates by column chromatography as shown in Table S1.
A series of pET28a plasmids encoding different regions of LigIII with an N-terminal his tag were constructed and verified by Sanger DNA sequencing. Expression of the recombinant proteins was induced in E. coli Rosetta 2 (R2L) cells with β-D-1-thiogalactopyranoside (IPTG). Cell pellets were resuspended in 50 mM NaH2PO4, 10 mM Tris-HCl pH 8.0, 300 mM NaCl, 0.5% Igepal, and cOmplete Mini EDTA-free Protease Inhibitor Cocktail tablets (Roche). After sonication, recombinant proteins were purified from cleared lysates by nickel affinity chromatography (HisTrap HP column, GE Healthcare). A series of pGEX-6p-1 plasmids encoding N-terminal GST fusions of full-length and truncated versions of TDP1 was constructed and verified by Sanger DNA sequencing. Expression of the recombinant proteins was induced in E. coli BL21 (DE3) cells with IPTG prior to purification of GST fusion proteins on a GSTrap HP (GE Healthcare) column according to the protocol from the company. The GST tag was removed from the GST- TDP1149–608 using the PreScission protease prior to SAXS analysis. All column chromatography was performed on an AKTA FPLC (GE Healthcare) using UNICORN 5.0 software.
Pull-down assays
His-tagged full length and truncated versions of LigIII were incubated with Ni-NTA beads in binding buffer (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 5% glycerol, 0.2% NP40, 1 mM benzamidine, and 0.2 mM PMSF) at 4 °C with rotation. After washing with binding buffer containing 30 mM imidazole, beads were incubated with full-length and truncated versions of TPD1 for 1 h at 4 °C. After washing with binding buffer containing 30 mM imidazole, the beads were resuspended in 4× SDS-PAGE sample buffer and then were heated at 100 °C for 5 min. Glutathione beads (Sigma Aldrich) were incubated with the GST-tagged protein in binding buffer containing 1× PBS, pH 7.5, 8% glycerol, 1 mM PMSF, and cOmplete Mini EDTA-free Protease Inhibitor Cocktail tablets (Roche). The same buffer was used to wash the GST protein-bound beads and the beads after the pull-down. Beads were resuspended in 4× SDS-PAGE sample buffer and then were heated at 100 °C for 5 min. Proteins released from the beads were separated by SDS-PAGE prior to detection by immunoblotting with the following antibodies; 6His (Abcam), GST (Abcam), TDP1 (Abcam), Anti-Flag-M2 (Sigma Aldrich).
Surface plasmon resonance
The interaction between LigIII and TDP1 was analyzed using OpenSPR (Nicoya Life Sciences Inc) according to the manufacturer’s guidelines. Running buffer contained 40 mM HEPES-NaOH, pH 7.5, 200 mM NaCl, 0.5% BSA, and 0.05% Tween-20. DNA LigIIIβ (50 μg/ml) was immobilized in channel 2 of the COOH sensor by two consecutive injections of 150 μl at a rate of 20 μl/min. XRCC1 (20 μg/ml) was then immobilized in channel 1 (reference channel) by two consecutive injections of 150 μl at a rate of 20 μl/min. Finally, any remaining active sites in both channels were inactivated by injection of blocking agent (Nicoya Life Sciences Inc) according to the manufacturer’s guidelines. Three different concentrations (3.66 nM, 11 nM, and 33 nM) of analytes were injected at 30 μl/min flow rate for each of the TDP1 variants (WT, S81E, and S81A). In between analyte injections, signal was allowed to return to a stable baseline before a surface regeneration injection (150 μl) of 40 mM HEPES-NaOH, pH 7.5, 600 mM NaCl at 130 μl/min. Sensorgram traces of the LigIII-TDP1 interactions were analyzed using the TraceDrawer software.
Mass spectrometry
After SDS-PAGE, the band corresponding to His-Flag-TDP1 purified from insect cells was detected by staining with Coomassie blue and then cut from the gel. After washing with 50% acetonitrile, the gel slice was sent to the Mass Spectrometry and Proteomics Resource Laboratory at Harvard University. Gel slices were incubated with 20 mM Tris-[2-carboxyethyl] phosphine in 25 mM triethylammonium bicarbonate (TEAB) for 45 min at 37oC for 45 min and then with iodoacetamide in 25 mM TEAB for 45 min a room temperature in the dark. After sequential washing with 200 μl 100 mM TEAB and acetonitrile, the gel slices were incubated with 20 μl 0.1% Rapi Gest reagent for 10 min and then dried in a SpeedVac. Proteins were digested with combinations of trypsin (0.02 μg/ml, cleavage at C-terminal side of Lys and Arg), LysC (0.02 μg/ml, cleavage at C-terminal side of Lys), Glu-C (0.02 μg/ml, cleavage at N-terminal side of Arg) and elastase (0.02 μg/ml, cleavage at C-terminal side of Ala, Val, Ser, Gly, Leu and Ile) at 37 °C overnight except for elastase (2.5 h). Peptides were separated and identified using Waters NanoAquity liquid chromatography system coupled to an Orbitrap Elite Hybrid Ion Trap-Orbitrap Mass Spectrometer (Thermo Fisher). Raw data was analyzed using Proteome Discoverer software (version 2.1.0.18, Thermo Scientific) and the human UniProtKB database (HUMAN2016_SPonly.fasta, 20,417 proteins) to identify peptide sequences. Two missed cleavages were permitted. The database search parameters were precursor peptide mass tolerance of 10 ppm set for parent ion search and mass tolerance of 0.6 Da was set for fragment ions. Carbamidomethyl was set as fixed modification to cysteine and deamidation of asparagine and glutamine as well as methionine oxidation were set as variable modifications. The search output was filtered to a false discovery rate of 1% calculated by dividing the number of false-positive peptides identified in the database by the number of total identified peptides. Mass spectrometry data is available at ftp://massive.ucsd.edu/MSV000087179/. Annotated mass-labeled MS/MS spectra of phosphopeptides are included in Supporting information (Fig. S2B).
Enzyme activity assay
All DNA oligonucleotides were from Integrated DNA Technologies (IDT) except for the oligonucleotide with a 3′-phosphotyrosine, which was from Midland Certified Reagent Company. To make the substrate for nick ligation, oligonucleotide 5′-CGC CAG GGT TCT GAG CAC AGT CAC GAC-3′ was 5′-end-labeled by T4-polynucleoide kinase (NEB) and annealed in a thermocycler, with two oligonucleotides, (5′-PO4-AGTAACACGACGGCCAGTGCTG-3′ and 5-CAGCACTGGCCGTCGTGTTACTGTCGTGACTGTGCTCAGAACCCTGGCG-3′) to generate a 49 nucleotide long duplex DNA substrate with a single ligatable nick. To make the substrate for TDP1, the 3′-phosphotyrosine modified oligo (5′ CGC CAG GGT TCT GAG CAC AGT CAC GAC-PO4-Tyr -3′) was end-labeled by T4-polynucleotide kinase (NEB) and then annealed with two oligonucleotides, (5′-PO4-AGTAACACGACGGCCAGTGCTG-3′ and 5-CAGCACTGGCCGTCGTGTTACTGTCGTGACTGTGCTCAGAACCCTGGCG-3′) to make a 49 nucleotide long nicked duplex DNA substrate with a 3′-phophotyrosine modification at the nick site. The buffer for nick ligation assays buffer contained 20 mM HEPES-NaOH pH7.5, 100 mM NaCl, 10 mM MgCl2, 2 mM ATP, and 0.1 mg/ml BSA. Assays measuring TDP1 activity utilized the same buffer except for the addition of 1 mM DTT and 0.01% Triton X-100.
SEC-MALS
Molecular mass estimation was carried out using a Superdex 200 Increase 10/300 GL gel filtration column operated with an AKTAmicro system (GE Healthcare) and connected to Dawn HELEOS II MALS and OptiLab T-rEX online refractive index detectors (Wyatt Technology). The system was equilibrated in 40 mM HEPES pH 7.5, 200 mM NaCl, 10% glycerol, and 0.1 mM TCEP prior to application of purified samples at 0.35 ml/min. The detectors were calibrated with BSA and data were processed using ASTRA Version 6.1.6.5 (Wyatt Technology). The gel filtration column was calibrated with protein standards of known molecular mass (Bio-Rad).
Negative-stain electron microscopy and image analysis
Gel-filtration-purified TDP1-LigIII170–755 and TDP1-LigIIIα complexes were each chemically cross-linked with 0.05% (v/v) glutaraldehyde in gel filtration buffer (40 mM HEPES pH 7.5, 200 mM NaCl, 10% glycerol) on ice for 5 min in the dark. The reactions were quenched by buffer exchange into low-salt buffer (40 mM HEPES pH 7.5, 50 mM NaCl, 0.1 mM TCEP) using Amicon Ultra centrifugal filters (Millipore Sigma). Cross-linking of complexes was confirmed by SDS-PAGE. Cross-linked complexes and gel-filtration-purified TDP1 were each diluted to ∼20 nM and adsorbed onto glow-discharged (Agar Scientific) carbon-coated copper grids (Electron Microscopy Sciences) for 1 min. Excess liquid was blotted off using Whatman No.1 filter paper and the grids were stained for 1 min with 1.5% uranyl formate (Electron Microscopy Sciences), followed by blotting and air drying. Micrographs were collected at room temperature using a FEI Tecnai T12 transmission electron microscopy operated at 120 keV and fitted with a LaB6 filament. The samples were imaged at 67,000× magnification (1.65 Å pixel size) using a CCD camera (FEI Eagle 4k x 4k) with defocus values between 0.5 and 2 μm. Images were acquired in low-dose mode using SerialEM (71). A total of 132, 199, and 336 micrographs were collected for TDP1, TDP1-LigIII170–755, and TDP1-LigIIIα, respectively. The Scipion framework (72) was used for all image analysis following a twofold downsampling of micrographs. Particles were selected automatically using a reference-free algorithm (73). Initial particle sets contained 26,000, 22,300, and 45,400 particles for TDP1, TDP1-LigIII170–755, and TDP1-LigIIIα, respectively. Errant picks and aggregated proteins were discarded through successive rounds of 2D classification using Xmipp’s CL2D (74). Initial 3D volumes were generated by means of a reference-free stochastic gradient descent program (75) using 6631 and 10,430 particles for TDP1-LigIII170–755 and TDP1-LigIIIα, respectively. 3D classification (76) began with the initial volume low-pass filtered to 60 Å and did not employ masks or symmetry. Particles belonging to similar majority classes were retained for 3D refinement of the principle class (3930 and 8124 particles for TDP1-LigIII170–755 and TDP1-LigIIIα, respectively). The TDP1-LigIII170–755 and TDP1-LigIIIα resolutions were calculated using the gold-standard Fourier shell correlation at the 0.5 criterion (77). UCSF Chimera (78) was used to dock crystal structures into the EM maps.
Small-angle X-ray scattering coupled with multiangle light scattering in line with size-exclusion chromatography
For SEC-SAXS-MALS experiments, 60 μl contained 10 mg/ml TDP1, 10 mg/ml of TDP1 treated with NEB Lambda Phosphatase (see above) in 200 mM NaCl, 20 mM Tris-HCl pH 7.5, 2% Glycerol, 2 mg/ml of TDP1149–608, 3 mg/ml of LigIIIα-6his.Flag-TDP1; 3 mg/ml of 6his-LigIII170–755-Flag-TDP1 in 200 mM NaCl and 40 mM HEPES pH 7.5.
SEC-SAXS-MALS data were collected at the ALS beamline 12.3.1 LBNL Berkeley, California (79, 80). The X-ray wavelength was set at λ = 1.127 Å and the sample-to-detector distance was 2100 mm resulting in scattering vectors, q, ranging from 0.01 Å−1 to 0.4 Å−1. The scattering vector is defined as q = 4πsinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20 °C and data was processed as described (43). Briefly, a SAXS flow cell was directly coupled with an online Agilent 1260 Infinity HPLC system using a Shodex KW802.5 column for TDP1 samples and KW804 column for LigIIIα-TDP1 complexes. The column was equilibrated with running buffer as indicated above with a flow rate of 0.5 ml/min. In total, 55 μl of each sample was run through the SEC and 3-s X-ray exposures were collected continuously during a 30-min elution. The SAXS frames recorded prior to the protein elution peak were used to subtract all other frames. The subtracted frames were investigated by radius of gyration (Rg) derived by the Guinier approximation I(q) = I(0) exp(−q2Rg2/3) with the limits qRg <1.5 (Fig. S11) (81). The elution peak was mapped by comparing the integral of ratios to background and Rg relative to the recorded frame using the program SCÅTTER. Nonuniform Rg values across an elution peak represent conformational variability of samples. Final merged SAXS profiles, derived by integrating multiple frames at the peak of the elution peak, were used for further analysis including Guinier plot, which determined aggregation free state (Fig. S11). The program SCÅTTER was used to compute the P(r) function. The distance r where P(r) approach zero intensity identifies the maximal dimension of the macromolecule (Dmax). P(r) functions were normalized based on the molecular mass of the assemblies as determined by SCÅTTER using volume of correlation Vc (82) (Table 1). The eluent was subsequently split 3:1 between SAXS line and a series of UV at 280 and 260 nm, MALS, quasi-elastic light scattering (QELS), and refractometer detector. MALS experiments were performed using an 18-angle DAWN HELEOS II light scattering detector connected in tandem to an Optilab refractive index concentration detector (Wyatt Technology). System normalization and calibration were performed with BSA using a 45 μl sample at 10 mg/ml in the same SEC running buffer and a dn/dc value of 0.19. The light scattering experiments were used to perform analytical-scale chromatographic separations for molecular mass determination of the main peaks in the SEC analysis. UV, MALS, and differential refractive index data was analyzed using Wyatt Astra 7 software to monitor the homogeneity of the sample across the elution peak complementary to the abovementioned SEC-SAXS signal validation (Fig. 5A).
Solution structure modeling
The model for full-length TDP1 was built based on the crystal structure of TDP1149–608 (PDB 1JY1) and by adding missing N- and C-terminal regions in MODELLER (83). To model conformation flexibility of the N-terminal region (residues 1–150), we performed conformational sampling using BILBOMD (54). The experimental SAXS profiles of TDP1 and dephosphorylated TDP1 were then compared to theoretical scattering curves of the atomistic models generated by BILBOMD (54) using FOXS (84, 85), followed by multistate model selection by MultiFoXS (48, 51).
The initial atomistic model of LigIII170–755-6hisFlag-TDP1 was built by connecting TDP1 atomistic model (Fig. 4) and LigIII170–755 domains (PDB 3L2P) by flexible linkers while preserving the relative position of domains determined by EM. Similarly, the initial model for LigIIIα-6his.Flag-TDP1 was built as above with the inclusion of the LigIII ZnF domain (PDB 1UW0) and BRCT homodimer (PDB 3PC8) and preserving the relative domain positions determined by EM. Minimal molecular dynamics simulations were performed on flexible regions in the models by the rigid body modeling strategy BILBOMD (54) in order to optimize conformational space of domains linkers alone or individual domains. The selection of multistate models was performed as described above for TDP1.
Cell culture and lentivirus particle production
Mouse embryonic fibroblast (MEF-wt and MEF-TDP1−/−-p53−/−) cells were a generous gift from Dr Peter McKinnon (St Jude Hospital). Cells were grown in DMEM/Ham’s F12 (1:1 Mixture) with L-glutamine, 10% fetal bovine serum and 1% Penicillin-Streptomycin. Nonessential amino acid (0.01%) and β mercaptoethanol (55 μM) were added immediately prior to use. cDNAs encoding TDP1-wt, TDP1149–608, TDP1-S81E, and TDP1-S81A were subcloned into the pLVX-EF1a-IRES-ZsGreen1 (Takara). Lentivirus particles were created in HEK293TN cells (HEK239TN; System Biosciences) using the pMDG envelope (Addgene) and psPAX2 (Addgene) packaging plasmids, along with derivatives of pLVX-EF1a-IRES-ZsGreen1. Virus particles were collected using Lenti-X-Concentrator (Takara) and used to transduce MEF-TDP1−/−-p53−/−cells.
Alkaline comet assay
After incubation with 25 μM CPT for 1 h, MEFs were trypsinized, collected by centrifugation, and then resuspended at 1 × 105 cells/ml in ice-cold PBS (Ca++ and Mg++ free). The cell suspension was mixed with 37 °C molten LMAgarose (Trevigen) at a ratio of 1:10 (v/v), and then 40 μl of the mixture was immediately pipetted onto each well of a CometSlide (20 well slide) (Trevigen). The alkaline comet assay was carried out according to the manufacturer’s instructions. DNA was stained with 50 μl of diluted SYBR Gold Nucleic Acid Gel Stain (1:10,000 SYBR Gold in TE buffer) that was placed onto each circle of dried agarose. The slides were viewed by an Olympus XI83 fluorescence microscope equipped with a DP72 camera and cellSens Dimension imaging software (Olympus America; ver. 1.17). The quantification done by image analysis using Comet Analysis Software Trevigen (version 1.2) was utilized to determine the comet tail moment, percentage of DNA in tail, and percentage of cells with tails. Images were manually reviewed to ensure appropriate selection of nuclei (size, roundness, shape, and direction of tail) and to exclude overlapping cells and staining or processing artifacts. The mean comet-tail moment was used to represent the result. The comet-tail moment represents the product of the fraction of cellular DNA in the comet tail and the tail length with higher values corresponding to a greater number of cellular DNA SSBs. At least 50 cells on each slide and a minimum three slides in each group were analyzed.
Data availability and accession numbers
Mass spectrometry raw data is available in the MassIVE database (ftp://massive.ucsd.edu/MSV000087179/). EM maps have been deposited in the EM databank with accession codes EMD-23299 (LigIII170–755-TDP1) and EMD-23301 (LigIIIα-TDP1). SAXS data and atomistic models of TDP1, dephosphorylated TDP1, LigIII170–755-TDP1, and LigIIIα-TDP1 complex have been deposited in the SASBDB with accession codes SASDJN2, SASDJQ2, SASDJP2, SASDJS2, SASDJR2.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank Dr Budnik and the staff at the Harvard Center for Mass Spectrometry for the uploading the raw data and information about the mass spectrometry analysis as well as the preparation of the annotated mass labeled MS/MS spectra of phosphopeptides. We are grateful to Dr Tom Ellenberger and Dr Peter J McKinnon for the plasmid constructs encoding LigIIIα domains and, the MEF-wt and MEF-TDP1−/−-p53−/− cells, respectively. We thank Dr Scott A Ness and the UNMCCC ATG Shared Resource for assistance with lentivirus expression and Dr Laurie Hudson for help with the comet assay.
Author contributions
I. R., M. H., J. M. P., J. A. T., and A. E. T. conceptualization; I. R., M. H., A. S., J. M. P., J. A. T., and A. E. T. formal analysis; J. M. P., J. A. T., and A. E. T. funding acquisition; I. R., M. H., A. S., and M.-S. T. investigation; J. M. P., J. A. T., and A. E. T. project administration; I. R., M. H., M.-S. T, and J. M. P. resources; A. E. T. supervision; I. R., M. H., A. S., M.-S. T., J. M. P., J. A. T., and A. E. T. writing-original draft; I. R., M. H., A. S., M.-S. T, J. M. P., J. A. T., and A. E. T. writing-review and editing.
Funding and additional information
This work was supported in part by National Institutes of Health (NIH) grants R01 ES012512 (A. E. T.), R35 CA220430 (J. A. T.), and the Structural Cell Biology of DNA Repair Program (P01 CA092584, A. E. T., J. M. P., J. A. T.) and a Discovery grant RGPIN-2015-05776 from the National Science and Engineering Research Council of Canada (J. M. P.). This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J. A. T. is also supported by Cancer Prevention Research Institute of Texas (CPRIT) grant RP180813, and a Robert A. Welch Chemistry Chair. Funding for the SIBYLS beamline at the Advanced Light Source was provided in part by the Offices of Science and Biological and Environmental Research, U.S. Department of Energy, under Contract DE-AC02-05BH11231.
Edited by Patrick Sung
Contributor Information
John A. Tainer, Email: jatainer@gmail.com.
Alan E. Tomkinson, Email: atomkinson@salud.unm.edu.
Supporting information
References
- 1.Olive P.L. The role of DNA single- and double-strand breaks in cell killing by ionizing radiation. Radiat. Res. 1998;150:S42–S51. [PubMed] [Google Scholar]
- 2.Vilenchik M.M., Knudson A.G. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldecott K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 4.Hanzlikova H., Kalasova I., Demin A.A., Pennicott L.E., Cihlarova Z., Caldecott K.W. The importance of poly(ADP-ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol. Cell. 2018;71:319–331.e313. doi: 10.1016/j.molcel.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannan W.J., Pederson D.S. Mechanisms and consequences of double-strand DNA break formation in chromatin. J. Cell Physiol. 2016;231:3–14. doi: 10.1002/jcp.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortes-Ledesma F., Aguilera A. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 2006;7:919–926. doi: 10.1038/sj.embor.7400774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 8.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., Martin N.M., Jackson S.P., Smith G.C., Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 9.Pommier Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 10.Takashima H., Boerkoel C.F., John J., Saifi G.M., Salih M.A., Armstrong D., Mao Y., Quiocho F.A., Roa B.B., Nakagawa M., Stockton D.W., Lupski J.R. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 11.El-Khamisy S.F., Saifi G.M., Weinfeld M., Johansson F., Helleday T., Lupski J.R., Caldecott K.W. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 12.Moreira M.C., Barbot C., Tachi N., Kozuka N., Uchida E., Gibson T., Mendonca P., Costa M., Barros J., Yanagisawa T., Watanabe M., Ikeda Y., Aoki M., Nagata T., Coutinho P. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat. Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 13.Ahel I., Rass U., El-Khamisy S.F., Katyal S., Clements P.M., McKinnon P.J., Caldecott K.W., West S.C. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 14.Hoch N.C., Hanzlikova H., Rulten S.L., Tetreault M., Komulainen E., Ju L., Hornyak P., Zeng Z., Gittens W., Rey S.A., Staras K., Mancini G.M., McKinnon P.J., Wang Z.Q., Wagner J.D. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature. 2017;541:87–91. doi: 10.1038/nature20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen J., Gilmore E.C., Marshall C.A., Haddadin M., Reynolds J.J., Eyaid W., Bodell A., Barry B., Gleason D., Allen K., Ganesh V.S., Chang B.S., Grix A., Hill R.S., Topcu M. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat. Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascal J.M. The comings and goings of PARP-1 in response to DNA damage. DNA Repair (Amst.) 2018;71:177–182. doi: 10.1016/j.dnarep.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das B.B., Huang S.Y., Murai J., Rehman I., Ame J.C., Sengupta S., Das S.K., Majumdar P., Zhang H., Biard D., Majumder H.K., Schreiber V., Pommier Y. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014;42:4435–4449. doi: 10.1093/nar/gku088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okano S., Lan L., Caldecott K.W., Mori T., Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M., Lu L.Y., Yang C.Y., Wang S., Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013;27:1752–1768. doi: 10.1101/gad.226357.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breslin C., Hornyak P., Ridley A., Rulten S.L., Hanzlikova H., Oliver A.W., Caldecott K.W. The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function. Nucleic Acids Res. 2015;43:6934–6944. doi: 10.1093/nar/gkv623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldecott K.W., McKeown C.K., Tucker J.D., Ljungquist S., Thompson L.H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caldecott K.W., Aoufouchi S., Johnson P., Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular 'nick-sensor' in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okano S., Lan L., Tomkinson A.E., Yasui A. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res. 2005;33:422–429. doi: 10.1093/nar/gki190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubota Y., Nash R.A., Klungland A., Schar P., Barnes D.E., Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: Interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehouse C.J., Taylor R.M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D.D., Weinfeld M., Caldecott K.W. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 26.Vidal A.E., Boiteux S., Hickson I.D., Radicella J.P. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S.Y., Murai J., Dalla Rosa I., Dexheimer T.S., Naumova A., Gmeiner W.H., Pommier Y. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs. Nucleic Acids Res. 2013;41:7793–7803. doi: 10.1093/nar/gkt483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Interthal H., Chen H.J., Champoux J.J. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flett F.J., Ruksenaite E., Armstrong L.A., Bharati S., Carloni R., Morris E.R., Mackay C.L., Interthal H., Richardson J.M. Structural basis for DNA 3'-end processing by human tyrosyl-DNA phosphodiesterase 1. Nat. Commun. 2018;9:24. doi: 10.1038/s41467-017-02530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi K., Kurahashi K., Gao R., Tsutakawa S.E., Tainer J.A., Pommier Y., Aihara H. Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2. Nat. Struct. Mol. Biol. 2012;19:1372–1377. doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortes Ledesma F., El Khamisy S.F., Zuma M.C., Osborn K., Caldecott K.W. A human 5'-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 32.Heideker J., Prudden J., Perry J.J., Tainer J.A., Boddy M.N. SUMO-targeted ubiquitin ligase, Rad60, and Nse2 SUMO ligase suppress spontaneous Top1-mediated DNA damage and genome instability. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiang S.C., Carroll J., El-Khamisy S.F. TDP1 serine 81 promotes interaction with DNA ligase IIIalpha and facilitates cell survival following DNA damage. Cell Cycle. 2010;9:588–595. doi: 10.4161/cc.9.3.10598. [DOI] [PubMed] [Google Scholar]
- 34.Das B.B., Antony S., Gupta S., Dexheimer T.S., Redon C.E., Garfield S., Shiloh Y., Pommier Y. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009;28:3667–3680. doi: 10.1038/emboj.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakshmipathy U., Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lakshmipathy U., Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28:3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das B.B., Dexheimer T.S., Maddali K., Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2010;107:19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi M.N., Carbone M., Mostocotto C., Mancone C., Tripodi M., Maione R., Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J. Biol. Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szczesny B., Brunyanszki A., Olah G., Mitra S., Szabo C. Opposing roles of mitochondrial and nuclear PARP1 in the regulation of mitochondrial and nuclear DNA integrity: Implications for the regulation of mitochondrial function. Nucleic Acids Res. 2014;42:13161–13173. doi: 10.1093/nar/gku1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simsek D., Furda A., Gao Y., Artus J., Brunet E., Hadjantonakis A.K., Van Houten B., Shuman S., McKinnon P.J., Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y., Katyal S., Lee Y., Zhao J., Rehg J.E., Russell H.R., McKinnon P.J. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brosey C.A., Tainer J.A. Evolving SAXS versatility: Solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr. Opin. Struct. Biol. 2019;58:197–213. doi: 10.1016/j.sbi.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammel M., Rashid I., Sverzhinsky A., Pourfarjam Y., Tsai M.S., Ellenberger T., Pascal J.M., Kim I.K., Tainer J.A., Tomkinson A.E. An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA ligase IIIalpha within a flexible DNA repair complex. Nucleic Acids Res. 2021;49:306–321. doi: 10.1093/nar/gkaa1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackey Z.B., Ramos W., Levin D.S., Walter C.A., McCarrey J.R., Tomkinson A.E. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellenberger T., Tomkinson A.E. Eukaryotic DNA ligases: Structural and functional insights. Annu. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cotner-Gohara E., Kim I.K., Hammel M., Tainer J.A., Tomkinson A.E., Ellenberger T. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry. 2010;49:6165–6176. doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Putnam C.D., Hammel M., Hura G.L., Tainer J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 48.Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016;44:W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rambo R.P., Tainer J.A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davies D.R., Interthal H., Champoux J.J., Hol W.G. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure. 2002;10:237–248. doi: 10.1016/s0969-2126(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 51.Schneidman-Duhovny D., Hammel M. Modeling structure and dynamics of protein complexes with SAXS profiles. Methods Mol. Biol. 2018;1764:449–473. doi: 10.1007/978-1-4939-7759-8_29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuneo M.J., Gabel S.A., Krahn J.M., Ricker M.A., London R.E. The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes. Nucleic Acids Res. 2011;39:7816–7827. doi: 10.1093/nar/gkr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mani R.S., Karimi-Busheri F., Fanta M., Caldecott K.W., Cass C.E., Weinfeld M. Biophysical characterization of human XRCC1 and its binding to damaged and undamaged DNA. Biochemistry. 2004;43:16505–16514. doi: 10.1021/bi048615m. [DOI] [PubMed] [Google Scholar]
- 54.Pelikan M., Hura G.L., Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammel M., Yu Y., Radhakrishnan S.K., Chokshi C., Tsai M.S., Matsumoto Y., Kuzdovich M., Remesh S.G., Fang S., Tomkinson A.E., Lees-Miller S.P., Tainer J.A. An intrinsically disordered APLF links Ku, DNA-PKcs, and XRCC4-DNA ligase IV in an extended flexible non-homologous end joining complex. J. Biol. Chem. 2016;291:26987–27006. doi: 10.1074/jbc.M116.751867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams G.J., Hammel M., Radhakrishnan S.K., Ramsden D., Lees-Miller S.P., Tainer J.A. Structural insights into NHEJ: Building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst.) 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammel M., Tainer J.A. X-ray scattering reveals disordered linkers and dynamic interfaces in complexes and mechanisms for DNA double-strand break repair impacting cell and cancer biology. Protein Sci. 2021 doi: 10.1002/pro.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pommier Y. Drugging topoisomerases: Lessons and challenges. ACS Chem. Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caldecott K.W., Tucker J.D., Stanker L.H., Thompson L.H. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nash R.A., Caldecott K.W., Barnes D.E., Lindahl T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- 61.Taylor R.M., Wickstead B., Cronin S., Caldecott K.W. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr. Biol. 1998;8:877–880. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 62.Pascal J.M., O'Brien P.J., Tomkinson A.E., Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 63.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Ioannes P., Malu S., Cortes P., Aggarwal A.K. Structural basis of DNA ligase IV-artemis interaction in nonhomologous end-joining. Cell Rep. 2012;2:1505–1512. doi: 10.1016/j.celrep.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montecucco A., Rossi R., Levin D.S., Gary R., Park M.S., Motycka T.A., Ciarrocchi G., Villa A., Biamonti G., Tomkinson A.E. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: Identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dutta A., Eckelmann B., Adhikari S., Ahmed K.M., Sengupta S., Pandey A., Hegde P.M., Tsai M.S., Tainer J.A., Weinfeld M., Hegde M.L., Mitra S. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 2017;45:2585–2599. doi: 10.1093/nar/gkw1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eckelmann B.J., Bacolla A., Wang H., Ye Z., Guerrero E.N., Jiang W., El-Zein R., Hegde M.L., Tomkinson A.E., Tainer J.A., Mitra S. XRCC1 promotes replication restart, nascent fork degradation and mutagenic DNA repair in BRCA2-deficient cell. NAR Cancer. 2020;2 doi: 10.1093/narcan/zcaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polo L.M., Xu Y., Hornyak P., Garces F., Zeng Z., Hailstone R., Matthews S.J., Caldecott K.W., Oliver A.W., Pearl L.H. Efficient single-strand break repair requires binding to both poly(ADP-ribose) and DNA by the central BRCT domain of XRCC1. Cell Rep. 2019;26:573–581.e575. doi: 10.1016/j.celrep.2018.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Plo I., Liao Z.Y., Barcelo J.M., Kohlhagen G., Caldecott K.W., Weinfeld M., Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst.) 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 70.Della-Maria J., Zhou Y., Tsai M.-S., Kuhnlein J., Carney J.P., Paull T.T., Tomkinson A.E. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIα/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J. Biol. Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 72.De la Rosa-Trevín J., Quintana A., Del Cano L., Zaldívar A., Foche I., Gutiérrez J., Gómez-Blanco J., Burguet-Castell J., Cuenca-Alba J., Abrishami V. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 2016;195:93–99. doi: 10.1016/j.jsb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 73.Kimanius D., Forsberg B.O., Scheres S.H., Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5 doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sorzano C., Bilbao-Castro J., Shkolnisky Y., Alcorlo M., Melero R., Caffarena-Fernández G., Li M., Xu G., Marabini R., Carazo J. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol. 2010;171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 76.Scheres S.H. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scheres S.H., Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 79.Dyer K.N., Hammel M., Rambo R.P., Tsutakawa S.E., Rodic I., Classen S., Tainer J.A., Hura G.L. High-throughput SAXS for the characterization of biomolecules in solution: A practical approach. Methods Mol. Biol. 2014;1091:245–258. doi: 10.1007/978-1-62703-691-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Classen S., Hura G.L., Holton J.M., Rambo R.P., Rodic I., McGuire P.J., Dyer K., Hammel M., Meigs G., Frankel K.A., Tainer J.A. Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the advanced light source. J. Appl. Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guinier A., Fournet F. Wiley Interscience; New York, NY: 1955. Small Angle Scattering of X-Rays. [Google Scholar]
- 82.Rambo R.P., Tainer J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 84.Schneidman-Duhovny D., Hammel M., Sali A. FoXS: A web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry raw data is available in the MassIVE database (ftp://massive.ucsd.edu/MSV000087179/). EM maps have been deposited in the EM databank with accession codes EMD-23299 (LigIII170–755-TDP1) and EMD-23301 (LigIIIα-TDP1). SAXS data and atomistic models of TDP1, dephosphorylated TDP1, LigIII170–755-TDP1, and LigIIIα-TDP1 complex have been deposited in the SASBDB with accession codes SASDJN2, SASDJQ2, SASDJP2, SASDJS2, SASDJR2.