Summary
In sexually reproducing animals, the oocyte contributes a large supply of RNAs that are essential to launch development upon fertilization. The mechanisms that regulate the composition of the maternal RNA contribution during oogenesis are unclear. Here, we show that a subset of RNAs expressed during the early stages of oogenesis is subjected to regulated degradation during oocyte specification. Failure to remove these RNAs results in oocyte dysfunction and death. We identify the RNA-degrading Super Killer complex and No-Go Decay factor Pelota as key regulators of oogenesis via targeted degradation of specific RNAs expressed in undifferentiated germ cells. These regulators target RNAs enriched for cytidine sequences that are bound by the polypyrimidine tract binding protein Half pint. Thus, RNA degradation helps orchestrate a germ cell-to-maternal transition that gives rise to the maternal contribution to the zygote.
Graphical Abstract
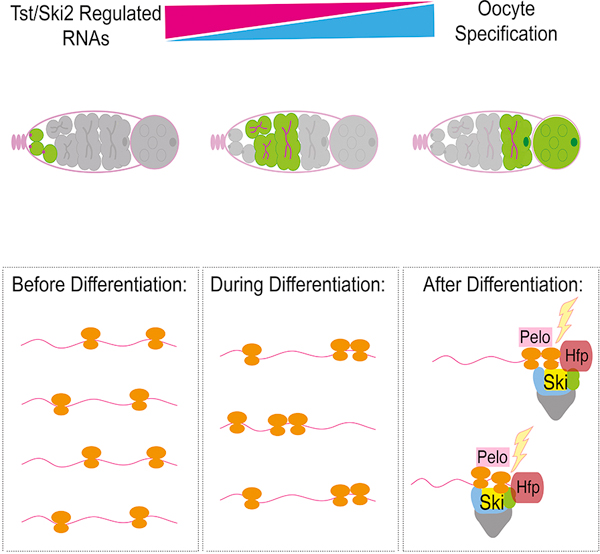
eToc blurb:
Blatt et al. find that a cohort of RNAs expressed in germ cells are targeted for regulated degradation mediated by components of the No Go Decay pathway when oocytes are specified. They find that this regulated RNA degradation is essential for proper oocyte development and fertility.
Introduction
A fertilized egg is totipotent, having the unique potential to differentiate into every cell lineage in the adult organism [1–3]. Across animals, RNA transcribed from 40–75% of all genes is deposited into the egg during oogenesis as a maternal contribution required for embryo development [4–6]. It is unlikely that every RNA synthesized during oogenesis is destined for the maternal contribution – RNAs that support oogenesis-specific functions, such as germline stem cell (GSC) self-renewal and differentiation, could be detrimental during embryogenesis. It is not known if such oogenesis-specific RNAs are targeted for elimination or what, if any, mechanisms ensure that only the appropriate RNAs are contributed to the egg. In Drosophila, oogenesis occurs in ovarioles beginning in germaria, which contain the GSCs and the GSC daughter cells (cystoblasts, CBs) that progressively differentiate into 16-cell cysts (Figure 1A–1B) [7–11]. In each cyst, the oocyte enlarges as it receives RNAs and proteins destined to become the maternal contribution from the remaining 15 nurse cells (Figure 1B) [12–18], but the eventual fate of the existing mRNAs that supported oocyte development is unclear.
Figure 1. Components of the Ski Complex are required in the germ line for oogenesis.
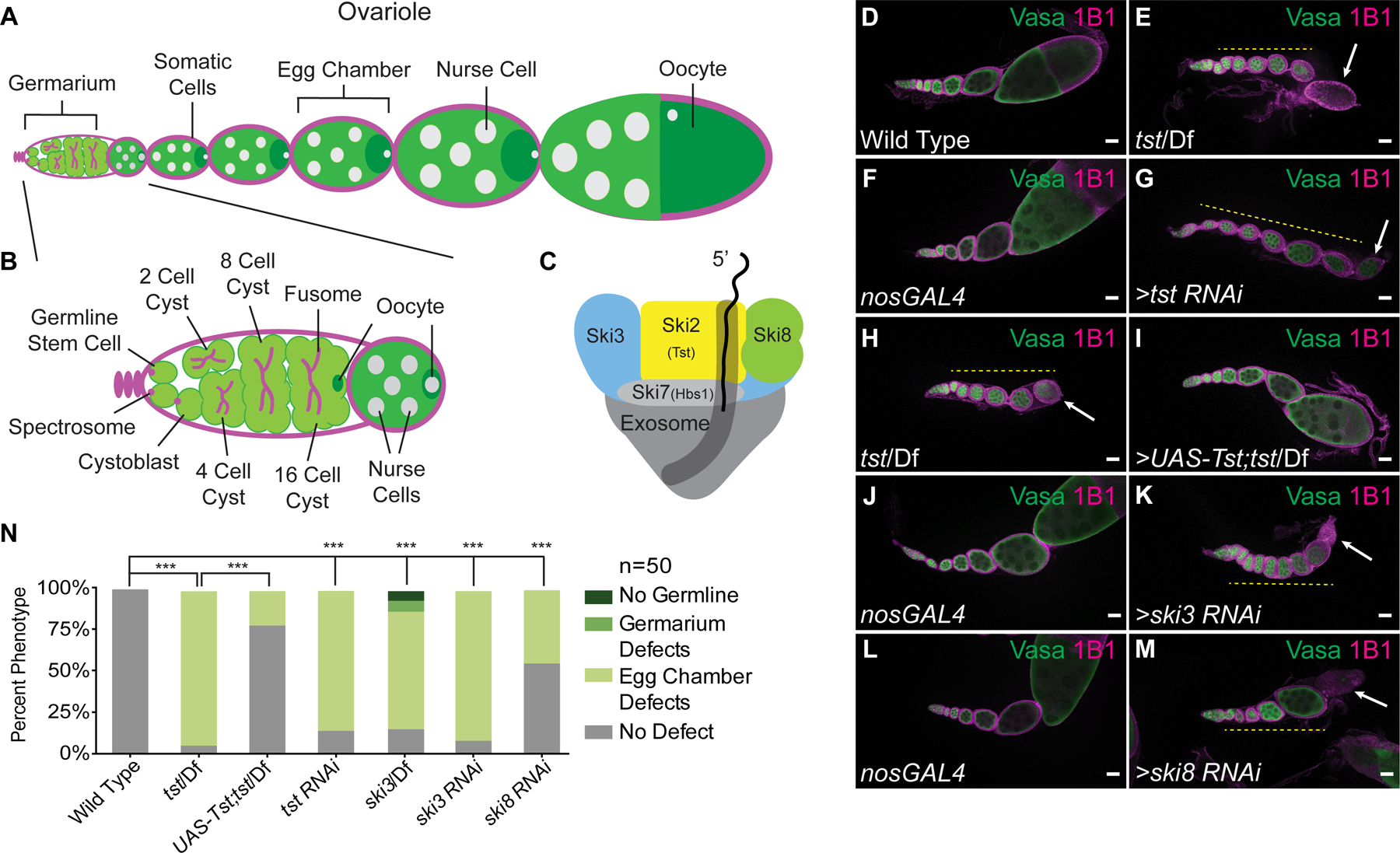
(A) Schematic of a Drosophila ovariole and (B) germarium. (C) The Ski complex is composed of Ski2 (Tst, yellow), Ski3 (blue), Ski8 (green), and Ski7 (Hbs1, light gray) threading mRNA into the exosome (dark gray). (D–M) Confocal images of ovarioles stained with Vasa (green) and 1B1 (magenta). (D) Adult WT ovarioles show normal egg chamber development. (E) Hypomorphic tst genomic mutant ovariole displaying egg chambers that do not grow in size (yellow dashed line) and a dying egg chamber (arrow). (F) nosGAL4 driver control ovariole shows WT morphology. (G) tst germline RNAi knockdown ovariole displaying egg chambers that do not grow in size (yellow dashed line) and a dying egg chamber (arrow). (H) tst genomic mutant ovariole (control for I) showing egg chamber defects (yellow dashed line) and dying egg chamber (white arrow) (I) tst genomic mutant ovariole expressing recombinant Tst protein in the germline rescues the egg chamber defects. (J–M) Germline RNAi knockdown for ski3 (K) and ski8 (M) also display egg chambers that do not grow in size (yellow dashed line) and dying egg chambers (arrow), whereas nosGAL4 driver controls (J, L) have WT egg chambers. (N) Quantification of oogenesis defect phenotypes (n=50 ovarioles per genotype, *** = p < 0.001, Chi-squared tests with Bonferroni correction, df=3). tst/Df and tst RNAi are not significantly different. Scale bars = 10µm. See also Figure S1.
Results
The Ski complex is required in the female germline for fertility
In a screen to identify novel regulators of oogenesis, we discovered that a component of the RNA-degradation-promoting Super Killer (Ski) complex (Figure 1C), Super Killer 2 (Ski2), called Twister (Tst) in Drosophila, is required for egg chamber growth and female fertility (Figure S1A) [19–22]. Wild type (WT) Drosophila ovarioles stained for Vasa (germ cells) and for 1B1 (somatic cell membranes) show the progression from the germarium to successively larger egg chambers (Figure 1D). In contrast, egg chambers failed to grow in tst mutant ovarioles (Figure 1D–1E, 1N) as well as upon germline RNAi depletion of tst (nanos-GAL4 >RNAi, Figure 1F–1G, 1N, S1B), but not when tst was depleted in the soma (traffic jam-GAL4 >RNAi, S1C-S1D) [23,24]. Egg chambers lacking tst expressed cleaved Caspase 3 at putative stages 6–7, suggesting that they undergo apoptosis (Figure S1E–S1F). tst mutant flies are otherwise viable, and successful oogenesis and viable egg production were restored in tst mutants by germline-specific expression of Tst protein (nanos-GAL4 driver, Figure 1H–1I, 1N, S1A). This maternally expressed tst is sufficient to allow for embryonic development, as eggs laid by rescued flies hatched at the same rate as WT control eggs (Figure S1G). Thus, Tst is required in the germline for proper oogenesis.
The Ski complex promotes 3’ to 5’ degradation of mRNAs [20]. In addition to SKI2 (Tst), the Ski complex consists of the scaffolding subunits SKI3 and SKI8, which are coupled to the exosome complex by SKI7 (Figure 1C) [20,21,25,26]. We found that ski3 (CG8777) mutant and germline depletion of ski3 and ski8 (CG3909) phenocopied tst mutants (Figure 1J–1N, S1H–S1J). The conserved GTPase, Hsp70 subfamily B Suppressor (HBS1), is thought to fulfill the role of SKI7 in Drosophila; however, female hbs1 mutants were previously found to be fertile, suggesting that SKI7/HBS1 is dispensable for Ski complex function in the female germline or acts redundantly with a yet-unidentified protein [27–29].
Defects in other mRNA degradation pathways do not elicit the same phenotype as loss of the Ski complex. Loss of exosome components that are required for either nuclear RNA degradation alone (rRNA-processing 6 (rrp6) and mRNA transport 4 (mtr4)) or both nuclear and cytoplasmic RNA degradation (chromosome disjunction 3 (dis3) and rrp40) resulted in earlier, germarium-stage defects and complete loss of the germline (Figure S1K–S1R). Defects to the cytoplasmic 5’ mRNA degradation pathway due to loss of xrn1 still produce late-stage egg chambers, though flies have reduced fecundity [30]. Thus, we conclude that the cytoplasmic 3’ mRNA degradation-promoting Ski complex is required specifically in the fly germ line for egg chamber growth and successful completion of oogenesis.
The Ski complex promotes degradation of a cohort of RNAs during oogenesis
Given the role of the Ski complex in exosome-mediated RNA degradation, we hypothesized that Tst promotes degradation of RNAs during oogenesis [21,31,32]. We performed RNA sequencing (RNA-seq) on ovaries of tst mutants and germline RNAi knockdown of tst and compared these samples to adult WT ovaries as well as ovaries from virgin flies (young WT) (Figure S2A–S2C). tst ovaries are both temporally and morphologically further developed than young WT ovaries, which contain only 2–3 early-stage egg chambers, compared to 6–7 egg chambers in tst ovarioles and up to 9–10 chambers in adult WT (Figure S2A–S2C) (see methods). RNA-seq indeed revealed that the tst ovary transcriptome closely resembles adult WT (Figure S2A); however, 296 genes are upregulated in ovaries lacking tst (Data S1A–B). Among these, 207 genes displayed >4-fold higher levels in germline tst RNAi compared to adult WT and are >2-fold upregulated in tst genomic mutant ovaries, including blanks and actin57B (act57B) (Figure 2A–2B). These genes likely encode transcripts strongly regulated by Tst in the germline.
Figure 2. Twister promotes degradation of a subset of transcripts concurrent with oocyte specification.
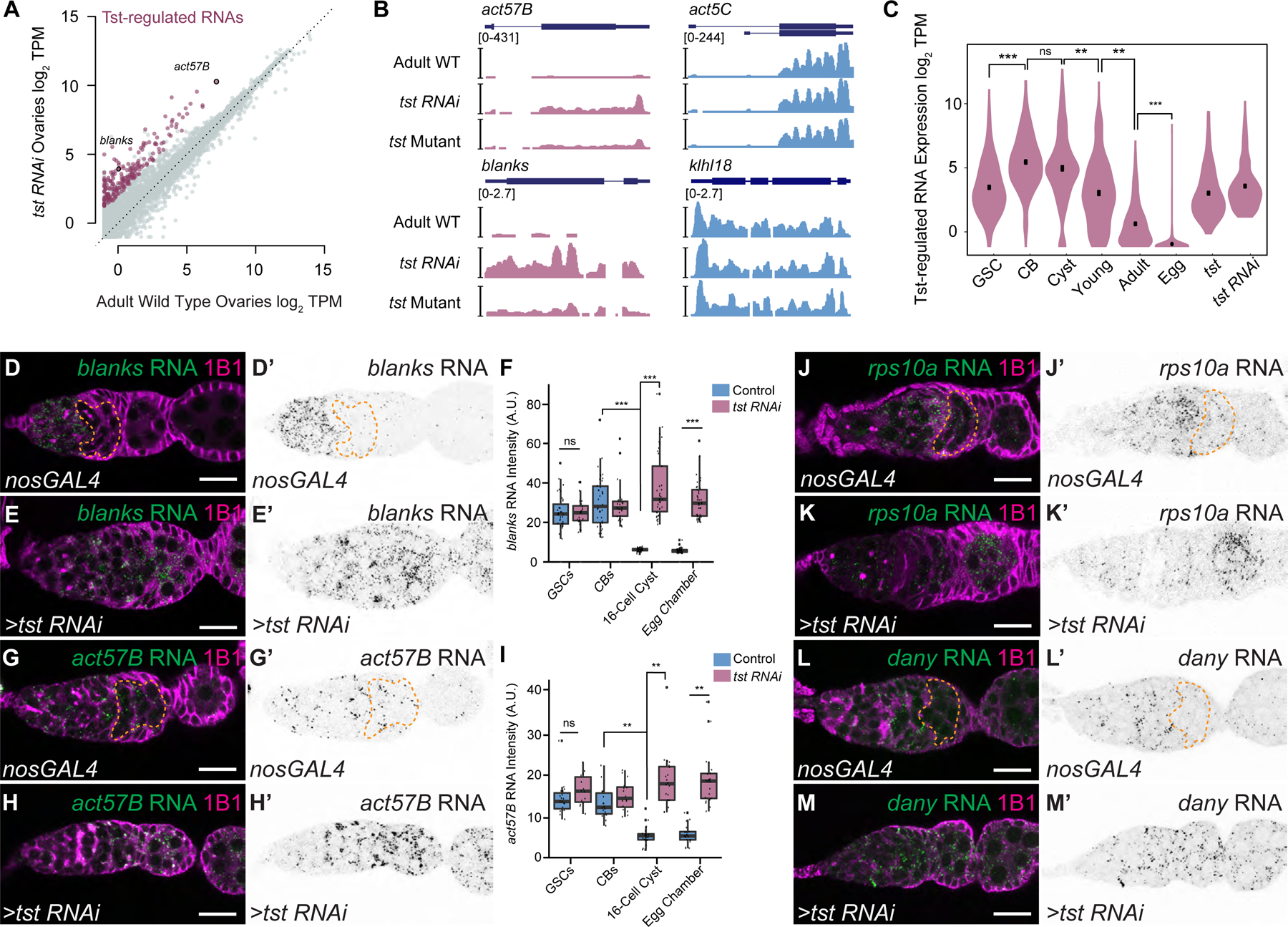
(A) RNA-seq biplot comparing adult WT and tst germline RNAi knockdown ovaries showing upregulated Tst-regulated RNAs (magenta). act57b and blanks are highlighted. TPM = transcripts per million. (B) RNA-seq coverage tracks of Tst-regulated genes act57B and blanks (magenta) and non-targets act5C and klhl18 (blue). (C) Violin plot of Tst-regulated RNAs (n=207) showing decrease in expression after differentiation and cyst stages. *** = p < 1e-25, Wilcoxon signed rank test with Bonferroni correction. GSC = germline stem cells, CB = cystoblasts. (D–H) Confocal images of in situ hybridizations probing for Tst-regulated RNAs blanks and act57B (green) and staining for 1B1 (magenta). (D’-H’) Corresponding in situ signals in grayscale. blanks RNA expression is restricted to undifferentiated cells and early cyst in a nosGAL4 driver control (dotted orange line) (D, D’) but is expanded to egg chambers in tst RNAi ovarioles (E, E’). (F) Quantification of blanks in situ signal in different cell types in the ovariole. *** = p < 0.001, Tukey’s post-hoc test (n=24–42 per cell type). Compared to nosGAL4 control (G, G’), act57B RNA expression is also expanded in tst RNAi (H, H’). (I) Quantification of act57B in situ signal in different cell types in the ovariole. ** = p < 0.01, Tukey’s post-hoc test (n=14–23 per cell type). Scale bars = 10µm. See Supplement for quantification for J–M’. A.U. = arbitrary units. See also Figure S2 and Data S1.
We considered whether the elevated levels of genes in tst samples could arise from the morphological differences between tst ovaries and adult WT, i.e. developmental arrest and loss of late-stage egg chambers that could enrich the tst RNA-seq samples for early-stage RNAs. Using previous published single-cell RNA sequencing (scRNA-seq) data from Drosophila ovaries [33], we were able to identify 18 genes specifically expressed during early oogenesis that would be expected to be over-represented in bulk RNA-seq samples that are enriched in early-stage egg chambers. However, only 1 out of 18 is >4-fold elevated in tst RNAi samples (rps10a), while genes such as vas, bam, and cona are not (Figure S2D). Conversely, maternal mRNAs that would be enriched in egg chambers such as polar granule component (pgc) and germ cell-less (gcl) are not decreased in tst ovaries compared to adult WT (Figure S2E, Data S1A–B). Thus, the transcriptome differences in tst ovaries are not simply due to morphological or developmental differences, and rather reflect specific regulation of a cohort of early-expressed RNAs during oogenesis.
To determine if the upregulation of Tst-regulated mRNAs is due to a defect in post-transcriptional regulation, we measured pre-mRNA levels of select Tst-regulated RNAs and nontargets by qRT-PCR and indeed found no significant difference between WT and tst germline RNAi flies (Figure S2F, p>0.05, Student’s t-test). We also determined that the changes of Tst-regulated RNA levels by RNA-seq are not due to changes in poly(A) tail length: by conducting qRT-PCR primed with random hexamers, we found that act57b, act42a, act87e, and blanks RNAs are indeed upregulated in tst samples (Figure S2G). Taken together, we find that tst promotes the post-transcriptional degradation of a distinct group of RNAs during oogenesis.
Ski complex mediated degradation occurs concurrent with oocyte specification
To characterize the timing of Tst-mediated RNA degradation during oogenesis, we profiled the expression of Tst-regulated RNAs in ovaries across an RNA-seq time course of oocyte development. We enriched for GSCs, CBs, and cysts using mutants (see methods) and compared these samples to germaria and early egg chambers (young WT ovaries), late-stage egg chambers (adult WT ovaries), and unfertilized eggs, which represent the maternal contribution [9,11,34–36]. Principal component analysis (PCA) of stage-enriched samples reveals the developmental trajectory of the oocyte transcriptome and places the tst mutant and tst RNAi transcriptomes close to the adult WT ovaries, far from the undifferentiated stages (Figure S2A). This suggests that Tst acts after GSC differentiation. Indeed, Tst-regulated RNAs decrease after oocyte differentiation in the cyst stages and are nearly absent in the egg (p < 1e- 215, Friedman test) (Figure 2C). This is pattern is not observed for non-targets (Figure S2H–S2I). In situ hybridization of the Tst-regulated RNAs such as blanks, act57B, rps10a, and dany demonstrated that these RNAs are present in the undifferentiated cells and early cyst stages but are degraded by the 16-cell cyst stage in WT when the oocyte is fully specified, in contrast to persistence throughout the egg chambers in tst germline RNAi (Figure 2D–2M’, S2J–S2K). To further validate when Tst-regulated RNAs are degraded, we also probed for proteins encoded by Tst-regulated blanks and actin RNAs. In WT, both Blanks and nuclear-Actins (detected by C4 staining) were highly expressed in GSCs and CBs but their expression is attenuated in the cysts, when the oocyte is specified. In contrast, upon loss of tst, protein expression of Blanks and Actin was expanded to cyst stages (Figure S2L–S2Q) [37,38]. Thus, Tst mediates the degradation of a cohort of RNAs concurrent with oocyte specification.
No Go decay pathway effector Pelota is required for degradation of Ski complex regulated RNAs
To investigate how specific transcripts are targeted by Tst, we considered the contribution of RNA surveillance pathways, which are known to direct RNAs to the Ski complex for degradation [39]. Nonsense mediated decay (NMD) and non-stop decay (NSD) are unlikely to be involved. In contrast to tst, ski3, and ski8 germline RNAi flies, germline mutant clones of the NMD pathway components up-frameshift 1 (Upf1), Upf2, and Upf3 do produce eggs, albeit with patterning defects [40]. We additionally looked for features in the RNAs that could trigger NMD or NSD. Most Tst-regulated RNAs do not encode introns in their 3’ untranslated regions (3’ UTR) (Figure S3A), nor show any evidence for aberrant splicing that would give rise to premature termination codons (Figure S3B), ruling out NMD [41–43]. NSD is triggered by ribosome read through into the 3’ UTR, but all Tst-regulated mRNAs are annotated transcripts that encode stop codons, suggesting that NSD is unlikely to be involved [44–46].
However, we did find evidence that no-go decay (NGD), which is activated when ribosomes stall on RNAs, was involved in the degradation of Tst-regulated RNAs. Pelota (Pelo/DOM34) is the critical effector protein of the NGD pathway that promotes recycling of stalled ribosomes on mRNAs [27,47,48]. Intriguingly, pelo mutants, like tst mutants, are homozygous viable but female sterile, and this role is germline specific [49]. pelo mutant egg chambers failed to grow and died mid-oogenesis, phenocopying tst mutant ovaries (Figure 3A–3C, S3C–S3E’). In addition, pelo mutants also lost GSCs, as previously described (Figure S3F–S3G) [49]. To test if pelo and tst co-regulate target RNAs, we performed RNA-seq on pelo mutant ovaries and found that 75% of genes upregulated upon the loss of tst were also upregulated >2-fold in pelo mutants (156/207, Figure 3D, S2A), including act57B. We also find that blanks is upregulated 1.9-fold in pelo mutants compared to control, just below a 2-fold cut-off. However, in situ hybridization for blanks RNA in pelo mutants showed expanded blanks mRNA expression, validating it as a bona fide target (Figure S3H–S3I’). These data suggest that pelo, a key component of the NGD pathway, promotes the degradation of a large fraction of Tst-regulated RNAs.
Figure 3. Twister-regulated RNAs are co-regulated by Pelota and exhibit an increased ribosome association concurrent with a decrease in RNA abundance.
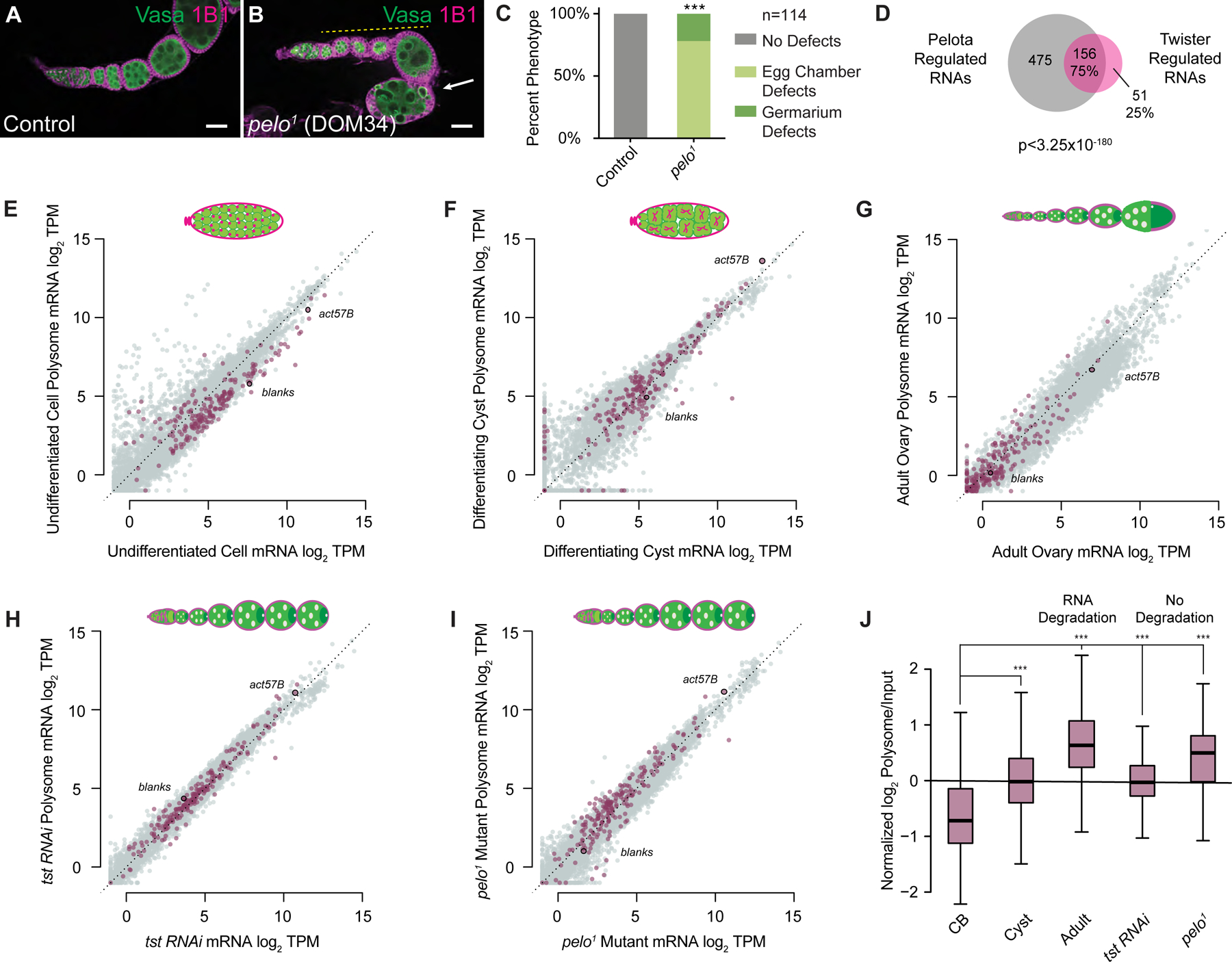
(A) Confocal image of a WT control and (B) pelo1 mutant ovariole stained with 1B1 (magenta) and Vasa (green) and indicating egg chambers that fail to grow (yellow dashed line) and subsequently die (arrow). Scale bars = 10µm. (C) Quantification of oogenesis phenotypes observed in pelo1 mutants (n=114 ovarioles, p < 0.001, Chi-squared tests, df=2). (D) Venn diagram illustrating overlap of Tst-regulated RNAs that are >2 fold upregulated upon loss of pelo (p < 3.25×10−180, Hypergeometric Test). (E–I) Biplots of poly(A)+ mRNA Input versus polysome associated mRNA showing Tst-regulated RNAs (magenta) and highlighting Tst-regulated blanks and act57B. In undifferentiated germ cells (E) and differentiating cysts (F), Tst-regulated RNAs show high abundance and ribosome association compared to adult WT (G). Tst-regulated RNAs also have high abundance and ribosome association in germline tst RNAi (H) and pelo1 (I) ovaries. (J) Box plots of normalized log2 polysome/input mRNA of Tst-regulated RNAs across the samples. Overall ribosome association increases during the transition from CB to cyst to adult. Ribosome association in tst RNAi and pelo1 ovaries, where Tst-regulated RNA degradation is not occurring, is elevated compared to CB, but comparable to cyst. *** = p < 0.001, Estimated marginal means post-hoc tests on repeated-measures ANOVA. See also Figure S3 and Data S1.
Pelota and Ski complex regulated RNAs exhibit signatures of ribosome stalling
To observe the translation dynamics of Tst-regulated RNAs, we conducted polysome profiling on ovaries enriched for different stages of oocyte development (Figure 3E–3G). We plotted the expression levels of RNA levels in the polysome RNA fraction (y-axis) compared to the total RNA transcriptome wide (x-axis). In undifferentiated CBs, Tst-regulated RNAs exhibit depressed polysome association (Figure 3E), but overall these RNAs appear to be translated, consistent with detection of Blanks protein in CBs (Figure S3J–S3K’). In differentiating cysts, polysome association of Tst-regulated RNAs increases compared to undifferentiated cells (Figure 3F). However, we did not observe robust Blanks protein in WT cysts (Figure S3K–S3K’, S3N), suggesting that ribosome engagement of Tst-regulated RNAs in WT cyst stages is not productive. Finally, in adult WT ovaries, Tst-regulated RNAs recapitulate the low expression we observed previously, but these RNAs exhibit elevated ribosome association as they are degraded (Figure 3G, 3J). In adult WT ovaries, consistent with the absence of blanks mRNA, we do not observe Blanks protein expression in the egg chambers (Figure S3K–S3L’, S3N).
Increased ribosome association is usually linked to increased RNA stability, but Tst-regulated RNAs showed an increase in ribosome association concomitant with their degradation (p < 2.2e-16, two-way repeated-measures ANOVA) (Figure 3E–G, J), suggesting the presence of stalled ribosomes [50]. As DOM34 (pelo) promotes recycling of stalled ribosomes, and SKI2 is required to both extract RNAs from stalled ribosomes and promote their degradation, we predicted that Tst-regulated RNAs would be associated with polysomes, but not degraded in tst and pelo mutants [51–53]. Indeed, we found that upon loss of tst or pelo, Tst-regulated RNAs remain highly expressed, associated with ribosomes, and present in egg chambers (Figure 3H–3J). However, in spite of the presence and association of blanks RNA with the polysomes, blanks RNA is not robustly translated in egg chambers lacking Tst or Pelo (Figure S3K–S3N). Taken together, these results suggest that engagement of ribosomes with Tst-regulated RNAs is unproductive prior to their degradation.
Polypyrimidine tracts are required for degradation Tst-regulated RNAs
Pelo-mediated degradation can be activated by features in the coding sequence that cause ribosome stalling, such as sub-optimal codons [54,55]. We found that Tst-regulated RNAs in fact have an elevated codon optimality compared to non-targets, as measured by the Codon Adaptation Index (CAI), suggesting sub-optimal codon frequency is not the trigger for degradation of Tst-regulated RNAs (Figure S4A) [56,57]. Instead, we identified a motif consisting of repeating, interspaced cytidine residues in the coding sequence (CDS), but not in the 5’UTRs or 3’UTRs, of 60% of Tst-regulated RNAs (124/207, Figure 4A, Data S2A–B), suggesting cytidine tracts might recruit Tst. To investigate this hypothesis, we compared three actin paralogs (act42A, act57B and act87E), which were upregulated upon the loss of tst, to a fourth, act5C, which was not upregulated. The actin coding sequences are highly similar (>84% nucleotide identity) and have similar CAIs (Figure S4B). A multiple sequence alignment revealed a repeating cytidine tract in the codon wobble position of act42A, act57B and act87E that is interrupted by purines in the non Tst-target act5C (Figure 4B). Disruption of the C-repeat motif correlated with the degree to which the expression of an Actin paralog is affected by loss of Tst: act57B with a strong C-repeat motif exhibits high up-regulation in tst ovaries compared to act5C, which contains purine substitutions and is unaffected by loss of tst, while act42A with one purine and one pyrimidine substitution was moderately up-regulated in tst ovaries (Figure 4B).
Figure 4. Twister-regulated RNAs are regulated by polypyrimidine rich sequence in their CDS and are regulated by Half Pint.
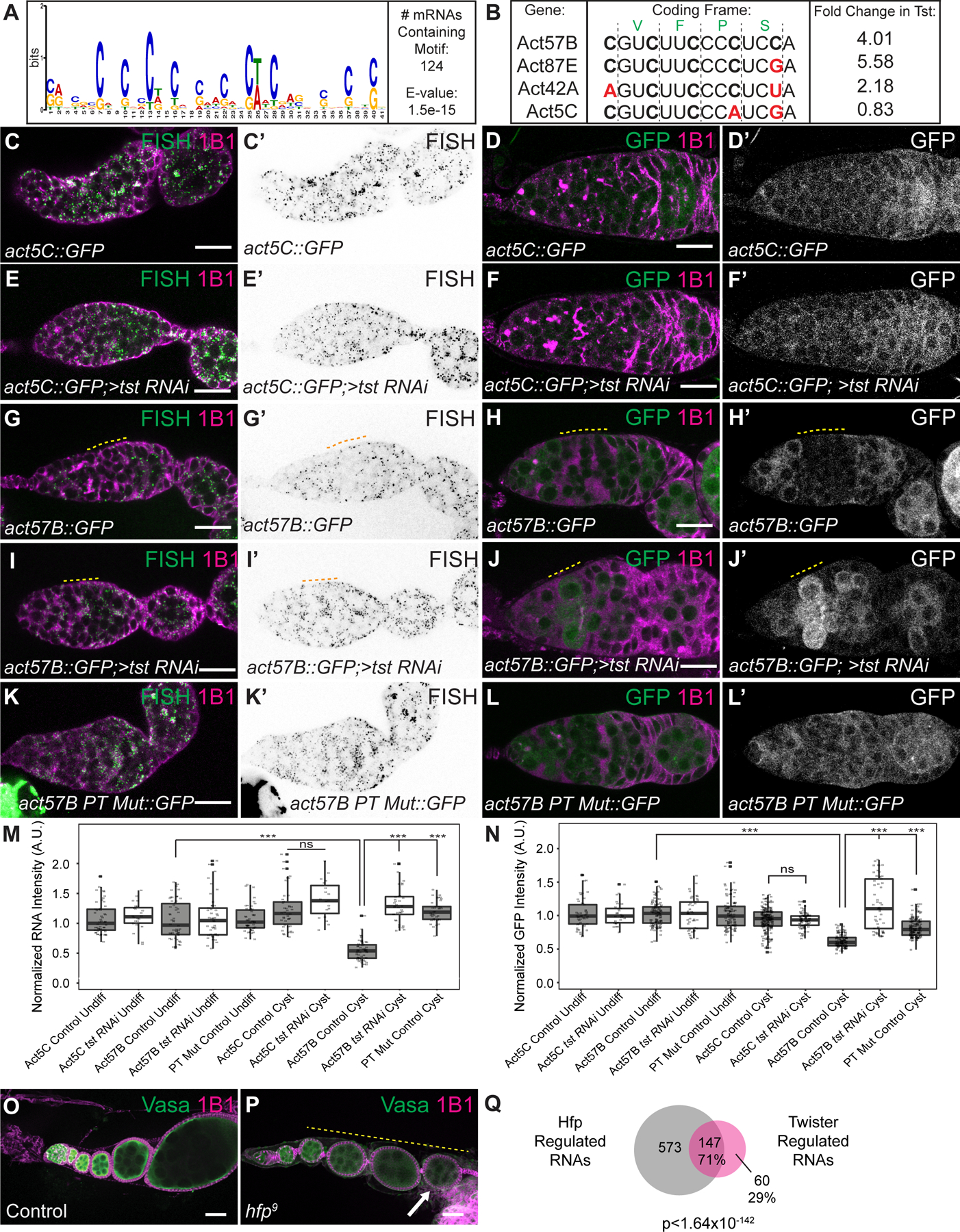
(A) Sequence logo of the polypyrimidine-rich motif identified by MEME to be enriched in the CDS of 124 Tst-regulated RNAs. (B) CDS alignment of Tst-regulated RNAs act57B and act87E, as well as act42A that is sensitive to tst, and non-target paralog act5C, showing divergent polypyrimidine-rich tracts (PTs). Black vertical lines indicate coding frame with amino acid symbols above. Fold-change increase in tst RNAi ovaries over WT is shown to the right. (C–L’) in situ hybridization and GFP fluorescence of reporters (green, grayscale) and 1B1 staining (magenta) in germaria. act5C::GFP in situ (C–C’) and GFP protein (D–D’) show uniform expression and do not change upon loss of tst (E–F’). act57B::GFP in situ (G–G’) and GFP protein (H–H’) expression are decreased in WT cyst stages (yellow dashed line) but expression is expanded into the cyst upon loss of tst (I–J’). act57B::GFP fusion with mutated PT shows ubiquitous expression in WT (K–L’). (M) Quantification of reporter RNA in situ intensity in undifferentiated cells and cyst stages (n=19–54 per cell type). *** = p < 0.001, Tukey’s post-hoc test. (N) Quantification of GFP intensity in undifferentiated cells and cyst stages (n=28–207 per cell type). *** = p < 0.001, Tukey’s post-hoc test. A.U. = arbitrary units. (O) Confocal image of control and (P) hfp9 mutant ovariole stained with 1B1 (magenta) and Vasa (green) showing egg chambers that do not grow in size (yellow dashed line) and dying egg chamber (arrow). (Q) Venn diagram of Tst-regulated RNAs and Hfp regulated RNAs indicating that Tst and Hfp share 71% (147/208) of their target RNAs (p < 1.64×10−142, Hypergeometric Test). Scale bars = 10µm. See also Figure S4, Data S1 and S2.
To evaluate the effect of these sequence differences in vivo, we built reporters with the GFP open reading frame fused to the CDS of non-target act5C or target act57B, as well as to a version of act57B with cytidine tracts mutated to match act5C (PT-mutant). We expressed each of these reporters under the control of the maternal germline promoter pgc, as well as the 3’UTR of K10, and 5’UTR of nos (Figure S4C–S4C’’), which are not translationally repressed [34,58–60]. We found no significant change to the levels of either act5C-GFP RNA or protein during oocyte specification, or upon loss of tst (Figure 4C–4F’, 4M–4N, p>0.05, two-way ANOVA). In contrast, the levels of both act57B-GFP RNA and protein were significantly reduced in WT cysts compared to undifferentiated cells (Figure 4G–4H’, 4M–4N, p<0.001, two-way ANOVA); we note that the reporter was re-expressed in the egg chambers, which may be due to the strong maternal germline promoter or additional layers of control on Tst-regulated RNAs. Strikingly, upon germline depletion of tst, the levels of act57B-GFP RNA and protein were strongly elevated in cysts (Figure 4I–4J’, 4M–4N). Expression of both RNA and protein from the act57B PT Mutant-GFP reporter were significantly higher in cysts compared to that of act57B-GFP (Figure 4K–4N, p<0.001, two-way ANOVA), matching act5C-GFP and demonstrating the importance of the cytidine tract in promoting destabilization of Tst-regulated RNAs during the cyst stages.
To identify the factor that recruits Tst to these cytidine tracts, we looked for polypyrimidine tract binding proteins (PTBs) expressed during oogenesis and found two: hephaestus (heph) and half pint (hfp), the homolog of human PUF60. While loss of heph does not phenocopy pelo and tst, loss of hfp partially phenocopied the oogenesis defects of pelo and tst, marked by egg chambers that fail to grow and mid-oogenesis death (Figure 4O–4P, S4D–S4G’) [61,62]. Consistent with previous reports, we found that Hfp is present in the nucleus, where it has been shown to regulate splicing, but we also observed it in the cytoplasm, suggesting it could affect RNA stability and translation (Figure S4H–S4I) [62]. To determine if Hfp was bound to Tst-regulated RNAs in vivo, we immunoprecipitated Hfp from young WT ovaries followed by qRT-PCR. We found that Tst-regulated RNAs such as act57B and rps10a were robustly associated with Hfp, whereas non-targets pgc and act5C were not (Figure S4J). To determine if Hfp also co-regulates Tst-regulated RNAs, we performed RNA-seq of hfp mutant ovaries. We found that 71% of the RNAs upregulated in tst-depleted ovaries were also upregulated >2-fold in hfp mutants, including act57B, act42A and act87E (148/207, Figure 4Q), whereas act5C was not upregulated in either mutant. Furthermore, 59% of Tst-regulated RNAs are upregulated in both pelo and hfp mutant ovaries, indicating a significant overlap of regulated RNAs between these three factors (Figure S4K, 122/207, p<9.84e-169, Hypergeometric test). We did not observe splicing defects of Tst-regulated RNAs in hfp mutants, ruling out mis-splicing as the reason for their upregulation (Figure S4L). To determine if Hfp acts in concert with Tst and Pelo, we created a fly carrying GFP-tagged Tst as well as HA-tagged Pelo, which we expressed in the germline with nosGAL4. We immunoprecipitated Pelo and probed for Tst and Hfp and found that these proteins interact (Figure S4M). Taken together, our data suggest that Hfp binding to a subset of Tst-regulated RNAs can elicit their degradation mediated by both Pelo and Tst by presumably modulating ribosome association.
Some Ski complex regulated genes are required for germline stem cell maintenance and differentiation, and their ectopic expression perturbs maintenance of oocyte fate
Having elucidated the mechanisms underlying how Tst-regulated RNAs are recognized for degradation, we sought to determine how correct temporal regulation of these RNAs contributes to oogenesis. We hypothesized that Tst-regulated targets are degraded at cyst stages because they have functions prior to or during differentiation and would be detrimental if expressed in the differentiated stages. To determine if there are functional classes of genes in Tst-regulated targets, we carried out gene ontology (GO) analysis and found Tst-regulated targets to be enriched in cytoskeleton components (p=1.03E-05), including act57B, act87E, and mlc1. Actin cytoskeleton is critical during cyst formation and progression [63]. We further assessed the functions of other Tst-regulated RNAs prior to their degradation. Individual depletion of 4 out of 50 Tst-regulated RNAs tested using germline-specific RNAi resulted in severe germline defects, including failure to develop past the germarium stage and complete loss of the germline (Figure S5A–S5E, Table S1). Thus, some Tst-regulated targets have critical function in the undifferentiated stages of oogenesis, though many do not seem to, notably blanks, as blanks mutant females are fertile [38].
To determine if ectopic expression of Tst-regulated mRNAs can lead to oogenesis defects, we overexpressed act57B alone in the germline utilizing nosGAL4 and a previously characterized UAS-act57B [64]. We chose act57B as it has previously been reported that overexpression of act57B results in female sterility, but the exact nature of the oogenesis defect was not known [64]. We stained ovaries overexpressing act57B in the germline and indeed observed egg chamber defects during mid-oogenesis (64%, n=50) (Figure S5F–S5H). Thus, our data suggest that some Tst-regulated targets have a functional role in the undifferentiated cells and need to be removed during oocyte specification to promote oogenesis.
Finally, to elucidate why ectopic persistence of Tst-regulated RNAs interferes with later oogenesis, we examined tst mutants for hallmarks of oogenesis defects. We did not find any changes in GSC differentiation or nurse-cell endocycling (Figure S5I–S5N) [11,17,65–68]. However, Egalitarian (Egl), a protein required to transport the maternal RNA contribution to the oocyte, was mislocalized (Figure 5A–5D’). While in WT, Egl always localized to the oocyte and persisted, in tst, pelo and hfp mutants, Egl localization was not maintained in later egg chambers (Figure 5A–5D’) [13,69]. While initial Egl localization to oocytes appeared to be normal, we cannot rule out subtle specification defects. Taken together, our data suggest that targeted RNA degradation is required for proper establishment of the maternal contribution and maintenance of oocyte fate. Thus, some Tst-regulated RNAs are required in undifferentiated cells for germline maintenance but are detrimental in subsequent stages to the transition to a mature oocyte.
Figure 5. Twister is required for maintaining oocyte fate.
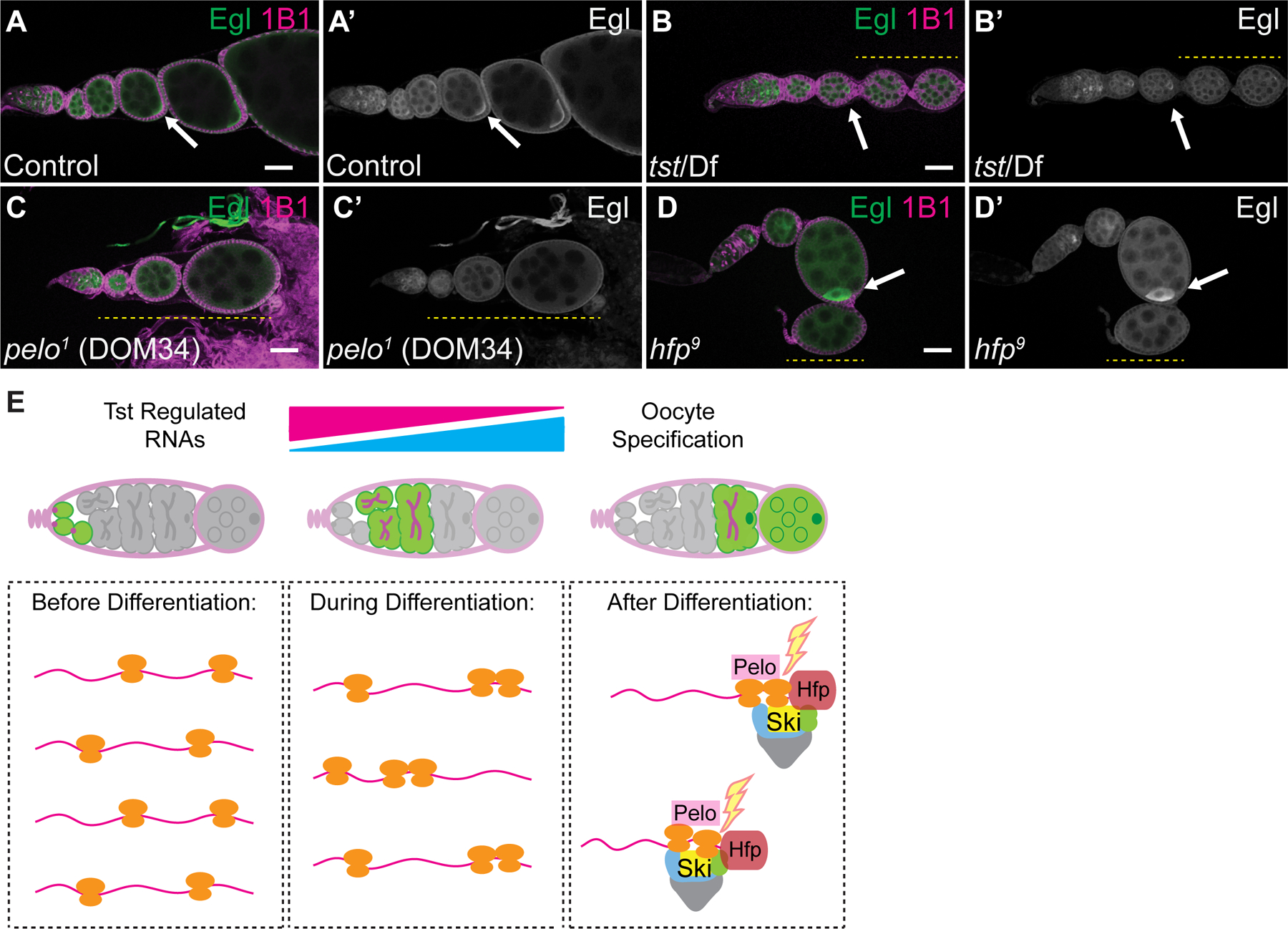
(A–A’) WT control, (B–B’) tst/Df, (C–C’) pelo1 and (D–D’) hfp9 mutant ovarioles stained with 1B1 (magenta) and Egl (green and grayscale) showing initial localization of Egl (arrow) and subsequent loss of Egl accumulation (yellow dashed line). Scale bars = 10µm. (E) In undifferentiated cells (left), Tst-regulated RNAs are highly expressed, yet lowly associated with ribosomes, and required for early oogenesis. During differentiation (center), ribosome association of Tst-regulated RNAs increases. After differentiation (right), during oocyte specification, Hfp protein binds in the CDS of Tst-regulated RNAs leading to targeting by Pelo, and the Ski complex. See also Figure S5 and Table S1.
Discussion
In conclusion, we find that specific RNAs expressed during the undifferentiated stages of oogenesis are degraded during oocyte specification, preventing them from being inherited as part of the maternal contribution, and that this turnover is mediated by the Ski complex and the NGD effector Pelo (Figure 5E). Aberrant persistence of these RNAs results in loss of oocyte maintenance and death of egg chambers. This suggests that precise curation of the maternal contribution is tightly coupled to successful egg production. Based on our observations, we propose that a germ cell-to-maternal transition (GMT) occurs during oocyte specification. We speculate that the GMT exists to enable both the transition from germ cell to oocyte identity and the establishment of the maternal RNA contribution to the embryo. mRNAs produced during oogenesis are subsequently cleared during the oocyte-to-embryo transition (OET) and the maternal-to-zygotic transition (MZT) to promote a zygotic identity [70–72]. Thus, RNA degradation bookends an oocyte’s fate, regulating both its inception and termination.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prashanth Rangan (Prangan@albany.edu).
Materials availability
Plasmids and transgenic flies generated during this study are maintained in our lab and available upon request.
Data and code availability
All RNA-Sequencing data generated during this study is available in the GEO repository under the Accession number: GSE166275. This study did not generate any code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Female Drosophila melanogaster flies were maintained in group housing at 25°C and utilized for these studies. Flies were grown at 25°C, fattened overnight on yeast, and dissected between 1–3 days post-eclosion. To achieve stage 9–10 ovarioles, adult WT flies were fattened on yeast overnight for 12–14 hrs.
Genotypes used to enrich specific stages of germline:
Germline Stem Cells: nosGAL4>UAS-tkv [9,34,73]. Cystoblasts: nosGAL4>bam RNAi [11,65,66]. Differentiating Cysts: nosGAL4>bam RNAi; hs-bam [35,74]. Female flies were heat shocked at 37° C for 2 hours, incubated at room temperature for 4 hours and heat shocked again for 2 hours. This was subsequently repeated the next day and flies were dissected. Young Wild Type: Female flies were collected and dissected within 2 hours of eclosion.
METHODS DETAILS
Dissection and Immunostaining
Flies were dissected in 1X PBS and samples were fixed for 10 minutes in 5% methanol-free formaldehyde [34]. Ovary samples were washed in 1 mL PBT (1X PBS, 0.5% Triton X-100, 0.3% BSA) 4 times for 7 minutes each. Primary antibodies were added in PBT and incubated at 4°C rotating overnight. Samples were washed 4 times for 7 minutes each in 1 mL PBT, and once in 1 mL PBT with 2% donkey serum for 15 minutes. Secondary antibodies were added in PBT with 4% donkey serum and incubated at room temperature for 2 hours. Samples were washed 4 times for 7 minutes each in 1 mL of 1X PBST (0.2% Tween 20 in 1x PBS) and incubated in Vectashield with DAPI for 30 minutes before mounting. The following primary antibodies were used: Mouse anti-1B1 (1:20), Rabbit anti-Vasa (1:1000), Chicken anti-Vasa (1:1000), Rabbit anti-GFP (1:2000), Rabbit anti-Blanks (1:1000), Mouse anti-Actin C4 (1:50), Rabbit anti-Cleaved Caspase3 (1:300), Rabbit anti-Egl (1:1000), Mouse anti-Hfp (1:25) [37,75–77]. The following secondary antibodies were used: Alexa 488, Cy3, and Cy5 were used at a dilution of 1:500.
Fluorescence Imaging
The ovary tissue samples were visualized under 10X dry, 20X dry and 40X oil objective lenses and images were acquired using a Zeiss LSM-710 confocal microscope. Confocal images were processed with ImageJ. A.U. The images were quantified using ImageJ with the Measurement function.
Generation of Transgenic Flies
The pCasper2 plasmid containing the pgc promoter, nos 5’UTR, eGFP CDS and K10 3’UTR was used as a backbone to generate Actin-GFP reporter constructs (Table S2) [34]. gBlocks (IDT) of the actin5C, actin57B and actin57B PT-Mutant CDSs were individually cloned upstream of GFP by digesting with SpeI. Constructs were ligated through Gibson Assembly, utilizing complementary overhangs between the CDS fragment and the pCasper2 backbone. Plasmid was then transformed into DH5α competent cells and plated on LB-Kan plates at 37°C overnight. Cells of individual colonies were propagated, and plasmid was purified. Injection of these plasmids into Drosophila embryos was conducted by BestGene Inc.
Gateway Cloning
The complete coding sequence of Tst was PCR amplified from cDNA to include flanking attB sites. BP recombination was carried out according to the manufacturer’s protocol using equimolar amounts (100 fmol) of the attB-PCR product and the pDONR entry clone plasmid (Table S2). Components were incubated in TE buffer with BP Clonase enzyme mix and reaction buffer at 25°C for one hour. 2 µg/µL Proteinase K was added to the reaction and incubated at 37°C for one hour. Plasmid was then transformed into DH5α competent cells and plated on LB-Kan plates at 37°C overnight. Cells of individual colonies were propagated, and plasmid was purified. LR recombination reaction was performed with the pPPW and pPGW destination vectors. Components were incubated in TE buffer with LR Clonase enzyme mix and reaction buffer at 25°C for one hour. 2 µg/µL Proteinase K was added to the reaction and incubated at 37°C for one hour. Plasmid was then transformed into XL10-Gold competent cells and plated on LB-Kan plates at 37°C overnight. Cells of individual colony samples were propagated, plasmid was purified and sequenced to verify insertion.
RNA Isolation and cDNA Synthesis
Ovaries were dissected in 1X PBS and homogenized in 50uL of TRIzol [34]. RNA was isolated by adding an additional 950 uL of TRIzol and 230uL of Chloroform with mixing. Samples were centrifuged at 13,000 rpm, 4°C for 15 minutes. Aqueous phase was transferred to a new tube, nucleic acids were precipitated using 1 mL of 100% ethanol, 50 µL of 3M Sodium Acetate and precipitated for >1 hour at −20°C. Samples were centrifuged at 13,000 rpm, 4°C for 20 minutes. Ethanol was decanted, pellet was washed with 70% ethanol and dried at room temperature for 10 minutes. Pellet was dissolved in 20 µL RNase free water and placed in a 42°C water bath for 10 minutes. Concentration of nucleic acid samples were measured on a spectrophotometer and treated with DNase according to the manufacturer’s protocol. cDNA was synthesized using SuperScript 2 and according to the manufacturer’s protocol. To reverse transcribe mRNAs, Oligo-dT was used from the SuperScript 2 kit. To reverse transcribe nascent pre-mRNAs, or mRNAs independent of poly(A) tail length, 150 ng of random hexamers per sample was used from the SuperScript 2 kit.
Quantitative Real Time-PCR (qRT-PCR)
1 µL of cDNA was amplified using 5µL of SYBR green Master Mix, 0.3 µL of 10µM of each reverse and forward primers in a 10 µL reaction [34]. The thermal cycling conditions consisted of 50°C for 2 minutes, 95°C for 10 minutes, 40 cycles at 95°C for 15 seconds, and 60°C for 60 seconds. The experiments were carried out in technical triplicate and minimum 2 biological replicates for each sample. rp49 gene was utilized as a control. To calculate fold change in mRNA levels to rp49 mRNA levels, average of the 2^∆Ct for the biological replicates was calculated. Error bars were plotted using standard error of the ratios. P-value was determined by a two sample Student’s t-test.
RNA-seq library preparation
Total RNA samples were run on a 1% agarose gel to assess sample integrity [78]. To generate mRNA-Seq libraries, total RNA was incubated with poly(A) selection beads. mRNA enriched sequencing libraries were made with the NEXTflex Rapid Directional RNAseq Kit and corresponding protocol. mRNA was fragmented at 95°C for 13 minutes to achieve ~300 bp fragments. The WT_1, Young_1, Tst_1 and Tst_RNAi_1 library samples are unstranded and were sequenced by Beijing Genomics Institute on a HiSeq4000. All other Library samples were sequenced with 75 bp single-end or paired-end mRNA by an Illumina NextSeq500, carried out by the Center for Functional Genomics (CFG).
EdU
Ovaries were dissected into Schneider’s Media and incubated in 10µM EdU solution rotating for one hour [79]. Samples were fixed in 3.7% formaldehyde in PBS, rotating for 30 minutes. Fixative was then aspirated and samples were washed with 1 mL PBS for 10 minutes and permeabilized in 1 mL of permeabilization solution (1% Triton X-100 in PBST) rotating for 20 minutes. Samples were then washed in 1 mL PBS rotating for 10 minutes. Click-iT reaction cocktail (PBS, CuSO4, Fluorescent dye azide and Reaction Buffer Additive) was made according to manufacturer’s directions and added to each sample. Tubes were protected from light and rotated at room temperature for 30 minutes. Samples were then washed once with 1 mL of Click-iT reaction rinse buffer and once with 1 mL PBS. Ovary samples were then transitioned to the immunostaining protocol.
Fluorescent in situ Hybridization
Ovaries were dissected in RNase free 1X PBS and fixed in 1 mL of 5% formaldehyde rotating for 10 minutes. Samples were then washed three times for five minutes each in PT buffer (PBS, 0.1% Triton X-100) and dehydrated in successive methanol washes for six minutes each (30%, 50%, 70%). A final 100% methanol wash was carried out for 12 minutes. Samples were equilibrated to PT buffer by conducting successive methanol washes for six minutes each (70%, 50%, 30%), followed by three PT washes of six minutes each. Ovaries were pre-hybridized for six minutes in 1 mL Wash buffer (10% Deionized Formamide, 2X SSC in RNase Free H2O). CALFluor590 tagged probes against GFP were used. Hybridization of probes was conducted at 32°C, covered for >16 hours. Samples were then washed six times in Wash buffer for 2 minutes per wash. Samples were then washed twice in 1 mL Wash buffer for 30 minutes at 30°C. Wash buffer was aspirated and incubated in Vectashield for 30 minutes before mounting. in situ experiments were repeated more than three times for control and experimental ovaries.
RNAScope™ Assay
We utilized a modified RNAscope procedure for Drosophila ovaries described previously [80]. Probes were designed and generated by Advanced Cell Diagnostics with specificity to target base pairs 29–1250 of blanks mRNA (NCBI accession number: NM_139709.2), base pairs 1196–1693 of actin57B mRNA (NM_079076.4), base pairs 213–1141 of dany mRNA (NM_166493.2), base pairs 2–599 of rps10a mRNA (NM_143319.4). Ovaries were dissected in RNase free 1X PBS and fixed in 1 mL of 5% formaldehyde rotating for 10 minutes. Samples were then washed three times for five minutes each in PT buffer (PBS, 0.1% Triton X-100) and dehydrated in successive methanol washes for six minutes each (30%, 50%, 70%). A final 100% methanol wash was carried out for 12 minutes. Samples were equilibrated to PT buffer by conducting successive methanol washes for six minutes each (70%, 50%, 30%), followed by three PT washes of six minutes each. Ovaries were pre-hybridized for six minutes in 1 mL of RNAScope Wash buffer. Hybridization of probes was conducted at 40°C, covered for >16 hours. Samples were then washed three times in RNAscope wash buffer for 5 minutes per wash, fixed in 4% formaldehyde in 1X PBS at room temperature for 10 minutes and washed in buffer three times for 5 minutes each. Ovaries were incubated in a successive series of amplifier solutions (Amp). Amp 1 for at least 45 minutes at 40°C, Amp 2 for 45 minutes at 40°C, Amp 3 for 45 minutes at 40°C, Amp 4 for 45 minutes at 40°C. After each Amp step ovaries were washed in wash buffer 5 times for 3 minutes each at room temperature. Samples were then washed in 1 mL PBT for 5 minutes and mounted in Vectashield. RNAscope experiments were repeated more than three times for control and, pelo, and tst RNAi ovaries.
Materials and reagents
Fly food was made according to previously published procedures, and filled narrow vials to approximately 12mL [34].
Immuno-Precipitation (IP) and RNA Immuno-Precipitation (RIP)
65 pairs of ovaries were dissected in 1X PBS [34]. After dissection, PBS was aspirated and 100 µl of RIPA buffer (50 mM Tris pH 8.0, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl, 1mM EDTA, 1 mM PMSF, 1 protease inhibitor pill per 50 ml) was added and the sample was homogenized. An additional 200 µl of RIPA buffer was added to the lysate and mixed. The lysate was then centrifuged at 13,000 rpm for 10 minutes at 4°C. The supernatant was transferred to a new tube. 10% of the cleared homogenate was set aside as input, 4X SDS buffer was added the sample was heated at 95°C for 5 minutes and stored at −20°C until Western analysis. An additional 10% of homogenate was used for RNA input, 100 µl of TriZol was added, mixed and this sample was stored in −80°C. 40% of homogenate was used for the IgG control and the remaining 40% was used for RIP. The following antibody was added to the lysate and incubated at 4°C for 3 hours; 1 µl of Rabbit anti-HA. Protein A Dynabeads were separated into 15 µl aliquots for each sample and washed four times in 400 µl of 1:10 diluted protease inhibitor-containing Net2 buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 10% NP-40) on a magnetic rack. The beads were then re-suspended in 100 µl of Net2 buffer. After lysate incubation 25 µl of washed beads was added to each sample and incubated overnight at 4°C. Beads were washed six times with 500 µl of 1:10 diluted Net2 buffer for 2 minutes each. Beads were then resuspended in 25 µl of Net2 buffer. An aliquot of 10 µl was used for Western Blot analysis. The remaining 15 µl was used for RNA extraction.
Subcellular Fractionation
50 adult Wild Type ovaries were dissected in 1X PBS and gently homogenized with 10–20 strokes of a plastic homogenizer in 100 µL hypotonic lysis buffer (10mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT). Homogenate was incubated on ice for 15 minutes. 50 µL of homogenate was aliquoted in a new tube, 4X SDS buffer was added, sample was boiled at 95°C for 5 minutes and stored in −20°C until use as total homogenate. The remaining homogenate was centrifuged for 10 minutes at 1000g. 50 µL of supernatant was collected 4X SDS buffer was added, sample was boiled at 95°C for 5 minutes and stored in −20°C until use as cytoplasmic fraction. The pellet was resuspended in high salt extraction buffer (20mM HEPES pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT) and centrifuged for 5 minutes at 20,000g. Supernatant was collected 4X SDS buffer was added, sample was boiled at 95°C for 5 minutes and stored in −20°C until use as total nuclear fraction.
Western Blot
Twenty wild-type size ovaries or 40 mutant size ovaries were dissected in 1X PBS [34]. After dissection, PBS was aspirated and 30 µl of NP-40 buffer with protease inhibitors added to the tissue and homogenized. The lysate was centrifuged at 13,000 rpm for 15 minutes at 4°C. Aqueous layer was transferred into a new tube while avoiding the top lipid layer. 1 µl of the protein extract was used to carry out a Bradford assay. 25 µg of protein was denatured with 4X Laemmli Sample Buffer and β-marcepthanol at 95°C for 5 minutes. The samples were loaded in a Mini-PROTEAN TGX 4–20% gradient SDS-PAGE gels and run at 110V for 1 hour. The proteins were then transferred to a 0.20 µm nitrocellulose membrane at 100V for 1 hour at 4°C. After transfer, the membrane was blocked in 5% milk in PBST for 2 hours at RT. The following antibodies were used: Mouse anti-Hfp (1:1000), Rabbit anti-Orb (1:1000), Mouse anti-Fibrillarin (1:25), Rabbit anti-HA (1:1000), Rabbit anti-GFP (1:1000). Primary antibody was prepared in 5% milk in PBST was added to the membrane and incubated at 4°C overnight. The membrane was then washed three times in 0.5% milk PBST. Anti-Rabbit HRP (1:10,000) or Anti-Mouse HRP (1:10,000) was prepared in 5% milk in PBST, and was added to the membrane and incubated at room temperature for 2 hours. The membrane was then washed 3 times in PBST. Bio-rad chemiluminescence ECL kit was used to image the membrane.
Egg Laying Test and Viability Assay
Newly eclosed flies were collected and fattened overnight on yeast. Assays were conducted in cages on apple juice plates containing 6 control or experimental females crossed to 4 adult Wild Type control males. Cages were maintained at 25°C and plates changed daily for egg counting. Analyses were performed on three consecutive days. Total number of eggs laid was counted and averaged. For viability assays, hatched larvae were counted 48 hours later and were assessed as a proportion of eggs hatched/eggs laid. Both control and experimental experiments were conducted in triplicate.
Polysome profiling and Polysome-Seq
30 Wild Type or 150 mutant ovary pairs were dissected in 1X PBS and immediately flash frozen on liquid nitrogen [34,81]. Samples were homogenized in lysis buffer and 20% of lysate was used as input for mRNA isolation and library preparation (as described above). Samples were loaded onto 10–45% CHX supplemented sucrose gradients in 9/16 × 3.5 PA tubes (Beckman Coulter, #331372). For samples used for protein isolation, 650 µg/mL of heparin was added to sucrose gradients. Gradients were spun at 35,000 x g in SW41 for 3 hours at 4°C. Gradients were fractionated with a Density Gradient Fractionation System (#621140007). RNA was extracted using acid phenol-chloroform and precipitated overnight. Pelleted RNA was resuspended in 20 µL water, treated with TURBO DNase and libraries were prepared as described above. For samples used for protein isolation, 4 volumes of methanol was added to each fraction and centrifuged at 13,000g for 5 minutes. Methanol was aspirated and pellet was dried before being resuspended in 1X Laemli SDS buffer.
MEME Analyses
The 5’UTR, CDS, 3’UTR and full transcript sequences of all 207 Tst-regulated target genes were individually analyzed by the MEME algorithm [82]. Discriminative mode analysis was conducted against 621 non-target gene sequences as background with default parameters. FIMO was conducted by entering a repeating “CNN” motif and conducting a search for this motif in the blanks CDS [83]. Motif logos, number of sites, and p-values all reported as produced by output of the program.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-seq analysis
Sequenced reads were aligned to the D. melanogaster genome (UCSC dm6 and FlyBase R6.01) using HISAT2 v2.0.5 [84]. Unambiguously mapping reads to RefSeq annotated mRNA and lincRNA were quantified using featureCounts v1.5.1 default parameters [85]. Genes with>=0.5 reads per million (RPM) in one of WT ovaries, tst mutant ovaries, tst RNAi ovaries, or young ovaries were retained for further analysis (N=9251 genes). Tst regulated mRNAs were classified as genes whose transcript-per-million (TPM) expression levels were >4-fold increased in the tst RNAi samples; a subset of these that are additionally >2-fold increased in the tst mutant samples were considered to be strong Tst regulated mRNAs (N=207 genes). DESeq2 was subsequently used to confirm significance of these targets (Data S1C–D)[86]. To curate a set of non-target genes to serve as a background set, we identified genes that differed <1.25 fold between WT and tst RNAi and selected the 3 non-targets with the most similar tst RNAi expression level to each Tst regulated mRNA, to yield a set of 621 non-targets. For polysome profiling samples, normalized ribosome occupancy was calculated as log2(polysome TPM / input TPM). A 0.5 pseudocount was added to each TPM value to prevent division by zero and taking the log of zero. To compare ribosome occupancy of Tst regulated mRNAs across different conditions relative to the global differences observed, we normalized Tst regulated mRNA ribosome occupancy by mean non-target occupancy, i.e. log2( (polysome_target TPM / input_target TPM) / average (polysome_non-target TPM / input_non-target TPM) ). Codon optimality index (CAI) was calculated for each gene relative to the codon frequencies in the top 100 expressed genes in WT ovaries according to the method of Sharp & Li, 1987 [56]. To measure Tst-target expression change over the developmental timecourse, we performed a Friedman test (non-parametric one-way ANOVA with repeated measures). Individual post-hoc tests between stages were performed using Wilcoxon signed rank tests with Bonferonni correction for (8 choose 2) = 28 comparisons. To measure changes in ribosome occupancy in the polysome profiling data, we performed a two-way repeated measures ANOVA modeling, per gene, the difference in ribosome occupancy (log2 polysome TPM / input TPM) versus classification as Twister target or non-target (occupancy ~ condition * target_type) using the lme function in the R package nlme (v3.1–149, Pinheiro & Bates). P value is from a likelihood test against a null model (occupancy ~ 1). Post-hoc tests between conditions given target type were performed using the R package emmeans (v1.5.2–1, Lenth et al). All RNA-Seq data utilized in this work are summarized in Data S1A–D, and raw reads are available under the following GEO accessions: GSE119458, GSE143728, and GSE166275.
Statistical Analyses
All statistical analyses were conducted in Excel or R as noted. The specific tests, sample, size, p-value and asterisks are displayed in the corresponding legends.
Supplementary Material
(A) Log2 Transcript Per Million (TPM) values and status of regulation by Tst, Pelo or Hfp for all Drosophila genes. RNA-Seq data sets include: adult WT (WT_1, 2), tst/Df (Tst_1, 2), tst RNAi (Tst_RNAi_1, 2), young WT (Young_1, 2), GSC enriched ovaries (GSC), undifferentiated CB enriched ovaries (CB), unfertilized Egg (Egg_pA), pelo1 heterozygous control (Pelo_Het_1, 2), pelo1 homozygous mutant (Pelo_1, 2), hfp9 heterozygous control (Hfp_Het_1, 2), hfp9 homozygous mutant (Hfp_1, 2), undifferentiated CB enriched ovary input (CB_Input), undifferentiated CB enriched ovary polysome RNA (CB_Psome), differentiating cyst enriched ovary input (Bam_hs_Input_1, 2), differentiating cyst enriched ovary polysome RNA (Bam_hs_Psome_1, 2), adult control input (Ctrl_Input_1, 2), adult control polysome RNA (Ctrl_Psome_1, 2), pelo1 homozygous mutant input (Pelo_Input_1, 2), pelo1 homozygous mutant polysome RNA (Pelo_Psome_1, 2), tst RNAi input (Tst_Input_1, 2), tst RNAi polysome RNA (Tst_Psome_1, 2). See methods for how specific stages were enriched. (B) RNA-Seq sample information, GEO identifiers, library properties, total reads and alignment statistics. (C) DESeq2 normalized log2 fold change (lfc), p value (pval), and adjusted p value (padj) for tst LOF vs WT, pelo vs WT, and hfp vs WT. (D) Sample comparisons used for DESeq2.
(A) MEME discriminative mode motif enrichment output of Tst-regulated gene names, starting position of the polypyrimidine rich motif, p-value and site within each CDS. (B) Find Individual Motif Occurrences (FIMO) output of the polypyrimidine rich motif (CNN) search in the blanks CDS.
List of Tst-regulated genes screened in the germline, BDSC stock number, and a description of the resulting phenotype.
Table S2. List of primers used in this study. Related to STAR methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | abCam | ab6556 |
| Mouse anti-1B1 | Developmental Studies Hybridoma Bank | Antibody Registry ID: 528070 |
| Rat monoclonal anti-HA high affinity | Roche Diagnostics | REF: 11867423001 |
| Rabbit polyclonal anti-Vasa | Rangan Lab | N/A |
| Chicken polyclonal anti-Vasa | Rangan Lab | N/A |
| Rabbit polyclonal anti-Blanks | Gift from Sontheimer Lab | N/A |
| Mouse polyclonal anti-Actin C4 | Sigma | MAB1501 |
| Rabbit polyclonal anti-Cleaved Caspase 3 | Cell Signalling | #96615 |
| Rabbit polyclonal anti-Egalitarian | Gift from Lehmann Lab | N/A |
| Mouse polyclonal anti-Half pint | Gift from Schüpbach Lab | N/A |
| Anti-rabbit Alexa 488 | Jackson ImmunoResearch Labs | Code:711–546-152 |
| Anti-mouse Cy3 | Jackson ImmunoResearch Labs | Code:715–546-150 |
| Anti-chicken Alexa 647 | Jackson ImmunoResearch Labs | Code:703–606-155 |
| Anti-rabbit HRP | Jackson ImmunoResearch Labs | Code:111–035-144 |
| Anti-mouse HRP | Jackson ImmunoResearch Labs | Code:115–035-003 |
| Rabbit Anti-Orb | Gift from Lehmann Lab | N/A |
| Rabbit Anti-Fibrillarin | abCam | ab166630 |
| Bacterial and Virus Strains | ||
| DH5α competent E. coli | New England Biolabs Inc. | #C25271 |
| XL-10 Gold Ultracompetent cells | Integrated Sciences | #200315 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde (Methanol Free), 10% Ultrapure | Polysciences Inc. | #04018–1 |
| Donkey Serum | Sigma-Aldrich | SKU: D9663 |
| Vectashield Antifade Mounting Medium with DAPI | Vector Laboratories | #H-1200 |
| Triton X-100 detergent | VWR | #97062–208 |
| Nonidet P-40 (NP-40) substitute | IBI Scientific | #9016–45-9 |
| Tween-20 detergent | VWR | #97062–332 |
| TRIzol | Invitrogen | #15596026 |
| Complete, EDTA-free Protease Inhibitor Cocktail Pill | Sigma-Aldrich | SKU: 11873580001 |
| Gibson Assembly Master Mix | New England Biolabs Inc. | E2611L |
| Restriction Endonuclease SpeI | New England Biolabs Inc. | R0133S |
| Gateway Clonase II | Invitrogen | #12535–029 |
| Schneider’s Drosophila Medium | Gibco | #21720024 |
| Dynabeads Protein A | Invitrogen | #10002D |
| Bradford reagent | Bio-Rad | #500–0205 |
| 4X Laemmli Sample Buffer | Bio-Rad | #161–0747 |
| SuperScript II | Invitrogen | 18064022 |
| Critical Commercial Assays | ||
| TURBO DNA-free Kit | Life Technologies | AM1907 |
| SYBR Green Master Mix | Applied Biosystems | #4367659 |
| NEXTflex Rapid Illumina DNA-Seq Library Prep Kit | BioO Scientific | NOVA-5138–11 |
| Click-iT EdU Cell Proliferation Kit | Invitrogen | C10337 |
| RNAScope Kit | Advanced Cell Diagnostics | #310091 |
| Mini-PROTEAN TGX 4–20% gradient SDS-PAGE gels | Bio-Rad | #456–1094 |
| Western ECL Substrate | Bio-Rad | #1705060 |
| Deposited Data | ||
| RNA-Seq Data | This paper | GEO: GSE166275 |
| RNA-Seq Data | [74] | GEO: GSE77294 |
| RNA-Seq Data | [34] | GEO: GSE119458 |
| RNA-Seq Data | [36] | GEO: GSE143728 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: RNAi for tst: y1 sc* v1 sev21; P{TRiP.HMC03796}attP40 | Bloomington Drosophila Stock Center | BDSC: 55647 |
| D. melanogaster: RNAi for CG8777 (Ski3): P{KK106325}VIE-260B | Vienna Drosophila Resource Center | VDRC: v100948 |
| D. melanogaster: bam Mutant: ry506 e1 bamΔ86/TM3, ryRK Sb1 Ser1 | Bloomington Drosophila Stock Center | BDSC: 5427 |
| D. melanogaster: UAS-tkv: w*; P{UAS-tkv.Q253D.Nb}3/TM3, Sb1 Ser1 | Bloomington Drosophila Stock Center | BDSC: 36536 |
| D. melanogaster: RNAi for CG3909 (Ski8): y1 sc* v1 sev21; P{TRiP.HMC04664}attP40 | Bloomington Drosophila Stock Center | BDSC: 57377 |
| D. melanogaster: RNAi for bam: y1 v1; P{TRiP.HMJ22155}attP40 | Bloomington Drosophila Stock Center | BDSC: 58178 |
| D. melanogaster: Heat shock bam: w1118; P{hs-bam.O}11d/TM3, Sb1 | Bloomington Drosophila Stock Center | BDSC: 24637 |
| D. melanogaster: nosGAL4 Driver: P{UAS-Dcr-2.D}1, w1118; P{GAL4-nos.NGT}40 | Bloomington Drosophila Stock Center | BDSC: 25751 |
| D. melanogaster: nosGAL4 Driver: UAS-Dcr2;nosGAL4;bamGFP | Lehmann Lab | N/A |
| D. melanogaster: nosGAL4 Driver: If/CyO;nosGAL4 | Lehmann Lab | N/A |
| D. melanogaster: nosGAL4 Driver: y1 w*; P{GAL4-nos.NGT}40 | Bloomington Drosophila Stock Center | BDSC: 4442 |
| D. melanogaster: tjGAL4 Driver: tjGAL4/CyO | Lehmann Lab | N/A |
| D. melanogaster: Wild type Control: y1 w1118 P{70FLP}3F | Bloomington Drosophila Stock Center | BDSC: 6420 |
| D. melanogaster: tst Mutant: w1118; Mi{ET1}tstMB10212/TM6C, Sb1 | Bloomington Drosophila Stock Center | BDSC: 29100 |
| D. melanogaster: hfp Mutant: hfp9,cu/TM2 | Schüpbach Lab | N/A |
| D. melanogaster: Hfp-HA: M{UAS-hfp.ORF.3xHA}ZH-86Fb | FlyORF | F000989 |
| D. melanogaster: Pelo-HA: M{UAS-pelo.ORF.3xHA.GW}ZH-86Fb | FlyORF | F003036 |
| D. melanogaster: CG8777 (Ski3) Mutant: y1 w*; Mi{MIC}CG8777MI02824 | Bloomington Drosophila Stock Center | BDSC: 35904 |
| D. melanogaster: CG8777 (Ski3) Deficiency: w[1118];Df(2R)ED1770,P{w[+mW.Scer\FRT.hs3]=3’.RS5+3.3’} ED1770/SM6a | Bloomington Drosophila Stock Center | BDSC: 9157 |
|
D. melanogaster: tst Deficiency: w[1118]; Df(3R)Exel9013/TM6B, Tb[1] |
Bloomington Drosophila Stock Center | BDSC: 7991 |
| D. melanogaster: pelo Mutant: pelo1/CyO | Bloomington Drosophila Stock Center | BDSC: 11757 |
| Oligonucleotides | ||
| Primers for qRT-PCR see Table S4 | This paper | N/A |
| Primers for Gateway Cloning see Table S4 | This paper | N/A |
| RNAScope Probe Against blanks | Advanced Cell Diagnostics | Base pairs 29–1250 NM_139709.2 |
| RNAScope Probe Against act57B | Advanced Cell Diagnostics | Base pairs 1196–1693 NM_079076.4 |
| RNAScope Probe Against dany | Advanced Cell Diagnostics | Base pairs 213–1141 NM_166493.2 |
| RNAScope Probe Against rps10a | Advanced Cell Diagnostics | Base pairs 2–599 NM_143319.4 |
| GFP RNA FISH probe labeled with CALFluor590 | [87] | N/A |
| Recombinant DNA | ||
| Plasmid: pCaSpeR2 P element transformation vector | Drosophila Genomics Resource Center | Stock Number: 1066 |
| Gateway Destination Vector Plasmid: pPWG | Drosophila Genomics Resource Center | Gateway 1 Collection |
| Gateway Destination Vector Plasmid: pPGW | Drosophila Genomics Resource Center | Gateway 1 Collection |
| Act5C Coding Sequence gBlock | Integrated DNA Technologies | NM_001297986.1 |
| Act57B Coding Sequence gBlock | Integrated DNA Technologies | NM_079076.4 |
| Act57B Mutant Coding Sequence gBlock | Integrated DNA Technologies | N/A |
| Gateway pDONR Vector | Invitrogen | #12536–017 |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| HISAT2 | [88] | https://ccb.jhu.edu/software/hisat2/index.shtml |
| DESeq2 | [86] | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| featureCounts | [85] | http://bioinf.wehi.edu.au/featureCounts/ |
| FIMO | [83] | https://meme-suite.org/meme/doc/fimo.html |
| MEME Suite | [89] | https://meme-suite.org/doc/overview.html. |
Highlights:
Ski complex mediated RNA degradation is required in the germline for fertility.
A subset of early oogenic RNAs are degraded concurrent with oocyte specification.
Early oogenic RNAs are degraded utilizing components of the No Go Decay pathway.
Degradation of early oogenic RNAs is required for maintenance of oocyte fate.
Inclusion and diversity.
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
Acknowledgements
We would like to thank all members of the Rangan Lab as well as Drs. Siekhaus D, Sano H, Juliano C, Belfort M, and Farrell J for discussion and comments on the manuscript. We would also like to thank the Schüpbach Lab for the Hfp antibody and mutant flies, the Sontheimer Lab for the Blanks antibody, and the Newbury Lab for flies and reagents. P.R. is funded by the NIH/NIGMS (R01GM111779–06 and R01GM135628–01). P.B. is funded by NIH (grant 1F31GM126784–01) and by the RNA Institute. M.T.L. was supported by the NIH/NIGMS (R35GM135099) and start-up funds from the Univ. of Pittsburgh.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- [1].Cinalli RM, Rangan P, Lehmann R. Germ cells are forever. Cell 2008;132:559–62. doi: 10.1016/j.cell.2008.02.003. [DOI] [PubMed] [Google Scholar]
- [2].Seydoux G, Braun RE. Pathway to Totipotency: Lessons from Germ Cells. Cell 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- [3].Reik W, Surani MA. Germline and Pluripotent Stem Cells. Cold Spring Harbor Perspectives in Biology 2015;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Walser CB, Lipshitz HD. Transcript clearance during the maternal-to-zygotic transition. Current Opinion in Genetics & Development 2011;21:431–43. doi: 10.1016/j.gde.2011.03.003. [DOI] [PubMed] [Google Scholar]
- [5].Laver JD, Marsolais AJ, Smibert CA, Lipshitz HD. Regulation and Function of Maternal Gene Products During the Maternal-to-Zygotic Transition in Drosophila. Current Topics in Developmental Biology 2015;113:43–84. doi: 10.1016/bs.ctdb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- [6].Lee MT, Bonneau AR, Giraldez AJ. Zygotic Genome Activation During the Maternal-to-Zygotic Transition. Annu Rev Cell Dev Biol 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lehmann R Germline stem cells: Origin and destiny. Cell Stem Cell 2012;10:729–39. doi: 10.1016/j.stem.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Spradling AC, de Cuevas M, Drummond-Barbosa D, Keyes L, Lilly M, Pepling M, et al. The Drosophila germarium: stem cells, germ line cysts, and oocytes. Cold Spring Harbor Symposia on Quantitative Biology 1997;62:25–34. [PubMed] [Google Scholar]
- [9].Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 1998;94:251–60. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- [10].Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature 2001;414:98–104. [DOI] [PubMed] [Google Scholar]
- [11].McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes & Development 1990;4:2242–51. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- [12].Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 2016;5:1–24. doi: 10.7554/eLife.16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol 2004;6:427–35. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- [14].Huynh J-R, St Johnston D. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Current Biology 2004;14:R438–49. [DOI] [PubMed] [Google Scholar]
- [15].Lantz V, Chang JS, Horabin JI, Bopp D, Schedl P. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes & Development 1994;8:598–613. doi: 10.1101/gad.8.5.598. [DOI] [PubMed] [Google Scholar]
- [16].Kugler J-M, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during drosophila oogenesis. Fly 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- [17].Spradling AC, Mahowald AP. Amplification of genes for chorion proteins during oogenesis in <em>Drosophila melanogaster</em>. Proc Natl Acad Sci USA 1980;77:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Navarro-Costa P, McCarthy A, Prudêncio P, Greer C, Guilgur LG, Becker JD, et al. Early programming of the oocyte epigenome temporally controls late prophase i transcription and chromatin remodelling. Nat Comms 2016;7. doi: 10.1038/ncomms12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Seago JE, Chernukhin IV, Newbury SF. The Drosophila gene twister, an orthologue of the yeast helicase SKI2, is differentially expressed during development. Mechanisms of Development 2001;106:137–41. doi: 10.1016/S0925-4773(01)00429-4. [DOI] [PubMed] [Google Scholar]
- [20].Anderson JS, Parker RP. The 39 to 59 degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 39 to 59 exonucleases of the exosome complex. Embo J 1998;17:1497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. Rna 2012;18:124–34. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schmidt C, Kowalinski E, Shanmuganathan V, Defenouillere Q, Braunger K, Heuer A, et al. The cryo-EM structure of a ribosome–Ski2-Ski3-Ski8 helicase complex. Science 2016;354:1431–3. doi: 10.1126/science.aaf7520. [DOI] [PubMed] [Google Scholar]
- [23].Tanentzapf G, Devenport D, Godt D, Brown NH. Integrin-dependent anchoring of a stem-cell niche. Nat Cell Biol 2007;9:1413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Doren MV, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Current Biology 1998;8:243–6. doi: 10.1016/S0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- [25].Halbach F, Reichelt P, Rode M, Conti E. The Yeast Ski Complex: Crystal Structure and RNA Channeling to the Exosome Complex. Cell 2013;154:814–26. doi: 10.1016/j.cell.2013.07.017. [DOI] [PubMed] [Google Scholar]
- [26].Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- [27].Becker T, Armache J-P, Jarasch A, Anger AM, Villa E, Sieber H, et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol 2011;18:715–20. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- [28].Yang F, Zhao R, Fang X, Huang H, Xuan Y, Ma Y, et al. The RNA surveillance complex Pelo-Hbs1 is required for transposon silencing in the Drosophila germline. EMBO Reports 2015;16:965–74. doi: 10.15252/embr.201540084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li Z, Yang F, Xuan Y, Xi R, Zhao R. Pelota-interacting G protein Hbs1 is required for spermatogenesis in Drosophila. Sci Rep 2019:1–14. doi: 10.1038/s41598-019-39530-6. [DOI] [PMC free article] [PubMed]
- [30].Nagarajan VK, Jones CI, Newbury SF, Green PJ. XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochimica Et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 2013;1829:590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. arXiv 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- [32].Halbach F. Structural and Functional Characterization of the Yeast Ski2-Ski3-Ski8 Complex 2013:1–126.
- [33].Jevitt A, Chatterjee D, Xie G, Wang X-F, Otwell T, Huang Y-C, et al. A single-cell atlas of adult Drosophila ovary identifies transcriptional programs and somatic cell lineage regulating oogenesis. PLoS Biol 2020;18:e3000538. doi: 10.1371/journal.pbio.3000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Flora P, Wong-Deyrup SW, Martin ET, Palumbo RJ, Nasrallah M, Oligney A, et al. Sequential Regulation of Maternal mRNAs through a Conserved cis-Acting Element in Their 3’ UTRs. CellReports 2018;25:3828–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ohlstein B, McKearin D. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development 1997;124:3651. [DOI] [PubMed] [Google Scholar]
- [36].Jia Ng SS, Zheng RT, Osman I, Pek JW. Generation of Drosophila sisRNAs by Independent Transcription from Cognate Introns. Iscience 2018;4:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kelpsch DJ, Groen CM, Fagan TN, Sudhir S, Tootle TL. Fascin regulates nuclear actin during Drosophila oogenesis. Molecular Biology of the Cell 2016;27:2965–79. doi: 10.1091/mbc.E15-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gerbasi VR, Preall JB, Golden DE, Powell DW, Cummins TD, Sontheimer EJ. Blanks, a nuclear siRNA/dsRNA-binding complex component, is required for Drosophila spermiogenesis. Pnas 2011;108:3204–9. doi: 10.1073/pnas.1009781108/-/DCSupplemental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schneider C, Tollervey D. Threading the barrel of the RNA exosome 2013:1–9. doi: 10.1016/j.tibs.2013.06.013. [DOI] [PMC free article] [PubMed]
- [40].Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. Rna 2011;17:624–38. doi: 10.1261/rna.2404211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Molecular Cell 2003;11:1405–13. doi: 10.1016/S1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- [42].Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nature Publishing Group 2015;16:665–77. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- [43].Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes & Development 1993;7:1737–54. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- [44].Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2013;2013:1–32. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hashimoto Y, Takahashi M, Sakota E, Nakamura Y. Nonstop-mRNA decay machinery is involved in the clearance of mRNA 5′-fragments produced by RNAi and NMD in Drosophila melanogaster cells. Biochemical and Biophysical Research Communications 2017;484:1–7. doi: 10.1016/j.bbrc.2017.01.092. [DOI] [PubMed] [Google Scholar]
- [46].Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay — Mechanisms of substrate mRNA recognition and degradation in mammalian cells. BBA - Gene Regulatory Mechanisms 2013;1829:612–23. [DOI] [PubMed] [Google Scholar]
- [47].Kobayashi K, Kikuno I, Kuroha K, Saito K, Ito K, Ishitani R, et al. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1alpha complex. Proc Natl Acad Sci USa 2010;107:17575–9. doi:1009598107 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jamar NH, Kritsiligkou P, Grant CM. The non-stop decay mRNA surveillance pathway is required for oxidative stress tolerance. Nucleic Acids Research 2017;45:6881–93. doi: 10.1093/nar/gkx306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xi R, Doan C, Liu D, Xie T. Pelota controls self-renewal of germline stem cells by repressing a Bam-independent differentiation pathway. Development 2005;132:5365–74. doi: 10.1242/dev.02151. [DOI] [PubMed] [Google Scholar]
- [50].Edri S, Tuller T. Quantifying the Effect of Ribosomal Density on mRNA Stability. PLoS ONE 2014;9:e102308. doi: 10.1371/journal.pone.0102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Guydosh NR, Green R. Dom34 Rescues Ribosomes in 3′ Untranslated Regions. Cell 2014;156:950–62. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Guydosh NR, Green R. Translation of poly(A) tails leads to precise mRNA cleavage. Rna 2017;23:749–61. doi: 10.1261/rna.060418.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zinoviev A, Ayupov RK, Abaeva IS, Hellen CUT, Pestova TV. Extraction of mRNA from Stalled Ribosomes by the Ski Complex. Molecular Cell 2020;77:1340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Simms CL, Yan LL, Zaher HS. Ribosome Collision Is Critical for Quality Control during No-Go Decay. Molecular Cell 2017;68:361–5. doi: 10.1016/j.molcel.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 2017. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed]
- [56].Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research 1987;15:1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Presnyak V, Alhusaini N, Chen Y-H, Martin S, Morris N, Kline N, et al. Codon optimality is a major determinant of mRNA stability. Cell 2015;160:1111–24. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Serano TL, Cheung H-K, Frank LH, Cohen RS. P element transformation vectors for studying Drosophila melanogaster oogenesis and early embryogenesis. Gene 1994;138:181–6. [DOI] [PubMed] [Google Scholar]
- [59].Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell 1992;71:301–13. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- [60].Gavis ER, Lehmann R. Translational regulation of nanos by RNA localization. Nature 1994;369:315–8. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- [61].McDermott SM, Davis I. Drosophila Hephaestus/polypyrimidine tract binding protein is required for dorso-ventral patterning and regulation of signalling between the germline and soma. PLoS ONE 2013;8:e69978–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Van Buskirk C, Trudi S. Half pint Regulates Alternative Splice Site Selection in Drosophila 2002:1–11. [DOI] [PubMed]
- [63].Chanet S, Huynh J-R. Collective Cell Sorting Requires Contractile Cortical Waves in Germline Cells. Current Biology 2020;30:4213–4. [DOI] [PubMed] [Google Scholar]
- [64].Roper K Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. Journal of Cell Science 2005;118:3937–48. doi: 10.1242/jcs.02517. [DOI] [PubMed] [Google Scholar]
- [65].Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Current Biology 2003;13:1786–91. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- [66].Chen D, Mckearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development (Cambridge, England) 2003;130:1159–70. doi: 10.1242/dev.00325. [DOI] [PubMed] [Google Scholar]
- [67].Lilly MA, Sptadling AC. The Drosophila endocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion. Genes & Development 1996;10:2514–26. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- [68].Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes & Development 1998;12:734–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes & Development 1997;11:423–35. doi: 10.1101/gad.11.4.423. [DOI] [PubMed] [Google Scholar]
- [70].Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 2002;8:323–31. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- [71].Tadros W, Houston SA, Bashirullah A, Cooperstock RL, Semotok JL, Reed BH, et al. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics 2003;164:989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Cabrera Quio LE, Schleiffer A, Mechtler K, Pauli A. Zebrafish Ski7 tunes RNA levels during the oocyte-to-embryo transition. bioRxiv 2020:2020.03.19.998716. [DOI] [PMC free article] [PubMed]
- [73].Xie T, Spradling AC. A Niche Maintaining Germ Line Stem Cells in the Drosophila Ovary. Science Reports 2000;290:328–30. [DOI] [PubMed] [Google Scholar]
- [74].McCarthy A, Sarkar K, Martin ET, Upadhyay M, James JR, Lin JM, et al. MSL3 coordinates a transcriptional and translational meiotic program in female Drosophila. bioRxiv 2019:2019.12.18.879874.
- [75].Wineland DM, Kelpsch DJ, Tootle TL. Multiple Pools of Nuclear Actin. Anat Rec 2018;301:2014–36. doi: 10.1002/ar.23964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gerbasi VR, Preall JB, Golden DE, Powell DW, Cummins TD, Sontheimer EJ. Blanks, a nuclear siRNA/dsRNA-binding complex component, is required for Drosophila spermiogenesis. Proceedings of the National Academy of Sciences 2011;108:3204–9. doi: 10.1073/pnas.1009781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Upadhyay M, Martino Cortez Y, Wong-Deyrup SW, Tavares L, Schowalter S, Flora P, et al. Transposon Dysregulation Modulates dWnt4 Signaling to Control Germline Stem Cell Differentiation in Drosophila. PLoS Genet 2016;12. doi: 10.1371/journal.pgen.1005918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].McCarthy A, Deiulio A, Martin ET, Upadhyay M, Rangan P. Tip60 complex promotes expression of a differentiation factor to regulate germline differentiation in female Drosophila. Molecular Biology of the Cell 2018;29:2933–45. doi: 10.1091/mbc.E18-06-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Flora P, Schowalter S, Wong-Deyrup S, DeGennaro M, Nasrallah MA, Rangan P. Transient transcriptional silencing alters the cell cycle to promote germline stem cell differentiation in Drosophila. Developmental Biology 2018;434:84–95. doi: 10.1016/j.ydbio.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang X, Page-McCaw A. Wnt6 maintains anterior escort cells as an integral component of the germline stem cell niche. Development (Cambridge, England) 2018;145. doi: 10.1242/dev.158527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Fuchs G, Diges C, Kohlstaedt LA, Wehner KA, Sarnow P. Proteomic analysis of ribosomes: translational control of mRNA populations by glycogen synthase GYS1. Journal of Molecular Biology 2011;410:118–30. doi: 10.1016/j.jmb.2011.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology 1994;2:28–36. [PubMed] [Google Scholar]
- [83].Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics 2011;27:1017–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Meth 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- [86].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Trcek T, Lionnet T, Shroff H, Lehmann R. mRNA quantification using single-molecule FISH in Drosophila embryos. Nat Protoc 2017;12:1326–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Meth 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research 2006;34:W369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Log2 Transcript Per Million (TPM) values and status of regulation by Tst, Pelo or Hfp for all Drosophila genes. RNA-Seq data sets include: adult WT (WT_1, 2), tst/Df (Tst_1, 2), tst RNAi (Tst_RNAi_1, 2), young WT (Young_1, 2), GSC enriched ovaries (GSC), undifferentiated CB enriched ovaries (CB), unfertilized Egg (Egg_pA), pelo1 heterozygous control (Pelo_Het_1, 2), pelo1 homozygous mutant (Pelo_1, 2), hfp9 heterozygous control (Hfp_Het_1, 2), hfp9 homozygous mutant (Hfp_1, 2), undifferentiated CB enriched ovary input (CB_Input), undifferentiated CB enriched ovary polysome RNA (CB_Psome), differentiating cyst enriched ovary input (Bam_hs_Input_1, 2), differentiating cyst enriched ovary polysome RNA (Bam_hs_Psome_1, 2), adult control input (Ctrl_Input_1, 2), adult control polysome RNA (Ctrl_Psome_1, 2), pelo1 homozygous mutant input (Pelo_Input_1, 2), pelo1 homozygous mutant polysome RNA (Pelo_Psome_1, 2), tst RNAi input (Tst_Input_1, 2), tst RNAi polysome RNA (Tst_Psome_1, 2). See methods for how specific stages were enriched. (B) RNA-Seq sample information, GEO identifiers, library properties, total reads and alignment statistics. (C) DESeq2 normalized log2 fold change (lfc), p value (pval), and adjusted p value (padj) for tst LOF vs WT, pelo vs WT, and hfp vs WT. (D) Sample comparisons used for DESeq2.
(A) MEME discriminative mode motif enrichment output of Tst-regulated gene names, starting position of the polypyrimidine rich motif, p-value and site within each CDS. (B) Find Individual Motif Occurrences (FIMO) output of the polypyrimidine rich motif (CNN) search in the blanks CDS.
List of Tst-regulated genes screened in the germline, BDSC stock number, and a description of the resulting phenotype.
Table S2. List of primers used in this study. Related to STAR methods.
Data Availability Statement
All RNA-Sequencing data generated during this study is available in the GEO repository under the Accession number: GSE166275. This study did not generate any code.


