Graphical abstract
Keywords: Ashwagandha, DNA damage, Withanone, Toxicity, Glutathione
Abbreviations: Win, withanone; cys, cysteine; DMEDA, dimethylethylenediamine; EDTA, ethylenediamine tetraacetic acid; GSH, glutathione; HDS, Herbal medicines and dietary supplements; LC–MS, liquid chromatography-mass spectrometry
Highlights
-
•
The widely used medicinal herb Withania somnifera (Ashwagandha) has been recently reported to cause liver damage.
-
•
Withanone is a major metabolite of Ashwagandha.
-
•
Withanone was found to cause DNA damage.
-
•
Withanone forms adducts with amines and thiols.
-
•
Withanone-mediated DNA damage has serious biological consequences.
Abstract
Withania somnifera, commonly known as Ashwagandha, is a medicinal plant used for thousands of years for various remedies. Extracts of Ashwagandha contain more than 200 metabolites, with withanone (win) being one of the major ones responsible for many of its medicinal properties. Recently, several cases of liver toxicity resulting from commercially available Ashwagandha products have been reported. The first report of Ashwagandha-related liver damage was from Japan, which was quickly resolved after drug-withdrawal. Later, similar cases of liver toxicity due to Ashwagandha consumption were reported from the USA and Iceland. Towards understanding the liver toxicity of Ashwagandha extracts, we studied win, a representative withanolide having toxicophores or structural alerts that are commonly associated with adverse drug reactions. We found that win can form non-labile adducts with the nucleosides dG, dA, and dC. Using various biochemical assays, we showed that win forms adducts in DNA and interfere with its biological property. Win also forms adducts with amines and this process is reversible. Based on the data presented here we concluded that win is detoxified by GSH but under limiting GSH levels it can cause DNA damage. The work presented here provides a potential mechanism for the reported Ashwagandha-mediated liver damage.
1. Introduction
In recent years, some consumers are using complementary and alternative medications (CAMs) to manage acute and chronic diseases due to perceived and reported adverse effects associated with some prescription drugs (Wooten, 2010, Kalgutkar et al., 2008, Troupin, 1996). An increasing trend in the use of herbal medicines and dietary supplements (HDS) has been noticed in the USA as well (Garcia-Cortes et al., 2016, Stickel and Shouval, 2015). This is because of the belief that herbal medicines are devoid of toxicity (Balunas and Kinghorn, 2005, Chin et al., 2006). HDS are considered as food by the FDA (USA) and, unlike prescription medicines, are assumed to be safe unless otherwise reported.
The Solanaceae family consists of agricultural crops, medicinal plants, spices, weeds, and ornamentals plants. Some commonly used food products from this family are tomatoes, potatoes, eggplant, bell and chili peppers, tomatillo, and tobacco. Many plants of this family contain interesting compounds that are either toxic or have therapeutic potential. Notable examples include solanine, scopolamine, nicotine, atropine, capsaicin, and withanolides (Balunas and Kinghorn, 2005, Choi et al., 2006). Withania somnifera (also known as Ashwagandha, Indian ginseng, or winter cherry) is a medicinal plant of the Solanaceae family that has been in use for thousands of years in India and its neighboring counties (Mukherjee et al., 2021). Extracts of Ashwagandha are utilized for remedies against inflammation, arthritis, diabetes, stress, asthma, and hypertension (Widodo et al., 2010, Widodo et al., 2007, Mishra et al., 2000). In addition, Ashwagandha extracts have antioxidant, neuroprotective, immunomodulatory, anti-cancer, and anti-viral properties (Widodo et al., 2010, Widodo et al., 2007, Mishra et al., 2000, Dar et al., 2016, Palliyaguru et al., 2016). Extracts from various parts of the plant contain more than 200 primary and secondary metabolites including alkaloids, flavanol glycosides, glycowithanolides, steroidal lactones (withanolides), sterols, and phenolics (Trivedi et al., 2017). To date, 12 alkaloids and 35 withanolides have been purified and characterized (Mirjalili et al., 2009, Atta-ur-Rahman et al., 1993, Choudary et al., 1996). Withanone (win) is one such withanolide that is found in significant amounts in Ashwagandha extracts (19 and 3 mg/g dry weight of leaves and roots, respectively) and is thought to be responsible for many of its medicinal properties (Rai et al., 2016, Kaul et al., 2016).
With the increase in the use of herbal supplements, HDS-induced liver injury and failure are becoming common (Garcia-Cortes et al., 2016, Stickel and Shouval, 2015, Navarro et al., 2017). A prospective study conducted by the drug-induced liver injury network (DILIN) in the USA has reported an increase in the proportion of DILI due to HDS from 7% in 2004–2005 to about 20% in 2010–2014 (Björnsson et al., 2020, Navarro et al., 2014). Similar trend (up to 27%) has been found in a retrospective study in Mainland China with Chinese traditional medicine and HDS. The majority of the reported cases of HDS-induced liver damage are associated with multi-ingredient mixtures, where identifying a single causative agent is extremely difficult (Björnsson et al., 2020). Ashwagandha, like most herbal supplements, is considered generally safe for consumption without considerable side effects (Ashwagandha, 2012). However, recently several cases of liver toxicity resulting from commercially available Ashwagandha products have been reported (Philips et al., 2020). The symptoms usually resolve with discontinued use and fatalities or chronic injuries are yet to be reported (Philips et al., 2020). Although impurities may be a cause for liver injury, there are some cases where the preparations were pure. Liver toxicity could also be due to extracts having higher concentrations compared to traditional use of Ashwagandha such as in a tonic or in a churna. In addition, toxicity may also result due to intake of more than recommended dose. In rats the no observed adverse effect level (NOAEL) of dry Ashwagandha extract was found to be 2000 mg/kg and the corresponding Human Equivalent Dose (HED) is ~325 mg/kg/day (Pires et al., 2020, Patel et al., 2016). The safety and toxicity profile of Ashwagandha extracts and/or the bioactive components therein have not been explored in detail. In a recent phase 1 clinical trial using a daily dose of 400 mg of dry ashwagandha extract, only grade 1 and 2 adverse events were observed (Pires et al., 2020). The first report of Ashwagandha-related liver damage was from Japan, which was quickly resolved after its withdrawal (Philips et al., 2020). Later, similar cases of liver toxicity due to Ashwagandha consumption were reported from the USA and Iceland (Philips et al., 2020).
Although Ashwagandha contains several metabolites, we specifically focused on the withanolide win, as a representative compound (Dar et al., 2015). Apart from being a major metabolite, win contains multiple electrophilic groups (two Michael acceptors and an epoxide) which are associated with adverse drug reactions (Vaishnavi et al., 2012). These functional groups, generally termed as toxicophores or structural alerts, are responsible for the toxicity and withdrawal of various drugs. Surprisingly though, there are some structural alerts containing drugs that are safe if properly used (Kalgutkar et al., 2008), a classic example being acetaminophen. Although acetaminophen is bioactivated to a reactive intermediate quinoneimine, it is efficiently detoxified by the endogenous protective system consisting of glutathione (GSH) (Kalgutkar, 2020, Obach et al., 2008). However, when the GSH system is overwhelmed due to drug overdose or reduced level of GSH, severe liver injury occurs. We have shown here and in detail elsewhere that win also forms adducts with GSH. Accordingly, we hypothesized that despite having structural alerts, the cellular detoxification system consisting of GSH renders Ashwagandha safe, when used in limited amounts. If used in excess or if the cellular GSH level is mitigated, electrophilic metabolites can cause toxicity. Herein, we provide strong evidence that win can cause alkylative DNA damage if not detoxified by GSH.
2. Experimental
2.1. Reagents and cell lines
Unless otherwise mentioned all reagents were of the highest purity available and obtained from Sigma Aldrich (now Merck, St. Louis, MO, USA); Agarose, ethidium bromide, catalase, ethanol, DMSO, NaCl, EDTA, Tris-Cl were obtained from HiMedia (Mumbai, India); DMEDA from Tokyo Chemical Industry (Tokyo, Japan); acetonitrile from JT Baker (Center Valley, PA, USA); glutathione from Sisco Research Lab. Pvt Ltd. (Mumbai, India); 2′-deoxyadenosine from Alfa Aesar (Haverhill, MA, USA); ethylamine and propylamine from Spectrochem Pvt Ltd. (Mumbai, India).
Cell lines authentication by STR profiling was outsourced to Lifecode Technologies (New Delhi, India).
2.2. DNA cleavage assays
In a typical DNA cleavage reaction, plasmid DNA (pUC19, 1 µg) was incubated with win (0–25 µM), catalase (250 µg/ml), desferal (500 µM) in potassium phosphate buffer (100 mM, pH 7.4), at 37 °C for 12 h. Following incubations, reactions were subjected to DMEDA (100 mM) workup for 2 h at 37 °C, quenched by addition of 5 µL of glycerol loading buffer, and then electrophoresed for 45 min at 80 V in 1% agarose gel (w/v) containing 0.5 µg/mL ethidium bromide. DNA was visualized and quantified using GE Image Quant LAS 500 gel documentation system (Massachusetts, USA). Strand breaks per plasmid DNA molecule (S) were calculated using the equation S = −ln f1, where f1 is the fraction of plasmid present as form I.
2.3. Formation of various win adducts
To form win adducts, win (100 µM) was incubated with deoxyguanosine (dG, 500 µM), deoxyadenosine (dA, 500 µM), deoxycytidine (dC, 500 µM), deoxythymidine (dT, 500 µM), inosine (I, 500 µM), or ethylamine (100 μM) in 100 mM potassium phosphate (pH 7.5) at 37 °C for 16 h.
2.4. Determination of stability and reversibility of win-ethylamine adduct
For determining the stability of win-ethylamine adduct, win (100 μM) and ethylamine (1 mM) in 100 mM potassium phosphate buffer pH 7.4 was incubated at 37 °C and analyzed using LCMS at various time intervals.
For reversibility assay, win-ethylamine adduct, synthesized as mentiond above, was HPLC purified (5.3–5.8 min). The purified win-ethylamine adduct was then incubated with calf thymus DNA (1 mM in bases) or propyl-amine (1 mM) at 37 °C and analyzed by LCMS at various time intervals.
2.5. Theoretical calculations
The mechanistic studies of the covalent adduct formation between EtNH2 and win were performed using Density Functional Theory (DFT). All the structures were optimized at M06-2X/6-31G* level of theory using the Gaussian 09 package (Zhao et al., 2018, Frisch et al., 2016). The optimizations were carried out in an implicit solvent using a CPCM solvent model, with parameters corresponding to water, which is the solvent used in the experimental studies (Cossi et al., 2003). The nature of stationary points was characterized by frequency calculations at the same level of theory. The single-point energy evaluations were done at the M06-2X/def2-TZVP level including the solvent (Weigend and Ahlrichs, 2005). The relative free energies were evaluated at 298.15 K.
2.6. Equilibrium dialysis
To determine the binding of win to DNA, equilibrium dialysis was performed. A solution containing 100 µM EDTA, 100 mM Potassium phosphate buffer (pH 7.5), 2 µM win, and ctDNA (1 mM in bps) was dialyzed against the same solution without the DNA at room temperature for 16 h. The dialysis membrane had a cutoff of 3.5 kDa. Following dialysis, the relative amount of win in the dialysate was measured by LC–MS and compared against no DNA control.
2.7. Formation of DNA adduct with calf thymus DNA
In a typical reaction, 200 μg of sonicated calf thymus DNA and win (200 µM) in 100 mM potassium phosphate buffer (pH 7.5) was incubated for 6 h at 37 °C. Following incubation, the DNA was precipitated with 70% ethanol and 0.3 M sodium acetate. The precipitated DNA was pelleted by centrifugation at 10,000×g, washed three times with cold 80% ethanol, and re-dissolved in 50 mM sodium acetate (pH 7.4) buffer containing 50 mM MgCl2. The DNA was digested with DNase (7 units), phosphodiesterase (0.01 units), benzonase (0.1 unit), and alkaline phosphatase (20 units) for 6 h at 37 °C. After digestion, the reaction mixture was quenched with cold acetonitrile (1:1), the protein was pelleted by centrifugation at 10,000×g for 20 min. The supernatant was dried under a stream of N2 and analyzed by LC-tandem MS.
2.8. Competition assay
To check the competition between DNA and GSH to form an adduct with win, win (20 μM), GSH (1 mM), and different concentration of calf thymus DNA (0.1–10 mM) in potassium phosphate buffer (100 mM, pH 7.4) was incubated at 37 °C for 5 h. The relative yield of the adducts was analyzed by LC–MS
2.9. LC–MS/MS detection and characterization of win and various win-adducts
An Agilent 6540 UHD Accurate-Mass Q-TOF LCMS System (Agilent Technologies, Santa Clara, CA, USA) with an Agilent UHPLC system was used for LC-tandem MS analysis. Phenomenex (Torrance, CA, USA) kinetex polar C18 column (Column A: 5 μm, 2.1 mm × 10 mm; Column B; 2.6 μm, 2.1 mm × 100 mm) was used for chromatography.
Win adducts were separated using solvent A (containing 0.1% HCO2H and water v/v) and solvent B (containing 0.1% HCO2H and CH3CN, v/v) following a gradient program with a flow rate of 300 μL min−1: 0–2 min, 95% A (v/v); 2.0–12.5 min, linear gradient to 100% B; 12.5–15.5 min, hold at 100% B (v/v); 15.5–16.0 min, linear gradient to 98% A (v/v); 16–20 min, hold at 98% A (v/v). The temperature of the column was maintained at 30 °C and samples (20 μL) were infused with an auto-sampler. For nucleoside adducts, the initial gradient was 98% A instead of 95%.
ESI conditions were as follows: gas temperature 325 °C, drying gas flow rate 8 l/min, nebulizer 35 psi, sheath gas temperature 300 °C, sheath gas flow rate 10 l/min, capillary voltage 2500 V, nozzle voltage 1000 V, capillary current 0.054 μA, chamber current 4.23 μA, fragmentor voltage 80 V, skimmer voltage 70 V. For MS/MS normalized collision energy of 30% was used.
2.10. Transformation
1 µg of pUC19 plasmid was incubated with 10 nM win in 100 mM potassium phosphate buffer pH 7.5 at 37 °C for 4 h. After incubation, DNA was precipitated with ethanol and sodium acetate. The precipitated plasmid DNA was washed and used for transformation into competent ampicillin-sensitive E. coli cells. Cells were transformed following standard protocol giving a heat shock at 42 °C. Because DNA damage can have significant effect on transformation, we introduced a correction factor for it (Huang et al., 2010). Transformed cells were initially incubated at 20 °C in LB media containing ampicillin for 5 h. This step was performed to allow only ampicillin-resistant cells to survive while preventing any cell growth/division. Following incubation, 100 μL of the culture was stained with DAPI to visualize live cells (Johnson and Criss, 2013). Live cells were counted under a microscope. The remaining culture was plated on LB-agar plates having ampicillin (100 μg/mL). The plates were incubated overnight and colonies counted. The number of colonies were corrected for difference in number of live cells plated after various treatments.
2.11. Transfection
30 µg of pGFP-N1 plasmid was incubated with 100 µM win and 100 mM potassium phosphate pH 7.5 at 37 °C for 3 h. After incubation DNA was precipitated with ethanol and sodium acetate. HEK-293 T cells were transfected with 10 µg of win-treated plasmid with lipofectamine 2000. Imaging was done at 10x in Leica DMi1 (Wetzlar, Germany) after 48 h.
2.12. Cell culture and cytotoxicity assays.
Cell lines HepG2 and MCF7 were acquired from National Center for Cell Science (Pune, Maharashtra, India) and MCF-10A was obtained from Dr. Carlos L. Arteaga at Southwestern Medical Center. Cells were grown in DMEM supplemented with 10% fetal bovine serum in a humidified CO2 incubator at 37 °C. All cell culture reagents were purchased from Thermo Fisher Scientific (Waltham, Massachusetts, USA).
Approx. 15,000 cells were plated in each well of transparent flat bottom 96 well plate. Treatment of win, with respective dosages, was started after cells attached overnight and continued for 72 h. At the end of experiment viability was measured either using Cell Titer Glo kit (Promega Corporation, Madison, Wisconsin, USA) as per manufacturer instructions or 0.5% crystal violet staining. Absorbance was measured at 595 nm.
3. Results and discussion
3.1. DNA cleavage assay to detect the formation of labile DNA adducts by win
Michael acceptors and epoxides are known to alkylate DNA, particularly N7 of guanine (Koivisto et al., 1999, Tang et al., 2011). Alkylation at N7G and N3A generate labile adducts that ultimately lead to DNA cleavage (Koivisto et al., 1999). DNA cleavage is known to have serious biological consequences including DNA double strand breaks, replication block, mutation and carcinogenesis (Wani et al., 2018a, Wani et al., 2018b). Therefore, we looked into the ability of win to cause DNA cleavage through the formation of labile DNA adducts. Formation of labile DNA adducts results in the generation of abasic sites which upon base (dimethylethylenediamine, DMEDA) workup results in DNA cleavage (Greenberg, 2014, Dahlmann et al., 2009). Thus, by measuring the extent of DNA cleavage in the plasmid DNA cleavage assay, (Wani et al., 2018b, Wani et al., 2017) we can determine the formation of labile DNA adducts by win. We found that there was no cleavage with increasing concentration of win, indicating that win does not form labile DNA adducts (Fig. S1, supporting information).
3.2. Win forms adduct with 2-deoxynucleoside
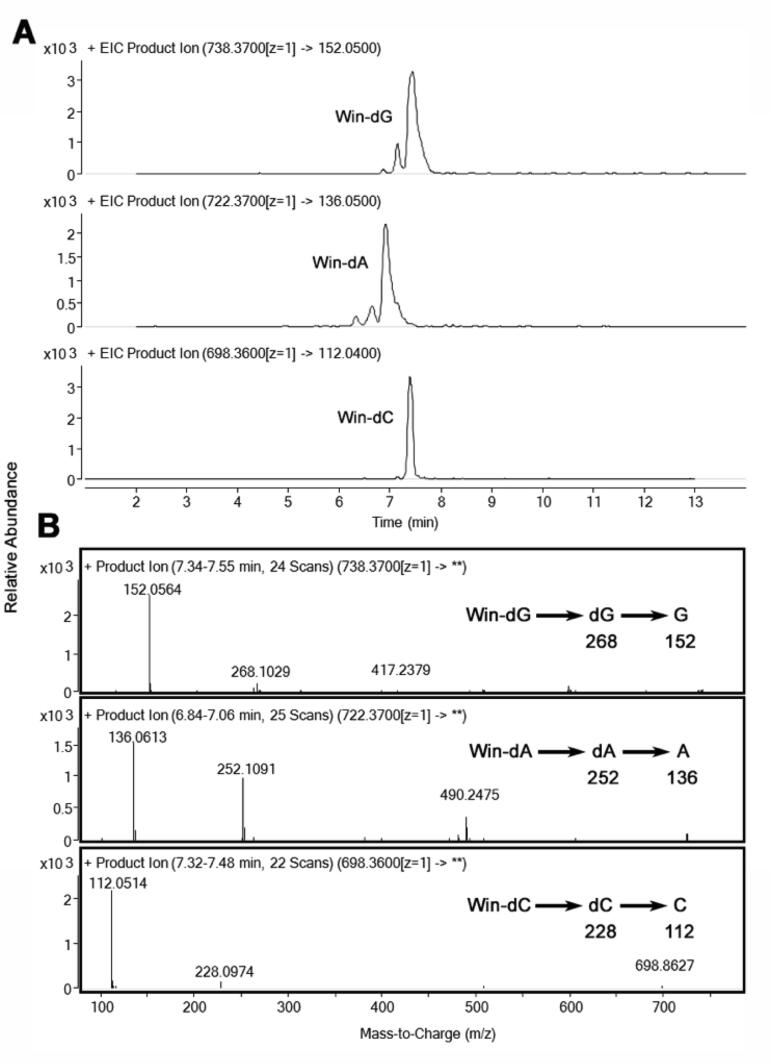
Although the DNA cleavage data indicated that win does not form labile DNA adducts, there still exists the possibility of forming non-labile adducts (Ewa and Danuta, 2017). Accordingly, we treated 2′-deoxynucleosides with win and looked for adducts using LC tandem MS. In accord with our expectation, we found that win forms adducts with dG, dA, and dC but not dT (observed [M + H]+ is 738.3695, 722.374, and 698.3668 and calculated 738.3709, 722.3760, and 698.3647 for dG, dA, and dC respectively, Fig. 1). Collision induced dissociation (CID) of the m/z 738.3695 (win-dG) peak produced fragments at m/z 622 (neutral loss of 116, which corresponds to the 2′-deoxyribose and is characteristic of nucleosides), 268 (dG) and 152 (G) (Fig. 1) (Chowdhury et al., 2013). The fragments are characteristics of a win-dG adduct. Similar results were obtained for the 722.374 (win-dA) and 698.3668 (win-dC) peaks as well (Fig. 1). Together, these results provide evidence that win forms an adduct with dG, dA, and dC. It is to be noted that in the LC-MS data the intensity of the parent ion peak is weak due to its fragmentation in the source, degradation during workup, or poor yield. Direct infusion of the reaction mixture showed a significant breakdown in the source itself.
Fig. 1.
Detection of win adducts of dG, dA, or dC. A) Win (100 μM) was treated with dG, dA or dC (500 μM) for 16 h at 37 °C in aqueous potassium phosphate buffer (100 mM, pH 7.4) and analyzed by LC-tandem MS. Adducts were detected using SRM. B) MS/MS spectra of the Win-dG, dA and dC adducts.
3.3. Characterization of the win-inosine adduct
Because we did not observe any adducts with dT, which unlike other nucleosides does not have any exocyclic amino group, we performed the alkylation reaction with inosine (I). Inosine has the same structure as that of dG without the exocyclic amino group and an additional 2′-hydroxy group. We found a peak at 7.2 min having m/z 739.3512 (calculated [M + H]+ 739.3549). This corresponds to the win-I adduct (Fig. S2, supporting information). CID of the m/z 739.3512 peak gave fragments at m/z 137 and 269, which corresponds to hypoxanthine and inosine (Fig. S2, supporting information). Together, the results indicated that non-labile nucleophilic sites in the purine ring of dG can participate in adduct formation with win. This observation is consistent with the presence of multiple peaks in the LC–MS chromatogram.
3.4. Win forms adducts with amines
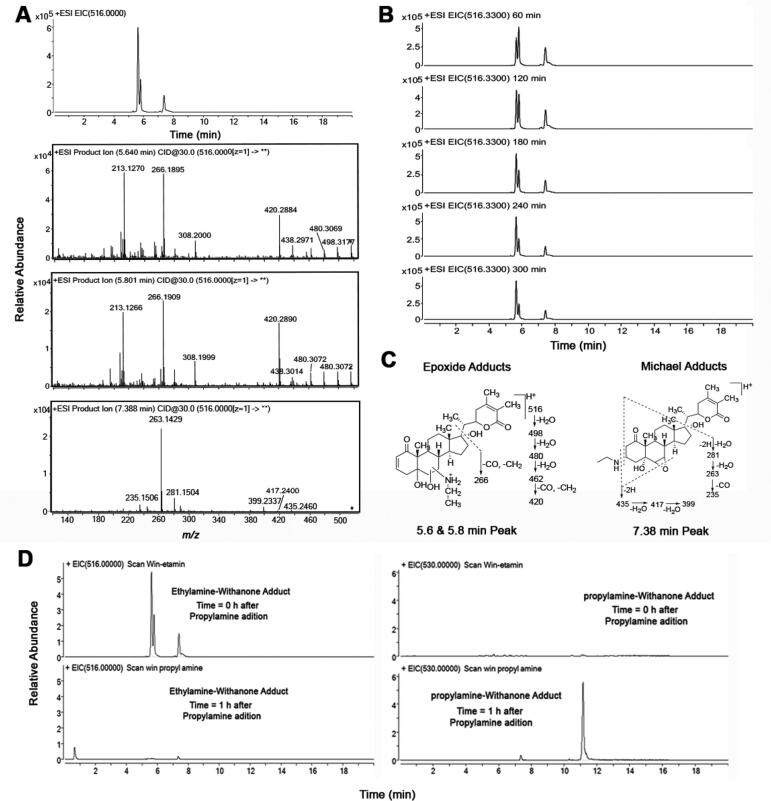
Because dG, dA, dC, and histones have primary amines that can act as nucleophiles, we performed assays to see if simple primary alkylamines will form adducts with win. Both Michael acceptors and epoxides are good electrophiles that can react with thiols, amines, and H2O. Because H2O is the most abundant nucleophile in a cell, we first checked the stability of win in pH 7.4 aqueous buffer using LC-MS. We found win to be stable in pH 7.4 aqueous buffer for 10 h (Fig. S3, supporting information). Although surprising, this stability of the epoxide is not unprecedented (Martin et al., 2000). We then looked into the ability of primary amines to alkylate win. When win was treated with ethylamine, which contains a single nucleophile (Scheme 1), three major peaks at 5.65, 5.8, and 7.4 min were obtained (Fig. 2). The obtained experimental [M + H]+ values (516.3339) were consistent with the calculated value (516.3320). CID of the three peaks gave two distinct fragmentation patterns, one for the 7.4 min peak and another for the 5.65- and 5.8-min peaks (Fig. 2A). Based on fragmentation analysis we concluded that the 7.4 min peak probably corresponds to the Michael adduct/s (R and/or S) and 5.65- and 5.8-min peaks to the two epoxide adducts (Fig. 2A and C).
Scheme 1.
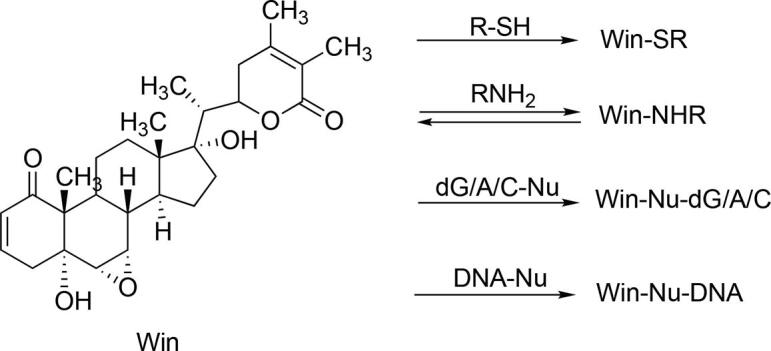
Reaction of win with various nucleophiles to form adducts.
Fig. 2.
Detection, stability, fragmentation analysis, and reversibility of win-ethylamine (win-NHEt) adducts. Win (100 μM) was treated with ethylamine (1 mM) for 0–6 h in aqueous potassium phosphate buffer (100 mM, pH 7.4) and analyzed by LC-tandem MS. A) LC-MS chromatogram showing the presence of win-NHEt adducts (m/z 516) along with the corresponding CID (MS/MS) of the respective adducts. B) LC-MS chromatogram showing the presence of win-NHEt adducts (m/z 516) at 60 min time interval. C) Fragmentation analysis of the Michael and epoxide adducts. D) LC-MS chromatograms showing the formation win-NHPr adducts following the addition of PrNH2 to the win-NHEt adducts.
Interestingly, the relative intensities of the peaks for the amine adducts changed over time. For ethylamine (EtNH2), the 7.4 and 5.8 min peaks were found to decrease with time while the relative intensity of the 5.65 min peak increased (Fig. 2B). This may be due to the adducts being reversible, results in the accumulation of the more thermodynamically stable epoxide adduct over time. To confirm if the adduct formation is reversible, we isolated the ethylamine adducts eluting at 5.65 and 5.8 min and treated with propylamine (the adduct at 7.4 min coelutes with win). The idea being that if the win-NHEt adducts are reversible, there should be generation of win from the purified win-NHEt adducts, which should then react with propylamine (PrNH2) to form new adducts. Thus, detection of win-NHPr adducts will provide evidence that the amine adducts of win are reversible. Consistent with our expectation, the formation of PrNH2 adduct/s of win was observed, indicating that the amine adducts of win are indeed reversible (Fig. 2D).
3.5. Thermodynamic stability of win-NHEt adducts determined by DFT calculation
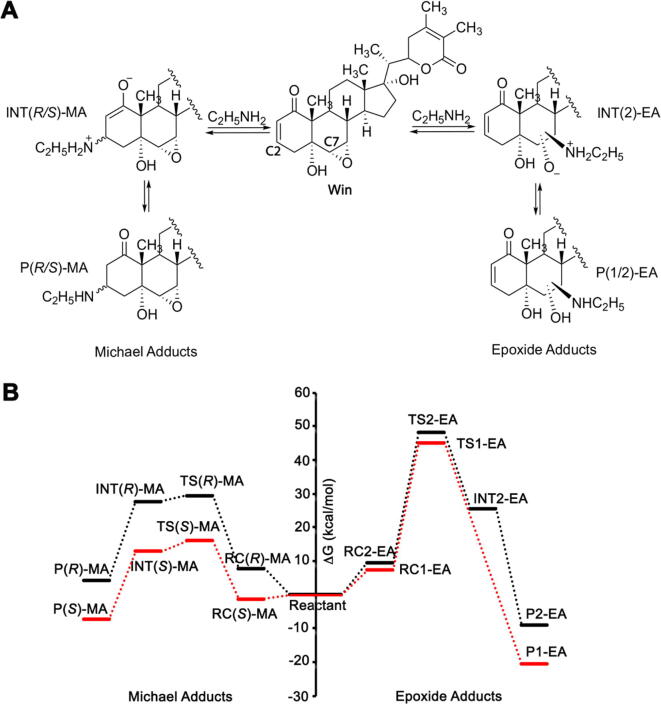
To gain additional insights on the reversibility and thermodynamic stability of the various win-NHEt adducts, Density Functional Theory (DFT) calculations were performed. Relative free energy (ΔG) calculations will provide additional evidence regarding the feasibility, reversibility, and thermodynamic stability of the different adducts formed between win and EtNH2. Based on the fragmentation pattern in the LC-MS/MS data, we focused on the adduct formation at the epoxide and the Michael acceptor groups (Fig. 3A). According to our proposed mechanism, attack of the amine to either of the epoxide carbon atoms will result in the formation of the zwitterion INT1-EA and INT2-EA (Fig. 3A). Intermolecular proton transfer will convert the intermediates to neutral adducts P1-EA and P2-EA. In the present work, we calculated the transition states for the steps leading to the formation of zwitterions. Interestingly, in the case of P1-EA, INT1-EA was not observed in our calculations possibly due to the direct conversion to the neutral product. Nucleophilic attack on the C2 atom of the Michael acceptor group can be either from the re or si face resulting in two diastereomers INT(R)-MA and INT(S)-MA, which following protonation will result in two final products, P(R)-MA and P(S)-MA.
Fig. 3.
A) Proposed reaction mechanism of win with EtNH2 showing the four possible covalent adducts. B) Free energy profiles showing all four possible pathways for the reaction of win with EtNH2. Relative free energies (ΔG) at 298.15 K were computed at the M06-2X/Def2-TZVP// M06-2X/6-31G* level of theory.
Our calculations indicate no thermodynamic stability for the P(R)-MA adduct while for the P(S)-MA adduct ΔG is −7.4 kcal/mol compared to that of the reactants (Fig. 3B). For the epoxide adducts, ΔG for P1-EA and P2-EA are −20.6 and −9.1 kcal/mol, respectively. Based on the calculated ΔG values, the formation of three win-NHEt adducts i.e. two epoxides (P1-EA and P2-EA) and one Michael [P(S)-MA] adduct is thermodynamically feasible. Thus, the results obtained from the theoretical studies are consistent with the observation of three peaks in the LC–MS data, out of which two were found to have a similar fragmentation pattern in the LC–MS/MS data. Hence, the LC–MS, LC–MS/MS and DFT data suggest that two peaks eluting at 5.65 and 5.8 min with similar fragmentation spectra are the epoxide adducts P1-EA and P2-EA while the 7.2 min peak corresponds to the Michael adduct P(S)-MA. Lower thermodynamics stability of the win-NHEt adducts compared to the win-SG and win-Cys adducts (communicated elsewhere, ΔG for irreversible win-SG and win-Cys adducts are greater than 50 kcal/mol) indicates the adduct formation reaction of win with the amine to be reversible under our experimental condition. This reversibility indeed explains the decrease of the 7.4 and 5.8 min peaks over time as seen in Fig. 3B. Thus, the Michael adduct P(S)-MA (peak at 7.2 min in Fig. 3B), which has a lower free energy barrier (~-7.4 kcal/mol) than for the epoxide adducts, although forms faster would convert to the thermodynamically most stable adduct P2-EA over time. Taken together the DFT calculations support the experimental observation that primary amines can react with win to form three different adducts and the reaction is reversible under the reaction conditions used here.
3.6. Equilibrium dialysis to detect DNA binding by win
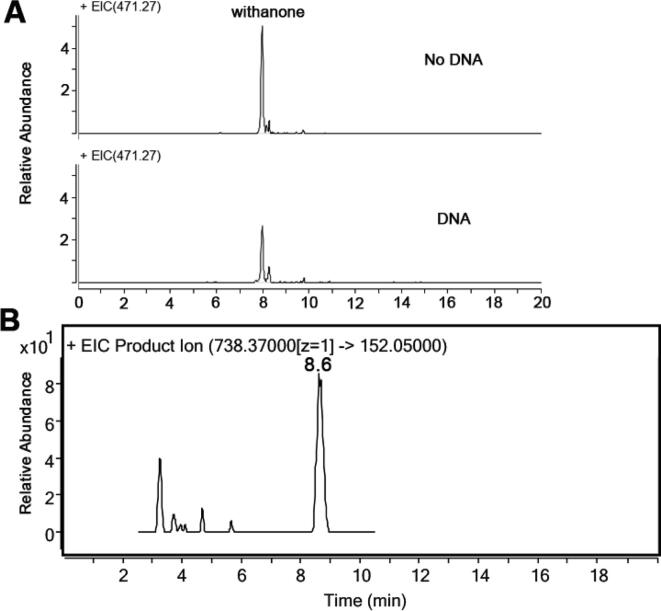
To determine if win will form adducts with DNA, we first performed an equilibrium dialysis experiment to detect the binding of win to DNA. ctDNA was taken in a dialysis bag and dialyzed against a solution of win for 16 h. Analysis of the solution outside the dialysis unit using LC-MS revealed that there is ~65% reduction in win concentration compared to a no DNA control (Fig. 4A). The results indicated that win binds to DNA.
Fig. 4.
DNA binding and detection of win-dG adduct from ctDNA treated with win. A) LC–MS chromatogram showing the presence of win in the dialysate. A solution containing 100 µM EDTA, 100 mM potassium phosphate buffer (pH 7.5), 2 µM win, and 1 mM ctDNA (in base pairs) was dialyzed against the same solution without the DNA at room temperature for 16 h. B) LC-MS/MS chromatogram showing the presence of win-dG adduct in ctDNA. 200 μg of sonicated ctDNA and win (200 µM) in 100 mM potassium phosphate buffer (pH 7.5) was incubated for 6 h at 37 °C. The DNA was precipitated, washed, digested, and analyzed by LC-MS/MS following the transition of m/z 738 → 152.
3.7. LC-tandem MS detection of DNA adducts of win
To confirm the formation of win-DNA adduct, we treated ctDNA with win, precipitated the DNA using 70% ethanol and 0.3 M sodium acetate, digested it to 2′-deoxynucleosides with nucleases and phosphatases (Chowdhury et al., 2014, Ahmed et al., 2020) and finally analyzed it using LC-tandem MS following the m/z 738 → 152 transition. As expected we did see a peak with a retention time of 8.6 min (Fig. 4B, the slight difference in retention time is probably due to variable time of usage of the columns used here). HRMS and CID of the ~8.6 min peak match well with the win-dG adduct obtained from dG. In a separate assay, we confirmed that win does not precipitate under the reaction condition used here (Fig. S4, supporting information).
3.8. Win forms DNA adducts in presence of amines and thiols
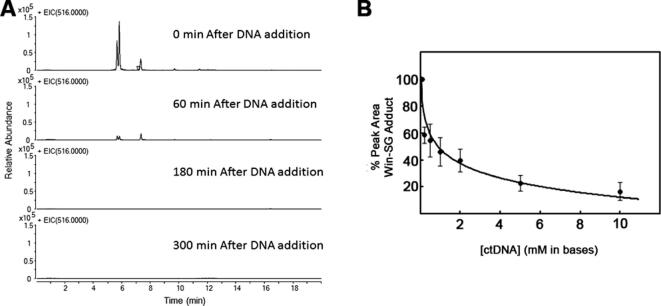
DNA in an eukaryotic cell is wrapped around histones to form chromatin. Histones are lysine and arginine-rich positively charged proteins. Because there are a lot of amines inside a cell, particularly in histones, it is important to check whether win can form DNA adducts in presence of primary amines. Accordingly, we first incubated win with EtNH2 (1 mM) for 1 h and then added ctDNA (1 mM in bases). The addition of ctDNA resulted in the disappearance of ~85% of the win-NHEt adducts within 1 h and >99% within 3 h (Fig. 5A). These results indicated that win can form DNA adducts in the presence of amines.
Fig. 5.
Formation of win-DNA adducts in the presence of amines and GSH. A) LC-MS extracted ion chromatogram showing the disappearance of the win-NHEt adducts following the addition of DNA over 300 mins. B) Competition between GSH (1 mM) and ctDNA for adduct formation with win. The graph represents the average of 2 independent data sets (N = 2) and error bars represent standard deviation.
Because there is a significant amount of the cellular protective nucleophile GSH (5 mM) inside a cell, we performed a competition experiment to determine if win would react with DNA in the presence of GSH. The idea was to determine if the cellular protective systems consisting of GSH would be able to protect the DNA from the potentially toxic alkylating effect of win (in the cytosol) and if DNA can compete with GSH for adduct formation (in the nucleus). We treated win with GSH (1 mM) and varying concentrations of DNA (0–10 mM in base pair) for 3 h. LC–MS analysis of the reaction mixture clearly showed that the yield of the win-SG adducts decreased with increasing concentration of DNA. When the concentration of GSH is similar to DNA (in bases), the win-SG adduct concentration is ~50% of the no DNA control. The results showed that win can form adducts with DNA in the presence of GSH. Hence, in the cytosol, where there is no DNA and the concentration of thiols are significant, win will form adducts with GSH and other thiols. (Fig. 5B). However, under limiting GSH condition (either due to depletion of GSH or use of excess win), win will escape the detoxification reaction in the cytosol and form adducts with DNA in the nucleus.
3.9. Win-treated plasmid DNA having the ampicillin resistance gene has reduced ability to confer ampicillin resistance phenotype in E. coli
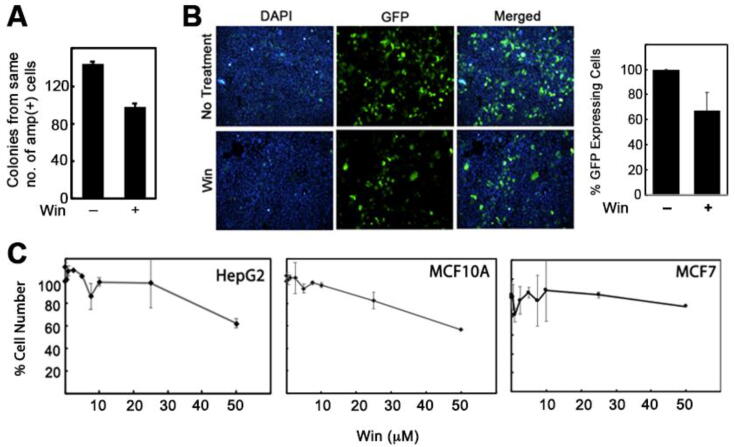
Finally, to determine if the win-DNA adducts have any biological consequence, we treated plasmid DNA harboring the ampicillin resistance gene with win followed by ethanol precipitation of the DNA. In a separate experiment, we have determined that win does not precipitate under the DNA precipitation conditions (Fig. S4, supporting information) used here. The precipitated plasmid DNA was washed and transformed into competent ampicillin-sensitive E. coli cells. Because DNA damage can have an effect on transformation efficiency, we introduced a correction factor. Transformed cells were initially incubated at 20 °C in LB media containing ampicillin for 5 h. This step was performed to allow only ampicillin-resistant (transformed) cells to survive while preventing any cell growth/division. Following incubation, an 100 μL aliquot of the culture was stained with DAPI to visualize live cells. Live cells were counted under a microscope. The remaining culture was plated on LB-agar plates having ampicillin (100 μg/mL). The plates were incubated overnight and colonies counted. The number of colonies were corrected for difference in number of live cells plated after various treatments. The results revealed that transformation of competent E. coli cells with plasmid DNA pre-treated with win produced ~60% fewer colonies than the no win pre-treatment control (Fig. 6A). These observations suggested that the win-DNA adducts caused either a block in DNA synthesis or mutations that rendered the ampicillin-resistance plasmid DNA ineffective.
Fig. 6.
Biological consequences of win exposure A) Ability of win-treated plasmid DNA bearing the ampicillin resistance factor to confer ampicillin resistance phenotype in transformed E. coli cells. B) Ability of win-treated plasmid DNA bearing the green fluorescence protein cDNA to confer green fluorescence phenotype in transfected HEK293T cells. C) Effects of increasing concentrations of win on hepatoma (HepG2), normal mammary epithelium (MCF10A), and mammary carcinoma (MCF-7) cell lines 72 h post-treatment. All graphs represent the average of 2 independent data sets (N = 2) and the error bars represent standard deviation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.10. Win-treated GFP cDNA containing plasmid DNA construct has reduced ability to confer green fluorescence phenotype in HEK-293 T cells
In an attempt to verify if win-DNA adduct affects gene expression in mammalian cells, we treated pGFP-N1 reporter plasmid with win, purified by ethanol purification and transiently transfected in SV40 T antigen transformed human embryonic kidney HEK-293 T cells. Imaging 48 h post-transfection revealed more than 30% decrease in reporter activity (as indicated by percent of GFP + cells) in cells transfected with win-treated DNA compared to untreated DNA transfected cells (Fig. 6B). There were no significant differences in total cell number (DAPI column in Fig. 6B) between DNA alone and win-DNA transfected cells.
3.11. Effect of win on cell survival and proliferation
We looked into the cytotoxicity of win using a cell proliferation assay. The cytotoxicity of win (0–50 μM) in both tumors (HepG2, MCF-7) and normal epithelial (MCF-10A) cell lines were measured. For all three cell lines used here, no change in percent cell number was observed till 20 μM of drug concentration (Fig. 6C). At 50 μM drug concentration, a 40%, reduction in cell number was observed for HepG2 and MCF-10A cell line while for MCF-7 10% reduction in cell number compared to DMSO control was observed (Fig. 6C). A review of the literature revealed that the amount of GSH in MCF-7 cells is significantly higher (8 μmol/mg protein) when compared to MCF-10A (90 nmol/mg protein), which might explain the reduced cytotoxicity in MCF-7 cells (Lewis-Wambi et al., 2008, Cheng et al., 2017).
4. Conclusions
The Ashwagandha metabolite win can form non-labile adducts with the nucleosides dG, dA, dC, and also with DNA. Win forms adducts with primary amines, although the process is reversible. Adduct formation occurs at both the electrophilic Michael acceptor and epoxide functional groups of win. The affinity of win for DNA is significantly more than amines. Win can also form adducts with GSH, indicating the involvement of possible detoxification pathways. Transformation and transfection assays with win-treated plasmid DNA revealed that the DNA lesions caused by win have serious biological consequences and may interfere with DNA transcription, replication and repair resulting in replication block, mutagenesis, apoptosis and cell death. The data presented here can be used to explain the recently reported liver damage resulting from the use of Ashwagandha. Because GSH can form adducts with win, it can potentially decrease Ashwagandha-induced genotoxicity. The data presented here led us to speculate that one of the potential cytotoxic pathways of win may involve concentrations of win that overwhelms the protective system of the cell or when the protective system involving GSH is suppressed. Further studies including mutational analysis, DNA repair, protein adduction will help us understand the occasional liver damage and toxicity caused by this medicinal herb and in turn the safe use of Ashwagandha.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported in part by the Department of Biotechnology (India), Ramalingaswami Re-entry fellowships [BT/RLF/RE-ENTRY/18/2013 to G.C., and BT/RLF/RE-ENTRY/35/2012 to A.C.] and Science and Engineering Research Board, Department of Science and Technology (India) early-career grant [ECR/2015/000197 to G.C. and ECR/2015/000198 to A.C.].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crtox.2021.02.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahmed N., Chakrabarty A., Guengerich F.P., Chowdhury G. Protective role of glutathione against peroxidenitrite-mediated DNA damage during acute inflammation. Chem. Res. Toxicol. 2020;33:2668–2674. doi: 10.1021/acs.chemrestox.0c00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwagandha LiverTox. National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda (MD): 2012. Clinical and Research Information on Drug-Induced Liver Injury. [Google Scholar]
- Atta-ur-Rahman S. Abbas, Dur-e-Shawar A.S., Jamal M.I. Choudhary. New withanolides from Withania spp. J. Nat. Prod. 1993;56:1000–1006. [Google Scholar]
- Balunas M.J., Kinghorn A.D. Drug discovery from medicinal plants. Life Sci. 2005;78(5):431–441. doi: 10.1016/j.lfs.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Björnsson H.K., Björnsson E.S., Avula B., Khan I.A., Jonasson J.G., Ghabril M., Hayashi P.H., Navarro V. Ashwagandha-induced liver injury: a case series from Iceland and the US Drug-Induced Liver Injury Network. Liver Int. 2020;40(4):825–829. doi: 10.1111/liv.14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.H., Tseng Y.M., Wu S.H., Tsai S.M., Tsai L.Y. Whey protein concentrate renders MDA-MB-231 cells sensitive to rapamycin by altering cellular redox state and activating GSK3beta/mTOR signaling. Sci. Rep. 2017;7(1):15976. doi: 10.1038/s41598-017-14159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin Y.W., Balunas M.J., Chai H.B., Kinghorn A.D. Drug discovery from natural sources. AAPS J. 2006;8(2):E239–253. doi: 10.1007/bf02854894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.K., Murillo G., Su B.N., Pezzuto J.M., Kinghorn A.D., Mehta R.G. Ixocarpalactone A isolated from the Mexican tomatillo shows potent antiproliferative and apoptotic activity in colon cancer cells. FEBS J. 2006;273(24):5714–5723. doi: 10.1111/j.1742-4658.2006.05560.x. [DOI] [PubMed] [Google Scholar]
- Choudary M.I., Abbas S., Jamal A.S., Atta-ur-Rahman Withania somnifera- a source of exotic withanolides. Heterocycles. 1996;42:555–563. [Google Scholar]
- Chowdhury G., Cho S.H., Pegg A.E., Guengerich F.P. Detection and characterization of 1,2-dibromoethane-derived DNA crosslinks formed with O(6) -alkylguanine-DNA alkyltransferase. Angew. Chem. Int. Ed. Engl. 2013;52(49):12879–12882. doi: 10.1002/anie.201307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury G., Shibata N., Yamazaki H., Guengerich F.P. Human cytochrome P450 oxidation of 5-hydroxythalidomide and pomalidomide, an amino analogue of thalidomide. Chem. Res. Toxicol. 2014;27(1):147–156. doi: 10.1021/tx4004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossi M., Rega N., Scalmani G., Barone V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J. Comput. Chem. 2003;24(6):669–681. doi: 10.1002/jcc.10189. [DOI] [PubMed] [Google Scholar]
- Dahlmann H.A., Vaidyanathan V.G., Sturla S.J. Investigating the biochemical impact of DNA damage with structure-based probes: abasic sites, photodimers, alkylation adducts, and oxidative lesions. Biochemistry. 2009;48(40):9347–9359. doi: 10.1021/bi901059k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar N.J., Hamid A., Ahmad M. Pharmacologic overview of Withania somnifera, the Indian Ginseng. Cell. Mol. Life Sci. 2015;72(23):4445–4460. doi: 10.1007/s00018-015-2012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar P.A., Singh L.R., Kamal M.A., Dar T.A. Unique medicinal properties of Withania somnifera: phytochemical constituents and protein component. Curr. Pharm. Des. 2016;22(5):535–540. doi: 10.2174/1381612822666151125001751. [DOI] [PubMed] [Google Scholar]
- Ewa B., Danuta M.S. Polycyclic aromatic hydrocarbons and PAH-related DNA adducts. J. Appl. Genet. 2017;58(3):321–330. doi: 10.1007/s13353-016-0380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., Li X., Caricato M., Marenich A., Bloino J., Janesko B.G., Gomperts R., Mennucci B., Hratchian H.P., Ortiz J.V., Izmaylov A.F., Sonnenberg J.L., Williams-Young D., Ding F., Lipparini F., Egidi F., Goings J., Peng B., Petrone A., Henderson T., Ranasinghe D., Zakrzewski V.G., Gao J., Rega N., Zheng G., Liang W., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Throssell K., Jr J.A.M., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Keith T., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Millam J.M., Klene M., Adamo C., Cammi R., Ochterski J.W., Martin R.L., Morokuma K., Farkas O., Foresman J.B., Fox D.J. Gaussian, Inc.; Wallingford CT: 2016. Gaussian 09, Revision D.01, Gaussian 09 suites. [Google Scholar]
- Garcia-Cortes M., Robles-Diaz M., Ortega-Alonso A., Medina-Caliz I., Andrade R.J. Hepatotoxicity by dietary supplements: a tabular listing and clinical characteristics. Int. J. Mol. Sci. 2016;17(4):537. doi: 10.3390/ijms17040537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M.M. Looking beneath the surface to determine what makes DNA damage deleterious. Curr. Opin. Chem. Biol. 2014;21:48–55. doi: 10.1016/j.cbpa.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.L., Li C.Z., Chen Z.R., He W., Zhou Y., Zhou Z.G., Liu S.W., Zhou C. Effects of UV-induced DNA damage on vector ligation and transformation into bacterial cells. Nan Fang Yi Ke Da Xue Xue Bao. 2010;30(1):111–113. [PubMed] [Google Scholar]
- Johnson M.B., Criss A.K. Fluorescence microscopy methods for determining the viability of bacteria in association with mammalian cells. J. Vis. Exp. 2013;79 doi: 10.3791/50729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalgutkar A.S. Designing around structural alerts in drug discovery. J. Med. Chem. 2020;63(12):6276–6302. doi: 10.1021/acs.jmedchem.9b00917. [DOI] [PubMed] [Google Scholar]
- Kalgutkar A.S., Fate G., Didiuk M.T., Bauman J. Toxicophores, reactive metabolites and drug safety: when is it a cause for concern? Expert Rev. Clin. Pharmacol. 2008;1(4):515–531. doi: 10.1586/17512433.1.4.515. [DOI] [PubMed] [Google Scholar]
- Kaul S.C., Ishida Y., Tamura K., Wada T., Iitsuka T., Garg S., Kim M., Gao R., Nakai S., Okamoto Y., Terao K., Wadhwa R. Novel methods to generate active ingredients-enriched Ashwagandha leaves and extracts. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0166945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivisto P., Kilpelainen I., Rasanen I., Adler I.D., Pacchierotti F., Peltonen K. Butadiene diolepoxide- and diepoxybutane-derived DNA adducts at N7-guanine: a high occurrence of diolepoxide-derived adducts in mouse lung after 1,3-butadiene exposure. Carcinogenesis. 1999;20(7):1253–1259. doi: 10.1093/carcin/20.7.1253. [DOI] [PubMed] [Google Scholar]
- Lewis-Wambi J.S., Kim H.R., Wambi C., Patel R., Pyle J.R., Klein-Szanto A.J., Jordan V.C. Buthionine sulfoximine sensitizes antihormone-resistant human breast cancer cells to estrogen-induced apoptosis. Breast Cancer Res. 2008;10(6):R104. doi: 10.1186/bcr2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H.J., Drescher M., Mulzer J. How stable are epoxides? A novel synthesis of epothilone B. Angew. Chem. Int. Ed. Engl. 2000;39(3):581–583. [PubMed] [Google Scholar]
- Mirjalili M.H., Bonfill M., Cusido R.M., Palazón J. Steroidal lactones from Withania somnifera, an ancient plant for novel medicine. Molecules. 2009;14:2373–2393. doi: 10.3390/molecules14072373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra L.C., Singh B.B., Dagenais S. Scientific basis for the therapeutic use of Withania somnifera (ashwagandha): a review. Altern. Med. Rev. 2000;5(4):334–346. [PubMed] [Google Scholar]
- Mukherjee Pulok K, Banerjee Subhadip, Biswas Sayan, Das Bhaskar, Kar Amit, Katiyar C K. Withania somnifera (L.) Dunal - Modern perspectives of an ancient Rasayana from Ayurveda. J Ethnopharmacol. 2021;264:113157. doi: 10.1016/j.jep.2020.113157. [DOI] [PubMed] [Google Scholar]
- Navarro V.J., Barnhart H., Bonkovsky H.L., Davern T., Fontana R.J., Grant L., Reddy K.R., Seeff L.B., Serrano J., Sherker A.H., Stolz A., Talwalkar J., Vega M., Vuppalanchi R. Liver injury from herbals and dietary supplements in the U.S. Drug-Induced Liver Injury Network. Hepatology. 2014;60(4):1399–1408. doi: 10.1002/hep.27317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro V.J., Khan I., Bjornsson E., Seeff L.B., Serrano J., Hoofnagle J.H. Liver injury from herbal and dietary supplements. Hepatology. 2017;65(1):363–373. doi: 10.1002/hep.28813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obach R.S., Kalgutkar A.S., Soglia J.R., Zhao S.X. Can in vitro metabolism-dependent covalent binding data in liver microsomes distinguish hepatotoxic from nonhepatotoxic drugs? An analysis of 18 drugs with consideration of intrinsic clearance and daily dose. Chem. Res. Toxicol. 2008;21(9):1814–1822. doi: 10.1021/tx800161s. [DOI] [PubMed] [Google Scholar]
- Palliyaguru D.L., Singh S.V., Kensler T.W. Withania somnifera: from prevention to treatment of cancer. Mol. Nutr. Food Res. 2016;60(6):1342–1353. doi: 10.1002/mnfr.201500756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.B., Rao N.J., Hingorani L.L. Safety assessment of Withania somnifera extract standardized for Withaferin A: acute and sub-acute toxicity study. J. Ayurveda Integr. Med. 2016;7(1):30–37. doi: 10.1016/j.jaim.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips C.A., Ahamed R., Rajesh S., George T., Mohanan M., Augustine P. Comprehensive review of hepatotoxicity associated with traditional Indian Ayurvedic herbs. World J. Hepatol. 2020;12(9):574–595. doi: 10.4254/wjh.v12.i9.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N., Gota V., Gulia A., Hingorani L., Agarwal M., Puri A. Safety and pharmacokinetics of Withaferin-A in advanced stage high grade osteosarcoma: a phase I trial. J. Ayurveda Integr. Med. 2020;11(1):68–72. doi: 10.1016/j.jaim.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, M., Jogee, P., Agarkar, G., d.S. CA, 2016. Anticancer activities of Withania somnifera: Current research, formulations, and future perspectives., Pharm. Biol. 54 189-197. [DOI] [PubMed]
- Stickel F., Shouval D. Hepatotoxicity of herbal and dietary supplements: an update. Arch. Toxicol. 2015;89(6):851–865. doi: 10.1007/s00204-015-1471-3. [DOI] [PubMed] [Google Scholar]
- Tang M.S., Wang H.T., Hu Y., Chen W.S., Akao M., Feng Z., Hu W. Acrolein induced DNA damage, mutagenicity and effect on DNA repair. Mol. Nutr. Food Res. 2011;55(9):1291–1300. doi: 10.1002/mnfr.201100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi M.K., Panda P., Sethi K.K., Jana S. Metabolite profiling in Withania somnifera roots hydroalcoholic extract using LC/MS, GC/MS and NMR Spectroscopy. Chem. Biodivers. 2017;14(3) doi: 10.1002/cbdv.201600280. [DOI] [PubMed] [Google Scholar]
- Troupin A.S. Dose-related adverse effects of anticonvulsants. Drug Saf. 1996;14(5):299–328. doi: 10.2165/00002018-199614050-00004. [DOI] [PubMed] [Google Scholar]
- Vaishnavi K., Saxena N., Shah N., Singh R., Manjunath K., Uthayakumar M., Kanaujia S.P., Kaul S.C., Sekar K., Wadhwa R. Differential activities of the two closely related withanolides, Withaferin A and Withanone: bioinformatics and experimental evidences. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0044419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani T.H., Chakrabarty A., Shibata N., Yamazaki H., Guengerich F.P., Chowdhury G. The dihydroxy metabolite of the teratogen thalidomide causes oxidative DNA damage. Chem. Res. Toxicol. 2017;30(8):1622–1628. doi: 10.1021/acs.chemrestox.7b00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani T.H., Surendran S., Jana A., Chakrabarty A., Chowdhury G. Quinone-based antitumor agent sepantronium bromide (YM155) causes oxygen-independent redox-activated oxidative DNA damage. Chem. Res. Toxicol. 2018;31(7):612–618. doi: 10.1021/acs.chemrestox.8b00094. [DOI] [PubMed] [Google Scholar]
- Wani T.H., Surendran S., Mishra V.S., Chaturvedi J., Chowdhury G., Chakrabarty A. Adaptation to chronic exposure to sepantronium bromide (YM155), a prototypical survivin suppressant is due to persistent DNA damage-response in breast cancer cells. Oncotarget. 2018;9(71):33589–33600. doi: 10.18632/oncotarget.26096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigend F., Ahlrichs R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. PCCP. 2005;7(18):3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- Widodo N., Kaur K., Shrestha B.G., Takagi Y., Ishii T., Wadhwa R., Kaul S.C. Selective killing of cancer cells by leaf extract of Ashwagandha: identification of a tumor-inhibitory factor and the first molecular insights to its effect. Clin. Cancer Res. 2007;13(7):2298–2306. doi: 10.1158/1078-0432.ccr-06-0948. [DOI] [PubMed] [Google Scholar]
- Widodo N., Priyandoko D., Shah N., Wadhwa R., Kaul S.C. Selective killing of cancer cells by Ashwagandha leaf extract and its component Withanone involves ROS signaling. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten J.M. Adverse drug reactions: part I. South Med. J. 2010;103(10):1025–1028. doi: 10.1097/SMJ.0b013e3181f0c866. quiz 1029. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Xia L., Liao X., He Q., Zhao M.X., Truhlar D.G. Extrapolation of high-order correlation energies: the WMS model. PCCP. 2018;20(43):27375–27384. doi: 10.1039/c8cp04973d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.