Abstract
Single-cell capillary electrophoresis mass spectrometry (CE-MS) is a promising platform to analyze cellular contents and probe cell heterogeneity. However, current single-cell CE-MS methods often rely on offline microsampling processes and may demonstrate low sampling precision and accuracy. We have recently developed an electrospray-assisted device, spray-capillary, for low-volume sample extraction. With the spray-capillary, low-volume samples (pL–nL) are drawn into the sampling end of the device, which can be used directly for CE separation and online MS detection. Here, we redesigned the spray-capillary by utilizing a capillary with a <15 μm tapered tip so that it can be directly inserted into single cells for sample collection and on-capillary CE-MS analysis. We evaluated the performance of the modified spray-capillary by performing single-cell microsampling on single onion cells with varying sample injection times and direct MS analysis or online CE-MS analysis. We have demonstrated, for the first time, online sample collection and CE-MS for the analysis of single cells. This application of the modified spray-capillary device facilitates the characterization and relative quantification of hundreds of metabolites in single cells.
Graphical Abstract
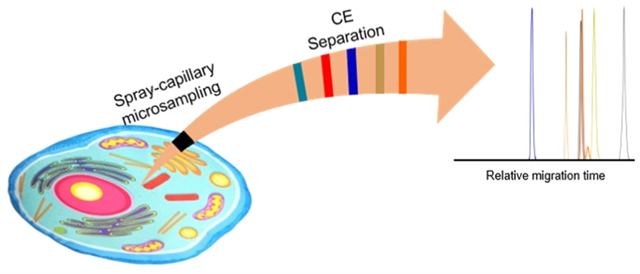
INTRODUCTION
Capillary electrophoresis mass spectrometry (CE-MS) has been one of the most promising techniques for single-cell analysis during the past decade1–6 due to inherent advantages such as low sample loading amounts (nL to μL level),7,8 high separation power,9,10 and high sensitivity with online MS detection.11,12 Many studies have successfully applied single-cell CE-MS approaches to address biological questions such as cell heterogeneity, cell signaling, and intrinsic cellular variation.4 However, there is still room for improvement of the overall performance of the single-cell CE-MS, especially from the perspective of developing efficient microsampling methods for CE-MS platforms.13
Microdissection-based microsampling methods have been routinely used for isolating single-cell samples followed by offline manual manipulation.14 This sampling method involves the isolation of the cell of interest from the tissue using surgical tools followed by further sample preparation steps. The Sweedler group has extensively applied microdissection methods for CE-MS analysis of single cells including the metabolite analysis of A. californica neuron cells, which demonstrated one of the best limits of detection (LODs) of neurotransmitter metabolites among their peers at that time.15 The same group later shared a detailed protocol of their single-cell CE-MS platform using microdissection-based microsampling and a sheath–liquid style interface for CE-MS.16 They further improved the performance of the platform by incorporating preconcentration approaches together with CE separation.17,18 Sun et al. performed single-cell proteomics using microdissection followed by CE with an electrokinetically pumped sheath–liquid interface on isolated blastomeres from different stages of Xenopus laevis embryos,19 and a similar strategy was used for metabolic profiling on kidney tissue.20 Although the microdissection-based microsampling methods are mature and straightforward, relatively lengthy offline sample preparation impedes the throughput of these methods.
Nanopipettes have been heavily used during ambient ionization MS microsampling for single-cell analysis.21–30 In these studies, nanopipettes (e.g., μm-scale tip diameter) were directly inserted into single cells and the cellular contents were analyzed by MS without further separation. Nanopipettes can also be coupled offline with CE-MS analysis of single cells. Nanopipettes (e.g., μm-scale tip diameter) can be directly inserted into single cells for microsampling of offline single-cell CE-MS analysis.5,16,31 The Nemes group has constructed a microprobe CE-MS platform for single-cell metabolomics, which consists of a pump-based nanopipette microsampling platform (microprobe) and a CE-MS platform with a sheath–liquid interface.32,33 They successfully used this platform to probe the cell heterogeneity of the left and right blastomeres in Xenopus laevis embryos (8-cell stage).34 In addition, they reported that microsampling using this platform permits in situ analysis of individual blastomeres, which allows for the monitoring of metabolites in the same blastomere at different stages of Xenopus laevis embryonic development.32 Kawai et al. utilized a similar platform for ultrasensitive single HeLa cell metabolomics, in which the vacuum driving force was manually provided by a syringe and a thin-walled sheathless interface was utilized for the CE-MS platform.35 Using Kawai’s platform, a total of 40 metabolites were identified and 20 amino acids were quantified. Further modifications to the nanopipette methods including electrophysiological methods such as the patch clamp have also been incorporated to improve the microsampling process.36 Meanwhile, other microsampling strategies (e.g., electroosmotic flow (EOF),37 microfluid chip,2,38,39 microjunction extraction40) that have been coupled with CE-MS may potentially be applied to single-cell analyses in the future.
The microsampling approaches used in the aforementioned platforms were generally coupled off-line with the CE-MS platform in the single-cell analysis; however, a microsampling technique capable of online coupling with CE-MS analysis may provide more sensitive and quantitative sampling for single cells. Therefore, our group has modified our spray-capillary device for direct microsampling of single cells followed by online CE-MS separation and analysis. The spray-capillary utilizes electrospray ionization (ESI) as the driving force for ultralow-volume sample handling (pL–nL) through a long capillary (e.g., 50 cm).41 The spray-capillary is capable of performing quantitative, low-volume sample injection (e.g., ∼15 pL/s injection flow rate) by utilizing the pressure difference produced when electrospray is formed at the ESI end of the spray-capillary. The spray-capillary can directly serve as a CE capillary for online separation and MS detection and eliminates sample loss that may occur during offline sample handling steps of previously reported microsampling methods. Here, we redesigned the sample inlet end of the spray-capillary so it can be directly inserted into a single cell (e.g., onion cell) for microvolume sample injection and CE-MS analysis. We investigated the sample injection flow rate and the performance of sample extraction from single onion cells using different sample injection times to control the amount of cellular material injected. We also performed online single-cell CE-MS analysis using the modified spray-capillary device. Overall, we demonstrated that the modified spray-capillary device can perform microsampling from single cells with on-capillary CE-MS analysis. To our knowledge, this represents the first online single-cell CE-MS platform.
EXPERIMENTAL SECTION
Chemicals and Reagents.
Red onion (Allium cepa) was purchased from a local supermarket. All chemicals such as formic acid, HPLC grade water, n-butanol, acetonitrile, hydrofluoric acid (HF), and standard metabolites (malic acid, phenylalanine, and glucose) were purchased from Sigma-Aldrich (St. Louis, MO) unless noted otherwise. The fused silica capillary with the tapered tip (SilicaTip, 50 cm length, 50 μm I.D., and 15 μm tip size) was purchased from New Objective (Littleton, MA).
Fabrication and Operation of the Modified Spray-Capillary Device.
A long pretapered-tip capillary purchased from New Objective was used to fabricate the modified spray-capillary. We used the tapered-tip end as the sample inlet, and the opposite end was fabricated for the MS inlet similarly to previously described.41 First, the MS end of the capillary was slightly trimmed using a Shortix capillary column cutter (Agilent, San Jose, CA). Next, the polymer coating of the MS end (∼3 cm) was removed using flame heating. The porous segment of the MS end was produced through chemical etching using HF while water was flowed through the column to prevent etching of the inner wall of the capillary.41,42 To introduce the water flow into the capillary through the small sample end (e.g., 15 μm tip) without damage, we inserted the tapered tip into a flow generator41 that used nitrogen gas to produce the flow through the modified spray-capillary. The water flow was adjusted to approximately 0.2 μL/min, and the exposed segment of the MS end was submerged in 48% HF for approximately 90 min to reduce the thickness of the capillary wall to 5–10 μm so that efficient electric contact through the porous wall to produce ESI was achieved. (Caution: HF should be handled properly in a ventilated chemical hood.) After etching, the capillary was thoroughly washed with water to remove residual HF. An inverted microscope was used throughout etching to monitor the thickness of the capillary. The sheathless interface for MS coupling was assembled by inserting the capillary through a PEEK tee union, which was then pushed through a short, stainless-steel tube (4 cm, 1/16 in. O.D., 0.04 in. I.D.) until about 1.5 cm of etched fused silica emerged from the metal tube, as detailed previously.41 The performance of the etched tip on the spray-capillary was evaluated primarily by its ability to produce stable electrospray. Briefly, a microscope was used to observe the formation of the electrospray and, for each etched tip, the injection flow rate was measured in triplicate to ensure reproducible microsampling.
Microsampling from Single Cells.
Onion cells were chosen as the model system because onion is relatively easy to handle and has been extensively used in previous studies.43–46 An inverted microscope (AmScope FMA050, CA) was used to monitor the microsampling process. The epidermal tissue was freshly peeled from the red onion and placed on a piece of parafilm. Two glass slides were used to fix the parafilm on the platform of the inverted microscope. Several drops of 0.1% FA were used to cover the target area to facilitate sample extraction. The modified spray-capillary was prefilled with column liquid (0.1% FA) until a droplet was observed at the tip. Then, the sample inlet end was moved to the targeted cell using an in-house assembled micromanipulator consisting of two lifting platforms, one probe holder, and one tube rack for the column liquid vials placed under the inverted microscope. The microsampling process was triggered by initiating the ESI voltage on the MS end (2–4 kV). Sample injection volume was controlled by varying the sample injection time. After the microsampling process was finished, the electrospray voltage was turned off and the sample inlet end was moved back to the column liquid. Between the analysis of individual samples, the spray-capillary device was flushed using 0.1% FA for 10 min. In addition, a blank run (column liquid) was included between sample analyses to ensure no significant contaminants were detectable from the previous sampling. New cells were probed for each sample injection. If the sample inlet becomes clogged with cell debris, the capillary can be flushed with organic solution (0.1% FA in ACN) at 40 psi or the sample inlet end can be sonicated in organic solution (50% MeOH in HPLC grade water).
Pressure-Based Sample Elution and MS Detection.
After single-cell sample injection using the modified spray-capillary, the same flow generator used in the etching step was utilized to apply pressure-based sample elution41 for the follow-up MS detection. Generally, for direct infusion analysis, the extracted cellular content was eluted at 3 psi (from an in-house nitrogen source). However, fine adjustments were made (2.5–5 psi) to maintain a stable electrospray as determined by monitoring the MS signal. 0.1% FA was used as the elution buffer. The MS end of the spray-capillary was used directly to produce the ESI to introduce the sample to an Orbitrap Elite mass spectrometer (Thermo Scientific, San Jose, CA) for MS analysis. The temperature of the inlet capillary was 275 °C. MS scans were acquired at a resolving power of 120 000 at m/z = 400. The AGC target was 1 × 106 with a maximum ion injection time of 1000 ms. The m/z scan range is 150–2000. All data files were collected in profile mode. Online MS/MS scans were acquired using ITMS with collisional induced dissociation (CID) at a normalized collision energy setting of 35%. The AGC for MS/MS was 3 × 104, and the max ion injection time was 50 ms with 3 microscans.
Single-Cell CE-MS Analysis.
The spray-capillary device was subjected to a preconditioning procedure before single-cell CE-MS analysis. The device was flushed using 1 M NaOH, HPLC grade water, and 0.1% FA in water (background electrolyte, BGE), consecutively. The device was flushed with each solvent for 20 min. Before single-cell microsampling, the spray-capillary was filled with BGE. The sample inlet was inserted into an intact onion cell, and the microsampling process was triggered by applying the ESI voltage (2–4 kV) on the MS end of the sprayc-apillary. After microsampling, the sample inlet end was moved to the sample vial containing BGE solution for online CE separation. A metal wire was inserted into the vial containing the sample inlet end of the 50 cm spray-capillary,41 and high voltage (15 kV) was applied to the metal wire to facilitate CE separation; 2.8 or 2.9 kV was used to produce electrospray at the MS end. (Note: to prevent electrical hazards, all the other conductive parts in the system were grounded as needed and proper warning signs were displayed.) The MS end of the spray-capillary was directly coupled in front of the Orbitrap Elite mass spectrometer inlet for MS analysis. The temperature of the inlet capillary was 275 °C. MS scans were acquired at the resolving power of 120 000 at m/z = 400. The AGC target was 1 × 106 with a maximum ion injection time of 1000 ms. The m/z scan range was 150–2000. All data files were collected in profile mode. Online MS/MS scans were acquired using ITMS with collisional induced dissociation (CID) at a normalized collision energy setting of 35%. The AGC for MS/MS was 3 × 104, and the max ion injection time was 50 ms with 3 microscans. After sample analysis, the spray-capillary device was flushed using the BGE buffer (0.1% FA). A blank run (BGE buffer) was included between sample analyses to ensure no significant contaminants were detectable from previous sampling as needed.
Data Analysis.
All raw MS data sets were converted to mzML files using MSConvert47 and processed using an in-house developed python software. For each spray-capillary MS analysis using pressure-based sample elution, all mass spectra collected during the elution window were summed, and peaks were extracted to produce a table of m/z values and intensities for each data set. To reduce random noise, all m/z values with an intensity lower than 500 were removed. For single-cell CE-MS analysis, similar mass feature lists were generated including elution time information.
The generated mass feature lists from different injection conditions (e.g., various injection times) were further compared using GraphPad Prism (San Diego, CA). A mass measurement accuracy of 10 ppm was used to determine shared m/z values from different injection conditions or different runs under the same injection conditions. Significantly changed features were characterized using volcano plots.
For putative metabolite identification, mass features were searched using METLIN,48 an online metabolite-searching tool (https://metlin.scripps.edu/landing_page.php?pgcontent=mainPage). The mass measurement accuracy was set to 10 ppm. An onion metabolite database (a total of 280 reported onion metabolites) was created by compiling previously reported onion metabolites49 and the human metabolome database (filtered with “onion metabolites”)50,51 for manual cross-checking of the metabolites putatively identified in this work to previous literature (Supplementary List 1).
RESULTS AND DISCUSSION
Performance Evaluation of the Modified Spray-Capillary Device.
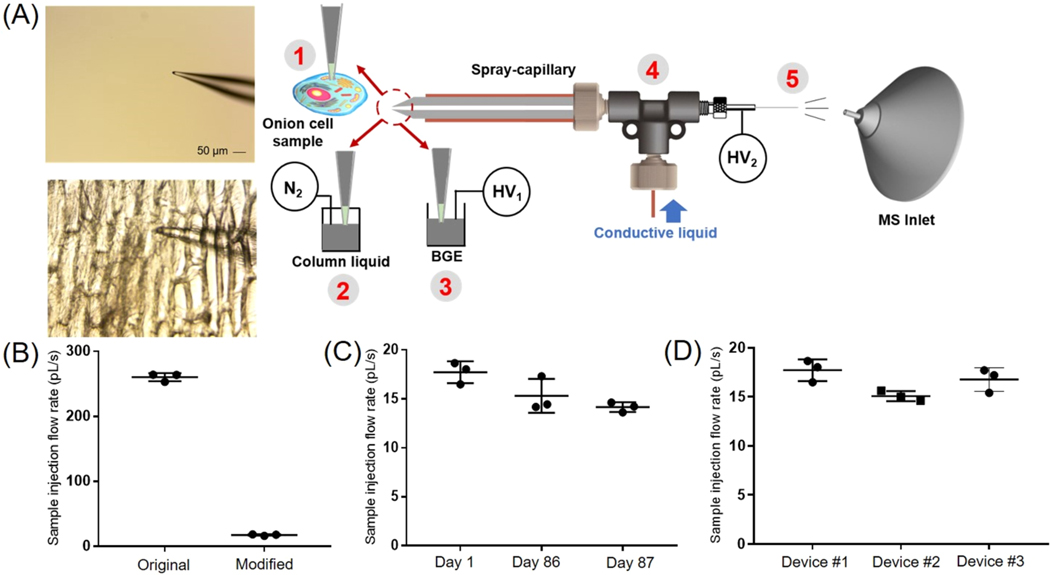
We first evaluated the sample injection flow rate of the modified spray-capillary device (Figure 1A) using a digital camera as previously described.41 The sample injection flow rate was likely to be altered from the original spray-capillary device due to the tapered tip at the sample inlet end (i.e., the inner diameter of the sample inlet tip decreased from 50 μm to less than 15 μm). In this study, H2O was used as the column liquid while an organic solution (90% n-butanol in water) was used to mimic the sample so that a clear boundary could be monitored and recorded using the camera. The microsampling process was filmed and used to calculate the sample injection flow rate (Movie S1). The average sample collection flow rate was 17.72 pL/s with an RSD value of 6.27%. This value is approximately 15-fold smaller than the original spray-capillary without the tapered tip41 (Figure 1B). Furthermore, we evaluated the durability of the device 86 days after fabrication and found that the performance of the modified spray-capillary device was relatively unchanged after ∼3 months (15.30 pL/s with an RSD value of 11.35%; Figure 1C). Likewise, the day-to-day reproducibility of the same spray-capillary was evaluated, and the average sample injection flow rate was similar on Day 1, Day 86, and Day 87.
Figure 1.
Single-cell MS analysis using the modified spray-capillary. (A) Schematic of the modified spray-capillary device for single-cell microsampling (A. cepa): The laser pulled sample inlet end (1) for penetrating single cells. The extracted cell content was either (2) directly analyzed by MS using an in-house built pressure generator or (3) used to perform online CE-MS analysis in the spray-capillary. The sheathless interface (4) with a porous segment (5) was utilized for ESI-based microsampling and MS coupling. (B) Sample injection flow rates were compared between the original spray-capillary and the modified spray-capillary. The day-to-day performance (C) and batch-to-batch reproducibility (D) were also evaluated. Error bars represent relative standard deviation.
It has been reported that different batches of nanopipette tips (sample inlet end) may result in inconsistent results for single-cell sampling, which can make reproducible sample handling challenging.22 To determine how batch-to-batch variation might affect the performance of the modified spray-capillary, we compared the microsampling reproducibility of three separate spray-capillaries. The average sample injection flow rates for these three devices are 17.72 pL/s (RSD = 6.27%), 15.07 pL/s (RSD = 3.43%), and 16.8 pL/s (RSD = 7.21%) (Figure 1D). The high reproducibility of our single-cell spray-capillary devices is likely due to the consistency of the commercial laser-pulled sampling ends (SilicaTip Emitter) purchased from New Objective.
Single-Cell Microsampling Coupled with MS Detection.
As detailed in the Experimental Section, we utilized an inverted microscope to monitor the single-cell sampling process. To avoid damaging the tip during the sampling process, we used a piece of parafilm to hold the freshly peeled onion epidermal tissue on top of a glass microscope slide that was fixed onto the tray of an inverted microscope. A proof-of-principle injection was performed in triplicate with three different onion cells. The single-cell microsampling process was triggered by initiating a 3 kV ESI voltage for 5 s on the MS end of the spray-capillary. After sample injection, the sample inlet end of the spray-capillary was inserted into the ESI buffer and the extracted cellular content was pressure eluted through the spray-capillary for online MS detection (Figure S1A). The average intensity of the total ion chromatograms (TICs) in the elution window was determined as the area under the curve (AUC), which was 1.45 × 108 with an RSD value of 10.69%. With the same elution pressure, the elution time of the single-cell components was highly reproducible (3.01 ± 0.13 min). The summed mass spectra for these three replicates are displayed in Figure S1B. In total, 1895 mass features were detected in these three replicate injections and 822 of these mass features were detected in all three runs.
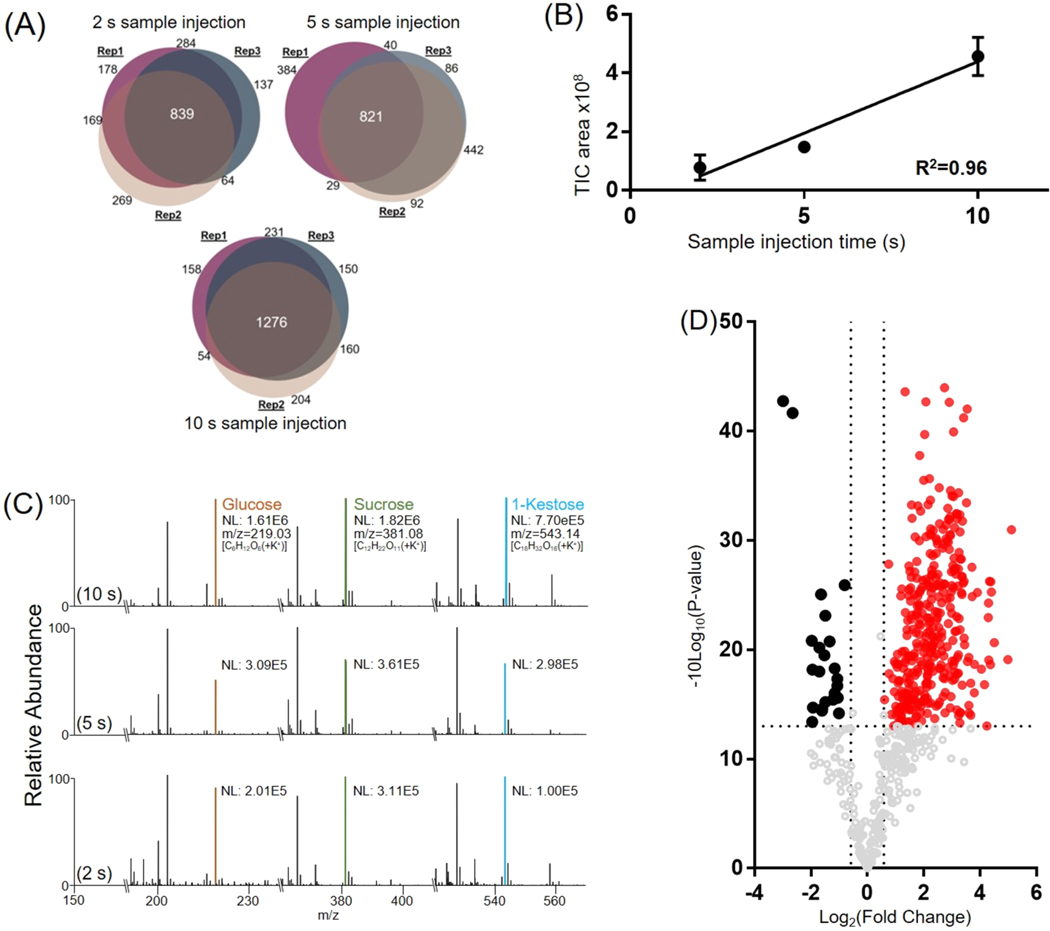
We evaluated the performance of the modified spray-capillary utilizing different sample injection times (2, 5, and 10 s) to determine if the device can be applied to relative quantification of single-cell metabolites (Figure 2). A 3 kV electrospray voltage was used to collect the sample for all runs, and experiments were performed in triplicate for each sample injection time. According to the previously measured sample injection flow rate, the estimated sample extraction volume is about 32–160 pL at 3 kV ESI voltage (average volume of onion epidermal cells is ∼30 nL52,53). We evaluated the reproducibility for each sample injection time using triplicate analysis. Using a 2 s sample collection duration, a total of 1939 mass features were detected; 837 mass features were detected in all three runs, and 1354 mass features were detected in at least two runs. At 5 s, a total of 1895 mass features were detected; 822 mass features were detected in all three runs, and 1330 mass features were detected in at least two runs. At 10 s, a total of 2248 mass features were detected; 1276 mass features were detected in all three runs, and 1706 mass features were detected in at least two runs (Figure 2A). Some runs appeared to be more similar than others performed under the same conditions demonstrating more similar mass feature identifications. This variation in reproducibility may be due to natural variation in onion cell contents and probing different regions of the cell. Still, we are encouraged by the relative metabolite quantification for single cells using our modified spray-capillary.
Figure 2.
Sample extraction from A. cepa single-cell samples using the modified spray-capillary device. (A) Venn diagrams comparing mass features (intensity > 500) in the elution window among triplicate trials for each sample injection time (2, 5, and 10 s). (B) Calibration curve demonstrating intensity as a function of sample injection time (n = 3 for each sample injection time). The error bars represent the relative standard deviation. (C) Representative mass spectra from different sample injection times (2, 5, and 10 s) labeled with putatively identified metabolites (NL: normalized intensity). (D) Volcano plot for comparing detected shared mass features between the 2 and 10 s sample injection times (red and black: fold change > 1.5, p value < 0.05).
We then studied the effect of sample injection time on the total amount of cellular material injected from a single cell. A good linear relationship (R2 = 0.96) was observed when correlating the area under the curves (AUCs) of the total ion chromatograms (TICs) to the sample injection time (Figure 2B), indicating that the amount of cellular material injected has linear proportionality with the sample injection time. We averaged the MS scans over the entire elution range for each injection time (Figure 2C) and found that the detected mass feature patterns were similar, but the intensity increased linearly with injection time. For example, simple sugars (e.g., monosaccharides or oligosaccharides) are commonly observed with high abundance in onion cells.49 Three high-intensity peaks that were observed in all MS spectra were confirmed to be monosaccharides or oligosaccharides that are known carbohydrates in onions. The detected m/z of 219.0281 is the potassium adduct ion of the monosaccharide (glucose or fructose). Its sodium adduct ion was also detected with high intensity. Similarly, the potassium adduct ions of a disaccharide (m/z = 381.0825, possibly sucrose) and a trisaccharide (m/z = 543.1560, possibly 1-kestose) were detected together with their sodium adduct ions. The intensities of these detected ions increased with increased injection time.
We further compared the intensities of all the shared mass features in the 2 and 10 s injection time runs using a volcano plot (Figure 2D) and found that a majority of the detected mass features (357 in 620) have significantly higher abundance (p value < 0.05 and fold change > 1.5) in the 10 s injection time run. This further suggests that a higher quantity of cellular material was injected into the spray-capillary with longer injection times. We note that some detected mass features have similar abundance or decreased abundance with longer injection time. To further evaluate the source of these mass features, the buffer was analyzed (n = 3) to detect any impurities that may be mistaken for onion metabolites (Figure S2). Unique mass features detected in the buffer were selected and compared with mass features detected in the onion sample. The volcano plot (Figure 2D) revealed 163 mass features that were not upregulated with increased injection volume. These 163 mass features were compared with the buffer control, and 122 (∼75%) mass features were detected in both the onion sample and the buffer control. This result confirmed that these mass features may originate from impurities in the buffer and instrumental noise.
Putative Metabolite Identification in Single Onion Cell Analysis.
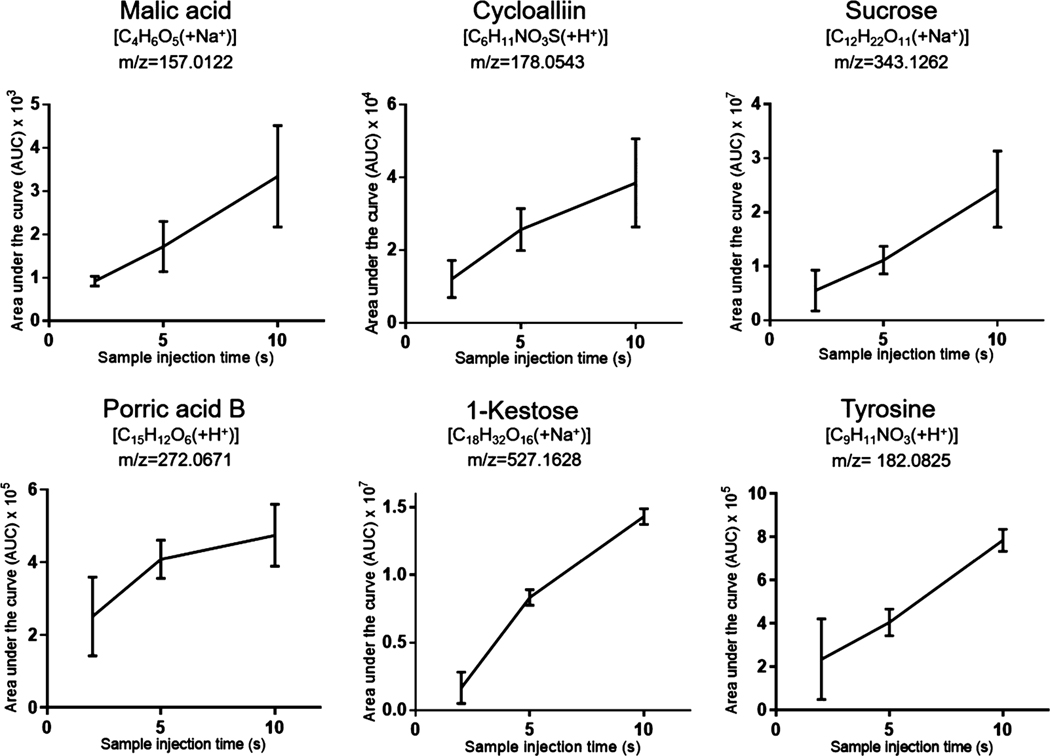
A total of 3247 mass features were detected from nine pressure elution runs (2, 5, and 10 s injection time, triplicate runs for each injection time). Among these mass features, 510 were detected in all 9 runs. We searched these 510 shared mass features using the METLIN48 database for putative metabolite identifications and found possible hits for 441 mass features. Among them, 80 putative METLIN identifications with low m/z values were confirmed by manual cross-checking with previously reported onion metabolites (Supplementary List 2). Also, among the 367 upregulated mass features between the 10 and 2 s injection time runs, 62 were confirmed to be onion metabolites by cross-checking with previous literature (Supplementary List 3). We selected 6 putatively identified metabolites that have been reported previously in onion samples (A. cepa) and plotted their detected intensities across different injection times (Figure 3). The amount of metabolite injected is proportional to the injection time as demonstrated by the linearity between peak intensity and injection time. The relatively large standard deviations may result from variation attributed to cell-to-cell heterogeneity. Our results suggested that the modified spray-capillary is capable of performing relative metabolite quantitation analysis at the single-cell level, which may provide better insights into biomarker characterization and cell-to-cell variation. Because some low-abundance metabolites may not be detected in shorter sample injection times (2 and 5 s), we also searched the 1276 mass features detected in the 10 s injection time trials using METLIN and putatively assigned 978 potential metabolites. 148 of these metabolites were also previously reported onion metabolites.
Figure 3.
Single-cell MS analysis (onion cells) with different sample injection times using the modified spray-capillary. Triplicate injections were performed for each injection time, and the error bars represent the relative standard deviation between the measurements.
Spray-Capillary-Based Single-Cell CE-MS Analysis.
We have previously demonstrated that the spray-capillary can directly serve as a CE separation capillary after microsampling for online separation and quantitative MS detection without additional devices. Here, we performed single-cell CE-MS analysis using the modified spray-capillary. An example CE-MS analysis of a single onion cell using the modified spray-capillary is shown in Figure 4 (3 kV injection ESI voltage and 10 s sample injection time, ∼160 pL of injected volume). Most of the detected metabolites were eluted between 3 and 16 min. Representative metabolites with their extracted ion electropherograms (EIEs) were selected to evaluate the CE separation performance. Many detected metabolites were efficiently separated with an intensity range from 1.89 × 103 to 7.81 × 105, demonstrating a good dynamic range of detection by the single-cell CE-MS analysis using the modified spray-capillary. A single-cell CE-MS experiment using our modified spray-capillary takes approximately 15–20 min.
Figure 4.
Example extracted ion electropherograms (EIEs) of putatively identified metabolites using spray-capillary-based single-cell (onion cells) CEMS analysis (NL: normalized intensity).
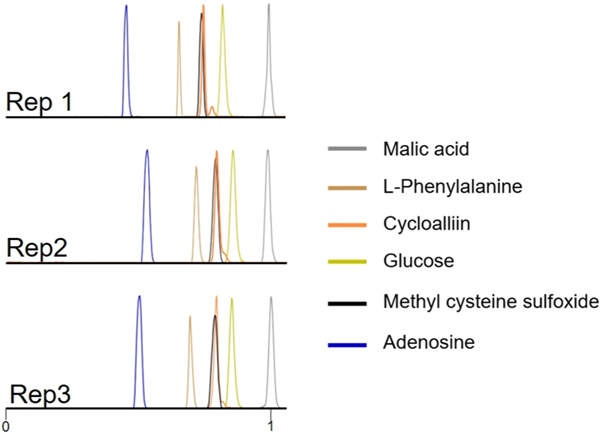
Figure 5 shows extracted EIEs from replicate trials using 3 kV as the electrospray voltage and 10 s as the single-cell sample injection time. Six putatively identified metabolites were evaluated in three individual single-cell CE-MS analyses on the basis of their relative migration times and measured intensities (Table S1). The migration times of all metabolites were normalized according to the migration time of malic acid in the corresponding run. The RSD of the relative migration times was less than 10% for each putatively identified metabolite. The measured peak areas of the same metabolites in different cells were also compared. RSDs of the peak areas for 5 of the 6 metabolites were less than 20%, which demonstrates the cell-to-cell reproducibility using the modified spray-capillary with online CE-MS analysis. However, a slightly higher variation of the peak area RSD of phenylalanine was observed (RSD = 35.3%), which may be attributed to cell-to-cell metabolite variation. Interestingly, a shoulder peak was repeatedly observed on the extracted ion electropherogram for cycloalliin (putatively assigned). It has been reported that the cycloalliin possesses a structural isomer54 (e.g., alliin, isoalliin) in onion, and the shoulder peak may be a result of the separation of the isomers.
Figure 5.
Representative extracted ion electropherograms (EIEs) of putatively identified metabolites in replicated single-cell CE-MS runs (malic acid [C4H6O5 + Na+], L-phenyalanine [C9H11NO2 + H+], cycloalliin [C6H11NO3S + H+], glucose [C6H12O6 + Na+], methylcysteine sulfoxide [C4H9NO3S + H+], and adenosine [C10H13N5O4 + H+]). The migration time of each metabolite was normalized with the migration time of malic acid.
In total, 2407 mass features were detected from replicate single-cell CE-MS experiments. Among them, 1620 were putatively assigned by searching METLIN and 163 were previously reported onion metabolites (Supplementary list 1 and 4). The mass features detected here that were not identified as onion metabolites may be the product of instrument or ambient chemical noise or impurities from the buffer solutions used to facilitate the microsampling and separation processes. We further compared the identification result between CE-MS analysis and direct infusion and observed 349 overlapped mass features. Among these 349 mass features, 324 were putatively assigned using METLIN. To confirm that the spray-capillary CE-MS device can be used to confidently identify metabolites, a selection of standard metabolites (phenylalanine, glucose, and malic acid) was analyzed, and the results were compared with the putative identification results from the onion cells. Relative migration times (normalized by the migration time of malic acid) are listed in Table S2. The relative migration time for the phenylalanine ion [C9H11NO2(+H+)] in the single onion cell CE-MS/MS analysis was 0.703 (RSD = 4.97%), which is comparable with the measured migration time for the standard phenylalanine sample, 0.754 (RSD = 0.80%). Similarly, the relative migration time for the measured glucose ion [C6H12O6(+Na+)] in the single onion cell CE-MS/MS analysis is 0.849 (RSD = 2.41%), and the measured migration time for the standard glucose sample is 0.872 (RSD = 1.18%). Additionally, the online CE-MS/MS fragmentation was used to confirm the identity of the metabolites in both the standard mixture and the onion cell (Figure S3). Additional MS/MS examples from the single-cell CE-MS/MS analysis are listed in Figure S4. The proof-of-principle experiments suggested that the CE migration time and online CE-MS/MS results can improve the identification confidence in single-cell spray-capillary-based CE-MS analysis.
Overall, our results demonstrate that metabolites can be reproducibly separated using the on-capillary CE-MS separation, and the single-cell metabolite profiling is similar between different single onion cells. Also, with the elimination of any intermediate steps between microsampling and sample injection for CE separation, the whole workflow has been largely simplified with minimized sample loss. The CE-MS provides separation of metabolites prior to MS detection and allows additional mass features to be detected with increased signal-to-noise ratios when compared with direct infusion experiments. However, some mass features were only detected in the direct infusion experiments. These compounds may bind more strongly to the column and may not elute within the detection window. In addition, a bare capillary was used in our proof-of-principle experiments, and some metabolites may adsorb to the negatively charged capillary wall, which may result in loss of signal.
CONCLUSION
Here, for the first time, we demonstrated a platform for online single-cell CE-MS analysis using the modified spray-capillary device. The simple and straightforward transition from microsampling using the modified spray-capillary device to CE separation largely eliminates potential contamination and/or sample loss introduced by intermediate or offline sample handling steps. The robustness of the proposed device was investigated by measuring sample injection flow rate directly after the device was fabricated and after ∼3 months. Batch-to-batch reproducibility was also studied by comparing the injection flow rate of three independently fabricated devices. Microsampling on single onion cells with MS detection was evaluated using various sample injection times, and single onion cell CE-MS analysis was successfully demonstrated. The modified spray-capillary device separated and putatively identified hundreds of metabolites; however, the system currently relies on manual operation, which requires caution throughout the process since the laser pulled tip (sample inlet end) as well as the MS end (porous interface) are both damage prone. Future improvements to spray-capillary-based single-cell CE-MS analysis may be made to improve CE separation performance, for example, the application of coating material (e.g., linear polyacrylamide, polyethylenimine)55,56 and the increase in capillary length or electric field strength.57 High quality coating processes can also help avoid the formation of unwanted adduct ions in the CE-MS analysis. Despite these weaknesses, the modified spray-capillary electrospray-assisted microsampling and CE-MS analysis platform was capable of metabolite analysis at the single-cell level. The throughput of this system can be elevated by automation and motorization. More importantly, with smaller laser pulled tips (8 μm or smaller), the simple design of the spray-capillary device may allow it to be applied to different types of single-cell samples (e.g., human cell lines), and the device holds great potential for the field of single-cell metabolomics.
Supplementary Material
ACKNOWLEDGMENTS
This work was partly supported by grants from NIH NIAID R01AI141625, NIH NIGMS R01GM118470, OCAST HR16125, and NIH NIAID 2U19AI062629.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c04624.
Supplementary List 1: onion metabolite database generated from previous reports; Supplementary Lists 2–4: putatively identified metabolites (XLSX)
Movie S1: microsampling process (MP4)
Triplicate sample extraction from A. cepa single cells; comparison between blank and onion sample; MS/MS spectra; CE separation; characterization of metabolites analysis; relative migration time (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.0c04624
The authors declare no competing financial interest.
Contributor Information
Lushuang Huang, Department of Chemistry and Biochemistry, University of Oklahoma, Norman, Oklahoma 73019, United States; .
Mulin Fang, Department of Chemistry and Biochemistry, University of Oklahoma, Norman, Oklahoma 73019, United States; .
Kellye A. Cupp-Sutton, Department of Chemistry and Biochemistry, University of Oklahoma, Norman, Oklahoma 73019, United States .
Zhe Wang, Department of Chemistry and Biochemistry, University of Oklahoma, Norman, Oklahoma 73019, United States; .
Kenneth Smith, Department of Arthritis and Clinical Immunology, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, United States.
Si Wu, Department of Chemistry and Biochemistry, University of Oklahoma, Norman, Oklahoma 73019, United States.
REFERENCES
- (1).Duncan KD; Fyrestam J; Lanekoff I Analyst 2019, 144 (3), 782–793. [DOI] [PubMed] [Google Scholar]
- (2).DeLaney K; Sauer CS; Vu NQ; Li LJ Molecules 2019, 24 (1), 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Yin L; Zhang Z; Liu Y; Gao Y; Gu J. Analyst 2019, 144 (3), 824–845. [DOI] [PubMed] [Google Scholar]
- (4).Evers TMJ; Hochane M; Tans SJ; Heeren RMA; Semrau S; Nemes P; Mashaghi A. Anal. Chem. 2019, 91 (21), 13314–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Onjiko RM; Moody SA; Nemes P. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (21), 6545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nemes P; Knolhoff AM; Rubakhin SS; Sweedler JV ACS Chem. Neurosci. 2012, 3 (10), 782–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lubeckyj RA; McCool EN; Shen XJ; Kou Q; Liu XW; Sun LL Anal. Chem. 2017, 89 (22), 12059–12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chen DY; Shen XJ; Sun LL Analyst 2017, 142 (12), 2118–2127. [DOI] [PubMed] [Google Scholar]
- (9).Gahoual R; Leize-Wagner E; Houze P; Francois YN Rapid Commun. Mass Spectrom. 2019, 33 (S1), 11–19. [DOI] [PubMed] [Google Scholar]
- (10).Gomes FP; Yates JR 3rd Mass Spectrom. Rev. 2019, 38 (6), 445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sun LL; Zhu GJ; Zhang ZB; Mou S; Dovichi NJ J. Proteome Res. 2015, 14 (5), 2312–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Faserl K; Sarg B; Kremser L; Lindner H. Anal. Chem. 2011, 83 (19), 7297–305. [DOI] [PubMed] [Google Scholar]
- (13).Altelaar AF; Heck AJ Curr. Opin. Chem. Biol. 2012, 16 (1–2), 206–13. [DOI] [PubMed] [Google Scholar]
- (14).Cecala C; Sweedler JV Analyst 2012, 137 (13), 2922–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lapainis T; Rubakhin SS; Sweedler JV Anal. Chem. 2009, 81 (14), 5858–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nemes P; Rubakhin SS; Aerts JT; Sweedler JV Nat. Protoc. 2013, 8 (4), 783–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Liu JX; Aerts JT; Rubakhin SS; Zhang XX; Sweedler JV Analyst 2014, 139 (22), 5835–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liao HW; Rubakhin SS; Philip MC; Sweedler JV Anal. Chim. Acta 2020, 1118, 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sun L; Dubiak KM; Peuchen EH; Zhang Z; Zhu G; Huber PW; Dovichi NJ Anal. Chem. 2016, 88 (13), 6653–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sanchez-Lopez E; Kammeijer GSM; Crego AL; Marina ML; Ramautar R; Peters DJM; Mayboroda OA Sci. Rep. 2019, 9 (1), 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Liu RM; Zhang GW; Sun M; Pan XL; Yang ZB Anal. Chim. Acta 2019, 1064, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yin R; Prabhakaran V; Laskin J. Anal. Chem. 2018, 90 (13), 7937–7945. [DOI] [PubMed] [Google Scholar]
- (23).Fujii T; Matsuda S; Tejedor ML; Esaki T; Sakane I; Mizuno H; Tsuyama N; Masujima T. Nat. Protoc. 2015, 10 (9), 1445–1456. [DOI] [PubMed] [Google Scholar]
- (24).Nakashima T; Wada H; Morita S; Erra-Balsells R; Hiraoka K; Nonami H. Anal. Chem. 2016, 88 (6), 3049–3057. [DOI] [PubMed] [Google Scholar]
- (25).Zhang XC; Wei ZW; Gong XY; Si XY; Zhao YY; Yang CD; Zhang SC; Zhang XR Sci. Rep. 2016, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang XC; Zang QC; Zhao HS; Ma XX; Pan XY; Feng JX; Zhang SC; Zhang RP; Abliz Z; Zhang XR Anal. Chem. 2018, 90 (16), 9897–9903. [DOI] [PubMed] [Google Scholar]
- (27).Esaki T; Masujima T. Anal. Sci. 2015, 31 (12), 1211–1213. [DOI] [PubMed] [Google Scholar]
- (28).Saha-Shah A; Green CM; Abraham DH; Baker LA Analyst 2016, 141 (6), 1958–65. [DOI] [PubMed] [Google Scholar]
- (29).Saha-Shah A; Karty JA; Baker LA Analyst 2017, 142 (9), 1512–1518. [DOI] [PubMed] [Google Scholar]
- (30).Xu MC; Pan RR; Zhu Y; Jiang DC; Chen HY Analyst 2019, 144 (3), 954–960. [DOI] [PubMed] [Google Scholar]
- (31).Morris CA; Friedman AK; Baker LA Analyst 2010, 135 (9), 2190–202. [DOI] [PubMed] [Google Scholar]
- (32).Onjiko RM; Portero EP; Moody SA; Nemes P. Anal. Chem. 2017, 89 (13), 7069–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Choi SB; Zamarbide M; Manzini MC; Nemes PJ Am. Soc. Mass Spectrom. 2017, 28 (4), 597–607. [DOI] [PubMed] [Google Scholar]
- (34).Onjiko RM; Morris SE; Moody SA; Nemes P. Analyst 2016, 141 (12), 3648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kawai T; Ota N; Okada K; Imasato A; Owa Y; Morita M; Tada M; Tanaka Y. Anal. Chem. 2019, 91 (16), 10564–10572. [DOI] [PubMed] [Google Scholar]
- (36).Aerts JT; Louis KR; Crandall SR; Govindaiah G; Cox CL; Sweedler JV Anal. Chem. 2014, 86 (6), 3203–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Duncan KD; Lanekoff I. Anal. Chem. 2019, 91 (12), 7819–7827. [DOI] [PubMed] [Google Scholar]
- (38).Li X; Zhao S; Hu H; Liu YM J. Chromatogr A 2016, 1451, 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kleparnik K. Electrophoresis 2013, 34 (1), 70–85. [DOI] [PubMed] [Google Scholar]
- (40).Comi TJ; Makurath MA; Philip MC; Rubakhin SS; Sweedler JV Anal. Chem. 2017, 89 (14), 7765–7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Huang L; Wang Z; Cupp-Sutton KA; Smith K; Wu S. Anal. Chem. 2020, 92 (1), 640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Moini M. Anal. Chem. 2007, 79 (11), 4241–4246. [DOI] [PubMed] [Google Scholar]
- (43).Shrestha B; Patt JM; Vertes A. Anal. Chem. 2011, 83 (8), 2947–55. [DOI] [PubMed] [Google Scholar]
- (44).Misra BB; Assmann SM; Chen S. Trends Plant Sci. 2014, 19 (10), 637–46. [DOI] [PubMed] [Google Scholar]
- (45).Shrestha B; Vertes A. Anal. Chem. 2009, 81 (20), 8265–71. [DOI] [PubMed] [Google Scholar]
- (46).Gong XY; Zhao YY; Cai SQ; Fu SJ; Yang CD; Zhang SC; Zhang XR Anal. Chem. 2014, 86 (8), 3809–3816. [DOI] [PubMed] [Google Scholar]
- (47).Kessner D; Chambers M; Burke R; Agus D; Mallick P. Bioinformatics 2008, 24 (21), 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Smith CA; O’Maille G; Want EJ; Qin C; Trauger SA; Brandon TR; Custodio DE; Abagyan R; Siuzdak G. Ther. Drug Monit. 2005, 27 (6), 747–751. [DOI] [PubMed] [Google Scholar]
- (49).Bottcher C; Krahmer A; Sturtz M; Widder S; Schulz H. Metabolomics 2017, 13 (4), 35. [Google Scholar]
- (50).Wishart DS; Feunang YD; Marcu A; Guo AC; Liang K; Vazquez-Fresno R; Sajed T; Johnson D; Li C; Karu N; Sayeeda Z; Lo E; Assempour N; Berjanskii M; Singhal S; Arndt D; Liang Y; Badran H; Grant J; Serra-Cayuela A; Liu Y; Mandal R; Neveu V; Pon A; Knox C; Wilson M; Manach C; Scalbert A. Nucleic Acids Res. 2018, 46 (D1), D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yannai S. Dictionary of Food Compounds with CD-ROM: Additives, Flavors, and Ingredients; Chapman & Hall/CRC: Boca Raton, FL, 2004; pp 1–1784. [Google Scholar]
- (52).Kafle K; Xi X; Lee CM; Tittmann BR; Cosgrove DJ; Park YB; Kim SH Cellulose 2014, 21 (2), 1075–1086. [Google Scholar]
- (53).Suslov D; Verbelen J-P; Vissenberg KJ Exp. Bot. 2009, 60 (14), 4175–4187. [DOI] [PubMed] [Google Scholar]
- (54).Breu W. Phytomedicine 1996, 3 (3), 293–306. [DOI] [PubMed] [Google Scholar]
- (55).Zhu G; Sun L; Dovichi NJ Talanta 2016, 146, 839–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Han X; Wang Y; Aslanian A; Fonslow B; Graczyk B; Davis TN; Yates JR 3rd J. Proteome Res. 2014, 13 (12), 6078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Onjiko RM; Portero EP; Moody SA; Nemes P. Anal. Chem. 2017, 89 (13), 7069–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.