Abstract
The impact of the human microbiome on health and disease is of utmost importance and has been studied intensively in recent years. Microbes promote immune system development and are essential to the production and absorption of nutrients for the host but are also implicated in disease pathogenesis. Particularly, bacterial biofilms have long been recognized as contributors to chronic infections and diseases in humans. However, our understanding of how the host responds to the presence of biofilms, specifically the immune response to biofilms, and how this contributes to disease pathogenesis is limited. This review aims to highlight what is known about biofilm formation and in vivo models available for the biofilm study. We critique the contribution of biofilms to human diseases, focusing on the lung diseases, cystic fibrosis and chronic obstructive pulmonary disease, and the gut diseases, inflammatory bowel disease and colorectal cancer.
Keywords: Immune response, biofilms, lung, gut
Introduction
The human body is home to a diverse community that spans most microbial kingdoms. The best-studied colonizers of the human body are bacteria, estimated to be ~3.8 x 1013 in total, with the highest density residing in the gastrointestinal tract1. Recent studies have only begun to unravel the intricate relationship(s) between this microbial community and its human host, informing us on how vital microbes are in health and in disease2,3.
Host-microbe interactions occur via individual planktonic bacteria or through complex polymicrobial communities, including a subset termed biofilms. Bacterial biofilms are implicated in several human infections including, but not limited to, periodontitis, endocarditis, otitis media, osteomyelitis, bacterial vaginosis, and device-associated infections (e.g., catheters)4-10. Because data on viruses, parasites and/or fungi in biofilm biology in human disease are limited, we will focus specifically on bacterial biofilms. This review highlights current knowledge about host responses and bacterial biofilms, focusing on biofilms described in cystic fibrosis (CF), chronic obstructive pulmonary disease (COPD), inflammatory bowel diseases (IBD), and colorectal cancer (CRC).
General Concepts: Bacterial Biofilms
As planktonic bacteria colonize a niche, they proliferate and adhere to a surface to form microcolonies (small aggregates of bacteria covered in a simple matrix). Microcolonies may progress to larger aggregates, called biofilms, classically defined in vitro as matrix-enclosed bacterial (and/or archaeal) populations adherent to each other and/or to surfaces or interfaces9. The bacterial-enclosing, self-secreted, hydrated matrix in vitro is composed of extracellular polymeric substances (EPS), which include polysaccharides, proteins, nucleic acids, lipids, and typically contributes >90% of the mass of biofilms11. When considering human mucosal-associated bacterial biofilms, the EPS may also include host proteins, mucopolysaccharides, nucleic acids and even whole host cells8. Biofilms are found throughout nature, can be adherent to biological and nonbiological surfaces, and allow for the exchange of nutrients with their environment9,12,13. Interestingly, biofilms appear to contribute to more than 80% of human soft and hard tissue infections9,10. Despite their prevalence in human infections, investigations defining how biofilms in vivo directly impact the human host remain limited.
Biofilm formation enables the survival and persistence of bacterial cells living in challenging environments, such as those that may be encountered in the human lung and gut. As such, the physiology and activity of bacteria within a biofilm greatly differs from that of planktonic bacteria4,6. For instance, mice inoculated with a slurry made from human biofilm-positive colon tumors displayed increased differential expression (RNA-seq) in genes related to sporulation, antibiotic synthesis pathways, and bacterial secretion systems, compared to mice inoculated with slurries from biofilm-negative colonoscopy biopsies14. Similarly, a meta-analysis of functional in silico predictions by PICRUSt using 16S rRNA amplicon sequence data comparing biofilm-positive and biofilm-negative human CRC tissues revealed an enrichment in the presence of genes related to peptidoglycan biosynthesis, cytoskeletal proteins, antimicrobial biosynthesis, and a decrease in genes related to flagellar assembly in biofilm-positive CRCs15. Importantly, biofilms on biological surfaces differ from those on nonbiological surfaces. For example, Streptococcus pyogenes biofilms exhibit 50% discordance in gene expression between tissue-associated and abiotic-associated biofilms16. Such changes in physiology occur, in part, due to inter-bacteria communication, referred to as quorum sensing, and lead to downregulation of genes and activities needed in their planktonic state, as well as upregulation of genes and activities needed for the adherent biofilm state4,6. In vivo, bacterial biofilm function is likely further impacted by the host response(s) to these microbial communities.
There are several advantages for bacteria embedded in biofilms, including a suitable environment for horizontal gene transfer, specifically of antimicrobial-resistance genes, making biofilm eradication by host defenses or antibiotics less effective. This evasion of anti-microbials is first impacted by the EPS, which serves as a protective barrier to the biofilm-embedded bacteria in a manner similar to how the mucus layer protects epithelia in the human body. The mixed bacterial populations--some viable and active, some dormant and stationary, and some resistant to antimicrobial agents-- of biofilms further promotes host-defense evasion. Biofilms facilitate preservation of persister bacteria, increasing the potential virulence of the biofilm community4,7,8,17. Persister bacteria are inactive phenotypic variants of bacteria that can survive in the presence of high levels of antibiotics. Evidence of this phenotype has largely been derived from in vitro data, and limited studies have demonstrated the phenotypic switch to persister cells in vivo18,19. Although biofilms are relatively stable growth environments, they are likely not permanent. Shear forces, such as those that may be experienced as luminal fluid moves through the gastrointestinal tract, can cause bacterial cell detachment followed by new niche colonization4,6. Exactly how (or if) the host overcomes biofilm formation and prevents long-term colonization of persister bacteria is unknown.
While the goal of this review is to discuss what is known about the relationship between biofilms and disease states in the lung and gut, it is important to note that not all biofilms are pathogenic. For example, in situ analyses of oral biofilm formation in healthy individuals detected a biphasic change in the biofilm-forming bacteria. These initial biofilm colonizers prevent direct contact of anaerobic bacteria that cause gingivitis. However, these anaerobic bacteria can begin to dominate after 48hrs. Thus, these results suggest that teeth should be brushed at least once within a 48hr period to prevent build-up of pro-gingivitis bacteria20. This scenario also suggests that biofilms may evolve from non-pathogenic to disease-initiating relatively quickly. As a second example, it has been suggested that bacteria form biofilms in the appendix, which may serve as a reservoir for commensal microbes to reseed the colon after an infection or antibiotic treatment21, however this evidence is still up for debate. Some studies also show that bacteria form biofilms on food particles passing through the gastrointestinal tract in order to facilitate particle breakdown, however there is debate on whether these attached bacterial communities are similar or different to unattached bacterial species present in the feces22-24.
Identification and Visualization of Biofilms
Bacterial biofilms were first observed by Antonie van Leeuwenhoek when investigating the differences between his teeth and another’s with poor oral hygiene7,25. In fact, much of the foundational work informing our understanding of biofilm formation, EPS molecules, as well as host responses to biofilms, has been performed in models of oral biofilms, characteristic of dental carries and periodontitis26, and in models of Staphylococcus aureus-related wound infections 27,28. Importantly, many of these studies use in vitro cultures of single microbes on abiotic surfaces; in vitro observations have been assumed to be occurring in vivo by the same organisms. Models of biofilms range from a variety of in vitro culturing systems, both static and dynamic, to ex vivo tissue studies, to invertebrate and mammalian models, and have been thoroughly reviewed by others26,29,30.
Identification and visualization of biofilms typically involve a variety of microscopy techniques, in combination with probes that target bacteria and EPS molecules. For example, early studies by Lawrence JR et al. used scanning confocal laser microscopy and viable negative fluorescence staining to visualize Pseudomonas aeruginosa, Pseudomonas fluorescens, and Vibrio parahaemolyticus biofilms in vitro31. This study found species-specific structural differences in biofilm formations; Pseudomonas biofilms, whether from P. aeruginosa or P. fluorescens, had higher cellular density at regions of attachment to glass slides and were less dense away from the attachment surface, while Vibrio biofilms were less dense at sites of attachment to glass slides and grew more dense away from the attachment site. Overall, they observed that biofilms were comprised of >70% extracellular matter, with only ~30% bacterial content31.
Lectins, or carbohydrate-binding proteins, have been used to visualize carbohydrate components of the EPS, such as mannopyranosyl- or glucopyranosyl-concanavalin A, or sialic acid- and N-acetyl-glucosamine-wheat germ agglutinin32,33. An important caveat is that, while lectins are useful to label biofilms developed under abiotic conditions, lectins do not distinguish microbial- vs. human-derived carbohydrates, and thus, cannot be used to determine the molecular origin of a biofilm matrix in vivo. Furthermore, lectins cannot differentiate between secreted carbohydrates and those attached to bacterial cell surfaces, and biofilms are defined classically by the presence of a secreted, non-attached EPS matrix. To overcome this lack of specificity, groups have generated bacteria expressing fluorescent EPS proteins primarily for use in in vitro studies. Methods to discriminate between specific host vs. bacterial EPS molecules in vivo have not yet been described for most bacterial biofilms. Thus, in vivo analyses of biofilm formation primarily utilize fluorescence, chromogenic and/or RNA- in situ hybridization (e.g., FISH, RNA-ISH) with oligonucleotide probes targeting the desired taxa, ranging from subspecies identification using specific genes, to kingdom level using conserved regions in the bacterial 16S rRNA gene (e.g., all-bacterial EUB338). These efforts are often complemented by scanning electron microscopy (SEM) to visualize the structure of the biofilms and their EPS 14,15,34-38.
Unique Features of Mucosal Biofilms in the Lung and Gut
Description of Lung and Colonic Mucus Layers
When considering biofilm locales, those found on mucosal surfaces, such as along the respiratory and gastrointestinal tracts, are of particular interest as mucosal tissues provide a multifaceted nutrient environment for biofilm formation. The mucus layers found in the lung and colon are comprised of polymeric mucin proteins that are heavily glycosylated. These mucins bind and trap water molecules to cover the epithelium and create a diffusion barrier that: 1) keeps the epithelial surface ‘clean’; and 2) protects the epithelium from hostile perturbations. In the normal, healthy lung, mucus bundles are swept across the epithelial surface by beating epithelial cilia to keep the epithelium ‘clean’. Under disease states such as CF and COPD, the sweeping function of the cilia is inhibited and the mucus bundles become attached to the epithelial surface, trapping bacteria that would normally be cleared. In contrast, as elegantly studied by the Hansson laboratory and others39-41, colonic epithelial cells lack beating cilia and are covered by two mucus layers, a non-attached, looser, outer mucus layer where bacteria often reside in a non-pathogenic state, and an inner, attached, stratified mucus layer that is mainly devoid of bacteria and limits contact between luminal contents and the epithelial surface. Under disease conditions in the gut, such as those in IBD and CRC, the inner mucus layer becomes penetrable and bacteria are able to establish direct contact with the underlying epithelium42 where they initiate host responses, leading to inflammation and other epithelial signaling that likely contribute to disease pathogenesis. When bacterial penetration of the inner stratified mucus layer meets certain criteria (see text below, Table 1), the term pathogenic biofilm becomes appropriate.
Table 1.
Comparison of mucosal biofilms in the lung and gut.
| Mucosal Biofilms | ||
|---|---|---|
| Definition: biofilms are matrix-enclosed bacterial populations adherent to each other and/or to surfaces or interfaces9. Thus, mucosal biofilms are biofilms adherent to mucosal surfaces. | ||
| Lung | Colon | |
| Bacterial composition | Single-species dominant, e.g. P. aeruginosa, NTHi | Polymicrobial, e.g. Bacteroidetes, Firmicutes, Proteobacteria, Fusobacteria |
| Extracellular matrix/EPS* components | Alginate (P. aeruginosa), lipooligosaccharide (NTHi) | Not described; likely very diverse and dependent on the bacterial composition |
| Bacterial quantification | CFU of cultured samples (in vitro), PNA-FISH (P. aeruginosa) | FISH (in vivo; Carnoy’s fixative is preferred over formalin as a more sensitive technique for biofilm detection): At ≥400X of a 10 X 10μm square field of a 5μm (500μm3) tissue section (=5x10−10 mL): 1 cell = 2X109 (bacteria/mL) 250 cells=1011 2,500 cells=1012 |
| Length spanning mucosa | Not determined | 50-200μM + within 1μM of epithelium |
| Verification of mucus invasion** | Electron microscopy, H&E, Gram stain, alcian blue | PAS, H&E, alcian blue, tissue autofluorescence, electron microscopy |
| References | 38,39 | 14, 15, 29-31 |
Abbreviations: EPS, extracellular polymeric substance; NTHi, non-typeable Hemophilus influenzae; CFU, colony-forming units; PNA-FISH, peptide nucleic acid fluorescent in situ hybridization; H&E, hematoxylin and eosin stain; PAS, Periodic acid-Schiff stain.
The mucus layers differ between the lung and colon mucosa; the lung has a single layer formed by mucus bundles, while the colon has an inner, sterile layer and an outer, non-sterile layer.
For biofilms to establish themselves in the lung and gut, they must overcome considerable, but still incompletely understood, host defense mechanisms. In both organs, shear forces from fluid flow by the airway surface liquid in the lung, and luminal contents in the gut, and rapid turnover of the host mucus and epithelium are major obstacles that limit extensive colonization and biofilm formation on these epithelial surfaces. Thus, the mucus layers in the lung and gut are designed to significantly limit invasive biofilm formation, and when these layers become dysfunctional, for example, due to altered host mechanisms or mucin-degrading bacteria, invasive bacterial biofilms can form. The biofilm-mucosa interface permits interactions between the adhesive biofilms and host epithelial proteins which lead to changes in both the microorganisms and the human host, often resulting in robust host responses and predicted disease worsening.
Defining Lung and Gut Mucosal Biofilms
There is a significant body of literature describing the presence of lung-associated bacterial biofilms using in vitro models as well as assessment of potential in vivo biofilms using patient samples43,44. Lung mucosal biofilms are unique because they are often dominated by a single bacterial species, such as P. aeruginosa or non-typeable Haemophilus influenzae (NTHi) and in contrast to multispecies colonic biofilms, have allowed for greater development of species-specific biofilm tools. For example, NTHi isolates cultured in vitro can be evaluated for biofilm formation by immune-transmission electron microscopy using a monoclonal antibody targeting a H. influenzae-specific lipooligosaccharide antigen45. Similarly, antibodies specific to alginate have been used to study biofilm-forming P. aeruginosa, as described by Bjarnsholt T et al46. These investigators examined paraformaldehyde-preserved autopsy tissues, explanted lungs, and sputum smears and cultures of CF patients using a combination of hematoxylin-eosin (H&E) staining, Gram and alcian-blue staining, and immunofluorescence imaging with P. aeruginosa-specific peptide nucleic acid (PNA) FISH probes and alginate-specific antibodies46. Using these techniques, they found that only mucoid P. aeruginosa aggregated in the respiratory bronchioles and alveoli prior to antibiotic treatment in autopsy samples. In contrast, in explanted lung samples taken after intensive antibiotic treatment, both mucoid and nonmucoid P. aeruginosa sputum aggregates were detected in the conductive zone, which includes the trachea, bronchi, and conducting bronchioles. The authors concluded that the conductive zone serves as a safe space where mucoid biofilms are protected from antibiotics and host immune defenses.
As per the canonical biofilm definition (bacterial aggregates enclosed in a secreted polymeric matrix and attached to an abiotic or biotic surface), biofilms have long been described in chronic CF and COPD infections. However, biofilms are rarely observed in direct contact with the airway mucosa, nor linked to mucosal function [likely due to infrequent acquisition of lung biopsies for in vivo study or difficulty with co-culturing bacteria with lung epithelial cells in vitro]. While we consider biofilms in direct contact with the mucosa (Table 1) as most likely to impact disease, it is important to note that the term “biofilm” is used differently in the CF and COPD literature, and often applied to bacterial aggregates from cultured sputum and BAL fluid samples assessed in vitro without direct demonstration of mucus-invasive lung mucosal biofilms and/or induction of functional mucosal changes.
In contrast to single-species dominant biofilms on lung mucosa, in vivo gut mucosal biofilms are complicated by their polymicrobial nature (i.e. more than one bacterial species) with species that display highly variable EPS molecules47. This concept arose from seminal studies by Swidsinski et al. in which bacterial group-specific FISH was performed on Carnoy’s-fixed human colon biopsies, along with H&E or alcian blue staining for mucus layer and epithelial visualization, to assess bacterial quantification and localization relative to the epithelial surface. Invasive biofilms were defined as carpets of bacteria that spanned at least 50μm and observed within 1 μm of the epithelium in the inner mucus layer. Biofilms contained at least 0.4 X 109 bacteria/mL within a 400μm3 tissue section (Table 1)34. Importantly, these structural criteria rigorously define colonic biofilms in contrast with the reductive description of lung biofilms as bacterial aggregates.
In an approach similar to Swidsinski et al34, the Sears laboratory defined colonic biofilms as a dense formation of at least 2x109 bacteria/mL that spanned at least 150-200μm and within 1μm of the underlying epithelium (Table 1). Invasion of the inner mucus layer was confirmed by periodic acid-Schiff (PAS) mucus staining of an adjacent tissue section from the same block14,15,35,36. Alternatively, both bacterial and mucus presence can be confirmed in parallel on a single slide by utilizing FISH probes and the inherent autofluorescence properties of the colon mucus layer. While colon bacterial- and host-specific EPS probes would be useful to directly test the host vs. bacterial origins of the EPS in colon biofilms, such tools are not available. However, polymicrobial biofilms detected on the human colon mucosa nearly always include classes of encapsulated organisms such as Bacteroidetes that produce surface polysaccharides that may also serve as EPS molecules (with Bacteroides fragilis alone displaying 8 capsular polysaccharides47). Thus, it is very probable that the invasive bacterial biofilms observed on human colon tissues are encased in a matrix that includes EPS from both bacterial and host origins. SEM images also help in detection of biofilm EPS presence35. Furthermore, based on the parameters described in Table 1, there is currently stronger evidence for bacterial biofilms on the gut mucosa compared to the lung mucosal surface.
Models of Pathogenic Biofilm Formation
There is an abundance of evidence showing the association of biofilms with human diseases10. However, whether bacterial biofilms cause disease, or result from the diseased state, is an ongoing field of research. Dongari-Bagtzoglou proposed two pathogenesis models that focus on host inflammatory responses. The first model proposes that mucosal biofilms induce a robust inflammatory response by the host, that leads to inflammation-associated tissue damage, and further biofilm proliferation. This type of model is proposed for P. aeruginosa-related biofilms in CF patients and will be described further below. In contrast, the second model of biofilm-related pathogenesis proposes that EPS molecules inhibit initial mucosal inflammation and phagocytic cell function, allowing the biofilm to propagate and stabilize8. For example, NTHi EPS phosphorylcholine decreases early inflammatory events to allow for biofilm outgrowth in models of otitis media48. Regardless, in both models, chronic inflammation and tissue damage ensue that benefit biofilm growth. Because bacteria:host interactions play a similar, and central, role in the pathophysiology of the lung and gut, and because of several other commonalities including the structure of their epithelium and protective mucus layers, herein, we chose to focus on biofilms in these two tissues. Prior reviews have discussed similarities between these two anatomical sites as contributing to common microbial and immune responses in their respective inflammatory diseases5,49-52. We will explore biofilm formation in select chronic diseases of the lung, CF and COPD, and of the gut, IBD and CRC. We will focus on characterization of lung and gut biofilms in human and experimental studies that probe inflammatory responses to biofilms and/or to disease-associated bacteria known to form biofilms in vitro.
Mucosal Biofilms in Lung Diseases
The lower respiratory tract is kept free of bacteria and inhaled particulate matter by the beating of epithelial cilia that sweep the airway surface liquid and mucus up towards the larynx42. Despite this, there is significant evidence suggesting the establishment of a diverse, but low bacterial biomass, lung microbiome, even in the normal host. There has been a recent upsurge in lung microbiome-related articles with the development of culture-independent techniques; according to PubMed (August 21, 2019), in 2010 only 16 papers were published on the lung microbiome, compared to 276 in 2018. Several studies have used 16S rRNA amplicon sequencing of healthy subjects, smokers, and/or those with respiratory diseases (e.g., asthma, COPD) to demonstrate that the phyla Bacteroidetes and Firmicutes dominate the healthy airway microbiome53-56. Some members of the normal respiratory tract microbiome may impact the pathogenesis of lung diseases. Since there has yet to be a clear definition of invasive biofilms in the lungs, we will review below the evidence of the host immune response to specific bacteria and their association with the lung diseases CF and COPD.
Cystic Fibrosis (CF) Biofilms
CF is an inherited genetic disease caused by mutations in the CF transmembrane conductance regulator (CFTR) gene, which encodes a Cl− channel expressed in epithelial tissues. The disease affects secretory tissues, such as those that secrete mucus, sweat, and digestive enzymes. In the lung, defunct CFTR results in a periciliary liquid layer that is not properly hydrated, and thus becomes very thick and viscous57. As the mucus builds up, it clogs the airways, compresses ciliary beating, impairing mucus clearance and facilitating persistent colonization of pathogenic bacteria that lead to chronic infections, inflammation, and eventually respiratory failure. Bacteria well-known to contribute to biofilm formation in vitro and to trigger infection in CF include S. aureus, H. influenzae, and P. aeruginosa, among others45,58. S. aureus and H. influenzae colonize CF patients early in life and induce the initial inflammatory environment, characterized by a robust neutrophil infiltration. P. aeruginosa is an aerobic, Gram-negative rod bacterium highly associated with late-stage CF disease. Mucoid P. aeruginosa produce a highly viscous polysaccharide called alginate that is thought to enable biofilm formation and evasion of host immune responses59. Doggett et al. showed that cultures of P. aeruginosa isolates from CF patients displayed a mucoid phenotype and induce biofilm formation58-60. However, successive sub-culturing of P. aeruginosa results in loss of the mucoid phenotype, suggesting the need of additional in vivo features to induce and maintain the mucoid characteristic61,62. Other studies have detected P. aeruginosa biofilms in vivo on patient tissues using alginate-specific probes46. In addition to studying CF patient samples, in vivo animal models of CF-biofilm infections have often used intratracheal or intranasal routes of infection with P. aeruginosa immobilized on agar-beads29,63.
P. aeruginosa biofilm-mediated infections in vivo are associated with leukocyte mobilization and bacterial-specific antibody production to clear the pathogen59,64-66. Multiple studies have established that P. aeruginosa triggers a predominant Th2 immune response in the human CF lung and in murine CF models64,67,68. CF patients chronically infected with P. aeruginosa have higher levels of interleukin (IL)-3, IL-4, and secreted immunoglobulin A (sIgA; Th2 markers) and lower interferon gamma (IFNγ; Th1 marker), compared to non-chronically infected patients65,69-71. Th2 responses have also been observed in vitro and in murine mouse models of CF in response to challenge with P. aeruginosa and/or its gene products72,73.
These studies suggest that approaches to enhance a Th1 intrapulmonary immune response could be beneficial to CF patients and help overcome P. aeruginosa biofilm-associated infections. The anti-inflammatory medication that has been most studied in people with CF is ibuprofen74. Other studies have assessed the effectiveness of inhaled corticosteroids which show a decrease in disease progression but with side-effects75. However, ongoing clinical trials are testing the action of additional anti-inflammatory drugs that, for example, activate the cannabinoid receptor type 2 (CB-2 agonist; JBT-101), inhibit the production of leukotriene B4 (CTX-4430) or inhibit neutrophil elastase (POL6014) among others76. Nonetheless, studies describing CF-related immune responses do not always assess biofilm formation and focus on targeting specific pathogens, such as P. aeruginosa, despite the implications of biofilms in inducing immune responses. Thus, the field would benefit by performing studies where biofilms and immune responses in the underlying epithelium are examined in parallel.
Chronic Obstructive Pulmonary Disease (COPD) Biofilms
Similar to CF, the chronic lung diseases that contribute to COPD, including emphysema, chronic bronchitis, and non-reversible asthma, result in reduced airflow, local and systemic inflammation, and several comorbidities58,77. Consistent with loss of the normal cilia-mediated mucus transport in the lung, proteomic analysis of BAL fluid from COPD patients and visualization of Carnoy’s-fixed CF and COPD lung tissues revealed a shift towards a more colonic-mucus-like environment, promoting more bacterial-epithelial contact in the stagnate mucus78. While the pathophysiology of COPD is very different from CF, the contribution of the lung microbiome appears to be a common denominator in these diseases, specifically the impact of NTHi and P. aeruginosa.
At baseline COPD airways in humans are highly inflamed, with reports of increased neutrophils, eosinophils, mast cells, dendritic cells, macrophages, and T and B lymphocytes79. Microbial infections, such as those induced by NTHi, can further exacerbate the inflammatory environments of COPD respiratory tissues. H. influenzae is a Gram-negative coccobacillus, and is the most common bacterium associated with COPD, colonizing 60% of COPD patients whereas P. aeruginosa only colonizes 6% of COPD patients58,79. NTHi can form biofilms that are resistant to host-clearance responses, but many NTHi biofilm observations are from in vitro studies and models of infection, such as otitis media in the chinchilla80,81. Thus, the relevance of NTHi biofilms to COPD in vivo is still up for debate. Nevertheless, to better understand the contribution of NTHi/biofilms to COPD pathogenesis, experimental models of COPD use mice treated with elastase or chronically exposed to cigarette smoke and then intratracheally injected with NTHi, resulting in morphological and functional changes similar to those seen in human COPD lungs29,82-84.
The data regarding NTHi biofilm formation, or any other bacteria, are sparse and therefore the relevance to lung (and/or systemic) immune responses in COPD remains to be established. NTHi has been shown to be associated with an imbalanced Th2/Th1 immune response79, including increased sIgA production85 and decreased Th1 responses86,87. In NTHi-infected COPD patients there are systemic increases in T regulatory cells, myeloid-derived suppressor cells, and exhausted effector T cells compared to healthy controls, with inhibited proliferation of NTHi-specific effector T cells88. Other studies have described innate immune responses to NTHi, including Toll-like Receptor (TLR)-2 and 4 activation, nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB)-dependent inflammatory responses79,89,90, and NOD-like receptor protein 3 (NLRP3) inflammasome activation91. Overall, the causative relationship between the mucosal immune response and the formation of pulmonary biofilms in COPD remains to be established.
Better characterization of pulmonary mucosal biofilms in patient samples and better delineation of the compositional and functional state of pulmonary mucosal biofilms would assist in understanding the pathological contribution of biofilms to CF and COPD pathogenesis.
Mucosal Biofilms in Gut Diseases
Physiologically, the gut is a hostile environment for biofilm formation because of the high turnover and shedding of the epithelium, the shear forces of luminal content movement down the intestinal tract, and the high viscosity of the mucus layer7. However, there is evidence that dysbiosis and an abnormal inflammatory environment predispose the colonic mucosa to biofilm formation6. As discussed earlier, colonic mucus is comprised of two distinct layers: a loose outer mucus layer full of bacteria41 and a stratified inner mucus layer, firmly attached to the underlying epithelium, devoid of bacteria39,40. The finely tuned crosstalk between the microbiota, epithelium, and immune system is critical for the maintenance of the mucosal architecture and the organization of the mucus layer92.
The recent description and characterization of biofilms in gut diseases, described hereafter, allowed for the striking observation of colonic biofilms in a significant proportion of specimens from patients with gut disorders including IBD and CRC compared to healthy controls 34,93,35. Whether the presence of colonic mucosal biofilms in the “healthy” human gut indicates disease onset is unknown as only cross-sectional human studies of colon mucosal biofilms presently exist. Longitudinal monitoring of patients harboring colonic biofilms will be necessary in order to evaluate their causative contribution to gut diseases.
Inflammatory Bowel Disease (IBD) Biofilms
IBD is classified as two chronic inflammatory disorders of the gastrointestinal tract, Crohn’s disease (CD) and ulcerative colitis (UC). CD can affect any region of the gut, while UC is chronic inflammation specifically in the colon. Most IBD studies have sought to unravel the relationships between the gut microbiota, aberrant immune responses, and host genetics94. IBD is characterized by disruption of the mucus layer and reduced epithelial barrier function that together permit excessive exposure of luminal bacteria to the host immune system42,94,95.
In the seminal study by Swidsinski et al.34, the spatial organization and bacterial composition of gut mucosal tissues (formalin- and Carnoy’s-fixed) were compared between patients with IBD, colitis, IBS and healthy controls using fluorescent bacterial-specific probes. Mucosal biofilms (at least 0.4x109 bacteria/mL, 50μm in length) were found in 95% of IBD patients and 95% of patients with self-limiting colitis compared to 35% of healthy control biopsies (n=20). The bacterial density of mucosal biofilms (Table 1) was higher in patients with IBD. UC bacterial density (0.26±0.31 x 1010 bacteria/mL), specifically, was ~35-fold less than CD (9.1±20.2 x 1010/mL; UC:), with even greater differences between CD and the other groups studied. Importantly, the integrity of invasive mucosal biofilms is poorly preserved by formalin fixation, and therefore a milder fixation procedure using Carnoy’s fixative better preserves the mucus layer to analyze mucosal biofilms. The Bacteroides fragilis group contributed to >60% of the biofilm mass in IBD patients, establishing B. fragilis group biofilms as a hallmark of IBD. It must be noted that the B. fragilis group probe96 targeted four Bacteroides species: B. fragilis, B. vulgatus, B. eggerthii, and B. thetaiotamicron; today, these species are detected independently using specific probes. In an additional study from Swidsinski et al. comparing the intestinal mucus barrier and bacterial infiltration between normal and inflamed colon, approximately 50% of the mucosa from UC patients was invaded by bacteria compared to <10% of the mucosa from healthy control biopsies93. Studies by Johansson et al.97 used several mouse models of colitis and reported similar invasion of the mucus by bacterial aggregates, although these were not formally quantitated as biofilms according to the criteria discussed in Table 1. They did observe significantly increased inflammation by histological analysis of the Il-10−/− mouse model of colitis in association with bacterial invasion of the mucus layer97. Overall, these studies in IBD patients and mouse models of colitis demonstrate the proinflammatory consequences of the disruption of the mucus layer and ensuing bacterial invasion.
Bacteroidetes members have been the prominent taxa identified in in vivo IBD mucosal biofilms to date. Whether or not the B. fragilis strains identified in IBD biofilms harbor the metalloprotease toxin gene, bft, is still unknown. However, adherent and invasive Escherichia coli (AIEC) may play a role as well since they are increased in CD and easily form biofilms in vitro. Several components of the AIEC-EPS have been studied for their immunogenicity, including cellulose, amyloid fibrils, and type 1 pili98. Studies using E. coli strains deficient in cellulose synthesis (ΔbcsA) resulted in reduced epithelial adhesion, and thus biofilm formation, in vitro on human colonic HT-29 cells, and in vivo in the ileum of BALB/c mice. The mutant E. coli strain resulted in a 28% inhibition of IL-8 production by HT-29 cells in vitro compared to the parent strain, suggesting that cellulose is important for adhesion to the epithelium, biofilm formation, and epithelial IL-8 production99. Further, mono-association of Il-10−/− mice with AIEC::ΔbcsA showed decreased Il17a transcription in the proximal colon 21 days post-infection, and decreased IL-17 production ex vivo by mesenteric lymph node cells, compared to mice infected with wild-type (WT) AIEC100.
Bacterial amyloid fibrils are components of biofilms produced by members of the phyla Bacteroidetes, Firmicutes, and Proteobacteria. Enterobacteriaceae family bacteria (phyla Proteobacteria) produce the amyloid fibril curli, which binds to TLR-1/2 and activates the NLRP3 inflammasome, leading to NFκB activation and Caspase 1 production. NFκB and Caspase 1 are responsible for the production of IL-18 and IL-1β, respectively101,102. Furthermore, ex vivo studies using purified curli fibrils lead to murine dendritic cell activation of Th17 differentiation and may contribute to pro-inflammatory responses in IBD patients103. Although not an EPS molecule and a component of E. coli biofilms, type 1 pili are essential for E. coli biofilm establishment and attachment to host cells, but results vary by type of in vitro surface104. The pili protein FimH has been shown to activate TLR-4, triggering innate immune responses and type 1 interferon signaling105,106. Altogether, these findings suggest that biofilms and EPS molecules from bacteria associated with IBD may have pro-inflammatory properties. Further studies in experimental models of colitis, as well as in IBD patient tissues, are needed to further establish their contribution to IBD pathogenesis.
Colorectal Cancer (CRC) Biofilms
Sporadic CRC is a major health burden both in the US and worldwide. Despite the success of screening colonoscopy in decreasing sporadic CRC in patients over the age of 50 in the United States, we are now seeing an alarming trend in early onset CRC (<50 years of age)107. The shift to presentation of CRC to earlier ages is making it ever more necessary to understand contributing factors, such as the microbiome and inflammation, as well as to identify novel biomarkers that facilitate detection of colon neoplasia.
A recent study by Dejea et al35 showed that sporadic CRC, not associated with IBD, may be linked to biofilm formation and pro-tumoral inflammation. They examined 36 CRC patients from the US to determine the spatial organization of colonic bacteria in comparison to healthy screening colonoscopy controls. Overall, 53% of CRC and adenomas (19/36) and 13% of healthy colonoscopy biopsies (15/120) displayed colon mucosal biofilms. 89% of right-sided tumors were biofilm-positive compared to only 12% of left-sided tumors; nearly all normal (non-tumor) distal mucosa from CRC patients were biofilm-positive if their respective tumors were biofilm-positive. Virtually all biofilms were associated with tissue-invasive bacteria and, therefore, established invasive microbial biofilms as a feature of right-sided (proximal colon) CRCs35. Biofilm-positive normal colon tissues distant to the tumors of CRC patients displayed loss of E-cadherin (consistent with disrupted colon barrier function), increased IL-6 expression, and overt Stat3 activation. These features are well-established as associated with human carcinogenesis and, therefore, suggest that colon mucosal biofilms may foster pro-carcinogenic inflammatory responses. Biofilm-positive healthy colonoscopy tissues also displayed disruption of E-cadherin and increased IL-6 expression, but Stat3 activation was not observed35. A follow-up study performed on a Malaysian CRC cohort confirmed that right-sided tumors were biofilm-positive; however, a higher portion (58%) of left-sided tumors were also biofilm-positive compared to the US CRC cohort.
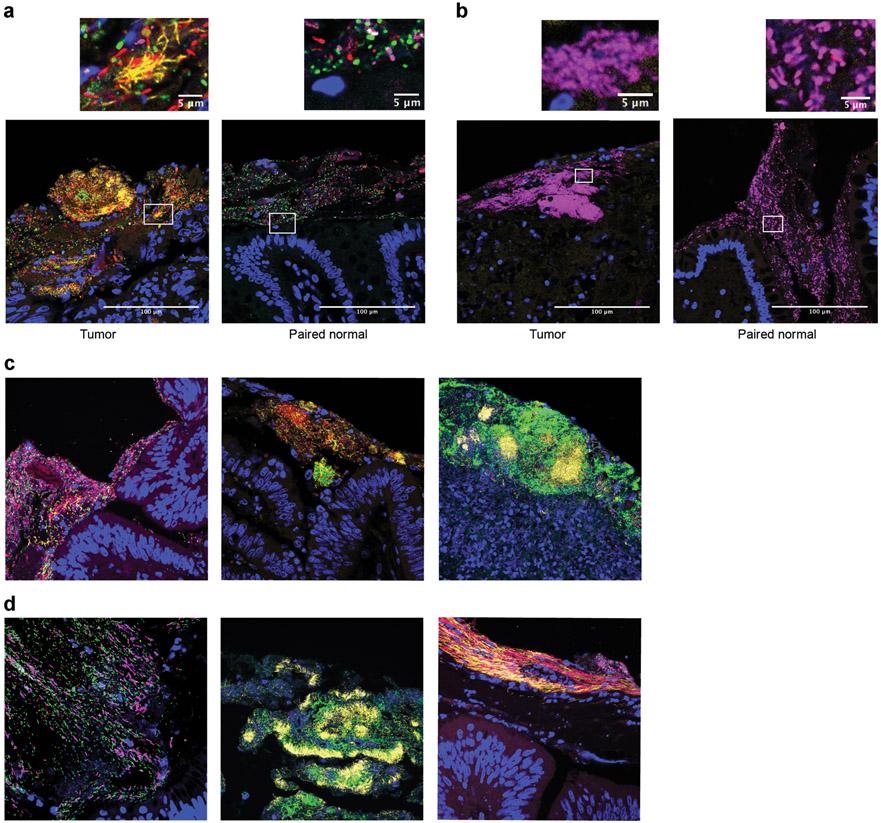
Multiplexed FISH staining of CRC biofilms with taxa-specific probes has revealed three major microbiological phenotypes: 1) polymicrobial (Lachnospiraceae, Bacteroidetes, and Proteobacteria) in 55% of patients (40/73), 2) polymicrobial with Fusobacteria blooms (40%; 29/73), and 3) Proteobacteria-dominant (5%; 4/73) biofilms (Figure 1)15,35. Elegant studies have identified intricate structures in other in vivo biofilms such as dental plaques, including “corncob” and “hedgehog” structures, reflecting an orderly assembly from initial attachment organisms, to bridging organisms, to late colonizers108. These intricate structures display gradients of nutrients/gases/sugars/etc108, similar to how mono-species colonies grown on an agar plate can have different zones of nutrient availability and anaerobicity. While the vast majority of invasive biofilms in the colon, thus far, have been characterized as having random organization (Fig. 1C, left), complex 3-dimensional structures can be observed including microcolonies of single species or mixed species within a larger, scattered biofilm (Fig. 1C, middle). Dense biofilms with embedded microcolonies are also frequently observed in necrotic tissues from tumor patients (Fig. 1C, right). Finally, in the outer mucus layer of these tumor biofilms, linear arrays of bacteria may be observed, likely due to shear flow of the luminal contents (Fig. 1D, left, middle, and right). In contrast, colonic biofilms in healthy hosts, thus far, have largely been defined as polymicrobial (Bacteroidetes/Lachnospiraceae)35. Complex structures such as the ones described above have yet to be observed. Whether this is due to differences in the tumor microenvironment, or reflect differences in the timing of bowel preparation, or the way that samples are procured (colonoscopy vs. surgery) is unknown.
Figure 1. Characterization of Colonic Biofilms.
Colon resections from a Malaysian CRC cohort were fixed in modified Carnoy’s (methacarn) and stained with oligonucleotide probes for Bacteroidetes (green), Lachnospiraceae (red), Fusobacteria (yellow), and Proteobacteria (magenta) as previously described (Drewes et al. 2017). Host nuclei are counterstained in DAPI. Images were captured at 40x by confocal microscopy. (A) A polymicrobial biofilm with Fusobacterial blooms from a tumor (left) and the paired normal biofilm without Fusobacteria (right). (B) A Proteobacteria-dominant biofilm from a tumor (left) and the paired normal biofilm (right). (C) Most CRC biofilms observed to date consist of scattered mixtures of bacteria (left), but complex 3D structures can be observed including microcolonies of specific species embedded within the larger biofilm (middle) or on necrotic tumor tissue (right). (D) In the outer mucus of tumors, distinct linear organization of the bacteria can sometimes be observed, potentially representing shear forces from the flow of gastrointestinal contents (left, center, and right images). Images in A and B were originally published in Drewes et al. 2017 NPJ Biofilms Microbiomes and have not been modified15.
Biofilms are described in familial adenomatous polyposis (FAP) patients36. FAP is a hereditary condition characterized by an APC mutation resulting in polyp and adenoma formation, an early stage of the adenoma-carcinoma sequence109. In contrast to the polymicrobial biofilms associated with sporadic CRC, FAP-associated biofilms were predominantly composed of enterotoxigenic B. fragilis (ETBF, positive for the bft toxin), and pks+ E. coli, both of which have intrinsic tumorigenic properties in mouse models of colon tumorigenesis110,111. Co-colonization of mice, including multiple intestinal neoplasia (Min) Apc+/− mice that model FAP, with ETBF and pks+ E. coli showed that both bacteria cooperate to trigger colon carcinogenesis. Colon tumor formation was dependent on expression of each organism’s virulence determinants, bft and pks, as well as ETBF-induced IL-17. IL-17 is a critical pro-inflammatory mediator contributing to colon tumorigenesis triggered by biofilms14, tumor progression112, and/or resistance to checkpoint blockade113 in human CRC. In this co-colonization model, mucus degradation by ETBF likely facilitated pks+ E. coli mucosal adherence. This resulted in increased DNA damage, that cooperated with bft-dependent IL-17 epithelial signaling, and initiated colon carcinogenesis and promotion of tumor growth. While these mice do not develop biofilms as observed in FAP patients, this experimental model provided insight into mechanisms of bacterial cooperation to promote colon tumorigenesis in humans36.
To test whether human colon mucosal biofilms are carcinogenic, or just a consequence of the changing tumor environment, bacterial slurries, made from mucosa collected from human biofilm-positive CRC and healthy biofilm-negative colonoscopy patients, were inoculated into germ-free (GF) and specific pathogen free (SPF) CRC-susceptible mice. Whereas mice do not normally develop biofilms, combined FISH and PAS staining of the mucus layer of GF murine proximal and distal colonic tissues, revealed robust invasive biofilm development 12 weeks after inoculation with biofilm-positive tissue slurries, but not with biofilm-negative biopsy slurries. Further, biofilm formation was observed as early as one week in GF WT mice inoculated with biofilm-positive human tumor slurries, but not with colonoscopy biofilm-positive or biofilm-negative samples. Importantly, all biofilm-positive tissue inocula, but not biofilm-negative biopsy inocula used as control, induced tumorigenesis in colonized mice. Unexpectedly, inoculation of biofilm-positive colonoscopy biopsy from cancer free individuals induced comparable tumorigenesis to biofilm-positive CRC inocula.
Histopathological analysis of GF mouse tissues showed overall similar levels of inflammation in mice inoculated with biofilm-positive slurries when compared to mice inoculated with biofilm-negative control biopsy slurries. Flow cytometry analyses of the colonic lamina propria leukocytes isolated from GF WT mice inoculated with human biofilm-positive or biofilm-negative biopsy slurries showed the recruitment of immunosuppressive myeloid cells and IL-17 production as early as one-week post-inoculation. Both immunosuppressive myeloid cells and IL-17 are commonly associated with colon carcinogenesis114. In contrast, biofilm-negative colonoscopy biopsy slurries triggered minimal inflammation. These findings provide direct evidence that human colonic biofilms alter the colonic mucosa architecture and inflammatory homeostasis, with mucus and tissue invasion of pro-tumorigenic bacteria likely leading to the induction of oncogenic and pro-inflammatory signaling that contribute to colon carcinogenesis. Future studies will focus on identifying biofilm-associated pro-carcinogenic and pro-inflammatory microbes.
Concluding Remarks and Future Directions
Our understanding of biofilm contributions to mucosal immune homeostasis and disease pathogenesis remains very limited. Experimental in vitro systems and in vivo mouse models are necessary to define biofilm microbial interactions and mechanisms of action, as well as to establish the causative link between biofilm and human disease. However, there is a significant gap between these experimental models and the complexity and intricacy of biological systems contributing to the formation of biofilms and their impact in vivo in the human lung or gut. Studies on biofilm-forming bacteria in CF and COPD are numerous but lack key studies examining direct evidence of mucosal biofilms in humans in vivo in relation to disease pathophysiology. Furthermore, our knowledge of gut mucosal biofilms remains insufficient, but nevertheless promising for understanding IBD and CRC pathogenesis. It will be critical to have access to biopsies from younger patient cohorts, increasingly susceptible to IBD and CRC, to better understand the relationship between their microbiome and disease risk.
Future studies should prioritize the detection and characterization of the compositional (microbial and EPS) and metabolic context of biofilms in order to enhance the understanding of their role in disease pathogenesis. Development of antibodies and probes targeting specific bacteria and their respective secreted EPS molecules will help. Application of uniform, rigorous criteria to in vivo bacterial biofilm detection would allow cross-comparison of biofilm biology across human tissues. Furthermore, technologies that can distinguish biofilm resident bacteria from their planktonic counterparts by analyzing differences in gene expression (quiescent vs. non-quiescent states) would assist in evaluating the role of biofilms in human disease. This can likely be accomplished by emerging novel technologies, such as meta-genomic, -transcriptomic, metabolic, and proteomic analyses. In addition to the development of new animal models, these technologies will improve our understanding of the interaction between host responses and biofilm structure/function in human diseases.
Acknowledgements
We’d like to acknowledge all the members of the Housseau and Sears laboratories for thoughtful discussions and their support. This work has also been supported by funds from the Cancer Research Institute/Fight CRC (to CLS, FH), the Bloomberg~Kimmel Institute for Cancer Immunotherapy, the Johns Hopkins School of Medicine and Department of Medicine, the Cancer Research UK’s Grand Challenge Initiative, the NCI HTAN U2CCA233291 (all to CLS), Swim Across America Grant (to FH), and National Institute of Health grants to JLD (National Cancer Institute K99 CA230192), FH (R01 CA203891), and CLS (R01 CA196845).
Footnotes
Conflict of interest: CLS reports research support from Bristol Myers Squibb, Merck, and Janssen Pharmaceuticals, Inc. FH participated in a grant review for Sanofi.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Sender R, Fuchs S & Milo R Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 14, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young VB The role of the microbiome in human health and disease: An introduction for clinicians. BMJ 356, j831 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Costerton JW & Stoodley P Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol 2, 95–108 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Dang AT & Marsland BJ Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 12, 843–850 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Tytgat HLP, Nobrega FL, van der Oost J & de Vos WM Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends Microbiol. 27, 17–25 (2019). [DOI] [PubMed] [Google Scholar]
- 7.DeVos WM PERSPECTIVE Microbial biofilms and the human intestinal microbiome. npj Biofilms Microbiomes 1, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dongari-Bagtzoglou A Mucosal biofilms: Challenges and future directions. Expert Rev. Anti. Infect. Ther 6, 141–144 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Costerton JW et al. Microbial biofilms. Annu Rev Microbiol 49, 711–745 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Jamal M et al. Bacterial biofilm and associated infections. J. Chinese Med. Assoc 81, 7–11 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Flemming HC & Wingender J The biofilm matrix. Nat. Rev. Microbiol 8, 623–633 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Costerton JW, Stewart PS & Greenberg EP Bacterial biofilms: A common cause of persistent infections. Science (80-. ). 284, 1318–1322 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Burmølle M, Ren D, Bjarnsholt T & Sørensen SJ Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 22, 84–91 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Tomkovich S, Jobin C & Sears CL Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest 129, 1699–1712 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drewes JL et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. npj Biofilms Microbiomes 3, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho KH & Caparon MG Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol 57, 1545–1556 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Balaban NQ, Merrin J, Chait R, Kowalik L & Leibler S Bacterial persistence as a phenotypic switch. Science (80-. ). 305, 1622–1625 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Helaine Sophie; Cheverton Angela; Watson Kathryn; Faure Laura; Matthews Sophie; Holden D Internalization of Salmonella by Macrophages Induces Formation of Nonreplicating Persisters. Science (80-. ). 343, 204–208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher RA, Gollan B & Helaine S Persistent bacterial infections and persister cells. Nat. Rev. Microbiol 15, 453–464 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Wake N et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. npj Biofilms Microbiomes (2016).doi: 10.1038/npjbiofilms.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Randal Bollinger R, Barbas AS, Bush EL, Lin SS & Parker W Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J. Theor. Biol (2007).doi: 10.1016/j.jtbi.2007.08.032 [DOI] [PubMed] [Google Scholar]
- 22.Macfarlane S & Macfarlane GT Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl. Environ. Microbiol (2006).doi: 10.1128/AEM.00754-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitch ECMW, Walker AW, Duncan SH, Holtrop G & Flint HJ Selective colonization of insoluble substrates by human faecal bacteria. Environ. Microbiol (2007).doi: 10.1111/j.1462-2920.2006.01186.x [DOI] [PubMed] [Google Scholar]
- 24.Walker AW et al. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ. Microbiol (2008).doi: 10.1111/j.1462-2920.2008.01717.x [DOI] [PubMed] [Google Scholar]
- 25.Zarco MF, Vess TJ & Ginsburg GS The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 18, 109–120 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Darrene LN & Cecile B Experimental Models of Oral Biofilms Developed on Inert Substrates: A Review of the Literature. Biomed Res. Int 2016, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bahamondez-Canas TF, Heersema LA & Smyth HDC Current status of in vitro models and assays for susceptibility testing for wound biofilm infections. Biomedicines 7, 1–31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moormeier DE & Bayles KW Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol 104, 365–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebeaux D, Chauhan A, Rendueles O & Beloin C From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2, 288–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabrilska RA & Rumbaugh KP Biofilm models of polymicrobial infection. Future Microbiol. 10, 1997–2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence JR, Korber DR, Hoyle BD, Costerton JW & Caldwell DE Optical sectioning of microbial biofilms. J. Bacteriol 173, 6558–6567 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leriche V, Sibille P & Carpentier B Use of an enzyme-linked lectinsorbent assay to monitor the shift in polysaccharide composition in bacterial biofilms. Appl. Environ. Microbiol 66, 1851–1856 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strathmann M, Wingender J & Flemming H Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. 50, 237–248 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Swidsinski A, Weber J, Loening-Baucke V, Hale LP & Lochs H Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol 43, 3380–3389 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dejea CM et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. U. S. A 111, 18321–18326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dejea CM et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science (80-. ). 359, 592–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macfarlane S, Woodmansey EJ & Macfarlane GT Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl. Environ. Microbiol 71, 7483–7492 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bullman S et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science (80-. ). 358, 1443–1448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson MEV et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U. S. A 105, 15064–15069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johansson MEV, Holmén Larsson JM & Hansson GC The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U. S. A 108, 4659–4665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansson GC Mucus and mucins in diseases of the intestinal and respiratory tracts. J. Intern. Med 285, 479–490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boisvert AA, Cheng MP, Sheppard DC & Nguyen D Microbial biofilms in pulmonary and critical care diseases. Ann. Am. Thorac. Soc (2016).doi: 10.1513/AnnalsATS.201603-194FR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bjarnsholt T The role of bacterial biofilms in chronic infections. APMIS (2013).doi: 10.1111/apm.12099 [DOI] [PubMed] [Google Scholar]
- 45.Starner TD, Zhang N, Kim GH, Apicella MA & McCray PB Haemophilus influenzae forms biofilms on airway epithelia: Implications in cystic fibrosis. Am. J. Respir. Crit. Care Med 174, 213–220 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bjarnsholt T et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol 44, 547–558 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Krinos CM et al. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414, 555–558 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Hong W et al. Phosphorylcholine decreases early inflammation and promotes the establishment of stable biofilm communities of nontypeable Haemophilus influenzae strain 86-028NP in a chinchilla model of otitis media. Infect. Immun 75, 958–965 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keely S, Talley NJ & Hansbro PM Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. 5, 7–18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tulic MK, Piche T & Verhasselt V Lung-gut cross-talk: Evidence, mechanisms and implications for the mucosal inflammatory diseases. Clin. Exp. Allergy 46, 519–528 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Wang H et al. Gut-lung crosstalk in pulmonary involvement with inflammatory bowel diseases. World J. Gastroenterol 19, 6794–6804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Budden KF et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol 15, 55–63 (2017). [DOI] [PubMed] [Google Scholar]
- 53.O’Dwyer DN, Dickson RP & Moore BB The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J. Immunol 196, 4839–4847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hilty M et al. Disordered microbial communities in asthmatic airways. PLoS One 5, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erb-Downward JR et al. Analysis of the lung microbiome in the ‘healthy’ smoker and in COPD. PLoS One 6, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morris A et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med 187, 1067–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao MC Physiology of Electrolyte Transport in the Gut: Implications for Disease. Compr. Physiol 9, 947–1023 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Fenker DE et al. A Comparison between Two Pathophysiologically Different yet Microbiologically Similar Lung Diseases: Cystic Fibrosis and Chronic Obstructive Pulmonary Disease. Int. J. Respir. Pulm. Med 5, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lyczak JB, Cannon CL & Pier GB Lung Infections Associated with Cystic Fibrosis. 15, 194–222 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doggett RG, Harrison GM, Stillwell RN & Wallis ES An atypical Pseudomonas aeruginosa associated with cystic fibrosis of the pancreas. J. Pediatr 68, 215–221 (1966). [Google Scholar]
- 61.Govan JRW, Fyfe JAM & McMillan C The instability of mucoid Pseudomonas aeruginosa: fluctuation test and improved stability of the mucoid form in shaken culture. J. Gen. Microbiol 110, 229–232 (1979). [DOI] [PubMed] [Google Scholar]
- 62.DeVries CA & Ohman DE Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol 176, 6677–6687 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffmann N et al. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr−/− mice. Antimicrob. Agents Chemother 51, 3677–3687 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moser C et al. Biofilms and host response – helpful or harmful. Apmis 125, 320–338 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Mauch RM et al. Secretory IgA response against Pseudomonas aeruginosa in the upper airways and the link with chronic lung infection in cystic fibrosis. Pathog. Dis 75, 10–13 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Khan T; Wagener J; Bost T; Martinez J; Accurso F; Riches D Early Pulmonary Inflammation in Infants with CF. Am J Respir Crit Care Med 151, 1075–1082 (1995). [DOI] [PubMed] [Google Scholar]
- 67.González JF, Hahn MM & Gunn JS Chronic biofilm-based infections: Skewing of the immune response. Pathog. Dis 76, 1–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yonker LM, Cigana C, Hurley BP & Bragonzi A Host-pathogen interplay in the respiratory environment of cystic fibrosis. J. Cyst. Fibros 14, 431–439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moser C et al. The immune response to chronic Pseudomonas aeruginosa lung infection in cystic fibrosis patients is predominantly of the Th2 tvpe. APMIS 108, 329–335 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Moser C et al. Serum concentrations of GM-CSF and G-CSF correlate with the Th1/Th2 cytokine response in cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. Apmis 113, 400–409 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Aanaes K et al. Secretory IgA as a diagnostic tool for Pseudomonas aeruginosa respiratory colonization. J. Cyst. Fibros 12, 81–87 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Moser C et al. Early immune response in susceptible and resistant mice strains with chronic Pseudomonas aeruginosa lung infection determines the type of T-helper cell response . Apmis 107, 1093–1100 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Bruscia EM et al. Macrophages directly contribute to the exaggerated inflammatory response in cystic fibrosis transmembrane conductance regulator−/−mice. Am. J. Respir. Cell Mol. Biol 40, 295–304 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lands LC & Stanojevic S Oral non-steroidal anti-inflammatory drug therapy for lung disease in cystic fibrosis. Cochrane Database Syst. Rev (2019).doi: 10.1002/14651858.CD001505.pub5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balfour-Lynn IM, Welch K & Smith S Inhaled corticosteroids for cystic fibrosis. Cochrane Database Syst. Rev (2019).doi: 10.1002/14651858.CD001915.pub6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cystic Fibrosis Clinical Trial Finder. at <https://www.cff.org/Trials/finder>
- 77.Hassett DJ, Borchers MT & Panos RJ Chronic obstructive pulmonary disease (COPD): Evaluation from clinical, immunological and bacterial pathogenesis perspectives. J. Microbiol 52, 211–226 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Fernández-Blanco JA et al. Attached stratified mucus separates bacteria from the epithelial cells in COPD lungs. JCI insight 3, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su YC, Jalalvand F, Thegerström J & Riesbeck K The interplay between immune response and bacterial infection in COPD: Focus Upon non-typeable Haemophilus influenzae. Front. Immunol 9, 1–26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jurcisek JA & Bakaletz LO Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol 189, 3868–3875 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swords WE Nontypeable Haemophilus influenzae biofilms: role in chronic airway infections. Front. Cell. Infect. Microbiol 2, 97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Serban KA & Petrache I Mouse models of COPD. In Methods Mol. Biol 1809, 379–394 (Humana Press Inc., 2018). [DOI] [PubMed] [Google Scholar]
- 83.Pang B et al. Lipooligosaccharides containing phosphorylcholine delay pulmonary clearance of nontypeable Haemophilus influenzae. Infect. Immun 76, 2037–2043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pang B, Hong W, West-Barnette SL, Kock ND & Swords WE Diminished ICAM-1 expression and impaired pulmonary clearance of nontypeable Haemophilus influenzae in a mouse model of chronic obstructive pulmonary disease/emphysema. Infect. Immun 76, 4959–4967 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Staples KJ et al. Relationships between mucosal antibodies, non-typeable Haemophilus influenzae (NTHi) infection and airway inflammation in COPD. PLoS One 11, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.King PT et al. Adaptive immunity to nontypeable Haemophilus influenzae. Am. J. Respir. Crit. Care Med 167, 587–592 (2003). [DOI] [PubMed] [Google Scholar]
- 87.King P et al. Effect of interferon gamma and CD40 ligation on intracellular monocyte survival of nontypeable Haemophilus influenzae. Apmis 116, 1043–1049 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Kalathil SG et al. T-regulatory cells and programmed death 1+ T cells contribute to effector T-Cell dysfunction in patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med 190, 40–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shuto T et al. Activation of NF-κB by nontypeable Hemophilus influenzae is mediated by toll-like receptor 2-TAK1-dependent NIK-IKKα/β-IκBα and MKK3/6-p38 MAP kinase signaling pathways in epithelial cells. Proc. Natl. Acad. Sci. U. S. A 98, 8774–8779 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X et al. Toll-Like Receptor 4 Mediates Innate Immune Responses to Haemophilus influenzae Infection in Mouse Lung . J. Immunol 168, 810–815 (2002). [DOI] [PubMed] [Google Scholar]
- 91.Rotta detto Loria J et al. Nontypeable Haemophilus Influenzae Infection Upregulates the NLRP3 Inflammasome and Leads to Caspase-1-Dependent Secretion of Interleukin-1β - A Possible Pathway of Exacerbations in COPD. PLoS One 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peterson LW & Artis D Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol 14, 141–153 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Swidsinski A et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut 56, 343–350 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maloy KJ & Powrie F Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011). [DOI] [PubMed] [Google Scholar]
- 95.van der Post S et al. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 1–10 (2019).doi: 10.1136/gutjnl-2018-317571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Franks AH et al. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol 64, 3336–3345 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johansson MEV et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63, 281–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ellerman M & Sartor RB Intestinal bacterial biofilms modulate mucosal immune responses. J. Immunol. Sci 2, 13–18 (2018). [PMC free article] [PubMed] [Google Scholar]
- 99.Monteiro C et al. Characterization of cellulose production in Escherichia coli Nissle 1917 and its biological consequences. Environ. Microbiol 11, 1105–1116 (2009). [DOI] [PubMed] [Google Scholar]
- 100.Ellermann M et al. Adherent-invasive Escherichia coli production of cellulose influences iron-induced bacterial aggregation, phagocytosis, and induction of colitis. Infect. Immun 83, 4068–4080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tükel Ç et al. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell. Microbiol 12, 1495–1505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rapsinski GJ et al. Toll-like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, curli. Infect. Immun 83, 693–701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nishimori JH et al. Microbial amyloids induce interleukin 17A (IL-17A) and IL-22 responses via toll-like receptor 2 activation in the intestinal mucosa. Infect. Immun 80, 4398–4408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lajhar SA, Brownlie J & Barlow R Correction: Characterization of biofilm-forming capacity and resistance to sanitizers of a range of E.coli O26 pathotypes from clinical cases and cattle in Australia (BMC Microbiology (2018) 18 (41) DOI: 10.1186/s12866-018-1182-z). BMC Microbiol. 18, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mossman KL et al. Cutting Edge: FimH Adhesin of Type 1 Fimbriae Is a Novel TLR4 Ligand. J. Immunol 181, 6702–6706 (2008). [DOI] [PubMed] [Google Scholar]
- 106.Ashkar AA, Mossman KL, Coombes BK, Gyles CL & Mackenzie R FimH adhesin of type 1 fimbriae is a potent inducer of innate antimicrobial responses which requires TLR4 and type 1 interferon signalling. PLoS Pathog. 4, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Virostko J, Capasso A, Yankeelov TE & Goodgame B Recent trends in the age at diagnosis of colorectal cancer in the US National Cancer Data Base, 2004-2015. Cancer (2019).doi: 10.1002/cncr.32347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Welch JLM, Rossetti BJ, Rieken CW, Dewhirst FE & Borisy GG Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U. S. A 113, E791–E800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leslie A, Carey FA, Pratt NR & Steele RJC The colorectal adenoma-carcinoma sequence. Br. J. Surg (2002).doi: 10.1046/j.1365-2168.2002.02120.x [DOI] [PubMed] [Google Scholar]
- 110.Wu S et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med 15, 1016–1022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arthur JC et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (80-. ). 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tosolini M et al. Clinical Impact of Different Classes of Infiltrating T Cytotoxic and Helper Cells (Th1, Th2, Treg, Th17) in Patients with Colorectal Cancer. Cancer Res. 71, 1263–1271 (2011). [DOI] [PubMed] [Google Scholar]
- 113.Llosa NJ et al. IMMUNOPATHOLOGIC STRATIFICATION OF COLORECTAL CANCER FOR CHECKPOINT BLOCKADE IMMUNOTHERAPY. (2019).doi: 10.1158/2326-6066.CIR-18-0927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Terzic′* J, Terzic′* T, Grivennikov S, Karin E & Karin M Inflammation and Colon Cancer. Gastroenterology 138, 2101–2114 (2010). [DOI] [PubMed] [Google Scholar]