Abstract
The goal of this study was to evaluate the accuracy, reproducibility, and efficiency of a 31P magnetic resonance spectroscopic fingerprinting (31P-MRSF) method for fast quantification of the forward rate constant of creatine kinase (CK) in mouse hindlimb. The 31P-MRSF method acquired spectroscopic fingerprints using interleaved acquisition of phosphocreatine (PCr) and γATP with ramped flip angles and a saturation scheme sensitive to chemical exchange between PCr and γATP. Parameter estimation was performed by matching the acquired fingerprints to a dictionary of simulated fingerprints generated from the Bloch-McConnell model. The accuracy of 31P-MRSF measurements was compared with the magnetization transfer (MT-MRS) method in mouse hindlimb at 9.4 T (n = 8). The reproducibility of 31P-MRSF was also assessed by repeated measurements. Estimation of the CK rate constant using 31P-MRSF (0.39 ± 0.03 s−1) showed a strong agreement with that using MT-MRS measurements (0.40 ± 0.05 s−1). Variations less than 10% were achieved with 2 min acquisition of 31P-MRSF data. Application of the 31P-MRSF method to mice subjected to an electrical stimulation protocol detected an increase in CK rate constant in response to stimulation-induced muscle contraction. These results demonstrated the potential of the 31P-MRSF framework for rapid, accurate, and reproducible quantification of the chemical exchange rate of CK in vivo.
Keywords: 31P spectroscopy, creatine kinase activity, magnetic resonance fingerprinting, magnetization transfer
1 |. INTRODUCTION
Creatine kinase (CK) is expressed at a high level in the heart, skeletal muscle, and brain.1,2 It catalyzes the exchange of a phosphate group between phosphocreatine (PCr) and adenosine triphosphate (ATP). In tissues with high and fluctuating metabolic demands, the CK system provides short-term ATP buffering when ATP demand exceeds mitochondria’s capacity of ATP generation via oxidative phosphorylation. This misbalance between ATP utilization and production may occur during normal physical activities such as exercise, or under pathological conditions such as ischemia and hypoxia. Hence, the CK system plays an important role in cellular metabolism by serving as the major energy reserve for maintaining constant ATP supply during metabolic perturbations. In addition, CK may act as an intracellular pH regulator by consuming protons produced in ATP hydrolysis.3 Mitochondrial CK located in the intermembrane space also plays a role in metabolic regulation by transporting adenosine diphosphate (ADP), a potent allosteric activator of glycolytic flux, from the cytosol into the mitochondria.4
Abnormal CK activity can disrupt the constant flow of energy and adversely impact tissue function. A decrease of ATP synthesis through CK was observed in patients with heart failure.5 Studies employing mouse models with CK gene deletion showed that CK knockout mice were more likely to develop left ventricular hypertrophy and dilatation.6 Impaired post-ischemic recovery of calcium homeostasis and contractile function were also observed in mouse hearts with CK knockout.7 On the other hand, mice overexpressing CK showed improved functional recovery after ischemia,8 as well as increased survival after aortic constriction surgery.9 In the skeletal muscle, a reduction in ATP during ischemia and more pronounced acidification were observed in the hindlimb of CK knockout mice.10 These studies provide direct evidence of the protective role of CK during metabolic perturbations and suggest CK as a promising therapeutic target.11
Phosphorus-31 (31P) magnetic resonance spectroscopy (MRS) employing magnetization transfer (MT) techniques provides the only methodology for in vivo quantification of CK activity.12–17 The technique entails measuring changes in the PCr signal after the saturation or inversion of the γATP signal. The forward rate constant of ATP synthesis via CK is determined by fitting the Bloch-McConnell model to the measured PCr signal as a function of the saturation or inversion time.18–20 Typically, a range of saturation or inversion times covering the entire dynamics of PCr signal changes is sampled to accurately measure the rate constant, thus leading to prolonged acquisition time. The small sizes of tissue masses in mice present additional challenges for quantifying CK activity because the signal-to-noise ratio (SNR) can be orders of magnitude lower.
Magnetic resonance fingerprinting (MRF) is a new framework of data acquisition that enables accurate parameter estimation at low SNR.21 In conjunction with model simulation and dictionary matching, MRF extracts a set of tissue parameters, e.g., relaxation times, from signals acquired at non-steady-state by constantly varying the sequence parameters. This approach offers significantly improved data acquisition efficiency, enabling accurate parameter estimation with drastically reduced scan time.21 Various MRF methods have been developed for fast and robust measurement of proton relaxation times in the brain,22–24 abdomen,25 heart,26 prostate,23 cartilage and bone,27 and cerebral vasculature.28 High-resolution MRF methods for preclinical applications in small animals are also emerging.29–31 Further, Kulpanovich and Tal have developed proton spectroscopic MRF (MRSF) methods for fast quantification of T1 and T2 relaxation times of several neural metabolites.32,33 The feasibility of MRF for fast quantification of chemical exchange saturation transfer (CEST) has also been explored.34,35
We recently explored the potential of using the MRF framework for rapid quantification of CK activity in vivo.36 A 31P-MRSF method was developed for measuring the CK rate constant in the skeletal muscle of rat hindlimb. The method is based on the balanced steady-state free precession (bSSFP) sequence with varying flip angles (FAs) for data acquisition. Frequency selective saturation pulses were interleaved with data acquisition to encode chemical exchange via CK. The method enabled accurate measurement of the CK rate constant in rat hindlimb with less than 5% variation in 20 s acquisition time. In the current study, we evaluated the utility of the 31P-MRSF method for rapid quantification of the CK rate constant in mouse hindlimb, which requires prohibitively longer acquisition time to gain adequate SNR by conventional MT-MRS methods. Measurement accuracy was validated using a conventional 31P MT-MRS method. Measurement efficiency was also compared between 31P-MRSF and the conventional MT-MRS method. Our results showed that 31P-MRSF achieved a measurement accuracy of less than 10% variation in 2 min acquisition time. Finally, the 31P-MRSF technique was applied to quantify changes in CK rate constant in response to muscle contraction induced by electrical stimulation.
2 |. MATERIALS AND METHODS
2.1 |. 31P-MRSF sequence and parameter matching
The 31P-MRSF method has been described in detail previously.36 A schematic diagram of the pulse sequence is shown in Figure 1. After a non-selective adiabatic inversion pulse, the sequence is followed by interleaved acquisition of PCr and γATP signals in 32 acquisition blocks (Figure 1A). In our current implementation, each block consisted of a train of 10 excitation pulses with alternating phase and a constant repetition time (TR) of 12.8 ms. A 4 ms frequency selective Gaussian pulse was used for spectrally selective excitation. The FA in each block was linearly ramped up and ramped down. The entire acquisition was organized into two 16-block segments with the maximal FA in each block varied sinusoidally (Figure 1B). Following each excitation, 7.7 ms of the free induction decay (FID) signal was collected with 256 data points, yielding a spectral resolution of 130 Hz. A 490 ms spectrally selective saturation block was applied between every two acquisition blocks. The saturation frequency was set at the resonant frequency of γATP in the second acquisition segment and at the frequency contralateral to γATP in the first segment. A total of 320 FID signals were collected in one fingerprint. The acquisition time of one fingerprint was 20 s.
FIGURE 1.
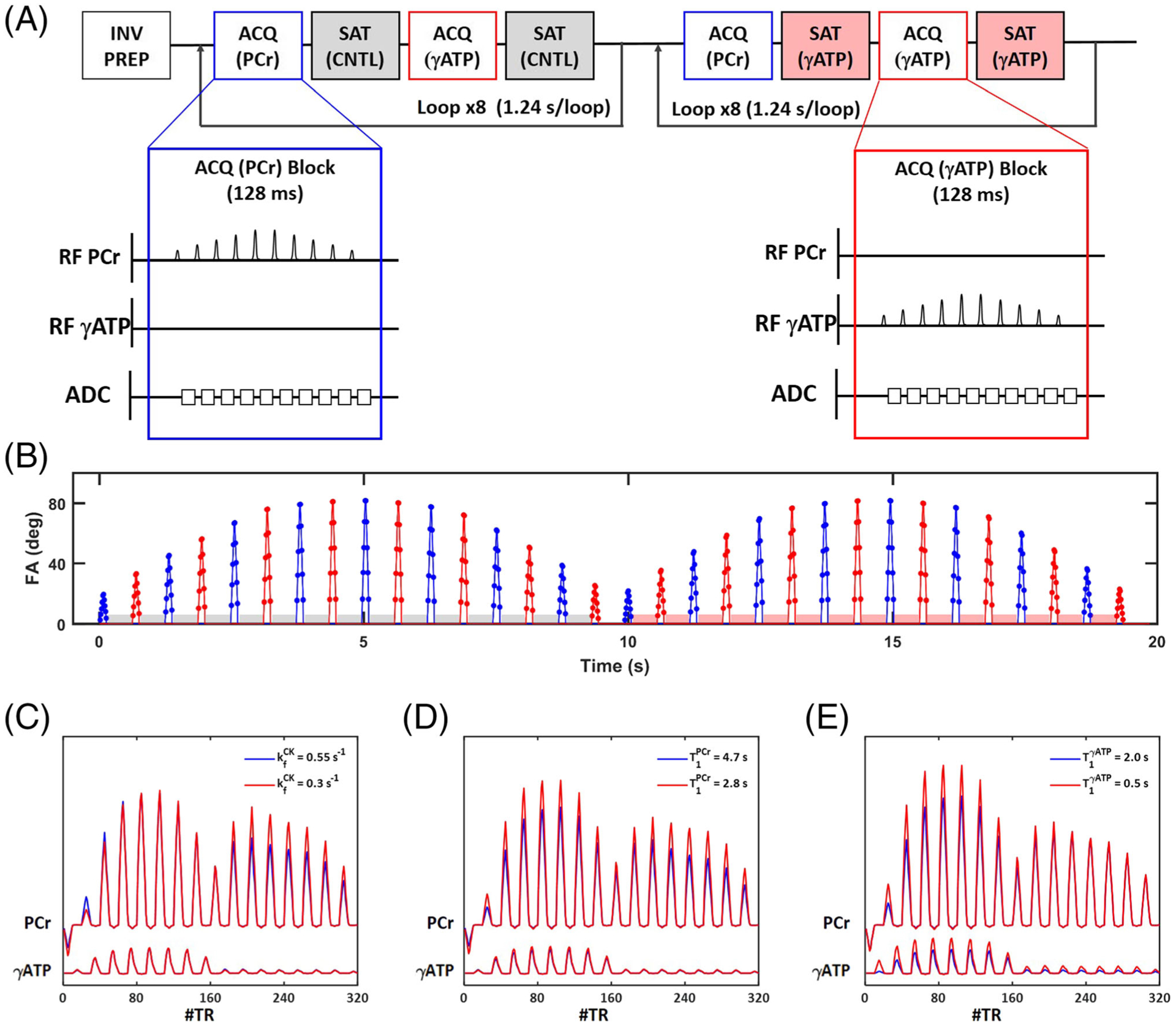
31P-MRSF pulse sequence. A, Schematic diagram of the pulse sequence. Blue and red insets show a single acquisition block of PCr and γATP, respectively. INV PREP, inversion preparation; ACQ (PCr) and ACQ (γATP), acquisition blocks for PCr and γATP, respectively; SAT (CNTL) and SAT (γATP), contralateral and γATP saturation blocks, respectively. B, Timing and nominal FAs of all excitation pulses. Blue and red colors indicate PCr and γATP excitation, respectively. Red and grey shaded boxes indicate γATP and contralateral saturation blocks, respectively. C-E. Simulated fingerprints with one of the matching parameters (from C to E: , , and , respectively) varied
A dictionary of fingerprints was constructed using an in-house-developed MATLAB-based simulator. The fingerprints were simulated by solving the modified Bloch-McConnell equation as described previously.36 A total of nine parameters were used in 31P-MRSF simulations (Table 1). To circumvent the computational challenge of matching all nine parameters, the 31P-MRSF sequence was designed to be relatively insensitive to the T1 relaxation time of γATP (), the T2 relaxation time of both PCr and γATP ( and ), and the linewidth (LW). Further, the resonant frequency of γATP (ωγATP) was constrained by the chemical shift difference between PCr and γATP (2.4 ppm). Hence, only four parameters were estimated from dictionary matching, including the forward CK rate constant (), the T1 relaxation time of PCr (), the resonance frequency of PCr (ωPCr), and the PCr-to-ATP ratio (). Table 1 also lists the parameter ranges and resolutions used for the current study on normal adult mice, as well as the values of the fixed parameters, which were obtained from pilot studies.
TABLE 1.
Parameters in fingerprint simulation
| Symbols | Unit | Parameter description | Range of simulation | Dictionary resolution |
|---|---|---|---|---|
| s−1 | forward CK rate constant | 0.2–0.6 | 0.005 | |
| ms | T1 relaxation time of PCr | 2000–5000 | 200 | |
| ms | T1 relaxation time of γATP | 800* | n.a. | |
| ms | T2 relaxation time of PCr | 120* | n.a. | |
| ms | T2 relaxation time of γATP | 16* | n.a. | |
| ωPCr | Hz | chemical shift of PCr | −15 to 15 | 3 |
| ωγATP | Hz | chemical shift of γATP | 2.4 ppm from PCr | n.a. |
| n.a. | PCr-to-ATP ratio | 2.0–6.0 | 0.2 | |
| LW | Hz | linewidth | 15* | n.a. |
Fixed parameter values obtained from pilot studies.
Figure 1C–E shows simulated fingerprints with one of the matching parameters varied. The impact of CK rate constant on signal evolution can be appreciated in Figures 1C, where the saturation of γATP in the second half of data acquisition induced more pronounced attenuation of the PCr signal with a larger CK rate constant (blue). Figure 1D shows the impact of on signal evolution. As expected, a shorter gave rise to faster signal recovery after the inversion pulse (red). Comparing Figure 1C and 1D, it can be appreciated that the first half of signal evolution encoded predominantly T1 relaxation time. However, the effect of inversion transfer was evident in Figure 1E, where chemical exchange between PCr and γATP accelerated the recovery of PCr signal when was shorter (red). These differential signal evolutions in a fingerprint enabled more robust estimation of the CK rate constant.
2.2 |. Conventional MT-MRS method
Parameter estimation using the conventional 31P MT-MRS method was also performed for validation and comparison. Specifically, nine spectra were acquired, including seven spectra acquired after γATP saturation with the saturation time varying from 0.4 to 7 s, a control spectrum acquired with 7 s contralateral saturation to correct for the spillover effects, and a conventional spectrum acquired without saturation. Acquisition of each spectrum used a TR of 16 s and a 90° hard pulse. The total acquisition time of a single dataset without signal averaging was approximately 150 s.
Post-processing of the MT-MRS data consisted of 15 Hz line broadening, Fourier transform, and phase and baseline correction as described previously.36 The PCr and γATP signals were quantified by fitting each peak to a Lorentzian curve and calculating the integral under the curve. Subsequently, the CK rate constant, T1 of PCr, and the steady-state magnetization of PCr were determined by fitting the experimental data to the explicit solution of the Bloch-McConnell equation. The spillover effects of γATP saturation were corrected using the method outlined by Kingsley and Monahan.37 The PCr-to-ATP ratio was calculated from the conventional spectrum.
2.3 |. In vivo study protocol
In vivo studies were performed on a horizontal 9.4 T preclinical scanner (Bruker BioSpin, Billerica, MA) to evaluate the accuracy and precision of 31P-MRSF measurements in mouse hindlimb. Male C57BL/6 mice 10–12 weeks of age (n = 8) were anesthetized with 1.5% isofluorane delivered via a nose cone. The mouse hindlimb was secured in a 10 mm 31P saddle coil (Figure 2A). The mouse cradle and the 31P coil were placed within a 1H volume coil at the isocenter of the scanner. The respiratory rate was maintained between 75 and 110 breaths per minute by manually adjusting the anesthesia level. Body temperature was maintained at above 35 °C via a feedback control system (SA Instruments, Stony Brook, NY). The animal protocol was approved by the Institutional Animal Care and Use Committee of Case Western Reserve University.
FIGURE 2.
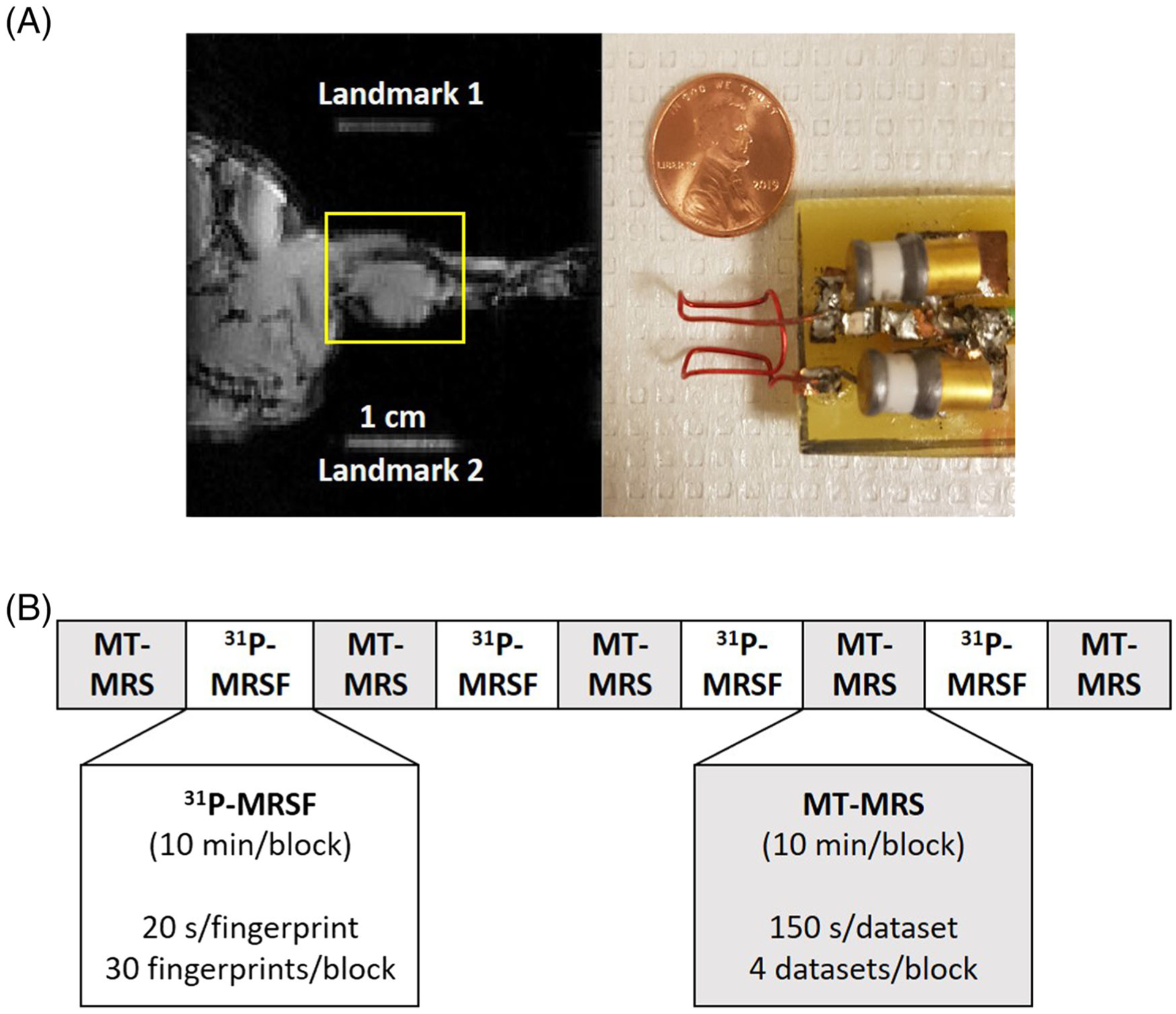
In vivo experimental settings. A, 31P coil and a sagittal view of mouse hindlimb. Two 1 cm segments of water-filled tubing placed on the edges of the cradle serve as the landmarks to demarcate the location of the 31P coil. The yellow box indicates the shimming volume. B, Data acquisition protocol. MT-MRS and 31P-MRSF data were acquired alternatingly in 10 min acquisition blocks
Following localized shimming and B1 power calibration, 31P-MRSF and MT-MRS datasets were acquired alternatingly in 10 min acquisition blocks (Figure 2B). In each acquisition block, 30 31P-MRSF fingerprints or 4 MT-MRS datasets were acquired. These acquisition blocks were repeated four times for 31P-MRSF and five times for MT-MRS. A total of 120 31P-MRSF fingerprints and 20 MT-MRS datasets were acquired in 90 min.
An additional seven C57BL/6 mice underwent an electrical stimulation protocol for evaluation of changes in CK activity after stimulation-induced muscle contraction. Two needle electrodes were inserted subcutaneously over the third lumbar vertebra and the greater trochanter, respectively. The electrodes were connected to a Grass stimulator (Grass Technologies, Warwick, RI). Muscle contraction was induced for 4 min by applying 3 Hz square wave pulses with 2 V amplitude and 1 ms pulse duration.38 31P-MRSF data were acquired before and 16 min after the stimulation.
2.4 |. Data analyses
All results are presented as mean ± standard deviation. The accuracy of parameter estimation by 31P-MRSF was evaluated by comparing 31P-MRSF measurements with MT-MRS, using data with different numbers of averages. The coefficient of variation (CV) was used to assess the precision of the estimated parameters. was used as an index of measurement efficiency. A two-tailed paired Student t-test was used to determine the significance of differences between the 31P-MRSF and the MT-MRS methods, as well as to compare estimated parameters before and after electrical stimulation. A p-value of less than 0.05 was considered statistically significant.
3 |. RESULTS
3.1 |. Fingerprint variations and matching residuals
Figure 3 shows representative 31P-MRSF fingerprints acquired with a single, 6, and 120 average(s), corresponding to acquisition times of 20 s, 2 min, and 40 min, respectively. The corresponding matched dictionary entries are also shown. Both the PCr and the ATP signals showed 16 lobes acquired in an alternating manner, with each lobe comprised of 10 data points acquired with ramped-up and ramped-down FAs. The amplitude of the first lobe in the PCr signal was negative due to the inversion preparation at the beginning of the pulse sequence. Because of γATP saturation during the second part of data acquisition, the last eight lobes of PCr signal showed a reduced amplitude compared with the first eight lobes, and the last eight lobes of the γATP signal were below the detection level, although simulated fingerprints showed a slight signal recovery during data acquisition.
FIGURE 3.
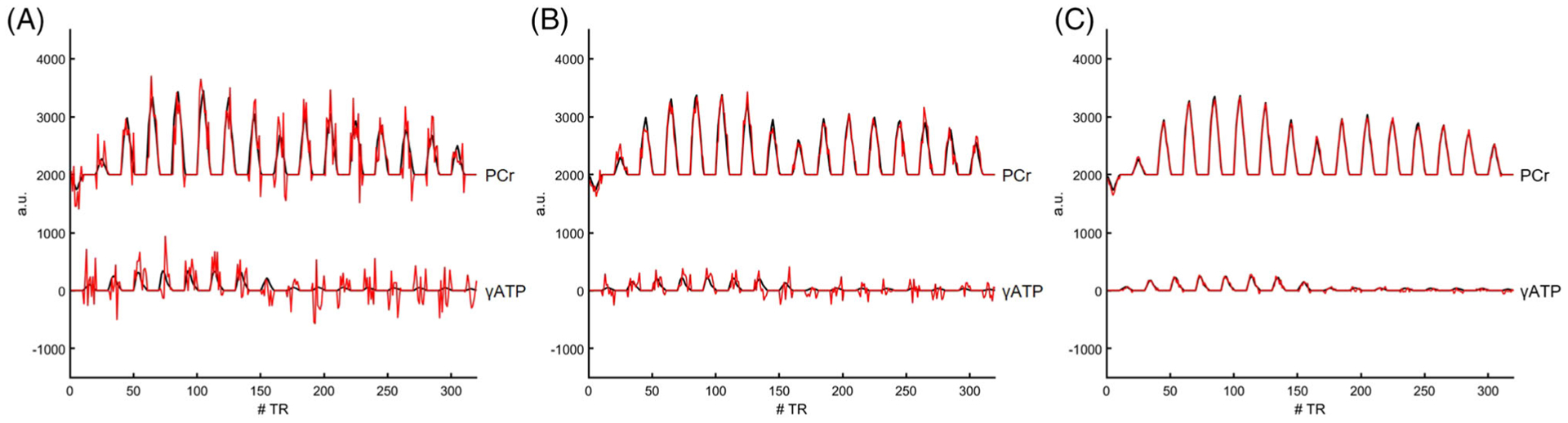
31P-MRSF fingerprints. Representative fingerprints (red) and the corresponding dictionary matches (black) from mouse hindlimb acquired with a single (A), 6 (B), and 120 (C) signal average(s), corresponding to acquisition times of 20 s, 2 min, and 40 min, respectively
The ranges of fingerprint variations with a single, 6, and 120 signal average(s), as well as the matching residuals calculated as the difference between the acquired and matched fingerprints, are shown in Figure 4. As expected, signal averaging improved the SNR of the acquired fingerprints, leading to reduced signal variations (Figure 4A) and decreased deviations from the matched dictionary entry (Figure 4B). Matching residuals for single-, 6-, and 120-average fingerprints were 975, 297, and 50, respectively. In general, performing six averages reduced signal variations and the matching residual by about 70%.
FIGURE 4.
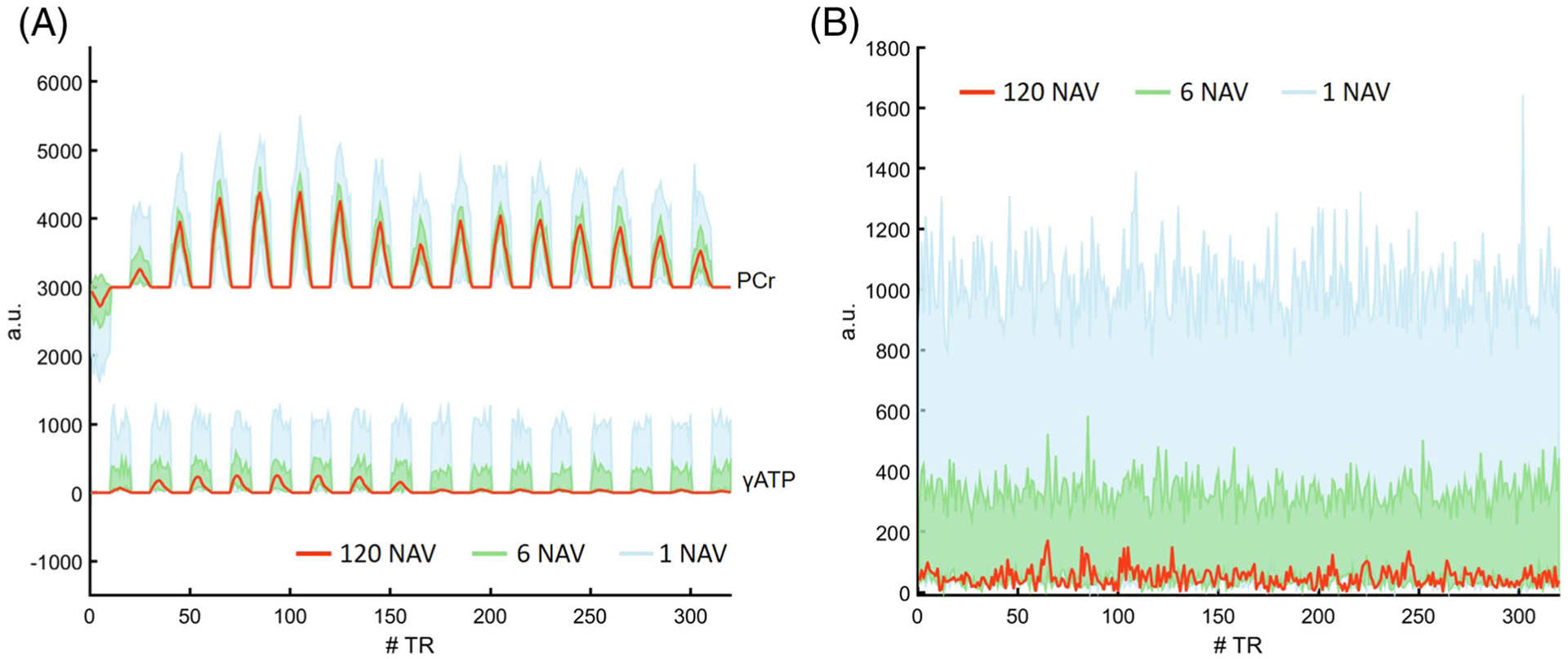
Fingerprint variations and matching residuals. A, Fingerprint acquired with 120 averages and the range of signal variations in fingerprints acquired with a single and six average(s), respectively. B, Matching residual of the 120-average fingerprint and the variations in matching residuals of single- and six-average fingerprints. Red curves represent the average fingerprint and the corresponding matching residual of all 120 fingerprints acquired during a scan session. Blue and green colors indicate fingerprints and matching residuals from a single and six signal average(s), respectively
3.2 |. Accuracy of parameter estimation by 31P-MRSF
Figure 5 shows a comparison of parameter estimation by 31P-MRSF and MT-MRS obtained by averaging all the signals acquired during a scan session. Representative 31P spectra from the MT-MRS acquisition are shown in Supplementary Figure S1. The two methods yielded similar measurements for the CK rate constant (0.39 ± 0.03 s−1 by 31P-MRSF versus 0.40 ± 0.05 s−1 by MT-MRS), T1 of PCr (3.27 ± 0.24 s versus 3.57 ± 0.43 s), and PCr-to-ATP ratio (3.69 ± 0.41 versus 3.86 ± 0.25). No statistically significant differences were detected by paired Student t-tests, suggesting unbiased parameter estimation by 31P-MRSF compared with the conventional MT-MRS method.
FIGURE 5.
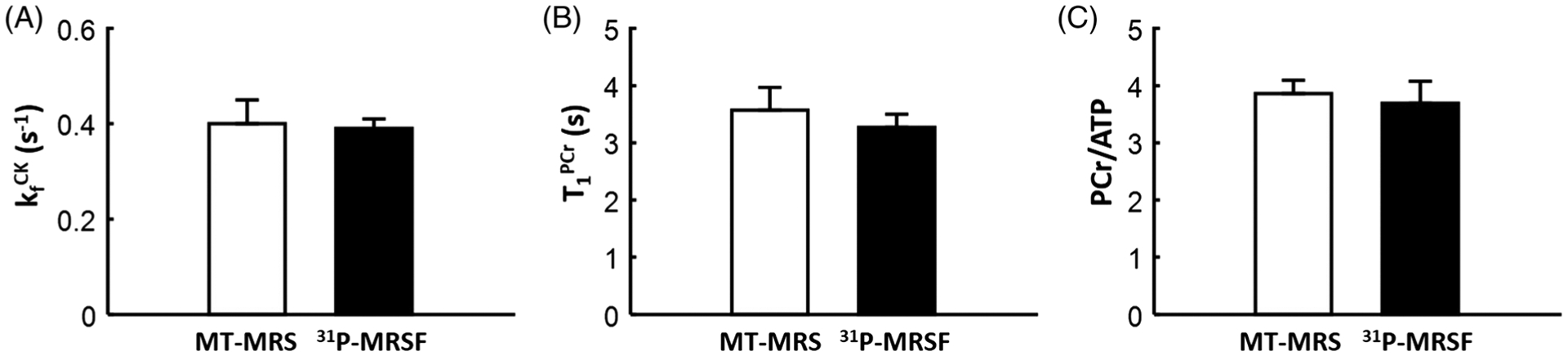
Comparison of parameter estimation by 31P-MRSF and MT-MRS. A, CK rate constant. B, T1 of PCr. C, PCr-to-ATP ratio
Figure 6 shows matched parameters and their ranges of variation as a function of signal averages. Although variations in matched parameters were larger with fewer signal averages due to the reduced SNR, the mean values of the estimated parameters showed minimal deviation from the estimated parameters obtained using more averages. For estimation of the CK rate constant, the average CV using a single average acquisition was 19.30 ± 4.68%, which decreased to 8.4 ± 2.7% with six averages. In general, less than 10% variation was achieved with six signal averages, corresponding to a 2 min acquisition time. These results suggest that unbiased parameter estimation can be achieved with relatively few signal averages.
FIGURE 6.
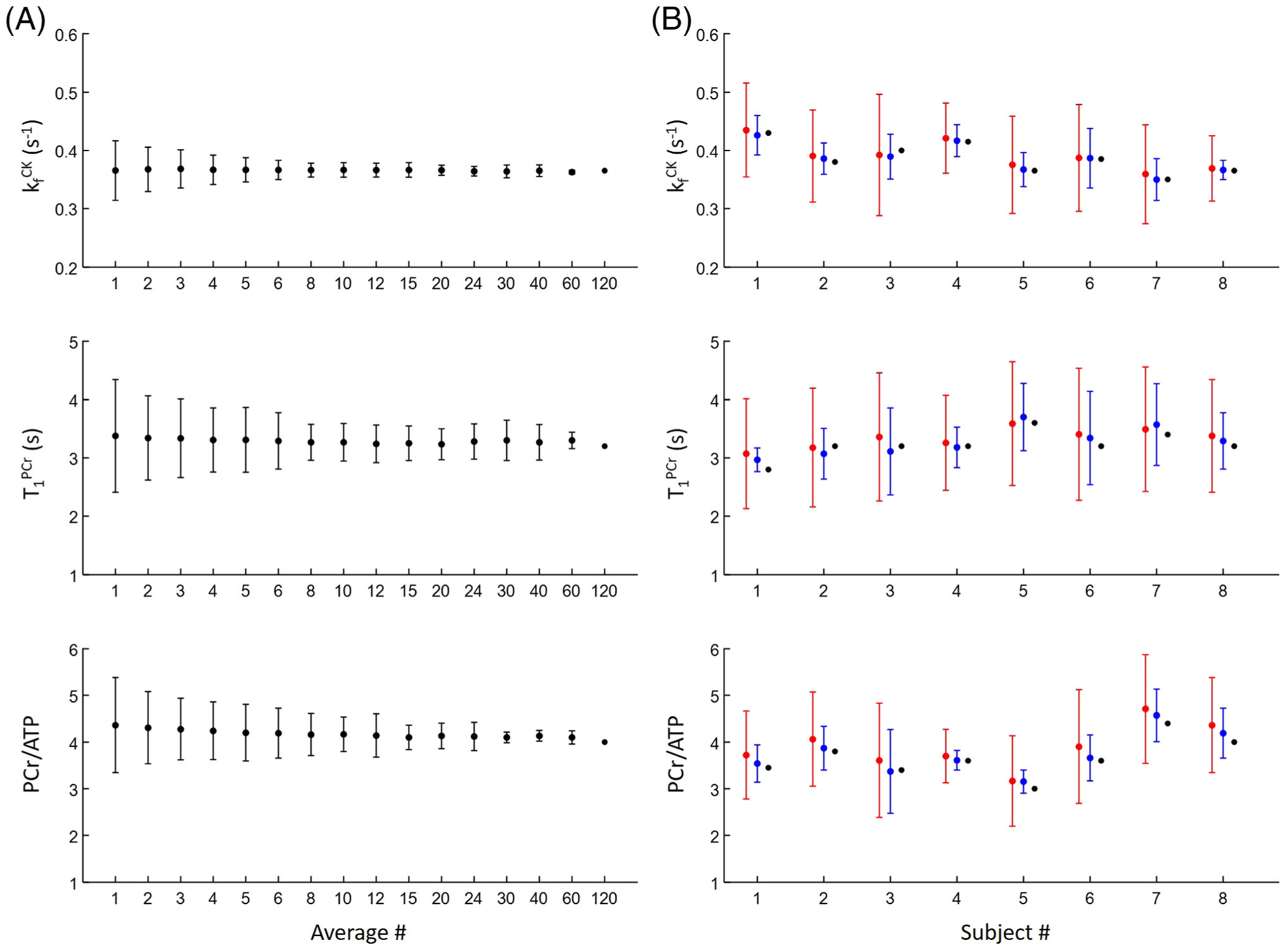
Comparison of parameter estimation using different numbers of signal averages. A, Parameter estimation in a single subject. B, Parameter estimation in all eight subjects using single (red), 6 (blue) and 120 (black) signal average(s)
3.3 |. Measurement efficiency
Figure 7A shows the time course of repeated measurements of the CK rate constant using MT-MRS and 31P-MRSF during a 90 min scan session. 31P-MRSF had higher measurement density than MT-MRS due to the difference in acquisition time of a single-average dataset (20 versus 150 s). Nevertheless, the 31P-MRSF measurements showed smaller variations compared with those of MT-MRS. The standard deviation of the CK rate constant measured by 31P-MRSF was in the range of 0.05 to 0.10 s−1 for all the animal subjects, while it varied from 0.10 to 0.98 s−1 in MT-MRS measurements. The mean CV of the CK rate constant estimated from single-average 31P-MRSF acquisition was about 60% lower than that from MT-MRS (19.30 ± 4.68% versus 49.09 ± 11.51%, p < 0.05, Figure 7B). Taking into account the more than 80% reduction in acquisition time, the acquisition efficiency of 31P-MRSF was six times higher than that of MT-MRS (Figure 7C).
FIGURE 7.
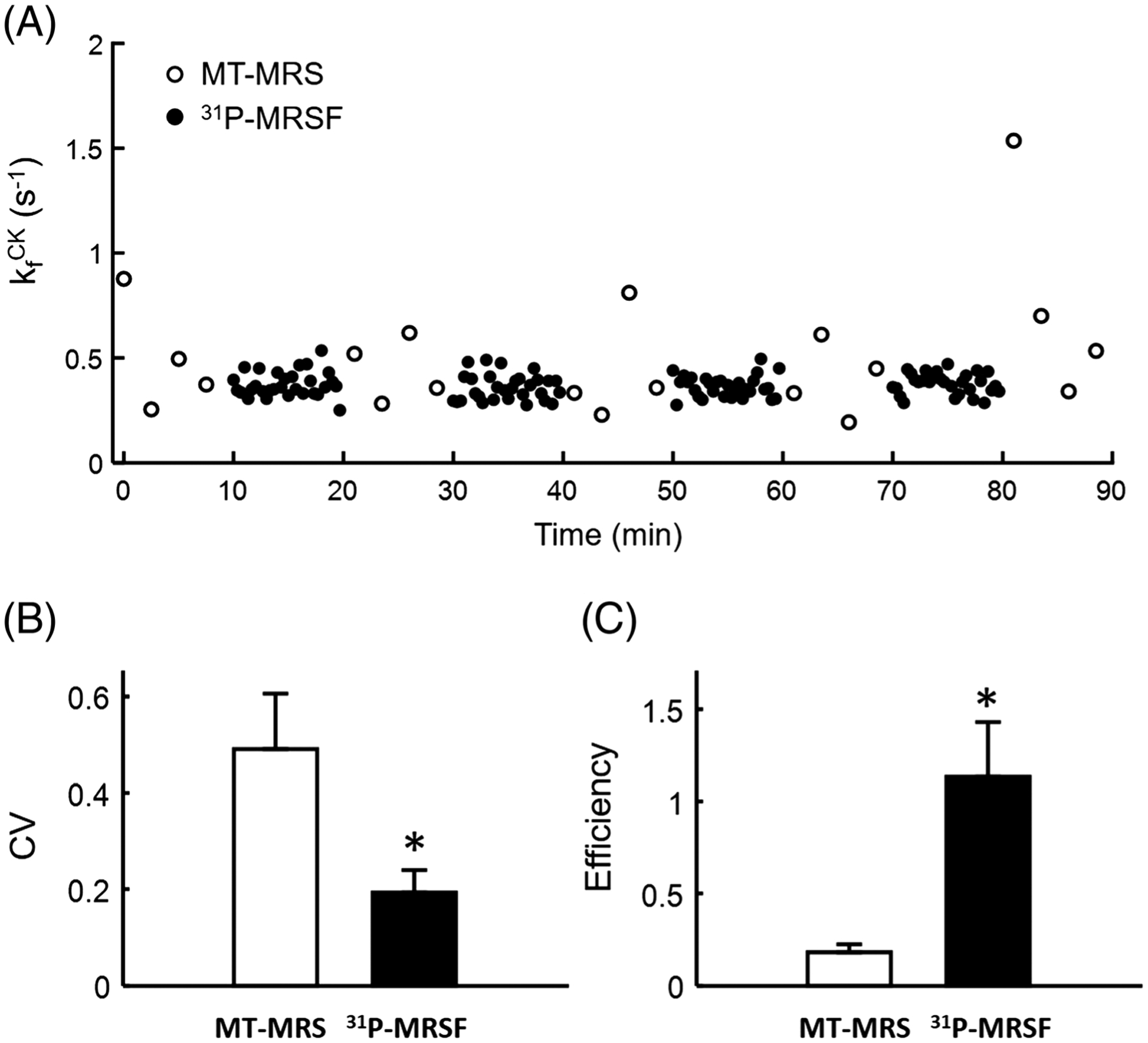
Reproducibility and efficiency. A, Repeated measurements of CK rate constant using MT-MRS and 31P-MRSF. B, Average CV. C, Average measurement efficiency. *p < 0.05
3.4 |. CK change induced by electrical stimulation
Table 2 shows estimated parameters from the 31P-MRSF fingerprints acquired before and after 4 min of electrical stimulation. The post-stimulation data were acquired 16 min after the stimulation to ensure that a new steady state was established. CK rate constant increased significantly after the electrical stimulation (p < 0.05). There was also a trend of decreased PCr-to-ATP ratio; however, no statistical difference was detected. T1 of PCr remained unaltered after the stimulation.
TABLE 2.
Estimated parameters before and after electrical stimulation
| Pre-stimulation | 0.32 ± 0.05 | 3.86 ± 0.22 | 3.51 ± 0.28 |
| Post-stimulation | 0.36 ± 0.05* | 3.69 ±0.16 | 3.49 ± 0.20 |
p < 0.05, pre-stimulation versus post-stimulation.
4 |. DISCUSSION
The role of CK in energy metabolism has long been recognized, particularly in organs with high and fluctuating energy requirements such as the heart, brain, and skeletal muscle.1–4 Mouse models with a gene knockout of either the cytosolic isoform or mitochondrial isoform of CK, or a combination of both, have been developed to investigate the consequences of loss in CK function.39–41 In skeletal muscle, it was shown that CK deficiency led to altered calcium handling, reduced muscle force development, slower relaxation, and impaired voluntary running capacity.41–43 On the other hand, mice overexpressing CK showed a faster force development and a trend of faster relaxation.44,45 Parallel to the effort of creating transgenic mouse models for investigating the CK system, MT-MRS methods were developed to quantify CK activity both in perfused organs and in vivo.12,46 With simultaneous recording of physiological functions during data acquisition, MT-MRS has greatly enhanced our understanding of the role of the CK system in regulating cellular metabolism and physiological responses. However, due to the small size of tissue masses in mice, very few MT-MRS studies have been performed on mice in vivo.8,9,39,40,44,47,48
In this study, we evaluated the utility of a novel 31P-MRSF method to quantify the CK rate constant in mouse hindlimb. The 31P-MRSF sequence used in the current study was modified from the sequence that has been validated in rat hindlimb previously.36 It was shown that a CV of less than 5% can be achieved in rat hindlimb with a single-average acquisition (20 s acquisition time). Similar to conventional MT-MRS methods, the 31P-MRSF method used spectrally selective saturation of γATP to encode chemical exchange via CK. The interleave of γATP saturation with a bSSFP-like acquisition further enhanced the MT effects by preserving the history of signal evolution throughout the data acquisition.49 In addition, the inversion recovery preparation at the beginning of fingerprint acquisition reduced the coupling of the CK rate constant and the T1 of PCr. These sequence design considerations enabled parameter estimation with improved reproducibility and accuracy. While both 31P-MRSF and conventional MT-MRS methods yielded similar measurements of the CK rate constant, T1 of PCr, and PCr-to-ATP ratio, the reproducibility of the parameters estimated using 31P-MRSF was significantly improved, with a 60% reduction in CV of single-average acquisitions. Combined with shortened acquisition time, the 31P-MRSF method led to a sixfold improvement in measurement efficiency.
Unlike conventional MT-MRS methods that use relatively long TR (>1 s) to acquire spectra with high spectral resolution (<5 Hz), the 31P-MRSF sequence used short TR (12.8 ms) to gain acquisition efficiency. As a result, only 7.7 ms of the FID signal was acquired, leading to a low spectral resolution of 130 Hz. Hence, the resonance peaks were largely compressed into a single spectral bin, which significantly simplified spectral quantification. While signal leakage to the neighboring spectral bins may occur in regions with poor B0 homogeneity (>50 Hz linewidth for PCr), this is unlikely to happen in mouse studies because of the small size of the tissue or organ to be characterized. However, spectral quantification may need to include more spectral bins when translating this technique to clinical applications on human subjects.
For the purposes of reducing matching time and computer memory requirements, only four out of nine parameters in the Bloch-McConnell model were matched. The values of T1 of γATP, T2 of both PCr and γATP, and linewidth were fixed in dictionary generation. Previously, we have performed simulation studies to evaluate the sensitivity of matched parameters to these fixed parameters.36 Our results indicated that quantification of the CK rate constant was insensitive to the linewidth and the T2 of both PCr and γATP, such that varying these parameters by 50% would lead to less than 5% deviation from its true value. While insensitivity to T1 of γATP is less ideal, a 25% variation will still give rise to only 5% matching error in the CK rate constant. On the other hand, the T1 of γATP is affected by multiple physical and chemical processes such as the cross-relaxation between γATP and βATP, and the chemical exchanges among ATP, PCr, and inorganic phosphate (Pi).20 As such, there may be physiological significance in measuring the T1 of γATP. In this case, expanding parameter matching to include the T1 of γATP may be considered. This will require adding another dimension to the current dictionary, which will significantly increase its size.
While our study demonstrated improved parameter estimation accuracy and efficiency of the 31P-MRSF method over the conventional MT-MRS method, the chosen sequence parameters are by no means optimized. Due to the large number of acquisition parameters used in the sequence, a theoretic framework and a computationally feasible method of sequence optimization are lacking. This is a major limitation of the current study, which needs to be addressed in the future. Another limitation of the current 31P-MRSF method is that the interleaved data acquisition scheme only acquires signal from PCr and γATP; signal evolution from Pi was not acquired. Fingerprint simulation used a two-compartment model that only involved exchange between PCr and ATP. As a result, Pi-to-ATP exchange was not evaluated in the current study, although γATP saturation also provides the opportunity of quantifying the Pi-to-ATP exchange rate.19 The low concentration of Pi is a significant challenge for quantitative assessment of the Pi-to-ATP exchange. Besides the need for acquiring the Pi signal with an adequate SNR, the expansion of a two-compartment model to a three-compartment model in dictionary generation and parameter matching also presents a computational challenge. How to improve the acquisition and computation efficiency for accurate assessment of the Pi-to-ATP exchange rate should be an important direction for future development of the 31P-MRSF technology. On the other hand, it is worth noting that the low concentration of Pi also enabled the use of a two-compartment model in the current study without compromising the accuracy of the estimated CK rate constant.36
Using the current 31P-MRSF method, we observed a significant increase in CK rate constant after stimulation-induced muscle contraction. Previously, we also observed an increased in CK rate constant in rats after an ischemia-reperfusion protocol.36 These results suggest a positive response of the CK system to metabolic perturbations that give rise to a transient misbalance between ATP production and demand. The strenuous muscle contraction induced by electrical stimulation elicits responses similar to that of acute exercise.50 The 31P-MRSF method provides an in vivo tool for investigating exercise physiology in widely used laboratory animals. The noninvasive nature of the 31P-MRSF method also enables the effects of chronic exercise to be monitored longitudinally.
In conclusion, the current study has demonstrated that 31P-MRSF allowed significantly increased accuracy and efficiency in measuring the CK rate constant in mouse hindlimb in vivo. It has paved the way for characterizing the CK system and energy metabolism in widely used laboratory animal models. Application of this technique to a large number of understudied mouse models of skeletal myopathies, such as muscular dystrophy,51 nemaline myopathy,52 and mitochondrial myopathy,53 will enhance our understanding of the role of the CK system in metabolic regulation and the pathogenesis of these diseases. The noninvasive nature of the technique and its high acquisition efficiency also provide potential for its clinical translation.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health R01-EB23704, R21-HL126215, F30-HL124894, and T32-EB007509. It made use of the High Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University.
Abbreviations:
- 31P
phosphorus-31
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- bSSFP
balanced steady-state free precession
- CEST
chemical exchange saturation transfer
- CK
creatine kinase
- CV
coefficient of variation
- FA
flip angle
- FID
free induction decay
- LW
linewidth
- MRF
magnetic resonance fingerprinting
- MRS
magnetic resonance spectroscopy
- MRSF
magnetic resonance spectroscopic fingerprinting
- MT
magnetization transfer
- PCr
phosphocreatine
- Pi
inorganic phosphate
- SNR
signal-to-noise ratio
- TR
repetition time
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Jacobus WE, Lehninger AL. Creatine kinase of rat heart mitochondria. Coupling of creatine phosphorylation to electron transport. J Biol Chem. 1973; 248(13):4803–4810. [PubMed] [Google Scholar]
- 2.Wallimann T, Wyss M, Brdiczka D, Nicolay K, Eppenberger HM. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the “phosphocreatine circuit” for cellular energy homeostasis. Biochem J. 1992;281:21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer RA, Sweeney HL, Kushmerick MJ. A simple analysis of the “phosphocreatine shuttle”. Am J Physiol. 1984;246(5 Pt 1):C365–C377. [DOI] [PubMed] [Google Scholar]
- 4.Schlattner U, Klaus A, Ramirez Rios S, Guzun R, Kay L, Tokarska-Schlattner M. Cellular compartmentation of energy metabolism: creatine kinase micro-compartments and recruitment of B-type creatine kinase to specific subcellular sites. Amino Acids. 2016;48(8):1751–1774. [DOI] [PubMed] [Google Scholar]
- 5.Weiss RG, Gerstenblith G, Bottomley PA. ATP flux through creatine kinase in the normal, stressed, and failing human heart. Proc Natl Acad Sci U S A. 2005;102(3):808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nahrendorf M, Spindler M, Hu K, et al. Creatine kinase knockout mice show left ventricular hypertrophy and dilatation, but unaltered remodeling post-myocardial infarction. Cardiovasc Res. 2005;65(2):419–427. [DOI] [PubMed] [Google Scholar]
- 7.Spindler M, Meyer K, Strömer H, et al. Creatine kinase-deficient hearts exhibit increased susceptibility to ischemia-reperfusion injury and impaired calcium homeostasis. Am J Physiol Heart Circ Physiol. 2004;287:1039–1045. [DOI] [PubMed] [Google Scholar]
- 8.Akki A, Su J, Yano T, et al. Creatine kinase overexpression improves ATP kinetics and contractile function in postischemic myocardium. Am J Physiol Heart Circ Physiol. 2012;303(7):H844–H852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta A, Akki A, Wang Y, et al. Creatine kinase-mediated improvement of function in failing mouse hearts provides causal evidence the failing heart is energy starved. J Clin Invest. 2012;122(1):291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.in’t Zandt HJA, Oerlernans F, Wieringa B, Heerschap A. Effects of ischemia on skeletal muscle energy metabolism in mice lacking creatine kinase monitored by in vivo 31P nuclear magnetic resonance spectroscopy. NMR Biomed. 1999;12:327–334. [DOI] [PubMed] [Google Scholar]
- 11.Cao F, Zervou S, Lygate CA. The creatine kinase system as a therapeutic target for myocardial ischaemia-reperfusion injury. Biochem Soc Trans. 2018; 46(5):1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown TR, Gadian DG, Garlick PB, Radda GK, Seeley PJ, Styles P. Creatine kinase activities in skeletal and cardiac muscle measured by saturation transfer NMR. In: Dutton PL, Leigh JS, Scarpa A, eds. Frontiers of Biological Energetics. New York: Academic; 1978:1341. [Google Scholar]
- 13.Bottomley PA, Hardy CJ. Mapping creatine kinase reaction rates in human brain and heart with 4 tesla saturation transfer 31P NMR. J Magn Reson. 1992;99:443–448. [Google Scholar]
- 14.Nicolay K, van Dorsten FA, Reese T, Kruiskamp MJ, Gellerich JF, van Echteld CJ. In situ measurements of creatine kinase flux by NMR. The lessons from bioengineered mice. Mol Cell Biochem. 1998;184(1/2):195–208. [PubMed] [Google Scholar]
- 15.Ren J, Sherry AD. Malloy CR. A simple approach to evaluate the kinetic rate constant for ATP synthesis in resting human skeletal muscle at 7 T. NMR Biomed. 2016;29(9):1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke WT, Robson MD, Neubauer S, Rodgers CT. Creatine kinase rate constant in the human heart measured with 3D-localization at 7 tesla. Magn Reson Med. 2017;78(1):20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parasoglou P, Xia D, Chang G, Convit A, Regatte RR. Three-dimensional mapping of the creatine kinase enzyme reaction rate in muscles of the lower leg. NMR Biomed. 2013;26(9):1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leibfritz D, Dreher W. Magnetization transfer MRS. NMR Biomed. 2001;14(2):65–76. [DOI] [PubMed] [Google Scholar]
- 19.Befroy DE, Rothman DL, Petersen KF, Shulman GI. 31P-magnetization transfer magnetic resonance spectroscopy measurements of in vivo metabolism. Diabetes. 2012;61(11):2669–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren J, Yang B. Sherry AD, Malloy CR. Exchange kinetics by inversion transfer: integrated analysis of the phosphorus metabolite kinetic exchanges in resting human skeletal muscle at 7 T. Magn Reson Med. 2014;73(4):1359–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn Reson Med. 2015;74(6):1621–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu AC, Badve C, Ponsky LE, et al. Development of a combined MR fingerprinting and diffusion examination for prostate cancer. Radiology. 2017;283(3):729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma D, Jiang Y, Chen Y, et al. Fast 3D magnetic resonance fingerprinting for a whole-brain coverage. Magn Reson Med. 2018;79(4):2190–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Jiang Y, Pahwa S, et al. MR fingerprinting for rapid quantitative abdominal imaging. Radiology. 2016;279(1):278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton JI, Jiang Y, Chen Y, et al. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn Reson Med. 2017;77(4):1446–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cloos MA, Assländer J, Abbas B, et al. Rapid radial T1 and T2 mapping of the hip articular cartilage with magnetic resonance fingerprinting. J Magn Reson Imaging. 2018;50(3):810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemasson B, Pannetier N, Coquery N, et al. MR vascular fingerprinting in stroke and brain tumors models. Sci Rep. 2016;6:37071–37081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Chen Y, Ma D, et al. Preclinical MR fingerprinting (MRF) at 7 T: effective quantitative imaging for rodent disease models. NMR Biomed. 2015; 28(3):384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Y, Wang CY, Anderson CE, et al. Fast magnetic resonance fingerprinting for dynamic contrast-enhanced studies in mice. Magn Reson Med. 2018;80(6):2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson CE, Wang CY, Gu Y, et al. Regularly incremented phase encoding-MR fingerprinting (RIPE-MRF) for enhanced motion artifact suppression in preclinical Cartesian MR fingerprinting. Magn Reson Med. 2018;79(4):2176–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulpanovich A, Tal A. The application of magnetic resonance fingerprinting to single voxel proton spectroscopy. NMR Biomed. 2018;31:e4001. [DOI] [PubMed] [Google Scholar]
- 33.Kulpanovich A, Tal A. What is the optimal schedule for multiparametric MRS? A magnetic resonance fingerprinting perspective. NMR Biomed. 2019; e4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen O, Huang S, McMahon MT, Rosen MS, Farrar CT. Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF). Magn Reson Med. 2018;80(6):2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, Han P, Zhou B, et al. Chemical exchange saturation transfer fingerprinting for exchange rate quantification. Magn Reson Med. 2018;80(4): 1352–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang CY, Liu Y, Huang S, Griswold MA, Seiberlich N, Yu X. 31P magnetic resonance fingerprinting for rapid quantification of creatine kinase reaction rate in vivo. NMR Biomed. 2017;30:e3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kingsley PB, Monahan WG. Corrections for off-resonance effects and incomplete saturation in conventional (two-site) saturation-transfer kinetic measurements. Magn Reson Med. 2000;43(6):810–819. [DOI] [PubMed] [Google Scholar]
- 38.Clifford B, Gu Y, Liu Y, et al. High-resolution dynamic 31P-MR spectroscopic imaging for mapping mitochondrial function. IEEE Trans Biomed Eng. 2020;67(10):2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Deursen J, Heerschap A, Oerlemans F, et al. Skeletal muscles of mice deficient in muscle creatine kinase lack burst activity. Cell. 1993;74(4): 621–631. [DOI] [PubMed] [Google Scholar]
- 40.van Deursen J, Ruitenbeek W, Heerschap A, Jap P, ter Laak H, Wieringa B. Creatine kinase (CK) in skeletal muscle energy metabolism: a study of mouse mutants with graded reduction in muscle CK expression. Proc Natl Acad Sci U S A. 1994;91(19):9091–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steeghs K, Benders A, Oerlemans F, et al. Altered Ca2+ responses in muscles with combined mitochondrial and cytosolic creatine kinase deficiencies. Cell. 1997;89(1):93–103. [DOI] [PubMed] [Google Scholar]
- 42.Dahlstedt AJ, Katz A, Westerblad H. Role of myoplasmic phosphate in contractile function of skeletal muscle: studies on creatine kinase-deficient mice. J Physiol. 2001;533(Pt 2):379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Momken I, Lechêne P, Koulmann N, et al. Impaired voluntary running capacity of creatine kinase-deficient mice. J Physiol. 2005;565(Pt 3):951–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brosnan MJ, Raman SP, Chen L, Koretsky AP. Altering creatine kinase isoenzymes in transgenic mouse muscle by overexpression of the B subunit. Am J Physiol Cell Physiol. 1993;264(1):C151–C160. [DOI] [PubMed] [Google Scholar]
- 45.Roman BB, Foley JM, Meyer RA, Koretsky AP. Contractile and metabolic effects of increased creatine kinase activity in mouse skeletal muscle. Am J Physiol Cell Physiol. 1996;270:236–245. [DOI] [PubMed] [Google Scholar]
- 46.Balaban RS, Kantor HL, Ferretti JA. In vivo flux between phosphocreatine and adenosine triphosphate determined by two-dimensional phosphorus NMR. J Biol Chem. 1983;258(21):12787–12789. [PubMed] [Google Scholar]
- 47.Nabuurs C, Huijbregts B, Wieringa B, Hilbers CW, Heerschap A. 31P saturation transfer spectroscopy predicts differential intracellular macromolecular association of ATP and ADP in skeletal muscle. J Biol Chem. 2010;285(51):39588–39596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kruiskamp MJ, van Vliet G, Nicolay K. 1H and 31P magnetization transfer studies of hindleg muscle in wild-type and creatine kinase-deficient mice. Magn Reson Med. 2000;43(5):657–664. [DOI] [PubMed] [Google Scholar]
- 49.Speck O, Scheffler K, Hennig J. Fast 31P chemical shift imaging using SSFP methods. Magn Reson Med. 2002;48(4):633–639. [DOI] [PubMed] [Google Scholar]
- 50.Kemp GJ, Ahmad RE, Nicolay K, Prompers JJ. Quantification of skeletal muscle mitochondrial function by 31P magnetic resonance spectroscopy techniques: a quantitative review. Acta Physiol. 2015;213:107–144. [DOI] [PubMed] [Google Scholar]
- 51.Whitmore C, Morgan J. What do mouse models of muscular dystrophy tell us about the DAPC and its components? Int J Exp Pathol. 2014;95(6): 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen MAT, Hardeman EC. Mouse models for thin filament disease. Adv Exp Med Biol. 2008;642:66–77. [DOI] [PubMed] [Google Scholar]
- 53.Wallace DC. Mouse models for mitochondrial disease. Am J Med Genet. 2001;106(1):71–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


