Abstract
Recent efforts to evaluate the heritability of the brain’s functional connectome have predominantly focused on static connectivity. However, evaluating connectivity changes across time can provide valuable insight about the inherent dynamic nature of brain function. Here, the heritability of Human Connectome Project resting-state fMRI data was examined to determine whether there is a genetic basis for dynamic fluctuations in functional connectivity. The dynamic connectivity variance, in addition to the dynamic mean and standard static connectivity, was evaluated. Heritability was estimated using Accelerated Permutation Inference for the ACE (APACE), which models the additive genetic (h2), common environmental (c2), and unique environmental (e2) variance. Heritability was moderate (mean h2: dynamic mean = 0.35, dynamic variance = 0.45, and static = 0.37) and tended to be greater for dynamic variance compared to either dynamic mean or static connectivity. Further, heritability of dynamic variance was reliable across both sessions for several network connections, particularly between higher-order cognitive and visual networks. For both dynamic mean and static connectivity, similar patterns of heritability were found across networks. The findings support the notion that dynamic connectivity is genetically influenced. The flexibility of network connections, not just their strength, is a heritable endophenotype that may predispose trait behavior.
Keywords: ACE, dynamic connectivity, heritability, resting state networks, static connectivity
Introduction
The combination of neuroimaging with genetic techniques is a powerful approach for understanding how individual differences in brain structure and function may be attributable in part to genetic variation (Meyer-Lindenberg and Weinberger 2006). Disentangling the relative effects of genes and environment on brain circuitry is crucial for understanding dynamics of both the healthy brain and those afflicted by neurologic or psychiatric disease, such as schizophrenia and Alzheimer’s Disease (Cannon et al. 2002; Jahanshad et al. 2013; Narayanan et al. 2015). These disorders have a strong genetic component and are associated with alterations in functional brain circuits. Imaging genetics studies, which explore the degree to which structural and functional features are influenced by genetic factors, have begun to identify heritable endophenotypes that are closely tied to the underlying neurobiology. A goal of imaging genetics is to develop precision medicine approaches, including improved characterization of subsyndromal or transdiagnostic biotypes, detection or prediction of early risk, and more refined treatments for mental health disorders.
To date, most imaging genetic studies focus on the heritability of structural phenotypes (Bartley et al. 1997; Sullivan et al. 2001; Thompson et al. 2001; Posthuma et al. 2002; Wright et al. 2002; Hulshoff Pol et al. 2006; Lenroot et al. 2009; Kremen et al. 2010; Rimol et al. 2010; Yoon et al. 2010; Joshi et al. 2011; Eyler et al. 2012; Chouinard-Decorte et al. 2014), such as cortical thickness and volume. These studies have indicated that heritability is regionally specific, with the highest heritability estimates in the prefrontal and temporal regions (Bartley et al. 1997; Thompson et al. 2001; Wright et al. 2002; Hulshoff Pol et al. 2006; Lenroot et al. 2009; Kremen et al. 2010; Rimol et al. 2010; Joshi et al. 2011; Chouinard-Decorte et al. 2014). One such study explored the effect of age on heritability of cortical thickness, reporting incomplete pleiotropy across age and suggesting that age-dependent gene expression contributes to cortical thickness (Chouinard-Decorte et al. 2014).
While early imaging genetic studies focused on structural indices, a growing body of research is examining the heritability of functional circuitry (Glahn et al. 2010; van den Heuvel et al. 2013; Korgaonkar et al. 2014; Moodie et al. 2014; Fu et al. 2015; Sinclair et al. 2015; Yang et al. 2016; Colclough et al. 2017; Ge et al. 2017; Sudre et al. 2017; Achterberg et al. 2018; Adhikari et al. 2018; Fox et al. 2018; Miranda-Dominguez et al. 2018). Most of these studies use resting-state functional magnetic resonance imaging (rsfMRI), a method that examines functional networks composed of spatially distributed regions in which spontaneous rsfMRI activity fluctuates together resulting in strong functional connectivity (FC) (Fox and Raichle 2007). These FC networks are commonly linked to both cognition and psychopathology. For example, network efficiency is associated with intelligence (Li et al. 2009; van den Heuvel et al. 2009) and abnormal network architecture has been found across a host of psychiatric disorders (Wang et al. 2010).
Imaging genetics studies using rsfMRI have consistently reported mild to strong heritability of global and network FC, with the majority of studies reporting mild to moderate values. A number of studies have found that heritability of the default mode network is especially strong, with genetic influence accounting for 15–42% of the variance within the network, significantly outweighing shared environmental contributions (Glahn et al. 2010; Korgaonkar et al. 2014; Colclough et al. 2017; Sudre et al. 2017; Miranda-Dominguez et al. 2018).
Traditionally, FC studies, including those utilizing imaging genetics, have focused on static measures of connectivity, in which connectivity reflects the correlation in activity between 2 regions across the entire scan. More recently, a growing body of evidence has shown that accounting for changes in FC across time can provide valuable insight into the dynamic nature of brain function (Hutchison et al. 2013; Calhoun et al. 2014; Lindquist et al. 2014; Lurie et al. 2020; Pervaiz et al. 2020). Changes in the strength and direction of network connections vary on the scale of seconds and minutes within a single rsfMRI scan (Chang and Glover 2010; Sakoglu et al. 2010; Kiviniemi et al. 2011; Handwerker et al. 2012; Jones et al. 2012), and such time-sensitive FC information is lost when FC reflects static correlation over the course of a testing session. By quantifying FC changes over time, dynamic FC preserves information about brain network dynamics which may contribute to phenotypic variation.
The utility of network dynamics in predicting trait behavior has been increasingly underscored in recent years (Chang and Glover 2010; Hutchison et al. 2013; Allen et al. 2014). Dynamic FC states reflect individual differences in clinical symptoms (Damaraju et al. 2014; Kucyi and Davis 2014; Elton and Gao 2015; Barber et al. 2018), and behavior (Barttfeld et al. 2015). It has been suggested that dynamic FC is a more reliable predictor of psychiatric disease, such as schizophrenia (Supekar et al. 2018), than static FC. Further, dynamic FC reflects on-going intrasubject fluctuations in behavioral performance, indicating momentary changes in attention and cognitive states (Thompson et al. 2013; Kucyi and Davis, 2014; Barttfeld et al., 2015). These findings suggest that examination of the heritability of network dynamics is warranted, although to date such studies have been limited. One study indicated that dynamic connectivity states were more similar between monozygotic (MZ) than dizygotic (DZ) twins, although heritability values were not explicitly estimated (Vidaurre et al. 2017).
Dynamic FC studies have revealed that both the magnitude of correlation coefficients and the variance (i.e., fluctuations) in the strength of those correlations over time both carry uniquely important information about brain network function (Thompson and Fransson 2015; Choe et al. 2017). For example, one study reported that approaches based on correlation magnitude were better suited to analyze within-network connectivity, while variance-based approaches were better suited for between-network connectivity analyses (Thompson and Fransson 2015). The dynamic conditional correlation (DCC) model is a time-series-based approach which is effective for estimating both mean correlation coefficients of networks as well as the variance in those correlations (Lebo and Box-Steffensmeier 2008). Recently, our understanding of dynamic FC variance has evolved from its initial designation as a source of noise to a putative index of brain flexibility and function (McIntosh et al. 2010; Garrett et al. 2013b; Tognoli and Kelso 2014; Nomi et al. 2017; Nomi et al. 2018). For example, patients with schizophrenia show less dynamic FC variance than healthy controls (Yu et al. 2015), individuals with autism spectrum disorders show altered variance (He et al. 2018; Guo et al. 2020), and dynamic variance predicts attention deficit hyperactivity disorder (ADHD) symptom severity (Nomi et al. 2018). Despite increasing interest in dynamic FC variance as biologically meaningful, there are no studies evaluating its heritability.
Examining twin pairs provides a valuable opportunity for understanding the heritability of brain network function. The Human Connectome Project (HCP) is a multimodal neuroimaging study that includes family structure for hundreds of sibling sets, including MZ twins, DZ twins, and nontwin siblings, as well as singletons. Here, we use DCC and the ACE model (Chen et al. 2014) to investigate heritability of network FC using both dynamic and static metrics in the HCP dataset. This is the first study to examine whether the variance of dynamic FC, a metric that reflects network flexibility, is itself heritable.
Materials and Methods
Participants
A total of 815 healthy adult participants from the HCP 900 Subjects Data Release (http://humanconnectome.org/documentation/S900) had complete data available for family structure, 4 resting state scans, and nuisance variables (age, gender, age2, age x gender, age2 x gender, handedness, race, ethnicity, and scan motion). The HCP contains a relatively high proportion of twins and/or sibling sets. For the current study sample, this included 81 MZ twins, 78 DZ twins, 103 nontwin sibling sets, and 113 unrelated participants. Families ranged in size from 2 to 6 siblings with some including both twins and nontwin siblings.
HCP Imaging Acquisition and Data Processing
The HCP rsfMRI protocol has been previously described in detail (Glasser et al. 2013; Smith et al. 2013; Van Essen et al. 2013). Four 14 min, 33 s rsfMRI runs—divided across 2 sessions on sequential days—were acquired on a Siemens connectome-Skyra 3 T scanner with 32-channel head coil (multiband acceleration factor of 8, TR = 0.72 s, 2 mm isotropic spatial resolution). The time-courses from the “Parcellation-Timeseries-Netmats (PTN) extensively processed resting-state fMRI dataset” were used to assess the heritability of network and edge-wise resting-state connectivity. This publicly released dataset was already processed, including artifact removal using ICA + FIX (Griffanti et al. 2014; Salimi-Khorshidi et al. 2014), temporal demeaning and variance normalization (Beckmann and Smith 2004), and data reduction using MELODIC for Incremental Group-PCA (Smith et al. 2014) and spatial Group-ICA (Beckmann and Smith 2004; Smith et al. 2013). The Group-ICA had been performed at several dimensionalities (i.e., 25, 50, 100, 200, 300 components). It was expected that the higher ICA dimensionalities would improve network connectivity estimates due to the more refined spatial identification of networks and due to the inclusion of a greater number of edges per network. For that reason, our primary analysis used variance component models to examine heritability with the 300-dimensional ICA. Follow-up analyses at other ICA dimensionalities were performed to assess the impact of ICA dimensionality on heritability estimates. An overview of the processing pipeline is shown in Figure 1.
Figure 1 .
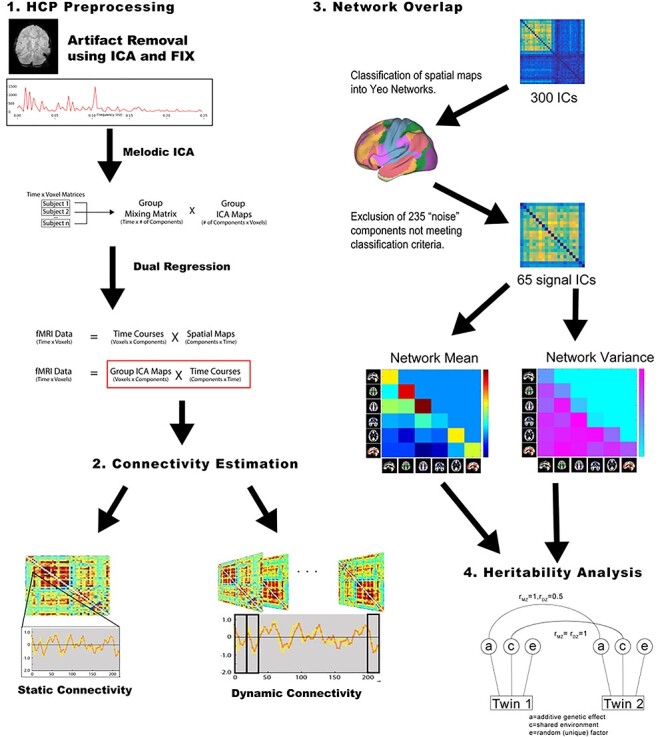
Schematic for data processing pipeline.
Network Identification
Our primary analysis examined heritability for mean within- and between-network connectivity estimates of the 300-dimensional ICA dataset. To assess the range of heritability values for the edge-wise connections, follow-up analyses were done on individual connections between all signal components. For the purposes of network classification and signal component identification, the volumetric MNI152 3D-space version of the Group-ICA component spatial maps was automatically labeled based on spatial overlap with the Yeo 7-network parcellation (Yeo et al. 2011) using previously defined criteria (Barber et al. 2018). This included thresholding the top 5% of voxels within each component map and then assigning the component to the network with the highest spatial overlap. Each component was thereby labeled as one of the 7 Yeo networks: Visual (VN), Somatomotor (SMN), Dorsal Attention (DAN), Ventral Attention (VAN), Limbic (LN), Frontoparietal (FPN), or Default Mode (DMN), or it was classified as “noise” if it had a low degree of network overlap (i.e., < 500 suprathreshold voxels or < 55% overlap with any one network). This resulted in 65 “signal” components (29 VN, 8 SMN, 3 DAN, 2 VAN, 8 FPN, and 15 DMN), which were used to compute connectivity. Since no components were classified as LN, only 6 networks were included for the network-wise analyses.
To determine whether this labeling method accurately characterized “noise” components, the ICA components were also labeled based purely on temporal characteristics of the component time-courses. This involved examination of the LF:HF power, based on a previous method (Allen et al. 2011). LF power was defined as the integral of power from 0.1 to 0.01 Hz, while HF power was defined as the power from 0.2 to 0.67 Hz. The latter was modified from that of Allen et al. (2011), who defined HF power as 0.15–0.25 Hz. Recent studies have determined that resting-state network signatures exist at higher frequencies than previously thought, so the current study increased the minimum frequency to 0.2 Hz and also set the maximum frequency to 0.67 Hz, due to the higher temporal resolution of the HCP multiband acquisition. To calculate the LF:HF power, the integral of power was first calculated within these prespecified ranges for each subject and each component timecourse. Each ICA component was then classified as signal if the mean LF:HF power across all subjects was greater than 2 (i.e., at least twice as much LF as HF power in the component), or was otherwise classified as noise. Comparing the new temporal classification with that of our previous component classification using the Yeo 7 network spatial overlap, we found 90% sensitivity and 89% specificity. Our previous spatial approach classified 65 out of 300 components as signal, while the new temporal approach classified 87 networks as signal. A total of 58 ICs were consistently classified as signal using both approaches and only 7 of the 65 components classified as signal using the Yeo spatial overlap approach were classified as noise using the temporal approach, suggesting that the previous spatial method does a relatively good job at distinguishing between signal and noise components.
The Yeo 7-network parcellation was used to label the ICs for the current study, rather than a finer-grained parcellation, since previous studies have shown that higher model order ICA tends to subdivide larger networks into smaller subnetworks (Kiviniemi et al. 2009; Smith et al. 2009; Abou-Elseoud et al. 2010; Abou Elseoud et al. 2011; Ray et al. 2013). Therefore, the 300-component high model order ICA is used to distinguish sub-networks characterized by each of the individual ICs. The coarse functional labeling is then used to group these components into larger-scale networks.
Dynamic and Static Connectivity Estimation
For both dynamic and static connectivity, primary ACE models examined heritability of network-wise connectivity and follow-up ACE models examined heritability of edge-wise connectivity. Network-wise estimation involved first computing edge-wise connectivity between all pairs of signal components and then averaging over those connections identified as a particular network pair. For the 6 Yeo networks that were included in the current study, this resulted in 21 network connections (i.e., 6 * 5/2 = 21), including both within- and between-network connections. For edge-wise connections between each of the 65 signal components, this resulted in 2080 connections (i.e., 65 * 64/2).
DCC was used to compute dynamic connectivity (https://github.com/canlab/Lindquist_Dynamic_Correlation) (Lindquist et al. 2014) (please refer to the Supplementary Information for a mathematical description of DCC). Briefly here, DCC is a multivariate volatility method in which the current conditional correlation is updated using a linear combination of past estimates of the conditional correlation and current observations. DCC estimates model parameters through quasi-maximum likelihood methods and therefore, unlike sliding window approaches, does not require an arbitrary window length to be set. Pairwise-dynamic connectivity values were obtained for every time-point of each participant’s 4 resting state runs. A matrix of 2080 edge-wise connectivity values (i.e., all pairwise connections between the 65 signal components) was generated at each of the 2400 time points for each scan session. A matrix of 21 network-wise connectivity values (i.e., all pairwise connections between 6 Yeo networks) was generated at each of the 2400 time points for each scan session. Dynamic connectivity was then summarized as the mean or variance across all time points within a run. Therefore, the DCC mean represented the strength of dynamic connectivity for a run, while the DCC variance represented the variability or flexibility of dynamic connectivity for a run. For each connection, static connectivity was the Pearson’s correlation coefficient between pairwise-signal component time-courses and therefore, represented standard FC across the run. As with the DCC measures, this was first computed within each run and then averaged over runs. ACE modeling was performed on the mean and variance of the dynamic connectivity and on the static connectivity for 21 network connections with follow-up heritability models done on the 2080 edge-wise connections.
Heritability of Dynamic Mean and Variance and Static Mean in Connectivity
Heritability analysis was performed using the Accelerated Permutation Inference for the ACE Model (APACE: (Chen et al. 2014; Colclough et al. 2017)). The ACE model of heritability is a variance decomposition of the additive genetic (A), common environmental (C), and unique environmental (E) variance, using family structure. Each APACE model included nuisance variables (age, gender, age2, age x gender, age2 x gender, handedness, race, ethnicity, and scan motion).
APACE speeds computation time by inferring the A, C, and E components from the squared differences of paired individuals (e.g., twin-pairs versus unrelated pairs). The parameters are then estimated using ordinary least squares regression and inference is done using the Likelihood Ratio Test (LRT). To assess whether the mean heritability across the 21 network connections is significantly greater than zero, permutation testing was performed by shuffling the relationship labels 1000 times and recomputing the LRT. Multiple comparison correction across the 21 network pairs was implemented using False Discovery Rate (FDR) in which the P value for each network pair was taken from the permutation-based empirical distribution. For the mean h2, c2, and e2 values across the 21 network pairs, APACE software computes the confidence intervals by bootstrapping 1000 times.
To determine the stability of the heritability findings, the APACE models were run on the 2 sessions, consisting of the 2 Day 1 runs and the 2 Day 2 runs, separately. In addition, to further assess the stability of the heritability findings, the APACE models were run using varying scan lengths. Scan length was examined in 2 ways: 1) consecutively and 2) averaged over runs. First, consecutive scan length was examined to determine the amount of time required for heritability estimates to stabilize. Second, scan length averaged over runs was examined since previous studies have found that FC values and heritability estimates are more reliable when pooling across multiple runs (Ge et al. 2017; Noble et al. 2017). For consecutive scan length, heritability was estimated across scan volumes in the order in which they were collected and included 300, 600, 900, 1200, 2400, 3600, or 4800 time points (i.e., corresponding to 3:36, 7:12, 10:48, 14:24, 28:48, 43:12, and 57:36 (min:s) of consecutive scanning). For scan length averaged over runs, heritability was estimated across the first 300, 600, 900, or 1200 time points for each run and then averaged over the 4 runs, resulting in 3.6, 7.2, 10.8, or 14.4 min of data per run (i.e., 14.4, 28.8, 43.2, or 57.6 min of data across the 4 runs).
Results
Heritability of Network Connectivity
Both dynamic (mean and variance) and static connectivity measures resulted in nonzero heritability values across all network pairs (Fig. S1). Heritability values for dynamic connectivity were within a similar range to that of more traditional static connectivity heritability values (Fig. 2). The heritability of the dynamic connectivity mean ranged from 0.21 to 0.53 (session 1, mean dynamic mean h2 = 0.39, P = 0.0070; session 2, mean dynamic mean h2 = 0.31, P = 0.012), which was within a similar range to the heritability of static connectivity (static h2 range = 0.05–0.53; session 1, mean static h2 = 0.38, P = 0.0060; session 2, mean static h2 = 0.37, P = 0.0030) and was comparable to that of previous studies examining the heritability of traditional static FC (Glahn et al. 2010; Yang et al. 2016; Ge et al. 2017; Adhikari et al. 2018). Likewise, the heritability of the dynamic connectivity variance ranged from 0.2 to 0.59 (session 1, mean dynamic variance h2 = 0.45, P = 0.0010; session 2, mean dynamic variance h2 = 0.45, P = 0.0010), which was also within a similar range to the heritability of static connectivity.
Figure 2 .
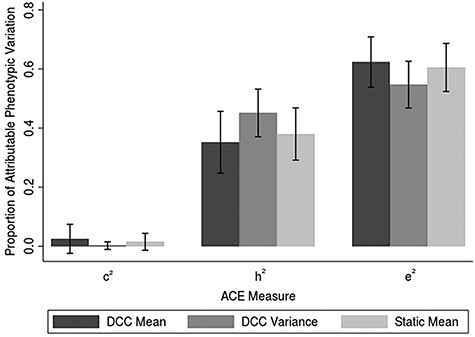
Relative contributions to connectivity measures. Bar graphs represent the averaged ACE measures across both sessions, with error bars reflecting standard deviation. For both sessions individually, heritability was higher for the DCC variance relative to DCC mean (session 1: t(20) = 2.20, P = 0.040; session 2: t(20) = 5.15, P = 4.88 × 10−5) and was higher for DCC variance relative to static connectivity (session 1: t(20) = 2.40, P = 0.026; session 2: t(20) = 2.87, P = 0.0095).Abbreviations: h2 = genetic variance, c2 = common environmental variance, e2 = unique environmental variance.
Examining Heritability across Network Pairs
Figure S1 displays the network-wise heritability values for the 3 connectivity metrics during each session. For both dynamic mean and static connectivity, only the heritability of the VN-DAN network connections was consistently significant for both sessions (dynamic mean, session 1: h2 = 0.45, FDR-corrected P value = 0.021; dynamic mean, session 2, h2 = 0.38, FDR-corrected P value = 0.029; static, session 1: h2 = 0.47, FDR-corrected P value = 0.042; static, session 2, h2 = 0.49, FDR-corrected P value = 0.021). For the variance of the dynamic connectivity, on the other hand, heritability was consistently significant for a number of network pairs. This included VN-SMN (session 1: h2 = 0.45, FDR-corrected P value = 0.031; session 2, h2 = 0.45, FDR-corrected P value = 0.021), VN-VAN (session 1: h2 = 0.55, FDR-corrected P value = 0.043; session 2, h2 = 0.46, FDR-corrected P value = 0.021), VN-FPN (session 1: h2 = 0.52, FDR-corrected P value = 0.013; session 2, h2 = 0.49, FDR-corrected P value = 0.021), VN-DMN (session 1: h2 = 0.50, FDR-corrected P value = 0.026; session 2, h2 = 0.47, FDR-corrected P value = 0.021), SMN-VAN (session 1: h2 = 0.36, FDR-corrected P value = 0.0070; session 2, h2 = 0.38, FDR-corrected P value = 0.021), and DAN-FPN (session 1: h2 = 0.50, FDR-corrected P value = 0.030; session 2, h2 = 0.53, FDR-corrected P value = 0.021).
Examining Heritability across Different Scan Lengths
The effect of scan length on heritability estimates was examined for consecutive time points as well as for time points averaged over runs. Figure 3 shows that for consecutive scan lengths of 300, 600, 900, 1200, 2400, 3600, and 4800 time points (i.e., 3:36, 7:12, 10:48, 14:24, 28:48, 43:12, and 57:36 (min:s)), relatively long scan epochs are needed before heritability estimates stabilize (~2400–3600 time points or ~ 28–43 min). This is the case for all 3 connectivity metrics; although, as has been consistently found in the current study, the DCC variability tends to result in higher heritability at all scan lengths, and requires fewer time points to stabilize compared to DCC mean and static connectivity. Figure 4 shows that for scan length averaged over runs, heritability values were generally consistent for scan lengths of 300, 600, 900, and 1200 across the 4 runs. For static connectivity, heritability values were lower at a scan length of 300, but tended to be consistent for scan length >300. For DCC measures, heritability values were stable across all scan lengths.
Figure 3 .
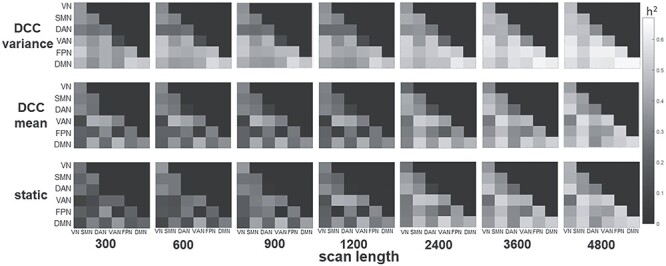
Heritability as a function of consecutive scan length. Connectivity values were computed for the first 300, 600, 900, 1200, 2400, 3600, or 4800 consecutive time points, resulting in 3.6, 7.2, 10.8, 14.4, 28.8, 43.2, or 57.6 min of data. The connectivity metrics were first computed over the consecutive scan length in each subject before performing APACE. Heritability based on consecutive scans required 2400–3600 time points (i.e., between 28 and 43 min of scanning) before the heritability measures became stable. This was the case for all 3 connectivity metrics although, as was consistently found in the current study, the DCC variability tended to result in higher heritability at all scan lengths, and required fewer time points to stabilize than DCC mean and static connectivity.
Figure 4 .
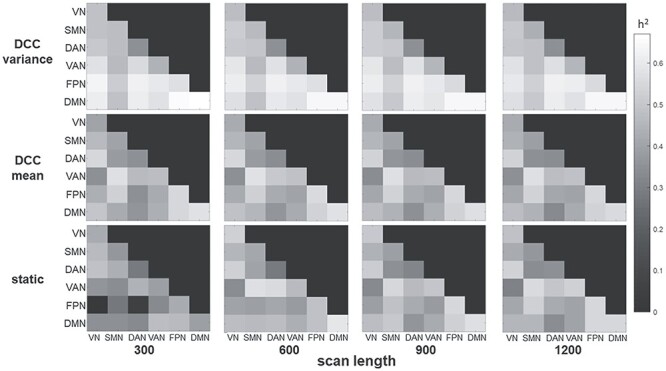
Heritability as a function of scan length averaged over 4 runs. Connectivity values were computed for the first 300, 600, 900, or 1200 time points of each run, resulting in 3.6, 7.2, 10.8, or 14.4 min of data per run (i.e., 14.4, 28.8, 43.2, or 57.6 min of data across the 4 runs). As with the primary analyses, the connectivity metrics were first computed within each run and then averaged across the 4 runs before performing APACE. Heritability values were generally stable across all scan lengths for the DCC metrics. Heritability values were tended to be lower for the scan length of 300 for static connectivity.
Examining Heritability across Dimensionalities
Table S1 displays the range and mean of the network-wise heritability values for sessions 1 and 2 at ICA dimensionalities of 25, 50, 100, 200, and 300 components. The heritability values did not change much across the ICA dimensionalities. While there was a slight increase in the heritability values for DCC variance from 25 to 300 components, this was mostly due to smaller values for ICA dimensionalities of less than 100 components. For DCC mean and static connectivity, this trend was less clear. Across all ICA dimensionalities, DCC variance tended to have higher heritability values than either DCC mean or static connectivity.
Examining Heritability of Individual Connections
Table S2 displays the range and mean of the individual connection heritability values of the 300-dimensional ICA for sessions 1 and 2. Due to the large number of individual connections, heritability was only examined for the primary 300-dimensional ICA results. We found that the heritability range tended to be larger and the mean heritability tended to be lower for individual connections, than for mean network connections. This may have been due to very low heritability for a few individual connections.
Higher Heritability for the Variance of Dynamic Connectivity
As found previously (Choe et al. 2017), the dynamic connectivity mean values were similar to the static FC values. This was true both for the actual connectivity values, as well as for the heritability. Further, the heritability of dynamic variance tended to be higher than either that of the dynamic mean or static connectivity. This trend was found across different dimensionalities (Table S1) as well as for the individual connections (Table S2). When the effect of scan length was considered, DCC variance also tended to have higher heritability values across all scan lengths (Fig. 3). To determine whether this trend was significant across the 21 network pairs for the 300-dimensional ICA primary heritability findings, we performed 2-tailed, paired t-tests which tested whether the heritability of dynamic variance was different from that of dynamic mean or of static connectivity across the 21 network pairs. We found that for both sessions, heritability tended to be higher for the dynamic variance as compared to dynamic mean (session 1: t(20) = 2.20, P = 0.040; session 2: t(20) = 5.15, P = 4.88 × 10−5) and also tended to be higher for dynamic variance as compared to static connectivity (session 1: t(20) = 2.40, P = 0.026; session 2: t(20) = 2.87, P = 0.0095).
Discussion
Our results provide novel and compelling evidence for the heritability of dynamic connectivity. Dynamic connectivity metrics were robustly heritable and the estimated heritability values for dynamic connectivity were within the range of static connectivity heritability values. Importantly, we found that the heritability of dynamic variance tended to be higher than that of either DCC mean or static connectivity, suggesting that genetic influence affects not only the strength but also the flexibility of network connectivity. Furthermore, a greater number of network-wise connections had robust heritability for DCC variance than for either DCC mean or static connectivity. Heritability of DCC variance was robust across both sessions for several VN, SMN, and higher-order network connections, whereas the heritability of DCC mean and static connectivity was more restricted and was only significant across both sessions for the VN-DAN connection.
Not only is this study the first to directly assess differences in heritability of dynamic and static connectivity, but also the first to establish the relatively greater heritability of DCC variance than DCC mean. Our results show robust and reliable heritability of DCC variance for VN network connections and also for connections with higher-order networks, in particular the FPN and DMN. Although variability in BOLD signal has traditionally been considered noise or a confounding factor, several recent studies suggest that signal variance may be important for optimal performance (McIntosh et al. 2010; Garrett et al. 2013a; Tognoli and Kelso 2014; Nomi et al. 2017). Increased variability in BOLD signal is associated with more efficient behavioral performance (Garrett et al. 2013a) and faster reaction times in attentional tasks (Garrett et al. 2011). Variability has also been identified as an important feature of networks, enabling the shift between integration and segregation (Tognoli and Kelso 2014). Given the implications of variance for optimal brain functioning, it is conceivable that network variance reflects genetic predisposition to cognitive phenotypes in both healthy individuals and those with psychiatric disorders.
Although dynamic connectivity, and the dynamic variance in particular, captures state changes in connectivity over time, the fact that dynamic variance is heritable suggests that it is an important contributor to trait-level individual differences in behavior. Further, it suggests that the degree to which particular network connections change over time plays an important role in circuit dynamics. This may reflect changes in network or global configurations, hormone and/or neurotransmitter concentrations, arousal, or the frequency of engagement of particular dynamic states. Thus, such state-level FC fluctuations may provide unique information regarding network dynamics.
Other recent studies using the HCP dataset have also reported robust heritability (ranging from 20 to 40% of the variance explained) for several functional networks, including the DMN, FPN, SMN, VN, SN, and attention networks (Ge et al. 2017; Adhikari et al. 2018; Miranda-Dominguez et al. 2018), with the DAN (Yang et al. 2016; Miranda-Dominguez et al. 2018) and VN (Yang et al. 2016) having some of the strongest genetic effects. Interestingly, 4 of the 6 significantly heritable network couplings identified in our analyses included the VN, and only the VN-DAN network coupling demonstrated robust heritability across both sessions. This heritability of the VN couplings is consistent with previous studies, which have reported that genetic influences are stronger for sensory networks, such as VN and sensorimotor, than for cognition-related networks (Fu et al. 2015).
Our findings should be considered with some limitations. First, although controlled for in our model, the twin/sibling groups in the HCP data differ in age, sex, and race. Future studies in samples with demographically matched groups could help to evaluate whether such demographic differences have differential effects on network heritability. Second, heritability could be examined in the context of other dynamic connectivity methods such as the identification of reoccurring dynamic connectivity states. Clustering algorithms may improve detection of genetic influences on connectivity patterns by classifying unique brain states. Future work could also expand upon our findings by including samples of extended families, rather than solely siblings and twins as we have explored here. Third, although we have employed a number of methods to reduce nuisance confounds (i.e., nuisance regression and group-level covariates), a number of uncontrolled state-level phenotypic variables (e.g., medication-use, hunger, fatigue, diurnal fluctuations, recent life events) are likely to have varying levels of influence on FC metrics and contribute to noise in ACE model estimates. The heritability estimates generated by the APACE model were generally consistent across both sessions; however, there were a few cases in which heritability was significant for one session, but not the other. Heritability studies, like most imaging studies examining individual differences, assume that functional differences reflect trait behavior, and therefore, can be profoundly influenced by uncontrolled state-level variables. Like previous studies, the current study found that reliability tended to be greater when pooling data across multiple sessions (Ge et al. 2017; Noble et al. 2017). Fourth, the LRT used in APACE will be mis-specified for variables that are not normally distributed. Using permutation testing improves Type I error control. However, the mis-specification can potentially negatively affect Type II error. Fifth, the current study used a high-order ICA approach and then classified the components into networks. While high-dimensional ICA has previously been shown to be effective in predicting behavioral utility (Pervaiz et al. 2020), it also assumes spatial independence of the ICA components. Other methods that relax this assumption may more accurately represent regions that change affiliations over time (Harrison et al. 2015; Bijsterbosch et al. 2019). Further, while we have shown that our network labeling method is fairly accurate in distinguishing signal and noise components, more accurate labeling methods may improve network heritability estimation. Finally, although these findings provide valuable insights as to the genetic contribution of network connectivity, they do not leverage molecular genetics to identify individual genes or gene networks that influenceFC.
Our results demonstrate the utility of examining the heritability of dynamic connectivity metrics and provide evidence that genetic influences are stronger for the variance of dynamic connectivity than for static and mean dynamic measures. This indicates that network flexibility, in addition to network strength, is an important contributor to trait-level differences in behavior. Establishing the heritability of functional networks has potentially far-reaching implications for understanding the etiology of disease. Many neurologic and psychiatric diseases, such as schizophrenia, autism spectrum disorder, Alzheimer’s, and ADHD, have a strong genetic component and are associated with aberrant function of brain circuits. Assessing the relationship of both dynamic mean and variance with genetics could help to identify endophenotypes. These endophenotypes may in turn illuminate the neurobiological etiologies of these disorders and aid in the development of novel therapeutics.
Notes
Conflict of Interest: The authors have no disclaimers.
Funding
This work was supported by the National Institutes of Mental Health (NIMH) R21 MH101506 (PI Karlsgodt) and R21 MH122886 (PI Barber).
Supplementary Material
Contributor Information
Anita D Barber, Division of Psychiatry Research, Zucker Hillside Hospital, Glen Oaks, New York, 11004, USA; Institute for Behavioral Science, The Feinstein Institutes for Medical Research, Manhasset, New York, 11030, USA; Department of Psychiatry, Zucker School of Medicine at Hofstra/Northwell, Hempstead, NY, 11549, USA.
Catherine E Hegarty, Department of Psychology, University of California, Los Angeles, 90095, USA.
Martin Lindquist, Department of Biostatistics, Johns Hopkins University, Baltimore, 21205, USA.
Katherine H Karlsgodt, Department of Psychology, University of California, Los Angeles, 90095, USA; Department of Psychiatry and Biobehavioral Sciences, University of California, Los Angeles, 90095, USA.
References
- Abou-Elseoud A, Starck T, Remes J, Nikkinen J, Tervonen O, Kiviniemi V. 2010. The effect of model order selection in group PICA. Hum Brain Mapp. 31:1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Elseoud A, Littow H, Remes J, Starck T, Nikkinen J, Nissila J, Timonen M, Tervonen O, Kiviniemi V. 2011. Group-ICA model order highlights patterns of functional brain connectivity. Front Syst Neurosci. 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achterberg M, Bakermans-Kranenburg MJ, van Ijzendoorn MH, van der Meulen M, Tottenham N, Crone EA. 2018. Distinctive heritability patterns of subcortical-prefrontal cortex resting state connectivity in childhood: a twin study. Neuroimage. 175:138–149. [DOI] [PubMed] [Google Scholar]
- Adhikari BM, Jahanshad N, Shukla D, Glahn DC, Blangero J, Fox PT, Reynolds RC, Cox RW, Fieremans E, Veraart J et al. 2018. Comparison of heritability estimates on resting state fMRI connectivity phenotypes using the ENIGMA analysis pipeline. Hum Brain Mapp. 39:4893–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. 2014. Trackgging whole-brain connectivity dynamics in the resting state. Cereb Cortex. 24:663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R et al. 2011. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci. 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber AD, Lindquist MA, DeRosse P, Karlsgodt KH. 2018. Dynamic functional connectivity states reflecting psychotic-like experiences. Biol Psychiatry Cogn Neurosci Neuroimaging. 3:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley AJ, Jones DW, Weinberger DR. 1997. Genetic variability of human brain size and cortical gyral patterns. Brain. 120(Pt 2):257–269. [DOI] [PubMed] [Google Scholar]
- Barttfeld P, Uhrig L, Sitt JD, Sigman M, Jarraya B, Dehaene S. 2015. Signature of consciousness in the dynamics of resting-state brain activity. Proc Natl Acad Sci U S A. 112:887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. 2004. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 23:137–152. [DOI] [PubMed] [Google Scholar]
- Bijsterbosch JD, Beckmann CF, Woolrich MW, Smith SM, Harrison SJ. 2019. The relationship between spatial configuration and functional connectivity of brain regions revisited. Elife. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Miller R, Pearlson G, Adali T. 2014. The chronnectome: time-varying connectivity networks as the next frontier in fMRI data discovery. Neuron. 84:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, van Erp TG, Glahn DC. 2002. Elucidating continuities and discontinuities between schizotypy and schizophrenia in the nervous system. Schizophr Res. 54:151–156. [DOI] [PubMed] [Google Scholar]
- Chang C, Glover GH. 2010. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 50:81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Smith S, Viding E, Nichols TE, editors. 2014. APACE: Accelerated Permutation Inference for the ACE Model., 20th Annual Meeting of the Organization for Human Brain Mapping; 2014 June 8–12. Germany: Hamburg. [Google Scholar]
- Choe AS, Nebel MB, Barber AD, Cohen JR, Xu Y, Pekar JJ, Caffo B, Lindquist MA. 2017. Comparing test-retest reliability of dynamic functional connectivity methods. Neuroimage. 158:155–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouinard-Decorte F, McKay DR, Reid A, Khundrakpam B, Zhao L, Karama S, Rioux P, Sprooten E, Knowles E, Kent JW Jr et al. 2014. Heritable changes in regional cortical thickness with age. Brain Imaging Behav. 8:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colclough GL, Smith SM, Nichols TE, Winkler AM, Sotiropoulos SN, Glasser MF, Van Essen DC, Woolrich MW. 2017. The heritability of multi-modal connectivity in human brain activity. Elife. 6:e20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju E, Allen EA, Belger A, Ford JM, McEwen S, Mathalon DH, Mueller BA, Pearlson GD, Potkin SG, Preda A et al. 2014. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. NeuroImage Clinical. 5:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton A, Gao W. 2015. Task-related modulation of functional connectivity variability and its behavioral correlations. Hum Brain Mapp. 36:3260–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler LT, Chen CH, Panizzon MS, Fennema-Notestine C, Neale MC, Jak A, Jernigan TL, Fischl B, Franz CE, Lyons MJ et al. 2012. A comparison of heritability maps of cortical surface area and thickness and the influence of adjustment for whole brain measures: a magnetic resonance imaging twin study. Twin Res Hum Genet. 15:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AS, Oler JA, Birn RM, Shackman AJ, Alexander AL, Kalin NH. 2018. Functional connectivity within the primate extended amygdala is heritable and associated with early-life anxious temperament. J Neurosci. 38:7611–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 8:700–711. [DOI] [PubMed] [Google Scholar]
- Fu Y, Ma Z, Hamilton C, Liang Z, Hou X, Ma X, Hu X, He Q, Deng W, Wang Y et al. 2015. Genetic influences on resting-state functional networks: a twin study. Hum Brain Mapp. 36:3959–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett DD, Kovacevic N, McIntosh AR, Grady CL. 2011. The importance of being variable. J Neurosci. 31:4496–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett DD, Kovacevic N, McIntosh AR, Grady CL. 2013a. The modulation of BOLD variability between cognitive states varies by age and processing speed. Cereb Cortex. 23:684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett DD, Samanez-Larkin GR, MacDonald SW, Lindenberger U, McIntosh AR, Grady CL. 2013b. Moment-to-moment brain signal variability: a next frontier in human brain mapping? Neurosci Biobehav Rev. 37:610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Holmes AJ, Buckner RL, Smoller JW, Sabuncu MR. 2017. Heritability analysis with repeat measurements and its application to resting-state functional connectivity. Proc Natl Acad Sci U S A. 114:5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, Curran JC, Olvera RL, Laird AR, Smith SM et al. 2010. Genetic control over the resting brain. Proc Natl Acad Sci U S A. 107:1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR et al. 2013. The minimal preprocessing pipelines for the human connectome project. Neuroimage. 80:105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L, Salimi-Khorshidi G, Beckmann CF, Auerbach EJ, Douaud G, Sexton CE, Zsoldos E, Ebmeier KP, Filippini N, Mackay CE et al. 2014. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. Neuroimage. 95:232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Duan X, Chen H, He C, Xiao J, Han S, Fan YS, Guo J, Chen H. 2020. Altered inter- and intrahemispheric functional connectivity dynamics in autistic children. Hum Brain Mapp. 41:419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Roopchansingh V, Gonzalez-Castillo J, Bandettini PA. 2012. Periodic changes in fMRI connectivity. Neuroimage. 63:1712–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SJ, Woolrich MW, Robinson EC, Glasser MF, Beckmann CF, Jenkinson M, Smith SM. 2015. Large-scale probabilistic functional modes from resting state fMRI. Neuroimage. 109:217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Chen Y, Jian T, Chen H, Guo X, Wang J, Wu L, Chen H, Duan X. 2018. Dynamic functional connectivity analysis reveals decreased variability of the default-mode network in developing autistic brain. Autism Res. 11:1479–1493. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RC, Baare WF, van Oel C, van Haren NE, Collins DL, Evans AC, Amunts K et al. 2006. Genetic contributions to human brain morphology and intelligence. J Neurosci 26:10235–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Della Penna S, Duyn JH, Glover GH, Gonzalez-Castillo J et al. 2013. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 80:360–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, Jack CR Jr, Saykin AJ, Green RC, Weiner MW et al. 2013. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc Natl Acad Sci U S A. 110:4768–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, Przybelski SA, Gregg BE, Kantarci K, Knopman DS et al. 2012. Non-stationarity in the "resting brain's" modular architecture. PLoS One. 7:e39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Lepore N, Joshi SH, Lee AD, Barysheva M, Stein JL, McMahon KL, Johnson K, de Zubicaray GI, Martin NG et al. 2011. The contribution of genes to cortical thickness and volume. Neuroreport. 22:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V, Starck T, Remes J, Long X, Nikkinen J, Haapea M, Veijola J, Moilanen I, Isohanni M, Zang YF et al. 2009. Functional segmentation of the brain cortex using high model order group PICA. Hum Brain Mapp. 30:3865–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V, Vire T, Remes J, Elseoud AA, Starck T, Tervonen O, Nikkinen J. 2011. A sliding time-window ICA reveals spatial variability of the default mode network in time. Brain Connect. 1:339–347. [DOI] [PubMed] [Google Scholar]
- Korgaonkar MS, Ram K, Williams LM, Gatt JM, Grieve SM. 2014. Establishing the resting state default mode network derived from functional magnetic resonance imaging tasks as an endophenotype: a twins study. Hum Brain Mapp. 35:3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, Franz CE, Lyons MJ, Pacheco J, Perry ME et al. 2010. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage. 49:1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucyi A, Davis KD. 2014. Dynamic functional connectivity of the default mode network tracks daydreaming. Neuroimage. 100:471–480. [DOI] [PubMed] [Google Scholar]
- Lebo MJ, Box-Steffensmeier JM. 2008. Dynamic conditional correlations in political science. Am J Polit Sci. 52:688–704. [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC, Giedd JN. 2009. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum Brain Mapp. 30:163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu Y, Li J, Qin W, Li K, Yu C, Jiang T. 2009. Brain anatomical network and intelligence. PLoS Comput Biol. 5:e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist MA, Xu Y, Nebel MB, Caffo BS. 2014. Evaluating dynamic bivariate correlations in resting-state fMRI: a comparison study and a new approach. Neuroimage. 101:531–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie DJ, Kessler D, Bassett DS, Betzel RF, Breakspear M, Kheilholz S, Kucyi A, Liegeois R, Lindquist MA, McIntosh AR et al. 2020. Questions and controversies in the study of time-varying functional connectivity in resting fMRI. Netw Neurosci. 4:30–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AR, Kovacevic N, Lippe S, Garrett D, Grady C, Jirsa V. 2010. The development of a noisy brain. Arch Ital Biol. 148:323–337. [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. 2006. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 7:818–827. [DOI] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Feczko E, Grayson DS, Walum H, Nigg JT, Fair DA. 2018. Heritability of the human connectome: a connectotyping study. Netw Neurosci. 2:175–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie CA, Wisner KM, MacDonald AW, 3rd. 2014. Characteristics of canonical intrinsic connectivity networks across tasks and monozygotic twin pairs. Hum Brain Mapp 35:5532–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan B, Ethridge LE, O'Neil K, Dunn S, Mathew I, Tandon N, Calhoun VD, Ruano G, Kocherla M, Windemuth A et al. 2015. Genetic sources of subcomponents of event-related potential in the dimension of psychosis analyzed from the B-SNIP study. Am J Psychiatry. 172:466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Spann MN, Tokoglu F, Shen X, Constable RT, Scheinost D. 2017. Influences on the test-retest reliability of functional connectivity MRI and its relationship with Behavioral utility. Cereb Cortex. 27:5415–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomi JS, Bolt TS, Ezie CEC, Uddin LQ, Heller AS. 2017. Moment-to-moment BOLD signal variability reflects regional changes in neural flexibility across the lifespan. J Neurosci. 37:5539–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomi JS, Schettini E, Voorhies W, Bolt TS, Heller AS, Uddin LQ. 2018. Resting-state brain signal variability in prefrontal cortex is associated with ADHD symptom severity in children. Front Hum Neurosci. 12:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervaiz U, Vidaurre D, Woolrich MW, Smith SM. 2020. Optimising network modelling methods for fMRI. Neuroimage. 211:116604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJ, Baare WF, Hulshoff Pol HE, Kahn RS, Boomsma DI. 2002. The association between brain volume and intelligence is of genetic origin. Nat Neurosci. 5:83–84. [DOI] [PubMed] [Google Scholar]
- Ray KL, McKay DR, Fox PM, Riedel MC, Uecker AM, Beckmann CF, Smith SM, Fox PT, Laird AR. 2013. ICA model order selection of task co-activation networks. Front Neurosci. 7:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimol LM, Panizzon MS, Fennema-Notestine C, Eyler LT, Fischl B, Franz CE, Hagler DJ, Lyons MJ, Neale MC, Pacheco J et al. 2010. Cortical thickness is influenced by regionally specific genetic factors. Biol Psychiatry. 67:493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoglu U, Pearlson GD, Kiehl KA, Wang YM, Michael AM, Calhoun VD. 2010. A method for evaluating dynamic functional network connectivity and task-modulation: application to schizophrenia. MAGMA. 23:351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi-Khorshidi G, Douaud G, Beckmann CF, Glasser MF, Griffanti L, Smith SM. 2014. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 90:449–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair B, Hansell NK, Blokland GA, Martin NG, Thompson PM, Breakspear M, de Zubicaray GI, Wright MJ, McMahon KL. 2015. Heritability of the network architecture of intrinsic brain functional connectivity. Neuroimage. 121:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Beckmann CF, Andersson J, Auerbach EJ, Bijsterbosch J, Douaud G, Duff E, Feinberg DA, Griffanti L, Harms MP et al. 2013. Resting-state fMRI in the human Connectome project. Neuroimage. 80:144–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A. 106:13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Hyvarinen A, Varoquaux G, Miller KL, Beckmann CF. 2014. Group-PCA for very large fMRI datasets. Neuroimage. 101:738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre G, Choudhuri S, Szekely E, Bonner T, Goduni E, Sharp W, Shaw P. 2017. Estimating the heritability of structural and functional brain connectivity in families affected by attention-deficit/hyperactivity disorder. JAMA Psychiat. 74:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A, Swan GE, Carmelli D. 2001. Heritability of hippocampal size in elderly twin men: equivalent influence from genes and environment. Hippocampus. 11:754–762. [DOI] [PubMed] [Google Scholar]
- Supekar K, Cai W, Krishnadas R, Palaniyappan L, Menon V. 2018. Dysregulated brain dynamics in a triple-network saliency model of schizophrenia and its relation to psychosis. Biol Psychiatry. 85:60–69. [DOI] [PubMed] [Google Scholar]
- Thompson GJ, Magnuson ME, Merritt MD, Schwarb H, Pan WJ, McKinley A, Tripp LD, Schumacher EH, Keilholz SD. 2013. Short-time windows of correlation between large-scale functional brain networks predict vigilance intraindividually and interindividually. Hum Brain Mapp. 34:3280–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam CG, Kaprio J, Khaledy M et al. 2001. Genetic influences on brain structure. Nat Neurosci. 4:1253–1258. [DOI] [PubMed] [Google Scholar]
- Thompson WH, Fransson P. 2015. The mean-variance relationship reveals two possible strategies for dynamic brain connectivity analysis in fMRI. Front Hum Neurosci. 9:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognoli E, Kelso JA. 2014. The metastable brain. Neuron. 81:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Kahn RS, Hulshoff Pol HE. 2009. Efficiency of functional brain networks and intellectual performance. J Neurosci. 29:7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, van Soelen IL, Stam CJ, Kahn RS, Boomsma DI, Hulshoff Pol HE. 2013. Genetic control of functional brain network efficiency in children. Eur Neuropsychopharmacol. 23:19–23. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, WU-MH C. 2013. The WU-Minn human Connectome project: an overview. Neuroimage. 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre D, Smith SM, Woolrich MW. 2017. Brain network dynamics are hierarchically organized in time. Proc Natl Acad Sci U S A. 114:12827–12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zuo X, He Y. 2010. Graph-based network analysis of resting-state functional MRI. Front Syst Neurosci. 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. 2002. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage. 17:256–271. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zuo XN, McMahon KL, Craddock RC, Kelly C, de Zubicaray GI, Hickie I, Bandettini PA, Castellanos FX, Milham MP et al. 2016. Genetic and environmental contributions to functional connectivity architecture of the human brain. Cereb Cortex. 26:2341–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon U, Fahim C, Perusse D, Evans AC. 2010. Lateralized genetic and environmental influences on human brain morphology of 8-year-old twins. Neuroimage. 53:1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Erhardt EB, Sui J, Du Y, He H, Hjelm D, Cetin MS, Rachakonda S, Miller RL, Pearlson G et al. 2015. Assessing dynamic brain graphs of time-varying connectivity in fMRI data: application to healthy controls and patients with schizophrenia. Neuroimage. 107:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


