Significance
Hepatocellular carcinoma (HCC), a type of liver cancer, has a poor 5-y survival rate and current therapies provide only marginal clinical benefits to most HCC patients. Therefore, new therapeutic approaches are needed for HCC treatment. Natural killer (NK) cells are cells of the innate immune system that can inhibit tumor development and progression. We find that pharmacological inhibition of EZH2 results in NK cell–mediated hepatic tumor growth inhibition in mice, which occurs, in part, due to the increased expression of the chemokine CXCL10, leading to increased NK cell migration. These results have implications for EZH2-dependent tumors, in which NK cell–mediated tumor clearance can be induced using EZH2 inhibitors.
Keywords: EZH2, hepatocellular carcinoma, NK cells, HDAC10, CXCL10
Abstract
Enhancer of zeste homolog 2 (EZH2) is a histone H3 lysine 27 methyltransferase that has been shown to function as an oncogene in some cancers. Previous reports have largely focused on the ability of EZH2 to regulate cell-intrinsic tumor regulatory pathways as its mechanism-of-oncogenic action. However, the role that EZH2-mediated immune suppression plays in its oncogenic activity is not fully known. In particular, the role of natural killer (NK) cells in EZH2-driven tumor growth remains incompletely understood. Here, we demonstrate that genetic or pharmacological inhibition of EZH2 induces reexpression of the chemokine CXCL10 in hepatic tumor cells. We find that histone deacetylase 10 (HDAC10) is necessary for EZH2 recruitment to the CXCL10 promoter, leading to CXCL10 transcriptional repression. Critically, CXCL10 is necessary and sufficient for stimulating NK cell migration, and EZH2’s ability to inhibit NK cell migration via CXCL10 suppression is conserved in other EZH2-dependent cancers. NK cell depletion in an immunocompetent syngeneic mouse model of hepatic tumorigenesis reverses the tumor inhibitory effects of an EZH2 inhibitor (GSK343), and inhibitor-mediated reexpression of CXCL10 is required for its tumor suppressive effects in the same mouse model. Collectively, these results reveal a decisive role for NK cells and CXCL10 in mediating the oncogenic function of EZH2.
Enhancer of zeste homolog 2 (EZH2) encodes a histone methyltransferase that constitutes the catalytic component of polycomb repressive complex 2 (PRC2). The gene silencing activity of PRC2 depends on its ability to trimethylate lysine 27 of histone H3 (H3K27me3), which occurs via the catalytic SET domain of the EZH2 subunit and at least two other PRC2 subunits, including suppressor of zeste 12 (SUZ12) and embryonic ectoderm development (EED) (1–3). In particular, the carboxyl-terminal domain of EED binds specifically to histone tails carrying trimethyl-lysine residues associated with repressive chromatin marks, which in turn, leads to allosteric activation of the methyltransferase activity of PRC2 (4).
The presence of gain-of-function oncogenic mutations in the EZH2 gene or EZH2 overexpression is observed in several cancer types, and in some cases, EZH2 also functions as an oncogene (5). These observations have led to the development of several potent and clinically efficacious EZH2 and EED inhibitors, many of which are already undergoing clinical testing for treating a wide variety of cancers (5). Based on the results of these clinical trials, one such EZH2 inhibitor, tazemetostat (Tazverik), has already been approved by the US Food and Drug Administration (US FDA) for treating follicular lymphoma and epithelioid sarcoma (6, 7). Collectively, these observations confirm that EZH2 is a druggable target with clinical value for cancer therapy.
The host immune response to cancer cells is a potent mechanism for tumor suppression. In this regard, cells of both the adaptive immune system (e.g., T cells and B cells) and the innate immune system (e.g., natural killer [NK] cells and macrophages) have been shown to play important roles in tumor initiation and progression (8). Therefore, a number of cancer therapeutic agents function to elicit tumor inhibitory immune responses that, in some cases, underpin their anticancer activity (9–11). Previous studies exploring the mechanisms of EZH2-mediated oncogenesis have largely focused on the cell-intrinsic mechanisms by which EZH2 regulates expression of genes that are necessary for cancer cell proliferation and survival to promote tumor development and progression (12, 13). However, the relative contribution of cell-extrinsic tumor regulatory pathways, including host immune response, relative to cell-extrinsic pathways, such as those that promote cell proliferation and survival, for mediating EZH2-driven oncogenesis, is not known.
We previously demonstrated that EZH2 promotes tumor development through a cell-extrinsic mechanism involving inhibition of the antitumor activity of NK cells (14). NK cells are components of the innate immune response that play a key role in eradicating infected and stress-damaged cells (15). These cells are also critical for inhibiting tumor initiation and progression (16, 17). There are two main mechanisms that regulate the antitumor activity of NK cells. These include the pathways that directly regulate NK cell–mediated tumor cell eradication, and those that control NK cell migration and recruitment to tumor sites, which are also necessary for their antitumor activity. The regulation of these NK cell activities is achieved, in part, by modulating the expression of NK cell–activating and repressing ligands on tumor cells (16). In our previous study, we demonstrated that EZH2 represses expression of the NK cell–activating ligand UL16 binding protein 1 (ULBP1) and showed that this ligand is necessary for NK cell–mediated inhibition of hepatic tumor cells in culture (14).
In addition, regulation of NK cell activities can occur through NK cell recruitment to tumor sites by the action of chemokines (18, 19), which are small cytokines that play vital roles in promoting immune cell migration, thereby regulating and controlling antitumor responses in the tumor microenvironment (20, 21). Tumor-derived chemokines in the tumor microenvironment play pivotal roles in controlling tumor growth and progression, in part through their ability to modulate immune responses directed against cancer cells (14, 21, 22). However, whether EZH2 modulates NK cell migration through a mechanism involving chemokines is not yet known. In this study, we investigated the role of chemokines in modulating EZH2-mediated NK cell migration and their role in mediating the oncogenic function of EZH2 by regulating NK cell activity. We demonstrate that EZH2 represses expression of the chemokine, CXC motif, ligand 10 (CXCL10) in a histone deacetylase 10 (HDAC10)–dependent manner. In addition, we demonstrate that both NK cells and the reexpression of CXCL10 in response to EZH2 inhibitor treatment are required for EZH2 inhibitor-induced tumor suppression in an immunocompetent syngeneic mouse model of hepatic tumor growth. Collectively, these results reveal a decisive role for NK cells and CXCL10 in EZH2 inhibition-mediated tumor suppression and highlight the importance of cell-extrinsic mechanisms, such as modulation of tumor growth inhibition by NK cells, in facilitating the oncogenic activities of EZH2.
Results
EZH2 Inhibition Increases Expression of CXCL10 in Hepatic Tumor Cells.
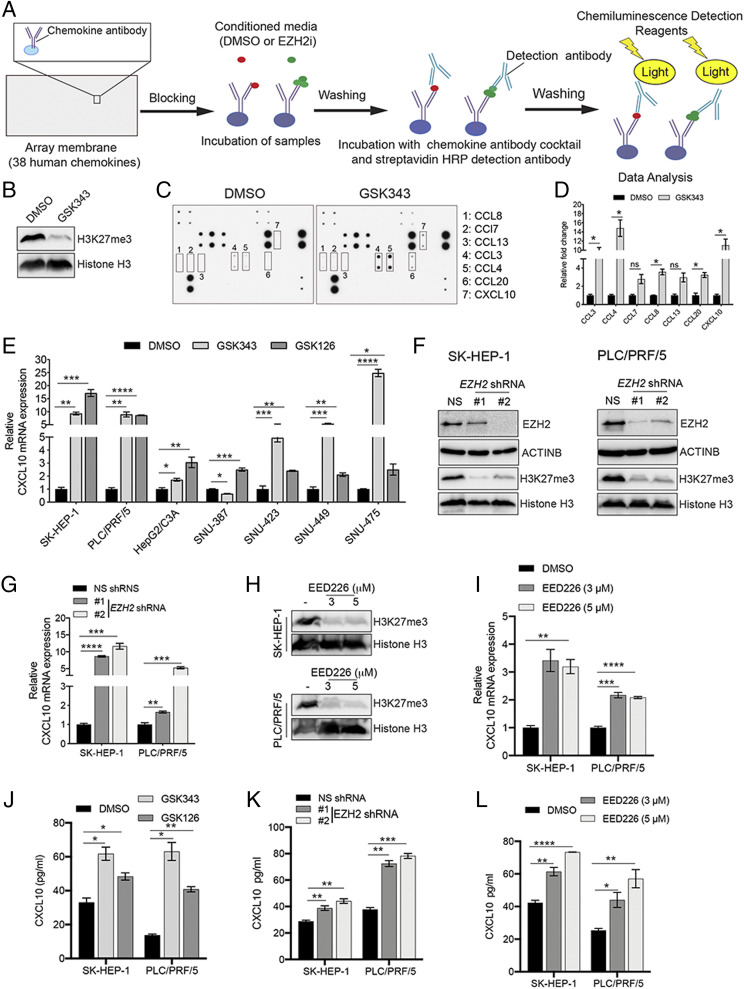
In a previous study, we showed that EZH2 inhibition results in the reexpression of natural killer group 2, member D (NKG2D) ligands in hepatic tumor cells, resulting in their increased inhibition by NK cells (14). Based on these findings and the previously documented roles for chemokines in the regulation of immune response against cancer cells, we asked whether EZH2 can regulate chemokine expression and thereby affect NK cell migration. To test this possibility and identify EZH2-regulated chemokines, the hepatic tumor cell line SK-HEP-1 was first treated with EZH2 inhibitor GSK343 or dimethyl sulfoxide (DMSO) for 72 h. Conditioned media were then collected and hybridized to human chemokine antibody array membranes, which can detect changes in expression of 38 human chemokines (Fig. 1A). We identified five chemokines that were significantly increased in response to pharmacological inhibition of EZH2 (Fig. 1 B–D). To further validate and generalize these results, we treated seven different hepatic tumor cell lines with two small molecule inhibitors of EZH2, GSK343 and GSK126, and measured mRNA expression levels of the five chemokines identified by chemokine array using qRT-PCR. We found that both GSK343 and GSK126 were able to induce expression of several chemokines in hepatic tumor cell lines. However, only CXCL10 was found to be consistently up-regulated in all hepatic tumor cell lines tested in response to both EZH2 inhibitors (Fig. 1E and SI Appendix, Fig. S1).
Fig. 1.
EZH2 and EED inhibition increases the expression of CXCL10 in hepatic tumor cells. (A) Schematic overview of human chemokine antibody array analysis to identify chemokines repressed by EZH2. (B) SK-HEP-1 cells were treated with GSK343 (3 μM) or DMSO control for 72 h, and immunoblot analysis was performed to measure trimethylation at lysine 27 of histone H3 (H3K27me3) and total histone H3 protein in whole-cell lysates. (C) SK-HEP-1 cells were treated with GSK343 (3 μM) or DMSO for 72 h. Conditioned media were collected and analyzed for expression of 38 different human chemokines using human chemokine antibody arrays. (D) Average fold change in expression of indicated chemokines, quantified from the image shown in C, using ImageJ software. (E) Relative CXCL10 mRNA expression was measured by qRT-PCR in SK-HEP-1 cells treated with GSK343 (3 µM) or GSK126 (2 µM) for 72 h compared to DMSO-treated cells. ACTINB expression was used for normalization. (F) Immunoblot analysis for the indicated proteins in SK-HEP-1 and PLC/PRF/5 cells expressing EZH2 shRNAs or nonspecific (NS) shRNA. (G) Relative CXCL10 mRNA expression measured by qRT-PCR in SK-HEP-1 and PLC/PRF/5 cells expressing EZH2 shRNA compared to NS shRNA-expressing cells. ACTINB expression was used for normalization. (H) Immunoblot analysis to measure H3K27me3 and histone H3 protein in SK-HEP-1 and PLC/PRF/5 cells treated with EED226 at the indicated concentrations for 72 h. (I) Relative CXCL10 mRNA expression measured by qRT-PCR in SK-HEP-1 and PLC/PRF/5 cells treated with EED226 at the indicated concentrations for 72 h compared to DMSO-treated cells. (J) CXCL10 levels measured by ELISA in supernatants from SK-HEP-1 and PLC/PRF/5 cells treated with GSK343 (3 µM), GSK126 (2 µM), or DMSO for 72 h. (K) CXCL10 levels measured by ELISA in supernatants from SK-HEP-1 and PLC/PRF/5 cells expressing EZH2 shRNAs or NS shRNA. (L) CXCL10 levels measured by ELISA in supernatants from SK-HEP-1 and PLC/PRF/5 cells treated with DMSO or EED226 at the indicated concentrations for 72 h. Data are presented as mean ± SEM; ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We then knocked down the expression of EZH2 using two sequence-independent short hairpin RNAs (shRNAs) and found that, as expected, EZH2 knockdown results in reduced expression of both EZH2 and the EZH2-associated histone mark, H3K27me3 (Fig. 1F). Additionally, in agreement with the results from EZH2 inhibitor treatment experiments, EZH2 knockdown also induces up-regulation of CXCL10 in hepatic tumor cell lines (Fig. 1G). To further determine whether repression of CXCL10 in response to EZH2 inhibition is due to PRC2 complex activity, we pharmacologically inhibited EED, another PRC2 complex protein that is required for EZH2 activity (23). We observed that similar to EZH2 inhibitors and EZH2 shRNAs, treatment with the EED inhibitor EED226 results in increased expression of CXCL10 (Fig. 1 H and I and SI Appendix, Fig. S2 A and B), indicating that PRC2 activity is required for CXCL10 repression.
Finally, to test whether CXCL10 protein levels correlate with mRNA levels, we treated SK-HEP-1 and PLC/PRF/5 cells with EZH2 inhibitors (GSK126 and GSK343) and measured CXCL10 protein expression in conditioned media using enzyme-linked immunosorbent assays (ELISAs). As expected, we found that CXCL10 protein levels are significantly increased in hepatic tumor cell lines treated with EZH2 inhibitors (Fig. 1J and SI Appendix, Fig. S2C). Similarly, increased CXCL10 protein levels are observed in conditioned media from hepatic tumor cells expressing an EZH2 shRNA (Fig. 1K) or after treatment with EED226 (Fig. 1L and SI Appendix, Fig. S2D). Collectively, these results show that CXCL10 is up-regulated in response to inhibition of EZH2 or EED, another component of the PRC2 complex, in hepatic tumor cells.
EZH2 Inhibition-Mediated Up-Regulation of CXCL10 Is Necessary for NK Cell Migration.
After identifying CXCL10 as a transcriptionally repressed target of EZH2, we next sought to understand the role of CXCL10 in regulation of NK cell function and hepatic tumor growth and progression. To this end, in order to first determine whether CXCL10 functions in a cell-intrinsic manner to regulate EZH2-mediated tumor growth, we knocked down CXCL10 expression in hepatic tumor cell lines (SI Appendix, Fig. S3 A and B) and measured their growth with or without GSK343 treatment using clonogenic assays. We found that knockdown of CXCL10 does not alter growth of hepatic tumor cells or affect the ability of GSK343 to inhibit hepatic tumor cell growth (SI Appendix, Fig. S3C). These results indicate that CXCL10 is not required for EZH2 inhibitor-mediated growth inhibition of hepatic tumor cells in cell culture, which rules out its role in a cell-intrinsic pathway.
We then asked whether CXCL10 knockdown affects the ability of NK cells to induce hepatic tumor cell cytotoxicity. To this end, hepatic tumor cells expressing CXCL10 shRNAs or a nonspecific shRNA control were treated with the EZH2 inhibitor GSK343. We then measured NK cell–mediated cytotoxicity and found that CXCL10 reexpression following EZH2 inhibitor treatment is not required for NK cell–mediated inhibition of hepatic tumor cells (SI Appendix, Fig. S3D). Similarly, ectopic expression of CXCL10 in hepatic tumor cells does not increase NK cell–mediated cytotoxicity against these cells (SI Appendix, Fig. S3E). Taken together, these results show that CXCL10 expression does not influence NK cell cytotoxicity against hepatic tumor cells.
NK cell recruitment to solid tumors is essential for direct contact between NK and cancer cells, and consequentially, NK cell–mediated cytotoxicity (24, 25). Previous studies have shown that chemokines directly influence NK cell recruitment to tumors, in part, by regulating NK cell migration (24, 25). Based on these findings, we next asked whether CXCL10 promotes NK cell migration to hepatic tumor cells. To this end, we determined whether conditioned media from hepatic tumor cells treated with EZH2 inhibitors (GSK343 or GSK126) can promote NK cell migration. For these experiments, two hepatic tumor cell lines—SK-HEP-1 and PLC/PRF/5—were treated with EZH2 inhibitors (GSK343 or GSK126) or a DMSO control. The conditioned media from treated cells were then collected and used to measure NK cell migratory activity (Fig. 2A). Our results show that conditioned media from SK-HEP-1 cells treated with EZH2 inhibitors significantly increased migration of NK cells, as compared to media from DMSO-treated cells (Fig. 2B). Consistent with the role of other PRC2 complex members in CXCL10 repression, increased NK cell migration is also observed with conditioned media from hepatic tumor cells treated with the EED inhibitor, EED226 (Fig. 2C).
Fig. 2.
EZH2 inhibition-mediated up-regulation of CXCL10 is necessary for NK cell migration. (A) Schematic overview of NK cell migration assays. (B) Average number of NK cells that migrated toward conditioned media collected from SK-HEP-1 and PLC/PRF/5 cells treated with EZH2 inhibitors GSK343 (3 µM) and GSK126 (2 µM) or DMSO for 72 h. (C) Average number of NK cells that migrated toward conditioned media collected from SK-HEP-1 and PLC/PRF5 cells treated with the EED inhibitor EED226 (5 µM) or DMSO for 72 h. (D) Average number of NK cells that migrated toward conditioned media collected from SK-HEP-1 and PLC/PRF/5 cells ectopically expressing CXCL10 or the vector control. (E) Average number of NK cells that migrated toward conditioned media collected from SK-HEP-1 and PLC/PRF/5 cells expressing CXCL10 shRNAs or NS shRNA control that were treated with either GSK343 (3 µM) or DMSO for 72 h. Data are presented as mean ± SEM; *P < 0.05, **P < 0.01, and ***P < 0.001.
To further determine whether CXCL10 can directly promote NK cell migration, we ectopically expressed CXCL10 in hepatic tumor cell lines (SK-HEP-1 and PLC/PRF/5) and measured NK cell migration. We found that conditioned media from CXCL10-expressing cells significantly enhanced NK cell migration, as compared to media from hepatic tumor cells containing the empty vector control (Fig. 2D). We then asked whether the EZH2 inhibitor-mediated increase in NK cell migration is dependent on CXCL10 reexpression. To this end, we knocked down expression of CXCL10 in hepatic tumor cell lines (SK-HEP-1 and PLC/PRF/5) and tested the effect on GSK343-mediated enhancement of NK cell migration. Our results show that CXCL10 knockdown leads to reduced NK cell migration toward media from GSK343-treated hepatic tumor cells, as compared to media from control cells expressing a nonspecific shRNA (Fig. 2E). Similar results were obtained when conditioned media from GSK343-treated hepatic tumor cells were treated with a CXCL10-neutralizing antibody (SI Appendix, Fig. S4A).
A previous report showed that the NK cell line NK-92MI expresses the CXCL10 receptor CXCR3 (26). Therefore, to further confirm the role of CXCL10 in mediating the effects of media from GSK343-treated cells in promoting NK cell migration, we knocked down expression of the CXCL10 receptor CXCR3 in the NK cell line NK-92MI using shRNAs (SI Appendix, Fig. S4 B and C). Similar to knockdown of CXCL10 in hepatic tumor cells, knockdown of CXCR3 in NK-92MI cells inhibits NK cell migration toward media from GSK343-treated hepatic tumor cells, relative to NK-92MI cells expressing a nonspecific shRNA (SI Appendix, Fig. S4D). Collectively, these results demonstrate that CXCL10 up-regulation resulting from EZH2 inhibition promotes NK cell migration, without influencing tumor cell growth in a cell-intrinsic manner or NK cell–mediated cytotoxicity in cell culture.
EZH2 Regulates CXCL10 Expression and NK Cell Migration in a HDAC10-Dependent Manner.
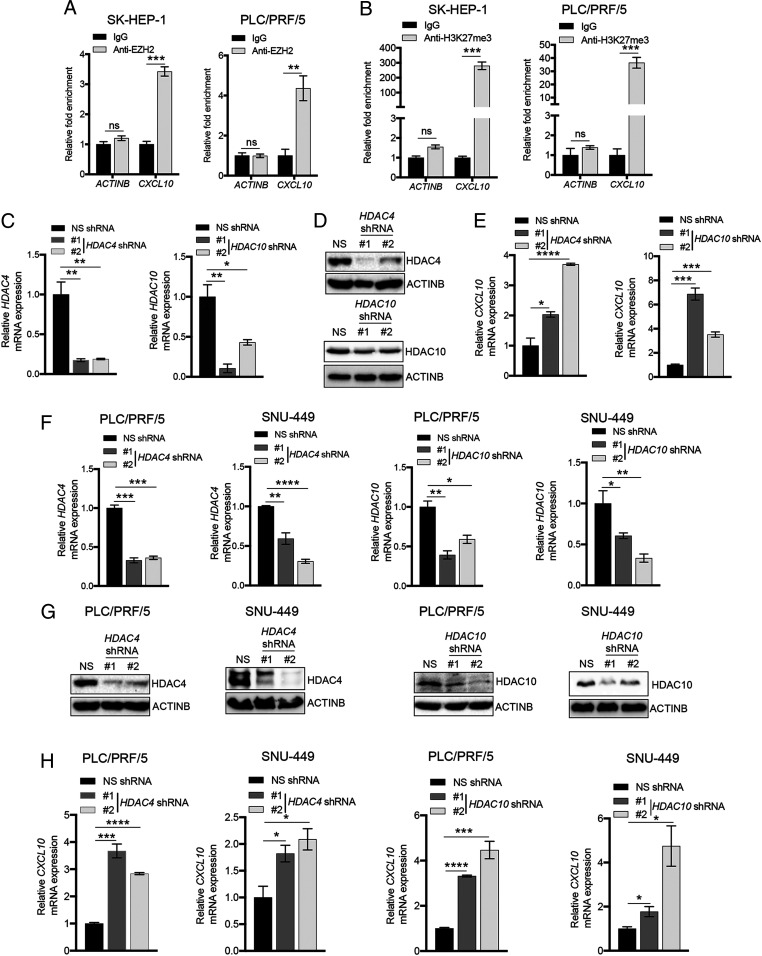
Based on our data showing that CXCL10 expression downstream of EZH2 regulates NK cell migration, we next sought to determine the mechanism underlying EZH2-mediated transcriptional repression of CXCL10. In particular, we first asked whether EZH2 can directly repress CXCL10 expression in hepatic tumor cell lines. To test this idea, we measured both EZH2 recruitment and the EZH2-associated H3K27me3 mark on the CXCL10 promoter using chromatin immunoprecipitation (ChIP) assays. Our results reveal that EZH2 is significantly enriched at the CXCL10 promoter, as compared to the promoter of the ACTINB gene (Fig. 3A). Similarly, the CXCL10 promoter contains increased levels of the H3K27me3 mark relative to the ACTINB promoter (Fig. 3B).
Fig. 3.
HDAC4 and HDAC10 are necessary for CXCL10 repression in hepatic tumor cells. (A) EZH2 recruitment to the CXCL10 and ACTINB promoters was measured in SK-HEP-1 and PLC/PRF/5 cells using ChIP assays. Relative fold enrichment of EZH2 on the CXCL10 and ACTINB promoters compared to IgG control is plotted for the indicated cell lines. (B) The H3K27me3 modification on the CXCL10 and ACTINB promoters was measured using ChIP assays. Relative fold enrichment compared to IgG control is plotted. (C) Relative mRNA expression of HDAC4 (Left) and HDAC10 (Right) measured by qRT-PCR in SK-HEP-1 cells expressing HDAC4 or HDAC10 shRNAs or nonspecific (NS) shRNA control. ACTINB expression was used for normalization. (D) Immunoblot analysis to measure the indicated proteins in SK-HEP-1 cells expressing NS, HDAC4, or HDAC10 shRNAs. (E) Relative CXCL10 mRNA expression measured by qRT-PCR in SK-HEP-1 cells expressing NS, HDAC4, or HDAC10 shRNAs. ACTINB expression was used for normalization. (F) Relative mRNA expression of HDAC4 and HDAC10 measured by qRT-PCR in PLC/PRF/5 and SNU-449 cells expressing NS, HDAC4, or HDAC10 shRNAs. ACTINB expression was used for normalization. (G) Immunoblot analysis to measure the indicated proteins in PLC/PRF/5 and SNU-449 cells expressing NS, HDAC4, or HDAC10 shRNAs. (H) Relative CXCL10 mRNA expression measured by qRT-PCR in PLC/PRF/5 and SNU-449 cells expressing NS, HDAC4, or HDAC10 shRNAs. ACTINB expression was used for normalization. Data are presented as the mean ± SEM; ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Previous studies have shown that EZH2-mediated transcriptional gene silencing is associated with increased promoter DNA methylation and/or reduced histone acetylation (27, 28). Therefore, to determine whether these modifications are also involved in CXCL10 regulation by EZH2, we treated hepatic tumor cells with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5Aza2dC), or the histone deacetylase inhibitor trichostatin A (TSA), and measured expression of CXCL10 mRNA. We detected robust induction of CXCL10 expression after treatment with TSA but only a modest increase after 5Aza2dC treatment (SI Appendix, Fig. S5A). These results suggest that a HDAC might be involved in CXCL10 repression in hepatic tumor cells. We therefore individually knocked down the expression of 18 known human HDACs in the SK-HEP-1 hepatic tumor cell line (Fig. 3 C and D and SI Appendix, Table S1 and Fig. S5B) and measured expression of CXCL10 mRNA, in order to determine which of these proteins are required for CXCL10 repression. We found that knockdown of six HDACs (i.e., HDAC4, HDAC7, HDAC10, SIRT1, SIRT2, and SIRT5) in SK-HEP-1 cells leads to increased expression of CXCL10 mRNA (Fig. 3E and SI Appendix, Fig. S5C). We then tested whether knockdown of these six HDACs can similarly promote increased expression of CXCL10 in two additional hepatic tumor cell lines (PLC/PRF/5 and SNU-449) (Fig. 3 F and G and SI Appendix, Fig. S5D). Our results show that only knockdown of HDAC4 and HDAC10 leads to increased mRNA expression of CXCL10 in both lines (Fig. 3H and SI Appendix, Fig. S5E), suggesting that these HDACs act as common repressors of CXCL10 expression in hepatic tumor cells.
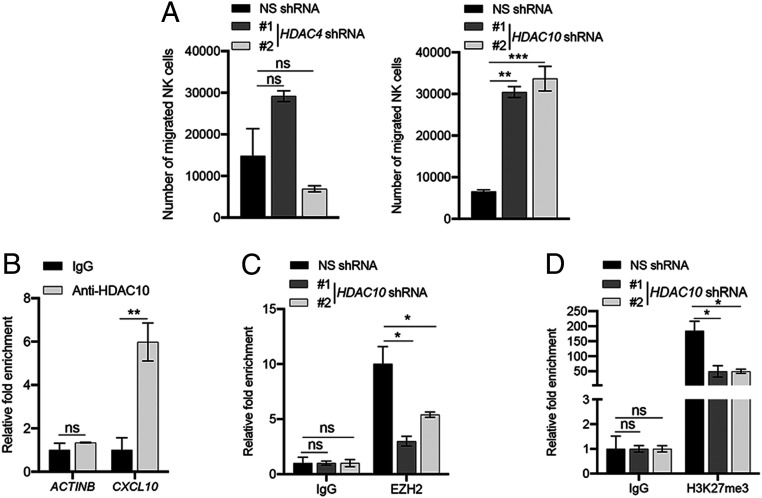
Next, we asked whether either HDAC4 or HDAC10, similar to EZH2, is involved in CXCL10-mediated NK cell migration. To test this, we individually knocked down expression of HDAC4 and HDAC10 in the SK-HEP-1 hepatic tumor cell line using shRNAs and measured NK cell migration toward conditioned media from these cells. We found that similar to EZH2 inhibition, knockdown of HDAC10 promotes enhanced NK cell migration, whereas HDAC4 knockdown does not significantly affect this process (Fig. 4A). We then determined whether HDAC10 can directly regulate CXCL10 expression by performing ChIP to measure recruitment of HDAC10 at the CXCL10 promoter. We found that HDAC10 is enriched on the CXCL10 promoter, as compared to the ACTINB promoter, in the hepatic tumor cell line SK-HEP-1 (Fig. 4B), suggesting that this protein directly regulates CXCL10 expression. We then measured both EZH2 recruitment and the H3K27me3 mark at the CXCL10 promoter in SK-HEP-1 cells expressing HDAC10 shRNAs to determine whether the increase in CXCL10 expression observed in HDAC-knockdown cells results from reduced EZH2 recruitment at the CXCL10 promoter. Notably, we observed significantly decreased recruitment of EZH2, as well as reduced levels of the H3K27me3 mark, on the CXCL10 promoter relative to the ACTINB promoter in hepatic tumor cell lines knocked down for HDAC10 (Fig. 4 C and D). However, coimmunoprecipitation experiments further revealed that HDAC10 does not directly interact with EZH2 or any other proteins in the PRC2 complex (SI Appendix, Fig. S6 A and B), indicating that HDAC10 is not part of this complex, but rather, specifically regulates EZH2 recruitment to the CXCL10 promoter. Collectively, our data show that knockdown of HDAC10 promotes increased expression of CXCL10 by inhibiting recruitment of EZH2 to the CXCL10 promoter, which ultimately leads to enhanced NK cell migration.
Fig. 4.
HDAC10 loss enhances NK cell migration, and HDAC10 is necessary for EZH2 recruitment to the CXCL10 promoter. (A) NK cell migration in response to conditioned media collected from 72-h culture of SK-HEP-1 cells expressing either nonspecific (NS), HDAC4, or HDAC10 shRNAs was measured. Average numbers of migrated NK cells are plotted. (B) HDAC10 recruitment to the CXCL10 and ACTINB promoters was measured in SK-HEP-1 cells using ChIP assays. Relative fold enrichment of HDAC10 on the CXCL10 and ACTINB promoters compared to IgG control is shown. (C) EZH2 recruitment to the CXCL10 and ACTINB promoters in SK-HEP-1 cells expressing NS or HDAC10 shRNAs was measured using ChIP assay. Relative fold enrichment of EZH2 on the CXCL10 and ACTINB promoters compared to IgG control under the indicated conditions is shown. (D) Levels of the H3K27me3 modification on the CXCL10 and ACTINB promoters was measured in SK-HEP-1 cells expressing NS or HDAC10 shRNAs using ChIP assays. Relative levels of the H3K27me3 mark on the CXCL10 and ACTINB promoters compared to IgG control under the indicated conditions are shown. Data are presented as the mean ± SEM; ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001.
EZH2-Mediated CXCL10 Repression and Its Effect on NK Cell Recruitment Are Conserved in Other EZH2-Dependent Cancers.
EZH2 has been shown to possess oncogenic activity in a variety of cancers, including diffuse large B cell lymphoma (DLBCL), breast cancer, and colon cancer (29–31). Therefore, we hypothesized that our observations on the role of EZH2 in hepatic tumor cells might be generalized to other EZH2-dependent cancer types. To test this idea, we used EZH2-mutant DLBCL cell lines (SU-DHL-10 and KARPASS-422) (32), EZH2-dependent breast cancer cell lines (MDA-MB-231 and MCF7), and colon cancer cell lines (HCT116 and HT29) for which EZH2 inhibition has been shown to inhibit growth (33, 34). We found that treatment of these cell lines with the EZH2 inhibitor GSK343 leads to decreased levels of the H3K27me3 mark at the CXCL10 promoter (SI Appendix, Fig. S7A), increased expression of CXCL10 mRNA (SI Appendix, Fig. S7B), and increased levels of CXCL10 protein in conditioned media (SI Appendix, Fig. S7C). Additionally, similar to hepatic tumor cells, conditioned media from breast cancer, colon cancer, and DLBCL cell lines treated with GSK343 significantly increased migration of NK cells relative to conditioned media from DMSO-treated cells (SI Appendix, Fig. S7D). Collectively, these results confirm that EZH2-mediated repression of CXCL10 and its role in NK cell migration is conserved in cells from other EZH2-dependent cancers.
EZH2 Inhibition-Induced Tumor Suppression Largely Depends on NK Cell Activity in a Syngeneic Immunocompetent Mouse Model of Hepatic Tumor Growth.
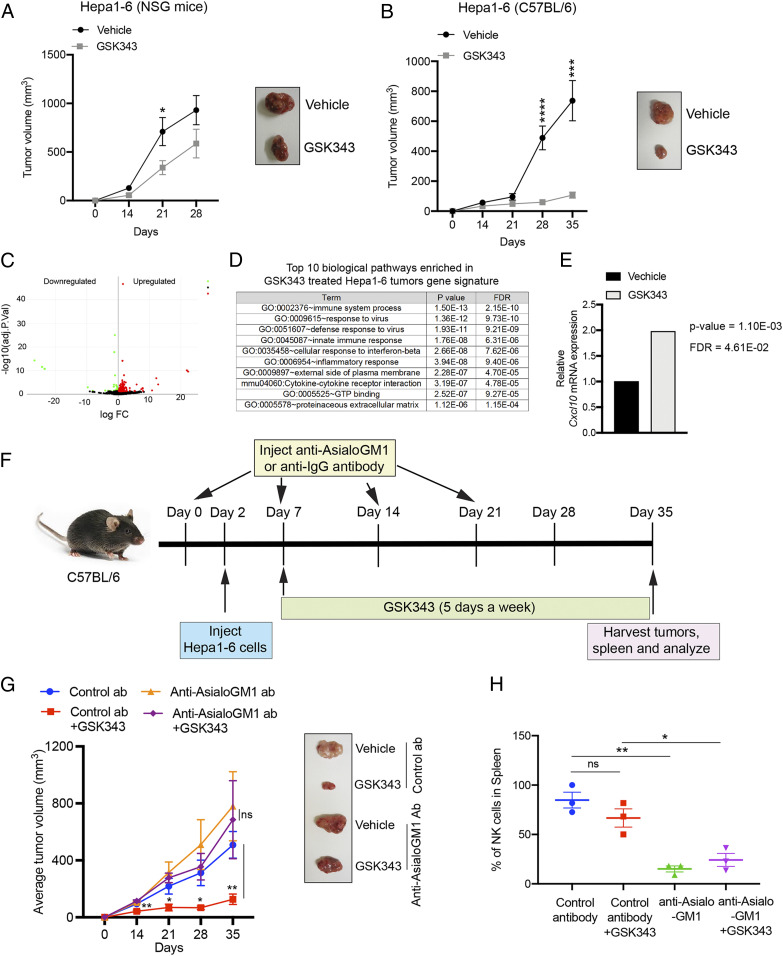
To determine the role of NK cells in EZH2 suppression-mediated tumor growth inhibition in vivo, we used the mouse hepatic tumor cell line Hepa1-6, which can form tumors in syngeneic C57BL/6 mice. For this model, Hepa1-6 cells were injected subcutaneously into either syngeneic C57BL/6 mice, which are immunocompetent and can mount antitumor immune responses, or NOD-scid IL2Rgammanull (NSG) immunodeficient mice that cannot mount antitumor immune responses. Tumor-bearing mice were then treated with either EZH2 inhibitor (GSK343) or vehicle control. We detected significantly increased inhibition of Hepa1-6 tumors in C57BL/6 mice (∼85% tumor inhibition) relative to tumors in NSG mice (∼37% tumor inhibition) after GSK343 treatment (Fig. 5 A and B). Analysis of vehicle- or GSK343-treated tumors using RNA-sequencing (RNA-seq) analysis identified significant changes in the mRNA expression levels of multiple genes, with 315 genes up-regulated in GSK343-treated Hepa1-6 tumors relative to those treated with vehicle control (Fig. 5C and SI Appendix, Fig. S8 and Table S2). Pathway analysis with genes up-regulated in GSK343-treated tumors revealed enrichment of immune regulatory pathways, including the innate immune response and cytokine–chemokine receptor interactions (Fig. 5D and SI Appendix, Table S3). Further, consistent with our findings in human hepatic tumor cell lines, RNA-seq of tumor-derived RNA also showed increased Cxcl10 mRNA expression in tumors treated with GSK343 (Fig. 5E).
Fig. 5.
NK cells are required for EZH2 inhibitor-mediated tumor suppression in a syngeneic mouse model of hepatic tumor growth. (A) Hepa1-6 cells (1 × 105) were injected subcutaneously into NSG mice (n = 8). After 5 d, mice were treated with 20 mg/kg GSK343 or vehicle control; treatment was administered 5 d/wk via i.p. injection for 28 d. Average tumor volumes at the indicated time points for vehicle- or GSK343-treated mice are plotted; representative tumors are shown at Right. (B) Hepa1-6 cells (3 × 106) were injected subcutaneously into C57BL/6 mice (n = 8). After 5 d, mice were treated with 20 mg/kg GSK343 or vehicle control; treatment was administered 5 d/wk via i.p. injection for 35 d. Average tumor volumes at the indicated time points for vehicle- or GSK343-treated mice are plotted; representative tumors are shown at Right. (C) Volcano plot showing genes that are differentially expressed in GSK343-treated Hepa1-6 tumors compared to vehicle-treated tumors. (D) The top 10 biological pathways enriched in gene signatures from GSK343-treated Hepa1-6 tumors. (E) Cxcl10 mRNA expression levels from RNA-seq analysis of Hepa1-6 tumors treated with GSK343 or vehicle control. (F) Schematic showing the timeline and experimental strategy for NK cell depletion assays. (G) Average tumor volumes (n = 10) for the indicated conditions are plotted; representative tumors are shown at Right. (H) Relative percentage (%) of NK cells in spleens from mice (n = 3) treated with indicated antibodies and either GSK343 (20 mg/kg) or vehicle control, 5 d/wk for 35 d. Data are presented as the mean ± SEM; ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Based on these results and our cell culture data, we then asked whether GSK343-mediated tumor suppression of Hepa1-6 tumors in syngeneic C57BL/6 mice is dependent upon NK cell activity. To this end, we used anti–Asialo-GM1 antibodies, which have been shown to effectively deplete NK cells in vivo (35, 36) (Fig. 5F); an isotype control IgG antibody was used as a control. We then treated mice with the EZH2 inhibitor GSK343 and observed significantly attenuated tumor growth inhibition in mice depleted for NK cells, as compared to control mice with normal NK cell levels (Fig. 5H). We further note that vehicle-treated tumors with or without NK cell depletion (treated with anti–Asialo-GM1 or IgG antibody, respectively) grew slightly faster, although this difference was not significant (Fig. 5G). Consistent with previous reports, we found that treatment with mouse anti–Asialo-GM1 antibodies effectively depletes NK cells in mice, as confirmed by analysis of the spleen (Fig. 5H). These results demonstrate that NK cells play a critical role in the in vivo tumor suppression that results from the inhibition of EZH2.
EZH2 Inhibition-Induced Reexpression of CXCL10 Is Required for Tumor Suppression in a Syngeneic Immunocompetent Mouse Model of Hepatic Tumor Growth.
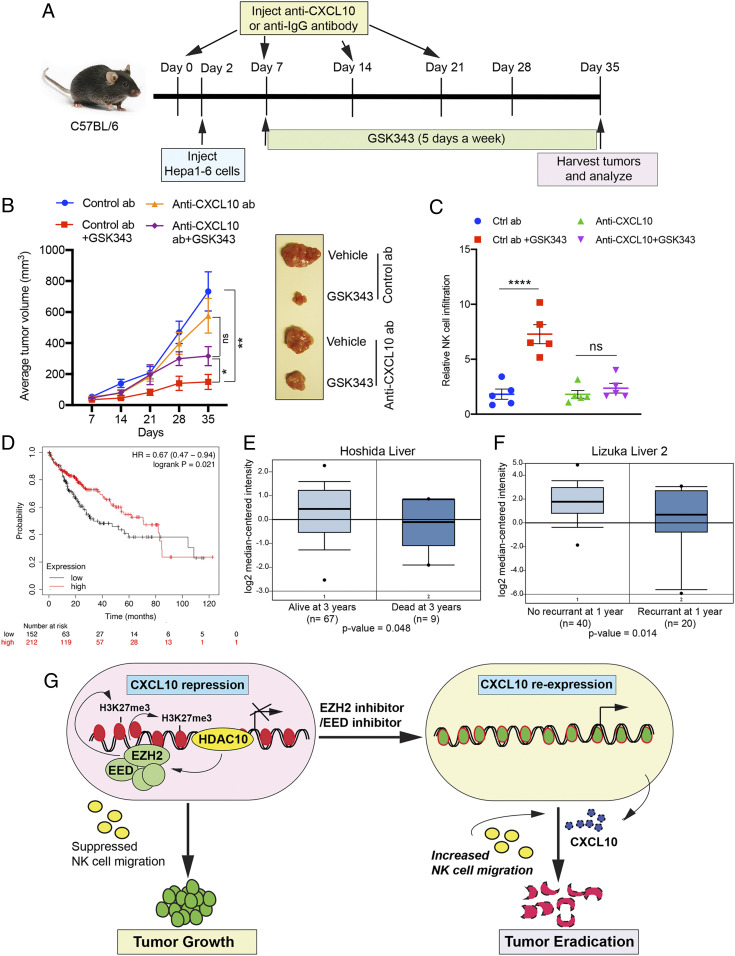
To further confirm the importance of CXCL10 in the oncogenic activity of EZH2 and elucidate the role that this chemokine plays in EZH2 inhibitor-mediated tumor suppression in vivo, we used the syngeneic mouse model of hepatic tumors generated from Hepa1-6 cells, as described above. To this end, we injected Hepa1-6 cells into syngeneic C57BL/6 mice and inhibited CXCL10 using a CXCL10-neutralizing antibody (Fig. 6A); control animals were injected with isotype control antibody. Mice were then treated with the EZH2 inhibitor GSK343 or vehicle control. Notably, we found that CXCL10 neutralization abolishes the tumor suppressive effects of GSK343, indicating that Cxcl10 repression by EZH2 is necessary for its oncogenic activity (Fig. 6B). Furthermore, we detected significantly reduced NK cell infiltration in tumors from CXCL10 antibody-treated mice relative to IgG antibody-treated mice after GSK343 treatment (Fig. 6C). These results are consistent with the proposed role of CXCL10 in promoting recruitment of NK cells to tumor sites.
Fig. 6.
EZH2 inhibitor-mediated reexpression of CXCL10 is necessary for tumor suppression in a syngeneic mouse model of hepatic tumor growth. (A) Schematic showing the timeline and experimental strategy for NK cell depletion experiments. (B) Average tumor volumes (n = 10) for the indicated conditions are plotted; representative tumors are shown at Right. (C) Relative percentage (%) of NK cell infiltration in Hepa1-6 tumors (n = 5) treated with control or Cxcl10-neutralizing antibodies, with or without GSK343 treatment. (D) Overall survival for hepatocellular carcinoma patients with higher or lower levels of CXCL10 mRNA expression. (E) Hoshida liver dataset showing CXCL10 expression in patients that are either alive or dead at year 3. (F) Lizuka liver 2 dataset showing CXCL10 expression in patients that are either alive or dead at year 1. (G) Model depicting proposed mechanism of CXCL10 regulation and its role in NK cell migration and consequential tumor growth inhibition. Data are presented as the mean ± SEM; ns, not significant; *P < 0.05, **P < 0.01, and ****P < 0.0001.
In addition, consistent with the critical role of CXCL10 in enhancing NK cell–mediated tumor growth inhibition, we further found that hepatocellular carcinoma patients with higher CXCL10 levels display longer overall survival, disease-free survival, and recurrence-free survival (Fig. 6 D–F). Collectively, our results identify CXCL10 as an important mediator of EZH2 oncogenic activity in vivo that functions by enhancing NK cell recruitment to tumor sites, leading to tumor suppression via NK cell–mediated tumor cell growth inhibition (Fig. 6G).
Discussion
EZH2 is mutated or overexpressed in several cancers, and in some of these, it can also function as an oncogene (32, 37–40). Increased EZH2 activity is generally thought to drive cancer development through the aberrant epigenetic silencing of tumor suppressor genes (41). Thus, the oncogenic function of this protein is, in large part, believed to arise from its ability to suppress genes that regulate cell-intrinsic tumor suppressor pathways (e.g., cell survival and cell proliferation regulatory genes) (12, 13). Here, however, we identify a decisive role for NK cells in mediating EZH2-driven oncogenesis in hepatic tumor cells, with likely implications in other EZH2-dependent tumor types. Specifically, we show that EZH2 inhibition leads to tumor suppression in hepatic tumor cells, primarily through the activation of NK cell–mediated antitumor responses. This activity is dependent on reexpression of the chemokine CXCL10, which is transcriptionally repressed by EZH2 in an HDAC10-dependent manner. Critically, reexpression of CXCL10, as a result of EZH2 inhibition, enhances NK cell migration and recruitment to tumor sites, leading to enhanced NK cell–mediated tumor growth inhibition (Fig. 6G).
In previous studies, we and others have shown that EZH2 plays an important role in immune suppression. This function arises from its ability to transcriptionally repress activating NK cell ligands on cancer cells, which in turn, blocks their tumor growth inhibitory activities (14, 42). EZH2 is also expressed in various immune cells and plays a number of diverse roles (43, 44). For example, genetic deletion of EZH2 in regulatory T cells (Tregs) leads to robust antitumor immunity (43). In contrast, EZH2 was found to be required for maximal antibody secretion by antibody-secreting cells, in part, due to its ability to regulate mitochondrial respiration, glucose metabolism, and the unfolded-protein response pathway (44). In NK cells, EZH2 has further been shown to regulate cellular differentiation and function through its histone methyltransferase activity (45).
Notably, however, the relative importance of EZH2-mediated suppression of NK cell antitumor activity for its ability to promote tumor growth has not been analyzed. Here, we find that EZH2 is necessary for the repression of several immune-modulatory chemokines. Chemokines are chemotactic cytokines that control immune cell migration (46). A variety of chemokines and their receptors are expressed by cancer cells and play important roles in cancer growth and progression (47–50). For example, CXCR4 is overexpressed in over 20 different tumor types and is required for tumor metastasis (47), including lymph node and distal organ metastasis (48–50).
In this study, we show that CXCL10 expression is repressed by EZH2 in hepatic tumor cells and other EZH2-dependent cancer types. CXCL10 has been shown to promote infiltration of T cells and NK cells into the tumor microenvironment (51, 52), and increased expression of CXCL10 enhances immune cell–mediated antitumor effects in ovarian cancer (53). Using hepatic tumor cells, we further demonstrate that EED, another component of the PRC2 complex, is also required for transcriptional repression of CXCL10. Based on our observation that treatment with an HDAC inhibitor can also promote reexpression of CXCL10 in hepatic tumor cells, we performed an unbiased screen with all human HDACs and identified HDAC10 as a major regulator of EZH2 recruitment to the CXCL10 promoter, leading to its transcriptional repression via the histone methyltransferase activity of EZH2. Importantly, we further show that all these regulators of CXCL10 (e.g., HDAC10, EZH2, and EED) are necessary for NK cell migration. Furthermore, consistent with the key role of CXCL10 in promoting NK cell migration, patients with hepatic tumors expressing higher levels of CXCL10 display better prognosis. These results are also in agreement with previous studies showing that CXCL10 expression positively corelates with tumor infiltration of NK cells and T cells in hepatic tumor patients and positively impacts overall survival (54).
To further confirm the validity of our cell culture findings in vivo, using a syngeneic mouse model of hepatic tumor growth, we demonstrate that both NK cell depletion and CXCL10 neutralization are sufficient to reverse the tumor suppressive effect of pharmacological EZH2 inhibition. Thus, we show a key role for CXCL10 and NK cells in mediating EZH2 inhibition-induced antitumor activity in vivo. We note, however, that in this study, we focused our efforts specifically on understanding the role of EZH2 in regulating NK cell function and NK cell migration and found this occurs in a CXCL10-dependent manner. However, we cannot rule out the possibility that other immune cells might also be important for the oncogenic function of EZH2 in hepatic tumor cells. For example, in a previous study, it was shown that EZH2 represses expression of programmed cell death-1 ligand 1 (PD-L1) (55), and inhibition of EZH2 in T cells has been shown to improve the efficacy of anti–CTLA-4 therapy (43). Here, we identify a decisive role for NK cells and CXCL10 in EZH2-mediated oncogenesis, and thus provide evidence for an EZH2-driven mechanism of evading NK cell–mediated tumor growth inhibition that promotes hepatic tumor growth. These findings also have implications for other EZH2-dependent cancers and may yield insights for new therapeutic strategies aimed at enhancing NK cell cytotoxic activity in these cancer types.
Materials and Methods
Cell Lines and Cell Culture.
SNU-387, SNU-423, SNU-449, SNU-475, HepG2/C3A, SK-HEP-1, and PLC/PRF/5 cell lines were purchased from the American Type Culture Collection (ATCC; liver cancel panel TCP-1011). HEK293T cells, breast cancer cell lines (MCF-7 and MDA-MB-231), DLBCL cell lines (KARPAS-422 and SU-DHL-10), colon cancer cell lines (HCT116 and HT29), and the Hepa1-6 mouse liver cancer cell line were also purchased from ATCC.
HepG2/C3A, SK-HEP-1, PLC/PRF/5, MCF-7, MDA-MB-231, HCT116, Hepa1-6, and HT29 cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) with high glucose and L-glutamine, supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich) and 1× penicillin/streptomycin (Invitrogen, Thermo Fisher Scientific). SNU-387, SNU-423, SNU-449, SNU-475, KARPAS-422, and SU-DHL-10 cells were grown in Roswell Park Memorial Institute (RPMI)-1640 media, supplemented with 10% FBS and penicillin/streptomycin. NK cells (NK-92MI cells) were purchased from ATCC and grown in Alpha-Minimum Essential Medium (Sigma-Aldrich; Cat. No. M4526) with 2-mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.2 mM inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid, 12.5% horse serum, and 12.5% FBS, but without ribonucleosides and deoxyribonucleosides.
Drug Treatments.
EZH2 inhibitors (GSK343 and GSK126) and EED inhibitor (EED226) were purchased from Selleck Chemical LLC; information on these inhibitors is provided in SI Appendix, Table S4. All inhibitors were dissolved in hybridoma-grade DMSO (Sigma-Aldrich) at a stock concentration of 10 mM. Cells were treated with the inhibitor drugs at the concentrations indicated in the figure legends or DMSO (in the amount administered with the highest drug concentration). For mouse studies, GSK343 was resuspended in anhydrous ethanol with moderate vortexing and then diluted in 1× phosphate-buffered saline (PBS), at a final concentration of 2% ethanol in 1× PBS. Additional details about these inhibitors are also provided in SI Appendix, Table S4.
Human Chemokine Array.
Human Chemokine Array C1 Kits (Cat. No. AAH-CHE-1) were purchased from RayBiotech, and these were performed according to the manufacturer’s instructions. Briefly, 2 × 105 SK-HEP-1 cells per well were plated into six-well plates in OPTI-MEM Reduced-Serum Medium (Invitrogen). After 36 h, cells were treated with either DMSO or EZH2 inhibitor GSK343 for 72 h. Cell culture supernatants (conditioned media) were then collected from DMSO- or GSK343-treated cells and concentrated using Centricon tubes (3-kDa cutoff) (Millipore Sigma). Array membranes were blocked in blocking buffer at room temperature (RT) for 1 h and membranes were incubated overnight at 4 °C with 1.5 mL of concentrated conditioned media collected from DMSO- or GSK343-treated cells. The next day, membranes were washed and incubated with 1 mL of primary biotin-conjugated antibody mixture provided by the kit at 4 °C overnight. Membranes were then washed and incubated with 2 mL of a 1:1,000 dilution of horseradish peroxidase (HRP)–conjugated streptavidin at RT for 2 h. After incubation, array membrane signals were detected using the SuperSignal West Chemiluminescent Substrate Kit (Pico or Femto, Thermo Fisher Scientific). Spot signal intensities were quantified using ImageJ software (NIH). Positive controls on each array membrane were used to normalize spot intensities across different array membranes.
NK Cell Migration Assay.
NK cell migration was measured as previously described (56). In brief, various cell lines, including those expressing CXCL10 shRNAs, HDAC4 and HDAC10 shRNAs, or nonspecific (NS) shRNA, were treated with EZH2 inhibitors (GSK343 or GSK126), EED inhibitor (EED226), or DMSO control for 72 h in OPTI-MEM Reduced-Serum Medium. Cell culture supernatants (conditioned media) were collected and concentrated using Centricon tubes (3 kDa). Concentrated culture supernatants were collected and added in the bottom chamber of a Corning Transwell system (6.5-mm diameter inserts and 5.0-μm pore size; Cat. No. 3421). In parallel, NK cells (NK-92MI) (2.5 × 105) were seeded in the upper Transwell chamber. Cells were incubated at 37 °C for 4 h, after which nonadherent cells from the bottom chamber were harvested and collected in fluorescence-activated cell sorting (FACS) tubes. A known number of fluorescent CountBright Absolute Counting Beads (Cat. No. C36950; Invitrogen) in a volume of 50 μL were added to the migrated NK cells in each tube, and 300 μL/well were analyzed by flow cytometry. The absolute number of NK cells in 300 μL was calculated using the following formula: A*(B/C), where A = number of NK cell events, B = the number of beads added to each culture, and C = number of bead events.
5Aza2dC and TSA Treatment.
SK-HEP-1 and PLC/PRF/5 cells were treated with 5 μM 5Aza2dC demethylating agent for 72 h, followed by treatment with 1 μM TSA for 12 h. The cells were then harvested in TRIzol reagent (Invitrogen) for total RNA isolation and analyzed for expression of CXCL10 mRNA by qRT-PCR analysis.
Chromatin Immunoprecipitation.
ChIP assays were performed in SK-HEP-1 or PLC/PRF/5 cell lines or in cells expressing NS or HDAC10 shRNA using the SimpleChIP Enzymatic Chromatin IP Kit (Cat. No. 9002S; Cell Signaling Technology), according to the manufacturer’s instructions, as previously described (14). Cell lysates were diluted in ChIP buffer, containing protease inhibitor mixture (Roche), and the samples were incubated with antibodies against EZH2, trimethyl H3K27 (Cell Signaling Technology), HDAC10 (Sigma-Aldrich), or control IgG (Cell Signaling Technology), followed by immobilization on Protein A/G Agarose Beads (Life Technologies, Thermo Fisher Scientific). The chromatin was eluted, and DNA was extracted using the included DNA purification columns. qPCR was then performed using CXCL10 promoter-specific primers, and relative fold change was calculated as the ratio of immunoprecipitated DNA to IgG-precipitated DNA. Primer sequences and antibodies used for ChIP assays are listed in SI Appendix, Table S4.
Mouse Experiments.
All animal experiments were conducted under the ethical guidelines and protocols approved by the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham. Male C57BL/6 or NSG mice were purchased from The Jackson Laboratory. For NSG mice, 1 × 105 Hepa1-6 cells were injected subcutaneously (s.c.), and for C57BL/6 mice, 3 × 106 Hepa1-6 cells were injected s.c. At 5 d postinjection, mice were treated with 20 mg/kg of the EZH2 inhibitor GSK343 via intraperitoneal (i.p.) injection; injections were administered 5 d a week until the end of the experiment.
For NK cell depletion, 100 μL of polyclonal Ultra-LEAF Purified anti–Asialo-GM1 (clone Poly21460, BioLegend) antibodies were injected i.p. into C57BL/6 mice once every week for 4 wk. As a control, 100 μg of Ultra-LEAF purified mouse IgG1, κ-isotype control antibody (BioLegend; clone MG1-45) were administrated i.p. once every week for 4 wk. For CXCL10 neutralization in vivo, 100 μg of CXCL10 antibodies (Cat. No. MA5-23774; Invitrogen) were injected i.p. into C57BL/6 mice once every week for 4 wk. As a control, 100 μg of Ultra-LEAF purified mouse IgG1, κ-isotype control antibodies (BioLegend; Clone MG1-45) were administrated i.p. once every week for 4 wk.
Isolation of RNA from Tumors.
Tumors were excised from mice, and necrotic areas were trimmed. Tumors were then cut into small pieces, and tissues were homogenized using the gentleMACS Octo dissociator. Total tumor RNA was extracted with TRIzol reagent (Invitrogen) and purified using RNeasy Mini Columns (QIAGEN) for RNA-seq analyses.
NK Cell Infiltration and Flow Cytometry Analysis.
Tumor or spleen dissociation was performed using the Tumor Dissociation Kit, Mouse (Cat. No. 130-096-730; Miltenyi Biotec), according to the manufacturer’s instructions. Briefly, tumors were excised, and necrotic areas were trimmed and excluded from the analysis. Tumors were cut into 3-mm pieces, and those from different areas of the tumor were added to MACS C tubes, containing RPMI medium with enzyme mix. Tissues were dissociated using a gentleMACS Octo dissociator with heaters. Cell suspensions were passed through a MACS SmartStrainer (70 μm) that was placed on a 15-mL tube to collect the single-cell suspensions. After several washes with FACS buffer (PBS with 2% FBS), cells were used for flow cytometry analysis. In brief, washed cells were incubated with Fc block (anti-mouse CD16/32 antibody) at a 1:100 dilution for 10 min. Cells were then stained with Alexa Fluor 488 anti-mouse NK-1.1 and APC (allophycocyanin) anti-mouse CD335 (NKp46) antibody mixtures from BioLegend for 30 min, washed again, and subjected to flow cytometry analysis. NK1.1 and CD335 (NKp46) double-positive infiltrated NK cells were measured in mouse tumors. FACS analyses were performed using a BD LSRFortessa (BD Biosciences). All FACS data were analyzed using FlowJo software. Antibodies used for FACS analysis are listed in SI Appendix, Table S4.
Statistical Analysis.
All experiments were conducted with at least three biological replicates. Results for individual experiments are expressed as the mean ± SEM. For analysis of tumor progression in mice, statistical assessment was performed using the area under the curve (AUC) method on GraphPad Prism, v.9.0 for Macintosh (GraphPad Software; http://www.graphpad.com). P values for all other experiments were calculated using the two-tailed unpaired Student’s t test in GraphPad Prism v.9.0 for Macintosh (GraphPad Software). P values <0.05 were considered statistically significant, with significance indicated as follows: ns, not significant; *P < 0.05; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Acknowledgments
M.R.G. and N.W. are supported by NIH Grant R01CA218008. N.W. is also supported by NIH Grant R01CA257046 and by funding from the Department of Defense (W81XWH1910480 and W81XWH-18-1-0069).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102718118/-/DCSupplemental.
Data Availability
RNA-seq data of this study have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE166553) (57). All additional data discussed in the paper are available in the main text and SI Appendix.
References
- 1.van Mierlo G., Veenstra G. J. C., Vermeulen M., Marks H., The complexity of PRC2 subcomplexes. Trends Cell Biol. 29, 660–671 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Cao R., et al., Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298, 1039–1043 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Laugesen A., Højfeldt J. W., Helin K., Role of the polycomb repressive complex 2 (PRC2) in transcriptional regulation and cancer. Cold Spring Harb. Perspect. Med. 6, a026575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margueron R., et al., Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim K. H., Roberts C. W., Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Italiano A., et al., Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: A first-in-human, open-label, phase 1 study. Lancet Oncol. 19, 649–659 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Positive results for tazemetostat in follicular lymphoma. Cancer Discov. 8, OF3 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez H., Hagerling C., Werb Z., Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 32, 1267–1284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller P., et al., Trastuzumab emtansine (T-DM1) renders HER2+ breast cancer highly susceptible to CTLA-4/PD-1 blockade. Sci. Transl. Med. 7, 315ra188 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Belvin M., Mellman I., Is all cancer therapy immunotherapy? Sci. Transl. Med. 7, 315fs48 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Triulzi T., et al., Early immune modulation by single-agent trastuzumab as a marker of trastuzumab benefit. Br. J. Cancer 119, 1487–1494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Souroullas G. P., et al., An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat. Med. 22, 632–640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zingg D., et al., The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 6, 6051 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Bugide S., Green M. R., Wajapeyee N., Inhibition of Enhancer of zeste homolog 2 (EZH2) induces natural killer cell-mediated eradication of hepatocellular carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 115, E3509–E3518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parham P., Guethlein L. A., Genetics of natural killer cells in human health, disease, and survival. Annu. Rev. Immunol. 36, 519–548 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Bugide S., Janostiak R., Wajapeyee N., Epigenetic mechanisms dictating eradication of cancer by natural killer cells. Trends Cancer 4, 553–566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morvan M. G., Lanier L. L., NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer 16, 7–19 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Grégoire C., et al., The trafficking of natural killer cells. Immunol. Rev. 220, 169–182 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris M. A., Ley K., Trafficking of natural killer cells. Curr. Mol. Med. 4, 431–438 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Balkwill F., Cancer and the chemokine network. Nat. Rev. Cancer 4, 540–550 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Nagarsheth N., Wicha M. S., Zou W., Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 17, 559–572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dangaj D., et al., Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell 35, 885–900.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Vlag J., Otte A. P., Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23, 474–478 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Cantoni C., et al., NK cells, tumor cell transition, and tumor progression in solid malignancies: New hints for NK-based immunotherapy? J. Immunol. Res. 2016, 4684268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castriconi R., et al., Molecular mechanisms directing migration and retention of natural killer cells in human tissues. Front. Immunol. 9, 2324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berahovich R. D., Lai N. L., Wei Z., Lanier L. L., Schall T. J., Evidence for NK cell subsets based on chemokine receptor expression. J. Immunol. 177, 7833–7840 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Viré E., et al., The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Gazin C., Wajapeyee N., Gobeil S., Virbasius C. M., Green M. R., An elaborate pathway required for Ras-mediated epigenetic silencing. Nature 449, 1073–1077 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCabe M. T., et al., EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Hirukawa A., et al., Targeting EZH2 reactivates a breast cancer subtype-specific anti-metastatic transcriptional program. Nat. Commun. 9, 2547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Böhm J., et al., Loss of enhancer of zeste homologue 2 (EZH2) at tumor invasion front is correlated with higher aggressiveness in colorectal cancer cells. J. Cancer Res. Clin. Oncol. 145, 2227–2240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morin R. D., et al., Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 42, 181–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song X., et al., Selective inhibition of EZH2 by ZLD1039 blocks H3K27 methylation and leads to potent anti-tumor activity in breast cancer. Sci. Rep. 6, 20864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fussbroich B., et al., EZH2 depletion blocks the proliferation of colon cancer cells. PLoS One 6, e21651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasai M., et al., In vivo effect of anti-asialo GM1 antibody on natural killer activity. Nature 291, 334–335 (1981). [DOI] [PubMed] [Google Scholar]
- 36.Zhou G., Juang S. W., Kane K. P., NK cells exacerbate the pathology of influenza virus infection in mice. Eur. J. Immunol. 43, 929–938 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Chase A., Cross N. C., Aberrations of EZH2 in cancer. Clin. Cancer Res. 17, 2613–2618 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Ernst T., et al., Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 42, 722–726 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Kleer C. G., et al., EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 100, 11606–11611 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varambally S., et al., The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Gan L., et al., Epigenetic regulation of cancer progression by EZH2: From biological insights to therapeutic potential. Biomark. Res. 6, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zingg D., et al., The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep. 20, 854–867 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Goswami S., et al., Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J. Clin. Invest. 128, 3813–3818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo M., et al., EZH2 represses the B cell transcriptional program and regulates antibody-secreting cell metabolism and antibody production. J. Immunol. 200, 1039–1052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin J., et al., Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 112, 15988–15993 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luster A. D., Chemokines – Chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 338, 436–445 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Chatterjee S., Behnam Azad B., Nimmagadda S., The intricate role of CXCR4 in cancer. Adv. Cancer Res. 124, 31–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Ordoñez A., et al., Breast cancer metastasis to liver and lung is facilitated by Pit-1-CXCL12-CXCR4 axis. Oncogene 37, 1430–1444 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Su L., et al., Differential expression of CXCR4 is associated with the metastatic potential of human non-small cell lung cancer cells. Clin. Cancer Res. 11, 8273–8280 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Smith M. C., et al., CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 64, 8604–8612 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Susek K. H., Karvouni M., Alici E., Lundqvist A., The role of CXC chemokine receptors 1-4 on immune cells in the tumor microenvironment. Front. Immunol. 9, 2159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wennerberg E., Kremer V., Childs R., Lundqvist A., CXCL10-induced migration of adoptively transferred human natural killer cells toward solid tumors causes regression of tumor growth in vivo. Cancer Immunol. Immunother. 64, 225–235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.K Au K., et al., CXCL10 alters the tumour immune microenvironment and disease progression in a syngeneic murine model of high-grade serous ovarian cancer. Gynecol. Oncol. 145, 436–445 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Chew V., et al., Chemokine-driven lymphocyte infiltration: An early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut 61, 427–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao G., et al., EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J. Immunother. Cancer 7, 300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chava S., Bugide S., Gupta R., Wajapeyee N., Measurement of natural killer cell-mediated cytotoxicity and migration in the context of hepatic tumor cells. J. Vis. Exp. 156, 1–7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bugide S., Wajapeyee N., Study the differential gene expression of HEPA1-6 tumors in C57/BL6 Mouse tumors treated with vehicle or EZH2 inhibitor GSK343. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166553. Deposited 10 February 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data of this study have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE166553) (57). All additional data discussed in the paper are available in the main text and SI Appendix.