Significance
Animal models and genome-scale metabolic models (GEMs) are two important tools for studying human diseases, but a proper integration of the two is lacking. To bridge them, we developed a reproducible and systematic framework by which the GEMs for major model animals are reconstructed and maintained. Due to the shared namespace, biological insights can be inferred from comparative analysis of the omics data from different animal models of the same disease. The drug targets and biomarkers derived from integrative metabolic analysis can thus be validated and translated to treatments and/or diagnostic methods. As an example, we demonstrated their utility in discovering potential biomarkers for early diagnosis of Alzheimer’s disease, where there is a lack of effective diagnostics and treatments.
Keywords: genome-scale model, animal model, Alzheimer’s disease, Aβ deposition, translational medicine
Abstract
Genome-scale metabolic models (GEMs) are used extensively for analysis of mechanisms underlying human diseases and metabolic malfunctions. However, the lack of comprehensive and high-quality GEMs for model organisms restricts translational utilization of omics data accumulating from the use of various disease models. Here we present a unified platform of GEMs that covers five major model animals, including Mouse1 (Mus musculus), Rat1 (Rattus norvegicus), Zebrafish1 (Danio rerio), Fruitfly1 (Drosophila melanogaster), and Worm1 (Caenorhabditis elegans). These GEMs represent the most comprehensive coverage of the metabolic network by considering both orthology-based pathways and species-specific reactions. All GEMs can be interactively queried via the accompanying web portal Metabolic Atlas. Specifically, through integrative analysis of Mouse1 with RNA-sequencing data from brain tissues of transgenic mice we identified a coordinated up-regulation of lysosomal GM2 ganglioside and peptide degradation pathways which appears to be a signature metabolic alteration in Alzheimer’s disease (AD) mouse models with a phenotype of amyloid precursor protein overexpression. This metabolic shift was further validated with proteomics data from transgenic mice and cerebrospinal fluid samples from human patients. The elevated lysosomal enzymes thus hold potential to be used as a biomarker for early diagnosis of AD. Taken together, we foresee that this evolving open-source platform will serve as an important resource to facilitate the development of systems medicines and translational biomedical applications.
Animal models have long been utilized as a fundamental tool for translational research in recapitulating phenotypic syndromes, clarifying underlying mechanisms, and translating biomedical discoveries into effective clinical treatments for human disease (1). Small rodents, including mouse (Mus musculus) and rat (Rattus norvegicus), account for 90% of the tens of millions of animals used annually in medical research (2, 3), and transgenic mice in particular are the most commonly used models for a plethora of human diseases including cancers, neurodegenerative dementia, diabetes, and many other metabolic disorders (4). In addition, with their unique anatomical and physiological features transgenic zebrafish (Danio rerio) and invertebrate models, such as the fruit fly (Drosophila melanogaster) and Nematoda worm (Caenorhabditis elegans), have been used for many years as inexpensive alternatives for studying human diseases through genetic manipulation of ortholog genes (5, 6). It is also important to note that nearly all the fundamental aspects of biology have been derived from the study of model organisms (7).
A genome-scale metabolic model (GEM) is a mathematical representation of the metabolism for an organism and it provides extensive gene–reaction–metabolite connectivity via two matrices: the S matrix for associating metabolites to reactions and the rxnGeneMat matrix associating reactions to corresponding enzymes and genes (8). Given that many human diseases, including cancer, type II diabetes, and many liver- and pancreas-related diseases can be attributed to metabolic disorders (9), human GEMs have been used to describe the metabolic conditions of specific tissues and cell types at the systems level with the integration of omics data (10–13). For the purpose of clarity, henceforth “GEM” is used here to refer to a computational metabolic model, and “model” refers to a transgenic animal developed for studying human disease.
The use of animal models together with GEMs poses an attractive approach to studying human disease. For example, mouse GEMs have previously been applied to investigate the influences of gut microbiota on host metabolism (14). To date, there have been a few existing GEMs for model animals, including MMR (14) and iMM1865 (15) for mouse, iRno (16) for rat, ZebraGEM (17, 18) for zebrafish, and iCEL1273 and iCEL1314 for worm (19, 20). However, these GEMs have not been developed and publicly curated to the same extent as that of yeast (21) and human (11). Their limited coverage in metabolic pathways and incompatible nomenclatures impede cross-species validation of biological discoveries and translational applications from animal models to human patients.
Recently, an open and version-controlled workflow has been introduced during the development of the most comprehensive yeast and human GEM, Yeast8 (21) and Human1 (11), respectively, which present high-quality templates to develop new GEMs in a systematic and reproducible manner. Databases such as MGD (22), FlyBase (23), ZFIN (24), and WormBase (25) that provide organism-specific annotation and human orthologs have been recently integrated into a centralized portal, the Alliance of Genome Resources (26), for consistent annotation and curation of gene ontology in relation to the human counterparts. Through channeling these reliable data sources we here present a unified GEM platform for mouse, rat, zebrafish, fruit fly, and worm. The derived GEMs (Mouse1, Rat1, Zebrafish1, Fruitfly1, and Worm1, respectively) were reconstructed from a robust modeling pipeline that combines both the orthology-based metabolic network and species-specific pathways. To validate this approach, we conducted an extensive GEM comparison and gene essentiality analysis using available experimental data and demonstrated that our GEMs generally outperform the previous ones. We also showcased the usefulness of Mouse1 in systems medicine discovery by performing integrative analysis of omics data from mouse models of Alzheimer’s disease (AD). We are confident that this versatile GEM platform covering all major model animals will greatly enhance the utilization of omics data from disease models in facilitating translational studies.
Results
GEM Reconstruction, Exploration, and Curation for Model Animals.
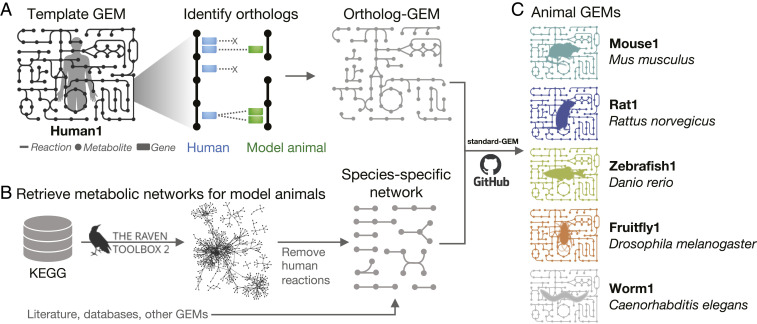
We developed a pipeline that combines various reliable data sources and generates a coherent collection of GEMs for major model animals, including mouse (M. musculus), rat (R. norvegicus), zebrafish (D. rerio), fruit fly (D. melanogaster), and Nematoda worm (C. elegans) (Fig. 1 and SI Appendix, Fig. S1). In this pipeline, the open-curated generic human GEM, Human1 (11), was used as a template. The well-annotated orthologs and paralogs associated from human to the five model organisms, as well as evaluating features bestForward, bestReverse, and methodCount (the number of different methods used in determining the orthologs), were retrieved from the Alliance Genomes databases using the stringent criteria provided (26). All one-to-one pairs were kept, while the one-to-multiple pairs were filtered with the following criteria: 1) exclude the pairs that are neither bestforward nor bestReverse and 2) only keep the pairs that are both the best froward and reverse hits. If steps 1 and 2 exclude all hits for a query gene, then we retrieved and kept the ortholog pair(s) with the highest methodCount. Using the RAVEN toolbox (27), the species-specific reactions and metabolites for each animal were extracted from the KEGG (Kyoto Encyclopedia of Genes and Genomes) database and manually inspected for integration (SI Appendix). Here, we report the first releases of these simulation-ready animal GEMs as Mouse1, Rat1, Zebrafish1, Fruitfly1 and Worm1, respectively (Fig. 1 and Dataset S1).
Fig. 1.
Genome-scale metabolic modeling for model animals. A reconstruction approach of combining (A) ortholog-GEMs derived from the Human1 template and (B) species-specific metabolic networks extracted from the KEGG database by the RAVEN package was used to obtain (C) the model animal GEMs that were deposited on GitHub according to the standard-GEM scheme (28).
These new GEMs were integrated into the accompanying web portal Metabolic Atlas (https://metabolicatlas.org/), which allows for interactive exploration and cross-species comparison of the GEMs through the GEM Browser, a tabular interface; the Map Viewer, a collection of manually curated two-dimensional maps and automated three-dimensional maps; and the Interaction Partners tool, a network view of gene-metabolite associations. Furthermore, the relational database (PostgreSQL) powering the Metabolic Atlas portal was replaced by a graph implementation (Neo4j), enabling new features such as GEM comparison via external identifiers. These improvements are released as Metabolic Atlas 2.0, which is open-source with the running website, graph database integration, upgraded three-dimensional viewer, and all data files publicly available (SI Appendix).
To facilitate open curation and continuous integration of biochemical knowledge from the research community, these GEMs are tightly integrated with GitHub (see Data Availability) in complying with the “standard-GEM” specifications (28), which defines a set of requirements and recommendations for versioning GEMs and structuring Git-based repositories.
GEM Comparison, Evaluation, and Validation.
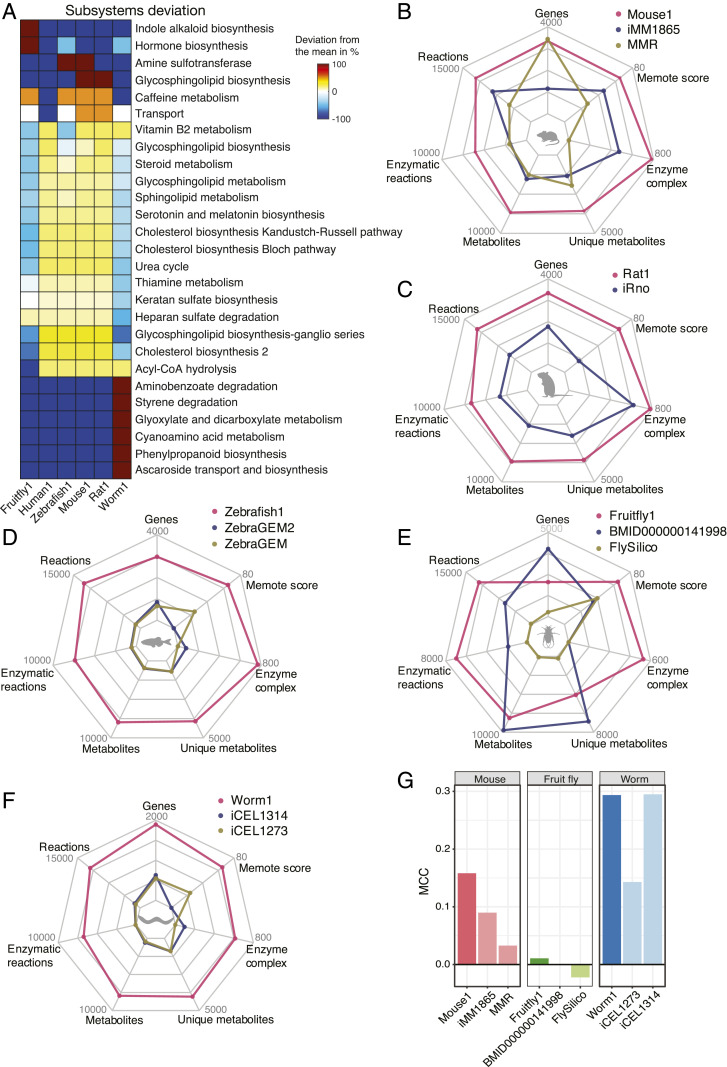
As generated by a coherent pipeline in which all primary identifiers belong to the same namespace, the new generic GEMs can be compared under a consistent scheme of subsystems (Fig. 2A). Despite the reduction in the total number of reactions, Worm1 contained new phenylpropanoid biosynthesis pathway reactions, and the hormone biosynthesis pathway in Fruitfly1 was augmented, after the integration of a species-specific metabolic network. Likewise, Mouse1 and Rat1 are capable of de novo synthesis of vitamin C (ascorbate), a metabolic feature retained in most rodent species but lost in humans (29). In addition, the primary identifiers of metabolites and reactions from the new GEMs were all provided with the MetaNetX identifiers (30) for convenient comparison and expansion.
Fig. 2.
Systematic comparison and evaluation of generic model animal GEMs. (A) Significantly altered subsystems with deviated reaction content between the newly generated GEMs and Human1. The color indicates the percent difference in the number of reactions within each subsystem for a GEM compared to the mean number of reactions in that subsystem across all GEMs. Radar plots showing the comparison in the numbers of reactions, metabolites, genes, and enzyme complexes that have a GPR with “and” relation, as well as benchmarking MEMOTE scores between the GEMs for (B) mouse, (C) rat, (D) zebrafish, (E) fruit fly, and (F) worm. (G) Evaluation of gene essentiality prediction performance among GEMs of mouse, fruit fly, and worm using the MCC, which scores the relative amount of true and false positive and negative predictions of gene essentiality.
To validate these newly developed GEMs, we conducted a systematic comparison with existing GEMs of the same species (Fig. 2 and Dataset S1). For each species, the newly generated GEMs obtained substantially expanded coverage in metabolic components of genes, reactions, and metabolites, as well as extended complex-subunit information (Fig. 2 B–F). An exception was the GEM BMID000000141998, which was produced from a fully automatic pipeline and included more genes and metabolites, but the quality of automatically generated GEMs is generally low due to lack of curation (31).
The quality of a GEM is often evaluated by predicting genes that are essential to the viability of the organism and comparing the predictions with experimental data. To validate our animal GEM reconstruction pipeline, a gene essentiality analysis was conducted for each species by comparing the predictive performance of new and previous GEMs (Fig. 2G and Dataset S2). In this analysis, the genes for a given GEM were individually knocked out in silico to identify those that are essential for biomass formation (cell viability). The prediction results were then compared with gene essentiality data retrieved from the Online Gene Essentiality (OGEE) database (32) to quantify the number of true and false positives and negatives. The Matthews correlation coefficient (MCC), a balanced metric of classification performance (33), was calculated for the GEMs of mouse, fruit fly, and worm, for which genome-scale experimental gene essentiality data were available (Dataset S2). Mouse1 demonstrated a substantial improvement in essentiality predictions (MCC = 0.158) compared to 0.09 and 0.03 for iMM1865 and MMR, respectively. Fruitfly1 also showed a better predictive capacity than FlySilico, a simple GEM covering only central carbon, amino acid, lipid, and carbohydrate metabolism (31). The automatically generated GEM BMID000000141998 predicted all genes as nonessential and therefore had an undefined MCC value (Fig. 2G). For the worm GEMs, Worm1 displayed improved prediction performance compared to iCEL1273 (19) and equivalent to the very recent iCEL1314, from which by incorporating the pathway for Ascaroside biosynthesis and transportation (20). This further highlights the importance of curation and more complete coverage of metabolism in obtaining systems-level insights.
The MEMOTE scores (34), which were estimated from a series of tests on stoichiometric consistency, mass-balanced and blocked reactions, associated annotations, and so on, were used for benchmarking. The new GEMs outperformed the existing GEMs for each species with regard to MEMOTE scores (Fig. 2 B–F).
Integrative Metabolic Analysis of AD Models Using Mouse1.
Due to many attractive features (small and easy to handle, prolific breeder, and amenable to genetic manipulation) of mice and high genomic similarity to human, mouse models exist for a substantial number of diseases (4). They are extremely useful in studying the conditions for which patient samples and experiments are unfeasible or unethical to obtain (e.g., neurodegenerative diseases). To date, there are ∼200 different mouse models that have been developed for understanding the mechanism of AD (35), which is the most prevalent dementia affecting millions of people worldwide (36).
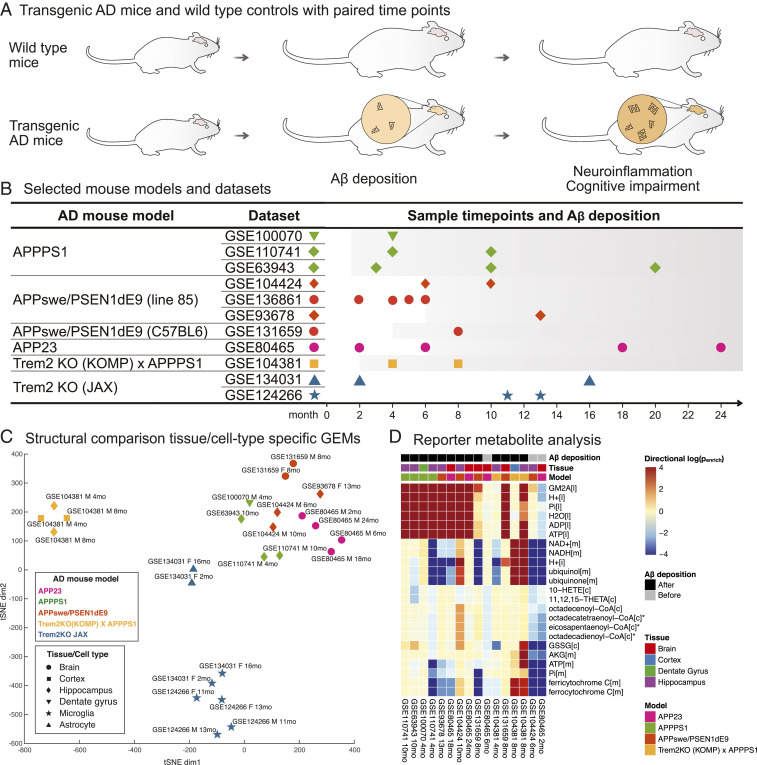
To demonstrate the utility of the GEMs developed using our framework, Mouse1 was used to investigate different AD models through omics data integration and gene set analysis (Fig. 3 and SI Appendix, Figs. S2–S6). Using a detailed search of the GEO database (SI Appendix), a total of 11 high-quality RNA-sequencing (RNA-seq) datasets including both transgenic mice and wild-type controls with paired time points were selected from six representative AD models (Fig. 3 A and B and Dataset S3). There were four amyloid precursor protein (APP) overexpression models that recapitulate the major AD pathology (i.e., the formation of intracellular plaques and tangles containing aggregated amyloid β (Aβ) peptide and hyperphosphorylated tau protein), as well as two Trem2KO models with a deleted Trem2 gene that had complete or partial absence of the Aβ deposition phenotype. To make the figures simple and easy to understand, the models APPswe/PSEN1dE9 (line 85) and APPswe/PSEN1dE9 (C57BL6) that have identical genotype but use different mouse strains were merged and the sample GEO IDs were consistently provided for clarification. Next, we applied the task-driven integrative network inference algorithm (tINIT) (37) to generate tissue- and cell-type-specific GEMs, which combine the metabolic network determined by the RNA-seq data of metabolic genes and the essential reactions required for all cell types (SI Appendix). A structural comparison showed that the GEMs from APP overexpression models are more homogeneous than those from Trem2KO models, regardless of tissue source, gender, and age (Fig. 3C and SI Appendix, Fig. S2). In contrast, the GEMs from Trem2KO model separated explicitly by cell types. GEMs from APP overexpression models were characterized by an enhanced coverage of pathways including cholesterol biosynthesis, beta oxidation of fatty acids, and amino acid metabolism but reduced coverage of biopterin metabolism (SI Appendix, Fig. S3). In addition, de novo synthesis of asparagine, cystine, and cysteine and energy-associated functions were reduced in Trem2KO model GEMs (SI Appendix, Fig. S4). Interestingly, the GEMs of the hybrid model that was developed by crossing the Trem2KO and APPPS1 transgenic mice presented different metabolic features from both ascendants in reaction content (Fig. 3C and SI Appendix, Fig. S2), subsystem coverage (SI Appendix, Fig. S3), and metabolic task performance (SI Appendix, Fig. S4). In summary, the comparison of these context-specific GEMs revealed distinct metabolism between the neuronal cells of APP overexpression and Trem2KO models.
Fig. 3.
RNA-seq data integration and gene set analysis using Mouse1. (A) Various AD models of transgenic mice have been developed to recapitulate the pathology of Aβ deposition and subsequent phenotypes of subsequent neuroinflammation and cognitive impairment. (B) RNA-seq datasets from transgenic and wild-type mice with paired time points were selected for studying the metabolic changes associated with AD progression. The colored symbols are used to depict the different datasets and their sampling time points in relation to the onset and progression of Aβ deposition, which is illustrated by a shaded background (Dataset S3). (C) Structural comparison of tissue/cell type-specific GEMs using t-distributed stochastic neighbor embedding (tSNE) analysis. (D) Reporter metabolite gene set analysis using Mouse1. The log-transformed Penrich value quantifies the significance of substantially up- (in positive values) or down-regulated (in negative values) gene sets between diseased and normal conditions. Subcellular compartment is indicated in brackets, in which l, m, and c refer to lysosome, mitochondrion, and cytosol, respectively. *The full names of the CoA metabolites are (6Z,9Z,12Z,15Z)-octadecatetraenoyl-CoA, (5Z,8Z,11Z,14Z,17Z)-eicosapentaenoyl-CoA, and (7Z)-octadecenoyl-CoA and (6Z,9Z)-octadecadienoyl-CoA, respectively.
Enrichment analysis conducted with various gene-set collections also showed distinct metabolic differences between APP overexpression and Trem2KO models (SI Appendix, Fig. S5). Among the APP overexpression models, substantial expression profile changes were observed along with Aβ deposition, including the up-regulation of pathways in apoptosis, signaling, cholesterol homeostasis, innate immune system, allograft rejection, inflammatory and interferon response, and a number of energy metabolism and neurogenerative diseases (SI Appendix, Fig. S5). To pinpoint the altered metabolic processes associated with Aβ deposition we performed comparative analysis of subsystems (SI Appendix, Fig. S6) and reporter metabolite gene sets (Fig. 3D). The oxidative phosphorylation, amino acid metabolism, fatty acid oxidation, and cholesterol metabolism were identified as prominently up-regulated during the progression of AD (SI Appendix, Fig. S6). This is consistent with the down-regulation of mitochondrial metabolite gene sets associated with energy metabolism in the AD models before the onset of Aβ deposition (Fig. 3D). The alteration patterns of these metabolic pathways were also consistent with that found in the previous enrichment analysis (SI Appendix, Fig. S5), as well as the tissue-specific GEM comparison (Fig. 3C and SI Appendix, Fig. S3). With gene–reaction–metabolite connectivity of GEMs, gene sets associated with each metabolite can be obtained and subjected to enrichment analysis that may reveal insights into network level alterations of given expression data. Notably, the gene sets of reporter metabolites (GM2A, H+, Pi, H2O, ADP, and ATP) associated with lysosome (Dataset S3) were uniformly enriched in the APP overexpression models after Aβ deposition but absent from the hybrid Trem2 KO (KOMP) × APPPS1 model (Fig. 3D).
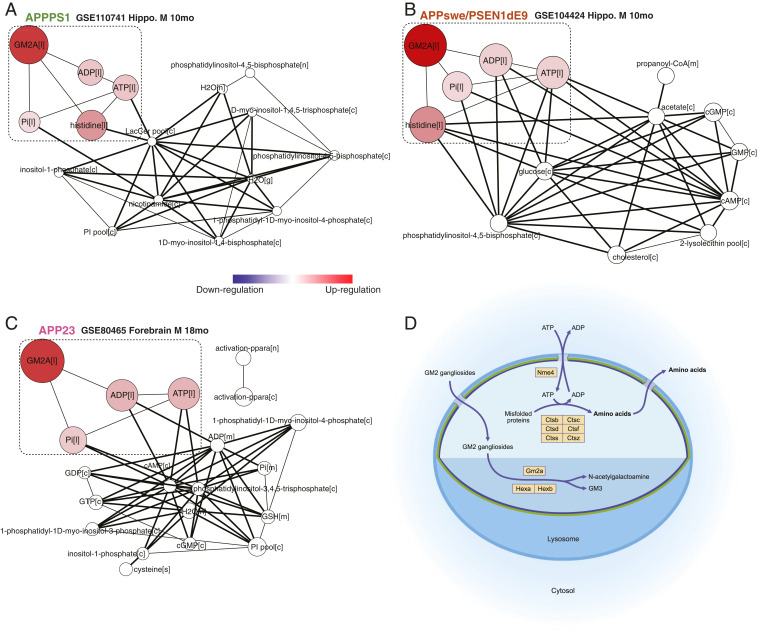
Aβ Accumulation Activates Gene Expression in Lysosomal Pathways of GM2 Gangliosides and Peptide Degradation.
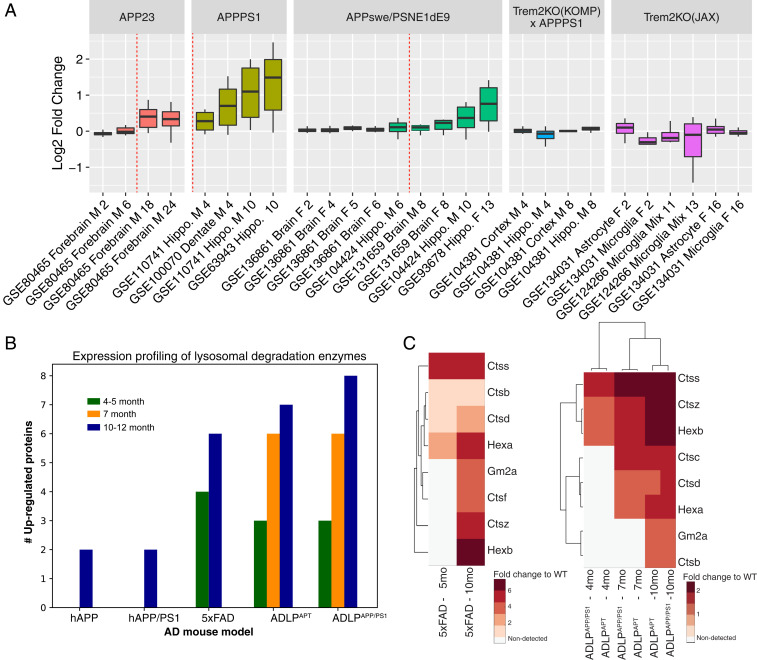
By integrative analysis of reporter metabolite gene sets with Mouse1 we discovered an up-regulated GM2A-centric lysosomal subnetwork shared between APP overexpression models (Fig. 4 and SI Appendix, Fig. S7). This subnetwork was not enriched in the data from Trem2KO models, as well as the conditions prior to Aβ plaque formation in APP overexpression models (SI Appendix, Fig. S8). It therefore appeared to be the signature response induced specifically by Aβ accumulation. By inspecting the expression pattern of the lysosomal genes across the collected AD models we confirmed their consistent up-regulation in APP overexpression models and only after the onset of Aβ deposition (Fig. 5A). Within the APP models, a higher magnitude of fold change of lysosomal genes was observed in APPPS1 compared to APP23 and APPswe/PSEN1dE9. Among the four time points from the APP23 model data (GSE80465), the up-regulation of the lysosomal genes peaked at 18 mo, whereas they appeared to slightly decrease in a later stage at 24 mo. In contrast, only marginal changes were observed in the APPswe/PSEN1dE9 (line 85) model (GSE136861), which were sampled with four time points (2, 4, 5, and 6 mo) when Aβ plaques have not been formed in the brain of transgenic mice (Dataset S3).
Fig. 4.
Integrative analysis of reporter metabolite gene sets with Mouse1. The up-regulation of a GM2A-centric subnetwork in lysosome is identified as a signature response to in APP overexpression models: (A) APPPS1, (B) APPswe/PSEN1dE9, and (C) APP23. This subnetwork, indicated by a dashed box, was found to be the most significant and consistent metabolic change to Aβ deposition. The nodes depict significantly changed reporter metabolite gene sets (cutoff: P < 0.002), each of which comprises all genes associated with reactions in which one metabolite is involved. The node color reflects directionality score that indicates the overall differential direction of the gene set, ranging from down- (blue) to up-regulation (red). The node sizes are proportional to the −log10p values of corresponding gene sets, and the edge width indicates the number of reactions shared by the metabolites. (D) The diagram depicts the lysosomal degradation pathways that were detected from the elevated mouse genes through integrative analysis.
Fig. 5.
Validation of elevated lysosomal enzymes at the RNA and protein level. (A) Log2 fold changes of lysosomal genes (Gm2a, Hexa, Hexb, Ctsb, Ctsc, Ctsd, Ctsf, Ctss, Ctsz, and Nme4) that are involved in GM2 ganglioside and peptide degradation. The data from each AD model are displayed in the order of sampling time, while the onset of Aβ deposition is indicated by a dashed red line. (B) Bar plot showing the number of significantly (P < 0.05) up-regulated lysosomal enzymes from five APP overexpression mouse models (hAPP, hAPP/PS1, 5xFAD, ADLPAPPPS1, and ADLPAPT) (Dataset S4). (C) Heat maps showing the fold changes of these significantly up-regulated lysosomal enzymes along AD progression.
Validation of Activated Lysosomal Degradation Pathways Using Proteomics Data from AD Models.
To validate the activation of the lysosomal degradation pathways at the protein level we reanalyzed proteomics datasets obtained from brain tissues of another five APP overexpression models: hAPP, hAPP/PS1, 5xFAD, ADLPADP, and ADLPAPP/PS1 (Fig. 5B and Dataset S3). Despite the different methods of mass spectrometric detection and quantification, a concerted up-regulation of the candidate lysosomal digestive enzymes, except Nme4, was observed among the differentially expressed proteins between transgenic versus wild-type mice at different time points (Fig. 5C). In 5xFAD, ADLPAPT, and ADLPAPP/PS1 models, AD progression was clearly accompanied by an increase of these lysosomal proteins in fold changes compared to the wild-type conditions. This suggests the potential application of their corresponding human orthologs, such as CTSS, as biomarkers for early diagnosis.
Investigation of Lysosomal Peptide Concentrations in Cerebrospinal Fluid Samples of AD Patients.
Since our approach ensured a consistent namespace and nomenclature between Human1 and new animal GEMs, this enables convenient translation of results from animal model experiments to human validation. Consequently, we sought to investigate the expression levels of these lysosomal enzymes in human samples. By analyzing a dataset from a recent study that quantified 51 targeted peptides in the cerebrospinal fluid samples from AD patients and healthy controls (38), we found that peptides from GM2A, CTSB, and CTSD were significantly enriched in AD patients compared to those from healthy individuals (SI Appendix, Fig. S9), supporting the association of altered lysosomal function in AD patients.
Discussion
In this study we presented a robust animal GEM development pipeline with which the most comprehensive GEMs (Mouse1, Rat1, Zebrafish1, Fruitfly1, and Worm1) for five major model animals were generated and maintained (Fig. 1 and SI Appendix, Fig. S1). With significantly expanded coverage of metabolic components compared to previously reported GEMs (Fig. 2 and Dataset S1), these new GEMs collectively serve as a coherent platform allowing systematic integration of high-throughput omics data from a wealth of disease models. These GEMs, capturing both the orthology-based metabolic network and species-specific pathways (Fig. 1) that can be interactively queried through the accompanying web portal Metabolic Atlas (11), will evolve as a continuously expanding knowledgebase for the study of animal metabolism.
We demonstrated a specific case of using Mouse1 through a systems biology approach with omics data integration (Fig. 3). There exists an abundance of rodent models developed by introducing AD risk genes (35) that include APP and presenilin 1 and 2 (PSEN1 and PSEN2), which have been determined to cause Aβ deposition in familial forms of AD (39), and other susceptible genes (e.g., TREM2) that were associated with sporadic AD (40, 41). Both APP overexpression and Trem2KO models, as well as the hybrid one Trem2KO × APPPS1 (42), were investigated to evaluate their metabolic features in terms of the different genetic backgrounds (Fig. 3 and SI Appendix, Figs. S2–S4). The illustration of model-specific changes revealed their distinct metabolism, supporting the claim that each model can only recapitulate partial AD pathologies (43). This further stresses the necessity of an integrative evaluation of different transgenic animals, and our GEM-based in silico analysis was shown to be very useful in characterizing various animal models of the same disease in their metabolic capacity and guiding downstream investigations toward targeted clinical application.
Aβ plaque formation increases with age in both transgenic mice and AD patients (6), making APP overexpression models useful tools in elucidating the temporal development of AD in human brain (35). Assisted by the comprehensive coverage of metabolic genes in Mouse1, a subnetwork of lysosomal degradation pathways was identified as a signature response to Aβ deposition across the APP overexpression models (Fig. 4 and SI Appendix, Fig. S7). The lysosomal pathways appeared to be activated immediately with the onset of Aβ deposition and sustained throughout the progression of AD in transgenic mice (Fig. 5). The increased transcription and translation of individual lysosomal components were previously detected in brains of AD and other neurodegenerative disease patients (44). This is consistent with the observation of reduced Aβ levels in the brain of transgenic mice after in vivo up-regulation of lysosomal activities (45). In particular, our findings address lysosomal genes hydrolyzing GM2 gangliosides and peptides (Fig. 4D), whose homeostasis in neurons is essential for brain maintenance (46). Gangliosides are abundant in brain tissue and constitute ∼10 to 12% of the lipid content in neuronal membranes (47). The content and composition of gangliosides decrease dramatically during aging (48). The GM2A, HEXA, and HEXB genes encode enzymes responsible for degradation of GM2 gangliosides, mutations obstructing this pathway induce neurodegenerative disorders, including Tay–Sachs disease, AB variant, and Sandhoff disease (49). Similarly, cysteine cathepsins CtsB/L have been demonstrated as essential in lysosomal degradation of Aβ peptides in mouse embryonic fibroblasts (50). In vitro screening also showed that CtsB and CtsL can proteolyze α-synuclein amyloid fibrils that are closely associated with neurodegeneration in parkinsonian disorders, while CtsD requires the assistance of anionic lipids for the hydrolysis (51). Collectively, a strong correlation between AD and endolysosomal activities has been established with a growing number of studies. Previously, genetic deficiency of lysosomal genes was speculated as the cause of endolysosomal and autophagic dysfunction that subsequently drives AD progression (52). Using a systematic analysis of AD models with and without Aβ plaques, however, our results suggests that up-regulation of lysosomal degradation pathways occurs downstream of Aβ deposition, such that elevated expression of lysosomal genes is a result of AD pathogenesis rather than a cause. Also, lysosomal dysfunction appears to be involved as a cascading effect of Aβ deposition and worth continued exploration.
Biomarkers for an early diagnosis are urgently needed for AD due to the lack of effective treatments (53). There are currently three well-established biomarkers of lower Aβ42 levels and higher levels of total and phosphorylated tau for AD diagnosis (54). By translating the metabolic alterations obtained from analyzing AD models with Mouse1 (Fig. 5), here we presented a collection of nine lysosomal enzymes as potential biomarkers, of which CSTD has been previously verified in cerebrospinal fluid (55) and plasma samples (56), while the others represent new candidates. Our statistical analysis showed that the peptides of CTSB, CTSD, and GM2A in cerebrospinal fluid samples of AD patients are significantly increased compared to those in healthy controls (SI Appendix, Fig. S9). In addition, the rapid elevation of expressed lysosomal genes following Aβ deposition in mouse models also suggests that these lysosomal enzymes may be up-regulated in the prodromal phase of AD (Fig. 5). This is in good agreement with the longitudinal analysis of plasma CSTD levels, which were sampled from the same patients at preclinical phase and after diagnosed as AD (with an interval of 1 to 10 y), and both were significantly elevated in comparison to that in healthy controls (56). Taken together, the presented lysosomal enzymes identified with altered expression by the approach of systems medicines constitute potential body fluid biomarkers that may be utilized for early-stage AD diagnosis.
In summary, we presented tissue- and time-dependent variations of AD mice and reported candidate biomarkers through integrative analysis of omics data with animal GEMs. By associating gene expression changes to subsystem and metabolite levels, this study demonstrated how results from model organism can be translated to clinical implications in human. Next, it is expected that the discovered disease-specific patterns and biomarkers could be inspected with large-scale comparative analysis of human datasets and body fluid samples for further verification. For quantitative flux predictions, we suggest contextualizing (e.g., tINIT) these GEMs with omics data and/or using the GECKO enzyme-constraint framework (57). Along with the rapidly growing omics data, we are confident that the set of animal GEMs will be a valuable platform assisting translational studies toward developing clinical treatments for a wide range of human diseases.
Materials and Methods
All the materials and methods are detailed in SI Appendix: generation, comparison, and evaluation of animal GEMs; development of Metabolic Atlas 2.0; RNA-seq data retrieval and differential expression analysis; gene set and network integrative analysis; proteomics data investigation; and statistical analyses. All GEM simulations were carried out in MATLAB using RAVEN toolbox (27) with the Gurobi solver (Gurobi Optimization, LLC).
Supplementary Material
Acknowledgments
We thank Leif Väremo, Francesco Gatto, Sinisa Bratulic, Rasool Saghaleyni, Fariba Roshanzamir, and Xin Chen for valuable discussions. The computations were performed on resources provided by the Swedish National Infrastructure for Computing at C3SE. This work was supported by the Knut and Alice Wallenberg Foundation and the Chalmers Foundation. H.Z. is a Wallenberg Scholar supported by grants from the Swedish Research Council (2018-02532), the European Research Council (681712), Swedish State Support for Clinical Research (ALFGBG-720931), the Alzheimer Drug Discovery Foundation (201809-2016862), and the UK Dementia Research Institute at University College London.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102344118/-/DCSupplemental.
Data Availability
The GEMs for Mouse1, Rat1, Zebrafish1, Fruitfly1, and Worm1 are available on GitHub at https://github.com/SysBioChalmers/Mouse-GEM, https://github.com/SysBioChalmers/Rat-GEM, https://github.com/SysBioChalmers/Zebrafish-GEM, https://github.com/SysBioChalmers/Fruitfly-GEM, and https://github.com/SysBioChalmers/Worm-GEM, respectively. The code for GEM development is available at https://github.com/SysBioChalmers/Human-GEM. The tissue- and cell-type-specific GEMs for transgenic mice and code for integrative analysis are available at https://github.com/SysBioChalmers/Mouse-GEM. All other data are available in supporting information.
References
- 1.Beck A. P., Meyerholz D. K., Evolving challenges to model human diseases for translational research. Cell Tissue Res. 380, 305–311 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Phillips K. A., et al., Why primate models matter. Am. J. Primatol. 76, 801–827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daneshian M., Busquet F., Hartung T., Leist M., Animal use for science in Europe. ALTEX 32, 261–274 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Rosenthal N., Brown S., The mouse ascending: Perspectives for human-disease models. Nat. Cell Biol. 9, 993–999 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Davis E. E., Katsanis N., Zebrafish: A Model System to Study the Architecture of Human Genetic Disease (Elsevier, 2017). [Google Scholar]
- 6.Dawson T. M., Golde T. E., Lagier-Tourenne C., Animal models of neurodegenerative diseases. Nat. Neurosci. 21, 1370–1379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fields S., Johnston M., Whither model organism research? Science (80-.) 307, 1885–1886 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Nielsen J., Systems biology of metabolism. Annu. Rev. Biochem. 86, 245–275 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Nielsen J., Systems biology of metabolism: A driver for developing personalized and precision medicine. Cell Metab. 25, 572–579 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Agren R., et al., Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLOS Comput. Biol. 8, e1002518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson J. L., et al., An atlas of human metabolism. Sci. Signal. 13, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis N. E., Abdel-Haleem A. M., The evolution of genome-scale models of cancer metabolism. Front. Physiol. 4, 237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yizhak K., Chaneton B., Gottlieb E., Ruppin E., Modeling cancer metabolism on a genome scale. Mol. Syst. Biol. 11, 817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mardinoglu A., et al., The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol. Syst. Biol. 11, 834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khodaee S., Asgari Y., Totonchi M., Karimi-Jafari M. H., iMM1865: A new reconstruction of mouse genome-scale metabolic model. Sci. Rep. 10, 6177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blais E. M., et al., Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nat. Commun. 8, 14250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bekaert M., Reconstruction of Danio rerio metabolic model accounting for subcellular compartmentalisation. PLoS One 7, e49903 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Steijn L., Verbeek F. J., Spaink H. P., Merks R. M. H., Predicting metabolism from gene expression in an improved whole-genome metabolic network model of Danio rerio. Zebrafish 16, 348–362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yilmaz L. S., Walhout A. J. M., A Caenorhabditis elegans genome-scale metabolic network model. Cell Syst. 2, 297–311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yilmaz L. S., et al., Modeling tissue-relevant Caenorhabditis elegans metabolism at network, pathway, reaction, and metabolite levels. Mol. Syst. Biol. 16, e9649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu H., et al., A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat. Commun. 10, 3586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bult C. J., et al., Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 47 (D1), D801–D806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin A., et al., FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 49, D899–D907 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruzicka L., et al., The Zebrafish Information Network: New support for non-coding genes, richer Gene Ontology annotations and the Alliance of Genome Resources. Nucleic Acids Res. 47, D867–D873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris T. W., et al., WormBase: A modern model organism information resource. Nucleic Acids Res. 48, D762–D767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agapite J., et al., Alliance of Genome Resources Portal: Unified model organism research platform. Nucleic Acids Res. 48, D650–D658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., et al., RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLOS Comput. Biol. 14, e1006541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anton M., ulfliebal, Wang H., MetabolicAtlas/Standard-GEM: Standard-GEM 0.5 (Version 0.5, Zenodo, 2021).
- 29.Nishikimi M., Kawai T., Yagi K., Guinea pigs possess a highly mutated gene for L-gulono-γ-lactone oxidase, the key enzyme for L-ascorbic acid biosynthesis missing in this species. J. Biol. Chem. 267, 21967–21972 (1992). [PubMed] [Google Scholar]
- 30.Moretti S., et al., MetaNetX/MNXref—Reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 44, D523–D526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schönborn J. W., Jehrke L., Mettler-Altmann T., Beller M., FlySilico: Flux balance modeling of Drosophila larval growth and resource allocation. Sci. Rep. 9, 17156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen W. H., Lu G., Chen X., Zhao X. M., Bork P., OGEE v2: An update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 45, D940–D944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matthews B. W., Comparison of the predicted and observed secondary structure of T4 phage lysozyme. BBA - Protein Struct. 405, 442–451 (1975). [DOI] [PubMed] [Google Scholar]
- 34.Lieven C., et al., MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 38, 272–276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ALZFORUM , AD research models. https://www.alzforum.org/research-models. Accessed 22 June 2020.
- 36.Nichols E., et al., Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 18, 88–106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agren R., et al., Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol. 10, 721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sjödin S., et al., Endo-lysosomal proteins and ubiquitin CSF concentrations in Alzheimer’s and Parkinson’s disease. Alzheimers Res. Ther. 11, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bekris L. M., Yu C. E., Bird T. D., Tsuang D. W., Genetics of Alzheimer disease. J. Geriatr. Psychiatry Neurol. 23, 213–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonsson T., et al., Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerreiro R., et al., TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368, 117–127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jay T. R., et al., TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212, 287–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verheijen J., Sleegers K., Understanding Alzheimer disease at the interface between genetics and transcriptomics. Trends Genet. 34, 434–447 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto A., Yue Z., Autophagy and its normal and pathogenic states in the brain. Annu. Rev. Neurosci. 37, 55–78 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Xiao Q., et al., Neuronal-targeted TFEB accelerates lysosomal degradation of app, reducing Aβ generation and amyloid plaque pathogenesis. J. Neurosci. 35, 12137–12151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frere S., Slutsky I., Alzheimer’s Disease: from firing instability to homeostasis network collapse. Neuron 97, 32–58 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Kolter T., Ganglioside Biochemistry. ISRN Biochem., 1–36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segler-Stahl K., Webster J. C., Brunngraber E. G., Changes in the concentration and composition of human brain gangliosides with aging. Gerontology 29, 161–168 (1983). [DOI] [PubMed] [Google Scholar]
- 49.Cachon-Gonzalez M. B., Zaccariotto E., Cox T. M., Genetics and therapies for GM2 gangliosidosis. Curr. Gene. Ther. 18, 68–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cermak S., et al., Loss of Cathepsin B and L leads to lysosomal dysfunction, NPC-Like cholesterol sequestration and accumulation of the key Alzheimer’s proteins. PLoS One 11, e0167428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGlinchey R. P., Lee J. C., Cysteine cathepsins are essential in lysosomal degradation of α-synuclein. Proc. Natl. Acad. Sci. U.S.A. 112, 9322–9327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whyte L. S., Lau A. A., Hemsley K. M., Hopwood J. J., Sargeant T. J., Endo-lysosomal and autophagic dysfunction: A driving factor in Alzheimer’s disease? J. Neurochem. 140, 703–717 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Dolgin E., How to defeat dementia. Nature 539, 156–158 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Olsson B., et al., CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: A systematic review and meta-analysis. Lancet Neurol. 15, 673–684 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Schwagerl A. L., et al., Elevated levels of the endosomal-lysosomal proteinase cathepsin D in cerebrospinal fluid in Alzheimer disease. J. Neurochem. 64, 443–446 (1995). [DOI] [PubMed] [Google Scholar]
- 56.Goetzl E. J., et al., Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 85, 40–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sánchez B. J., et al., Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEMs for Mouse1, Rat1, Zebrafish1, Fruitfly1, and Worm1 are available on GitHub at https://github.com/SysBioChalmers/Mouse-GEM, https://github.com/SysBioChalmers/Rat-GEM, https://github.com/SysBioChalmers/Zebrafish-GEM, https://github.com/SysBioChalmers/Fruitfly-GEM, and https://github.com/SysBioChalmers/Worm-GEM, respectively. The code for GEM development is available at https://github.com/SysBioChalmers/Human-GEM. The tissue- and cell-type-specific GEMs for transgenic mice and code for integrative analysis are available at https://github.com/SysBioChalmers/Mouse-GEM. All other data are available in supporting information.