Significance
Parasitic plants like witchweed cause huge losses in crop yield in Africa. A key part to the success of witchweed is to start its life cycle upon sensing small molecules called strigolactones, which are exuded from roots of host plants into the soil. Witchweed sense host-derived strigolactones through receptors called HTLs. It is thought that the evolutionary origin of HTLs is a receptor called KAI2 in nonparasitic plants, which can respond to different small molecules such as karrikins. By making three changes in the protein sequence of KAI2, this hybrid receptor can now sense both strigolactones and karrikins. These results help in understanding how receptors can evolve to sense different signals and can lead to solutions for combating pesky witchweed.
Keywords: receptor evolution, plant hormone, strigolactones, karrikins, plants
Abstract
Uncovering the basis of small-molecule hormone receptors’ evolution is paramount to a complete understanding of how protein structure drives function. In plants, hormone receptors for strigolactones are well suited to evolutionary inquiries because closely related homologs have different ligand preferences. More importantly, because of facile plant transgenic systems, receptors can be swapped and quickly assessed functionally in vivo. Here, we show that only three mutations are required to turn the nonstrigolactone receptor, KAI2, into a receptor that recognizes the plant hormone strigolactone. This modified receptor still retains its native function to perceive KAI2 ligands. Our directed evolution studies indicate that only a few keystone mutations are required to increase receptor promiscuity of KAI2, which may have implications for strigolactone receptor evolution in parasitic plants.
Plants and animals use small-molecule hormones to drive growth and development, and for this reason, considerable effort is placed on understanding how hormone receptors perceive their ligands (1, 2). Most inquiries are tuned to identify receptor amino acids that contribute to ligand specificity using “loss-of-function” approaches, in which an amino acid is replaced with a chemically inert moiety, usually an alanine, to assess the binding of its ligand (3–5). The less explored “gain-of-function” approach involves swapping amino acids between related receptors that respond to different ligands (6, 7). Gain-of-function approaches have the potential to not only answer questions about receptor–ligand relationships but also can give insights into how receptors evolve. As selection drives changes in the receptor sequence to recognize new ligands, important questions regarding this molecular process can be posed: Is this process gradually occurring through many amino acid changes or are only a few keystone substitutions required? When receptors evolve the ability to recognize new ligands, do they lose the ability to bind ancestral ligands? Are there amino acid changes that stabilize a receptor, permitting other mutations to accumulate, thereby generating multiple paths of receptor evolution? Gleaning this type of information through gain-of-function approaches, however, has challenges (8). First, amino acids are not discrete entities acting in isolation but are a sea of interdependent residues. As a result, swapping amino acids into a new context can often result in a nonfunctional protein because of negative epistasis. Secondly, depending on the number of potential swaps, the number of variants can quickly scale up, limiting analysis to simple in vitro assays rather than more complex functional in vivo analyses.
With this said, the perception of the plant hormone strigolactone (SL) may be a useful system to understanding the evolution of small-molecule receptors. The root parasite Striga hermonthica (Striga), for example, uses a group of α/β hydrolases designated HYPOSENSITIVE TO LIGHT (ShHTL) to perceive host-derived SLs, allowing the parasite to coordinate its germination with the lifecycle of a host plant (9). By contrast, the related ShHTL receptor, HYPOSENSITIVE TO LIGHT/KARRIKIN INSENSITIVE 2 (KAI2), which is also involved in germination of the model plant Arabidopsis thaliana (Arabidopsis), is not considered an SL receptor (10). Plants only produce SLs with a 2′R enantiomeric configuration of the D-ring, and this enantiomer is not recognized by KAI2 (SI Appendix, Fig. S1) (11, 12). In Arabidopsis, (2′R)-SLs are recognized by a more distantly related α/β hydrolase, DWARF14, a receptor that is involved primarily in vegetative growth (13, 14). KAI2 does respond to a collection of smoke-derived butenolides collectively called karrikins (KARs) and, strangely, is activated by nonnatural (2′S)-SL enantiomers (SI Appendix, Fig. S1) (10, 12, 15). This has led to suggestions that KAI2 receptors regulate germination by perceiving an unidentified, butenolide-based ligand, designated KAI2-ligand (KL) (16). Because parasitic lifestyles are derived, this implies that ShHTL parasitic receptors evolved the ability to perceive (2′R)-SLs from ancestor KAI2 receptors that bind KL.
Structural analysis of KL and SL receptors generally conclude that ShHTL receptors have larger binding pockets, allowing for greater choice to an array of (2′R)-SLs, a trait thought to increase Striga’s host range (9, 17, 18). Also, some changes, such as the substitution of leucine with phenylalanine at position 153 of ShHTL7, are thought to improve ShHTL interactions with downstream partners (19). These conclusions, however, are based solely on in vitro experiments because Striga is an experimentally intractable genetic system. Fortunately, ShHTL receptors function in Arabidopsis mutants deficient in KAI2 function and under specific conditions confer seed germination sensitivity to SLs (20). Furthermore, model plants like Arabidopsis, unlike their animal counterparts, are easily transformed, making high-throughput analysis at the whole-organism level feasible (21). This cross-species functionality means that homologous substitution mutagenesis between KL-based KAI2 and SL-based ShHTL receptors can be performed to probe evolutionary questions of ligand sensitivity, specificity, and promiscuity in an in vivo context.
Here, we systematically substituted key amino acids from the highly responsive SL receptor, ShHTL7, into the homologous positions in KAI2 and assayed their ability to sense SLs in vivo. We show, using both yeast- and plant-based assays, that only three amino acid changes were required to repurpose KAI2 into an SL receptor that recognizes (2′R)-SL enantiomers. The emergent properties of this chimeric KAI2 receptor does not appear to require the loss of KL perception. Finally, one ShHTL7 substitution appears to play a stabilizing role, which may allow other amino acid changes. Structural and molecular dynamic (MD) modeling indicates that the substitution of three active site amino acids from ShHTL7 into KAI2 produces a larger, more flexible binding pocket that better accommodates SLs including (2′R)-SL enantiomers, likely resulting in better interactions with downstream signaling partners. The short mutational path converting KAI2 into an SL receptor may have implications to why Striga coopted this regulator to be a germination sensor of host-derived SLs.
Results
Identifying Amino Acids Involved in SL Binding.
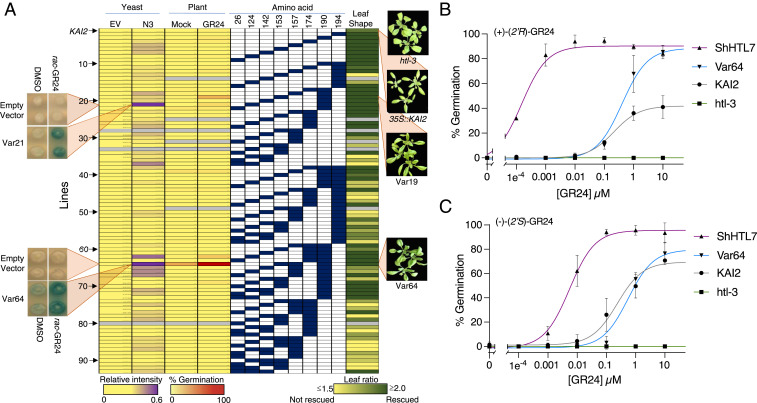
KAI2 and ShHTL receptors share β‐sheet cores flanked by α‐helices, with their ligand-binding pockets covered by a V-shaped cap (SI Appendix, Fig. S2A) (22–24). Within the binding pocket is a catalytic triad of amino acids that is involved in ligand hydrolysis (9, 25). To identify important amino acid differences between a KL and an SL receptor, we compared amino acid sequences within the ligand-binding pocket of KAI2 to the Striga SL receptor, ShHTL7. Since the larger ShHTL7-binding pocket can accommodate (2′R)-SLs, we focused on ShHTL7 residues that were less bulky and/or more polar than their homologous counterparts in KAI2 (SI Appendix, Fig. S2 B and C). Eight ShHTL7 residues (Tyr26, Leu124, Thr142, Leu153, Thr157, Tyr174, Thr190, and Cys194) were identified and substituted into the equivalent homologous position in KAI2 in all single, double, and triple combinations to generate 92 chimeric KAI2 gene variants (Fig. 1A).
Fig. 1.
Identification of KAI2 variants that perceives SL. (A) Var1 is the KAI2 control. (Columns 1 and 2) Increased purple shading represents an increased Y2H interaction intensity on rac-GR24 relative to the DMSO control for KAI2 chimeric variants queried against the N3 MAX2 fragment (N3) and empty vector control (EV) (SI Appendix, Figs. S3–S5). Representative colonies for Var27 (Tyr124Leu and Gly190Thr) and Var64 (Trp153Leu, Phe157Thr, and Gly190Thr) are shown in the Left Insets. (Columns 3 and 4) Mean germination percentages of three independent transgenics expressing individual KAI2 chimeric variants on PAC (20 µM) in the absence or presence of 1 µM rac-GR24. (Columns 5 to 12) Graphic representation of 93 KAI2 variants. Blue boxes represent the amino acid position in KAI2, where an ShHTL7 amino acid is substituted. (Column 13) Degree of rescue of the htl-3 leaf-shape phenotype by each KAI2 chimeric variant based on quantified leaf-shape ratios (SI Appendix, Figs. S7 and S8). Colors are from 0 (yellow) to 100% rescued (dark green). Gray indicates that the variant was not available. Representative images of htl-3, 35S::KAI2, Var19, and Var64 plants are shown to the right of the heatmap. (B) Effective concentrations of (+)-(2′R)-GR24 required to germinate 50% of htl-3, 35S::KAI2, 35S::ShHTL7, and Var64 transgenic seeds on 20 µM PAC. (C) Effective concentrations of(−)-(2′S)-GR24 required to germinate 50% of htl-3, 35S::KAI2, 35S::ShHTL7, and Var64 transgenic seeds on 20 µM PAC. All points represents three independent biological replicates. Bar = SD.
Three Substitutions Change KAI2 Responsiveness to SL.
To systematically test the SL responsiveness of 92 chimeric KAI2 receptors, we developed two in vivo high-throughput assays. KAI2 interacts with its signaling partner, MORE AXILLARY MERISTEMS 2 (MAX2), making this interaction a good candidate for a yeast two-hybrid (Y2H) assay (26). KAI2, however, autoactivates in Y2H assays in some conditions, and MAX2 is also unstable in yeast (17, 27). Therefore, we developed Y2H growth conditions in which KAI2 did not autoactivate (see Materials and Methods) and fragmented MAX2 into nine constructs (N1 to N9) to identify a fragment that produced a positive Y2H signal in the presence of an SL (SI Appendix, Fig. S3). Although wild-type KAI2 (Var1) and the chimeric receptor variants (Var2 to Var93) showed no or poor interactions with most MAX2 fragments, the N3 fragment interacted with some chimeric receptors to varying degrees, in the presence of a racemic mixture (2′R and 2′S) of the artificial SL, rac-GR24 (SI Appendix, Fig. S4A and Fig. 1A). Based on a cutoff of four SDs above the mean interaction intensity, we identified seven variants (Var21, Var37, Var62, Var64, Var65, Var66, and Var67) that showed rac-GR24–dependent interactions with the N3 fragment of MAX2 (SI Appendix, Fig. S4B). Six of these variants contained an ShHTL7 amino acid substitution at position 190, and four variants contained substitutions in positions 124 or 157, suggesting that these amino acids influence the rac-GR24–dependent interactions in yeast (SI Appendix, Fig. S4C). The identification of amino acids positioned at 124, 157, and 190 is consistent with in vitro studies that demonstrate the importance of these amino acids for SL binding and downstream partner recognition (17, 19).
Parallel to Y2H assays, we evaluated the in vivo function of 87 chimeric KAI2 receptors by transforming them into Arabidopsis deficient for KAI2 function (htl-3) (26). Using an SL-dependent germination assay involving GA-depleted seed, we found one variant, Var64 (Trp153Leu, Phe157Thr, and Gly190Thr), that germinated well upon rac-GR24 addition (Fig. 1A). Subsequent analysis of three independent transgenic lines (64A, 64B, and 64C) indicated that these lines have nanomolar sensitivity to rac-GR24 (SI Appendix, Fig. S6A). Moreover, Var64 transgenic lines all showed low-KAI2 expression compared to a KAI2-expressing line, indicating that the increased SL sensitivity of these lines was not simply due to increased KAI2 expression (SI Appendix, Fig. S6B). Finally, we found KUF1, a gene that is induced by both SL and KAR signaling, had increased expression in the Var64 line (SI Appendix, Fig. S6C).
The preferences of ShHTL7 and KAI2 for enantiomeric forms of SLs suggested that we should test Var64 lines for GR24 enantiomeric specificity. ShHTL7 receptors responded to both isomers but preferred (+)-(2′R)-GR24 by two orders of magnitude. This flexibility most likely reflects the large binding pocket of this parasitic receptor (18). By contrast, KAI2 receptors favor (−)-(2′S)-GR24, and in our experiments, the germination of Arabidopsis seed harboring these receptors reflected these preferences (Fig. 1B). Var64 seed germination showed similar KAI2 sensitivity to (−)-(2′S)-GR24, but this variant also displayed a marked increase in germination in response to (+)-(2′R)-GR24 (Fig. 1B). Thus, substituting three KAI2 amino acids (153, 157, and 190) with their ShHTL7 counterparts relaxes receptor specificity to now recognize (+)-(2′R)-GR24 without loss of (−)-(2′S)-GR24 recognition. Notably, Var64 was identified by both our yeast- and plant-based SL assays. This convergence suggests that doing less time-consuming yeast interaction assays is an ideal preliminary screen to guide more laborious plant-based assays.
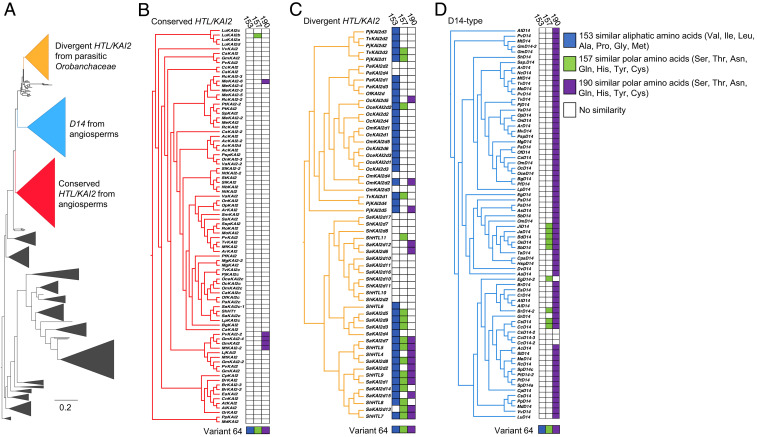
The identification of Leu153, Thr157, and Thr190 as having roles in SL perception encouraged us to determine the prevalence of structurally related amino acids to these positions in KAI2 proteins across a large collection of land plants (Fig. 2A) (28). We found that clades of receptors not expected to bind SLs (conserved) were mostly devoid of ShHTL7-related amino acids (Fig. 2B). By contrast, clades of receptors that were expected to bind SLs (divergent) were highly populated with ShHTL7-related residues (Fig. 2C). Divergent KAI2 proteins were particularly enriched for Leu153 or hydrophobic amino acids at this position, and proteins more closely related to ShHTL7 possessed similar amino acids at positions Thr157 and Thr190. Within the divergent group, the lone clade of proteins depleted of ShHTL7 amino acid identities were most closely related to receptors ShHTL10 and ShHTL11 (e.g., SaKAI2d2, SaKAI2d10, SaKAI2d11, SaKAI2d16, ShKAI2d10, and ShKAI2d11), which do not have a role in SL-dependent germination, as assayed in Arabidopsis (21). Interestingly, applying the same analysis to D14-type SL receptors that have vegetative roles in plant development found that these receptors were mostly enriched for Thr190 conservation (Fig. 2D). Together, the distribution of ShHTL7 amino acids across land plants suggests that our in vivo screening identified important amino acids involved in turning KAI2 to an SL receptor.
Fig. 2.
Conservation of Var64 amino acids in land plants. (A) Phylogenetic tree built using KAI2/DLK/DDK/D14 sequences from land plants and rooted using KAI2 sequences from hornworts. Sequences within each of the three colored clades were investigated in more detail for their similarity with variant 64 at positions 153, 157, and 190. (B) KAI2 divergent sequences from parasitic Orobanchaceae. (C) Conserved KAI2 sequences from angiosperms. (D) Conserved D14 sequences from angiosperms. Colored boxes beside the clades indicate amino acid similarity to its counterpart in Var64 at positions 153 (blue), 157 (green), and 190 (purple).
Var64 Binds and Hydrolyses Naturally Occurring (2′R) SL Enantiomers.
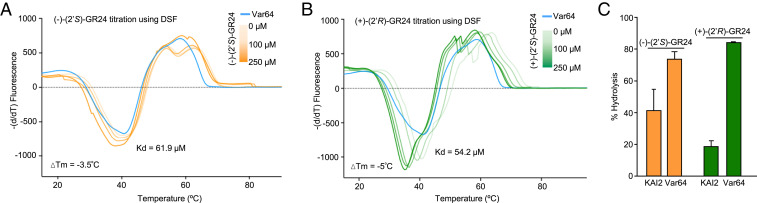
To biochemically characterize Var64, we performed differential scanning fluorimetry (DSF), which monitors receptor–ligand interactions by shifts in protein melting temperatures (Tm). Previous DSF analysis of KAI2 showed that Tm decreases and fluorescence broadens with increasing (−)-(2′R)-GR24 concentrations, but these shifts were not detected in the presence of (+)-(2′R)-GR24 (29). We observed decreasing Tm shifts with Var64 protein in the presence of increasing (−)-(2′R)-GR24, and these shifts allowed us to calculate an SL binding Kd of 61.9 µM (Fig. 3A). In contrast to KAI2, (+)-(2′R)-GR24 also resulted in decreasing Tm shifts in Var64 protein, resulting in a Kd of 54.2 µM (Fig. 3B). Next, we directly measured receptor hydrolase activity by monitoring intact GR24 depletion in the presence of either enantiomer. KAI2 protein hydrolyzed (−)-(2′S)-GR24, but under our conditions, this receptor showed some ability to also hydrolyze (+)-(2′R)-GR24 (Fig. 3C). Var64 protein also hydrolyzed (−)-(2′S)-GR24 and, more importantly, hydrolyzed (+)-(2′R)-GR24 at a rate four times higher than KAI2 (Fig. 3C). In summary, consistent with our germination data, Var64 binds and hydrolyzes both GR24 enantiomers, with a preference for (+)-(2′R)-GR24.
Fig. 3.
Var64 binds both (2′S)- and (2′R)-GR24 enantiomers (A and B), Tm curves Var64 protein preincubated with (−)-(2′S)-GR24 or (+)-(2′R)-GR24. A first derivative of the change in fluorescence due to gradual protein melting was plotted against Tm. These curves were used to calculate inflection points of fluorescence versus Tm curves, as well as Kd values. Each line represents the average protein melt curve for three replicate samples. (C) Quantification of hydrolysis of GR24 enantiomers by liquid chromatography–mass spectrometry (LC-MS). The rate of hydrolysis was monitored in triplicate by production of the ABC-ring–hydrolyzed product by LC-MS.
Var64 Responds to KARs.
Since the identity of KL is unknown, direct assessment of its binding to variant receptors cannot be performed. In Arabidopsis, however, kai2 leaves show shape defects thus rescue of this mutant phenotype by variant KAI2 receptors is an indirect readout of endogenous KL binding (30). Under our growth conditions, length-to-width ratios of htl-3 rosette leaves were less than wild type (≤1.85 versus ≥1.92), and this difference was used to quantify the degree to which chimeric variants complement htl-3 leaf-shape defects (SI Appendix, Fig. S7). Generally, higher-order mutant lines showed less rescue of the htl-3 leaf phenotypes than lower-order mutants, which could reflect a loss of endogenous KL recognition (Fig. 1A). This pattern, however, may also reflect increasing negative epistasis, as newly introduced amino acids pile up, resulting in nonfunctional proteins (8). We noticed, however, that triple substitution variant lines with an ShHTL7 amino acid at position 190 complemented htl-3 leaf defects more frequently, compared to other triple mutants or even double mutants lacking this substitution (Fig. 1A). Related to this, Thr190 was found in six of the seven constructs that allowed positive interactions in our Y2H assay (SI Appendix, Fig. S4). Deep sequencing analysis suggests that some amino acid substitutions can be neither positive nor negative in terms of protein function but simply stabilize proteins to allow for a larger number of substitutions to be tolerated during evolution (31). Perhaps, threonine at position 190 permits alternative mutational paths for ligand recognition. In this context, the fact that Thr190 was highly conserved in D14 clades may mean that this amino acid stabilized substitutions that encouraged SL binding to this receptor (Fig. 2D). With this said, Var64 lines, which contain a Thr190 substitution, fully complemented htl-3 leaf defects, suggesting that this chimeric receptor retained an ability to recognize KL (Fig. 1C). Consistent with this, GA-depleted Var64 seed germinated on KAR2 to similar levels seen in KAI2 seed, again indicating that this chimeric variant retains a capacity to recognize natural KAI2 ligands (SI Appendix, Fig. S9).
Var64 Structural Analysis.
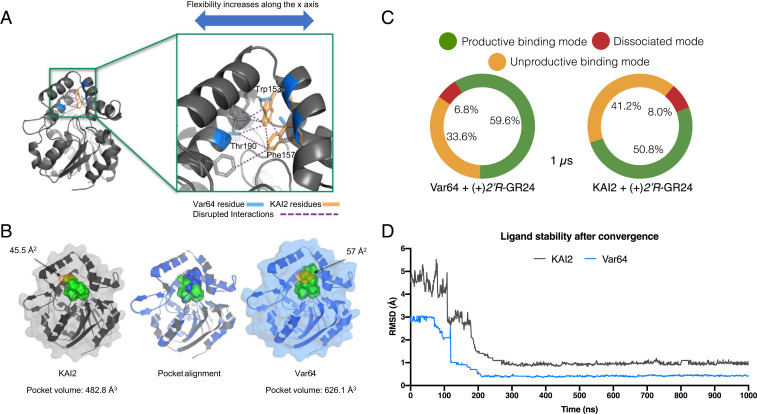
Structural comparisons of a Var64 protein model to a KAI2 crystallographic structure (Protein Data Bank [PDB] ID: 4IH1) suggests that replacing KAI2 aromatic residues Trp153 and Phe157 with Leu and Thr disrupted several hydrogen bond interactions that would occur between the α-B, α-C, and α-D helices of the lid domain (Fig. 4A). This change increased the pocket mouth area from 45.5 to 57 Å2, which would allow for larger substrates to access the receptor active site. These disruptions also increased the overall pocket solvent–accessible volume by ∼140 Å3 (Fig. 4 A and B). Larger binding pockets would increase ligand accessibility, and to explore this, we modeled (+)-(2′R)-GR24 and (−)-(2′S)-GR24 ligand–receptor interactions, quantifying the number of interactions generated between KAI2, Var64, and ShHTL7 with their ligands. ShHTL7 and Var64 showed 11 amino acid interactions with (+)-(2′R)-GR24, with a 64% overlap of interacting residues (SI Appendix, Fig. S10). By contrast, ShHTL7, KAI2, and Var64 docked with (−)-(2′S)-GR24–presented interactions with 7 to 8 residues, with KAI2 showing only a 45% overlap with ShHTL7 (SI Appendix, Fig. S10). These results explain, at least partially, why ShHTL7 and Var64 have an increased affinity for (+)-(2′R)-GR24 versus KAI2. In summary, substituting Leu153, Thr157, and Thr190 into KAI2 would, in principle, relax the lid covering the active site. Increasing lid flexibility would translate into increased binding pocket volume and could allow better interaction of the receptor with downstream partners (19). The recent in vitro demonstration that Thr157 dramatically increases the affinity of the ShHTL7 receptor for MAX2 is consistent with this view.
Fig. 4.
Var64-derived amino acids create an SL-binding domain. (A) The substitution of KAI2 aromatic residues for ShHTL7 equivalents results in the disruption of several interactions (purple dashed lines) along the x-axis of the protein. This disruption translates into increased motility of the lid domain (Movie S1), consistent with the receptor’s new ability to accommodate SL molecules. (B) Pocket alignment of KAI2 and Var64 receptors reveals an 11.5 Å larger pocket mouth (orange shading) and a 29.7% increased pocket volume in the Var64 receptor. The pocket mouths are shown in orange shading, with width shown in Å2. (C) A summary of the three most common binding states between Var64, KAI2, and 2′R-GR24. Binding modes of ligand–protein complexes were determined by 1-µs MD simulations of Var64 and KAI2 receptors. The Var64–(2′R)-GR24 complex was found in a productive binding mode 59.6% of the time, while the same state was found only 50% of the time for the KAI2 complex. (D) Calculation of the magnitude change in the pose of the ligand throughout the MD simulation. Both proteins stabilized the ligand after 0.1-µs simulation, but its position on the KAI2 pocket is compromised briefly before final convergence, as the interaction is less stable.
Var64 Receptor Dynamics.
Unlike traditional structural docking analysis, which give static snapshots of protein structure, MD simulations take into account the temporal evolution and velocities of all of the atoms of a protein, averaged over a specific period of time, to give a more realistic, atomic-level detail of the protein. MD simulations of Var64 and KAI2 receptors in both the absence and presence of (+)-(2′R)-GR24 were conducted in triplicate 1-µs simulations to evaluate the dynamics of apoprotein– and ligand–receptor complexes. Using the top binding pose, based on the lowest Gibbs free energy provided by two docking engines, the fluctuation of the ligand positions throughout the simulation were measured, particularly after convergence was reached (SI Appendix, Tables S1 and S2). We quantitatively distinguish between three ligand-binding states, based on the length range of any bonds produced between KAI2 homologs and a GR24 molecule (SI Appendix, Table S3). A productive mode was defined by the close proximity (≤5.0 Å) of the A-ring moiety of (+)-(2′R)-GR24 to the receptor, with its surface area mostly covering the active site of the protein. An unproductive mode was where the ligand is further away from the pocket opening (5.1 to 7.0 Å) and not interacting with the active site. And finally, a dissociated mode was where the ligand is far from the protein (≥7.5 Å), and interactions were very unlikely. Using these criteria, (+)-(2′R)-GR24 was in a productive binding mode with the Var64 receptor in almost 60% (±2.1%) of the simulation time versus only 50% (±1.8%) for KAI2 (Fig. 4C). Furthermore, when docked on KAI2, (+)-(2′R)-GR24 spent ∼8% (±3.3%) of the simulation time in a dissociated mode versus 6.8% (±2.9%) for Var64. Finally, we calculated the magnitude of location change of the position of (+)-(2′R)-GR24 on the two receptors using the rms module. For a productive ligand–receptor interaction, minimal fluctuation is expected, as the ligand is tightly bound until it is metabolized or released. The simulations showed that the position of (+)-(2′R)-GR24 in KAI2 fluctuates between 2.8 and 5.6 Å (average fluctuation using three replicates), once initially bound to the protein (Fig. 4D). By contrast, this oscillation is greatly reduced in the Var64 protein, confirming the ability of the new variant to form a stable interaction with an SL enantiomer (Fig. 4D).
Visualization of the MD simulations revealed that the motility of the α-E loop of unbound Var64 is increased, consistent with increased flexibility of the receptor’s active site and the expansion of the cavity depth and volume (Movie S1). The trajectories also show three main changes between bound and unbound receptors. First, hydrophobicity of the binding domain of Var64 was slightly reduced throughout the simulation after binding (+)-(2′R)-GR24. Second, (+)-(2′R)-GR24 appeared to be more attracted by the polar ShHTL7 Thr190 in Var64 than the KAI2 Gly190. This substitution would explain the higher ligand stability provided by Var64. Lastly, a conformational change of the α-D helix of Var64 upon (+)-(2′R)-GR24 binding was detected, and previous studies have suggested conformation changes in α-D helix that encourage interactions with downstream partners (Movie S2).
Interestingly, Var19 (Trp153Leu and Gly190Thr), which contains a subset of Var64 amino acids, showed the next highest level of rac-GR24–dependent germination and also showed the rescue of the htl-3 leaf-shape phenotype (Fig. 1C and SI Appendix, Fig. S11). We performed MD analysis (0.5-µs simulations) on Var19 protein, and trajectories showed a slightly decreased flexibility of the lid domain across the horizontal plane (Movie S3). This is because the Phe157 amino acid of Var19 formed at least three additional interactions with Thr190 and neighboring residues. Consequently, Var19 pocket mouth area was 51 Å2, which is ∼6 Å smaller than that of Var64 (Movie S3). Var19 and Var64 lines differ only at position 157, and in vitro analysis suggests that in ShHTL7 this amino acid is key for MAX2 partner interactions, resulting in hypersensitivity to SL (19). Furthermore, studies in Lotus indicate that this position may also be important in the differentiation of GR24 2′-enantiomers by KAI2 (32). Whatever the case, these results again are consistent with receptor flexibility being a key component in the SL perception and their interaction with signaling partners (33, 34).
Discussion
Structure-guided mutational analysis is an effective approach to dissecting how a protein’s sequence contributes to biochemical activity but elucidation of how these biochemical properties evolve is more experimentally challenging (8). From an evolutionary perspective, loss-of-function approaches are not necessarily informative, and often experiments on protein evolution are not performed in an in vivo context, which allows proteins to interact with other cellular constituents and processes. In this study, we used a gain-of-function approach with active site amino acid–targeted substitutions, informed by structural and evolutionary considerations, to determine if a nonparasitic plant orphan receptor, KAI2, that is incapable of recognizing natural SL ligands harnessed by parasitic plants, could be converted into a receptor that recognizes such ligands. Importantly, we assayed these receptor variants using yeast- and Arabidopsis-based systems to evaluate receptor function under in vivo conditions.
Our approach unambiguously determined that only three amino acid substitutions were necessary to convert KAI2 into a functional SL receptor. By introducing smaller, more polar amino acids into KAI2, we produced a protein with a more flexible pocket and lid, which affects receptor function in two ways. First, it increases the pocket volume to accommodate natural SL conformations, like that of (2′R)-GR24. Second, it increases the overall elasticity of the protein, which may allow better interactions with signaling partners (19, 33, 34). Our in vivo findings also contribute to discussions concerning the role of gradual versus keystone mutations in small-molecule receptor evolution. Animal studies using ancestral gene reconstructions involving steroid receptors show that as few as two mutations can switch ligand specificity (35). Our results now extend the importance of keystone mutations to plant hormone receptor evolution.
General models of hormone evolution suggest that promiscuous ancestral forms evolve specificity so as to prevent deleterious ligand cross-talk (36). Three lines of evidence in this study, however, suggest that a path to SL perception in KAI2 does not necessarily require the loss of responsiveness to previous ligands. First, our variant receptor complements the KAI2 loss-of-function phenotype, suggesting it still recognizes KL. Second, variant receptor seeds are sensitive to (2′S)-GR24 enantiomers, a known substrate for KAI2. Finally, variant lines respond to the KARs. The ability of variant receptors to respond to both ancestral as well as derived ligands supports the notion that SL perception can evolve through minimal specificity rather than by a narrowing of specificity. Minimal specificity model receptors as being adequate but not overly so to distinguish endogenous ligands (37). This “just enough” specificity explains why receptors often show sensitivity to drug structures unrelated to their natural ligands. The femtomolar responsiveness of ShHTL7 to an artificial agonist with little similarity to SLs supports a sufficient promiscuity model (38).
SL hydrolysis is maintained in Var64 protein, supporting a connection between hydrolysis and signaling. The directed evolution to create Var64, however, most likely does not reflect the natural progression by which KAI2 genes evolve. Our KAI2 variants were not under the constraints seen in natural environments that are important for fitness. We understand that monitoring KAI2 function using the output of Arabidopsis germination is superior to in vitro based experiments but even our system certainly misrepresents the frequency of beneficial mutations relative to natural evolution. With this said, the occurrence of amino acids biochemically similar to the ShHTL7 residues at the 153, 157, and 190 positions within divergent KAI2 receptors suggests that these residues may play roles in SL perception. Finally, directed protein-engineering studies tell us that enzymes usually possess minor nonselective activities that are often enhanced by laboratory evolution (39, 40). If true, KAI2 may possess a to-date, undetected low level of responsiveness to 2′R enantiomers. In vitro, KAI2 appears to bind (2′S)-GR24, but in vivo, it can perceive (2′R)-GR24 only when the SL receptor, D14, is defective (12, 41). Possibly, the absence of D14 uncovers the latent binding ability of KAI2 for 2′R enantiomers, as it is free from competition for (2′R)-SL substrates.
The role of KAI2, in germination together with its ability to perceive small molecules, makes it easy to understand why this gene was coopted during the evolution of Striga parasitism. Obligate parasites like Striga not only require a cue to signal proximity to a host but a long-term survival strategy in the absence of a host. Seeds are an attractive solution to establishing long-term viability in the absence of a host, as dormant seeds can survive long periods under a variety of environmental conditions. An unintended consequence of using dormant seeds as the protective state in the absence of a host is that they easily contaminate soil and are difficult to purge from farmer’s fields. For these reasons, Striga has contaminated over 20 African nations and is the largest biological impediment to food security on the continent (42). Understanding the steps by which KAI2 evolved to become an SL receptor will give insights not only into the role of this key regulator of the Striga lifecycle but will also help define the chemical space of this receptor, which should open leads to chemical solutions to this African scourge.
Materials and Methods
Germination Assays.
Germination assays were performed on 1/2 Murashige and Skoog (MS) minimal agar medium. Stocks of rac-GR24 and GA3 were prepared in DMSO, and paclobutrazol (PAC) was prepared in ethanol. Each stock was diluted 1:1,000 in 1/2 MS to a final solvent percentage of 0.1 (volume/volume). For germination assays, the seeds were at least 1 mo old. All seeds were surface sterilized with 70% (volume/volume) ethanol. For germination assays, ∼50 seeds were used per assay. After seeds were plated on agar, they were stratified for 4 d at 4 °C and then placed under continuous white light at 25 to 26 °C. After 7 d under continuous white light, germination was scored using radicle emergence. GR24 dose–response curves and effective concentration values were generated with SigmaPlot 11.0.
Y2H Assays.
The constructs encoding AtKAI2 chimeric variants were synthesized and cloned into pENTR SD-D-TOPO vectors (Invitrogen) by the US Department of Energy’s (DOE) Joint Genomics Institute. Y2H experiments were performed as published (43). Constructs for the 90 variants, representing all 1-, 2-, and 3-substitution variants, were cloned into pEG202 vectors for expression in the “bait” orientation, consisting of an N-terminal fusion to the LexA DNA-binding domain using Invitrogen Gateway LR Clonase II. LR reaction products were transformed into Escherichia coli DH5-α cells. Expression vectors were transformed into RFY206 A-type mating strain yeast, containing the pSH18-34 vector and the reporter gene LacZ. Full-length MAX2 and MAX2 fragments were in the prey vector, pJG4-5, and transformed into yeast strain, EGY48. Each query was tested on reporter plates containing 0.1% (volume/volume) DMSO, as well as plates containing 50 µM racemic mixture of rac-GR24. Plates were incubated at 30 °C and scanned at 3 d.
Quantification of Interaction Strengths.
Scanned images of reporter plates at 72 h postpinning were quantified using ImageJ software (44). Colonies were highlighted using the ellipse selection tool, and mean gray values for colonies were used to determine colony shading intensity for each query on DMSO and rac-GR24. Two colonies were scored, and the mean was calculated for the final interaction strength for each variant-MAX2. Mean gray values on SL conditions for each variant were subtracted from those recorded for DMSO conditions, and the resulting values were log2 transformed. These log2-fold changes in interaction strength between the rac-GR24 and DMSO treatments were used as a measure of the SL dependence of each interaction.
Identification of Homozygous KAI2 Variant Overexpression Lines.
KAI2 chimeric variant constructs were cloned into pGWB612 vectors, and Agrobacterium-mediated transformation was used to transform the constructs into Arabidopsis htl-3 lines. Homozygous lines from three independent insertion events were identified. In the event that no homozygote lines were identified for a given insertion event, the line showing the highest proportion of resistant seedlings was chosen as a representative line. It was not possible to make even variant lines. SL-dependent germination assays were performed as published (20).
Quantification of Leaf Shape.
Ellipses were used to approximate leave shape using ImageJ, and major axis/minor axis ratio of the ellipse represented the narrowness of the leaf. Three independent transformed lines of 2-wk, two-rosette leaf, 6 to 8, were used. We used normal distribution to model the distribution of major/minor ratio among the natural population of wild type and htl-3 using data from controls to graph the curve. The probability density function of a normal distribution is given as , in which μ and σ are the mean and SD of the population. Sample mean (x) and SD (s) from controls were used to approximate that of the corresponding population.
Phylogenetic Reconstruction and Amino Acid Analysis.
A total of 568 sequences (coding sequence; CDS), encoding members of the D14/HTL/KAI2/DDK -hydrolase superfamily, were used. Duplicate sequences were identified and removed by a combination of basic local alignment search tool analyses (BLAST) and manual inspection. To simplify the phylogenetic tree, charophyte and lycophyte sequences were also removed, leaving a final dataset of 472 nonredundant CDSs that were aligned using multiple sequence comparison by log-expectation (MUSCLE) version v3.8.31. The alignment was trimmed using Gblocks version 0.91b with the following parameters: −t = d, −b3 = 10, −b4 = 3, −b5 = h, −d = y, which retained 700 phylogenetically informative positions and was used to build a phylogenetic tree using randomized axelerated maximum likelihood (version 8) according to the general time reversible model (GTR+GAMMA+I) with the rapid bootstrap option set and 200 bootstrap replicates. The tree was rooted with five sequences from hornworts (45). The tree was inspected and nodes collapsed using FigTree (version 1.4.4). Three clades of sequences were identified for further analysis: divergent HTL/KAI2d from parasitic Orobanchaceae, conserved HTLs/KAI2 from angiosperms, and D14 proteins from angiosperms. For each of these clades, the CDSs were extracted, translated to their corresponding amino acid sequences, and aligned to the KAI2 sequence from A. thaliana (AT4G37470.1) and variant 64 using MUSCLE. The amino acid alignments for each clade of sequences were then parsed using a custom Python script to determine the amino acid similarity of each sequence at amino acid positions 153, 157, and 190 of variant 64.
Protein Expression and Purification.
Protein CDSs for Var64 (codon optimized) and KAI2 were cloned into pMCSG53 and pET28-SBP-TEV, respectively, and transformed into BL21-Gold (DE3). Protein expression was induced using 1 mM IPTG shaken at 200 rpm overnight at 17 °C. Both proteins were purified using a Ni-affinity column and by fast protein liquid chromatography against a final elution buffer consisting of 0.15 M NaCl, 10 mM HEPES pH 7.0, and 1 mM TCEP. Fresh protein preps were used for subsequent biochemical assays.
Plant Total Protein Extraction and Western Blot Analysis.
Approximately 50 seedlings were flash frozen, beaten with glass beads, and dissolved in 60 µL distilled water. A total of 20 µg protein was separated in a 15% SDS-polyacrylamide gel electrophoresis gel and blotted onto a PVDF membrane. Primary antibody (anti-FLAG produced in mouse, Sigma) was added (1:10,000) in 3% skim milk and incubated for 30 min at room temperature. After five 3-min washes with phosphate-buffered saline with Tween, the membrane was incubated with 1:10,000 of secondary antibody (anti-mouse IgG HRP-linked anti-FLAG, Cell Signaling Technology) for 1 h at room temperature. The membrane was incubated for 10 min with Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific).
GR24 Hydrolysis Quantification by Liquid Chromatography–Mass Spectrometry.
A total of 5 µM of either Var64 or KAI2 was incubated in triplicate with 10 µg (+)2′R GR24 or (−)2′S GR24 at room temperature for 12 h. After incubation, the reactions were quenched with 2 µL trifluoroacetic acid to denature the protein. The reactions were then centrifuged at maximum speed for 5 min at 4 °C. Liquid chromatography–mass spectrometry analysis of GR24 and the potential hydrolysis products was performed using a Dionex Ultimate 3000 ultra-high performance liquid chromatography system and a Q-Exactive mass spectrometer equipped with a heated-electrospray ionization II probe from Thermo Scientific and controlled by Thermo XCalibur 4.1 software. LC separation was conducted on a Hypersil Gold C18 column (50 × 2.1 mm, 1.9-µm particle size, Thermo Scientific), equipped with a guard column, at 40 °C. Solvent A was 0.1% formic acid (in water), and solvent B was 0.1% formic acid (in methanol) at a flowrate of 0.3 mL/min. Autosampler temperature was maintained at 8 °C, and injection volume was 10 µL. The gradient was 0 to 1 min: 95% A; 1 to 7 min: linear gradient to 2% A, 98% B; 10 to 10.5 min: 2% A, 98% B; and 10.5 to 15 min: 95% A. Data collection was done in positive and negative ionization mode with a scan range m/z 100 to 1,500, resolution 140,000 at 1 Hz, AGC target of 3e6 ions, and a maximum injection time of 250 ms.
DSF Titrations.
DSF experiments were performed on a BioRad CFX96 Real-Time PCR Detection System using the fluorescence resonance energy transfer channel for an excitation (450 to 490 nm) and emission (560 to 580 nm) wavelengths. Samples were prepared in triplicate using 5 µg protein and increasing concentrations of ligand in a buffer containing 10 mM Tris Cl pH 6.8 and 200 mM NaCl. Following a 30-min incubation with the ligand, 0.5 mM Sypro Orange was added and used as the reporter dye. After a 5-min equilibration at 5 °C, samples were heat denatured using a linear 5 to 95 °C gradient at a rate of 0.5 °C with 20-s intervals. Protein unfolding was monitored by detecting changes in Sypro Orange fluorescence. The inflection points of fluorescence versus temperature curves, as well as Kd values, were determined using the DSF fitting program (46).
Protein Structural Analyses and Docking.
The structures of Var64 and Var19 were modeled using the Phyre2 web portal for protein modeling, based on the crystallographic structure of KAI2 (PDB ID: 4IH1). All static structures were analyzed using PyMOL for educational use version 2.4.0 and the University of California, San Francisco (UCSF) Chimera version 1.14. Protein cavities and mouths were measured using CAVER Analyst 2.0. The docking of GR24 enantiomers was performed using SwissDock and AutoDock. The top binding pose was selected based on the lowest Gibbs free energy and inspected using Chimera viewer to verify that no clashes with the protein would be generated. Minimal differences were observed between the most energetically favored binding states obtained by the two docking engines, so further analysis was carried out with the SwissDock models, as these reflected the lowest free energy values on average. To further validate docking results, a structural alignment of the docked complexes and the crystallographic structure of rice D14-GR24 (PDB ID:5DJ5) was performed, showing minimal deviations on the pose of the docked ligands (quantified in rmsd Å), which confirms that the selected binding poses were adequate (SI Appendix, Table S1). Individual amino acid–ligand contacts were analyzed using LigPlot.
MD Simulations.
MD simulations for the KAI2–Var64 complexes with (+)2′R-GR24 were performed in triplicate 1-µs cycles (0.5 µs for Var19) using the Groningen machine for chemical simulations (GROMACS) (version 4.6.1) simulation package (47). The starting coordinates used were obtained from the crystallographic structure of KAI2 (PDB ID: 4IH1) and the models described in Protein Structural Analyses and Docking. All crystallographic water molecules were removed from the protein file. The substrate (GR24) was placed in the active site, as described in Protein Structural Analyses and Docking, and validated using the crystallographic structure of D14-GR24. The force field used was CHARMM36 (48). Amino acids were assumed to be in their standard protonation state at pH 7.0. Hydrogens were added to the .pdb files using the molecule editor Avogadro (49). Ligand topologies were generated using the CGenFF server and validated by the server’s penalty score system to ensure that atom types, bond connectivity, and charges were all below a penalty score of 10. The structures were solvated using the pdb2gmx and solvate modules (transferable intermolecular potential with three points water for solvent configuration) and placed in the center of a dodecahedral unit cell (dimensions = 8.51 × 8.51 × 6.31 Å), at least 1 nm from the box edge, filled with Simple Point Charge water molecules extending to at least 8 Å from the complex atoms. The solvated systems were neutralized by adding Na+ and Cl− ions using the genion module.
The systems were initially minimized using 1,000 steps steepest descent, followed by conjugate gradient minimization, for a total maximum number of minimization steps of 50,000. To obtain the NVT ensembles, the minimized systems were heated from 100 to 300 K over 0.04 ns and then for another 0.06 ns for Tm equilibration; Tm was maintained using the Velocity Rescale Thermostat. To obtain NPT ensembles, the system was equilibrated for 0.1 ns at constant pressure using the modified Berendsen barostat for pressure coupling with isotropic position scaling, with a reference pressure of 1 bar. Continuing from NPT, the equilibrated systems were exposed to a fully unrestrained production simulation of 1 µs (0.5 µs for Var19). All H-bonds were constrained with the linear constraint solver constraint algorithm; long-range electrostatics were calculated with Particle Mesh Ewald using a timestep of 2 fs throughout the simulation. Finally, the molecules’ coordinates were recentered within the unit cell. The number and distance of hydrogen bonds were calculated using the H-bond module, while the rms module was used to quantify the location change of the ligand. Small differences between each simulation replicate were quantified (SI Appendix, Table S2); the average rmsd of the ligand after convergence and the average potential energy at the end of each simulation replicate did not vary significantly.
All simulations were analyzed using the modules mentioned in this section. To determine the gradient of potential interactions, the structures of KAI2 homologs in complex with SL molecules (intact, intermediate, or antagonist) were analyzed with the Protein–Ligand Interaction Profiler web tool to quantify the length of any bonds produced between protein and SL (50). In summary, the length for all bonds was found to be in the range of 2.32 to 5.41 Å (SI Appendix, Table S3).
The trajectories of the replicates with the lowest potential energy for each simulation were visualized using the Visual MD package (version 1.9.4) using PyMOL, the UCSF Chimera package, and Final Cut Pro (version 10.5.1 for macOS Catalina) for graphics production (51). All simulations were carried out in the Cedar compute and Cedar graphics processing unit servers of the Compute Canada Database.
Supplementary Material
Acknowledgments
We thank the US DOE Joint Genome Institute DNA Synthesis program for the synthesis of KAI2 variants used in this study. The work conducted by the US DOE Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract DE-AC02-05CH11231. This research was enabled in part by support provided by Compute Ontario (https://www.computeontario.ca/) and Compute Canada (http://www.computecanada.ca). A.A.-S. was partially funded by the Mexican National Council of Science and Technology and by a Mitacs Globalink Graduate Scholarship. This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (06752), an NSERC Accelerator Supplement (507992), an NSERC Research Tools and Instruments Grant (00356), a New Frontiers in Research Fund (NFRFE-2018-00118) and a France–Canada Research Fund awarded to S.L., and an NSERC Discovery Grant (04298) awarded to P.M.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103175118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Evans R. M., Mangelsdorf D. J., Nuclear receptors, RXR, and the Big Bang. Cell 157, 255–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lumba S., Cutler S., McCourt P., Plant nuclear hormone receptors: A role for small molecules in protein-protein interactions. Annu. Rev. Cell Dev. Biol. 26, 445–469 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Morrison K. L., Weiss G. A., Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 5, 302–307 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Howlader M. T. H., et al., Alanine scanning analyses of the three major loops in domain II of Bacillus thuringiensis mosquitocidal toxin Cry4Aa. Appl. Environ. Microbiol. 76, 860–865 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao L.-H., et al., Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25, 1219–1236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobilka B. K., et al., Chimeric alpha 2-,beta 2-adrenergic receptors: Delineation of domains involved in effector coupling and ligand binding specificity. Science 240, 1310–1316 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Cunningham B. C., Jhurani P., Ng P., Wells J. A., Receptor and antibody epitopes in human growth hormone identified by homolog-scanning mutagenesis. Science 243, 1330–1336 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Hochberg G. K. A., Thornton J. W., Reconstructing ancient proteins to understand the causes of structure and function. Annu. Rev. Biophys. 46, 247–269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuchiya Y., et al., Parasitic plants. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Scaffidi A., et al., Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165, 1221–1232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneyama K., et al., Which are the major players, canonical or non-canonical strigolactones? J. Exp. Bot. 69, 2231–2239 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Flematti G. R., Scaffidi A., Waters M. T., Smith S. M., Stereospecificity in strigolactone biosynthesis and perception. Planta 243, 1361–1373 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Obando M., Ligerot Y., Bonhomme S., Boyer F.-D., Rameau C., Strigolactone biosynthesis and signaling in plant development. Development 142, 3615–3619 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Waldie T., McCulloch H., Leyser O., Strigolactones and the control of plant development: Lessons from shoot branching. Plant J. 79, 607–622 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Waters M. T., Scaffidi A., Flematti G. R., Smith S. M., The origins and mechanisms of karrikin signalling. Curr. Opin. Plant Biol. 16, 667–673 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Conn C. E., Nelson D. C., Evidence that KARRIKIN-INSENSITIVE2 (KAI2) Receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 6, 1219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y., et al., Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat. Commun. 9, 3947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toh S., et al., Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., et al., Molecular basis for high ligand sensitivity and selectivity of strigolactone receptors in Striga. Plant Physiol. 185, 1411–1428 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunsick M., et al., SMAX1-dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat. Plants 6, 646–652 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Kagiyama M., et al., Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Guo Y., Zheng Z., La Clair J. J., Chory J., Noel J. P., Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 8284–8289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shahul Hameed U., et al., Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor. EMBO Rep. 19, e45619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Saint Germain A., et al., An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 12, 787–794 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toh S., Holbrook-Smith D., Stokes M. E., Tsuchiya Y., McCourt P., Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem. Biol. 21, 988–998 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Yao J., et al., An allelic series at the KARRIKIN INSENSITIVE 2 locus of Arabidopsis thaliana decouples ligand hydrolysis and receptor degradation from downstream signalling. Plant J. 96, 75–89 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Conn C. E., et al., Plant evolution. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349, 540–543 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Waters M. T., et al., A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27, 1925–1944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters M. T., et al., Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Starr T. N., Thornton J. W., Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carbonnel S., et al., Lotus japonicus karrikin receptors display divergent ligand-binding specificities and organ-dependent redundancy. PLoS Genet. 16, e1009249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bürger M., et al., Structural basis of karrikin and non-natural strigolactone perception in Physcomitrella patens. Cell Rep. 26, 855–865.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H. W., et al., Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners. J. Biol. Chem. 295, 4181–4193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harms M. J., et al., Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. Proc. Natl. Acad. Sci. U.S.A. 110, 11475–11480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tawfik D. S., Messy biology and the origins of evolutionary innovations. Nat. Chem. Biol. 6, 692–696 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Eick G. N., Colucci J. K., Harms M. J., Ortlund E. A., Thornton J. W., Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 8, e1003072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uraguchi D., et al., A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science 362, 1301–1305 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Bloom J. D., Arnold F. H., In the light of directed evolution: Pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. U.S.A. 106 (suppl. 1), 9995–10000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khersonsky O., Tawfik D. S., Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Villaécija-Aguilar J. A., et al., SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet. 15, e1008327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker C., Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag. Sci. 65, 453–459 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Lumba S., et al., A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Dev. Cell 29, 360–372 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bythell-Douglas R., et al., Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol. 15, 52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai N., Roder H., Dickson A., Karanicolas J., Isothermal analysis of thermofluor data can readily provide quantitative binding affinities. Sci. Rep. 9, 2650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abraham M. J., et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 48.Huang J., et al., CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanwell M. D., et al., Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 4, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salentin S., Schreiber S., Haupt V. J., Adasme M. F., Schroeder M., PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 43 W443–W447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.