Abstract
Establishing cell-type specific gene expression programs relies on the action of distal enhancers, cis-regulatory elements that can activate target genes over large genomic distances - up to Mega-bases away. How distal enhancers physically relay regulatory information to target promoters has remained a mystery. Here we review the latest developments and insights on promoter-enhancer communication mechanisms revealed by live-cell, real-time single-molecule imaging approaches.
Introduction
Enhancers are cis-regulatory elements that activate target promoters over large genomic distances, in a time- and tissue- dependent manner. Enhancers contain binding sites for sequence specific transcription factors (TFs) [1], and specific chromatin modifications such as H3K4me1 and H3K27ac [2, 3]. Enhancer-bound TFs and chromatin adapter molecules recruit several important regulatory factors (RFs): co-activators, chromatin remodelers and epigenetic regulators [2]. The recruited RFs act to modulate the activity of the transcription machinery at the target gene promoter and shape the nascent RNA production kinetics. How exactly this long-range communication takes place has not yet been understood in detail. Several physical communication mechanisms have been proposed: DNA looping [4], tracking[5, 6], linking [7], and mobilization to nuclear compartments such as transcription “factories” [8–11]. Looping has been the prevailing model that has dominated our thinking of how enhancers communicate with distal promoters (e.g. [12–15]). However whether such putative loops are stable or transient, how close and how frequently enhancers come with target promoters, and the mechanisms by which enhancer-associated regulatory complexes control the transcription cycle of the target promoter have not been elucidated.
Recent breakthrough technological developments in live-cell optical nanoscopy, single-molecule tracking, and improved fluorescent probes for intracellular labeling, as well as in genome editing tools and fast-acting chemical probes, have enabled direct visualization of promoter-enhancer communication mechanisms. New pictures of promoter-enhancer communication are emerging that further refine ideas from early classical models and also incorporate a more detailed understanding of the nano-scale nuclear environment of active transcription sites in live cells.
Visualizing spatio-temporal relations between enhancers and promoters in live cells
How close and how often do enhancers come to target genes? Although fluorescence in-situ hybridization techniques had addressed the spatial relationships between different genomic loci [16–19], tracking the dynamics of enhancer-promoter (P-E) communication required the ability to tag and visualize specific genomic loci in live cells. One of the first studies of this kind [20] used time-lapse confocal microscopy to probe 3D physical distances between P-E with respect to nascent RNA production at single-cell level in live Drosophila embryos. It focused on even-skipped (eve) gene, one of the essential pair-rule genes regulating segment pattern formation. The authors coupled genome editing with 3-color imaging approaches to study how the endogenous even-skipped (eve) enhancers activated a synthetic reporter gene integrated ~150 kb away from the endogenous eve locus. The reporter cassette was engineered to harbor an eve promoter and a homie insulator element, the latter facilitating self-pairing with an endogenous homie near the eve locus. The authors tracked the positions of the enhancers and target reporter gene, as well as simultaneously monitor reporter gene activity, in real-time. A DNA labeling system (ParS/ParB) [21] was used to visualize the position of the reporter gene, while an RNA labeling system (PP7/PCP) [22] was used to visualize the nascent RNA produced by the reporter. An orthogonal RNA labeling method (MS2/MCP) [23] was used to tag the endogenous eve RNA as a proxy of the endogenous eve enhancers, which are located within ~10 kb from the eve promoter.
The results of this study showed strong correlation between physical proximity with gene activity: when transcription is OFF, P-E distances were ~700 nm r.m.s. This distance decreased to ~340 nm r.m.s. right before transcription switches ON. Similarly, transcription switching from ON to OFF was accompanied by increased P-E distances. Interestingly, more detailed analysis revealed that mere physical proximity after homie-homie pairing was not sufficient to activate the reporter gene. In the paired state, transcription activity was associated with additional conformational changes, which further compacted the locus and brought enhancer-promoter in closer proximity, from ~385 nm to ~330 nm.
A similar result was also observed for enhancer-promoter communication in trans, during transvection between homologous chromosomes in Drosophila [24]. The authors inserted reporter transgenes on the two chromosomes: one allele harbored a PP7 reporter with an upstream snail (sna) enhancer, while the other allele harbored an identical MS2 reporter but without an enhancer. Pairing was facilitated by gypsy insulator elements placed upstream. A fraction of nuclei show coordinated transcription bursts of PP7 and MS2, indicating co-regulated transcription of the two reporters from the same sna enhancer. The location of the PP7 and MS2 RNA probes provides an approximate measurement of the distances between the shared enhancer and the target genes. In this case promoter-enhancer communication in trans is associated with proximity of ~280 nm. Further results with different configurations of co-regulated PP7 and MS2 reporters in cis and trans indicates that in this setting promoter-enhancer communication generally involved distances in the order of 200-300 nm.
Embryonic stem cells (ESCs) are another biological system where promoter-enhancer spatio-temporal relationships and nascent transcription kinetics were simultaneously visualized. A study of the Sox2 locus in mouse ESCs (mESCs) [25] focused on activation of Sox2 by a distal cluster of enhancers, located ~100 kb downstream. The authors developed a modular system for inserting genome tags based on knock-in of transposase landing pads, with subsequent insertion of Tetracycline and Cumate operator arrays. P-E distance measurements reveal that the Sox2 promoter and its distal enhancer are separated by ~300 nm on average.
By also integrating an MS2 reporter to visualize real-time activity of Sox2, the authors discover that Sox2 exhibits short intermittent transcription bursts, where the nascent RNA switches between ON and OFF states on the time-scale of minutes. Interestingly, no temporal correlation (e.g. increased spatial proximity during or preceding a burst) between transcription activity and P-E distances was observed.
The observations in the Sox2 locus initially appear to be at odds with the results of the eve reporter, where changes in P-E distances result in immediate and robust activation of transcription. Although different P-E pairs might have different minimum distance requirements for transcription activation, the major distance changes associated with eve activation involved homie pairing that presumably reflects formation of a topological “loop” domain. Such a domain might be constitutively formed and always present at Sox2 in mESCs. It remains possible that any more subtle and transient conformational changes associated with on-off transitions during bursts might have been missed in work of [25]. The observations in Sox2 are entirely consistent with a picture where any P-E contacts involved in initiating a burst are very transient and/or there is a long and variable time-lag between E-P contacts and appearance of nascent RNA that washes out any correlations.
Limitations due to the modest spatiotemporal resolution of the imaging modalities used and the sizes of the tags acting as proxies for promoter and enhancer location might have resulted in missing very transient (seconds) and subtle (~10’s of nm) conformational changes that accompany P-E communication. Despite imperfections, an important picture emerges from these live-cell imaging studies: in both Drosophila and mESCs, enhancers and promoters appear to not associate through persistent molecular contact (e.g. 10’s of nanometers distance). Proximity ligation methods rely on close contact between fragments to be joined by ligase, but only provide averages over populations of cells and it is not straightforward to quantify distances from sequence reads distributions. Although technical limitations for directly measuring molecular contacts by optical means still remain, the imaging data suggest that such events might occur very infrequently and only transiently. Rather, transcription activity is associated with only approximate proximity, of ~200 nm on average. This picture, also suggested by certain FISH experiments in fixed cells (e.g. [26]), is inconsistent with looping models that posit P-E frequently residing in close proximity, held together by molecular-scale bridges.
Visualizing nanoscale organization of enhancer-associated regulatory factors in live cells
Regulatory information is likely relayed by interactions between enhancer-associated factors and components of the transcription machinery at the promoter. A key question then is, if enhancers and promoters are separated by ~200 nm distances, without stable molecular complexes bridging the two, how do such interactions physically take place? Alternative models postulate that enhancer-promoter communication is achieved via approximate proximity, with enhancer-bound factors propagating towards the promoter (such as the classical linking and tracking models). A similar but physically distinct mechanism posits that due to the increased high local concentration of factors recruited at the vicinity of the enhancer, individual molecules might only have to diffuse through a small intervening space to interact with the Pol II machinery at the promoter [27]. An extension of the idea that enhancers work by increasing the local concentration of transcriptional regulators is a class of models that postulate formation of specialized transcriptional compartments or activating local environments [28] in the vicinity of the transcription site.
Although such ideas have been somewhat speculative, early immunofluorescence of RNA Polymerase II [11, 29] and TFs [30] had already suggested inhomogeneous nuclear distributions, although it was hard to know how fixation and antibody staining might have perturbed the native structures. Recently, in one of the earliest observations of RFs in live cells, the authors created mice with knock-in of mCherry in the endogenous Cdk9 gene [31]. Intriguingly, fluorescent Cdk9 is not uniform throughout the nucleus, but accumulates in clusters. By using FRAP measurements, the authors further showed that these stable clusters reflect steady-state accumulation of multiple molecules that rapidly exchange – residence times of Cdk9 molecules into the clusters are merely a few seconds. More recently, several more RFs have been shown to form nuclear clusters that are also stabilized by dynamic interactions, including coactivators [32–34], as well as a variety of TFs [35–40]. When directly imaged by confocal or lattice light-sheet microscopy such clusters appear as stable structures, persisting for minutes to hours (e.g. [31, 35, 41]). Temporal analyses of single-molecule blinking events concluded that ~100 molecules of Dendar2-tagged Pol II [42] and Mediator [33] can also transiently accumulate in sub-diffraction regions but disperse quickly - within seconds – after cluster formation. Given recent improvements in 4D resolution and detection sensitivity of single-gene and single-molecule imaging methods [43–45] it will be interesting to re-examine Pol II and RF spatio-temporal dynamics in different live-cell settings, using advanced techniques. Interestingly, the short residence times into clusters also parallels the short residence times of TFs on nuclear chromatin seen by single-particle tracking techniques (e.g. [46–48], reviewed in [49]).
The discoveries of clusters of regulatory factors throughout the nucleus of live cells was a significant and important advance, however it was hard to relate clustering to transcription of specific gene loci and test whether clustering relates to transcription activity. Before detailed mechanisms could be dissected, at least four key requirements needed to be met: 1) locating the target gene of interest in 3D nuclear space; 2) simultaneously detecting its transcription activity; 3) detecting down to single molecules of Pol II and RFs engaged at the target gene; 4) rapidly perturbing the system and analyzing in real-time the effects of perturbations as they happen at the transcription site, to capture primary effects and dissect cause-and-effect relations.
A recent work reported this type of single-molecule/single-gene imaging of transcription in live cells [44]. The authors developed new single-molecule optical nanoscopy techniques that enabled zooming into specific gene loci in live cells and imaging, tracking and quantifying Pol II and RFs with down to single-molecule resolution. To visualize the endogenous protein factors, SNAP-tag was knocked into the endogenous genes. The authors focused on Pou5f1 (Oct4) and Nanog, two genes encoding TFs important for the pluripotent state and which are controlled by clusters of distal enhancers. Intriguingly, they discovered that key enhancer-associated RFs – pTEFb, Mediator, Brd4, and Sox2 – form small, sub-diffraction clusters in the vicinity (~100-200 nm) of the transcription site. These small RF clusters contain up to 20 molecules of each RF and are spatially separated in the nanometer scale from clusters of ~5-10 elongating Pol II molecules, which colocalize to <100 nm from the MS2-tagged nascent RNA. Similar RF clusters were also later observed in the Sox2 locus [45]. Intriguingly, transcription “factories” observed by electron microscopy [29, 50] have average diameters of ≤100 nm, similar to the clusters of nascent RNA and Pol II observed by single-molecule nanoscopy at single-genes in live cells. The small sub-diffraction RF clusters observed using the highly sensitive imaging methods in the vicinity of specific gene loci in live mESCs [44, 45] are distinct from much larger clusters (~14 clusters per cell that are >300 nm in size and contain 100’s of molecules for e.g. Pol II and the Mediator complex [33]) that do not co-localize with active genes and can be readily observed in other regions of the nucleus. The few large Pol II and Mediator clusters per cell that were originally seen with less sensitive optical methods [33] appear to represent just the tip of the ice-berg of a range of distinct types of RF clusters formed throughout the nucleus of mESCs (Figure 1).
Figure 1. Sub-diffraction clusters of RNA Pol II and RFs at the Pou5f1 transcription site discovered by optical nanoscopy.
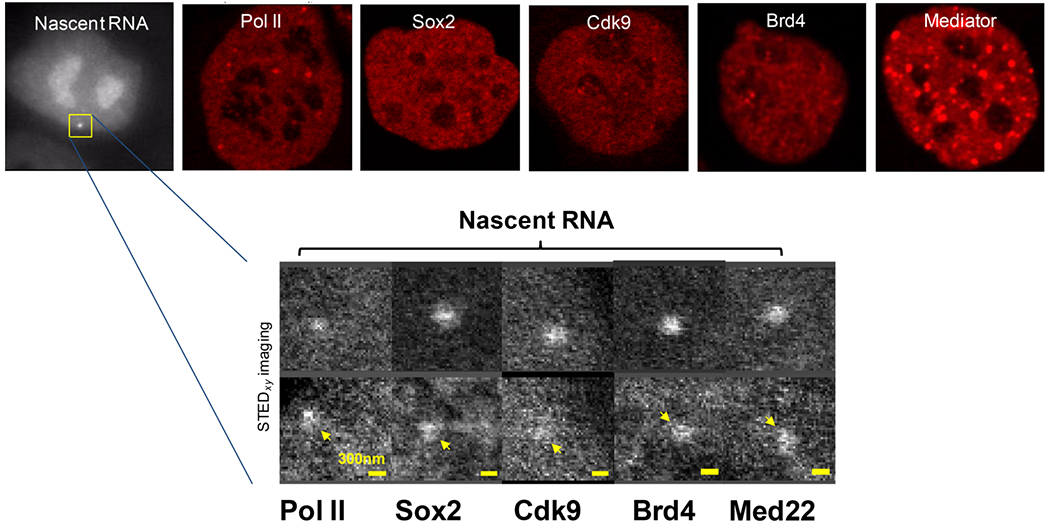
Top row: whole nucleus live-cell images, showing the Pou5f1 transcription site (Nascent RNA visualized using MCP-mNeonGreen binding to 24xMS2 stem-loops), as well as RNA Polymerase II (Rpb1 subunit), Sox2, Cdk9, Brd4 and Mediator (Med22 subunit), visualized using SNAP-tag knock-ins and a SiR dye. Bottom row shows zoomed-in regions of the vicinity of the transcription site in live-cells, showing small (5-20 molecules), sub-diffraction clusters of Pol II and RFs. Top row: A.P. and J.L., unpublished data; bottom row: reproduced, with permission, from [44].
Control of promoter kinetics by focal accumulation of enhancer-associated RFs
The observation that different types of RFs form sub-diffraction clusters in the vicinity of active genes gives rise to a couple of questions: what are the interdependencies of the different RFs and their respective activities? How does RF clustering relate to nascent transcription activity? To address these questions, the authors used fast-acting small-molecule chemical inhibitors: A-485, targeting the p300/CBP Histone Acetyltransferase (HAT) activity; JQ1, competing with Brd4 binding to acetylated chromatin; Flavopiridol, targeting the kinase Cdk9 that controls promoter-proximal pause release of Pol II into productive elongation. At the Pou5f1 locus, there is a hierarchy of activities: at the top of this hierarchy, a positive feedback loop between Sox2 binding, recruitment of p300/CBP, and reinforcement of Sox2 accumulation by p300/CBP HAT activity sets up Sox2 clustering and a establishes the acetylated enhancer chromatin state. Multiple acetylated targets then enable binding of multiple Brd4 molecules, which form a platform for recruitment of multiple pTEFb/Cdk9, as well as Mediator molecules. Finally, the high local concentration of clustered pTEFb/Cdk9 controls the rate of release of Pol II into elongation and the production of nascent RNA.
Interestingly, for Nanog, which exhibits intermittent transcriptional bursting (on-off switching) on the minute time-scale [51], clustering of Brd4 molecules appears to control the burst frequency (burst initiation rate), with smaller effects on burst amplitude and almost no effect on burst duration. The discovery that clustering of an enhancer-associated RF (Brd4) controls bursting frequency is a step forward towards elucidating the exact molecular mechanisms of bursting, a still enigmatic phenomenon. Modulation of bursting frequency by distal enhancers appears to be a more prevalent phenomenon, as seen by real-time imaging for Drosophila enhancers [52], as well as from genome-wide analysis of single-cell RNA sequencing experiments [53], that correlate enhancer acetylation levels and inferred bursting frequencies. Interestingly, pools of shared clustered RFs had been hypothesized to initiate coordinated bursting of two linked promoters by the same enhancer [54]. Strikingly, a shared or partially overlapping pool of clustered Brd4 molecules between the two sister chromatids was observed during simultaneous bursting of the two sister chromatids, for Nanog and Sox2. It will be interesting to see if a similar arrangement underlies coordinated bursting and co-regulation of different genes, both is cis and in trans [24, 52, 54–56].
Relating focal RF accumulation and underlying genome topologies
What are the mechanisms for forming the discovered clusters at the vicinity of important developmental genes? Focal accumulation of multiple TFs and chromatin regulators could reflect clustered DNA and chromatin binding sites, either due to the arrangement of enhancer clusters in the 1D genome, or via juxtaposition of multiple distal enhancers in 3D. An alternative model suggests that weak, multivalent protein-protein interactions between low-complexity domains (LCDs) might create a unique environment, possibly even a distinct physical phase/state (i.e. a “condensate”) that encompasses the transcription site. Dissecting the relative importance of these mechanisms required measuring the spatial relationships of target genes and distal enhancers with high resolution, as well as detailed analysis of the molecular interactions that are responsible for recruiting key RFs into such clusters.
A recent study [45] sheds more light into the mechanisms of Sox2 and Brd4 clustering in mESCs. By measuring the ability of various point and truncation mutants, the authors discovered that recruitment of Sox2 and Brd4 into clusters throughout the nucleus, as well as at the Pou5f1 locus, is dependent on specific molecular recognition of Sox DNA motifs and acetylated lysine targets, respectively. Neither Sox2 nor Brd4 LCDs were needed for efficient incorporation into clusters, and LCD-LCD interactions alone were not sufficient for efficient recruitment into clusters. These observations strongly suggested that the underlying mechanism for Sox2 and Brd4 cluster formation is clustering of multiple DNA and chromatin binding sites in the vicinity of the transcription site.
To better understand the possible enhancers that underlie RF clustering, the same study then focused on measuring the proximity of various distal genomic locations from the extended locus relative to the MS2-tagged transcription site. The authors leveraged improved sensitivity and resolution imaging, capable of detecting down to a few molecules of chromatin bound RFs, to image dCas9-Halo bound to the DNA. This approach bypassed the need for exogenous tags (such as bulky TetO arrays), while also achieving ~1 kb resolution with down to 12 unique gRNAs. dCas9-Halo tagged distal enhancers were observed close to target gene, within the same distances as clustered Sox2 and Brd4 (<200 nm). High frequency of pair-wise interactions suggests that the extended locus adopts configurations that often juxtapose multiple distal enhancers with the target gene. Such enhancer cluster “super-clusters” are likely underlying the formation of Sox2 and Brd4 clusters in mESCs.
Local activating environments with accumulated RFs and underlying clustering of enhancers could be a general feature of transcription regulation. The Ultrabithorax (Ubx) TF in Drosophila [40] was seen inhomogeneously distributed in live cell nuclei, while a DNA binding deficient Ubx mutant protein exhibited uniform nuclear distribution, without incorporating into clusters. Although the authors could not observe real-time transcription in live cells, using RNA FISH and immunofluorescence, they observe that actively transcribed shavenbaby (svb) locus is embedded in a ~250 nm region of high local Ubx concentration. Interestingly, ectopic svb enhancers also embedded into the same Ubx microenvironment as the endogenous svb locus, suggesting specialized local environments enable transcription by related co-localized enhancers.
Towards elucidating physical mechanisms of promoter-enhancer communication
The compartment and local “environment” hypothesis appears to be broadly consistent with the experimental observations of enhancers and promoters separated by a few hundred nanometers and residing within a local environment of focal RF accumulation at active transcription sites. A specific variant of such models, the “hub” or “condensate” model [57], envisioned that clusters of multiple RFs, held together by LCD-LCD interactions and spanning a ~300-400 nm volume that bridges enhancer and promoter. This model was originally supported by the propensity of LCDs of Pol II and RFs to drive droplet formation in vitro [32, 58], the formation of large nuclear puncta when LCDs are overexpressed and artificially multimerized in live cells [59], as well as the observation of a few (~14 per cell) large clusters formed by endogenous Mediator and Pol II in mESCs that exhibited apparent “droplet-fusion” characteristics [33]. More detailed analysis with single-molecule resolution at specific gene loci shows that 1) RF clusters forming within the vicinity of single gene loci such as Sox2, Pou5f1 and Nanog are much smaller, while the few large clusters of Mediator and Pol II do not spent significant time in the vicinity of these genes [44, 45]; 2) RF recruitment into clusters does not involve LCD-LCD interactions but is predominantly through specific recognition of DNA and chromatin sites and 3D clustering of multiple distal enhancers into “super-clusters” [45]. Overall, these results point to a more nuanced physical P-E communication mechanism than originally put forth by “condensate” models.
Two proposed models (Figure 2) of how the promoter senses high local RF concentrations created within the enhancer cluster “super-clusters” could help establish a conceptual framework for further studies of physical P-E communication mechanisms. The two models are differentiated by whether RFs remain almost exclusively bound to their cognate DNA and chromatin binding sites or whether RFs are free to explore the intervening P-E space, possibly in a facilitated fashion. The first model (“dynamic promoter-enhancer contact model”) posits that RFs within the cluster remain mostly bound to their targets at the enhancer, and the promoter senses the high local RF concentration through dynamic/transient interactions with the individual enhancers, that bring individual RF molecules and the transcription machinery at molecular proximity. This view is supported by single-locus tracking experiments, showing exploration of nuclear space by cis-elements and active genes consistent with an (unconstraint) movement of the underlying chromatin polymer [44, 60]. Additional active processes (e.g. by cohesin-mediated loop extrusion), provided that they can synergize rather than interfere with other genomic interactions mechanisms (e.g. TF-TF association, DNA looping), could further facilitate short-range interactions, changing the search dimensionality from 1D to 3D. Cohesin- and TF-mediated interactions might also be transient, given the residence times on chromatin seen by single-molecule tracking, ranging from minutes for cohesin [61] to seconds for TFs [35, 46, 62]. The second model (“local RF trapping model”) envisions that the promoter does not come in direct contact with the individual enhancers, remaining at ~100-200 nm separations. Rather approximate proximity of multiple enhancers creates a local environment where individual RFs are kinetically trapped. Local trapping could be mediated by “avidity” effects, due to the large local concentration of binding sites [41]. Intriguingly, the movements of certain TFs in the nucleus have also been shown to exhibit behaviors consistent with local trapping, due to interactions with RNAs [63] or LCDs [35, 62]. RF clusters at specific genomic loci nucleate from a scaffold of underlying binding sites on chromatin, and do not abide by pure LLPS rules, but likely other model with more complex and still unresolved biophysical properties. It will be of great interest to study the movement of individual RFs in the vicinity of specific genes, and relatively specific tagged enhancers. We also note that in cases where RFs are recruited into clusters through binding to specific sites, local trapping processes (e.g. through LCD-LCD interactions) would not be expected to be main drivers for partitioning and enrichment of RFs into clusters. Although the majority of RF molecules within the cluster might be bound to specific sites at any given time, Local trapping might nonetheless be crucial for P-E communication: a minority of molecules, possibly down to a single molecule at any given point, that dissociate from their cognate sites might use local trapping mechanisms to exploring the local environment created by clustered enhancers and enhancer-associated RFs, to find and interact with the transcription machinery at the promoter.
Figure 2. Proposed physical models for enhancer-promoter communication.
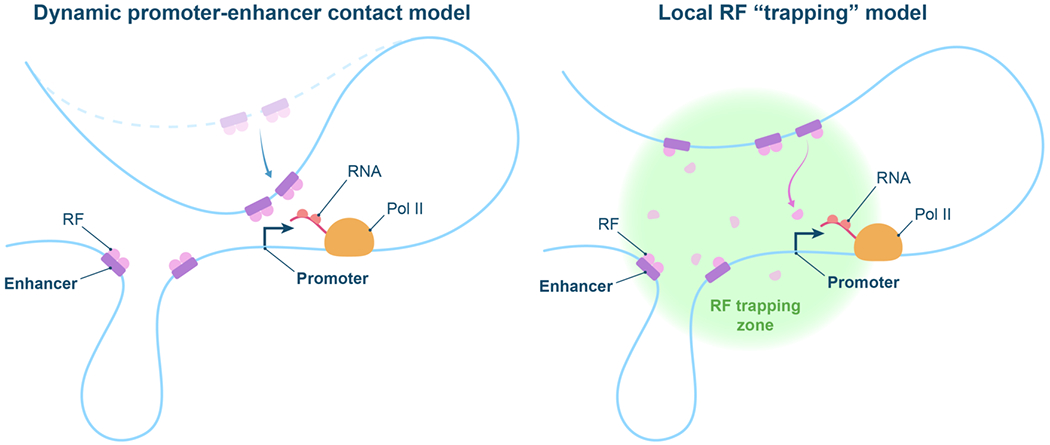
Dynamic P-E contact model: RFs stay mostly bound to cognate DNA and chromatin sites at the enhancers; RF interactions with the transcription machinery at the promoter are then facilitated by dynamic movements of the chromatin polymer that bring promoter and individual enhancers in molecular proximity. Local RF trapping model: P-E are staying at approximate proximity (100-200nm) while single RF molecules can dissociate from their DNA/chromatin sites and explore the local environment of the clustered enhancers, possibly through constrained motion, searching for the transcription machinery at the promoter.
Conclusions and Outlook
The last several years have seen a tremendous progress in our abilities to probe long-range gene regulation by distal enhancers by imaging single-molecules in live cells. New emerged methods that can image track and quantify nascent transcription, genomic interactions and regulatory factor dynamics, at specific gene loci, hold great promise for increasing our mechanistic understanding of promoter-enhancer communication and the interplay between genome organization and transcription. Further developments in terms of spatio-temporal resolution and detection sensitivity [43, 44, 64, 65] will enable delving deeper into the physical properties of the nanoscale environment created by clustered enhancers and accumulated RFs. We anticipate that single-particle tracking at ~1 msec temporal and <10 nm 3D spatial resolution will directly track individual RF molecules and individual constituent enhancers (e.g. tagged with single dCas9 or other programmable DNA probes) in the vicinity of the transcription site and relatively to the promoter, enabling discriminating between fundamentally distinct communication models and providing further insights.
Better defining the roles of the different RFs in facilitating P-E communication and transcription activation is also a key area where new breakthroughs are expected. Coupling real-time imaging methods with acute perturbations (recently reviewed in [66]), e.g. using small-molecule chemical inhibitors or targeted protein degradation further enhances our abilities to capture primary events that happen at the transcription site and probe cause-and-effect relations. We are envisioning several key developments in the near future, specifically delineating the detailed roles of key genome architectural factors that are proposed to facilitate genomic interactions and promoter-enhancer communication. These include cohesin, proposed to extrude chromatin loops [67, 68], as well as TFs proposed to self-associate [63, 69–71], possibly bringing or holding together distal genomic loci. Given that cohesin rings, postulated to enclose chromatin strands, are ~50 nm in diameter [72, 73], some close contacts might be expected. Evaluating cohesin-mediated contacts by live-cell single-molecule imaging of tagged genomic loci will provide new insights, particularly for genes that show reduced P-E contacts and reduced transcription activity upon cohesin depletion [74]. Finally, genomics [75] and imaging [76] studies suggest antagonistic relations between segregaton id transcriptionally active and inactive chromatin compartments and topological domains formed by cohesin and CTCF. It will be interesting to see how different factors and activities shape the search process and stability of different types of genomic interactions, and how these structures guide P-E communication.
Targeted perturbation experiments, together with improved imaging and genome tagging resolution will also be crucial in defining what constitutes a productive genomic interaction, specifically the length-scales at which P-E proximity mediated by architectural and putative looping factors results in transcription activation. A further important future task is to quantify the detailed relation between P-E contact frequency and transcriptional output. Apparent conundrum in the field is how topological structures such as TADs constrain promoter-enhancer communication. Interactions across TAD boundaries are only 2-fold different than within boundaries, seen both by proximity ligation approaches [77, 78] and by imaging [17]. Furthermore TADs do not exist as defined structures in individual cells but arise as statistical structures after averaging populations of cells [16, 18, 78]. Teasing out detailed quantitative relations between P-E proximity/contact frequency and promoter activity will be important to understand the roles of TADs in defining the dynamic range of P-E communication and the mechanisms of insulation.
Perspectives.
Importance of the field. How distal enhancers specify gene expression programs remains a key question. Shedding light into this decades-old problem required new tools that enabled zooming into the crowded and compartmentalized nuclear milieu, to visualize nascent transcription, genomic interactions and regulatory factor dynamics, in real-time, at specific gene loci, in live cells.
Current thinking. New insights from dynamic single-molecule imaging in live cells show that in certain settings enhancers cluster in 3D, to scaffold the formation of local nano-scale environments (~100-200 nm) characterized by high local concentration of important transcription regulatory proteins. This focal RF accumulation controls key steps in the transcription cycle of target promoters embedded in such local nano-environments.
Future directions. Presently the roles of the various regulatory and genome architectural factors in creating these local environments, as well as the detailed physical mechanisms of how the transcription machinery at the promoter senses this high local concentration of accumulated RFs in the vicinity of active genes are not well understood. Future increases in spatio-temporal resolution of single-molecule nanoscopy approaches, together with rapid perturbations using chemical tools, will shed new light into these important questions.
Funding Information
This work was supported by a NYSTEM Postdoctoral Training Award (C32599GG; J.L.), the MSKCC Functional Genomics Initiative (GC-242240; A.P.), the MSKCC Center for Epigenetics Research and the Metropoulos Family Foundation (A.P.), a National Cancer Institute Grant (P30 CA008748), and the National Institute of General Medical Sciences of NIH (1R01GM135545-01 and 1R21GM134342-01; A.P.).
Footnotes
Declaration of competing interests
The authors declare no competing interests.
References
- 1.Spitz F and Furlong EEM, Transcription factors: from enhancer binding to developmental control. Nature reviews Genetics, 2012. 13(9): p. 613–26. [DOI] [PubMed] [Google Scholar]
- 2.Long HK, Prescott SL, and Wysocka J, Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell, 2016. 167(5): p. 1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. , Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics, 2007. 39(3): p. 311–8. [DOI] [PubMed] [Google Scholar]
- 4.Carter D, Chakalova L, Osborne CS, Dai Y.-f., and Fraser P, Long-range chromatin regulatory interactions in vivo. Nature genetics, 2002. 32(4): p. 623–6. [DOI] [PubMed] [Google Scholar]
- 5.Blackwood EM and Kadonaga JT, Going the distance: a current view of enhancer action. Science, 1998. 281(5373): p. 60–3. [DOI] [PubMed] [Google Scholar]
- 6.Hatzis P and Talianidis I, Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell, 2002. 10(6): p. 1467–77. [DOI] [PubMed] [Google Scholar]
- 7.Bulger M and Groudine M, Looping versus linking: toward a model for long-distance gene activation. Genes Dev, 1999. 13(19): p. 2465–77. [DOI] [PubMed] [Google Scholar]
- 8.Papantonis A and Cook PR, Transcription factories: genome organization and gene regulation. Chemical reviews, 2013. 113(11): p. 8683–705. [DOI] [PubMed] [Google Scholar]
- 9.Cook PR and Marenduzzo D, Transcription-driven genome organization: a model for chromosome structure and the regulation of gene expression tested through simulations. Nucleic acids research, 2018. 46(19): p. 9895–9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francastel C, Schubeler D, Martin DI, and Groudine M, Nuclear compartmentalization and gene activity. Nature reviews Molecular cell biology, 2000. 1(2): p. 137–43. [DOI] [PubMed] [Google Scholar]
- 11.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, et al. , Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet, 2004. 36(10): p. 1065–71. [DOI] [PubMed] [Google Scholar]
- 12.Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, et al. , Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell, 2014. 158(4): p. 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, et al. , Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell, 2012. 148(1-2): p. 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 2014. 159(7): p. 1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenfelder S and Fraser P, Long-range enhancer-promoter contacts in gene expression control. Nature reviews Genetics, 2019. 20(8): p. 437–455. [DOI] [PubMed] [Google Scholar]
- 16.Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, et al. , Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell, 2014. 157(4): p. 950–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn EH, Pegoraro G, Brandao HB, Valton AL, Oomen ME, Dekker J, et al. , Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell, 2019. 176(6): p. 1502–1515 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bintu B, Mateo LJ, Su JH, Sinnott-Armstrong NA, Parker M, Kinrot S, et al. , Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science, 2018. 362(6413). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williamson I, Lettice LA, Hill RE, and Bickmore WA, Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development (Cambridge, England), 2016. 143(16): p. 2994–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, and Gregor T, Dynamic interplay between enhancer-promoter topology and gene activity. Nat Genet, 2018. 50(9): p. 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Germier T, Sylvain A, Silvia K, David L, and Kerstin B, Real-time imaging of specific genomic loci in eukaryotic cells using the ANCHOR DNA labelling system. Methods, 2018. 142: p. 16–23. [DOI] [PubMed] [Google Scholar]
- 22.Larson DR, Zenklusen D, Wu B, Chao JA, and Singer RH, Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science, 2011. 332(6028): p. 475–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, and Long RM, Localization of ASH1 mRNA particles in living yeast. Molecular cell, 1998. 2(4): p. 437–45. [DOI] [PubMed] [Google Scholar]
- 24.Heist T, Fukaya T, and Levine M, Large distances separate coregulated genes in living Drosophila embryos. Proc Natl Acad Sci U S A, 2019. 116(30): p. 15062–15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, et al. , Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, et al. , Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol Cell, 2019. 76(3): p. 473–484 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, and Bejerano G, Enhancers: five essential questions. Nature reviews Genetics, 2013. 14(4): p. 288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiter F, Wienerroither S, and Stark A, Combinatorial function of transcription factors and cofactors. Current opinion in genetics & development, 2017. 43: p. 73–81. [DOI] [PubMed] [Google Scholar]
- 29.Iborra FJ, Pombo A, Jackson DA, and Cook PR, Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci, 1996. 109 ( Pt 6): p. 1427–36. [DOI] [PubMed] [Google Scholar]
- 30.van Steensel B, Brink M, van der Meulen K, van Binnendijk EP, Wansink DG, de Jong L, et al. , Localization of the glucocorticoid receptor in discrete clusters in the cell nucleus. J Cell Sci, 1995. 108 ( Pt 9): p. 3003–11. [DOI] [PubMed] [Google Scholar]
- 31.Ghamari A, van de Corput MP, Thongjuea S, van Cappellen WA, van Ijcken W, van Haren J, et al. , In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev, 2013. 27(7): p. 767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. , Coactivator condensation at super-enhancers links phase separation and gene control. Science, 2018. 361(6400). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, et al. , Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science, 2018. 361(6400): p. 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma L, Gao Z, Wu J, Zhong B, Xie Y, Huang W, et al. , Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Molecular cell, 2021. [DOI] [PubMed] [Google Scholar]
- 35.Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, et al. , Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science (New York, N Y ), 2018. 361(6400). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. , Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell, 2018. 175(7): p. 1842–1855 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, et al. , Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nature structural & molecular biology, 2019. 26(3): p. 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, et al. , Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nature cell biology, 2019. 21(12): p. 1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mir M, Reimer A, Haines JE, Li XY, Stadler M, Garcia H, et al. , Dense Bicoid hubs accentuate binding along the morphogen gradient. Genes Dev, 2017. 31(17): p. 1784–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai A, Muthusamy AK, Alves MR, Lavis LD, Singer RH, Stern DL, et al. , Nuclear microenvironments modulate transcription from low-affinity enhancers. elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, et al. , 3D imaging of Sox2 enhancer clusters in embryonic stem cells. elife, 2014. 3: p. e04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, et al. , Real-time dynamics of RNA polymerase II clustering in live human cells. Science, 2013. 341(6146): p. 664–7. [DOI] [PubMed] [Google Scholar]
- 43.Cao B, Wang G, Li J, and Pertsinidis A, 3D Interferometric Lattice Light-Sheet Imaging. BioRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Dong A, Saydaminova K, Chang H, Wang G, Ochiai H, et al. , Single-Molecule Nanoscopy Elucidates RNA Polymerase II Transcription at Single Genes in Live Cells. Cell, 2019. 178(2): p. 491–506 e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Hsu A, Hua Y, Wang G, Cheng L, Ochiai H, et al. , Single-gene imaging links genome topology, promoter-enhancer communication and transcription control. Nature structural & molecular biology, 2020. 27(11): p. 1032–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, et al. , Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell, 2014. 156(6): p. 1274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, et al. , Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell, 2016. 165(3): p. 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gebhardt JCM, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, et al. , Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nature methods, 2013. 10(5): p. 421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z and Tjian R, Visualizing transcription factor dynamics in living cells. J Cell Biol, 2018. 217(4): p. 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pombo A, Hollinshead M, and Cook PR, Bridging the resolution gap: Imaging the same transcription factories in cryosections by light and electron microscopy. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society, 1999. 47(4): p. 471–80. [DOI] [PubMed] [Google Scholar]
- 51.Ochiai H, Sugawara T, Sakuma T, and Yamamoto T, Stochastic promoter activation affects Nanog expression variability in mouse embryonic stem cells. Scientific reports, 2014. 4: p. 7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukaya T, Lim B, and Levine M, Enhancer Control of Transcriptional Bursting. Cell, 2016. 166(2): p. 358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larsson AJM, Johnsson P, Hagemann-Jensen M, Hartmanis L, Faridani OR, Reinius B, et al. , Genomic encoding of transcriptional burst kinetics. Nature, 2019. 565(7738): p. 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim B, Heist T, Levine M, and Fukaya T, Visualization of Transvection in Living Drosophila Embryos. Mol Cell, 2018. 70(2): p. 287–296 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, Monahan K, et al. , Enhancer interaction networks as a means for singular olfactory receptor expression. Cell, 2014. 159(3): p. 543–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Di Giammartino DC, Kloetgen A, Polyzos A, Liu Y, Kim D, Murphy D, et al. , KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nature cell biology, 2019. 21(10): p. 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furlong EEM and Levine M, Developmental enhancers and chromosome topology. Science, 2018. 361(6409): p. 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, et al. , Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell, 2013. 155(5): p. 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, et al. , Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell, 2018. 175(6): p. 1481–1491 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu B, Swigut T, Spencley A, Bauer MR, Chung M, Meyer T, et al. , Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science, 2018. 359(6379): p. 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hansen AS, Pustova I, Cattoglio C, Tjian R, and Darzacq X, CTCF and cohesin regulate chromatin loop stability with distinct dynamics. elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia DA, Johnson TA, Presman DM, Fettweis G, Wagh K, Rinaldi L, et al. , An intrinsically disordered region-mediated confinement state contributes to the dynamics and function of transcription factors. Molecular cell, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen AS, Hsieh TS, Cattoglio C, Pustova I, Saldana-Meyer R, Reinberg D, et al. , Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol Cell, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang GS, Hauver J, Thomas Z, Darst SA, and Pertsinidis A, Single-Molecule Real-Time 3D Imaging of the Transcription Cycle by Modulation Interferometry. Cell, 2016. 167(7): p. 1839-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balzarotti F, Eilers Y, Gwosch KC, Gynna AH, Westphal V, Stefani FD, et al. , Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science, 2017. 355(6325): p. 606–612. [DOI] [PubMed] [Google Scholar]
- 66.Jaeger MG and Winter GE, Fast-acting chemical tools to delineate causality in transcriptional control. Molecular cell, 2021. [DOI] [PubMed] [Google Scholar]
- 67.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, and Mirny LA, Formation of Chromosomal Domains by Loop Extrusion. Cell Rep, 2016. 15(9): p. 2038–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nuebler J, Fudenberg G, Imakaev M, Abdennur N, and Mirny LA, Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci U S A, 2018. 115(29): p. E6697–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beagan JA, Duong MT, Titus KR, Zhou L, Cao Z, Ma J, et al. , YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res, 2017. 27(7): p. 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, et al. , YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell, 2017. 171(7): p. 1573–1588 e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, et al. , Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell, 2012. 149(6): p. 1233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huis in ‘t Veld PJ, Herzog F, Ladurner R, Davidson IF, Piric S, Kreidl E, et al. , Characterization of a DNA exit gate in the human cohesin ring. Science (New York, N Y ), 2014. 346(6212): p. 968–72. [DOI] [PubMed] [Google Scholar]
- 73.Gruber S, Haering CH, and Nasmyth K, Chromosomal cohesin forms a ring. Cell, 2003. 112(6): p. 765–77. [DOI] [PubMed] [Google Scholar]
- 74.Liu NQ, Maresca M, van den Brand T, Braccioli L, Schijns MMGA, Teunissen H, et al. , WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nature genetics, 2021. 53(1): p. 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, et al. , Two independent modes of chromatin organization revealed by cohesin removal. Nature, 2017. 551(7678): p. 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gu B, Comerci CJ, McCarthy DG, Saurabh S, Moerner WE, and Wysocka J, Opposing Effects of Cohesin and Transcription on CTCF Organization Revealed by Super-resolution Imaging. Molecular cell, 2020. 80(4): p. 699–711. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, et al. , Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature, 2013. 502(7469): p. 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Flyamer IM, Gassler J, Imakaev M, Brandao HB, Ulianov SV, Abdennur N, et al. , Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature, 2017. 544(7648): p. 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]


