Abstract
Chromatin ‘readers’ are central interpreters of the epigenome that facilitate cell-specific transcriptional programs and are therapeutic targets in cancer and inflammation. The Speckled Protein (SP) family of chromatin ‘readers’ in humans consists of SP100, SP110, SP140, and SP140L. SPs possess functional domains (SAND, PHD, bromodomain) that dock to DNA or post-translationally modified histones and a caspase activation and recruitment domain (CARD) to promote multimerization. Mutations within immune expressed SPs associate with numerous immunological diseases including Crohn’s disease, multiple sclerosis, chronic lymphocytic leukemia, veno-occlusive disease with immunodeficiency, as well as Mycobacterium tuberculosis infection, underscoring their importance in immune regulation. In this review, we posit that SPs are central chromatin regulators of gene silencing that establish immune cell identity and function.
SPs as Chromatin Regulators of Innate and Adaptive Immunity
SPs were first described as autoantigens in primary biliary cholangitis (see Glossary) [1–7] and as components of promyelocytic leukemia (PML) nuclear bodies (NBs) in human cell lines [4,8,9]. The SP family comprises SP100, SP110, SP140, and SP140-like protein (SP140L), which are all characterized by the presence of a nuclear localization signal (NLS) and multiple functional domains that implicate these proteins as chromatin ‘readers’ [10] (Figure 1, Key Figure). A rare SAND domain (named after proteins that contain it: SP100, Aire, NucP41/P75, and DEAF) interacts with DNA directly [11] or may promote protein–protein interactions [12], a plant homeodomain (PHD) reads histone methylation, and a bromodomain (BRD) reads histone acetylation [13]. SPs also share a CARD, previously characterized as a SP100-like domain or a homogeneously staining region (HSR) domain that may induce SP homo or heteromultimerization (Figure 1).
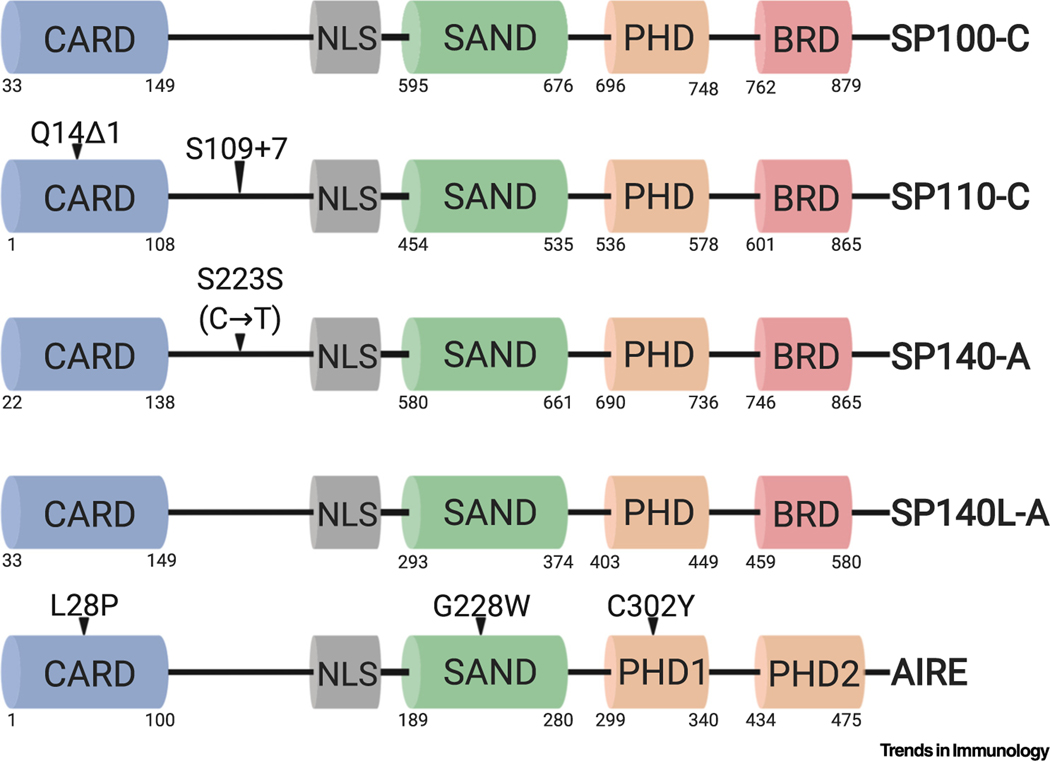
Figure 1. Functional Domains and Disease-Associated Mutations in Human Speckled Proteins (SPs) and Autoimmune Regulator (Aire).
SPs and Aire express highly similar caspase activation and recruitment domains (CARDs), Sp100, Aire, NucP41/75, DEAF-1 shared domains (SANDs), and plant homeodomains (PHDs). The SP family PHD is similar to Aire’s PHD1. Aire contains two unique PHDs and lacks a bromodomain (BRD). Highlighted are representative mutations associated with immunological diseases. Depicted are full-length SP isoforms: SP100 isoform C (SP100-C), SP110 isoform C (SP110-C), SP140 isoform A (SP140-A), and SP140-like (SP140L) isoform A (SP140L-A). Protein domains are depicted with corresponding amino acid residue positions underneath. The synonymous mutation depicted in SP140 is a cytidine-to-thymidine nucleotide mutation. Δ1, one-nucleotide deletion; +7, seven-nucleotide insertion. This figure was created using BioRender (https://biorender.com/).
The SP genetic locus (Figure 2) experienced duplication or insertion events throughout evolution. As a result, the locus is highly repetitive and contains pseudogenes, previously making the creation of mouse genetic models difficult. Although all functional protein domains are highly conserved, there is only approximately 45% amino acid identity between mouse and human homologs, necessitating the need for both mouse and human studies (Figure 2). SP140L is not expressed in mice as it recently evolved in primates [1] (Figure 2). Furthermore, human SPs have greater alternative isoform usage than mouse (Figure 3), but the effect of alternative splicing on SP function remains unexplored.
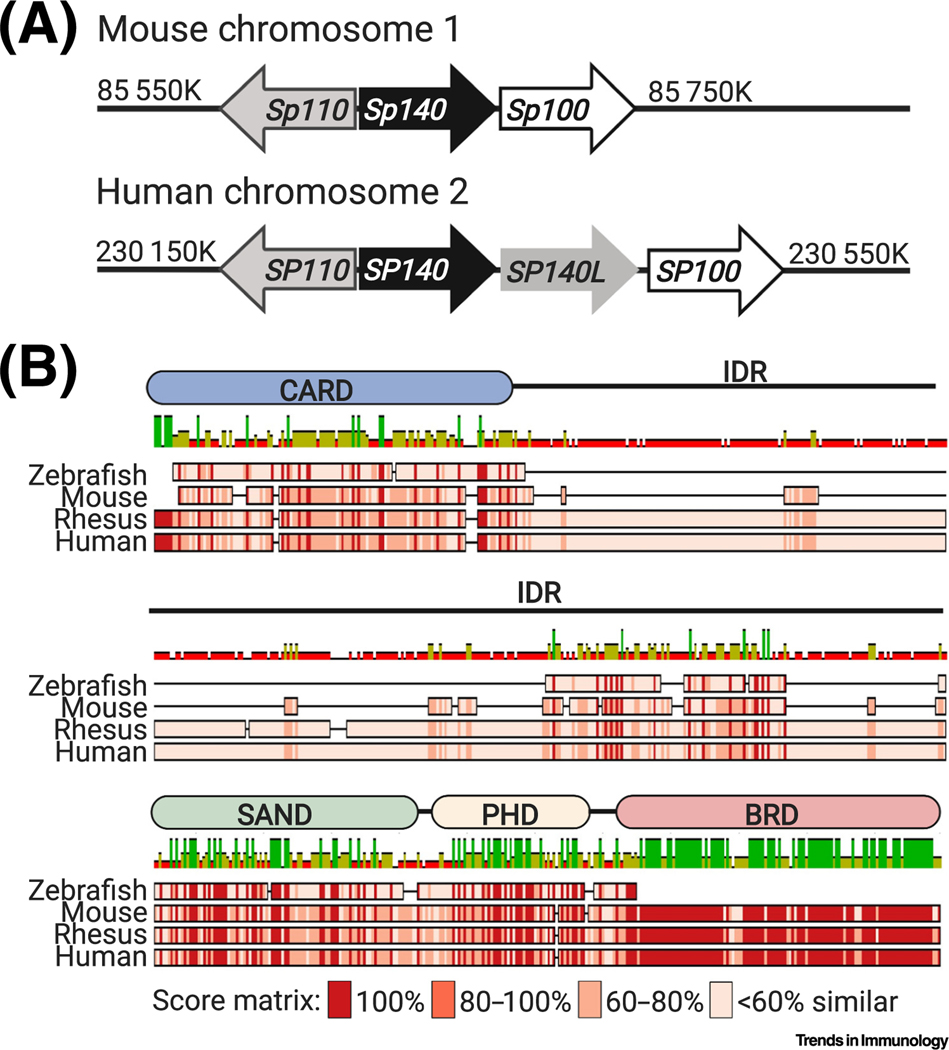
Figure 2. Genomic Organization of Speckled Protein (SP) Genes and Evolution of SP140.
(A) Schematic of SP genes on human chromosome 2 and Sp genes on mouse chromosome 1. (B) Here, we use SP140 as a model protein of the SP family to show the high degree of evolution in these proteins and the importance of studying this family in both mouse and human. Mouse Sp140, human SP140, rhesus monkey SP140, and zebrafish SP140 amino acid sequences were aligned using ClustalW, which determined that there was 34.7% pairwise identity between mouse and human at the amino acid level. Sequence similarity was determined using the Blosum62 score matrix and a threshold of 1. A schematic of human SP140 with functional domains is shown above the alignment. The intrinsically disordered region (IDR) is indicated on the human schematic but is mostly absent in the mouse. Abbreviations: BRD, bromodomain; CARD, caspase activation and recruitment domain; PHD, plant homeodomain; SAND, Sp100, Aire, NucP41/75, DEAF-1 shared domain.
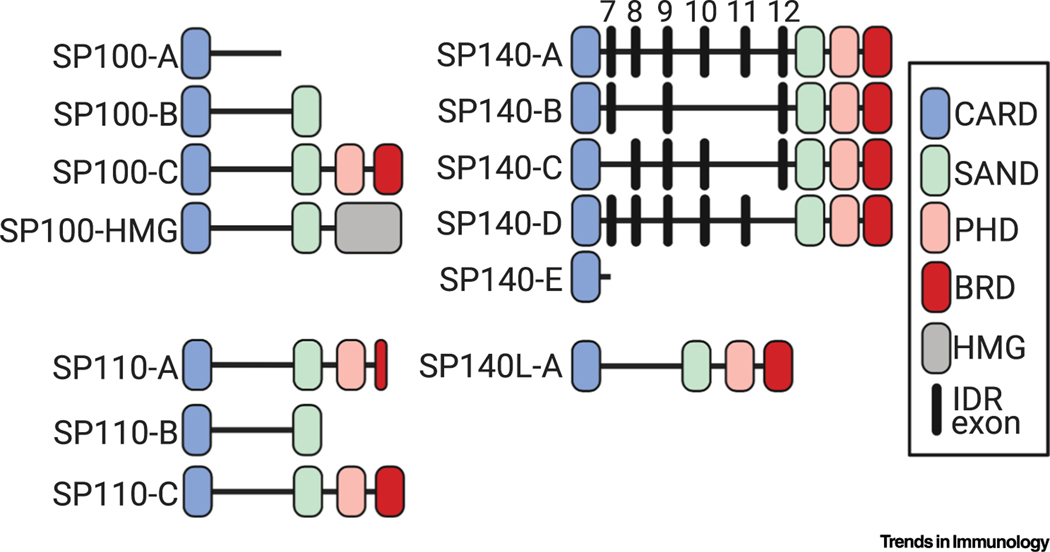
Figure 3. Outline of All Known Human Isoforms of Speckled Proteins (SPs).
Functional domains are color coded and indicated in the key: CARD, caspase activation and recruitment domain; SAND, Sp100, Aire, NucP41/75, DEAF-1 shared domain; PHD, plant homeodomain; BRD, bromodomain; HMG, high-mobility group protein domain. SP110 isoform A (SP110-A) contains a truncated BRD. In the SP140 alignment, most isoforms differ by exon usage in the intrinsically disordered region (IDR). Numbering above these IDR exons corresponds to exon numbering in full-length human SP140 isoform A (SP140-A). SP140L, SP140-like.
Both innate and adaptive immune cell lineages as well as non-immune cells express SP100 and SP110 [4], but SP140 is exclusively expressed in immune cells [14] (Figure 4). Human SP110 and SP140 were initially shown to be highly expressed in peripheral blood leukocytes and spleen [4,5,8]. As evidenced by RNA-seq data, all SPs are highly expressed in developing and mature B cells, activated CD4+ and CD8+ T cells, and myeloid lineages [14] (Figure 4). Moreover, SPs are interferon (IFN)-stimulated genes (ISGs) [6,14–17]. Of clinical relevance, SP110 and SP140 mutations associate with a number of heritable and acquired human immune disorders, such as Crohn’s disease (CD) [18,19], multiple sclerosis (MS) [20], susceptibility to infection [21–25], and immunodeficiency [26]. These disease associations underscore the vital role of the SP family for homeostatic immune cell identity and function (Table 1).
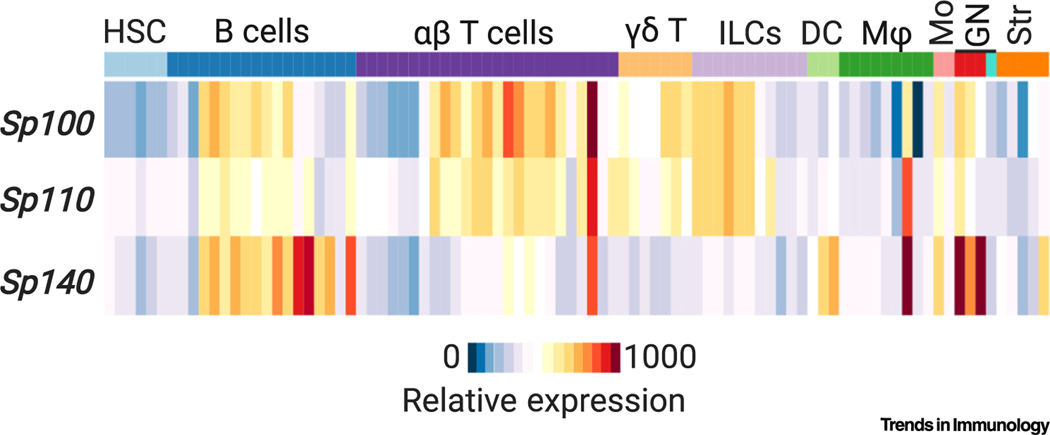
Figure 4. Expression of Speckled Proteins (SPs) in Murine Immune Cell Subsets.
Sp100, Sp110, and Sp140 expression in indicated mouse leukocytes based on RNA-seq data (Immgen.org)i. Abbreviations; DC, dendritic cell; GN, granulocyte; HSC, hematopoietic stem cell; ILCs, innate lymphoid cells; Mφ, macrophage; Mo, monocyte.
Table 1.
Summary of Disease Associations with SPs and Airea
| Gene | Human disease associations | Mouse disease associations | Refs |
|---|---|---|---|
| SP100 | Autoantigen in primary biliary cholangitis | Unknown | [3] |
| SP110 | VODI Mycobacterium tuberculosis (TB) |
Sst1S mice: ↑MTB (type I IFN and IL-1Ra dependent) ↑Listeria monocytogenes ↑Chlamydia pneumoniae |
[21–26,40,42–46] |
| SP140 | Autoantigen in primary biliary cholangitis MS CD CLL |
Hematopoietic Sp140 shRNA knockdown mice: ↑DSS-induced colitis Sst1S and Sp140−/− mice: ↑MTB (type I IFN and IL-1Ra dependent) ↑Listeria monocytogenes ↑Legionella pneumophila |
[14,18–20,27,30,40–46,91] |
| SP140L | Autoantigen in primary biliary cholangitis | Unknown | [1] |
| Aire | APS-1 |
Aire−/− mice: Multiorgan autoimmune disease |
[92–94] |
DSS, dextran sulfate sodium; B6, C57BL/6. The Sst1S allele derived from C3HeB/FeJ (C3H) mice contains mutations that confer loss of Sp110 [40] and Sp140 [41] expression. Most recent work implicates Sp140 as the causative gene for MTB susceptibility in mice [41]. Upward arrows denote increased disease associations.
The SP family has recently garnered interest for its role in chromatin and transcriptional regulation in immune cells during homeostasis, immunity to bacterial and viral infections, and the development of inflammatory diseases or immunodeficiency. Here, we discuss the nuclear localization of SPs and their chromatin interactions that orchestrate immune cell identity and function. It is also pertinent to note that SPs share high homology with autoimmune regulator (Aire) (Figure 1). We thus highlight advances in our understanding of Aire as a transcriptional regulator and draw parallels to inform future SP family studies.
Mutations in SPs Associate with Immunodeficiency, Inflammatory Diseases, and Infectious Disease in Humans
Patients with the autoimmune disease primary biliary cholangitis produce autoantibodies to SP100, SP140, and SP140L [1–3,27]. Human SP110 loss-of-function mutations associate with an immunodeficiency of the adaptive immune system: veno-occlusive disease with immunodeficiency (VODI) [26,28,29]. Furthermore, genome-wide association studies (GWASs) have identified human SP110 SNPs that associate with Mycobacterium tuberculosis (MTB) infection depending on disease subtype and genetic background [21–25]. The SP110 polymorphisms associated with MTB are unique from the insertions and deletions that lead to VODI, and the result of the MTB-associated polymorphisms on SP110 function is unknown. GWASs have also identified loss-of-function SNPs in SP140 that associate with three acquired immunological diseases: CD [18,19], chronic lymphocytic leukemia (CLL) [30], and MS [20] (Figure 1 and Table 1). Finally, many viruses have evolved mechanisms to inhibit SP function in PML-NBs, organelles associated with viral gene repression [31], suggesting that SPs mediate protective viral defense mechanisms. Therefore, it will be interesting to determine whether SP mutations that associate with inflammatory diseases are protective in the context of virus infection.
SP110 Mutations Result in VODI
Hepatic VODI is an autosomal-recessive primary immunodeficiency with a typical disease onset before age 6 months. VODI is associated with combined T and B cell immunodeficiencies, hypogammaglobulinemia, a lack of lymph node germinal centers and tissue plasma cells, and hepatic vascular occlusion and fibrosis [26]. VODI patients exhibit normal numbers of circulating mature B cells; however, they lack plasma cells and memory B cells [26]. In addition, VODI patients harbor reduced numbers of memory T cells in circulation and low T cell cytokine expression compared with healthy individuals [26]. Fine-mapping genetic studies of families afflicted by VODI identified early exon deletions and insertions in SP110 that lead to protein truncation [26,28,29] (Figure 1). Sp110 is expressed throughout adaptive immune cell lineages (Figure 4), so it may influence adaptive immunity via cell-intrinsic mechanisms. Whether mice that lack Sp110 have adaptive immune cell defects similar to those of patients carrying SP110 mutations is unknown. Nonetheless, despite the essential responsibility of SP110 for adaptive immune cell development and function, the mechanistic role of SP110 as a transcriptional regulator remains to be fully characterized.
SP140 SNPs Associate with CD, MS, and CLL
The top human SP140 SNPs identified by GWASs that associate with CD [18,19], MS [20], and CLL [30] are intronic but in perfect linkage disequilibrium (LD) with rs28445040, a synonymous SNP in exon 7 (Figure 1). Exon 7 lies in the intrinsically disordered region (IDR) (Box 1) that is exclusive to human SPs and is alternatively spliced into five protein-coding SP140 mRNA isoforms. The presence of the rs28445040 mutation in minigene assays led to the exclusion of exon 7 during splicing [32]. In agreement with this work, peripheral blood mononuclear cells (PBMCs) from CD patients homozygous for SP140 risk alleles (SNP+/+) exhibited upregulation of one SP140 isoform (SP140-C) (Figure 3) – devoid of exons 7 and 11 – in addition to down-regulation of all other SP140 isoforms, relative to SP140 expression in sex- and age-matched controls lacking SP140 SNPs (SNP−/−) [14]. SP140 SNP+/+ cells also displayed a pronounced loss of SP140 protein and a truncation in the remaining protein as shown by immunoblot for endogenous SP140. Expression of SP110, which shares a divergent promoter with SP140, was unchanged [14]. Thus, closely linked SP140 SNPs associated with CD, MS, and CLL all resulted in a loss of SP140 expression.
Box 1. IDRs in Phase-Separated Proteins.
SP140, and possibly other SPs, contain an IDR that is characteristic of proteins that are capable of liquid–liquid phase separation. Rather than tight ‘lock-and-key’ protein–protein interactions, IDRs mediate weak and flexible protein–protein interactions [95,96]. IDRs have evolved in higher organisms to perform a wider variety of tasks [97]. The IDR of SP140 developed in primates and is absent in mouse Sp140. Not only is phase separation crucial for signal transduction events in immune cells [98,99], but phase separation of transcriptional machinery also plays an important role during gene regulation [96,100]. Considering that a disease-associated mutation is found in the IDR of SP140 and may alter alternative splicing of the IDR, it will be of interest to determine the role of this mutation in SP140 phase separation, PML-NB localization, and transcriptional regulation.
The precise immunological outcome of reduced SP140 expression that contributes to three complex immune diseases is an active area of investigation. Hematopoietic depletion of Sp140 via inducible short hairpin (sh)RNA-mediated knockdown exacerbated intestinal inflammation in mice, thereby validating the GWAS association of SP140 with CD [14]. Moreover, SNP+/+ PBMCs from CD patients exhibited a suppressed innate immune signature that stratified them from other CD patients not carrying SP140 variants, as evidenced by unbiased RNA-seq and measurement of cytokine secretion upon Toll-like receptor (TLR) stimulation [14]. Thus, SP140 is likely to control intestinal homeostasis through the regulation of innate immune responses. Many inflammatory bowel disease (IBD)-associated genes (NOD2, AIM2, ATG16L1, GPR65) are essential for innate immunity and their loss or variation results in impaired barrier defense, decreased cytokine production, and an inability to maintain a gut microbial balance that ultimately enhances intestinal inflammation [33,34]. Future investigations will need to determine the causality of SP140 mutations in MS and CLL, the precise cell types affected, and whether there is a common mechanism by which SP140 loss drives pathology in three pathologically distinct immune diseases. Emerging insight implicates innate immune function and dysbiosis in the commensal ecosystem as risk factors for not only IBD [34] but also MS [35–38]. Furthermore, an association with CLL suggests that SP140 function in B cells is paramount for immune homeostasis. Despite a lack of evidence for the role of humoral immunity in driving MS pathogenesis, B cell depletion has therapeutic efficacy in MS patients [39]. Furthermore, many IBD-associated risk alleles (STAT4, BACH2, PRDM1) are enriched in B cells [34], but the pathogenic role of B cells in IBD remains a matter of debate [33]. Given the multifactorial nature of IBD, MS, and CLL, SP140 mutations will nonetheless constitute one of many factors that ultimately lead to disease susceptibility.
Role of SPs in Intracellular Pathogen Infections
Genetic Determinant of Intracellular Bacteria Resistance in Mice
The Super-susceptibility to tuberculosis (Sst1) locus, a mouse chromosome 1 region of approximately 50 genes including all SP genes and pseudogenes, was discovered as a genetic determinant of host susceptibility to MTB infection in C3HeB/FeJ (C3H) mice [40]. The Sst1 susceptible (Sst1s) allele derived from C3H mice confers loss of Sp110 and Sp140, whereas Sp100 is unaffected [40,41]. Expression of the Sst1s allele in mice leads to exacerbated MTB infection and rapid death, as well as necrotic lung inflammation during infection with virulent MTB (Table 1) [40,42,43]. Besides MTB infection, Listeria monocytogenes [44] and Chlamydia pneumoniae [45] burdens were also exacerbated in infected Sst1S mice. Reconstitution of Sst1S mice with a Sp110 (also known as Ipr1) transgene reduced the bacterial burdens of MTB and L. monocytogenes in lung and in bone marrow-derived macrophages (BMDMs) [40], suggesting that Sp110 mediates protection against intracellular bacterial infections. However, recent work demonstrated that Sp110−/− mice exhibited unaltered MTB bacterial burdens and survival rates compared with B6 wild-type (WT) controls [41]. By contrast, Sp140−/− mice were more susceptible to MTB, L. monocytogenes, and Legionella pneumophila infection [41]. By utilizing newly generated knockout mice, this recent study indicated that Sp140 might be the key MTB resistance gene in the Sst1 locus (Table 1).
Conditional deletions in mice are required to definitively determine the cell types involved in Sp110- or Sp140-mediated resistance to intracellular pathogen infections. However, recent work corroborates a likely role of the Sst1 locus in the type I IFN response; specifically, BMDMs derived from Sst1S mice or Sp140−/− mice upregulated IFN-β expression in vitro after tumor necrosis factor (TNF) stimulation [41,46] or a C. pneumoniae infection model [45], relative to WT cells. Furthermore, MTB-infected Sst1S and Sp140−/− mice exhibited upregulated lung IFN-β expression in vivo relative to WT mice [41,46]. Enhanced IFN production in Sst1S mice contributed to increased MTB susceptibility via the induction of Il1rn, which encodes interleukin (IL)-1 receptor antagonist (IL-1Ra). Blocking the IFN-α/β receptor, heterozygous deficiency of Il1rn, or antibody-mediated neutralization of IL-1Ra reversed bacterial burdens in the lungs of MTB-infected mice compared with Sst1S mice or mice that received PBS injections [46]. This observation is in line with extensive work suggesting that elevated type I IFN signaling is deleterious during MTB infection in mice and humans, as previously reviewed [47]. Thus, Sp140 expression is protective during intracellular bacterial infections by negatively regulating the induction of the type I IFN response [41]. Considering that these findings implicate Sp140 in type I IFN regulation, it is possible that Sp140 loss of function may be beneficial during viral defense – a concept that certainly merits further investigation.
SPs in PML-NB and Virus Defense Mechanisms
SPs each contain an NLS, so this family of proteins resides primarily in the nucleus. Confocal immunofluorescence has demonstrated that human SP100, SP110, and SP140 can localize to PML-NBs in human cell lines via endogenous or ectopic expression [4,5,8,9]. This subnuclear localization of SPs to PML-NBs may be dependent on cell type, cell cycle, or experimental conditions. The role of PML-NBs as sites of DNA and RNA virus replication restriction has been previously reviewed [31]. Many viruses have thus evolved proteins that target PML-NB components directly, including PML and SP100 [48–55]. For example, coexpression of human cytomegalovirus (HCMV) protein IE1 or the herpes simplex virus type 1 (HSV-1) protein ICP0 with SP100 in human cell lines resulted in reduced SUMOylation and proteasomal degradation of SP100 [49,56,57]. Of note, SP140 also contains SUMOylation sites [58] and SUMO-interacting motifs (SIMs); however, the importance of these features has not been investigated in the context of PML-NB localization or viral defense.
Ultimately, degradation of SP100 by viral proteins or experimental knockdown of SP100 in human cells results in enhanced viral gene expression and viral replication [49,59–61]. These results suggest that SP100 association with PML-NBs can restrict DNA virus replication, but it is unknown whether SP100 directly regulates viral gene transcription or acts via host DNA- or chromatin-binding capabilities. SP100 may bind to unmethylated CpG viral DNA [62] via its SAND domain [62,63] and contains a heterochromatin protein 1 alpha (HP1-α)-binding domain [61], suggesting that SP100 may act as a viral transcriptional repressor by binding to HP1-α and maintaining heterochromatin.
Finally, a co-immunoprecipitation study demonstrated that human SP140 interacted with the HIV-1 protein Vif in an infected HeLa cell line that expressed CD4 and SP140 [15], suggesting that SP140 might also play roles during viral infection, which remain to be assessed. Moreover, infection of HeLa-CD4 cells with HIV-1 resulted in dispersal of SP140 from the nucleus to the cytoplasm [15], consistent with other studies showing cytoplasmic localization of PML-NB components after HIV-1 infection [64,65]. Like HSV-1, HCMV, and other viruses, presumably, HIV-1 may have evolved effector proteins to interrupt the normal function of antiviral PML-NBs. Given that SPs are ISGs, whether SPs can restrict viruses by regulating viral transcription, host immune cell transcription, or both needs clarification. Furthermore, the relationship between SP localization to PML-NBs and their function as chromatin readers remains to be resolved.
SPs Maintain Gene Silencing for Cell Identity and Function
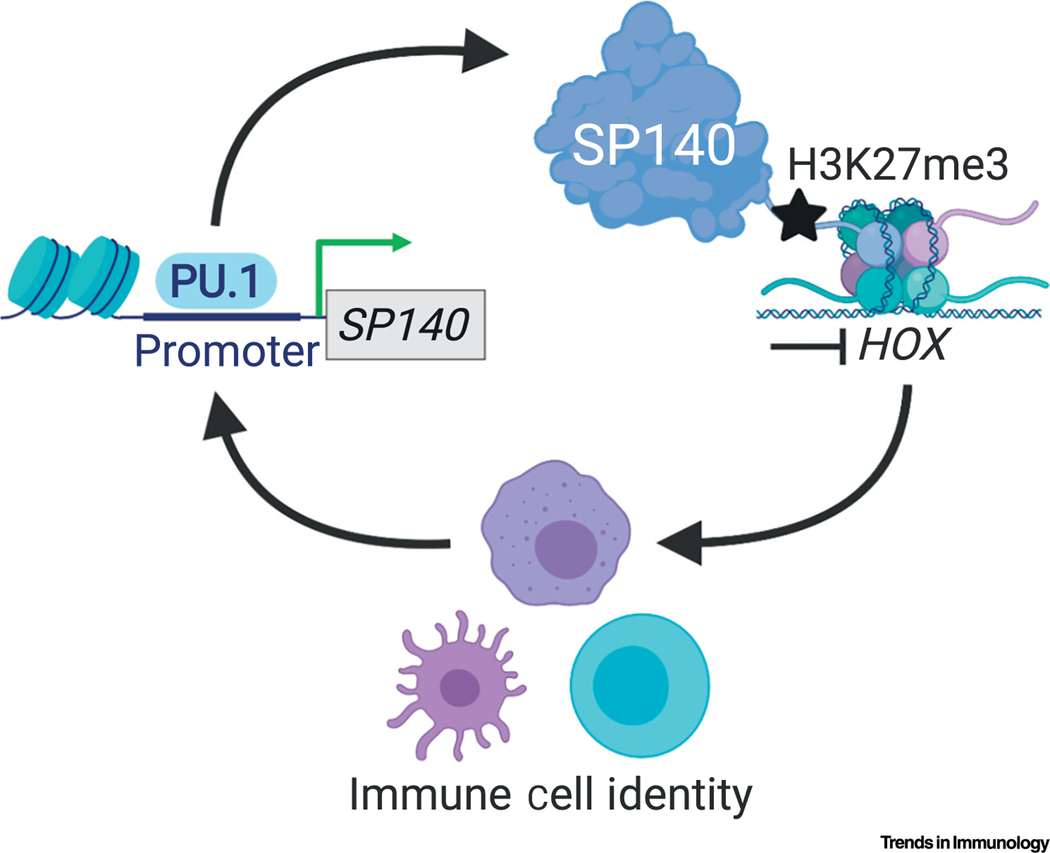
An nderstanding of the mechanism by which SPs regulate host transcription has emerged from recent work that identified SP140 as an essential orchestrator of macrophage transcriptional programs [14]. By occupying and repressing lineage-inappropriate genes, such as HOX. OLIG and FOX genes in human macrophages, SP140 ultimately maintains macrophage cell identity [14] (Figure 5). Using ChIP-seq, the majority of endogenous SP140 in primary human macrophages occupied promoters [14]. Furthermore, SP140 preferentially occupied heterochromatic regions with low chromatin accessibility and low transcriptional activity, indicated by high histone H3 lysine 27 trimethylation (H3K27me3) and low histone H3 lysine 4 trimethylation (H3K4me3), in human macrophages [14]. Given that siRNA-mediated knockdown of SP140 in human macrophages resulted in increased chromatin accessibility and upregulated expression of normally silenced lineage-inappropriate genes, such as HOX genes that were occupied by SP140 [14], SP140 appears to predominantly be a repressor of gene expression. Knockdown of SP140 in mouse or human macrophages resulted in severely impaired cytokine responses to microbial ligands as determined by RNA-seq and qPCR. This was deemed to be dependent on elevated expression of silenced lineage-inappropriate genes, such HOX, as ChIP-seq revealed that SP140 did not directly occupy cytokines in human macrophages, and double knockdown of SP140 and HOXA9 in human macrophages restored cytokine expression [14]. Thus, regulation of proinflammatory cytokines by SP140 is likely to occur through indirect mechanisms that regulate heterochromatin and cell state (Figure 5). PBMCs from CD patients with SP140 SNPs have been shown to exhibit reduced expression of many macrophage surface receptors compared with healthy controls and are hyporesponsive to both TLR4 and TLR3 activation in vitro [14]. SP140 is also abundantly expressed in B cells (Figure 4) and suppression of HOX expression is a key determinant for B cell development [66]; therefore, it is feasible that SP140 might also control B cell identity through similar repressive mechanisms. Nevertheless, these possibilities warrant further investigation.
Figure 5. Mechanism of Action to date for Speckled Protein 140 (SP140) for Immune Cell Identity and Function.
Lineage-defining transcription factors (TFs) for mature macrophages (such as PU.1) drive SP140 transcription. SP140 in turn docks to repressed chromatin marked by H3K27me3 and maintains the silencing of lineage-inappropriate genes, such as HOX, OLIG, and FOX, for the preservation of cell identity and function.
SPs and Aire Have Divergent Roles in Transcriptional Regulation
SPs and Aire are highly homologous proteins (Figure 1). Thus, expansive work on the structurally similar protein Aire may offer suggestions for how SPs are recruited to chromatin, interact with protein complexes, and regulate transcription. Mass spectrometry and many other experiments in Aire-expressing HEK293T cells, in addition to ChIP-seq and assay for transposase-accessible chromatin sequencing (ATAC-seq) studies of mouse medullary thymic epithelial cells (mTECs), have culminated in a model whereby Aire is recruited to superenhancers and interacts with topoisomerase 1 (TOP1); this in turn promotes the recruitment of DNA-damage response proteins (e.g., DNA-PK), topoisomerase 2 (TOP2), and other transcriptional regulators (e.g., RNA-pol II) to stabilize enhancer RNA (eRNA) transcription, depletes nucleosomes, and relieves DNA torsional stress [67,68]. Ultimately, Aire localization and protein complex formation at superenhancers promotes transcription elongation at peripheral tissue antigen (PTA) transcription start sites (TSSs) via chromosomal looping [67,69,70] and the localization of Aire specks (PML negative) to the nuclear matrix [71].
Although SPs share high sequence homology with Aire, they may have opposite modes of action. SP140 appears to lock chromatin in an inactive state, particularly at TSSs, to prevent the expression of silenced lineage-inappropriate genes in macrophages [14] (Figure 5). Conversely, Aire promotes transcription, particularly by activating superenhancers [67]. These differences in function may involve defined reading of post-translationally modified histones by SPs and Aire. However, localization to PML-NBs may also distinguish their function, since, unlike SP proteins [4,9], Aire does not localize to PML-NBs [71], which are known sites of transcriptional repression [31]. How other SPs compare with SP140 in terms of chromatin associations and transcriptional regulation is unclear and further work is necessary to determine whether SPs and Aire associate with similar or distinct protein complexes.
SP CARD for Multimerization
Chromatin ‘readers’ contain structurally diverse protein domains that precisely recognize and dock to covalent modifications of histones, DNA, or transcription factors. SP family members contain the unique combination of a CARD, SAND, PHD, and BRD (Figure 1). The N termini of SPs contain a previously termed ‘HSR’ domain that contains homology to the CARD domain of other proteins, including Aire (Figure 1 and Figure 6). The role of SP CARD domains is a nascent area of investigation, but the field may be able to hypothesize its function based on Aire and other CARD-containing proteins demonstrating a role for the CARD in forming large protein complexes. The CARD domain of Aire is a conserved region where missense mutations are present in some autoimmune polyendocrine syndrome type 1 (APS-1) patients [72,73]. Mutations, such as L28P (Figure 1), in the CARD domain of Aire prevent homodimerization and punctate formation of Aire, as well as PTA transcription [72,74]. The L28 residue is positioned in the hydrophobic core of the CARD domain; hence, a mutation here may affect protein stabilization [72]. However, other mutations that occur on the outside surface of the CARD tertiary structure may affect protein–protein interactions [72]. Further work is necessary to determine whether SP CARDs are involved in homodimerization and it is unknown whether SP family members can heterodimerize if they are coexpressed. Furthermore, it will be important to determine whether individuals that are heterozygous for disease-associated SP mutations have impaired immune cell function, considering that one protein molecule may ‘poison’ dimer functionality similar to Aire dominant-negative mutations [75,76].
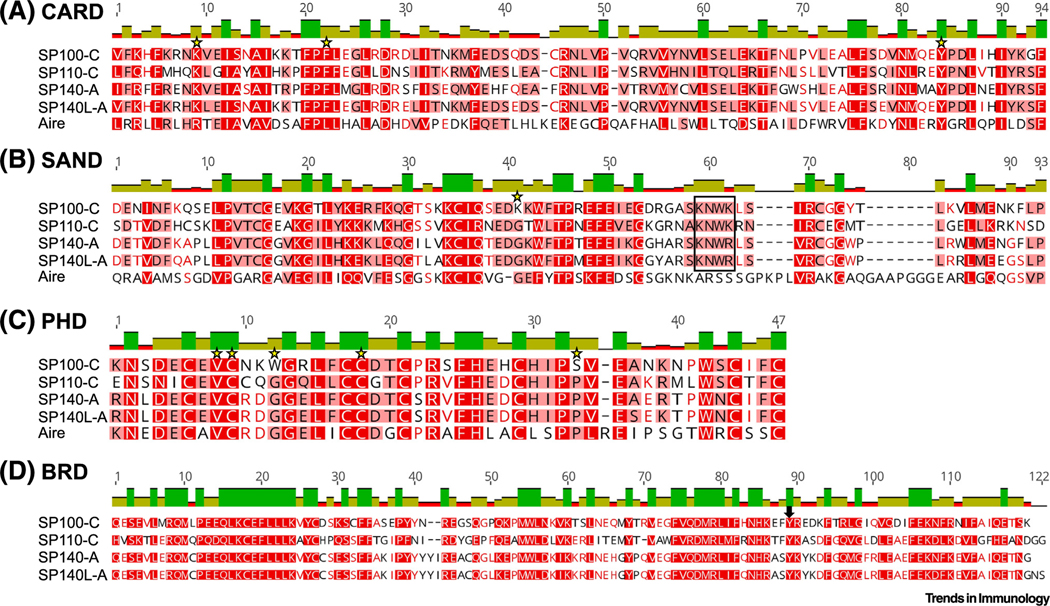
Figure 6. Alignments of Functional Domains of Human Speckled Protein (SP) Isoforms and Autoimmune Regulator (Aire).
Sequences were aligned in Geneious using ClustalW. Red shading of residue and bar graphs above sequences corresponds to percentage identity at each residue. Stars indicate conserved residues that are mutated in Aire in patients with autoimmune polyendocrine syndrome type 1 (APS-1). In (B), a black box indicates the KDWK-like DNA-binding motif [11]. In (D), the arrow points to the position where SPs lack a conserved asparagine residue previously reported to anchor bromodomains to histone tail acetyl groups via hydrogen bonds [13]. Abbreviations: BRD, bromodomain; CARD, caspase activation and recruitment domain; PHD, plant homeodomain; SAND, Sp100, Aire, NucP41/75, DEAF-1 shared domain; SP140L, SP140-like protein; SP100-C, SP100 isoform C.
SPs Contain the Rare SAND DNA-Binding Domain
The SAND domain (named after a few proteins that contain it: Sp100, Aire, NucP41/P75, and DEAF-1) was originally identified as a gene encoded by the yeast Saccharomyces cerevisiae. Later, it was also found in proteins expressed in Caenorhabditis elegans, Drosophila melanogaster, and the plant Arabidopsis thaliana [77,78]. For example, the SAND domain-containing protein ULT1 functions as a chromatin remodeling factor that activates genes in plants [78]. Moreover, DEAF-1 regulates innate immunity in Drosophila via the activation of Toll pathway target genes [79,80]. Human NUDR, Drosophila DEAF-1, and rat GMEB were shown to bind to DNA in a sequence-dependent manner via a conserved KDWK amino acid motif in their SAND domains [81–84]. Likewise, mouse and human SPs contain a similar KNWK motif (Figure 6), and the human SP100-B SAND domain can bind double-stranded DNA via the positively charged KNWK fold [11]. Unlike SPs, the Aire SAND domain does not contain a KDWK-like motif and is not thought to bind DNA but rather, directs protein–protein interactions [12,75]. Nevertheless, it is possible that SP SANDs are also needed for protein–protein interactions.
Chromatin Reader Domains: PHD and BRD
SP functional protein domains include a PHD that reads histone methylation and a BRD that reads histone acetylation [13] (Figure 1). An early study that solved the structure of the human SP140 PHD determined that it did not bind to H3K4me0 or an array of modified or unmodified histone peptides [85]. However, a later study showed that the expression of SP140’s full C terminus comprising the PHD and BRD enabled binding to H3K4me0 [86]. Similarly, Sp100-C was able to bind to H3K4me0 as well as to H3K9me3, H3T3ph, and H3S10ph and was crystallized with the heterochromatin-associated H3K9me3 peptide [86]; this suggested that SP PHDs enable docking to inactive chromatin regions. Of note, SP family members contain one PHD domain that is similar in sequence to the PHD1 of Aire (Figure 6) that was also shown to preferentially bind to unmethylated H3K4me0 in a histone peptide microarray and peptide pulldown assays [86–90]. Furthermore, the two sides of Aire’s folded PHD1 seem to have different functions of H3K4me0 recognition versus protein–protein interactions, as determined by the location of loss-of-function mutations in APS-1 patients [89].
In addition to binding H3K4me0, the PHD of human SP140 was recently shown to mediate SUMOylation of the adjacent BRD [58]. Canonical BRDs bind acetylated histones and the BRD of SP140 was shown to bind promiscuously to acetylated H3 or H4 histone peptides [13], but human SP100-C showed minimal binding to acetylated H3 or H4 peptides [86]. All SP BRDs lack a canonical tyrosine residue found in most BRD-containing proteins, rendering this BRD more atypical (Figure 6). Furthermore, since SP BRDs can be SUMOylated [58], their BRD may also be necessary for interaction with PML-NBs. Thus, it possible that the BRD of SPs may not be utilized for docking to histone acetylation that is normally found at active and open chromatin but rather, for localization to PML-NBs that are sites of repression. However, this hypothesis will evidently require formal testing.
Concluding Remarks
In summary, the SP family represents a novel class of chromatin regulators that mediate gene silencing with designated roles in immune cells. All SPs are associated with autoimmune, inflammatory, or infectious diseases in humans, underscoring their essential role in maintaining immune homeostasis and proper functional responses to pathogens. Nonetheless, we have only begun to understand the precise role of these proteins in the immune system: SP100 may mediate host defense mechanisms against DNA virus infections, SP110 or SP140 play roles in restricting intracellular pathogen infections, and SP140 orchestrates macrophage transcriptional programs to maintain cellular identity. Knowledge of their role in adaptive immune cells, in which they are abundant, is currently completely lacking. Although SPs share high homology, individual SPs are likely to play distinct (and perhaps nonredundant) roles in transcriptional regulation by virtue of their unique expression profiles and association with distinct diseases. However, currently there are limited mouse models of individual SPs due to both the infancy of the field and the repetitive nature of SP loci. Nonetheless, the low homology of mouse and human SPs does emphasize the need to examine human SPs. Furthermore, it will be necessary to consider the role of various SP mRNA spliced isoforms (see Outstanding Questions). Importantly, studying the mechanisms of SPs in immune cells will provide an understanding of how epigenetic alterations can contribute to immune disorders and will aid in the design of new putative therapies for those diseases associated with SP loss. Furthermore, given the success of targeting more ubiquitous chromatin ‘reader’ proteins for therapeutic benefit, immune-expressed SP proteins may offer novel and more refined therapeutic avenues for taming hyperactive immune responses through disruption of immune cell states.
Outstanding Questions.
What is the function of individual speckled proteins (SPs) in regulating immune cell identity and function? Is there redundancy between SP family members in terms of functionality?
Given that SPs are ISGs and localize to promyelocytic (PML) nuclear bodies (NBs), what is the role of SPs in antiviral defense?
What is the role of SPs in the adaptive immune system?
What are the mechanisms by which individual SPs regulate chromatin dynamics and gene transcription, and which proteins associate with SPs during these events?
What is the function of SP caspase activation and recruitment (CARD) domains?
How does the unique combination of a SAND, a plant homeodomain, and a bromodomain contribute to SP function?
What is the relationship of SPs to AIRE? If AIRE is expressed in cells outside the thymus, does AIRE interact or compete with SPs?
Does the human SP140 intrinsically disordered region (IDR) mediate liquid–liquid phase separation of SP140 and how does alternative splicing of the IDR alter function?
Is the alternative splicing of SPs differentially regulated in various cell lineages and between cell states?
Do SPs localize to PML-NBs in all immune cells? How does PML-NB localization affect the ability of SPs to regulate host chromatin organization and transcription?
How does SP140 dysfunction lead to diseases as diverse as CD, CLL, and MS?
Do SPs mediate homeostatic interactions between innate immunity at mucosae or skin and commensal microbes?
Are SPs druggable like other chromatin readers? What chromatin reader domain should be targeted?
By virtue of their localization to PML-NBs, SPs have been implicated in transcriptional repression of host or viral genomes. The function of SP140 was recently elucidated showing that it occupies heterochromatin and represses lineage-inappropriate genes such as HOX to maintain macrophage identity in mice and humans.
Highlights.
The speckled protein (SP) family of chromatin ‘readers’ in humans comprises SP100, SP110, SP140, and SP140L.
The mouse SP family comprises Sp100, Sp110, and Sp140 and is only ~45% homologous to human SPs at the amino acid level, necessitating both mouse and human studies of SPs.
SPs are highly expressed in innate and adaptive immune cells, with SP140 being immune restricted. SPs are also interferon (IFN)-stimulated genes (ISGs).
Mutations in human SP140 associate with three immunological diseases: Crohn’s disease, chronic lymphocytic leukemia, and multiple sclerosis. Mutations in human SP110 associate with veno-occlusive disease with immunodeficiency (VODI). Mouse SPs can act as determinants of resistance to intracellular pathogen infections such as Mycobacterium tuberculosis.
The SP family members possess a SAND domain, a plant homeodomain (PHD), and a caspase activation and recruitment domain (CARD) that share high homology with these domains found in autoimmune regulator (Aire); these domains suggest that SPs can bind to DNA directly, ‘read’ histone methylation status, and multimerize, respectively. In addition, SPs contain a bromodomain (BRD) that may read histone acetylation status.
Human SPs localize to promyelocytic leukemia (PML) nuclear bodies (NBs) in human cell lines – dynamic nuclear protein aggregates interspersed between chromatin that can measure up to 2 μm in diameter. The presence of an intrinsically disordered region (IDR) exclusively in human SPs suggest that they may phase separate.
Acknowledgments
We apologize to members of the community whose studies were not cited herein because of space constraints. This study was supported by National Institutes of Health (NIH) grant R01DK119996 (to K.L.J).
Glossary
- Assay for transposase-accessible chromatin sequencing (ATAC-seq)
technique for determining genome-wide chromatin accessibility characterized by a lack of nucleosome occupancy
- Autoimmune polyendocrine syndrome type 1 (APS-1)
multiorgan autoimmune disease caused by Aire mutations; characterized by autoimmune infiltrates and autoantibodies in multiple endocrine organs
- Autoimmune regulator (Aire)
transcriptional regulator of PTA genes in medullary thymic epithelial cells
- ChIP-seq
technique for identifying genome-wide DNA-binding sites of proteins
- Chronic lymphocytic leukemia (CLL)
type of cancer that usually develops in older adults; characterized by the abnormal expansion of a clonal population of B cells
- Crohn’s disease (CD)
type of IBD; causes chronic inflammation in the gastrointestinal tract
- Histone H3 lysine 4 trimethylation (H3K4me3)
associated with active or poised promoters
- Histone H3 lysine 27 trimethylation (H3K27me3)
associated with transcriptionally repressed regions
- Interferon (IFN)-stimulated genes (ISGs)
genes whose expression is induced by type I, II, or III IFN signaling
- Linkage disequilibrium (LD)
SNPs in LD will segregate together in individuals during population dynamics
- Multiple sclerosis (MS)
autoimmune disease in which T cells react to selfmyelin antigens and cause inflammation in the central nervous system as well as destruction of the protective myelin sheath covering nerve fibers, ultimately resulting in neurological deficits
- Peripheral tissue antigen (PTA)
normally expressed in a specific peripheral organ; ectopic expression in medullary thymic epithelial cells allows thymic T cell selection
- Primary biliary cholangitis
previously known as primary biliary cirrhosis; autoimmune disease of the liver
- Promyelocytic leukemia (PML) nuclear bodies (NBs)
nuclear organelles regulated by a variety of stimuli such as viral infection and DNA damage; implicated in a range of cellular processes, including higher-order chromatin organization and gene repression; viewed as specks via confocal immunofluorescence microscopy and dependent on PML expression
- SUMOylation
post-translational modification that stabilizes PML-NBs
- Superenhancers
long chromatin regions densely occupied by general and cell-type-specific transcriptional factors that drive high-level expression of target genes
- Toll-like receptors (TLRs)
family of surface and endosomal pattern-recognition receptors that detect microbial products; TLR3 senses double-stranded RNA; TLR4 senses bacterial lipopolysaccharide (LPS)
- Topoisomerases
enzymes that cleave and reseal DNA strands to relieve topological strain from DNA supercoils
- Type I interferon (IFN)
subgroup of cytokines including IFN-α and IFN-β. They function to prevent viral replication in cells
- Unmethylated CpG DNA
‘CpG’ is shorthand for the occurrence of a cytosine linked, through a phosphate bond, to a guanine. In humans, 70–80% of cytosines in CpGs are methylated, which silences genes. Unmethylated CpGs are often a signature of pathogen-associated DNA
- Veno-occlusive disease with immunodeficiency (VODI)
autosomal-recessive primary immunodeficiency with typical disease onset before 6 months of age
Footnotes
Resources
References
- 1.Saare M. et al. (2015) SP140L, an evolutionarily recent member of the SP100 family, is an autoantigen in primary biliary cirrhosis. J. Immunol. Res 2015, 526518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szostecki C. et al. (1990) Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J. Immunol 145, 4338–4347 [PubMed] [Google Scholar]
- 3.Szostecki C. et al. (1987) Autoimmune sera recognize a 100 kD nuclear protein antigen (sp-100). Clin. Exp. Immunol 68, 108–116 [PMC free article] [PubMed] [Google Scholar]
- 4.Bloch DB et al. (1996) Identification and characterization of a leukocyte-specific component of the nuclear body. J. Biol. Chem 271, 29198–29204 [DOI] [PubMed] [Google Scholar]
- 5.Dent AL et al. (1996) LYSP100-associated nuclear domains (LANDs): description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood 88, 1423–1426 [PubMed] [Google Scholar]
- 6.Grötzinger T. et al. (1996) A highly amplified mouse gene is homologous to the human interferon-responsive Sp100 gene encoding an autoantigen associated with nuclear dots. Mol. Cell. Biol 16, 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadereit S. et al. (1993) Molecular cloning of two new interferon-induced, highly related nuclear phosphoproteins. J. Biol. Chem 268, 24432–24441 [PubMed] [Google Scholar]
- 8.Bloch DB et al. (2000) Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol. Cell. Biol 20, 6138–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloch DB et al. (1999) Structural and functional heterogeneity of nuclear bodies. Mol. Cell. Biol 19, 4423–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta S. and Jeffrey KL (2015) Beyond receptors and signaling: epigenetic factors in the regulation of innate immunity. Immunol. Cell Biol 93, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottomley MJ et al. (2001) The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol 8, 626–633 [DOI] [PubMed] [Google Scholar]
- 12.Waterfield M. et al. (2014) The transcriptional regulator Aire coopts the repressive ATF7ip–MBD1 complex for the induction of immunotolerance. Nat. Immunol 15, 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filippakopoulos P. et al. (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta S. et al. (2017) Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140. Sci. Immunol 2, eaag3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madani N. et al. (2002) Implication of the lymphocyte-specific nuclear body protein Sp140 in an innate response to human immunodeficiency virus type 1. J. Virol 76, 11133–11138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadler M. et al. (1995) Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11, 2565–2573 [PubMed] [Google Scholar]
- 17.Guldner HH et al. (1992) IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J. Immunol 149, 4067–4073 [PubMed] [Google Scholar]
- 18.Jostins L. et al. (2012) Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke A. et al. (2010) Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet 42, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International Multiple Sclerosis Genetics Consortium (IMSGC) et al. (2013) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet 45, 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox GJ et al. (2014) Polymorphisms of SP110 are associated with both pulmonary and extra-pulmonary tuberculosis among the Vietnamese. PLoS One 9, e99496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tosh K. et al. (2006) Variants in the SP110 gene are associated with genetic susceptibility to tuberculosis in West Africa. Proc. Natl. Acad. Sci. U. S. A 103, 10364–10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abhimanyu et al. (2011) Genetic association study suggests a role for SP110 variants in lymph node tuberculosis but not pulmonary tuberculosis in north Indians. Hum. Immunol 72, 576–580 [DOI] [PubMed] [Google Scholar]
- 24.Babb C. et al. (2007) SP110 polymorphisms are not associated with pulmonary tuberculosis in a South African population. Hum. Genet 121, 521–522 [DOI] [PubMed] [Google Scholar]
- 25.Leu J-S et al. (2017) SP110b controls host immunity and susceptibility to tuberculosis. Am. J. Respir. Crit. Care Med 195, 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roscioli T. et al. (2006) Mutations in the gene encoding the PML nuclear body protein Sp110 are associated with immunodeficiency and hepatic veno-occlusive disease. Nat. Genet. 38, 620–622 [DOI] [PubMed] [Google Scholar]
- 27.Granito A. et al. (2010) PML nuclear body component Sp140 is a novel autoantigen in primary biliary cirrhosis. Am. J. Gastroenterol 105, 125–131 [DOI] [PubMed] [Google Scholar]
- 28.Wang T. et al. (2012) Hepatic veno-occlusive disease with immunodeficiency (VODI): first reported case in the U.S. and identification of a unique mutation in Sp110. Clin. Immunol 145, 102–107 [DOI] [PubMed] [Google Scholar]
- 29.Cliffe ST et al. (2012) Clinical, molecular, and cellular immunologic findings in patients with SP110-associated veno-occlusive disease with immunodeficiency syndrome. J. Allergy Clin. Immunol 130, 735–742.e6 [DOI] [PubMed] [Google Scholar]
- 30.Sillé FCM et al. (2012) Post-GWAS functional characterization of susceptibility variants for chronic lymphocytic leukemia. PLoS One 7, e29632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scherer M. and Stamminger T. (2016) Emerging role of PML nuclear bodies in innate immune signaling. J. Virol 90, 5850–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matesanz F. et al. (2015) A functional variant that affects exon-skipping and protein expression of SP140 as genetic mechanism predisposing to multiple sclerosis. Hum. Mol. Genet 24, 5619–5627 [DOI] [PubMed] [Google Scholar]
- 33.Uhlig HH and Powrie F. (2018) Translating immunology into therapeutic concepts for inflammatory bowel disease. Annu. Rev. Immunol 36, 755–781 [DOI] [PubMed] [Google Scholar]
- 34.Graham DB and Xavier RJ (2020) Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 578, 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berer K. et al. (2017) Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. U. S. A 114, 10719–10724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cekanaviciute E. et al. (2017) Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U. S. A 114, 10713–10718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.International Multiple Sclerosis Genetics Consortium (2019) Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duscha A. et al. (2020) Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell 180, 1067–1080.e16 [DOI] [PubMed] [Google Scholar]
- 39.Häusser-Kinzel S. and Weber MS (2019) The role of B cells and antibodies in multiple sclerosis, neuromyelitis optica, and related disorders. Front. Immunol 10, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan H. et al. (2005) Ipr1 gene mediates innate immunity to tuberculosis. Nature 434, 767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji DX et al. (2020) Role of the transcriptional regulator SP140 in resistance to bacterial infections via repression of type I interferons. bioRxiv. Published online January 8, 2020. 10.1101/2020.01.07.897553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kramnik I. et al. (2000) Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A 97, 8560–8565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pichugin AV et al. (2009) Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. Am. J. Pathol 174, 2190–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyartchuk V. et al. (2004) The host resistance locus sst1 controls innate immunity to Listeria monocytogenes infection in immunodeficient mice. J. Immunol 173, 5112–5120 [DOI] [PubMed] [Google Scholar]
- 45.He X. et al. (2013) The sst1 resistance locus regulates evasion of type I interferon signaling by Chlamydia pneumoniae as a disease tolerance mechanism. PLoS Pathog. 9, e1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji DX et al. (2019) Type I interferon-driven susceptibility to Mycobacterium tuberculosis is mediated by IL-1Ra. Nat. Microbiol 4, 2128–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreira-Teixeira L. et al. (2018) Type I interferons in tuberculosis: foe and occasionally friend. J. Exp. Med 215, 1273–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Everett RD et al. (2013) The replication defect of ICP0-null mutant herpes simplex virus 1 can be largely complemented by the combined activities of human cytomegalovirus proteins IE1 and pp71. J. Virol 87, 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tavalai N. et al. (2011) Evidence for a dual antiviral role of the major nuclear domain 10 component Sp100 during the immediate-early and late phases of the human cytomegalovirus replication cycle. J. Virol 85, 9447–9458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y-E et al. (2011) Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J. Virol 85, 11928–11937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walters MS et al. (2010) The RING finger domain of varicella-zoster virus ORF61p has E3 ubiquitin ligase activity that is essential for efficient autoubiquitination and dispersion of Sp100-containing nuclear bodies. J. Virol 84, 6861–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Full F. et al. (2012) Herpesvirus saimiri antagonizes nuclear domain 10-instituted intrinsic immunity via an ORF3-mediated selective degradation of cellular protein Sp100. J. Virol 86, 3541–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desbois C. et al. (1996) Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science 273, 951–953 [DOI] [PubMed] [Google Scholar]
- 54.Szekely L. et al. (1996) The Epstein–Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J. Virol 70, 2562–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoppe A. et al. (2006) Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J. Virol 80, 3042–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chelbi-Alix MK and de The H. (1999) Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18, 935–941 [DOI] [PubMed] [Google Scholar]
- 57.Müller S. and Dejean A. (1999) Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol 73, 5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zucchelli C. et al. (2019) Sp140 is a multi-SUMO-1 target and its PHD finger promotes SUMOylation of the adjacent bromodomain. Biochim. Biophys. Acta 1863, 456–465 [DOI] [PubMed] [Google Scholar]
- 59.Lee H-R et al. (2004) Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol 78, 6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Everett RD et al. (2008) Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol 82, 2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berscheminski J. et al. (2014) Sp100 isoform-specific regulation of human adenovirus 5 gene expression. J. Virol 88, 6076–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isaac A. et al. (2006) SP100B, a repressor of gene expression preferentially binds to DNA with unmethylated CpGs. J. Cell. Biochem 98, 1106–1122 [DOI] [PubMed] [Google Scholar]
- 63.Negorev DG et al. (2006) Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J. Virol 80, 8019–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dutrieux J. et al. (2015) PML/TRIM19-dependent inhibition of retroviral reverse-transcription by Daxx. PLoS Pathog. 11, e1005280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kahle T. et al. (2015) TRIM19/PML restricts HIV infection in a cell type-dependent manner. Viruses 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang M. et al. (2018) Transcription factor Hoxb5 reprograms B cells into functional T lymphocytes. Nat. Immunol 19, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bansal K. et al. (2017) The transcriptional regulator Aire binds to and activates super-enhancers. Nat. Immunol 18, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abramson J. et al. (2010) Aire’s partners in the molecular control of immunological tolerance. Cell 140, 123–135 [DOI] [PubMed] [Google Scholar]
- 69.Meredith M. et al. (2015) Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat. Immunol 16, 942–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brennecke P. et al. (2015) Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat. Immunol 16, 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akiyoshi H. et al. (2004) Subcellular expression of autoimmune regulator is organized in a spatiotemporal manner. J. Biol. Chem 279, 33984–33991 [DOI] [PubMed] [Google Scholar]
- 72.Ferguson BJ et al. (2008) AIRE’s CARD revealed, a new structure for central tolerance provokes transcriptional plasticity. J. Biol. Chem 283, 1723–1731 [DOI] [PubMed] [Google Scholar]
- 73.Anderson MS and Su MA (2016) AIRE expands: new roles in immune tolerance and beyond. Nat. Rev. Immunol 16, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guha M. et al. (2017) DNA breaks and chromatin structural changes enhance the transcription of autoimmune regulator target genes. J. Biol. Chem 292, 6542–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Su MA et al. (2008) Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J. Clin. Invest 118, 1712–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oftedal BE et al. (2015) Dominant mutations in the autoimmune regulator AIRE are associated with common organ-specific autoimmune diseases. Immunity 42, 1185–1196 [DOI] [PubMed] [Google Scholar]
- 77.Cottage A. et al. (2004) Molecular characterisation of the SAND protein family: a study based on comparative genomics, structural bioinformatics and phylogeny. Cell. Mol. Biol. Lett 9, 739–753 [PubMed] [Google Scholar]
- 78.Carles CC and Fletcher JC (2009) The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 23, 2723–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reed DE et al. (2008) DEAF-1 regulates immunity gene expression in Drosophila. Proc. Natl. Acad. Sci. U. S. A 105, 8351–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuttenkeuler D. et al. (2010) A large-scale RNAi screen identifies Deaf1 as a regulator of innate immune responses in Drosophila. J. Innate Immun 2, 181–194 [DOI] [PubMed] [Google Scholar]
- 81.Oshima H. et al. (1995) The factor binding to the glucocorticoid modulatory element of the tyrosine aminotransferase gene is a novel and ubiquitous heteromeric complex. J. Biol. Chem 270, 21893–21901 [DOI] [PubMed] [Google Scholar]
- 82.Gross CT and McGinnis W. (1996) DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO J. 15, 1961–1970 [PMC free article] [PubMed] [Google Scholar]
- 83.Christensen J. et al. (1999) Two new members of the emerging KDWK family of combinatorial transcription modulators bind as a heterodimer to flexibly spaced PuCGPy half-sites. Mol. Cell. Biol 19, 7741–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huggenvik JI et al. (1998) Characterization of a nuclear Deformed epidermal autoregulatory factor-1 (DEAF-1)-related (NUDR) transcriptional regulator protein. Mol. Endocrinol 12, 1619–1639 [DOI] [PubMed] [Google Scholar]
- 85.Zucchelli C. et al. (2014) Structure of human Sp140 PHD finger: an atypical fold interacting with Pin1. FEBS J. 281, 216–231 [DOI] [PubMed] [Google Scholar]
- 86.Zhang X. et al. (2016) Multifaceted histone H3 methylation and phosphorylation readout by the plant homeodomain finger of human nuclear antigen Sp100C. J. Biol. Chem 291, 12786–12798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Org T. et al. (2008) The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chignola F. et al. (2009) The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res. 37, 2951–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaetani M. et al. (2012) AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome. Nucleic Acids Res. 40, 11756–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koh AS et al. (2008) Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc. Natl. Acad. Sci. U. S. A 105, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Di Bernardo MC et al. (2008) A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet 40, 1204–1210 [DOI] [PubMed] [Google Scholar]
- 92.Anderson MS et al. (2002) Projection of an immunological self shadow within the thymus by the Aire protein. Science 298, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 93.Finnish-German APECED Consortium (1997) An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet 17, 399–403 [DOI] [PubMed] [Google Scholar]
- 94.Nagamine K. et al. (1997) Positional cloning of the APECED gene. Nat. Genet 17, 393–398 [DOI] [PubMed] [Google Scholar]
- 95.Shin Y. and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 [DOI] [PubMed] [Google Scholar]
- 96.Hnisz D. et al. (2017) A phase separation model for transcriptional control. Cell 169, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao A. et al. (2018) Evolution of weak cooperative interactions for biological specificity. Proc. Natl. Acad. Sci. U. S. A 115, E11053–E11060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Su X. et al. (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Du M. and Chen ZJ (2018) DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sabari BR et al. (2018) Coactivator condensation at superenhancers links phase separation and gene control. Science 361, eaar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]