Abstract
Most currently used conventional influenza vaccines are based on 1940s technology. Advances in vaccine immunogen design and delivery emerging over the last decade promise new options for improving influenza vaccines. In addition, new technologies for immune profiling provide better-defined immune correlates of protection and precise surrogate biomarkers for vaccine evaluations. Major technological advances include single-cell analysis, high-throughput antibody discovery, next-generation sequencing of antibody gene transcripts, antibody ontogeny, structure-guided immunogen design, nanoparticle display, delivery and formulation options, and better adjuvants. In this review, we provide our prospective outlook for improved influenza vaccines in the foreseeable future.
PAST, PRESENT, AND FUTURE INFLUENZA VACCINES
The discovery of the causative agent of swine influenza was reported in 1931 by Shope (Shope 1931) and was quickly followed by the identification of human influenza virus by Smith et al. in 1933 (Smith et al. 1933). Woodruff and Goodpasture made another major virology breakthrough in 1931 when they documented a virus cultivation method utilizing fertilized chick embryos (Goodpasture et al. 1931). In 1936, Smorodintseff developed the first attenuated influenza virus vaccine using embryonated eggs, and soon after, this approach was further refined by Francis and Salk (Francis and Salk 1942). Notably, Salk's development of an effective polio vaccine in 1952 was possible because of his experience with influenza vaccine research. The first egg-based influenza vaccine was licensed in 1945, and it astonishingly remains as the basis for the majority of influenza vaccines produced today. In 2007, the European Medicines Agency (EMA) approved the first non-egg-derived influenza vaccine for human use, made in the Madin–Darby canine kidney (MDCK) cells (Optaflu), and 5 years later the same vaccine (renamed Flucelvax) was approved by the Food and Drug Administration (FDA) in the United States. Although the initial method of virus production differs, these cell culture–derived vaccines are ultimately processed in the same way as egg-based vaccines (i.e., both yield split-virus vaccines). In 2013, the FDA approved another new vaccine that is conceptually different from the conventional split-virus (or whole-virion) vaccines. This vaccine (Flublok) was based on recombinant hemagglutinin (HA) proteins produced in insect cells. Its effectiveness in humans is similar (if not superior) to egg-based conventional vaccines and is not affected by the egg-adapted HA mutations that might be acquired during productive virus replication in eggs, which may alter immunogenicity and vaccine efficacy. This licensure of an HA subunit protein-based influenza vaccine was groundbreaking and paved the way for other recombinant protein-based vaccine approaches.
Immunization practices vary widely worldwide, but annual influenza vaccination is routinely recommended for everyone older than 6 mo living in the United States, especially for the elderly, pregnant women, young children, and health-care providers (https://www.cdc.gov/flu/prevent/vaccinations.htm). This is an elaborate system that has become a $4 billion per year global market and involves biannual influenza strain selection recommendations by the World Health Organization (WHO) advisory group based on the data from the WHO Global Influenza Surveillance and Response System (www.who.int/influenza/vaccines/virus/recommendations/en). Influenza virus strains anticipated to be prevalent in the upcoming season are selected for inclusion in the next version of influenza vaccines to be manufactured for the commercial use. This typically includes the selection of four viruses (H1N1 and H3N2 subtype influenza A, and two lineages of influenza B). There are several competing manufacturers with licensed products who race to produce the expected number of needed doses between February and September for the Northern hemisphere (offset 6 mo for the Southern hemisphere) either by growing viruses in eggs or in cell lines or producing recombinant proteins from insect cells infected by engineered baculovirus-encoding respective HAs. The vaccine products include live-attenuated vaccines, inactivated virus–based vaccines (split vaccine), or hemagglutinin protein containing vaccines that are monodispersed or aggregated as rosettes. A primary problem with current approaches is that the efficacy against medically attended influenza illness is only ∼60% in years when selected strains are matched to epidemic strains, and only ∼10% in years when there is significant antigenic drift in circulating strains (www.cdc.gov/flu/vaccines-work/past-seasons-estimates.html) (Lloyd et al. 1986; Belongia et al. 2009, 2011, 2016; Griffin et al. 2011; Treanor et al. 2012; Ohmit et al. 2014; McLean et al. 2015; Ferdinands et al. 2019). Moreover, the time required to make new vaccines based on novel strains is often too long to adequately respond to influenza outbreaks caused by strains with pandemic potential (Broadbent and Subbarao 2011). Given the current state of influenza vaccines and the existing technological opportunities for vaccine innovation, there is a large ongoing effort to identify new approaches for vaccine design that could achieve broadly protective, longer lasting immunity against influenza.
The concept of a universal influenza vaccine is attractive and gaining attention, but what this means in practice has many different interpretations. A vaccine given once in childhood that would provide lifelong protection to all current and future strains of influenza is highly unlikely to be achievable. However, there are different degrees of universality that could extend immunity either across strains within a subtype, across subtypes within influenza A virus groups (either group 1 or 2 defined by HA subtype), across both influenza A virus groups, and/or across all influenza A and B viruses. Another component desired for a universal influenza vaccine is greater durability of protective immunity. Achieving broad universal immunity for even one year at a time by a single vaccine formulation (reaching efficacy levels of >75% against all circulating strains on a given year) would be a huge advance and significantly reduce manufacturing costs. The ultimate goal, however, would be to design a vaccine capable of inducing broad immunity for several years at a time. Because there are several recent comprehensive reviews of universal vaccines in development, this review will not list every feasible approach being studied. Instead, their development will be approached from a conceptual viewpoint. From a vaccine development perspective, it is useful to define the intended purpose for a particular approach rather than categorize or list the immunological goals for universality. We have divided influenza vaccine product types into five major categories with distinct practical purposes: (1) high-performance seasonal vaccines would compete with and upgrade existing conventional vaccines; (2) supraseasonal vaccines are intended to substantially improve upon and replace current vaccines by providing consistent year-to-year efficacy without annual reformulation over two or more seasons; (3) vaccines for pandemic preparedness implies there is a product in a vial that can be deployed immediately in the case of a future outbreak; (4) vaccines for pandemic response would be products that could be rapidly produced in response to a specific threat; and (5) an ultimate universal vaccine would be an advanced supraseasonal vaccine that could protect individuals from all existing and future strains including noncirculating viruses at the time of administration for at least 3–5 yr.
BIOLOGICAL CHALLENGES
The virology and immunology of influenza has been described in other reviews (Fodor and te Velthuis 2020; tenOever 2020; Topham et al. 2020; Wu and Wilson 2020), but some of the features relevant to vaccine development will be briefly reviewed. The goal of influenza immunization is to prevent disease caused by two genera in the Orthomyxoviridae family, influenza A and B. Orthomyxoviruses have a segmented, negative-sense, single-stranded RNA genome, and influenza A and B have eight gene segments that encode at least 11 characterized proteins (Bouvier and Palese 2008). Their tissue tropism is largely based on α2,3-linked or α2,6-linked sialoside receptor dependence for entry, and species tropism is based on the distribution of cells expressing those receptors and the ability to escape intracellular restriction factors including innate immune sensors (Rogers and Paulson 1983; Nicholls et al. 2008; de Graaf and Fouchier 2014; Villalón-Letelier et al. 2017; Long et al. 2019). Effective immunity would interfere with either viral attachment and entry mediated by the HA surface protein, virus shedding and spread through interference with neuraminidase (NA) function, or rapid clearance of virus-infected cells with minimal immunopathology or dysfunction in airway physiology. The major viral defense mechanism against the host's preexisting immunity appears to be its genetic plasticity, which allows HA and NA to remain functional despite extensive antigenic variation (Kosik and Yewdell 2019). The antigenic variation arises from a diverse sequence space generated by an error-prone viral polymerase, is multifactorial, and is primarily driven by immunological pressure from antibodies that select for escape mutations, mutations that modulate receptor-binding kinetics, and/or epistatic mutations that alter functional features of HA and NA or interplay of the two proteins (Yewdell 2011). The antigenic variation results in antigenic drift, which occurs continuously in circulating influenza subtypes, limited principally by virus replication fitness and transmissibility. In addition, because of the segmented genome and extensive zoonotic reservoir in nature, influenza genomic segments can be reassorted when cells are co-infected with two or more viruses that might result in new influenza strains with exotic HA and/or NA subtypes for which little preexisting immunity is present in the general population. This process is termed antigenic shift and in humans is limited primarily by viral fitness to replicate within the host or spread within the population. This complex multilevel capacity for antigenic diversity and immune escape makes it challenging to design vaccines that will be capable of inducing effective immunity against future unforeseen influenza viruses (Taubenberger and Kash 2010). Therefore, effective universal influenza immunity has to target minimally variant sequences and structures critical for virus propagation for which alterations would result in a high cost of virus fitness. In principle, these selected sites of vulnerability should not be subject to the continuous antigenic drift and shift that occur in an unpredictable manner.
Another major biological challenge for influenza immunization is that most people of >2 yr of age have some level of preexisting immunity from prior exposure to influenza infection and/or vaccination. They have been imprinted through the establishment of memory B-cell and T-cell repertoires that shape the landscape of subsequent responses to similar antigens and alter responses to new antigenic determinants on new viruses (Francis 1960; Fonville et al. 2014; Monto et al. 2017; Guthmiller and Wilson 2018). Therefore, improved influenza vaccines (especially those dependent on antibodies to HA and NA) need to precisely maneuver responses to intended target sites of vulnerability to avoid the potentially negative impact of preexisting immunity.
ANTIGEN TARGETS FOR INFLUENZA VACCINES
HA and NA as described above are important targets for neutralizing antibodies (nAbs). HA is a trimeric class 1 fusion glycoprotein and has been studied in great detail. It was the first viral glycoprotein crystalized with a published structure (Wilson et al. 1981). It is critical for viral attachment and entry into cells and is the primary target for vaccine-induced protective immunity. HA engages sialosides on host glycoproteins and glycolipids through its receptor-binding site located on the membrane distal, globular head domain (Skehel and Wiley 2000; de Graaf and Fouchier 2014). Antibodies to this domain compete with receptor binding and therefore block viral attachment and ability to infect cells. This antibody-mediated inhibition of receptor-binding is the basis of hemagglutination inhibition (HAI) assay, and the HAI titer has been used as a surrogate for protective immunity and for licensure. There are well-characterized antigenic sites on the globular head domain for both H1 and H3 subtype HAs; however, they do not share the same nomenclature in the respective subtypes (Gerhard et al. 1981; Wiley et al. 1981; Wilson et al. 1981). Although the head domain contains extremely neutralization-sensitive epitopes, these epitopes are under strong immune pressure and readily acquire mutations that evade antibody recognition without compromising receptor-binding properties (Wu and Wilson 2017). Indeed, influenza virus occasionally acquires mutations in HA during vaccine manufacturing using embryonated eggs, leading to antigenic mismatch and lower vaccine effectiveness (Raymond et al. 2016; Zost et al. 2017; Garretson et al. 2018). The membrane proximal stem domain of HA is much more conserved than the head domain (Wu and Wilson 2017). The stem domain is responsible for the membrane fusion process and undergoes a dramatic conformational change at low pH in the late endosomal environment (Russell 2014). Antibodies directed toward this domain inhibit the function of the viral fusion machinery that is required for entry of the nucleocapsid into the cytoplasm and infection of the host cell (Corti et al. 2017). Because the stem domain is largely conserved within given HA subtypes and to a lesser extent across subtypes, antibodies to this domain, particularly those targeting the hydrophobic groove surrounding the Trp21HA2 residue, exhibit neutralizing breadth against multiple subtype viruses within influenza A group 1 subtypes (Okuno et al. 1993; Throsby et al. 2008; Ekiert et al. 2009; Sui et al. 2009), group 2 subtypes (Ekiert et al. 2011; Friesen et al. 2014), or across both group 1 and group 2 subtypes (Corti et al. 2011; Nakamura et al. 2013; Joyce et al. 2016; Kallewaard et al. 2016; Lang et al. 2017). Although the stem domain appears to be an appealing vaccine target, eliciting antibody response with such broad neutralizing activity has been challenging (Nabel and Fauci 2010). First, the stem domain is immunologically subdominant. In the presence of the head domain, immune responses directed to the stem are outcompeted by the head-directed response (Ellebedy et al. 2014; Andrews et al. 2015; Tan et al. 2019). Second, the native stem domain by itself is not suitable as a vaccine immunogen because the stem is metastable in the absence of the head domain (Lu et al. 2014; Impagliazzo et al. 2015; Yassine et al. 2015). The latter issue has been solved by structure-guided protein engineering approaches (Impagliazzo et al. 2015; Yassine et al. 2015; Corbett et al. 2019). The resulting stabilized stem-based immunogens are further discussed in the Vaccines for Pandemic Preparedness section. Other conserved HA regions of vulnerability are also regarded as improved vaccine targets, inspired by highly neutralizing human nAbs targeting the receptor-binding site (Whittle et al. 2011; Ekiert et al. 2012; Lee et al. 2014; Schmidt et al. 2015; Shen et al. 2017) and the lateral patch or the vestigial esterase domain of the head (Dreyfus et al. 2012; Iba et al. 2014; Chai et al. 2017; Raymond et al. 2018; Kanekiyo et al. 2019b). Although the precise mechanism of action remains uncertain, broadly cross-reactive yet nonneutralizing antibodies targeting the trimer interface of the head are also highly protective in a mouse model (Lee et al. 2016; Bangaru et al. 2019; Watanabe et al. 2019).
NA is a type II membrane protein and forms homotetramers on the virus surface (Air 2012). It is the second most abundant viral surface protein and is found as unevenly distributed clusters surrounded by HA trimers on the virion (Murti and Webster 1986; Harris et al. 2006). Each NA protomer consists of a membrane-anchored amino-terminal stalk domain and the globular domain containing the catalytic site that recognizes and cleaves sialic acids from glycans (Air and Laver 1989). The name NA implies this protein has neuraminidase activity and has been shown to facilitate egress of virions from infected cells (Air 2012; de Vries et al. 2020). Inhibition of this process either by small molecules or antibodies results in virus sequestration on the host cell membrane, leading to inefficient viral spread. NA also has sialic acid–binding properties (Laver et al. 1984; Mögling et al. 2017) and, in principle, can rescue the host cell attachment function of HA if needed for initial binding. Analogous to HA, NA has also been under immune pressure and undergone evolution over time (Kosik and Yewdell 2019). Although NA has been largely overlooked as a potential immunogen to elicit protective influenza immunity until recently (Wohlbold and Krammer 2014; Krammer et al. 2018; Eichelberger and Monto 2019), there is growing evidence to support the idea that antibody-mediated immunity to NA could confer broad protection or positively modulate disease progression/severity (Schulman et al. 1968; Murphy et al. 1972; Monto and Kendal 1973; Gilbert et al. 2019; Ng et al. 2019; Weiss et al. 2019). Furthermore, antibody-mediated inhibition of its role in facilitating virus release and shedding could reduce transmission and spread within the population. In particular, the discovery of broadly cross-reactive protective human monoclonal antibodies (mAbs) against NA has revitalized the NA-based universal influenza vaccine efforts (Chen et al. 2018; Stadlbauer et al. 2019). One such antibody isolated from an individual infected with H3N2 recognizes all influenza A virus NA subtypes as well as influenza B NAs by precisely mimicking the atomic interactions made by the sialic acid ligand and, hence, inhibiting the catalytic activity of NA (Stadlbauer et al. 2019). The identification of this antibody has served as a proof of principle that NA could contribute to universal influenza immunity and should be considered for inclusion in future vaccine approaches. Although the current influenza vaccine products, except the recombinant HA-based products, contain NA, the quantity and quality of NA in the vaccines are not actively monitored, regulated, and standardized. Over the last several decades, significant progress has been made in defining the structures of all known NA subtypes from both human influenza A and B viruses (Laver and Valentine 1969; Colman et al. 1983; Varghese et al. 1983; Colman and Ward 1985; Burmeister et al. 1992; McAuley et al. 2019). However, producing antigenically genuine and structurally stable NA protein has not been extensively explored, and delivery of an effective immunogen will likely require additional structural manipulation, protein engineering, and establishment of potency assays for quality assurance.
M2 is a tetrameric type III membrane protein and functions as an ion channel represented on the viral membrane although in low abundance. It is analogous to the small hydrophobic protein (SH) of respiratory syncytial virus. Vaccine prototypes based on the highly conserved ectodomain have been evaluated for both viruses and have advanced into clinical evaluation (Langley et al. 2018; Saelens 2019), but to date have not progressed beyond phase 2 trials (Kolpe et al. 2017; Mezhenskaya et al. 2019). Antibodies to M2 ectodomain (M2e) mediate protection through Fc functions and do not have in vitro neutralizing activity (Neirynck et al. 1999; Grandea et al. 2010; El Bakkouri et al. 2011). Whether delivery of a M2e-based vaccine constructs will be sufficiently immunogenic to establish broad immunity that is more effective than that induced by natural infection remains to be proven in humans. One of the challenges with this type of vaccine is shared by products based on HA stem immunogens, because conventional HAI activity cannot be measured and used as a test of potency or as a benchmark for licensure. Therefore, clinical development will likely require large field trials and will need to demonstrate added value to conventional vaccine approaches.
Nucleoprotein (NP) and other internal viral proteins could also be included in vaccines to promote T-cell responses. Townsend et al. (Townsend and Skehel 1982, 1984; Townsend et al. 1986) first described the importance of CD8+ T-cell epitopes from internal viral proteins for broad recognition of virus-infected cells using influenza NP. Internal proteins tend to be more conserved than HA and NA and have been proposed as the basis for influenza vaccine candidates based on CD8+ T-cell-mediated immunity (Rimmelzwaan et al. 2007). Although there is indirect evidence that CD8+ T cells directed to conserved epitopes confer some protection in humans (McMichael et al. 1983; Sridhar et al. 2013; Wang et al. 2015) and there is ample evidence for protection in animal models, significant efficacy against influenza disease may depend on the localization of effector cells. Achieving robust tissue resident memory CD8+ T cells (Trm) and/or CD4+ T effector cells in the airways will require delivery of vaccine antigens into lung tissue which may pose technical challenges and alter the safety profile of the prototype vaccine.
TARGET PRODUCT PROFILES FOR NEXT-GENERATION INFLUENZA VACCINES
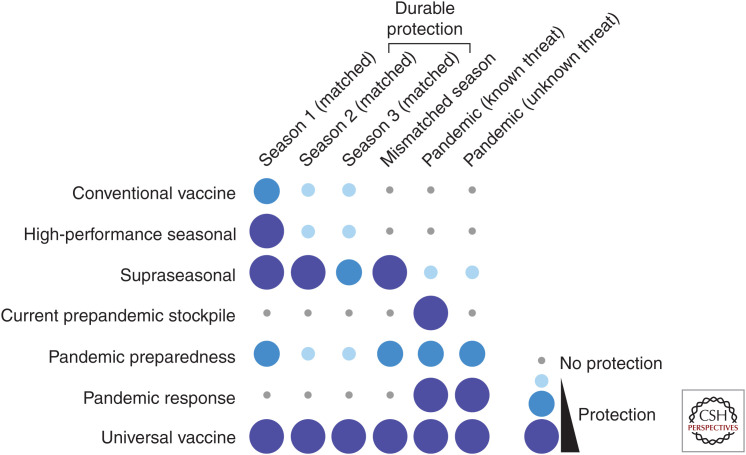
There is a growing consensus that the time is right for identifying disruptive approaches that will significantly improve influenza vaccine efficacy and there is a need to develop vaccine approaches that would be accessible to a larger fraction of the world's population. A practical and critical step in development involves defining the purpose and the desired properties of the vaccine product. Articulating the clinical and public health needs for a vaccine is the first step in this process. Based on desirable target product profiles, we propose five classifications for next-generation influenza vaccines below (Fig. 1).
Figure 1.
Visualized summary of target product profile of current and improved influenza vaccines. Conventional vaccines (current influenza vaccines) confer protection against antigenically matched seasonal influenza viruses. Vaccine efficacy varies year by year. An example of high-performance seasonal vaccines would be one that offers better protective immunity to antigenically matched seasonal influenza viruses, even though they may not provide durable multiyear protection nor protection against mismatched or pandemic viruses. An example of a supraseasonal vaccine would be one that provides consistent protection against antigenically drifted (mismatched) viruses and offers multiyear protection beyond a single influenza season, but may have limited efficacy against pandemic strains. Prepandemic stockpile vaccines (conventional) would work only if the pandemic virus is closely related to the stockpile vaccine strain. An example of a vaccine for pandemic preparedness would be one that provides broad protection against both seasonal and pandemic viruses, although the protective efficacy may not be as high as with strain-matched vaccines. This would allow options for immediate deployment during outbreak with a partially effective intervention. Vaccines for pandemic response would offer potent protection against pandemic viruses as these vaccines would be produced by platform manufacturing technologies using specific sequences from the emerging virus. Universal vaccines are expected to provide robust protection against both seasonal and pandemic viruses over multiple years.
High-Performance Seasonal Vaccines
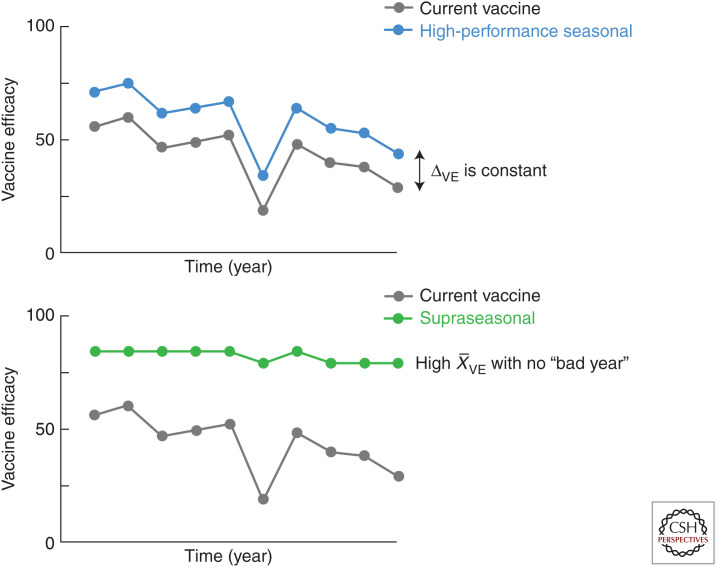
Several approaches have been attempted or suggested in order to improve the current conventional vaccines. Increasing the vaccine dose, especially for elderly subjects, has been shown to improve effectiveness (DiazGranados et al. 2014; Wilkinson et al. 2017; Lee et al. 2018). In addition, adding adjuvants improves efficacy (Frey et al. 2003, 2014; Domnich et al. 2017; Wilkins et al. 2017) but may also increase the risk of adverse events, which has been controversial in some cases (Ahmed et al. 2014). Mutations associated with adaptation to growth in eggs have been shown to diminish immunogenicity against circulating strains (Raymond et al. 2016; Zost et al. 2017; Garretson et al. 2018); moving toward vaccines derived from recombinant methods or from virus grown in mammalian cells would avoid this problem. Another approach would be to measure and standardize the amount of NA in current vaccines. The presence of NA-specific antibodies has been associated with reduced illness in humans (Gilbert et al. 2019; Ng et al. 2019; Weiss et al. 2019), has the capacity to limit viral propagation (Chen et al. 2018; Stadlbauer et al. 2019), and may also play a role in reducing transmission by reducing the viral shedding from infected cells (Schulman 1969). Real-time sequence analysis (www.nextstrain.org/flu) and increasing bioinformatic capability would contribute to more accurate predictions of upcoming circulating strains. It would also improve vaccine efficacy by reducing the chance of antigenic mismatch of strains selected for vaccines with circulating strains and, thus, remediate the seasons with extremely low vaccine efficacy. There is a lower bar for the technological advances and regulatory requirements needed for these types of improvements, so the investment needed would be smaller compared to most of the approaches listed below. However, potential gains would also be limited (Fig. 2).
Figure 2.
Expected vaccine efficacy profile of improved seasonal influenza vaccines. Comparison of theoretical vaccine efficacy between the current vaccine and an example of high-performance seasonal vaccine (top). Although average vaccine efficacy over time () is higher for high-performance seasonal vaccines compared to conventional vaccines, the difference in vaccine efficacy (ΔVE) between vaccine modalities for each year is consistent and, hence, vaccine efficacy will fluctuate depending on accuracy of antigenic match between vaccine and circulating strains. Theoretical vaccine efficacy of supraseasonal vaccines (bottom). Unlike high-performance seasonal and conventional vaccines, supraseasonal vaccines would provide consistent vaccine efficacy year to year so would remain high with no fluctuation. Supraseasonal vaccines would not be significantly affected by antigenic mismatch between vaccine and circulating strains, and, therefore, they may not need annual updates of vaccine components and reformulation.
Supraseasonal Vaccines
The concept of a supraseasonal vaccine is that efficacy could be maintained from season to season without the need for annual reformulation and perhaps immunization. This type of vaccine would be expected to have superior breadth of coverage and efficacy compared to currently available vaccines and would eventually replace current vaccines (Fig. 2). It would have robust efficacy against drifted strains but not necessarily against pandemic strains (i.e., antigenically shifted viruses). Even if it required annual administration, it would not require annual reformulation and manufacturing. Such a vaccine would likely need to induce immune responses to invariant sites of vulnerability of the virus. This could occur by amplifying the response to selected epitopes known to be conserved like the sites on the HA stem or head including the receptor-binding site, the lateral patch, the vestigial esterase site, and the trimer interface (Wu and Wilson 2020), the NA catalytic site (Stadlbauer et al. 2019), and the M2e (Grandea et al. 2010) or by inducing effector T cells, especially if they are tissue resident memory cells localized in the lower airway and lung (Van Braeckel-Budimir and Harty 2017). Another approach would be to prevent induction of antibodies to sites that have narrower specificity. Immunodominance of responses that have limited breadth occurs for multiple reasons. Greater physical access of B cells to the membrane distal head domain of HA compared to the stem results in one plausible reason for immunodominance of the variable HA head. Another cause of immunodominance could be differences in lymphocyte precursor frequencies either inherent or established during early life infection or vaccination. Immunological imprinting has been shown to affect subsequent immunity to heterosubtypic influenza viruses on a HA group-specific bias (Gostic et al. 2016). For example, if early imprinting with H1N1 (or other group 1 viruses) expanded predominantly VH1-69 lineages in the B-cell response to the HA stem epitope, later exposure to an H3N2 or other group 2 virus may have lower capacity to drive evolution of B-cell lineages known to gain cross-neutralizing activity against both group 1 and group 2 viruses, such as VH6-1 and VH1-18 lineages. One approach for avoiding immunodominance is to design vaccine antigens that facilitate expansion of B cells that recognize subdominant yet vulnerable sites with the potential for greater breadth. An alternative approach is to co-display heterotypic antigens on a single continuous surface such as nanoparticles. This reduces the frequency of responses intolerant to variability and increases the chances for amplifying B cells that tolerate antigenic variation and, hence, subvert the immunodominance (Kanekiyo et al. 2019b). It may also be possible to accumulate greater breadth of antibody-mediated protection by using cocktails of immunogens or serial immunization with multiple specificities in an attempt to generate a greater diversity of memory precursors. However, serial exposure of patients to different viral strains during natural infection and repeated vaccination has not resulted in broad immunity and has in fact sometimes been associated with reduced immunity to divergent strains (McLean et al. 2014; Belongia et al. 2017).
Vaccines for Pandemic Preparedness
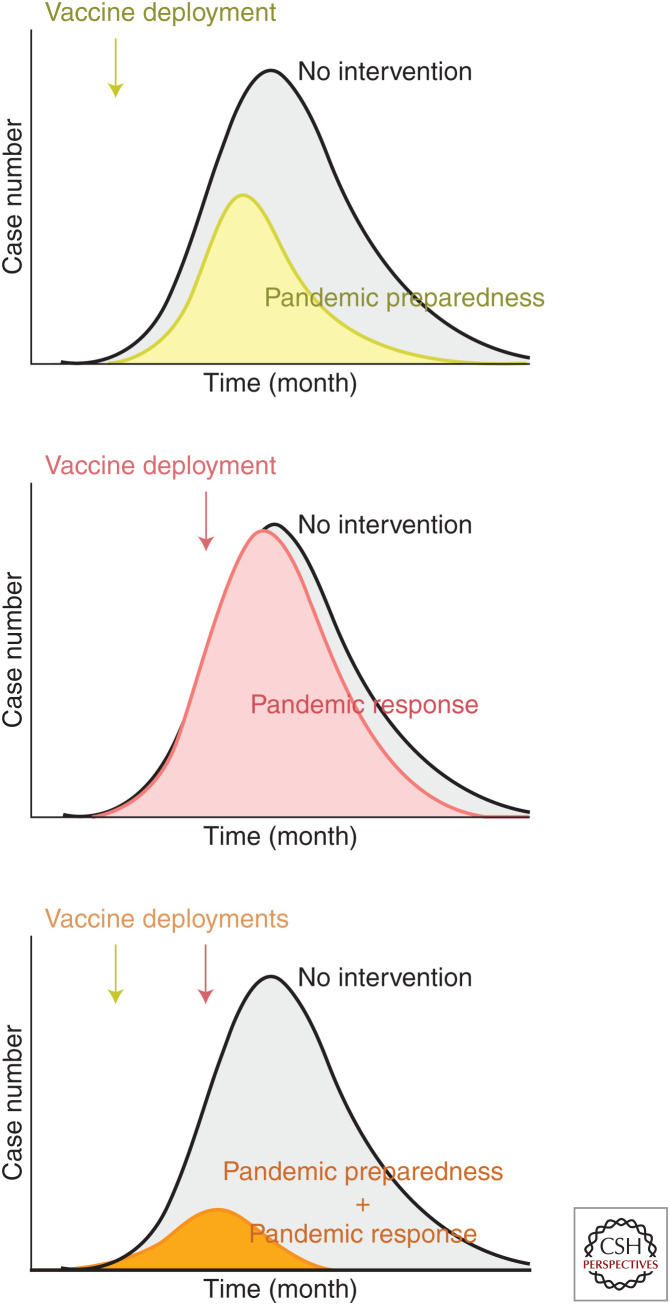
The concept of pandemic preparedness encompasses many facets of surveillance and medical care systems. There is a consensus among researchers, medical professionals, and public health officials that we are not adequately prepared for a pandemic catastrophe like the one that occurred in 1918–1919 (Taubenberger et al. 2019). Even for industrialized countries preparedness would at the very least require improved surveillance and response systems including a more robust distribution infrastructure for preexisting vaccines and therapeutics than currently exists. Prepandemic vaccine supplies would need to be stockpiled and accessible for immediate deployment to effectively interrupt transmission cycles. Modeling has shown that early deployment of a partially effective vaccine is more effective than delayed deployment of a more efficacious vaccine (Feng et al. 2011; Larson and Teytelman 2012). The emergence of avian H5N1 and H7N9 influenza viruses that could infect humans led to the manufacturing of U.S. prepandemic stockpiles of pandemic vaccines produced with conventional technology. These stockpiled doses were ready for rapid deployment in case of a pandemic caused by these subtypes. However, the number of doses was limited and after a few years, the antigenicity of prevalent avian strains evolved away from the available vaccine, making it a less dependable prepandemic countermeasure. Therefore, vaccines for pandemic preparedness that could be immediately available for a pandemic event would need to be periodically replaced (an expensive proposition) or designed with a long shelf life and able to elicit broad immunity while not being affected by antigenic drift and shift. Candidate vaccines that best fit this product profile are those that target conserved epitopes like the HA stem, the NA catalytic domain, or M2e antigens that would induce antibody-based protection or proteins that would induce cross-reactive T-cell responses. None of these approaches have yet been shown to be efficacious in controlled field trials, but are in early phase development and should continue to advance to answer questions about the degree of cross-protection achieved. For example, if HA stem-based vaccines using either chimeric molecules with exotic HA head domains (Krammer et al. 2013; Bernstein et al. 2020), headless HA stem trimers delivered as soluble proteins (Impagliazzo et al. 2015), or displayed on nanoparticles (Yassine et al. 2015; Corbett et al. 2019) were shown to be efficacious, then these products could be produced and stockpiled and be available for deployment against any emerging influenza A virus threat. This may also provide time to produce a strain-matched vaccine that would be expected to have higher efficacy (Fig. 3). Inherent to this concept is the idea that effective immunity would be achieved rapidly from a single vaccine administration.
Figure 3.
Potential scenario of an influenza pandemic with different countermeasure options. In the case of an influenza pandemic caused by a new virus with no antigenically matched stockpile vaccines, vaccines for pandemic preparedness and for pandemic response or a combination of the two vaccines could make a substantial impact on the epidemic curve and reduce the cumulative health burden. Vaccines for pandemic preparedness could be deployed as soon as the outbreak spreads locally and is recognized to be caused by a pandemic strain, so medical providers and at-risk population can be immunized before the pandemic peaks (top). Although vaccine efficacy for this type of vaccine may not be optimal, it will provide time for mass production of strain-matched vaccines (pandemic response). Relying on deployment of strain-matched vaccines alone as a pandemic response may not be sufficient as it may not be feasible to produce enough doses in time to substantially impact the epidemic (middle). This situation will be mitigated as newer technologies for rapid vaccine production such as mRNA-based modalities become available. Sequential deployment of vaccines for pandemic preparedness and pandemic response would be ideal to contain pandemic with minimum burden taking into account the importance of both speed and specificity (bottom).
Vaccines for Pandemic Response
Platform technologies for antigen design and manufacturing would be needed to achieve large-scale production of a vaccine after the pandemic strain emerges, in the shortest time frame. This type of response is not easily achieved using conventional vaccine approaches as demonstrated by the 2009 H1N1 pandemic. Even though the manufacturing process was initiated during early spring when the dominant strain emerged, the product was not available until after the peak of the epidemic in early fall (Broadbent and Subbarao 2011; Skowronski et al. 2011). Had there been a partially effective vaccine targeting conserved epitopes as described above for immediate use, it could have been followed by rapid manufacturing of vaccine constructs specific for the pandemic strain (Fig. 3). This could potentially be achieved using egg-grown virus, but the supply of eggs may be limiting in this setting. Recombinant HA and/or NA proteins derived from pandemic strain sequences could also be produced in insect or mammalian cells either as soluble proteins or displayed on nanoparticles (Kanekiyo et al. 2013; Georgiev et al. 2018; Marcandalli et al. 2019). The advantage of nanoparticles is that the orderly array of antigens can effectively cluster B-cell receptors and activate B cells to proliferate and mature (Bachmann et al. 1993). This would reduce the need to formulate the protein-based vaccine with an adjuvant. In a pandemic setting, simplifying the supply chain and using the minimal components is more likely to be scalable and less likely to encounter bottlenecks. Genetic delivery of the vaccine antigen is an attractive platform manufacturing technology for a pandemic response. RNA-based delivery, in particular, looks promising because it typically requires a lower dose than DNA-based delivery and can potentially be rapidly manufactured. A major advantage of genetic delivery is the ability to use the same platform and manufacturing protocol for multiple vaccine variants, and the regulatory requirements would be reduced compared to recombinant vector or recombinant protein approaches. RNA-delivered immunogens would also have the capacity for inducing more robust CD8+ T-cell responses than protein-based vaccines in addition to the capacity to induce antibody responses.
Universal Influenza Vaccines
As noted before, there are many degrees of universality. For the purpose of this review, the definition of a universal influenza vaccine is one that can induce protective immunity against all influenza A and B viruses. The efficacy would be consistent from year to year, ideally for more than 3 years, with an at least 75% reduction in medically attended lower respiratory tract disease or hospitalization. This means the vaccine would not need annual reformulation and would maintain immunity against drifted seasonal and pandemic viruses. It would be distinguished from a supraseasonal vaccine by inducing greater coverage of pandemic subtypes and protective immunity from a single injection lasting more than 1 year. Therefore, a universal influenza vaccine would have better efficacy than currently available vaccines when matched with prevalent strains, which implies a greater potency or more favorable anatomic location for induction of protective antibody and T-cell responses. It would also need to have greater breadth of immunity to be effective in subsequent years without reformulation. Ideally, this could be accomplished with a single immunization every 3–5 yr or longer, but even if the vaccine required annual administration, it would represent a major advance. The types of products and technologies on the near horizon may need to be combined to achieve anything close to the aforementioned universal protection criteria, but there are currently many opportunities to make incremental and potentially breakthrough advances.
CONCLUDING REMARKS: TECHNOLOGY-DRIVEN OPPORTUNITIES AND REMAINING CHALLENGES
A confluence of technological advances over the last decade has given rise to new vaccine design options. These technologies have all contributed to an era of precision vaccinology in which it is possible to conceive influenza immunogens in different ways. In particular, the ability to define and manipulate atomic level details of HA structure and topology including antibody-liganded dynamic conformations has allowed the exact mapping of antigenic supersites and identification of new conserved targets for future vaccines (for review, see Wu and Wilson 2020). The ability to structurally define new sites of vulnerability has been amplified by the ability to rapidly isolate and characterize numerous human mAbs (for review, see Crowe 2018). Sequence analysis of individual B cells sorted by their ability to recognize conserved antigenic supersites combined with bioinformatic tools has identified several convergent multidonor B-cell lineages with desirable functional antibody properties that could serve as molecular end points for vaccine evaluation (Joyce et al. 2016; Andrews et al. 2017; Andrews and McDermott 2018; Crowe 2018). Advances in computational biology and protein engineering have enabled the generation of self-assembling nanoparticles that can display geometrically defined arrays of homotypic or heterotypic antigens have provided new options for organizing, delivering, and presenting immunogens to the immune system (for review, see Kanekiyo et al. 2019a). Particularly, combining structure-based antigen design with customized nanoparticle display has opened new possibilities for inducing supranormal levels of vaccine-elicited immunity (Marcandalli et al. 2019). New capabilities for platform manufacturing have provided potential avenues for rapidly converting to vaccines for pandemic response and reduce uncertainties related to the development process and safety of new products (Graham et al. 2018). New insights into lymph node and B-cell biology and better understanding of adjuvants as Toll-like receptor (TLR) agonists are also contributing to more potent and durable vaccine-induced immunity (Mesin et al. 2016; Andrews and McDermott 2018; Del Giudice et al. 2018). These advances have ushered in a new era of precision vaccinology and have made it technically possible to substantially improve influenza vaccines. They have also introduced new challenges for clinical development and regulatory pathways. There will be complex business and public health decisions about how to replace existing products with new products without having gaps in availability. Importantly, there are still a number of biological questions to solve that not only apply to influenza but to other respiratory diseases. The recent breakthroughs in structural biology, protein engineering, antibody ontogeny, and B-cell biology have been opportunistically applied to achieve systemic antibody-mediated protection from lower airway infection. Although there have been recent advances in our understanding of Fc-mediated functions with capacity for clearing virus-infected cells (Bournazos and Ravetch 2017; Boudreau and Alter 2019), our ability to control events if viruses evade antibody neutralization and infect substantial numbers of airway cells is limited. There is still a lot to be learned regarding the pathogenesis of airway damage and remodeling, T-cell-mediated viral clearance, and direct induction of secretory IgA and other factors unique to mucosal immunity. Achieving optimal protection of the airways from the pathophysiological consequences of respiratory viral infections may require new methods for targeted delivery of immunogens to specific tissues and induction of local B-cell and T-cell effector functions.
Influenza is a persistent threat to human health and can potentially cause devastating pandemics that can compromise societal infrastructure and stability. Therefore, it is incumbent on politicians, scientists, public health officials, and industry to use the tools of precision vaccinology to develop fit-for-purpose products that can induce protective immunity from seasonal, drifted, and pandemic strains while working toward a truly universal influenza vaccine. We have outlined several specific unmet needs requiring innovation and application of new approaches and trust that the technologies described will provide solutions for preventing influenza disease.
ACKNOWLEDGMENTS
The authors thank Drs. John Mascola, Karin Bok, Jeffrey Boyington, and Sarah Andrews (VRC) for helpful comments and discussion. This work was supported by the intramural research program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (M.K. and B.S.G.).
This article has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this subject collection.
- Ahmed SS, Schur PH, MacDonald NE, Steinman L. 2014. Narcolepsy, 2009 A(H1N1) pandemic influenza, and pandemic influenza vaccinations: what is known and unknown about the neurological disorder, the role for autoimmunity, and vaccine adjuvants. J Autoimmun 50: 1–11. 10.1016/j.jaut.2014.01.033 [DOI] [PubMed] [Google Scholar]
- Air GM. 2012. Influenza neuraminidase. Influenza Other Respir Viruses 6: 245–256. 10.1111/j.1750-2659.2011.00304.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Air GM, Laver WG. 1989. The neuraminidase of influenza virus. Proteins 6: 341–356. 10.1002/prot.340060402 [DOI] [PubMed] [Google Scholar]
- Andrews SF, McDermott AB. 2018. Shaping a universally broad antibody response to influenza amidst a variable immunoglobulin landscape. Curr Opin Immunol 53: 96–101. 10.1016/j.coi.2018.04.009 [DOI] [PubMed] [Google Scholar]
- Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, et al. 2015. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 7: 316ra192. 10.1126/scitranslmed.aad0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SF, Joyce MG, Chambers MJ, Gillespie RA, Kanekiyo M, Leung K, Yang ES, Tsybovsky Y, Wheatley AK, Crank MC, et al. 2017. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol 2: eaan2676. 10.1126/sciimmunol.aan2676 [DOI] [PubMed] [Google Scholar]
- Bachmann MF, Rohrer UH, Kundig TM, Burki K, Hengartner H, Zinkernagel RM. 1993. The influence of antigen organization on B cell responsiveness. Science 262: 1448–1451. 10.1126/science.8248784 [DOI] [PubMed] [Google Scholar]
- Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, et al. 2019. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177: 1136–1152.e18. 10.1016/j.cell.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belongia EA, Kieke BA, Donahue JG, Greenlee RT, Balish A, Foust A, Lindstrom S, Shay DK, Marshfield Influenza Study G. 2009. Effectiveness of inactivated influenza vaccines varied substantially with antigenic match from the 2004–2005 season to the 2006–2007 season. J Infect Dis 199: 159–167. 10.1086/595861 [DOI] [Google Scholar]
- Belongia EA, Kieke BA, Donahue JG, Coleman LA, Irving SA, Meece JK, Vandermause M, Lindstrom S, Gargiullo P, Shay DK. 2011. Influenza vaccine effectiveness in Wisconsin during the 2007–08 season: comparison of interim and final results. Vaccine 29: 6558–6563. 10.1016/j.vaccine.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Belongia EA, Simpson MD, King JP, Sundaram ME, Kelley NS, Osterholm MT, McLean HQ. 2016. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis 16: 942–951. 10.1016/S1473-3099(16)00129-8 [DOI] [PubMed] [Google Scholar]
- Belongia EA, Skowronski DM, McLean HQ, Chambers C, Sundaram ME, De Serres G. 2017. Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev Vaccines 16: 723–736. 10.1080/14760584.2017.1334554 [DOI] [PubMed] [Google Scholar]
- Bernstein DI, Guptill J, Naficy A, Nachbagauer R, Berlanda-Scorza F, Feser J, Wilson PC, Solórzano A, Van der Wielen M, Walter EB, et al. 2020. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect Dis 20: 80–91. 10.1016/S1473-3099(19)30393-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau CM, Alter G. 2019. Extra-neutralizing FcR-mediated antibody functions for a universal influenza vaccine. Front Immunol 10: 440. 10.3389/fimmu.2019.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, Ravetch JV. 2017. Fcγ receptor function and the design of vaccination strategies. Immunity 47: 224–233. 10.1016/j.immuni.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM, Palese P. 2008. The biology of influenza viruses. Vaccine 26(Suppl 4): D49–D53. 10.1016/j.vaccine.2008.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent AJ, Subbarao K. 2011. Influenza virus vaccines: lessons from the 2009 H1N1 pandemic. Curr Opin Virol 1: 254–262. 10.1016/j.coviro.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister WP, Ruigrok RW, Cusack S. 1992. The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J 11: 49–56. 10.1002/j.1460-2075.1992.tb05026.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai N, Swem LR, Park S, Nakamura G, Chiang N, Estevez A, Fong R, Kamen L, Kho E, Reichelt M, et al. 2017. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat Commun 8: 14234. 10.1038/ncomms14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, et al. 2018. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173: 417–429.e10. 10.1016/j.cell.2018.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman PM, Ward CW. 1985. Structure and diversity of influenza virus neuraminidase. Curr Top Microbiol Immunol 114: 177–255. [DOI] [PubMed] [Google Scholar]
- Colman PM, Varghese JN, Laver WG. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303: 41–44. 10.1038/303041a0 [DOI] [PubMed] [Google Scholar]
- Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, Myers SI, Tsybovsky Y, Wheatley AK, Schramm CA, et al. 2019. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. MBio 10: e02810-18. 10.1128/mBio.02810-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333: 850–856. 10.1126/science.1205669 [DOI] [PubMed] [Google Scholar]
- Corti D, Cameroni E, Guarino B, Kallewaard NL, Zhu Q, Lanzavecchia A. 2017. Tackling influenza with broadly neutralizing antibodies. Curr Opin Virol 24: 60–69. 10.1016/j.coviro.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JE Jr. 2018. Is It possible to develop a “universal” influenza virus vaccine? Potential for a universal influenza vaccine. Cold Spring Harb Perspect Biol 10: a029496. 10.1101/cshperspect.a029496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf M, Fouchier RAM. 2014. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 33: 823–841. 10.1002/embj.201387442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Giudice G, Rappuoli R, Didierlaurent AM. 2018. Correlates of adjuvanticity: a review on adjuvants in licensed vaccines. Semin Immunol 39: 14–21. 10.1016/j.smim.2018.05.001 [DOI] [PubMed] [Google Scholar]
- de Vries E, Du W, Guo H, de Haan CAM. 2020. Influenza A virus hemagglutinin-neuraminidase-receptor balance: preserving virus motility. Trends Microbiol 28: 57–67. 10.1016/j.tim.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiazGranados CA, Dunning AJ, Kimmel M, Kirby D, Treanor J, Collins A, Pollak R, Christoff J, Earl J, Landolfi V, et al. 2014. Efficacy of high-dose versus standard-dose influenza vaccine in older adults. N Engl J Med 371: 635–645. 10.1056/NEJMoa1315727 [DOI] [PubMed] [Google Scholar]
- Domnich A, Arata L, Amicizia D, Puig-Barberà J, Gasparini R, Panatto D. 2017. Effectiveness of MF59-adjuvanted seasonal influenza vaccine in the elderly: a systematic review and meta-analysis. Vaccine 35: 513–520. 10.1016/j.vaccine.2016.12.011 [DOI] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337: 1343–1348. 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberger MC, Monto AS. 2019. Neuraminidase, the forgotten surface antigen, emerges as an influenza vaccine target for broadened protection. J Infect Dis 219: S75–S80. 10.1093/infdis/jiz017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger M-A, Friesen RHE, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324: 246–251. 10.1126/science.1171491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RHE, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJWM, Brandenburg B, et al. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333: 843–850. 10.1126/science.1204839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, et al. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489: 526–532. 10.1038/nature11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bakkouri K, Descamps F, De Filette M, Smet A, Festjens E, Birkett A, Van Rooijen N, Verbeek S, Fiers W, Saelens X. 2011. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J Immunol 186: 1022–1031. 10.4049/jimmunol.0902147 [DOI] [PubMed] [Google Scholar]
- Ellebedy AH, Krammer F, Li G-M, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, et al. 2014. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci 111: 13133–13138. 10.1073/pnas.1414070111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Towers S, Yang Y. 2011. Modeling the effects of vaccination and treatment on pandemic influenza. AAPS J 13: 427–437. 10.1208/s12248-011-9284-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinands JM, Gaglani M, Martin ET, Middleton D, Monto AS, Murthy K, Silveira FP, Talbot HK, Zimmerman R, Alyanak E, et al. 2019. Prevention of influenza hospitalization among adults in the United States, 2015–2016: results from the US hospitalized adult influenza vaccine effectiveness network (HAIVEN). J Infect Dis 220: 1265–1275. 10.1093/infdis/jiy723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Fodor E, te Velthuis. 2019. Structure and function of the influenza virus transcription and replication machinery. Cold Spring Harb Perspect Med 10.1101/cshperspect.a038398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, Xue L, Jones TC, Le NMH, Pham QT, et al. 2014. Antibody landscapes after influenza virus infection or vaccination. Science 346: 996–1000. 10.1126/science.1256427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis TJ. 1960. On the doctrine of original antigenic sin. Proc Am Philos Soc 104: 572–578. [Google Scholar]
- Francis T Jr, Salk JE. 1942. A simplified procedure for the concentration and purification of influenza virus. Science 96: 499–500. 10.1126/science.96.2500.499 [DOI] [PubMed] [Google Scholar]
- Frey S, Poland G, Percell S, Podda A. 2003. Comparison of the safety, tolerability, and immunogenicity of a MF59-adjuvanted influenza vaccine and a non-adjuvanted influenza vaccine in non-elderly adults. Vaccine 21: 4234–4237. 10.1016/S0264-410X(03)00456-0 [DOI] [PubMed] [Google Scholar]
- Frey SE, Reyes MRA-DL, Reynales H, Bermal NN, Nicolay U, Narasimhan V, Forleo-Neto E, Arora AK. 2014. Comparison of the safety and immunogenicity of an MF59®-adjuvanted with a non-adjuvanted seasonal influenza vaccine in elderly subjects. Vaccine 32: 5027–5034. 10.1016/j.vaccine.2014.07.013 [DOI] [PubMed] [Google Scholar]
- Friesen RHE, Lee PS, Stoop EJM, Hoffman RMB, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, et al. 2014. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci 111: 445–450. 10.1073/pnas.1319058110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garretson TA, Petrie JG, Martin ET, Monto AS, Hensley SE. 2018. Identification of human vaccinees that possess antibodies targeting the egg-adapted hemagglutinin receptor binding site of an H1N1 influenza vaccine strain. Vaccine 36: 4095–4101. 10.1016/j.vaccine.2018.05.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Joyce MG, Chen RE, Leung K, McKee K, Druz A, Van Galen JG, Kanekiyo M, Tsybovsky Y, Yang ES, et al. 2018. Two-component ferritin nanoparticles for multimerization of diverse trimeric antigens. ACS Infect Dis 4: 788–796. 10.1021/acsinfecdis.7b00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard W, Yewdell J, Frankel ME, Webster R. 1981. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290: 713–717. 10.1038/290713a0 [DOI] [PubMed] [Google Scholar]
- Gilbert PB, Fong Y, Juraska M, Carpp LN, Monto AS, Martin ET, Petrie JG. 2019. HAI and NAI titer correlates of inactivated and live attenuated influenza vaccine efficacy. BMC Infect Dis 19: 453. 10.1186/s12879-019-4049-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpasture EW, Woodruff AM, Buddingh GJ. 1931. The cultivation of vaccine and other viruses in the chorio-allantoic membrane of chick embryos. Science 74: 371–372. 10.1126/science.74.1919.371 [DOI] [PubMed] [Google Scholar]
- Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. 2016. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354: 722–726. 10.1126/science.aag1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BS, Mascola JR, Fauci AS. 2018. Novel vaccine technologies: essential components of an adequate response to emerging viral diseases. J Am Med Assoc 319: 1431–1432. 10.1001/jama.2018.0345 [DOI] [PubMed] [Google Scholar]
- Grandea AG III, Olsen OA, Cox TC, Renshaw M, Hammond PW, Chan-Hui PY, Mitcham JL, Cieplak W, Stewart SM, Grantham ML, et al. 2010. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc Natl Acad Sci 107: 12658–12663. 10.1073/pnas.0911806107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin MR, Monto AS, Belongia EA, Treanor JJ, Chen Q, Chen J, Talbot HK, Ohmit SE, Coleman LA, Lofthus G, et al. 2011. Effectiveness of non-adjuvanted pandemic influenza A vaccines for preventing pandemic influenza acute respiratory illness visits in 4 U.S. communities. PLoS One 6: e23085. 10.1371/journal.pone.0023085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmiller JJ, Wilson PC. 2018. Harnessing immune history to combat influenza viruses. Curr Opin Immunol 53: 187–195. 10.1016/j.coi.2018.05.010 [DOI] [PubMed] [Google Scholar]
- Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci 103: 19123–19127. 10.1073/pnas.0607614103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, Wakiyama M, Shirouzu M, Okada J, Okuno Y, et al. 2014. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol 88: 7130–7144. 10.1128/JVI.00420-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RMB, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, et al. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349: 1301–1306. 10.1126/science.aac7263 [DOI] [PubMed] [Google Scholar]
- Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, et al. 2016. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 166: 609–623. 10.1016/j.cell.2016.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, et al. 2016. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 166: 596–608. 10.1016/j.cell.2016.05.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M, Wei CJ, Yassine HM, McTamney PM, Boyington JC, Whittle JRR, Rao SS, Kong WP, Wang L, Nabel GJ. 2013. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499: 102–106. 10.1038/nature12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M, Ellis D, King NP. 2019a. New vaccine design and delivery technologies. J Infect Dis 219: S88–S96. 10.1093/infdis/jiy745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, et al. 2019b. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat Immunol 20: 362–372. 10.1038/s41590-018-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpe A, Schepens B, Fiers W, Saelens X. 2017. M2-based influenza vaccines: recent advances and clinical potential. Expert Rev Vaccines 16: 123–136. 10.1080/14760584.2017.1240041 [DOI] [PubMed] [Google Scholar]
- Kosik I, Yewdell JW. 2019. Influenza hemagglutinin and neuraminidase: yin–yang proteins coevolving to thwart immunity. Viruses 11: 346. 10.3390/v11040346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Pica N, Hai R, Margine I, Palese P. 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87: 6542–6550. 10.1128/JVI.00641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, Wilson PC, Compans RW, Skountzou I, Monto AS. 2018. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? MBio 9: e02332-17. 10.1128/mBio.02332-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S, Xie J, Zhu X, Wu NC, Lerner RA, Wilson IA. 2017. Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in VH1-69 antibody orientation on the HA stem. Cell Rep 20: 2935–2943. 10.1016/j.celrep.2017.08.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley JM, MacDonald LD, Weir GM, MacKinnon-Cameron D, Ye L, McNeil S, Schepens B, Saelens X, Stanford MM, Halperin SA. 2018. A respiratory syncytial virus vaccine based on the small hydrophobic protein ectodomain presented with a novel lipid-based formulation is highly immunogenic and safe in adults: a first-in-humans study. J Infect Dis 218: 378–387. 10.1093/infdis/jiy177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RC, Teytelman A. 2012. Modeling the effects of H1N1 influenza vaccine distribution in the United States. Value Health 15: 158–166. 10.1016/j.jval.2011.07.014 [DOI] [PubMed] [Google Scholar]
- Laver WG, Valentine RC. 1969. Morphology of the isolated hemagglutinin and neuraminidase subunits of influenza virus. Virology 38: 105–119. 10.1016/0042-6822(69)90132-9 [DOI] [PubMed] [Google Scholar]
- Laver WG, Colman PM, Webster RG, Hinshaw VS, Air GM. 1984. Influenza virus neuraminidase with hemagglutinin activity. Virology 137: 314–323. 10.1016/0042-6822(84)90223-X [DOI] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. 2014. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5: 3614. 10.1038/ncomms4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee CH, Lavinder JJ, et al. 2016. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med 22: 1456–1464. 10.1038/nm.4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JKH, Lam GKL, Shin T, Kim J, Krishnan A, Greenberg DP, Chit A. 2018. Efficacy and effectiveness of high-dose versus standard-dose influenza vaccination for older adults: a systematic review and meta-analysis. Expert Rev Vaccines 17: 435–443. 10.1080/14760584.2018.1471989 [DOI] [PubMed] [Google Scholar]
- Lloyd PE, Schacher S, Kupfermann I, Weiss KR. 1986. Release of neuropeptides during intracellular stimulation of single identified Aplysia neurons in culture. Proc Natl Acad Sci 83: 9794–9798. 10.1073/pnas.83.24.9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Mistry B, Haslam SM, Barclay WS. 2019. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17: 67–81. 10.1038/s41579-018-0115-z [DOI] [PubMed] [Google Scholar]
- Lu Y, Welsh JP, Swartz JR. 2014. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci 111: 125–130. 10.1073/pnas.1308701110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcandalli J, Fiala B, Ols S, Perotti M, de van der Schueren W, Snijder J, Hodge E, Benhaim M, Ravichandran R, Carter L, et al. 2019. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell 176: 1420–1431.e17. 10.1016/j.cell.2019.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Gilbertson BP, Trifkovic S, Brown LE, McKimm-Breschkin JL. 2019. Influenza virus neuraminidase structure and functions. Front Microbiol 10: 39. 10.3389/fmicb.2019.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean HQ, Thompson MG, Sundaram ME, Meece JK, McClure DL, Friedrich TC, Belongia EA. 2014. Impact of repeated vaccination on vaccine effectiveness against influenza A(H3N2) and B during 8 seasons. Clin Infect Dis 59: 1375–1385. 10.1093/cid/ciu680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean HQ, Thompson MG, Sundaram ME, Kieke BA, Gaglani M, Murthy K, Piedra PA, Zimmerman RK, Nowalk MP, Raviotta JM, et al. 2015. Influenza vaccine effectiveness in the United States during 2012–2013: variable protection by age and virus type. J Infect Dis 211: 1529–1540. 10.1093/infdis/jiu647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael AJ, Gotch FM, Noble GR, Beare PA. 1983. Cytotoxic T-cell immunity to influenza. N Engl J Med 309: 13–17. 10.1056/NEJM198307073090103 [DOI] [PubMed] [Google Scholar]
- Mesin L, Ersching J, Victora GD. 2016. Germinal center B cell dynamics. Immunity 45: 471–482. 10.1016/j.immuni.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezhenskaya D, Isakova-Sivak I, Rudenko L. 2019. M2e-based universal influenza vaccines: a historical overview and new approaches to development. J Biomed Sci 26: 76. 10.1186/s12929-019-0572-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mögling R, Richard MJ, Vliet SVD, van Beek R, Schrauwen EJA, Spronken MI, Rimmelzwaan GF, Fouchier RAM. 2017. Neuraminidase-mediated haemagglutination of recent human influenza A(H3N2) viruses is determined by arginine 150 flanking the neuraminidase catalytic site. J Gen Virol 98: 1274–1281. 10.1099/jgv.0.000809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto AS, Kendal AP. 1973. Effect of neuraminidase antibody on Hong Kong influenza. Lancet 301: 623–625. 10.1016/S0140-6736(73)92196-X [DOI] [PubMed] [Google Scholar]
- Monto AS, Malosh RE, Petrie JG, Martin ET. 2017. The doctrine of original antigenic sin: separating good from evil. J Infect Dis 215: 1782–1788. 10.1093/infdis/jix173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BR, Kasel JA, Chanock RM. 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med 286: 1329–1332. 10.1056/NEJM197206222862502 [DOI] [PubMed] [Google Scholar]
- Murti KG, Webster RG. 1986. Distribution of hemagglutinin and neuraminidase on influenza virions as revealed by immunoelectron microscopy. Virology 149: 36–43. 10.1016/0042-6822(86)90084-X [DOI] [PubMed] [Google Scholar]
- Nabel GJ, Fauci AS. 2010. Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat Med 16: 1389–1391. 10.1038/nm1210-1389 [DOI] [PubMed] [Google Scholar]
- Nakamura G, Chai N, Park S, Chiang N, Lin Z, Chiu H, Fong R, Yan D, Kim J, Zhang J, et al. 2013. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe 14: 93–103. 10.1016/j.chom.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, Fiers W. 1999. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med 5: 1157–1163. 10.1038/13484 [DOI] [PubMed] [Google Scholar]
- Ng S, Nachbagauer R, Balmaseda A, Stadlbauer D, Ojeda S, Patel M, Rajabhathor A, Lopez R, Guglia AF, Sanchez N, et al. 2019. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat Med 25: 962–967. 10.1038/s41591-019-0463-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls JM, Chan RWY, Russell RJ, Air GM, Peiris JSM. 2008. Evolving complexities of influenza virus and its receptors. Trends Microbiol 16: 149–157. 10.1016/j.tim.2008.01.008 [DOI] [PubMed] [Google Scholar]
- Ohmit SE, Thompson MG, Petrie JG, Thaker SN, Jackson ML, Belongia EA, Zimmerman RK, Gaglani M, Lamerato L, Spencer SM, et al. 2014. Influenza vaccine effectiveness in the 2011–2012 season: protection against each circulating virus and the effect of prior vaccination on estimates. Clin Infect Dis 58: 319–327. 10.1093/cid/cit736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67: 2552–2558. 10.1128/JVI.67.5.2552-2558.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond DD, Stewart SM, Lee J, Ferdman J, Bajic G, Do KT, Ernandes MJ, Suphaphiphat P, Settembre EC, Dormitzer PR, et al. 2016. Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat Med 22: 1465–1469. 10.1038/nm.4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, Harrison SC. 2018. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci 115: 168–173. 10.1073/pnas.1715471115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan GF, Fouchier RAM, Osterhaus ADME. 2007. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol 18: 529–536. 10.1016/j.copbio.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127: 361–373. 10.1016/0042-6822(83)90150-2 [DOI] [PubMed] [Google Scholar]
- Russell CJ. 2014. Acid-induced membrane fusion by the hemagglutinin protein and its role in influenza virus biology. Curr Top Microbiol Immunol 385: 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens X. 2019. The role of matrix protein 2 ectodomain in the development of universal influenza vaccines. J Infect Dis 219: S68–S74. 10.1093/infdis/jiz003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC. 2015. Viral receptor-binding site antibodies with diverse germline origins. Cell 161: 1026–1034. 10.1016/j.cell.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman JL. 1969. The role of antineuraminidase antibody in immunity to influenza virus infection. Bull World Health Organ 41: 647–650. [PMC free article] [PubMed] [Google Scholar]
- Schulman JL, Khakpour M, Kilbourne ED. 1968. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J Virol 2: 778–786. 10.1128/JVI.2.8.778-786.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Chen J, Li R, Zhang M, Wang G, Stegalkina S, Zhang L, Chen J, Cao J, Bi X, et al. 2017. A multimechanistic antibody targeting the receptor binding site potently cross-protects against influenza B viruses. Sci Transl Med 9: eaam5752. 10.1126/scitranslmed.aam5752 [DOI] [PubMed] [Google Scholar]
- Shope RE. 1931. Swine influenza: III. Filtration experiments and etiology. J Exp Med 54: 373–385. 10.1084/jem.54.3.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69: 531–569. 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- Skowronski DM, Janjua NZ, De Serres G, Hottes TS, Dickinson JA, Crowcroft N, Kwindt TL, Tang P, Charest H, Fonseca K, et al. 2011. Effectiveness of AS03 adjuvanted pandemic H1N1 vaccine: case-control evaluation based on sentinel surveillance system in Canada, autumn 2009. BMJ 342: c7297. 10.1136/bmj.c7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W, Andrewes CH, Laidlaw PP. 1933. A virus obtained from influenza patients. Lancet 222: 66–68. 10.1016/S0140-6736(00)78541-2 [DOI] [Google Scholar]
- Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, Bean T, Barclay W, Deeks JJ, Lalvani A. 2013. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 19: 1305–1312. 10.1038/nm.3350 [DOI] [PubMed] [Google Scholar]
- Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, Strohmeier S, Yu W, Nachbagauer R, Mudd PA, et al. 2019. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 366: 499–504. 10.1126/science.aay0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16: 265–273. 10.1038/nsmb.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HX, Jegaskanda S, Juno JA, Esterbauer R, Wong J, Kelly HG, Liu Y, Tilmanis D, Hurt AC, Yewdell JW, et al. 2019. Subdominance and poor intrinsic immunogenicity limit humoral immunity targeting influenza HA stem. J Clin Invest 129: 850–862. 10.1172/JCI123366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7: 440–451. 10.1016/j.chom.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Kash JC, Morens DM. 2019. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci Transl Med 11: eaau5485. 10.1126/scitranslmed.aau5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.tenOever B. 2019. Synthetic virology: building viruses to better understand them. Cold Spring Harb Perspect Med 10.1101/cshperspect.a038703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LLM, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3: e3942. 10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Topham DJ, DeDiego ML, Nogales A, Sangster MY, Sant A. 2019. Immunity to influenza infection in humans. Cold Spring Harb Perspect Med 10.1101/cshperspect.a038729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend AR, Skehel JJ. 1982. Influenza A specific cytotoxic T-cell clones that do not recognize viral glycoproteins. Nature 300: 655–657. 10.1038/300655a0 [DOI] [PubMed] [Google Scholar]
- Townsend AR, Skehel JJ. 1984. The influenza A virus nucleoprotein gene controls the induction of both subtype specific and cross-reactive cytotoxic T cells. J Exp Med 160: 552–563. 10.1084/jem.160.2.552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend AR, Rothbard J, Gotch FM, Bahadur G, Wraith D, McMichael AJ. 1986. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell 44: 959–968. 10.1016/0092-8674(86)90019-X [DOI] [PubMed] [Google Scholar]
- Treanor JJ, Talbot HK, Ohmit SE, Coleman LA, Thompson MG, Cheng PY, Petrie JG, Lofthus G, Meece JK, Williams JV, et al. 2012. Effectiveness of seasonal influenza vaccines in the United States during a season with circulation of all three vaccine strains. Clin Infect Dis 55: 951–959. 10.1093/cid/cis574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Braeckel-Budimir N, Harty JT. 2017. Influenza-induced lung Trm: not all memories last forever. Immunol Cell Biol 95: 651–655. 10.1038/icb.2017.32 [DOI] [PubMed] [Google Scholar]
- Varghese JN, Laver WG, Colman PM. 1983. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 303: 35–40. 10.1038/303035a0 [DOI] [PubMed] [Google Scholar]
- Villalón-Letelier F, Brooks AG, Saunders PM, Londrigan SL, Reading PC. 2017. Host cell restriction factors that limit influenza A infection. Viruses 9: 376. 10.3390/v9120376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wan Y, Qiu C, Quiñones-Parra S, Zhu Z, Loh L, Tian D, Ren Y, Hu Y, Zhang X, et al. 2015. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8+ T cells. Nat Commun 6: 6833. 10.1038/ncomms7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, et al. 2019. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 177: 1124–1135.e16. 10.1016/j.cell.2019.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss CD, Wang W, Lu Y, Billings M, Eick-Cost A, Couzens L, Sanchez JL, Hawksworth AW, Seguin P, Myers CA, et al. 2019. Neutralizing and neuraminidase antibodies correlate with protection against influenza during a late season A/H3N2 outbreak among unvaccinated military recruits. Clin Infect Dis ciz1198. 10.1093/cid/ciz1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci 108: 14216–14221. 10.1073/pnas.1111497108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289: 373–378. 10.1038/289373a0 [DOI] [PubMed] [Google Scholar]
- Wilkins AL, Kazmin D, Napolitani G, Clutterbuck EA, Pulendran B, Siegrist CA, Pollard AJ. 2017. AS03- and MF59-adjuvanted influenza vaccines in children. Front Immunol 8: 1760. 10.3389/fimmu.2017.01760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K, Wei Y, Szwajcer A, Rabbani R, Zarychanski R, Abou-Setta AM, Mahmud SM. 2017. Efficacy and safety of high-dose influenza vaccine in elderly adults: a systematic review and meta-analysis. Vaccine 35: 2775–2780. 10.1016/j.vaccine.2017.03.092 [DOI] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289: 366–373. 10.1038/289366a0 [DOI] [PubMed] [Google Scholar]
- Wohlbold TJ, Krammer F. 2014. In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses 6: 2465–2494. 10.3390/v6062465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Wilson IA. 2017. A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J Mol Biol 429: 2694–2709. 10.1016/j.jmb.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wu NC, Wilson IA. 2020. Influenza hemagglutinin structures and antibody recognition. Cold Spring Harb Perspect Med 10: a038778. 10.1101/cshperspect.a038778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong W-P, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21: 1065–1070. 10.1038/nm.3927 [DOI] [PubMed] [Google Scholar]
- Yewdell JW. 2011. Viva la revolución: rethinking influenza A virus antigenic drift. Curr Opin Virol 1: 177–183. 10.1016/j.coviro.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost SJ, Parkhouse K, Gumina ME, Kim K, Diaz Perez S, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE. 2017. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci 114: 12578–12583. 10.1073/pnas.1712377114 [DOI] [PMC free article] [PubMed] [Google Scholar]