Abstract
Treatment of high-risk neuroblastoma typically incorporates multiagent chemotherapy, surgery, radiation therapy, autologous stem cell transplantation, immunotherapy, and differentiation therapy. The discovery of activating mutations in ALK receptor tyrosine kinase (ALK) in ∼8% of neuroblastomas opens the possibility of further improving outcomes for this subset of patients with the addition of ALK inhibitors. ALK inhibitors have shown efficacy in tumors such as non-small-cell lung cancer and anaplastic large cell lymphoma in which wild-type ALK overexpression is driven by translocation events. In contrast, ALK mutations driving neuroblastomas are missense mutations in the tyrosine kinase domain yielding constitutive activation and differing sensitivity to available ALK inhibitors. We describe a case of a patient with relapsed, refractory, metastatic ALK F1174L-mutated neuroblastoma who showed no response to the first-generation ALK inhibitor crizotinib but had a subsequent complete response to the ALK/ROS1 inhibitor lorlatinib. The patient's disease relapsed after 13 mo of treatment. Sequencing of cell-free DNA at the time of relapse pointed toward a potential mechanism of acquired lorlatinib resistance: amplification of CDK4 and FGFR1 and a NRAS Q61K mutation. We review the literature regarding differing sensitivity of ALK mutations found in neuroblastoma to current FDA-approved ALK inhibitors and known pathways of acquired resistance. Our report adds to the literature of important correlations between neuroblastoma ALK mutation status and clinical responsiveness to ALK inhibitors. It also highlights the importance of understanding acquired mechanisms of resistance.
Keywords: neuroblastoma
INTRODUCTION
Neuroblastoma is the most common extracranial solid tumor in children and most often arises from precursor sympathoadrenal cells of the neural crest (Stiller and Parkin 1992). Successful treatment of patients with high-risk neuroblastoma remains a major challenge, and metastatic neuroblastoma is a leading cause of childhood cancer–related death, despite the use of multimodal dose–intensive chemotherapy, surgery, radiation therapy, 131I-metaiodobenzylguanidine (MIBG) therapy, immunotherapeutic strategies, and autologous hematopoietic stem cell transplantation (Cohn et al. 2009; Smith et al. 2010; Park et al. 2013). Genomic amplification of MYCN, chromosome alterations of 1p deletion, 11q aberration, and unbalanced gain of 17q are associated with poor prognosis in neuroblastoma (Attiyeh et al. 2005; Bagatell et al. 2009; Schleiermacher et al. 2012). The discovery that ∼8% of neuroblastomas harbor activating mutations in ALK receptor tyrosine kinase (ALK) opened the possibility of using ALK inhibitors as new treatment strategies (Chen et al. 2008; George et al. 2008; Janoueix-Lerosey et al. 2008; Mossé et al. 2008; De Brouwer et al. 2010; Bresler et al. 2014). Although translocation events up-regulate wild-type (WT) ALK expression in ALK-driven tumors such as anaplastic large cell lymphoma (ALCL) (Tsuyama et al. 2017) or non-small-cell lung cancer (NSCLC) (Choi et al. 2010), ALK mutations in neuroblastoma most often occur in the tyrosine kinase domain and result in its constitutive activation. Eighty-five percent of ALK mutations in neuroblastoma are found in one of three hotspot residues in the tyrosine kinase domain—amino acids F1174, F1245, and R1275 (Pugh et al. 2013). These mutations inherently confer variable resistance to clinically available ALK inhibitors, with F1174 mutations reported as the most resistant to the first-generation ALK inhibitor crizotinib. Here we present a case of a child with relapsed, refractory, metastatic ALK F1174L-mutated neuroblastoma, treated with the third-generation ALK /ROS1 inhibitor lorlatinib, who achieved a durable complete response lasting 13 mo. Upon subsequent relapse, next-generation sequencing revealed potential resistance mechanisms that had arisen in the tumor.
RESULTS
The patient initially presented at 37 mo of age with a large facial mass, and initial imaging and diagnostic studies led to a diagnosis of metastatic, high-risk neuroblastoma. She had no significant past medical history, and no pertinent family history suggestive of an underlying cancer predisposition syndrome. Biopsy of the facial mass yielded tumor cells that stained positive for chromogranin, synaptophysin, and neuron-specific enolase. The stains for CD45, CD99, cytokeratin, myogenin, and S100 were negative. The International Neuroblastoma Pathology Classification was poorly differentiated, unfavorable histology. Initial sites of MIBG-avid disease included a large 7.7-cm left facial mass, a 2.8-cm right parietal bone metastasis, and a 1.5-cm mass centered in the right laminae of the T7 and T8 vertebrae (Fig. 1A,B) with a corresponding Curie score of 3 (Yanik et al. 2013). Bilateral posterior iliac crest bone marrow biopsies showed almost complete replacement of normal hematopoiesis with a uniform population of small round blue cells consistent with neuroblastoma. Initial molecular diagnostic studies revealed no evidence of MYCN amplification, no loss of heterozygosity (LOH) of Chr 1p, and no unbalanced LOH at Chr 11q. The DNA ploidy index was 1.385 (favorable). Urine catecholamine to urine creatinine ratios were significantly elevated for both vanillylmandelic acid (VMA) and homovanillic acid (HVA).
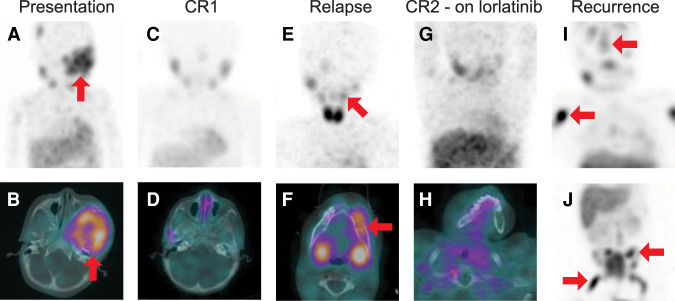
Figure 1.
Serial 131I-metaiodobenzylguanidine (MIBG) single-photon emission computed tomography (SPECT)/computed tomography (CT) studies. (A) Baseline maximum intensity projection (MIP) image of the head, neck, and chest and (B) SPECT/CT image through the skull base demonstrate bone metastatic disease including a large, radiotracer-avid skull base lesion (red arrows). (C) MIP and (D) axial SPECT/CT through the skull base following initial therapy show complete resolution of the radiotracer-avid metastatic disease. (E) MIP and (F) axial SPECT/CT through the mandible at the time of recurrence demonstrate multifocal radiotracer-avid bone metastases, including a lesion in the left mandibular body (red arrows). The MIP is obliqued in order to project the mandibular lesion away from physiologic uptake in the salivary glands. (G) MIP and (H) axial SPECT/CT through the mandible after therapy shows complete resolution of radiotracer-avid disease. (I,J) MIP images of the head, neck and chest (I) and of the abdomen and pelvis (J) demonstrate extensive new bone metastases. Representative lesions in the calvarium, right humerus, left iliac, and right femur are demarcated with red arrows.
Initial therapy included induction chemotherapy with six cycles of chemotherapy according to the schema of Children's Oncology Group (COG) Study ANBL0532, including autologous stem cell collection. Using International Neuroblastoma Response Criteria (INRC) (Park et al. 2017), a partial response (PR) was achieved, with complete response (CR) of the thoracic spine primary tumor, PR of metastatic sites with measurable tumor remaining in the face and parietal bone (Curie score of 2). Minimal disease (MD) remained in the bone marrow. A course of therapeutic MIBG resulted in a continued PR with CR of the facial tumor but residual MIBG avidity in the parietal bone, MD in the bone marrow, and a Curie score of 1. Consolidation therapy included a single autologous hematopoietic stem cell transplant using a busulfan/melphalan preparative regimen. The patient was treated with post-transplant external beam radiation to the left facial tumor (3060 cGy), parietal bone (3060 cGy), and thoracic spine (2160 cGy). Restaging postradiation therapy showed an overall CR (CR 1, Fig. 1C,D). Postconsolidation therapy with the anti-disialoganglioside (anti-GD2) monoclonal antibody dinutuximab, cytokines, and isotretinoin was completed on COG study ANBL0032. She remained in complete remission for the next 34 mo.
First Relapse
At the age of 7 yr, our patient relapsed with MIBG-avid masses under the left mandible and overriding the calvarium (Curie score of 2), as well as bilateral bone marrow involvement (Fig. 1E,F). We treated with dinutuximab, irinotecan, and temozolomide according to the COG study ANBL1221 Arm B (Mody et al. 2017). After anaphylaxis to dinutuximab, several cycles of irinotecan and temozolomide were given without dinutuximab and resulted in stable disease (SD) by INRC. Another course of therapeutic 131I-MIBG resulted in a PR with shrinkage of mandible and metastatic sites, MD in the bone marrow, symptom palliation, and clinical stability for the next 4 mo. At 9 mo after initial detection of relapse, progressive disease was detected in the mandible and calvarium along with multiple new bony metastatic sites in the thoracic and cervical vertebrae.
Next-generation sequencing on the relapsed tumor using a 125-gene commercial cancer sequencing panel had been performed revealing an ALK F1174L mutation (27% variant allele frequency [VAF]) and a non-hotspot PIK3CA mutation (C692F frameshift—26% VAF), see Table 1. Tumor purity was estimated to be 40% in the sequenced sample with DNA from peripheral blood sequenced to clarify somatic mutations. No mutations in TP53 or other reported neuroblastoma-associated genes, including ATRX, NRAS, or PTPN1, were identified (Pugh et al. 2013). As the patient's family opted against traveling for a clinical trial, ALK-targeted therapy was initiated to treat the progressive disease with the first-generation ALK inhibitor crizotinib (240 mg/m2/dose given twice daily), together with the standard cytotoxic chemotherapy regimen of cyclophosphamide (250 mg/m2/dose days 1–5) and topotecan (0.75 mg/m2/dose days 1–5) as per COG study ADVL1212 Arm C (Greengard et al. 2020). The mandible tumor progressed through this regimen during cycle 1 and palliative radiation therapy (3400 cGy) was given to the mandible to prevent airway obstruction. A final attempt with cytotoxic chemotherapy included high-dose ifosfamide, carboplatin, and etoposide (ICE) with autologous stem cell rescue (Kushner et al. 2013). Imaging following the mandible radiation and ICE chemotherapy showed stable disease persisting in the mandible, calvarium, and cervical and thoracic spine. With complications of the cytotoxic chemotherapy and prolonged treatment course, the family declined further cytotoxic chemotherapy and enrolled in hospice care. With the U.S. Federal Drug Administration (FDA) approval of lorlatinib for patients whose tumors had progressed on a first-generation ALK inhibitor, we attempted another systemic ALK-specific targeted therapy with daily dosing of lorlatinib (95 mg/m2/dose, continuous). Over the following months, symptoms, including jaw pain, fatigue, anorexia, and weight loss, completely resolved. Whole-body MIBG scan performed 7 mo after initiating lorlatinib showed CR (CR2, Fig. 1G,H). The only significant side effect of the drug was grade 2 hypercholesterolemia (total cholesterol 332–393 mg/dL).
Table 1.
Mutations in relapsed tumor detected by targeted gene sequencing panel
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP/dbVar ID | Genotype (hetero zygous/homozygous) | ClinVar ID |
|---|---|---|---|---|---|---|---|---|
| ALK | Chr 2:29220829 (GRCh38.p12) | NC_000002.12:g.29220829G > C | NP_004295.2: p.Phe1174Leu | Missense | Substitution | rs863225281 | VAF 27% | VCV000217852.1 |
| PIK3CA | Chr 3:179221045 (GRCh38.p12) | NC_000003.12: g.179221045del | NP_006209.2: p.Cys692Phefs*8 | Single-base pair deletion | Frameshift | VAF 26% | VCV000988765.1 |
(VAF) Variant allele frequency.
Second Relapse
After 13 mo on lorlatinib, our patient presented with focal back pain, and imaging revealed a mass infiltrating the T10 vertebral body and surrounding soft tissue. An MIBG scan showed diffuse, widespread avidity with Curie score of 10 (Fig. 1I,J). As the family opted for only palliative measures at this point, surgical biopsy of the relapsed disease sites was not performed. A “liquid biopsy” (sequencing cell-free tumor DNA (cfDNA) from blood) using a 74-gene commercial sequencing assay was obtained at this time in an attempt to identify a potential etiology for lorlatinib resistance (Odegaard et al. 2018). The ALK F1174L mutation was again present (31.1% cfDNA) with the same PIK3CA mutation (C692F frameshift (37.5% cfDNA) and was now accompanied by previously undetected amplifications of CDK4 (plasma copy number 17.6) and FGFR1 (plasma copy number 14.4) genes (see Methods for details of interpretation) and a new hotspot missense mutation in NRAS (Q61K, 0.4% cfDNA) (Table 2). Our patient's family elected to discontinue disease-directed therapy at that time, and she died from progressive disease 3 mo later.
Table 2.
Genomic changes detected in cfDNA after relapse during lorlatinib therapy
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP/dbVar ID | Genotype | ClinVar ID |
|---|---|---|---|---|---|---|---|---|
| ALK | Chr 2:29220829 (GRCh38.p12) | NC_000002.12:g.29220829G > C | NP_004295.2:p.Phe1174Leu | Missense | Substitution | rs863225281 | 31.1% cfDNA | VCV000217852.1 |
| PIK3CA | Chr 3:179221045 (GRCh38.p12) | NC_000003.12:g.179221045del | NP_006209.2: p.Cys692Phefs*8 | Single-base pair deletion | Frameshift | 37.5% cfDNA | VCV000988765.1 | |
| NRAS | Chr 1:114713909 (GRCh38.p12) | NC_000001.11:g.114713909G > T | NP_002515.1: p.Gln61Lys | Missense | Substitution | rs863225281 | 0.4% cfDNA | VCV000073058.5 |
| CDK4 | Chr 12:57,747,255–57,752,919 (GRCh38.p13) | NC_000012.12:57,747,255–57,752,919 | NP_000066.1 | Gene amplification | Plasma copy number: 17.6 | |||
| FGFR1 | Chr 8: 38,411,143–38,468,635 (GRCh38.p13) | NC_000008.11:38,411,143–38,468,635 | NP_001167534.1 | Gene amplification | Plasma copy number 14.4 |
(cfDNA) Cell-free tumor DNA.
DISCUSSION
We report here a case of relapsed ALK F1174L mutation–driven neuroblastoma resistant to the first-generation ALK inhibitor crizotinib combined with chemotherapy, but sensitive to the third-generation ALK/ROS1 inhibitor lorlatinib, resulting in a 13-mo remission. Sequencing of cfDNA at the time of relapse pointed toward potential ALK-independent mechanisms of acquired lorlatinib resistance—amplification of CDK4 and FGFR1, and a NRAS Q61K mutation.
ALK mutations have been identified in 6%–10% of neuroblastomas comprising both somatic missense alterations and inherited germline mutations (Chen et al. 2008; George et al. 2008; Janoueix-Lerosey et al. 2008; Mossé et al. 2008; Bresler et al. 2014; Tolbert et al. 2017). Of these, mutation at R1275 is the most common (43% of ALK mutant tumors) followed by F1174 (30%) and F1245 (12%) (Bresler et al. 2014). The remaining 15% of ALK mutations also localize to the tyrosine kinase (TK) domain of ALK. In studies of patients with germline ALK mutations, very high penetrance is observed in those harboring a R1275Q mutation. Only one case each of germline mutation at F1174 and F1245 have been reported accompanied by severe neurodevelopmental deficits (de Pontual et al. 2011). In addition to these activating missense mutations, copy-number gains and rare high-level amplification of ALK can also be seen in neuroblastoma (17% of a large neuroblastoma cohort and 4% of the high-risk patients) (Caren et al. 2008; Bresler et al. 2014). Activating mutation events and copy-number variations appear to be mutually exclusive but either case portends a worse event-free and overall survival (Bresler et al. 2014). Early implementation of effective ALK inhibition strategies could improve clinical outcomes and is being studied in ongoing clinical trials (further detailed below).
ALK tyrosine kinase inhibitors (TKI) targeting ALK mutations have been evaluated in both preclinical and clinical studies of neuroblastoma with findings summarized in Table 3. The first-generation ALK inhibitor crizotinib demonstrated remarkable clinical efficacy in patients with ALK-fusion positive NSCLC and ALCL; however, neuroblastoma harboring ALK mutations has not been similarly sensitive to crizotinib (Bresler et al. 2014). In preclinical studies, neuroblastoma cell lines with ALK R1275Q, L1198F, and I1170N mutations are sensitive to crizotinib, whereas those with ALK F1174L/C/V/I, I1171N, L1196M, L1152P, C1156Y, and G1269A mutations exhibit a relative resistance (Guan et al. 2016). Crizotinib induced a complete and sustained tumor suppression in ALK R1275Q-mutant neuroblastoma xenografts, whereas only partial inhibition of growth was seen in those driven by ALK F1174L (Bresler et al. 2011). To date, data regarding the correlation between specific ALK mutations, response to crizotinib, and clinical outcomes have been sparse. Data from neuroblastoma patients with known ALK mutation status treated on the COG trial ADVL0912 (pediatric phase I study of crizotinib monotherapy) showed one CR and one prolonged SD, both in patients with germline ALK R1275Q. In contrast, two patients whose tumors harbored somatic ALK R1275Q had progressive disease. In the same trial, four patients with the ALK F1174L mutation were treated, resulting in one patient with SD and three others with progressive disease (Mossé et al. 2013). Primary resistance is a major concern treating patients with ALK F1174L-driven neuroblastoma with single agent crizotinib, and thus approaches using combinations with chemotherapy are being tested. Crizotinib combined with cyclophosphamide and topotecan achieved complete tumor regressions in ALK-mutant neuroblastoma xenograft models (F1245C and F1174L), which are resistant to crizotinib alone (Krytska et al. 2016). Despite these preclinical data, our patient's cancer progressed through crizotinib combined with topotecan and cyclophosphamide. Combining chemotherapy with crizotinib has been tested in a pilot pediatric clinical trial (COG trial ADVL1212) (Greengard et al. 2020) to determine a recommended phase 2 dose. Five neuroblastoma patients enrolled on the trial with one achieving a CR (received crizotinib in combination with vincristine and doxorubicin) and another experiencing prolonged stable disease (received crizotinib with topotecan and cyclophosphamide). The ALK genomic alteration status (mutation or amplification) of the five neuroblastoma patients was not reported, thus limiting interpretation of the clinical response data. The ongoing COG trial for high-risk neuroblastoma ANBL1531 (NCT03126916) should yield more definitive data correlating ALK mutation status with clinical responses to crizotinib. For patients with tumors harboring ALK aberrations, crizotinib is being added to up-front induction chemotherapy and continued during consolidation, postconsolidation, and continuation therapy. Tumor and blood samples to determine ALK mutation status and monitor cell-free DNA during therapy will provide a robust data set to better understand the therapeutic role of crizotinib in ALK-mutated neuroblastoma.
Table 3.
Mutation-specific responses to ALK tyrosine kinase inhibitors in neuroblastoma
| Crizotinib |
Ceritinib |
Alectinib |
Brigatinib |
Lorlatinib | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | IC50 (nM) | Sens. | Ref. | IC50 (nM) | Sens. | Ref. | IC50 (nM) | Sens. | Ref. | IC50 (nM) | Sens. | Ref. | IC50 (nM) | Sens. | Ref. |
| Preclinical reports | |||||||||||||||
| F1174L | 130 ∼ 898 | R | 1–7 | 117 | R | 4,8 | 44 ∼ 62 | S | 4,10,11 | 55 ± 21 | S | 4 | 1.2 ∼ 26.6 | S | 2,5 |
| F1174C/V/I | 238 ∼ 274 | R | 2,4 | 109 ∼ 121 | R | 4 | 31 ∼ 46 | S | 4 | 58 ∼ 64 | S | 4 | 4.9 ± 2.6 | S | 2 |
| F1245C/V | 75 ∼ 814 | I/R | 1,2,5,7 | R | 8 | 0.8 ∼ 21.8 | S | 2,5 | |||||||
| R1275Q | 84 ∼ 320 | I/R | 3,5 | S | 8 | S | 11 | 6.1 ∼ 16.8 | S | 5 | |||||
| G1128A | 65 ∼ 138 | I/R | 1,7 | ||||||||||||
| I1170N/S | 76 ∼ 77 | S | 1 | ||||||||||||
| I1171N | 157 ∼ 532 | R | 1,4 | 119 ± 42 | S | 4 | 724 ± 130 | R | 4 | 124 ± 36 | S | 4 | |||
| R1192P | 65 ∼ 125 | S/R | 1,2,7 | 0.6 | S | 2 | |||||||||
| L1196M | 521 ∼ 589 | R | 1,4 | 67 ± 17 | S | 4 | 133 ± 55 | S | 4,11 | 41 ± 14 | S | 4 | |||
| Y1278S | 113 | R | 1 | ||||||||||||
| M1166R | 76.1 ± 2.4 | S | 1 | ||||||||||||
| Crizotinib |
Ceritinib |
Alectinib |
|||||||||||||
| Mutation | Clinical response |
Ref. | Clinical response |
Ref. | Clinical response |
Ref. | |||||||||
| Clinical reports | |||||||||||||||
| F1174L | SD, PD | 9 | |||||||||||||
| F1245C | PD | 9 | CR | 12 | |||||||||||
| R1275Q | CR, SD, PD | 9 | |||||||||||||
| Y1278S | PD | 9 | |||||||||||||
| I1171T | CR | 13 | |||||||||||||
(Sens.) Sensitivity as reported in cited references, (S) sensitive, (I) intermediate, (R) resistant, (CR) complete response, (PD) progressive disease, (SD) stable disease, (IC50) half-maximal inhibitory concentration.
References: 1 (Bresler et al. 2014), 2 (Guan et al. 2016), 3 (Bresler et al. 2011), 4 (Zhang et al. 2016), 5 (Infarinato et al. 2016), 6 (Sasaki et al. 2010), 7 (Schönherr et al. 2011), 8 (Wood et al. 2017), 9 (Mossé et al. 2013), 10 (Lu et al. 2017), 11 (Sakamoto et al. 2011), 12 (Heath et al. 2018), 13 (Guan et al. 2018).
The sensitivity of ALK mutations in neuroblastoma to second- and third-generation ALK TKI has been characterized. Although ceritinib and alectinib both demonstrate activity against ALK-mutant cell lines as single agents, alectinib has improved efficacy compared to ceritinib in the inhibition of the ALK F1174L mutation (Zhang et al. 2016). Brigatinib has shown activity against a wide range of ALK mutations in neuroblastoma, including many mutations that mediate resistance to crizotinib, ceritinib, and alectinib (Shaw et al. 2016). Brigatinib recently received FDA approval as frontline therapy for ALK-positive NSCLC, following demonstration of superior progression-free survival compared to crizotinib (Camidge et al. 2018). Clinical experience using brigatinib in neuroblastoma patients has not been reported. Lorlatinib, a third-generation inhibitor of ALK and ROS1, was developed to overcome crizotinib resistance and demonstrates activity both in vitro and in neuroblastoma xenograft models harboring F1174L and F1245C ALK mutations (Johnson et al. 2014; Guan et al. 2016; Infarinato et al. 2016; Shaw et al. 2016). Although the clinical activity of lorlatinib in patients with neuroblastoma is not yet known, a phase II study in patients with ALK-positive NSCLC treated with lorlatinib showed promising efficacy in patients with ALK mutations (Shaw et al. 2019). This study led to its FDA approval for patients with NSCLC as second-line therapy for those who have experienced progression on alectinib or ceritinib or as a third-line therapy following progression on crizotinib and a second-line ALK inhibitor. To begin to establish the role of these next-generation ALK inhibitors in neuroblastoma, a pediatric phase I trial, sponsored by the New Approaches to Neuroblastoma Therapy (NANT) Consortium, of lorlatinib alone or in combination with cyclophosphamide and topotecan in patients with ALK-driven relapsed or refractory neuroblastoma is currently enrolling patients (NANT 2015-02, NCT03107988). Detailed molecular testing of the tumors being treated in this trial will also provide necessary data to correlate response and ALK mutation status. Based on promising data showing clinical activity of lorlatinib against the broad range of ALK mutations present in neuroblastoma, COG has discussed substituting lorlatinib for crizotinib as the ALK inhibitor given to patients with ALK aberrations on the ongoing COG ANBL1531 clinical trial.
Although ALK-targeted therapies have dramatically improved clinical outcomes in multiple cancers, the development of treatment-emergent resistance remains a significant hurdle. Both ALK-dependent and ALK-independent mechanisms of resistance have been described. ALK-dependent resistance mechanisms include development of a point mutation in the TK domain in ALK-fusion or ALK-amplified tumors that initially harbor WT ALK TK domains. Second-site compound mutations in the ALK TK domain have been observed in patients treated with crizotinib (Doebele et al. 2012; Katayama et al. 2012), alectinib (Katayama et al. 2014), ceritinib (Friboulet et al. 2014), and lorlatinib (Yoda et al. 2018), and resistance-conferring activity of these mutations has been validated in cellular systems. To our knowledge, secondary resistance compound ALK mutations have not yet been reported in neuroblastoma patients. Some hypothesize that sequential treatment with first-generation and then later-generation ALK inhibitors may potentiate the accumulation of compound mutations in ALK (Yoda et al. 2018). As the role of ALK inhibitors expands in neuroblastoma, examples of this resistance mechanism may emerge. Multiple pathways resulting in ALK-independent resistance have been detailed including activation of EGFR (Yamada et al. 2012), HER2 (Wilson et al. 2015), IGF1R (Lovly et al. 2014), and KIT (Katayama et al. 2012), as well as mutations in KRAS (Doebele et al. 2012). Yoda et al. reported a variety of co-occurring mutations specifically associated with lorlatinib resistance, of which TP53 mutation was the most common. Other mutations included NRAS, BRCA1/2, APC, MAP3K1, BRAF, and CDKN2A/B copy-number variants (Yoda et al. 2018). Specific to neuroblastoma, bypass signaling pathways such as activation of AXL (Debruyne et al. 2016), PIM1 (Trigg et al. 2019), EGFR, and ERBB4 (Redaelli et al. 2018) have been reported in association with ALK TKI resistance. As the ALK F1174L mutation persisted in our patient after relapse on lorlatinib, ALK-independent mechanisms were the likely drivers of resistance. We conjecture, as have others (Yoda et al. 2018), that use of later-generation ALK inhibitors such as lorlatinib may predispose to ALK-independent resistance mechanisms in ALK-mutated neuroblastoma, an area that deserves further scientific investigation. Larger studies such as NANT 2015-02 and ANBL1531 hold promise to provide crucial information to understand the development of ALK resistance in neuroblastoma.
Analysis of cfDNA in our patient at the time of progression on lorlatinib demonstrated potential ALK-independent resistance mechanisms, including CDK4 and FGFR1 amplifications, and the NRAS Q61K mutation. As lorlatinib-resistant tumor sites could not be biopsied because of the patient's clinical condition, further validation and analysis (e.g., determination of gene copy numbers in the tumor) of these additional genetic changes was not feasible. Mutations in all three of these genes have been described in neuroblastoma and were more prevalent in relapsed neuroblastoma tumors compared to tumors sampled at diagnosis (Padovan-Merhar et al. 2016). CDK4 acts as a positive cell cycle regulator, and its activation has been implicated in the tumorigenesis of neuroblastoma and represents a therapeutic target (Rihani et al. 2015). Fibroblast growth factor receptors (FGFRs) activate downstream MEK-ERK signaling and thereby regulate cell survival and proliferation. FGFRs can also be up-regulated in response to TKI such as ALK inhibitors via STAT3 activation (Lee et al. 2014). Amplification of FGFR1 is found in several cancer types and novel inhibitors of this receptor family are being developed (Babina and Turner 2017). Mutations activating the RAS-ERK signaling pathway are associated with resistance to conventional chemotherapies in patients with relapsed neuroblastoma, suggesting that inhibition of this pathway might be clinically beneficial (Eleveld et al. 2015).
Although the exact interplay of these specific genomic events in conferring resistance to lorlatinib in our patient's neuroblastoma cannot be ascertained, their co-occurrence suggests that combination targeted therapies may be more effective in such tumors. The combination of the ALK inhibitor ceritinib and CDK4/6 inhibitor ribociclib demonstrated potent cytotoxicity in ALK F1174L and F1245C-mutant cell line and patient-derived xenograft models of relapsed or refractory neuroblastoma (Wood et al. 2017) and is now being investigated in an ongoing clinical trial (NCT02780128). FGFR1/2 amplification or activating mutations, however, promote resistance to CDK4/6 inhibition (Formisano et al. 2019) and SHP2 inhibition (Lu et al. 2020), suggesting the possibility that a CDK4/6 inhibitor, or other relevant combinations, may not have benefited our patient after her second relapse. FGFR signaling activation is a common event mediating the adaptive response to downstream signaling inhibition. TKI and/or MEK inhibition leads to loss of feedback activation of FGFR and STAT3 signaling (Lee et al. 2014), and in KRAS mutant lung cancer, FGFR1 signaling emerged as a prominent mechanism of adaptive resistance to trametinib, which was sensitive to dual MEK /FGFR inhibition (Manchado et al. 2016). Perhaps for our patient and others, the combination of effective ALK inhibition with FGFR inhibitors might have enhanced antitumor activity, a strategy that has not to our knowledge been explored clinically.
When targeting tumor resistance mechanisms, tumor heterogeneity cannot be ignored. We suspect the ALK F1174L mutation was uniformly present when our patient initially relapsed as all her sites of disease completely regressed while on lorlatinib. However, deep sequencing of the initial relapsed tumor to determine and confirm clonality was not performed. The cfDNA analysis after her second relapse definitively indicated genomic heterogeneity in the sites of disease with different levels of the genomic alterations detected from the blood. Padovan-Merhar et al. (2016) detailed DNA sequencing analysis of different relapse sites in neuroblastoma patients and demonstrated striking heterogeneity including examples of loss of previously identified mutations, accumulation of new mutations, and further amplification events. Tumor whole-exome sequencing and cfDNA studies in a cohort of neuroblastoma patients also identified significant heterogeneity not only in relapsed disease but also at the time of initial diagnosis (Chicard et al. 2018). Although the cfDNA analysis from our patient pointed to potential resistance mechanisms that represented potential targets for therapy, it is impossible to know which resistance mechanisms might be dominant drivers in the various sites of relapsed disease. Tumor heterogeneity remains a significant obstacle for both cytotoxic and targeted therapy approaches.
Our case highlights the potential utility of liquid biopsies for genomic testing of cfDNA in patients treated with targeted therapy. The significant benefit in our patient was identifying new molecular alterations at the time of relapse without requiring a morbid biopsy. In retrospect, there may have been much to learn had we used a cfDNA assay to monitor for treatment-emergent resistance during the period of lorlatinib treatment. Advances in the field of liquid biopsies for pediatric tumors were recently reviewed (Van Paemel et al. 2020). In neuroblastoma patients, MYCN amplification, ALK mutations, and whole-chromosome and segmental chromosomal abnormalities have been detected from cfDNA (Kurihara et al. 2015; Van Roy et al. 2017; Klega et al. 2018). Liquid biopsy–based monitoring of therapy response in pediatric cancer patients has been evaluated in some small studies and appears promising to monitor therapy or predict relapse (Chicard et al. 2018; Su et al. 2020). Chicard et al. utilized cfDNA to identify clones resistant to treatment in neuroblastoma patients and discover potential targetable mutations (Chicard et al. 2018). We hypothesize that serial monitoring of cfDNA will have significant utility in patients like ours receiving targeted therapies in which emergent resistance is expected. This hypothesis will be tested in larger ongoing prospective trials of patients with neuroblastoma with correlative aims analyzing cfDNA throughout therapy (COG ANBL1531 NCT03126916, NANT 2015-02 NCT03107988, NCT02546453).
In summary, we describe a patient with relapsed, refractory neuroblastoma harboring the ALK F1174L mutation resistant to the first-generation ALK inhibitor crizotinib combined with chemotherapy in whom lorlatinib induced a durable complete remission of 13 mo. However, as is often the case with targeted therapeutic approaches, resistance, potentially mediated by new genomic alterations including CDK4 and FGFR1 amplification and NRAS mutation, led to disease recurrence. Our case provides an example of clinical benefit made possible by the development of next-generation ALK inhibitors but also highlights the need for increased understanding of mechanisms of acquired resistance. We propose that molecular monitoring during therapy may guide rational combination multidrug approaches to overcome and prevent resistance.
METHODS
Next-generation sequencing of the relapsed tumor was conducted using the CancerSELECT 125 panel commercially available from Personal Genome Diagnostics, which evaluates a total of 125 cancer-associated genes for detection of single-nucleotide variants (SNVs), small indels, amplifications, translocations, and microsatellite instability. Sequencing was performed using DNA isolated from both formalin-fixed paraffin-embedded tissue tumor (relapsed tumor from mandibular mass with estimated 40% pathological tumor purity) and peripheral blood specimens to identify somatic mutations. Sequences were aligned to Human Reference genome hg19. A complete list of genes included in the assay and sequencing coverage can be found in Supplemental Table S1. Details regarding the sequencing and bioinformatics analysis pipeline were previously published (Jones et al. 2015).
At the time of second relapse on lorlatinib, cfDNA was assessed with the Guardant360 CDx assay (Guardant Health, Inc.). Guardant360 sequenced 74 cancer-associated genes to identify somatic alterations. cfDNA was extracted from plasma, enriched for targeted regions, and sequenced using the Illumina platform and hg19 as the reference genome as previously described (Lanman et al. 2015; Odegaard et al. 2018). All exons are sequenced in some genes; only clinically significant exons are sequenced in other genes. The types of genomic alterations detected by Guardant360 include SNVs, gene amplifications, fusions, short insertions/deletions, and splice site–disrupting events. Gene amplification events are reported as the average copy number for the detected gene in the circulating cfDNA. This copy number is dependent on both tumor fraction of the total cfDNA and the magnitude of the tumor amplification. Thus, the reported plasma copy number is not the same as the number of gene amplification events that could be measured directly by fluorescence in situ hybridization of tumor tissue or DNA sequencing of a tumor sample. Guardant360 reports a semiquantitative scale for amplification events. For our patient, amplification of CDK4 (plasma copy number 17.6) is reported as “high” (amplification magnitude is greater than the 50th percentile for CDK4 amplifications detected by Guardant360) and FGFR1 (plasma copy number 14.4) is reported as “high” (amplification magnitude is greater than the 90th percentile for FGFR1 amplifications detected by Guardant360). The 74 genes analyzed in the assay include AKT1, ALK, APC, AR, ARAF, ARID1A, ATM, BRAF, BRCA1, BRCA2, CCND1, CCND2, CCNE1, CDH1, CDK12, CDK4, CDK6, CDKN2A, CTNNB1, DDR2, EGFR, ERBB2, ESR1, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, GATA3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KIT, KRAS, MAP2K1, MAP2K2, MAPK1, MAPK3, MET, MLH1, MPL, MTOR, MYC, NF1, NFE2L2, NOTCH1, NPM1, NRAS, NTRK1, NTRK3, PDGFRA, PIK3CA, PTEN, PTPN11, RAF1, RB1, RET, RHEB, RHOA, RIT1, ROS1, SMAD4, SMO, STK11, TERT, TP53, TSC1, and VHL. As previously published, the average sequencing coverage for the gene regions analyzed is 15,000× (Odegaard et al. 2018).
ADDITIONAL INFORMATION
Data Deposition and Access
Sequencing data for the CancerSELECT 125 panel (Personal Genome Diagnostics, Baltimore, Maryland) and the cfDNA assay from the Guardant360 CDx assay (Guardant Health, Inc.) were not made available for public distribution. The novel variant in PIK3CA detected in both the CancerSELECT 125 panel and the Guardant360 CDx cfDNA assay can be found on the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) under accession number VCV000988765.1.
Ethics Statement
Written consent was obtained from the patient's guardian at the time of submission for clinical testing with targeted next-generation DNA sequencing of the tumor and cfDNA. Further verbal consent from the patient's guardian was obtained and documented to publish nonidentifying details about the patient's specific diagnosis, clinical course, and care for advancement of clinical–academic practice.
Author Contributions
All authors wrote and edited the manuscript. S.P.R. composed the figure. M.D.M., A.R.C., and B.H.L. provided clinical care.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Olena M. Vaske
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Attiyeh EF, London WB, Mossé YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, et al. 2005. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med 353: 2243–2253. 10.1056/NEJMoa052399 [DOI] [PubMed] [Google Scholar]
- Babina IS, Turner NC. 2017. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer 17: 318–332. 10.1038/nrc.2017.8 [DOI] [PubMed] [Google Scholar]
- Bagatell R, Beck-Popovic M, London WB, Zhang Y, Pearson AD, Matthay KK, Monclair T, Ambros PF, Cohn SL, International Neuroblastoma Risk Group. 2009. Significance of MYCN amplification in international neuroblastoma staging system stage 1 and 2 neuroblastoma: a report from the International Neuroblastoma Risk Group database. J Clin Oncol 27: 365–370. 10.1200/JCO.2008.17.9184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler SC, Wood AC, Haglund EA, Courtright J, Belcastro LT, Plegaria JS, Cole K, Toporovskaya Y, Zhao H, Carpenter EL, et al. 2011. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci Transl Med 3: 108ra114. 10.1126/scitranslmed.3002950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler SC, Weiser DA, Huwe PJ, Park JH, Krytska K, Ryles H, Laudenslager M, Rappaport EF, Wood AC, McGrady PW, et al. 2014. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 26: 682–694. 10.1016/j.ccell.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camidge DR, Kim HR, Ahn M-J, Yang JC-H, Han J-Y, Lee J-S, Hochmair MJ, Li JY-C, Chang G-C, Lee KH, et al. 2018. Brigatinib versus crizotinib in ALK-positive non-small-cell lung cancer. N Eng J Med 379: 2027–2039. 10.1056/NEJMoa1810171 [DOI] [PubMed] [Google Scholar]
- Caren H, Abel F, Kogner P, Martinsson T. 2008. High incidence of DNA mutations and gene amplifications of the ALK gene in advanced sporadic neuroblastoma tumours. Biochem J 416: 153–159. 10.1042/BJ20081834 [DOI] [PubMed] [Google Scholar]
- Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, et al. 2008. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 455: 971–974. 10.1038/nature07399 [DOI] [PubMed] [Google Scholar]
- Chicard M, Colmet-Daage L, Clement N, Danzon A, Bohec M, Bernard V, Baulande S, Bellini A, Deveau P, Pierron G, et al. 2018. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin Cancer Res 24: 939–949. 10.1158/1078-0432.CCR-17-1586 [DOI] [PubMed] [Google Scholar]
- Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, et al. 2010. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 363: 1734–1739. 10.1056/NEJMoa1007478 [DOI] [PubMed] [Google Scholar]
- Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, et al. 2009. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 27: 289–297. 10.1200/JCO.2008.16.6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brouwer S, De Preter K, Kumps C, Zabrocki P, Porcu M, Westerhout EM, Lakeman A, Vandesompele J, Hoebeeck J, Van Maerken T, et al. 2010. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res 16: 4353–4362. 10.1158/1078-0432.CCR-09-2660 [DOI] [PubMed] [Google Scholar]
- Debruyne DN, Bhatnagar N, Sharma B, Luther W, Moore NF, Cheung NK, Gray NS, George RE. 2016. ALK inhibitor resistance in ALKF1174L-driven neuroblastoma is associated with AXL activation and induction of EMT. Oncogene 35: 3681–3691. 10.1038/onc.2015.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pontual L, Kettaneh D, Gordon CT, Oufadem M, Boddaert N, Lees M, Balu L, Lachassinne E, Petros A, Mollet J, et al. 2011. Germline gain-of-function mutations of ALK disrupt central nervous system development. Hum Mutat 32: 272–276. 10.1002/humu.21442 [DOI] [PubMed] [Google Scholar]
- Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, Kondo KL, Linderman DJ, Heasley LE, Franklin WA, et al. 2012. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non–small cell lung cancer. Clin Cancer Res 18: 1472–1482. 10.1158/1078-0432.CCR-11-2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleveld TF, Oldridge DA, Bernard V, Koster J, Colmet Daage L, Diskin SJ, Schild L, Bentahar NB, Bellini A, Chicard M, et al. 2015. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet 47: 864–871. 10.1038/ng.3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formisano L, Lu Y, Servetto A, Hanker AB, Jansen VM, Bauer JA, Sudhan DR, Guerrero-Zotano AL, Croessmann S, Guo Y, et al. 2019. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun 10: 1373. 10.1038/s41467-019-09068-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, Michellys PY, Awad MM, Yanagitani N, Kim S, et al. 2014. The ALK inhibitor ceritinib overcomes crizotinib resistance in non–small cell lung cancer. Cancer Discov 4: 662–673. 10.1158/2159-8290.CD-13-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RE, Sanda T, Hanna M, Fröhling S, Luther W, Zhang J, Ahn Y, Zhou W, London WB, McGrady P, et al. 2008. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 455: 975–978. 10.1038/nature07397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greengard E, Mossé YP, Liu X, Minard CG, Reid JM, Voss S, Wilner K, Fox E, Balis F, Blaney SM, et al. 2020. Safety, tolerability and pharmacokinetics of crizotinib in combination with cytotoxic chemotherapy for pediatric patients with refractory solid tumors or anaplastic large cell lymphoma (ALCL): a Children's Oncology Group phase 1 consortium study (ADVL1212). Cancer Chemother Pharmacol 86: 829–840. 10.1007/s00280-020-04171-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J, Tucker ER, Wan H, Chand D, Danielson LS, Ruuth K, El Wakil A, Witek B, Jamin Y, Umapathy G, et al. 2016. The ALK inhibitor PF-06463922 is effective as a single agent in neuroblastoma driven by expression of ALK and MYCN. Dis Model Mech 9: 941–952. 10.1242/dmm.024448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J, Fransson S, Siaw JT, Treis D, Van den Eynden J, Chand D, Umapathy G, Ruuth K, Svenberg P, Wessman S, et al. 2018. Clinical response of the novel activating ALK-I1171T mutation in neuroblastoma to the ALK inhibitor ceritinib. Cold Spring Harb Mol Case Stud 4: a002550. 10.1101/mcs.a002550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath JA, Campbell MA, Thomas A, Solomon B. 2018. Good clinical response to alectinib, a second generation ALK inhibitor, in refractory neuroblastoma. Pediatr Blood Cancer 65: e27055. 10.1002/pbc.27055 [DOI] [PubMed] [Google Scholar]
- Infarinato NR, Park JH, Krytska K, Ryles HT, Sano R, Szigety KM, Li Y, Zou HY, Lee NV, Smeal T, et al. 2016. The ALK/ROS1 inhibitor PF-06463922 overcomes primary resistance to crizotinib in ALK-driven neuroblastoma. Cancer Discov 6: 96–107. 10.1158/2159-8290.CD-15-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, et al. 2008. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455: 967–970. 10.1038/nature07398 [DOI] [PubMed] [Google Scholar]
- Johnson TW, Richardson PF, Bailey S, Brooun A, Burke BJ, Collins MR, Cui JJ, Deal JG, Deng YL, Dinh D, et al. 2014. Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain exposure and broad-spectrum potency against ALK-resistant mutations. J Med Chem 57: 4720–4744. 10.1021/jm500261q [DOI] [PubMed] [Google Scholar]
- Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, Shukla M, Chesnick B, Kadan M, Papp E, et al. 2015. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 7: 283ra253. 10.1126/scitranslmed.aaa7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, Jessop NA, Wain JC, Yeo AT, Benes C, et al. 2012. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med 4: 120ra117. 10.1126/scitranslmed.3003316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama R, Friboulet L, Koike S, Lockerman EL, Khan TM, Gainor JF, Iafrate AJ, Takeuchi K, Taiji M, Okuno Y, et al. 2014. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res 20: 5686–5696. 10.1158/1078-0432.CCR-14-1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klega K, Imamovic-Tuco A, Ha G, Clapp AN, Meyer S, Ward A, Clinton C, Nag A, Van Allen E, Mullen E, et al. 2018. Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis Oncol 2018: PO.17.00285. 10.1200/PO.17.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krytska K, Ryles HT, Sano R, Raman P, Infarinato NR, Hansel TD, Makena MR, Song MM, Reynolds CP, Mossé YP. 2016. Crizotinib synergizes with chemotherapy in preclinical models of neuroblastoma. Clin Cancer Res 22: 948–960. 10.1158/1078-0432.CCR-15-0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara S, Ueda Y, Onitake Y, Sueda T, Ohta E, Morihara N, Hirano S, Irisuna F, Hiyama E. 2015. Circulating free DNA as non-invasive diagnostic biomarker for childhood solid tumors. J Pediatr Surg 50: 2094–2097. 10.1016/j.jpedsurg.2015.08.033 [DOI] [PubMed] [Google Scholar]
- Kushner BH, Modak S, Kramer K, Basu EM, Roberts SS, Cheung NK. 2013. Ifosfamide, carboplatin, and etoposide for neuroblastoma: a high-dose salvage regimen and review of the literature. Cancer 119: 665–671. 10.1002/cncr.27783 [DOI] [PubMed] [Google Scholar]
- Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, Collisson EA, Divers SG, Hoon DSB, Kopetz ES, et al. 2015. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 10: e0140712. 10.1371/journal.pone.0140712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. 2014. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26: 207–221. 10.1016/j.ccr.2014.05.019 [DOI] [PubMed] [Google Scholar]
- Lovly CM, McDonald NT, Chen H, Ortiz-Cuaran S, Heukamp LC, Yan Y, Florin A, Ozretić L, Lim D, Wang L, et al. 2014. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 20: 1027–1034. 10.1038/nm.3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Guan S, Zhao Y, Yu Y, Woodfield SE, Zhang H, Yang KL, Bieerkehazhi S, Qi L, Li X, et al. 2017. The second-generation ALK inhibitor alectinib effectively induces apoptosis in human neuroblastoma cells and inhibits tumor growth in a TH-MYCN transgenic neuroblastoma mouse model. Cancer Lett 400: 61–68. 10.1016/j.canlet.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Liu C, Huynh H, Le TBU, LaMarche MJ, Mohseni M, Engelman JA, Hammerman PS, Caponigro G, Hao H-X. 2020. Resistance to allosteric SHP2 inhibition in FGFR-driven cancers through rapid feedback activation of FGFR. Oncotarget 11: 265–281. 10.18632/oncotarget.27435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Weissmueller S, Morris JP 4th, Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, et al. 2016. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534: 647–651. 10.1038/nature18600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody R, Naranjo A, Van Ryn C, Yu AL, London WB, Shulkin BL, Parisi MT, Servaes S-EN, Diccianni MB, Sondel PM, et al. 2017. Irinotecan–temozolomide with temsirolimus or dinutuximab in children with refractory or relapsed neuroblastoma (COG ANBL1221): an open-label, randomised, phase 2 trial. Lancet Oncol 18: 946–957. 10.1016/S1470-2045(17)30355-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, et al. 2008. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455: 930–935. 10.1038/nature07261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossé YP, Lim MS, Voss SD, Wilner K, Ruffner K, Laliberte J, Rolland D, Balis FM, Maris JM, Weigel BJ, et al. 2013. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children's Oncology Group phase 1 consortium study. Lancet Oncol 14: 472–480. 10.1016/S1470-2045(13)70095-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, Fairclough SR, Zill OA, Sikora M, Mokhtari R, et al. 2018. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res 24: 3539–3549. 10.1158/1078-0432.CCR-17-3831 [DOI] [PubMed] [Google Scholar]
- Padovan-Merhar OM, Raman P, Ostrovnaya I, Kalletla K, Rubnitz KR, Sanford EM, Ali SM, Miller VA, Mossé YP, Granger MP, et al. 2016. Enrichment of targetable mutations in the relapsed neuroblastoma genome. PLoS Genet 12: e1006501. 10.1371/journal.pgen.1006501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JR, Bagatell R, London WB, Maris JM, Cohn SL, Mattay KK, Hogarty M, Committee COGN. 2013. Children's Oncology Group's 2013 blueprint for research: neuroblastoma. Pediatr Blood Cancer 60: 985–993. 10.1002/pbc.24433 [DOI] [PubMed] [Google Scholar]
- Park JR, Bagatell R, Cohn SL, Pearson AD, Villablanca JG, Berthold F, Burchill S, Boubaker A, McHugh K, Nuchtern JG, et al. 2017. Revisions to the International Neuroblastoma Response Criteria: a consensus statement from the National Cancer Institute clinical trials planning meeting. J Clin Oncol 35: 2580–2587. 10.1200/JCO.2016.72.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. 2013. The genetic landscape of high-risk neuroblastoma. Nat Genet 45: 279–284. 10.1038/ng.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redaelli S, Ceccon M, Zappa M, Sharma GG, Mastini C, Mauri M, Nigoghossian M, Massimino L, Cordani N, Farina F, et al. 2018. Lorlatinib treatment elicits multiple on- and off-target mechanisms of resistance in ALK-driven cancer. Cancer Res 78: 6866. 10.1158/0008-5472.CAN-18-1867 [DOI] [PubMed] [Google Scholar]
- Rihani A, Vandesompele J, Speleman F, Van Maerken T. 2015. Inhibition of CDK4/6 as a novel therapeutic option for neuroblastoma. Cancer Cell Int 15: 76. 10.1186/s12935-015-0224-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami TA, Oikawa N, Tsukuda T, Ishii N, Aoki Y. 2011. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell 19: 679–690. 10.1016/j.ccr.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L, Gray NS, Wilner K, Christensen JG, Demetri G, et al. 2010. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res 70: 10038–10043. 10.1158/0008-5472.CAN-10-2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiermacher G, Mosseri V, London WB, Maris JM, Brodeur GM, Attiyeh E, Haber M, Khan J, Nakagawara A, Speleman F, et al. 2012. Segmental chromosomal alterations have prognostic impact in neuroblastoma: a report from the INRG project. Br J Cancer 107: 1418–1422. 10.1038/bjc.2012.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönherr C, Ruuth K, Yamazaki Y, Eriksson T, Christensen J, Palmer RH, Hallberg B. 2011. Activating ALK mutations found in neuroblastoma are inhibited by crizotinib and NVP-TAE684. Biochem J 440: 405–413. 10.1042/BJ20101796 [DOI] [PubMed] [Google Scholar]
- Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Bergqvist S, Brooun A, Burke BJ, Deng YL, Liu W, Dardaei L, et al. 2016. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med 374: 54–61. 10.1056/NEJMoa1508887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Solomon BJ, Besse B, Bauer TM, Lin CC, Soo RA, Riely GJ, Ou SI, Clancy JS, Li S, et al. 2019. ALK resistance mutations and efficacy of lorlatinib in advanced anaplastic lymphoma kinase-positive non-small-cell lung cancer. J Clin Oncol 37: 1370–1379. 10.1200/JCO.18.02236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O'Leary M, Smith FO, Reaman GH. 2010. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol 28: 2625–2634. 10.1200/JCO.2009.27.0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller CA, Parkin DM. 1992. International variations in the incidence of neuroblastoma. Int J Cancer 52: 538–543. 10.1002/ijc.2910520407 [DOI] [PubMed] [Google Scholar]
- Su Y, Wang L, Jiang C, Yue Z, Fan H, Hong H, Duan C, Jin M, Zhang D, Qiu L, et al. 2020. Increased plasma concentration of cell-free DNA precedes disease recurrence in children with high-risk neuroblastoma. BMC Cancer 20: 102. 10.1186/s12885-020-6562-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert VP, Coggins GE, Maris JM. 2017. Genetic susceptibility to neuroblastoma. Curr Opin Genet Dev 42: 81–90. 10.1016/j.gde.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigg RM, Lee LC, Prokoph N, Jahangiri L, Reynolds CP, Amos Burke GA, Probst NA, Han M, Matthews JD, Lim HK, et al. 2019. The targetable kinase PIM1 drives ALK inhibitor resistance in high-risk neuroblastoma independent of MYCN status. Nat Commun 10: 5428. 10.1038/s41467-019-13315-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyama N, Sakamoto K, Sakata S, Dobashi A, Takeuchi K. 2017. Anaplastic large cell lymphoma: pathology, genetics, and clinical aspects. J Clin Exp Hematop 57: 120–142. 10.3960/jslrt.17023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Paemel R, Vlug R, De Preter K, Van Roy N, Speleman F, Willems L, Lammens T, Laureys G, Schleiermacher G, Tytgat GAM, et al. 2020. The pitfalls and promise of liquid biopsies for diagnosing and treating solid tumors in children: a review. Eur J Pediatr 179: 191–202. 10.1007/s00431-019-03545-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roy N, Van Der Linden M, Menten B, Dheedene A, Vandeputte C, Van Dorpe J, Laureys G, Renard M, Sante T, Lammens T, et al. 2017. Shallow whole genome sequencing on circulating cell-free DNA allows reliable noninvasive copy-number profiling in neuroblastoma patients. Clin Cancer Res 23: 6305–6314. 10.1158/1078-0432.CCR-17-0675 [DOI] [PubMed] [Google Scholar]
- Wilson FH, Johannessen CM, Piccioni F, Tamayo P, Kim JW, Van Allen EM, Corsello SM, Capelletti M, Calles A, Butaney M, et al. 2015. A functional landscape of resistance to ALK inhibition in lung cancer. Cancer Cell 27: 397–408. 10.1016/j.ccell.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AC, Krytska K, Ryles HT, Infarinato NR, Sano R, Hansel TD, Hart LS, King FJ, Smith TR, Ainscow E, et al. 2017. Dual ALK and CDK4/6 inhibition demonstrates synergy against neuroblastoma. Clin Cancer Res 23: 2856–2868. 10.1158/1078-0432.CCR-16-1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Takeuchi S, Nakade J, Kita K, Nakagawa T, Nanjo S, Nakamura T, Matsumoto K, Soda M, Mano H, et al. 2012. Paracrine receptor activation by microenvironment triggers bypass survival signals and ALK inhibitor resistance in EML4-ALK lung cancer cells. Clin Cancer Res 18: 3592–3602. 10.1158/1078-0432.CCR-11-2972 [DOI] [PubMed] [Google Scholar]
- Yanik GA, Parisi MT, Shulkin BL, Naranjo A, Kreissman SG, London WB, Villablanca JG, Maris JM, Park JR, Cohn SL, et al. 2013. Semiquantitative mIBG scoring as a prognostic indicator in patients with stage 4 neuroblastoma: a report from the Children's Oncology Group. J Nucl Med 54: 541–548. 10.2967/jnumed.112.112334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda S, Lin JJ, Lawrence MS, Burke BJ, Friboulet L, Langenbucher A, Dardaei L, Prutisto-Chang K, Dagogo-Jack I, Timofeevski S, et al. 2018. Sequential ALK inhibitors can select for lorlatinib-resistant compound ALK mutations in ALK-positive lung cancer. Cancer Discov 8: 714–729. 10.1158/2159-8290.CD-17-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Anjum R, Squillace R, Nadworny S, Zhou T, Keats J, Ning Y, Wardwell SD, Miller D, Song Y, et al. 2016. The potent ALK inhibitor brigatinib (AP26113) overcomes mechanisms of resistance to first- and second-generation ALK inhibitors in preclinical models. Clin Cancer Res 22: 5527–5538. 10.1158/1078-0432.CCR-16-0569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data for the CancerSELECT 125 panel (Personal Genome Diagnostics, Baltimore, Maryland) and the cfDNA assay from the Guardant360 CDx assay (Guardant Health, Inc.) were not made available for public distribution. The novel variant in PIK3CA detected in both the CancerSELECT 125 panel and the Guardant360 CDx cfDNA assay can be found on the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) under accession number VCV000988765.1.