Abstract
The vertebrate genome contains an endogenous retrovirus that has been inherited from the past millions of years. Although approximately 8% of human chromosomal DNA consists of sequences derived from human endogenous retrovirus (HERV) fragments, most of the HERVs are currently inactive and non-infectious due to recombination, deletions, and mutations after insertion into the host genome. Several studies suggested that Human endogenous retroviruses (HERVs) factors are significantly related to certain cancers. However, only limited studies have been conducted to analyze the expression of HERV derived elements at protein levels in certain cancers. Herein, we analyzed the expression profiles of HERV-K envelope (Env) and HERV-R Env proteins in eleven different kinds of cancer tissues. Furthermore, the expression patterns of both protein and correlation with various clinical data in each tissue were analyzed. The expressions of both HERV-K Env and HERV-R Env protein were identified to be significantly high in most of the tumors compared with normal surrounding tissues. Correlations between HERV Env expressions and clinical investigations varied depending on the HERV types and cancers. Overall expression patterns of HERV-K Env and HERV-R Env proteins were different in every individual but a similar pattern of expressions was observed in the same individual. These results demonstrate the expression profiles of HERV-K and HERV-R Env proteins in various cancer tissues and provide a good reference for the association of endogenous retroviral Env proteins in the progression of various cancers. Furthermore, the results elucidate the relationship between HERV-Env expression and the clinical significance of certain cancers.
Keywords: Cancer, Env, Expression pattern, HERV
INTRODUCTION
Even if most human endogenous retroviruses (HERVs) are known as endogenous and a trace of ancient viruses, recent studies have provided evidence that some viral proteins are still expressed and produce viral particles (1). Various research groups have demonstrated that between 7 and 30% of all HERV sequences in the genome are transcriptionally active (1-4). Several groups have suggested the relationship between HERV-derived genes and tumor progression and there have been various hypotheses concerning the potential contribution of gene expression to oncogenesis. HERV-K derived genes are significantly up-regulated at the RNA level in lymphoma, breast cancer, prostate cancer (5), melanoma (6), pancreatic cancer (7), and ovarian cancer (8). HERV-R derived genes have also been reported to be related to lymphoma (9).
Although the expression of HERV-derived elements may play an important role in normal physiology and cancer development, studies at the protein level are limited because of the absence of validated antibodies. In recent studies, a few antibodies for HERV-K Env and HERV-R Env have been confirmed to specifically detect each antigen (8, 10).
We previously screened the HERV-R Env protein expression profile in both adult human organs and tumors and focused on the expression profile of HERV-R Env in colorectal cancers using tissue microarrays (10, 11). Also, the HERV-K Env protein expression pattern was screened in various normal and cancer tissue microarrays (12). In this study, expressions of HERV-K Env and HERV-R Env proteins were analyzed at a grand scale involving eleven different cancer tissue microarrays containing 60 different cancer tissues of different individuals in each slide. The clinical data including gender, age, tumor stage, metastatic potential, and other conditions were also considered and analyzed to elucidate the relationship between the expression pattern of HERV-K Env and HERV-R Env proteins and cancer progression.
Furthermore, the expression pattern, co-localization, and cor-relation of HERV-K and HERV-R Env protein in the same tissue were analyzed. Our results provide a good guideline to study the expression patterns of HERV-K Env and HERV-R Env proteins and support further studies to elucidate the relationship between HERV derived proteins and the progression of certain cancers.
RESULTS
Antibody specificity
Although the specificity of antibodies has been validated in previous studies (8, 10, 12, 13-17), we confirmed the results by constructing HERV-K Env and HERV-R Env over-expressing cells. Over-expressions of HERV-K Env and HERV-R Env proteins were detected in transfected cells thereby confirming the antibody specificities for both HERV-K Env and HERV-R Env proteins (Supplementary Fig. 1A). We also conducted immunofluorescence and immunohistochemical analysis with HERV-K or HERV-R Env over-expressing cells. HERV-K Env and HERV-R Env proteins were stained specifically in transfected cells thereby confirming the antibody specificities for both HERV-K and HERV-R Env proteins in both immunofluorescence and immunohistochemical staining (Supplementary Fig. 1B). To confirm the antibodies’ cross-reactivity, HERV-K and HERV-R Env proteins were over-expressed by transfection of expression vectors and blotted with each antibody. HERV-K Env exhibited stronger staining in HERV-K Env over-expressing cells than HERV-R Env over-expressing cells when stained with HERV-K Env antibody (Supplementary Fig. 1C). HERV-R Env was also stained stronger in HERV-R over-expressing cells than HERV-K Env over-expressing cells when stained with HERV-R antibody. We performed HERV-K Env and HERV-K Env knockout for the antibody test (Supplementary Fig. 1D). These results demonstrate the specificity of both the antibodies with no cross-reactivity to each other.
General expression patterns of HERV-K Env and HERV-R Env proteins
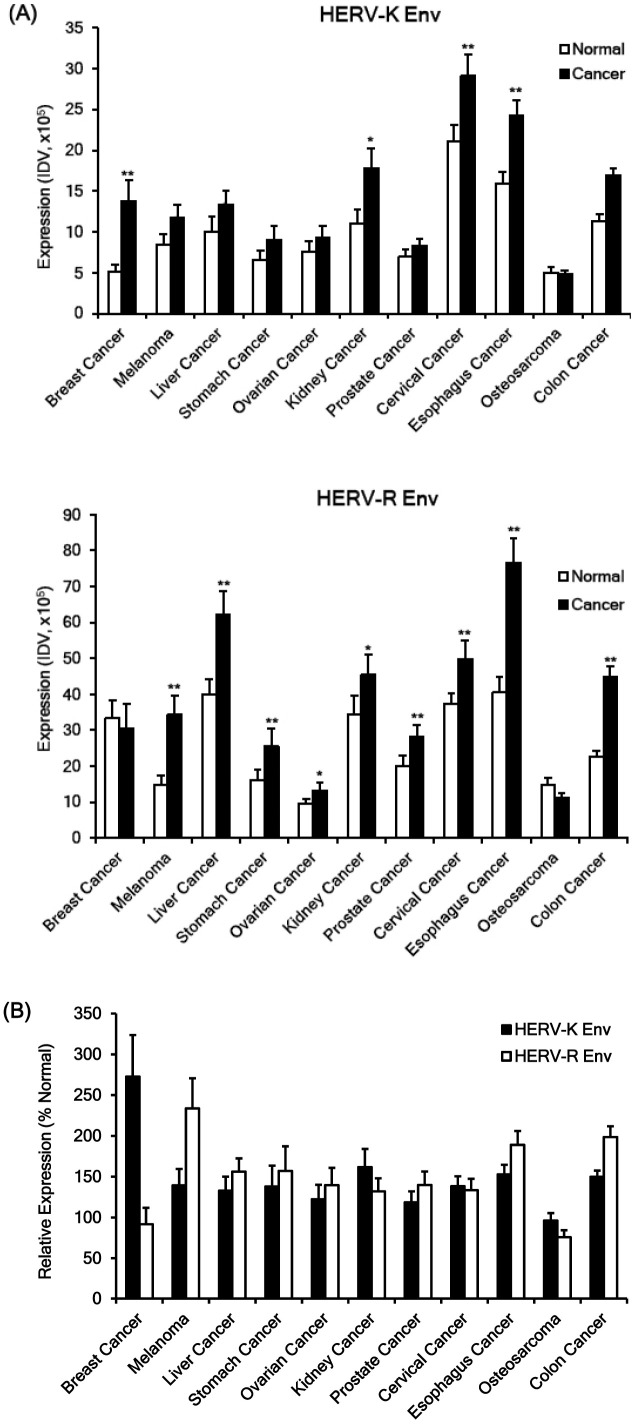
The expressions of HERV-K Env and HERV-R Env protein were significantly higher in tumor tissues compared with normal surrounding tissues in almost all types of tumors (Fig. 1). The expression of HERV-K Env was specifically high in breast cancer, melanoma, kidney cancer, prostate cancer, cervical cancer, esophagus cancer, and colon cancer. The expression of HERV-R Env protein was specifically high in melanoma, liver cancer, stomach cancer, ovarian cancer, cervical cancer, esophagus cancer, and colon cancer. However, only osteosarcoma showed weak to moderate expressions of HERV-K Env and HERV-R Env proteins (Fig. 1A, B). The relative expression of HERV-K Env and HERV-R Env to the normal surrounding tissues (%Normal) was usually similar in different tumors except breast cancer and melanoma. HERV-K Env protein demonstrated much higher expression than HERV-R Env in breast cancer whereas HERV-R demonstrated much higher expression than HERV-K Env in melanoma (Fig. 1B). All raw data of immunofluorescence staining in 11 types of cancers are shown in Supplementary Fig. 2.
Fig. 1.
Expressions of HERV-K Env and HERV-R Env proteins in normal and tumor tissues. The expressions of HERV-K Env and HERV-R Env proteins were analyzed in tissue arrays of eleven different cancers; breast cancer, melanoma, liver cancer, stomach cancer, ovarian cancer, kidney cancer, prostate cancer, cervical cancer, esophagus cancer, osteosarcoma, and colon cancer by immunofluorescence analysis and the expressions were measured by Integrated Density Value (IDV) of fluorescence with image analysis program. The expressions of HERV-K Env and HERV-R Env proteins in cancer were compared with the expression of the two proteins in the surrounding normal tissues. (A) Expressions of HERV-K Env and HERV-R Env proteins in various cancers and surrounding normal tissues. (B) Comparison of relative expressions of HERV-K Env and HERV-R Env proteins in various cancer tissues. Relative expressions of HERV-K Env and HERV-R Env were compared with the expression of the two proteins in the surrounding normal tissues (% Normal). *P < 0.05, **P < 0.01.
Analysis based on clinical data
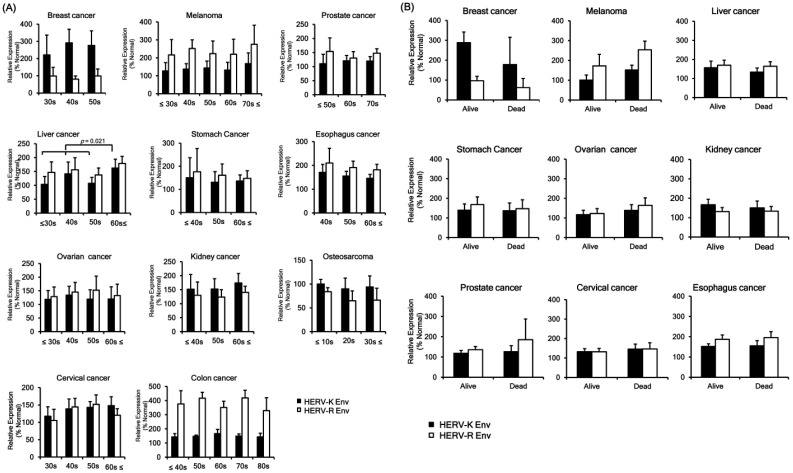
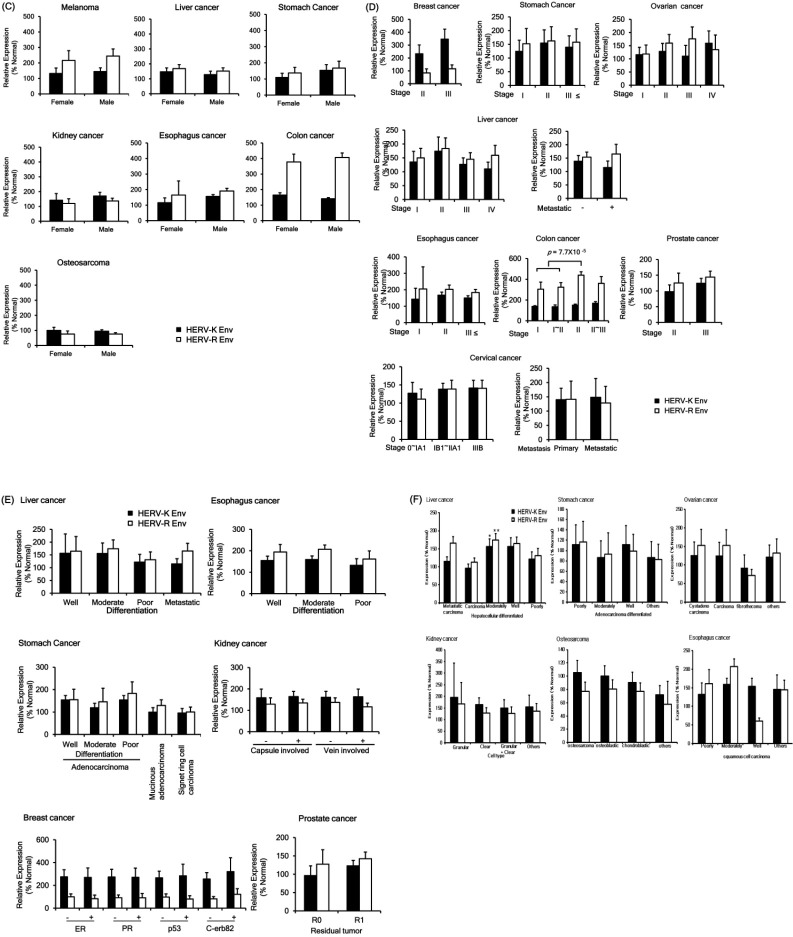
The relationship between HERVs envelope protein expression and clinical data of various cancers were analyzed. The expression of HERV-K Env protein was up-regulated in the old-age group (over 60 years) than the young age group (below 60 years) of liver cancer patients (Fig. 2A). However, HERV-K Env and HERV-R Env protein levels demonstrated no significant change among different age groups. Expressions of HERV-K Env and HERV-R Env proteins with survival prognosis showed a different pattern in certain tumors. Both HERV-K Env and HERV-R Env were downregulated in the dead group in breast cancer but up-regulated in the dead group in melanoma (Fig. 2B). However, both the patterns did not show a significant difference due to an insufficient number of patients, and further studies are needed to identify the significant relationship with a validated number of patient samples. The protein levels of HERV-K Env and HERV-R Env were not significantly changed upon survival prognosis in other cancers. HERV-K Env and HERV-R Env proteins levels were also not significantly changed with gender (Fig. 2C) and tumor stage in almost all the cancers (Fig. 2D). However, HERV-R Env protein expression was significantly increased in the tumor stage Ⅱ group of colon cancer patients (Fig. 2D). HERV-K Env protein expression was higher in the stage III group of breast cancer compared with stage II but the change was not statistically significant. Other factors including differentiation level, types of tumor, capsule or vein involvement, tumor markers, and residual tumor were not related to HERV-K Env or HERV-R Env proteins expression patterns (Fig. 2E, F). We also analyzed the correlations between the clinical data and the intensity of HERV Env protein expression in normal tissues; however, there was no significant difference in terms of age, gender, etc.
Fig. 2.
Expressions of HERV-K Env and HERV-R Env proteins according to clinical factors. (A) Expressions of HERV-K Env and HERV-R Env proteins according to age group. (B) Expressions of HERV-K Env and HERV-R Env proteins according to the survival prognosis. (C) Expressions of HERV-K Env and HERV-R Env proteins according to gender. (D) Expressions of HERV-K Env and HERV-R Env proteins according to the stage of the tumor. (E) Expressions of HERV-K Env and HERV-R Env proteins according to other factors including differentiation, cancer type, and other markers or characteristics. The expression patterns of HERV-K Env and HERV-R Env proteins were compared based on the degree of differentiation in the liver, eso-phagus, and stomach cancer. The expression patterns of HERV-K Env and HERV-R Env proteins were also compared based on capsule involvement, tumor markers, and residual type in kidney cancer, breast cancer, and prostate cancer, respectively. (F) The expression patterns of HERV-K Env and HERV-R Env proteins were compared based on the tumor type in liver cancer, stomach cancer, ovarian cancer, kidney cancer, osteosarcoma, and esophagus cancer. Relative expressions of HERV-K Env and HERV-R Env were compared with the expressions in the surrounding normal tissues (% Normal).
Correlation between HERV-K Env and HERV-R Env expressions
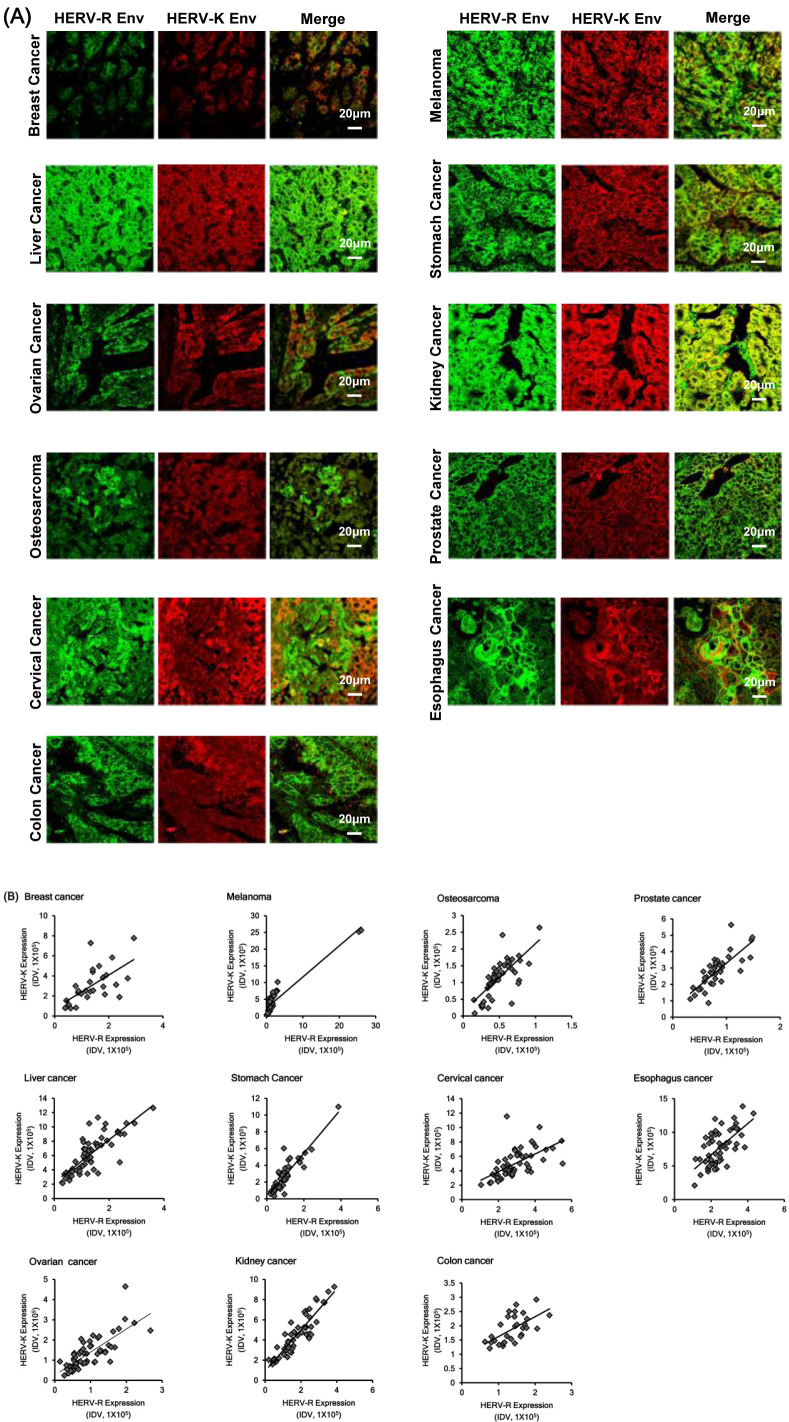
Overall expression patterns of HERV-K Env and HERV-R Env proteins were significantly up-regulated in various cancers but the expression patterns differed in each individual. Interestingly, the expression patterns of HERV-K Env and HERV-R Env proteins exhibited a significant correlation with each other in each individual. In addition, the localization of HERV-K and HERV-R Env protein in the same patient was highly merged and showed similar expression patterns (Fig. 3A). To confirm the correlation between HERV-K Env and HERV-R Env protein expressions, we calculated the correlation coefficient and significance between the expression of both the proteins. Expressions of HERV-K Env and HERV-R Env protein were extremely correlated with each other and showed significantly similar expression patterns in all 11 types of cancers and their surrounding normal tissues (Supplementary Table 2). Correlation between HERV-K and HERV-R expression patterns was significantly high in all the cancer types thereby suggesting the possibility that overall epigenomic regulation may be consistent and demonstrating the association in each individual. Fig. 3B also shows the correlation between HERV-K Env and HERV-R Env protein expressions in each patient. The analysis of the relationship between HERV-K Env and HERV-R Env protein expressions reveals significant relations in various cancers.
Fig. 3.
Correlation between HERV-K Env and HERV-R Env protein expressions in various cancers. (A) Co-localized expression of HERV-K Env and HERV-R Env proteins in various cancers. (B) Correlation between HERV-K and HERV-R Env protein expressions in various cancers. The analysis of the relationship between HERV-K Env and HERV-R Env protein expressions revealed significant relationships in various cancers.
DISCUSSION
HERVs related genetic elements constitute approximately 8% of the human genome and are distributed in multiple loci on human chromosomes (18). Despite the existence of a lot of HERV elements genes in human genes, they have been neglected as junk DNA. Accumulating evidence suggests HERV derived elements may be related to certain physiological and pathological processes (19-22). For example, HERV-W Env proteins have been reported to be involved in placental development (23, 24). Another study showed that HERVs are linked to rheumatic disease thereby aiding in the discovery of novel therapeutic agents and even cures for these chronic diseases (25). Furthermore, recent studies also indicate that HERVs related elements are significantly related to certain cancers.
HERV-H and HERV-E related elements are over-expressed in leukemia (26) and prostate carcinoma cell lines (27), respectively. The endogenous retroviral particles of HERV-K have also been found in cancer cells and they demonstrate similarity to exogenous cancer-inducing retroviruses, thereby suggesting that these ancient viruses are possibly associated with malignant transformation (15, 17, 28). The abnormal expression of HERV-K seems to trigger pathological processes that lead to melanoma onset but also contributes to the morphological and functional cellular modifications implicated in melanoma maintenance and progression (29). The prevalence of HML-2 pol, gag and env mRNA, and Gag and Env proteins has also been well established to be related to melanoma (16, 30, 31). HERV-K Env expression has also been reported to be significantly higher in most breast cancer tissues than in normal breast tissues (32). Although HERV-related proteins appear to have significant roles in normal physiology and cancers, few studies have investigated the expression pattern and localization of HERV-related proteins in normal tissues and cancer tissues. To elucidate the relationship between HERV related proteins and certain cancers, numerous studies have been conducted with various normal and cancer tissue microarrays with HERV-K and HERV-R Env antibodies (12). We selected both types of HERV Env proteins because antibodies for both proteins ensure validated results through quality control experiments. However, the previous studies have a limitation concerning the number of samples to validate the uniformity and individual difference of certain cancers. Therefore, we measured the expression levels of HERV-K and HERV-R Env protein in certain cancers with each type of cancer tissue array including around 60 different patients’ tissue samples. However, this study is associated with certain limitations because each type of tissue microarray was purchased from company and different factors including the ethnic group, location of patients, etc. were limited. The expression patterns of HERVs Env protein in each individual can be different depending on various factors so further studies with a large number of patients are necessitated. Despite the limitation of this study, there exist no studies to date on the determination of the total HERVs Env protein expression profiles in various tumors and analysis with clinical data. Therefore, this study provides a good basis for future studies to identify the role of HERVs Env protein in cancer progression with clinical significance. Furthermore, the data also suggests the possibility that HERVs Env proteins can be used as a diagnostic marker of cancer.
The mechanism of the correlation between the two proteins can be suggested based on different possibilities. The expression of HERV factors was usually regulated by epigenetic control and the correlation can be related to the methylation condition of each patient. Another mechanism to explain the correlation between the two proteins is the possibility of being controlled by the same transcription factor. HERV-K and HERV-R belong to the same HERV family, and there is a possibility that their factors will be suppressed or activated under the same regulation. To elucidate the exact mechanism of the correlation between the two proteins, further studies are necessitated.
MATERIALS AND METHODS
Antibody specificity
To test antibody specificities, we cloned full-length HERV-K env (1.7 Kb, Gene bank accession No.: DQ112095) and HERV-R env (1.8 Kb, Gene bank accession No.: NM_001007253) genes to pcDNA3.1 animal cell expression vector (Thermo Fisher Scientific, Waltham, MA, USA), respectively. HERV-K env and HERV-R env over-expression vectors were transfected into MDA-MB231 breast cancer cells using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Total cell lysates and cultured cells on chamber slides were analyzed by Western blot analysis and immunostaining, respectively.
Tissue microarray
Tissue microarray slides of certain tumors including breast cancer, melanoma, liver cancer, stomach cancer, ovarian cancer, kidney cancer, prostate cancer, cervical cancer, esophagus cancer, and osteosarcoma were purchased from SuperBioChips (SuperBioChips Laboratories, Seoul, Korea). Tissue microarray slides of colon cancer tissue were purchased from Abcam (Cambridge, UK). Around sixty normal surrounding tissues (Normal) or tumors, surgical specimens of different individuals were put on one tissue microarray slide, respectively. No other personal data except gender and age were provided. Clinical data including age, gender, tumor markers, tumor stage, survivals, tumor type, and differentiation status were supplied from the manufacturer’s datasheet. All clinical information of tissue microarray patients is described in supplementary clinical data.
Immunostaining
Immunofluorescence analysis was conducted as described previously (13). Tissue microarray slides of certain tumors were deparaffinized and hydrated before staining. For antigen retrieval, slides were immersed in citrate buffer (0.01 M, pH 6.0) and heated twice in a microwave (700 W) for 10 min. Slides were permeabilized by incubation in 10% normal serum in PBS for 1 h to block nonspecific antibodies. Slides were stained with antibody to HERV-K (1:300 dilution, HERM-1811-5, Austral Biologicals CA, San Ramon, USA) and HERV-R (1:100 dilution, ab116723, Abcam, Cambridge, UK) for overnight at 4°C. After washing thrice in PBS for 5 min, slides were incubated with secondary antibodies including Alexa Fluor 546 anti-mouse and Alexa Fluor 488 anti-rabbit antibody (Invitrogen, Carlsbad, CA, USA). Fluorescence was determined using a confocal laser-scanning microscope (LSM510 META, ZEISS, Jena, Germany). Confocal images were analyzed using AlphaEase FC image analysis software (Randburg, SA, USA). For immunohistochemistry, after antibody incubation, the color reaction was developed with Dako’s EnVision System (DAKO, Carpinteria, CA, USA).
Construction of HERV-K and HERV-R env knockout cell lines
HERV-K and HERV-R env knockout cell lines were constructed by the CRISPR-Cas9 system. Guide RNA for each env gene and CRISPR-Cas9 system were co-transfected into DLD-1 colorectal cancer cell line and stable cell lines were selected by hygromycin at a concentration of 100 μg/ml.
Statistical analysis
Descriptive statistics were used for describing patient characteristics. A two-tailed P-value < 0.05 was considered to indicate statistical significance. The most intensely stained area in the confocal microscope field of view was chosen and the fluorescence intensity was measured by image analysis software, FluorChem HD2 (San Jose, CA, USA). The intensity was used to calculate the mean values and 95% confidence intervals (CIs). Each value of cancer tissue was normalized by the average value of normal tissue. Statistical significance of differences among the groups was determined using a two-tailed Student’s t-test. The fluorescence intensity was read by image analysis software, and mean values and 95% confidence intervals (CIs) were calculated. The statistical significance of differences among the groups was determined using a two-tailed Student’s t-test. The correlation between HERV-K and HERV-R protein expressions was calculated by statistical analysis with SPSS software (IBM, Armonk, NY, USA).
Supplemental Materials
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation Grant funded by the Korean Government (NRF-2016R1D1A3 B01007444).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
References
- 1.Contreras-Galindo R, Kaplan MH, Leissner P, et al. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol. 2008;82:9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conley AB, Piriyapongsa J, Jordan IK. Retroviral promoters in the human genome. Bioinformatics. 2008;24:1563–1567. doi: 10.1093/bioinformatics/btn243. [DOI] [PubMed] [Google Scholar]
- 3.Oja M, Peltonen J, Blomberg J, Kaski S. Methods for estimating human endogenous retrovirus activities from EST databases. BMC Bioinformatics. 2007;8 Suppl 2:S11. doi: 10.1186/1471-2105-8-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perot P, Mugnier N, Montgiraud C, et al. Microarray-based sketches of the HERV transcriptome landscape. PLoS One. 2012;7:e40194. doi: 10.1371/journal.pone.0040194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishida T, Obata Y, Ohara N, et al. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun. 2008;8:15. [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn S, Ugurel S, Hanschmann KM, et al. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res Hum Retroviruses. 2008;24:717–723. doi: 10.1089/aid.2007.0286. [DOI] [PubMed] [Google Scholar]
- 7.Li M, Radvanyi L, Yin B, et al. Downregulation of Human endogenous retrovirus type K (HERV-K) viral env RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin Cancer Res. 2017;23:5892–5911. doi: 10.1158/1078-0432.CCR-17-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang-Johanning F, Rycaj K, Plummer JB, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst. 2012;104:189–210. doi: 10.1093/jnci/djr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kewitz S, Staege MS. Expression and regulation of the endogenous retrovirus 3 in Hodgkin's lymphoma cells. Front Oncol. 2013;3:179. doi: 10.3389/fonc.2013.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SH, Kang YJ, Jo JO, et al. Elevation of human ERV3-1 env protein expression in colorectal cancer. J Clin Pathol. 2014;67:840–844. doi: 10.1136/jclinpath-2013-202089. [DOI] [PubMed] [Google Scholar]
- 11.Rhyu DW, Kang YJ, Ock MS, et al. Expression of human endogenous retrovirus env genes in the blood of breast cancer patients. Int J Mol Sci. 2014;15:9173–9183. doi: 10.3390/ijms15069173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo JO, Kang YJ, Ock MS, et al. Expression profiles of HERV-K Env protein in normal and cancerous tissues. Genes Genom. 2016;38:91–107. doi: 10.1007/s13258-015-0343-9. [DOI] [Google Scholar]
- 13.Yun KY, Ko EJ, Kim HY, et al. Long interspersed element-1 open reading frame 1 protein expression profiles in ovarian cancers. Genes Genom. 2017;39:1157–1162. doi: 10.1007/s13258-017-0589-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Rycaj K, Geng S, et al. Expression of human endogenous retrovirus type K envelope protein is a novel candidate prognostic marker for human breast cancer. Genes Cancer. 2011;2:914–922. doi: 10.1177/1947601911431841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krone B, Grange JM. Melanoma, Darwinian medicine and the inner world. J Cancer Res Clin Oncol. 2010;136:1787–1794. doi: 10.1007/s00432-010-0949-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 17.Kang YJ, Jo JO, Ock MS, et al. Human ERV3-1 env protein expression in various human tissues and tumours. J Clin Pathol. 2014;67:86–90. doi: 10.1136/jclinpath-2013-201841. [DOI] [PubMed] [Google Scholar]
- 18.Kurth R, Bannert N. Beneficial and detrimental effects of human endogenous retroviruses. Int J Cancer. 2010;126:306–314. doi: 10.1002/ijc.24902. [DOI] [PubMed] [Google Scholar]
- 19.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A. 2004;101 Suppl 2:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol. 2010;20:246–253. doi: 10.1016/j.semcancer.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Freimanis G, Hooley P, Ejtehadi HD, et al. A role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: investigating mechanisms of pathogenesis. Clin Exp Immunol. 2010;160:340–347. doi: 10.1111/j.1365-2249.2010.04110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong BH, Lee YJ, Carp RI, Kim YS. The prevalence of human endogenous retroviruses in cerebrospinal fluids from patients with sporadic Creutzfeldt-Jakob disease. J Clin Virol. 2010;47:136–142. doi: 10.1016/j.jcv.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Blond JL, Lavillette D, Cheynet V, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/JVI.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mi S, Lee X, Li X, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 25.Tugnet N, Rylance P, Roden D, Trela M, Nelson P. Human endogenous retroviruses (HERVs) and autoimmune rheumatic disease: is there a link? Open Rheumatol J. 2013;7:13–21. doi: 10.2174/1874312901307010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindeskog M, Blomberg J. Spliced human endo-genous retroviral HERV-H env transcripts in T-cell leukaemia cell lines and normal leukocytes: alternative splicing pattern of HERV-H transcripts. J Gen Virol. 1997;78(Pt 10):2575–2585. doi: 10.1099/0022-1317-78-10-2575. [DOI] [PubMed] [Google Scholar]
- 27.Wang-Johanning F, Frost AR, Jian B, et al. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 28.Kaufmann S, Sauter M, Schmitt M, et al. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J Gen Virol. 2010;91:1494–1502. doi: 10.1099/vir.0.014241-0. [DOI] [PubMed] [Google Scholar]
- 29.Cegolon L, Salata C, Weiderpass E, Vineis P, Palu G, Mastrangelo G. Human endogenous retroviruses and cancer prevention: evidence and prospects. BMC Cancer. 2013;13:4. doi: 10.1186/1471-2407-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiche J, Pauli G, Ellerbrok H. Differential expression of human endogenous retrovirus K transcripts in primary human melanocytes and melanoma cell lines after UV irradiation. Melanoma Res. 2010;20:435–440. doi: 10.1097/CMR.0b013e32833c1b5d. [DOI] [PubMed] [Google Scholar]
- 31.Serafino A, Balestrieri E, Pierimarchi P, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res. 2009;315:849–862. doi: 10.1016/j.yexcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 32.Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.