Abstract
Background
Considering the clinical and genetic characteristics, acute lymphoblastic leukemia (ALL) is a rather heterogeneous hematological neoplasm for which current standard diagnostics require various analyses encompassing morphology, immunophenotyping, cytogenetics, and molecular analysis of gene fusions and mutations. Hence, it would be desirable to rely on a technique and an analytical workflow that allows the simultaneous analysis and identification of all the genetic alterations in a single approach. Moreover, based on the results with standard methods, a significant amount of patients have no established abnormalities and hence, cannot further be stratified.
Methods
We performed WTS and WGS in 279 acute lymphoblastic leukemia (ALL) patients (B-cell: n = 211; T-cell: n = 68) to assess the accuracy of WTS, to detect relevant genetic markers, and to classify ALL patients.
Results
DNA and RNA-based genotyping was used to ensure correct WTS-WGS pairing. Gene expression analysis reliably assigned samples to the B Cell Precursor (BCP)-ALL or the T-ALL group. Subclassification of BCP-ALL samples was done progressively, assessing first the presence of chromosomal rearrangements by the means of fusion detection. Compared to the standard methods, 97% of the recurrent risk-stratifying fusions could be identified by WTS, assigning 76 samples to their respective entities. Additionally, read-through fusions (indicative of CDKN2A and RB1 gene deletions) were recurrently detected in the cohort along with 57 putative novel fusions, with yet untouched diagnostic potentials. Next, copy number variations were inferred from WTS data to identify relevant ploidy groups, classifying an additional of 31 samples. Lastly, gene expression profiling detected a BCR-ABL1-like signature in 27% of the remaining samples.
Conclusion
As a single assay, WTS allowed a precise genetic classification for the majority of BCP-ALL patients, and is superior to conventional methods in the cases which lack entity defining genetic abnormalities.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-08635-5.
Keywords: Patient classification, Whole transcriptome sequencing, Fusion transcript calling, Gene expression profiling
Background
Transcriptome sequencing, usually gene expression arrays, has been a well-established diagnostic tool to characterize and quantify gene expression profiles and to detect fusion transcripts for many years. The development of RNA sequencing (RNA-Seq), including polyA-selected and whole transcript sequencing (WTS), has made it possible to broaden the analytical spectrum to study multiple transcriptional events (e.g., chimeric transcripts, isoform switching, expression, etc.) with a single approach. Compared to the expression arrays, RNA-Seq offers single base pair resolution, and considerably less background noise, providing hence, a relatively unbiased analysis of the transcriptome. Although guidelines have been established to allow for precise and context-dependent data analysis [1], no gold standard exists for any of the preprocessing steps or the downstream analyses. Hence, integrating RNA-Seq into the necessarily rigid quality standards of clinical diagnostic workflows is challenging. However, the multifaceted output of the assay can greatly benefit clinical diagnostics as indicated by various studies [2–4] and reviews [5, 6].
Acute lymphoblastic leukemia (ALL) is a heterogeneous hematological neoplasm when considering the clinical and genetic characteristics [7]. The World Health Organization (WHO) recognizes nine different sub-entities within the BCP-ALL with recurrent genetic abnormalities [8], including 4 groups characterized by the specific translocations, which result in the formation of aberrant chimeric transcripts detectable by RNA-Seq fusion calling (BCR-ABL1, KMT2A-rearranged, ETV6-RUNX1, TCF3-PBX1). Additional entities are characterized by abnormalities in chromosome numbers, or by partial amplifications: BCP-ALLs with hyperdiploidy, hypodiploidy or intrachromosomal amplification of chromosome 21 (iAMP21). More recently, further distinct ALL subtypes, including BCR-ABL1-like and ETV6-RUNX1-like ALL, were identified based on their gene expression profiles [9–11]. BCR-ABL1-like was included in the WHO classification of 2017, as a provisional entity based on treatment and prognostic implications that are associated with this high-risk subtype [8].
Currently, the diagnosis of ALL patients requires various analyses encompassing morphology, immunophenotyping, molecular analysis of gene fusions and mutations, and detection of numerical and structural abnormalities based on chromosomal banding analysis (CBA) and fluorescence in situ hybridization (FISH) [12]. With WTS parallel analysis of gene expression profiles, fusion transcripts and copy number changes becomes feasible, leading to an in-depth characterization of a patients’ genetic profile as a basis for disease classification based on the data set of a single approach.
Nevertheless, studies that comprehensively assessed all the transcriptional events in a clinical setting are scarce. In the current study, we performed detailed WTS analysis in 279 patients with newly diagnosed ALL of B- and T-lineage, to explore the complete diagnostic potential of WTS for the genetic characterization of ALL and its applicability in routine practice.
Methods
Patients and samples
Two hundred seventy nine patients with newly diagnosed ALL, sent to MLL Leukemia Laboratory between 03/2006–01/2017 for diagnostic work-up, were selected based on sample availability for WTS and WGS. ALL diagnosis was established based on morphology, immunophenotype, and cytogenetics, as previously published [13–15]. The cohort comprised 115 female (41%) and 164 male (59%) patients, with a median age of 49 years (range 0.1–93 years) at diagnosis (Additional file 3: Table S1). The patients showed B-cell precursor (BCP-ALL; n = 211) or T-cell precursor immunophenotype (T-ALL; n = 68). For WTS analysis 64 (45% female, 55% male), healthy individuals were sequenced as controls.
CBA, FISH, array-CGH
CBA was performed for all 279 cases as previously described [13]. Classification of chromosomal aberrations and karyotypes was performed according to the ISCN 2016 guidelines [16]. The FISH probes used in diagnostic work-up were selected based on recommendations, aberrations detected in CBA, and the availability of probes. Array-CGH analyses were carried out for 123 cases (4x180K microarray slides, Agilent Technologies, Santa Clara, CA). The design was based on UCSC hg19 (NCBI Build 37, February 2009).
Library preparation, sequencing, and data preprocessing
Library preparation was done as previously described [17]. In brief, genomic DNA and total RNA were extracted from lysed cell pellet of diagnostic bone marrow (n = 196) or peripheral blood (n = 83). Two hundred fifty ng of high-quality RNA were used as input for the TruSeq Total Stranded RNA kit (Illumina, San Diego, CA, USA). WGS libraries were prepared from 1 μg of DNA with the TruSeq PCR free library prep kit (Illumina). For WTS, 101 bp paired-end reads were produced on a NovaSeq 6000 system with a median yield of 68 million cluster per sample. WGS libraries were sequenced on a NovaSeq 6000 or HiSeqX instrument with 90x coverage and 150 bp paired-end sequences (Illumina). FASTQ generation was performed applying Illumina’s bcl2fastq software (v2.20). Using BaseSpace’s RNA-seq Alignment app (v2.0.1) with default parameters, reads were mapped with STAR aligner (v2.5.0a, Illumina) to the human reference genome hg19 (RefSeq annotation). Reads from WGS libraries were aligned to the human reference genome (GRCh37, Ensembl annotation) using the Isaac aligner (version 03.16.02.19).
DNA and RNA-based genotyping
The haplotype caller was used to identify the variant allele frequencies of 50 single nucleotide polymorphisms (SNP) [18], following the best practice guidelines of GATK4 [19]. The allele concordance score, defined as the ratio between the number of identical alleles and the total number of alleles, was computed for all pairwise comparisons to identify the best WGS match to each WTS profile. WTS coverage of the chromosomal region of the various SNPs were assessed by samtools [20] depth command. Only the SNPs with at least 5 reads in a patients’ WTS data, were used for the comparisons.
Structural variant and copy number variant detection on WGS data
For WGS, no sample specific normal tissue was available. A sequencing-platform and gender specific genomic DNA from a mixture of multiple anonymous donors (Promega, Fitchburg, WI, USA) was used as a normal in a tumor/unmatched normal workflow to call structural variants (SV; aberrations with > 50 bp in size) with Manta (v0.28.0). For somatic copy number variations (CNV), GATK4 was used following the Broad’s recommended best practices with a panel of normals. Specific gene deletions (IKZF1, CDKN2A, RB1) were identified by matching SV and CNV calls within the respective region.
Gene expression analysis
Estimated read counts per gene were obtained from Cufflinks 2 (version 2.2.1). Non expressed genes were filtered out (< 2 counts). Raw counts were normalized by applying the Trimmed mean of M-values method from the edgeR package [21], producing log2 CPM values. t-SNE plots were generated with the R package Rtsne (https://github.com/jkrijthe/Rtsne). Circos plots were generated with RCircos [22]. Venn diagrams were produced with BioVenn [23]. The remaining plots were generated with ggplot2 (ref. [24]). For BCR-ABL1-like expression analysis the median expression profiles of selected genes from 40 BCR-ABL1 positive and 65 BCR-ABL1 negative cases were used as references. Classification was done based on the minimal Euclidean distance.
Fusion calling on WTS data
Arriba (https://github.com/suhrig/arriba), STAR-Fusion [25], and MANTA [26] were selected for fusion calling. All the algorithms were used with default settings, except for STAR-Fusion --min_FFPM, which was set to zero, to include all candidate fusion transcripts independent of estimated expression. Fusions were only considered for further analysis, if they were called by at least two callers, could be confirmed by WGS, and were not detected in control samples. Putative novel fusions were queried against the Mitelman Database of Chromosome Aberrations and Gene Fusions (https://mitelmandatabase.isb-cgc.org/) and ChimerDB [27].
Copy number inference on WTS data
The copy number states of the autosomes were inferred from raw gene counts with the ‘import-rna’ option of the software package CNVkit [28]. The obtained results were further filtered, and only the calls with a weight > 15 were considered. Individual calls were aggregated per chromosome. A copy number state was considered as aberrant, if the log2 value was > 0.15. Samples with > 3 copy number changes (chromosome gains or losses) were selected as potential low hypodiploidv/near triploidy and high hyperdiploidy cases. Samples with either loss of ≥5 chromosomes or the specific loss of chromosomes 3, 7, 13, and 17, were assigned to the low hypodiploid/near-triploid group. Cases were categorized as high hyperdiploid if at least 2 of the following chromosomes were gained: 4, 6, 10, 14, 17, 18, and 21.
Selected small nucleotide variant analysis
The WTS data was evaluated for SNVs in CRLF2, DUX4, JAK2, KRAS, NRAS, PAX5 and TP53. Variants were called with the Isaac Variant Caller (version 2.3.13) and only the passed variants with a matching call in WGS data were included. For the WGS data, a gender-matched reference DNA was used for unmatched normal variant calling with Strelka2 (version 2.4.7).
Results
SNP profiles verify correct WGS/WTS pairing
A recently established SNP panel for both DNA and RNA-based genotyping [18], was used to identify potential sample mix-ups and/or contaminations between the WGS and WTS samples. The allele concordance score (ranging from 0 to 1; Patients & Methods) was used to identify the best matching WGS sample for each WTS profile. For 278/279 cases, the best matching WGS sample belonged to the same patient as the WTS sample with a minimal allele concordance score of 0.81 (Additional file 4: Table S2). However, for one of the samples, a substantial number of SNPs showed divergent VAFs between the WTS and WGS datasets, resulting in the elimination of the patients’ dataset.
Gene expression reliably segregates BCP-ALL from T-ALL patients
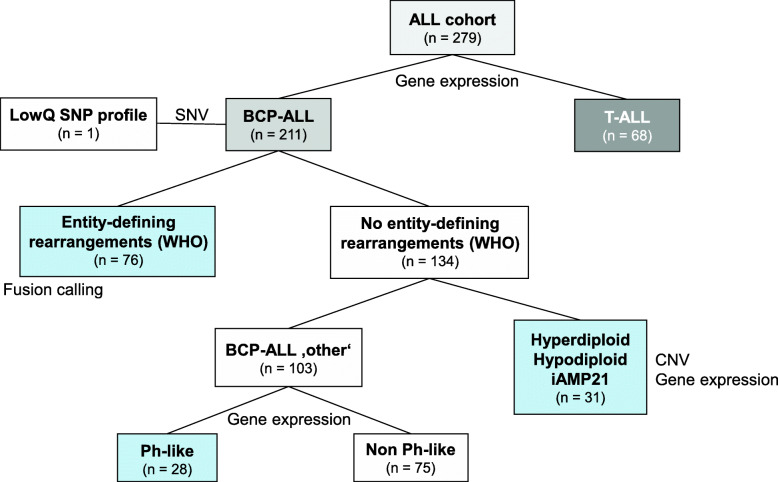
The samples were classified by WTS data following the classification tree depicted in Fig. 1. The initial classification step comprised the assignment of the samples to either the T, or the B lineage. As expected, the gene expression data could be used to reliably differentiate between BCP-ALL and T-ALL samples, based on the expression levels of 14 described markers (Additional file 1: Fig. S1A, Additional file 5: Table S3) [29]. Both lineages comprise different subtypes, characterized by the expression of various differentiation markers, and thereby defining the maturation state. Mapping the sample subtype classification (immunophenotyping data, Additional file 3: Table S1) to the two groups showed that the clusters within the groups fitted loosely to these subtypes (Additional file 1: Fig. S1B). However, further subclassification based on gene expression data, is rather challenging, and only for CD10 (common B-ALL) and CD1A (thymic T-ALL) discriminative power of the expression data could be detected (Additional file 1: Fig. S1C). The CD10 and CD1A expression values obtained from WTS correlated well with the percentage of positive cells obtained from immunophenotyping (R2 = 0.75; Additional file 1: Fig. S1D).
Fig. 1.
Classification tree. Design of the stepwise classification approach and distribution of the patients. First, DNA and RNA-based genotyping (SNV) was used to ensure correct WTS-WGS pairing, excluding one sample with low concordance. Gene expression analysis was used to distinguish between BCP-ALL or the T-ALL group. Subclassification of BCP-ALL samples was done progressively, assessing first the presence of entity-defining rearrangements (WHO) by the means of fusion detection. Next, copy number variations were inferred from WTS data to identify relevant ploidy groups. Lastly, gene expression profiling was used to identify BCR-ABL1-like signatures. ALL: Acute lymphoblastic leukemia; BCP-ALL: B-cell precursor acute lymphoblastic leukemia; CNV: copy number variation; iAMP21: intrachromosomal amplification of chromosome 21; LowQ: low quality; Ph: BCR-ABL1; SNP: single nucleotide polymorphism; T-ALL: T-cell acute lymphoblastic leukemia; WHO: world health organization
Fusion calling identifies subgroup defining rearrangements with high accuracy
Following the segregation of the samples into the two lineages, the BCP-ALL samples are further subclassified by the identification of recurrent risk-stratifying gene fusions. The median of fusions per patient was 1 (range 0–8). In total, 100 unique fusion transcripts were called in the BCP-ALL cohort. Fourteen fusion transcripts occurred recurrently, while 86 fusion transcripts could only be detected in a single patient. 56% of these fusion transcripts involved genes located on the same chromosome (48/86; intra-chromosomal fusion) and 44% were caused by structural rearrangements between two different chromosomes (38/86; inter-chromosomal fusion). Based on the results from CBA, 41 BCR-ABL1, 23 KMT2A-AFF1, 5 ETV6-RUNX1, and 4 TCF3-PBX1 fusions were detected in the cohort. The fusion calling based on WTS data identified 97% of these fusions (Table 1) with no false positives. In addition, WTS detected three other known fusion partners of KMT2A (MLLT10, MLLT1, and USP2), each in a different case, but missed one KMT2A-EPS15 fusion, assigning 76/211 BCP-ALL samples to their respective subgroups. For 15% of these samples an additional fusion transcript was called. Except for WDR37-TBRG1, which co-occurred with the reciprocal of the KMT2A-MLLT10 fusion transcript and involved the same chromosomes, but with breakpoints further apart, all of the additional fusion transcripts were intra-chromosomal (Additional file 6: Table S4).
Table 1.
Comparison of detected fusion transcripts by RNA-Seq to expected translocations by chromosome banding analysis (CBA)
| CBA | RNA-Seq | RNA-Seq/CBA [%] | |
|---|---|---|---|
| BCR-ABL1 | 41 | 40 | 98 |
| ETV6-RUNX1 | 5 | 4 | 80 |
| KMT2A-AFF1 | 23 | 23 | 100 |
| TCF3-PBX1 | 4 | 4 | 100 |
Broadening the spectra of fusion transcripts
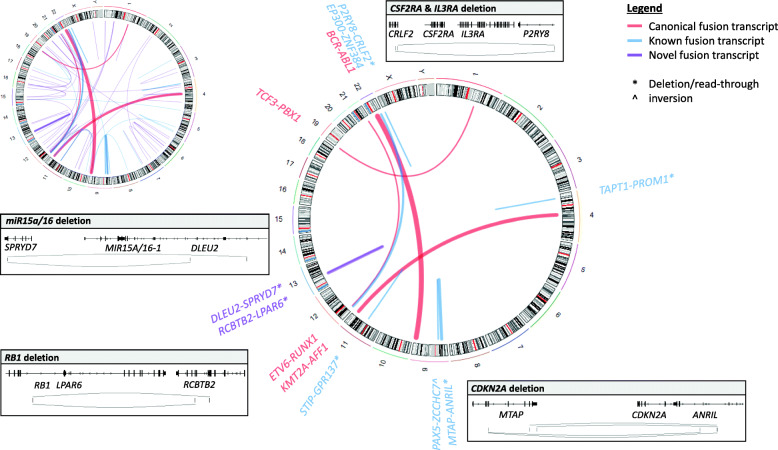
In addition to the subtype defining rearrangements, among the BCP-ALLs we identified well characterized fusions involving ZNF384 (n = 8), PAX5 (n = 3), and two fusion transcripts containing NUTM1: BRD9-NUTM1, which has been described in infant ALLs [30], and the novel fusion CHD4-NUMT1. We also identified one case with an EBF1-PDGFRB fusion, which arose from an interstitial 5q33 deletion (WGS data), and another, with a TCF3-HLF fusion transcript. Known fusion transcripts in the T-ALL cohort mainly involved MLLT10 (n = 3) and genes encoding for proteins of the nuclear pore complex (n = 5). Interestingly, we also identified recurrent read-through events, such as MTAP-ANRIL (n = 15), RCBTB2-LPAR6 (n = 12), P2RY8-CRLF2 (n = 3) and DLEU2-SPRYD7 (n = 3) in both groups. Even if the fusions themselves are most likely not biologically active, MTAP-ANRIL has been detected in melanoma patients in association with the deletion of the tumor suppressor genes CDKN2A/B [31], RCBTB2-LPAR6 indicates a partial RB1 loss as part of a larger deletion [32], and DLEU2-SPRYD7 indicates the deletion of the miR-15a/16–1 cluster (Fig. 2). The deletions were confirmed by WGS SV and CNV calls in the respective patients. Since WTS is not limited to the detection of already known chimeric transcripts, we also identified in total 57 putative novel fusion transcripts (Additional file 6: Table S4). Although the potential therapeutic consequences and functions are yet to be determined, multiple genes associated with cancer or implied in non-hematologic malignancies were found as fusion partner genes in our dataset (e.g. CHD4, HOXA7, FOXO3).
Fig. 2.
Fusion landscape of BCP-ALL. The Circos plots depict the spectra of identified fusion transcripts. Top left: all fusion transcripts; right: recurrent fusion transcripts. The line width of the central links correlates with the frequency of the fusion transcript. The colors represent the status of the fusion transcripts: Red - canonical, blue - known, purple - novel. * indicate deletion/read-through events, ^ inversions. Rectangles show fusion breakpoints of selected deletion/read-through events accompanied with gene annotations
CNV calling based on WTS data identifies relevant ploidy groups
For the remaining 134 BCP-ALLs with no subtype defining rearrangements, CNV calling was performed based on the WTS data to identify relevant ploidy groups for further subclassification (Fig. 1). Here, the CNVkit algorithm was used to identify patients with high hyperdiploidy or low hypodiploidy/near-triploidy (Patients and Methods). The algorithm correctly identified 17 (94%) low hypodiploid/near-triploid ALLs and 12 (80%) high hyperdiploid ALLs as defined by WGS, arrayCGH, and FISH (Additional file 7: Table S5). One case was misclassified as high hyperdiploid, but is most likely a near-triploid ALL, according to the WGS data. The algorithm missed 1 hypodiploid/near-triploid ALL with low blast count (20%), 3 high hyperdiploid ALL, 1 near haploid ALL, and 1 iAMP21 ALL. However, the resolution of the algorithm might be too low to reliably detect iAMP21. We thus, analyzed the expression of DYRK1A and CHAF1B that have recently been associated with iAMP21-positive ALLs [33]. The expression of both genes was indeed heightened in the iAMP21 case (Additional file 2: Fig. S2A). Based on this classification, 103 (49%) BCP-ALL samples of our cohort have no established abnormalities and are further referred to as BCP-ALL ‘other’.
BCR-ABL1-like signature identification by WTS
A compilation of the various published gene lists [34–36] was used to test for their ability to differentiate between BCR-ABL1 positive and BCR-ABL1 negative cases in our cohort. A final list of 26 genes with the highest variation between BCR-ABL1 positive and BCR-ABL1 negative cases and the reference profiles of 41 BCR-ABL1 positive and 65 BCR-ABL1 negative cases (Additional file 8: Table S6) were used to classify the 103 BCP-ALL ‘other’ cases into the BCR-ABL1-like and non BCR-ABL1-like groups, based on minimal distance. Twenty eight cases were classified as BCR-ABL1-like and the remaining 75, as non BCR-ABL1-like. Recently, a targeted RNA-Seq panel of 38 genes was published to identify adult BCP-ALL pts. with BCR-ABL1-like characteristics [37]. The application of this gene panel identified 30 BCR-ABL1-like cases, of which 28 (93%) were concordant with the group classification, by the list of 26 genes. Hence, the concordant subset of 28 samples was assigned to the BCR-ABL1-like subtype (Additional file 9: Table S7).
Characteristics of BCR-ABL1-like and non BCR-ABL1-like cases
There were no significant differences in baseline characteristics such as age, gender, and ALL phenotype between BCR-ABL1-like and non BCR-ABL1-like patients (Additional file 3: Table S1), but for an elevation of white blood cell counts in BCR-ABL1-like patients (59.03 × 109/L vs 25.18 × 109/L, P = 0.025). Among the BCP-ALL ‘other’ cases, CRLF2 showed a clear bimodal expression (Additional file 2: Fig. S2B), with a significant higher CRLF2 expression in the BCR-ABL1-like group, as compared to the non BCR-ABL1-like cases (logFC 5.17, P < 0.0001). The high CRLF2 expression could either be linked to the occurrence of a CRLF2 rearrangement or CRLF2 mutations. Only one case with a high CRLF2 expression was assigned to the non BCR-ABL1-like group. In addition, a significant enrichment of JAK2 mutations (mainly c.2047A > G) could be observed in the BCR-ABL1-like group (42% vs 0%, P < 0.001), whereas the non BCR-ABL1-like group carried a higher proportion of NRAS/KRAS (28% vs 4%, P = 0.007), PAX5 (c.239C > G, 8% vs 0%, P = 0.12) and TP53 (8% vs 0%, P = 0.12) mutations (Additional file 9: Table S7). Fusions involving PAX5, CRLF2, and tyrosine kinases were exclusively found in the BCR-ABL1-like group. All samples with detected NUTM1, HLF, and ZNF384 fusion transcripts were assigned to the non BCR-ABL1-like group and, hence, could be further subclassified based on these genetic alterations. WGS data showed that 34% of the BCP-ALL ‘other’ cases harbored a deletion in IKZF1, and as expected, these deletions were significantly more common in the BCR-ABL1-like group (61% vs 24%, P < 0.001). A similar trend could be observed for RB1 deletions (WGS data, 18% vs 4%, P = 0.019). In contrast, deletions of the tumor-suppressor gene CDKN2A (WGS data) were fairly common amongst both groups (32% vs 44%), and were not enriched in BCR-ABL1-like or non BCR-ABL1-like cases (Additional file 9: Table S7).
A multi-modal approach is superior to a classification based on gene expression profiles alone
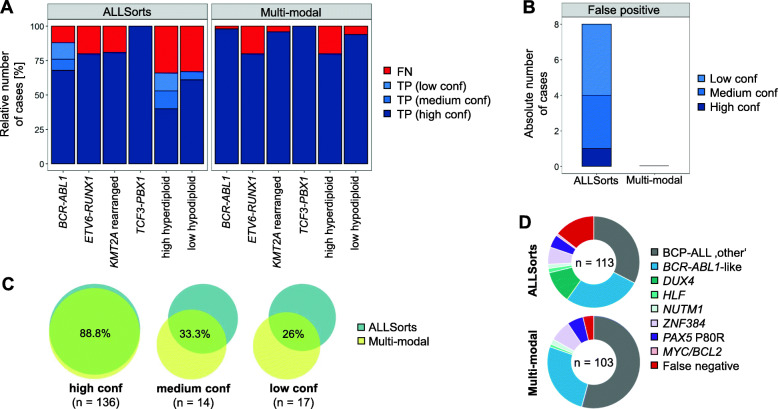
Most genetic alterations in ALLs are also associated with specific gene expression profiles, providing the basis for expression-based classification approaches such as ALLsorts (https://github.com/Oshlack/AllSorts). Hence, for BCP-ALL patients, we compared our results from the multi-modal approach to the ALLSorts classifier (see Patients & Methods; Additional file 10: Table S8). The ALLSorts classifier returns a matrix with per sample probabilities for each subtype. For the comparison, only the highest subtype probability was considered for each sample. The ALLSorts predictions were grouped into; unclassified (= BCP-ALL ‘other’; probability < 50%), low confidence (50–80% probability), medium confidence (80–90% probability), and high confidence (probability > 90%) calls.
For the fusion transcript and ploidy based WHO subgroups, the ALLSorts classifier achieved an overall accuracy of 86%, compared to 97% of our stepwise approach. The ploidy groups had the highest number of false negative calls, and less than 50% of the high hyperdiploid cases were called with high confidence by ALLSorts (Fig. 3A). Although the single iAMP21 case could be identified by gene expression as mentioned above, it was not identified as such, by ALLSorts. The ALLSorts classifier also made 8 false positive calls with different confidence levels, compared to zero false positive calls of the fusion calling (Fig. 3B). It was also evident that the overlap of assigned class labels between ALLSorts and the multi-method approach dropped from 89 to 26% with decreasing probability values (Fig. 3C). Due to the higher number of false negative calls, ALLSorts assigned more cases to the BCP-ALL ‘other’ group compared to the multi-modal approach (113 vs 103 Fig. 3D). The approaches agreed on 26 of the BCR-ABL1-like cases, while ALLSorts misclassified 3 BCR-ABL1 cases as BCR-ABL1-like. ALLSorts classified 15 patient profiles as DUX4 rearranged (Fig. 3D). However, neither DUX4 fusion transcripts nor DUX4 expression (WTS data) or the IGH-DUX4 structural variants (WGS data), could be identified in those cases. Nevertheless, compared to BCP-ALL ‘other’ cases not classified as DUX4 rearranged, an overexpression of DUX4 target genes such as PCDH17 (logFC 7.51, P-value < 0.0001), PDGFRA (logFC 5.65, P-value < 0.0001), and AGAP1 (logFC 5.52, P-value < 0.0001) could be observed. ALLSorts correctly identified 6 samples with a PAX5 c.239C > G mutation. However, one case of PAX5 c.239C > G was missed, and in one case the additional high hyperdiploidy was not detected. Both cases were correctly identified by the stepwise approach. ALLSorts correctly identified all cases harboring a ZNF384 or NUTM1 fusion transcript and one case with a HLF fusion transcript as detected by the multi-modal approach. One case was labeled as a MYC/BCL2 double-hit BCP-ALL by ALLSorts, but solely carried a MYC translocation (WGS data).
Fig. 3.
Performance comparison between the multi-modal approach and ALLSorts. Accuracy comparison for BCP-ALL patients harboring entity-defining fusion transcripts or abnormalities in chromosome number (a) and the number of false positive calls (b). c Overlap in label class for different confidence levels (low: 50–80% probability; medium: 80–90% probability; high: > 90% probability). d Breakdown of the identified BCP-ALL ‘other’ cases by both approaches. Conf: confidence; FN: false negative; TP: true positive
Discussion
Genetic aberrations in ALL are structurally diverse, and currently detected by a variety of diagnostic assays. The aim of this study was to compile a diagnostic workflow to establish whole transcriptome RNA sequencing as a reliable, comprehensive, and efficient assay for ALL diagnostics. We demonstrated that typical genetic alterations can be identified with high accuracy, while at the same time the unbiased assessment of the transcriptome also allows the identification of potentially new targets in patients, where these genetic aberrations are absent. Our results further suggest that careful selection of the algorithms for each molecular type is beneficial for accurate sample classification.
We demonstrated that samples could efficiently be classified in a stepwise approach (Fig. 1). As previously shown [38], BCP-ALLs were characterized by a homogenous CD19 gene expression, whereas T-ALLs could be identified by CD3D and CCR9 expression. Multiple entity-defining fusion transcripts are known in BCP-ALLs (BCR-ABL1, KMT2A-AFF1, TCF3-PBX1, ETV6-RUNX1), and a reliable detection is mandatory for every diagnostic workflow. The applied fusion calling procedure identified 97% of the fusions, as detected by gold standard techniques, which is in line with previous RNA-Seq studies in pediatric ALL cohorts that reported detection rates between 91 and 97% [3, 39–41]. The number of true positive calls can be increased by considering the overlap of different callers [42], while simultaneously reducing the number of false negative ones. In this study, we used three different algorithms, with only 68% of the risk-stratifying fusion events being called by all the three algorithms (Additional file 6: Table S4), advocating the combined approach. Two fusion transcripts were missed most likely due to the low fusion transcripts expression, which are very difficult to detect [3, 40] and cannot be rescued by this approach. Here, only a greater sequencing depth could solve this issue.
Beside subtype defining rearrangements, other previously described translocations could be identified, including 8 rearrangements involving ZNF384, which were recently described to constitute a new molecular subtype of BCP-ALL ‘other’ with a good response to prednisone and conventional chemotherapy [43–45]. Our study showed that WTS is especially beneficial to identify cytogenetically cryptic events (e.g. EP300-ZNF384, PAX5 fusion transcripts, SET-NUP214, etc) and unknown or divers fusion partners, in an unbiased and cost-effective way. In addition, we identified multiple recurrent read-through events indicative of gene deletions, frequent in ALL (e.g. CDKN2A/B, RB1, MIR15A/16–1), which were exclusively called by STAR-Fusion and arriba. To the best of our knowledge, two of these events have not been described before, whereas the MTAP-ANRIL fusion has been identified in a melanoma cohort, in the context of CDKN2A/B deletions [31]. CDKN2A/B deletions have been associated with poor prognosis and it has been suggested to declare them as an additional B-ALL subgroup [46]. Moreover, WTS identified 57 putative novel fusions with the majority occurring only in a single patient; similar to the findings in a study of pediatric ALL [39]. As all these fusions were detected only once, the putative role in ALL pathogenesis and their diagnostic and prognostic potential has to be determined by combining data from several studies. However, fusion transcripts involving genes with a large number of pseudogenes (e.g. DUX4) or highly variable genomic regions (e.g. IGH gene locus) are still challenging to detect with most fusion calling algorithms but this might be improved by their continuous optimization.
Estimating abnormalities involving the chromosome number, plays a major role in ALL classification and prognostication. While ALL with high hyperdiploidy is associated with a favorable prognosis, ALL with low hypodiploidy shows a poor outcome [15, 47]. Due to the interplay of multiple regulating factors, inferring copy number changes from WTS data is rather challenging [48]. In our study, the determination of ploidy groups had the highest error rate, missing 5 cases, compared to the ones from WGS, arrayCGH, and FISH. However, CBA missed 4 low hypodiploid/near-triploid cases due to low in vitro proliferation, which were identified based on WTS data and confirmed by WGS. The resolution of the applied algorithm was too low to identify the iAMP21 case, or to reliably detect single gene deletions. While in the case of iAMP21 the gene expression could be used for the classification, the same did not hold true for gene deletions, as mentioned in a previous study [3]. Here, the analysis of isoforms and differential transcript usage might provide the needed insights, but these analyses were out of the scope of this work. In addition, a larger set of iAMP21-positive cases is needed to proof the validity of CHAF1B and DYRK1A gene expression as biomarkers for the presence of iAMP21, since our cohort included only one such case.
BCR-ABL1-like ALLs are one of the most relevant new subgroups due to the potential benefit for treatment with tyrosine kinase inhibitors, as further underlined by the poor outcome for this ALL subtype on conventional treatment strategies [49]. False negative results are rare, and the actual risk and clinical impact in such cases is unknown [50]. Various gene lists have been published in the literature [34–36] for gene expression profiling, with only partial overlapping between the lists and the resulting classification. For WTS we identified a list of 26 genes to identify BCR-ABL1-like cases. The majority of the genes (65%) were also present in the recently published list of a targeted RNA-Seq panel [18], resulting in an overlap of 93% in classification results. We only characterized 13% of our BCP-ALL cohort as BCR-ABL1-like cases, which is below the typical range of 24–33% [37, 51, 52]. However, 67% of the patients from our BCP ALL cohort fall into the adult or elderly age group, and it has been shown that the frequency of BCR-ABL1-like cases declines with age, with an incidence just of between 7 and 20% for adults and elderly patients [53, 54].
In line with published data, cases with CRLF2 rearrangements and IKZF1 deletions were significantly more common in BCR-ABL1-like cases [49, 55]. One case with an IGH-CRLF2 rearrangement and high CRLF2 expression was not classified as BCR-ABL1-like ALL. In pediatric ALL it has been reported that 5 to 10% of patients with CRLF2-rearranged ALL have distinctly different gene-expression profiles without the kinase-activated signature [49]. In the BCR-ABL1-like subgroup we also identified 3 patients harboring a P2RY8-CRLF2 fusion, which is associated with poor prognosis in children [56], and 3 patients with fusions involving PAX5. Cases harboring PAX5 or CRLF2 fusions have been proposed as an independent subgroup in BCP-ALL [2]. 42% of BCR-ABL1-like cases carried a JAK2 mutation, which is comparable to previous studies that reported 27–57% of mutated JAK2 [37, 51, 52]. Among the non BCR-ABL1-like subgroup, we identified various cases with PAX5 (c.239C > G) mutations, along with cases harboring ZNF384, HLF or NUTM rearrangements, all of which have been recently identified as new BCP ALL subgroups [2, 45]. The ALLSorts algorithm also identified a DUX4 transcriptional signature in 15 cases, but no indication for DUX4 fusion transcripts could be identified in them, as these fusions were predominantly described in pediatric and AYA ALL (adolescent and young adults) [57], and our cohort included only a small number of young patients. However, it is well known that fusion transcripts involving DUX4 are difficult to detect with standard fusion calling pipelines and gene expression profiling might be superior in these instances.
Further comparison between the gene expression profile based ALLSorts classifier and our stepwise approach, showed a good concordance for high confidence calls (Fig. 3C). However, our approach applies optimized algorithms for the different molecular types, resulting in an overall more precise classification, with superior performance for the identification of ploidy groups and a reduced number of false positive calls.
Conclusion
In summary, our study demonstrates that WTS can be used to reliably classify ALL patients with a single assay and is superior to conventional methods in cases which lack entity-defining genetic abnormalities. With the decrease in sequencing costs, the integration of WTS in routine diagnostics of ALL patients seems feasible, however, requiring the definition of standardized quality parameters and data analysis workflows to enable reproducibility and comparability between laboratories.
Supplementary Information
Additional file 1: Figure S1. Gene expression of the B-cell and T-cell lineage. t-SNE plot of gene expression of selected marker genes (a-c, perplexity: 25). d) Correlation between CD10/CD1A gene expression and CD10+/CD1A+ cells as determined by immunophenotyping. Colors correspond to lineage, immunophenotypic subtype or expression as indicated by the plot legends.
Additional file 2: Figure S2. Expression of selected genes. a) CHAF1B and DYRK1A expression of BCP-ALL patients (n = 104) without risk-stratifying fusions or abnormal chromosome number. b) CRLF2 expression of BCP-ALL ‘other’ patients (n = 103).
Additional file 3: Table S1. Patient data.
Additional file 4: Table S2. Allele concordance scores between WTS and WGS SNP profiles.
Additional file 5: Table S3. Gene expression of known lineage markers.
Additional file 6: Table S4. List of called fusion transcripts in both lineages.
Additional file 7: Table S5. Copy number variations called by CNVkit for WTS data.
Additional file 8: Table S6. Expression of BCR-ABL1-like signature genes.
Additional file 9: Table S7. Characteristics of BCR-ABL1-like and non BCR-ABL1-like cases.
Additional file 10: Table S8. Comparison to ALLSorts algorithm.
Acknowledgements
The authors would like to thank all co-workers at the MLL Munich Leukemia Laboratory for their dedicated work. The authors would also like to thank all the physicians involved in patient care, data collection and providing the samples.
Abbreviations
- ALL
Acute lymphoblastic leukemia
- AYA
Adolescent and young adults
- CBA
Chromosomal banding analysis
- CNV
Copy number variation
- FISH
Fluorescence in situ hybridization
- iAMP21
Intrachromosomal amplification of chromosome 21
- SNP
Single nucleotide polymorphism
- SV
Structural variant
- WGS
Whole genome sequencing
- WHO
World Health Organization
- WTS
Whole transcriptome sequencing
Authors’ contributions
WW designed the study, WW, RS, AS and CH interpreted the data, WW wrote the manuscript. AS and CH were responsible for chromosome banding and FISH analyses, WW and MM for molecular and bioinformatic analyses, WK for immunophenotyping and TH for cytomorphologic analyses. All authors read and contributed to the final version of the manuscript.
Funding
RS received grant support from Torsten Haferlach Leukämiediagnostik Stiftung, Munich, Germany.
Availability of data and materials
All data analysed during this study are included in this published article and its supplementary information files. Additional datasets are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and the Ethics Committee of the Bavarian Medical Association for the use of archived DNA/RNA samples and associated clinical information. The clinical data were retrieved, and the samples were collected and analysed with the endorsement of the Ethics Committee of the Bavarian Medical Association. All patients had given a written informed consent, to the use of genetic and clinical data according to the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
CH, WK, and TH declare part ownership of Munich Leukemia Laboratory (MLL). WW, AS, MM are employed by the MLL. RS received grant support from Torsten Haferlach Leukämiediagnostik Stiftung, Munich, Germany.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17(1):13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J-F, Dai YT, Lilljebjörn H, Shen SH, Cui BW, Bai L, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A. 2018;115(50):E11711–E11720. doi: 10.1073/pnas.1814397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown LM, et al. The application of RNA sequencing for the diagnosis and genomic classification of pediatric acute lymphoblastic leukemia. Blood Adv 2020;4(5):930–942. Blood Adv. 2020;4:1217. doi: 10.1182/bloodadvances.2020001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arindrarto W, Borràs DM, de Groen RAL, van den Berg RR, Locher IJ, van Diessen SAME, et al. Comprehensive diagnostics of acute myeloid leukemia by whole transcriptome RNA sequencing. Leukemia. 2020;35(1):47–61. doi: 10.1038/s41375-020-0762-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet. 2016;17(5):257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marco-Puche G, Lois S, Benítez J, Trivino JC. RNA-seq perspectives to improve clinical diagnosis. Front Genet. 2019;10:1152. doi: 10.3389/fgene.2019.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullighan CG, Downing JR. Global genomic characterization of acute lymphoblastic leukemia. Semin Hematol. 2009;46(1):3–15. doi: 10.1053/j.seminhematol.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Den Boer ML, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10(2):125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7(1):11790. doi: 10.1038/ncomms11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoelzer D, Bassan R, Dombret H, Fielding A, Ribera JM, Buske C, et al. Acute lymphoblastic leukaemia in adult patients: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(suppl 5):v69–v82. doi: 10.1093/annonc/mdw025. [DOI] [PubMed] [Google Scholar]
- 13.Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U, et al. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia. 2002;16(1):53–59. doi: 10.1038/sj.leu.2402329. [DOI] [PubMed] [Google Scholar]
- 14.Haferlach T, Schoch C, Löffler H, Gassmann W, Kern W, Schnittger S, et al. Morphologic dysplasia in de novo acute myeloid leukemia (AML) is related to unfavorable cytogenetics but has no independent prognostic relevance under the conditions of intensive induction therapy: results of a multiparameter analysis from the German AML cooperative group studies. J Clin Oncol. 2003;21(2):256–265. doi: 10.1200/JCO.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Mühlbacher V, Zenger M, Schnittger S, Weissmann S, Kunze F, Kohlmann A, et al. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%: characterization of low Hypodiploid ALL. Genes Chromosomes Cancer. 2014;53(6):524–536. doi: 10.1002/gcc.22163. [DOI] [PubMed] [Google Scholar]
- 16.McGowan-Jordan J, Simons A, Schmid M. ISCN 2016: an international system for human cytogenomic nomenclature (2016) Basel: S Karger AG; 2016. [Google Scholar]
- 17.Stengel A, Shahswar R, Haferlach T, Walter W, Hutter S, Meggendorfer M, et al. Whole transcriptome sequencing detects a large number of novel fusion transcripts in patients with AML and MDS. Blood Adv. 2020;4(21):5393–5401. doi: 10.1182/bloodadvances.2020003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousefi S, et al. A SNP panel for identification of DNA and RNA specimens. BMC Genomics. 2018;19(1):90. doi: 10.1186/s12864-018-4482-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poplin R, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017. 10.1101/201178.
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14(1):244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9(1):488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickham H. Ggplot2: elegant graphics for data analysis. 1. New York: Springer; 2010. [Google Scholar]
- 25.Haas BJ, et al. STAR-fusion: fast and accurate fusion transcript detection from RNA-Seq. bioRxiv. 2017. 10.1101/120295.
- 26.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 27.Jang YE, Jang I, Kim S, Cho S, Kim D, Kim K, et al. ChimerDB 4.0: an updated and expanded database of fusion genes. Nucleic Acids Res. 2020;48(D1):D817–D824. doi: 10.1093/nar/gkz1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiaretti S, Zini G, Bassan R. Diagnosis and subclassification of acute lymphoblastic leukemia. Mediterr J Hematol Infect Dis. 2014;6(1):e2014073. doi: 10.4084/mjhid.2014.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson AK, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47(4):330–337. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie H, Rachakonda PS, Heidenreich B, Nagore E, Sucker A, Hemminki K, et al. Mapping of deletion breakpoints at the CDKN2A locus in melanoma: detection of MTAP-ANRIL fusion transcripts. Oncotarget. 2016;7(13):16490–16504. doi: 10.18632/oncotarget.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwab CJ, Chilton L, Morrison H, Jones L, al-Shehhi H, Erhorn A, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98(7):1081–1088. doi: 10.3324/haematol.2013.085175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov Öfverholm I, Zachariadis V, Taylan F, Marincevic-Zuniga Y, Tran AN, Saft L, et al. Overexpression of chromatin remodeling and tyrosine kinase genes in iAMP21-positive acute lymphoblastic leukemia. Leuk Lymphoma. 2020;61(3):604–613. doi: 10.1080/10428194.2019.1678153. [DOI] [PubMed] [Google Scholar]
- 34.Harvey RC, et al. Development and validation of a highly sensitive and specific gene expression classifier to prospectively screen and identify B-precursor acute lymphoblastic leukemia (ALL) patients with a Philadelphia chromosome-like (“Ph-like” or “BCR-ABL1-like”) signature for therapeutic targeting and clinical intervention. Blood. 2013;122:826. doi: 10.1182/blood.V122.21.826.826. [DOI] [Google Scholar]
- 35.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371(11):1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiaretti S, Messina M, Grammatico S, Piciocchi A, Fedullo AL, di Giacomo F, et al. Rapid identification ofBCR/ABL1-like acute lymphoblastic leukaemia patients using a predictive statistical model based on quantitative real time-polymerase chain reaction: clinical, prognostic and therapeutic implications. Br J Haematol. 2018;181(5):642–652. doi: 10.1111/bjh.15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sánchez R, Ribera J, Morgades M, Ayala R, Onecha E, Ruiz-Heredia Y, et al. A novel targeted RNA-Seq panel identifies a subset of adult patients with acute lymphoblastic leukemia with BCR-ABL1-like characteristics. Blood Cancer J. 2020;10(4):43. doi: 10.1038/s41408-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiaretti, et al. Gene expression profiles of B-lineage adult acute lymphocytic leukemia reveal genetic patterns that identify lineage derivation and distinct mechanisms of transformation. Clin Cancer Res. 2005;11(20):7209–7219. doi: 10.1158/1078-0432.CCR-04-2165. [DOI] [PubMed] [Google Scholar]
- 39.Marincevic-Zuniga Y, Dahlberg J, Nilsson S, Raine A, Nystedt S, Lindqvist CM, et al. Transcriptome sequencing in pediatric acute lymphoblastic leukemia identifies fusion genes associated with distinct DNA methylation profiles. J Hematol Oncol. 2017;10(1):148. doi: 10.1186/s13045-017-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winters JL, Davila JI, McDonald AM, Nair AA, Fadra N, Wehrs RN, et al. Development and verification of an RNA sequencing (RNA-seq) assay for the detection of gene fusions in tumors. J Mol Diagn. 2018;20(4):495–511. doi: 10.1016/j.jmoldx.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Schieck M, Lentes J, Thomay K, Hofmann W, Behrens YL, Hagedorn M, et al. Implementation of RNA sequencing and array CGH in the diagnostic workflow of the AIEOP-BFM ALL 2017 trial on acute lymphoblastic leukemia. Ann Hematol. 2020;99(4):809–818. doi: 10.1007/s00277-020-03953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas BJ, Dobin A, Li B, Stransky N, Pochet N, Regev A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20(1):213. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gocho Y, et al. A novel recurrent EP300-ZNF384 gene fusion in B-cell precursor acute lymphoblastic leukemia. Leukemia. 2015;29(12):2445–2448. doi: 10.1038/leu.2015.111. [DOI] [PubMed] [Google Scholar]
- 44.Qian M, Zhang H, Kham SKY, Liu S, Jiang C, Zhao X, et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017;27(2):185–195. doi: 10.1101/gr.209163.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, et al. ZNF384 -related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 2017;102(1):118–129. doi: 10.3324/haematol.2016.151035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Kuang P, Liu T. Prognostic significance of CDKN2A/B deletions in acute lymphoblastic leukaemia: a meta-analysis. Ann Med. 2019;51(1):28–40. doi: 10.1080/07853890.2018.1564359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45(3):242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talevich E, Shain AH. CNVkit-RNA: copy number inference from RNA-sequencing data. bioRxiv. 2018. 10.1101/408534.
- 49.Harvey RC, Tasian SK. Clinical diagnostics and treatment strategies for Philadelphia chromosome-like acute lymphoblastic leukemia. Blood Adv. 2020;4(1):218–228. doi: 10.1182/bloodadvances.2019000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frisch A, Ofran Y. How I diagnose and manage Philadelphia chromosome-like acute lymphoblastic leukemia. Haematologica. 2019;104(11):2135–2143. doi: 10.3324/haematol.2018.207506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jain N, Roberts KG, Jabbour E, Patel K, Eterovic AK, Chen K, et al. Ph-like acute lymphoblastic leukemia: a high-risk subtype in adults. Blood. 2017;129(5):572–581. doi: 10.1182/blood-2016-07-726588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen IM, et al. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol. 2017;35(4):394–401. doi: 10.1200/JCO.2016.69.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herold T, Baldus CD, Gökbuget N. Ph-like acute lymphoblastic leukemia in older adults. N Engl J Med. 2014;371(23):2235. doi: 10.1056/NEJMc1412123. [DOI] [PubMed] [Google Scholar]
- 54.Tasian SK, Hurtz C, Wertheim GB, Bailey NG, Lim MS, Harvey RC, et al. High incidence of Philadelphia chromosome-like acute lymphoblastic leukemia in older adults with B-ALL. Leukemia. 2017;31(4):981–984. doi: 10.1038/leu.2016.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coccaro N, Anelli L, Zagaria A, Specchia G, Albano F. Next-generation sequencing in acute lymphoblastic leukemia. Int J Mol Sci. 2019;20(12):2929. doi: 10.3390/ijms20122929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palmi C, Vendramini E, Silvestri D, Longinotti G, Frison D, Cario G, et al. Poor prognosis for P2RY8-CRLF2 fusion but not for CRLF2 over-expression in children with intermediate risk B-cell precursor acute lymphoblastic leukemia. Leukemia. 2012;26(10):2245–2253. doi: 10.1038/leu.2012.101. [DOI] [PubMed] [Google Scholar]
- 57.Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet. 2016;48(5):569–574. doi: 10.1038/ng.3535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Gene expression of the B-cell and T-cell lineage. t-SNE plot of gene expression of selected marker genes (a-c, perplexity: 25). d) Correlation between CD10/CD1A gene expression and CD10+/CD1A+ cells as determined by immunophenotyping. Colors correspond to lineage, immunophenotypic subtype or expression as indicated by the plot legends.
Additional file 2: Figure S2. Expression of selected genes. a) CHAF1B and DYRK1A expression of BCP-ALL patients (n = 104) without risk-stratifying fusions or abnormal chromosome number. b) CRLF2 expression of BCP-ALL ‘other’ patients (n = 103).
Additional file 3: Table S1. Patient data.
Additional file 4: Table S2. Allele concordance scores between WTS and WGS SNP profiles.
Additional file 5: Table S3. Gene expression of known lineage markers.
Additional file 6: Table S4. List of called fusion transcripts in both lineages.
Additional file 7: Table S5. Copy number variations called by CNVkit for WTS data.
Additional file 8: Table S6. Expression of BCR-ABL1-like signature genes.
Additional file 9: Table S7. Characteristics of BCR-ABL1-like and non BCR-ABL1-like cases.
Additional file 10: Table S8. Comparison to ALLSorts algorithm.
Data Availability Statement
All data analysed during this study are included in this published article and its supplementary information files. Additional datasets are available from the corresponding author on reasonable request.