Abstract
Background
Usual vulvar intraepithelial neoplasia (uVIN) is a premalignancy caused by persistent infection with high-risk types of human papillomavirus (HPV), mainly type 16. Even though different treatment modalities are available (eg, surgical excision, laser evaporation or topical application of imiquimod), these treatments can be mutilating, patients often have recurrences and 2%–8% of patients develop vulvar carcinoma. Therefore, immunotherapeutic strategies targeting the pivotal oncogenic HPV proteins E6 and E7 are being explored to repress carcinogenesis.
Method
In this phase I/II clinical trial, 14 patients with HPV16+ uVIN were treated with a genetically enhanced DNA vaccine targeting E6 and E7. Safety, clinical responses and immunogenicity were assessed. Patients received four intradermal HPV-16 E6/E7 DNA tattoo vaccinations, with a 2-week interval, alternating between both upper legs. Biopsies of the uVIN lesions were taken at screening and +3 months after last vaccination. Digital photography of the vulva was performed at every check-up until 12 months of follow-up for measurement of the lesions. HPV16-specific T-cell responses were measured in blood over time in ex vivo reactivity assays.
Results
Vaccinations were well tolerated, although one grade 3 suspected unexpected serious adverse reaction was observed. Clinical responses were observed in 6/14 (43%) patients, with 2 complete responses and 4 partial responses (PR). 5/14 patients showed HPV-specific T-cell responses in blood, measured in ex vivo reactivity assays. Notably, all five patients with HPV-specific T-cell responses had a clinical response.
Conclusions
Our results indicate that HPV-16 E6/E7 DNA tattoo vaccination is a biologically active and safe treatment strategy in patients with uVIN, and suggest that T-cell reactivity against the HPV oncogenes is associated with clinical benefit.
Trial registration number
NTR4607.
Keywords: immunogenicity, vaccine, adaptive immunity, cytokines
Introduction
Usual vulvar intraepithelial neoplasia (uVIN), also known as vulvar high-grade squamous intraepithelial lesions, is a premalignant chronic skin disorder of the vulva and associated with a persistent infection with high risk types of human papillomavirus (HPV), mainly HPV type 16.1–3 Spontaneous regression is rare, restricted to 1%–2% of women, and progression to vulvar cancer is observed in 2%–8% of cases.4–8 Current treatment strategies are laser ablation, local excision or topical treatment with the toll-like receptor (TLR) 7-ligand imiquimod. Since patients frequently suffer from recurrent disease, different sequential therapies are often applied over the years.5 7–9 Multiple surgical treatments can however be mutilating, and induce psychosexual dysfunction.10 11 Also, topical treatment with imiquimod is associated with side effects such as pruritus and pain.12 In order to avoid the need for debilitating treatments, and prevent relapses and potential malignant transformation, new therapeutic strategies should be explored with a final goal to eradicate transformed, oncoprotein E6 and E7 expressing epithelial cells.
Infection with high-risk genotypes of HPV leads to the expression of the oncogenic HPV proteins E6 and E7. Together, E6 and E7 drive cellular immortalization and maintain the transformed phenotype during tumor progression.13–15 The E6 and E7 oncoproteins are continuously expressed in transformed cells, consequently enabling presentation of E6 and E7 epitopes by the transformed cells and creating the opportunity for T-cell recognition. Notably, patients with persistent uVIN often have dysfunctional HPV16-specific T-cell responses,16–18 suggesting that immune stimulating therapies that induce or enhance functional HPV16-specific T-cell responses may lead to clinical benefit.
In line with this notion, several HPV-vaccination studies targeting E6 and/or E7 have been performed with some promising immunological and clinical responses, confirming the suitability of the target proteins. Strategies that have been studied included genetic vaccines (DNA/RNA/virus/bacterial), protein-based, peptide-based or dendritic cell-based vaccines.19–22 To date, these vaccines have not found their way to clinical practice because of little efficacy, high production costs, or cumbersome production processes like dendritic cell-based vaccines which requires a personalized cell product. Also upscaling the cell expansion protocol for adoptive transfer can be complicated and troublesome.
DNA vaccination forms an attractive approach for the induction of cellular immune responses, as these vaccines are easy to produce, very stable, relatively cheap and do not suffer from the drawback of pre-existing immunity or induction of antivector immunity, as is the case for most viral vectors.23 24 Since subcutaneous administration with adjuvant of peptide-based therapeutic HPV-vaccines can cause significant adverse events (such as local skin swelling)21 we focused on improving the administration route and optimization of immunogenicity of the vaccine. Therefore, we developed a DNA vaccination strategy based on DNA tattoo vaccination, which demonstrated a 10–100 fold increase in vaccine specific T-cell responses as compared with classical intramuscular DNA vaccination when tested in non-human primates.25
Recently, we performed for the first time a phase I clinical trial using the E7 directed DNA vaccine tetanus toxin fragment C (TTFC)-E7SH, which was delivered using the tattooing technique in patients with uVIN.26 This DNA vaccine was well tolerated and the tattoo-induced skin damage was completely reversible. However, no induction of E7 directed CD8+ responses nor clinical responses could be observed.26
The aim of the current study is to improve the immunological response and monitor clinical outcome in patients with uVIN. Therefore, we developed a novel DNA vaccine that can be administered by DNA tattoo vaccination.27 Since targeting both E6 and E7 has been reported to have a synergistic effect on HPV infection control,26 28 both oncogenes are targeted in this new format. With the combined novel DNA vaccines sig-HELP-E6SH-KDEL and sig-HELP-E7SH-KDEL (further referred to as HPV-16 E6/E7 DNA tattoo vaccine), we aim to increase the immunogenicity towards E6 and E7 by inducing CD4+ helper T cells and including signals for enhanced endoplasmic reticulum targeting and retention. Here, we describe the results of a phase I/II clinical trial in which we evaluated the toxicity, clinical response and immunogenicity of this HPV-16 E6/E7 DNA tattoo vaccination in patients with uVIN.
Materials and methods
Patients
Fourteen female patients with histology and PCR proven HPV16+ uVIN lesions were included between January 2017 and December 2019. Patients needed to have adequate bone marrow function, renal function and liver function. Exclusion criteria were pregnancy/lactation, active infectious disease, autoimmune disease or immunodeficiency. Other exclusion criteria were use of oral anticoagulant drugs or an indication of severe cardiac, respiratory or metabolic disease. Furthermore, patients could not participate if the uVIN was treated with another modality within 6 weeks prior to enrolment, if patients were treated before with therapeutic HPV vaccines, or if patients participated in a study with another investigational drug (for different indications than uVIN) within 30 days prior to enrolment. Patient characteristics are shown in table 1.
Table 1.
Baseline characteristics of the study population
| Patient no. | Age | Multi/unifocal | Symptoms | Smoker | Previous treatment(s) | First diagnosis uVIN | Lesion size (cm²) |
| 1 | 51 | Uni | Pruritus | No | Laser, LE (2×), imiquimod | 2012 | 1.4 |
| 2 | 64 | Multi | Pruritus | Former smoker (stopped in 2016) | Laser, imiquimod | 2015 | 1.3 |
| 3 | 55 | Multi | Pruritus | Smoker | None | 2017 | 0.6 |
| 4 | 37 | Multi | None | Former smoker (stopped in 2017) | LE | 2013 | 3.5 |
| 5 | 65 | Uni | Pain | Former smoker (stopped in 1998) | None | 2017 | 3.5 |
| 6 | 69 | Uni | None | Former smoker | Laser (2×), imiquimod | 1996 | 0.9 |
| 7 | 46 | Multi | None | Smoker | LE (3×) | 2010 | 3.7 |
| 8 | 45 | Uni | None | Former smoker (stopped in 2018) | Imiquimod | 2018 | 3.8 |
| 9 | 41 | Multi | Pruritus | Smoker | Laser (3×), LE (3×), imiquimod (3×) | 2005 | 36 |
| 10 | 50 | Multi | Pruritus | Smoker | LE (3×), laser (6×), imiquimod | 1993 | 6.8 |
| 11 | 46 | Multi | None | Smoker | Laser (2×), imiquimod | 2016 | 1.7 |
| 12 | 61 | Multi | Pruritus, pain | Former smoker (stopped in 1995) | Laser, LE, imiquimod | 2003 | 3.5 |
| 13 | 29 | Multi | None | Smoker | Imiquimod | 2019 | 0.7 |
| 14 | 36 | Multi | Pruritus, pain | Smoker | Laser | 2017 | 2.0 |
All patients were diagnosed with human papillomavirus (HPV) type 16, but patient #10 had a coinfection with HPV type 56 and patient #13 had a coinfection with HPV type 40.
LE, local excision; uVIN, usual vulvar intraepithelial neoplasia.
Vaccine composition
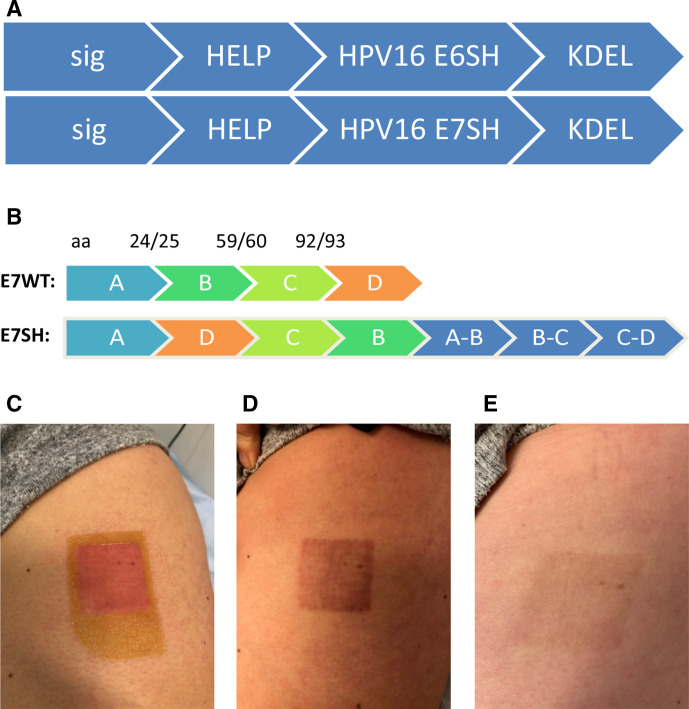
The HPV-16 E6/E7 DNA vaccine comprises of sig-HELP-E6SH-KDEL and sig-HELP-E7SH-KDEL, which are plasmid DNA constructs of 4814 and 5240 base pairs, respectively (figure 1A). In this plasmid, the cytomegalovirus promoter drives the continuous expression of E6SH and E7SH. To prevent toxicity and protect against the transforming properties of E6 and E7, coding sequences were rearranged (‘shuffled’). To prevent loss of potential immunogenic epitopes, sequences flanking the positions where the coding sequence was cut were added 3′ from the coding regions (figure 1B). The HPV-16 E6/E7 DNA vaccine includes three CD4 helper sequences: antigenic epitopes of the negative factor protein from HIV (39 bp),29 the P30 epitope derived from tetanus toxin (63 bp)30 and the universal synthetic, non-natural pan human leukocyte antigen (HLA) DR epitope (39 bp).31 By only inserting the relevant CD4 epitopes, and not the full protein domains, the risk of antigenic competition and skewing of the CD8+ T cell response towards the helper epitopes was minimized. The C-terminal KDEL amino acid sequence was included to achieve endoplasmic reticulum targeting and retention, resulting in higher immunogenicity.24 32
Figure 1.
pUMC3 sig-HELP-E6SH-KDEL and pUMVC3 sig-HELP-E7SH-KDEL plasmids used in this trial and administered by tattoo vaccination. (A) Schematic representation of the therapeutic region of the plasmid, including three helper sequences: synthetic epitope pan human leukocyte antigen (HLA) DR epitope (39 bp), negative factor from HIV (39 bp) and P30 from tetanus toxin (63 bp) for CD4 help. Sig and KDEL for improved endoplasmatic reticulumtargeting and retention, resulting in better antigen uptake by dentritic cellss, enhanced processing and presentation. (B) To prevent toxicity, E6 and E7 coding sequences were shuffled. Splice sites are added at the back of the construct so no potential immunogenic epitopes are lost. (C) Picture of the patients’ skin immediately after vaccination with human papillomavirus (HPV)-16 E6/E7 tattoo vaccination. (D) Picture of the skin 2 weeks after vaccination. (E) Picture of the skin 6 months after last vaccination, demonstrating hardly any visible tissue scar remains.
For the manufacturing of both vaccines, a standard Good manufacturing practice production process was followed as described earlier.33 Resulting DNA vaccines were formulated as a lyophilized powder for solution for intradermal injection, using sucrose as stabilizer.33 Just before administration, 1 mg of sig-HELP-E6SH-KDEL was reconstituted with 0.4 mL water for injection and mixed with 1 mg reconstituted sig-HELP-E7SH-KDEL to obtain 2 mg of the combined HPV-16 E6/E7 DNA tattoo vaccine at a concentration of 5 mg/mL. For each of the four subsequent vaccinations, 2 mg of the combined HPV-16 E6/E7 DNA tattoo vaccine was used.
Study design
This was a single center, non-randomized phase I/II study, consisting of two cohorts. In the first cohort, five patients were treated, followed by an interim analysis that assessed vaccine immunogenicity. Since the criteria for continuation after interim analysis were met (a twofold increase in the T cell response compared with baseline in ≥2 out of 5 patients), an additional 9 patients were enrolled. Patients in both cohorts were treated identically. The primary objective of this trial was to study the systemic HPV-specific immune response of patients with HPV16+ uVIN that received the HPV-16 E6/E7 DNA tattoo vaccine. Secondary objectives were the safety and clinical responses. However, to improve the readability of the paper, we will first discuss our clinical findings, followed by the immunogenicity data.
The HPV-16 E6/E7 DNA tattoo vaccine was applied topically on the skin of the upper legs (close to a regional lymph node area) on days 0, 14, 28 and 42 and administered into the skin using a permanent make-up tattoo device (Derm.MT GmbH, Berlin, Germany). Patients received 2 mg of vaccine injected over a skin surface of 16 cm². Prior to tattoo vaccination, the skin area was treated with an epilating cream (Veet; Reckitt Benckiser Healthcare B.V., Hoofddorp, The Netherlands). Vaccination at day 28 was administered to the same area as vaccination at day 0, and vaccination at day 42 was administered to the same area as vaccination at day 14. Patients were observed during 1 hour after tattooing. Peripheral blood mononuclear cell (PBMC) isolation was performed at day 0 and day 28 before vaccine administration, and at follow-up on day 56 and day 84. A biopsy of the uVIN was taken before treatment and 3 months after the last vaccination. Patients were seen for follow-up after 3, 6, 9 and 12 months after last vaccination with evaluation of the vulvar lesions including photography and measurement of the size of the lesion(s).
Safety and toxicity
The Common Terminology Criteria for Adverse Events (CTCAE) V.4.03 was used for the assessment of adverse events. All patients that received at least one vaccine dose were included in the evaluation of safety. Vital signs were measured at baseline and at all visiting days. Hematology and biochemistry tests were performed before inclusion, and at days 0, 28, 56 and 84. Unacceptable toxicity was defined as an adverse event of the following types for which the relation to the study treatment was likely or not assessable: non-hematological toxicity of grade 3 or higher, hematological toxicity grade 4, neutropenia grade 3 plus fever, or non-reversible neurotoxicity of grade >2. In case unacceptable toxicity occurred in more than 30% of patients, the study would be discontinued. Local toxicity was scored as CTCAE ‘injection site reaction’.
Clinical responses
Lesions were examined and the size was measured bi-dimensionally by an experienced gynecologist and another member of the study team. Drawings were made on a vulvoscopy form in the medical record. Furthermore, the lesions were monitored by digital photography. The total area (in mm²) of the lesions was determined using ImageJ. A complete response (CR) was defined as a complete disappearance of the lesion(s) and a partial response (PR) defined as at least 50% regression of the lesion. A patient was classified as a non-responder (NR) if lesion size was reduced by less than 50% compared with the original lesion size, or in case of progressive disease.
Immune monitoring
To assess systemic induction of HPV E6 and E7 specific T cells, PBMCs were collected at baseline (day 0) and at days 28, 56, and 84 after the first HPV-16 E6/E7 DNA tattoo vaccination. PBMCs were isolated from fresh heparinized blood samples by Ficoll density-gradient centrifugation and cryopreserved until further use.
Presence and magnitude of HPV E6 and E7 specific T-cell responses were determined by co-culture of T cells with autologous antigen presenting cells (APCs) loaded with long overlapping peptides for 6 hours (adapted protocol based on method described by Samuels et al26). To obtain peptide loaded APCs, PBMCs were thawed and plated in 24 well plates at a concentration of 0.3–1.5*106 cells/mL in T cell mixed media (20% Roswell Park Memorial Institute/ 80% AIM-V medium) without serum. Monocytes were separated by plate adherence, and the non-adherent cells were harvested to be used as T cell input in the co-culture. Monocytes were peptide loaded in T cell mixed media with 800 U/mL granulocyte-macrophage colony-stimulating factor (Invitrogen/Thermo Fisher Scientific, California, USA) with five different peptide pools. Long overlapping peptides covering the entire E6 protein were split over pool 1 and 2, long overlapping peptides covering the entire E7 protein were combined in pool 3. Pool 4 consisted of all epitopes that arose as a consequence of shuffling E6 and E7 proteins. The full amino acid sequences of the long overlapping peptides from these four pools are listed in online supplemental table 1. Pool 5 consisted of a set of 32 viral epitopes covering multiple HLA-alleles, and served as a positive control to assess immune competence (ICE peptide pool, U-CyTech biosciences, Utrecht, The Netherlands). Because these were short peptides that could be directly presented without processing, the ICE peptide pool was loaded onto the APCs for only 1.5 hours prior to the start of cocultures. An unloaded APC condition was taken along in order to determine the background reactivity. Five hours after peptide loading, monocytes were cultured overnight in the presence of 25 µg/mL poly(I:C) (InvivoGen, California, USA), to generate monocyte-derived APCs. The previously harvested non-adherent T cells were rested overnight in T cell mixed media without serum or cytokines. After overnight incubation, peptide loaded APCs were washed and T cells were added, alongside the CD107a antibody. After 1 hour, 0.7 µL/mL Golgistop and 1 µL/mL Golgiplug were added to each well (BD Biosciences, USA), and cultures were continued for an additional 5 hours. Subsequently, T cells were harvested and stained for surface markers and intracellular cytokines and analyzed by multiparametric flow cytometry (antibody panel listed in online supplemental table 2). Acquisition of cells was performed using an LSR II flowcytometer (BD Biosciences). Flow cytometry standard files were analyzed using FlowJo software (FlowJo_V.10.6.1).
jitc-2021-002547supp002.pdf (80.2KB, pdf)
Immunological responses were assessed by measuring intracellular cytokine production (interferon gamma (IFNγ), tumor necrosis factor alpha (TNFα) and interleukin 2 (IL-2)) and the degranulation marker CD107a (lysosomal-associated membrane protein-1). Gates were placed based on the negative population with the highest mean fluoresence intensity and consistent for stimulated and unstimulated conditions. Patients were considered an immunological responder when the frequency of positive cells for one or more readout molecules exceeded that of the unloaded APC control by at least a factor two at any time point. In addition, the magnitude of the response should be greater than 0.1% from respectively the CD4+—or CD8+ T-cell parent population. A T-cell response was considered vaccine induced, when the response was not yet present at baseline.
Blood counts by hemocytometer
Routine blood counts were measured with a hemocytometer at the Clinical Chemistry Department at the Netherlands Cancer Institute. Blood was analyzed on the Xn2000 system (Sysmex). Lymphocyte, neutrophil, eosinophil and monocyte counts were extracted and analyzed from the patient records by the involved study team.
Statistical analysis
For sample size calculation, an optimal Simons two-stage design was implemented, aimed to exclude an immunological response rate of 30% and targeting a response rate of 60%. With α=0.1 and power=80%, five patients had to be enrolled in the first stage and the vaccine-induced immune response had to be observed in at least two patients to continue to the next stage (second cohort of n=9).
Patients were included in the evaluation of HPV-specific immune responses if they had received at least two doses of the vaccine, and if blood samples were drawn at baseline and at least two during therapy. Fishers exact test was used to test whether responding patients had significantly more immunological responses ex vivo compared with non-responding patients.
Blood counts were compared between responders (CR and PR) and NRs using the non-parametric two-tailed Mann-Whitney U test. Paired analysis of the same patient over two time points was performed using the Wilcoxon signed rank test. P<0.05 is * and p<0.01 is **.
Results
Safety and toxicity
Thirteen patients received all four vaccinations and one patient received only two vaccinations due to adverse reactions. All adverse events are listed in table 2. The patient (patient #12) who had to discontinue vaccination was diagnosed (by biopsy of a skin eruption) with a Stevens-Johnson syndrome grade 3, 2 weeks after the second vaccination. Although she presented with similar symptoms earlier that year during imiquimod treatment and before she received the first vaccination, an effect of the vaccination could not be excluded, and this event was therefore reported as a suspected unexpected serious adverse reaction (SUSAR). This patient fully recovered from the SUSAR, within 4 months after last vaccination all skin lesions had disappeared. Other patients did not have treatment-related adverse events higher than grade 1 (table 2). Pruritus at the injection site after vaccination was the most commonly observed adverse event (36%). A picture of the injected skin immediately after vaccination, 2 weeks after vaccination and 6 months after vaccination is shown in figure 1C–E.
Table 2.
Overview of treatment-related adverse events
| Toxicity | Grade | Related | No of patients |
| Steven Johnsons Syndrome | 3 | Unlikely | 1 |
| Pruritus | 1 | Definitely | 5 |
| Injection site reaction | 1 | Definitely | 3 |
| Fatigue | 1 | Possibly | 3 |
| Influenza-like symptoms | 1 | Possibly | 3 |
| Dizziness | 1 | Possibly | 2 |
| Dysgeusia | 1 | Possibly | 2 |
| Local infection after skin biopsy | 1 | Definitely | 1 |
| Hot flushes | 1 | Possibly | 1 |
| Pain of skin | 1 | Possibly | 1 |
Grades according to the Common Terminology Criteria for Adverse Events V.4.03.
Observation of clinical responses after HPV-vaccination in patients with uVIN lesions
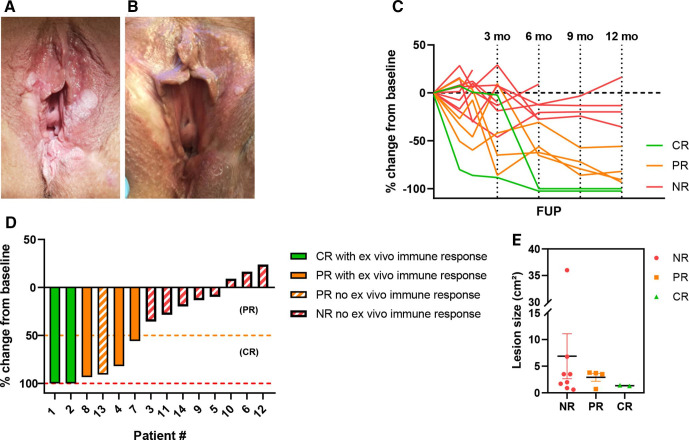
In the first cohort, we included five patients. In this cohort, a CR was observed in two patients and a PR was seen in one patient (figure 2). Both CRs were seen after 6 months of follow-up and the PRs after 3 months. The uVIN lesions did not recur after a CR had been observed for the duration of follow-up (12 months after the last vaccination). Patient #3 showed no clinical response and was treated with laser evaporation 2 years after vaccination. Patient #5 showed no response and started with imiquimod treatment 3 months after the last vaccination. Clinical responses after the start of new treatments were not taken into account in this study. In the second cohort, nine patients were included. In this second cohort three patients showed a durable PR during follow-up. An example of a patient showing a PR is shown in figure 2A, B. The biopsies of the vulva at 3 months follow-up showed uVIN in all of the vaccinated patients. This correlates with the clinical observation that CRs were first seen at 6 months after vaccination. Six patients showed no clinical response. One patient (patient #11) was diagnosed with microinvasive vulvar cancer after 6 months of follow-up for which a local excision was performed. Patient #10 underwent laser treatment. Patient #12 underwent laser excision after 84 days of follow-up. Patient #14 showed no response. An overview of all clinical responses is given in figure 2C, D. In figure 2E, the pretreatment size of the lesions per group (NR, PR, CR) is illustrated. The patients with the biggest lesion size (#9 and #10) were both NRs. These two patients also had received most previous treatments before inclusion in this study, as shown in table 1.
Figure 2.
clinical response data of cohort 1 and 2. (A) Usual vulvar intraepithelial neoplasia (uVIN) lesions visible at screening visit. (B) Partial response of uVIN lesions visible at follow-up +12 months after vaccination with human papillomavirus-16 E6/E7 tattoo vaccination. (C) Overview of uVIN lesion size changes (as percentage change compared with baseline) during follow-up. (D) Waterfall plot showing percentage change of uVIN lesion at last follow-up compared with baseline lesion size (=lesion size at screening). (E) Lesion size before therapy per response category. Complete responders are depicted in green, partial responders in orange and non-responders in red. CR, complete response; NR, non-responder; PR, partial response.
Phenotypic characterization of systemic T cells
Patient PBMCs from baseline samples, as well as from ~day 28,~56, and ~84 after primary vaccination were subjected to basic phenotypic characterization, as determined by multiparameter flow cytometry (see online supplemental figure 1 for gating strategy). Programmed cell death protein 1 (PD-1) expression on systemic T cells was overall low (<0.4%) and did not show any directionality in terms of response prediction or evaluation (see online supplemental figure 2A). We also did not uncover an increase in PD-1 expression in CD4+ and CD8+ T cells on vaccination. The absence of PD-1 expression on circulating T cells does not necessarily reflect expression levels of PD-1 on T cells infiltrating the uVIN lesions. No differences between responders and NRs could be found in the differentiation state of T cells based on the surface marker expression of CD45RA and CCR7 (see online supplemental figure 2B).
jitc-2021-002547supp001.pdf (2.6MB, pdf)
Systemic HPV-16 specific T-cell responses
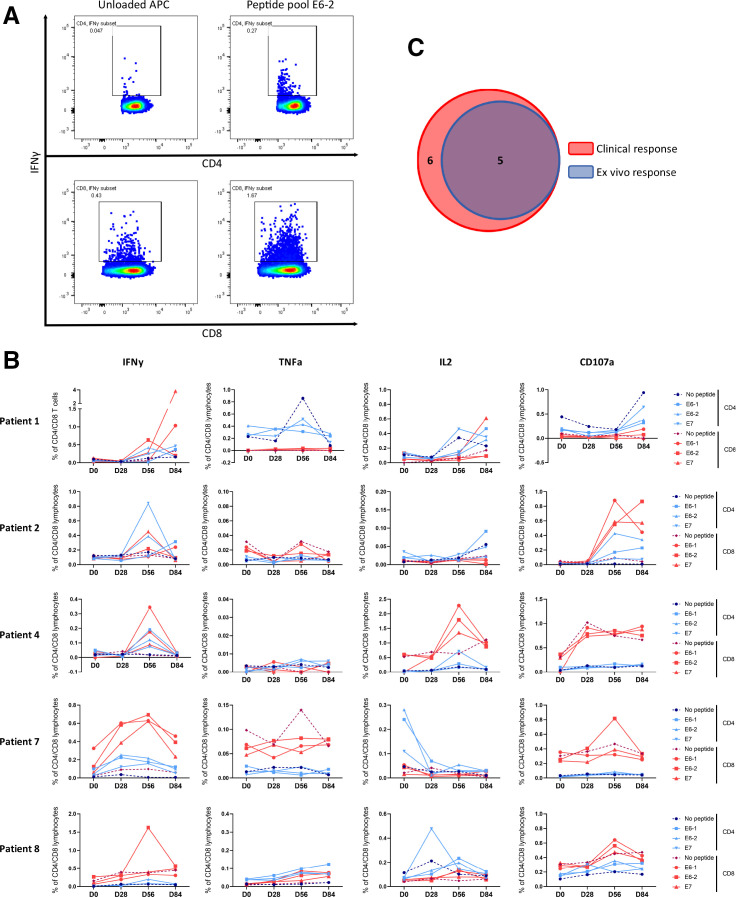
The same PBMCs used for phenotypic characterization of T cells were also used to monitor systemic immune responses against the HPV16 E6 and E7 oncoproteins. A patient was considered an immunological responder if the percentage of positive CD4+ and/or CD8+ T cells for one or more of the measured molecules (IFNγ, TNFα, IL-2 and CD107a) was greater than 0.1% and at least two times higher than the background. Furthermore, a response was considered vaccine induced when it was not yet present at baseline. To illustrate an ex vivo immune response, expression of IFNγ in CD4+ and CD8+ T cells in the presence or absence of stimulation with peptide-loaded APCs from an immunological responder (patient #8) are displayed in figure 3A. T-cell responses against E6-1, E6-2 and E7 peptide pool compared with unloaded APCs of all immunological responders are presented in figure 3B. Table 3 provides an overview of the CD4+ and CD8+ T-cell responses against E6-1, E6-2 and E7 peptide pools from all patients, depicted as the fold change over the unloaded APC background.
Figure 3.
ex vivo reactivity data. (A) Example of an immunological responder (patient #8) at day 56, in which you can appreciate a cloud of interferon gamma (IFNγ) producing CD4 and CD8 cells, that also meets the fold increase over background requirement. (B) Time course of all immunological responders. T cell responses against E6-1, E6-2 and E7 peptide pool are depicted. The dashed line represents the ‘no peptide’ control to visualize background reactivity. Time courses of IFNγ, tumor necrosis factor alpha (TNFα), interleukin 2 (IL-2) and CD107a production for all patients are displayed in online supplemental figure 4). (C) Venn diagram visualizing the overlap between clinical responders (6/14) and immunological responders (5/14) (Fishers exact test, p=0.003). APC, antigen presenting cell.
Table 3.
Overview of immunological responses against E6 and E7 peptide pools
| Patient | Ex vivo | Clinical | E6-1 | E6-2 | E7 | |||||||||||
| Response | Response | IFNy | TNFa | IL2 | CD107a | IFNy | TNFa | IL2 | CD107a | IFNy | TNFa | IL2 | CD107a | |||
| 1 | Yes | CR | CD4 | D0 | <0.1% | 1.1 | <0.1% | 0.4 | <0.1% | 1.8 | 1.3 | 0.4 | <0.1% | 1.2 | 1.2 | 0.4 |
| D56 | <0.1% | 0.4 | 0.4 | 0.7 | 3.4 | 0.5 | 0.3 | 0.9 | 2.0 | 0.6 | 1.4 | 0.7 | ||||
| CD8 | D0 | <0.1% | n.d. | n.d | <0.1% | <0.1% | n.d. | n.d. | <0.1% | 1.5 | n.d. | n.d. | <0.1% | |||
| D56 | <0.1% | n.d. | <0.1% | <0.1% | 8.7 | n.d. | <0.1% | <0.1% | 3.8 | n.d. | 2.0 | 0.0 | ||||
| 2 | Yes | CR | CD4 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | 0.8 | <0.1% | <0.1% | <0.1% |
| D56 | 0.7 | <0.1% | <0.1% | 15.9 | 2.3 | <0.1% | <0.1% | 39.9 | 4.9 | <0.1% | <0.1% | <0.1% | ||||
| CD8 | D0 | 0.9 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | 0.2 | |||
| D56 | 0.8 | <0.1% | <0.1% | 9.5 | 1.7 | <0.1% | <0.1% | 5.9 | 3.4 | <0.1% | <0.1% | 6.3 | ||||
| 3 | No | NR | CD4 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | 1.1 | <0.1% | <0.1% | <0.1% | 1.0 | <0.1% |
| D56 | <0.1% | <0.1% | 1.0 | <0.1% | <0.1% | <0.1% | 0.9 | <0.1% | <0.1% | <0.1% | 1.0 | <0.1% | ||||
| CD8 | D0 | 1.2 | <0.1% | 0.6 | 0.8 | 0.9 | <0.1% | 1.0 | 1.2 | 0.7 | <0.1% | 1.0 | 0.8 | |||
| D56 | <0.1% | <0.1% | 1.0 | 1.7 | <0.1% | <0.1% | 1.0 | 1.0 | <0.1% | <0.1% | 1.0 | 0.9 | ||||
| 4 | Yes | PR | CD4 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% |
| D56 | 10.1 | <0.1% | 1.6 | 1.8 | 6.4 | <0.1% | 0.9 | 1.2 | <0.1% | <0.1% | 4.0 | <0.1% | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | 1.2 | 1.2 | <0.1% | <0.1% | 1.1 | 0.9 | |||
| D56 | 24.2 | <0.1% | 3.6 | 1.0 | 12.4 | n.d. | 2.8 | 1.1 | <0.1% | <0.1% | 2.1 | 1.0 | ||||
| 5 | No | NR | CD4 | D0 | <0.1% | <0.1% | <0.1% | 0.9 | <0.1% | <0.1% | <0.1% | 0.9 | <0.1% | <0.1% | <0.1% | <0.1% |
| D56 | <0.1% | <0.1% | <0.1% | 1.2 | <0.1% | <0.1% | <0.1% | 0.9 | <0.1% | <0.1% | <0.1% | 0.7 | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | |||
| D56 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | ||||
| 6 | No | NR | CD4 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% |
| D56 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | |||
| D56 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | ||||
| 7 | Yes, | PR | CD4 | D0 | <0.1% | <0.1% | 5.2 | <0.1% | <0.1% | <0.1% | 6.0 | <0.1% | <0.1% | <0.1% | 2.4 | <0.1% |
| Vaccine enhanced, | D56 | 22.1 | <0.1% | <0.1% | <0.1% | 27.3 | <0.1% | <0.1% | <0.1% | 19.8 | <0.1% | <0.1% | <0.1% | |||
| not induced | CD8 | D0 | 13.8 | <0.1% | <0.1% | 1.2 | 5.3 | <0.1% | <0.1% | 0.8 | <0.1% | <0.1% | <0.1% | 0.8 | ||
| D56 | 6.4 | <0.1% | <0.1% | 0.7 | 7.0 | <0.1% | <0.1% | 1.7 | 6.3 | <0.1% | <0.1% | 0.8 | ||||
| 8 | Yes | PR | CD4 | D0 | <0.1% | <0.1% | <0.1% | 1.4 | <0.1% | <0.1% | <0.1% | 1.7 | <0.1% | <0.1% | <0.1% | 1.4 |
| D56 | <0.1% | <0.1% | 2.3 | 1.5 | 3.2 | <0.1% | 1.9 | 1.8 | <0.1% | <0.1% | 1.4 | 1.0 | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | 0.8 | 1.7 | <0.1% | <0.1% | 0.9 | 0.7 | <0.1% | <0.1% | 1.1 | |||
| D56 | 0.9 | <0.1% | 2.6 | 1.4 | 4.3 | <0.1% | 2.9 | 1.2 | 1.1 | <0.1% | <0.1% | 1.0 | ||||
| 9 | No | NR | CD4 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% |
| D56 | <0.1% | <0.1% | <0.1% | 1.0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | |||
| D56 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | 0.9 | <0.1% | <0.1% | <0.1% | <0.1% | ||||
| 10 | No | NR | CD4 | D0 | <0.1% | <0.1% | <0.1% | 0.6 | <0.1% | 1.1 | <0.1% | 1.1 | <0.1% | <0.1% | <0.1% | 0.7 |
| D56 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | 1.2 | <0.1% | <0.1% | <0.1% | <0.1% | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | 0.7 | <0.1% | <0.1% | <0.1% | 1.2 | <0.1% | <0.1% | <0.1% | 0.8 | |||
| D56 | <0.1% | <0.1% | <0.1% | 1.2 | <0.1% | <0.1% | <0.1% | 0.9 | <0.1% | <0.1% | <0.1% | 1.1 | ||||
| 11 | No | NR | CD4 | D0 | <0.1% | <0.1% | 1.7 | <0.1% | <0.1% | <0.1% | 0.7 | <0.1% | <0.1% | <0.1% | 1.0 | 1.8 |
| D56 | <0.1% | <0.1% | <0.1% | 1.0 | <0.1% | <0.1% | 0.9 | 1.0 | <0.1% | <0.1% | 0.6 | 0.9 | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | 0.9 | 1.1 | <0.1% | <0.1% | 1.0 | 1.1 | <0.1% | <0.1% | 1.1 | |||
| D56 | 1.1 | <0.1% | <0.1% | 0.9 | 1.2 | <0.1% | <0.1% | 0.9 | 1.1 | <0.1% | <0.1% | 1.0 | ||||
| 12 | No | NR | CD4 | D0 | <0.1% | <0.1% | <0.1% | 0.9 | <0.1% | <0.1% | 1.0 | 0.9 | <0.1% | <0.1% | 1.7 | 0.7 |
| D56 | <0.1% | <0.1% | <0.1% | 1.2 | <0.1% | <0.1% | 1.4 | 1.6 | <0.1% | <0.1% | 1.0 | 1.3 | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | 1.0 | <0.1% | <0.1% | <0.1% | 1.1 | <0.1% | <0.1% | <0.1% | <0.1% | |||
| D56 | 0.7 | <0.1% | <0.1% | 0.9 | 0.7 | <0.1% | 1.8 | 0.9 | 0.5 | <0.1% | <0.1% | 0.9 | ||||
| 13 | No | PR | CD4 | D0 | <0.1% | <0.1% | <0.1% | 1.8 | <0.1% | <0.1% | <0.1% | 1.8 | <0.1% | <0.1% | <0.1% | <0.1% |
| D56 | <0.1% | <0.1% | <0.1% | 1.9 | <0.1% | <0.1% | <0.1% | 1.7 | <0.1% | <0.1% | <0.1% | <0.1% | ||||
| CD8 | D0 | <0.1% | <0.1% | <0.1% | 0.7 | <0.1% | 1.0 | <0.1% | 1.0 | <0.1% | <0.1% | <0.1% | <0.1% | |||
| D56 | <0.1% | 0.8 | <0.1% | 0.7 | <0.1% | 0.6 | <0.1% | 0.9 | <0.1% | <0.1% | <0.1% | 1.2 | ||||
| 14 | No | NR | CD4 | D0 | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% | <0.1% |
| D56 | <0.1% | <0.1% | <0.1% | 1.9 | <0.1% | <0.1% | <0.1% | 1.2 | <0.1% | <0.1% | <0.1% | <0.1% | ||||
| CD8 | D0 | <0.1% | <0.1% | 0.6 | 0.8 | <0.1% | <0.1% | 1.0 | 0.7 | <0.1% | <0.1% | <0.1% | <0.1% | |||
| D56 | <0.1% | <0.1% | <0.1% | 0.8 | <0.1% | <0.1% | <0.1% | 0.9 | <0.1% | <0.1% | 1.1 | 0.6 | ||||
Numbers represent the fold change over background (unloaded APCs). n.d.: no positive cells detected. Fold changes greater than two are highlighted in green. <0.1% indicates that the fraction of positive cells was less than 0.1% from its parent (CD4+ or CD8+ T cells).
CR, complete response; IFNγ, interferon gamma; IL2, interleukin 2; NR, no response; PR, partial response; TNFa, tumor necrosis factor alpha.
The peak of the immunological response in blood was mostly detected at day 56; 2 weeks after the boost vaccination. From the 14 patients treated in this study, five showed an ex vivo immunological response (36% immunological response rate). Four of these immunological responses were not detected at baseline, and one response showed a substantial increase after vaccination (figure 3B, patient #7 IFNγ). Furthermore, 4 out of 5 of these responses could still be detected at day 84, over a month after the last vaccination that was given at day 42 (namely in patient #1, #2, #7 and #8).
The effector molecule measured in the response varied between patients, but IFNγ was the dominant effector molecule (4/5). Interestingly, both CD4+ and CD8+ T-cell reactivity against all peptide pools was observed (figure 3B). In all immunological responders (5/5), the response could be detected in both the CD4+ and CD8+ T cell compartments. A Boolean gating strategy was applied to distinguish single, double and triple producing T cells (combinations of IFNγ, TNFα and IL2), with or without coexpression of degranulation marker CD107a in responding patients. T cells predominantly produced one cytokine (online supplemental figure 3), indicating suboptimal functionality of the T cells.34 35 Time course graphs showing the IFNγ, TNFα, IL-2 and CD107a responses against E6-1, E6-2 and E7 peptide pool of all patients (including the NRs) can be found in online supplemental figure 4.
As described in the method section, E6 and E7 coding sequences needed to be shuffled for safety reasons. To assess the immunogenicity of the junction sites of the shuffled proteins, all possible epitopes covering those regions were taken along in a separate pool during the ex vivo immune reactivity assays. In online supplemental figure 5, reactivity from CD4+ and CD8+ T cells against the shuffle points is depicted at day 0 and day 56. CD8+ T cells from patient #1 and patient #8 (both responders) produced IFNγ on stimulation with the shuffle point peptide pool. For patient #8, shuffle point reactivity seemed vaccine induced and for patient #1 the reactivity was also found in the baseline samples, possibly indicating cross reactivity towards another epitope. The magnitude of the response against the shuffle points was occasionally higher than the magnitude of the response against E6 and E7 epitopes. We do not know the exact reason for this, although we could speculate that this is due to differences in antigen processing and/or presentation between patients. In general, we do not see common reactivity against the shuffle point epitopes and it is important to note that no ‘on target, of lesion’ toxicity was observed in any of the patients.
Reactivity against the ICE peptide pool consisting of 32 viral epitopes covering multiple HLA-alleles was tested to assess differences in immune competence between responding and non-responding patients (see online supplemental figure 6). In total, CD8+ T cells from baseline samples of 9/14 patients produced cytokines on culturing with ICE peptide loaded APCs and no CD4 reactivity was measured against the ICE peptide pool (see online supplemental figure 6). As a positive control, we took along four healthy donors, which were all responsive towards the ICE peptide pool (see online supplemental figure 6). Also, all patient samples produced high amounts of cytokines after phorbol 12-myristate 13-acetate/ionomycin stimulation (data not shown).
Correlation between T-cell reactivity against the HPV oncogenes and clinical benefit
Notably, all patients who showed ex vivo HPV E6 or E7 specific T-cell responses also experienced clinical benefit from the vaccine (figure 3C). In contrast, such HPV E6 or E7 specific T-cell responses were completely absent in clinical NRs (0/8). For one out of six patients that showed a clinical response, no ex vivo immune reactivity could be determined (table 3 and online supplemental figure 4, patient #13). Collectively, these findings demonstrate that there is a strong correlation between the induction of immune reactivity and clinical response (Fischer’s exact test, p=0.003).
Lymphocyte counts in relation to clinical response
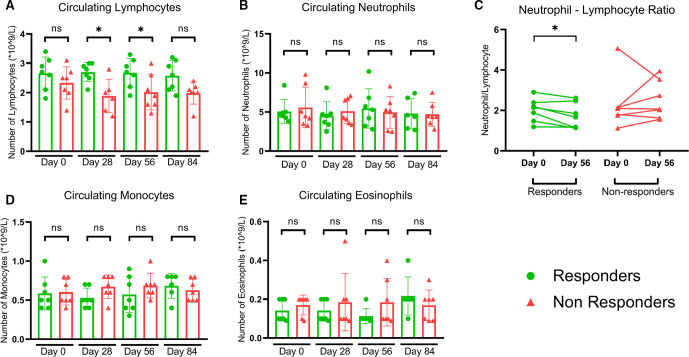
Regular blood cell counts were established at matching time points with the PBMC isolation for ex vivo reactivity assays. At baseline, no statistically significant differences in number of circulating lymphocytes, neutrophils, monocytes and eosinophils could be found between responding and non-responding patients (shown in figure 4). During the vaccinations at day 28, as well as at the peak of the response at day 56, lymphocyte counts were significantly higher in responding patients than in non-responding patients (figure 4A). Systemic neutrophil to lymphocyte ratio was decreased at the peak of the response compared with baseline in responding patients. No significant change in neutrophil to lymphocyte ratio over time was found in non-responding patients (figure 4C). The number of circulating neutrophils, monocytes and eosinophils remained similar for responders and NRs and unaltered compared with baseline levels (figure 4B, D, E).
Figure 4.
Systemic blood counts reveal differences between responders and non-responders during- and post vaccination. (A) Significantly reduced number of circulating lymphocytes in non-responding patients, compared with responding patients. Two tailed Mann-Whitney test D28 p=0.015, D56 p=0.048. (B) No statistically significant differences detected between responding and non-responding patients at any time point in neutrophils. (C) Responding patients show a decreased neutrophil to lymphocyte ratio at the peak the immune response compared with baseline. No significant changes in neutrophil to lymphocyte ratio in non-responding patients. Wilcoxon matched-pairs signed rank test p=0.0469. No statistically significant differences detected between responding and non-responding patients at any time point in circulating (D) monocytes and (E) eosinophils.
Despite a variety of treatment modalities for patients suffering from uVIN, these patients are often confronted with recurrent disease and are at risk to progress to invasive disease. In this study, we have used a therapeutic HPV-16 E6/E7 DNA tattoo vaccine comprizing of sig-HELP-E6SH-KDEL and sig-HELP-E7SH-KDEL. In mice, this DNA vaccine has shown to be much more immunogenic than the variants with other helper cassettes (such as TTFC) that were previously used in the clinic.24 32 This is the first clinical trial using this optimized DNA vaccine targeting the HPV oncoproteins E6 and E7 in patients with uVIN.
Several HPV-vaccination studies targeting E6 and/or E7 have been performed with varying results. Intramuscular TA-cervical intraepithelial neoplasia (CIN) (fusion protein HPV16 E6E7L2) administration preceded by local imiquimod application has been studied, with a clinical response rate of 63% in patients with uVIN, but all patients in this study displayed moderate (n=5, 26%), or severe (n=14, 74%) side effects.22 TA-HPV, a recombinant vaccinia virus, encoding modified HPV 16 and 18 E6 and E7, has also been successfully applied in uVIN and vaginal intraepithelial neoplasia patients. This was resulting in both a potent clinical responses (8/18 and 5/12, respectively) and immunological responses (13/18 and 6/10, respectively).16 36 However, the use of live vaccinia virus limits the broad application of this therapy. In trials investigating subcutaneously administered HPV16 E6 and E7 synthetic long peptides (SLP), clinical responses were observed after 12 months in 52%–79% of women with uVIN.21 37 However, grade 1 and grade 2 side effects were reported at very high frequencies and were probably linked to the use of the Montanide ISA51. In our trial, no adverse events higher than grade 1 were reported, (apart from one patient with a grade 3 SUSAR that was probably unrelated as symptoms had occurred before the first vaccination) and at much lower frequencies, suggesting that HPV-16 E6/E7 DNA tattoo vaccination is safe to use. This difference in toxicity and tolerability can likely be explained by the fact that we used the tattoo technique, and no adjuvant or other initial treatment modality such as imiquimod was used in our trial. Since subcutaneous administration of a therapeutic HPV peptide vaccine with adjuvant can cause significant adverse events (such as local skin swelling), we focused on improving the administration route and optimization of immunogenicity of the vaccine.
Our data indicate a 43% clinical response rate. A clinically durable and ongoing CR was seen in 14% of the patients. PRs were observed in 29% of patients and were ongoing at the time of most recent follow-up. Importantly, unlike other treatment modalities (eg, laser ablation, surgical excision or imiquimod application) in which up to one-third of patients show a recurrence,9 38 none of the responders in our study had recurrences or increasing lesion size over time. A likely explanation for this difference is that our vaccination strategy targets the cause of the disease, that is, HPV16 E6 and E7 expressing cells, and this is underlined by the fact that 83% of the responding patients showed a clear E6/E7 specific T-cell response in their blood. However, recurrences often occur over 1 year after treatment in this patient group and follow-up period in this study was only 12 months. Future studies have to point out whether the recurrence of uVIN is maintained more than a year after therapeutic HPV-vaccination. Furthermore, no HPV-testing at the end of follow-up was performed, which would be interesting to incorporate in follow-up studies to confirm the successful clearance of the virus at the uVIN lesions after vaccination.
Although responses were durable in our study, CR rates were still low (2/14). Therefore, we would like to advocate the combination of our vaccine with for instance immune checkpoint inhibitors such as (locally administrated) anti PD-(L)1 or TLR_agonists, such as poly (I:C) (TLR3 agonist) or Imiquimod (TLR7 agonist). Besides this, it might be beneficial for patients with large uVIN lesions to first decrease lesion size (eg, by laser or topical therapy), before administering our vaccine, because patients with largest uVIN lesion size at baseline did not show any response to vaccination in this trial. However, since the sample size in this study was quite small, future studies have to reveal whether this effect will still be observed. Interestingly though, Kenter et al also reported that lesions were smaller in the CR group after E6 and E7 synthetic long-peptide vaccination in patients with uVIN.21
Systemic immunological HPV-specific T-cell responses were found in both the CD4+ as well as the CD8+ compartment. These responses were either vaccine induced (4/5) or vaccine enhanced (1/5). Interestingly, five out of six patients with complete or PRs showed systemic HPV-specific T-cell responses in ex vivo assays. Likewise, patients without a clinical response did not show an HPV-specific T-cell response ex vivo. Previous HPV targeting vaccines, in the same patient group, observed a similar relationship. Both Kenter et al and van Poelgeest et al reported a correlation between (the magnitude of) the ex vivo response and the clinical outcome of the patients after vaccination with HPV16 E6 and E7 SLPs21 37 However, in a study evaluating the effect of a TA-HPV vaccine against E6 and E7, ex vivo responsiveness to the vaccine vector was confirmed in all patients, there was no relation with clinical benefit.20 The differences between clinical and immunological responses between our study and previous studies could be explained by a different study design, different vaccine, different patient group and a different technique used to identify ex vivo immune responses.
At baseline, the number of circulating lymphocytes, neutrophils, monocytes and eosinophils did not differ statistically significant between responders and NRs. On treatment, NRs had statistically significant fewer circulating lymphocytes than responders, which could potentially be a reflection of a less competent immune system.
Future experiments will tell whether responders will have relatively higher numbers of VIN lesion infiltrating lymphocytes compared with NRs, and what potential immunosuppressive mechanisms in the lesions might have hampered a T-cell response in the non-responding patients.
Follow-up studies should be performed to determine the effects of this vaccination strategy in a larger cohort of patients with uVIN, as well as patients with other intraepithelial neoplasia caused by HPV 16, such as anal intraepithelial neoplasia, penile intraepithelial neoplasia and CIN. Patients with HPV-16 and HPV-18 CIN2/3 have already shown to respond to other types of DNA vaccination targeting E6 and E7 proteins.39
In conclusion, we found in this phase I/II clinical trial that HPV-16 E6/E7 DNA tattoo vaccination for the treatment of HPV16 positive uVIN is a safe and immunologically effective strategy. Interestingly, in five out of six clinically responding patients, E6/E7 specific CD4+ and CD8+ T cell reactivity could be detected in blood samples. Such responses were not observed in patients without a clinical response. Therefore, HPV-16 E6/E7 DNA tattoo could possibly be a clinically meaningful treatment strategy in patients with uVIN.
Acknowledgments
The authors would like to thank Thomas Lanigan from the University of Michigan for kindly supplying us with the pUMVC3 backbone. The authors would also like to thank Renate de Boer, Maaike van Zon and Edith Vermeij for their help in manufacturing of DNA vaccines, and the QC and QA department of the Pharmacy of The Netherlands Cancer Institute- Antoni van Leeuwenhoek for the generous support.
Footnotes
NAMB, JR, JHvdB and NEvT contributed equally.
Contributors: NAMB constructed the plasmids and produced DNA vaccines, designed and performed immune monitoring experiments, interpreted data and cowrote the manuscript. JR vaccinated patients, collected and interpreted clinical data and co-wrote the manuscript. MvB involved in patient care. HJMAAZ involved in patient care. SS involved in writing the study protocol. MvR vaccinated patients and collected clinical data. BN and JB supervised DNA production. TS conceived the project, designed plasmids, interpreted data and reviewed the manuscript. JH conceived the project, designed plasmids and interpreted data. KDV interpreted data and reviewed the manuscript. TDdG interpreted data. ESJ interpreted data. GGK conceived the project, interpreted data. JHvdB conceived the project, interpreted data, designed plasmids.
Funding: We thank the Rational molecular Assessment Innovative Drug selection (RAIDs) consortium (http://www.raids-fp7.eu). This trial is part of the RAIDs project and received funding from the European Union’s Seventh Program for Research, Technological Development, and Demonstration (Grant No. 304810). Additional funding was obtained through the Louise Vehmeijer Foundation, The Netherlands. Part of the salary costs were provided by Oncode Institute, The Netherlands.
Competing interests: JB is (part time) employee of Modra Pharmaceuticals and stockholder in Modra Pharmaceuticals. (not related to the manuscript). TS is advisor for Adaptive Biotechnologies, Allogene Therapeutics, Merus, Neogene Therapeutics, and Scenic Biotech; is a recipient of research support from Merck KgaA; is a stockholder in Allogene Therapeutics, Merus, Neogene Therapeutics and Scenic Biotech; and is venture partner at Third Rock Ventures, all not related to the current work. JH is advisor to Achilles Therapeutics, Bristol-Myers Squibb, BioNTech USA, Ipsen, Gadeta, Immunocore, MSD, Merck Serono, Molecular Partners, Neogene Therapeutics, Novartis, Pfizer, Roche/Genentech, Sanofi, Seattle Genetics, Third Rock Ventures, is stock holder in Neogene Therapeutics, and is a recipient of grant or research support from Bristol-Myers Squibb, MSD, Novartis and BioNTech USA. KDV reports research funding from Roche and is consultant for Third Rock Ventures, all outside the scope of this work. TDdG has served as advisor to TILT Biotherapeutics, LAVA Therapeutics, Macrophage Pharma and DCPrime, is a recipient of a grant from Idera Pharmaceuticals, and is co-founder and shareholder of LAVA Therapeutics. JHvdB is a recipient of grant or research support from BioNTech USA and Astra Zeneca, and is currently employed at CellPoint B.V.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The study was approved by the Central Committee on Research Involving Human Subjects (In Dutch: Centrale Commissie Mensgebonden Onderzoek; CCMO) in The Hague, the Netherlands (Number NL46637.000.13) and registered at trialregister.nl (NTR4607). All study protocols were conducted in accordance with the ICH Harmonised Tripartite Guideline for Good Clinical Practice and the principles of the Declaration of Helsinki. All patients provided written informed consent before enrolment.
References
- 1.De Vuyst H, Clifford GM, Nascimento MC, et al. Prevalence and type distribution of human papillomavirus in carcinoma and intraepithelial neoplasia of the vulva, vagina and anus: a meta-analysis. Int J Cancer 2009;124:1626–36. 10.1002/ijc.24116 [DOI] [PubMed] [Google Scholar]
- 2.Serrano B, de Sanjosé S, Tous S, et al. Human papillomavirus genotype Attribution for HPVs 6, 11, 16, 18, 31, 33, 45, 52 and 58 in female anogenital lesions. Eur J Cancer 2015;51:1732–41. 10.1016/j.ejca.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 3.Srodon M, Stoler MH, Baber GB, et al. The distribution of low and high-risk HPV types in vulvar and vaginal intraepithelial neoplasia (VIN and vain). Am J Surg Pathol 2006;30:1513–8. 10.1097/01.pas.0000213291.96401.48 [DOI] [PubMed] [Google Scholar]
- 4.Fehr MK, Baumann M, Mueller M, et al. Disease progression and recurrence in women treated for vulvovaginal intraepithelial neoplasia. J Gynecol Oncol 2013;24:236–41. 10.3802/jgo.2013.24.3.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones RW, Rowan DM, Stewart AW. Vulvar intraepithelial neoplasia: aspects of the natural history and outcome in 405 women. Obstet Gynecol Dec;106:1319–26. 10.1097/01.AOG.0000187301.76283.7f [DOI] [PubMed] [Google Scholar]
- 6.McNally OM, Mulvany NJ, Pagano R, et al. Vin 3: a clinicopathologic review. Int J Gynecol Cancer 2002;12:490–5. 10.1136/ijgc-00009577-200209000-00014 [DOI] [PubMed] [Google Scholar]
- 7.Satmary W, Holschneider CH, Brunette LL, et al. Vulvar intraepithelial neoplasia: risk factors for recurrence. Gynecol Oncol 2018;148:126–31. 10.1016/j.ygyno.2017.10.029 [DOI] [PubMed] [Google Scholar]
- 8.van Seters M, van Beurden M, de Craen AJM. Is the assumed natural history of vulvar intraepithelial neoplasia III based on enough evidence? A systematic review of 3322 published patients. Gynecol Oncol 2005;97:645–51. 10.1016/j.ygyno.2005.02.012 [DOI] [PubMed] [Google Scholar]
- 9.Wallbillich JJ, Rhodes HE, Milbourne AM, et al. Vulvar intraepithelial neoplasia (VIN 2/3): comparing clinical outcomes and evaluating risk factors for recurrence. Gynecol Oncol 2012;127:312–5. 10.1016/j.ygyno.2012.07.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Likes WM, Stegbauer C, Tillmanns T, et al. Correlates of sexual function following vulvar excision. Gynecol Oncol 2007;105:600–3. 10.1016/j.ygyno.2007.01.027 [DOI] [PubMed] [Google Scholar]
- 11.Thuesen B, Andreasson B, Bock JE. Sexual function and somatopsychic reactions after local excision of vulvar intra-epithelial neoplasia. Acta Obstet Gynecol Scand 1992;71:126–8. 10.3109/00016349209007969 [DOI] [PubMed] [Google Scholar]
- 12.van Seters M, van Beurden M, ten Kate FJW, et al. Treatment of vulvar intraepithelial neoplasia with topical imiquimod. N Engl J Med 2008;358:1465–73. 10.1056/NEJMoa072685 [DOI] [PubMed] [Google Scholar]
- 13.Sher Y-P, Lee C, Liu S-Y, et al. A therapeutic vaccine targeting HPV E6/E7 with intrinsic Toll-like receptor 2 agonist activity induces antitumor immunity. Am J Cancer Res 2018;8:2528–37. [PMC free article] [PubMed] [Google Scholar]
- 14.Tan S, de Vries EGE, van der Zee AGJ, et al. Anticancer drugs aimed at E6 and E7 activity in HPV-positive cervical cancer. Curr Cancer Drug Targets 2012;12:170–84. 10.2174/156800912799095135 [DOI] [PubMed] [Google Scholar]
- 15.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 2010;10:550–60. 10.1038/nrc2886 [DOI] [PubMed] [Google Scholar]
- 16.Davidson EJ, Boswell CM, Sehr P, et al. Immunological and clinical responses in women with vulval intraepithelial neoplasia vaccinated with a vaccinia virus encoding human papillomavirus 16/18 oncoproteins. Cancer Res 2003;63:6032–41. [PubMed] [Google Scholar]
- 17.de Vos van Steenwijk PJ, Piersma SJ, Welters MJP, et al. Surgery followed by persistence of high-grade squamous intraepithelial lesions is associated with the induction of a dysfunctional HPV16-specific T-cell response. Clin Cancer Res 2008;14:7188–95. 10.1158/1078-0432.CCR-08-0994 [DOI] [PubMed] [Google Scholar]
- 18.van Poelgeest MIE, van Seters M, van Beurden M, et al. Detection of human papillomavirus (HPV) 16-specific CD4+ T-cell immunity in patients with persistent HPV16-induced vulvar intraepithelial neoplasia in relation to clinical impact of imiquimod treatment. Clin Cancer Res 2005;11:5273–80. 10.1158/1078-0432.CCR-05-0616 [DOI] [PubMed] [Google Scholar]
- 19.Chabeda A, Yanez RJR, Lamprecht R, et al. Therapeutic vaccines for high-risk HPV-associated diseases. Papillomavirus Res 2018;5:46–58. 10.1016/j.pvr.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson EJ, Faulkner RL, Sehr P, et al. Effect of TA-CIN (HPV 16 L2E6E7) booster immunisation in vulval intraepithelial neoplasia patients previously vaccinated with TA-HPV (vaccinia virus encoding HPV 16/18 E6E7). Vaccine 2004;22:2722–9. 10.1016/j.vaccine.2004.01.049 [DOI] [PubMed] [Google Scholar]
- 21.Kenter GG, Welters MJP, Valentijn ARPM, et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N Engl J Med 2009;361:1838–47. 10.1056/NEJMoa0810097 [DOI] [PubMed] [Google Scholar]
- 22.Daayana S, Elkord E, Winters U, et al. Phase II trial of imiquimod and HPV therapeutic vaccination in patients with vulval intraepithelial neoplasia. Br J Cancer 2010;102:1129–36. 10.1038/sj.bjc.6605611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet 2008;9:776–88. 10.1038/nrg2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oosterhuis K, van den Berg JH, Schumacher TN, et al. DNA vaccines and intradermal vaccination by DNA tattooing. Curr Top Microbiol Immunol 2012;351:221–50. 10.1007/82_2010_117 [DOI] [PubMed] [Google Scholar]
- 25.Verstrepen BE, Bins AD, Rollier CS, et al. Improved HIV-1 specific T-cell responses by short-interval DNA tattooing as compared to intramuscular immunization in non-human primates. Vaccine 2008;26:3346–51. 10.1016/j.vaccine.2008.03.091 [DOI] [PubMed] [Google Scholar]
- 26.Samuels S, Marijne Heeren A, Zijlmans HJMAA, et al. Hpv16 E7 DNA tattooing: safety, immunogenicity, and clinical response in patients with HPV-positive vulvar intraepithelial neoplasia. Cancer Immunol Immunother 2017;66:1163–73. 10.1007/s00262-017-2006-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oosterhuis K, Aleyd E, Vrijland K, et al. Rational design of DNA vaccines for the induction of human papillomavirus type 16 E6- and E7-specific cytotoxic T-cell responses. Hum Gene Ther 2012;23:1301–12. 10.1089/hum.2012.101 [DOI] [PubMed] [Google Scholar]
- 28.Peng S, Tomson TT, Trimble C, et al. A combination of DNA vaccines targeting human papillomavirus type 16 E6 and E7 generates potent antitumor effects. Gene Ther 2006;13:257–65. 10.1038/sj.gt.3302646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pancré V, Georges B, Angyalosi G, et al. Novel promiscuous HLA-DQ HIV Nef peptide that induces IFN-gamma-producing memory CD4+ T cells. Clin Exp Immunol 2002;129:429–37. 10.1046/j.1365-2249.2002.01934.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panina-Bordignon P, Tan A, Termijtelen A, et al. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol 1989;19:2237–42. 10.1002/eji.1830191209 [DOI] [PubMed] [Google Scholar]
- 31.Alexander J, Sidney J, Southwood S, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity 1994;1:751–61. 10.1016/S1074-7613(94)80017-0 [DOI] [PubMed] [Google Scholar]
- 32.Ahrends T, Spanjaard A, Pilzecker B, et al. CD4+ T Cell Help Confers a Cytotoxic T Cell Effector Program Including Coinhibitory Receptor Downregulation and Increased Tissue Invasiveness. Immunity 2017;47:848–61. 10.1016/j.immuni.2017.10.009 [DOI] [PubMed] [Google Scholar]
- 33.Quaak SGL, van den Berg JH, Toebes M, et al. GMP production of pDERMATT for vaccination against melanoma in a phase I clinical trial. Eur J rm Biopharm 2008;70:429–38. 10.1016/j.ejpb.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 34.Kannanganat S, Ibegbu C, Chennareddi L, et al. Multiple-cytokine-producing antiviral CD4 T cells are functionally superior to single-cytokine-producing cells. J Virol 2007;81:8468–76. 10.1128/JVI.00228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almeida JR, Price DA, Papagno L, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med 2007;204:2473–85. 10.1084/jem.20070784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldwin PJ, van der Burg SH, Boswell CM, et al. Vaccinia-expressed human papillomavirus 16 and 18 E6 and E7 as a therapeutic vaccination for vulval and vaginal intraepithelial neoplasia. Clin Cancer Res 2003;9:5205–13. [PubMed] [Google Scholar]
- 37.van Poelgeest MIE, Welters MJP, Vermeij R, et al. Vaccination against oncoproteins of HPV16 for noninvasive Vulvar/Vaginal lesions: lesion clearance is related to the strength of the T-cell response. Clin Cancer Res 2016;22:2342–50. 10.1158/1078-0432.CCR-15-2594 [DOI] [PubMed] [Google Scholar]
- 38.Mahto M, Nathan M, O'Mahony C. More than a decade on: review of the use of imiquimod in lower anogenital intraepithelial neoplasia. Int J STD AIDS 2010;21:8–16. 10.1258/ijsa.2009.009309 [DOI] [PubMed] [Google Scholar]
- 39.Trimble CL, Morrow MP, Kraynyak KA, et al. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2B trial. Lancet 2015;386:2078–88. 10.1016/S0140-6736(15)00239-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-002547supp002.pdf (80.2KB, pdf)
jitc-2021-002547supp001.pdf (2.6MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. All data relevant to the study are included in the article or uploaded as supplementary information.