Abstract
Health organizations worldwide have warned that we are on the cusp of a “post-antibiotic era,” necessitating new approaches to combat antibiotic resistant infections. One such approach is the development of antibiotic adjuvants, which have little or no inherent antibiotic activity at their active concentrations but instead potentiate the activity of antibiotics against antibiotic-resistant bacteria. Recently, we demonstrated that meridianin D, a natural product originally reported to have activity against Staphylococcus aureus and Mycobacterium tuberculosis, possesses the ability to reverse colistin resistance in colistin resistant bacteria. As most natural product screens typically involve screening for only certain activities (anticancer, antiviral, and antimicrobial are typical), we posited that the meridianin D discovery was not unique and there are potentially many natural products that have adjuvant activity. To explore this, the National Cancer Institute (NCI) Natural Product Library Set IV was screened for adjuvant activity using four classes of antibiotics (β-lactams, aminoglycosides, macrolides, and polymyxins) against three bacterial pathogens (methicillin-resistant Staphylococcus aureus (MRSA), Acinetobacter baumannii, and Klebsiella pneumoniae). Sixteen compounds suppressed β-lactam resistance in MRSA, five of which effected a 16-fold reduction in the oxacillin minimum inhibitory concentration (MIC). Two natural products effectively suppressed aminoglycoside resistance in both of the Gram-negative species tested, and no hits were observed with macrolides. In contrast, a larger number of natural product adjuvants were identified when screening against colistin-resistant strains of A. baumannii and K. pneumoniae. Nine compounds reduced the colistin MIC to its breakpoint or lower (up to a 1024-fold reduction). Clorobiocin, novobiocin, and prodigiosin were most effective, reducing the colistin MIC in K. pneumoniae strain B9 to 2 μg/mL at concentrations as low as 0.625, 2.5, and 1.25 μm, respectively. Restored sensitivity to colistin with these compounds does not appear to coincide with known mechanisms of colistin resistance.
Keywords: Klebsiella pneumoniae, Acinetobacter baumannii, prodigiosin, novobiocin, colistin resistance, antibiotic adjuvant
Graphical Abstract
Multidrug resistant (MDR) bacteria have been a developing public health issue for decades. The Centers for Disease Control and Prevention (CDC) estimate 2.8 million antibiotic-resistant infections occur in the US every year, of which 35,000 are lethal.1 Many of these lethal infections are caused by members of a group commonly referred to as ESKAPE pathogens, which include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.2,3 Infections caused by these bacteria are of particular concern to immunocompromised individuals, including those with cancer or HIV/AIDS. After years of widespread antibiotic use, these bacteria have developed resistance to many classes of antibiotics (in some cases all), necessitating the investigation into alternative therapies to circumvent resistance mechanisms and to continue to effectively treat MDR bacterial infections.4
The identification and development of antibiotic adjuvants represent one such alternative. These small molecules do not exhibit antibacterial properties on their own at their active adjuvant concentrations but preserve or enhance the efficacy of a drug when used in combination with antibiotics.5,6 Mechanisms by which they accomplish this can include increasing uptake of an antibiotic,7,8 inhibiting antibiotic modification,9,10 inhibiting target modification,11,12 or inhibiting the efficiency of efflux pumps.13 A well-known antibiotic/adjuvant combination, amoxicillin/clavulanic acid (marketed as Augmentin), is in common clinical use.14,15
Approximately 75% of antibiotics in use today were originally derived from naturally produced compounds, particularly secondary metabolites produced by fungi or bacteria, such as those of the genus Streptomyces.16–18 Furthermore, approximately half of the new chemical entities introduced between 1981 and 2002 were either natural products or derived from/inspired by natural products.19,20 Given their chemical diversity and the similarity of targets across species, natural products have been a productive source of leads for novel drug therapies.21
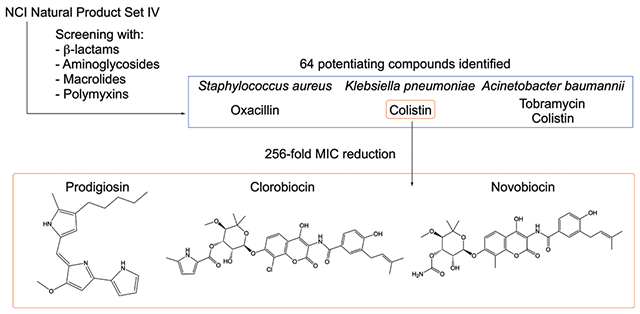
The National Cancer Institute (NCI) Developmental Therapeutics Program has accumulated a repository of natural products from various plant, marine, and microbial samples for the purpose of identifying scaffolds or derivatives of natural products to treat a variety of diseases.22 Libraries of natural products have traditionally been screened for anticancer, antiviral, or antibacterial activities. Recently, we demonstrated that meridianin D, an indolic marine natural product originally documented to have activity against S. aureus23 and Mycobacterium tuberculosis,24 also acts as an adjuvant to break colistin resistance in colistin-resistant bacteria.25 On the basis of this discovery and natural products rarely being screened for adjuvant activity, we hypothesized that current collections of purified natural products or extracts may contain previously undisclosed adjuvant activity. Herein, we report that multiple compounds from the NCI natural products repository Natural Products Set IV (plate numbers 13160330 and 13160331) act as effective antibiotic adjuvants in multiple ESKAPE pathogens with varying antibiotic resistance profiles. They show particular promise in combatting colistin resistance.
RESULTS
Natural Products Effectively Lower Minimum Inhibitory Concentrations of Multiple Classes of Antibiotics against Both Gram-Positive and Gram-Negative Pathogens.
A screen to identify adjuvants in the NCI Natural Product Library Set IV (hereafter referred to as Set IV) was conducted with three ESKAPE pathogens (S. aureus, A. baumannii, and K. pneumoniae) and four different antibiotic classes. The Gram-positive pathogen methicillin-resistant S. aureus (MRSA strain ATCC BAA-1556) was screened first for resensitization to the β-lactam antibiotic oxacillin. At 20 μM, 16 natural products reduced the minimum inhibitory concentration (MIC) of oxacillin by at least 4-fold (Table S1). Of these, five further reduced the oxacillin MIC 16-fold, bringing the required dosage for growth inhibition to 2 μg/mL. These compounds include: (plate 13160330) E12, michell-amine B diacetic acid salt; F13, cryptopimaric acid; G9, CAS 32986-79-1; (plate 13160331) I11, sirodesmin A; L16, variabiline dihydrochloride (Figure 1).
Figure 1.
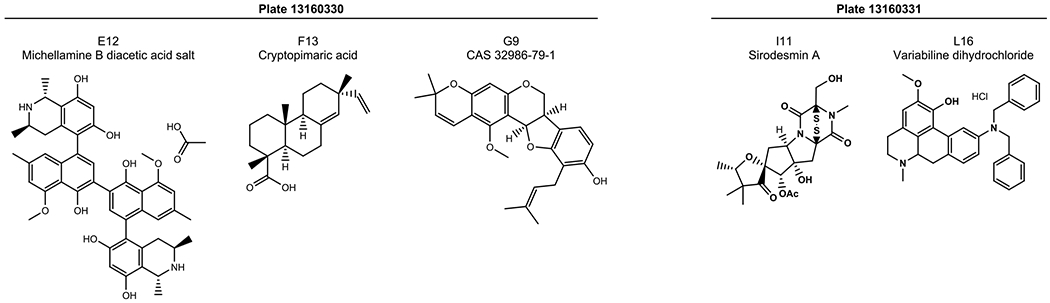
Natural products that resensitize MRSA 1556 to oxacillin. Michellamine B diacetic acid salt (E12, plate 13160330), cryptopimaric acid (F13, plate 13160330), CAS 32986-79-1 (G9, plate 13160330), sirodesmin A (I11, plate 13160331), and variabiline dihyrdrochloride (L16, plate 13160331) were all able to lower the MIC of oxacillin 16-fold in MRSA 1556. Natural products were tested at 20 μM.
Next, we focused on screening for adjuvant activity with Gram-negative bacteria given the paucity of new antibiotics that have been developed for these bacteria. K. pneumoniae strain ATCC BAA-2146 (KP2146) was screened for potentiation of clarithromycin and meropenem; A. baumannii strain 5075 (AB5075) was screened with tobramycin in addition to both of the aforementioned antibiotics. At 20 μM, adjuvants were identified only in AB5075 in combination with tobramycin. Two compounds lowered the tobramycin MIC 8-fold (Figure 2): J12 (plate 13160330, ricinoleic acid sodium salt) and L13 (plate 13160331, epirubicin). Though additional compounds were identified as preventing bacterial growth in initial screening with tobramycin, subsequent counter-screening for stand-alone toxicity showed them to have antibiotic activity at 20 μM. Compounds identified as having antibiotic activity can be found in Table S2 (plate 13160330) and Table S3 (plate 13160331).
Figure 2.
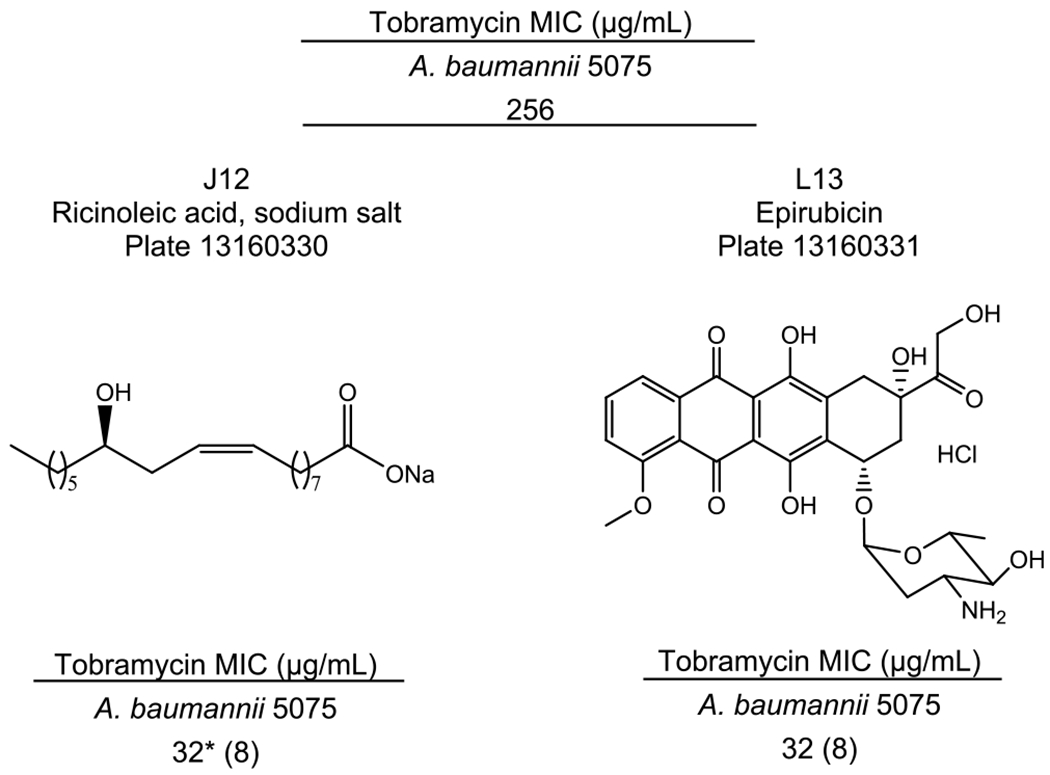
Resensitization of A. baumannii 5075 to tobramycin. Ricinoleic acid sodium salt (J12, plate 13160330) and epirubicin (L13, plate 13160331) both lowered the MIC of tobramycin 8-fold in AB5075. Natural products were tested at 20 μM. Fold change as compared to the control antibiotic MIC is expressed in parentheses. * indicates the lowest antibiotic concentration tested.
After observing so few hits in Gram-negative pathogens, we additionally screened Set IV for compounds exhibiting activity with colistin, a polymyxin generally regarded as the antibiotic of last resort for the treatment of MDR Gram-negative infections. With colistin, we saw a dramatic increase in the number of compounds displaying adjuvant activity (Table S4). In colistin-susceptible AB5075, 31 natural products in Set IV reduced the colistin MIC at least 4-fold, from 1 μg/mL down to 0.25 μg/mL. The lowest MIC was reduced 16-fold, by plate 13160331 compound H6 (dianemycin). Fourteen compounds also reduced the colistin MIC by 4-fold in the colistin-susceptible KP2146. Three compounds further lowered the MIC 16-fold, all of which are located on plate 13160331: H6, dianemycin; J4, scopafungin; N15, variabiline monohydrochloride (Figure 3).
Figure 3.
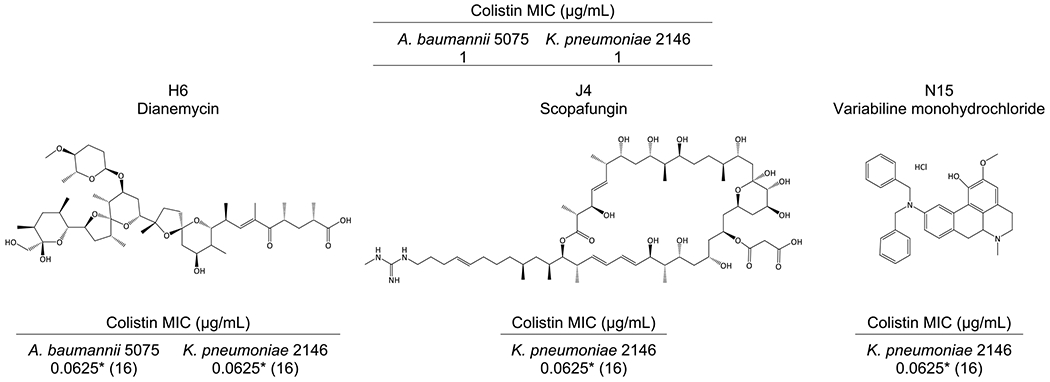
Structures and colistin MICs of the most effective compounds in colistin-susceptible strains of A. baumannii and K. pneumoniae. All compounds were located on plate 13160331. Dianemycin lowered the colistin MIC 16-fold in both AB5075 and KP2146, from 1 μg/mL to 0.0625 μg/mL. Scopafungin and variabiline monohydrochloride had the same activity in KP2146. Natural products were tested at 20 μM. Fold change as compared to the control antibiotic MIC is expressed in parentheses. * indicates the lowest antibiotic concentration tested.
Following this observed adjuvant activity, the screen with colistin was conducted in two colistin-resistant strains. A. baumannii 4106 (AB4106) is highly resistant to colistin via mutations in pmrAB, which lead to modifications of the lipopolysaccharide,26 registering an MIC of 1024 μg/mL; 23 of the compounds from Set IV lowered that MIC at least 4-fold, and three compounds reduced the colistin MIC 512-fold, down to the breakpoint concentration of 2 μg/mL. These compounds were all located on plate 13160331: D7, streptovaricin G; G7, rifamycin, 3-[(dimethylamino)methyl]-(9CI); H8, borrelidin (Figure 4, Table 1). When screened against the colistin-resistant K. pneumoniae B9 (KPB9), which expresses Ara4N modifications of LPS,27 the number of compounds with adjuvant activity doubled compared to the colistin-sensitive strain. Thirty compounds reduced the colistin MIC from 512 to 128 μg/mL (a 4-fold change); nine compounds further reduced the MIC to 2 μg/mL (plate 13160330: G20, prodigiosin; L9, nigericin sodium salt; plate 13160331: D7, streptovaricin G; G7, rifamycin, 3-[(dimethylamino)methyl]-(9CI); H8, borrelidin; H10, pepleomycin sulfate; I8, withanolide E; K12, stubomycin; L8, clorobiocin), and two compounds lowered the colistin MIC to 1 μg/mL (plate 13160330: F16, streptovaricin C; N5, carbomycin) (Figure 4, Table 1). Three compounds potentiated colistin in all four strains tested (plate 13160331: F6, rhodomycin A; F8, aclarubicin; H6, dianemycin); however, the activity of these compounds is modest.
Figure 4.
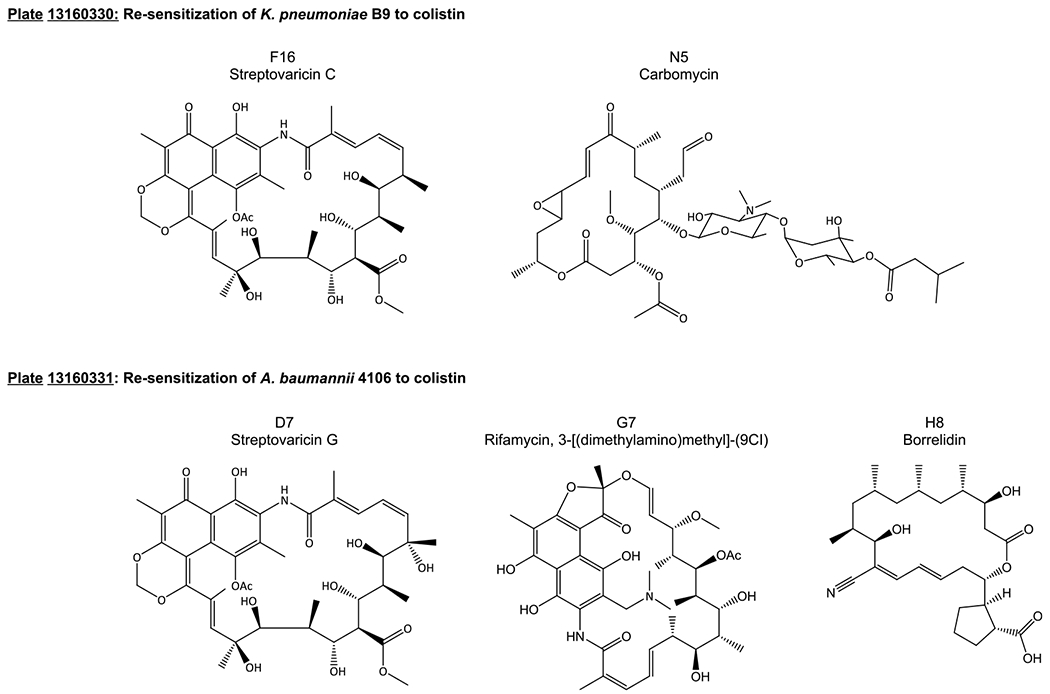
Structures of compounds that lowered colistin MICs most effectively in KPB9 (top) and AB4106 (bottom). Streptovaricin C and carbomycin lowered the KPB9 colistin MIC from 512 to 1 μg/mL. Streptovaricin G, rifamycin, 3-[(dimethylamino)methyl]-(9CI), and borrelidin lowered the AB4106 colistin MIC from 1024 to 2 μ/mL.
Table 1.
Potentiation of Colistin against Colistin-Susceptible and -Resistant Strains of A. baumannii and K. pneumoniaea
| colistin MIC (μg/mL) |
|||||
|---|---|---|---|---|---|
| compound name | A. baumannii 5075 | A. baumannii 4106 | K. pneumoniae 2146 | K pneumoniae B9 | |
| control | 1 | 1024 | 1 | 512 | |
| Plate 13160330 | |||||
| F16 | streptovaricin C | 1b (512) | |||
| G20 | prodigiosin | 0.125 (8) | 0.25 (4) | 2 (256) | |
| L9 | nigericin sodium salt | 0.125 (8) | 64 (16) | 2 (256) | |
| N5 | carbomycin | 0.25 (4) | 64 (16) | 1b (512) | |
| Plate 13160331 | |||||
| D7 | streptovaricin G | 0.125 (8) | 2b (512) | 2b (256) | |
| G7 | rifamycin, 3-[(dimethylamino)methyl]-(9CI) | 0.25 (4) | 2b (512) | 2b (256) | |
| H8 | borrelidin | 0.25 (4) | 2b (512) | 2b (256) | |
| H10 | pepleomycin sulfate | 0.125 (8) | 4 (256) | 2b (256) | |
| I8 | withanolide E | 0.25 (4) | 2b (256) | ||
| K12 | stubomycin | 64 (16) | 2b (256) | ||
| L8 | clorobiocin | 2b (256) | |||
Natural products were tested at 20 μM. Fold change as compared to the control antibiotic MIC is expressed in parentheses. A full list of compounds that lowered colistin MICs at least 4-fold can be found in Table S4.
Indicates the lowest antibiotic concentration tested.
Dose responses were then measured for natural products that demonstrated the highest potentiation of colistin. The colistin concentration was held constant at the MIC value achieved with each respective natural product at 20 μM, and the concentration of natural product was then serially lowered from 20 μM to determine the lowest effective concentration. The most activity was seen when compounds were screened against KPB9, where more than half of the compounds identified were still active at or below 1.25 μM (16-fold lower than the initial screen concentration of 20 μM) (Table 2). The Set IV natural products that lowered the colistin MIC the furthest with the lowest concentration of compound were: streptovaricin C; prodigiosin; streptovaricin G; rifamycin, 3-[(dimethylamino)methyl]-(9CI); withanolide E; clorobiocin.
Table 2.
Lowest Effective Concentration of Natural Products with Adjuvant Activity
| bacterial strain | colistin MIC (μg/mL) | plate # | well ID | compound name | lowest active concentration (μM) | colistin concentration (μg/mL) |
|---|---|---|---|---|---|---|
| A. baumannii 5075 | 1 | 13160331 | H6 | dianemycin | 10 | 0.0625 |
| A. baumannii 4106 | 1024 | 13160331 | D7 | streptovaricin G | 10 | 2 |
| G7 | rifamycin, 3-[(dimethylamino) methyl]-(9CI) | 10 | 2 | |||
| H8 | borrelidin | 10 | 2 | |||
| K. pneumoniae 2146 | 1 | 13160331 | H6 | dianemycin | 20 | 0.0625 |
| J4 | scopafungin | 20 | 0.0625 | |||
| N15 | variabiline monohydrochloride | 20 | 0.0625 | |||
| K. pneumoniae B9 | 512 | 13160330 | F16 | streptovaricin C | 1.25 | 2 |
| G20 | prodigiosin | 1.25 | 2 | |||
| L9 | nigericin sodium salt | 10 | 2 | |||
| N5 | carbomycin | 10 | 2 | |||
| 13160331 | D7 | streptovaricin G | 0.3125 | 2 | ||
| G7 | rifamycin, 3-[(dimethylamino) methyl]-(9CI) | 1.25 | 2 | |||
| H8 | borrelidin | 15 | 2 | |||
| H10 | pepleomycin sulfate | 15 | 2 | |||
| I8 | withanolide E | 0.625 | 2 | |||
| K12 | stubomycin | 10 | 2 | |||
| L8 | clorobiocin | 0.625 | 2 |
Compounds Similar to Active Natural Product Adjuvants Maintain Activity.
On the basis of the activity observed with prodigiosin, rifamycin, 3-[(dimethylamino)-methyl]-(9CI), and clorobiocin, three structurally related compounds (obatoclax, rifampicin, and novobiocin; Figure 5) were also screened for their ability to potentiate colistin (Table 3). Obatoclax, a synthetic drug similar in structure to prodigiosin (G20, 13160330), was less active than prodigiosin in A. baumannii but maintained similar activity in colistin-resistant K. pneumoniae. Rifampicin, which is structurally similar to the rifamycin derivative (G7, 13160331), showed better activity with colistin in both AB4106 and KPB9 but was also more toxic, with MICs 4–16-fold below that of G7. Novobiocin, structurally related to clorobiocin (L8, 13160331), required higher concentrations for activity in KPB9 but was far more active than clorobiocin in AB strains. While clorobiocin did not show any activity in AB4106, novobiocin potentiated colistin in this strain as well as four additional colistin-resistant strains of AB tested (Table S5). Fractional inhibitory concentration indices (FICI) were calculated for each combination; aside from prodigiosin in AB4106, all combinations have FICI < 0.5, indicating synergy between colistin and each compound. The addition of fetal bovine serum to the potentiation assay did have a slight effect on activity of the combination of colistin with novobiocin and prodigiosin, increasing the required concentration of compound by up to 4-fold (Table S6).
Figure 5.
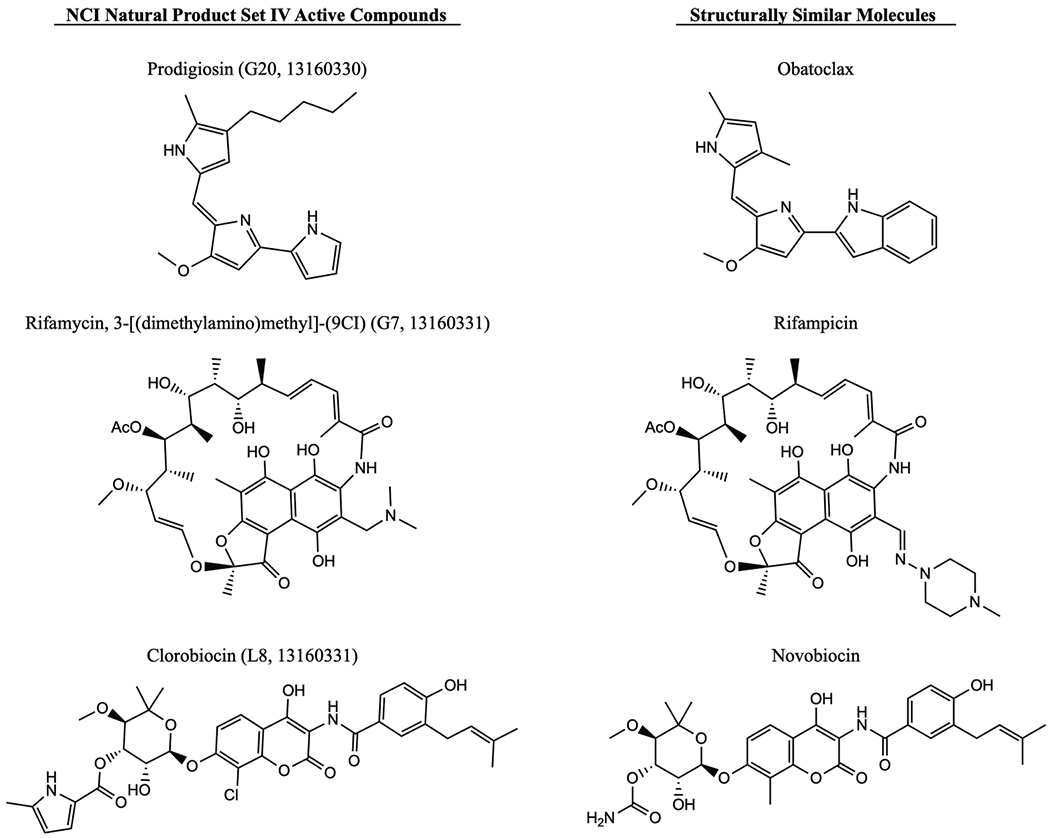
Structures of active compounds from NCI Natural Product Set IV (left) and molecules with similar structures screened for activity with colistin (right).
Table 3.
Colistin Potentiation Activity of Active Natural Products and Structurally Similar Molecules in Colistin-Resistant A. baumannii and K. pneumoniae
| A. baumannii | K pneumoniae | ||
|---|---|---|---|
| lowest effective concentration | bacterial strain | 4106 | B9 |
| colistin MICa | 1024 | 512 | |
| prodigiosin MIC (μM) | 10 | >160 | |
| prodigiosin (μM) | 5 | 1.25 | |
| colistin MICa (fold-reduction) | 4 (256) | 2 (256) | |
| fractional inhibitory concentration index (FICI) | 0.504 | 0.008b | |
| obatoclax MIC (μM) | >160 | >160 | |
| lowest effective concentration | obatoclax (μM) | 20 | 1.25 |
| colistin MICa (fold-reduction) | 4 (256) | 2 (256) | |
| FICI | 0.066 | 0.008 | |
| rifamycin, 3-[(dimethylamino) methyl]-(9CI) MIC (μM) | >20 | 100 | |
| lowest effective concentration | rifamycin, 3-[(dimethylamino) methyl]-(9CI) (μM) | 10 | 0.3125 |
| colistin MICa (fold-reduction) | 2 (512) | 2 (256) | |
| FICI | 0.252 | 0.007 | |
| rifampicin MIC (μM) | 5 | 20 | |
| lowest effective concentration | rifampicin (μM) | 0.3125 | 0.156 |
| colistin MICa (fold-reduction) | 2 (512) | 8 (64) | |
| FICI | 0.064 | 0.023 | |
| clorobiocin (μM) | >20 | 50 | |
| lowest effective concentration | clorobiocin (μM) | NAc | 0.625 |
| colistin MICa (fold-reduction) | NA | 2 (256) | |
| FICI | 0.016 | ||
| novobiocin MIC (μM) | 20 | >160 | |
| lowest effective concentration | novobiocin (μM) | 2.5 | 2.5 |
| colistin MICa (fold-reduction) | 4 (256) | 2 (256) | |
| FICI | 0.129 | 0.012 |
Colistin MICs are presented in μg/mL. The numbers in parentheses indicate the fold-reduction from the original colistin MIC of each strain.
When the MIC of the compound is above the highest tested concentration, FICI is approximated using double the highest tested concentration (e.g., when MIC > 160 μg/mL, FICI is approximated using 320 μg/mL).
NA indicates no activity in the strain.
The activity of select compounds in additional colistin-resistant strains can be found in Table S5. Checkerboard assays for prodigiosin and novobiocin in combination with colistin, and associated FICI, can be found in Figures S1 and S2 and Table S8, respectively.
Colistin Potentiation Is Not Related to Membrane Permeability or Reversal of Lipid A Modifications.
Given the activity and commercial availability, novobiocin and prodigiosin were chosen for preliminary mechanism of action (MOA) studies using KPB9 as a representative bacterial strain. Colistin has been previously shown to potentiate Gram-positive selective antibiotics in strains containing the mobile colistin resistance plasmid mcr-1,28 while novobiocin and clorobiocin have been reported to sensitize bacteria to colistin by accelerating LPS turnover.29 We were curious as to which compound (colistin or natural products identified in the screen) was sensitizing bacteria and which was inhibiting growth or whether they acted synergistically. We first evaluated the effects of cotreatment or pretreatment with colistin, novobiocin, or prodigiosin on planktonic growth in KPB9. Novobiocin, at half its MIC (4× lowest effective concentration), inhibited growth by 57% after 14 h; prodigiosin did not inhibit growth. Only in simultaneous combination with colistin did either novobiocin or prodigiosin prevent bacterial growth (Figure S3). Treatment with PMBN in place of colistin did not show the same inhibitory effect, indicating that the low concentration of colistin is not permeabilizing the outer membrane to enhance antibiotic effects of novobiocin or prodigiosin (Table S7). We then evaluated membrane integrity via the BacLight assay. Neither compound alone increased membrane permeability, though membrane integrity was compromised when cells were treated with either compound in combination with colistin (Figure 6). Adding prodigiosin at its lowest effective concentration (1.25 μM) marginally increased membrane permeability by an average of 11%. Novobiocin at its lowest effective concentration did not, on average, affect membrane permeability. The lethal combinations of colistin with prodigiosin or novobiocin significantly increased membrane permeability, though not to the level of a lethal concentration of colistin.
Figure 6.
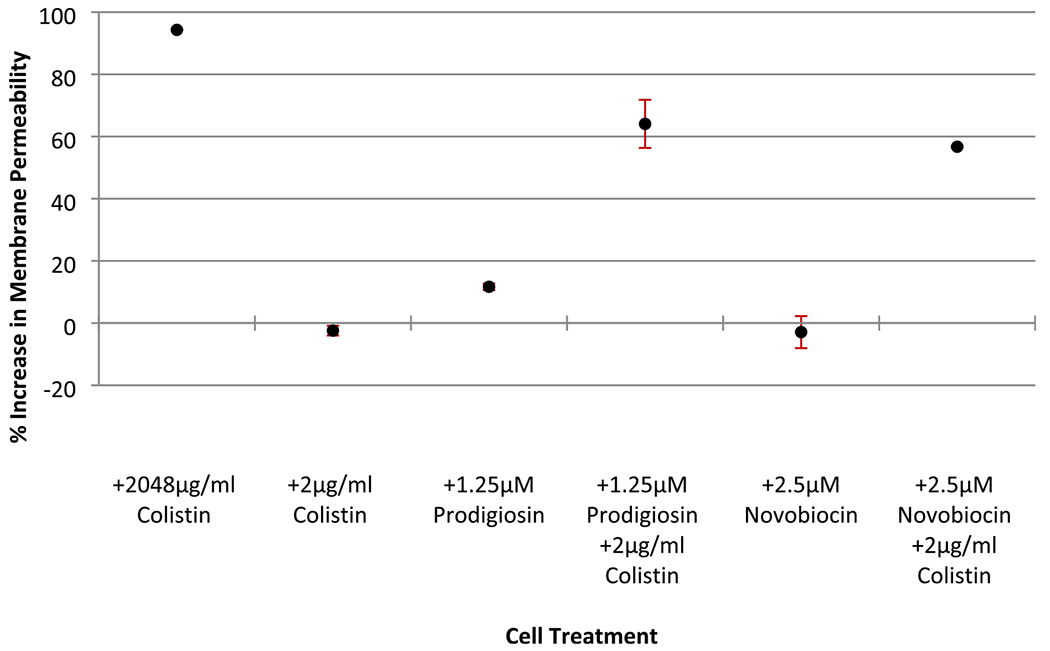
Membrane permeability of K. pneumoniae B9 does not change with treatment of 2 μg/mL colistin or compound alone but increases significantly under lethal combination conditions. The percent increase in membrane permeability was calculated by comparing SYTO9: propidium iodide fluorescence ratios of treated cells to untreated cells. Error bars represent standard error of at least two independent biological samples from experiments conducted on separate days. Note: treatment conditions “+2048 μg/mL colistin” and “+2.5 μM novobiocin + 2 μg/mL colistin” have standard error below 1%.
Colistin resistance is associated with modifications to the lipid A anchor of the lipopolysaccharide (LPS) component of the outer membrane, such as the addition of l-4-amino-arabinose or phosphoethanolamine.27,30–33 Such modifications reduce the net negative charge of LPS, reducing the affinity to cationic polypeptides like colistin. Several studies have shown that these modifications are mediated by the pmrAB or phoPQ regulatory systems,31,34–36 including in colistin-resistant K. pneumoniae.27,37–39 We conducted RT-qPCR analysis to determine if treatment with prodigiosin or novobiocin significantly altered the expression of these genes (Figure 7). Though prodigiosin slightly upregulated expression of phoQ and pmrA (approximately 2- and 4-fold, respectively), it did not significantly change the expression of other genes in the respective systems. The addition of colistin with prodigiosin followed the same trend. Treatments with novobiocin, either alone or with colistin, had no major effect on the expression of these genes. These results are in agreement with previous reports that link novobiocin to increased LPS transport and enhanced polymyxin activity.29,40 As resensitization to colistin by other small molecules has been shown to coincide with reversal of lipid A modification,12,41 we complemented the RT-qPCR data with MALDI-ToF analysis of lipid A from KPB9 treated with prodigiosin or novobiocin and found no significant changes in l-4-aminoarabinose or phosphoethanolamine content (Figure S4).
Figure 7.
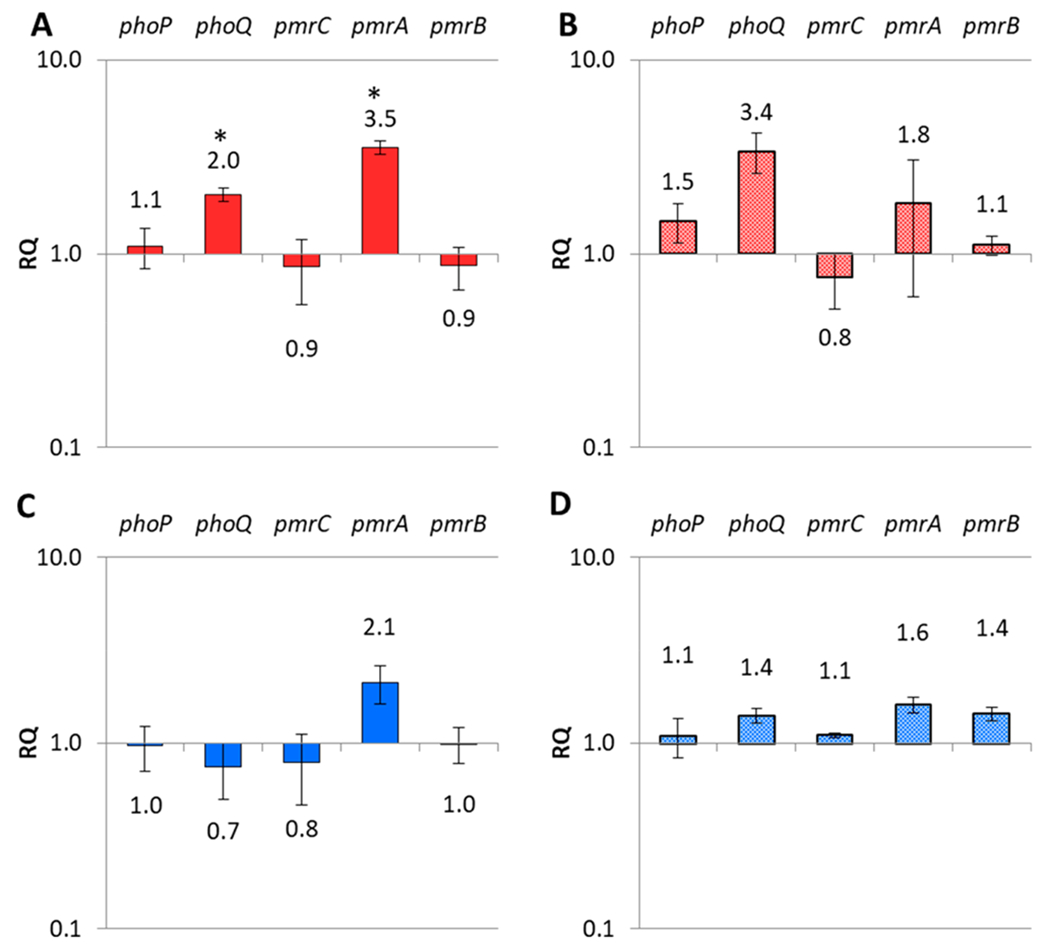
Variation in gene expression for the two component system phoPQ and the pmrCAB operon in K. pneumoniae B9 upon treatment with (A) 5 μM prodigiosin, (B) 1.25 μM prodigiosin with 2 μg/mL colistin, (C) 10 μM novobiocin, and (D) 2.5 μM novobiocin with 2 μg/mL colistin. All relative quantification (RQ) is calculated using the 12S rRNA rpsL as the reference gene. Error bars represent standard error of ΔΔCt values across 6 reactions in 2 independent samples. *p < 0.05 by Student’s t test comparing ΔCt of test reactions and control reactions, indicating statistically significant differences upon treatment with 5 μM prodigiosin.
Combination of Prodigiosin and Colistin Shows an Increase in Reactive Oxygen Species.
Prodigiosin and polymyxins have both been reported to cause bacterial cell death via reactive oxygen species (ROS) generation.42–44 On the basis of these reports, we also evaluated the presence of ROS using the fluorescent dye 2′,7′-dichlorofluorescin diacetate (DCFH-DA) in cells treated with prodigiosin, novobiocin, or either in combination with colistin (Figure 8). Compared to untreated KPB9 cells, ROS increased marginally after treatment with 1.25 μM prodigiosin (11%) or 2.5 μM novobiocin (5%). The most striking result, however, was the increase in ROS seen upon treatment with both prodigiosin and colistin: compared to an untreated control, ROS increased exponentially. While colistin and prodigiosin each individually elevated ROS by approximately 10%, together they produced increases in ROS up to 1656%. An increase of this magnitude was not observed with the lethal combination of novobiocin/colistin nor with hydrogen peroxide combined with colistin, indicating specificity to the combination of prodigiosin and colistin. This also indicates ROS is not increasing simply due to the onset of cell death, as novobiocin and hydrogen peroxide combined with colistin also prevent cell growth but without such drastic changes in ROS (Figure S5).
Figure 8.
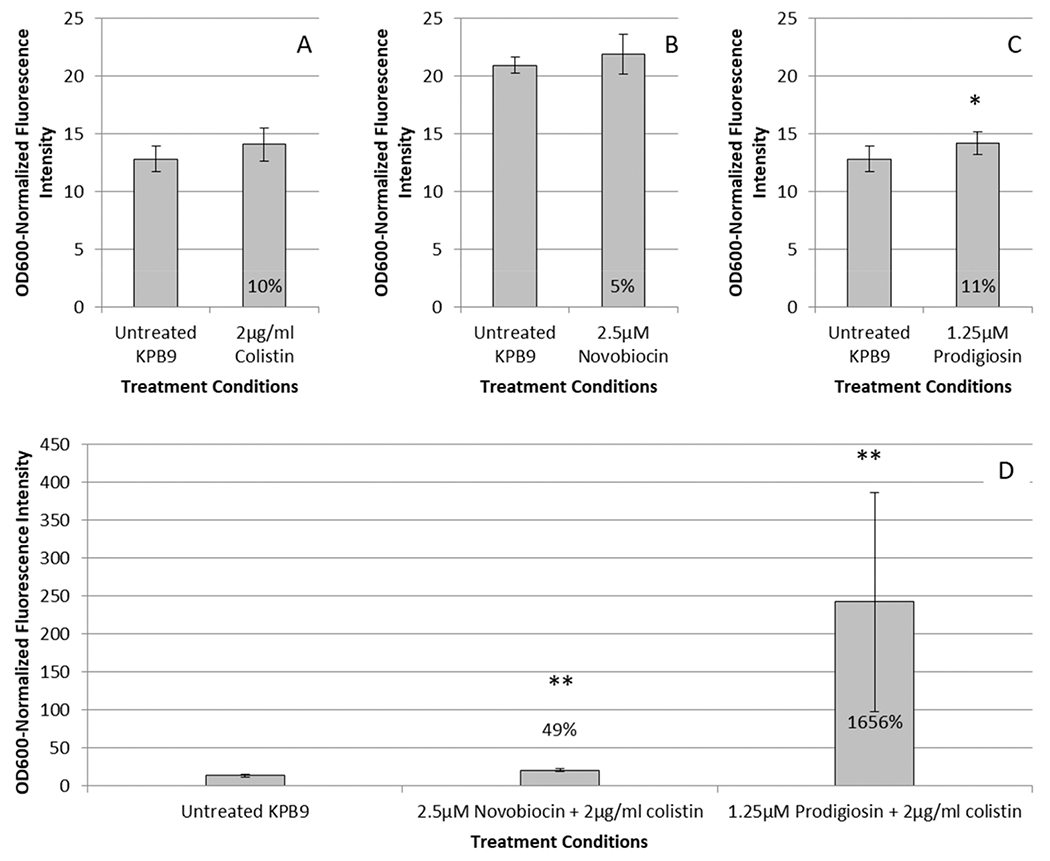
Average OD600 normalized ROS fluorescence intensity compared to untreated cells when treated with (A) 2 μg/mL colistin, (B) novobiocin, (C) prodigiosin, or (D) prodigiosin or novobiocin in combination with 2 μg/mL colistin. Percentages shown indicate the average increase in fluorescence; * indicates a p-value < 0.06, and ** indicates a p-value < 0.005 compared to untreated KPB9 subjected to the fluorescent dye treatment. Fluorescence of DCFH-DA was normalized by the OD600 of the well to account for potential differences due to growth rates; measurements were obtained simultaneously. Error bars indicate standard deviation of five measurements in each of six independent biological samples using the same stock solution of DCFH-DA.
DISCUSSION
We have identified a number of natural products from the NCI Natural Products Set IV with adjuvant activity against A. baumannii and K. pneumoniae. Out of the 419 compounds contained in Set IV, 64 showed the ability to potentiate at least one of the antibiotics tested, lowering the MIC at least 4-fold without showing inherent toxicity at 20 μM (a hit rate of 15.3% overall and 12.9% for Gram-negative bacteria). Of the antibiotics tested, the highest number of adjuvants was identified with colistin. Colistin has been used as a drug of last resort due to nephrotoxicity and neurotoxicity. However, resistance to colistin is becoming more frequent, especially with the plasmid-borne mcr genes being disseminated into the human pathogen pool. Prodigiosin and novobiocin were both able to potentiate colistin down to the breakpoint concentration in highly colistin resistant strains of A. baumannii and K. pneumoniae. Both were particularly active in K. pneumoniae strain B9, only requiring low concentrations to effectively lower the colistin MIC.
The activity observed for the combination of novobiocin (or clorobiocin) and colistin was not unexpected, as similar synergy has been shown between novobiocin and polymyxins in multiple Gram-negative pathogens, including sensitive strains of A. baumannii and K. pneumoniae29 and an Escherichia coli strain harboring the mcr-1 plasmid.28 We have added to these observations by establishing that novobiocin also potentiates the activity of colistin against both A. baumannii and K. pneumoniae in clinical isolates with chromosomally encoded colistin resistance. Novobiocin has been shown to increase the transport of LPS to the outer membrane, decreasing the membrane permeability while also increasing the activity of polymyxins. We have further established that activity is not related to the reversal of lipid A modification nor the generation of reactive oxygen species.
Prodigiosin has shown antibacterial, antimalarial, antifungal, anticancer, and apoptotic properties, but its mode of action as an antibacterial agent remains unknown.45–47 Some studies have pointed to the membrane as a potential target,43,44 with a recent simulation study suggesting that prodigiosin localizes in the lipid bilayer and alters the local architecture, potentially allowing certain molecules into the cell without majorly affecting membrane integrity.48 We do qualitatively observe the interaction of prodigiosin with the membrane, as the characteristic red coloring of prodigiosin remains in the bacterial pellet after treatment, even after multiple washes. Additionally, our findings agree that membrane integrity is somewhat compromised, as we observe a very modest 11.2% increase in membrane permeability that is similar to what was previously reported for prodigiosin-treated E. coli.43 Prodigiosin has been connected to increased intracellular reactive oxygen species and SOS response.43,44,49 The combination of prodigiosin and colistin results in a large increase in ROS (upward of 1656-fold) not seen in other lethal combinations tested here. Such a large increase may underpin the activity of this combination, but more experimentation is required to fully elucidate the mechanism of action.
CONCLUSION
Here, we have shown that established libraries of natural compounds contain a number of previously undisclosed antibiotic adjuvants. In screening the NCI Natural Products Set IV and related compounds, novobiocin and prodigiosin were found to be particularly effective at breaking colistin resistance. Novobiocin is used clinically and, so, may be a good candidate to develop for therapeutic use with colistin; prodigiosin has been shown to cause genetic damage in human cell lines,50 and thus, less toxic derivatives would be needed before consideration for therapeutic use. The activity does not appear to correspond with known mechanisms of colistin resistance, with both compounds showing minimal effects on the expression of the phoPQ or pmrAB regulatory systems and displaying no indication of reversing lipid A modifications. Large increases in ROS, however, may explain the activity of the effective combination of colistin and prodigiosin. Together, this could point to a new line of investigation toward breaking colistin resistance.
METHODS
Natural Product Library Maintenance.
The National Cancer Institute (NCI) Natural Product Library Set IV was obtained from the NCI Chemotherapeutic Agents Repository via Fisher BioServices. Compounds in Set IV were received as 10 mM solutions (20 μL per compound) distributed across two 384-well polypropylene microtiter plates (plate 13160330 and plate 13160331, replicate 175). Compounds in Set IV have a purity of >90% by evaporative light scattering detector (ELSD), and the major peak has correct mass ion, according to the NCI library standards. Stocks were diluted to 1 mM and transferred to 96-well plates (4 per original plate, distributed by quadrants thusly: A1-H12, A13-H24, I1-P12, and I13-P24). All compound plates were wrapped in Parafilm and stored at −80 °C. 1 mM working stocks were thawed for 30 min before use in potentiation assays.
Bacterial Strains and Growth Conditions.
Bacterial strains and their minimum inhibitory concentrations with regard to tested antibiotics used in this study can be found in Table 4. Acinetobacter baumannii and Klebsiella pneumoniae strains were routinely grown on LB Lennox 1.5% (w/v) agar plates from glycerol stocks every 2 weeks. For use in biological assays, these strains were grown in cation-adjusted Mueller Hinton broth (CAMHB, BD). Methicillin-resistant Staphylococcus aureus (MRSA) was routinely plated from glycerol stock on tryptic soy agar plates (1.5% agar (w/v)). Broths were grown in Mueller Hinton broth (MHB).
Table 4.
Bacterial Strains and Minimum Inhibitory Concentrations (MIC)
| species | strain | antibiotic; MIC (μg/mL) |
|---|---|---|
| Acinetobacter baumannii | 5075 | tobramycin; 256 |
| meropenem; 32 | ||
| clarithromycin; 32 | ||
| colistin; 1 | ||
| 4106 | colistin; 1024 | |
| 3941 | colistin; 512 | |
| 3942 | colistin; 512 | |
| 4112 | colistin; 1024 | |
| 4119 | colistin; 1024 | |
| Klebsiella pneumoniae | 2146 | clarithromycin; 1024 |
| meropenem; 256 | ||
| colistin; 1 | ||
| B9 | colistin; 512 | |
| C3 | colistin; 256 | |
| Methicillin-resistant Staphylococcus aureus | 1556 | tobramycin; 32 |
Minimum Inhibitory Concentration (MIC) Determination.
MICs were determined following the Clinical and Laboratory Standards Institute (CLSI) guidelines. Bacterial strains were always cultured in duplicate. Strains were grown in 3 mL of appropriate media (K. pneumoniae and A. baumannii in CAMHB; MRSA in MHB) for 6 h at 37 °C with shaking at 200 rpm and then diluted to 5 × 105 CFU in fresh media. Subcultures were aliquoted and dosed with antibiotic or natural product. Each compound was serially diluted in 100 μL of subculture in a 96-well plate and incubated at 37 °C overnight without shaking. A Synergy HTX Multimode plate reader was used to determine the optical density (OD600) of cell growth in each well. The lowest dilution with <10% growth as compared to a positive control was recorded as the MIC. MICs were repeated regularly to ensure consistent levels of antibiotic resistance in each strain as well as to ensure the potency of new antibiotic stock.
Antibiotic Potentiation Assay.
Potentiation assays were completed as previously described.51,52 Bacterial cells were grown in 3 mL of media for 6 h at 37 °C with shaking at 200 rpm and then diluted to 5 × 105 CFU in fresh media. For initial screening of the NCI natural product plates 13160330 and 13160331, the appropriate antibiotic was added to a concentration equal to 1/4 of the determined MIC, as described in Table 1. 98 μL of cell culture/antibiotic mix was dispensed into sterile U-bottom 96-well plates (Costar). Each well was dosed with 2 μL of 1 mM natural product in DMSO for a final concentration of 20 μM. Plates were covered and incubated without shaking for 18 h at 37 °C. A control plate containing the cell culture/antibiotic mix without natural product additions (positive control) and fresh media (negative control) was included for each set of screenings. A Synergy HTX Multimode plate reader was used to determine the OD600 of each well; the negative control was subtracted from each reading, and the OD600 was compared to the positive control. Less than 10% growth compared to the positive control was recorded as significant growth inhibition.
Natural product compounds exhibiting 90% growth inhibition in combination with 1/4 MIC of antibiotic were further evaluated for inherent antibacterial activity. Assays were conducted and evaluated as before but without the addition of antibiotic. Once the natural product was determined not to be toxic at 20 μM, resensitization assays were repeated with breakpoint antibiotic concentrations. If the combination of natural product and lower antibiotic concentration was still effective at inhibiting growth, dose responses were completed with regard to the natural product and the antibiotic to determine the lowest effective concentrations.
Polymyxin B Nonapeptide Permeabilization.
To help determine if colistin was permeabilizing outer membranes to novobiocin or prodigiosin, MICs of prodigiosin and novobiocin were evaluated with A. baumannii 5075 and 4106 and K. pneumoniae 2146 and B9 in the presence of the cationic polypeptide polymyxin B nonapeptide (PMBN; Sigma). Growth media was supplemented with 2, 4, or 8 μg/mL PMBN, and compounds of interest were serially diluted from 20 μM to determine any changes in MIC.
Effects of Serum on Combination Efficacies.
To determine the effect of serum on the efficacy of prodigiosin or novobiocin with colistin, effective concentrations of prodigiosin or novobiocin were evaluated with a fixed 2 μg/mL colistin. Growth medium was supplemented with 5%, 10%, or 20% (v/v) fetal bovine serum (FBS). Prodigiosin and novobiocin were serially diluted from 20 μM. Fold change was determined on the basis of a 0% (v/v) FBS control.
Growth Curves.
Bacterial strains included in growth curves were grown in CAMHB for 6 h at 37 °C, diluted to 5 × 105 CFU, and aliquoted into a round-bottom 96-well plate. Wells were supplemented with selected compounds and/or antibiotic. A Synergy HTX Multimode plate reader set to 37 °C was used to read OD600 every 20 min for 14 h. The plate was shaken for 5 min before each reading.
Analysis of Lipid A Modification.
Lipid A was analyzed for modification as previously described.41 K. pneumoniae B9 was cultured in 5 mL of CAMHB for 18 h at 37 °C and then subcultured to 1 × 106 CFU in 120 mL of fresh CAMHB. Subcultures were split and left untreated or dosed with novobiocin or prodigiosin at 4× the minimum effective concentration, but below the compound MIC, and incubated for 8 h. Each concentration was completed with biological duplicates. Cells were pelleted for 15 min at 4000 rpm and 4 °C. The supernatant was discarded, and the pellet was washed with 5 mL of endotoxin free water. The cells were pelleted again, and the supernatant was discarded. Pellets were stored at −80 °C until processed for analysis.
Thawed pellets were resuspended in 70% isobutyric acid/1 M ammonium hydroxide (5:3 vol/vol) and incubated at 100 °C for 1 h. Samples were centrifuged (2000g, 15 min). Supernatant was added to equal volumes of endotoxin-free water, frozen on dry ice, and lyophilized. Dried samples were washed with methanol and centrifuged (2000g, 15 min). Lipid A was solubilized in chloroform/methanol/water (3:1.5:0.25 vol/vol/vol). 1 μL of sample was spotted on a steel reusable MALDI plate with 1 μL of norharmane matrix (10 mg/mL in 2:1 C/M). Samples were analyzed on a Bruker Microflex in negative ion mode. Data were processed with flexAnalysis software.
Reverse-Transcriptase Quantitative PCR (RT-qPCR) Analysis.
Bacterial strains were incubated at 37 °C overnight in 5 mL of CAMHB and subcultured 1:20 in fresh media in 15 mL conical tubes. Subcultures were incubated for 2 h at 37 °C, at which point novobiocin, prodigiosin, or a sublethal combination of either with colistin was added to the subculture. Cultures were incubated for another 2 h. Cells were centrifuged (4000 rpm, 20 min, 4 °C), and the supernatant was discarded. RNA was isolated from the bacterial pellet with TRIzol reagent (ThermoFisher), following the manufacturer’s instructions except for an additional wash with 75% ethanol before solubilizing the RNA pellet.
Solubilized RNA was treated with DNase (Turbo DNA-free Kit, Invitrogen) following the manufacturer’s instructions for undiluted samples. Digestion was completed in 2 steps: 2 μL of DNase was added to the sample, and it was incubated at 37 °C for 30 min; another 2 μL of DNase was added before an additional 30 min incubation. DNase treated samples were transferred to new 1.5 mL centrifuge tubes. RNA was quantified using a Take3 plate on a BioTek Synergy HTX multimode plate reader. Samples were stored at −20 °C overnight.
Changes in expression for genes associated with colistin resistance were evaluated by RT-qPCR, including pmrCAB and phoPQ. Primers used in this study can be found in Table 5. RT-qPCR was conducted with the Power SYBR Green RNA-to-CT 1-Step Kit (ThermoFisher) according to the manufacturer’s instructions using an Applied Biosystems StepOne instrument. 12S rRNA gene rpsL was used as the reference gene in all experiments. Relative standard curves were conducted to ensure linear amplification before completing comparative Ct experiments. Reactions were performed in triplicate on two independent biological samples.
Table 5.
Primers Used in RT-qPCR Gene Expression Studies
| primer name | sequence 5′–3′ | source |
|---|---|---|
| KP pmrC forward | GCGTGATGAATATCCTCACCA | 39 |
| KP pmrC reverse | CACGCCAAAGTTCCAGATGA | |
| KP pmrA forward | GATGAAGACGGGCTGCATTT | |
| KP pmrA reverse | ACCGCTAATGCGATCCTCAA | |
| KP pmrB forward | TGCCAGCTGATAAGCGTCTT | |
| KP pmrB reverse | TTCTGGTTGTTGTGCCCTTC | |
| KP phoP forward | GCGTCACCACCTCAAAGTTC | |
| KP phoP reverse | GGCGATATCCGGGAGATGTT | |
| KP phoQ forward | CTCAAGCGCAGCTATATGGT | |
| KP phoQ reverse | TCTTTGGCCAGCGACTCAAT | |
| KP rpsL forward | CCGTGGCGGTCGTGTTAAAGA | |
| KP rpsL reverse | GCCGTACTTGGAGCGAGCCTG |
Evaluation of the Production of Reactive Oxygen Species (ROS).
2′,7′-Dichlorofluorescin diacetate fluorescent dye (DCFH-DA, Calbiochem) was used to measure changes in general intracellular ROS upon treatment with prodigiosin, novobiocin, colistin, and combinations thereof. 20 mM stocks of DCFH-DA were prepared in DMSO and stored, protected from light, at −20 °C in 400 μL aliquots to reduce freeze/thaw cycles. Bacterial strains were incubated in 10 mL of CAMHB at 37 °C and 200 rpm overnight. Cultures were split and diluted 1:2 in fresh media. Cells were either left untreated or dosed with one of the following conditions: 0.15% H2O2 (positive control), 2 μg/mL colistin, 0.07% H2O2 with 2 μg/mL colistin, 2.5 μM or 1.25 μM prodigiosin, 1.25 μM prodigiosin with 2 μg/mL colistin, 2.5 μM novobiocin, or 2.5 μM novobiocin with 2 μg/mL colistin. Cultures were incubated in the dark at 37 °C and 240 rpm for 4 h. 2 mL of treated culture was centrifuged (15 000 rcf, 6 min) and washed with 1× PBS. The pellets were suspended in 1 mL of 100 μM DCFH-DA in 1× PBS and incubated in the dark at 37 °C for 1.5 h. Cells were pelleted (15 000 rcf, 6 min) and washed with 1× PBS before resuspension in fresh CAMHB. Cells recovered for 1 h at 37 °C. 100 μL of cell suspension was aliquoted into black-walled, clear-bottomed 96-well plates. Fluorescence was measured on a HTX Synergy Multimode plate reader with excitation at 485/20 nm and emission at 528/20 nm. Each treatment was measured 5 times in 6 separate biological samples. CAMHB without cells was measured for potential background fluorescence, and 100 μM DCFH-DA in PBS was measured to track spontaneous fluorescence of the dye. p-values were determined by the Student’s t test with the Tukey Method of eliminating outliers.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH National Institute of Allergy and Infectious Disease (RO1AI136904).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00259.
NCI Natural Product Library Set IV with MRSA and oxacillin, antibiotic activities of Set IV compounds at 20 μM full list of compounds active with colistin in tested A. baumannii and K. pneumoniae strains, additional activities in colistin-resistant A. baumannii and K. pneumoniae strains, supplementation with fetal bovine serum, supplementation with polymyxin B nonapeptide, checkerboard assays and FICI for A. bamaunnii 4106 and K. pneumoniae B9, growth curves, Lipid A compositions, and ROS fluorescence assay of K. pneumoniae B9 cells treated with hydrogen peroxide and colistin (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsinfecdis.0c00259
The authors declare the following competing financial interest(s): Dr. C. Melander is a cofounder of Agile Sciences, a biotechnology company seeking to commercialize antibiotic adjuvants.
Contributor Information
Anne E. Mattingly, Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana 46556, United States
Karlie E. Cox, Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana 46556, United States
Richard Smith, Department of Microbial Pathogenesis, University of Maryland-Baltimore, Baltimore, Maryland 21201, United States.
Roberta J. Melander, Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana 46556, United States
Robert K. Ernst, Department of Microbial Pathogenesis, University of Maryland-Baltimore, Baltimore, Maryland 21201, United States
Christian Melander, Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana 46556, United States.
REFERENCES
- (1).CDC (2019) Antibiotic Resistance Threats in the United States, 2019, U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- (2).Mulani MS, Kamble EE, Kumkar SN, Tawre MS, and Pardesi KR (2019) Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol 10, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Pendleton JN, Gorman SP, and Gilmore BF (2013) Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther 11, 297–308. [DOI] [PubMed] [Google Scholar]
- (4).Ventola CL (2015) The antibiotic resistance crisis: part 1: causes and threats. P T 40, 277–283. [PMC free article] [PubMed] [Google Scholar]
- (5).Gill EE, Franco OL, and Hancock REW (2015) Antibiotic Adjuvants: Diverse Strategies for Controlling Drug-Resistant Pathogens. Chem. Biol. Drug Des 85, 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Melander RJ, and Melander C (2017) The Challenge of Overcoming Antibiotic Resistance: An Adjuvant Approach? ACS Infect. Dis 3, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Stowe SD, Thompson RJ, Peng L, Su Z, Blackledge MS, Draughn GL, Coe WH, Johannes E, Lapham VK, Mackenzie J, Melander C, and Cavanagh J (2015) Membrane-permeabilizing activity of reverse-amide 2-aminoimidazole antibiofilm agents against Acinetobacter baumannii. Curr. Drug Delivery 12, 223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Stokes JM, MacNair CR, Ilyas B, French S, Cote JP, Bouwman C, Farha MA, Sieron AO, Whitfield C, Coombes BK, and Brown ED (2017) Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol 2, 17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Reading C, and Cole M (1977) Clavulanic acid: a beta-lactamase-inhiting beta-lactam from Streptomyces clavuligerus. Antimicrob. Agents Chemother 11, 852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).King AM, Reid-Yu SA, Wang W, King DT, De Pascale G, Strynadka NC, Walsh TR, Coombes BK, and Wright GD (2014) Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature 510, 503–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Boudreau MA, Fishovitz J, Llarrull LI, Xiao Q, and Mobashery S (2015) Phosphorylation of BlaR1 in Manifestation of Antibiotic Resistance in Methicillin-Resistant Staphylococcus aureus and Its Abrogation by Small Molecules. ACS Infect. Dis 1 , 454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Harris TL, Worthington RJ, Hittle LE, Zurawski DV, Ernst RK, and Melander C (2014) Small Molecule Down-regulation of PmrAB Reverses Lipid A Modification and Breaks Colistin Resistance. ACS Chem. Biol 9, 122–127. [DOI] [PubMed] [Google Scholar]
- (13).Lamut A, Peterlin Masic L, Kikelj D, and Tomasic T (2019) Efflux pump inhibitors of clinically relevant multidrug resistant bacteria. Med. Res. Rev 39, 2460–2504. [DOI] [PubMed] [Google Scholar]
- (14).Brogden RN, Carmine A, Heel RC, Morley PA, Speight TM, and Avery GS (1981) Amoxycillin/clavulanic acid: a review of its antibacterial activity, pharmacokinetics and therapeutic use. Drugs 22, 337–62. [DOI] [PubMed] [Google Scholar]
- (15).McLeod DC, Smith BR, and LeFrock JL (1985) Amoxicillin-potassium clavulanate: a novel beta-lactamase inhibitor. Drug Intell Clin Pharm. 19, 415–20. [DOI] [PubMed] [Google Scholar]
- (16).Nothias LF, Knight R, and Dorrestein PC (2016) Antibiotic discovery is a walk in the park. Proc. Natl. Acad. Sci. U. S. A 113, 14477–14479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Watve MG, Tickoo R, Jog MM, and Bhole BD (2001) How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol 176, 386–390. [DOI] [PubMed] [Google Scholar]
- (18).de Lima Procopio RE, da Silva IR, Martins MK, de Azevedo JL, and de Araujo JM (2012) Antibiotics produced by Streptomyces. Braz. J. Infect. Dis 16, 466–471. [DOI] [PubMed] [Google Scholar]
- (19).Newman DJ, Cragg GM, and Snader KM (2003) Natural products as sources of new drugs over the period 1981–2002. J. Nat. Prod 66, 1022–1037. [DOI] [PubMed] [Google Scholar]
- (20).Koehn FE, and Carter GT (2005) The evolving role of natural products in drug discovery. Nat. Rev. Drug Discovery 4, 206–220. [DOI] [PubMed] [Google Scholar]
- (21).Harvey AL (2007) Natural products as a screening resource. Curr. Opin. Chem. Biol 11, 480–484. [DOI] [PubMed] [Google Scholar]
- (22).Thornburg CC, Britt JR, Evans JR, Akee RK, Whitt JA, Trinh SK, Harris MJ, Thompson JR, Ewing TL, Shipley SM, Grothaus PG, Newman DJ, Schneider JP, Grkovic T, and O’Keefe BR (2018) NCI Program for Natural Product Discovery: A Publicly-Accessible Library of Natural Product Fractions for High-Throughput Screening. ACS Chem. Biol 13, 2484–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bharate SB, Yadav RR, Khan SI, Tekwani BL, Jacob MR, Khan IA, and Vishwakarma RA (2013) Meridianin G and its analogs as antimalarial agents. MedChemComm 4, 1042–1048. [Google Scholar]
- (24).Yadav RR, Khan SI, Singh S, Khan IA, Vishwakarma RA, and Bharate SB (2015) Synthesis, antimalarial and antitubercular activities of meridianin derivatives. Eur. J. Med. Chem 98, 160–9. [DOI] [PubMed] [Google Scholar]
- (25).Huggins WM, Barker WT, Baker JT, Hahn NA, Melander RJ, and Melander C (2018) Meridianin D Analogues Display Antibiofilm Activity against MRSA and Increase Colistin Efficacy in Gram-Negative Bacteria. ACS Med. Chem. Lett 9, 702–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lesho E, Yoon E-J, McGann P, Snesrud E, Kwak Y, Milillo M, Onmus-Leone F, Preston L, Clair K, Nikolich M, Viscount H, Wortmann G, Zapor M, Grillot-Courvalin C, Courvalin P, Clifford R, and Waterman PE (2013) Emergence of Colistin-Resistance in Extremely Drug-Resistant Acinetobacter baumannii Containing a Novel pmrCAB Operon During Colistin Therapy of Wound Infections. J. Infect. Dis 208, 1142–1151. [DOI] [PubMed] [Google Scholar]
- (27).Leung LM, Cooper VS, Rasko DA, Guo QL, Pacey MP, McElheny CL, Mettus RT, Yoon SH, Goodlett DR, Ernst RK, and Doi Y (2017) Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother 72, 3035–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).MacNair CR, Stokes JM, Carfrae LA, Fiebig-Comyn AA, Coombes BK, Mulvey MR, and Brown ED (2018) Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun 9, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Mandler MD, Baidin V, Lee J, Pahil KS, Owens TW, and Kahne D (2018) Novobiocin Enhances Polymyxin Activity by Stimulating Lipopolysaccharide Transport. J. Am. Chem. Soc 140, 6749–6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Scott AJ, Oyler BL, Goodlett DR, and Ernst RK (2017) Lipid A structural modifications in extreme conditions and identification of unique modifying enzymes to define the Toll-like receptor 4 structure-activity relationship. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1862, 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, and Miller SI (1998) PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol 27, 1171–82. [DOI] [PubMed] [Google Scholar]
- (32).Simpson BW, and Trent MS (2019) Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol 17, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Klein G, and Raina S (2019) Regulated Assembly of LPS, Its Structural Alterations and Cellular Response to LPS Defects. Int. J. Mol. Sci 20, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, and Hancock RE (2011) The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother 55, 3743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Gunn JS, and Miller SI (1996) PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol 178, 6857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, and Hoiby N (2012) PmrB Mutations Promote Polymyxin Resistance of Pseudomonas aeruginosa Isolated from Colistin-Treated Cystic Fibrosis Patients. Antimicrob. Agents Chemother 56, 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Cheng HY, Chen YF, and Peng HL (2010) Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci 17, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Cannatelli A, D’Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, and Rossolini GM (2013) In Vivo Emergence of Colistin Resistance in Klebsiella pneumoniae Producing KPC-Type Carbapenemases Mediated by Insertional Inactivation of the PhoQ/PhoP mgrB Regulator. Antimicrob. Agents Chemother 57, 5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Jayol A, Nordmann P, Brink A, and Poirel L (2015) Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother 59, 2780–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).May JM, Owens TW, Mandler MD, Simpson BW, Lazarus MB, Sherman DJ, Davis RM, Okuda S, Massefski W, Ruiz N, and Kahne D (2017) The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport. J. Am. Chem. Soc 139, 17221–17224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Barker WT, Martin SE, Chandler CE, Nguyen TV, Harris TL, Goodell C, Melander RJ, Doi Y, Ernst RK, and Melander C (2017) Small molecule adjuvants that suppress both chromosomal and mcr-1 encoded colistin-resistance and amplify colistin efficacy in polymyxin-susceptible bacteria. Bioorg. Med. Chem 25, 5749–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Sampson TR, Liu X, Schroeder MR, Kraft CS, Burd EM, and Weiss DS (2012) Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother 56, 5642–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Danevcic T, Vezjak MB, Zorec M, and Stopar D (2016) Prodigiosin - A Multifaceted Escherichia coli Antimicrobial Agent. PLoS One 11, e0162412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Darshan N, and Manonmani HK (2016) Prodigiosin inhibits motility and activates bacterial cell death revealing molecular biomarkers of programmed cell death. AMB Express 6, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Perez-Tomas R, Montaner B, Llagostera E, and Soto-Cerrato V (2003) The prodigiosins, proapoptotic drugs with anticancer properties. Biochem. Pharmacol 66, 1447–52. [DOI] [PubMed] [Google Scholar]
- (46).Stankovic N, Senerovic L, Ilic-Tomic T, Vasiljevic B, and Nikodinovic-Runic J (2014) Properties and applications of undecylprodigiosin and other bacterial prodigiosins. Appl. Microbiol. Biotechnol 98, 3841–58. [DOI] [PubMed] [Google Scholar]
- (47).Williamson NR, Fineran PC, Gristwood T, Chawrai SR, Leeper FJ, and Salmond GP (2007) Anticancer and immunosuppressive properties of bacterial prodiginines. Future Microbiol. 2, 605–18. [DOI] [PubMed] [Google Scholar]
- (48).Ravindran A, Anishetty S, and Pennathur G (2020) Molecular dynamics of the membrane interaction and localisation of prodigiosin. J. Mol. Graphics Modell 98, 107614. [DOI] [PubMed] [Google Scholar]
- (49).Hage-Hulsmann J, Grunberger A, Thies S, Santiago-Schubel B, Klein AS, Pietruszka J, Binder D, Hilgers F, Domrose A, Drepper T, Kohlheyer D, Jaeger KE, and Loeschcke A (2018) Natural biocide cocktails: Combinatorial antibiotic effects of prodigiosin and biosurfactants. PLoS One 13, No. e0200940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lins JC, De Melo ME, Do Nascimento SC, and Adam ML (2015) Differential genomic damage in different tumor lines induced by prodigiosin. Anticancer research 35, 3325–3332. [PubMed] [Google Scholar]
- (51).Minrovic BM, Jung D, Melander RJ, and Melander C (2018) New Class of Adjuvants Enables Lower Dosing of Colistin Against Acinetobacter baumannii. ACS Infect. Dis 4, 1368–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Hubble VB, Hubbard BA, Minrovic BM, Melander RJ, and Melander C (2019) Using Small-Molecule Adjuvants to Repurpose Azithromycin for Use against Pseudomonas aeruginosa. ACS Infect. Dis 5, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


