Abstract
The histone H3 family in animals and plants includes replicative H3 and nonreplicative H3.3 variants. H3.3 preferentially associates with active transcription, yet its function in development and transcription regulation remains elusive. The floral transition in Arabidopsis (Arabidopsis thaliana) involves complex chromatin regulation at a central flowering repressor FLOWERING LOCUS C (FLC). Here, we show that H3.3 upregulates FLC expression and promotes active histone modifications histone H3 lysine 4 trimethylation (H3K4me3) and histone H3 lysine 36 trimethylation (H3K36me3) at the FLC locus. The FLC activator FRIGIDA (FRI) directly mediates H3.3 enrichment at FLC, leading to chromatin conformation changes and further induction of active histone modifications at FLC. Moreover, the antagonistic H3.3 and H2A.Z act in concert to activate FLC expression, likely by forming unstable nucleosomes ideal for transcription processing. We also show that H3.3 knockdown leads to H3K4me3 reduction at a subset of particularly short genes, suggesting the general role of H3.3 in promoting H3K4me3. The finding that H3.3 stably accumulates at FLC in the absence of H3K36me3 indicates that the H3.3 deposition may serve as a prerequisite for active histone modifications. Our results reveal the important function of H3.3 in mediating the active chromatin state for flowering repression.
Establishment of the permissive chromatin state at floral repressors requires histone variant H3.3.
Introduction
Histone variants are related but functional distinct proteins in the same histone family (Talbert and Henikoff, 2017). The incorporation of histone variants could generate profound impacts on nucleosome property and chromatin function (Jiang and Berger, 2017; Talbert and Henikoff, 2017; Borg et al., 2021). The histone H3 family consists of three major variants: canonical H3.1/H3.2, H3.3, and CenH3/CENP-A (Hake and Allis, 2006; Jiang and Berger, 2017). Though H3.1 and H3.3 are distinguished by only a few amino acids, they acquired distinct expression patterns and deposition modes. The S phase-specific H3.1 is deposited during DNA replication via histone chaperone complex CHROMATIN ASSEMBLY FACTOR-1 (CAF1; Smith and Stillman, 1989), while H3.3 is expressed throughout the cell cycle and replication-independently incorporated into the chromatin by HISTONE REGULATORY HOMOLOG A (HIRA) complex, ALPHA THALASSEMIA MENTAL RETARDATION SYNDROME X-LINKED (ATRX)-DAXX and DEK-domain containing protein (DEK; Tagami et al., 2004; Goldberg et al., 2010; Sawatsubashi et al., 2010; Talbert and Henikoff, 2017). Another histone chaperone ANTI-SILENCING FUNCTION 1 (ASF1) cooperates with both CAF1 and HIRA in the H3.1 and H3.3 deposition (Tagami et al., 2004). Although H3.1 plays an essential role in the chromatin assembly of doubled genome during DNA replication, the function of H3.3 is complex and remains undetermined. In both animals and plants, the lack of H3.3 causes defects in development (Hodl and Basler, 2009; Sakai et al., 2009; Szenker et al., 2012; Jang et al., 2015; Wollmann et al., 2017), demonstrating its important role in multicellular eukaryotes.
On the genome, H3.3 is associated with actively transcribed genes and gene regulatory elements (Wirbelauer et al., 2005; Goldberg et al., 2010; Szenker et al., 2011; Stroud et al., 2012; Wollmann et al., 2012). However, H3.3 is nonessential for most of the transcriptional events in Drosophila (Drosophila melanogaster; Hodl and Basler, 2009) and it is largely interchangeable with the replicative H3 (Sakai et al., 2009). In Arabidopsis (Arabidopsis thaliana), only a small number of H3.3-enriched genes show transcriptional defects when H3.3 levels are reduced, indicating that H3.3 may not be directly required for transcription (Wollmann et al., 2017). Other studies using different organisms and materials have shown that H3.3 modulates both active and repressive histone modifications (Banaszynski et al., 2013; Wollmann et al., 2017; Martire et al., 2019; Armache et al., 2020), adding to the complexity of the H3.3 function. Therefore, the role of H3.3 on transcription and chromatin modifications remains to be clarified. Besides its potential regulatory roles in histone modifications, H3.3 actively interplays with another histone variant H2A.Z. H2A.Z is associated with both gene activation and repression, likely depending on its genic localization and the chromatin context (Raisner et al., 2005; Jarillo and Pineiro, 2015; Chang et al., 2020). The coexistence of H3.3 and H2A.Z destabilizes nucleosomes (Fan et al., 2002; Jin and Felsenfeld, 2007), and H3.3 globally prevents the accumulation of H2A.Z at the 3′ gene end in Arabidopsis (Wollmann et al., 2017). However, the interaction between H3.3 and H2A.Z in the context of transcription regulation remains to be investigated.
Arabidopsis floral transition involves complex regulations at the chromatin level, and many of them act on a key floral repressor FLOWERING LOCUS C (FLC; He, 2012; Bao et al., 2020). In addition to FLC, the Arabidopsis genome encodes five FLC homologs, FLOWERING LOCUS M (FLM)/MADS AFFECTING FLOWERING 1 (MAF1)–MAF5, which act together with FLC to repress the floral transition (Ratcliffe et al., 2003; Scortecci et al., 2003; Kim and Sung, 2010; Gu et al., 2013). FLC activation requires permissive histone modifications, such as H3K4me3 and H3K36me3 (Xu et al., 2008, 2020; Jiang et al., 2009). H3K4me3 at FLC and its homologs MAF4 and MAF5 is catalyzed by the evolutionarily conserved COMPASS-like complex, which includes WDR5a, ASH2R, RBL, and a histone methyltransferase (Miller et al., 2001; Jiang et al., 2009, 2011, 2018; Ding et al., 2012). EARLY FLOWERING IN SHORT DAYS (EFS)/SET DOMAIN GROUP 8 (SDG8) is a major methyltransferase catalyzing H3K36 di- and trimethylation in Arabidopsis. In efs mutant, H3K36me2 and H3K36me3 are reduced at genome-wide level including the FLC, MAF4, and MAF5 loci (Xu et al., 2008). The chromatin remodeling complex SWR1 (SWR1-C), which mediates H2A.Z deposition, also activates the expression of FLC, MAF4, and MAF5 (Noh and Amasino, 2003; Deal et al., 2007). The disruption of SWR1-C leads to reduced FLC transcripts (Deal et al., 2007), suggesting a positive role of H2A.Z in FLC expression.
The expression of FLC is highly elevated in late-flowering Arabidopsis winter annual accessions by a FRIGIDA (FRI) containing protein complex (FRI-C). FRI-C interacts with several chromatin modifiers including COMPASS-like and EFS, and enhances their binding at the FLC locus (Jiang et al., 2009; Ko et al., 2010; Choi et al., 2011; Li et al., 2018; Luo and He, 2020). Hence, active histone modifications at FLC are elevated by FRI-C, resulting in strong FLC induction and flowering inhibition. In addition to promoting active histone modifications, FRI-C stimulates the FLC 5′ to 3′ gene looping (Li et al., 2018). Active FLC transcription per se does not regulate gene loop formation (Li et al., 2018), indicating that other mechanisms are responsible for the FLC 5′ to 3′ gene looping.
The presence of multiple chromatin modifications and their vigorous interactions at FLC makes it a paradigm for the understanding of chromatin-based plant gene regulation (He, 2012). Here, we show that H3.3 stimulates the expression of FLC and its homologs and promotes the active histone modifications at their loci. FRI directly interacts with the H3.3 chaperone HIRA and elevates H3.3 deposition toward the FLC 3′ region. The enriched H3.3 facilitates FLC 3′ end interaction with the 5′ end, promoting active histone modifications around the transcription start site (TSS) of FLC. Moreover, though H3.3 antagonizes H2A.Z, both of them are essential for FLC activation, and the loss of either one compromises FLC expression. In addition to FLC and its homologs, H3.3 is required for H3K4me3 at a subset of especially short genes. Our findings reveal the important function of H3.3 in the regulation of the active chromatin state, which enhances FLC transcription and floral repression.
Results
H3.3 represses the floral transition in Arabidopsis
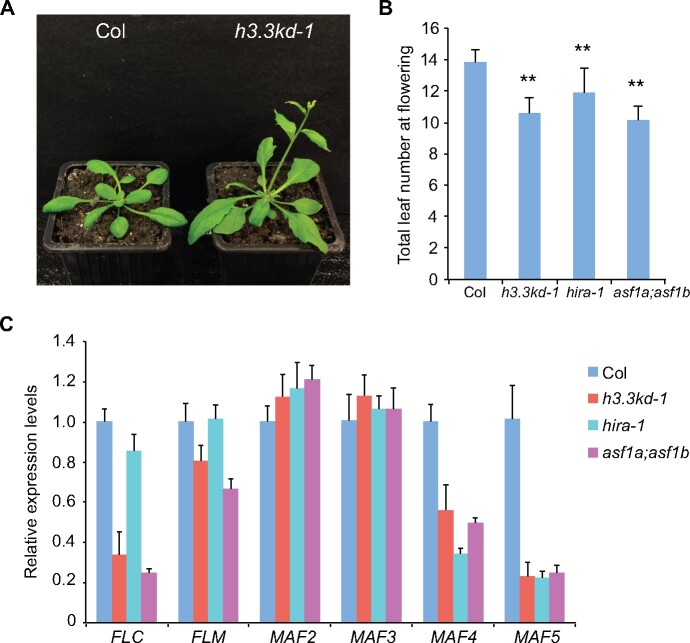
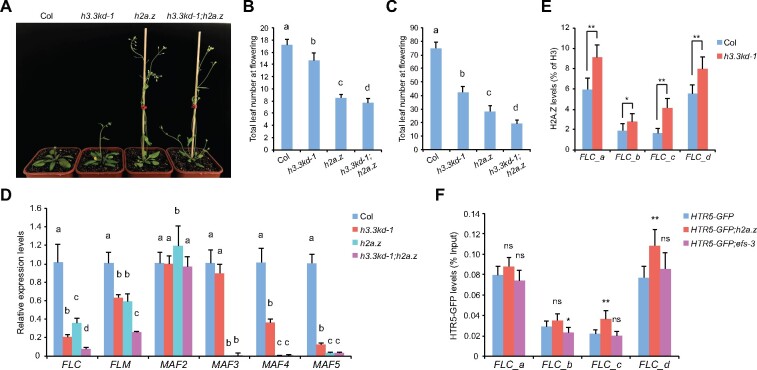
A previous study has shown that H3.3 knockdown (h3.3kd) led to defects in leaf development and fertility (Wollmann et al., 2017). To further elucidate the function of H3.3 in plant development, we examined the flowering behavior of h3.3kd. h3.3kd lines h3.3kd-1 and h3.3kd-3 flowered earlier than the wild-type (WT) Columbia (Col; Figure 1, A and B;Supplemental Figure S1A). In Arabidopsis, the floral transition is repressed by FLC and its homologs. An examination of their expression by reverse transcription quantitative polymerase chain reaction (RT-qPCR) revealed that transcripts levels of FLC, MAF4, MAF5 and to a lesser extent FLM were reduced in h3.3kd lines (Figure 1C;Supplemental Figure S1B; Wollmann et al., 2017), consistent with their early flowering phenotypes. H3.3 is deposited by the conserved HIRA complex (Nie et al., 2014; Duc et al., 2015). Moreover, two functional redundant ASF1 homologs ASF1a and ASF1b were identified in Arabidopsis (Zhu et al., 2011). Similar to h3.3kd, hira and asf1a;asf1b mutants showed early flowering phenotype and reduced expression of FLC, MAF4, and MAF5 (Figure 1, B and C). The mild flowering phenotype and FLC reduction in hira mutant is likely due to the functional redundancy from other H3.3 deposition pathways mediated by ATRX or DEK (Sawatsubashi et al., 2010; Duc et al., 2017; Wang et al., 2018). As both h3.3kd-1 and h3.3kd-3 lines had similar flowering phenotypes and gene expression changes, we used h3.3kd-1 for further analyses.
Figure 1.
H3.3 represses flowering and activates the expression of FLC and its homologs. A, The flowering phenotype of h3.3kd-1 grown in long days. B, The flowering time of indicated lines grown in long days. The total number of primary rosette and cauline leaves at flowering were counted; 15–20 plants were scored for each line. Values are means ± sd. Statistical significance relative to Col was determined by two-tailed Student’s t test (**P<0.01). C, Relative transcripts of FLC and its homologs determined by RT-qPCR. TUB2 was used as an endogenous control. Values are means ± sd of three biological repeats.
H3.3 promotes active histone modifications at FLC and its homologs
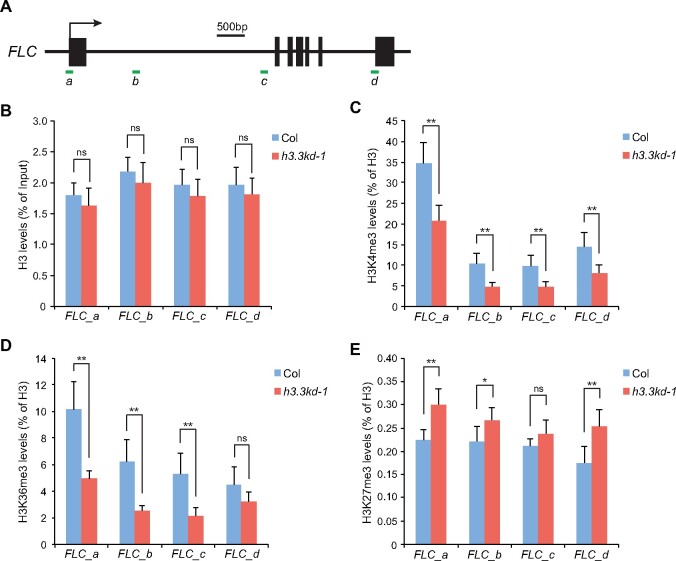
Previous RNA-seq results in h3.3kd have shown that only a small portion of H3.3-enriched genes were misregulated, suggesting that the loss of H3.3 per se might not be enough to alter transcription activity (Wollmann et al., 2017). The lack of H3.3 could reduce nucleosome density at the chromatin, which may affect transcription activity. We examined nucleosome enrichment at FLC, MAF4, and MAF5 by profiling total H3 levels using chromatin immunoprecipitation (ChIP). The H3 levels were largely maintained with minimal reduction in h3.3kd (Figure 2, A and B;Supplemental Figure S2, A and B). It is possible that in h3.3kd, H3.1 compensates for the nucleosome deficit at FLC, MAF4, and MAF5 loci.
Figure 2.
H3.3 promotes active histone modifications at FLC. A, Schematic structure of FLC. Filled boxes represent exons; an arrow indicates the transcription start site. ChIP examined regions are indicated by green lines. B–E, H3 (B), H3K4me3 (C), H3K36me3 (D), and H3K27me3 (E) levels at FLC determined by ChIP. The amounts of immunoprecipitated DNA fragments were quantified by qPCR and subsequently normalized to input DNA (B) or H3 antibody-precipitated DNA (C–E). Values are means ± sd of three biological repeats. Statistical significance was determined by two-tailed Student’s t test (**P<0.01; *P<0.05; ns, not significant, P>0.05).
We searched for other chromatin state changes that could contribute to gene expression defects induced by H3.3 knockdown. FLC activation requires active histone modifications including H3K4me3 and H3K36me3, both are highly enriched around the TSS of FLC (Yang et al., 2014; Li et al., 2016). We thus analyzed H3K4me3 and H3K36me3 levels at the FLC locus in h3.3kd. Considering the slight difference in H3 enrichment between WT and h3.3kd, histone modification levels were normalized to the H3 levels. H3K4me3 and H3K36me3 were reduced at FLC in h3.3kd (Figure 2, C and D), showing that H3.3 is required for their deposition at the FLC chromatin. Similarly H3K4me3 and H3K36me3 levels were significantly decreased at the TSS of MAF4 and MAF5 loci (Supplemental Figure S2, C and D). The FLC locus is also enriched with the repressive histone H3 lysine 27 trimethylation (H3K27me3; Jiang et al., 2008; Zhou et al., 2018). We observed a moderate increase of H3K27me3 in h3.3kd compared to WT, especially at the FLC 5′ and 3′ gene ends (Figure 2E). This could be due to the antagonism between H3K27me3 and active histone modifications at FLC (Jiang et al., 2008; Yang et al., 2014), or the loss of H3.3-induced H3.1 accumulation, which closely associates with and facilitates methylation at H3K27 (Stroud et al., 2012; Jacob et al., 2014; Jiang and Berger, 2017). Together, these results suggest that H3.3 promotes active histone modifications at FLC, MAF4, and MAF5.
FRI-mediated FLC activation requires H3.3
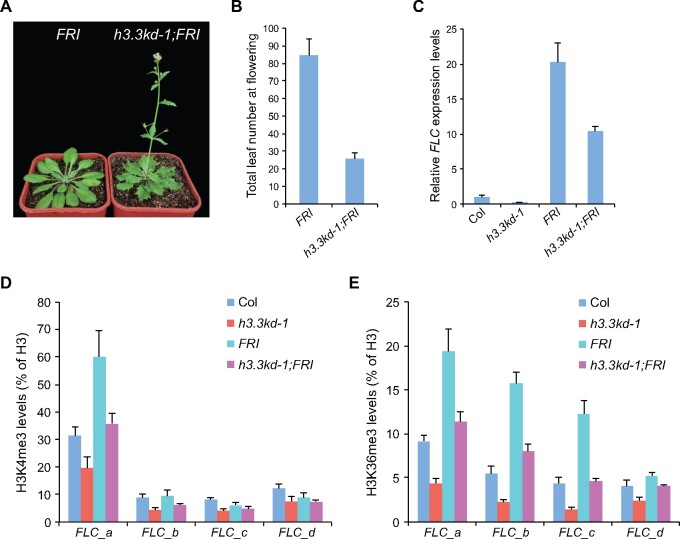
In winter-annual Arabidopsis accessions, FRI elevates FLC expression to levels that inhibit flowering before winter (Johanson et al., 2000; Michaels et al., 2003). To evaluate the role of H3.3 in FRI-mediated FLC upregulation, we introduced a functional FRI allele into h3.3kd (Lee et al., 1994), and observed that h3.3kd strongly suppressed the late-flowering phenotype of FRI (Figure 3, A and B). The suppression of the late-flowering phenotype by h3.3kd is likely due to the reduction of FLC expression, hence, we examined its expression in h3.3kd-1;FRI. Indeed, elevated FLC expression levels in FRI were reduced upon H3.3 knockdown (Figure 3C). Autonomous pathway genes such as FVE and FLOWERING LOCUS D (FLD) repress FLC expression, the loss of FVE or FLD causes delayed flowering due to FLC upregulation (He et al., 2003; Ausin et al., 2004). We further crossed h3.3kd with fve-4 and fld-3 mutants; similarly the late flowering phenotypes and upregulated FLC transcripts of fve-4 and fld-3 were suppressed by h3.3kd (Supplemental Figure S3). Therefore, H3.3 is required for FLC activation induced by FRI and autonomous pathway mutations.
Figure 3.
FRI-induced FLC upregulation and active histone modifications require H3.3. A, The flowering phenotype of h3.3kd-1;FRI grown in long days. B, The flowering time of indicated lines grown in long days. The total number of primary rosette and cauline leaves at flowering were counted; 10 and 16 plants were scored for FRI and h3.3kd-1;FRI, respectively. Values are means ± sd. C, Relative transcripts of FLC determined by RT-qPCR. TUB2 was used as an endogenous control. Values are means ± sd of three biological repeats. D, E, H3K4me3 (D) and H3K36me3 (E) levels at FLC determined by ChIP. The amounts of immunoprecipitated DNA fragments were quantified by qPCR, and subsequently normalized to H3 antibody-precipitated DNA. Values are means ± sd of two biological repeats.
FRI acts in FRI-C, which recruits and stabilizes chromatin modifiers at the FLC locus, and thus promotes active histone modifications including H3K4me3 and H3K36me3 at FLC (Choi et al., 2011; Li et al., 2018). We found that FRI-induced H3K4me3 and H3K36me3 at FLC were strongly suppressed by h3.3kd (Figures 2A; 3, D and E). These results demonstrate that H3.3 is essential for FRI-mediated establishment of active histone modifications at FLC.
Enhanced H3.3 deposition mediated by FRI promotes FLC gene loop formation
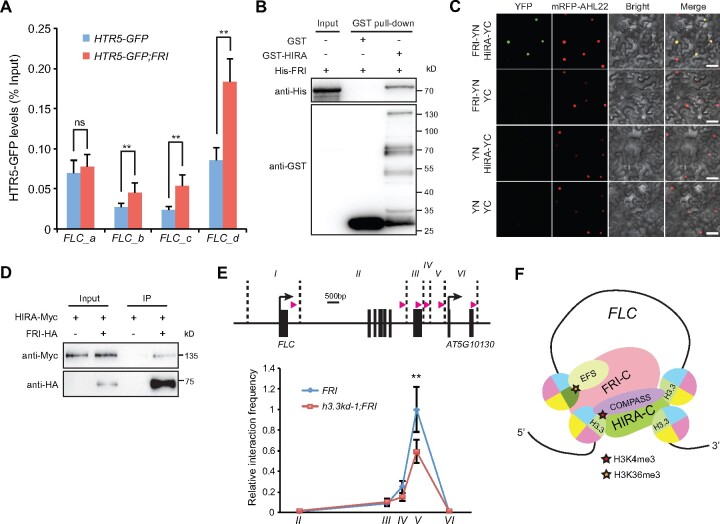
To investigate whether FRI regulates H3.3 deposition at the FLC locus, we combined an HTR5-GFP (HTR5 is one of the three H3.3 coding genes in Arabidopsis) reporter line with the functional FRI and performed ChIP experiment. Due to the lack of specific antibody against plant H3.3, the levels of HTR5-GFP at chromatin were used as an indication for H3.3 deposition levels (Wollmann et al., 2012). The enrichment of HTR5-GFP was enhanced by FRI especially toward the FLC 3′ region (Figures 2A, 4A), while the overall protein abundance of HTR5-GFP was not affected by FRI (Supplemental Figure S4A). Thus, FRI induces H3.3 deposition at the FLC chromatin.
Figure 4.
FRI-enriched H3.3 deposition promotes FLC 5′ to 3′ looping. A, HTR5-GFP enrichment levels at FLC in indicated lines. The amounts of immunoprecipitated DNA fragments were quantified by qPCR and normalized to input DNA. Values are means ± sd of three biological repeats. Statistical significance was determined by two-tailed Student’s t test (**P<0.01; ns, P>0.05). B, Pull-down assay of glutathione S-transferase (GST)-HIRA with His-FRI. C, Physical interaction of FRI-NYFP (YN) with HIRA-CYFP (YC) in N. benthamiana leaf cells analyzed by BiFC. mRFP-fused AHL22 (AT-hook motif nuclear-localized protein 22) signals were used to label nuclei (Meng et al., 2019). Scale bars: 50 μm. D, Co-IP assay of HIRA with FRI. Total proteins were extracted from Arabidopsis protoplasts coexpressing HIRA-Myc and FRI-HA. E, Quantitative 3C of the FLC 5′ to 3′ loop formation using region I as the anchor region in indicated lines. In the up graph, BamHI and BglII cutting sites are indicated by the vertical dashed lines, pink arrow indicates the position and direction of the primer used in each region. The interaction frequency was normalized to that of region I to region V in FRI, which was set as 1.0. Values are means ± sd of three biological repeats. Statistical significance was determined by two-tailed Student’s t test (**P<0.01). F, A possible function for H3.3 in FRI-induced active histone modifications and gene looping at FLC. FRI-C enhances the H3.3 deposition toward the FLC 3′ end. The enriched H3.3 may stabilize the binding of histone modifiers such as COMPASS-like at the spatially nearby 5′ and 3′ ends, and/or stimulate the activity of histone modifiers, resulting in increased gene loop formation and induced active histone modifications predominantly at the 5′ end.
To explore how FRI enriches H3.3 at FLC, we tested the interactions between FRI-C and the H3.3 chaperones. Direct interaction of FRI with HIRA was detected by pull-down assay (Figure 4B). Bimolecular fluorescence complementation (BiFC) experiment further confirmed that FRI directly associates with HIRA (Figure 4C). Moreover, co-immunoprecipitation (Co-IP) assay was performed with FRI-HA and HIRA-Myc coexpressed Arabidopsis protoplasts. HIRA-Myc was copurified with FRI-HA but not with the HA antibody-coupled beads (Figure 4D). Together, these results indicate that FRI-C directly enriches H3.3 deposition at FLC by interacting with its chaperone HIRA complex.
In plants, H3K4me3 and H3K36me3 are accumulated at the TSS of transcribed genes (Sequeira-Mendes et al., 2014; Liu et al., 2019). A similar pattern was observed at the FLC locus (Figure 2, A, C, and D; Yang et al., 2014; Li et al., 2016). However, H3.3 is generally enriched around the 3′ gene end (Stroud et al., 2012; Wollmann et al., 2012). Moreover, though FRI enhances H3K4me3 and H3K36me3 levels mainly around the FLC 5′ region, the elevated deposition of H3.3 mediated by FRI is more predominant toward the gene body and 3′ end (Figures 2A; 3, D and E; 4A). At the FLC locus, a peak of H3.3 was also detected at TSS (Figures 2A, 4A). The H3.3 enrichment at both gene ends is a signature chromatin state for moderately expressed genes with a gene loop structure (Liu et al., 2016). The 5′ and 3′ of FLC form a gene loop and this structure is enhanced by FRI (Crevillen et al., 2013; Li et al., 2018). We thus performed a chromosome conformation capture (3C) experiment to examine the impact of H3.3 on 5′ to 3′ looping. Interestingly, the loss of H3.3 impaired loop formation at FLC (Figure 4E). Thus, H3.3 facilitates FLC 5′ to 3′ loop formation (Figure 4F).
H3.3 and H2A.Z are both required for FLC activation but oppose each other at the FLC chromatin
Activation of FLC and its homologs also depends on the chromatin remodeling complex SWR1-C that deposits H2A.Z. Disruption of SWR1-C led to downregulated FLC, MAF4, and MAF5 expression and accelerated flowering (Noh and Amasino, 2003; Deal et al., 2007), presumably due to the lack of H2A.Z at their chromatin. To directly address the impact of H2A.Z on flowering gene expression, we used a near-null h2a.z mutant defective in all three H2A.Z coding genes, which similarly showed early flowering phenotype (Figure 5, A–C; Coleman-Derr and Zilberman, 2012). In addition to FLC, MAF4, and MAF5, the loss of H2A.Z led to the reduced expression of FLM and MAF3 (Figure 5D). Thus, our results confirmed that H2A.Z activates FLC and its homologs.
Figure 5.
H3.3 and H2A.Z oppose each other at the FLC chromatin but coactivate FLC expression. A, The flowering phenotype of indicated lines grown in long days. B and C, The flowering time of indicated lines grown in long days (B) and short days (C). The total number of primary rosette and cauline leaves at flowering were counted. For (B), 15–18 plants were scored for each line. For (C) 9 plants were scored for each line. Values are means ± sd. The significance of differences was tested using one-way ANOVA with Tukey’s test (P<0.05), different letters indicate statistical significance. D, Relative transcripts of FLC and its homologs determined by RT-qPCR. TUB2 was used as an endogenous control. Values are means ± sd of three biological repeats. The significance of differences at each gene was tested using one-way ANOVA with Tukey’s test (P<0.05), different letters indicate statistical significance. E, H2A.Z levels at FLC determined by ChIP. The amounts of immunoprecipitated DNA fragments were quantified by qPCR, and subsequently normalized to H3 antibody-precipitated DNA. Values are means ± sd of three biological repeats. Statistical significance was determined by two-tailed Student’s t test (*P<0.05; **P<0.01). F, HTR5-GFP enrichment levels at FLC in indicated lines. The amounts of immunoprecipitated DNA fragments were quantified by qPCR and normalized to input DNA. Values are means ± sd of three biological repeats. Statistical significance relative to HTR5-GFP was determined by two-tailed Student’s t test (*P<0.05; **P<0.01; ns, P>0.05).
We sought to investigate how H3.3 and H2A.Z, both of them required for the activation of FLC, interact at its loci. H2A.Z is enriched at both the 5′ and 3′ of FLC (Figures 2A, 5E), consistent with a similar pattern reported previously (Deal et al., 2007). Interestingly, H3.3 knockdown enhanced H2A.Z occupancy at FLC (Figure 5E), despite reduced FLC expression in h3.3kd. To examine H3.3 accumulation in the absence of H2A.Z, HTR5-GFP enrichment was quantified by ChIP-qPCR after it was introduced into h2a.z mutant. A slight induction of HTR5-GFP levels was detected toward the FLC 3′ end (Figures 2A, 5F). This is not due to the increased expression of HTR5-GFP, as its protein levels were rather decreased in h2a.z for unknown reasons (Supplemental Figure S4B). We also confirmed that FLC expression was reduced in HTR5-GFP;h2a.z (Supplemental Figure S4C). Thus, although both are activators of FLC, H3.3 and H2A.Z antagonize each other at the FLC locus.
We next tested the genetic relationship of H3.3 and H2A.Z in flowering by crossing h3.3kd with h2a.z. H3.3 knockdown in h2a.z further enhanced the reduction of especially FLC and FLM transcripts (Figure 5D), and concomitantly accelerated flowering in both long-day and short-day conditions (Figure 5, B and C). Together, these results suggest close cooperation of H3.3 and H2A.Z on FLC activation and the presence of both H3.3 and H2A.Z is required to maintain FLC at a transcription active state in Col, while lacking of either one (even when the enrichment of the other one is increased) leads to a reduction in FLC expression.
H3K36me3 is not required for H3.3 accumulation at FLC
The above results suggest that H3.3 regulates chromatin modifications at FLC. To investigate the impact of chromatin modifications on H3.3 deposition, we examined H3.3 enrichment with mutants defective in chromatin modifications. EFS deposits H3K36me3 at FLC to stimulate its expression. We measured HTR5-GFP accumulation at FLC in efs mutant, in which FLC expression is reduced (Supplemental Figure S4C). The HTR5-GFP protein expression levels and its enrichment at FLC were comparable to that in WT (Figure 5F;Supplemental Figure S4B), suggesting that H3K36me3 is not required for H3.3 deposition.
H3.3 knockdown affects H3K4me3 at a subset of loci
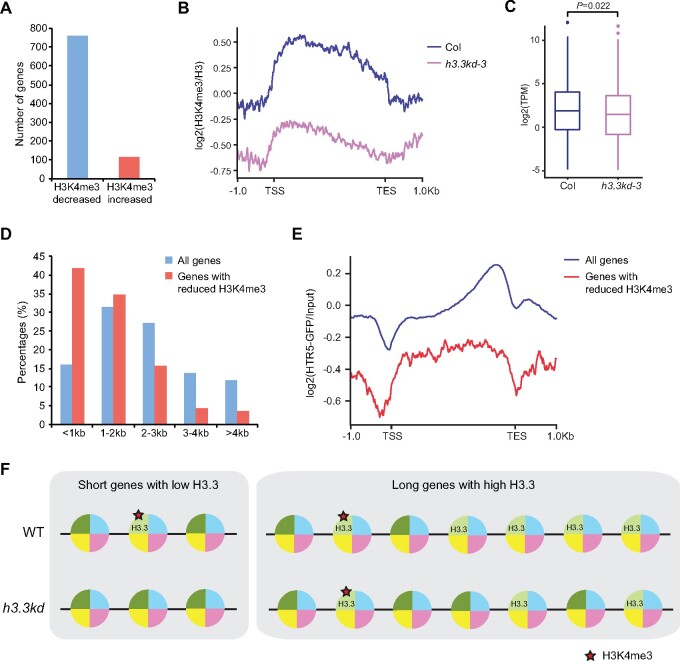
A previous study has shown that H3.3 knockdown had little impact on H3K4me3 globally when all the genes were evaluated, while H3K36me3 enrichment was affected especially at misexpressed genes upon the loss of H3.3 (Wollmann et al., 2017). To further investigate the impact of H3.3 at H3K4me3 in detail, we analyzed the published ChIP-seq data generated using h3.3kd-3 seedlings to identify genes with altered H3K4me3 levels. ChIP-seq detected less H3K4me3 at FLC, MAF4, and MAF5 in h3.3kd-3, confirming that H3.3 is required for H3K4me3 at these loci (Supplemental Figure S5A). The lack of H3.3 led to strong H3K4me3 reductions at 761 loci (>1.5-fold change), while 116 loci gained H3K4me3 in h3.3kd (Figure 6, A and B;Supplemental Table S1). In line with the role of H3K4me3 in gene activation, genes with reduced H3K4me3 showed lower expression levels in h3.3kd (Figure 6C). Notably, the H3K4me3 is broadly distributed at these genes, albeit it is generally enriched around the TSS (Figure 6B). Such a distribution pattern resembles H3K4me3 enrichment at genes with short length (Li et al., 2016). We thus examined the length of genes that displayed H3K4me3 reduction in h3.3kd and found that they were indeed enriched with short genes (Figure 6D). H3.3 distribution was further analyzed by profiling chromatin occupancy of HTR5-GFP in seedlings using ChIP-seq. Like H3K4me3, H3.3 tended to occupy the whole body of short genes (Supplemental Figure S5B). Moreover, genes with reduced H3K4me3 in h3.3kd had less H3.3 accumulation compared with the average level (Figure 6E), suggesting that the H3K4me3 is more vulnerable at genes where H3.3 could be nearly depleted when a limited amount of it is available in h3.3kd (Figure 6F).
Figure 6.
H3K4me3 is reduced at a subset of short genes in h3.3kd. A, The number of H3K4me3 decreased and increased genes in h3.3kd. B, Normalized ChIP-seq profiles of H3K4me3 enrichment in Col and h3.3kd-3 over genes with reduced H3K4me3 in h3.3kd-3. TSS: transcription start site, TES: transcription end site. C, RNA-seq determined expression profiles of genes with reduced H3K4me3 in h3.3kd-3. The P-value is based on the Mann–Whitney U test. The RNA-seq data were generated in a previous study (Wollmann et al., 2017). D, Gene length distribution of all genes and genes with reduced H3K4me3 in h3.3kd-3. E, Normalized ChIP-seq profiles of HTR5-GFP enrichment in Col over all genes and genes with reduced H3K4me3 in h3.3kd-3. F, A possible explanation for the predominant loss of H3K4me3 at short genes with low levels of H3.3 in h3.3kd. At their loci, limited supply of H3.3 in h3.3kd may lead to a near-complete depletion of H3.3, which likely serves as a substrate for H3K4me3 or directly regulates H3K4me3 at nearby nucleosomes, resulting in strong H3K4me3 reduction. At long genes with high levels of H3.3, a considerable amount of H3.3 could be kept at their loci in h3.3kd, which is enough to maintain H3K4me3.
Discussion
On the Arabidopsis genome, H3.3 preferentially associates with active genes, but the function of H3.3 on transcription and chromatin regulation is still not clear. The knockdown of H3.3 causes leaf serration, reduced fertility, and misexpression of particularly responsive genes (Wollmann et al., 2017). The early flowering phenotype and reduced expression of FLC and its homologs in h3.3kd indicate that H3.3 also regulates the floral transition (Figure 1;Supplemental Figure S1). The loss of H3.3 may directly affect transcription as H3.3-containing nucleosomes are less stable (Jin and Felsenfeld, 2007). However, the majority of the H3.3-enriched genes expressed normally upon H3.3 knockdown, suggesting that the loss of H3.3 itself may not directly or be significant enough to affect transcription (Wollmann et al., 2017). Our results show that H3.3 promotes the deposition of active histone modifications at FLC, MAF4, and MAF5 (Figure 2;Supplemental Figure S2). Moreover, H3K4me3 reduction induced by H3.3 knockdown is coupled with reduced gene transcription (Figure 6C). It is possible that decreased active histone modifications, together with the lack of H3.3 deposition, contribute to the reduced expression of FLC and its homologs in h3.3kd.
In the absence of FRI, H3.3 is enriched at both FLC 5′ and 3′ ends (Figure 4A). In winter annuals, FRI directly associates with HIRA and stimulates H3.3 deposition at FLC gene body and 3′ end (Figure 4, A–D), and elevated H3.3 facilitates FRI-induced 5′ end enrichment of H3K4me3 and H3K36me3 at FLC (Figure 3, D and E). We show that H3.3 promotes the FLC 5′ to 3′ looping (Figure 4E), which may explain how 3′-enriched H3.3 enhances active histone modifications at the 5′ end. The COMPASS-like complex binds both the FLC 5′ and 3′ ends and is required for FRI-induced loop formation. In addition, H3K4me3 at the FLC 3′ end is moderately induced by FRI (Li et al., 2018). H3.3 accumulation at the 3′ end may enhance the activity of histone modifiers and/or stabilize their binding at the spatially nearby 5′ and 3′ ends, resulting in increased active histone modifications and gene loop formation (Figure 4F). It has been reported that genes with a loop structure tend to carry H3.3 at both gene ends (Liu et al., 2016), a pattern similar to that at FLC. Therefore, it is of great interest to further investigate the general function of H3.3 at genes with a loop structure.
We found that H3.3 knockdown has minimal effects on H3K4me3 levels at long genes, where H3K4me3 is enriched at the 5′ end; while H3.3 is highly accumulated at the 3′ end (Supplemental Figure S5B; Li et al., 2016). In contrast, H3K4me3 is strongly reduced in h3.3kd at particularly short genes, at which H3.3 and H3K4me3 share similar distribution patterns and localize across the whole genic region (Figure 6B;Supplemental Figure S5B; Li et al., 2016), suggesting that H3.3 may serve as a substrate for H3K4me3 or directly regulate H3K4me3 at nearby nucleosomes. This is in line with the reduction of H3K4me3 in h3.3kd at the 5′ end of FLC, a long gene (∼6 kb) but accumulates H3.3 around both TSS and TES. We also noted that genes with strong H3K4me3 reductions in h3.3kd carry less H3.3 than average (Figure 6E). h3.3kd plants show mild developmental defects, whereas h3.3 knockout lines are not viable, indicating that a substantial amount of H3.3 is still available in h3.3kd (Wollmann et al., 2017). It is possible that at genes with little H3.3, the lack of H3.3 supply caused by H3.3 knockdown could lower its levels below a critical threshold essential for the maintenance of H3K4me3, yet genes with high H3.3 levels may still carry enough H3.3 for H3K4me3 deposition (Figure 6F). Further studies using materials with stronger H3.3 impairment would help to provide a more comprehensive view of the function of H3.3 in histone modifications and chromatin regulation in plants.
Our study shows that although H3K36me3 requires H3.3, the deposition of H3.3 at FLC is independent of H3K36me3 (Figure 5F). Hence, H3.3 incorporation is likely the prerequisite of H3K36me3. Recent studies in animals have shown that the phosphorylation at Serine31, a specific amino acid on H3.3, directly impacts H3K36me3 and H3K27ac in-cis (Armache et al., 2020; Sitbon et al., 2020). Plant H3.3 proteins contain a specific threonine at residue 31, which might undergo phosphorylation. It is of note that the alanine at position 31 on H3.1 is specifically recognized by ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5 (ATXR5) and ATXR6, two plant-specific methyltransferases preferentially catalyzing H3K27me1 on H3.1 (Jacob et al., 2009, 2014). The A31 residue on plant H3.1 is also required for the H3K27me3 maintenance, mediated specifically by H3.1 (Jiang and Berger, 2017). The distinct amino acids on H3.1 and H3.3 may stimulate or exclude specific enzymes, and thus enable H3.1 and H3.3 to serve as favored substrates for different types of histone modifications. Alternatively, H3.3 may attract specific binding partners and engage the configuration of chromatin structure suitable for active histone modifications.
Nucleosomes containing both H3.3 and H2A.Z are unstable (Jin and Felsenfeld, 2007). At Arabidopsis gene loci, H3.3 overall opposes H2A.Z accumulation especially at the 3′ gene end (Wollmann et al., 2017). Our results confirmed the antagonism between H3.3 and H2A.Z at FLC (Figure 5, E and F). H2A.Z is often associated with transcriptional repression, but at the FLC locus it activates its expression. Interestingly, it has been reported that the H2A.Z abundance at FLC is not positively associated with FLC expression levels, rather the less FLC is expressed, the higher H2A.Z is enriched (Deal et al., 2007). This is in line with our observations that H3.3 knockdown induces H2A.Z accumulation but leads to decreased FLC expression. These results suggest that in Col, the presence of both H3.3 and H2A.Z is required for the FLC activation while lacking either one compromises FLC expression. Hence, balanced H3.3 and H2A.Z deposition may help to smooth the transcription process and establish a competent state for FLC transcription. H3.3 and H2A.Z are both enriched at the FLC 5′ and 3′ ends (Figures 4A; 5, E and F). It is tempting to speculate that at FLC, H3.3 and H2A.Z co-exist in the same nucleosomes, which are highly sensitive to disruption. In contrast, the loss of either H2A.Z or H3.3 increases the nucleosome stability and transcription blockage at FLC, resulting in a less competent state for transcription.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) h3.3kd lines (Wollmann et al., 2017), h2a.z (Coleman-Derr and Zilberman, 2012), FRI-Col (Lee et al., 1994), fve-4 (Kim et al., 2004), fld-3 (He et al., 2003), flc-3 (Michaels and Amasino, 1999), efs-3 (Kim et al., 2005), and HTR5-GFP (Wollmann et al., 2012) were described previously. Since in h2a.z (hta8; hta9; hta11), the hta8 mutant was derived from the Ws (Wassilewskija) background, h2a.z was backcrossed with Col three times prior to any analyses. Plants were grown in long days (16 h light/8 h dark) or short days (8 h light/16 h dark) at ∼22°C.
RNA analysis
Total RNAs from 10-day-old seedlings were extracted with Minibest plant RNA extraction kit (Takara). Reverse transcription was performed using TransScript one-step gDNA removal and cDNA synthesis supermix (TransGen). RT-qPCR was carried out on an Applied Biosystems QuantStudio 6 Flex Real-Time PCR System using TransStart top green qPCR supermix (TransGen). TUB2 was used as an endogenous control for normalization. Primers used for amplification are listed in Supplemental Table S2.
For the analysis of published RNA-seq data (Wollmann et al., 2017), reads were mapped to the Arabidopsis genome (TAIR10) using Hisat2 (Kim et al., 2019). Reads per gene were counted by HTseq (Anders et al., 2015), and transcripts per million values were generated using R.
ChIP-qPCR
ChIP experiments were carried out using sonicated chromatin extracted from 10-day-old seedlings as previously described (Jiang and Berger, 2017). Immunoprecipitations were performed with anti-H3 (Abcam, ab1791), anti-H3K4me3 (Abcam, ab8580), anti-H3K36me3 (Abcam, ab9050), anti-H3K27me3 (Millipore, 07-449), and anti-H2A.Z (Yelagandula et al., 2014) antibodies. ChIP with HTR5-GFP-related lines were conducted with GFP-trap agarose beads (Chromotek, gta-20). The amounts of immunoprecipitated DNA were quantified by quantitative real-time PCR (qPCR). Primers used for amplification are specified in Supplemental Table S2.
ChIP-seq analysis
To analyze genome-wide H3K4me3 changes in h3.3kd, published ChIP-seq datasets were downloaded (Wollmann et al., 2017), and reads were aligned to the Arabidopsis genome (TAIR10) with Botiew2 (Langmead and Salzberg, 2012). Reads were filtered for duplicated reads by using Picard MarkDuplicates (https://github.com/broadinstitute/picard). For data visualization, bigwig coverage files of H3K4me3 relative to H3 (log2 ratio) were generated using deepTools utility bamCoverage with a bin size of 10 bp (Ramirez et al., 2014). To identify differential H3K4me3 enriched genes, the average scores for H3K4me3/H3 over the whole gene body were calculated with deepTools utility multiBigwigSummary, genes with H3K4me3/H3 ratio more than one in Col were retained for further analysis and differential genes were called by requiring more than 1.5-fold difference in h3.3kd compared with Col. Average ChIP-seq profiles were generated using deepTools utility plotProfile.
To profile HTR5-GFP enrichment in 10-day-old seedlings, materials were ground with liquid nitrogen into fine powder and fixed with 1% (v/v) formaldehyde, followed by nuclei extraction and micrococcal nuclease (Sigma, N5386) digestion to generate mononucleosomes as previously described (Jiang and Berger, 2017). Immunoprecipitation was performed with GFP-trap agarose beads (Chromotek, gta-20). ChIP-recovered DNA was subjected to library preparation with VAHTS universal DNA library prep kit for Illumina (Vazyme, ND607) according to manufacturer’s instruction and sequenced with HiSeq 2500 to generate paired-end 150-bp reads. Adapter trimming was performed and low quality reads were filtered with fastp (Chen et al., 2018). Reads were mapped to the Arabidopsis genome (TAIR10) with Botiew2 (Langmead and Salzberg, 2012), and filtered for duplicated reads by using Picard MarkDuplicates (https://github.com/broadinstitute/picard). For data visualization, bigwig coverage files of HTR5-GFP relative to input (log2 ratio) were generated using deepTools utility bamCoverage with a bin size of 10 bp (Ramirez et al., 2014). Average ChIP-seq profiles were generated using deepTools utility plotProfile.
Pull-down assay
Full-length coding regions of HIRA and FRI were cloned into pGEX-5X-2 and pRSETA for the production of GST-HIRA and His-FRI proteins respectively. Proteins were expressed using Escherichia coli BL21 (DE3) by IPTG induction. GST-HIRA and His-FRI were mixed together with glutathione-agarose resin (GE Healthcare) in pull-down buffer (50-mM Tris pH 7.5, 150-mM NaCl, 1-mM EDTA, 0.5% Nonidet P-40, protease inhibitor cocktail) at 4°C overnight. After washing four times with pull-down buffer, proteins retained on the beads were eluted by boiling with sodium dodecyl sulfate (SDS) loading buffer, separated by SDS–polyacrylamide gel electrophoresis, and detected with anti-GST (Sigma, G7781) and anti-His (CWBio, CW0286) antibodies. Primers used for generating constructs are listed in Supplemental Table S2.
Western blot
To analyze HTR5-GFP expression, total proteins extracted from 10-day-old seedlings were transferred to a 0.2-μm nitrocellulose membrane and detected with anti-GFP (TransGen, HT801) and anti-Actin (CWBio, CW0264) antibodies.
Bimolecular fluorescence complementation
The full-length coding sequences of FRI and HIRA were cloned into pEarleyGate201-YN and pEarleyGate202-YC vectors (Lu et al., 2010) via gateway technology (Thermo Fisher Scientific) and subsequently transformed into Agrobacterium tumefaciens strain GV3101. The Agrobacterium strains harboring YN, YC, and mRFP-AHL22 were coinfiltrated into Nicotiana benthamiana plant leaves. YFP and mRFP signals were observed 2–3 days after infiltration using a Zeiss confocal laser scanning microscope. For detecting YFP signals, excitation was performed at 514 nm (2% laser intensity), and emission was collected at 519–579 nm (gain: 972). For detecting mRFP signals, excitation was performed at 561 nm (2% laser intensity), and emission was collected at 578–650 nm (gain: 700). Primers used for generating constructs are listed in Supplemental Table S2.
Co-IP assay
To express tag-fused FRI and HIRA, CaMV 35S promoter was first inserted into pGWB513 and pGWB516 (Nakagawa et al., 2007), subsequently the full-length coding sequences of FRI and HIRA except stop codon were inserted to fuse with HA and Myc in pGWB513 and pGWB516 respectively. Protoplast transformation was performed as previously described (Cheng et al., 2015). After transformation, protoplasts were kept overnight and subjected to total protein extraction with Co-IP buffer (50-mM HEPES, pH 7.5, 150-mM KCl, 1-mM EDTA, 1-mM DTT, 0.3% Triton X-100, protease inhibitor cocktail). After centrifugation, the supernatant was incubated with anti-HA beads (Thermo scientific, 88836) for overnight. Tagged proteins were detected with anti-Myc (Easybio, BE2011) and anti-HA (CST, 3724) antibodies. Primers used for generating constructs are listed in Supplemental Table S2.
3C experiment
3C experiment was performed as previously described (Louwers et al., 2009; Crevillen et al., 2013; Li et al., 2018). Briefly, nuclei extracted from freshly collected 10-day-old seedlings were fixed with 2% formaldehyde and subsequently lysed with 0.2% SDS at 65°C for 30 min. SDS was sequestered with 2% Triton X-100 at 37°C for 1 h. BamHI and BglII were used to digest chromatin overnight and were inactivated with 1.6% SDS. After the sequestration of SDS with 1% Triton X-100, ligation was performed at 16°C for 5 h with T4 DNA ligase, followed by reverse crosslink and DNA recovery.
Relative interaction frequencies were quantified and calculated as described previously (Crevillen et al., 2013; Li et al., 2018). Amplifications of a loading control region that does not contain restriction sites were used to normalize DNA concentration between samples. Primer efficiency was normalized by amplification with a control template DNA consists of equal amounts of all possible ligation products, which was generated by ligating a BamHI and BglII cut plasmid containing an 11-kb FLC genomic fragment (Li et al., 2018). Values are averages of three biological replicates. Primers used for amplification were adopted from a previous study (Li et al., 2018).
Statistical analysis
Statistical significance was determined with two-tailed Student’s t test, one-way Analysis of Variance (ANOVA) with Tukey’s test or Mann–Whitney U test.
Data availability
The published H3K4me3 and RNA-seq datasets were downloaded from GEO GSE96873. The ChIP-seq data of HTR5-GFP in seedlings are available in the GEO under accession number GSE167384.
Accession numbers
FLC (AT5G10140), FLM/MAF1 (AT1G77080), MAF2 (AT5G65050), MAF3 (AT5G65060), MAF4 (AT5G65070), MAF5 (AT5G65080), HIRA (AT3G44530), ASF1a (AT1G66740), ASF1b (AT5G38110), FRI (AT4G00650), FVE (AT2G19520), FLD (AT3G10390), EFS/SDG8 (AT1G77300), HTR5 (AT4G40040).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Flowering phenotype and gene expression analyses in h3.3kd lines.
Supplemental Figure S2 . Chromatin modification changes at MAF4 and MAF5 in h3.3kd-1.
Supplemental Figure S3 . h3.3kd-1 represses the late flowering phenotype of fve-4 and fld-3.
Supplemental Figure S4 . HTR5-GFP and FLC expression in HTR5-GFP-related lines.
Supplemental Figure S5 . ChIP-seq analysis of H3K4me3 and HTR5-GFP distribution.
Supplemental Table S1 . List of genes at which H3K4me3 changed more than 1.5-fold in h3.3kd-3.
Supplemental Table S2 . Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Hui Li and Mande Xue (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for their assistance with the experiments.
D.J. conceived the research. F.Z., T.Z., and D.J. performed the experiments. F.Z., H.Z., and D.J. analyzed the data. Z.L. provided the experimental materials. D.J. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Danhua Jiang (dhjiang@genetics.ac.cn).
Funding
This work was supported by the National Key R&D Program of China Grant (grant no. 2019YFA0903903), the Strategic Priority Research Program of the Chinese Academy of Sciences (Precision Seed Design and Breeding, grant no. XDA24020303) and the National Natural Science Foundation of China (grant no. 31970527).
Conflict of interest statement. None declared.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache A, Yang S, de Paz AM, Robbins LE, Durmaz C, Cheong JQ, Ravishankar A, Daman AW, Ahimovic DJ, Klevorn T, et al. (2020) Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 583: 852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Wen DC, Dewell S, Whitcomb SJ, Lin MY, Diaz N, Elsasser SJ, Chapgier A, Goldberg AD, Canaani E, et al. (2013) Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155: 107–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Hua C, Shen L, Yu H (2020) New insights into gibberellin signaling in regulating flowering in Arabidopsis. J Integr Plant Biol 62: 118–131 [DOI] [PubMed] [Google Scholar]
- Borg M, Jiang D, Berger F (2021) Histone variants take center stage in shaping the epigenome. Curr Opin Plant Biol 61: 101991. [DOI] [PubMed] [Google Scholar]
- Chang YN, Zhu C, Jiang J, Zhang H, Zhu JK, Duan CG (2020) Epigenetic regulation in plant abiotic stress responses. J Integr Plant Biol 62: 563–580 [DOI] [PubMed] [Google Scholar]
- Chen SF, Zhou YQ, Chen YR, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Li JF, Niu Y, Zhang XC, Woody OZ, Xiong Y, Djonovic S, Millet Y, Bush J, McConkey BJ, et al. (2015) Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521: 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D (2012) Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillen P, Sonmez C, Wu Z, Dean C (2013) A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J 32: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Ndamukong I, Xu ZS, Lapko H, Fromm M, Avramova Z (2012) ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PloS Genet 8: e1003111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc C, Benoit M, Detourne G, Simon L, Poulet A, Jung M, Veluchamy A, Latrasse D, Le Goff S, Cotterell S, et al. (2017) Arabidopsis ATRX modulates H3.3 occupancy and fine-tunes gene expression. Plant Cell 29: 1773–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc C, Benoit M, Le Goff S, Simon L, Poulet A, Cotterell S, Tatout C, Probst AV (2015) The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J 81: 707–722 [DOI] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ (2002) The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol 9: 172–176 [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XF, Le C, Wang YZ, Li ZC, Jiang DH, Wang YQ, He YH (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun 4:1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Allis CD (2006) Histone H3 variants and their potential role in indexing mammalian genomes: the "H3 barcode hypothesis. Proc Natl Acad Sci USA 103: 6428–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y (2012) Chromatin regulation of flowering. Trends Plant Sci 17:556–562 [DOI] [PubMed] [Google Scholar]
- He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754 [DOI] [PubMed] [Google Scholar]
- Hodl M, Basler K (2009) Transcription in the absence of histone H3.3. Curr Biol 19: 1221–1226 [DOI] [PubMed] [Google Scholar]
- Jacob Y, Bergamin E, Donoghue MT, Mongeon V, LeBlanc C, Voigt P, Underwood CJ, Brunzelle JS, Michaels SD, Reinberg D, et al. (2014) Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343: 1249–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD (2009) ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CW, Shibata Y, Starmer J, Yee D, Magnuson T (2015) Histone H3.3 maintains genome integrity during mammalian development. Genes Dev 29: 1377–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Pineiro M (2015) H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. Plant J 83: 96–109 [DOI] [PubMed] [Google Scholar]
- Jiang D, Berger F (2017) DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 357: 1146–1149 [DOI] [PubMed] [Google Scholar]
- Jiang D, Berger F (2017) Histone variants in plant transcriptional regulation. Biochim Biophys Acta Gene Regul Mech 1860: 123–130 [DOI] [PubMed] [Google Scholar]
- Jiang D, Gu X, He Y (2009) Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21: 1733–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Kong NC, Gu X, Li Z, He Y (2011) Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet 7: e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Wang S, Jiang H, Cheng B, Wu K, Ding Y (2018) The COMPASS-like complex promotes flowering and panicle branching in rice. Plant Physiol 176: 2761–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin CY, Felsenfeld G (2007) Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sung S (2010) The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc Natl Acad Sci USA 107: 17029–17034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Kim HJ, Lee MH, Moon J, Lee I, et al. (2004) A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet 36: 167–171 [DOI] [PubMed] [Google Scholar]
- Kim SY, He YH, Jacob Y, Noh YS, Michaels S, Amasino R (2005) Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Mitina I, Tamada Y, Hyun Y, Choi Y, Amasino RM, Noh B, Noh YS (2010) Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J 29: 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM (1994) The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J 6: 903–909 [Google Scholar]
- Li Z, Jiang D, Fu X, Luo X, Liu R, He Y (2016) Coupling of histone methylation and RNA processing by the nuclear mRNA cap-binding complex. Nat Plants 2: 16015. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang D, He Y (2018) FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat Plants 4: 836–846 [DOI] [PubMed] [Google Scholar]
- Liu B, Liu Y, Wang B, Luo Q, Shi J, Gan J, Shen WH, Yu Y, Dong A (2019) The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment. Nat Commun 10: 2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Wang C, Wang G, Becker C, Zaidem M, Weigel D (2016) Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res 26: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers M, Splinter E, van Driel R, de Laat W, Stam M (2009) Studying physical chromatin interactions in plants using Chromosome Conformation Capture (3C). Nat Protocols 4: 1216–1229 [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang XR, Tian G, Wang F, Liu KD, Nguyen V, Kohalmi SE, Keller WA, Tsang EWT, Harada JJ, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Luo X, He Y (2020) Experiencing winter for spring flowering: a molecular epigenetic perspective on vernalization. J Integr Plant Biol 62: 104–117 [DOI] [PubMed] [Google Scholar]
- Martire S, Gogate AA, Whitmill A, Tafessu A, Nguyen J, Teng YC, Tastemel M, Banaszynski LA (2019) Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat Genet 51:941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng YY, Wang ZY, Wang YQ, Wang CN, Zhu BT, Liu H, Ji WK, Wen JQ, Chu CC, Tadege M, et al. (2019) The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula. Plant Cell 31: 2751–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A (2001) COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nie X, Wang H, Li J, Holec S, Berger F (2014) The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol Open 3: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD (2005) Histone variant H2A.Z marks the 5 ' ends of both active and inactive genes in euchromatin. Cell 123: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T (2014) deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Schwartz BE, Goldstein S, Ahmad K (2009) Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol 19: 1816–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, et al. (2010) A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev 24: 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci K, Michaels SD, Amasino RM (2003) Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol Biol 52: 915–922 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Araguez I, Peiro R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C (2014) The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon D, Boyarchuk E, Dingli F, Loew D, Almouzni G (2020) Histone variant H3.3 residue S31 is essential for Xenopus gastrulation regardless of the deposition pathway. Nat Commun 11: 1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B (1989) Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58: 15–25 [DOI] [PubMed] [Google Scholar]
- Stroud H, Otero S, Desvoyes B, Ramirez-Parra E, Jacobsen SE, Gutierrez C (2012) Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 5370–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenker E, Lacoste N, Almouzni G (2012) A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep 1: 730–740 [DOI] [PubMed] [Google Scholar]
- Szenker E, Ray-Gallet D, Almouzni G (2011) The double face of the histone variant H3.3. Cell Res 21: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S (2017) Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Biol 18: 115–126 [DOI] [PubMed] [Google Scholar]
- Wang H, Jiang D, Axelsson E, Lorkovic ZJ, Montgomery S, Holec S, Pieters B, Al Temimi AHK, Mecinovic J, Berger F (2018) LHP1 interacts with ATRX through plant-specific domains at specific loci targeted by PRC2. Mol Plant 11: 1038–1052 [DOI] [PubMed] [Google Scholar]
- Wirbelauer C, Bell O, Schubeler D (2005) Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev 19: 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H, Holec S, Alden K, Clarke ND, Jacques PE, Berger F (2012) Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet 8: e1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H, Stroud H, Yelagandula R, Tarutani Y, Jiang D, Jing L, Jamge B, Takeuchi H, Holec S, Nie X, et al. (2017) The histone H3 variant H3.3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol 18: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Liu Q, Chen G, Yan Z, Hu H (2020) Aldehyde dehydrogenase ALDH3F1 involvement in flowering time regulation through histone acetylation modulation on FLOWERING LOCUS C. J Integr Plant Biol 62: 1080–1092 [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28: 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Howard M, Dean C (2014) Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24: 1793–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelagandula R, Stroud H, Holec S, Zhou K, Feng S, Zhong X, Muthurajan UM, Nie X, Kawashima T, Groth M, et al. (2014) The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JX, Liu ZW, Li YQ, Li L, Wang B, Chen S, He XJ (2018) Arabidopsis PWWP domain proteins mediate H3K27 trimethylation on FLC and regulate flowering time. J Integr Plant Biol 60: 362–368 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Weng MJ, Yang Y, Zhang C, Li Z, Shen WH, Dong AW (2011) Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J 66: 443–455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published H3K4me3 and RNA-seq datasets were downloaded from GEO GSE96873. The ChIP-seq data of HTR5-GFP in seedlings are available in the GEO under accession number GSE167384.