Abstract
An indispensable feature of episodic memory is our ability to temporally piece together different elements of an experience into a coherent memory. Hippocampal time cells—neurons that represent temporal information—may play a critical role in this process. Although these cells have been repeatedly found in rodents, it is still unclear to what extent similar temporal selectivity exists in the human hippocampus. Here, we show that temporal context modulates the firing activity of human hippocampal neurons during structured temporal experiences. We recorded neuronal activity in the human brain while patients of either sex learned predictable sequences of pictures. We report that human time cells fire at successive moments in this task. Furthermore, time cells also signaled inherently changing temporal contexts during empty 10 s gap periods between trials while participants waited for the task to resume. Finally, population activity allowed for decoding temporal epoch identity, both during sequence learning and during the gap periods. These findings suggest that human hippocampal neurons could play an essential role in temporally organizing distinct moments of an experience in episodic memory.
SIGNIFICANCE STATEMENT Episodic memory refers to our ability to remember the what, where, and when of a past experience. Representing time is an important component of this form of memory. Here, we show that neurons in the human hippocampus represent temporal information. This temporal signature was observed both when participants were actively engaged in a memory task, as well as during 10-s-long gaps when they were asked to wait before performing the task. Furthermore, the activity of the population of hippocampal cells allowed for decoding one temporal epoch from another. These results suggest a robust representation of time in the human hippocampus.
Keywords: human electrophysiology, human hippocampus, sequence learning, temporal coding, time cells
Introduction
Creating episodic memories requires linking distinct events of an experience with temporal fidelity. The brain must represent the temporal flow and order of events, and glue them together in the correct sequential order. Time cells in the hippocampus and adjacent structures might play an essential role in this temporal organization of memory (Hasselmo, 2009; Eichenbaum, 2014; Howard et al., 2014). In rodents, time cells signal changing temporal contexts in a variety of paradigms (Manns et al., 2007; Pastalkova et al., 2008; MacDonald et al., 2011, 2013; Kraus et al., 2013, 2015). They fire at successive moments of time during a fixed interval, and the activity of the population of time cells covers the entire time interval (Pastalkova et al., 2008). More recently, another class of ramping cells in the lateral entorhinal cortex has been discovered. Ramping cells show slowly rising or decaying activity with time, over a range of time scales. Temporal epoch identity could be decoded from the firing activity of the population of cells (Tsao et al., 2018).
Temporal coding has also been observed in neuronal activity patterns in the human hippocampus. For instance, neuronal activity in the human medial temporal lobe shows gradual changes over time in memory tasks (Howard et al., 2012; Folkerts et al., 2018). The recall of a particular item is accompanied by the reinstatement of its initial temporal representation (Gelbard-Sagiv et al., 2008; Howard et al., 2012; Folkerts et al., 2018). More recently, single neurons have also been shown to be modulated by time, akin to time cells in rodents, during encoding and retrieval in a free recall memory task (Umbach et al., 2020).
In the current study, we asked if human hippocampal neurons represent temporal information during sequential order learning. A large body of work in animals and humans has shown that the hippocampus is essential for remembering the temporal order of sequential events (Eichenbaum, 2013). For example, in humans, the hippocampus is activated when subjects recall the order of objects, and conversely, patients with hippocampal damage have trouble in temporal order judgements (Spiers et al., 2001; Ekstrom and Bookheimer, 2007). In animals, rats with hippocampal damage are impaired at remembering the sequential order of odors (Fortin et al., 2002). Given the importance of the hippocampus in sequence order learning and temporal order judgements, we tested whether human hippocampal neurons represented temporal information while participants learned the order of a sequence of items. We tested for temporal modulation of hippocampal activity in two experiments (1) during sequence learning (SL: Fig. 1A) and (2) during empty gap periods inserted in the task during which participants passively waited for the sequence to resume (Fig. 1B). Note that in these gap periods, any potential temporal information is not driven by external stimuli or events but rather represents inherent changes in the patients' moment-to-moment experience. We report that human hippocampal neurons fire at successive moments during these structured time periods, both while subjects actively monitor a sequence as well as during empty temporal gaps between events.
Figure 1.
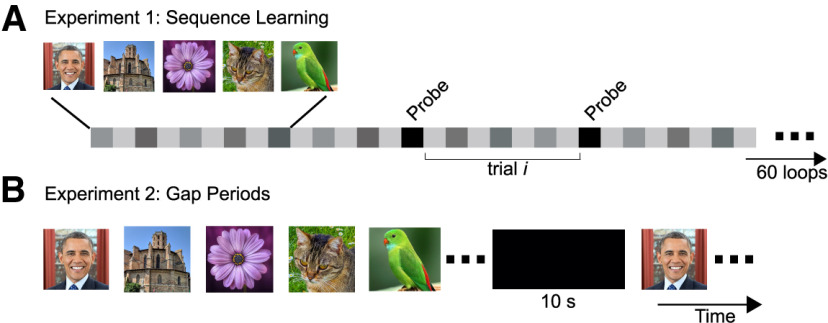
Experimental design. A, In the sequence learning experiments, participants saw a sequence of images in a fixed order, and were asked to learn the sequence order. The stimulus sequence consisted of five to seven image periods (image number fixed per session and determined by the availability of the patient) separated by ISI periods. Each image was presented for 1.5 s followed by an ISI of 0.5 s. The sequence was repeated for 60 loops. 20% of the time, a probe event occurred (black squares) during which participants had to decide which of two choice images was the correct one at the current position in the sequence. The probe events occurred at random positions of the sequence. After the probe event, the sequence resumed. In our main analysis, we consider time periods that occurred between two consecutive probe events as the trials of interest. As shown, each postprobe trial consisted of several image and ISI periods (gray squares). B, The design of experiment 2 was similar to that of experiment 1 except for the insertion of 10-s-long gap periods (black rectangle) during sequence learning. These gap periods occurred periodically (see below, Materials and Methods). During the gap periods, the sequence stopped, and patients were presented with a blank screen. They were asked to simply wait until the sequence resumed.
Materials and Methods
In this study, human epileptic patients performed two sequence learning tasks while single neuron activity was recorded from microelectrodes implanted in the hippocampus (Fig. 2). We quantified the influence of time on the firing activity of individual neurons using a stepwise general linear model (GLM), as has previously been used in the rodent literature (MacDonald et al., 2011; Tsao et al., 2018). In this GLM, a predictor variable is included in the model only if it is found to significantly improve the prediction of the response variable (see below).
Figure 2.
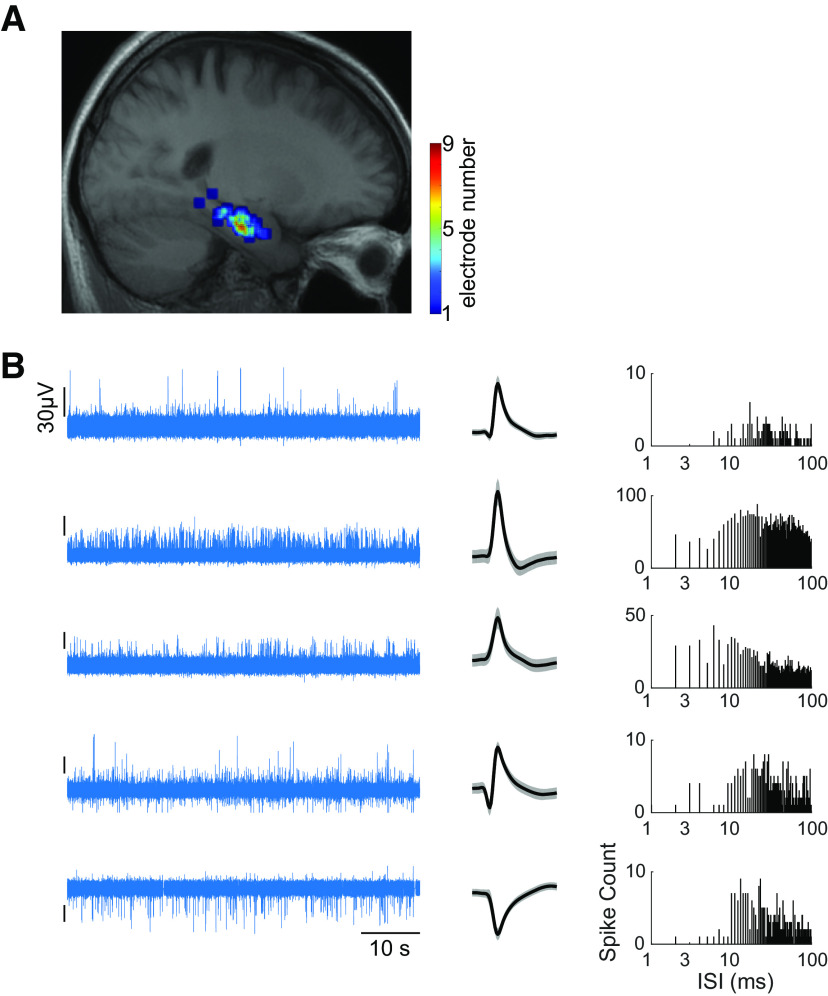
A, Electrode locations shown on a sagittal slice of the average MRI of patients registered to the Montreal Neurological Institute brain template. B, Bandpass-filtered (300–3000 Hz) signal from five different channels (left), mean waveforms recorded on these channels (middle), and the corresponding distributions of interspike intervals (right). The black vertical tick marks on the left plots indicate a scale of 30 uV.
Patients
Nine patients of either sex participated in the first experiment, and six patients of either sex participated in the second experiment. The patients were diagnosed with pharmacologically intractable epilepsy and were undergoing epileptological evaluation at the Amsterdam University Medical Center. Patients were implanted with chronic depth electrodes for 7–10 d to localize the seizure focus for possible surgical resection (Fried et al., 1997; Engel et al., 2005). All surgeries were performed by Johannes C. Baayen and Sander Idema. The Medical Ethics Committee at the medical center approved the studies. The electrode locations were based entirely on clinical criteria and were evaluated based on the presurgical planned trajectories on the basis of structural MRI scans. For each electrode the planned trajectory was adjusted to ensure that the tip of the macroelectrode was at least 3 mm within the body of the hippocampus. The clinical team aimed for microwires that extended ∼2–3 mm from the tip of the macroelectrode. The accuracy of the implantation was always checked using a computerized tomography scan coregistered to the MRI. We only included electrodes that were within a 3 mm deviation from the target (based on visual confirmation). Each electrode contained eight microwires (Behnke-Fried electrodes, Ad-Tech Medical) from which we recorded multiunit activity, and a ninth microwire that served as a local reference. The signal from the microwires was recorded using a 64-channel Neuralynx system, filtered between 1 and 9000 Hz, sampled at 32KHz. On average, each patient was implanted with 34 ± 11.8 microwires [range = (16, 48)]. Participants sat in their hospital room at the Epilepsy Monitoring Unit and performed the experimental sessions on a laptop computer.
Spike detection and sorting
Spike detection and sorting were performed with Wave_clus (Quiroga et al., 2004). Briefly, the data were bandpass filtered between 300 and 3000 Hz, and spikes were detected with an automatic amplitude threshold (Reddy et al., 2015). Spike sorting was performed with a wavelet transform that extracted the relevant features of the spike waveform. Clustering was performed using a superparamagnetic clustering algorithm. Clusters were visually reviewed by Leila Reddy for (1) the mean spike shape and its variance, (2) the ratio between the spike peak value and the noise level, (3) the inter spike interval distribution of each cluster, (4) the presence of a refractory period, and (5) the similarity of each cluster to other clusters from the same microwire. Based on manual inspection of these criteria, clusters were retained, merged, or discarded.
Experimental design and statistical analyses
Behavioral Task
In experiment 1, sequence learning, the patients performed a total of 31 SL sessions (Fig. 1A). In each SL session, participants were presented with a sequence of 5–7 images (image number determined as a function of the difficulty level and the availability of the patient). The images were always presented in a predetermined order so that a given image, A, predicted the identity of the next image, B, and so on. Subjects were asked to remember the order of the images in the sequence. Each image was presented for 1.5 s (image period) followed by an interstimulus interval (ISI) period of 0.5 s. The sequence was repeated continually 60 times, resulting in experimental sessions of 10 min for five-image sequences and 14 min for seven-image sequences, not including time spent by the subject to respond on probe events. On a random 20% of image periods, the sequence stopped, and participants were presented with probe events. In these probe events, instead of being presented with the next image of the sequence, subjects were shown two images side by side and asked to decide (by pressing one of two keys on the keyboard) which of the two was the next image in the sequence. After the subjects responded, the sequence resumed.
From the point of view of the subject, the probe events were salient moments of an otherwise repetitive experiment because the probes stopped the sequence and tested subjects on their learning of the sequence order. Thus, we considered sequence segments between two consecutive probe events as our trials of interest, that is, structured, temporal experiences between two salient markers. We asked whether hippocampal neurons tracked time in this interval. In control analyses described below, we verified these results with respect to other time periods in the experiment.
In experiment 2, sequence learning with temporal gaps (Fig. 1B), six new patients performed eight sessions of a second SL experiment. This second experiment followed the design of the first SL experiment described above, except for the following modifications: After a fixed number of repeats of the sequence, a 10-s-long empty gap interval was presented. During these gap intervals, participants were presented with a black screen, without any stimulus input, and were asked to simply wait until the sequence started again. For three participants these gap intervals occurred after every five repeats of the sequence (resulting in six gap intervals in the experiment), whereas for the remaining three participants, these gap intervals occurred after every two repeats of the sequence (resulting in 15 gap intervals in the experiment). The sequence was repeated only 30 times instead of 60 times.
In the nine patients who performed the first experiment, we recorded from 429 neurons in the hippocampus, and in the six patients who performed the second experiment, we recorded 96 hippocampal units.
Time cell identification with a general linear model (experiment 1)
Time cell identification was performed with a GLM as in previous studies (MacDonald et al., 2011; Tsao et al., 2018). The firing activity of each neuron was modeled as a function of time, image identity, and whether the temporal period corresponded to an image or ISI period.
For the purposes of the GLM, as described above (Fig. 1A), we defined trials as segments of the sequence between two consecutive probe events (number of sequence segments or trials between two consecutive probe events across sessions: mean ± SEM = 73.6 ± 2.4). We made this choice because (1) as explained previously, these probe events were the most salient events of the experiment, and (2) if we simply consider time = 0 as the start of each five to seven image sequence, time in the sequence is directly confounded by image identity because the sequence order is fixed. By redefining time = 0 as the time at which the sequence restarted after the probe events, we avoided this confound because time is not confounded with image identity with respect to probe events (the sequence segment after the probe is random since the probe events occurred at random moments). In control analyses we also considered different temporal intervals for determining time cells.
Each of the postprobe trials consisted of several image and ISI periods that regularly followed each other (Fig. 1A). The median number of image and ISI periods in a trial was seven, corresponding to a median trial length of 6.5 s. For all subsequent analyses, only the first 6.5 s of each trial were included in the GLM (i.e., trials shorter than this duration were excluded from the GLM). Thus, each trial between probe events was a well-structured temporal interval during which the sequence progressed according to its fixed order. The average number of trials included in the GLM was 49.7 ± 0.8 [mean ± SD, range = (25, 64)].
In the GLM, the firing activity vector (Y) on each trial contained the average firing rates for each period of the trial, with no smoothing or additional preprocessing. Y was modeled as a function of three variables: image identity in each period, time of the period [i.e., time of the midpoint of the period with respect to trial onset/probe offset; time varied between 0s and 6.5s], and whether the period corresponded to an image or an ISI event. A linear factor for time assumes that time cells either show a ramping up or a ramping down of firing activity during the trial. To also include the possibility of cells having a preferred time not just at the beginning and end of trials but also at intermediate points, we included a quadratic time term (i.e., t2, a parabola-shaped function; for this purpose, time was recentered to the middle of the trial and thus varied between −3.25 and 3.25 s). The firing activity (Y) was modeled as follows:
The GLM analysis was performed using the MATLAB stepwiselm function, including the variables image/ISI, imageID, Time, and Time2, in a linear model, with a constant term as the baseline model, the sum of squared error (SSE) criterion (PEnter = 0.05), and other default parameters. The variables image/ISI and imageID were entered as categorical variables, and the time variables were continuous variables. Stepwise regression systematically tests the variance explained by adding and removing variables from a linear model based on their statistical significance in explaining the response variable. Note that the order in which regressors are entered into the stepwise linear model does not affect its outcome. Time cells were defined as cells for which the time terms (i.e., Time and/or Time2) were added by the stepwiselm function (PEnter < 0.05).
Statistical testing was performed for each time cell with permutation testing in which the firing rates were shuffled with respect to the task design, and the stepwise regression repeated. This procedure was repeated 1000 times per cell. For each cell, pactual was the p value returned by the regression analysis for the real data. pactual was compared with the distribution of p values returned from the shuffling procedure. For cells where both the time and time2 terms were significant the p value corresponded to the smaller of the corresponding p values. We defined pshuffle as the proportion of shuffles that produced a smaller p value than pactual. Only cells with pshuffle < 0.05 were ultimately considered time cells in this analysis (Umbach et al., 2020).
As a separate test to confirm our classification of time cells, different from the stepwise regression test, we performed a likelihood ratio test to compare the log likelihood values of a restricted linear model that included all terms except the time terms, and a full model which also included the two time terms.
Time cell identification with a general linear model (experiment 2)
The 10 s gap intervals of experiment 2 were epoched into 500 ms nonoverlapping windows, and, as above, a stepwise regression analysis was performed. The firing activity (Y) in each epoch was modeled as a function of time in the epoch (Time and a quadratic time term Time2). All other parameters in this analysis were identical to those described for experiment 1. The firing activity (Y) was modeled as follows
Control analyses for defining trial periods and time cells
In the main analysis of experiment 1, a trial was defined with respect to the probe events (i.e., the sequence segment that occurred between two consecutive probe events). We performed several additional analyses for identifying time cells; in each case permutation testing was performed for each time cell as described above. The results of the control analyses are summarized in Table 1.
Table 1.
The number of time cells identified in the control analyses and the overlap between time cells identified in the main analysis and the control analyses
| Control Analysis | Number of time cells identified (% of overlap with time cells from the main analysis) | Concern |
|---|---|---|
| Likelihood ratio test | 138 (84.7%) | Identification of time cells is method dependent |
| Stepwise GLM on ISI periods only | 106 (76.4%) | Identification of time cells is dependent on image information |
| Stepwise GLM excluding first ISI | 65 (81.5%) | Identification of time cells is dependent on the probe events |
| Stepwise GLM with trials defined with respect to sequence onset | 69 (30.4%) | Identification of time cells is dependent on the temporal interval |
| N-way ANOVA | 101(88.1%) | Identification of time cells is method dependent |
| Stepwise GLM excluding the quadratic time term | 114 (88.7%) | Identification of ramping cells |
First, time cells were identified when the first period after the probe event was excluded from the GLM. Second, time cells were identified when the GLM analysis was performed on only the ISI periods. In this control, the Y vector contained the firing rates in the ISI periods, and the regressor matrix X consisted of the time factors and an image identity factor (i.e., the identity of the image following the ISI period, to account for image-specific anticipatory responses that can be observed in the ISI periods during sequence learning (Reddy et al., 2015)). Third, trials were redefined as sequence segments with respect to the onset of each repetition of the sequence. Note that in this case, time selectivity can be confounded by image selectivity (as the same stimulus sequence repeats identically in every loop); however, the MATLAB stepwiselm function that we used for determining time selectivity could disentangle the potential contributions of the time and image ID variables because it systematically tests for the addition and removal of each variable in significantly explaining the response variable. Nonetheless, to avoid any ambiguity in interpretation, we elected to present time selectivity with respect to probe events as our main analysis, as it precludes this potential confound. Fourth, time cells were also identified in a control analysis that used an N-way ANOVA to test for an interaction between firing rates and time, as in Umbach et al., 2020. For each trial we computed the firing rate within each ISI and image period. We tested for temporal modulation of firing activity with predictors time bin, image identity, and image/ISI. As in Umbach et al., 2020, cells with a significant main effect of time (i.e., pactual < 0.05) from this procedure were passed on for statistical testing. Statistical testing was performed with permutation testing in which the firing rates were shuffled with respect to the task design, and the ANOVA was repeated on the shuffled data. As explained previously, only potential time cells with pshuffle < 0.05 were ultimately considered time cells in this analysis. Finally, to identify ramping cells (Tsao et al., 2018), we repeated the stepwise GLM approach as in the main analysis but with the exclusion of the quadratic time term. A summary of the control analyses is shown in Table 1.
Statistical test for the number of time cells
Statistical significance for the number of time cells identified by the GLM analysis was evaluated using a permutation test. For the permutation test, in experiment 1, surrogate data were created by randomly shuffling the image and ISI periods on each trial. For experiment 2 surrogate data were created by randomly shuffling epoch time. The stepwise regression was then performed on this surrogate data on 106 iterations. The proportion of surrogates that had a higher number of time cells was p < 10−6 in all analyses (in other words, none of the surrogates ever had a higher number of time cells).
Heat maps cross-validation and statistical test
The original heat maps (Fig. 3B; see Fig. 5B) were constructed by sorting the time cells according to the latency of peak firing and using this sorted order to plot the firing rate of each cell over the time interval. However, a heat map generated from random data, sorted and plotted according to the maximum value of each entry, will also show a similar well-organized pattern. Thus, to ensure the reliability of these maps, we performed a cross-validation analysis on the real heat maps to verify their reproducibility.
Figure 3.
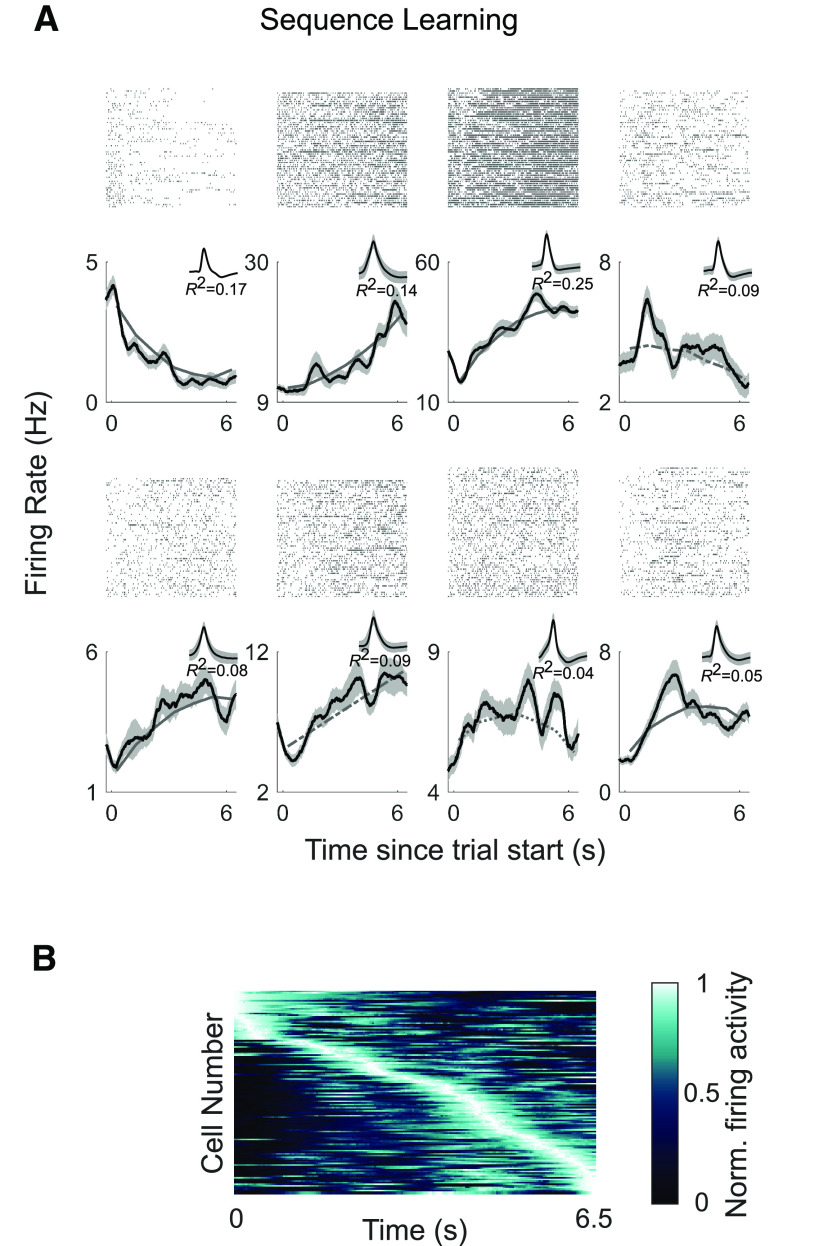
Time cells fire at specific moments during sequence learning. A, Raster plots (top) and poststimulus time histograms (bottom) are shown for eight example time cells. The x-axis corresponds to time of the median trial length (6.5 s; see above, Materials and Methods). The black line is the average firing activity, and the shaded area corresponds to the SE of the mean across trials. The gray lines show the model fit (solid gray line for cells that were classified as time cells according to the inclusion of the linear and quadratic terms, dashed gray line for cells classified based on the linear term alone, and stippled gray line for cells classified based on the quadratic term alone). Insets, The waveforms and the R2 of the model for each cell. B, Firing activity of the population of time cells (N = 128) identified as being significantly modulated by time (i.e., time and/or time2) in the sequence learning experiment. Each row shows the firing activity for an individual time cell, averaged across trials. The x-axis corresponds to time of the median trial length. The neurons are sorted by the latency of the maximum firing rate.
Figure 5.
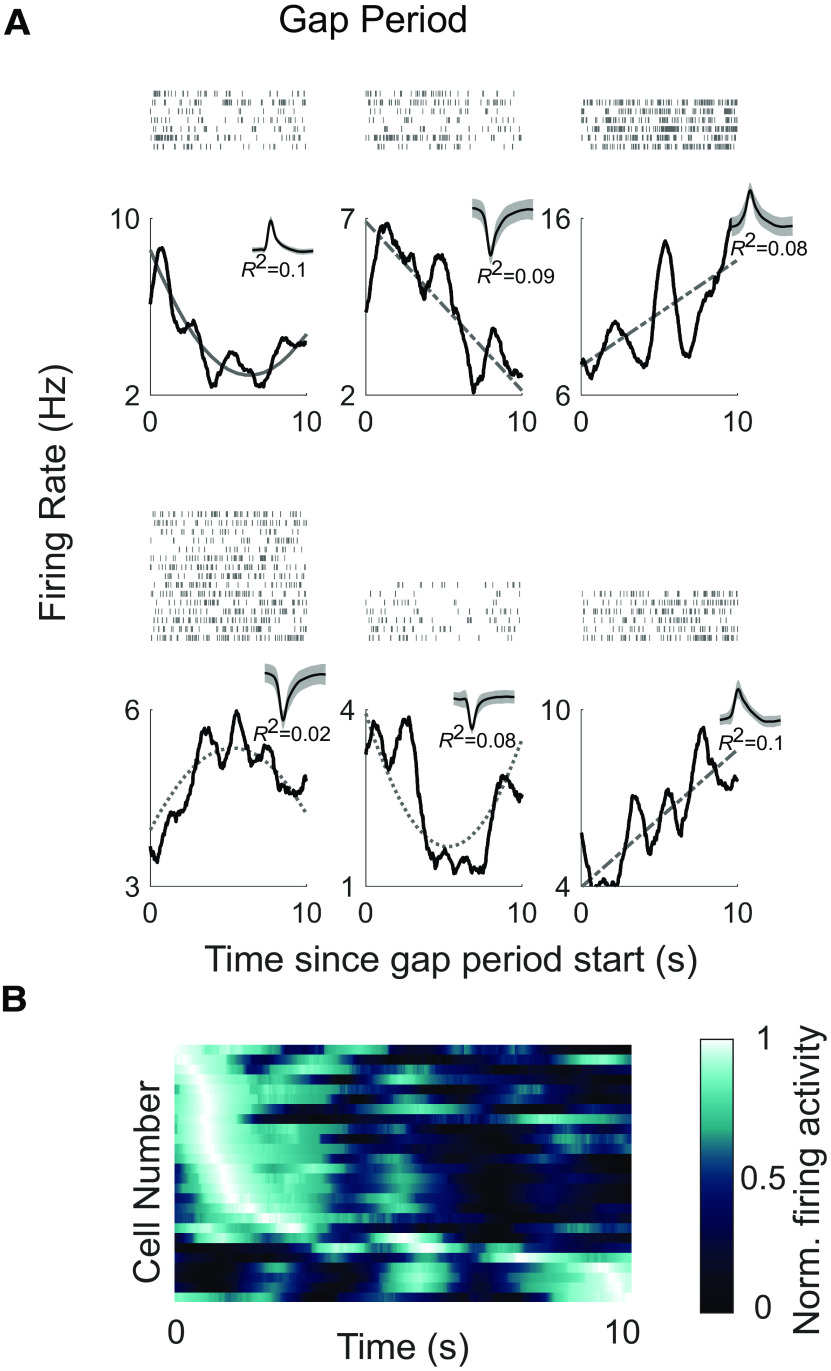
Time cells fire at specific moments during the gap periods. A, Raster plots (top) and poststimulus time histograms (bottom) are shown for six example time cells. The x-axis corresponds to the 10 s duration of the gap period. The gray lines show the model fit (solid gray line for cells that were classified as time cells according to the inclusion of the linear and quadratic terms, dashed gray line for cells classified based on the linear term alone, and stippled gray line for cells classified based on the quadratic term alone). Insets, The waveforms and the R2 of the model for each cell. B, Firing activity of time cells in the temporal gap experiment (N = 26). Each row shows the firing activity for an individual cell averaged across the gap periods. The x-axis corresponds to the 10 s gap period. The neurons are sorted by the latency of the maximum firing rate.
For the cross-validated heat maps, the order in which the cells were plotted was determined from the latencies on a random half of the data, and the firing rates were plotted for the remaining half of the data. If the cells had no true temporal preference, the peak latency on the second half should be unrelated to the latency measured on the first half, and no meaningful ordering of cells should appear. This cross-validation procedure was repeated 1000 times, and the resulting heat maps were averaged across cross-validations to generate cross-validated maps. We quantified the reliability of the heat maps by performing a nonparametric permutation test. We first computed the Spearman correlation pointwise between the average cross-validated heat map and the original heat map, resulting in the correlation measure corrorig,cv. To simulate the null hypothesis that the cells do not have a reliable time preference, on each cross-validation iteration we generated 105 surrogate heat maps by randomizing the cell order obtained from the first half of the data (instead of determining their order based on peak firing time over the first half), and generating a heat map with this random order for the second half of the data. Each of these 105 surrogate heat maps was correlated with the original heat map (corrorig,surr). After averaging across the 1000 cross-validations, none of the 105 surrogates yielded a correlation value corrorig,surr higher than the real correlation corrorig,cv, that is, p < 10−5.
Population pattern analysis
For this analysis, the population size was 429 neurons for experiment 1 and 96 neurons for Experiment 2.
In experiment 1, sequence learning, the population pattern analysis was performed on the image periods. The population pattern analysis was based on a pairwise comparison of image periods, thus, chance performance for all analyses described below is 50%.
As mentioned above, each trial (i.e., the sequence segment between two consecutive probe events) consisted of a varying number of image periods (Fig. 1A). In the population pattern analysis, our goal was to discriminate temporal period identity (e.g., image period 1 versus image period 2). Pairwise discrimination performance was evaluated for different numbers of temporal periods from the start of the trial (ranging from two to five; i.e., discriminating between the first two image periods after the probe event, the first three image periods, and so on). To ensure that performance was not driven by an unequal representation of the different images in the different periods, we created a balanced dataset for the population pattern analysis. In this balanced dataset we included the subset of trials per cell that ensured that each image was equally present in each period (see Fig. 6A). The balanced dataset only included cells for which a minimum number of trials had been recorded. The minimum number of trials was the smallest number so that at least 300 cells were included in the analysis.
Figure 6.
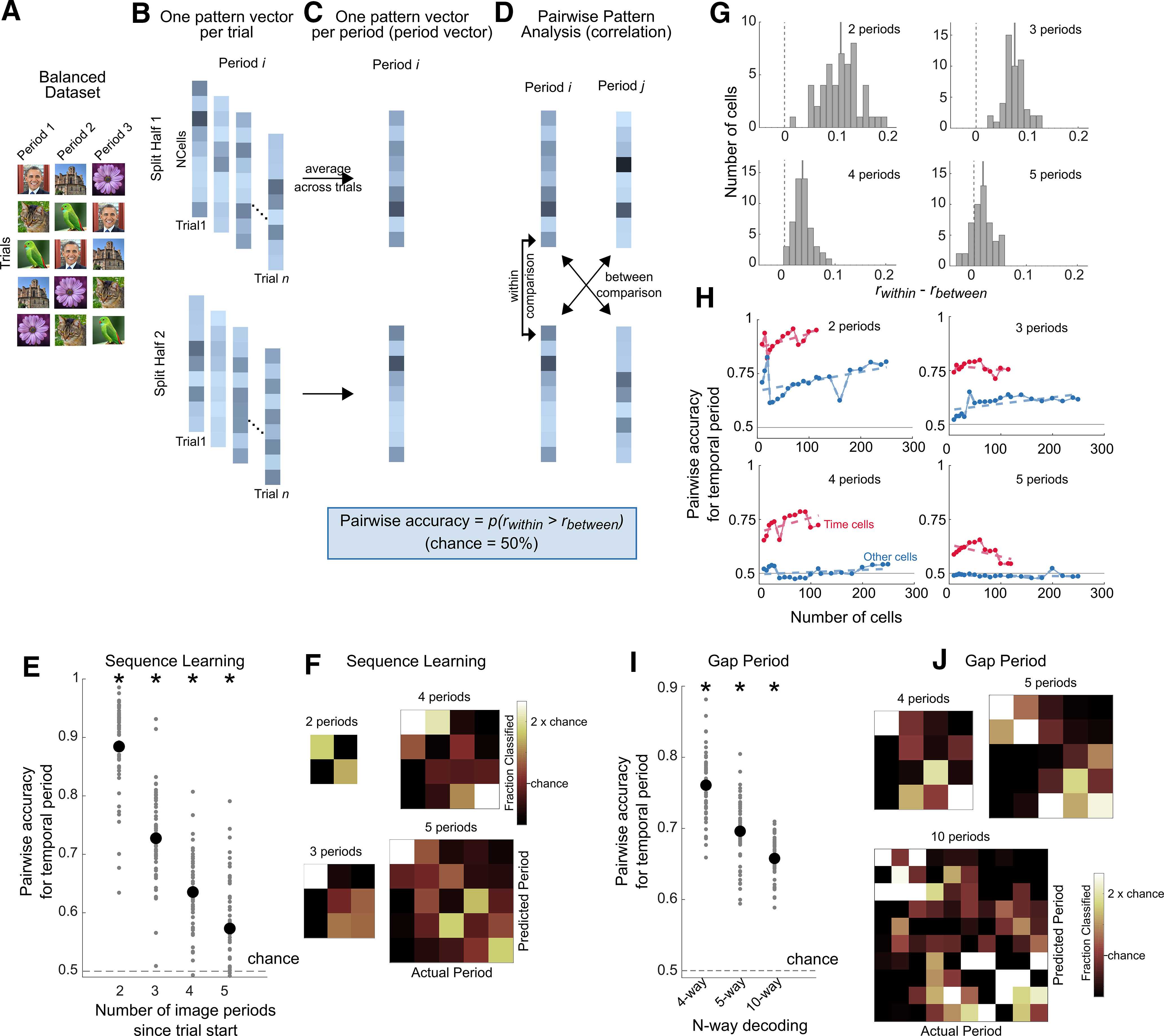
Population pattern analysis. In experiment 1, the population pattern analysis was performed on image periods (e.g., discriminating image period 1 vs image period 2). In experiment 2, the population pattern analysis was performed on the 10 s gap interval. Because this decoding method was based on pairwise comparisons, chance performance is always 50%. A, For experiment 1, a balanced dataset was created by selecting a subset of trials so that each image was equally present in each period across trials (see above, Materials and Methods). B, Population pattern analysis procedure. The trials of the balanced dataset (experiment 1), or the gap intervals (experiment 2) were split into two halves (repeated on 200 iterations; see above, Materials and Methods). In each half of the dataset, and for each period, the firing activity of the population of cells was arranged into a pattern vector for each trial. C, An average pattern vector of population firing activity was obtained for each image period by averaging across trials (period vectors). D, Pairwise discrimination was performed on the period vectors across the two halves of the dataset. For all pairs of periods, the Pearson's correlation was computed across the two halves, and the same-period comparisons (within comparisons) were compared with the different-period (between) comparisons. Pairwise accuracy was the proportion of within comparison correlations (rwithin) that were higher than the between comparison correlations (rbetween) (Haxby et al., 2001; Pereira et al., 2018). E, Population pattern analysis accuracies. Pairwise accuracy for temporal period identity during the sequence learning experiment using the split-half procedure described in A–D (population size = 429 neurons). The x-axis shows the number of image periods that the classifier was tested on (i.e., discrimination between the first two image periods, the first three image periods, and so on from the start of the trial). Pairwise accuracy (mean ± SE) for discriminating the first two periods = 88.4 ± 8%, the first three periods = 72.7 ± 8%, the first four periods = 63.5 ± 6%, the first five periods = 57.3 ± 9%. The black dots correspond to the mean accuracy across the 50 iterations for creating the balanced dataset, and the gray dots show the distribution of accuracies obtained across iterations. Asterisks denote significance based on a t test against chance, (p < 10−3). As the analysis is based on pairwise comparisons, chance performance is always 50%. F, Decoding errors mainly occurred for predicting neighboring temporal periods. Confusion matrices during sequence learning when discriminating the first two temporal periods, the first three temporal periods, and so on. G, Distributions of the difference of (rwithin) and (rbetween) for decoding of the first two image periods, the first three image periods, and so on. The solid gray line corresponds to the mean of the distribution. H, Pairwise accuracy for size-matched populations of time cells (in red) and other (nontime) cells (in blue), for decoding of the first two image periods, the first three image periods, and so on. The dashed lines correspond to a linear fit through the data. The horizontal gray line corresponds to chance performance (50%). I, Population pattern analysis performance during the temporal gap experiment (population size = 96 neurons). The 10 s gap periods were split into 4, 5, or 10 discrete periods, and discrimination of temporal period identity was computed using the procedure shown in B–D. Pairwise accuracy was significantly above chance for all comparisons [t test against chance (0.5), p < 10−3]. Pairwise accuracy, mean ± SD = 76.0 ± 4.7% for four-way decoding, 69.6 ± 4.6% for five-way decoding, and 65.7 ± 3.1% for 10-way decoding. The black dots correspond to the mean accuracy across the 50 iterations for creating the balanced dataset, and the gray dots show the distribution of accuracies obtained across iterations. Asterisks denote significance based on a t test against chance (p < 10−3). Because the analysis is based on pairwise comparisons, chance performance is always 50%. J, Confusion matrix for the gap experiment for different epoch lengths.
For the two-way discrimination (period 1 vs period 2), the balanced dataset required 42 trials, and 397 cells were included; for a three-way discrimination (period 1 vs period 2 vs period 3) it required 30 trials and included 303 cells; for a four-way discrimination it required 12 trials and included 320 cells; and for a five-way discrimination, it required 6 trials and included 367 cells. Discrimination of temporal period identity in this balanced dataset was thus not influenced by an imbalance in the number of presentations of each image across temporal periods. Note that the balanced dataset did not include the first ISI period after the probe, and decoding was thus not influenced by the offset of the probe events. As mentioned below, trial selection for creating the balanced dataset was repeated on 50 iterations, and the population pattern analysis was performed on each iteration.
The population pattern analysis was performed using a split-half approach (Haxby et al., 2011; Pereira et al., 2018) on the firing rates of the balanced dataset (see Fig. 6B–D). The trials in the balanced dataset were randomly split into two halves, and in each half the firing activity of the population of neurons was arranged into vectors per period and per trial (see Fig. 6B). These vectors were averaged across trials, yielding a population pattern vector for each period (period vectors) in each half of the dataset (see Fig. 6C). To quantify decoding or discrimination performance, we pairwise correlated the period vectors in one-half of the dataset with the period vectors in the other half, and used the pairwise correlation values to measure the percentage of correct classification (see Fig. 6D). To be more precise, for each period we computed the correlation for this period across the two halves of the dataset (within comparison, rwithin), and compared it to the correlation with a different period in the other half of the dataset (between comparison, rbetween). Decoding was correct if the within comparison was larger than the between comparison (see Fig. 6G; Haxby et al., 2001). This procedure was repeated for all pairs of periods, and pairwise decoding accuracy was the proportion of correct comparisons. Because this procedure was based on pairwise comparisons, chance performance was 50% for all analyses. Feature normalization was performed on the dataset for this analysis by performing a z score on the data for each cell along the periods dimension. Feature normalization was performed on the whole dataset based on the mean and SD measured in the training half of the dataset (to avoid leakage of information from the training half to the test half during the split-half cross-validation).
Note that in the split-half approach the training data (one half of the data) and testing data (the other half of the data) are independent by construction.
To increase reliability, the population pattern analysis was performed over several iterations: (1) trial selection for the balanced dataset was repeated 50 times; (2) on each of these 50 iterations, the dataset was randomly split into two halves 200 times. The reported mean decoding accuracy was the average decoding performance across the 50 iterations for creating the balanced dataset. The SD was computed across the 50 balanced datasets of the mean decoding accuracy across the 200 split halves. Statistical significance was computed with a t test against chance (0.5) across the 50 iterations for creating the balanced dataset (all p < 10−3).
To compare decoding performance for time cells and nontime cells (other cells), and to determine how population size affects decoding accuracy, we performed the pairwise pattern analysis separately for time cells and other cells, and for different population sizes. Size-matched populations were generated by randomly subsampling the population of cells. For each population size, the same procedure described above was repeated on 20 balanced datasets and 50 split halves.
In experiment 2, sequence learning with temporal gaps, for the population pattern analysis of the gap intervals, each gap interval was split into 4, 5, or 10 discrete periods, and discrimination of temporal period identity was performed using a split-half approach (50 iterations). The procedure was the same as the one described above for experiment 1 (see Fig. 6); and as above, chance performance was 50% because the method was based on pairwise comparisons. Feature normalization was performed on the dataset by performing a z score on the data. Statistical significance was tested using a t test against chance performance (0.5) across the 50 split-half iterations. As some subjects had 6 gap periods, and others had 15 gap periods, on each of the 50 iterations for splitting the data a random set of 6 gap periods was chosen from the datasets that contained 15 gap periods.
Results
Behavioral task and number of units
To determine whether human hippocampal neurons are modulated by the temporal context, we recorded from hippocampal neurons in human patients who performed a sequence learning task (Fig. 1A,B). We performed two independent sequence-learning experiments in two groups of patients implanted with intracranial microelectrodes. In experiment 1, we recorded the activity of 429 hippocampal neurons in 9 patients, and in experiment 2 we recorded from 96 hippocampal neurons in a new group of 6 patients.
In both experiments, the patients were presented with a fixed number of images (five to seven, depending on the patients' availability) in a predefined order (Fig. 1A,B) and were asked to learn the sequence order (Reddy et al., 2015). The sequence was repeated continually 60 times. Experiment 2 was similar to experiment 1 except for the periodic insertion of 10-s-long gap periods during the experiment. During these gap periods, the sequence stopped, and the patients had to wait until the sequence resumed (Fig. 1B).
The duration of each image period in the sequence was 1.5 s, and each image period was followed by an ISI of 0.5 s. On a random 20% of image periods, subjects were probed on their learning of the image order. During these probe events, the sequence momentarily stopped, and subjects were presented with two images from the sequence. Their task was to report which of the two images was the correct one at the current sequence position. The sequence then resumed until the next probe event. Participants rapidly learned the sequence order and achieved >90% performance on probe trials within the first six sequence presentations (Reddy et al., 2015). From the point of view of the subjects, the probe events were salient moments of the experiment because they stopped the ongoing sequence and tested learning.
Human hippocampal neurons are modulated by time during sequence learning
Time cells have been characterized as neurons whose activity is modulated by temporal context within a well-defined time window. Our experiment design lent itself to identifying time cells because the task consisted of a structured image sequence that occurred in a fixed and predictable time interval. In the time domain, our experiment involved three distinct time lines: (1) experiment time running from the beginning to the end of the experiment, (2) sequence time with respect to the start of each iteration of the sequence, and (3) probe time running between consecutive probe events. We first focus on probe time because, as described above, the probe events were salient moments from the point of view of the participants. Furthermore, focusing on probe time allowed us to decouple time from image identity as the postprobe trials consisted of varying segments of image and ISI periods.
We defined trials as segments of the experiment that occurred between two consecutive probe events in experiment 1 (Fig. 1A). These trials were of a relatively fixed duration (median trial length = 6.5 s), and consisted of a sequence of image and ISI periods. To identify time cells, we examined whether the firing activity of hippocampal neurons was modulated by time. Previous studies have identified time cells using a variety of frameworks such as fitting a Gaussian function to firing activity (Park et al., 2014; Salz et al., 2016), a one-way ANOVA of time and firing rate (Umbach et al., 2020), or a GLM factoring in the influence of time and other experimental factors on firing activity (MacDonald et al., 2011; Tsao et al., 2018). In the current study we elected to use the stepwise GLM method established by Tsao et al. (2018) because it allows us to identify time cells while also measuring the influence of the other experimental parameters on hippocampal responses.
In the stepwise GLM framework used here, a predictor variable is included in the model only if it is found to significantly improve the prediction of the response variable (firing rate). For each neuron, we modeled the firing rate in each image/ISI period of the trial sequence with different potential predictors: image identity, period type (i.e., image or ISI period), and two time terms (a linear and a quadratic time term). A linear combination of the linear and quadratic terms allowed for detecting maximal/minimal firing at all points in the time window of interest. Time cells were identified as those in which one or both of the time terms was selected for inclusion in the stepwise linear model. To determine how likely it is that the temporal modulation of firing rates exhibited by each time cell arose because of chance, we performed a shuffling procedure. Separately, statistical significance for the number of time cells was evaluated using a permutation test in which the image and ISI periods on each trial were randomly shuffled on 106 iterations, and the stepwise GLM was performed on each iteration (see above, Materials and Methods).
We identified a significant number of hippocampal neurons (30%) that were modulated by time during sequence learning (128 of 429 neurons passed the shuffling test, and this number was more than expected by chance based on a permutation test, p < 10−6; 106 of these neurons were located in the anterior hippocampus and 22 in the posterior hippocampus). Individual examples of time cells are shown in Figure 3A. The reliability of time cells was evaluated by comparing the regressor values for the time variables on odd/even trials, or the first/second half of trials (Fig. 4A,B). One hundred and seven time cells were modulated by the linear time term, 62 by the quadratic time term, and 41 by both time terms. Of the linearly modulated cells, 32.7% decreased their firing rates and 67.3% increased their firing rates (Fig. 4F). To test for the presence of ramping cells (Tsao et al., 2018), we reran the analysis with a model that excluded the quadratic time term. In this analysis, 27% of hippocampal neurons (114/429) were significantly modulated by the linear time term, with a 94% overlap with the linearly modulated cells identified by the full model. This proportion of ramping cells is similar to a previous study that reported 34.5% of ramping cells in the human hippocampus (Umbach et al., 2020).
Figure 4.
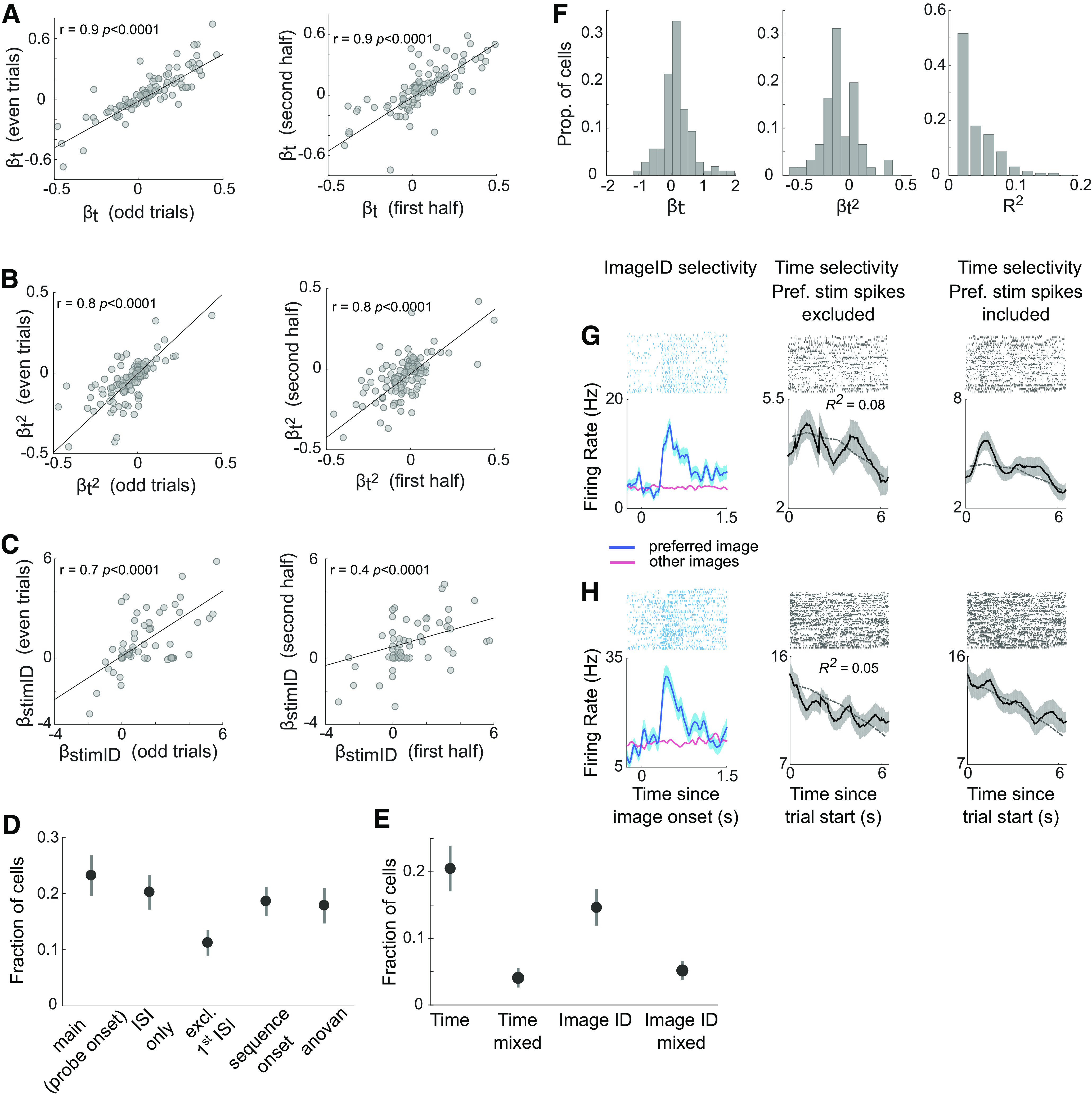
Time cell stability and proportion of time cells identified in control analyses. A–C, Stability of time cells (A, B) and image responsive cells (C) across trials. The trials were split into odd/even trials, or first/second half of trials. The β coefficients for the time and time2 regressors are plotted for the time cells (A, B) and for the imageID regressor for the image responsive cells (C) in each subset of trials. D, Fraction of cells that were modulated by time in the main analysis (i.e., with respect to the offset of the probe event), when time cells were identified on only the ISI periods (ISI only), when the first ISI period was excluded (excl. First ISI), when time cells were identified with respect to the onset of each iteration of the sequence (sequence onset), and with an N-way ANOVA (anovan). E, Fraction of cells that were exclusively modulated by time (Time), modulated by time and another variable (Time mixed, e.g., Time and ImageID), exclusively modulated by image type (ImageID), and modulated by image type and another variable (ImageID mixed). The dots and vertical lines correspond to the mean and SE of the mean across recording sessions respectively (N = 31 sessions). F, Distribution of β coefficients for the linear and quadratic time terms, and R2 values for cells with significant time predictors. G, H, Examples of two cells that showed mixed selectivity for the factors of ImageID and Time. Left, These two cells were modulated by the factor of ImageID. The raster plots and poststimulus time histograms are aligned to the onset of each image in the sequence. The blue curve is the response to the image in the sequence that elicited the highest response (preferred image). The red curve is the mean response to the other images in the sequence. The blue shaded area is the SE of the mean across trials of image presentation. Right, The same two cells were also modulated by the factor of time. The raster plots and poststimulus time histograms are now aligned to trial onset (i.e., the offset of the probe events). The format of the panels on the right is the same as in Figure 3A. For visualization purposes only, and to better disentangle the effects of Time and ImageID, the spikes corresponding to the preferred images have been excised from the middle panels. The gray lines show the model fit (these cells were classified as time cells based on the linear term).
In a separate analysis to confirm our classification of neurons as time cells, we performed a likelihood ratio test to compare the log likelihood values of a restricted linear model that included all terms except the time terms, and a full model that also included the time terms. Time cells were identified as cells that had a significantly higher log likelihood value according to the likelihood ratio test (p < 0.05; 138 cells identified). Eighty-five percent of cells based on this method overlapped with the population identified by the stepwise regression method (Table 1).
We conducted a number of control analyses to verify that the temporal modulation of firing rates was observed under different analysis parameters (Table 1; Fig. 4D). Time cells were identified when the GLM was performed on the firing rates of only the ISI periods (106 cells, permutation test, p < 10−6) and when excluding the first ISI period after the probe (65 cells, permutation test, p < 10−6). Time cells were also detected when trials were redefined as sequence segments with respect to the onset of each repetition of the sequence (i.e., instead of with respect to the onset of the probe events as in the main analysis). In this analysis, we identified 69 neurons that were significantly modulated by the time variables (permutation test, p < 10−6). Finally, time cells were also detected based on an ANOVA (101 cells) as in (Umbach et al., 2020).
Thus, human hippocampal neurons represent a changing temporal context while participants are actively engaged in memorizing the order of a sequence of events. Previous studies have shown that when considered at the population level, the firing activity of time cells covers the duration of a given temporal epoch (Pastalkova et al., 2008; MacDonald et al., 2011). Likewise, across all participants we observed neuronal peak firing at successive moments in time, and when each cell was ordered by its preferred moment of firing, population activity spanned the temporal window (Fig. 3B). For consistency with previous studies, we illustrate these data as a population-level heat map. Although population-level heat maps are primarily used for display purposes, it can be informative to test the reliability of these as random data sorted by peak value could also generate well-organized heat maps. To test the reliability of our heat maps we performed a cross-validation analysis in which the preferred time of firing for each cell was determined in one-half of the data, and the consistency of time preference was measured in the second half of the data. Statistical significance of the cross-validated heat maps was evaluated using a permutation method (see above, Materials and Methods) in which cross-validated and surrogate (randomly permuted) heat maps were correlated with the original heat map. The proportion of surrogates that had a higher correlation than the cross-validated data was p < 10−5, supporting the notion that temporal preference was reliable in our neuronal population.
An advantage of the GLM-based approach used in the present study is that it allowed us to tease apart the influence of different experimental factors on the firing activity of hippocampal neurons (Fig. 4C–E,G,H). The GLM analysis quantified the influence of stimulus presence (i.e., image periods vs ISI periods), image identity, and time in the trial. A considerable number of neurons was modulated exclusively by time (111 cells) or image identity (50 cells), but we also found neurons selective for a combination of these factors (17 neurons for time and another factor; 17 neurons for image identity and another factor).
Internally generated time selectivity in human hippocampal neurons
The results from experiment 1 demonstrate that human hippocampal neurons are modulated by the temporal context of an explicit task. Do human hippocampal neurons also represent the temporal structure of an experience in the absence of external inputs or an overt task?
We performed a second experiment in six new patients, who performed a different version of the sequence-learning task (Fig. 1B). In this new task, participants learned the sequence order as before, but every so often the sequence stopped for 10 s, and participants waited until the sequence resumed. The participants had no stimulus input during these 10 s gap periods—they were presented with a blank screen. We isolated 96 hippocampal neurons in this second experiment and used a GLM approach to determine whether human hippocampal neurons represent temporal information during the gap periods. As before, significance testing was based on a permutation test in which we repeated the GLM 106 times after shuffling the data (see above, Materials and Methods).
During the gap periods, 26 hippocampal neurons (27% of cells; Fig. 5A) were significantly modulated by time (more than expected by chance based on a permutation test, p < 10−6), whereas while the patients were engaged in sequence learning, 13 neurons were time selective. Only three neurons encoded temporal information during both the task period and the gap period, suggesting that the recruitment of temporally sensitive cells can change with task demands or behavior (Pastalkova et al., 2008; MacDonald et al., 2011; Tsao et al., 2018; Umbach et al., 2020). During the gap periods, the time of peak firing in the population occurred at successive moments, and population activity spanned the entire 10 s interval (Fig. 5B). As in experiment 1, the reliability of the heat map was statistically verified with a cross-validation procedure (permutation test p < 10−5; see above, Materials and Methods). As reported in rodents, there was a stronger representation of earlier time points (Salz et al., 2016). To summarize, even in the absence of visual input or an overt task, the firing activity of hippocampal neurons is inherently modulated by a changing temporal context.
Hippocampal population activity encodes temporal information
In the rodent brain, time information is signaled explicitly in individual neurons and can also be gleaned from population-level dynamics of time-selective and non-time-selective cells (Tsao et al., 2018). Is time information also reflected in the population activity of human hippocampal neurons? To address this question, we performed a population pattern analysis of image period identity in the sequence-learning sessions (Fig. 6).
In experiment 1, each trial of the experiment consisted of a sequence of image periods between two consecutive probes, and the goal of the population pattern analysis was to determine whether hippocampal population dynamics reflected the temporal identity of each image period (e.g., discriminate image period 1 vs image period 2). To ensure that decoding or discrimination performance was not driven by an unequal representation of the different images in the different image periods, we created a balanced dataset in which accurate decoding of the temporal identity of each image period cannot arise from merely decoding spurious image information (Fig. 6A). Decoding performance was based on a pairwise comparison method (Fig. 6B–D) as in Haxby et al. (2001) and Pereira et al. (2018); hence, chance performance was 50% for all comparisons.
Discrimination performance was evaluated for different numbers of image periods from the start of the trial (ranging from two to five, i.e., discriminating between the first two image periods after the probe event, the first three image periods and so on). High decoding accuracy for temporal period identity was observed for discriminating all image periods (Fig. 6E; mean ± SD of decoding accuracy for discrimination of the first two periods, 88.4 ± 8%, t(49) = 34.8, p < 0.001; of the first three periods, 72.7 ± 8%, t(49) = 20.1, p < 0.001; of the first four periods, 63.5 ± 6%, t(49) = 14.9, p < 0.001; and of the first five periods, 57.3 ± 9%, t(49) = 5.6, p < 0.001). Decoding errors occurred primarily for neighboring periods (Fig. 6F). Decoding accuracy could not be biased by the offset of the probe events because the first ISI period after the probe event was excluded in the population pattern analysis. Thus, hippocampus population dynamics uniquely represented each temporal period.
To compare how temporal information is represented for time cells versus nontime cells (other cells), and to determine how population size affects decoding accuracy, we performed the pairwise pattern analysis separately for time cells and other cells and for size-matched cell populations. As expected, decoding performance was consistently higher for time cells, but nontime cells also showed significant decoding, suggesting that these cells encode temporal information but possibly in a form that is not detected when modeled with a combination of linear and quadratic time terms or when only present in the population code (Fig. 6H).
Temporal epoch information was also present in population-level dynamics during the gap periods. The gap periods were split into 4, 5 or 10 discrete epochs and a discrimination of temporal epoch identity was performed. A population pattern analysis during these gap periods revealed a significant representation of time information at the level of the overall population, and decoding errors mainly occurred for neighboring epochs (Fig. 6I,J). High decoding accuracy for temporal epoch identity was observed for different temporal epoch sizes (mean accuracy ± SD for four-way decoding, 76.0 ± 4.7%, t(49) = 38.9, p < 0.001; for five-way decoding, 69.6 ± 4.6%, t(49) = 29.9, p < 0.001; and for 10-way decoding = 65.4 ± 3.1%, t(49) = 36.3, p < 0.001). Decoding accuracy during the gap periods could not have been driven by external visual input or overt behavior; rather the high decoding accuracy reflects an internally generated temporal context signal represented in the population of neurons.
Discussion
In this study we report that human hippocampal neurons represent temporal information as subjects progress through a sequence of events and during empty gap periods in the sequence. Time cells responded successively at different moments of the task, and together, the activity of these neurons covered the entire task period. Population-level activity allowed for successful decoding of temporal epoch identity.
Temporal information is represented at different time scales in the hippocampus and neighboring brain regions. In rodents, hippocampal time cells typically show sharp tuning for particular moments of a fixed interval (Pastalkova et al., 2008; MacDonald et al., 2011), whereas in the lateral entorhinal cortex, temporally sensitive neurons show a more gradual ramping of firing activity (Tsao et al., 2018). In humans, population activity gradually drifts over a period of minutes (Folkerts et al., 2018), but single neurons can also show punctuated time-cell-like firing patterns as we show here and as previously reported (Umbach et al., 2020). The relationship between a gradually changing temporal context versus a more precise representation of time in the human hippocampus remains an open question for future work. Task demands such as free recall versus precise temporal order judgments and the resolution at which temporal information is relevant at the behavioral level may influence the temporal precision of human time cells.
Rodent time cells display flexibility in temporal coding. For instance, they exhibit scalar coding of time so that cells that are active later in a time window fire for longer periods. Further, just like place cells remap, time cells have been observed to retime: cells can change temporal preferences within the same recording session when the temporal structure of the experience is changed (MacDonald et al., 2011). For our patients, the image sequence was generally fixed across sessions, and patients typically performed only one experimental session per day. We were thus unable to test whether temporal preferences change when the image order of the sequence changes or when the same sequence is repeated after a short interval. Observations of retiming in rodent time cells raise intriguing possibilities for future investigations of temporal coding in the human hippocampus.
Perhaps most relevant to the current study are recent findings of Umbach et al. (2020). That study reported time cell activity in the human medial temporal lobe during a free recall memory task. Hippocampal neurons encoded temporal information during the task, and entorhinal cortex neurons showed ramping activity. Unlike our task design, which consisted of a structured and predictable sequence of items that repeated continually for 10–15 min, the free recall task in Umbach et al. consisted of two distinct phases, one in which words were presented in a structured list, and another that consisted of a period of free recall. Both studies report a significant number of time cells, although we report a higher proportion (30%) compared with Umbach et al.'s study (15%). This difference cannot solely be because of the different methodology used in the two studies for detecting time cells (stepwise regression versus a one-way ANOVA) because we identified ∼24% of time cells when using an ANOVA as our model. Instead, it seems likely that the different proportions of time cells could arise from differences in task structure or behavioral requirements. For instance, the hippocampus is essential for remembering the temporal order of sequential events (Eichenbaum, 2013), and it is possible that temporal order learning engages a larger population of cells for representing time information. The study by Umbach et al. also showed that time cell activity was relevant to subsequent performance on a memory test. Our study did not directly test for the relationship between time cell firing and behavioral performance as the task was expressly designed to be easy for patients to perform. Thus, we did not have enough error trials to correlate time cell activity with performance on the sequence memory task. Nevertheless, despite these differences in task design and methodology, both the study by Umbach et al. and our study report a significant proportion of hippocampal neurons that are modulated by time, suggesting that a temporal code may be a general property of the human hippocampus network.
The temporal modulation of time cells in our study was also observed during empty 10 s gap periods in which patients were not presented with any visual input or required to perform an explicit task. Time cells were observed to fire at successive moments in these blank periods. Temporal modulation during these gap periods could not have been driven by external events; rather, they appear to represent an evolving temporal signal as a result of changes in the patients' experience during this time of waiting. Related to this point, temporal coding in the hippocampus and the lateral entorhinal cortex has been shown to change with behavior and task demands (MacDonald et al., 2013; Tsao et al., 2018). The design of experiment 2 allowed us to directly examine whether cells that were modulated by time during sequence learning were also recruited during the gap periods when the patients did not overtly perform a task. We found that different subsets of cells were active during these two task periods, with a small overlap of cells that were active in both periods. These results suggest a context-dependent recruitment of cells for the representation of temporal information and raise interesting questions for future research of how the encoding of temporal information in single cells and at the population level is affected when switching between tasks or experiences.
We found evidence for time modulation in various control analyses that considered the trial structure (i.e., sequence time vs probe time), or trial contents (e.g., with or without image information). Interestingly, we found that excluding the first ISI period of each trial from the analysis resulted in an important difference in the number of time cells identified (Table 1). This finding, along with the heat maps shown in Figures 3B and 5B, suggests that a large number of cells show firing rate changes shortly after the probe, similar to reports in rodents and humans of a stronger representation of temporal information early in the trial (Salz et al., 2016; Umbach et al., 2020). Importantly, however, this early representation of temporal information did not compromise population-level representations of elapsed time as our decoding analyses, performed on image periods only, showed robust temporal information at the population level.
In rodents, time cells and place cells do not uniquely represent temporal and spatial information respectively. Rather, medial temporal lobe neurons can be influenced by various experimental factors, including the stimulus-related, spatial and temporal facets of an experience (Komorowski et al., 2009; Tsao et al., 2018). For example, in rodents it has been reported that place cells can code for distance, time, or visual cues (Ravassard et al., 2013; Eichenbaum, 2014; Acharya et al., 2016), and time cells can encode spatial or stimulus information (Eichenbaum, 2014; Tsao et al., 2018). Similarly, we found that human time cells also encoded sensory information about the presence or absence of a stimulus and the identity of the stimulus. Such multidimensional representations could play a critical role in episodic memory mechanisms in which the what, where, and when elements of an experience are bound together into a coherent memory.
The phenomenon of subjective mental time travel is a cornerstone of episodic memory (Tulving, 2002). Central to our experience of reliving the past is our ability to vividly recall specific events that occurred at a specific place and in a specific temporal order. Time cells in rodents and humans, and other temporally sensitive populations of neurons, support theoretical frameworks that suggest that temporal context information plays an important role in memory mechanisms in the hippocampus (Howard et al., 2014, 2015). Our results provide further evidence that human hippocampal neurons represent the flow of time in an experience.
Footnotes
This work was supported by grants from the French Agence Nationale de la Recherche (ANR-12-JSH2-0004-01 and ANR-18-CE37-0007-01), Artificial and Natural Intelligence Toulouse Institute Research Chair (ANR-19-PI3A-0004), the Fyssen Foundation, and the Université Paul Sabatier (Bonus Qualité Recherche, 2009, and Appel à Projets de Recherche Labellisés, 2013), to L.R. A Dutch Research Council Onderzoekstalent Grant awarded to J.K.P and M.W.S and a Dutch Research Agenda Startimpuls Grant awarded to M.W.S. We thank Pieter Roelfsema for support.
The authors declare no competing financial interests.
References:
- Acharya L, Aghajan ZM, Vuong C, Moore JJ, Mehta MR (2016) Causal influence of visual cues on hippocampal directional selectivity. Cell 164:197–207. 10.1016/j.cell.2015.12.015 [DOI] [PubMed] [Google Scholar]
- Eichenbaum H (2013) Memory on time. Trends Cogn Sci 17:81–88. 10.1016/j.tics.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H (2014) Time cells in the hippocampus: a new dimension for mapping memories. Nat Rev Neurosci 15:732–744. 10.1038/nrn3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom AD, Bookheimer SY (2007) Spatial and temporal episodic memory retrieval recruit dissociable functional networks in the human brain. Learn Mem 14:645–654. 10.1101/lm.575107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AK, Moll CK, Fried I, Ojemann GA (2005) Invasive recordings from the human brain: clinical insights and beyond. Nat Rev Neurosci 6:35–47. 10.1038/nrn1585 [DOI] [PubMed] [Google Scholar]
- Folkerts S, Rutishauser U, Howard MW (2018) Human episodic memory retrieval is accompanied by a neural contiguity effect. J Neurosci 38:4200–4211. 10.1523/JNEUROSCI.2312-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin NJ, Agster KL, Eichenbaum HB (2002) Critical role of the hippocampus in memory for sequences of events. Nat Neurosci 5:458–462. 10.1038/nn834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried I, MacDonald KA, Wilson CL (1997) Single neuron activity in human hippocampus and amygdala during recognition of faces and objects. Neuron 18:753–765. 10.1016/s0896-6273(00)80315-3 [DOI] [PubMed] [Google Scholar]
- Gelbard-Sagiv H, Mukamel R, Harel M, Malach R, Fried I (2008) Internally generated reactivation of single neurons in human hippocampus during free recall. Science 322:96–101. 10.1126/science.1164685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmo ME (2009) A model of episodic memory: mental time travel along encoded trajectories using grid cells. Neurobiol Learn Mem 92:559–573. 10.1016/j.nlm.2009.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P (2001) Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science 293:2425–2430. 10.1126/science.1063736 [DOI] [PubMed] [Google Scholar]
- Haxby JV, Guntupalli JS, Connolly AC, Halchenko YO, Conroy BR, Gobbini MI, Hanke M, Ramadge PJ (2011) A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron 72:404–416. 10.1016/j.neuron.2011.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MW, Viskontas IV, Shankar KH, Fried I (2012) Ensembles of human MTL neurons “jump back in time” in response to a repeated stimulus. Hippocampus 22:1833–1847. 10.1002/hipo.22018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MW, MacDonald CJ, Tiganj Z, Shankar KH, Du Q, Hasselmo ME, Eichenbaum H (2014) A unified mathematical framework for coding time, space, and sequences in the hippocampal region. J Neurosci 34:4692–4707. 10.1523/JNEUROSCI.5808-12.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MW, Shankar KH, Aue WR, Criss AH (2015) A distributed representation of internal time. Psychol Rev 122:24–53. 10.1037/a0037840 [DOI] [PubMed] [Google Scholar]
- Komorowski RW, Manns JR, Eichenbaum H (2009) Robust conjunctive item-place coding by hippocampal neurons parallels learning what happens where. J Neurosci 29:9918–9929. 10.1523/JNEUROSCI.1378-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus BJ, Robinson RJ 2nd, White JA, Eichenbaum H, Hasselmo ME (2013) Hippocampal “time cells”: time versus path integration. Neuron 78:1090–1101. 10.1016/j.neuron.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus BJ, Brandon MP, Robinson RJ 2nd, Connerney MA, Hasselmo ME, Eichenbaum H (2015) During running in place, grid cells integrate elapsed time and distance run. Neuron 88:578–589. 10.1016/j.neuron.2015.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald CJ, Lepage KQ, Eden UT, Eichenbaum H (2011) Hippocampal “time cells” bridge the gap in memory for discontiguous events. Neuron 71:737–749. 10.1016/j.neuron.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald CJ, Carrow S, Place R, Eichenbaum H (2013) Distinct hippocampal time cell sequences represent odor memories in immobilized rats. J Neurosci 33:14607–14616. 10.1523/JNEUROSCI.1537-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns JR, Howard MW, Eichenbaum H (2007) Gradual changes in hippocampal activity support remembering the order of events. Neuron 56:530–540. 10.1016/j.neuron.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IM, Meister ML, Huk AC, Pillow JW (2014) Encoding and decoding in parietal cortex during sensorimotor decision-making. Nat Neurosci 17:1395–1403. 10.1038/nn.3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastalkova E, Itskov V, Amarasingham A, Buzsáki G (2008) Internally generated cell assembly sequences in the rat hippocampus. Science 321:1322–1327. 10.1126/science.1159775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F, Lou B, Pritchett B, Ritter S, Gershman SJ, Kanwisher N, Botvinick M, Fedorenko E (2018) Toward a universal decoder of linguistic meaning from brain activation. Nat Commun 9:963. 10.1038/s41467-018-03068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga RQ, Nadasdy Z, Ben-Shaul Y (2004) Unsupervised spike detection and sorting with wavelets and superparamagnetic clustering. Neural Comput 16:1661–1687. 10.1162/089976604774201631 [DOI] [PubMed] [Google Scholar]
- Ravassard P, Kees A, Willers B, Ho D, Aharoni DA, Cushman J, Aghajan ZM, Mehta MR (2013) Multisensory control of hippocampal spatiotemporal selectivity. Science 340:1342–1346. 10.1126/science.1232655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy L, Poncet M, Self MW, Peters JC, Douw L, van Dellen E, Claus S, Reijneveld JC, Baayen JC, Roelfsema PR (2015) Learning of anticipatory responses in single neurons of the human medial temporal lobe. Nat Commun 6:8556. 10.1038/ncomms9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz DM, Tiganj Z, Khasnabish S, Kohley A, Sheehan D, Howard MW, Eichenbaum H (2016) Time Cells in Hippocampal Area CA3. J Neurosci 36:7476–7484. 10.1523/JNEUROSCI.0087-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers HJ, Burgess N, Hartley T, Vargha-Khadem F, O'Keefe J (2001) Bilateral hippocampal pathology impairs topographical and episodic memory but not visual pattern matching. Hippocampus 11:715–725. 10.1002/hipo.1087 [DOI] [PubMed] [Google Scholar]
- Tsao A, Sugar J, Lu L, Wang C, Knierim JJ, Moser MB, Moser EI (2018) Integrating time from experience in the lateral entorhinal cortex. Nature 561:57–62. 10.1038/s41586-018-0459-6 [DOI] [PubMed] [Google Scholar]
- Tulving E (2002) Episodic memory: from mind to brain. Annu Rev Psychol 53:1–25. 10.1146/annurev.psych.53.100901.135114 [DOI] [PubMed] [Google Scholar]
- Umbach G, Kantak P, Jacobs J, Kahana M, Pfeiffer BE, Sperling M, Lega B (2020) Time cells in the human hippocampus and entorhinal cortex support episodic memory. Proc Natl Acad Sci U S A 117:28463–28474. 10.1073/pnas.2013250117 [DOI] [PMC free article] [PubMed] [Google Scholar]