Abstract
Cell survival, tissue integrity and organismal health depend on the ability to maintain functional protein networks even under conditions that threaten protein integrity. Protection against such stress conditions involves the adaptation of folding and degradation machineries, which help to preserve the protein network by facilitating the refolding or disposal of damaged proteins. In multicellular organisms, cells are permanently exposed to stress resulting from mechanical forces. Yet, for long time mechanical stress was not recognized as a primary stressor that perturbs protein structure and threatens proteome integrity. The identification and characterization of protein folding and degradation systems, which handle force‐unfolded proteins, marks a turning point in this regard. It has become apparent that mechanical stress protection operates during cell differentiation, adhesion and migration and is essential for maintaining tissues such as skeletal muscle, heart and kidney as well as the immune system. Here, we provide an overview of recent advances in our understanding of mechanical stress protection.
Keywords: autophagy, chaperones, mechanobiology, proteostasis, signal transduction
Subject Categories: Autophagy & Cell Death; Post-translational Modifications, Proteolysis & Proteomics; Protein Biosynthesis & Quality Control
Cells are constantly subjected to mechanical stress. This review discusses how chaperone and protein degradation systems handle force‐unfolded proteins to preserve protein homeostasis in mechanically stressed cells and tissues.
Glossary
- ABD
actin‐binding domain
- AJ
adherens junction
- CASA
chaperone‐assisted selective autophagy
- CIM
critical illness myopathy
- CMA
chaperone‐mediated autophagy
- ECM
extracellular matrix
- FA
focal adhesion
- FAK
focal adhesion kinase
- HSP
heat shock protein
- ICU
intensive care unit
- Ig
immunoglobulin
- mTORC1
mechanistic target of rapamycin complex I
- PDZ
PSD‐95/Dlg1/ZO‐1 domain
- SD
slit diaphragm
- sHSP
small heat shock protein
- TPR
tetratricopeptide repeat
- TSC
tuberous sclerosis complex
- UCS
UNC‐45/CRO1/She4p domain
- UnDOx
unfolded domain oxidation
- UPS
ubiquitin–proteasome system
Introduction
In a living organism, the integrity of the cellular protein network is under constant threat. Increased temperature, the generation of reactive oxygen species or the accumulation of aberrant proteins lead to heat, oxidative and proteotoxic stress, which shifts the conformational equilibrium of proteins towards partially unfolded and misfolded states prone to aggregation (Richter et al, 2010; Dahl et al, 2015; Sala et al, 2017; Hipp et al, 2019). Protein aggregation can interfere with essential cellular processes causing severe pathology including neurodegeneration and dementia (Douglas & Dillin, 2010; Choe et al, 2016; Woerner et al, 2016; Hipp et al, 2019). To protect the proteome against stressful conditions, the cell employs protein folding and degradation systems. These highly regulated systems recognize partially unfolded and misfolded conformers and promote refolding or, in case of terminal damage, mediate disposal (Fig 1A) (Buchberger et al, 2010; Balchin et al, 2016; Cohen‐Kaplan et al, 2016; Dikic, 2017; Klimek et al, 2017; Nillegoda et al, 2018). Key players in stress protection are molecular chaperones and their regulatory cochaperones. They sense protein unfolding based on the exposure of hydrophobic surfaces that are otherwise buried in the native structure (Kim et al, 2013b; Dahiya & Buchner, 2019; Rosenzweig et al, 2019). Chaperone binding prevents aggregation and directs the non‐native client onto specific folding or degradation pathways. Protein degradation can be mediated by the ubiquitin–proteasome system (UPS) or selective autophagy (Cohen‐Kaplan et al, 2016; Dikic, 2017; Höhfeld & Hoppe, 2018). In both cases, degradation is usually initiated by the attachment of a ubiquitin‐derived degradation signal onto the client, based on the activity of client‐specific ubiquitin conjugation systems (Fig 1A) (Khaminets et al, 2016; Gatica et al, 2018). Depending on the selected degradation pathway, the generated degradation signal is recognized either by ubiquitin receptors in the proteasome or by autophagic ubiquitin adaptors that facilitate an association of the ubiquitylated client with phagophore membranes leading to autophagosome formation and client degradation in autolysosomes. An extensive body of work has elucidated the adaptation of cellular folding and degradation machineries in response to heat, oxidative and proteotoxic stress (Richter et al, 2010; Vihervaara & Sistonen, 2014; Dahl et al, 2015; Sala et al, 2017; Nillegoda et al, 2018; Dahiya & Buchner, 2019; Hipp et al, 2019; Rosenzweig et al, 2019). In contrast, the impact of mechanical stress on these machineries became only recently apparent, when force‐regulated proteostasis systems were identified and shown to be essential for cell and tissue homeostasis (Janiesch et al, 2007; Arndt et al, 2010; Gazda et al, 2013; Ulbricht et al, 2013, 2015; Kathage et al, 2017; Rinschen et al, 2017b; Donkervoort et al, 2020).
Figure 1. Protecting the proteome against mechanical stress relies on protein folding and degradation systems that recognize force‐unfolded proteins.

(A) Stress protection systems comprise chaperone/cochaperone complexes and specialized E2/E3 ubiquitin conjugation systems, which recognize unfolded proteins under stress conditions to facilitate their refolding or their degradation by the proteasome and autophagic/lysosomal pathways. Ubiquitylation can be assisted by molecular chaperones or can proceed through a direct recognition of unfolded proteins by quality control E3 ubiquitin ligases. Stress‐induced signalling pathways regulate the activity of the involved folding and degradation systems. (B) Cellular systems are permanently exposed to a wide variety of mechanical signals. After recognition and transmission through cell–cell and cell–matrix contacts as well as cytoskeletal systems, these signals induce a variety of specific cell responses. (C) Mechanical forces can trigger the unfolding of mechanosensory proteins such as talin, which links integrin‐containing adhesion complexes in the plasma membrane (PM) to the actin cytoskeleton (ECM—extracellular matrix).
Mechanical stress is a highly relevant physiological stimulus
Cells in multicellular organisms are permanently exposed to mechanical forces, which are either passively applied or generated inside the cell (Discher et al, 2009; Irianto et al, 2016; Hu et al, 2017; Wolfenson et al, 2019) (Fig 1B). Indeed, the ability to generate and respond to mechanical forces is a basic requirement for the development, life and survival of organisms (Alon & Dustin, 2007; Johnson et al, 2007; Moore et al, 2010; DuFort et al, 2011; Hoffman et al, 2011; Mammoto et al, 2013; Yusko & Asbury, 2014). To stand and walk, to take a breath, or to pump blood through the body depends on force generation in skeletal and cardiac muscle. Cells in the skin, the cardio‐vascular system and the kidney must withstand mechanical forces under touch and during blood circulation and filtration. Immune cells are subjected to mechanical forces when they attach to blood vessel walls upon exit from the vasculature and during migration to sites of infection and inflammation. Finally, the division, migration, functionality and differentiation of cells rely on their ability to sense the mechanical properties of their surrounding such as tissue elasticity and matrix stiffness (Engler et al, 2006; Gilbert et al, 2010; Hersch et al, 2013; Wolfenson et al, 2019). Impairment of these processes results in diverse diseases, including myopathies, heart and kidney failure, leukocyte adhesion deficiencies and cancer (Etzioni, 2007; DuFort et al, 2011; Hoffman et al, 2011; Yang et al, 2015; Ayad et al, 2019; Maurer & Lammerding, 2019).
The actin cytoskeleton is centrally involved in generating and responding to mechanical forces (Small & Resch, 2005; Johnson et al, 2007; Luo et al, 2013; Schiller & Fässler, 2013; Gautel & Djinović‐Carugo, 2016; Schiffhauer et al, 2016). Comprised of globular actin molecules, which polymerize into a dynamic filament network, it provides shape and mechanical stability to cells, generates mechanical force and drives cell migration. Actin‐anchoring sites are of particular importance for force generation and force transduction. At the plasma membrane, proteins of the integrin and cadherin families together with their interaction partners form cell adhesions that link the actin cytoskeleton to the extracellular matrix and neighbouring cells (Fig 1B and C) (Schiller & Fässler, 2013; Irianto et al, 2016; Schiffhauer et al, 2016). At the nucleo‐cytoskeletal interface, the actin cytoskeleton contacts the nuclear envelope to impact genome functions (Simon & Wilson, 2011; Isermann & Lammerding, 2013; Graham & Burridge, 2016; Cho et al, 2017; Nava et al, 2020). Furthermore, actin filaments are anchored to the constituents of the blood filtration barrier at the slit diaphragm in the kidney (Huber & Benzing, 2005; New et al, 2014), and in skeletal and cardiac muscle, actin anchoring is mediated by Z‐discs, which limit the minimal contractile units of muscle, the sarcomeres (Fig 2) (Frank et al, 2006). Mechanical forces, externally applied through blood pressure and movement or internally generated through actomyosin contraction, accumulate at anchoring sites leading to a high stress concentration.
Figure 2. The UNC‐45‐containing chaperone system mediates the folding and assembly of myosin in muscle sarcomeres.
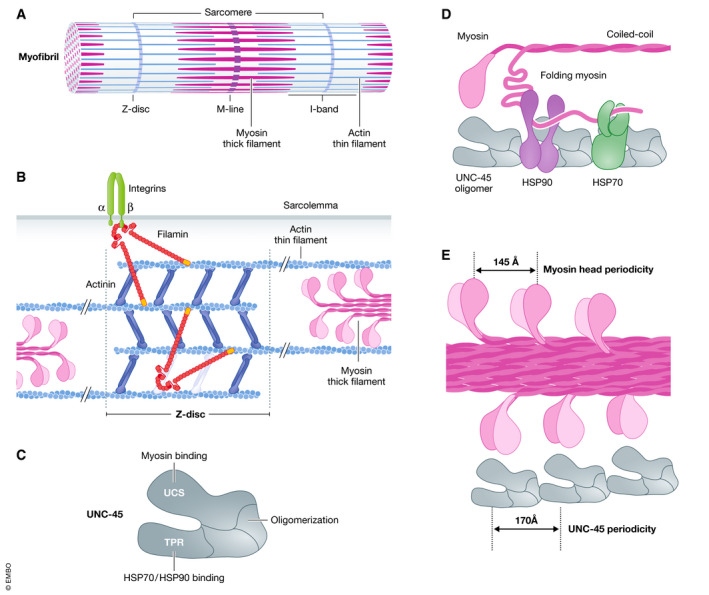
(A) The sarcomere represents the smallest contractile unit of striated muscles. It is repetitively arranged in tubular myofibrils, numerous bundles of which form the muscle fibre. Z‐discs limit the sarcomere on both sides and mediate the anchoring of actin thin filaments. Myosin thick filaments are intercalated between the actin filaments and are connected at the M‐line. The I‐band is the region that contains exclusively actin filaments. (B) Actin and filamin crosslink actin filaments within the Z‐disc. In addition, filamin interacts with integrin molecules in the sarcolemma. (C) Schematic representation of the domain structure of the cochaperone UNC‐45. UCS, UNC‐45/CRO1/She4p domain and TPR, tetratricopeptide repeat. (D) Oligomeric UNC‐45 coordinates the activity of HSP70 and HSP90 chaperone proteins during the folding and assembly of myosin filaments. (E) The UNC‐45 oligomer provides a molecular scaffold for enforcing the regular spacing of folded myosin head domains in the myosin thick filament.
Force‐induced protein unfolding
Many constituents of actin‐anchoring sites undergo force‐induced unfolding (Hu et al, 2017; Saini & Discher, 2019). One such protein is vinculin, which links the actin cytoskeleton to integrin complexes at cell–matrix adhesions (focal adhesions, FAs) and to cadherin complexes at cell–cell junctions (adherens junctions, AJs) (Atherton et al, 2016; Goldmann, 2016). Mechanical force disrupts auto‐inhibitory interactions between the head and tail domains of vinculin and in this way stimulates binding to other anchoring components and actin filaments (Atherton et al, 2016). At FAs, vinculin interacts with talin, which is also a mechanosensitive protein (Fig 1C) (Austen et al, 2015; Ringer et al, 2017; Lemke et al, 2019; Fischer et al, 2021). Remarkably, mechanical forces do not just induce a domain rearrangement, but instead trigger the loss of tertiary and secondary structural elements within talin (del Rio et al, 2009; Yao et al, 2014a, 2016). Talin possesses an amino‐terminal integrin binding domain connected to a rod‐like structure formed by 13 α‐helical bundles, the last of which contacts actin. When talin is stretched in cells, tensile forces are exerted linearly along the molecule (Fig 1C) (Ringer et al, 2017). The mechanical properties of talin domains were analysed in biochemical experiments using magnetic tweezers, total internal reflection fluorescence and atomic force microscopy (del Rio et al, 2009; Yao et al, 2014a, 2016). These experiments demonstrated that 12 out of the 13 helical bundles undergo force‐induced unfolding, leading to a disruption of helix–helix interactions and the complete unfolding of individual helices. In turn, buried vinculin binding sites become exposed. Talin unfolding thus contributes to the transduction of mechanical signals. Importantly, the force required for the unfolding of different helical bundles varies between 5 and 20 pN (Yao et al, 2016). This suggests that the talin rod acts as a force buffer. Integrin–actin contacts are maintained by talin over a significant force range through the subsequent unfolding of single helical bundles (Fig 1C).
Another proposed mechanosensor and interactor of various FA proteins (i.e. vinculin and FAK) is the stretch‐sensitive adaptor protein p130Cas (Sawada et al, 2006). Following early FA maturation, p130Cas is recruited to FAs, unfolds due to applied tension and exposes up to 15 formerly hidden phosphorylation sites (Sawada et al, 2006). Phosphorylated p130Cas engages in different mechanosensitive signalling cascades, in agreement with a role of the protein as a central hub for force transmission. Moreover, the interaction of p130Cas with vinculin has been proposed to stabilize an open conformation of vinculin, thereby promoting talin binding and FA stability (Janoštiak et al, 2014). Finally, p130Cas has been shown to be important for actin reorientation in response to cyclic mechanical stretch (Niediek et al, 2012).
Importantly, mechanical forces are transduced not only at the cell–matrix interface but also at the cell–cell contacts through AJs. AJs contain α‐catenin as a main mechanosensor (Huveneers & de Rooij, 2013; Yao et al, 2014b; Ladoux et al, 2015). α‐catenin physically couples the cadherin complex, the core component of AJs, to the actin cytoskeleton. Loss of α‐catenin function disrupts epithelial integrity, the ability of cells to undergo coordinated cell movements and the ability to reorient actin fibres under cyclic strain (Noethel et al, 2018). α‐catenin comprises three evolutionary conserved regions: an N‐terminal region that binds to β‐catenin, a central M‐region and a C‐terminal actin‐binding domain (ABD). Opening of the ABD under tension results in enforced actin binding and facilitates dimerization of the ABD to promote actin bundling (Buckley et al, 2014; Ishiyama et al, 2018). In addition, also the M‐region of α‐catenin undergoes a conformational change when stretched to reveal a formerly hidden binding site for vinculin (Yonemura et al, 2010; Choi et al, 2012; Yao et al, 2014b). The recruitment of vinculin is thought to strengthen the AJ‐actin link and seems to enhance the mechanosensitivity of α‐catenin (Noethel et al, 2018).
A function as mechanotransducer and force buffer was also assigned to filamin, which binds actin along stress fibres, at focal adhesions and at the sarcomeric Z‐disc (Fig 2B) (Frank et al, 2006; Ehrlicher et al, 2011; Nakamura et al, 2011, 2014). Filamin is a large homodimeric protein comprised of two rods with terminal actin and integrin binding sites, respectively, on opposing ends. This domain arrangement enables the protein to crosslink and anchor actin filaments. The filamin rod is formed by 24 immunoglobulin (Ig) domains, some of which engage in pairwise interactions (Nakamura et al, 2011). Single‐molecule mechanical measurements demonstrated that the Ig domain pair 20‐21 of filamin functions as an auto‐inhibited force‐activatable mechanosensor (Lad et al, 2007; Rognoni et al, 2012). In the inhibited state, the amino‐terminal ß‐strand of domain 20 folds onto domain 21. Mechanical force disrupts this interaction, leading to rod extension and the exposure of binding sites in domain 21 for integrins and other partner proteins. Molecular dynamics simulation suggests that also other Ig domains of filamin are sensitive to mechanical force (Kesner et al, 2010). Thereby, unfolding of the amino‐terminal ß‐strand seems to represent a common initial step, resulting in metastable states similar to those observed upon thermal unfolding (Kesner et al, 2010).
The gigantic myofilament protein titin (Mr: ˜3,800 kDa) spans the muscle sarcomere from the Z‐disc to the M‐band and confers elasticity and passive force under stretch (Linke & Hamdani, 2014). Titin regions localized in the I‐band adjacent to the Z‐disc function as molecular springs that extend and condense as the muscle stretches and contracts. At low forces, regions containing tandemly arranged Ig domains elongate due to the straightening of interdomain linkers, whereas at higher forces, intrinsically disordered structures are extended (Linke & Hamdani, 2014; Mártonfalvi et al, 2014).
As already mentioned above, talin unfolding is observed at forces between 5 and 20 pN (Yao et al, 2016). Unfolding of the Ig domain 20–21 pair of filamin is triggered at forces between 2 and 4 pN (Rognoni et al, 2012), and conformational transitions of titin occur upon stretching with forces as low as 5 pN (Mártonfalvi et al, 2014). To put this into perspective, one should note that the contraction of a single actin–myosin unit generates a force of 3–4 pN (Finer et al, 1994). Thus, cytoskeleton components constantly undergo unfolding reactions under physiological conditions. Indeed, studies with cultured cells illustrate the prevalence and significance of force‐induced protein unfolding in the cellular context. In one approach, the accessibility of cysteine residues was monitored (Johnson et al, 2007). Cysteines are often buried within tertiary and/or quaternary structure, and their force‐induced exposure is therefore a readout for a significant loss of structural elements. Red blood cells were subjected to fluid shear stress to determine the force‐induced unfolded proteome, and also adherent mesenchymal stem cells were analysed, in which traction forces are generated based on actomyosin contraction (Johnson et al, 2007). In both cell types, mechanical stress caused unfolding of the actin‐associated scaffolding protein spectrin, confirming previous biochemical studies on the mechanosensitive properties of the cytoskeleton protein (Rief et al, 1999; Altmann et al, 2002; Johnson et al, 2007). Moreover, in adherent mesenchymal stem cells traction forces resulted in the unfolding of non‐muscle myosin and filamin (Johnson et al, 2007). The latter was also at the focus of another study, in which a fluorescence resonance energy transfer (FRET)‐based sensor was constructed to visualize the disruption of the Ig domain 20–21 pair of filamin in adherent mammalian cells (Nakamura et al, 2014). Maximal opening of filamin occurred predominantly at the cell edge and in protruding areas following pharmacological stimulation of cell spreading and migration.
Taken together, the described findings demonstrate that cells of a multicellular organism are challenged continuously by mechanical protein unfolding. Even in the absence of externally applied force, the tension generated inside adherent and migrating cells is sufficient to alter the conformation of abundant cytoskeleton proteins, leading to a significant loss of structural elements. This sets the stage for a critical involvement of protein folding and degradation machineries, which recognize force‐unfolded proteins during mechanical stress protection.
Proteostasis machineries involved in mechanical stress protection
Molecular chaperones are defined by the ability to bind and stabilize unfolded proteins (see Fig 1A) (Hartl et al, 2011; Dahiya & Buchner, 2019; Rosenzweig et al, 2019). This enables them to assist in the folding, sorting and degradation of proteins, often in cooperation with regulating cochaperones. Small heat shock proteins (sHSPs), members of the HSP70 and HSP90 chaperone families as well as diverse cochaperones of HSP70 and HSP90, have been linked to mechanical stress protection (Arndt et al, 2010; Sarparanta et al, 2012; Gazda et al, 2013; Ulbricht et al, 2013; Adriaenssens et al, 2017; Jacko et al, 2020).
The HSP90 chaperone system is essential for muscle assembly in worms, fish, mice and men (Janiesch et al, 2007; Du et al, 2008; Gaiser et al, 2011; Donlin et al, 2012; Gazda et al, 2013; Echeverría et al, 2016). In zebrafish, for example, HSP90α is abundantly expressed in skeletal muscle and its knockdown results in paralysed animals with massive perturbation of the myofibrillar organization of skeletal muscle (Etard et al, 2007; Du et al, 2008). The chaperone apparently exerts essential functions in the assembly of muscular structures during development. Yet, with respect to a possible involvement in mechanical stress protection, it is necessary to investigate the maintenance of muscle following, for example, a depletion of HSP90 isoforms in adult organisms. In fact, such an approach was performed in the nematode C. elegans. Adult worms were treated with silencing RNAs directed against the HSP90 homolog DAF‐21, which shows strong expression in body wall muscle cells (Gaiser et al, 2011). Depletion severely reduced motility and caused a profound disruption of muscle structure. The findings demonstrate that HSP90 is required for both the initial assembly of skeletal muscles and their maintenance when force is generated. Furthermore, maintaining muscle structure and activity relies on the cooperation of HSP90 with several of its cochaperones, including the HSP70/HSP90 organizing protein HOP/STI1, AHA1 and p23, which regulate the ATP‐regulated client binding cycle of HSP90, as well as UNC‐45 (Barral et al, 1998; Frumkin et al, 2014; Donkervoort et al, 2020).
What are clients of the HSP90 chaperone system under mechanical stress? Linke and coworkers showed that HSP90 binds to the N2‐A region of cardiac and skeletal titin isoforms (Donlin et al, 2012; Voelkel et al, 2013). This region of titin is intrinsically unstructured and its extension under force contributes to the elasticity of the myofilament protein (Linke & Hamdani, 2014). Abrogating HSP90 recruitment to this region impaired skeletal and cardiac muscle activity in zebrafish and led to severe disorganization of the sarcomeric I‐band, where the N2‐A region is located (Donlin et al, 2012; Voelkel et al, 2013). Thus, HSP90 exerts an essential chaperone function on mechanosensitive domains of titin.
Myosin is another client of HSP90. The chaperone cooperates with HSP70 and its cochaperone UNC‐45 to facilitate myosin folding and to organize sarcomere architecture (Etard et al, 2007; Wohlgemuth et al, 2007; Gazda et al, 2013). The sarcomeric repeat is a supra‐molecular structure, in which thin (actin) and thick (myosin) filaments and their associated proteins are arranged in a precise order that is required to link the formation of actin–myosin cross‐bridges with filament gliding and muscle contraction (Fig 2A and B) (Geeves & Holmes, 2005; Henderson et al, 2017). The C. elegans UNC‐45 protein is the founding member of the conserved UCS (UNC‐45/CRO1/She4p) domain family and orchestrates the activity and assembly of various myosin subtypes (Fig 2C) (Price et al, 2002; Etard et al, 2007; Wohlgemuth et al, 2007; Kim et al, 2008; Gazda et al, 2013). Mutations in UNC‐45 were initially shown to induce myosin disorganization and severe motility defects (Venolia & Waterston, 1990; Barral et al, 1998, 2002; Price et al, 2002). In Drosophila, zebrafish and Xenopus, UNC‐45 homologs are essential for muscle development and maintenance under stress (Etard et al, 2007, 2008; Wohlgemuth et al, 2007; Melkani et al, 2011). In zebrafish, for example, an UNC‐45 homolog plays a role in the formation of myocyte attachment complexes that transmit forces from the contractile apparatus to the extracellular matrix, ensuring mechanical stability of muscles during contraction (Myhre et al, 2014). In mammals, two UNC‐45 homologs exist, indicating an evolutionary conserved requirement for myosin‐specific cochaperones (Price et al, 2002). UNC‐45A represents a broadly expressed isoform, which is necessary for actomyosin bundling and plays a key role in cell adhesion and migration, promoting the proliferation and invasiveness of cancer cells (Bazzaro et al, 2007; Guo et al, 2011; Lee & Kumar, 2016). On the other hand, UNC‐45B is predominantly expressed in striated muscles. Indeed, abnormal UNC‐45B function is associated with severe muscle defects resulting in skeletal and cardiac myopathies (Janiesch et al, 2007; Donkervoort et al, 2020).
UNC‐45 is able to interact with HSP70 and HSP90 simultaneously and to form multimeric linear assemblies (Fig 2D) (Gazda et al, 2013). It thus provides a multisite‐docking platform to assemble myosin filaments in a precisely defined pattern into thick myofilaments and eventually myofibrils (Fig 2E). Importantly, this highly conserved chaperone system is required not only for sarcomere formation during development but also for its maintenance in the contracting muscle (Kim et al, 2008; Gaiser et al, 2011; Pokrzywa & Hoppe, 2013; Donkervoort et al, 2020). In response to stress or damage to the myofibre, HSP90 and UNC‐45 dissociate from the Z‐disc and transiently associate with myosin (Etard et al, 2008), indicating their essential involvement in assembly processes under mechanical stress.
In single‐molecule experiments using atomic force microscopy, UNC‐45 was shown to bind to the myosin motor domain upon force‐induced unfolding and to prevent aberrant protein interactions of the unfolded domain (Kaiser et al, 2012). Thus, stabilization of force‐unfolded myosin appears to contribute to the observed essential activity of UNC‐45 in contracting muscle.
Coping with mechanical stress does not only depend on the induction of chaperone‐based folding and assembly machineries, but damaged proteins also need to be disposed to maintain the integrity of the proteome. Indeed, selective ubiquitin‐dependent autophagy emerged in recent years as a degradation pathway of major importance in mechanical stress protection. First evidence was provided when a mechanically induced interaction of titin with the autophagic ubiquitin adaptor NBR1 was described (Lange et al, 2005). The adaptor protein participates in the selective autophagic removal of pathologic protein aggregates and peroxisomes in non‐muscle cells (Kirkin et al, 2009; Odagiri et al, 2012; Deosaran et al, 2013). Accordingly, the force‐induced recruitment of NBR1 to titin might localize the adaptor at the cytoskeleton for the autophagic disposal of mechanically damaged proteins. Many studies illustrate the importance of autophagy for muscle maintenance (Sandri, 2010). Deletion of key autophagy factors in transgenic animal models leads to a progressive deterioration of muscle structure and activity under mechanical stress, which is accompanied by the accumulation of protein aggregates (Tanaka et al, 2000; Raben et al, 2008; Masiero et al, 2009; He et al, 2012; Kim et al, 2013a). However, observed pathologies were usually attributed to impaired glucose and energy metabolism in the absence of autophagy (Sandri, 2010; He et al, 2012; Kim et al, 2013a). An essential involvement of autophagy in the removal of mechanically unfolded and damaged cytoskeleton proteins became apparent with the functional characterization of the cochaperone BAG3 (Arndt et al, 2010).
BAG3 can interact simultaneously with members of the HSP70 chaperone family and with small heat shock proteins, such as HSPB8 (also known as HSP22) (Carra et al, 2008; Gamerdinger et al, 2009, 2011; Arndt et al, 2010; Crippa et al, 2016; Ganassi et al, 2016; Judge et al, 2017; Rauch et al, 2017) (Fig 3A). Small heat shock proteins form oligomeric chaperone complexes that maintain non‐native proteins in a state competent for processing by other proteostasis factors (Mainz et al, 2015; Ungelenk et al, 2016; Dahiya & Buchner, 2019). BAG3‐mediated chaperone coupling allows to coordinate the holding function of small heat shock proteins with the chaperone function of HSP70 proteins (Rauch et al, 2017). In addition, the BAG3 chaperone complex can contain the HSP70‐associated ubiquitin ligase CHIP, which mediates the ubiquitylation of chaperone‐bound client proteins to initiate their disposal (Fig 3A) (Arndt et al, 2010). When ubiquitylation occurs in association with the BAG3 chaperone complex, CHIP generates a signal for autophagic degradation. It triggers the recruitment of the autophagic ubiquitin adaptor SQSTM1/p62 to stimulate the engulfment of the BAG3 chaperone complex by autophagosome precursor membranes, so‐called phagophores, leading to client degradation in autolysosomes (Fig 3B) (Gamerdinger et al, 2009; Arndt et al, 2010). In muscle cells, autophagosome formation is further facilitated through cooperation of BAG3 with the actin‐associated protein SYNPO2, which links the BAG3 chaperone complex to a phagophore membrane fusion machinery (Ulbricht et al, 2013). The BAG3‐mediated autophagy pathway was termed chaperone‐assisted selective autophagy (CASA) to distinguish it from chaperone‐mediated autophagy (CMA) (Arndt et al, 2010; Kaushik & Cuervo, 2012). In contrast to CASA, CMA does not involve autophagosome formation but instead depends on a chaperone‐mediated direct transfer of client proteins across the lysosomal membrane for degradation (Kaushik & Cuervo, 2012).
Figure 3. The BAG3 chaperone complex initiates a force‐induced autophagy pathway in contracting muscle.
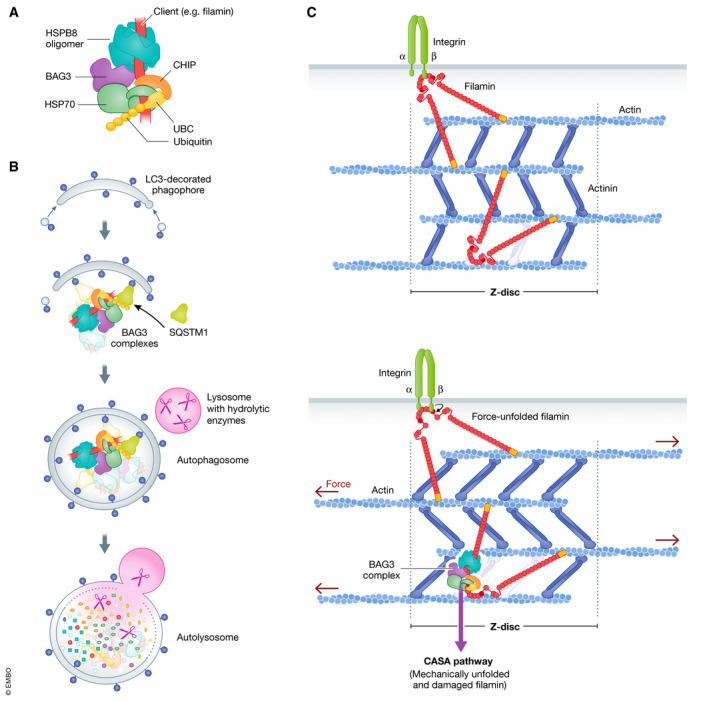
(A) BAG3 links chaperones of the HSP70 family to small heat shock proteins such as HSPB8. The HSP70‐associated ubiquitin ligase CHIP can associate with the BAG3 chaperone complex to ubiquitylate a bound client protein in cooperation with a ubiquitin conjugation enzyme (UBC). (B) Enclosure of the BAG3 chaperone complex by phagophore membranes is facilitated by the autophagic ubiquitin adaptor SQSTM1, leading to autophagosome formation and ultimately to degradation in autolysosomes. (C) Actin–myosin contraction causes the force‐induced unfolding of the actin‐crosslinking protein filamin at the Z‐disc of the muscle sarcomere. Mechanically unfolded and damaged forms of filamin are recognized by the BAG3 complex and directed onto the CASA pathway for disposal.
BAG3 was initially shown to participate in the autophagic degradation of aggregation‐prone, pathologic proteins in neuronal cells and later on also found to be involved in the turnover of the microtubule organizing protein tau in neuronal processes, thus providing an essential proteostasis function in the nervous system (Carra et al, 2008; Gamerdinger et al, 2009, 2011; Crippa et al, 2010; Lei et al, 2015). However, BAG3‐deficient animals and patients carrying BAG3 mutant alleles display a profound pathology in skeletal and cardiac muscle, characterized by a progressive deterioration of sarcomeric structures under mechanical stress (Homma et al, 2006; Selcen et al, 2009; Arndt et al, 2010; Arimura et al, 2011; Claeys et al, 2013; Ruparelia et al, 2014; Konersman et al, 2015; Quintana et al, 2016; Fang et al, 2017; Meister‐Broekema et al, 2018; Schänzer et al, 2018; Kimura et al, 2021). Moreover, a decline in BAG3‐mediated sarcomere turnover contributes to the development of heart failure, the leading cause of mortality in the industrialized world (Martin et al, 2021). Ultimately, the mechanosensitive actin‐crosslinking protein filamin was identified as a target of CASA (Arndt et al, 2010; Ulbricht et al, 2013). In an unbiased biochemical assay using isolated cytoskeleton from differentiated murine muscle cells and purified CASA components, BAG3 stimulated the release of filamin from the cytoskeleton, which can be considered as a requirement for subsequent sorting to the lysosome and autophagic degradation (Arndt et al, 2010). BAG3 and its chaperone partners HSP70 and HSPB8 bind to the mechanosensitive region of filamin comprising Ig domains 19–21 (see above) and direct mechanically unfolded and damaged forms onto the CASA pathway for disposal (Fig 3C) (Arndt et al, 2010; Klimek et al, 2017). Indeed, filamin forms aggregates in skeletal and cardiac muscle when the CASA pathway is compromised under pathological conditions (Arimura et al, 2011; Ruparelia et al, 2016; Fang et al, 2017; Kimura et al, 2021; Martin et al, 2021), illustrating the relevance of CASA for myofilament homeostasis in contracting muscle.
Recognition of mechanically unfolded filamin by the CASA machinery seems to be facilitated by LDB3 (Pathak et al, 2021). Like SYNPO2, LDB3 is a cytoskeleton‐associated PDZ domain protein and acts as a multivalent interaction hub. It directly binds to the mechanosensitive region of filamin and also contacts HSP70. Similar to findings for BAG3, mutations in LDB3 cause skeletal and cardiac myopathy in animal models and patients (Cassandrini et al, 2021; Pathak et al, 2021). Pathology is accompanied by the formation of protein aggregates containing filamin, HSP70, HSPB8, BAG3 and ubiquitin, suggesting an impairment of CASA‐mediated quality control upon expression of non‐functional LDB3 (Pathak et al, 2021).
The HSP70 cochaperone DNAJB6 has been detected in association with BAG3, HSPB8 and CHIP in rat muscle (Sarparanta et al, 2012). DNAJB6 is a critical sensor of cellular stress, which contributes to the regulation of stress responses (Klaips et al, 2020). Mutations in DNAJB6 cause a form of limb‐girdle muscular dystrophy, characterized by a progressive weakness and wasting of arm and leg muscles, consistent with a maintenance and stress protection function of DNAJB6 (Ruggieri et al, 2016). Strikingly, disease‐associated mutations of DNAJB6 were shown to abrogate the ability of the cochaperone to bind to and stabilize unfolded proteins (Sarparanta et al, 2012). Because DNAJ‐type cochaperones help to load clients onto HSP70 (Faust et al, 2020), it is conceivable that DNAJB6 may assist in the initial recognition of at least a subset of client proteins by the BAG3 machinery. However, physiological relevant clients of DNAJB6 in the context of mechanical stress protection remain to be identified.
An expansion of the CASA client repertoire may also be achieved through cooperation with different small heat shock proteins. In addition to HSPB8, BAG3 can bind HSPB1 (HSP27), HSPB5 (αB‐crystallin) and HSPB6 (HSP20) (Rauch et al, 2017). Mutations in HSPB1 and HSPB5 have been linked to different forms of muscle weakness (Adriaenssens et al, 2017), and phosphorylation of HSPB5 at serine‐59, which increases the chaperone activity of HSPB5 (Peschek et al, 2013), is induced in human leg muscle following resistance exercise (Jacko et al, 2020). Moreover, HSPB1 and HSPB5 were shown to engage in a force‐induced association with mechanosensitive elements of sarcomeric titin, which prevents titin stiffening and aggregation in stretched sarcomeres (Kötter et al, 2014). Binding of BAG3 to HSPB1 and HSPB5 may thus provide access to specific client proteins during mechanical stress protection. Of note, HSPB1 interacts with SQSTM1/p62 and regulates autophagosome formation (Haidar et al, 2019), pointing to an active role of HSPB1 in the disposal of unfolded client proteins. Small heat shock proteins apparently emerge as key players in mechanical stress protection.
In a recent study, Martin et al (2021) monitored the BAG3‐mediated release of proteins from the cytoskeleton of cardiomyocytes by mass spectrometry and identified several sarcomere proteins as potential new clients of the CASA pathway, including the mechanosensitive protein spectrin. Quality control functions of BAG3 under mechanical stress thus seem to extend beyond filamin homeostasis.
Protein triage under mechanical stress
What determines the fate of a force‐unfolded mechanosensory protein? How is a decision reached between spontaneous or chaperone‐assisted refolding and chaperone‐assisted degradation? To answer these questions, it appears helpful to look at current concepts regarding protein triage in the proteostasis field. A prevalent view invokes a kinetic partitioning of proteins between free and chaperone‐bound states to facilitate de novo folding and to ensure conformational flexibility required for function (Kim et al, 2013b). According to this concept, chaperones would bind to force‐unfolded domains of mechanosensors engaged in the transduction of mechanical signals, when the rate of domain refolding is slower than the rate of chaperone binding. Transient interaction with chaperones would prevent aggregation and reduce the concentration of free folding intermediates at force‐bearing and force‐generating sites. Slow refolding kinetics may result in repeated recognition of unfolded mechanosensors by chaperone complexes and may ultimately lead to the recruitment of degradation factors. Dynamic low affinity interactions between mechanosensors, chaperones, degradation‐inducing cochaperones, chaperone‐associated ubiquitin ligases and ubiquitin receptors would shift the balance towards degradation. In such a scenario, a clear distinction between “productive” force‐induced unfolding during mechanotransduction and “unproductive” mechanical damage would be difficult to draw. Instead, folding and refolding kinetics of mechanosensitive domains would determine functionality, chaperone recognition and initiation of degradation.
In case of the giant muscle protein titin, reversible oxidation was recently shown to reduce the protein's elasticity through preferential modification of mechanosensitive Ig domains in the I‐band region of the muscle sarcomere (Loescher et al, 2020). This process, termed unfolded domain oxidation (UnDOx), increases cardiac stiffness under mechanical force and oxidative stress. By increasing the aggregation propensity of the unfolded and oxidized Ig domains, homotypic interactions between titin molecules are promoted to control myocardial stiffness. Of note, HSP90 and the autophagic ubiquitin adaptor NBR1 bind to the I‐band region of titin during quality control (Lange et al, 2005; Donlin et al, 2012), and the small heat shock proteins HSPB1 and HSPB5 recognize unfolded Ig domains of titin to suppress titin stiffening (Kötter et al, 2014). UnDOx may thus facilitate recognition by proteostasis factors and may influence the kinetic partitioning of titin.
Reversible prolyl isomerization regulates the refolding kinetics of the mechanosensitive Ig 20–21 domain pair in the BAG3 client filamin (Rognoni et al, 2014). While disruption of the 20–21 interdomain interaction occurs at forces around 4 pN, higher forces lead to a cis‐trans isomerization of a proline switch in domain 20 causing domain unfolding. The trans state persists up to minutes even when force is reduced, leaving the domain unstable for a prolonged time.
The studies by Loescher et al and Rognoni et al reveal molecular events that alter the folding and refolding kinetics of mechanosensitive domains, which in turn affect protein stability and chaperone recognition. Oxidation and force‐induced isomerization could thus be decisive steps during protein triage. Yet, both studies emphasize the relevance of the observed processes for the regulation of the mechanical properties of the cytoskeleton. Transient population of unstable, aggregation‐prone states of mechanosensors under surveillance of chaperones and other proteostasis factors appears to be an integral part of the adaptation of cells and tissues to mechanical forces.
Balancing transcription, translation and autophagy under mechanical stress
The BAG3 chaperone system mediates the degradation of filamin under mechanical stress. Yet, BAG3 also contributes to maintaining filamin homeostasis by stimulating the synthesis of new filamin molecules through interactions with components of the Hippo and mTOR signalling pathways, respectively (Ulbricht et al, 2013; Kathage et al, 2017). The Hippo pathway (named after the kinase Hippo involved in this pathway in Drosophila) is a mechanoresponsive signalling pathway that controls the expression of extracellular matrix components and cytoskeleton proteins including filamin (Dupont et al, 2011; Yu & Guan, 2013; Wackerhage et al, 2014). Hippo signalling proteins that are contacted by BAG3 include LATS1, LATS2, AMOT and AMOTL1 (Ulbricht et al, 2013; Taipale et al, 2014). In their activated state, these proteins interact with the transcriptional coactivators YAP and TAZ, which results in coactivator retention in the cytoplasm to attenuate target gene expression in the absence of a mechanical stimulus (Wackerhage et al, 2014; Yusko & Asbury, 2014). BAG3 interacts with the Hippo signalling proteins in a manner competitive with YAP/TAZ binding, causing YAP/TAZ release and translocation into the nucleus for the activation of target gene expression, including filamin (Ulbricht et al, 2013). BAG3‐activated transcription of filamin allows to compensate the disposal of mechanically damaged filamin molecules and to maintain the cytoskeleton under mechanical stress.
Protein synthesis is stimulated by BAG3 through spatial regulation of the mTOR kinase complex I (mTORC1) (Kathage et al, 2017). mTORC1 represents a master regulator of anabolic and catabolic processes in eukaryotic cells, whose activity is controlled by the inhibitory tuberous sclerosis complex (TSC) (Laplante & Sabatini, 2012; Betz & Hall, 2013; Menon et al, 2014). In adherent smooth muscle cells, BAG3 recruits the TSC complex to actin stress fibres by binding to the complex subunit TSC1 (Kathage et al, 2017). TSC recruitment results in an inhibition of mTORC1 along stress fibres, which is essential for the initiation of CASA at sites of mechanical protein unfolding and damage. At the same time, TSC recruitment and sequestration by BAG3 attenuates mTORC1 inhibition throughout the remaining cytoplasm, causing an upregulation of protein translation (Kathage et al, 2017). The BAG3‐mediated spatial regulation of mTORC1 apparently allows for a simultaneous induction of autophagy and protein synthesis in response to mechanical stress.
Mechanical stress signalling
The expression of many proteostasis factors is elevated when non‐native proteins accumulate to increase the folding and degradation capacity of the cell. A key component in this regulatory circuitry is the heat shock factor 1 (HSF1) (Vihervaara & Sistonen, 2014; Li et al, 2017; Naidu & Dinkova‐Kostova, 2017; Barna et al, 2018; Klaips et al, 2020). Identified for the first time in heat shocked fruit flies (which explains the name), HSF1 has emerged as a general stress protective transcription factor that upregulates the expression of folding and degradation proteins in response to diverse stress stimuli (Vihervaara & Sistonen, 2014; Barna et al, 2018). In the repressed state, monomeric HSF1 is complexed with chaperone proteins, i.e. HSP70 and HSP90. When non‐native proteins accumulate upon cell stress, chaperones dissociate from HSF1 to stabilize non‐native clients. In turn, HSF1 trimerizes and activates target gene expression (Li et al, 2017). Importantly, also mechanical stress triggers HSF1 activation (Li et al, 2013; Ulbricht et al, 2013). This leads to an elevated expression of many proteins that were discussed above to contribute to mechanical stress protection, including core components of the CASA machinery such as HSP70, BAG3, SQSTM1 and ubiquitin (Franceschelli et al, 2008; Barna et al, 2018). Accordingly, an HSF1‐ and BAG3‐dependent mechanotransduction pathway was proposed for the conversion of mechanical forces into biochemical and cell biological responses (Ulbricht et al, 2013). On this pathway, force‐unfolded mechanosensory proteins are recognized by chaperone proteins, causing HSF1 derepression and activation. The resultant increase of the cellular concentration of BAG3 and its partner proteins facilitates degradation of mechanically damaged client proteins through CASA. At the same time, increased engagement of BAG3 with Hippo signalling proteins activates YAP/TAZ‐dependent gene expression and stimulates protein synthesis through TSC complex sequestration and mTORC1 activation (Ulbricht et al, 2013). Balancing of transcription, translation and degradation is required to maintain the cytoskeleton under mechanical stress.
Recent work also reveals an unexpected link between HSF2 and mechanotransduction (Joutsen et al, 2020). Similar to HSF1, HSF2 is activated upon proteostasis imbalance, following proteasome inhibition for example. In this situation, HSF2‐dependent gene expression is required to maintain cadherin‐mediated cell–cell adhesions and to ensure cell survival (Joutsen et al, 2020). The findings suggest that HSF2 could contribute to the transcriptional control of mechanical stress protection.
In addition to regulated gene expression, posttranslational modifications, in particular phosphorylation and dephosphorylation events, contribute to the induction and adjustment of mechanical stress protection systems. Mechanical forces activate diverse signalling pathways governed by multiple kinases and phosphatases to tightly control contractile functions and to maintain the cytoskeleton. For example, mechanical stress imposed by acute exercise was shown to activate the extracellular signal‐regulated kinases ERK1 and ERK2, Jun N‐terminal kinase (JNK) and p38 mitogen‐activated protein kinase (p38, MAPK) (Kumar et al, 2002; Coffey et al, 2006; Hoffman et al, 2015; Nelson et al, 2019). Hypertrophy‐stimulating resistance training activates the protein kinases Akt and mTOR (Léger et al, 2006), and the Akt pathway is involved in the adaptation of skeletal muscle fibres to changes in workload (Bodine et al, 2001), resulting in an increase in fibre size (Rommel et al, 2001). Further kinases implicated in controlling protective and adaptive processes to mechanical stress in muscle cells include protein kinase C (PKC), calmodulin‐dependent protein kinase II (CaMKII), p21‐activated kinase 1 (PAK1) and AMP‐activated protein kinase (AMPK) (Egan & Zierath, 2013; Hoffman et al, 2015; Nelson et al, 2019). However, the lack of detailed knowledge about kinase–substrate and phosphatase–substrate relationships remains a major bottleneck in decoding signalling networks under mechanical stress. Only a few global phosphoproteomic studies of skeletal muscle exist to date. Phosphosites identified in proteins from non‐exercised skeletal muscle and/or differentiated skeletal muscle cells without fully assembled sarcomeres were used to predict the involved kinases (Højlund et al, 2009; Hou et al, 2010; Drexler et al, 2012; Kettenbach et al, 2015). However, activities of many kinases drastically change in response to acute exercise as demonstrated by quantitative phosphoproteomics of human skeletal muscle (Hoffman et al, 2015; Nelson et al, 2019). Recently, a mouse myotube model was established to study force‐regulated signalling processes (Reimann et al, 2017). Murine C2 or C2C12 skeletal muscle cells were differentiated to mature myotubes and their contraction was triggered by electrical pulse stimulation (EPS), followed by quantitative phosphoproteomics with a focus on Z‐disc and Z‐disc‐associated proteins. Z‐discs have been proposed as nodal points of myocyte signalling (Pyle & Solaro, 2004; Frank et al, 2006). Since Z‐discs limit the force‐generating sarcomeres, help to connect myofibrils to each other and further link the contractile apparatus to the sarcolemma and the extracellular matrix, they are in an ideal position to sense mechanical strain and orchestrate signalling processes (see Fig 2). Most importantly, the establishment of the Z‐disc protein phosphorylation landscape identified clustered multisite phosphorylation in the CASA client filamin and core CASA components including BAG3, HSPB8 and SYNPO2 (Reimann et al, 2017). Moreover, force‐induced and phosphorylation‐regulated interactions of filamin with HSPB1 and the degradation factor FILIP1, respectively, were recently described (Collier et al, 2019; Reimann et al, 2020). The findings indicate that interactions, localization, dynamics and/or functions of mechanosensitive proteins and force‐regulated proteostasis factors are modulated by reversible phosphorylation under mechanical stress.
In Drosophila, the serine/threonine kinase NUAK interacts with Starvin, the fly orthologue of BAG3 (Brooks et al, 2020). Starvin and NUAK mutant flies exhibit muscle degeneration and muscle contraction defects, and similar phenotypes are also observed following a depletion of Drosophila HSC70 and the autophagic membrane marker ATG8 (Arndt et al, 2010; Brooks et al, 2020). Moreover, muscle degeneration is accompanied by the accumulation of protein aggregates that contain filamin. NUAK thus seems to exert a central regulatory function in controlling CASA activity in fly muscles. It remains to be investigated how NUAK is regulated in response to muscle contraction and mechanical stress and whether NUAK orthologues exert a similar function in mammals.
Strikingly, phosphoproteomics of human exercised muscle and EPS‐stimulated murine myoblasts revealed several conserved sites in BAG3 that are subjected to dephosphorylation in response to mechanical force (Hoffman et al, 2015; Reimann et al, 2017). This suggests an essential involvement of phosphatases in BAG3 regulation. However, involved enzymes and functional consequences of BAG3 dephosphorylation remain to be established. Notably, the protein phosphatase 5 (PP5) is a direct binding partner of HSP70 and HSP90 (Connarn et al, 2014; Oberoi et al, 2016) and acts as an HSP90‐activated phosphatase at a mechanosensitive region of titin to modulate titin elasticity and myofilament‐associated signalling pathways (Krysiak et al, 2018). The work provides a first conceptual framework to explain the involvement of phosphatases in mechanical stress protection.
Signalling pathways not only activate stress protection systems but can also contribute to the shutdown of these systems, when necessary. Indeed, the Hippo network kinase STK38 was recently identified as a CASA inhibitor, which binds BAG3 to abrogate the interplay of the cochaperone with HSPB8 and SYNPO2 during the initiation of autophagy (Klimek et al, 2019). STK38 is activated upon starvation or when mechanical signals cease during cell detachment (Joffre et al, 2015; Bettoun et al, 2016), and STK38 activation stimulates BAG3 binding (Klimek et al, 2019). The resultant CASA inhibition may increase the availability of commonly used autophagy factors for the execution of other autophagy pathways, i.e. starvation and detachment induced autophagy, which might be more important under these conditions for cell survival.
Mechanical stress protection in the kidney
The previous chapters highlighted the relevance of mechanical stress protection for muscle maintenance. It is important to note, however, that mechanical stress protection is not limited to skeletal and cardiac muscle but represents a universal process. To illustrate this, we will discuss stress protection systems in the kidney and in immune cells in more detail.
Ultrafiltration in the kidney is a necessary physiological function for maintaining organismal homeostasis. The human kidney generates 180 l of primary urine each day in the 1 million glomeruli of the renal filtration unit (Fig 4A). This three‐layered filtration unit is formed by endothelia, the glomerular basement membrane and podocytes enwrapping the glomerular tuft from the outside (Fig 4B). The highly sophisticated ultrastructural architecture of podocytes consists of primary and secondary interdigitating processes covering the outer surface of glomerular capillaries (Brinkkoetter et al, 2013). Podocyte processes are connected to the underlying glomerular basement membrane by cell‐specific focal adhesion/cytoskeleton interactions (Sachs & Sonnenberg, 2013; Lennon et al, 2014) and to neighbouring cells through a highly specialized membrane‐like cell junction called the slit diaphragm (Benzing, 2004; Grahammer et al, 2013; Endlich et al, 2017). Failure of the filtration barrier becomes apparent with the key symptom of proteinuria—the loss of protein into the urine. Genetic causes for proteinuria affect genes almost exclusively coding for proteins of the focal adhesion/cytoskeleton interface or the slit diaphragm protein complex (Perico et al, 2016; Schell & Huber, 2017). The slit diaphragm contains an evolutionarily conserved mechanosensitive ion channel super‐complex, comprised of the homodimeric podocin proteins and the ion channel TRPC6 (Fig 4C) (Benzing, 2004; Huber et al, 2006; Huber et al, 2007; Hagmann et al, 2014). In addition, the Ig superfamily members nephrin and Neph1, and signalling adapters such as PI3 kinase, Nck1/2 and Crk are part of the super‐complex (Huber et al, 2003; Höhne et al, 2011; New et al, 2014; Grahammer et al, 2016). More recently identified members of this complex include the mechanosensitive cytoskeletal adapter and CASA client filamin (Venkatareddy et al, 2011), and the autophagy‐regulating protein C1QBP (Rinschen et al, 2016). Mutations in virtually all of these proteins are a major cause of genetic forms of proteinuria indicating that integrity of this protein complex is essential for kidney function (Perico et al, 2016).
Figure 4. Podocytes withstand high mechanical forces at the kidney filtration barrier.

(A) Schematic representation of the kidney glomerulus. Blood is filtered across the walls of the glomerular capillaries lined with podocytes. The filtrate enters the urinary space and exits through the proximal tubule. (B) The glomerular filtration barrier is formed by a fenestrated endothelium, the glomerular basement membrane and podocytes. Podocytes possess long foot processes that wrap around the capillaries, leaving slit diaphragms (Sd) available for blood filtration. (C) At the slit diaphragm, foot processes are connected by a mechanosensitive protein complex containing nephrin, Neph1 and podocin. The complex is linked to TRPC6 ion channels, diverse kinases and the actin cytoskeleton. Filamin mediates actin anchoring and crosslinking at the slit diaphragm.
Due to their location, the podocytes are constantly subjected to mechanical stress resulting from the blood pressure (Endlich & Endlich, 2012; Brinkkoetter et al, 2013). The terminally differentiated podocytes have to withstand this mechanical stress to maintain a physiological kidney function, despite an only limited capacity for self‐renewal (Assady et al, 2019). Podocytes thus rely on adaptive mechanisms that allow repair in response to mechanical damage to prevent their loss by detachment (Kriz & Lemley, 2015). Adaptation might be especially important in disease states with glomerular hypertension and hyperfiltration, when mechanical forces increase even further (Helal et al, 2012; Srivastava et al, 2018). Indeed, by combining quantitative analyses with advanced mathematical modelling, it could be demonstrated that the permeability of the filtration barrier is modulated through compression of the capillary wall. Disease‐associated morphological alterations of podocytes lead to reduced compressive forces that counteract filtration pressure, thereby rendering the filtration barrier leaky (Butt et al, 2020). This work proves an active role of the podocyte in counteracting filtration pressure to sustain the filtration barrier.
Considering the presence of the CASA client filamin in the podocin super‐complex (Fig 4C) and the key role of BAG3‐dependent proteostasis mechanisms for filamin homeostasis in muscle cells, it appears likely that BAG3 also represents an important mechanical stress protection factor in podocytes. In support of this hypothesis, the entire CASA complex, including BAG3, HSPB8, HSP70, the E3 ubiquitin ligase CHIP and the ubiquitin adaptor SQSTM1, was detected in an unbiased proteomic screen for proteins enriched in podocytes (Rinschen et al, 2018). In addition, phosphoproteome studies revealed multiple phosphorylation sites in BAG3, CHIP and filamin in primary podocytes, which are altered during podocyte differentiation (Rinschen et al, 2014, 2016). Induction of filamin expression represents an early response upon filtration barrier failure and knockdown of filamin in Drosophila reduces filtration capacity, pointing to key role of the BAG3 client in determining the mechanical properties of the filtration barrier (Koehler et al, 2020). Of note, the central podocyte actin‐cytoskeleton protein synaptopodin has been shown to play a role in BAG3‐mediated autophagy in neurons (Ji et al, 2019). Recent evidence also suggests that a YAP‐mediated mechanoresponse is required for maintaining podocyte integrity throughout lifetime (Keyvani Chahi et al, 2016; Schwartzman et al, 2016) and that YAP is an essential mediator of mechanical activation of podocytes (Rinschen et al, 2017a). Finally, mutations in the CASA component SYNPO2 were recently shown to contribute to monogenic nephrotic syndrome (Mao et al, 2020). Taken together, the data strongly suggest that mechanical stress protection systems are present and functional in podocytes and might be conserved, at least in part, between podocytes and muscle cells.
Mechanical stress protection in the immune system
The immune system may be viewed as a complex cell system, which is not embedded in “static” tissue architectures (Huse, 2017). This is even true for immune organs such as lymph nodes, which have a highly dynamic content of motile cells (Miller et al, 2002). In mammals, most immune cells originate in the bone marrow (Schulz et al, 2012). Precursor cells, together with already mature immune cells, exit the bone marrow via the bloodstream and are transported to their destination, namely resting or inflamed tissues. In addition, T cells recirculate between blood and lymphatic organs, that is they are cycling between blood‐borne and tissue‐borne states. Thus, a universal property of immune cells is their capacity to exit the circulation and to enter tissues (Fig 5). The ability to sense and respond to mechanical cues is essential for the development, activation, differentiation and expansion of immune cells (Zhang et al, 2020). In the blood, immune cells are subjected to the flow forces generated by the heartbeat, which gives rise to blood pressure. When immune cells enter tissues, they need to adhere to the endothelia of the destination organ and to traverse the respective endothelial barriers. This process, also called extravasation, has been extensively studied and led to a widely accepted model, according to which immune cells utilize two major classes of adhesion molecules, termed integrins and selectins (Ley et al, 2007). Both play essential roles in the recruitment of such cells to endothelial membranes at so‐called post‐capillary venules and subsequently to the underlying tissues. Several laboratories have shown that haemodynamic, flow‐mediated shear forces (or shear stresses) play an essential role in these processes (Shamri et al, 2005; Alon & Dustin, 2007; Alon & Ley, 2008). Flow forces alter the regulation and possibly the mechanism of extravasation, which involves two‐dimensional crawling and transmigration of immune cells between endothelial cells (paracytosis) or through their cell bodies (transcytosis) (Fig 5A) (Carman et al, 2007). Haemodynamic forces were also shown to be relevant for the motile behaviour of non‐classical monocytes, which move on barrier membranes of capillaries, larger veins and arteries via intraluminal crawling, as such cells constantly patrol in these blood vessels (Auffray et al, 2007). Finally, also the intraluminal crawling of T cells has been shown to be affected by shear stress (Valignat et al, 2013).
Figure 5. Mechanical forces govern the adhesion, migration and activity of immune cells.

(A) Immune cells, such as leukocytes, are subjected to variety of mechanical forces when they leave the blood stream and migrate through sites of infection and inflammation. (B) Schematic representation of the integrin machinery that operates in immune cells. The transduction of mechanical signals between intracellular processes and the extracellular matrix (ECM) via integrin heterodimers requires a two‐way communication system that employs cytoskeletal adapters (e.g. talin, kindlin, vinculin), kinases (e.g. FAK, Src) and mediators of Rho‐GTPase signalling. Integrin engagement with extracellular ligands affects motility and cell fate decisions, including survival, proliferation and differentiation via downstream signalling cascades. PM, plasma membrane.
Upon tissue entry, immune cells are further subjected to multiple forces or have to generate forces, respectively. Leukocytes migrate through a diverse array of three‐dimensional environments, and their motility mechanisms highly depend on the cell type and on the surrounding matrix (Huse, 2017; Gauthier & Roca‐Cusachs, 2018; Kechagia et al, 2019). Several classes of molecules have been shown to regulate functional force transduction in leukocytes. This includes several integrin adhesion receptors, which can bind either to cellular ligands or to extracellular matrix ligands such as collagen or fibronectin receptors (Fig 5B). Ligand binding of the integrin adhesome is dynamically regulated by conformational switching between active and inactive states (Horton et al, 2015; Li & Springer, 2017). Forces generated by the cell during crawling or extracellular forces applied by the surrounding tissues are transduced to the effector machinery of the actin cytoskeleton (Huse, 2017). Finally, an array of intracellular signalling molecules, including Rho, Rap and Arf‐GTPases (Reiner & Lundquist, 2018), and the cytoskeletal adapters talin and kindlin (Goult et al, 2018; Rahikainen et al, 2019) regulate both adhesion and cytoskeletal dynamics (Fig 5B).
Of note, the stimulation of adhesion and spreading of human Jurkat T lymphoblasts induces the expression of BAG3 and filamin and results in increased filamin turnover (Ulbricht et al, 2013). Moreover, inhibition of autophagy strongly impairs the adhesion, spreading and migration of Jurkat cells and attenuates the resistance of lymphoblasts and human primary leukocytes to shear stress (Ulbricht et al, 2013). These findings point to an essential role of the CASA machinery and the autophagic disposal of force‐unfolded proteins in mechanically stressed immune cells. Future studies will have to reveal the link between force‐regulated signalling pathways and BAG3‐dependent proteostasis mechanisms in these cells.
Always stressed
We use the term “mechanical stress” throughout this review. Per definition, the term “stress” refers to a state of increased strain or tension resulting from exceptionally adverse or demanding circumstances. Yet, in multicellular organisms, mechanical forces of a scale sufficient to disturb the structure of mechanosensitive proteins are not exceptional but ever‐present and affect diverse types of cells and tissues, as outlined above. Recent results, however, revealed an even more unanticipated and fundamental role of mechanical stress protection systems in the determination of cell behaviour. The identification of BAG3, HSPB8 and SQSTM1 as essential factors for cell division points to a very general function of the CASA pathway in all higher eukaryotic cells (Fuchs et al, 2015; Varlet et al, 2017; Luthold et al, 2020). In dividing human HeLa and HEK 293T cells, BAG3 is hyperphosphorylated at mitotic entry and localizes to centrosomal regions. Depletion of BAG3 and HSPB8 severely affects spindle orientation in mitotic cells due to disorganized actin‐rich retraction fibres, and similar phenotypic aberrations are observed upon depletion of the autophagic ubiquitin adaptor SQSTM1, which cooperates with BAG3 and HSPB8 during CASA (Fuchs et al, 2015; Luthold et al, 2020). Furthermore, the BAG3/HSPB8 machinery was also found to be essential for the accurate disassembly of the actin‐based contractile ring during cytokinesis, and alterations in this process triggered by the depletion of the CASA components could be alleviated by a pharmacological induction of autophagy (Varlet et al, 2017). Although the relevant CASA targets during mitosis remain to be identified, the CASA‐mediated autophagic disposal of cytoskeleton proteins seems to be of critical importance during cell division at stages when profound changes in cell tension occur.
Relevance for human physiology and pathophysiology
The inactivation or deregulation of mechanical stress protection factors causes severe pathology in humans. For example, mutations in BAG3 give rise to skeletal muscle myopathies and cardiomyopathies (Claeys et al, 2013; Fang et al, 2017; Kimura et al, 2021), a decline of BAG3 activity contributes to heart failure (Martin et al, 2021), and mutation‐induced functional impairment of UNC‐45 leads to skeletal muscle myopathy and ovarian and breast cancer (Bazzaro et al, 2007; Janiesch et al, 2007; Donkervoort et al, 2020). Advancing our knowledge about mechanical stress protection systems in humans will thus be instrumental for the diagnosis of diverse diseases and may pave the road for therapeutic intervention using, for example, proteostasis modulators, such as the autophagy inducing compound rapamycin. In addition, such knowledge might provide means to sustain the fitness and increase the performance of healthy individuals.
It is generally accepted that performance and health in human beings is critically dependent on muscle mass and strength, especially in the elderly (McLeod et al, 2016). Mechanical strain induced by resistance exercise is a necessary cellular stimulus to trigger anabolic signalling (Gehlert et al, 2015) and to induce protein synthesis and muscle growth (Mayhew et al, 2009; Damas et al, 2015). However, muscle stimulation by resistance exercise in athletes or in patients during rehabilitation programmes needs to be repeated over several weeks to achieve significant muscle growth and strength gain (Bird et al, 2005). Yet, resistance exercise also causes the force‐induced unfolding of sarcomeric proteins and muscle damage (Proske & Morgan, 2001; Ulbricht et al, 2015). Thus, increased muscle anabolism has to be accompanied by an induction of mechanoprotective systems to ensure a coordinated adaptation to mechanical stress in exercised muscle (Arndt et al, 2010; Ulbricht et al, 2015). In fact, strenuous resistance exercise leads to an immediate induction of the CASA machinery in human leg muscle, which will then continue at least for the next 24 h to degrade mechanically unfolded and damaged filamin and possibly other mechanosensory proteins (Ulbricht et al, 2015). Moreover, repeated resistance exercise during several weeks of training causes an elevated expression of CASA components, which limits muscular damage during a subsequent bout of exercise (Ulbricht et al, 2015). CASA apparently acts as a central adaptation mechanism in human muscle, responsive to acute physical exercise and repeated mechanical stimulation. The BAG3‐mediated regulation of mTORC1, which activates protein synthesis in response to mechanical stress (Kathage et al, 2017), might also contribute to the increase of muscle mass following exercise (Goodman, 2014; Jacobs et al, 2014).
The BAG3‐interacting small heat shock protein HSPB5 is rapidly phosphorylated at serine‐59 in human skeletal muscle after strenuous resistance exercise, and the extent of phosphorylation correlates with the intensity and number of muscle contractions (Jacko et al, 2019, 2020). Furthermore, HSPB5 phosphorylation declines during repeated exercise and re‐increases upon exercise following training cessation (Jacko et al, 2020), suggesting rapid adaptation and de‐adaptation in response to the timing of mechanical stimulation. Elucidating the relationship between exercise parameters, alterations in mechanical stress protection systems and muscle damage and growth will be important for establishing optimized training programmes for athletes and individuals who need to build up muscle during rehabilitation.
It is interesting to consider not only situations with increased mechanical stress such as resistance exercise, but also conditions when mechanical signals cease, for example in immobilized and ventilated patients in intensive care units (ICU), the number of which dramatically increased during the COVID pandemic. Life‐saving treatment of ICU patients often involves periods of mechanical ventilation, sedation and complete mechanical silencing. Under these conditions, up to 30% of patients develop critical illness myopathy (CIM), characterized by a massive loss of muscular myosin (Friedrich et al, 2018). The resultant muscle weakness can be as severe as causing a complete quadriplegia and represents a major obstacle for the recovery of affected ICU patients. Elaborate animal models established the complete mechanical silencing as a predominant factor in the development of CIM, which causes aberrant autophagic flux, increased levels of stress protection factors including HSP70, SQSTM1 and HSPB5, and altered expression of components of the ubiquitin–proteasome system in affected muscle (Ochala et al, 2011; Llano‐Diez et al, 2019). As outlined above, myosin is a critical client of the UNC‐45/HSP90 chaperone machinery and, indeed, also this machinery is present at elevated levels in animal models of CIM (Llano‐Diez et al, 2019). UNC‐45 protein levels need to be precisely regulated to ensure the progression of myosin folding (Hoppe et al, 2004; Janiesch et al, 2007). Consequently, an increase of UNC‐45 expression in CIM patients might directly contribute to the collapse of myosin homeostasis. In addition to myosin, also levels of the mechanosensitive protein titin are reduced in muscles of CIM patients and titin loss results in Z‐disc disassembly and sarcomere disintegration (Swist et al, 2020).
Although more research is required to understand the molecular alterations that cause critical illness myopathy, available data already illustrate that mechanical silencing has a profound impact on the proteostasis network and causes drastic changes in mechanical stress protection systems. The altered homeostasis of mechanosensitive client proteins is apparently incompatible with cell and tissue maintenance. A tight interplay between mechanical stimulation and proteome maintenances is further supported by the findings that passive mechanical loading as well as a pharmacological induction of proteostasis factors, for example based on the administration of the chaperone co‐inducer BGP‐15, alleviates aspects of CIM pathology (Salah et al, 2016; Llano‐Diez et al, 2019). It seems that our cells and tissues are not just mechanically stressed all the time, but that this stress is necessary for fitness and survival by establishing a functional proteostasis network.
Concluding remarks
Mechanobiology and proteostasis research are booming areas in biochemistry, cell biology and biomedicine. However, until recently, there has been very little exchange between these areas. Mechanobiologists wanted to identify and understand force‐generating, force‐bearing and force‐transducing structures, but usually overlooked that force‐induced protein unfolding poses a problem in cells, coped with by providing specialized protein folding and degradation systems. Proteostasis researchers, on the other hand, focussed their attention on cellular and organismal responses to heat, oxidative and proteotoxic stress, but largely ignored mechanical forces as an ever‐present major threat for protein integrity and proteome maintenance. The work described here illustrates that mechanobiology and proteostasis research need to be integrated to understand cell and tissue homeostasis in the presence of mechanical forces. Future studies should provide a comprehensive picture of the force‐sensitive proteome and the protein folding and degradation systems necessary for its maintenance. Elucidating involved proteostasis systems and signalling networks may ultimately provide the means to treat diseases caused by dysregulated mechanical stress protection ranging from myopathies to renal failure and immune deficiencies.
In need of answers.
-
What are the constituents of the force‐unfolded proteome and mechanical stress protection systems?
State‐of‐the‐art proteomic approaches of cells and tissues subjected to mechanical forces of defined intensity should be performed in future studies to identify the force‐unfolded proteome and the proteostasis systems that are involved in maintaining this part of the proteome.
-
What determines the recognition and processing of mechanosensitive domains by protein quality control systems?
Single‐molecule studies have revealed force‐induced unfolding events in mechanosensors. It will be necessary to include chaperones and other proteostasis factors in such studies to elucidate how the dynamic unfolding of mechanosensitive domains during mechanotransduction is linked to the surveillance by protein quality control systems. Analysis should also include posttranslational modifications of mechanosensors and proteostasis factors that alter folding and interaction kinetics.
-
Which kinase/phosphatase networks control mechanical stress protection and what are their targets?
Our knowledge about the signalling pathways that operate under mechanical stress and their critical targets is still limited. The further development of techniques for elucidating phosphoproteomes coupled to force stimulation of cells, tissues and model organisms will be important to establish regulatory networks.
-
Are there common and tissue‐specific mechanical stress protection systems?
For obvious reasons, the analysis of mechanical stress protection initially focussed on muscle. A comparative analysis of different tissues will be required to elucidate common principles and more specific responses. This may pave the road for a tissue‐specific pharmacological intervention to treat diverse diseases associated with a dysregulation of mechanical stress protection.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
Work in the authors' laboratories was supported by a grant of the Deutsche Forschungsgemeinschaft DFG FOR 2743. Research of B.W. and M.K. was further supported by the Excellence Strategy (CIBSS—EXC‐2189), and J.H. received additional funding from the European Space Agency (ESA‐CORA‐GBF‐2018‐005). Open Access funding enabled and organized by Projekt DEAL.
EMBO reports (2021) 22: e52507.
See the Glossary for abbreviations used in this article.
References
- Adriaenssens E, Geuens T, Baets J, Echaniz‐Laguna A, Timmerman V (2017) Novel insights in the disease biology of mutant small heat shock proteins in neuromuscular diseases. Brain 140: 2541–2549 [DOI] [PubMed] [Google Scholar]
- Alon R, Dustin ML (2007) Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen‐presenting cells. Immunity 26: 17–27 [DOI] [PubMed] [Google Scholar]
- Alon R, Ley K (2008) Cells on the run: shear‐regulated integrin activation in leukocyte rolling and arrest on endothelial cells. Curr Opin Cell Biol 20: 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann SM, Grünberg RG, Lenne P‐F, Ylänne J, Raae A, Herbert K, Saraste M, Nilges M, Hörber JKH (2002) Pathways and intermediates in forced unfolding of spectrin repeats. Structure 10: 1085–1096 [DOI] [PubMed] [Google Scholar]
- Arimura T, Ishikawa T, Nunoda S, Kawai S, Kimura A (2011) Dilated cardiomyopathy‐associated BAG3 mutations impair Z‐disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum Mutat 32: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK et al (2010) Chaperone‐assisted selective autophagy is essential for muscle maintenance. Curr Biol 20: 143–148 [DOI] [PubMed] [Google Scholar]
- Assady S, Benzing T, Kretzler M, Skorecki KL (2019) Glomerular podocytes in kidney health and disease. Lancet 393: 856–858 [DOI] [PubMed] [Google Scholar]
- Atherton P, Stutchbury B, Jethwa D, Ballestrem C (2016) Mechanosensitive components of integrin adhesions: role of vinculin. Exp Cell Res 343: 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C, Fogg D, Garfa M, Elain G, Join‐Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, Geissmann F (2007) Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317: 666–670 [DOI] [PubMed] [Google Scholar]
- Austen K, Ringer P, Mehlich A, Chrostek‐Grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M, Grashoff C (2015) Extracellular rigidity sensing by talin isoform‐specific mechanical linkages. Nat Cell Biol 17: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayad NME, Kaushik S, Weaver VM (2019) Tissue mechanics, an important regulator of development and disease. Philos Trans R Soc B Biol Sci 374: 20180215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchin D, Hayer‐Hartl M, Hartl FU (2016) In vivo aspects of protein folding and quality control. Science 353: aac4354 [DOI] [PubMed] [Google Scholar]
- Barna J, Csermely P, Vellai T (2018) Roles of heat shock factor 1 beyond the heat shock response. Cell Mol Life Sci 75: 2897–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Bauer CC, Ortiz I, Epstein HF (1998) Unc‐45 mutations in Caenorhabditis elegans implicate a CRO1/She4p‐like domain in myosin assembly. J Cell Biol 143: 1215–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF (2002) Role of the myosin assembly protein UNC‐45 as a molecular chaperone for myosin. Science 295: 669–671 [DOI] [PubMed] [Google Scholar]
- Bazzaro M, Santillan A, Lin Z, Tang T, Lee MK, Bristow RE, Shih I‐M, Roden RBS (2007) Myosin II co‐chaperone general cell UNC‐45 overexpression is associated with ovarian cancer, rapid proliferation, and motility. Am J Pathol 171: 1640–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing T (2004) Signaling at the slit diaphragm. J Am Soc Nephrol 15: 1382–1391 [DOI] [PubMed] [Google Scholar]
- Bettoun A, Joffre C, Zago G, Surdez D, Vallerand D, Gundogdu R, Sharif AAD, Gomez M, Cascone I, Meunier B et al (2016) Mitochondrial clearance by the STK38 kinase supports oncogenic Ras‐induced cell transformation. Oncotarget 7: 44142–44160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Hall MN (2013) Where is mTOR and what is it doing there? J Cell Biol 203: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird SP, Tarpenning KM, Marino FE (2005) Designing resistance training programmes to enhance muscular fitness: a review of the acute programme variables. Sport Med 35: 841–851 [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ et al (2001) Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3: 1014–1019 [DOI] [PubMed] [Google Scholar]
- Brinkkoetter PT, Ising C, Benzing T (2013) The role of the podocyte in albumin filtration. Nat Rev Nephrol 9: 328–336 [DOI] [PubMed] [Google Scholar]
- Brooks D, Naeem F, Stetsiv M, Goetting SC, Bawa S, Green N, Clark C, Bashirullah A, Geisbrecht ER (2020) Drosophila NUAK functions with Starvin/BAG3 in autophagic protein turnover. PLoS Genet 16: e1008700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T (2010) Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell 40: 238–252 [DOI] [PubMed] [Google Scholar]
- Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR (2014) Cell adhesion. The minimal cadherin‐catenin complex binds to actin filaments under force. Science 346: 1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt L, Unnersjö‐Jess D, Höhne M, Edwards A, Binz‐Lotter J, Reilly D, Hahnfeldt R, Ziegler V, Fremter K, Rinschen MM et al (2020) A molecular mechanism explaining albuminuria in kidney disease. Nat Metab 2: 461–474 [DOI] [PubMed] [Google Scholar]
- Carman CV, Sage PT, Sciuto TE, de la Fuente MA, Geha RS, Ochs HD, Dvorak HF, Dvorak AM, Springer TA (2007) Transcellular diapedesis is initiated by invasive podosomes. Immunity 26: 784–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, Lambert H, Landry J (2008) HspB8 chaperone activity toward poly(Q)‐containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem 283: 1437–1444 [DOI] [PubMed] [Google Scholar]
- Cassandrini D, Merlini L, Pilla F, Cenni V, Santi S, Faldini C, Santorelli FM, Sabatelli P (2021) Protein aggregates and autophagy involvement in a family with a mutation in Z‐band alternatively spliced PDZ‐motif protein. Neuromuscul Disord 31: 44–51 [DOI] [PubMed] [Google Scholar]
- Cho S, Irianto J, Discher DE (2017) Mechanosensing by the nucleus: from pathways to scaling relationships. J Cell Biol 216: jcb.201610042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe Y‐J, Park S‐H, Hassemer T, Körner R, Vincenz‐Donnelly L, Hayer‐Hartl M, Hartl FU (2016) Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531: 191–195 [DOI] [PubMed] [Google Scholar]
- Choi H‐J, Pokutta S, Cadwell GW, Bobkov AA, Bankston LA, Liddington RC, Weis WI (2012) αE‐catenin is an autoinhibited molecule that coactivates vinculin. Proc Natl Acad Sci USA 109: 8576–8581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys KG, Fardeau M, Olivé M, Kley RA, Goldfarb LG, Claeys KG, Fardeau M (2013) Myofibrillar myopathies. Handb Clin Neurol 113: 1337–1342 [DOI] [PubMed] [Google Scholar]
- Coffey VG, Zhong Z, Shield A, Canny BJ, Chibalin AV, Zierath JR, Hawley JA (2006) Early signaling responses to divergent exercise stimuli in skeletal muscle from well‐trained humans. FASEB J 20: 190–192 [DOI] [PubMed] [Google Scholar]
- Cohen‐Kaplan V, Livneh I, Avni N, Cohen‐Rosenzweig C, Ciechanover A (2016) The ubiquitin‐proteasome system and autophagy: coordinated and independent activities. Int J Biochem Cell Biol 79: 403–418 [DOI] [PubMed] [Google Scholar]
- Collier MP, Reid Alderson T, De Villiers CP, Nicholls D, Gastall HY, Allison TM, Degiacomi MT, Jiang H, Mlynek G, Fürst DO et al (2019) HspB1 phosphorylation regulates its intramolecular dynamics and mechanosensitive molecular chaperone interaction with filamin C. Sci Adv 5: eaav8421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connarn JN, Assimon VA, Reed RA, Tse E, Southworth DR, Zuiderweg ERP, Gestwicki JE, Sun D (2014) The molecular chaperone Hsp70 activates protein phosphatase 5 (PP5) by binding the tetratricopeptide repeat (TPR) domain. J Biol Chem 289: 2908–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa V, D’Agostino VG, Cristofani R, Rusmini P, Cicardi ME, Messi E, Loffredo R, Pancher M, Piccolella M, Galbiati M et al (2016) Transcriptional induction of the heat shock protein B8 mediates the clearance of misfolded proteins responsible for motor neuron diseases. Sci Rep 6: 22827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa V, Sau D, Rusmini P, Boncoraglio A, Onesto E, Bolzoni E, Galbiati M, Fontana E, Marino M, Carra S et al (2010) The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). Hum Mol Genet 19: 3440–3456 [DOI] [PubMed] [Google Scholar]
- Dahiya V, Buchner J (2019) Functional principles and regulation of molecular chaperones. Adv Protein Chem Struct Biol 114: 1–60 [DOI] [PubMed] [Google Scholar]
- Dahl J‐U, Gray MJ, Jakob U (2015) Protein quality control under oxidative stress conditions. J Mol Biol 427: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas F, Phillips S, Vechin FC, Ugrinowitsch C (2015) A review of resistance training‐induced changes in skeletal muscle protein synthesis and their contribution to hypertrophy. Sport Med 45: 801–807 [DOI] [PubMed] [Google Scholar]
- Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott‐Schwartz J et al (2013) NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 126: 939–952 [DOI] [PubMed] [Google Scholar]
- Dikic I (2017) Proteasomal and autophagic degradation systems. Annu Rev Biochem 86: 193–224 [DOI] [PubMed] [Google Scholar]
- Discher D, Dong C, Fredberg JJ, Guilak F, Ingber D, Janmey P, Kamm RD, Schmid‐Schönbein GW, Weinbaum S (2009) Biomechanics: cell research and applications for the next decade. Ann Biomed Eng 37: 847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkervoort S, Kutzner CE, Hu Y, Lornage X, Rendu J, Stojkovic T, Baets J, Neuhaus SB, Tanboon J, Maroofian R et al (2020) Pathogenic variants in the myosin chaperone UNC‐45B cause progressive myopathy with eccentric cores. Am J Hum Genet 107: 1078–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin Lt, Andresen C, Just S, Rudensky E, Pappas Ct, Kruger M, Jacobs Ey, Unger A, Zieseniss A, Dobenecker M‐w et al (2012) Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev 26: 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Dillin A (2010) Protein homeostasis and aging in neurodegeneration. J Cell Biol 190: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HCA, Ruhs A, Konzer A, Mendler L, Bruckskotten M, Looso M, Günther S, Boettger T, Krüger M, Braun T (2012) On marathons and sprints: an integrated quantitative proteomics and transcriptomics analysis of differences between slow and fast muscle fibers. Mol Cell Proteomics 11: M111.010801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Li H, Bian Y, Zhong Y (2008) Heat‐shock protein 90alpha1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc Natl Acad Sci USA 105: 554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuFort CC, Paszek MJ, Weaver VM (2011) Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol 12: 308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S et al (2011) Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183 [DOI] [PubMed] [Google Scholar]
- Echeverría PC, Briand P‐A, Picard D (2016) A remodeled Hsp90 molecular chaperone ensemble with the novel cochaperone Aarsd1 is required for muscle differentiation. Mol Cell Biol 36: 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, Zierath JR (2013) Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab 17: 162–184 [DOI] [PubMed] [Google Scholar]
- Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP (2011) Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478: 260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endlich K, Kliewe F, Endlich N (2017) Stressed podocytes—mechanical forces, sensors, signaling and response. Pflügers Arch ‐ Eur J Physiol 469: 937–949 [DOI] [PubMed] [Google Scholar]
- Endlich N, Endlich K (2012) The challenge and response of podocytes to glomerular hypertension. Semin Nephrol 32: 327–341 [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126: 677–689 [DOI] [PubMed] [Google Scholar]
- Etard C, Behra M, Fischer N, Hutcheson D, Geisler R, Strähle U (2007) The UCS factor Steif/Unc‐45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev Biol 308: 133–143 [DOI] [PubMed] [Google Scholar]
- Etard C, Roostalu U, Strähle U (2008) Shuttling of the chaperones Unc45b and Hsp90a between the A band and the Z line of the myofibril. J Cell Biol 180: 1163–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzioni A (2007) Leukocyte adhesion deficiencies: molecular basis, clinical findings, and therapeutic options. Immune Mediat Dis Theory Ther 601: 51–60 [DOI] [PubMed] [Google Scholar]
- Fang Xi, Bogomolovas J, Wu T, Zhang W, Liu C, Veevers J, Stroud MJ, Zhang Z, Ma X, Mu Y et al (2017) Loss‐of‐function mutations in co‐chaperone BAG3 destabilize small HSPs and cause cardiomyopathy. J Clin Invest 127: 3189–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust O, Abayev‐Avraham M, Wentink AS, Maurer M, Nillegoda NB, London N, Bukau B, Rosenzweig R (2020) HSP40 proteins use class‐specific regulation to drive HSP70 functional diversity. Nature 587: 489–494 [DOI] [PubMed] [Google Scholar]
- Finer JT, Simmons RM, Spudich JA (1994) Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature 368: 113–119 [DOI] [PubMed] [Google Scholar]
- Fischer LS, Klingner C, Schlichthaerle T, Strauss MT, Böttcher R, Fässler R, Jungmann R, Grashoff C (2021) Quantitative single‐protein imaging reveals molecular complex formation of integrin, talin, and kindlin during cell adhesion. Nat Commun 12: 919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschelli S, Rosati A, Lerose R, De Nicola S, Turco MC, Pascale M (2008) bag3 gene expression is regulated by heat shock factor 1. J Cell Physiol 215: 575–577 [DOI] [PubMed] [Google Scholar]
- Frank D, Kuhn C, Katus HA, Frey N (2006) The sarcomeric Z‐disc: a nodal point in signalling and disease. J Mol Med 84: 446–468 [DOI] [PubMed] [Google Scholar]
- Friedrich O, Diermeier S, Larsson L (2018) Weak by the machines: muscle motor protein dysfunction ‐ a side effect of intensive care unit treatment. Acta Physiol 222: e12885 [DOI] [PubMed] [Google Scholar]
- Frumkin A, Dror S, Pokrzywa W, Bar‐Lavan Y, Karady I, Hoppe T, Ben‐Zvi A (2014) Challenging muscle homeostasis uncovers novel chaperone interactions in Caenorhabditis elegans . Front Mol Biosci 1: 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M, Luthold C, Guilbert SM, Varlet AA, Lambert H, Jetté A, Elowe S, Landry J, Lavoie JN (2015) A role for the chaperone complex BAG3‐HSPB8 in actin dynamics, spindle orientation and proper chromosome segregation during mitosis. PLOS Genet 11: e1005582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser AM, Kaiser CJO, Haslbeck V, Richter K (2011) Downregulation of the Hsp90 system causes defects in muscle cells of Caenorhabditis elegans . PLoS One 6: e25485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C (2009) Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J 28: 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C (2011) BAG3 mediates chaperone‐based aggresome‐targeting and selective autophagy of misfolded proteins. EMBO Rep 12: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G et al (2016) A surveillance function of the HSPB8‐BAG3‐HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell 63: 796–810 [DOI] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, Klionsky DJ (2018) Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20: 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M, Djinović‐Carugo K (2016) The sarcomeric cytoskeleton: from molecules to motion. J Exp Biol 219: 135–145 [DOI] [PubMed] [Google Scholar]
- Gauthier NC, Roca‐Cusachs P (2018) Mechanosensing at integrin‐mediated cell–matrix adhesions: from molecular to integrated mechanisms. Curr Opin Cell Biol 50: 20–26 [DOI] [PubMed] [Google Scholar]
- Gazda L, Pokrzywa W, Hellerschmied D, Löwe T, Forné I, Mueller‐Planitz F, Hoppe T, Clausen T (2013) The myosin chaperone UNC‐45 is organized in tandem modules to support myofilament formation in C. elegans . Cell 152: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Holmes KC (2005) The molecular mechanism of muscle contraction. Adv Protein Chem 71: 161–193 [DOI] [PubMed] [Google Scholar]
- Gehlert S, Suhr F, Gutsche K, Willkomm L, Kern J, Jacko D, Knicker A, Schiffer T, Wackerhage H, Bloch W (2015) High force development augments skeletal muscle signalling in resistance exercise modes equalized for time under tension. Pflugers Arch Eur J Physiol 467: 1343–1356 [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM (2010) Substrate elasticity regulates skeletal muscle stem cell self‐renewal in culture. Science 329: 1078–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann WH (2016) Role of vinculin in cellular mechanotransduction. Cell Biol Int 40: 241–256 [DOI] [PubMed] [Google Scholar]
- Goodman CA (2014) The role of mTORC1 in regulating protein synthesis and skeletal muscle mass in response to various mechanical stimuli. Rev Physiol Biochem Pharmacol 166: 43–95 [DOI] [PubMed] [Google Scholar]
- Goult BT, Yan J, Schwartz MA (2018) Talin as a mechanosensitive signaling hub. J Cell Biol 217: 3776–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DM, Burridge K (2016) Mechanotransduction and nuclear function. Curr Opin Cell Biol 40: 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahammer F, Schell C, Huber TB (2013) The podocyte slit diaphragm ‐ From a thin grey line to a complex signalling hub. Nat Rev Nephrol 9: 587–598 [DOI] [PubMed] [Google Scholar]
- Grahammer F, Wigge C, Schell C, Kretz O, Patrakka J, Schneider S, Klose M, Arnold SJ, Habermann A, Bräuniger R et al (2016) A flexible, multilayered protein scaffold maintains the slit in between glomerular podocytes. JCI Insight 1: e86177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Chen D, Fan Z, Epstein HF (2011) Differential turnover of myosin chaperone UNC‐45A isoforms increases in metastatic human breast cancer. J Mol Biol 412: 365–378 [DOI] [PubMed] [Google Scholar]
- Hagmann H, Kuczkowski A, Ruehl M, Lamkemeyer T, Brodesser S, Horke S, Dryer S, Schermer B, Benzing T, Brinkkoetter PT (2014) Breaking the chain at the membrane: paraoxonase 2 counteracts lipid peroxidation at the plasma membrane. FASEB J 28: 1769–1779 [DOI] [PubMed] [Google Scholar]
- Haidar M, Asselbergh B, Adriaenssens E, De Winter V, Timmermans JP, Auer‐Grumbach M, Juneja M, Timmerman V (2019) Neuropathy‐causing mutations in HSPB1 impair autophagy by disturbing the formation of SQSTM1/p62 bodies. Autophagy 15: 1051–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer‐Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332 [DOI] [PubMed] [Google Scholar]
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q et al (2012) Exercise‐induced BCL2‐regulated autophagy is required for muscle glucose homeostasis. Nature 481: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helal I, Fick‐Brosnahan GM, Reed‐Gitomer B, Schrier RW (2012) Glomerular hyperfiltration: definitions, mechanisms and clinical implications. Nat Rev Nephrol 8: 293–300 [DOI] [PubMed] [Google Scholar]
- Henderson CA, Gomez CG, Novak SM, Mi‐Mi L, Gregorio CC (2017) Overview of the muscle cytoskeleton. In Comprehensive physiology, pp 891–944. Hoboken, NJ: John Wiley & Sons, Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch N, Wolters B, Dreissen G, Springer R, Kirchgessner N, Merkel R, Hoffmann B (2013) The constant beat: cardiomyocytes adapt their forces by equal contraction upon environmental stiffening. Biol Open 2: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Kasturi P, Hartl FU (2019) The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol 20: 421–435 [DOI] [PubMed] [Google Scholar]
- Hoffman BD, Grashoff C, Schwartz MA (2011) Dynamic molecular processes mediate cellular mechanotransduction. Nature 475: 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N, Parker B, Chaudhuri R, Fisher‐Wellman K, Kleinert M, Humphrey S, Yang P, Holliday M, Trefely S, Fazakerley D et al (2015) Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise‐regulated kinases and AMPK substrates. Cell Metab 22: 922–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld J, Hoppe T (2018) Ub and down: ubiquitin exercise for the elderly. Trends Cell Biol 28: 512–522 [DOI] [PubMed] [Google Scholar]
- Höhne M, Lorscheider J, von Bardeleben A, Dufner M, Scharf MA, Gödel M, Helmstädter M, Schurek E‐M, Zank S, Gerke P et al (2011) The BAR domain protein PICK1 regulates cell recognition and morphogenesis by interacting with Neph proteins. Mol Cell Biol 31: 3241–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højlund K, Bowen BP, Hwang H, Flynn CR, Madireddy L, Geetha T, Langlais P, Meyer C, Mandarino LJ, Yi Z (2009) In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC−ESI−MS/MS. J Proteome Res 8: 4954–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma S, Iwasaki M, Shelton GD, Engvall E, Reed JC, Takayama S (2006) BAG3 deficiency results in fulminant myopathy and early lethality. Am J Pathol 169: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Cassata G, Barral JM, Springer W, Hutagalung AH, Epstein HF, Baumeister R (2004) Regulation of the myosin‐directed chaperone UNC‐45 by a novel E3/E4‐multiubiquitylation complex in C. elegans . Cell 118: 337–349 [DOI] [PubMed] [Google Scholar]
- Horton ER, Byron A, Askari JA, Ng DHJ, Millon‐Frémillon A, Robertson J, Koper EJ, Paul NR, Warwood S, Knight D et al (2015) Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol 17: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Cui Z, Xie Z, Xue P, Wu P, Chen X, Li J, Cai T, Yang F (2010) Phosphoproteome analysis of rat L6 myotubes using reversed‐phase C18 prefractionation and titanium dioxide enrichment. J Proteome Res 9: 777–788 [DOI] [PubMed] [Google Scholar]
- Hu X, Margadant FM, Yao M, Sheetz MP (2017) Molecular stretching modulates mechanosensing pathways. Protein Sci 26: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TB, Benzing T (2005) The slit diaphragm: a signaling platform to regulate podocyte function. Curr Opin Nephrol Hypertens 14: 211–216 [DOI] [PubMed] [Google Scholar]
- Huber TB, Hartleben B, Kim J, Schmidts M, Schermer B, Keil A, Egger L, Lecha RL, Borner C, Pavenstädt H et al (2003) Nephrin and CD2AP associate with phosphoinositide 3‐OH kinase and stimulate AKT‐dependent signaling. Mol Cell Biol 23: 4917–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TB, Schermer B, Muller RU, Hohne M, Bartram M, Calixto A, Hagmann H, Reinhardt C, Koos F, Kunzelmann K et al (2006) Podocin and MEC‐2 bind cholesterol to regulate the activity of associated ion channels. Proc Natl Acad Sci USA 103: 17079–17086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber TB, Schermer B, Benzing T (2007) Podocin organizes ion channel‐lipid supercomplexes: implications for mechanosensation at the slit diaphragm. Nephron Exp Nephrol 106: e27–e31 [DOI] [PubMed] [Google Scholar]
- Huse M (2017) Mechanical forces in the immune system. Nat Rev Immunol 17: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S, de Rooij J (2013) Mechanosensitive systems at the cadherin‐F‐actin interface. J Cell Sci 126: 403–413 [DOI] [PubMed] [Google Scholar]
- Irianto J, Pfeifer CR, Xia Y, Discher DE (2016) SnapShot: mechanosensing matrix. Cell 165: 1820e1‐1820.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann P, Lammerding J (2013) Nuclear mechanics and mechanotransduction in health and disease. Curr Biol 23: R1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N, Sarpal R, Wood MN, Barrick SK, Nishikawa T, Hayashi H, Kobb AB, Flozak AS, Yemelyanov A, Fernandez‐Gonzalez R et al (2018) Force‐dependent allostery of the α‐catenin actin‐binding domain controls adherens junction dynamics and functions. Nat Commun 9: 5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacko D, Bersiner K, Hebchen J, De Marées M, Bloch W, Gehlert S (2019) Phosphorylation of B‐crystallin and its cytoskeleton association differs in skeletal myofiber types depending on resistance exercise intensity and volume. J Appl Physiol 126: 1607–1618 [DOI] [PubMed] [Google Scholar]
- Jacko D, Bersiner K, Schulz O, Przyklenk A, Spahiu F, Höhfeld J, Bloch W, Gehlert S (2020) Coordinated alpha‐crystallin B phosphorylation and desmin expression indicate adaptation and deadaptation to resistance exercise‐induced loading in human skeletal muscle. Am J Physiol Cell Physiol 319: C300–C312 [DOI] [PubMed] [Google Scholar]
- Jacobs BL, Goodman CA, Hornberger TA (2014) The mechanical activation of mTOR signaling: an emerging role for late endosome/lysosomal targeting. J Muscle Res Cell Motil 35: 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmüller H, Cassata G, Krause S, Hoppe T (2007) The ubiquitin‐selective chaperone CDC‐48/p97 links myosin assembly to human myopathy. Nat Cell Biol 9: 379–390 [DOI] [PubMed] [Google Scholar]
- Janoštiak R, Brábek J, Auernheimer V, Tatárová Z, Lautscham LA, Dey T, Gemperle J, Merkel R, Goldmann WH, Fabry B et al (2014) CAS directly interacts with vinculin to control mechanosensing and focal adhesion dynamics. Cell Mol Life Sci 71: 727–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Tang M, Zeidler C, Höhfeld J, Johnson GV (2019) BAG3 and SYNPO (synaptopodin) facilitate phospho‐MAPT/Tau degradation via autophagy in neuronal processes. Autophagy 15: 1199–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffre C, Dupont N, Hoa L, Gomez V, Pardo R, Gonçalves‐Pimentel C, Achard P, Bettoun A, Meunier B, Bauvy C et al (2015) The Pro‐apoptotic STK38 kinase is a new beclin1 partner positively regulating autophagy. Curr Biol 25: 2479–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CP, Tang H‐Y, Carag C, Speicher DW, Discher DE (2007) Forced unfolding of proteins within cells. Science 317: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutsen J, Da Silva AJ, Luoto JC, Budzynski MA, Nylund AS, de Thonel A, Concordet J‐P, Mezger V, Sabéran‐Djoneidi D, Henriksson E et al (2020) Heat shock factor 2 protects against proteotoxicity by maintaining cell‐cell adhesion. Cell Rep 30: 583–597.e6 [DOI] [PubMed] [Google Scholar]
- Judge LM, Perez‐Bermejo JA, Truong A, Ribeiro AJS, Yoo JC, Jensen CL, Mandegar MA, Huebsch N, Kaake RM, So P‐L et al (2017) A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight 2: e94623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CM, Bujalowski PJ, Ma L, Anderson J, Epstein HF, Oberhauser AF (2012) Tracking UNC‐45 chaperone‐myosin interaction with a titin mechanical reporter. Biophys J 102: 2212–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathage B, Gehlert S, Ulbricht A, Lüdecke L, Tapia VE, Orfanos Z, Wenzel D, Bloch W, Volkmer R, Fleischmann BK et al (2017) The cochaperone BAG3 coordinates protein synthesis and autophagy under mechanical strain through spatial regulation of mTORC1. Biochim Biophys Acta Mol Cell Res 1864: 62–75 [DOI] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM (2012) Chaperone‐mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol 22: 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechagia JZ, Ivaska J, Roca‐Cusachs P (2019) Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol 20: 457–473 [DOI] [PubMed] [Google Scholar]
- Kesner BA, Ding F, Temple BR, Dokholyan NV (2010) N‐terminal strands of filamin Ig domains act as a conformational switch under biological forces. Proteins 78: 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Sano H, Keller SR, Lienhard GE, Gerber SA (2015) SPECHT ‐ single‐stage phosphopeptide enrichment and stable‐isotope chemical tagging: quantitative phosphoproteomics of insulin action in muscle. J Proteomics 114: 48–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyvani Chahi A, Martin CE, Jones N (2016) Nephrin suppresses hippo signaling through the adaptor proteins Nck and WTIP. J Biol Chem 291: 12799–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Behl C, Dikic I (2016) Ubiquitin‐dependent and independent signals in selective autophagy. Trends Cell Biol 26: 6–16 [DOI] [PubMed] [Google Scholar]
- Kim J, Löwe T, Hoppe T (2008) Protein quality control gets muscle into shape. Trends Cell Biol 18: 264–272 [DOI] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim Y‐N, Kim SS, Kim DH, Hur KY, Kim HK et al (2013a) Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med 19: 83–92 [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer‐Hartl M, Ulrich Hartl F (2013b) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82: 323–355 [DOI] [PubMed] [Google Scholar]
- Kimura K, Ooms A, Graf‐Riesen K, Kuppusamy M, Unger A, Schuld J, Daerr J, Lother A, Geisen C, Hein L et al (2021) Overexpression of human BAG3P209L in mice causes restrictive cardiomyopathy. Nat Commun 12: 3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou Y‐S, Bjørkøy G, Nunn JL, Bruun J‐A, Shvets E, McEwan DG, Clausen TH, Wild P et al (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Klaips CL, Gropp MHM, Hipp MS, Hartl FU (2020) Sis1 potentiates the stress response to protein aggregation and elevated temperature. Nat Commun 11: 6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimek C, Jahnke R, Wördehoff J, Kathage B, Stadel D, Behrends C, Hergovich A, Höhfeld J (2019) The Hippo network kinase STK38 contributes to protein homeostasis by inhibiting BAG3‐mediated autophagy. Biochim Biophys Acta Mol cell Res 1866: 1556–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimek C, Kathage B, Wördehoff J, Höhfeld J (2017) BAG3‐mediated proteostasis at a glance. J Cell Sci 130: 2781–2788 [DOI] [PubMed] [Google Scholar]
- Koehler S, Kuczkowski A, Kuehne L, Jüngst C, Hoehne M, Grahammer F, Eddy S, Kretzler M, Beck BB, Höhfeld J et al (2020) Proteome analysis of isolated podocytes reveals stress responses in glomerular sclerosis. J Am Soc Nephrol 31: 544–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konersman CG, Bordini BJ, Scharer G, Lawlor MW, Zangwill S, Southern JF, Amos L, Geddes GC, Kliegman R, Collins MP (2015) BAG3 myofibrillar myopathy presenting with cardiomyopathy. Neuromuscul Disord 25: 418–422 [DOI] [PubMed] [Google Scholar]
- Kötter S, Unger A, Hamdani N, Lang P, Vorgerd M, Nagel‐Steger L, Linke WA (2014) Human myocytes are protected from titin aggregation‐induced stiffening by small heat shock proteins. J Cell Biol 204: 187–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz W, Lemley KV (2015) A potential role for mechanical forces in the detachment of podocytes and the progression of CKD. J Am Soc Nephrol 26: 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysiak J, Unger A, Beckendorf L, Hamdani N, von Frieling‐Salewsky M, Redfield MM, dos Remedios CG, Sheikh F, Gergs U, Boknik P et al (2018) Protein phosphatase 5 regulates titin phosphorylation and function at a sarcomere‐associated mechanosensor complex in cardiomyocytes. Nat Commun 9: 262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Chaudhry I, Reid MB, Boriek AM (2002) Distinct signaling pathways are activated in response to mechanical stress applied axially and transversely to skeletal muscle fibers. J Biol Chem 277: 46493–46503 [DOI] [PubMed] [Google Scholar]
- Lad Y, Kiema T, Jiang P, Pentikäinen OT, Coles CH, Campbell ID, Calderwood DA, Ylänne J (2007) Structure of three tandem filamin domains reveals auto‐inhibition of ligand binding. EMBO J 26: 3993–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoux B, Nelson WJ, Yan J, Mège RM (2015) The mechanotransduction machinery at work at adherens junctions. Integr Biol 7: 1109–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B et al (2005) The kinase domain of titin controls muscle gene expression and protein turnover. Science 308: 1599–1603 [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149: 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kumar S (2016) Actomyosin stress fiber mechanosensing in 2D and 3D. F1000Res 5: 2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léger B, Cartoni R, Praz M, Lamon S, Dériaz O, Crettenand A, Gobelet C, Rohmer P, Konzelmann M, Luthi F et al (2006) Akt signalling through GSK‐3β, mTOR and Foxo1 is involved in human skeletal muscle hypertrophy and atrophy. J Physiol 576: 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Brizzee C, Johnson GVW (2015) BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiol Aging 36: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke SB, Weidemann T, Cost A‐L, Grashoff C, Schnorrer F (2019) A small proportion of Talin molecules transmit forces at developing muscle attachments in vivo. PLOS Biol 17: e3000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon R, Randles MJ, Humphries MJ (2014) The importance of podocyte adhesion for a healthy glomerulus. Front Endocrinol 5: 160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley K, Laudanna C, Cybulsky MI, Nourshargh S (2007) Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol 7: 678–689 [DOI] [PubMed] [Google Scholar]
- Li J, Labbadia J, Morimoto RI (2017) Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol 27: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Springer TA (2017) Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc Natl Acad Sci USA 114: 4685–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Y, Cui L, Wang J, Pang X, Lai Y, Yao Y, Liu X, Li Y (2013) Mechanical stretch changes coronary artery fibroblasts function by upregulating HSF1 protein expression. Int J Biol Macromol 59: 105–110 [DOI] [PubMed] [Google Scholar]
- Linke WA, Hamdani N (2014) Gigantic business: titin properties and function through thick and thin. Circ Res 114: 1052–1068 [DOI] [PubMed] [Google Scholar]
- Llano‐Diez M, Fury W, Okamoto H, Bai Y, Gromada J, Larsson L (2019) RNA‐sequencing reveals altered skeletal muscle contraction, E3 ligases, autophagy, apoptosis, and chaperone expression in patients with critical illness myopathy. Skelet Muscle 9: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loescher CM, Breitkreuz M, Li Y, Nickel A, Unger A, Dietl A, Schmidt A, Mohamed BA, Kötter S, Schmitt JP et al (2020) Regulation of titin‐based cardiac stiffness by unfolded domain oxidation (UnDOx). Proc Natl Acad Sci USA 117: 24545–24556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Mohan K, Iglesias PA, Robinson DN (2013) Molecular mechanisms of cellular mechanosensing. Nat Mater 12: 1064–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthold C, Varlet A‐A, Lambert H, Bordeleau F, Lavoie JN (2020) Chaperone‐assisted mitotic actin remodeling by BAG3 and HSPB8 involves the deacetylase HDAC6 and its substrate cortactin. Int J Mol Sci 22: 142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainz A, Peschek J, Stavropoulou M, Back KC, Bardiaux B, Asami S, Prade E, Peters C, Weinkauf S, Buchner J et al (2015) The chaperone αB‐crystallin uses different interfaces to capture an amorphous and an amyloid client. Nat Struct Mol Biol 22: 898–905 [DOI] [PubMed] [Google Scholar]
- Mammoto T, Mammoto A, Ingber DE (2013) Mechanobiology and developmental control. Annu Rev Cell Dev Biol 29: 27–61 [DOI] [PubMed] [Google Scholar]
- Mao Y, Schneider R, van der Ven PFM, Assent M, Lohanadan K, Klämbt V, Buerger F, Kitzler TM, Deutsch K, Nakayama M et al (2020) Recessive mutations in SYNPO2 as a candidate of monogenic nephrotic syndrome. Kidney Int Rep 6: 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TG, Myers VD, Dubey P, Dubey S, Perez E, Moravec CS, Willis MS, Feldman AM, Kirk JA (2021) Cardiomyocyte contractile impairment in heart failure results from reduced BAG3‐mediated sarcomeric protein turnover. Nat Commun 12: 2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mártonfalvi Z, Bianco P, Linari M, Caremani M, Nagy A, Lombardi V, Kellermayer M (2014) Low‐force transitions in single titin molecules reflect a memory of contractile history. J Cell Sci 127: 858–870 [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M (2009) Autophagy is required to maintain muscle mass. Cell Metab 10: 507–515 [DOI] [PubMed] [Google Scholar]
- Maurer M, Lammerding J (2019) The driving force: nuclear mechanotransduction in cellular function, fate, and disease. Annu Rev Biomed Eng 21: 443–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew DL, Kim JS, Cross JM, Ferrando AA, Bamman MM (2009) Translational signaling responses preceding resistance training‐mediated myofiber hypertrophy in young and old humans. J Appl Physiol 107: 1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod M, Breen L, Hamilton DL, Philp A (2016) Live strong and prosper: the importance of skeletal muscle strength for healthy ageing. Biogerontology 17: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister‐Broekema M, Freilich R, Jagadeesan C, Rauch JN, Bengoechea R, Motley WW, Kuiper EFE, Minoia M, Furtado GV, van Waarde MAWH et al (2018) Myopathy associated BAG3 mutations lead to protein aggregation by stalling Hsp70 networks. Nat Commun 9: 5342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkani GC, Bodmer R, Ocorr K, Bernstein SI (2011) The UNC‐45 chaperone is critical for establishing myosin‐based myofibrillar organization and cardiac contractility in the drosophila heart model. PLoS One 6: e22579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD (2014) Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156: 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Wei SH, Parker I, Cahalan MD (2002) Two‐photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 296: 1869–1873 [DOI] [PubMed] [Google Scholar]
- Moore SW, Roca‐Cusachs P, Sheetz MP (2010) Stretchy proteins on stretchy substrates: the important elements of integrin‐mediated rigidity sensing. Dev Cell 19: 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhre JL, Hills JA, Jean F, Pilgrim DB (2014) Unc45b is essential for early myofibrillogenesis and costamere formation in zebrafish. Dev Biol 390: 26–40 [DOI] [PubMed] [Google Scholar]
- Naidu SD, Dinkova‐Kostova AT (2017) Regulation of the mammalian heat shock factor 1. FEBS J 284: 1606–1627 [DOI] [PubMed] [Google Scholar]
- Nakamura F, Song M, Hartwig JH, Stossel TP (2014) Documentation and localization of force‐mediated filamin A domain perturbations in moving cells. Nat Commun 5: 4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH (2011) The filamins: organizers of cell structure and function. Cell Adh Migr 5: 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava MM, Miroshnikova YA, Biggs LC, Whitefield DB, Metge F, Boucas J, Vihinen H, Jokitalo E, Li X, García Arcos JM et al (2020) Heterochromatin‐driven nuclear softening protects the genome against mechanical stress‐induced damage. Cell 181: 800–817.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ME, Parker BL, Burchfield JG, Hoffman NJ, Needham EJ, Cooke KC, Naim T, Sylow L, Ling NX, Francis D et al (2019) Phosphoproteomics reveals conserved exercise‐stimulated signaling and AMPK regulation of store‐operated calcium entry. EMBO J 38: e102578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- New LA, Martin CE, Jones N (2014) Advances in slit diaphragm signaling. Curr Opin Nephrol Hypertens 23: 420–430 [DOI] [PubMed] [Google Scholar]
- Niediek V, Born S, Hampe N, Kirchgessner N, Merkel R, Hoffmann B (2012) Cyclic stretch induces reorientation of cells in a Src family kinase‐ and p130Cas‐dependent manner. Eur J Cell Biol 91: 118–128 [DOI] [PubMed] [Google Scholar]
- Nillegoda NB, Wentink AS, Bukau B (2018) Protein disaggregation in multicellular organisms. Trends Biochem Sci 43: 285–300 [DOI] [PubMed] [Google Scholar]
- Noethel B, Ramms L, Dreissen G, Hoffmann M, Springer R, Rübsam M, Ziegler WH, Niessen CM, Merkel R, Hoffmann B (2018) Transition of responsive mechanosensitive elements from focal adhesions to adherens junctions on epithelial differentiation. Mol Biol Cell 29: 2317–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi J, Dunn DM, Woodford MR, Mariotti L, Schulman J, Bourboulia D, Mollapour M, Vaughan CK (2016) Structural and functional basis of protein phosphatase 5 substrate specificity. Proc Natl Acad Sci USA 113: 9009–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochala J, Gustafson A‐M, Diez ML, Renaud G, Li M, Aare S, Qaisar R, Banduseela VC, Hedström Y, Tang X et al (2011) Preferential skeletal muscle myosin loss in response to mechanical silencing in a novel rat intensive care unit model: underlying mechanisms. J Physiol 589: 2007–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odagiri S, Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K (2012) Autophagic adapter protein NBR1 is localized in Lewy bodies and glial cytoplasmic inclusions and is involved in aggregate formation in α‐synucleinopathy. Acta Neuropathol 124: 173–186 [DOI] [PubMed] [Google Scholar]
- Pathak P, Blech‐Hermoni Y, Subedi K, Mpamugo J, Obeng‐Nyarko C, Ohman R, Molloy I, Kates M, Hale J, Stauffer S et al (2021) Myopathy associated LDB3 mutation causes Z‐disc disassembly and protein aggregation through PKCα and TSC2‐mTOR downregulation. Commun Biol 4: 355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perico L, Conti S, Benigni A, Remuzzi G (2016) Podocyte‐actin dynamics in health and disease. Nat Rev Nephrol 12: 692–710 [DOI] [PubMed] [Google Scholar]
- Peschek J, Braun N, Rohrberg J, Christiane Back K, Kriehuber T, Kastenmüller A, Weinkauf S, Buchner J (2013) Regulated structural transitions unleash the chaperone actièity of αb‐crystallin. Proc Natl Acad Sci USA 110: E3780–E3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrzywa W, Hoppe T (2013) Chaperoning myosin assembly in muscle formation and aging. Worm 2: e25644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MG, Landsverk ML, Barral JM, Epstein HF (2002) Two mammalian UNC‐45 isoforms are related to distinct cytoskeletal and muscle‐specific functions. J Cell Sci 115: 4013–4023 [DOI] [PubMed] [Google Scholar]
- Proske U, Morgan DL (2001) Muscle damage from eccentric exercise: mechanism, mechanical signs, adaptation and clinical applications. J Physiol 537: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle WG, Solaro RJ (2004) At the crossroads of myocardial signaling. Circ Res 94: 296–305 [DOI] [PubMed] [Google Scholar]
- Quintana MT, Parry TL, He J, Yates CC, Sidorova TN, Murray KT, Bain JR, Newgard CB, Muehlbauer MJ, Eaton SC et al (2016) Cardiomyocyte‐specific human Bcl2‐associated anthanogene 3 P209L expression induces mitochondrial fragmentation, Bcl2‐associated anthanogene 3 haploinsufficiency, and activates p38 signaling. Am J Pathol 186: 1989–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P (2008) Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet 17: 3897–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahikainen R, Ohman T, Turkki P, Varjosalo M, Hytonen VP (2019) Talin‐mediated force transmission and talin rod domain unfolding independently regulate adhesion signaling. J Cell Sci 132: jcs226514 [DOI] [PubMed] [Google Scholar]
- Rauch JN, Tse E, Freilich R, Mok S‐A, Makley LN, Southworth DR, Gestwicki JE (2017) BAG3 Is a modular, scaffolding protein that physically links heat shock protein 70 (Hsp70) to the small heat shock proteins. J Mol Biol 429: 128–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann L, Schwäble AN, Fricke AL, Mühlhäuser WWD, Leber Y, Lohanadan K, Puchinger MG, Schäuble S, Faessler E, Wiese H et al (2020) Phosphoproteomics identifies dual‐site phosphorylation in an extended basophilic motif regulating FILIP1‐mediated degradation of filamin‐C. Commun Biol 3: 253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann L, Wiese H, Leber Y, Schwäble AN, Fricke AL, Rohland A, Knapp B, Peikert CD, Drepper F, van der Ven PFM et al (2017) Myofibrillar Z‐discs are a protein phosphorylation hot spot with protein kinase C (PKCα) modulating protein dynamics. Mol Cell Proteomics 16: 346–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner DJ, Lundquist EA (2018) Small GTPases. WormBook 2018: 1–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J (2010) The heat shock response: life on the verge of death. Mol Cell 40: 253–266 [DOI] [PubMed] [Google Scholar]
- Rief M, Pascual J, Saraste M, Gaub HE (1999) Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J Mol Biol 286: 553–561 [DOI] [PubMed] [Google Scholar]
- Ringer P, Weißl A, Cost AL, Freikamp A, Sabass B, Mehlich A, Tramier M, Rief M, Grashoff C (2017) Multiplexing molecular tension sensors reveals piconewton force gradient across talin‐1. Nat Methods 14: 1090–1096 [DOI] [PubMed] [Google Scholar]
- Rinschen MM, Gödel M, Grahammer F, Zschiedrich S, Helmstädter M, Kretz O, Zarei M, Braun DA, Dittrich S, Pahmeyer C et al (2018) A Multi‐layered quantitative in vivo expression atlas of the podocyte unravels kidney disease candidate genes. Cell Rep 23: 2495–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinschen MM, Grahammer F, Hoppe A‐K, Kohli P, Hagmann H, Kretz O, Bertsch S, Höhne M, Göbel H, Bartram MP et al (2017a) YAP‐mediated mechanotransduction determines the podocyte’s response to damage. Sci Signal 10: eaaf8165 [DOI] [PubMed] [Google Scholar]
- Rinschen MM, Hoppe A‐K, Grahammer F, Kann M, Völker LA, Schurek E‐M, Binz J, Höhne M, Demir F, Malisic M et al (2017b) N‐degradomic analysis reveals a proteolytic network processing the podocyte cytoskeleton. J Am Soc Nephrol 28: 2867–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinschen MM, Schroeter CB, Koehler S, Ising C, Schermer B, Kann M, Benzing T, Brinkkoetter PT (2016) Quantitative deep mapping of the cultured podocyte proteome uncovers shifts in proteostatic mechanisms during differentiation. Am J Physiol Cell Physiol 311: C404–C417 [DOI] [PubMed] [Google Scholar]
- Rinschen MM, Wu X, König T, Pisitkun T, Hagmann H, Pahmeyer C, Lamkemeyer T, Kohli P, Schnell N, Schermer B et al (2014) Phosphoproteomic analysis reveals regulatory mechanisms at the kidney filtration barrier. J Am Soc Nephrol 25: 1509–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A, Perez‐Jimenez R, Liu R, Roca‐Cusachs P, Fernandez JM, Sheetz MP (2009) Stretching single talin rod molecules activates vinculin binding. Science 323: 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni L, Möst T, Žoldák G, Rief M (2014) Force‐dependent isomerization kinetics of a highly conserved proline switch modulates the mechanosensing region of filamin. Proc Natl Acad Sci USA 111: 5568–5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni L, Stigler J, Pelz B, Ylänne J, Rief M (2012) Dynamic force sensing of filamin revealed in single‐molecule experiments. Proc Natl Acad Sci USA 109: 19679–19684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ (2001) Mediation of IGF‐1‐induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol 3: 1009–1013 [DOI] [PubMed] [Google Scholar]
- Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B (2019) The Hsp70 chaperone network. Nat Rev Mol Cell Biol 20: 665–680 [DOI] [PubMed] [Google Scholar]
- Ruggieri A, Saredi S, Zanotti S, Pasanisi MB, Maggi L, Mora M (2016) DNAJB6 myopathies: focused review on an emerging and expanding group of myopathies. Front Mol Biosci 3: 63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruparelia AA, Oorschot V, Vaz R, Ramm G, Bryson‐Richardson RJ (2014) Zebrafish models of BAG3 myofibrillar myopathy suggest a toxic gain of function leading to BAG3 insufficiency. Acta Neuropathol 128: 821–833 [DOI] [PubMed] [Google Scholar]
- Ruparelia AA, Oorschot V, Ramm G, Bryson‐Richardson RJ (2016) FLNC myofibrillar myopathy results from impaired autophagy and protein insufficiency. Hum Mol Genet 25: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Sachs N, Sonnenberg A (2013) Cell‐matrix adhesion of podocytes in physiology and disease. Nat Rev Nephrol 9: 200–210 [DOI] [PubMed] [Google Scholar]
- Saini K, Discher DE (2019) Forced unfolding of proteins directs biochemical cascades. Biochemistry 58: 4893–4902 [DOI] [PubMed] [Google Scholar]
- Sala AJ, Bott LC, Morimoto RI (2017) Shaping proteostasis at the cellular, tissue, and organismal level. J Cell Biol 216: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah H, Li M, Cacciani N, Gastaldello S, Ogilvie H, Akkad H, Namuduri AV, Morbidoni V, Artemenko KA, Balogh G et al (2016) The chaperone co‐inducer BGP‐15 alleviates ventilation‐induced diaphragm dysfunction. Sci Transl Med 8: 350ra103 [DOI] [PubMed] [Google Scholar]
- Sandri M (2010) Autophagy in skeletal muscle. FEBS Lett 584: 1411–1416 [DOI] [PubMed] [Google Scholar]
- Sarparanta J, Jonson PH, Golzio C, Sandell S, Luque H, Screen M, McDonald K, Stajich JM, Mahjneh I, Vihola A et al (2012) Mutations affecting the cytoplasmic functions of the co‐chaperone DNAJB6 cause limb‐girdle muscular dystrophy. Nat Genet 44: 450–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin‐Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP (2006) Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127: 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schänzer A, Rupp S, Gräf S, Zengeler D, Jux C, Akintürk H, Gulatz L, Mazhari N, Acker T, Van Coster R et al (2018) Dysregulated autophagy in restrictive cardiomyopathy due to Pro209Leu mutation in BAG3. Mol Genet Metab 123: 388–399 [DOI] [PubMed] [Google Scholar]
- Schell C, Huber TB (2017) The evolving complexity of the podocyte cytoskeleton. J Am Soc Nephrol 28: 3166–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffhauer ESS, Luo T, Mohan K, Srivastava V, Qian X, Griffis ERR, Iglesias PAA, Robinson DNN (2016) Mechanoaccumulative elements of the mammalian actin cytoskeleton. Curr Biol 26: 1473–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller HB, Fässler R (2013) Mechanosensitivity and compositional dynamics of cell‐matrix adhesions. EMBO Rep 14: 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Perdiguero EG, Chorro L, Szabo‐Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SEW, Pollard JW et al (2012) A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science 335: 86–90 [DOI] [PubMed] [Google Scholar]
- Schwartzman M, Reginensi A, Wong JS, Basgen JM, Meliambro K, Nicholas SB, D’Agati V, McNeill H, Campbell KN (2016) Podocyte‐specific deletion of yes‐associated protein causes FSGS and progressive renal failure. J Am Soc Nephrol 27: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcen D, Muntoni F, Burton BK, Pegoraro E, Sewry C, Bite A, Engel AG (2009) Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol 65: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamri R, Grabovsky V, Gauguet J‐M, Feigelson S, Manevich E, Kolanus W, Robinson MK, Staunton DE, von Andrian UH, Alon R (2005) Lymphocyte arrest requires instantaneous induction of an extended LFA‐1 conformation mediated by endothelium‐bound chemokines. Nat Immunol 6: 497–506 [DOI] [PubMed] [Google Scholar]
- Simon DN, Wilson KL (2011) The nucleoskeleton as a genome‐associated dynamic ‘network of networks’. Nat Rev Mol Cell Biol 12: 695–708 [DOI] [PubMed] [Google Scholar]
- Small JV, Resch GP (2005) The comings and goings of actin: coupling protrusion and retraction in cell motility. Curr Opin Cell Biol 17: 517–523 [DOI] [PubMed] [Google Scholar]
- Srivastava T, Hariharan S, Alon US, McCarthy ET, Sharma R, El‐Meanawy A, Savin VJ, Sharma M (2018) Hyperfiltration‐mediated injury in the remaining kidney of a transplant donor. Transplantation 102: 1624–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swist S, Unger A, Li Y, Vöge A, von Frieling‐Salewsky M, Skärlén Å, Cacciani N, Braun T, Larsson L, Linke WA (2020) Maintenance of sarcomeric integrity in adult muscle cells crucially depends on Z‐disc anchored titin. Nat Commun 11: 4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, Larsen B, Choi H, Berger B, Gingras AC, Lindquist S (2014) A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 158: 434–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann‐Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P (2000) Accumulation of autophagic vacuoles and cardiomyopathy in LAMP‐2‐deficient mice. Nature 406: 902–906 [DOI] [PubMed] [Google Scholar]
- Ulbricht A, Eppler FJ, Tapia VE, Van Der Ven PFM, Hampe N, Hersch N, Vakeel P, Stadel D, Haas A, Saftig P et al (2013) Cellular mechanotransduction relies on tension‐induced and chaperone‐assisted autophagy. Curr Biol 23: 430–435 [DOI] [PubMed] [Google Scholar]
- Ulbricht A, Gehlert S, Leciejewski B, Schiffer T, Bloch W, Höhfeld J (2015) Induction and adaptation of chaperone‐assisted selective autophagy CASA in response to resistance exercise in human skeletal muscle. Autophagy 11: 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungelenk S, Moayed F, Ho C‐T, Grousl T, Scharf A, Mashaghi A, Tans S, Mayer MP, Mogk A, Bukau B (2016) Small heat shock proteins sequester misfolding proteins in near‐native conformation for cellular protection and efficient refolding. Nat Commun 7: 13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valignat M‐P, Theodoly O, Gucciardi A, Hogg N, Lellouch AC (2013) T lymphocytes orient against the direction of fluid flow during LFA‐1‐mediated migration. Biophys J 104: 322–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlet AA, Fuchs M, Luthold C, Lambert H, Landry J, Lavoie JN (2017) Fine‐tuning of actin dynamics by the HSPB8‐BAG3 chaperone complex facilitates cytokinesis and contributes to its impact on cell division. Cell Stress Chaperones 22: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatareddy M, Cook L, Abuarquob K, Verma R, Garg P (2011) Nephrin regulates lamellipodia formation by assembling a protein complex that includes Ship2. Filamin and Lamellipodin. PLoS One 6: e28710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L, Waterston RH (1990) The unc‐45 gene of Caenorhabditis elegans is an essential muscle‐affecting gene with maternal expression. Genetics 126: 345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Sistonen L (2014) HSF1 at a glance. J Cell Sci 127: 261–266 [DOI] [PubMed] [Google Scholar]
- Voelkel T, Andresen C, Unger A, Just S, Rottbauer W, Linke WA (2013) Lysine methyltransferase Smyd2 regulates Hsp90‐mediated protection of the sarcomeric titin springs and cardiac function. Biochim Biophys Acta Mol Cell Res 1833: 812–822 [DOI] [PubMed] [Google Scholar]
- Wackerhage H, Del Re DP, Judson RN, Sudol M, Sadoshima J (2014) The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal 7: re4–re4 [DOI] [PubMed] [Google Scholar]
- Woerner Ac, Frottin F, Hornburg D, Feng Lr, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer Kf, Hartl Fu et al (2016) Cytoplasmic protein aggregates interfere with nucleo‐cytoplasmic transport of protein and RNA. Science 351: 173–176 [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SL, Crawford BD, Pilgrim DB (2007) The myosin co‐chaperone UNC‐45 is required for skeletal and cardiac muscle function in zebrafish. Dev Biol 303: 483–492 [DOI] [PubMed] [Google Scholar]
- Wolfenson H, Yang B, Sheetz MP (2019) Steps in mechanotransduction pathways that control cell morphology. Annu Rev Physiol 81: 585–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zhang X, Guo Y, Meng F, Sachs F, Guo J (2015) Mechanical dynamics in live cells and fluorescence‐based force/tension sensors. Biochim Biophys Acta Mol Cell Res 1853: 1889–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J, Hoffman BD, Crocker JC, Bershadsky AD, Balaban NQ et al (2014a) Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep 4: 259–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Goult BT, Klapholz B, Hu X, Toseland CP, Guo Y, Cong P, Sheetz MP, Yan J (2016) The mechanical response of talin. Nat Commun 7: 11966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Qiu W, Liu R, Efremov AK, Cong P, Seddiki R, Payre M, Lim CT, Ladoux B, Mège R‐M et al (2014b) Force‐dependent conformational switch of α‐catenin controls vinculin binding. Nat Commun 5: 4525 [DOI] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M (2010) alpha‐Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 12: 533–542 [DOI] [PubMed] [Google Scholar]
- Yu F‐X, Guan K‐L (2013) The Hippo pathway: regulators and regulations. Genes Dev 27: 355–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko EC, Asbury CL (2014) Force is a signal that cells cannot ignore. Mol Biol Cell 25: 3717–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kim TH, Thauland TJ, Li H, Majedi FS, Ly C, Gu Z, Butte MJ, Rowat AC, Li S (2020) Unraveling the mechanobiology of immune cells. Curr Opin Biotechnol 66: 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]


