SUMMARY
Glucocorticoids (GCs) are widely used as anti-inflammatory drugs, but their long-term use has severe metabolic side effects. Here, by treating multiple individual adipose stem cell-derived adipocytes and induced pluripotent stem cell-derived hepatocytes with the potent GC, dexamethasone (Dex), we uncovered cell-type-specific and individual-specific GC-dependent transcriptomes and glucocorticoid receptor (GR) cistromes. Individual-specific GR binding could be traced to single nucleotide polymorphisms (SNPs) that altered the binding motifs of GR or its cooperating factors. We also discovered another set of genetic variants that modulated Dex response through affecting chromatin accessibility or chromatin architecture. Several SNPs that altered Dex-regulated GR binding and gene expression controlled Dex-driven metabolic perturbations. Remarkably, these genetic variations were highly associated with increases in serum glucose, lipids, and body mass in subjects on GC therapy. Knowledge of the genetic variants that predispose individuals to metabolic side effects allows for a precision medicine approach to the use of clinically-relevant GCs.
In Brief:
Applying functional genomics to individual stem cell-derived adipocytes and hepatocytes Hu and colleagues reveal natural genetic variations that modulate and determine the metabolic side effects of a clinically-relevant glucocorticoid.
INTRODUCTION
Glucocorticoids (GCs) are steroid hormones that play an important role in regulating diverse physiologic processes including energy homeostasis and the immune response (Patel et al., 2014; Sacta et al., 2016). Clinically-relevant GCs are highly effective and widely prescribed for the treatment of severe inflammation, but their use causes many metabolic side effects (Barnes, 2011; Desmet and De Bosscher, 2017). GCs function by targeting the glucocorticoid receptor (GR) in multiple tissues, including adipose and liver (Sacta et al., 2016). Excess GCs cause a severe pathology referred to as Cushing’s syndrome, which refers to a constellation of central obesity with variable hyperglycemia, dyslipidemia and hepatic steatosis (Mazziotti et al., 2011). GCs exert complex effects on lipid metabolism in adipose tissues, generating glycerol and fatty acids, which provide substrate and energy for hepatic gluconeogenesis (Chen et al., 1999; Yu et al., 2010). GCs also directly regulate lipogenic gene expression by working with other factors, such as insulin (Gathercole et al., 2011). In the liver, GC-activated GR has been shown to bind the promoter regions of PCK1 and G6PC, whose protein products catalyze the rate-limiting and final step of gluconeogenesis, respectively (Imai et al., 1993; She et al., 2000). This enhances hepatic glucose production, leading to diabetes in individuals treated with GCs (Brenner et al., 2020; Desmet and De Bosscher, 2017). GCs also augment insulin-stimulated lipogenesis and reduce lipolysis in the liver, contributing to hypertriglyceridemia and hepatic steatosis observed in people with Cushing’s disease and those undergoing exogenous GC therapy (Dolinsky et al., 2004; Rockall et al., 2003; Sholter and Armstrong, 2000).
In addition to the use of GCs to treat inflammatory conditions, such as rheumatoid arthritis, systemic lupus erythematosus and inflammatory bowel disease, they are frequently included in chemotherapeutic regimens (Pufall, 2015). Moreover, the powerful GR ligand dexamethasone (Dex) is now widely used to treat people with advanced COVID-19 (Group et al., 2020); yet metabolic dysfunction increases the risk and severity of COVID infection (Zhu et al., 2020). Thus, a better understanding of the underlying mechanisms of GC response in the liver and adipose tissue at the individual-specific level could help precision medicine approach to guide therapy and minimize unwanted side effects.
Precision medicine holds great promise for disease prevention and treatment, since individuals differ in their genetics, epigenetics and environment (Costantino et al., 2018; Shastry, 2002). Genetic variations in the form of single nucleotide polymorphisms (SNPs) are statistically associated with complex diseases and modulate the individual’s response to drugs, pathogen and other environmental stimuli (Groner and Brown, 2017; Lee et al., 2014; Matsa et al., 2016). The majority of SNPs identified by genome-wide association studies (GWASs) are located in non-coding regions of the genome and are thus likely to serve as regulatory regions that affect the openness of chromatin and the genomic binding of sequence-specific transcription factors (TFs) that regulate nearby gene expression, or modulate distal gene expression through affecting chromatin architecture (Deplancke et al., 2016; Waszak et al., 2015). There is a great interest in developing appropriate research platforms to study how genetic variants modulate drug responses, considering the timing, cost, availability and safety of treatment with a given medication (Visscher et al., 2017). Individual-specific stem cell-derived cell types provide a useful platform for studying the individual response to drugs (Hu et al., 2019; Matsa et al., 2016).
Here, we focused on individual stem cell-derived adipocytes and hepatocytes, the cell types responsible for much of the metabolic toxicity of GC therapy, to determine how genetic variations modulate human responses to Dex. We found that individual-derived adipocytes and hepatocytes show cell type-specific GR genomic occupancy and Dex responses, some of which are also individual-specific. Genetic variations transform DNA recognition motifs for GR or its cooperating TFs, or alter chromatin accessibility and/or architecture that result in the individual-specific drug response. Remarkably, genetic variations controlling the Dex-responsiveness of genes crucial for hepatocyte or adipocyte metabolism proved to be powerful predictors of metabolic disturbances in individuals receiving GC therapy.
RESULTS
Individual-specific response to Dex in human stem cell-derived adipocytes and hepatocytes
To better understand the mechanism by which Dex produces adverse metabolic effects in people treated with the drug, we generated adipocytes from human adipose stem cells (hASCs) and hepatocytes from human induced pluripotent stem cells (hiPSCs). Specifically, we isolated ASCs from the subcutaneous abdominal adipose tissue of healthy subjects undergoing elective abdominoplasty (Shan et al., 2017). Mature adipocytes were generated from hASCs in well-defined adipogenic differentiation medium for 2 weeks as previously described (Hu et al., 2019), cultured in maintenance medium for 1 week and then incubated with 1.5% BSA medium for an additional 2 days to wash out the potential influence of Dex that was included in the adipogenic and maintenance medium (Figure 1A). Eight individual adipocytes showing similar adipogenic differentiation efficiency according to the morphology and adipocyte maker genes expression were used to model the Dex response (Figures S1A, S1B and S1C). Meanwhile, we also successfully induced eleven hiPSC lines to differentiate into hepatocyte-like cells (HLCs) in a step-wise manner as previously described but omitting hydrocortisone in the maturation stage (Figure 1B) (Pashos et al., 2017). Hepatocyte marker genes Albumin (ALB) and HNF4 were expressed at comparable levels in all eleven hiPSC-derived HLCs based on immunostaining and western blot analysis (Figures S1D and S1E). Thus we have generated multiple individual-specific adipocytes and HLCs with similar properties from human stem cells as models to study Dex responses.
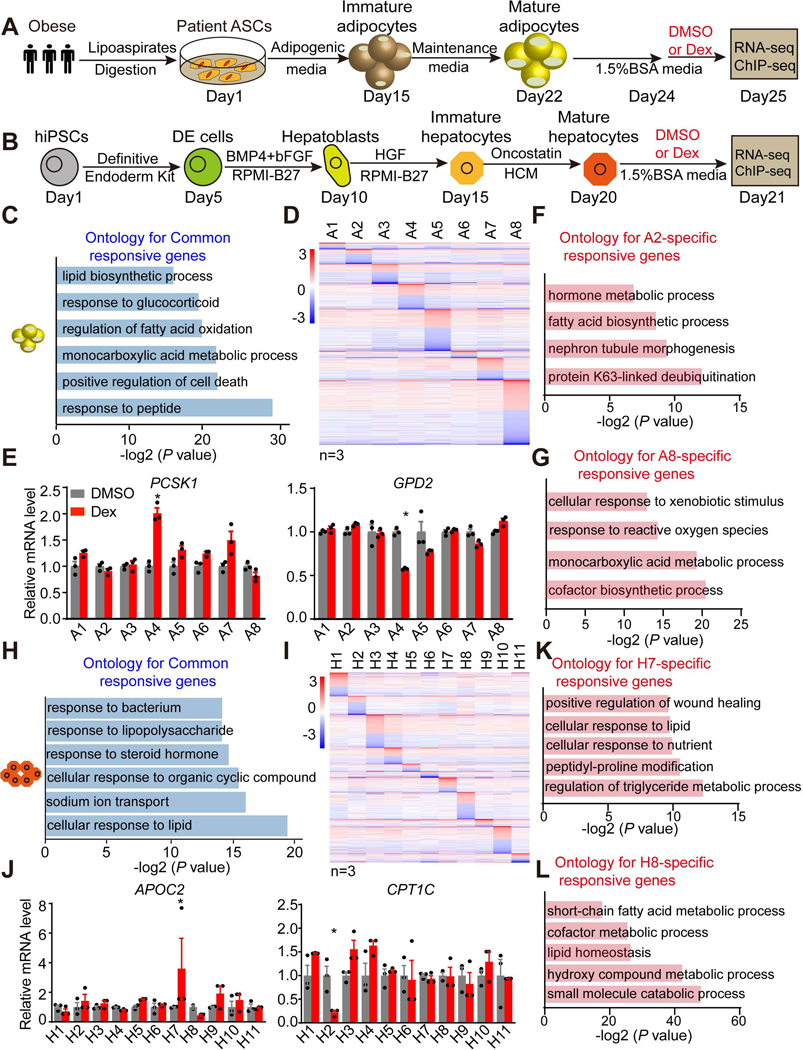
Figure 1. Individual-specific Dex responsiveness in individual stem cell-derived adipocytes and HLCs.
(A and B) Scheme of the adipogenic (A) and hepatic (B) differentiation procedure and DMSO or Dex (1 μM) treatment.
(C) Gene ontology for Dex-responsive genes in adipocytes from all eight individuals.
(D) Heatmap of individual-specific responsive genes that are significantly regulated by Dex in the adipocytes from only one individual. The color bar indicates log2(fold change) in Dex vs DMSO.
(E) mRNA expression of individual A4-specific upregulated gene PCSK1 and downregulated gene GPD2 in adipocytes from eight individuals, normalized to DMSO, as measured by RNA-seq.
(F and G) Gene ontology for A2-specific Dex-responsive genes (F) and A8-specific Dex-responsive genes (G).
(H) Gene ontology for Dex-responsive genes in HLCs from all eleven individuals.
(I) Heatmap of individual-specific responsive genes that are significantly regulated by Dex in only one individual’s HLCs. The color bar indicates log2(fold change) in Dex vs DMSO.
(J) mRNA expression of individual H7-specific upregulated gene APOC2 and H2-specific downregulated gene CPT1C in the HLCs of eleven individuals, normalized to DMSO, as measured by RNA-seq.
(K and L) Gene ontology for H7-specific Dex-responsive genes (K) and H8-specific Dex-responsive genes (L).
*P < 0.05, by Student’s t-test (E and J).
See also Figures S1–S3.
We first examined hASC-derived adipocytes treated with Dex for 24 hrs beginning at day 24 (Figure 1A). Reassuringly, three biological replicates of adipocytes from each individual treated with DMSO or Dex generally clustered together (Figures S1F and S1G). The transcriptomes showed larger inter-individual variation while replicates from the same individual were more highly correlated than that from different individuals (Figure S1H), adding confidence to our ability to detect individual-specific drug responses. Indeed, we found 696 genes that were regulated by Dex in all eight individual adipocytes (329 upregulated, 367 downregulated) (Figure S2A). Gene ontology (GO) and Gene Set Enrichment Analysis (GSEA) showed that, as expected, the genes commonly regulated by Dex were enriched for pathways including response to glucocorticoid, lipid biosynthetic processes and insulin signaling pathways (Figures 1C and S2B). For example, the classic GR-target gene FKBP5 was commonly induced by Dex while UCP2 was commonly repressed by Dex (Figures S2C and S2D). Intriguingly, besides the 696 commonly regulated genes, we also identified 1494 genes that were specifically responsive to Dex in adipocytes from only one individual (Figures 1D and S2E), which was not due to the variation of basal gene expression (Figure S2F). Moreover, there were 2466 genes that respond to Dex in adipocytes from multiple individuals. Subject A8 had the greatest number of individual-specific Dex-responsive genes (Figures 1D and S2G). Of note, two metabolic genes PCSK1 and GPD2 were uniquely induced or repressed by Dex in a individual-specific manner (Figure 1E). Gene ontology analysis revealed that fatty acid biosythesis pathway and other metabolic pathways were enriched in individual A2- and A8-specific responsive genes (Figures 1F and 1G). In contrast, we also noted that there were many genes unresponsive to Dex treatment in adipocytes derived from only one of the eight individuals (Figures S2H and S2I). These individual-specific responses were not attributable to differential GR (NR3C1) expression (Figure S2J). These data indicate that individual-derived adipocytes with different genetic backgrounds exhibit variable responses to Dex.
Exogenous GC administration in results in hyperglycemia in humans, largely due to uncontrolled hepatic gluconeogenesis (Desmet and De Bosscher, 2017). To test how hepatocytes derived from different iPSCs respond to Dex, we treated the iPSC-derived HLCs for 24 hrs (Figure 1B). Principle Component Analysis (PCA) revealed that biological replicates from the same individual and the same treatment condition nearly always clustered together (Figures S3A and S3B). Consistently, inter-individual variations were larger than that of replicates from the same individual (Figure S3C). 138 genes that were commonly regulated in all eleven HLCs were, as expected, enriched in pathways reflecting cellular response to lipid, response to steroid hormone, and glucose homeostasis (Figures 1H, S3D and S3E). For example, angiopoietin-like 4 (ANGPTL4), which regulates glucose homeostasis (Gusarova et al., 2018), was induced by Dex in all eleven HLCs (Figure S3F). The eleven individual HLCs exhibited a individual-specific response to Dex (Figure S3G), which was not due to variation in basal gene expression (Figure S3H). We were particularly interested in genes that responded to Dex in only one of eleven HLCs (Figures 1I and S3I), such as the two lipid metabolic genes Apolipoprotein C2 (APOC2) and Carnitine Palmitoyltransferase 1C (CPT1C) that were uniquely induced in H7 or repressed in H2 by Dex treatment, respectively (Figure 1J). Gene ontology analysis revealed that many lipid metabolic pathways were enriched in individual H7- and H8-specific responsive genes (Figures 1K and 1L). Conversely, a few genes were not responsive to Dex in HLCs from only one of the eleven individuals, such as Carbamoyl-Phosphate Synthase 1 (CPS1) (Figures S3J and S3K). These individual-specific responses were not attributable to differential GR (NR3C1) expression (Figure S3L). Furthermore, to examine if the 24hrs Dex treatment will induce indirect targets, secondary/tertiary responses, we picked some top Dex-responsive genes and examined their responses to Dex at different treatment times. The data showed that most genes responded to Dex similarly at 6h and 24h treatment, although some genes such as FKBP5 and MAOA exhibited attenuated or enhanced Dex responses upon prolonged Dex treatment, respectively (Figure S3M). Overall, these studies reveal the two cell types that are critical for glucose and lipid metabolism differentially respond to Dex treatment in different subjects.
Individual adipocytes and HLCs show individual-specific genomic occupancy of GR
GCs modulate target gene expression by binding to GR, causing its translocation to the nucleus where it binds to the genome in a sequence-specific manner (Syed et al., 2020). In adipocytes, we found that a 6hr Dex treatment induced GR translocation to the nucleus (Figure S4A). We performed chromatin immunoprecipitation sequencing (ChIP-seq) for GR in these individual stem cell-derived adipocytes that showed comparable GR expression level (Figure S4B), which further confirmed the requirement of GC treatment for GR genomic binding (Figure S4C). ChIP-seq analysis of all eight individual stem cell-derived adipocytes identified 11904 to 31362 GR peaks in the eight Dex-treated adipocytes, mainly located in the intronic and intergenic regions of the genome and highly enriched for the classic GR DNA binding motif, as expected (Figures 2A and S4D). GSEA analysis revealed that these GR peaks were enriched for pathways that include respond to glucocorticoid, insulin receptor signaling pathway and lipid metabolism (Figure S4E).
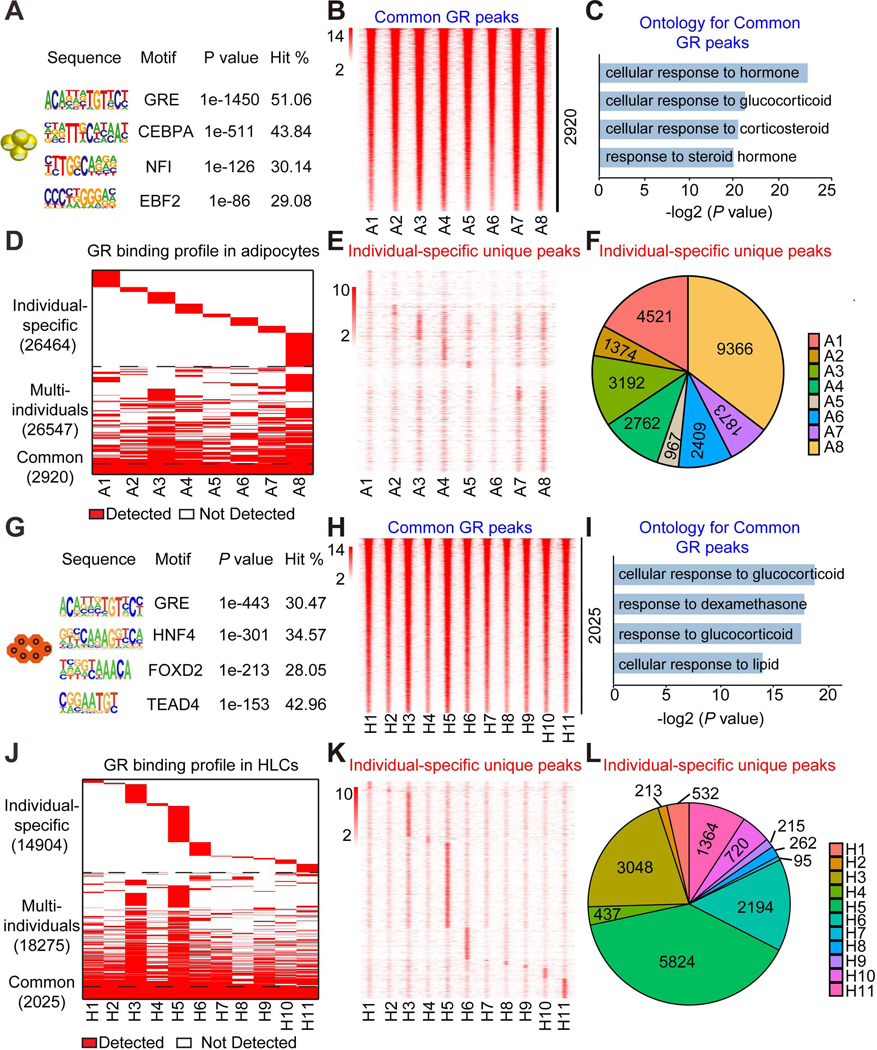
Figure 2. Differential Dex-induced genomic binding of GR in individual stem cell-derived adipocytes and HLCs.
(A) Consensus motif logos for the top-scoring motif families found in GR binding regions in adipocytes treated with Dex.
(B) Heatmap of common GR peaks that are detected similarly in adipocytes from all eight individuals. The color bar indicates the scale of normalized tag counts.
(C) Gene ontology for the nearest genes of GR binding sites that are detected similarly in adipocytes from eight individuals.
(D) Overall GR binding profile in adipocytes from eight individuals.
(E) Heatmap of individual-specific unique peaks that are specifically unique in adipocytes from only one individual.
(F) Proportion of individual-specific GR peaks that are specifically detected in adipocytes from only one individual.
(G) Consensus motif logos for the top-scoring motif families found in GR binding regions in HLCs treated with Dex.
(H) Heatmap of common GR peaks that are detected similarly in HLCs from all eleven individuals. The color bar indicates the scale of normalized tag counts.
(I) Gene ontology for the nearest genes of GR binding sites that are detected similarly in HLCs from all eleven individuals.
(J) Overall GR binding profile in HLCs from all eleven individuals.
(K) Heatmap of individual-specific unique peaks that are specifically unique in HLCs from only one individual.
(L) Proportion of individual-specific GR peaks that are specifically detected in HLCs from only one individual.
See also Figures S4–S5.
Similar studies were performed in HLCs. We first compared the genomic binding of GR after 6hrs and 24hrs of Dex treatment. The data revealed that the 6hr Dex treatment induced stronger genomic occupancy of GR (Figures S4F and S4G), consistent with previous findings in transformed cell lines (McDowell et al., 2018). After 6hrs, Dex treatment again led to the nuclear translocation of GR (Figure S4H), which was expressed at comparable levels in all eleven HLCs (Figure S4I). GR ChIP-seq showed that GR genomic binding reproducibly required Dex on a genome-wide scale (Figure S4J). Analysis of eleven Dex-treated HLCs identified 7485 to 24983 GR peaks, mainly located in the intronic and intergenic regions of the genome (Figure S4K). The genes near these GR peaks were enriched for pathways that included gluconeogenesis and annotated glucocorticoid responses (Figure S4L). These data demonstrate the robustness of the GR cistromes generated in adipocytes and HLCs treated with Dex.
We next compared the results across the individual-derived adipocytes and HLCs. Among the eight individual adipocyte samples, we detected 2920 shared sites with comparable binding intensity (Figure 2B). The genes near these common sites were highly enriched for functional annotation related to response to glucocorticoid (Figure 2C). Remarkably, in addition to the common GR binding sites, we also noted genomic occupancy of GR that was individual-specific (Figures 2D). For each individual studied, we found that some GR-binding sites were specifically gained (Figures 2E, 2F and S5A). Interestingly, subject A8 had the greatest number of individual-specific GR binding sites (Figures 2E and 2F), consistent with this individual having the most unique Dex-responsive gene regulation in the transcriptome analysis. In contrast, we also identified some GR binding sites that were absent in only one of eight individual adipocytes (Figure S5B), such as individual A5 whose adipocytes had 628 GR binding sites with an average intensity much lower than others (Figure S5C).
Parallel GR ChIP-seq analysis of all eleven HLCs revealed the GR peaks highly enriched for the canonical GR-binding motif (Figure 2G) and 2025 GR peaks with similar intensity across individuals, whose nearby genes were enriched for pathways including response to glucocorticoid (Figures 2H and 2I). Strikingly, similar to the findings in adipocytes, GR genomic binding also exhibited individual-specificity (Figure 2J), with some GR binding sites uniquely gained in HLCs from only one individual (Figures 2K, 2L and S5D), while other GR binding sites were specifically lost in only one of eleven individual HLCs (Figures S5E and S5F). Overall, the peak intensity of common peaks was higher than that of individual-specific GR peaks (Figures S5G and S5H). Together, these data revealed that a large fraction of genomic GR binding is different among individual adipocytes and HLCs.
Individual-specific GR binding drives differential Dex responses
We next tested whether the differential genomic GR binding drove the individual-specific Dex responses in adipocytes (Figure S6A). Indeed, Dex-regulated genes were more likely associated with GR binding sites than random genes (Figure 3A). GR binding sites were highly enriched in Dex-responsive genes (Figure S6B). Stronger GR binding promoted higher Dex response, and vice versa (Figures 3B and 3C). Individual-specific Dex-responsive genes were much more likely to be regulated by individual-specific GR binding sites than common GR binding sites (Figure S6C).
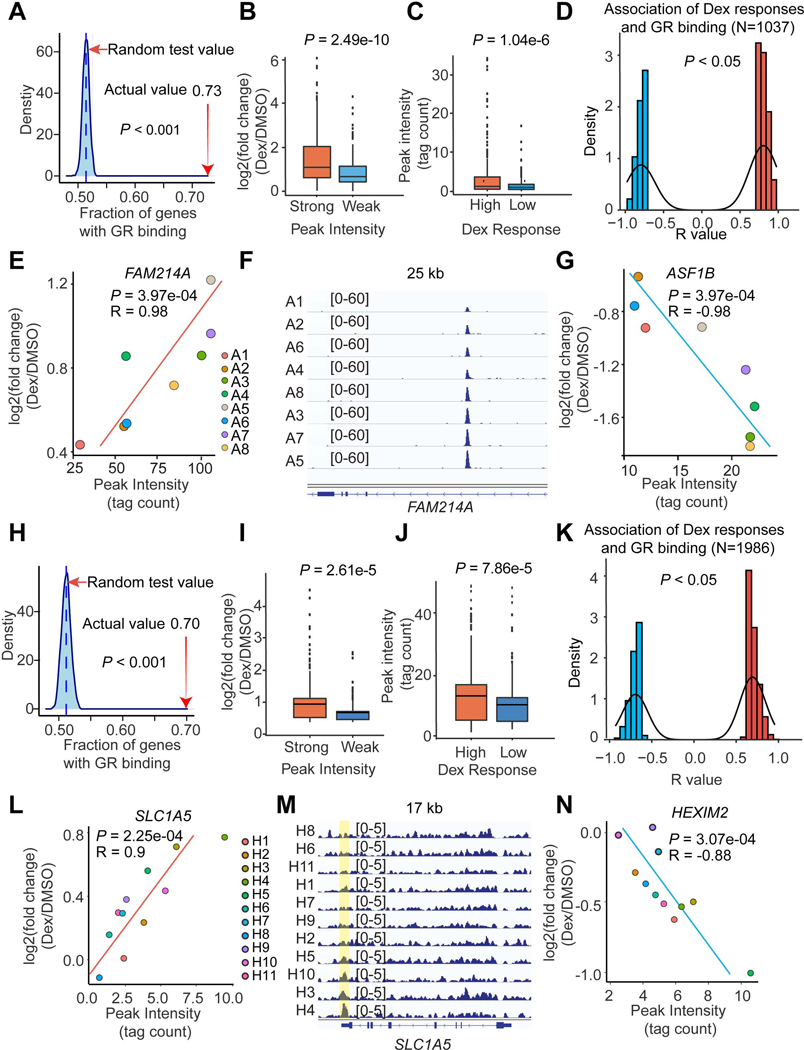
Figure 3. Differential GR binding drives individual-specific Dex responses.
(A and H) Dex-responsive genes are enriched in GR peaks in adipocytes (A) and HLCs (H) based on a random test. The percentage of Dex-responsive genes that have GR binding sites within 10 kb is indicated with a red arrow.
(B and I) The Dex responses near strong (Top 500 peaks) and weak (Bottom 500 peaks) GR binding sites in adipocytes (B) and HLCs (I).
(C and J) The GR binding intensities near high (Top 500 genes) and low (Bottom 500 genes) Dex-responsive genes in adipocytes (C) and HLCs (J).
(D and K) The number of gene-peak pairs in which gene responses to Dex are tightly associated with the intensities of GR bindings within 200 kb in adipocytes (D) and HLCs (K).
(E - G) Two example genes FAM214A (E) and ASF1B (G) whose responsiveness to Dex are positively or negatively associated with GR binding intensity in adipocytes. The GR-ChIP tracks at FAM214A locus were shown (F).
(L - N) Two example genes SLC1A5 (L) and HEXIM2 (N) whose responsiveness to Dex are positively or negatively associated with GR binding intensity in HLCs. The GR-ChIP tracks at SLC1A5 locus were shown (M).
The P values were calculated by permutation test (A and H), Student’s t-test (B, C, I and J) or R function cor.test (D, E, G, K, L and N).
See also Figures S6.
Combining the gene response and GR binding intensity in all eight individual adipocytes, we identified 1037 gene-peak pairs in which gene responses to Dex were related to the intensity of their nearby GR binding sites (Figure 3D). For example, the induction of FAM214A gene by Dex was strongly positively correlated with nearby GR binding intensity (Figures 3E and 3F), while ASF1B gene expression was tightly but inversely correlated with GR binding intensity (Figures 3G and S6D). The same analysis was also performed in HLCs (Figure S6E). Dex-regulated genes tended to have GR binding sites nearby (Figures 3H and S6F), with the binding intensity strongly correlating with the magnitude of the Dex response (Figures 3I and 3J). Individual-specific Dex-responsive genes were much more likely to be near individual-specific GR binding sites (Figure S6G). In total, 1986 gene-peak pairs demonstrated a close association between Dex response amplitude and GR binding intensity (Figure 3K), including SLC1A5 (Figures 3L and 3M) and HEXIM2 (Figures 3N and S6H) whose regulation by Dex was positively or negatively related to GR binding intensity, respectively. Collectively, these data indicate that individual-specific responses to Dex are determined by individual differences in genomic occupancy of GR in adipocytes and HLCs.
Cell type-specific Dex responses are related to differential GR binding
The functions of GCs are not only pleiotropic, including regulation of cell growth and development, metabolism, and inflammation, but also tissue-specific (Quatrini and Ugolini, 2020). Comparison of the adipocyte and HLC transcriptomes revealed that only a small fraction of genes that were Dex-regulated in at least half of the individuals overlapped between the two cell types (Figure 4A). The Dex-regulated genes in both individual-derived adipocytes and HLCs were enriched for cellular responses to lipid and lipid biosynthetic process (Figure 4B). We observed clear cell type-specific Dex responses (Figure 4A), which were not attributed to the differential basal gene expression profiles between adipocytes and HLCs (Figure S7A). GO analysis also showed that adipocyte-specific responsive genes were enriched for regulation of cell differentiation, ossification, and lipid metabolic processes (Figure S7B), while HLC-specific Dex-responsive genes were enriched for amino acid metabolic processes, regulation of hormone levels, and fatty acid biosynthetic processes (Figure S7C). Of note, Dex regulated many more genes in adipocytes than in HLCs, which may be due to the higher GR expression in adipocytes than in HLC (Figure S7D). Consistent with the small amount of common Dex-regulated genes, we also noted that GR binding sites detected in at least half of each individual population were rarely shared between adipocytes and HLCs (Figure 4C).
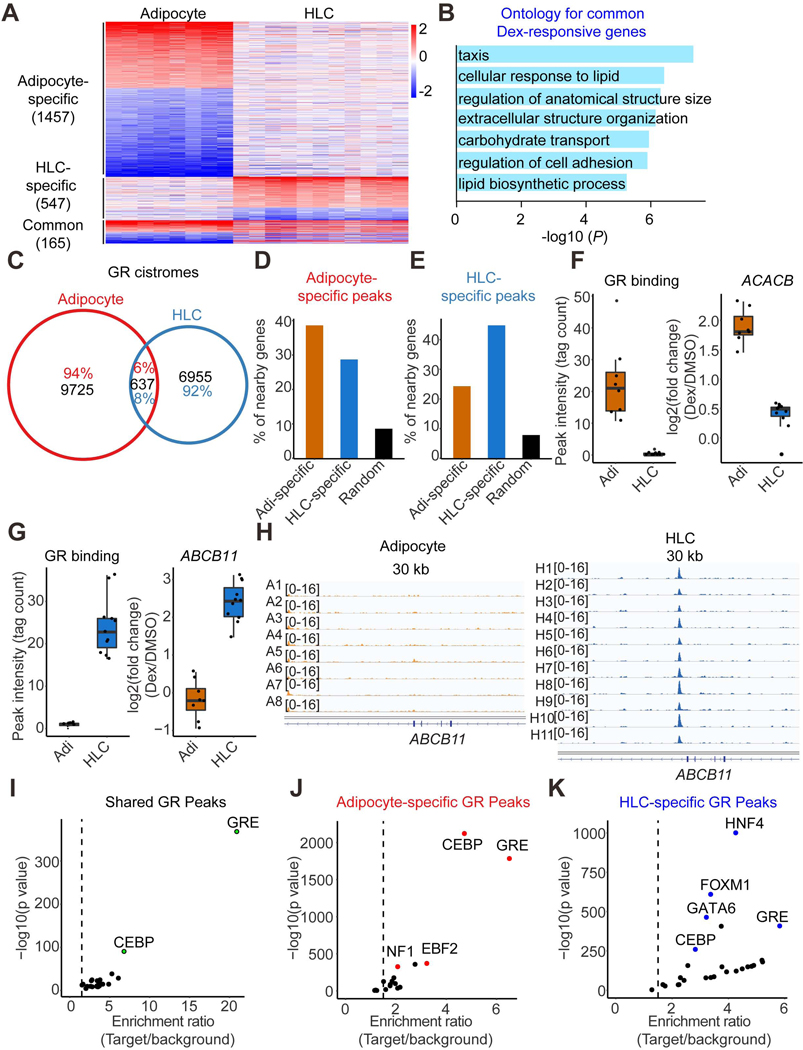
Figure 4. Cell type-specific Dex responses and GR genomic binding.
(A) Heatmap demonstrating the Dex-responsive genes in at least half of the individual adipocytes or HLCs. The color bar indicates log2(fold change) in Dex vs DMSO.
(B) Gene ontology for Dex-responsive genes observed in both adipocytes and HLCs.
(C) Venn diagram demonstrating the GR genomic binding in at least half of the individual adipocytes and HLCs.
(D and E) The percentage of adipocyte-specific Dex-responsive genes, HLCs-specific Dex-responsive genes or random genes that have nearby adipocyte-specific GR binding sites (D) or HLCs-specific GR binding sites (E).
(F) One example gene ACACB that’s induced by Dex in adipocytes only have higher nearby GR binding intensity in adipocytes compared to that in HLCs. Boxplots show median as a horizontal line, interquartile range as a box.
(G) One example gene ABCB11 that’s induced by Dex in HLCs only have higher nearby GR binding intensity in HLCs compared to that in adipocytes. Boxplots show median as a horizontal line, interquartile range as a box.
(H) GR ChIP-seq tracks at ABCB11 locus in adipocytes and HLCs.
(I-K) De novo motif analysis for common (I), adipocyte-specific (J) and HLC-specific (K) GR peaks.
See also Figures S7.
We next addressed the question of whether cell type-specific GR binding explained cell type-specific Dex responses. Approximately 39% of adipocyte-specific Dex-responsive genes had adipocyte-specific GR binding sites nearby, which was higher than that of HLC-specific Dex-responsive genes or random genes (Figure 4D). Similarly, ~45 percent of HLC-specific Dex-responsive genes were driven by HLC-specific GR binding sites nearby, which was higher than that of adipocyte-specific Dex-responsive genes or random genes (Figure 4E). For example, the Acetyl-CoA Carboxylase Beta gene (ACACB), which is important for fatty acid metabolism, was specifically induced by Dex in adipocytes, with much higher GR binding nearby in adipocytes than in HLCs (Figures 4F and S7E). Conversely, ATP Binding Cassette Subfamily B Member 11 (ABCB11) was specifically induced by Dex in HLCs, with stronger GR binding at this locus in HLCs than in adipocytes (Figures 4G and 4H). By contrast, the classic GR target gene FKBP5 was induced by Dex in both adipocytes and HLCs, with high intensity of GR binding at this locus (Figure S7F). These data suggest that cell type-specific Dex responses are mainly controlled by cell type-specific GR genomic binding.
To explore the underlying mechanisms driving the cell type-specific Dex response and GR binding, we performed de novo motif analysis for common and cell type-specific GR peaks. As expected, the GR peaks shared between adipocyte and HLCs were significantly enriched for GR- and CEBP-binding motifs (Figure 4I). By contrast, adipocyte-specific GR peaks were enriched for motifs shown to be involved in adipocyte lineage determination, such as NF1 and EBF (Hiraike et al., 2017; Rajakumari et al., 2013) (Figure 4J). Similarly, hepatocyte lineage determining factors HNF4 and Forkhead Box family members were enriched in HLC-specific GR binding sites (Figure 4K). These data suggest that lineage-determining TFs cooperate with GR to regulate adipocyte- and hepatocyte-specific Dex responses.
Genetic variations determine individual-specific Dex responsiveness
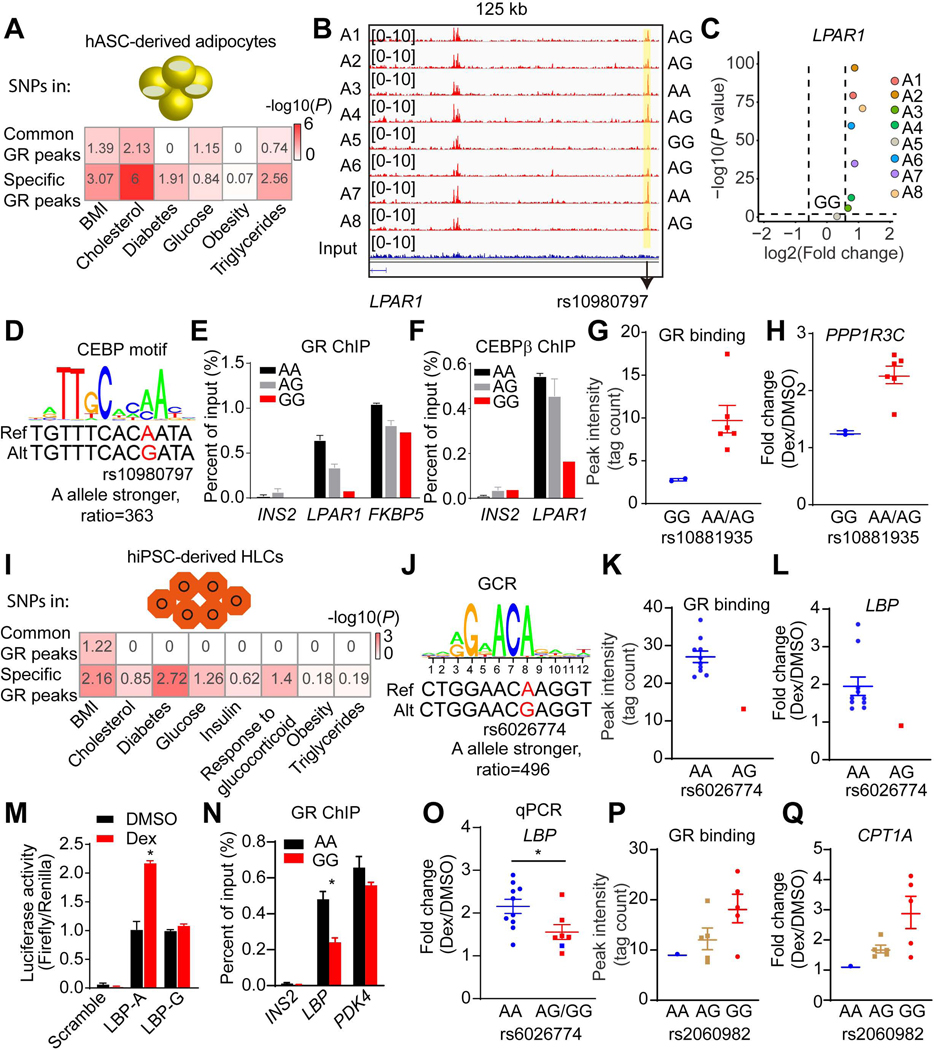
To determine whether the individual-specific GR peaks were associated with complex metabolic traits and diseases, we obtained GWAS summary statistics for metabolic disorders and traits (Buniello et al., 2019) and performed enrichment analysis, as previously reported (Zhang et al., 2020). In adipocytes, individual-specific GR binding sites were significantly enriched for SNPs associated with metabolic traits, including Body Mass Index (BMI), cholesterol, diabetes and triglycerides levels, while common GR binding peaks exhibited much weaker enrichment (Figure 5A). Systematic integration of SNPs from dbSNP150 with the individual-specific genes and individual-specific GR peaks identified 288 SNPs located in individual-specific GR binding sites and affected the motif of GR or a cooperating TF such as CEBP, PPARγ, FOX, AP1 or NFI.
Figure 5. Genetic variants determine GR function and Dex responses.
(A and I) Enrichment of genetic variants associated with metabolic disorders in common or specific GR peaks in adipocytes (A) and in HLCs (I). The number is - log10 (P), which is also reflected by color.
(B) Visualization of an A5-specific absent peak region (yellow box) at LPAR1 locus across individual adipocytes. The black arrow indicates the position of rs10980797.
(C) Scatter plot showing the LPAR1 gene is not responsive to Dex in only individual A5 adipocytes.
(D) The putative effect of rs10980797 on CEBP motif.
(E and F) GR (E) and CEBPβ (F) ChIP-qPCR for LPAR1, FKBP5, and INS2 in eight individual adipocytes (2 A/A, 5 A/G and 1 G/G). Data are expressed as mean ± SEM.
(G and H) Differential GR binding near PPP1R3C locus (G) and Dex response of PPP1R3C (H) between different genotypes of rs10881935 across individual adipocytes. Data are expressed as mean ± SEM.
(J) The putative effect of rs6026774 on GR motif.
(K and L) Differential GR binding near LBP locus (K) and Dex response of LBP (L) between different genotypes of rs6026774 across individual HLCs. Data are expressed as mean ± SEM.
(M) The activities of luciferase reporters with the different alleles for rs6026774 and control reporter PGL4.24 in hepG2 cell lines treated with DMSO or Dex. Data are expressed as mean ± SEM.
(N) GR ChIP-qPCR for LBP, PDK4, and INS2 in HLCs (9 A/A and 2 G/G). Data are expressed as mean ± SEM.
(O) mRNA expression of LBP was examined by qPCR in 17 individual HLCs. Blue dots represent repeated experiment of individuals H1-H11. Red dots represent new HLCs (4 A/G, 2 G/G). Data are expressed as mean ± SEM fold change due to Dex.
(P and Q) Differential GR binding near CPT1A locus (P) and Dex response of CPT1A (Q) between different genotypes of rs2060982 across individual HLCs. Data are expressed as mean ± SEM.
*P < 0.05, by Student’s t-test (M, N and O).
See also Figures S8.
To validate the concept that genetic variations determine individual-specific Dex-responsiveness by modulating the genomic binding of GR, we focused on SNP rs10980797 in the vicinity of Lysophosphatidic Acid Receptor 1 (LPAR1) gene. Individual A5 was uniquely homozygous for the minor G allele (GG) (minor allele frequency [MAF], G = 0.33, 1000 Genomes), while all of the other seven individuals had at least one A allele, with 2 being AA. Moreover, GR binding was much weaker in the genomic region spanning rs10980797 in this individual than all 7 others (Figure 5B). Consistent with this being a functional binding site, Dex induced LPAR1 gene expression in adipocytes from every individual except individual A5 (Figure 5C). Mechanistically, rs10980797 does not affect the GR binding motif, but has a major effect on a motif favored by CEBP, a TF that has been shown to cooperate with GR at the genome (Grontved et al., 2013); the score of agreement with the CEBP consensus motif was 363-fold greater for the A allele than for the G allele (Figure 5D). Indeed, GR and CEBPβ displayed much greater binding in this region of the LPAR1 locus in adipocytes from individuals with at least one A allele than from G/G individuals (Figures 5E and 5F), while GR binding was genotype independent on an unrelated genomic region (FKBP5). Similarly, another SNP rs10881935 that is 5kb downstream of the PPP1R3C gene also showed genotype-dependent GR binding and Dex response (Figures 5G and 5H), with an A allele favoring GR binding and Dex response, possibly by affecting a FOXC2 motif (Figure S8A).
A similar bioinformatics pipeline was used to study how genetic variation controls GR function and Dex response in HLCs, with the advantage of available genotype information of these eleven iPSC lines (Figure S8B) (Pashos et al., 2017). We first demonstrated that individual-specific GR binding sites in HLCs were significantly enriched for SNPs associated with metabolic traits including BMI, diabetes, and glucocorticoid response based on the GWAS catalog (Figure 5I). 534 SNPs located in individual-specific GR binding sites and affected Dex response by altering the motif of GR or a cooperating TF. Notably, SNP rs6026774, which markedly affects a GR binding motif (Figure 5J) near the Lipopolysaccharide Binding Protein (LBP) gene, controlled GR binding and LBP gene induction by Dex. HLCs from individual H8, of the AG genotype, had much weaker GR binding and Dex response compared to the ten other individual HLCs with the AA genotype (Figures 5K, 5L and S8C).
The regulatory function of rs6026774 was assessed using luciferase reporter assays in the human liver-derived HepG2 cell line. Consistent with the endogenous A allele supporting GR binding and Dex-responsiveness of the native LBP gene, the A allele supported Dex-activation of a reporter gene whereas the G allele did not (Figure 5M). Moreover, independent GR ChIP-qPCR data revealed much greater binding in this region of the LBP locus in HLCs from individuals with AA allele than from GG individuals (Figure 5N), while GR binding was genotype independent on an unrelated genomic region (PDK4). Further validating the transcriptional importance of rs6026774, the requirement of the AA genotype for Dex induction of the LBP gene was replicated in the original 11 HLCs lines as well HLCs from 6 additional iPSC lines (4 AG, 2 GG) (Figure 5O). Moreover, we identified another SNP, rs2060982, ~1kb downstream of the CPT1A gene that exhibited genotype-dosage effects on GR binding (Figure 5P) and Dex responsiveness of the mRNA expression (Figure 5Q). Together, these results demonstrate that genetic variation modulates GR genomic occupancy and Dex responses in both adipocytes and HLCs.
To further explore the tight relationship of GR occupancy and Dex-responsiveness of nearby genes, we performed allele-specific analysis and identified 3795 SNPs located in imbalanced GR binding sites, which exhibited a cis-effect on GR binding (Figure S8D). These allele-imbalanced SNPs were enriched in promoter regions, enhancer regions and eQTLs in liver tissues from GTEx (Figure S8E) (Consortium, 2015), suggesting they are functional in gene regulation. Moreover, these allele-imbalanced SNPs were associated with the metabolic traits BMI and cholesterol (Figure S8F). 950 allele-imbalanced SNPs were associated with allele-selective Dex responses by affecting the motif of GR or its cooperating TFs. rs13038139 was of particular interest because it is in a GR binding site ~18 kb downstream from the PCK1 gene which encodes phosphoenolpyruvate carboxykinase (PEPCK), the rate-limiting enzyme in hepatic gluconeogenesis (She et al., 2000). 5 individual HLCs were heterozygous for this SNP, with ~6-fold greater binding at the C allele as determined by deep sequencing of the ChIP-seq products (Figure S8G). Another SNP, rs1042531, was heterozygous in two of the individuals and located in the last exon of the PCK1 gene, with the G allele (37% frequency) in cis with the C allele of rs13038139 (Machiela and Chanock, 2015). Deep sequencing of the PCK1 transcripts revealed no difference in the absence of Dex, but about a 3-fold excess of the G allele in the Dex-treated HLCs (Figure S8G). The imbalanced GR binding and gene expression was further confirmed by the SNaPshot assay (Figure S8H). These data demonstrate a cis-acting relationship between the GR binding site and the associated PCK1 transcript.
Genetic variations control Dex responsiveness by affecting chromatin accessibility and architecture
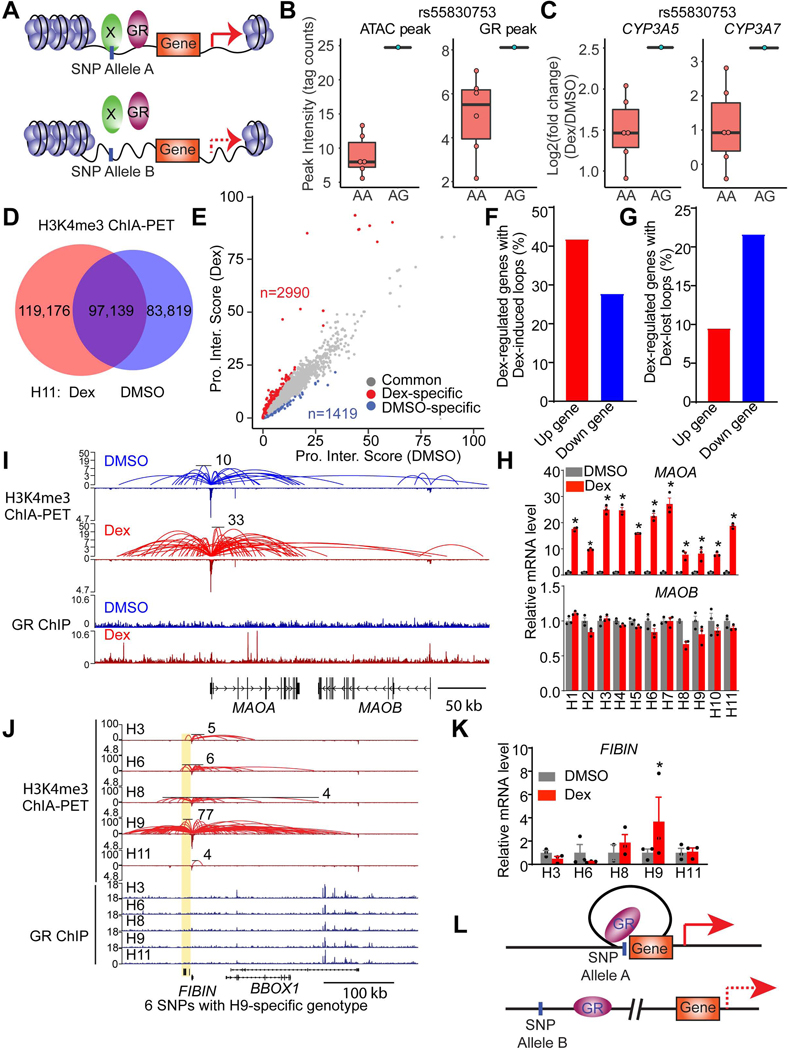
We noted that while ~50% of individual-specific GR binding sites contained SNPs within 100 bp of the center of the peak, nearly half did not. To explain individual-specific GR binding differences in the absence of genetic variation at the site of binding, we hypothesized that SNPs could also modulate chromatin accessibility and architecture to indirectly modulate GR binding at a distance, as has been suggested for other sequence-specific TFs (Behera et al., 2018; Deplancke et al., 2016; John et al., 2011; Waszak et al., 2015). To test this, we performed ATAC-seq in eight iPSC-derived HLCs. We detected 42,457 to 87,424 peaks, which were mainly located in the intronic and intergenic regions of the genome (Figure S9A) and highly enriched for the binding motifs of hepatic lineage factors FOXA1 and HNF4 as well as CTCF (Figure S9B), suggesting a faithful ATAC-seq dataset. Of note, all 8 HLCs shared 26,409 ATAC peaks (Figures S9C and S9D) but also exhibited individual-specific differences (Figures S9E and S9F).
1030 SNPs were noted to differentially affect chromatin accessibility near sites of variable GR binding and Dex-responsive gene expression (Figure 6A). For example, SNP rs55830753 (with genotype information available in 7 of 8 HLCs) is located 1.3 kb from a variable GR binding site that is 13kb upstream of CYP3A5 and 7kb downstream of CYP3A7. HLCs carrying the rs55830753 AG genotype showed high chromatin accessibility at this genomic site and increased GR binding intensity at the nearby site (Figures 6B and S9G). Remarkably, both the CYP3A5 and CYP3A7 genes were highly induced specifically in HLCs with the rs55830753 AG genotype (Figure 6C). These data suggest that the SNPs modulate GR genomic occupancy and function by affecting chromatin accessibility at a distance from the site of GR binding.
Figure 6. Genetic variations modulate GR function through affecting chromatin accessibility and architecture.
(A) Diagram depicting SNPs modulate gene response to Dex by affecting chromatin accessibility.
(B and C) rs55830753 affects ATAC-seq (left of B) signal and GR binding (right of B) near CYP3A5 and CYP3A7 locus and modulates their response to Dex (C).
(D) Venn diagram demonstrating the numbers of H3K4me3-mediated loops in DMSO- or Dex-treated H11 HLCs.
(E) Scatter plot revealing the genes whose promoter interaction score is 2 fold higher in Dex or DMSO group. Promoter interaction score represents the total PET number of each gene promoter.
(F and G) The percentage of Dex-responsive genes with Dex-specific interactions (F) or DMSO-specific interactions (G).
(H) mRNA expression of MAOA and MAOB in HLCs from eleven individuals, normalized to DMSO, as measured by RNA-seq. Data are expressed as mean ± SEM.
(I) Visualization of promoter-enhancer interactions (Top) and GR binding (Bottom) at MAOA and MAOB locus in DMSO or Dex-treated H11 HLCs. The scale of Y-axis of ChIA-PET is a log scale.
(J) Visualization of promoter-enhancer interactions (Top) and GR binding (Bottom) at FIBIN locus in five Dex-treated HLCs. Yellow box indicates genomic regions with H9-specifc promoter-enhancer loops and H9-specific genotypes. The scale of Y-axis of ChIA-PET is a log scale.
(K) mRNA expression level of FIBIN in HLCs from five individuals, normalized to DMSO, as measured by RNA-seq. Data are expressed as mean ± SEM.
(L) Diagram depicting the mechanisms of SNPs that modulate gene response to Dex by affecting the generation of loops.
*P < 0.05, by Student’s t-test (H and K).
See also Figures S9.
To examine how genetic variations impact enhancer-promoter looping to modulate gene expression, we performed Chromatin Interaction Analysis with Paired-End Tag (ChIA-PET) sequencing for a mark of active transcription, histone 3 lysine 4 trimethylation (H3K4me3) (Kouzarides, 2007; Li et al., 2017). We first identified 119,176 loops that were gained in individual H11 Dex-treated HLCs compared to DMSO-treated controls (Figure 6D). Enhancer-promoter interaction scores were assigned for each gene as described in the Methods section, and we noted 2,990 genes that exhibited increased enhancer-promoter interaction scores in Dex-treated HLCs than that in DMSO-treated HLCs (Figure 6E). Conversely, 1,419 genes decreased enhancer-promoter interaction scores in Dex-treated HLCs. Interestingly, ~42% of Dex-induced genes showed increased enhancer-promoter interaction after Dex treatment, in contrast, only ~28% of Dex-repressed genes gained enhancer-promoter interactions (Figure 6F). Similarly, ~22% of Dex-repressed genes showed decreased enhancer-promoter interaction after Dex treatment, whereas only ~9% of Dex-induced genes lost enhancer-promoter interactions (Figure 6G). This suggests that the transcriptional response to Dex is controlled by changes in enhancer-promoter interaction. For example, MAOA and MAOB are located near each other in the genome, yet MAOA was strongly induced by Dex while MAOB was not (Figure 6H). Remarkably, and consistent with the gene regulation, strong enhancer-promoter loops were seen at the MAOA but not the MAOB locus (Figure 6I), although we also found that the MAOA locus showed strong GR peak gain compared to MAOB after Dex treatment.
We then performed H3K4me3 ChIA-PET in five iPSC-derived HLCs treated with Dex, and identified 4872 SNPs located at the anchor of individual-specific loops linked to individual-specific Dex-responsive genes. For example, 6 SNPs with individual H9-specific genotype were noted at the anchor of loops markedly enriched in the HLCs of individual H9 near the FIBIN gene (Figure 6J). There is GR binding at this locus, though not at the loop anchor and of similar magnitude in all 5 individual HLCs (Figure 6J). These SNPs do not appear to alter the motifs for GR or cooperating TFs, but one SNP, rs7111860, alters the binding motif for CTCF (Figure S9H), which may cause altered chromatin architecture at the FIBIN locus. The FIBIN gene was uniquely induced by Dex in HLCs from individual H9 (Figure 6K), suggesting that the individual-specific loops facilitated the ability of Dex-bound GR activating the FIBIN gene by creating a favorable architecture that was not present in the other individual HLCs (Figure 6L). These data indicate that genetic variations can modulate GR function by affecting chromatin architecture.
Genetic variations altering GC responsiveness in adipocytes and HLCs predict metabolic adverse effects of drug treatment in the clinic
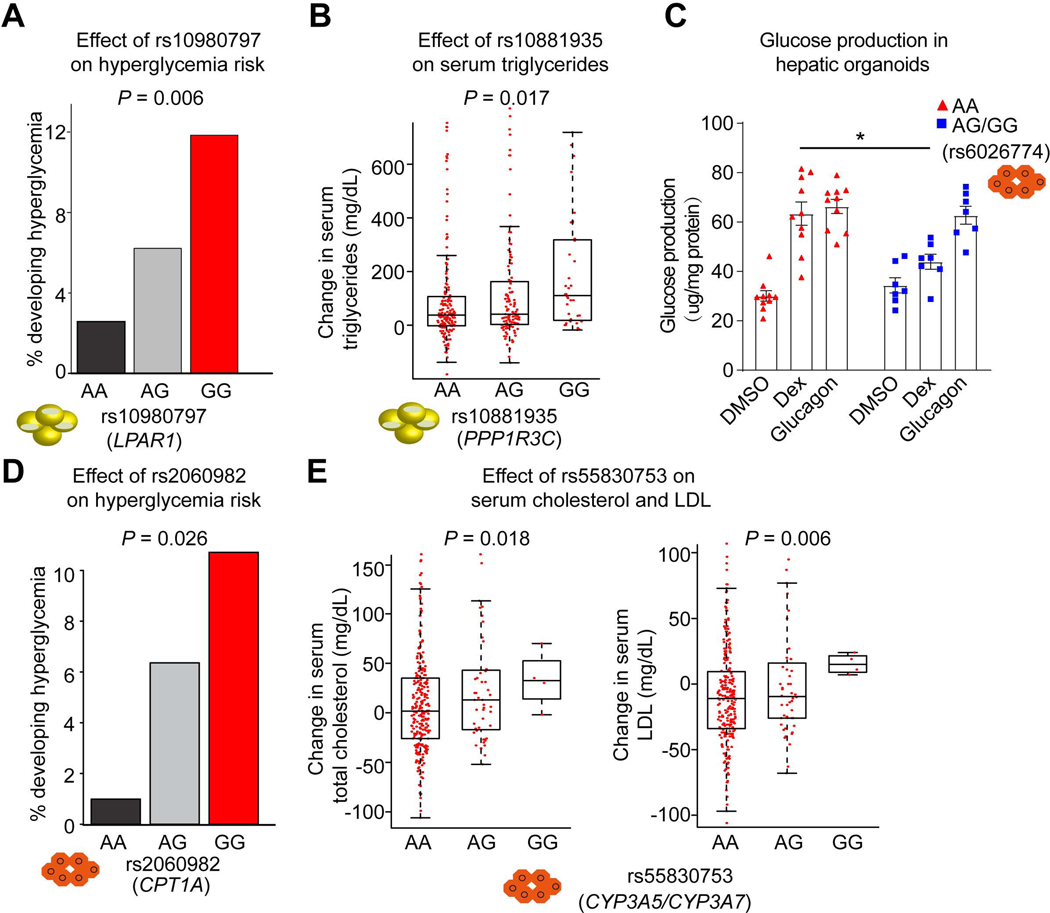
We next explored the clinical relevance of several SNPs that affect GR function and Dex response in adipocytes and HLCs. In GWASs, the A allele of rs10980797 that controls Dex-responsiveness of the LPAR1 gene (Figure 5C) was associated with BMI-adjusted waist-hip ratio (p = 5 × 10−20) (Pulit et al., 2019). We then evaluated the relationship between this SNP and metabolic parameters in 387 children with acute lymphoblastic leukemia (ALL) who received dexamethasone as part of their chemotherapy (Kawedia et al., 2011; Ramsey et al., 2017). Overall, 6% of children with ALL developed hyperglycemia (defined as blood glucose level >160mg/dL) among those treated with Dex. Notably, we found that homozygosity for the G allele of rs10980797 was associated with a nearly 5-fold increase in the percentage of individuals who developed hyperglycemia compared to AA individuals (Figure 7A). AG heterozygotes had an intermediate risk, indicative of a SNP-dosage effect (Figure 7A). While the mechanism is not clear, LPAR1 has previously been shown to regulate glucose uptake in adipocytes (Yea et al., 2008).
Figure 7. Genetic variants determine metabolic side effects of GC therapy.
(A) Percentage of pediatric individuals carrying each genotype of rs10980797 becoming hyperglycemic (>160 mg/dL) during treatment. 117 AA, 194 AG and 76 GG children with ALL who received chemotherapy with glucocorticoids.
(B) The change of triglyceride level after Dex treatment (Continuation Week8 – Week7) in individuals with ALL carrying different genotypes at rs10881935. Boxplots show median as a horizontal line, interquartile range as a box.
(C) Glucose production in hepatic organoids with different genotypes at rs6026774 treated with DMSO, Dex and Glucagon. 10 AA (red) and 7 AG or GG (blue) iPSC-derived hepatic organoids. Data are expressed as mean ± SEM. *P < 0.05 (Student’s t-test).
(D) Percentage of pediatric individuals with ALL carrying each genotype of rs2060982 becoming hyperglycemic (>160 mg/dL). 112 AA, 173 AG and 102 GG.
(E) The change of total cholesterol level (left) and LDL level (right) after Dex treatment (Continuation week8 – week7) in individuals with ALL carrying different genotypes at rs55830753.
The P value was calculated by multiple logistic regression analysis (A, B, D and E). See also Figures S10.
Dex treatment can also result in increased serum triglyceride level (Brotman et al., 2005). We assessed the impact of genetic variation at rs10881935, where the A allele favored GR binding and Dex-induction of PPP1R3C (Figure 5H). PPP1R3C-deficient mice have increased levels of circulating triglycerides (Crosson et al., 2003), and thus we predicted that the A allele would be protective. Indeed, the therapy-induced increase in serum triglyceride level was blunted in AA relative to GG with AG heterozygotes being in between (Figure 7B).
We next investigated whether rs6026774, which controls the LBP induction by Dex in HLCs (Figure 5L), had any relation to the adverse effects of Dex in the clinic. In mice, knockdown of LBP in the liver improved glucose homeostasis (Molinaro et al., 2020). Dex is known to increase hepatic glucose production (Desmet and De Bosscher, 2017), a major contributor to blood glucose levels, and thus we first tested the effect of SNP rs6026774 on Dex-modulation of glucose metabolism, employing a hepatic organoid model that is more mature and metabolically active than the HLCs used earlier (Ouchi et al., 2019; Shinozawa et al., 2020). 17 iPSC lines (rs6026774 genotypes: 10 AA, 5 AG and 2 GG) were differentiated into hepatic organoids and measured glucose production. As the A allele controls GR binding and induction of LBP by Dex (Figures 5K and 5L), we predicted that the G allele should protect against Dex-induced glucose production. Glucose production in AA hepatic organoids was stimulated ~2-fold both by Dex and by glucagon, a canonical regulator of hepatic glucose output (Habegger et al., 2010); whereas, by contrast, glucose production by iPSC-derived hepatic organoids derived from individuals with the AG and GG genotype showed a blunted response to Dex, while the glucagon response was approximately equal to that of the AA genotype (Figure 7C).
We next explored whether SNP rs6026774 modulates the hyperglycemic effects of GC in the clinic. Unfortunately, rs6026774 was not successfully imputed in the pediatric ALL cohort. As an alternative, we examined the electronic records of 167 adult subjects at the Hospital of the University of Pennsylvania for which SNP information was available and who had normal blood glucose levels (<160mg/dL) prior to beginning GC treatment with a variety of regimens for disparate medical conditions and did not take insulin during steroid treatment. Interestingly, blood glucose levels increased to a greater degree after GC treatments in individuals with the AA SNP compared to those with the GG SNP (Figure S10A), which is consistent with the protective effect of the G allele seen in the hepatic organoids.
We were able to assess rs2060982, the SNP that regulates CPT1A responsiveness to Dex (Figure 5Q), in the treated ALL children cohort. CPT1 is the rate-limiting step in fatty acid oxidation, and inhibition of CPT1 has been reported to reduce glucose production by the liver (Conti et al., 2011). Thus, given that in individual-derived HLCs the GG genotype conferred Dex-induction of CPT1A, individuals with the AA SNP would be predicted to be further protected from Dex-induced hyperglycemia. Indeed, the A allele was significantly associated with decreased risk of hyperglycemia in a dosage-dependent manner (Figure 7D).
Allele-specific analysis also revealed that the C allele of rs13038139 was associated with allele-selective induction of PCK1 by Dex in HLCs (Figure S8G). In the Dex-treated children with ALL, individuals with the CC SNP exhibited greater increases in BMI than that of individuals with the TT SNP (Figure S10B), which was supported by previous studies (Chang et al., 2008; Zhao et al., 2008). Moreover, SNP rs55830753, identified earlier as a genetic modulator of GR function and the Dex responses of CYP3A5 and CYP3A7 genes by affecting chromatin accessibility (Figure 6B and 6C), was associated with changes in total cholesterol and LDL levels after Dex treatment in children with ALL (Figure 7E). Thus, distinct SNPs affecting GR function in different tissues underly adverse metabolic consequences of GC treatment. Further, we looked at GWAS results focused on treatment outcome (“any relapses”) and early treatment response of remission/induction therapy in the children with ALL in a St. Jude cohort (Yang et al., 2011; Yang et al., 2009). Only the rs10881935_G allele was associated with a better outcome using “any relapse” as the dependent variable (P = 0.0173) (Figure S10C). Considering multiple testing, this result is suggestive and will need to be confirmed by future studies.
DISCUSSION
GWASs have been effective at identifying alleles that confer disease risk and drug response, but are time-consuming and costly due to the need for recruiting large cohorts of individuals and performing extensive genotyping and phenotyping (Visscher et al., 2017). As most GWAS-identified SNPs are in non-coding regions, it has recently been more widely accepted that many of these sites of natural genetic variation are affecting TF binding and enhancer function (Deplancke et al., 2016). Even with this recognition, GWASs often fail to provide insight into the TFs, as well as the cell types, most affected. Here, by studying adipocytes and hepatocytes derived from individual stem cells using genomic and epigenomic approaches, we have demonstrated predictable, individual-specific GC effects on gene expression and metabolism. Genetic variations were associated with Dex response in a cell-type-specific manner by affecting GR binding or interrupting chromatin accessibility and/or architecture. Remarkably, SNPs identified in this way effectively predict dysmetabolic side-effects of clinical GC use.
Previous studies have shown that genetic variations modulate gene expression through affecting genomic binding of TFs, but surprisingly, only a small fraction of SNPs can explain differential TF binding by directly interrupting the motif of the studied TFs, while the majority of variable genomic binding of TFs events appear to be attributed to mechanisms other than SNPs in the cognate motifs (Gosselin et al., 2014; Hu et al., 2019; Soccio et al., 2015). Consistently, in this study, GR motif-altering SNPs only accounted for a minority of individual-specific GR binding locations in adipocytes and hepatocytes. Genetic variation can also affect TF binding on chromatin through altering the motifs of its cooperating or collaborating factors (Deplancke et al., 2016; Soccio et al., 2015), which was supported by the finding that many SNPs affect genomic binding of GR by interfering with the motifs for CEBP, Forkhead Box family members and other TFs which function cooperatively or collaboratively with GR on chromatin (Grontved et al., 2013; Waddell et al., 2008).
SNPs can also function to alter chromatin accessibility and/or architecture. A previous study noted that the majority of PU.1 DNA binding variation in human lymphoblastoid cell lines was driven by SNPs located distally to the PU.1 binding region with a median distance between 20 and 30 kb from the transcribed region (Waszak et al., 2015). Similarly, by performing ATAC-seq and ChIA-PET on hepatocytes from multiple individuals, we noted numerous genes whose GR binding and Dex-dependent expression are associated with SNPs regulating chromatin accessibility and architecture. Intriguingly, we also uncovered SNPs that affect Dex-responsive gene expression, apparently due to changes in chromatin accessibility and architecture, with or without altering GR binding. Thus, natural genetic variation modulates individual-specific GC action by a multitude of decipherable mechanisms.
Previous studies have indicated that GCs exhibit cell-type-specific functions in humans (Gross and Cidlowski, 2008). For example, the transcriptional responses to GC are quite different between human primary hematopoietic and non-hematopoietic cells (Franco et al., 2019). Consistent with this, we found very little overlap between the Dex-regulated transcriptomes of human adipocytes and hepatocytes, and thus individual-specific SNPs controlling Dex-responsiveness function largely in a cell-specific manner. This is largely driven by cell type-specific GR genomic binding. Analysis of the sequence motifs reveals that cell-type-specific TFs cooperate with GR, likely by serving pioneering, lineage-specifying roles (Heinz et al., 2015; Zaret et al., 2016). Other possible mechanisms for cell-type-specific GR binding and function include different GR isoforms and post-translational modification, which potentially regulate different sets of genes (Cohen and Steger, 2017).
The models of stem cell-derived adipocytes and hepatocytes that we have derived from individuals may also be valuable for investigators searching for selective GR-modulating ligands with anti-inflammatory actions but without untoward metabolic effects. Analogous selective estrogen receptor modulators have been of enormous clinical utility in the treatments of breast cancer, osteoporosis and menopausal-related estrogen deficiency (Patel and Bihani, 2018). Selective GR modulating compounds have had less clinical success (Cole, 2006; Safy et al., 2017), in part because of the lack of translation of non-human models, a problem which our human systems would overcome. In addition, the advantage of using iPSCs is that multiple cell types can be generated with the same genetic background. Notably, beyond metabolic cell types, including adipocytes and hepatocytes that are reported here, the stem cells have the potential to be differentiated into immune cell types, such as macrophages and lymphocytes, to dissect both the theraputic effects of and metabolic side effects of selective GR ligands. It would also be interesting to determine if genes and metabolic processes altered in a individual-specific manner by Dex are differentially affected by other GR-modulators.
Previous studies on the role of genetics in modulating the response to GCs have largely focused on the GR itself, including rare individuals with GC resistance due to loss-of-function GR mutations (Bachmann et al., 2007) as well as more subtle exonic variations associated with GC responsiveness, such as ER22/23EK, which reduces GC sensitivity (Russcher et al., 2005) and N363S, which is associated with hypersensitivity to GCs (van Rossum and Lamberts, 2004). Further, the Pharmacogenomics Knowledge Base includes some non-coding SNPs that have been associated with the efficacy and adverse effects of Dex, but the underlying biological mechanisms have not been determined (Karol et al., 2016). Our genome-wide pipeline and integrative analysis of individual stem cell-derived adipocytes and hepatocytes models may allow for a more systematic identification of additional non-coding SNPs that affect Dex responses by affecting GR binding and/or chromatin accessibility and architecture.
Consistent with our findings, a previous study identified several enhancer SNPs that regulate GR-mediated gene expression using a massive Dex-responsive reporter activity assay (Johnson et al., 2018). Another study revealed wide gene expression variation following GR activation and found that the presumed enhancer and long distance chromatin regions modulate GR target gene expression in lymphoblastoid cell lines derived from multiple individuals (Maranville et al., 2013).The value of our approach is clear from our finding that several SNPs controlling gene responses to GC in human adipocytes and/or hepatocytes have clinical impact on increases in serum glucose, lipids and BMI, which are the most concerning metabolic side effects of GC therapy. Future studies in larger, well-phenotyped populations may be even more powerful to uncover synergistic effects of two or more SNPs that predict which individuals will develop metabolic adverse effects. In addition, natural language processing of electronic health records should expand the phenotypic range of future studies by searching for terms like “central obesity” and “Cushingoid” (Nieman, 2015).
In sum, our studies of individual-derived adipocytes and hepatocytes have revealed cell type-specific and individual-specific GC responses, setting the stage for identifying people who will have the most severe metabolic adverse drug effects. More generally, our “biased” approach of focusing on the TF targeting by a known drug, and the cell types most likely to be causing the adverse effects of the drug target, allowed for the discovery of risk alleles. The success of this approach, using stem cell-derived mature differentiated cell types from a few different individuals, suggests a way forward to personalized and precision approaches to the treatment of cancer and immune diseases with GCs.
Limitations of study
GR activation is required for hepatocyte maturation and adipogenic differentiation. It is possible that Dex could affect adipocyte and/or HLC differentiation, although we have taken several steps to limit this concern, such as omitting hydrocortisone from the HCM kit and only adding Oncostatin M to promote hepatocyte maturation, treating the mature adipocytes with 1.5% BSA medium without Dex for an additional 2 days to wash the effect for Dex, and choosing adipocytes and HLCs with similar differentiation efficiencies. Nevertheless, the variability in differentiation may affect the individual magnitude of Dex responses. Another control could be to study mature adipocytes from the same biopsied material used to prepare pre-adipocytes. Also, although our studies strongly suggest the regulatory role of SNPs by reporter assay and allele-specific analysis, it would be valuable to prove the causal role of the SNPs by generating isogenic iPSCs. Future studies should investigate the inter-individual genomic and gene expression variation of mature fat cell depots from clinical studies of subjects with annotated phenotypic responses to glucocorticoids. Finally, our examination of the clinical relevance of implicated SNPs in children with ALL is limited by the relatively small size of the population, and may not be relevant to adults.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mitchell A. Lazar, M.D., Ph.D. (lazar@pennmedicine.upenn.edu).
Materials Availability
The iPSC lines used in this study are from the publicly available WiCell Research Institute depository for the NHLBI Next Generation Genetic Association Studies Consortium collection. The hASC lines used in this study are kindly provided by Dr. Ivona Percec, which are available from the lead contact upon request.
Data and Code Availability
The accession number for the sequencing data reported in this paper is GEO: GSE163061.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study Cohorts
The patient setting, treatment strategy and clinical data collection were described as previously (Kawedia et al., 2011; Ramsey et al., 2017). This study was approved by the St. Jude Institutional Review Board, and informed consent was obtained from subjects who were 18 years old and, in the case of younger individuals, from their parents or guardians, in accordance with the Declaration of Helsinki. The clinical trial number is NCT00137111. In brief, children with newly diagnosed ALL (n = 498) were enrolled. 394 children have measured lab values. The children age is from 1.0 to 18.8 years with a median age 5.4 years, and 54.5% children are male. After a common remission induction and consolidation phase, dexamethasone at 8 mg/m2 per day (low-risk arm) and 12 mg/m2 per day (standard-risk/high-risk arm) was given orally for 5 days at weeks 1, 4, and 9 of continuation therapy. During both reinductions I and II (weeks 7–9, weeks 17–19), all patients received dexamethasone at 8 mg/m2 per day orally for 7 to 8 days. We collected heparinized blood samples on day 15 of consolidation and day 1 of week 2 (or week 5), week 7, week 8 (after 7 continuous days of dexamethasone), and week 12 of the continuation phase.
We also examined the electronic records of participants of the Penn Medicine Biobank. The University of Pennsylvania Institutional Review Board approved the study, and subjects consented to the use of genetic and clinical data for the purposes of research. 167 participants whose SNP information was available and who had normal random blood glucose levels (<160 mg/dL) prior to beginning high dose GC treatment with a variety of medical conditions and did not admit insulin during GC treatment were included for this analysis. The average age of participants is 62.3 ± 14.6 years, and 54.0% participants are male.
Human iPSC lines
Human iPSC lines are selected based on their differentiation ability into hepatocytes from a cohort of iPSC lines generated in our previous study (Pashos et al., 2017). 58% iPSC lines used in this study were generated from male subjects. The University of Pennsylvania Human Subjects Research Institutional Review Board approved the study protocol for the generation of these iPSC lines, and all subjects gave written informed consent for study participation. iPSCs were cultured in mTeSR medium at 37 °C with 5% O2 and 5% CO2.
Primary hASCs
Subcutaneous abdominal adipose tissues were excised from consented healthy female subjects undergoing elective abdominoplasty (University of Pennsylvania IRB approval Protocol number 812150). ASCs were isolated from fat tissue samples according to a standard collagenase protocol (Shan et al., 2017). The ASCs were maintained in DMEM medium at 37 °C with 5% CO2 and passaged 2–3 times before adipogenic differentiation.
METHOD DETAILS
Adipogenic differentiation of hASCs
hASCs at passage P2-P3 were cultured in DMEM medium. Confluent hASCs were then transferred into adipogenic medium (DMEM with 10% FBS, 1% penicillin/streptomycin, dexamethasone (1 μM), IBMX (0.5 mM), indomethacin (0.2mM), Insulin (10 μg/ml) and rosiglitazone (1 μM)) for 14 days. Then, cells were further cultured in maintenance medium (DMEM with 10% FBS, 1% penicillin/streptomycin, dexamethasone (1 μM), IBMX (0.5 mM), indomethacin (0.2 mM) and Insulin (10 μg/ml)) another 7 days. Before treatment with Dex, the cells were cultured with 1.5% BSA/DMEM medium for 2 days. We selected eight subject adipocytes with similar adipogenic differentiation efficiency based on morphology and gene expression from a pool of 23 individual ASCs as a celluar model system to study glucocorticoid response.
Differentiation of hiPSCs into HLCs
The differentiation of hiPSCs into HLCs were performed as described previously with minor modification (Pashos et al., 2017). Briefly, we treated hiPSC at 30–40% confluence 24 hrs after plating for differentiation with Definitive Endoderm Kit from day 1 to day 4 following manufacturer’s instruction. Then, the cells were transferred into RPMI-B27 (made with 2% B-27 Supplement) supplemented with BMP-4 (20 ng/mL) and FGF basic (10 ng/mL) for 5 days in 5% oxygen/5% CO2. Subsequently, RPMI-B27 supplemented with recombinant human HGF (20 ng/mL) were applied to the cells for 5 days in 5% oxygen/5% CO2. Finally, the cells were treated with HCM Hepatocyte Culture Medium (Lonza) without EGF and hydrocortisone and supplemented with recombinant human oncostatin M (20 ng/mL) for 5 days in ambient oxygen/5% CO2, yielding mature HLCs.
Immunostaining
For immunostaining, cells plated on glass coverslips were fixed by 4% PFA solution for 10 min at room temperature. Then cells were incubated in blocking buffer (1% bovine serum albumin in PBS) with 0.5% Triton X-100 for 30 min at RT. Afterwards, samples were incubated with primary antibodies (GR, Cell Signaling Technology, 1:100 dilution; HNF4, 1:100 dilution; Albumin, 1:100 dilution) at 4 °C overnight and then with appropriate fluorescent probe-conjugated secondary antibodies for 1 h at RT. Images were captured with fluorescence microscope or Leica TCS SP8.
Western blot and gene expression analysis
For western blotting, HLCs were washed with cold PBS and lysed with Laemmli’s sample buffer, and then cell lysates were separated on SDS-PAGE, transferred onto nitrocellulose membrane and blotted with indicated primary antibodies. The membrane was detected by secondary antibody conjugated to HRP.
For gene expression analysis, total RNA samples were collected with RNeasy kit (QIAGEN) according to manufacturer’s instructions. The RNA for each reaction was reverse-transcribed to cDNA using High-Capacity cDNA Reverse Transcription Kit. Quantitative real-time PCR was subsequently conducted with specific primers and Power SYBR Green PCR Master Mix (Applied Biosystems). The relative expression levels were normalized against the internal control (HPRT). Primers used were listed in Supplement information, Table 1.
RNA-seq and data processing
Total RNA samples were prepared with RNeasy kit (QIAGEN) according to manufacturer’s instructions. More than 2 ug RNA from three biological replicates were sent to Novogene (CA) for library preparation (Illumina) and sequencing at pair-end at 150bp read length on Novaseq 6000. Raw reads were trimmed using fastp v0.19.5 to remove reads with low quality, too short, or too many N. Trimmed reads were then aligned to human reference genome (hg38) using Hisat2 v2.1 with default parameters. Only unique mapped reads extracted by SAMtools v1.9 were considered for downstream analyses. Quantification of gene annotated in GRCh38 v94 from Ensembl database was estimated using StringTie v1.3.4. Genes with normalized expression value, fragments per kilobase of exon per million reads mapped (FPKM), larger than 1 in at least one sample were considered. To assess the reproducibility of replicates, PCA analysis was performed, and three biological replicates of adipocytes from each individual treated with DMSO or Dex generally clustered together. Pearson correlation between replicates from each individual was also calculated. The correlation coefficients were all larger than 0.99 among replicates. To calculate Pearson correlation coefficients for inter-samples, average expression of the replicates for each gene was calculated, following by the calculation of Pearson correlation coefficient between each two individuals based on the average expression values.
Read counts that were measured for each gene using featureCounts v1.5.1 were used as the input to DESeq2 for differential expression analysis with adjusted p value (Benjamini Hochberg) < 0.01 and fold change ≥ 1.5. Genes that were deferentially expressed in only one individual were defined as individual-specific responsive genes. GSEA analysis, GO and KEGG enrichment analysis were performed using clusterProfiler. For GSEA analysis, genes ranked by average log2(fold change) in Dex vs DMSO across all individuals. For heatmaps showing individual-specific and common Dex-responsive genes in Figure S2E and S3D, triplicate results were combined for this analysis following a published method (Wang et al., 2020).
ChIP
Mature adipocytes and HLCs were treated with Dex for 6h. DMSO-treated cells were included as control. ChIP assays were performed in two biological replicates as described previously (Hu et al., 2019). Mature adipocytes and HLCs were crosslinked in 1% formaldehyde for 15 min and then quenched with 1/20 volume of 2.5 M glycine solution for 5 min. Soluble chromatin was sonicated into small fragments and then was incubated with anti-GR antibody (CST) or anti-CEBPβ antibody (Santa-Cruz). The pulled DNA was decrosslinked overnight at 65°C in SDS buffer (50 mM Tris-HCL, 10 mM EDTA, 1% SDS at pH 8), and DNA was purified using phenol/chloroform/isoamyl alcohol. Precipitated DNA was analyzed by qPCR or sequencing.
ChIP-seq and data processing
ChIP experiments were performed independently on hASC-derived adipocytes and iPSC-derived HLCs. The library preparation for ChIP-seq follows the guide provided by Illumina. ChIP-seq libraries were sequenced single-end at 50bp read length on Illumina HiSeq2000 by the Functional Genomics Core of the Penn Diabetes Research Center or sequenced pair-end at 150bp read length on Illumina HiSeq X by Novogene (CA). For ChIP-seq in adipocytes, we downsampled the number of read pairs to 40M seperately for each sample. ChIP-seq raw reads were trimmed using fastp v0.19.5. Trimmed reads were then aligned to human reference genome (hg38) using Bowtie2 v2.3.0 with default parameters. SAMtools v1.9 was used to extract unique mapped reads and remove duplicated reads. Biological replicates from the same individual were pooled together following by peak calling and de novo motif analyses using HOMER v4.9 with default parameters. The overlap among biological replicates was calculated before pooling the replicates for downstream analysis. Peaks detected in only one individual were defined as individual-specific peaks. Peaks detected in all individuals were defined as common GR peaks. GSEA analysis, GO and KEGG enrichment analysis based on the nearest genes were performed using clusterProfiler. For GSEA analysis, genes ranked by average intensity of their counterpart peaks across all individuals.
Integrated analysis of Dex-responsive genes and GR cistromes
GR binding sites were paired with Dex-responsive genes within 200KB. Then the Pearson correlations between peaks and genes were calculated based on the intensities of GR peaks and Dex responses of genes across individuals. To check if individual-specific GR binding sites are likely to regulate individual-specific Dex responsive genes, a random test was performed by shuffling the genomic region label for Dex responsive genes for 1000 times. RP score for each Dex responsive gene were calculated using GR bindings as the input using the regulatory potential (RP) model decribed as previously (Wang et al., 2016).
To identify motif-altering SNPs that control GR function and Dex responses, protocol described as previously with minor modification (Hu et al., 2019) was used. Briefly, peak-gene pairs in which peaks contain a SNP were considered. For HLCs, genotype data came from our previous study (Pashos et al., 2017). And for adipocytes, genotypes were predicted based on GR ChIP-seq data. Briefly, reads spanning the interesting SNPs were extracted, following by the extraction of the nucleotides locating at the same position of the SNPs. Only reads whose depth above 5 were considered. Next, individuals were divided into different groups based on the genotypes. For each SNP, the difference of peak intensities (or gene response) between different groups was required larger than 1.5 fold. Only peak-gene pairs in which group with higher peak intensities having stronger Dex response were considered. Then, the hg38 reference sequences including 20 bp on either side of candidate SNPs were retrieved, as well as the sequences of the SNPs with alternative allele. FIMO algorithm in MEME suite with p value < 0.001 was performed to examine if these SNPs alter the motifs of GR or its cooperating factors. The SNPs with position weight at a position weighted matrixes (PWMs) larger than 0.3 and with the ratio of the position weights between two alleles of the SNP larger than 3 were considered.
Allele-specific analysis
To identify allele-specific binding (ASB) SNPs, read counts for each allele were calculated using perl scripts based on bam files of ChIP-seq data. To maximize the power to detect ASB SNPs, reads from all heterozygous individuals were pooled for each SNP, and only SNPs with read counts > 6 were considered. The p-values were calculated based on binomial test using binom.test from the R package, and FDR correction were applied using R function p.adjust(x,method = “fdr”). Only SNPs with FDR < 0.1 were considered. To examine if the nearby GR target genes also show allele-selective response to Dex, the same strategy was applied to RNA-seq data of target genes containing heterozygous SNPs. To analyze the enrichment of ASB SNPs in promoters, enhancers, liver eQTL and metabolic traits-related SNPs, protocol described previously (Zhang et al., 2020) was used. Briefly, the enrichment p values were calculated using binom.test from the R package. The fold enrichment was calculated using the formula described previously (Zhang et al., 2020). Promoter is defined as upstream 2 kb and downstream 2 kb of TSS annotated in GRCh38 v94 from Ensembl database. And enhancer regions were defined as H3K27ac peaks in hepatocyte without overlapping with promoter and exon regions annotated in GRCh38 v94 from Ensembl database. eQTL in liver tissue was download from GTEx (Consortium, 2015), and the metabolic traits-related SNPs came from GWAS catalog (Buniello et al., 2019).
Cell types-specific Dex-responsive genes and GR cistromes
Only high confident Dex-responsive genes and GR cistromes were considered, in which genes that are Dex-responsive and GR peaks that are detected in at least half of the individuals were considered. De novo motif analyses for cell type-specific GR cistromes were performed using HOMER v4.9. Gene ontology analyses for cell type-specific Dex responsive genes were performed using clusterProfiler.
ATAC-seq and data processing
ATAC-seq experiments were performed as described previously (Corces et al., 2016). In brief, 100,000 hiPSC-derived HLCs were resuspended in 50 μl of transposase mixture (25 μl of 2× TD buffer, 2.5 μl of TDE1, 0.5 μl of 1% digitonin, and 22 μl of nuclease-free water) (Illumina). Transposition reactions were incubated at 37 °C for 30 min in a Thermo Mixer with agitation at 300 rpm. Transposed DNA was purified using a QIAGEN MinElute Reaction Cleanup kit. Transposed fragments were subjected to library preparation following previous protocol (Buenrostro et al., 2015). ATAC-seq libraries were sequenced using paired-end, dual-index sequencing on a NextSeq 500 instrument.
ATAC-seq raw reads were trimmed using fastp v0.19.5. Next, trimmed reads were aligned to human reference genome (hg38) using Bowtie2 v2.3.0 with parameter --very-sensitive. Reads mapped to mitochondria were ignored. SAMtools v1.9 was applied to extract unique mapped reads and remove duplicated reads. Peak calling was performed using Genrich (https://github.com/jsh58/Genrich#contact), which was developed by John M. Gaspar in the Informatic Group with an alternative analysis mode only for ATAC-seq, and also recommended in the latest ATAC-seq Guidelines (https://informatics.fas.harvard.edu/atac-seq-guidelines.html#overview). Peaks detected in only one individual were defined as individual-specific peaks.
Integrated analysis of Dex-responsive genes and ATAC-seq data
ATAC-seq peaks were paired with Dex-responsive genes within 200KB. To identify motif-altering SNPs that control chromatin accessibility and Dex responses, similar pipeline described above was used. Briefly, ATAC peak-gene pairs in which peaks contain a SNP were considered. Next, individuals were divided into different groups based on the genotypes. For each SNP, the difference of ATAC peak intensities (or gene response) between different groups was larger than 1.5 fold. Only ATAC peak-gene pairs in which group with higher peak intensities having stronger Dex response were considered. We further filtered the ATAC peak-gene pairs by associated them with GR binding that’s 2 kb from ATAC peaks and exhibited similar pattern of binding as ATAC peaks.
H3K4me3 in-situ ChIA-PET
The H3K4me3 in-situ ChIA-PET was performed according to the previously described protocol (Li et al., 2017) with minor modifications. Briefly, 5 million human iPSC-derived HLCs were harvested and crosslinked with 1% formaldehyde for 20 mins at room temperature. Then cells were quenched by 0.125 M glycine for 10 mins at room temperature. Then, the cells were fixed again by 1.5 mM EGS for 45 mins at 37 °C and quenched by 0.125 M glycine for 10 mins at room temperature. The dual-crosslinked cells were lysed and digested with AluI enzyme (NEB). Digested DNA ends were A-tailed and proximal ligated with biotinylated bridge linker overnight (Tang et al., 2015). The ligated chromatin was sheared by sonication and pull-down by H3K4me3 antibody (Millipore). Immunoprecipitated DNA was subjected to Tn5 (Illumina) tagmentation, M280 Dynabeads (Thermo Fisher Scientific) streptavidin pull-down and PCR amplification (Illumina). PCR products were subjected to size-selection and pair-end sequencing on Illumina HiSeq X platform.
ChIA-PET data processing
ChIA-PET data was processed by ChIA-PIPE tools (Lee et al., 2020). Briefly, ChIA-PET paired-end reads were first removed bridge linker and split into 3 category tags: none-linker containing tags, one-end tags and pair-end tags (PETs). Tags longer than 18 bp were used to mapped to human genome (hg38) by Burrows–Wheeler alignment (BWA). Duplicated PETs are filtered and uniquely non-redundant PETs were classified into inter-chromosomal PETs, intra-chromosomal PETs (two tags distance >=8 kb) and self-ligated PETs (two tags distance <8 kb). Multiple intra-chromosomal PETs whose respective ends were within 1 kb were then clustered together. PET count of each PET cluster represents the interaction strength. Loops with PETs ≥3 were considered for downstream analysis. Loops detected in only one individual were defined as individual-specific Loops. H3K4me3-marked Promoter Interaction Score (PIS) was defined by the following steps: each promoter (extended TSS from −2.5 to +2.5 kb) was overlapped with all PET clusters, only ones which have at least 1 anchor overlapped with promoters would be kept. PIS was calculated by summing all PET counts of these overlapped clusters within each promoter and normalized by total intra-chromosomal PET number. ChIA-PET data was visualized by a customized karyoploteR package (Gel and Serra, 2017).
To identify SNP that control chromatin loops and gene responses, loops with one anchor spanning a SNP and the other anchor overlapped with gene promoter were considered, and similar pipeline described above was used. Briefly, individuals were divided into different groups based on the genotypes. For each SNP, the difference of loop intensities (or gene response) between different groups was larger than 1.5 fold. Only loop-gene pairs in which group with higher loop intensities having stronger Dex response were considered.
Luciferase reporter assay
249bp human DNA fragments encompassing SNP rs6026774 reference or alternative allele were synthesized from Integrated DNA Technologies. They were cloned into the XhoI and BglII restriction sites of the pGL4.24 luciferase reporter (Promega) and sequence-verified. Transient transfections of HepG2 cells were performed in 24-well plates, n = 4 wells per condition, using Lipofectamine 3000 (Invitrogen) to add 400 ng of pGL4 luciferase reporter and 2 ng of renilla luciferase for normalization. One day later, the cells were treated with 1 μM Dex for 24 hours. The Dual-Luciferase Kit (Promega) was used to measure luciferase activities on a Synergy HT plate reader (Biotek).
SNaPshot assay
Genotyping were carried out with SNaPshot Multiplex Kit (Applied Biosystem) according to manufacturer’s instructions. Briefly, each region of interest flanking the SNP was PCR amplified, and 5 μL of PCR product was purified by ExoSAP-IT reagent (Affmetrix). Primer extension was performed by adding 1 μL of purified PCR product to a mix of 2.5 μL SNaPshot reagent, 1 μL water and 0.2 pmol extension primer for 25 cycles on Thermocycler. To remove unincorporated fluorescent dNTPs post-extension, each reaction was incubated 1U of Calf Intestinal Phosphatase (New England BioLabs). Samples were sequenced by the Penn DNA Sequencing Facility on an ABI 3730, and genotypes were identified manually using Peak Scanner Software (Life Technologies).
Glucose production assay
Human hepatic organoids from 17 human iPSC lines were generated exactly following the previous protocol (Ouchi et al., 2019). The glucose production assay was performed as previously (Zou et al., 2018). Briefly, each well of hepatic organoids from 6 well plates at day 23 were transferred to one well of 24 well ultra-low attachment plate, and cultured with complete HCM (Lonza HCM bullet kit, except no EGF, supplemented with 100 nM Dex, 20 ng/mL oncostatin M, and 20 ng/mL HGF) with 10% Matrigel for 3 days. Then, hepatic organoids were washed with PBS and starved in GOM buffer (118 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 25 mM NaHCO3, 1.2 mM CaCl2, pH= 7.4) plus 0.1% BSA for 6 hrs. After starvation, hepatic organoids were treated in glucose-free media supplemented with 1 μM Dex or 100 nM Glucagon, 1x P/S, 10 mM Lactate and 1 mM Pyruvate for 24 hrs. Glucose concentration in cell culture media was determined by Glucose Colorimetric Detection Kit (ThermoFisher Scientific) following the manufacturer’s instruction. Glucose production rate was normalized to protein content.
Clinical analysis
387 children with ALL with available phenotype and genotype data were included for this analysis. Hyperglycemia was defined according to National Cancer Institute Common Terminology Criteria for Adverse Events, Version 3.0 (Grade 2 or above, blood glucose level >160 mg/dL) during the treatment course. Genotypes were coded as 0, 1 or 2, representing number of minor alleles and were tested for additive effect for their association with phenotypes. Multiple logistic regression was used in the analysis of hyperglycemia, which was coded as a binary variable. The effects of genotypes on changes in lipid measures, and BMI were evaluated using analysis of covariance based on a general linear model using the measures at a later timepoint (week 8) as the dependent variable and the measures at baseline (week 7, or at diagnosis for BMI) as covariates. Genetic ancestry, age, gender and risk arm were included in all models as covariates. For therapeutic effect analysis, we looked at GWAS results that we have previously published on treatment outcome (“any relapses”) and early treatment response (minimal residual disease during [mrd_day19] or at the end of [mrd_day46] of remission/induction therapy (Yang et al., 2011; Yang et al., 2009). For “any relapse”, we used cox-proportional hazard regression (n = 475), adjusting for covariates include race and treatment risk group; For “MRD D19”, we used ordinal regression (n = 465), treating dependent variable MRD as three levels: <0.01% vs. 0.01–1% vs. >= 1%, adjusting for race; For “MRD D46”, we used logistic regression (n = 469), <0.01% vs. >= 0.01%, adjusting for race.
To test the association of rs6026774 with glucose level after GC treatment, we examined the electronic records of participants of the Penn Medicine Biobank with available genotype data. 167 participants whose SNP information was available and who had normal random blood glucose levels (<160 mg/dL) prior to beginning high dose GC treatment with a variety of medical conditions and did not admit insulin during GC treatment were included for this analysis. High dose GC therapy was defined as dexamethasone ≥2 mg or equivalent doses of prednisone, methylprednisolone, or hydrocortisone. The change of glucose level was calculated by comparing the maximal glucose level within 6 days post GC treatment to that before GC treatment.
Genotyping
For the clinical study at St. Jude Children’s Research Hospital, single nucleotide polymorphism (SNP) genotyping was performed using the Affymetrix Gene Chip Human Mapping Array, using 500K or 6.0 arrays (Affymetrix) (Ramsey et al., 2017). Imputation was performed using Michigan Imputation Server using 1000genome population data as reference. SNPs with imputation r2 > 0.6 and following Hardy-Weinberg equilibrium > 0.05 were included in the analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Error bars represent the standard error to mean (SEM), and statistical significance was determined by unpaired two-tailed Student t-test or multiple logistic regression analysis using GraphPad Prism software or R software; a P-value of ≤ 0.05 was considered significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GR | Cell Signaling Technology | Cat# 3660S; RRID: AB_11179215 |
| GR | Proteintech | Cat# 24050-1-AP; RRID: AB_2813890 |
| Albumin | Bethyl Laboratories | Cat# A80-229A; RRID: AB_67018 |
| HNF4 | Thermo Fisher Scientific | Cat# MA1199; RRID: AB_2633309 |
| Histone H3K4me3 | Millipore | Cat# 04-745; RRID: AB_1163444 |
| CEBPβ | Santa Cruz | Cat# sc-7962; RRID: AB_626772 |
| Chemicals, peptides, and recombinant proteins | ||
| mTeSR 1 | STEMCELL Technologies | Cat# 05850 |
| Gelt rex | Thermo Fisher Scientific | Cat# A1413302 |
| Matrigel | Corning | Cat# 356237 |
| Accutase | Innovative Cell Technologies | Cat# AT-104 |
| STEMdiff Definitive Endoderm Kit | STEMCELL Technologies | Cat# 05110 |
| RPMI 1640 | Thermo Fisher Scientific | Cat# 22400105 |
| Advanced DMEM/F12 | Thermo Fisher Scientific | Cat# 12634010 |
| B-27 Supplement | Thermo Fisher Scientific | Cat# 17504044 |
| N-2 Supplement | Thermo Fisher Scientific | Cat# 17502048 |
| Knockout Serum Replacement | Thermo Fisher Scientific | Cat# 10828-028 |
| GlutaMAX | Thermo Fisher Scientific | Cat# 35050061 |
| HCM BulletKit | Lonza | Cat# CC-3198 |
| Human BMP-4 | Pe protech | Cat# 120-05ET |
| Human FGF-basic | Pe protech | Cat# 100-18B |
| Human HGF | Pe protech | Cat# 100-39H |
| Recombinant Human Oncostatin M | R&D Systems | Cat# 8475-OM-050 |
| Human/Murine/Rat Activin A | Pe protech | Cat# 120-14E |
| Recombinant Human FGF-4 | Peprotech | Cat# 100-31 |
| DMEM | Thermo Fisher Scientific | Cat# 11995065 |
| DMEM/F12 | Thermo Fisher Scientific | Cat# 11330032 |
| DMEM, no glucose | Thermo Fisher Scientific | Cat# 11966025 |
| Fetal bovine serum | Atlanta Biologicals | Cat# S11150 |
| Insulin solution human | Sigma | Cat# 19278 |
| 3-lsobutyl-1-methylxanthine | Sigma | Cat# 15879 |
| Dexamethasone | Sigma | Cat# D4902 |
| Indomethacin | Sigma | Cat# 17378 |
| Rosiglitazone | Sigma | Cat# R2408 |
| Y27632 | Selleckchem | Cat# S1049 |
| CHIR99021 | Selleckchem | Cat# S1263 |
| Retinoic acid | Sigma | Cat# R2625 |
| Glucagon | Sigma | Cat# G2044 |
| Lactate | Sigma | Cat# L1750 |
| Sodium Pyruvate | Thermo Fisher Scientific | Cat# 11360070 |
| Paraformaldehyde Solution, 4% | Affymetrix | Cat# 19943 1 LT |
| cOmplete, EDT-free Pro tease Inhibitor Cocktail | Roche | Cat# 11873580001 |
| Chloroform | Sigma | Cat# C2432-500ML |
| RNase A (DNase and Protease Free) | Fermentas Life Sciences | Cat# EN0531 |
| Phenol/Chloroform/ Isoamyl Alcohol | Fisher Scientific | Cat# BP1753I-400 |
| ChIP DNA Clean & ConcentratorCapped column | Zymo Research | Cat# D5205 |
| DNA LoBind Microcentrifuge Tube | Eppendorf | Cat# 30108.051 |
| Deoxynucleotide (dNTP) Solution Mix | New England Biolabs | Cat# N0447S |
| Phusion Hot Start II DNA Polymerase | Thermo Fisher Scientific | Cat# F-549L |
| UltraPure Glycogen | Thermo Fisher Scientific | Cat# 10814010 |
| T4 DNA Polymerase | New England Biolabs | Cat# M0203S |
| Klenow Fragment | New England Biolabs | Cat# M0212S |
| Klenow DNA Polymerase | New England Biolabs | Cat# M0210S |
| T4 Polynucleotide Kinase | New England Biolabs | Cat# M0201S |
| EGS | Thermo Fisher Scientific | Cat# 21565 |
| Alul | New England Biolabs | Cat# R0137L |
| Tn5 | Illumina | Cat# 20034197 |
| Human Liver Whole Tissue Lysate | Novus Biologicals | Cat# NB820-59232 |
| Human NR3C1 knockout HeLa cell lysate | Abeam | Cat# ab257009 |
| Critical commercial assays | ||
| RNeasy Mini Kit | QIAGEN | Cat# 74106 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat# 4368813 |
| Power SYBR Green PCR Master Mix | Applied Biosystems | Cat# 4367659 |
| Agilent high sensitivity DNA assay | Agilent | Cat# 5067-4626 |
| M280 Dynabeads | Thermo Fisher Scientific | Cat# 11206D |
| Agencourt AM Pure XP beads | Beckman Coulter | Cat# A63881 |
| Nextera DNA Library Prep Kit | lllumina | Cat# FC-121-1031 |
| MinElute Reaction Cleanup Kit | QIAGEN | Cat# 28204 |
| NEBNext High-Fidelity 2X PCR Master Mix | New England Biolabs | Cat# M0541S |
| Dual-Luciferase Reporter Assay System | Promega | Cat# E1910 |
| Glucose Colorimetric Detection Kit | Thermo Fisher Scientific | Cat# EIAGLUC |
| SNaPshot Multiplex Kit | Thermo Fisher Scientific | Cat# 4323159 |
| Deposited data | ||
| RNA-seq, GR ChlP-seq, H3K4me3 ChlA-PET, and ATAC-seq | This study | GEO: GSE163061 |
| Genotype data | Pashos et al., 2017 | dbGaP: phs001341.v1.p1 |
| Oligonucleotides | ||
| See Table S1 | N/A | N/A |
| Software and algorithms | ||
| Fastp 0.19.5 | Chen et al., 2018 | https://github.com/OpenGene/fastp |
| BWA 0.7.17 | Li and Durbin, 2009 | http://biobwa.sourceforge.net/ |
| Bowtie 2.3.0 | Langmead and Salzberg, 2012 | http://bowtiebio.sourceforge.net/bowtie2/manual.shtml |
| SAMtools 1.9 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Bedtools 2.26.0 | Quinlan and Hall, 2010 | http://bedtools.readthedocs.org/en/latest/ |
| Homer v4.9 | Heinz et al., 2010 | http://homer.salk.edu/homer/ |
| R 3.3.2 | N/A | https://www.r-project.org/ |
| StringTie 1.3.4 | Pertea et al., 2015 | https://ccb.jhu.edu/software/stringtie/ |
| Hisat2 2.1 | Kim et al., 2015 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| IGV 2.4 | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv |
| MEME SUITE 4.12.0 | Grant et al., 2011 | http://meme-suite.org/ |
| featureCounts v1.5.1 | Liao et al., 2014 | http://bioinf.wehi.edu.au/featureCounts/ |
| DESeq2 1.26.0 | Love et al., 2014 | http://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html |
| clusterProfile 3.14.3 | Yu et al., 2012 | https://guangchuangyu.github.io/software/clusterProfiler/ |
| ChIA -PIPE v1.0 | Lee et al., 2020 | https://github.com/TheJacksonLaboratory/ChlA-PIPE |
HIGHLIGHTS.
Human adipocytes and hepatocytes responses to glucocorticoids (GCs) were tested
Individual-specific genomic binding by GR determined these responses
Genetic variations control GR function and GC response
Genetic variations predict individual adverse metabolic effects of GC therapy
ACKNOWLEDGEMENTS
We thank all other members of the Lazar lab for technical support and valuable discussions. We thank Dr. Ivona Percec for generously providing us with human adipose stem cells. We thank Deepti Abbey for technical help with the hepatocyte differentiation from iPSCs. We thank Joseph Park for extracting genotype information from Penn Medicine Biobank funded by a gift from the Smilow family, the National Center for Advancing Translational Sciences of the National Institutes of Health under CTSA Award Number UL1TR001878. We thank Wenli Yang for providing the whole genome sequencing data of iPSC line SV20 (H11). We thank the Functional Genomics Core of the Penn Diabetes Research Center (P30 DK19525) for next-generation sequencing. This work was supported by the Cox Medical Institute, the JPB Foundation (to M.A.L.), ALSAC (to M.V.R. and C.-H.P) as well as by National Institutes of Health grants (R01-DK049780 to M.A.L.; R01-CA142665 and P50 GM115279 to M.V.R.; CA021765 to C.-H.P.; K23-HL145121 to J.R.W.; K01-DK125602 to D.G.). W.H. was supported by American Diabetes Association Training grant #1–18-PDF-132.
M.A.L. is an advisory board member for Pfizer Inc., and consultant to Flare Therapeutics, Madrigal Pharmaceuticals, and Novartis. M.V.R. receives investigator-initiated research support from Servier. D.J.R. is an advisory board member for Alnylam, Novartis, Pfizer, and Verve and a co-founder of Staten Biotechnology. C.H.P is an advisory board member for Adaptive Biotechnology, Inc, is on Data Monitoring Committee for Novartis, and has received honorarium from Amgen and Erytech.
Footnotes
DECLARATION OF INTERESTS
The remaining authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bachmann PS, Gorman R, Papa RA, Bardell JE, Ford J, Kees UR, Marshall GM, and Lock RB (2007). Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res 67, 4482–4490. [DOI] [PubMed] [Google Scholar]
- Barnes PJ (2011). Glucocorticosteroids: current and future directions. Br J Pharmacol 163, 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera V, Evans P, Face CJ, Hamagami N, Sankaranarayanan L, Keller CA, Giardine B, Tan K, Hardison RC, Shi J, et al. (2018). Exploiting genetic variation to uncover rules of transcription factor binding and chromatin accessibility. Nat Commun 9, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner LN, Mercader JM, Robertson CC, Cole J, Chen L, Jacobs SBR, Rich SS, and Florez JC (2020). Analysis of Glucocorticoid-Related Genes Reveal CCHCR1 as a New Candidate Gene for Type 2 Diabetes. J Endocr Soc 4, bvaa121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman DJ, Girod JP, Garcia MJ, Patel JV, Gupta M, Posch A, Saunders S, Lip GY, Worley S, and Reddy S. (2005). Effects of short-term glucocorticoids on cardiovascular biomarkers. J Clin Endocrinol Metab 90, 3202–3208. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–21 29 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. (2019). The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47, D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TJ, Lee WJ, Chang HM, Lee KC, and Chuang LM (2008). Expression of subcutaneous adipose tissue phosphoenolpyruvate carboxykinase correlates with body mass index in nondiabetic women. Metabolism 57, 367–372. [DOI] [PubMed] [Google Scholar]
- Chen X, Iqbal N, and Boden G. (1999). The effects of free fatty acids on gluconeogenesis and glycogenolysis in normal subjects. J Clin Invest 103, 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DM, and Steger DJ (2017). Nuclear Receptor Function through Genomics: Lessons from the Glucocorticoid Receptor. Trends Endocrinol Metab 28, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TJ (2006). Glucocorticoid action and the development of selective glucocorticoid receptor ligands. Biotechnol Annu Rev 12, 269–300. [DOI] [PubMed] [Google Scholar]
- Consortium GT (2015). Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti R, Mannucci E, Pessotto P, Tassoni E, Carminati P, Giannessi F, and Arduini A. (2011). Selective reversible inhibition of liver carnitine palmitoyl-transferase 1 by teglicar reduces gluconeogenesis and improves glucose homeostasis. Diabetes 60, 644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino S, Libby P, Kishore R, Tardif JC, El-Osta A, and Paneni F. (2018). Epigenetics and precision medicine in cardiovascular patients: from basic concepts to the clinical arena. Eur Heart J 39, 4150–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson SM, Khan A, Printen J, Pessin JE, and Saltiel AR (2003). PTG gene deletion causes impaired glycogen synthesis and developmental insulin resistance. J Clin Invest 111, 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Alpern D, and Gardeux V. (2016). The Genetics of Transcription Factor DNA Binding Variation. Cell 166, 538–554. [DOI] [PubMed] [Google Scholar]
- Desmet SJ, and De Bosscher K. (2017). Glucocorticoid receptors: finding the middle ground. J Clin Invest 127, 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky VW, Douglas DN, Lehner R, and Vance DE (2004). Regulation of the enzymes of hepatic microsomal triacylglycerol lipolysis and re-esterification by the glucocorticoid dexamethasone. Biochem J 378, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco LM, Gadkari M, Howe KN, Sun J, Kardava L, Kumar P, Kumari S, Hu Z, Fraser IDC, Moir S, et al. (2019). Immune regulation by glucocorticoids can be linked to cell type-dependent transcriptional responses. J Exp Med 216, 384–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gathercole LL, Morgan SA, Bujalska IJ, Hauton D, Stewart PM, and Tomlinson JW (2011). Regulation of lipogenesis by glucocorticoids and insulin in human adipose tissue. PLoS One 6, e26223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gel B, and Serra E. (2017). karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 3088–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, et al. (2014). Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, and Brown M. (2017). Role of steroid receptor and coregulator mutations in hormone-dependent cancers. J Clin Invest 127, 1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grontved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, and Hager GL (2013). C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J 32, 1568–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross KL, and Cidlowski JA (2008). Tissue-specific glucocorticoid action: a family affair. Trends Endocrinol Metab 19, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group RC, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, et al. (2020). Dexamethasone in Hospitalized Patients with Covid-19 - Preliminary Report. N Engl J Med.
- Gusarova V, O’Dushlaine C, Teslovich TM, Benotti PN, Mirshahi T, Gottesman O, Van Hout CV, Murray MF, Mahajan A, Nielsen JB, et al. (2018). Genetic inactivation of ANGPTL4 improves glucose homeostasis and is associated with reduced risk of diabetes. Nat Commun 9, 2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habegger KM, Heppner KM, Geary N, Bartness TJ, DiMarchi R, and Tschop MH (2010). The metabolic actions of glucagon revisited. Nat Rev Endocrinol 6, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, and Glass CK (2015). The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraike Y, Waki H, Yu J, Nakamura M, Miyake K, Nagano G, Nakaki R, Suzuki K, Kobayashi H, Yamamoto S, et al. (2017). NFIA co-localizes with PPARgamma and transcriptionally controls the brown fat gene program. Nat Cell Biol 19, 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Jiang C, Guan D, Dierickx P, Zhang R, Moscati A, Nadkarni GN, Steger DJ, Loos RJF, Hu C, et al. (2019). Patient Adipose Stem Cell-Derived Adipocytes Reveal Genetic Variation that Predicts Antidiabetic Drug Response. Cell Stem Cell 24, 299–308 e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai E, Miner JN, Mitchell JA, Yamamoto KR, and Granner DK (1993). Glucocorticoid receptor-cAMP response element-binding protein interaction and the response of the phosphoenolpyruvate carboxykinase gene to glucocorticoids. J Biol Chem 268, 5353–5356. [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, and Stamatoyannopoulos JA (2011). Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Barrera A, McDowell IC, D’Ippolito AM, Majoros WH, Vockley CM, Wang X, Allen AS, and Reddy TE (2018). Human genome-wide measurement of drug-responsive regulatory activity. Nat Commun 9, 5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol SE, Mattano LA Jr., Yang W, Maloney KW, Smith C, Liu C, Ramsey LB, Fernandez CA, Chang TY, Neale G, et al. (2016). Genetic risk factors for the development of osteonecrosis in children under age 10 treated for acute lymphoblastic leukemia. Blood 127, 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawedia JD, Kaste SC, Pei D, Panetta JC, Cai X, Cheng C, Neale G, Howard SC, Evans WE, Pui CH, et al. (2011). Pharmacokinetic, pharmacodynamic, and pharmacogenetic determinants of osteonecrosis in children with acute lymphoblastic leukemia. Blood 117, 2340–2347; quiz 2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- Lee B, Wang J, Cai L, Kim M, Namburi S, Tjong H, Feng Y, Wang P, Tang Z, Abbas A, et al. (2020). ChIA-PIPE: A fully automated pipeline for comprehensive ChIA-PET data analysis and visualization. Sci Adv 6, eaay2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, Imboywa SH, Chipendo PI, Ran FA, Slowikowski K, et al. (2014). Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343, 1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Luo OJ, Wang P, Zheng M, Wang D, Piecuch E, Zhu JJ, Tian SZ, Tang Z, Li G, et al. (2017). Long-read ChIA-PET for base-pair-resolution mapping of haplotype-specific chromatin interactions. Nat Protoc 12, 899–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, and Chanock SJ (2015). LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranville JC, Baxter SS, Witonsky DB, Chase MA, and Di Rienzo A. (2013). Genetic mapping with multiple levels of phenotypic information reveals determinants of lymphocyte glucocorticoid sensitivity. Am J Hum Genet 93, 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsa E, Burridge PW, Yu KH, Ahrens JH, Termglinchan V, Wu H, Liu C, Shukla P, Sayed N, Churko JM, et al. (2016). Transcriptome Profiling of Patient-Specific Human iPSC-Cardiomyocytes Predicts Individual Drug Safety and Efficacy Responses In Vitro. Cell Stem Cell 19, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotti G, Gazzaruso C, and Giustina A. (2011). Diabetes in Cushing syndrome: basic and clinical aspects. Trends Endocrinol Metab 22, 499–506. [DOI] [PubMed] [Google Scholar]
- McDowell IC, Barrera A, D’Ippolito AM, Vockley CM, Hong LK, Leichter SM, Bartelt LC, Majoros WH, Song L, Safi A, et al. (2018). Glucocorticoid receptor recruits to enhancers and drives activation by motif-directed binding. Genome Res 28, 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro A, Koh A, Wu H, Schoeler M, Faggi MI, Carreras A, Hallen A, Backhed F, and Caesar R. (2020). Hepatic expression of lipopolysaccharide-binding protein (Lbp) is induced by the gut microbiota through Myd88 and impairs glucose tolerance in mice independent of obesity. Mol Metab 37, 100997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman LK (2015). Cushing’s syndrome: update on signs, symptoms and biochemical screening. Eur J Endocrinol 173, M33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi R, Togo S, Kimura M, Shinozawa T, Koido M, Koike H, Thompson W, Karns RA, Mayhew CN, McGrath PS, et al. (2019). Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab 30, 374–384 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, et al. (2017). Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell 20, 558–570 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel HK, and Bihani T. (2016). Chromatin Scanning by Dynamic Binding of Pioneer Factors. Mol Cell 62, 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R, Williams-Dautovich J, and Cummins CL (2014). Minireview: new molecular mediators of glucocorticoid receptor activity in metabolic tissues. Mol Endocrinol 28, 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufall MA (2015). Glucocorticoids and Cancer. Adv Exp Med Biol 872, 315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, Yengo L, Ferreira T, Marouli E, Ji Y, et al. (2019). Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet 28, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini L, and Ugolini S. (2020). New insights into the cell- and tissue-specificity of glucocorticoid actions. Cell Mol Immunol. [DOI] [PMC free article] [PubMed]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, and Seale P. (2013). EBF2 determines and maintains brown adipocyte identity. Cell Metab 17, 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey LB, Pounds S, Cheng C, Cao X, Yang W, Smith C, Karol SE, Liu C, Panetta JC, Inaba H, et al. (2017). Genetics of pleiotropic effects of dexamethasone. Pharmacogenet Genomics 27, 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockall AG, Sohaib SA, Evans D, Kaltsas G, Isidori AM, Monson JP, Besser GM, Grossman AB, and Reznek RH (2003). Hepatic steatosis in Cushing’s syndrome: a radiological assessment using computed tomography. Eur J Endocrinol 149, 543–548. [DOI] [PubMed] [Google Scholar]
- Russcher H, Smit P, van den Akker EL, van Rossum EF, Brinkmann AO, de Jong FH, Lamberts SW, and Koper JW (2005). Two polymorphisms in the glucocorticoid receptor gene directly affect glucocorticoid-regulated gene expression. J Clin Endocrinol Metab 90, 5804–5810. [DOI] [PubMed] [Google Scholar]
- Sacta MA, Chinenov Y, and Rogatsky I. (2016). Glucocorticoid Signaling: An Update from a Genomic Perspective. Annu Rev Physiol 78, 155–180. [DOI] [PubMed] [Google Scholar]
- Safy M, de Hair MJH, Jacobs JWG, Buttgereit F, Kraan MC, and van Laar JM (2017). Efficacy and safety of selective glucocorticoid receptor modulators in comparison to glucocorticoids in arthritis, a systematic review. PLoS One 12, e0188810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Roberts C, Kim EJ, Brenner A, Grant G, and Percec I. (2017). Transcriptional and Cell Cycle Alterations Mark Aging of Primary Human Adipose-Derived Stem Cells. Stem Cells 35, 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry BS (2002). SNP alleles in human disease and evolution. J Hum Genet 47, 561–566. [DOI] [PubMed] [Google Scholar]
- She P, Shiota M, Shelton KD, Chalkley R, Postic C, and Magnuson MA (2000). Phosphoenolpyruvate carboxykinase is necessary for the integration of hepatic energy metabolism. Mol Cell Biol 20, 6508–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozawa T, Kimura M, Cai Y, Saiki N, Yoneyama Y, Ouchi R, Koike H, Maezawa M, Zhang RR, Dunn A, et al. (2020). High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell-Derived Organoids. Gastroenterology. [DOI] [PMC free article] [PubMed]
- Sholter DE, and Armstrong PW (2000). Adverse effects of corticosteroids on the cardiovascular system. Can J Cardiol 16, 505–511. [PubMed] [Google Scholar]
- Soccio RE, Chen ER, Rajapurkar SR, Safabakhsh P, Marinis JM, Dispirito JR, Emmett MJ, Briggs ER, Fang B, Everett LJ, et al. (2015). Genetic Variation Determines PPARgamma Function and Anti-diabetic Drug Response In Vivo. Cell 162, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed AP, Greulich F, Ansari SA, and Uhlenhaut NH (2020). Anti-inflammatory glucocorticoid action: genomic insights and emerging concepts. Curr Opin Pharmacol 53, 35–44. [DOI] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. (2015). CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 163, 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum EF, and Lamberts SW (2004). Polymorphisms in the glucocorticoid receptor gene and their associations with metabolic parameters and body composition. Recent Prog Horm Res 59, 333–357. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, and Yang J. (2017). 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet 101, 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, and Bodine SC (2008). The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab 295, E785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zang C, Xiao T, Fan J, Mei S, Qin Q, Wu Q, Li X, Xu K, He HH, et al. (2016). Modeling cis-regulation with a compendium of genome-wide histone H3K27ac profiles. Genome Res 26, 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng Y, Li J, Yu Y, Zhang W, Song M, Liu Z, Min Z, Hu H, Jing Y, et al. (2020). Single-Cell Transcriptomic Atlas of Primate Ovarian Aging. Cell 180, 585–600 e519. [DOI] [PubMed] [Google Scholar]
- Waszak SM, Delaneau O, Gschwind AR, Kilpinen H, Raghav SK, Witwicki RM, Orioli A, Wiederkehr M, Panousis NI, Yurovsky A, et al. (2015). Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell 162, 1039–1050. [DOI] [PubMed] [Google Scholar]
- Yang JJ, Cheng C, Devidas M, Cao X, Fan Y, Campana D, Yang W, Neale G, Cox NJ, Scheet P, et al. (2011). Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet 43, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Cheng C, Yang W, Pei D, Cao X, Fan Y, Pounds SB, Neale G, Trevino LR, French D, et al. (2009). Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA 301, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yea K, Kim J, Lim S, Park HS, Park KS, Suh PG, and Ryu SH (2008). Lysophosphatidic acid regulates blood glucose by stimulating myotube and adipocyte glucose uptake. J Mol Med (Berl) 86, 211–220. [DOI] [PubMed] [Google Scholar]
- Yu CY, Mayba O, Lee JV, Tran J, Harris C, Speed TP, and Wang JC (2010). Genome-wide analysis of glucocorticoid receptor binding regions in adipocytes reveal gene network involved in triglyceride homeostasis. PLoS One 5, e15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Lerner J, and Iwafuchi-Doi M. (2016). Chromatin Scanning by Dynamic Binding of Pioneer Factors. Mol Cell 62, 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhang H, Zhou Y, Qiao M, Zhao S, Kozlova A, Shi J, Sanders AR, Wang G, Luo K, et al. (2020). Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science 369, 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Xiao P, Guo Y, Liu YJ, Pei YF, Yang TL, Pan F, Chen Y, Shen H, Zhao LJ, et al. (2008). Bivariate genome linkage analysis suggests pleiotropic effects on chromosomes 20p and 3p for body fat mass and lean mass. Genet Res (Camb) 90, 259–268. [DOI] [PubMed] [Google Scholar]
- Zhu L, She ZG, Cheng X, Qin JJ, Zhang XJ, Cai J, Lei F, Wang H, Xie J, Wang W, et al. (2020). Association of Blood Glucose Control and Outcomes in Patients with COVID-19 and Pre-existing Type 2 Diabetes. Cell Metab 31, 1068–1077 e1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Liu Q, Meng L, Zhou J, Da C, Wu X, Jiang L, Shou J, and Hua H. (2018). Chemical genetic-based phenotypic screen reveals novel regulators of gluconeogenesis in human primary hepatocytes. NPJ Genom Med 3, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequencing data reported in this paper is GEO: GSE163061.