Abstract
Ndr/Lats kinases bind Mob coactivator proteins to form complexes that are essential and evolutionarily conserved components of “Hippo” signaling pathways, which control cell proliferation and morphogenesis in eukaryotes. All Ndr/Lats kinases have a characteristic N-terminal regulatory (NTR) region that binds a specific Mob cofactor: Lats kinases associate with Mob1 proteins, and Ndr kinases associate with Mob2 proteins. To better understand the significance of the association of Mob protein with Ndr/Lats kinases and selective binding of Ndr and Lats to distinct Mob cofactors, we determined crystal structures of Saccharomyces cerevisiae Cbk1NTR–Mob2 and Dbf2NTR–Mob1 and experimentally assessed determinants of Mob cofactor binding and specificity. This allowed a significant improvement in the previously determined structure of Cbk1 kinase bound to Mob2, presently the only crystallographic model of a full length Ndr/Lats kinase complexed with a Mob cofactor. Our analysis indicates that the Ndr/LatsNTR–Mob interface provides a distinctive kinase regulation mechanism, in which the Mob cofactor organizes the Ndr/Lats NTR to interact with the AGC kinase C-terminal hydrophobic motif (HM), which is involved in allosteric regulation. The Mob-organized NTR appears to mediate association of the HM with an allosteric site on the N-terminal kinase lobe. We also found that Cbk1 and Dbf2 associated specifically with Mob2 and Mob1, respectively. Alteration of residues in the Cbk1 NTR allows association of the noncognate Mob cofactor, indicating that cofactor specificity is restricted by discrete sites rather than being broadly distributed. Overall, our analysis provides a new picture of the functional role of Mob association and indicates that the Ndr/LatsNTR–Mob interface is largely a common structural platform that mediates kinase–cofactor binding.
Graphical Abstract
Hippo signaling systems are widespread in the eukaryotic world, playing roles in cell proliferation control, tissue development, and cell morphogenesis.1 These pathways have a deeply conserved signaling core in which upstream Ste20-family Mst/hippo kinases phosphorylate key C-terminal activating sites of Ndr/Lats kinases that belong to the AGC superfamily of protein kinases (e.g., PKA, -B, or -C). The Ndr/Lats kinases fall into Ndr and Lats subfamilies that, while closely related, are distinct from one another in amino acid sequence and cellular function from yeast to animals (reviewed in refs 2-4). In animals, for example, Lats-related kinases inhibit YAP/TAZ-family transcriptional coactivators driving genes that promote cell cycle entry and apoptosis resistance; this system links cell proliferation to tissue organization.1,2,5,6 Animal Ndr kinases play separate and incompletely understood roles in cell proliferation and morphogenesis of polarized structures, notably organization of neurites.1-4,6 The Ndr/Lats kinases have a distinctive segment immediately N-terminal to their kinase domains generally termed the NTR (N-terminal regulatory) region. This NTR region binds to the “Mob” coactivator protein, an association that appears to be essential for kinase function.2,7,8 In the budding yeast Saccharomyces cerevisiae, where Mob coactivators were discovered, the paralogous Lats-related kinases Dbf2 and Dbf20 bind the Mob1 protein, forming a central component of a hippo pathway system called the mitotic exit network (MEN) that controls cytokinesis and the transition from M phase to G19-20 (reviewed in refs 21 and 22). The budding yeast Ndr-subfamily kinase Cbk1 associates with the Mob2 coactivator and functions in a different pathway that controls the final stage of cell separation and polarized cell growth called the Regulation of Ace2 and Morphogenesis (RAM) network. Notably, the MEN and RAM network control distinct processes that are important for late cell division and morphogenesis, with no evidence of functional overlap of Dbf2/20 and Cbk1.16,17,19,23-25 Cbk1–Mob1 or Dbf2/20–Mob2 complexes do not appear to form,26-30 despite the simultaneous presence of all of the proteins in the cytosol, suggesting a mechanism that enforces kinase–coactivator association specificity. Studies of homologous Schizosaccharomyces pombe Sid2(Lats)–Mob1 and Orb6(Ndr)–Mob2 complexes further indicate highly specific Ndr/Lats–Mob interactions,31 despite the highly conserved nature of both the Mob cofactor and the kinase NTR across the entire Ndr/Lats family.32 While the Mob coactivator is required for kinase function, poor resolution in previous crystal structures of the Cbk1–Mob2 interface gave only limited information about the role of Mob binding.33
In this study, we collected higher-resolution crystallographic data to obtain a better structural model of the Mob2–Cbk1 interface. In the light of this new structure, we updated the NTR in the full Cbk1–Mob2 complex33 and found that this region adopts a V-shaped helical hairpin similar to those of the human Lats1–Mob134,35 and Ndr–Mob136 complexes. Furthermore, our improved structure highlights a critical role for Mob binding in positioning the hydrophobic motif (HM) of the kinase. Ndr-family kinases contain a C-terminal hydrophobic motif (HM) that upon phosphorylation of a critical threonine residue by an upstream kinase activates the enzyme in vivo.37 The Ndr/Lats group is unique among AGC kinases: apart from HM phosphorylation and autoactivation at their activation loop (AL), they depend on Mob cofactor binding. The new structure combined with modeling reveals that the phosphorylated Thr-743 in Cbk1’s HM becomes proximal to a conserved Arg in the NTR upon Mob binding. Mob-driven orientation in this region likely drives optimal positioning of the kinase’s otherwise flexible αC, a component critical for kinase activation.
The higher-resolution structure of the Cbk1–Mob2 complex allowed the comparison of the kinase–coactivator interface with that of the Dbf2–Mob1 complex to better understand the mechanism of specificity. We found that a short motif in the Mob structure that differs between Mob1 and Mob2 strongly contributes to the molecular recognition between the kinase and the cofactor; mutation of this motif permits noncognate complex formation and perturbs the binding affinity of the cognates.
RESULTS
The Cbk1NTR–Mob2 Complex Contains Conserved Structural Motifs.
The mechanism by which binding of the Mob cofactor to Ndr-family kinases controls kinase function and kinase–cofactor binding specificity is not well-understood. We set out to determine the crystal structure of the Cbk1NTR–Mob2 complex. As we were unable to stably express monomeric Mob2 in an Escherichia coli overexpression system, we engineered Mob2 V148C Y153C, termed zinc-binding Mob2 in this work, which recapitulates a zinc-binding motif found in most metazoan Mob2 orthologs as well as in S. cerevisiae Mob1 (Figure S1). We found this change stabilized Mob2 and allowed suitable E. coli expression for biochemistry. Using this construct, we determined the structure of zinc-binding Mob245-287 bound to Cbk1NTR (251–351) to 2.8 Å resolution (Table 1 and Figure 1A).
Table 1.
Crystallographic Data Collection and Refinement
| Dbf2NTR–Mob1 | Cbk1NTR–Mob2 | Cbk1–Mob2– pepSsd1 |
|
|---|---|---|---|
| Data Collection | |||
| wavelength (Å) | 0.978560 | 0.978720 | 1.0000 |
| space group | P6122 | P41212 | C121 |
| cell dimensions | |||
| a, b, c (Å) | 108.28, 108.28, 134.76 | 126.23, 126.27, 49.34 | 138.43, 79.99, 117.59 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | 90.0, 117.6, 90.0 |
| resolution (Å) | 46.9–3.5 (3.59–3.50) | 39.9–2.8 (2.87–2.80) | 44.60–3.15 (3.23–3.15) |
| CC1/2 | 99.0 (79.1) | 100.0 (67.0) | 99.6 (80.4) |
| Rmerge | 54.1 (259.2) | 10.0 (152.4) | 16.8 (130.4) |
| mean I/σI | 9.34 (1.49) | 21.79 (2.21) | 9.37 (2.05) |
| completeness (%) | 99.9 (100) | 99.8 (100) | 99.7 (99.7) |
| redundancy | 24.60 (25.83) | 14.16 (14.74) | 7.73 (7.82) |
| no. of reflections | 6385 (447) | 10323 (750) | 19888 (1455) |
| Refinement | |||
| Rwork, Rfree | 0.2292, 0.2631 | 0.2490, 0.2838 | 0.2310, 0.2983 |
| no. of atoms | |||
| protein | 2024 | 2196 | 4715 |
| ligand/ion | 17 | 1 | 31 |
| B-factor | |||
| protein | 88.3 | 92.7 | 98.8 |
| ligand | 139.1 | 114.7 | 92.6 |
| Ramachandran plot (%) | |||
| favored | 93.5 | 93 | 76 |
| allowed | 5 | 4 | 23 |
| outliers | 1.6 | 3 | 1 |
| rotamer outliers | 1.85 | 15 | 21.5 |
| RMSD | |||
| bond lengths (Å) | 0.010 | 0.004 | 0.011 |
| bond angles (°) | 0.75 | 0.70 | 1.35 |
Figure 1.
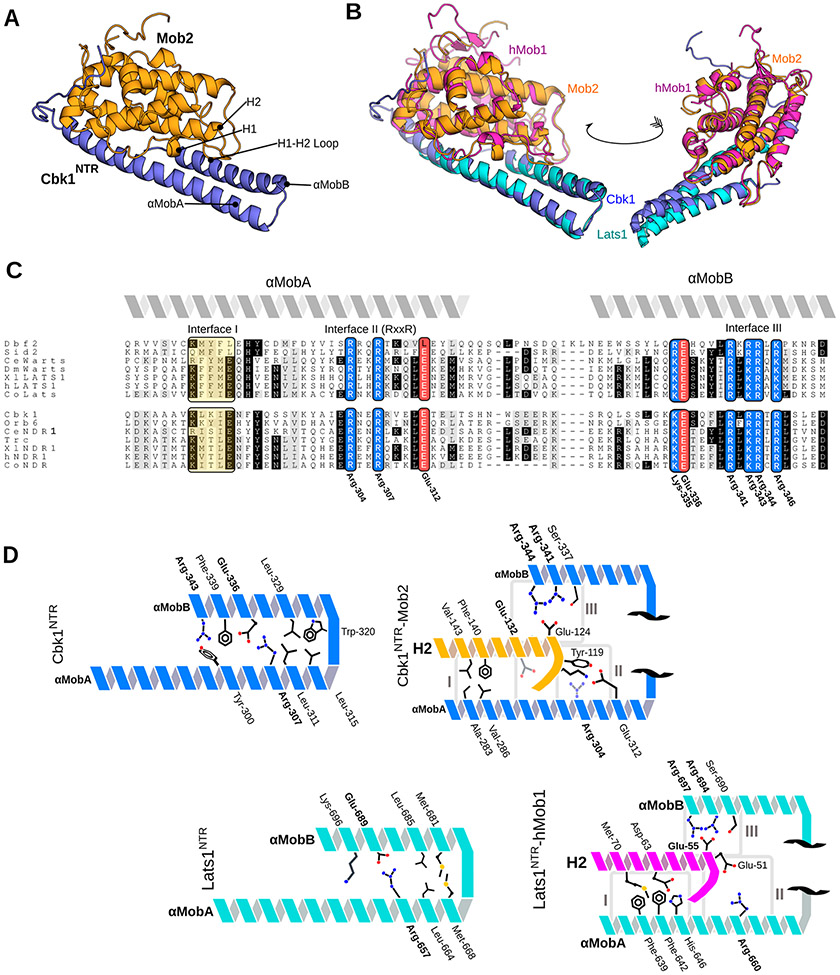
Crystal structure of the Cbk1NTR–Mob2 complex that highlights conserved structural motifs at the Ndr/Lats–Mob interface. (A) Cbk1NTR (residues 251–351, blue) complexed with zinc-binding Mob2 (residues 45–287, orange). (B) Cbk1NTR–Mob2 (blue/orange) overlaid with Lats1NTR–hMob1 (PDB entry 5B5W, cyan/purple). (C) Alignment of the Ndr/Lats NTR regions across eukaryotes. Abbreviations: Ce, Caenorhabditis elegans; Dm/Wts, Drosophila melanogaster; Xl, Xenopus laevis; Co, Capsaspora owczarzaki. Basic residues are colored blue, acidic residues red, and hydrophobic regions yellow. Interfaces I–III equivalent to those described in the Lats1–hMob1 structure are labeled. Conserved residues shown in Cbk1 are labeled. (D) Cbk1NTR–Mob2 (top) and Lats1NTR–Mob1 interaction architecture. The NTR is comprised of two Mob-binding helices: αMobA and αMobB. Conserved residues are labeled in bold.
The NTR forms a bihelical conformation similar to that observed in LatsNTR– and NdrNTR–Mob1 structures; an overlay with a previously determined Lats1NTR–hMob1 [Protein Data Bank (PDB) 5B5W] crystal structure demonstrates substantial similarity, with a root-mean-square deviation (RMSD) of 1.082 Å (Figure 1B). The new Cbk1NTR–Mob2 complex highlighted structurally conserved features of the Ndr/Lats–Mob interface, which could be mapped to three distinct regions (interfaces I–III). Regions previously observed to be responsible for binding are highly conserved within a variety of eukaryotes (Figure 1C), with interfaces II and III showing similarity in the alignment. This includes conserved Cbk1 residues Arg-304 and -307 (RxxR, at interface II), which have a geometry similar to that of Lats1 Arg-657 and -660.34,35 A number of residues are responsible for both cohesion between the two helical regions of the Cbk1 NTR (αMobA and αMobB; αMob refers to those helical kinase NTR regions that interface with the Mob cofactor) (Figure 1D, left) and adhesion to the Mob2 cofactor, specifically helix H2 (Figure 1D, right).
The Dbf2–Mob1 Structure Is Similar to the Lats–Mob1 Structure.
While a majority of the yeast Mob1’s biochemical properties have been derived from temperature sensitive mutations discovered in Mob1,15-17 the interaction interface with Dbf2 remains poorly characterized. To more completely understand this interaction and compare it with those of the Cbk1 and Lats1 homologues, we determined the structure of Mob179-314 bound to Dbf2NTR (85–173) to 3.5 Å resolution (Figure 2A). As in Cbk1NTR–Mob2, a variety of molecular contacts cohere the Dbf2 αMob helices as well as the Dbf2–Mob1 interaction, many of them similar to those of Cbk1NTR–Mob2 (compare Figure 2B with Figure 1D).
Figure 2.
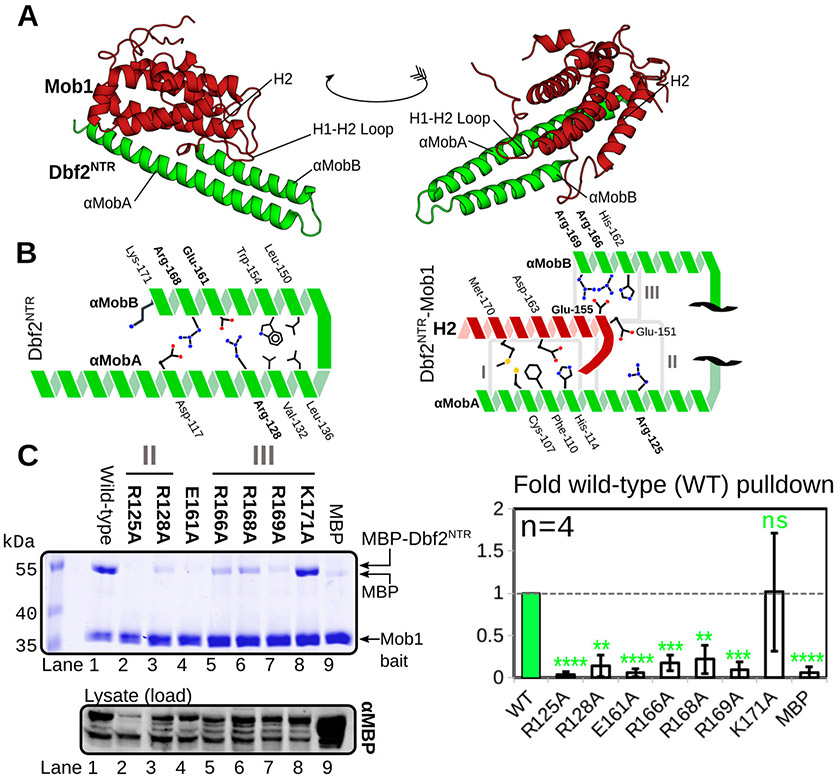
Dbf2 interacts with Mob1 through conserved residues. (A) Crystal structure of Dbf2NTR–Mob1 with conserved motifs labeled: Dbf2, green; Mob1, dark red. (B) Interaction architecture of interacting residues within the Dbf2 NTR helices αMobA and αMobB (left) and between Dbf2 NTR and Mob1 (right). Conserved residues are labeled in bold. (C) MBP-fused Dbf2 NTR (residues 85–173) and His6-fused Mob1 (residues 79–314) co-expressed in E. coli followed by co-purification of the cognate binding proteins with nickel resin. Samples were subjected to SDS–PAGE and stained with Coomassie. The bottom panel represents Western blots of the lysate as a loading control. For quantification (right), the MBP-Dbf2 NTR Coomassie signal was normalized to the input anti-MBP signal and divided by the bait Mob1 Coomassie signal. The resulting ratios were normalized to the wild-type input (WT) and averaged over four replicates (n = 4). Error bars represent the standard deviation: ns, p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; or ****p < 0.0001, based on a two-tailed t test (n = 4).
We next verified the structure by performing interaction assays with point mutants in conserved residues identified in the crystal structures. Unfortunately, Dbf2 and Cbk1 NTRs were highly prone to aggregation when expressed alone, even when fused to maltose-binding protein (MBP). SEC-MALS of purified MBP fusions demonstrated extensive oligomerization, prohibiting direct interaction assays (Figure S2). To test binding, we co-expressed Dbf2NTR and Mob1 from separate plasmids in E. coli with His6-fused Mob179-314. His6-Mob1 was purified with nickel resin, and the ability of the kinase NTR to co-purify was examined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie staining. This MBP–Dbf2NTR and Mob1 interaction assay demonstrated that Dbf2 Arg-125, Arg-166, and Arg-169 contribute strongly to the interaction, as predicted by the crystal structure (Figure 2C, lanes 2, 5, and 7). Mutation of non-Mob-binding Arg-128, Glu-161, and Arg-168 either ablated or abolished the interaction (Figure 2C, lanes 3, 4, and 6), indicating the integrity of the NTR is crucial to formation of the complex similar to the intra-αMob interaction between Lats1 Arg-660 and Glu-689.34 Interestingly, Lys-171 does not participate in Mob1 binding despite strong charge conservation across the eukaryotic world (Figure 2C, lane 8). Thus, conserved residues in the Dbf2–Mob1 structure participate in binding.
Ndr/Lats–Mob Complexes Contain Extensively Conserved Regions.
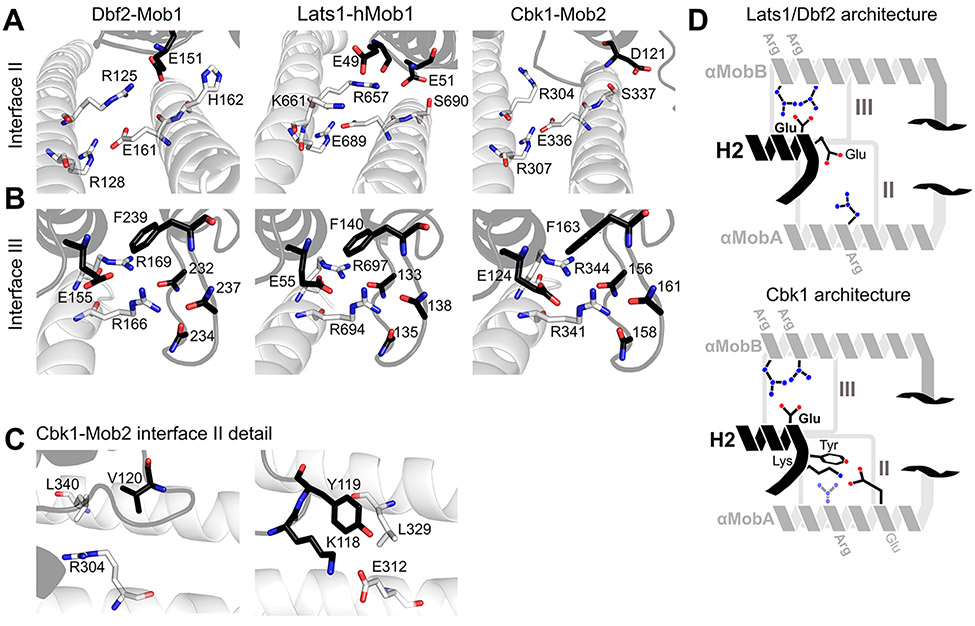
Overall, we found both yeast complexes to be conformationally similar to previously published structures of mammalian Mob1/Lats134,35 and Mob1/Ndr2.36 Both Cbk1NTR and Dbf2NTR form a two-helix bundle (αMobA and αMobB), and helix 2 (H2) of the Mob protein is incorporated into a cleft formed between the two αMob helices. First, we examined a highly conserved interface (interface II) that holds the αMobA and αMobB helices together (Figure 3A). Similar to Lats–Mob1, Dbf2 Arg-125 and Arg-128 protrude from αMobA to hold Glu-161 on αMobB (Figure 3A, left panel) akin to interface II in the Mob1–Lats1NTR structure (Figure 3A, center panel).34 Dbf2 Arg-125 interacts with Mob1 Glu-151, which is conserved in the Mob1 coactivator subfamily,15 and with Dbf2 αMobB His-162 (Figure 3A, left panel). Similarly, the Cbk1–Mob2 structure reveals a salt bridge between Arg-307 and Glu-336 (Figure 3A, right panel).
Figure 3.
Conserved regions cohere Ndr/LatsNTR–Mob complexes. (A) Comparison of interface II from Lats1NTR–hMob1 (PDB entry 5B5W, center panel) with Cbk1NTR–Mob2 (right) and Dbf2NTR–Mob1 (left). Highlighted residues shown as sticks are important for the interaction. (B) Comparisons of interface III from Lats1NTR–hMob1 (center), Cbk1NTR–Mob2 (right), and Dbf2NTR–Mob1 (left). (C) More detailed view of interface II of the Cbk1NTR–Mob2 complex showing the orientation of Cbk1 Arg-304 in relation to Leu-340 and Mob2 Val-120 (left) and Cbk1 Glu-312 and Mob2 Lys-118 and Tyr-119 (right). The interaction between Lys-118/Tyr-119 of Mob2 and Glu-312/Leu-329 of Cbk1 is distinct from those of Lats and Dbf2–Mob1 complexes. (D) Generalized architecture of Dbf2/Lats1–Mob1 in relation to that of Cbk1–Mob2. Conserved binding interfaces II and III as shown in panels A–C are boxed. Basic residues are colored blue, and acidic residues red. Mob is colored black, and the Ndr/Lats kinase NTR is colored gray.
Also similar to Mob1–Lats1,34 several highly conserved basic residues form an interface (interface III) to stabilize the Mob–NTR interaction (Figure 3B). Dbf2 Arg-166 and Arg-169 contact Mob1 Glu-155 (Figure 3B, left panel). It was shown that Lats–Mob1 shows a similar organization with the conserved Lats Arg-694 and Arg-697 interacting with Mob1 Glu-55 (Figure 3B, center panel).34 Similarly, Cbk1 Arg-341 and Arg-344 form an electrostatic interaction with Mob2 Glu-124 (Figure 3B, right panel). Taken together, these data demonstrate how the geometry of the Mob1–NTR interaction at interface III is strongly conserved; Lats1, Dbf2, and Cbk1 bind their cognate Mobs in a nearly identical fashion, especially in interface III, with some differences in interface II described below.
While Cbk1 Arg-307 on αMobA forms a bridge to Glu-336 on αMobB similar to the interhelical cohesion between Lats1 Arg-657 and Glu-689 (see Figure 1D), Cbk1 Arg-304 does not interact with Mob in the crystal structure as Lats1 Arg-660 and Dbf2 Arg-125 do (see Figures 1D and 2B), instead being sterically pushed toward the center of Mob2 by the hydrophobic Val-120 and Cbk1 Leu-340 (Figure 3C, left panel). As a replacement for this interaction, Mob2 Lys-118 and Tyr-119 extend from the Mob H1–H2 loop and contact Cbk1 Glu-312, forming a salt bridge and a hydrogen bond, respectively, and laying in a pocket formed at the intersection of the two αMob helices (Figure 3C, right panel). Glu-312 is not conserved in Dbf2 (see Figure 1C) but is found in other known Ndr/Lats-subfamily kinases in metazoans. Thus, Cbk’s interaction with Mob2 partially occurs through an interaction distinct from the Lats1–Dbf2 interaction (Figure 3D).
Budding Yeast Mob1 and Mob2 Exhibit Intrinsic Binding Specificity for Their Cognate Kinases.
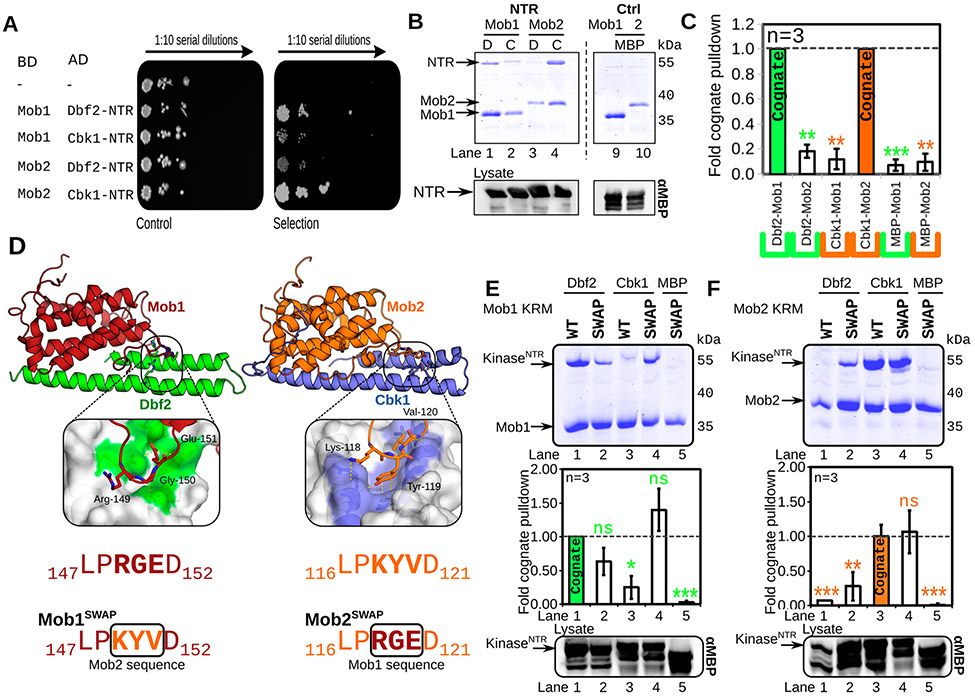
Evidence from S. pombe suggests yeast kinases Sid2 (SpDbf2) and Orb6 (SpCbk1) bind Mob1 and Mob2 to form the cognate Sid2–Mob1 and Orb6–Mob2 complexes, respectively, without cross-interaction.31,38 While genetic evidence supports a similar organization in S. cerevisiae, this has never been demonstrated biochemically. We therefore sought to examine the specificity of binding of Dbf2 and Cbk1 to Mob1 and Mob2, respectively. First, we used the yeast two-hybrid assay to probe Dbf2–Mob1 and Cbk1 NTR–Mob2 interaction. This genetic assay showed that cognate Dbf2NTR–Mob1 and Cbk1NTR–Mob2 interaction is stronger in vivo compared to noncognate ones, as expected (Figure 4A). Next, we examined kinase NTR–Mob interaction specificity in biochemical binding assays. To test binding, we co-expressed Dbf2–Mob1 and Cbk1–Mob2 interaction from separate plasmids in E. coli with His6-fused Mob179-278. His6-Mob was purified with nickel resin, and the ability of the kinase NTR to co-purify was examined by SDS–PAGE and Coomassie stain. We found cognate Dbf2–Mob1 and Cbk1–Mob2 complexes co-purified (Figure 4B,C, lanes 1 and 4), while the noncognate Dbf2–Mob2 complex did not (Figure 4B,C, lane 3). Cbk1 showed a weak interaction with Mob1 (Figure 4B,C, lane 2). Taken together, these data support the idea that limited cross-interaction occurs between the noncognate complexes, suggesting Dbf2–Mob1 and Cbk1–Mob2 act as the dominating complexes.
Figure 4.
Ndr/Lats–Mob binding is specific and controlled through a three-residue motif. (A) Mob–NTR interaction specificity is maintained in vivo. Yeast two-hybrid test of kinase–NTR complexes. Selection: yeast cells containing binding domain (BD)–Mob and activation domain (AD)–NTR fragments on dropout agar serially diluted. Growth on this “Selection” agar plate is indicative of binding between BD and AD constructs (BD, DNA-binding domain construct; AD, transcriptional activation domain construct; −, empty plasmid). Viability/growth “Control” plate: single AD construct transformants grown on nonselective YPD agar plates. (B) Dbf2 and Cbk1 form specific complexes with Mob1 and Mob2, respectively, in vitro. MBP-fused Dbf2 NTR (85–173), labeled as “D”, and Cbk1NTR (251–351), labeled as “C”, were co-expressed with His6-fused Mob1 and Mob2 V148C Y153C in E. coli, isolated via nickel chromatography, separated via SDS–PAGE, and stained with Coomassie. The dotted line demarcates a division in the gel. Bottom panels show Western blots of the E. coli lysate using anti-MBP antibody (αMBP). NTR, intact MBP–NTR fusion. (C) Quantification of panel B. The Coomassie signal from prey Dbf2–Cbk1 was normalized to the amount in the total lysate and divided by the total bait signal. Three individual replicates were performed. Samples were normalized to the “cognate” Dbf2–Mob1 (green) or Cbk1–Mob2 (orange) signals [ns, p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; or ****p < 0.0001, based on a two-tailed t test (n = 3)]. (D) Diagram of wild-type (WT) and “swapped” Mob cofactors. The kinase restrictor motif (KRM), a short, three-amino acid motif at the interaction interface (purple) between Mob and the kinase NTR, was mutated to reflect the paralogous sequence in Mob1 and Mob2. Mob1SWAP was mutated to possess the Mob2 sequence (KYV); Mob2SWAP has the KRM mutated to the Mob1 sequence (RGE). (E and F) Mob1SWAP and Mob2SWAP mutants gain the ability to bind noncognate kinases. in the top panels, wild-type (WT) or His6-fused swapped (SWAP) Mob1 (E) or Mob2 (F) was co-expressed with MBP-fused Dbf2 or Cbk1 NTR in E. coli, isolated via nickel chromatography, separated via SDS–PAGE, and stained with Coomassie. The middle panels show the quantification of the top panels as in panel B. The bottom panels show Western blots of the E. coli lysate using the anti-MBP antibody (αMBP). KinaseNTR, intact MBP–NTR fusion.
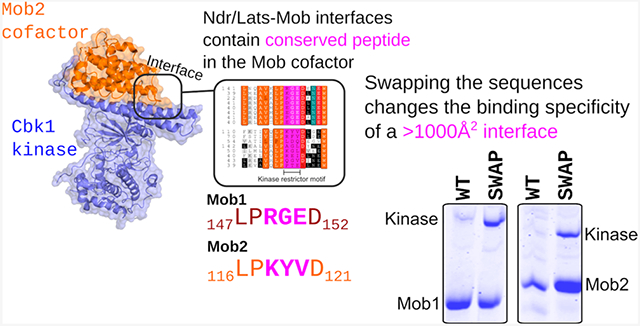
The Mob Kinase Restrictor Motif (KRM) Constrains the Binding of Kinase to a Single Cofactor.
Despite differences in sequence, all crystallized Ndr/Lats kinases interact with Mob through a conserved loop connecting the two first two helices of Mob (H1 and H2) (Figure 4D).15,34,35 Alignment of the H1 and H2 loops of yeast and human Mob reveals a highly conserved sequence (LP)xxx(D,N) that separates Mob1 and Mob2 subfamilies (Figure S1). In yeast Mob1, the conserved Pro-148 is hypothesized to stabilize the H1–H2 loop,15 with the solvent-exposed Asp-152 initiating helix 2. Mutations in Pro-148 lead to a loss of Mob1 function.15 Importantly, the three variable residues are subfamily-specific, with xGE representing Mob1 and Mob2 having no strongly conserved alignment (Figure S1). We named this tripeptide the kinase restrictor motif (KRM) and posited it strengthens or otherwise affects kinase–Mob binding specificity.
We sought to determine the role of this motif in the kinase–Mob interaction. Mob1 and Mob2 with sequences swapped between proteins were co-expressed with stabilized MBP fusions of Dbf2NTR and Cbk1NTR. The Mob1SWAP (Mob1 R149K G150Y E151V) mutant bound its cognate Dbf2 weakly and the noncognate Cbk1 with greater affinity when compared with those of the wild type (Figure 4E). The Mob2SWAP (Mob2 K118R Y119G V120E) chimera bound Dbf2, an interaction not reported in S. pombe, in S. cerevisiae, or in human Lats1 (Figure 4F).8,38,39 Thus, exchange of this region between Mob cofactors can confer or enhance their ability to bind noncognate Ndr/Lats kinases.
The Higher-Resolution Structure of Cbk1–Mob2 Reveals Mob Binding Properly Positions the Kinase HM Motif.
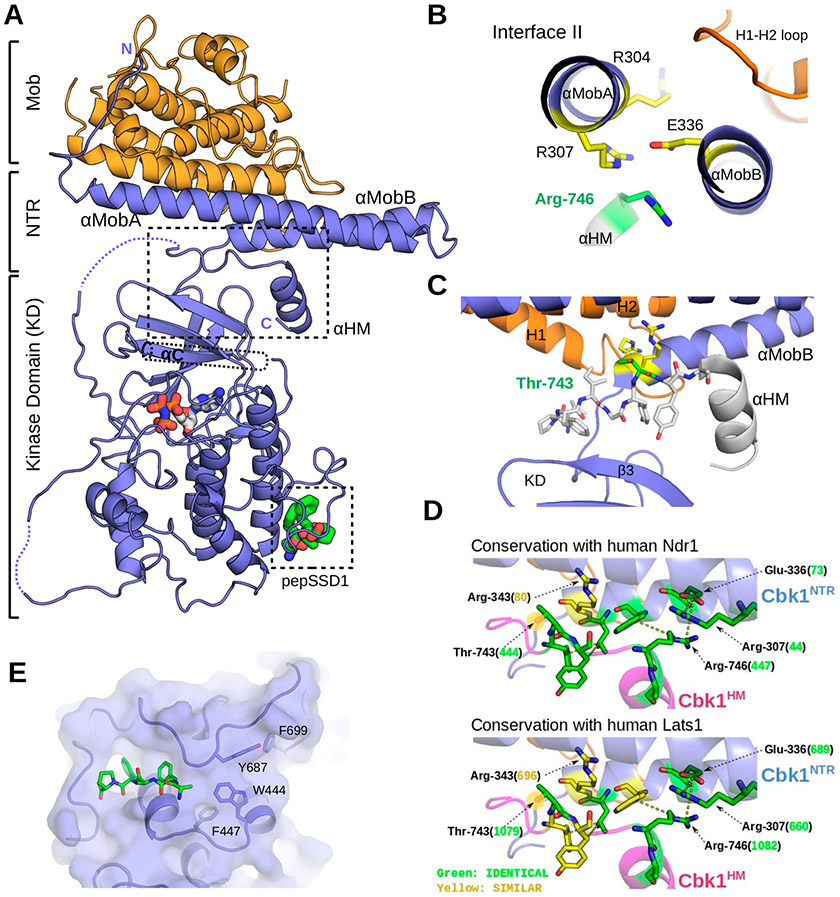
On the basis of the higher-resolution Cbk1NTR–Mob2 structure, we revisited our crystallographic analysis of the full length Cbk1–Mob2 complex.33 We improved the electron density map of the Cbk1–Mob2 complex, which enabled us to correct inaccuracies in the lower-resolution model, such as the conformation of the DFG motif important in kinase activation and the NTR of Mob2, particularly αMobA, which was not interpreted as being α-helical in the old model (Figure 5 and Figure S3). Notably, enhancement of the Cbk1–Mob2 structure resolves the C-terminal extension of Cbk1’s hydrophobic motif (HM), providing a new view of the conformation of this important region involved in allosteric activation of the kinase.37 We found that Cbk1’s C-terminal HM regulatory region is helical (αHM) in the structure, making contact with residues in Cbk1’s αMobB and the core of the kinase domain (Figure 5A). In addition, we found that Arg-746 of Cbk1’s αHM interacts with Glu-336 between the αMob helices, potentially explaining notable phosphorylation-independent interaction of the HM with the kinase as we described previously (Figure 5B).33 The Cbk1 HM falls next to residues forming interface II between Cbk1NTR and the H1–H2 loop from Mob2. Additionally, our revised model shows Thr-743 in the HM is positioned proximal to the conserved Arg-343 and -344 of the NTR (Figure 5C). This suggests an activating phosphorylation at Thr-743 could hypothetically provide an electrostatic interaction that stabilizes the HM conformation, which in turn drives a disorder-to-order transition of the αB–αC region, similar to what had been shown for AKT/PKB40 (Figure 6).
Figure 5.
Revised Cbk1–Mob2 crystal structure that reveals Ndr structural components conserved in Ndr1/Lats1. (A) The crystal structure of the inactive Cbk1–Mob2 complex was remodeled on the basis of the new Cbk1NTR–Mob2 structure. In the new model, the coactivator binds to two N-terminal helices (αMobA and αMobB) that together with the top of the kinase domain (β3) form a binding slot for the AGC kinase hydrophobic motif (HM). The C-terminal part of this motif also adopts a helical conformation (αHM). (B) Cbk1’s hydrophobic motif (HM) binds residues in interface II of the Cbk1NTR. The panel shows the view along the axis of αMobA/B and highlights Arg-746 and critical residues at Cbk1NTR–Mob interface II (Arg-307, Arg-304, and Glu-336). (C) The phosphorylation site on the HM (Thr-743, green) is next to Arg-343 and Arg-344 (yellow) from αMobB, hinting about the importance of Mob coactivator binding in the allosteric phosphoregulation of the Ndr/Lats kinase domain. The HM is shown in stick representation (737-LPFIGYTY-744) or as an α-helical cartoon (745-SRFDYLTKRKNAL-756) in gray. (D) Residues in the interacting region between the HM and the Cbk1NTR are conserved in human Ndr1/Lats1. Green residues are identical; yellow residues are chemically similar. Numbers in parentheses indicate the human Ndr/Lats residue orthologous to the indicated Cbk1 amino acid (black). (E) Crystallographic model rebuilding also improved the overall quality of the electron density and allowed the Ssd1 docking peptide to be traced (green sticks).
Figure 6.
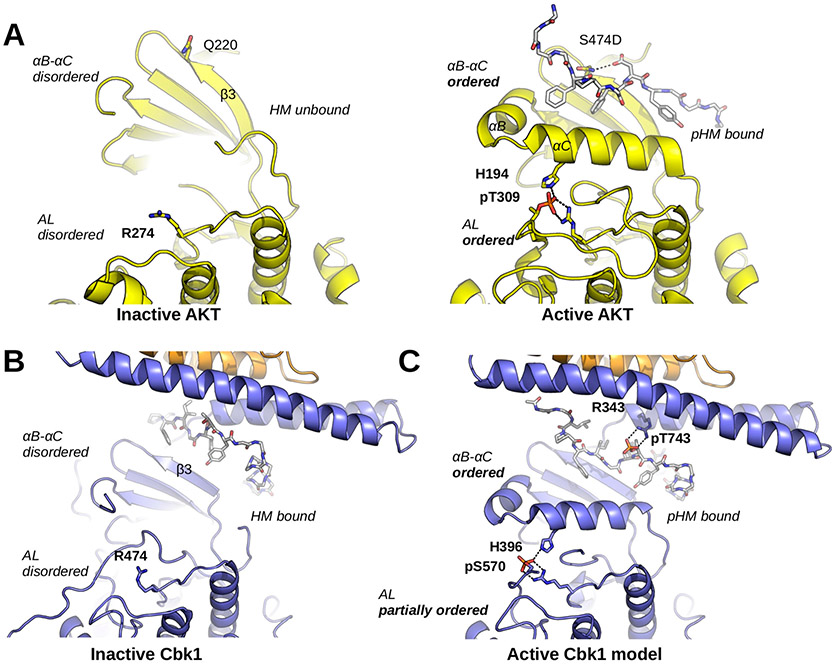
Cbk1 activation: the role of HM phosphorylation and Mob binding. (A) The activation of AKT/PKB involves a disorder-to-order transition at the αB–αC segment, the activation loop (AL), and the HM. Q220 coordinates the phosphorylated HM (here shown with the S474D phosphomimicking mutation; PDB entry 1O6K). The phosphorylated activation loop (AL; phosphoThr-309) is coordinated by Arg-274 and His-194 (from αC). (B) The inactive Cbk1 structure shows a similar structure apart from the position of its HM. The inactive crystal structure of the Cbk1–Mob2 complex shows that the HM is bound to the groove formed by the Mob-bound NTR and β3 from Cbk1. (C) For activation, the ordering of helix αC in Cbk1 similar to that of AKT/PKB is likely required (MD model). This latter may be supported by interactions formed by phospho-HM and phospho-AL. The activation loop phosphosite (AKT/PKB, Thr-309; Cbk1, Ser-570) is coordinated by an arginine residue from the kinase HRD motif (AKT/PKB, Arg-274; Cbk1, Arg-474) and by another residue from helix αC (AKT/PKB, His-194; Cbk1, His-396). The hydrophobic motif phosphosite (AKT/PKB, Ser-474; Cbk1, Thr-743) in most AGC kinases is coordinated by a single residue from β3 (AKT/PKB, Gln-220). In the case of Cbk1, an arginine residue (Arg-343) from the Mob-bound αMobB coordinates the HM phosphosite while β3 may not be involved. For both AKT/PKB and Cbk1, phospho-HM coordination (by Gln-220-pSer-474 or by Arg-343-pThr-743) brings hydrophobic residues much closer to αC and probably facilitates its ordering. This activation model suggests that coactivator binding is essential not only in HM binding but also in the precise coordination of the kinase’s active state when it is phosphorylated. AKT/PKB is colored yellow, and both HM regions are shown with gray sticks.
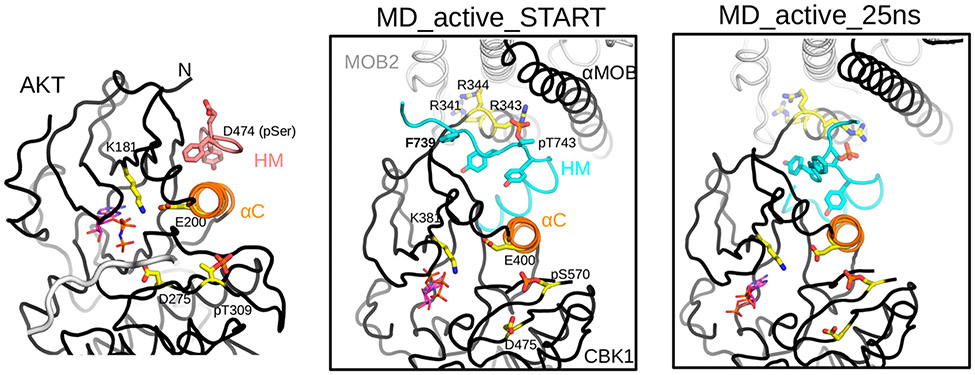
Helix C (αC) plays an important role in protein kinase activity regulation by affecting ATP binding; moreover, for AGC kinases, HM phosphorylation may directly influence αC flexibiliy or conformation.40 This critical region, however, is disordered in the nonphosphorylated Cbk1–Mob2 crystal structure, as seen for another AGC kinase, AKT/PKB. In AKT/PKB, αC becomes ordered upon HM phosphorylation and this promotes the proper positioning of ATP for substrate phosphorylation and makes the activation loop (AL) ordered. We used the active AKT/PKB crystallographic model to build αB–αC and the corresponding AL region of the Cbk1 kinase domain. This model was then phosphorylated at the HM phosphosite (p-T743) and at the activation loop (p-S570) in silico and subjected to energy minimization to obtain a biochemically feasible starting model for MD simulations (Figure S4). This latter showed that phospoThr-743–Arg-343 binding may hold the HM fixed in the binding slot between the NTR and the N-terminal kinase lobe (N-lobe) of Cbk1, if the αC from Cbk1 were ordered (Figure 7). Indeed, MD simulation showed that the salt bridge between phosphoThr-743 (pT743) and Arg-343 is stable during the time course of the simulation (25 ns), suggesting that this interaction may serve as an anchor around which the HM, particularly the bulky tyrosine or phenylalanine side chains, may pivot and thus push the αC more toward the core of the kinase N-lobe (Figure 7, right panel). We postulate that Cbk1 activation may mechanistically proceed similarly to AKT/PKB activation, but the former also depends on Mob2 binding as the cofactor holds the NTR in position for HM and particularly for phosphorylated HM binding. According to this, Ndr/Lats kinases substitute the classical HM-binding pocket in AGC kinases located on the top of the N-lobe (such as in AKT/PKB, and termed the PIF pocket) with a unique HM-binding slot that is buttressed between the NTR and the kinase N-lobe.
Figure 7.
Mechanistic model of activation of the Cbk1–Mob2 complex by the HM and activation loop phosphorylation. The crystal structure of active AKT/PKB (PDB entry 1O6K) highlights the central role of αC in assembling a catalytically competent AGC kinase active site. Glu-200 on αC together with Lys-181 from the N-lobe of the kinase orients the phosphate moiety of ATP (shown in sticks where carbon atoms are colored magenta) and aligns it for substrate phosphorylation involving Asp-275. (The backbone of the substrate peptide bound in the substrate-binding pocket is colored gray.) Activated HM binding in the PIF pocket and activation loop (AL) phosphorylation at Thr-309 buttress αC and promote the proper ATP alignment. The middle panel shows the structural model of the activated Cbk1–Mob2 complex (MD_active_START). The model, which had αC built on the basis of the active AKT/PKB crystallographic model shown in the left panel (but lacked long flexible regions invisible in the crystal structure, e.g., the linker connecting the HM to the kinase domain core or most of the long AL), was created using the revised crystal structure of the nonphosphorylated Cbk1–Mob2 complex. The panel shows the energy-minimized phosphorylated Cbk1–Mob2 structural model and highlights the same key residues in the AGC kinase domain as shown in the AKT/PKB structure on the left. Note that Arg-343 is well-positioned to coordinate phosphoThr-743 on the HM, and this together with phosphoSer570 at the AL affects salt bridge formation between Glu-400 and Lys-381, which in turn could influence active site assembly as in AKT/PKB. The right panel shows the active Cbk1–Mob2 complex after a 25 ns molecular dynamics simulation (MD_active_25 ns).
Our revised model also provided additional insight into the substrate binding of the Cbk1 kinase. Full length Cbk1–Mob2 was crystallized in the presence of a small docking peptide from a Cbk1 substrate, Ssd1, but electron density corresponding to this peptide could not be traced in the old crystal structure.33 Our improved map, however, showed additional electron density close to the predicted peptide-binding groove (Figure 5E). The observed density occurred close to Phe-447, Trp-444, and Tyr-687, mutation of which interfered with peptide binding, and was distant from Phe-699, a mutation that did not affect binding.33 These data reveal the region on Cbk1 required for docking motif binding.
Strikingly, all major interacting amino acids we identified in Cbk1’s NTR and HM are either identical or highly similar at corresponding sites in human Lats1 and Ndr1 (Figure 5D). The crucial Arg-343 of Cbk1 is charge-conserved in human Lats and Ndr kinases at a similar position in NTR–Mob1 structures. This would position a positively charged side chain above the key HM phosphorylation site (Cbk1 Thr-743, Ndr1 Thr-444, and Lats1 Thr-1079) with additional conserved salt bridges forming between Cbk1 Glu-336 and Arg-746. Notably, the Lats equivalent of Cbk1 Glu-336, Glu-689, is crucial for interaction with Mob1 in humans and is also conserved.35 Taken together, the revised Cbk1–Mob2 structure highlights an extensive electrostatic network among the residues of the HM, NTR, and Mob, and evolutionary sequence conservation of these suggests that a similar regulatory mechanism likely present in other hippo pathway components across the eukaryotic world.
DISCUSSION
Dbf2 and Cbk1 are conserved AGC kinases in the Ndr/Lats family and are evolutionarily related to their human counterparts. The conserved arginines on the αMobA and αMobB helices of Dbf2, Cbk1, Lats, and Ndr contain a conserved RxxR motif (Figure 1C). Dbf2/Lats uses this motif to bind its Mob factor, whereas Cbk1 does not appear to. While the second arginine appears to electrostatically cohere the αMob helices, the function of Cbk1’s Arg-304 remains unknown, as it does not bind Mob2. However, Cbk1 Arg-304 may have some role in stabilizing the HM-binding site in the kinase–coactivator complex as Cbk1 HM Arg-746 interacts electrostatically with the conserved Glu-336 that is coordinated by the RxxR of the Cbk1 NTR (Figure 5B). Mob1 binds Dbf2 RxxR (involving Arg-125 and Arg-128) directly to form interface II (Figures 2B and 3A), suggesting roles for the cofactor in kinase regulation through modulation of the HM-binding site if the structures are similar.
Comparing the kinase restrictor motifs (KRMs) of eukaryotic Mob1 and Mob2 subfamilies may reveal similarities in cofactor function. Mob1’s high degree of conservation in this region is consistent with its generally conserved role in binding Lats-subfamily kinases and in cell cycle exit. Mob2 is more diverse, potentially reflecting the larger variety of functions performed by the Ndr subfamily of kinases in eukaryotic systems. Human Mob2 may interact with Ndr through Arg-48 and Glu-49 of its KRM. In contrast, Drosophila Mob2, which contains Ala/Gly in place of Arg/Glu, may not utilize the motif to bind Tricornered, the Drosophila Ndr. Such large differences within the normally conserved Ndr–Mob families may explain the difficulty in abolishing the human Ndr1–Mob2 interaction through point mutations at conserved residues in previous work.39
In our revised structure, the conserved arginines in Cbk1’s Mob2-binding NTR (RxxR) associate with Arg-746 in the kinase’s C-terminal regulatory hydrophobic motif (HM) (Figure 5B,D). The position of the Mob2 KRM directly covering the highly conserved HM-binding site in Cbk1 suggests a role for regulation (Figures 4D and 5B). As the hMob1/Mob1 KRM directly binds the homologous domain through Glu-51/151 (Figure 3A,D), we hypothesize Mob1 modulates the conformation of the putative Lats/Dbf2 HM-binding region and subsequently controls the kinase.
CONCLUSIONS
Our analysis here indicates that the Ndr/Lats NTR–Mob interface is a common structural platform through which kinase–cofactor binding is mediated; however, amino acid variations in key positions contribute to subgroup- and organism-specific differences. While the first Mob crystal structures suggested that a conserved and acidic surface was responsible for interactions with Ndr/Lats through the kinase N-terminus, the reality may be more complex. Some Mob cofactors may bind and activate their cognate kinases through a conserved set of charged residues; others may bind using distal regions and use these residues for activation only, or for alternative functions of Mob.
MATERIALS AND METHODS
Cloning of DNA Constructs.
We were unable to stably express and purify monomeric Mob2 in an E. coli overexpression system. Therefore, to produce suitable protein for crystallization and biochemistry, we engineered Mob2 V148C Y153C, which recapitulates a zinc-binding motif found in most metazoan Mob2 orthologs as well as in S. cerevisiae Mob1. Unless otherwise noted, interaction data and the Cbk1NTR–Mob2 crystal structure were obtained using this mutant.
For expression of Cbk1NTR–Mob2 for crystallography, the modified bicistronic pBH4 vector containing His6-Cbk1 (Cbk1 NTR, residues 251–351) and zinc-binding GST-Mob2 V148C Y153C (residues 45–287) were expressed from a single T7 promoter. The expression construct was made by cloning the cDNA corresponding to Cbk1 residues 251–756 and Mob2 residues 45–287 into the backbone, and then Cbk1 Phe-352 was mutated to a stop codon using the QuickChange technique (Agilent). His6-Cbk1NTR is 5′ to GST-Mob2 in this vector; both open reading frames contain separate ribosome-binding sites. Val-148 and Tyr-153 were mutated to cysteine using the same technique. For expression of Dbf2NTR–Mob1 for crystallography, a GST-less form of the bicistronic vector used for Cbk1NTR–Mob2 was utilized. His6-Dbf2NTR (residues 85–173) was cloned into the 5′ cistron, and tag-less Mob179-314 was cloned into the 3′ cistron. For vectors used for pull-downs, Dbf2 (85–173) and Cbk1 (251–352) were cloned into pMAL-c2x (New England Biolabs) and fused to an N-terminal MBP tag. S. cerevisiae Mob1 (79–314) and zinc-binding Mob2 V148C Y153C (45–278) were cloned into pET28a (EMD Biosciences).
Expression and Purification of Proteins.
To purify the zinc-binding Cbk1NTR–Mob2 V148C Y153C complex, protein was overexpressed in BL21(DE3) RIL (Agilent) at 37 °C in Terrific Broth (TB) containing 40 μM zinc chloride. After growth to mid-log phase, IPTG was added to a final concentration of 0.2 mM and expressed overnight at 37 °C. Cell pellets were lysed by sonication and purified using nickel chromatography, and tags cleaved using tobacco etch virus (TEV) protease overnight on wet ice. Cleaved protein was further purified using a hand-poured SP-Sepharose (GE) column and eluted with a gradient from 50 to 2000 mM NaCl, exchanged into 20 mM HEPES (pH 7.4) and 300 mM NaCl using size exclusion chromatography, and concentrated to 55 mg/mL.
To purify the Dbf2NTR–Mob1 complex, a bicistronic vector containing His6-Dbf2NTR (85–173) and untagged Mob1 (79–314) expressed from a single T7 promoter was cloned and overexpressed in BL21 (DE3). Mob1 (79–314) contained the N-terminal amino acid sequence ENLYFQGS as a byproduct of cloning. TB cultures containing 40 μM zinc chloride were inoculated, grown to mid-log phase, induced with IPTG to 0.2 mM, and expressed overnight at 24 °C. Cell pellets were lysed by sonication. Protein was purified using nickel chromatography and cleaved overnight on wet ice using TEV protease. Residual TEV and nickel-binding contaminants were reabsorbed using nickel resin, and the final protein was then exchanged into 100 mM sodium phosphate (pH 7.4), 50 mM KCl, and 100 mM l-arginine HCl using size exclusion chromatography and concentrated to 30 mg/mL. l-Arginine was present throughout the purification to prevent oligomerization of the complex and did not interfere with nickel chromatography.
Crystallization of Cbk1NTR–Mob2, Dbf2NTR–Mob1, and Cbk1–Mob2–pepSsd1 Complexes.
Ndr/LatsNTR–Mob crystals were obtained at 22.3 °C. Cbk1NTR–Mob2 was crystallized using the hanging-drop method; the well solution containing 500 μL of 100 mM MES/imidazole (pH 6.5), 30 mM CaCl2, 30 mM MgCl2, 12.5% (w/v) PEG1000, 12.5% (w/v) PEG3350, and 12.5% (v/v) MPD produced crystals. Dbf2NTR–Mob1 was crystallized using the microbatch method under 200 μL of mineral oil (Fisher BioReagents); crystals were obtained in 100 mM Bicine/Tris (pH 8.5), 30 mM LiCl, 30 mM NaCl, 30 mM KCl, 30 mM RbCl, 12.5% (w/v) PEG1000, 12.5% (w/v) PEG3350, and 12.5% (v/v) MPD. All crystals were frozen in liquid nitrogen without the addition of further cryoprotectant. Cbk1NTR–Mob2 and Dbf2NTR–Mob1 diffracted to 2.80 and 3.50 Å, respectively. Data were processed with XDS,41 and the structures of Cbk1NTR–Mob2 and Dbf2NTR–Mob1 complexes were determined by molecular replacement with PHASER.42 The MR search identified single NTR–Mob complexes with a searching model of Lats1–hMob1 (PDB entry 5BRK). Structure refinement was carried out using PHENIX,43 and structure remodeling and building was performed in Coot.44
Crystallization of the Cbk1–Mob2–pepSsd1 complex was identical to that previously published.33 To increase the redundancy of our crystallographic data and to maximize the gained experimental information, we merged two independent data sets collected on this complex (Table 1). This helped to increase the resolution range to 3.15 Å, meaning an additional ~15% experimental data on top of duplicated multiplicity. Data were collected at 100 K on the PXIII beamlines of the Swiss Light Source (Villigen, Switzerland) and at the Advanced Photon Source (Argonne, IL).
Pull-Down Experiments.
Dbf2–Mob1 and Cbk1–Mob2 constructs were co-transformed into BL21(DE3) or BL21(DE3) RIL, grown in TB at 37 °C for 2 h, induced with 0.2 mM IPTG, and grown overnight at 18 or 24 °C. Cell pellets were lysed by sonication, and raw protein was measured by a Bio-Rad Protein Assay. Lysates were normalized, incubated with nickel resin (Qiagen), washed twice with 20 mM imidazole, and eluted in 300 mM imidazole. Purified protein was separated using 15% SDS–PAGE gels and visualized using an Odyssey infrared imager (LI-COR Biosciences).
Immunoblotting.
Bacterial cell lysates normalized in raw protein concentration were separated via 15% SDS–PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes. Blots were blocked using Odyssey Blocking Buffer (LI-COR Biosciences) for 30 min and probed with mouse αMBP or rabbit αHis6 antibodies for 1 h at room temperature. Membranes were washed three times with TBS-T, probed with fluorophore-conjugated IRDye 680LT goat anti-mouse or IRDye 800CW goat anti-Rabbit antibody (LI-COR Biosciences) for 30 min, and washed three more times with TBS-T. Blots were imaged using an Odyssey infrared imager.
Yeast Two-Hybrid Assay.
Plasmids containing Mob179-314 and Mob145-278 were fused to Gal-binding domains and transformed into the Y2H Gold strain. THe Dbf2 and Cbk1 NTR were fused to the Gal activation domain and transformed into Y187. Serial dilutions of cultures were grown on –Leu –Trp –His triple dropout medium until colonies were visible. The dropout agar plates contained 10 mM 3-amino-1,2,4-triazole (3-AT) to repress spurious activation.
Molecular Dynamics Simulations.
Energy minimization, MD simulations were carried out as described earlier but using the revised Cbk1–Mob2 crystal structure presented in this study.33 Because αC of Cbk1 was not resolved in the crystal structure, we used homology modeling based on the structure of activated AKT/PKB to build this region.40 Furthermore, the activation loop segment with the Ser-570 autophosphorylation site as well as the DFG loop of the Cbk1–Mob2 complex was remodeled to make it adopt a similar structure as it had formerly been observed in other activated AGC kinases. The presence of long disordered loops (which could not be traced in the Cbk1–Mob2 crystal structure, e.g., AL and the linker that connects the HM to the kinase domain core) prevented fully unrestrained MD simulations (see Figure S4A). For example, (1) biochemically feasible modeling of the critical αC segment became possible only if main chain restraints were applied, and (2) trajectories were too sensitive to differences in the starting model as the outcome varied if the full kinase–cofactor structure was allowed to move freely. Therefore, the ends of unstructured loops, the Mg·ATP, most of the Cbk1 kinase domain, and Mob2 had to be restrained. Therefore, we applied heavy atom position restraints on those parts of the complex that did not directly seem to be involved in forming the active state in GROMACS version 4.5.5,45 but NDR/LATS kinase-specific (e.g., NTR, HM, and the C-terminal part of the activation loop harboring an autoregulatory phosphorylation site) as well as universally important kinase regions relevant for activity (e.g., αC, DFG, and HRD loops) were allowed to move freely. Main chain atom restraints were also needed to keep the homology-modeled αC and the small αB region helical (see Figure S4B).
Structure Deposition.
The crystallographic models of the Dbf2NTR-Mob1, Cbk1NTR-Mob2, and revised Cbk1–Mob2–pepSsd1 complexes have been deposited as PDB entries 5NCN, 5NCM, and 5NCL, respectively.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Kyle Schneider for the Cbk1–Mob2 expression plasmids. The authors thank Jonathan Hsu for the bicistronic plasmid backbone. The authors acknowledge a grant of computer time from the CSCS Swiss National Supercomputing Centre and the NIIF Hungarian National Information Infrastructure Development Institute. The authors acknowledge staff and instrumentation support from the Structural Biology Facility at Northwestern University, the Robert H. Lurie Comprehensive Cancer Center of Northwestern University, and NCI CCSG P30 CA060553. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corp. and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Funding
This work was supported by the National Institutes of Health (Grant 5R01GM084223-08) and by the Hungarian National Research, Development and Innovation Office (NN 114309 and KKP 126963). The work of C.H. and M.B. was supported by the Hungarian National Research, Development and Innovation Office (K123836) and the János Bolyai Research Scholarship of the Hungarian Academy of Sciences. The authors acknowledge the grant of computer time from the Governmental Information Technology Development Agency (KIFÜ) of Hungary. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.9b01096.
Figures S1-S4 (PDF)
Contributor Information
Benjamin W. Parker, Department of Biochemistry, University of Illinois Urbana-Champaign, Urbana, Illinois 61801, United States.
Gergo Gogl, Institute of Organic Chemistry, Research Center for Natural Sciences, 1117 Budapest, Hungary; Equipe Labellisee Ligue 2015, Department of Integrated Structural Biology, Institut de Genetique et de Biologie Moleculaire et Cellulaire (IGBMC), INSERM U1258/CNRS UMR 7104/Universite de Strasbourg, F-67404 Illkirch, France.
Mónika Bálint, Department of Pharmacology and Pharmacotherapy, Medical School, University of Pécs, 7624 Pécs, Hungary.
Csaba Hetényi, Department of Pharmacology and Pharmacotherapy, Medical School, University of Pécs, 7624 Pécs, Hungary.
Attila Reményi, Institute of Organic Chemistry, Research Center for Natural Sciences, 1117 Budapest, Hungary.
Eric L. Weiss, Molecular Biosciences, Northwestern University, Evanston, Illinois 60208, United States
REFERENCES
- (1).Meng Z, Moroishi T, and Guan K-L (2016) Mechanisms of Hippo pathway regulation,. Genes Dev. 30 (1), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hergovich A, Stegert MR, Schmitz D, and Hemmings BA (2006) NDR kinases regulate essential cell processes from yeast to humans,. Nat. Rev. Mol. Cell Biol 7 (4), 253–264. [DOI] [PubMed] [Google Scholar]
- (3).Emoto K (2011) The growing role of the Hippo-NDR kinase signalling in neuronal development and disease. J. Biochem 150 (2), 133–141. [DOI] [PubMed] [Google Scholar]
- (4).Hergovich A (2016) The Roles of NDR Protein Kinases in Hippo Signalling,. Genes 7 (5), 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhao B, et al. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control,. Genes Dev. 21 (21), 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hergovich A, and Hemmings BA (2012) Hippo signalling in the G2/M cell cycle phase: Lessons learned from the yeast MEN and SIN pathways,. Semin. Cell Dev. Biol 23 (7), 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bichsel SJ, Tamaskovic R, Stegert MR, and Hemmings BA (2004) Mechanism of Activation of NDR (Nuclear Dbf2-related) Protein Kinase by the hMOB1 Protein,. J. Biol. Chem 279 (34), 35228–35235. [DOI] [PubMed] [Google Scholar]
- (8).Bothos J, Tuttle RL, Ottey M, Luca FC, and Halazonetis TD (2005) Human LATS1 Is a Mitotic Exit Network Kinase,. Cancer Res. 65 (15), 6568–6575. [DOI] [PubMed] [Google Scholar]
- (9).Yoshida S, and Toh-E A (2001) Regulation of the localization of Dbf2 and mob1 during cell division of saccharomyces cerevisiae,. Genes Genet. Syst 76 (2), 141–147. [DOI] [PubMed] [Google Scholar]
- (10).Frenz LM, Lee SE, Fesquet D, and Johnston LH (2000) The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis,. J. Cell Sci 113 (19), 3399–3408. [DOI] [PubMed] [Google Scholar]
- (11).Mah AS, Jang J, and Deshaies RJ (2001) Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex,. Proc. Natl. Acad. Sci. U. S. A 98 (13), 7325–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Visintin R, and Amon A (2001) Regulation of the Mitotic Exit Protein Kinases Cdc15 and Dbf2,. Mol. Biol. Cell 12 (10), 2961–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rock JM, et al. (2013) Activation of the Yeast Hippo Pathway by Phosphorylation-Dependent Assembly of Signaling Complexes. Science 340 (6134), 871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Luca FC, and Winey M (1998) MOB1, an Essential Yeast Gene Required for Completion of Mitosis and Maintenance of Ploidy,. Mol. Biol. Cell 9 (1), 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Stavridi ES, et al. (2003) Crystal Structure of a Human Mob1 Protein: Toward Understanding Mob-Regulated Cell Cycle Pathways,. Structure 11 (9), 1163–1170. [DOI] [PubMed] [Google Scholar]
- (16).Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, and Winey M (2001) Saccharomyces cerevisiae Mob1p Is Required for Cytokinesis and Mitotic Exit,. Mol. Cell. Biol 21 (20), 6972–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Komarnitsky SI, et al. (1998) DBF2 Protein Kinase Binds to and Acts through the Cell Cycle-Regulated MOB1 Protein,. Mol. Cell. Biol 18 (4), 2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mah AS, et al. (2005) Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening,. BMC Biochem. 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Johnston LH, Eberly SL, Chapman JW, Araki H, and Sugino A (1990) The product of the Saccharomyces cerevisiae cell cycle gene DBF2 has homology with protein kinases and is periodically expressed in the cell cycle.,. Mol. Cell. Biol 10 (4), 1358–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Toyn JH, and Johnston LH (1994) The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition.,. EMBO J. 13 (5), 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hotz M, and Barral Y (2014) The Mitotic Exit Network: new turns on old pathways,. Trends Cell Biol. 24 (3), 145–152. [DOI] [PubMed] [Google Scholar]
- (22).Bardin AJ, and Amon A (2001) MEN and SIN: what’s the difference?,. Nat. Rev. Mol. Cell Biol 2 (11), 815–826. [DOI] [PubMed] [Google Scholar]
- (23).Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, and Luca FC (2002) The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor,. J. Cell Biol 158 (5), 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, and Deshaies RJ (2009) Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis,. J. Cell Biol 184 (4), 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Brace J, Hsu J, and Weiss EL (2011) Mitotic Exit Control of the Saccharomyces cerevisiae Ndr/LATS Kinase Cbk1 Regulates Daughter Cell Separation after Cytokinesis,. Mol. Cell. Biol 31 (4), 721–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Nelson B, et al. (2003) RAM: A Conserved Signaling Network That Regulates Ace2p Transcriptional Activity and Polarized Morphogenesis,. Mol. Biol. Cell 14 (9), 3782–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Voth WP, Olsen AE, Sbia M, Freedman KH, and Stillman DJ (2005) ACE2, CBK1, and BUD4 in Budding and Cell Separation,. Eukaryotic Cell 4 (6), 1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Jansen JM, Barry MF, Yoo CK, and Weiss EL (2006) Phosphoregulation of Cbk1 is critical for RAM network control of transcription and morphogenesis,. J. Cell Biol 175 (5), 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Schneper L, Krauss A, Miyamoto R, Fang S, and Broach JR (2004) The Ras/Protein Kinase A Pathway Acts in Parallel with the Mob2/Cbk1 Pathway To Effect Cell Cycle Progression and Proper Bud Site Selection,. Eukaryotic Cell 3 (1), 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Mrkobrada S, Boucher L, Ceccarelli DFJ, Tyers M, and Sicheri F (2006) Structural and Functional Analysis of Saccharomyces cerevisiae Mob1,. J. Mol. Biol 362 (3), 430–440. [DOI] [PubMed] [Google Scholar]
- (31).Hou M-C, Wiley DJ, Verde F, and McCollum D (2003) Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression,. J. Cell Sci 116 (1), 125–135. [DOI] [PubMed] [Google Scholar]
- (32).Hergovich A (2011) MOB control: Reviewing a conserved family of kinase regulators,. Cell. Signalling 23 (9), 1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gógl G, et al. (2015) The Structure of an NDR/LATS Kinase-Mob Complex Reveals a Novel Kinase-Coactivator System and Substrate Docking Mechanism,. PLoS Biol. 13 (5), No. e1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ni L, Zheng Y, Hara M, Pan D, and Luo X (2015) Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling,. Genes Dev. 29 (13), 1416–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kim S-Y, Tachioka Y, Mori T, and Hakoshima T (2016) Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway. Sci. Rep 6, 28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kulaberoglu Y, et al. (2017) Stable MOB1 interaction with Hippo/MST is not essential for development and tissue growth control. Nat. Commun 8, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Pearce LR, Komander D, and Alessi DR (2010) The nuts and bolts of AGC protein kinases,. Nat. Rev. Mol. Cell Biol 11 (1), 9–22. [DOI] [PubMed] [Google Scholar]
- (38).Hou M-C, Guertin DA, and McCollum D (2004) Initiation of Cytokinesis Is Controlled through Multiple Modes of Regulation of the Sid2p-Mob1p Kinase Complex,. Mol. Cell. Biol 24 (8), 3262–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kohler RS, Schmitz D, Cornils H, Hemmings BA, and Hergovich A (2010) Differential NDR/LATS Interactions with the Human MOB Family Reveal a Negative Role for Human MOB2 in the Regulation of Human NDR Kinases,. Mol. Cell. Biol 30 (18), 4507–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yang J, Cron P, Good VM, Thompson V, Hemmings BA, and Barford D (2002) Crystal structure of an activated Akt/Protein Kinase B ternary complex with GSK3-peptide and AMP-PNP,. Nat. Struct. Biol 9 (12), 940–944. [DOI] [PubMed] [Google Scholar]
- (41).Kabsch W (2010) XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr 66 (Part 2), 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software,. J. Appl. Crystallogr 40 (Part 4), 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Adams PD, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structuresolution,. Acta Crystallogr., Sect. D: Biol. Crystallogr 66 (Part 2), 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot,. Acta Crystallogr., Sect. D: Biol. Crystallogr 66 (Part4), 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Pronk S, et al. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29 (7), 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.